- 1Anhui Provincial Key Laboratory of Microbial Pest Control, Anhui Agricultural University, Hefei, China
- 2School of Plant Protection, Anhui Agricultural University, Hefei, China
- 3School of Transportation Engineering, Anhui Sanlian University, Hefei, China
The APSES family is a unique family of transcription factors with a basic helix-loop-helix structure (APSES: Asm1p, Phd1p, Sok2p, Efg1p, and StuAp), which are key regulators of cell development and sporulation-related processes. However, the functions of the APSES family of genes in the entomopathogenic fungus Metarhizium robertsii have not been reported. Here, we report the identification and characterization of the MrStuA gene, a member of the APSES family, in M. robertsii. The selected gene was identified as StuA in M. robertsii (MrStuA) because the gene product contains two conserved sequences, an APSES-type DNA-binding domain and a KilA DNA-binding domain, and has the highest homology with the StuA in the C-II clade of the APSES family. We found that the number of conidia produced by the ΔMrStuA strain was 94.45% lower than that in the wild type. Additionally, in the mutant, the conidia displayed an elongated shape, the sporulation was sparse and the phialide were slender. In addition, transcription levels of two central regulators of asexual development, AbaA and WetA, were significantly reduced in the mutant; furthermore, the transcription levels of other sporulation related genes, such as Mpk, Phi, Med, Aco, Flu, and FlbD, also decreased significantly. We also show that the median lethal time (LT50) of the mutant increased by 19%. This increase corresponded with a slower growth rate and an earlier conidia germination time compared to that of the wild strain. However, the resistance of the mutant to chemicals or physical stressors, such as ultraviolet radiation or heat, was not significantly altered. Our results indicate that in M. robertsii, MrStuA may play a crucial role in regulating sporulation as well as virulence, germination, and vegetative growth. This study improves our understanding of the impact of the transcription factor StuA on sporulation processes in filamentous fungi and provides a basis for further studies aimed at improving sporulation efficiency of these fungi for use as a biocontrol agent.
Introduction
Transcription factors are an important component of the signal transduction pathway, which regulates the expression of the target gene(s) to further regulate the growth and differentiation of cells (Shelest, 2008). There are several exclusive transcription factor families in fungi, including the TEA/ATTS and APSES families. The transcriptional regulatory proteins in the APSES family belong to the bHLH class of transcription factors. There are five types of proteins in the APSES family, Asm1, Phd1, Sok2, Efg1, and StuA; these proteins play important roles in mycelial growth, sporulation, and pathogenicity (Twumasi-Boateng et al., 2009). In recent years, a large number of APSES family proteins have been identified, especially in filamentous fungi. There are five members identified in most fungi, including Aspergillus fumigatus and A. nidulans; meanwhile, other fungi, such as Magnaporthe oryzae and Cryptococcus neoformans, have fewer members (between two and four) (Nishimura et al., 2009).
A. nidulans StuA was the first transcriptional factor with a basic helix-loop-helix structure found in the APSES family (Ballance et al., 1983). The gene, which has a 3-kb-long promoter that is regulated by the most prominent regulator BrlA, is responsible for the spatial expression of the AbaA transcription factor (Wu and Miller, 1997). This protein plays an important role in the structure and cell morphogenesis during the sexual and asexual phases of reproduction in A. nidulans. However, the transcription factors in the APSES family have different functions in different species. For example, they regulate attachment-mediated infection in M. grisea (Nishimura et al., 2000); yeast-mycelial two-phase transformation in Wangiella dermatitidis; secondary metabolites in A. fumigates (Twumasi-Boateng et al., 2009); bisexual dimorphism, sporulation, and pathogenic development in Ustilago maydis (Garcia-Pedrajas et al., 2010); and conidia development, pathogenicity, and secondary metabolites in Fusarium graminearum (Lysoe et al., 2011).
The STUA protein (65.01 kDa) in A. nidulans contains two conserved fragments: an APSES-type DNA-binding domain and a KILA (N-terminal/APSES-type HTH) DNA-binding domain (Twumasi-Boateng et al., 2009). The two conserved domains regulate the expression of related genes through interactions with the STREs sequence of the promoter region in the AbaA gene, thus impacting the conidiation of strains. For example, a spatial rearrangement of the conidiophores and a small amount of visible conidia are observed in the A. nidulans knockout mutant of StuA; meanwhile, no normal conidiophore structure has been found in a Penicillium oxalicum StuA mutant. The StuA gene has two transcription sites, StuAα and StuAβ. StuAα activates the first intron in StuAβ. Both of the sites have long 5′ leaders and mini open reading frames (ORFs) that code regulatory functions of StuA at a translational level.
M. robertsii (formerly known as M. anisopliae) is an important insect pathogen that is commonly used as a biological control agent (Zhou et al., 2012). Spores are the main reproductive organs of fungi as well as the main component of fungal pesticides. The bottleneck limiting the use of M. robertsii as a biopesticide is the low sporulation rate. Thus, a greater understanding of the function of sporulation-related genes will help make the application of strains as fungicides feasible.
There are some existing reports on sporulation-related genes in the genus Metarhizium. The molecular mechanism of the sporulation process is complicated in M. anisopliae and other entomopathogenic fungi (Clarkson and Charnley, 1996). Two sporulation pathways involving many regulatory genes, such as Flu, G-flbs, and FadA, co-regulate sporulation in M. robertsii; this process is similar to that of A. nidulans. The Cag8 gene, which is homologous with the FlbA gene in A. nidulans, is required for sporulation. This requirement has been confirmed by showing that a knockout strain cannot produce conidia (Timberlake, 1980). Using RNA interference (RNAi), a micro-circulation sporulation gene Pky was found to produce more varied shapes of conidia (Xu et al., 2011).
The mechanisms of sporulation in A. nidulans and Neurospora crassa have been well studied (Park and Yu, 2012). Blasting the sequences of StuA and other sporulation related genes from A. nidulans, such as BrlA, AbaA, WetA, FlbA, FluG, AcoB, FlbC, FlbD, MedA, PhiA, SteC, MpkA, FadA, HymA, and Sak A, show that homologous genes also exist in M. robertsii. Thus, it is speculated that the molecular mechanism of sporulation in M. robertsii may be similar to that of A. nidulans. However, the functions of the APSES family of proteins in the entomopathogenic fungi M. robertsii have not been reported. We generated a MrStuA knockout mutant (ΔMrStuA) and found that StuA may play a crucial regulatory role in sporulation, virulence, germination, and vegetative growth in M. robertsii. This study deepened our understanding of the impact of the StuA-like transcription factor on sporulation in filamentous fungi.
Materials and Methods
Fungal Culture
The wild-type (WT) strain M. robertsii ARSEF23 (ATCC No. MYA-3075) was obtained from the Anhui Provincial Key Laboratory of Microbial Pest Control (RCEF5501). To obtain conidia, the WT strain, a MrStuA knockout strain (ΔMrStuA), and a complemented strain (C’) were grown on potato dextrose agar (PDA; 200 g of potato, 20 g of dextrose, 20 g of agar and 1,000 ml of water) at 25°C for 7–8 days. Subsequently, conidial suspensions (0.05% v/v Tween 80) were obtained by filtering the cultures through sterile non-woven fabrics to remove mycelia. The conidial suspensions were inoculated onto PDA medium overlain with sterile cellophane paper using spreaders and incubated at 25°C. Colony phenotype assays were performed on solid PDA, 1/4 Sabouraud dextrose agar with yeast extract (1/4 SDAY; 10 g of glucose, 2.5 g of yeast extract powder, 2.5 g of peptone, 20 g of agar, and 1,000 ml of water) and SDAY.
The host E. coli Trans 10 was purchased from the TransGen Biotech Corporation (China). The plasmid pDHt-bar/ben and Agrobacterium tumefaciens AGL-1 were donated by Chengshu Wang (Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences).
Peptide Domain and Homology Analysis
The amino acid sequences of APSES proteins were retrieved from NCBI1. The conserved domains were predicted using InterProScan 5.0. We identified the protein structure of the ARSEF 23 APSES transcription factor (XP_007819177) (Figure 1A) and found that this protein has high homology with the APSES family of proteins from a diverse range of fungi.
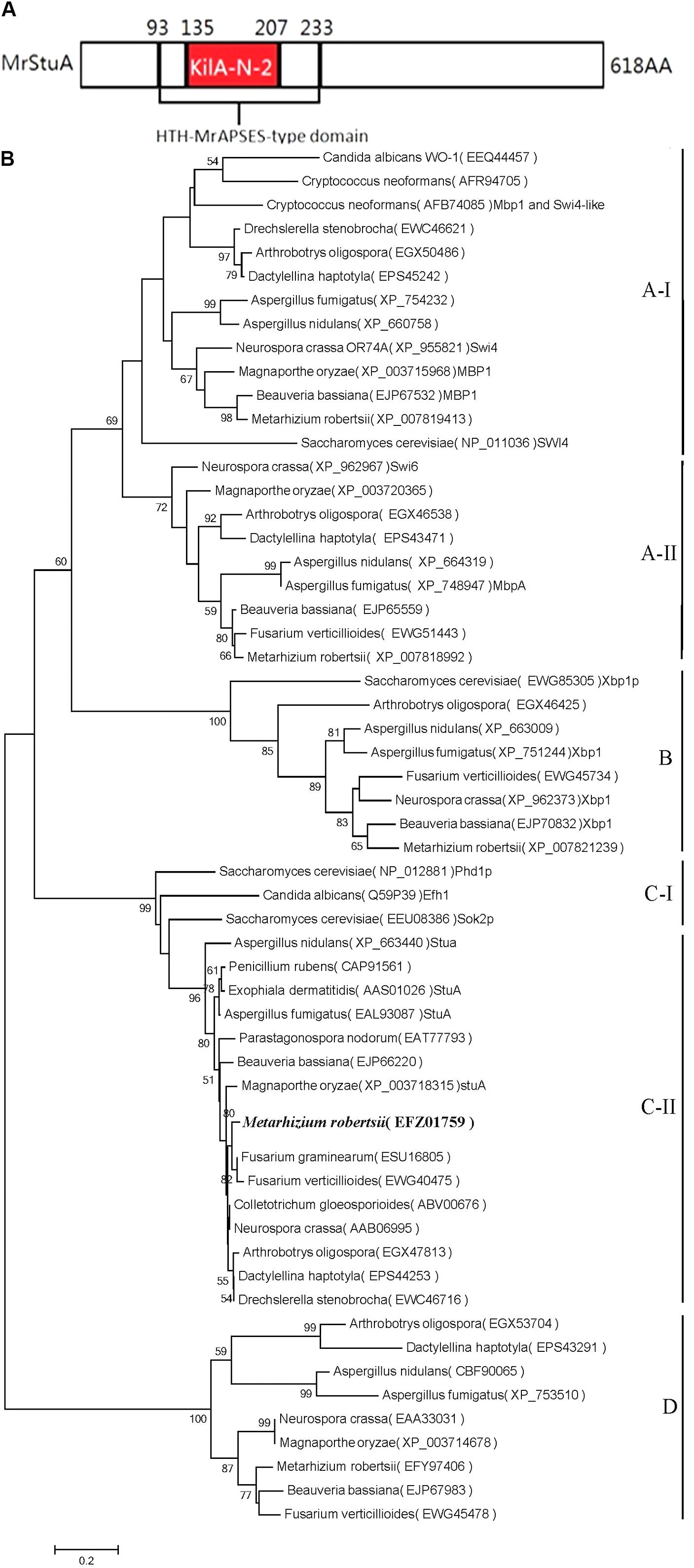
FIGURE 1. Bioinformatics analysis of the MrStuA protein. (A) Peptide domain analysis of the APSES transcription factor from M. robertsii. (B) A phylogenetic tree based on the APSES-type DNA-binding domains from different fungi. Bootstrap values (expressed as percentages of 1000 replications) greater than 50% are indicated on the tree. Numbers below the nodes indicate the bootstrap values. The bar markers indicate genetic distances, which are proportional to the number of amino acid substitutions.
Multiple Alignment of the APSES-Type DNA-Binding Domain and Construction of a Phylogenetic Tree
The conserved amino acid sequences of the APSES domains (IPR003163) were aligned using Clustal W parameters (Supplementary Figure S1). MEGA version 5.1 was used to construct a neighbor-joining (NJ) tree with bootstrap analysis using 1,000 replicates. In total, 58 APSES domains from different fungi were identified for phylogenetic analysis. The neighbor-joining tree was submitted to TreeBase (Figure 1B) (Study Accession URL2).
Construction of a MrStuA Knockout Mutant, ΔMrStuA
To characterize the functions of the MrStuA in M. robertsii, a gene replacement mutant was generated using a dominant selectable marker gene, bar. The ΔMrStuA deletion mutant was constructed by replacing the MrStuA gene regions with the bar gene cassette. A complementation strain was obtained by transforming pben-MrStuA into the ΔMrStuA strain. Based on the gene sequences of APSES (MAA_02988) transcription factors in M. robertsii ARSEF 23, the primers MrStuA-F and MrStuA-R (Supplementary Table S1) were designed to amplify the MrStuA fragment. The PCR amplification product was purified, cloned, and sequenced.
A MrStuA deletion vector, pDHt-bar-StuA-Del, was constructed by inserting two flanking sequences of MrStuA into the left and right sides of bar (a kanamycin resistance gene) in the pDHt-bar vector using the primers MrStuAupF+MrStuAupR (Supplementary Table S1). A 1,008 bp upstream flanking sequence of MrStuA was amplified from M. roberstii genomic DNA and was inserted into the EcoR I-Pst I sites of the pDHt-bar vector to generate a plasmid, pDHt-bar-StuA-upstream. Subsequently, a 1,285 bp downstream flanking sequence of MrStuA was amplified from M. roberstii genomic DNA using the primers MrStuAdownF+MrStuAdownR (Supplementary Table S1). This sequence was inserted into the XbaI site of the pDHt-bar-StuA-upstream vector to generate the pDHt-bar-StuA-Del plasmid. Finally, a 3,267 bp fragment containing the StuA- upstream- bar- StuA- downstream cassette was obtained through PCR amplification with the MrStuAupF+MrStuAdownR primers from the pDHt-bar-StuA-Del vector. The resulting PCR product was purified and used for A. tumefaciens-mediated transformation. The ΔMrStuA strain was constructed using Agrobacterium-mediated fungal transformation (ATMT) and inserted by homologous recombination as previously described (Duan et al., 2009).
All recombinant strains were confirmed by PCR and reverse transcription-PCR (RT-PCR). PCR analysis indicated that a 1,050-bp fragment corresponding to the partial MrStuA gene was present in the WT and C’ strains, but not in the ΔMrStuA strain. A partial bar gene fragment (814-bp) was present in the ΔMrStuA and C’ strains, and a partial Ben gene fragment (328-bp) was present in the C’ strain.
Construction of the Complement Plasmid for ΔMrStuA
The MrStuA complement plasmid ΔMrStuA::MrStuA was constructed using the backbone of the pDHt-ben vector. The full-length MrStuA (3,528 bp including the promoter region) was amplified from the genomic DNA of the WT strain using cpMrStuAupF+cpMrStuAupR primers (Supplementary Table S1). This sequence was subsequently cloned into the Spe I-Xba I site of pDHt-ben to create the pDHt-ben-ΔMrStuA::MrStuA plasmid. Transformation of pDHt-ben-ΔMrStuA::MrStuA with the full length of MrStuA was performed as previously described using carb (50 μg/mL) and kan (50 μg/mL) as selective agents. Transformants were co-cultured with the ΔMrStuA strain. A portion (1,050 bp) of the MrStuA sequence was amplified using MrStuAF+MrStuAR primers (Supplementary Table S1) to obtain the C’ strain.
Morphological Observations at the Microscopic Level
The WT, ΔMrStuA, and C’ strains were grown on PDA medium at 25°C for 3 days to determine hyphal growth and conidiation. Fresh mycelia and conidia were taken from the edges of colonies of each strain and stained with lactophenol cotton blue. Micromorphology was examined using an Olympus optical microscope (BX51, Olympus Co, Tokyo, Japan) and measured using the Measurement Tool program in the DP2-BSW software.
Sporulation Capacity Tests
Fresh conidia from the WT, ΔMrStuA, and C’ strains were collected after being cultured at 25°C for 8 days. The conidia were suspended, and 30 μl of each suspension was inoculated onto solid PDA (35 mm disposable plastic petri dish) for incubation at 25°C for 12 days (Liu et al., 2013; Xiong et al., 2013). The quantity of conidia in each plate was counted using a hemacytometer, after which the number of conidia per unit area was calculated. There were three replicates for each strain and all assays were repeated in triplicate.
Color, Morphology, and Growth of Colonies
The WT, ΔMrStuA, and C’ strains were grown on PDA, SDAY, and 1/4SDAY plates, respectively, at 25°C for 7–8 days. Conidial suspensions (107 conidia/mL) were prepared for each strain to inoculate a new plate (1 μl of suspension/plate). These plates were cultured at 25°C for 12 days. Each strain was inoculated on 3–5 plates. All assays were repeated in triplicate. The morphology and growth status of the colonies were observed and measured every 48 h starting on the fourth day of culture. The diameter of each colony was measured by the cross method, and all assays were repeated in triplicate.
Changes in Germination Patterns
Conidia were harvested and suspended in Tween 80 solution (0.05% v/v). Then, 30 μL of the filtered suspension (106 conidia/mL) was resuspended in 30 μL of GM (0.2% trehalose, 0.25% peptone, and 0.25% yeast extract powder) liquid medium and incubated at 25°C for 16–22 h. Germination was observed at 400× magnification after the conidia were placed on the medium. All assays were repeated in triplicate.
Chemical Stress Challenges
To examine chemical stress tolerance, the mycelium of the WT, ΔMrStuA, and C’ strains were cut off from YEB medium by perforations (Chen et al., 2015a). The mycelium of each strain were grown on solid PDA and PDA supplemented with 2 μg/mL sodium dodecyl sulfate (SDS), 2 μg/mL Congo red (CR), 1 mol/L NaCl, 2 μg/mL carbendazim, 2 μg/mL menadione, and 2 mmol/L H2O2. Then, the mycelium were incubated at 25°C for 10 days. Morphological changes in the colonies were observed and measured. All assays were repeated in triplicate.
UV Radiation and Heat Shock Assay
Thirty microliters of conidial suspensions (106 conidia/mL) were placed on a glass slide and dried on a sterile table to eliminate moisture. The UV light wavelength of the hybrid furnace was set to UV 254 nm, Time1, Energy2. Germination liquid (GM) was added after UV irradiation, and samples were cultured at 25°C for 16–24 h. At the same time, the filtered suspensions were transferred to Eppendorf tubes and treated in a 42°C water bath for 1 h. Aliquots of the suspensions (30 μL) were inoculated onto PDA medium (35 mm disposable plastic petri dish) and cultured at 25°C for 16–24 h for examination of conidium germination. The data were recorded every 2 h. The inhibition rates were then analyzed (Rangel et al., 2006; Chen et al., 2015b).
Virulence Assays
To investigate the effects of MrStuA on fungal virulence, insect bioassays were conducted using wax moths. Conidia harvested from cultures grown on PDA medium were applied topically by immersing the wax moths for 1 min in an aqueous suspension (107 conidia/mL). Each treatment had three replicates with 15 insects each, and mortality was recorded every 12 h to calculate the median lethal time (LT50).
Real-Time Quantitative Reverse Transcription PCR
Hyphal and conidia were harvested from cultures grown on solid PDA for 60 h (initial period of conidia formation). Total RNA was extracted from the samples using the Trizol reagent (Invitrogen, Foster City, CA, United States). cDNA was synthesized from 1 μg of total RNA using Moloney murine leukemia virus reverse transcriptase (TaKaRa, Dalian, China) according to the manufacturer’s instructions. This cDNA was used as the template for qRT-PCR with specific primer pairs (Supplementary Table S2). The qRT-PCR was performed using the SYBR Green (TAKARA) detection method (CFX96TM Real-Time System, Bio-Rad Laboratories, Hercules, CA, United States). All PCR reactions were performed in triplicate. The 2-ΔΔCT method (Livak and Schmittgen, 2001) was used to calculate relative expression levels, and the glyceraldehyde 3-phosphate dehydrogenase gene (primer pair gpdF/gpdR) was used as an internal control for each sample. The data are expressed as the mean ± standard deviation (SD) for three independent experiments.
Results
Summary of the Gene StuA in Metarhizium robertsii
An analysis of the M. robertsii genome database revealed that the putative StuA transcription factor gene consists of 2,074 nucleotides with two introns and three exons (427 bp, 94 bp, and 1,336 bp) and is predicted to encode a protein with 618 amino acids. The protein contains two conserved sequences, the APSES-type DNA-binding domain (IPR003163, 93rd–208th amino acid) and the KILA, N-terminal/APSES-type HTH, DNA-binding domain (IPR018004, 135th–207th amino acid, Figure 1A). The APSES core domain in fungi contains 73 amino acid residues consisting of six beta-folded and two pairs of alpha helices. The amino acid sequence from M. robertsii putative StuA had 65%, 43%, and 43% homologies with those of F. graminearum, A. nidulans, and P. chrysogenum, respectively.
Based on the phylogenetic tree (Figure 1B), the selected APSES family proteins were divided into four clades (A, B, C, and D), each of which has differently conserved amino acid sequences. The M. robertsii putative StuA transcription factor gene (XP_007819177) is in the C clade, which also includes the StuA genes from F. graminearum, A. nidulans, P. chrysogenum, and Beauveria bassiana. Within the clade, the M. robertsii gene is most closely related to the homologous genes from F. graminearum and F. verticillioides and is most distinct from that of B. bassiana. Based on the conserved domains, homology with other genes, and similarity within the phylogenetic tree, the gene was identified as the StuA transcription factor and was named MrStuA.
MrStuA Is Required for Sporulation
There were numerous morphological differences between the colonies of the ΔMrStuA strain and those of the WT and C’ strains. The ΔMrStuA strain formed white colonies with only a few conidia; meanwhile, the WT and C’ strains produced dark green colonies with many green conidia (Figure 2A). On PDA medium, the ΔMrStuA strain yielded 94.45% fewer conidia than did the WT strain, and the mutant was nearly totally unable to produce conidia (Figure 2B). Microscopic observations showed that the conidium shape, sporulation structure, and phialides were elongated, sparser, and more slender, respectively, in the mutant compared to those in the wild type. In the wild-type and C’ strains, the conidia column contained many parallel conidial chains; however, this feature was not present in the mutant. A length–width ratio was calculated for conidia to better determine the variety in conidia shape. This ratio revealed that the ΔMrStuA strain (ratio = 3.64 ± 0.36) had conidia that were significantly more slender (40%) than those of the WT strain (ratio = 2.6 ± 0.36) (P < 0.01) (Figure 2D).
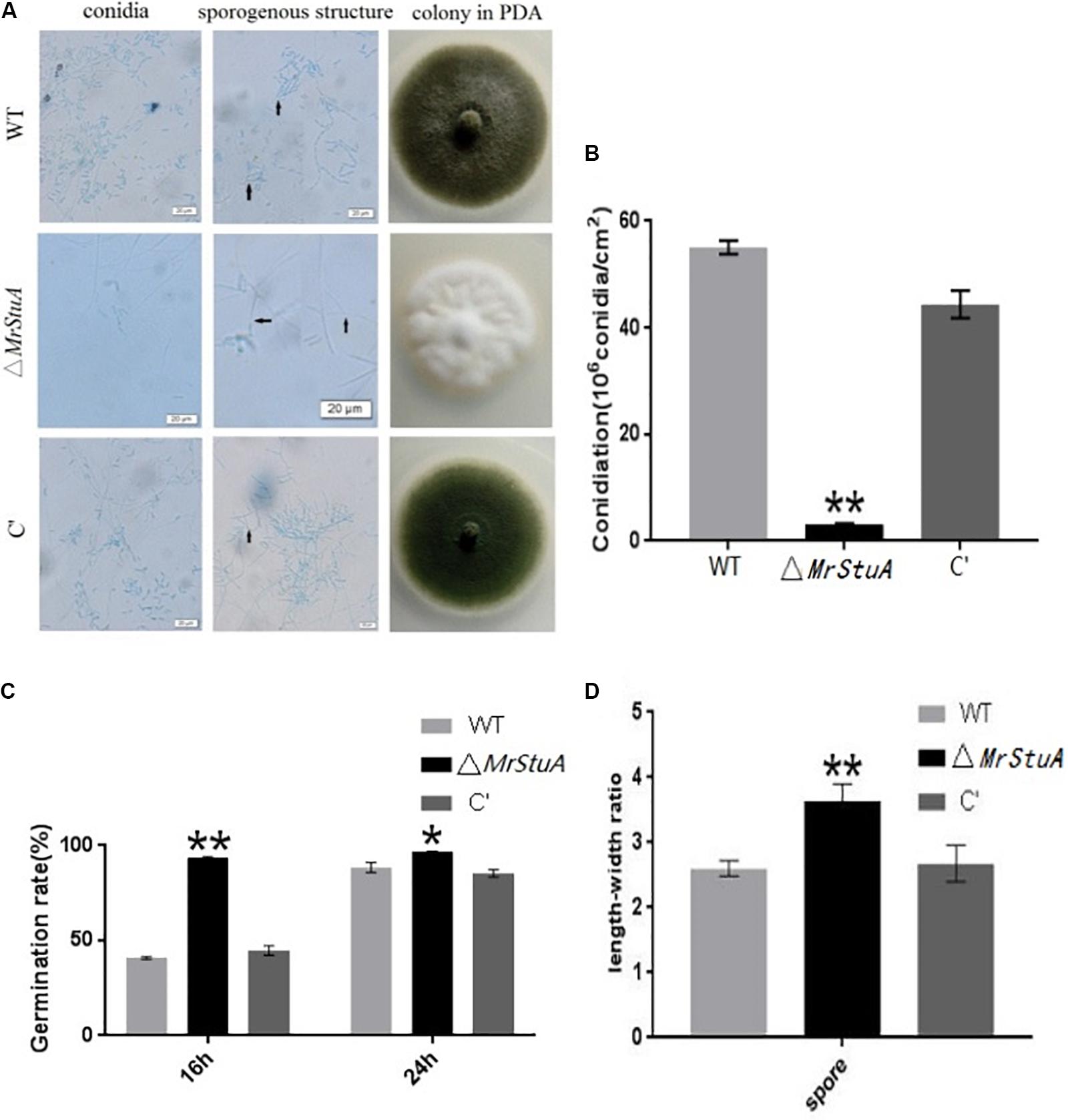
FIGURE 2. Changes in conidia capacity, conidium shape, conidiophore structure, and germination time between the wild type and mutant strains. (A) Morphology of the conidia, hypha and conidiophore structures, and colonies for the WT, ΔMrStuA, and C’ strains. (B) Conidia numbers per growth area (cm2) for the WT, ΔMrStuA, and C’ strains. (C) Germination rate of conidia for the WT, ΔMrStuA, and C’ strains. (D) The aspect ratio of the conidia for the WT, ΔMrStuA, and C’ strains. Student’s t-test: ∗p < 0.05, ∗∗p < 0.01.
After 16 h, the germination rate of the ΔMrStuA strain was more than 90%; meanwhile, the germination rates of the WT and C’ strains were less than 50%. However, there were no differences in germination rate between strains at 24 h (Figure 2C). Thus, it appears that germination occurs sooner in the mutant strain than that in the WT or C’ strains.
Deletion of MrStuA led to changes in sporulation capability, conidia shape, conidiophore structure, and germination time; thus, it appears that MrStuA is required for sporulation.
Expression of Sporulation Related Genes in ΔMrStuA
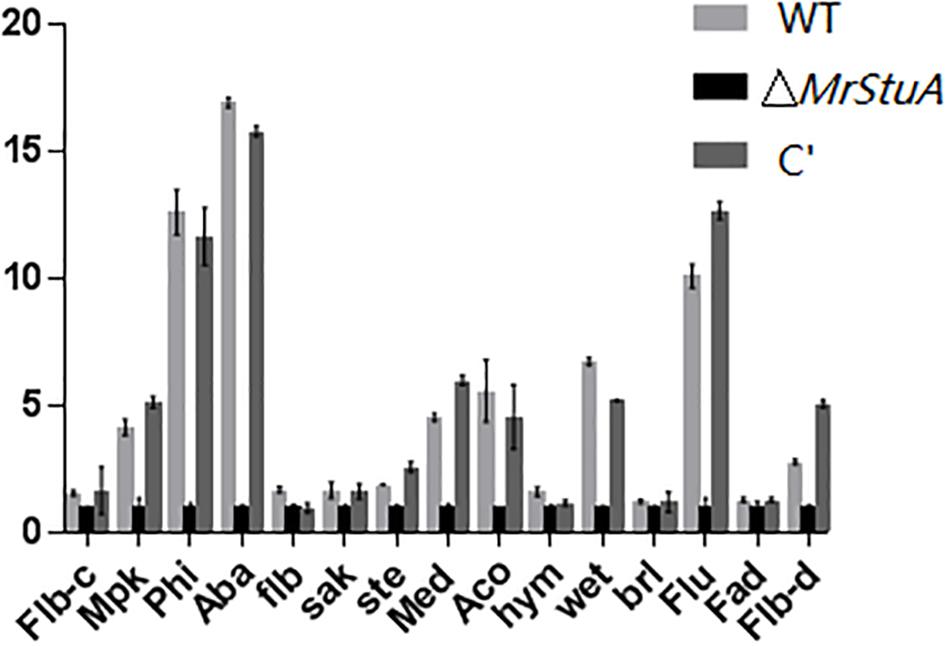
We monitored the mRNA levels of genes involved in conidiogenesis in M. robertsii by qRT-PCR, including the FlbA, BrlA, FluG, AcoB, AbaA, FlbC, FlbD, MedA, PhiA, WetA, SteC, MpkA, FadA, HymA, and SakA genes. The Brl A, Aba A, and Wet A genes are pivotal to the central regulatory pathway of asexual sporulation in A. nidulans. The transcription levels of two of these pivotal sporulation-related genes, AbaA and WetA, were significantly downregulated in ΔMrStuA compared to those in the WT and C’ strains (p < 0.01) (Figure 3). In addition, the transcription levels of other sporulation related genes, including MpkA, PhiA, MedA, AcoB, FluG, and FlbD, were also downregulated. Meanwhile, the expression levels of other genes, such as FlbA, BrlA, FlbC, SteC, FadA, HymA, and SakA, were not altered in the ΔMrStuA mutant. Collectively, the results indicate that the MrStuA gene regulates two key transcription factors, AbaA and WetA, that in turn impact a series of downstream sporulation-related genes at the molecular level. Therefore, MrStuA is necessary for normal sporulation in M. robertsii.
Effect of MrStuA Function on Other Phenotypic Traits
Morphological observations showed differences between ΔMrStuA colonies and those of the WT and C’ strains. On solid PDA, SDAY, and 1/4SDAY medium, the ΔMrStuA mutant produced white colonies, while the WT and C’ strains produced dark green colonies (Figure 4A). The radial growth of ΔMrStuA was significantly reduced compared to that of the WT and C’ strains (p < 0.05) under all growth conditions (Figure 4B).
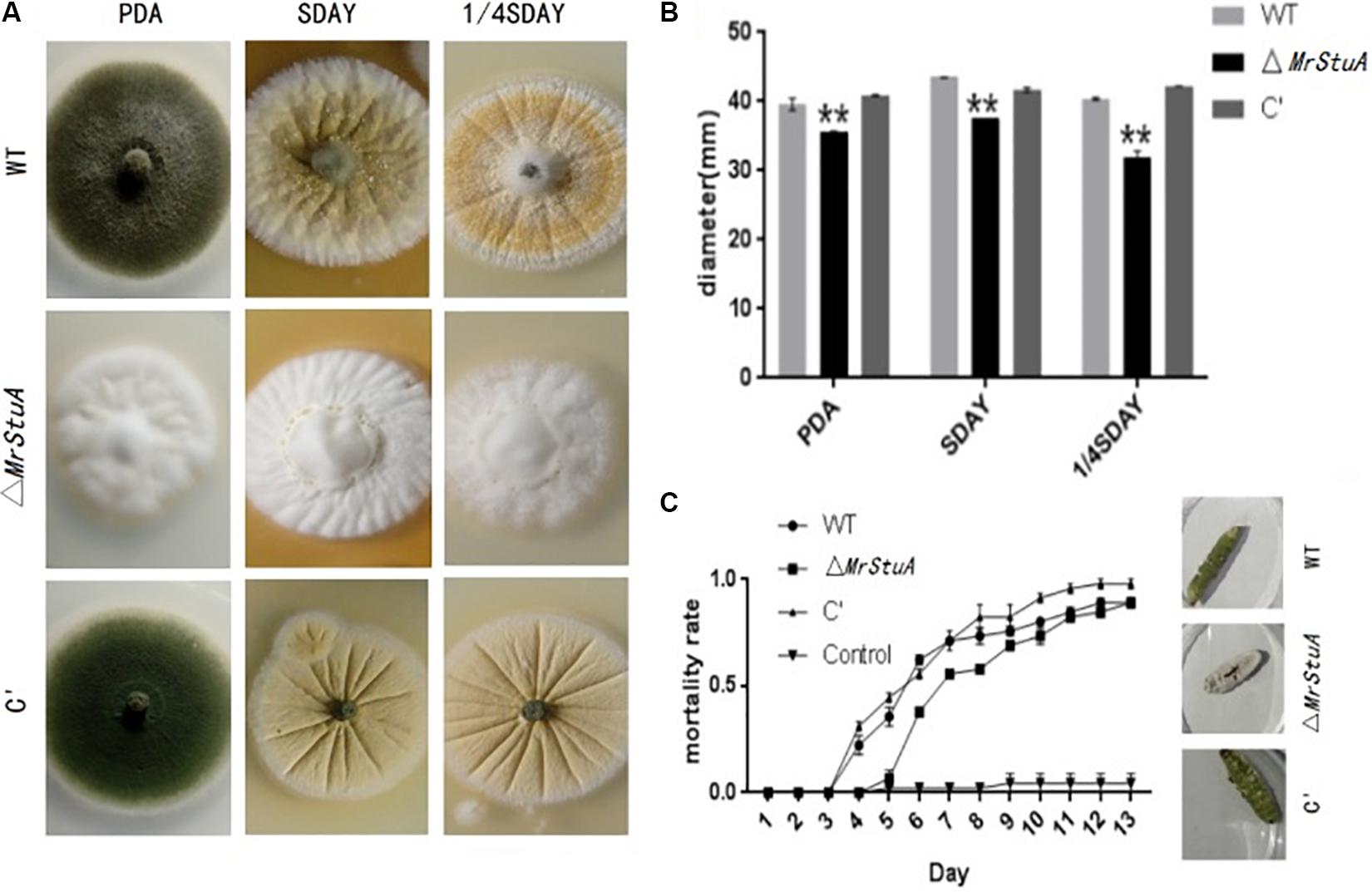
FIGURE 4. Effect of the MrStuA gene on other phenotypic traits. (A) Colonies from each strain grown on solid PDA, SDAY, and 1/4SDAY. (B) Radial growth rate of each strain on solid PDA, SDAY, and 1/4SDAY. (C) Virulence curves of each strain. Error bars are standard error of at least three independent experiments. Student’s t-test: ∗p < 0.05, ∗∗p < 0.01.
In the UV treated group, the relative germination rates of the WT, ΔMrStuA, and C’ strains at 24 h were 62.55%, 63.38%, and 66.06%, respectively (Supplementary Figure S4). Under the 42°C water-bath treatment condition, the relative germination rates of the WT, ΔMrStuA, and C’ strains at 24 h were 76.77%, 81.95%, and 81.8%, respectively (Supplementary Figure S5). There were no significant differences in the tolerance of the germination of conidia to UV or heat treatments. Furthermore, there were no differences in the growth diameters of the WT, ΔMrStuA, and C’ strains on all chemical stress medium (Supplementary Figures S2, S3).
The ΔMrStuA strain exhibited reduced virulence compared to WT and C’ strains based on infection bioassays performed on the greater wax moth (Galleria mellonella). The ΔMrStuA strain (LT50 = 8.07 ± 0.68 d) took a longer time (19%) to kill the wax moths than the WT strain did (LT50 = 6.78 ± 0.77 d) (P < 0.05) (Figure 4C).
There were also differences in sporulation of the WT, ΔMrStuA, and C’ strains on host cadavers. The dead greater wax moths infected with the three M. robertsii strains were incubated at 25°C at 100% relative humidity for a week. Those killed by the WT and C’ strains produced numerous green spores; however, there were only white mycelia and a few conidia on the surface of host cadavers killed by the ΔMrStuA strain (Figure 4C).
Our results indicate that MrStuA may be involved in virulence and vegetative growth but has no impact on resistance to heat, UV, or chemical stress.
Discussion
Proteins in the APSES family are regulatory transcription factors that exist ubiquitously in fungi. The functions of these proteins are related to sporulation, development, and the tolerance to unfavorable conditions (Doedt et al., 2004; Nishimura et al., 2009). StuA is a common member of Clade C-II of the APSES family; the function of StuA varies across different fungi. For example, StuA regulates sexual/asexual sporulation in A. nidulans, appressorium-mediated infection in M. oryzae (Doedt et al., 2004), two-phase conversion in W. dermatitidis (Wang and Szaniszlo, 2007), and secondary metabolites in A. fumigatus (Grosse et al., 2008). We found that in M. robertsii, MrStuA may play a crucial regulatory role in sporulation as well as minor roles in regulating virulence, germination, and vegetative growth.
The M. robertsii APSES transcription factor (EFZ01759) was identified as a C-II clade transcription factor based on a phylogenetic tree of APSES family gene data from different fungi. Some APSES family proteins are divided into the A-I, A-II, B, C-I, C-II, and D clades; at the same time, other APSES family genes, including XP_007819413, XP_007818991, XP_007821139, and EFY97406 from M. robertsii, belong to the A-I, A-II, B, and D clades, respectively. Thus, all five APSES protein family members are present in M. robertsii, demonstrating the richness and diversity of transcription factors of this family within M. robertsii. These APSES family proteins may play important roles in regulating the growth, differentiation, and infection processes in entomopathogenic fungi, especially for sporulation-related processes.
We found that the conidial yield from the ΔMrStuA mutant was 94.45% lower than the yield from the control strains. This observation is consistent with results from A. nidulans and P. oxalicum, as well as those from other fungi (Park and Yu, 2012). Microscopy revealed that the morphology of the conidia and sporulation structures of the mutant strains were significantly altered compared with those of the WT strains in some fungi (Mirabito et al., 1989; Prade and Timberlake, 1993). We found that in the M. robertsii mutant, the conidia became elongated, the sporulation became sparse, and the phialide became slender. However, the knockout of this gene in A. nidulans resulted in shortened conidiophores, reduced conidiophores, missing metulae and phialide, and a “stunted” phenotype. It has also been reported that there were no conidiophore structures when StuA was deleted in P. oxalicum. In addition, transcription levels of two central regulators of asexual development, AbaA and WetA, were significantly reduced in the ΔMrStuA mutant. These results indicate that MrStuA plays a regulatory role in some key genes, which in turn impact sporulation ability, conidia morphology, and sporulation structure. Thus, the gene MrStuA is required for sporulation.
The mutant strain has a significantly early germination time indicating that the StuA gene may be involved in regulating the germination process in M. robertsii. In A. nidulans, it has been reported that the StuA protein may regulate the temporal expression of genes (Adams et al., 1998). Thus, we speculate that this gene can impact the temporal expression of related genes.
The virulence of the mutant was significantly decreased, and the ΔMrStuA strain only produced a few conidia on the surface of host cadavers. The sporulation ability on host cadavers is also an important factor when determining insect virulence. In M. acridum, virulence decreased when a sporulation-related gene was knocked out; this effect is mainly due to the increased immunity of the host insect or the impact on its ability to form appressoria (Zhang et al., 2017). Thus, we conclude that the MrStuA gene is related to the virulence of M. robertsii.
In summary, the knock-out of the MrStuA gene led to significant changes in M. robertsii sporulation at both a phenotypic and molecular level; the deletion impacted virulence, germination, and vegetative growth. Thus, the MrStuA gene is involved in both infection and vegetative growth and is required for the sporulation process. It remains unclear how MrStuA regulates downstream genes, and further studies should focus on sporulation regulation pathways.
Author Contributions
BH and ZW conceived and designed the study and edited the manuscript. WY wrote the manuscript, conducted the experiments, and analyzed the data. HW, QS, and LQ did a part of the experiments. BH supervised the project. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (Grant numbers: 2017YFD0200400 and 2017YFD0201202) and the National Natural Science Foundation of China (Grant numbers: 31772226, 31471821, and 31572060).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01208/full#supplementary-material
Footnotes
References
Adams, T. H., Wieser, J. K., and Yu, J. H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54.
Ballance, D. J., Buxton, F. P., and Turner, G. (1983). Transformation of Aspergillus nidulans by the orotidine-5′-phosphate decarboxylase gene of Neurospora crassa. Biochem. Biophys. Res. Commun. 112, 284–289. doi: 10.1016/0006-291x(83)91828-4
Chen, Y. X., Duan, Z. B., Chen, P. L., Shang, Y. F., and Wang, C. S. (2015a). The Bax inhibitor MrBI-1 regulates heat tolerance, apoptotic-like cell death, and virulence in Metarhizium robertsii. Sci. Rep. 5:11. doi: 10.1038/srep10625
Chen, Y. X., Feng, P., Shang, Y. F., Xu, Y. J., and Wang, C. S. (2015b). Biosynthesis of non-melanin pigment by a divergent polyketide synthase in Metarhizium robertsii. Fungal Genet. Biol. 81, 142–149. doi: 10.1016/j.fgb.2014.10.018
Clarkson, J. M., and Charnley, A. K. (1996). New insights into the mechanisms of fungal pathogenesis in insects. Trends Microbiol. 4, 197–203. doi: 10.1016/0966-842x(96)10022-6
Doedt, T., Krishnamurthy, S., Bockmuhl, D. P., Tebarth, B., Stempel, C., Russell, C. L., et al. (2004). APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell 15, 3167–3180. doi: 10.1091/mbc.E03-11-0782
Duan, Z. B., Shang, Y. F., Gao, Q., Zheng, P., and Wang, C. S. (2009). A phosphoketolase Mpk1 of bacterial origin is adaptively required for full virulence in the insect-pathogenic fungus Metarhizium anisopliae. Environ. Microbiol. 11, 2351–2360. doi: 10.1111/j.1462-2920.2009.01961.x
Garcia-Pedrajas, M. D., Baeza-Montanez, L., and Gold, S. E. (2010). Regulation of Ustilago maydis dimorphism, sporulation, and pathogenic development by a transcription factor with a highly conserved APSES domain. Mol. Plant Microbe Interact. 23, 211–222. doi: 10.1094/mpmi-23-2-0211
Grosse, C., Heinekamp, T., Kniemeyer, O., Gehrke, A., and Brakhage, A. A. (2008). Protein kinase a regulates growth, sporulation, and pigment formation in Aspergillus fumigatus. Appl. Environ. Microbiol. 74, 4923–4933. doi: 10.1128/aem.00470-08
Liu, Q., Ying, S. H., Li, J. G., Tian, C. G., and Feng, M. G. (2013). Insight into the transcriptional regulation of Msn2 required for conidiation, multi-stress responses and virulence of two entomopathogenic fungi. Fungal Genet. Biol. 54, 42–51. doi: 10.1016/j.fgb.2013.02.008
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lysoe, E., Pasquali, M., Breakspear, A., and Kistler, H. C. (2011). The transcription factor Fgstuap influences spore development, pathogenicity, and secondary metabolism in Fusarium graminearum. Mol. Plant Microbe Interact. 24, 54–67. doi: 10.1094/mpmi-03-10-0075
Mirabito, P. M., Adams, T. H., and Timberlake, W. E. (1989). Interactions of three sequentially expressed genes control temporal and spatial specificity in Aspergillus development. Cell 57, 859–868. doi: 10.1016/0092-8674(89)90800-3
Nishimura, M., Fukada, J., Moriwaki, A., Fujikawa, T., Ohashi, M., Hibi, T., et al. (2009). Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Biosci. Biotechnol. Biochem. 73, 1779–1786. doi: 10.1271/bbb.90146
Nishimura, M., Hayashi, N., Jwa, N. S., Lau, G. W., Hamer, J. E., and Hasebe, A. (2000). Insertion of the LINE retrotransposon MGL causes a conidiophore pattern mutation in Magnaporthe grisea. Mol. Plant Microbe Interact. 13, 892–894. doi: 10.1094/mpmi.2000.13.8.892
Park, H. S., and Yu, J. H. (2012). Genetic control of asexual sporulation in filamentous fungi. Curr. Opin. Microbiol. 15, 669–677. doi: 10.1016/j.mib.2012.09.006
Prade, R. A., and Timberlake, W. E. (1993). The Aspergillus nidulans brlA regulatory locus consists of overlapping transcription units that are individually required for conidiophore development. EMBO J. 12, 2439–2447.
Rangel, D. E. N., Butler, M. J., Torabinejad, J., Anderson, A. J., Braga, G. U. L., Day, A. W., et al. (2006). Mutants and isolates of Metarhizium anisopliae are diverse in their relationships between conidial pigmentation and stress tolerance. J. Invertebr. Pathol. 93, 170–182. doi: 10.1016/j.jip.2006.06.008
Shelest, E. (2008). Transcription factors in fungi. FEMS Microbiol. Lett. 286, 145–151. doi: 10.1111/j.1574-6968.2008.01293.x
Timberlake, W. E. (1980). Developmental gene regulation in Aspergillus nidulans. Dev. Biol. 78, 497–510. doi: 10.1016/0012-1606(80)90349-8
Twumasi-Boateng, K., Yu, Y., Chen, D., Gravelat, F. N., Nierman, W. C., and Sheppard, D. C. (2009). Transcriptional profiling identifies a role for brla in the response to nitrogen depletion and for stua in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 8, 104–115. doi: 10.1128/ec.00265-08
Wang, Q., and Szaniszlo, P. J. (2007). WdStuAp, an APSES transcription factor, is a regulator of yeast-hyphal transitions in Wangiella (Exophiala) dermatitidis. Eukaryot. Cell 6, 1595–1605. doi: 10.1128/ec.00037-07
Wu, J., and Miller, B. L. (1997). Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulating stunted gene expression. Mol. Cell. Biol. 17, 6191–6201. doi: 10.1128/mcb.17.10.6191
Xiong, C. H., Xia, Y. L., Zheng, P., and Wang, C. S. (2013). Increasing oxidative stress tolerance and subculturing stability of Cordyceps militaris by overexpression of a glutathione peroxidase gene. Appl. Microbiol. Biotechnol. 97, 2009–2015. doi: 10.1007/s00253-012-4286-7
Xu, F., Peng, G. X., and Xia, Y. X. (2011). Cloning and analysis by RNAi of a pyruvate kinase gene expressed during microcycle conidiation in Metarhizium anisopliae. Mycosystema 38, 380–382.
Zhang, J., Jin, K., and Xia, Y. X. (2017). Contributions of beta-tubulin to cellular morphology, sporulation and virulence in the insect-fungal pathogen, Metarhizium acridum. Fungal Genet. Biol. 103, 16–24. doi: 10.1016/j.fgb.2017.03.005
Keywords: virulence, conidium capacity, transcription factor, asexual development gene, Metarhizium robertsii
Citation: Yang W, Wu H, Wang Z, Sun Q, Qiao L and Huang B (2018) The APSES Gene MrStuA Regulates Sporulation in Metarhizium robertsii. Front. Microbiol. 9:1208. doi: 10.3389/fmicb.2018.01208
Received: 30 March 2018; Accepted: 17 May 2018;
Published: 05 June 2018.
Edited by:
Sibao Wang, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Sheng-hua Ying, Zhejiang University, ChinaChangbin Chen, Institut Pasteur of Shanghai (CAS), China
Copyright © 2018 Yang, Wu, Wang, Sun, Qiao and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Huang, Ymh1YW5nQGFoYXUuZWR1LmNu
 Wenjing Yang1
Wenjing Yang1 Bo Huang
Bo Huang