
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci., 29 September 2020
Sec. Marine Fisheries, Aquaculture and Living Resources
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.561402
This article is part of the Research TopicFish Nutrition, Metabolism and PhysiologyView all 20 articles
This study was designed to evaluate the effect of dietary lipid levels on the mRNA transcripts of lipid metabolic regulatory genes. Nine combinations of experimental diets containing graded lipid (80, 100, and 120 g/kg) and protein (450, 500, and 550 g/kg) levels were fed to the Clarias magur (Indian walking catfish) larvae from 14 to 35 dph (day post hatching). All the lipolytic genes, such as pancreatic triacylglycerol lipase (PL), lipoprotein lipase (LPL), and bile salt-activated lipase (BAL), and genes for long-chain polyunsaturated fatty acid (LC-PUFA) biosynthetic enzymes like fatty acyl desaturase-2 (FADS2), fatty acyl desaturase-5 (FADS5), and elongase (ELOV) were expressed in a wide range of tissues. A high abundance of mRNA transcript levels of lipolytic genes was detected in the intestine and liver, and similarly, desaturases and elongase were predominantly found to be expressed in the liver, brain, and intestine. Among the diets, a significantly high expression of both lipolytic and LC-PUFA biosynthetic genes were observed at 8% dietary lipid level. The mRNA expression of all the studied genes was down-regulated at 12% dietary lipid contents. Hence, the present study concludes that the efficient nutrient utilization and the lipid metabolic pathway occur at the optimum dietary lipid level of 8% in C. magur larvae.
Dietary lipids are considered to be the important source of energy and the essential fatty acids for the fish among all the nutrients besides acting as the carriers of fat soluble vitamins (Liu et al., 2018). Dietary lipids also have the property of protein sparing action where the optimum levels of lipid inclusion in the diets results in the proficient utilization of dietary protein for improved growth performance (Cho and Kaushik, 1990). The high dietary lipid level might lead to lipid accumulation in the body which should not mislead to a pathological condition but could be a strategy in the natural environment of fish in order to survive during the periods of starvation (Kjaer et al., 2009; Han et al., 2011). However, in cultured conditions where fishes are fed continuously, fat accumulation in the body could be a problem and may affect the feed intake and the lipid metabolic enzymes of the fish (Martino et al., 2002; Du et al., 2008).
A plethora of reports revealed the impact of dietary lipid levels on the lipid metabolic enzymes of fish (Alvarez et al., 2000; Wang et al., 2005; Zheng et al., 2010). The high dietary lipid beyond the optimal level leads to negative impact on the metabolic enzymes like lipoprotein lipase (LPL), pancreatic lipase (PL), and hepatic lipase (HL) during the early larval development of Darkbarbel catfish (Zheng et al., 2010). Li et al. (2016) also concluded that dietary lipid level had a great impact on the lipid metabolism related genes in orange-spotted grouper.
Several lipases belonging to the same family are responsible for the lipolytic breakdown of dietary lipids. LPL, PL, and BAL have a significant level of structural resemblance and perform distinct functions in lipid metabolism (Wong and Schotz, 2002). The LPL plays a crucial role in the metabolism of plasma lipoproteins, and its expression is regulated by the physiological, nutritional, and developmental state of animals in a tissue-specific manner (Tavangar et al., 1992; Wang et al., 2016). On the other hand, PL and BAL are also important lipases secreted from pancreas which are also essential for the metabolic breakdown of dietary lipids. PL is substrate specific and leads to the breakdown of triacylglycerol (TAG) into diacylglycerol (DAG), monoacylglycerol (MAG), and eventually the free fatty acids. Whereas, BAL catalyzes the lysis of carboxyl ester bonds of phospholipids, fat-soluble vitamin esters, and cholesterol esters in addition to triglycerides and has been revealed to be the most important digestive lipase in teleosts (Gjellesvik et al., 1992; Wang and Hartsuck, 1993; González-Félix et al., 2018).
Freshwater fishes have the ability to desaturate and elongate linolenic (18:3n-3) and linolenic acid (18:2n-6) to produce the n-3 and n-6 LC-PUFA, respectively with the help of desaturase and elongase enzymes (Leonard et al., 2004; Zheng et al., 2004; Bell and Tocher, 2009). The biosynthesis of LC-PUFA is initiated by the insertion of double bonds at selected positions, which is catalyzed by the fatty acid desaturases (FADS), more specifically FADS6 and FADS5 (Sprecher, 1981). The desaturase FADS-6 enzyme is encoded by the gene fatty acid desaturase-2 (FADS2) and is mainly responsible for the rate-limiting step of fatty acid biosynthesis of arachidonic acid (Nakamura and Nara, 2004; Gol et al., 2018). The biosynthesis of highly unsaturated fatty acids like EPA (eicosapentaenoic acid) requires FADS6 desaturase-mediated desaturation of 18:3n-3 to produce 18:4n-3 and elongation catalyzed by elongase followed by a desaturation by FADS5 (Cook, 1996). However, the synthesis of DHA (docosahexaenoic acid) requires two additional steps viz. desaturation catalyzed by Δ6 desaturase and a peroxisomal chain shortening step (Sprecher et al., 1995).
Clarias magur is an important food fish of aquacultural importance with good growth potential and excellent nutritional profile due to its low fat (1%), high protein (15%), and high iron (710 mg/100 g) contents (Hossain et al., 2006; Argungu et al., 2013). The aim of the present study was to investigate the effect of dietary lipid levels on the lipid breakdown and fatty acid synthetic pathways at the molecular level and to determine the optimal amount of lipids to be incorporated in the diets of C. magur larvae.
A dietary strategy was designed containing graded levels of lipids with constant protein contents among all the treatments to elucidate their effect on the mRNA expression of lipid metabolism-related genes of C. magur from 14 to 35 dph (days post hatching). During the rearing period, different water quality parameters like dissolved oxygen, temperature, and pH were observed to be in the optimum ranges of 6–8 mg L–1, 28–30°C and 7.8–8.2, respectively. The experiment conducted from the 14 dph stage was based on the inference from our previous study (Mir et al., 2019), where the C. magur was found to efficiently utilize dietary lipids from 13 dph onward. Nine experimental diets were prepared with combination of three lipids (8, 10, and 12%) and three protein levels (55, 50, and 45%). The protein contents were decreased from 55 to 45% in all treatments (T1, T2, and T3), since the requirement of dietary protein reduces as the fish grows with age. The experimental design and the dietary combinations followed are mentioned in Table 1. The feeding trial was carried to evaluate the effect of the experimental diets on the relative mRNA expression of lipid metabolism-related genes viz. LPL, PL, BAL, fatty acyl desaturase-2 (FADS2), fatty acyl desaturase-5 (FADS5), and fatty acyl elongase (ELOV).
The experimental diets were prepared by using several commercially available feed ingredients (Table 2). Soya protein hydrolysate (SPH) and fish protein hydrolysate (FPH) were procured from New Alliance Dye Chem Pvt. Ltd. (Mumbai, India) and Jeevan chemical Pvt. Ltd. (Mumbai, India), respectively. Nine experimental diets of variable lipid and protein contents were formulated and prepared in the Fish Nutrition and Feed Technology Laboratory of ICAR-Central Institute of Fisheries Education. The diets were composed of 45% crude protein (CP) and 8% lipid level (CL)-F1; 50% CP 8% CL (F2); 55% CP 8% CL (F3); 45% CP 10% CL (F4); 50% CP 10% CL (F5); 55% CP 10% CL (F6); 45% CP 12% CL (F7); 50% CP 12% CL (F8); and 55% CP with 12% CL (F9) as displayed in Table 2.
All the selected ingredients were accurately weighed and thoroughly mixed with adequate addition of water, except soya lecithin, vitamin C, oil, and vitamin-mineral mix. The dough of the mixture was steam-cooked for 15 min in autoclave and then cooled to room temperature. Additives like butylated hydroxyl toluene (BHT) dissolved in oil, premix of vitamin-mineral mix, and vitamin C along with betaine and choline chloride were mixed thoroughly to the dough for homogenous distribution of the respective additives. The dough was then formed into pellets by a pelletizer with 1 mm diameter die size and later transferred to a spheronizer. The moist pellets were air-dried for some time and then kept in a hot air oven at 40°C until the moisture level reduces to 10–12%. The dried pellets were crumbled to small size particulate feeds and were packed in airtight zipper bags to be used for the feeding trial.
The collection of samples was done randomly in the morning before the feeding of fishes. Gene expression analysis was carried out by sampling six different pools of larvae from the three experimental larval rearing tanks each stocked with 500 larvae. The samples were taken at 21 dph (35 larvae numbers), 28 dph (30 larvae), and 35 dph (19 larvae) in all the three treatments for analysis after being euthanized with ice-cold water. Moreover, the tissues such as liver, brain, stomach, intestine, muscle, and gill were collected from adult fishes in triplicates for basal distribution of the selected genes.
Total RNA was extracted from the whole larval samples taken at the abovementioned time points of the study, using TrizolTM reagent (Invitrogen, United States) as per manufacturer’s instructions. The genomic DNA was removed from the extracted RNA using DNase I (Thermo Scientific, United States) treatment following our previously published study (Mir et al., 2018b). The concentration of RNA as well as the purity was determined by NanoDrop spectrophotometer (Thermo Scientific, United States) at an optical density of 260/280. The cDNA were synthesized from DNAse-treated RNA using the RevertAidTM First Strand cDNA Synthesis kit (Thermo Scientific, United States) with Oligo(dT) primer as per the protocol of manufacturer.
The fatty acyl desaturases selected in the present study were first cloned as no such earlier sequence of these genes was available from the GenBank. Primers from the sequences of closely related fish species were designed for FADS2 and FADS5 genes (Table 3), using Gene Runner software (v. 4.0.9.62), for sequencing in C. magur. Amplification reaction was carried out by using a 20-μL reaction volume under the conditions as follows: initial denaturation or pre-heating at 94°C for 4 min, followed by 35 cycles at 94°C for 30 s, annealing at 57°C (FADS2) and 59°C (FADS5) for 30 s, 72°C for 40 s and a post extension of 10 min at 72°C by using 96-well PCR System (Takara, Japan). The amplified PCR products with expected band sizes were excised from gel, cloned into the pTZ57R/T vector (Thermo Scientific, United States), then sequenced and confirmed by NCBI BLAST analysis.
The sequence similarity of the FADS2 and FADS5 putative proteins was evaluated by using the BLASTP algorithm of the NCBI. The phylogenetic analyses of both the sequences were done with sequences of other fishes and mammals deposited in NCBI GenBank, using the MEGA 7.0 software.
Primers for qPCR analysis were listed in Table 4 and were designed from the already submitted sequences to the GenBank. The PCR efficiency (E) of primers was determined by using the formula: E (%) = (10–1/slope−1) × 100. The primers with efficiency in the ranges of 95–105% were selected for quantitative gene expression analysis (Kubista et al., 2006). The mRNA transcript levels of respective genes were carried out in a LightCycler® Real-time PCR detection system (Roche, United States). Beta actin (β-actin) was selected as internal control for the expression analysis of lipid metabolism related genes as per our previous report (Mir et al., 2018a).

Table 4. Primers used for the qRT-PCR analysis of lipolytic and long-chain fatty acid biosynthesis related genes.
The reaction volume (25 μL) comprising of 12.5 μL of MaximaTM SYBR Green qPCR master mix (Thermo Scientific, United States), 1 μL (10 pM) of each forward and reverse primer, 1 μL (100 ng) of cDNA, and 9.5 μL nuclease-free water was used for the amplification in qPCR. Each sample was divided into two wells each with 10-μL volume. The real-time PCR program followed was pre-heating of 10 min at 95°C, followed by denaturation of 40 cycles at 95°C for 20 s, annealing at 58°C (PL gene), 59°C (LPL gene), 59°C (BAL gene), 60°C (FADS2 gene), 57°C (FADS5 gene) and 59°C (ELOV gene) for 30 s and final extension of 30 s at 72°C. The qPCR analysis was done by following the 2–ΔCt method (Livak and Schmittgen, 2001).
One and two-way ANOVA was used for the analysis of data at P < 0.05 significance level. Shapiro-Wilk’s test was used for evaluating the normality, and equality of variances was determined by Levene’s test before carrying out ANOVA. SPSS 22.0 software was used to conduct all the statistical analysis (SPSS Inc., United States).
The growth of the larvae on average wet weight basis was assessed in all the treatments and was observed to be significantly high at 8% dietary lipid level as shown in our previous report (Mir et al., 2020).
Nucleotide sequences of 612 and 507 bp were acquired from C. magur for FADS2 and FADS5 genes, respectively (Figure 1). The size of the putative proteins were 204 (FADS2) and 169 (FADS5) amino acid residues. The respective sequences were later submitted to the GenBank database under the accession numbers MG654483.1 (FADS2) and MG654484.1 (FADS5).
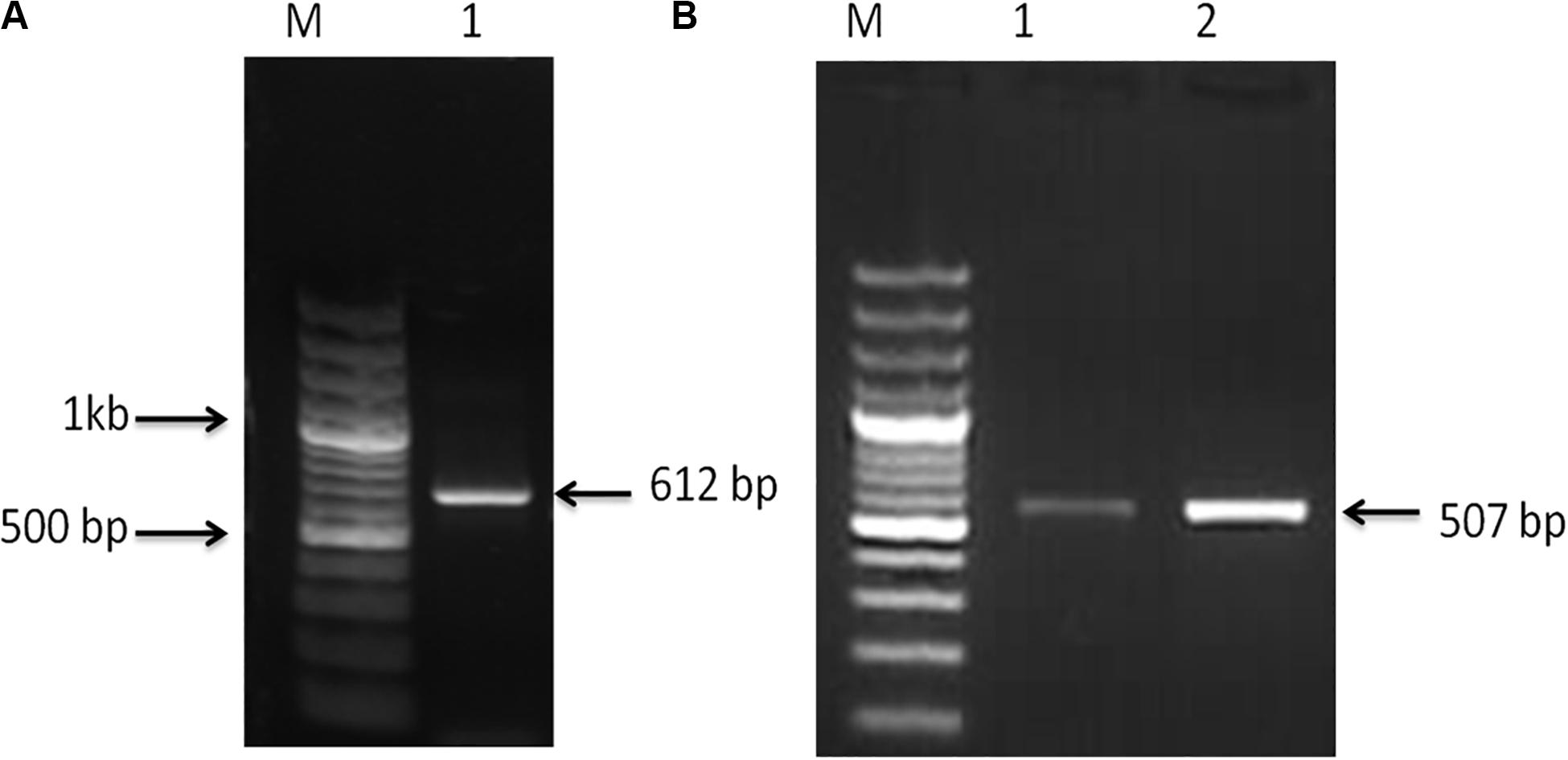
Figure 1. (A) PCR amplified product, Lane M; 100 bp plus ladder (Fermentas, United States) and Lane 1 fatty acid desaturase2 (FADS2) amplified product (612 bp). (B) PCR amplified product, Lane M; 100 bp plus ladder (Fermentas, United States) and, Lanes 1 and 2; fatty acid desaturase5 (FADS5) amplified product (507 bp).
The protein sequence of C. magur FADS2 showed 65–95% similarity with other fish species and around 57–62% with mammals using protein BLAST of NCBI. On the other hand, FADS5 putative protein of C. magur displayed 67–89% similarity with other fish and 64–75% with mammalian species.
Phylogenetic neighbor joining tree analysis of C. magur FADS2 showed more similarity with other catfish like Thai magur, pangas, and marine catfishes and, hence, were clustered in one clade (Figure 2). Similarly, the FADS5 putative protein exhibited higher similarity with other marine catfish (Figure 3).
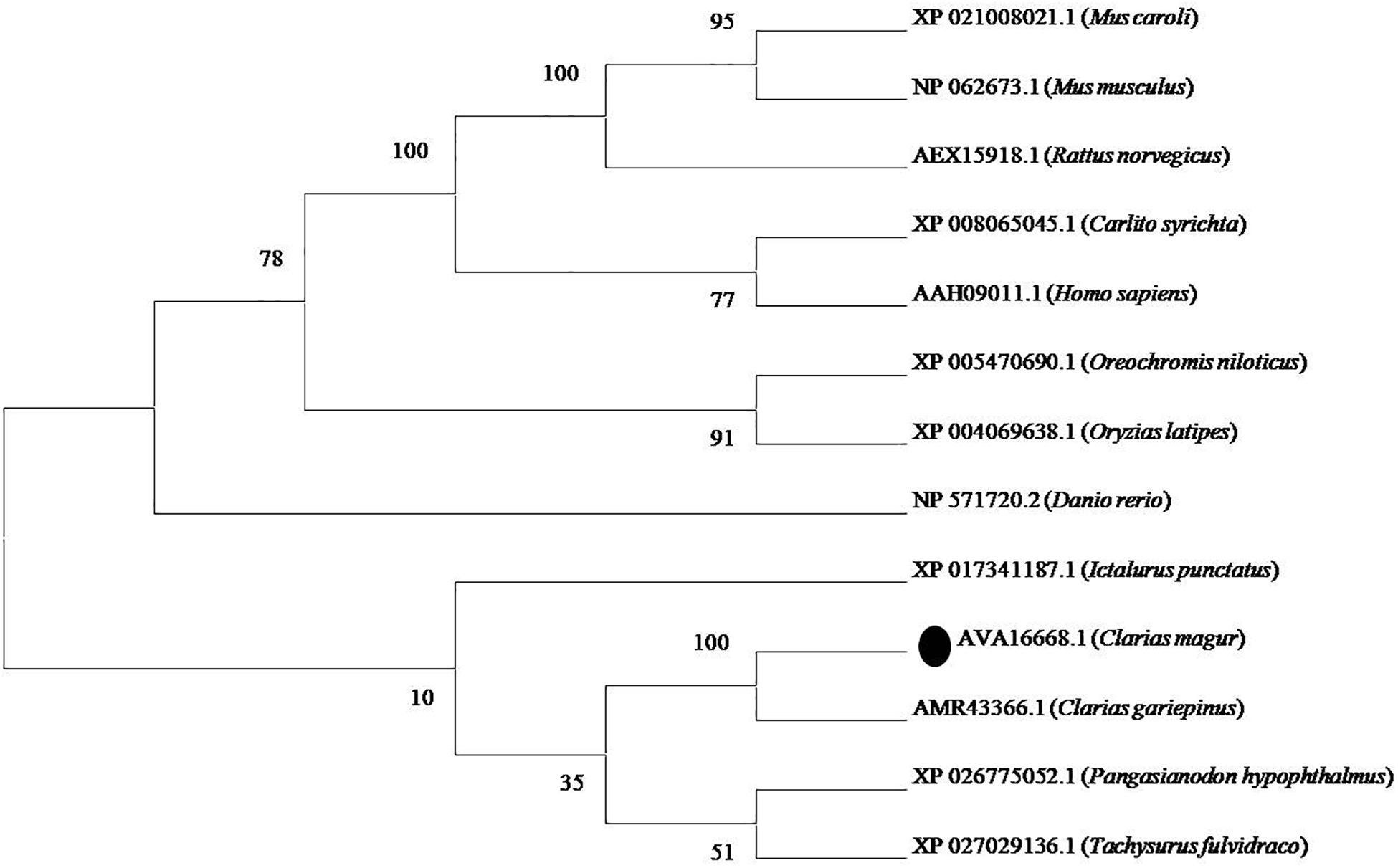
Figure 2. Phylogenetic analysis of C. magur FADS2 putative protein was done using MEGA 7.0 software. Phylogenetic relationship with other piscine and selected mammalian FADS2 proteins. The evolutionary tree was formed using the neighbor-joining method and a bootstrap analysis with 1000 replicates was used to assess the strength of nodes in the tree. Accession numbers are given in the brackets.
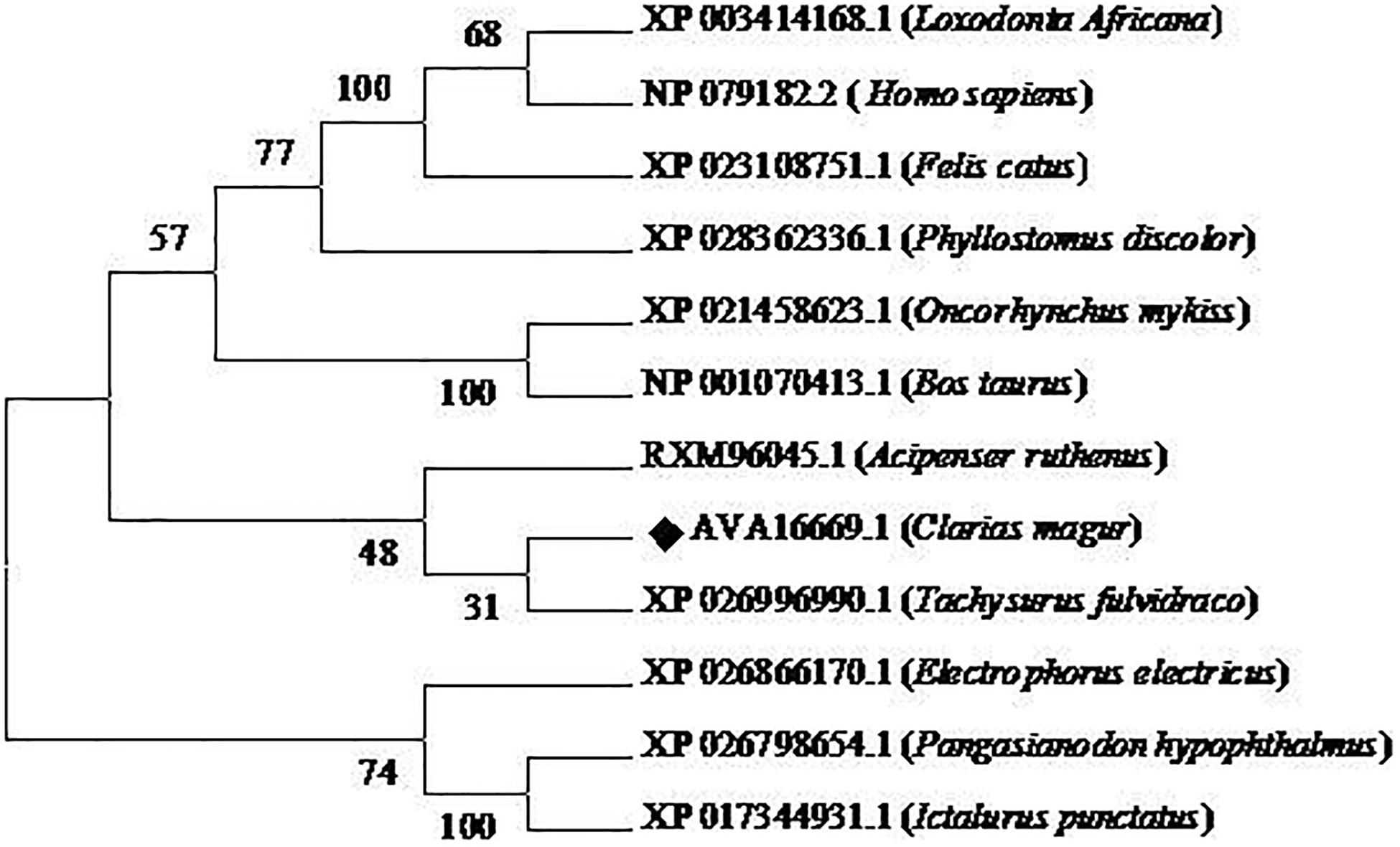
Figure 3. Phylogenetic analysis of C. magur FADS5 putative protein was done using MEGA 7.0 software. Phylogenetic relationship with other piscine and selected mammalian FADS5 proteins. The evolutionary tree was formed using the neighbor-joining method and a bootstrap analysis with 1000 replicates was used to assess the strength of nodes in the tree. Accession numbers are given in the brackets.
Quantitative real-time PCR analysis showed that all the selected genes related to lipolysis were broadly distributed and their mRNA expression varied among the tissues (Figure 4). The significantly high level of LPL mRNA expression was observed in intestine and liver followed by muscle, and least transcript level was found in gill and stomach. PL gene showed higher expression in intestine followed by liver, and significantly low levels were observed in brain, stomach, muscle, and gill tissues. In addition, BAL transcript levels were significantly high in intestine followed by liver and the rest of studied tissues.
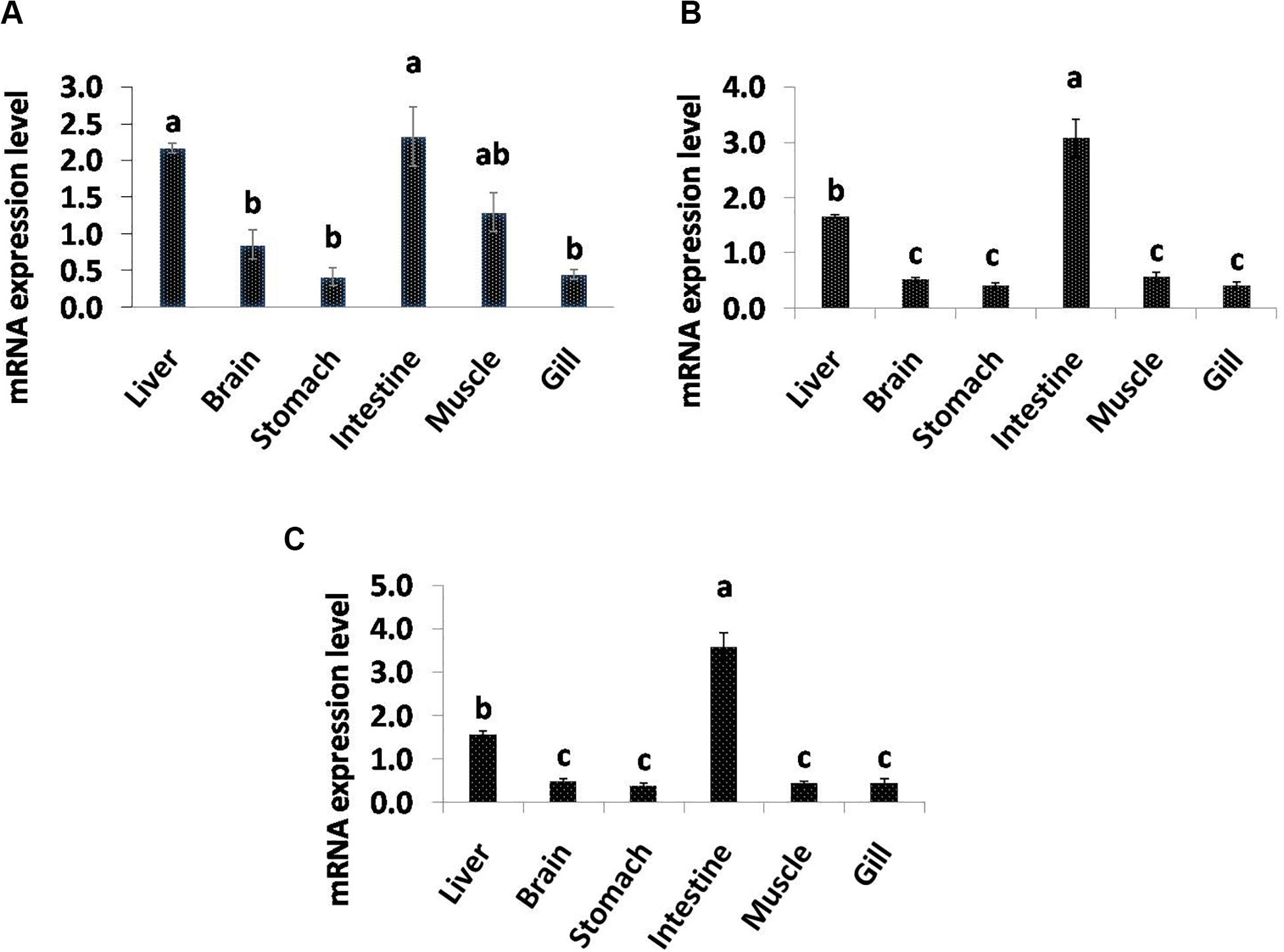
Figure 4. Quantitative analysis of the relative mRNA expression levels of LPL (A), PL (B) and BAL (C) in different tissues of C. magur. Data (mean ± SEM, n = 3) were expressed relative to expression of β-actin gene. Groups with different letters were significantly (P < 0.05) different.
In the present study, qPCR analysis of the desaturase and elongase genes displayed a variable pattern of expression among different tissues (Figure 5). C. magur FADS2 exhibited a significantly high level of mRNA expression in liver followed by brain, intestine, stomach, muscle, and gill. Furthermore, FADS5 transcript level was found to be highest in liver followed by brain and intestine among the other tissues. However, the expression of ELOV gene was high in the brain, liver, and intestine with no significant difference, while a least transcript level was observed in the rest of the tissues studied (stomach, muscle, and gill).
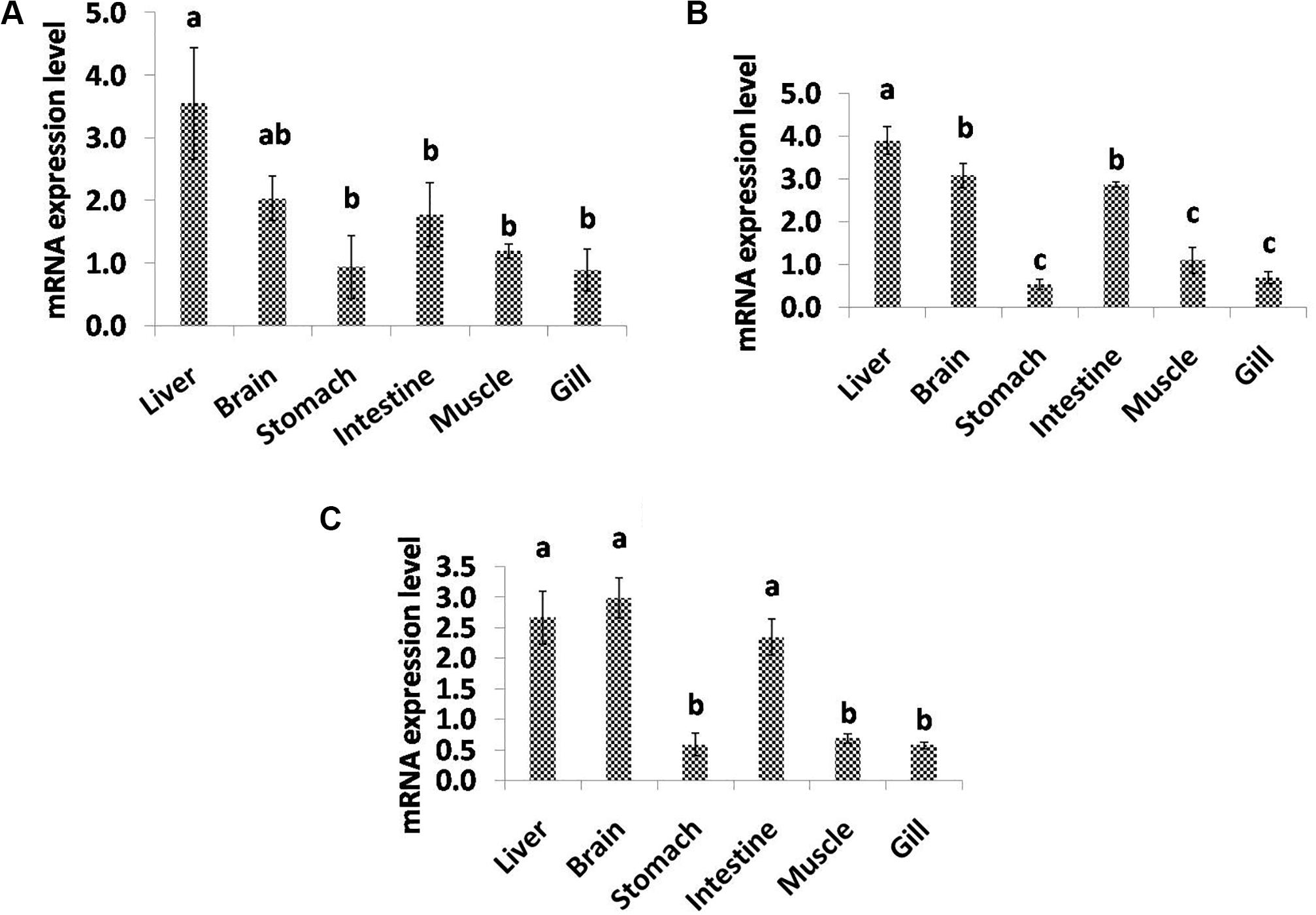
Figure 5. Quantitative analysis of the relative mRNA expression levels of FADS2 (A), FADS5 (B), and ELOV (C) in different tissues of C. magur. Data (mean ± SEM, n = 3) were expressed relative to expression of β-actin gene. Groups with different letters were significantly (P < 0.05) different.
The mRNA expression of lipolytic genes in response to graded dietary lipid levels was assessed in whole larval samples of C. magur at different time points as displayed in Table 5. Significant difference was observed among the relative expression of lipolytic genes between all the combinations of lipid and age groups within the treatments. The level of LPL mRNA expression decreased significantly (P < 0.05) from 21 to 35 dph in the treatments from T1 to T3. PL gene did not exhibit a significant (P > 0.05) difference in the mRNA expression during 21 and 28 dph in T1 and T2 treatments. In T1, the relative expression further declined in 35 dph, whereas in T2, it increased significantly during 35 dph among all the other groups. However, a significantly (P < 0.05) low expression level was observed in T3 group at all the time points. Similarly, the mRNA expression of BAL gene was found to be significantly high during the 21 dph of T1 treatment among all the other groups. Although there was no significant difference between T1 and T2 but, the expression level declined from 21 to 35 dph in both the treatments. Like the other two lipolytic genes, BAL gene showed a low expression level in the treatment T3 (12% lipid level).
The individual effect of lipid and age revealed that 10% lipid level exhibits a significantly high level of LPL expression compared to the other two levels and 21 and 28 dph time points did not show any significant difference and was higher than 35 dph. Lipid has a significant influence on the mRNA expression pattern of PL and BAL. There was no significant difference in the level of PL gene expression in 8 and 10% lipid levels and was observed to be higher than 12% lipid level. Age of the larvae did not display any significant effect on the mRNA expression pattern of PL gene. The BAL gene expression was found to be significantly high at 8% lipid level followed by 10 and 12%. Like the PL gene, the age has no significant effect on the expression of BAL gene. The insignificant difference due to age might be also due to level of protein as the particular age represents a specific dietary protein level as mentioned in the experimental design. The lipolytic genes studied in the present study did not exhibit any interaction effect between lipid and age.
The regulation of expression of different genes related to fatty acid synthesis in response to the feeding graded levels of dietary lipid during different time points was evaluated and is shown in Table 6. The expression of all the genes vary in the three treatments with significantly low expression level in T3 compared to T1 and T2. Dietary lipid showed a significant effect on the mRNA transcript level of FADS2, FADS5, and ELOV with high expression at 8% lipid content than 10 and 12% levels. So far as effect of age is concerned, the mRNA expression of FADS2 gene exhibited a significant increasing trend from 21 to 35 dph. However, there was a significantly higher expression of FADS5 at 35 dph and with no significant difference at 21 and 28 dph age groups.
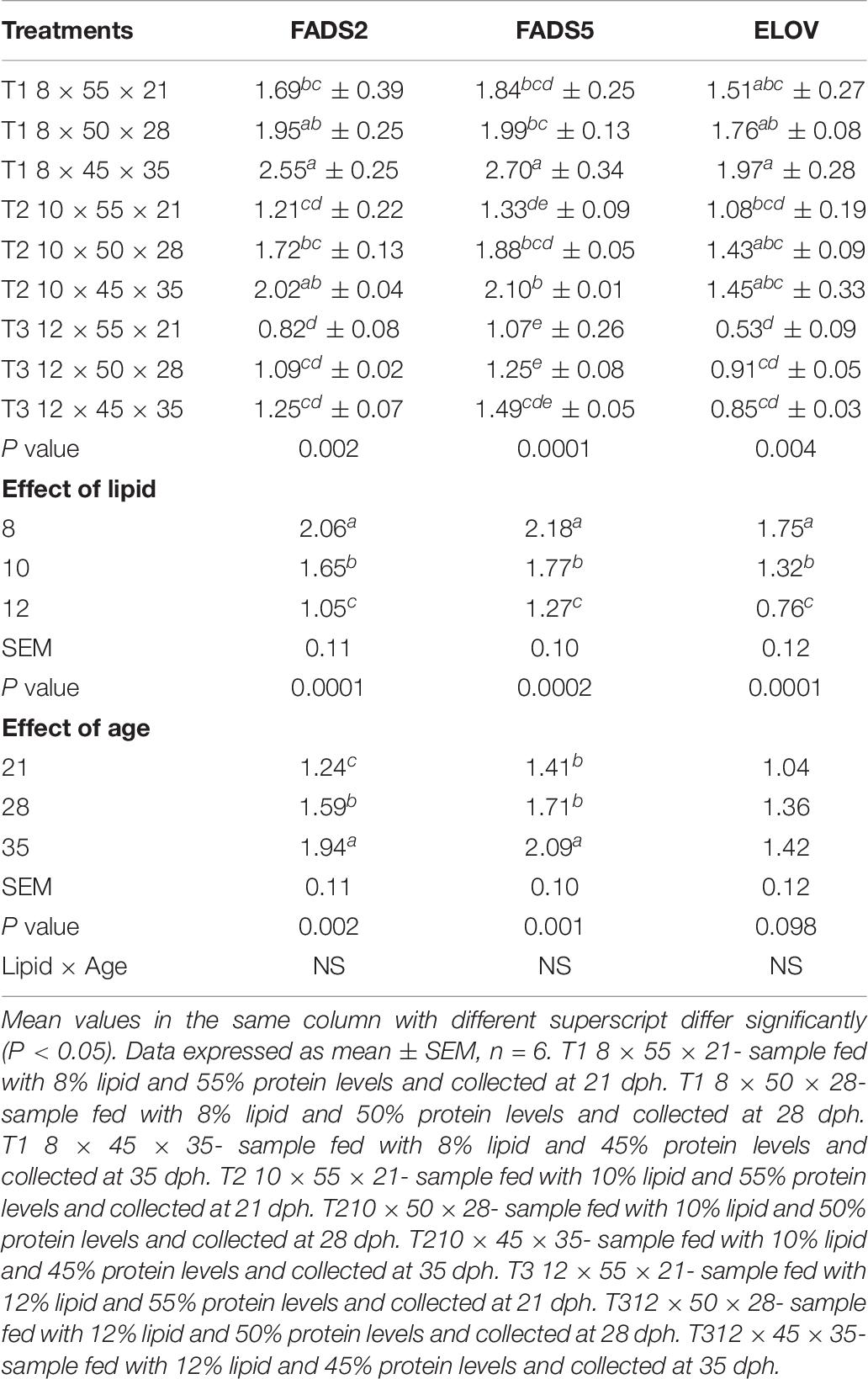
Table 6. Effects of dietary lipid levels and age on the expression of fatty acid synthesis related genes of C. magur.
A variable level of mRNA expression patterns of ELOV gene were observed in different dietary treatments across the age groups. The mRNA transcript level was significantly (P < 0.05) high at 8% dietary lipid level, and it was declined as the content of lipid was increased in the experimental diets. Unlike the desaturases studied in the present study, the expression of ELOV was not affected by age, and there was no significant difference.
Lipid nutrition in fish has attracted the researcher’s interest in particular with the diversity in requirements and lipid metabolism, such as the mechanism and pathways of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis among different species (Bell and Tocher, 2009). Fatty acid desaturases and elongases form the important enzymes for biosynthesis of LC-PUFA especially EPA, DHA, and AA. Among the desaturases, Δ5 and Δ6 fatty acid desaturases form key enzymes which lack in marine fish along with the elongases (Sargent et al., 2002; Tocher, 2003). However, delta-6-desaturase enzyme is encoded by the fatty acid desaturase-2 (FADS2) gene, which is responsible for the rate-limiting step in the biosynthesis of LC-PUFA (Gol et al., 2018). Delta 5-Desaturase (FADS5) is a fatty acid metabolic enzyme which is responsible for the biosynthesis of EPA and DHA. FADS5 uses eicosatetraenoic acid (ETA, 20:4n-3) as a substrate and leads to the insertion of a double bond for the synthesis of EPA. On the other hand, lipolysis is also an important biochemical pathway responsible for the breakdown of lipids to make them usable for energy and several body functions. Dietary lipids have a great impact on the biosynthetic and lipolytic pathway of fish that varies with the type of species. In the present study, the key desaturases (FADS2 and FADS5) were characterized partially in order to elucidate the pattern of expression in freshwater fish C. magur fed with variable level of lipids. The putative proteins of FADS2 and FADS5 showed more similarity to other catfish species as shown in the phylogenetic neighbor joining tree analysis, which infer that genes are highly conserved among the catfishes.
Tissue distribution studies revealed a variable pattern of expression among the tissues taken in the present study. The lipolytic genes (LPL, PL, and BAL) primarily exhibited a high relative expression in intestine. The high level of expression of lipolytic genes in the intestine infers the maturation of gut tissues responsible for the secretion of respective enzymes. The high expression of lipolytic enzymes in the intestine was also observed in the previous study in rohu and sardine fish (Nayak et al., 2003). In contrast to our study, LPL mRNA expression was significantly found high in liver in earlier reports on rainbow trout and gilthead sea bream (Lindberg and Olivecrona, 2002; Saera-Vila et al., 2005). On the other hand, desaturases and elongase also exhibited a variable pattern of expression among the different tissues. The higher transcript levels of FADS2 and FADS5 were observed in liver followed by brain and intestine among all the tissues, whereas significantly high ELOV mRNA expression was found in brain followed by liver and intestine. Aliyu-Paiko et al. (2013) also reported the highest expression of desaturases and elongase in liver and brain of Channa striata. The study is also corroborated by Zheng et al. (2009), with high transcript level in liver and brain followed by intestine of cobia (Rachycentron canadum). Similarly, a widespread expression pattern of desaturase and elongase was observed in the liver and brain of rainbow trout, Oncorhynchus mykiss (Seiliez et al., 2001). This indicates the liver has the central role in lipid metabolism, where fatty acids are being synthesized and modified. The higher expression of desaturases and elongase in the brain and liver depicted the importance of LC-PUFA particularly DHA in neural tissue development as suggested by Sargent et al. (2002).
Dietary lipid levels significantly influence the mRNA expression of lipolytic genes. It was observed that 8 and 10% lipid level exhibited high the expression level of LPL, PL, and BAL genes compared to 12% lipid level. However, there was no significant difference between 8 and 10% in the mRNA abundance of LPL and PL, whereas, 8 and 10% lipid-fed groups varied in the case of BAL expression. This suggests that there is increased utilization of lipids at 8 and 10% dietary lipid levels during larval development. Further, it revealed that BAL was more responsive to dietary differences in lipid than LPL and PL. It was reported that LPL mRNA expression level increased first and reached a plateau with increasing dietary lipid level from 5.8 to 19.9% in darkbarbel catfish larvae (Zheng et al., 2010), which is in agreement with our study. Li et al. (2016) also found that the LPL mRNA level of orange-spotted grouper larvae increased first from 5.94 to 13.3% then remained constant and later declined at 21.87% dietary lipid level. However, the mRNA expression of LPL in different tissues like adipose tissue, liver, and muscle of Megalobrama amblycephala was found to be high at 12% dietary lipid levels (Li et al., 2013). This may be due to the different optimum dietary requirement of lipids which varies from species to species. Similarly, PL mRNA expression was also found to decrease as the dietary lipid level increased from 13 to 23% in sea bass (Cahu et al., 2003). Zheng et al. (2010) also reported that PL activity decreased as the lipid content of diets was increased in dark barbell catfish larvae. This decrease in activity can be due to the nutritional regulation at transcriptional level. Bile salt-activated lipase gene expression was found to be significantly higher at 8% level and decreased further as the lipid level was increased. It is clearly indicated that dietary lipid level affects the expression of BAL, which is reported to be the common lipase in teleosts (Patton et al., 1977). However, Hoehne-Reitan et al. (2001) stated that the bile salt dependent lipase (BSDL) level of turbot larvae was not significantly influenced by the lipid level of the prey, which is a contrast statement with respect to our results. Péres et al. (1996), on the other hand suggested that the expression of digestive enzymes was affected by diet composition at the transcriptional and translational levels. A decrease in the efficiency or activity of digestive enzymes along with a growth depressing effect due to high dietary lipid levels was reported by Morais et al. (2007).
The temporal variation in expression levels of lipolytic genes was evaluated, and it was found that only LPL displayed significant difference in the relative mRNA levels. The significant decrease in the LPL gene expression can be correlated with the results of our previous study (Mir et al., 2019), where it showed a declining trend after 16 dph. Whether the effect on the lipolytic gene expression was due to age or difference in dietary protein level could not be differentiated as the three time points represent the samples fed with different protein levels. However, Cai et al. (2017) suggested that the mRNA expression levels of key lipases were mainly controlled by age in early stage and by the exogenous feeding in later stages during the ontogenetic development of large yellow croaker larvae.
In the present study, the regulation of biosynthesis of LC-PUFA involving FADS2, FADS5, and ELOV in C. magur fed with graded level of dietary lipid varies significantly. The biosynthetic pathway of LC-PUFA was inhibited as the dietary lipid levels were increased from 8 to 12%, as depicted by the down-regulation of desaturases and elongase genes. The down-regulation could be due to enhanced concentration of LC-PUFA in the treatments with higher dietary lipid levels. The study is supported by several previous reports in freshwater fish and mammals (Christiansen et al., 1991; Izquierdo et al., 2008; Ren et al., 2012; Xu et al., 2014; Nayak et al., 2018). The increasing pattern of FADS expression during the developmental ontogeny of C. magur from 21 to 35 dph suggested the enhanced capacity of LC-PUFA biosynthesis as the larvae grows. Similar increase in the FADS expression during larval development was also reported in Channa striata and zebrafish (Monroig et al., 2009; Tan et al., 2010; Jaya-Ram et al., 2011).
The study suggested that the efficient nutrient utilization and the metabolism of lipids occurs at an optimum dietary lipid level of 8%. Based on our results, feeding C. magur larvae with a diet containing a high lipid level (12%) significantly reduces its efficiency in the breakdown of complex lipids and the biosynthetic pathway of LC-PUFA. Hence, the present study can be used as a model for the optimization of dietary lipids and with the potential to synthesize the long-chain PUFA’s at a suitable lipid level without affecting the growth of the fish. Furthermore, the effect of dietary lipids on fatty acid composition needs to be studied in future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The care and treatment of animals used in this study were in accordance with the guidelines of the CPCSEA [(Committee for the Purpose of Control and Supervision of Experiments on Animals), Ministry of Environment & Forests (Animal Welfare Division), Govt of India] on care and use of animals in scientific research. The study was approved by the Board of Studies and authorities of the Fish Nutrition, Biochemistry and Physiology division of ICAR-Central Institute of Fisheries Education (Deemed University), Mumbai.
IM: methodology set-up, sample collection, analysis, data generation and validation, and manuscript writing. PS: supervision, manuscript review and editing, and scientific suggestions. IB: sample collection and analysis. JD: concept design, data interpretation and presentation, and manuscript writing. DA: manuscript review and editing. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are thankful to Director and Vice-Chancellor, ICAR-Central Institute of Fisheries Education, Mumbai, India, for their kind support and providing necessary facilities for conducting this work.
Aliyu-Paiko, M., Jaya-Ram, A., Chong, A., Hashim, R., and Amuzat, A. O. (2013). Estimation of tissue distribution of mRNA transcripts for desaturase and elongase enzymes in Channa striata (Bloch, 1793) fingerlings using PCR technique. Taiwania 58, 112–118.
Alvarez, M., Diez, A., Lopez-Bote, C., Gallego, M., and Bautista, J. (2000). Short-term modulation of lipogenesis by macronutrients in rainbow trout (Oncorhynchus mykiss) hepatocytes. Br. J. Nutr. 84, 619–628. doi: 10.1017/S0007114500001951
Argungu, L., Christianus, A., Amin, S., Daud, S., Siraj, S., and Aminur Rahman, M. (2013). Asian catfish Clarias batrachus (Linnaeus, 1758) getting critically endangered. Asian J. Anim. Vet. Adv. 8, 168–176. doi: 10.3923/ajava.2013.168.176
Bell, M. V., and Tocher, D. R. (2009). “Biosynthesis of polyunsaturated fatty acids in aquatic ecosystems: general pathways and new directions,” in Lipids in Aquatic Ecosystems, eds M. Kainz, M. Brett, and M. Arts (New York, NY: Springer), 211–236. doi: 10.1007/978-0-387-89366-2_9
Cahu, C. L., Infante, J. L. Z., and Barbosa, V. (2003). Effect of dietary phospholipid level and phospholipid: neutral lipid value on the development of sea bass (Dicentrarchus labrax) larvae fed a compound diet. Br. J. Nutr. 90, 21–28. doi: 10.1079/BJN2003880
Cai, Z., Xie, F., Mai, K., and Ai, Q. (2017). Molecular cloning and genetic ontogeny of some key lipolytic enzymes in large yellow croaker larvae (Larimichthys crocea R.). Aquac. Res. 48, 1183–1193. doi: 10.1111/are.12960
Cho, C., and Kaushik, S. (1990). “Nutritional energetics in fish: energy and protein utilization in rainbow trout (Salmo gairdneri),” in Aspects of Food Production, Consumption and Energy Values, ed. G. H. Bourne (Basel: Karger Publishers), 132–172. doi: 10.1159/000417529
Christiansen, E. N., Lund, J. S., Rørtveit, T., and Rustan, A. C. (1991). Effect of dietary n- 3 and n- 6 fatty acids on fatty acid desaturation in rat liver. Biochim. Biophys. Acta LipidsLipid Metab. 1082, 57–62. doi: 10.1016/0005-2760(91)90299-W
Cook, H. W. (1996). “Fatty acid desaturation and chain elongation in eukaryote,” in Biochemistry of Lipids, Lipoproteins and Membranes, eds D. E. Vance and J. E. Vance (Amsterdam: Elsevier), 129–152. doi: 10.1016/S0167-7306(08)60512-8
Du, Z., Clouet, P., Huang, L., Degrace, P., Zheng, W., He, J., et al. (2008). Utilization of different dietary lipid sources at high level in herbivorous grass carp (Ctenopharyngodon idella): mechanism related to hepatic fatty acid oxidation. Aquac. Nutr. 14, 77–92. doi: 10.1111/j.1365-2095.2007.00507.x
Gjellesvik, D., Lombardo, D., and Walther, B. (1992). Pancreatic bile salt dependent lipase from cod (Gadus morhua): purification and properties. Biochim. Biophys. Acta LipidsLipid Metab. 1124, 123–134. doi: 10.1016/0005-2760(92)90088-D
Gol, S., Pena, R. N., Rothschild, M. F., Tor, M., and Estany, J. (2018). A polymorphism in the fatty acid desaturase-2 gene is associated with the arachidonic acid metabolism in pigs. Sci. Rep. 8, 1–9. doi: 10.1038/s41598-018-32710-w
González-Félix, M. L., Gatlin, D. M., Perez-Velazquez, M., Webb, K., García-Ortega, A., and Hume, M. (2018). Red drumSciaenopsocellatus growth and expression of bile salt-dependent lipase in response to increasing dietary lipid supplementation. Fish Physiol. Biochem. 44, 1319–1331. doi: 10.1007/s10695-018-0523-z
Han, C. Y., Wen, X. B., Zheng, Q. M., and Li, H. B. (2011). Effects of dietary lipid levels on lipid deposition and activities of lipid metabolic enzymes in hybrid tilapia (Oreochromis niloticus× O. aureus). J. Anim. Physiol. Anim. Nutr. 95, 609–615. doi: 10.1111/j.1439-0396.2010.01091.x
Hoehne-Reitan, K., Kjørsvik, E., and Reitan, K. (2001). Bile salt-dependent lipase in larval turbot, as influenced by density and lipid content of fed prey. J. Fish Biol. 58, 746–754. doi: 10.1111/j.1095-8649.2001.tb00526.x
Hossain, Q., Hossain, M. A., and Parween, S. (2006). Artificial breeding and nursery practices of Clarias batrachus (Linnaeus, 1758). Sci. World 4, 32–37.
Izquierdo, M., Robaina, L., Juárez-Carrillo, E., Oliva, V., Hernández-Cruz, C. M., and Afonso, J. M. (2008). Regulation of growth, fatty acid composition and delta 6 desaturase expression by dietary lipids in gilthead seabream larvae (Sparus aurata). Fish Physiol. Biochem. 34, 117–127. doi: 10.1007/s10695-007-9152-7
Jaya-Ram, A., Ishak, S. D., Enyu, Y.-L., Kuah, M.-K., Wong, K.-L., and Shu-Chien, A. C. (2011). Molecular cloning and ontogenic mRNA expression of fatty acid desaturase in the carnivorous striped snakehead fish (Channa striata). Comparat. Biochem. Physiol. Part A Mol. Integrat. Physiol. 158, 415–422. doi: 10.1016/j.cbpa.2010.11.018
Kjaer, M. A., Vegusdal, A., Berge, G. M., Galloway, T. F., Hillestad, M., and Krogdahl, Å (2009). Characterisation of lipid transport in Atlantic cod (Gadus morhua) when fasted and fed high or low fat diets. Aquaculture 288, 325–336. doi: 10.1016/j.aquaculture.2008.12.022
Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., et al. (2006). The real-time polymerase chain reaction. Mol. Aspects Med. 27, 95–125. doi: 10.1016/j.mam.2005.12.007
Leonard, A. E., Pereira, S. L., Sprecher, H., and Huang, Y. (2004). Elongation of long-chain fatty acids. Prog. Lipid Res. 43, 36–54. doi: 10.1016/S0163-7827(03)00040-7
Li, S., Mai, K., Xu, W., Yuan, Y., Zhang, Y., Zhou, H., et al. (2016). Effects of dietary lipid level on growth, fatty acid composition, digestive enzymes and expression of some lipid metabolism related genes of orange-spotted grouper larvae (Epinephelus coioides H.). Aquac. Res. 47, 2481–2495. doi: 10.1111/are.12697
Li, X.-F., Jiang, G.-Z., Qian, Y., Xu, W.-N., and Liu, W.-B. (2013). Molecular characterization of lipoprotein lipase from blunt snout bream Megalobrama amblycephala and the regulation of its activity and expression by dietary lipid levels. Aquaculture 416, 23–32. doi: 10.1016/j.aquaculture.2013.08.020
Lindberg, A., and Olivecrona, G. (2002). Lipoprotein lipase from rainbow trout differs in several respects from the enzyme in mammals. Gene 292, 213–223. doi: 10.1016/S0378-1119(02)00680-7
Liu, K., Liu, H., Chi, S., Dong, X., Yang, Q., and Tan, B. (2018). Effects of different dietary lipid sources on growth performance, body composition and lipid metabolism-related enzymes and genes of juvenile golden pompano, Trachinotus ovatus. Aquac. Res. 49, 717–725. doi: 10.1111/are.13502
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martino, R. C., Cyrino, J. E. P., Portz, L., and Trugo, L. C. (2002). Effect of dietary lipid level on nutritional performance of the surubim, Pseudoplatystoma coruscans. Aquaculture 209, 209–218. doi: 10.1016/S0044-8486(01)00738-4
Mir, I. N., Srivastava, P. P., Bhat, I. A., Muralidhar, A. P., Gireesh-Babu, P., Varghese, T., et al. (2018a). Reference gene selection for quantitative real-time RT-PCR normalization in Clarias magur at different larval developmental stages. Indian J. Anim. Sci. 88, 380–382.
Mir, I. N., Srivastava, P. P., Bhat, I. A., Muralidhar, A. P., Varghese, T., Gireesh-Babu, P., et al. (2018b). Expression and activity of trypsin and pepsin during larval development of Indian walking catfish (Clarias magur). Aquaculture 491, 266–272.
Mir, I. N., Srivastava, P. P., Bhat, I. A., Dar, S. A., Sushila, N., Varghese, T., et al. (2019). Expression and activity of key lipases during larval development of Indian walking catfish (Clarias magur). J. Exp. Zool. B Mol. Dev. Evol. 332, 149–157. doi: 10.1002/jez.b.22861
Mir, I. N., Srivastava, P. P., Bhat, I. A., Jaffar, Y. D., Sushila, N., Sardar, P., et al. (2020). Optimal dietary lipid and protein level for growth and survival of catfish Clarias magur larvae. Aquaculture 520:734678. doi: 10.1016/j.aquaculture.2019.734678
Monroig, Ó, Rotllant, J., Sánchez, E., Cerdá-Reverter, J. M., and Tocher, D. R. (2009). Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1791, 1093–1101. doi: 10.1016/j.bbalip.2009.07.002
Morais, S., Conceição, L., Rønnestad, I., Koven, W., Cahu, C., Infante, J. Z., et al. (2007). Dietary neutral lipid level and source in marine fish larvae: effects on digestive physiology and food intake. Aquaculture 268, 106–122. doi: 10.1016/j.aquaculture.2007.04.033
Nakamura, M. T., and Nara, T. Y. (2004). Structure, function, and dietary regulation of Δ6, Δ5, and Δ9 desaturases. Annu. Rev. Nutr. 24, 345–376. doi: 10.1146/annurev.nutr.24.121803.063211
Nayak, J., Viswanathan Nair, P., Ammu, K., and Mathew, S. (2003). Lipase activity in different tissues of four species of fish: rohu (Labeo rohita Hamilton), oil sardine (Sardinella longiceps Linnaeus), mullet (Liza subviridis Valenciennes) and Indian mackerel (Rastrelliger kanagurta Cuvier). J. Sci. Food Agric. 83, 1139–1142. doi: 10.1002/jsfa.1515
Nayak, M., Saha, A., Pradhan, A., Samanta, M., Mohanty, T. K., and Giri, S. S. (2018). Influence of dietary lipid levels on growth, nutrient utilization, tissue fatty acid composition and desaturase gene expression in silver barb (Puntius gonionotous) fingerlings. Comparat. Biochem. Physiol. Part B Biochem. Mol. Biol. 226, 18–25. doi: 10.1016/j.cbpb.2018.08.005
Patton, J. S., Warner, T. G., and Benson, A. (1977). Partial characterization of the bile salt-dependent triacylglycerol lipase from the leopard shark pancreas. Biochim. Biophys. Acta LipidsLipid Metab. 486, 322–330. doi: 10.1016/0005-2760(77)90028-90025
Péres, A., Cahu, C., Infante, J. Z., Le Gall, M., and Quazuguel, P. (1996). Amylase and trypsin responses to intake of dietary carbohydrate and protein depend on the developmental stage in sea bass (Dicentrarchus labrax) larvae. Fish Physiol. Biochem. 15, 237–242. doi: 10.1007/BF01875574
Ren, H. T., Yu, J. H., Xu, P., and Tang, Y. K. (2012). Influence of dietary fatty acids on muscle fatty acid composition and expression levels of Δ6 desaturase-like and Elovl5-like elongase in common carp (Cyprinus carpio var. Jian). Comparat. Biochem. Physiol. Part B Biochem. Mol. Biol. 163, 184–192. doi: 10.1016/j.cbpb.2012.05.016
Saera-Vila, A., Calduch-Giner, J. A., Gómez-Requeni, P., Médale, F., Kaushik, S., and Pérez-Sánchez, J. (2005). Molecular characterization of gilthead sea bream (Sparus aurata) lipoprotein lipase. Transcriptional regulation by season and nutritional condition in skeletal muscle and fat storage tissues. Comparat. Biochem. Physiol. Part B 142, 224–232. doi: 10.1016/j.cbpb.2005.07.009
Sargent, J. R., Tocher, D. R., and Bell, J. G. (2002). “The lipids,” in Fish Nutrition, eds J. E. Halver and R. W. Hardy (San Diego, CA: Elsevier), 181–257. doi: 10.1016/B978-012319652-1/50005-50007
Seiliez, I., Panserat, S., Kaushik, S., and Bergot, P. (2001). Cloning, tissue distribution and nutritional regulation of a Δ6-desaturase-like enzyme in rainbow trout. Comparat. Biochem. Physiol. Part B Biochem. Mol. Biol. 130, 83–93. doi: 10.1016/S1096-4959(01)00410-419
Sprecher, H. (1981). Biochemistry of essential fatty acids. Prog. Lipid Res. 20, 13–22. doi: 10.1016/0163-7827(81)90009-90006
Sprecher, H., Luthria, D. L., Mohammed, B., and Baykousheva, S. P. (1995). Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. J. Lipid Res. 36, 2471–2477. doi: 10.1201/9781439831939.ch2
Tan, S.-H., Chung, H.-H., and Shu-Chien, A. C. (2010). Distinct developmental expression of two elongase family members in zebrafish. Biochem. Biophys. Res. Commun. 393, 397–403. doi: 10.1016/j.bbrc.2010.01.130
Tavangar, K., Murata, Y., Patel, S., Kalinyak, J. E., Pedersen, M. E., Goers, J. F., et al. (1992). Developmental regulation of lipoprotein lipase in rats. Am. J. Physiol. Endocrinol. Metab. 262, E330–E337. doi: 10.1152/ajpendo.1992.262.3.E330
Tocher, D. R. (2003). Metabolism and functions of lipids and fatty acids in teleost fish. Rev. Fish. Sci. 11, 107–184. doi: 10.1080/713610925
Wang, C. S., and Hartsuck, J. A. (1993). Bile salt-activated lipase. A multiple function lipolytic enzyme. Biochim. Biophys. Acta 1166, 1–19. doi: 10.1016/0005-2760(93)90277-G
Wang, J. T., Liu, Y. J., Tian, L. X., Mai, K. S., Du, Z. Y., Wang, Y., et al. (2005). Effect of dietary lipid level on growth performance, lipid deposition, hepatic lipogenesis in juvenile cobia (Rachycentron canadum). Aquaculture 249, 439–447. doi: 10.1016/j.aquaculture.2005.04.038
Wang, M., Xu, D., Liu, K., Yang, J., and Xu, P. (2016). Molecular cloning and expression analysis on LPL of Coilianasus. Gene 583, 147–159. doi: 10.1016/j.gene.2016.02.017
Wong, H., and Schotz, M. C. (2002). The lipase gene family. J. Lipid Res. 43, 993–999. doi: 10.1194/jlr.R200007-JLR200
Xu, H., Dong, X., Ai, Q., Mai, K., Xu, W., Zhang, Y., et al. (2014). Regulation of tissue LC-PUFA contents, Δ6 fatty acyl desaturase (FADS2) gene expression and the methylation of the putative FADS2 gene promoter by different dietary fatty acid profiles in Japanese seabass (Lateolabrax japonicus). PLoS One 9:e0087726. doi: 10.1371/journal.pone.0087726
Zheng, K., Zhu, X., Han, D., Yang, Y., Lei, W., and Xie, S. (2010). Effects of dietary lipid levels on growth, survival and lipid metabolism during early ontogeny of Pelteobagrus vachelli larvae. Aquaculture 299, 121–127. doi: 10.1016/j.aquaculture.2009.11.028
Zheng, X., Ding, Z., Xu, Y., Monroig, O., Morais, S., and Tocher, D. R. (2009). Physiological roles of fatty acyl desaturases and elongases in marine fish: characterisation of cDNAs of fatty acyl Δ6 desaturase and elovl5 elongase of cobia (Rachycentron canadum). Aquaculture 290, 122–131. doi: 10.1016/j.aquaculture.2009.02.010
Zheng, X., Tocher, D. R., Dickson, C. A., Bell, J. G., and Teale, A. J. (2004). Effects of diets containing vegetable oil on expression of genes involved in highly unsaturated fatty acid biosynthesis in liver of Atlantic salmon (Salmo salar). Aquaculture 236, 467–483. doi: 10.1016/j.aquaculture.2004.02.003
Keywords: dietary lipid, lipolytic, desaturase, elongase, transcript, Clarias magur
Citation: Mir IN, Srivastava PP, Bhat IA, Dar JY and Agarwal D (2020) Expression Profiling of Lipolytic and Long-Chain Fatty Acid Biosynthesis Genes in Catfish Larvae Fed With Graded Lipid Levels. Front. Mar. Sci. 7:561402. doi: 10.3389/fmars.2020.561402
Received: 12 May 2020; Accepted: 31 August 2020;
Published: 29 September 2020.
Edited by:
Chunnuan Zhang, Henan University of Science and Technology, ChinaReviewed by:
Li Li, Ocean University of China, ChinaCopyright © 2020 Mir, Srivastava, Bhat, Dar and Agarwal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ishfaq Nazir Mir, aXNoZmFxbWlyMTBAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.