- 1Westerdijk Fungal Biodiversity Institute, Utrecht, Netherlands
- 2Department of Dermatology, Shanghai Changzheng Hospital, Second Military Medical University, Shanghai, China
- 3Shanghai Key Laboratory of Molecular Medical Mycology, Shanghai Institute of Medical Mycology, Shanghai Changzheng Hospital, Second Military Medical University, Shanghai, China
- 4Department of Medical Mycology/Invasive Fungi Research Center, School of Medicine, Mazandaran University of Medical Sciences, Sari, Iran
- 5Student Research Committee, Mazandaran University of Medical Sciences, Sari, Iran
- 6Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, Netherlands
Candida auris, C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii are closely related and highly multidrug resistant yeast pathogens. The high cost and low accuracy of current diagnostics may underestimate their prevalence, especially in medical resource-limited regions. In this study, we used 172 C. auris stains and its relatives and 192 other fungal strains to establish and validate a novel multiplex end-point PCR. A prospective and a retrospective clinical screenings using this assay were further performed in China and Iran respectively. We identified the first isolate of C. pseudohaemulonii in China and the first isolate of C. haemulonii in Iran from 821 clinical isolates in total, without any false positive. Animal models of C. auris and C. haemulonii were established for validation. The overall positive rates of the assay for mice blood and tissue were 28.6 and 92.9%, respectively. Compared with previously developed assays, our assay is more available and affordable to the developing countries, and may contribute to a better understanding of the epidemiology of C. auris and its relatives in these regions.
Introduction
The evolving epidemiology of non-albicans Candida species and other emerging opportunistic pathogenic yeasts contributes to an increasing morbidity and mortality globally (Miceli et al., 2011; Fang et al., 2017). Among them, Candida auris and its medically important relatives in the Metschnikowiaceae clade, e.g., C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii, are well known as highly multidrug resistant pathogens that can lead to both superficial and deep-seated infections (Cendejas-Bueno et al., 2012). C. auris appears to be the most notable species at present due to its outbreak potential and high mortality rate (30–50%) (Calvo et al., 2016; de Almeida et al., 2016). After the first isolate from the external ear canal of a Japanese inpatient in 2009, a number of severe C. auris cases occurred subsequently in 17 countries from five continents (Chowdhary et al., 2017; Mohsin et al., 2017; Arauz et al., 2018). Meanwhile, other phylogenetic related species (C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii) share similar characteristics of C. auris and are also emerging worldwide (Sugita et al., 2006; Ramos et al., 2015; Boatto et al., 2016; de Almeida et al., 2016; Fang et al., 2016; Kumar et al., 2016).
Besides the antifungal profile and outbreak characteristics, clinical intervention for C. auris, C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii infections are mainly challenged by diagnostic difficulties. Firstly, the most commonly used identification systems, e.g., VITEK and API-20C AUX, are time-consuming and tend to misidentify C. auris as other yeasts, such as C. haemulonii, C. sake, C. famata, Rhodotorula glutinis, Saccharomyces cerevisiae (Chowdhary et al., 2017). Secondly, more specific diagnostic methods (e.g., sequence-, RT-PCR- and MALDI-TOF MS- based) require high-cost equipment and trained technicians (Girard et al., 2016; Kordalewska et al., 2017). Thirdly, although there are some in-house developed detective systems, such as the updated MALDI-TOF MS library for C. auris, there is no FDA approved one that can be used in the clinic till now (Mizusawa et al., 2017).
The high cost, low accuracy and unavailability of current diagnostics tools may underestimate the global prevalence of these pathogens, especially in medically resource-limited regions (such as the Africa and Southeast Asia). Consequently, this may lead to the fact that nearly all the reported cases were from developed countries (such as United States, United Kingdom, and Germany) or medically developed cities in the low-income regions (such as New Delhi, India) (Kumar et al., 2016; Chowdhary et al., 2017; Rudramurthy et al., 2017).
Herein, the aim of our study is to establish a multiplex assay that can identify C. auris, C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii. Simplicity and affordability of our assay allow the researchers in developing countries to exploit our multiplex PCR assay as a robust screening tool.
Materials and Methods
Fungal Isolates
Reference strains for the tetraplex PCR validation comprised C. auris (n = 138), C. haemulonii (n = 26), C. duobushaemulonii (n = 6), and C. pseudohaemulonii (n = 2) (Table 1). Human genomic DNA (Sigma–Aldrich, St. Louis, MO, United States) and 192 other fungal strains ranging from the closest to distant related species were used for specificity testing (Table 2). All the samples were obtained from the collection department of Westerdijk Fungal Biodiversity Institute in the form of freeze-dried powder. Subsequently the strains were incubated on Glucose Yeast Extract Peptone Agar plates at 25°C for 48 h, and subsequently pure colonies were confirmed by MALDI-TOF MS (Bruker, Billerica, MA, United States) and LSU sequencing (Vlek et al., 2014; Stielow et al., 2015).
DNA Sample Preparation
DNA extraction from pure colonies of yeast cells was performed using the CTAB method as previously described (Gupta et al., 2004). DNA was purified from blood and tissue (kidney) samples using the animal tissue and blood DNA isolation kit (DENAzist Asia, Mashhad, Iran) according to the manufacturer’s instructions.
Primer Design and PCR Amplification
The 26s rDNA sequence were obtained from our own in-house database or from NCBI Nucleotide Database1 and searched for species-specific and universal regions for primer design. Kordalewska et al. (2017) classified 47 C. auris isolates from India, Pakistan, Venezuela, Japan, and South Africa into four clades using Genome-wide SNP-based phylogenetic analyses, which indicated there is high genetic heterogeneity within C. auris globally. Considering this and also the large number of new cases after 2016, we designed the primers for C. auris by analyzing more isolates from a wider range. For the reverse specific primer for C. auris, we analyzed the 26S rDNA sequence of 233 isolates from Kuwait, Japan, Korea, Pakistan, South Africa, Venezuela, India, Malaysia, Israel, United States, and Oman. 137 sequences were done by ourselves and the rest were downloaded from Genbank. By alignment, genetic heterogeneity was easily found in 26S rDNA. Accordingly, a region which is stable within C. auris, and specific to other species was chosen for reverse primer. The primer system used in this assay were shown in Figure 1, which contained one universal forward primer (Uni-F: 5′-GAACGCACATTGCGCCTTGG-3′) and four species-specific primers (C. auris: Au-R, 5′-TCCAAAGGACTTGCCTGCT-3′; C. duobushaemulonii: Du-R, 5′-GTAGACTTCGCTGCGGATATGTTA-3′; C. haemulonii: Ha-R, 5′-ATTGCGCCAGCATCCTTATTG-3′; C. pseudohaemulonii: Ps-R, 5′-GCACCCGATGCTGACAGTCTAC-3′). Primers were synthesized by Integrated DNA Technologies, Coralville, IA, United States.
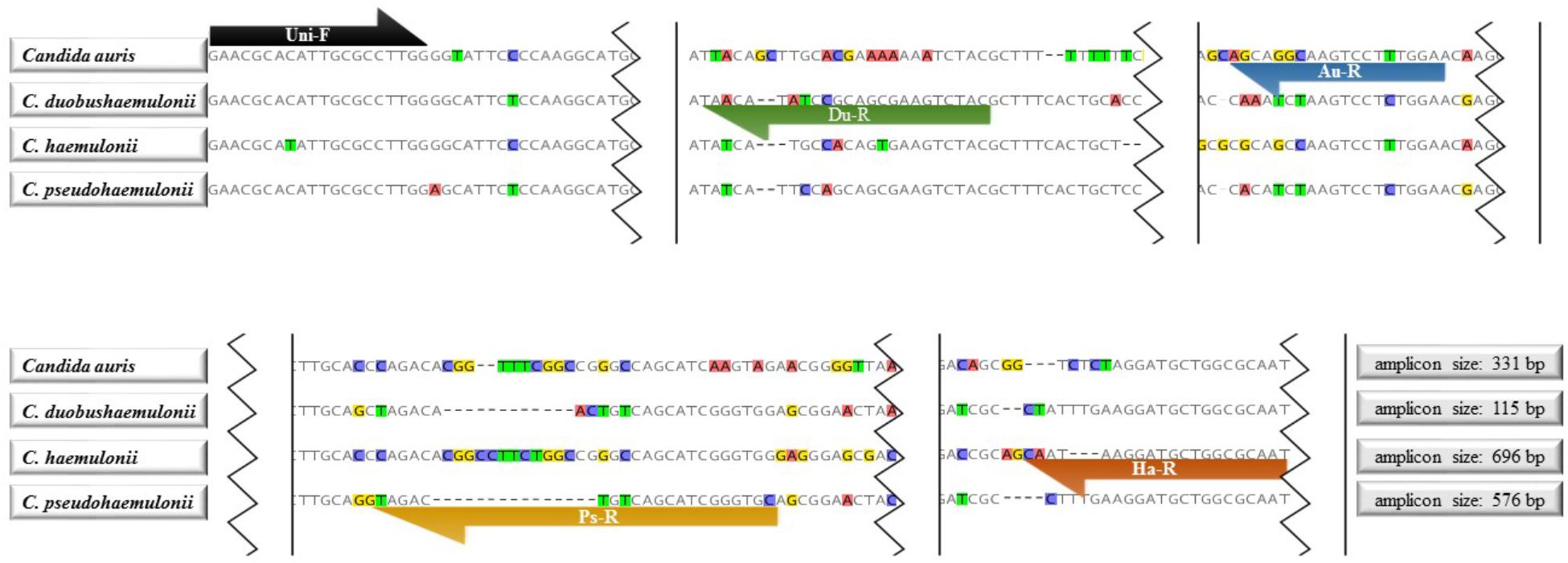
FIGURE 1. Primer sequences and positions used in the tetraplex PCR. Uni-F: Universal forward primer for Candida auris, C. duobushaemulonii, C. haemulonii, and C. pseudohaemulonii; Du-R: C. duobushaemulonii specific reverse primer; Au-R: C. auris specific reverse primer; Ps-R: C. pseudohaemulonii specific reverse primer; Ha-R: C. haemulonii specific reverse primer.
A total volume of 50 μl PCR mixture containing 5 μl of buffer, 1.5 mM magnesium chloride, 2.5 units of Taq polymerase enzyme (BIO-21040, BioLine Company, London, United Kingdom), 0.2 mM dNTP (BIO-39043, BioLine Company, London, United Kingdom), 5 pM Uni-F primer, 5 pM Du-R, 2 pM Ps-R, 3 pM Au-R, and 10 pM Ha-R, 1 μl template DNA or single colony pick (≈1 mm3), and 38.3 μl Mili-Q water (Millipore Corporation, Billerica, MA, United States) was used for amplification of DNA from target species.
The PCR was performed using a 2720 thermal cycler (Applied Biosystems, Waltham, MA, United States), and the PCR program consisted of 5 min pre-denaturation at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 62°C, 30 s at 72°C and 8 min at 72°C as final extension.
Validation via Animal Model
When certain infected human samples are not easily available for diagnostic validation, animal model can be an alternative (Bialek et al., 2001). To evaluate its potential use for infected human blood or tissue, animal models infected by the most important pathogens in Metschnikowiaceae clade, i.e., C. auris and C. haemulonii, were developed for diagnostic purpose. All procedures related to animal experiments were approved by the ethics and research committee of Mazandaran University of Medical Sciences, Sari, Iran.
Candida auris (CBS 10913, type strain), C. haemulonii (CBS 5149, type strains) and C. albicans (ATCC 29008, control) were cultured twice at 35°C for 48 h on Sabouraud dextrose agar (SDA, Difco). All isolates were subcultured in brain heart infusion broth and incubated at 37°C with shaking at 150 rpm. Supernatants were carefully removed and washed twice in sterile phosphate buffered saline (PBS). After centrifugation, the cells were washed in PBS and a hemocytometer was used to count the yeast cell. The yeast cell concentration was adjusted to an inoculum size of 5 × 105 yeast/ml. The viable counts of isolates were confirmed by 10-fold serially diluting the cell suspension on SDA plates.
Immunocompetent female ICR (CD-1 specific pathogen- free) mice with a mean weight of 22 g (purchased from Royan Institute, Tehran, Iran) were used in the study. Animals were housed at the accredited Animal Experimentation Facility in standard cages, received sterilized food and were monitored daily (Fakhim et al., 2018). Seven mice were allocated into each of the three mice groups infected with C. auris. C. haemulonii and C. albicans. Infection was induced in the mice for each group with an inoculum of 5 × 105 CFU/mouse in a volume of 0.2 ml into the lateral tail vein. No immunosuppressive scheme was used. Mice were checked daily and euthanized when symptoms of disseminated infection were detected. Any animal that had more than one of the criteria including, decreased activity, hunched posture, torticollis or barrel rolling, inability to eat or drink and hypothermia was humanly euthanized by intracardiac puncture under general anesthesia (Conti et al., 2014). Tissue sections were also stained with hematoxylin and eosin (H&E) for microscopic examination. Tissue (kidney) and blood samples were recovered under aseptic conditions. Tissue samples were homogenized in sterile saline and 100 μl of homogenate was cultured on SDA at 35°C to confirm Candida infection and determine the colony forming units (CFU) were determine (Fakhim et al., 2018). All yeast cells cultures were identified by our tetraplex PCR. In addition, 100 μl of each EDTA-blood sample was cultured on SDA plates. The rest of the homogenate and blood samples were used for tetraplex PCR test.
Application in Clinical Setting
A single-center prospective clinical screening for C. auris, C. duobushaemulonii, C. haemulonii and C. pseudohaemulonii was performed during 2017.05.01 to 2017.11.01 in Shanghai Changzheng Hospital, China. The newly developed multiplex PCR system worked as a supplementary tool to BD BACTECTM FX40 Instrument (Becton, Dickinson and Company, Franklin Lakes, NJ, United States), CHROMagarTM Candida (CHROMagar Microbiology, Paris, France) and microscopic examination to finding infection cases by the targeted species. In order to further test the specificity of our assay using more clinical isolates, we used a wide range of sample types for screening, including blood, sputum, feces, bronchoalveolar lavage fluid and oral swab. We test all the clinical yeast isolates that are identified by CHROMagarTM Candida as non-albicans Candida species. As soon as the clinical culture is obtained by conventional diagnostic methods, single colony (≈1 mm3) is tested by this multiplex PCR system. DNA extraction and ITS sequencing are done as well to confirm the accuracy of the assay in the clinical settings as described before (Stielow et al., 2015).
Two hundred and fifty five clinical non-albicans Candida isolates (identified by CHROMagarTM Candida) from Mazandaran University of Medical Sciences, Iran, were used as a retrospective screening sample in this study. The isolates were collected from blood, sputum, feces, oral swab and vaginal samples during 2016.1–2017.6. Multiplex PCR were performed, and MALDI-TOF and LSU sequencing were done as well to confirm the accuracy of the assay in the clinical settings (Vlek et al., 2014; Stielow et al., 2015).
All the tests were approved by the ethics and research committees of Shanghai Changzheng Hospital and Mazandaran University of Medical Sciences.
Results
Analytical Validation
Figure 2 showed that four amplicons exhibited major bands that could be differentiated from each other. The gel figure for all the strains of C. auris and its relatives tested in this study was shown in Supplementary Figure S1. Sequencing results of PCR amplicons were 100% matched with the targeted species. Human genomic DNA and 192 other fungal species were tested, and no cross-reactivity was found (100% specificity).
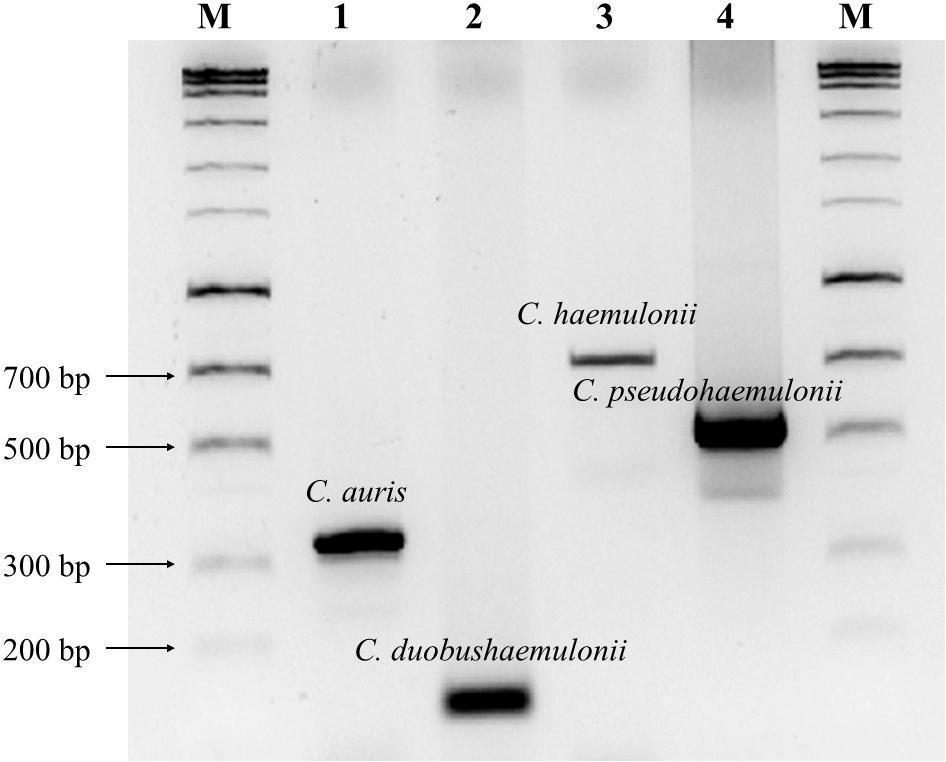
FIGURE 2. Agarose gel electrophoresis (2% agarose) of PCR amplicons. Lanes 1–4 are C. auris (CBS 10913 type, 331 bp), C. duobushaemulonii (CBS 7798 type, 115 bp), C. haemulonii (CBS 5149 type, 696 bp) and C. pseudohaemulonii (CBS 10004 type, 576 bp), respectively. Lane M, HyperLadderTM 100 bp (Bioline, London, United Kingdom).
Animal Model Testing
Figure 3 showed the detailed information of the animal model testing.
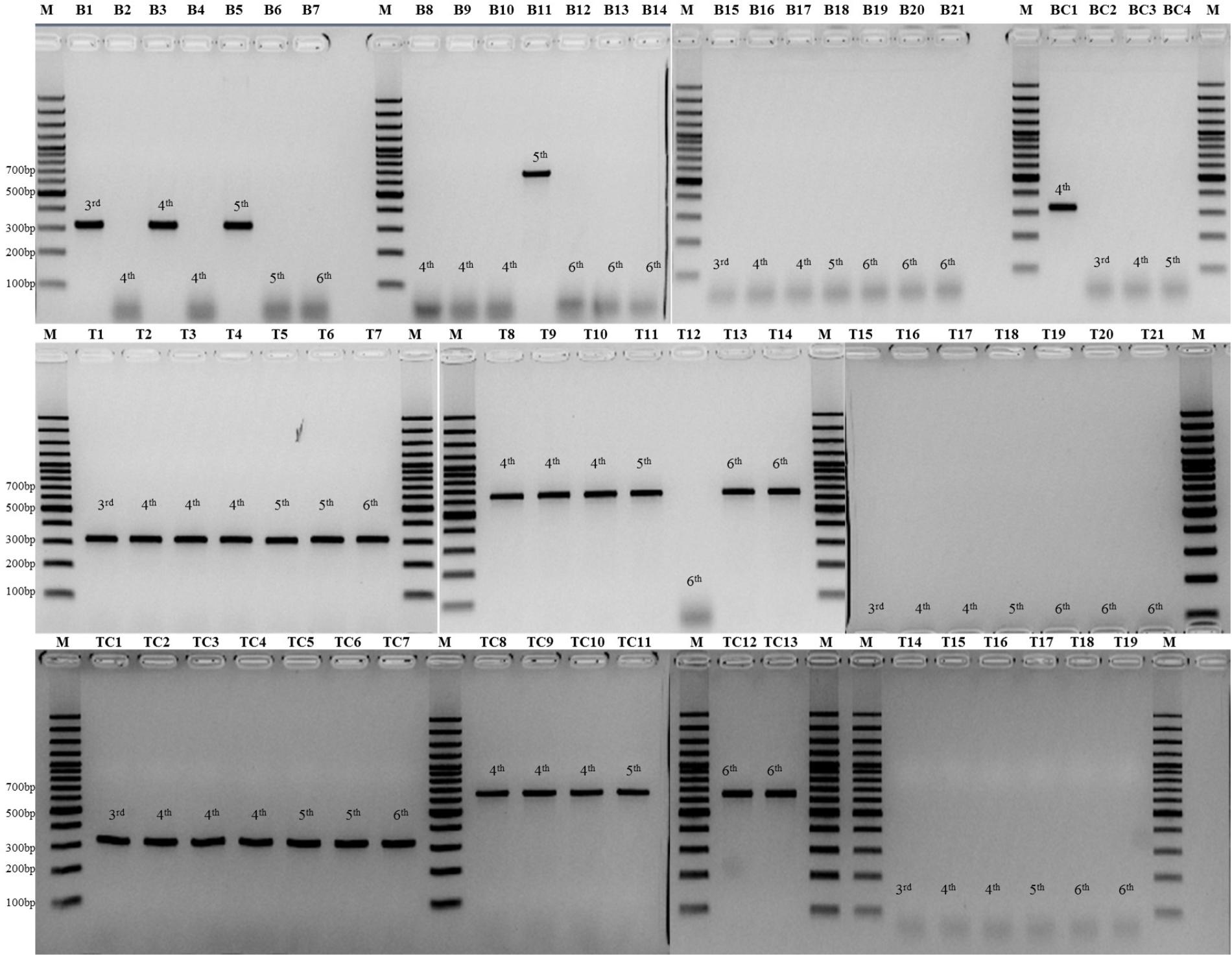
FIGURE 3. Lanes B1–B7 are PCR results of blood sample from seven mice infected with C. auris; Lanes B8-B14 are PCR results of blood sample from seven mice infected with C. haemulonii; Lanes B15-B21 are PCR results of blood sample from seven mice infected with C. albicans; Lane BC1–4 are PCR results of four cultured strains from blood samples of all the 21 mice; Lanes T1–T7 are PCR results of kidney tissue sample from seven mice infected with C. auris; Lanes T8–T14 are PCR results of kidney tissue sample from seven mice infected with C. haemulonii; Lanes T15–T21 are PCR results of kidney tissue sample from seven mice infected with C. albicans; Lane TC1–7 are PCR results of seven cultured strains from kidney samples of mice infected with C. auris; Lane TC8–13 are PCR results of six cultured strains from kidney samples of mice infected with C. haemulonii; Lane TC14–19 are PCR results of six cultured strains from kidney samples of mice infected with C. albicans; Lane M, GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher, Waltham, MA, United States).
Application of the tetraplex PCR in blood showed a higher overall positive rate (4 of 14, 28.6%) than blood culture positive rate (1 of 14, 7.1%) in animal models of C. auris and C. haemulonii. Specifically, three C. auris (3/7, on the 3rd, 4th, and 5th day post-infection (PI)) and one C. haemulonii (1/7, on the 5th day, PI) infected mice showed positive PCR results from blood, without cross-reactivity in C. albicans group (0/7). Culture results were positive only in one blood samples of C. auris (1/7, on the 4th day, PI), negative in all C. haemulonii infected mice (0/7), and positive in three C. albicans infected mice (3/7, on the 3rd, 4th, and 5th day PI), with CFUs ranging from 1 to 6 colonies in 100 μl.
The overall positive PCR rate of mice kidney samples was 92.8% (13/14) among C. auris and C. haemulonii, without cross-reactivity in C. albicans group (0/7). Among the kidney samples from 21 mice examined by culture, 19 were positive due to C. auris (7/7), haemulonii (6/7), and C. albicans (6/7) with CFUs ranging from 8.3 × 10 to 4.3 × 104 colonies in 100 μl (mean 3.5 × 103 colonies).
Clinical Validation
Applying our assay in a prospective (566 clinical isolates) and a retrospective cohorts (255 clinical isolates), revealed one C. haemulonii isolate (S49AF, accession number: KY112738) from Iran and one C. haemulonii isolate (SCZ90793, accession number: MG637448), one C. duobushaemulonii isolate (SCZ91445, accession number: MG963993) and one C. pseudohaemulonii isolate (SCZ90800, accession number: MG242063) from China. Specifically, the Chinese C. pseudohaemulonii was isolated from sputum of a 65-year-old male who suffered from lower esophageal resection (Shanghai, China). To our knowledge this the first isolate of C. pseudohaemulonii from China. Whereas the Iranian C. haemulonii isolate was recovered from toenail sample of a 36-year-old diabetic male in 2016 (Shiraz, Iran), and also was the first C. haemulonii isolate from Iran.
Discussion
Achieving a timely and accurate diagnosis of infections caused by C. auris and its phylogenetic relatives is of great difficulty in clinical settings (Mizusawa et al., 2017). Nearly all the C. auris isolates were misdiagnosed as C. haemulonii, C. sake, C. famata, Rhodotorula glutinis, Saccharomyces cerevisiae by clinical commercial methods such as Vitek-2 YST ID system, API-20C AUX, AuxaColor 2, BD Phoenix, and MicroScan (Chowdhary et al., 2017). Studies also showed that C. pseudohaemulonii failed to be distinguished from C. haemulonii by these methods (Kim et al., 2009). All the identifications from the biochemical assays should be confirmed by sequencing. The Centers for Disease Control of America speculated that the unavailability of appropriate diagnostic method underestimate its prevalence in many countries (Sarma and Upadhyay, 2017), which includes medical resource-limited regions. Hence, we developed a novel end-point tetraplex PCR system in a low-cost, rapid and accurate format. This novel assay has been validated by a large number of CBS/clinical isolates, diagnostic animal models and clinical application. We hope this assay can help to understand the global clinical impact of C. auris and its relatives, especially in the under-served and poor regions.
Our new end-point tetraplex PCR was designed to identify and differentiate the most medically important and related species to C. auris. Recently, two real-time PCR based assays for C. auris were developed (Kordalewska et al., 2017; Leach et al., 2017). Compared with the real-time PCR method, our end-point PCR is non-automated and labor-intensive. The electrophoresis step of our assay normally required 45 min, which is more time-consuming than real-time PCRs. Additionally, since the nature of end-point PCR, our assay is unable to quantify the fungal load of host. However, the advantages of our assay over theirs are also obvious.
Kordalewska et al. (2017) established the first SYBR Green real-time PCR system for C. auris, C. duobushaemulonii, C. haemulonii, and C. lusitaniae; While Leach et al. (2017) developed a probe-based singleplex real-time PCR only for C. auris. Considering the high misdiagnostic rate between C. auris, C. haemulonii, C. duobushaemulonii, and C. pseudohaemulonii, and higher resistance of C. pseudohaemulonii to AMB than C. auris, C. haemulonii, C. duobushaemulonii, and C. lusitaniae (Kim et al., 2009; Shin et al., 2012), we used a multiplex strategy for this assay and included C. pseudohaemulonii instead of C. lusitaniae. Among the selected species, C. haemulonii initially was known as a multi-antifungal resistant pathogen since the first case report in 1984, and it can cause outbreaks and infect immunocompetent individuals (Loosova et al., 2001; Khan et al., 2007). Subsequently, C. haemulonii was recognized as a complex species and reclassified as C. haemulonii, C. duobushaemulonii, and C. haemulonii var. vulnera (Cendejas-Bueno et al., 2012). C. auris and C. pseudohaemulonii were described in 2009 and 2006, respectively. Both of them were multi-antifungal resistant pathogens and the latter is spreading across the world at an accelerated pace (Sugita et al., 2006; Chowdhary et al., 2017). Correct species-level identification for the four species is needed due to the clinical importance and heterogeneity in terms of taxonomy, spreading prevalence and antifungal susceptibility profile.
Our multiplex PCR system is the first assay for C. auris that has been validated by animal model testing. The overall PCR positive rates of mice blood and tissue were 28.6 and 92.9%, respectively, which showed slightly higher performance than culture based identification. And PCR/culture from tissue samples showed higher value than those from blood for diagnosing infections caused by these pathogens. Moreover, our new assay is much faster than culture based identification. PCR directly from tissue/blood only requires 3 h from sample preparation to result read-out; while culture based identification is more time-consuming (normally 1–3 days).
Our new assay is also featured by its low cost and potential to be used in medical resource-limited settings. Due to the poor economic condition, lack of basic medical facilities and the relatively poor population health, the developing countries were reported to have a much higher risk of infections than the developed ones (Pittet et al., 2008). Among the 17 countries reported with C. auris prevalence, seven are the developing ones (Oman, Venezuela, Colombia, South Africa, Kenya, Pakistan, and India). Considering this, our new assay was established to help to extend the global knowledge of the epidemiology of these important and emerging pathogens, especially for medical resource-limited countries. Compared with MALDI-TOF and real-time PCR cycler, end-point PCR cycler used in this assay is more portable and much cheaper ($130–$4,000 per cycler) (Wong et al., 2015). For clinical validation, we used isolates from two clinical settings that can represent different situations of medical care in developing countries (Shanghai, China and Sari, Iran). In Shanghai, specific diagnostic tools such as MALDI-TOF and sequencing are available to most hospitals. However, since China is a developing country, there are still a large number of patients that cannot afford these tests. While in Iran, to our knowledge, there is no MALDI-TOF and sequencing machine. There was only one epidemiology study related to C. haemulonii and C. duobushaemulonii in China and no epidemiology data is available in Iran (Hou et al., 2016). Hence, we hold the prevalence of C. auris and its close related relatives are underestimated due to the lack of low-cost and accurate assays in both countries. By using this assay, we successfully found the first clinical isolates of C. haemulonii in Iran and first clinical isolates of C. pseudohaemulonii in China. However, no C. auris isolate was found in both cohorts.
Conclusion
The novel end-point tetraplex PCR for C. auris and its phylogenetic relatives is rapid and 100% specific. Clinical validation and animal model testing further proved its clinical performance. This assay only requires the simple devices of end-point PCR cycler and electrophoresis instrument, showing its availability and affordability to most of the developing countries, and can contribute to a better understanding of the clinical occurrence of C. auris and its relatives.
Ethics Statement
This study was carried out in accordance with the recommendations of Guide for the Care and Use of Laboratory Animals, committee of the Update of the Guide of the Care and Use of Laboratory Animals. The protocol was approved by the Ethics and Research Committee of Mazandaran University of Medical Sciences, Sari, Iran (No. 2321).
Author Contributions
AA and WF participated in primer design, PCR optimization, data collection, and drafted the manuscript. WP, FH, and TB participated in designing this study and revising the manuscript. HB and AV participated in animal model testing and collecting Iranian clinical isolates. WJ participated in clinical validation in China. All authors contributed to the writing of the final manuscript.
Funding
This work was supported by the Major National R&D Projects of the National Health Department (2018ZX10101003), European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No. 642095, National Natural Science Foundation of China (31770161), Shanghai Science and Technology Committee (Grant Nos. 14DZ2272900 and 14495800500).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01119/full#supplementary-material
FIGURE S1 | Gel figure for all the strains of C. auris and its relatives tested in this study.
Footnotes
References
Araúz, A. B., Caceres, D. H., Santiago, E., Armstrong, P., Arosemena, S., Ramos, C., et al. (2018). Isolation of Candida auris from nine patients in Central America: importance of accurate diagnosis and susceptibility testing. Mycoses 61, 44–47. doi: 10.1111/myc.12709
Bialek, R., Fischer, J., Feucht, A., Najvar, L. K., Dietz, K., Knobloch, J., et al. (2001). Diagnosis and monitoring of murine histoplasmosis by a nested PCR assay. J. Clin. Microbiol. 39, 1506–1509. doi: 10.1128/jcm.39.4.1506-1509.2001
Boatto, H. F., Cavalcanti, S. D., Del Negro, G. M., Girao, M. J., Francisco, E. C., Ishida, K., et al. (2016). Candida duobushaemulonii: an emerging rare pathogenic yeast isolated from recurrent vulvovaginal candidiasis in Brazil. Mem. Inst. Oswaldo Cruz 111, 407–410. doi: 10.1590/0074-02760160166
Calvo, B., Melo, A. S., Perozo-Mena, A., Hernandez, M., Francisco, E. C., Hagen, F., et al. (2016). First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J. Infect. 73, 369–374. doi: 10.1016/j.jinf.2016.07.008
Cendejas-Bueno, E., Kolecka, A., Alastruey-Izquierdo, A., Theelen, B., Groenewald, M., Kostrzewa, M., et al. (2012). Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 50, 3641–3651. doi: 10.1128/JCM.02248-12
Chowdhary, A., Sharma, C., and Meis, J. F. (2017). Candida auris: a rapidly emerging cause of hospital-acquired multidrug-resistant fungal infections globally. PLoS Pathog. 13:e1006290. doi: 10.1371/journal.ppat.1006290
Conti, H. R., Huppler, A. R., Whibley, N., and Gaffen, S. L. (2014). Animal models for candidiasis. Curr. Protoc. Immunol. 105, 11–17. doi: 10.1002/0471142735.im1906s105
de Almeida, J. N. Jr., Assy, J. G., Levin, A. S., Del Negro, G. M., Giudice, M. C., Tringoni, M. P., et al. (2016). Candida haemulonii complex species. Emerg. Infect. Dis. 22, 561–563. doi: 10.3201/eid2203.151610
Fakhim, H., Vaezi, A., Dannaoui, E., Chowdhary, A., Nasiry, D., Faeli, L., et al. (2018). Comparative virulence of Candida auris with Candida haemulonii, Candida glabrata and Candida albicans in a murine model. Mycoses 61, 377–382. doi: 10.1111/myc.12754
Fang, S. Y., Wei, K. C., Chen, W. C., Lee, S. J., Yang, K. C., Wu, C. S., et al. (2016). Primary deep cutaneous candidiasis caused by Candida duobushaemulonii in a 68-year-old man: the first case report and literature review. Mycoses doi: 10.1111/myc.12540 [Epub ahead of print].
Fang, W., Zhang, L., Liu, J., Denning, D. W., Hagen, F., Jiang, W., et al. (2017). Tuberculosis/cryptococcosis co-infection in China between 1965 and 2016. Emerg. Microbes Infect. 6:e73. doi: 10.1038/emi.2017.61
Girard, V., Mailler, S., Chetry, M., Vidal, C., Durand, G., van Belkum, A., et al. (2016). Identification and typing of the emerging pathogen Candida auris by matrix-assisted laser desorption/ionisation time of flight mass spectrometry. Mycoses 59, 535–538. doi: 10.1111/myc.12519
Gupta, A. K., Boekhout, T., Theelen, B., Summerbell, R., and Batra, R. (2004). Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J. Clin. Microbiol. 42, 4253–4260. doi: 10.1128/JCM.42.9.4253-4260.2004
Hou, X., Xiao, M., Chen, S. C., Wang, H., Cheng, J. W., Chen, X. X., et al. (2016). Identification and antifungal susceptibility profiles of Candida haemulonii species complex clinical isolates from a multicenter study in China. J. Clin. Microbiol. 54, 2676–2680. doi: 10.1128/jcm.01492-16
Khan, Z. U., Al-Sweih, N. A., Ahmad, S., Al-Kazemi, N., Khan, S., Joseph, L., et al. (2007). Outbreak of fungemia among neonates caused by Candida haemulonii resistant to amphotericin B, itraconazole, and fluconazole. J. Clin. Microbiol. 45, 2025–2027. doi: 10.1128/JCM.00222-07
Kim, M. N., Shin, J. H., Sung, H., Lee, K., Kim, E. C., Ryoo, N., et al. (2009). Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin. Infect. Dis. 48, e57–e61. doi: 10.1086/597108
Kordalewska, M., Zhao, Y., Lockhart, S. R., Chowdhary, A., Berrio, I., and Perlin, D. S. (2017). Rapid and accurate molecular identification of the emerging multidrug resistant pathogen Candida auris. J. Clin. Microbiol. 55, 2445–2452. doi: 10.1128/JCM.00630-17
Kumar, A., Prakash, A., Singh, A., Kumar, H., Hagen, F., Meis, J. F., et al. (2016). Candida haemulonii species complex: an emerging species in India and its genetic diversity assessed with multilocus sequence and amplified fragment-length polymorphism analyses. Emerg. Microbes Infect. 5:e49. doi: 10.1038/emi.2016.49
Leach, L., Zhu, Y., and Chaturvedi, S. (2017). Development and validation of a real-time PCR assay for rapid detection of Candida auris from surveillance samples. J. Clin. Microbiol. 56:e01223-17. doi: 10.1128/jcm.01223-17
Loosova, G., Jindrak, L., and Kopacek, P. (2001). Mortality caused by experimental infection with the yeast Candida haemulonii in the adults of Ornithodoros moubata (Acarina: argasidae). Folia Parasitol. 48, 149–153. doi: 10.14411/fp.2001.023
Miceli, M. H., Diaz, J. A., and Lee, S. A. (2011). Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151. doi: 10.1016/S1473-3099(10)70218-8
Mizusawa, M., Miller, H., Green, R., Lee, R., Durante, M., Perkins, R., et al. (2017). Can multidrug-resistant Candida auris be reliably identified in clinical microbiology laboratories? J. Clin. Microbiol. 55, 638–640. doi: 10.1128/jcm.02202-16
Mohsin, J., Hagen, F., Al-Balushi, Z. A. M., de Hoog, G. S., Chowdhary, A., Meis, J. F., et al. (2017). The first cases of Candida auris candidaemia in Oman. Mycoses 60, 569–575. doi: 10.1111/myc.12647
Pittet, D., Allegranzi, B., Storr, J., Bagheri Nejad, S., Dziekan, G., Leotsakos, A., et al. (2008). Infection control as a major world health organization priority for developing countries. J. Hosp. Infect. 68, 285–292. doi: 10.1016/j.jhin.2007.12.013
Ramos, L. S., Figueiredo-Carvalho, M. H., Barbedo, L. S., Ziccardi, M., Chaves, A. L., Zancope-Oliveira, R. M., et al. (2015). Candida haemulonii complex: species identification and antifungal susceptibility profiles of clinical isolates from Brazil. J. Antimicrob. Chemother. 70, 111–115. doi: 10.1093/jac/dku321
Rudramurthy, S. M., Chakrabarti, A., Paul, R. A., Sood, P., Kaur, H., Capoor, M. R., et al. (2017). Candida auris candidaemia in Indian ICUs: analysis of risk factors. J. Antimicrob. Chemother. 72, 1794–1801. doi: 10.1093/jac/dkx034
Sarma, S., and Upadhyay, S. (2017). Current perspective on emergence, diagnosis and drug resistance in Candida auris. Infect. Drug Resist. 10, 155–165. doi: 10.2147/idr.s116229
Shin, J. H., Kim, M. N., Jang, S. J., Ju, M. Y., Kim, S. H., Shin, M. G., et al. (2012). Detection of amphotericin B resistance in Candida haemulonii and closely related species by use of the Etest, Vitek-2 yeast susceptibility system, and CLSI and EUCAST broth microdilution methods. J. Clin. Microbiol. 50, 1852–1855. doi: 10.1128/JCM.06440-11
Stielow, J. B., Levesque, C. A., Seifert, K. A., Meyer, W., Iriny, L., Smits, D., et al. (2015). One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35, 242–263. doi: 10.3767/003158515x689135
Sugita, T., Takashima, M., Poonwan, N., and Mekha, N. (2006). Candida pseudohaemulonii Sp. Nov., an amphotericin B-and azole-resistant yeast species, isolated from the blood of a patient from Thailand. Microbiol. Immunol. 50, 469–473. doi: 10.1111/j.1348-0421.2006.tb03816.x
Vlek, A., Kolecka, A., Khayhan, K., Theelen, B., Groenewald, M., Boel, E., et al. (2014). Interlaboratory comparison of sample preparation methods, database expansions, and cutoff values for identification of yeasts by matrix-assisted laser desorption ionization-time of flight mass spectrometry using a yeast test panel. J. Clin. Microbiol. 52, 3023–3029. doi: 10.1128/JCM.00563-14
Keywords: end-point PCR, Candida auris, multi-drug resistance, animal model, clinical validation, molecular diagnosis
Citation: Arastehfar A, Fang W, Badali H, Vaezi A, Jiang W, Liao W, Pan W, Hagen F and Boekhout T (2018) Low-Cost Tetraplex PCR for the Global Spreading Multi-Drug Resistant Fungus, Candida auris and Its Phylogenetic Relatives. Front. Microbiol. 9:1119. doi: 10.3389/fmicb.2018.01119
Received: 23 February 2018; Accepted: 11 May 2018;
Published: 29 May 2018.
Edited by:
Dominique Sanglard, Université de Lausanne, SwitzerlandReviewed by:
Alix Thérèse Coste, Centre Hospitalier Universitaire Vaudois (CHUV), SwitzerlandGuillermo Quindós, University of the Basque Country (UPV/EHU), Spain
Copyright © 2018 Arastehfar, Fang, Badali, Vaezi, Jiang, Liao, Pan, Hagen and Boekhout. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiwei Jiang, amlhbmd4dWVqaWFuZ3dlaUBxcS5jb20= Weihua Pan, cGFud2VpaHVhQHNtbXUuZWR1LmNu
†These authors have contributed equally to this work.
 Amir Arastehfar
Amir Arastehfar Wenjie Fang
Wenjie Fang Hamid Badali
Hamid Badali Afsane Vaezi5
Afsane Vaezi5 Weiwei Jiang
Weiwei Jiang Wanqing Liao
Wanqing Liao Weihua Pan
Weihua Pan Ferry Hagen
Ferry Hagen