- 1Institute of Psychiatry and Neuroscience, Xinxiang Medical University, XinXiang, China
- 2Priority Area Asthma and Allergy, Research Center Borstel, Airway Research Center North, Members of the German Center for Lung Research, Borstel, Germany
Rheumatoid diseases are a group of systemic autoimmune diseases which affect multiple organs with largely unknown etiology. In the past decade, long non-coding RNAs (lncRNAs) have emerged as important regulators of biological processes and contribute deeply to immune cell development and immune responses. Substantial evidences have been accumulated showing that LncRNAs involved in the pathogenesis of the rheumatoid diseases, including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE) and Sjögren's syndrome (SS). In this review, we summarize literature combined with bioinformatics methods to analyze the unique and common lncRNAs patterns in rheumatoid diseases and try to reveal the important function of lncRNAs in RA, SLE and SS.
Introduction
Autoimmune diseases are disorders caused by the break of the immune tolerance to autoantigens which is marked by emerging of autoreactive T cells and autoantibodies. Autoimmune diseases can be categorized into two groups based on organ-specificity. One group is organ-specific autoimmune disease that affects particular organs, and the other group is systemic autoimmune diseases that affect multiple organs (1). Rheumatoid diseases are a group of systemic autoimmune diseases, including rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), Sjogren's syndrome (SS), polymyositis and dermatomyositis etc. (1). Rheumatoid diseases often share some common clinical and immunological features. For example, SS is closely related to other two rheumatoid diseases, RA and SLE (2). SS develop alone as primary SS (pSS), while it is referred to as secondary Sjogren's syndrome(2ndSS) when it occurs with other autoimmune diseases. The 2ndSS always coexist with SLE and RA, wherein 9–19% SLE patients (3, 4) and 4–31% RA patients (5, 6) develop the 2ndSS. In terms of immunological features, RA, SLE, and SS are all characterized with anti-nuclear antibodies (ANA) and rheumatoid factor (RF) (7). In addition, anti-Ro/ SSA and anti-La/SSB autoantibodies which are diagnostic markers for SS also can be detected in both SLE and RA, although with relatively low prevalence (8). Besides clinical and immunological features, the three rheumatoid diseases also share some common features at the molecule level. Analyses of gene expression profiles of peripheral blood mononuclear cell (PBMC) demonstrate that RA, SLE and SS share some common gene expression profiles and biological processes (9, 10). Finally, the three rheumatoid diseases also share some susceptibility genes, such as PTPN22 (11), STAT4 (12) and IRF5 (13–15). Taken together, the common features among RA, SLE, and SS in clinical, immunological and molecule levels suggest that they share some causes in their pathogenesis.
Long non-coding RNAs (lncRNAs) are molecules longer than 200 nucleotides in length (16). LncRNAs are RNAs with little or no protein-coding capacity (17). Since the first lncRNA discovered in the early 1990s (18), around 172,216 transcripts have been annotated in the human according to the NONCODE database (http://www.noncode.org/analysis.php) (17). As small RNA sponging and molecular scaffolds, lncRNAs play an important role in regulation of gene expression through multiple mechanisms (19–22). lncRNAs are actively involved in the regulation of many biological processes, including development and function of immune cells. Recently, a body of evidence has been accumulated showing that lncRNAs are involved in the pathogenesis of rheumatoid diseases, such as SS, SLE and RA (23–25).
In this review, we aim to provide some scientific insights into the role of lncRNAs in rheumatoid diseases. We summarize the recent progresses and discuss the role of LncRNAs in the pathogenesis of RA, SLE, and SS, and highlight those lncRNAs associated with multiple rheumatoid diseases. In addition, by comparing rheumatoid disease-associated proteins retrieved from Coremine Medical (http://www.coremine.com/medical/#search) with proteins regulated by lncRNA associated with rheumatoid diseases, we try to explore the mechanism behind the role of lncRNAs in RA, SLE, and SS.
Long Non-Coding RNAs Function and Mechanisms
According to the criterion defined by GENCODE (26), lncRNAs are subclassified into antisense lncRNA, sense lncRNA, bidirectional lncRNA, and long intergenic noncoding RNAs (lincRNA) (27). Since the discovery of the first lncRNA (18), our knowledge about lncNRAs has been considerably increased. LncRNAs interact with RNA or DNA through complementary base-paring, and interact with protein via direct interaction (28, 29). Interactions between lncRNA with RNA, DNA, and proteins enable lncRNAs to regulate gene expression at multiple levels, including transcription, posttranscription, translation, post-translation, and epigenetic modification.
Transcriptional Regulation
LncRNAs can mediate both cis- and trans-regulation of transcription. Acting as enhancer associated RNAs (e-RNAs) and affect neighboring intra-chromosomal genes in cis-regulation manner (30). E-RNAs are transcribed from enhancer regions harboring specific chromatin states such as H3K4me1 and H3K27ac (31). E-RNAs can regulate neighboring genes expression through recruiting transcription factors or complexes and guiding them to the specific region of target genes (32). Also, lncRNAs can interact with the transcription factors to form the preinitiation complex and thus either activate or inhibit gene transcription (33, 34). Another group of lncRNAs called ncRNA-activating are able to recruit the transcriptional co-activator mediator to promote chromatin looping of neighboring genes, resulting in the transcriptional activation of those genes (35). Finally, the process of transcription of lncRNA is also capable to affect adjacent genes transcription (36).
Apart from cis-regulation, lncRNAs also regulate genes expression in trans-acting manner. For example, lncRNA 7SK negatively regulates transcription elongation factor P-TEFb complex by interacting with high mobility group protein A1 (HMGA1) (37, 38) and lncRNA B2 acts as brakes which hold back the advancement of RNA polymerase II (39, 40).
Post-Transcriptional Regulation
LncRNAs play a role in post-transcriptional regulation by influence the pre-mRNA procession and mRNA stability (36). Alternative splicing of pre-mRNA is an important event in the eukaryotes gene transcription, which provides diverse transcripts. For example, interaction between nuclear enriched abundant transcript 1 (NEAT1) and serine/arginine-rich splicing factor 5 (SRp40) influences the function of splicing factors that act on splicing of the PPARγ2 gene during adipogenesis (41). Metastasis associated lung adenocarcinoma transcript 1 (MALAT1, also known as NEAT2) is another lncRNA contributing to the pre-mRNA splicing. Previous studies suggest that the interaction between MALAT1 and serine/arginine splicing factors act a pivotal part in cancer progression (42), synapse formation (43), and growth signal responsive genes expression (44).
LncRNAs also regulate mRNA stability via multiple ways. First, lncRNAs can act as a sponge to absorb microRNA and thus regulate the stability of target mRNA (45). For example, Linc-MD1 affects skeletal muscle differentiation and disease though sponging miR-133 and miR135 (46). In addition, lncRNAs are also able to regulate mRNA stability by interacting with the 3′UTR or the coding region of target mRNAs. A good example is half-STAU1-binding site RNAs (1/2-sbsRNAs), a group of lncRNAs which form imperfect base-pairs with an Alu element in the 3′ UTR of a target mRNA and another Alu element in a 1/2-sbsRNAs, and this formation is required for the STAU1-mediated messenger RNA decay (45). Another example is lncRNA maternally expression gene 3 (MEG3) which recruit polypyrimidine tract-binding protein 1 (PTBP1) to bind to the coding region of small heterodimer partner (SHP) mRNA and subsequently decay the mRNA (47). Interestingly, in a feedback-regulatory fashion, SHP inhibits cAMP response element-binding protein (CREB) mediated expression of MEG3 (47), suggesting a crosstalk between lncRNA and mRNA.
Translational Regulation
By interacting with the apparatus of translation, lncRNAs enhance or suppress gene expression at the translational level. For example, Ubiquitin carboxy-terminal hydrolase L1 antisense RNA 1 (UCHL1-AS1) interacts with polysomes and thus promotes the translation of UCHL1 mRNA (48). And non-coding RNA activated by DNA damage (NORAD) maintains genomic stability by sequestering PUMILIO protein, which repress the stability and translation of the target mRNA (49). By interacting with translation initiation factor 4E (eIF4E), growth arrest-specific transcript 5 (GAS5) inhibit the translation of c-Myc mRNA (50). Finally, LncMyod, a lncRNA encoded next to the Myod gene directly binds to IGF2-mRNA-binding protein 2 (IMP2) and negatively regulates IMP2-mediated gene translation such as c-Myc (51).
Post-Translational Regulation
Post-translational modification of protein refers to the process of adding or removing chemical components on protein, as well as the process of protein folding and degradation (52). Phosphorylation and acetylation of a protein usually is associated with its activation status, while ubiquitination of a protein means protein degradation. LncRNAs are involved in the regulation of post-translational modification of proteins at multiple levels. For example, lncPRESS1, a p53-regulated transcript, interacts physically with SIRT6 and prevents SIRT6-mediated histone H3K56 and H3K9 deacetylation, therefore maintaining the pluripotency of human embryonic stem cells (53). Another example is lncRNAs associated with breast cancer brain metastases (lnc-BM), which binds to JH2 domain of JAK2 protein thus mediate oncostatin-M and signal transducer and activator of transcription 3 (STAT3) phosphorylation by increasing JAK2 kinase activity (54). Furthermore, lnc-DC, a lncRNA exclusively expressed in human conventional DCs, regulates the phosphorylation of STAT3 by direct binding to STAT3 and prevent its dephosphorylation by SHP1 (55).
Besides function in abovementioned transcriptional, post-transcriptional, translational and post-translation levels, lncRNAs regulate gene expression via some other mechanisms. LncRNAs can affect epigenetic regulation by altering the DNA methylation (56), histone modifications (57) and genetic imprinting (58). In addition, some lncRNAs containing short open reading frames are translated into functional micro-peptides which might carry their function in the regulation of gene expression (59). Taken together, lncRNAs regulates biological processes interacts by interacting with other molecules such as miRNA, DNA, and protein, which is an important mechanism for maintaining life function.
Long Non-Coding RNA in Rheumatoid Disease
Given the important role of lncRNAs in regulating gene expression, it is conceivable to speculate that lncRNAs participates in various physiological and pathological processes of rheumatoid diseases. Using a variety of methods, such as microarray, quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) and whole transcriptome sequencing, the researchers demonstrated that many lncRNAs are differentially expressed in rheumatoid patients and healthy controls, and this differential expression is associated with disease characteristics. These results suggest that lncRNAs play a role in developing of Rheumatoid disease (60). Therefore, exploration of the function of lncRNAs will help us clarify the mechanism of rheumatoid disease and provide new diagnostic markers and therapeutic targets.
LncRNAs in Rheumatoid Arthritis
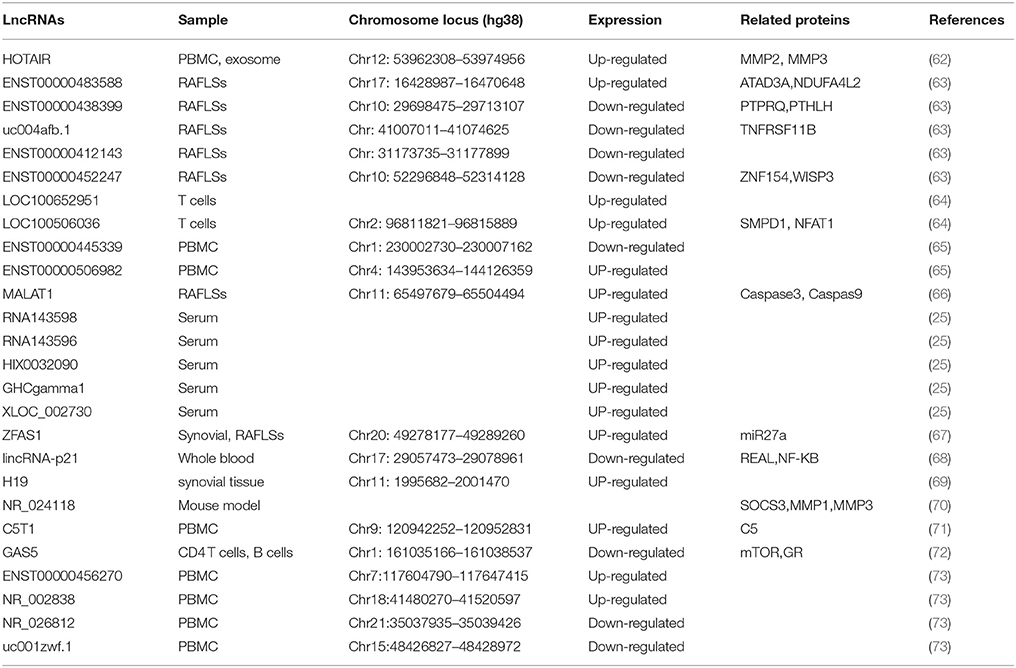
The pathogenesis of RA is complex and involves the interaction of host factors (e.g., genetic susceptibility and sex hormones) with environmental players (e.g., bacterial or viral infection) (61). So far, 27 lncRNAs have been implicated to play a role in RA (Table 1).
The first lncRNA implicated in RA is H19. In 2003, Stuhlmueller et al. reported that H19 expression was significantly higher in the synovial tissues (ST) from patients with RA and osteoarthritis (OA) than that in normal control or individual with joint trauma (69). In the cultured synovial fibroblasts (SFBs), the H19 expression is low but can be strongly induced by serum starvation. Interestingly, this starvation-induced increase of H19 expression in RA-SFBs is significantly higher than in OA-SFBs and control SFBs (69). Significant over expression of H19 RNA and its increased sensitivity to starvation/cytokine regulation in RA implicate a role of H19 in the pathogenesis of RA.
Since upregulation of ZFAS1 has been observed in cancers and promotes cell migration and invasion (74–76), Ye et al. investigated the role of this lncRNA in RA. They found that ZFAS1 expression level was increased in synovial tissue from patients with RA compared with that in controls (67). Their study further proved that lncRNA ZFAS1 promoted the migration and invasion of RA FLS is a miR-27a-dependent manner (67).
Another lncRNA associated in RA is growth arrest-specific 5 (GAS5) which acts as a potent repressor of the glucocorticoid receptor (GR) through its RNA “glucocorticoid response element (GRE)” (77). In 2016, Mayama and colleagues examined GAS5 levels in multiple immune related diseases (72). They found the expression level of GAS5 was significantly reduced in the CD4+ T cells and B cells from patients with RA compared with that in controls, Mayama et al. (72).
LncRNA C5T1 is located in a genomic region comprising a RA risk locus TRAF1-C5 (71). C5T1 expression is positively correlated with C5 mRNA, and knockdown of C5T1 specifically decreases the level of C5 mRNA, suggesting that C5T1 positively regulates C5 expression (71). Since C5 is overexpressed in inflamed joint of patients with RA (78) and C5-deficient mice are resistant to the development of RA model (79), it is conceivable that C5T1, as the C5 gene regulator, is associated with RA.
T cells play important roles in the pathogenesis of RA (80). To investigate the role of lncRNAs in RA patients, Lu et al. determine 10 potentials aberrantly expressed lncRNAs in T cells from 39 RA patients and 17 health cohort by RT-PCR (64). Among the 10 candidate lncRNAs, LOC100652951 and LOC100506036 are up-regulated. Interestingly, treated with tumor necrosis factor (TNF) antagonists could decrease LOC100652951 expression level. In addition, knockdown of LOC100506036 by siRNA could inhibit the production of the interferon gamma and decrease the expression level of nuclear factor of activated T cells (NFAT) and sphingomyelin phosphodiesterase 1 (SMPD1) (80). Their results indicate that LOC100652951 may be involved in the production of cytokines and LOC100506036 may contribute to the inflammatory response in RA.
Compared with detecting candidate lncRNAs, determination of the expression profile of lncRNAs by microarray, and subsequent validation with quantitative RT-PCR is a high throughput strategy for identification of disease-related lncRNAs. In 2015, Song et al. analyzed the expression profile of 83 lncRNAs in PBMC and blood exosome from 28 RA patients and 10 controls (62). Compared with healthy controls, the expression level of HOTAIR is increased in both PBMC and blood exosome of RA patients. Functional study shows that HOTAIR contributes to the migration of activated macrophage as well as the MMP2 and MMP13 activation (62). This indicates that HOTAIR might involve in the inflammation and the dissolution of bone and cartilage matrix in RA (62).
Using a biochip capable of detecting 40,173 lncRNAs, Yuan et al. analyzed the expression profiles of lncRNA in three pairs of samples (73). They identified 2,099 lncRNAs which were differentially expressed in PBMC between RA and controls. With PBMC from 36 RA patients and 24 healthy controls, the authors verified four significantly differentially expressed lncRNAs. ENST00000456270 and NR_002838 were up-regulated, whereas NR_026812 and uc001zwf.1 were down-regulated in patients with RA as compared to controls (73). Furthermore, the expression level of ENST00000456270 strongly correlates with the level of IL-6 and TNF-α in serum and the simplified Disease Activity Index (SDAI) of the RA patients (73).
Another study determining the expression profile of lncRNA in PMBC from RA patient was performed by a Chinese research group. Through a microarray capable of detecting 30,586 human lncRNA, Luo and colleagues determined the expression profile of lncRNAs in PBMC from 10 patients with RA and 10 well-matched health controls (65). The results revealed that 139 lncRNAs were significantly differentially expressed between RA patients and controls. The authors further validated the two most significantly deregulated lncRNAs, ENST00000445339 and ENST00000506982 using quantitative RT-PCR in the 24 patients with RA and 24 controls.
Fibroblast-like synovial cells (FLSs), one of the key effector cells in RA synovium, have attracted increasing attention (81). To explore lncRNAs expression pattern in the FLSs of RA, Zhang et al. determined the of lncRNAs profiles in FLSs of 10 RA patients and 10 patients with trauma as controls (63). Among the 30,586 lncRNAs detected by the microarray, 135 were differentially expressed in FLS between RA group and controls. By quantitative RT-PCR, the authors confirmed four differentially expressed lncRNAs, including ENST00000483588 which was up-regulated in RA FLSs as well as ENST00000438399, uc004afb.1, and ENST00000452247 which were down-regulated in RA FLSs as compared with controls. Notably, the expression level of ENST00000483588 is positively correlated with C-reactive protein (CRP) level and the simplified Disease Activity Index (SDAI) score (r = 0.79, P < 0.01). In addition, four lncRNAs show good diagnostic value for RA with the area under ORC curve ranging from 0.85 to 0.97 (63).
Besides cells, serum samples also be used for identification of RA-related lncRNAs. To identify lncRNAs associated with RA, Xu et al. investigated the expression profile of lncRNAs in serum samples from 3 RA patients and 3 health controls by microarray and then verified the interested lncRNAs in 43 RA patients and 40 healthy controls by quantitative RT-PCR (25). This study has identified 5 significantly up-regulated lncRNAs in RA as compared with controls, including RNA143598, RNA143596, HIX0032090, IGHCgamma1 and XLOC_002730. Moreover, these lncRNAs are positively correlated with some immunological and clinical features of RA, including rheumatoid factor (RF), erythrocyte sedimentation rate (ESR), anti-cyclic citrullinated peptide (anti-CCP) antibody and disease course (25).
Besides lncRNAs which are differentially expressed and functionally related with disease pathogenesis, some other lncRNAs have been implicated in RA because they are associated with therapeutic efficacy in the disease. Quercetin, a free oxygen radical scavenger (82), is effective in the management of RA (83). In vitro, Quercetin decreases the viability and promotes the apoptosis of FLS from patients with RA (84). Interestingly, treated with quercetin induces the expression of lncRNAs MALAT1 and knockdown of MALAT1 decreases the expression of caspase-3 and caspase-9 thus inhibits the FLS apoptosis induced by quercetin. Therefore, this study suggests that MALAT1 is involved in the quercetin-induced apoptosis of FLS and thus MALAT1 maybe have a therapeutic efficacy in RA. Zicao (purple gromwell) is a traditional Chinese herbal medicine, its major active component is Shikonin (66). Shikonin possesses anti-inflammatory property and can effective reduce the incidence and severity of RA in a collagen-induced arthritis mouse model. These results suggest that Shikonin is a good candidate of RA protective medicine (85). In the CIA model, Shikonin treatment increases the expression of lncRNAs NR_024118 in the joint of diseased mice through increasing the acetylation of H3 in the promoter of the lncRNAs (70). The knockdown of NR_024118 could reverse the effects of shikonin on proinflammatory cytokines and MMPs, suggesting that shikonin exerts it therapeutic effect in mouse model of RA via lncRNA-NR024118. Another lncRNA implicated in the therapeutic efficacy in RA is LincRNA-p21, one of lncRNAs induced in p53-mediated DNA damage response (68). Methotrexate (MTX) is an anchor therapy for the management of RA. In RA patients, MTX treatment increases the expression of lincRNA-p21 and decreases the level of p65 (RelA) phosphorylation compared with untreated RA patients (86), Interestingly, the basal levels of lincRNA-p21RA is reduced, while basal levels of RelA increased in RA patients (86). In addition, T cells from RA patients have deficiencies in DNA damage response as indicated by upregulation of DNA-PKcs (87). This suggests that MTX treatment might act through upregulating the lincRNA-p21 expression and thus prevent DNA damage and apoptosis in T cells. This note is supported by in vitro finding that MTX treatment induces the expression level of lincRNA-p21 through a DNA-PKcs-dependent mechanism in primary T cells or Jurkat cells (86).
LncRNAs in Systemic Lupus Erythematosus
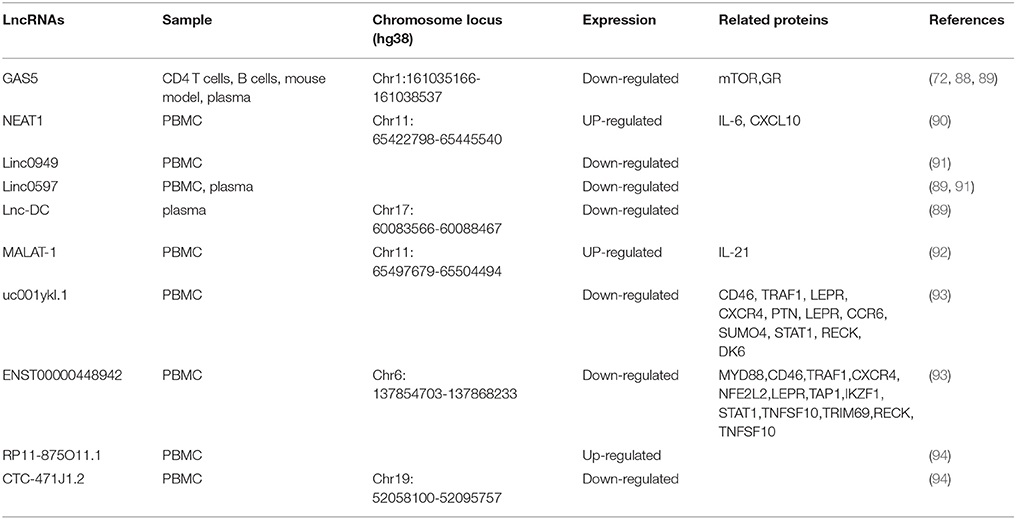
SLE Patient usually presents highly heterogeneous in pathogenesis and disease features, which makes it difficult to understand the etiology of SLE. In recent years, emerging evidence has demonstrated that lncRNAs are involved in the pathogenesis of SLE, which brings new insights into SLE research (Table 2).
As in RA, the expression level of GAS5 is also decreased in the patients with SLE compared to control (72), indicating that GAS5 is associated with SLE. The association between GAS5 and SLE has been further supported by some other evidence. In BXSB mice, a spontaneous mouse model for SLE, GAS5 gene is located within a disease-susceptibility interval and carries multiple polymorphism which may account for the disease-related gene expression profile, suggesting that it is a candidate susceptibility gene in this SLE model (88). Furthermore, a recent study performed by Wu et al. demonstrated that the plasma level of GAS5 in SLE patients is significantly lower than that in healthy controls (89). Besides GAS5, Wu et al. also identified two other SLE-associated lncRNAs, lnc-DC and linc0597. Lin-DC, an intergenic lncRNAs, is exclusively expressed in human conventional DCs (55). Lnc-DC bound directly to STAT3 in the cytoplasm and promote the activation of STAT3. Knockdown of lnc-DC impaired DC differentiation in vitro and in vivo and reduced capacity of DCs to stimulate T cell activation (55). Compared with controls, the expression level of lnc-DC is significantly decreased in the plasma from patients with SLE (89). Furthermore, compared with SLE without nephritis, the lnc-DC expression level is significantly increased in lupus nephritis, making lnc-DC a promising marker distinguishing the two subgroups of SLE. In contrast to GAS5 and lnc-DC, the expression of linc0597 is significantly increased in plasma of SLE patients as compared with controls (89).
The association between linc0597 and SLE is confirmed by another study in which Wu et al. examined the expression levels of 4 immune-related lncRNAs in PBMC from 102 SLE patients and 76 healthy controls (91). They found that the expression levels of linc0949 and linc0597 were significantly decreased in SLE patients compared with those in controls. Furthermore, correlation analysis has demonstrated that linc0949 is negatively correlated with disease activity and positively correlated with complement component C3. In addition, the level of linc0949 is negatively associated with lupus nephritis and cumulative organ damage (91).
LncRNAs MALAT1 is associated with RA (84) and involved in the development and metastasis of cancer (95). To explore the role of MALAT1 in the pathogenesis of SLE, Yang et al. analyzed the expression of MALAT1 in PBMC from 39 SLE patients and 45 matched normal controls (92). They found that MALAT1 was abnormally increased in the patients with SLE and predominantly expressed in monocytes. In monocytes of patients with SLE, silencing MALAT1 significantly reduced the expression of IL-21 (92), an important cytokine in the pathogenesis of SLE. Furthermore, this study has also demonstrated that MALAT-1 exerts its detrimental effects by regulating silent information regulator 1 (SIRT1) signaling. Nuclear enriched abundant transcript 1 (NEAT1), a lncRNA often colocalized with MALAT1, has also been implicated in SLE. In 2016, Zhang et al. detected the level of NEAT1 in the PBMC from 39 SLE patients and 50 normal controls (90). They found that the NEAT1 level was significantly increased in SLE patients and the expression level of NEAT1 was positively correlated with disease activity of SLE (90). In both human monocyte cell line and primary monocytes, LPS or pam3cks4 stimulation could increase the expression of NEAT1. In addition, silencing of NEAT1 significantly reduced the expression of a group of chemokines and cytokines such as IL-6, CXCL10, etc., mainly through affection the late MAPK pathways, especially the phosphorylation of JNK and ERK.
High-throughput method such as microarray also apply to identify disease associated lncRNAs in SLE. In 2017, Li et al. analyzed the expression profiles of lncRNAs in T cells from SLE patients and healthy controls (93). Using quantitative RT-PCR, the authors verified that two lncRNAs uc001yk1.1 and ENST00000448942 were significantly downregulated in SLE patients with compared to controls. Moreover, the expression level of ENST00000448942 is correlated with anti-Sm antibodies and ESR (erythrocyte sedimentation rate), whereas the expression level of uc0011yk1.1 is correlated with ESR and C-reactive protein (93). Another study determining expression profile of lncRNAs was performed by Luo et al. through PBMC from SLE patients and controls (94). The results indicated that 8,868 lncRNAs (3,657 upregulated and 5,211 downregulated) were differentially expressed in SLE samples compared with the control group (94). By quantitative RT-PCR, the authors verified the upregulation of RP11-875O11.1 and the down-regulation of CTC-471J1.2 (94).
LncRNAs in Sjögren's Syndrome
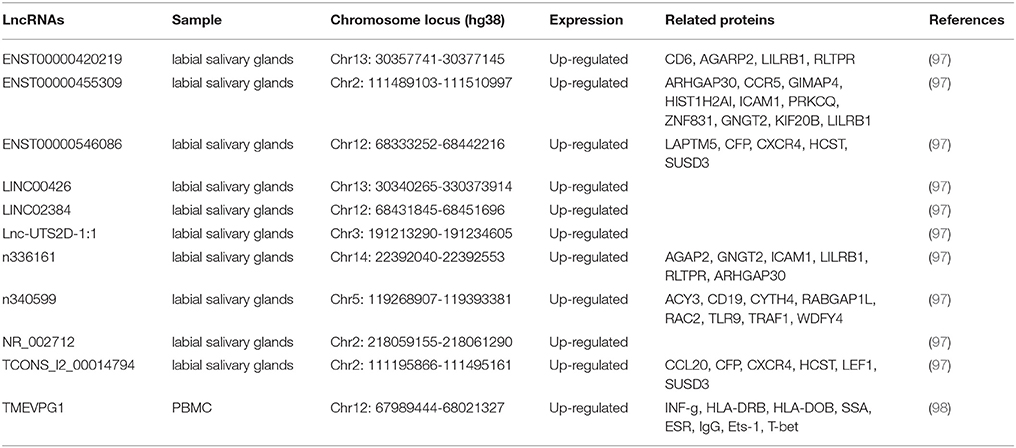
SS is a systemic autoimmune disease featured by dysfunction of exocrine glands (96). So far, 11 lncRNAs have been suggested to be associated with SS (Table 3).
LncRNA TMEVPG1 [also named Ifng-AS1 and Nettoie Salmonella pas Theiler's (NeST)] is located on the DNA stands opposite to interferon gamma (IFNG) coding gene and it can promote the transcription of IFNG as an enhancer (99). TMEVPG1 is expressed predominately in T cells (CD4 and CD8) and NK cells (100). In 2016, Wang and colleagues reported that the expression level of TMEVPG1 is increased in CD4+ T cells in SS patients compared with that in controls (98). Moreover, the expression of TMEVPG1 is positively correlated with levels of anti-SSA antibody, erythrocyte sedimentation rate (ESR), total IgG amount and the proportion of Th1 cells. In addition, in vitro study showed that the knockdown of TMEVPG1 could decrease the proportion of Th1 cells in CD4+ T cells from patients (98).
To identify whether the dysfunction of lncRNAs is involved in the pathogenesis of pSS, Shi et al. analyzed the expression profile of lncRNAs in labial salivary glands of patients with pSS and controls by microarray (97). Using a chip with a total capacity of 63,431 lncRNAs, They found 1243 lncRNAs were differentially expressed in pSS patients compared with controls, including 890 upregulated and 353 downregulated lncRNAs. By quantitative RT-PCR, the authors validated eight upregulated lncRNAs, named ENST00000420219.1, ENST00000455309.1, n336161, NR_002712, ENST00000546086.1, Lnc-UTS2D-1:1, n340599 and TCONS_l2_00014794 (97). The authors further analyzed the correlation between those eight lncRNAs and the clinical characteristics of pSS. The results revealed strong correlations between these lncRNAs and pSS characteristics, including β2 microglobulin, erythrocyte sedimentation rate (ESR), rheumatoid factor (RF), amount of IgA and IgM, visual analog scale (VAS) of parotid swelling and VAS of dry eyes (97).
A Role of LncRNAs in RA, SLE, and SS
As mentioned in the introduction, RA, SLE, and SS share many common clinical features at immunological and molecular levels (60). However, the molecular mechanisms behind the common features among rheumatoid diseases are not well understood. Association between lncRNAs and rheumatoid diseases might shed some new light on our understanding the pathogenesis of rheumatoid diseases.
Among the tens of lncRNAs involved in RA, SLE and SS, those associated with multiple diseases are special interesting since lncRNAs involved in the pathogenesis of multiple diseases would provide a hint for interpreting common features of rheumatoid diseases. Although lncRNAs implicated in RA, SLE, or SS, none of them is shared in all three rheumatoid diseases. However, one lncRNA, TMEVPG1, has been implicated in SLE and SS, and two lncRNAs, MALAT1 and GAS5, have been implicated in RA and SLE (Figure 1A).
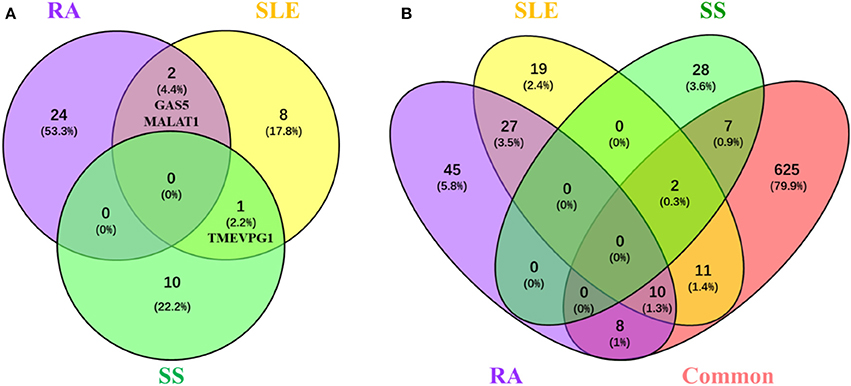
Figure 1. LncRNAs and their related proteins inplicated in RA, SLE, and pSS. (A) Venn diagram of the lncRNA implicated in RA, SLE, and pSS. Lists of lncRNAs that implicated in RA, SLE, and SS, respectively, were submitted to Venny webset (http://bioinfogp.cnb.csic.es/tools/venny/index.html) to analyze lncRNAs shared by multiple diseases. The number and percentage of lncRNAs in each fraction are depicted in the corresponding fraction. LncRNAs implicated in at least two diseases are depicted. RA, Rheumatoid arthritis; SLE, systemic lupus erythematosus; SS, Sjogren syndrome. (B) Venn diagram of the proteins interacted with lncRNAs that are implicated in RA, SLE, and SS. Using lncRInter websver (http://bioinfo.life.hust.edu.cn/lncRInter/), we obtained the proteins related with lncRNAs. For those lncRNAs, did not record in the lncRInter, we got its related proteins by searching literature manually. We also obtain 663 proteins which common to pathological basic of RA, SLE, and pSS by searching coremine database (http://coremine.com/medical/#search). The list of proteins interacted with lncRNAs that are implicated in RA, SLE, and pSS, respectively, and the 663 common proteins were submitted to Venny webset (http://bioinfogp.cnb.csic.es/tools/venny/index. html) to analyze lncRNA-related proteins shared by multiple diseases. The number and percentages of proteins in each fraction are depicted in the corresponding fraction. LncRNA-related proteins associated with at least two diseases are depicted. RA, Rheumatoid arthritis; SLE, systemic lupus erythematosus; SS, Sjogren syndrome.
It is conceivable that lncRNAs affect the development of rheumatoid disease via regulating the expression disease related proteins. To explore the possible mechanisms of lncRNA and related proteins in rheumatoid diseases, we collected 663 proteins have been suggested involved in the common pathological progress of RA, SLE and SS by either retrieving them from the coremine database (http://coremine.com/medical/#search) or manually from literature. Notably, 18 out of 90 proteins related to lncRNAs implicated in RA, 23 out of 69 proteins related to lncRNAs implicated in SLE and 9 out of 37 proteins related to lncRNAs implicated in SS (Figure 1B).
We speculate that lncRNAs might contribute to the pathogenesis of rheumatoid diseases through regulating those proteins. Thus, we generated an interaction network using MALAT1, NEAT1, TEMVP1, GAS5, and lnc-DC with their related proteins to demonstrate that lncRNAs might contribute to the pathogenesis of rheumatoid diseases at multiple steps which interact with each other (Figure 2).
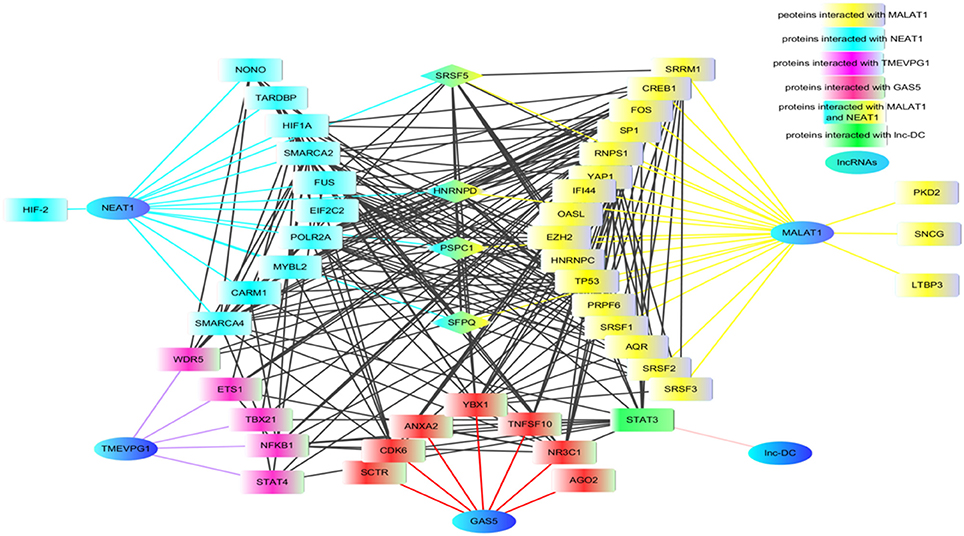
Figure 2. The proteins network that interacted with lncRNAs. Using lncRInter websever (http://bioinfo.life.hust.edu.cn/lncRInter/), we obtained the proteins related with lncRNAs. For those lncRNAs, did not record in the lncRInter, we got its related proteins by searching literature manually. A list of proteins interacted with lncRNAs that are implicated in RA, SLE, and pSS, respectively, were submitted to STRING website to analyze the proteins shared by multiple lncRNAs. The results from STRING were further modified by software cytoscape (version 3.6). In the network, the proteins SRSF5, HNENPD, PSPC1 and SFPQ shared by lncRNAs MALAT1 and NEAT1. The protein NONO, SFPQ, and PSPC1 are components of paraspeckle which was the key element of HDP-RNP. The color blue indicates lncRNAs, the color yellow indicates proteins related with MALAT1, the color wathet blue indicated proteins interacted with NEAT1, the color wathet blue and yellow indicates the proteins shared by MALAT1 and NEAT1, the color red indicated proteins interacted with GAS5, the color purple indicated proteins related with TMEVP1, the color green indicated proteins interacted with lnc.,-DC. SRSF5, serine and arginine rich splicing factor 5; HNENPD, heterogeneous nuclear ribonucleoprotein D; PSPC1, paraspeckle component 1; SFPQ, splicing factor proline and glutamine rich; NONO, non-POU domain-containing octamer-binding protein; HDP-RNP, HEXIM1-DNA-PK-paraspeckle components-ribonucleoprotein complex.
Based on the function and interaction of lncRNA-related proteins, here we propose a hypothetical model for the role of lncRNAs in pathogenesis of RA, SLE and SS, with a focus in TMEVPG1, MALAT1, GAS5, NEAT1, Lnc-DC, and C5T1 (Figure 3). TMEVPG1 was first identified in mice in a susceptibility locus to Theiler's virus (99). The transcription of TMEVPG1 is regulated by transcription factors STAT4 and T-bet and TMEVPG1 acting cooperator with these transcription factors positively regulates the IFN-γ expression (101). Compared with matched controls, TMEVPG1 is upregulated in the PBMC of both SS (98) and SLE (93) patients. IFNs, particularly type I IFNs, signature is a molecular feature for both in SS and SLE, which suggest that IFNs play an essential role in the two diseases (102). With regards to the IFN-γ regulated by TMEVPG1, there is an increasing body of evidence that IFN-γ plays an important role in developing of SLE and SS (103). On one hand, IFN-γ is the feature cytokine of the Th1 cells which has been demonstrated to play a role in the SLE and SS (104). On the other hand, IFN-γ could induce some chemokines, such as IFN-γ inducible 10kd protein (IP-10 or CXCL10) and CXCL9, which have important functions in attracting T cells to the target tissues in SS (105) and SLE (106). Therefore, TMEVPG1 might contribute to the development of SLE and SS through affecting Th1 cells and chemokines. In addition, since both SS (107) and SLE (108) are characterized by IFN gene signature, TMEVPG1 might also contribute to the diseases via acting on IFN-related signaling pathways.
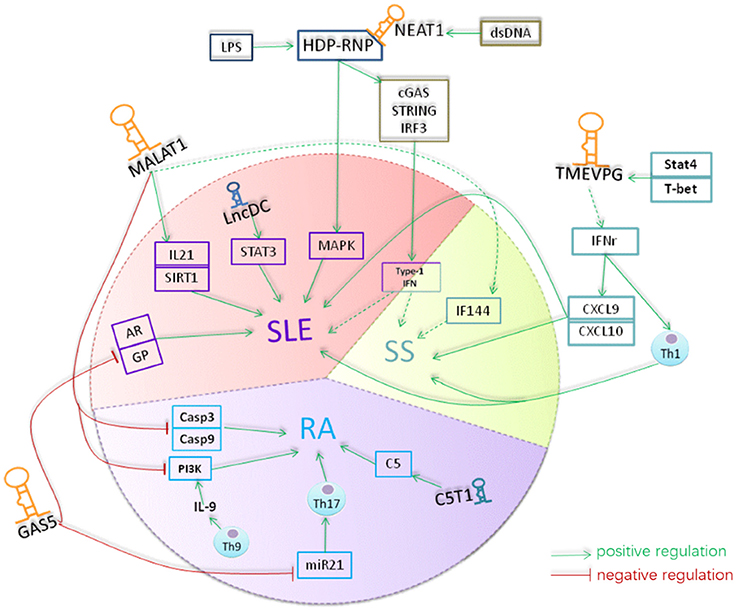
Figure 3. A hypothetical model for the role of lncRNAs in rheumatoid diseases. Lnc-DC was down-regulated in the plasma of SLE. Lnc-DC regulates the expression of STAT3 which plays a crucial role in Th17 differentiation. The downregulation of lnc-DC in SLE might regulate Th17 cell differentiation and thus involved in the pathogenesis of SLE. MALAT1 increases the expression of caspase3 and caspase9 and promotes the apoptosis of RAFLS cells in RA. MALAT1 is involved in the regulation of expression of E2F1 and thus the cell growth and apoptosis. Also, MALAT1 can inhibit the activity of PI3K and thus suppress the proliferation of RAFLS in RA. Th9 cells can produce IL-9 which promoting the activity of PI3K in RAFLs. The expression level of MALAT1 was increased in SLE and its expression was correlated with the expression of IL-21 and SIRT1 signaling in SLE. MALAT1 can affect the expression of IFI44 which was identified as one of the classifier genes to differentiate pSS from healthy controls. NEAT1 was up-regulated in monocyte of SLE patients with unknown mechanism. NEAT1 targeted to MAKP pathway in SLE. TMEVPG can regulate the expression of INF-γ which inducing the expression of CXCL9 and CXCL10 and thus involve in the pathogenesis of SLE and pSS. Gas5 is down-regulated in CD4 T and B cells of patients with RA and patients with SLE. mammalian target of rapamycin (mTOR)can induce the accumulation of GAS5 during growth arrest of cells. GAS5 can induce cell cycler arrest at the G0/G1 phase by inhibiting the transcription of gene such as glucocorticoid receptor and androgen. GAS5 also acted as microRNA sponge to absorb and quench microRNA like miR-21 which is essentially involved in promoting Th17 cell differentiation and contribute to pathogenesis of many autoimmune diseases. lncRNA C5T1, located in a susceptibility locus of RA, positively regulate the expression of C5 which has been suggested to play an essential role in pathogenesis of RA and its animal models. The yellow color indicates lncRNAs, the blue color indicates the molecular interacted with lncRNAs, and the red color indicated the cells that lncRNAs function. The dash arrows indicate the interaction was predicted and the solid arrows indicates the interaction were confirmed by related assay.
In the patients with SLE, MALAT1 was predominantly expressed in monocytes with an abnormally increased expression level (92). MALAT1 is upregulated in FLSs of RA patients compared with corresponding controls. MALAT1 is also involved in the regulation of E2F1 expression, cell growth and apoptosis (109). In FLS from RA, the unregulated MALAT1 is associated with increased cell apoptosis, and the knockdown of MALAT1 inhibits the apoptosis of FLS by suppressing the expression of caspase-3 and caspase-9 (84), suggesting that MALAT1 contribute to the pathogenesis of RA through regulating apoptosis. In SLE, the upregulated expression of MALAT1 is positively related with SIRT1 signaling and the expression of IL-21 (92). Therefore, although MALAT1 is upregulated in both SLE and RA, it might contribute to the disease development via different mechanism. Of note, it has been shown that MALAT1 regulates interferon-induced protein 44 (IFI44) expression (110), a molecule which has been suggested to be a classifier gene to distinguish SS from healthy controls (110). Therefore, it would be interesting to evaluate the association between MALAT1 and SS. LncRNA NEAT1 colocalizes with MALAT1 to many transcription start sites. NEAT1 and MALAT1 are independent but complementary in functions (111). The expression level of NEAT1 was significantly increased in the monocytes from patients with SLE compared with controls, and the expression level of NEAT positively correlates with the clinical disease activity in SLE patient (90). NEAT1 was first identified in the mouse infected with Rabies virus (112), and the subsequent studies showed that it plays an import role in innate immune response (113). For example, NEAT1 can affect the activity of MAPK signaling thereby participates the inflammatory process mediated by TLR-4 (90). In addition, lncNRAs NEAT1 together with hexamethylene bis-acetamide-inducible protein 1 (HEXIM1), DNA-PK subunits, paraspeckle proteins and ribonucleoprotein complex can form a HEXIM1-DNA-PK-paraspeckle components-ribonucleoprotein (HDP-RNP) complex which plays key roles in DNA-mediated innate immune response through the cGAS-STING-IRF3 pathway (114). Therefore, NEAT might contribute to SS and SLE by regulating innate immune responses.
The expression of lncRNA GAS5 is significantly down regulated in both CD4+ T and B cells from RA and SLE patients compared with controls. It has been reported that GAS5 is accumulated during growth arrest induced by serum starvation or suppression of mammalian target of rapamycin (mTOR) (115). Moreover, it is known that GAS5 can induce cell cycler arrest at the G0/G1 phase by inhibiting the transcription of gene such as glucocorticoid receptor and androgen (77, 115), suggesting GAS5 might contribute to the pathogenesis of RA and SLE by regulating cell arrest. In addition, GAS5 also acted as microRNA sponge to absorb and quench microRNA like miR-21 (116). Since miR-21 is essentially involved in promoting Th17 cell differentiation and contribute to pathogenesis of many autoimmune diseases, the down regulated GAS5 might contribute to RA and SLE through uncontrolled miR-21 and Th17 cells (117) (Figure 3).
Another lncRNA of interest is lnc-DC which is exclusively expressed in DC (55). Lnc-DC is down-regulated in the plasma of SLE and associated with the presence of lupus nephritis (89). In the cytoplasm, lnc-DC directly binds to STAT3, thus promoting STAT3 phosphorylation by preventing the interaction between STAT3 and SHP1 (55). Therefore, lnc-DC might contribute to SLE development by regulating DC differentiation. It is reported that incomplete differentiation of pluripotent progenitor cells contributes to rheumatic diseases pathogenesis (118). Thus, lncRNAs which impaired the differentiation of pluripotent progenitor cells such as HOXA transcript antisense RNA, myeloid-specific 1(HOTAIRM1) (119) and H19 (120) may also contribute to the pathology of rheumatic disease.
Finally, lncRNA C5T1 has been implicated in RA because it is located in a susceptibility locus of RA (71). C5T1 positively regulate C5 expression. Complement activation has been suggested to play an essential role in pathogenesis of RA (79). Therefore, C5T1 might play a role in the pathogenesis of RA by regulating the complement system.
Those results indicate the relation of lncRNAs and rheumatic diseases pathogenesis, but further researches on animal model or clinical samples needed to confirm the role of lncRNAs in rheumatic diseases.
Conclusions
LncRNAs plays an important role in gene regulation network during rheumatoid diseases development,thus lncRNAs may become an important target to reveal disease mechanisms and therapy. Recently, the role of lncRNAs in rheumatoid diseases has been extensively investigated. A large number of lncRNAs have been identified to be differentially expressed in rheumatoid diseases as compared with corresponding controls. However, it need to be mentioned that differential expression of lncRNAs is not by default a sign for contribution of them in rheumatic disease pathogenesis. Besides abnormal expression, more evidence are required for the identification of lncRNAs contributing to rheumatoid disease, such as association with clinical characteristics or therapeutic efficacy. Moreover, the role of lncRNA need to be validated and investigated in animal models of rheumatoid diseases.
In this review, we summarized 27, 10, and 11 lncRNAs have been implicated in RA, SLE, and pSS respectively. We discussed some possible mechanisms of these lncRNAs in detail based on published literature. By sharing lncRNAs and related proteins network analysis, we specially found that multiple components of the HDP-RNP pathway were involved in the disease, suggesting that lncRNAs regulated innate immune responses could have important roles in rheumatoid diseases.
In conclusion, accumulated evidence suggests that lncRNAs contribute to the pathogenesis of rheumatoid diseases in the past decade. Further identification of novel disease-related lncRNAs and exploration of the role of them will help us to understand the pathogenesis. We believe that the disease-related functional lncRNAs would be promising diseases biomarkers and therapeutic targets. The development of new RNA delivery entities like nanoparticles (121) and modifications such as CRISPRi (122) will promote the lncRNA-based therapeutics for a broader range of disease in the future.
Author Contributions
JZ, XY, and YG wrote the manuscript. SL prepared manuscript figure. JZ and ZZ edited the manuscript. All authors approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SG and handling Editor declared their shared affiliation.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No.81571593), the start-up packages from Institute of Psychiatry and Neurology, Xinxiang Medical University, and the Deutsche Forschungsgemeinschaft, German Center for Lung Research (DZL).
References
1. Lehuen A, Marshak-Rothstein A. Organ-specific and systemic autoimmune diseases. Curr Opin Immunol. (2013) 25:667–9. doi: 10.1016/j.coi.2013.10.014
2. Huizinga TW, Grondal G. Shared symptoms in rheumatic diseases: a blessing or a curse? Arthritis Rheum. (2009) 60:2547–9. doi: 10.1002/art.24746
3. Gilboe IM, Kvien TK, Uhlig T, Husby G. Sicca symptoms and secondary Sjogren's syndrome in systemic lupus erythematosus: comparison with rheumatoid arthritis and correlation with disease variables. Ann Rheum Dis. (2001) 60:1103–9. doi: 10.1136/ard.60.12.1103
4. Menelaos N. Manoussakis HMM. In: Autoimmune Diseases, 4th Edn. Manhattan, NY: Academic Press (2006)
5. Uhlig T, Kvien TK, Jensen JL, Axell T. Sicca symptoms, saliva and tear production, and disease variables in 636 patients with rheumatoid arthritis. Ann Rheum Dis. (1999) 58:415–22. doi: 10.1136/ard.58.7.415
6. Mattey DL, Gonzalez-Gay MA, Hajeer AH, Dababneh A, Thomson W, Garcia-Porrua C, et al. Association between HLA-DRB1*15 and secondary Sjogren's syndrome in patients with rheumatoid arthritis. J Rheumatol. (2000) 27:2611–6.
7. Birtane M, Yavuz S, Tastekin N. Laboratory evaluation in rheumatic diseases. World J Methodol. (2017) 7:1–8. doi: 10.5662/wjm.v7.i1.1
8. Franceschini F, Cavazzana I. Anti-Ro/SSA and La/SSB antibodies. Autoimmunity (2005) 38:55–63. doi: 10.1080/08916930400022954
9. Song GG, Kim JH, Seo YH, Choi SJ, Ji JD, Lee YH. Meta-analysis of differentially expressed genes in primary Sjogren's syndrome by using microarray. Hum Immunol. (2014) 75:98–104. doi: 10.1016/j.humimm.2013.09.012
10. Wang L, Wu LF, Lu X, Mo XB, Tang ZX, Lei SF, et al. Integrated analyses of gene expression profiles digs out common markers for Rheumatic diseases. PLoS ONE (2015) 10:e0137522. doi: 10.1371/journal.pone.0137522
11. Zheng J, Ibrahim S, Petersen F, Yu X. Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun. (2012) 13:641–52. doi: 10.1038/gene.2012.46
12. Zheng J, Yin J, Huang R, Petersen F, Yu X. Meta-analysis reveals an association of STAT4 polymorphisms with systemic autoimmune disorders and anti-dsDNA antibody. Hum Immunol. (2013) 74:986–92. doi: 10.1016/j.humimm.2013.04.034
13. Graham RR, Kozyrev SV, Baechler EC, Reddy MVPL, Plenge RM, Bauer JW, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. (2006) 38:550–5. doi: 10.1038/ng1782
14. Miceli-Richard C, Comets E, Loiseau P, Puechal X, Hachulla E, Mariette X. Association of an IRF5 gene functional polymorphism with Sjogren's syndrome. Arthritis Rheum. (2007) 56:3989–94. doi: 10.1002/art.23142
15. Garnier S, Dieude P, Michou L, Barbet S, Tan A, Lasbleiz S, et al. IRF5 rs2004640-T allele, the new genetic factor for systemic lupus erythematosus, is not associated with rheumatoid arthritis. Ann Rheum Dis. (2007) 66:828–31. doi: 10.1136/ard.2006.061390
16. Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. (2012) 3:219. doi: 10.3389/fgene.2012.00219
17. Hassan MQ, Tye CE, Stein GS, Lian JB. Non-coding RNAs: epigenetic regulators of bone development and homeostasis. Bone (2015) 81:746–56. doi: 10.1016/j.bone.2015.05.026
18. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. (1990) 10:28–36. doi: 10.1128/MCB.10.1.28
19. Wilusz JE, Freier SM, Spector DL. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell (2008) 135:919–32. doi: 10.1016/j.cell.2008.10.012
20. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell (2009) 136:629–41. doi: 10.1016/j.cell.2009.02.006
21. Xiong XD, Ren X, Cai MY, Yang JW, Liu X, Yang JM. Long non-coding RNAs: an emerging powerhouse in the battle between life and death of tumor cells. Drug Resist Updat. (2016) 26:28–42. doi: 10.1016/j.drup.2016.04.001
22. Kitagawa M, Kotake Y, Ohhata T. Long non-coding RNAs involved in cancer development and cell fate determination. Curr Drug Targets (2012) 13:1616–21. doi: 10.2174/138945012803530026
23. Li Z, Li X, Jiang C, Qian W, Tse G, Chan MTV, et al. Long non-coding RNAs in rheumatoid arthritis. Cell Prolif. (2018) 51:e12404. doi: 10.1111/cpr.12404
24. Sandhya P, Joshi K, Scaria V. Long noncoding RNAs could be potential key players in the pathophysiology of Sjogren's syndrome. Int J Rheum Dis. (2015) 18:898–905. doi: 10.1111/1756-185X.12752
25. Xu D, Jiang Y, Yang L, Hou X, Wang J, Gu W, et al. Long noncoding RNAs expression profile and functional networks in rheumatoid arthritis. Oncotarget (2017) 8:95280–92. doi: 10.18632/oncotarget.20036.
26. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. (2012) 22:1760–74. doi: 10.1101/gr.135350.111
27. Jalali S, Jayaraj GG, Scaria V. Integrative transcriptome analysis suggest processing of a subset of long non-coding RNAs to small RNAs. Biol Direct. (2012) 7:25. doi: 10.1186/1745-6150-7-25
28. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature (2012) 482:339–46. doi: 10.1038/nature10887
29. Rutenbergschoenberg M, Sexton AN, Simon MD. The properties of long noncoding RNAs that regulate chromatin. Ann Rev Genomics Hum Genet. (2016) 17:69–94. doi: 10.1146/annurev-genom-090314-024939
30. Ounzain S, Pedrazzini T. The promise of enhancer-associated long noncoding RNAs in cardiac regeneration. Trends Cardiovasc Med. (2015) 25:592–602. doi: 10.1016/j.tcm.2015.01.014
31. Ounzain S, Pedrazzini T. Super-enhancer lncs to cardiovascular development and disease. Biochim Biophys Acta (2016) 7:24. doi: 10.1016/j.bbamcr.2015.11.026
32. Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature (2011) 472:120–4. doi: 10.1038/nature09819
33. Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. (1996) 10:2657–83. doi: 10.1101/gad.10.21.2657
34. Sengupta S, Mitra S, Bhakat KK. Dual regulatory roles of human AP-endonuclease (APE1/Ref-1) in CDKN1A/p21 expression. PLoS ONE (2013) 8:e68467. doi: 10.1371/journal.pone.0068467
35. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature (2013) 494:497–501. doi: 10.1038/nature11884
36. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature (2016) 539:452–5. doi: 10.1038/nature20149
37. Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature (2001) 414:317–22. doi: 10.1038/35104575
38. Eilebrecht S, Benecke BJ, Benecke AG. Latent HIV-1 TAR Regulates 7SK-responsive P-TEFb target genes and targets cellular immune responses in the absence of Tat. Genomics Proteomics Bioinformatics (2017) 15:313–23. doi: 10.1016/j.gpb.2017.05.003
39. Zovoilis A, Cifuentes-Rojas C, Chu HP, Hernandez AJ, Lee JT. Destabilization of B2 RNA by EZH2 activates the stress response. Cell (2016) 167:1788–802e13. doi: 10.1016/j.cell.2016.11.041
40. Espinoza CA, Goodrich JA, Kugel JF. Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA (2007) 13:583–96. doi: 10.1261/rna.310307
41. Cooper DR, Carter G, Li P, Patel R, Watson JE, Patel NA. Long Non-Coding RNA NEAT1 Associates with SRp40 to temporally regulate PPARgamma2 splicing during adipogenesis in 3T3-L1 Cells. Genes (2014) 5:1050–63. doi: 10.3390/genes5041050
42. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell (2010) 39:925–38. doi: 10.1016/j.molcel.2010.08.011
43. Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. (2010) 29:3082–93. doi: 10.1038/emboj.2010.199
44. Wilusz JE. Long noncoding RNAs: re-writing dogmas of RNA processing and stability. Biochim Biophys Acta (2016) 1859:128–38. doi: 10.1016/j.bbagrm.2015.06.003
45. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature (2014) 505:344–52. doi: 10.1038/nature12986
46. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell (2011) 147:358–69. doi: 10.1016/j.cell.2011.09.028
47. Zhang L, Yang Z, Trottier J, Barbier O, Wang L. Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay. Hepatology. (2017) 65:604–15. doi: 10.1002/hep.28882
48. Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature (2012) 491:454–7. doi: 10.1038/nature11508
49. Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, et al. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell (2016) 164:69–80. doi: 10.1016/j.cell.2015.12.017
50. Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS ONE (2014) 9:e107016. doi: 10.1371/journal.pone.0107016
51. Gong C, Li Z, Ramanujan K, Clay I, Zhang Y, Lemire-Brachat S, et al. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell (2015) 34:181–91. doi: 10.1016/j.devcel.2015.05.009
52. Liu J, Qian C, Cao X. Post-translational modification control of innate immunity. Immunity (2016) 45:15–30. doi: 10.1016/j.immuni.2016.06.020
53. Jain AK, Xi Y, McCarthy R, Allton K, Akdemir KC, Patel LR, et al. LncPRESS1 Is a p53-Regulated LncRNA that safeguards pluripotency by disrupting SIRT6-mediated de-acetylation of histone H3K56. Mol Cell (2016) 64:967–81. doi: 10.1016/j.molcel.2016.10.039
54. Wang S, Liang K, Hu Q, Li P, Song J, Yang Y, et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J Clin Invest. (2017) 127:4498–15. doi: 10.1172/JCI91553
55. Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, et al. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science (2014) 344:310–3. doi: 10.1126/science.1251456
56. Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature (2013) 503:371–6. doi: 10.1038/nature12598
57. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science (2010) 329:689–93. doi: 10.1126/science.1192002
58. Sasaki H, Ishihara K, Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J Biochem. (2000) 127:711–5. doi: 10.1093/oxfordjournals.jbchem.a022661
59. Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, et al. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature (2017) 541:228–32. doi: 10.1038/nature21034
60. Tang Y, Zhou T, Yu X, Xue Z, Shen N. The role of long non-coding RNAs in rheumatic diseases. Nat Rev Rheumatol. (2017) 13:657–69. doi: 10.1038/nrrheum.2017.162
61. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. New Engl J Med. (2011) 365:2205–19. doi: 10.1056/NEJMra1004965
62. Song J, Kim D, Han J, Kim Y, Lee M, Jin EJ. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin Exp Med. (2015) 15:121–6. doi: 10.1007/s10238-013-0271-4
63. Zhang Y, Xu YZ, Sun N, Liu JH, Chen FF, Guan XL, et al. Long noncoding RNA expression profile in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Res Ther. (2016) 18:227. doi: 10.1186/s13075-016-1129-4
64. Lu MC, Yu HC, Yu CL, Huang HB, Koo M, Tung CH, et al. Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol Res. (2016) 64:576–83. doi: 10.1007/s12026-015-8756-8
65. Luo Q, Xu C, Li X, Zeng L, Ye J, Guo Y, et al. Comprehensive analysis of long non-coding RNA and mRNA expression profiles in rheumatoid arthritis. Exp Ther Med. (2017) 14:5965–73. doi: 10.3892/etm.2017.5284
66. Chen X, Oppenheim J, Howard OM. Shikonin, a component of antiinflammatory Chinese herbal medicine, selectively blocks chemokine binding to CC chemokine receptor-1. Int Immunopharmacol. (2001) 1:229–36. doi: 10.1016/S1567-5769(00)00033-3
67. Ye Y, Gao X, Yang N. LncRNA ZFAS1 promotes cell migration and invasion of fibroblast-like synoviocytes by suppression of miR-27a in rheumatoid arthritis. Human Cell (2018) 31:14–21. doi: 10.1007/s13577-017-0179-5
68. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell (2010) 142:409–19. doi: 10.1016/j.cell.2010.06.040
69. Stuhlmuller B, Kunisch E, Franz J, Martinez-Gamboa L, Hernandez MM, Pruss A, et al. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am J Pathol. (2003) 163:901–11. doi: 10.1016/S0002-9440(10)63450-5
70. Yang KY, Chen DL. Shikonin inhibits inflammatory response in rheumatoid arthritis synovial fibroblasts via lncRNA-NR024118. Evid Based Complement Alternat Med. (2015) 2015:631737. doi: 10.1155/2015/631737
71. Messemaker TC, Frank-Bertoncelj M, Marques RB, Adriaans A, Bakker AM, Daha N, et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. (2016) 17:85–92. doi: 10.1038/gene.2015.54
72. Mayama T, Marr AK, Kino T. Differential Expression of Glucocorticoid Receptor Noncoding RNA Repressor Gas5 in Autoimmune and Inflammatory Diseases. Horm Metab Res. (2016) 48:550–7. doi: 10.1055/s-0042-106898
73. Yuan M, Wang S, Yu L, Qu B, Xu L, Liu L, et al. Long noncoding RNA profiling revealed differentially expressed lncRNAs associated with disease activity in PBMCs from patients with rheumatoid arthritis. PLoS ONE (2017) 12:e0186795. doi: 10.1371/journal.pone.0186795
74. Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. (2015) 75:3181–91. doi: 10.1158/0008-5472.CAN-14-3721
75. Nie F, Yu X, Huang M, Wang Y, Xie M, Ma H, et al. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget (2017) 8:38227–38. doi: 10.18632/oncotarget.9611
76. Fang C, Zan J, Yue B, Liu C, He C, Yan D. Long non-coding ribonucleic acid zinc finger antisense 1 promotes the progression of colonic cancer by modulating ZEB1 expression. J Gastroenterol Hepatol. (2017) 32:1204–11. doi: 10.1111/jgh.13646
77. Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. (2010) 3:ra8. doi: 10.1126/scisignal.2000568
78. Cooke TD, Hurd ER, Jasin HE, Bienenstock J, Ziff M. Identification of immunoglobulins and complement in rheumatoid articular collagenous tissues. Arthritis Rheum. (1975) 18:541–51. doi: 10.1002/art.1780180603
79. Wang Y, Kristan J, Hao L, Lenkoski CS, Shen Y, Matis LA. A role for complement in antibody-mediated inflammation: C5-deficient DBA/1 mice are resistant to collagen-induced arthritis. J Immunol. (2000) 164:4340–7. doi: 10.4049/jimmunol.164.8.4340
80. Gizinski AM, Fox DA. T cell subsets and their role in the pathogenesis of rheumatic disease. Curr Opin Rheumatol. (2014) 26:204–10. doi: 10.1097/BOR.0000000000000036
81. Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. (2010) 233:233–55. doi: 10.1111/j.0105-2896.2009.00859.x
82. Hollman PC, vd Gaag M, Mengelers MJ, van Trijp JM, de Vries JH, Katan MB. Absorption and disposition kinetics of the dietary antioxidant quercetin in man. Free Rad Biol Med. (1996) 21:703–7. doi: 10.1016/0891-5849(96)00129-3
83. Natarajan V, Krithica N, Madhan B, Sehgal PK. Formulation and evaluation of quercetin polycaprolactone microspheres for the treatment of rheumatoid arthritis. J Pharm Sci. (2011) 100:195–205. doi: 10.1002/jps.22266
84. Pan F, Zhu L, Lv H, Pei C. Quercetin promotes the apoptosis of fibroblast-like synoviocytes in rheumatoid arthritis by upregulating lncRNA MALAT1. Int J Mol Med. (2016) 38:1507–14. doi: 10.3892/ijmm.2016.2755
85. Kim YO, Hong SJ, im SV. The efficacy of shikonin on cartilage protection in a mouse model of rheumatoid arthritis. Korean J Physiol Pharmacol. (2010) 14:199–204. doi: 10.4196/kjpp.2010.14.4.199
86. Spurlock CF III, Tossberg JT, Matlock BK, Olsen NJ, Aune TM. Methotrexate inhibits NF-kappaB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. (2014) 66:2947–57. doi: 10.1002/art.38805
87. Shao L, Goronzy JJ, Weyand CM. DNA-dependent protein kinase catalytic subunit mediates T-cell loss in rheumatoid arthritis. EMBO Mol Med. (2010) 2:415–27. doi: 10.1002/emmm.201000096
88. Haywood ME, Rose SJ, Horswell S, Lees MJ, Fu G, Walport MJ, et al. Overlapping BXSB congenic intervals, in combination with microarray gene expression, reveal novel lupus candidate genes. Genes Immun. (2006) 7:250–63. doi: 10.1038/sj.gene.6364294
89. Wu GC, Li J, Leng RX, Li XP, Li XM, Wang DG, et al. Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus. Oncotarget (2017) 8:23650–63. doi: 10.18632/oncotarget.15569
90. Zhang F, Wu L, Qian J, Qu B, Xia S, La T, et al. Identification of the long noncoding RNA NEAT1 as a novel inflammatory regulator acting through MAPK pathway in human lupus. J Autoimmun. (2016) 75:96–104. doi: 10.1016/j.jaut.2016.07.012
91. Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B, et al. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res Ther. (2015) 17:131. doi: 10.1186/s13075-015-0632-3
92. Yang H, Liang N, Wang M, Fei Y, Sun J, Li Z, et al. Long noncoding RNA MALAT-1 is a novel inflammatory regulator in human systemic lupus erythematosus. Oncotarget (2017) 8:77400–6. doi: 10.18632/oncotarget.20490
93. Li LJ, Zhao W, Tao SS, Li J, Xu SZ, Wang JB, et al. Comprehensive long non-coding RNA expression profiling reveals their potential roles in systemic lupus erythematosus. Cell Immunol. (2017) 319:17–27. doi: 10.1016/j.cellimm.2017.06.004
94. Luo Q, Li X, Xu C, Zeng L, Ye J, Guo Y, et al. Integrative analysis of long non-coding RNAs and messenger RNA expression profiles in systemic lupus erythematosus. Mol Med Rep. (2017) 17:3489–96. doi: 10.3892/mmr.2017.8344
95. Fang Z, Zhang S, Wang Y, Shen S, Wang F, Hao Y, et al. Long non-coding RNA MALAT-1 modulates metastatic potential of tongue squamous cell carcinomas partially through the regulation of small proline rich proteins. BMC Cancer. (2016) 16:706. doi: 10.1186/s12885-016-2735-x
96. Gaubitz M. Epidemiology of connective tissue disorders. Rheumatology (Oxford) (2006) 45(Suppl 3):iii3–4. doi: 10.1093/rheumatology/kel282
97. Shi H, Cao N, Pu Y, Xie L, Zheng L, Yu C. Long non-coding RNA expression profile in minor salivary gland of primary Sjogren's syndrome. Arthritis Res Ther. (2016) 18:109. doi: 10.1186/s13075-016-1005-2
98. Wang J, Peng H, Tian J, Ma J, Tang X, Rui K, et al. Upregulation of long noncoding RNA TMEVPG1 enhances T helper type 1 cell response in patients with Sjogren syndrome. Immunol Res. (2016) 64:489–96. doi: 10.1007/s12026-015-8715-4
99. Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell (2013) 152:743–54. doi: 10.1016/j.cell.2013.01.015
100. Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by Th1 cells. J Immunol. (2012) 189:2084–8. doi: 10.4049/jimmunol.1200774
101. Collier SP, Henderson MA, Tossberg JT, Aune TM. Regulation of the Th1 genomic locus from Ifng through Tmevpg1 by T-bet. J Immunol. (2014) 193:3959–65. doi: 10.4049/jimmunol.1401099
102. Ronnblom L, Eloranta ML. The interferon signature in autoimmune diseases. Curr Opin Rheumatol. (2013) 25:248–53. doi: 10.1097/BOR.0b013e32835c7e32
103. Pollard KM, Cauvi DM, Toomey CB, Morris KV, Kono DH. Interferon-gamma and systemic autoimmunity. Discov Med. (2013) 16:123–31.
104. Struyf NJ, Snoeck HW, Bridts CH, De Clerck LS, Stevens WJ. Natural killer cell activity in Sjogren's syndrome and systemic lupus erythematosus: stimulation with interferons and interleukin-2 and correlation with immune complexes. Ann Rheum Dis. (1990) 49:690–3. doi: 10.1136/ard.49.9.690
105. Cha S, Brayer J, Gao J, Brown V, Killedar S, Yasunari U, et al. A dual role for interferon-gamma in the pathogenesis of Sjogren's syndrome-like autoimmune exocrinopathy in the nonobese diabetic mouse. Scand J Immunol. (2004) 60:552–65. doi: 10.1111/j.0300-9475.2004.01508.x
106. Elia G. Interferon-gamma-inducible chemokines in systemic lupus erythematosus. La Clin Ter. (2015) 166:e41–6. doi: 10.7417/T.2015.1808
107. Li H, Ice JA, Lessard CJ, Sivils KL. Interferons in Sjögren's syndrome: genes, mechanisms, and effects. Front Immunol. (2013) 20:290. doi: 10.3389/fimmu.2013.00290
108. Hagberg J, Rönnblom N.L. Systemic lupus erythematosus–a disease with A dysregulated type I interferon system. Scand J Immunol. (2015). 82:199–207. doi: 10.1111/sji.12330
109. Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell (2011) 147:773–88. doi: 10.1016/j.cell.2011.08.054
110. Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, et al. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA (2012) 18:738–51. doi: 10.1261/rna.028639.111
111. West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, et al. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell (2014) 55:791–802. doi: 10.1016/j.molcel.2014.07.012
112. Saha S, Murthy S, Rangarajan PN. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol. (2006) 87(Pt 7):1991–5. doi: 10.1099/vir.0.81768-0
113. Geng H, Tan XD. Functional diversity of long non-coding RNAs in immune regulation. Genes Dis. (2016) 3:72–81. doi: 10.1016/j.gendis.2016.01.004
114. Morchikh M, Cribier A, Raffel R, Amraoui S, Cau J, Severac D, et al. HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol Cell. (2017) 67:387-99e5. doi: 10.1016/j.molcel.2017.06.020
115. Pickard MR, Williams GT. Molecular and cellular mechanisms of action of tumour suppressor GAS5 LncRNA. Genes (2015) 6:484–99. doi: 10.3390/genes6030484
116. Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. (2014) 32:1628–35. doi: 10.1002/jor.22718
117. Wang S, Wan X, Ruan Q. The MicroRNA-21 in autoimmune diseases. Int J Mol Sci. (2016) 17:864. doi: 10.3390/ijms17060864
118. Wang L, Wang FS, Gershwin ME. Human autoimmune diseases: a comprehensive update. J Int Med. (2015) 278:369–95. doi: 10.1111/joim.12395
119. Zhang X, Weissman SM, Newburger PE. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. (2014) 11:777–87. doi: 10.4161/rna.28828
120. Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, et al. Maternal imprinting at the H19–Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature (2013) 500:345–9. doi: 10.1038/nature12303
121. Miele E, Spinelli GP, Di Fabrizio E, Ferretti E, Tomao S, Gulino A. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. Int J Nanomed. (2012) 7:3637–57. doi: 10.2147/IJN.S23696
Keywords: rheumatoid diseases, long non-coding RNAs, rheumatoid arthritis, systemic lupus erythematosus (SLE), Sjögren's syndrome
Citation: Gao Y, Li S, Zhang Z, Yu X and Zheng J (2018) The Role of Long Non-coding RNAs in the Pathogenesis of RA, SLE, and SS. Front. Med. 5:193. doi: 10.3389/fmed.2018.00193
Received: 05 April 2018; Accepted: 11 June 2018;
Published: 03 July 2018.
Edited by:
George Bertsias, University of Crete, GreeceReviewed by:
Efstathia K. Kapsogeorgou, National and Kapodistrian University of Athens, GreeceAggelos Banos, Biomedical Research Foundation of the Academy of Athens, Greece
Spiros Georgakis, University of Crete, Greece
Copyright © 2018 Gao, Li, Zhang, Yu and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinhua Yu, eGluaHVheXVAZnotYm9yc3RlbC5kZQ==
Junfeng Zheng, MTcxMDEzQHh4bXUuZWR1LmNu
 Yunzhen Gao1
Yunzhen Gao1 Xinhua Yu
Xinhua Yu