
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Med. , 07 August 2020
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 7 - 2020 | https://doi.org/10.3389/fmed.2020.00444
This article is part of the Research Topic Coronavirus Disease (COVID-19): Pathophysiology, Epidemiology, Clinical Management and Public Health Response View all 400 articles
COVID-19, a disease induced by SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus-2), has been the cause of a worldwide pandemic. Though extensive research works have been reported in recent days on the development of effective therapeutics against this global health crisis, there is still no approved therapy against SARS-CoV-2. In the present study, plant-synthesized secondary metabolites (PSMs) have been prioritized to make a review focusing on the efficacy of plant-originated therapeutics for the treatment of COVID-19. Plant metabolites are a source of countless medicinal compounds, while the diversity of multidimensional chemical structures has made them superior to treat serious diseases. Some have already been reported as promising alternative medicines and lead compounds for drug repurposing and discovery. The versatility of secondary metabolites may provide novel antibiotics to tackle MDR (Multi-Drug Resistant) microbes too. This review attempted to find out plant metabolites that have the therapeutic potential to treat a wide range of viral pathogens. The study includes the search of remedies belonging to plant families, susceptible viral candidates, antiviral assays, and the mode of therapeutic action; this attempt resulted in the collection of an enormous number of natural therapeutics that might be suggested for the treatment of COVID-19. About 219 plants from 83 families were found to have antiviral activity. Among them, 149 plants from 71 families were screened for the identification of the major plant secondary metabolites (PSMs) that might be effective for this pandemic. Our investigation revealed that the proposed plant metabolites can serve as potential anti- SARS-CoV-2 lead molecules for further optimization and drug development processes to combat COVID-19 and future pandemics caused by viruses. This review will stimulate further analysis by the scientific community and boost antiviral plant-based research followed by novel drug designing.
Coronaviruses comprise a group of large, enveloped, positive-sensed, single-stranded RNA viruses that damage the respiratory tract of mammals including humans, bats, and other animals, leading to infections in the respiratory tract (1–5). The Coronavirus disease 2019 (COVID-19), initially called 2019 novel coronavirus (2019-nCoV), is an agile respiratory disease caused by a novel coronavirus primarily detected in Wuhan, China (6, 7). Now, it has spread to 216 countries and caused the death of more than 0.5 million people worldwide and was declared as a pandemic by the World Health Organization (WHO) (8, 9). Seven types of human coronaviruses have been reported so far, including HCoV-OC43, HCoV-229E, HCoV-HKU1, HCoV-NL63, severe acute respiratory syndrome (SARS)-CoV, Middle East respiratory syndrome (MERS-CoV), and 2019-novel coronavirus nCoV (10). Among them, MERS-CoV, SARS-CoV, and nCoV have taken the concern of scientists worldwide. In 2003, the severe acute respiratory syndrome (SARS) outbreak occurred in Guangdong (southern China) (6, 11) which infected 8,000 people and resulted in 800 deaths in 26 countries. Only a decade later, another coronavirus has attacked the world and caused another devastating outbreak, MERS, which infected 2,494 people and caused the deaths of 858 worldwide (12, 13). However, the COVID-19 pandemic caused by SARS CoV-2 resulted in remarkable levels of morbidity and mortality all over the world. Initially China, followed by the USA, Italy, France, Iran, Spain, Russia, Turkey, and the UK became hotspots for SARS CoV-2. The virus hotspot has now moved to Latin America and, at this time, Brazil, Mexico, and Peru are the new hotspots of SARS CoV-2. The important aspects of the pathobiology, a viral response phase, and a hyperbolic host response phase are linked with the morbidity and mortality in COVID-19 patients (14). However, the increased cytokine levels (IL-6, IL-10, and TNF-α), lymphopenia (in CD4+ and CD8+ T cells), and decreased IFN-γ expression in CD4+ T cells are the more risky and possibly life-threatening events related to severe COVID-19 (15–17). The infection rate of COVID-19 is increasing gradually but scientists have not been able to suggest any specific drug, vaccine, or any other certified therapeutic agents against SARS-CoV-2, which consequently leads to the significant morbidity and mortality.
On the other hand, plants have been essential to human welfare for their uses as therapeutics since ancient times (18, 19). According to the WHO, about 80% of the world's population depends on medicinal plants or herbs to fulfill their medicinal needs (20–22). A significant amount of antiviral compounds produced from numerous kinds of plants have been used in many studies (23–25). Researchers all around the world are screening therapeutic drugs from existing antiviral plant secondary metabolites (PSMs) and are also trying to find novel compounds from medicinal plants [(26–159); Supplementary Table 1] to avert this global crisis. Plant metabolites can halt the activity of enzymes involved in the replication cycle of CoVs including papain-like protease and 3CL protease, halt the fusion of the S protein of coronaviruses and ACE2 of the host, and also inhibit cellular signaling pathways (123, 144, 160). Screening from existing PSMs, researchers have been trying to find novel compounds from medicinal plants to prevent numerous diseases, including COVID-19 (Supplementary Table 1). Therefore, the current manuscript aims to describe potential metabolites from plant sources that have antiviral properties that might be aligned for the alternative approach against COVID-19. Hence, understanding the structure, life cycle, pathogenicity, cell signaling, epidemiology of the recently emerging virus, drug targets, and drug discovery process have become very important issues to find specific/effective therapeutics.
In December 2019, SARS CoV-2, one of the most devastating viral outbreaks since SARS CoV and MERS, originated from Wuhan city seafood market in China (161–163). The virus was found to be transmitted through close contact with infected people or through exposure to coughing, sneezing, and respiratory droplets (164, 165). It has already been reported to have spread to 216 countries and caused more than 0.5 million deaths. Brazil is now the new hotspot for SARS CoV-2 after the USA, Russia, France, Italy, Germany, Spain, and the UK, where more than 11 million people are infected (166, 167).
The pleomorphic or spherical shaped SARS COV-2 has a single-stranded RNA genome of 26.4–31.7 kb in length and a crown-like glycoproteins on its surface (168–173). It is more similar to SARS CoV (over 80%) than MERS (174, 175). However, the RNA genome of CoV-2 is considered as one of the largest genomes compared to those of other RNA viruses (176, 177). The largest open reading frame, ORF1ab, encodes non-structural proteins while the remaining ORFs encode four structural proteins, namely the envelope glycoprotein or spike protein (S), envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein. The S protein mediates attachment to the host cell while the E protein is involved in virus assembly, membrane permeability of the host cell, and virus-host cell interaction. The M protein is known as a central organizer for the coronavirus assembly and the nucleocapsid (N) protein is usually involved in the processing of helical ribonucleocapsid complex, including some accessory proteins (172, 178). Six types of mutations are found in the genome of SARS CoV-2 while three mutations have been reported in orf 1ab gene, two mutation in S gene, and the final one in the orf 7b and orf 8 (174, 175). Proteomic analysis revealed that SARS CoV-2 is vastly homologous to SARS CoV but two proteins, orf 8 and orf 10, are not homologous to SARS CoV (175). To complete its life cycle, SARS CoV-2 passes into the human body through the nose, mouth, or eyes and then attaches itself to the receptor-binding domain (RBD) using the surface glycoprotein (Spike-protein) of the virion which tries to attach with the hACE2 receptor (179, 180). The entry mechanism of SARS CoV-2 depends on cellular transmembrane serine protease 2 (TMPRSS2) and furin, along with viral receptor ACE2 (180–182). However, after the fusion of the SARS CoV-2 virion particle with the host cell membrane, the envelope and capsid part of the virus are removed. The virus releases its genetic material (RNA) into the host cell cytoplasm and acts as mRNA for the translation from ORF1a and ORF1ab to produce pp1a and pp1ab polypeptides (169, 183). Subsequently, chymotrypsin-like protease (3CLpro) slices these polypeptides into 16 non-structural proteins (NSPs) that are responsible for replication and transcription (184). Then, infected cells produce proteins when they become hijacked by SARS CoV-2. In this situation, the immune system supports the assembly of SARS CoV-2 into new copies of virion particles (185, 186). Freshly synthesized viral nucleic acids and proteins then assemble into the lumen of the ERGIC (Endoplasmic Reticulum Golgi Intermediate Compartment) and leave the cells through exocytosis [(187, 188); Figure 1]. Infected cells release virions and infect other human cells.
SARS-CoV-2 viral infection can be divided into three stages: the asymptomatic period, non-severe symptomatic period, and the severe infection stage (17, 189). SARS CoV-2 patients are reported to have a significant amount of cytokines and chemokines; the levels of cytokines are especially highly increased in patients admitted to ICUs (Intensive Care Unit) (190, 191). These significantly high levels are what results in a patient reaching a critical stage. However, the main mediator of SARS CoV-2, the spike glycoprotein, is found in two conformations (192) and the enzyme 3CLpro of SARS-CoV-2 share a 99.02% sequence identity with 3CLpro of SARS-CoV, which is also highly similar to bat SARS CoV 3CLpro (193). SARS CoV-2 binds to the host cell receptor with a higher affinity than SARS CoV (194). SARS CoV-2 has shown some strategic alteration with the substrate-binding site of bat SARS CoV-2 and 12 point-mutations are found in SARS CoV-2 compared to SARS CoV. Mutations disrupt the significant hydrogen bonds and modify the receptor binding site (RBS) of SARS-CoV-2 3CLpro. However, the occurrence of recurrent mutations can lead to new strains with alterations in virulence, which one of the reasons discovering a suitable vaccine to combat SARS CoV-2 is challenging (175, 195).
A fundamental therapeutic approach to treat multi-viral infections is the interruption of human host-virus interactions (17). The major structural proteins of SARS CoV-2 can be obvious targets for drugs designed against COVID-19. In addition, 16 non-structural proteins (NSPs) can also be considered (169). However, the manifestation of recurrent recombination events is a major hindrance to develop SARS CoV-2 specific vaccines/drugs (176). Up-to-date studies revealed that, though SARS-CoV-2 and SARS-CoV identify a similar receptor (ACE2) in humans (194, 196), there is a noteworthy variation in the antigenicity between SARS-CoV and SARS-CoV-2 which has significance on the development of therapeutic options against this rapidly emerging virus (197). The SARS-CoV-2 spike protein exhibits a higher affinity to the ACE2 receptor in comparison to SARS-CoV, but hACE2 showed a lower binding affinity to RBD (Receptor Binding Domain) of SARS COV-2 when compared to SARS CoV (194, 198). The two most paramount enzymes of SARS CoV-2, proprotein convertase furin- potentiates cell fusion and serine protease TMPRSS2, are responsible for S-protein activation and are propitious drug targets for the treatment of COVID (180, 194, 199).
Though extensive research works are being continued for the development of effective vaccines or drug compounds against SARS-CoV-2, efficacious therapeutics have not yet been attained (200). Moreover, interferon therapies, monoclonal antibodies, oligonucleotide-based therapies, peptides, small-molecule drugs, and vaccines, are regarded as some strategic approaches for controlling or preventing COVID-19 (201, 202). Existing drugs can be used as the first-line treatment for coronavirus outbreaks, but this is not the ultimate solution to eradicate the disease (203). Therefore, the development of therapeutic drugs for the treatment of the COVID-19 outbreak have gathered considerable attention. Scientists from different fields are trying to figure out the way to develop therapeutics. However, experimental implications of drug recombination might be both expensive and time-consuming, whereas computational evaluation may bring about testable hypotheses for systematic drug recombination (174).
Though there are approved, repurposed drugs currently in clinical use, there is still an urgent need for specific antiviral therapeutics and vaccines (199). Bioengineered and vectored antibodies and therapies based on cytokines and nucleic acid which target virus gene expression have been found as promising to treat coronavirus infections (204). For example, the repurposing drugs, including favipiravir, remdesivir, lopinavir, ritonavir, nebulized α-interferon, chloroquine, hydroxychloroquine, ribavirin, and interferon (IFN), have been shown to be effective for the treatment of COVID-19. Apart from this, some therapeutics are in clinical trials, such as peptide vaccine (mRNA-1273) (198) and antibody therapies (205). Recently, plasma therapy showed promising results for COVID-19 treatment (206, 207). But, application of these synthetic drugs are not efficient as they exhibit adverse direct or indirect side effects [(208–220); Table 1]. In addition, scientists all around the world are trying to find out some prominent drug and multi-epitope vaccine candidates against this deadly virus using various kinds of immuno-informatics approaches (221, 222). Therefore, the urgent need for safe, effective, and inexpensive therapies/drugs with negligible side effects against COVID-19 is imperative.
PSMs are a source of natural antiviral compounds that could be an effective option, as most of them are safer and more cost-effective compared to orthodox drugs (223), though some PSMs are toxic too. The dependency on and popularity of plant-based drugs are increasing day by day (224). Due to the presence of multiple compounds in crude plant extracts, it can be either beneficial or not, depending on the amounts used each time; if properly regulated, better activity might be shown. It was also found that crude extracts can target multiple sites at a time in a virion particle (225). However, this is yet to be tested against SARS-CoV-2. PSMs can affect the disruption of cell membrane functions and structures (226), interference with intermediary metabolisms (227), interruption of DNA/RNA synthesis and function (228), interruption of normal cell communication (quorum sensing) (229), and the induction of coagulation of cytoplasmic constituents (230). Different kinds of plant metabolites act against SARS CoV (Supplementary Table 1). Plant-based products affect several key events in the pathogenic process. For example, curcumin is effective for its antineoplastic, anti-proliferative, anti-aging, anti-inflammatory, anti-angiogenic, antiviral and anti-oxidant effects, and can regulate redox status, protein kinases, transcription factors, adhesion molecules, and cytokines in the human body (231). In silico analysis revealed that anti-SARS CoV PSMs could be one of the most valuable drug targets against SARS CoV-2 [(232); Table 2]. A huge amount of plant metabolites have remained unexplored due to the extensive process of isolation of the target compound. Now, various types of modern techniques have been developed for the isolation of lead compounds from crude extracts including maceration, percolation, decoction, reflux extraction, soxhlet extraction, pressurized liquid extraction, supercritical fluid extraction, ultrasound assisted extraction, microwave-assisted extraction, pulsed electric field extraction, enzyme assisted extraction, hydro distillation, and steam distillation (179). These techniques can lead us to find out novel anti-SARS CoV-2 compounds earlier than traditional techniques. In addition, plant metabolomics are used as a tool for the discovery of novel drugs from plant resources (262, 263).
Plants produce diversified low molecular weight PSMs to protect them from different herbivores and microbes (264). Before the discovery of allopathic drugs, these leading natural sources were extensively used for treating several kinds of human diseases (265, 266). Due to the increased resistance of microbial pathogens against allopathic drugs, researchers have now returned to natural resources, focusing especially on plant metabolites, to find out lead compounds to fight against human pathogens (175). Moreover, about 35% of the global medicine market (which accounts for 1.1 trillion US dollars) have been shared by medicinal products prepared using natural plants or herbs (265). Investigations are undergoing for the finding of novel and modern drugs from numerous herbal preparations to fight against this microbial resistance war. Many similarities have been found between SARS CoV and SARS CoV-2 (both of them belong to beta family, containing the same genetic material-RNA, and using the same receptor for viral attachment-ACE2, with an 86% identity and 96% similarity of genome, with almost the same pathogenesis). Thus, previously reported antiviral plant metabolites for SARS CoV can be considered as emerging drug candidates for COVID-19. Right now, the setbacks arising from viral infection around the world have placed budget constraints on researchers trying to discover effective antiviral drugs. However, some PSMs have already shown anti-SARS CoV activity where other antiviral activities are also reported (Supplementary Table 1). These results suggest that there is a scope to find alternative medicines and specific compounds. So, plants could be a vital resource in the fight against COVID-19. Our study suggests that around 76 natural metabolites from different plant species can be efficiently active against COVID-19 (Table 3 and Supplementary Figure 1).
A wide variety of antiviral compounds were found from 219 medicinal plants (26–159) belonging to 83 plant families (Supplementary Table 1). First and foremost are polyphenols, which contain multiple phenolic rings, and are classified as phenols, flavonoids, lignans, hydroxycinnamic acid, stilbenes, and hydroxybenzoic acid (267). We found polyphenols in numerous plants (Table 4) which exerted antiviral activity (269–271) against a wide range of viruses including HIV-1, HIV-2, HSV-1, HSV-2, Influenza virus, Dengue virus, HBV, HCV, Infectious bronchitis virus (IBV), Murbarg virus, Ebola virus, Newcastle disease virus (NDV), Poliomyelitis-1 virus, Lentivirus, and Coronavirus. Polyphenols work against coronaviruses using diverse mechanisms including actuating or inhibiting cellular signaling pathways or halting papain-like protease (PLpro) and 3-chymotripsin-like protease (3CLpro) enzyme (269, 272). Some polyphenol compounds (30-(3-methylbut-2-enyl)-30, 4-hydroxyisolonchocarpin, broussochalcone A, 4,7-trihydroxyflavane, broussochalcone B, papyriflavonol A, kazinol A, kazinol B, kazinol F, kazinol J, and broussoflavan A) isolated from Broussonetia papyrifera showed promising activity against SARS CoV. Higher efficiency against PLpro as observed by these compounds though activity against Mpro or 3CLpro is not up to the mark. Specially, papyriflavonol A possesses impressive activity against SARS CoV (IC50 3.7, l M) (272). In silico analysis revealed that polyphenols can inhibit SARS CoV-2 Mpro and RdRp effectively (273, 274). In our study, we have found another widely distributed, low molecular weight phenolic compound named as a flavonoid which showed strong antiviral activity against SARS CoV, Influenza virus, HBV, HSV, HCV, HIV, Dengue virus, Simian virus, Human rotavirus, Bovine viral diarrhea virus, Poliomyelitis-1 virus, Vesicular stomatitis virus (VSV), and Newcastle disease virus (NDV) (Table 4). Flavonoid type compounds, such as apigenin and quercetin, showed activity against SARS CoV virion particles through the inhibition of Mpro enzymes with an IC50 of 38.4 ± 2.4 μM and 23.8 μM, respectively (144, 150, 275). According to in silico analysis, flavonoid compounds can terminate the activity of Mpro of SARS CoV-2 (276, 277).
Alkaloids are another class of natural organic compounds which are classified into several groups based on their heterocyclic ring, such as tropanes, pyrrolidines, isoquinoline purines, imidazoles, quinolizidines, indoles, piperidines, and pyrrolizidines (278). Alkaloids are very promising against HIV-1, HSV-1, HSV-2, DNV, VSV, Influenza virus, and Newcastle disease virus (NDV) (Table 4). Different kinds of alkaloids showed anti-SARS activity including emetine, Ipecac, Macetaxime, tylophorine, and 7-methoxy cryptopleurine, through the inhibition of protease enzyme, RNA synthesis, and protein synthesis (244, 279). In addition, some alkaloids act against SARS CoV as a nucleic acid intercalating agent such as tetrandrine, fangchinoline, cepharanthine, and lycorine through degrading nucleic acids and inhibiting spike and nucleocapsid proteins (280). Virtual screening analysis revealed that 10-Hydroxyusambarensine and Cryptoquindoline—two alkaloid compound isolated from African medicinal plants showed anti-SARS CoV and anti-SARS CoV-2 activity through inhibition of their Mpro (256). Chloroquine, a derivative of alkaloid, is found to be active against anti-SARS CoV-2 (281). So, some PSMs as alkaloids can be alternative drug targets for COVID-19 (280).
Another class of PSMs, saponins (amphipathic glycosides), are found ubiquitously in plants which showed antiviral activities against Newcastle disease virus (NDV), Simian (SA-11) virus, Murine norovirus (MNV) and Feline calicivirus (FCV), RSV, VSV, HSV-1,HSv-2, HIV-1, Epstein–Barr virus (EBV), (SA-11) and human (HCR3) rotaviruses, Influenza virus, and Dengue virus (Table 4). Plants produce five carbon isoprene derived terpenes which are the largest and most diverse group of PSM. They are classified by monoterpenes, diterpenes, triterpenes, sesterterpenes, hemi terpenes, and sesquiterpenes (282). They exhibited antiviral activity against Bovine viral diarrhea virus, HSV-1, Poliovirus type 2 (PV-2) and vesicular stomatitis virus (VSV), Dengue virus serotype-1 (DENV-1), Influenza A and B viruses, HIV-1, HIV-2, SIV mac 251, and SARS-CoV (Table 4). Ten diterpenes, two sesquiterpenes, and two triterpenes showed anti-SARS activity with IC50 of 3–10 μM (283). In silico analysis also revealed that terpene Ginkgolide A can strongly inhibit SARS CoV-2 protease enzyme (284). Carbohydrates, mainly classified as monosaccharides, disaccharides, polysaccharides, and oligosaccharides (282), are found as antiviral agent against Human rotavirus, Influenza A virus, HSV-1, HSV-2, Herpes simplex virus (HSV), RSV, Coxsackie B3 Virus, and VSV [(285); Table 4]. Acyclovir is an FDA (Food and Drug Administration) approved antiviral drug which is obtained from Carissa edulis (Supplementary Table 1). It is mainly used for herpes simplex virus, chickenpox, and shingles. The group basis structure of some major compounds can be found in Table 5.
Drug discovery from plant metabolites refers to the extraction and purification of active ingredients from conventional cures. Natural plant products comprise complicated chemical structures which differ according to their numerous species. There are several classes of PSMs which are responsible for the biological activities of herbal medicines. PSMs exert their actions on molecular targets that differ from one case to the other. These targets may be enzymes, mediators, transcription factors, or even nucleic acids (286). Good knowledge of the chemical composition of plants leads to a better understanding of their possible and specific medicinal value. Drug discovery and development have become a wide interdisciplinary field over recent decades and many factors are involved in the successful evolution from a bioactive compound into a potential drug [(287, 288); Figure 2]. When existing methods with advanced technologies are applied, it can lead to a modern revelation of drugs, benefitting medicinal purposes (223, 289). The development of modern technologies has streamlined the screening of natural products in discovering new drugs. Research for drug discovery must create robust and prudent lead molecules, which is progressed from a screening hit to a drug candidate through structural elucidation and structure recognizable proof available from high throughput technology like GC–MS, NMR, IR, HPLC, and HPTLC. Utilizing these advanced technologies gives us an opportunity to perform research in screening novel molecules employing a computer program and database to set up common items as a major source for drug discovery. It finally leads to lead structure discovery. Powerful new technologies are revolutionizing natural herbal drug discovery (223). Steps associated with the drug discovery process from natural resources is illustrated (Figure 3).
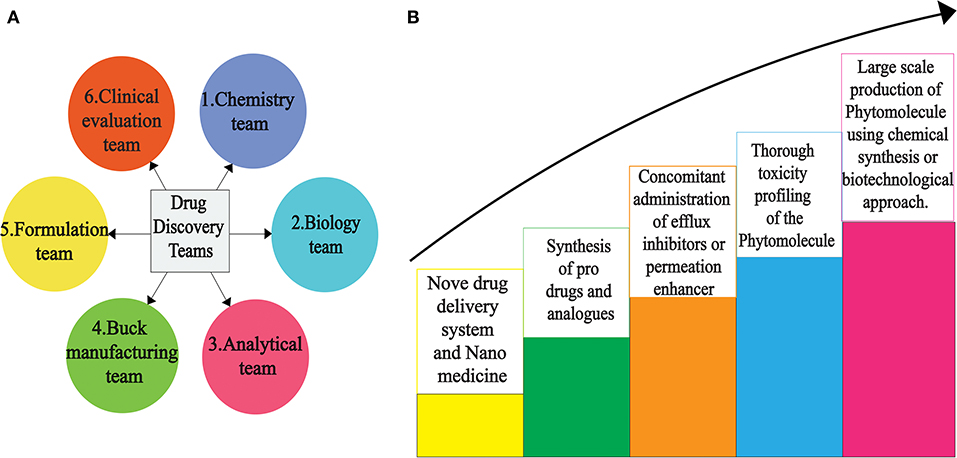
Figure 2. Scientific teams (A) to overcome various hurdles for successful novel drug discovery (B) from PSMs.
However, several factors involving the conversion of a desirable compound into a valuable drug candidate include availability, bioavailability, intellectual property, and the strong pharmacokinetic profile of the compound (268, 290). Sometimes researchers find great bioactivity of a plant-derived compound in in vitro analysis but unfortunately, the desired compound becomes ineffectual under in vivo conditions (291). In vivo is a very crucial step to move to animal trials or subsequent clinical trials. Even if the compound shows promising activity in in vivo assay but it can still become ineffective in animal model trials due to a poor pharmacokinetic profile (292). Under in vivo condition, the target compound remains in direct contact with cells, while in animal models the compound moves to various stages where it might lose its bioactivity (292). For example, despite curcumin having promising antioxidant, anticancer, anti-inflammatory, and antimicrobial activities, it has not been released as a drug yet due to its poor bioavailability (292). Another propitious drug candidate, epigallocatechin gallate (EGCG), showed antioxidant, antihypertensive, anticancer, antimicrobial, and anti-inflammatory activity (293, 294) but unfortunately, it has also failed to obtain drug designation due to the same reason mentioned for curcumin (292).
To remedy these problems, researchers around the world are working to develop new approaches. Changing the administration route might increase the bioavailability of a compound. For example, the bioavailability of an anti-inflammatory compound, andrographolide, is increased when it is administered intravenously instead of through oral administration (295). Other methods to enhance the bioavailability of target compound include using drug delivery systems, the nano-formulation of a drug, using adjuvant systems, or altering structural analogs (220, 296). Furthermore, the modification of pharmacokinetic profiles of compounds like absorption, distribution, metabolism, and excretion can escalate its probability as drug candidate (268). Indeed, there is an urgent need for specific protocols for invention of novel bioactive compounds and for this purpose it is very crucial for related organizations, companies, and agencies, including the World Health Organization (WHO), Food and Drug Administration (FDA), European Medicines Agency (EMA), World Trade Organization (WTO), International Conference on Harmonization (ICH), World Intellectual Property Organization (WIPO), biotech companies, pharmaceutical pharmaceuticals companies, and several other companies and agencies, to work together. However, plant-originated therapeutics need to be taken under consideration against SARS-CoV-2 as they have already shown promising hopes for different critical conditions caused by deadly pathogens.
The seven major drug targets of SARS CoV-2 were described before (176). Similarly, screening of PSMs for drug establishment by molecular docking is efficient in terms of time and cost. Even the development of vaccines through computational biology was found to be effective for previous severe viruses like MERS using animal models, target antigens, and probable vaccine candidates (181). But still, there exists a lack of a complete review for PSMs as alternative drug therapeutics. Our review aims at establishing PSMs as a strong and safe candidate for the treatment of SARS CoV-2. Through suggesting probable antiviral plant metabolites or screening, druggability analysis of plant metabolites against SARS-CoV-2 has become a time-saving practice (280, 297). Without establishing a drug development pipeline that includes clinical trials, these suggested candidate PSMs will end up only in journal publications or be shelved as herbal formulations on a supermarket store as a traditional medicine and will never be a modern drug. Undoubtedly, the plant an underutilized source of novel bioactive compounds and is one of the hotspots to fight against this microbial resistance war. The decrypting of PSMs is not increasing so much in comparison to the number of metabolites produced from plants. A biotechnological approach can offer a desired amount of secondary metabolites in a rapid and eco-friendly way against SARS-CoV-2 (298). In addition, plant metabolomics are now used as a tool for discovery of novel drugs from plant resources (299). Characterization of genes and proteins involved in secondary metabolic pathways are also very crucial to understand. Therefore, omics approaches (transcriptomics, proteomics, and metabolomics) have paramount importance in food research and drug discovery (300, 301) for human welfare. Genetic modifications for engineering plant metabolites can be helpful for reaching a specific drug. Quality control of natural products is also very important. So, laboratory support, skilled manpower, and funding is also very important for drug discovery from natural resources.
Scientists all around the world are trying to discover the most effective antiviral drug to combat SARS CoV-2. In this situation, our study accentuated some plant secondary metabolites that showed prominent antiviral activity against coronaviruses through impeding the main machinery used in their pathogenesis and replication cycle. The in vitro, in vivo, and in silico investigations revealed numerous plant-derived compounds with promising anti- SARS CoV and anti- SARS CoV-2 activity [Table 2; (179, 219, 222, 233–261, 297)]. Plants are a dramatically underutilized source of bioactive compounds with a broad spectrum of antiviral activities. Some Chinese traditional plant formulations have been reported as being anti- SARS CoV-2 and this formulation is also provided in COVID patients (302, 303). We reported here on 219 plants which act against a wide range of DNA/RNA viruses, but the plant PSMs that showed promising activity against SARS CoV and MERS might be a desired drug candidate against SARS CoV-2. So, this review gathered all antiviral plants in a single platform to facilitate laboratory-based research for the development of novel drug/molecular therapeutics to overcome this and future pandemic situations. The world is facing a serious health crisis, and it needs an effective solution to combat the burning flame of COVID-19. Researchers are trying to find an effective way to overcome this situation, and the present study could help them to think with a new dimension by using the knowledge from the databases based on the plant metabolites (304, 305). Finally, advanced and rapid acting extraction, purification, and characterization techniques used for plant metabolites as well as multidisciplinary expertise and funding are very essential for novel drug discovery.
Articles were selected and identified by searching specific keywords and journal citations for each section of a manuscript. Related peer reviewed scientific journal articles were screened from different journal depositories after reviewing abstracts and original data.
The authors initiated this project to facilitate the research on molecular therapeutics from plant sources as an immediate action in response to the COVID-19 pandemic situation.
FB and MH designed the project. FB prepared the first draft. FB, SH, TR, and MH have investigated the data and completed the manuscript. All authors have read through the manuscript and approved it for submission and publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful to Mr. Md. Fayezur Rahman (MSS in Economics, Comilla University) for drawing the figures as authors expected. Our sincere gratitude to Mr. Mohammad Saifullah (MA in English, University of Chittagong) for proofreading of the manuscript. We are grateful to the editors and reviewers for their valuable comments during the review process to improve the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2020.00444/full#supplementary-material
Supplementary Figure 1. Different plant families showing antiviral properties. (Each portion of the pie chart describes a specific Family alongside its total number of plants that have antiviral properties).
Supplementary Table 1. List of secondary metabolites found from medicinal plants.
1. Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. (2005) 287:1–30. doi: 10.1007/3-540-26765-4_1
2. Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. (2020) 395:565–74. doi: 10.1016/S0140-6736(20)30251-8
3. Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. (2007) 38:281–97. doi: 10.1051/vetres:2006055
4. Ismail MM, Tang Y, Saif YM. Pathogenicity of turkey coronavirus in turkeys and chickens. Avian Dis. (2003) 47:515–22. doi: 10.1637/5917
5. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. (2015) 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1
6. Lan Jun, Jiwan Ge, Jinfang Yu, Sisi Shan, Huan Zhou, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. (2020) 581:215–20 doi: 10.1101/2020.02.19.956235
7. Chen Y, Peng H, Wang L, Zhao Y, Zeng L, Gao H, et al. Infants born to mothers with a new coronavirus (COVID-19). Front Pediatr. (2020) 8:104. doi: 10.3389/fped.2020.00104
8. WHO. Director-General's Opening Remarks at the Media Briefing on COVID-19. (2020). Available online at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19 (accessed May 15, 2020).
9. WHO. Director-General's Opening Remarks at the Media Briefing on COVID-19. (2020). Available online at: https://www.who.int/dg/speeches/detail/who-director-general-sopening-remarks-at-the-media-briefing-on-covid-19 (accessed April 5, 2020).
10. Kanne JP. Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: Key points for the radiologist. Radiology. (2020) 295:16–7. doi: 10.1148/radiol.2020200241
11. Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen KY. Coronaviruses-drug discovery and therapeutic options. Nat Rev Drug Discov. (2016) 15:327–47. doi: 10.1038/nrd.2015.37
12. Heymann DL, Rodier G. Global surveillance, national surveillance, and SARS. Emerg Infect Dis. (2004) 10:173–5. doi: 10.3201/eid1002.031038
13. Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. (2012) 367:1814–20. doi: 10.1056/NEJMoa1211721
14. Shetty R, Ghosh A, Honavar SG, Khamar P, Sethu S. Therapeutic opportunities to manage COVID-19/SARS-CoV-2 infection: present and future. Indian J Ophthalmol. (2020) 68:693–702. doi: 10.4103/ijo.IJO_639_20
15. Hirano T, Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. (2020) 52:731–3. doi: 10.1016/j.immuni.2020.04.003
16. Pedersen SF, Ho YC. SARS-CoV-2: a storm is raging. J Clin Invest. (2020) 130:2202–5. doi: 10.1172/JCI137647
17. Coperchini F, Chiovato L, Croce L, Magri F, Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. (2020) 53:25–32. doi: 10.1016/j.cytogfr.2020.05.003
18. Moses T, Goossens A. Plants for human health: greening biotechnology and synthetic biology. J Exp Bot. (2017) 68:4009–11. doi: 10.1093/jxb/erx268
19. Schaal B. Plants and people: our shared history and future. Plants People Planet. (2019) 1:14–9. doi: 10.1002/ppp3.12
20. World Health Organization. WHO Global Report on Traditional and Complementary Medicine 2019. World Health Organization (2019).
21. Hoque Haque MI, Chowdhury ABMA, Shahjahan M, Harun MGD. Traditional healing practices in rural Bangladesh: a qualitative investigation. BMC Complement Altern Med. (2018) 18:62. doi: 10.1186/s12906-018-2129-5
22. Ozioma E, Okaka A. Herbal medicines in African traditional medicine. In: Ozioma EJ and Chinwe OAN, editors. Herbal Medicine. Intratech open (2019). p. 1–25.
23. Jassim SAA, Naji MA. Novel antiviral agents: a medicinal plant perspective. J Appl Microbiol. (2003) 95:412–27. doi: 10.1046/j.1365-2672.2003.02026.x
24. Hussain W, Haleem KS, Khan I, Tauseef I, Qayyum S, Ahmed B, et al. Medicinal plants: a repository of antiviral metabolites. Future Virol. (2017) 12:299–308. doi: 10.2217/fvl-2016-0110
25. Özçelik B, Kartal M, Orhan I. Cytotoxicity, antiviral and antimicrobial activities of alkaloids, flavonoids, and phenolic acids. Pharm Biol. (2011) 49:396–402. doi: 10.3109/13880209.2010.519390
26. Pick A, Ling K, Khoo BF, Seah CH, Foo KY, Cheah RK. Inhibitory activities of methanol extracts of Andrographis Paniculata and Ocimum Sanctum against Dengue-1 virus. In: International Conference on Biological, Environment & Food Engineering. Bali (2014). p. 6.
27. Namazi R, Zabihollahi R, Behbahani M, Rezae A. Inhibitory activity of Avicennia marina, a medicinal plant in persian folk medicine, against HIV and HSV. Iran J Pharm Res.. (2013) 12:435–43.
28. Zhou B, Yang Z, Feng Q, Liang X, Li J, Zanin M, et al. Aurantiamide acetate from baphicacanthus cusia root exhibits anti-inflammatory and anti-viral effects via inhibition of the NF-κB signaling pathway in influenza A virus-infected cells. J Ethnopharmacol. (2017) 199:60–7. doi: 10.1016/j.jep.2017.01.038
29. Castillo-Maldonado I, Moreno-Altamirano MMB, Serrano-Gallardo LB. Anti-dengue serotype-2 activity effect of Sambucus nigra leaves-and flowers-derived compounds. Virol Res Rev. (2017) 1:1–5. doi: 10.15761/VRR.1000117
30. Chen C, Zuckerman DM, Brantley S, Sharpe M, Childress K, Hoiczyk E, et al. Sambucus nigra extracts inhibit infectious bronchitis virus at an early point during replication. BMC Vet Res. (2014) 10:24. doi: 10.1186/1746-6148-10-24
31. Andleeb R, Ashraf A, Muzammil S, Naz S, Asad F, Ali T, et al. Analysis of bioactive composites and antiviral activity of Iresine herbstii extracts against Newcastle disease virus in ovo. Saudi J Biol Sci. (2020) 27:335–40. doi: 10.1016/j.sjbs.2019.10.002
32. Szlμvik L, Minμrovits J, Forgo P. Alkaloids from Leucojum vernum and antiretroviral activity of Amaryllidaceae alkaloids. Planta Medica. (2004) 70:871–3. doi: 10.1055/s-2004-827239
33. Lopes RC, Oliveira DB, Costa SS, Miranda MMFS, Romanos MT V, Santos NSO, et al. In vitro anti-rotavirus activity of some medicinal plants used in Brazil against diarrhea. J Ethnopharmacol. (2005) 99:403–7. doi: 10.1016/j.jep.2005.01.032
34. Reichling J, Neuner A, Sharaf M, Harkenthal M, Schnitzler P. Antiviral activity of Rhus aromatica (fragrant sumac) extract against two types of herpes simplex viruses in cell culture. Pharmazie. (2009) 64:538–41.
35. Modi M, Nutan, Pancholi B, Kulshrestha S, Rawat AKS, Malhotra S, et al. Anti-HIV-1 activity, protease inhibition and safety profile of extracts prepared from Rhus parviflora. BMC Complement Altern Med. (2013) 13:158. doi: 10.1186/1472-6882-13-158
36. Nocchi SR, Companhoni MVP, De Mello JCP, Dias Filho BP, Nakamura CV, Carollo CA, et al. Antiviral activity of crude hydroethanolic extract from Schinus terebinthifolia against herpes simplex virus type 1. Planta Med. (2017) 83:509–18. doi: 10.1055/s-0042-117774
37. Park JY, Ko JA, Kim DW, Kim YM, Kwon HJ, Jeong HJ, et al. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J Enzyme Inhib Med Chem. (2016) 31:23–30. doi: 10.3109/14756366.2014.1003215
38. Tolo FM, Rukunga GM, Muli FW, Njagi ENM, Njue W, Kumon K, et al. Anti-viral activity of the extracts of a Kenyan medicinal plant Carissa edulis against Herpes simplex virus. J Ethnopharmacol. (2006) 104:92–9. doi: 10.1016/j.jep.2005.08.053
39. Bonvicini F, Lianza M, Mandrone M, Poli F, Gentilomi GA, Antognoni F. Hemidesmus indicus (L.) R. Br. Extract inhibits the early step of herpes simplex type 1 and type 2 replication. New Microbiol. (2018) 41:187–94.
40. Rittà M, Marengo A, Civra A, Lembo D, Cagliero C, Kant K, et al. Antiviral activity of a Arisaema tortuosum leaf extract and some of its constituents against herpes simplex virus type 2. Planta Med. (2020) 86:267–75. doi: 10.1055/a-1087-8303
41. Lee JS, Ko E, Hwang HYESUK, Lee Y, Kwon Y, Kim M, et al. Antiviral activity of ginseng extract against respiratory syncytial virus infection. Int J Mol Med. (2014) 34:183–90. doi: 10.3892/ijmm.2014.1750
42. Lee MH, Lee B, Jung J, Cheon D, Kim K, Choi C. Antiviral effect of Korean red ginseng extract and ginsenosides on murine norovirus and feline calicivirus as surrogates for human norovirus. J Ginseng Res. (2011) 35:429–35. doi: 10.5142/jgr.2011.35.4.429
43. Choi J, Jin Y, Lee H, Oh TW, Yim N, Cho W. Protective effect of Panax notoginseng root water extract against influenza a virus infection by enhancing antiviral interferon-mediated immune responses and natural killer cell activity. Front Immunol. (2017) 8:1542. doi: 10.3389/fimmu.2017.01542
44. Glatthaar-Saalmüller B, Fal AM, Schönknecht K, Conrad F, Sievers H, Saalmüller A. Antiviral activity of an aqueous extract derived from Aloe arborescens Mill. against a broad panel of viruses causing infections of the upper respiratory tract. Phytomedicine. (2015) 22:911–20. doi: 10.1016/j.phymed.2015.06.006
45. Zandi K, Zadeh MA, Sartavi K, Rastian Z. Antiviral activity of Aloe vera against herpes simplex virus type 2 : an in vitro study. Afr J Biotechnol. (2007) 6:1770203. doi: 10.5897/AJB2007.000-2276
46. Moradi M, Rafieian-kopaei M, Karimi A. A review study on the effect of Iranian herbal medicines against in vitro replication of herpes simplex virus. Avicenna J Phytomed. (2016) 6:506–15.
47. Soltan MM, Zaki AK. Antiviral screening of forty-two Egyptian medicinal plants. J Ethnopharmacol. (2009) 126:102–7. doi: 10.1016/j.jep.2009.08.001
48. Patocka J, Navratilova Z. Achillea fragrantissima : pharmacology review. Clin Oncol. (2019) 4:1601.
49. Visintini M, Redko F, Muschietti L, Campos R, Martino V, Cavallaro LV. in vitro antiviral activity of plant extracts from Asteraceae medicinal plants. Virol J. (2013) 10:245. doi: 10.1186/1743-422X-10-245
50. Derksen A, Kühn J, Hafezi W, Sendker J, Ehrhardt C, Ludwig S, et al. Antiviral activity of hydroalcoholic extract from Eupatorium perfoliatum L. against the attachment of influenza A virus. J Ethnopharmacol. (2016) 188:144–52. doi: 10.1016/j.jep.2016.05.016
51. Lani R, Hassandarvish P, Chiam CW, Moghaddam E, Chu JJH, Rausalu K, et al. Antiviral activity of silymarin against chikungunya virus. Sci Rep. (2015) 5:11421. doi: 10.1038/srep11421
52. Benassi-Zanqueta É, Marques CF, Valone LM, Pellegrini BL, Bauermeister A, Ferreira ICP, et al. Evaluation of anti-HSV-1 activity and toxicity of hydroethanolic extract of Tanacetum parthenium (L.) Sch.Bip. (Asteraceae). Phytomedicine. (2019) 55:249–54. doi: 10.1016/j.phymed.2018.06.040
53. Rehman S, Ijaz B, Fatima N, Aun S. Sciencedirect therapeutic potential of Taraxacum officinale against HCV NS5B polymerase : in-vitro and in silico study. Biomed Pharmacother. (2016) 83:881–91. doi: 10.1016/j.biopha.2016.08.002
54. He W, Han H, Wang W, Gao B. Anti-influenza virus effect of aqueous extracts from dandelion. Virol J. (2011) 8:538. doi: 10.1186/1743-422X-8-538
55. Rothan HA, Zulqarnain M, Ammar YA, Tan EC, Rahman NA, Yusof R. Screening of antiviral activities in medicinal plants extracts against dengue virus using dengue NS2B-NS3 protease assay. Trop Biomed. (2014) 31:286–96.
56. Mgole S, Pieters L, David O, Apers S, Vingerhoets R, Cos P, et al. Screening of some Tanzanian medicinal plants from Bunda District for antibacterial, antifungal and antiviral activities. J Ethnopharmacol. (2008) 119:58–66. doi: 10.1016/j.jep.2008.05.033
57. Al-thobaiti SA, Zeid IMA. Phytochemistry and pharmaceutical evaluation of Balanites aegyptiaca : an overview. J Exp Biol Agric Sci. (2018) 6:453–65 doi: 10.18006/2018.6(3).453.465
58. Cho WK, Kim H, Choi YJ, Yim NH, Yang HJ, Ma JY. Epimedium koreanum Nakai water extract exhibits antiviral activity against porcine epidermic diarrhea virus in vitro and in vivo. Evid Based Complement Altern Med. (2012) 2012:985150. doi: 10.1155/2012/985151
59. Park JY, Jeong HJ, Kim JH, Kim YM, Park SJ, Kim D, et al. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol Pharm Bull. (2012) 35:2036–42. doi: 10.1248/bpb.b12-00623
60. Tung NH, Kwon HJ, Kim JH, Ra JC, Ding Y, Kim JA, et al. Anti-influenza diarylheptanoids from the bark of Alnus japonica. Bioorganic Med Chem Lett. (2010) 20:1000–3. doi: 10.1016/j.bmcl.2009.12.057
61. Lin CW, Tsai FJ, Tsai CH, Lai CC, Wan L, Ho TY, et al. Anti-SARS coronavirus 3C-like protease effects of Isatis indigotica root and plant-derived phenolic compounds. Antiviral Res. (2005) 68:36–42. doi: 10.1016/j.antiviral.2005.07.002
62. Chen F, Yang L, Huang Y, Chen Y, Sang H, Duan W, et al. Isocorilagin, isolated from Canarium album (Lour.) Raeusch, as a potent neuraminidase inhibitor against influenza A virus. Biochem Biophys Res Commun. (2020) 523:183–9. doi: 10.1016/j.bbrc.2019.12.043
63. César GZJ, Alfonso MGG, Marius MM, Elizabeth EM, Ángel CBM, Maira HR, Guadalupe CLM, et al. Inhibition of HIV-1 reverse transcriptase, toxicological and chemical profile of Calophyllum brasiliense extracts from Chiapas, Mexico. Fitoterapia. (2011) 82:1027–34. doi: 10.1016/j.fitote.2011.06.006
64. Ibrahim AK, Youssef AI, Arafa AS, Ahmed SA. Anti-H5N1 virus flavonoids from Capparis sinaica Veill. Nat Prod Res. (2013) 27:2149–53. doi: 10.1080/14786419.2013.790027
65. Ghazal EA, Khamis IMA, Elhaw MHM. Chemical constituents of Capparis sinaica Veill. plant and its antimicrobial effects. Middle East J Appl Sci. (2015) 5:411–22.
66. Lam S-Z, Ng T-B. A protein with antiproliferative, antifungal and HIV-1 reverse transcriptase inhibitory activities from caper (Capparis spinosa) seeds. Phytomedicine. (2009) 16:444–50. doi: 10.1016/j.phymed.2008.09.006
67. Callies O, Bedoya LM, Beltra M, Mun A, Obrego P, Osorio AA, et al. Isolation, structural modification, and HIV inhibition of pentacyclic lupane-type triterpenoids from Cassine xylocarpa and Maytenus cuzcoina. J Nat Prod. (2015) 78:1045–55. doi: 10.1021/np501025r
68. Inoue R. Orally administered Salacia reticulata extract reduces h1n1 influenza clinical symptoms in murine lung tissues putatively due to enhanced natural killer cell activity. Front Immunol. (2016) 7:115. doi: 10.3389/fimmu.2016.00115
69. Rebensburg S, Helfer M, Schneider M, Koppensteiner H, Eberle J, Schindler M, et al. Potent in vitro antiviral activity of Cistus incanus extract against HIV and Filoviruses targets viral envelope proteins. Sci Rep. (2016) 6:20394. doi: 10.1038/srep20394
70. Javed T, Ashfaq UA, Riaz S, Rehman S, Riazuddin S. In-vitro antiviral activity of Solanum nigrum against Hepatitis C Virus. Virol J. (2011) 8:26. doi: 10.1186/1743-422X-8-26
71. Alcami J, Bermejo P. Phytomedicine Ellagitannins from Tuberaria lignosa as entry inhibitors of HIV. Phytomedicine. (2010) 17:69–74. doi: 10.1016/j.phymed.2009.08.008
72. Mushi NF, Mbwambo ZH, Innocent E, Tewtrakul S. Activities of aqueous ethanolic extracts from Combretum adenogonium Steud. Ex A. Rich (Combretaceae). BMC Complement Altern Med. (2012) 12:163. doi: 10.1186/1472-6882-12-163
73. Liu M, Katerere DR, Gray AI, Seidel V. Phytochemical and antifungal studies on Terminalia mollis and Terminalia brachystemma. Fitoterapia. (2009) 80:369–73. doi: 10.1016/j.fitote.2009.05.006
74. Lavoie S, Côté I, Pichette A, Gauthier C, Ouellet M, Nagau-Lavoie F, et al. Chemical composition and anti-herpes simplex virus type 1 (HSV-1) activity of extracts from Cornus canadensis. BMC Complement Altern Med. (2017) 17:2. doi: 10.1186/s12906-017-1618-2
75. Hsieh CF, Chen YL, Lin CF, Ho JY, Huang CH, Chiu CH, et al. An extract from Taxodium distichum targets hemagglutinin- and neuraminidase-related activities of influenza virus in vitro. Sci Rep. (2016) 6:36015. doi: 10.1038/srep36015
76. Xu H, Ma Y, Huang X, Geng C, Wang H. Bioactivity-guided isolation of anti-hepatitis B virus active sesquiterpenoids from the traditional Chinese medicine : Rhizomes of Cyperus rotundus. J Ethnopharmacol. (2015) 171:131–40. doi: 10.1016/j.jep.2015.05.040
77. Danciu C, Muntean D, Alexa E, Farcas C, Oprean C, Zupko I, et al. Phytochemical characterization and evaluation of the antimicrobial, antiproliferative and pro-apoptotic potential of Ephedra alata Decne. hydroalcoholic extract against the MCF-7 breast cancer cell line. Molecules. (2018) 24:13. doi: 10.3390/molecules24010013
78. Churqui MP, Lind L, Thörn K, Svensson A, Savolainen O, Aranda KT, et al. Extracts of Equisetum giganteum L and Copaifera reticulate Ducke show strong antiviral activity against the sexually transmitted pathogen herpes simplex virus type 2. J Ethnopharmacol. (2018) 210:192–7. doi: 10.1016/j.jep.2017.08.010
79. Shamsabadipour S, Ghanadian M, Saeedi H, Reza Rahimnejad M, Mohammadi-Kamalabadi M, Ayatollahi SM, et al. Triterpenes and steroids from Euphorbia denticulata Lam. with anti-herpes symplex virus activity. Iran J Pharm Res. (2013) 12:759–67.
80. Gyuris A, Szlávik L, Minárovits J, Vasas A, Molnár J, Hohmann J. Antiviral activities of extracts of Euphorbia hirta L. against HIV-1, HIV-2 and SIVmac251. In Vivo. (2008) 23:429–32.
81. Jiang C, Luo P, Zhao Y, Hong J, Morris-Natschke SL, Xu J, et al. Carolignans from the aerial parts of Euphorbia sikkimensis and their anti-HIV activity. J Nat Prod. (2016) 79:578–83. doi: 10.1021/acs.jnatprod.5b01012
82. Shoji M, Woo SY, Masuda A, Win NN, Ngwe H, Takahashi E, et al. Anti-influenza virus activity of extracts from the stems of Jatropha multifida Linn. Collected in Myanmar. BMC Complement Altern Med. (2017) 17:76. doi: 10.1186/s12906-017-1612-8
83. Ghoke SS, Sood R, Kumar N, Pateriya AK, Bhatia S, Mishra A, et al. Evaluation of antiviral activity of Ocimum sanctum and Acacia arabica leaves extracts against H9N2 virus using embryonated chicken egg model. BMC Complement Altern Med. (2018) 18:174. doi: 10.1186/s12906-018-2238-1
84. Makau JN, Watanabe K, Mohammed MMD, Nishida N. Antiviral activity of peanut (Arachis hypogaea L.) skin extract against human influenza viruses. J Med Food. (2018) 21:777–84. doi: 10.1089/jmf.2017.4121
85. Knipping K, Garssen J, van't Land B. An evaluation of the inhibitory effects against rotavirus infection of edible plant extracts. Virol J. (2012) 9:137. doi: 10.1186/1743-422X-9-137
86. Fahmy NM, Al-Sayed E, Moghannem S, Azam F, El-Shazly M, Singab AN. Breaking down the barriers to a natural antiviral agent: antiviral activity and molecular docking of Erythrina speciosa extract, fractions, and the major compound. Chem Biodivers. (2020) 17:2. doi: 10.1002/cbdv.201900511
87. Donalisio M, Cagno V, Civra A, Gibellini D, Musumeci G, Rittà M, et al. The traditional use of Vachellia nilotica for sexually transmitted diseases is substantiated by the antiviral activity of its bark extract against sexually transmitted viruses. J Ethnopharmacol. (2018) 213:403–8. doi: 10.1016/j.jep.2017.11.039
88. Nutan SK, Modi M, Dezzutti CS, Kulshreshtha S, Rawat AKS, Srivastava SK, et al. Extracts from Acacia catechu suppress HIV-1 replication by inhibiting the activities of the viral protease and Tat. Virol J. (2013) 10:309. doi: 10.1186/1743-422X-10-309
89. Karimi A, Rafieian-kopaei M, Moradi M. Anti -herpes simplex virus type-1 activity and phenolic content of crude ethanol extract and four corresponding fractions of Quercus brantii L Acorn. J Evid Based Complement Altern Med. (2016) 22:455–61. doi: 10.1177/2156587216676421
90. Rafieian-kopaei M, Saeedi M, Asgari S, Karimi A, Moradi M-T. Antiviral activity of Quercus persica L.: high efficacy and low toxicity. Adv Biomed Res. (2013) 2:36. doi: 10.4103/2277-9175.109722
91. Choi JG, Kim YS, Kim JH, Chung HS. Antiviral activity of ethanol extract of Geranii Herba and its components against influenza viruses via neuraminidase inhibition. Sci Rep. (2019) 9:12132. doi: 10.1038/s41598-019-48430-8
92. Helfer M, Koppensteiner H, Schneider M, Rebensburg S, Forcisi S, Müller C, et al. The root extract of the medicinal plant Pelargonium sidoides is a potent HIV-1 attachment inhibitor. PLoS ONE. (2014) 9:e87487. doi: 10.1371/journal.pone.0087487
93. Michaelis M, Doerr HW, Cinatl J. Investigation of the influence of EPs® 7630, a herbal drug preparation from Pelargonium sidoides, on replication of a broad panel of respiratory viruses. Phytomedicine. (2011) 18:384–6. doi: 10.1016/j.phymed.2010.09.008
94. Haasbach E, Hartmayer C, Hettler A, Sarnecka A, Wulle U, Ehrhardt C, et al. Antiviral activity of Ladania067, an extract from wild black currant leaves against influenza a virus in vitro and in vivo. Front Microbiol. (2014) 5:171. doi: 10.3389/fmicb.2014.00171
95. Theisen LL, Erdelmeier CAJ, Spoden GA, Boukhallouk F, Sausy A, Florin L, et al. Tannins from Hamamelis virginiana bark extract: characterization and improvement of the antiviral efficacy against influenza, a virus and human papillomavirus. PLoS ONE. (2014) 9:e88062. doi: 10.1371/journal.pone.0088062
96. Erdelmeier CAJ, Cinatl J, Rabenau H, Doerr HW, Biber A, Koch E. Antiviral and antiphlogistic activities of Hamamelis virginiana bark. Planta Med. (1996) 62:241–5. doi: 10.1055/s-2006-957868
97. Chen SG, Leu YL, Cheng ML, Ting SC, Liu CC, Wang S Der, et al. Anti-enterovirus 71 activities of Melissa officinalis extract and its biologically active constituent rosmarinic acid. Sci Rep. (2017) 7:12264. doi: 10.1038/s41598-017-12388-2
98. Schnitzler P, Schuhmacher A, Astani A, Reichling J. Melissa officinalis oil affects infectivity of enveloped herpesviruses. Phytomedicine. (2008) 15:734–40. doi: 10.1016/j.phymed.2008.04.018
99. Tang LIC, Ling APK, Koh RY, Chye SM, Voon KGL. Screening of anti-dengue activity in methanolic extracts of medicinal plants. BMC Complement Altern Med. (2012) 12:3. doi: 10.1186/1472-6882-12-3
100. Chiu LCM, Zhu W, Ooi VEC. A polysaccharide fraction from medicinal herb Prunella vulgaris downregulates the expression of herpes simplex virus antigen in Vero cells. J Ethnopharmacol. (2004) 93:63–8. doi: 10.1016/j.jep.2004.03.024
101. Brindley MA, Widrlechner MP, McCoy JA, Murphy P, Hauck C, Rizshsky L, et al. Inhibition of lentivirus replication by aqueous extracts of Prunella vulgaris. Virol J. (2009) 6:8. doi: 10.1186/1743-422X-6-8
102. Ma F-W, Kong S-Y, Tan H-S, Wu R, Xia B, Zhou Y, et al. Structural characterization and antiviral effect of a novel polysaccharide PSP-2B from Prunellae Spica. Carbohydr Polym. (2016) 152:699–709. doi: 10.1016/j.carbpol.2016.07.062
103. Mancini DAP, Torres RP, Pinto JR, Mancini-Filho J. Inhibition of DNA virus: Herpes-1 (HSV-1) in cellular culture replication, through an antioxidant treatment extracted from rosemary spice. Brazilian J Pharm Sci. (2009) 45:127–33. doi: 10.1590/S1984-82502009000100016
104. Shi H, Ren K, Lv B, Zhang W, Zhao Y, Tan RX, et al. Baicalin from Scutellaria baicalensis blocks respiratory syncytial virus (RSV) infection and reduces inflammatory cell infiltration and lung injury in mice. Sci Rep. (2016) 6:35851. doi: 10.1038/srep35851
105. Cao YG, Hao Y, Li ZH, Liu ST, Wang LX. Antiviral activity of polysaccharide extract from Laminaria japonica against respiratory syncytial virus. Biomed Pharmacother. (2016) 84:1705–10. doi: 10.1016/j.biopha.2016.10.082
106. Yarmolinsky L, Zaccai M, Ben-shabat S, Mills D, Huleihel M. Antiviral activity of ethanol extracts of Ficus binjamina and Lilium candidum in vitro. (2009) 26:307–13. doi: 10.1016/j.nbt.2009.08.005
107. Tsai YC, Hohmann J, El-Shazly M, Chang LK, Dankó B, Kúsz N, et al. Bioactive constituents of Lindernia crustacea and its anti-EBV effect via Rta expression inhibition in the viral lytic cycle. J Ethnopharmacol. (2020) 250:112493. doi: 10.1016/j.jep.2019.112493
108. Boff L, Silva IT, Argenta DF, Farias LM, Alvarenga LF, Pádua RM. Strychnos pseudoquina A. St. Hil.: a Brazilian medicinal plant with promising in vitro antiherpes activity. J Appl Microbiol. (2016) 121:1519–29. doi: 10.1111/jam.13279
109. Arunkumar J, Rajarajan S. Study on antiviral activities, drug-likeness and molecular docking of bioactive compounds of Punica granatum L. to Herpes simplex virus - 2 (HSV-2). Microb Pathog. (2018) 118:301–9. doi: 10.1016/j.micpath.2018.03.052
110. Haidari M, Ali M, Ward Casscells S, Madjid M. Pomegranate (Punica granatum) purified polyphenol extract inhibits influenza virus and has a synergistic effect with oseltamivir. Phytomedicine. (2009) 16:1127–36. doi: 10.1016/j.phymed.2009.06.002
111. Fang C, Chen S, Wu H, Ping Y, Lin C. Honokiol, a Lignan biphenol derived from the magnolia tree, inhibits Dengue virus type 2 infection. Viruses. (2015) 7:4894–910. doi: 10.3390/v7092852
112. Baatartsogt T, Bui VN, Trinh DQ, Yamaguchi E, Gronsang D, Thampaisarn R, et al. High antiviral effects of hibiscus tea extract on the H5 subtypes of low and highly pathogenic avian influenza viruses. J Vet Med Sci. (2016) 78:1405–11. doi: 10.1292/jvms.16-0124
113. Sood R, Raut R, Tyagi P, Pareek PK, Barman TK, Singhal S, et al. Cissampelos pareira Linn: Natural source of potent antiviral activity against all four Dengue virus serotypes. PLoS Negl Trop Dis. (2015) 9:e0004255. doi: 10.1371/journal.pntd.0004255
114. Yarmolinsky L, Huleihel M, Zaccai M, Ben-shabat S. Potent antiviral flavone glycosides from Ficus benjamina leaves. Fitoterapia. (2012) 83:362–7. doi: 10.1016/j.fitote.2011.11.014
115. Aref HL, Gaaliche B, Fekih A. in vitro cytotoxic and antiviral activities of Ficus carica latex extracts. Nat Prod Res. (2011) 25:310–9. doi: 10.1080/14786419.2010.528758
116. Ghosh M, Civra A, Rittà M, Cagno V, Mavuduru SG, Awasthi P, et al. Ficus religiosa L. bark extracts inhibit infection by herpes simplex virus type 2 in vitro. Arch Virol. (2016) 161:3509–14. doi: 10.1007/s00705-016-3032-3
117. Huang NC, Hung WT, Tsai WL, Lai FY, Lin YS, Huang MS, et al. Ficus septica plant extracts for treating Dengue virus in vitro. Peer J. (2017) 5:e3448. doi: 10.7717/peerj.3448
118. Sa S, Abdullahi H. Antifungal activity and phytochemical analysis of Ficus sycomorus leaf extract on malassezia glubosa. Adv Plants Agric Res. (2018) 8:432–6. doi: 10.15406/apar.2018.08.00362
119. Batiha GES, Alkazmi LM, Wasef LG, Beshbishy AM, Nadwa EH, Rashwan EK. Syzygium aromaticum L. (myrtaceae): traditional uses, bioactive chemical constituents, pharmacological and toxicological activities. Biomolecules. (2020) 10:202. doi: 10.3390/biom10020202
120. Benzekri R, Bouslama L, Papetti A, Hammami M, Smaoui A, Limam F. Anti HSV-2 activity of Peganum harmala (L.) and isolation of the active compound. Microb Pathog. (2018) 114:291–8. doi: 10.1016/j.micpath.2017.12.017
121. Li J, Yang X, Huang L. Anti-Influenza virus activity and constituents characterization of Paeonia delavayi extracts. Molecules. (2016) 21:1133. doi: 10.3390/molecules21091133
122. Ho JY, Chang HW, Lin CF, Liu CJ, Hsieh CF, Horng JT. Characterization of the anti-influenza activity of the Chinese herbal plant Paeonia lactiflora. Viruses. (2014) 6:1861–75. doi: 10.3390/v6041861
123. Cho JK, Curtis-Long MJ, Lee KH, Kim DW, Ryu HW, Yuk HJ, et al. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorganic Med Chem. (2013) 21:3051–7. doi: 10.1016/j.bmc.2013.03.027
124. Lv J, Yu S, Wang Y, Wang D, Zhu H, Cheng R, et al. Anti-hepatitis B virus norbisabolane sesquiterpenoids from Phyllanthus acidus and the establishment of their absolute configurations using theoretical calculations. J Org Chem. (2014) 79:5432–47. doi: 10.1021/jo5004604
125. Ravikumar YS, Ray U, Nandhitha M, Perween A, Raja Naika H, Khanna N, et al. Inhibition of hepatitis C virus replication by herbal extract: Phyllanthus amarus as potent natural source. Virus Res. (2011) 158:89–97. doi: 10.1016/j.virusres.2011.03.014
126. Tan WC, Jaganath IB, Manikam R, Sekaran SD. Evaluation of antiviral activities of four local Malaysian phyllanthus species against herpes simplex viruses and possible antiviral target. Int J Med Sci. (2013) 10:1817–29. doi: 10.7150/ijms.6902
127. Zhang X, Yang LM, Liu GM, Liu YJ, Zheng CB, Lv YJ, et al. Potent anti-HIV activities and mechanisms of action of a pine cone extract from Pinus yunnanensis. Molecules. (2012) 17:6916–29. doi: 10.3390/molecules17066916
128. Hsu WC, Chang SP, Lin LC, Li CL, Richardson CD, Lin CC, et al. Limonium sinense and gallic acid suppress hepatitis C virus infection by blocking early viral entry. Antiviral Res. (2015) 118:139–47. doi: 10.1016/j.antiviral.2015.04.003
129. Chavan RD, Shinde P, Girkar K, Madage R, Chowdhary A. Assessment of Anti-Influenza activity and hemagglutination inhibition of Plumbago indica and Allium sativum extracts. Pharmacognosy Res. (2016) 8:105–11. doi: 10.4103/0974-8490.172562
130. Xiong HR, Luo J, Hou W, Xiao H, Yang ZQ. The effect of Emodin, an anthraquinone derivative extracted from the roots of Rheum tanguticum, against herpes simplex virus in vitro and in vivo. J Ethnopharmacol. (2011) 133:718–23. doi: 10.1016/j.jep.2010.10.059
131. Su M, Li Y, Leung KT, Cen Y, Li T, Chen R, et al. Antiviral activity and constituent of Ardisia Chinensis Benth against coxsackie B3 virus. Phytother Res. (2006) 20:634–9. doi: 10.1002/ptr.1912
132. Hossan MS, Fatima A, Rahmatullah M, Khoo TJ, Nissapatorn V, Galochkina AV, et al. Antiviral activity of Embelia ribes Burm. f. against influenza virus in vitro. Arch Virol. (2018) 163:2121–31. doi: 10.1007/s00705-018-3842-6
133. Hung TC, Jassey A, Lin CJ, Liu CH, Lin CC, Yen MH, et al. Methanolic extract of rhizoma coptidis inhibits the early viral entry steps of hepatitis C virus infection. Viruses. (2018) 10:669. doi: 10.3390/v10120669
134. Kim HB, Lee CY, Kim SJ, Han JH, Choi KH. Medicinal herb extracts ameliorate impaired growth performance and intestinal lesion of newborn piglets challenged with the virulent porcine epidemic diarrhea virus. J Anim Sci Technol. (2015) 57:33. doi: 10.1186/s40781-015-0065-1
135. Shin WJ, Lee KH, Park MH, Seong BL. Broad-spectrum antiviral effect of Agrimonia pilosa extract on influenza viruses. Microbiol Immunol. (2010) 54:11–9. doi: 10.1111/j.1348-0421.2009.00173.x
136. Bisignano C, Mandalari G, Smeriglio A, Trombetta D, Pizzo MM, Pennisi R, et al. Almond skin extracts abrogate HSV-1 replication by blocking virus binding to the cell. Viruses. (2017) 9:178. doi: 10.3390/v9070178
137. Ratnoglik SL, Aoki C, Sudarmono P, Komoto M, Deng L, Shoji I, et al. Antiviral activity of extracts from Morinda citrifolia leaves and chlorophyll catabolites, pheophorbide a and pyropheophorbide a, against hepatitis C virus. Microbiol Immunol. (2014) 58:188–94. doi: 10.1111/1348-0421.12133
138. Pratheeba T, Taranath V, Sai Gopal DVR, Natarajan D. Antidengue potential of leaf extracts of Pavetta tomentosa and Tarenna asiatica (Rubiaceae) against dengue virus and its vector Aedes aegypti (Diptera: Culicidae). Heliyon. (2019) 5:e02732. doi: 10.1016/j.heliyon.2019.e02732
139. Brijesh S, Daswani P, Tetali P, Antia N, Birdi T. Studies on the antidiarrhoeal activity of Aegle marmelos unripe fruit : validating its traditional usage. BMC Complement Altern Med. (2009) 12:47. doi: 10.1186/1472-6882-9-47
140. Apriyanto DR, Aoki C, Hartati S, Hanafi M, Kardono LBS, Arsianti A, et al. Anti-hepatitis C virus activity of a crude extract from longan (Dimocarpus longan Lour.) leaves. Jpn J Infect Dis. (2016) 69:213–20. doi: 10.7883/yoken.JJID.2015.107
141. Song JH, Ahn JH, Kim SR, Cho S, Hong EH, Kwon BE, et al. Manassantin B shows antiviral activity against coxsackievirus B3 infection by activation of the STING/TBK-1/IRF3 signalling pathway. Sci Rep. (2019) 9:9413. doi: 10.1038/s41598-019-45868-8
142. Wang GW, Hu WT, Huang BK, Qin LP. Illicium verum: a review on its botany, traditional use, chemistry and pharmacology. J Ethnopharmacol. (2011) 136:10–20. doi: 10.1016/j.jep.2011.04.051
143. Abdelgawad AAM. Tamarix nilotica (Ehrenb) bunge: a review of phytochemistry and pharmacology. J Microb Biochem Technol. (2017) 9:544–53. doi: 10.4172/1948-5948.1000340
144. Ryu YB, Jeong HJ, Kim JH, Kim YM, Park JY, Kim D, et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CLpro inhibition. Bioorganic Med Chem. (2010) 18:7940–7. doi: 10.1016/j.bmc.2010.09.035
145. Karamese M, Aydogdu S, Karamese SA, Altoparlak U. Preventive effects of a major component of green tea, Epigallocathechin-3-Gallate, on Hepatitis-B virus DNA replication. Asian Pac J Cancer Prev. (2015) 16:40199–202. doi: 10.7314/APJCP.2015.16.10.4199
146. Xu J, Wang J, Deng F, Hu Z, Wang H. Green tea extract and its major component epigallocatechin gallate inhibits hepatitis B virus in vitro. Antiviral Res. (2008) 78:242–9. doi: 10.1016/j.antiviral.2007.11.011
147. Dai J, Tao H, Min Q, Zhu Q. Anti-hepatitis B virus activities of friedelolactones from Viola diffusa Ging. Phytomedicine. (2015) 22:724–9. doi: 10.1016/j.phymed.2015.05.001
148. Kwon HJ, Kim HH, Yoon SY, Ryu YB, Chang JS, Cho KO, et al. in vitro inhibitory activity of Alpinia katsumadai extracts against influenza virus infection and hemagglutination. Virol J. (2010) 7:307. doi: 10.1186/1743-422X-7-307
149. Thuy BTP, My TTA, Hai NTT, Hieu LT, Hoa TT, Thi Phuong Loan H, et al. Investigation into SARS-CoV-2 resistance of compounds in garlic essential oil. ACS Omega. (2020) 5:8312–8320. doi: 10.1021/acsomega.0c00772
150. Kim DW, Seo KH, Curtis-Long MJ, Oh KY, Oh JW, Cho JK, et al. Phenolic phytochemical displaying SARS-CoV papain-like protease inhibition from the seeds of Psoralea corylifolia. J Enzyme Inhib Med Chem. (2014) 29:59–63. doi: 10.3109/14756366.2012.753591
151. Li SY, Chen C, Zhang HQ, Guo HY, Wang H, Wang L, et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antiviral Res. (2005) 67:18–23. doi: 10.1016/j.antiviral.2005.02.007
152. Ulasli M, Gurses SA, Bayraktar R, Yumrutas O, Oztuzcu S, Igci M, et al. The effects of Nigella sativa (Ns), Anthemis hyalina (Ah) and Citrus sinensis (Cs) extracts on the replication of coronavirus and the expression of TRP genes family. Mol Biol Rep. (2014) 41:1703–11. doi: 10.1007/s11033-014-3019-7
153. Ryu YB, Park SJ, Kim YM, Lee JY, Seo WD, Chang JS, et al. SARS-CoV 3CLpro inhibitory effects of quinone-methide triterpenes from Tripterygium regelii. Bioorganic Med Chem Lett. (2010) 20:1873–76. doi: 10.1016/j.bmcl.2010.01.152
154. Loizzo MR, Saab AM, Tundis R, Statti GA, Menichimi F, Lampronti D, et al. Phytochemical analysis and in vitro antiviral activities of the essential oils of seven Lebanon species. Chem Biodivers. (2008) 5:461–70. doi: 10.1002/cbdv.200890045
155. Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. (2020) 10:766–88. doi: 10.1016/j.apsb.2020.02.008
156. Wen CC, Shyur LF, Jan JT, Liang PH, Kuo CJ, Arulselvan P, et al. Traditional Chinese medicine herbal extracts of Cibotium barometz, Gentiana scabra, Dioscorea batatas, Cassia tora, and Taxillus chinensis inhibit SARS-CoV replication. J Tradit Complement Med. (2011) 1:41–50. doi: 10.1016/S2225-4110(16)30055-4
157. Zhuang M, Jiang H, Suzuki Y, Li X, Xiao P, Tanaka T, et al. Procyanidins and butanol extract of Cinnamomi Cortex inhibit SARS-CoV infection. Antiviral Res. (2009) 82:73–81. doi: 10.1016/j.antiviral.2009.02.001
158. Ho TY, Wu SL, Chen JC, Li CC, Hsiang CY. Emodin blocks the SARS coronavirus spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res. (2007) 74:92–101. doi: 10.1016/j.antiviral.2006.04.014
159. Luo W, Su X, Gong S, Qin Y, Liu W, Li J, et al. Anti-SARS coronavirus 3C-like protease effects of Rheum palmatum L. extracts. Biosci Trends. (2009) 3:124–6.
160. Yang C, Lee Y, Hsu H, Shih C, Chao Y, Lee S. Targeting coronaviral replication and cellular JAK2 mediated dominant NF- κB activation for comprehensive and ultimate inhibition of coronaviral activity. Sci Rep. (2017) 7:4105. doi: 10.1038/s41598-017-04203-9
161. Adhikari SP, Meng S, Wu Y, Mao Y, Ye R, Wang Q, et al. Novel Coronavirus during the early outbreak period: epidemiology, causes, clinical manifestation and diagnosis, prevention and control. Infect Dis Poverty. (2020) 9:29. doi: 10.1186/s40249-020-00646-x
162. Nickbakhsh S, Ho A, Marques DFP, McMenamin J, Gunson RN, Murcia PR. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of Coronavirus disease 2019. J Infect Dis. (2020) 222:17–25. doi: 10.1093/infdis/jiaa185
163. Wu D, Wu T, Liu Q, Yang Z. The SARS-CoV-2 outbreak: what we know. Int J Infect Dis. (2020) 94:44–8. doi: 10.1016/j.ijid.2020.03.004
164. Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. (2020) 382:1199–207. doi: 10.1056/NEJMoa2001316
165. Phan LT, Nguyen TV, Luong QC, Nguyen TV, Nguyen TV, Nguyen HT, et al. Importation and human-to-human transmission of a novel Coronavirus in Vietnam. N Engl J Med. (2020) 382:872–4. doi: 10.1056/NEJMc2001272
166. Cots JM, Alós J, Bárcena M, Boleda X. COVID-19 in Brazil: “So what?”. Lancet. (2020) 395:1461. doi: 10.1016/S0140-6736(20)31095-3
167. COVID-19 Dashboard by the Center for Systems Science Engineering (CSSE) at Johns Hopkins University (JHU). Available online at: https://coronavirus.jhu.edu/map.html (accessed July 5, 2020).
168. Yang H, Bartlam M, Rao Z. Drug design targeting the main protease, the Achilles Heel of coronaviruses. Curr Pharm Des. (2006) 12:4573–90. doi: 10.2174/138161206779010369
169. Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect. (2020). doi: 10.1016/j.jmii.2020.03.022. [Epub ahead of print].
170. Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. (2020) 11:1620. doi: 10.1038/s41467-020-15562-9
171. Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. (2020) 176:140742. doi: 10.1016/j.antiviral.2020.104742
172. Liang Y, Wang M, Chien C, Yarmishyn AA. Highlight of immune pathogenic response and hematopathologic effect in SARS-CoV, MERS-CoV and SARS-Cov-2 infection. Front Immunol. (2020) 11:1022. doi: 10.3389/fimmu.2020.01022
173. Chen B, Tian E, He B, Tian L, Han R, Wang S, et al. Overview of lethal human coronaviruses. Sig Transduct Target Ther. (2020) 5:89. doi: 10.1038/s41392-020-0190-2
174. Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. (2020) 6:14. doi: 10.1038/s41421-020-0153-3
175. Xu J, Zhao S, Teng T, Abdalla AE, Zhu W, Xie L, et al. Systematic comparison of two animal-to-human transmitted human Coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. (2020) 12:244. doi: 10.3390/v12020244
176. Prajapat M, Sarma P, Shekhar N, Avti P, Sinha S, Kaur H, et al. Drug targets for corona virus: a systematic review. Indian J Pharmacol. (2020) 52:56–65. doi: 10.4103/ijp.IJP_115_20
177. Rabaan AA, Al-Ahmed SH, Haque S, Sah R, Tiwari R, Malik YS, et al. SARS-CoV-2, SARS-CoV, and MERS-COV: a comparative overview. Le Infez Med. (2020) 28:174–84.
178. Tang X, Wu C, Li X, Song Y, Yao X, Wu X, et al. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. (2020) 3:nwaa036. doi: 10.1093/nsr/nwaa036
179. Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. (2020) 46:586–90. doi: 10.1007/s00134-020-05985-9
180. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 2:52. doi: 10.1016/j.cell.2020.02.052
181. Skariayachan S, Challapilli SB, Pacckirisamy S, Kumargowda ST, Sridhar VS. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutics interventions for Middle East respiratory syndrome coronavirus infections. Front Microbiol. (2019) 10:569. doi: 10.3389/fmicb.2019.00569
182. Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J Adv Res. (2020) 24:91–8. doi: 10.1016/j.jare.2020.03.005
183. Rampersad S, Tennant P. Replication and expression strategies of viruses. Viruses. (2018) 2018:55–82. doi: 10.1016/B978-0-12-811257-1.00003-6
184. Wang H, Xue S, Yang H, Chen C. Recent progress in the discovery of inhibitors targeting coronavirus proteases. Virol Sin. (2016) 31:24–30. doi: 10.1007/s12250-015-3711-3
185. Cao W, Li T. COVID-19: towards understanding of pathogenesis. Cell Res. (2020) 30:367–9. doi: 10.1038/s41422-020-0327-4
186. Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Oude Munnink BB, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. (2020) 368:1012–5. doi: 10.1126/science.abb7314
187. Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. (2020) 92:418–23. doi: 10.1002/jmv.25681
188. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. (2020) 181:914–21. doi: 10.1016/j.cell.2020.04.011
189. Shi Y, Wang Y, Shao C, Huang J, Gan J, Huang X, et al. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. (2020) 27:1451–4. doi: 10.1038/s41418-020-0530-3
190. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
191. Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. (2020) 109:102433. doi: 10.1016/j.jaut.2020.102433
192. Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome. J Microbiol Immunol Infect. (2020) 53:425–35. doi: 10.1016/j.jmii.2020.04.015
193. Tahir ul Qamar M, Alqahtani SM, Alamri MA, Chen LL. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. (in press) doi: 10.20944/preprints202002.0193.v1
194. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. (2020) 581:221–4. doi: 10.1038/s41586-020-2179-y
195. Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. (2014) 281:4085–96. doi: 10.1111/febs.12936
196. Zeng Q, Langereis MA, van Vliet ALW, Huizinga EG, de Groot RJ. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci USA. (2008) 105:9065–9. doi: 10.1073/pnas.0800502105
197. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. (2020) 1843–44. doi: 10.1001/jama.2020.3786
198. Saxena SK, Kumar S, Maurya VK, Sharma R, Dandu HR, Bhatt MLB. Current insight into the novel coronavirus disease 2019 (COVID-19). Nat Public Health Emerg Collect. (2020) 2019:1–8. doi: 10.1007/978-981-15-4814-7_1
199. Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, et al. Research and development on therapeutic agents and vaccines for covid-19 and related human coronavirus diseases. ACS Cent Sci. (2020) 6:315–31. doi: 10.1021/acscentsci.0c00272
200. Salata C, Calistri A, Parolin C, Palù G. Coronaviruses: a paradigm of new emerging zoonotic diseases. Pathog Dis. (2020) 77:ftaa006. doi: 10.1093/femspd/ftaa006
201. Wilder-smith A, Chiew CJ, Lee VJ. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect Dis. (2020) 20:e102–7. doi: 10.1016/S1473-3099(20)30129-8
202. Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov. (2020) 19:149–50. doi: 10.1038/d41573-020-00016-0
203. Andersen PI, Ianevski A, Lysvand H, Vitkauskiene A, Oksenych V, Bjørås M, et al. Discovery and development of safe-in-man broad-spectrum antiviral agents. Int J Infect Dis. (2020) 93:268–76. doi: 10.1016/j.ijid.2020.02.018
204. Liu M, Yu Q, Xiao H, Yi Y, Cheng H, Putra DF, et al. Antiviral activity of Illicium verum Hook. f. extracts against grouper iridovirus infection. J Fish Dis. (2020) 43:531–40. doi: 10.1111/jfd.13146
205. Fuzimoto AD, Isidoro C. The antiviral and the coronavirus-host protein pathways inhibiting properties of herbs and natural compounds - additional weapons in the fight against the COVID-19 pandemic? J Tradit Complement Med. (2020) 10:405–19. doi: 10.1016/j.jtcme.2020.05.003
206. Chen L, Xiong J, Bao L, Shi Y. Convalescent plasma as a potential therapy for COVID-19. Lancet Infect Dis. (2020) 20:398–400. doi: 10.1016/S1473-3099(20)30141-9
207. Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. (2020) 20:269–70. doi: 10.1038/s41577-020-0308-3
208. Vankadari N. Arbidol: a potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. Int J Antimicrob Agents. (2020) 28:105998. doi: 10.1016/j.ijantimicag.2020.105998
209. Khan SA, Zia K, Ashraf S, Uddin R, Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J Biomol Struct Dyn. (2020) 2020:1–10. doi: 10.1080/07391102.2020.1751298
210. Delang L, Abdelnabi R, Neyts J. Favipiravir as a potential countermeasure against neglected and emerging RNA viruses. Antiviral Res. (2018) 153:85–94. doi: 10.1016/j.antiviral.2018.03.003
211. Wang M, Cao R, Zhang L, Yang X, Liu J, Xu M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. (2020) 30:269–71. doi: 10.1038/s41422-020-0282-0
212. Caly L, Druce JD, Catton MG, Jans DA, Wagstaff KM. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. (2020) 178:3–6. doi: 10.1016/j.antiviral.2020.104787
213. Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, et al. A trial of lopinavir-ritonavir in adults hospitalized with severe covid-19. N Engl J Med. (2020) 382:1787–99. doi: 10.1056/NEJMc2008043
214. Elmezayen AD, Al-Obaidi A, Sahin AT, Yelekçi K. Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J Biomol Struct Dyn. (2020) 2020:1–13. doi: 10.1080/07391102.2020.1758791
215. Muralidharan N, Sakthivel R, Velmurugan D, Gromiha MM. Computational studies of drug repurposing and synergism of lopinavir, oseltamivir and ritonavir binding with SARS-CoV-2 protease against COVID-19. J Biomol Struct Dyn. (2020) 2020:1–6. doi: 10.1080/07391102.2020.1752802
216. Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov Ther. (2020) 14:58–60. doi: 10.5582/ddt.2020.01012
217. Sarma P, Sekhar N, Prajapat M, Avti P, Kaur H, Kumar S, et al. In-silico homology assisted identification of inhibitors of RNA binding against 2019-nCoV N-protein (N terminal domain). J Biomol Struct Dyn. (2020) 2020:1–9. doi: 10.1080/07391102.2020.1753580
218. Baron SA, Devaux C, Colson P, Raoult D, Rolain JM. Teicoplanin: an alternative drug for the treatment of COVID-19? Int J Antimicrob Agents. (2020) 55:105944. doi: 10.1016/j.ijantimicag.2020.105944
219. Enmozhi SK, Raja K, Sebastine I, Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn. (2020) 2020:1–7. doi: 10.1080/07391102.2020.1760136
220. Peng S, Li Z, Zou L, Liu W, Liu C, McClements DJ. Enhancement of curcumin bioavailability by encapsulation in Sophorolipid-coated nanoparticles: an in vitro and in vivo study. Agric Food Chem. (2018) 66:1488–97. doi: 10.1021/acs.jafc.7b05478
221. Wu R, Wang L, Kuo HCD, Shannar A, Peter R, Chou PJ, et al. An update on current therapeutic drugs treating COVID-19. Curr Pharmacol Rep. (2020) 11:1–15. doi: 10.1007/s40495-020-00216-7
222. Maurya VK, Kumar S, Bhatt MLB, Saxena SK. Therapeutic development and drugs for the treatment of COVID-19. Nat Public Health Emerg Collect. (2020) 2019:109–26. doi: 10.1007/978-981-15-4814-7_10
223. Koparde AA, Doijad RC, Magdum CS, Koparde AA. Natural products in drug discovery. In: Doijad RC, editor. Pharmacognosy - Medicinal Plants. Rijeka: IntechOpen. (2019). p. 14. doi: 10.5772/intechopen.82860
224. Ekor M. The growing use of herbal medicines: issues relating to adverse reactions and challenges in monitoring safety. Front Neurol. (2014) 4:177. doi: 10.3389/fphar.2013.00177
225. Ganjhu RK, Mudga PP, Maity H, Dowarha D, Devadiga S, Nag S, et al. Herbal plants and plant preparations as a remedial approach for viral diseases. Virus Dis. (2015) 26:225–36. doi: 10.1007/s13337-015-0276-6
226. Chitemerere TA, Mukanganyama S. Evaluation of cell membrane integrity as a potential antimicrobial target for plant products. BMC Complement Altern Med. (2014) 14:278. doi: 10.1186/1472-6882-14-278
227. Anandhi D, Srinivasan PT, Kumar GP, Jagatheesh S. Original research article DNA fragmentation induced by the glycosides and flavonoids from C. coriaria. Int J Curr Microbiol App Sci. (2014) 3:666–73.
228. Zhao X, Zhao F, Zhong N. Quorum sensing inhibition and anti-biofilm activity of traditional Chinese medicine. In: El-Samragy Y, editor. Food Safety - Some Global Trends. Itratech Open (2018). p. 14. doi: 10.5772/intechopen.74658
229. Radulovic NS, Blagojevic PD, Stojanovic-Radic ZZ, Stojanovic NM. Antimicrobial plant metabolites: structural diversity and mechanism of action. Curr Med Chem. (2013) 20:932–52. doi: 10.2174/0929867311320070008
230. Mogosanu GD, Grumezescu AM, Huang KS, Bejenaru LE, Bejenaru C. Prevention of microbial communities: novel approaches based natural products. Curr Pharm Biotechnol. (2015) 16:94–111. doi: 10.2174/138920101602150112145916
231. Anand U, Jacobo-Herrera N, Altemimi A, Lakhssassi N. A comprehensive review on medicinal plants as antimicrobial therapeutics: potential avenues of biocompatible drug discovery. Metabolites. (2019) 9:258. doi: 10.3390/metabo9110258
232. Zhang D, Wu K, Zhang X, Deng S, Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. (2020) 18:152–8. doi: 10.1016/j.joim.2020.02.005
233. Kiran G, Karthik L, Shree Devi MS, Sathiyarajeswaran P, Kanakavalli K, Kumar KM, et al. In silico computational screening of Kabasura Kudineer - official Siddha Formulation and JACOM against SARS-CoV-2 spike protein. J Ayurveda Integr Med. (in press) doi: 10.1016/j.jaim.2020.05.009
234. Subbaiyan A, Ravichandran K, Singh SV, Sankar M, Thomas P, Dhama K, et al. In silico molecular docking analysis targeting SARS-CoV-2 spike protein and selected herbal constituents. J Pure Appl Microbiol. (2020) 14:989–98. doi: 10.22207/JPAM.14.SPL1.37
235. Nivetha R, Bhuvaragavan S, Janarthanan S. Inhibition of multiple SARS-CoV-2 proteins by an antiviral biomolecule, seselin from Aegle marmelos deciphered using molecular docking analysis. Res Sq. (2020) 2020:v1. doi: 10.21203/rs.3.rs-31134/v1
236. Basu A, Sarkar A, Maulik U. Computational approach for the design of potential spike protein binding natural compounds in SARS- CoV2. Pharmacodynamics. (2020). doi: 10.21203/rs.3.rs-33181/v1. [Epub ahead of print].
237. Krishnasamy R, Anand T, Baba M, Bharath MV, Phuntsho J, Arunachalam D, et al. In silico analysis of active compounds from siddha herbal infusion of Ammaiyar Koondhal Kudineer (Akk) against SARS-CoV-2 spike protein and its ACE2 receptor complex. SSRN Online J. (2020). doi: 10.2139/ssrn.3578294. [Epub ahead of print].
238. Pandit M, Latha N. In silico studies reveal potential antiviral activity of phytochemicals from medicinal plants for the treatment of COVID-19 infection. Res Sq. (2020). doi: 10.21203/rs.3.rs-22687/v1. [Epub ahead of print].
239. Pandeya KB, Ganeshpurkar A, Kumar Mishra M. Natural RNA dependent RNA polymerase inhibitors: molecular docking studies of some biologically active alkaloids of Argemone mexicana. Med Hypotheses. (2020) 2020:109905. doi: 10.1016/j.mehy.2020.109905
240. Aanouz I, Belhassan A, El-Khatabi K, Lakhlifi T, El-ldrissi M, Bouachrine M. Moroccan medicinal plants as inhibitors against SARS-CoV-2 main protease: computational investigations. J Biomol Struct Dyn. (2020) 2020:1–9. doi: 10.1080/07391102.2020.1758790
241. Bhardwaj VK, Singh R, Sharma J, Rajendran V, Purohit R, Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. (2020) 2020:1–11. doi: 10.1080/07391102.2020.1766572
242. Serseg T, Benarous K, Yousfi M. Hispidin and Lepidine E: two natural compounds and folic acid as potential inhibitors of 2019-novel coronavirus main protease (2019-nCoVMpro), molecular docking and SAR study. Curr Comput Aided Drug Des. (2020). doi: 10.2174/1573409916666200422075440. [Epub ahead of print].
243. Kumar A, Choudhir G, Shukla SK, Sharma M, Tyagi P, Bhushan A, et al. Identification of phytochemical inhibitors against main protease of COVID-19 using molecular modeling approaches. J Biomol Struct Dyn. (2020) 2020:1–11. doi: 10.21203/rs.3.rs-31210/v1
244. Islam MT, Sarkar C, El-Kersh DM, Jamaddar S, Uddin SJ, Shilpi JA, et al. Natural products and their derivatives against coronavirus: a review of the non-clinical and pre-clinical data. Phyther Res. (2020). doi: 10.1002/ptr.6700
245. Sayed AM, Khattab AR, AboulMagd AM, Hassan HM, Rateb ME, Zaid H, et al. Nature as a treasure trove of potential anti-SARS-CoV drug leads: a structural/mechanistic rationale. RSC Adv. (2020) 10:19790–802. doi: 10.1039/D0RA04199H
246. Sepay N, Sepay N, Al Hoque A, Mondal R, Halder UC, Muddassir M. In silico fight against novel coronavirus by finding chromone derivatives as inhibitor of coronavirus main proteases enzyme. Struct Chem. (2020) 2020:1–10. doi: 10.1007/s11224-020-01537-5
247. Umesh U, Kundu D, Selvaraj C, Singh SK, Dubey VK. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J Biomol Struct Dyn. (2020) 2020:1–9. doi: 10.1080/07391102.2020.1763202
248. Singh A, Mishra A. Leucoefdin a potential inhibitor against SARS CoV-2 M. J Biomol Struct Dyn. (2020) 2020:1–6. doi: 10.1080/07391102.2020.1777903
249. Gupta MK, Vemula S, Donde R, Gouda G, Behera L, Vadde R. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn. (2020) 2020:1–11. doi: 10.1080/07391102.2020.1751300
250. Joshi RS, Jagdale SS, Bansode SB, Shankar SS, Tellis MB, Pandya VK, et al. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J Biomol Struct Dyn. (2020) 2020:1–6. doi: 10.20944/preprints202004.0068.v2
251. Joshi T, Joshi T, Sharma P, Mathpal S, Pundir H, Bhatt V, et al. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur Rev Med Pharmacol Sci. (2020) 24:4529–36. doi: 10.26355/eurrev_202004_21036
252. Salman S, Shah FH, Idrees J, Idrees F, Velagala S, Ali J, et al. Virtual screening of immunomodulatory medicinal compounds as promising anti-SARS-COV-2 inhibitors. Future Virol. (2020). doi: 10.2217/fvl-2020-0079. [Epub ahead of print].
253. Wahedi HM, Ahmad S, Abbasi SW. Stilbene-based natural compounds as promising drug candidates against COVID-19. J Biomol Struct Dyn. (2020) 2020:1–10. doi: 10.1080/07391102.2020.1762743
254. Abdelli I, Hassani F, Bekkel Brikci S, Ghalem S. In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from Western Algeria. J Biomol Struct Dyn. (2020) 2020:1–14. doi: 10.1080/07391102.2020.1763199
255. Murugan NA, Pandian CJ, Jeyakanthan J. Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J Biomol Struct Dyn. (2020) 2020:1–12. doi: 10.1080/07391102.2020.1777901
256. Gyebi GA, Ogunro OB, Adegunloye AP, Ogunyemi OM, Afolabi SO. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): an in silico screening of alkaloids and Terpenoids from African medicinal plants. J Biomol Struct Dyn. (2020) 20201–13. doi: 10.1080/07391102.2020.1764868
257. Sampangi-Ramaiah MH, Vishwakarma R, Shaanker RU. Molecular docking analysis of selected natural products from plants for inhibition of SARS-CoV-2 main protease. Curr Sci. (2020) 118:1087–92.
258. Kumar V, Dhanjal JK, Bhargava P, Kaul A, Wang J, Zhang H, et al. Withanone and withaferin-A are predicted to interact with transmembrane protease serine 2 (TMPRSS2) and block entry of SARS-CoV-2 into cells. J Biomol Struct Dyn. (2020) 2020:1–13. doi: 10.1080/07391102.2020.1775704
259. Borkotoky S, Banerjee M. A computational prediction of SARS-CoV-2 structural protein inhibitors from Azadirachta indica (Neem). J Biomol Struct Dyn. (2020) 2020:1–7. doi: 10.1080/07391102.2020.1774419
260. Elfiky AA. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J Biomol Struct Dyn. (2020) 2020:1–10. doi: 10.21203/rs.3.rs-22458/v1
261. Ahmad S, Abbasi HW, Shahid S, Gul S, Abbasi SW. Molecular docking, simulation and MM-PBSA studies of nigella sativa compounds: a computational quest to identify potential natural antiviral for COVID-19 treatment. J Biomol Struct Dyn. (2020) 2020:1–9. doi: 10.1080/07391102.2020.1775129
262. Saito K. Phytochemical genomics-a new trend. Curr Opin Plant Biol. (2013) 16:373–80. doi: 10.1016/j.pbi.2013.04.001
263. Mathur S, Hoskins C. Drug development: lessons from nature. Biomed Rep. (2017) 6:612–4. doi: 10.3892/br.2017.909
264. Wink M. Plant secondary metabolites modulate insect behavior-steps toward addiction? Front Physiol. (2018) 9:364. doi: 10.3389/fphys.2018.00364
265. Ebenezer KS, Manivannan R, Punniyamoorthy A, Tamilselvan C. Plant secondary metabolites of antiviral properties a rich medicinal source for drug discovery: a mini review. J Drug Deliv Ther. (2019) 9:161–7. doi: 10.22270/jddt.v9i5.3471
266. Calixto JB. The role of natural products in modern drug discovery. An Acad Bras Cienc. (2019) 91:20190105. doi: 10.1590/0001-3765201920190105
267. Hardman WE. Diet components can suppress inflammation and reduce cancer risk. Nutr Res Prac.t. (2014) 8:233–40. doi: 10.4162/nrp.2014.8.3.233
268. Atanasov AG, Waltenberger B, Pferschy-Wenzig EM, Linder T, Wawrosch C, Uhrin P, et al. Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol Adv. (2015) 33:1582–614. doi: 10.1016/j.biotechadv.2015.08.001
269. Annunziata G, Sanduzzi Zamparelli M, Santoro C, Ciampaglia R, Stornaiuolo M, Tenore GC, et al. May polyphenols have a role against coronavirus infection? An overview of in vitro evidence. Front Med. (2020) 7:240. doi: 10.3389/fmed.2020.00240
270. Chowdhury P, Sahuc M, Rouillé C, Rivière C, Bonneau N, Vandeputte A, et al. Theaflavins, polyphenols of black tea, inhibit entry of hepatitis C virus in cell culture. PLoS ONE. (2018) 13:11. doi: 10.1371/journal.pone.0198226
271. Vázquez-Calvo Á, Oya N, Martín-Acebes MA, Garcia-Moruno E, Saiz J. Antiviral properties of the natural polyphenols delphinidin and epigallocatechin gallate against the flaviviruses West Nile virus, Zika virus, and dengue virus. Front Microbiol. (2017) 8:1314. doi: 10.3389/fmicb.2017.01314
272. Park JY, Yuk HJ, Ryu HW, Lim SH, Kim KS, Park KH, et al. Evaluation of polyphenols from Broussonetia papyrifera as coronavirus protease inhibitors. J Enzyme Inhib Med Chem. (2017) 32:504–12. doi: 10.1080/14756366.2016.1265519
273. Abdelmageed MI, Abdelmoneim AH, Mustafa MI, Elfadol NM, Murshed NS, Shantier SW, et al. Design of a multiepitope-based peptide vaccine against the E protein of human COVID-19: an immunoinformatics approach. BioMed Res Int. (2020) 2020:2683286. doi: 10.1101/2020.02.04.934232
274. Ghosh R, Chakraborty A, Biswas A, Chowdhuri S. Evaluation of green tea polyphenols as novel coronavirus (SARS CoV-2) main protease (Mpro) inhibitors – an in silico docking and molecular dynamics simulation study. J Biomol Struct Dyn. (2020) 1–11. doi: 10.1080/07391102.2020.1779818
275. Jo S, Kim S, Shin DH, Kim MS. Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem. (2020) 35:145–51. doi: 10.1080/14756366.2019.1690480
276. Peterson L. In silico molecular dynamics docking of drugs to the inhibitory active site of SARS-CoV-2 protease and their predicted toxicology and ADME. SSRN Electron J. (2020). doi: 10.26434/chemrxiv.12155523.v1
277. Owis A, Marwa EH, Dalia EA, Omar A, Usama A, Mohamed K. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. (2020) 10:19570–75. doi: 10.1039/D0RA03582C
278. Thawabteh A, Juma S, Bader M, Karaman D, Scrano L, Bufo SA, et al. The biological activity of natural alkaloids against herbivores, cancerous cells and pathogens. Toxins. (2019) 11:656–83. doi: 10.3390/toxins11110656
279. Kim DE, Min JS, Jang MS, Lee JY, Shin YS, Park CM, et al. Natural bis-benzylisoquinoline alkaloids-tetrandrine, fangchinoline, and cepharanthine, inhibit human coronavirus oc43 infection of mrc-5 human lung cells. Biomolecules. (2019) 9:696. doi: 10.3390/biom9110696
280. Wink M. Potential of DNA intercalating alkaloids and other plant secondary metabolites against SARS-CoV-2 causing COVID-19. Diversity. (2020) 12:175. doi: 10.3390/d12050175
281. Colson P, Rolain J, Roult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents. (2020) 55:105923. doi: 10.1016/j.ijantimicag.2020.105923
282. Hussein RA, El-Anssary AA. Plants secondary metabolites: the key drivers of the pharmacological actions of medicinal plants. Herb Med. (2019) doi: 10.5772/intechopen.76139
283. Wen CC, Kuo YH, Jan JT, Liang PH, Wang SY, Liu HG, et al. Specific plant terpenoids and lignoids possess potent antiviral activities against severe acute respiratory syndrome coronavirus. J Med Chem. (2007) 50:4087–95. doi: 10.1021/jm070295s
284. Álvarez DM, Castillo E, Duarte LF, Arriagada J, Corrales N, Farías MA, et al. Current antivirals and novel botanical molecules interfering with herpes simplex virus infection. Front Microbiol. (2020) 11:139. doi: 10.3389/fmicb.2020.00139
285. Sun Z, Yu C, Wang W, Yu G, Zhang T, Zhang L, et al. Aloe polysaccharides inhibit Influenza A virus infection—a promising natural anti-flu drug. Front Microbiol. (2018) 9:2338. doi: 10.3389/fmicb.2018.02338
286. Mera IFG, Falconí DEG, Córdova VM. Secondary metabolites in plants: main classes, phytochemical analysis and pharmacological activities. Rev Bionatura. (2019) 4:1000–9. doi: 10.21931/RB/2019.04.04.11
287. Strovel J, Sittampalam S, Coussens NP, Hughes M, Inglese J, Kurtz A, et al. Early Drug Discovery and Development Guidelines: for Academic Researchers, Collaborators, and Start-Up Companies. Assay Guidance Manual. Bethesda, MD: Eli Lilly & Company and the National Center for Advancing Translational Sciences (2004). p. 1–35.
288. Krüger A, Gonçalves Maltarollo V, Wrenger C, Kronenberger T. ADME profiling in drug discovery and a new path paved on silica. In: Gaitonde V, editor. Drug Discovery and Development - New Advances. Itratech open (2020). p. 32. doi: 10.5772/intechopen.86174
289. Seca AML, Pinto DCGA. Biological potential and medical use of secondary metabolites. Medicines. (2019) 6:66. doi: 10.3390/medicines6020066
290. Dias DA, Urban S, Roessner U. A historical overview of natural products in drug discovery. Metabolites. (2012) 2:303–36. doi: 10.3390/metabo2020303
291. Anand U, Nandy S, Mundhra A, Das N, Pandey DK, Dey A. A review on antimicrobial botanicals, phytochemicals and natural resistance modifying agents from Apocynaceae family: possible therapeutic approaches against multidrug resistance in pathogenic microorganisms. Drug Resist Update. (2020) 51:100695. doi: 10.1016/j.drup.2020.100695
292. Anwar N, Teo Y, Joash T. The role of plant metabolites in drug discovery: current challenges and future perspectives. In: Swamy, MK, Akhtar, MS, editors. Natural Bio-active Compounds, Volume 2: Chemistry, Pharmacology and Health Care Practices. New York, NY: Springer Publications (2019). p. 25–51. doi: 10.1007/978-981-13-7205-6_2
293. Gonzalez-Alfonso JL, Peñalver P, Ballesteros AO, Morales JC, Plou FJ. Effect of α-Glycosylation on the stability, antioxidant properties, toxicity, and neuroprotective activity of (-)-Epigallocatechin gallate. Front Nutr. (2019) 6:30. doi: 10.3389/fnut.2019.00030
294. Sharifi-Rad M, Pezzani R, Redaelli M, Zorzan M, Imran M, Khalil AA, et al. Preclinical pharmacological activities of Epigallocatechin-3-gallate in signaling pathways: an update on cancer. Molecules. (2020) 25:467. doi: 10.3390/molecules25030467
295. Banerjee A, Czinn SJ, Reiter RJ, Blanchard TG. Crosstalk between endoplasmic reticulum stress and anti-viral activities: a novel therapeutic target for COVID-19. Life Sci. (2020) 255:117842. doi: 10.1016/j.lfs.2020.117842
296. Karade PG, Jadhav NR. Colon targeted curcumin microspheres laden with ascorbic acid for bioavailability enhancement. J Microencapsul. (2018) 35:372–80. doi: 10.1080/02652048.2018.1501111
297. Azim KF, Ahmed SR, Banik A, Khan MMR, Deb A, Somana SR. Screening and druggability analysis of some plant metabolites against SARS-CoV-2: an integrative computational approach. Inform Med Unlocked. (2020) 20:100367. doi: 10.1016/j.imu.2020.100367
298. Capell T, Twyman RM, Armario NV, Ma JKC, Schillberg S, Christou P. Potential applications of plant biotechnology against SARS-CoV-2. Trends Plant Sci. (2020) 25:635–43. doi: 10.1016/j.tplants.2020.04.009
299. Nakabayashi R, Saito K. Metabolomics for unknown plant metabolites. Anal Bioanal Chem. (2013) 405:5005–11. doi: 10.1007/s00216-013-6869-2
300. Li D Halitschke R Baldwin IT Gaquerel E. Information theory tests critical predictions of plant defense theory for specialized metabolism. Sci Adv. (2020) 6:eaaz0381. doi: 10.1126/sciadv.aaz0381
301. Bhuiyan FR, Campos NA, Swennen R, Carpentier S. Characterizing fruit ripening in plantain and Cavendish bananas: a proteomics approach. J Proteomics. (2020) 214:103632. doi: 10.1016/j.jprot.2019.103632
302. Runfeng L, Yunlong H, Jicheng H, Weiqi P, Qinhai M, Yongxia S, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol Res. (2020) 156:104761. doi: 10.1016/j.phrs.2020.104761
303. Cyranoski D. China is promoting coronavirus treatments based on unproven traditional medicines. Nature. (2020). doi: 10.1038/d41586-020-01284-x. [Epub ahead of print].
304. Nguyen-Vo TH, Nguyen L, Do N, Nguyen TN, Trinh K, Cao H, et al. Plant metabolite databases: from herbal medicines to modern drug discovery. J Chem Inf Model. (2020) 60:1101–10. doi: 10.1021/acs.jcim.9b00826
Keywords: medicinal plants, secondary metabolites, antiviral activities, natural therapeutics/alternative medicine, drug discovery, COVID-19
Citation: Bhuiyan FR, Howlader S, Raihan T and Hasan M (2020) Plants Metabolites: Possibility of Natural Therapeutics Against the COVID-19 Pandemic. Front. Med. 7:444. doi: 10.3389/fmed.2020.00444
Received: 14 April 2020; Accepted: 06 July 2020;
Published: 07 August 2020.
Edited by:
Zisis Kozlakidis, International Agency for Research on Cancer (IARC), FranceReviewed by:
Tauqeer Hussain Mallhi, Al Jouf University, Saudi ArabiaCopyright © 2020 Bhuiyan, Howlader, Raihan and Hasan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Farhana Rumzum Bhuiyan, ZmFyaGFuYS5ib3RAY3UuYWMuYmQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.