- 1Department of Basic Pathology, Federal University of Paraná, Curitiba, Brazil
- 2Department of Genetics, Federal University of Paraná, Curitiba, Brazil
- 3Department of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, Lexington, KY, United States
- 4Center for Pharmaceutical Research and Innovation, College of Pharmacy, University of Kentucky, Lexington, KY, United States
- 5Department of Biology, Federal University of Mato Grosso do Sul, Campo Grande, Brazil
Endophytic actinomycetes from medicinal plants produce a wide diversity of secondary metabolites (SM). However, to date, the knowledge about endophytes from Brazil remains scarce. Thus, we analyzed the antimicrobial potential of 10 actinomycetes isolated from the medicinal plant Vochysia divergens located in the Pantanal sul-mato-grossense, an unexplored wetland in Brazil. Strains were classified as belonging to the Aeromicrobium, Actinomadura, Microbacterium, Microbispora, Micrococcus, Sphaerisporangium, Streptomyces, and Williamsia genera, through morphological and 16S rRNA phylogenetic analyzes. A susceptibility analysis demonstrated that the strains were largely resistant to the antibiotics oxacillin and nalidixic acid. Additionally, different culture media (SG and R5A), and temperatures (28 and 36°C) were evaluated to select the best culture conditions to produce the active SM. All conditions were analyzed for active metabolites, and the best antibacterial activity was observed from metabolites produced with SG medium at 36°C. The LGMB491 (close related to Aeromicrobium ponti) extract showed the highest activity against methicillin-resistant Staphylococcus aureus (MRSA), with a MIC of 0.04 mg/mL, and it was selected for SM identification. Strain LGMB491 produced 1-acetyl-β-carboline (1), indole-3-carbaldehyde (2), 3-(hydroxyacetyl)-indole (4), brevianamide F (5), and cyclo-(L-Pro-L-Phe) (6) as major compounds with antibacterial activity. In this study, we add to the knowledge about the endophytic community from the medicinal plant V. divergens and report the isolation of rare actinomycetes that produce highly active metabolites.
Introduction
Endophytes are microorganisms that inhabit the internal tissues of plants without causing any negative effects, and actinomycetes isolated from plants have been widely studied due their ability to produce active metabolites (Kim et al., 2000; Zhao et al., 2011; Kadiri et al., 2014; Golinska et al., 2015; Savi et al., 2015a,b). Actinomycetes have been used for drug discovery for more than five decades, producing more than 10,000 bioactive compounds. Of these ~75% are produced by Streptomyces, the by far mostly explored actinomycete genus. The remaining 25% bioactive compounds were isolated from “rare actinomycetes”, i.e., actinomycetes isolated in lower frequency than Streptomyces (Rong and Huang, 2012; Tiwari and Gupta, 2012). Since, the rare actinomycetes are an underexplored group, the use of these organisms, and their compounds have gained great importance in drug discovery programs (Rong and Huang, 2012; Tiwari and Gupta, 2012), mainly to combat infections caused by resistant microorganisms. The widespread use of broad-spectrum antibiotics has created a strong selective pressure, resulting in survival, and spread of resistant bacteria (Davies and Davies, 2010). The increase in bacterial resistance is a major concern for public health (Ventola, 2015). Unfortunately, many pharmaceutical companies have reduced or eliminated their search for new antibiotics, due to economic reasons, exasperating the problem further (Borrero et al., 2014). In order to find microorganisms with potential to produce active metabolites our group has been searching endophytic microorganisms from medicinal plants located in underexplored environments, such as the Brazilian wetland regions (Savi et al., 2015a,b; Hokama et al., 2016; Peña et al., 2016; Santos et al., 2016; Tonial et al., 2017). The Brazilian Pantanal is the largest wetland in the world, and it is characterized by two seasons: flooding and the dry. Hence, the Pantanal has developed a peculiar biological diversity regarding its fauna and flora (Alho, 2008). According to Arieira and Cunha (2006), only 5% of the species of plants of the Pantanal can survive the stress caused by drought and flood periods. Among them is the medicinal plant Vochysia divergens, which is commonly used in form of syrups and teas for the treatment of colds, coughs, fever, pneumonia, and other diseases (Pott et al., 2004). In a study carried out with endophytes from V. divergens, Savi et al. (2015a) identified actinomycetes able to produce highly active metabolites. However, the study was performed with a small number of isolates, and the diversity of V. divergens remained little explored. Thus, the focus of this study is to identify endophytic actinomycetes from the medicinal plant V. divergens and to assay their secondary metabolites, dependent on different culture conditions, against clinical pathogens associated with antibiotic resistance.
Materials and Methods
Sample Collection
V. divergens leaves with no marks or injuries were collected from 21 plants located in the Pantanal sul-mato-grossense/Brazil, specifically in two regions of the Pantanal of Miranda, Abobral (19°30′09.5″S, 57°02′32.2″W) and São Bento (19°28′53.9″S, 57°02′36.9″W).
Isolation of Actinomycetes
The leaves from V. divergens were subjected to surface sterilization according to a protocol described by Petrini (1986). The leaves were fragmented (8 × 8 mm) and deposited on petri dishes containing starch casein agar (SCA) (Mohseni et al., 2013), with nalidixic acid (50 μg/mL) and cycloheximide (50 μg/mL). The plates were incubated at 28°C for 30 days, and were examined daily for the presence of colonies. The actinomycetes isolates were deposited in the Laboratório de Genética de Microrganismos (LabGeM) culture collection, Federal University of Paraná, Brazil (http://www.labgem.ufpr.br/).
Identification
Morphological Analysis
Four different culture media were used to access the macro-morphological characteristics, ISP2—Agar yeast-malt extract; ISP3—Oat Agar; ISP4—Agar Starch and inorganic salts; ISP5—Glycerol Asparagine Agar (Shirling and Gottlieb, 1966). The isolates were streaked on the plates and incubated at 28°C for 21 days. The characteristics evaluated were growth rate, the formation and color of aerial spore mass and substrate mycelia.
Molecular Taxonomy
Total genomic DNA was extracted from 3 day old cultures using the method described by Raeder and Broda (1985). Partial sequence of the 16S rRNA gene was amplified using primers 9F (5′GAGTTTGATCCTGGCTCAG3′) and 1541R (5′AAGGAGGTGATCCAGCC3′), as described by Savi et al. (2016). The PCR product was purified using Exo1 and FastAP enzymes (GE Healthcare, USA), and sequenced using the BigDye® Terminator v3.1 Kit. The products were purified with SephadexG50 and submitted to an ABI3500® automated sequencer (Applied Biosystems, Foster City, CA, USA). Consensus sequences were analyzed and aligned using Mega 6.0 (Tamura et al., 2013) and BioEdit, and compared to sequences available in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/). Type strain sequences were found through search in the List of Prokaryotic Names with Standing Nomenclature database (http://www.bacterio.net/). All sequences obtained were deposited in the GenBank, the accession numbers are listed in Table 1. For Bayesian inference analysis, a Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with posterior probabilities using MrBayesv3.2.6 x86 (Ronquist et al., 2011). GRT evolutionary model was determined using the Akaike Information Criterion (AIC) in R software (R Core Team, 2017) and the phangorn package (Schliep, 2011). Comparisons of sequences with respect to their percentile similarity were estimated using the R software (R Core Team, 2017) and the pegas package (Paradis, 2010).
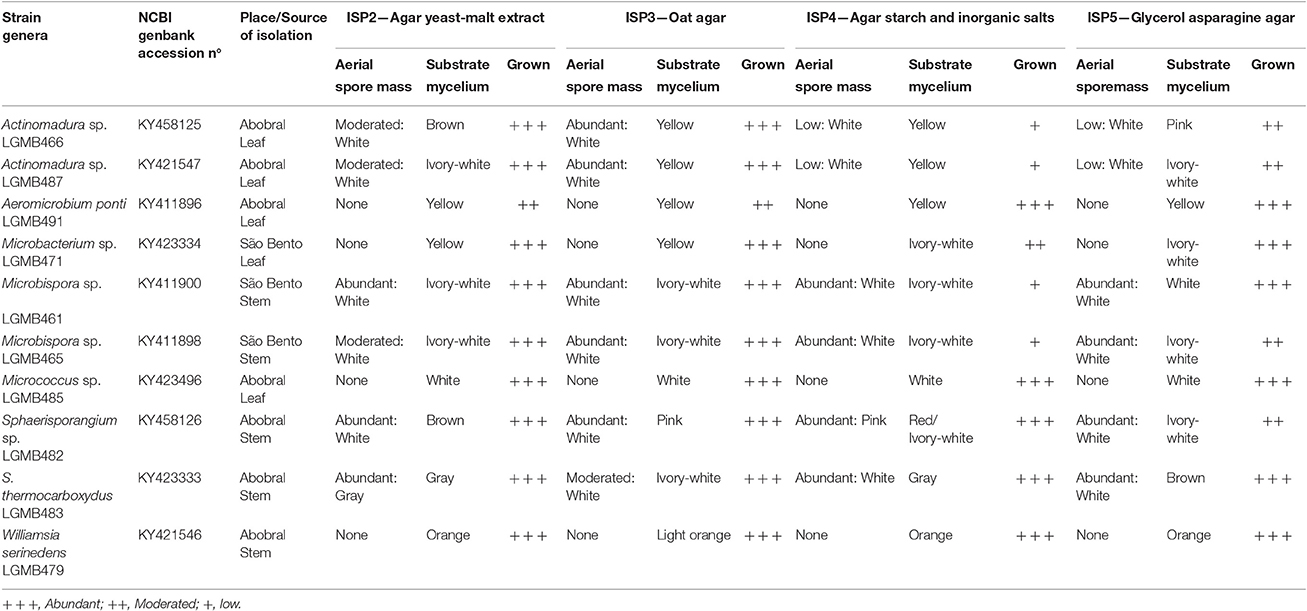
Table 1. Identification, place, and source of isolation and morphological characteristic of endophytic actinomycetes isolates, morphological characteristics 21 days after inoculation in four different culture media at 28°C.
Antibiotic Sensitivity
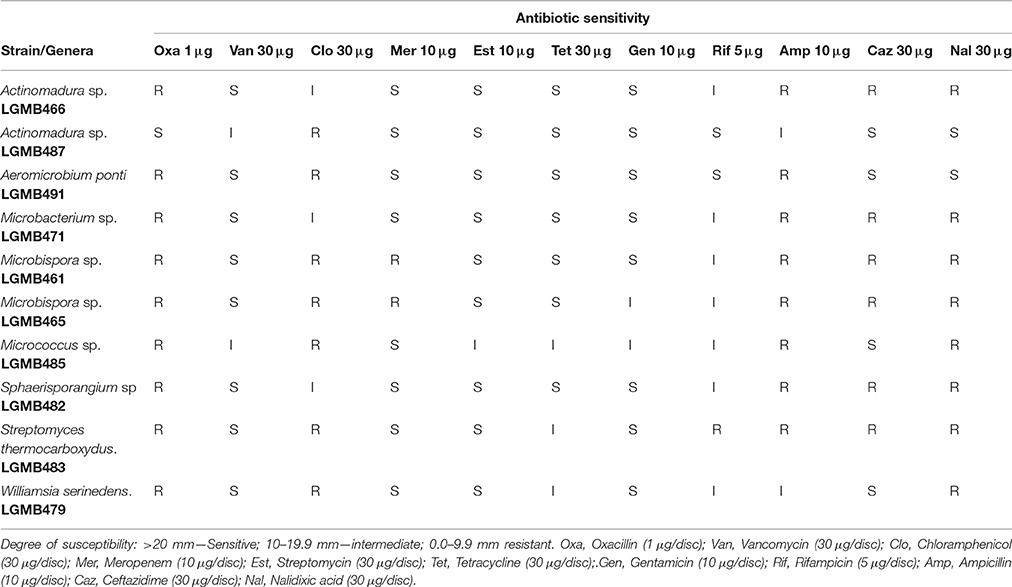
The susceptibility of the endophytes to 11 antibiotics, oxacilin (a penicillin), vancomycin (a glycopeptide), chloramphenicol (an amphionicol), meropenem (a carbapenem), streptomycin (an aminoglycoside), tetracycline (a tetracycline), gentamicin (another aminoglycoside), rifampicin (a macrolactam), ampicillin (another penicillins), ceftazidime (a third generation cephalosporin), and nalidixic acid (a quinolone) were evaluated as described by Passari et al. (2015). The analysis was performed considering the isolate sensitive (S) with an inhibition zone > 20 mm, intermediate (I) with an inhibition zone of 10–19.9 mm and resistant (R), if the inhibition zone was between 0.0–9.9 mm (Williams et al., 1989).
Biological Activity
Screening of Culture Conditions
Isolates were inoculated in 50 mL of SG medium (Shaaban et al., 2011), incubated for 3 days at 36°C and 180 rpm. Subsequently, 1 mL from the pre-culture was inoculated in SG and R5A media (100 mL) (Fernandez et al., 1998), and incubated for 10 days at two different temperatures, 28 and 36°C, and 180 rpm. The culture was filtered-off on Whatmann 4 filters, the water fraction was extracted with EtOAc (3 × 100 mL). The combined organics were evaporated in vacuo at 40°C and diluted in methanol at 10 mg/mL.
Antibacterial Activity–Disk Diffusion Assays
The antibacterial activity of crude extracts and the isolated compounds 1–9 was evaluated against methicillin-sensitive Staphylococcus aureus (MSSA) (ATCC 25923), methicillin-resistant S. aureus (MRSA) (BACHC-MRSA), Pseudomonas aeruginosa (ATCC 27853), Candida albicans (ATCC 10231), Acinetobacter baumannii (BACHC-ABA), Klebsiella pneumoniae, the producer of the enzyme KPC (K. pneumoniae carbapenemase) (BACHC-KPC), Stenotrophomonas maltophilia (BACHC-SMA), and Enterobacter cloacae a producer of the enzyme VIM (Verona integron-encoded metallo-β-lactamase) (BACHC-VIM). The bacteria were cultivated for 12 h at 37°C, and diluted according to the McFarland standard 0.5 scale. Each test organism was streaked on a sterile Mueller-Hinton agar plate with a cotton swab. Extracts were aliquoted in 100 μg amounts per 6 mm sterile filter disc. The discs were placed on plates and incubated for 24 h at 37°C. The diameter halos were measured in millimeters. As a positive control, a disc with a standard antibiotic with activity against each of the bacteria was used, and pure methanol was used as negative control (CLSI, 2015; Savi et al., 2015b).
MIC–Minimum Inhibitory Concentration and MBC–Minimum Bactericidal Concentration
Extracts from strain LGMB491 that showed high antibacterial activity were selected to determine the minimum inhibitory concentration. The MIC of extracts against the clinical pathogens was performed as described by Ostrosky et al. (2008) and CLSI. The minimum bactericidal concentration was determined as described by Soltani and Moghaddam (2014).
Statistical Analyses
The statistical analysis was performed using analysis of variance (ANOVA) to compare extract effects to their respective controls. We also performed Post-hoc tests using Tukey's honest significant difference. All tests premises were fulfilled; the significance level used was 0.05 (α).
Large-Scale Fermentation, Extraction and Isolation
A large-scale fermentation (10 L) of strain LGMB491 was performed using SG culture medium at 36°C for 10 days. The culture was subjected to extraction with EtOAc (3 × v/v), and the combined organic layers were evaporated in vacuo at 40°C to yield 653 mg of crude extract. The crude extract was subjected to reverse phase C18 column chromatography (20 × 8 cm, 250 g), eluted with a gradient of H2O-MeOH (100:0–0:100) to produce fractions FI-FV. The single fractions were subjected to HPLC and Sephadex LH-20 (MeOH; 1 × 20 cm) purifications to yield compounds 1–9 in pure form (Figure 9, Figure S9). NMR spectra were measured using a Varian (Palo Alto, CA) Vnmr 400 (1H, 400 MHz; 13C, 100 MHz) spectrometer, δ-values were referenced to the respective solvent signals (CD3OD, δH 3.31 ppm, δC 49.15 ppm; DMSO-d6, δH 2.50 ppm, δC 39.51 ppm). HPLC-MS analyses were accomplished using a Waters (Milford, MA) 2695 LC module (Waters Symmetry Anal C18, 4.6 × 250 mm, 5 μm; solvent A: H2O/0.1% formic acid, solvent B: CH3CN/0.1% formic acid; flow rate: 0.5 mL min−1; 0–4 min, 10% B; 4–22 min, 10–100% B; 22–27 min, 100% B; 27–29 min, 100–10% B; 29–30 min, 10% B). HPLC analyses were performed on an Agilent 1260 system equipped with a photodiode array detector (PDA) and a Phenomenex C18 column (4.6 × 250 mm, 5 μm; Phenomenex, Torrance, CA). Semi-preparative HPLC was accomplished using Phenomenex (Torrance, CA) C18 column (10 × 250 mm, 5 μm) on a Varian (Palo Alto, CA) ProStar Model 210 equipped with a photodiode array detector and a gradient elution profile (solvent A: H2O, solvent B: CH3CN; flow rate: 5.0 mL min−1; 0–2 min, 25% B; 2–15 min, 25–100% B; 15–17 min, 100% B; 17–18 min, 100–25% B; 18–19 min, 25% B). All solvents used were of ACS grade and purchased from the Pharmco-AAPER (Brookfield, CT). Size exclusion chromatography was performed on Sephadex LH-20 (25–100 μm; GE Healthcare, Piscataway, NJ).
Results
Isolation of Endophytic Actinomycetes
From 2,988 fragments analyzed, 10 endophytic actinomycetes were isolated (Table 1), thus the isolation frequency was 0.34%. From the 10 isolates, 70% (n = 7) were isolated from the Abobral, and 30% (n = 3) from the São Bento region. Five isolates were obtained from stems, and five from leaf tissues of the plant (Table 1).
Morphological Identification
A great macro-morphological diversity was observed, with white, ivory-white, pink, brown, gray, orange, and yellow colony colors. Most of isolates showed abundant to moderate growth after 21 days of incubation, and six isolates showed abundant to moderate spore formation on ISP2 and ISP3 media. Isolates LGMB461 and LGMB465 showed high morphological similarity, and probably represent the same species (Table 1).
Molecular Analysis
Using a BLAST analysis in the GenBank database, the isolates were classified as eight genera: Aeromicrobium, Williamsia, Microbacterium, Sphaerisporangium, Micrococcus, Microbispora, Actinomadura, and Streptomyces. Each genus was analyzed in a separate phylogenetic tree based on Bayesian inference.
Actinomadura (LGMB466 and LGMB487)
The alignment consisted of strains LGMB466 and LGMB487, 55 type strains representative of Actinomadura genus, and Streptomyces glauciniger (AB249964) as out group taxa. The analysis comprises of 1,402 characters, 1,011 of these were conserved, 124 were parsimony informative and 131 were uninformative. Strains LGMB466 and LGMB487 showed high similarity among themselves (98.86%), and in the phylogenetic analysis these isolates did not cluster with any species from the Actinomadura genus (Figure 1, Table S1), and probably represent a new species.
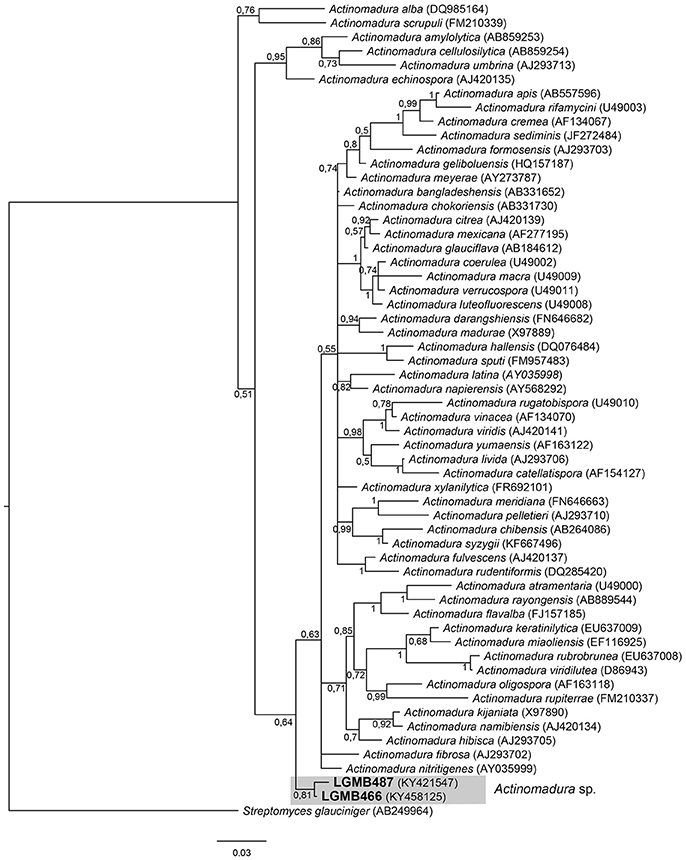
Figure 1. Bayesian phylogenic tree based on 16S rRNA gene of LGMB466, LGMB487, and the 53 type strain of Actionomadura genus. Values on the node indicate Bayesian posterior probabilities. The species Streptomyces glauciniger was used as out group. Scale bar indicates the number of substitutions per site.
Aeromicrobium (LGMB491)
Strain LGMB491 was aligned with all type strains from the Aeromicrobium genus (12 species), and Nocardioides albus (X53211) was used as out group taxa. The alignment consisted of 1,336 characters, 1,164 of these were conserved, 89 were parsimony informative and 68 were uninformative. Based on this phylogenetic analysis, strain LGMB491 is close related to Aeromicrobium ponti (Figure 2), sharing high sequence similarity, 99.25 % (Table S2).
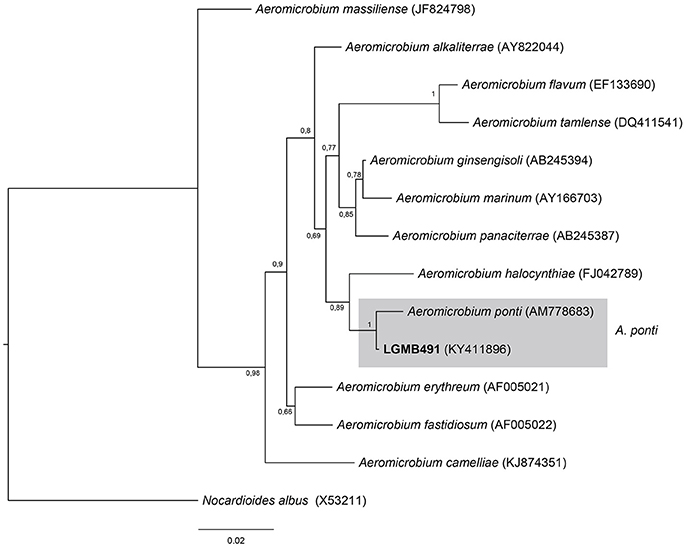
Figure 2. Bayesian phylogenic tree based on 16S rRNA gene of LGMB491 and the 12 type strain of Aeromicrobium genus. Values on the node indicate Bayesian posterior probabilities. The species Nocardioides albus was used as out group. Scale bar indicates the number of substitutions per site.
Microbacterium (LGMB471)
Strain LGMB471 was aligned with type strains from the Microbacterium genus, and Agrococcus jenensis (X92492) as out group taxa. The alignment comprised of 1,314 characters, of those 721 conserved sites, 122 were parsimony informative, and 57 uninformative. In the phylogenetic tree, isolate LGMB471 ended up in a single branch related to species Microbacterium liquefaciens, Microbacterium maritypicum, Microbacterium oxydans, Microbacterium luteolum, Microbacterium saperdae, and Microbacterium paraoxydans (Figure 3, Table S3).
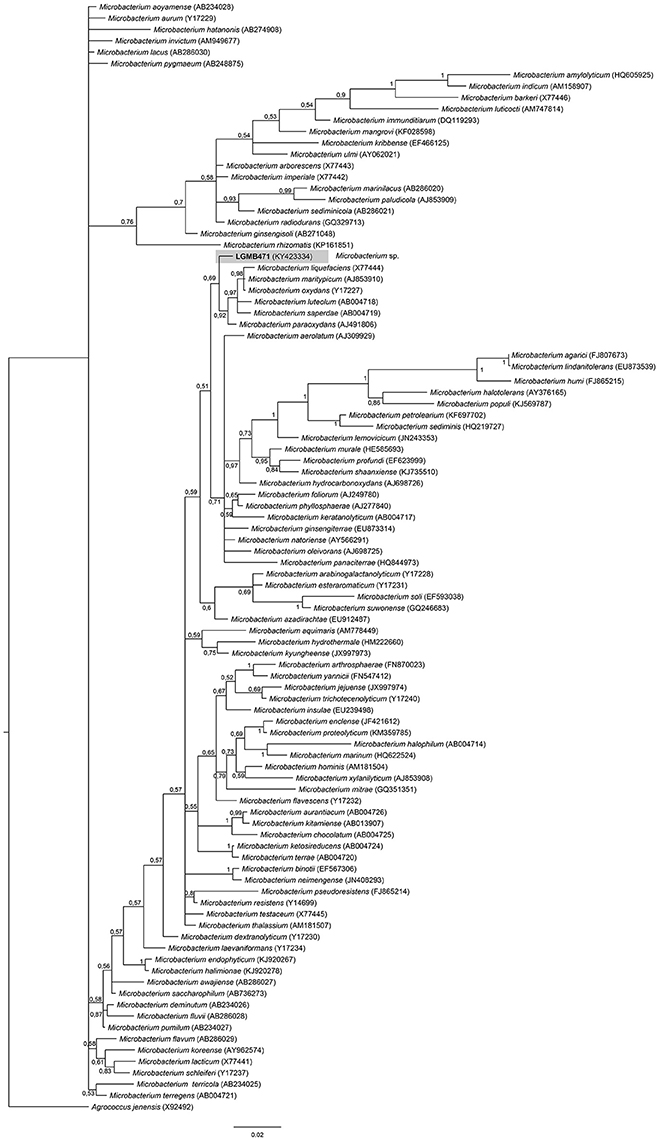
Figure 3. Bayesian phylogenic tree based on 16S rRNA gene of LGMB471 and the 94 type strain of Microbacterium genus. Values on the node indicate Bayesian posterior probabilities. The species Agrococcus jenensis was used as out group. Scale bar indicates the number of substitutions per site.
Microbispora (LGMB461 and LGMB465)
The analysis comprises of strains LGMB461 and LGMB465, 10 species accepted in Microbispora genus, and the isolates previously reported as Microbispora sp.1, Microbispora sp.2, and Microbispora sp.3 (Savi et al., 2016). Actinomadura echinospora (AJ420135) was used as out group taxa. The alignment consists of 1,371 characters, 1,309 of these were conserved, 33 were parsimony informative, and 29 uninformative. In the phylogenetic analysis strains LGMB461 and LGMB465 presented similarity with Microbispora sp.1 (LGMB259) with 99.84 and 100% f similarity, respectively (Figure 4, Table S4).
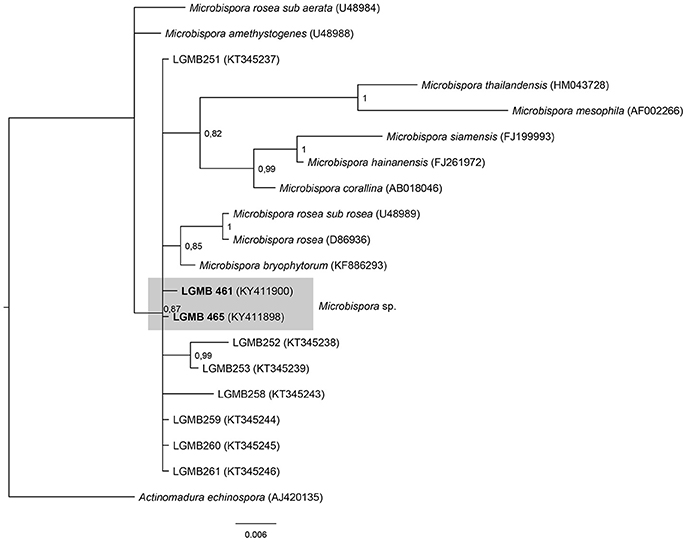
Figure 4. Bayesian phylogenic tree based on 16S rRNA gene of LGMB461, LGMB465, the 10 type strain of Microbispora genus, and 7 strains previously reported by Savi et al. (2016). Values on the node indicate Bayesian posterior probabilities. The species Citricoccus parietis was used as out group. Scale bar indicates the number of substitutions per site.
Micrococcus (LGMB485)
The Bayesian analysis comprised of all Micrococcus type strains, strain LGMB485 and Citricoccus parietis (FM9923367) as out group taxa (Figure 5). The alignment consisted of 1,340 characters with 452 conserved sites, nine were parsimony informative and 19 uninformative. Since the sequences were very similar (Table S5) and the alignment had only nine parsimony informative sites, a species designation cannot be assigned, and isolate LGMB485 was identified as Micrococcus sp.
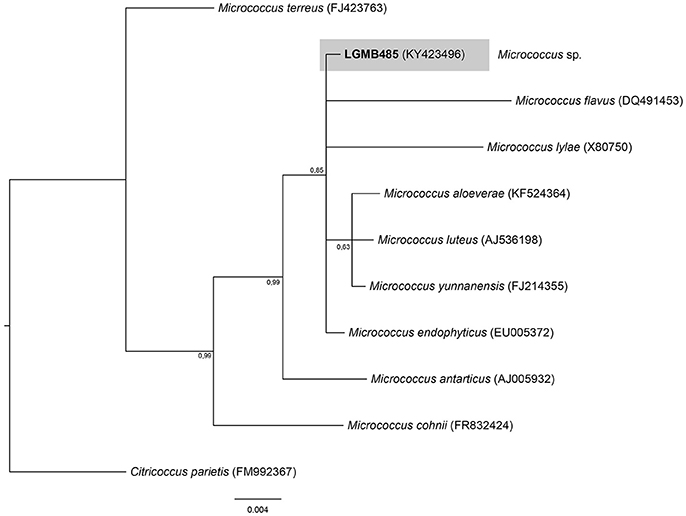
Figure 5. Bayesian phylogenic tree based on 16S rRNA gene of LGMB485 and the 9 type strain of Micrococcus genus. Values on the node indicate Bayesian posterior probabilities. The species Citricoccus parietis was used as out group. Scale bar indicates the number of substitutions per site.
Sphaerisporangium (LGMB482)
For the Bayesian analysis, the sequence from LGMB482 was aligned with strains of the Sphaerisporangium genus, and Actinomadura madurae (X97889) was used as out group taxa. The alignment consisted of 1,320 characters, 886 of these were conserved, 51 were parsimony informative and 47 were uninformative. Strain LGMB482 is closely related to S. melleum AB208714 (99.4% similarity) and S. viridalbum X89953 (97.89% similarity), however, it is in an isolated branch and may represent a new species of the Sphaerisporangium genus (Figure 6, Table S6).
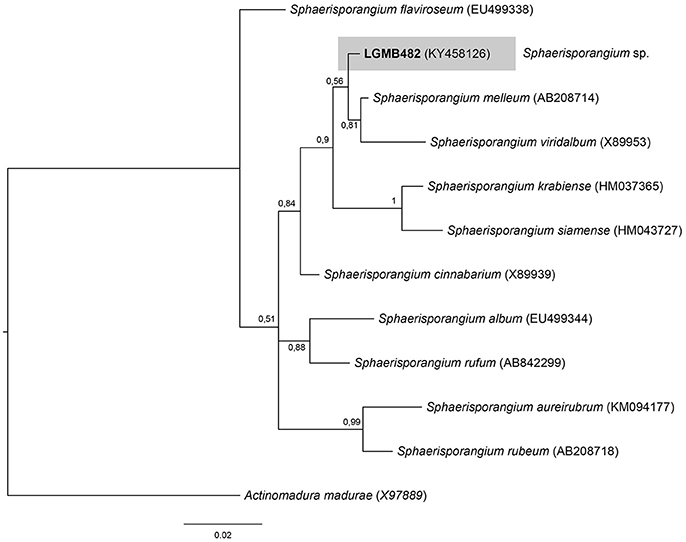
Figure 6. Bayesian phylogenic tree based on 16S rRNA gene of LGMB482 and the 10 type strain of Sphaerisporangium genus. Values on the node indicate Bayesian posterior probabilities. The species Actinomadura madurae was used as out group. Scale bar indicates the number of substitutions per site.
Streptomyces (LGMB483)
The phylogenetic analysis was performed using 23 type strains closely related with LGMB483; including Streptomyces albus subsp. albus (X53163) as out group taxa. The alignment consisted of 1,391 characters, with 1,291 conserved sites, 45 were parsimony informative, and 39 uninformative. In the phylogenetic tree, isolate LGMB483 grouped with Streptomyces thermocarboxydus, sharing 99.86% of similarity (Figure 7, Table S7), and thus we suggest this isolate may belongs to this species.
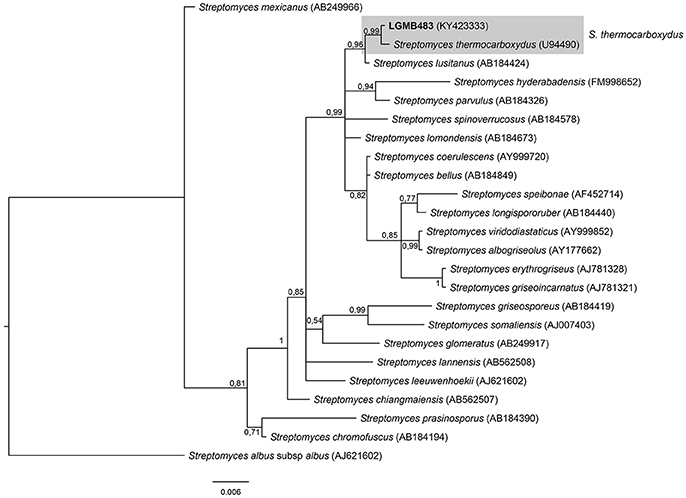
Figure 7. Bayesian phylogenic tree based on 16S rRNA gene of LGMB483 and the 33 type strain of Streptomyces genus. Values on the node indicate Bayesian posterior probabilities. The species Streptomyces albus subsp.albus was used as out group. Scale bar indicates the number of substitutions per site.
Williamsia (LGMB479)
The analysis consists of 11 sequences, including all type strains of the Williamsia genus, the strain LGMB479, and Mycobacterium tuberculosis (X58890) was used as out group taxa. The alignment comprises of 1,346 characters, of these 1,185 were conserved, 81 were parsimony informative and 56 were uninformative. Strain LGMB479 was in the same clade with Williamsia serinedens (AM283464) (Figure 8) and share 99.85% sequence similarity (Table S8), and may belongs to this species.
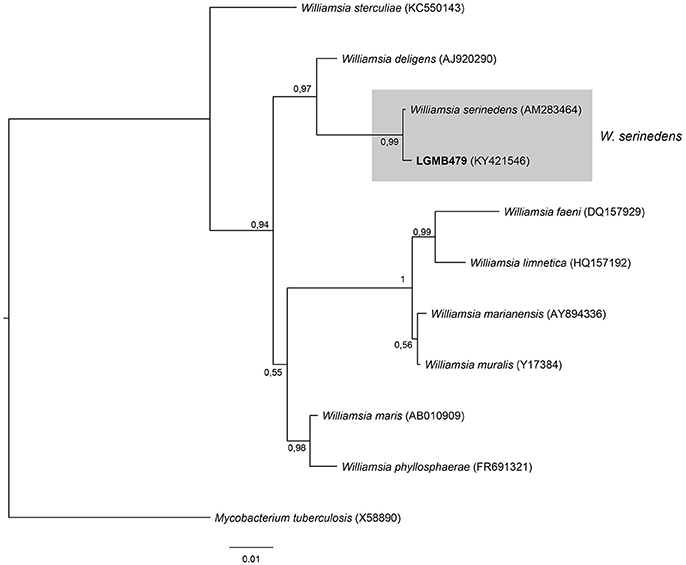
Figure 8. Bayesian phylogenic tree based on 16S rRNA gene of LGMB479 and the 9 type strain of Willamsia genus. Values on the node indicate Bayesian posterior probabilities. The species Mycobacterium tuberculosis was used as out group. Scale bar indicates the number of substitutions per site.
Antibiotic Sensitivity Test
In order to characterize the susceptibility profiles of the endophytes, 11 antibiotics with different mechanisms-of-action were utilized. Isolates were susceptible to vancomycin (80% sensitive and 20% intermediate), streptomycin (90% sensitive and 10% intermediate), tetracycline (70% sensitive and 30% intermediate), and gentamicin (80% sensitive and 20% intermediate). The two isolates of Microbispora sp. (LGMB461 and LGMB465) showed resistance to meropenem, and 90% of the isolates showed resistance to oxacillin, and nalidixic acid (Table 2).
Antibacterial Activity of Crude Extracts
All strains and culture conditions analyzed produced active extracts (Table 3, Table S9), however, the extract from LGMB491 (close related to A. ponti) cultured in SG medium at 36°C showed great antibacterial activity against S. aureus (22 mm) and MRSA (19.8 mm), and moderate activity against others clinical pathogens (Table 3, Figures S1–S8). The MIC and MBC of extract from LGMB491 against S. aureus and methicillin-resistant S. aureus were 0.02, and 0.04 mg/mL, respectively, and the MBC was 5 mg/mL for both bacteria (Table 4). In addition, the crude extract from LGMB491 had an MIC of 0.63 mg/mL against gram-negative bacteria associated with antibiotic resistance, K. pneumoniae KPC, S. maltophilia, and E. cloacae VIM, and a MIC of 0.31 mg/mL against A. baumannii and P. aeruginosa, respectively (Table 4).
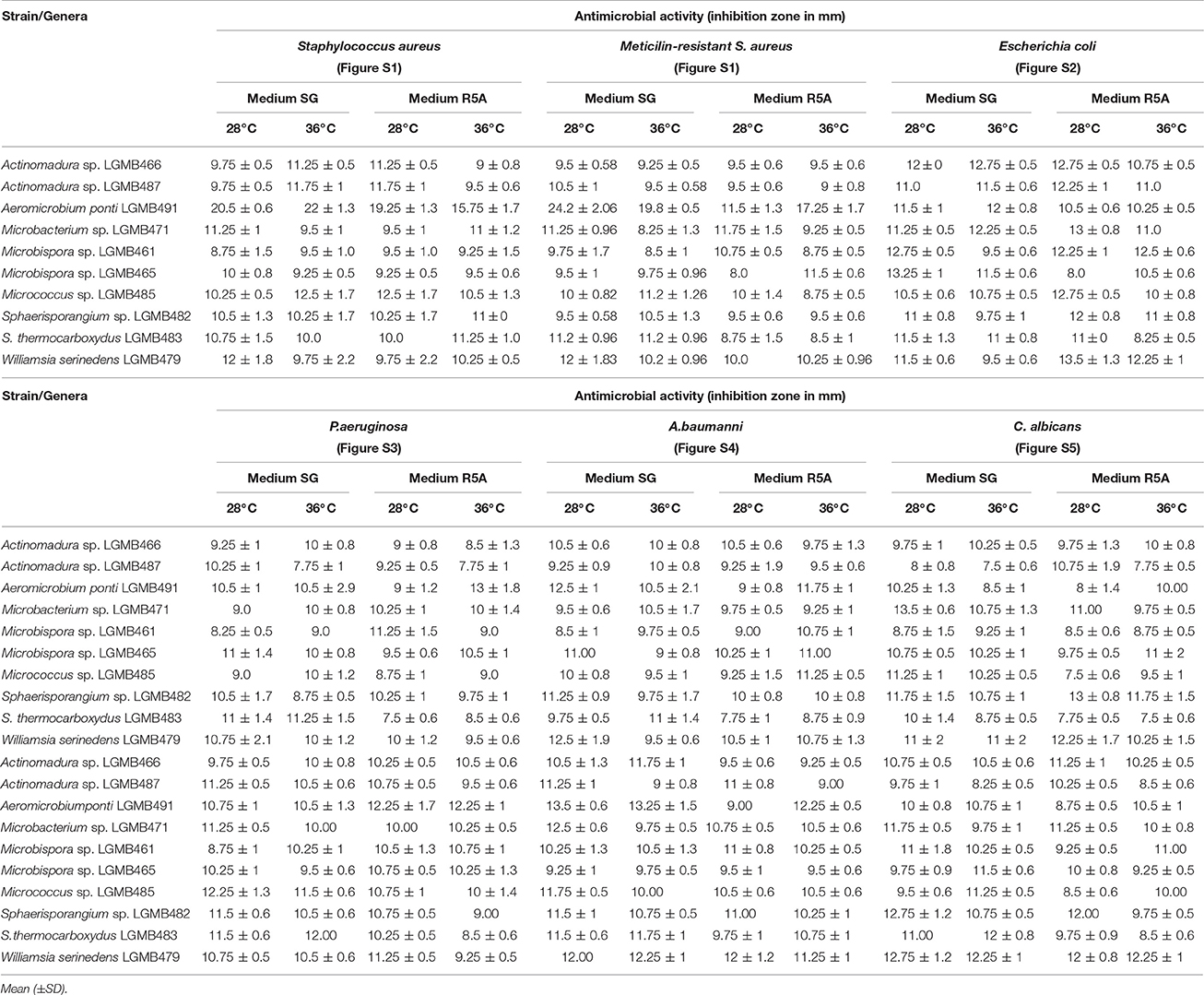
Table 3. Antibacterial activity of the extracts from endophytic actinomycetes in two culture media (SG and R5A) and two temperatures (28°C and 36°C) against Clinical pathogens.
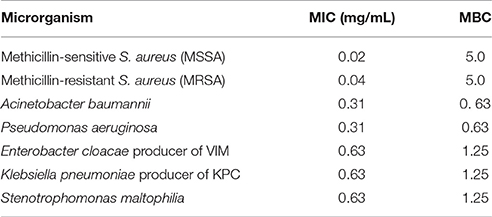
Table 4. Minimum Inhibitory and Minimum Bactericidal Concentrations of the extract from strain Aeromicrobium ponti LGMB491.
Structure Determination of Secondary Metabolites from Strain LGMB491
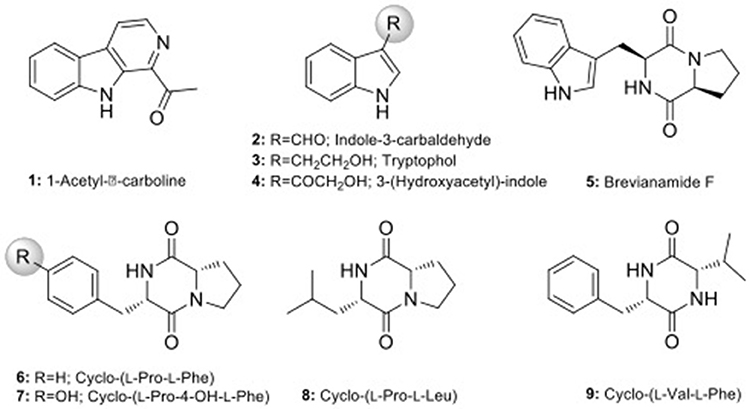
Scale-up fermentation of strain LGMB491 (10 L) using SG medium, followed by extraction afforded 653 mg of crude extract. Fractionation, isolation and purification of the obtained extract using various chromatographic techniques resulted in compounds 1–9 in pure forms (Figure S9). Thorough analyses of the HPLC/UV, ESIMS and NMR spectroscopy data (Figure S10–S43), and by comparison with literature data (Laatsch, 2012), the compounds were identified as 1-acetyl-β-carboline (1) (Shaaban et al., 2007; Savi et al., 2015b), indole-3-carbaldehyde (2) (Zendah et al., 2012; Savi et al., 2015b), tryptophol (3) (Rayle and Purves, 1967), 3-(hydroxyacetyl)-indole (4) (Zendah et al., 2012), brevianamide F (5) (Shaaban, 2009), cyclo-(L-Pro-L-Phe) (6) (Barrow and Sun, 1994), cyclo-(L-Pro-L-Tyr) (7) (Barrow and Sun, 1994), cyclo-(L-Pro-L-Leu) (8) (Yan et al., 2004), and cyclo-(L-Val-L-Phe) (9) (Pickenhagen et al., 1975) (Figure 9). In order to determine the compounds responsible for the biological activity observed for the crude extract of strain LGMB491, we evaluated the antibacterial activity of compounds 1–9 against S. aureus and methicillin-resistant S. aureus. 1-Acetyl-β-carboline (1) showed an equivalent activity as the antibiotic methicillin against S. aureus, however, different from this antibiotic, compound 1 also showed activity against MRSA (Table 5). In addition, compounds 2, 4–6 also showed moderate activity against both MSSA and MRSA.
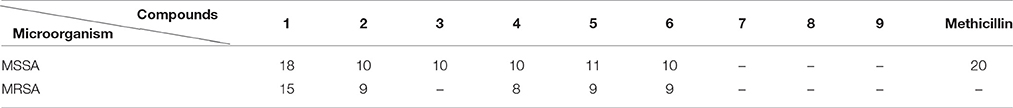
Table 5. Inhibition zone (mm) growth of methicillin-sensitive Staphylococcus aureus (MSSA) and methicillin-resistant S. aureus (MRSA) of compounds 1–9 (100 μg/disk).
Discussion
Endophytes Isolation and Identification
Actinomycetes from medicinal plants are the source of several secondary metabolites with biological activity (Qin et al., 2015; Savi et al., 2015b), and their metabolites may be associated with the medicinal properties of the plant host (Kusari et al., 2013; Santos et al., 2015). We explored the endophytes from the medicinal plant V. divergens, in order to catalog the species richness and biological properties. A low frequency of isolation (0.34%), compared with the isolation of terrestrial actinomycetes, was observed, in agreement with literature data (Passari et al., 2015). However, despite the lower isolation frequency a higher richness of genera was observed (Passari et al., 2015; Saini et al., 2016). We reported for the first time the isolation of strains close related to the species A. ponti (LGMB491) and Williamsia serinedens (LGMB479) as endophytes. A. ponti was originally isolated from seawater (Lee and Lee, 2008), and has been found in this environment (Jiang et al., 2010; Claverias et al., 2015). W. serinedens was first isolated from an oil-contaminated soil sample and it is common isolated from different types of soil (Yassin et al., 2007). In addition, species S. thermocarboxydus was isolated from soil (Kim et al., 2000), and was recently described as endophyte from a medicinal plant in India (Passari et al., 2015). Based on the 16S rRNA phylogenetic analysis we suggest that strains LGMB471 and LGMB482 may represent new species within the Microbacterium and Sphaerisporangium genera, respectively (Figures 3, 6), and isolates LGMB466 and LGMB487 seem to be a new species within the Actinomadura genus (Figure 1). Isolates LGMB461 and LGMB465 belong to genus Microbispora, and showed high sequence similarity with strains belonging to Microbispora sp.1 group, previously isolated from V. divergens (Savi et al., 2016). However, sequencing others genes than 16S rRNA, and DNA-DNA hybridization would be required for species description (Meyers, 2014). Microbacterium, Sphaerisporangium, and Micrococcus species are common associated with medicinal plants in different regions, and climate conditions (Kim et al., 2000; Kamil et al., 2014; Xing et al., 2015). However, none of these has been isolated from wetland regions. Savi et al. (2015a) performed the first report about actinomycetes from the medicinal plant V. divergens. However, despite the higher number of isolates, the authors then just identified three genera as endophytes from this plant, Microbispora, Micromonospora, and Streptomyces. In addition to those genera previously mentioned (Microbispora and Streptomyces) we isolated species belonging to Actinomadura, Aeromicrobium, Microbacterium, Sphaerisporangium, Micrococcus, and Williamsia (Figures 1–8), thereby significantly increasing the knowledge regarding endophytes from V. divergens.
Antibiotic Sensitivity Assay
In order to characterize the susceptibility profile as well as to suggest antibiotics to be used in actinomycete isolation, we evaluated the susceptibility profile of endophytes. We detected significant resistance to antibiotics oxacillin and nalidixic acid, only strain Actinomadura LGMB487 was sensitive to both compounds (Table 2). Nalidixic acid is the antibiotic used to inhibit bacterial growth during actinomycete isolation, however, even with the use of this compound, the presence of contaminating bacteria was common (Baskaran et al., 2011; Kadiri et al., 2014). Therefore, based on the high resistance to oxacillin observed in this study, we suggest the use of this antibiotic to inhibit bacterial growth during the isolation of actinomycetes. Strains LGMB466 and LGMB487, both characterized as Actinomadura sp., showed a complete different sensitivity pattern: strain LGMB487 was resistant only to chloramphenicol, and LGMB466 showed resistance to four antibiotics, and intermediate resistant to chloramphenicol, and rifampicin, suggesting that the resistance profile of isolates is not associated with the intrinsic factors of Actinomadura genus. The resistance observed in these strains can result from the presence of plasmids, which contributes to the well-known problem of antibiotic resistance (Wintersdorff et al., 2016). In addition, vancomycin, streptomycin, tetracycline, and gentamicin were previously reported from actinomycetes (Gonzalez and Spencer, 1998; Chopra and Roberts, 2001; Levine, 2006; Zumla et al., 2013), however, all strains evaluated here showed some sensitivity level to these antibiotics, which suggest that these compounds are not present as secondary metabolites from our isolates.
Biological Activity and Secondary Metabolites Identification
All isolates and conditions analyzed produced active secondary metabolites, ratios superior than observed in previous studies (Higginbotham and Murphy, 2010; Passari et al., 2015; Tonial et al., 2016), suggesting the high biotechnological potential of the evaluated strains. This may be related to the culture conditions used to obtain the secondary metabolites. Extracts from LGMB491 (close related to A. ponti) showed great activity against MRSA, with inhibition zones higher than caused by vancomycin, the clinical antibiotic used for the treatment of this resistant bacterium (Table 3). In addition, extracts from strain LGMB491 also had considerable MIC, and MBC values against S. aureus, MRSA, K. pneumoniae KPC, S. maltophilia, A. baumannii, P. aeruginosa, and E. cloacae VIM. These data suggest the presence of metabolites with broad spectrum activity (Smith et al., 2011). Compounds with broad spectrum activity are required to treat multidrug resistant pathogens, such as MRSA, S. maltophilia, P. aeruginosa, and A. baumannii (Bonomo and Szabo, 2006; Çıkman et al., 2016), bacteria that are considered one of the most urgent issues in modern healthcare (Paulus et al., 2017). Therefore, due to the good activity observed, and the absence of studies about metabolites with biological activity from A. ponti species, we decided to characterize the major compounds produced by strain LGMB491. From the nine secondary metabolites isolated, 1-acetyl-β-carboline (1) turned out to be the compound responsible for the antibacterial activity of the LGMB491 extract. The compound displayed high activity against the MRSA (Table 5). β-carbolines are normally isolated from plants with a large spectrum of biological activity (Lee et al., 2013). Savi et al. (2015b) reported the production of four β-carbolines by the Microbispora sp. 1 also isolated from V. divergens. The authors isolated as the major metabolite the compound 1-vinyl-β-carboline-3-carboxylic acid, and attributed the vinyl chain as the likely responsible structural feature causing the antibacterial activity of this natural product. However, 1-acetyl-β-carboline (1), found during this study, showed also high biological activity, which is unlikely associated with the acetyl chain in position 1. Several studies demonstrated great activity of compound 1 against MRSA, and suggest the use of this compound for an effective treatment of this resistant bacterium (Shin et al., 2010; Lee et al., 2013). In addition to 1-acetyl-β-carboline (1), compounds 2–6 displayed moderate antibacterial activity, and may act synergistically with compound (1), contributing for the activity observed. Brevianamide F (5), an alkaloid, was isolated for the first time from Penicillium brevicompactum (Birsh and Wright, 1969), and has nematocidal (Shiomi and Omura, 2004), anti-inflammatory (Rand et al., 2005), and antibacterial activity against methicillin-sensitive and resistant S. aureus (Kumar et al., 2014; Alshaibani et al., 2016). Cyclo-(L-Pro-L-Phe) (6) is a diketopiperazine, i.e., a member of these cyclic dipeptides commonly isolated from microorganisms that have been associated with antimicrobial activity, and plant growth regulation (Zhang et al., 2013; Kalinovskaya et al., 2017). Interesting, several diketopiperazines, including cyclo-(L-Pro-L-Phe), were previously isolated from Aspergillus fumigatus from a soil sample of the Pantanal, and showed high antibacterial activity against S. aureus (Furtado et al., 2005), which supports the idea of synergism of the compounds produced by strain LGMB491. The indoles isolated from strain LGMB491 are commonly produced by plants and endophytic microorganism (Braga et al., 2016). 3-(Hydroxyacetyl)-indole (4) showed a broad-spectrum antibacterial activity against methicillin-resistant S. aureus, and against vancomycin-sensitive or resistant Enterococci, attributed to disruption of cell membrane (Sung and Lee, 2007). In plants, indole-3-carbaldehyde (3) is associated with the innate immunity to microbial pathogen infections (Stahl et al., 2016). This compound was also produced by Microbispora sp. 1 previously isolated from the medicinal plant V. divergens (Savi et al., 2015b). Some studies suggested that indole compounds play an important role in plant-microorganism interaction and plant defense (Gamir et al., 2012; Lin and Xu, 2013; Jeandet et al., 2014).
Conclusion
In this study, we increased the knowledge regarding the endophytic community of the medicinal plant V. divergens, through the isolation of rare actinomycetes, some of which were never described as endophytes. We identified for the first time some secondary metabolites produced by one strain close related to the species A. ponti, and demonstrated that this species is able to produce indoles, β-carbolines, brevianamide, and diketopiperazines. Future studies to evaluate the potential of these compounds in animal models are required to better understand the potential of compound 1-acetyl-β-carboline as an alternative to treat MRSA infections. Our results indicate that actinomycetes from V. divergens have biotechnological potential as producer of bioactive compounds.
Author Contributions
All the authors contributed to the experimental design of the work; as well as to the acquisition, analysis, and interpretation of the obtained results; moreover, all the authors contributed to the writing and the critical revision of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Fundação Araucária de Apoio e Desenvolvimento Científico e Tecnológico do Paraná—Brazil, grant 441/2012—23510 to CG, CNPq-Brazil grant 486016/2011-0 to CG, and CAPES-Brazil—grant to DS. It was also supported in part by the University of Kentucky College of Pharmacy, the University of Kentucky Markey Cancer Center and the National Center for Advancing Translational Sciences (UL1TR001998), and by NIH grants CA 091091 and GM 105977 as well as an Endowed University Professorship in Pharmacy to JR.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01642/full#supplementary-material
References
Alho, C. J. (2008). Biodiversity of the Pantanal: response to seasonal flooding regime and to environmental degradation. Braz. J. Biol. 68, 957–966. doi: 10.1590/S1519-69842008000500005
Alshaibani, M. M., Jalil, J., Sidik, N. M., Edrada-Ebel, R., and Zin, N. M. (2016). Isolation and characterization of cyclo-(tryptophanylprolyl) and chloramphenicol from Streptomyces sp. SUK 25 with antimethicillin-resistant Staphylococcus aureus activity. Drug Des. Dev. Ther. 31, 1817–1827. doi: 10.2147/DDDT.S101212
Arieira, J., and Cunha, C. N. (2006). Fitossociologia de uma floresta inundavelmonodominante de Vochysia divergens Pohl (Vochysiaceae), no Pantanal Norte, MT, Brasil. Acta Bot. Bras. 20, 569–580. doi: 10.1590/S0102-33062006000300007
Barrow, C. J., and Sun, H. H. (1994). Spiroquinazoline, a novel substance P inhibitor with a New carbon skeleton, isolated from Aspergillus flavipes. J. Nat. Prod. 57, 471–476. doi: 10.1021/np50106a005
Baskaran, R., Vijayakumar, R., and Mohan, P. (2011). Enrichment method for the isolation of bioactive actinomycetes from mangrove sediments of Andaman Islands India. Malays. J. Microbiol. 7, 26–32. doi: 10.21161/mjm.24410
Birsh, A., and Wright, J. (1969). The Brevianamides: a new class of fungal alkaloid. J. Chem. Soc. D. 12, 644–645.
Bonomo, R. A., and Szabo, D. (2006). Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin. Infect. Dis. 43, 49–56. doi: 10.1086/504477
Borrero, N. V., Bai, F., Perez, C., Duong, B. Q., Rocca, J. R., Jin, S., et al. (2014). Phenazine antibiotic inspired discovery of potent bromophenazine antibacterial agents against Staphylococcus aureus and Staphylococcus epidermidis. Org. Biomol. Chem. 12, 881–886. doi: 10.1039/C3OB42416B
Braga, R. M., Dourado, M. N., and Araujo, W. L. (2016). Microbial interactions: ecology in a molecular perspective. Braz. J. Microbiol. 47, 86–98. doi: 10.1016/j.bjm.2016.10.005
Chopra, I., and Roberts, M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65, 232–260. doi: 10.1128/MMBR.65.2.232-260.2001
Çıkman, A., Parlak, M., Bayram, Y., Güdücüoğlu, H., and Berktaş, M. (2016). Antibiotics resistance of Stenotrophomonas maltophilia strains isolated from various clinical species. Afr. Health Sci. 16, 149–152. doi: 10.4314/ahs.v16i1.20
Claverias, F. P., Undabarrena, A., Gonzalez, M., Seeger, M., and Camara, B. (2015). Culturable diversity and antimicrobial activity of Actinobacteria from marine sediments in Valparaiso bay, Chile. Front. Microbiol. 6:737. doi: 10.3389/fmicb.2015.00737
CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. CLSI document M100–S25. Wayne, PA: Clinical and Laboratory Standards Institute.
Davies, J., and Davies, D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433. doi: 10.1128/MMBR.00016-10
Fernandez, E., Weibach, U., Reillo, C. S., Brana, A. F., Mendez, C., Rohr, J., et al. (1998). Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 180, 4929–4937.
Furtado, N. A. J. C., Pupo, M. T., Carvalho, I., Campo, V. L., Duarte, M. C. T., and Bastos, J. K. (2005). Diketopiperazines produced by an Aspergillus fumigatus Brazilian strain. J. Braz. Chem. Soc. 16, 1448–1453. doi: 10.1590/S0103-50532005000800026
Gamir, J., Pastor, V., Cerezo, M., and Flors, V. (2012). Identification of indole-3-carbolylic acid as mediator of priming against Plectosphaerella cucumerina. Plant Physiol. Bioch. 61, 169–179. doi: 10.1016/j.plaphy.2012.10.004
Golinska, P., Wypij, M., Agarkar, G., Rathod, D., Dahm, H., and Rai, M. (2015). Endophytic actinobacteria of medicinal plants: diversity and bioactivity. Ant. Van Leew 108, 267–289. doi: 10.1007/s10482-015-0502-7
Gonzalez, L. S., and Spencer, J. P. (1998). Aminoglycosides: a practical review. Am. Fam. Phys. 58, 1811–1820.
Higginbotham, S. J., and Murphy, C. D. (2010). Identification and characterization of Streptomyces sp. isolate exhibiting activity against methicillin-resistant Staphylococcus aureus. Microb. Res. 165, 82–86. doi: 10.1016/j.micres.2008.12.004
Hokama, Y., Savi, D. C., Assad, B., Aluizio, R., Gomes-Figueiredo, J., Adamoski, D., et al. (2016). “Endophytic fungi isolated from Vochysia divergens in the pantanal, mato grosso do sul: diversity, phylogeny and biocontrol of Phyllosticta citricarpa,” in Endophytic Fungi: Diversity, Characterization and Biocontrol, 4th Edn., ed E. Hughes (Hauppauge, NY: Nova Publishers), 1–25.
Jeandet, P., Hebrard, C., Deville, M. A., Cordelier, S., Dorey, S., Aziz, A., et al. (2014). Deciphering the role of phytoalexins in plant-microorganism interaction and human health. Molecules 19, 18033–18056. doi: 10.3390/molecules191118033
Jiang, H., Lin, R., Chen, L., Lin, H., Nie, Y., and Lian, Y. (2010). Actinobacterial diversity of marine sediment samples from Chile. Wei Sheng Wu XueBao 50, 862–869.
Kadiri, S. K., Yarla, N. S., and Vidavalur, S. (2014). Screening and isolation of antagonistic actinobacteria associated with marine sponges from Indian coast. J. Microb. Bioch. Technol. 8, 1–3. doi: 10.4172/1948-5948.S8-003
Kalinovskaya, N. I., Romanenko, L. A., and Kalinovsky, A. I. (2017). Antibacterial low-molecular-weight compounds produced by the marine bacterium Rheinheimera japonica KMM 9513T. Antonie Van Leeuwenhoek. 110, 719–726. doi: 10.1007/s10482-017-0839-1
Kamil, I., Gencbay, T., Zedmir-Kocak, F., and Cil, E. (2014). Molecular identification of different actinomycetes isolated from east black sea region plateau soil by 16S rDNA gene sequencing. Afric. J. Microbiol. 8, 878–887. doi: 10.5897/AJMR2013.6174
Kim, B. S., Surk, S. S., and Hwang, B. K. (2000). Structure elucidation and antifungal activity of an anthracycline antibiotic, Daunomycin, isolated from Actinomadura roseola. J. Agric. Food Chem. 48, 1875–1881. doi: 10.1021/jf990402u
Kumar, S. N., Mohandas, C., and Nambisan, B. (2014). Purification, structural elucidation and bioactivity of tryptophan containing diketopiperazines, from Comamonas testosterone associated with a rhabditidentomopathogenic nematode against major human-pathogenic bacteria. Peptides 53, 48–58. doi: 10.1016/j.peptides.2013.09.019
Kusari, S., Pandey, S. P., and Spiteller, M. (2013). Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochem 91, 81–87. doi: 10.1016/j.phytochem.2012.07.021
Lee, D. S., Eom, S. H., Jeong, S. Y., Shin, H. J., Je, J. Y., Lee, E. W., et al. (2013). Anti-methicillin-resistant Staphylococcus aureus (MRSA) substance from the marine bacterium Pseudomonas sp. UJ-6. Environ. Toxicol. Pharmacol. 35, 171–177. doi: 10.1016/j.etap.2012.11.011
Lee, D. W., and Lee, S. D. (2008). Aeromicrobium ponti sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 58, 987–991. doi: 10.1099/ijs.0.65575-0
Lin, L., and Xu, X. (2013). Indole-3-acetic acid production by Endophytic streptomyces sp. En-1 isolated from medicinal plants. Curr. Microbiol. 67, 209–217. doi: 10.1007/s00284-013-0348-z
Meyers, P. R. (2014). Gyrase subunit B amino acid signatures for the actinobacterial family Streptosporangiaceae. Syst. Appl. Microbiol. 4, 252–260. doi: 10.1016/j.syapm.2013.12.002
Mohseni, M., Norouzi, H., Hamedi, J., and Roohi, A. (2013). Screening of antibacterial producing actinomycetes from sediments of the Caspian Sea. Int. J. Mol. Cell. Med. 2, 64–71.
Ostrosky, E. A., Mirian, K. M., Lima, M. E. L., Kaneko, T. M., Nishikawa, S. O., and Freitas, B. R. (2008). Divulgação da Concentração Mínima Inibitória (CMI) de plantas medicinais. Rev. Bras. Farmacogn. 18, 301–307. doi: 10.1590/S0102-695X2008000200026
Paradis, E. (2010). Pegas: an R package for population genetics with an integrated-molecular approach. Bioinformatics 26, 419–420. doi: 10.1093/bioinformatics/btp696
Passari, A. K., Mishra, V. K., Saikia, R., Gupta, V. K., and Singh, B. P. (2015). Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 6:273. doi: 10.3389/fmicb.2015.00273
Paulus, C., Rebets, Y., Tokovenko, B., Nadmid, S., Terekhova, L. P., Myronovskyi, M., et al. (2017). New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP 131-18. Nature 7:42382. doi: 10.1038/srep42382
Peña, L. C., Jung, L. F., Savi, D. C., Servienski, A., Aluizio, R., Goulin, E. H., et al. (2016). A Muscodor strain isolated from Citrus sinensis and its production of volatile organic compounds inhibiting Phyllosticta citricarpa. J. Plant Dis. Protect. 124, 349–360. doi: 10.1007/s41348-016-0065-5
Petrini, O. (1986). “Taxonomy of endophytic fungi of arial plant tissues,” in Microbiology of Phyllosphere, eds N. J. Fokkema and J. V. Heuvel (Cambridge, UK: University Press), 175–187.
Pickenhagen, W., Dietrich, P., Keil, B., Polonsky, J., Nouaille, F., and Lederer, E. (1975). Identification of the bitter principle of cocoa. Helv. Chim. Acta 58, 1078–1086. doi: 10.1002/hlca.19750580411
Pott, A., Pott, V. J., and Sobrinho, A. A. B. (2004). “Plantas uteis a sobrevivência no Pantanal,” in IV Simpósio sobre Recurso Naturais e Sócio-Econômico do Pantanal (Corumbá, MS), 1–16.
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.R-project.org/
Qin, S., Miao, Q., Feng, W. W., Wang, Y., Zhu, X., Xing, K., et al. (2015). Biodiversity and plant growth promoting traits of culturable endophytic actinobacteria associated with Jatrophacurcas, L. growing in Panxi dry-hot valley soil. App. Soil Ecol. 93, 47–55. doi: 10.1016/j.apsoil.2015.04.004
Raeder, U., and Broda, P. (1985). Rapid preparation of DNA from filamentous fungi. Lett. App. Microbiol. 1, 17–20. doi: 10.1111/j.1472-765X.1985.tb01479.x
Rand, T. G., Giles, S., Flemming, J., Miller, J. D., and Puniani, E. (2005). Inflammatory and cytotoxic responses in mouse lugs exposed to purified toxins from building isolated Penicillium brevicompactum Dierckyx and P. chrysogenum Thom. Toxicol. Sci. 87, 213–222. doi: 10.1093/toxsci/kfi223
Rayle, D. L., and Purves, W. K. (1967). Isolation and identification of Indole-3-Ethanol (Tryptophol) from cucumber seedlings. Plant Physiol. 42, 520–524. doi: 10.1104/pp.42.4.520
Rong, X., and Huang, Y. (2012). Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. App. Microbiol. 35, 7–18. doi: 10.1016/j.syapm.2011.10.004
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., and Darling, A. (2011). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Saini, P., Gangwar, M., Kalia, A., Singh, N., and Narang, D. (2016). Isolation of endophytic actinomycetes from Syzygiumcumini and their antimicrobial activity against human pathogens. J. App. Nat. Sci. 8, 416–422.
Santos, I. P., Silva, L. C. N., Silva, M. V., Araujo, J. M., Cavalcanti, M. S., and Lima, V. L. M. (2015). Antibacterial activity of endophytic fungi from leaves of Indigofera sufruticosa Miller (Fabaceae). Front. Microbiol. 6, 1–7. doi: 10.3389/fmicb.2015.00350
Santos, P. J. C., Savi, D. C., Gomes, R. R., Goulin, E. H., Senkiv, C. C., Tanaka, F. A. O., et al. (2016). Diaporthe endophytica and D. terebinthifolii from medicinal plants for biological control of Phyllosticta citricarpa. Microbiol. Res. 186, 153–160. doi: 10.1016/j.micres.2016.04.002
Savi, D. C., Aluizio, R., Galli-Terasawa, L., Kava, V., and Glienke, C. (2016). 16S-gyrB-rpoB multilocus sequence analysis for species identification in the genus Microbispora. Ant. van Leeuw 109, 801–815. doi: 10.1007/s10482-016-0680-y
Savi, D. C., Haminiuk, C. W. I., Sora, G. T. S., Adamoski, D. M., Kensiki, J., Winnischofer, S. M. B., et al. (2015a). Antitumor, antioxidant and antibacterial activities of secondary metabolites extracted by endophytic actinomycetes isolated from Vochysia divergens. Int. J. Pharm. Chem. Biol. Sci. 5, 347–356.
Savi, D. C., Shaaban, K. A., Vargas, N., Ponomareva, L. V., Possiede, Y. M., Thorson, J. S., et al. (2015b). Microbispora sp. LGMB259 endophytic actinomycete isolated from Vochysia divergens (Pantanal, Brazil) producing B-carbolines and indoles with biological activity. Cur. Microbiol. 70, 345–354. doi: 10.1007/s00284-014-0724-3
Schliep, K. P. (2011). phangorn: phylogenetic analysis in R. Bioinformatics 27, 592–593. doi: 10.1093/bioinformatics/btq706
Shaaban, K. A. (2009). Nafisamycin, Cyclization Product of a New Enediyne, P.recursor, Highly Cytotoxic, M.ansouramycins, Karamomycins Possessing a Novel Heterocyclic, S.keleton, and Further Unusual Secondary Metabolites from Terrestrial and Marine Bacteria. Ph.D. thesis, University of Göttingen.
Shaaban, K. A., Srinivasan, S., Kumar, R., Damodaran, C., and Rohr, J. (2011). Landomycins, P.-W., cytotoxic angucyclines from Streptomyces cyanogenus S-136. J. Nat. Prod. 74, 2–11. doi: 10.1021/np100469y
Shaaban, M., Schroder, D., Shaaban, K. A., Helmke, E., Grun-Wollny, I., Wagner-Dobler, I., et al. (2007). Flazin, perlolyrin, and other β-carbolines from marine-derived Bacteria. Rev. Latinoamer. Quím. 35, 58–67.
Shin, H. J., Lee, H. S., and Lee, D. S. (2010). The synergistic antibacterial activity of 1-acetyl-beta-carboline and beta-lactams against methicillin-resistant Staphylococcus aureus (MRSA). J. Microbiol. Biotechnol. 20, 501–505. doi: 10.4014/jmb.0910.10019
Shiomi, K., and Omura, S. (2004). Antiparasitic agents produced by microorganisms. Proc. Jpn. Acad. Ser. 80, 245–258. doi: 10.2183/pjab.80.245
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Smith, P. A., Roberts, T. C., and Romesberg, F. E. (2011). Broad spectrum antibiotic activity of the Arylomycin natural products is masked by natural target mutations. Chem. Biol. 17, 1223–1231. doi: 10.1016/j.chembiol.2010.09.009
Soltani, J., and Moghaddam, M. S. H. (2014). Diverse and bioactive endophyticAspergilli inhabit Cupressaceae plant family. Arch. Microbiol. 196, 635–644. doi: 10.1007/s00203-014-0997-8
Stahl, E., Bellwon, P., Huber, S., Schlaeppi, K., Bernsdorff, F., Vallat-Michel, A., et al. (2016). Regulatory and functional aspects of Indolic metabolism in plant systemic acquired resistance. Mol. Plant. 9, 662–681. doi: 10.1016/j.molp.2016.01.005
Sung, W. S., and Lee, D. G. (2007). In vitro antimicrobial activity and the mode of action of Indole-3-Carbinol against human pathogenic microorganisms. Biol. Pharm. Bull. 30, 1865–1869. doi: 10.1248/bpb.30.1865
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2013). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tiwari, K., and Gupta, R. K. (2012). Rare actinomycetes: a potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 32, 108–132. doi: 10.3109/07388551.2011.562482
Tonial, F., Gomes, R. R., Gomes-Figueiredo, J., Savi, D. C., Maia, B. H. L. S., and Glienke, C. (2016). Influence of culturing conditions on bioprospecting and antimicrobial potential of endophytic fungi from Schinus terebinthifolious. Curr. Microbiol. 71, 1–11. doi: 10.1007/s00284-015-0929-0
Tonial, F., Maia, B. H. L. N. S., Savi, D. C., Aparecida, V., Gomes, R. R., and Glienke, C. (2017). Biological activity of Diaporthe terebinthifolii extracts against Phyllosticta citricarpa. FEMS Microbiol. Lett. 1, 1–17. doi: 10.1093/femsle/fnx026
Williams, S. T., Sharpe, M. E., and Holt, J. G. (1989). Bergey's Manual of Systematic Bacteriology, Vol. 4. Baltimore, MD: Williams and Wilkins.
Wintersdorff, C. J. H. V., Penders, J., Niekerk, J. M. V., Mills, N., Majumder, S., Alphen, L. B. V., et al. (2016). Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front. Microbiol. 7:173. doi: 10.3389/fmicb.2016.00173
Xing, J., Liu, C., Zhang, Y., He, H., Zhou, Y., Li, L., et al. (2015). Sphaerisporangium dianthi sp. nov., an endophytic actinomycete isolated from a root of Dianthus chinensis L. Anton. van Leeuw 107, 9–14. doi: 10.1007/s10482-014-0298-x
Yan, P.-S., Song, Y., Sakuno, E., Nakajima, H., Nakagawa, H., and Yabe, K. (2004). Cyclo(L-Leucyl-L-Prolyl) produced by Achromobacter xylosoxidans inhibits aflatoxin production by Aspergillus parasiticus. Appl. Environm. Microbiol. 70, 7466–7473. doi: 10.1128/AEM.70.12.7466-7473.2004
Yassin, A. F., Young, C. C., Lai, W. A., Hupfer, H., Arun, A. B., Shen, F. T., et al. (2007). Williamsia serinedens sp. nov., isolated from an oil-contaminated soil. Int. J. Syst. Evol. Microbiol. 57, 558–561. doi: 10.1099/ijs.0.64691-0
Zendah, I., Shaaban, K. A., Helmke, E., Maier, A., Fiebig, H., and Laatsch, H. (2012). Barakacin: A thiazolyl-indole alkaloid isolated from a ruminal Pseudomonas sp. Z. Naturforsch. 67, 417–420. doi: 10.5560/znb.2011-0277
Zhang, Q., Wang, S. Q., Tang, H. Y., Li, X. J., Zhang, L., Xiao, J., et al. (2013). Potential allelopathic indole diketopiperazines produced by the plant endophytic Aspergillus fumigatus using the one strain-many compounds method. J. Agric. Food Chem. 61, 11447–11452. doi: 10.1021/jf403200g
Zhao, K., Penttinen, P., Guan, T., Xiao, J., Chen, Q., Xu, J., et al. (2011). The diversity and anti-microbial activity of endophytic actinomycetes isolated from medicinal plants in Panxi Plateau, China. Curr. Microbiol. 62, 182–190. doi: 10.1007/s00284-010-9685-3
Keywords: actinomycetes, endophytes, Vochysia divergens, pantanal, MRSA, secondary metabolites
Citation: Gos FMWR, Savi DC, Shaaban KA, Thorson JS, Aluizio R, Possiede YM, Rohr J and Glienke C (2017) Antibacterial Activity of Endophytic Actinomycetes Isolated from the Medicinal Plant Vochysia divergens (Pantanal, Brazil). Front. Microbiol. 8:1642. doi: 10.3389/fmicb.2017.01642
Received: 12 May 2017; Accepted: 14 August 2017;
Published: 06 September 2017.
Edited by:
Elizabeth M. H. Wellington, University of Warwick, United KingdomReviewed by:
D. Ipek Kurtboke, University of the Sunshine Coast, AustraliaAtte Von Wright, University of Eastern Finland, Finland
Copyright © 2017 Gos, Savi, Shaaban, Thorson, Aluizio, Possiede, Rohr and Glienke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jürgen Rohr, anJvaHIyQGVtYWlsLnVreS5lZHU=
Chirlei Glienke, Y2hnbGllbmtlQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work.
 Francielly M. W. R. Gos
Francielly M. W. R. Gos Daiani C. Savi
Daiani C. Savi Khaled A. Shaaban
Khaled A. Shaaban Jon S. Thorson3,4
Jon S. Thorson3,4 Yvelise M. Possiede
Yvelise M. Possiede Chirlei Glienke
Chirlei Glienke