- 1Institute of Environmental Microbiology, College of Resources and Environment, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Life Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 3College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
- 4J. Craig Venter Institute, La Jolla, CA, United States
Rho GTPases are signaling macromolecules that are associated with developmental progression and pathogenesis of Fusarium graminearum. Generally, enzymatic activities of Rho GTPases are regulated by Rho GTPase guanine nucleotide exchange factors (RhoGEFs). In this study, we identified a putative RhoGEF encoding gene (FgBUD3) in F. graminearum database and proceeded further by using a functional genetic approach to generate FgBUD3 targeted gene deletion mutant. Phenotypic analysis results showed that the deletion of FgBUD3 caused severe reduction in growth of FgBUD3 mutant generated during this study. We also observed that the deletion of FgBUD3 completely abolished sexual reproduction and triggered the production of abnormal asexual spores with nearly no septum in ΔFgbud3 strain. Further results obtained from infection assays conducted during this research revealed that the FgBUD3 defective mutant lost its pathogenicity on wheat and hence, suggests FgBud3 plays an essential role in the pathogenicity of F. graminearum. Additional, results derived from yeast two-hybrid assays revealed that FgBud3 strongly interacted with FgRho4 compared to the interaction with FgRho2, FgRho3, and FgCdc42. Moreover, we found that FgBud3 interacted with both GTP-bound and GDP-bound form of FgRho4. From these results, we subsequently concluded that, the Rho4-interacting GEF protein FgBud3 crucially promotes vegetative growth, asexual and sexual development, cell division and pathogenicity in F. graminearum.
Introduction
Fusarium head blight (FHB), a disease of wheat and barley is mainly caused by the filamentous ascomycete Fusarium graminearum (teleomorph Gibberella zeae) (Goswami and Kistler, 2004). Besides causing huge yield and economic losses, the FBH fungus also secretes a variety of harmful mycotoxins, including, deoxynivalenol (DON), zearalenone and T-2 toxin, into infested grains (Desiardins et al., 1996; Bottalico and Perrone, 2002; Proctor et al., 2002; Goswami and Kistler, 2004; Starkey et al., 2007).
Collectively, Rho GTPases are a small group of GTPases in the Ras GTPase superfamily. They switch between active and inactive form by binding to GTP and GDP respectively. The activation of Rho GTPases by guanine nucleotide-exchange factors (GEFs) transforms it from GDP binding state to GTP binding state (Iden and Collard, 2008). In the fungal kingdom, Rho GTPases including Rho1 and Cdc42 and their corresponding GEF activators have been well studied in budding yeast. For instance, research has shown that in Saccharomyces cerevisiae three RhoGEFs; Rom1, Rom2 and Tus1 play coordinated roles in activating Rho1 which is an essential GTPase associated within the cell wall integrity pathway, actin ring assembly and cytokinesis processes (Ozaki et al., 1996; Manning et al., 1997; Schmidt et al., 1997; Schmelzle et al., 2002; Yoshida et al., 2006, 2009). Loss of Tus1 or Rom1 alone resulted in only subtle phenotypes whereas loss of Rom2 caused cell lysis at high temperatures (Schmelzle et al., 2002; Lesage et al., 2005; Hillenmeyer et al., 2008). Rom1 and Rom2 perform overlapping functions, hence, loss of both of genes had a lethal effect and caused cell lysis at all temperatures (Ozaki et al., 1996). Contrary to previous suggestions that Cdc24 functions as the sole RhoGEF for Cdc42, current investigations interestingly identified Bud3, a protein containing a putative GTP-binding motif and a Dbl homology (DH) domain (also known as RhoGEF domain), as additional RhoGEF for Cdc42 and further proceeded to show that Bud3 functions as Cdc42 GEF during early G1 phase in budding yeast (Sloat et al., 1981; Adams et al., 1990; Chenevert et al., 1994; Zheng et al., 1994; Kang et al., 2014). Previous studies also identified Bud3 homologues in Neurospora crassa and Aspergillus nidulans as Rho4 GEFs (Justa-Schuch et al., 2010; Si et al., 2010). Insights gained from studies conducted in yeast and filamentous fungi showed that some of these Rho GTPases, i.e., Rho1 in the budding yeast, could be activated by more than one GEF (Justa-Schuch et al., 2010; Krause et al., 2012; Kang et al., 2014). Additional evidence also confirmed that a given GEF could regulate two different Rho GTPases. For example, it has been shown that Cdc24, besides operating as GEF for Cdc42, also activated GEF for Rac1 in N. crassa (Araujo-Palomares et al., 2011). Normally, activated Rho GTPases interact with downstream effectors on membranes to regulate signal transduction pathways (Iden and Collard, 2008). For instance, FgRac1 specifically interacted with downstream targets including FgCla4 and FgNoxR to regulate asexual and sexual development in F. graminearum, respectively (Zhang et al., 2013, 2016). In some plant pathogens, Rho GTPases were not only important for polarity growth, sexual and asexual reproduction, cytokinesis but also pathogenesis (Zheng et al., 2007, 2009; Chen et al., 2008; Harris, 2011; Kwon et al., 2011; Nesher et al., 2011; Zhang et al., 2013). Up to now, the functions of RhoGEFs and their relationship with different Rho GTPases in plant pathogens are still less studied. Therefore, it is important to identify their roles in fungal development and pathogenesis process in plant pathogens.
In our previous study, we identified six Rho GTPases in F. graminearum, and showed that all six Rho GTPases were associated with development and pathogenesis of F. graminearum in varying manner (Zhang et al., 2013). For example, we demonstrated that deletion of FgCdc42 caused serious impairment in, growth, conidiation, sexual development and rendered resultant mutant strains non-pathogenic, while a deletion of FgRho2 only caused a slight reduction in growth and virulence (Zhang et al., 2013). However, the influence of GEF proteins on regulatory activities of Rho GTPases in F. graminearum has not been reported. In this study, we identified six putative RhoGEF proteins in F. graminearum by homology alignment. We further characterized functions of a Rho4 interacting RhoGEF protein, FgBud3, and found it was not only important for vegetative growth, reproduction and pathogenicity, but also for cell division in F. graminearum.
Materials and Methods
Strains, Media and Growth Condition
Conidia of the FgBUD3 deletion mutants, ΔFgbud3-1 and ΔFgbud3-6, were stored in 20% glycerol solution at -80°C. Complete medium (CM) and synthetic low-nutrient agar (SNA) medium was used for mycelial growth assays and conidiation assays as previously described (Zheng et al., 2012). Strains grown on CM plates supplemented with 0.1 mg/mL Calcofluor White (CFW) were used to test the sensitivity against cell-wall-disrupting agents (Jiang et al., 2011). Sexual reproduction assays were performed on carrot agar (CA) medium in accordance with a previously described experimental protocol (Bowden and Leslie, 1999; Zheng et al., 2013).
Generation of the FgBUD3 Deletion Mutants and the Complementary Strain
To generate an FgBUD3 deletion vector construct, we first use primer pairs AF-BUD3/AR- BUD3 and BF- BUD3/BR- BUD3 (Supplementary Table S1) to amplify the upstream and downstream fragments of the FgBUD3 gene from the genome of F. graminearum wild-type strain PH-1, the resulting amplicons were cloned by linking upstream and downstream sequences of hygromycin-resistance gene in a pCX62 vector respectively. The protoplasting buffer [0.5 g driselase (D9515, Sigma-Aldrich, Inc.), 0.1 g lysing enzymes (L1412, Sigma-Aldrich, Inc.) and 20 mL 1 M KCl solution] was used for protoplast preparation of F. graminearum. The details of the protoplast preparation and fungal transformation were described in an established protocol (Hou et al., 2002). Hygromycin-resistant transformants were screened by PCR and RT-PCR with primer pair OF- BUD3/OR- BUD3 and further verified by Southern blot.
For complementation of the FgBUD3 deletion, a fragment amplified by primer pairs CF-BUD3/CR-BUD3 (Supplementary Table S1) was co-transformed with a geneticin-resistant gene fragment into the protoplast of the FgBUD3 deletion mutant (ΔFgbud3-1). Geneticin-resistant transformants were screened by PCR and RT-PCR.
Infection and DON Production Assays
For flowering wheat heads (Bainong 979) infection, conidia were collected from 7-day-old SNA plates and resuspended in sterile distilled water to a concentration of 2 × 105 conidia/ml. The middle spikelet of wheat flowers was inoculated with 10 μl of the conidial suspension as described (Gale et al., 2002; Wang et al., 2012). Autoclaved rice grains were inoculated with conidia and cultured for 2 weeks and assayed for DON toxin as described (Seo et al., 1996). Ergosterol levels were used to normalize DON content per fungal mass. The DON production level was also detected by the DON Plate Kit (Shenzhen Finder Biotech Co., Ltd.), 104 conidia of each strain were grown in 1.5 ml liquid TBIA culture (Zhang et al., 2016). Fifty microliter of 8-old-day cultures or cultures with 10-fold or 1000-fold dilutions were used for DON detection following the manipulation instructions of kit. The thorough dried mycelia weight of each strain in the cultures was used to normalize the DON production level.
Staining and Microscopy Observation
Conidia or mycelia were stained with 10 μg/mL CFW (Rasmussen and Glass, 2007) and 5 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) (Seong et al., 2008) for septa and nuclei observation, respectively. An Olympus BX51 Microscope was used to perform microscopic observations. The morphology of aerial hypha, conidia and germinated conidia of different strains were observed under light microscopy. The septa and nuclei observation was performed under UV microscopy. The microscopy images were used to observe the morphology of conidia and hypha and to calculate the quantity of septa and nuclei. We randomly chose more than 100 conidia of each strain to calculate the length and width of conidia and the quantity of septa or nuclei.
Yeast Two-Hybrid Assay
Yeast two-hybrid assay was performed using the MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech). FgRho4 ORF amplicon was amplified with primer pairs BDF-RHO4/BDR-RHO4 from cDNA of PH-1 with the site mutation C267S to ensure that the FgRho4 protein cannot be prenylated and was thus soluble (Chen et al., 2008). The amplicon was cloned into the pGBKT7 vector to create the vector BD-FgRho4. Similar methods were used to construct the vectors BD-FgRho1, BD-FgRho2, BD-FgRho3, BD-FgRac1 and BD-FgCdc42. Template BD-FgRho4 vector and two primers CAF and CAR (Supplementary Table S1) were used to generate a constitutively active (G64V) mutation with primers containing the substitution of the glycine (G64V) of FgRho4 with valine. Primers DNF and DNR (Supplementary Table S1) were used to generate dominant negative mutations, the (D172A) mutation with primers containing the substitution of the aspartic acid (D172A) with alanine. The resultant vectors were named BD-FgRho4 (CA) and BD-FgRho4 (DN). A partial FgBUD3 ORF including RhoGEF domain region was amplified from cDNA of PH-1 with primer pairs ADF-BUD3/ADR-BUD3, and the amplicon was cloned into pGADT7 vector to create AD-FgBud3 vector. The resultant bait and prey vectors were confirmed by sequencing and were co-transformed in pairs into the yeast strain AH109 (Schiestl and Gietz, 1989). The plasmid pairs, pGBKT7-53 and pGADT7 and pGBKT7- Lam and pGADT7-T, served as the positive and the negative control, respectively. The isolation and confirmation of transformants were described previously (Chen et al., 2008).
Results
Identification of RhoGEF Proteins in F. graminearum
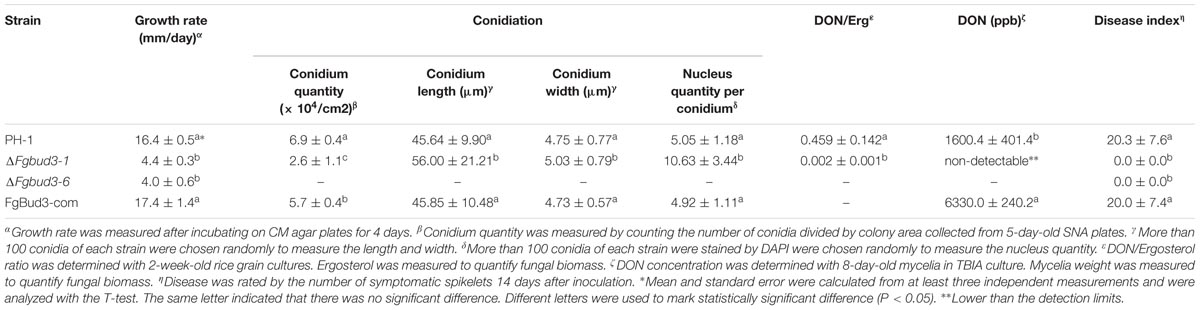
We identified six proteins harboring RhoGEF domains in the F. graminearum genome database1 encoded by, FGSG_08572, FGSG_08568, FGSG_01266, FGSG_11886, FGSG_05273, and FGSG_10511. These proteins differed in lengths ranging from 582 to 1704 amino acids. In addition to the RhoGEF domain, four of these proteins (FGSG_08572, FGSG_8568, FGSG_11886, and FGSG_10511) contained additional domains, which include, BAR, PB1, CDC24 domains and thus, suggested these four proteins may assume diverse functions in F. graminearum (Figure 1A). Subsequent alignments performed with amino acid sequences of these six putative F. graminearum RhoGEFs recorded very low homology between the respective GEF proteins. However, domain alignment results showed that seven residues in two of the motifs identified are only conserved in the RhoGEF domain sequences of the six proteins (Figure 1B). These results suggested these seven amino acids might be important for the function of RhoGEF proteins.
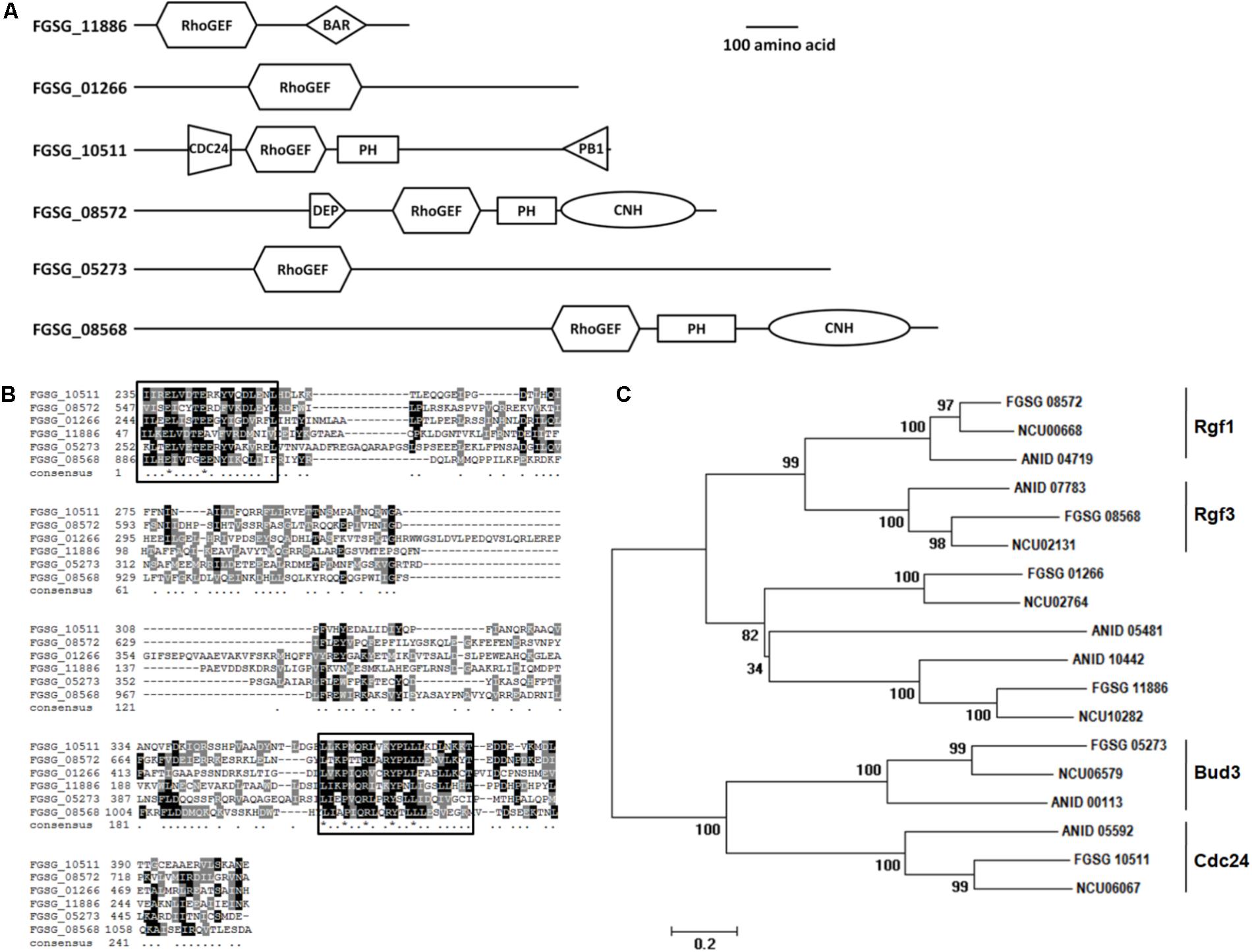
FIGURE 1. Fusarium graminearum encodes six putative RhoGEF proteins. (A) The domain distribution of six F. graminearum RhoGEFs. (B) The RhoGEF domain amino acid sequences of six F. graminearum RhoGEFs were aligned. Residues conserved in all proteins were highlighted in black or gray shaded. The black frames showed the conserved regions. (C) Phylogenetic comparison of six F. graminearum RhoGEFs with other fungal RhoGEFs. The amino acid sequences of RhoGEFs from N. crassa (NCU), A. nidulans (ANID) and F. graminearum (FGSG) were analyzed by the Clustal X 1.83 program to create a progressive alignment dendrogram. The branch lengths were proportional to the mean number of differences per residue along each branch. Accession number of each protein was shown as follows: FGSG_08572, XP_011320178.1; NCU00668, XP_965808.3; ANID_04719, XP_662323.1; ANID_07783, XP_681052.1; FGSG_08568, XP_011320182.1; NCU02131, XP_011392943.1; FGSG_01206, XP_011317046.1; NCU02764, XP_964259.2; ANID_05481, XP_663085.1; ADID_10442, XP_661358.1; FGSG_11886, XP_011316798.1; NCU10282, XP_001728403.1; FGSG_05273, XP_011323784.1; NCU06579, XP_961939.3; ANID_00113, XP_657717.1; ANID_05592, XP_663196.1; FGSG_10511, XP_011319503.1; NCU06067, XP_959830.3.
We also initiated a comparative homology search by comparing these F. graminearum RhoGEFs proteins identified in this study with RhoGEF proteins previously reported in N. crassa and A. nidulans (Justa-Schuch et al., 2010; Si et al., 2010; Araujo-Palomares et al., 2011; Richthammer et al., 2012). This search found homologs of F. graminearum RhoGEF proteins in these two fungi. Maximum likelihood analysis of these RhoGEF proteins revealed six independent lineages for each RhoGEF member in N. crassa, A. nidulans, and F. graminearum (Figure 1C). From these integrated analyses, we successfully identified four F. graminearum RhoGEF proteins in accordance with their homologs in N. crassa and A. nidulans and named them FgBud3, FgRgf3, FgRgf1, and FgCdc24, which encoded by FGSG_05273, FGSG_08568, FGSG_08572, and FGSG_10511, respectively (Justa-Schuch et al., 2010; Si et al., 2010; Araujo-Palomares et al., 2011; Richthammer et al., 2012). One of these F. graminearum RhoGEFs, FgBud3, was further characterized in this study.
The Deletion of FgBUD3 Exerted Serious Adverse Effect on Vegetative Growth of ΔFgbud3 Strain
FgBud3 contains 1477 amino acids and a RhoGEF domain at the N-terminus (Figure 1B). To study the function of FgBud3, we generated FgBUD3 deletion mutants by replacing the ORF of FgBUD3 with a hygromycin resistance gene as a selectable marker in F. graminearum and confirmed the generated gene deletion by PCR, RT-PCR and Southern blot assay (Supplementary Figure S1).
Two of the FgBUD3 deletion mutants (ΔFgbud3-1 and ΔFgbud3-6) and the wild-type strain were grown on CM plates for 4 days. The FgBUD3 deletion mutants grew much slower than the wild-type (Table 1 and Figure 2A). Compared with the dense aerial hypha produced by the wild-type strain PH-1, ΔFgbud3-1 only produced tiny aerial hypha (Figure 2A). We further generated a complemented strain (FgBud3-com) by reintroducing the native FgBUD3 into ΔFgbud3-1 strain and normal vegetative growth phenotype was observed (Table 1 and Figure 2A). The FgBUD3 deletion mutant grew very poorly on CM plates containing 100 μg/mL CFW or 1.5 mg/mL Congo red. The sensitivity of ΔFgbud3-1 to CFW or Congo red is higher than sensitivity of PH-1 and FgBud3-com (Figure 2B). Moreover, many protoplasts could be observed after mycelia lysed after incubation in protoplasting buffer for 1 h (data not shown) in the FgBUD3 deletion mutant. In contrast, much fewer protoplasts of the wild-type strain could be observed under the same treatment, suggesting the FgBUD3 deletion mutant was highly sensitive to cell wall damaging agents. These results suggested FgBud3 was involved in polarity growth and cell wall integrity.
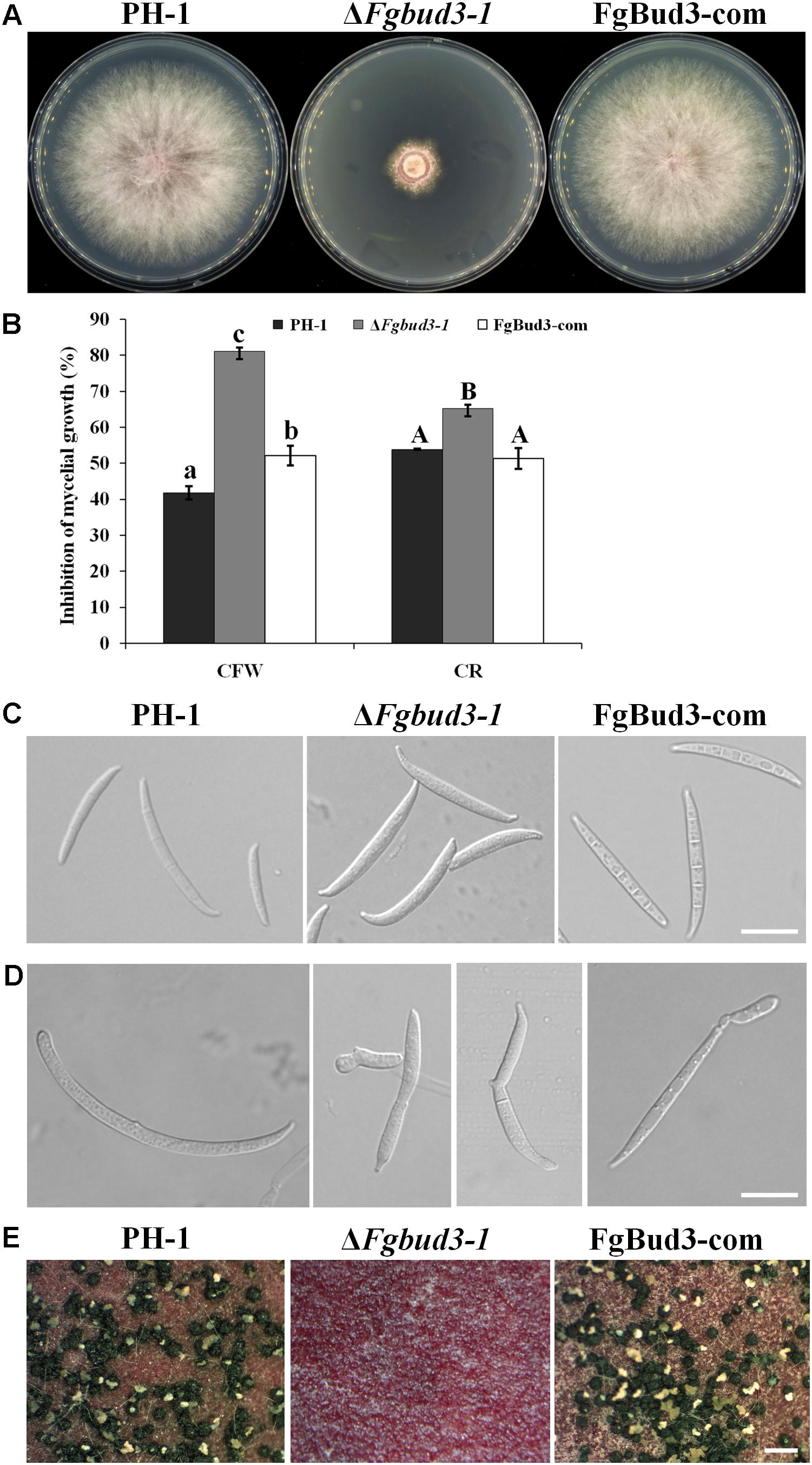
FIGURE 2. The hyphal growth, asexual and sexual developmental defects of the FgBUD3 deletion mutant. (A) The hyphal growth of strains grown on CM plates. (B) Sensitivity of each strain to 100 μg/mL CFW and 1.5 mg/mL CR (Congo red). Different letters were used to mark statistically significant difference (P < 0.05). (C) Conidia produced by PH-1, ΔFgbud3-1 and FgBud3-com with normal Fusarium-shape. Bar = 20 μm. (D) Conidia produced by the FgBUD3 deletion mutant with different kinds of abnormal shape. Bar = 20 μm. (E) The sexual development defect of the FgBUD3 deletion mutant. Bar = 500 μm. PH-1, wild-type strain; ΔFgbud3-1, gene FgBUD3 deletion mutants; FgBud3-com, complemented strain of FgBUD3 deletion mutant.
FgBud3 Is Required for Conidiogenesis and Sexual Reproduction
We incubated the FgBUD3 deletion mutant and the wild-type strain on SNA plates for 5 days, and observed that the mutant produced fewer conidia than the wild-type strain (Table 1). We also noticed that the average conidium size of ΔFgbud3-1 was larger than PH-1 and FgBud3-com; many conidia of ΔFgbud3-1 were deformed in shape, for example, some these conidia conjugated with the phialide cell (Table 1 and Figures 2C,D). From these results, we inferred that FgBud3 is involved in the regulation of the conidia morphogenesis process in F. graminearum.
Our investigations further showed that deletion of FgBUD3 in the homothetic fungus, F. graminearum completely abolished perithecia production in the ΔFgbud3-1 strain cultured on CA plates after induction, indicating that FgBud3 is essential for sexual reproduction in F. graminearum (Figure 2E).
FgBud3 Is Involved in Cell Division
Deletion of BUD3 in A. nidulans and N. crassa resulted in defective septum formation (Justa-Schuch et al., 2010; Si et al., 2010). In accordance with this report, we hypothesized that FgBud3 might play a similar role in F. graminearum. To test this hypothesis, we stained conidia of the FgBUD3 deletion mutant and wild-type strain with CFW. Corresponding results obtained from these assays showed that most conidia produced by ΔFgbud3-1 lacked septa while the majority of conidia produced by the wild-type strain possessed 3–5 septa (Figures 3A,B). In some rare cases, we observed 1–2 septa in some conidia produced by the deletion mutant (Figures 3A,B, 4A). In addition, we used DAPI to perform conidia nuclei staining. Results from these examinations revealed that conidia produced by the ΔFgbud3-1 strain contain more nuclei than conidia obtained from the wild-type or complemented strain (Table 1 and Figure 4A). Similar results were observed in fresh hypha germinating from conidia after incubation in CM culture for 6 h (Figures 3C, 4B). Moreover, protoplasts from ΔFgbud3-1 generated in protoplasting buffer were much bigger and harbored more nuclei than protoplasts of the wild-type or the complemented strain (Figure 4C). These results indicated the FgBud3 play key role in cell division related processes.
FIGURE 3. The septum defective phenotype of the FgBUD3 deletion mutant. (A) Conidia of PH-1, ΔFgbud3-1 and FgBud3-com were stained with 1 μg/ml CFW and examined by microscopy under DIC or UV light. (B) Ratio of conidia with different septal quantity of each strain. (C) Hypha of each strain were stained and examined. The arrows indicate septa. Bar = 20 μm.
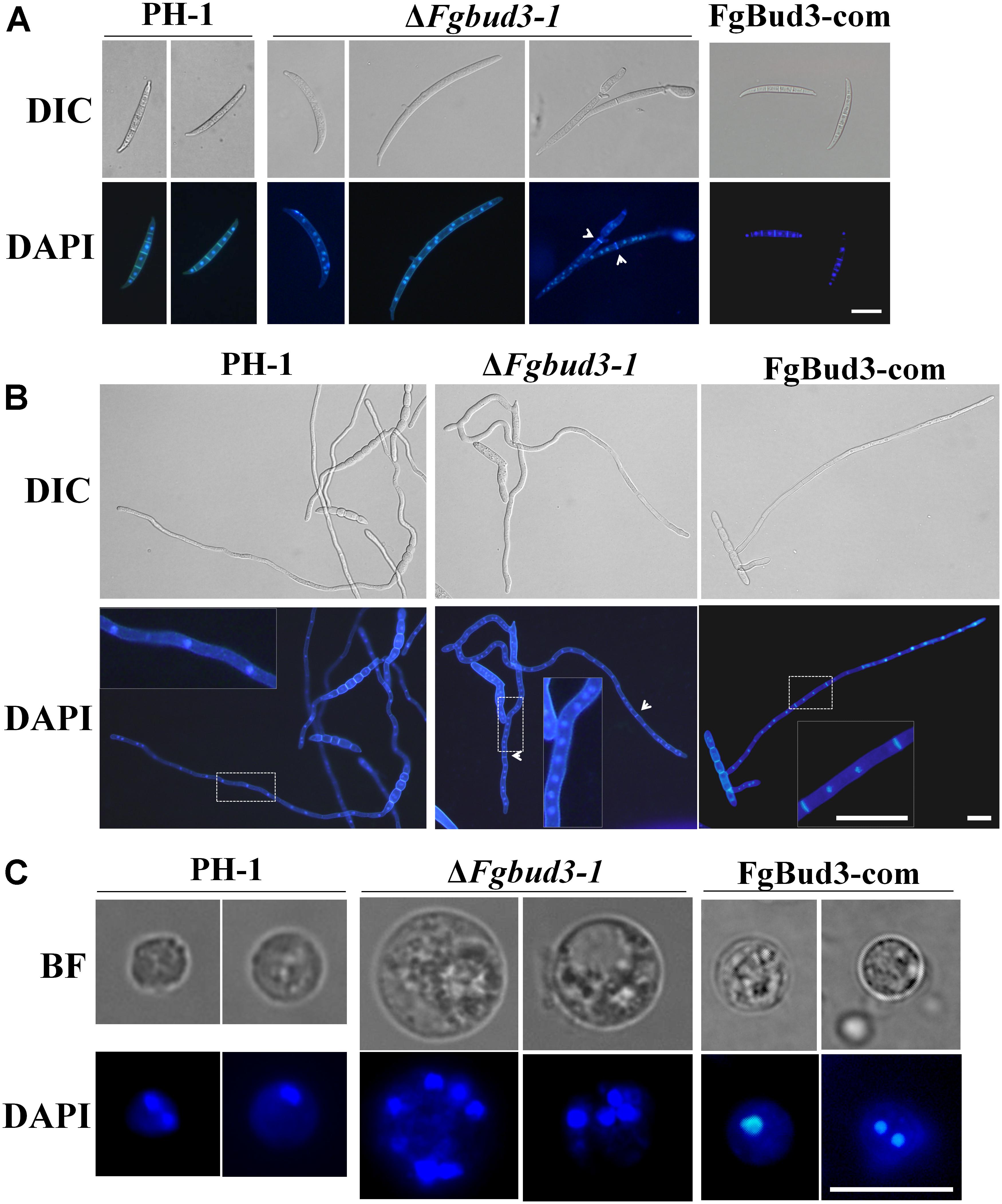
FIGURE 4. The nuclear division phenotype of the FgBUD3 deletion mutant. Conidia (A) or hyphae (B) of PH-1, ΔFgbud3-1 and FgBud3-com were stained with 4′,6-diamidino-2-phenylindole (DAPI) and examined by microscopy under DIC or UV light. The arrows indicate septa. Bar = 20 μm. (C) Protoplasts of each strain were stained and examined. Bar = 10 μm.
FgBud3 Is Important for Pathogenicity and DON Production
We inoculated a spikelet in the middle of wheat heads with conidia of the FgBUD3 deletion mutants, wild-type strain or the complemented strain. After 14 days, serious symptoms were observed in wheat heads inoculated with conidia obtained from the wild-type strain or the complemented strain (Table 1 and Figure 5). On the contrarily, no symptoms were observed in wheat head spikelets inoculated with conidia harvested from the ΔFgbud3-1 strain, indicating that FgBud3 is required for pathogenicity of F. graminearum (Table 1 and Figure 5). Furthermore, we monitored the level of DON generated in the ΔFgbud3-1 strain compared to the wild-type strain in rice grains and TBIA culture. DON production assessment results showed that, the level of DON generated in both rice grains and TBIA mudium inoculated with ΔFgbud3-1 strain was at a significantly lower than that inoculated with wild-type strain, suggesting that FgBud3 played an important role in regulating DON production F. graminearum (Table 1).

FIGURE 5. Infection assays with flowering wheat heads. Wheat heads infected with conidia of PH-1, ΔFgbud3-1 and FgBud3-com. The arrows indicate inoculated spikelets.
FgBud3 Interacts With Both GDP-Bound and GTP-Bound FgRho4
Bud3-homologs were identified as Rho4GEFs in both N. crassa and A. nidulans, FgBud3 could play a similar role in F. graminearum (Justa-Schuch et al., 2010; Si et al., 2010). As a putative Rho4GEF, FgBud3 would be predicted to interact with an inactivated (GDP-bound) FgRho4 to replace GTP for GDP. Thus, a yeast two-hybrid assay was performed between RhoGEF domain of FgBud3 and different states of FgRho4. The result indicated the FgBud3 RhoGEF domain interacted with wild-type FgRho4 and both GTP-bound (FgRho4-CA) and GDP-bound FgRho4 (FgRho4-DN) (Figure 6A). Furthermore, we performed another yeast two-hybrid assay to determine if FgBud3 interacted with other Rho GTPases in F. graminearum. The result indicated the FgBud3 RhoGEF domain interacted not only with FgRho4 but also with FgRho2, FgRho3 and FgCdc42. However, the interaction between the FgBud3 RhoGEF domain and FgRho4 was stronger than other interactions (Figure 6B).
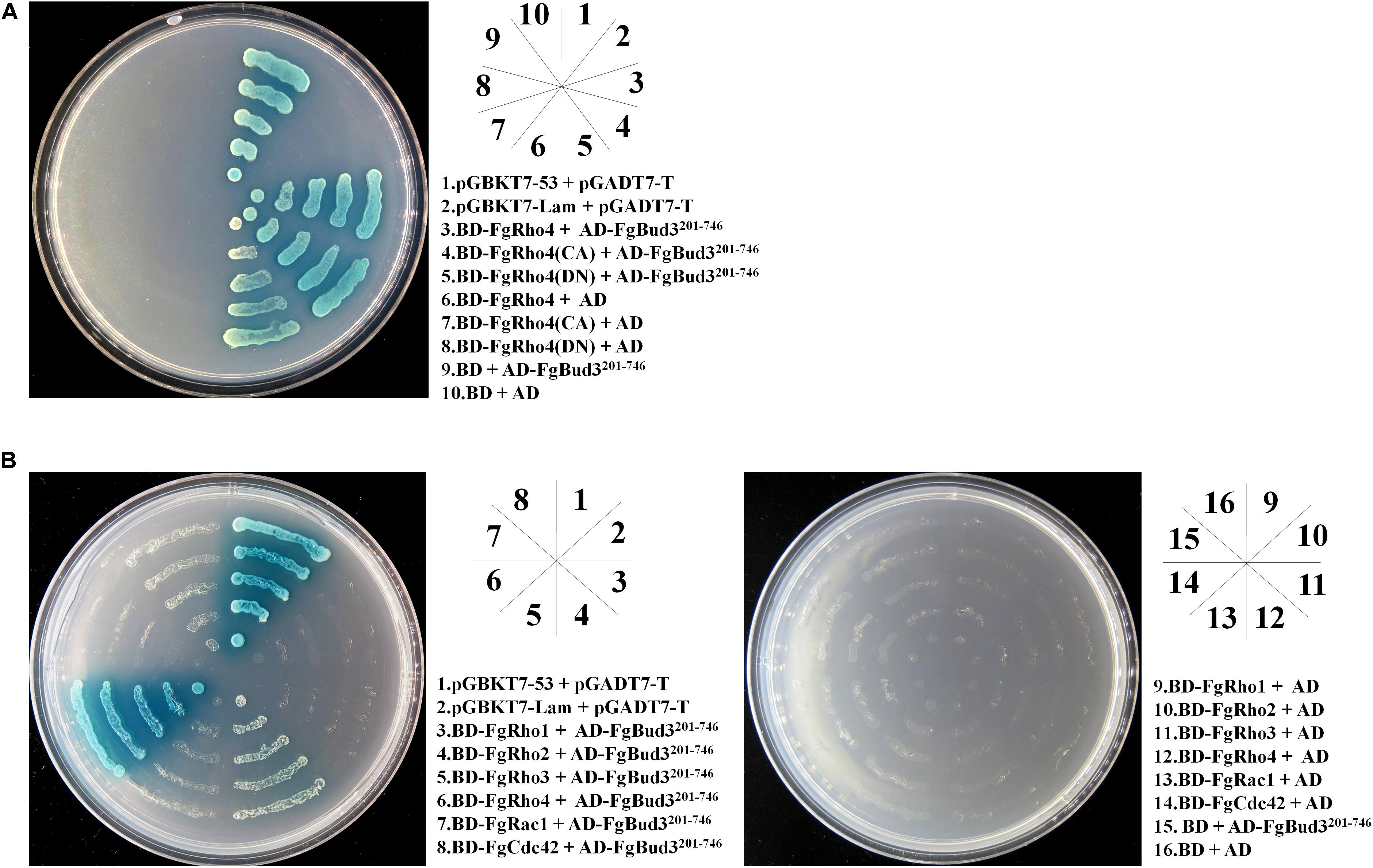
FIGURE 6. FgBud3 interacts with FgRho4. (A) Yeast two-hybrid assay with FgRho4 (3) or FgRho4-CA (4) or FgRho4-DN (5) as the bait and FgBud3201-746 (3–5) as the prey. (B) Yeast two-hybrid assay with each FgRho GTPase (3–8) as the bait and FgBud3201-746 (3–8) as the prey. Twenty microliter yeast cells (107 cell/mL) of each yeast transformants were grown on the SD-Leu-Trp-His-Ade plates were assayed for α-galactosidase activity. The interaction of pGBKT7-53 and pGADT7-T (1) was used as the positive control. The other interactions were used as the negative control.
Discussion
The exchange of GDP for GTP is required for the activation of Rho GTPases and RhoGEF proteins are responsible for this process. Rho GTPases in F. graminearum were shown to be important to fungal development and/or pathogenesis, but their activators, RhoGEF proteins, had not been reported yet (Zhang et al., 2013). In this study, we identified six putative RhoGEF proteins in F. graminearum. In N. crassa, a RhoGEF, Cdc24, regulated two Rho GTPases (Cdc42 and Rac1) while Rho4 was activated by two RhoGEFs (Bud3 and Rgf3) (Justa-Schuch et al., 2010; Araujo-Palomares et al., 2011). It suggested the relationship between RhoGEFs and Rho GTPases was not “one RhoGEF to one Rho GTPase.” This pattern may be also occurring in another filamentous fungus, F. graminearum. Homologs of Cdc24, Bud3, Rgf3 and Rgf1 in N. crassa and A. nidulans play important roles in polarity growth, conidiation and/or septation suggesting F. graminearum RhoGEFs could play similar roles (Justa-Schuch et al., 2010; Si et al., 2010; Araujo-Palomares et al., 2011; Richthammer et al., 2012). In addition, they could have some different functions, e.g., a function in pathogenesis, as compared to homologs in N. crassa and A. nidulans. Therefore, we characterized the functions of one of the four RhoGEF proteins, FgBud3, in F. graminearum. Our results further demonstrated FgBud3 is involved in multiple processes, such as polarity growth, conidiogenesis, sexual reproduction, cell division and pathogenicity.
Bud3-homolog has been first studied in a plant pathogen in our study, and the result indicated FgBud3 was essential for pathogenicity in F. graminearum (Table 1 and Figure 5). The serious growth defect is one of the reasons accounting for pathogenicity defects displayed by the FgBUD3 deletion mutant as shown in many other mutants of F. graminearum with growth defects, such as deletion mutants of some transcription factors or kinases (Son et al., 2011; Wang et al., 2011). The deletion of these types of genes usually does not only exert serious effect on growth, but also interferes with cell wall sensitivity, triggers reduction in DON production and/or renders respective deletion mutants non-pathogenic. Although, the growth of FgRho2 deletion mutant was indistinguishable from the wild-type strain, it however, displayed attenuated virulence and compromised cell wall sensitivity (Zhang et al., 2013). The deletion mutants of some TRI genes such as TRI6 and TRI10 did not display any DON production and subsequently almost no virulence without having any other defects suggesting this toxin was a key factor for pathogenesis (Seong et al., 2009). Thus, it comes as no surprise that the FgBUD3 deletion mutant with sensitive cell wall and almost no DON production is also non-pathogenic.
The FgBUD3 deletion mutant showed multiple defects compared to wild-type, and the complemented strain FgBud3-com was successful to recover these defects. Interestingly, FgBud3-com produced four times higher DON than wild-type PH-1. We can reason that the high DON production level of the complemented strain was due to the random insertion of FgBUD3 in the complemented strain which could cause a different expression pattern of FgBUD3 compare to the wild-type.
Depletion of FgBud3 resulted in serious defects in aerial hyphal growth and abnormal conidiation suggesting FgBud3 is required for polarity growth and maintenance. Fungal Rho GTPases are well known for regulating polarity growth and maintenance (Harris, 2011). In F. graminearum, loss of FgRHO4 caused serious conidiogenesis defects such as producing many abnormal conidia, and some conidia could even be generated on other conidia (Zhang et al., 2013). FgRac1 and FgCdc42, two other Rho GTPases, were also shown to be important for polarity growth and maintenance in that their corresponding gene deletion mutant led to a hyper-branching phenotype and an abnormal conidia shape, respectively (Zhang et al., 2013). The PAK kinase FgCla4, a downstream target of both FgCdc42 and FgRac1, was also required for polarity growth (Zhang et al., 2013). These results suggested FgBud3 might be involved in activating at least one of these Rho GTPases to regulate polarity growth by interacting with their downstream targets. In the Rho GTPase family, Rho4 is well conserved and regulates septation in filamentous fungi (Rasmussen and Glass, 2005, 2007; Justa-Schuch et al., 2010; Si et al., 2010; Kwon et al., 2011; Zhang et al., 2013). In F. graminearum, FgRho4 is required for both septum formation and nuclear division (Zhang et al., 2013). FgBud3, a putative RhoGEF, exerted similar functions, not only on septum formation and nuclear division but also on vegetative growth, cell wall integrity, conidiogenesis, sexual reproduction and pathogenesis in a manner similar to FgRho4. These results strongly imply FgBud3 to be a Rho4GEF for FgRho4. A yeast two hybrid assay was performed to investigate the interaction between FgBud3 and FgRho4. Unfortunately, the interaction was negative. We supposed it was because the large size of the FgBud3 protein (1477 amino acids) and it was difficult to make a correct protein folding in yeast. Therefore, we used the RhoGEF domain to replace the full length of FgBud3 as a prey in the yeast two hybrid assay. The result revealed the interaction between an inactivated form (GDP-bound) of FgRho4 and FgBud3 indicating FgBud3 was a GEF of FgRho4 (Figure 6A).
In N. crassa, the RhoGEF Cdc24 can activate two Rho GTPases, Rac1 and Cdc42 (Araujo-Palomares et al., 2011). Thus, we hypothesized that FgBud3 might interact with other Rho GTPases to regulate their downstream targets in F. graminearum. To test this hypothesis, one more yeast two hybrid assay was exerted and the result showed the RhoGEF domain of FgBud3 also interacted with FgRho2, FgRho3, and FgCdc42, though the intensity of these interactions were weaker than the interaction between FgBud3 and FgRho4 (Figure 6B). In N. crassa, Bud3 was a specific RhoGEF of Rho4 (Justa-Schuch et al., 2010). If the pattern was also happening in F. graminearum, it seemed the weak interactions between partial of FgBud3 and other Rho GTPases, e.g., FgRho3, may not be accurate enough due to the compromised specificity of FgBud3. However, the interaction results were still reliable, because the RhoGEF domain of FgBud3 did not interact with FgRho1 and FgRac1 (Figure 6B). The recent result showed Bud3 activated Cdc42 to establish a proper growth site in budding yeast (Kang et al., 2014), suggesting FgBud3 could also be an activator of FgCdc42 in F. graminearum. Rac1 is a Rho GTPase that closely related to Cdc42, which is not appeared in budding yeast but filamentous fungi. Besides, Rac1 and Cdc42 share overlapping functions in some filamentous fungi (Virag et al., 2007; Araujo-Palomares et al., 2011; Kwon et al., 2011; Zhang et al., 2013). Surprisingly, FgCdc42 but not FgRac1 can interact with FgBud3 (Figure 6B). Both FgCDC42 deletion mutant and FgRAC1 deletion mutant revealed serious growth and conidiation defects (Zhang et al., 2013). However, deletion of FgCDC42 but not FgRAC1 led to conidia with a serious abnormal shape which is a similar phenotype of a FgBUD3 deletion mutant (Zhang et al., 2013). It implied FgBud3 also activated FgCdc42 to regulate morphogenesis of conidia. Rho1 was an upstream regulator of the well known cell wall integrity MAPK pathway in budding yeast (Levin, 2005). FgBud3 did not interact with FgRho1 indicating FgBud3 regulated cell wall integrity not through the potential FgRho1-MAPK pathway in F. graminearum. FgRho2 and FgRho4 were important to cell wall integrity (Zhang et al., 2013). The interaction between FgBud3 and FgRho2 or FgRho4 suggested FgBud3 could be a GEF of both FgRho2 and FgRho4 to regulate cell wall integrity.
As we have mentioned above, the FgBUD3 deletion mutant showed almost the same phenotype as the FgRHO4 deletion mutant, however, we still found a difference in septum formation between them. In rare cases, conidia with one or even two septa were generated in the FgBUD3 deletion mutant but not in the FgRHO4 deletion mutant (Figures 3A,B, 4A) (Zhang et al., 2013). One possible reason behind this difference could be attributed to a residual natural capacity in the exchange of GDP to GTP without activation by RhoGEF proteins. The other possible reason could be the existence of another putative RhoGEF protein for FgRho4. In N. crassa, two Rho4-specific Rho GEF proteins, Bud3 and Rgf3, are required at different stages of the septation process (Justa-Schuch et al., 2010). We thus speculate that a Rgf3-homolog (gene number, FGSG_08568) could be the other FgRho4 GEF and might also contribute to septum formation in F. graminearum. RhoGEF interacts with GDP-bound Rho GTPase to perform the exchange GDP for GTP. In this study, FgBud3 did not only interact with GDP-bound but also with GTP-bound FgRho4. These findings implied different Rho4GEF guided the activated FgRho4 to different targets that may be important for different functions. RhoGEFs usually contain various domains that may be involved in interactions with different effectors to direct Rho GTPase downstream signaling (Mertens et al., 2003). FgRgf3 was predicted to have two additional domains when compared to FgBud3 indicating FgRgf3 may play a number of different roles (Figure 1). Future work is needed to further characterize the role of FgRgf3 and determine whether it is also a Rho4 RhoGEF involved in some other functions in F. graminearum.
Author Contributions
CZ, GW, HL, CR, and ZW: conceived and designed the experiments. CZ, ZL, DH, LS, and HY: performed the experiments. CZ and ZL: analyzed the data. CZ: wrote the paper. CZ, HL, CR, and ZW: originated research leading up to this paper and provided guidance and review.
Funding
This work was supported by the Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University, the General Financial Grant from the China Postdoctoral Science Foundation under Grant No. 2017M612110, and NSFC Grant 31770123.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Justice Norvienyeku and Mr. Xie Dang (Fujian Agriculture and Forestry University, China) for helpful discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01209/full#supplementary-material
FIGURE S1 | Generation of the FgBUD3 deletion mutants. (A) FgBUD3 gene locus and gene replacement construct. The FgBUD3 and hph genes are marked with empty and black arrows, respectively. P, Pst I. (B) Genomic DNA isolated from mycelia of PH-1, and some transformants were subjected to PCR using primer pairs OF/OR and UAF/H853 marked in (A). (C) Total RNA samples isolated from mycelia of PH-1, the FgBUD3 deletion mutant and the complemented strain were subjected to RT-PCR using the FgBUD3 gene-specific primer QF/QR (Supplementary Table S1), ACTIN gene was amplified as positive control. (D) DNA gel blots of restriction enzymes marked in (A) digested genomic DNA were hybridized with probe marked in (A). PH-1, wild-type strain; (ΔFgbud3, gene FgBUD3 deletion mutants; Fgbud3-com, complemented strain of FgBUD3 deletion mutant.
TABLE S1 | PCR primers used in this study.
Footnotes
References
Adams, A. E., Johnson, D. I., Longnecker, R. M., Sloat, B. F., and Pringle, J. R. (1990). CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111, 131–142. doi: 10.1083/jcb.111.1.131
Araujo-Palomares, C. L., Richthammer, C., Seiler, S., and Castro-Longoria, E. (2011). Functional characterization and cellular dynamics of the CDC-42 - RAC - CDC-24 module in Neurospora crassa. PLoS One 6:e27148. doi: 10.1371/journal.pone.0027148
Bottalico, A., and Perrone, G. (2002). Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. Eur. J. Plant Pathol. 108, 611–624. doi: 10.1023/A:1020635214971
Bowden, R. L., and Leslie, J. F. (1999). Sexual recombination in Gibberella zeae. Phytopathology 89, 182–188. doi: 10.1094/PHYTO.1999.89.2.182
Chen, J., Zheng, W., Zheng, S., Zhang, D., Sang, W., Chen, X., et al. (2008). Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea. PLoS Pathog. 4:e1000202. doi: 10.1371/journal.ppat.1000202
Chenevert, J., Valtz, N., and Herskowitz, I. (1994). Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics 136, 1287–1296.
Desiardins, A. E., Proctor, R. H., Bai, G. H., Mccormick, S. P., Shaner, G., Buechley, G., et al. (1996). Reduced virulence of trichothecene-nonproducing mutants of Gibberella zeae in wheat field tests. Mol. Plant Microbe Interact. 9, 775–781. doi: 10.1094/MPMI-9-0775
Gale, L. R., Chen, L. F., Hernick, C. A., Takamura, K., and Kistler, H. C. (2002). Population analysis of Fusarium graminearum from wheat fields in eastern China. Phytopathology 92, 1315–1322. doi: 10.1094/Phyto.2002.92.12.1315
Goswami, R. S., and Kistler, H. C. (2004). Heading for disaster: Fusarium graminearum on cereal crops. Mol. Plant Pathol. 5, 515–525. doi: 10.1111/j.1364-3703.2004.00252.x
Harris, S. D. (2011). Cdc42/Rho GTPases in fungi: variations on a common theme. Mol. Microbiol. 79, 1123–1127. doi: 10.1111/j.1365-2958.2010.07525.x
Hillenmeyer, M. E., Fung, E., Wildenhain, J., Pierce, S. E., Hoon, S., Lee, W., et al. (2008). The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365. doi: 10.1126/science.1150021
Hou, Z. M., Xue, C. Y., Peng, Y. L., Katan, T., Kistler, H. C., and Xu, J. R. (2002). A mitogen-activated protein kinase gene (MGV1) in Fusarium graminearum is required for female fertility, heterokaryon formation, and plant infection. Mol. Plant Microbe Interact. 15, 1119–1127. doi: 10.1094/Mpmi.2002.15.11.1119
Iden, S., and Collard, J. G. (2008). Crosstalk between small GTPases and polarity proteins in cell polarization. Nat. Rev. Mol. Cell Biol. 9, 846–859. doi: 10.1038/nrm2521
Jiang, J., Liu, X., Yin, Y., and Ma, Z. (2011). Involvement of a velvet protein FgVeA in the regulation of asexual development, lipid and secondary metabolisms and virulence in Fusarium graminearum. PLoS One 6:e28291. doi: 10.1371/journal.pone.0028291
Justa-Schuch, D., Heilig, Y., Richthammer, C., and Seiler, S. (2010). Septum formation is regulated by the RHO4-specific exchange factors BUD3 and RGF3 and by the landmark protein BUD4 in Neurospora crassa. Mol. Microbiol. 76, 220–235. doi: 10.1111/j.1365-2958.2010.07093.x
Kang, P. J., Lee, M. E., and Park, H. O. (2014). Bud3 activates Cdc42 to establish a proper growth site in budding yeast. J. Cell Biol. 206, 19–28. doi: 10.1083/jcb.201402040
Krause, S. A., Cundell, M. J., Poon, P. P., Mcghie, J., Johnston, G. C., Price, C., et al. (2012). Functional specialisation of yeast Rho1 GTP exchange factors. J. Cell Sci. 125, 2721. doi: 10.1242/jcs.100685
Kwon, M. J., Arentshorst, M., Roos, E. D., van den Hondel, C. A., Meyer, V., and Ram, A. F. (2011). Functional characterization of Rho GTPases in Aspergillus niger uncovers conserved and diverged roles of Rho proteins within filamentous fungi. Mol. Microbiol. 79, 1151–1167. doi: 10.1111/j.1365-2958.2010.07524.x
Lesage, G., Shapiro, J., Specht, C. A., Sdicu, A. M., Menard, P., Hussein, S., et al. (2005). An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet 6:8. doi: 10.1186/1471-2156-6-8
Levin, D. E. (2005). Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69, 262–291. doi: 10.1128/MMBR.69.2.262-291.2005
Manning, B. D., Padmanabha, R., and Snyder, M. (1997). The Rho-GEF Rom2p localizes to sites of polarized cell growth and participates in cytoskeletal functions in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 1829–1844. doi: 10.1091/mbc.8.10.1829
Mertens, A. E., Roovers, R. C., and Collard, J. G. (2003). Regulation of Tiam1–Rac signalling. FEBS Lett. 546, 11–16. doi: 10.1016/S0014-5793(03)00435-6
Nesher, I., Minz, A., Kokkelink, L., Tudzynski, P., and Sharon, A. (2011). Regulation of pathogenic spore germination by CgRac1 in the fungal plant pathogen Colletotrichum gloeosporioides. Eukaryot. Cell 10, 1122–1130. doi: 10.1128/EC.00321-10
Ozaki, K., Tanaka, K., Imamura, H., Hihara, T., Kameyama, T., Nonaka, H., et al. (1996). Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15, 2196–2207.
Proctor, R. H., Desjardins, A. E., Mccormick, S. P., Plattner, R. D., Alexander, N. J., and Brown, D. W. (2002). Genetic analysis of the role of trichothecene and fumonisin mycotoxins in the virulence of Fusarium. Eur. J. Plant Pathol. 108, 691–698. doi: 10.1007/978-94-010-0001-7_12
Rasmussen, C. G., and Glass, N. L. (2005). A Rho-type GTPase, rho-4, is required for septation in Neurospora crassa. Eukaryot. Cell 4, 1913–1925. doi: 10.1128/EC.4.11.1913-1925.2005
Rasmussen, C. G., and Glass, N. L. (2007). Localization of RHO-4 indicates differential regulation of conidial versus vegetative septation in the filamentous fungus Neurospora crassa. Eukaryot. Cell 6, 1097–1107. doi: 10.1128/EC.00050-07
Richthammer, C., Enseleit, M., Sanchez-Leon, E., März, S., Heilig, Y., Riquelme, M., et al. (2012). RHO1 and RHO2 share partially overlapping functions in the regulation of cell wall integrity and hyphal polarity in Neurospora crassa. Mol. Microbiol. 85, 716–733. doi: 10.1111/j.1365-2958.2012.08133.x
Schiestl, R. H., and Gietz, R. D. (1989). High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16, 339–346. doi: 10.1007/BF00340712
Schmelzle, T., Helliwell, S. B., and Hall, M. N. (2002). Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell Biol. 22, 1329–1339. doi: 10.1128/MCB.22.5.1329-1339.2002
Schmidt, A., Bickle, M., Beck, T., and Hall, M. N. (1997). The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell 88, 531–542. doi: 10.1016/S0092-8674(00)81893-0
Seo, J. A., Kim, J. C., Lee, D. H., and Lee, Y. W. (1996). Variation in 8-ketotrichothecenes and zearalenone production by Fusarium graminearum isolates from corn and barley in Korea. Mycopathologia 134, 31–37. doi: 10.1007/BF00437050
Seong, K. Y., Pasquali, M., Zhou, X., Song, J., Hilburn, K., Mccormick, S., et al. (2009). Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Mol. Microbiol. 72, 354–367. doi: 10.1111/j.1365-2958.2009.06649.x
Seong, K. Y., Zhao, X., Xu, J. R., Güldener, U., and Kistler, H. C. (2008). Conidial germination in the filamentous fungus Fusarium graminearum. Fungal Genet. Biol. 45, 389–399. doi: 10.1016/j.fgb.2007.09.002
Si, H., Justa-Schuch, D., Seiler, S., and Harris, S. D. (2010). Regulation of septum formation by the Bud3-Rho4 GTPase module in Aspergillus nidulans. Genetics 185, 165–176. doi: 10.1534/genetics.110.114165
Sloat, B. F., Adams, A., and Pringle, J. R. (1981). Roles of the CDC24 gene product in cellular morphogenesis during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 89, 395–405. doi: 10.1083/jcb.89.3.395
Son, H., Seo, Y. S., Min, K., Park, A. R., Lee, J., Jin, J. M., et al. (2011). A phenome-based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum. PLoS Pathog. 7:e1002310. doi: 10.1371/journal.ppat.1002310
Starkey, D. E., Ward, T. J., Aoki, T., Gale, L. R., Kistler, H. C., Geiser, D. M., et al. (2007). Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 44, 1191–1204. doi: 10.1016/j.fgb.2007.03.001
Virag, A., Lee, M. P., Si, H., and Harris, S. D. (2007). Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol. Microbiol. 66, 1579–1596. doi: 10.1111/j.1365-2958.2007.06021.x
Wang, C., Zhang, S., Hou, R., Zhao, Z., Zheng, Q., Xu, Q., et al. (2011). Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum. PLoS Pathog. 7:e1002460. doi: 10.1371/journal.ppat.1002460
Wang, G. H., Wang, C. F., Hou, R., Zhou, X. Y., Li, G. T., Zhang, S. J., et al. (2012). The AMT1 arginine methyltransferase gene is important for plant infection and normal hyphal growth in Fusarium graminearum. PLoS One 7:e38324. doi: 10.1371/journal.pone.0038324
Yoshida, S., Bartolini, S., and Pellman, D. (2009). Mechanisms for concentrating Rho1 during cytokinesis. Gene. Dev. 23, 810–823. doi: 10.1101/gad.1785209
Yoshida, S., Kono, K., Lowery, D. M., Bartolini, S., Yaffe, M. B., Ohya, Y., et al. (2006). Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science 313, 108–111. doi: 10.1126/science.1126747
Zhang, C., Lin, Y., Wang, J., Wang, Y., Chen, M., Norvienyeku, J., et al. (2016). FgNoxR, a regulatory subunit of NADPH oxidases, is required for female fertility and pathogenicity in Fusarium graminearum. FEMS Microbiol. Lett. 363:fnv223. doi: 10.1093/femsle/fnv223
Zhang, C., Wang, Y., Wang, J., Zhai, Z., Zhang, L., Zheng, W., et al. (2013). Functional characterization of Rho family small GTPases in Fusarium graminearum. Fungal Genet. Biol. 61, 90–99. doi: 10.1016/j.fgb.2013.09.001
Zheng, Q., Hou, R., Zhang, J. Y., Ma, J. W., Wu, Z. S., Wang, G. H., et al. (2013). The MAT locus genes play different roles in sexual reproduction and pathogenesis in Fusarium graminearum. PLoS One 8:e66980. doi: 10.1371/journal.pone.0066980
Zheng, W., Chen, J., Liu, W., Zheng, S., Zhou, J., Lu, G., et al. (2007). A Rho3 homolog is essential for appressorium development and pathogenicity of Magnaporthe grisea. Eukaryot. Cell 6, 2240–2250. doi: 10.1128/EC.00104-07
Zheng, W., Zhao, X., Xie, Q., Huang, Q., Zhang, C., Zhai, H., et al. (2012). A conserved homeobox transcription factor Htf1 is required for phialide development and conidiogenesis in Fusarium species. PLoS One 7:e45432. doi: 10.1371/journal.pone.0045432
Zheng, W., Zhao, Z., Chen, J., Liu, W., Ke, H., Zhou, J., et al. (2009). A Cdc42 ortholog is required for penetration and virulence of Magnaporthe grisea. Fungal Genet. Biol. 46, 450–460. doi: 10.1016/j.fgb.2009.03.005
Keywords: Fusarium graminearum, RhoGEF, FgBUD3, septum formation, pathogenicity
Citation: Zhang C, Luo Z, He D, Su L, Yin H, Wang G, Liu H, Rensing C and Wang Z (2018) FgBud3, a Rho4-Interacting Guanine Nucleotide Exchange Factor, Is Involved in Polarity Growth, Cell Division and Pathogenicity of Fusarium graminearum. Front. Microbiol. 9:1209. doi: 10.3389/fmicb.2018.01209
Received: 15 December 2017; Accepted: 17 May 2018;
Published: 07 June 2018.
Edited by:
Angel Medina, Cranfield University, United KingdomReviewed by:
Susan Schlimpert, John Innes Centre (JIC), United KingdomSomayeh Dolatabadi, Westerdijk Fungal Biodiversity Institute, Netherlands
Copyright © 2018 Zhang, Luo, He, Su, Yin, Wang, Liu, Rensing and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Liu, ZmphdWxpdWhvbmdAMTYzLmNvbQ== Christopher Rensing, cmVuc2luZ0BmYWZ1LmVkdS5jbg== Zonghua Wang, d2FuZ3poQGZhZnUuZWR1LmNu; em9uZ2h1YXdAMTYzLmNvbQ==
 Chengkang Zhang
Chengkang Zhang Zenghong Luo3
Zenghong Luo3 Guo Wang
Guo Wang Hong Liu
Hong Liu Christopher Rensing
Christopher Rensing Zonghua Wang
Zonghua Wang