- 1Faculty of Law, Queensland University of Technology, Brisbane, QLD, Australia
- 2Griffith Law School, Faculty of Arts, Education and Law, Griffith University, Nathan, QLD, Australia
- 3Department of Life Sciences, Natural History Museum, London, United Kingdom
- 4London Centre for Neglected Tropical Disease Research, London, United Kingdom
- 5Department of Chemistry, School of Natural and Computing Sciences, University of Aberdeen, Aberdeen, United Kingdom
Negotiations are underway for a new treaty for the conservation and sustainable use of biodiversity of areas beyond national jurisdiction under the United Nations Convention on the Law of the Sea. Points of contention in the negotiation concern marine genetic resources and questions of monetary and/or non-monetary benefit sharing arising from their use. Tracing the origin of marine genetic resources used in scientific research, development and commercialization may offer the evidence needed to prove they came from areas beyond national jurisdiction and that benefit sharing is owed. Traceability is complex and multidisciplinary: involving legal, scientific and informatics considerations. We look at different traceability approaches within national jurisdictions and how these might provide lessons for the proposed treaty, using one of the few case studies available to trace the commercial development of a marine genetic resource from areas beyond national jurisdiction. We discuss this case study in relation to existing legal frameworks including the Nagoya Protocol and other systems based on open sharing of information and materials, including existing scientific practice. We conclude that a well-designed traceability system tailored to the unique geographical, political and jurisdictional characteristics of areas beyond national jurisdiction could lead to more equitable outcomes for the sharing of benefits from the use of marine genetic resources. Our key recommendations are that any traceability mechanism needs to be light touch, integrated with existing systems such as bioinformatics databases and not impose additional burdens on scientific users. Systems should be designed to improve scientific knowledge of ocean biodiversity to allow better conservation measures to be developed. If treaty negotiators engage commercial sectors to find workable policy solutions for the draft treaty that promote greater transparency and data sharing from these sectors, there may be a greater chance for traceability mechanisms to support benefit sharing.
Introduction
Countries are negotiating a new treaty framework for the conservation and sustainable use of marine biological diversity in areas beyond national jurisdiction (the BBNJ Agreement). Key areas of debate in the treaty negotiations are: (1) how to regulate the collection and use of these marine genetic resources (MGRs) that are not within the sovereign control of any country; (2) whether to require monetary as well as non-monetary benefits from their use and if so; (3) how to share any benefits with other countries, particularly developing countries who may have limited technological capabilities to undertake this work (UN, 2019). Being able to point to evidence that a final product (e.g., a pharmaceutical or cosmetic) incorporates MGRs of ABNJ is a key step for establishing that benefit sharing is owed from the use of the original MGR. Although traceability mechanisms are essential for establishing this connection and have been explored in the context of genetic resource access and benefit sharing (ABS) (Rohden et al., 2020), there is relatively little research about how they may work in practice for the BBNJ Agreement (see Humphries et al., 2020). Understanding what ‘traceability’ means varies between international ABS frameworks and national laws. Using a case study of AbyssineTM, a cosmetic product incorporating products derived from an MGR of ABNJ, this paper aims to explore the practical operation of four key traceability approaches and offers insights into how they may work in the context of the BBNJ agreement.
The BBNJ Agreement proposes to address implementation and governance gaps in the United Nations Convention on the Law of the Sea (UNCLOS) concerning the conservation and sustainable use of BBNJ (UN, 2012). The UN’s mandate is to conduct four negotiating sessions for parties to agree on the framework for four key elements concerning biodiversity – (1) marine genetic resources (MGRs), including questions on the sharing of benefits; (2) area-based management tools, including marine protected areas; (3) environmental impact assessments; and (4) capacity-building and the transfer of marine technology. The November 2019 Draft Text includes provisions on access and benefit sharing (ABS) of MGRs and traditional knowledge associated with MGRs (UN, 2019)1. Section “BBNJ Proposed Approach to Traceability in the Draft Text” outlines the current proposed approach to traceability in the Draft Text.
During text negotiations, there has been little agreement between countries about how to practically achieve traceability in the BBNJ context. In the closing remarks of the third negotiating session on the Draft Text in 2019, the Facilitator of the MGR element outlined the differing country positions about whether a traceability system is even desirable or feasible,
With regard to monitoring, generally, further discussions could clarify how to balance the need for transparency in the utilization of marine genetic resources of areas beyond national jurisdiction with the need to avoid creating disincentives for marine scientific research. Two divergent perspectives were noticeable. One perspective emphasized the need for a robust track-and-trace mechanism and consequently offered proposals on who would be in charge of monitoring, the activities that would be subject to monitoring, and how monitoring would be carried out, including whether it would be through the clearing-house mechanism, a scientific and technical body, an obligatory notification system or a combination of those mechanisms. Another perspective questioned the feasibility and desirability of a monitoring mechanism that would include the use of identifiers, notifications by databases, repositories and gene banks, and the submission of periodic status reports by proponents of marine scientific research in areas beyond national jurisdiction (UN, 2019, p. 7).
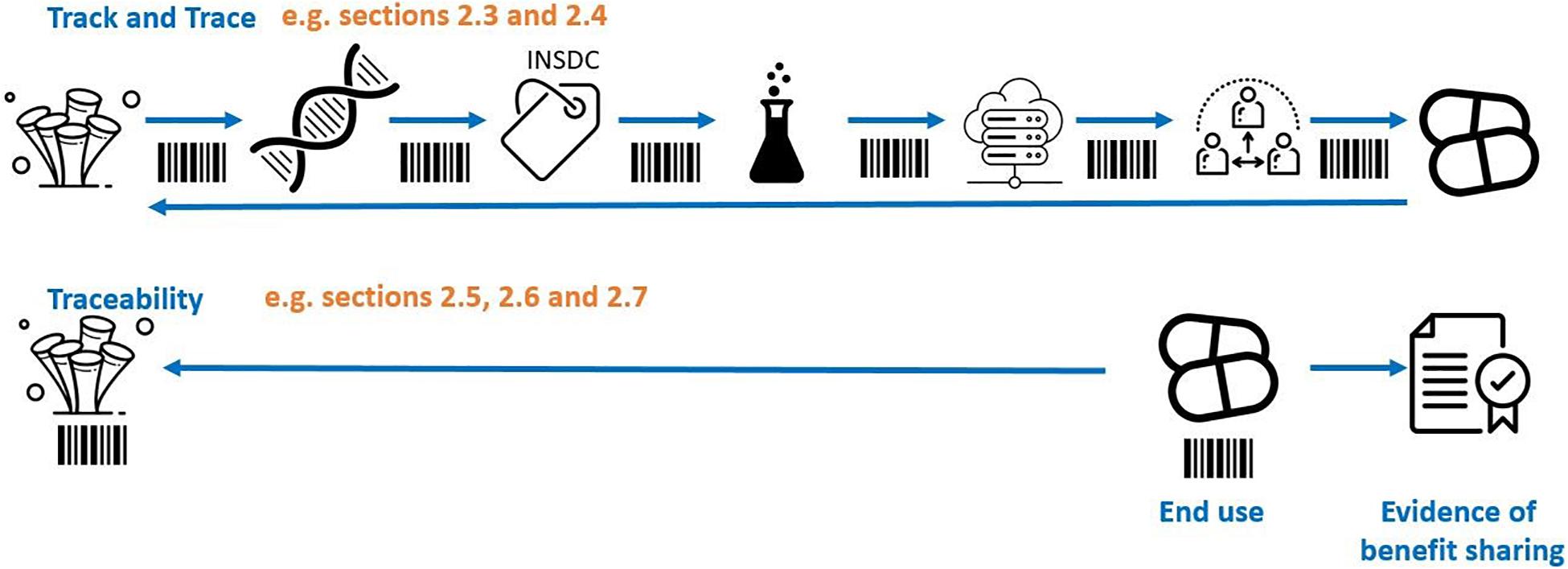
Various traceability mechanisms have been proposed in the treaty discussion to date, for example ‘track and trace.’ Conceptually it is helpful to visualize ‘track and trace’ methods of traceability as akin to a parcel tracking system that records the movements of a parcel at every point along the chain of custody (Figure 1). In the context of BBNJ, this would require reporting obligations and monitoring infrastructure (e.g., identifiers, reports) to link subsequent uses and benefit sharing back to the original collection of potentially very large numbers of MGRs from areas beyond national jurisdiction covering over 60% of the planet’s oceans (Humphries et al., 2020). The ‘track and trace’ approach to ABS is outlined in sections “Traceability Infrastructure Under CBD and Nagoya Protocol” and “Track and Trace Options.” Other traceability mechanisms are more akin to an automobile recall system where it is not necessary to track the location of a car at every moment (Humphries et al., 2020; Figure 1). In this case, traceability is only triggered by an event (e.g., an engine problem) at which time the software will piece together the ownership details through connected databases (e.g., car registration systems). MGR traceability options using this approach are outlined in sections “Multilateral (Contractual) Traceability Options” and “End-Product/End-User Traceability Options.” Section “Existing Open-Access Traceability Options” highlights existing open access systems and section “Combined Traceability approaches” outlines traceability options using a combination of traceability approaches.
Figure 1. ‘Track and trace’ showing tracing throughout the chain of custody versus tracing origin from the end product/user (simplified example). Adapted from Humphries et al. (2020). The images are from the Noun Project.
Identifiers are an essential component of traceability, which allow this connectivity between databases. For identifiers to be valid, i.e., provide stable links (in science generally referred to as ‘unique’ or ‘persistent’), they need to meet certain criteria: resolvability, persistence, authority and uniqueness (Guralnick et al., 2015). Digital Object Identifiers (DOIs) for example fulfill these requirements. Such identifiers are in widespread usage in biology, such as globally unique identifiers or GUIDS (which meet the above criteria), allocated to individual specimen records (reviewed in Rabone et al., 2019). In contrast, current identifiers in legal contexts are unlikely to meet these requirements or provide stable links between systems (see section “Track and Trace Options”).
When considering the scope of what is to be traced, there is little agreement in the November 2019 Draft Text about whether information associated with genetic resources should be included in the governance and traceability arrangements (UN, 2019, p. 6). BBNJ physical materials and information are different entities, but intrinsically linked, given that information originates from a sample, and the scientific value of a sample equates to its associated data. In terms of physical sample, a marine organism collected from an ABNJ may contain biochemicals that can be used in generating a commercial product or process, such as a novel pharmaceutical, or an enzyme that can be used in an industrial process. In some cases, it is the associated information generated from a sample that is utilized for commercial applications, rather than the physical sample itself. The use of sequence data from MGR samples (DNA/RNA sequences and possibly protein sequences: see Houssen et al., 2020) to generate products and processes relies heavily on available sequence information in large databases (i.e., INSDC, the International Nucleotide Sequence Database Collaboration). These databases contain billions of sequences (DNA/RNA/proteins), some of which have been annotated with experimentally confirmed functions or putative functions based on theoretical predictions. Using sequence information is more straightforward for the generation of proteins and enzymes than small molecule metabolites, e.g., potential pharmaceuticals. Inherent in the idea of traceability of MGR is the assumption that a link between the MGR and a final product can be made. In some cases this may be possible (and where the ‘final product’ is not a commercial product but a scientific publication this is generally the case) but this becomes very difficult when it comes to derivatives (e.g., proteins/chemical compounds). Some forums have attempted to address this to a certain extent e.g., intellectual property (see section “End-Product/End-User Traceability Options”) and some multilateral systems (see section “Multilateral (Contractual) Traceability Options”). However, the scope of what is actually being traced is a fundamental policy question that is missing from the ILBI discussion about how to implement monitoring in practice. To simplify the analysis of traceability mechanisms that relate to information associated with genetic resources, this paper will consider the implications for nucleotide sequence data (NSD) as a subset of information comprised of DNA and RNA sequence data and annotations (Houssen et al., 2020).
There is limited data showing the actual value and volume of commercial products that incorporate MGRs of ABNJ (Broggiato et al., 2018). On the one hand this may indicate traceability tools are necessary to link MGR origins with benefit sharing under the ILBI. On the other hand, it may raise questions about whether a potential traceability mechanism is commensurate with the scale of activity or even necessary. This paper explores the feasibility and practical considerations of traceability options using one of the few documented examples of MGRs collected from ABNJ being incorporated into a cosmetic product for commercial sale – the case of Alteromonas macleodii subsp. fijiensis biovar deepsane (the ‘Deepsane’ strain).
There are many reported uses of MGRs in cosmetics and other personal care products (Martins et al., 2014). Most of these are derived from organisms collected within national jurisdiction, but one, marketed as the face cream ‘AbyssineTM’ contains materials derived from organisms collected in ABNJ. A French Cruise owned and conducted by L’Institut Français de Recherche pour l’Exploitation de la Mer (IFREMER) in 1987 (‘Hydronaut’ cruise) collected the worm Alvinella pompejana at a depth of 2625 m from a hydrothermal vent located on the East Pacific Rise, from which a microorganism was isolated. Researchers at IFREMER carried out the basic research on the microorganism (16S rRNA sequence, phylogenetic and chemical analyses) that was found to produce an unusual exopolysaccharide, which provides protection to a cell. The bacterium (HYD657) isolated from the worm, later identified as Alteromonas macleodii subsp. fijiensis biovar deepsane based on its biological characteristics and 16S rRNA2 sequence (Cambon-Bonavita et al., 2002) and entered this genetic sequence into the European Molecular Biology Laboratory (EMBL) database. IFREMER holds a patent (see Table 1) for the method of producing a low-molecular-weight sulfated polysaccharide derivative (‘Deepsane’) from the microorganism that could be used in cosmetics and pharmaceuticals. Later research defined the full biological activity of this material and determined its complete chemical structure (Le Costaouëc et al., 2012). Commercial research was conducted by Lanatech, which patented a skin-care application of the material. The material was produced commercially by Lucas Meyers, later purchased by the cosmetic ingredients company Unipex. Cosmetics incorporating the material and marketed under the AbyssineTM brand are promoted to soothe sensitive skin against irritation caused by chemical, mechanical and UV-B exposure.

Table 1. Information obtained from a combination of the Cruise report,1 Cambon-Bonavita et al. (2002), GenBank2.
Table 1 identifies the key events concerning the collection, research, development, and commercialization that are relevant for the traceability option analysis outlined in this article.
For each of the traceability options examined in section “Traceability Policy Options and Implications,” we briefly outline examples of legal and policy frameworks and identify key considerations for effective implementation of the option in the BBNJ context using the AbyssineTM Deepsane case study. In our analysis we do not suggest those involved in Deepsane case acted contrary to their obligations at the time of access or use. Instead, we analyze how traceability mechanisms could work in future for other cases, using the facts of this case. Section “Policy Recommendations” makes policy recommendations for traceability options in the BBNJ context followed by a conclusion in section “Conclusion.”
Traceability Policy Options and Implications
BBNJ Proposed Approach to Traceability in the Draft Text
The November 2019 Draft Text outlines proposed obligations for access and utilization of MGRs, the sharing of benefits, monitoring and intellectual property concerning MGRs. Countries have not yet agreed about which body would conduct monitoring of the utilization of MGRs of ABNJ. Proposed obligations for collecting MGRs of ABNJ include a requirement to provide the secretariat with information about the collection, including geographical coordinates, and to deposit samples in collections and data in open source platforms such as databases and gene banks [draft article 10(2)]. Negotiating countries have agreed there should be benefit sharing obligations (draft article 11) but disagreement remains about the modalities of benefit sharing (UN, 2019).
Options for monitoring under draft article 13 include the proposed BBNJ clearing house mechanism, a scientific and technical body, an electronic notification system managed by the BBNJ secretariat and existing international institutions. Countries disagree about traceability infrastructure that States Parties must incorporate in their legislative and other measures (UN, 2019). Options in the Draft Text include assigning an identifier to MGRs collected in ABNJ and those in ex situ and in silico conditions3, obligations for notifying a BBNJ body when MGRs are accessed and obligations for researchers to submit periodic status reports and research findings to a BBNJ body. The use of the term ‘identifier’ in the Draft Text appears to refer to a legal identifier (used for administrative purposes), not a unique identifier associated with a specimen (generally used for scientific purposes) that is retrievable and persistent (see section “Existing Open-Access Traceability Options” below). However, the meaning and use of the term in the text is not clear, potentially causing confusion for some delegations. The Draft Text lacks a shared understanding of: (a) the type of identifier it proposes; (b) the capabilities or limits of legal and scientific identifiers; and (c) how they might interact (or not) in a practical sense within databases and between countries’ existing traceability mechanisms. A key area of disagreement in the negotiations is whether the proposed agreement should include intellectual property provisions (UN, 2019). Options relevant to traceability in the Draft Text include requiring States Parties to take domestic measures to ensure that applicants or users of patents on inventions incorporating MGRs of ABNJ disclose the origin of the MGRs [draft article 12(3)].
Figure 2 outlines the proposed monitoring infrastructure at the various phases of collection and use of physical materials and NSD. This figure illustrates that the primary burden for monitoring is on the initial researcher and not downstream users. In relation to the Deepsane example, under the Draft Text approach, the initial researcher IFREMER would have ongoing reporting obligations, an obligation to assign an identifier to samples and information and an obligation to deposit duplicate samples in an ex situ facility. However, it is unclear how far these obligations would extend to IFREMER to ensure subsequent users must also comply with these traceability measures. Downstream users such as Lucas Meyers would have no traceability obligations under the current proposed Draft Text model, obligations would only potentially arise if they patented an invention incorporating BBNJ materials (possible disclosure of origin obligations, see Table 1). In the case of US Patent 8,598,142, the inventors and assignees are all French based so there appears to be no traceability obligation on the entity that commercializes the product incorporating MGRs from ABNJ in this case.
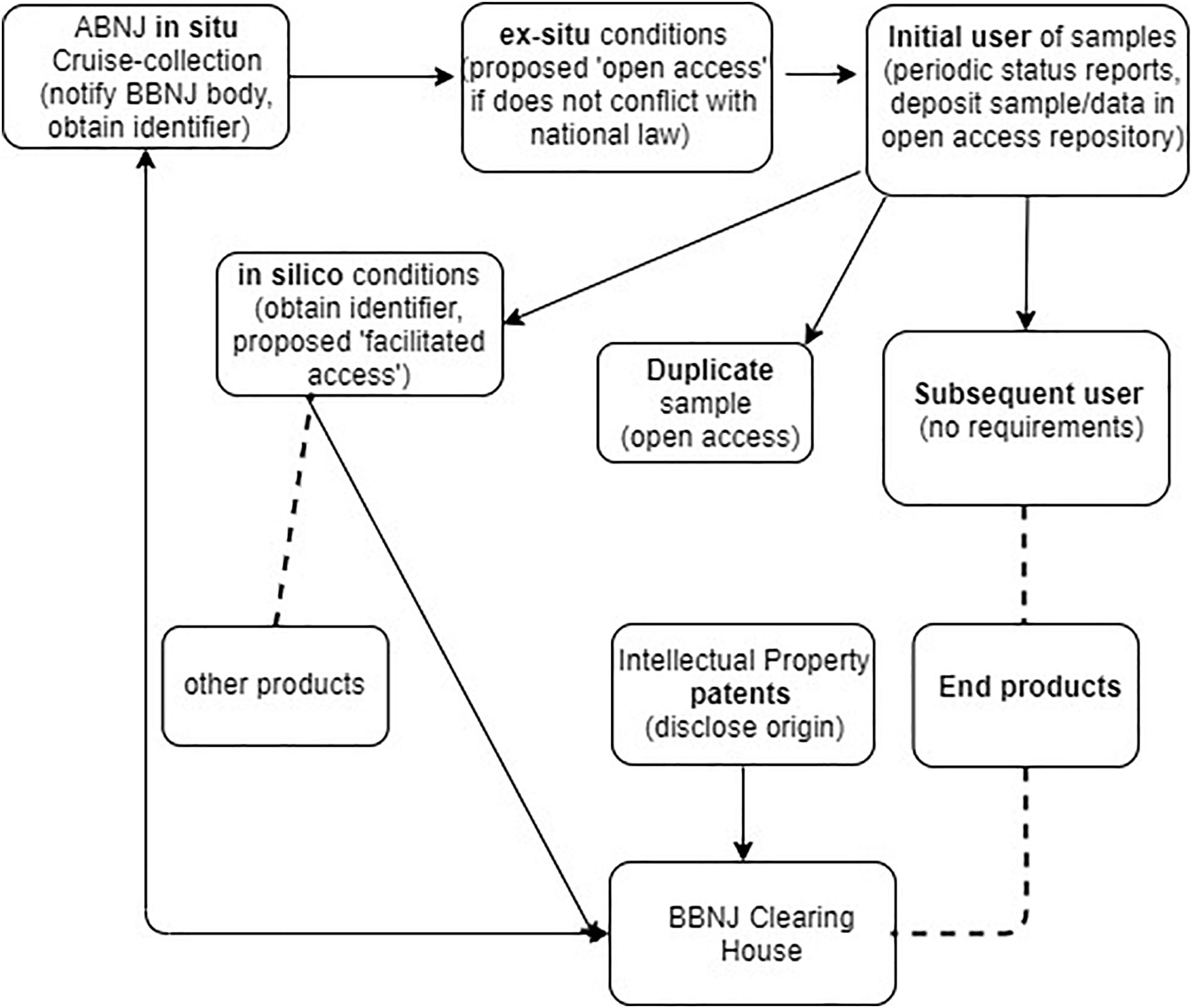
Figure 2. November 2019 Draft Text proposed obligations in relation to proposed requirements for traceability of MGR, e.g., requirement for deposit of samples/data in an open access repository or disclosure of origin for any patent arising from a product developed from an MGR. Arrows show movement/requirements for MGR samples or data, dashed line shows break in traceability.
Traceability Infrastructure Under CBD and Nagoya Protocol
Under the Convention on Biological Diversity (CBD) and the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from Their Utilization to the Convention on Biological Diversity (Nagoya Protocol), countries can exercise their sovereign rights to implement a traceability system for their genetic resources and associated knowledge that suits their national interests. The Nagoya Protocol sets up a minimum standard of a monitoring and compliance framework for countries to help ensure that genetic resources utilized within their jurisdiction have been accessed in accordance with prior informed consent (PIC), and that mutually agreed terms (MAT) have been established (articles 15-18). The traceability elements primarily relate to Internationally Recognized Certificates of Compliance (IRCC), the checkpoint mechanism, the ABS Clearing House (ABSCH) and User Measures outlined below. Any infrastructure developed for the ILBI would need to account for this infrastructure to avoid duplication and ensure consistency with national CBD approaches to traceability.
The Internationally Recognized Certificates of Compliance (IRCC), the checkpoint mechanism and the ABSCH4 is infrastructure designed to capture evidence of compliance with PIC and establishment of MAT in accordance with national laws. The IRCCs are usually in the form of an access permit5 but they may be in another form6. IRCCs must contain certain non-confidential information, including details of issuing authority, the provider and user, the unique identifier of the certificate (known as the ‘ABSCH Unique Identifier’), the subject matter of resources covered by the certificate, confirmation that PIC was obtained and that MAT was established and the type of use allowed under the authorization (NP article 17). Each IRCC is issued with an ABSCH Unique Identifier, which is attached to the certificate and not assigned to the actual genetic resources that are the subject of the authorization. Amendments to the records can be tracked by adding a revision number to the ABSCH Unique Identifier7. As at March 24, 2021, there are 2142 IRCCs recorded on the ABSCH site, but there is little information about the movement or physical location of the genetic resources after the ABSCH Unique Identifier is assigned.
To assist with monitoring movements within and between jurisdictions, Parties to the Protocol must designate one or more checkpoints to collect or receive relevant information (including the IRCC) on PIC, MAT, and the source and use of genetic resources (article 17). These checkpoints may relate to the collection or use of materials or information at any stage of the research, development, innovation, pre-commercialization, or commercialization phases. This has resulted in a range of checkpoints that vary according to national law, including intellectual property offices, pharmaceutical regulatory agencies and public research bodies, universities and museums8. Many countries have not yet designated checkpoints and those that do are mainly departments, organizations and institutions that handle the movement of physical samples and not NSD.
User Measure obligations tackle movements of materials and knowledge across borders. The Protocol requires States Parties to establish User Measures, namely to: (a) adopt measures to provide for compliance with provider country PIC and MAT requirements; (b) enforce user compliance with these measures; and (c) cooperate with other countries to address alleged violation of provider country measures (article 15). A user country that waives its own right to require PIC may still be subject to these obligations to comply with provider country requirements (Greiber et al., 2012). These obligations are duties to confirm that users have complied with PIC at the time of access and not an obligation on countries to apply provider country laws and remedies (Morgera et al., 2014). There are very few examples of countries with User Measures embedded in their ABS legislation (e.g., Uganda9 and Malaysia10). The European Union’s due diligence framework essentially operates as a User Measure, where EU Member States must ensure that before someone can use genetic resources and traditional knowledge in their jurisdiction, they first need to demonstrate they were accessed in accordance with the legislation of the providing country11.
‘Track and Trace’ Options
The benefit of using a ‘track and trace’ approach is that it helps to provide evidence linking the benefits from the use of a particular resource to the original access within a country. This section uses the Deepsane example to illustrate that unless the relevant materials fall within scope of ABS laws and all countries along the research and development continuum for the particular resource have traceability infrastructure and obligations similar to those under the Nagoya Protocol framework (section “Traceability Infrastructure Under CBD and Nagoya Protocol” above), then it is difficult if not impossible to ‘track and trace’ the movement of ABNJ MGRs and associated information once they leave the possession of the original collectors of the samples.
The Nagoya Protocol’s infrastructure theoretically helps countries that have implemented ‘track and trace’ systems to know where the resources/knowledge are at any given time and how they are being used. At the national level, this may also be assisted by reporting, change of use and transfer provisions, which significantly vary in practice between countries. Reporting obligations range from sending regular and multiple reports to the government on the progress of research by users, such as in Vietnam12, to requirements for personal record keeping such as in Australia13. Many countries such as Zambia14 require users to reapply for government and/or community permission when they seek to change the use of the resources or knowledge, such as from non-commercial to commercial research. There are several approaches that countries take to track the transfer of genetic materials to third parties or transfers of materials out of the country. Some countries such as India15 only allow transfers with government permission in addition to the original access requirements. Countries such as Kenya have prescribed material transfer agreement and reporting processes for sending resources outside the country with strict use conditions16. Other countries like Ethiopia require letters from international users’ home countries that they will enforce Ethiopia’s ABS (including reporting) obligations (Mulesa and Westengen, 2020). There is little published information about whether or how countries with ABS laws manage the usage of identifiers to trace the materials and/or associated information under their monitoring frameworks within and between jurisdictions in practice.
In the case of Deepsane, France might have claimed jurisdiction over the genetic resources because they were collected by a French research organization on a French owned ship. Although States have no sovereign rights of ABNJ resources in situ, once they are landed on board a vessel, they fall within the jurisdiction of the flag state (Humphries et al., 2020). If the ship was registered in another jurisdiction, that State might raise a competing claim for jurisdiction over the resources. France’s ABS law came into effect in 2016 and would not have applied to the Deepsane collection (HYD 657). If a similar collection were to occur today, French law excludes genetic resources collected from areas outside its national jurisdiction, indicating that MGRs collected from ABNJ (even those held in French ex situ collections) are not subject to ABS and its traceability arrangements17. However, being part of the European Union means that users of genetic resources in France are subject the EU’s Due Diligence system so that France must ensure compliance with the ABS legislation of a provider Party to the Nagoya Protocol of resources accessed after 201418. In the Deepsane case, the resources were collected in 1987 so the EU Due Diligence procedures again would not apply and there is no avenue for ‘track and trace’ under this infrastructure.
There might also be competing claims to benefits from the use of MGRs from other jurisdictions involved in the movement of the samples and information, for example the port where the samples were first landed after collection from ABNJ. The question comes down to the scope of subject matter that is captured under national laws. The CBD’s framework offers States Parties flexibility to include within their ABS genetic resources for which they are the country of origin or the country where they have gained their distinctive characteristics (article 2). States have interpreted this in a variety of ways ranging from only including native genetic resources within scope of their measures (e.g., Australia19), to including those that are simply present in their jurisdiction, and not only those for which they are the country of origin (e.g., Malaysia20). Some countries such as Uganda exclude resources merely transiting through its jurisdiction21. In the Deepsane case, the materials came from ABNJ to Mexico. Mexico has ABS measures but not a comprehensive legally binding ABS framework (Jefferson et al., 2018)22, which would not have applied to the Deepsane collection in 1987. For future ABNJ collections, whether ABS laws and ‘track and trace’ requirements apply to transit jurisdictions, it may come down to whether the samples are deemed to have ‘landed’ in the port country (or simply remained on the vessel before returning home) and whether they were in scope and accessed, used or transferred within the meaning of the relevant ABS law of the port country.
Another jurisdiction along the chain of custody may be ex situ facilities to which duplicate samples are sent. For example, in the Deepsane case, a duplicate sample was sent in 1995 to a facility in France for the purposes of satisfying patentability requirements23 (Table 1). There may have been other samples (not on the public record) sent to facilities in other countries that might claim jurisdiction over them. Whether ex situ materials are captured by ABS laws varies considerably between countries. Some laws do not apply to ex situ resources in private facilities on private land, while others do (e.g., Australia24). As French ABS obligations do not appear to apply in the Deepsane case (see above), there would be no legal obligation for subsequent users to make public the movement of the samples to other facilities around the world. When the resource is subsequently developed in a country where it does fall within scope, then ABS obligations may be triggered.
Where MGRs of ABNJ are captured under national ABS laws and traceability mechanism, there are several infrastructure challenges for maintaining evidence of the chain of custody of MGRs of ABNJ through a ‘track and trace’ system similar to that under the Nagoya Protocol’s infrastructure. Challenges for physical materials include the reliance on users to update the government about the use and movement of the resources, when sometimes it is more economically viable to pay a fine for failure to report. The international checkpoint system is designed to catch unauthorized movements and uses but the system is currently ad hoc with many gaps for undetected movements. As regular information updates to the ABSCH depend on the goodwill or priorities of Parties to the Nagoya Protocol, often the information about resource/knowledge uses and movements and IRCCs is out of date or unavailable. User Measures under the Nagoya Protocol framework only ensure that MAT have been established and do not cover compliance with the terms or content of a benefit sharing agreement to ensure that the share of the benefits from the use of the materials or knowledge with the provider is fair and equitable (Young and Tvedt, 2017). However, the Protocol obliges all parties ‘to ensure that an opportunity to seek recourse is available under their legal system, consistent with applicable jurisdictional requirements, in cases of disputes arising from mutually agreed terms’ [article 18(2)].
These challenges are magnified in the case of information associated with MGRs. It is difficult to prove the origin and provider of the information for the purposes of sharing the benefits of the information’s use, particularly when the information is used separately from the physical materials and entered into an open-access database system (see section “Existing Open-Access Traceability Options”). A CBD fact-finding study in 2020 found that the only system where it is practically possible to link an accession number of an NSD entry to an access permit for the purposes of traceability is the ABSCH Unique Identifier and link generated by an IRCC published on the ABS Clearing House website (Rohden et al., 2020). Whereas it will be possible to have a link from the metadata of the NSD to the IRCC, the reverse may be more difficult. This is because numbers of sequences (NSD) from a given genetic resource may increase and linking these from the IRCC to the metadata of the NSD will require constant updates to the IRCC. The CBD study authors could not find an example of a linkage between an IRCC and NSD entry and suggest the reason is the relative novelty of the system. Tracing categories of information that are broader than NSD such as contextual or ecological information is beyond current technical capabilities for ‘track and trace’ and have been excluded from the working definition of DSI (Digital Sequence Information) by the AHTEG (Scholz et al., 2020; CBD AHTEG report, 2020).
For this kind of ‘track and trace’ system to work in BBNJ, due diligence evidencing compliance with ABS measures and an update to the ABSCH Unique Identifier would be required under national laws at every step (from collection to final product) by the next user in every jurisdiction they move through. Blockchain is attracting more attention as a potential platform for recording objects, providers, users, and ABS terms and conditions across national jurisdictions (Scholz et al., 2020), for example through the use of smart contracts. These contracts may take many forms but are generally a ‘computerized program between two or more parties, whose conditions are defined in advance and stored in the blockchain, and which can be automatically executed or enforced, without the need for a third party’ (Carron and Botteron, 2019 p. 106). The provider country however must have the technological capacity to interact with the blockchain infrastructure, such as creating, accessing, and approving smart contracts as a basis for linking subsequent users to the original terms and conditions of access (Scholz et al., 2020). Blockchain technology could enable ‘track and trace’ approaches but could equally be used for other traceability approaches outlined below. If BBNJ negotiators or implementers consider blockchain as a tool for MGR regulation, they might weigh the benefits of traceability with the energy intensive requirements of blockchain and its appropriateness for an environmental treaty (Mignon, 2019; Leape et al., 2020). Scholz et al. (2020) have also pointed out that major upfront investment would be required for such technology development, which is difficult to justify given existing traceability of NSD through the INSDC infrastructure; and potential issues futureproofing for a rapidly evolving technology. As a general point, maintenance of both databases (e.g., INDSC and biodiversity databases25) and physical collections have substantial (long-term) cost implications (Rabone et al., 2019; Collins et al., 2020; Rohden et al., 2020), which should be considered in the development of any potential traceability system. The cost of developing a new, stand-alone global traceability system should be balanced against the more likely successful and more cost-effective approach of building on, and improving, current scientific collections and databases.
If BBNJ negotiators are considering extending the CBD’s ‘track and trace’ model to MGRs collected within ABNJ, then the infrastructure and processes would need to be compatible with those within national jurisdiction for the reasons outlined above. Figure 3 illustrates using the Deepsane case that while there may be evidence of movement of samples and information from the time of collection, there is no legal means for tracking and tracing the materials back to BBNJ (e.g., to the BBNJ Clearing House Mechanism). This is because firstly French ABS law (including its monitoring and due diligence obligations) would not apply and there is no obligation to trace movement of the materials from the initial researchers to subsequent users such as Lucas Meyers and other organizations that developed the compound into the AbyssineTM product, apparently also in France. Secondly, under the Draft Text proposed approach (see Figure 1) the use of collected samples in ex situ conditions such as those in the Collection Nationale de Culture de Microorganismes (France) would be ‘open,’ resulting in a break in the chain of custody – a chain on which the ‘track and trace’ approach depends.
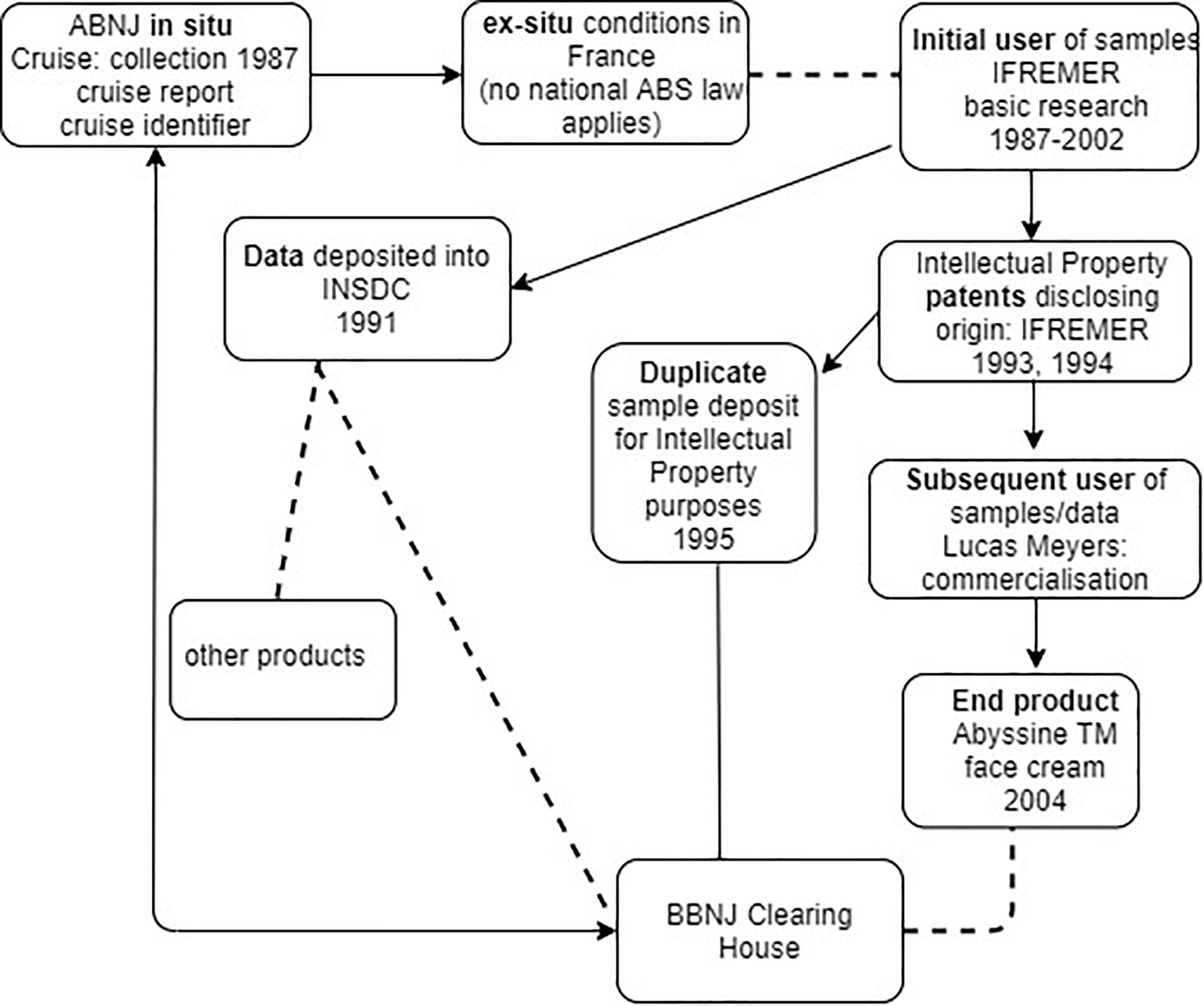
Figure 3. Traceability in practice including the commercialization phase using the Deepsane/Abyssine example (not shown - patents were also filed in 1998 by Lanatech, subsequently acquired by Lucas Meyers). Arrows show movement of data, dashed line a break in track and trace, and plain line is possible back-track of information.
Multilateral (Contractual) Traceability Options
While the Nagoya Protocol’s traceability infrastructure largely relates to tracking whether PIC has been obtained and MAT have been established in accordance with national law (but not whether a user has complied with the terms of the agreement, see section “Traceability Infrastructure Under CBD and Nagoya Protocol”), contractual mechanisms can accompany the movement of materials and associated information and knowledge across jurisdictions and be used for traceability purposes. This section identifies how standardized material transfer agreements (MTAs) (including data transfer agreements) and other contractual mechanisms attach terms and conditions to the transfer of genetic resources/information and the role these may play in ABS multilateral traceability mechanisms.
MTAs have the potential to put the burden of compliance with ABS obligations on end users of the biological resources and/or knowledge. This is because contracts can include clauses that manage the conditions of subsequent uses or transfers of the resources/knowledge to third parties, if allowed under the contract (Young and Tvedt, 2017). MTAs of ex situ collections of genetic resources are increasingly incorporating ABS obligations as part of their terms and conditions26. Some MTAs require the recipient to exercise due diligence to ascertain that the materials and information have been accessed and used in accordance with PIC and MAT requirements27. The MTAs can include specific benefit sharing obligations and clear consequences if the recipient breaches these obligations (Von Kries et al., 2013). Standardized agreements are common in the biotechnology sector and create legal certainty for transfer of materials, intellectual property, and evidence of provenance for physical materials (Rourke, 2019).
With machine readable technology, it is also possible to incorporate contractual obligations for specific information associated with genetic resources. For example, Scholz et al. propose the use of a standard creative commons licence as an option for an ABS tool (Scholz et al., 2020). This would be a legally binding agreement requiring people who are submitting DSI to a database to enter a standardized license identifier into a metadata field linked to an online version of the license. Their paper proposes four standardized licenses (to be determined at an international level, e.g., through a Public/Private Partnership) with different levels of information and benefit sharing. DSI users would bear responsibility for keeping track of the licenses associated with the DSI they are using, which would be less onerous for a standardized license than case-by-case ABS conditions. This builds on creative commons licenses (already in widespread use in science for publishing research findings) and depends on machine readable technology and interoperability of databases28.
Traceability approaches using Standard MTAs in combination with other traceability infrastructure include multilateral ABS systems for plant genetic resources for food and agriculture and certain viruses. The International Treaty for Plant Genetic Resources for Food and Agriculture (Plant Treaty) explained briefly below uses a Standard Material Transfer Agreement (SMTA) that does not require the provider to track single accessions once they have afforded access to them [article 12(3)(b)] but instead passes obligations to update movements onto subsequent users. The World Health Organization’s (WHO) Pandemic Influenza Preparedness Framework for the Sharing of Influenza Viruses and Access to Vaccines and other Benefits (PIP Framework) explained below has a system of two SMTAs, identifiers and other traceability mechanisms that trace the movement of H5N1 influenza viruses.
The Plant Treaty creates a multilateral system of ABS for a specific selection of plant genetic resources used for food and agriculture but not other purposes such as pharmaceuticals (article 10). The purpose is to enable other Treaty Members to access plant genetic resources for research, breeding, conservation and training, subject to benefit sharing arrangements intended to benefit farmers, especially those from income poor countries, who conserve and sustainably use the resources [article 13(3)]. The terms and conditions of transfer must be under the SMTA [article 12(4)], which is a bilateral contract between providers (e.g., CGIAR collections) and recipient institutions, and not a bilateral contract with the original provider countries. Providers of multilateral materials have no obligation to track individual accessions but they must provide with the material all available passport data (Lawson et al., 2018)29 and, subject to applicable law, any other associated available non-confidential descriptive information (article 12(3)(b)(c) and SMTA article 5). SMTA binds non-parties to the agreement through a clause that requires recipients to only transfer the materials under the terms of the SMTA through a new MTA and to notify the Governing Body of the transfer [SMTA article 6(5)(a)]. Central to the traceability framework is the Global Information System (GLIS) that the Governing Body set up to allow information about plant materials to be collected, made available and shared (article 17.1). GLIS is used as an information hub that directs users to other databases such as the System-Wide Information Network for Genetic Resources (SINGER) and Genetic Resources Information Network (GRIN) (Lawson et al., 2019).
The PIP Framework is a multilateral system for the sharing of H5N1 and other influenza viruses with human pandemic potential, access to vaccines and sharing other benefits (article 2) that is not dependent on ABS obligations under a treaty. Under this system, samples, sequence data and analyses must be shared through the Global Influenza Surveillance and Response System (GISRS), which is an international network of influenza laboratories that conduct virus surveillance, assess pandemic risks and assist in preparedness measures30. This framework has two SMTAs of relevance to its traceability system: SMTA 1 is between WHO (a laboratory in the GISRS) and the depositing country’s (WHO recognized) laboratory which contributes the physical samples to the multilateral system; and SMTA 2 is between WHO and pharmaceutical companies that want to access the sample for various purposes (article 5.4.2). Transfers to subsequent users must be under the same terms and conditions as those provided in the SMTAs (Annex 1 article 4). The PIP Framework encourages information sharing in publicly accessible databases and requires WHO GISRS laboratories to submit genetic sequence data to databases in a timely manner consistent with the SMTA (Annex 4 principle 9)31. The PIP Framework’s Influenza Virus Traceability Mechanism is a public access database maintained by WHO that records the receipt and transfer of PIP biological materials into, within and to parties outside the WHO GISRS (Annex 4 principle 8). It enables anyone to track the transfers of materials and the results of analyses and tests carried out with them32.
There are challenges with applying a multilateral traceability model similar to the Plant Treaty or PIP Framework to the BBNJ context. These include defining the scope of ABNJ MGRs that would fall within the multilateral system. The Plant Treaty applies to specific crop species, while the PIP framework applies to a specific virus strain in contrast to countless species that may be collected from 60% of the world’s oceans (Humphries et al., 2020). Kamau (2013); Halewood (2014) argue that the Plant Treaty notification process is burdensome and has practical difficulties for attempting to distinguish public domain or government-controlled resources from those that are not. Consequently, notifications are low. Technical and administrative burdens may account for some of the parties’ reluctance to report (Reichman et al., 2015). To date the process of negotiating PIP Framework SMTA2 agreements has been lengthy and complex, resulting in a limited number of concluded agreements, often with details of the benefit sharing agreements unavailable to the public (Rourke, 2019).
In the Deepsane case, we could not find information about any MTAs used for the transfer of materials to subsequent users nor mention of identifiers for the relevant materials. However, if they exist, machine-readable publicly available MTAs could contribute to the information network relevant for ABS traceability. If these MTAs are standardized, there is a greater likelihood of user uptake through cost and time reductions compared with ABS negotiations on a case-by-case basis33. SMTAs may not on their own be sufficient for effective MGR of ABNJ traceability but they offer one means for monitoring the movement of materials and certain information. BBNJ negotiators might learn from the experiences of the very different PIP and Plant Treaty (contractual) multilateral systems and may find a hybrid model that suits the unique environment and geo-political considerations of ABNJ.
End-Product/End-User Traceability Options
In contrast to ‘track and trace,’ emerging registration systems relevant to genetic resource/information use offer an approach to traceability that does not require every movement to be traced between users and subsequent users. Instead, they require downstream users to report products or activities at which time certain disclosure, reporting and/or benefit sharing obligations are triggered. The section below explores two examples of traceability options that target the end users or end products of genetic resource use.
The first example is an end-user registration system where end-users rather than the initial researchers (if they are different from the end users) engage with the traceability system. While the Deepsane materials do not fall within scope of any end-product traceability systems, Brazil’s ABS measures offer insight into how this might work in practice. In 2017, Brazil changed from a system requiring PIC and MAT at the time of access to (collection of) its genetic resources to a registration mechanism controlling the economic exploitation of products or reproductive materials arising from their use (da Silva and de Oliveira, 2018). To be granted a Certificate of Lawful Access, a user must register on the National System for Genetic Heritage and Associated Traditional Knowledge Management (SisGen) before a specified event such as commercialization, intellectual property applications and public disclosure of results (Brazil Government, 2019). Upon Notification of a Finished Product on SisGen, parties negotiate benefit sharing arrangements, which can be monetary (e.g., contribution to a fund) or non-monetary (e.g., capacity building) (Brazil Government, 2019).
A second example is an end-product traceability system using an Intellectual Property disclosure of origin framework that is partially proposed as an option in the Draft Text. This proposes obligations on parties to take measures that ensure patent applicants that use MGRs of ABNJ disclose the origin of the MGRs they use, and if they do not, they are ‘not approved’ (draft article 12). Patents prevent others from using a patented product or invention without the permission of a patent holder34. From the perspective of technology poor countries, there are concerns of an inequitable distribution of benefits from the use of MGRs of ABNJ to users in technologically rich countries (Broggiato et al., 2018). The Draft Text intellectual property provisions (see section “BBNJ Proposed Approach to Traceability in the Draft Text” above) aim to trigger disclosure at or before the time of commercialization of product or process inventions that incorporate MGRs.
There are other forms of intellectual property that could help to link end-products with ABNJ such as trademarks and geographical indications. Geographical indications identify a ‘good’ as originating from a particular location “where a given quality, reputation or other characteristic of the good is essentially attributable to its geographical origin.”35 Examples include Champagne and Roquefort cheese in France. This form of intellectual property provides protection (often through a trademark or other mark) against use by a third party of a product that does not conform to the relevant standard (e.g., content, production method). These are normally associated with territories within national jurisdiction but ILBI negotiators might consider the implications of this form of intellectual property or a similar system for identifying origin of MGR products. On the other hand, this type of system might lead to inequitable sharing of benefits since an owner of a geographical indication mark has the right to only allow other producers to use it, so long as they comply with the relevant standards. Negotiators have used the broad term intellectual property rights (instead of patents which they seem to be targeting) in the Draft Text traceability option, which may inadvertently introduce more layers of red tape through other forms of private rights connected with MGRs, including copyright, database protection and trademarks. Given the emphasis on private rights inherent in intellectual property, any system here is likely to be much less fit-for-purpose than one based on for example current science practice, which already emphasizes open access and publishing of data (including collection/site information, i.e., origin of MGR).
In the Deepsane case, there appears to be a myriad of French, European, Japanese and US patents relating to the sample collected from ABNJ in 1987 (some examples in Table 1), however it would require extensive investigation of commercial-in-confidence materials to determine which of these led to the final AbyssineTM product. Most of the patents refer to process inventions for producing a low-molecular-weight sulfated polysaccharide derivative from the microorganism for a variety of applications36. Since intellectual property is administered and enforced under national laws, determining the extent to which the end-product incorporates MGRs of ABNJ would require an investigation in each country where a patent may have been claimed. In the case of Deepsane, this is assisted through other sources such as publications (e.g., Cambon-Bonavita et al., 2002) and end-product marketing (e.g., AbyssineTM). However, the Treaty could propose a requirement under national laws to add the specimen identifier of an MGR from ABNJ to patent claims and then link patent databases to patent checkpoints and/or other infrastructure that can alert the proposed BBNJ Clearing House to its existence. What is currently missing is an emphasis on transparency and accessibility of patent data, where databases are numerous, but disparate and poorly integrated.
Assuming technological developments are able to link information, the benefits of this end-product approach to traceability is that intellectual property systems are well established in most countries, offering a ready-made source for information sharing. For many years, a group of biodiversity provider countries have been seeking amendments to the Agreement on Trade Related Aspects of Intellectual Property Rights (TRIPS) to require a minimum standard in national patent laws for disclosure of origin (WTO, 2011). While these calls have been unsuccessful, national ABS or patent laws such as those of Malaysia, Vietnam and India are increasingly requiring origin disclosure when applying for or registering patents (WIPO, 2020). Despite these efforts, a 2016 study found that 90% of patent applications do not provide information on origin (Hammond, 2016). Without all or at least the key countries where inventions using MGRs are patented or commercialized, the loopholes that could be exploited by end-users targeting countries without disclosure systems would seriously undermine traceability. Further, there would need to be some common standards across countries for determining to what extent a product or process incorporates the MGRs and/or associated information before disclosure is required (e.g., whether the genetic trait is merely present or functional; Humphries, 2015). Many of the current national examples only require origin to be disclosed and not whether PIC was obtained and whether benefits were shared with the correct provider (WIPO, 2020). Many national laws requiring disclosure of origin for patentability purposes only relate to materials for which they are the country of origin, which is likely to exclude BBNJ materials. At best, tapping into existing Intellectual Property mechanisms may only be one ad hoc source of information for piecing together the movements of MGRs of ABNJ.
Existing Open-Access Traceability Options
The CBD/Nagoya Protocol do not mention open access but the BBNJ Draft Text proposes that States Parties must take the necessary measures “to ensure that ex situ access to [MGRs]…is free and open” [draft article 10(3)]. On the other hand, it proposes that access to MGRs in silico “is facilitated” [draft article 10(4)] without explaining what facilitation would entail. This section explores examples of the use of open-access databases to trace the movements of NSD under INSDC and materials and chemical compounds in the US National Cancer Institute Developmental Therapeutics Program (US-NCI). It concludes with some implications of open-access traceability options for the BBNJ agreement.
INSDC is the existing fundamental mechanism for access, sharing and traceability of NSD. This database consortium consists of a collaboration between GenBank (of NCBI), the European Nucleotide Archive- ENA, and the DNA Data Bank of Japan (DDBJ). These databases receive approximately 10 billion data requests a year (WiLDSI report, 2020)37. Open access is embedded in the fabric of the INSDC databases and their usage. In genetic/genomic research, publishing of data on these databases is ubiquitous. Database accession numbers29 are allocated to sequences upon submission and represent a unique identifier for any sequence within the INSDC system and is therefore key to traceability. These numbers are cited in resulting publications; also as machine readable links in some cases (see ABYSSLINE case study in Rabone et al., 2019). INSDC therefore is fundamental to traceability and open access in biological research, and as such plays a critical role in supporting the reproducibility of science. For the Deepsane example, the original accession (16S rRNA sequence of strain HYD657) was added to European Molecular Biology Laboratory (EMBL), accession number AJ319537 (Cambon-Bonavita et al., 2002). The accession numbers of the other sequences used in phylogenetic analysis of the strain are also listed in the paper, and similarly, any subsequent usage of the 16S rRNA sequence for phylogenetics (or other analyses) will cite this accession number in a publication. The EMBL entry includes the raw sequence, the publication data, and links to the same entry in other related databases, and records that the organism was “isolated from deep-sea hydrothermal vent.”
Current traceability is most evident at the individual specimen level, represented by records in biodiversity databases, such as the Ocean Biodiversity Information System (OBIS). For MGR collections in museums for example, specimens are allocated a unique identifier when accessioned into a collection and a database record is created. This identifier can then be traced through these databases. Traceability at collections or bulk specimen lot29 level (i.e., all specimens from a given sampling event29) can be more challenging. For example, all collections from a given cruise may be housed at different institutions, and while bulk specimen lots may have entries on an institution database they are generally not published online (Rabone et al., 2019; Collins et al., 2020). Work is underway in the biodiversity informatics community to try to capture the ‘gray’ data of unsorted collections, for example in publishing bulk specimen lots on OBIS, and development of both collections-level data descriptions (Thessen et al., 2019) and of existing collections registries, e.g., GRSciColl (Schindel et al., 2016). Traceability could be strengthened by further developing linkages between INSDC and biodiversity databases, i.e., linking the specimen record that a sequence was derived from and the corresponding INSDC record. More fundamentally, specimens with data published on INSDC should be archived in natural history collections by default (Droege et al., 2019; Rabone et al., 2019). In general, improved practices of ‘citation’ of physical specimens/samples in the science community are needed (Thessen et al., 2019).
In terms of sample sharing, natural history collections have long been available to the science community (Rabone et al., 2019), with traceability through registration/accession numbers, referenced in associated publications (and INSDC entries as above). Organizations such as the Global Genome Biodiversity Network (GGBN) facilitate sharing of genomic collections held by their partners (Droege et al., 2014). SMTAs have also been developed for biological collections by the science community38. For traceability of permit information, the Global Genome Biodiversity Network (GGBN) has developed data standards which allows for the capture of MTA, PIC, and MAT information with the sample/collection and therefore traceability of compliance (Droege et al., 2016). General usage and development of global data standards in biological research such as DarwinCore for biodiversity data (Wieczorek et al., 2012)39 and MIxS for sequence data (Yilmaz et al., 2011) has been recently reviewed in Rabone et al. (2019). These global standards are critical to ensure data are FAIR (findable, accessible, interoperable, and reusable) and therefore support traceability. Considering sample sharing for potential commercialization, the US-NCI has a collection of ca. 100,000 plant and marine organism extracts sourced internationally under its ‘Letter of Collection.’40 The extracts containing mixtures of chemical compounds, but no genetic materials, are made available to participating scientists under an MTA41, which protect the rights of the source country, participating scientists and the US-NCI. The open repository mechanism starts with identification of active extracts for which materials are provided to the partner carrying out biological activity testing, and if activity is identified, work on the isolation of bioactives can proceed with regular updates to the NCI. Any development is not tracked by the NCI, but the MTA requires (in addition to giving due credit to the source country and NCI and quoting the NCI sample identifier in publications) the participating scientists to negotiate and sign a benefit sharing agreement with the source country before the commercial sale of any products arising from this work. This unique identifier must be passed on to any subsequent users. The US-NCI open repository therefore uses a sample identifier, together with user compliance, to trace materials from extract to commercial product.
To apply this approach to the Deepsane example, if commercial utilization is likely to occur, the user would be obliged to notify the ex situ collection (or clearing house) with the identifier of the original material leading to the product and commit to sharing a percentage of profits with a central fund. This approach requires user compliance by the researchers and due diligence by downstream users. Using gatekeepers such as national funding agencies and academic journals would ensure that the majority of users of MGR from ABNJ could comply with this light-touch system (see Figure 4).
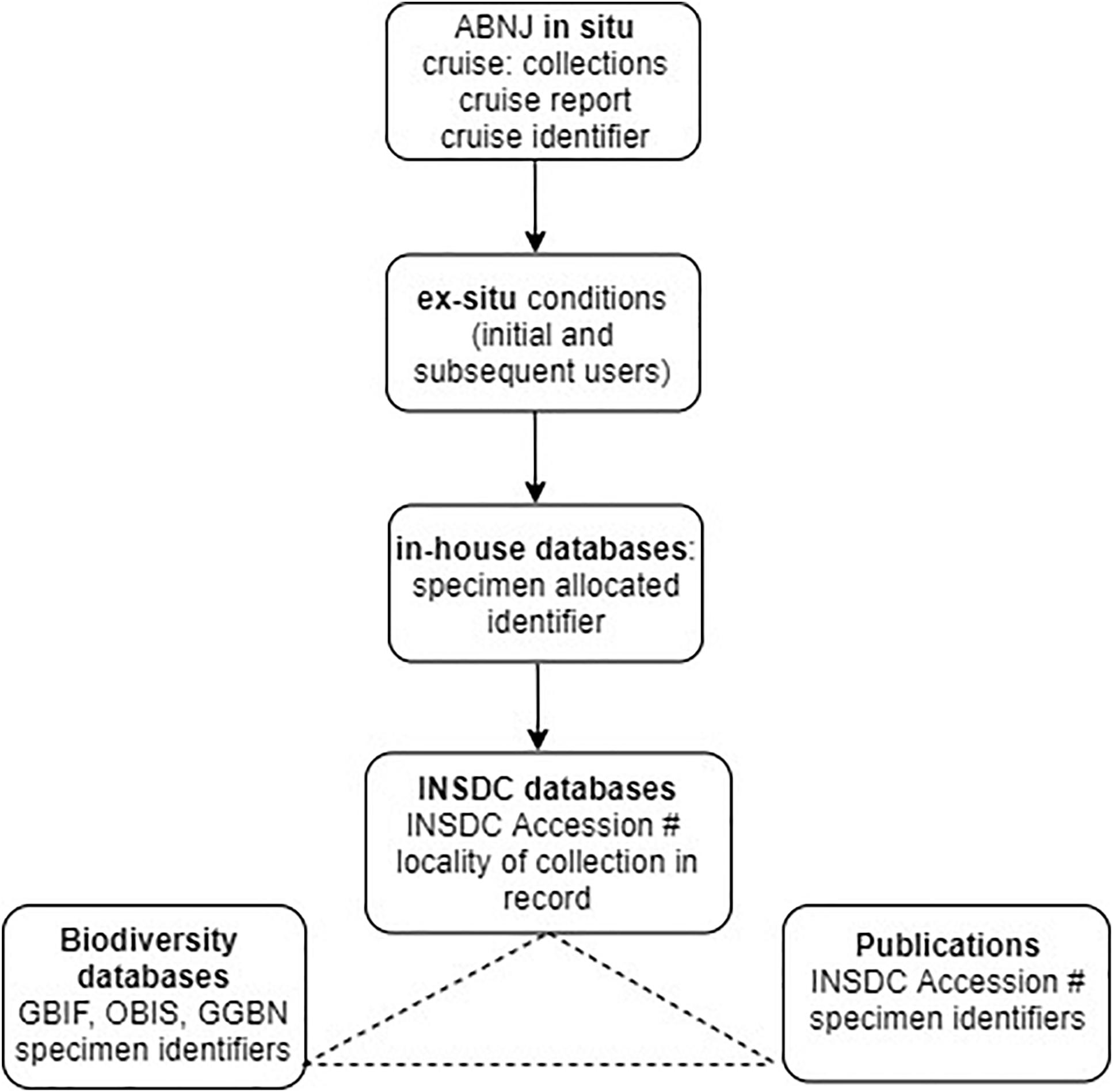
Figure 4. Existing traceability in science highlighting usage of identifiers and linkages between databases. Cruise identifier refers to the existing system of cruise codes allocated to any deep-sea cruise, INSDC accession # refers to accession numbers allocated to sequence data submissions/records in INSDC databases which represent a unique identifier within this system (see Glossary). Specimen identifiers refers to the unique identifiers used in biodiversity databases such as OBIS, e.g., globally unique identifiers, or GUIDS (section “Existing Open-Access Traceability Options”). Potential traceability links between GenBank, publications and biodiversity databases are shown as linkages and integration between some databases/publications exist but not in all cases but are not comprehensive.
Combined Traceability Approaches
Biodiversity beyond national jurisdiction policy makers may decide to move away from the current one-size-fits all approach to MGR traceability and instead adopt a combination of traceability approaches and infrastructure with standard options for a subsequent user depending on the activity or resource (see Figure 5). Such options might include open access, contribution to a BBNJ Fund, engagement with a purpose based multilateral mechanism (e.g., for BBNJ conservation) and/or BBNJ Capacity Building and Technology Transfer (e.g., ‘needs based’42) or an end product/user model as outlined in section “End-Product/End-User Traceability Options” above. Another option is a subscription or tax model, requiring payments (outside the ABS transaction) either by users accessing the resources (e.g., PIP Framework) or an impost on contracting parties (e.g., Norway seed sales tax) (Lawson et al., 2019). This section explores two options that have combined approaches to traceability.
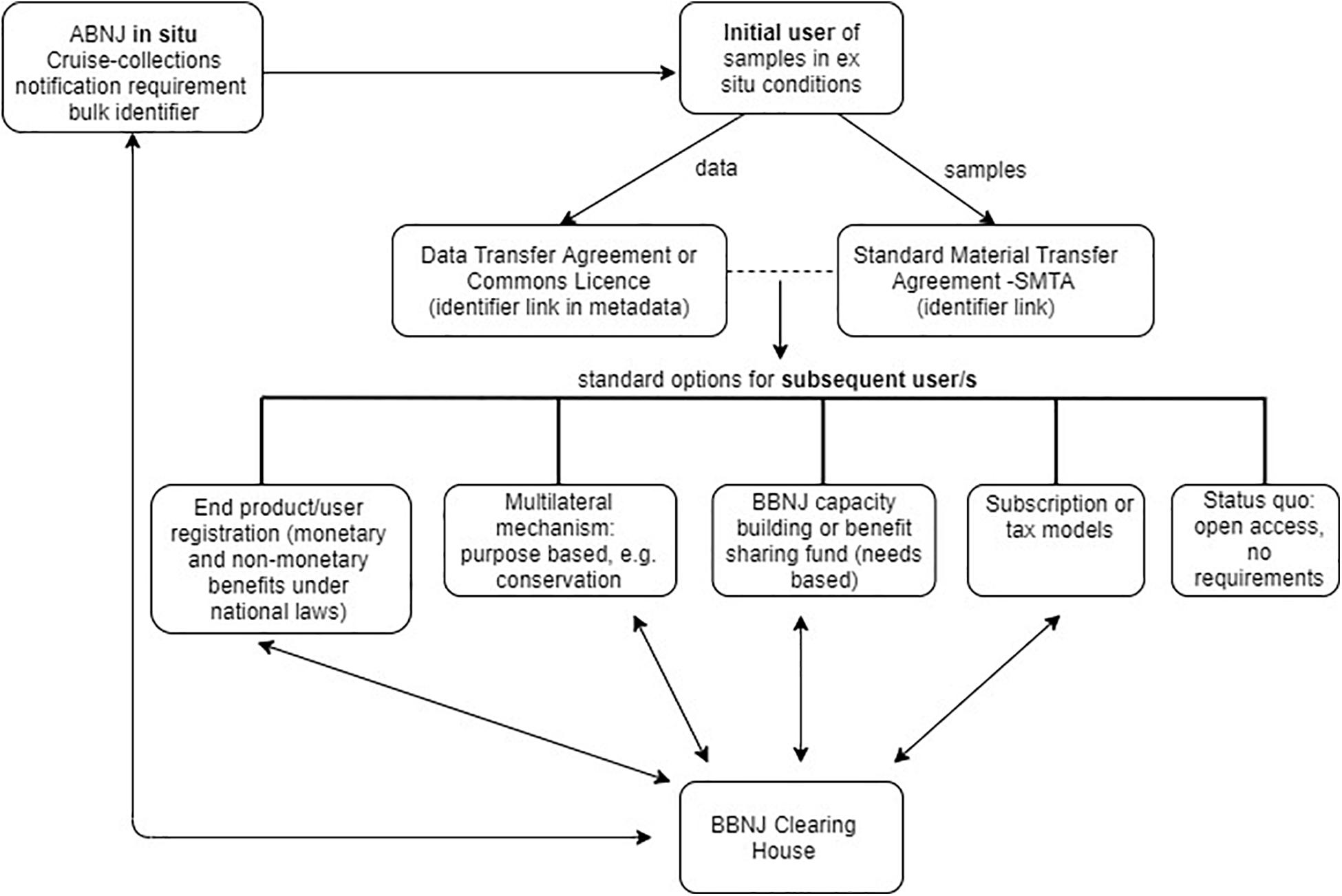
Figure 5. Combined approaches to traceability, showing potential standard options for subsequent users, such as end/product/user registration, multilateral mechanism- purpose based (e.g., for conservation), BBNJ capacity building or benefit sharing fund (needs-based), subscription or tax models, and status quo – i.e., open access, no requirements. Arrow shows flow of information.
OPEN Traceability Option
The Mare Geneticum concept provides a series of building blocks, based on good scientific practice, toward a fair and effective regime to regulate benefit sharing of MGR from ABNJ (Broggiato et al., 2018). Within this, a traceability system called ‘Obligatory Prior Electronic Notification’ (OPEN) is proposed which provides facilitated and conditional access to MGR from ABNJ. Before a collection of MGRs is made, a minimal dataset is recorded in the OPEN system (for instance including information on the collector, geographical area, dates, research aims, type of sampling) with a commitment to make materials collected available openly in a biorepository and to update the information in the OPEN at certain milestones starting with provision of sample data upon return from the collection cruise. An identifier is associated with each sample collected and this will allow determination of its provenance and provide legal certainty for subsequent users, upon change of use and at different milestones which may include taxonomic identification, generating DNA sequences, testing for function/activity, publication, patent application and (potential) eventual commercial application. Benefit sharing upon commercial income being generated is proposed to be a fixed percentage by sector (e.g., pharmaceutical, biotechnology, and nutrition) to help simplify negotiations. Other (non-monetary) benefits to be shared are access to the collected materials themselves, and to make sample and sequence data openly available. This system requires due diligence by subsequent users to ensure they have legal certainty by obtaining the unique identifier, without the need for an onerous ‘track and trace’ system as discussed in Section “Track and Trace Options.”
Tiered Approach Traceability Option
Humphries et al. (2020) propose a tiered ‘activities’ approach to MGR governance in ABNJ as an alternative to the ‘one size fits all’ approach currently under negotiation. In relation to the traceability elements, the authors raise a range of options including a user-driven and transparent web-based traceability platform similar to the PIP Framework’s Influenza Virus Traceability Mechanism (see section “Multilateral (Contractual) Traceability Options”). Before undertaking activities in ABNJ, a researcher would obtain a cruise identifier (as is common practice), receive information on ILBI requirements and register on a Capacity Building Database. After samples are collected and investigated in a laboratory, the researcher would register on an information hub. They suggest a Facilitated Information and Sample Sharing Hub (FISSH) could offer electronic reporting of the movement and uses of samples and data using a system of Bulk collection identifiers. Instead of issuing unique identifiers at collection, which may be impractical (see section “BBNJ Proposed Approach to Traceability in the Draft Text”), the Bulk collection identifier is associated with a given sampling event29 and linked to all resulting collections on a given cruise. When the samples are identified on shore, the FISSH could link to the Bulk identifier with any unique identifiers for specific samples and resulting sequences issued by other repositories and databases as they move through research, development and commercialization phases (Humphries et al., 2020). Further investigation into this idea could include the practicalities of assigning a Bulk Identifier for different collection methods (e.g., trawling or single core sample), different events (the whole cruise or a single sampling event) and different locations (sampling within and beyond national jurisdiction on one cruise). The authors suggest an End-User Due Diligence Approach to benefit sharing (see section “End-Product/End-User Traceability Options” above), including market based, research and private sector incentives for user engagement, especially for using information separately from the physical materials. In this way the onus is on those doing commercial development, and not (unnecessarily) burdensome to those doing non-commercial biodiversity research.
Many aspects of these combined approaches are similar to the open access in current usage in science (see Section “Existing Open-Access Traceability Options”). A unique identifier accompanies a sample using a due diligence approach, but the challenge is how to associate this with any results or data generated from the sample. This may include taxonomic information, nucleotide sequence data, and small molecule derivative information, making a connected network of information on the MGR from ABNJ. The system again relies on due diligence by the initial and downstream users. Different levels of requirements in the OPEN and Tiered Approach mean different potential burdens on the users, and different levels of data deposition either in centralized or connected databases. It makes sense to deposit data in the relevant open-access database such as the INSDC databases for nucleotide sequences, taxonomic data in WoRMS, and specimen records in biodiversity databases such as OBIS. Given existing practices in science of publishing collections records and usage of identifiers at various levels: cruise/sampling event/specimen and sequence identifiers (see section “Existing Open-Access Traceability Options”), integration with existing processes and databases would be preferable to development of new systems. Requirements for improved traceability, e.g., a BBNJ bulk identifier, could be supported by the proposed BBNJ Clearing House in the Draft Text or other body responsible for information exchange.
The main differences between ‘track and trace’ and traceability systems are highlighted in Figure 6 and Table 2. The complexity and likely associated costs of full ‘track and trace’ systems means that these are less desirable. More realistic are systems relying on end user compliance or hybrid systems that build on current good scientific practice and are likely to be more cost effective, will benefit ocean science to a greater extent, and may meet with greater user acceptance.
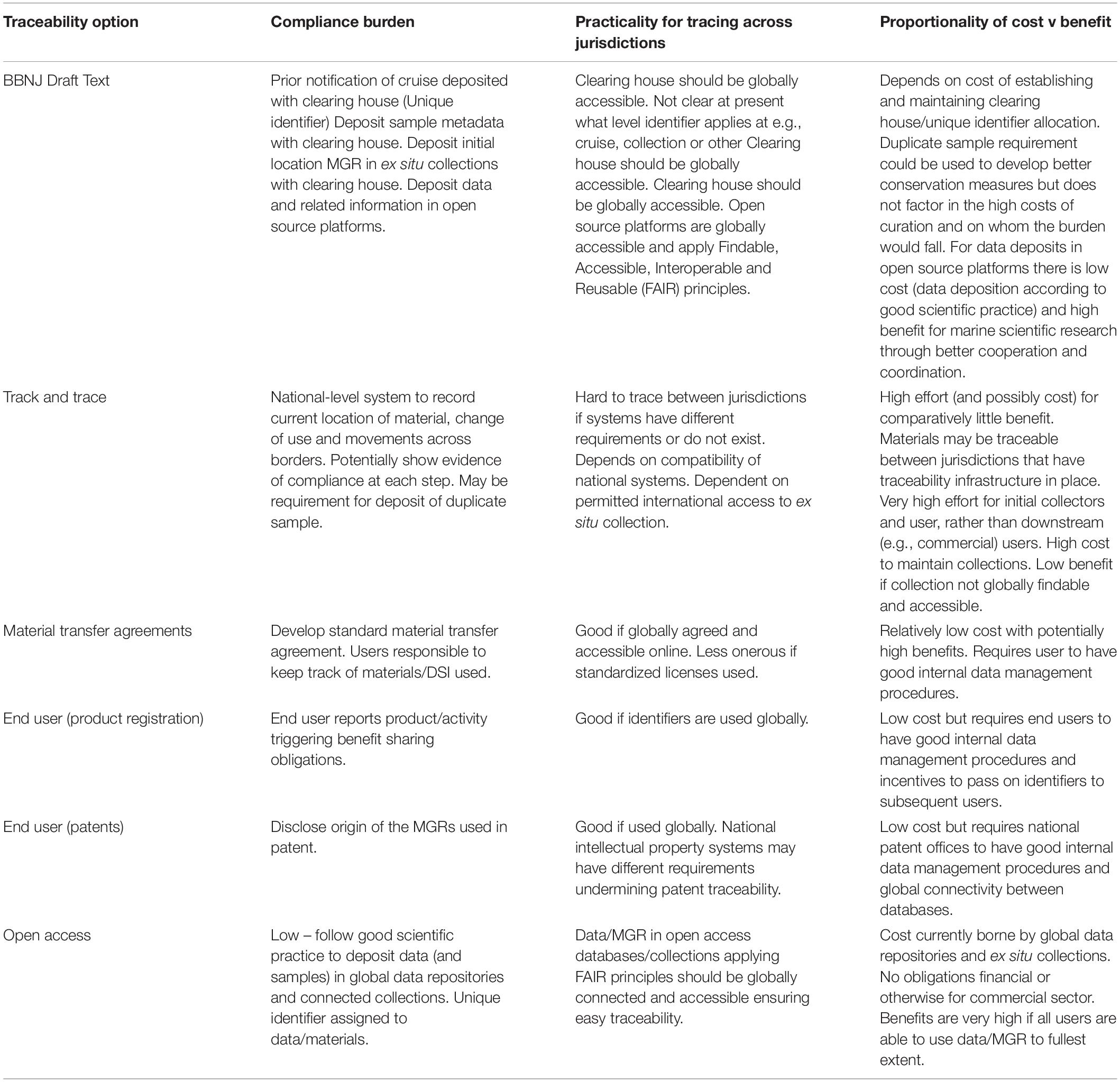
Table 2. A summary of the compliance burden, practicality and cost/benefit proportionality for the different traceability options discussed.
Policy Recommendations
As with all international framework agreements, States Parties will have the flexibility to implement their proposed BBNJ Agreement obligations to suit their national circumstances. However, with increasing digitization of genetic resources in open source databases beyond the jurisdiction of any one nation (Lawson et al., 2019) and the multi-national characteristics of ABNJ research cruises and collaborations (Rabone et al., 2019), policy-makers could consider designing digital traceability arrangements for a connected world. The key question to be addressed is whether full traceability is feasible at all, and if so, what the costs would be of an all-encompassing global traceability system. Benefits of a truly global system, if feasible, are that material might be traceable in multiple jurisdictions and throughout the potentially discontinuous development pipeline. The alternative is multiple ad hoc national systems that don’t interact well, potentially losing sight of the development of a product as it travels between jurisdictions and up the commercial value chain. Below we summarize our recommendations based on the foregoing discussions:
Recommendation 1
It is essential that any system is light touch and does not impede marine scientific research with additional bureaucratic burdens. Most marine scientific research is basic research, with commercial applications far downstream. Marine scientific research is essential to allow us to understand the basic biology, ecology and chemistry of the oceans, permitting the development of better conservation measures. The ideal system will benefit marine scientific research, making it more coordinated and connected, thus ensuring adoption/buy-in from the scientists involved.
Recommendation 2
Any system developed should learn from, and build on, good scientific practice so that duplication does not occur. In the Deepsane/Abyssine example, the strain was allocated an identifier (HYD-657) which (while not meeting current criteria for unique identifiers, section “Existing Open-Access Traceability Options”) was good scientific practice for the period and crucially allowed its progress in the scientific and patent literature to be traced over decades. Similarly, specimens in an ex situ or data repository will generally receive a globally unique identifier so BBNJ requirements could adapt to existing infrastructure and scientific practice, without duplicating requirements. Usage of persistent/unique identifiers is a common feature in the global data ecosystem and like usage of data standards, ensures data is FAIR (Findable, Accessible, Interoperable, Reusable). The Draft Text or subsequent protocols/guidelines could recognize that a model of traceability already exists in INSDC, biodiversity databases and scientific publications. It could seek to support these systems as far as possible and acknowledge their centrality to biological research. These databases are critical to the functioning biological research community and continued open access to their data holdings must be assured including through underwriting long-term funding.
Recommendation 3
Traceability and reporting requirements should be placed on the eventual end users and not the scientists. Scientists should follow best practice guidelines for data and sample management including recording relevant identifiers in databases/publications and archiving of samples in natural history collections including those where associated sequences are published (Droege et al., 2019; Rabone et al., 2019). If the Deepsane/Abyssine example had required benefit sharing this would be possible as the identifier is quoted in the patents, so an end user compliance mechanism might be feasible. This would require buy-in from stakeholders which may be possible via market-based (e.g., certification), funding-based (e.g., grant incentives) or corporate (e.g., Global Compact) incentives (Humphries et al., 2020). Given that AbyssineTM refers to the BBNJ origin in its name, product information and advertising materials, it is a good example of how end-users could engage with the proposed BBNJ frameworks if it has some form of incentive to declare the origin of MGRs at a defined stage of research like the Brazilian approach. The alternative may be a very complex and costly global full ‘track and trace’ system which may not be desirable or even feasible.
Recommendation 4
In addition to incentives (‘carrots’), enforcement measures (‘sticks’) may be required to encourage end users to engage with the system. Humphries et al. (2020) suggest that ‘stick’ measures might include an alert back to the proposed ABNJ Clearing House, for example through the existing CBD checkpoint infrastructure (see section “Traceability Infrastructure Under CBD and Nagoya Protocol”) for products that have information (e.g., an identifier) associated with a species linked to an ABNJ Bulk Identifier. If the Deepsane case was to occur today, French ABS laws might not be triggered but the ILBI could still require alerts from its checkpoints43. For this to be workable, infrastructure between the Nagoya Protocol and the ILBI need to be compatible for a coherent system.
Recommendation 5
There is a need for increased transparency in the commercial phase regarding movement and use of resources and data. For the Deepsane/Abyssine example, relevant information was available about the scientific research, but it was much harder to get information about the commercial research phases because information held in private databases for commercial objectives are not well integrated or connected. This could be solved to a certain extent by clarifying origin of material and information in the patent claim and labeling on eventual products. For scientific reasons, location of collection (with GPS coordinates) is a standard field in the INSDC databases, although recording of these data could be improved (reviewed in Rabone et al., 2019). The World Intellectual Property Organization (WIPO) is moving to allow machine readable NSD, meaning that sequences included in patents can be automatically linked to their INSDC database entries, and linked to origin if specified in the record44. Such a model could allow origin of material to be specified indirectly in patents. In addition, some information from MTAs should be made publicly available insofar as this does not break confidentiality. An arms-length body that gathers anonymized information might encourage industry to share information on the size of its business and profits under general headings such as ‘pharmaceuticals,’ ‘enzymes,’ or ‘cosmetics’ amongst others without sharing details of the exact products. Some information regarding origin such as whether the MGR was obtained from an EEZ or ABNJ would segregate the data in a useful way. The proposed arms-length body could aggregate the data to give an estimate of the overall size of the blue biotechnology market under these headings and show how much is from the EEZ and how much from ABNJ, all without industry divulging commercially sensitive information.
Recommendation 6
There is a need to clarify the extent of the commercial enterprise around MGR from ABNJ and how fast it is expected to grow in the future. For instance, the Deepsane/Abyssine example may have remained overlooked until now, but extensive analysis is still required to fully explore the size of current and future biotechnology based on BBNJ. Most surveys to date have focused on healthcare products, but we need to find hidden uses of marine biotechnology such as enzymes, agriculture, personal care products and nutritional products. Whereas scientific literature and patent searches may yield a large amount of information, many fields such as cosmetics and nutrition may not use patents to protect their products and will remain undiscoverable. Even profits made from particular BBNJ-derived products may be confidential and a mechanism may need to be developed to obtain this information without divulging commercially sensitive information. On the question of whether it is feasible or even desirable to develop a new traceability system, there is a need for an independent and evidence-based assessment of the extent of commercial benefits from MGRs of ABNJ and a cost-benefit analysis of the potential implications of traceability requirements for scientific progress in technology-rich and technology-poor countries, in light of realistic expectations of commercial benefits.
Conclusion
The fourth IGC of the BBNJ process has been postponed until August 2021. The intersessional period has given delegates and civil society ample time to consider and informally discuss the more complex issues in the process. We hope that our discussion and recommendations in this paper will add to the evidence needed to develop a workable traceability approach that benefits marine scientific research and ocean conservation. We have illustrated through a concrete example that a full ‘track and trace’ system is not workable or even desirable. Evidence from the current implementation of the Nagoya Protocol at a national level shows that such systems are often non-existent or incomplete and would not allow any level of traceability at an international level.
Any additional traceability requirements imposed by the ILBI will need to be carefully assessed with consultation and input from marine science and bioinformatics communities and other stakeholders and it is vital that additional administrative burdens are minimized and any that arise are borne by commercial end-users rather than initial researchers. Improved traceability that builds on current good practice will benefit science by making data and materials widely available in a consistent and coordinated fashion. These measures could benefit the baseline biodiversity research that is needed to develop the most effective conservation measures in ABNJ. Our case study shows that good scientific practice makes traceability possible through the scientific process even though the collections were made several decades ago. However, traceability either slows considerably or stops when a product enters commercialization. The commercial sector therefore needs to be engaged in finding workable solutions for the Draft Text to be more transparent and share vital data that will make traceability possible, following on from current scientific practice that encourages openness. This could include measures such as sharing of MTAs, the use of direct or indirect measures (including incentives) to disclose the source of MGRs in products or patents and initiatives to gather anonymized information about the commercial use of MGRs from ABNJ that may otherwise be commercially sensitive. The availability of commercial data showing how many products based on MGR from ABNJ exist or are being developed would indicate the size and growth of the global market and assist in assessing whether a potential traceability mechanism is commensurate with the scale of activity. A landscape analysis of usage of identifiers both within science and in the commercial phase in a BBNJ context is another crucial step for an effective traceability system.
Drawing from the Deepsane example, the seven policy recommendations in this paper essentially urge an approach that is a light touch, builds on existing traceability mechanisms including proper usage of identifiers and places obligations on end-users through incentives and transparent measures that encourage commercial buy-in. The feasibility and desirability of developing a traceability system should be based on comprehensive evidence, particularly the implications of any legal requirements on publicly funded science. Even well-intentioned legal requirements may have unintended consequences. Importantly, negotiators of the ILBI have the benefit of learning from the challenges with national implementation of traceability under the Nagoya Protocol framework, which cannot effectively cope with advances in scientific practice, such as the use of digital sequence information. A well-designed traceability system tailored to the unique geographical, political and jurisdictional characteristics of ABNJ could lead to more equitable outcomes for the sharing of benefits from the use of MGRs. Ultimately it will create a greater depth of knowledge of the oceans and biodiversity, enabling their conservation and sustainable use by current and future generations.
Author Contributions
FH conceptualized the study and produced the initial draft. MJ produced the tables. FH and MR created the figures. All authors contributed to the writing and editing process.
Funding
The open access fee was funded by the Griffith University Queensland.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Aurélie Lécolier and Sylvia Colliec Jouault at IFREMER for information on Deepsane; Marjo Vierros for providing information hydrothermal vent patents from ABNJ, and Charles Lawson for providing comments on the manuscript. The cartoon figures used in Figures 1, 6 are from the Noun Project (DNA by Vectors Point, coral by Nook Fulloption, liquid by Smalllike, database by Flatart, third party by Priyanka, drug by Adindar, label by AB Designs, certificate by Libertetstudio, aeroplane by Deepz, car by Adrien Coquet, dolly by Vectorstall, box by IconMark, factory by Iconsphere, and barcode by Adned Kadri).
Glossary
ABNJ, Areas Beyond National Jurisdiction; ABS, Access and Benefit Sharing; ABSCH, Access and Benefit Sharing Clearing House (the CBD clearing house mechanism); ABSCH Unique Identifier, the identifier allocated by the ABSCH Clearing House, unique within that system; AN, Accession number (specifically for INSDC database entries): these represent a unique identifier for a record within the INSDC system; BBNJ, Biodiversity Beyond National Jurisdiction; BBNJ Bulk Identifier, a collection level or bulk specimen lot identifier (see ‘bulk specimen lot’ below); Bulk specimen lot, mixed and unsorted sample/specimen lots, generally containing different species but from one sampling event for example all the organisms from a trawl are preserved and stored in one container, for future identification/analysis; CBD, Convention on Biological Diversity; DOI, Digital object identifier; DDBJ, DNA Data Bank of Japan; DSI, Digital Sequence Information; ENA, European Nucleotide Archive; EMBL, European Molecular Biology Laboratory; GGBN, Global Genome Biodiversity Network; GISRS, Global Influenza Surveillance and Response System; GUID, Globally Unique Identifier; IFREMER, L’Institut Français de Recherche pour l’Exploitation de la Mer; ILBI, International Legally Binding instrument; IRCC, Internationally Recognized Certificates of Compliance; INSDC, International Nucleotide Sequence Database Collaboration; MAT, Mutually Agreed Terms; MGR, Marine Genetic Resource; MTA, Material Transfer Agreement; Nagoya Protocol, Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from Their Utilization to the Convention on Biological Diversity; NCBI, National Center for Biotechnology Information; NSD, Nucleotide Sequence Data; OBIS, Ocean Biodiversity Information System; Passport data, information required under Plant Treaty article 12.3(c), which is ‘essentially identifying information that allows the correct material to be properly exchanged (Lawson et al., 2018); PIC, Prior Informed Consent; PIP, Pandemic Influenza Preparedness Framework for the Sharing of Influenza Viruses and Access to Vaccines and other Benefits; Plant Treaty, International Treaty for Plant Genetic Resources for Food and Agriculture; Sampling event, the collection or sampling at a given locality and timepoint, for example a box core sample recovered from the sea floor, or a trawl recovered from the water column would be considered a discrete sampling event; SMTA, Standard Material Transfer Agreement; TRIPS, Agreement on Trade Related Aspects of Intellectual Property Rights; UNCLOS, United Nations Convention on the Law of the Sea; WHO, World Health Organization; WIPO, World Intellectual Property Organization.
Footnotes
- ^ Exploration of BBNJ Agreement provisions on traditional knowledge is beyond the scope of this paper: see Mulalap et al. (2020) and Vierros et al. (2020).
- ^ 16S ribosomal RNA sequence, GenBank Accession: AJ319537.1.
- ^ ‘In silico’ is undefined but concerns information associated with the physical materials.
- ^ See https://absch.cbd.int/
- ^ See https://absch.cbd.int/search/nationalRecords?schema=absPermit
- ^ For example, India has an ABS E-filing system where upon approval of access, instead of issuing a permit, the authority enters into an agreement with the applicant, which serves as the equivalent of an IRCC http://absefiling.nic.in/NBA/login/auth; Indian Government (2017) Interim National Report on the Implementation of the Nagoya Protocol (November 2, 2017).
- ^ https://absch.cbd.int/about/faqs
- ^ https://absch.cbd.int/search/nationalRecords?schema=absCheckpoint
- ^ National Environment (Access to Genetic Resources and Benefit Sharing) Regulations 2005 (section 23).
- ^ Access to Biological Resources and Benefit Sharing Act 2017 (sections 12 and 15).
- ^ European Union regulation No.511/2014 of the European Parliament and of the Council on compliance measures for users from the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization in the Union [article 4(1)].
- ^ Law on Biodiversity 2008 No. 20/2008/QH12 (article 60); Decree No. 59/2017/ND-CP on the Management of Access To Genetic Resources and The Sharing of Benefits Arising From Their Utilization article 25.
- ^ Environment Protection and Biodiversity Conservation Regulation 2000 (Cth) section 8A.19.
- ^ Protection of Traditional Knowledge, Genetic Resources and Expressions of Folklore Act 2016 [section 36(h)].
- ^ Biological Diversity Act in 2002 section 20(2).
- ^ Environmental, Management and Coordination Act (Conservation of Biological Diversity and Resources, Access to Genetic Resources and Benefit Sharing) Regulation 2006 section 18.
- ^ ‘Les ressources génétiques prélevées en dehors du territoire national et des zones sous souveraineté ou juridiction française’ Loi n 2016-1087 du 8 aout 2016 pour la reconquete de la biodiversite, de la nature et des paysages (1) Titer V: Accés aux ressources genetiqués et partage juste et equitable des avantages article L412.5 (II)(1)(b). Other countries more explicitly state genetic resources from ABNJ do not fall within their obligations e.g., Republic of Korea Act on Access to and Utilization of Genetic Resources and Benefit-Sharing 2017 article 3.2 (unofficial translation).
- ^ European Union regulation No.511/2014 of the European Parliament and of the Council on compliance measures for users from the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization in the Union article 2(4).
- ^ Environment Protection and Biodiversity Conservation Regulation 2000 (Cth) regulation 8A.03(1).
- ^ Access to Biological Resources and Benefit Sharing Act 2017 section 5(1).
- ^ National Environment (Access to Genetic Resources and Benefit Sharing) Regulations 2005 article 4.
- ^ E.g., National Strategy on Biodiversity (“ENBioMex”) 2016–2030 and a range of national laws that might apply to research and species collections; see Jefferson et al. (2018).
- ^ Under the terms of the Budapest Treaty on the International Recognition of the Deposit of Microorganisms for the Purposes of Patent Procedure https://www.wipo.int/treaties/en/registration/budapest/
- ^ Australia’s Commonwealth legislation Environment Protection and Biodiversity Conservation Regulation 2000 (Cth) does not apply to private facilities whereas Australia’s Northern Territory legislation does – Biological Resources Act 2006 (NT).
- ^ Key biodiversity databases include GBIF (Global Biodiversity Information Facility), www.gbif.org; the Ocean Biodiversity Information System OBIS, obis.org; and Global Genome Biodiversity Facility, GGBN, www.ggbn.org (Droege et al., 2014).
- ^ E.g., clause 7 of European Culture Collection’s Organization Material Transfer Agreement 2009, ‘If the RECIPIENT desires to use the MATERIAL or MODIFICATIONS for COMMERCIAL PURPOSE(S), it is the responsibility of the RECIPIENT, in advance of such use, to negotiate in good faith the terms of any benefit sharing with the appropriate authority in the country of origin of the MATERIAL, as indicated by the COLLECTION’s documentation.’ https://www.eccosite.org
- ^ E.g., MTAs from Belgian Co-ordinated Collections of Microorganisms; available at: http://bccm.belspo.be/services/mta
- ^ Creative commons licenses cover over “1.4 billion species occurrence records submitted by 1,612 publishing institutions” (Scholz et al., 2020, p. 27).
- ^ See Glossary of Terms.
- ^ https://www.who.int/influenza/pip/virus_sharing/en/
- ^ At least 6 databases contain influenza genetic sequence data: https://www.who.int/influenza/pip/GSD_EN_3Apr2018.pdf
- ^ https://extranet.who.int/ivtm/
- ^ Some countries such as Uganda have standard ABS terms and conditions embedded in their MTA requirements under national ABS laws – see National Environment (Access to Genetic Resources and Benefit Sharing) Regulations 2005 sections 12 and 16.
- ^ See Agreement on Trade Related Aspects of Intellectual Property Rights (TRIPS) article 5.
- ^ TRIPS article 22.
- ^ E.g., US Patent 8,598,142.
- ^ This estimate doesn’t include downloads of the entire database (for further information see WiLDSi Report, 2020).
- ^ Consortium of European Taxonomic Facilities (CETAF) have developed a SMTA and provide best practice guidance: https://cetaf.org/services/natural-science-collections-and-access-and-benefit-sharing
- ^ DarwinCore is a data standard maintained by the Taxonomic Databases Working Group (TDWG) https://dwc.tdwg.org/
- ^ A comprehensive agreement between the source country and the US-NCI committing it to ‘conservation of biological diversity, as well as to policies of fair and equitable collaboration and compensation in interacting with the Source Countries participating in the collection programs’. https://dtp.cancer.gov/organization/npb/docs/NCI_LOC_NPCA_Aug 2015.pdf; https://dtp.cancer.gov/organization/npb/introduction.htm
- ^ https://dtp.cancer.gov/organization/npb/docs/template_MTA_NPB_June2015.pdf
- ^ See closing statement of the third negotiating session section VI paragraph 10 – UN (2019).
- ^ The Ministère de l’Enseignement supérieur, de la Recherche et de l’Innovation (at the stage of external funding of research projects) and the Ministère de la Transition Ecologique et Solidaire (in charge of requests for access to genetic resources and traditional knowledge and sharing benefits from their use in France): https://absch.cbd.int/countries/FR
- ^ https://www.wipo.int/standards/en/pdf/03-26-01.pdf
References
Brazil Government (2019). Brazil’s position on DSI (Notification 2019-012). Available online at: https://www.cbd.int/abs/DSI-views/2019/Brazil-DSI.pdf (accessed date 03 June 2019)
Broggiato, A., Vanagt, T., Lallier, L. E., Jaspars, M., Burton, G., and Muyldermans, D. (2018). Mare geneticum: balancing governance of marine genetic resources in international waters. Int. J. Mar. Coastal Law 33, 3–33. doi: 10.1163/15718085-13310030
Cambon-Bonavita, M. A., Raguenes, G., Jean, J., Vincent, P., and Guezennec, J. (2002). A novel polymer produced by a bacterium isolated from a deep-sea hydrothermal vent polychaete annelid. J. Appl. Microbiol. 93, 310–315. doi: 10.1046/j.1365-2672.2002.01689.x
Carron, B., and Botteron, V. (2019). “How smart can a contract be?,” in Blockchains, Smart Contracts, Decentralised Autonomous Organisation and the Law, eds D. Kraus, T. Obrist, and O. Hari (Cheltenham: Edward Elgar).
Collins, J., Rabone, M., Vanagt, T., Amon, D. J., Gobin, J., and Huys, I. (2020). Strengthening the Global Network for Sharing of Marine Biological Collections: Recommendations for a New Agreement for Biodiversity Beyond National Jurisdiction. ICES J Mar Sci 2020:fsaa227. doi: 10.1093/icesjms/fsaa227
da Silva, M., and de Oliveira, D. R. (2018). The new Brazilian legislation on access to the biodiversity (Law 13,123/15 and Decree 8772/16). Braz. J. Microbiol. 49, 1–4. doi: 10.1016/j.bjm.2017.12.001
Droege, G., Barker, K., Astrin, J. J., Bartels, P., Mackenzie-Dodds, J., et al. (2014). The global genome biodiversity network (GGBN) data portal. Nucleic Acids Res. 42, D607–D612. doi: 10.1093/nar/gkt928
Droege, G., Barker, K., Seberg, O., Coddington, J. A., Benson, E., Berendsohn, W. G., et al. (2016). The global genome biodiversity network (GGBN) data standard specification. Database 2016:baw125. doi: 10.1093/database/baw125
Droege, G., Karsch-Mizrachi, I., Barker, K., Coddington, J., and Seberg, O. (2019). Towards Linked Open Molecular Data: Recommendations for researchers, collections, infrastructures and publishers. BISS 3:e37204. doi: 10.3897/biss.3.37204
Greiber, T., Pena Moreno, S., Ahren, M., Nieto Carrasco, J., Kamau, E. C., Cabrera Medaglia, J., et al. (2012). An Explanatory Guide to the Nagoya Protocol on Access and Benefit-Sharing (No. 83). Switzerland: IUCN.
Guralnick, R. P., Cellinese, N., Deck, J., Pyle, R. L., Kunze, J., Penev, L., et al. (2015). Community Next Steps for Making Globally Unique Identifiers Work for Biocollections Data. ZooKeys 494, 133–154. doi: 10.3897/zookeys.494.9352
Halewood, M. (2014). International efforts to pool and conserve crop genetic resources in times of radical legal change. Intellectual property rights: Legal and economic challenges for development. The Initiative for Policy Dialogue Series. Oxford: Oxford University Press.
Hammond, E. (2016). More Patent Claims on Genetic Resources of Secret Origin: An Update on Disclosure of Origin in Patent Applications Under the Budapest Treaty Biodiversity, Knowledge and Rights Series No. 4. Penang: Third World Network.
Houssen, W., Sara, R., and Jaspars, M. (2020). Digital Sequence Information on Genetic Resources: Concept, Scope and Current Use. Ad Hoc Technical Expert Group on Digital Sequence Information on Genetic Resources. 29 January 2020. CBD/DSI/AHTEG/2020/1/3. Available online at: https://www.cbd.int/doc/c/fef9/2f90/70f037ccc5da885dfb293e88/dsi-ahteg-2020-01-03-en.pdf
Humphries, F. (2015). Shellfish Patents Krill Experimentation: Defences for Sharing Patented Aquatic Genetic Materials. EIPR 37, 210–224.
Humphries, F., Gottlieb, H. M., Laird, S., Wynberg, R., Lawson, C., Jaspars, M., et al. (2020). A tiered approach to the marine genetic resource governance framework under the proposed UNCLOS agreement for biodiversity beyond national jurisdiction (BBNJ). Mar. Policy 122:103910. doi: 10.1016/j.marpol.2020.103910
Jefferson, D. J., Fraire, I. S., and Beltrán-Morales, L. F. (2018). “Intellectual Property and the Governance of Plant Genetic Resources in Mexico: Trends and Implications for Research and Innovation,” in Mexican Natural Resources Management and Biodiversity Conservation, eds I. Azuz-Adeath and A. Cuevas Corona (Cham: Springer), 131–148. doi: 10.1007/978-3-319-90584-6_6
Kamau, E. C. (2013). 17 The multilateral system of the International Treaty on Plant Genetic Resources for Food and Agriculture. Common pools of genetic resources: Equity and innovation in international biodiversity law. London: Earthscan-Routledge.
Lawson, C., Burton, H., and Humphries, F. (2018). The important place of information in the evolving legal and policy framework for the conservation and sustainable use of the world’s plant genetic resources for food and agriculture’. Eur. Intellect. Prop. Rev. 40, 243–259.
Lawson, C., Humphries, F., and Rourke, M. (2019). The future of information under the CBD, Nagoya Protocol, Plant Treaty, and PIP Framework. J. World Intellect. Prop. 22, 103–119. doi: 10.1111/jwip.12118
Lawson, C., Wraith, J., and Pickering, C. M. (2019). Regulating wild collected orchids? The CBD, Nagoya Protocol and CITES overlaps. Environ. Plan. Law J. 36, 339–361.
Le Costaouëc, T., Cérantola, S., Ropartz, D., Ratiskol, J., Sinquin, C., Colliec-Jouault, S., et al. (2012). Structural data on a bacterial exopolysaccharide produced by a deep-sea Alteromonas macleodii strain. Carbohydr. Polym. 90, 49–59. doi: 10.1016/j.carbpol.2012.04.059
Leape, J., Abbott, M., and Sakaguchi, H. (2020). Technology, Data and New Models for Sustainably Managing Ocean Resources. Washington, DC: World Resources Institute.
Martins, A., Vieira, H., Gaspar, H., and Santos, S. (2014). Marketed Marine Natural Products in the Pharmaceutical and Cosmeceutical Industries: Tips for Success. Mar. Drugs 12, 1066–1101. doi: 10.3390/md12021066
Mignon, V. (2019). “Blockchains – perspectives and challenges,” in Blockchains, Smart Contracts, Decentralised Autonomous Organisation and the Law, eds D. Kraus, T. Obrist, and O. Hari (Cheltenham: Edward Elgar).
Morgera, E., Tsioumani, E., and Buck, M. (2014). Unraveling the Nagoya Protocol: a commentary on the Nagoya Protocol on access and benefit-sharing to the convention on biological diversity. Leiden: Martinus Nijhoff Publishers.
Mulalap, C. Y., Frere, T., Huffer, E., Hviding, E., Paul, K., Smith, A., et al. (2020). Traditional knowledge and the BBNJ instrument. Mar. Policy 122:104103. doi: 10.1016/j.marpol.2020.104103
Mulesa, T. H., and Westengen, O. T. (2020). Against the grain? A historical institutional analysis of access governance of plant genetic resources for food and agriculture in Ethiopia. J. World Intellect. Prop. 23, 82–120. doi: 10.1111/jwip.12142
Rabone, M., Horton, T., Harden-Davies, H., Collins, J. A., Zajderman, S., and Horton, et al. (2019). Access to Marine Genetic Resources (MGR): raising awareness of best-practice through a new agreement for Biodiversity Beyond National Jurisdiction (BBNJ). Front. Mar. Sci. 6:520. doi: 10.3389/fmars.2019.00520
Reichman, J. H., Uhlir, P. F., and Dedeurwaerdere, T. (2015). Governing digitally integrated genetic resources, data, and literature: global intellectual property strategies for a redesigned microbial research commons. Cambridge, MA: Cambridge University Press.
Rohden, F., Dröge, G., and Scholz, A. H. (2020). Combined study on Digital Sequence Information (DSI) in public and private databases and traceability. Ad Hoc Technical Expert Group on Digital Sequence Information on Genetic Resources. 31 January 2020. CBD/DSI/AHTEG/2020/1/4. Available online at: https://www.cbd.int/doc/c/1f8f/d793/57cb114ca40cb6468f479584/dsi-ahteg-2020-01-04-en.pdf
Rourke, M. (2019). Access by Design, Benefits if Convenient: A Closer Look at the Pandemic Influenza Preparedness Framework’s Standard Materials Transfer Agreements. Milbank Q. 97, 91–112. doi: 10.1111/1468-0009.12364
Schindel, D., Miller, S., Trizna, M., Graham, E., and Crane, A. (2016). The global registry of biodiversity repositories: a call for community curation. Biodivers. Data J. 4:e10293. doi: 10.3897/BDJ.4.e10293
Scholz, A. H., Hillebrand, U., Freitag, J., Cancio, I., Dos S. Ribeiro, C., Haringhuizen, G., et al. (2020). Finding Compromise on ABS and DSI in the CBD: Requirements and Policy Ideas from a Scientific Perspective, Bundesministerium für Bildung und Forschung, BMBF, WiLDSI. Available online at: https://www.dsmz.de/fileadmin/user_upload/Collection_allg/Final_WiLDSI_White_Paper_Oct7_2020.pdf
Thessen, A. E., Woodburn, M., Koureas, D., Paul, D., Conlon, M., Shorthouse, D. P., et al. (2019). Proper Attribution for Curation and Maintenance of Research Collections: Metadata Recommendations of the RDA/TDWG Working Group. Data Sci. J. 18:54. doi: 10.5334/dsj-2019-054
UN (2012). United Nations General Assembly Resolution 66/231, Resolution Adopted by the General Assembly On 24 December 2011. 66th Session, Agenda item 76(a). UN Doc A/RES/66/231, 2012. New York, NY: UN.
UN (2019). Statement by the President of the Conference at the Closing of the Third Session, Intergovernmental Conference on an Internationally Legally Binding Instrument under the United Nations Convention on the Law of the Sea on the Conservation and Sustainable Use of Marine Biological Diversity of Areas Beyond National Jurisdiction. Third Session, 13 September 2019, UN Doc. A/CONF.232/2019/10. New York, NY: UN.
Vierros, M. K., Harrison, A. L., Sloat, M. R., Crespo, G. O., Moore, J. W., Govan, H., et al. (2020). Considering indigenous peoples and local communities in governance of the global ocean commons. Mar. Policy 119:104039. doi: 10.1016/j.marpol.2020.104039
Von Kries, C., Broggiato, A., Dedeurwaerdere, T., and Winter, G. (2013). MICRO B3 Model Agreement on Access to Marine Microorganisms and Benefit Sharing (MICRO B3 WP8, version 1.0, 10 June 2013, updated 17 December 2013). Available online at: https://www.microb3.eu/sites/default/files/pdf/MICRO_B3_ABS_model_agreement_17122013%20explanatory%20notes.pdf (accessed date 17 December 2013)
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., et al. (2012). Darwin core: an evolving community-developed biodiversity data standard. PLoS One 7:e29715. doi: 10.1371/journal.pone.0029715
WIPO (2020). Key questions on patent disclosure requirements for genetic resources and traditional knowledge, 2nd edition. Geneva: WIPO.
WTO (2011). ‘Draft Decision to Enhance Mutual Supportiveness Between the Trips Agreement and the Convention on Biological Diversity’ World Trade Organization (WTO document TN/C/W/59 of 19 April 2011). Geneva: WTO.
Yilmaz, P., Kottmann, R., Field, D., Knight, R., Cole, J. R., Amaral-Zettler, L., et al. (2011). Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420. doi: 10.1038/nbt.1823
Keywords: marine genetic resources, biodiversity beyond national jurisdiction, traceabilily, benefit sharing, INSDC, open access
Citation: Humphries F, Rabone M and Jaspars M (2021) Traceability Approaches for Marine Genetic Resources Under the Proposed Ocean (BBNJ) Treaty. Front. Mar. Sci. 8:661313. doi: 10.3389/fmars.2021.661313
Received: 30 January 2021; Accepted: 31 March 2021;
Published: 27 April 2021.
Edited by:
Elizabeth Mendenhall, University of Rhode Island, United StatesReviewed by:
Arianna Broggiato, European Commission, BelgiumNengye Liu, Macquarie University, Australia
Copyright © 2021 Humphries, Rabone and Jaspars. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fran Humphries, fran.humphries@qut.edu.au
 Fran Humphries
Fran Humphries Muriel Rabone
Muriel Rabone Marcel Jaspars
Marcel Jaspars