- 1Department of Medical Microbiology, School of Medicine, Nanchang University, Nanchang, China
- 2Key Laboratory of Tumor Pathogenesis and Molecular Pathology, School of Medicine, Nanchang University, Nanchang, China
Outer membrane vesicles (OMVs) are valued for their unique, convenient, and amendable functions. They are flexible, controllable nanoparticles, which can be modified in different ways, for use in a wide array of applications like adjuvants, vaccines, drug presentation, and fluorescence tracking. Attempts have been made to alter the nanoparticles in liposomes and bacterial ghosts using traditional methods. However, it has not been highly successful owing to certain unavoidable disadvantages. OMVs have unique advantages and are considered novel platforms for heterologous protein presentation. Although previous reviews have described this to some extent, owing to the rapid development in this field, there is a need for the regular update of the reviews. This review focuses on few novel applications of engineered OMVs, and redefines the significance of OMVs as novel multifunctional delivery platforms. Additionally, this review also supplements and provides an update on the OMV transformation methods.
Introduction
Outer membrane vesicle (OMV), naturally secreted by Gram-negative bacteria, is a spherical nanostructure with a diameter of 30–200 nm (Figure 1) (Liu et al., 2016). Bacteria can produce OMVs in solid and liquid media, or in biofilms and intracellular infections under in vitro conditions (Unal et al., 2011; Klimentová and Stulík, 2014; Caruana and Walper, 2020). The generation of OMVs is considered a stress response. Various stress factors, such as temperature, nutrient consumption, and exposure to harmful chemicals, may cause accumulation and aggregation of misfolded proteins in the periplasm. Packing these pressure products to the outer membrane and releasing them, OMVs are formed (Klimentová and Stulík, 2014). Quantitative analysis showed that Escherichia coli packages ~0.2–0.5% of outer membrane and periplasmic protein into OMV (Kesty and Kuehn, 2004). Approximately 1% of the outer membrane material could be incorporated into the vesicles of Pseudomonas aeruginosa, while the logarithmic phase culture of Neisseria meningitidis incorporates 8–12% of its total protein and endotoxin into vesicles (Devoe and Gilchrist, 1973; Bauman and Kuehn, 2006). Most of the components of OMVs are lipopolysaccharide (LPS), glycerophospholipids, and proteins including periplasmic proteins (Ellis and Kuehn, 2010; Underhill and Goodridge, 2012). The various components on OMVs serve different functions. LPS of OMVs gives its adjuvant function, which is useful for vaccine formulation, and some other antigen components that can be used for vaccine development. In addition, other components on OMVs also play roles that cannot be ignored. OMVs have been found to function as a multifaceted delivery system with intra- and inter-species interactions through membrane proteins (Ellis and Kuehn, 2010). The OMVs produced by Mycobacterium tuberculosis carries the iron-carrier mold, mycomycin, which transports this essential nutrient over long distances in a hostile host environment (Prados–Rosales et al., 2014). Regarding the interspecies interactions of OMVs, multiple studies have indicated the use of OMVs by many bacterial pathogens to deliver virulent determinants to host cells, at proximal and distal sites, thereby impairing the host defense system and promoting infection. In contrast, OMVs from various mucosal pathogens show numerous pathogen-associated molecular patterns including LPS, lipoproteins, DNA, and RNA. Their interaction with the pattern recognition receptors (PRR) on host epithelial cells stimulates the production of cytokines and chemokines on the mucosal epithelial surface, their interactions with immune cells, and regulates pathology in a variety of ways (Yoon, 2016). The virulence factors contained in OMVs can be distributed over long distances, and also transferred onto the plasma membranes of the host cells (Ellis and Kuehn, 2010).
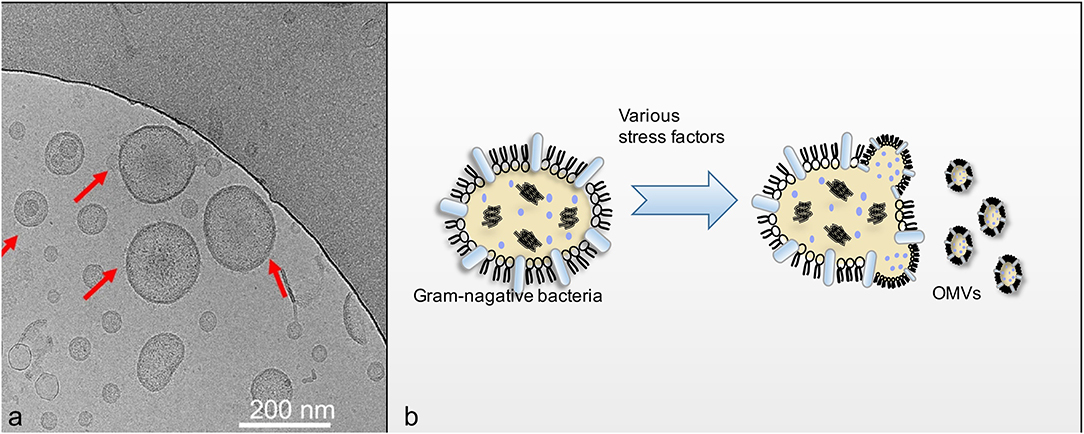
Figure 1. (a) Showed that Cryo-EM imaging of OMVs (Liu et al., 2016). OMVs derived from the flagellin-deficient S.typhimurium were visualized using cryo-EM. The red arrows indicate the visible OMVs; (b) showed schematic diagram of OMVs secretion process.
Schulz et al. (2018) characterized the morphology of OMVs by electron microscopy and found that OMVs are biocompatible with epithelial cells and differentiated macrophages (Schulz et al., 2018). They showed low endotoxin activity compared to control samples, indicating low acute inflammation potential. At the same time, OMVs also showed good biodegradability, and it has been proven that OMV in the lysosome can be completely degraded after 48 h of endocytosis (Kim et al., 2012). Bioengineering can be used to increase OMV output, and may be used in conjunction with specific production processes to obtain a large number of well-defined, stable, and unified OMV particle vaccine products (van der Pol et al., 2015). Commercially available N. meningitidis serotype B OMV vaccine indicates the use of OMVs for disease prevention and treatment (Mirlashari et al., 2010; Collins, 2011; Santos et al., 2012). Currently, research has been focused on the use of OMVs in vaccine transformation platforms; different heterologous proteins can be efficiently added to achieve the use of such nanoparticles in multiple applications (Chen et al., 2010; Bartolini et al., 2013; Alves et al., 2015).
Although previous reviews have described OMVs in vaccine transformation platforms, since this is a rapidly growing area of research, the reviews need to be updated frequently (Gerritzen et al., 2017). This review mainly describes the applications of the most recently engineered OMVs, and gives an update on few novel engineering methods. Additionally, the advantages and disadvantages of the different engineering methods have been compared, and few prospective applications of the engineered OMVs have been described.
Purification, the Key Step in OMV Usage in Vaccine Applications
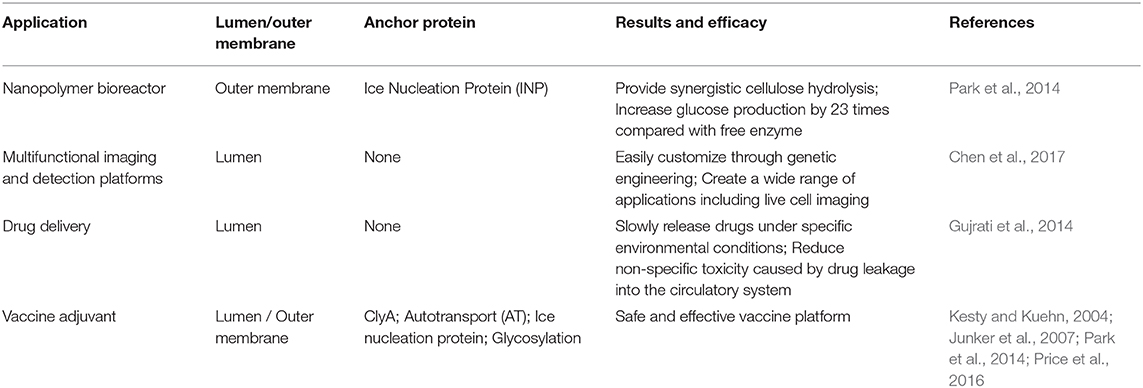
Some bacterial nanoparticles have gradually gaining interest and are being developed with antigen-related functions. However, compared with OMV, the production process is more complicated and expensive, and it is difficult to ensure its safety in living organisms (Table 1) (Carita et al., 2018; Hu et al., 2019; Lee et al., 2019; Sánchez et al., 2020). Presently, there have been a variety of bacterial OMVs used in vaccine production attempts, such as E. coli, P. aeruginosa, Shigella, Salmonella, Helicobacter pylori (H. pylori), Campylobacter jejuni, Borrelia burgdorferi, Vibrio, and Neisseria spp. (Bauman and Kuehn, 2006; Chatterjee and Chaudhuri, 2011; Elmi et al., 2012; Schwechheimer and Kuehn, 2015; Turner et al., 2015; Elhenawy et al., 2016). In the making of vaccines of different strains, some components on OMVs (such as LPS) could affect the safety of the vaccine. Purification can remove some unnecessary or harmful components, thereby making OMVs more in line with needs. Purification is the first and the most important step in the use of OMVs and impurities can have a large impact, resulting in extremely unfavorable on experimental outcomes. Thus, the purification of OMVs using traditional methods has been summarized, and the latest approaches for the purification of OMVs using genetic modifications have been elaborated.
Purification of OMVs Using Traditional Methods
Traditional methods of OMV purification include ultracentrifugation, density gradient separation, precipitation, and gel filtration techniques (Kataoka et al., 2014; Klimentová and Stulík, 2014; Cecil et al., 2016; Kim et al., 2017a,b). Density gradient centrifugation and gel filtration are commonly used methods for OMV purification (Kulp and Kuehn, 2010), while ultracentrifugation and precipitation are the primary methods to obtain OMVs. Although ultracentrifugation methods have been widely used in the preparation of OMVs, this method has certain shortcomings. The primary problem is the complexity of the operation process. It is time consuming, and requires skilled labor, owing to the complexity of the process. In addition, the high cost of the purification equipment makes it difficult to commercialize. Although sedimentation is relatively inexpensive, it also has certain unavoidable disadvantages. The preparation of the salt solution required for precipitation is difficult, and the steps are complicated. Also, the proteins on the OMVs are easily damaged, thereby allowing for a negative impact on experimental results (Qing et al., 2019). Additionally, considering the impact of LPS on the experiment, researchers have attempted certain special methods. Initially, detergent was added during the extraction process to attenuate LPS. However, studies have shown that LPS is a potential endotoxin, and acts as a powerful adjuvant for specific antigens (Mitra et al., 2013). Consequently, the complete removal of LPS from OMVs was required, rather than LPS attenuation with detergent, which proved unsuitable for the betterment of vaccine performance (Mitra et al., 2016). Therefore, researchers proposed the use of EDTA extraction (chelating agent) to obtain high quality OMVs, by accurately controlling the pH and collection point (van de Waterbeemd et al., 2012). However, this method demands stringent extraction conditions, and has a complex operation process. If the purification methods do not meet the research requirements, the development and application of OMVs in the biomedical field will be limited. Therefore, the development of a simpler and more feasible method is very crucial.
Genetic Engineering Used for Attenuating Toxicity of OMVs
Generally, major pathogenic Gram-negative bacteria can secrete OMVs and interact with the host through OMVs (Kuehn and Kesty, 2005). Although OMVs can be obtained by extraction with detergents, the improper use of chemicals can result in the loss of lipoproteins and enzymes, and their activity, which impairs the ability of OMVs to stimulate cross-protective immunity. In order to obtain OMVs with low toxicity and high safety, the genetic modification, and purification of OMVs has been proposed. Genetic modification is primarily aimed at the attenuation of the OMVs, and improvement of its yield. An established method is the knock out of the msbB gene, which regulates LPS in bacteria. The inactivation of msbB leads to a significant decrease in the level of LPS endotoxin (Kim et al., 2009; Ranallo et al., 2010). This method has been widely used in the preparation of OMVs (Leitner et al., 2015). Latest research has combined a new modification scheme with the genetic modification of the msbB gene, and has helped to obtain good results. At the same time, the Dutch vaccine research institute found that the deletion of the lpxL1 gene attenuated LPS toxicity while retaining the adjuvant activity required for the immune response (van der Ley et al., 2001; Fisseha et al., 2005; Koeberling et al., 2008). Engineered OMVs obtained from Bordetella pertussis expressing the pagL gene of Bordetella bronchiseptica, had lower endotoxin activity and better adjuvant performance. Attenuated OMVs are considered as prospective candidates in the development of novel vaccines targeting multiple antigens (Asensio et al., 2011).
Compared to the traditional methods, genetically engineered bacterial strains not only possess exclusively attenuated LPS, but also have the ability to induce pathogen-specific immune responses to OMVs, which is more suitable for the non-endotoxin conversion of OMVs (Shim et al., 2017). These pivotal genes attracted attention, and their knockout lead to a decrease in immunogenicity. Consequently, researchers have gradually started to explore more novel methods such as using non-pathogenic symbiotic bacteria as the source of OMVs, which can overcome the risks of gene knockout and render a certain degree of safety (Carvalho et al., 2019). Now, OMVs can be mass-produced through optimized purification methods, and for reference, Gerritzen et al. (2019) has summarized various purification methods of OMV (Gerritzen et al., 2019).
Genetic Engineering in the Increasing Production of OMVs
The Tol-Pal system continues to attract attention since its functioning affects the OMV production (Shrivastava et al., 2017). The five proteins of the Tol-Pal system includes three inner membrane proteins (TolA, TolQ, and TolR), a periplasmic protein (TolB), and a peptidoglycan-related lipoprotein (Pal) associated with the outer membrane proteins (Derouiche et al., 1995). The tolB mutant shows increased OMV release in many bacteria like E. coli, Salmonella typhimurium, and H. pylori (Deatherage et al., 2009; Nevermann et al., 2019). Interestingly, tol mutants can often be introduced into the bacteria of interest by means of electrotransportation (Turner et al., 2015). Additionally, the rmpM gene deletion mutant was found to have a loosely attached outer membrane, which increased OMV release (Maharjan et al., 2016). Theoretically, the combination of the lpxL1 and rmpM mutations can address toxicity and resolve issues associated with the detergent-free OMV vaccines. Both gene mutations have been reported to have beneficial effects, making the detergent-free method a better alternative to the traditional detergent-based OMV purification methods (van de Waterbeemd et al., 2010). With gradual improvement in genetically engineered OMVs, multiple genes have been modified for the purification of OMVs. For example, few studies have attempted to incorporate a non-natural histidine amino acid. The sequence (His-tag) has been used to purify the bacterial OMVs by affinity chromatography (Alves et al., 2015). Additionally, genetic engineering has also been used to produce a high-yield of bacteria lacking the OmpA gene, which has been widely used in OMV engineering (Moon et al., 2012; Kulp et al., 2015).
Studies in E. coli have shown that the stability of the envelope comes from three major cross-links: OmpA, LppAB, and the Tol-Pal complex (Mizushima, 1984; Yeh et al., 2010; Park et al., 2012). When there is a decrease in these cross-links, the yield of OMV increases, and vice versa. Thus, S. typhimurium mutants lacking OmpA, LppAB, Pal, TolA, or TolB show increased OMV production (Deatherage et al., 2009). Similarly, gene deletion can have a negative impact on immunogenicity. For example, the TolA gene can increase the resistance of S. typhimurium to bile (Lahiri et al., 2011). The tolR gene encodes proteins with exercise-related functions in S. typhimurium and E. coli (Santos et al., 2014). Deletion of these genes will inevitably have a negative impact on the function of OMVs, like reduced immunogenicity. Consequently, the question of balancing yield and the immune efficacy remains unsolved.
Update on the Application of OMVs as Antigen Presentation Platforms Using Genetic Engineering
Increasing evidence exists for the delivery of antigens by OMVs, to proximal host cells and distal host cells, which in turn stimulates the immune system and triggers immune response in the host (Shen et al., 2012). The antigens in OMVs can be distributed over long distances and are not damaged by dynamic physical and biochemical stresses. In addition, they can be transferred to the plasma membrane of the host cell (Sharpe et al., 2011; Yoon et al., 2011). Moreover, some transmembrane proteins on OMVs can be used as anchoring proteins, and play a role in presenting heterologous proteins in engineering transformation. To design a safer and a more efficient antigen delivery system, the naturally occurring OMVs have been modified by genetic engineering to obtain a multifunctional antigen delivery platform (Kim et al., 2009). By summarizing existing transformation methods, we hope to highlight the systematic processes of engineering OMVs (Figure 2).
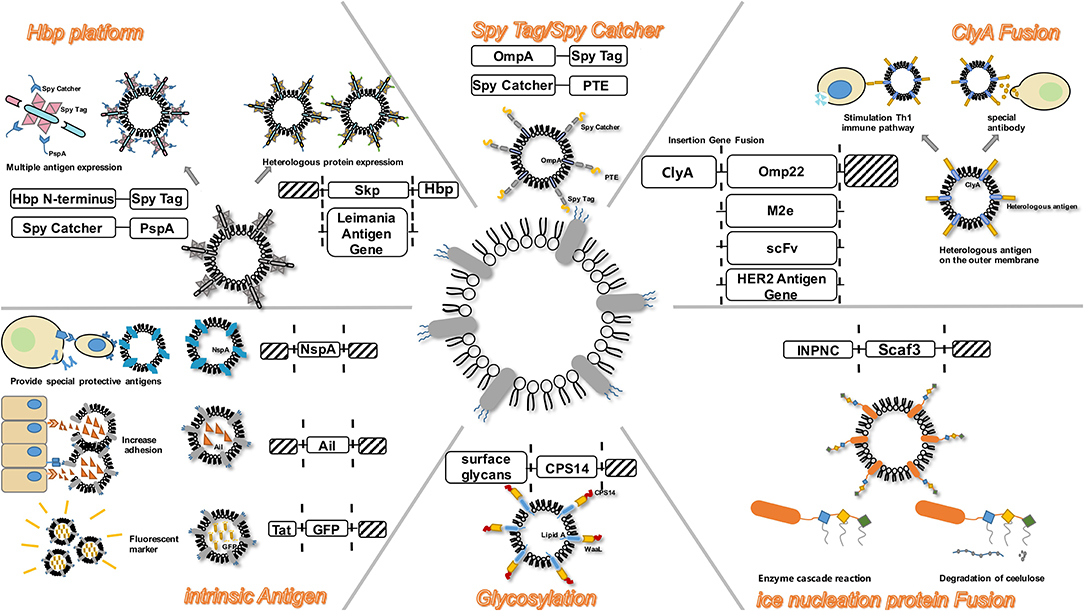
Figure 2. OMV serves as a versatile transformation platform. Engineering methods include: ClyA Fusion; Spy Tag/Spy Catcher; Hbp platform; intrinsic antigen; Glycosylation; Ice nucleation protein Fusion.
Presentation of Heterologous Proteins
The modification of OMVs, in terms of the presentation of heterologous antigens, is an established method. Since this modification alters the communication between the cells and the bacteria to a certain extent, it affects the functionality of OMVs as vaccines. Most of the heterologous antigens are heterologous proteins.
Intrinsic Antigens
The expression of intrinsic antigens in OMVs is widely used, and is closely associated with the development of vaccines. The symbiotic bacteria Neisseria gonorrhoeae and N. meningitidis share a tendency to overproduce the bacterial outer membrane, leading to the formation and release of OMVs during growth. OMVs from the meningococcal and commensal intima have been reported to function as a vaccine against meningococcal disease (Mirlashari et al., 2010). The nspA gene has been shown to be highly conserved in a variety of meningococcal strains, and has been extensively studied as a potential antigen for vaccines (Lewis et al., 2010, 2012; Echenique–Rivera et al., 2011; Giuntini et al., 2011). However, this method is difficult to commercialize. Previous studies have reported that recombinant E. coli was not suitable for the expression of other meningococcal OMV vaccine components, owing to its ability to genetically up-regulate certain outer membrane proteins in meningococcus, and separately engineer OMVs (O'dwyer et al., 2004). Ail is an outer membrane adhesin and invasin from Yersinia enterocolitica. Following genetic engineering, the OMVs of the E. coli DH5α strain carrying Ail can be internalized in the eukaryotic cells. The presence of Ail enabled non-invasive strains to invade the CHO cells (Kesty and Kuehn, 2004).
Moreover, green fluorescent protein (GFP) fused with the Tat signal sequence, was secreted into the periplasm by the double arginine transporter (Tat) in the E. coli DH5α strain. Pronase-resistant fluorescence was detected in the vesicles of the Tat-GFP transformed strain, demonstrating the presence of GFP in intact vesicles (Kesty and Kuehn, 2004). The inclusion of GFP cargo increased the vesicle density, but did not bring about any morphological changes in the vesicles. More than 60 membrane proteins have been detected in the OMVs (Molloy et al., 2000; Tokuda and Matsuyama, 2004; Lee et al., 2007). These proteins continue to be identified as anchoring proteins in the engineered OMVs. With further study of OMVs, more proteins with important functions will be identified.
Fusion of ClyA
Cytosolic A (ClyA), a 34-kDa cytosolic protein encoded by the ClyA gene (also known as sheA and hlyE), is located on the K-12 chromosome of E. coli (Westermark et al., 2000). Lately, researchers have favored the genetic fusion of ClyA. ClyA acts as a leading sequence and aids in the localization of heterologous proteins in the outer membranes where OMVs are secreted, resulting in the display of functional proteins on the surface of the OMVs (Kim et al., 2008). The gene for the heterologous protein expresses a recombinant outer membrane protein by fusion with the ClyA gene. Since ClyA is anchored to the outer membrane, the heterologous antigen can be expressed in an extracellular manner. Experiments on the heterologous protein presentation of ClyA-OMVs have been gradually extended. For example, expression of the Omp22 protein using the E. coli DH5α strain and the Fv fragment of the presenting antibody, the HER2 antibody, and the influenza virus M2e antibody, and so on (Kesty and Kuehn, 2004; Kim et al., 2008; Gujrati and Sangyong, 2014; Garrett Rappazzo et al., 2016; Huang et al., 2016).
Vesicles produced by bacterial pathogens promote the direct delivery of antigens into the host cells. In this regard, the recombinant ClyA-OMV increases the antigen delivery efficiency by 8-fold (Wai et al., 2003). Further research shows that the protein fusion on ClyA has different effects. Fusion with the N-terminal of ClyA produces heterologous antigens that are inconsistent, while C-terminal fusions always produce well-displayed proteins which retain their biological functions (Kim et al., 2008). This may be due to the fact that the C-terminus of ClyA is closer to the outer surface compared to the distance between the N-terminus of ClyA and the outer membrane, and is therefore more likely to extend into the extracellular environment (Tzokov et al., 2006; Eifler et al., 2014). Additionally, the physiological state of ClyA also affects its localization in the OMVs. According to experiments, the redox state of ClyA is pivotal in regulating its localization in the OMVs (Kim et al., 2008).
AT Platform
Autotransport (AT) is a large virulence factor (usually over 100 kDa) secreted by Gram-negative bacteria. The AT system is simple, and is hypothesized to carry all information concerning the inner periplasm, and the outer membrane translocation of the protein itself. AT is synthesized as a large precursor protein containing three domains: first, the N-terminal signal peptide, which targets the protein to the Sec translocator and initiates the transfer through the intima; second, the passenger domain; third, the C-terminal beta-domain, which integrates into the outer membrane and plays a crucial role in passenger translocation into the extracellular space (Eifler et al., 2012). The AT system has been used as a transport vehicle for many heterologous fusion partners in Gram-negative bacteria (Kjaergaard et al., 2000; Junker et al., 2019). In most cases, variants of the Neisseria IgA protease and the endogenous E. coli AT AIDA-I are used to engineer antigens specifically for surface display (Junker et al., 2007). Two model antigens of Leishmania were successfully expressed using this method (Schroeder and Aebischer, 2009). Recently, the skp protein of ETEC has been successfully loaded into OMVs through AT, which fused with the glutathione-S-transferase (GST) epitope and combined with cholera toxin to prepare an ideal vaccine (Hays et al., 2018). OspA has been extensively studied as a vaccine component against Lyme disease (Wressnigg et al., 2014; Comstedt et al., 2015). The OspA antigen has been successfully expressed in OMVs but has not been exposed on the surface of OMVs. The improved method uses a meningococcal surface protein, fHbp, as the anchor protein. By connecting OspA to the N-terminus of fHbp, the strong immune response elicited by the engineered OMVs was successfully detected (Salverda et al., 2016; Zhang et al., 2016). Ovalbumin (OVA) hemoglobin protease has also been successfully expressed on the surface of engineered OMVs through the AT platform, and has been shown to have the ability to induce antigen-specific CD8 + T cell responses (Schetters et al., 2019). Unlike ClyA, the AT system can successfully present multiple antigens using a slightly different approach. Using the crystal structure of the AT passenger, five passenger side domains were selected and replaced with the M. tuberculosis antigen ESAT6 sequentially, while the β-helix core structure (β-stem) remained intact (Otto et al., 2005). The resulting Hbp-ESAT6 chimera was efficiently and stably secreted into the medium of E. coli. The Hbp-ESAT6 display variant was constructed by disrupting the cleavage site between the passenger and the β-domain, which maintains cell correlation and promotes efficient surface exposure of ESAT6 (Eifler et al., 2012). Compared to the ClyA fusion system, the AT system has multiple sites for the insertion of heterologous sequences that can simultaneously bind multiple antigens. This enables the formation of a multivalent recombinant live vaccine. This is of high value because multivalent vaccines are a key requirement for the prevention of infectious diseases such as tuberculosis (Aagaard et al., 2011). Importantly, since the passenger side domains have Hbp function, their replacement with antigenic proteins automatically eliminates potential toxic effects, making the proposed Hbp platform safe for antigen delivery (van Bloois et al., 2011). Thus, the AT system has been found to be more efficient, convenient, and safer than the engineered OMVs associated with ClyA.
Spy Tag/Spy Catcher System
The SpyCatcher is a genetically encoded protein designed to spontaneously form a covalent bond with its peptide partner SpyTag (Brune et al., 2016). Studies have demonstrated the ability of this bioconjugate system to deliver antigens, which can be linked to an antigen delivery system via common cell-linking proteins (Alves et al., 2015). It has been found that this system can be used as a modification of the autotransport system (Jong et al., 2010). In the AT system, although antigen delivery has been successfully accomplished, their density on the OMVs has been found to be affected (Jong et al., 2010). The SpyCatcher/SpyTag protein ligation technology can enzymatically attach the antigen to the high-density Hbp in OMVs, without being hampered by the membrane environment, as verified by recent experiments. This protein ligation system is based on the fibronectin of Streptococcus pyogenes, and forms an intramolecular isopeptide bond between the adjacent lysine and aspartate residues by an autocatalytic mechanism (Zakeri et al., 2012). The domain was split into a 13 amino acid (AA) peptide called the SpyTag, and a 138-AA fragment called the SpyCatcher, to create a SpyCatcher/SpyTag pair for coupling with heterologous proteins and autotransporters. When fused to different proteins, the SpyTag and the SpyCatcher recombine with the domain following mixing and form stable intermolecular covalent bonds under certain conditions (Fierer et al., 2014). Additionally, a similar structure, the orthogonal SnoopTag/SnoopCatcher system, can also couple multiple antigenic modules to Hbp by a sequential ligation strategy without affecting antigenic display (Veggiani et al., 2016). Spy ligation can be easily performed by gene fusion to the end of the Hbp display vector, and directional loading onto the OMVs. OMV display technology, combined with protein linkages, constitutes a plug-and-play system designed for customized antigen delivery. As a “mediating” protein system, the Spy Tag/Spy Catcher can also link other anchoring proteins such as OmpA, to deliver the corresponding antigen. This has been verified in the presentation of the active enzyme phosphodiesterase (PTE) (Alves et al., 2015). Therefore, the Spy Tag/Spy Catcher system can be used for modifying anchoring proteins, allowing engineered OMVs to perform antigen delivery more efficiently.
Ice Nucleation Protein
To display multiple functional proteins on OMVs, a trivalent scaffold (Scaf3) plasmid was constructed to express the protein of interest (Tsai et al., 2009). Park et al. (2014) used an enzyme scaffold to express the protein in bacteria, and immobilized it on the outer membrane using a truncated ice nucleation protein anchor. The plasmid pINP was constructed by amplification using a truncated ice nucleation protein gene from pPNC20 (Bae et al., 2002). High levels of INP-scaf3 were successfully detected on engineered E. coli OMVs, by binding Scf3 to INP by primers (Park et al., 2014). This provides a simple and convenient way to express different enzymes on OMVs. The assembled enzyme complex not only retains full activity, but also leads to the hydrolysis of cellulose, 23 times faster than the conventional enzymes. This method can thus be used as a simple platform for the efficient functioning of multiple enzymes as nano-biocatalysts (Park et al., 2014). The flexibility to design scaffolds using various specific binding domains allows for the display of unlimited number of functional proteins on OMVs.
Similar to the AT system, the enzyme scaffold is also a high-performance system that can simultaneously load multiple heterologous proteins. However, the AT system has certain inevitable shortcomings, like a truncated passenger structure used in the loading of multiple antigens (Jong et al., 2010). Similar to the AT system, the enzyme scaffold also binds the target protein in an orderly manner, forming a multi-functional system independently. Therefore, it is expected to demonstrate more applications.
Glycosylation Linkage
Glycosylation-modified OMVs (geOMVs), as seen in Streptococcus pneumoniae capsules and C. jejuni N-linked glycans, have been shown to produce a good immune response (Price et al., 2016). The geOMV platform utilizes the loose glycan structure of the O-antigen ligase. Specifically, they are delivered to the lipid A-core molecule in the inner membrane (Kaniuk et al., 2004; Han et al., 2012). First, glycans were customized using the initial glycosyltransferase, WecA, and a specific glycosyltransferase encoded on a plasmid. The flip enzyme Wzx subsequently moved the glycans to the periplasmic surface of the inner membrane, and the enzyme Wzy polymerized the glycan subunits into a high-molecular-weight form of the surface glycans. The WaaL ligase then attached the glycan to the lipid A-core molecule. Finally, these customized LPS molecules were transported to the outer membrane and flipped into the extracellular space through the Lpt protein complex (Kaniuk et al., 2004; Simpson et al., 2015). When bacteria formed the OMVs, LPS as well as proteins and lipids were packaged into OMVs, resulting in the formation of mature geOMVs. The S. pneumoniae capsules were expressed in E. coli cells lacking the relevant O antigen, and the relevant plasmid was constructed and introduced into E. coli to secrete the corresponding OMVs. The immunological potency was confirmed using animal experiments (Price et al., 2016). Glycosylated conjugate vaccines are valuable. However, technical challenges exist with respect to the attachment of glycan chains (Jones, 2005; Wacker et al., 2006; Bottomley et al., 2012). The glycol-engineered E. coli-expressing OMVs can express the glycans required for binding to the vaccine, and thus can complement the current conjugate vaccines, particularly for serotypes corresponding to glycan structures that have not been resolved. The studies of glycans were less than those of proteins, lipids, and nucleic acids (Williams et al., 2018). The successful validation of glycosylated OMVs provides an inspiring platform for future engineered glycosylation efforts (Liang et al., 2014; Shimoda et al., 2017).
Other Applications of OMVs Using Genetic Engineering
In addition to presenting antigens for vaccine application, the transformation of OMVs has now embraced more functions, including drug delivery, bioimaging, nanogenerators, and new adjuvants (Table 2). OMVs are being developed for more applications due to their flexible retrofit characteristics.
Drug Delivery
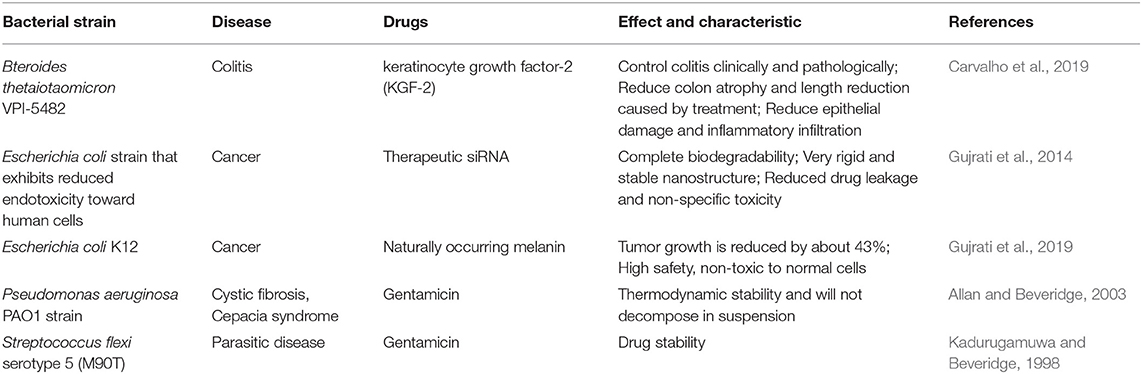
In addition to serving as a platform for surface antigen presentation, the function of OMVs to encapsulate small molecule particles in the lumen for presentation is also of interest. Recent studies have found that OMVs have a great potential in the transport of small molecule drugs (Table 3). The method of loading drugs into the lumen of OMVs by electroporation has been proven. For relatively hydrophobic and positively charged small molecule drugs that easily interact with lipophilic membranes, passive diffusion can be used. However, for hydrophilic molecules, the bilayer lipid membrane may constitute an obstacle, in which case electroporation may be the method of choice (Gujrati et al., 2014) (Figure 3). Although OMVs can only encapsulate about 15% of KSP-siRNA, the amount is sufficient to exert cytotoxicity. This electrotransport method is convenient and easy to implement, and has been widely used in loading small molecular substances (Turner et al., 2015). Recently, scientists have tried to use bioengineering methods to successfully upload cancer treatment drugs on OMVs. OMVs from the non-pathogenic commensal bacteria were used to deliver keratinocyte growth factor 2 (KGF-2), a small molecule with the potential to treat inflammatory bowel disease (Baumgart and Sandborn, 2007). Adding intact KGF-2 OMVs to epithelial cell culture promoted cell proliferation and accelerated wound healing, in a safe manner (Carvalho et al., 2019). Melanin, a specific agent for photothermia therapy in cancer treatment, was presented through OMVs, and almost all the cancer cells in the animal model were destroyed (Gujrati et al., 2019) (Figure 4). Another loading method is the incorporation of drugs into OMVs during their biological generation through parental bacteria. Further to cancer treatment, drug delivery by OMVs has also been confirmed. Allan and Beveridge (2003) have demonstrated that the incorporation of gentamicin during the growth of P. aeruginosa PAO1 strains produced gentamicin-containing OMVs, which can deliver drugs to the target, Burkholderia cepacia (Allan and Beveridge, 2003). This was also confirmed in Shigella flexneri OMVs (Kadurugamuwa and Beveridge, 1998).
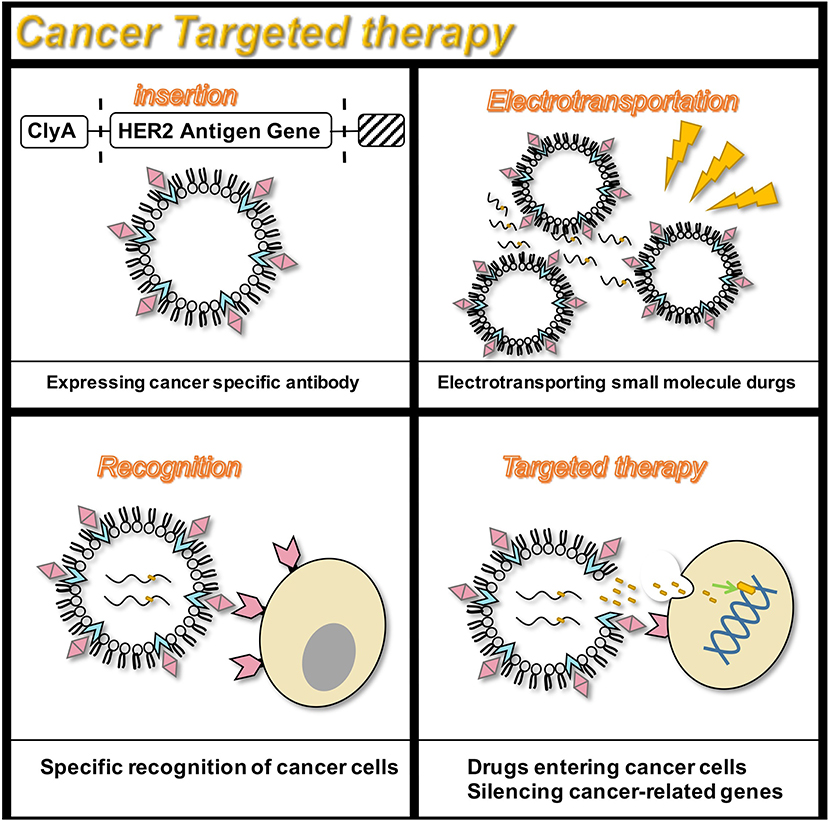
Figure 3. Targeted therapy for the HER2 receptor. OMVs are genetically engineered to carry siRNA that silence cancer genes, and specifically target cancer cells that express the HER2 antigen to exert anti-cancer effects.
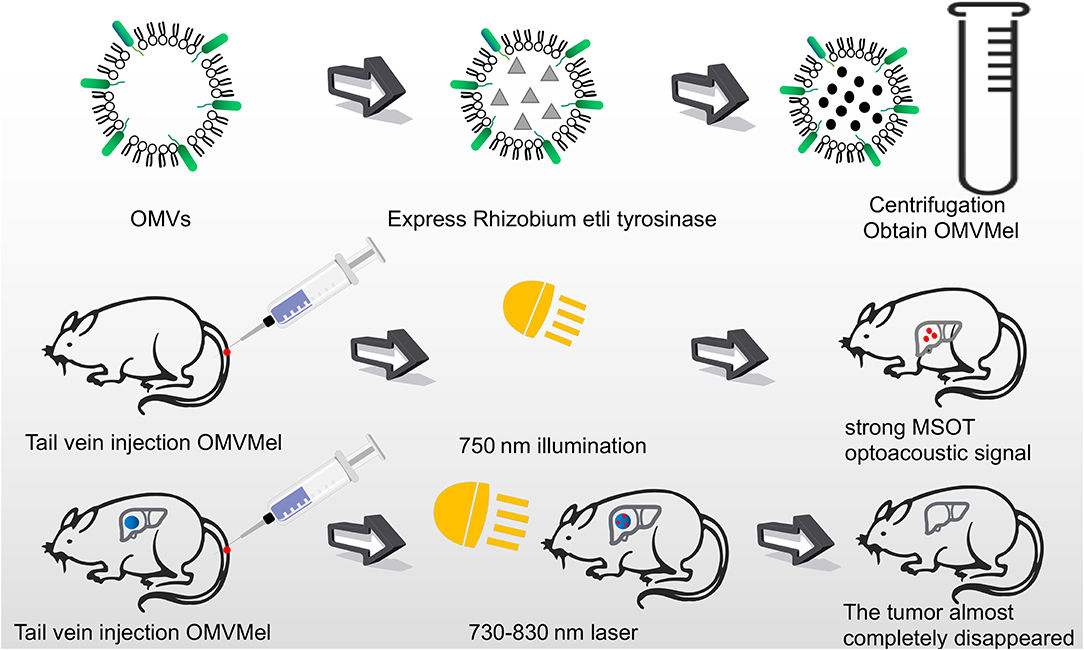
Figure 4. OMVs loaded with melanin were injected into mice through the tail vein. After irradiating at 750 nm, a strong photoacoustic signal of MSOT was generated. The tumor cells were almost completely killed by laser irradiation at 730–830 nm for several minutes.
As a drug presentation tool, OMVs have the advantage of being small, stable, well-tolerated, and highly specific. OMVs can slowly release drugs under specific Physiological conditions, reducing non-specific toxicity caused by drug leakage into the circulatory system. In addition, this slow-release property can provide sufficient time for siRNA to reach its target cancer cells in areas with abnormal blood supply (Gujrati et al., 2014). OMVs have thermodynamic stability and will not decompose in suspension, while ordinary liposomes have the disadvantages of drug leakage and easy degradation of circulation. Although the packaging efficiency is not ideal, it is sufficient to exert drug efficacy.
Multifunctional Imaging and Detection Platforms
Complex, multi-antigen-bound engineered OMVs have begun to evolve and grow. Based on this technology, scientists have proposed a “one-pot synthesis” model (Chen et al., 2017). According to the mutual binding between proteins, the nano-luciferase (Nluc) and the Z domain recruits antibodies which exists in OMVs at the same time (Tashiro et al., 1997). Experiments have confirmed the feasibility of imaging cancer cells by multifunctional OMVs (Ferreira et al., 2006). Currently, multifunctional OMVs can be customized with ease through genetic engineering, creating an almost unlimited combination of “capture” and “report,” for a wide range of applications including live cell imaging (Chen et al., 2017) (Figure 5). At the same time, the photoacoustic imaging method has proven to be feasible to produce encapsulated biopolymer-melanin OMV (OMV Mel), using bacterial strains expressing the tyrosinase transgenes. The OMV Mel is generated under near-infrared light, and is suitable for imaging applications. The strong photoacoustic signal non-invasively monitors tumor-associated OMV Mel distribution in vivo (Gujrati et al., 2019). This experiment represents the rationale on the use of bioengineered OMVs as effective alternatives to synthetic particles in photoacoustic imaging, for image enhancement and photo-thermal applications (Figure 4).
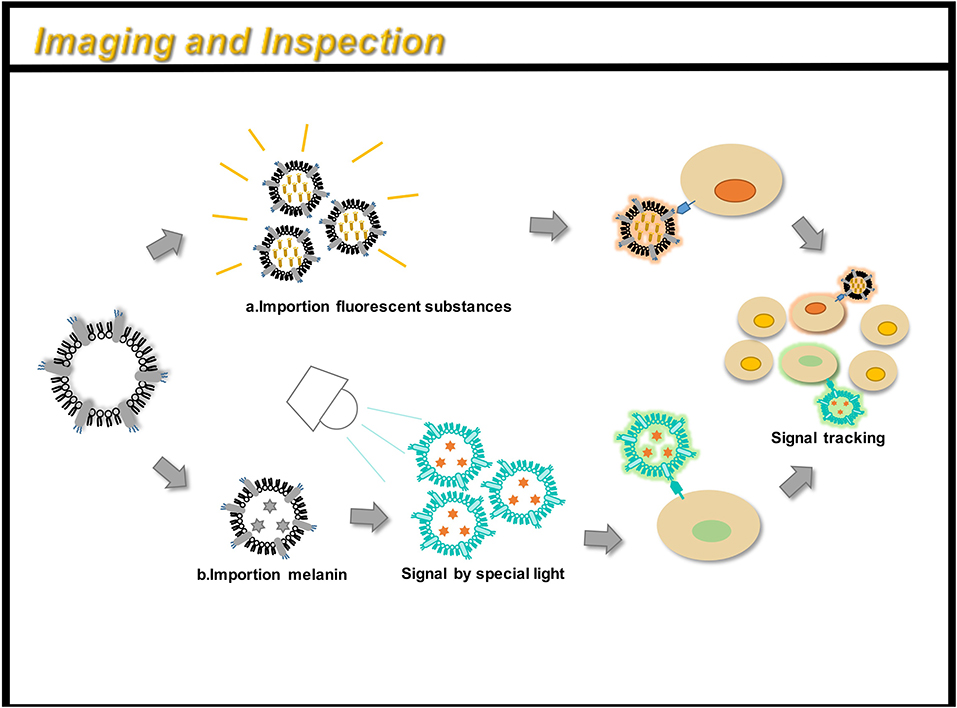
Figure 5. OMVs can be engineered for immunoimaging and cell detection. There are two main methods: (a) directly loading a fluorescent substance; (b) indirectly loading a small molecular substance (such as melanin), and a signal can be obtained under a specific light. By taking OMV preparation, you can track the path of action of the drug in the body and monitor its efficacy.
OMVs have been reported to possess excellent sensitivity. When used in thrombin detection, the detection limit of thrombin by OMVs was found to be 0.5 nm (Chen et al., 2017). Additionally, the preparatory method is simple, and a high yield of OMVs can be successfully obtained with ease only through a simple fermentation process. Furthermore, since more than 60 membrane-anchoring proteins have been detected in OMVs, a variety of modified enzymes can be presented by OMVs. Therefore, OMVs can flexibly modify electrochemical-based detection or colorimetric detection of enzymes. Owing to these advantages, OMVs can be applied in immunoassays, in vivo imaging, and cancer cell detection.
Nanopolymer Bioreactor
Multi-step enzyme pathways play a key role in cell metabolism. Enzyme cascade reactions are highly complex systems. A key feature of a highly efficient enzymatic pathway is the synergistic and spatial organization of the enzyme to ensure sequential transformation of the substrate (Agapakis et al., 2012). OMV is relatively stable in the surrounding environment and can be manufactured cost-effectively, and is considered to have the potential of a bioreactor. A novel study by Park et al. (2014) identified a single protein scaffold based on the Coh-Doc interaction, which can be used to sequentially assemble multiple enzymes onto the surface of OMVs, thereby resulting in a nanoreactor capable of complex biocatalysis (Park et al., 2014) (Figure 6). By genetically engineering enzyme scaffolds into OMVs, scientists have succeeded in obtaining an enzyme system with an enzyme cascade. Recently, the expression of organophosphate hydrolase in E. coli OMVs has also been successful, and engineered OMVs have enhanced thermal stability and pH stability (Su et al., 2017). After 15 reaction cycles, at least 83% of the OMVs initial activity is retained. After 40 days, about 20–30% of initial activity is retained (Su et al., 2017). In addition, phosphotriesterase was displayed in OMVs through the outer membrane porin OmpA of E. coli (Alves et al., 2018).
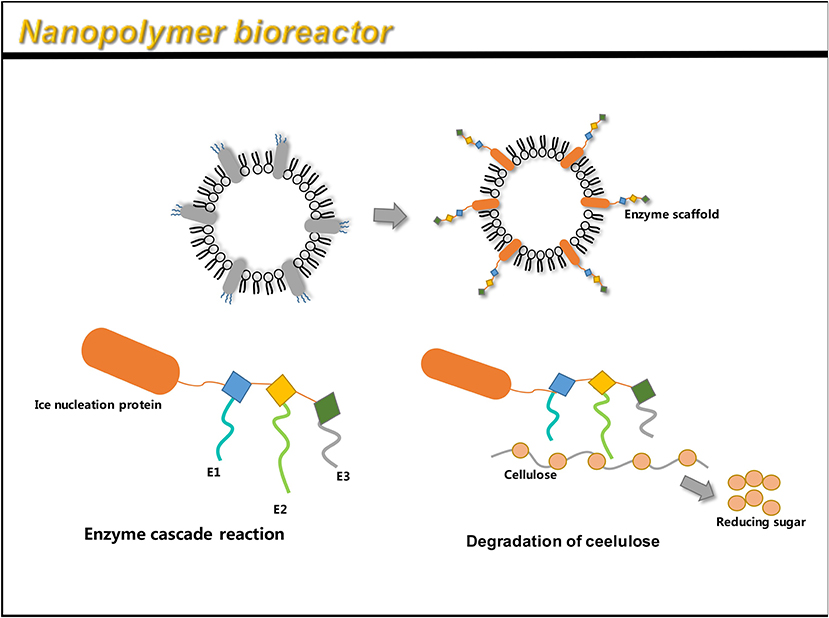
Figure 6. OMV can be linked to an enzyme scaffold through gene functions to connect three enzyme molecules that can complete a complete set of reactions. Through the action of enzymes, cellulose is successfully hydrolyzed to reduce sugars.
In recent decades, various methods have been developed to create artificial multi-enzyme complexes, including OMVs, polyhydroxyalkanoate (PHA) particles, virus-like particles (VLP), enzyme-derived nanoparticles (EZPs), and magnetosomes (Parlane et al., 2017; Yan et al., 2017; Wilkerson et al., 2018; Schmid-Dannertand and López-Gallego, 2019). However, in terms of its wider application, there are still some obstacles. For example, due to steric hindrance, the encapsulation rate of large proteins is low, and the substrate cannot fully enter the enzyme assembly and the vesicle shell (Wong et al., 2020). These nanoparticles are difficult to challenge for long-term storage and have poor biocompatibility. The enzyme system expressed through the outer membrane vesicles has been proven stable for long-term storage and has the potential to be used as a carrier for industrial production of enzyme systems.
As a Vaccine Adjuvant
An adjuvant is defined as a molecule that aids an antigen in eliciting an immune response. Adjuvants improve the immune response in people with poor immune response, expand cross-protection, induce a shift in the specific T cell-assisted response, and reduce the amount of antigen required for vaccine development (Maisonneuve et al., 2014). OMVs have been considered as potential vaccine adjuvants since their inception (Figure 7). However, due to the toxicity of wild-type LPS, such as flagellin, lipoprotein, and other OMPs, they may trigger allergic reactions such as excessive inflammation (Arigita et al., 2005; Thompson et al., 2005). Therefore, the OMV endotoxins are removed manually following production. Another disadvantage is that the LPS-deficient OMVs generally exhibit lower immunogenicity compared to the immunogenicity of the wild-type OMVs containing wild-type LPS (Gnopo et al., 2017). Therefore, the appropriate modification of LPS is necessary to obtain an optimal balance between low-toxicity and high-immunogenicity. The latest method of LPS modification involves the synthetic assembly or genetic modification of bacteria (Arenas et al., 2010). Further, OMVs with LpxL gene deletion and PagL gene expression have been proven to possess good adjuvant ability and show significantly better abilities compared to that of the wild type (Arenas et al., 2010; van de Waterbeemd et al., 2010; Asensio et al., 2011). Further comparison has revealed that the PagL gene expression possesses higher endotoxic activity compared to that of the LpxL gene, which was also supported by animal studies. The PagL gene is considered to have a better adjuvant effect (Asensio et al., 2011).
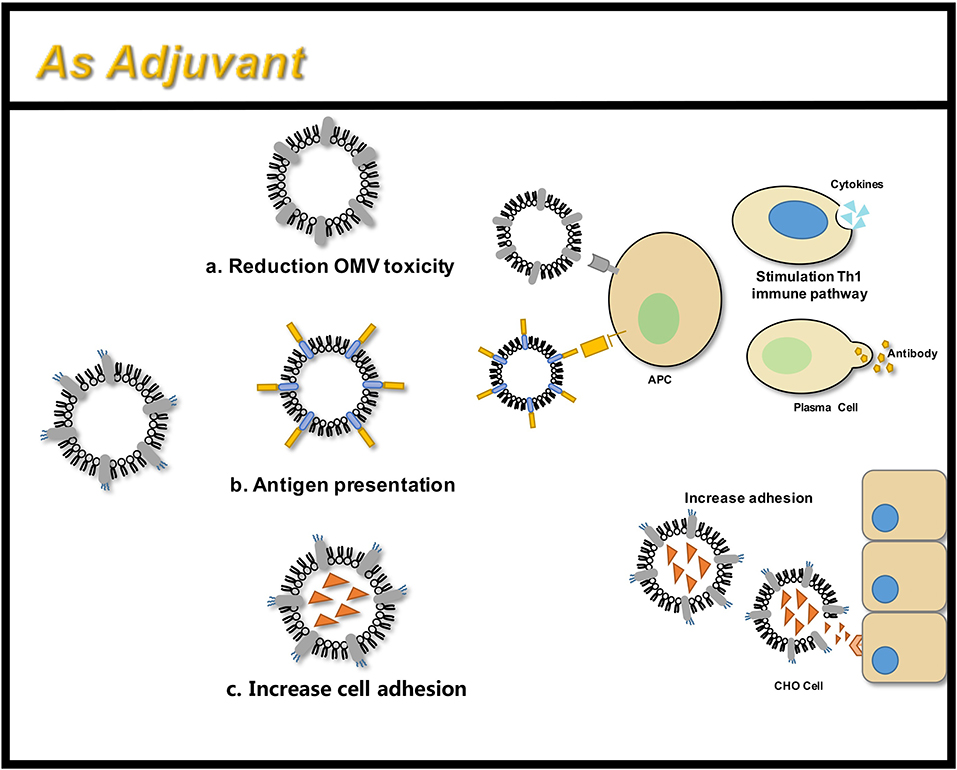
Figure 7. OMVs function as an adjuvant through three main areas: (a) attenuated antigen stimulation; (b) presentation of heterologous antigens; (c) increased adhesion to tissue cells.
Recent studies have shown that the adjuvant ability of engineered OMVs continues to attract the attention of researchers. Engineered OMVs carrying the Ail antigen have been shown to increase adhesion to the CHO cells, and non-invasive vesicles have been modified to invade the CHO cells (Kesty and Kuehn, 2004). Recent developments in vaccine research involve the use of more antigen profiles, expressing mutant forms of HlaH35L, SpAKKAA, LukE, LukD/E, one of the two components of the interleukin Csa1A, and the iron binding protein, FhuD2, on meningococcal OMVs, have proven to possess good immunogenicity (Falugi et al., 2013; Gasanov et al., 2013; Spaan et al., 2017). The OMVs produced in Salmonella carrying pneumococcal protein antigens have shown promising results in a murine model (Kuipers et al., 2015). This allows glycans and proteins to synergistically enhance the immunogenicity of vaccines (Price et al., 2016). In contrast, the currently existing and the most successful antimicrobial vaccine glycoconjugates face multiple problems like complex preparation, slow development, insufficient safety, instability, and batch-to-batch variability. In addition, glycoconjugates do not contain multiple serotypes. OMVs can be genetically modified with flexibility and ease to produce geOMVs with the perfect balance between reduced toxicity and optimal adjuvant properties.
Future Research Directions
Compared to conventional adjuvants, OMVs show a clear advantage in terms of future development (Tan et al., 2018). As a platform for the convenient implementation of genetic modification, OMVs have received attention in the field of precision medicine, in addition to a wide range of vaccine applications (Wang et al., 2019). For example, OMVs have become a potential choice for the targeted delivery of cancer drugs. As an easily engineered nanoparticle, OMVs can transport a variety of small molecules with different characteristics, thereby involving in important applications such as immunoimaging, enzyme-linked reaction polymers, etc. Further research will help to unveil the undiscovered aspects and future applications of the engineered OMVs.
Targeted Cancer Therapy Delivery Platform
Targeted therapy continues to receive attention owing to its fewer side effects. Bio-nanocarriers derived from a variety of natural resources like pathogens and mammalian cells have been identified to be well-suited for drug delivery (Yoo et al., 2011), and nanomaterials have been widely studied as drug delivery vehicles in the treatment of cancer (Petros and DeSimone, 2010; Shi et al., 2010). Designing OMVs to target specific cell types makes them an attractive treatment option. In addition, engineering OMVs offers several advantages. The outer membrane protects the OMV contents such as chemotherapeutic agents, from protease or nuclease degradation, thereby increasing the circulating half-life of the drug. Recent studies have successfully designed tumor-targeted OMVs carrying an anti-tumor siRNA (Gujrati and Sangyong, 2014; Gujrati et al., 2014). The transmembrane receptor HER2, chosen as the cell-specific target, was found to be overexpressed in a range of cancers, including breast, ovarian, and gastric cancers (Weaver and Cleveland, 2005). By binding HER2 antibody to ClyA, OMVs targeting cancer cells were obtained. In vitro studies by Gujrati et al. (2014) have demonstrated the ability of the siRNA-loaded OMVs to deliver siRNA in ovarian and breast cancer cell lines, subsequently inducing cancer cell death (Gujrati et al., 2014). Multiple forms of cancer treatment have been proven feasible. By presenting melanin, a substance with good light-to-heat conversion rate, a treatment can be achieved that can kill tumor cells by selectively irradiating the melanin-containing tissues, thereby inducing local heating (Liu et al., 2013). Animal studies have shown that almost all tumor cells in the body were eliminated. Furthermore, this photothermal therapy monitors the effect of local treatment by photoacoustic imaging (Gujrati et al., 2019). Additionally, recent studies have shown that engineered OMVs, which transport OVA hemoglobin protease using the AT system, have a mediatory function in the induction of antigen-specific CD8+ T cell response (Schetters et al., 2019). CD8+ T cells control viral replication and tumor growth. This property of OMVs also hints at its powerful potential in cancer treatment.
OMVs deliver heterologous proteins thus, acting as tools for delivering targeted drugs. Such vesicles are expected to be further developed in the near future. The advent of nanobiotechnology, and advances in genomics, immunology, microbiology, and vaccine needs, alone and in combination, have made OMVs a novel choice for precision therapies and targeted-drug therapies.
Multifunctional Delivery Platform
In addition to the widely known function of antigen presentation, OMVs have been used in a wide array of applications. For example, OMVs can be used as bioreactors for enzyme cascade reactions. By carrying anchor proteins which load multiple heterologous proteins such as ice nucleating proteins, OMVs can combine multiple related enzymes to complete a more complex set of biological reactions (Park et al., 2014). Additionally, engineered OMVs have also been used in the field of immune bioimaging. OMVs can be directly tagged with fluorescent substances that can be easily detected, such as GFP, and perform functions such as in vivo imaging and remote tracking (Chen et al., 2017). Indirect detection methods have also been confirmed. OMVs carrying melanin generate strong photoacoustic signals suitable for imaging applications under near-infrared light (Gujrati et al., 2019). This method has also proven to be therapeutically effective. For rendering OMVs in various platforms, secure performance is a pre-requisite. In addition to existing methods for attenuating gene knockouts, the use of non-pathogenic symbiotic bacteria as a source of OMVs has also received attention (Carvalho et al., 2019). Reconstruction of these OMVs can easily produce engineered OMVs with high safety.
With further study of OMVs, it has been shown that this bio-nanoparticle has a variety of functions not limited to vaccine adjuvants. Relying on flexible genetic modifications, OMVs perform multiple functions by carrying small molecules with multiple functions. OMVs are expected to demonstrate more applications in the fields of immunology, diagnostics, and so on.
Conclusion
OMVs have attracted attention as a new engineering platform for vaccine development. Through research on the engineering of OMVs, we find that the applications of OMVs are not limited to vaccines. As a convenient bio-nanoparticle, OMV has the potential to function as a multi-functional delivery platform, and thus can be applied in many fields such as immunology, clinical medicine, diagnostics, and biochemistry. However, there are still some problems in the application of OMVs: (i) the toxicity of LPS on OMVs is still a major concern, especially when applied in the human body; (ii) in biological engineering, OMVs load heterologous proteins and the efficiency of small molecular substances is not high; (iii) in large-scale production, the size of OMVs is not easy to control, and the difference between batches is large. At the same time, the lack of understanding of the exact assembly mechanism of OMVs leads to poor programmability of the particles. In future research, attention should be paid to other problems of modifying OMVs such as: (i) increasing the load rate; (ii) improving safety and reduce costs when large-scale production is required; and (iii) delineating the mechanism of OMVs germination for a better understanding of the mechanism of its functions. This review focuses on some of the latest advances in the application of engineered OMVs such as drugs for cancer treatment, immune imaging, etc. It is anticipated that the functions of OMVs may be better manipulated through genetic engineering in subsequent studies.
Author Contributions
RL wrote the manuscript. QL and RL revised the manuscript for integrity and accuracy. QL approved the final version of the manuscript and takes responsibility for its content. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (31760261), the Science and Technology Research Project of Jiangxi Provincial Education Department (60224), and Key Research and Development Projects of Jiangxi Natural Science Foundation (20192BBG70067).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
OMVs, Outer membrane vesicles; LPS, lipopolysaccharide; PRR, pattern recognition receptors; GFP, green fluorescent protein; ClyA, Cytosolic A; AT, Autotransport; GST, glutathione-S-transferase; OVA, Ovalbumin; β-stem, β-helix core structure; AA, amino acid; PTE, phosphodiesterase; Scaf3, trivalent scaffold; geOMVs, Glycosylation-modified OMVs; KGF-2, keratinocyte growth factor 2; Nluc, nano-luciferase; OMV Mel, biopolymer-melanin OMV.
References
Aagaard, C., Hoang, T., Dietrich, J., Cardona, P.-J., Izzo, A., Dolganov, G., et al. (2011). A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat. Med. 17, 189–194. doi: 10.1038/nm.2285
Agapakis, C. M., Boyle, P. M., and Silver, P. A. (2012). Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat. Chem. Biol. 8, 527–535. doi: 10.1038/nchembio.975
Allan, N. D., and Beveridge, T. J. (2003). Gentamicin delivery to Burkholderia cepacia group IIIa strains via membrane vesicles from Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47, 2962–2965. doi: 10.1128/AAC.47.9.2962-2965.2003
Alves, N. J., Moore, M., Johnson, B. J., Dean, S. N., Turner, K. B., Medintz, I. L., et al. (2018). Environmental decontamination of a chemical warfare simulant utilizing a membrane vesicle-encapsulated phosphotriesterase. ACS Appl. Mater. Interfaces 10, 15712–15719. doi: 10.1021/acsami.8b02717
Alves, N. J., Turner, K. B., Daniele, M. A., Oh, E., Medintz, I. L., and Walper, S. A. (2015). Bacterial nanobioreactors-directing enzyme packaging into bacterial outer membrane vesicles. ACS Appl. Mater. Interfaces 7, 24963–24972. doi: 10.1021/acsami.5b08811
Arenas, J., van Dijken, H., Kuipers, B., Hamstra, H. J., Tommassen, J., and van der Ley, P. (2010). Coincorporation of LpxL1 and PagL mutant lipopolysaccharides into liposomes with Neisseria meningitidis opacity protein: influence on endotoxic and adjuvant activity. Clin. Vaccine Immunol. 17, 487–495. doi: 10.1128/CVI.00423-09
Arigita, C., Luijkx, T., Jiskoot, W., Poelen, M., Hennink, W. E., Crommelin, D. J. A., et al. (2005). Well-defined and potent liposomal meningococcal B vaccines adjuvated with LPS derivatives. Vaccine 23, 5091–5098. doi: 10.1016/j.vaccine.2005.06.001
Asensio, C. J. A., Gaillard, M. E., Moreno, G., Bottero, D., Zurita, E., Rumbo, M., et al. (2011). Outer membrane vesicles obtained from Bordetella pertussis tohama expressing the lipid A deacylase PagL as a novel acellular vaccine candidate. Vaccine 29, 1649–1656. doi: 10.1016/j.vaccine.2010.12.068
Bae, W., Mulchandani, A., and Chen, W. (2002). Cell surface display of synthetic phytochelatins using ice nucleation protein for enhanced heavy metal bioaccumulation. J. Inorg. Biochem. 88, 223–227. doi: 10.1016/S0162-0134(01)00392-0
Bartolini, E., Ianni, E., Frigimelica, E., Petracca, R., Galli, G., Scorza, F. B., et al. (2013). Recombinant outer membrane vesicles carrying chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J. Extracell. Vesicles 2:20181. doi: 10.3402/jev.v2i0.20181
Bauman, S. J., and Kuehn, M. J. (2006). Purification of outer membrane vesicles from Pseudomonas aeruginosa and their activation of an IL-8 response. Microb. Infect. 8, 2400–2408. doi: 10.1016/j.micinf.2006.05.001
Baumgart, D. C., and Sandborn, W. J. (2007). Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet 369, 1641–1657. doi: 10.1016/S0140-6736(07)60751-X
Bottomley, M. J., Serruto, D., Sáfadi, M. A. P., and Klugman, K. P. (2012). Future challenges in the elimination of bacterial meningitis. Vaccine 30, B78–B86. doi: 10.1016/j.vaccine.2011.12.099
Brune, K. D., Leneghan, D. B., Brian, I. J., Ishizuka, A. S., Bachmann, M. F., Draper, S. J., et al. (2016). Plug-and-display: decoration of virus-like particles via isopeptide bonds for modular immunization. Sci. Rep. 6:19234. doi: 10.1038/srep19234
Carita, A. C., Eloy, J. O., Chorilli, M., Lee, R. J., and Leonardi, G. R. (2018). Advances, and perspectives in liposomes for cutaneous drug delivery. Curr. Med. Chem. 25, 606–635. doi: 10.2174/0929867324666171009120154
Caruana, J. C., and Walper, S. A. (2020). Bacterial membrane vesicles as mediators of microbe - microbe and microbe - host community interactions. Front. Microbiol. 11:432. doi: 10.3389/fmicb.2020.00432
Carvalho, A. L., Fonseca, S., Miquel-Clopés, A., Cross, K., Kok, K.-S., Wegmann, U., et al. (2019). Bioengineering commensal bacteria-derived outer membrane vesicles for delivery of biologics to the gastrointestinal and respiratory tract. J. Extracell. Vesicles 8:1632100. doi: 10.1080/20013078.2019.1632100
Cecil, J. D., O'Brien-Simpson, N. M., Lenzo, J. C., Holden, J. A., Chen, Y.-Y., et al. (2016). Differential responses of pattern recognition receptors to outer membrane vesicles of three periodontal pathogens. PLoS ONE 11:e0151967. doi: 10.1371/journal.pone.0151967
Chatterjee, D., and Chaudhuri, K. (2011). Association of cholera toxin with vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett. 585, 1357–1362. doi: 10.1016/j.febslet.2011.04.017
Chen, D. J., Osterrieder, N., Metzger, S. M., Buckles, E., Doody, A. M., DeLisa, M. P., et al. (2010). Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc. Natl. Acad. Sci. U.S.A. 107, 3099–3104. doi: 10.1073/pnas.0805532107
Chen, Q., Rozovsky, S., and Chen, W. (2017). Engineering multi-functional bacterial outer membrane vesicles as modular nanodevices for biosensing and bioimaging. Chem. Commun. 53, 7569–7572. doi: 10.1039/C7CC04246A
Collins, B. S. (2011). Gram-negative outer membrane vesicles in vaccine development. Discov. Med. 12, 7–15.
Comstedt, P., Hanner, M., Schüler, W., Meinke, A., Schlegl, R., and Lundberg, U. (2015). Characterization and optimization of a novel vaccine for protection against lyme borreliosis. Vaccine 33, 5982–5988. doi: 10.1016/j.vaccine.2015.07.095
Deatherage, B. L., Lara, J. C., Bergsbaken, T., Barrett, S. L. R., Lara, S., and Cookson, B. T. (2009). Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72, 1395–1407. doi: 10.1111/j.1365-2958.2009.06731.x
Derouiche, R., Bénédetti, H., Lazzaroni, J. C., Lazdunski, C., and Lloubès, R. (1995). Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J. Biol. Chem. 270, 11078–11084. doi: 10.1074/jbc.270.19.11078
Devoe, I. W., and Gilchrist, J. E. (1973). Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J. Exp. Med. 138, 1156–1167. doi: 10.1084/jem.138.5.1156
Echenique–Rivera, H., Muzzi, A., Del Tordello, E., Seib, K. L., Francois, P., Rappuoli, R., et al. (2011). Transcriptome analysis of Neisseria meningitidis in human whole blood and mutagenesis studies identify virulence factors involved in blood survival. PLoS Pathog. 7:e1002027. doi: 10.1371/journal.ppat.1002027
Eifler, N., Vetsch, M., Gregorini, M., Ringler, P., Chami, M., Philippsen, A., et al. (2012). A structurally informed autotransporter platform for efficient heterologous protein secretion and display. Microbiol. Cell. Fact. 11:85. doi: 10.1186/1475-2859-11-85
Eifler, N., Vetsch, M., Gregorini, M., Ringler, P., Chami, M., Philippsen, A., et al. (2014). Cytotoxin ClyA from Escherichia coli assembles to a 13-meric pore independent of its redox-state. EMPO J. 25, 2652–2661. doi: 10.1038/sj.emboj.7601130
Elhenawy, W., Bording–Jorgensen, M., Valguarnera, E., Haurat, M. F., Wine, E., and Feldman, M. F. (2016). LPS remodeling triggers formation of outer membrane vesicles in salmonella. mBio 7, e00940–e00916. doi: 10.1128/mBio.00940-16
Ellis, T. N., and Kuehn, M. J. (2010). Virulence and immunomodulatoary roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74, 81–94. doi: 10.1128/MMBR.00031-09
Elmi, A., Watson, E., Sandu, P., Gundogdu, O., Mills, D. C., Inglis, N. F., et al. (2012). Campylobacter jejuni outer membrane vesicles play an important role in bacterial interactions with human intestinal epithelial cells. Infect. Immun. 80, 4089–4098. doi: 10.1128/IAI.00161-12
Falugi, F., Kim, H. K., Missiakas, D. M., and Schneewind, O. (2013). Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 4:e00575. doi: 10.1128/mBio.00575-13
Ferreira, C. S. M., Matthews, C. S., and Missailidis, S. (2006). DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers. Tumour Biol. 27, 289–301. doi: 10.1159/000096085
Fierer, J. O., Veggiani, G., and Howarth, M. (2014). SpyLigase peptide-peptide ligation polymerizes affibodies to enhance magnetic cancer cell capture. Proc. Natl. Acad. Sci. U.S.A. 111, E1176–E1181. doi: 10.1073/pnas.1315776111
Fisseha, M., Chen, P., Brandt, B., Kijek, T., Moran, E., and Zollinger, W. (2005). Characterization of native outer membrane vesicles from lpxL mutant strains of Neisseria meningitidis for use in parenteral vaccination. Infect. Immun. 73, 4070–4080. doi: 10.1128/IAI.73.7.4070-4080.2005
Garrett Rappazzo, C., Watkins, H. C., Guarino, C. M., Chau, A., Lopez, J. L., DeLisa, M. P., et al. (2016). Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine 34, 1252–1258. doi: 10.1016/j.vaccine.2016.01.028
Gasanov, A. G., Mamedbeili, E. G., Guseinov, N. S., Ayubov, I. G., and Gasanov Kh, I. (2013). Dienes from the C5 pyrolyzate fraction in thermal and catalytic [4 + 2]-cycloaddition reactions. Petroleum Chem. 53, 54–58. doi: 10.1134/S0965544113010052
Gerritzen, M. J. H., Martens, D. E., Wijffels, R. H., van der Poland, L., and Stork, M. (2017). Bioengineering bacterial outer membrane vesicles as vaccine platform. Biotechnol. Adv. 35, 565–574. doi: 10.1016/j.biotechadv.2017.05.003
Gerritzen, M. J. H., Salverda, M. L. M., Martens, D. E., Wijffels, R. H., and Stork, M. (2019). Spontaneously released Neisseria meningitidis outer membrane vesicles as vaccine platform: production and purification. Vaccine 37, 6978–6986. doi: 10.1016/j.vaccine.2019.01.076
Giuntini, S., Reason, D. C., and Granoff, D. M. (2011). Complement-mediated bactericidal activity of anti-factor H binding protein monoclonal antibodies against the meningococcus relies upon blocking factor H binding. Infect. Immun. 79, 3751–3759. doi: 10.1128/IAI.05182-11
Gnopo, Y. M. D., Watkins, H. C., Stevenson, T. C., DeLisa, M. P., and Putnam, D. (2017). Designer outer membrane vesicles as immunomodulatory systems - reprogramming bacteria for vaccine delivery. Adv. Drug Deliv. Rev. 114, 132–142. doi: 10.1016/j.addr.2017.05.003
Gujrati, V., Kim, S., Kim, S.-H., Min, J. J., Choy, H. E., Kim, S. C., et al. (2014). Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano 8, 1525–1537. doi: 10.1021/nn405724x
Gujrati, V., Prakash, J., Malekzadeh–Najafabadi, J., Stiel, A., Klemm, U., Mettenleiter, G., et al. (2019). Bioengineered bacterial vesicles as biological nano-heaters for optoacoustic imaging. Nat. Commun. 10:1114. doi: 10.1038/s41467-019-09034-y
Gujrati, V. B., and Sangyong, J. (2014). Bioengineered bacterial outer membrane vesicles: what is their potential in cancer therapy? Nanomedicine 9, 933–935. doi: 10.2217/nnm.14.56
Han, W., Wu, B., Li, L., Zhao, G., Woodward, R., Pettit, N., et al. (2012). Defining function of lipopolysaccharide O-antigen ligase WaaL using chemoenzymatically synthesized substrates. J. Biol. Chem. 287, 5357–5365. doi: 10.1074/jbc.M111.308486
Hays, M. P., Houben, D., Yang, Y., Luirink, J., and Hardwidge, P. R. (2018). Immunization with Skp delivered on outer membrane vesicles protects mice against enterotoxigenic Escherichia coli challenge. Front. Cell. Infect. Microbiol. 8:132. doi: 10.3389/fcimb.2018.00132
Hu, J., Zuo, J., Chen, Z., Fu, L., Lv, X., Hu, S., et al. (2019). Use of a modified bacterial ghost lysis system for the construction of an inactivated avian pathogenic Escherichia coli vaccine candidate. Vet. Microbiol. 229, 48–58. doi: 10.1016/j.vetmic.2018.12.020
Huang, W., Wang, S., Yao, Y., Xia, Y., Yang, X., Li, K., et al. (2016). Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci. Rep. 6:37242. doi: 10.1038/srep37242
Jones, C. (2005). Vaccines based on the cell surface carbohydrates of pathogenic bacteria. An. Acad. Bras. Cienc. 77, 293–324. doi: 10.1590/S0001-37652005000200009
Jong, W. S. P., Saurí, A., and Luirink, J. (2010). Extracellular production of recombinant proteins using bacterial autotransporters. Curr. Opin. Biotechnol. 21, 646–652. doi: 10.1016/j.copbio.2010.07.009
Junker, M., Besingi, R. N., and Clark, P. L. (2007). The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiol. Mol. Biol. Rev. 71, 600–619. doi: 10.1128/MMBR.00011-07
Junker, M., Besingi, R. N., and Clark, P. L. (2019). Vectorial transport, and folding of an autotransporter virulence protein during outer membrane secretion. Mol. Microbiol. 96:333a. doi: 10.1016/j.bpj.2008.12.1677
Kadurugamuwa, J. L., and Beveridge, T. J. (1998). Delivery of the non-membrane-permeative antibiotic gentamicin into mammalian cells by using Shigella flexneri membrane vesicles. Antimicrob. Agents Chemother. 42, 1476–1483. doi: 10.1128/AAC.42.6.1476
Kaniuk, N. A., Vinogradov, E., and Whitfield, C. (2004). Investigation of the structural requirements in the lipopolysaccharide core acceptor for ligation of O antigens in the genus salmonella: WaaL “ligase” is not the sole determinant of acceptor specificity. J. Biol. Chem. 279, 36470–36480. doi: 10.1074/jbc.M401366200
Kataoka, M., Yamaoka, A., Kawasaki, K., Shigeri, Y., and Watanabe, K. (2014). Extraordinary denaturant tolerance of keratinolytic protease complex assemblies produced by meiothermus ruber H328. Appl. Microbiol. Biotechnol. 98, 2973–2980. doi: 10.1007/s00253-013-5155-8
Kesty, N. C., and Kuehn, M. J. (2004). Incorporation of heterologous outer membrane and periplasmic proteins into Escherichia coli outer membrane vesicles. J. Biol. Chem. 279, 2069–2076. doi: 10.1074/jbc.M307628200
Kim, J.-Y., Doody, A. M., Chen, D. J., Cremona, G. H., Shuler, M. L., Putnam, D., et al. (2008). Engineered bacterial outer membrane vesicles with enhanced functionality. J. Mol. Biol. 380, 51–66. doi: 10.1016/j.jmb.2008.03.076
Kim, O. Y., Hong Dinh, N. T., Park, H. T., Choi, S. J., Hong, K., and Gho, Y. S. (2017a). Bacterial protoplast-derived nanovesicles for tumor targeted delivery of chemotherapeutics. Biomaterials 113, 68–79. doi: 10.1016/j.biomaterials.2016.10.037
Kim, O. Y., Park, H. T., Hong Dinh, N. T., Choi, S. J., Lee, J., Kim, J. H., et al. (2017b). Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 8, 626–626. doi: 10.1038/s41467-017-00729-8
Kim, S., Kim, D., Jung, H. H., Lee, I.-H., Kim, J. I., Suh, J.-Y., et al. (2012). Bio-inspired design and potential biomedical, applications of a novel class of high-affinity peptides. Angew. Chem. Int. Ed. Engl. 51, 1890–1894. doi: 10.1002/anie.201107894
Kim, S.-H., Kim, K.-S., Lee, S.-R., Kim, E., Kim, M.-S., Lee, E.-Y., et al. (2009). Structural modifications of outer membrane vesicles to refine them as vaccine delivery vehicles. Biochim. Biophys. Acta 1788, 2150–2159. doi: 10.1016/j.bbamem.2009.08.001
Kjaergaard, K., Schembri, M. A., Hasman, H., and Klemm, P. (2000). Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182, 4789–4796. doi: 10.1128/JB.182.17.4789-4796.2000
Klimentová, J., and Stulík, J. (2014). Methods of isolation and purification of outer membrane vesicles from gram-negative bacteria. Microbiol. Res. 170, 1–9. doi: 10.1016/j.micres.2014.09.006
Koeberling, O., Seubert, A., and Granoff, D. M. (2008). Bactericidal antibody responses elicited by a meningococcal outer membrane vesicle vaccine with overexpressed factor H–binding protein and genetically attenuated endotoxin. J. Infect. Dis. 198, 262–270. doi: 10.1086/589308
Kuehn, M. J., and Kesty, N. C. (2005). Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19, 2645–2655. doi: 10.1101/gad.1299905
Kuipers, K., Daleke-Schermerhorn, M. H., Jong, W. S. P., ten Hagen-Jongman, C. M., van Opzeeland, F., Simonetti, E., et al. (2015). Salmonella outer membrane vesicles displaying high densities of pneumococcal antigen at the surface offer protection against colonization. Vaccine 33, 2022–2029. doi: 10.1016/j.vaccine.2015.03.010
Kulp, A., and Kuehn, M. J. (2010). Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64, 163–184. doi: 10.1146/annurev.micro.091208.073413
Kulp, A. J., Sun, B., Ai, T., Manning, A. J., Orench–Rivera, N., Schmid, A. K., et al. (2015). Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS ONE 10:e0139200. doi: 10.1371/journal.pone.0139200
Lahiri, A., Ananthalakshmi, T. K., Nagarajan, A. G., Ray, S., and Chakravortty, D. (2011). TolA mediates the differential detergent resistance pattern between the salmonella enterica subsp. enterica serovars typhi and typhimurium. Microbiology 157, 1402–1415. doi: 10.1099/mic.0.046565-0
Lee, E.-Y., Bang, J. Y., Park, G. W., Choi, D.-S., Kang, J. S., Kim, H.-J., et al. (2007). Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7, 3143–3153. doi: 10.1002/pmic.200700196
Lee, Y., Cho, I. J., Choi, S. Y., and Lee, S. Y. (2019). Systems metabolic engineering strategies for non-natural microbial polyester production. Biotechnol. J. 14:e1800426. doi: 10.1002/biot.201800426
Leitner, D. R., Lichtenegger, S., Temel, P., Zingl, F. G., Ratzberger, D., Roier, S., et al. (2015). A combined vaccine approach against Vibrio cholerae and ETEC based on outer membrane vesicles. Front. Microbiol. 6:823. doi: 10.3389/fmicb.2015.00823
Lewis, L. A., Carter, M., and Ram, S. (2012). The relative roles of factor H binding protein, neisserial surface protein A, and lipooligosaccharide sialylation in regulation of the alternative pathway of complement on meningococci. J. Immunol. 188, 5063–5072. doi: 10.4049/jimmunol.1103748
Lewis, L. A., Ngampasutadol, J., Wallace, R., Reid, J. E. A., Vogel, U., and Ram, S. (2010). The meningococcal vaccine candidate neisserial surface protein A (NspA) binds to factor H and enhances meningococcal resistance to complement. PLoS Pathog. 6:e1001027. doi: 10.1371/journal.ppat.1001027
Liang, Y., Eng, W. S., Colquhoun, D. R., Dinglasan, R. R., Graham, D. R., and Mahal, L. K. (2014). Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. J. Biol. Chem. 289, 32526–32537. doi: 10.1074/jbc.M114.606269
Liu, Q., Yi, J., Liang, K., Hu, B., Zhang, X., Curtiss, R. III., et al. (2016). Outer membrane vesicles from flagellin-deficient Salmonella enterica serovar Typhimurium induce cross-reactive immunity and provide cross-protection against heterologous Salmonella challenge. Sci. Rep. 6:34776. doi: 10.1038/srep34776
Liu, Y., Ai, K., Liu, J., Deng, M., He, Y., and Lu, L. (2013). Dopamine-melanin colloidal nanospheres: an efficient near-infrared photothermal therapeutic agent for in vivo cancer therapy. Adv. Mater. 25, 1353–1359. doi: 10.1002/adma.201204683
Maharjan, S., Saleem, M., Feavers, I. M., Wheeler, J. X., Care, R., and Derrick, J. P. (2016). Dissection of the function of the RmpM periplasmic protein from Neisseria meningitidis. Microbiology 162, 364–375. doi: 10.1099/mic.0.000227
Maisonneuve, C., Bertholet, S., Philpott, D. J., and De Gregorio, E. (2014). Unleashing the potential of NOD- and Toll-like agonists as vaccine adjuvants. Proc. Natl. Acad. Sci. U.S.A. 111, 12294–12299. doi: 10.1073/pnas.1400478111
Mirlashari, M. R., Høiby, E. A., Holst, J., and Lyberg, T. (2010). Outer membrane vesicles from Neisseria meningitidis. Cytokine 110, 193–204. doi: 10.1034/j.1600-0463.2002.100301.x
Mitra, S., Chakrabarti, M. K., and Koley, H. (2013). Multi-serotype outer membrane vesicles of shigellae confer passive protection to the neonatal mice against shigellosis. Vaccine 31, 3163–3173. doi: 10.1016/j.vaccine.2013.05.001
Mitra, S., Sinha, R., Mitobe, J., and Koley, H. (2016). Development of a cost-effective vaccine candidate with outer membrane vesicles of a tolA-disrupted shigella boydii strain. Vaccine 34, 1839–46. doi: 10.1016/j.vaccine.2016.02.018
Mizushima, S. (1984). Post-translational modification and processing of outer membrane prolipoproteins in Escherichia coli. Mol. Cell. Biochem. 60, 5–15. doi: 10.1007/BF00226297
Molloy, M. P., Herbert, B. R., Slade, M. B., Rabilloud, T., Nouwens, A. S., Williams, K. L., et al. (2000). Proteomic analysis of the Escherichia coli outer membrane. Eur. J. Biochem. 267, 2871–2881. doi: 10.1046/j.1432-1327.2000.01296.x
Moon, D. C., Choi, C. H., Lee, J. H., Choi, C.-W., Kim, H.-Y., Park, J. S., et al. (2012). Acinetobacter baumannii outer membrane protein a modulates the biogenesis of outer membrane vesicles. J. Microbiol. 50, 155–160. doi: 10.1007/s12275-012-1589-4
Nevermann, J., Silva, A., Otero, C., Oyarzún, D. P., Barrera, B., Gil, F., et al. (2019). Identification of genes involved in biogenesis of outer membrane vesicles (OMVs) in salmonella enterica serovar typhi. Front. Microbiol. 10:104. doi: 10.3389/fmicb.2019.00104
O'dwyer, C. A., Reddin, K., Martin, D., Taylor, S. C., Gorringe, A. R., Hudson, M. J., et al. (2004). Expression of heterologous antigens in commensal Neisseria spp.: preservation of conformational epitopes with vaccine potential. Infect. Immun. 72, 6511–6518. doi: 10.1128/IAI.72.11.6511-6518.2004
Otto, B. R., Sijbrandi, R., Luirink, J., Oudega, B., Heddle, J. G., Mizutani, K., et al. (2005). Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J. Biol. Chem. 280, 17339–17345. doi: 10.1074/jbc.M412885200
Park, J. S., Lee, W. C., Yeo, K. J., Ryu, K.-S., Kumarasiri, M., Hesek, D., et al. (2012). Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J. 26, 219–228. doi: 10.1096/fj.11-188425
Park, M., Sun, Q., Liu, F., DeLisa, M. P., and Chen, W. (2014). Positional assembly of enzymes on bacterial outer membrane vesicles for cascade reactions. PLoS ONE 9:e97103. doi: 10.1371/journal.pone.0097103
Parlane, N. A., Gupta, S. K., Rubio–Reyes, P., Chen, S., Gonzalez–Miro, M., Wedlock, D. N., et al. (2017). Self-assembled protein-coated polyhydroxyalkanoate beads: properties and biomedical applications. ACS Biomater. Sci. Eng. 3, 3043–3057. doi: 10.1021/acsbiomaterials.6b00355
Petros, R. A., and DeSimone, J. M. (2010). Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug Discov. 9, 615–627. doi: 10.1038/nrd2591
Prados–Rosales, R., Weinrick, B. C., Piqué, D. G., Jacobs, W. R. Jr., Casadevall, A., and Rodriguez, M. G. (2014). Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J. Bacteriol. 196, 1250–1256. doi: 10.1128/JB.01090-13
Price, N. L., Goyette–Desjardins, G., Nothaft, H., Valguarnera, E., Szymanski, C. M., Segura, M., et al. (2016). Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci. Rep. 6:24931. doi: 10.1038/srep24931
Qing, G., Gong, N., Chen, X., Chen, J., Zhang, H., Wang, Y., et al. (2019). Natural and engineered bacterial outer membrane vesicles. Biophys. Rep. 5, 184–198. doi: 10.1007/s41048-019-00095-6
Ranallo, R. T., Kaminski, R. W., George, T., Kordis, A. A., Chen, Q., Szabo, K., et al. (2010). Virulence, inflammatory potential, and adaptive immunity induced by Shigella flexneri msbB mutants. Infect. Immun. 78, 400–412. doi: 10.1128/IAI.00533-09
Salverda, M. L. M., Meinderts, S. M., Hamstra, H.-J., Wagemakers, A., Hovius, J. W. R., van der Ark, A., et al. (2016). Surface display of a borrelial lipoprotein on meningococcal outer membrane vesicles. Vaccine 34, 1025–1033. doi: 10.1016/j.vaccine.2016.01.019
Sánchez, J. M., López-Laguna, H., Álamo, P., Serna, N., Sánchez-Chardi, A., Nolan, V., et al. (2020). Artificial inclusion bodies for clinical development. Adv. Sci. 7:1902420. doi: 10.1002/advs.201902420
Santos, S., de Arauz, L. J., Baruque–Ramos, J., Lebrun, I., Carneiro, S. M., Barreto, S. A., et al. (2012). Outer membrane vesicles (OMV) production of Neisseria meningitidis serogroup B in batch process. Vaccine 30, 6064–6069. doi: 10.1016/j.vaccine.2012.07.052
Santos, T. M. A., Lin, T.-Y., Rajendran, M., Anderson, S. M., and Weibel, D. B. (2014). Polar localization of Escherichia coli chemoreceptors requires an intact Tol-Pal complex. Mol. Microbiol. 92, 985–1004. doi: 10.1111/mmi.12609
Schetters, S. T. T., Jong, WSP, Horrevorts, S. K., Kruijssen, L. J. W., Engels, S., Stolk, D., et al. (2019). Outer membrane vesicles engineered to express membrane-bound antigen program dendritic cells for cross-presentation to CD8+ T cells. Acta Biomater. 91, 248–257. doi: 10.1016/j.actbio.2019.04.033
Schmid-Dannertand, C., and López-Gallego, F. (2019). Advances and opportunities for the design of self-sufficient and spatially organized cell-free biocatalytic systems. Curr. Opin. Chem. Biol. 49, 97–104. doi: 10.1016/j.cbpa.2018.11.021
Schroeder, J., and Aebischer, T. (2009). Recombinant outer membrane vesicles to augment antigen-specific live vaccine responses. Vaccine 27, 6748–6754. doi: 10.1016/j.vaccine.2009.08.106
Schulz, E., Goes, A., Garcia, R., Panter, F., Koch, M., Müller, R., et al. (2018). Biocompatible bacteria-derived vesicles show inherent antimicrobial activity. J. Control. Release 290, 46–55. doi: 10.1016/j.jconrel.2018.09.030
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Sharpe, S. W., Kuehn, M. J., and Mason, K. M. (2011). Elicitation of epithelial cell-derived immune effectors by outer membrane vesicles of nontypeable Haemophilus influenzae. Infect. Immun. 79, 4361–4369. doi: 10.1128/IAI.05332-11
Shen, Y., Torchia, M. L. G., Lawson, G. W., Karp, C. L., Ashwell, J. D., and Mazmanian, S. K. (2012). Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12, 509–520. doi: 10.1016/j.chom.2012.08.004
Shi, J., Votruba, A. R., Farokhzad, O. C., and Langer, R. (2010). Nanotechnology in drug delivery and tissue engineering: from discovery to applications. Nano Lett. 10, 3223–3230. doi: 10.1021/nl102184c
Shim, S.-M., Song, E.-J., Song, D., Lee, T.-Y., Kim, D.-J., Nam, J.-H., et al. (2017). Nontoxic outer membrane vesicles efficiently increase the efficacy of an influenza vaccine in mice and ferrets. Vaccine 35, 3741–3748. doi: 10.1016/j.vaccine.2017.05.053
Shimoda, A., Tahara, Y., Sawada, S.-I., Sasaki, Y., and Akiyoshi, K. (2017). Glycan profiling analysis using evanescent-field fluorescence-assisted lectin array: importance of sugar recognition for cellular uptake of exosomes from mesenchymal stem cells. Biochem. Biophys. Res. Commun. 491, 701–707. doi: 10.1016/j.bbrc.2017.07.126
Shrivastava, R., Jiang, X., and Chng, S.-S. (2017). Outer membrane lipid homeostasis via retrograde phospholipid transport in Escherichia coli. Mol. Microbiol. 106, 395–408. doi: 10.1111/mmi.13772
Simpson, B. W., May, J. M., Sherman, D. J., Kahne, D., and Ruiz, N. (2015). Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Philos. Trans. R Soc. Lond. B Biol. Sci. 370, 209–211. doi: 10.1098/rstb.2015.0029
Spaan, A. N., van Strijp, J. A. G., and Torres, V. J. (2017). Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 15, 435–447. doi: 10.1038/nrmicro.2017.27
Su, F.-H., Tabañag, I. D. F., Wu, C.-Y., and Tsai, S.-L. (2017). Decorating outer membrane vesicles with organophosphorus hydrolase and cellulose binding domain for organophosphate pesticide degradation. Chem. Eng. J. 308, 1–7. doi: 10.1016/j.cej.2016.09.045
Tan, K., Li, R., Huang, X., and Liu, Q. (2018). Outer membrane vesicles: current status, and future direction, of these novel vaccine adjuvants. Front. Microbiol. 9:783. doi: 10.3389/fmicb.2018.00783
Tashiro, M., Tejero, R., Zimmerman, D. E., Celda, B., Nilsson, B., and Montelione, G. T. (1997). High-resolution solution NMR structure of the Z domain of staphylococcal protein A. J. Mol. Biol. 272, 573–590. doi: 10.1006/jmbi.1997.1265
Thompson, B. S., Chilton, P. M., Ward, J. R., Evans, J. T., and Mitchell, T. C. (2005). The low-toxicity versions of LPS, MPL® adjuvant and RC529, are efficient adjuvants for CD4+ T cells. J. Leukoc. Biol. 78, 1273–1280. doi: 10.1189/jlb.0305172
Tokuda, H., and Matsuyama, S. (2004). Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 1693, 5–13. doi: 10.1016/j.bbamcr.2004.02.005
Tsai, S.-L., Oh, J., Singh, S., Chen, R., and Chen, W. (2009). Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 75, 6087–6093. doi: 10.1128/AEM.01538-09
Turner, L., Praszkier, J., Hutton, M. L., Steer, D., Ramm, G., Kaparakis–Liaskos, M., et al. (2015). Increased outer membrane vesicle formation in a Helicobacter pylori tolB mutant. Helicobacter 20, 269–283. doi: 10.1111/hel.12196
Tzokov, S. B., Wyborn, N. R., Stillman, T. J., Jamieson, S., Czudnochowski, N., Artymiuk, P. J., et al. (2006). Structure of the hemolysin E (HlyE, ClyA, and SheA) channel in its membrane-bound form. J. Biol. Chem. 281, 23042–23049. doi: 10.1074/jbc.M602421200
Unal, C. M., Schaar, V., and Riesbeck, K. (2011). Bacterial outer membrane vesicles in disease and preventive medicine. Semin. Immunopathol. 33, 395–408. doi: 10.1007/s00281-010-0231-y
Underhill, D. M., and Goodridge, H. S. (2012). Information processing during phagocytosis. Nat. Rev. Immunol. 12, 492–502. doi: 10.1038/nri3244
van Bloois, E., Winter, R. T., Kolmar, H., and Fraaije, M. W. (2011). Decorating microbes: surface display of proteins on Escherichia coli. Trends Biotechnol. 29, 79–86. doi: 10.1016/j.tibtech.2010.11.003
van de Waterbeemd, B., Streefland, M., van der Ley, P., Zomer, B., van Dijken, H., Martens, D., et al. (2010). Improved OMV vaccine against Neisseria meningitidis using genetically engineered strains and a detergent-free purification process. Vaccine 28, 4810–4816. doi: 10.1016/j.vaccine.2010.04.082
van de Waterbeemd, B., Streefland, M., van Keulen, L., van den Ijssel, J., de Haan, A., Eppink, M. H., et al. (2012). Identification and optimization of critical process parameters for the production of NOMV vaccine against Neisseria meningitidis. Vaccine 30, 3683–3690. doi: 10.1016/j.vaccine.2012.03.028
van der Ley, P., Steeghs, L., Hamstra, H. J., ten Hove, J., Zomer, B., and van Alphen, L. (2001). Modification of lipid A biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect. Immun. 69, 5981–5990. doi: 10.1128/IAI.69.10.5981-5990.2001
van der Pol, L., Stork, M., and van der Ley, P. (2015). Outer membrane vesicles as platform vaccine technology. Biotechnol. J. 10, 1689–1706. doi: 10.1002/biot.201400395
Veggiani, G., Nakamura, T., Brenner, M. D., Gayet, R. V., Yan, J., Robinson, C. V., et al. (2016). Programmable polyproteams built using twin peptide superglues. Proc. Natl. Acad. Sci. U.S.A. 113, 1202–1207. doi: 10.1073/pnas.1519214113
Wacker, M., Feldman, M. F., Callewaert, N., Kowarik, M., Clarke, B. R., Pohl, N. L., et al. (2006). Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc. Natl. Acad. Sci. U.S.A. 103, 7088–7093. doi: 10.1073/pnas.0509207103
Wai, S. N., Lindmark, B., Söderblom, T., Takade, A., Westermark, M., Oscarsson, J., et al. (2003). Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial clya cytotoxin. Cell 115, 25–35. doi: 10.1016/S0092-8674(03)00754-2
Wang, S., Gao, J., and Wang, Z. (2019). Outer membrane vesicles for vaccination and targeted drug delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 11:e1523. doi: 10.1002/wnan.1523
Weaver, B. A. A., and Cleveland, D. W. (2005). Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell 8, 7–12. doi: 10.1016/j.ccr.2005.06.011
Westermark, M., Oscarsson, J., Mizunoe, Y., Urbonaviciene, J., and Uhlin, B. E. (2000). Silencing and activation of ClyA cytotoxin expression in Escherichia coli. J. Bactorial. 182, 6347–6357. doi: 10.1128/JB.182.22.6347-6357.2000
Wilkerson, J. W., Yang, S.-O., Funk, P. J., Stanley, S. K., and Bundy, B. C. (2018). Nanoreactors: strategies to encapsulate enzyme biocatalysts in virus-like particles. N. Biotechnol. 44, 59–63. doi: 10.1016/j.nbt.2018.04.003
Williams, C., Royo, F., Aizpurua–Olaizola, O., Pazos, R., Boons, G.-J., Reichardt, N.-C., et al. (2018). Glycosylation of extracellular vesicles: current knowledge, tools and clinical perspectives. J. Extracell. Vesicles 7:1442985. doi: 10.1080/20013078.2018.1442985
Wong, J. X., Ogura, K., Chen, S., and Rehm, B. H. A. (2020). Bioengineered polyhydroxyalkanoates as immobilized enzyme scaffolds for industrial applications. Front. Bioeng. Biotechnol. 8:156. doi: 10.3389/fbioe.2020.00156
Wressnigg, N., Barrett, P. N., Pöllabauer, E.-M., O'Rourke, M., Portsmouth, D., Schwendinger, M. G., et al. (2014). A Novel multivalent OspA vaccine against Lyme borreliosis is safe and immunogenic in an adult population previously infected with Borrelia burgdorferi sensu lato. Clin. Vaccine Immunol. 21, 1490–1499. doi: 10.1128/CVI.00406-14
Yan, L., Da, H., Zhang, S., López, V. M., and Wang, W. (2017). Bacterial magnetosome and its potential application. Microbiol. Res. 203, 19–28. doi: 10.1016/j.micres.2017.06.005
Yeh, Y.-C., Comolli, L. R., Downing, K. H., Shapiro, L., and McAdams, H. H. (2010). The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J. Bacteriol. 192, 4847–4858. doi: 10.1128/JB.00607-10
Yoo, J.-W., Irvine, D. J., Discher, D. E., and Mitragotri, S. (2011). Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat. Rev. Drug Discov. 10, 521–535. doi: 10.1038/nrd3499
Yoon, H. (2016). Bacterial outer membrane vesicles as a delivery system for virulence regulation. J. Microbiol. Biotechnol. 26, 1343–7. doi: 10.4014/jmb.1604.04080
Yoon, H., Ansong, C., Adkins, J. N., and Heffron, F. (2011). Discovery of salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infect. Immun. 79, 2182–2192. doi: 10.1128/IAI.01277-10
Zakeri, B., Fierer, J. O., Celik, E., Chittock, E. C., Schwarz–Linek, U., Moy, V. T., et al. (2012). Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 109, E690–E697. doi: 10.1073/pnas.1115485109
Keywords: genetic engineering, multifunctional delivery platform, outer membrane vesicles (OMVs), LPS, ClyA, AT
Citation: Li R and Liu Q (2020) Engineered Bacterial Outer Membrane Vesicles as Multifunctional Delivery Platforms. Front. Mater. 7:202. doi: 10.3389/fmats.2020.00202
Received: 28 January 2020; Accepted: 03 June 2020;
Published: 10 July 2020.
Edited by:
Huanan Wang, Dalian University of Technology, ChinaReviewed by:
David Wibowo, Griffith University, AustraliaMichihiro Nakamura, Yamaguchi University, Japan
Copyright © 2020 Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiong Liu, cWlvbmdsaXVAbmN1LmVkdS5jbg==
 Ruizhen Li
Ruizhen Li Qiong Liu
Qiong Liu