
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 20 June 2018
Sec. Microbial Physiology and Metabolism
Volume 9 - 2018 | https://doi.org/10.3389/fmicb.2018.01322
This article is part of the Research Topic Tuning Metabolic Efficiency Through Flavin-based Electron Bifurcation View all 6 articles
Reduction of the disulfide of coenzyme M and coenzyme B (CoMS–SCoB) by heterodisulfide reductases (HdrED and HdrABC) is the final step in all methanogenic pathways. Flavin-based electron bifurcation (FBEB) by soluble HdrABC homologs play additional roles in driving essential endergonic reactions at the expense of the exergonic reduction of CoMS–SCoM. In the first step of the CO2 reduction pathway, HdrABC complexed with hydrogenase or formate dehydrogenase generates reduced ferredoxin (Fdx2-) for the endergonic reduction of CO2 coupled to the exergonic reduction of CoMS–SCoB dependent on FBEB of electrons from H2 or formate. Roles for HdrABC:hydrogenase complexes are also proposed for pathways wherein the methyl group of methanol is reduced to methane with electrons from H2. The HdrABC complexes catalyze FBEB-dependent oxidation of H2 for the endergonic reduction of Fdx driven by the exergonic reduction of CoMS–SCoB. The Fdx2- supplies electrons for reduction of the methyl group to methane. In H2- independent pathways, three-fourths of the methyl groups are oxidized producing Fdx2- and reduced coenzyme F420 (F420H2). The F420H2 donates electrons for reduction of the remaining methyl groups to methane requiring transfer of electrons from Fdx2- to F420. HdrA1B1C1 is proposed to catalyze FBEB-dependent oxidation of Fdx2- for the endergonic reduction of F420 driven by the exergonic reduction of CoMS–SCoB. In H2- independent acetotrophic pathways, the methyl group of acetate is reduced to methane with electrons derived from oxidation of the carbonyl group mediated by Fdx. Electron transport involves a membrane-bound complex (Rnf) that oxidizes Fdx2- and generates a Na+ gradient driving ATP synthesis. It is postulated that F420 is reduced by Rnf requiring HdrA2B2C2 catalyzing FBEB-dependent oxidation of F420H2 for the endergonic reduction of Fdx driven by the exergonic reduction of CoMS–SCoB. The Fdx2- is recycled by Rnf and HdrA2B2C2 thereby conserving energy. The HdrA2B2C2 is also proposed to play a role in Fe(III)-dependent reverse methanogenesis. A flavin-based electron confurcating (FBEC) HdrABC complex is proposed for nitrate-dependent reverse methanogenesis in which the oxidation of CoM-SH/CoB-SH and Fdx2- is coupled to reduction of F420. The F420H2 donates electrons to a membrane complex that generates a proton gradient driving ATP synthesis.
Methane-producing archaea (methanogens) are terminal organisms of anaerobic microbial food chains decomposing complex organic matter in Earth’s anaerobic biosphere which includes the lower intestinal tract of humans, the hind gut of termites, the rumen of animals, natural wetlands and rice paddies. As such, methanogens are an essential link in the global carbon cycle (Figure 1). In step 1, CO2 is incorporated into biomass by photosynthetic plants and microbes. In oxygenated environments, O2-respiring microbes oxidize the biomass producing CO2 that re-enters the carbon cycle (step 2). A significant fraction of the biomass enters anaerobic biospheres where it is converted to CO2 and CH4 by microbial food chains comprised of at least four metabolic groups (steps 3–6). The fermentative group digests the complex biomass producing acetate, H2, and CO2 along with other volatile fatty acids (step 3) that are oxidized to acetate plus either formate or H2 (step 4) by syntrophic acetogens. The CO2-reducing methanogen group reduces CO2 to CH4 with electrons derived from oxidation of H2 or formate (step 5). This group forms symbioses with the acetogens that supply H2 or formate the methanogens metabolize to concentrations thermodynamically favorable for the acetogens in a process termed interspecies electron transfer (ISET) (Sieber et al., 2009). The acetate-utilizing (acetoclastic) methanogen group converts the methyl group to CH4 and the carbonyl group to CO2 (step 6). A portion of the CH4 is oxidized to CO2 (step 7) by the anaerobic oxidation of methane (AOM) proposed to involve the reversal of methanogenic pathways. The CO2 and remaining CH4 escapes into oxygenated zones where O2-respiring methanotrophic microbes oxidize CH4 to CO2 (step 8), closing the carbon cycle. As a greenhouse gas, methane is nearly 20-fold more potent than CO2; thus, the aerobic and anaerobic oxidation of CH4 plays an important role in controlling Earth’s climate (Valentine, 2002; Rhee et al., 2009).
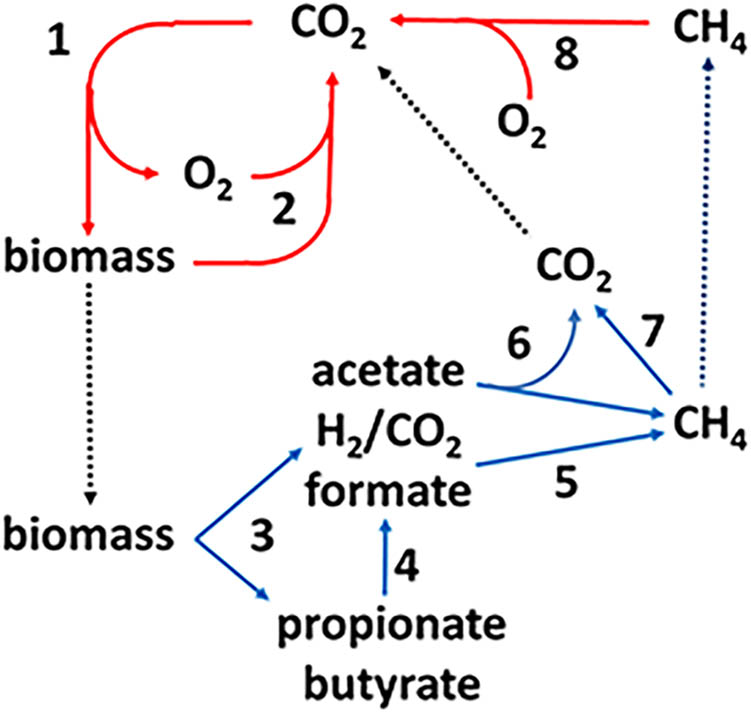
FIGURE 1. The global carbon cycle. Solid lines indicate aerobic (red) and anaerobic (blue) steps in the cycle and dotted lines indicate transfer of material between aerobic and anaerobic environments. (1) Photosynthesis, (2) aerobic decomposition, (3) fermentation, (4) syntrophic acetogenesis, (5) CO2 reduction to methane with electrons derived from formate or H2, (6) acetotrophic methanogenesis, (7) anerobic oxidation of methane, and (8) aerobic oxidation of methane.
Electron transport is much less understood than the comprehensive biochemical understanding of carbon transformations in methanogenic and reverse methanogenic pathways. Herein is reviewed the current understanding of electron transport with a focus on the role of flavin-based electron bifurcation (FBEB) and confurcation (FBEC).
As the name implies, this group only produces CH4 by reducing CO2, primarily with electrons from oxidation of H2 or formate. The pathway (Figure 2) is the subject of reviews (Liu and Whitman, 2008; Thauer et al., 2008; Ferry, 2010). The first step is reduction of CO2 to formyl-methanofuran (CHO-MF) catalyzed by formylmethanofuran dehydrogenase (Fwd or Fmd) (Wagner et al., 2016). The reaction is endergonic and dependent on reduced ferredoxin (Fdx2-). The formyl group of CHO-MF is transferred to tetrahydromethanopterin (H4MPT) and reduced to yield CH3 - H4MPT. Most methanogens contain H4MPT, whereas Methanosarcina species contain the functionally equivalent tetrahydrosarcinapterin (H4SPT). The electron donor is reduced coenzyme F420 (F420H2) generated from H2 or formate by F420-dependent hydrogenases (Fru, Frc, Frh) or formate dehydrogenase (Fdh) (Thauer et al., 2010). Coenzyme F420 is an obligate two-electron carrier donating or accepting a hydride. The methyl group of CH3 - H4MPT is transferred to coenzyme M (HS-CoM) catalyzed by methyltransferase (Mtr) to generate CH3 - SCoM. This exergonic reaction is linked to translocation of Na+ outside the membrane generating a gradient (high outside). Without cytochromes in obligate CO2 reducing methanogens, this is the only mechanism generating an ion gradient that drives ATP synthesis (Thauer et al., 2008). Methyl-SCoM methylreductase (Mcr) catalyzes the reductive demethylation of CH3 - SCoM to CH4 involving coenzyme B (HS-CoB) accompanied by formation of CoMS-SCoB. Heterodisulfide reductase (HdrABC) reduces the disulfide bond with electrons supplied from the oxidation of 2H2 or 2HCO2H (E∘′ ∼-420 mV) catalyzed by F420-independent hydrogenase or Fdh. The exergonic reduction of CoMS-SCoB (E∘′ = –140 mV) drives the endergonic reduction of CO2 (E∘′ = –500 mV) in the first step (Figure 2) via FBEB by HdrABC (Buckel and Thauer, 2013).
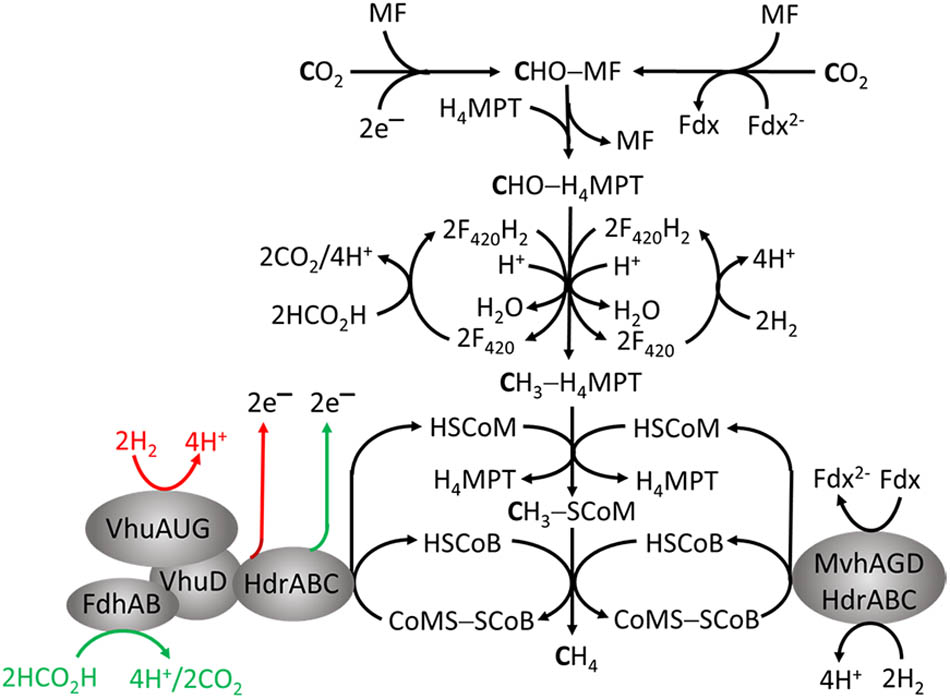
FIGURE 2. Roles for flavin-based electron bifurcation (FBEB) by heterodisulfide reductases in the formate- and H2-dependent CO2 reduction pathway of methanogenesis in Methanothermobacter marburgensis and Methanococcus maripaludis. The red and green arrows represent the FBEB-dependent generation of electrons by HdrABC originating from oxidation of H2 and HCO2H, respectively, for reduction of CO2 in the first step of the pathway.
Two variations are proposed for FBEB of obligate CO2-reducing methanogens (Figure 2). FBEB in strictly hydrogenotrophic Methanothermobacter marburgensis involves the soluble MvhAGD:HdrABC for which the crystal structure of the heterododecameric complex from Methanothermococcus thermolithotrophicus supports a proposed mechanism (Figure 3; Thauer et al., 2008; Kaster et al., 2011; Buckel and Thauer, 2013; Wagner et al., 2017). H2 is oxidized at the catalytic [NiFe] center of MvhA with transfer of electrons to the [2Fe-2S] cluster of MvhD mediated by the [4Fe-4S] clusters of MvhA and MvhG. The bifurcating FAD of HdrA sequentially accepts two electrons from the [2Fe-2S] cluster that contrasts with other characterized FBEB enzymes for which a hydride is donated to FAD (Lubner et al., 2017; Peters and Lubner, 2017). Any of three conformational changes are proposed to overcome the >30 Å distance observed in the crystal structure that otherwise would prohibit electron transfer between the [2Fe-2S] cluster of MvhD and FAD of HdrA. At this juncture the electrons from reduced FAD (FADH-) bifurcate into a high-potential and a low-potential electron. The high-potential electron from FADH- is transported via [4Fe-4S] clusters of HdrA and HdrC to the active-site non-cubane [4Fe-4S] clusters of HdrB where CoMS-SCoB is reduced. The low-potential electron of the resulting semiquinone radical (FADH) is transported via [4Fe-4S] clusters (HA3, HA5, and HA6) of HdrA to Fdx. It is proposed that a conformational change overcomes the 21.5 Å distance in the crystal structure that would otherwise prohibit electron transfer between HA3 and HA5 (Figure 3). The structure reveals residues adjacent to the isoalloxazine ring of FAD proposed to achieve the low-potential neutral FADH radical and a postulated anionic semiquinone (FAD-) intermediate during reduction of FAD. The process occurs twice yielding HSCoM, HSCoB, and Fdx2- that donates electrons to Fwd reducing CO2 to CHO-MF (Figure 2). Generation of Fdx2- by the membrane-bound energy-converting hydrogenase Eha (Figure 4) of obligate CO2 reducing methanogens serves an anaplerotic role and validates the essentiality of FBEB (Lie et al., 2012).
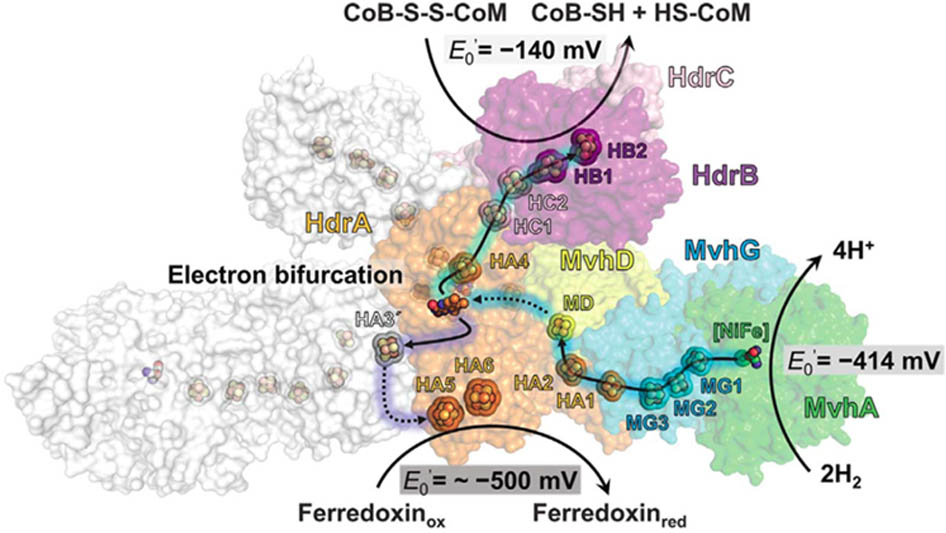
FIGURE 3. Proposed electron-transfer pathway in the heterodisulfide reductase [NiFe]–hydrogenase complex (HdrABC-MvhAGD) from Methanothermococcus thermolithotrophicus. MvhA (green), MvhG (cyan), MvhD (yellow), HdrA (orange), HdrB (purple), HdrC (light pink). The solid arrows indicate the electron-transfer pathway composed of iron-sulfur clusters at distances less than 13.5 Å. The dashed arrows correspond to a hypothetical electron-transfer pathway with distances longer than 15 Å between the redox centers. Reproduced by permission (Wagner et al., 2017).
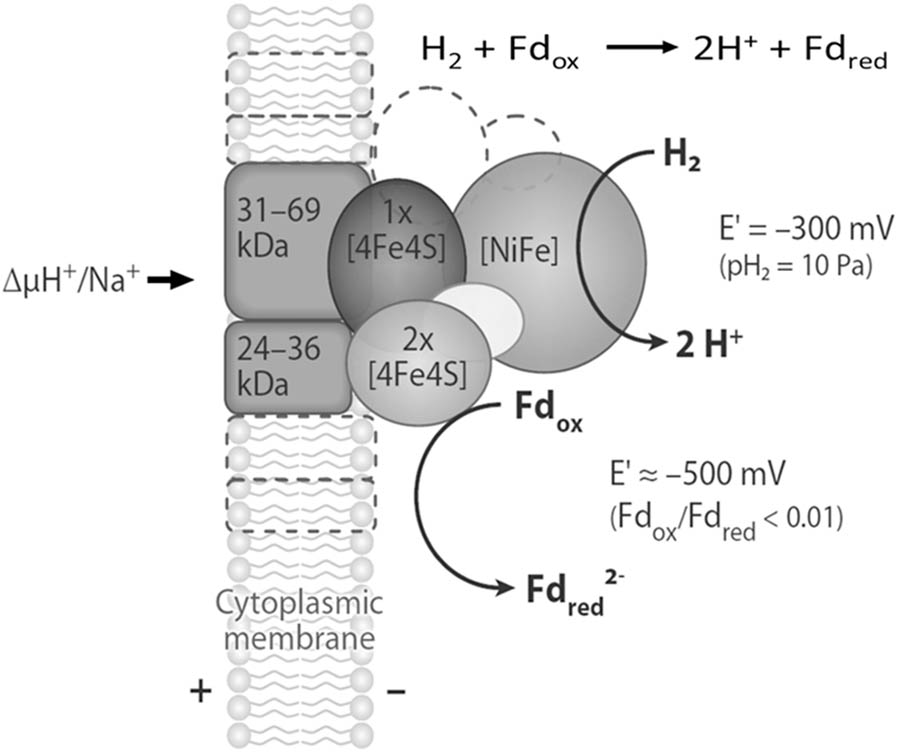
FIGURE 4. Schematic representation of the structure and function of the energy-converting [NiFe]-hydrogenases EchA-F, EhaA-T, EhbA-Q, and MbhA-N found in methanogenic archaea. The energy-converting hydrogenase EchA-F is composed only of the six conserved core subunits, which are shown in shades of gray. The energy-converting hydrogenases EhaA-T, EhbA-Q, and MbhA-N also contain several hydrophobic and hydrophilic subunits of unknown function. These subunits are symbolized by areas with dashed boundaries. Abbreviations: Fdox, oxidized ferredoxin (Fdx); Fdred2-, two-electron-reduced ferredoxin (Fdx2-). Adapted from (Thauer et al., 2010).
In contrast to strictly hydrogenotrophic M. marburgensis, Methanococcus maripaludis utilizes either H2 or formate as electron donors for reduction of CO2 to CH4 requiring FBEB mechanisms for each substrate (Figure 2). A protein complex isolated from formate-grown cells contains HdrABC, F420-non-reducing selenocysteine-containing hydrogenase (Vhu), Fdh, and the tungsten-containing Fwd (Costa et al., 2010). This result lead to the conclusion that Fdh oxidizes formate with direct transfer of electrons to HdrABC without first producing H2 by a F420-dependent formic hydrogenlyase system and then oxidation of the H2 by the F420-independent MvhAGD hydrogenase as in FBEB by M. marburgensis. Direct transfer without participation of H2 as an intermediate is supported by robust growth with formate, although not H2, for a mutant deleted of genes encoding subunits of the selenocysteine-containing (vhu) and cysteine-containing (vhc) F420-independent hydrogenases associated with HdrABC. However, the mutant retained the vhuD and vhcD genes homologous to mvhD of M. marburgensis obscuring potential roles for VhuD and VhcD. When grown under conditions where both Fdh and Vhu are expressed, the enzymes compete for binding to VhuD, and are fully functional and bound to VhuD (Costa et al., 2013). Further, Fdh co-purifies with VhuD in the absence of other hydrogenase subunits. It was concluded that VhuD, also containing a [2Fe-2S] cluster, functions analogous to MvhD by mediating direct electron transfer from Vhu or Fdh to HdrABC (Figure 2; Costa et al., 2013). The mechanism for transfer of electrons from HdrABC to Fwd is unknown although likely mediated by Fdx as for M. marburgensis (Costa et al., 2010). Not reported is biochemical validation of electron bifurcation by the proposed complex as was shown for the MvhAGD:HdrABC and HdrA2B2C2 complexes of M. marburgensis and Methanosarcina acetivorans (Kaster et al., 2011; Yan et al., 2017). Nonetheless, an in silico genome-scale metabolic reconstruction of M. maripaludis indicates the organism is unable to grow without the energy-conserving complex (Richards et al., 2016).
In addition to supplying Fdx2- for the first step in the CO2-reduction pathway of methanogenesis, it is proposed that energy-conserving FBEB is instrumental for growth of Methanocella conradii when concentrations of H2 are exceptionally low (Liu et al., 2014). A transcription unit comprised of genes encoding Fwd, HdrABC and MvhD is up regulated in M. conradii grown syntrophically in co-culture with H2-producing acetogens utilizing propionate and butyrate (Liu et al., 2014). Thus, it is proposed that an electron bifurcating MvhD/HdrABC/Fwd complex is essential for syntrophic growth with low concentrations of H2. As M. conradii encodes MvhGA remote from the up regulated transcription unit, the mechanism by which H2 is oxidized and electrons transferred to HdrABC is unknown. Interestingly, obligate CO2-reducing methanogens of the order Methanomicrobiales are missing genes encoding MvhA and MvhG but encode MvhD and HdrABC (Browne et al., 2016). These methanogens could form an MvhD/HdrABC complex associated with energy-converting hydrogenases EchA–F, EhaA–T, or MbhA–N dependent on ion gradients to supply Fdx2- for reduction of CO2 to CHO-MF, although reduction of CoMS-SCoB would be energy consuming. Thus, it is proposed that these methanogens substitute MvhA and MvhG with FrhA and FrhG of the F420-reducing hydrogenase (FrhABG) contained in all methanogens without cytochromes (Thauer et al., 2010; Gilmore et al., 2017). In this way, FrhAG would be present in an FrhAG/MvhD/HdrABC complex with the potential for FBEB of H2 that generates the Fdx2- required for reduction of CO2 to CHO-MF (Figure 5) with the added advantage of conserving energy. In this scenario, the energy-converting hydrogenases play a role in only providing Fdx2- for biosynthesis (Major et al., 2010). However, the possibility of an FrhABG/MvhD/HdrABC complex (Figure 5) cannot be ruled out at this juncture. Inclusion of the F420-binding FrhB subunit invokes electron transport dependent on FrhABG producing F420H2 for which the electron pair is bifurcated by MvhD/HdrABC reducing Fdx and CoMS-SCoB analogous to the HdrA2B2C2 of M. acetivorans (Vitt et al., 2014; Yan et al., 2017). However, it is unknown which FBEB pathway is physiologically relevant.
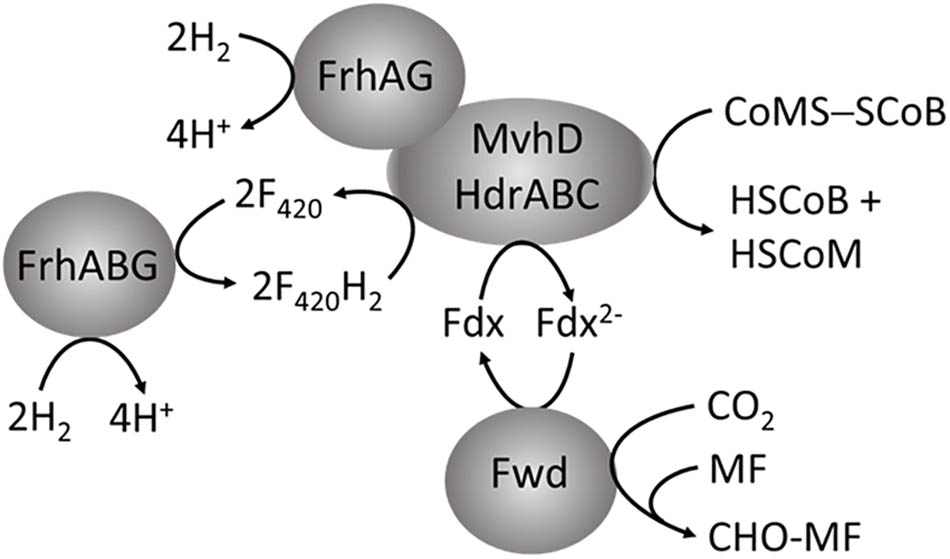
FIGURE 5. Proposed role for FBEB by the heterodisulfide reductase in obligate CO2-reducing methanogens of the order Methanomicrobiales. MF, methanofuran; Fdx, ferredoxin.
Methanogens from the order Methanosarcinales grow and produce CH4 with methyl-containing substrates (methanol, methylamines, and methyl sulfides) (Liu and Whitman, 2008). A few also grow by reducing CO2 with H2. Unlike obligate CO2 reducers, these methanogens contain cytochromes and generate a proton gradient dependent on electron transport involving hydrogenases (Thauer et al., 2008). Like obligate CO2 reducers, Fdx2- is required to supply electrons to Fwd/Fmd catalyzing reduction of CO2 to CHO-MF; however, Fdx2- is generated independent of FBEB by the membrane-bound energy-converting Ech hydrogenase driven by the proton gradient (Figure 4).
The methylotrophic pathway of the order Methanosarcinales involves transfer of substrate methyl groups to HSCoM forming a CH3-SCoM pool of which one-fourth of the methyl groups are oxidized to CO2 via reversal of the CO2-reduction pathway to supply F420H2 and Fdx2- required for reductive demethylation of the remaining three-fourths CH3-SCoM to CH4 (Figure 6). The F420H2 is oxidized by a membrane-bound complex (Fpo) that donates electrons to a quinone-like electron carrier (methanophenazine, MP) coupled to generation of a proton gradient. It is proposed that Fdx2- is re-oxidized by reducing F420 although the mechanism is unknown. The production of CH4 from CH3-SCoM is similar to obligate CO2 reducing methanogens involving HSCoB and Mcr with the exception of the membrane-bound heterodisulfide reductase (HdrDE) that reduces CoMS-SCoB to the sulfhydryl forms of the cofactors. Electrons are supplied to HdrDE by MPH2 with the scalar translocation of protons contributing to the proton gradient that drives ATP synthesis. However, the genomes of all sequenced Methanosarcinales also contain genes encoding the HdrABC homologs HdrA1B1C1 and HdrA2B2C2 (Buan and Metcalf, 2010). HdrA1B1C1 is elevated during methylotrophic growth of M. acetivorans for which the ΔhdrA1B1C1 mutant strain is growth impaired. Thus, a role in methylotrophic growth is proposed wherein FBEB by HdrA1B1C1 reduces F420 and CoMS-SCoB with electron pairs donated by two Fdx2- generated in the oxidation of CHO-MF (Figure 6), thereby allowing energy conservation via Fpo (Buan and Metcalf, 2010).
Methanosphaera stadtmanae, isolated from the human gut, is also a methylotrophic methanogen reducing the methyl group of methanol to CH4 although belonging to the order Methanobacteriales that do not contain cytochromes necessary for electron-transport coupled proton translocation that drives ATP synthesis. The genome also lacks a complete gene set necessary for reversal of the CO2 reduction pathway and therefore requires the oxidation of H2 to supply electrons for reductive demethylation of CH3-SCoM to CH4 (Fricke et al., 2006). A scheme is proposed that includes FBEB of H2 by an MvhADG:HdrABC complex to explain the finding that ATP synthesis is driven by an ion gradient (Figure 7; Sparling et al., 1993; Thauer et al., 2008). The Fdx2- produced donates electrons to the membrane-bound energy-converting Ehb complex that generates a Na+ gradient driving ATP synthesis and regenerates H2 recycled for FBEB by the MvhADG:HdrABC complex. A similar FBEB/H2 cycling scheme is proposed for a sixth class of methanogens, ‘Candidatus Methanofastidiosa,’ based on metagenome-derived draft genomes that also lack cytochromes and genes encoding enzymes for reversal of the CO2 reducing pathway (Nobu et al., 2016). However, this class is restricted to reducing methylthiols with H2. A seventh order, the Methanomassiliicoccales, also grows by reducing the methyl groups of methylotrophic substrates with H2. Analyses of several genomes show this class also lacks cytochromes and genes required for reversal of the CO2 reducing pathway (Borrel et al., 2014; Kroninger et al., 2015; Lang et al., 2015). Unlike M. stadtmanae and the ‘Candidatus Methanofastidiosa’ class, genes encoding the membrane-bound energy-converting complexes are absent and genes encoding an Fpo-like complex and the HdrD subunit of HdrDE are present. Figure 8 shows the pathway proposed for Methanomassiliicoccus luminyensis. The Fpo complex oxidizes Fdx2- generated via FBEB of H2 by the MvhADG:HdrABC complex. HdrD accepts electrons from Fpo and reduces CoMS-SCoB coupled to generation of a H+ gradient. Roles for involvement of the Ech1 and Ech2 hydrogenases are ruled out based on low abundance of transcripts and low membrane-bound hydrogenase activity (Kroninger et al., 2015). Thus, CoMS-SCoB is essential for both FBEB and the terminal electron acceptor which is distinct from that proposed for M. stadtmanae and the ‘Candidatus Methanofastidiosa’ class which involves H2 cycling (Figure 7).
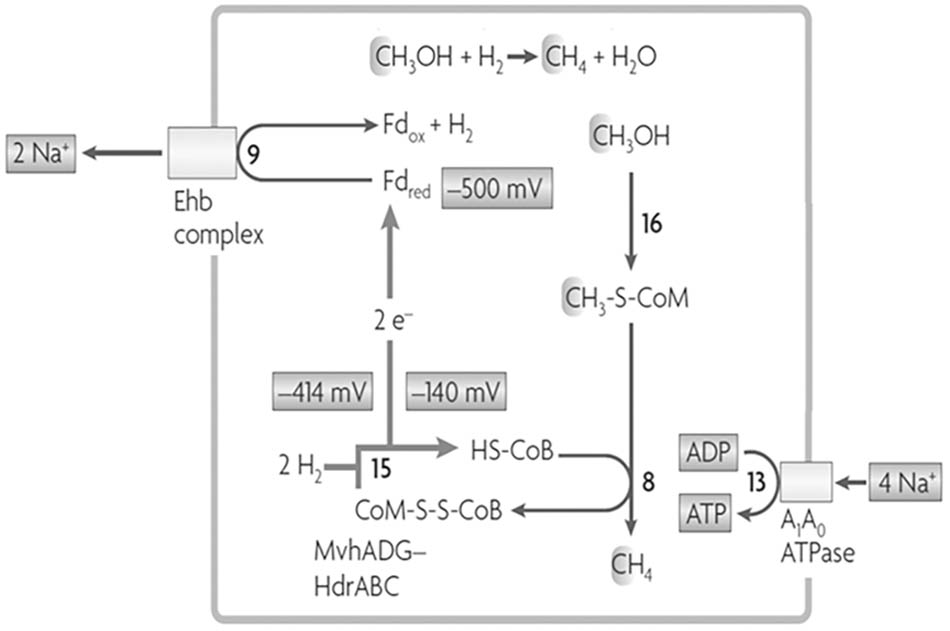
FIGURE 7. Proposed role for FBEB by heterodisulfide reductase in Methanosphaera stadtmanae growing with methanol and H2. Reactions 9 and 15 are coupled by FBEB. The redox potentials are standard potentials at pH 7.0. Fdox, oxidized ferredoxin (Fdx); Fdred, two-electron-reduced ferredoxin (Fdx2-). Adapted from (Thauer et al., 2008).
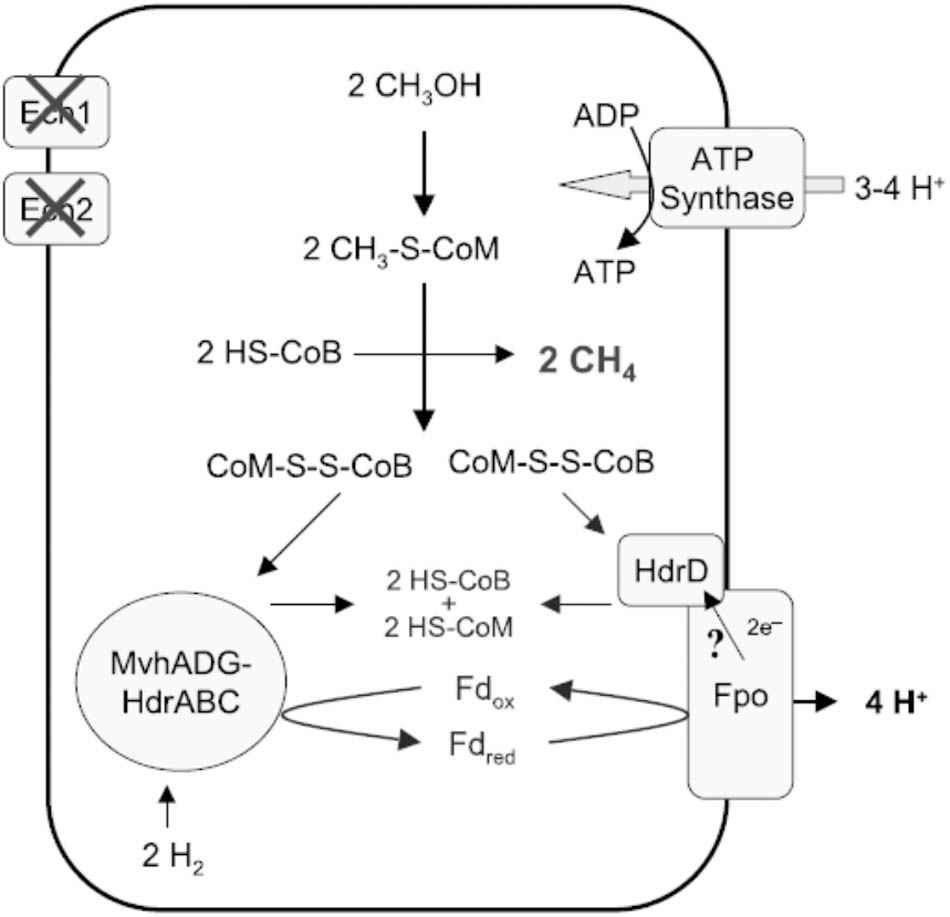
FIGURE 8. Proposed role for FBEB by heterodisulfide reductase in energy-conservation by Methanomassiliicoccus luminyensis. Ech1 and Ech2 are covered with crosses to indicate non-involvement in generating the proton gradient driving ATP synthesis (Kroninger et al., 2015). The question mark indicates a reaction not experimentally verified, although includes 2H+ pumped by the Fpo complex and another 2H+ translocated scalar via reduction and re-oxidation of methanophenazine. Fdox, oxidized ferredoxin (Fdx); Fdred, two-electron-reduced ferredoxin (Fdx2-). Reproduced by permission (Kroninger et al., 2015).
Methanosarcina and Methanosaeta are the only described genera of acetotrophic methanogens that are the subject of recent reviews (Ferry, 2013; Schlegel and Muller, 2013; Welte and Deppenmeier, 2014; Ferry, 2015). Most biochemical investigations have involved Methanosarcina species. M. acetivorans is a model for species that do not metabolize H2 which constitute the majority of Methanosarcina species (Figure 9A). Acetate is converted to acetyl-CoA at the expense of one ATP followed by cleavage of the C-C and C-S bonds yielding a methyl group that is transferred to H4SPT and a carbonyl group that is oxidized to CO2 with transfer of electrons to Fdx. The methyl group of CH3-H4SPT is transferred to HS-CoM followed by reductive demethylation of CH3S-CoM to methane involving reactions common to all methanogenic pathways. The Mtr complex catalyzes the exergonic methyl transfer coupled to generation of a Na+ gradient. The reduced Fdx2- is electron donor to the Na+-pumping Rnf complex that donates electrons to cytochrome c that is the electron donor to MP (Wang et al., 2011). As in the methylotrophic pathway, HdrDE oxidizes MPH2 and reduces CoMS-SCoB with scalar translocation of H+ that generates a gradient. The multisubunit Na+/H+ antiporter Mrp adjusts the Na+/H+ ratio optimal for the ATP synthase which is dependent on both Na+ and H+ gradients (Schlegel et al., 2012a; Jasso-Chavez et al., 2013, 2016).
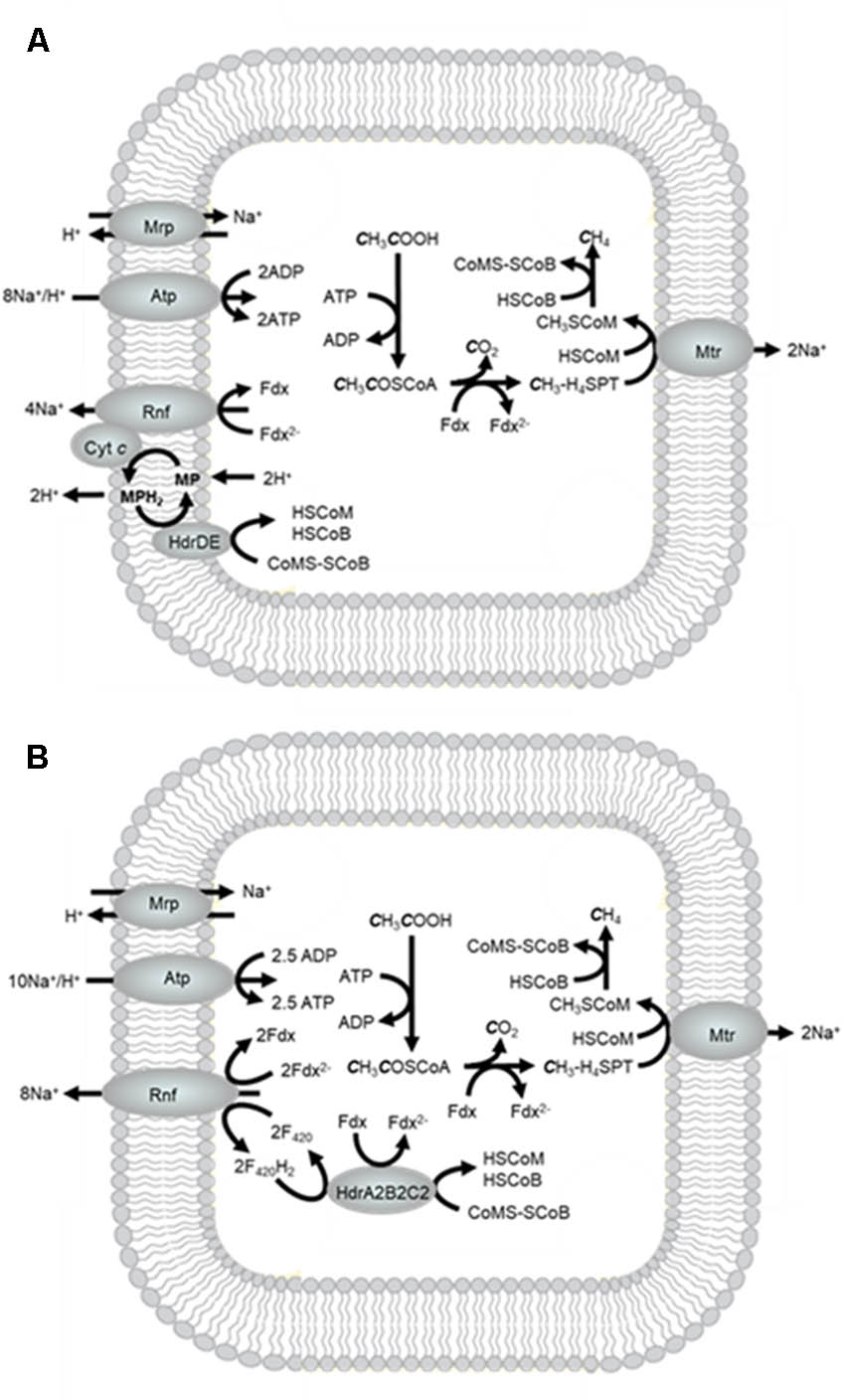
FIGURE 9. Acetotrophic pathway of Methanosarcina acetivorans. (A) Pathway based on the current biochemical understanding of electron transport. (B) Pathway showing electron transport based on the postulated role for FBEB by HdrA2B2C2 (Buckel and Thauer, 2018a).
When switched from growth with methanol to growth with acetate, M. acetivorans up regulates an electron bifurcating heterodisulfide reductase (HdrA2B2C2) that oxidizes F420H2 (Em = -380 mV) and reduces Fdx (Em = -520 mV) driven by reduction of CoMS-SCoB (Em = -140 mV) (Yan et al., 2017). A role has been proposed for HdrA2B2C2 dependent on reduction of NAD-like coenzyme F420 (F420) by the Rnf complex analogous to Fdx-dependent reduction of NAD+ by homologous Rnf complexes from the domain Bacteria (Figure 9B; Buckel and Thauer, 2018a,b). In this way, Fdx reduced by HdrA2B2C2 is re-oxidized by Rnf thereby supplementing the translocation of Na+. The Na+ gradient formed by Rnf and Mtr could be exchanged with H+ by Mrp to adjust the Na+/H+ ratio optimal for ATP synthesis. The process generates more ATP than electron transport involving MP and HdrDE (Figure 9). However, it is reported that HdrDE is essential for acetotrophic growth suggesting the possibility of both electron transport pathways oxidizing Fdx2- and reducing CoMS-SCoB (Buan and Metcalf, 2010). Having alternate electron transport pathways with different thermodynamic efficiencies could provide the cell with options for responding to fluctuations in available free energy proportional to levels of acetate in the environment. Indeed, the conversion of acetate to CH4 and CO2 provides only a marginal amount of energy available for growth (ΔG∘′ = -36 kJ/CH4) that requires cells to maximize the thermodynamic efficiency.
A genome-wide analysis of Methanosaeta thermophila revealed genes encoding enzymes catalyzing carbon transformation reactions in the pathway of acetate to CH4 similar to Methanosarcina species (Smith and Ingram-Smith, 2007). However, genes encoding the Rnf complex are absent in the genome of Methanosaeta suggesting an unknown alternative electron transport pathway and mechanism for energy conservation.
It is postulated that AOM is accomplished by a reversal of methanogenic pathways based on environmental metagenomic and metatranscriptomic analyses of sediments (Hallam et al., 2004; McGlynn, 2017; Timmers et al., 2017). Although discovered nearly four decades ago, the unavailability of pure cultures prevented biochemical investigations of AOM. However, M. acetivorans is capable of “trace methane oxidation” (TMO) defined as reverse methanogenesis during net CH4 production from growth substrates (Moran et al., 2005, 2007; Timmers et al., 2017). More recently, methanotrophic growth dependent on reduction of Fe(III) was documented for M. acetivorans (Soo et al., 2016). Figure 10 illustrates the reverse methanogenesis pathway proposed for M. acetivorans based on a biochemical understanding of Fe(III)-dependent mechanisms driving endergonic reactions and energy conservation essential for methanotrophic growth (Yan et al., 2018). It is remarkably similar to the pathway proposed for anaerobic methanotrophic archaea (ANME) based on metagenomic and transcriptomic analyses of uncultured Methanosarcinales sp. ANME-2a (Wang et al., 2014).
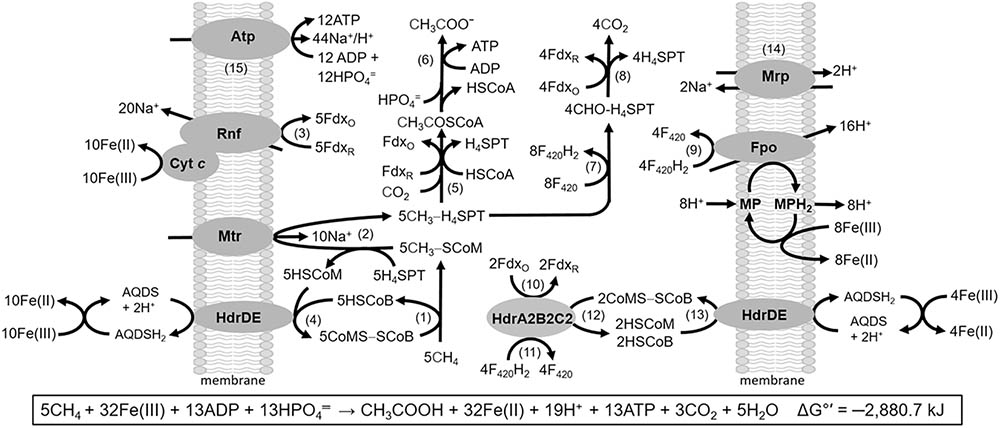
FIGURE 10. Pathway proposed for Fe(III)-dependent methane oxidation and conservation of energy by M. acetivorans. Enzymes not discussed in the text: CO dehydrogenase/acetyl CoA synthase (Rxn. 5); acetate kinase and phosphotransacetylase (Rxn. 6); coenzyme F420 (F420)-dependent methylene-H4SPT reductase, F420-dependent methylene-H4SPT dehydrogenase, methenyl-H4SPT cyclohydrolase, formylmethanofuran:H4SPT formyltransferase (Rxn. 7); formylmethanofuran dehydrogenase (Fwd) (Rxn. 8). MP, methanophenazine; AQDS, anthraquinone-2,6-disulfonate; FdxO, oxidized ferredoxin (Fdx); FdxR, two-electron-reduced ferredoxin (Fdx2-). Reproduced by permission (Yan et al., 2018).
The M. acetivorans pathway is a reversal of established acetate-utilizing and CO2-reducing methanogenic pathways (Li et al., 2005, 2006, 2007; Lessner et al., 2006; Ferry, 2008; Wang et al., 2011; Schlegel et al., 2012b; Welte and Deppenmeier, 2014). Methane is oxidized by Mcr (Rxn. 1) with the methyl group of CH3-SCoM transferred to H4SPT by Mtr (Rxn. 2) representing the reversal of reactions common to all methanogenic pathways. The HdrDE oxidizes HSCoM and HSCoB coupled to reduction of Fe(III) that regenerates CoMS-SCoB (Rxn. 4). Removal of HSCoM, HSCoB, and CH3-SCoM products by HdrDE and Mtr drives the endergonic oxidation of CH4 by Mcr. The endergonic methyl transfer producing CH3-H4SPT is driven with the Na+ gradient generated by the Rnf/cytochrome c complex catalyzing the highly exergonic oxidation of Fdx2- and reduction of Fe(III) (Rxn. 3). Fdx2- is also utilized in reduction of CO2 that supplies the carbonyl group for condensation with the methyl group of CH3-H4SPT producing acetate (Rxns. 5 and 6). Fdx2- and F420H2 are generated in reversal of the CO2 reduction pathway (Rxns. 7 and 8). F420H2 is oxidized by the Fpo complex (Rxn. 9) with transfer of electrons to MP and Fe(III) coupled to generation of a H+ gradient. The H+ gradient, together with the Na+ gradient, drives ATP synthesis assisted by the Mrp antiporter that optimizes the H+/Na+ ratio optimal for the ATP synthase dependent on both H+ and Na+ (Rxns. 14 and 15) (Schlegel et al., 2012a; Jasso-Chavez et al., 2013). The reverse methanogenesis pathway is remarkably similar to that proposed for uncultivated Methanosarcinales sp. ANME-2a present in marine sediments that perform AOM It is also proposed that Fdx2- is generated by HdrA2B2C2 previously shown to oxidize F420H2 and reduce Fdx coupled to reduction of CoMS-SCoB via energy-conserving FBEB (Rxns. 10–12) (Yan et al., 2017). The HSCoM and HSCoB produced are oxidized by HdrDE coupled to the reduction of Fe(III) regenerating CoMS-SCoB (Rxn. 13). This proposed role would be essential in the environment where low availability of Fe(III) limits the generation of Na+ and H+ gradients by the Rnf and Fpo complexes. In this scenario, the Fdx2- produced by HdrA2B2C2 is used to reduce CO2 for the synthesis of acetate and ATP by substrate level phosphorylation (Rxns. 5 and 6). Notably, the Methanosarcinales sp. ANME-2a metagenome encodes HdrA2, HdrB2, and HdrC2 homologs with 59, 72, and 59% identities (Supplementary Figure S1) consistent with a role in reverse methanogenesis by ANME.
Unlike the HdrA1B1C1 of M. acetivorans and the HdrABC of obligate CO2-reducing methanogens, the C-terminal domain of HdrA2 extends with sequences homologous to MvhD (Yan et al., 2017). Although the function of this fused MvhD is unknown, HdrA2 homologs are ubiquitous in acetotrophic and methylotrophic species of the order Methanosarcinales suggesting important functions. Remarkably, HdrA2 and HdrBC homologs are present in non-methanogenic species of the domain Bacteria signaling diverse functions.
A metagenomics-based metabolic model of electron transport is proposed for the nitrate-dependent reverse methanogenesis by Methanoperedens-like ANME (Figure 11). Apart from the F420H2, HSCoM/HSCoB, and Fdx2- generated by reverse methanogenesis, the model contrasts with the Fe(III)-dependent pathway of M. acetivorans (Arshad et al., 2015). Foremost, the genome encodes a Rieske-type protein, cytochromes c and b, and a nitrate reductase that reduces nitrate to nitrite with reduced menaquinone (MQH2) generated by a F420H2 dehydrogenase (Fqo) that combine to generate a proton gradient driving ATP synthesis. Reduction of MQ is also accomplished by oxidation of HSCoM/HSCoB with HdrDE. An energy-converting hydrogenase homolog (Ech) is proposed to oxidize Fdx2- and contribute to the proton gradient although the fate of produced H2 is unknown. Alternatively, Fdx2- is the electron donor to a flavin-based electron confurcating complex comprised of an HdrABC homolog oxidizing HSCoM/HSCoB and donating electrons to the F420-dependent hydrogenase subunit (FrhB) proposed to oxidize Fdx2- and reduce F420. Energy is conserved in the confurcation reaction rather than lost as heat should FrhB alone oxidize Fdx2- and reduce F420 (ΔE∘′ = 120 mV). The genome of “Candidatus Methanoperedens nitroreducens” encodes a homolog of M. acetivorans HdrA2 which presents the possibility of an HdrA2B2C2 homolog catalyzing the confurcation reaction (Berger et al., 2017; Yan et al., 2017).
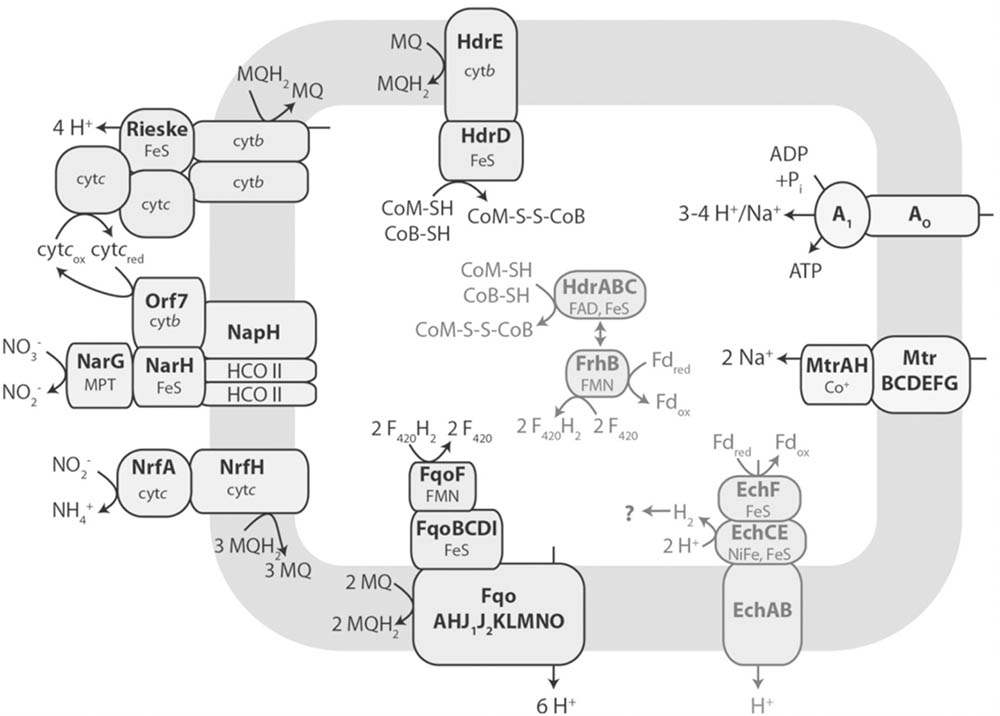
FIGURE 11. Metagenomics-based model of electron transport in reverse methanogenesis by Methanoperedens. Key: Fdred, reduced ferredoxin (Fdx2-); Fdox, oxidized ferredoxin (Fdx); Fqo, F420H2 dehydrogenase complex; MQ, menaquinone; MQH2, menaquinol; Hdr, heterodisulfide reductase; cyt b, cytochrome b; cytc, cytochrome c; Nar, nitrate reductase complex complex; Nrf, nitrite reductase; Ech, ferredoxin-dependent hydrogenase complex; FrhB, F420-binding subunit of F420-dependent hydrogenase; Mtr, methyltransferase complex; A1AO, ATP synthase; HCO II, heme copper oxidase subunit-like proteins; FMN, flavin mononucleotide; FAD, flavin adenine dinucleotide; MPT, molybdopterin; NiFe, nickel-iron center. Reproduced by permission (Arshad et al., 2015).
Methanogenic and reverse methanogenic pathways are proposed to involve FBEB or FBEC in electron transport that also serve as mechanisms of energy conservation. However, there is a significant lack of understanding requiring further investigation.
(1) Biochemical confirmation of FBEB is needed for the several proposed complexes other than that shown for the purified MvhADG:HdrABC of M. marburgensis and HdrA2B2C2 of M. acetivorans.
(2) A more detailed understanding of the FBEB mechanism of HdrABC is needed. The crystal structure of MvhADG:HdrABC from M. thermolithotrophicus has provided a guide for experiments to address questions of electron gating and stabilization of reduced flavin intermediates. The ability to produce the catalytically active recombinant HdrA2B2C2 of M. acetivorans, combined with the crystal structure of MvhADG:HdrABC, provides a foundation for genetic approaches generating enzyme variants that will facilitate a detailed understanding of FBEB.
(3) Validation is needed for the proposed role of HdrA1B1C1 in the methylotrophic pathway of M. acetivorans and related methylotrophic methanogens; in particular, the proposal that HdrA1B1C1 of M. acetivorans oxidizes F420H2 in analogy to that shown for HdrA2B2C2. Also worthy of investigation are the uncharacterized HdrA2B2C2 homologs in the order Methanosarcinales and the domain Bacteria.
(4) Investigations are in order to determine the mechanism by which H2 is oxidized and electrons are delivered to the proposed MvhD/HdrABC and MvhD/HdrABC/Fwd complexes of methanogens in the orders Methanocellales and Methanomicrobiales.
(5) The proposed roles for FBEB and FBEC in reverse methanogenesis pathways require validation via analyses of deletion mutants.
ZY performed the research. JF wrote the manuscript.
This work was supported by the Department of Energy Advanced Research Projects Agency-Energy #0881-1525 to JF (70%); the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through grant DE-FG02-95ER20198 (20%); the Person endowment to JF (10%).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01322/full#supplementary-material
Arshad, A., Speth, D. R., De Graaf, R. M., Op Den Camp, H. J., Jetten, M. S., and Welte, C. U. (2015). A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like Archaea. Front. Microbiol. 6:1423. doi: 10.3389/fmicb.2015.01423
Berger, S., Frank, J., Dalcin Martins, P., Jetten, M. S. M., and Welte, C. U. (2017). High-quality draft genome sequence of “Candidatus Methanoperedens sp.” strain BLZ2, a nitrate-reducing anaerobic methane-oxidizing archaeon enriched in an anoxic bioreactor. Genome Announc. 5:e01159-17. doi: 10.1128/genomeA.01159-17
Borrel, G., Parisot, N., Harris, H. M., Peyretaillade, E., Gaci, N., Tottey, W., et al. (2014). Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15:679. doi: 10.1186/1471-2164-15-679
Browne, P., Tamaki, H., Kyrpides, N., Woyke, T., Goodwin, L., Imachi, H., et al. (2016). Genomic composition and dynamics among Methanomicrobiales predict adaptation to contrasting environments. ISME J. 11, 87–99. doi: 10.1038/ismej.2016.1104
Buan, N. R., and Metcalf, W. W. (2010). Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol. Microbiol. 75, 843–853. doi: 10.1111/j.1365-2958.2009.06990.x
Buckel, W., and Thauer, R. K. (2013). Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim. Biophys. Acta 1827, 94–113. doi: 10.1016/j.bbabio.2012.07.002
Buckel, W., and Thauer, R. K. (2018a). Flavin-based electron bifurcation, a new mechanism of biological energy coupling. Chem. Rev. 8, 3862–3886. doi: 10.1021/acs.chemrev.7b00707
Buckel, W., and Thauer, R. K. (2018b). Flavin-Based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD(+) (Rnf) as electron acceptors: a historical review. Front. Microbiol. 9:401. doi: 10.3389/fmicb.2018.00401
Costa, K. C., Lie, T. J., Xia, Q., and Leigh, J. A. (2013). VhuD facilitates electron flow from H2 or formate to heterodisulfide reductase in Methanococcus maripaludis. J. Bacteriol. 195, 5160–5165. doi: 10.1128/JB.00895-13
Costa, K. C., Wong, P. M., Wang, T., Lie, T. J., Dodsworth, J. A., Swanson, I., et al. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U.S.A. 107, 11050–11055. doi: 10.1073/pnas.1003653107
Ferry, J. G. (2008). “Acetate-based methane production,” in Bioenergy, eds J. D. Wall, C. S. Harwood, and A. Demain (Washington, DC: ASM press), 155–170.
Ferry, J. G. (2010). How to make a living exhaling methane. Annu. Rev. Microbiol. 64, 453–473. doi: 10.1146/annurev.micro.112408.134051
Ferry, J. G. (2013). “Acetate: A key intermediate during the anaerobic degradation of organic matter,” in Acetate: Versatile Building Block of Biology and Chemistry, ed. D. A. Sanders (Hauppauge, NY: Nova Science), 1–14.
Ferry, J. G. (2015). Acetate metabolism in anaerobes from the domain Archaea. Life 5, 1454–1471. doi: 10.3390/life5021454
Fricke, W. F., Seedorf, H., Henne, A., Kruer, M., Liesegang, H., Hedderich, R., et al. (2006). The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon Is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 188, 642–658. doi: 10.1128/JB.188.2.642-658.2006
Gilmore, S. P., Henske, J. K., Sexton, J. A., Solomon, K. V., Seppala, S., Yoo, J. I., et al. (2017). Genomic analysis of methanogenic archaea reveals a shift towards energy conservation. BMC Genomics 18:639. doi: 10.1186/s12864-017-4036-4
Hallam, S. J., Putnam, N., Preston, C. M., Detter, J. C., Rokhsar, D., Richardson, P. M., et al. (2004). Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462. doi: 10.1126/science.1100025
Jasso-Chavez, R., Apolinario, E. E., Sowers, K. R., and Ferry, J. G. (2013). MrpA functions in energy conversion during acetate-dependent growth of Methanosarcina acetivorans. J. Bacteriol. 195, 3987–3994. doi: 10.1128/JB.00581-13
Jasso-Chavez, R., Diaz-Perez, C., Rodriguez-Zavala, J. S., and Ferry, J. G. (2016). Functional role of MrpA in the MrpABCDEFG Na+/H+ antiporter complex from the Archaeon Methanosarcina acetivorans. J. Bacteriol. 199, 1–13. doi: 10.1128/JB.00662-16
Kaster, A. K., Moll, J., Parey, K., and Thauer, R. K. (2011). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U.S.A. 108, 2981–2986. doi: 10.1073/pnas.1016761108
Kroninger, L., Berger, S., Welte, C., and Deppenmeier, U. (2015). Evidence for the involvement of two different heterodisulfide reductases in the energy conserving system of Methanomassiliicoccus luminyensis. FEBS J. 283, 472–483. doi: 10.1111/febs.13594
Lang, K., Schuldes, J., Klingl, A., Poehlein, A., Daniel, R., and Brune, A. (2015). New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of Candidatus methanoplasma termitum. Appl. Environ. Microbiol. 81, 1338–1352. doi: 10.1128/AEM.03389-14
Lessner, D. J., Li, L., Li, Q., Rejtar, T., Andreev, V. P., Reichlen, M., et al. (2006). An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U.S.A. 103, 17921–17926. doi: 10.1073/pnas.0608833103
Li, L., Li, Q., Rohlin, L., Kim, U., Salmon, K., Rejtar, T., et al. (2007). Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J. Proteome Res. 6, 759–771. doi: 10.1021/pr060383l
Li, Q., Li, L., Rejtar, T., Karger, B. L., and Ferry, J. G. (2005). The proteome of Methanosarcina acetivorans. Part II, comparison of protein levels in acetate- and methanol-grown cells. J. Proteome Res. 4, 129–136. doi: 10.1021/pr049831k
Li, Q., Li, L., Rejtar, T., Lessner, D. J., Karger, B. L., and Ferry, J. G. (2006). Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188, 702–710. doi: 10.1128/JB.188.2.702-710.2006
Lie, T. J., Costa, K. C., Lupa, B., Korpole, S., Whitman, W. B., and Leigh, J. A. (2012). Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc. Natl. Acad. Sci. U.S.A. 109, 15473–15478. doi: 10.1073/pnas.1208779109
Liu, P., Yang, Y., Lu, Z., and Lu, Y. (2014). Response of a rice paddy soil methanogen to syntrophic growth as revealed by transcriptional analyses. Appl. Environ. Microbiol. 80, 4668–4676. doi: 10.1128/AEM.01259-14
Liu, Y., and Whitman, W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 1125, 171–189. doi: 10.1196/annals.1419.019
Lubner, C. E., Jennings, D. P., Mulder, D. W., Schut, G. J., Zadvornyy, O. A., Hoben, J. P., et al. (2017). Mechanistic insights into energy conservation by flavin-based electron bifurcation. Nat. Chem. Biol. 6, 655–659. doi: 10.1038/nchembio.2348
Major, T. A., Liu, Y., and Whitman, W. B. (2010). Characterization of energy conserving hydrogenase B in Methanococcus maripaludis. J. Bacteriol. 192, 4022–4030. doi: 10.1128/JB.01446-09
McGlynn, S. E. (2017). Energy metabolism during anaerobic methane oxidation in ANME archaea. Microbes Environ. 32, 5–13. doi: 10.1264/jsme2.ME16166
Moran, J. J., House, C. H., Freeman, K. H., and Ferry, J. G. (2005). Trace methane oxidation studied in several Euryarchaeota under diverse conditions. Archaea 1, 303–309. doi: 10.1155/2005/650670
Moran, J. J., House, C. J., Thomas, B., and Freeman, K. H. (2007). Products of trace methane oxidation during nonmethyltrophic growth by Methanosarcina. J. Geophys. Res. 112, G02011–G02011. doi: 10.1029/2006JG000268
Nobu, M. K., Narihiro, T., Kuroda, K., Mei, R., and Liu, W. T. (2016). Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 10, 2478–2487. doi: 10.1038/ismej.2016.33
Peters, J. W., and Lubner, C. (2017). Electron bifurcation makes the puzzle pieces fall energetically into place in methanogenic energy conservation. Chembiochem 18, 2295–2297. doi: 10.1002/cbic.201700533
Rhee, T. S., Kettle, A. J., and Andreae, M. O. (2009). Methane and nitrous oxide emissions from the ocean: a reassessment using basin-wide observations in the Atlantic. J. Geophys. Res. 114:D12304. doi: 10.1029/2008JD011662
Richards, M. A., Lie, T. J., Zhang, J., Ragsdale, S. W., Leigh, J. A., and Price, N. D. (2016). Exploring hydrogenotrophic methanogenesis: a genome scale metabolic reconstruction of Methanococcus maripaludis. J. Bacteriol. 198, 3378–3390. doi: 10.1128/JB.00571-16
Schlegel, K., Leone, V., Faraldo-Gomez, J. D., and Muller, V. (2012a). Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc. Natl. Acad. Sci. U.S.A. 109, 947–952. doi: 10.1073/pnas.1115796109
Schlegel, K., and Muller, V. (2013). Evolution of Na+ and H+ bioenergetics in methanogenic archaea. Biochem. Soc. Trans. 41, 421–426. doi: 10.1042/BST20120294
Schlegel, K., Welte, C., Deppenmeier, U., and Muller, V. (2012b). Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279, 4444–4452. doi: 10.1111/febs.12031
Sieber, J. R., Mcinerney, M. J., Plugge, C. M., Schink, B., and Gunsalus, R. P. (2009). “Methanogenesis: syntrophic metabolism,” in Microbiology of Hydrocarbons, Oils, Lipids, ed. K. N. Timmis (Heidelberg: Springer-Verlag), 337–355.
Smith, K. S., and Ingram-Smith, C. (2007). Methanosaeta, the forgotten methanogen? Trends Microbiol. 7, 150–155.
Soo, V. W., Mcanulty, M. J., Tripathi, A., Zhu, F., Zhang, L., Hatzakis, E., et al. (2016). Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell Fact. 15:11. doi: 10.1186/s12934-015-0397-z
Sparling, R., Blaut, M., and Gottschalk, G. (1993). Bioenergetic studies of Methanosphaera stadtmanae, an obligate H2-methanol utilising methanogen. Can. J. Microbiol. 39, 742–748. doi: 10.1139/m93-109
Thauer, R. K., Kaster, A. K., Goenrich, M., Schick, M., Hiromoto, T., and Shima, S. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu. Rev. Biochem. 79, 507–536. doi: 10.1146/annurev.biochem.030508.152103
Thauer, R. K., Kaster, A. K., Seedorf, H., Buckel, W., and Hedderich, R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. doi: 10.1038/nrmicro1931
Timmers, P. H., Welte, C. U., Koehorst, J. J., Plugge, C. M., Jetten, M. S., and Stams, A. J. (2017). Reverse methanogenesis and respiration in methanotrophic Archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237
Valentine, D. L. (2002). Biogeochemistry and microbial ecology of methane oxidation in anoxic environments: a review. Anton. Van Leeuwenhoek 81, 271–282. doi: 10.1023/A:1020587206351
Vitt, S., Ma, K., Warkentin, E., Moll, J., Pierik, A., Shima, S., et al. (2014). The F420-reducing [NiFe]-hydrogenase complex from Methanothermobacter marburgensis, the first X-ray structure of a group 3 family member. J. Mol. Biol. 426, 2813–2826. doi: 10.1016/j.jmb.2014.05.024
Wagner, T., Ermler, U., and Shima, S. (2016). The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters. Science 354, 114–117. doi: 10.1126/science.aaf9284
Wagner, T., Koch, J., Ermler, U., and Shima, S. (2017). Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction. Science 357, 699–703. doi: 10.1126/science.aan0425
Wang, M., Tomb, J. F., and Ferry, J. G. (2011). Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 11:165. doi: 10.1186/1471-2180-11-165
Wang, F. P., Zhang, Y., Chen, Y., He, Y., Qi, J., Hinrichs, K. U., et al. (2014). Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J. 8, 1069–1078. doi: 10.1038/ismej.2013.212
Welte, C., and Deppenmeier, U. (2014). Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim. Biophys. Acta 1837, 1130–1147. doi: 10.1016/j.bbabio.2013.12.002
Yan, Z., Joshi, P., Gorski, C. A., and Ferry, J. G. (2018). A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat. Commun. 9:1642. doi: 10.1038/s41467-018-04097-9
Keywords: archaea, heterodisulfide reductase, methane, ferredoxin, hydrogen, acetate, formate
Citation: Yan Z and Ferry JG (2018) Electron Bifurcation and Confurcation in Methanogenesis and Reverse Methanogenesis. Front. Microbiol. 9:1322. doi: 10.3389/fmicb.2018.01322
Received: 03 April 2018; Accepted: 30 May 2018;
Published: 20 June 2018.
Edited by:
Michael W. Adams, University of Georgia, United StatesReviewed by:
Wolfgang Buckel, Philipps University of Marburg, GermanyCopyright © 2018 Yan and Ferry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James G. Ferry, amdmM0Bwc3UuZWR1
†Present address: Zhen Yan, Department of Molecular, Cellular and Developmental Biology, University of Michigan, Ann Arbor, MI, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.