- Department of Biotechnology, Faculty of Bioresource Sciences, Akita Prefectural University, Akita, Japan
Isoleucine 2-epimerase (ILEP) is a novel branched-chain amino acid racemase isolated from Lactobacillus buchneri. In this study, we examined production of free D-branched-chain amino acids such as D-valine, D-leucine, and D-allo-isoleucine, using lactic acid bacteria carrying homologs to ILEP. Twelve selected strains of lactic acid bacteria were grown at optimal growth temperatures and accumulation of D-branched-chain amino acids in the medium was monitored in exponential, early stationary, and stationary phases. To analyze the D-branched-chain amino acids, enantiomers in the medium were initially converted into diastereomers using pre-column derivatization with o-phthaldialdehyde plus N-isobutyryl-L-cysteine. The resultant fluorescent isoindole derivatives were analyzed on an octadecylsilyl stationary phase using ultra-high performance liquid chromatography. The analyses revealed that the seven following lactic acid bacteria carrying homologs showing 53–60% amino acid sequence identity to the L. buchneri ILEP accumulate D-branched-chain amino acids: Lactobacillus fermentum and Weissella paramesenteroides produce D-valine, D-leucine, and D-allo-isoleucine; Lactobacillus reuteri, Leuconostoc mesenteroides subsp. mesenteroides, and Leuconostoc gelidum subsp. gasicomitatum accumulate D-leucine and D-allo-isoleucine; and Lactobacillus vaginalis and Leuconostoc pseudomesenteroides produce D-allo-isoleucine. These results suggest that D-branched-chain amino acids are produced by a variety of lactic acid bacteria species, particularly those carrying homologs to the ILEP.
Introduction
D-Amino acids, enantiomers of L-amino acids, play key roles as components of the peptidoglycan cell wall of bacteria. Within bacterial cell walls, D-alanine (D-Ala), and D-glutamate (D-Glu) are often the most common D-amino acids (Hancock, 1960; Schleifer and Kandler, 1972). In some lactic acid bacteria, which have a peptidoglycan type A4α or A4β, the peptidoglycans contain D-aspartate (D-Asp) in addition to D-Ala and D-Glu (Schleifer and Kandler, 1972; Bellais et al., 2006; Veiga et al., 2006). These D-amino acids are primarily produced from the corresponding L-enantiomers by enzymes such as alanine racemase (EC 5.1.1.1), glutamate racemase (EC 5.1.1.3), and aspartate racemase (EC 5.1.1.13). In our earlier study, marked accumulation of D-branched-chain amino acids (D-BCAAs) such as D-valine (D-Val), D-leucine (D-Leu), and D-allo-isoleucine (D-allo-Ile) was observed in the growth medium of the lactic acid bacterium Lactobacillus otakiensis. The enzyme activity responsible was detected as branched-chain amino acid racemase (BCAAR) activity including leucine racemase, valine racemase, and isoleucine 2-epimerase activities in L. otakiensis cells. On the basis of the N-terminal amino acid sequence of the purified enzyme, a gene encoding a homolog of this racemase from L. otakiensis was identified in the genome of Lactobacillus buchneri (Mutaguchi et al., 2013b). Although this gene is annotated as a γ-aminobutyrate aminotransferase (GABA-AT), the gene product enzyme expressed in Escherichia coli showed BCAAR activity, but not GABA-AT activity. The recombinant BCAAR, which is pyridoxal 5′-phosphate (PLP)-dependent, preferentially epimerizes between L-isoleucine (L-Ile) and D-allo-Ile, and also racemizes nonpolar amino acids including Val and Leu. This enzyme was therefore designated isoleucine 2-epimerase (ILEP) and assigned an identical EC number (EC 5.1.1.21). ILEP is a novel enzyme that catalyzes the racemization of BCAAs as its main substrate and seems to be involved in the production of D-BCAAs in lactic acid bacteria. In fact, homologous genes whose deduced amino acid sequence shows more than 30% identity with that of the L. buchneri ILEP are widely distributed among lactic acid bacteria. Elucidation the actual ability of D-BCAAs production by lactic acid bacteria carrying the ILEP homologous gene is extremely informative to reveal the physiological functions of D-BCAAs in lactic acid bacteria, thereby leading to the application of the amino acids production. In this study, therefore, we investigated the levels of D-BCAAs in growth media and the ILEP activity in cells of 12 strains of lactic acid bacteria carrying the ILEP homologous gene.
Materials and Methods
Materials
N-Isobutyryl-L-cysteine (NIC) and o-phthaldialdehyde (OPA) were, respectively, purchased from Sigma-Aldrich Corp. LLC (St. Louis, MO, USA) and Nacalai Tesque Inc. (Kyoto, Japan). MRS medium was obtained from Becton, Dickinson and Co. (Franklin Lakes, NJ, USA). D-Amino acid oxidase from porcine kidney and catalase from bovine liver were purchased from Sigma.
Microorganisms and Growth Conditions
Twelve strains of lactic acid bacteria used for this study and database information of their ILEP homologous gene are presented in Table 1. All strains were obtained from the Japan Collection of Microorganisms (JCM, Tsukuba, Japan) and the National Institute of Technology and Evaluation Biological Resource Center (NBRC, Tokyo, Japan). These strains were aerobically cultured in their recommended media at their respective optimum cultivation temperatures (Table 1). The recommended medium for Oenococcus oeni JCM 6125, 143 Leuconostoc oenos medium, was prepared according to the recipe published on the JCM website (http://www.jcm.riken.jp/cgi-bin/jcm/jcm_grmd?GRMD=143).
Preparation of Culture Media for D-Amino Acid Analyses
For sequential analysis of the D-amino acid content of the culture medium conditioned by each of the lactic acid bacteria, sample solutions were prepared as follows. Each of strains was cultured to the stationary phase in 5 mL of the recommended medium. Aliquots (1 mL) of the culture medium were centrifuged (10,000 × g for 1 min at 4°C), and the supernatants were filtered through a centrifugal filter 3 K device (Amicon Ulta 0.5 mL, Merck Millipore, Darmstadt, Germany). The prepared samples were stored at −20°C until use.
Analysis of L-Amino and D-Amino Acids Using UHPLC
D- and L-Amino acids in sample solutions were derivatized with OPA and NIC (Brückner et al., 1994) using an OPA-NIC solution prepared by dissolving 10 mg of OPA and 10 mg of NIC in 1 mL of methanol. The reaction mixture (250 μL) for the derivatization contained 5 μL of amino acid sample, 10 μL of OPA-NIC solution, and 35 μL of 0.4 M borate-sodium hydroxide buffer (pH 10.4). After derivatization for 2 min at 15°C in the dark, an aliquot (1 μL) of the reaction mixture was introduced into an ultra-high performance liquid chromatography (UHPLC) system (Nexera X2; Shimadzu Corp., Kyoto, Japan). The diastereoisomeric derivatives of amino acids formed with OPA-NIC were applied to a TSKgel ODS-12OH 2.0 mm I.D. × 15 cm column (Tosoh Corp., Tokyo, Japan) in the UHPLC. The UHPLC system consisted of a system controller CBM-20A, a solvent delivery unit LC-30AD, an HPLC column oven CTO-20AC, an autosampler SIL-30AC and a fluorescence detector RF-20Axs (all apparatus from Shimadzu Corp.). The excitation and emission wavelengths for fluorescent detection of the diastereoisomeric amino acid derivatives were, respectively, 230 and 445 nm. The data were processed using Labstations (Shimadzu). The system was operated at a flow rate of 0.25 mL/min at 25°C. The UHPLC gradient system for analysis of OPA-NIC derivatives (A = 50 mM sodium acetate, pH 5.9, B = methanol, and C = acetonitrile) was 12.2–22.8% B and 2.0–3.8% C for 6 min, 22.8% B and 3.8% C for 4 min, 22.8–33.4% B and 3.8–5.6% C for 2 min, 33.4% B and 5.6% C for 2 min, 33.4–35.5% B and 5.6–5.9% C for 0.25 min, 35.5% B and 5.9% C for 1.25 min, 35.5–41.4% B and 5.9–6.9% C for 0.5 min, 41.4% B and 6.9% C for 5 min, 41.4–49.3% B and 6.9–8.2% C for 3 min, 49.3% B and 8.2% C for 3.5 min, and 49.3–12.2% B and 8.2–2.0% C for 0.01 min. The peak area and retention time were, respectively, used for amino acid quantification and identification. Using OPA-NIC derivatization, 35 kinds of amino acids were analyzed simultaneously: D-form and L-form of Asp, Glu, Ala, Val, Leu, asparagine (Asn), glutamine (Gln), serine (Ser), methionine (Met), tryptophan (Trp), tyrosine (Tyr), phenylalanine (Phe), arginine (Arg), histidine (His), lysine (Lys), L-Ile, D-allo-Ile, L-threonine (L-Thr), D-allo-threonine (D-allo-Thr), and glycine (Gly). For this study, analysis was conducted with emphasis on D-BCAAs.
D-Amino Acid Oxidase Treatment
Bacterial medium samples usually contain any unidentified amino compound that gives the same retention time-peak as a D-amino acid. Therefore, the peaks of D-amino acids, such as D-branched-chain amino acids, were confirmed by the reduction in fluorescence intensity elicited by treatment with D-amino acid oxidase (DAO) from Sus scrofa (Tosa et al., 1974). The DAO has very broad substrate specificity, oxidizing 12 D-amino acids including D-branched-chain amino acids (D'Aniello et al., 1993). Treatment with DAO removed the 12 susceptible D-amino acids from the medium samples, causing the respective peaks to be smaller than those obtained with the corresponding untreated samples. The DAO treatment was conducted as described in an earlier report (Mutaguchi et al., 2013a).
Preparation of Cell Extract for BCAAR Assay
For determination of BCAAR activity in lactic acid bacteria cells, cell extract was prepared as follows. Each of 12 strains was cultured until the end of exponential phase in 37.5 mL of recommended medium, and the cells were collected by centrifugation. The harvested cells were washed twice with 150 mM NaCl, suspended in 1 mL of 50 mM sodium-phosphate buffer (pH 7.2), disrupted using a multi-bead shocker (Yasui Kikai, Osaka, Japan), and centrifuged (10,000 × g for 30 min at 4°C). The resultant supernatant was used as the cell extract.
Determination of BCAAR Activity in the Bacterial Cells
BCAAR activity was assayed by measuring the amount of D-BCAAs resulted from L-BCAAs during the epimerization or racemization reaction. The reaction mixture (500 μL), consisting of 200 mM sodium-phosphate buffer (pH 7.0), 50 mM substrate (L-Val, L-Leu, or L-Ile), 0.1 mM PLP and 50 μL of the cell extract, was incubated at the same temperature as the optimum growth temperature of the strain. The reaction was stopped by addition of 125 μL of 50% trichloroacetic acid (TCA) to the reaction mixture when less than 20% of the substrate was consumed. The maximum reaction time was up to 24 h. After incubation for 5 min at 25°C, the mixture was centrifuged (10,000 × g for 15 min at 4°C). An aliquot (500 μL) of the supernatant was neutralized by addition of 300 μL of 1 M NaOH. The amount of product, D-BCAA, in the solution was measured using UHPLC as described above.
Protein Determination
Protein concentration was determined using the method described by Bradford (Bradford, 1976). Bovine serum albumin was used as the standard.
Results
Analysis of the D-BCAAs in the Culture Medium of 12 Strains of Lactic Acid Bacteria
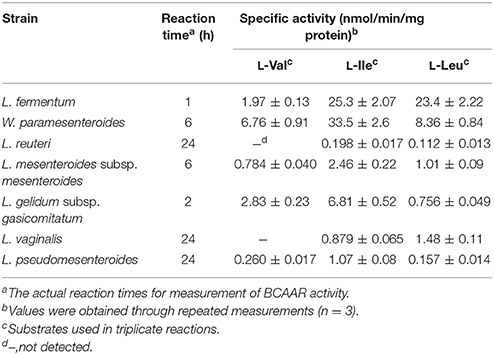
Each of 12 strains of lactic acid bacteria carrying an L. buchneri ILEP homologous gene was cultured until the stationary phase, and accumulation of D-BCAAs in the culture medium was investigated. For D-Val, D-allo-Ile, and D-Leu, the UHPLC peaks were identified using retention times and confirmed to be derived from D-amino acids by DAO treatment. The results were the following: Lactobacillus furmentum and Weissella paramesenteroides produced D-Val, D-allo-Ile, and D-Leu (Figures 1A,B); Lactobacillus reuteri, Leuconostoc mesenteroides subsp. mesenteroides, and Leuconostoc gelidum subsp. gasicomitatum produced D-allo-Ile and D-Leu (Figures 1C–E); Lactobacillus vaginalis and Leuconostoc pseudomesenteroides produced only D-allo-Ile (Figures 1F,G). However, no D-BCAA was produced by Lactobacillus ruminis, Streptococcus mutans, Enterococcus malodoratus, Oenococcus oeni, nor Lactobacillus plantarum subsp. plantarum.
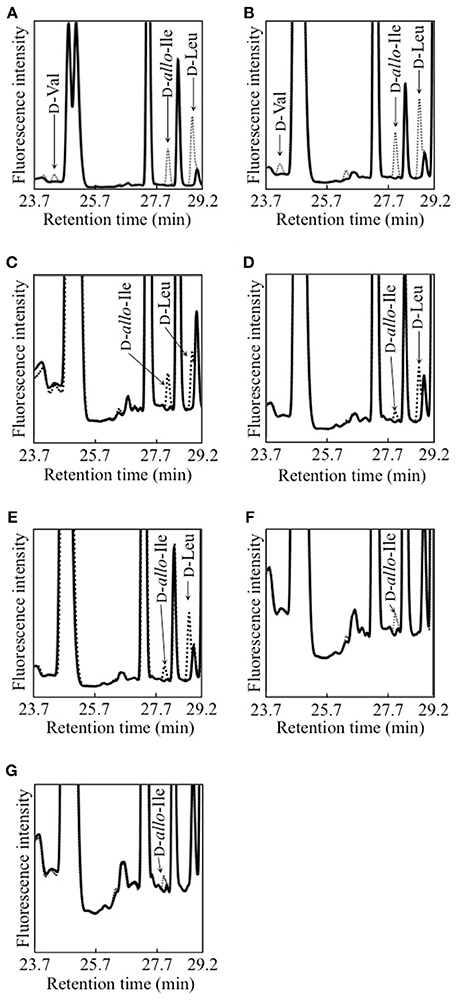
Figure 1. Detection of D-BCAAs in culture media of 7 strains of lactic acid bacteria. (A), L. fermentum; (B) W. paramesenteroides; (C), L. reuteri; (D), L. mesenteroides subsp. mesenteroides; (E), L. gelidum subsp. gasicomitatum; (F), L. vaginalis; (G), L. pseudomesenteroides. Dotted and solid lines show chromatograms of samples before and after DAO treatment, respectively.
Subsequently, the D-BCAA concentrations in the culture media were measured in the middle and end of exponential phase and in the stationary phase for the D-BCAA producing seven strains. The results are portrayed in Figure 2 and D-BCAAs accumulated by the lactic acid bacteria are summarized in Table 2. The levels of D-Val, D-allo-Ile, and D-Leu produced by L. fermentum and W. paramesenteroides continued to increase until their stationary phase (Figures 2A,B). The levels of D-BCAAs produced by L. reuteri, L. mesenteroides subsp. mesenteroides, L. gelidum subsp. gasicomitatum, L. vaginalis, and L. pseudomesenteroides, however, increased until the end of exponential phase (Figures 2C–G). Results show that the remarkable increases in the D-BCAAs were occurred in the exponential phase and the D-BCAA levels continued to be almost constant through out the stationary phase.
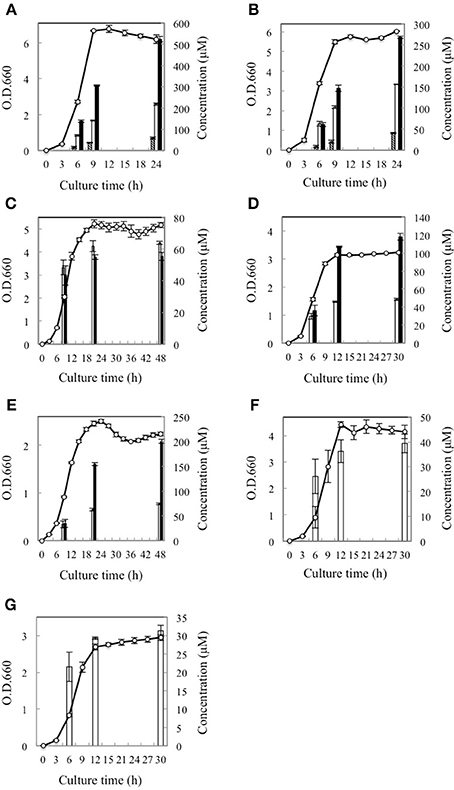
Figure 2. Time course of D-BCAA accumulation in culture media of 7 strains of lactic acid bacteria. (A), L. fermentum; (B), W. paramesenteroides; (C), L. reuteri; (D), L. mesenteroides subsp. mesenteroides; (E), L. gelidum subsp. gasicomitatum; (F), L. vaginalis; (G), L. pseudomesenteroides. Solid bars, empty bars, and diagonal lines bars, respectively, show concentrations of D-Leu, D-allo-Ile, and D-Val. Cell concentrations (open circles) were assayed at O.D.600.
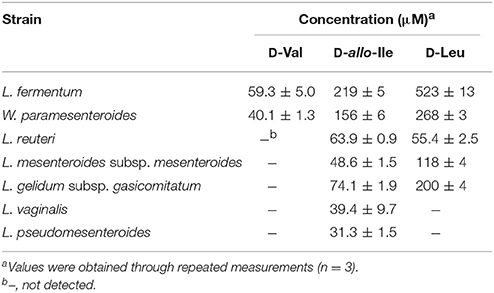
Table 2. D-BCAA concentrations in the culture media in the stationary phase for the D-BCAA producing strains.
Determination of BCAAR Activity in the Bacterial Cells
In order to confirm that D-BCAAs accumulated in the culture media were derived from racemization or epimerization of BCAAs in the bacterial cells tested, BCAAR activity in the cells was determined using cell extracts of the 12 strains of lactic acid bacteria. Results show that BCAAR activity, which resulted in the accumulation of D-BCAA in culture medium, was detected in the cells of seven strains (Table 3). In addition, racemase activity for D-BCAAs not accumulated in the culture medium was also detected in the four strains: leucine racemase activity in L. vaginalis and L. pseudomesenteroides; valine racemase activity in L. mesenteroides subsp. mesenteroides, L. gelidum subsp. gasicomitatum, and L. pseudomesenteroides. However, no BCAAR activity was detected in the cell extracts of L. ruminis, S. mutans, E. malodoratus, O. oeni, nor L. plantarum subsp. plantarum after the incubation of reaction mixture for 24 h.
Discussion
In this study, the production of D-BCAAs such as D-Val, D-allo-Ile, and D-Leu by 12 strains of lactic acid bacteria carrying a homologous gene to L. buchneri ILEP was investigated. Amino acid sequences of these homologous genes show more than 30% identity with that of the L. buchneri ILEP. These homologous genes are annotated as putative aminotransferases such as acetylornithine aminotransferase (AO-TA) and GABA-AT (Table 1). Specifically examining the phylogenetic tree based on the deduced amino acid sequences of the homologous genes, three clades were readily apparent (Figure 3). The first clade included homologs showing 30% identity with the L. buchneri ILEP. For strains carrying homologs included in the clade, neither accumulation of D-BCAAs in the growth medium nor BCAAR activity in the cell extract was detected. According to the genome database, the homologous gene of L. ruminis is annotated as a putative GABA-AT gene, with some γ-aminobutyrate metabolism-related genes lying around it. The homologous genes of S. mutans and E. malodoratus are annotated as a putative AO-AT gene, with some genes possibly related to acetylornithine metabolism around them. It is, therefore, considered that enzymes expressed from these homologous genes in L. ruminis, S. mutans, and E. malodoratus might be responsible for a function as an aminotransferase, but not as an ILEP.
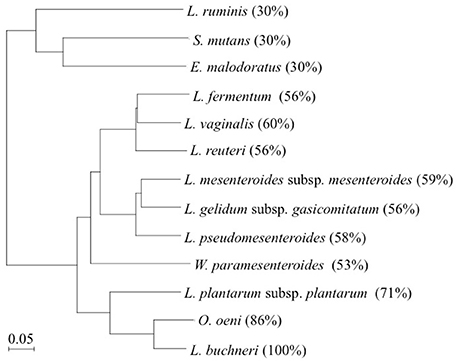
Figure 3. Phylogenetic tree of L. buchneri ILEP with ILEP homologs in 12 lactic acid bacteria. Numbers in parentheses denote identity with L. buchneri ILEP based on the amino acid sequence.
Homologs showing between 53 and 60% identity with the L. buchneri ILEP were included in the second clade, and all the strains in this clade produced some of D-BCAAs and showed BCAAR activity in the cells. In addition, each of homologous genes in this clade neighbors a putative amino acid permease gene in each genome without any other neighboring genes. These facts suggest that enzymes expressed from these homologous genes belonging to the clade played a role in the same manner as L. buchneri ILEP, and the permease might be involved in D-BCAA production.
The third clade, to which L. buchneri ILEP belonged, consists of homologs conserved in genomes of O. oeni and L. plantarum subsp. plantarum. The homologs in the two strains showed more than 70% identity with the L. buchneri ILEP. In addition, the members in this clade also accompany a putative amino acid permease gene in the genome as observed in the second clade homologs. Despite the high identity, however, O. oeni and L. plantarum subsp. plantarum failed to produce D-BCAAs in the growth media and showed no BCAAR activity was detected in the cell extracts. Hayashi et al. reported that L-Ile epimerization by L. buchneri ILEP proceeds through abstraction of the α-hydrogen from the substrate by Lys280, whereas Asp222 serves as the catalytic residue adding an α-hydrogen to the quinonoid intermediate to form D-allo-Ile (Hayashi et al., 2017). Figure 4 shows the amino acid sequence alignment among ILEP homologs of the two bacteria, L. fermentum, and L. buchneri to reveal that the amino acid residues corresponding to Lys280 and Asp222 of L. buchneri ILEP were well conserved in the homologs of O. oeni (Lys280 and Asp222) and L. plantarum subsp. plantarum (Lys281 and Asp223), similarly in those of L. fermentum. In addition, most of substrate-binding residues and cofactor-binding residues of L. buchneri ILEP (Hayashi et al., 2017) were also conserved among the homologs. It is therefore crucial to clarify the reason for failure of D-BCAA production by the two lactic acid bacteria through analysis of the transcription levels of the ILEP homologous genes together with enzymatic characterization of the recombinant protein.

Figure 4. Multiple alignment of amino acid sequences of L. buchneri ILEP with ILEP homologs. L. buc, L. buchneri; O. oen, O. oeni; L. pla, L. plantarum subsp. plantarum; L. fer, L. fermentum. Lys residues abstracting α-hydrogen of L-form of the substrate and Asp residues adding α-hydrogen to the quinonoid intermediate are surrounded by lines. The substrate-binding and cofactor-binding residues are, respectively, marked by filled circles and open circles.
Lam et al. reported that Vibrio cholerae produced several D-amino acids including D-Leu and D-Val from the end of its logarithmic growth phase to the stationary phase. The bacterium used these D-amino acids for regulating cell wall remodeling during its stationary phase (Lam et al., 2009). In contrast, however, for the seven lactic acid bacteria used in this study, the production and accumulation of D-BCAAs started simultaneously with the start of the bacterial growth, and concentrations of D-BCAAs in the cultures increased along with the bacterial growth. To elucidate physiological functions of D-BCAAs in lactic acid bacteria, it is thus necessary to discuss the relation between D-BCAA production and growth of lactic acid bacteria.
Recently, many novel functions of bacterial D-amino acids have been reported except for those as peptidoglycan building blocks. For instance, Sasabe et al. reported that enterobacterial D-amino acids such as D-Ala, D-Asp, and D-Glu regulate gut microbiota of host mammalian via DAO from the host mammalian (Sasabe et al., 2016). Moreover, as described above, Vibrio cholerae are considered to use several D-amino acids for regulating cell wall remodeling during their stationary phase (Lam et al., 2009). These reports commonly show that free D-amino acids secreted from one bacterial cell directly or indirectly affect other bacterial cells. Free D-BCAAs produced by lactic acid bacteria might also have effects on other bacterial cells of the same or different species present in the surrounding environment. Among the lactic acid bacteria producing D-BCAAs examined for this study, L. fermentum, L. vaginalis, and L. reuteri are human indigenous bacteria isolated from mouth, vagina, and intestine, respectively (Lerche and Reuter, 1962; Embley et al., 1989; Dellaglio et al., 2004). The results of this study, therefore, indicate that D-BCAAs from these lactic acid bacteria might be related to human health through their influences on microbiota in human tissues. Furthermore, many kinds of D-amino acids including D-BCAAs are known to be much sweeter than the corresponding L-amino acids (Solms et al., 1965; Schiffman et al., 1981; Kawai et al., 2012). This study showed that lactic acid bacteria isolated from foods such as L. mesenteroides subsp. mesenteroides, L. pseudomesenteroides, L. gelidum subsp. gasicomitatum, and W. paramesenteroides (Collins et al., 1993; Björkroth et al., 2000) produced D-BCAAs, which might affect the taste of foods.
Although little has been clarified about free D-BCAAs production and functions in all organisms, this study shows D-BCAAs production by several lactic acid bacteria that are closely related to human life and indicates the importance of further studies focusing on the function of the amino acids in human health and food quality.
Author Contributions
YM planned this research and did all the practical experiment of this research. KK and IK contributed conception of this study and advised about organization of this manuscript.
Funding
This work was supported by Department of Biotechnology, Faculty of Bioresource Sciences, Akita Prefectural University, 2015–2017.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bellais, S., Arthur, M., Dubost, L., Hugonnet, J. E., Gutmann, L., van Heijenoort, J., et al. (2006). Aslfm, the D-aspartate ligase responsible for the addition of D-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J. Biol. Chem. 281, 11586–11594. doi: 10.1074/jbc.M600114200
Björkroth, K. J., Geisen, R., Schillinger, U., Weiss, N., De Vos, P., Holzapfel, W. H., et al. (2000). Characterization of Leuconostoc gasicomitatum sp. nov., associated with spoiled raw tomato-marinated broiler meat strips packaged under modified-atmosphere conditions. Appl. Environ. Microbiol. 66, 3764–3772. doi: 10.1128/AEM.66.9.3764-3772.2000
Bradford, M. M. (1976). A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Brückner, H., Haasmann, S., Langer, M., Westhauser, T., and Wittner, R. (1994). Liquid chromatographic determination of D- and L-amino acids by derivatization with o-phthaldialdehyde and chiral thiols: applications with reference to biosciences. J. Chromatogr. A 666, 259–273. doi: 10.1016/0021-9673(94)80388-9
Collins, M. D., Samelis, J., Metaxopoulos, J., and Wallbanks, S. (1993). Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 75, 595–603. doi: 10.1111/j.1365-2672.1993.tb01600.x
D'Aniello, A., Vetere, A., and Petrucelli, L. (1993). Further study on the specificity of D-amino acid oxidase and of D-aspartate oxidase and time course for complete oxidation of D-amino acids. Comp. Biochem. Physiol. 105B, 731–734. doi: 10.1016/0305-0491(93)90113-J
Dellaglio, F., Torriani, S., and Felis, G. E. (2004). Reclassification of Lactobacillus cellobiosus Rogosa et al. 1953 as a later synonym of Lactobacillus fermentum Beijerinck 1901. Int. J. Syst. Evol. Microbiol. 54, 809–812. doi: 10.1099/ijs.0.02947-0
Embley, T. M., Faquir, N., Bossart, W., and Collins, M. D. (1989). Lactobacillus vaginalis sp. nov. from the human vagina. Int. J. Syst. Bacteriol. 39, 368–370. doi: 10.1099/00207713-39-3-368
Hancock, R. (1960). The amino acid composition of the protein and cell wall of Staphylococcus aureus. Biochim. Biophys. Acta. 37, 42–46. doi: 10.1016/0006-3002(60)90076-7
Hayashi, J., Mutaguchi, Y., Minemura, Y., Nakagawa, N., Yoneda, K., Ohmori, T., et al. (2017). Crystal structure of the novel amino-acid racemase isoleucine 2-epimerase from Lactobacillus buchneri. Acta. Crystallogr. D Struct. Biol. 73, 428–437. doi: 10.1107/S2059798317005332
Kawai, M., Sekine-Hayakawa, Y., Okiyama, A., and Ninomiya, Y. (2012). Gustatory sensation of L- and D-amino acids in humans. Amino Acids 43, 2349–2358. doi: 10.1007/s00726-012-1315-x
Lam, H., Oh, D. C., Cava, F., Takacs, C. N., Clardy, J., de Pedro, M. A., et al. (2009). D-Amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555. doi: 10.1126/science.1178123
Lerche, M., and Reuter, G. (1962). Das Vorkommen aerob wachsender grampositiver Stäbchen des Genus Lactobacillus Beijerinck im Darminhalt erwachsener Menschen. Zentralbl. Bakteriol. I. Abt. Orig. 185, 446–481.
Mutaguchi, Y., Ohmori, T., Akano, H., Doi, K., and Ohshima, T. (2013a). Distribution of D-amino acids in vinegars and involvement of lactic acid bacteria in the production of D-amino acids. Springerplus 2:691. doi: 10.1186/2193-1801-2-691
Mutaguchi, Y., Ohmori, T., Wakamatsu, T., Doi, K., and Ohshima, T. (2013b). Identification, purification, and characterization of a novel amino acid racemase, isoleucine 2-epimerase, from Lactobacillus species. J. Bacteriol. 195, 5207–5215. doi: 10.1128/JB.00709-13
Sasabe, J., Miyoshi, Y., Rakoff-Nahoum, S., Zhang, T., Mita, M., Davis, B. M., et al. (2016). Interplay between microbial d-amino acids and host d-amino acid oxidase modifies murine mucosal defence and gut microbiota. Nat. Microbiol. 1:16125. doi: 10.1038/nmicrobiol.2016.125
Schiffman, S. S., Sennewwald, K., and Gagnon, J. (1981). Comparison of taste qualities and thresholds of D- and L-amino acids. Physiol. Behav. 27, 51–59. doi: 10.1016/0031-9384(81)90298-5
Schleifer, K. H., and Kandler, O. (1972). Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477.
Solms, J., Vuataz, L., and Egli, R. H. (1965). The taste of L- and D-amino acids. Experientia 21, 692–694. doi: 10.1007/BF02138474
Tosa, T., Sano, R., and Chibata, I. (1974). Immobilized D-amino acid oxidase preparation, some enzymatic properties, and potential uses. Agr. Biol. Chem. 38, 1529–1534. doi: 10.1080/00021369.1974.10861362
Keywords: D-amino acid, branched-chain amino acid, epimerase, isoleucine 2-epimerase, lactic acid bacteria, racemase
Citation: Mutaguchi Y, Kasuga K and Kojima I (2018) Production of D-Branched-Chain Amino Acids by Lactic Acid Bacteria Carrying Homologs to Isoleucine 2-Epimerase of Lactobacillus buchneri. Front. Microbiol. 9:1540. doi: 10.3389/fmicb.2018.01540
Received: 30 March 2018; Accepted: 20 June 2018;
Published: 13 July 2018.
Edited by:
Tohru Yoshimura, Nagoya University, JapanReviewed by:
Mamoru Wakayama, Ritsumeikan University, JapanAtte Von Wright, University of Eastern Finland, Finland
Copyright © 2018 Mutaguchi, Kasuga and Kojima. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuta Mutaguchi, y-muta@akita-pu.ac.jp
 Yuta Mutaguchi
Yuta Mutaguchi Kano Kasuga
Kano Kasuga