- 1Faculty of Science, Sydney School of Veterinary Science, The University of Sydney, Camperdown, NSW, Australia
- 2Department of Biological Sciences, Macquarie University, North Ryde, NSW, Australia
Anthropogenic activities and pollution are impacting marine environments globally. As a consequence, increasing numbers of human-associated phylotypes of Escherichia coli, an indicator of fecal contamination, have been found in both aquatic environments and marine mammals considered sentinels for marine health. The objective of this study was to determine the presence and diversity of E. coli in pups of three species of free-ranging pinnipeds in Australia. Fecal samples (n = 963) were collected between 2016 and 2019 from Australian sea lion (Neophoca cinerea), Australian fur seal (Arctocephalus pusillus doriferus) and long-nosed fur seal (Arctocephalus forsteri) pups from eight breeding colonies extending along the Southern Australian coast. E. coli were isolated from 842 (87.3%) samples and molecular screening was applied to assign isolates to E. coli phylotypes and sub-types. The human associated E. coli phylotype B2 was the most frequently isolated in all species at seven of the eight colonies, with 73.7% of all E. coli isolates belonging to this phylotype. Phylotype distribution did not differ significantly within or across species, breeding colonies or breeding seasons. Analysis of B2 isolates into sub-types showed a significant difference in sub-type distribution across breeding seasons at two colonies (Seal Rocks and Cape Gantheaume). The predominance of the B2 phylotype could indicate that all colonies are exposed to similar levels of anthropogenic pollution. This widespread occurrence of the human-associated E. coli phylotypes highlights the imperative for ongoing monitoring and surveillance of microbes in both the marine environment and sentinel species.
Introduction
The contamination of the marine environment with atypical microorganisms as a result of anthropogenic pollution poses unknown risks to both marine wildlife and the wider ecosystem (Oates et al., 2012; Baily et al., 2015). Fecal coliforms such as Escherichia coli are used globally as indicators of anthropogenic fecal contamination and as a measure of pollution of waterways and coastal ecosystems (Beversdorf et al., 2007; Schaefer et al., 2011; Ahmed et al., 2016). E. coli is a commensal species colonizing the intestinal tract of birds and mammals (Gordon and Cowling, 2003; Guenther et al., 2011). However, some strains of E. coli have virulence traits that render them opportunistic pathogens that can cause a range of diseases in multiple host species, including urinary tract infections, neonatal meningitis and septicemia (Russo and Johnson, 2000; Mora et al., 2009; Clermont et al., 2011). There are eight E. coli lineages (A, B1, B2, C, D, E, F, clade I), termed phylotypes, that differ in ecological niches, life histories, host affinities and pathogenic abilities; phylotypes B2 and D are commonly isolated from and associated with humans (Gordon and Cowling, 2003; Gordon et al., 2008). Many clinically relevant strains of E. coli that have the potential to cause extraintestinal disease in humans, domestic animals and birds belong to phylotype B2 (Johnson and Russo, 2002; Clermont et al., 2014).
Marine mammals are considered sentinels of ocean health with marine mammal health status providing insights into the health of the wider marine ecosystem (Bossart, 2010). Escherichia coli has been isolated from a number of aquatic mammal species including pinnipeds (Stoddard et al., 2008, 2009; Wallace et al., 2013; Delport et al., 2015; Power et al., 2016; Fulham et al., 2018; Mora et al., 2018), cetaceans (Schaefer et al., 2011; Melendez et al., 2019) and river otters (Oliveira et al., 2018). Despite this, little is known about the host and environmental factors that influence the diversity and prevalence of E. coli in marine mammals. Furthermore, the intestinal population of E. coli in mammals differs between individuals of the same species (Guenther et al., 2011). As a result, studies that focus on one species at a single time point are unlikely to accurately represent the diversity of E. coli within a species (Schierack et al., 2008, 2009; Leser and Mølbak, 2009).
In the Southern hemisphere, E. coli has been isolated from multiple pinniped species including southern elephant seals (Mirounga leonina), Weddell seals (Leptonychotes weddellii), Antarctic fur seals (Arctocephalus gazella) and Australian sea lions (Neophoca cinerea) (Hernandez et al., 2007; Delport et al., 2015; Power et al., 2016; Fulham et al., 2018; Mora et al., 2018). These studies have characterized E. coli isolated from free-ranging individuals with a majority of samples collected from adults and a predominance of phylotype B2 identified; the prevalence of the B2 phylotype ranging from 49% (Fulham et al., 2018) to 65% (Power et al., 2016). Comparisons of E. coli prevalence and phylotype distribution in captive and free-ranging adult N. cinerea (Delport et al., 2015) determined a higher prevalence in captive individuals; however, in both captive and free-ranging animals, the human-associated phylotype B2 was the most frequently identified (Delport et al., 2015). A higher E. coli prevalence and differing trends in phylotype distribution was described in free-ranging N. cinerea pups from two colonies in South Australia (Fulham et al., 2018). Of the two colonies, phylotype B2 was more prevalent in samples from the colony closer to anthropogenic sources of pollution than the colony located further from human habitation (Fulham et al., 2018). It is unknown whether host or environmental factors could be contributing to the differences in phylotype distribution observed in E. coli isolated from free-ranging N. cinerea pups. For this reason, further investigations are required to understand the distribution and diversity of E. coli in marine mammal species in Australian waters, including other pinniped species such as the Australian fur seal (Arctocephalus pusillus doriferus) and long-nosed fur seal (Arctocephalus forsteri) where the prevalence and diversity of E. coli has not been reported previously.
The three pinniped species, N. cinerea, A. p. doriferus, and A. forsteri, inhabit numerous offshore colonies along the Australian coast from Western Australia to Tasmania (Kirkwood and Goldsworthy, 2013). All three species were subjected to commercial sealing practices during the nineteenth and twentieth centuries which decimated their numbers (Ling, 1999). Population recovery of each species from historical harvesting has differed greatly, with numbers of A. p. doriferus and A. forsteri steadily increasing since the 1960s and 1970s (Shaughnessy et al., 2015; McIntosh et al., 2018). In contrast, N. cinerea have undergone continual decline; the species is listed as endangered on the IUCN Red list (Goldsworthy et al., 2015). The reasons underlying these differing rates of population recovery are likely complex and multi-factorial and include differing life histories, breeding strategies, geographical distribution, extent of fisheries interactions and disease (Goldsworthy et al., 2009; Shaughnessy et al., 2011; Marcus et al., 2015). The geographical range of these species frequently overlap, with some species breeding at the same sites. This geographical proximity provides an unparalleled opportunity for comparative investigations of E. coli to better understand the role of host and environmental factors on E. coli prevalence and diversity.
The objective of this study was to characterize the diversity of E. coli in three species of free-ranging pinniped pups in Southern Australia, establishing base line data for future comparative studies and to assess potential effects of anthropogenic impacts.
Materials and Methods
Study Sites
Fecal swabs (n = 963) were collected from eight pinniped breeding colonies across multiple breeding seasons between 2016 and 2019 (Figure 1 and Table 1). Of the 963 fecal swabs collected, a total of n = 401 fecal swabs were from N. cinerea pups, n = 400 from A. p. doriferus pups and n = 162 from A. forsteri pups. Samples collected from N. cinerea pups at Seal Bay in 2016 and Dangerous Reef 2017 were collected and analyzed as part of a previous study (Fulham et al., 2018). Samples were collected across two breeding seasons for all sites except N. cinerea at Dangerous Reef and A. p. doriferus at Deen Maar Island, Cape Bridgewater and The Skerries. The proximity to human settlements differs for each colony; Dangerous Reef and Olive Island are the most remote locations, located approximately 33 and 15 km offshore, respectively, while Seal Rocks and Deen Maar Island are located less than 10 km offshore (Figure 1).
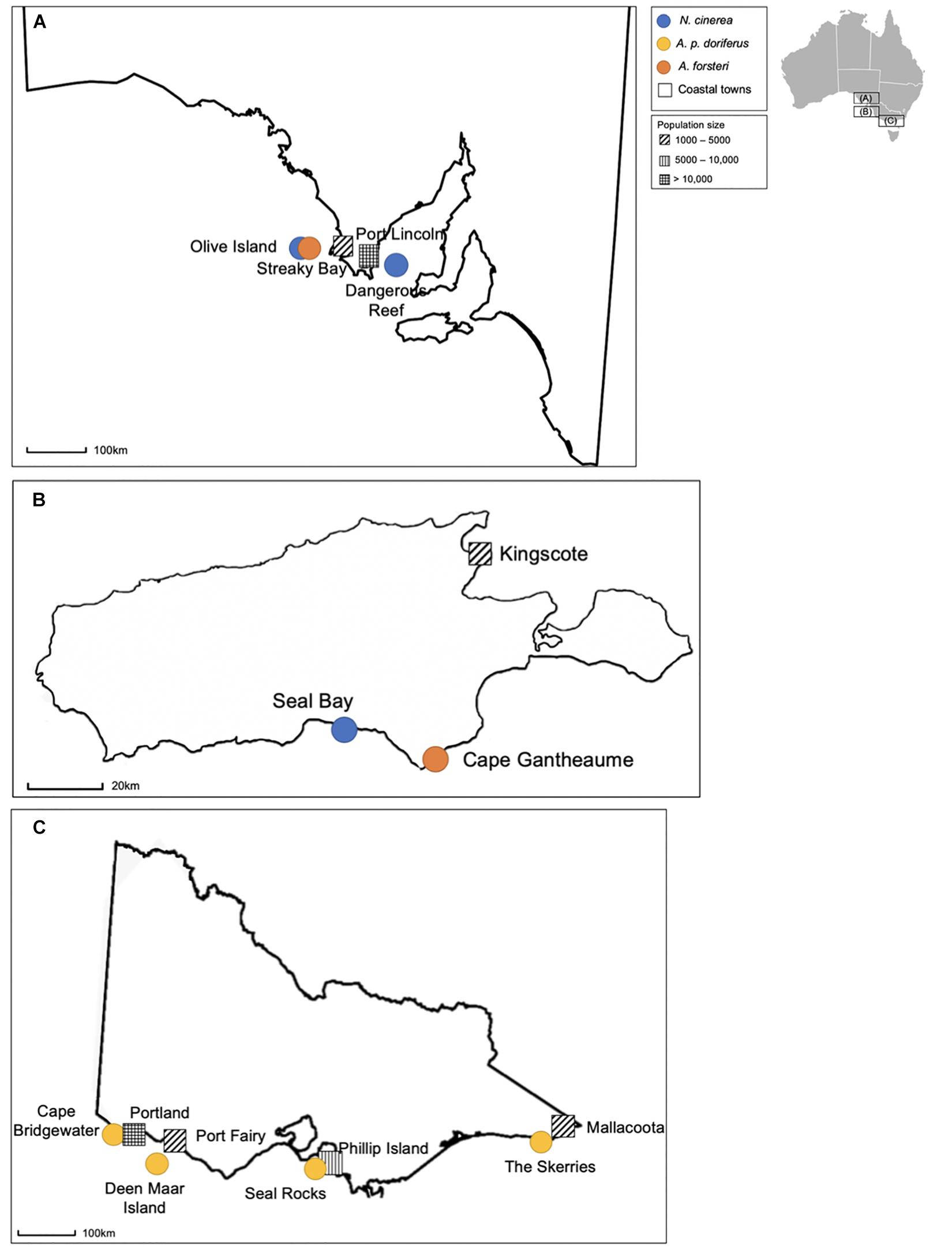
Figure 1. Location of breeding colonies and species sampled at each colony in South Australia (A), on Kangaroo Island (B), and in Victoria (C), with closest coastal town and approximate population size. Colonies sampled in South Australia include Olive Island, Dangerous Reef and two colonies on Kangaroo Island (Seal Bay and Cape Gantheaume). The remaining colonies, Cape Bridgewater, Deen Maar Island, Seal Rocks and the Skerries are located in Victoria.
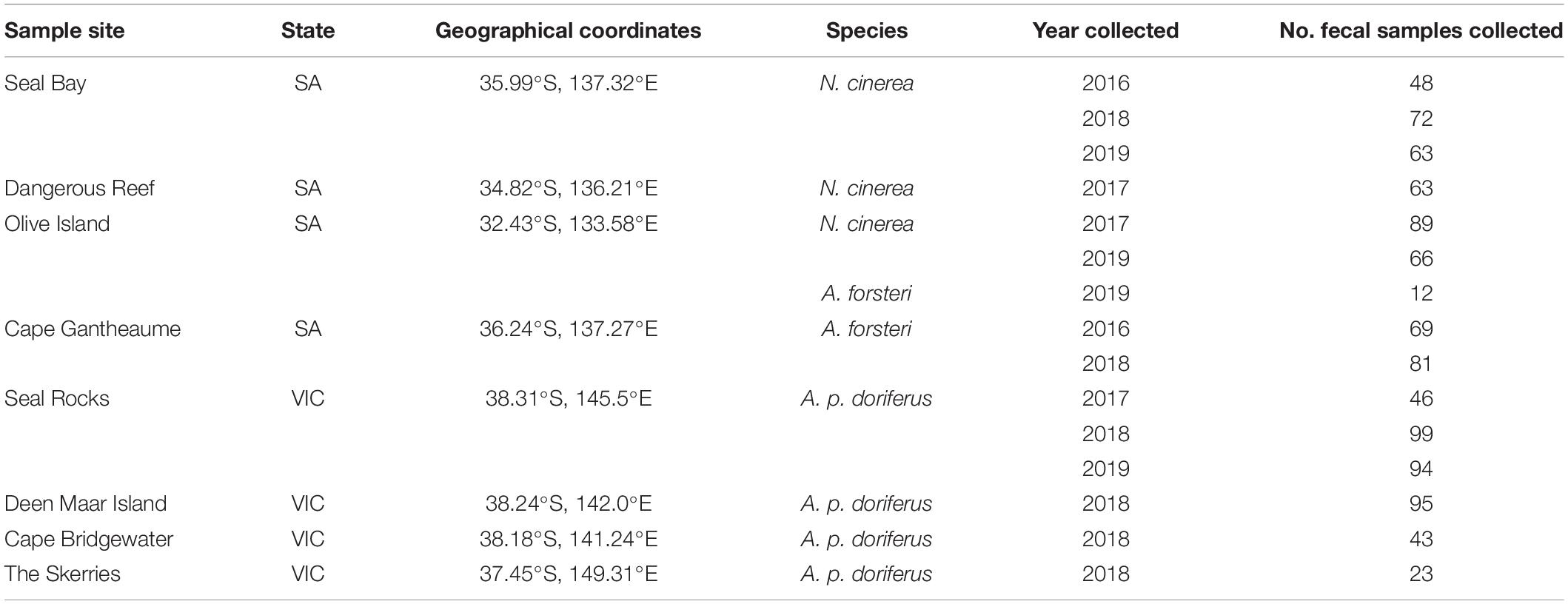
Table 1. Geographical location for each breeding colony and total number of fecal samples collected each year at each breeding colony.
Sample Collection
Pinniped pups, ranging in age from 3 to 6 weeks for A. forsteri and A. p. doriferus and 2 to 6 weeks for N. cinerea, were captured by hand as part of ongoing health investigations and restrained in canvas pup bags with breathing holes for the duration of capture and sample collection(Fulham et al., 2018). Pups were sampled when mothers were absent. Fecal samples were collected by inserting a sterile swab (Copan, Brescia, Italy) covered by a lubricated sheath directly into the rectum or by swabbing a fecal sample passed by the pup during capture and restraint. Fecal swabs were then sub-sampled into Sterile FecalSwabTM (Copan, Brescia, Italy). All swabs were refrigerated at 4°C until culture, usually within 7 days of collection. All sampling methods for N. cinerea and A. forsteri were approved by the Animal Ethics Committee at the University of Sydney (Protocol Nos. 2014/726 and 2017/1260); sampling methods for A. p. doriferus were approved by the Phillip Island Nature Park Animal Ethics Committee (Protocol No. 2.2016).
E. coli Culture, Isolation, and Preservation
Fecal swab media was inoculated onto Chromocult® coliform agar (Merck, Millipore, Australia) and cultured plates incubated at 37°C for 24 h. The E. coli colonies were identified by morphological features – being round, and dark blue-violet. Single colonies were selected and sub-cultured onto Chromocult® coliform agar plates to obtain pure cultures. To preserve cultures, pure E. coli isolates were grown in Luria Bertani (LB) broth (5 mL) at 37°C for 24 h and 500 μL of broth culture was combined with 500 μL 70% glycerol in 1.0 mL cryovials and samples were stored at −80°C.
Extraction of DNA From Preserved Bacteria
DNA was extracted using a boil preparation method. Preserved bacteria (40 μL) was inoculated into LB broth (150 μL) and incubated at 37°C for 24 h. Broth cultures were centrifuged for 5 min at 4000 rpm (Eppendorf 5810 R, rotor: A-4-62), the supernatant was removed and the bacterial pellet was re-suspended in molecular grade water (40 μL). Samples were then heated for 5 min at 95°C and centrifuged for 5 min at 4000 rpm (Eppendorf 5430 R, rotor: FA-45-24-11-HS). Lysates were stored in 1.5 mL microcentrifuge tubes at −30°C until PCR analysis.
Phylotyping of E. coli Isolates and Phylotype-Specific PCRs
To assign E. coli isolates to a phylotype, lysates were analyzed using a quadruplex PCR protocol following the methodology of Clermont et al. (2013). Isolates were assigned to a phylotype based on the presence or absence of four genes: ChuA, yjaA, TspE4.C2, and arpA. Isolates identified as A/C and D/E were further analyzed using two phylotype-specific PCRs. To assign isolates to phylotypes A or C, a singleplex PCR was conducted using the primers trpAgpC.1 and trpAgpC.2 with internal control primers trpBA.f and trpBA.r (Clermont et al., 2013). To assign isolates to phylotypes D or E, a singleplex PCR was conducted using primers ArpAgpE.f and ArpAgpE.r with internal control primers trpBA.f and trpBA.r (Clermont et al., 2013). All phylotyping PCRs were performed using GoTaq® Green Master Mix (Promega, Madison, United States) and included controls representing each phylotype (Power et al., 2016), and a negative control containing PCR water.
All reactions described above were resolved using gel electrophoresis (2% agarose w/v) conducted at 100 V for 30 min in TBE (Tris, boric acid, ethylenediaminetetraacetic acid) with SYBR safe gel stain (Invitrogen, Mulgrave, Australia). Product sizes were approximated against a HyperLadderII 50 bp DNA marker (Bioline, Sydney, Australia).
E. coli Phylotype B2 Sub-Typing
Isolates classified as B2 were further analyzed using two multiplex PCR panels to assign isolates to one of nine sub-groups using primers described by Clermont et al. (2014). Minor modifications were made to the PCR protocol by altering the annealing temperatures for both panels. Samples were assigned to sub-groups based on the presence of the following fragments: putP, pabB, trpA, trpA, polB, dinB, icd, aes (IX), and aes (X). The chuA gene was included as an internal control. Multiplex panel 1 tested for sub-groups II, III, VI, VII, and IX, and multiplex panel 2 tested for sub-groups I, VI, V, and X. The PCRs for panels 1 and 2 were performed using GoTaq® Green 2X (Promega, Madison, United States) with the addition of MgCl2 to increase the concentration to 2.0 mM MgCl2. The following PCR conditions were used: initial denaturation at 94°C for 4 min; 35 cycles at 94°C for 5 s, 59°C for 20 s (panel 1) or 58°C for 20 s (Panel 2), 72°C for 20 s, 72°C for 5 min and held at 4°C. Isolates were either assigned one of the nine sub-groups or considered unassigned. All reactions were resolved using gel electrophoresis using methods described in section “Phylotyping of E. coli Isolates and Phylotype-Specific PCRs”.
Statistical Analysis
RStudio (V 1.2.5042, Boston, MA, United States) software was utilized for all statistical analyses. A one-way ANOVA was used to compare E. coli prevalence across breeding seasons within each species. A generalized linear model was employed to analyze the relationship between the following factors: phylotype and sub-type prevalence, colony location, species, and breeding season. Normality of data was tested using the Shapiro-Wilk test. Factors were tested and included in the model based on AIC values. Statistical significance was determined when p < 0.05.
Results
Prevalence of E. coli in Pinniped Pups
E. coli was detected in 842/963 (87.3%) fecal samples collected from N. cinerea, A. p. doriferus, and A. forsteri pups. The total prevalence of E. coli varied across pinniped species and sampling sites (Table 2). E. coli prevalence was highest in A. p. doriferus (88.7%), followed by A. forsteri (87.0%) and N. cinerea (86.2%). There was no significant difference in E. coli prevalence across sites (p = 0.442), species (p = 0.564) or breeding season (p = 0.293).
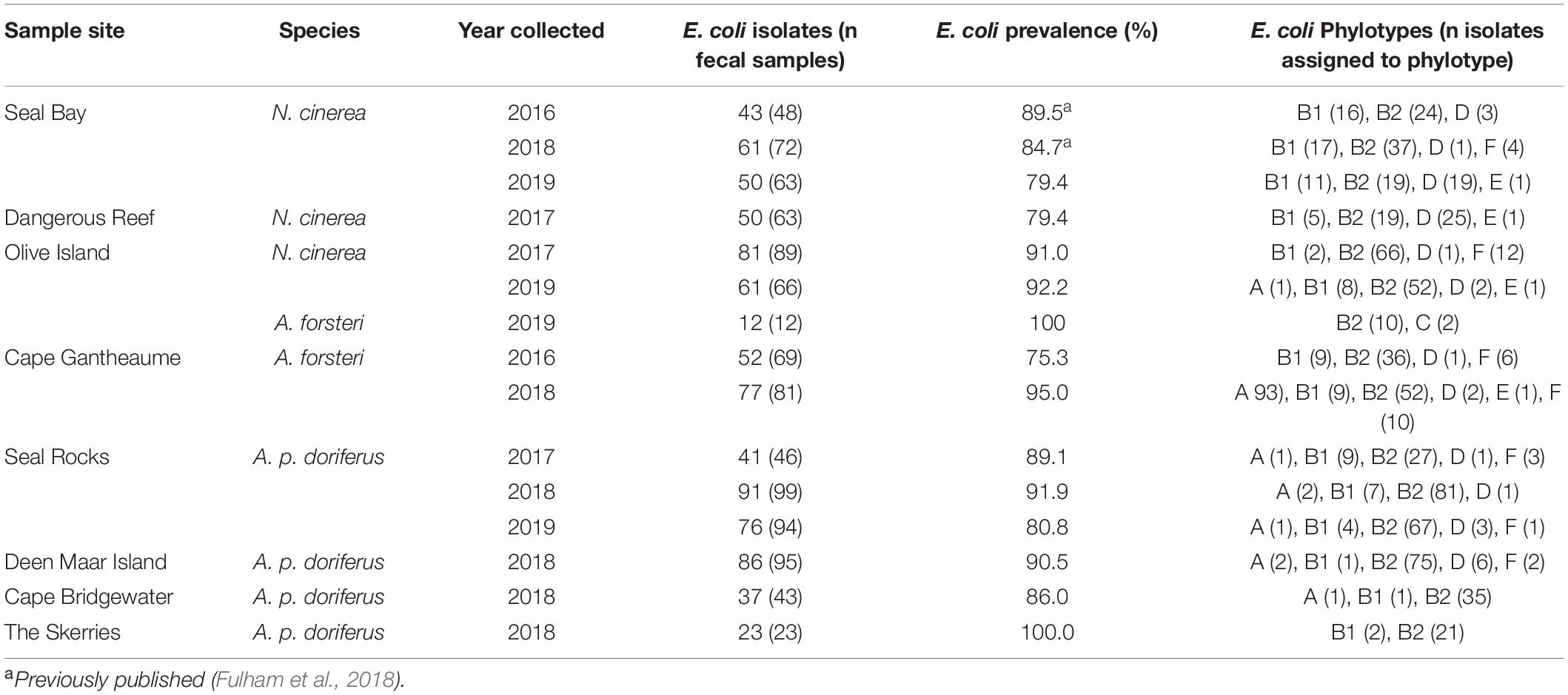
Table 2. E. coli isolates and prevalence of E. coli in pinniped pups at all eight colonies sampled for each breeding season.
Distribution of E. coli Phylotypes in Pinniped Pups
Phylotyping of E. coli isolates showed only minor differences in distribution across the three species (Figure 2). There was no significant difference in E. coli phylotypes across species (p = 0.055), sampling sites (p = 0.437) or breeding season at colonies sampled over multiple breeding seasons; Seal Bay (p = 0.272), Olive Island (p = 0.199), Seal Rocks (p = 0.880), and Cape Gantheaume (p = 0.076) (Figure 3). The B2 phylotype was most frequently isolated from all samples with 73.7% of all E. coli isolates identified as B2, followed by B1, D, F, A, E, and C.
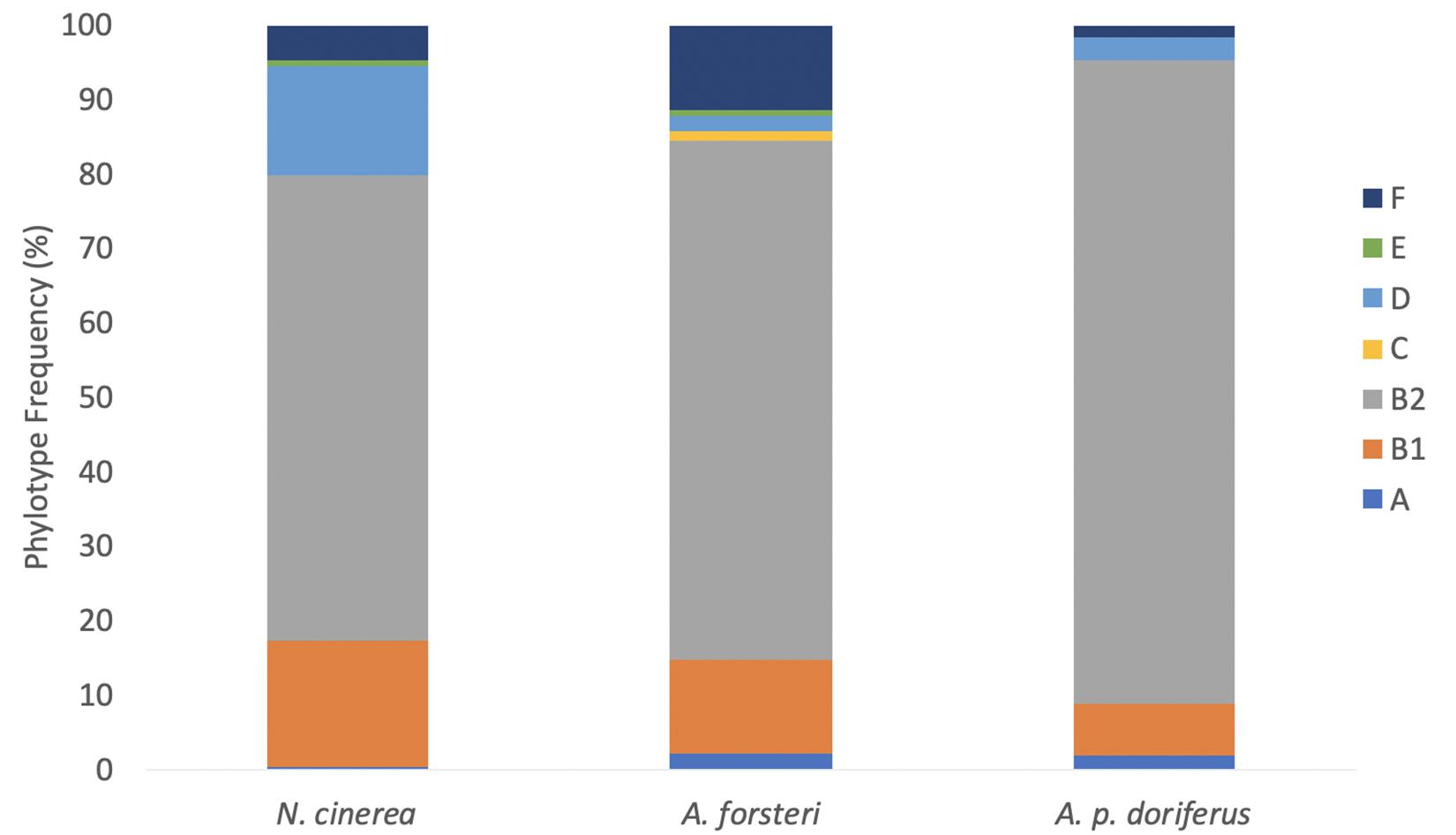
Figure 2. Distribution of E. coli phylotypes in each species across all breeding seasons (2016–2019).
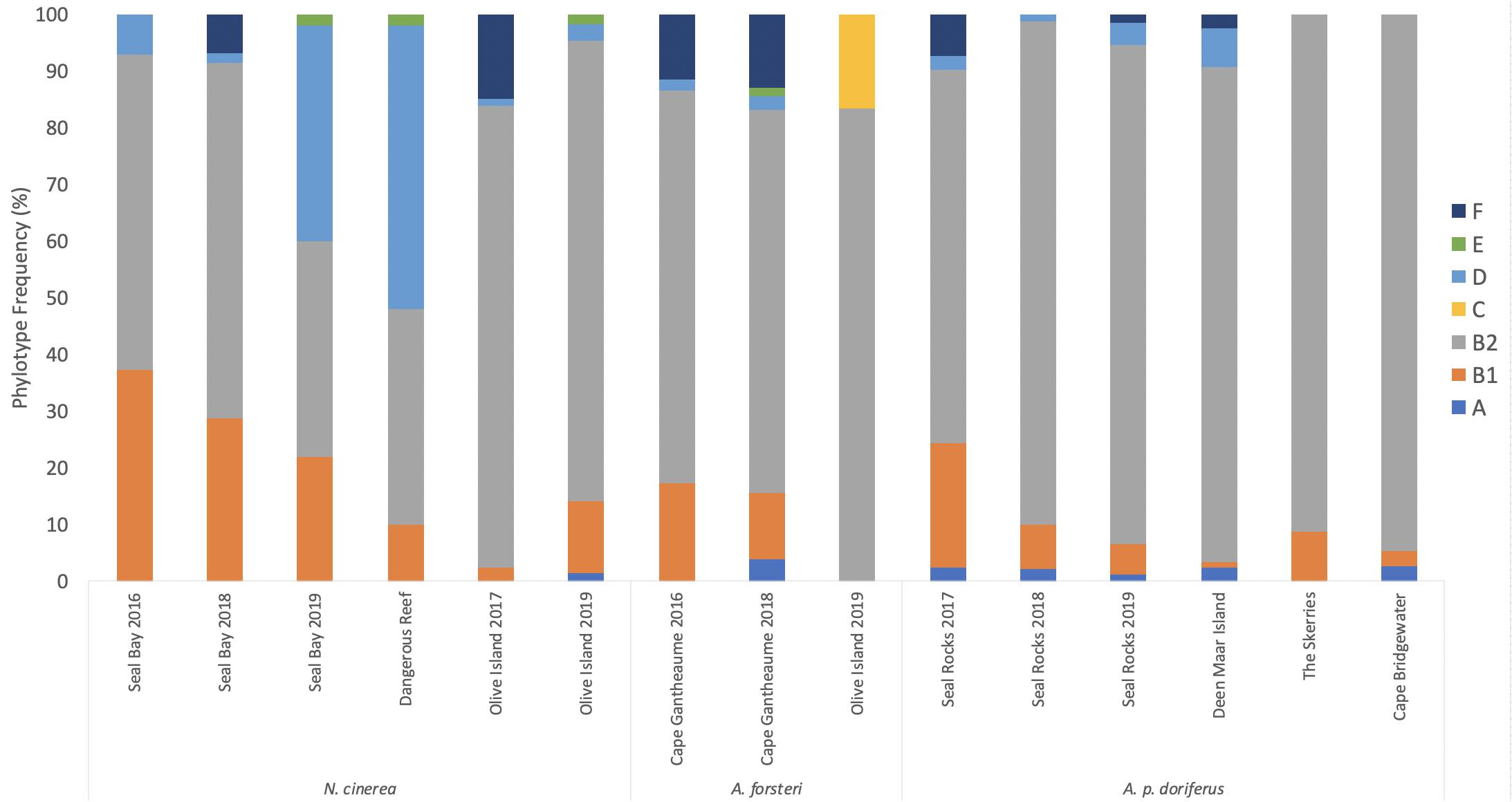
Figure 3. Distribution of E. coli phylotypes across breeding season for each pinniped species across the eight sampling sites between 2016 and 2019.
In N. cinerea, A. forsteri, and A. p. doriferus the most frequently identified phylotype was B2, with 62.5, 86.4, and 69.5% of isolates assigned to this phylotype, respectively (Table 2 and Figure 3). There were no E. coli isolates from A. p. doriferus pups assigned to phylotype E. Isolates from A. forsteri pups were the most diverse with seven phylotypes identified and phylotype C was only found in E. coli isolates from A. forsteri pups at Olive Island.
Distribution of B2 Sub-Types in Pinniped Pups
Isolates from N. cinerea, A. forsteri, and A. p. doriferus pups that were assigned to the B2 phylotype were further analyzed and assigned to one of nine B2 sub-types. There was no significant difference in B2 sub-type distribution across sampling sites (p = 0.768) or species (p = 0.121) (Figure 4). At Seal Bay and Olive Island there was no significant difference in B2 sub-type distribution across breeding seasons (p = 0.483 and p = 0.098, respectively) (Figure 5). There was a significant difference in sub-type distribution across breeding seasons at Seal Rocks (p = 0.046) and Cape Gantheaume (p < 0.001). At Seal Rocks there was a decrease in the frequency of sub-types II, IV, VI, VII, IX, and X between seasons, while the frequency of sub-type III increased (Figure 4). The significant difference seen at Cape Gantheaume was due to an increase in sub-types II, V, VI, and IX and a decrease in the frequency of sub-types III, IV, and VII.
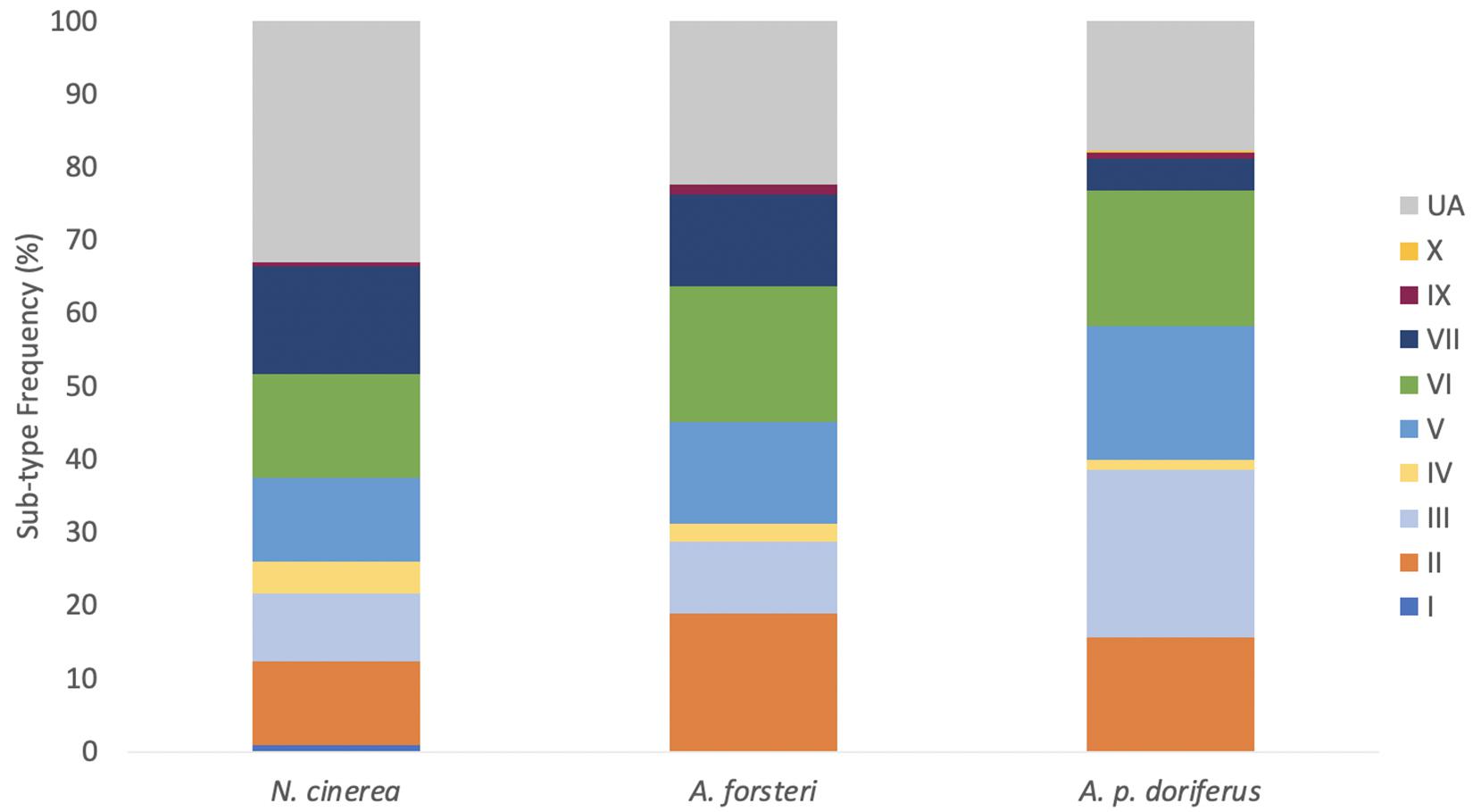
Figure 4. Combined distribution of E. coli B2 sub-types for each species across breeding seasons sampled (2016–2019). Samples that could not be assigned to a sub-type were considered “unassigned” (UA). All nine sub-groups were identified in at least one of the three species.
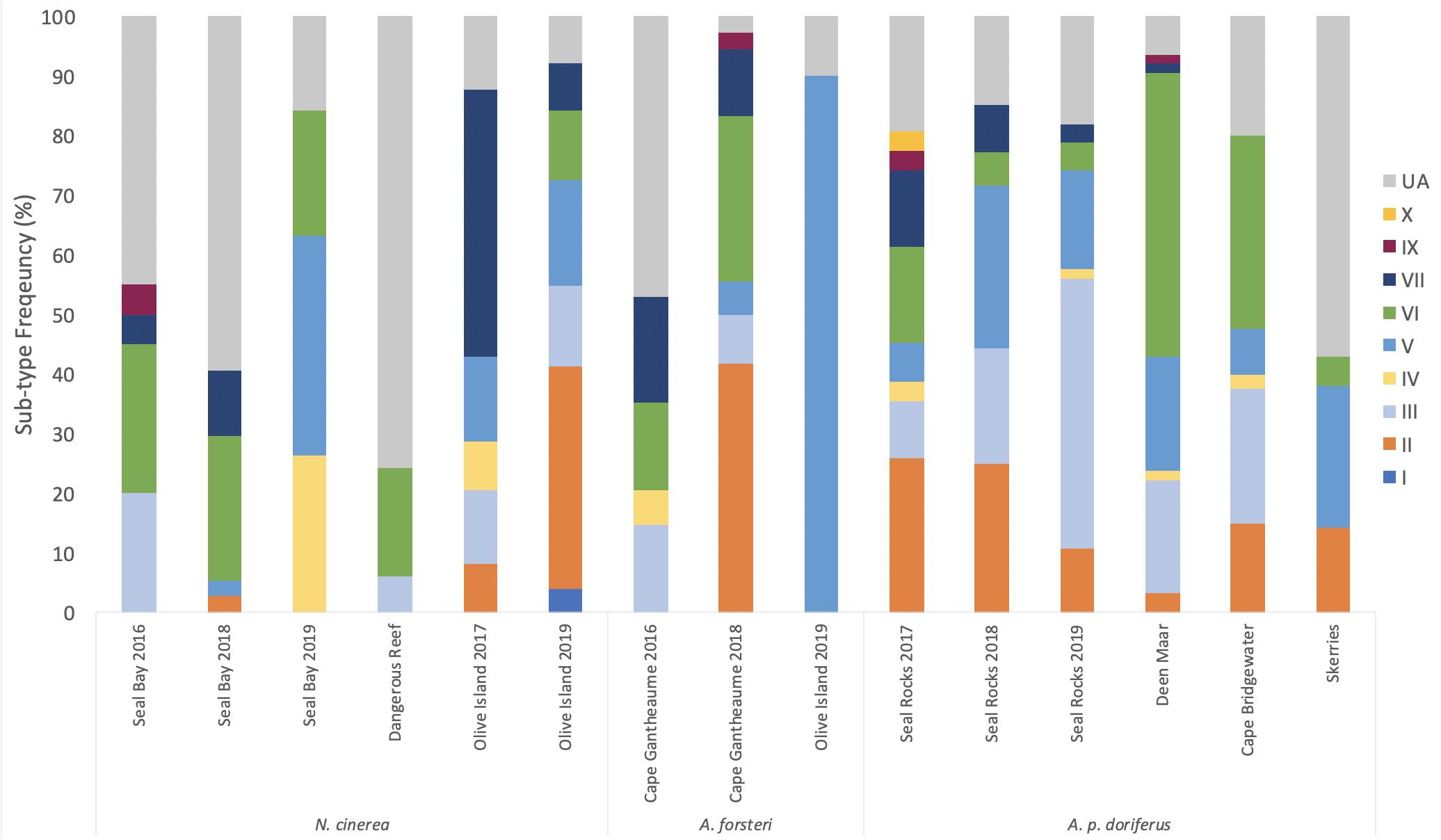
Figure 5. Distribution of E. coli B2 sub-types for each breeding season in each species across the eight sampling sites. Samples that could not be assigned were classed as “unassigned” (UA). Sub-type I was only identified in N. cinerea and sub-group X was only detected in A. p. doriferus pups.
Sub-type distribution differed slightly between species with sub-type I only detected in a single N. cinerea pup sampled at Olive Island; sub-type X was only detected in one A. p. doriferus pup sampled at Seal Rocks. The most frequently isolated sub-type across all species was VI, followed by III, V, II, VII, IV, IX, I, and X (Figure 4). A total of 23.6% of B2 isolates could not be assigned (UA) to a sub-type.
Discussion
This study reports the prevalence and diversity of E. coli isolated from free-ranging N. cinerea, A. p. doriferus, and A. forsteri pups at eight breeding colonies in Australia, finding no significant difference in E. coli prevalence or the distribution of phylotypes across species, colonies or breeding seasons.
This is the first investigation of the comparative presence and distribution of E. coli in free-ranging A. p. doriferus and A. forsteri pups. The prevalence of E. coli was similar across all three species and supports previous reports in Antarctic pinnipeds including Antarctic fur seals, Southern elephant seals and Weddell seals (Power et al., 2016; Mora et al., 2018), and captive adult N. cinerea (Delport et al., 2015). Previous studies have suggested that E. coli is uncommon in marine mammals (Johnson et al., 1998; Hernandez et al., 2007) with a higher prevalence associated with proximity to humans (Stoddard et al., 2008; Delport et al., 2015). In this study the differences in proximity to humans at each breeding colony and the high prevalence of E. coli observed in N. cinerea, A. p. doriferus, and A. forsteri pups could potentially indicate that proximity to humans is not the only factor contributing to E. coli prevalence in marine mammals.
The B2 phylotype was most frequently identified from pups of all three species, with 73.7% of all E. coli isolates assigned to this phylotype. This is the highest frequency of the B2 phylotype identified in pinnipeds in Australia; in free-ranging N. cinerea, B2 made up 67% of all isolates in adults (Delport et al., 2015). It has been suggested that strains belonging to the B2 phylotype are well adapted to the intestinal environment of mammals (Gordon and Cowling, 2003; Nowrouzian et al., 2006), which could be a contributing factor to the high frequency identified in this study. The B2 phylotype was the most frequently isolated phylotype at seven of the eight breeding colonies during every breeding season sampled. It was previously hypothesized that the difference in phylotype distribution observed in free-ranging N. cinerea pups was due to colony location (Fulham et al., 2018), however, the results from this study indicate that geographical location and proximity to humans, is not a contributing factor to E. coli diversity in N. cinerea, A. p. doriferus, or A. forsteri pups.
The prevalence of E. coli in free-ranging pinniped pups is likely diet related; E. coli is a lactose fermenter and as the sampled pups were feeding solely on milk, high E. coli prevalence is not unexpected (Fulham et al., 2018). In adult N. cinerea, E. coli prevalence from free-ranging individuals was significantly lower (7.7%) compared to captive individuals (84%) suggesting that E. coli does not occur naturally in free-ranging adult N. cinerea (Delport et al., 2015). Marine wildlife species that forage in coastal or nearshore waters are at greater risk of exposure to pathogens in fecal bacteria as a result of anthropogenic pollution compared to species further removed from anthropogenic influence (Oates et al., 2012). In Antarctic pinnipeds, it was hypothesized that the presence of human-associated E. coli could be due to the geographic mobility of the animals sampled (Mora et al., 2018). Free-ranging N. cinerea, A. p. doriferus, and A. forsteri adults occupy large geographical ranges (Shaughnessy et al., 2011, 2015; McIntosh et al., 2018), encountering a variety of environments and environmental conditions that could influence their exposure to anthropogenic pollution. In other pinniped species, there is evidence of maternal transmission of gut microbes (Nelson et al., 2013b), however, it is unknown whether the acquisition of E. coli in pinniped pups in this study is environmental or through maternal transfer. Understanding the prevalence of E. coli and phylotype diversity in free-ranging adults could provide valuable insights into the factors that contribute to the acquisition of E. coli and the trends in E. coli prevalence and phylotype distribution seen in pups.
Studies investigating the transfer of E. coli between humans and wildlife species are limited to terrestrial environments. Generally, E. coli isolated from wildlife species that share habitats and have higher levels of interactions with humans and livestock were genetically similar to E. coli isolated from humans (Goldberg et al., 2007; Rwego et al., 2008a,b). This genetic similarity suggests E. coli transmission between these species in shared habitats is likely a result of indirect contact through contaminated environmental sources rather than direct contact (Goldberg et al., 2007). For example, E. coli isolated from the feces of banded mongoose (Mungos mungo) was genetically similar to E. coli isolated from human fecal waste in their environment, highlighting the importance of indirect routes of transmission (Pesapane et al., 2013). These results suggest that the acquisition of human-associated bacteria by wildlife species is linked to fecal contamination of the environment.
Coastal environments can be contaminated by fecal pollution through a number of different sources including sewage and storm water runoff from agricultural, urban and commercial land (Crain et al., 2009; Pandey et al., 2014). The population size and density of people in coastal towns will influence the amount of bacterial contamination of the environment from runoff, with higher density resulting in higher levels of fecal bacteria (Pandey et al., 2014). The population size of towns closest to the pinniped breeding colonies sampled in this study varied, however, the prevalence of E. coli did not differ across colonies or seasons, suggesting that the contribution of human population density to trends in E. coli prevalence observed requires further investigation. Previous studies have determined that E. coli isolates from wildlife species occupying habitats in close association with humans or that are exposed to fecal pollution are more likely to belong to phylotype B2 than those isolated from wildlife living in isolation from humans (Gordon and Cowling, 2003). E. coli isolates belonging to B2 and D phylotypes have been found in treated sewage, suggesting a greater ability to survive treatment processes (Anastasi et al., 2010). The survival of these phylotypes in wastewater coupled with increasing pollution of coastal ecosystems presents a higher risk of transfer of human-associated bacteria into the marine environment. The predominance of the B2 phylotype across the three pinniped species studied could therefore suggest that all species are exposed to similar levels of anthropogenic pollution. This could be important for pup health, given that strains belonging to this phylotype are associated with extraintestinal disease in humans (Dale and Woodford, 2015).
Ocean currents and tides are also potential factors contributing to the high prevalence of E. coli in pinnipeds. The main source of ocean pollution is of terrestrial origin (Robinson et al., 2017) and as a consequence, coastal areas in close proximity to populated areas are exposed to higher levels of anthropogenic pollution (Partelow et al., 2015). The marine environment, being connected over longer timescales (Jönsson and Watson, 2016) has greater connectivity compared to terrestrial environments, with fewer physical barriers between areas. Consequently, the pathways of water that flow along coastlines can influence the dispersal of anthropogenic pollutants. Wastewater effluent is a known source of human-associated E. coli phylotypes (de Stoppe et al., 2017) and E. coli that originate from wastewater have an enhanced capacity to survive in the marine environment, able to survive for several days outside of a host (Rozen and Belkin, 2001). In addition, E. coli can attach to particles in the water column, facilitating movement in the marine environment (Mallin et al., 2000). This enhanced survival and attachment to particles in the water column could result in prolonged environmental persistence of E. coli, facilitating acquisition by marine wildlife. The similarity in E. coli distribution across species and breeding colonies in this study could be indicative of the high degree of connectivity of marine ecosystems (Crain et al., 2009; Robinson et al., 2017) or similar anthropogenic pollution across this large geographical range.
The identification of B2 sub-types I, II and IX are of particular interest given that clonal complexes that belong to these sub-types are commonly associated with disease in avian species and humans (Clermont et al., 2014; Riley, 2014). ST131 and ST95 strains belonging to sub-types I and IX, respectively, are associated with disease caused by extraintestinal pathogenic E. coli (ExPEC) (Dale and Woodford, 2015). These sub-types or strains have previously been identified in Antarctic pinnipeds (Power et al., 2016; Mora et al., 2018). Assignment of E. coli isolates to strains was not undertaken in this study, however, given the presence of B2 sub-types associated with pathogenic strains, further analysis of B2 isolates will assist our understanding of the potential risks posed to pup health by their presence.
It is also important to consider that E. coli is a very small component of the intestinal microbiota in pinnipeds (Nelson et al., 2013a,b; Delport et al., 2016). However, the ease with which E. coli can be cultured and characterized has resulted in E. coli being commonly used for monitoring fecal contamination of marine environments (Beversdorf et al., 2007). For this reason, it can be a useful indicator of the diversity of E. coli phylotypes present in marine mammals, and a relatively high prevalence may suggest greater exposure to anthropogenic pollution. Sampling of substrate and water surrounding breeding colonies and comparing those E. coli phylotypes with diversity seen in pinniped pups could provide useful insights into potential sources of environmental contamination. Investigating the presence of specific markers through microbial source tracking could also be utilized to determine the origin of E. coli found in both wildlife species and contaminated environments.
The presence of bacteria and protozoa that are associated with humans should be explored to further understand potential pathogen transmission from anthropogenic sources into the marine environment. Toxoplasma gondii, Campylobacter spp. and Salmonella spp. have been isolated from marine mammals inhabiting coastal environments, including gray seals (Halichoerus grypus), Californian sea otters (Enhydra lutris nereis) and northern elephant seals (Stoddard et al., 2005; Oates et al., 2012; Shapiro et al., 2012; Baily et al., 2015). Campylobacter spp. isolated from H. grypus were genetically similar to isolates commonly found in agricultural and human sources, demonstrating the dissemination of a human pathogen into the marine environment (Baily et al., 2015). Similar investigations in Australian pinniped species could enable the identification of the source and dissemination of anthropogenic microbial pollution into the Australian marine environment.
In this study, the predominance of the human associated B2 phylotype and similarity of E. coli prevalence and phylotype diversity seen across species, colonies and breeding seasons could indicate that all colonies are exposed to similar levels of anthropogenic pollution. This widespread occurrence of human associated phylotypes highlights the need for ongoing monitoring and surveillance of microbes in both the marine environment and sentinel species, particularly those with potential pathogenicity for marine mammals. Future investigations should focus on whether E. coli is an atypical bacterium in these pinniped species and determine its reliability as an indicator of marine pollution.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Committee at the University of Sydney and Phillip Island Nature Parks.
Author Contributions
MF, RG, and MP contributed to conceptualization and design of the study. MF and RG collected samples from pinniped pups. MF completed laboratory analysis of samples and data analysis. MF drafted the manuscript. RG and MP participated in revising the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Collection and analysis of samples was funded by the Ecological Society of Australia through the provision of the Holsworth Wildlife Research Endowment and the Sydney School of Veterinary Science at University of Sydney through the Late Dorothy Minchin Bequest. Field work support was provided by the Hermon Slade Foundation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff at Seal Bay, Kangaroo Island, Department for Environment and Water (DEW), South Australia and Rebecca McIntosh and staff at Phillip Island Nature Parks (PINP), Victoria for field assistance and logistical support; Simon Goldsworthy and South Australian Research and Development Institute (SARDI) for field assistance. We also thank Scott Lindsay, Shannon Taylor, and Matthew Gray for assistance in sample collection; Daniel Russell and Fiona McDougall, Macquarie University, for laboratory assistance; Victorian Fisheries Authorities, T-cat Charters, Seatec Marine services and Darren Guidera for marine charters. Sample collection was made possible through the collaborative support of DEW, PINP, and SARDI.
References
Ahmed, W., Sidhu, J. P. S., Smith, K., Beale, D. J., Gyawali, P., and Toze, S. (2016). Distributions of fecal markers in wastewater from different climatic zones for human fecal pollution tracking in Australian surface waters. Appl. Environ. Microbiol. 82, 1316–1323. doi: 10.1128/AEM.03765-15
Anastasi, E. M., Matthews, B., Gundogdu, A., Vollmerhausen, T. L., Ramos, N. L., Stratton, H., et al. (2010). Prevalence and persistence of Escherichia coli strains with uropathogenic virulence characteristics in sewage treatment plants. Appl. Environ. Microbiol. 76, 5882–5886. doi: 10.1128/AEM.00141-10
Baily, J. L., Méric, G., Bayliss, S., Foster, G., Moss, S. E., Watson, E., et al. (2015). Evidence of land-sea transfer of the zoonotic pathogen Campylobacter to a wildlife marine sentinel species. Mol. Ecol. 24, 208–221. doi: 10.1111/mec.13001
Beversdorf, L. J., Bornstein-Forst, S. M., and McLellan, S. L. (2007). The potential for beach sand to serve as a reservoir for Escherichia coli and the physical influences on cell die-off. J. Appl. Microbiol. 102, 1372–1381. doi: 10.1111/j.1365-2672.2006.03177.x
Bossart, G. D. (2010). Marine mammals as sentinel species for oceans and human health. Vet. Pathol. 48, 676–690. doi: 10.1177/0300985810388525
Clermont, O., Christenson, J. K., Daubié, A.-S., Gordon, D. M., and Denamur, E. (2014). Development of an allele-specific PCR for Escherichia coli B2 sub-typing, a rapid and easy to perform substitute of multilocus sequence typing. J. Microbiol. Methods 101, 24–27. doi: 10.1016/j.mimet.2014.03.008
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clermont, O., Olier, M., Hoede, C., Diancourt, L., Brisse, S., Keroudean, M., et al. (2011). Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect. Genet. Evol. 11, 654–662. doi: 10.1016/j.meegid.2011.02.005
Crain, C. M., Halpern, B. S., Beck, M. W., and Kappel, C. V. (2009). Understanding and managing human threats to the coastal marine environment. Ann. N. Y. Acad. Sci. 1162, 39–62. doi: 10.1111/j.1749-6632.2009.04496.x
Dale, A. P., and Woodford, N. (2015). Extra-intestinal pathogenic Escherichia coli (ExPEC): disease, carriage and clones. J. Infect. 71, 615–626. doi: 10.1016/j.jinf.2015.09.009
de Stoppe, N. C., Silva, J. S., Carlos, C., Sato, M. I. Z., Saraiva, A. M., Ottoboni, L. M. M., et al. (2017). Worldwide phylogenetic group patterns of Escherichia coli from commensal human and wastewater treatment plant isolates. Front. Microbiol. 8:2512. doi: 10.3389/fmicb.2017.02512
Delport, T. C., Harcourt, R. G., Beaumont, L. J., Webster, K. N., and Power, M. L. (2015). Molecular detection of antibiotic-resistance determinants in Escherichia coli isolated from the endangered Australian sea lion (Neophoca cinerea). J. Wildl. Dis. 51, 555–563. doi: 10.7589/2014-08-200
Delport, T. C., Power, M. L., Harcourt, R. G., Webster, K. N., and Tetu, S. G. (2016). Colony location and captivity influence the gut microbial community composition of the Australian sea lion (Neophoca cinerea). Appl. Environ. Microbiol. 82, 3440–3449. doi: 10.1128/AEM.00192-16
Fulham, M., Power, M., and Gray, R. (2018). Comparative ecology of Escherichia coli in endangered Australian sea lion (Neophoca cinerea) pups. Infect. Genet. Evol. 62, 262–269. doi: 10.1016/j.meegid.2018.05.002
Goldberg, T. L., Gillespie, T. R., Rwego, I. B., Wheeler, E., Estoff, E. L., and Chapman, C. A. (2007). Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol. Conserv. 135, 511–517. doi: 10.1016/j.biocon.2006.10.048
Goldsworthy, S. D., Gales, N. J., and Chilvers, B. L. (2015). Neophoca Cinerea. IUCN Red List Threat. Species 2015 e.T14549A45228341. Gland: IUCN.
Goldsworthy, S. D., McKenzie, J., Shaughnessy, P. D., McIntosh, R. R., Page, B., and Campbell, R. (2009). An Update of the Report: Understanding the Impediments to the Growth of Autralian Sea Lion Populations. SARDI Publ. Number F2008/00847-1 SARDI Reearch Report Series. No. 297. Urrbrae, SA: South Australian Research and Development Institute
Gordon, D. M., Clermont, O., Tolley, H., and Denamur, E. (2008). Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 10, 2484–2496. doi: 10.1111/j.1462-2920.2008.01669.x
Gordon, D. M., and Cowling, A. (2003). The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149, 3575–3586. doi: 10.1099/mic.0.26486-0
Guenther, S., Ewers, C., and Wieler, L. H. (2011). Extended-spectrum Beta-Lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2:246. doi: 10.3389/fmicb.2011.00246
Hernandez, J., Prado, V., Torres, D., Waldenström, J., Haemig, P. D., and Olsen, B. (2007). Enteropathogenic Escherichia coli (EPEC) in Antarctic fur seals Arctocephalus gazella. Polar Biol. 30, 1227–1229. doi: 10.1007/s00300-007-0282-2
Johnson, J. R., and Russo, T. A. (2002). Extraintestinal pathogenic Escherichia coli: “The other bad E coli.”. J. Lab. Clin. Med. 139, 155–162. doi: 10.1067/mlc.2002.121550
Johnson, S. P., Nolan, S., and Gulland, F. M. (1998). Antimicrobial susceptibility of bacteria isolated from pinnipeds stranded in central and northern California. J. Zoo Wildl. Med. 29, 288–294.
Jönsson, B. F., and Watson, J. R. (2016). The timescales of global surface-ocean connectivity. Nat. Commun. 7:11239. doi: 10.1038/ncomms11239
Kirkwood, R., and Goldsworthy, S. (2013). Fur Seals and Sea Lions. Collingwood, VIC: CSIRO PUBLISHING.
Leser, T. D., and Mølbak, L. (2009). Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ. Microbiol. 11, 2194–2206. doi: 10.1111/j.1462-2920.2009.01941.x
Ling, J. K. (1999). Exploitation of fur seals and sea lions from Australian, New Zealand and adjacent subantarctic islands during the eighteenth, nineteenth and twentieth centuries. Aust. Zool. 31, 323–350. doi: 10.7882/AZ.1999.036
Mallin, M. A., Williams, K. E., Esham, E. C., and Lowe, R. P. (2000). Effect of human development on bacteriological water quality in coastal watersheds. Ecol. Appl. 10, 1047–1056. doi: 10.1890/1051-07612000010
Marcus, A. D., Higgins, D. P., and Gray, R. (2015). Health assessment of free-ranging endangered Australian sea lion (Neophoca cinerea) pups: Effect of haematophagous parasites on haematological parameters. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 184, 132–143. doi: 10.1016/j.cbpa.2015.02.017
McIntosh, R. R., Kirkman, S. P., Thalmann, S., Sutherland, D. R., Mitchell, A., Arnould, J. P. Y., et al. (2018). Understanding meta-population trends of the Australian fur seal, with insights for adaptive monitoring. PLoS One 13:e0200253. doi: 10.1371/journal.pone.0200253
Melendez, D., Roberts, M. C., Greninger, A. L., Weissman, S., No, D., Rabinowitz, P., et al. (2019). Whole-genome analysis of extraintestinal pathogenic Escherichia coli (ExPEC) MDR ST73 and ST127 isolated from endangered southern resident killer whales (Orcinus orca). J. Antimicrob. Chemother. 74:2176. doi: 10.1093/jac/dkz159
Mora, A., García-Peña, F., Alonso, M., Pedraza-Diaz, S., Ortega-Mora, L., Garcia-Parraga, D., et al. (2018). Impact of human-associated Escherichia coli clonal groups in Antarctic pinnipeds: presence of ST73, ST95, ST141 and ST131. Sci. Rep. 8:4678. doi: 10.1038/s41598-018-22943-0
Mora, A., López, C., Dabhi, G., Blanco, M., Blanco, J. E., Alonso, M. P., et al. (2009). Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 9:132. doi: 10.1186/1471-2180-9-132
Nelson, T. M., Rogers, T. L., and Brown, M. V. (2013a). The gut bacterial community of mammals from marine and terrestrial habitats. PLoS One 8:e83655. doi: 10.1371/journal.pone.0083655
Nelson, T. M., Rogers, T. L., Carlini, A. R., and Brown, M. V. (2013b). Diet and phylogeny shape the gut microbiota of Antarctic seals: a comparison of wild and captive animals. Environ. Microbiol. 15, 1132–1145. doi: 10.1111/1462-2920.12022
Nowrouzian, F. L., Adlerberth, I., and Wold, A. E. (2006). Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 8, 834–840. doi: 10.1016/j.micinf.2005.10.011
Oates, S. C., Miller, M. A., Byrne, B. A., Chouicha, N., Hardin, D., Jessup, D., et al. (2012). Epidemiology and potential land-sea transfer of enteric bacteria from terrestrial to marine species in the Monterey Bay region of California. J. Wildl. Dis. 48, 654–668. doi: 10.7589/0090-3558-48.3.654
Oliveira, M., Freire, D., and Pedroso, N. M. (2018). Escherichia coli is not a suitable fecal indicator to assess water fecal contamination by otters. Braz. J. Biol. 78, 155–159. doi: 10.1590/1519-6984.167279
Pandey, P. K., Kass, P. H., Soupir, M. L., Biswas, S., and Singh, V. P. (2014). Contamination of water resources by pathogenic bacteria. AMB Express 4:51. doi: 10.1186/s13568-014-0051-x
Partelow, S., von Wehrden, H., and Horn, O. (2015). Pollution exposure on marine protected areas: A global assessment. Mar. Pollut. Bull. 100, 352–358. doi: 10.1016/j.marpolbul.2015.08.026
Pesapane, R., Ponder, M., and Alexander, K. A. (2013). Tracking pathogen transmission at the human–wildlife interface: banded mongoose and Escherichia coli. Ecohealth 10, 115–128. doi: 10.1007/s10393-013-0838-2
Power, M. L., Samuel, A., Smith, J. J., Stark, J. S., Gillings, M. R., and Gordon, D. M. (2016). Escherichia coli out in the cold: dissemination of human-derived bacteria into the Antarctic microbiome. Environ. Pollut. 215, 58–65. doi: 10.1016/j.envpol.2016.04.013
Riley, L. W. (2014). Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect 20, 380–390. doi: 10.1111/1469-0691.12646
Robinson, J., New, A. L., Popova, E. E., Srokosz, M. A., and Yool, A. (2017). Far-field connectivity of the UK’s four largest marine protected areas: Four of a kind? Earth’s Futur. 5, 475–494. doi: 10.1002/2016EF000516
Rozen, Y., and Belkin, S. (2001). Survival of enteric bacteria in seawater. FEMS Microbiol. Rev. 25, 513–529. doi: 10.1111/j.1574-6976.2001.tb00589.x
Russo, T. A., and Johnson, J. R. (2000). Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181, 1753–1754. doi: 10.1086/315418
Rwego, I. B., Gillespie, T. R., Isabirye-Basuta, G., and Goldberg, T. L. (2008a). High rates of Escherichia coli transmission between livestock and humans in rural Uganda. J. Clin. Microbiol. 46, 3187–3191. doi: 10.1128/JCM.00285-08
Rwego, I. B., Isabirye-Basuta, G., Gillespie, T. R., and Goldberg, T. L. (2008b). Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park. Uganda. Conserv. Biol. 22, 1600–1607. doi: 10.1111/j.1523-1739.2008.01018.x
Schaefer, A. M., Bossart, G. D., Mazzoil, M., Fair, P. A., and Reif, J. S. (2011). Risk factors for colonization of E. coli in atlantic bottlenose dolphins (Tursiops truncatus) in the Indian River Lagoon. Florida. J. Environ. Public Health 2011:597073. doi: 10.1155/2011/597073
Schierack, P., Romer, A., Jores, J., Kaspar, H., Guenther, S., Filter, M., et al. (2009). Isolation and characterization of intestinal Escherichia coli clones from wild boars in Germany. Appl. Environ. Microbiol. 75:695. doi: 10.1128/AEM.01650-08
Schierack, P., Walk, N., Ewers, C., Wilking, H., Steinrück, H., Filter, M., et al. (2008). ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E.coli in a small piglet group. Environ. Microbiol. 10, 1742–1751. doi: 10.1111/j.1462-2920.2008.01595.x
Shapiro, K., Miller, M., and Mazet, J. (2012). Temporal association between land-based runoff events and California sea otter (Enhydra lutris nereis) protozoal mortalities. J. Wildl. Dis. 48, 393–404. doi: 10.7589/0090-3558-48.2.394
Shaughnessy, P. D., Goldsworthy, S. D., Hamer, D. J., Page, B., and McIntosh, R. R. (2011). Australian sea lions Neophoca cinerea at colonies in South Australia: Distribution and abundance, 2004 to 2008. Endanger. Species Res. 13, 87–98. doi: 10.3354/esr00317
Shaughnessy, P. D., Goldsworthy, S. D., and Mackay, A. I. (2015). The long-nosed fur seal (Arctocephalus forsteri) in South Australia in 2013–14: abundance, status and trends. Aust. J. Zool. 63:101. doi: 10.1071/ZO14103
Stoddard, R. A., Atwill, E. R., Conrad, P. A., Byrne, B. A., Jang, S., Lawrence, J., et al. (2009). The effect of rehabilitation of northern elephant seals (Mirounga angustirostris) on antimicrobial resistance of commensal Escherichia coli. Vet. Microbiol. 133, 264–271. doi: 10.1016/j.vetmic.2008.07.022
Stoddard, R. A., Atwill, E. R., Gulland, F. M. D., Miller, M. A., Dabritz, H. A., Paradies, D. M., et al. (2008). Risk factors for infection with pathogenic and antimicrobial-resistant fecal bacteria in northern elephant seals in California. Public Health Rep. 123, 360–370. doi: 10.1177/003335490812300316
Stoddard, R. A., Gulland, M. D. F., Atwill, E. R., Lawrence, J., Jang, S., and Conrad, P. A. (2005). Salmonella and Campylobacter spp. in northern elephant seals. California. Emerg. Infect. Dis. 11, 1967–1969. doi: 10.3201/eid1112.050752
Keywords: Escherichia coli, pinnipeds, wildlife, anthropogenic pollution, human-associated bacteria
Citation: Fulham M, Power M and Gray R (2020) Diversity and Distribution of Escherichia coli in Three Species of Free-Ranging Australian Pinniped Pups. Front. Mar. Sci. 7:571171. doi: 10.3389/fmars.2020.571171
Received: 10 June 2020; Accepted: 19 August 2020;
Published: 09 September 2020.
Edited by:
Michele Thums, Australian Institute of Marine Science (AIMS), AustraliaReviewed by:
Peter Douglas Shaughnessy, South Australian Museum, AustraliaTiffanie Maree Nelson, Deakin University, Australia
Andrew Damon Lowther, Norwegian Polar Institute, Norway
Copyright © 2020 Fulham, Power and Gray. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachael Gray, cmFjaGFlbC5ncmF5QHN5ZG5leS5lZHUuYXU=
 Mariel Fulham
Mariel Fulham Michelle Power
Michelle Power Rachael Gray
Rachael Gray