- Department of Dermatology, Kagoshima University Graduate School of Medical and Dental Sciences, Kagoshima, Japan
The diagnosis of early stage cutaneous T-cell lymphoma is often difficult, particularly in mycosis fungoides (MF), because the clinical presentation, histological findings, and laboratory findings of MF resemble those of inflammatory skin diseases such as atopic dermatitis, psoriasis, and parapsoriasis en plaque. Furthermore, MF sometimes occurs with or after these inflammatory skin diseases. The current diagnostic criteria heavily rely on clinical impressions along with assessments of T cell clonality. To make a diagnosis of early-stage MF, the detection of a malignant clone is critical. T cell receptor (TCR) gene rearrangements have been detected by southern blotting or polymerase chain reaction for this purpose, but the results of these methods are insufficient. High-throughput TCR sequencing has provided insights into the complexities of the immune repertoire. Accordingly, his technique is more sensitive and specific than current methods, making it useful for the detection of early lesions and monitoring responses to therapy.
Introduction
Cutaneous T cell lymphomas (CTCLs) comprise a heterogeneous class of non-Hodgkin lymphomas that are derived from skin-tropic T cells. Mycosis fungoides (MF), the most prevalent type of primary CTCL, accounts for almost half of all cases (1–3). MF is clinically characterized by erythematous patches, plaques, or skin tumors, and is can be associated with lymph node, blood, and internal organ involvement. More than two-thirds of MF patients are in early stage at first presentation (3–5). MF often starts as an unspecific erythema, similar to many inflammatory skin diseases.
Histopathologically, MF can be characterized by the epidermotropic proliferation of small- to medium-sized pleomorphic lymphocytes forming intraepidermal collections, also called Pautrier's microabscesses. This microabscess is considered the histopathological hallmark of disease, but it is only observed in <20% of early MF cases (6). These microabscesses are also usually recognized as epidermotropic atypical lymphocyte infiltration without spongiosis, although spongiotic variants of MF have been reported (7, 8). Morphologic characterization of early-stage MF might show non-specific findings (9), because skin lesions are infiltrated by large numbers of non-malignant memory T cells, often making it impossible to distinguish malignant T cell clones from activated benign infiltrating T cells based on histopathology (6). Clinical and histopathological algorithms have been developed to aid early diagnosis (10), but the specificity and sensitivity of these algorithms for early diagnosis in individual patients are by no means established. A definitive diagnosis can only be made based on careful clinicopathological correlations (9).
Early stages of MF can resemble benign inflammatory skin disorders (11, 12) like chronic dermatitis including atopic dermatitis (AD), psoriasis, and parapsoriasis en plaque (PEP), among others. AD is a common chronic inflammatory skin disorder that has a T-helper (Th) 2 type-dominant phenotype, skin-barrier dysfunction, and pruritus (13). In contrast AD is an inflammatory disorder, and its pathophysiology is similar to that of AD. Mycosis fungoides, characterized as a Th2-type disease (14, 15), is frequently linked to eosinophilia and high serum immunoglobulin E levels. Although affected skin and peripheral blood T-cells express a Th1 cytokine profile during early-stage MF (16, 17), chemokines expressed in MF lesional skin, such as CCL17, CCL11, and CCL26, are supposed to induce a Th2 milieu in MF (18).
Barrier dysfunction is also observed in MF (19). Lower levels of skin moisture, with increased transepidermal water loss, have been observed in the lesional skin of CTCL, compared to that in normal skin. CTCL lesional skin also displays decreased levels of filaggrin and loricrin mRNAs compared to those in normal skin, which has also been demonstrated for AD. Pruritus is often present in MF patients (20) and constitutes one of the most disturbing symptoms for patients (21). Therefore, it is occasionally difficult to clinically differentiate MF from AD. The coexistence of MF and AD in patients was also reported in several studies (22, 23).
Psoriasis is a common, chronic inflammatory skin disorder defined by thickened, red, scaly plaques with systemic inflammation. The relationship between MF and psoriasis is sometimes difficult to determine, as there is often significant overlap in terms of pathological and clinical observations, particularly in early stages (24). Psoriasis and MF have the abnormal function of T cells as a common symptom. Psoriasis was recognized as a Th1 disease, although recent data suggest that it might be a Th17 disease (25). MF during preliminary stages also exhibits a Th1 phenotype (26); moreover, Krejsgaard et al. (27) reported that malignant T cells from CTCL lesions produce IL-17. Therefore, many early MF cases are misdiagnosed as psoriasis, whereas another group of cases occur in which the two diseases coexist and/or psoriasis develops into MF. The prevalence of psoriasis among patients with MF was found to be higher than that estimated for the general population (24, 28) and patients with psoriasis have an elevated risk of lymphomas including CTCL (29, 30). In addition to the common symptoms, immunosuppressive agents might also promote MF development in patients with psoriasis (31).
PEP is a chronic, inflammatory skin disorder, closely resembling early-stage MF. Clinically, PEP consists of persistent, scaly, well-demarcated erythematous lesions. Pathologically, it is associated with superficial lymphocyte infiltration with various degree of epidermotropism (32). More than 30% patients with large plaque parapsoriasis develop pathologically-confirmed MF (32), and therefore, PEP is often an early manifestation of MF. However, the individual clinical course might determine the difference between early-stage MF and PEP.
Because of difficulties associated with differential diagnosis, MF often remains undiagnosed for years. Accordingly, the average time from the appearance of lesions to a definitive diagnosis was to be estimated 3–6 years (33, 34).
T-cell Receptor Clonality is an Important Criterion in MF Diagnosis
Diagnosing T-cell malignancies is often hampered by difficulties in distinguishing neoplastic T cells from reactive T cells based on conventional morphological and immunopathological criteria (35). Skin lesions of MF patients are infiltrated by many non-malignant memory T cells, and thus, it is often impossible to distinguish a malignant T cell clone from activated, benign, infiltrating T cells by histopathology, particularly for early-stage lesions (10).
T-cell receptor (TCR) gene configurations are thought to be the most promising marker to identify malignant T-cell proliferation (36). TCRs are cell-surface protein heterodimers that are expressed on T cells and comprise either α and β chains or γ and δ chains. Immature T-cells rearrange their TCR genes during maturation in the thymus, then mature as either αβ or γδ lineage T-cells (37). TCRs are unique to individual T cell clones. Malignant cells in MF express αβ TCRs. The gene segments that encode variable (V), diversity (D) (β and δ chains only), joining (J), and constant (C) domains of the TCR protein exist as multiple unique sets (38). Since TCR genes are rearranged during thymic T-cell development (Figure 1) but not in mature T-cells, a peripheral T-cell lymphoma clone of malignant cells should only have a single TCR gene sequence. TCR genes are rearranged polyclonally in normal and reactively proliferating T cells as rearrangements are random, whereas neoplastic cells contain identically-rearranged TCR genes. Similarly, molecular analysis of such rearrangements can be useful to differentiate MF from benign skin conditions (39). Half of patients with large plaque parapsoriasis were reported to have TCR monoclonality in the lesional skin (40), and thus, the detection of monoclonality in a skin lesion generally suggests CTCL including MF, rather than inflammatory skin disorders.
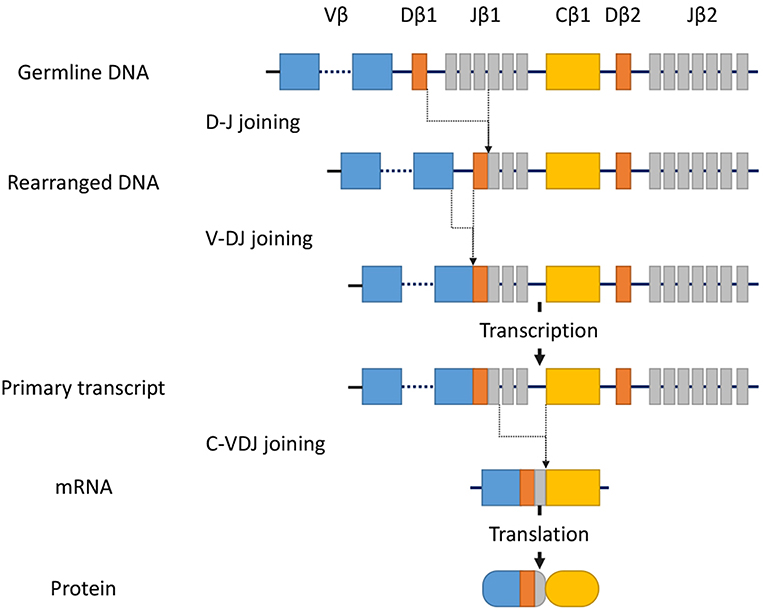
Figure 1. Somatic recombination of the TCRβ gene. Rearrangement begins with D-J recombination followed by subsequent V-DJ recombination. The segments recombine randomly to generate TCR diversity. After transcription, intervening sequences are spliced, generating the TCRβ chain transcript with V, D, J, and C region segments. Finally, transcripts are translated into protein. In contrast, the TCRα gene lacks the D-segment, and its rearrangement starts with V-J recombination.
Conventional Methods for Detecting T-cell Clonality
Initially, Southern blotting was used to determine TCR gene rearrangement clonality (41). This technique can detect clonal T-cell populations in most T-cell lymphomas without prior amplification, but has several disadvantages including low sensitivity and the requirement for large amounts of fresh frozen tissue. Therefore, since the effectiveness of Southern blotting in the diagnostic work-up of MF was reported (42–44) in the early 1990s, more sensitive polymerase chain reaction (PCR)-based methods have been developed. PCR amplification of rearranged TCR gamma genes using genomic DNA as the template was reported to permit the detection of clonal T cells with a sensitivity of 0.1–1% from a background population of polyclonal T-cells (45). Conventional agarose gel electrophoresis of the PCR products often fails to reliably differentiate polyclonal from monoclonal TCR junctions (46), because the narrow size range of the PCR products makes multiple bands appear as single bands. Therefore, PCR amplification with subsequent denaturing gradient polyacrylamide gel electrophoresis and gel scanning (47, 48) or the Biomed GeneScan analysis of flat or capillary polyacrylamide gels (49, 50) has been used as a diagnostic assay for clonality in CTCL patients.
Despite these technical advances, current methods for TCR clonality are still sometimes insufficient for a definitive diagnosis, particularly at early stages, because early lesions often do not contain sufficient numbers of clonal T-cells (51–53). These non-quantitative tests have significant false negative and positive rates for MF (50, 54) and are particularly unreliable for early-stage MF (55).
High-Throughput Sequencing Technologies for Diagnosing MF
Recent improvements in assays to assess T cell clonality have been achieved based on next-generation high-throughput sequencing (NGS) technologies. By sequencing the third complementarity-determining regions (CDR3s) of genes encoding TCRβ and TCRγ, the number of individual T cell clones present in a sample, the relative proportions of specific clones, and the CDR3 region sequences of each clone can be quantified (56, 57). Likewise, NGS represents a superior method to diagnose CTCL through the precise identification of malignant T cell clones (58–60). This technique is more sensitive than previous techniques for the detection of clonality (59, 60). Further, NGS-based methods allow the clinician to follow specific clones when monitoring disease recurrence and progression (60). Furthermore, TCR sequencing has clarified that neoplastic cells in some MF lesions might be as few as 1% of the total population of T cells (59). These data clearly explain the difficulties encountered in the histopathological assessment of early-stage MF. In contrast, the frequencies of the most dominant T cell clones range from 1 to 10% in most cases of inflammatory skin disorders such as psoriasis and eczematous dermatitis, among others, whereas the frequency of the most dominant T cell clones adjusted by total nucleated cells could distinguish MF from inflammatory skin disorders (59). Therefore, PEP often demonstrates TCR rearrangement, and NGS-based TCR gene analysis might overcome difficulties in distinguishing PEP and early-stage MF T-cell repertoires.
Most MF cases present as early-stage, typically with a chronic, indolent clinical course. Greater than 80% of patients with early-stage disease will present with an indolent life-long course, free of disease progression, independent of the treatment modality (5). For many years, most patients will also exhibit short-term clinical response associated with recurrent disease, as well as a normal life expectancy in the majority of cases. Furthermore, the limited efficacy associated with chemotherapy has been discussed in retrospective studies (61, 62), making it clear that potentially toxic and aggressive therapies should be avoided (63, 64). However, a small number of early-stage cases will progress. Since advanced-stage patients have poor prognoses, the early identification of high-risk subpopulations is important.
Using NGS technologies, de Masson et al. (55) demonstrated that an enhanced proportion of a malignant T cell clone in the skin is strongly correlated with reductions in progression-free and overall survival for patients with CTCL, and particularly for patients with early-stage MF with a T2 distribution. Further, based on high throughput DNA sequencing of the TCRβ gene, a tumor clone frequency of >25% was found to be a strong predictor of disease progression and poor survival for MF patients with disease limited to the skin.
In summary, evidence for TCR clonality from any method is strong evidence for malignancy. However, it is not conclusive, because benign conditions have also been associated with clonal T-cell populations, such as reactive or autoimmune conditions (65, 66).
Conclusion
NGS can be used to assess TCR clonality with superior sensitivity compared to current methods and is useful to diagnose early stage MF. Moreover, this technique permits the tracking of specific clones across different time points or in multiple lesions for a more accurate diagnosis of MF recurrence or progression (55, 59, 60).
Author Contributions
KF conceived the concept and wrote the manuscript. TK edited and improved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Editage (www.editage.com) for English language editing.
References
1. Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. (2005) 105:3768–85. doi: 10.1182/blood-2004-09-3502
2. Bradford PT, Devesa SS, Anderson WF, Toro JR. Cutaneous lymphoma incidence patterns in the United States: a population-based study of 3884 cases. Blood. (2009) 113:5064–73. doi: 10.1182/blood-2008-10-184168
3. Hamada T, Iwatsuki K. Cutaneous lymphoma in Japan: a nationwide study of 1733 patients. J Dermatol. (2014) 41:3–10. doi: 10.1111/1346-8138.12299
4. Assaf C, Gellrich S, Steinhoff M, Nashan D, Weisse F, Dippel E, et al. Cutaneous lymphomas in Germany: an analysis of the Central Cutaneous Lymphoma Registry of the German Society of Dermatology (DDG). J Dtsch Dermatol Ges. (2007) 5:662–8. doi: 10.1111/j.1610-0387.2007.06337.x
5. Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. J Clin Oncol. (2010) 28:4730–9. doi: 10.1200/JCO.2009.27.7665
6. Massone C, Kodama K, Kerl H, Cerroni L. Histopathologic features of early (patch) lesions of mycosis fungoides: a morphologic study on 745 biopsy specimens from 427 patients. Am J Surg Pathol. (2005) 29:550–60. doi: 10.1097/01.pas.0000153121.57515.c6
7. Shapiro PE, Pinto FJ. The histologic spectrum of mycosis fungoides/Sezary syndrome (cutaneous T-cell lymphoma). A review of 222 biopsies, including newly described patterns and the earliest pathologic changes. Am J Surg Pathol. (1994) 18:645–67. doi: 10.1097/00000478-199407000-00001
8. Shamim H, Johnson EF, Gibson LE, Comfere N. Mycosis fungoides with spongiosis: a potential diagnostic pitfall. J Cutan Pathol. (2019). doi: 10.1111/cup.13477. [Epub ahead of print].
9. Cerroni L. Mycosis fungoides-clinical and histopathologic features, differential diagnosis, and treatment. Semin Cutan Med Surg. (2018) 37:2–10. doi: 10.12788/j.sder.2018.002
10. Pimpinelli N, Olsen EA, Santucci M, Vonderheid E, Haeffner AC, Stevens S, et al. Defining early mycosis fungoides. J Am Acad Dermatol. (2005) 53:1053–63. doi: 10.1016/j.jaad.2005.08.057
11. Elmer KB, George RM. Cutaneous T-cell lymphoma presenting as benign dermatoses. Am Fam Phys. (1999) 59:2809–13.
12. Nashan D, Faulhaber D, Stander S, Luger TA, Stadler R. Mycosis fungoides: a dermatological masquerader. Br J Dermatol. (2007) 156:1–10. doi: 10.1111/j.1365-2133.2006.07526.x
13. Kabashima K. New concept of the pathogenesis of atopic dermatitis: interplay among the barrier, allergy, and pruritus as a trinity. J Dermatol Sci. (2013) 70:3–11. doi: 10.1016/j.jdermsci.2013.02.001
14. Dummer R, Kohl O, Gillessen J, Kagi M, Burg G. Peripheral blood mononuclear cells in patients with nonleukemic cutaneous T-cell lymphoma. Reduced proliferation and preferential secretion of a T helper-2-like cytokine pattern on stimulation. Arch Dermatol. (1993) 129:433–6. doi: 10.1001/archderm.129.4.433
15. Guenova E, Watanabe R, Teague JE, Desimone JA, Jiang Y, Dowlatshahi M, et al. TH2 cytokines from malignant cells suppress TH1 responses and enforce a global TH2 bias in leukemic cutaneous T-cell lymphoma. Clin Cancer Res. (2013) 19:3755–63. doi: 10.1158/1078-0432.CCR-12-3488
16. Asadullah K, Docke WD, Haeussler A, Sterry W, Volk HD. Progression of mycosis fungoides is associated with increasing cutaneous expression of interleukin-10 mRNA. J Invest Dermatol. (1996) 107:833–7. doi: 10.1111/1523-1747.ep12330869
17. Asadullah K, Haeussler A, Sterry W, Docke WD, Volk HD. Interferon gamma and tumor necrosis factor alpha mRNA expression in mycosis fungoides progression. Blood. (1996) 88:757–8.
18. Sugaya M. Chemokines and cutaneous lymphoma. J Dermatol Sci. (2010) 59:81–5. doi: 10.1016/j.jdermsci.2010.05.005
19. Suga H, Sugaya M, Miyagaki T, Ohmatsu H, Kawaguchi M, Takahashi N, et al. Skin barrier dysfunction and low antimicrobial peptide expression in cutaneous T-cell lymphoma. Clin Cancer Res. (2014) 20:4339–48. doi: 10.1158/1078-0432.CCR-14-0077
20. Beynon T, Selman L, Radcliffe E, Whittaker S, Child F, Orlowska D, et al. 'We had to change to single beds because I itch in the night': a qualitative study of the experiences, attitudes and approaches to coping of patients with cutaneous T-cell lymphoma. Br J Dermatol. (2015) 173:83–92. doi: 10.1111/bjd.13732
21. Sampogna F, Frontani M, Baliva G, Lombardo GA, Alvetreti G, Di Pietro C, et al. Quality of life and psychological distress in patients with cutaneous lymphoma. Br J Dermatol. (2009) 160:815–22. doi: 10.1111/j.1365-2133.2008.08992.x
22. Mehrany K, El-Azhary RA, Bouwhuis SA, Pittelkow MR. Cutaneous T-cell lymphoma and atopy: is there an association? Br J Dermatol. (2003) 149:1013–7. doi: 10.1111/j.1365-2133.2003.05551.x
23. Onsun N, Kural Y, Su O, Demirkesen C, Buyukbabani N. Hypopigmented mycosis fungoides associated with atopy in two children. Pediatr Dermatol. (2006) 23:493–6. doi: 10.1111/j.1525-1470.2006.00291.x
24. Nikolaou V, Marinos L, Moustou E, Papadavid E, Economidi A, Christofidou E, et al. Psoriasis in patients with mycosis fungoides: a clinicopathological study of 25 patients. J Eur Acad Dermatol Venereol. (2017) 31:1848–52. doi: 10.1111/jdv.14365
25. Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. (2014) 32:227–55. doi: 10.1146/annurev-immunol-032713-120225
26. Papadavid E, Economidou J, Psarra A, Kapsimali V, Mantzana V, Antoniou C, et al. The relevance of peripheral blood T-helper 1 and 2 cytokine pattern in the evaluation of patients with mycosis fungoides and Sezary syndrome. Br J Dermatol. (2003) 148:709–18. doi: 10.1046/j.1365-2133.2003.05224.x
27. Krejsgaard T, Ralfkiaer U, Clasen-Linde E, Eriksen KW, Kopp KL, Bonefeld CM, et al. Malignant cutaneous T-cell lymphoma cells express IL-17 utilizing the Jak3/Stat3 signaling pathway. J Invest Dermatol. (2011) 131:1331–8. doi: 10.1038/jid.2011.27
28. Donigan JM, Snowden C, Carter JB, Kimball AB. The temporal association between cutaneous T-cell lymphoma and psoriasis: implications for common biologic processes. J Eur Acad Dermatol Venereol. (2016) 30:e31–2. doi: 10.1111/jdv.13281
29. Morales MM, Olsen J, Johansen P, Kaerlev L, Guenel P, Arveux P, et al. Viral infection, atopy and mycosis fungoides: a European multicentre case-control study. Eur J Cancer. (2003) 39:511–6. doi: 10.1016/S0959-8049(02)00773-6
30. Gelfand JM, Shin DB, Neimann AL, Wang X, Margolis DJ, Troxel AB. The risk of lymphoma in patients with psoriasis. J Invest Dermatol. (2006) 126:2194–201. doi: 10.1038/sj.jid.5700410
31. Nikolaou V, Papadavid E, Economidi A, Marinos L, Moustou E, Karampidou K, et al. Mycosis fungoides in the era of antitumour necrosis factor-alpha treatments. Br J Dermatol. (2015) 173:590–3. doi: 10.1111/bjd.13705
32. Väkevä L, Sarna S, Vaalasti A, Pukkala E, Kariniemi AL, Ranki A. A retrospective study of the probability of the evolution of parapsoriasis en plaques into mycosis fungoides. Acta Derm Venereol. (2005) 85:318–23. doi: 10.1080/00015550510030087
33. van Doorn R, Van Haselen CW, van Voorst Vader PC, Geerts ML, Heule F, de Rie M, et al. Mycosis fungoides: disease evolution and prognosis of 309 Dutch patients. Arch Dermatol. (2000) 136:504–10. doi: 10.1001/archderm.136.4.504
34. Scarisbrick JJ, Quaglino P, Prince HM, Papadavid E, Hodak E, Bagot M, et al. The PROCLIPI international registry of early-stage mycosis fungoides identifies substantial diagnostic delay in most patients. Br J Dermatol. (2019) 181:350–7. doi: 10.1111/bjd.17258
35. Witzens M, Mohler T, Willhauck M, Scheibenbogen C, Lee KH, Keilholz U. Detection of clonally rearranged T-cell-receptor gamma chain genes from T-cell malignancies and acute inflammatory rheumatic disease using PCR amplification, PAGE, and automated analysis. Ann Hematol. (1997) 74:123–30. doi: 10.1007/s002770050269
36. Bertness V, Kirsch I, Hollis G, Johnson B, Bunn PA Jr. T-cell receptor gene rearrangements as clinical markers of human T-cell lymphomas. N Engl J Med. (1985) 313:534–8. doi: 10.1056/NEJM198508293130902
37. Robey E, Fowlkes BJ. The alpha beta versus gamma delta T-cell lineage choice. Curr Opin Immunol. (1998) 10:181–7. doi: 10.1016/S0952-7915(98)80247-1
38. Chitgopeker P, Sahni D. T-cell receptor gene rearrangement detection in suspected cases of cutaneous T-cell lymphoma. J Invest Dermatol. (2014) 134:1–5. doi: 10.1038/jid.2014.73
39. Bergman R. How useful are T-cell receptor gene rearrangement studies as an adjunct to the histopathologic diagnosis of mycosis fungoides? Am J Dermatopathol. (1999) 21:498–502. doi: 10.1097/00000372-199910000-00019
40. Simon M, Flaig MJ, Kind P, Sander CA, Kaudewitz P. Large plaque parapsoriasis: clinical and genotypic correlations. J Cutan Pathol. (2000) 27:57–60. doi: 10.1034/j.1600-0560.2000.027002057.x
41. Weiss LM, Hu E, Wood GS, Moulds C, Cleary ML, Warnke R, et al. Clonal rearrangements of T-cell receptor genes in mycosis fungoides and dermatopathic lymphadenopathy. N Engl J Med. (1985) 313:539–44. doi: 10.1056/NEJM198508293130903
42. Dosaka N, Tanaka T, Fujita M, Miyachi Y, Horio T, Imamura S. Southern blot analysis of clonal rearrangements of T-cell receptor gene in plaque lesion of mycosis fungoides. J Invest Dermatol. (1989) 93:626–9. doi: 10.1111/1523-1747.ep12319746
43. Amagai M, Hayakawa K, Amagai N, Kobayashi K, Onodera Y, Shimizu N, et al. T cell receptor gene rearrangement analysis in mycosis fungoides and disseminated lymphocytoma cutis. Dermatologica. (1990) 181:193–6. doi: 10.1159/000247922
44. Lynch JW Jr, Linoilla I, Sausville EA, Steinberg SM, Ghosh BC, Nguyen DT, et al. Prognostic implications of evaluation for lymph node involvement by T-cell antigen receptor gene rearrangement in mycosis fungoides. Blood. (1992) 79:3293–9.
45. Bourguin A, Tung R, Galili N, Sklar J. Rapid, nonradioactive detection of clonal T-cell receptor gene rearrangements in lymphoid neoplasms. Proc Natl Acad Sci USA. (1990) 87:8536–40. doi: 10.1073/pnas.87.21.8536
46. Kneba M, Bolz I, Linke B, Bertram J, Rothaupt D, Hiddemann W. Characterization of clone-specific rearrangement T-cell receptor gamma-chain genes in lymphomas and leukemias by the polymerase chain reaction and DNA sequencing. Blood. (1994) 84:574–81.
47. Wood GS, Tung RM, Haeffner AC, Crooks CF, Liao S, Orozco R, et al. Detection of clonal T-cell receptor gamma gene rearrangements in early mycosis fungoides/Sezary syndrome by polymerase chain reaction and denaturing gradient gel electrophoresis (PCR/DGGE). J Invest Dermatol. (1994) 103:34–41. doi: 10.1111/1523-1747.ep12389114
48. Theodorou I, Delfau-Larue MH, Bigorgne C, Lahet C, Cochet G, Bagot M, et al. Cutaneous T-cell infiltrates: analysis of T-cell receptor gamma gene rearrangement by polymerase chain reaction and denaturing gradient gel electrophoresis. Blood. (1995) 86:305–10.
49. Assaf C, Hummel M, Dippel E, Goerdt S, Muller HH, Anagnostopoulos I, et al. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferations by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood. (2000) 96:640–6.
50. Ponti R, Fierro MT, Quaglino P, Lisa B, Paola F, Michela O, et al. TCRgamma-chain gene rearrangement by PCR-based GeneScan: diagnostic accuracy improvement and clonal heterogeneity analysis in multiple cutaneous T-cell lymphoma samples. J Invest Dermatol. (2008) 128:1030–8. doi: 10.1038/sj.jid.5701109
51. Fivenson DP, Hanson CA, Nickoloff BJ. Localization of clonal T cells to the epidermis in cutaneous T-cell lymphoma. J Am Acad Dermatol. (1994) 31(5 Pt 1):717–23. doi: 10.1016/S0190-9622(94)70231-4
52. Ponti R, Quaglino P, Novelli M, Fierro MT, Comessatti A, Peroni A, et al. T-cell receptor gamma gene rearrangement by multiplex polymerase chain reaction/heteroduplex analysis in patients with cutaneous T-cell lymphoma (mycosis fungoides/Sezary syndrome) and benign inflammatory disease: correlation with clinical, histological and immunophenotypical findings. Br J Dermatol. (2005) 153:565–73. doi: 10.1111/j.1365-2133.2005.06649.x
53. Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome): part I. Diagnosis: clinical and histopathologic features and new molecular and biologic markers. J Am Acad Dermatol. (2014) 70: 205 e201–216; quiz 221-202. doi: 10.1016/j.jaad.2013.07.049
54. Sandberg Y, Heule F, Lam K, Lugtenburg PJ, Wolvers-Tettero IL, van Dongen JJ, et al. Molecular immunoglobulin/T- cell receptor clonality analysis in cutaneous lymphoproliferations. Experience with the BIOMED-2 standardized polymerase chain reaction protocol. Haematologica. (2003) 88:659–70.
55. de Masson A, O'Malley JT, Elco CP, Garcia SS, Divito SJ, Lowry EL, et al. High-throughput sequencing of the T cell receptor beta gene identifies aggressive early-stage mycosis fungoides. Sci Transl Med. (2018) 10:eaar5894. doi: 10.1126/scitranslmed.aar5894
56. Weng WK, Armstrong R, Arai S, Desmarais C, Hoppe R, Kim YH. Minimal residual disease monitoring with high-throughput sequencing of T cell receptors in cutaneous T cell lymphoma. Sci Transl Med. (2013) 5:214ra171. doi: 10.1126/scitranslmed.3007420
57. Kirsch IR, Watanabe R, O'Malley JT, Williamson DW, Scott LL, Elco CP, et al. TCR sequencing facilitates diagnosis and identifies mature T cells as the cell of origin in CTCL. Sci Transl Med. (2015) 7:308ra158. doi: 10.1126/scitranslmed.aaa9122
58. Sufficool KE, Lockwood CM, Abel HJ, Hagemann IS, Schumacher JA, Kelley TW, et al. T-cell clonality assessment by next-generation sequencing improves detection sensitivity in mycosis fungoides. J Am Acad Dermatol. (2015) 73:228–36 e222. doi: 10.1016/j.jaad.2015.04.030
59. Hughes CF, Khot A, McCormack C, Lade S, Westerman DA, Twigger R, et al. Lack of durable disease control with chemotherapy for mycosis fungoides and Sezary syndrome: a comparative study of systemic therapy. Blood. (2015) 125:71–81. doi: 10.1182/blood-2014-07-588236
60. Hanel W, Briski R, Ross CW, Anderson TF, Kaminski MS, Hristov AC, et al. A retrospective comparative outcome analysis following systemic therapy in Mycosis fungoides and Sezary syndrome. Am J Hematol. (2016) 91:E491–5. doi: 10.1002/ajh.24564
61. Trautinger F, Knobler R, Willemze R, Peris K, Stadler R, Laroche L, et al. EORTC consensus recommendations for the treatment of mycosis fungoides/Sezary syndrome. Eur J Cancer. (2006) 42:1014–30. doi: 10.1016/j.ejca.2006.01.025
62. Wilcox RA. Cutaneous T-cell lymphoma: 2017 update on diagnosis, risk-stratification, and management. Am J Hematol. (2017) 92:1085–102. doi: 10.1002/ajh.24876
63. Plaza JA, Morrison C, Magro CM. Assessment of TCR-beta clonality in a diverse group of cutaneous T-Cell infiltrates. J Cutan Pathol. (2008) 35:358–65. doi: 10.1111/j.1600-0560.2007.00813.x
64. Nakasone Y, Kumagai K, Matsubara R, Shigematsu H, Kitaura K, Suzuki S, et al. Characterization of T cell receptors in a novel murine model of nickel-induced intraoral metal contact allergy. PLoS ONE. (2018) 13:e0209248. doi: 10.1371/journal.pone.0209248
65. Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. (2009) 114:4099–107. doi: 10.1182/blood-2009-04-217604
Keywords: mycosis fungoides, early stage, T-cell receptor, rearrangement, next-generation sequencing
Citation: Fujii K and Kanekura T (2019) Next-Generation Sequencing Technologies for Early-Stage Cutaneous T-Cell Lymphoma. Front. Med. 6:181. doi: 10.3389/fmed.2019.00181
Received: 26 June 2019; Accepted: 29 July 2019;
Published: 13 August 2019.
Edited by:
Taku Fujimura, Tohoku University, JapanReviewed by:
Toshihisa Hamada, Takamatsu Red Cross Hospital, JapanTomomitsu Miyagaki, The University of Tokyo, Japan
Copyright © 2019 Fujii and Kanekura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kazuyasu Fujii, a2F6ZnVqaWkmI3gwMDA0MDttMi5rdWZtLmthZ29zaGltYS11LmFjLmpw
 Kazuyasu Fujii
Kazuyasu Fujii Takuro Kanekura
Takuro Kanekura