- 1Department of Molecular, Cellular and Biomedical Sciences, School of Marine Science and Ocean Engineering, University of New Hampshire, Durham, NH, United States
- 2School of Marine Science and Ocean Engineering, University of New Hampshire, Durham, NH, United States
The climate change related decline of shallow (<30 m) coral reef ecosystems has been driven by the mortality of scleractinian corals caused primarily by the phenomenon known as “coral bleaching.” But despite pervasive phase shifts and macroalgal dominance on many coral reefs, some coral species have persisted. One of those species is Montastraea cavernosa which has been categorized as resilient to a range of biotic and abiotic stressors. In order to understand the mechanism(s) of resistance in this coral, we present the results of a thermal stress and ocean acidification (OA) experiment on M. cavernosa, both its brown and orange color morphs, representing conditions predicted by the Representative Concentration Pathway (RCP) 6.0 scenario in the year 2100. We assessed the community response of the prokaryotic microbiome, the photophysiological response of the endosymbiotic Symbiodiniaceae and the molecular responses of critical pathways in the host by quantifying transcript abundances of genes encoding fluorescent proteins, heat shock proteins, antioxidant enzymes and regulators of apoptosis. After a 12 d acclimatization experiment, no visible bleaching was observed in any treatment, and the excitation pressure on photosystem II of the symbiotic Symbiodiniaceae showed no effects of the independent or interactive effects of thermal stress and OA, while only minor, but significant, changes in the prokaryotic microbiome were observed when exposed to RCP 6.0 predicted OA conditions. At the end of the experiment, the host heat shock protein 90 showed an increase in transcript abundance under the combined effects of thermal stress and OA compared to high temperatures alone, but these treatment groups were not significantly different from treatments under normal temperatures. While Bax, an activator of apoptosis, was significantly higher under thermal stress alone compared to control samples. Taken together, M. cavernosa, exhibits ecological stability over time and this may be based on its physiological persistence, resistance and resilience when experimentally exposed to the ecologically realistic RCP 6.0 climate model predictions.
Introduction
The worldwide decline of coral reefs has been attributed to a variety of natural and anthropogenic stressors (Hoegh-Guldberg et al., 2007, 2017; Hughes et al., 2018), and we are now in a new epoch of rapid ecological change known as the Anthropocene (Williams et al., 2019). However, climate change and specifically ocean warming (OW) have contributed to multiple incidences of coral bleaching in recent years (Donner et al., 2005; Hoegh-Guldberg et al., 2017; Eakin et al., 2019; Fordyce et al., 2019). In addition, the effect of ocean acidification (OA) on calcification and community composition is believed to be a serious threat to coral reefs (Hoegh-Guldberg et al., 2007, 2017; Edmunds et al., 2016), and the interaction between the effects of elevated OW and OA is an area of continuing concern (Pandolfi et al., 2011; Kroeker et al., 2013; Boyd et al., 2014; Harborne et al., 2017). These issues are of particular concern in the Caribbean Basin, where 70% of the coral reefs are already listed as threatened, critically endangered, or have undergone a phase shift to algal or soft coral dominated coral reefs (Norström et al., 2009; Dudgeon et al., 2010). The multiple stressors associated with climate change (e.g., Boyd et al., 2014) represent a “tipping point” for these coral reefs that are already severely impacted by hurricanes, overfishing, eutrophication, coastal development, hypoxic events and invasions by non-native species (Hughes et al., 2003, 2017; Lesser, 2004; Hoegh-Guldberg et al., 2007, Hoegh-Guldberg et al., 2017; Lesser and Slattery, 2011; Altieri et al., 2017).
The most significant consequence of climate change on coral reefs, however, is the phenomenon of “bleaching” in scleractinian corals where the primary cause is exposure to thermal stress and/or high irradiances of solar radiation, both its visible and ultraviolet components (Banaszak and Lesser, 2009). Subsequently, this leads to photooxidative stress in the host tissues and symbiotic dinoflagellates in the Family Symbiodiniaceae, followed by expulsion of the algae into the environment (Lesser, 2006, 2011a,b, 2019). The loss of these photosynthetic endosymbionts, or a change in their community composition (i.e., shuffling) to more resistant communities, can be a significant energetic cost to the host (Muscatine, 1990; Lesser, 2013), a strong selective pressure on corals (Baker et al., 2008), and an important influence on the resilience of corals to future climate change (Nyström et al., 2000; Anthony et al., 2015; Suggett et al., 2017). Recent studies have suggested that evolutionary adaptations can occur in corals over short temporal scales (Bay and Palumbi, 2014; Palumbi et al., 2014; Matz et al., 2018; Thomas et al., 2018; DeCarlo et al., 2019; Wright et al., 2019), but under current climate predictions it is clear that there will be “winners” and “losers” at the species level (Loya et al., 2001; Somero, 2010). Community composition changes, however, will also occur (van Woesik et al., 2011) and members of the community who were initially winners will become losers under prolonged, or repeated, exposures to environmental stress (Grottoli et al., 2014; Cacciapaglia and van Woesik, 2015).
Most severe bleaching occurs in the first 15 m depth on a coral reef (e.g., Baird et al., 2018) where irradiances are high (e.g., Lesser, 2000) and experimental evidence has supported a significant, and interactive, role for solar radiation in the bleaching phenomenon (Lesser et al., 1990; Brown et al., 2002; Bhagooli and Hidaka, 2004; Lesser and Farrell, 2004; Lesser, 2010, 2011a,b). Both the host and dinoflagellate symbionts respond by differential expression of stress genes that include heat shock proteins, enzymatic antioxidants, fluorescent proteins and regulators of apoptosis, autophagy or necrosis (Lesser and Farrell, 2004; Baird et al., 2009; Lesser, 2011a,b), or by switching to symbionts with increased organismal functionality (i.e., temperature compensated photophysiology) under environmental stress (Suggett et al., 2017; Lesser, 2019). One way to identify coral phenotypes resistant to environmental stress is to use molecular markers known to respond to multiple environmental stressors (Downs et al., 2000; Tchernov et al., 2004; Kenkel et al., 2011, 2014; Jin et al., 2016; Louis et al., 2017). We now know that the differential expression of genes involved in upstream pathways that respond to multiple environmental stressors such as quenching the formation of reactive oxygen (ROS) and nitrogen species (RNS), and the activation of downstream pathways related to coral bleaching including apoptosis, autophagy and necrosis (Lesser, 2006, 2011a,b; Weis, 2008), should be successful candidates to identify both resistant and susceptible phenotypes. These genes are highly conserved and have been identified in numerous cnidarians, including scleractinian corals with photoautotrophic endosymbionts in the Symbiodiniaceae (Bhattacharya et al., 2016).
In the Caribbean most species of coral have declined in abundance and percent cover over the past 40 years due to multiple factors that has resulted in a pervasive phase shift from coral to macroalgae cover (Jackson et al., 2014; de Bakker et al., 2016). Some species, such as Montastraea cavernosa, have continued to be a consistently small, and variable, component in many of these communities across the Caribbean on decadal time scales (Jackson et al., 2014). M. cavernosa occurs in three color morphs, cyan, orange and brown/green which express a range of fluorescent proteins as well as fluorescent pigments associated with their symbiotic cyanobacteria (Kelmanson and Matz, 2003; Lesser et al., 2004; Kao et al., 2007; Jarett et al., 2017). In the Caribbean, shallow populations of M. cavernosa have been categorized based on several phenotypic traits including sensitivity to bleaching, disease susceptibility and mortality, and found to be highly resistant to these biotic and abiotic stressors (Fitt and Warner, 1995; Smith et al., 2013). M. cavernosa also exhibits high resilience, described as a decrease in bleaching and mortality, in the face of repeated bleaching events (Gintert et al., 2018). Quantifying the underlying traits responsible for the ecological and evolutionary phenotypes that lead to the success of populations of M. cavernosa under current, and potentially future, environmental stress is essential (sensu Edmunds et al., 2014).
Here we present the results of an experiment on the Caribbean coral M. cavernosa that exposes both its brown and orange color morphs to OW and OA representing predictions from the RCP 6.0 climate model (Rogelj et al., 2012; Intergovernmental Panel on Climate Change [IPCC], 2014). Both physiological and molecular biomarkers of the host, its Symbiodiniaceae community and its prokaryotic microbiome were used to evaluate their response to future climate change scenarios that represent conditions coral reefs are likely to experience, based on realistic estimates of coal reserves and their use, as well as the absence of any climate change mitigation policies, which suggest that RCP 8.5 represent conditions are unrealistic (Hausfather and Peters, 2020).
Materials and Methods
Sampling, Experimental Conditions, Design and Statistical Analysis
Samples of both the brown (n = 16) and orange (n = 16) morphs of M. cavernosa were collected from South Perry Reef on Lee Stocking Island in the Bahamas (23°47′0.03″N, 76°6′5.14″W) at a depth of ∼15 m in August of 2011. All samples represented individual ramets that were collected by chiseling a piece of coral (=ramet) from individual colonies (=genet) in the field. Coral ramets ranged in size from 7.62 to 12.7 cm base diameter and were randomly selected and placed in different treatment groups. These corals were then acclimatized (n = 4 brown and n = 4 orange samples for each treatment group) in ambient seawater pressure of carbon dioxide (pCO2) and temperature for 3 days supplied to corals in individual aquaria (8 L) and placed in a raceway system of flowing seawater while being exposed to daily, full spectrum, solar radiation (∼13 h of light) under neutral density cloth. After 3 days, the seawater for each individual aquarium was supplied from header tanks (n = 2) that passed through a coarse sand filter allowing all corals to access ambient levels of food at ∼1.0 L h–1 for 12 d, a moderate duration experiment as defined by McLachlan et al. (2020), in a fully orthogonal matrix design of CO2 concentrations and temperature conditions.
At the time of the experiment CO2 concentrations that reflected current pCO2 (∼390 μatm) conditions and the A2 climate model (pCO2 of ∼800 μatm) scenario, as well as the current and predicted summer OW from that model (Intergovernmental Panel on Climate Change [IPCC], 2007) were used. The resulting conditions were pCO2 ∼390 μatm; (resulting pH: 8.07 ± 0.05 [SD]) and pCO2 = 800 μatm; (resulting pH: 7.81 ± 0.12 [SD]). Within each pCO2 treatment, corals were also acclimatized to current temperatures (mean 29.2 ± 0.9 [SD]°C; Range 27.3–30.1°C), or elevated temperatures (mean 31.4 ± 1.07 [SD]°C; Range 29.3–33.4°C). Both the temperature and pH values are within the predictions for RCP 6.0 (Rogelj et al., 2012; Intergovernmental Panel on Climate Change [IPCC], 2014). The mean differences in the abiotic factors were significant for both OA (t-test; two-tailed, P < 0.001) and temperature (t-test; two-tailed, P < 0.001). All measured as well as predicted carbonate chemistry values (e.g., pHT values) for each treatment group are presented in Supplementary Table 1.
The seawater pH values for these conditions were maintained by bubbling, using air stones, one header tank with CO2 and one with ambient air. pH was held constant using the pH-stat approach with WTW 3310 pH meters (accuracy = ±0.005 pH capability). The mean high temperature treatments were maintained using JBJ titanium heaters (accuracy = ±0.5°C) and monitored using HOBO Water Temperature Pro v2 data loggers (Onset Corp.). The experiment tested four treatments (n = 4 brown and n = 4 orange morphs per treatment); ambient T°C and pH (AmbNorm), ambient T°C and low pH (AmbLow), elevated T°C and normal pH (HiNorm), and elevated T°C and low pH (HiLow) for each color morph. The maximum irradiance of photosynthetically active radiation (PAR; 400–700 nm) under the neutral density cloth, measured with a LI-COR LI-192 cosine corrected underwater quantum sensor, was 450–500 μmol quanta m–2 s–1 representing the downwelling irradiance of the site and depth of collection at midday (e.g., Lesser and Gorbunov, 2001). All treatments, including color morph, and time effects and their interaction, were tested using ANOVA with post hoc multiple comparison testing (i.e., Tukey’s HSD), or repeated measures ANOVA, as needed.
Carbonate Chemistry
Independent seawater samples (n = 3) were collected from the experimental aquaria at the end of the experiment and analyzed for total alkalinity (TA) (μmol kg seawater–1), total CO2 (TCO2) (μmol kg seawater–1) and the partial pCO2 (μatm) at the University of New Hampshire Ocean Process Analysis Laboratory. TA was analyzed using an Apollo SciTech AS-A2 automated analyzer, which employs the Gran end-point titration procedure using 0.1 N HCl to pH 4.5 with a precision of 0.1%. The initial pH for the titration was measured on the sample using a Thermo Orion combination electrode (precision = ±0.027 pH units). TCO2 samples were analyzed immediately after opening the sample bottle. Using a digital syringe, a small amount of sample (0.5 ml) was acidified with 10% phosphoric acid and the evolved CO2 subsequently measured with a LICOR 6262 non-dispersive infrared gas analyzer. The precision of these measurements averaged ∼0.1% (range 0.05–0.5%). pCO2 (±4 μatm) was measured using a three-chamber equilibrator and a LiCor 840A non-dispersive infrared CO2 gas analyzer. Certified reference materials were used to generate standard curves and to ensure the precision of all measurements (Dickson et al., 2007). The pHT and Ω Ca of each sample were then calculated using CO2calc software (Robbins et al., 2010) using the temperature of each treatment, a salinity of 37.0 ppt, and the inorganic carbon dissociation constants from Mehrbach et al. (1973) as refitted by Dickson and Millero (1987). Given that the CO2calc software only required two of three of the directly measured carbonate parameters, TA and pO2 were chosen as inputs and the calculated TCO2 used as a check against the directly measured TCO2.
Active Fluorescence
The quantum yield of photosystem II (PSII) fluorescence was measured using a pulse amplitude modulated (PAM) diving fluorometer (Walz, Inc.) at noon and at 8–10 PM, equivalent to the maximum and minimum irradiances for the beginning (T0) and end (T12) of the experiment. The in vivo fluorescence of chlorophyll varies between a minimum yield, Fo, and maximum yield, Fm. The difference between Fo and Fm fluorescent yields (Fm-Fo) is the variable fluorescence, Fv and the ratio Fv/Fm or ΔF/Fm′ is the maximum (i.e., dark adapted) and steady state (i.e., in the light), respectively, is the quantum yield of PSII fluorescence (Gorbunov et al., 2001; Warner et al., 2010). The PAM fluorometer utilized was a blue LED version (470 nm excitation, emission > 630 nm) with the fiber-optic probe held 1 cm from the coral in a perpendicular orientation. A saturation pulse of 0.8 s at 3000 μmol quanta m–2 s–1 was used to record maximal fluorescence (Fm) at a gain setting of 6, and all coral measurements (n = 3 for each sample in each treatment) were taken at the same instrument settings. All quantum yield measurements are ratios and therefore a priori not normally distributed, so these measurements were transformed (Log + 1) prior to analysis and back transformed for presentation. Measurements were taken at the beginning (T0) and end (T12) of the experiment and analyzed for treatment effects; color morph and time of day (i.e., midday or midnight) were each treated as an independent factor using a multi-factor, repeated measures, ANOVA with interaction for the analysis of the data.
Additionally, the fluorescence measurements were used to assess the level of excitation over PSII defined as Qm = 1−[(ΔF/Fm′)/(Fv/Fm)] sensu Iglesias-Prieto et al. (2004). These data are also not a priori normally distributed and were transformed (Arcsine) prior to analysis and back transformed for presentation.
Metagenetic Analysis of 16S rRNA Genes
At the end of the 12 d experiment, samples of coral were preserved in both DNA preservation buffer (Seutin et al., 1991) and RNALater (Qiagen), transferred to the University of New Hampshire frozen, and maintained at −80°C until processed. Samples preserved in DNA buffer from each treatment; AmbNorm (n = 8), AmbLow (n = 7), HiNorm (n = 7), and HiLow (n = 8) were thawed, and rinsed with sterile 1× phosphate buffered saline (PBS) using an airbrush at a distance of 10–20 cm for ∼2 min. Subsequently, pressurized air from a Husky Pistol Grip Blow Gun fitted to a SCUBA cylinder was used to remove coral tissues into 10 ml of sterile 1× PBS. The tip was wiped with 70% ethanol followed by sterile 1× PBS after each coral sample was processed. The tissue slurry was homogenized for 30 s at medium speed using a tissue homogenizer which was run in 70% ethanol followed by sterile 1× PBS between each sample. Symbiodiniaceae and skeletal debris were pelleted (400 × g, 5 min, 4°C) and 1.5 ml of the tissue supernatant pipetted off into each of two 2 ml cryovials in DNA buffer (Seutin et al., 1991) for microbial analysis and stored at −20°C while the pellet was also stored in 1.5 ml of DNA buffer.
DNA was extracted from both coral supernatants and pellets using a modified CTAB (cetyltrimethylammonium bromide) protocol as follows. Lysis took place in the presence of 600 μl 2 × CTAB (0.4 M NaCl, 50 mM Na2 EDTA, pH 8.0) and 1.2 μl 2-mercaptoethanol. Mechanical lysis was achieved by the addition of one sterilized steel carbide bead (Qiagen) and 10 min of shaking at 50 Hz in a Tissue-Lyzer (Qiagen). Each sample was then incubated with 5 μl of Proteinase K (20 mg ml–1 stock in 10% SDS) at 55°C for 4 h. After incubation, samples were centrifuged at 2,000 × g for 1 min to pellet any debris and the liquid portion of the sample transferred to a new microcentrifuge tube. An equal volume of chloroform: isoamyl alcohol (24:1) was added and mixed by inversion. The aqueous phase was isolated by centrifugation for 10 min at 15,000 × g and transferred to a new microcentrifuge tube and mixed with an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1). The aqueous phase was again recovered by centrifugation for 10 min at 15,000 × g and rinsed with an equal volume of chloroform: isoamyl alcohol (24:1). Following a final centrifugation, the aqueous phase was recovered, and DNA precipitated with 2 × volumes of 100% cold ethanol and incubated for 1 h at −20°C. DNA was pelleted by centrifugation for 20 min at 20,000 × g at 4°C and the supernatant was subsequently discarded. Pellets were washed with 70% ethanol, pelleted by centrifugation for 20 min at 20,000 × g at 4°C and then dried at 45°C for 20 min in a SpeedVac to remove residual ethanol. Pellets were then rehydrated in 50 ul nuclease-free MilliQ water.
DNA was amplified using the polymerase chain reaction (PCR) with primer sets targeting the universal prokaryotic 16S rRNA gene. Degenerate primers designed to amplify the hypervariable region V3–V4 of the 16S rRNA gene were used and included a forward primer 515F (5′- GTGYCAGCMGCCGCGGTAA; Parada et al., 2016) and reverse primer 806R (5′- GGACTACN- VGGGTWTCTAAT; Apprill et al., 2015). Fluidigm linker sequences CS1 (5′- ACACTGACGACATGGTTC- TACA) and CS2 (5′- TACGGTAGCAGAGACTTGGTCT) were added to the 5′ end of both forward and reverse primers to facilitate Illumina MiSeq. The 16S rRNA gene PCR consisted of a 25 μl reaction with 12.5 μl AmpliTaq Gold 360 Master Mix (Applied Biosystems), 1.0 μl GC-enhancer, 0.5 μl 515F (10 μM) and 0.5 μl 806R (10 μM), 2.0 μl of DNA template (40–60 ng) and 8.5 μl nuclease free water (Integrated DNA Technologies, Coralville, Iowa). Reactions were performed using the following protocol: initial denaturation for 10 min at 95°C, 30 cycles of 95°C for 45 s, 50°C for 60 s, and 72°C for 90 s, followed by a 10 min extension at 72°C. The PCR products were then electrophoresed on a 1% agarose gel. For all DNA extractions both mock extractions (reagent controls) and PCR negative controls (no DNA) were conducted. The PCR products of these extractions, and samples, were run out on electrophoresis gels before sequencing. Even though mock extractions and negative controls showed no bands after PCR, a selection of these samples was submitted for 16S rRNA amplicon sequencing. These sequencing reactions did not reveal significant contamination (<500 reads per sample) and were excluded from downstream analysis because of insufficient reads to analyze with statistical confidence.
Sequencing and Bioinformatics
The 16S rRNA PCR amplicons containing Fluidigm linkers were sequenced on an Illumina MiniSeq System employing V2 chemistry (2 × 150 bp reads) at the University of Illinois at Chicago (UIC) Research Resources Center’s Sequencing Core. Amplicon sequence variants (ASVs) were inferred and tabulated across samples using DADA2 (Callahan et al., 2016). Briefly: raw reads were trimmed of the initial 20 bp to remove residual primer and then truncated beyond the first instance of quality scores below 3 (“truncQ = 2”). The maximum expected error during denoising (“maxEE”) was 2 and 5 for forward and reverse reads, respectively. The error model was built from the first 100M bases and inspected using “plotErrors.” Denoised reads were then merged and chimeric contigs discarded using “mergePairs” and “removeBimeraDenovo,” respectively. Taxonomic ranks were assigned to the inferred ASVs using the SILVA ribosomal reference database release 132, using DADA2 function “assignTaxonomy.” Raw 16S rRNA reads were submitted to the NCBI Sequence Read Archive under BioProject accession number PRJNA590192.
Analyses of the prokaryote communities was conducted using PhyloSeq functions in R (McMurdie and Holmes, 2013). All ASVs assigned as “chloroplast” were removed prior to analysis. Any samples recovering fewer than 10,000 total reads were omitted to ensure sufficient sequence coverage before normalization (Supplementary Figure 1). Samples were further filtered to exclude ASVs detected in in only one sample or amounting to less than 10 occurrences across all samples after assessment of ASV prevalence and abundance. Samples were then rarefied to even read depths to normalize for sequencing effort. Alpha diversities and evenness were estimated using the “estimate_richness” and “evenness” functions which calculate several common indices. Samples were ordinated based on Bray-Curtis distance using principal coordinates analysis (“method = PCoA”). In order to assess prokaryotic compositional differences at the class taxonomic level, the rarefied ASV count table was consolidated by rank using the “tax_glom” function. Raw counts were transformed to center log ratios using the “transform” function (“transform = CLR”) from the R “microbiome” package (Lahti et al., 2017). A global test of compositional differences among coral color morph, and treatments were then tested using PERMANOVA on sample Aitchison distances (Gloor et al., 2017) with the “adonis” function from the R package “vegan” (Oksanen et al., 2019). Subsequently, any significant global analysis was followed-up with post hoc tests of pairwise compositional differences among treatments and/or color morphs using PERMANOVA with Bonferroni correction of P-values for multiple tests using the “p.adjust” function in R. Global tests for the effect of experimental treatments and color morph on the abundance of dominant prokaryotic classes (i.e., classes comprising ≥ 1% of reads) were conducted using ANOVA and Tukey’s HSD for post hoc treatment comparisons.
In this study we were unable to amplify the DNA isolated in the samples collected at the end of the experiment for Symbiodiniaceae ITS2. Given that the same population of corals from the same reef on Lee Stocking Island in the Bahamas were used in the experiment described above as previously analyzed in Jarett et al. (2017), a re-analysis of those Symbiodiniaceae Sanger sequences from brown (n = 3) and orange (n = 3) morphs of M. cavernosa was undertaken using the internal transcribed spacer 2 (ITS2) data from Jarett et al. (2017). Cloned products previously amplified with ITS2 primers from Apprill and Gates (2007) were Sanger sequenced as described by Jarett et al. (2017) and re-analyzed here based on the latest taxonomic revisions of LaJeunesse et al. (2018). Symbiodinaceae taxonomy was assigned to each ITS2 OTU by BLASTn query against a curated Symbiodiniaceae ITS2 database provided in the SymITS21 pipeline known as SymTyper (Cunning et al., 2015) to identify top sequence matches with similarities ≥ 97%.
Analyses of the Symbiodinaceae community variation represented in the cloned PCR products from Jarett et al. (2017) was accomplished using the PhyloSeq functions in R (McMurdie and Holmes, 2013). To reduce the conflation of inter- and intragenomic ITS2 variation (Arif et al., 2014), diversity analyses compared abundances for each of the OTUs clustered at 97% similarity. Alpha diversities were estimated using the Shannon richness index. Samples were ordinated based on Bray-Curtis distance using principal coordinates analysis (method = “PCoA”). In order to assess compositional differences between color morphs, the OTU count table was consolidated by best ITS2 hit using the “tax_glom” function. Compositional differences between color morphs were then tested using PERMANOVA on sample Aitchison distances (Gloor et al., 2017) with the “adonis” function from the R package “vegan” (Oksanen et al., 2019).
Predicted Function of 16S rRNA Communities Using PICRUSt2
Predicted functional capacity was derived from 16S rRNA abundances using PICRUSt2 v2.1.0–b2. Briefly, 16S rRNA ASVs inferred by DADA2 were aligned with HMMER (Eddy, 2008) and placed in the provided reference tree using EPA–ng and GAPPA (Barbera et al., 2019). Gene family presence were predicted for 16S rRNA as well as KEGG functions (i.e., EC and KO accessions) using Hidden State Prediction (castor; Louca and Doebeli, 2018) based on 16S rRNA ASV abundances and phylogenetic proximity to reference taxa with available genomes. To minimize error in predicted functional capacity based on gene presence due to poor matches to available genomes, ASVs receiving Nearest-Sequenced-Taxon-Index (NSTI) scores above two were discarded from subsequent analyses. Abundances of biological pathways encoded by prokaryotic microbiomes were then predicted using MinPath (Ye and Doak, 2009). Abundances of KEGG accessions corresponding to metabolic processes of interest were tested for treatment effects using ANOVA and Tukey’s HSD for post hoc treatment comparisons.
RNA Extraction
For coral samples in RNALater, coral tissue equaling approximately 0.5 cm2 with its calcium carbonate skeleton was added to 1 ml Qiazol (Qiagen) containing a 5 mm steel bead (Qiagen) and homogenized for 2 min at 50 hz. Samples were clarified by centrifuging at 12,000 × g for 10 min at 4°C. The supernatant was saved and incubated for 5 min at room temperature. To each sample 200 μl chloroform was added and shaken vigorously for 15 s before incubating at room temperature for 3 min and centrifuged for 15 min at 12,000 × g in 4°C. The upper aqueous layer was saved and 500 μl 70% ethanol was added and the sample was mixed by pipetting. Half of the sample was added to a RNeasy mini spin column (Qiagen), centrifuged for 15 s at 8,000 × g. The process was repeated for the remaining sample. The column was washed in 700 μl Buffer RW1 and centrifuged for 15 s at 8,000 × g. An additional wash using 500 μl Buffer RPE was centrifuged for 15 s at 8,000 × g. The column was dried by centrifuging for 2 min at 8,000 × g and RNA was eluted by adding 30 μl RNase-free water before centrifuging for 1 min at 8,000 × g. A second elution was performed by adding the flow through back to the column and centrifuging for 1 min at 8,000 × g.
cDNA Synthesis and Real-Time PCR
RNA was treated with TURBO DNase (Thermo Fisher) per manufacturer’s directions followed by cDNA generation with ProtoScript® II First Strand cDNA Synthesis Kit (New England BioLabs) per manufacturer’s directions using Random Primers and the Easy Protocol. Reactions for Real-time PCR included Luna® Universal qPCR Master Mix (New England BioLabs), used per the manufacturer’s instructions. Genes for fluorescent protein gene amplification and quantification included: Gp1 (cyan), Gp3 (green), Gp4, (photoconvertible GFP to RFP) and β-tubulin as the housekeeping gene (Supplementary Table 2; Kao et al., 2007). Primers for the stress response genes Mn superoxide dismutase (MnSOD), Cu/Zn superoxide dismutase (Cu/Zn SOD), Catalase, heat shock protein 70 (HSP 70) and heat shock protein (HSP 90) were developed along with the regulators of apoptosis Bax, Bak, and Bcl-2. These primers were constructed by querying the Acropora millepora database using primer sequences from Skutnik (2016) and blasting them against the NCBI database, and by using the Genbank ID for specific genes (i.e., MnSOD and HSP 70) from Császár et al. (2009) and blasting those sequences against the NCBI database. After obtaining partial or full-length CDs for each gene, they were blasted against the 20-coral database3 used in Bhattacharya et al. (2016). Sequences identified from this database were compiled and conserved regions used for primer development (PrimerQuest, Integrated DNA Technologies) and testing. These primers were used with EF1 as the housekeeping gene (Supplementary Table 2).
Primer location and size were optimized (i.e., similar Tm) to allow primers to be used with the same thermocycler protocol. All primers were blasted to the NCBI database to ensure that they match the target gene. Primer working concentrations were optimized for each reaction (Supplementary Table 2), and a melting curve analysis was done on each product. No primer dimers were observed and technical replicates (n = 3) were done for each sample reaction. Quantitative Real-time PCR was performed using an Applied Biosystems 7300 Fast Real-Time PCR System (Thermo Fisher) using the thermoprofile of 95°C for 10 min, and 40 cycles of 95°C for 10 s and 60°C for 20 s using 1 ng μg–1 cDNA per reaction. Relative expression was calculated from the PCR efficiencies of the target and housekeeping gene, and the crossing point deviation of an unknown versus control sample as described by Pfaffl (2001). Housekeeping genes exhibited no significant differences in expression across all treatments.
Results
Active Fluorescence
The effects of both OW and OA, and color morph on the quantum yield of PSII fluorescence of the Symbiodiniaceae symbionts of M. cavernosa was assessed using both dark-adapted Fv/Fm and steady state ΔF/Fm′ quantum yields measured at noon and night with a PAM fluorometry (Table 1). No visible signs of “bleaching” were observed in experimental corals and Fv/Fm measurements were generally higher, overall, than measurements of ΔF/Fm′. There were no effects for treatment (ANOVA: F3,4 = 0.184, P = 0.9) or color morph (ANOVA: F1,52 = 0.17, P = 0.68) or their interaction (ANOVA: F3,52 = 1.29, P = 0.29) when the correct variance component estimate for error from the repeated measures assessment of time is used. This suggests that for the quantum yields of PSII fluorescence either no damage to PSII, or increased abilities across all treatments to repair any PSII damage, was occurring (Table 1). For the maximum excitation pressure (i.e., Qm values) there were also no effect of treatment (ANOVA: F3,4 = 0.153, P = 0.92) or color morph (ANOVA: F1,52 = 0.196, P = 0.66) or their interaction (ANOVA: F3,52 = 0.53, P = 0.67) (Table 1).
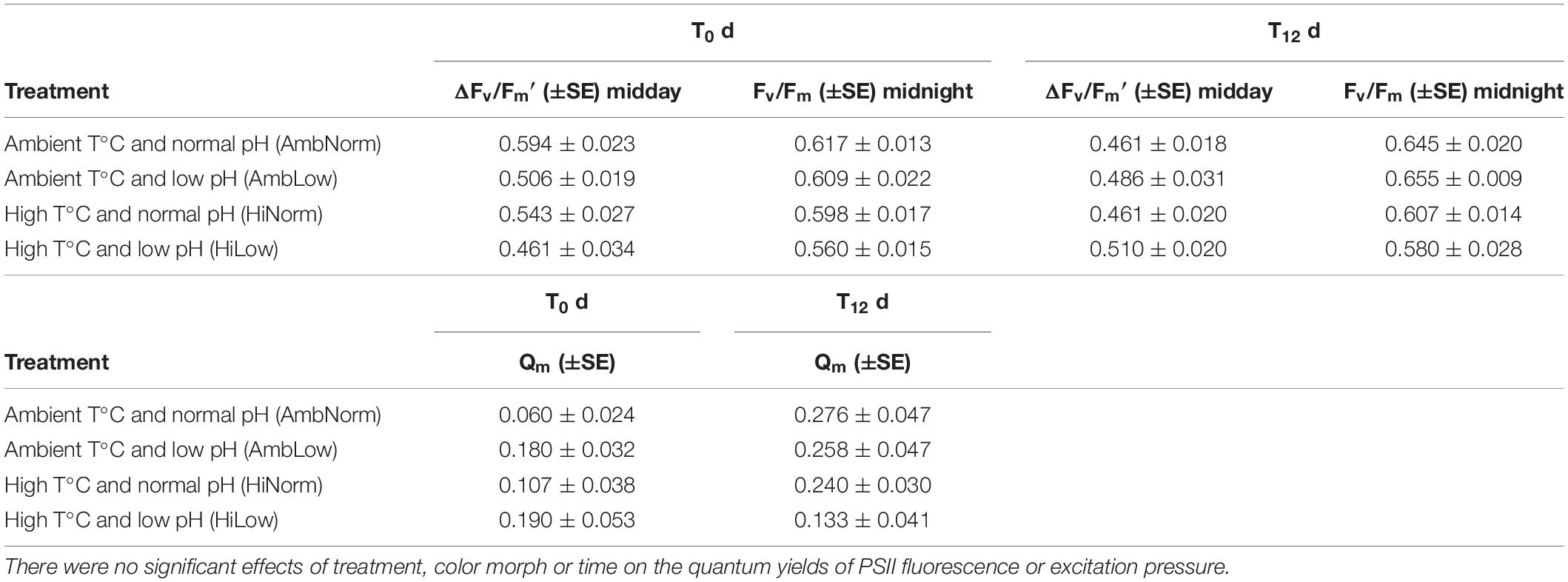
Table 1. Values for the quantum yield of PSII fluorescence and the excitation pressure (Qm) over PSII at the beginning (T0) and end (T12) of the experiment.
Metagenetic Analysis of 16S rRNA
Samples of the orange (n = 14), and brown morphs of M. cavernosa (n = 16) across all treatments were successfully sequenced and analyzed. A total of 543,317 16S rRNA MiniSeq read pairs were recovered after initial merging and quality trimming with DADA2 and before normalization. After the removal of reads assigned to chloroplasts, 490,880 read pairs remained and ranged from 10,556 to 21,422 reads per sample (average 15,449 ± 3,099) of 253 bp per read. A total of 456 unique ASVs were initially recovered, but any ASVs unique to a single sample or with fewer than 10 total observations were removed from the dataset, leaving a total of 190 unique ASVs for subsequent analyses.
Using the rarefied ASV count table (Supplementary Table 3) classified to the class level, those classes that consisted of greater than 1% of the relative abundance (30 classes) the 16S rRNA prokaryotic community showed significant compositional differences at the class level (Figure 1A) for treatment effects (PERMANOVA: F3,21 = 2.00, R2 = 0.184, P = 0.028) but not color morph (PERMANOVA: F1,21 = 1.79, P = 0.108). Samples of M. cavernosa exposed to lower pHs exhibited distinct 16S rRNA communities compared to all other treatments (Figures 1A,B). Post hoc multiple comparisons on the effect of treatment detected significant differences between AmbNorm and AmbLow treatments (PERMANOVA: F1,13 = 2.51, R2 = 0.162, P = 0.017) (Figure 1B) and between HiNorm and AmbLow treatments (PERMANOVA: F1,11 = 3.22, R2 = 0.23, P = 0.014). Significant post hoc differences between treatments for classes of prokaryotes occurred only for the abundance of the taxa Clostridia and Deltaproteobacteria. Clostridia increased in response to thermal stress (ANOVA: F7,21 = 6.97, P = 0.002; Tukey’s HSD: AmbNorm versus HiNorm, P = 0.03) while Deltaproteobacteria showed a global effect of treatment (ANOVA F7,21 = 2.9, P = 0.028) with only the HiNorm versus AmbLow comparisons being significantly different (Tukey’s HSD: P = 0.046) (Figure 1B and Supplementary Table 4). Additionally, as a result of the analyses above we conducted a a posteriori analysis of pH alone by placing all samples in a normal or low pH category. The results of this analysis revealed the presence of 23 classes of prokaryotes in the two groups (Supplementary Figure 2). An ANOVA on the abundance in each class showed 10 of these classes were significantly different between normal and low pH treatments (Supplementary Table 5). Notably, the abundance of Melainabacteria, a non-photosynthetic, nitrogen fixing class of bacteria related to the Cyanobacteria decreased, as did the Gammaproteobacteria.
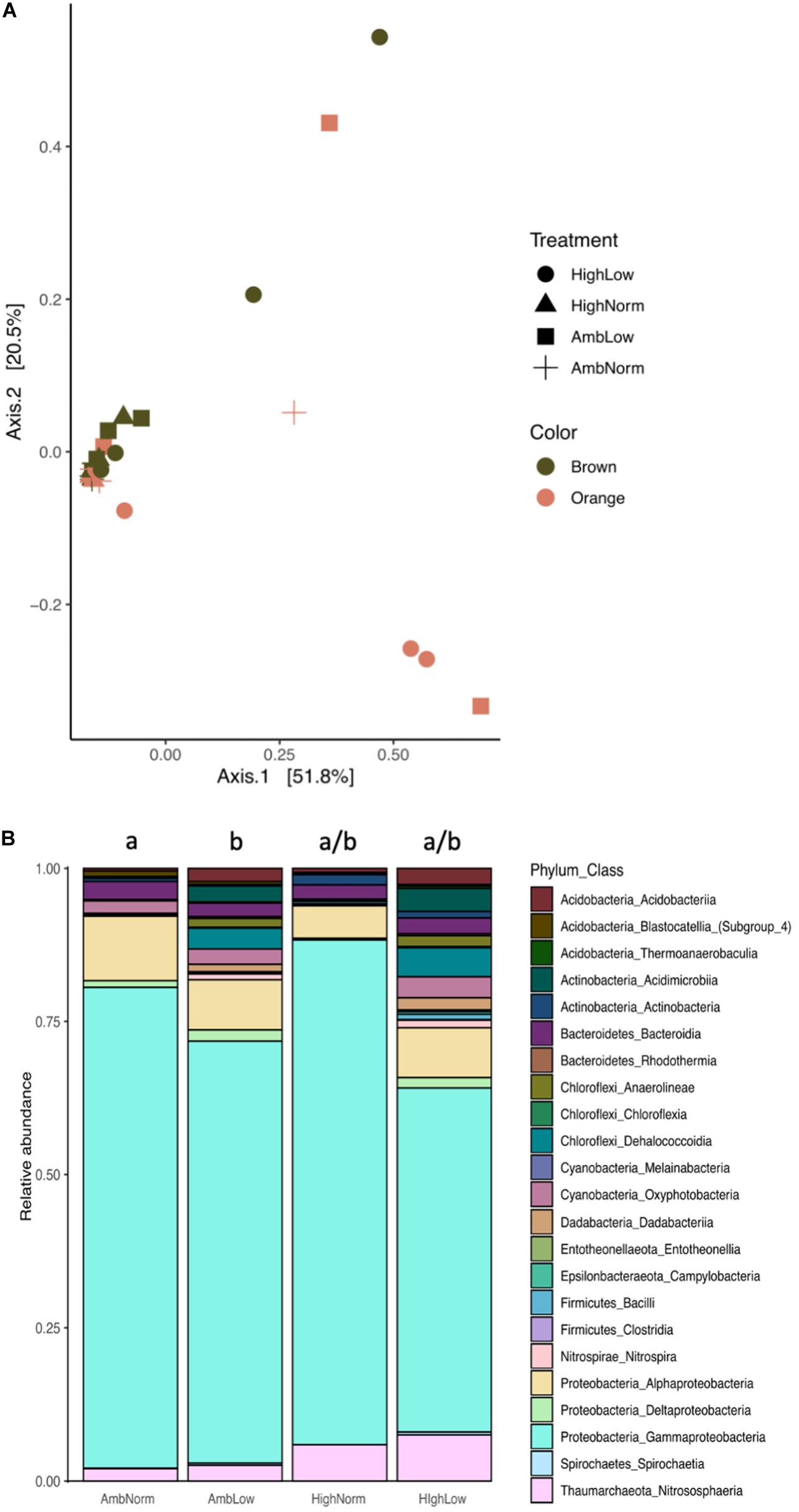
Figure 1. Prokaryotic communities of Montastraea cavernosa. (A) Multidimensional analysis of the b–diversity estimates using Bray–Curtis distance matrices at the class level for each treatment group and color morph. (B) Average relative abundance of ASVs consolidated to class level for each treatment group and color morph. Common superscripts indicate treatment groups not significantly different from each other (Tukey’s HSD, P > 0.05).
Additionally, given that the orange morphs of M. cavernosa are known to contain large populations of cyanobacteria (Lesser et al., 2004; Jarett et al., 2017) we examined the cyanobacterial composition and abundance between orange and brown morphs. For overall compositional differences both treatment (PERMANOVA: F3,28 = 4.78, R2 = 0.355, P = 0.015) and color (PERMANOVA: F1,28 = 4.79, R2 = 0.118, P = 0.04) were significant. Specifically, orange morphs showed an increase in the relative abundance of Oxyphotobacteria that was driven by the Order Synechococcales (ANOVA: F1,24 = 5.33, P = 0.03).
For the prokaryotic communities the predicted functional capacity analysis using PICRUSt2 revealed that those genes involved in carbon metabolism were affected significantly by treatment and/or color morph. For the tricarboxylic cycle (TCA), aconitate hydratase (acnA [K01681]) was significantly affected by the interaction between treatment and color morph (ANOVA: F3,21 = 3.10, P = 0.049) with multiple-comparison testing showing significant predicted gene enrichment (Tukey’s HSD, P = 0.022) in the AmbNorm brown versus AmbNorm orange morph. For glycolysis, the glyceraldehyde-3-phosphate dehydrogenase (gapA [K00134]) gene showed a significant interaction between treatment and color morph (ANOVA: F3,21 = 3.48, P = 0.034) with multiple-comparison testing showing significant predicted gene enrichment (Tukey’s HSD, P = 0.024) in the AmbNorm brown versus AmbNorm orange morph. For both the metabolism of pyruvate and glyoxylate, the gene for malate synthetase (glcB [K01638]) showed a significant effect of color morph (ANOVA: F1,21 = 4.92, P = 0.038) with multiple-comparison testing showing significant predicted gene enrichment (Tukey’s HSD, P = 0.049) in the AmbNorm brown morph versus AmbNorm orange morph.
Similarly, there were significant treatment and color morph effects on specific predicted functional capacity for genes involved in nitrogen cycling. For ammonification the urease gene (ureC [K01428]) showed a significant effect of color morph (ANOVA: F1,21 = 4.92, P = 0.038) with multiple-comparison testing showing significant predicted gene enrichment (Tukey’s HSD, P = 0.05) in the AmbNorm brown morph versus AmbNorm orange morph. Genes for denitrification, nitrate reductase (narG [K00370]), showed a significant effect of color morph (ANOVA: F1,21 = 4.92, P = 0.038) but multiple-comparison testing showed no significant differences in the predicted gene enrichment (Tukey’s HSD, P = 0.060) for the AmbNorm brown morph versus AmbNorm orange morph. Genes for nitrogen fixation, nifH [K02588] and nifD [K02586], both showed a significant interaction between treatment and color morph (ANOVA: F3,21 = 6.4, P = 0.003 and ANOVA: F3,21 = 5.3, P = 0.007, respectively), with multiple-comparison testing showing a significant predicted functional capacity (Tukey’s HSD, P = 0.023 and P = 0.032, respectively) in the AmbNorm orange morph versus AmbNorm brown morph.
For the orange and brown morphs of M. cavernosa, the 159 cloned sequences for the Symbiodiniaceae ITS2 resulted in 39 unique OTUs clustering at 97% similarity all matching published genotypes in the genus Cladocopium (Clade C). All of the 39 OTUs best-matched one of 12 possible NCBI accessions. There were no significant differences in the diversity of Symbiodiniaceae between orange and brown color morphs of Symbiodiniaceae (Shannon diversity, ANOVA: F1,5 = 0.97, P = 0.381). Coral samples were ordinated based on Bray-Curtis distances and using principal coordinates analysis (Figure 2A) the Symbiodiniaceae ITS2 community diversity showed no significant compositional differences between orange and brown morphs of M. cavernosa (PERMANOVA: F1,5 = 0.434, R2 = 0.098, P = 0.6). There were also no compositional differences between color morphs based on top-hit assignments (PERMANOVA: F1,5 = 4.41, R2 = 0.524, P = 0.1). Most Symbiodiniaceae OTUs (≥85%) from both color morphs at the beginning of the experiment belong to the species Cladocopium sp. (Phylotype C3) with the remaining 15% or less represented by Cladocopium goreaui (Phylotype C1) (Figure 2B).
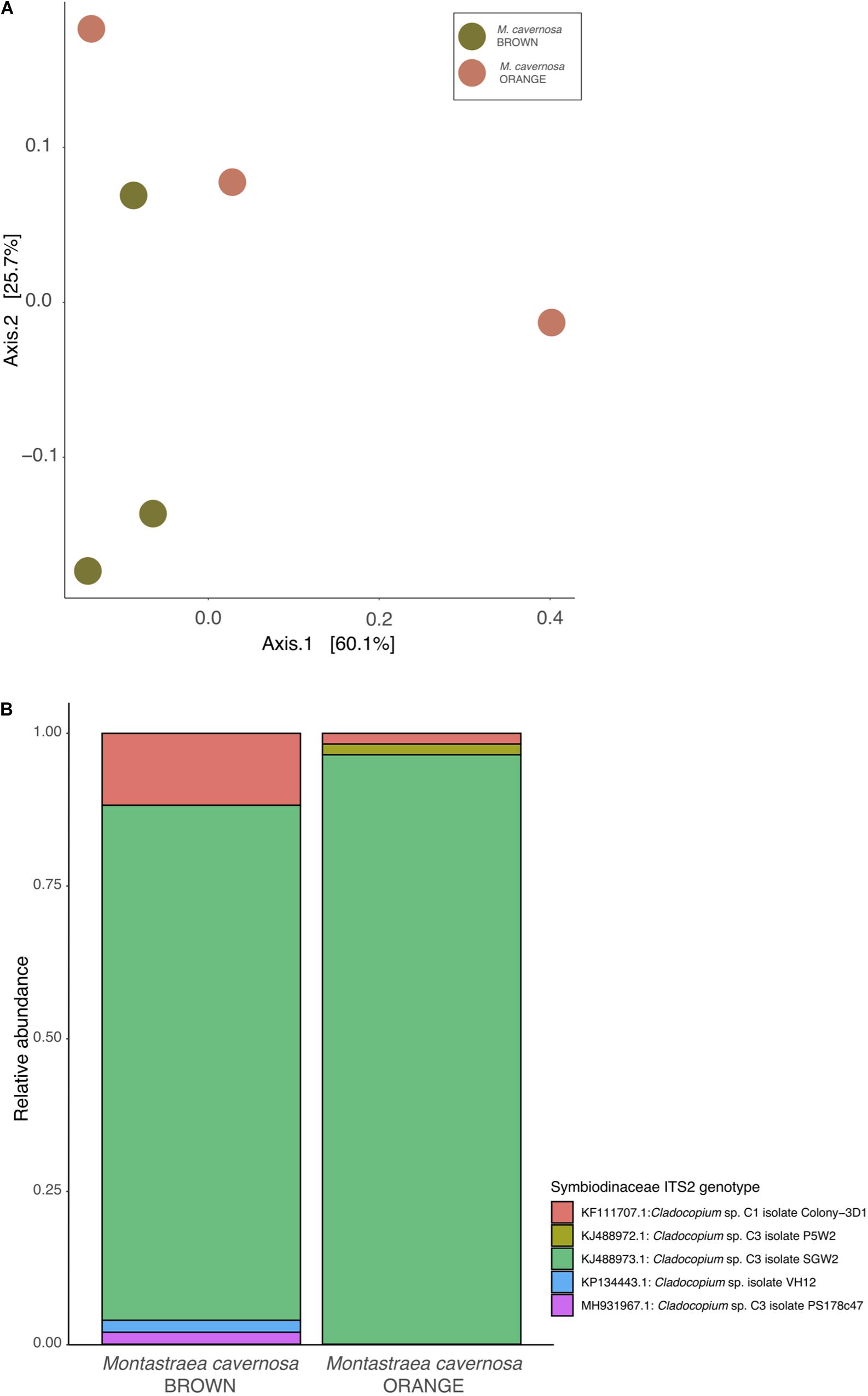
Figure 2. Symbiodiniaceae communities of Montastraea cavernosa color morphs. (A) Non-metric MDS plot of Symbiodiniaceae diversity across treatment group and color morph. (B) Average relative abundances of Symbiodiniaceae OTUs, clustered to 97%. ITS2 variant clusters are colored by closest match in the curated Symbiodiniaceae SymTyper database.
Relative Gene Expression Using qPCR
At the end of the experiment the differential expression of cyan (Gp1), green (Gp3) and photoconvertible (Gp4) fluorescent proteins, relative to the housekeeping gene, for the orange and brown color morphs of M. cavernosa showed no significant effect of color morph and treatment, or their interaction (ANOVA: cyan, F7,25 = 0.353, P = 0.918; green, F7,25 = 0.844, P = 0.566; photoconvertible, F7,25 = 0.182, P = 0.986) (Figure 3A). For the antioxidant genes there were no significant effects of color morph or treatment on Catalase (ANOVA: F7,28 = 1.138, P = 0.378) or MnSOD (ANOVA: F7,28 = 0.527, P = 0.805) transcript abundance while for Cu/Zn SOD there was a significant effect of treatment (ANOVA: F3,28 = 3.183, P = 0.045). Multiple comparison testing, however, showed no significant differences (Figure 3B) between any treatments (Tukey’s HSD, P > 0.05). Given these results a Dunnett’s Method multiple comparison test was done where treatment groups are compared against an identified control group (i.e., AmbNorm) which resulted in a significant difference between the AmbNorm and HighNorm treatments (Dunnett’s Method, P = 0.032). For those genes involved in the heat shock response HSP 70 showed no effect of treatment or color morph (ANOVA: F7,28 = 0.907, P = 0.520). At the end of the experiment HSP 90 showed a significant effect of treatment (ANOVA: F3,28 = 3.463, P = 0.035) with multiple comparison testing showing the HiLow treatment with the greatest expression of HSP 90 transcripts (Figure 3C), but these were not significantly different from treatments under normal temperatures.
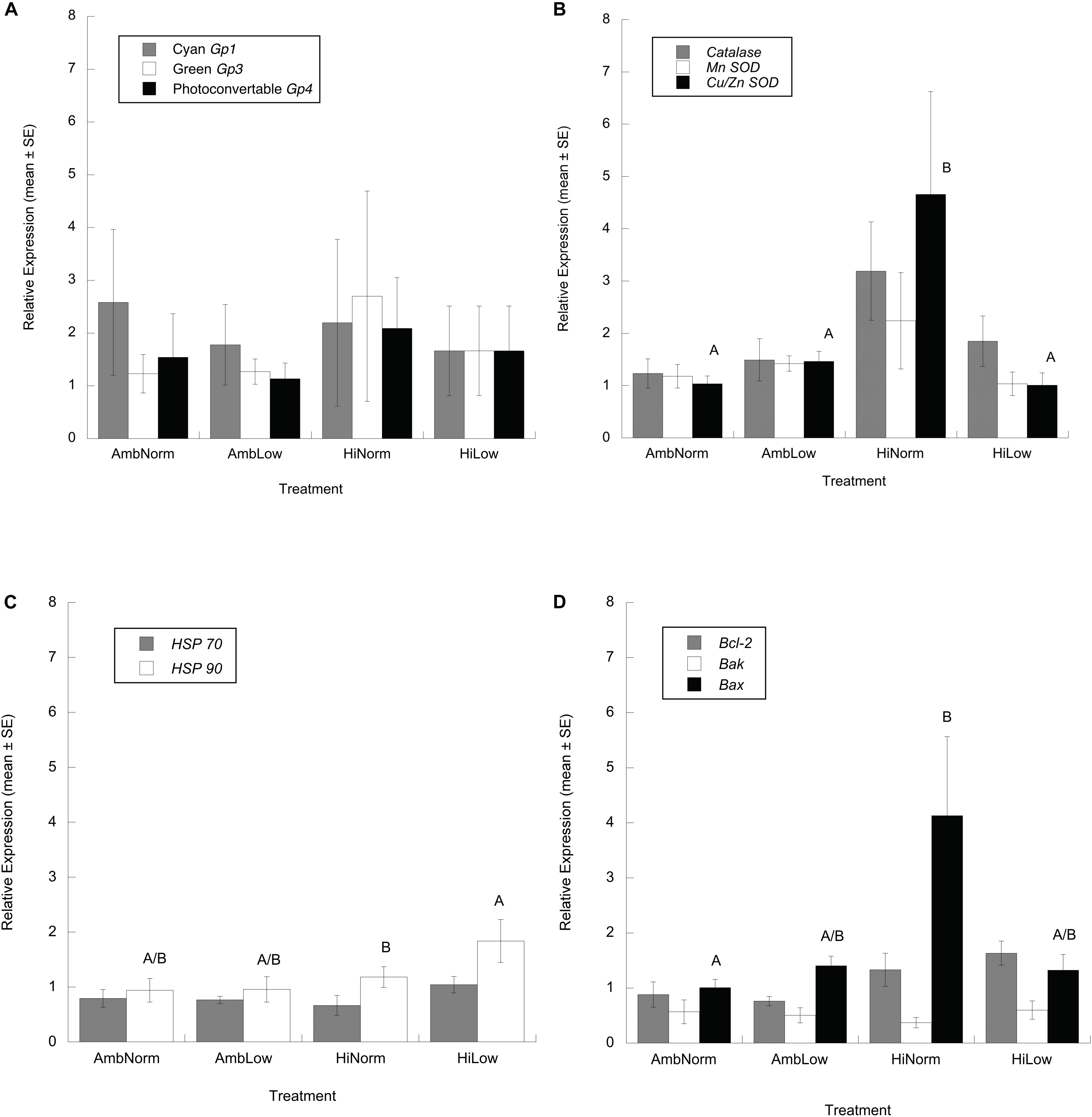
Figure 3. Relative expression ratios, or fold changes, of target and housekeeping gene transcripts (mean ± SE) for Montastraea cavernosa host as determined by qPCR. (A) Fluorescent proteins. (B) Antioxidant enzymes. (C) Heat shock proteins. (D) Regulators of apoptosis. Common superscripts indicate treatment groups not significantly different from each other (Tukey’s HSD, P > 0.05, panels C,D and Dunnett’s Method, P > 0.05, panel B).
Lastly, for the regulators of apoptosis Bcl-2 showed a significant effect of treatment (ANOVA: F3,28 = 3.101, P = 0.049) but multiple comparison testing showed no significant differences between any treatments (Tukey’s HSD, P > 0.05). As above, a Dunnett’s Method multiple comparison test was done where treatment groups are compared against an identified control group which resulted in no significant treatment differences detected (Dunnett’s Method, P > 0.05). For Bak, a significant effect of color morph was detected (ANOVA: F1,28 = 10.940, P = 0.003) with orange morphs expressing ∼2.5 time more transcripts than brown morphs. Bax expression showed a significant effect of treatment (ANOVA: F3,28 = 3.722, P = 0.027) with multiple comparison testing showing significantly higher expression of Bax transcripts in the HiNorm treatment group (Tukey’s HSD, P < 0.05) compared to control samples (Figure 3D).
Discussion
On a global scale both elevated OW and OA is proceeding unabated and changing the framework and biological communities of coral reefs in unprecedented ways (Hughes et al., 2003, 2017; McClenachan et al., 2017; Skirving et al., 2019). In particular, the coral reefs of the Caribbean basin have been affected not only by global climate change but other factors such as eutrophication and overfishing (Hughes et al., 2003, 2017). The observed degradation and phase shifts on coral reefs over time underscores the notion that sustaining, or improving, coral reef resilience and recovery is going to be very difficult (Hughes et al., 2010). The experiment presented here was based on the RCP climate model predictions, specifically the RCP 6.0 model (Rogelj et al., 2012; Intergovernmental Panel on Climate Change [IPCC], 2014) for temperature exposures. Additionally, this experiment used ecologically realistic irradiances of PAR, unlike most bleaching experiments that utilize sub-saturating irradiances, based on measurements from the site of collection for these corals (Lesser and Gorbunov, 2001), but were, in fact, exposed to higher irradiances of UVR with its known adverse effects on corals (Banaszak and Lesser, 2009). The general resistance of this coral to these experimental conditions is evident, but it is also possible that the moderate duration of the experiment (sensu McLachlan et al., 2020) was an insufficient timeframe to elicit a change in the parameters measured here, and that longer duration experiments would be needed to fully describe the response of this coral to RCP 6.0 conditions.
We suggest that given the speed that coral reef degradation is currently occurring, it seems to lack any ecological realism to conduct experiments under predicted RCP 8.5 predictions, commonly known as the “business as usual” scenario (Hausfather and Peters, 2020). Based on current models we are quickly surpassing RCP 2.6 conditions, and most coral reef ecosystems would be functionally extinct under RCP 8.5 conditions. Fortunately, the RCP 8.5 scenario actually appears to be unlikely to occur (Hausfather and Peters, 2020), such that we should be looking at the more plausible effects of RCP 4.5 and 6.0 on corals, and coral reefs, and the potential for coral adaptation and acclimatization to those scenarios (Somero, 2010; Bay et al., 2017). The conditions described here are representative of those predicted by the RCP 6.0 (climate model for the year 2100).
Quantifying the physiological impacts of the two primary drivers of coral reef degradation, OW and OA, at the organismal level (Rodolfo-Metalpa et al., 2011; Putnam et al., 2017), is essential in order to make predictions for marine populations, communities, and ultimately ecosystem services in the future (Doney et al., 2009, 2012; Anthony et al., 2011; Logan et al., 2014). Understanding the physiological responses of OW and OA is also essential for guiding the screening of naturally occurring coral phenotypes for potential rearing and explanting for restoration efforts (sensu Morikawa and Palumbi, 2019). Studies that have examined the resistance of corals to predicted changes in OW and OA have resulted in variable results reflecting the inter- and intraspecific phenotypic plasticity of corals and their Symbiodiniaceae symbionts (Hoadley et al., 2015; Bahr et al., 2016; Castillo et al., 2018; Cole et al., 2018; Davies et al., 2018). For Symbiodiniaceae, the diversity in phenotypic characters related to the stress response is significant (Díaz-Almeyda et al., 2018; Lesser, 2019), and recent studies, including those on M. cavernosa, have shown that “symbiont shuffling” to stress-tolerant Symbiodiniaceae (i.e., Durusdinium trenchi [Phylotype D1a]) can occur after exposure to thermal stress (Silverstein et al., 2015, 2017; Cunning et al., 2018). The change to, or addition of, D. trenchi symbionts to the Symbiodiniaceae community potentially adds resistance to future thermal perturbation (Silverstein et al., 2015). While some studies incorporating both long and short-exposures of corals to combined OW and OA conditions have revealed little effect of OA on gene expression or the functional diversity of their Symbiodiniaceae (Baghdasarian et al., 2017; Davies et al., 2018), other studies on the functional responses of corals in response to OW and OA have shown significant effects where OA lowers the thermal response threshold of the coral (e.g., Anthony et al., 2008) when using ecologically relevant irradiances (sensu Lesser, 2019).
For the Symbiodiniaceae community of M. cavernosa used in these experiments, the conservative taxonomic identification of Cladocopium sp. (Phylotype C3) as the dominant species in these experimental corals, as well as the source populations (Jarett et al., 2017) is consistent with other work (e.g., Silverstein et al., 2015) on this species. Despite harboring thermally sensitive Cladocopium sp. (Phylotype C3) this population of M. cavernosa has a long history of exposure to temperatures at, or above, 30°C during late Summer (Fitt et al., 2000; Smith, 2001; Manzello et al., 2009), and irradiances typical of shallow coral reefs. It therefore seems reasonable that one working hypothesis for the resistance and resilience of M. cavernosa to OW and OA is the selection for resistant Cladocopium sp. (Phylotype C3) phenotypes over time in the absence of any detectable D. trenchi symbionts. Previous work, on the same population of M. cavernosa, has shown that the Symbiodiniaceae communities have robust non-photochemical quenching (qN) capabilities that allowed these corals to maintain high rates of photosynthesis throughout the day without any evidence of photodamage (Lesser and Gorbunov, 2001). Those results are consistent with the lack of treatment effects on the quantum yields of, and the excitation pressure on, PSII observed here. This suggests that the community of Symbiodiniaceae in M. cavernosa have either enhanced PSII repair capabilities, down-regulation of PSII reaction centers or no significant photodamage (Gorbunov et al., 2001; Takahashi et al., 2004).
In this experiment there were significant treatment effects observed on the prokaryotic microbiome but for only 2 of 24 classes (Clostridia and Deltaproteobacteria, Supplementary Table 4). For the control treatments for the orange and brown morphs (i.e., AmbNormBr and AmbNormOR) the 16S rRNA sequence composition is similar to previous descriptions for this population of M. cavernosa (Jarett et al., 2017) where the primary difference is the ∼5% of reads assigned to the Class Oxyphotobacteria (Phylum Cyanobacteria) that increases in orange morphs in the low pH treatments (Figure 1B). Different coral species appear to harbor different “core” prokaryotic communities where differences in the host and Symbiodiniaceae communities may strongly influence the community structure of their prokaryotic microbiome (Bourne et al., 2016; Hernandez-Agreda et al., 2017; Morrow et al., 2018). The prokaryotic microbiome of corals can be sensitive to changes in their environment, especially those changes associated with climate change such as OW and OA (Bourne et al., 2016; Morrow et al., 2018). Many studies have shown shifts in community structure and changes in functional attributes such as the biogeochemical cycling of essential nutrients (e.g., nitrogen), or changes in the community where opportunistic microorganisms can cause stochastic transitions to unstable community states, dysbiosis and disease (Lesser et al., 2007; Bourne et al., 2016; Zaneveld et al., 2017; Morrow et al., 2018). The effects of OW and OA, and their interaction, have been shown to vary between sensitive (=losers) and resistant (=winners) corals (Webster et al., 2016; Grottoli et al., 2018). Similar to the results for resistant corals, M. cavernosa showed no significant overall changes in its prokaryotic microbiome indicating stability in the face of predicting changes in OW and OA. However, the relative abundance of Class Oxyphotobacteria, Order Synechococcales increases in orange morphs in those treatments experiencing lower pHs suggesting that increasing pCO2, as a result of the lower pH, has a significant positive effect on these cyanobacteria as reported for pelagic species (e.g., Levitan et al., 2007).
When the potential range of functional characteristics of the prokaryotic microbiome were examined using a predictive metagenomic approach (i.e., PICRUSt2) there were genes associated with carbon metabolism that showed no effect of OW or OA, but were predicted to be significantly enriched based on color morph with brown morphs always exhibiting greater enrichment. Similarly, nitrogen cycle genes associated with ammonification and denitrification showed no effects of OW or OA but were predicted to be significantly enriched in brown versus orange morphs. Genes for nitrogen fixation, however, were predicted to be significantly enriched in orange morphs as previously shown in a study using the qPCR analysis of nifH genes (Jarett et al., 2017). These differences are consistent with both the known genetic differences between the color morphs of M. cavernosa and the presence of nitrogen fixing bacteria in orange morphs of M. cavernosa (Jarett et al., 2017). Based on the PICRUSt2 analysis there were, however, no differences overall for carbon and nitrogen metabolism between the color morphs.
For the host compartment of M. cavernosa the quantitative analysis of transcript abundance (i.e., qPCR) showed no significant differences between color morphs of M. cavernosa in the expression of fluorescent protein transcripts. Cyan (Gp1), Green (Gp3) and photoconvertible (Gp4) transcripts were highly variable and were not significantly affected by exposure to OW or OA conditions (Figure 3A). Green fluorescent protein (GFP) can quench superoxide radicals (O2–) (Bou-Abdallah et al., 2006), as can other chromoprotein representatives of fluorescent proteins (Dove et al., 2006), and can also scavenge hydrogen peroxide (H2O2), a product of the dismutation of O2– (Palmer et al., 2009). The modest antioxidant activity of fluorescent proteins may well be effective because they occur in such high concentrations in corals and other symbiotic cnidarians (Mazel et al., 2003; Dove, 2004; Leutenegger et al., 2007), making it an important contributor to the overall antioxidant defenses in corals.
In addition to fluorescent proteins the expression of HSP 70 and HSP 90 was quantified. Heat shock proteins are highly conserved molecular chaperones that interact with cellular proteins during normal metabolism and in response to environmental stress by targeting proteins for degradation and removal from the cell (Feder and Hofmann, 1999). In addition, heat shock proteins are now known to be regulators of the cell cycle and apoptosis (Beere and Green, 2001; Calderwood and Gong, 2016). As in most metazoans, there are multiple heat shock proteins in the coral host tissues that respond to OW stress (Robbart et al., 2004; Fitt et al., 2009; Kvitt et al., 2016), as do their endosymbiotic Symbiodiniaceae (Rosic et al., 2011), but not OA conditions (Nakamura et al., 2012). For M. cavernosa neither OW or OA independently caused an increase in the expression of HSP 70 transcripts but exposure to the combined effects of OW and OA did result in an increase in HSP 90 transcript abundance compared to OW alone, but these were not significantly different from treatments under normal temperatures (Figure 3C).
The principal cause of bleaching in corals and other symbiotic cnidarians is the creation of hyperoxic conditions, caused by photosynthetically produced oxygen, acting synergistically with ecologically relevant irradiances of solar radiation, especially UVR, and thermal stress to produce ROS in both host tissues and Symbiodiniaceae beyond their capacity to quench these products of oxygen metabolism (Lesser, 2006, 2011a,b). Given the important role of light in the generation of photooxidative stress the increased tissue thickness, with its unique optical properties for M. cavernosa, might modulate the amount of photoinhibitory light reaching the endosymbionts (Wangpraseurt et al., 2012; Scheufen et al., 2017). Nonetheless, the production of ROS is inevitable, even in the absence of thermal stress, as it is directly related to pO2 (Jamieson et al., 1986). As a result, the endosymbiotic Symbiodiniaceae contribute significant amounts of photogenerated O2– and H2O2 (Lesser, 1996, 2019; Suggett et al., 2008; Saragosti et al., 2010; McGinty et al., 2012), and both host cells (Perez and Weis, 2006) and Symbiodiniaceae also produce nitric oxide (NO⋅) (Bouchard and Yamasaki, 2008). Nitric oxide synthase (NOS) activity in corals and sea anemones (Morrall et al., 2000; Trapido-Rosenthal et al., 2005) suggest that NO⋅ production, and subsequently reactive RNS, including the highly reactive peroxynitrite radical (ONOO–), are produced and are considered important contributors to redox stress and initiation of coral bleaching (Weis, 2008). Despite these known effects of OW and OA under ecologically realistic irradiances for many coral species and their symbionts, for M. cavernosa only Cu/Zn SOD was significantly elevated, and only when corals were exposed to OW. The transcript abundance of all other treatment groups was not significantly different from the control group. In order to understand whether the constitutive expression of antioxidant genes in the host compartment of M. cavernosa contributed to its resilience to OW and OA a comparative study with multiple coral species is required.
Photooxidative stress has been proposed as a mechanism by which multiple environmental stressors, either independently or interactively, cause damage to the endosymbiotic Symbiodiniaceae and the host simultaneously (Lesser and Farrell, 2004), which leads to apoptosis that manifests itself as coral bleaching (Lesser, 2006, 2011a,b). Cells exposed to oxidative stress, or UVR, can also incur DNA damage (Lesser and Farrell, 2004; Fitt et al., 2009) which if not repaired results in a cellular cascade of molecular events causing apoptosis via a highly conserved intrinsic pathway (Bhattacharya et al., 2016; Kvitt et al., 2016). The tumor suppressor gene, p53, is upregulated in response to DNA damage in corals (Lesser and Farrell, 2004) and coordinates expression of downstream genes including highly conserved anti-apoptotic Bcl-2, and the pro-apoptotic genes, Bak and Bax (Ainsworth et al., 2008, 2011, 2016; Kvitt et al., 2011, 2016; Pernice et al., 2011). Here, we observed that only for the pro-apoptotic gene Bax, transcripts were significantly elevated in the HiNorm treatment for M. cavernosa. Current models for apoptosis in corals suggest that host cell populations are represented by cells where apoptosis is suppressed and in others it is promoted (Ainsworth et al., 2011; Tchernov et al., 2011). Therefore, the observed increases in the pro-apoptotic Bax gene are not completely unexpected even for corals that have not bleached, show high functional integrity in the photosynthetic apparatus of their Symbiodiniaceae and low levels of oxidative stress as indicated by their relative expression of antioxidant genes.
When M. cavernosa is exposed to RCP 8.5 predictions for OW and OA a ∼40–50% decrease in rates of calcification occurs (Okazaki et al., 2017). Given the resistance to bleaching and mortality by M. cavernosa shown in this experiment, there is the potential to withstand RCP 6.0 effects on calcification, but that is likely to change significantly if the environment approaches RCP 8.5 conditions. Taken together, under near-term predicted climate change conditions of both OW and OA the phenotype of M. cavernosa, both its orange and brown morphs, is consistent with resistance to these climate change stressors. Given the recent history of multiple worldwide bleaching events (Eakin et al., 2019; Fordyce et al., 2019), M. cavernosa is a member of the Caribbean coral community whose abundance has been fairly consistent through time (e.g., Jackson et al., 2014), in spite of the overall low coral biodiversity in the Caribbean. The molecular phenotype described above for M. cavernosa from the Bahamas may also represent a strategy of “front-loading” based on previous exposure to OW and OA (sensu Barshis et al., 2013) where the transcripts or proteins of critical genes have already been upregulated in order to respond to future stress events. Given the degree of population differentiation observed in M. cavernosa between geographically separated sites (Brazeau et al., 2013) it is also possible that this population of M. cavernosa is uniquely resistant to the effects of OW and OA and the observed effects from this experiment are not generalizable to other populations of M. cavernosa.
The ecological demographics for M. cavernosa and the results presented here suggest that this coral is persistent, resistant and resilient to past and near future OW and OA conditions. This occurs in the absence of a highly variable environment where resilience to climate change stressors often occurs (Barshis et al., 2013), or in extreme environments where strong directional selection for thermally resistant phenotypes can also occur (Hume et al., 2014; Parker et al., 2020). This makes M. cavernosa a climate change winner in the near-term and reinforces the need to look at both the independent and interactive effects of OW and OA not only at the organismal level, but from a life-history perspective as well (Byrne and Przeslawski, 2013). In addition to studies that incorporate both realistic OW and OA conditions we must also include relevant irradiance, flow and nutrient conditions to fully understand a coral’s potential response to future increases in environmental stress.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Sequence Read Archive under BioProject accession number PRJNA590192.
Author Contributions
ML, JJ, and CF designed and performed the research. ML, MT, MP, and KM analyzed the samples and data. ML, MT, and MP wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Support for this research was provided by the NSF Biological Oceanography program (OCE-1231468 and 1437054) and the NOAA Ocean Exploration Program to ML. All research and sample export/import were done in compliance with the applicable laws of the Bahamas and the United States.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the staff of the Caribbean Marine Research Center, Lee Stocking Island, Bahamas and Marc Slattery, Elizabeth Kintzing, and Cole Easson for general support, help with sample collection and maintenance of the experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00728/full#supplementary-material
Footnotes
- ^ https://github.com/jrcunning/SymITS2
- ^ https://github.com/picrust/picrust2/wiki
- ^ http://comparative.reefgenomics.org/
References
Ainsworth, T. D., Heron, S. F., Ortiz, J. C., Mumby, P. J., Grech, A., Ogawa, D., et al. (2016). Climate change disables coral bleaching protection on the Great Barrier Reef. Science 352, 338–342. doi: 10.1126/science.aac7125
Ainsworth, T. D., Hoegh-Guldberg, O., Heron, S. F., Skirving, W. J., and Leggat, W. (2008). Early cellular changes are indicators of pre-bleaching thermal stress in the coral host. J. Exp. Mar. Biol. Ecol. 364, 63–71. doi: 10.1016/j.jembe.2008.06.032
Ainsworth, T. D., Wasmund, K., Ukani, L., Seneca, F., Yellowlees, D., Miller, D., et al. (2011). Defining the tipping point, A complex cellular life/death balance in corals in response to stress. Sci. Rep. 1:160.
Altieri, A. H., Harrison, S. B., Seemann, J., Collin, R., Diaz, R. J., and Knowlton, N. (2017). Tropical dead zones and mass mortalities on coral reefs. Proc. Natl. Acad. Sci. U.S.A. 114, 3660–3665. doi: 10.1073/pnas.1621517114
Anthony, K. R. N., Kline, D. I., Diaz-Pulido, G., Dove, S., and Hoegh-Guldberg, O. (2008). Ocean acidification causes bleaching and productivity loss in coral reef builders. Proc. Natl. Acad. Sci. U.S.A. 105, 17442–17446. doi: 10.1073/pnas.0804478105
Anthony, K. R. N., Marshall, P. A., Abdulla, A., Beeden, R., Bergh, C., Black, R., et al. (2015). Operationalizing resilience for adaptive coral reef management under global environmental change. Glob. Change Biol. 21, 48–61. doi: 10.1111/gcb.12700
Anthony, K. R. N., Maynard, J. A., Diaz-Pulido, G., Mumby, P. J., Marshall, P. A., Cao, L., et al. (2011). Ocean acidification and warming will lower coral reef resilience. Glob. Change Biol. 17, 1798–1808. doi: 10.1111/j.1365-2486.2010.02364.x
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Apprill, A. M., and Gates, R. D. (2007). Recognizing diversity in coral symbiotic dinoflagellate communities. Mol. Ecol. 16, 1127–1134. doi: 10.1111/j.1365-294x.2006.03214.x
Arif, C., Daniels, C., Bayer, T., Banguera-Hinestroza, E., Barbrook, A., Howe, C. J., et al. (2014). Assessing Symbiodinium diversity in scleractinian corals via next-generation sequencing-based genotyping of the ITS2 rDNA region. Mol. Ecol. 23, 4418–4433. doi: 10.1111/mec.12869
Baghdasarian, G., Osberg, A., Mihora, D., Putnam, H., Gates, R. D., and Edmunds, P. J. (2017). Effects of temperature and pCO2 on population regulation of Symbiodinium spp. in a tropical reef coral. Biol. Bull. 232, 123–139. doi: 10.1086/692718
Bahr, K. D., Jokiel, P. L., and Rodgers, K. S. (2016). Relative sensitivity of five Hawaiian coral species to high temperature and high-pCO2 conditions. Coral Reefs 35, 729–738. doi: 10.1007/s00338-016-1405-4
Baird, A. H., Bhagooli, R., Ralph, P. J., and Takahashi, S. (2009). Coral bleaching: the role of the host. Trends Ecol. Evol. 24, 16–20. doi: 10.1016/j.tree.2008.09.005
Baird, A. H., Madin, J. S., Álvarez-Noriega, M., Fontoura, L., Kerry, J. T., Kuo, C.-Y., et al. (2018). A decline in bleaching suggests that depth can provide a refuge from global warming in most coral taxa. Mar. Ecol. Prog. Ser. 603, 257–264. doi: 10.3354/meps12732
Baker, A. C., Glynn, P. W., and Riegl, B. (2008). Climate change and coral reef bleaching: an ecological assessment of long-term impacts, recovery trends and future outlook. Estuar. Coast. Shelf Sci. 80, 435–471. doi: 10.1016/j.ecss.2008.09.003
Banaszak, A. T., and Lesser, M. P. (2009). Effects of solar ultraviolet radiation on coral reef organisms. Photochem. Photobiol. Sci. 8, 1276–1294. doi: 10.1039/b902763g
Barbera, P., Kozlov, A. M., Czech, L., Morel, B., Darriba, D., Flouri, T., et al. (2019). EPA-ng: massive parallel evolutionary placement of genetic sequences. Syst. Biol. 68, 365–369. doi: 10.1093/sysbio/syy054
Barshis, D. J., Ladner, J. T., Oliver, T. A., Seneca, F. O., Traylor-Knowles, N., and Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Bay, R. A., and Palumbi, S. R. (2014). Multilocus adaptation associated with heat resistance in reef-building corals. Curr. Biol. 24, 2952–2956. doi: 10.1016/j.cub.2014.10.044
Bay, R. A., Rose, N. H., Logan, C. A., and Palumbi, S. A. (2017). Genomic models predict succesful coral adatation if future warming rates are reduced. Sci. Adv. 3:e1701413. doi: 10.1126/sciadv.1701413
Beere, H. M., and Green, D. R. (2001). Stress management- heat shock protein-70 and the regulation of apoptosis. Trends Cell Biol. 11, 6–10. doi: 10.1016/s0962-8924(00)01874-2
Bhagooli, R., and Hidaka, M. (2004). Photoinhibition, bleaching susceptibility and mortality in two scleractinian corals, Platygyra ryukyuensis and Stylophora pistillata,in response to thermal and light stress. Comp. Biochem. Physiol. A 137, 547–555. doi: 10.1016/j.cbpb.2003.11.008
Bhattacharya, D., Agrawal, S., Aranda, M., Baumgarten, S., Belcaid, M., Drake, J. L., et al. (2016). Comparative genomics explains the evolutionary success of reef-forming corals. eLife 5:e13288.
Bou-Abdallah, F., Chasteen, N. D., and Lesser, M. P. (2006). Quenching of superoxide radicals by green fluorescent protein. Biochim. Biophys. Acta 1760, 1690–1695. doi: 10.1016/j.bbagen.2006.08.014
Bouchard, J. N., and Yamasaki, H. (2008). Heat stress stimulates nitric oxide production in Symbiodinium microadriaticum: a possible linkage between nitric oxide and the coral bleaching phenomenon. Plant Cell Physiol. 49, 641–652. doi: 10.1093/pcp/pcn037
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Boyd, P. W., Lennartz, S. T., Glover, D. M., and Doney, S. C. (2014). Biological ramifications of climate-change-mediated oceanic multi-stressors. Nat. Clim. Chang. 5, 71–79. doi: 10.1038/nclimate2441
Brazeau, D. A., Lesser, M. P., and Slattery, M. (2013). Genetic structure in the coral, Montastraea cavernosa: assessing genetic differentiation among and within mesophotic reefs. PLoS One 8:e65845. doi: 10.1371/journal.pone.0065845
Brown, B. E., Downs, C. A., Dunne, R. P., and Gibb, S. W. (2002). Exploring the basis of thermotolerance in the reef coral Goniastrean aspera. Mar. Ecol. Prog. Ser. 242, 119–129. doi: 10.3354/meps242119
Byrne, M., and Przeslawski, R. (2013). Multistressor impacts of warming and acidification of the ocean on marine invertebrates’ life histories. Integr. Comp. Biol. 53, 582–596. doi: 10.1093/icb/ict049
Cacciapaglia, C., and van Woesik, R. (2015). Reef-coral refugia in a rapidaly changing ocean. Glob. Change Biol. 21, 2272–2282. doi: 10.1111/gcb.12851
Calderwood, S. K., and Gong, J. (2016). Heat shock proteins promote cancer: it’s a protection racket. Trends Biochem. 41, 311–323. doi: 10.1016/j.tibs.2016.01.003
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Castillo, K. D., Ries, J. B., Bruno, J. F., and Westfield, I. T. (2018). The reef-building coral Siderastrea siderea exhibits parabolic responses to ocean acidification and warming. Proc. R. Soc. B 281:20141856. doi: 10.1098/rspb.2014.1856
Cole, C., Finch, A. A., Hintz, C., Hintz, K., and Allison, N. (2018). Effects of seawater pCO2 and temperature on calcification and productivity in the coral genus Porites spp.: an exploration of potential interaction mechanisms. Coral Reefs 37, 471–481. doi: 10.1007/s00338-018-1672-3
Császár, N. B. M., Seneca, F. O., and van Oppen, M. J. H. (2009). Variation in antioxidant gene expression in the scleractinian coral Acropora millepora under laboratory thermal stress. Mar. Ecol. Prog. Ser. 392, 93–102. doi: 10.3354/meps08194
Cunning, R., Silverstein, R. N., and Baker, A. C. (2018). Symbiont shuffling linked to differential photochemical dynamics of Symbiodinium in three Caribbean reef corals. Coral Reefs 37, 145–152. doi: 10.1007/s00338-017-1640-3
Cunning, R., Yost, D. M., Guarinello, M. L., Putnam, H. M., and Gates, R. D. (2015). Variability of Symbiodinium communities in waters, sediments, and corals of thermally distinct reef pools in American Samoa. PLoS One 10:e0145099. doi: 10.1371/journal.pone.0145099
Davies, S. W., Ries, J. B., Marchetti, A., and Castillo, K. D. (2018). Symbiodinium functional diversity in the coral Siderastria siderea is influenced by thermal stress and reef environment, but not ocean acidification. Front. Mar. Sci. 5:150. doi: 10.3389/fmars.2018.00150
de Bakker, D. M., Meesters, E. H., Bak, R. P. M., Nieuwland, G., and van Duyl, F. C. (2016). Long-term shifts in coral communities on shallow to deep reef slopes of Curacao and Bonaire: Are there any winners? Front. Mar. Sci. 3:247. doi: 10.3389/fmars.2016.00247
DeCarlo, T. M., Harrison, H. B., Gajdzik, L., Alaguarda, D., Rodolfo-Metalpa, R., D’Olivo, J., et al. (2019). Acclimatization of massive reef-building corals to consecutive heatwaves. Proc. R. Soc. B 286:20190235. doi: 10.1098/rspb.2019.0235
Díaz-Almeyda, E. M., Prada, C., Ohdera, A. H., Moran, H., Civitello, D. J., Iglesias-Prieto, R., et al. (2018). Intraspecific and interspecific variation in thermotolerance and photoacclimation in Symbiodinium dinoflagellates. Proc. R. Soc. B 284:20171767. doi: 10.1098/rspb.2017.1767
Dickson, A. G., and Millero, F. J. A. (1987). Comparison of the equilibrium-constants for the dissociation of carbonic-acid in seawater media. Deep Sea Res. A 34, 1733–1743. doi: 10.1016/0198-0149(87)90021-5
Dickson, A. G., Sabine, C. L., and Christian, J. R. (2007). Guide to Best Practices for Ocean CO2 Measurements. Sidney: North Pacific Marine Science Organization.
Doney, S. C., Fabry, V. J., Feely, R. A., and Kleypas, J. A. (2009). Ocean acidification: the other CO2 problem. Annu. Rev. Mar. Sci. 1, 69–92.
Doney, S. C., Ruckelshaus, M., Duffy, E. J., Barry, J. P., Chan, F., English, C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37.
Donner, S. D., Skirving, W. J., Little, C. M., Oppenheimer, M., and Hoegh-Guldberg, O. (2005). Global assessment of coral bleaching and required rates of adaptation under climate change. Glob. Change Biol. 11, 2251–2265. doi: 10.1111/j.1365-2486.2005.01073.x
Dove, S. (2004). Scleractinian corals with photoprotective host pigments are hypersensitive to thermal bleaching. Mar. Ecol. Prog. Ser. 272, 99–116. doi: 10.3354/meps272099
Dove, S., Hoegh-Gulderg, O., and Lesser, M. (2006). All-protein chromophores isolated from corals, quench superoxide radicals. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 143:S132.
Downs, C. A., Mueller, E., Phillips, S., Fauth, J. E., and Woodley, C. M. (2000). A molecular biomaker system for assessing the health of coral (Montastraea faveolata) during heat stress. Mar. Biotechnol. 2, 533–544. doi: 10.1007/s101260000038
Dudgeon, S. R., Aronson, R. B., Bruno, J. F., and Precht, W. F. (2010). Phase shifts and stable states on coral reefs. Mar. Ecol. Prog. Ser. 413, 201–216. doi: 10.3354/meps08751
Eakin, M. C., Sweatman, H. P. A., and Brainard, R. E. (2019). The 2014-2017 global-scale bleaching event: insights and impacts. Coral Reefs 38, 539–545. doi: 10.1007/s00338-019-01844-2
Eddy, S. R. (2008). A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 4:e1000069. doi: 10.1371/journal.pcbi.1000069
Edmunds, P. J., Burgess, S. C., Putnam, H. M., Baskett, M. L., Bramanti, L., Fabina, N. S., et al. (2014). Evaluating the causal basis of ecological success within the scleractinia: an integral projection model approach. Mar. Biol. 161, 2719–2734. doi: 10.1007/s00227-014-2547-y
Edmunds, P. J., Comeau, S., Lantz, C., Andersson, A., Briggs, C., Cohen, A., et al. (2016). Integrating the effects of ocean acidification across functional scales on tropical coral reefs. Bioscience 66, 350–362. doi: 10.1093/biosci/biw023
Feder, M. E., and Hofmann, G. E. (1999). Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282. doi: 10.1146/annurev.physiol.61.1.243
Fitt, W., Gates, R., Hoegh-Guldberg, O., Grottoli, A., Gomez, M., Fisher, P., et al. (2009). Response of two species of Indo-Pacific corals, Porites cylindrica and Stylophora pistillata, to thermal stress: the host does matter in determining the tolerance of corals to bleaching. J. Exp. Mar. Biol. Ecol. 373, 102–110. doi: 10.1016/j.jembe.2009.03.011
Fitt, W. K., McFarland, F. K., Warner, M. E., and Chilcoat, G. C. (2000). Seasonal patterns of tissue biomass and densities of symbiotic dinoflagellates in reef corals and relation to coral bleaching. Limnol. Oceanogr. 45, 677–685. doi: 10.4319/lo.2000.45.3.0677
Fitt, W. K., and Warner, M. E. (1995). Bleaching patterns of four species of Caribbean reef corals. Biol. Bull. 189, 298–307. doi: 10.2307/1542147
Fordyce, A. J., Ainsworth, T. D., Heron, S. F., and Leggat, W. (2019). Marine heatwave hotspots in coral reef environments: physical drivers, ecophysiological outcomes, and impact upon structural complexity. Front. Mar. Sci. 6:498. doi: 10.3389/fmars.2019.00498
Gintert, B. E., Manzello, D. P., Enochs, I. C., Kolodziej, G., Carlton, R., Gleason, A. C. R., et al. (2018). Marked annual coral bleaching reselience of an inshore patch reef in the Florida Keys: a nugget of hope, aberrance, or last man standing? Coral Reefs 37, 533–547. doi: 10.1007/s00338-018-1678-x
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V., and Egozcue, J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2017.02224
Gorbunov, M. Y., Kolber, Z. S., Lesser, M. P., and Falkowski, P. G. (2001). Photosynthesis and photoprotection in corals. Limnol. Oceanogr. 46, 75–85. doi: 10.4319/lo.2001.46.1.0075
Grottoli, A. G., Martins, P. D., Wilkins, M. J., Johnston, M. D., Warner, M. E., Cai, W.-J., et al. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS One 13:e0191156. doi: 10.1371/journal.pone.0191156
Grottoli, A. G., Warner, M. E., Levas, S. J., Aschaffenburg, M. D., Schoepf, V., McGinley, M., et al. (2014). The cumulative impact of annual coral bleaching can turn some coral species winners into losers. Glob. Change Biol. 20, 3823–3833. doi: 10.1111/gcb.12658
Harborne, A. R., Rogers, A., Bozec, Y.-M., and Mumby, P. J. (2017). Multiple stressors and the functioning of coral reefs. Annu. Rev. Mar. Sci. 9, 445–468. doi: 10.1146/annurev-marine-010816-060551
Hausfather, Z., and Peters, G. P. (2020). Emissions-the ‘business as usual’ story is misleading. Nature 577, 618–620. doi: 10.1038/d41586-020-00177-3
Hernandez-Agreda, A., Gates, R. D., and Ainsworth, T. D. (2017). Defining the core microbiome in corals’ microbial soup. Trends Microbiol. 25, 125–140. doi: 10.1016/j.tim.2016.11.003
Hoadley, K. D., Pettay, D. T., Grottoli, A. G., Cai, W.-J., Melman, T. F., Schoepf, V., et al. (2015). Physiological response to elevated temperature and pCO2 varies across four Pacific coral species: understanding the unique host + symbiont response. Sci. Rep. 5:18371.
Hoegh-Guldberg, O., Mumby, P. J., Hooten, A. J., Steneck, R. S., Greenfield, P., Gomez, E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742.
Hoegh-Guldberg, O., Poloczanska, E. S., Skirving, W., and Dove, S. (2017). Coral reef ecosystems under climate change and ocean acidification. Front. Mar. Sci. 4:158. doi: 10.3389/fmars.2017.00158
Hughes, T. P., Anderson, K. D., Connolly, S. R., Heron, S. F., Kerry, J. T., Lough, J. M., et al. (2018). Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359, 80–83. doi: 10.1126/science.aan8048
Hughes, T. P., Baird, A. H., Bellwood, D. R., Card, M., Connolly, S. R., Folke, C., et al. (2003). Climate change, human impacts, and the resilience of coral reefs. Science 301, 929–933. doi: 10.1126/science.1085046
Hughes, T. P., Barnes, M. L., Bellwood, D. R., Cinner, J. E., Cumming, G. S., Jackson, J. B. C., et al. (2017). Coral reefs in the Anthropocene. Nature 546, 82–90.
Hughes, T. P., Graham, N. A., Jackson, J. B., Mumby, P. J., and Steneck, R. S. (2010). Rising to the challenge of sustaining coral reef resilience. Trends Ecol. Evol. 25, 633–642. doi: 10.1016/j.tree.2010.07.011
Hume, B. C. C., D’Angelo, C., Smith, E. G., Stevens, J. R., Burt, J., and Wiedenmann, J. (2014). Symbiodinium thermophilum sp. nov., a thermotolerant symbiotic alga prevalent in corals of the world’s hottest sea, the Persian/Arabian Gulf. Sci. Rep. 5:8562.
Iglesias-Prieto, R., Beltrán, V. H., LaJeunesse, T. C., Reyes-Bonilla, H., and Thomé, P. E. (2004). Different algal symbionts explain the vertical distribution of dominant reef corals in the eastern Pacific. R. Soc. Lond. B 271, 1757–1763. doi: 10.1098/rspb.2004.2757
Intergovernmental Panel on Climate Change [IPCC] (2007). “Climate change 2007: the physical science basis,” in Contribution of Working Group I to the 4th Assessment Report of the Intergovernmental Panel on Climate Change, eds S. Solomon, D. Qin, M. Manning, et al. (Cambridge: Cambridge University Press), 996.
Intergovernmental Panel on Climate Change [IPCC] (2014). in Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds Core Writing Team, R. K. Pachauri, and L. A. Meyer (Geneva: IPCC), 151.
Jackson, J. B. C., Donovan, M. K., Cramer, K. L., and Lam, V. V. (eds) (2014). Status and Trends of Caribbean Coral Reefs: 1970-2012. Gland: IUCN.
Jamieson, D., Chance, B., Cadenas, E., and Boveris, A. (1986). The relation of free radical production to hyperoxia. Annu. Rev. Physiol. 48, 703–719. doi: 10.1146/annurev.ph.48.030186.003415
Jarett, J. K., MacManes, M. D., Morrow, K. M., Pankey, M. S., and Lesser, M. P. (2017). Comparative genomics of color morphs in the coral Montastraea cavernosa. Sci. Rep. 7:16039.
Jin, Y. K., Lundgren, P., Lutz, A., Raina, J.-P., Howells, E. J., Paley, A. S., et al. (2016). Genetic markers for antioxidant capacity in a reef-building coral. Sci. Adv. 2:e1500842. doi: 10.1126/sciadv.1500842
Kao, H. T., Sturgis, S., DeSalle, R., Tsai, J., Davis, D., Gruber, D. F., et al. (2007). Dynamic regulation of fluorescent proteins from a single species of coral. Mar. Biotechnol. 9, 733–746. doi: 10.1007/s10126-007-9025-1
Kelmanson, I. V., and Matz, M. V. (2003). Molecular basis and evolutionary origins of color diversity in Great Star coral Montastraea cavernosa (Scleractinia: Faviida). Mol. Biol. Evol. 20, 1125–1133. doi: 10.1093/molbev/msg130
Kenkel, C. D., Aglyamova, G., Alamaru, A., Bhagooli, R., Capper, R., Cunning, R., et al. (2011). Development of gene expression markers of acute heat-light stress in reef building corals of the genus Porites. PLoS One 6:e26914. doi: 10.1371/journal.pone.0026914
Kenkel, C. D., Sheridan, C., Leal, M. C., Bhagooli, R., Castillo, K. D., Kurata, N., et al. (2014). Diagnostic gene expression biomakers of coral thermal stress. Mol. Ecol. Resour. 14, 667–678. doi: 10.1111/1755-0998.12218
Kroeker, K. J., Kordas, R. L., Crim, R., Hendriks, I. E., Ramajo, L., Singh, G. S., et al. (2013). Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob. Change Biol. 19, 1884–1896. doi: 10.1111/gcb.12179
Kvitt, H., Rosenfeld, H., and Tchernov, D. (2016). The regulation of thermal stress induced apoptosis in corals reveals high similarities in gene expression and function to higher animals. Sci. Rep. 6:30359.
Kvitt, H., Rosenfeld, H., Zandbank, K., and Tchernov, D. (2011). Regulation of apoptotic pathways by Stylophora pistillata (Anthozoa: Pocilloporidae) to survive thermal stress and bleaching. PLoS One 6:e28665. doi: 10.1371/journal.pone.0028665
Lahti, L., Shetty, S., Blake, T., and Salojarvi, J. (2017). Tools for Microbiome Analysis in R. Version 1.9.1. Available online at: http://microbiome.github.com/microbio
LaJeunesse, T. C., Parkinson, J. E., Gabrielson, P. W., Jeong, H. J., Reimer, J. D., Voolstra, C. R., et al. (2018). Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580. doi: 10.1016/j.cub.2018.07.008
Lesser, M. P. (1996). Exposure of symbiotic dinoflagellates to elevated temperatures and ultraviolet radiation causes oxidative stress and photosynthesis. Limnol. Oceanogr. 41, 271–283. doi: 10.4319/lo.1996.41.2.0271
Lesser, M. P. (2000). Depth-dependent photoacclimatization to solar ultraviolet radiation in the Caribbean coral Montastraea faveolata. Mar. Ecol. Prog. Ser. 192, 137–151. doi: 10.3354/meps192137
Lesser, M. P. (2004). Experimental Biology of coral reef ecosystems. J. Exp. Mar. Biol. Ecol. 300, 217–252. doi: 10.1016/j.jembe.2003.12.027
Lesser, M. P. (2006). Oxidative stress in marine environments: biochemistry and physiological ecology. Annu. Rev. Physiol. 68, 253–278. doi: 10.1146/annurev.physiol.68.040104.110001
Lesser, M. P. (2010). Interactions between stressors on coral reefs: analytical approaches, re-analysis of old data, and different conclusions. Reply to Dunne (2010). Coral Reefs 29, 615–619. doi: 10.1007/s00338-010-0625-2
Lesser, M. P. (2011a). “Coral bleaching: causes and mechanisms,” in Coral Reefs: An Ecosystem in Transition, eds Z. Dubinsky and N. Stambler (Amsterdam: Springer), 405–419. doi: 10.1007/978-94-007-0114-4_23
Lesser, M. P. (2011b). “Oxidative stress in tropical marine ecosystems,” in Oxidative Stress in Aquatic Ecosystems, eds D. Abele, J. P. Vázquez-Medina, and T. Zenteno-Savín (Chichester: John Wiley & Sons, Ltd), 9–19.
Lesser, M. P. (2013). Using energetic budgets to assess the effects of environmental stress on coral: Are we measuring the right things? Coral Reefs 32, 25–33. doi: 10.1007/s00338-012-0993-x
Lesser, M. P. (2019). Phylogenetic signature of light and thermal stress for the endosymbiotic dinoflagellates of corals (Family Symbiodiniaceae). Limnol. Oceanogr. 64, 1852–1863. doi: 10.1002/lno.11155
Lesser, M. P., Bythell, J. C., Gates, R. D., Johnstone, R. W., and Hoegh-Guldberg, O. (2007). Are infectious diseases really killing corals? J. Exp. Mar. Biol. Ecol. 346, 36–44. doi: 10.1016/j.jembe.2007.02.015
Lesser, M. P., and Farrell, J. H. (2004). Exposure to solar radiation increases damage to both host tissues and algal symbionts of corals during thermal stress. Coral Reefs 23, 367–377. doi: 10.1007/s00338-004-0392-z
Lesser, M. P., and Gorbunov, M. Y. (2001). Diurnal and bathymetric changes in chlorophyll fluorescence yields of reef corals measured in situ with a fast repetition rate fluorometer. Mar. Ecol. Prog. Ser. 212, 69–77. doi: 10.3354/meps212069
Lesser, M. P., Mazel, C. H., Gorbunov, M. Y., and Falkowski, P. G. (2004). Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science 305, 997–1000. doi: 10.1126/science.1099128
Lesser, M. P., and Slattery, M. (2011). Phase Shift to Algal Dominated Communities at Mesophotic Depths Associated with Lionfish (Pterois volitans) Invasion on a Bahamian Coral Reef. Biol. Invasions 13, 1855–1868. doi: 10.1007/s10530-011-0005-z
Lesser, M. P., Stochaj, W. R., Tapley, D. W., and Shick, J. M. (1990). Bleaching in coral reef anthozoans: effects of irradiance, ultraviolet radiation, and temperature on the activities of protective enzymes against active oxygen. Coral Reefs 8, 225–232. doi: 10.1007/bf00265015
Leutenegger, A., D’Angelo, C., Matz, M. V., Denzel, A., Oswald, F., Salih, A., et al. (2007). It’s cheap to be colorful. Anthozoans show a slow turnover of GFP-like proteins. FEBS J. 274, 2496–2505.
Levitan, O., Rosenber, G., Setlik, I., Setlikova, E., Klepetar, J., Prasil, O., et al. (2007). Elevated CO2 enhances nitrogen fixation and growth in the marine cyanobacterium Trichodesmium. Glob. Change Biol. 13, 531–538.
Logan, C. A., Dunne, J. P., Eakin, C. M., and Donner, S. D. (2014). Incorporating adaptive responses into future projections of coral bleaching. Glob. Change Biol. 20, 125–139.
Louca, S., and Doebeli, M. (2018). Efficient comparative phylogenetics on large trees. Bioinformatics 34, 1053–1055.
Louis, Y. D., Bhagooli, R., Kenkel, C. D., Baker, A. C., and Dyall, S. D. (2017). Gene expression biomarkers of heat stress in scleractinian corals: promises and limitations. Comp. Biochem. Physiol. C 191, 63–77.
Loya, Y., Sakai, K., Yamazato, K., Nakano, Y., Sambali, H., and van Woesik, R. (2001). Coral bleaching: the winners and the losers. Ecol. Lett. 4, 122–131.
Manzello, D., Warner, M., Stabenau, E., Hendee, J., Lesser, M., and Jankulak, M. (2009). Remote monitoring of chlorophyll fluorescence in two reef corals during the 2005 bleaching event at Lee Stocking Island, Bahamas. Coral Reefs 28, 209–214.
Matz, M. V., Treml, E. A., Aglyamova, G. V., and Bay, L. K. (2018). Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral. PLoS Genet. 14:e1007220. doi: 10.1371/journal.pgen.1007220
Mazel, C. H., Lesser, M. P., Gorbunov, M. Y., Barry, T. M., Farrell, J. H., Wyman, K. D., et al. (2003). Green-fluorescent proteins in Caribbean corals. Limnol. Oceanogr. 48, 402–411.
McClenachan, L., O’Connor, G., Neal, B. P., Pandolfi, J. M., and Jackson, J. B. C. (2017). Ghost reefs: nautical charts document large spatial scale of coral reef loss over 240 years. Sci. Adv. 3:e1603155.
McGinty, E. S., Pieczonka, J., and Mydlarz, L. D. (2012). Variations in reactive oxygen release and antioxidant activity in multiple Symbiodinium types in response to elevated temperature. Microb. Ecol. 64, 1007–1012.
McLachlan, R. H., Price, J. T., Solomon, S. L., and Grottoli, A. G. (2020). Thirtyn years of coral heat-stress experiments: a review of methods. Coral Reefs 39, 885–902.
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Mehrbach, C., Culberso, C., Hawley, J., and Pytkowic, R. (1973). Measurement of apparent dissociation constants of carbonic-acid in seawater at atmospheric pressure. Limnol. Oceanogr. 18, 897–907.
Morikawa, M. K., and Palumbi, S. R. (2019). Using naturally occurring climate reselient corals to construct bleaching-resistant nurseries. Proc. Natl. Acad. Sci. U.S.A. 116, 10586–10591.
Morrall, C. E., Galloway, T. S., Trapido-Rosenthal, H. G., and Depledge, M. H. (2000). Characterization of nitric oxide synthase activity in the tropical sea anemone Aiptasia pallida. Comp. Biochem. Physiol. B 125, 483–491.
Morrow, K. M., Muller, E., and Lesser, M. P. (2018). “How does the coral microbiome cause, respond to, or modulate the bleaching process?” in Coral Bleaching, Springer International Publishing, Vol. 233, eds M. J. H. van Oppen and J. M. Lough (Cham: Springer), 153–188.
Muscatine, L. (1990). “The role of symbiotic algae in carbon and energy flux in coral reefs,” in Ecosystems of the World, ed. Z. Zubinsky (Amsterdam: Elsevier), 75–87.
Nakamura, M., Morita, M., Kurihara, H., and Mitarai, S. (2012). Expression of hsp70, hsp90 and hsf1 in the reef coral Acropora digitifera under prospective acidified conditions over the next several decades. Biol. Open 1, 75–81.
Norström, A. V., Nyström, M., Lokrantz, J., and Folke, C. (2009). Alternative states on coral reefs: beyond coral–macroalgal phase shifts. Mar. Ecol. Prog. Ser. 376, 295–306.
Nyström, M., Folke, C., and Moberg, F. (2000). Coral reef disturbance and resilience in a human-dominated environment. Trends Ecol. Evol. 15, 413–417.
Okazaki, R. R., Towle, E. K., van Hooidonk, R., Mor, C., Winter, R. N., Piggot, A. M., et al. (2017). Species-specific responses to climate change and community composition determine future calcification rates of Florida Keys reefs. Glob. Change Biol. 23, 1023–1035.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., Minchin, O. R., et al. (2019). vegan: Community Ecology Package. R package version 2.4-3. Available online at: https://CRAN.R-project.org/package=vegan
Palmer, C. V., Modi, C. K., and Mydlarz, L. D. (2009). Coral fluorescent proteins as antioxidants. PLoS One 4:e7298. doi: 10.1371/journal.pone.0007298
Palumbi, S. R., Barshis, D. J., Traylor-Knowles, N., and Bay, R. A. (2014). Mechanisms of reef coral resistance to future climate change. Science 344, 895–898.
Pandolfi, J. M., Connolly, S. R., Marshall, D. J., and Cohen, A. L. (2011). Projecting coral reef futures under global warming and ocean acidification. Science 333, 418–422.
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414.
Parker, K. E., Ward, J. O., Eggleston, E. M., Federov, E., Parkinson, J. E., Dahlgren, C. P., et al. (2020). Characterization of a thermally tolerant Orbicella faveolata reef in Abaco, The Bahamas. Coral Reefs 39, 675–685.
Perez, S., and Weis, V. (2006). Nitric oxide and cnidarian bleaching: an eviction notice mediates breakdown of a symbiosis. J. Exp. Biol. 209, 2804–2810.
Pernice, M., Dunn, S. R., Miard, T., Dufour, S., Dove, S., and Hoegh-Guldberg, O. (2011). Regulation of apoptotic mediators reveals dynamic responses to thermal stress in the reef building coral Acropora millepora. PLoS One 6:e16095. doi: 10.1371/journal.pone.0016095
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45.
Putnam, H. M., Barott, K. L., Ainsworth, T. D., and Gates, R. D. (2017). The vulnerability and resilience of reef-building corals. Curr. Biol. 27, R528–R540.
Robbart, M. L., Peckol, P., Scordills, S. P., Curran, H. A., and Brown-Saracino, J. (2004). Population recovery and differential heat shock protein expression for the corals Agaricia agaricites and A. tenuifolia in Belize. Mar. Ecol. Prog. Ser. 283, 151–160.
Robbins, L. L., Hansen, M. E., Kleypas, J. A., and Meylan, S. C. (2010). CO2calc—A User-Friendly Seawater Carbon Calculator for Windows, Max OS X, and iOS (iPhone): U.S. Geological Survey Open-File Report 1280. Reston, VA: U.S. Geological Survey.
Rodolfo-Metalpa, R., Houlbréque, F., Tambutté, É., Boisson, F., Baggini, C., Patti, F. P., et al. (2011). Coral and mollusk resistance to ocean acidification adversely affected by warming. Nat. Clim. Change 1, 308–312.
Rogelj, J., Meinshausen, M., and Knutti, R. (2012). Global warming under old and new scenarios using IPCC climate sensitivity estimates. Nat. Clim. Change 2, 248–253.
Rosic, N. N., Pernice, M., Dove, S., Dunn, S., and Hoegh-Guldberg, O. (2011). Gene expression profiles of cytosolic heat shock proteins Hsp and Hsp90 from symbiotic dinoflagellates in response to thermal stress: possible implications for coral bleaching. Cell Stress Chaperones 16, 69–80.
Saragosti, E., Tchernov, D., Katsir, A., and Shaked, Y. (2010). Extracellular production and degradation of superoxide in the coral Stylophora pistillata and cultured Symbiodinium. PLoS One 5:e12508. doi: 10.1371/journal.pone.0012508
Scheufen, T., Iglesias-Prieto, R., and Enríquez, S. (2017). Changes in the number of symbionts and Symbiodinium cell pigmentation modulate differentially coral light absorption and photosynthetic performance. Front. Mar. Sci. 4:309. doi: 10.3389/fmars.2017.00309
Seutin, G., White, B. N., and Boag, P. T. (1991). Preservation of avian blood and tissue samples for DNA analyses. Can. J. Zool. 69, 82–90.
Silverstein, R. N., Cunning, R., and Baker, A. C. (2015). Change in algal symbiont communities after bleaching, not prior heat exposure, increases heat tolerance of reef corals. Glob. Change Biol. 21, 236–249.
Silverstein, R. N., Cunning, R., and Baker, A. C. (2017). Tenacious D: Symbiodinium in clade D remain in reef corals at both high and low temperature extremes despite impairment. J. Exp. Biol. 220, 1192–1196.
Skirving, W. J., Heron, S. F., Marsh, B. L., Liu, G., De La Cour, J. L., Geiger, E. F., et al. (2019). The relentless march of mass coral bleaching: a global perspective of changing heat stress. Coral Reefs 38, 547–557.
Skutnik, J. (2016). Examining the Effect of Climate Change on the Upper Mesophotic Coral Montastrea cavernosa (Linnaeus 1767). Master’s thesis, Grand Valley State University, Allendale, MI, 103.
Smith, N. P. (2001). Weather and hydrographic conditions associated with coral bleaching: Lee Stocking Island, Bahamas. Coral Reefs 20, 415–422.
Smith, T. B., Brandt, M. E., Calnan, J. M., Nemeth, R. S., Blondeau, J., Kadison, E., et al. (2013). Convergent mortality responses of Caribbean coral species to seawater warming. Ecosphere 4:art87.
Somero, G. N. (2010). The physiology of climate change: how potentials for acclimatization and genetic adaptation will determine ‘winners’ and ‘losers’. J. Exp. Biol. 213, 912–920.
Suggett, D. J., Warner, M. E., and Leggat, W. (2017). Symbiotic dinoflagellate functional diversity mediates coral survival under ecological crisis. Trends Ecol. Evol. 32, 735–745.
Suggett, D. J., Warner, M. E., Smith, D. J., Davey, P., Hennige, S., and Baker, N. R. (2008). Photosynthesis and production of hydrogen peroxide by Symbiodinium (Pyrrhophyta) phylotypes with different thermal tolerances. J. Phycol. 44, 948–956.
Takahashi, S., Nakamura, T., Sakamizu, M., van Woesik, R., and Yamasaki, H. (2004). Repair machinery of symbiotic photosynthesis as the primary target of heat stress for reef-building coral. Plant Cell Physiol. 45, 251–255.
Tchernov, D., Gorbunov, M. Y., de Vargas, C., Yadav, S. N., Milligan, A. J., Häggblom, M., et al. (2004). Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proc. Natl. Acad. Sci. U.S.A. 101, 13531–13535.
Tchernov, D., Kvitt, H., Haramaty, L., Bibby, T. S., Gorbunov, M. Y., Rosenfeld, H., et al. (2011). Apoptosis and the selective survival of host animals following thermal bleaching in zooxanthellate corals. Proc. Natl. Acad. Sci. U.S.A. 24, 9905–9909.
Thomas, L., Rose, N. H., Bay, R. A., López, E. H., Morikawa, M. K., Ruiz-Jones, L., et al. (2018). Mechanisms of thermal tolerance in reef-building corals across a fine-grained environmental mosaic: lessons from Ofu, American Samoa. Front. Mar. Sci. 4:434. doi: 10.3389/fmars.2017.00434
Trapido-Rosenthal, H., Zielke, S., Owen, R., Buxton, L., Boeing, B., Bhagooli, R., et al. (2005). Increased zooxanthellae nitric oxide synthase activity is associated with coral bleaching. Biol. Bull. 208, 3–6.
van Woesik, R., Sakai, K., Ganase, A., and Loya, Y. (2011). Revisiting the winners and the losers a decade after coral bleaching. Mar. Ecol. Prog. Ser. 434, 67–76.
Wangpraseurt, D., Larkum, A. W. D., Ralph, P. J., and Kühl, M. (2012). Light gradients and optical microniches in coral tissues. Front. Microbiol. 3:316. doi: 10.3389/fmicb.2012.00316
Warner, M. E., Lesser, M. P., and Ralph, P. (2010). “Chlorophyll fluorescence in reef building corals,” in Chlorophyll a Fluorescence in Aquatic Sciences: Methods and Applications, eds D. Suggett, O. Prasil, and O. Borowitzka (Cham: Springer), 209–222.
Webster, N. S., Negri, A. P., Botté, E. S., Laffy, P. W., Flores, F., Noonan, S., et al. (2016). Host-associated coral reef microbes respond to cumulative pressures of ocean warming and ocean acidification. Sci. Rep. 6: 19324.
Weis, V. M. (2008). Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. J. Exp. Biol. 211, 3059–3066.
Williams, G. J., Graham, N. A. J., Jouffray, J.-P., Norström, A. V., Nyström, M., Gove, J. M., et al. (2019). Coral reef ecology in the Anthropocene. Funct. Ecol. 33, 1014–1022.
Wright, R. M., Mera, H., Kenkel, C. D., Nayfa, M., Bay, L. K., and Matz, M. V. (2019). Positive genetic associations among fitness traits support evolvability of a reef-building coral under multiple stressors. Glob. Change Biol. 25, 3294–3304.
Ye, Y., and Doak, T. G. (2009). A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol. 5:e1000465. doi: 10.1371/journal.pcbi.1000465
Keywords: coral reefs, climate change, thermal stress, ocean acidification, apoptosis, coral bleaching
Citation: Lesser MP, Jarett JK, Fiore CL, Thompson MM, Pankey MS and Macartney KJ (2020) A New “Business as Usual” Climate Scenario and the Stress Response of the Caribbean Coral Montastraea cavernosa. Front. Mar. Sci. 7:728. doi: 10.3389/fmars.2020.00728
Received: 15 March 2020; Accepted: 11 August 2020;
Published: 16 September 2020.
Edited by:
Stefano Goffredo, University of Bologna, ItalyReviewed by:
Claudia Pogoreutz, University of Konstanz, GermanyEslam O. Osman, Pennsylvania State University (PSU), United States
Chris Langdon, University of Miami, United States
Copyright © 2020 Lesser, Jarett, Fiore, Thompson, Pankey and Macartney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael P. Lesser, bXBsQHVuaC5lZHU=
†Present address: Jessica K. Jarett, AnimalBiome, Oakland, CA, United States; Cara L. Fiore, Department of Biology, Appalachian State University, Boone, NC, United States
 Michael P. Lesser
Michael P. Lesser Jessica K. Jarett
Jessica K. Jarett Cara L. Fiore
Cara L. Fiore Megan M. Thompson
Megan M. Thompson M. Sabrina Pankey
M. Sabrina Pankey Keir J. Macartney1
Keir J. Macartney1