- 1Department of Microbiology, National Taiwan University College of Medicine, Taipei, Taiwan
- 2Department of Pediatrics, Chang Gung Children’s Hospital, Chang Gung Memorial Hospital, College of Medicine, Chang Gung University, Taoyuan, Taiwan
- 3Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan
Mycobacterium marinum is a close relative of Mycobacterium tuberculosis that can cause systemic tuberculosis-like infections in ectotherms and skin infections in humans. Sliding motility correlates with biofilm formation and virulence in most bacteria. In this study, we used a sliding motility assay to screen 2,304 transposon mutants of M. marinum NTUH-M6885 and identified five transposon mutants with decreased sliding motility. Transposons that interrupted the type VII secretion system (T7SS) ESX-1-related genes, espE (mmar_5439), espF (mmar_5440), and eccA1 (mmar_5443), were present in 3 mutants. We performed reverse-transcription polymerase chain reaction to verify genes from mmar_5438 to mmar_5450, which were found to belong to a single transcriptional unit. Deletion mutants of espE, espF, espG (mmar_5441), and espH (mmar_5442) displayed significant attenuation regarding sliding motility and biofilm formation. M. marinum NTUH-M6885 possesses a functional ESX-1 secretion system. However, deletion of espG or espH resulted in slightly decreased secretion of EsxB (which is also known as CFP-10). Thus, the M. marinum ESX-1 secretion system mediates sliding motility and is crucial for biofilm formation. These data provide new insight into M. marinum biofilm formation.
Introduction
Mycobacterium marinum is a non-tuberculous photochromogenic mycobacterium. It is a close relative of Mycobacterium tuberculosis, and also causes systemic tuberculosis-like infections in ectotherms (Solomon et al., 2003; Hagedorn and Soldati, 2007; Alibaud et al., 2011). Researchers first isolated M. marinum from saltwater fish in the 1920s and identified it as a human pathogen in the 1950s (Riera et al., 2015). This species usually occurs in warm saltwater, freshwater, and poikilothermic animals; fish, frogs, and amphibians are its main natural hosts. It grows best at a temperature of 25–35°C (McArdle et al., 2016). The prevalence of M. marinum infections in humans has risen in recent years because of the increasing popularity of home aquariums (Riera et al., 2015). M. marinum infections most commonly occur on the skin, especially the extremities, because of the low-temperature requirements of the bacteria for growth (Oh et al., 2015). An M. marinum skin infection is referred to as an aquarium granuloma, swimming pool granuloma, or fish tank granuloma (Solomon et al., 2003; Meijer, 2015; Sette et al., 2015).
M. marinum infection usually occurs following contact with an infected animal or handling of contaminated aquariums or water. It can also occur as an opportunistic infection, primarily in immune-deficient patients, such as those with human immunodeficiency virus (HIV)/acquired immunodeficiency syndrome (AIDS) (Ren et al., 2007; Rombouts et al., 2010). Clinicians characterize the stages of M. marinum infections as the initial stage (type I), in which there are single or multiple skin papules or nodules; advanced stage (type II), in which there are granulomas; and severe stage (type III), in which immunosuppressed patients experience tenosynovitis, arthritis, or osteomyelitis (Riera et al., 2015). The diagnosis of M. marinum infection is often delayed, due to its low prevalence or because there are few specific clinical signs and symptoms. Thus, many patients initially receive incorrect diagnoses and inappropriate treatments.
Many mycobacteria can spread on a surface by sliding (also called “growth-powered passive surface translocation”), which is driven by the outward pressure of cell growth. This process, which does not require flagella (Martínez et al., 1999), occurs due to surfactants, which decrease the surface tension and allow the spread of cells from their origin (Kearns, 2010). Several studies have indicated that lipooligosaccharides (LOSs), glycolpeptidolipids (GPLs), phthiocerol dimycocerosates (PDIMs), and phenolic glycolipids (PGLs) on the outer surface of mycobacteria have important functions in sliding motility (Ren et al., 2007; Tatham et al., 2012; Pang et al., 2013; Mohandas et al., 2016).
Sliding motility allows the colony diameter of non-swarming bacteria to increase during prolonged incubation (Kearns, 2010). Mycobacteria are prototypical non-flagellated microorganisms that spread slowly in a uniform monolayer due to sliding motility (Shi et al., 2011; Maya-Hoyos et al., 2015). Furthermore, sliding movements and biofilm formation facilitate diffusion and colonization by mycobacteria (Pang et al., 2013).
A biofilm is a thin and slimy film of one or more species of bacteria that adhere to each other and/or a solid surface and it can help to increase the virulence of the bacterial species. Biofilm formation may be considered a survival “strategy” for bacteria. There are several consecutive stages of biofilm formation: reversible attachment, irreversible attachment, mature biofilm formation, and dispersion. A biofilm consists of bacteria and matrix material, which includes extracellular polymeric substances, such as polysaccharides, lipids, membrane vesicles, and nucleic acids (Sousa et al., 2015; Toyofuku et al., 2015). Bacteria in biofilms have significantly enhanced resistance to antibiotics and the human immune system (Shi et al., 2011). Biofilm formation is highly related to sliding motility in Mycobacterium spp. (Nessar et al., 2011; Mohandas et al., 2016). Previous studies indicated that Mycobacterium smegmatis with defects in biofilm formation also have impaired sliding motility (Recht and Kolter, 2001). In addition, Ghosh et al. (2013) reported that the ability of M. smegmatis to form biofilms declined as sliding motility declined. Many other species of mycobacteria, including Mycobacterium avium and Mycobacterium fortuitum, are well-known to produce biofilms, although the ability of M. marinum to form biofilms remains largely unknown (Ren et al., 2007).
There has been an increased number of cases of M. marinum infection due to the increasing popularity of home aquariums. However, there is little knowledge about the pathogenic mechanism of this species in humans. Moreover, sliding motility is highly associated with biofilm formation in mycobacteria. In this study, we aimed to identify genes that have roles in sliding motility in M. marinum.
Materials and Methods
Bacteria Strains
M. marinum NTUH-M6885 is a strain that was clinically isolated at the National Taiwan University Hospital. It was cultured in 7H9 medium supplemented with 10% oleic acid/albumin/dextrose/catalase (OADC), 0.5% glycerol, and 0.05% Tween-80 at 32°C (Tan et al., 2006; Chen et al., 2015). Escherichia coli DH10B was grown in Luria broth (LB). When required for the experiments, the antibiotic hygromycin was used at a concentration of 50 mg/L for M. marinum and 100 mg/L for E. coli.
Generation of M. marinum Transposon Mutant Library
M. smegmatis mc2155 was used to propagate the TM4-derived conditionally replicating phage phAE94 [which was a kind gift from Dr. William R. Jacobs, Jr., Howard Hughes Medical Institute, New York, NY, United States (Bardarov et al., 1997)]. This phage carries the kanamycin-resistance transposon Tn5367 (Shin et al., 2006). We followed the procedures used in previous studies (Rybniker et al., 2003; Chen et al., 2015) to promote phAE94 infection of M. marinum NTUH-M6885 cells.
Screening for M. marinum Transposon Mutants With Decreased Sliding Motility
Sliding motility mutations were screened for using a sliding agar plate (7H9, 6% glycerol and 0.3% agarose) (Mohandas et al., 2016). An aliquot of 1 μL bacteria culture (in the stationary phase) was dropped onto these agar plates (24- or 6-wells) and cultured at 32°C for 9–11 days. To confirm the sliding defect in transposon and deletion mutants, 1 μL bacteria culture adjusted to an optical density at 600 nm (OD600) of 1 was dropped onto 6-well plates and cultured at 32°C for 7 days. The diameters of the sliding areas were then measured.
Identification of Transposon Mutants by Semi-Random Polymerase Chain Reaction (PCR)
The insertion sites of Tn5367 in the transposon mutants were identified by semi-random PCR and DNA sequencing as previously described (Chun et al., 1997; Choi et al., 2001; Shin et al., 2006; Chen et al., 2015). Supplementary Table 1 shows the primers used in these experiments.
Construction of Deletion Mutants
Gene-deleted fragments were constructed using primers listed in Supplementary Table 1. The Hygr-lacZ-sacB cassette of the pGOAL19 plasmid (Addgene plasmid #20190, Cambridge, MA, United States) was digested with ScaI and the gene-deleted fragments were then cloned into the ScaI site of the digested plasmid. We then transformed the constructed plasmid into M. marinum NTUH-M6885 by electroporation (BTX®, ECM 630 Electroporation System, MA, United States) under the following conditions: 2500 mV, 1,000 Ω, and 25 μF (Larsen et al., 2007). Following the procedure of Parish and Stoker (2000) and Chen et al. (2015), M. marinum unmarked deletion mutants (with in-frame deletions) were obtained after two rounds of homologous recombination. After recovering from the electroporation, each transformed M. marinum mutant was cultured on 7H11 with 50 mg/L hygromycin for 2 weeks. The single colony was then subcultured on 7H11 with 100 mg/L X-gal and 50 mg/L hygromycin for 1 week to select the single cross-over transformants (blue colonies). The blue colonies were selected and cultured on 7H11 with 2% sucrose and 100 mg/L X-gal for 1 week to obtain the deletion mutants (white colonies).
5′-Rapid Amplification of cDNA Ends (RACE)
The 5′-RACE procedure was performed using a SMARTerTM RACE cDNA Amplification Kit1 (CA, United States). This kit includes SMARTer II A oligonucleotides and SMARTScribe Reverse Transcriptase. This allowed isolation of the complete 5′ sequence of our target transcript from the total RNA.
Construction of Complemented Strains
After isolating the complete 5′ sequence of the target operon, we amplified and connected the promoter region and target genes using overlap- or inverse-PCR, with primers listed in Supplementary Table 1. The complementation fragments were then cloned into a blunted HindIII-site of pMN437 (which was a kind gift from Dr. Michael Niederweis, University of Alabama at Birmingham, Birmingham, AL, United States) (Steinhauer et al., 2010). Subsequently, we transformed the complementation plasmid into deletion mutants.
Growth Curve
Cultures were inoculated with fresh precultures to an OD600nm of 0.1. Bacterial growth was monitored spectrophotometrically, and colony counts were determined every day.
Biofilm Formation
M. marinum was cultured in 7H9 medium (without Tween-80) with OADC, and with shaking at 100 rpm (Pang et al., 2012; Mohandas et al., 2016). The cell concentration was adjusted to an OD600nm of 0.01 in round-bottom 96-well polypropylene cell culture plates (costar® 3879, New York, NY, United States) at 32°C, and the cells were then cultured for 3 weeks. After 3 weeks, the medium was removed. We stained the biofilm with 200 μL 1% crystal violet (CV) for 10 min (Trivedi et al., 2016). We then removed the CV and washed the biofilm twice with 1× phosphate-buffered saline (PBS). Subsequently, we extracted the CV from the biofilm using 99.5% EtOH and assessed the biofilm formation by measuring OD490nm.
Confocal Laser Scanning Microscopy
M. marinum was cultured on cover slides with 7H9 medium (without Tween-80) with OADC, and with shaking at 100 rpm for 2 weeks. The bacteria were then washed with PBS before fixing in formalin. The biofilm was observed with a Leica TCS SP5 confocal laser scanning microscope (Leica, Wetzlar, Germany) and three-dimensional images were analyzed using Volocity software (version 6.0.1, MA, United States).
Western Blotting
Bacteria were cultured in 7H9 medium for 4 days (to mid-log phase). We then removed the culture medium and washed the bacteria with Sauton’s medium. The bacteria were then cultured for 4 days at 32°C (Gao et al., 2004). Culture filtrates collected by centrifugation and filtration through 0.22 μm-pore-size polyethersulfone filter were condensed using a concentrator (Amicon, NJ, United States) with a 3-kDa cut-off. Next, 10 μg proteins were loaded on 15% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel. The proteins were detected using antibodies against 10 kDa culture filtrate protein (CFP-10; which is also known as EsxB), Ag85B (Ag85B protein levels was used for normalization), and heat-shock protein 65 (Hsp-65; also called grOEL; Hsp-65 protein levels was used for normalization). The details regarding the antibodies were are follows: anti-CFP-10 antibody (Abcam, ab45074, 1:5000), anti-Ag85B antibody (Abcam, ab43019, 1:3000), and anti-Hsp-65 antibody (grOEL; Abcam, ab20519, 1:200).
Two-Dimensional Thin-Layer Chromatography (2D-TLC)
The extraction and 2D-TLC analysis of polar lipids was based on established procedures used in our previous study (Chen et al., 2015) and in a study by Burguiere et al. (2005). First, the polar lipids were extracted from M. marinum grown on 7H11 agar plates. Second, lipids were examined using TLC aluminum sheets (Merck, Summit, NJ, United States). Third, the LOS signals were visualized by spraying the plates with ceric ammonium molybdate [CAM; 24 g (NH4)6Mo7O24⋅4H2O, 0.5 g ammonium cerium nitrate, 500 mL H2O, and 28 mL H2SO4], followed by gentle charring of the plates (Chen et al., 2015).
Statistical Analysis
All the data were from three independent experiments and they are presented as means ± standard deviations (SDs). We estimated the statistical significance of the differences using one-way analysis of variance (ANOVA) or two-tailed Student’s t-tests using GraphPad Prism software (version 5.01, La Jolla, CA, United States).
Results
Screening an M. marinum Transposon Library for Sliding Motility
We first constructed a transposon library of M. marinum NTUH-M6885 containing 2,304 mutants to screen for genes associated with sliding motility. We then randomly selected 16 of these mutants and used semi-random PCR and sequencing to characterize them. The results showed that these 16 mutants had unique transposon insertion sites (Supplementary Table 2), indicating that the library had good diversity.
Next, we screened the 2,304 mutants for sliding motility using 24-well sliding agar plates. We then re-examined and identified 13 mutants with defects in sliding motility using 6-well sliding agar plates. The wild-type strain slid to the edges of the culture well (3.5 cm in diameter), whereas the 13 mutants had sliding distances <2 cm (Supplementary Figure 1). Finally, we reperformed the same experiment using an equal quantity of bacteria cultured for 7 days (OD600 = 1), and we found that only five mutants had sliding motility defects (Supplementary Figure 2).
Role of the Early Secreted Antigen 6 kDa (ESAT-6) Secretion System 1 (ESX-1) Genes in Sliding Motility
We identified the disrupted genes of the five transposon mutants with sliding motility defects using semi-random PCR and DNA sequencing (Table 1). Two mutants harbored insertions within tetR and phoU, which both belong to putative regulators. The other three transposon mutants—23-B1 (mmar_5439), 8-C7 (upstream of mmar_5440), and 22-B5 (mmar_5443)—had disruptions in the gene cluster encoding the type VII secretion system ESX-1 (Figures 1A,B). Previous studies named mmar_5439 as espE, mmar_5440 as espF, and mmar_5443 as eccA1 (Bitter et al., 2009; Sani et al., 2010). Among these gene candidates, we focused on the ESX-1 cluster containing espE, espF, and eccA1. The products of all three of these genes are supposed to be displayed at the cell surface but their functions regarding sliding motility have not been characterized in M. marinum. These initial results suggested that ESX-1-related genes could be involved in sliding motility.
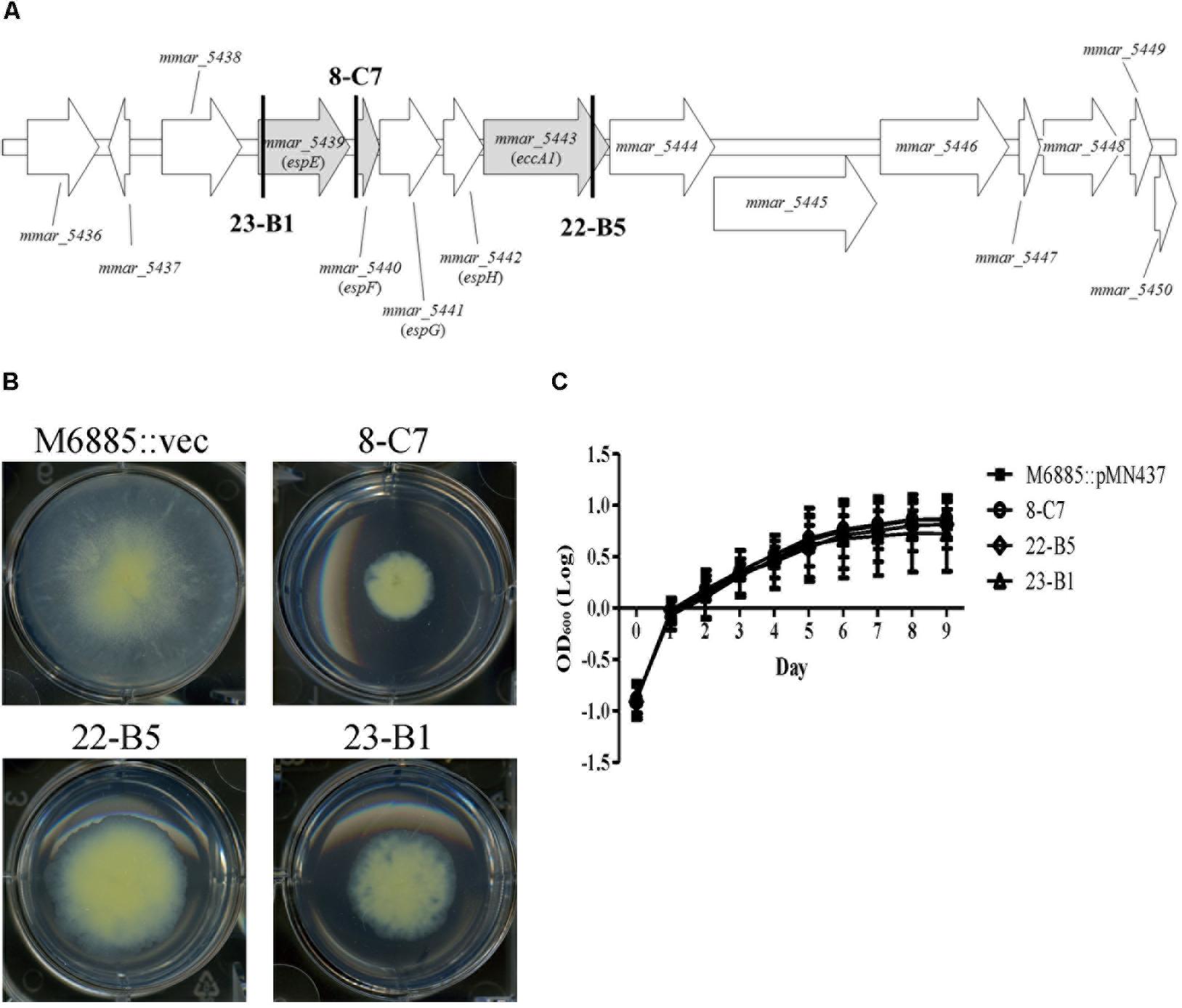
FIGURE 1. Transposon disruption of three ESX-1-related genes significantly impairs sliding motility but not growth. (A) Transposon insertion sites within the ESX-1 locus. The three disrupted genes (gray) were espE (mmar_5439), espF (mmar_5440), and eccA1 (mmar_5443). The bold vertical lines indicate the transposon insertion sites. (B) Sliding motility of the three transposon mutants (8-C7, 22-B5, and 23-B1). Aliquots of 1 μL bacteria culture (OD600 = 1) were added to 6-well sliding agar plates and cultured at 32°C for 1 week. (C) Bacteria were cultured in 7H9 medium supplemented with 10% oleic acid/albumin/dextrose/catalase (OADC), 0.5% glycerol, and 0.05% Tween-80. The growth of different strains was assessed based on OD600nm. Here and below, all data are from three independent experiments and presented as means ± SDs with one-way ANOVA.
Detection of Growth Rates in Transposon Mutants With Sliding Defects
The presence of a surfactant in the culture medium and a high bacterial growth rate can increase sliding motility (Kearns, 2010). Thus, it is necessary to determine whether the impaired sliding of the transposon mutants resulted from a growth defect. Recovery of bacterial counts (data not shown) and monitoring changes in OD600nm over time was carried out to measure the growth rates of the wild-type strain and its three transposon mutants (23-B1, 8-C7, and 22-B5). Based on assays in general culture medium (7H9 with 10% OADC, 0.5% glycerol, and 0.05% Tween-80) or sliding broth medium (7H9 with 0.5% glycerol), the growth rates of the three transposon mutants showed no significant differences compared to the corresponding growth of the wild-type strain (Figure 1C and Supplementary Figure 3A). Thus, the sliding defects of these three mutants were not due to defects in growth.
Transcriptional Units and Sequence Alignments of the ESX-1 Gene Cluster
Reverse-transcription (RT)-PCR to determine the transcriptional units of the ESX-1 gene cluster was performed with total RNA from M. marinum NTUH-M6885 as the template and primer pairs that hybridize within two consecutive genes. Total RNA without RT was used as the negative control, to exclude genomic DNA contamination. Figures 2A,B showed positive results for the junctions of genes from mmar_5438 to mmar_5450. In addition, Supplementary Figures 4A,B show the longer amplified PCR products containing two or three adjacent genes. The PCR products with expected sizes were also confirmed by sequencing. These results indicate that the ESX-1 gene cluster contains 13 genes that belong to a single transcriptional unit. This operon starts with mmar_5438 and ends with mmar_5450.
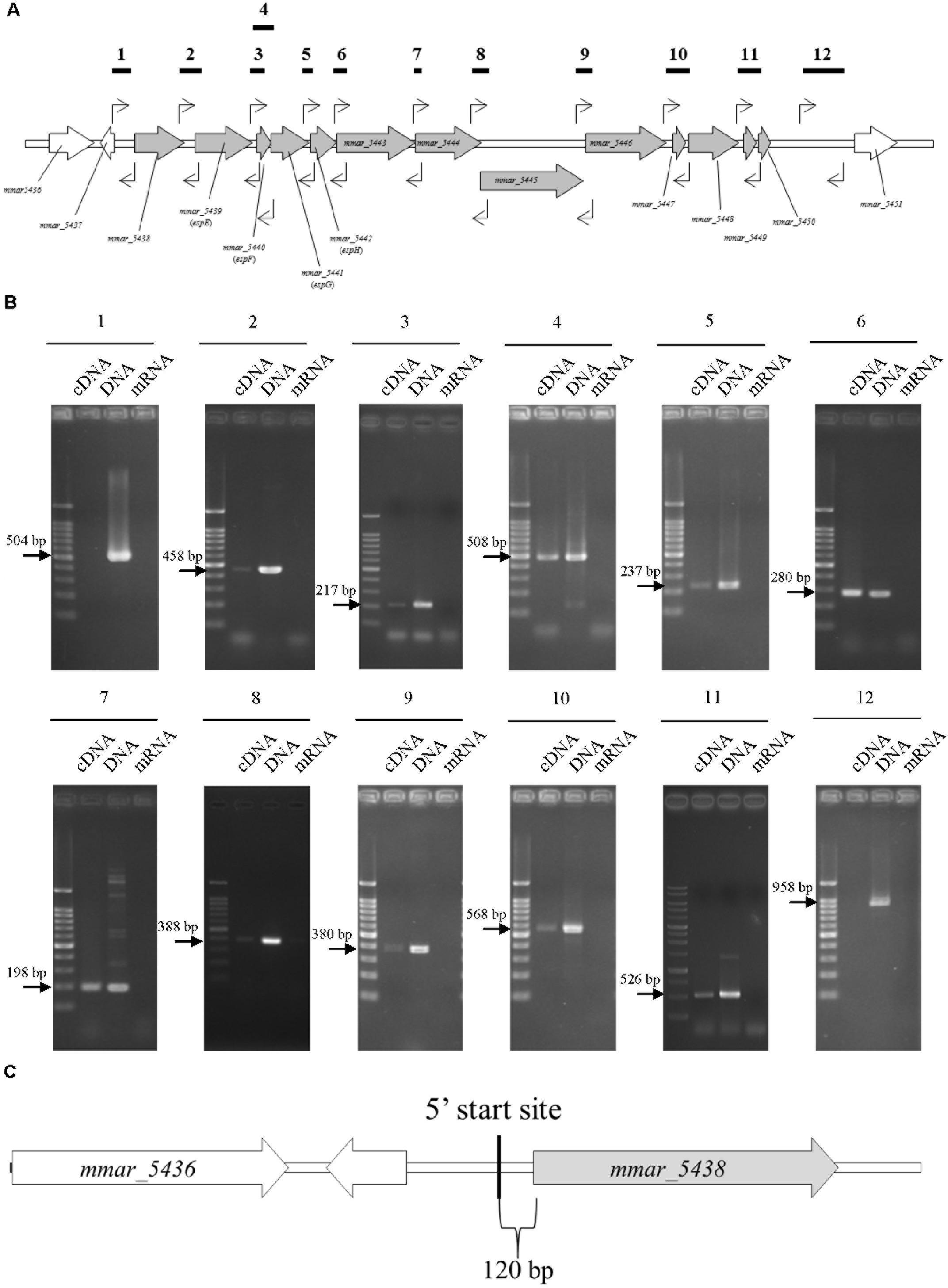
FIGURE 2. The sliding-related operon involves mmar_5438 to mmar_5450, and its 5′start site is 120 bp upstream of mmar_5438. The primer pairs were: 1, 5437-38 gap-F/5437-38 gap-R; 2, 5438-39 gap-F/5438-39 gap-R; 3, 5439-40 gap-F/5439-40 gap-R; 4, 5439-40 gap-F/5440-41 gap R; 5, 5441-42 gap-F/5441-42 gap-R; 6, 5442-43 gap-F/5442-43 gap-R; 7, 5443-44 gap-F/5443-44 gap-R; 8, 5444-45 gap-F/5444-45 gap-R; 9, 5445-46 gap-F/5445-46 gap-R; 10, 5446-47 gap-F/5447-48 gap-R; 11, 5448-49 gap-F/5449-50 gap-R; 12, 5450-51 gap-F/5450-51 gap-R2. The primer list is shown in Supplementary Table 1. (A) Primer recognition sites. The bold horizontal lines indicate amplified gene fragments, and arrows indicate primers. (B) Data showing that the sliding-related operon involves mmar_5438 to mmar_5450. Each gel is independent and not cropped from different parts of the same gel. cDNA: cDNA of M. marinum NTUH-M6885 (template); DNA: DNA of M. marinum NTUH-M6885 (positive control); mRNA: mRNA of M. marinum NTUH-M6885 (negative control, to exclude genomic DNA contamination). (C) The 5′ start site begins 120 bp upstream of mmar_5438 (gray). The bold vertical line indicates the 5′ start site, detected using a SMARTerTM RACE cDNA Amplification Kit.
Next, we identified the 5′ start site of this operon using a SMARTerTM RACE cDNA Amplification Kit. The result showed that the transcriptional start site of the ESX-1 operon was located 120 bp upstream of mmar_5438 (Figure 2C). Furthermore, we determined the full DNA sequence of this operon in M. marinum NTUH-M6885 using next-generation sequencing. Analysis of the nucleotide sequences of the ESX-1 cluster from M. marinum NTUH-M6885 (National Center for Biotechnology Information [NCBI] accession number: MF034931) revealed 99% sequence similarity compared with the cluster from the M. marinum M strain.
Roles of espE, espF, espG, and espH Genes in Sliding Motility
The genes, espE (mmar_5439), espF (mmar_5440), and eccA1 (mmar_5443) are all located in the same operon. To further characterize whether the specific genes of the ESX-1 cluster play roles in sliding motility, unmarked deletion mutants of espE, espF, and eccA1 and also of espG (mmar_5441) and espH (mmar_5442), were constructed. The deletions of espE, espF, espG, espH, and eccA1 were validated by PCR using two primer pairs targeted to the deleted genes and their flanking regions (Supplementary Figure 5). Figures 3A,B show that the sliding motility of each unmarked deletion mutant, except for the eccA1 mutant (∆eccA1), was dramatically reduced compared with the sliding motility of the wild-type strain. The motility defects of the gene deletion mutants were significantly restored by complementation with the corresponding gene (Figures 3C,D). The growth rates of these deletion mutants were not significantly different compared to the growth rate of the wild-type strain (Supplementary Figure 3B). However, the sliding motility of the espG-complemented strain was only partially restored by complementation with the plasmid containing the espG gene. As the ESX-1 operon contains the promoter for 13 genes, the results imply that proper regulation of espG gene expression might require cis-elements within the operon. Thus, these results suggest that espE, espF, espG, and espH have important roles in sliding motility in M. marinum NTUH-M6885.
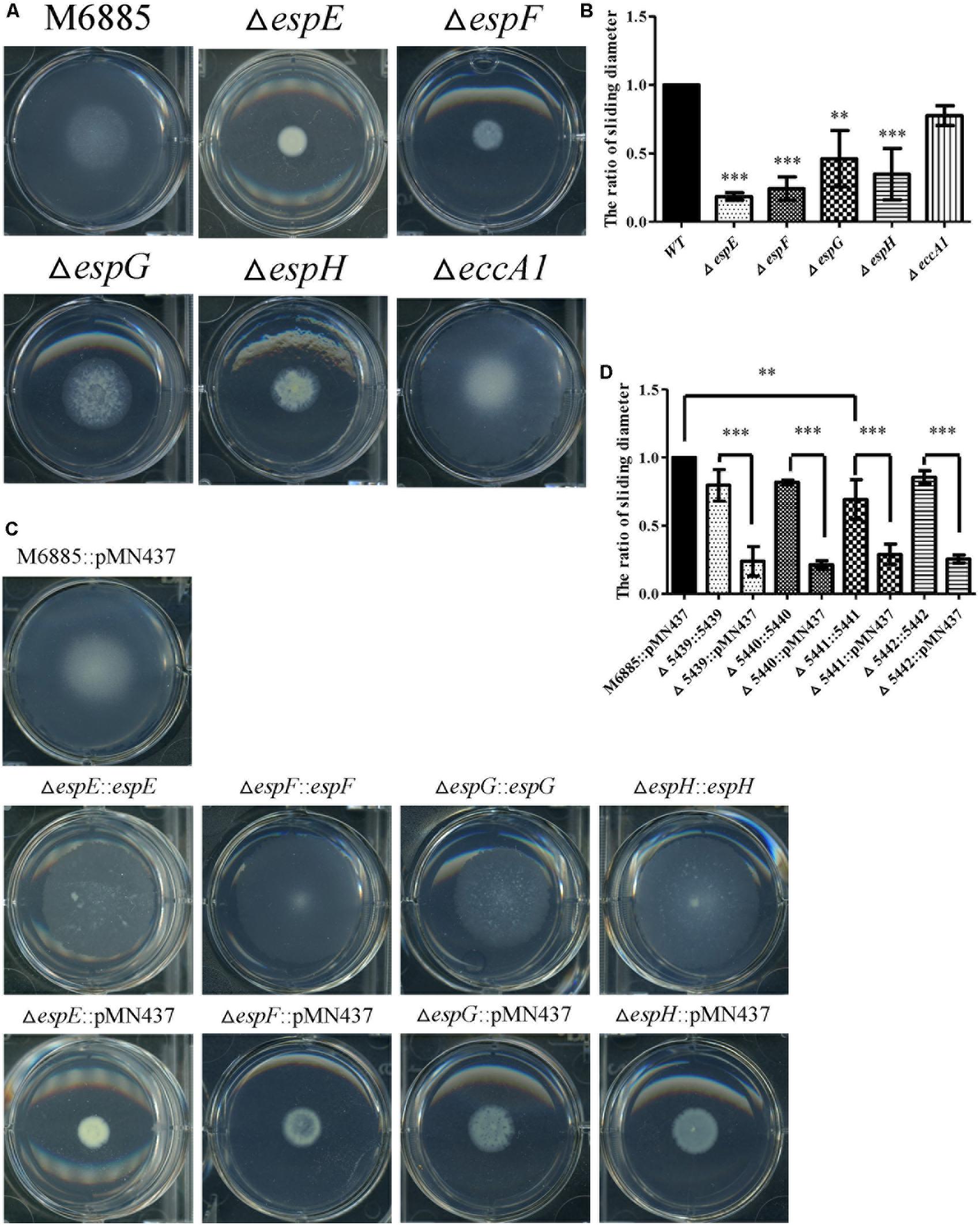
FIGURE 3. Four mutants (espE, espF, espG, and espH) have significantly impaired sliding motility. (A) Sliding motility of M. marinum deletion mutants (∆espE, ∆espF, ∆espG, ∆espH, and ∆eccA1) and M. marinum NTUH-M6885 (wild-type) on sliding agar plates. (B) Quantification of sliding diameters of the 6 strains shown in (A). (C) Sliding motility of complemented strains relative to M. marinum NTUH-M6885 harboring pMN437 on sliding plates. (D) Quantification of sliding diameters of the nine strains in (C). The quantification of the sliding diameters was used for normalization, with the wild-type diameter being assigned a value of 1. Means and SDs from three independent experiments were calculated with one-way ANOVA (∗∗p < 0.01, ∗∗∗p < 0.001).
Roles of espE, espF, espG, and espH Genes in Biofilm Formation
Previous studies indicated that sliding motility correlates with biofilm formation in Mycobacterium spp. (Nessar et al., 2011; Sousa et al., 2015; Mohandas et al., 2016). To discern whether the ESX-1-related genes contribute to biofilm formation, we compared the biofilm formation of the deletion mutants with that of the wild-type strain. After 3 weeks of culturing in general culture medium without Tween-80, the deletion mutants exhibited decreased biofilm formation compared with the wild-type strain. All complemented strains produced biofilms similar to that of the wild-type strain (Figure 4). To further confirm the role of these ESX-1 genes in biofilm formation, three-dimensional biofilms were formed on cover slides and they were analyzed using confocal laser scanning microscopy (Figure 5). The biofilms of these deletion mutants were thinner and more scattered than those of the wild-type strain, and complementation significantly restored biofilm formation. These data indicate that espE, espF, espG, and espH are required for biofilm formation in M. marinum.
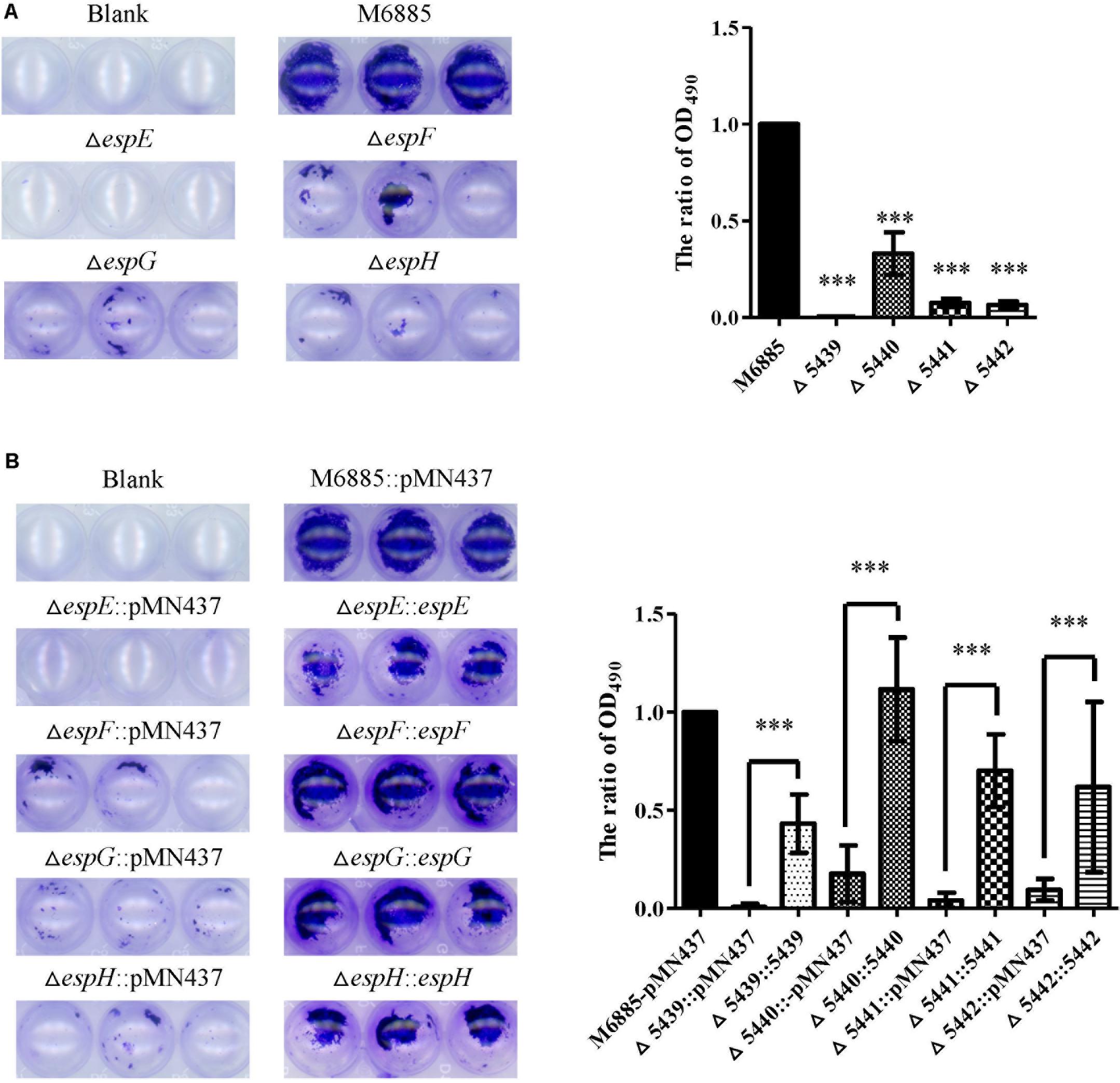
FIGURE 4. The espE, espF, espG, and espH genes also function in biofilm formation. Cells were stained with 1% CV and OD490nm was measured after 10 min. (A) Biofilm formation by the deletion mutants (∆espE, ∆espF, ∆espG, and ∆espH) and M. marinum NTUH-M6885 (wild-type), and quantitation of these results in all four strains (right). The quantification of biofilm was used for normalization, with the wild-type diameter being assigned a value of 1. All deletion mutants were compared with the wild-type strain. Means and SDs from three independent experiments from triplicates were calculated with one-way ANOVA (∗∗∗p < 0.001). (B) Biofilm formation in complemented strains, and quantification of these results in all 9 strains (right). The quantification of biofilm was used for normalization, with the wild-type diameter being assigned a value of 1. All deletion mutants were compared with the wild-type strain and their corresponding complemented strain. Means and SDs from three independent experiments from triplicates were calculated with one-way ANOVA (∗∗∗p < 0.001).
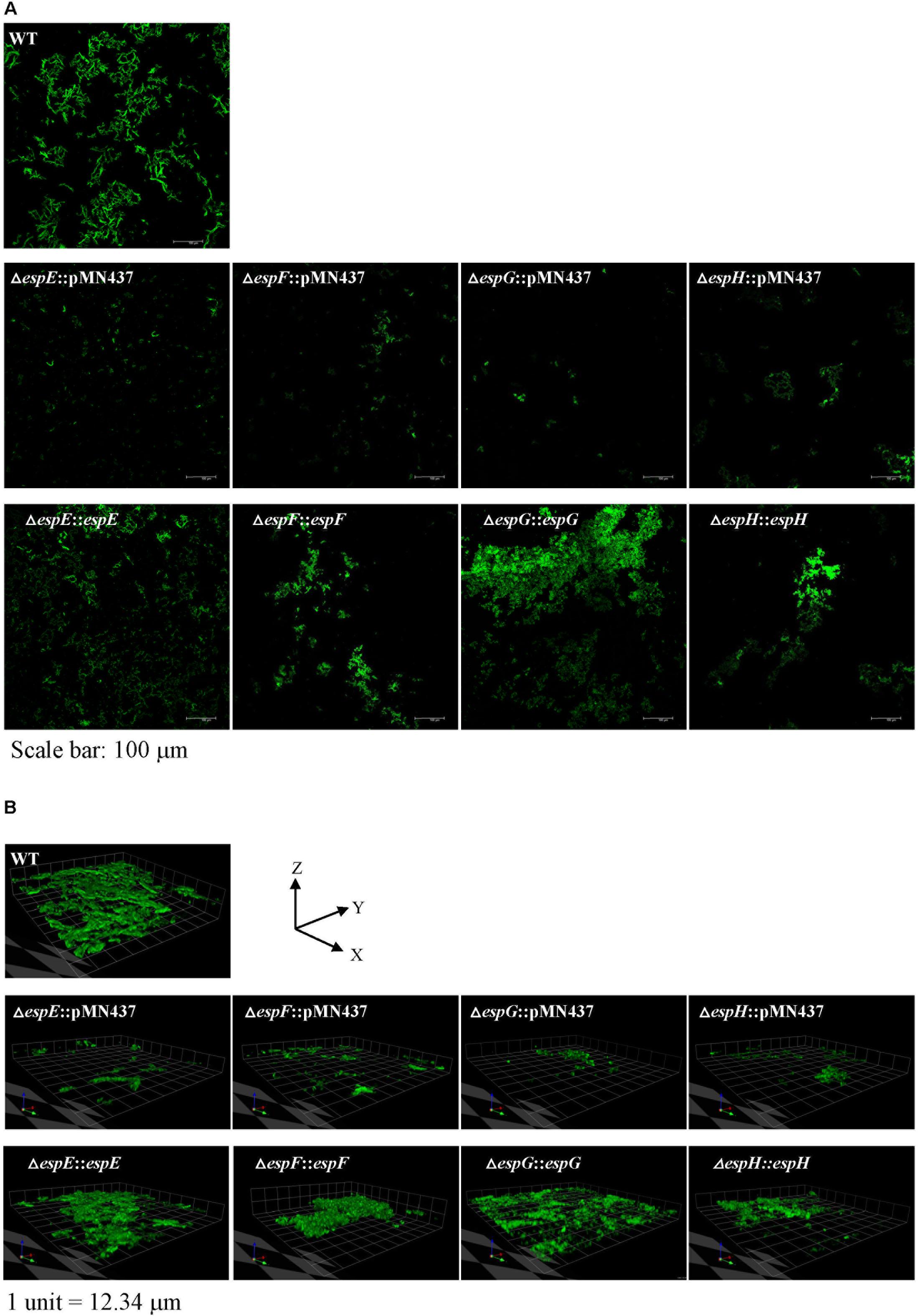
FIGURE 5. Visualization of biofilm formation on cover slides with confocal laser scanning microscopy (CLSM). To further analyze the structure of the biofilms, CLSM was performed using bacteria carrying pMN437, which harbors gfp. (A) Microscopic image at 20× magnification. Scale bar = 100 μm. (B) 3D imaging with a Z-stack of biofilm structure.1 unit = 12.34 μm.
EsxB Secretion in Δ espE, Δ espF, Δ espG, and Δ espH Mutants
According to previous studies, both EsxA (ESAT-6) and EsxB (CFP-10) are indispensable components of the ESX-1 secretion system (Ates et al., 2016; Wong, 2017) and they are considered to be indicators of the ESX-1 system with secretion function (Gao et al., 2004; Brodin et al., 2006; Champion et al., 2014). To investigate whether M. marinum NTUH-M6885 had a functional ESX-1 system, EsxB secretion from the wild-type strain was detected by western blotting. Cell filtrate of M. tuberculosis H37Rv was used as a positive control (Supplementary Figure 6). The M. marinum NTUH-M6885 wild-type strain and also all of the Esx-1 gene deletion mutants accumulated EsxB protein in their cell lysates. However, deletion of espE, espF, espG, and espH led to 0.35-, 0.59-, 0.82-, and 0.73-fold lower secretion of EsxB protein than the secretion of the wild-type strain. These results indicate that wild-type M. marinum NTUH-M6885 could secrete EsxB protein. There was a substantial decrease in secretion for espG and espH deletion mutants, and a slight decrease for espE and espF deletion mutants.
Discussion
The M. marinum NTUH-M6885 transposon mutant library was screened, and 5 mutants (8-C7, 12-B1, 14-A7, 22-B5, and 23-B1) that had defective sliding motility were identified. 12-B1 and 14-A7 had an interruption in tetR and phoU, respectively. Both genes are putative regulators and they might regulate the other downstream effector genes for sliding motility directly or indirectly. Further studies are needed to understand whether the regulation of tetR or phoU is critical for sliding motility in M. marinum. 22-B5 had a transposon insertion in the eccA1 gene and showed significantly decreased sliding motility; however, the sliding motility of the eccA1 gene deletion strain was similar to that of the wild-type strain. Several studies have reported that the insertion of transposable elements influences the expression of nearby genes (Wei and Cao, 2016) (which is known as the polar effect). The expression levels of genes both upstream and downstream of insertion sites can be affected (Cheng et al., 2015). Quantification of mRNA indicated that the expression of neighboring genes (espE, espF, and espG) was reduced in strain 22-B5 (Supplementary Figure 7). Thus, the sliding defect in strain 22-B5 was due to a polar effect. The virulence of the eccA1 transposon mutants in previous studies could be fully complemented (Gao et al., 2004; Joshi et al., 2012), indicating that eccA1 affects virulence through a mechanism unrelated to the effect on sliding ability. The discrepancy might be due to the virulence of M. marinum not being caused by sliding ability.
A previous study showed that EccA1 plays a regulatory role in mycolic acid synthesis in M. marinum (Joshi et al., 2012). In addition, alterations in mycolic acid synthesis and composition can affect biofilm formation in M. smegmatis (Ojha et al., 2005). In the present study, the deletion of eccA1 in M. marinum did not significantly affect sliding motility and biofilm formation (data not shown), indicating that alterations in the mycolic acid synthesis in the eccA1 mutant do not correlate with sliding motility and biofilm formation in M. marinum.
The mmar_5438-mmar_5450 gene cluster of M. marinum and the Rv3863-Rv3875 gene cluster of M. tuberculosis share 79% identity based on nucleotide sequence analysis. Bottai et al. (2011) previously revealed the operon structures of the ESX-1 gene cluster in M. tuberculosis, showing that espE, espF-espG, and espH-eccA1 belong to three independent operons. However, our results demonstrated that the operon structure of the ESX-1 loci in M. marinum involves a single operon. Therefore, we speculate that the ESX-1 gene cluster in different mycobacteria species might have different sequence alignments and operon structures.
Previous research indicated that EspE and EspF are major cell surface proteins in M. marinum (Sani et al., 2010; van der Woude et al., 2012). EspG proteins are cytosolic chaperones that specifically interact with heterodimeric PE-PPE substrates (Ates et al., 2016) and EspH is required for substrate export (EsxA and EsxB) (Gao et al., 2004; McLaughlin et al., 2007; Champion et al., 2014). In addition, a previous study showed that a defect in M. marinum espE led to the formation of smooth colonies (Carlsson et al., 2009). In agreement with this finding, we also found morphological changes in ∆espE, ∆espF, ∆espG, and ∆espH mutants and smooth colonies were observed (data not shown). Several studies have reported that morphological change correlates with reduced sliding motility (Maya-Hoyos et al., 2015; Lee et al., 2017). These might be the reasons why the ∆espE and ∆espF mutants had more severe defects in sliding motility and biofilm formation.
espE encodes the major surface-associated protein in M. marinum (Sani et al., 2010). The deletion of espE resulted in the most profoundly reduced sliding motility and biofilm formation abilities. To further investigate whether the surface properties of the espE mutant were changed, the surface charge and hydrophobicity of the wild-type and ∆espE strains were determined by using a Zetasizer Nano ZS (Malvern, Malvern, United Kingdom) (Kundu et al., 2017) and a bacterial adhesion to solvent assay (Miyamoto et al., 2004; Xu et al., 2009). Our results showed that the surface charge and hydrophobicity of the espE mutant were not significantly different compared with the wild-type values (data not shown). The LOS biosynthesis of ∆espE was assessed by 2D-TLC and was similar to that of the wild-type strain (Supplementary Figure 8). Thus, we suppose that the decreased sliding ability and biofilm formation of the espE mutant were not due to the alterations of the surface charge, hydrophobicity, or LOS biosynthesis.
Our results indicate that the deletion of espE, espF, espG, and espH resulted in extremely decreased sliding motility and biofilm formation. The incomplete restoration exhibited by complemented strains might be due to different expression levels of complemented genes carried by the plasmids, which was reported previously (Karnholz et al., 2006; Lin et al., 2006). M. marinum NTUH-M6885 could secrete EsxB protein and had a functional ESX-1 system. In addition, our data show that espE, espF, espG, and espH mutants accumulated EsxB protein in the cytosol and had reduced EsxB protein in the cell filtrates when compared with the wild-type strain (Supplementary Figure 6B). These results are consistent with those of a previous study that indicated that an M. marinum espE transposon mutant could not secrete EsxB protein and accumulated it in the cytosol (Carlsson et al., 2009). EsxB plays roles in cell membrane lysis and virulence (Unnikrishnan et al., 2017). Several previous studies showed that the secretion of the EsxA-EsxB heterodimer is co-dependent on the secretion of the EspA-EspC heterodimer, suggesting that the EspA-EspC heterodimer has an important function in ESX-1 secretion (Abdallah et al., 2007; Ates and Brosch, 2017; Wong, 2017). Additionally, the signal motifs of EspE and EspF were similar to those of EspA and EspC based on an analysis using the structural homology server PHYRE2 (Solomonson et al., 2015). Sliding motility and biofilm formation correlate with virulence in Mycobacterium spp. (Schorey and Sweet, 2008; Pang et al., 2013). Esp proteins regulate substrate export, which might be involved in sliding motility and biofilm formation.
The ESX-1 system is one of the major groups of T7SS (Bottai et al., 2011; Korotkova et al., 2014) and its role in pathogenesis has been reported in M. tuberculosis (Unnikrishnan et al., 2017; Wong, 2017) and M. marinum (Gao et al., 2004; Cardenal-Munoz et al., 2017; Unnikrishnan et al., 2017). Previous studies reported that espG and espH affect M. marinum virulence by influencing cytolysis, cytotoxicity, growth in macrophages, and spreading (Gao et al., 2004; McLaughlin et al., 2007). Bottai et al. (2011) indicated that espF and espG1 were associated with virulence during M. tuberculosis infection (Brodin et al., 2006) and M. tuberculosis can survive in bone marrow-derived macrophages. The present study is the first study to demonstrate the contribution of the ESX-1 system to sliding motility and biofilm formation in M. marinum.
In summary, we demonstrated that espE, espF, espG, and espH genes are critical for sliding motility and biofilm formation in M. marinum. These genes, which are located in the T7SS ESX-1 operon, are important virulence factors in M. marinum.
Author Contributions
T-LL and J-TW designed the research, discussed the analysis, and revised the paper. L-YL and Y-YC prepared materials and performed experiments. L-YL, Y-YC, T-LL, and P-FH analyzed the data. L-YL, T-LL, and P-FH wrote the main text of the manuscript. All the authors reviewed and approved the final version of the manuscript.
Funding
This study was supported by the Ministry of Science and Technology, National Taiwan University, National Taiwan University Hospital of Taiwan, and Liver Disease Prevention and Treatment Research Foundation of Taiwan.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Feng-Ling Yang and Dr. Shih-Hsiung Wu (Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan) for their kind help with the lipid extraction.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01160/full#supplementary-material
Footnotes
References
Abdallah, A. M., Gey Van Pittius, N. C., Champion, P. A., Cox, J., Luirink, J., Vandenbroucke-Grauls, C. M., et al. (2007). Type VII secretion–mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–891. doi: 10.1038/nrmicro1773
Alibaud, L., Rombouts, Y., Trivelli, X., Burguiere, A., Cirillo, S. L., Cirillo, J. D., et al. (2011). A Mycobacterium marinum TesA mutant defective for major cell wall-associated lipids is highly attenuated in Dictyostelium discoideum and zebrafish embryos. Mol. Microbiol. 80, 919–934. doi: 10.1111/j.1365-2958.2011.07618.x
Ates, L. S., and Brosch, R. (2017). Discovery of the type VII ESX-1 secretion needle? Mol. Microbiol. 103, 7–12. doi: 10.1111/mmi.13579
Ates, L. S., Houben, E. N., and Bitter, W. (2016). Type VII secretion: a highly versatile secretion system. Microbiol. Spectr. 4:1. doi: 10.1128/microbiolspec.VMBF-0011-2015
Bardarov, S., Kriakov, J., Carriere, C., Yu, S., Vaamonde, C., Mcadam, R. A., et al. (1997). Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 94, 10961–10966. doi: 10.1073/pnas.94.20.10961
Bitter, W., Houben, E. N., Bottai, D., Brodin, P., Brown, E. J., Cox, J. S., et al. (2009). Systematic genetic nomenclature for type VII secretion systems. PLoS Pathog. 5:e1000507. doi: 10.1371/journal.ppat.1000507
Bottai, D., Majlessi, L., Simeone, R., Frigui, W., Laurent, C., Lenormand, P., et al. (2011). ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J. Infect. Dis. 203, 1155–1164. doi: 10.1093/infdis/jiq089
Brodin, P., Majlessi, L., Marsollier, L., De Jonge, M. I., Bottai, D., Demangel, C., et al. (2006). Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect. Immun. 74, 88–98. doi: 10.1128/IAI.74.1.88-98.2006
Burguiere, A., Hitchen, P. G., Dover, L. G., Kremer, L., Ridell, M., Alexander, D. C., et al. (2005). LosA, a key glycosyltransferase involved in the biosynthesis of a novel family of glycosylated acyltrehalose lipooligosaccharides from Mycobacterium marinum. J. Biol. Chem. 280, 42124–42133. doi: 10.1074/jbc.M507500200
Cardenal-Munoz, E., Arafah, S., Lopez-Jimenez, A. T., Kicka, S., Falaise, A., Bach, F., et al. (2017). Mycobacterium marinum antagonistically induces an autophagic response while repressing the autophagic flux in a TORC1- and ESX-1-dependent manner. PLoS Pathog. 13:e1006344. doi: 10.1371/journal.ppat.1006344
Carlsson, F., Joshi, S. A., Rangell, L., and Brown, E. J. (2009). Polar localization of virulence-related Esx-1 secretion in mycobacteria. PLoS Pathog. 5:e1000285. doi: 10.1371/journal.ppat.1000285
Champion, M. M., Williams, E. A., Pinapati, R. S., and Champion, P. A. (2014). Correlation of phenotypic profiles using targeted proteomics identifies mycobacterial esx-1 substrates. J. Proteome Res. 13, 5151–5164. doi: 10.1021/pr500484w
Chen, Y. Y., Yang, F. L., Wu, S. H., Lin, T. L., and Wang, J. T. (2015). Mycobacterium marinum mmar_2318 and mmar_2319 are responsible for lipooligosaccharide biosynthesis and virulence toward Dictyostelium. Front. Microbiol. 6:1458. doi: 10.3389/fmicb.2015.01458
Cheng, C., Nair, A. D., Jaworski, D. C., and Ganta, R. R. (2015). Mutations in Ehrlichia chaffeensis causing polar effects in gene expression and differential host specificities. PLoS One 10:e0132657. doi: 10.1371/journal.pone.0132657
Choi, K. P., Bair, T. B., Bae, Y. M., and Daniels, L. (2001). Use of transposon Tn5367 mutagenesis and a nitroimidazopyran-based selection system to demonstrate a requirement for fbiA and fbiB in coenzyme F(420) biosynthesis by Mycobacterium bovis BCG. J. Bacteriol. 183, 7058–7066. doi: 10.1128/JB.183.24.7058-7066.2001
Chun, K. T., Edenberg, H. J., Kelley, M. R., and Goebl, M. G. (1997). Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13, 233–240. doi: 10.1002/(SICI)1097-0061(19970315)13:3<233::AID-YEA88>3.0.CO;2-E
Gao, L. Y., Guo, S., Mclaughlin, B., Morisaki, H., Engel, J. N., and Brown, E. J. (2004). A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53, 1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x
Ghosh, S., Indi, S. S., and Nagaraja, V. (2013). Regulation of lipid biosynthesis, sliding motility, and biofilm formation by a membrane-anchored nucleoid-associated protein of Mycobacterium tuberculosis. J. Bacteriol. 195, 1769–1778. doi: 10.1128/JB.02081-12
Hagedorn, M., and Soldati, T. (2007). Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell Microbiol. 9, 2716–2733. doi: 10.1111/j.1462-5822.2007.00993.x
Joshi, S. A., Ball, D. A., Sun, M. G., Carlsson, F., Watkins, B. Y., Aggarwal, N., et al. (2012). EccA1, a component of the Mycobacterium marinum ESX-1 protein virulence factor secretion pathway, regulates mycolic acid lipid synthesis. Chem. Biol. 19, 372–380. doi: 10.1016/j.chembiol.2012.01.008
Karnholz, A., Hoefler, C., Odenbreit, S., Fischer, W., Hofreuter, D., and Haas, R. (2006). Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J. Bacteriol. 188, 882–893. doi: 10.1128/JB.188.3.882-893.2006
Kearns, D. B. (2010). A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8, 634–644. doi: 10.1038/nrmicro2405
Korotkova, N., Freire, D., Phan, T. H., Ummels, R., Creekmore, C. C., Evans, T. J., et al. (2014). Structure of the Mycobacterium tuberculosis type VII secretion system chaperone EspG5 in complex with PE25-PPE41 dimer. Mol. Microbiol. 94, 367–382. doi: 10.1111/mmi.12770
Kundu, P., Dutta, D., and Kumar Das, A. (2017). The alpha1beta1 region is crucial for biofilm enhancement activity of MTC28 in Mycobacterium smegmatis. FEBS Lett. 591, 3333–3347. doi: 10.1002/1873-3468.12823
Larsen, M. H., Biermann, K., Tandberg, S., Hsu, T., and Jacobs, W. R. Jr. (2007). Genetic manipulation of Mycobacterium tuberculosis. Curr. Protoc. Microbiol. 6:Chapter 10:Unit 10A.2. doi: 10.1002/9780471729259.mc10a02s6
Lee, S. Y., Kim, H. Y., Kim, B. J., Kim, H., Seok, S. H., Kim, B. J., et al. (2017). Effect of amikacin on cell wall glycopeptidolipid synthesis in Mycobacterium abscessus. J. Microbiol. 55, 640–647. doi: 10.1007/s12275-017-6503-7
Lin, T. L., Shun, C. T., Chang, K. C., and Wang, J. T. (2006). Isolation and characterization of a competence operon associated with transformation and adhesion in Helicobacter pylori. Microbes Infect. 8, 2756–2765. doi: 10.1016/j.micinf.2006.08.006
Martínez, A. N., Torello, S., and Kolter, R. (1999). Sliding motility in Mycobacteria. J. Bacteriol. 181, 7331–7338.
Maya-Hoyos, M., Leguizamon, J., Marino-Ramirez, L., and Soto, C. Y. (2015). Sliding motility, biofilm formation, and glycopeptidolipid production in Mycobacterium colombiense strains. Biomed. Res. Int. 2015:419549. doi: 10.1155/2015/419549
McArdle, A., Nolan, M., Fleming, C., Tuite, H., Markham, T., and Kelly, J. L. (2016). Mycobacterium marinum hand infection. Br. J. Plast. Surg. 86, 1066–1067.
McLaughlin, B., Chon, J. S., Macgurn, J. A., Carlsson, F., Cheng, T. L., Cox, J. S., et al. (2007). A mycobacterium ESX-1-secreted virulence factor with unique requirements for export. PLoS Pathog. 3:e105. doi: 10.1371/journal.ppat.0030105
Meijer, A. H. (2015). Protection and pathology in TB: learning from the zebrafish model. Semin. Immunopathol. 38, 261–273. doi: 10.1007/s00281-015-0522-4
Miyamoto, Y., Mukai, T., Takeshita, F., Nakata, N., Maeda, Y., Kai, M., et al. (2004). Aggregation of mycobacteria caused by disruption of fibronectin-attachment protein-encoding gene. FEMS Microbiol. Lett. 236, 227–234. doi: 10.1111/j.1574-6968.2004.tb09651.x
Mohandas, P., Budell, W. C., Mueller, E., Au, A., Bythrow, G. V., and Quadri, L. E. (2016). Pleiotropic consequences of gene knockouts in the phthiocerol dimycocerosate and phenolic glycolipid biosynthetic gene cluster of the opportunistic human pathogen Mycobacterium marinum. FEMS Microbiol. Lett. 363:fnw016. doi: 10.1093/femsle/fnw016
Nessar, R., Reyrat, J. M., Davidson, L. B., and Byrd, T. F. (2011). Deletion of the mmpL4b gene in the Mycobacterium abscessus glycopeptidolipid biosynthetic pathway results in loss of surface colonization capability, but enhanced ability to replicate in human macrophages and stimulate their innate immune response. Microbiology 157, 1187–1195. doi: 10.1099/mic.0.046557-0
Oh, H. W., Youn, S. H., Kim, M. S., Na, C. H., Jang, S. J., Kim, C. K., et al. (2015). Mycobacterium marinum infection on the face diagnosed by polymerase chain reaction amplification and direct sequencing. Ann. Dermatol. 27, 639–641. doi: 10.5021/ad.2015.27.5.639
Ojha, A., Anand, M., Bhatt, A., Kremer, L., Jacobs, W. R. Jr., and Hatfull, G. F. (2005). GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123, 861–873. doi: 10.1016/j.cell.2005.09.012
Pang, J. M., Layre, E., Sweet, L., Sherrid, A., Moody, D. B., Ojha, A., et al. (2012). The polyketide Pks1 contributes to biofilm formation in Mycobacterium tuberculosis. J. Bacteriol. 194, 715–721. doi: 10.1128/JB.06304-11
Pang, L., Tian, X., Pan, W., and Xie, J. (2013). Structure and function of mycobacterium glycopeptidolipids from comparative genomics perspective. J. Cell Biochem. 114, 1705–1713. doi: 10.1002/jcb.24515
Parish, T., and Stoker, N. G. (2000). Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146(Pt 8), 1969–1975. doi: 10.1099/00221287-146-8-1969
Recht, J., and Kolter, R. (2001). Glycopeptidolipid acetylation affects sliding motility and biofilm formation in Mycobacterium smegmatis. J. Bacteriol. 183, 5718–5724. doi: 10.1128/JB.183.19.5718-5724.2001
Ren, H., Dover, L. G., Islam, S. T., Alexander, D. C., Chen, J. M., Besra, G. S., et al. (2007). Identification of the lipooligosaccharide biosynthetic gene cluster from Mycobacterium marinum. Mol. Microbiol. 63, 1345–1359. doi: 10.1111/j.1365-2958.2007.05603.x
Riera, J., Conesa, X., Pisa, J., Moreno, J., Siles, E., and Novell, J. (2015). Septic arthritis caused by Mycobacterium marinum. Arch. Orthop. Trauma Surg. 136, 131–134. doi: 10.1007/s00402-015-2358-8
Rombouts, Y., Elass, E., Biot, C., Maes, E., Coddeville, B., Burguiere, A., et al. (2010). Structural analysis of an unusual bioactive N-acylated lipo-oligosaccharide LOS-IV in Mycobacterium marinum. J. Am. Chem. Soc. 132, 16073–16084. doi: 10.1021/ja105807s
Rybniker, J., Wolke, M., Haefs, C., and Plum, G. (2003). Transposition of Tn5367 in Mycobacterium marinum, using a conditionally recombinant mycobacteriophage. J. Bacteriol. 185, 1745–1748. doi: 10.1128/JB.185.5.1745-1748.2003
Sani, M., Houben, E. N., Geurtsen, J., Pierson, J., De Punder, K., Van Zon, M., et al. (2010). Direct visualization by cryo-EM of the mycobacterial capsular layer: a labile structure containing ESX-1-secreted proteins. PLoS Pathog. 6:e1000794. doi: 10.1371/journal.ppat.1000794
Schorey, J. S., and Sweet, L. (2008). The mycobacterial glycopeptidolipids: structure, function, and their role in pathogenesis. Glycobiology 18, 832–841. doi: 10.1093/glycob/cwn076
Sette, C. S., Wachholz, P. A., Masuda, P. Y., Da Costa Figueira, R. B., De Oliveira Mattar, F. R., et al. (2015). Mycobacterium marinum infection: a case report. J. Venom. Anim. Toxins Incl. Trop. Dis. 21:7. doi: 10.1186/s40409-015-0008-9
Shi, T., Fu, T., and Xie, J. (2011). Polyphosphate deficiency affects the sliding motility and biofilm formation of Mycobacterium smegmatis. Curr. Microbiol. 63, 470–476. doi: 10.1007/s00284-011-0004-4
Shin, S. J., Wu, C. W., Steinberg, H., and Talaat, A. M. (2006). Identification of novel virulence determinants in Mycobacterium paratuberculosis by screening a library of insertional mutants. Infect. Immun. 74, 3825–3833. doi: 10.1128/IAI.01742-05
Solomon, J. M., Leung, G. S., and Isberg, R. R. (2003). Intracellular replication of Mycobacterium marinum within Dictyostelium discoideum: efficient replication in the absence of host coronin. Infect. Immun. 71, 3578–3586. doi: 10.1128/IAI.71.6.3578-3586.2003
Solomonson, M., Setiaputra, D., Makepeace, K. A. T., Lameignere, E., Petrotchenko, E. V., Conrady, D. G., et al. (2015). Structure of EspB from the ESX-1 type VII secretion system and insights into its export mechanism. Structure 23, 571–583. doi: 10.1016/j.str.2015.01.002
Sousa, S., Bandeira, M., Carvalho, P. A., Duarte, A., and Jordao, L. (2015). Nontuberculous mycobacteria pathogenesis and biofilm assembly. Int. J. Mycobacteriol. 4, 36–43. doi: 10.1016/j.ijmyco.2014.11.065
Steinhauer, K., Eschenbacher, I., Radischat, N., Detsch, C., Niederweis, M., and Goroncy-Bermes, P. (2010). Rapid evaluation of the mycobactericidal efficacy of disinfectants in the quantitative carrier test EN 14563 by using fluorescent Mycobacterium terrae. Appl. Environ. Microbiol. 76, 546–554. doi: 10.1128/AEM.01660-09
Tan, T., Lee, W. L., Alexander, D. C., Grinstein, S., and Liu, J. (2006). The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol. 8, 1417–1429. doi: 10.1111/j.1462-5822.2006.00721.x
Tatham, E., Sundaram Chavadi, S., Mohandas, P., Edupuganti, U. R., Angala, S. K., Chatterjee, D., et al. (2012). Production of mycobacterial cell wall glycopeptidolipids requires a member of the MbtH-like protein family. BMC Microbiol. 12:118. doi: 10.1186/1471-2180-12-118
Toyofuku, M., Tashiro, Y., Hasegawa, Y., Kurosawa, M., and Nomura, N. (2015). Bacterial membrane vesicles, an overlooked environmental colloid: biology, environmental perspectives and applications. Adv. Colloid Interface Sci. 226, 65–77. doi: 10.1016/j.cis.2015.08.013
Trivedi, A., Mavi, P. S., Bhatt, D., and Kumar, A. (2016). Thiol reductive stress induces cellulose-anchored biofilm formation in Mycobacterium tuberculosis. Nat. Commun. 7:11392. doi: 10.1038/ncomms11392
Unnikrishnan, M., Constantinidou, C., Palmer, T., and Pallen, M. J. (2017). The enigmatic Esx proteins: looking beyond mycobacteria. Trends Microbiol. 25, 192–204. doi: 10.1016/j.tim.2016.11.004
van der Woude, A. D., Sarkar, D., Bhatt, A., Sparrius, M., Raadsen, S. A., Boon, L., et al. (2012). Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. J. Biol. Chem. 287, 20417–20429. doi: 10.1074/jbc.M111.336461
Wei, L., and Cao, X. (2016). The effect of transposable elements on phenotypic variation: insights from plants to humans. Sci. China Life Sci. 59, 24–37. doi: 10.1007/s11427-015-4993-2
Wong, K. W. (2017). The role of ESX-1 in Mycobacterium tuberculosis pathogenesis. Microbiol. Spectr. 5:3. doi: 10.1128/microbiolspec.TBTB2-0001-2015
Keywords: type VII secretion system, Mycobacterium marinum, ESX-1, sliding motility, biofilm formation
Citation: Lai L-Y, Lin T-L, Chen Y-Y, Hsieh P-F and Wang J-T (2018) Role of the Mycobacterium marinum ESX-1 Secretion System in Sliding Motility and Biofilm Formation. Front. Microbiol. 9:1160. doi: 10.3389/fmicb.2018.01160
Received: 23 February 2018; Accepted: 14 May 2018;
Published: 30 May 2018.
Edited by:
George Grant, University of Aberdeen, United KingdomReviewed by:
Siouxsie Wiles, University of Auckland, New ZealandMeera Unnikrishnan, University of Warwick, United Kingdom
Copyright © 2018 Lai, Lin, Chen, Hsieh and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Town Wang, d2FuZ2p0QG50dS5lZHUudHc=
 Li-Yin Lai1
Li-Yin Lai1 Jin-Town Wang
Jin-Town Wang