
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Med. , 19 September 2018
Sec. Infectious Diseases: Pathogenesis and Therapy
Volume 5 - 2018 | https://doi.org/10.3389/fmed.2018.00262
 Raphaël M. Zellweger1
Raphaël M. Zellweger1 Buddha Basnyat2,3,4
Buddha Basnyat2,3,4 Poojan Shrestha2
Poojan Shrestha2 Krishna G. Prajapati5
Krishna G. Prajapati5 Sabina Dongol2
Sabina Dongol2 Paban K. Sharma5
Paban K. Sharma5 Samir Koirala2
Samir Koirala2 Thomas C. Darton1,6
Thomas C. Darton1,6 Christine Boinett1
Christine Boinett1 Corinne N. Thompson1
Corinne N. Thompson1 Guy E. Thwaites1,3
Guy E. Thwaites1,3 Stephen Baker1,3,7*
Stephen Baker1,3,7* Abhilasha Karkey2,3,4
Abhilasha Karkey2,3,4A comprehensive longitudinal understanding of the changing epidemiology of the agents causing bacteraemia and their AMR profiles in key locations is crucial for assessing the progression and magnitude of the global AMR crisis. We performed a retrospective analysis of routine microbiological data from April 1992 to December 2014, studying the time trends of non-Salmonella associated bacteraemia at a single Kathmandu healthcare facility. The distribution of aetiological agents, their antimicrobial susceptibility profiles, and the hospital ward of isolation were assessed. Two hundred twenty-four thousand seven hundred forty-one blood cultures were performed over the study period, of which, 30,353 (13.5%) exhibited growth for non-contaminant bacteria. We observed a significant increasing trend in the proportion of MDR non-Salmonella Enterobacteriaceae (p < 0.001), other Gram-negative organisms (p = 0.006), and Gram-positive organisms (p = 0.006) over time. Additionally, there was an annual increasing trend in the proportion of MDR organisms in bacteria-positive blood cultures originating from patients attending the emergency ward (p = 0.006) and the outpatient department (p = 0.006). This unique dataset demonstrates that community acquired non-Salmonella bacteraemia has become an increasingly important cause of hospital admission in Kathmandu. An increasing burden of bacteraemia associated with MDR organisms in the community underscores the need for preventing the circulation of MDR bacteria within the local population.
Bacteraemia continues to be a common cause of febrile illness worldwide, and is associated with substantial morbidity and mortality (1, 2). The spectrum of organisms causing bacteraemia is variable and highly setting-specific (2–4). The disease severity and outcome of bacteraemia can also vary significantly, and are related to multiple factors, including the genetic composition of the infecting bacteria, susceptibility of the host, and the clinical management of the patient (5–7).
In recent years, there have been substantial changes in the aetiological agents of bacteraemia and their associated antimicrobial susceptibility profiles (5, 6, 8). The rapid administration of an effective antimicrobial is imperative for the clinical management of bacteraemia; however increasing antimicrobial resistance (AMR) restricts treatment options. The sustained usage of broad-spectrum antimicrobials has become associated with the emergence, selection, and circulation of invasive bacterial pathogens exhibiting AMR to many empirically prescribed antimicrobials (9). Therefore, a comprehensive longitudinal understanding of the changing epidemiology of the agents causing bacteraemia and their AMR profiles in specific locations is crucial for assessing the scale of the global AMR crisis, revising rational management strategies, and improving treatment guidelines (10, 11).
In Nepal, febrile disease is a common reason for seeking medical attention (12). Like in many other low-income countries, much of the febrile disease burden in Nepal is associated with community-acquired infections. Invasive Salmonella serovars Typhi and Paratyphi A, now with resistance against fluoroquinolones, remain the organisms most frequently isolated from the blood of febrile patients in the Kathmandu Valley (12–14). However, invasive Salmonella are less adept at acquiring resistance to multiple antimicrobials than other indicator organisms and may not reflect general trends in AMR (15). Furthermore, Salmonella Typhi and Paratyphi A are well established and well described pathogens in the Kathmandu Valley, but little has been reported for non-Salmonella associated bacteraemia. To investigate the epidemiology and AMR profile of agents causing bacteraemia in this setting, we performed a retrospective analysis on 23 years of data regarding the etiology of non-Salmonella associated bacteraemia at Patan hospital, a major general hospital in the Kathmandu Valley.
This study used anonymised data, originating from the microbiology laboratory of Patan Hospital in Kathmandu, consisting of culture result, isolated organism, ward in which the blood sample was taken, and associated AMR profile. This study was, therefore, a component of the routine surveillance measures for infection control at Patan Hospital and local ethical approval and individual written informed consent were not required. However, a written permission, for access and analysis of the data, was sought and obtained from the Hospital.
This study was a retrospective analysis of routine microbiology laboratory results of all blood cultures taken at Patan Hospital between April 1992 and December 2014. It is one of three general hospitals in the greater metropolitan area of Kathmandu. The hospital had a capacity of 138 beds in 1992, 350 beds in 2014, a current capacity of 592 beds, and provides emergency and elective services to outpatients (~200,000 outpatient visits per year) and inpatients, 90% of which live in Lalitpur Sub Metropolitan City LSMC.
Systematic criteria concerning which patients had a blood culture performed were not used during the course of the study. However, a blood culture was generally performed on patients in whom a bacterial infection was suspected on the basis of a fever (>38°C) or evidence of sepsis on the basis of the presence of two or more of the following criteria: fever (>38°C) or sub-normal temperature (<36°C), tachycardia, tachypnoea, elevated white cell count (>12,000 cells/mm3), or depressed white cell count (<4,000 cells/mm3). Review of the microbiological standard operating procedures over the analysis time period suggest no substantive changes in the application of these criteria. Bacteraemia was defined as isolation of at least one clinically relevant pathogen from one blood culture drawn from a patient with an indicative clinical syndrome. Organisms of the same species with the same antimicrobial susceptibility profile isolated from the same patient (matching hospital ID number) were removed from analysis.
The majority of blood cultures were performed manually by inoculating 3–5 mL (pediatric patients) or 5–8 mL (adult patients) of blood into 30–50 mL of media containing tryptone soya broth and 0.05% sodium polyanetholesulfonate. An automated BACTEC (Becton Dickinson, MD, USA) system was introduced for cultures on blood taken from those aged <14 years from 2005 onwards. Blood samples from children were inoculated into BACTEC Peds plus bottles following the manufacturers recommendations (Becton Dickinson, MD, USA). All inoculated blood bottles were incubated at 37°C and examined daily over 7 days for bacterial growth, or until flagged in the automated system. For all conventional bottles, irrespective of whether turbidity developed or not, the inoculated broth was sub-cultured and Gram stained on sixth day of incubation. Organisms isolated from the primary cultures were sub-cultured onto sheep blood agar and chocolate agar. Plates were incubated at 37°C for 48 h. Organisms were identified by standard methods including API20E identification kits (Bio-Mérieux, Craponne, France) and specific antisera where possible. Coagulase negative staphylococci were reported only for neonates as pathogens and as contaminants for the rest of the population. Gram-negative rods that were not identifiable through biochemical tests were reported as Gram-negative rods.
Antimicrobial susceptibility testing was performed at time of isolation by the modified Kirby-Bauer disc diffusion method. Zone size interpretations were initially performed following the guidelines provided in the antimicrobial disc packaging; from 2003 onwards, susceptibilities were inferred by following the appropriate CLSI guidelines. A range of antimicrobials were tested, this was largely dependent on the period of isolation (Table S1).
For analysis purposes, resistant and intermediate organisms were grouped as “non-susceptible.” Multi-drug resistance (MDR) was defined as “acquired non-susceptibility to at least one agent in three or more antimicrobial categories” (16). The categories used in this study are described in Table S2. As current practice, laboratory results reporting resistance to an antimicrobial for which an organism was intrinsically resistant were not taken into account when generating the MDR profile. Intrinsic resistance were determined according to the 2014 Clinical and Laboratory Standards Institute (CLSI) guidelines (Table S3) (17). Increasing or decreasing trends in isolated organisms and AMR profiles were measured using Cox and Stuart test for trends (R-package “randtrends”). Davies' test (R-package “segmented”) was used to detect a non-constant regression parameter (i.e., change in slope) when the percentage of MDR was regressed on years. All analyses were performed using the statistical software R version 3.3.2 (18).
From April 1992 to December 2014, a total of 224,741 blood cultures were performed at Patan Hospital, of which 173,892 (77.4%) were culture negative, and a further 20,410 (9.1%) contained a contaminant organism. A non-contaminant fungal pathogen was identified in 86 blood samples, the remaining 30,353 (13.5%) blood cultures exhibited growth for non-contaminant bacteria (Table 1). The most frequently isolated organisms were Salmonella Typhi and Salmonella Paratyphi A, associated with 44.8% (13,592/30,353) and 20.0% (6,057/30,353) of all bacteria-positive blood cultures, respectively (Table S4).
The most frequently isolated non-Salmonella organisms were (in decreasing frequency) Enterobacter spp., Acinetobacter spp., Coagulase negative Staphylococci, Escherichia coli, Klebsiella spp. (including both K. pneumoniae and K. oxytoca), Staphylococcus aureus, Non-haemolytic Streptococci, and Streptococcus pneumoniae (Table S4). Between 1992 and 2014, the total number of blood cultures performed increased, but the proportion of blood samples exhibiting bacterial growth showed a decreasing trend (p < 0.001; Cox and Stuart test) (Figure S1).
The annual distribution of the aetiological agents associated with bacteraemia, stratified by non-Salmonella Enterobacteriaceae, other Gram-negative organisms, and Gram-positive organisms (19), is shown in Figure 1. The absolute annual counts of Enterobacteriaceae and Gram-positive organisms showed an increasing trend from 1992 to 2014 (p < 0.001, Cox and Stuart test). The proportion of non-Salmonella Enterobacteriaceae isolated from 1992 to 2014 additionally exhibited an increasing annual trend (p < 0.001, Cox and Stuart test) (Figure S2). No such trend was observed for other Gram-negative organisms, which exhibited a peak in 2007.
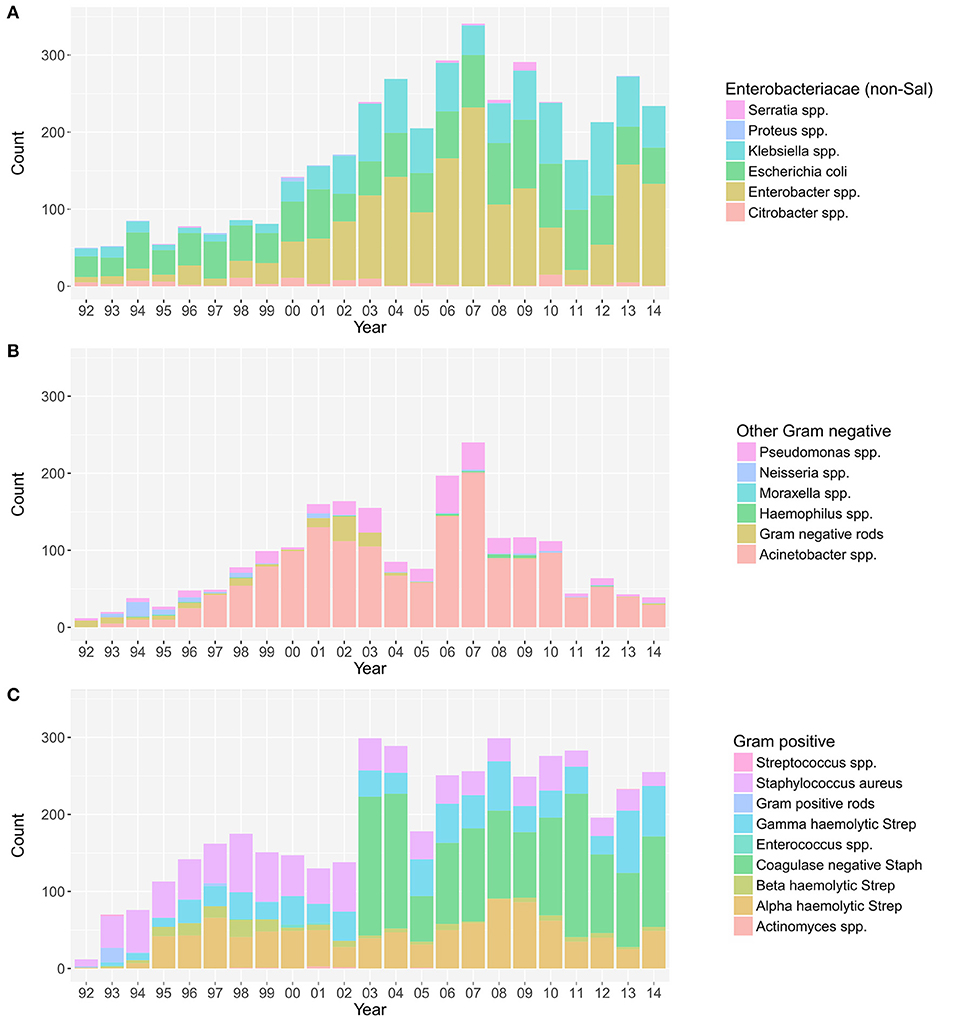
Figure 1. The annual distribution of the aetiological agents associated with bacteraemia. The distribution of non-Salmonella Enterobacteriaceae (A), other Gram-negative organisms (B), and Gram-positive organisms (C) isolated from positive blood cultures in this single healthcare facility between April 1992 and December 2014.
Over the study period we identified a significant increasing trend in the proportion of MDR non-Salmonella Enterobacteriaceae (p < 0.001; Cox and Stuart test), other Gram-negative organisms (p = 0.006), and Gram-positive organisms (p = 0.006) isolated (Figure 3 and Figure S3). The proportion of MDR organisms was regressed on time to assess the annual increase in the proportion of MDR organisms. We additionally assessed the presence of breakpoints in the regression to identify any specific years that were associated with a major change in the rate of the increase in MDR percentage. For the non-Salmonella Enterobacteriaceae and other Gram-negative organisms, there was no breakpoint in the regression plots, signifying a constant increase in the proportion of MDR organisms of 1.5 and 1.9% per year, respectively (Figure 3). Conversely, the Gram-positive organisms exhibited a breakpoint in 2010/2011, indicating acceleration in the annual increase of the proportion of MDR organisms from 2011 onwards (1992–2010: 1.26% annual increase in the proportion of MDR organisms, 2011–2014: 7.61% annual increase).
On investigating the time trends for MDR and non-MDR organisms we identified three distinct periods during which: (i) the number of isolated non-MDR organisms was greater than the number of MDR organisms (1992–2000, MDR/non-MDR <1), (ii) the number of isolated non-MDR organisms and MDR organisms was approximately equal (2000–2010, MDR/non-MDR≈1), and (iii) the number of MDR organisms was greater than the number of non-MDR organisms (2010–2014, MDR/non-MDR > 1) (Figure 2).
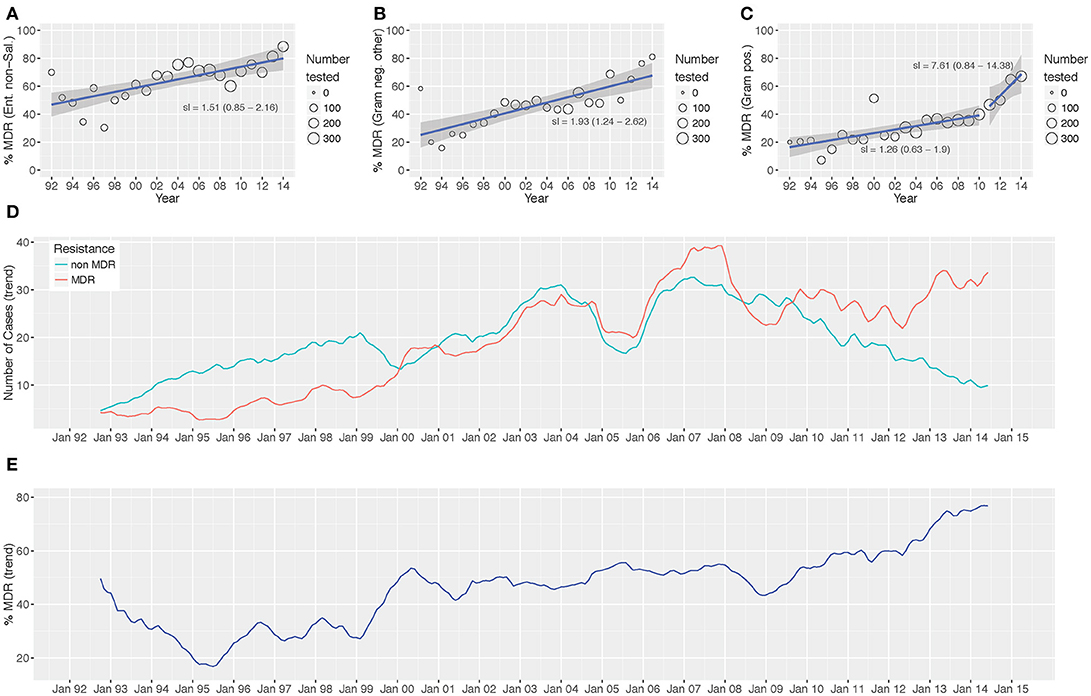
Figure 2. Antimicrobial resistance trends from 1992 to 2014. The proportion of MDR Enterobacteriaceae non-Salmonella (A), other Gram-negative organisms (B), and Gram-positive organisms (C) isolated between 1992 and 2014. Time trends of MDR vs. non-MDR isolates [(D) count and (E) distribution]. Linear trend lines (blue) and 95% CI for the regression lines (shaded) are overlaid; the slopes of the regression line (with 95% CI) are indicated (A-C) two regression lines are present in (C) to highlight the “breakpoint” in the slope.
Over the investigative period, the majority of positive blood cultures originated from patients attending the emergency department (3,808/10,496; 36.3%), the pediatric ward (2,217/10,496; 21.1%), and the outpatient department (1,528/10,496; 14.6%). The yearly distribution of wards from where the various groups of organisms were isolated is shown in Figure 3. The proportion of blood cultures positive with Enterobacteriaceae from patients attending the emergency department (in comparison to other wards in the hospital) showed an increasing trend between 1992 and 2014 (p = 0.03; Cox and Stuart test).
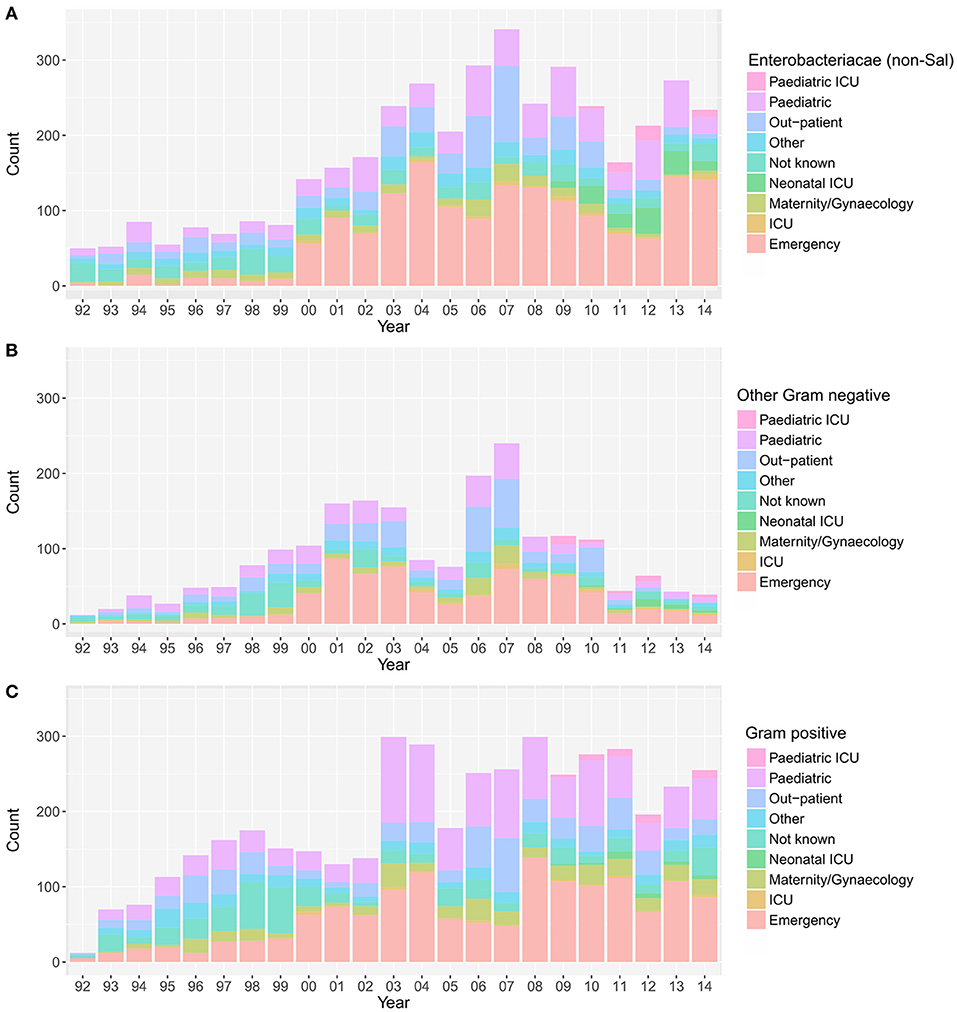
Figure 3. The annual distribution of wards in which the positive blood samples originated. Plots showing the hospital wards from which the positive blood samples originated between April 1992 and December 2014; Enterobacteriaceae non-Salmonella (A), other Gram-negative organisms (B), and Gram-positive organisms (C). Maternity, birthing, post-partum and gynecology wards were grouped under “maternity/gynecology,” pediatric and nursery wards were grouped under “pediatric,” and orthopedic, private, medical, and surgical wards were grouped under “other”.
Finally, we assessed the relative distribution of MDR and non-MDR organisms in different hospital wards (Figure S4). Over the entire period, for both the MDR and the non-MDR organisms, the majority of organisms were isolated from patients attending the emergency department (MDR: 36.4%; 1,938/5,319, non-MDR: 36.3%; 1,859/5,127), followed by the pediatric ward (MDR: 22.7%; 1,206/5,319, non-MDR: 19.6%; 1,004/5,127), and the out-patient department (MDR: 12.5%; 663/5,319, non-MDR: 16.7%; 857/5,127). The number of bacteriologically positive cultures originating from the emergency department increased throughout the study period. There was an increasing trend in the proportion of MDR organisms in all bacteria-positive cultures in the emergency ward (p = 0.006, Cox and Stuart test), the pediatric department (p = 0.006, Cox and Stuart test), and the outpatient department (p = 0.006, Cox and Stuart test).
In this retrospective analysis of bacteraemia from a single hospital in Kathmandu, we report a 13.5% bacterial isolation rate from blood cultures, which is comparable with other bacteraemia studies from Nepal and elsewhere (5, 20–22). Similarly, the array of isolated organisms was similar to those reported in other studies investigating the etiology of bloodstream infections (2, 3, 22–25), and childhood septicaemia (5, 11, 25, 26). The most common Gram-negative, non-Salmonella organisms to be isolated from blood cultures within this population were Enterobacter spp., followed by Acinetobacter spp., E. coli and Klebsiella spp., a distribution that has been observed previously (3, 5, 23, 27). The dominance of Enterobacter spp. in this setting was unexpected, but there have been recent observations regarding the increasing presence of Enterobacter spp. bacteraemia (28). This escalation is cause for concern, as this bacterial genus presents particular challenges for the selection of optimal antimicrobial therapy due to the presence of chromosomally encoded AmpC beta-lactamases (25). Among the Gram-positive organisms (excluding Coagulase negative Staphylococci, which were reported for neonates only), S. aureus, Non-haemolytic Streptococci and S. pneumoniae were the most commonly isolated pathogens from blood cultures; this is also a common array of organisms associated with bacteraemia (5, 8, 24, 29). None of these bacteria were covered by any vaccine schedule in the country during the period of analysis.
Over the study period, there was a notable increase in bacteraemia cases originating from the emergency department, and a corresponding increase in the proportion of MDR organisms originating from patients in the emergency and outpatient departments, both of which receive patients directly from the community. This observation suggests that community-acquired bacteraemia caused by MDR pathogens is increasing and becoming a major cause of hospital admissions. These data also demonstrated a hospital-wide increase in the proportion of MDR Enterobacteriaceae, other Gram-negative organisms, and Gram-positive organisms isolated over the analysis period. In fact, from 2010, MDR isolates represented more than half of the non-Salmonella bacteria isolated from blood in this location. These data detail a worrying increase in MDR Enterobacter spp., Klebsiella spp., and E. coli, circulating both within the community and the hospital with the capacity to cause bloodstream infections. The increasing significance of Klebsiella pneumoniae, an organism that has already caused major outbreaks in this hospital, raises specific concerns given its ability to be a reservoir of AMR genes that can be transferred to other Enterobacteriaceae (30).
The precise reason(s) for the increase in the proportion of MDR organisms being isolated from patients in the community is unclear; we speculate that this increase is associated with an increasing population and ease of access to antimicrobials. Nepal is a landlocked country located between India and China and Kathmandu in the central valley is the capital city. Therefore, this location naturally acts as major nucleus within South Asia, with an elevated likelihood of antimicrobial resistant organisms being imported from neighboring countries. Furthermore, immigration into the capital city has increased dramatically over the last 25 years, both as a consequence of the civil war (1996–2006) and economic migration from rural areas. This increase in population has placed substantial strains on the healthcare system and the city infrastructure, making pathogen transmission more likely. Access to antimicrobials in the community without prescription and the increasing empirical use of latter generation broad spectrum antimicrobials in healthcare facilities has potentially created an increasing problem. This scenario has likely made Kathmandu a hub for the circulation of MDR organisms in the community and healthcare facilities.
The strength of this analysis is the aggregation of 23 years of data from the same healthcare facility. These longitudinal data provide a rare opportunity to observe and document changes in the epidemiology and the AMR trends of organisms associated with bacteraemia. With the exception of a recent 9-year study of bloodstream infections in Bangladesh (21), we are unaware of similar longitudinal data from other locations in South Asia with such large numbers of blood cultures and MDR time trends. However, some limitations need to be considered. Over the prolonged period, the consistency of microbiological testing (blood culture and antimicrobial susceptibility testing) needs to be ensured to allow for a meaningful comparison of trends. It is difficult, however, to wholly eliminate any changes in microbiological practices or hospital organization over time. For example, the sudden increase in the number of Coagulase negative Staphylococci recorded from 2003 was associated with the opening of the neonatal intensive care unit. A further limitation was the absence of demographic and clinical information in the data, thus preventing our ability to associate a specific group of organisms or change in AMR profile with patient demographics and/or clinical outcome. Similarly, treatment data and information regarding antimicrobial use prior to blood culture were not available. Lastly, as this analysis was conducted using routine hospital data, these data may not precisely reflect the epidemiology of bacteraemia in the community. However, it is important to note that the majority of positive blood cultures originated from individuals attending hospital directly from the community. Similarly, the results of this study are indicative of the situation in the Kathmandu Valley and may not reflect the situation in the rest of South Asia.
Our work illustrates worrying changes in epidemiology and AMR of bacteraemia within a healthcare facility in Kathmandu and highlights the increasing significance of non-Salmonella Enterobacteriaceae and Gram-positive bacteria over the last two and a half decades. This study documents the rise of AMR and MDR bloodstream pathogens in Nepal, demonstrating that MDR is not only a concern for hospital-acquired infections, but is rapidly becoming a significant problem for organisms circulating in the community. Our study will serve as a unique reference for improving clinical care, establishing treatment guidelines, and gaining a better understanding of the changing epidemiology of AMR pathogens.
BB, PoS, SD, CT, SB, and AK original study design. PoS, KP, SD, PKS, SK, and AK collected the data for the study. BB, TD, GT, and AK provided data interpretation. RZ, PoS, CB, CT, SB, and AK performed the analysis. RZ, SB, and AK drafted the manuscript. RZ, BB, PoS, KP, SD, PKS, SK, TD, CB, CT, GT, SB, and AK approved the final version of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We wish to acknowledge the support provided Patan hospital for allowing us to access and analyze these data. This project was funded by the Wellcome Trust of Great Britain (106158/Z/14/Z). SB is a Sir Henry Dale Fellow, jointly funded by the Wellcome Trust and the Royal Society (100087/Z/12/Z). AK is funded as leadership fellows through the Oak Foundation (OCAY-15-547). Data was digitized through funding by the Global Antimicrobial Resistance Partnership (GARP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2018.00262/full#supplementary-material
1. Prasad N, Murdoch DR, Reyburn H, Crump JA, Rangel J. Etiology of severe febrile illness in low- and middle-income countries: a systematic review. PLoS ONE (2015) 10:e0127962. doi: 10.1371/journal.pone.0127962
2. Luzzaro F, Ortisi G, Larosa M, Drago M, Brigante G, Gesu G. Prevalence and epidemiology of microbial pathogens causing bloodstream infections: results of the OASIS multicenter study. Diagn Microbiol Infect Dis. (2011) 69:363–9. doi: 10.1016/j.diagmicrobio.2010.10.016
3. Deen J, von Seidlein L, Andersen F, Elle N, White NJ, Lubell Y. Community-acquired bacterial bloodstream infections in developing countries in south and southeast Asia: a systematic review. Lancet Infect Dis. (2012) 12:480–7. doi: 10.1016/S1473-3099(12)70028-2
4. Wilson J, Elgohari S, Livermore DM, Cookson B, Johnson A, Lamagni T, et al. Trends among pathogens reported as causing bacteraemia in England, 2004-2008. Clin Microbiol Infect. (2011) 17:451–8. doi: 10.1111/j.1469-0691.2010.03262.x
5. Murdoch DR. Microbiological patterns in sepsis: what happened in the last 20 years? Int J Antimicrob Agents (2009) 34:S5–8. doi: 10.1016/S0924-8579(09)70557-6
6. Karchmer AW. Bloodstream infections: the problem and the challenge. Int J Antimicrob Agents (2009) 34:S2–4. doi: 10.1016/S0924-8579(09)70556-4
7. Nasa P, Juneja D, Singh O, Dang R, Singh A. An observational study on bloodstream extended-spectrum beta-lactamase infection in critical care unit: incidence, risk factors and its impact on outcome. Eur J Intern Med. (2012) 23:192–5. doi: 10.1016/j.ejim.2011.06.016
8. Rice LB. Antimicrobial resistance in gram-positive bacteria. Am J Med. (2006) 119:S11–9. doi: 10.1016/j.amjmed.2006.03.012
9. Allison MG, Heil EL, Hayes BD. Appropriate Antibiotic Therapy. Emerg Med Clin North Am. (2017) 35:25–42. doi: 10.1016/j.emc.2016.08.003
10. Lamba M, Sharma R, Sharma D, Choudhary M, Maheshwari RK. Bacteriological spectrum and antimicrobial susceptibility pattern of neonatal septicaemia in a tertiary care hospital of North India. J Matern Neonatal Med. (2016) 29:3993–8. doi: 10.3109/14767058.2016.1152251
11. Ansari S, Nepal HP, Gautam R, Shrestha S, Neopane P, Rimal B, et al. Childhood septicaemia in Nepal: documenting the bacterial etiology and its susceptibility to antibiotics. Int J Microbiol. (2014) 2014:452648. doi: 10.1155/2014/452648
12. Murdoch DR, Woods CW, Zimmerman MD, Dull PM, Belbase RH, Keenan AJ, et al. The etiology of febrile illness in adults presenting to Patan hospital in Kathmandu, Nepal. Am J Trop Med Hyg. (2004) 70:670–5. doi: 10.4269/ajtmh.2004.70.670
13. Maskey AP, Basnyat B, Thwaites GE, Campbell JI, Farrar JJ, Zimmerman MD. Emerging trends in enteric fever in Nepal: 9124 cases confirmed by blood culture 1993-2003. Trans R Soc Trop Med Hyg. (2008) 102:91–5. doi: 10.1016/j.trstmh.2007.10.003
14. Karkey A, Thompson CN, Tran Vu Thieu N, Dongol S, Le Thi Phuong T, Voong Vinh P, et al. Differential epidemiology of Salmonella Typhi and Paratyphi A in Kathmandu, Nepal: a matched case control investigation in a highly endemic enteric fever setting. PLoS Negl Trop Dis. (2013) 7:e2391. doi: 10.1371/journal.pntd.0002391
15. Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine (2015) 33(Suppl 3):C21–9. doi: 10.1016/j.vaccine.2015.03.102
16. Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
17. CLSI. Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement, CLSI document M100-S24. Clinical and Laboratory Standards Institute (2014).
18. Team R. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. (2012). Available onlne at: http://cran.r-project.org
19. Reddy EA, Shaw A V, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis. (2010) 10:417–32. doi: 10.1016/S1473-3099(10)70072-4
20. Mehl A, Åsvold BO, Lydersen S, Paulsen J, Solligård E, Damås JK, et al. Burden of bloodstream infection in an area of Mid-Norway 2002-2013: a prospective population-based observational study. BMC Infect Dis. (2017) 17:205. doi: 10.1186/s12879-017-2291-2
21. Ahmed D, Nahid MA, Sami AB, Halim F, Akter N, Sadique T, et al. Bacterial etiology of bloodstream infections and antimicrobial resistance in Dhaka, Bangladesh, 2005–2014. Antimicrob Resist Infect Control (2017) 6:2. doi: 10.1186/s13756-016-0162-z
22. Pandey S, Raza S, Bhatta CP. The aetiology of the bloodstream infections in the patients who presented to a tertiary care teaching hospital in kathmandu, Nepal. J Clin Diagn Res. (2013) 7:638–41. doi: 10.7860/JCDR/2013/4752.2871
23. Stryjewski ME, Boucher HW. Gram-negative bloodstream infections. Int J Antimicrob Agents (2009) 34:S21–5. doi: 10.1016/S0924-8579(09)70561-8
24. Nickerson EK, West TE, Day NP, Peacock SJ, Xu YC, Chen MJ. Staphylococcus aureus disease and drug resistance in resource-limited countries in south and east Asia. Lancet Infect Dis. (2009) 9:130–5. doi: 10.1016/S1473-3099(09)70022-2
25. Harris PNA, Peri AM, Pelecanos AM, Hughes CM, Paterson DL, Ferguson JK. Risk factors for relapse or persistence of bacteraemia caused by Enterobacter spp.: a case-control study. Antimicrob Resist Infect Control (2017) 6:14. doi: 10.1186/s13756-017-0177-0
26. Kebede HK, Gesesew HA, Woldehaimanot TE, Goro KK, Namusisi O, Mukanga D. Antimicrobial use in pediatric patients in a teaching hospital in Ethiopia. PLoS ONE (2017) 12:e0173290. doi: 10.1371/journal.pone.0173290
27. van der Mee-Marquet NL, Blanc DS, Gbaguidi-Haore H, Dos Santos Borges S, Viboud Q, Bertrand X, et al. Marked increase in incidence for bloodstream infections due to Escherichia coli, a side effect of previous antibiotic therapy in the elderly. Front Microbiol. (2015) 6:646. doi: 10.3389/fmicb.2015.00646
28. Al-Hasan MN, Lahr BD, Eckel-Passow JE, Baddour LM. Temporal trends in Enterobacter species bloodstream infection: a population-based study from 1998–2007. Clin Microbiol Infect. (2011) 17:539–45. doi: 10.1111/j.1469-0691.2010.03277.x
29. Rasmussen RV, Fowler VG Jr, Skov R, Bruun NE. Future challenges and treatment of Staphylococcus aureus bacteremia with emphasis on MRSA. Future Microbiol. (2011) 6:43–56. doi: 10.2217/fmb.10.155
Keywords: bloodstream infections, bacteraemia, blood culture, surveillance, community acquired, antimicrobial resistance, enterobacteriaceae, Nepal
Citation: Zellweger RM, Basnyat B, Shrestha P, Prajapati KG, Dongol S, Sharma PK, Koirala S, Darton TC, Boinett C, Thompson CN, Thwaites GE, Baker S and Karkey A (2018) Changing Antimicrobial Resistance Trends in Kathmandu, Nepal: A 23-Year Retrospective Analysis of Bacteraemia. Front. Med. 5:262. doi: 10.3389/fmed.2018.00262
Received: 02 May 2018; Accepted: 30 August 2018;
Published: 19 September 2018.
Edited by:
Alessandro Cassini, European Centre for Disease Prevention and Control, SwedenReviewed by:
Sergey Eremin, World Health Organization, SwitzerlandCopyright © 2018 Zellweger, Basnyat, Shrestha, Prajapati, Dongol, Sharma, Koirala, Darton, Boinett, Thompson, Thwaites, Baker and Karkey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stephen Baker, c2Jha2VyQG91Y3J1Lm9yZw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.