- 1Department of Agricultural Biotechnology, Center for Food Safety and Toxicology, Seoul National University, Seoul, South Korea
- 2Food-borne Pathogen Omics Research Center (FORC), Seoul National University, Seoul, South Korea
- 3Department of Life Science, Sogang University, Seoul, South Korea
- 4Department of Applied Chemistry & Biological Engineering, Ajou University, Suwon, South Korea
- 5Department of Food Science and Biotechnology, Kyung Hee University, Yongin, South Korea
- 6Department of Animal Resources Science, Dankook University, Cheonan, South Korea
- 7Department of Animal Science and Biotechnology, Seoul National University, Seoul, South Korea
- 8Department of Food Science and Technology, Chungnam National University, Daejeon, South Korea
- 9Department of Life Science, Multidisciplinary Genome Institute, Hallym University, Chuncheon, South Korea
Vibrio parahaemolyticus can cause gastrointestinal illness through consumption of seafood. Despite frequent food-borne outbreaks of V. parahaemolyticus, only 19 strains have subjected to complete whole-genome analysis. In this study, a novel strain of V. parahaemolyticus, designated FORC_022 (Food-borne pathogen Omics Research Center_022), was isolated from soy sauce marinated crabs, and its genome and transcriptome were analyzed to elucidate the pathogenic mechanisms. FORC_022 did not include major virulence factors of thermostable direct hemolysin (tdh) and TDH-related hemolysin (trh). However, FORC_022 showed high cytotoxicity and had several V. parahaemolyticus islands (VPaIs) and other virulence factors, such as various secretion systems (types I, II, III, IV, and VI), in comparative genome analysis with CDC_K4557 (the most similar strain) and RIMD2210633 (genome island marker strain). FORC_022 harbored additional virulence genes, including accessory cholera enterotoxin, zona occludens toxin, and tight adhesion (tad) locus, compared with CDC_K4557. In addition, O3 serotype specific gene and the marker gene of pandemic O3:K6 serotype (toxRS) were detected in FORC_022. The expressions levels of genes involved in adherence and carbohydrate transporter were high, whereas those of genes involved in motility, arginine biosynthesis, and proline metabolism were low after exposure to crabs. Moreover, the virulence factors of the type III secretion system, tad locus, and thermolabile hemolysin were overexpressed. Therefore, the risk of foodborne-illness may be high following consumption of FORC_022 contaminated crab. These results provided molecular information regarding the survival and pathogenesis of V. parahaemolyticus FORC_022 strain in contaminated crab and may have applications in food safety.
Introduction
Vibrio parahaemolyticus is curved rod-shape, Gram-negative halophilic bacterium with a single polar flagellum and is abundant in estuarine environments and various seafood, including shellfish, oysters, clams, cockles, crabs, and shrimps (Joseph et al., 1982; Newton et al., 2014; Rodgers et al., 2014; Xu et al., 2014; Malcom et al., 2015). This bacterium causes seafood borne gastroenteritis, and its infection is usually associated with the consumption of raw or undercooked seafood (Daniels et al., 2000; Letchumanan et al., 2016). Recently, the occurrence of food borne disease and outbreaks due to seafood contamination with V. parahaemolyticus have increased significantly around the world (Letchumanan et al., 2015a,b). The symptoms include watery diarrhea, vomiting, nausea, abdominal cramps, septicemia and even death (Hazen et al., 2015). To understand the pathogenicity of V. parahaemolyticus, several studies have analyzed the association of its virulence factors with food poisoning (Haendiges et al., 2015; Li et al., 2017). The complete genomes of 19 strains of V. parahaemolyticus have been sequenced and are currently available from the National Center for Biotechnology Information (NCBI) database1.
Vibrio parahaemolyticus strains isolated from patients hospitalized due to food-borne illness harbor multiple virulence genes, including thermostable direct hemolysin (tdh) and TDH-related hemolysin (trh) (Kishishita et al., 1992). These genes are indicators of V. parahaemolyticus pathogenicity and elicit enterotoxic effects on human intestinal cells (Ottaviani et al., 2012; Ludeke et al., 2015). For example, V. parahaemolyticus KCTC 2471 (=ATCC 33844 = CDC strain KC 824), which harbored tdh gene as major virulence factor, caused food poisoning in Japan (Baumann et al., 1971; Hossain et al., 2013). However, several strains isolated from clinical samples were found to be negative for tdh and trh genes, suggesting that these strains may carry some other virulence factors (Park et al., 2004a; Lynch et al., 2005; Velazquez-Roman et al., 2012). Genes related to type III secretion, tight adhesion locus (tad locus), and hemolysin have been suggested as possible contributors to V. parahaemolyticus pathogenicity (Tomich et al., 2007; Caburlotto et al., 2010). Various secretion systems (types I, II, III, IV, and VI) are conserved in Gram-negative bacteria and are known to play a role in pathogenicity of V. parahaemolyticus by mediating the transportation of virulence-related proteins across the bacterial membrane (Pallen et al., 2003; Delepelaire, 2004; Park et al., 2004b; Abendroth et al., 2009; Wang et al., 2015). In addition, V. parahaemolyticus islands (VPaI), which are located in pathogenic islands in V. parahaemolyticus, are thought to play a role in the pathogenicity of this organism (Hurley et al., 2006). Infection of human cell lines by V. parahaemolyticus lacking tdh and trh genes also results in significant cytotoxicity (Broberg et al., 2011; Ritchie et al., 2012). Therefore, major virulence factors of V. parahaemolyticus have still not been identified at the genomic level.
About 28% of outbreaks by V. parahaemolyticus attributed to a single food commodity are due to aquatic products, such as crustaceans and shellfish (Wu et al., 2014). Crabs are a major source of V. parahaemolyticus outbreaks among aquatic products in northeast Asian due to improper cooking and wound infection from mishandling (Rodgers et al., 2014). However, no reports have described whole-genome sequences of V. parahaemolyticus isolated from contaminated crab; therefore, no information is available regarding the major virulence factors of V. parahaemolyticus obtained from crabs. Furthermore, information regarding the expression of virulence factors of V. parahaemolyticus in contaminated food is limited. Because crabs are a popular source of seafood in Asia, analyses of whole-genome sequences and the transcriptome of V. parahaemolyticus isolated from crab products are necessary to investigate the potential risks of foodborne illnesses from contaminated products.
In this study, we performed complete whole-genome sequencing of V. parahaemolyticus FORC_022 strain, isolated from soy sauce marinated swimming crabs in South Korea, and compared with complete sequences of this species from the public database. In addition, the transcriptome of this strain was analyzed to elucidate the potential pathogenicity of this strain in crabs using an artificial contact experiment. These results provide a better understanding of the pathogenicity of V. parahaemolyticus in crab-based foods and could facilitate the prevention of foodborne illness.
Materials and Methods
Isolation, Growth Conditions and Morphology
FORC_022 strain was isolated from soy sauce marinated crabs by the Jeollanam-do Institute of Health and Environment, South Korea. The strain was cultivated aerobically at 30°C in modified Luria-Bertani medium supplemented with 1% (w/v) NaCl for 12 h (Sakazaki, 1983).
The cells were negatively stained with uranyl acetate for 1 min and were then observed using a transmission electron microscopy (JEM-2100; JEOL, Tokyo, Japan) at 200 kV. The FORC_022 strain was a curved, rod-shaped bacterium that was 1.5–2.0 μm in length and 0.6–0.8 μm in width with a single polar flagellum (Supplementary Figure S1).
PCR amplification was performed to determine serotype of FORC_022 strain using genetic markers of O-serotypes specific genes and toxRS sequences unique to the pandemic O3:K6 serotype of V. parahaemolyticus (Supplementary Figure S2) (Matsumoto et al., 2000; Nasu et al., 2000; Chen et al., 2012; Akther et al., 2016).
Cytotoxicity Test
Cytotoxicity of the FORC_022 strain was evaluated by measuring the activity of cytoplasmic lactate dehydrogenase (LDH) released from INT-407 human epithelial cells (ATCC, Manassas, VA, United States) after the plasma membrane was damaged. INT-407 cells were grown in minimum essential medium containing 1% (v/v) fetal bovine serum (MEMF; Gibco-BRL, Gaithersburg, MD, United States) in 96-well culture dishes (Nunc, Roskilde, Denmark) as described previously (Kim et al., 2014). Cells (2 × 104 cells/well) were infected with FORC_022 or KCTC 2471 (control) at various multiplicities of infection (MOI) for 4 h. The MOI is the ratio for the number of bacterial cells to the number of epithelial cells. V. parahaemolyticus KCTC 2471 strain, which was isolated from patients of food poisoning in Japan, was used as control in LDH assay (Baumann et al., 1971). The LDH activity in the supernatants was determined using a cytotoxicity detection kit (Roche, Mannheim, Germany).
Genome Sequencing and Annotation
Genomic DNA was extracted from cultured strains using a DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, United States) according to the manufacturer’s protocol. Contamination of pure cultured strain was verified using 16S rRNA sequencing, and taxonomic identification was conducted by phylogenetic tree analysis using MEGA6 (Tamura et al., 2013). The phylogenetic tree was constructed using the neighbor-joining method with 1,000 bootstrap replicates. Genome sequencing was conducted at ChunLab, Inc. (Seoul, South Korea) using hybrid sequencing of Illumina MiSeq (Illumina, San Diego, CA, United States) and a PacBio RS II system (Pacific Biosciences, Menlo Park, CA, United States) according to the manufacturers’ protocols. Raw sequences obtained from PacBio RS II were assembled by PacBio SMRT Analysis ver. 2.3.0 software (Pacific Biosciences), and raw sequences from Illumina MiSeq were assembled by CLC Genomics Workbench ver. 7.5.1 (CLC bio, Aarhus, Denmark). The hybrid assembly of generated contigs from both systems was performed using the CodonCode Aligner (CodonCode, Co., Dedham, MA, United States).
Open reading frames (ORFs) and annotations were predicted by the Rapid Annotation using Subsystem Technology (RAST) server (Aziz et al., 2008) and GeneMarkS program (Besemer et al., 2001). The ribosome binding sites were predicted using RBSfinder (J. Craig Venter Institute, Rockville, MD, United States). Subsequent predictions of the functions of ORFs and their conserved protein domains were carried out using InterProScan 5 (Jones et al., 2014) and COG-based WebMGA programs (Wu et al., 2011). The circular genome maps were drawn using the GenVision program (DNASTAR, Madison, WI, United States). The putative virulence factors of FORC_022 were characterized using BLAST against the Virulence Factor Database2 (Chen et al., 2005).
Comparative Genome Analysis
The genome tree was used to determine the closest strain to FORC_022 among completely sequenced V. parahaemolyticus strains (CDC_K4557, ATCC 17802, BB22OP, FDAARGOS_191, RIMD 2210633, 10329, FORC_008, FORC_018, FORC_014, FORC_006, MAVPQ, MAVP-Q, UCM-V493, FORC_023, CHN25, MAVP-R, FORC_004, and FDA_R31) based on ANI values (Supplementary Figure S3). The ANI values were obtained from whole-genome sequences of strains using the JSpecies program (Richter and Rossello-Mora, 2009) by comparing sequences fragmented into 1,020-bp sections based on BLAST analysis. The genome tree was constructed using the R program (3.4.4). A comparative genome analysis between FORC_022 and CDC_K4557 was conducted with the Artemis Comparison Tool (ACT) (Carver et al., 2005). Pangenome analysis of FORC_022 with CDC_K4557 and RIMD 2210633 was conducted using GView Server3.
Artificial Contact With Swimming Crab and RNA Extraction From the FORC_022 Strain
To identify the potential survival mechanism and pathogenicity of FORC_022 contaminating swimming crab during handling or cooking, we artificially exposed crab to the FORC_022 strain. FORC_022 was grown to mid-log phase (A600 of 0.8) in V. fischeri minimal medium containing glycerol and then exposed to crab for 4 h. The V. fischeri minimal medium containing glycerol was used for artificial seawater medium for Vibrio (Cao et al., 2012; Kim et al., 2013). FORC_022 strain without exposure to crabs in the same medium was also prepared as a negative control. These experiments were performed in triplicate. The culture was filtrated with a syringe, sterilized gauze and a vacuum filter with Whatman no.1 filter paper (Whatman International Ltd., Maidstone, England). Subsequently, the filtered product was transferred to 50 mL falcon tubes (SPL, Kyungki, South Korea) and centrifuged at 5,000 × g and 4°C for 10 min. The pellets were resuspended in 0.5 mL cold diethyl phosphorocyanidated-treated phosphate-buffered saline after centrifugation, and the solutions were then mixed with 1 mL RNAprotect Bacteria Reagent (Qiagen). Total RNAs were isolated using a miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. DNA contaminations were removed using TURBO DNase (AMbion, Austin, TX, United States), and extracted RNAs were then cleaned up using an RNeasy MinElute Cleanup kit (Qiagen). The extracted RNA was verified with an Agilent 2100 Bioanalyzer with Agilent RNA 6000 Nano reagents (Supplementary Table S1) (Agilent Technologies, Waldbronn, Germany).
Transcriptome Analysis
To sequence the RNA, mRNA was enriched from extracted total RNA by depleting rRNAs by using a Ribo-ZeroTM rRNA Removal kit (Epicentre, Madison, WI, United States). The cDNA library was constructed from enriched mRNA using a TruSeq Stranded mRNA Sample Preparation kit (Illumina) following the manufacturer’s instructions. The quality of cDNA libraries was determined using an Agilent 2100 Bioanalyzer (Agilent Technologies). Strand-specific paired-ended 100-nucleotide reads from each cDNA library were obtained using HiSeq2500 (Illumina). For biological replication, two libraries were separately constructed and sequenced from RNAs isolated from two independently filtered culture supernatants of FORC_022.
Reads obtained from RNA sequencing were mapped to the FORC_022 reference genome using CLC Genomics Workbench ver. 7.5.1 (CLC Bio). The relative transcript abundance was measured by reads per kilobase of transcript per million mapped sequence reads (Mortazavi et al., 2008). Genes with two or greater fold change with p-values < 0.01 were considered differentially expressed in samples using the CLRNASeqTM program (ChunLab). Transcriptome analysis and visualization of virulence gene data were carried out using Gitools (Perez-Llamas and Lopez-Bigas, 2011).
Quantitative Real Time PCR (qRT-PCR)
Extracted RNA was converted to cDNA for qRT-PCR using an iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, United States). Real-time PCR of cDNA was performed using a Chromo 4 Real-time PCR Detection system (Bio-Rad) with SYBR Green I (Kim et al., 2013). The primers used in this study are listed in Supplementary Table S2. Relative expression levels of specific transcripts were calculated using the 16S rRNA and recA expression level as an internal reference for normalization (Ma et al., 2015). The amplification efficiencies and stability of reference genes were calculated using Delta CT method in Bio-Rad CFX Manager software ver. 3.1 (Supplementary Table S3). qRT-PCR data are presented as mean ± standard deviation of three independent experiments. The differences between groups were determined using two-tailed t-tests in SigmaPlot software (ver. 12.0).
Sequence Deposit in Public Databases
The whole genome sequence of V. parahaemolyticus FORC_022 was deposited in the GenBank of NCBI4 under accession numbers CP013248, CP013249 and CP013250 for chromosomes I, II, and a plasmid, respectively. Transcriptome analysis data of virulence factors in V. parahaemolyticus FORC_022 was deposited in the NCBI Sequence Read Archive (SRA) under the accession number SRP135619.
Results and Discussion
General Genome Features
The FORC_022 strain was isolated from a soy sauce marinated crab and identified as a species of V. parahaemolyticus by phylogenetic tree analysis of the 16S rRNA gene (Supplementary Figure S4). The genome of FORC_022 consisted of two circular chromosomes and a plasmid with 5,379,414 bp with 45.25% GC content (Supplementary Table S4). Chromosome I consisted of 3,397,828 bp with 45.20% GC content and contained 3,066 predicted ORFs, 119 tRNA genes, and 34 rRNA genes. Moreover, 2,423 ORFs (79.02%) were predicted to be functional, and 643 ORFs (17.71%) were predicted to encode hypothetical proteins. Chromosome II consisted of 1,879,989 bp with 45.37% GC content and contained 1,685 predicted ORFs, 14 tRNA genes, and three rRNA genes. Among the ORFs, 1,322 (78.46%) were predicted to be functional, and 363 (21.54%) were predicted to encode hypothetical proteins. The plasmid pFORC22 consisted of 100,597 bp with 44.49% GC content containing 107 predicted ORFs. Of these, 41 ORFs (38.32%) were predicted to be functional, and 66 ORFs (61.68%) were predicted to encode hypothetical proteins. Genome maps of chromosomes and a plasmid are shown in Supplementary Figure S5.
Pathogenesis and Virulence Factors
The cytotoxicity of FORC_022 was compared with KCTC 2471 (=ATCC 33844 = CDC strain KC 824) strain as a positive control, which was isolated from a food poisoning patient (Baumann et al., 1971; Kim et al., 2012; Hossain et al., 2013). The cytotoxicity of FORC_022 was higher than that of KCTC 2471 strain in lactate dehydrogenase (LDH) release assays (Figure 1). However, FORC_022 did not encode major virulence factors such as tdh and trh genes, which are correlates with the Kanagawa phenomenon and induce significant cytotoxicity (Kishishita et al., 1992; Raghunath, 2014). Most strains of V. parahaemolyticus usually contain tdh and trh genes, which have been shown to be associated with pathogenicity, leading to clinical outbreaks (DePaola et al., 2000, 2010; Broberg et al., 2011; Ludeke et al., 2015). The TDH and TRH toxins produced by V. parahaemolyticus can invade the host and disrupt its membrane leading to hemolysis and cytotoxicity (Casandra et al., 2013). However, recent studies reported that V. parahaemolyticus isolated from clinical sources induced cytotoxicity without tdh and/or trh genes (Broberg et al., 2011). In addition, mutant strains of Δtdh/Δtrh show significant cytotoxicity, suggesting that other virulence factors of V. parahaemolyticus may be involved in its pathogenicity (Xu et al., 1994; Park et al., 2004b; Lynch et al., 2005; Pazhani et al., 2014). Consistent with this finding, our results showed that V. parahaemolyticus FORC_022 isolated from a soy sauce marinated crab caused cytotoxicity in human cell lines without the tdh and trh genes. This result indicated that the FORC_022 strain could contain other virulence factors. To identify virulence factors of FORC_022 related to cytotoxicity, the virulence factors were searched for known virulence factors in the Virulence Factor Database (Chen et al., 2005) and annotated information for the FORC_022 genome (Supplementary Table S5).
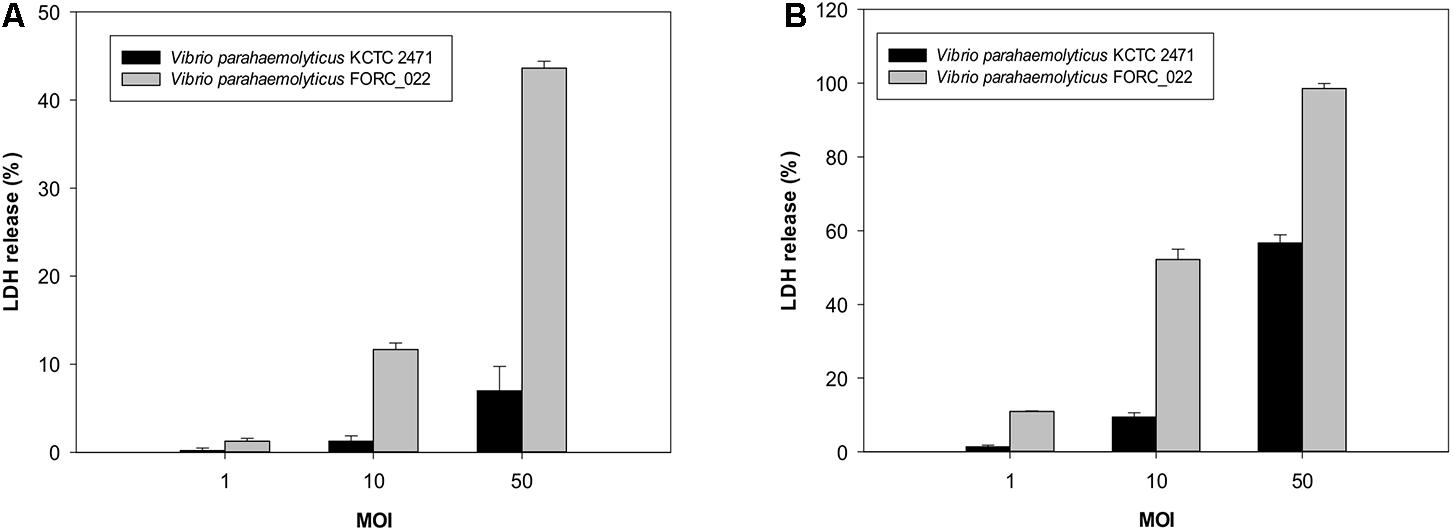
FIGURE 1. Cytotoxicity analyses of the FORC_022 strain were compared with the KCTC 2472 strain by measuring the activity of cytoplasmic lactate dehydrogenase (LDH). INT-407 cells were infected with FORC_022 or KCTC 2471 at various multiplicities of infection (MOIs) for (A) 2 h and (B) 3 h. Cytotoxicity was determined as the percentage of LDH leakage using the amount of LDH from the cells that were completely lysed by 2% Triton X-100. Error bars represent the standard errors of the means.
Several ORFs encoding various hemolysin genes (FORC22_0344, FORC22_0414, FORC22_1716, FORC22_2576, and FORC22_3059 in chromosome I; FORC22_3287, FORC22_3317, and FORC22_3346 in chromosome II) were detected; these genes may be responsible for the virulence of the FORC_022 strain. In addition, various secretion systems including types I, II, III, IV, and VI (T1SS, T2SS, T3SS, T4SS, and T6SS) were detected in the genome sequence of the FORC_022 strain. Type I secretion systems were detected from FORC22_0395, FORC22_1432, FORC22_1615-1620, and FORC22_2067 in chromosome I and FORC22_4335, and FORC22_4733-4736 in chromosome II; these secretion systems are required for the secretion of repeat-in-toxin (RTX; the major virulence factor of V. cholerae) (Boardman et al., 2007). Type II secretion systems were detected from FORC22_2454 to FORC22_2462 in chromosome I and from FORC22_3787 to FORC22_3862 in chromosome II; these secretion systems are required for secretion of the cholera toxin (the major virulence factor of V. cholerae) (Abendroth et al., 2009). Type III secretion systems, which serve several pathogenic functions, such as apoptosis and autophagy (Ono et al., 2006), were detected in a region of chromosome I (FORC22_1641-1687). T3SS has been shown to be involved in the cytotoxicity of V. parahaemolyticus in eukaryotic cells (Park et al., 2004b; Kodama et al., 2007). Moreover, T3SS2 effectors are translocated into the host cell membrane to cause enterotoxicity in colon epithelial cells. Type IV secretion systems, which are associated with effector protein injection machinery by disrupting the actin cytoskeleton or by inducing cell death pathways in host immune cells (Ham et al., 2011), were detected in the region from FORC22_2454 to FORC22_2462 in chromosome I and from FORC22_3787 to FORC22_3862 in chromosome II. Type VI secretion systems act as toxin proteins by delivering bacterial proteins into eukaryotic cells and causing cell death (Costa et al., 2015); these regions were detected from FORC22_1401 to FORC22_1409 in chromosome I and FORC22_4148 to FORC22_4166 in chromosome II. In addition, V. parahaemolyticus has two icmF family genes (icmF1 and icmF2) in type VI secretion systems, which contribute to pathogenicity including adhesion to host epithelial cells and cytotoxicity (Yu et al., 2012). The icmF1 was detected at FORC22_1405 and the icmF2 was detected at 1,086,583–1,086,627 position in chromosome I. Therefore, the FORC_022 may have the potential to induce pathogenesis through these virulence factors. Further studies are necessary to verify the effects of these secretion systems on disease onset.
Iron uptake from host cells can play a key role in survival of V. parahaemolyticus leading to its pathogenicity. Previous study reported that iron loss caused by V. parahaemolyticus affects the integrity of heme protein and leads to host cell death (Hurley et al., 2006). FORC_022 included enterobactin receptors (detected at the FORC22_2639 region on chromosome I and FORC22_3685 region on chromosome II), heme receptors (FORC22_4014 and FORC22_4477 regions on chromosome II), and iron ABC transport (FORC22_3679 to FORC22_3682 on chromosome II) (Supplementary Table S5). These iron uptake-related genes may affect the survival of FORC_022 in host cells.
Comparative Genome Analysis of FORC_022
The genome tree of FORC_022 with other completely sequenced V. parahaemolyticus strains (CDC_K4557, ATCC 17802, BB22OP, FDAARGOS_191, RIMD 2210633, 10329, FORC_008, FORC_018, FORC_014, FORC_006, MAVPQ, MAVP-Q, UCM-V493, FORC_023, CHN25, MAVP-R, FORC_004, and FDA_R31) was obtained based on ANI values (Supplementary Figure S3), and the general features of genomes were compared (Table 1). The highest ANI values (98.54%) were detected for FORC_022 and CDC_K4557 strain (Table 1), the latter of which was isolated from the stool of a patient in Louisiana in 2007 and was submitted to the Centers for Disease Control and Prevention (CDC) (Morrison et al., 2012). Significantly different regions between FORC_022 and CDC_K4557 were detected ranging from positions 1,720,159 to 1,726,760 (FORC22_1562 to FORC22_1576) in chromosome I (Figure 2A and Supplementary Figure S6A). These regions were only detected in the FORC_022 strain and contained the accessory cholera enterotoxin (FORC22_1570) and zona occludens toxin (FORC22_1571), which are associate with colonization and pathogenesis (Tacket et al., 1993; Mukhopadhyay et al., 1995). Accessory cholera enterotoxin contributes to intestinal secretion and diarrhea by stimulating Ca2+-dependent Cl-/HCO3- symporters (Trucksis et al., 2000), whereas zona occludens toxin weakens intestinal tight junctions, leading to body fluid secretion into the intestinal lumen (Fasano et al., 1997). Another different region was detected in chromosome II (ranging from FORC22_3757 to FORC22_3830; positions 750,924-830,901; Figure 2B and Supplementary Figure S6B). The tad locus was detected only in FORC_022 (ranging from FORC22_3784 to FORC22_3797) and has been shown to be related to biofilm formation, colonization, and pathogenicity of strains, thereby leading to several diseases in both humans and animals (Tomich et al., 2007; Morrison et al., 2012). Although the level of cytotoxicity in human cell lines cannot be directly compared between FORC_022 and CDC_K4557 due to unavailability of CDC_K4557, the presence of additional virulence factors may contribute to the cytotoxicity of FORC_022 in various human cells lines. CDC_K4557 did not harbor other virulence factors, such as the tad locus, which mediates the formation of biofilms and facilitates survival by utilizing nutrients from the host and protecting the organism from host immune surveillance detected in Pasteurella multocida and Yersinia ruckeri (Fuller et al., 2000; Fernandez et al., 2004). Although the role of the tad locus in pathogenicity has not yet been clarified, our data demonstrated the existence of the tad locus in V. parahaemolyticus. These results indicated that FORC_022 may be pathogenic to humans since its genome contains several putative virulence factors.
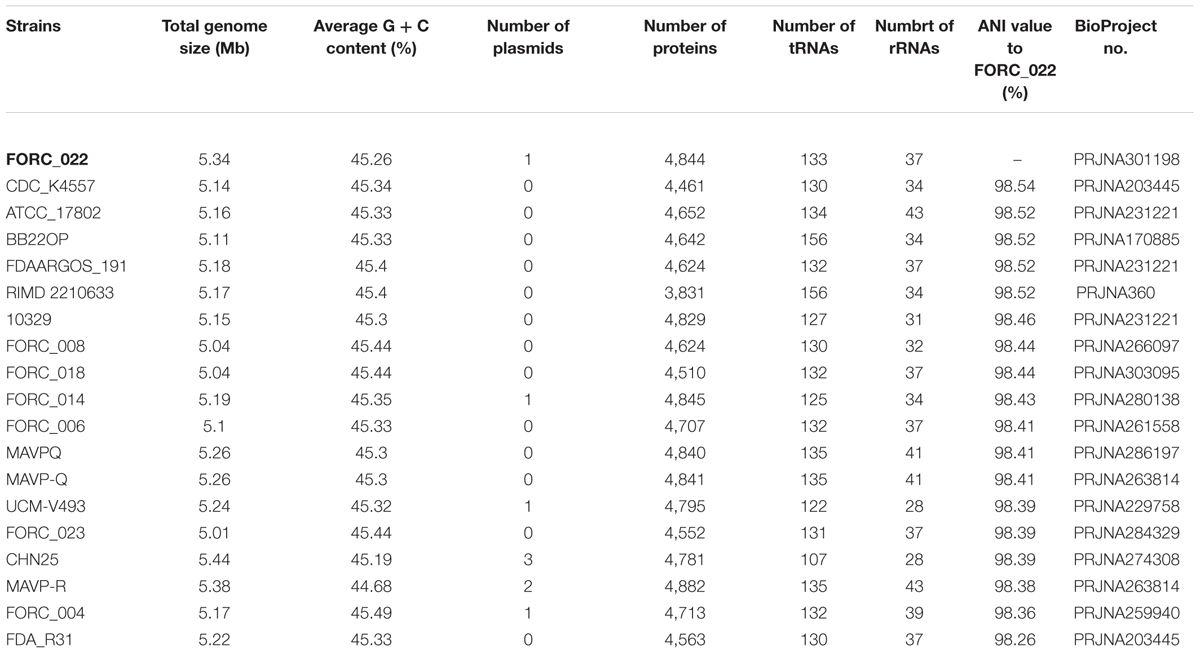
TABLE 1. Genome features of completely sequenced Vibrio parahaemolyticus strains and average nucleotide identity (ANI) values to FORC_022.
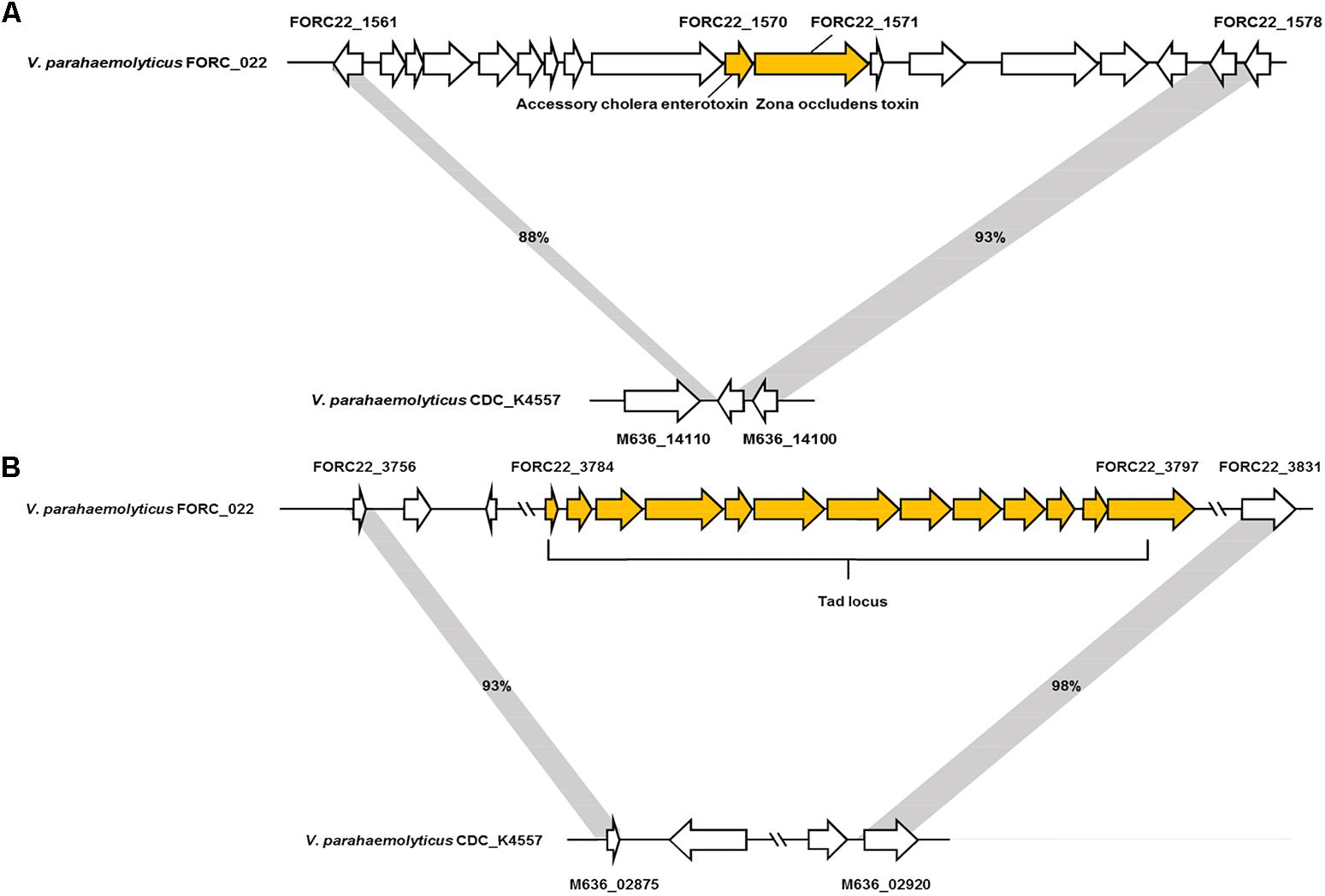
FIGURE 2. Two non-homologous regions between FORC_022 and CDC_K4557 strains. (A) Different regions detected in chromosome I and (B) chromosome II of FORC_022.
VPaIs in FORC_022 and CDC_K4557 were identified by BLAST search against RIMD 2210633 strain, a clinical isolate used to identify genomic islands that could be markers for pandemic clones (Table 2) (Hurley et al., 2006). Four VPaIs were detected in the FORC_022 genome, as follows: VPaI-1 (FORC22_0350-FORC22_0352, FORC22_0358-FORC22_0360, FORC22_0362, FORC22_0365-FORC22_0368, and FORC22_0372-FORC22_0373), VPaI-3 (FORC22_1007-FORC22_1009 and FORC22_1034-FORC22_1041), VPaI-6 (FORC22_4366), and VPaI-7 (FORC22_4405 and FORC22_4406). VPaI-1 was reported to be unique to the pandemic group of V. parahaemolyticus strains isolated after 1995 (Hurley et al., 2006). VPaI-3 has been shown to encode integrase, signal transduction histidine kinase, helicase, methyl accepting chemotaxis protein, AcrBDF family protein as well as hypothetical proteins, and VPaI-6 encode putative virulence genes, such as hydrolases, cytotoxin integrase, and colicins. Broberg et al. (2011) also demonstrated that T3SS2 is encoded by the VPaI region, which is now referred to as VPaI-7. Our results showed that FORC_022 harbored T3SS and VPal-7 as well, accompanied by cytotoxicity in colon epithelial cells, implying that T3SS may be a major virulence factor in the FORC_022 strain. In addition, different virulence factors were detected in pangenome analysis of FORC_022, CDC_K4557, and RIMD22106333 (Figure 3). Three virulence factors of virulence-associated E, Type II/IV secretion systems, and Type IV pilin PilA were detected only in FORC_022. Therefore, VPaIs and additional virulence factors in FORC_022 may play important roles in the pathogenicity of this strain.
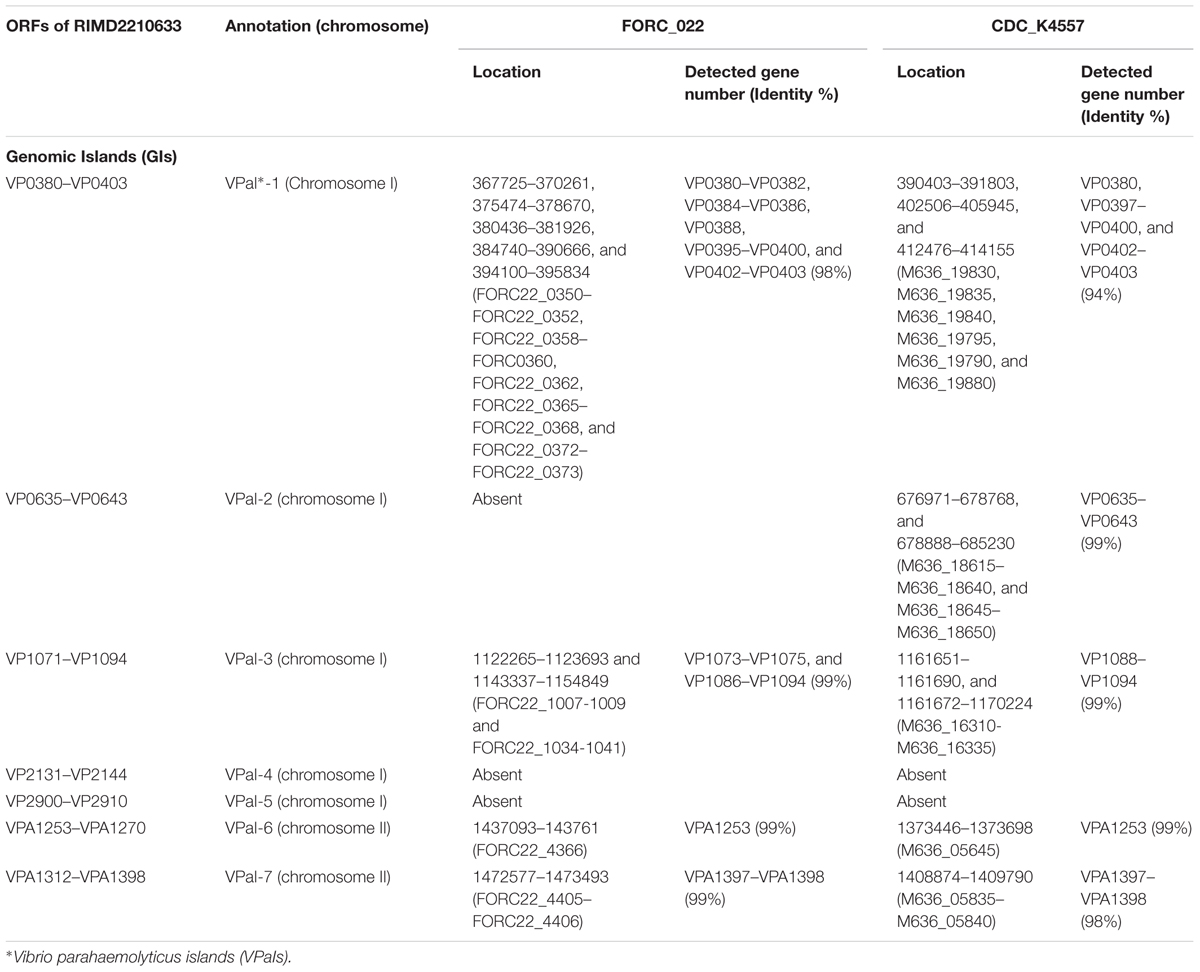
TABLE 2. Genomic islands (GIs) of V. parahaemolyticus FORC_022 and CDC_K4557, predicted by comparing the GIs in V. parahaemolyticus RIMD2210633.
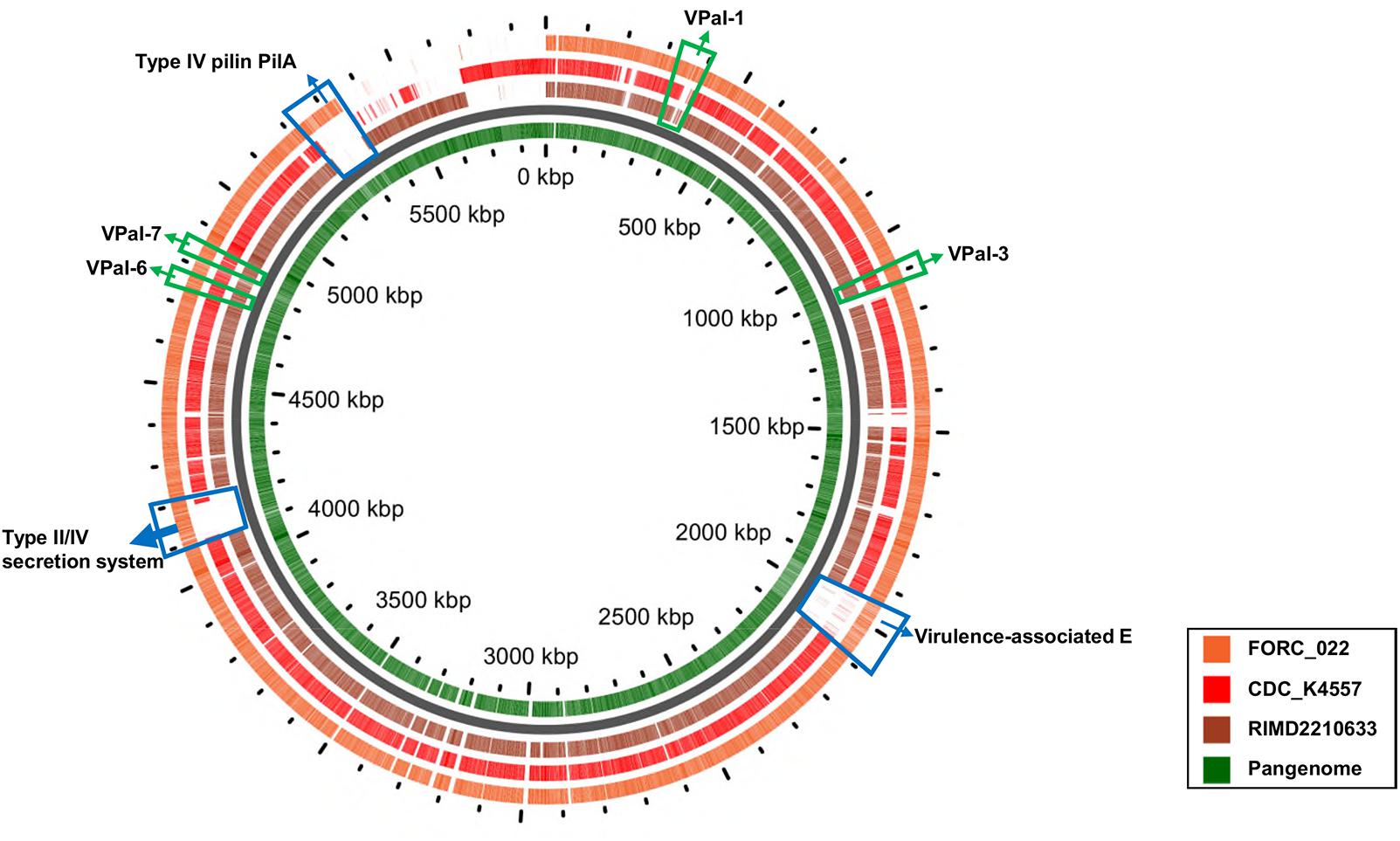
FIGURE 3. A pangenome analysis between three V. parahaemolyticus strains. The central circle belongs to the main bacterial strain, and the outer circles belong to other strains. The innermost circle shows the pangenome (green color), and the outer circles indicate the V. parahaemolyticus strains FORC_022 (orange), CDC_K4557 (red), and RIMD2210633 (dark brown).
Differentially Expressed Genes in FORC_022 After Infection of Crabs
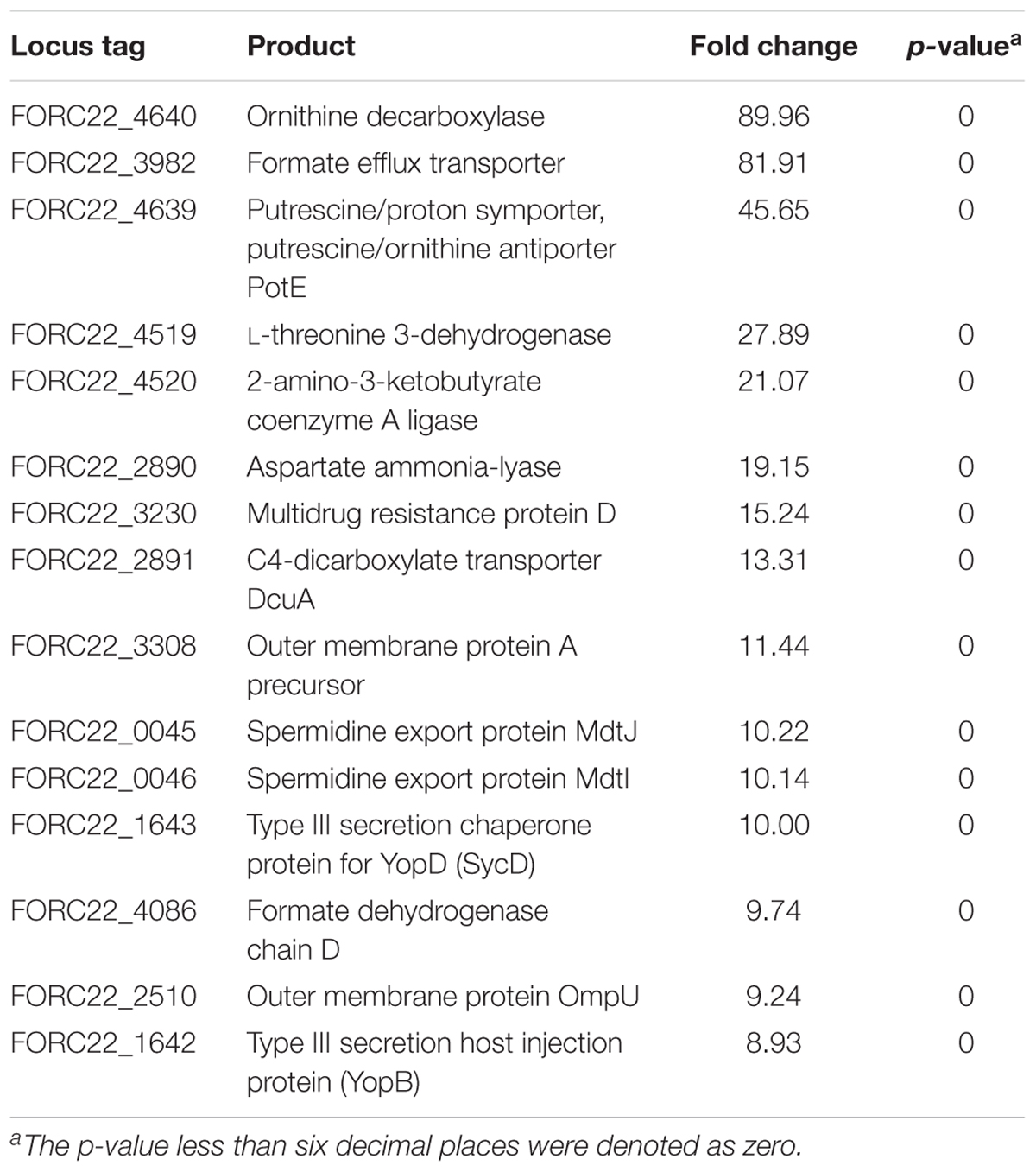
To compare gene expression in strains with or without contact with crabs, FORC_022 was exposed to washed swimming crabs in inoculation with minimal medium (mimic artificial seawater) and incubated for 4 h. The crabs tended to decompose after 6 h; thus, we analyzed samples after 4 h of incubation. Numerous genes were differentially expressed with significance in volcano plot (p-value < 0.01, two fold threshold; Supplementary Figure S7). A total of 1,283 genes were found to be differentially expressed between strains with or without contact with crabs (650 and 633 genes were upregulated and downregulated, respectively). Differently expressed genes were clustered into functionally related groups using the WebMGA server5 with the FORC_022 genome as the reference database (Figure 4). The top 15 overexpressed genes (over 5-fold change) from FORC_022 exposed to crabs are summarized in Table 3, and the top 15 down-regulated genes are presented in Supplementary Table S6.
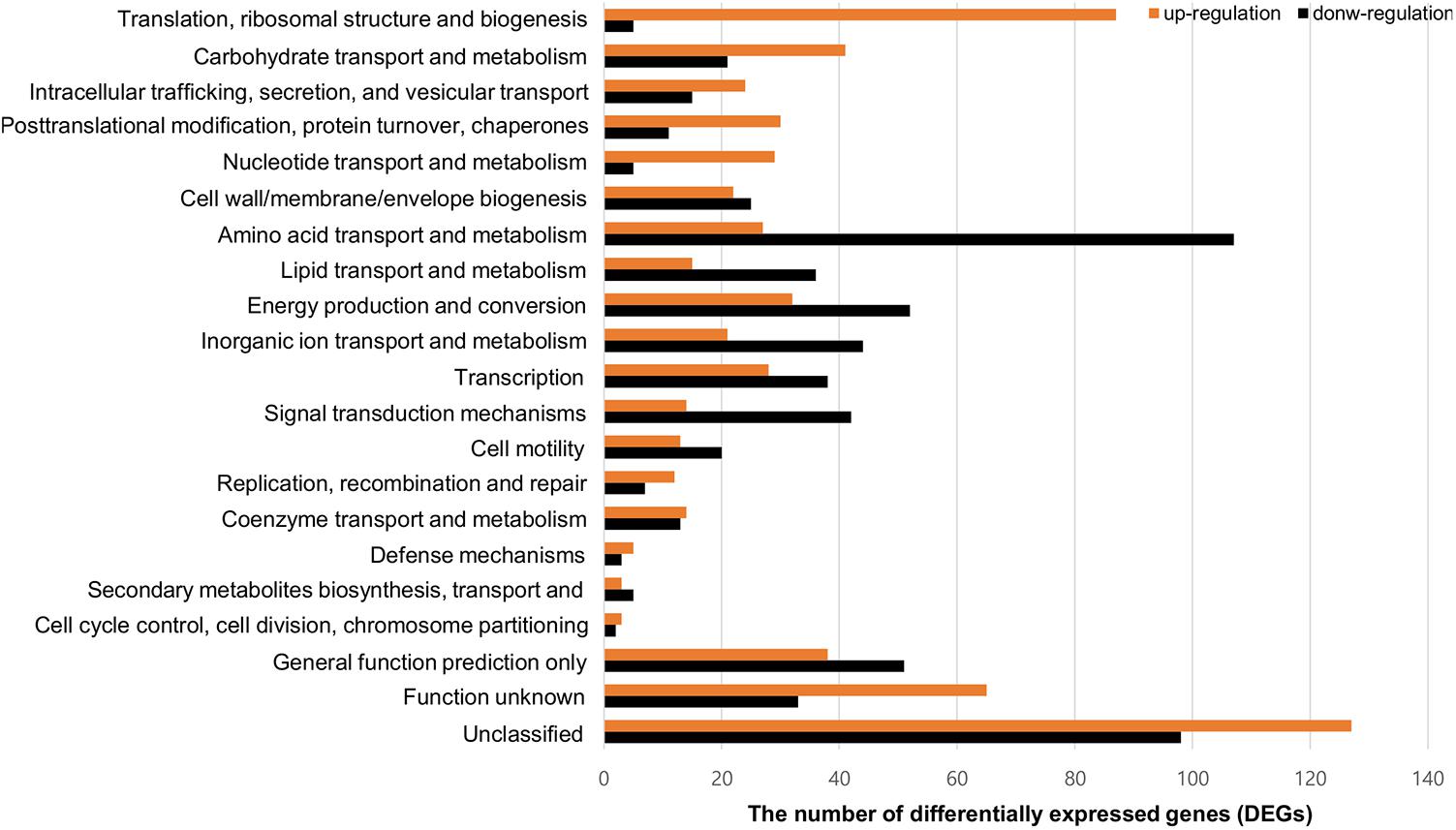
FIGURE 4. Functional analysis of differentially expressed gene (>2-fold change) in the FORC_022 strain exposed to crabs. Functional categories were determined by the Clusters of Orthologous Groups (COG) database. Genes upregulated (orange) and downregulated (black) after exposure to crabs are shown.
The expression levels of genes related to amino acid transport and metabolism and lipid transport and metabolism were lower in the contact strain than in the strain not exposed to crabs, whereas the expression levels of genes related to carbohydrate transport and metabolism were higher in strains after contact with crabs. Genes related to biofilm formation such as tad locus, capsular polysaccharide (CPS), and lipopolysaccharide (LPS) (Yildiz and Visick, 2009), were overexpressed in the strain exposed to crabs. Therefore, FORC_022 could form biofilm to enhance its growth and survival in crabs, establishing a reservoir. To verify the overexpressed genes related to biofilm formation, we performed quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR; Supplementary Figure S8). The amplified efficiencies for qRT-PCR were between 80.9 and 163.2%, and the efficiency curves were found to be linear with correlation coefficients (R2) ranging from 0.919 to 0.999. In addition, virulence factors, such as the type III secretion system, which exhibited cytotoxic activity toward human cells and was related to inflammatory diarrhea and septicemia (Caburlotto et al., 2010; Tamura et al., 2013), were also overexpressed in the strain following contact with crabs (Table 3).
A comparison of expressed virulence genes between strains with and without contacted with crabs is presented in the form of a heatmap (p-value < 0.01; Figure 5A). This result showed that genes related to virulence factors, such as the EPS type II secretion system, type III secretion system, MSHA type IV pilus, thermolabile hemolysin (tlh), and heme receptors, were overexpressed in the contacted strain (Supplementary Table S7). The transcription levels of type III secretion system and tight adhesion genes were confirmed by real-time PCR using the same RNA extracts used for transcriptome analysis (Figures 5B,C). The levels detected by real-time PCR were consistent with the transcriptome results. These results suggested that the potential risk of foodborne illness by ingestion of contaminated crab with FORC_022 could be high due to its virulence factors.
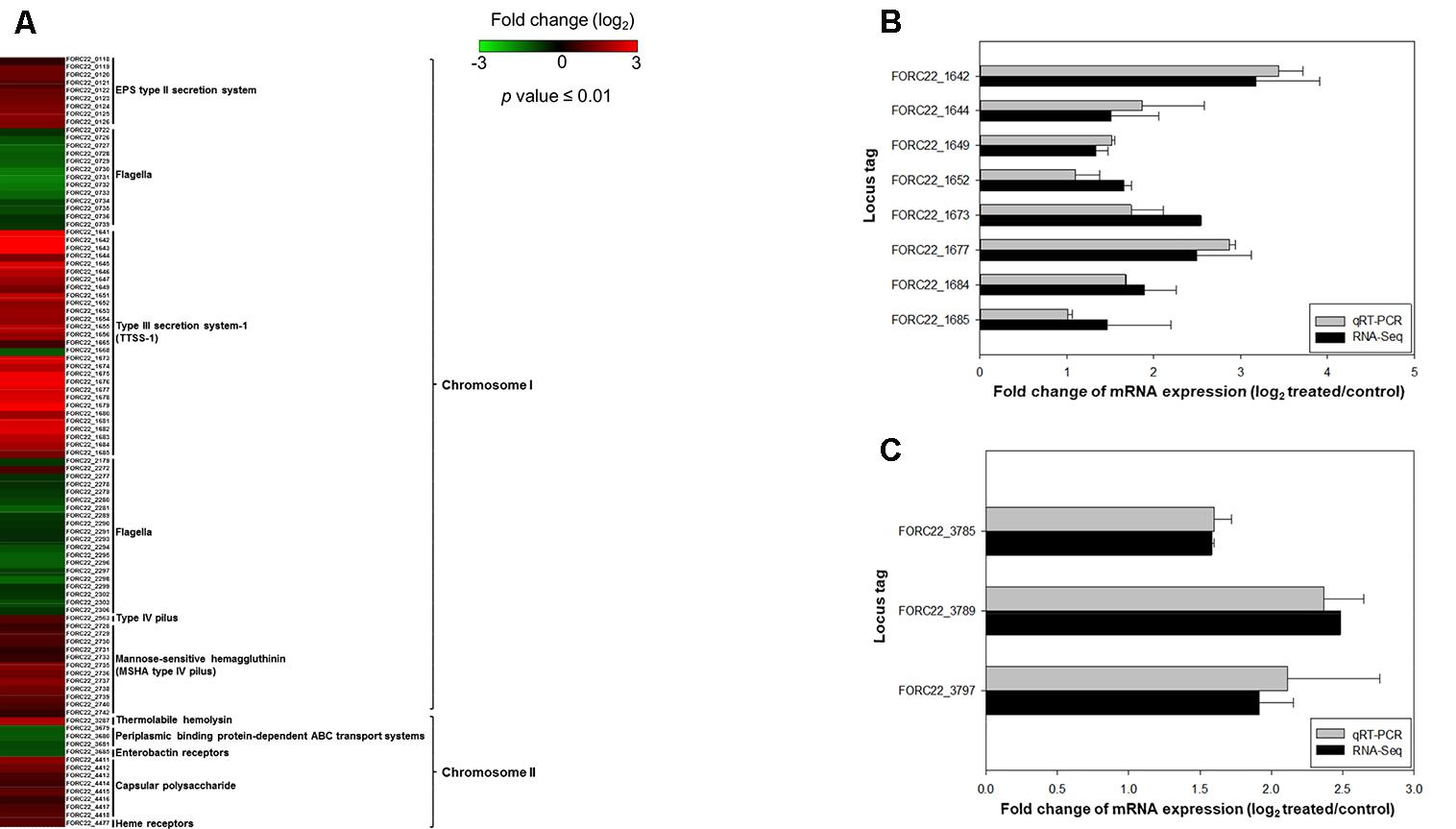
FIGURE 5. The relatively expression of virulence genes after exposure to crabs (p-value ≤ 0.01). (A) Fold changes of virulence genes in chromosomes I and II in heatmap. (B) Relative expression of the type III secretion system was determined by qRT-PCR. (C) Relative gene expression of the tight adhesion gene was determined. Error bars represent standard errors of the means from two independent experiments.
Conclusion
In the present study, we analyzed the complete genome sequences of V. parahaemolyticus FORC_022 strain isolated from soy sauce marinated crabs in South Korea. The genome of FORC_022 did not harbor tdh and trh genes, which are major virulence factors in clinically isolated V. parahaemolyticus. However, the cytotoxicity of FORC_022 was higher than the food poisoning causing strain (KCTC 2471), suggesting that other virulence factors may play a role in the pathogenesis of this infection. FORC_022 had additional virulence factors, such as accessory cholera enterotoxin, zona occludens toxin, and tad locus, compared with CDC_K4557 (the strain most closely related to FORC_022). In addition, the pandemic O3:K6 serotype specific gene (toxRS) and VPaI-1 were detected in FORC_022. Expression levels of adherence-, carbohydrate transporter-, and biofilm formation-related genes increased simultaneously after FORC_022 was exposed to crabs (Table 3). Therefore, this strain could survive and grow on crabs. In addition, several virulence factors, including the type III secretion system, tad locus, and thermolabile hemolysin, were overexpressed (Figure 5); this could lead to a higher risk of infection and pathogenicity (Morrison et al., 2012; Tamura et al., 2013). Although all of results in present study were analyzed by genomic data and gene expression, the genomic and transcriptomic results of FORC_022 provided new insights into our understanding of the molecular mechanisms mediating the survival and pathogenesis of V. parahaemolyticus in the crab products. Further studies including immunology analysis and animal model experiments are necessary to verify the pathogenicity of FORC_022 in present study.
Author Contributions
K-HL, SR, HC, J-HL, HBK, HK, HJ, SC, and B-SK designed the study. HC, BL, and B-SK analyzed the sequencing data. HC, BL, and EN performed the experiments. HC, K-HL, SR, HY, J-HL, HBK, HK, HJ, SC and B-SK discussed the results and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Ministry of Food and Drug Safety, South Korea in 2015 (14162MFDS972), and by the National Research Foundation of Korea, funded by the Ministry of Science, ICT, and Future Planning (2017R1E1A1A01074639 to SC).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01504/full#supplementary-material
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/genome/genomes/691
- ^ http://www.mgc.ac.cn/VFs/main.htm
- ^ https://server.gview.ca/
- ^ http://www.ncbi.nlm.nih.gov/
- ^ http://weizhong-lab.ucsd.edu/metagenomic-analysis/
References
Abendroth, J., Kreger, A. C. Hol, Wim G. J., and Hol, W. G. (2009). The dimer formed by the periplasmic domain of EpsL from the Type 2 secretion system of Vibrio parahaemolyticus. J. Struct. Biol. 168, 313–322. doi: 10.1016/j.jsb.2009.07.022
Akther, F., Neogi, S. B., Chowdhury, W. B., Sadique, A., Islam, A., Akhter, M. Z., et al. (2016). Major tdh+ Vibrio parahaemolyticus serotype changes temporally in the bay of Bengal estuary of Bangladesh. Infect. Genet. Evol. 41, 153–159. doi: 10.1016/j.meegid.2016.04.003
Aziz, R. K. Bartels, D., Best, A., DeJongh, M., Disz, T., Edwards, R. A., and et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Baumann, P., Baumann, L., and Mandel, M. (1971). Taxonomy of marine bacteria: the genus Beneckea. J. Bacteriol. 107, 268–294.
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Boardman, B. K., Meehan, B. M., and Fullner Satchell, K. J. (2007). Growth phase regulation of Vibrio cholerae RTX toxin export. J. Bacteriol. 189, 1827–1835. doi: 10.1128/JB.01766-06
Broberg, C. A., Calder, T. J., and Orth, K. (2011). Vibrio parahaemolyticus cell biology and pathogenicity determinants. Microbes Infect. 13, 992–1001. doi: 10.1016/j.micinf.2011.06.013
Caburlotto, G., Lleo, T. M., Hilton, T., Huq, A., Colwell, R. R., and Kaper, J. B. (2010). Effect on human cells of environmental Vibrio parahaemolyticus strains carrying type III secretion system 2. Infect. Immun. 78, 3280–3287. doi: 10.1128/IAI.00050-10
Cao, X., Studer, S. V., Wassarman, K., Zhang, Y., Ruby, E. G., and Miyashiro, T. (2012). The novel sigma factor-like regulatior RpoQ controls luminescence, chitinase activity, and motility in Vibrio fischeri. MBio 3:e00285-11. doi: 10.1128/mBio.00285-11
Carver, T. J., Rutherford, K. M., Berriman, M., Rajandream, M. A., Barrell, B. G., and Parkhill, J. (2005). ACT: the artemis comparison Tool. Bioinformatics 21, 3422–3423. doi: 10.1093/bioinformatics/bti553
Casandra, K., Gutierrez, W., Klein, S. L., and Lovell, H. R. (2013). High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl. Environ. Microbiol. 79, 2247–2252. doi: 10.1128/AEM.03792-12
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Chen, M., Guo, D., Wong, H., Zhang, X., Liu, F., Chen, H., et al. (2012). Development of O-serogroup specific PCR assay for detection and identification of Vibrio parahaemolyticus. Int. J. Food Microbiol. 159, 122–129. doi: 10.1016/j.ijfoodmicro.2012.08.012
Costa, T. R., Felisberto-Rodrigues, C., Catarina, F. R., Meir, A., Prevost, M. S., Redzej, A., et al. (2015). Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 13, 343–359. doi: 10.1038/nrmicro3456
Daniels, N. A., MacKinnon, L., Bishop, R., Altekruse, S., Ray, B., Hammond, R. M. (2000). Vibrio parahaemolyticus infections in the United States, 1973-1998. J. Infect. Dis. 181, 1661–1666. doi: 10.1086/315459
Delepelaire, P. (2004). Type I secretion in gram-negative bacteria. Biochim. Biophys. Acta 1694, 149–161. doi: 10.1016/j.bbamcr.2004.05.001
DePaola, A., Jones, J. L., Woods, J., Burkhardt, W., Calci, K. R., Krantz, J. A., et al. (2010). Bacterial and viral pathogens in live oysters: 2007 United States market survey. Appl. Environ. Microbiol. 76, 2754–2768. doi: 10.1128/AEM.02590-09
DePaola, A., Kaysner, C. A., Bowers, J., and Cook, D. W. (2000). Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998). Appl. Environ. Microbiol. 66, 4649–4654. doi: 10.1128/AEM.66.11.4649-4654.2000
Fasano, A., Uzzau, S., Fiore, C., and Margaretten, K. (1997). The enterotoxic effect of zonula occludens toxin on rabbit small intestine involves the paracellular pathway. Gastroenterology 112, 839–846. doi: 10.1053/gast.1997.v112.pm9041245
Fernandez, L., Marquez, I., and Guijarro, J. A. (2004). Identification of specific in vivo-induced (ivi) genes in Yersinia ruckeri and analysis of ruckerbactin, a catecholate siderophore iron acquisition system. Appl. Environ. Microbiol. 70, 5199–5207. doi: 10.1128/AEM.70.9.5199-5207.2004
Fuller, T. E., Kennedy, M. J., and Lowery, D. E. (2000). Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29, 25–38. doi: 10.1006/mpat.2000.0365
Haendiges, J., Timme, R., Allard, M. W., Myers, R. A., Brown, E. W., and Gonzalez-Escalona, N. (2015). Characterization of Vibrio parahaemolyticus clinical strains from Maryland (2012-2013) and comparisons to a locally and globally diverse V. parahaemolyticus strains by whole-genome sequence analysis. Front. Microbiol. 6:125. doi: 10.3389/fmicb.2015.00125
Ham, H., Sreelatha, A., and Orth, K. (2011). Manipulation of host membranes by bacterial effectors. Nat. Rev. Microbiol. 9, 635–646. doi: 10.1038/nrmicro2602
Hazen, T. H., Lafon, P. C., Garrett, N. M., Lowe, T. M., Silberger, D. J., Rowe, L. A., et al. (2015). Insights into the environmental reservoir of pathogenic Vibrio parahaemolyticus using comparative genomics. Front. Microbiol. 6:204. doi: 10.3389/fmicb.2015.00204
Hossain, M. T., Kim, Y. O., and Kong, I. S. (2013). Multiplex PCR for the detection and differentiation of Vibrio parahaemolyticus strains using the groEL, tdh and trh genes. Mol. Cell. Probes 27, 171–175. doi: 10.1016/j.mcp.2013.04.001
Hurley, C. C., Quirke, A., Reen, F. J., and Boyd, E. F. (2006). Four genomic islands that mark post-1995 pandemic Vibrio parahaemolyticus isolates. BMC Genomics 7:104. doi: 10.1186/1471-2164-7-104
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., McWilliam, H., and et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Joseph, S. W., Colwell, R. R., and Kaper, J. B. (1982). Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 10, 77–124. doi: 10.3109/10408418209113506
Kim, J. H., Jun, J. W., Choresca, C. H., Shin, S. P., Han, J. E., and Park, S. C. (2012). Complete genome sequence of a novel marine Siphovirus, pVp-1, infecting Vibrio parahaemolyticus. J. Virol. 86, 7013–7014. doi: 10.1128/JVI.00742-12
Kim, S., Bang, Y. J., Lim, J. G., Oh, M. H., and Choi, S. H. (2014). Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Mol. Microbiol. 93, 992–1009. doi: 10.1111/mmi.12712
Kim, S. M., Park, J. H., Lee, H. S., Kim, W. B., Ryu, J. M., Han, H. J., et al. (2013). LuxR homologue SmcR is essential for Vibrio vulnificus pathogenesis and biofilm detachment, and its expression is induced by host cells. Infect. Immun. 81, 3721–3730. doi: 10.1128/IAI.00561-13
Kishishita, M., Matsuoka, N., Kumagai, K., Yamasaki, S., Takeda, Y., and Nishibuchi, M. (1992). Sequence variation in the thermostable direct hemolysin-related hemolysin (trh) gene of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 58, 2449–2457.
Kodama, T., Rokuda, M., Park, K. S., Cantarelli, V. V., Matsuda, S., Iida, T., and et al. (2007). Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell. Microbiol. 9, 2598–2609. doi: 10.1111/j.1462-5822.2007.00980.x
Letchumanan, V., Chan, K. G., Pusparajah, P., Saokaew, S., Duangjai, A., Goh, B. H. et al. (2016). Insights into bacteriophage application in controlling Vibrio species. Front. Microbiol. 7:1114. doi: 10.3389/fmicb.2016.01114
Letchumanan, V., Pusparajah, P., Tan, L. T. H., Yin, W. F., Lee, L. H., and Chan, K. G. (2015a). Occurrence and antibiotic resistance of Vibro parahaemolyticus from shellfish in Selangor, Malaysia. Front. Microbiol. 6:1417. doi: 10.3389/fmicb.2015.01417
Letchumanan, V., Yin, W. F., Lee, L. H., and Chan, K. G. (2015b). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Li, H., Tang, R., Lou, Y., Cui, Z., Chen, W., Hong, Q., et al. (2017). A comprehensive epidemiological research for clinical Vibrio parahaemolyticus in Shanghai. Front. Microbiol. 8:1043. doi: 10.3389/fmicb.2017.01043
Ludeke, C. H., Kong, N., Weimer, B. C., Fischer, M., and Jones, J. L. (2015). Complete genome sequences of a clinical isolate and an environmental isolate of Vibrio parahaemolyticus. Genome Announc. 3:e00216-15. doi: 10.1128/genomeA.00216-15
Lynch, T., Livingstone, S., Buenaventura, E., Fedwick, J., Buret, A. G., Graham, D., and et al. (2005). Vibrio parahaemolyticus disruption of epithelial cell tight junctions occurs independently of toxin production. Infect. Immun. 73, 1275–1283. doi: 10.1128/IAI.73.3.1275-1283.2005
Malcom, T. T. H., Cheah, Y. K., Radzi, C. W. J. W. M., Kasim, F. A., Kantilal, H. K., John, T. Y. H., et al. (2015). Detection and quantification of pathogenic Vibrio parahaemolyticus in shellfish by using multiplex PCR and loop-mediated isothermal amplification assay. Food Control 47, 664–671. doi: 10.1016/j.foodcont.2014.08.010
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., and et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585.
Ma, Y. J., Sun, X., Xu, X., Zhao, Y., Pan, Y., Hwang, C., et al. (2015). Investigation of reference genes in Vibrio parahaemolyticus for gene expression analysis using quantitative RT-PCR. PLoS One 10:e0144362. doi: 10.1371/journal.pone.0144362
Morrison, S. S., Williams, T., Cain, A., Froelich, B., Taylor, C., Baker-Austin, C., et al. (2012). Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One 7:e37553. doi: 10.1371/journal.pone.0037553
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mukhopadhyay, A. K., Saha, P. K., Garg, S., Bhattacharya, S. K., Shimada, T., Takeda, T., et al. (1995). Distribution and virulence of Vibrio cholerae belonging to serogroups other than O1 and O139: a nationwide survey. Epidemiol. Infect. 114, 65–70. doi: 10.1017/S0950268800051918
Nasu, H., Iida, T., Suqahara, T., Yamaichi, Y., Park, K. S., Yokoyama, K., et al. (2000). A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38, 2156–2161.
Newton, A. E., Garrett, N., Stroika, S. G., Halpin, J. L., Turnsek, M., and Mody, R. K. (2014). Increase in Vibrio parahaemolyticus infections associated with consumption of Atlantic Coast shellfish-2013. Morb. Mortal. Wkly. Rep. 63, 335–336.
Ono, T., Park, K. S., Ueta, M., Iida, T., and Honda, T. (2006). Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect. Immun. 74, 1032–1042. doi: 10.1128/IAI.74.2.1032-1042.2006
Ottaviani, D., Leoni, F., Serra, R., Serracca, L., Decastelli, L., Rocchegiani, E., et al. (2012). Nontoxigenic Vibrio parahaemolyticus strains causing acute gastroenteritis. J. Clin. Microbiol. 50, 4141–4143. doi: 10.1128/JCM.01993-12
Pallen, M. J., Chaudhuri, R. R., and Henderson, I. R. (2003). Genome analysis of secretion systems. Curr. Opin. Microbiol. 6, 519–527. doi: 10.1016/j.mib.2003.09.005
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Iida, T., and Honda, T. (2004a). Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol. Immunol. 48, 313–318. doi: 10.1111/j.1348-0421.2004.tb03512.x
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Iida, T., and Honda, T. (2004b). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004
Pazhani, G. P., Bhowmik, S. K., Ghosh, S., Guin, S., Dutta, S., Rajendran, K., et al. (2014). Trends in the epidemiology of pandemic and non-pandemic strains of Vibrio parahaemolyticus isolated from diarrheal patients in Kolkata, India. PLoS Negl. Trop. Dis. 8:e2815. doi: 10.1371/journal.pntd.0002815
Perez-Llamas, C., and Lopez-Bigas, N. (2011). Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One 6:e19541. doi: 10.1371/journal.pone.0019541
Raghunath, P. (2014). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5, 2010–2013. doi: 10.3389/fmicb.2014.00805
Ritchie, J. M., Rui, H., Zhou, X., Iida, T., Kodoma, T., Ito, S., et al. (2012). Inflammation and disintegration of intestinal villi in an experimental model for Vibrio parahaemolyticus-induced diarrhea. PLoS Pathog. 8:e1002593. doi: 10.1371/journal.ppat.1002593
Rodgers, C., Parveen, S., Chigbu, P., Jacobs, J., Rhodes, M., Harter-Dennis, J., et al. (2014). Prevalence of Vibrio parahaemolyticus, and Vibrio vulnificus in blue crabs (Callinectes sapidus), seawater and sediments of the Maryland Coastal Bays. J. Appl. Microbiol. 117, 1198–1209. doi: 10.1111/jam.12608
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Tacket, C. O., Losonsky, G., Nataro, J. P., Cryz, S. J., Edelman, R., Fasano, A., et al. (1993). Safety and immunogenicity of live oral cholera vaccine candidate CVD 110, a delta ctxA delta zot delta ace derivative of El Tor Ogawa Vibrio cholerae. J. Infect. Dis. 168, 1536–1540. doi: 10.1093/infdis/168.6.1536
Tomich, M., Planet, P. J., and Figurski, D. H. (2007). The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375. doi: 10.1038/nrmicro1636
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Trucksis, M., Conn, T. L., Wasserman, S. S., and Sears, C. L. (2000). Vibrio cholerae ACE stimulates Ca(2+)-dependent Cl(-)/HCO(3)(-) secretion in T84 cells in vitro. Am. J. Physiol. Cell Physiol. 279, C567–C577. doi: 10.1152/ajpcell.2000.279.3.C567
Velazquez-Roman, J., León-Sicairos, N., Flores-Villaseñor, H., Villafaña-Rauda, S., and Canizalez-Romen, A. (2012). Association of pandemic Vibrio parahaemolyticus O3:K6 present in the coastal environment of Northwest Mexico with cases of recurrent diarrhea between 2004 and 2010. Appl. Environ. Microbiol. 78, 1794–1803. doi: 10.1128/AEM.06953-11
Wang, R., Zhong, Y., Gu, X., Yuan, J., Saeed, A. F., and Wang, S. (2015). The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 6:144. doi: 10.3389/fmicb.2015.00144
Wu, S., Zhu, Z., Fu, L., Niu, B., and Li, W. (2011). WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12:444, doi: 10.1186/1471-2164-12-444
Wu, Y., Wen, J., Ma, Y., Ma, X., and Chen, Y. (2014). Epidemiology of foodborne disease outbreaks caused by Vibrio parahaemolyticus, China, 2003-2008. Food Control 46, 197–202. doi: 10.1016/j.foodcont.2014.05.023
Xu, M., Yamamoto, K., Honda, T., and Ming, X. (1994). Construction and characterization of an isogenic mutant of Vibrio parahaemolyticus having a deletion in the thermostable direct hemolysin-related hemolysin gene (trh). J. Bacteriol. 176, 4757–4760. doi: 10.1128/jb.176.15.4757-4760.1994
Xu, X., Wu, Q., Zhang, J., Cheng, J., Zhang, S., and Wu, K. (2014). Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Control 46, 81–85. doi: 10.1016/j.foodcont.2014.04.042
Yildiz, F. H., and Visick, K. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118. doi: 10.1016/j.tim.2008.12.004
Keywords: Vibrio parahaemolyticus, whole genome sequencing, genomic comparison, transcriptome, virulence factors, crab, FORC_022
Citation: Chung HY, Lee B, Na EJ, Lee K-H, Ryu S, Yoon H, Lee J-H, Kim HB, Kim H, Jeong HG, Kim B-S and Choi SH (2018) Potential Survival and Pathogenesis of a Novel Strain, Vibrio parahaemolyticus FORC_022, Isolated From a Soy Sauce Marinated Crab by Genome and Transcriptome Analyses. Front. Microbiol. 9:1504. doi: 10.3389/fmicb.2018.01504
Received: 03 April 2018; Accepted: 18 June 2018;
Published: 06 July 2018.
Edited by:
Jennifer Ronholm, McGill University, CanadaReviewed by:
Adrian Canizalez-Roman, Autonomous University of Sinaloa, MexicoLearn-Han Lee, Monash University Malaysia, Malaysia
Copyright © 2018 Chung, Lee, Na, Lee, Ryu, Yoon, Lee, Kim, Kim, Jeong, Kim and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bong-Soo Kim, YmtpbTc5QGhhbGx5bS5hYy5rcg== Sang H. Choi, Y2hvaXNoQHNudS5hYy5rcg==
 Han Y. Chung
Han Y. Chung Byungho Lee
Byungho Lee Eun J. Na
Eun J. Na Kyu-Ho Lee
Kyu-Ho Lee Sangryeol Ryu
Sangryeol Ryu Hyunjin Yoon
Hyunjin Yoon Ju-Hoon Lee
Ju-Hoon Lee Hyeun B. Kim
Hyeun B. Kim Heebal Kim
Heebal Kim Hee G. Jeong2,8
Hee G. Jeong2,8 Bong-Soo Kim
Bong-Soo Kim Sang H. Choi
Sang H. Choi