
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mater. , 18 September 2018
Sec. Colloidal Materials and Interfaces
Volume 5 - 2018 | https://doi.org/10.3389/fmats.2018.00057
This article is part of the Research Topic Biointerfacial Science of Supported Lipid Membranes: A Nano-Perspective View all 7 articles
Accurate detection and sensing of bacteria are becoming increasingly important not only in microbiology but in a variety of fields including medicine, food, public health, and environmental science. However, even new rapid methods are not convenient enough. Here, we describe a simple and efficient label-free bacterial detection system using the polydiacetylene (PDA) liposomes immobilized on the 3D polydimethylsiloxane (PDMS) micropillars. Our system utilizes the colorimetric response of amine functionalized PDA vesicles to surfactin, a bacterial cyclic lipopeptide commonly released by Gram-positive Bacillus species as an antibiotic. To improve the sensitivity of PDA vesicles to surfactin by increasing the number and surface area of immobilized vesicles, the PDA vesicles were immobilized on the micropillar structure to give a hierarchical 3D PDA vesicle structure. For the fabrication of the 3D micropillar structure, polydimethylsiloxane (PDMS) was used to overcome the limitations imposed by silicon-based fabrication. In contrast to the 2D-PDA-PDMS system, which could only hardly detect the presence of 500 μM surfactin, the 3D-PDA-PDMS system could efficiently detect the presence of 5 μM surfactin and the initial presence of 4 × 101 cells/ml of Bacillus subtilis NCIB3610, which actively produces surfactin. Furthermore, bacterial strains that are known to produce no surfactin, such as Staphylococcus aureus Newman, Escherichia coli DH5α, and Pseudomonas aeruginosa PA14 were not detected by our system suggesting that the 3D-PDA-PDMS system is highly specific to surfactin but not to other chemicals produced by bacteria. Taken together, our results suggest that the 3D-PDA-PDMS system can sensitively and selectively be used for the high throughput detection and screening of biotechnologically important surfactin-producing bacterial strains.
Many bacteria are considered to be harmful pathogens in several plants and animals including human beings (Duc et al., 2005; DuPont, 2007; Apetroaie-Constantin et al., 2009). However, certain bacterial strains are used as materials for genetic engineering and an industrial workhorse for the economic production of numerous enzymes, proteins, and antibiotics (Darken et al., 1960; Konings et al., 1997; Clardy et al., 2009). B. subtilis as a model of Gram-positive bacteria have been intensively characterized in a variety of ways for the microbiological research as well as biotechnology (Härtig and Jahn, 2012; Thwaite and Atkins, 2012). B. subtilis can synthesize and secrete a well-known biosurfactant surfactin. The surfactin produced by various strains of B. subtilis is recognized as a cyclic heptapeptide bound to the carboxyl and hydroxyl groups of a 13–15 carbon fatty acid (Peypoux et al., 1999; Zhu et al., 2014). Importantly, the surfactin has much potential in a variety of commercial applications such as strong surface activity (Grangemard et al., 2001; Yoneda, 2001; Razafindralambo et al., 2004), hemolytic activity (Deleu et al., 2003), and antimicrobial/fungal activities (Liu et al., 2012). Thus, it is worth developing sensing materials to detect B. subtilis by using the surfactin as an indirect mediator.
The technologies for the purpose of bacterial detection can be classified into conventional methods and newly employed technologies. The conventional methods such as specimen culture and colony counting, which are sensitive and selective enough but often have long processing times (5–7 days), are labor-intensive and require trained personnel (Fournier et al., 2013; Ahmed et al., 2014). More recently, rapid detection method technologies have been developed such as polymerase chain reaction (PCR) technology, immunoassays (ELISA) and immunomagnetic separation method. PCR is an advent technique of genetic analysis with an extreme sensitivity (Burnham and Carroll, 2013; Croxen et al., 2013). Yet, incorrect pairing and sample matrix inhibition in PCR reactions may lead to false positive results, and it is possible that a genetically mutated strain could escape the matching process even after correct probing (Espy et al., 2006; Fernández-No et al., 2011). The ELISA method only requires extra labels of antibodies, whereas the immunomagnetic separation method often needs a combination of other methods. Critically, these techniques are generally time-consuming and costly due to the requirements of particular types of equipment and reagents. New types of detection methods have been made possible through biosensors.
Biosensors is a rapid and cost-effective method that combine biomaterial compounds with a physicochemical transducer with the ability to obtain a measurement with minimal perturbation of the sample (Deisingh and Thompson, 2004; Ahmed et al., 2014). The recognition of the biomaterials in biosensor can be achieved either by using a label or label-free systems. Labels have been essential in bioanalytical research; however, this technique has several drawbacks such as only providing an endpoint reading, noncontinuous monitoring, and multiple washing stages, thereby reducing throughput effectivity and increasing the cost. Due to these considerations, the development of label-free biosensors that can reduce assay cost and complexity while providing quantitative information with high throughput effectivity is an important research goal.
Polydiacetylene (PDA)-based sensor as label-free bacteria detection has been intensively investigated due to its unique stimulus-induced significant color-changing properties from brilliant blue to red (Silbert et al., 2006; Scindia et al., 2007; Meir et al., 2008; Lee et al., 2016; Park et al., 2016). The fact that such a visible color change occurs in response to a variety of environmental perturbations including temperature, heat, mechanical stress, pH, and solvents lead to a nonspecific colorimetric response. Therefore, specifically designed ligands have been introduced as the terminal group of PDA for detecting specific types of bacterial strains with high sensitivity, such as glycine terminated PDA for colorimetric signaling of bacterial pore-forming toxin (Ma and Cheng, 2005), peptide functionalized PDA for detecting bacterial lipopolysaccharide (Wu et al., 2011), aptamer-based PDA for Escherichia coli O157:H7 detection (Wu et al., 2012), amine functionalized PDA for detecting surfactin from B. subtilis NCIB3610 (Park et al., 2016) and a EuIII-EDTA functionalized PDA for recognizing calcium dipicolinate (DPA), a major component of Bacillus anthrax endospores (Seo et al., 2017). However, the majority of the PDA-based sensors that have been reported were prepared in the form of a liposome in aqueous solution or thin film as two dimensional (2D) structures (Silbert et al., 2006; Scindia et al., 2007; Meir et al., 2008; Pires et al., 2011; Park et al., 2016; Seo et al., 2017).
The high selectivity of amine functionalized PDA solutions toward surfactin has been reported in a number of works (Deleu et al., 2003; Bouffioux et al., 2007; Park et al., 2016). The interaction of amine functionalized PDA has been tested with various targets including the various surfactants (anion and cation), simple anions, and fatty acids. The simple anions, short-chain fatty acids, and cation surfactants did not trigger colorimetric changes in PDA liposomes. Considering the pKb values of the terminal amine group on the PDA, it is expected that ammonium cation and electrostatic interaction with the negative charges of surfactin occurs (Park et al., 2016). Furthermore, the fatty acid chain of surfactin also can enhance the binding affinity with carbon backbones in PDA by van der Waals interaction and due to its amphiphilic nature, it can penetrate into the charged bilayer. (Hupfer et al., 1983; Deleu et al., 2003; Bouffioux et al., 2007; Park et al., 2016).
A label-free bacteria sensor for detecting bacteria producing surfactin by using amine functionalized PDA was introduced by (Park et al., 2016). However, by embedding the PDA with the medium agar, the PDA platform possesses several drawbacks such as poor stability, low sensitivity, and the inability for precise quantitative analysis. Therefore, to overcome the problems associated with PDA embedded systems, in this study PDA was immobilized in the form of vesicles. Our previous study reported the immobilization of PDA liposomes onto hierarchical 3D networked nanostructures to overcome the intrinsic limitations of the 2D system and affords a significant increase in the sensitivity of PDA (Lee et al., 2016). However, this sensor was fabricated on the micropillar silicon substrate, which also has a limitation. The hardness of silicon poses limits to its broad application in the biosensor (Zhou et al., 2010). Other problems are the high cost of the fabrication process and the involvement of dangerous chemicals (Moschou and Tserepi, 2017). These limitations motivated the development of other chip materials that can be easily fabricated and are compatible with broader biological applications.
In this article, we developed a PDA vesicle-based sensor on PDMS micropillars (3D-PDA-PDMS) for detecting B. subtilis. In order to develop immobilized polydiacetylene vesicles on polydimethylsiloxane micropillars (3D-PDA-PDMS), amine functionalized diacetylene monomers PCDA-EDEA and PCDA-EDA were prepared. The mixture of PCDA-EDEA and PCDA-EDA, which has different lengths of amine ligands and has gaps along the length of its hydrophilic exterior, can ensure a better adherence and reaction (Kim et al., 2005; Park et al., 2016). The assembled vesicles of PCDA-EDEA and PCDA-EDA were formed with the molar ratio 1:1, which is the optimal mixture ratio, as reported elsewhere (Kim et al., 2005). The chromatic change in PDA caused by the surfactin was used as an indirect sensing method to readily detect the presence of bacteria and a high throughput screening tool for certain bacterial strains that produce surfactin. Polydimethylsiloxane (PDMS) was used to fabricate a micropillar template by controlling the diameter, height, and gap of the pillars.
10,12 pentacosadynoic acid (PCDA) was purchased from GFS chemicals. Sylgard 184 elastomer kit was purchased from Dow Corning and 3-(2-aminoethylamino)propyldimethoxymethylsilane(AEAPDMS) was purchased from Junsei. 2,2'- (ethylenedioxy)bis-(ethylamine; EDEA), ethylenediamine (EDA), octadecyltrichlorosilane (OTS), glutaraldehyde (GA) 25% in H2O and surfactin from B. subtilis, 98% (HPLC) were purchased from Sigma Aldrich.
NCIB3610 (Purchased from Korean Agricultural Culture Collection, No.10854) and CU1065 (Ollinger et al., 2006) were used as surfactin-producing and -defective strains of B. subtilis, respectively. Two other Bacillus genus strains, B. cereus ATCC11778 (KCTC1012) and Bacillus licheniformis ATCC14580 (KCTC1918), were purchased from the Korean Collection for Type Cultures. For the test of Gram-negative bacterium which does not produce surfactin, E. coli DH5α (Novagen) strain was used. The Staphylococcus aureus Newman (Ji et al., 2015) and Pseudomonas aeruginosa PA14 (Heo et al., 2010) strains were used as representative pathogens of Gram-positive and Gram-negative bacteria, respectively. All of the strains were routinely grown in Lysogenic Broth (LB) with a shaking incubator (210 rpm/37°C) for the designated incubation times as described in the results and discussion section. Bacterial growth was monitored by measuring absorbance at 600 nm using UV-Vis spectrophotometer (Shimadzu, Japan).
The PDMS micropillars were fabricated using the two-step replica method (Supplementary Figure 1). A three-dimensional pillar silicon wafer was used as the master mold. The uncured PDMS mixture (base: curing agent, 10:1 w/w) was poured on the master mold and cured at 80°C for an hour. After cooling down the process, the hole patterned-PDMS was detached from the master mold. Before undergoing second step molding, the hole patterned-PDMS mold was exposed to UV-ozone cleaner (Nippon Laser, Japan) for 10 min to decrease the hydrophobicity for further modification. The UV-ozone treated hole patterned-PDMS molds were treated with OTS in an ethanol solution for 30 min under nitrogen atmosphere. The substrate was washed several times with ethanol. The uncured PDMS mixture (base: curing agent, 10:1 w/w) was poured on the OTS treated hole patterned-PDMS mold and cured at 80°C for an hour. After the cooling down process, the micropillar PDMS was peeled off from the hole patterned-PDMS mold.
The amine-terminated diacetylene monomers PCDA-EDEA and PCDA-EDA were synthesized by employing standard procedures (Kim et al., 2003). A mixture of EDEA and EDA terminated diacetylene monomers were dissolved in chloroform in a test tube. The solvent was evaporated by a stream of N2 gas, and deionized water was added to the test tube to give the desired concentration of lipid (1 mM). The resultant suspension was sonicated (Fisher, USA) for 15 min at 80°C followed by filtering for removing dispersed lipid aggregates by using a 0.8 μm filter and cooled at 4°C overnight.
The surface modification process is shown in Figure 1. The PDMS substrates were set in a UV-Ozone cleaner (Nippon Laser, Japan) for 10 min to form hydroxyl groups. The substrates were silanized in a 4.2 mM of AEAPDMS in anhydrous ethanol (Daejung, Korea) for 3 h at room temperature. The substrates were rinsed five times with anhydrous ethanol and dried in the nitrogen atmosphere inside the glove box. The AEAPDMS modified substrates were incubated in a solution containing 2.5% v/v glutaraldehyde in deionized water for 5 hrs. The substrates were washed with deionized water to remove unreacted reagents and further dried under a stream of nitrogen. The surface morphology of AEAPDMS and GA modified substrate was identified by atomic force microscope (Park Systems, Korea) with NCHR non-contact tip, and the thickness of the resulted layer was measured by an ellipsometer (Rudolph Research, USA). The GA formation on the quartz substrate was confirmed by using a UV/Vis/NIR spectrometer (PerkinElmer, USA). The liposome was dried in air for 3 h. The aldehyde-modified substrates were placed in a diacetylene lipid vesicles solution for 1 h followed by rinsing with deionized water and dried under a stream of nitrogen. The immobilized diacetylene lipid vesicles were irradiated under UV light (1 mW cm−2) with a wavelength of 254 nm for 1 min. The morphology and size of the immobilized PDA liposome were analyzed by scanning electron microscope (Hitachi, Japan) operated at a beam energy of 15 kV. The immobilized PDA liposome was coated with platinum before the imaging process.
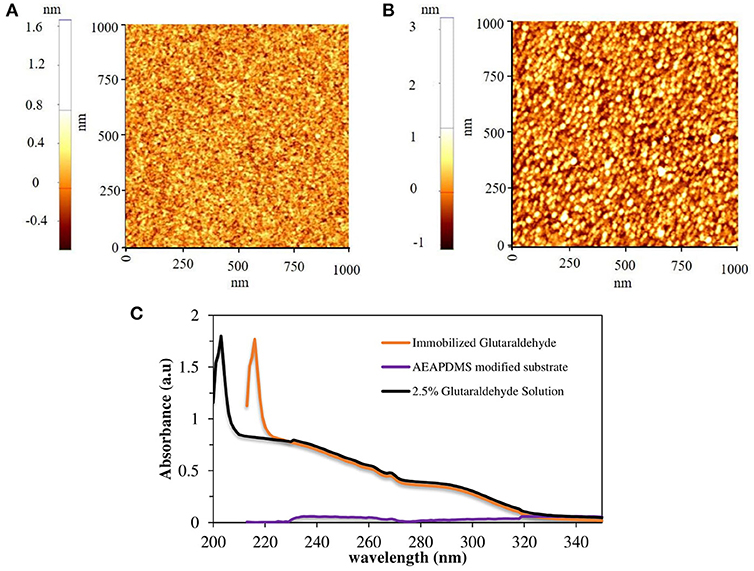
Figure 1. AFM images of (A) AEAPDMS modified substrate and (B) glutaraldehyde modified substrate, (C) UV-Vis spectra of glutaraldehyde solutions and glutaraldehyde modified quartz substrate.
In order to quantify the amount of surfactin produced from bacterial cells by measuring the fluorescence, cell-free culture media were directly incubated with immobilized PDA vesicles. To recover the supernatants after cell cultures, the cells were discarded by centrifugation with 15,000 × g for 5 min. After immersing the immobilized PDA vesicles in the supernatants for 1 h at 25°C, the immobilized PDA vesicles were washed with ethanol and deionized water. The fluorescence microscopy images were taken from the optical microscope (Olympus, Japan) and analyzed by using ImageJ (National Institute of Health, USA). To determine the detection limit of surfactin by immobilized PDA vesicles, a dose-dependent test was performed using various starting cultures with different initial concentrations of bacterial cells from 4 × 107 to 4 × 101 cells/ml (assuming that OD 600 = 1 correspond to 8 × 108 cells/ml, Park et al., 2016). The selectivity of the immobilized PDA vesicles to the surfactin was tested using seven different strains of bacteria: B. subtilis NCIB3610, B. subtilis CU1065, B. cereus ATCC11778, B. licheniformis ATCC14580, S. aureus Newman, P. aeruginosa PA14, and E. coli DH5α. The initial inoculum with a cell density of 4 × 107 cells/ml was cultured for 24 h. The surfactin standard curve was generated using fluorescence signals obtained from the 2D or 3D-PDA-PDMS immersed in different concentrations of the standard surfactin ranging from 500 nM to 1 mM.
The cell-free media supernatants were recovered as described above for fluorescence measurement. The concentration of surfactin in the supernatants was determined using an HPLC-UV/Vis quantification method. The analysis was performed with standard high-pressure liquid chromatography (HPLC) (Agilent, Germany) equipped with a reverse phase column (C18 150 × 4.6 mm, 120 A, Hyperclone, USA) at 30°C. An isocratic method was performed with 80% acetonitrile and 20% 3.8 mM trifluoroacetic acid for 25 min. The peptide bonds of the surfactin were detected at a wavelength of 210 nm.
The routine procedure for immobilizing the assembled vesicles of PCDA-EDEA and PCDA-EDA is by using glutaraldehyde (GA) on a solid support with a surface amine functional group (Kim et al., 2005). In this study, dialkoxysilane monomers (AEAPDMS) were used to generate the monolayer and amine functionalization on the surface of PDMS for uniform immobilization of glutaraldehyde. In order to obtain a preliminary clue for ensuring the formation of a monolayer on the sensor template, 2D silicon substrate, and quartz substrate were used for surface characterization. The surface properties of the amine-modified and GA modified substrate were characterized by using an ellipsometer and AFM. The measured thickness of the AEAPDMS monolayer on the Si substrate after 3 h of modification was 11 ± 1 Å, and the average thickness of GA was about 8 Å which is in agreement with its theoretical thickness (Salem et al., 2010). Figures 1A,B showed 1.0 × 1.0 μm AFM images of the AEAPDMS modified and GA modified Si substrate, respectively. The non-contact mode AFM images did not show any aggregated multilayer formation after AEAPDMS modification with the RMS value of 0.18 nm, and it was similar to RMS value after GA modification. Based on these results, the resultant layer of GA is regarded as a monolayer. However, a successful surface modification on the sensor template cannot be decided just by looking at the morphology, to confirm a successful GA modification on an AEAPDMS-modified substrate. Figure 1C shows the UV-visible spectrum of GA solution and immobilized GA on the quartz substrate where the GA has sharp characteristic absorption at 203 nm, which has been assigned to the π-π* transition of the carbonyl group (Tang et al., 2016) whereas the immobilized GA on the quartz substrate has similar sharp spectral peaks as that of the GA solution at 216 nm with a redshift of 13 nm. A successful modification of the sensor template with the GA molecules can offer the reproducibility and uniformity of immobilized diacetylene vesicles.
A reliable biosensor platform requires structure uniformity and stability of the receptor on the sensor template. Since the diacetylene is a colloidally dispersed liposome, it can be formed as a film or vesicle during the immobilization process (Carpick et al., 2000; Teschke and de Souza, 2002; Kim et al., 2006). However, the vesicle form has been reported to be more sensitive than the film form (Song et al., 2002; Davis et al., 2014; Lee et al., 2016). Figures 2A,B shows the SEM and fluorescence images of immobilized PDA on the 2D PDMS substrate in film form, while Figures 2C,D shows the SEM and fluorescence images of the immobilized PDA on the 2D PDMS in vesicle form at approximately 30-80 nm in size which is in agreement with previous study (Park et al., 2016). The fluorescence images were obtained after heat treatment (100°C, 1 min). The heat treatment induces a blue to the red transition of the PDA since the blue phase PDA is non-fluorescent. As presented in Figures 2B,D, the fluorescence intensity of the vesicle form immobilized PDA is two times higher than the film form, which is in agreement with a previous report (Song et al., 2002). The vesicle form of the PDA is presumed as a spherical form that has a higher surface area than the film form of the PDA. Since there is a proportional relationship between the fluorescence intensity and surface area, the vesicle form emitted a higher intensity compared to the film form (Reimhult et al., 2002; Stoner et al., 2016). Therefore, in this study, we selected immobilized PDA in vesicle form to develop a PDA-based label-free bacteria sensor.
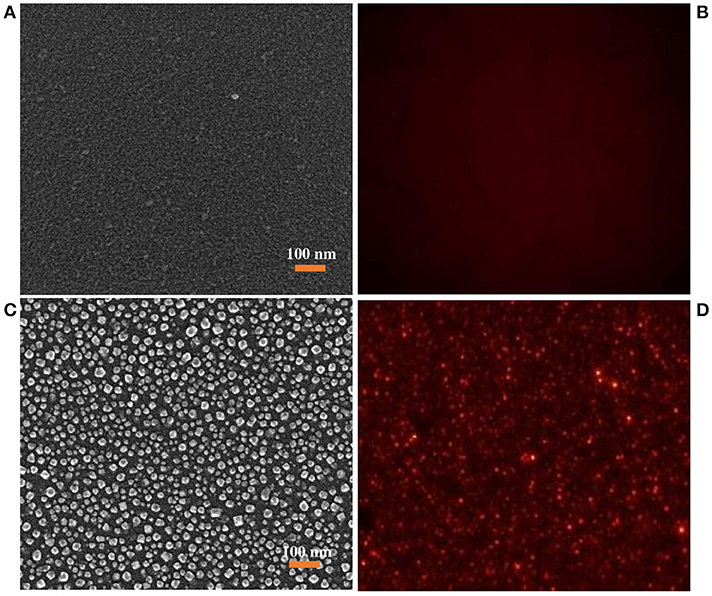
Figure 2. (A) SEM images of immobilized PDA liposome in film structure, (B) corresponding fluorescence images of Immobilized PDA liposome in film structure obtained after heating treatment, (C) SEM images of immobilized PDA liposome in vesicle structure, (D) corresponding fluorescence images of Immobilized PDA liposome in vesicle structure obtained after heating treatment.
The immobilized PDA vesicles on PDMS micropillars (3D-PDA-PDMS) were also prepared under the same conditions used for the 2D PDMS substrate. The SEM images of fabricated PDMS micropillars show height, gap and diameter around 5, 4 and 2 μm respectively as presented in Supplementary Figure 2. As presented in Figure 3A and Supplementary Figure 3, the PDA vesicles were also immobilized uniformly on the 3D PDMS micropillars substrate. For proving a successful immobilization of PDA vesicles on the 3D PDMS micropillars substrate, fluorescence images (Figures 3B,C) and UV-visible spectra (Figure 3D) of the PDA after 0.1 mM surfactin incorporation were obtained. The fluorescence image of the top view (Figure 3B) and the side view (Figure 3C) prove the efficiency of the PDA immobilization and UV-visible spectra. Figure 3D presents a significant UV absorption shift from the blue phase (645 nm) to the red phase (545 nm) after 0.1 mM surfactin incorporation.
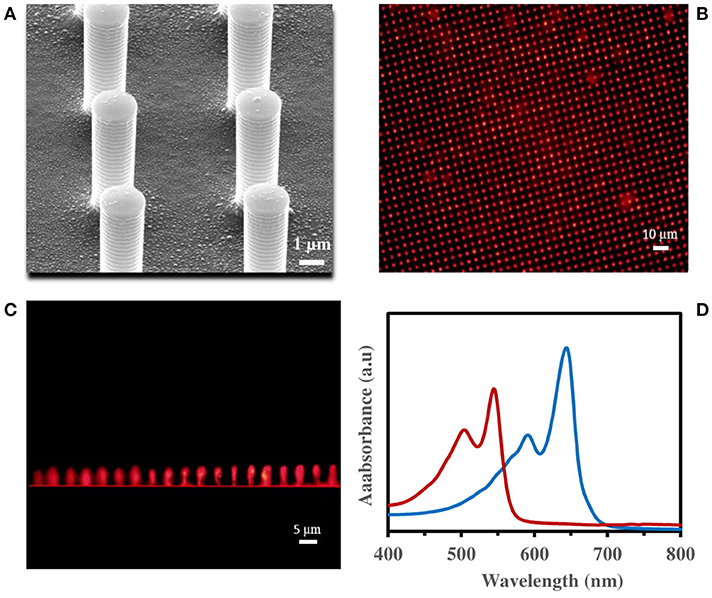
Figure 3. (A) SEM images of immobilized PDA vesicles on PDMS micropillars, fluorescence microscopic image of PDA immobilized on PDMS micropillars from (B) top view, (C) side view, (D) visible spectra of immobilized PDA vesicles on quartz substrate, before (blue line) and after (red line) 1 h incubation in 0.1 mM surfactin.
Figures 4A,B illustrate the surfactin based label-free bacterial sensor platform using 3D-PDA-PDMS. Thus, in pursuance of a feasible sensor platform using immobilized PDA vesicles, the 2D and 3D-PDA-PDMS were exposed to various concentrations of surfactin. As shown in Figure 5A, the fluorescence signals from the 2D-PDA-PDMS were hardly observable until 500 μM of surfactin whereas a red fluorescence intensity from 3D-PDA-PDMS was significantly enhanced with increasing concentration of surfactin in a range from 5 μM to 1 mM (Figure 5B). Figure 5C displays a calibration curve which consists of plots of fluorescence intensity as a function of the surfactin concentration at log scale. The calibration curve was used for quantitative estimation of the surfactin by a linear regression model for curve-fitting analysis. The R-squared of the experimental data of the 3D-PDA-PDMS was 0.99. The detection limit of the 2D and 3D-PDA-PDMS were found to be 500 and 5 μM, respectively. Thus, an increase in the sensitivity of at least 100 times was obtained with the 3D over the 2D-PDA-PDMS. These results clarify our previous study conducted using 3D networked PDA for quantification of α-cyclodextrin (Lee et al., 2016) where the large increase in the sensitivity of the 3D-PDA system over 2D-PDA system was not only due to the increment of the template surface area but was also caused by the significant increase in the target molecule accessibility to interact with immobilized PDA. Moreover, the immobilized PDA on the 2D system tends to form an aggregate that prevents the accessibility of the target molecules whereas the 3D system increases the accessibility of the target molecules through the ordered pathways of the 3D templates. The reproducibility of the 3D-PDA-PDMS was evaluated by repeatedly assaying using the coefficient of variation (CV, n = 3). The CV value of the 3D-PDA-PDMS was calculated to be 0.78%. A small CV value calculated at < 2% for this method indicates the precision of the developed platform. In addition, the accuracy of the 3D-PDA-PDMS was observed by the standard addition method. The % recovery values for the accuracy of the 3D-PDA-PDMS on the quantification of the surfactin was calculated to be 96.8%. This complies with the % recovery values for a unit concentration of 0.5 mM. Therefore, the results of the developed platform show a good accuracy of surfactin quantification.
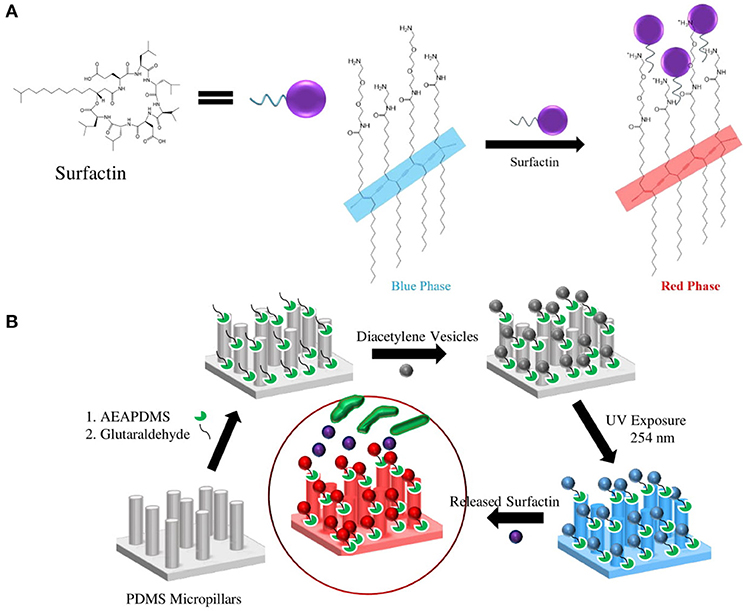
Figure 4. Schematic of (A) PDA-surfactin interaction through the ionic interaction between the hydrophilic head of surfactin and amine group of PDA and fatty acid chain of surfactin with carbon backbones in PDA, (B) procedure of surface modifications and immobilization of PCDA on the PDMS micropillars.
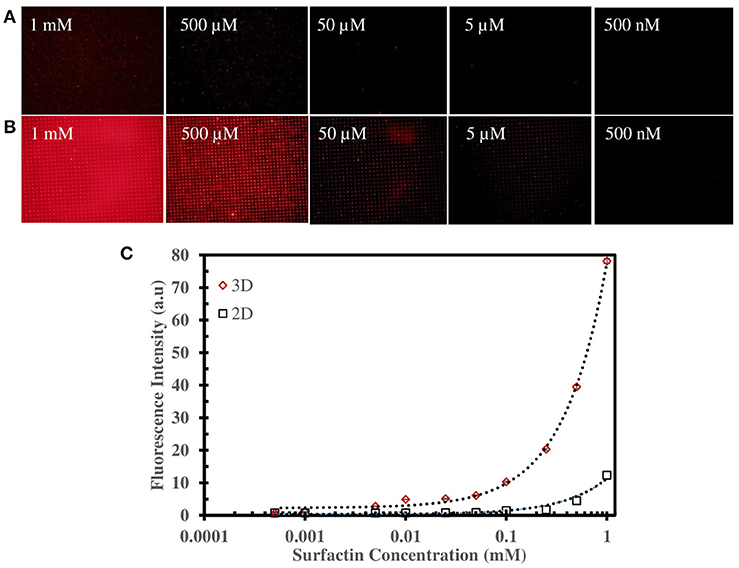
Figure 5. Fluorescence microscopic image of (A) 2D-PDA-PDMS, (B) 3D-PDA-PDMS after exposure to various concentration of surfactin, (C) plot of fluorescence intensity of PDA sensors in 2D and 3D-PDA-PDMS as a function of surfactin concentration in log scale.
To investigate the sensing ability of our platform as a label-free bacterial sensor, we first examined the surfactin-induced chromatic response using two different Gram-positive B. subtilis strains (the NCIB3610 strain as a normal surfactin-producer and the CU1065 strain as a surfactin-defective producer), as described previously. For quantitative evaluation and comparison of surfactin production during bacterial growth, both strains were cultivated in the growth medium. Cell-free media were obtained after 0, 4, 8, 12, and 24 h (Figure 6). The CU1065 strain was likely to reach a stationary phase around 12 h, while the NCIB3610 strain was able to continuously grow for 24 h, indicating that CU1065 achieved the maximum growth yield earlier than that of the NCIB3610 strain. Consistent with previous studies, our data also show that the production levels of surfactin have a correlation with the growth pattern where the maximum production of the surfactin occurs at the end of the exponential growth phase (Kim et al., 1997; de Oliveira et al., 2013; Mubarak et al., 2017).
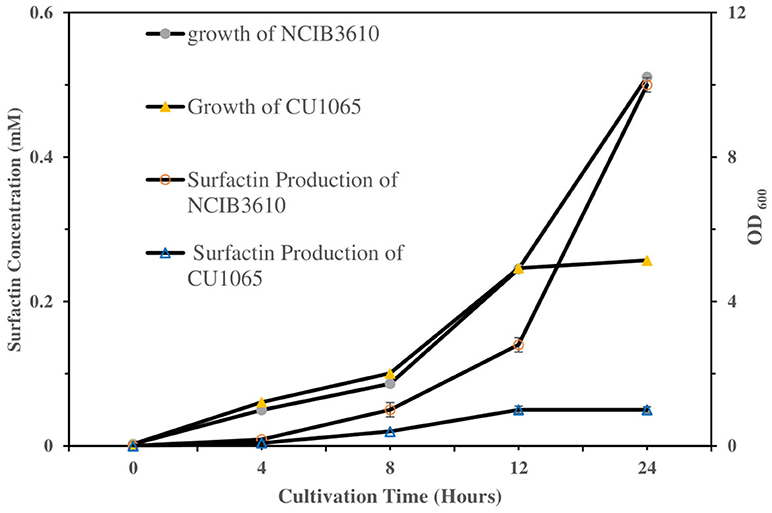
Figure 6. Comparison of the bacterial growth and surfactin production of NCIB3610 and CU1065 strains as a function of the cultivation time. Each of supernatant samples was analyzed by HPLC.
Next, to evaluate our platform as an effective bacterial detection platform, we determined the detection limit of the initial bacteria concentration by 3D-PDA-PDMS. We prepared various bacterial solutions of NCIB3610 and CU1065 strains with different initial cell concentrations ranging from 4 × 107 to 4 × 101 cells/ml. The sensor platforms were exposed to each media supernatant after 24 h of cultivation, and the corresponding fluorescence intensities were measured. As shown in Figures 7A,B, even in the groups where the initial concentration was 4 × 101 cells/ml (for both NCIB3610 and CU1065), distinguishable changes of fluorescence intensity were observed compared to the control group (no cell). Furthermore, the initial cell concentrations of the surfactin-producing NCIB3610 strain displayed a strong linear positive correlation with the concentrations of surfactin (which was calculated based on the standard curve as shown in Figure 5C) produced by live cells in growth media (Figure 7C). In order to confirm our 3D-PDA-PDMS based surfactin quantification method, the data deduced in Figure 7C was further verified by direct comparison with the results obtained from the HPLC quantification method (Table 1). The results from our 3D-PDA-PDMS method were in good agreement with those using the HPLC method since the calculated t (tcal) were less than table t (ttab) with ttab of 2.45 (df = 6 and α = 0.05).
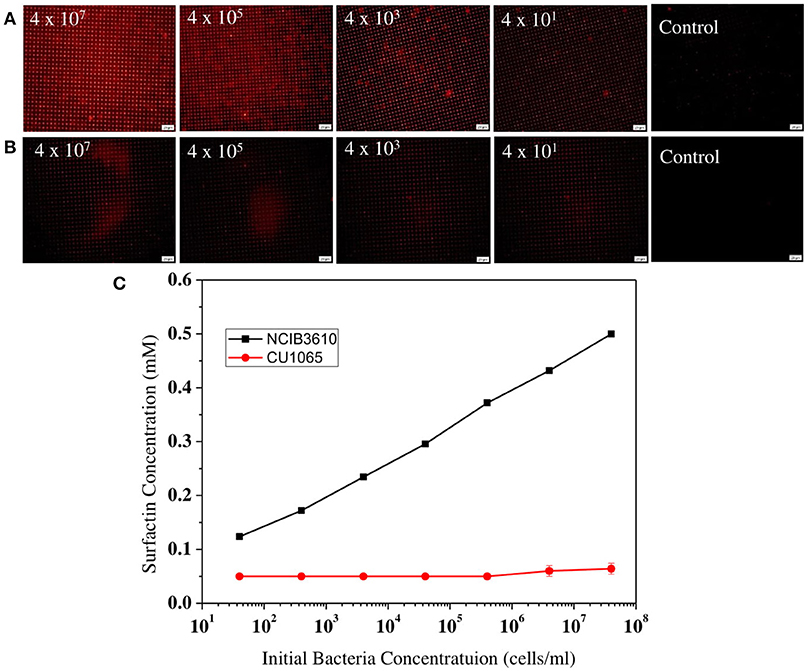
Figure 7. Comparison of the surfactin-detecting sensitivities between B. subtilis NCIB3610 and B. subtilis CU1065. The fluorescence images of sensor platform after exposed to media supernatants of (A) NCIB3610 strain and (B) CU1065 strain, (C) Plot of 3D-PDA-PDMS measurement of surfactin production levels depending of the initial cell concentrations.
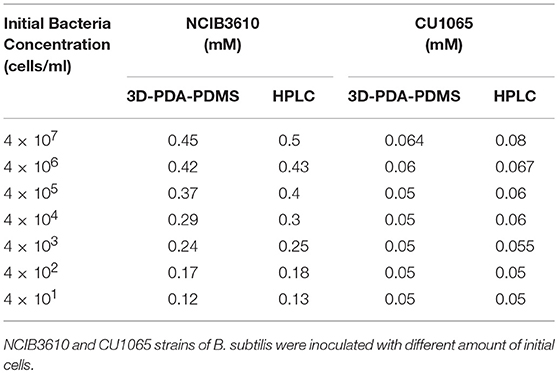
Table 1. Quantification and comparison of surfactin levels measured by using the immobilized PDA lipid vesicles and HPLC method.
As described above, despite the similar cell concentration, our 3D-PDA-PDMS platform exhibited a significant difference in color changes between surfactin-producing NCIB3610 and surfactin-defective CU1065 strains. This observation strongly suggests that other chemicals, except for surfactin, produced by bacteria are not efficiently sensed by our 3D-PDA-PDMS platform. To further investigate the specificity of the 3D-PDA-PDMS platform to surfactin, five additional well-characterized bacteria were selected: B. licheniformis ATCC14580 [close relative to B. subtilis and surfactin-positive (El-sheshtawy et al., 2016)], B. cereus ATCC11778 (surfactin-lacking Bacillus strain), S. aureus Newman (surfactin-lacking Gram-positive pathogenic bacterium), P. aeruginosa PA14 (surfactin-lacking Gram-negative pathogenic bacterium), and E. coli DH5α (surfactin-lacking Gram-negative model bacterium). After cultivation of the cells for 24 h, their media supernatants were then exposed to 3D-PDA-PDMS, followed by observation of fluorescence images. As expected, only our 3D-PDA-PDMS incubated with cultured media from B. licheniformis exhibited a notable color change, whereas no remarkable change was observed for the other tested strains (Figure 8). Since there was no significant difference in the growth yield between the surfactin-producers and non-surfactin-producers, the color change detected by our 3D-PDA-PDMS seems to be mainly due to the presence of surfactin rather than the other chemicals produced by the cells.
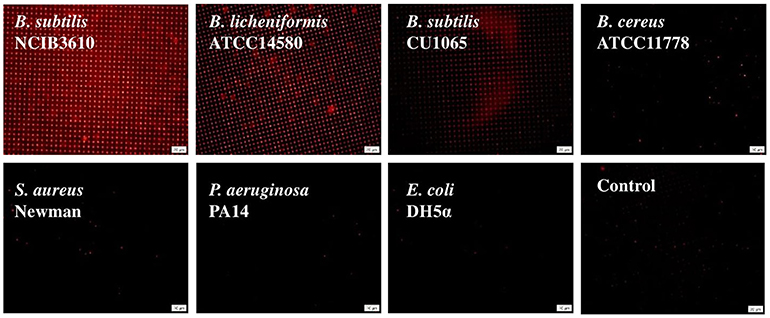
Figure 8. Fluorescence images of sensor platform after exposed to the media supernatant of different bacterial strains which were grown in LB for 24 h.
We developed a sensitive and reproducible 3D-PDA-PDMS for a surfactin-based label-free bacteria sensor by utilizing the chromatic response of immobilized amine functionalized PDA. The surface of amine-modified PDMS was uniformly modified with glutaraldehyde for facilitating PDA immobilization. The potential applications of 3D-PDA-PDMA as a feasible bacterial sensor was investigated through quantitative analysis of surfactin. The sensitivity of 3D-PDA-PDMS was approximately 100 times higher than that of the 2D-PDA-PDMS. The sensitivity enhancement of the 3D-PDA-PDMS was caused by the increment of total binding sites and the facile accessibility of the target molecules. The 3D-PDA-PDMS system can detect the presence of 5 μM surfactin and the initial presence of 4 × 101 cells/ml of Bacillus subtilis NCIB3610 which actively produce surfactin. Furthermore, the bacterial strains known to produce no surfactin were not detected by 3D-PDA-PDMS. It is therefore suggested that 3D-PDA-PDMS is highly specific to surfactin. The sensing ability of the 3D-PDA-PDMS was assessed through a comparison with the standard HPLC method. The results of the surfactin concentration produced by bacteria measured by 3D-PDA-PDMS were in agreement with those measured by HPLC. In summary, the 3D-PDA-PDMS can be used for sensitive and selective high throughput detection and screening of surfactin-producing bacterial strains.
FJ proposed the idea, and FJ performed the experiments with J-HK. JoK provided the diacetylene vesicles. HL and J-WL conceived the study, revised the manuscript, and supervised the work. JuK contributed to data analysis and manuscript review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Research Foundation (NRF) grants funded by the Ministry of Education (2012R1A6A1029029) and the Ministry of Science, ICT and Future Planning (IITP-2018-2015-0-00390). This work was also supported by the Ministry of Finance of the Republic Indonesia through Endowment Fund for Education (LPDP). Funding was also provided by the grant (FA2386-15-1-4081) from the AFOSR/AOARD, USA.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmats.2018.00057/full#supplementary-material
Ahmed, A., Rushworth, J. V., Hirst, N. A., and Millner, P. A. (2014). Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 27, 631–646. doi: 10.1128/CMR.00120-13
Apetroaie-Constantin, C., Mikkola, R., Andersson, M. A., Teplova, V., Suominen, I., Johansson, T., et al. (2009). Bacillus subtilis and B. mojavensis strains connected to food poisoning produce the heat stable toxin amylosin. J. Appl. Microbiol. 106, 1976–1985. doi: 10.1111/j.1365-2672.2009.04167.x
Bouffioux, O., Berquand, A., Eeman, M., Paquot, M., Dufrêne, Y. F., Brasseur, R., et al. (2007). Molecular organization of surfactin–phospholipid monolayers, Effect of phospholipid chain length and polar head. Biochim. Biophys. Acta 1768, 1758–1768. doi: 10.1016/j.bbamem.2007.04.015
Burnham, C.-A. D., and Carroll, K. C. (2013). Diagnosis of clostridium difficile infection: an ongoing conundrum for clinicians and for clinical laboratories. Clin. Microbiol. Rev. 26, 604–630. doi: 10.1128/CMR.00016-13
Carpick, R. W., Mayer, T. M., Sasaki, D. Y., and Burns, A. R. (2000). Spectroscopic ellipsometry and fluorescence study of thermochromism in an ultrathin poly(diacetylene) film: reversibility and transition kinetics. Langmuir 16, 4639–4647. doi: 10.1021/la991580k
Clardy, J., Fischbach, M. A., and Currie, C. R. (2009). The natural history of antibiotics. Curr. Biol. 19, R437–R441. doi: 10.1016/j.cub.2009.04.001
Croxen, M. A., Law, R. J., Scholz, R., Keeney, K. M., Wlodarska, M., and Finlay, B. B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880. doi: 10.1128/CMR.00022-13
Darken, M. A., Berenson, H., Shirk, R. J., and Sjolander, N. O. (1960). Production of tetracycline by streptomyces aureofaciens in synthetic media. Appl. Microbiol. 8, 46–51.
Davis, B. W., Burris, A. J., Niamnont, N., Hare, C. D., Chen, C. Y., Sukwattanasinitt, M., et al. (2014). Dual-mode optical sensing of organic vapors and proteins with Polydiacetylene (PDA)-embedded electrospun nanofibers. Langmuir 30, 9616–9622. doi: 10.1021/la5017388
de Oliveira, D. W. F., França, Í. W. L., Félix, A. K. N., Martins, J. J., Giro, M. E., Melo, V. M., et al. (2013). Kinetic study of biosurfactant production by Bacillus subtilis LAMI005 grown in clarified cashew apple juice. Coll. Surf. B Biointer. 101, 34–43. doi: 10.1016/j.colsurfb.2012.06.011
Deisingh, A. K., and Thompson, M. (2004). Biosensors for the detection of bacteria. Can. J. Microbiol. 50, 69–77. doi: 10.1139/w03-095
Deleu, M., Bouffioux, O., Razafindralambo, H., Paquot, M., Paquot, H., Thonart, P., et al. (2003). Interaction of surfactin with membranes: a computational approach. Langmuir 19, 3377–3385. doi: 10.1021/la026543z
Duc, L. H., Dong, T. C., Logan, N. A., Sutherland, A. D., Taylor, J., Cutting, S. M., et al. (2005). Cases of emesis associated with bacterial contamination of an infant breakfast cereal product. Int. J. Food Microbiol. 102, 245–251. doi: 10.1016/j.ijfoodmicro.2004.11.022
DuPont, H. L. (2007). The growing threat of foodborne bacterial enteropathogens of animal origin. Clin. Infect. Dis. 45, 1353–1361. doi: 10.1086/522662
El-sheshtawy, H. S., Aiad, I., Osman, M. E., Abu-Elnasr, A. A., and Kobisy, A. S. (2016). Production of biosurfactants by Bacillus licheniformis and Candida albicans for application in microbial enhanced oil recovery. Egypt. J. Petrol. 25, 293–298. doi: 10.1016/j.ejpe.2015.07.018
Espy, M. J., Uhl, J. R., Sloan, L. M., Buckwalter, S. P., Jones, M. F., Vetter, E. A., et al. (2006). Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19, 165–256. doi: 10.1128/CMR.19.1.165-256.2006
Fernández-No, I. C., Guarddon, M., Böhme, K., Cepeda, A., Calo-Mata, P., and Barros-Velázquez, J. (2011). Detection and quantification of spoilage and pathogenic Bacillus cereus, Bacillus subtilis and Bacillus licheniformis by real-time PCR. Food Microbiol. 28, 605–610. doi: 10.1016/j.fm.2010.10.014
Fournier, P.-E., Drancourt, M., Colson, P., Rolain, J. M., La Scola, B., Raoult, D., et al. (2013). Modern clinical microbiology: new challenges and solutions. Nat. Rev. Microbiol. 11:574. doi: 10.1038/nrmicro3068
Grangemard, I., Wallach, J., Maget-Dana, R., and Peypoux, F. (2001). Lichenysin. Appl. Biochem. Biotechnol. 90, 199–210. doi: 10.1385/ABAB:90:3:199
Härtig, E., and Jahn, D. (2012). “Chapter Five - Regulation of the Anaerobic Metabolism in Bacillus subtilis,” in Advances in Microbial Physiology, ed R. K. Poole (Cambridge: Academic Press), 195–216. doi: 10.1016/B978-0-12-394423-8.00005-6
Heo, Y. J., Chung, I. Y., Cho, W. J., Lee, B. Y., Kim, J. H., Choi, K. H., et al. (2010). The Major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J. Bacteriol. 192, 381–390. doi: 10.1128/JB.00980-09
Hupfer, B., Ringsdorf, H., and Schupp, H. (1983). Liposomes from polymerizable phospholipids. Chem. Phys. Lipids 33, 355–374. doi: 10.1016/0009-3084(83)90028-2
Ji, C.-J., Kim, J.-H., Won, Y.-B., Lee, Y.-E., Choi, T.-W., Ju, S.-Y., et al. (2015). Staphylococcus aureus PerR is a hypersensitive hydrogen peroxide sensor using iron-mediated histidine oxidation. J. Biol. Chem. 290, 20374–20386. doi: 10.1074/jbc.M115.664961
Kim, H.-S., Yoon, B.-D., Lee, C.-H., Suh, H. H., Katsuragi, T.et al., et al. (1997). Production and properties of a lipopeptide biosurfactant from Bacillus subtilis C9. J. Ferment. Bioeng. 84, 41–46. doi: 10.1016/S0922-338X(97)82784-5
Kim, J.-M., Lee, Y. B., Yang, D. H., Lee, J.-S., Lee, G. S., Ahn, D. J., et al. (2005). A polydiacetylene-based fluorescent sensor chip. J. Am. Chem. Soc. 127, 17580–17581. doi: 10.1021/ja0547275
Kim, J. M., Ji, E. K., Woo, S. M., Lee, H., and Ahn, DJ. (2003). Immobilized polydiacetylene vesicles on solid substrates for use as chemosensors. Adv. Mater. 15, 1118–1121. doi: 10.1002/adma.200304944
Kim, J. M., Lee, Y. B., Chae, S. K., and Ahn, D. J. (2006). Patterned color and fluorescent images with polydiacetylene supramolecules embedded in poly(vinyl alcohol) films. Adv. Funct. Mater. 16, 2103–2109. doi: 10.1002/adfm.200600039
Konings, W. N., Lolkema, J. S., Bolhuis, H., van Veen, H. W., Poolman, B., Driessen, A. J., et al. (1997). The role of transport processes in survival of lactic acid bacteria, Energy transduction and multidrug resistance. Antonie Van Leeuwenhoek 71, 117–128. doi: 10.1023/A:1000143525601
Lee, S., Lee, J., Lee, D. W., Kim, J. M., and Lee, H. (2016). A 3D networked polydiacetylene sensor for enhanced sensitivity. Chem. Commun. 52, 926–929. doi: 10.1039/C5CC08566G
Liu, X., Ren, B., Gao, H., Liu, M., Dai, H., Song, F., et al. (2012). Optimization for the production of surfactin with a new synergistic antifungal activity. PLoS ONE 7:e34430. doi: 10.1371/journal.pone.0034430
Ma, G., and Cheng, Q. (2005). Vesicular polydiacetylene sensor for colorimetric signaling of bacterial pore-forming Toxin. Langmuir 21, 6123–6126. doi: 10.1021/la050376w
Meir, D., Silbert, L., Volinsky, R., Kolusheva, S., Weiser, I., and Jelinek, R. (2008). Colorimetric / fluorescent bacterial sensing by agarose-embedded lipid / polydiacetylene films. J. Appl. Microbiol. 104, 787–795. doi: 10.1111/j.1365-2672.2007.03614.x
Moschou, D., and Tserepi, A. (2017). The lab-on-PCB approach: tackling the [small mu ]TAS commercial upscaling bottleneck. Lab Chip 17, 1388–1405. doi: 10.1039/C7LC00121E
Mubarak, M. Q. E., Jufri, S. H. M., Zahar, S. M. S. N. S., Sahaid, M., Hamid, A. A., and Isa, M. H. M. (2017). Kinetics of surfactin production by Bacillus subtilis in a 5 l stirred-tank bioreactor. Sains Malays 46, 1541–1548. doi: 10.17576/jsm-2017-4601-24
Ollinger, J., Song, K. B., Antelmann, H., Hecker, M., and Helmann, J. D. (2006). Role of the Fur regulon in iron trans-port in Bacillus subtilis. J. Bacteriol. 188, 3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006
Park, J., Ku, S. K., Seo, D., Hur, K., Jeon, H., Shvartsman, D., et al. (2016). Label-free bacterial detection using polydiacetylene liposomes. Chem. Commun. 52, 10346–10349. doi: 10.1039/C6CC03116A
Peypoux, F., Bonmatin, J. M., and Wallach, J. (1999). Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 51, 553–563. doi: 10.1007/s002530051432
Pires, A. C. d., da Silva, L. H. M., da Silva, M. d. C. H., De Almeida, M. V., Le Hyaric, M., et al. (2011). A colorimetric biosensor for the detection of foodborne bacteria. Sens Actuat. B 153, 17–23. doi: 10.1016/j.snb.2010.09.069
Razafindralambo, H., Thonart, P., and Paquox, M. (2004). Dynamic and equilibrium surface tensions of surfactin aqueous solutions. J. Surfactants Deterg. 7, 41–46. doi: 10.1007/s11743-004-0286-x
Reimhult, E., Höök, F., and Kasemo, B. (2002). Vesicle adsorption on SiO2 and TiO2: dependence on vesicle size. J. Chem. Phys. 117, 7401–7404. doi: 10.1063/1.1515320
Salem, M., Mauguen, Y., and Prangé, T. (2010). Revisiting glutaraldehyde cross-linking: the case of the Arg–Lys intermolecular doublet. Acta Crystallogr. Sect. F 66, 225–228. doi: 10.1107/S1744309109054037
Scindia, Y., Silbert, L., Volinsky, R., Kolusheva, S., and Jelinek, R. (2007). Colorimetric detection and fingerprinting of bacteria by glass-supported lipid/polydiacetylene films. Langmuir 23, 4682–4687. doi: 10.1021/la0636208
Seo, H., Singha, S., and Ahn, K. H. (2017). Ratiometric fluorescence detection of anthrax biomarker with EuIII-EDTA functionalized mixed poly(diacetylene) liposomes. Asian J. Org. Chem. 6:1257–1263. doi: 10.1002/ajoc.201700158
Silbert, L., Ben Shlush, I., Israel, E., Porgador, A., Kolusheva, S., and Jelinek, R. (2006). Rapid chromatic detection of bacteria by use of a new biomimetic polymer sensor. Appl. Environ. Microbiol. 72, 7339–7344. doi: 10.1128/AEM.01324-06
Song, X., Wang, H. L., Shi, J., Park, J. W., and Swanson, B. I. (2002). Conjugated polymers as efficient fluorescence quenchers and their applications for bioassays. Chem. Mater. 14, 2342–2347. doi: 10.1021/cm011681f
Stoner, S. A., Duggan, E., Condello, D., Guerrero, A., Turk, J. R., Narayanan, P. K., et al. (2016). High sensitivity flow cytometry of membrane vesicles. Cytometry 89, 196–206. doi: 10.1002/cyto.a.22787
Tang, S., Ma, W., Xie, G., Su, Y., and Jiang, Y. (2016). Acetylcholinesterase-reduced graphene oxide hybrid films for organophosphorus neurotoxin sensing via quartz crystal microbalance. Chem. Phys. Lett. 660, 199–204. doi: 10.1016/j.cplett.2016.08.025
Teschke, O., and de Souza, E. F. (2002). Liposome structure imaging by atomic force microscopy: verification of improved liposome stability during adsorption of multiple aggregated vesicles. Langmuir 18, 6513–6520. doi: 10.1021/la025689v
Thwaite, J. E., and Atkins, H. S. (2012). “21 - Bacillus: Anthrax; food poisoning A2 - Greenwood, David,” in Medical Microbiology 18th Edn, M. Barer, R. Slack, and W. Irving W. (Edinburgh: Churchill Livingstone), 237–244. doi: 10.1016/B978-0-7020-4089-4.00036-6
Wu, J., Zawistowski, A., Ehrmann, M., Yi, T., and Schmuck, C. (2011). Peptide functionalized polydiacetylene liposomes act as a fluorescent turn-on sensor for bacterial lipopolysaccharide. J. Am. Chem. Soc. 133, 9720–9723. doi: 10.1021/ja204013u
Wu, W., Zhang, J., Zheng, M., Zhong, Y., Yang, J., Zhao, Y., et al. (2012). An aptamer-based biosensor for colorimetric detection of Escherichia coli O157:H7. PLoS ONE 7:e48999. doi: 10.1371/journal.pone.0048999
Yoneda, T. (2001). Surfactin sodium salt : an excellent biosurfactant for cosmetics. J. Cosmet. Sci. 52, 153–154.
Zhou, J., Ellis, A. V., and Voelcker, N. H. (2010). Recent developments in PDMS surface modification for microfluidic devices. Electrophoresis 31, 2–16. doi: 10.1002/elps.200900475
Keywords: biointerfaces, label-free sensor, liposome, polydiacetylene, surfactin, bacteria sensor, polydimethylsiloxane
Citation: Jannah F, Kim J-H, Lee J-W, Kim J-M, Kim J-M and Lee H (2018) Immobilized Polydiacetylene Lipid Vesicles on Polydimethylsiloxane Micropillars as a Surfactin-Based Label-Free Bacterial Sensor Platform. Front. Mater. 5:57. doi: 10.3389/fmats.2018.00057
Received: 23 April 2018; Accepted: 29 August 2018;
Published: 18 September 2018.
Edited by:
Nam-Joon Cho, Nanyang Technological University, SingaporeReviewed by:
Joshua A. Jackman, Stanford University, United StatesCopyright © 2018 Jannah, Kim, Lee, Kim, Kim and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiwon Lee, aGFpd29uQGhhbnlhbmcuYWMua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.