- 1Centre for Inflammation, Centenary Institute, Sydney, NSW, Australia
- 2Faculty of Science, University of Technology Sydney, Sydney, NSW, Australia
- 3Discipline of Pharmacy, Graduate School of Health, University of Technology Sydney, Sydney, NSW, Australia
- 4Priority Research Centre for Healthy Lungs, Hunter Medical Research Institute, The University of Newcastle, Newcastle, NSW, Australia
- 5Faculty of Medicine, Thoracic Research Centre, The University of Queensland, Brisbane, QLD, Australia
- 6Department of Thoracic Medicine, The Prince Charles Hospital, Brisbane, QLD, Australia
- 7Woolcock Institute of Medical Research, University of Sydney, Sydney, NSW, Australia
- 8School of Medicine, Western Sydney University, Sydney, NSW, Australia
- 9St George and Sutherland Clinical School, University of New South Wales, Sydney, NSW, Australia
Respiratory diseases such as asthma, chronic obstructive pulmonary disease (COPD), lung fibrosis, and lung cancer, pose a huge socio-economic burden on society and are one of the leading causes of death worldwide. In the past, culture-dependent techniques could not detect bacteria in the lungs, therefore the lungs were considered a sterile environment. However, the development of culture-independent techniques, particularly 16S rRNA sequencing, allowed for the detection of commensal microbes in the lung and with further investigation, their roles in disease have since emerged. In healthy individuals, the predominant commensal microbes are of phylum Firmicutes and Bacteroidetes, including those of the genera Veillonella and Prevotella. In contrast, pathogenic microbes (Haemophilus, Streptococcus, Klebsiella, Pseudomonas) are often associated with lung diseases. There is growing evidence that microbial metabolites, structural components, and toxins from pathogenic and opportunistic bacteria have the capacity to stimulate both innate and adaptive immune responses, and therefore can contribute to the pathogenesis of lung diseases. Here we review the multiple mechanisms that are altered by pathogenic microbiomes in asthma, COPD, lung cancer, and lung fibrosis. Furthermore, we focus on the recent exciting advancements in therapies that can be used to restore altered microbiomes in the lungs.
Introduction
The lungs are vital organs that facilitate gas exchange during respiration. They are highly susceptible to disease risk factors such as exposure to air pollution, occupational exposure to toxins, cigarette smoke and infections. Hence, the burden of chronic lung diseases (CLDs) is rising (1–4). Lung diseases are the second leading cause of death worldwide (after cardiovascular disease) and account for more than 10% of disability-adjusted life years (5). Asthma, chronic obstructive pulmonary disease (COPD), idiopathic pulmonary fibrosis (IPF), and lung cancer are the most prevalent CLDs globally.
Briefly, asthma and COPD are chronic inflammatory diseases with different pathophysiology. The former is characterized by repeated episodic symptoms like shortness of breath and wheezing upon exposure to allergen, while the latter presents with chronic inflammation and irreversible airflow limitation (6–11). Around 334 million individuals suffer from asthma alone and the global prevalence of the disease has increased by 12.6% between 1990 and 2015 (12). Likewise, with a 44.2% increase in incidence, COPD is now the third leading cause of death worldwide (12, 13).
IPF is a progressive lung disease with a median survival rate of 3–5 years after diagnosis (14–16). Recent studies show that the rate of IPF is ~3–9 per 100,000 for Europe and North America, with limited data available on global prevalence (17, 18). It is characterized by progressive fibrosis of the lungs with an unknown cause leading to impaired forced expiratory volume, vital capacity and mortality in patients with an advanced stage of disease (19).
Lung cancer is one of the most commonly diagnosed cancers worldwide. Lung cancer is categorized into two major subtypes; 85% are non-small cell lung cancers (NSCLCs) and 15% are small cell lung cancers (20). Currently, lung cancer accounts for 18.4% of total cancer related deaths and therefore imparts a huge socio-economic burden on society (21). Furthermore, the prevalence of lung cancer is expected to rise due to the continued increase in risk factors such as smoking.
The causes of asthma, COPD, IPF and lung cancer are multifactorial and influenced by both genetics as well as the environment. Some of the known causes include long-term exposure to environmental and occupational hazards such as allergens, cigarette smoke, air pollution, asbestos, silica, coal dust, beryllium, hard metals, and radiation treatments (22, 23). These environmental factors along with genetic predisposition together drive the development and progression of lung disease.
Despite the increasing burden of these lung diseases, current treatment options are limited. In asthma, β2 agonists and inhaled corticosteroids remain as the first-line treatments, however 5–25% of patients fail to respond to steroids (6, 24, 25). Likewise, there is no effective therapy for COPD that stops or reverses the progressive nature of the disease and current disease management strategies primarily aim to treat associated symptoms (3, 26). For IPF, international guidelines for the management and treatment are lacking, with patients often limited to the prescription of anti-fibrotic drugs (nintedanib and pirfenidone) and broad spectrum antibiotics (azithromycin and polymyxin) (27, 28). For lung cancer, the overall 5-year survival rate was <5% for NSCLC until the last decade when the advent of immunotherapies raised this to 20–30% (29–31). Unfortunately, only 21% of NSCLC patients and 3.7% of small cell lung cancer patients are eligible for immunotherapy (32). Hence, the development of better treatment strategies is urgently needed, which can be achieved through improved understanding of the pathogenesis of CLDs.
There is growing interest in understanding host-microbiome interactions and manipulating the lung microbiome by targeting specific microbes or microbial products such as toxins and metabolites as potential novel treatments. There have been major recent advances in the understanding of the molecular and cellular mechanisms of CLDs, and new research is continuously revealing that many of these processes are driven by interactions with the respiratory microbiome (6, 33, 34). Metagenomics is the investigation of the collective genomic material in an environment (35). DNA libraries are prepared from samples such as bronchoalveolar lavage fluid (BALF) and tracheal aspirates, sequenced using Miseq (next generation sequencing technology) and analyzed using metagenomics tools. This approach provides a broad scale analysis of microbial species, their distribution, metabolic properties and impact on disease. Several factors influence bacterial persistence in the lower respiratory tract including; oxygen gradients, nutrient availability, temperature, pH and surfactants such as phosphatidylcholine-containing lipids (primarily dipalmitoyl phosphatidylcholine) and sphingomyelins (36). Furthermore, metagenomics can be used for molecular diagnostics and surveillance of pathogens in diseases (37).
One of the key characteristics used to assess the microbiome is diversity: the number of different species (richness) and the abundance of different species (evenness). Diversity of bacterial species can be measured within samples, which is known as α-diversity (using Simpson or Shannon index) or between samples, which is known as β-diversity (using principal coordinate analysis) (38). Bacterial taxa found in healthy individuals include Bacteroidetes and Firmicutes comprising of Veillonella, Lachnospira, and Rothia that have been detected in nasopharyngeal swabs, oral washes or bronchoalveolar lavage (39). A microbial imbalance, known as dysbiosis, has been associated with the development of several diseases including lung diseases (40).
Recent studies have highlighted the modulatory role of the microbiome in inflammation and inflammatory diseases. The term “microbiome” refers to the aggregate of all the commensal microbiota present on or in a particular host, including viruses, fungi, bacteria, archaea, and protozoa (1). The human microbiome is highly diverse, with distinct microbial profiles at different anatomical sites, and has been implicated in disease. For instance, it has been well established that that microbiome in the gut plays a distinct role in the pathogenesis of diseases such as irritable bowel syndrome, ulcerative colitis, allergy (e.g., atopic dermatitis) and obesity (41–45). Accordingly, recent research suggests that the resident lung microbiome is altered during the development of lung diseases, which allows for the colonization of pathogenic bacteria. Several bacterial species such as Legionella (46), Escherichia coli, H. influenzae, S. pneumoniae, Enterobacter spp. (47), and Moraxella spp. (48) can initiate lung inflammation. Until recently, the lungs of healthy individuals were considered to be sterile as culture-dependent techniques were unable to detect the presence of bacteria. However, with the development of culture-independent techniques, microbes of different phyla and genera have been found in the lungs and are associated with lung disease (49). The alteration in respiratory microbiomes during CLDs and how they are affected by challenges such as indoor/outdoor air pollution, biomass/cigarette smoke, and pathogenic infections are being recently elucidated. The immunological crosstalk between gut and lung, termed as “gut-lung axis” can impact the health of the lung as microbiome in lung are altered in CLDs. This involve activation of numerous factor such as inflammasomes and microRNAs (4). It is now well-established that the primary microbiome phyla residing in healthy lungs include Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria and others (49). Pathogenic microbes, such as Pseudomonas aeruginosa and Haemophilus influenzae, have the capacity to trigger different immune responses. Thus, it may be beneficial to examine the role of an altered lung microbiome in the pathogenesis and progression of lung diseases (1). Accordingly, in this review, we will discuss recent updates regarding the role of the lung microbiome in the context of COPD, asthma, IPF and lung cancer.
Lung Microbiome and Asthma
Asthma affects >330 million people worldwide and is a major healthcare issue. The annual increase in asthma incidence was 1.4% for children and 2.1% for adults between 2011 and 2015 (24, 50). It is a complex and heterogeneous inflammatory disease with variable phenotypes. Classically, asthma is characterized by increased type 2 immune responses and eosinophilic inflammation (51). However, recent studies have linked elevated type 1/17 immune responses with non-eosinophilic (i.e., neutrophilic) inflammation especially in adults with moderate to severe asthma (24, 51, 52). Interestingly, exogenous factors including respiratory viral and bacterial infections, a high-fat diet and/or obesity, air pollution and cigarette smoke exposure are associated with severe asthma in adults as well as with exacerbations of the disease (24, 53–56).
Unfortunately, 5–10% of asthmatics do not respond to steroid therapies and are more likely to have severe disease. Severe steroid-resistant (SSR) asthma is a condition where asthmatic patients do not respond to mainstay corticosteroid therapies (57). SSR is associated with non-eosinophilic endotypes of the disease, including neutrophilic asthma, and involves activation of innate immune responses, particularly those mediated by Toll-like receptor (TLR)2 and TLR4 responses, and NLRP3 (nucleotide-binding oligomerisation domain-like receptor family, pyrin domain-containing 3) inflammasome/interleukin (IL)-1β responses (11, 25, 56, 58–60). The lung microbiome has been linked SSR asthma. Recently, Durack et al., showed that variations in bronchial microbiome composition is associated with the immunological, steroid-responsiveness and clinical features of asthma. Patients with asthma had an enrichment of Haemophilus, Neisseria, Fusobacterium, Porphyromonas and Sphingomonodaceae, and a depletion of Mogibacteriaceae and Lactobacillales. The study also showed distinct changes in specific microbial members after fluticasone treatment, demonstrating that steroid treatment can alter the microbiome which may influence steroid-responsiveness (61). Accordingly, researchers have shown that pathogenic microbes such as Haemophilus spp. and Chlamydia infections induce the development of SSR experimental asthma when combined with mild to moderate asthma models (62–67). Therefore, a thorough understanding of the lung microbiome may be essential to understand the immunological differences between asthma endotypes and how to treat them.
Several studies have characterized the bacterial profile of the lung microbiome in asthma. In asthma there is an elevated abundance of Haemophilus influenzae, Streptococcus pneumoniae, Staphylococcus aureus, and Moraxella catarrhalis in nasopharyngeal swabs, compared to healthy controls. While frequently detected as part of a normal microbiome, these bacteria are known pathogens that can cause infectious exacerbations (35). Furthermore, across two studies, Huang et al., showed that Actinobacteria can have a high abundance in patients with severe asthma, which correlate with disease outcomes, including elevated sputum leukocytes and eosinophils in bronchial biopsies (68), and the abundance of Comamonadaceae, Sphingomonadaceae, and Oxalobacteraceae in asthma patients correlates with airway hyperreactivity (69). Denner et al., showed that bacterial α-diversity in endobronchial brushings of asthmatic subjects corresponds with eosinophil numbers in lavage as well as forced expiratory volume in 1 s (FEV1) (70). The alterations in lower airway microbiomes of patients with asthma also correlated with FEV1 as a biomarker of airflow obstruction. Patient with asthma with FEV1 < 60% had low α-diversity but high β-diversity, low abundance of Firmicutes, Bacteroidetes, and Actinobacteria and potential pathogenic genus Streptococcus and the commensal genera Veillonella and Prevotella, compared to asthma patients with an FEV1 > 80% (70). Overall, these studies demonstrate that the lung microbiome has an impact on patient outcomes in asthma.
In addition to patient outcomes, the lung microbiome is associated with systemic and bronchial markers of inflammation (71). In the lower airway microbiome of patient with severe asthma, pathogenic microbes Moraxella catarrhalis, Haemophilus spp., and Streptococcus spp. are predominate and their presence correlates with cytokine levels (IL-8), eosinophilia, and neutrophilia (72, 73). S. pneumoniae is another pathogenic microbe that is associated with asthma, but conflicting reports are available regarding its influence. In a mouse model of allergic airway disease, it was reported that mice infected with non-lethal S. pneumoniae as neonates before sensitization and challenge with ovalbumin (OVA) had increased interleukin-17A (IL-17A) production, elevated neutrophil recruitment and Th17 cells (74). However, we previously showed that exposure to non-pathogenic S. pneumoniae, through either infection, killed bacteria or its components, can protect against allergic airway disease in adult mice through the modulation of dendritic cells, natural killer cells, and the induction of regulatory T cells (Tregs) (75–79). It would be interesting to continue this work and examine the role of microbial dysbiosis on the development of SSR asthma.
Microbial metabolites, such as short chain fatty acids (SCFAs) acetate, propionate and butyrate, regulate the physiology and immune response in humans (80). SCFAs are produced by the gut microbiome via fermentation of dietary fibers and are distributed across the body through the bloodstream. SCFAs are utilized by the body to provide energy or as signaling molecules (81, 82). SCFAs inhibit histone deacetylases (HDACs) and promote anti-inflammatory cell phenotypes (i.e., neutrophils) that maintain homeostasis, and suppress nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) and tumor necrosis factor-α (TNF-α) production (80, 83, 84). Cait et al., identified SCFAs and the depletion of SCFA-producing bacteria as a mechanistic link between the microbiome and asthma susceptibility or severity. Exacerbation of OVA-induced allergic lung inflammation by vancomycin treatment was due to the lack of gut microbiome populations responsible for producing SCFAs, particularly butyrate. This work highlighted the potential of probiotics as a novel therapy for asthma (85). Hougee et al., utilizing OVA induced mouse model of asthma reported that oral administration of bacteria Bifidobacterium breve M-16V and Lactobacillus plantarum NumRes8 reduced the eosinophilic inflammation, OVA-specific IgE levels, and cytokines production (86). Likewise, the protective effects of administration of Lactobacillus rhamnosus GG and Lactobacillus paracasei L9 have been reported against the birch pollen-induced allergic asthma and urban airborne particular matter 2.5 μm-induced enhancement of airway hyperresponsiveness in the mouse model of asthma (87, 88). Huang et al., evaluated the effect of Lactobacillus aracasei, Lactobacillus fermentum or their combination on immune biomarkers, clinical severity and quality of life in 160 children aged 6–18 years of age with asthma in a double-blind, prospective, randomized, placebo-controlled trial. Serum IFN-γ, IL-4, IgE and TNF-α levels were evaluated for immune biomarkers, but only levels of IgE decreased significantly in the combination treatment group. Combined L. aracasei and L. fermentum also significantly improved quality of life. All intervention groups had lower levels of Clostridium compared to controls (89). This suggests that colonization by other microorganisms, and increasing gut microbial diversity, are potentially other protective factors against asthma (90). However, further studies are required to better understand the role of the microbiome in asthma and to define the potential for therapeutic manipulation.
Lung Microbiome and COPD
The prevalence of COPD is increasing, and its management is a formidable challenge to healthcare systems. There is growing evidence that the lung microbiome is altered during the course of the disease and contributes to COPD pathogenesis (91–93). Hilty et al., provided early evidence of the presence of dysbiosis in COPD. 16S rRNA analysis was performed on DNA collected from swabs from the nose and oropharynx, and bronchial brushings from the left upper lung lobe of COPD patients and healthy controls. Sequencing showed that COPD patients have a distinct microbiome in their lungs compared to healthy individuals. Specifically, pathogenic Proteobacteria (Haemophilus spp.) were more common in COPD than Bacteroidetes, with Prevotella spp. being especially reduced (94). Subsequent studies showed the predominance of Firmicutes, Bacteriodetes, Proteobacteria, Fusobacteria, and Actinobacteria in healthy individuals, in contrast to pathogenic Haemophilus, Streptococcus, Klebsiella, Pseudomonas, and Moraxella in COPD patients (91–93, 95, 96). We propose that the lung microbiome is dynamic and transient, with specific taxa being present at different stages of disease progression and impacted by clinical factors such as age, smoking, medications, or seasonal changes (33, 34).
Cigarette smoking is a major risk factor for many lung diseases. Hence, researchers have investigated the effects on the lung microbiome. A growing number of reports indicate that smoking alters the oral and respiratory bacterial microbiome (97–99). Limited studies have been carried out in the context of COPD. Erb-Downward et al., assessed BALF samples from smokers with COPD, healthy smokers and non-smokers and showed notable β-diversity (i.e., differences in bacterial evenness) between the cohorts which is likely caused by outgrowth of bacteria within the population, rather than colonization of different bacteria (91). However, contradictory results were found in a subsequent study by Sze et al., utilizing lung tissue samples from non-smokers, smokers without COPD, and COPD patients where there was low diversity between non-smokers and healthy smokers, and a high β-diversity compared to COPD (smokers & non-smokers) (100). The disparity in outcomes may be due to differences in samples used (BALF vs. lung tissue) and small sample size. Nevertheless, these studies suggest that cigarette smoke is associated with alterations in lung bacterial microbiomes in healthy smokers and COPD patients. Whether these changes in bacterial populations are associated with the development and progression of COPD still needs to be determined.
In addition to the impact of cigarette smoke on bacterial populations, a recent study reported that cigarette smoke alters the lung virome. In BALF samples from smokers and non-smokers, bacterial communities were found to be similar among the two groups; however, significant differences were found in the lung virome, which correlated with levels of IL-8 and arachidonic acid. Both IL-8 and arachidonic acid are involved in COPD pathogenesis. The former is a potent neutrophil attractant and the pro-inflammatory effects of the latter have been reported in COPD (101, 102). In smokers, bacteriophages infecting Prevotella, Actinomyces, Aeromonas, Capnocytophaga, Haemophilus, Rodoferax, and Xanthomonas were increased, while Enhydrobacter and Morganella bacteriophages were dominated in non-smokers (103). Future studies are required to address: (1) which microbiome populations (bacterial, viral, fungal) in the lung are influenced by smoking, and; (2) how changes in the microbiome contribute to the susceptibility and progression of COPD.
Furthermore, smoking can indirectly effect the lung microbiome by altering the extra-cellular matrix (ECM) (104–107). H. influenzae, S. pneumoniae, Pseudomonas, and Moraxella are frequent pathogenic colonizers in COPD patients (108). During non-typeable H. influenzae (NTHi) infections, the ECM proteins fibronectin, laminin, collagen IV, and vitronectin play critical roles in colonization. Briefly, NTHi binds to the host vitronectin via bacterial protein E resulting in subsequent intracellular invasion. Further, high affinity of Hap (one of the bacterial adhesins) has been reported for fibronectin, laminin, and collagen IV, which eventually aid NTHi invasion (109, 110). Likewise, during lung infections by Streptococcus, Pseudomonas, and Moraxella, laminin, collagen I, V, and VI are involved in adhesion and invasion (111). Since the levels of ECM proteins (fibronectin/fibrinogen/collagen/laminins/fibulin-1c) are altered in patients with COPD, relationship between smoking, pathogenic microbes and ECM need to be explored (107, 112–114).
Research investigating the mechanism by which the lung microbiome can influence the COPD-linked immune response is in its infancy. Studies have shown pathogenic microbes cause lung inflammation in healthy individuals (115–118). The immune-stimulatory capacity of Gram-negative pathogenic bacteria associated with COPD (Haemophilus spp. & Moraxella) is significantly higher than Gram-negative commensal bacteria (Prevotella spp.) (119). Further, poor oral health is an important risk factor for inflammatory lung diseases (120). Studies have shown that microaspiration of oral taxa (particularly Veillonella and Prevotellla) is associated with elevated Th17 lymphocytes in lungs (115, 121). Thus, dysbiosis in the COPD lung may be associated with COPD exacerbations (122). Epithelial cells and alveolar macrophages form the first line of defense against pathogens. They express pattern recognition receptors that recognize pathogen-associated molecular patterns to initiate the innate immune response. H. influenzae interacts with TLR2 and activates NF-κB (the master regulator of pro-inflammatory response) via translocation dependent and independent pathways. In the translocation dependent pathway, activation of NF-κB inducing kinase (NIK)–IKKα/β complex leads to KiBα phosphorylation and there is degradation. The NF-κB translocation-independent pathway occurs through activation of MKK3/6–p38 mitogen-activated protein (MAP) kinase pathway (123, 124). In addition to TLR2, TLR4 induced NF- κB activation has also been reported in context of H. influenzae (125, 126). Like H. influenzae, P. aeruginosa is recognized by pattern recognition receptors. P. aeruginosa activates NF-κB via TLRs through both TIRAP-dependent and independent pathways. Additionally, this bacterium is involved in the activation of MAPK cascades (127). Thus, pathogenic microbes induce inflammatory responses that are part of the natural defense against foreign pathogens. However, in COPD patients, excessive, persistent and chronic inflammation is a major driver of disease pathogenesis. The activation of TLRs by the presence of pathogenic microbes can further aggravate the inflammatory state in the lungs of COPD patients (128). This eventually results in tissue damage and the production of damage-associated molecular patterns that further activate inflammatory pathways, leading to a vicious cycle of pathogenesis (129). Our recent study utilizing TLR2- and TLR4-deficient mice showed that TLR2 and TLR4 play opposing roles in COPD pathogenesis. TLR2 inhibition increased COPD features in an experimental mouse model, while these features were reduced in the absence of TLR4 (128). Thus, modulation of TLRs appear to be a potential strategy to suppress COPD and bacteria-induced exacerbations. However, future studies examining the relationship between TLRs expression, bacterial load, and lung inflammation area required. Further, the protective role of TLR-2 and 4 through activation of innate immune system should not be ignored.
Additionally, studies have been carried out for the analysis of microbiome dynamics associated with COPD exacerbations. Wang et al., and Mayheve et al., utilizing sputum samples from the COPD subjects reported that Moraxella abundance is increased during COPD exacerbation compared to stable COPD subjects. However, Haemophilus abundance did not show significant change between stable and exacerbated COPD individuals. Both Haemophilus and Moraxella are known to form biofilms that protect them from antibiotics and immune cells, thus their persistence is associated with repeated bacterial exacerbations (122, 130). It is important to mention here that different sampling procedures like BALF, sputum, lung tissue explants represent microbiome from different respiratory regions. Hence, future studies utilizing BALF and lung tissue samples should be carried out to study microbiome dynamics with different stages of COPD. Further, thresholds should be determined to identify significant bacterial over-representations for all sample types.
Thus, a limited number of studies have explored the role of lung dysbiosis in COPD but it is difficult to draw any conclusions. Although they are valuable for our understanding, there are issues with existing studies such as the use of different samples (BALF vs. lung tissues), limited numbers of participants, low biomass and potential for contamination. Further studies are required to elucidate the role of the lung microbiome in the pathogenesis and progression of COPD. Additionally, studies evaluating the impact of TLR-agonists should be performed.
Lung Microbiome and Lung Cancer
The main causative factors in lung cancer are exposure to carcinogens, cigarette smoke, toxic compounds in the environment/industry, chronic airway inflammation driven by pathogenic infections, and fibrosis/scarring from co-morbid lung disease (30, 131). The lung microbiome may be altered by these causative factors and has been linked to lung cancer progression, phenotype and severity.
Research has shown that specific species/genera/phyla of bacteria are associated with lung cancer progression; specifically, Helicobacter pylori (132), Acidovorax temporans (133), Cyanobacteria (134), Actinobacteria, Bacteroidetes, Proteobacteria, Firmicutes (135), Cytomegalovirus (136), Thermus, Legionella, Megasphaera, Veillonella (137), Capnocytophaga, Neisseria and Selenomonas (138). The specific pathogenic mechanisms of microbiome-mediated lung cancer progression are not widely known include the effects of bacterial toxins such as lipopolysaccharide (LPS) and inflammatory cytokine release by immune cells. The LPS of H. pylori can stimulate the production of pro-inflammatory factors including TNF-α, IL-1, and IL-6. These inflammatory mediators promote chronic lung diseases such as COPD and bronchitis that are followed by lung cancer (133). Various other bacterial toxins also have crucial roles in tumor initiation and progression. Cytolethal distending toxin (CDT), Bacteroides fragilis toxin and cytotoxic necrotizing factor-1 can damage DNA repair machinery leading to tumorigenesis (133, 139). The toxin microcystin from Cyanobacteria was associated with diminished CD36 and upregulation of poly ADP ribose polymerase 1 (PARP1) levels in silico, which was confirmed in human lung epithelial carcinoma cells (A427) in vitro and in human adenocarcinoma samples (134). Similarly, heat-inactivated E. coli stimulated TLR4 and induced NSCLC metastasis and adhesion in vivo. These effects were specifically induced through extracellular signal-regulated kinase (ERK)1/2 and p38 mitogen-activated protein kinases (MAPK) pathways (140).
Some bacterial species directly promote the development of lung cancer while some are found in lung cancer patients after tumor has progressed. A pooled meta-analysis study suggests that H. pylori promotes lung cancer progression by 3.24-fold compared to controls (without known history of H. pylori infection and lung cancer) (141). Another meta-analysis suggests that previous lung disease with pneumonia and Mycobacterium tuberculosis could significantly increase lung cancer risk (142). Similarly, chronic infections with Chlamydia pneumoniae are also associated with increased risk in male patients aged ≤55 years (143). D'Journo et al., investigated the association of the lung microbiome and cancer through RT-PCR of 16S rRNA genes of bacteria in lung cancer patients. Microbiological examination was carried out in lung sections of patients selected for major lung surgery. This study showed that Cytomegalovirus were abundant in the non-cancer distal airway and parenchyma of lung sections (144). Similarly, Lee et al., investigated the lung microbiome in the BALF samples from lung cancer patients as well as those with benign mass-like lesions. 16S rRNA sequencing of BALF samples showed that two phyla (TM7 and Firmicutes) were more abundant in lung cancer. Also, the genera Veillonella and Megasphaera were comparatively higher in lung cancer patients, suggesting that dysbiosis may be a biomarker for lung cancer (137).
The association of the commensal microbiome with development of lung cancer was further elaborated in a nested case control study of 4,336 subjects with lung cancer and 10,000 controls. This study was performed with subjects 40–84 years of age to find links between the use of antibiotics and cancer risk. The relative risk of lung cancer was 2.52 (95% CI, 2.25–2.83) in subjects receiving ≥10 antibiotics compared to controls with no antibiotics. The higher relative risk was probably because of inflammatory conditions due to frequent infection and alterations in lung microbiomes in patients taking antibiotics (145).
The lung microbiome is critical in maintaining immune homeostasis in the airway mucosa. Alterations in the normal lung microenvironment can have deleterious effects including tumor progression. An in vivo mouse model study showed that antibiotic treatment makes mice more prone to the formation of engrafted Lewis lung carcinoma and B16/F10 melanoma, resulting in increased tumor multiplicity and sizeable tumor foci in the lungs and reduced mean survival. Treatment resulted in the malfunction of γδT17 cell responses in the lungs, leading to more invasive tumor progression. Restoration of altered immune surveillance in antibiotic treated mice could be achieved by adding IL-17 producing γδT cells and by supplementation with IL-17. The promotion of lung cancer development by commensal bacteria is mediated by activating lung-resident γδT cells to provoke inflammation (146). This recent discovery in mice introduced a new cancer-promoting mechanism for microbiome-activated γδT17 cells in lung cancer, highlighting the complex balance between inflammation and immune surveillance in the lung. Collectively, these studies highlight the significance of commensal bacteria in strengthening the immune response and protecting against lung cancer (147).
In addition to lung cancer progression, the lung microbiome can be linked to cancer phenotype. More specifically, the distribution of bacterial species can vary between different lung cancer subtypes. Comamonas, Rhodoferax, Acidovorax, Polarmonas, and Klebsiella are often observed in squamous cell carcinoma (SCC) but not adenocarcinoma (133). Apopa et al., investigated the microbiome composition in lung squamous cell carcinoma, adenocarcinoma and healthy control tissues and found that the phylum Cyanobacteria was persistent in adenocarcinoma samples (134). The potential association between salivary microbiome and lung cancer occurrence was evaluated in subjects with lung adenocarcinoma and squamous cell carcinoma. Deep sequencing analysis of the salivary microbiome of these lung cancers were compared with healthy controls, which showed that the genera Veillonella, Capnocytophaga, Neisseria, and Selenomonas were remarkably higher in subjects with cancer compared to controls (138). A recent clinical study of 143 lung cancer patients and 33 healthy controls revealed the presence of a distinct lung microbiome in the cancer group compared to controls. Particularly, in patients with squamous cell carcinoma, Acidovorax temporans was detected in tumor tissues by fluorescent in situ hybridization. This was further confirmed with 16S rRNA analysis where Acidovorax dominated in squamous cell carcinoma with mutations in the TP53 gene, but was not observed in adenocarcinoma (133).
The characterization of lung microbiomes in lung cancer patients show heterogeneity among various stages of disease. Analysis of seven and 151 patients with stage IV and I-IIIA stage lung cancer, respectively, showed that the genus Thermos was more abundant in stage IV (46). Huang et al., analyzed the differential taxonomy of squamous cell carcinoma with (SCC_M1) or without metastasis (SCC_M0) and lung adenocarcinoma with (AD_M1) or without metastasis (AD_M1). In stage I-III, the phylum Firmicutes and genera Veillonella, Megasphaera, Actinomyces and Arthrobacter were increased in AD_M0 compared to SCC_M0. In stage IV, the genera Capnocytophaga and Rothia were decreased in AD_M1 compared with SCC_M1. Interestingly, no difference was observed in these 6 genera among former smoking and non-smoking groups indicating that smoking was not responsible for the variation in the differential genera (148).
Recently, researchers have began focusing on opportunities to treat lung cancer by manipulating the lung microbiome. Targeting the microbiome and their products (toxin/metabolites) could be a promising approach as adjunct therapies or alone as suggested by various in vivo and in vitro studies (149). Oral administration of Enterococcus hirae and Barnesiella intestinihominis enhanced the therapeutic efficacy of cyclophosphamide and increased the survival in lung cancer patients. Bacteria specific memory Th1 cell immune responses selectively predicted the longer progression free survival in patients with advanced lung cancer (150). Another in vivo study in mice evaluated the efficacy of an antitumor vaccine prepared from B. subtilis B-7025 in experimental models of solid sarcoma 37 (S37) and metastatic Lewis lung carcinoma. The vaccine was prepared on the basis of lectines from B. subtilis B-7025 and its synergetic effects tested with concurrent administration of a probiotic mixture of Enterococcus faecium K-50 and Saccharomyces cerevisiae 14K, compared to vaccine alone. Combination therapy resulted in potent synergistic anti-tumor effects in S37-bearing mice. Moreover, there was 2 to 2.5-fold inhibition of metastasis of Lewis lung carcinoma compared to vaccine alone (151). Similarly, an in vitro study in a lung cancer cell line (SK-MES-1) showed probiotics Lactococcus lactis KC24 and NK34 from Korean fermented food Kimchi resulted in 86.53 ± 0.96% and 96.71 ± 0.00% reduction in lung cancer cell viability, respectively (152, 153). Taken together, these data show that targeting the microbiome or its metabolites could be a novel way to manage lung cancer progression. However, more in vitro and in vivo studies are essential to validate and explain the evidence-based beneficial roles of probiotics in lung cancer.
Lung Microbiome and IPF
Recently, a potential role of microbiomes in IPF has been reported (154, 155). Specifically, it was shown that changes in bacterial diversity were related to disease progression. However, how the microbiome influences respiratory function and how it can be used as a clinical marker in IPF remains to be elucidated. Some potential roles of the microbiome in the progression of IPF have been reported (156). Dysbiosis and the colonization of pathogenic bacteria can cause epithelial cell injury and activate immune responses leading to pro-inflammatory and pro-fibrotic cascades resulting in structural changes in the lungs (157). Current therapies for IPF are limited and glucocorticoids or immunosuppressive agents are associated with increased risk of death and hospitalization. Fortunately, antibiotics can improve quality of life and reduce mortality suggesting that the microbiome may have a role in the pathogenesis of the disease (158–160).
Interestingly, studies have reported that BALF from IPF patients have changes in the lung microbiome due to changes in the abundance of the phyla Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria. Specifically, there was a decrease in diversity due to an increase in Firmicutes and a decrease in Proteobacteria (161). This loss of diversity correlated with IPF symptoms including low forced expiratory and vital capacity, high serum surfactant protein-D and lactate dehydrogenase (both clinical biomarkers of IPF progression). The decrease of lung bacterial diversity correlated with pro-inflammatory and pro-fibrotic cytokines in the alveoli in IPF patients, suggesting that loss of diversity may have some impact on pathogenesis (156).
A higher bacterial burden has been observed in patients with IPF who had minor allele at the promoter of the mucin 5B (MUC5B), a gene that encodes a mucin family of protein which composes mucus, suggesting that the bacterial burden and MUC5B mutation are linked mechanistically (154, 162–165). This suggests that MUC5B may play a key role in IPF pathogenesis. Moreover, the development and progression of IPF has been associated with single nucleotide polymorphisms in microbially linked genes such as Toll-interacting proteins, which operate as an adaptor protein for TLRs that mediate microbe-host interactions (165, 166). Some studies found that BALF neutrophilia and expression levels of genes involved in host defense response (NLRC4, PGLYRP1, MMP9, DEFA4) as well as encoding antibacterial peptides (SLPI and CAMP) were increased suggesting a correlation with both increased bacterial burden and disease progression in IPF (167). The absence of secretory leukocyte protease inhibitor (SLPI), a host protein implicated in IPF, leads to the impairment of collagen gene expression in a mouse model of bleomycin-induced lung fibrosis (168). This suggests that there is a potential mechanistic link between the microbiome and lung fibrosis. During IPF exacerbations, patients have increased numbers of neutrophils in BALF due to increased bacterial burden, suggesting that the microbiome might play a causative role in exacerbations (169). Overall, these studies suggest that genetics and transcriptomic alterations in innate immune response genes are all associated with increases in bacterial load in IPF and its progression. This increase in bacterial burden is associated with increased epithelial cell damage, which leads to impaired lung function (170, 171). Interestingly, study demonstrate that fibroblasts obtained from IPF patients who had an altered microbiome or increased microbial load showed enhanced expression of the α-smooth muscle actin gene, which is a marker of lung fibrosis (172). Moreover, it has been reported that lung dysbiosis induces pro-inflammatory cytokines, including IL-17B and TNF-α, that cooperatively activate lung epithelial cells leading to inflammation and fibrosis (160), suggesting that there is a strong mechanistic link between the microbiome and fibrogenesis.
Despite these studies demonstrating the association between the microbiome and fibrogenesis, key questions remain unanswered. Firstly, is microbial diversity or the abundance of specific species responsible for the development and progression of IPF. Secondly, can the lung microbiome serve as a biomarker for the development and progression of IPF. These questions remain challenging and require further study.
Discussion and Future Perspectives
Numerous studies have shown important roles for the lung microbiome in both the maintenance of respiratory homeostasis and pathogenesis of CLDs (1, 4, 34). Thus, there is an exciting opportunity to target the lung microbiome for the treatment of CLDs. Even though microbiome modulation as a therapy for CLDs is currently limited, antibiotics, anti-inflammatory agents, probiotics and diet can help significantly in reducing disease exacerbations (33, 173, 174). Importantly, proper analysis and understanding the spectrum of the effects of the lung microbiome creates new avenues for the development of targeted therapies as well as prognostic and diagnostic markers for CLDs (34).
The universal adoption of culture independent techniques including 16s rRNA gene sequencing and now metagenomics has advanced the study of the lung microbiome in recent years. However, caution is needed when applying these techniques as laboratory contamination from reagents can impact the analysis of microbiome sequencing (175, 176). Indeed, contamination can heavily compromise the consistency of microbiome data, specifically in low microbiome biomass samples such as the lungs. A study investigating the extent of contamination attributed by consumables such as the PCR master mix and DNA extraction kits during 16S rRNA gene sequencing revealed that the master mix was a prime source. This was supported by a significantly reduced blank signal and improved precision by enzymatic removal of contamination (177). Thus, there is a need to optimize and standardize methods of DNA extraction from various sample types when sequencing the microbiome.
It is well known that there is a close relationship between the lung microbiome and inflammation and innate immune responses (178–181). LPS is a classical bacterial component that can induce inflammation in the lungs, which activates innate immune responses in humans and mice (182). LPS acts via TLR4 and induces proinflammatory signaling in COPD as well as in asthma (183–185). Other bacterial components like peptidoglycans and lipoteichoic acids in Gram-positive bacteria, act synergistically to induce lung inflammation by neutrophilic influx and by release of IL-6 (186–188). Likewise, outer membrane vesicles derived from lung commensal microbes (Bacteroides and Prevotella) can stimulate IL-17B production through TLR-MyD88 adaptor signaling to promote IPF (160).
The main therapy for asthma includes anti-inflammatory corticosteroids, which are generally administered in combination with short- or long-acting β2-adrenoceptor agonists (bronchodilators) (24, 189). Although combination therapies improve symptoms by suppressing type 2 cytokine responses, inflammation and by dilating the airways, they possess nonspecific anti-inflammatory activity and have long term adverse side effects if administered at higher doses (6). In asthma, the increased load and alterations to the composition of the lung microbiome is correlated with elevated eosinophil and neutrophil numbers in the bronchi, and IL-1β, IL-6, IL-17A, IL-8, IL-12, and TNF-α levels in sputum (190). A healthy microbiome during the early stage of life plays a key role in lung development, reducing the possibility of the development of asthma (191), providing impetus for giving probiotics and prebiotics in early life to improve the microbiome (192). Pre-clinical investigations into the therapeutic potential of pro- and prebiotics have shown that altering the microbiome can influence overall immunity and reduce allergic inflammation and sensitization, suggesting the protective effect of pre- and probiotics for asthma (193). Furthermore, macrolide-based antibiotics have been studied as a long-term treatment option to decrease airway inflammation and hypersensitiveness (194). In a landmark study by Gibson et al., azithromycin treatment led to decreased bacterial burden in the lung and gut, which reduced asthma exacerbations (195). In investigations of the lung microbiome with corticosteroid treatment, Goleva et al., showed that there are specific Gram-negative bacteria which trigger corticosteroid resistance via factor-b–associated kinase-1 (TAK1)/MAPK activation (196). Overall, the use of pre- and probiotic supplements, macrolide antibiotics and corticosteroids can reduce asthma exacerbations by decreasing harmful bacterial load (194).
Likewise, for IPF, various reports suggest that targeting the microbiome can increase patients' quality of life (15). In an observational pilot study in 14 advanced IPF patients, treatment with the combination of the anti-viral drug ganciclovir for 2 weeks (5 mg/kg twice daily) and prednisolone reduced disease progression in eight patients as shown by improvement in forced vital capacity (197). This study suggests that there might be a viral co-factors contributing to IPF progression that can be further validated through randomized controlled trials (197). Similarly, co-trimoxazole, a combination of trimethoprim and sulfamethoxazole bactericidal drugs, reduced respiratory infections and mortality, suggesting the use of antibiotics may be beneficial as a combination therapy along with standard drugs for IPF (198). Additionally, there are two ongoing clinical trials with antibiotic treatment in IPF patients. In one, the macrolide azithromycin is being used to treat cough and improve lung function1. In the second, a conjugated therapy of antibiotics co-trimoxazole and doxycycline is being tested in IPF patients2.
In COPD, bronchoscopy sampling from the lower respiratory tract showed that 54% of exacerbations are due to bacterial infections, which shows the potential of anti-bacterial agents (199). In a longitudinal clinical study, antibiotic treatments reduced harmful bacteria, whereas corticosteroids led to increased pathogenic bacteria levels, which clearly shows the effectiveness of antibiotic treatment in COPD (200). A randomized clinical trial conducted by Clancy et al., found that an oral non-typeable H. influenzae vaccine did not protect patients with COPD against exacerbations although patients were not stratified for H. infleunzae carriage (201). Similarly, a recent systematic review indicated that there was no reduction in the severity and frequency of acute exacerbations after oral administration of H. influenzae vaccine in people with chronic bronchitis and COPD (202). However, more positive results may be achieved if patients were stratified and treated according to whether they were H. influenzae positive. Apart from the conventional therapy with inhaled corticosteroids, long-acting muscarinic antagonist and long-acting β-agonist, antibiotics and anti-inflammatory treatment helps in overcoming COPD exacerbations. However, studies are required to define their effects on the lung microbiome (203).
Pre-clinical and clinical studies suggest that associations of the microbiome with lung cancer and further studies explaining the nature of these interactions are needed (204). Detection of specific bacteria like Veillonella and Capnocytophaga by 16S sequencing in saliva samples from lung cancer patients confirm alterations in the microbiome in lung cancer (138). Furthermore, immunotherapy with monoclonal antibodies with immune checkpoint inhibitors like programmed cell death (PD)-1 and targeting its ligand PD-L1 is used in many metastatic cancers as a therapy (205). Interestingly, the use of antibiotics in lung cancer patients resulted in reduced PD-1 response due to dysbiosis which clearly shows the role of microbiome in lung cancer (206). Until now, there are no reports of people using any specific treatments to target the microbiome during lung cancer, which may be a potential therapeutic and diagnostic tool.
Overall, targeting the lung microbiome and reverting dysbiosis may prove beneficial in CLDs. The use of probiotics and antibiotics appear beneficial (Figure 1), but robust cause and effect is lacking, and it is essential that treatment studies and translational research are performed to understand and apply therapeutic interventions. These then need to be formally tested in clinical trials and then implemented into clinical practice.
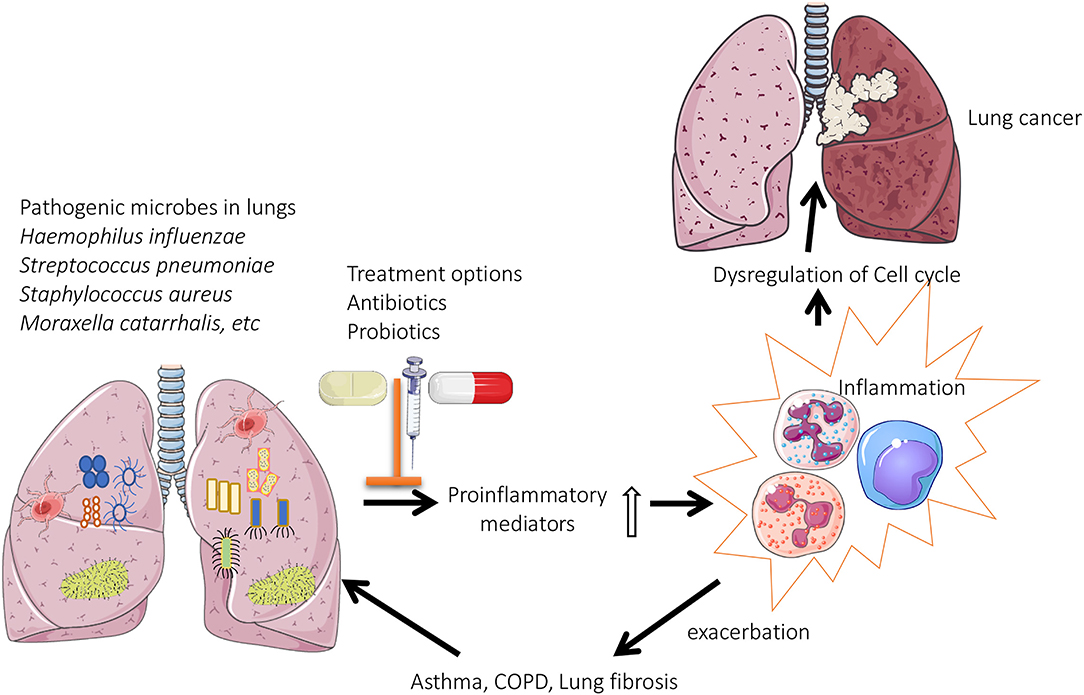
Figure 1. Role of lung microbiome in the pathogenesis of chronic lung diseases: Pathogenic microbes are predominately present in the lungs of patients with chronic respiratory diseases. This results in enhanced production of proinflammatory mediators, which eventually leads to oxidative stress induced dysregulation of cell cycle (lung cancer) or exacerbations associated with asthma/COPD.
Author Contributions
KP and VD have contributed equally to this manuscript. KP, VD, and PH design the conceptual outline of the review. KP, VD, VP, IG, RW, and VM wrote the review. SS, KB, NH, MB, KD, AV, MK-C, and IY provided critical feedback and proof-read the manuscript. All authors contributed to the article and approved the submission version.
Funding
PH is funded by a Fellowship and grants from the National Health and Medical Research Council (NHMRC) of Australia (1175134), the University of Technology Sydney, Lung Foundation of Australia and Cancer Council of NSW, the International Association for the Study of Lung Cancer and the Maitland Cancer Appeal, and the Rainbow Foundation/Hunter Medical Research Institute. KP was supported by a Prevent Cancer Foundation (PCF) and the International Association for the Study of Lung Cancer (IASLC) foundation fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge University of Technology Sydney for their support.
Footnotes
References
1. Shukla SD, Budden KF, Neal R, Hansbro PM. Microbiome effects on immunity, health and disease in the lung. Clin Transl Immunology. (2017) 6:e133. doi: 10.1038/cti.2017.6
2. Keely S, Talley NJ, Hansbro PM. Pulmonary-intestinal cross-talk in mucosal inflammatory disease. Mucosal Immunol. (2012) 5:7–18. doi: 10.1038/mi.2011.55
3. Jones B, Donovan C, Liu G, Gomez HM, Chimankar V, Harrison CL, et al. Animal models of COPD: What do they tell us? Respirology. (2017) 22:21–32. doi: 10.1111/resp.12908
4. Chotirmall SH, Gellatly SL, Budden KF, Mac Aogain M, Shukla SD, Wood DL, et al. Microbiomes in respiratory health and disease: an Asia-Pacific perspective. Respirology. (2017) 22:240–50. doi: 10.1111/resp.12971
5. E, Forum of International Respiratory Societies. The Global Impact of Respiratory Disease. 2nd ed. Sheffield: European Respiratory Society (2017).
6. Hansbro PM, Kaiko GE, Foster PS. Cytokine/anti-cytokine therapy - novel treatments for asthma? Br J Pharmacol. (2011) 163:81–95. doi: 10.1111/j.1476-5381.2011.01219.x
7. Hansbro PM, Hamilton MJ, Fricker M, Gellatly SL, Jarnicki AG, Zheng D, et al. Importance of mast cell Prss31/transmembrane tryptase/tryptase-gamma in lung function and experimental chronic obstructive pulmonary disease and colitis. J Biol Chem. (2014) 289:18214–27. doi: 10.1074/jbc.M114.548594
8. Kim TM, Paudel KR, Kim DW. Eriobotrya japonica leaf extract attenuates airway inflammation in ovalbumin-induced mice model of asthma. J Ethnopharmacol. (2019) 253:112082. doi: 10.1016/j.jep.2019.112082
9. Sethi GS, Dharwal V, Naura AS. Poly(ADP-Ribose)polymerase-1 in lung inflammatory disorders: a review. Front Immunol. (2017) 8:1172. doi: 10.3389/fimmu.2017.01172
10. Starkey MR, Jarnicki AG, Essilfie AT, Gellatly SL, Kim RY, Brown AC, et al. Murine models of infectious exacerbations of airway inflammation. Curr Opin Pharmacol. (2013) 13:337–44. doi: 10.1016/j.coph.2013.03.005
11. Kim RY, Pinkerton JW, Gibson PG, Cooper MA, Horvat JC, Hansbro PM. Inflammasomes in COPD and neutrophilic asthma. Thorax. (2015) 70:1199–201. doi: 10.1136/thoraxjnl-2014-206736
12. G.B.D. Collaborators CRD. Global, regional, national deaths. prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir Med. (2017) 5:691–706. doi: 10.1016/S2213-2600(17)30293-X
13. Vogelmeier CF, Criner GJ, Martinez FJ, Anzueto A, Barnes PJ, Bourbeau J, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am J Respir Crit Care Med. (2017) 195:557–82. doi: 10.1164/rccm.201701-0218PP
14. Ley B, Collard HR, King TE Jr. Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2011) 183:431–40. doi: 10.1164/rccm.201006-0894CI
15. Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. (2011) 183:788–824. doi: 10.1164/rccm.2009-040GL
16. Nathan SD, Shlobin OA, Weir N, Ahmad S, Kaldjob JM, Battle E, et al. Long-term course and prognosis of idiopathic pulmonary fibrosis in the new millennium. Chest. (2011) 140:221–9. doi: 10.1378/chest.10-2572
17. Hutchinson J, Fogarty A, Hubbard R, McKeever T. Global incidence and mortality of idiopathic pulmonary fibrosis: a systematic review. Eur Respir J. (2015) 46:795–806. doi: 10.1183/09031936.00185114
18. Richeldi L, Collard HR, Jones MG. Idiopathic pulmonary fibrosis. Lancet. (2017) 389:1941–52. doi: 10.1016/S0140-6736(17)30866-8
19. du Bois RM, Weycker D, Albera C, Bradford WZ, Costabel U, Kartashov A, et al. Forced vital capacity in patients with idiopathic pulmonary fibrosis: test properties and minimal clinically important difference. Am J Respir Crit Care Med. (2011) 184:1382–9. doi: 10.1164/rccm.201105-0840OC
20. Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. (2015) 16:e165–72. doi: 10.1016/S1470-2045(14)71180-5
21. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
22. Hubbard R, Lewis S, Richards K, Johnston I, Britton J. Occupational exposure to metal or wood dust and aetiology of cryptogenic fibrosing alveolitis. Lancet. (1996) 347:284–9. doi: 10.1016/S0140-6736(96)90465-1
23. Taskar VS, Coultas DB. Is idiopathic pulmonary fibrosis an environmental disease? Proc Am Thorac Soc. (2006) 3:293−8. doi: 10.1513/pats.200512-131TK
24. Hansbro PM, Kim RY, Starkey MR, Donovan C, Dua K, Mayall JR, et al. Mechanisms and treatments for severe, steroid-resistant allergic airway disease and asthma. Immunol Rev. (2017) 278:41–62. doi: 10.1111/imr.12543
25. Wang W, Li JJ, Foster PS, Hansbro PM, Yang M. Potential therapeutic targets for steroid-resistant asthma. Curr Drug Targets. (2010) 11:957–70. doi: 10.2174/138945010791591412
26. Kerstjens HAM, Upham JW, Yang IA. Airway pharmacology: treatment options and algorithms to treat patients with chronic obstructive pulmonary disease. J Thorac Dis. (2019) 11:S2200–9. doi: 10.21037/jtd.2019.10.57
27. Kreuter M, Polke M, Walsh S, Collard HR, Chaudhuri N, Avdeev S, et al. A global perspective on acute exacerbation of idiopathic pulmonary fibrosis (AE-IPF): results from an international survey. Eur Respiratory Soc. (2018) 52:OA542. doi: 10.1183/13993003.congress-2018.OA542
28. Glassberg MK. Overview of idiopathic pulmonary fibrosis, evidence-based guidelines, and recent developments in the treatment landscape. Am J Manag Care. (2019) 25:S195–203.
29. Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non-small cell lung cancer: a review. JAMA. (2019) 322:764–74. doi: 10.1001/jama.2019.11058
30. Eapen MS, Hansbro PM, Larsson-Callerfelt AK, Jolly MK, Myers S, Sharma P, et al. Chronic obstructive pulmonary disease and lung cancer: underlying pathophysiology and new therapeutic modalities. Drugs. (2018) 78:1717–40. doi: 10.1007/s40265-018-1001-8
31. Caramori G, Ruggeri P, Mumby S, Ieni A, Lo Bello F, Chimankar V, et al. Molecular links between COPD and lung cancer: new targets for drug discovery? Expert Opin Ther Targets. (2019) 23:539–53. doi: 10.1080/14728222.2019.1615884
32. Haslam A, Prasad V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw Open. (2019) 2:e192535. doi: 10.1001/jamanetworkopen.2019.2535
33. Budden KF, Gellatly SL, Wood DL, Cooper MA, Morrison M, Hugenholtz P, et al. Emerging pathogenic links between microbiota and the gut-lung axis. Nat Rev Microbiol. (2017) 15:55–63. doi: 10.1038/nrmicro.2016.142
34. Budden KF, Shukla SD, Rehman SF, Bowerman KL, Keely S, Hugenholtz P, et al. Functional effects of the microbiota in chronic respiratory disease. Lancet Respir Med. (2019) 7:907–20. doi: 10.1016/S2213-2600(18)30510-1
35. Thomas T, Gilbert J, Meyer F. Metagenomics - a guide from sampling to data analysis. Microb Inform Exp. (2012) 2:3. doi: 10.1186/2042-5783-2-3
36. Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The microbiome and the respiratory tract. Annu Rev Physiol. (2016) 78:481–504. doi: 10.1146/annurev-physiol-021115-105238
37. Takeuchi S, Kawada J-I, Horiba K, Okuno Y, Okumura T, Suzuki T, et al. Metagenomic analysis using next-generation sequencing of pathogens in bronchoalveolar lavage fluid from pediatric patients with respiratory failure. Sci Rep. (2019) 9:12909. doi: 10.1038/s41598-019-49372-x
38. Wagner BD, Grunwald GK, Zerbe GO, Mikulich-Gilbertson SK, Robertson CE, Zemanick ET, et al. On the use of diversity measures in longitudinal sequencing studies of microbial communities. Front Microbiol. (2018) 9:1037. doi: 10.3389/fmicb.2018.01037
39. Abdel-Aziz MI, Vijverberg SJH, Neerincx AH, Kraneveld AD, A.H. Maitland-van der Zee, The crosstalk between microbiome and asthma: exploring associations and challenges. Clin Exp Allergy. (2019) 49:1067−86. doi: 10.1111/cea.13444
40. Hufnagl K, Pali-Scholl I, Roth-Walter F, Jensen-Jarolim E. Dysbiosis of the gut and lung microbiome has a role in asthma. Semin Immunopathol. (2020) 42:75–93. doi: 10.1007/s00281-019-00775-y
41. Mohajeri MH, Brummer RJM, Rastall RA, Weersma RK, Harmsen HJM, Faas M, et al. The role of the microbiome for human health: from basic science to clinical applications. Eur J Nutr. (2018) 57:1–14. doi: 10.1007/s00394-018-1703-4
42. Sundin J, Rangel I, Fuentes S, Heikamp-de Jong I, Hultgren-Hornquist E, de Vos WM, et al. Altered faecal and mucosal microbial composition in post-infectious irritable bowel syndrome patients correlates with mucosal lymphocyte phenotypes and psychological distress. Aliment Pharmacol Ther. (2015) 41:342–51. doi: 10.1111/apt.13055
43. Hamer HM, Jonkers DM, Vanhoutvin SA, Troost FJ, Rijkers G, de Bruine A, et al. Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin Nutr. (2010) 29:738–44. doi: 10.1016/j.clnu.2010.04.002
44. Cox LM, Blaser MJ. Pathways in microbe-induced obesity. Cell Metab. (2013) 17:883–94. doi: 10.1016/j.cmet.2013.05.004
45. Moro G, Arslanoglu S, Stahl B, Jelinek J, Wahn U, Boehm G. A mixture of prebiotic oligosaccharides reduces the incidence of atopic dermatitis during the first six months of age. Arch Dis Child. (2006) 91:814–9. doi: 10.1136/adc.2006.098251
46. Yu G, Gail MH, Consonni D, Carugno M, Humphrys M, Pesatori AC, et al. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. (2016) 17:163. doi: 10.1186/s13059-016-1021-1
47. Laroumagne S, Salinas-Pineda A, Hermant C, Murris M, Gourraud PA, Do C, et al. [Incidence and characteristics of bronchial colonisation in patient with lung cancer: a retrospective study of 388 cases]. Rev Mal Respir. (2011) 28:328–35. doi: 10.1016/j.rmr.2010.05.020
48. Wong JL, Evans SE. Bacterial pneumonia in patients with cancer: novel risk factors and management. Clin Chest Med. (2017) 38:263–77. doi: 10.1016/j.ccm.2016.12.005
49. Faner R, Sibila O, Agusti A, Bernasconi E, Chalmers JD, Huffnagle GB, et al. The microbiome in respiratory medicine: current challenges and future perspectives. Eur Respir J. (2017) 49:1602086. doi: 10.1183/13993003.02086-2016
50. Genuneit J, Seibold AM, Apfelbacher CJ, Konstantinou GN, Koplin JJ, La Grutta S, et al. Overview of systematic reviews in allergy epidemiology. Allergy. (2017) 72:849–56. doi: 10.1111/all.13123
51. Hansbro PM, Scott GV, Essilfie AT, Kim RY, Starkey MR, Nguyen DH, et al. Th2 cytokine antagonists: potential treatments for severe asthma. Expert Opin Investig Drugs. (2013) 22:49–69. doi: 10.1517/13543784.2013.732997
52. Foster PS, Maltby S, Rosenberg HF, Tay HL, Hogan SP, Collison AM, et al. Modeling TH 2 responses and airway inflammation to understand fundamental mechanisms regulating the pathogenesis of asthma. Immunol Rev. (2017) 278:20–40. doi: 10.1111/imr.12549
53. Kim RY, Rae B, Neal R, Donovan C, Pinkerton J, Balachandran L, et al. Elucidating novel disease mechanisms in severe asthma. Clin Transl Immunol. (2016) 5:e91. doi: 10.1038/cti.2016.37
54. Hansbro PM, Beagley KW, Horvat JC, Gibson PG. Role of atypical bacterial infection of the lung in predisposition/protection of asthma. Pharmacol Ther. (2004) 101:193–210. doi: 10.1016/j.pharmthera.2003.10.007
55. Kim RY, Horvat JC, Pinkerton JW, Starkey MR, Essilfie AT, Mayall JR, et al. MicroRNA-21 drives severe, steroid-insensitive experimental asthma by amplifying phosphoinositide 3-kinase-mediated suppression of histone deacetylase 2. J Allergy Clin Immunol. (2017) 139:519–32. doi: 10.1016/j.jaci.2016.04.038
56. Kim RY, Pinkerton JW, Essilfie AT, Robertson AAB, Baines KJ, Brown AC, et al. Role for NLRP3 inflammasome-mediated, IL-1beta-dependent responses in severe, steroid-resistant asthma. Am J Respir Crit Care Med. (2017) 196:283–97. doi: 10.1164/rccm.201609-1830OC
57. Wadhwa R, Dua K, Adcock IM, Horvat JC, Kim RY, Hansbro PM. Cellular mechanisms underlying steroid-resistant asthma. Eur Respir Rev. (2019) 28:190096. doi: 10.1183/16000617.0096-2019
58. Barnes PJ, Adcock IM. Glucocorticoid resistance in inflammatory diseases. Lancet. (2009) 373:1905–17. doi: 10.1016/S0140-6736(09)60326-3
59. Pinkerton JW, Kim RY, Robertson AAB, Hirota JA, Wood LG, Knight DA, et al. Inflammasomes in the lung. Mol Immunol. (2017) 86:44–55. doi: 10.1016/j.molimm.2017.01.014
60. Hirota JA, Gold MJ, Hiebert PR, Parkinson LG, Wee T, Smith D, et al. The nucleotide-binding domain, leucine-rich repeat protein 3 inflammasome/IL-1 receptor I axis mediates innate, but not adaptive, immune responses after exposure to particulate matter under 10 mum. Am J Respir Cell Mol Biol. (2015) 52:96–105. doi: 10.1165/rcmb.2014-0158OC
61. Durack J, Lynch SV, Nariya S, Bhakta NR, Beigelman A, Castro M, et al. Blood Institute's, Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J Allergy Clin Immunol. (2017) 140:63–75. doi: 10.1016/j.jaci.2016.08.055
62. Essilfie AT, Horvat JC, Kim RY, Mayall JR, Pinkerton JW, Beckett EL, et al. Macrolide therapy suppresses key features of experimental steroid-sensitive and steroid-insensitive asthma. Thorax. (2015) 70:458–67. doi: 10.1136/thoraxjnl-2014-206067
63. Essilfie AT, Simpson JL, Dunkley ML, Morgan LC, Oliver BG, Gibson PG, et al. Combined Haemophilus influenzae respiratory infection and allergic airways disease drives chronic infection and features of neutrophilic asthma. Thorax. (2012) 67:588–99. doi: 10.1136/thoraxjnl-2011-200160
64. Essilfie AT, Simpson JL, Horvat JC, Preston JA, Dunkley ML, Foster PS, et al. Haemophilus influenzae infection drives IL-17-mediated neutrophilic allergic airways disease. PLoS Pathog. (2011) 7:e1002244. doi: 10.1371/journal.ppat.1002244
65. Kaiko GE, Phipps S, Hickey DK, Lam CE, Hansbro PM, Foster PS, et al. Chlamydia muridarum infection subverts dendritic cell function to promote Th2 immunity and airways hyperreactivity. J Immunol. (2008) 180:2225–32. doi: 10.4049/jimmunol.180.4.2225
66. Horvat JC, Starkey MR, Kim RY, Phipps S, Gibson PG, Beagley KW, et al. Early-life chlamydial lung infection enhances allergic airways disease through age-dependent differences in immunopathology. J Allergy Clin Immunol. (2010) 125:617–25:625 e1–5 e6. doi: 10.1016/j.jaci.2009.10.018
67. Starkey MR, Essilfie AT, Horvat JC, Kim RY, Nguyen DH, Beagley KW, et al. Constitutive production of IL-13 promotes early-life Chlamydia respiratory infection and allergic airway disease. Mucosal Immunol. (2013) 6:569–79. doi: 10.1038/mi.2012.99
68. Huang YJ, Nariya S, Harris JM, Lynch SV, Choy DF, Arron JR, et al. The airway microbiome in patients with severe asthma: associations with disease features and severity. J Allergy Clin Immunol. (2015) 136:874–84. doi: 10.1016/j.jaci.2015.05.044
69. Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Blood Institute's Asthma Clinical Research, Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. (2011) 127:372–81 e1–3. doi: 10.1016/j.jaci.2010.10.048
70. Denner DR, Sangwan N, Becker JB, Hogarth DK, Oldham J, Castillo J, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. (2016) 137:1398–405 e3. doi: 10.1016/j.jaci.2015.10.017
71. Durack J, Huang YJ, Nariya S, Christian LS, Ansel KM, Beigelman A, et al. Blood Institute's, Bacterial biogeography of adult airways in atopic asthma. Microbiome. (2018) 6:104. doi: 10.1186/s40168-018-0487-3
72. Zhang Q, Cox M, Liang Z, Brinkmann F, Cardenas PA, Duff R, et al. Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS ONE. (2016) 11:e0152724. doi: 10.1371/journal.pone.0152724
73. Green BJ, Wiriyachaiporn S, Grainge C, Rogers GB, Kehagia V, Lau L, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS ONE. (2014) 9:e100645. doi: 10.1371/journal.pone.0100645
74. Yang B, Liu R, Yang T, Jiang X, Zhang L, Wang L, et al. Neonatal Streptococcus pneumoniae infection may aggravate adulthood allergic airways disease in association with IL-17A. PLoS ONE. (2015) 10:e0123010. doi: 10.1371/journal.pone.0123010
75. Preston JA, Essilfie AT, Horvat JC, Wade MA, Beagley KW, Gibson PG, et al. Inhibition of allergic airways disease by immunomodulatory therapy with whole killed Streptococcus pneumoniae. Vaccine. (2007) 25:8154–62. doi: 10.1016/j.vaccine.2007.09.034
76. Preston JA, Thorburn AN, Starkey MR, Beckett EL, Horvat JC, Wade MA, et al. Streptococcus pneumoniae infection suppresses allergic airways disease by inducing regulatory T-cells. Eur Respir J. (2011) 37:53–64. doi: 10.1183/09031936.00049510
77. Thorburn AN, Brown AC, Nair PM, Chevalier N, Foster PS, Gibson PG, et al. Pneumococcal components induce regulatory T cells that attenuate the development of allergic airways disease by deviating and suppressing the immune response to allergen. J Immunol. (2013) 191:4112–20. doi: 10.4049/jimmunol.1201232
78. Thorburn AN, Foster PS, Gibson PG, Hansbro PM. Components of Streptococcus pneumoniae suppress allergic airways disease and NKT cells by inducing regulatory T cells. J Immunol. (2012) 188:4611–20. doi: 10.4049/jimmunol.1101299
79. Thorburn AN, O'Sullivan BJ, Thomas R, Kumar RK, Foster PS, Gibson PG, et al. Pneumococcal conjugate vaccine-induced regulatory T cells suppress the development of allergic airways disease. Thorax. (2010) 65:1053–60. doi: 10.1136/thx.2009.131508
80. Usami M, Kishimoto K, Ohata A, Miyoshi M, Aoyama M, Fueda Y, et al. Butyrate and trichostatin A attenuate nuclear factor kappaB activation and tumor necrosis factor alpha secretion and increase prostaglandin E2 secretion in human peripheral blood mononuclear cells. Nutr Res. (2008) 28:321–8. doi: 10.1016/j.nutres.2008.02.012
81. Panth N, Abbott KA, Dias CB, Wynne K, Garg ML. Differential effects of medium- and long-chain saturated fatty acids on blood lipid profile: a systematic review and meta-analysis. Am J Clin Nutr. (2018) 108:675–87. doi: 10.1093/ajcn/nqy167
82. Anand S, Mande SS. Diet, microbiota and gut-lung connection. Front Microbiol. (2018) 9:2147. doi: 10.3389/fmicb.2018.02147
83. Vinolo MA, Rodrigues HG, Hatanaka E, Sato FT, Sampaio SC, Curi R. Suppressive effect of short-chain fatty acids on production of proinflammatory mediators by neutrophils. J Nutr Biochem. (2011) 22:849–55. doi: 10.1016/j.jnutbio.2010.07.009
84. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. (2014) 20:159–66. doi: 10.1038/nm.3444
85. Cait A, Hughes MR, Antignano F, Cait J, Dimitriu PA, Maas KR, et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. (2018) 11:785–95. doi: 10.1038/mi.2017.75
86. Hougee S, Vriesema AJ, Wijering SC, Knippels LM, Folkerts G, Nijkamp FP, et al. Oral treatment with probiotics reduces allergic symptoms in ovalbumin-sensitized mice: a bacterial strain comparative study. Int Arch Allergy Immunol. (2010) 151:107–17. doi: 10.1159/000236000
87. Spacova I, Petrova MI, Fremau A, Pollaris L, Vanoirbeek J, Ceuppens JL, et al. Intranasal administration of probiotic Lactobacillus rhamnosus GG prevents birch pollen-induced allergic asthma in a murine model. Allergy. (2019) 74:100–10. doi: 10.1111/all.13502
88. Wang X, Hui Y, Zhao L, Hao Y, Guo H, Ren F. Oral administration of Lactobacillus paracasei L9 attenuates PM2.5-induced enhancement of airway hyperresponsiveness and allergic airway response in murine model of asthma. PLoS ONE. (2017) 12:e0171721. doi: 10.1371/journal.pone.0171721
89. Huang CF, Chie WC, Wang IJ. Efficacy of lactobacillus administration in school-age children with asthma: a randomized, placebo-controlled trial. Nutrients. (2018) 10:1678. doi: 10.3390/nu10111678
90. Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. (2014) 44:842–50. doi: 10.1111/cea.12253
91. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE. (2011) 6:e16384. doi: 10.1371/journal.pone.0016384
92. Zakharkina T, Heinzel E, Koczulla RA, Greulich T, Rentz K, Pauling JK, et al. Analysis of the airway microbiota of healthy individuals and patients with chronic obstructive pulmonary disease by T-RFLP and clone sequencing. PLoS ONE. (2013) 8:e68302. doi: 10.1371/journal.pone.0068302
93. Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS ONE. (2012) 7:e47305. doi: 10.1371/journal.pone.0047305
94. Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS ONE. (2010) 5:e8578. doi: 10.1371/journal.pone.0008578
95. Wu D, Hou C, Li Y, Zhao Z, Liu J, Lu X, et al. Analysis of the bacterial community in chronic obstructive pulmonary disease sputum samples by denaturing gradient gel electrophoresis and real-time PCR. BMC Pulm Med. (2014) 14:179. doi: 10.1186/1471-2466-14-179
96. Murphy TF, Brauer AL, Grant BJ, Sethi S. Moraxella catarrhalis in chronic obstructive pulmonary disease: burden of disease and immune response. Am J Respir Crit Care Med. (2005) 172:195–9. doi: 10.1164/rccm.200412-1747OC
97. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. I. Lung VMP. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. (2013) 187:1067–75. doi: 10.1164/rccm.201210-1913OC
98. Zhang R, Chen L, Cao L, Li KJ, Huang Y, Luan XQ, et al. Effects of smoking on the lower respiratory tract microbiome in mice. Respir Res. (2018) 19:253. doi: 10.1186/s12931-018-0959-9
99. Huang C, Shi G. Smoking and microbiome in oral, airway, gut and some systemic diseases. J Transl Med. (2019) 17:225. doi: 10.1186/s12967-019-1971-7
100. Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. (2012) 185:1073–80. doi: 10.1164/rccm.201111-2075OC
101. Rutting S, Papanicolaou M, Xenaki D, Wood LG, Mullin AM, Hansbro PM, et al. Dietary omega-6 polyunsaturated fatty acid arachidonic acid increases inflammation, but inhibits ECM protein expression in COPD. Respir Res. (2018) 19:211. doi: 10.1186/s12931-018-0919-4
102. Barnes PJ. Mediators of chronic obstructive pulmonary disease. Pharmacol Rev. (2004) 56:515–48. doi: 10.1124/pr.56.4.2
103. Gregory AC, Sullivan MB, Segal LN, Keller BC. Smoking is associated with quantifiable differences in the human lung DNA virome and metabolome. Respir Res. (2018) 19:174. doi: 10.1186/s12931-018-0878-9
104. Ghosh A, Coakley RD, Ghio AJ, Muhlebach MS, Esther CR Jr, et al. Chronic E-cigarette use increases neutrophil elastase and matrix metalloprotease levels in the lung. Am J Respir Crit Care Med. (2019) 200:1392–401. doi: 10.1164/rccm.201903-0615OC
105. Vogel ER, VanOosten SK, Holman MA, Hohbein DD, Thompson MA, Vassallo R, et al. Cigarette smoke enhances proliferation and extracellular matrix deposition by human fetal airway smooth muscle. Am J Physiol Lung Cell Mol Physiol. (2014) 307:L978–86. doi: 10.1152/ajplung.00111.2014
106. Chen L, Ge Q, Tjin G, Alkhouri H, Deng L, Brandsma CA, et al. Effects of cigarette smoke extract on human airway smooth muscle cells in COPD. Eur Respir J. (2014) 44:634–46. doi: 10.1183/09031936.00171313
107. Liu G, Cooley MA, Jarnicki AG, Hsu AC, Nair PM, Haw TJ, et al. Fibulin-1 regulates the pathogenesis of tissue remodeling in respiratory diseases. JCI Insight. (2016) 1:e86380. doi: 10.1172/jci.insight.86380
108. Simpson JL, Baines KJ, Horvat JC, Essilfie AT, Brown AC, Tooze M, et al. COPD is characterized by increased detection of Haemophilus influenzae, Streptococcus pneumoniae and a deficiency of Bacillus species. Respirology. (2016) 21:697–704. doi: 10.1111/resp.12734
109. Ikeda M, Enomoto N, Hashimoto D, Fujisawa T, Inui N, Nakamura Y, et al. Nontypeable Haemophilus influenzae exploits the interaction between protein-E and vitronectin for the adherence and invasion to bronchial epithelial cells. BMC Microbiol. (2015) 15:263. doi: 10.1186/s12866-015-0600-8
110. Fink DL, Green BA, St Geme JW III. The Haemophilus influenzae Hap autotransporter binds to fibronectin, laminin, collagen IV. Infect Immun. (2002) 70:4902–7. doi: 10.1128/IAI.70.9.4902-4907.2002
111. Singh B, Fleury C, Jalalvand F, Riesbeck K. Human pathogens utilize host extracellular matrix proteins laminin and collagen for adhesion and invasion of the host. FEMS Microbiol Rev. (2012) 36:1122–80. doi: 10.1111/j.1574-6976.2012.00340.x
112. Hussell T, Lui S, Jagger C, Morgan D, Brand O. The consequence of matrix dysfunction on lung immunity and the microbiome in COPD. Eur Respir Rev. (2018) 27:180032. doi: 10.1183/16000617.0032-2018
113. Kranenburg AR, Willems-Widyastuti A, Moori WJ, Sterk PJ, Alagappan VK, de Boer WI, et al. Enhanced bronchial expression of extracellular matrix proteins in chronic obstructive pulmonary disease. Am J Clin Pathol. (2006) 126:725–35. doi: 10.1309/JC477FAEL1YKV54W
114. Annoni R, Lancas T, Yukimatsu Tanigawa R, de Medeiros Matsushita M, de Morais Fernezlian S, Bruno A, et al. Extracellular matrix composition in COPD. Eur Respir J. (2012) 40:1362–73. doi: 10.1183/09031936.00192611
115. Segal LN, Clemente JC, Tsay JC, Koralov SB, Keller BC, Wu BG, et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat Microbiol. (2016) 1:16031. doi: 10.1038/nmicrobiol.2016.31
116. Hakansson AP, Orihuela CJ, Bogaert D. Bacterial-host interactions: physiology and pathophysiology of respiratory infection. Physiol Rev. (2018) 98:781–811. doi: 10.1152/physrev.00040.2016
117. Shen Y, Chen L, Wang M, Lin D, Liang Z, Song P, et al. Flagellar hooks and hook protein flge participate in host microbe interactions at immunological level. Sci Rep. (2017) 7:1433. doi: 10.1038/s41598-017-01619-1
118. Aguilera ER, Lenz LL. Inflammation as a modulator of host susceptibility to pulmonary influenza, pneumococcal, and co-infections. Front Immunol. (2020) 11:105. doi: 10.3389/fimmu.2020.00105
119. Larsen JM, Musavian HS, Butt TM, Ingvorsen C, Thysen AH, Brix S. Chronic obstructive pulmonary disease and asthma-associated Proteobacteria, but not commensal Prevotella spp., promote Toll-like receptor 2-independent lung inflammation and pathology. Immunology. (2015) 144:333–42. doi: 10.1111/imm.12376
120. Gaeckle NT, Pragman AA, Pendleton KM, Baldomero AK, Criner GJ. The oral-lung axis: the impact of oral health on lung health. Respir Care. (2020) 65:1211–20. doi: 10.4187/respcare.07332
121. Segal LN, Alekseyenko AV, Clemente JC, Kulkarni R, Wu B, Gao Z, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. (2013) 1:19. doi: 10.1186/2049-2618-1-19
122. Mayhew D, Devos N, Lambert C, Brown JR, Clarke SC, Kim VL, et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax. (2018) 73:422–30. doi: 10.1136/thoraxjnl-2017-210408
123. Su YC, Jalalvand F, Thegerstrom J, Riesbeck K. The interplay between immune response and bacterial infection in COPD: focus upon non-typeable haemophilus influenzae. Front Immunol. (2018) 9:2530. doi: 10.3389/fimmu.2018.02530
124. Shuto T, Xu H, Wang B, Han J, Kai H, Gu XX, et al. Activation of NF-kappa B by nontypeable Hemophilus influenzae is mediated by toll-like receptor 2-TAK1-dependent NIK-IKK alpha /beta-I kappa B alpha and MKK3/6-p38 MAP kinase signaling pathways in epithelial cells. Proc Natl Acad Sci USA. (2001) 98:8774–9. doi: 10.1073/pnas.151236098
125. Di Stefano, Ricciardolo FLM, Caramori G, Adcock IM, Chung KF, Barnes PJ, et al. Bronchial inflammation and bacterial load in stable COPD is associated with TLR4 overexpression. Eur Respir J. (2017) 49:1602006. doi: 10.1183/13993003.02006-2016
126. Hughes BM, Burton CS, Reese A, Jabeen MF, Wright C, Willis J, et al. Pellino-1 regulates immune responses to haemophilus influenzae in models of inflammatory lung disease. Front Immunol. (2019) 10:1721. doi: 10.3389/fimmu.2019.01721
127. McIsaac SM, Stadnyk AW, Lin TJ. Toll-like receptors in the host defense against Pseudomonas aeruginosa respiratory infection and cystic fibrosis. J Leukoc Biol. (2012) 92:977–85. doi: 10.1189/jlb.0811410
128. Haw TJ, Starkey MR, Pavlidis S, Fricker M, Arthurs AL, Nair PM, et al. Toll-like receptor 2 and 4 have opposing roles in the pathogenesis of cigarette smoke-induced chronic obstructive pulmonary disease. Am J Physiol Lung Cell Mol Physiol. (2018) 314:L298–317. doi: 10.1152/ajplung.00154.2017
129. Zuo L, Lucas K, Fortuna CA, Chuang CC, Best TM. Molecular regulation of toll-like receptors in asthma and COPD. Front Physiol. (2015) 6:312. doi: 10.3389/fphys.2015.00312
130. Wang Z, Bafadhel M, Haldar K, Spivak A, Mayhew D, Miller BE, et al. Lung microbiome dynamics in COPD exacerbations. Eur Respir J. (2016) 47:1082–92. doi: 10.1183/13993003.01406-2015
131. Shiels MS, Albanes D, Virtamo J, Engels EA. Increased risk of lung cancer in men with tuberculosis in the alpha-tocopherol, beta-carotene cancer prevention study. Cancer Epidemiol Biomarkers Prev. (2011) 20:672–8. doi: 10.1158/1055-9965.EPI-10-1166
132. GonzAlez I, Araya P, Rojas A. Helicobacter pylori infection and lung cancer: new insights and future challenges. Zhongguo Fei Ai Za Zhi. (2018) 21:658–62. doi: 10.3779/j.issn.1009-3419.2018.09.03
133. Greathouse KL, White JR, Vargas AJ, Bliskovsky VV, Beck JA, von Muhlinen N, et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. (2018) 19:123. doi: 10.1186/s13059-018-1501-6
134. Apopa PL, Alley L, Penney RB, Arnaoutakis K, Steliga MA, Jeffus S, et al. PARP1 is up-regulated in non-small cell lung cancer tissues in the presence of the cyanobacterial toxin microcystin. Front Microbiol. (2018) 9:1757. doi: 10.3389/fmicb.2018.01757
135. Gomes S, Cavadas B, Ferreira JC, Marques PI, Monteiro C, Sucena M, et al. Profiling of lung microbiota discloses differences in adenocarcinoma and squamous cell carcinoma. Sci Rep. (2019) 9:12838. doi: 10.1038/s41598-019-49195-w
136. Wang YC, Wang NC, Lin JC, Perng CL, Yeh KM, Yang YS, et al. Risk factors and outcomes of cytomegalovirus viremia in cancer patients: a study from a medical center in northern Taiwan. J Microbiol Immunol Infect. (2011) 44:442–8. doi: 10.1016/j.jmii.2011.04.004
137. Lee SH, Sung JY, Yong D, Chun J, Kim SY, Song JH, et al. Characterization of microbiome in bronchoalveolar lavage fluid of patients with lung cancer comparing with benign mass like lesions. Lung Cancer. (2016) 102:89–95. doi: 10.1016/j.lungcan.2016.10.016
138. Yan X, Yang M, Liu J, Gao R, Hu J, Li J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res. (2015) 5:3111–22.
139. Nesic D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature. (2004) 429:429–33. doi: 10.1038/nature02532
140. Chow SC, Gowing SD, Cools-Lartigue JJ, Chen CB, Berube J, Yoon HW, et al. Gram negative bacteria increase non-small cell lung cancer metastasis via Toll-like receptor 4 activation and mitogen-activated protein kinase phosphorylation. Int J Cancer. (2015) 136:1341–50. doi: 10.1002/ijc.29111
141. Zhuo WL, Zhu B, Xiang ZL, Zhuo XL, Cai L, Chen ZT. Assessment of the relationship between Helicobacter pylori and lung cancer: a meta-analysis. Arch Med Res. (2009) 40:406–10. doi: 10.1016/j.arcmed.2009.05.002
142. Brenner DR, McLaughlin JR, Hung RJ. Previous lung diseases and lung cancer risk: a systematic review and meta-analysis. PLoS ONE. (2011) 6:e17479. doi: 10.1371/journal.pone.0017479
143. Kocazeybek B. Chronic Chlamydophila pneumoniae infection in lung cancer, a risk factor: a case-control study. J Med Microbiol. (2003) 52:721–6. doi: 10.1099/jmm.0.04845-0
144. D'Journo XB, Bittar F, Trousse D, Gaillat F, Doddoli C, Dutau H, et al. Molecular detection of microorganisms in distal airways of patients undergoing lung cancer surgery. Ann Thorac Surg. (2012) 93:413–22. doi: 10.1016/j.athoracsur.2011.09.049
145. Zhang H, Garcia Rodriguez LA, Hernandez-Diaz S. Antibiotic use and the risk of lung cancer. Cancer Epidemiol Biomarkers Prev. (2008) 17:1308–15. doi: 10.1158/1055-9965.EPI-07-2817
146. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via gammadelta T cells. Cell. (2019) 176:998–1013 e16. doi: 10.1016/j.cell.2018.12.040
147. Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, et al. Microbiota modulate tumoral immune surveillance in lung through a gammadeltaT17 immune cell-dependent mechanism. Cancer Res. (2014) 74:4030–41. doi: 10.1158/0008-5472.CAN-13-2462
148. Huang D, Su X, Yuan M, Zhang S, He J, Deng Q, et al. The characterization of lung microbiome in lung cancer patients with different clinicopathology. Am J Cancer Res. (2019) 9:2047−63.
149. Sharma A, Viswanath B, Park YS. Role of probiotics in the management of lung cancer and related diseases: an update. J Funct Foods. (2018) 40:625–33. doi: 10.1016/j.jff.2017.11.050
150. Daillere R, Vetizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, et al. Enterococcus hirae and barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. (2016) 45:931–43. doi: 10.1016/j.immuni.2016.09.009
151. Tanasienko OA, Cheremshenko NL, Titova GP, Potebnya MG, Gavrilenko MM, Nagorna SS, et al. Elevation of the efficacy of antitumor vaccine prepared on the base of lectines from B. subtilis B-7025 upon its combined application with probiotics in vivo. Exp Oncol. (2005) 27:336–8.
152. Lee NK, Han KJ, Son SH, Eom SJ, Lee SK, Paik HD. Multifunctional effect of probiotic Lactococcus lactis KC24 isolated from kimchi. LWT Food Sci Technol. (2015) 64:1036–41. doi: 10.1016/j.lwt.2015.07.019
153. Han KJ, Lee NK, Park H, Paik HD. Anticancer and anti-inflammatory activity of probiotic Lactococcus lactis NK34. J Microbiol Biotechnol. (2015) 25:1697–701. doi: 10.4014/jmb.1503.03033
154. Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, et al. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2014) 190:906–13. doi: 10.1164/rccm.201403-0541OC
155. Han MK, Zhou Y, Murray S, Tayob N, Noth I, Lama VN, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. Lancet Respir Med. (2014) 2:548–56. doi: 10.1016/S2213-2600(14)70069-4
156. O'Dwyer DN, Ashley SL, Gurczynski SJ, Xia M, Wilke C, Falkowski NR, et al. Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am J Respir Crit Care Med. (2019) 199:1127–38. doi: 10.1164/rccm.201809-1650OC
157. Molyneaux PL, Maher TM. Respiratory microbiome in IPF: cause, effect, or biomarker? Lancet Respir Med. (2014) 2:511–3. doi: 10.1016/S2213-2600(14)70088-8
158. Maher TM, Wells AU, Laurent GJ. Idiopathic pulmonary fibrosis: multiple causes and multiple mechanisms? Eur Respir J. (2007) 30:835–9. doi: 10.1183/09031936.00069307
159. Idiopathic Pulmonary Fibrosis Clinical Research Network, Raghu G, Anstrom KJ, King TE Jr, Lasky JA, Martinez FJ, et al. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. (2012) 366:1968–77. doi: 10.1056/NEJMoa1113354
160. Yang D, Chen X, Wang J, Lou Q, Lou Y, Li L, et al. Dysregulated lung commensal bacteria drive interleukin-17B production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity. (2019) 50:692–706 e7. doi: 10.1016/j.immuni.2019.02.001
161. Takahashi Y, Saito A, Chiba H, Kuronuma K, Ikeda K, Kobayashi T, et al. Impaired diversity of the lung microbiome predicts progression of idiopathic pulmonary fibrosis. Respir Res. (2018) 19:34. doi: 10.1186/s12931-018-0736-9
162. Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. (2011) 364:1503–12. doi: 10.1056/NEJMoa1013660
163. Roy MG, Livraghi-Butrico A, Fletcher AA, McElwee MM, Evans SE, Boerner RM, et al. Muc5b is required for airway defence. Nature. (2014) 505:412–6. doi: 10.1038/nature12807
164. Helling BA, Gerber AN, Kadiyala V, Sasse SK, Pedersen BS, Sparks L, et al. Regulation of MUC5B expression in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. (2017) 57:91–9. doi: 10.1165/rcmb.2017-0046OC
165. Oldham JM, Ma SF, Martinez FJ, Anstrom KJ, Raghu G, Schwartz DA, et al. TOLLIP, MUC5B, and the response to N-acetylcysteine among individuals with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2015) 192:1475–82. doi: 10.1164/rccm.201505-1010OC
166. Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. (2013) 1:309–17. doi: 10.1016/S2213-2600(13)70045-6
167. Molyneaux PL, Willis-Owen SAG, Cox MJ, James P, Cowman S, Loebinger M, et al. Host-microbial interactions in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2017) 195:1640–50. doi: 10.1164/rccm.201607-1408OC
168. Habgood AN, Tatler AL, Porte J, Wahl SM, Laurent GJ, John AE, et al. Secretory leukocyte protease inhibitor gene deletion alters bleomycin-induced lung injury, but not development of pulmonary fibrosis. Lab Invest. (2016) 96:623–31. doi: 10.1038/labinvest.2016.40
169. Molyneaux PL, Cox MJ, Wells AU, Kim HC, Ji W, Cookson WO, et al. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir Res. (2017) 18:29. doi: 10.1186/s12931-017-0511-3
170. Huang Y, Ma SF, Espindola MS, Vij R, Oldham JM, Huffnagle GB, et al. Microbes are associated with host innate immune response in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. (2017) 196:208−19. doi: 10.1164/rccm.201607-1525OC
171. Xu MY, Porte J, Knox AJ, Weinreb PH, Maher TM, Violette SM, et al. Lysophosphatidic acid induces alphavbeta6 integrin-mediated TGF-beta activation via the LPA2 receptor and the small G protein G alpha(q). Am J Pathol. (2009) 174:1264–79. doi: 10.2353/ajpath.2009.080160
172. S. Holm Nielsen, Willumsen N, Leeming DJ, Daniels SJ, Brix S, Karsdal MA, et al. Serological assessment of activated fibroblasts by alpha-smooth muscle actin (alpha-SMA): a noninvasive biomarker of activated fibroblasts in lung disorders. Transl Oncol. (2019) 12:368–74. doi: 10.1016/j.tranon.2018.11.004
173. Shukla SD, Shastri MD, Chong WC, Dua K, Budden KF, Mahmood MQ, et al. Microbiome-focused asthma management strategies. Curr Opin Pharmacol. (2019) 46:143–9. doi: 10.1016/j.coph.2019.06.003
174. Chellappan DK, Yee LW, Xuan KY, Kunalan K, Rou LC, Jean LS, et al. Targeting neutrophils using novel drug delivery systems in chronic respiratory diseases. Drug Dev Res. (2020) 81:419–36. doi: 10.1002/ddr.21648
175. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. (2014) 12:87. doi: 10.1186/s12915-014-0087-z
176. Dickson RP, Erb-Downward JR, Freeman CM, McCloskey L, Falkowski NR, Huffnagle GB, et al. Bacterial topography of the healthy human lower respiratory tract. mBio. (2017) 8:1–12. doi: 10.1128/mBio.02287-16
177. Stinson LF, Keelan JA, Payne MS. Identification and removal of contaminating microbial DNA from PCR reagents: impact on low-biomass microbiome analyses. Lett Appl Microbiol. (2019) 68:2–8. doi: 10.1111/lam.13091
178. Mendez R, Banerjee S, Bhattacharya SK, Banerjee S. Lung inflammation and disease: a perspective on microbial homeostasis and metabolism. IUBMB Life. (2019) 71:152–65. doi: 10.1002/iub.1969
179. Li KJ, Chen ZL, Huang Y, Zhang R, Luan XQ, Lei TT, et al. Dysbiosis of lower respiratory tract microbiome are associated with inflammation and microbial function variety. Respir Res. (2019) 20:272. doi: 10.1186/s12931-019-1246-0
180. Clemente JC, Manasson J, Scher JU. The role of the gut microbiome in systemic inflammatory disease. BMJ. (2018) 360:j5145. doi: 10.1136/bmj.j5145
181. Slingerland AE, Schwabkey Z, Wiesnoski DH, Jenq RR. Clinical evidence for the microbiome in inflammatory diseases. Front Immunol. (2017) 8:400. doi: 10.3389/fimmu.2017.00400
182. Poroyko V, Meng F, Meliton A, Afonyushkin T, Ulanov A, Semenyuk E, et al. Alterations of lung microbiota in a mouse model of LPS-induced lung injury. Am J Physiol Lung Cell Mol Physiol. (2015) 309:L76–83. doi: 10.1152/ajplung.00061.2014
183. Knobloch J, Chikosi SJ, Yanik S, Rupp J, Jungck D, Koch A. A systemic defect in Toll-like receptor 4 signaling increases lipopolysaccharide-induced suppression of IL-2-dependent T-cell proliferation in COPD. Am J Physiol Lung Cell Mol Physiol. (2016) 310:L24–39. doi: 10.1152/ajplung.00367.2014
184. Starkhammar M, Kumlien Georen S, Dahlen SE, Cardell LO, Adner M. TNFalpha-blockade stabilizes local airway hyperresponsiveness during TLR-induced exacerbations in murine model of asthma. Respir Res. (2015) 16:129. doi: 10.1186/s12931-015-0292-5
185. Lu YC, Yeh WC, Ohashi PS. LPS/TLR4 signal transduction pathway. Cytokine. (2008) 42:145–51. doi: 10.1016/j.cyto.2008.01.006
186. Ohshima Y, Ko HL, Beuth J, Roszkowski K, Roszkowski W. Biological properties of staphylococcal lipoteichoic acid and related macromolecules. Zentralbl Bakteriol. (1990) 274:359–65. doi: 10.1016/S0934-8840(11)80693-6
187. Leemans JC, Heikens M, van Kessel KP, Florquin S, van der Poll T. Lipoteichoic acid and peptidoglycan from Staphylococcus aureus synergistically induce neutrophil influx into the lungs of mice. Clin Diagn Lab Immunol. (2003) 10:950–3. doi: 10.1128/CDLI.10.5.950-953.2003
188. Leemans JC, Vervoordeldonk MJ, Florquin S, van Kessel KP, van der Poll T. Differential role of interleukin-6 in lung inflammation induced by lipoteichoic acid and peptidoglycan from Staphylococcus aureus. Am J Respir Crit Care Med. (2002) 165:1445–50. doi: 10.1164/rccm.2106045
189. Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids–new mechanisms for old drugs. N Engl J Med. (2005) 353:1711–23. doi: 10.1056/NEJMra050541
190. Wang YH, Wills-Karp M. The potential role of interleukin-17 in severe asthma. Curr Allergy Asthma Rep. (2011) 11:388–94. doi: 10.1007/s11882-011-0210-y
191. Huang YJ. Asthma microbiome studies and the potential for new therapeutic strategies. Curr Allergy Asthma Rep. (2013) 13:453–61. doi: 10.1007/s11882-013-0355-y
192. Rauch M, Lynch SV. The potential for probiotic manipulation of the gastrointestinal microbiome. Curr Opin Biotechnol. (2012) 23:192–201. doi: 10.1016/j.copbio.2011.11.004
193. Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol. (1997) 159:1739–45.
194. Amayasu H, Yoshida S, Ebana S, Yamamoto Y, Nishikawa T, Shoji T, et al. Clarithromycin suppresses bronchial hyperresponsiveness associated with eosinophilic inflammation in patients with asthma. Ann Allergy Asthma Immunol. (2000) 84:594–8. doi: 10.1016/S1081-1206(10)62409-X
195. Gibson PG, Yang IA, Upham JW, Reynolds PN, Hodge S, James AL, et al. Effect of azithromycin on asthma exacerbations and quality of life in adults with persistent uncontrolled asthma (AMAZES): a randomised, double-blind, placebo-controlled trial. Lancet. (2017) 390:659–68. doi: 10.1016/S0140-6736(17)31281-3
196. Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. (2013) 188:1193–201. doi: 10.1164/rccm.201304-0775OC
197. Egan JJ, Adamali HI, Lok SS, Stewart JP, Woodcock AA. Ganciclovir antiviral therapy in advanced idiopathic pulmonary fibrosis: an open pilot study. Pulm Med. (2011) 2011:240805. doi: 10.1155/2011/240805
198. Shulgina L, Cahn AP, Chilvers ER, Parfrey H, Clark AB, Wilson EC, et al. Treating idiopathic pulmonary fibrosis with the addition of co-trimoxazole: a randomised controlled trial. Thorax. (2013) 68:155–62. doi: 10.1136/thoraxjnl-2012-202403
199. Sethi S, Murphy TF. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N Engl J Med. (2008) 359:2355–65. doi: 10.1056/NEJMra0800353
200. Huang YJ, Sethi S, Murphy T, Nariya S, Boushey HA, Lynch SV. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J Clin Microbiol. (2014) 52:2813–23. doi: 10.1128/JCM.00035-14
201. Clancy RL, Dunkley M. Acute exacerbations in COPD and their control with oral immunization with non-typeable haemophilus influenzae. Front Immunol. (2011) 2:7. doi: 10.3389/fimmu.2011.00007
202. Teo E, Lockhart K, Purchuri SN, Pushparajah J, Cripps AW, van Driel ML. Haemophilus influenzae oral vaccination for preventing acute exacerbations of chronic bronchitis and chronic obstructive pulmonary disease. (2017) 2017:CD010010. doi: 10.1002/14651858.CD010010.pub3
203. Horita N, Goto A, Shibata Y, Ota E, Nakashima K, Nagai K, et al. Long-acting muscarinic antagonist (LAMA) plus long-acting beta-agonist (LABA) versus LABA plus inhaled corticosteroid (ICS) for stable chronic obstructive pulmonary disease (COPD). Cochrane Database Syst Rev. (2017) 2:CD012066. doi: 10.1002/14651858.CD012066.pub2
204. Maddi A, Sabharwal A, Violante T, Manuballa S, Genco R, Patnaik S, et al. The microbiome and lung cancer. J Thorac Dis. (2019) 11:280–91. doi: 10.21037/jtd.2018.12.88
205. Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. (2018) 359:1366–70. doi: 10.1126/science.aar6918
Keywords: asthma, chronic obstructive pulmonary disease, lung fibrosis, lung cancer, microbiome
Citation: Paudel KR, Dharwal V, Patel VK, Galvao I, Wadhwa R, Malyla V, Shen SS, Budden KF, Hansbro NG, Vaughan A, Yang IA, Kohonen-Corish MRJ, Bebawy M, Dua K and Hansbro PM (2020) Role of Lung Microbiome in Innate Immune Response Associated With Chronic Lung Diseases. Front. Med. 7:554. doi: 10.3389/fmed.2020.00554
Received: 27 February 2020; Accepted: 03 August 2020;
Published: 18 September 2020.
Edited by:
Anne Hilgendorff, Ludwig Maximilian University of Munich, GermanyReviewed by:
Jeffrey Louis Curtis, University of Michigan School of Medicine, United StatesBenjamin G. Wu, New York University School of Medicine, United States
Copyright © 2020 Paudel, Dharwal, Patel, Galvao, Wadhwa, Malyla, Shen, Budden, Hansbro, Vaughan, Yang, Kohonen-Corish, Bebawy, Dua and Hansbro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip M. Hansbro, cGhpbGlwLmhhbnNicm9AdXRzLmVkdS5hdQ==
†These authors have contributed equally to this work
 Keshav Raj Paudel
Keshav Raj Paudel Vivek Dharwal
Vivek Dharwal Vyoma K. Patel
Vyoma K. Patel Izabela Galvao1,2
Izabela Galvao1,2 Vamshikrishna Malyla
Vamshikrishna Malyla Sj Sijie Shen
Sj Sijie Shen Mary Bebawy
Mary Bebawy Kamal Dua
Kamal Dua Philip M. Hansbro
Philip M. Hansbro