- 1Data Analytics Unit, bioMérieux, La Balme-les-Grottes, France
- 2Strategic Intelligence, Business Development Direction, bioMérieux, Marcy-l’Étoile, France
Staphylococcus aureus is an opportunistic pathogen of animals and humans that is capable of both colonizing and infecting its eukaryotic host. It is frequently detected in the clinical microbiology routine laboratory. S. aureus is capable of acquiring antibiotic resistance traits with ease and, given its rapid global dissemination, resistance to meticillin in S. aureus has received extensive coverage in the popular and medical press. The detection of meticillin-resistant versus meticillin-susceptible S. aureus (MRSA and MSSA) is of significant clinical importance. Detection of meticillin resistance is relatively straightforward since it is defined by a single determinant, penicillin-binding protein 2a’, which exists in a limited number of genetic variants carried on various Staphylococcal Cassette Chromosomes mec. Diagnosis of MRSA and MSSA has evolved significantly over the past decades and there has been a strong shift from culture-based, phenotypic methods toward molecular detection, especially given the close correlation between the presence of the mec genes and phenotypic resistance. This brief review summarizes the current state of affairs concerning the mostly polymerase chain reaction-mediated detection of MRSA and MSSA in either the classical laboratory setting or at the point of care. The potential diagnostic impact of the currently emerging whole genome sequencing (WGS) technology will be discussed against a background of diagnostic, surveillance, and infection control parameters. Adequate detection of MSSA and MRSA is at the basis of any subsequent, more generic antibiotic susceptibility testing, epidemiological characterization, and detection of virulence factors, whether performed with classical technology or WGS analyses.
Introduction
Detection of infectious agents and their diseases is performed through a wide array of diagnostic methodologies. These range from in silico methods assessing host’ susceptibility to colonization and infection (Suh et al., 2018) to direct or indirect detection of the pathogen itself. The latter tests are collectively known as in vitro diagnostics (IVD) and their execution requires a qualified laboratory environment and highly educated technicians and (clinical) microbiologists. Next to the laboratory-based tests there are also more simple formats that should be safe to use outside of the laboratory by trained non-professionals at the point-of-need (PoN) or point-of-care (PoC). The more popular diagnostic tools are increasingly molecular in nature, having speed, specificity, and sensitivity superior to those of more classical, culture-based technologies. Molecular tests are based on different principles of which direct hybridization is among the most ancient. In addition, several different nucleic acid amplification technologies (NAATs) have been implemented [see Muldrew (2009) for a review]. Post-amplification processing often includes DNA fragment analysis and/or sequencing. Such tests are mostly aiming at detection and identification of disease-invoking bacterial species. Of note, primary detection and identification of micro-organisms are obvious pre-requisites to their further epidemiological characterization or research into their resistance and virulence characteristics. Complete diagnostic data sets can then be used for curing patients or for prevention of cross infection and infection control.
In the current era of multi-to-pan antibiotic resistance, there is an increasing interest in microbial tests that detect antibiotic resistance, one of the current medical scourges (Okeke et al., 2011; Kelly et al., 2016). Although phenotypic analysis is our heritage, optimal molecular diagnosis should allow for the simultaneous detection, identification, and genetic antibiotic susceptibility testing (AST) of infectious agents. DNA sequences at the species level and at the level of resistance genes can be amplified at the same time in the same assay using the same clinical material as source (see Figure 1 for a conceptual explanation). Indirect AST results in the detection of resistance markers and should lead to targeted treatment on the basis of the presumed activity of the product for which only the gene was detected. When innovative analytical techniques such as mass spectrometry, liquid chromatography, and nucleic acid sequencing are included advices on treatment may become more encompassing and cover-all.
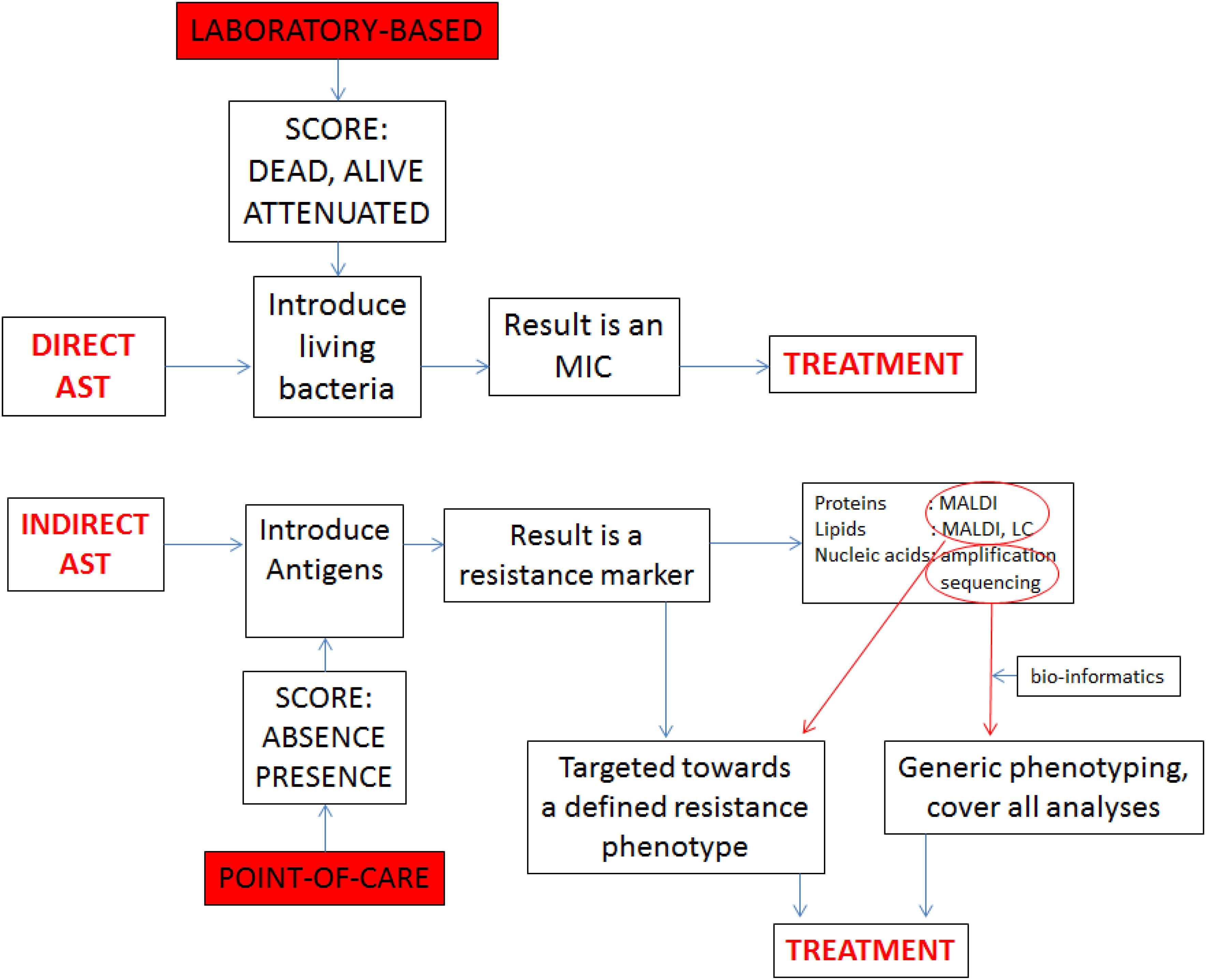
FIGURE 1. Comparison between direct AST and indirect AST, showing the difference between the laboratory based and PoC protocols. Whereas the laboratory exercise generates precise minimum inhibitory concentrations (MICs), the indirect approach generates markers that need to be associated with therapeutic modalities through bio-informatics.
Here, we will focus on the detection of meticillin resistance in the bacterial species Staphylococcus aureus as a model for the recent evolution of and the huge value of AST in clinical care. Meticillin-resistant S. aureus (MRSA) generates twice as much mortality than methicillin-susceptible S. aureus (MSSA) (Turnidge et al., 2009) and rapid molecular diagnostics has already been shown to reduce hospital stay and costs associated with MRSA infection (Brown and Paladino, 2010). Detecting both MSSA and MRSA is important since it guides therapeutic interventions with optimal antibiotics (Liu et al., 2011). Successful molecular diagnostic tests were developed by individual researchers (in house technology) but also by the IVD industry. In addition, quantification of the absolute number of bacterial cells in a clinical specimen is also important since different bacterial titers may be involved in colonization versus infection of human individuals. We review nucleic acid-based tests that have been made recently available for the detection of MSSA and MRSA. When such tests are correctly used they do facilitate subsequent studies into the epidemiology, evolution, and spread of both MSSA and MRSA. Detailed assessment of resistance to a wider spectrum of antimicrobial agents can be performed and implementation of enhanced infection control becomes an option.
Culture-Based Detection of MSSA and MRSA
There is no way of discussing molecular detection of MSSA and MRSA without briefly sketching the pre- and peri-molecular diagnostic landscape. Traditionally, staphylococcal colonization, and infection were diagnosed using culture-based technologies. These either employ generic, highly fertile culture media coupled to downstream bacterial species identification or species-specific enrichment media containing S. aureus selective components such as elevated salt concentrations. Addition of chromogenic compounds in the medium helps to identify S. aureus on the basis of colony morphology and color (Perry, 2017). Further taxonomic classification and identification of S. aureus can be done via agglutination assays or (commercially available) biochemical reactivity (e.g., API strips). Using combinations of simple phenotypic tests has been shown to allow for the adequate distinction of MSSA and MRSA (Verroken et al., 2016; Lüthje et al., 2017; Rees and Barr, 2017; Ábrók et al., 2018). Recent immunochromatographic methods such as the BinaxNOW (Alere, Scarborough, ME, United States) and the Clearview Exact PBP2a assay (Alere, Scarborough, ME, United States) have acquired a good position in the clinical laboratory given their modest price, rapidity, and good sensitivity and specificity (e.g., Kong et al., 2014). Still, modern laboratories now consider matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI ToF MS) as Gold Standard for staphylococcal identification (Bernardo et al., 2002; Szabados et al., 2010; Zhu et al., 2015). Distinguishing MRSA from MSSA using MALDI ToF MS is controversial with positive reports (Ueda et al., 2015; Rhoads et al., 2016; Sogawa et al., 2017) alternating with more negative ones (Du et al., 2002; Goldstein et al., 2013). Recent papers document the successful distinction between MSSA and MRSA, even including characterization of different MRSA clones (Østergaard et al., 2015; Zhang et al., 2015; Camoez et al., 2016). The main issue with these studies is that MALDI TOF MS detects proteins and that it is claimed that the mecA protein is produced in small amounts, often impossible to detect even by targeted mass spectrometry methods. So there is a significant risk that MALDI TOF MS will detect surrogate markers that also distinguish MRSA from MSSA. These may be markers of clonality rather than methicillin resistance and for this reason the collection of (preferably epidemiologically non-related) strains is of key importance. However, the overall impression is that MALDI ToF MS can be useful in the field of bacterial epidemiology but it will certainly not provide a universal “typing” solution. Advanced MS methods, including for instance electron spray ionization (ESI) MS, may bring more universal solutions but these methods are too cumbersome, too time-consuming and too expensive at this stage (Charretier et al., 2015). Although MS essentially provides molecular testing, albeit at the protein level, NAATs still provide the best tool for distinguishing MSSA and MRSA.
Short Introduction into Molecular Technologies
Using polymerase chain reaction (PCR), nucleic acid sequence-based amplification (NASBA), recombinase polymerase amplification (RPA), loop-mediated isothermal amplification (LAMP) and other systems, minute amounts of DNA can be amplified and detected using a variety of technological formats (Almassian et al., 2013; Yan et al., 2014). All of these methods have been shown to be useful in the detection of infectious agents and both PCR and several non-PCR tests have been commercialized (e.g., the LAMP-based Eazyplex test, see Henares et al., 2017). New methods surface regularly (e.g., those methods employing the specificity, sensitivity of enzymes involved in CRISPCas-mediated bacterial immunity to bacteriophages, see Gootenberg et al., 2017, 2018, for more details), several of them allowing genetic AST, and this import of new testing formats including their automatization will continue in the years to come.
There is a set of technologies that will undoubtedly have a huge impact on microbial detection and characterization: next generation sequencing (NGS) leading to the elucidation of the primary structure of complete bacterial chromosome sequences. Multiple elegant whole genome sequencing (WGS) NGS technologies have been developed three of which are currently commercialized and well-accessible to the diagnostic laboratory. Companies such as Illumina (San Diego, CA, United States), PacBio (Menlo Park, CA, United States), and Oxford Nanopore (Oxford, United Kingdom) provide exemplary methodologies suited for WGS. Further technical and usage detail on these methods will not be provided here but can be easily accessed through various recent reviews (Miyamoto et al., 2014; Quainoo et al., 2017; Rossen et al., 2017). NGS will find its way into the clinical microbiology routine laboratory over the years to come where it will fill in important niches in the rapid detection of pathogens and their epidemiological, antibiotic resistance, and virulence characteristics, possibly directly from clinical specimens. NGS will allow parallel sequencing of host DNA and define the host’s susceptibility to certain diseases. High throughput sequencing of RNA will allow for more precise expression monitoring via transcriptomics.
Realistically speaking though, we currently dispose of two main techniques for distinction between MSSA and MRSA: those targeting specific diagnostic signature sequences and those that characterize entire chromosomes and then depend on bio-informatic analyses to highlight the presence of the same sequence motifs used by the specific methods (Figure 1).
Targeted Genetic Detection of MSSA and MRSA
It needs to be realized that for the molecular detection of MRSA there should be a differentiation between screening tests (for carriage) and hard-core diagnostic tests for infection (Osiecki, 2010; Trouillet-Assant et al., 2013). Both tests have different requirements for sensitivity, specificity, costs, and speed. Screening may not need high-speed but must be focused on specific detection of high-rate carriers (Bode et al., 2010). A major hurdle to developing molecular MRSA-specific tests is the fact that the gene encoding meticillin resistance occurs in other staphylococcal species as well. A solution to this issue is the inclusion of species-specific assays in the amplification reaction (e.g., targeting the nuc or femA genes). The second hurdle is that the mec gene is present in four variants (mecA, mecC, and the more recently discovered mecB (Gómez-Sanz et al., 2015; Becker et al., 2018) and mecD (Schwendener et al., 2017)) and these reside in a growing number of genetic islands (Kolenda et al., 2017); mec genes are embedded in various Staphylococcal Cassette Chromosome mec (SCCmec) for which more than 10 different types have already been identified (Hill-Cawthorne et al., 2014; Kaya et al., 2018). Molecular tests for rapid discrimination of SCCmec types continue to be developed though their use is mostly of epidemiological rather than clinical value (Brukner et al., 2013). The mecC variant, mostly found in livestock associated MRSA and sharing about 70% sequence homology with mecA, was discovered as recently as 2011 (García-Álvarez et al., 2011). Finally, rapidity of a test can be affected by whether or not a test is (semi-)quantitative and performed in real-time or not (Verhoeven et al., 2012). The need for real-time testing differs per clinical application but in case of sepsis detection, for instance, speed is of utmost importance (Frye et al., 2012). Modern MRSA detection obviously is part of multiplexed, syndrome-oriented diagnostic testing (Blaschke et al., 2012; Ramanan et al., 2017).
A variety of experimental testing formats has been suggested for targeted MRSA detection but most of which have not reached the diagnostic market (yet). Table 1 reviews the status of a significant number of current PCR tests and highlights a domain of importance: the next generation routine-applicable tests may very well-originate from this pool of potentially high throughput tools.
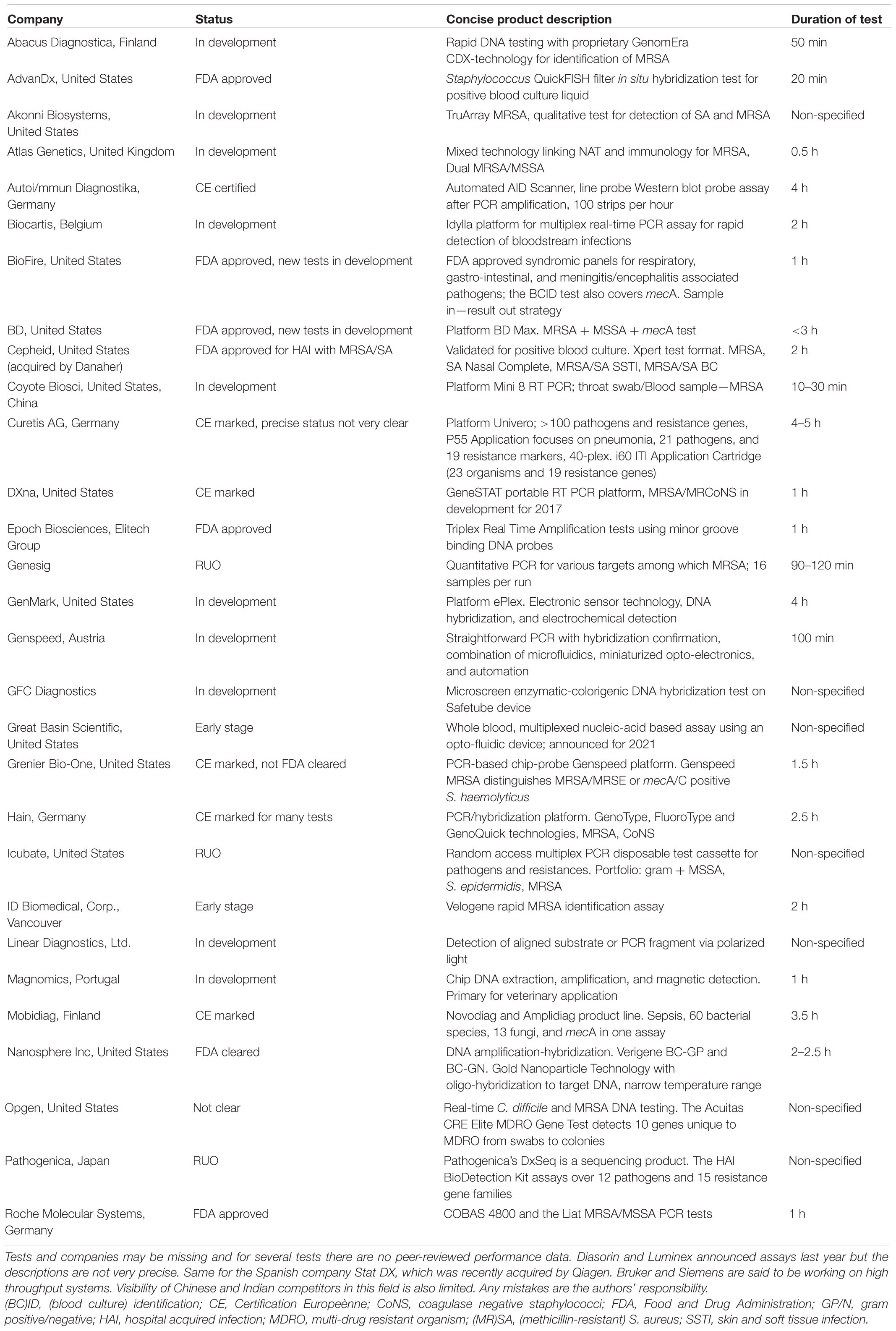
TABLE 1. Global review of future and commercial PCR tests for meticillin-resistant and -susceptible strains of Staphylococcus aureus.
Some of the technologies are worth mentioning separately given the fact that they can be considered extremely elegant from an experimental design point of view. Digital droplet PCR for instance was shown to be sensitive and rapid and it has to be realized that instruments allowing in house development of droplet-based PCR tests are already (commercially) available (Luo et al., 2017). Nanowires are attractive because of size, relatively low costs, and speed of the assay and broad applicability of the technology which, in addition, is easy to multiplex (Ibarlucea et al., 2017). Similarly, using albumin stabilized fluorescent gold nanoclusters as selective probes, MSSA and MRSA can be reliably distinguished (Chan and Chen, 2012). Sensitivity and specificity of these often still quite experimental tests are usually good and offer a positive perspective on future developments in this field (Bakthavatchalam et al., 2017). Note that Table 1 highlights the post-PCR use of array technology, filter in situ hybridization, minor groove binding DNA probes, and magnetic capturing as additional clever read-out methods.
Laboratory-Based vs. PoC Testing
Classical testing for microbial pathogens usually leads to amplification of viable cells. This requires the use of specialized laboratories where employees and the community outside of the lab are protected from infection through specific control measures. This has often blocked PoC test development and deployment. Now, with the possibility to detect pathogens by amplifying non-infectious components of such pathogens the door toward out-of-laboratory testing has been opened wide. Miniaturized tools have been developed that are based on microfluidics (Yeh et al., 2017), LAMP combined with cellulose-based nucleic acid binding paper (Bearinger et al., 2011), isothermal amplification tests (Toley et al., 2015) but also based upon labeling- and amplification-free techniques (Corrigan et al., 2013). With such technologies in mind it was established that PoC testing for MSSA and MRSA was among the priorities when remote and even disaster testing was due (Brock et al., 2010; Kost et al., 2012). Clearly, tools for bedside diagnostics are available that allow for in-department infection control and outbreak management.
In PoC testing both technical and clinical aspects are of key importance. Technical requirements are largely covered by the WHO ASSURED criteria. The acronym lists affordability, sensitivity, specificity, user-friendliness, rapidity and robustness, no need for complicated equipment, and providing solutions that can be easily delivered to end users. If all these requirement are met in a single test (which at this stage is non-existent) then clinical applicability is essentially global. However, if a test would, for example, be too expensive then application in developing economies would essentially be blocked. MRSA/MSSA PoC tests would be particularly useful for rapid assessment of (nasal) carriage for infection control, whereas screening for staphylococcal wound infection and respiratory infection would also have strong added value.
The first PoC MRSA projects have been published. Leone et al. (2013) did an intensive care-based study into the use of MSSA/MRSA detection in patients with ventilator-associated pneumonia. They showed that with a negative predictive value of 99.8% PoC testing efficiently excluded the presence of MRSA among the patients. They did warn that the reliability of this type of testing is dependent on the local prevalence of MRSA carriage. In an orthopedic readmission study it was shown that the Cepheid Xpert MRSA with its 75% sensitivity in this groups of patients with complicated problems performed quite well (Parcell and Phillips, 2014). Screening more than 10,000 patients at admission for detection of MRSA carriage was very efficient as well (Wu et al., 2017). Although the list of publications is relatively short, it is clear that detection of MSSA/MRSA at the PoC fulfills a real medical need. In case of epidemic spread of MRSA clones rapid and sensitive detection are key and in many cases the use of PoC testing allows for accelerated testing in comparison with more conventional laboratory assays. Speed really is the key to high throughput surveillance and subsequent rapid infection control.
Genomic Detection of MSSA and MRSA
Single genomic molecules can be captured to microscopic beads, which are equipped with biotinylated probes to which streptavidin-complexed galactosidase binds and which facilitates the detection of sub-femtomolar concentrations of specific DNA molecules. This Single Molecule Array tests (developed by Quanterix Corporation, Lexington, MA, United States) has been adapted for the detection of MRSA as well (Song et al., 2013). Beyond capturing and detecting “full” genomes, there is now also the option to have a staphylococcal genome sequenced de novo and in toto. Thousands of MSSA and MRSA strains have been subjected to genome sequencing and the software that allows for post-sequencing detection of the mec genes is available (Gordon et al., 2014). Current and well-known software packages for such purposes include CLC Bio (Qiagen, Hilden, Germany), Seqsphere (Ridom, Münster, Germany), and Bionumerics (Applied Maths, bioMérieux, St. Martens Lathem, Belgium). Hence genomic characterization of MRSA is feasible and with the rise in sequencing directly from clinical specimens the impact of direct detection of MRSA will change significantly over the years to come (Lefterova et al., 2015). However, in order to be applicable in routine high-throughput clinical laboratories the technology needs to be quicker, less expensive with data that should be easy to interpret preferably in a (semi-)quantitative fashion. Obviously, genome sequencing provides the ultimate tool for epidemiological typing of MRSA and MSSA (Quainoo et al., 2017) and many studies where NGS has been exploited to define epidemiological patterns of spreading of MRSA have been published before (Tewhey et al., 2012; Price et al., 2013; Stone et al., 2013; Kong et al., 2016; Ward et al., 2016; Planet et al., 2017; and references therein).
Concluding Remarks
Whereas classical detection and speciation of staphylococci has improved significantly upon the introduction of MALDI ToF MS in the diagnostic laboratory, molecular tests, mostly based on specific gene amplification, are still required for the rapid distinction between MSSA and MRSA. The availability of WGS and NGS has now opened up alternative avenues for the detection of resistance genes, the mec-variants included. The near future will bring (genome) sequencing and comprehensive software packages allowing for the unequivocal bio-informatic AST of MSSA and MRSA using WGS, even for non-bio-informaticians. The position of PoC testing in all of this is still poorly defined and needs to be clarified. Inclusion of additional patient data beyond laboratory results is an important additive to the optimization of PoC testing (Yoshioka et al., 2018). In conclusion, molecular testing for MRSA has been accepted by the diagnostic community and is performing well. New technology will challenge the molecular tests and there will be fierce clinical, commercial, and academic completion before full acceptation of the new wave of genomic testing formats.
Author Contributions
AB and OR: conceived, wrote and illustrated the manuscript. Data for the table was assembled using aimed searches in relevant databases and publicly available information in a variety of corporate websites and communications between company employees and the authors. Omissions in the table are for the responsibility of the authors only.
Funding
Research in the laboratory of AB has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie (Grant Agreement No. 675412) [New Diagnostics for Infectious Diseases (ND4ID)].
Conflict of Interest Statement
The authors are employees of bioMérieux, a company designing, developing, and selling diagnostic assays in the field of infectious diseases. Opinions and conclusions phrased in the current text are the author’s, not necessarily equaling the formal bioMérieux policies.
References
Ábrók, M., Lázár, A., Szécsényi, M., Deák, J., and Urbán, E. (2018). Combination of MALDI-TOF MS and PBP2’ latex agglutination assay for rapid MRSA detection. J. Microbiol. Methods 144, 122–124. doi: 10.1016/j.mimet.2017.11.021
Almassian, D. R., Cockrell, L. M., and Nelson, W. M. (2013). Portable nucleic acid thermocyclers. Chem. Soc. Rev. 42, 8769–8798. doi: 10.1039/c3cs60144g
Bakthavatchalam, Y. D., Nabarro, L. E., and Veeraraghavan, B. (2017). Evolving rapid methicillin-resistant Staphylococcus aureus detection: cover all the bases. J. Glob. Infect. Dis. 9, 18–22. doi: 10.4103/0974-777X.199997
Bearinger, J. P., Dugan, L. C., Baker, B. R., Hall, S. B., Ebert, K., Mioulet, V., et al. (2011). Development and initial results of a low cost, disposable, point-of-care testing device for pathogen detection. IEEE Trans. Biomed. Eng. 58, 805–808. doi: 10.1109/TBME.2010.2089054
Becker, K., van Alen, S., Idelevich, E. A., Schleimer, N., Seggewiß, J., Mellmann, A., et al. (2018). Plasmid-encoded transferable mecB-mediated methicillin resistance in Staphylococcus aureus. Emerg. Infect. Dis. 24, 242–248. doi: 10.3201/eid2402.171074
Bernardo, K., Pakulat, N., Macht, M., Krut, O., Seifert, H., Fleer, S., et al. (2002). Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics 2, 747–753. doi: 10.1002/1615-9861(200206)2:6<747::AID-PROT747>3.0.CO;2-V
Blaschke, A. J., Heyrend, C., Byington, C. L., Fisher, M. A., Barker, E., Garrone, N. F., et al. (2012). Rapid identification of pathogens from positive blood cultures by multiplex polymerase chain reaction using the FilmArray system. Diagn. Microbiol. Infect. Dis. 74, 349–355. doi: 10.1016/j.diagmicrobio.2012.08.013
Bode, L. G., Kluytmans, J. A., Wertheim, H. F., Bogaers, D., Vandenbroucke-Grauls, C. M., Roosendaal, R., et al. (2010). Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N. Engl. J. Med. 362, 9–17. doi: 10.1056/NEJMoa0808939
Brock, T. K., Mecozzi, D. M., Sumner, S., and Kost, G. J. (2010). Evidence-based point-of-care tests and device designs for disaster preparedness. Am. J. Disaster Med. 5, 285–294.
Brown, J., and Paladino, J. A. (2010). Impact of rapid methicillin-resistant Staphylococcus aureus polymerase chain reaction testing on mortality and cost effectiveness in hospitalized patients with bacteraemia: a decision model. Pharmacoeconomics 28, 567–575. doi: 10.2165/11533020-000000000-00000
Brukner, I., Oughton, M., Giannakakis, A., Kerzner, R., and Dascal, A. (2013). Significantly improved performance of a multitarget assay over a commercial SCCmec-based assay for methicillin-resistant Staphylococcus aureus screening: applicability for clinical laboratories. J. Mol. Diagn. 15, 577–580. doi: 10.1016/j.jmoldx.2013.04.009
Camoez, M., Sierra, J. M., Dominguez, M. A., Ferrer-Navarro, M., Vila, J., and Roca, I. (2016). Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 22, .e1–.e161. doi: 10.1016/j.cmi.2015.10.009
Chan, P. H., and Chen, Y. C. (2012). Human serum albumin stabilized gold nanoclusters as selective luminescent probes for Staphylococcus aureus and methicillin-resistant Staphylococcus aureus. Anal. Chem. 84, 8952–8956. doi: 10.1021/ac302417k
Charretier, Y., Dauwalder, O., Franceschi, C., Degout-Charmette, E., Zambardi, G., Cecchini, T., et al. (2015). Rapid bacterial identification, resistance, virulence and type profiling using selected reaction monitoring mass spectrometry. Sci. Rep. 5:13944. doi: 10.1038/srep13944
Corrigan, D. K., Schulze, H., Henihan, G., Hardie, A., Ciani, I., Giraud, G., et al. (2013). Development of a PCR-free electrochemical point of care test for clinical detection of methicillin resistant Staphylococcus aureus (MRSA). Analyst 138, 6997–7005. doi: 10.1039/c3an01319g
Du, Z., Yang, R., Guo, Z., Song, Y., and Wang, J. (2002). Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 74, 5487–5491. doi: 10.1021/ac020109k
Frye, A. M., Baker, C. A., Rustvold, D. L., Heath, K. A., Hunt, J., Leggett, J. E., et al. (2012). Clinical impact of a real-time PCR assay for rapid identification of staphylococcal bacteremia. J. Clin. Microbiol. 50, 127–133. doi: 10.1128/JCM.06169-11
García-Álvarez, L., Holden, M. T., Lindsay, H., Webb, C. R., Brown, D. F., Curran, M. D., et al. (2011). Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11, 595–603. doi: 10.1016/S1473-3099(11)70126-8
Goldstein, J. E., Zhang, L., Borror, C. M., Rago, J. V., and Sandrin, T. R. (2013). Culture conditions and sample preparation methods affect spectrum quality and reproducibility during profiling of Staphylococcus aureus with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Lett. Appl. Microbiol. 57, 144–150. doi: 10.1111/lam.12092
Gómez-Sanz, E., Schwendener, S., Thomann, A., Gobeli Brawand, S., and Perreten, V. (2015). First Staphylococcal cassette chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus caseolyticus isolate from a canine infection. Antimicrob. Agents Chemother. 59, 4577–4583. doi: 10.1128/AAC.05064-14
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444. doi: 10.1126/science.aaq0179
Gootenberg, J. S., Abudayyeh, O. O., Lee, J. W., Essletzbichler, P., Dy, A. J., Joung, J., et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. doi: 10.1126/science.aam9321
Gordon, N. C., Price, J. R., Cole, K., Everitt, R., Morgan, M., Finney, J., et al. (2014). Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J. Clin. Microbiol. 52, 1182–1191. doi: 10.1128/JCM.03117-13
Henares, D., Brotons, P., Buyse, X., Latorre, I., de Paz, H. D., and Muñoz-Almagro, C. (2017). Evaluation of the eazyplex MRSA assay for the rapid detection of Staphylococcus aureus in pleural and synovial fluid. Int. J. Infect. Dis. 59, 65–68. doi: 10.1016/j.ijid.2017.04.013
Hill-Cawthorne, G. A., Hudson, L. O., El Ghany, M. F., Piepenburg, O., Nair, M., Dodgson, A., et al. (2014). Recombinations in staphylococcal cassette chromosome mec elements compromise the molecular detection of methicillin resistance in Staphylococcus aureus. PLoS One 9:e101419. doi: 10.1371/journal.pone.0101419
Ibarlucea, B., Rim, T., Baek, C. K., de Visser, J. A. G. M., Baraban, L., and Cuniberti, G. (2017). Nanowire sensors monitor bacterial growth kinetics and response to antibiotics. Lab Chip 17, 4283–4293. doi: 10.1039/c7lc00807d
Kaya, H., Hasman, H., Larsen, J., Stegger, M., Johannesen, T. B., Allesøe, R. L., et al. (2018). SCCmecFinder, a web-based tool for typing of Staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3:e00612-17. doi: 10.1128/mSphere.00612-17
Kelly, R., Zoubiane, G., Walsh, D., Ward, R., and Goossens, H. (2016). Public funding for research on antibacterial resistance in the JPIAMR countries, the European Commission, and related European Union agencies: a systematic observational analysis. Lancet Infect. Dis. 16, 431–440. doi: 10.1016/S1473-3099(15)00350-3
Kolenda, C., Dupieux, C., Decousser, J. W., Larsen, A. R., Pichon, B., Holmes, M., et al. (2017). Comparison of automated antimicrobial susceptibility testing systems to detect mecC-positive methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 55, 3554–3556. doi: 10.1128/JCM.01150-17
Kong, H., Tong, L., Zhang, W., Fu, Y., and Li, X. (2014). Combined use of the BinaxNOW Staphylococcus aureus test with the Clearview PBP2a assay for the early detection of methicillin-resistant S. aureus from positive blood cultures. Diagn. Microbiol. Infect. Dis. 78, 226–228. doi: 10.1016/j.diagmicrobio.2013.11.018
Kong, Z., Zhao, P., Liu, H., Yu, X., Qin, Y., Su, Z., et al. (2016). Whole-genome sequencing for the investigation of a hospital outbreak of MRSA in China. PLoS One 11:e0149844. doi: 10.1371/journal.pone.0149844
Kost, G. J., Mecozzi, D. M., Brock, T. K., and Curtis, C. M. (2012). Assessing point-of-care device specifications and needs for pathogen detection in emergencies and disasters. Point Care 11, 119–125. doi: 10.1097/POC.0b013e31825a25cb
Lefterova, M. I., Suarez, C. J., Banaei, N., and Pinsky, B. A. (2015). Next-generation sequencing for infectious disease diagnosis and management: a report of the association for molecular pathology. J. Mol. Diagn. 17, 623–634. doi: 10.1016/j.jmoldx.2015.07.004
Leone, M., Malavieille, F., Papazian, L., Meyssignac, B., Cassir, N., Textoris, J., et al. (2013). Routine use of Staphylococcus aureus rapid diagnostic test in patients with suspected ventilator-associated pneumonia. Crit. Care 17:R170. doi: 10.1186/cc12849
Liu, C., Bayer, A., Cosgrove, S. E., Daum, R. S., Fridkin, S. K., Gorwitz, R. J., et al. (2011). Clinical practice guidelines by the infectious diseases society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52, e18–e55. doi: 10.1093/cid/ciq146
Luo, J., Li, J., Yang, H., Yu, J., and Wei, H. (2017). Accurate detection of methicillin-resistant Staphylococcus aureus in mixtures by use of single-bacterium duplex droplet digital PCR. J. Clin. Microbiol. 55, 2946–2955. doi: 10.1128/JCM.00716-17
Lüthje, P., Pranada, A. B., Carruthers-Lay, D., Desjardins, M., Gaillot, O., Wareham, D., et al. (2017). Identification of microorganisms grown on chromogenic media by MALDI-TOF MS. J. Microbiol. Methods 136, 17–20. doi: 10.1016/j.mimet.2017.03.001
Miyamoto, M., Motooka, D., Gotoh, K., Imai, T., Yoshitake, K., Goto, N., et al. (2014). Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes. BMC Genomics 15:699. doi: 10.1186/1471-2164-15-699
Muldrew, K. L. (2009). Molecular diagnostics of infectious diseases. Curr. Opin. Pediatr. 21, 102–111. doi: 10.1097/MOP.0b013e328320d87e
Okeke, I. N., Peeling, R. W., Goossens, H., Auckenthaler, R., Olmsted, S. S., de Lavison, J. F., et al. (2011). Diagnostics as essential tools for containing antibacterial resistance. Drug Resist. Updat. 14, 95–106. doi: 10.1016/j.drup.2011.02.002
Osiecki, J. (2010). MRSA Screening: Culture or Molecular? Available at: www.advanceweb.com
Østergaard, C., Hansen, S. G., and Møller, J. K. (2015). Rapid first-line discrimination of methicillin resistant Staphylococcus aureus strains using MALDI-TOF MS. Int. J. Med. Microbiol. 305, 838–847. doi: 10.1016/j.ijmm.2015.08.002
Parcell, B. J., and Phillips, G. (2014). Use of Xpert MRSA PCR point-of-care testing beyond the laboratory. J. Hosp. Infect. 87, 119–121. doi: 10.1016/j.jhin.2014.04.002
Perry, J. D. (2017). A decade of development of chromogenic culture media for clinical microbiology in an era of molecular diagnostics. Clin. Microbiol. Rev. 30, 449–479. doi: 10.1128/CMR.00097-16
Planet, P. J., Narechania, A., Chen, L., Mathema, B., Boundy, S., Archer, G., et al. (2017). Architecture of a species: phylogenomics of Staphylococcus aureus. Trends Microbiol. 25, 153–166. doi: 10.1016/j.tim.2016.09.009
Price, J., Gordon, N. C., Crook, D., Llewelyn, M., and Paul, J. (2013). The usefulness of whole genome sequencing in the management of Staphylococcus aureus infections. Clin. Microbiol. Infect. 19, 784–789. doi: 10.1111/1469-0691.12109
Quainoo, S., Coolen, J. P. M., van Hijum, S. A. F. T., Huynen, M. A., Melchers, W. J. G., van Schaik, W., et al. (2017). Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin. Microbiol. Rev. 30, 1015–1063. doi: 10.1128/CMR.00016-17
Ramanan, P., Bryson, A. L., Binnicker, M. J., Pritt, B. S., and Patel, R. (2017). Syndromic panel-based testing in clinical microbiology. Clin. Microbiol. Rev. 31:e00024-17. doi: 10.1128/CMR.00024-17
Rees, J. C., and Barr, J. R. (2017). Detection of methicillin-resistant Staphylococcus aureus using phage amplification combined with matrix-assisted laser desorption/ionization mass spectrometry. Anal. Bioanal. Chem. 409, 1379–1386. doi: 10.1007/s00216-016-0070-3
Rhoads, D. D., Wang, H., Karichu, J., and Richter, S. S. (2016). The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn. Microbiol. Infect. Dis. 86, 257–261. doi: 10.1016/j.diagmicrobio.2016.08.001
Rossen, J. W. A., Friedrich, A. W., Moran-Gilad, J. and ESCMID study group for genomic and molecular diagnostics (ESGMD) (2017). Practical issues in implementing whole-genome-sequencing in routine diagnostic microbiology. Clin. Microbiol. Infect. 24, 355–360. doi: 10.1016/j.cmi.2017.11.001
Schwendener, S., Cotting, K., and Perreten, V. (2017). Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Sci. Rep. 7:43797. doi: 10.1038/srep43797
Sogawa, K., Watanabe, M., Ishige, T., Segawa, S., Miyabe, A., Murata, S., et al. (2017). Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus using MALDI-TOF mass spectrometry. Biocontrol Sci. 22, 163–169. doi: 10.4265/bio.22.163
Song, L., Shan, D., Zhao, M., Pink, B. A., Minnehan, K. A., York, L., et al. (2013). Direct detection of bacterial genomic DNA at sub-femtomolar concentrations using single molecule arrays. Anal. Chem. 85, 1932–1939. doi: 10.1021/ac303426b
Stone, M. J., Wain, J., Ivens, A., Feltwell, T., Kearns, A. M., and Bamford, K. B. (2013). Harnessing the genome: development of a hierarchical typing scheme for meticillin-resistant Staphylococcus aureus. J. Med. Microbiol. 62(Pt 1), 36–45. doi: 10.1099/jmm.0.049957-0
Suh, H. J., Park, W. B., Jung, S.-I., Song, K.-H., Kwak, Y. G., Kim, K.-H., et al. (2018). A risk scoring system for predicting methicillin resistance in community-onset Staphylococcus aureus bacteremia in Korea. Microb. Drug Resist. 24, 556–562. doi: 10.1089/mdr.2017.0236
Szabados, F., Woloszyn, J., Richter, C., Kaase, M., and Gatermann, S. (2010). Identification of molecularly defined Staphylococcus aureus strains using matrix-assisted laser desorption/ionization time of flight mass spectrometry and the Biotyper 2.0 database. J. Med. Microbiol. 59(Pt 7), 787–790. doi: 10.1099/jmm.0.016733-0
Tewhey, R., Cannavino, C. R., Leake, J. A., Bansal, V., Topol, E. J., Torkamani, A., et al. (2012). Genetic structure of community acquired methicillin-resistant Staphylococcus aureus USA300. BMC Genomics 13:508. doi: 10.1186/1471-2164-13-508
Toley, B. J., Covelli, I., Belousov, Y., Ramachandran, S., Kline, E., Scarr, N., et al. (2015). Isothermal strand displacement amplification (iSDA): a rapid and sensitive method of nucleic acid amplification for point-of-care diagnosis. Analyst 140, 7540–7549. doi: 10.1039/c5an01632k
Trouillet-Assant, S., Rasigade, J. P., Lustig, S., Lhoste, Y., Valour, F., Guerin, C., et al. (2013). Ward-specific rates of nasal cocolonization with methicillin-susceptible and -resistant Staphylococcus spp. and potential impact on molecular methicillin-resistant Staphylococcus aureus screening tests. J. Clin. Microbiol. 51, 2418–2420. doi: 10.1128/JCM.00491-13
Turnidge, J. D., Kotsanas, D., Munckhof, W., Roberts, S., Bennett, C. M., Nimmo, G. R., et al. (2009). Staphylococcus aureus bacteraemia: a major cause of mortality in Australia and New Zealand. Med. J. Aust. 191, 368–373.
Ueda, O., Tanaka, S., Nagasawa, Z., Hanaki, H., Shobuike, T., and Miyamoto, H. (2015). Development of a novel matrix-assisted laser desorption/ionization time-of-flight mass spectrum (MALDI-TOF-MS)-based typing method to identify meticillin-resistant Staphylococcus aureus clones. J. Hosp. Infect. 90, 147–155. doi: 10.1016/j.jhin.2014.11.025
Verhoeven, P. O., Grattard, F., Carricajo, A., Lucht, F., Cazorla, C., Garraud, O., et al. (2012). Quantification by real-time PCR assay of Staphylococcus aureus load: a useful tool for rapidly identifying persistent nasal carriers. J. Clin. Microbiol. 50, 2063–2065. doi: 10.1128/JCM.00157-12
Verroken, A., Defourny, L., le Polain de Waroux, O., Belkhir, L., Laterre, P. F., Delmée, M., et al. (2016). Clinical impact of MALDI-TOF MS identification and rapid susceptibility testing on adequate antimicrobial treatment in sepsis with positive blood cultures. PLoS One 11:e0156299. doi: 10.1371/journal.pone.0156299
Ward, M. J., Goncheva, M., Richardson, E., McAdam, P. R., Raftis, E., Kearns, A., et al. (2016). Identification of source and sink populations for the emergence and global spread of the East-Asia clone of community-associated MRSA. Genome Biol. 17:160. doi: 10.1186/s13059-016-1022-0
Wu, P. J., Jeyaratnam, D., Tosas, O., Cooper, B. S., and French, G. L. (2017). Point-of-care universal screening for meticillin-resistant Staphylococcus aureus: a cluster-randomized cross-over trial. J. Hosp. Infect. 95, 245–252. doi: 10.1016/j.jhin.2016.08.017
Yan, L., Zhou, J., Zheng, Y., Gamson, A. S., Roembke, B. T., Nakayama, S., et al. (2014). Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 10, 970–1003. doi: 10.1039/c3mb70304e
Yeh, E. C., Fu, C. C., Hu, L., Thakur, R., Feng, J., and Lee, L. P. (2017). Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 3:e1501645. doi: 10.1126/sciadv.1501645
Yoshioka, N., Deguchi, M., Hagiya, H., Yoshida, H., Yamamoto, N., Hashimoto, S., et al. (2018). Available, bed-sided, comprehensive (ABC) score to a diagnosis of Methicillin-resistant Staphylococcus aureus infection: a derivation and validation study. BMC Infect. Dis. 18:19. doi: 10.1186/s12879-017-2919-2
Zhang, T., Ding, J., Rao, X., Yu, J., Chu, M., Ren, W., et al. (2015). Analysis of methicillin-resistant Staphylococcus aureus major clonal lineages by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS). J. Microbiol. Methods 117, 122–127. doi: 10.1016/j.mimet.2015.08.002
Keywords: Staphylococcus aureus, MSSA, MRSA, molecular testing, point of care, next generation sequencing, whole genome sequences
Citation: van Belkum A and Rochas O (2018) Laboratory-Based and Point-of-Care Testing for MSSA/MRSA Detection in the Age of Whole Genome Sequencing. Front. Microbiol. 9:1437. doi: 10.3389/fmicb.2018.01437
Received: 25 March 2018; Accepted: 11 June 2018;
Published: 29 June 2018.
Edited by:
Richard Vernon Goering, Creighton University School of Medicine, United StatesReviewed by:
Yan Q. Xiong, UCLA David Geffen School of Medicine, United StatesPaul Douglas Fey, University of Nebraska Medical Center, United States
Copyright © 2018 van Belkum and Rochas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alex van Belkum, alex.vanbelkum@biomerieux.com
 Alex van Belkum
Alex van Belkum Olivier Rochas2
Olivier Rochas2