
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 07 July 2020
Sec. Marine Molecular Biology and Ecology
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.00554
 Md Asaduzzaman1*
Md Asaduzzaman1* Md Abdul Wahab2
Md Abdul Wahab2 Md Moshiur Rahman3
Md Moshiur Rahman3 Mariom4
Mariom4 Md Nahiduzzaman2
Md Nahiduzzaman2 Md Jalilur Rahman2
Md Jalilur Rahman2 Barun Kanti Roy1
Barun Kanti Roy1 Michael J. Phillips5
Michael J. Phillips5 Li Lian Wong6*
Li Lian Wong6*Migration of an anadromous fish to heterogeneous environment continuously enforces a selective pressure that incorporates a wide range of life-history strategies by which individuals adapt to the prevailing conditions. Therefore, we used the landmark-based morphometric truss network method and nextRAD genotyping-based putatively adaptive SNP loci dataset to know how the anadromous Hilsa shad (Tenualosa ilisha) morpho-genetically adapt to the heterogeneous habitats across their migratory routes by investigating 300 individuals, collected from nine strategic sampling sites covering sea, estuary, and upstream freshwater rivers. Different multivariate and clustering analyses revealed that the riverine populations were morphometrically wider (broad type) than the estuarine and marine populations (slender type). In the case of riverine population, the north-western turbid population (the Padma and Jamuna rivers) had wider body depth than the north-eastern clear water population (the Meghna river). The linear model and spatial multivariate analyses further revealed that the outcomes of morphometric dataset were in complete concordance with the results of putatively adaptive SNP loci dataset for different Hilsa shad populations. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis of the 36 genes, which are encoded by the putative adaptive SNP loci, supported the presence of multiple genes involved in the growth, metabolism, homeostasis and osmoregulation related functions. Several non-mutually exclusive hypotheses were attributed to explain the observation of continuum differentiation at both phenotypes and genotypes: (i) the genetic variation largely determines the morphometric discrimination, (ii) the interactive evolutionary processes and salinity predominantly contribute to the morphogenetic difference between the marine-estuarine and the freshwater riverine populations, (iii) environmental heterogeneity largely influences the genotypes leading to the phenotypic plasticity, and (iv) the local environmental heterogeneity may contribute to the morphogenetic divergence between the riverine populations. Finally, we concluded that the genetic adaptation, phenotypic plasticity and interactive ecological and evolutionary consequences jointly determine the morphogenetic divergence of Hilsa shad. The interaction of all of these forces and their relative strength in heterogeneous environments, however, made it rather challenging to determine the most probable selective pressure, which has shaped the Hilsa shad morphogenetic divergence across their diverse migratory habitats.
Over the last few decades, many innovative studies have revealed that environmental changes have influenced the morpho-genetic alterations in both plant and animal kingdom (Bradshaw and Holzapfel, 2006; Borges, 2008; Hoffmann et al., 2015). Evidences from different aquatic organisms and fishes have suggested that aquatic animals possess special abilities to swiftly adapt to the changing environments (Lee and Gelembiuk, 2008; Vera et al., 2016). Migration of an anadromous fish from marine to freshwater environment incorporates a wide range of life-history strategies by which individuals adapt to the selective pressure driven by the different scales of temporal and spatial environmental heterogeneity (reviewed in Bernatchez, 2016). Therefore, anadromous fishes offer good model systems to investigate how the fish cope with and respond to the selective pressure driven by the environmental heterogeneity (Bernatchez, 2016; Tamario et al., 2019). As a short-term response, anadromous fish may acclimate to the changing environmental conditions through phenotypic plasticity, i.e., by expressing special adaptive phenotypes (Bernatchez, 2016). However, the long-term and ultimate response to the environmental heterogeneity is to adapt to the existing environmental conditions, which infers evolutionary (genetic) changes in response to the shifting conditions (Bernatchez, 2016; Tamario et al., 2019). In general, the phenotypic discriminations are largely determined by the genetic differences and are affected by the interaction of ecological and evolutionary systems in response to the spatio-temporal heterogenic environmental conditions (Tamario et al., 2019). Given such complex adaptation strategies of anadromous fish, there is a need for a better understanding about the pattern of morpho-genetic variation in response to their migratory behavior in heterogeneous environments. The major overarching question for the scientists over many years is how the anadromous fish respond to the environmental heterogeneity and cope with the changing environmental conditions when they migrate from the marine waters to the freshwater rivers for breeding and/or feeding purposes.
The Hilsa shad (Tenualosa ilisha) is an anadromous fish and its marine distribution ranges from the Persian Gulf to the Arabian Sea (west coast of India) and the Bay of Bengal (Blaber, 2000; Arai and Amalina, 2014). Along the Bay of Bengal region, they migrate from the open sea to the upstream Indo-Gangetic and Brahmaputra freshwater riverine network for breeding and nursing of the juveniles, and then return to the original habitats (Islam et al., 2016; Rahman et al., 2018; Hossain et al., 2019). They are the most dominating food fish in the countries within the Bay of Bengal region (BOBLME, 2012; Rahman et al., 2018), which contributed around 44% of the captured fish production, 10.5% of the total annual fish production, and over 1% of the total gross domestic product of Bangladesh (Department of Fisheries [DOF], 2018) and to a limited extent in both India and Myanmar (BOBLME, 2012; Rahman et al., 2018; Hossain et al., 2019). As an anadromous fish, Hilsa shad migrates between water bodies with different salinities, turbidities, food resources, and other ecological factors (Hossain et al., 2016), which may result in divergence in morphological traits and reproductive strategies among the populations for local adaptation as reported in other anadromous fish species (Palstra et al., 2007; Paez et al., 2010). As described in other studies (Kawecki and Ebert, 2004; Bernatchez, 2016), the migrating population of Hilsa shad may evolve in these heterogenic environmental conditions for thousands of years that have shaped distinct genetic composition, and local adaptation of genetically based phenotypic traits among populations for superior fitness. Earlier works have consistently revealed that the morpho-genetic variation of fish due to the environmentally driven local adaptation contributes to more stable populations and reduced their risk of extinction (Forsman and Wennersten, 2016; Bernatchez, 2016; Des Roches et al., 2018; Tamario et al., 2019). Although the annual production of this species has been increased and reached to more than 0.5 million tons in recent years due to higher marine catch, their production from the upstream freshwater rivers has alarmingly decreased (BOBLME, 2012; Food and Agricultural Organization [FAO], 2017; Department of Fisheries [DOF], 2018). The upstream migrating population are under threats because of habitat modification by siltation and upstream dams constructed on its migratory course, fragmentations and destructions of breeding and nursery habitats, pollutions, climatic variability, and overexploitations of juveniles and broods (BOBLME, 2012; Miah, 2015; Dutton et al., 2018; Hossain et al., 2019). Apart from the instantaneous negative impacts linked with declining Hilsa shad populations in the upstream rivers, these may in turn affect the size-structure, recruitment and dynamics of the populations as stated in other fish species (Kuparinen and Merilä, 2007; Lowerre-Barbieri et al., 2017). Each of the Hilsa shad ecotypes adapted to the local environment may, therefore, need a unique management scheme to enable their conservation because of environmental changes, anthropogenic disturbance, habitat modification, and overexploitation (Asaduzzaman et al., 2020). Therefore, a thorough understanding about how Hilsa shad populations have morpho-genetically varied and adapted in the face of environmental changes along their migratory routes is crucial for their conservation, management and sustainable exploitation. Morphological studies of fishes are important in the viewpoints of ecology, conservation, evolution and stock management (AnvariFar et al., 2011). Therefore, the morphometric discrimination of anadromous Hilsa shad among different migratory habitats and ecological gradients would be applicable to investigate short-term environmental variation (Tzeng, 2004b; Dwivedi, 2019a, b), and this could provide a basis for stock structure (Cadrin, 2000; Tzeng, 2004a). Among the various methods, the morphometric study using landmark-based truss network system is a quantitative and effective method to illustrate the comprehensive information about the fish shape (Cavalcanti et al., 1999). This illustration applies a precise morphological landmarks that encompasses across the entire fish and depicts geo-morphometric, which stances no limitation to indicate the variation and localization of shape changes (Sen et al., 2011). Therefore, this landmark-based truss network method has been used in this study to discriminate the morphometric shape of the Hilsa shad population along the ecological gradients of their migratory habitats.
While a significant number of studies have demonstrated morphometric changes in natural populations to the local environmental adaptation, evidence is required to demonstrate that the species may also genetically evolve to these heterogenic environmental conditions. In contrast to morphometric study, the genetic discrimination of anadromous fish populations from heterogenic environments is imponderable because of their large population size, increased rate of gene flow and high connection among the divergent populations (Hess et al., 2012; Carreras et al., 2016; Asaduzzaman et al., 2020). However, the recent development of next-generation sequencing (NGS) based nextRAD method is widely used to genotype huge numbers of single nucleotide polymorphisms (SNPs) loci in numerous individuals at a comparatively cheaper cost to expose the molecular basis of local adaptation for anadromous fish. After finalizing the SNPs loci datasets through subsequent filtering process, various outlier tests can be applied as a means of discovering putatively adaptive markers to genetically discriminate anadromous fish population in response to their spatial environmental heterogeneity (Hess et al., 2012; Drinan et al., 2018).
Earlier studies of Hilsa shad population structure in Bangladesh waters have used morphometric (Quddus et al., 1984; Rahman et al., 1997) and neutral marker-based genetic approaches (Rahman and Naevdal, 2000; Salini et al., 2004; Ahmed et al., 2004; Mazumder and Alam, 2009; Brahmane et al., 2013; Behera et al., 2015), but given inconclusive results. Subsequently, our recent studies have applied the nextRAD approaches to resolve the earlier inconclusive outcomes on their population genetic structure throughout their diverse migratory environments (Asaduzzaman et al., 2019, 2020). Although the population genetic structure has been resolved recently, the degree of morphometric discrimination of Hilsa shad population in response to environmental heterogeneity along their migratory routes in Bangladesh waters has remained obscure until now. Moreover, it is unknown whether the morphological phenotypic variation of Hilsa shad corresponds with their fine-scale genetic divergence for local adaptation in spatial environmental heterogeneity. Therefore, the morpho-genetic variation of anadromous Hilsa shad was compared through different multivariate approaches by using putatively adaptive SNP loci dataset, which were previously identified by applying different outlier approaches (Asaduzzaman et al., 2020), and 22 truss networks morphometric distances on the same fish collected from the spatial heterogenic habitats.
During the commencement of southwest monsoon (July–August), a total of 300 individuals of T. ilisha were collected from the nine fixed sites of five important habitats that they mostly use during migration, which include sea, estuary, and major freshwater river systems in Bangladesh (Figure 1). Based on the annual variations of the water quality parameters, these habitats showed environmental heterogeneity in terms of salinity and suspended sediment concentrations (Supplementary Table S1). Hilsa shad were caught by the government permitted gillnets (average length was 1179.3 ± 917 m and mesh size was 6.5 ± 1.6 cm) with hiring the local fishermen net and mechanized boat facilities. For the present study, a total of 60 individuals from the marine habitat (two collection sites), 30 individuals from estuarine habitat (one collection site), 60 individuals from the Meghna and its upper tributary rivers (two collection sites), 60 individuals from the Jamuna river (two collection sites), and 90 individuals from Padma river (two collection sites) were collected. Except the estuarine habitats, T. ilisha were collected from both the lower and upper part of each habitat. Immediately after collection, each fish was marked with an individual identification number and then a portion of the dorsal fin was taken and preserved in absolute ethanol for the nextRAD genotyping sequencing. Initially, all the collected Hilsa shad samples were grouped according to their collection habitats, i.e., (1) sea, (2) estuary, (3) the Meghna river, (4) the Jamuna river, and (5) the Padma river. Subsequently, these five strategic habitats were further divided into three categories based on the salinity levels of the collection sites such as (1) marine (salinity 30.3–33.4 ppt), (2) estuarine (salinity 2.5 to 20.6 ppt) and (3) riverine habitats (salinity < 1 ppt). Furthermore, all riverine habitats were divided into two categories based on the annual variation of suspended sediment concentration (Barua, 1990; Allison, 1998; Sarker et al., 2003). These include (1) the western turbid rivers which consist of the Jamuna and Padma rivers (SSC-190 to 1600 mg/L) and (2) the eastern clear rivers which consist of the Meghna and its upper tributary the Surma-Kushiara rivers (20–750 mg/L). Hilsa shad does not have the endangered or protected status, and do not require the collection permits for non-commercial purposes in the sampling locations except during the banning periods of 22 days of their peak spawning season. However, the non-surgical tissue sampling was conducted in accordance with the animal care protocol as approved by the Chattogram Veterinary and Animal Sciences University’s Animal Care and Biosafety Committee, Bangladesh.
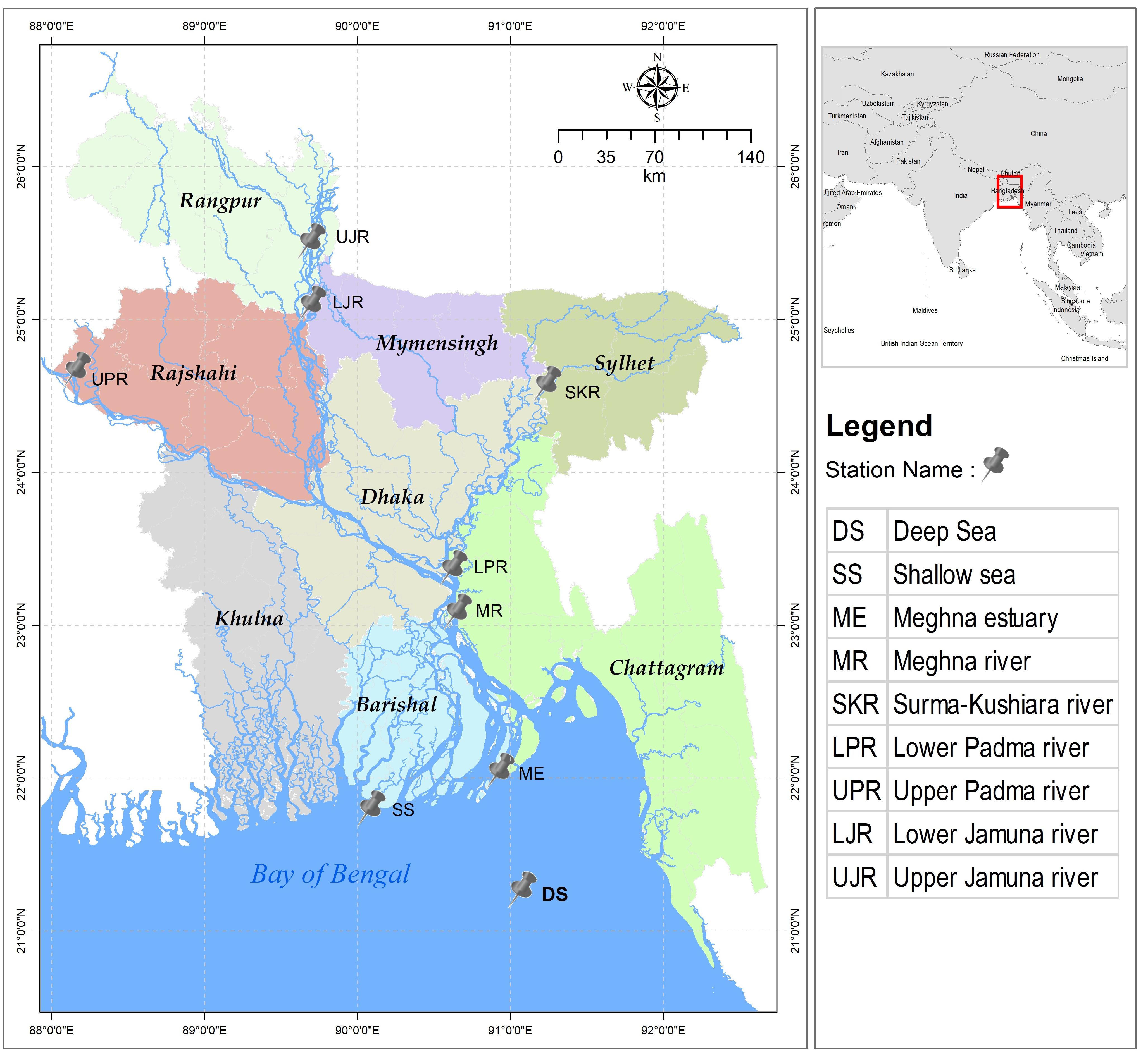
Figure 1. Map indicating the nine sampling sites of five heterogenous habitats of the anadromous Hilsa shad Tenualosa ilisha in Bangladesh across its diverse migratory habitats, including the Bay of Bengal (SS and DS), the Meghna estuary (ME), the Meghna river and its upper tributary (MR and SKR), the Padma river (LPR and UPR) and the Jamuna river (LJR and UJR).
Immediately after collection, a clear photograph of each fish was taken under a standard lighting condition by placing the fish on a laminated graph paper. A DSLR digital camera (Canon EOS Kiss X6i) was used to take the photographs from a fixed distance of 36 cm. The image of each fish also included an identification number so that the subsequent analyses of morphometric measurement can be blindly performed for all collection sites. The raw uncompressed images were imported into the SigmaScan Pro 5.0 software for the measurement of standard length and twelve landmarks delineating 22 truss distances of all fish (Supplementary Table S2 and Figure 2). For all the collection sites, a fixed ratio of female and male (6:4) Hilsa shad were used for all collection sites.
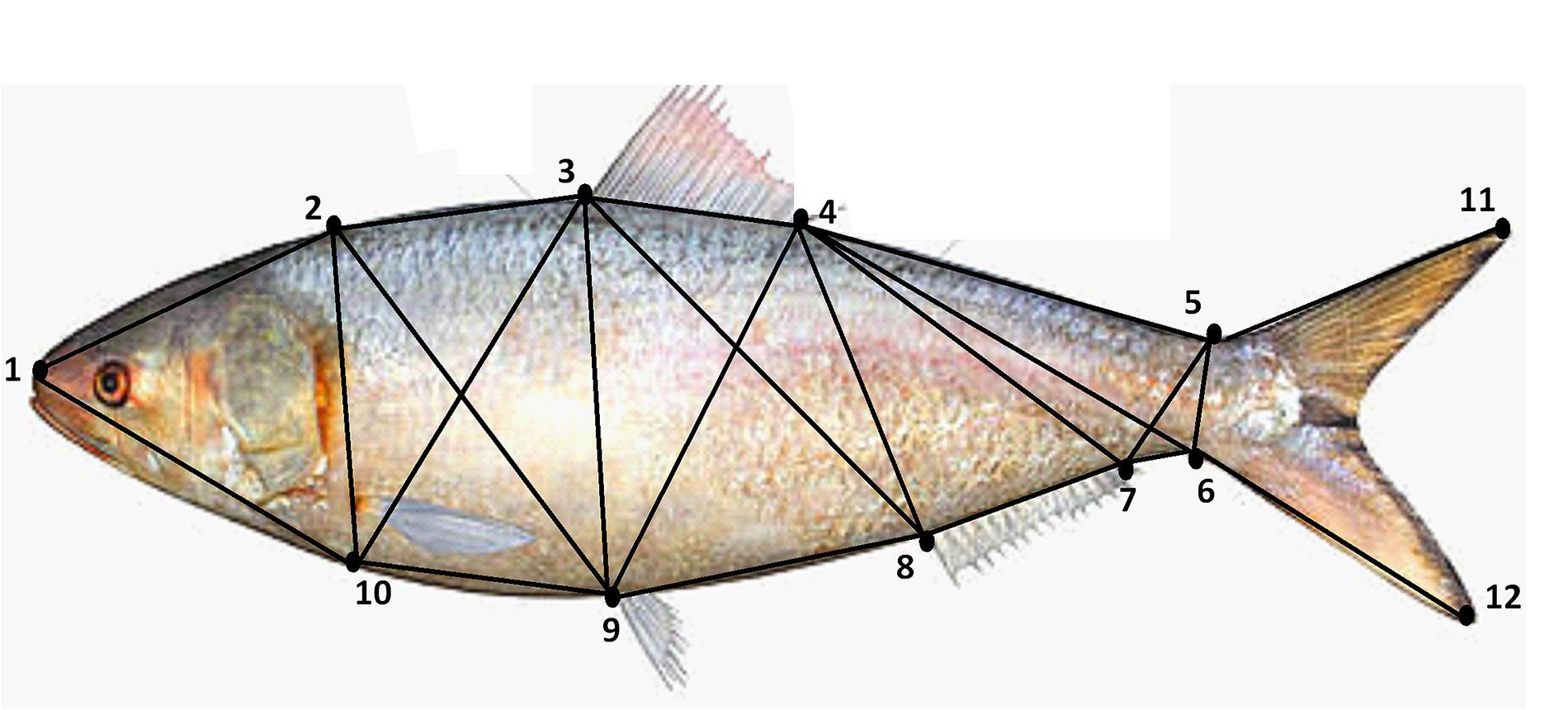
Figure 2. Location of the landmarks used for the morphometric shape analysis of Hilsa shad Tenualosa ilisha. In the study, 12 landmark points delineating 22 truss morphometric distances were superimposed on the body for shape analysis.
For ensuring the morphological variations were due to the body shape differences but not to the relative sizes of the fish, the size effects of the Hilsa shad samples were eliminated from the dataset before executing different statistical approaches. Therefore, size-dependent variations from these 22 truss morphological measurements were eliminated by employing an algometric method followed by Elliot et al. (1995):
Where, Madj is the size adjusted measurement of different morphometric length, M is the original measurement of different morphometric lengths, Ls is the overall mean of the standard length for all Hilsa shad fishes from all samples in each analysis, Lo is the standard length of the Hilsa shad, and b is the estimated value for each character from the observed data as the slope of the regression of log M on log Lo using all Hilsa shad fish from both groups. The size adjusted values derived from the allometric method were further validated by employing the significance testing of the correlation between standard length and the transformed variables (Turan, 1999).
The R version 3.6.1 (R Development Core Team, 2019) was used for performing all analyses of morphometric variation among different habitats. The “car” package (Fox and Weisberg, 2011) was used for performing the univariate analysis of variance (ANOVA) and Tukey multiple comparison test was subsequently done for each morphometric character using the “multcomp” package (Hothorn et al., 2008). The ANOVA model showed that all 22 landmark-based truss morphometric distances significantly differed to varying degrees among the nine collection sites of five heterogenous habitats (Supplementary Table 3). Therefore, all of these 22 truss morphometric distances were used to obtain the consistent outcomes from the multivariate (PCA, LDFA, CVA, and CA) analyses. Under these circumstances, the N:P ratio was 13.64 (300/22) that revealed Hilsa shad samples size were adequate for stable outcomes of multivariate analyses. Moreover, suitability of the morphometric datasets for multivariate analyses were further confirmed by the Kaiser–Meier–Olkin (KMO) test and the Bartlett’s Test of Sphericity. The KMO test measures whether the partial correlation among variables is sufficiently high for determining the sampling adequacy tests (Yakubu and Okunsebor, 2011). The value of KMO statistics ranges between 0 and 1, and the values more than 0.6 are acceptable (AnvariFar et al., 2011; Yakubu and Okunsebor, 2011). On the other hand, the Bartlett’s Sphericity test hypothesized that the values of the correlation matrix equals zero and the statistically significant outcomes (P < 0.05) is acceptable. In the present study, the outcomes of the Bartlett’s Test of Sphericity was found to be significant (P ≤ 0.01) and the overall matrix value of KMO test was 0.88. The outcomes of the Bartlett’s and KMO test recommended that the sampled data is adequate and appropriate for applying the multivariate factor analysis procedure (AnvariFar et al., 2011).
In order to confirm the existence of morphometric variation among different habitats, different multivariate approaches including Linear Discriminant Function Analysis (LDFA), the Principal Component Analysis (PCA), Canonical Variates Analysis (CVA), and cluster analysis (CA) using Euclidean distance method were employed in the present study. The PCAs were executed by using the ‘FactoMineR’ package (Sebastien et al., 2008). Furthermore, the contributions of variables to principal components (PCs) were also examined to determine which truss lengths were mostly varied among the different habitats and ecological groups of Hilsa shad. Only the first and second PCs (PC1 and PC2) were considered in this study as they explained most of the variability. The LDFA and CVA were performed by using ‘MASS’ package (Venables and Ripley, 2002). All the PCA, LDFA, and CVA plots were made using the ‘ggplot2’ package (Wickham, 2009). As a complement, cluster analysis was performed by using different morphometric distances among the individuals of different habitats (Veasey et al., 2001). Euclidean distance as a measure of dissimilarity and the UPGMA (unweighted pair group method with arithmetical average) as the clustering algorithm were adopted in the clustering analysis approach. The cluster analysis was done using the ‘dendextend’ package (Galili, 2015). The correlations among the geographic distances of the collection sites and morphometric lengths were tested and plotted using the “PerformanceAnalytics” packages (Peterson and Carl, 2019).
The detailed methodology of nextRAD genotyping, sequence assembly, filtering and SNP discovery of these Hilsa shad fishes were described in our previous study (Asaduzzaman et al., 2020). Briefly, DNA were isolated by using Promega DNA purification system (Promega, Madison, WI, United States) in accordance with the manufacturer’s protocol. DNA quantifications were performed by using the Quant-it kit (Life Technologies, Foster City, CA) and the real-time PCR fluorescence measurements of double stranded DNA. For the preparation of nextRAD genotyping-by-sequencing libraries, genomic DNA was fragmented with Nextera DNA Flex Library Prep Kit (illumina, San Diego, California, United States) at the SNPsaurus (SNPsaurus, LLC, United States). Then, an Illumina HiSeq 4000 with eight lanes of 150 bp reads (University of Oregon, United States) was used for the sequencing the nextRAD libraries. The BBDuk in custom scripts of SNPsaurus, LLC (BBMap tools, Eugene, OR, United States) was used for trimming of sequencing reads. The callVariants (BBMap tools) method was used for performing the genotype calling. Filtering of the vcf was performed for removing the alleles with a population frequency of <3%. Loci which had more than 2 alleles in a sample (suggesting collapsed paralogs) and heterozygous in all samples were removed. After these filtering process, data for 26,718 SNPs loci existing within a catalog of 92, 721 consensus nextRAD tagged sequences of 150 bases each were remained in the original shadstringent.vcf file. In the subsequent filtering process, less than 5% overall minor allele frequency, less than 80% completeness of data among samples and complex SNPs with more than two alleles were removed. Finally, a total of 15, 453 individual SNP loci remained in the dataset after executing all filtering steps.
In our previous study, different clustering analyses using the 15,453 putative SNP loci and 15,379 neutral loci displayed very little genetic discrimination among various collection sites of Hilsa shad (Asaduzzaman et al., 2020). Therefore, only 74 outlier SNP loci, which were identified as putatively adaptive by the FST Outflank approach were used to explore whether phenotypic variation in the body shape of Hilsa shad samples corresponds with their fine-scale genetic divergence for local adaptation. Principal component analysis (PCA) of these putatively adaptive SNP loci among different habitats and subsequent ecological groupings were conducted using the ‘adegenet’ R package (Jombart, 2008), while the LDFA and CVA analysis was done using the ‘MASS’ package (Venables and Ripley, 2002). All the PCA, LDFA, and CVA plots were made using the ‘ggplot2’ package (Wickham, 2009). Using a pairwise distance matrix for all collection sites, an isolation by distance analysis was performed by the ‘adegenet’ R package for the outlier dataset. Using an Edward’s genetic distance matrix and a physical distance matrix between the collection sites, significance testing for the isolation by distance analysis was conducted through a Mantel test with 9999 iterations (also using the ‘adegenet’ package). Neighbor-joining trees were generated using these outlier datasets by adopting Nei’s genetic distance method using ‘adegenet’ R package.
To identify the genes encoded within the adaptive SNP loci, a BLASTN search program was applied for each putatively adaptive SNP locus with all sequences available in the NCBI non-redundant database (word size = 11; mismatch scores = 2 and −3; and maximum e-value = 1 × 10–5). BLASTN search program of the SNP flanking sequences showed that the total of 36 putatively adaptive SNP loci were observed within the protein coding region (Asaduzzaman et al., 2020). In this study, gene annotations (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyses1 were performed using a web-based software, Enrichr2 to know the functional distribution of the encoded genes. GO terms having a p-value < 0.05 were selected as significantly enriched by the encoded genes.
Different multivariate and cluster analyses initially indicated that morphogenetic divergence of Hilsa shad population from different habitats overlapped among each other, and was mostly discriminated by the salinity gradients of their diverse migratory habitats (Supplementary Figures S1, S2). To determine which morphometric distances were most efficiently discriminated by the salinity gradients, the contributions of different variables to principal components (PCs) were investigated using morphometric dataset. The PCA extracted four significant and cross-validated PCs having eigenvalues > 1, all of which together accounted for 61.97% of the variation in the original dataset (Supplementary Table S4). Interestingly, the PC1 (27.21% variability) was dominated by the different body depth related morphometric distances such as D2-9 (0.93), D2-10 (0.87), D3-8 (0.81), D3-9 (0.95), D3-10 (0.84), D4-8 (0.83), and D4-9 (0.88), while the PC2 (13.73% variability) was mostly dominated by the body length related morphometric distances such as D2-3 (0.66), D3-4 (-0.72), D4-5 (0.53), D4-6 (0.57), D5-6 (-0.52), and D8-9 (0.52) (Figure 3A and Supplementary Table S4). Similarly, the LDFA analysis revealed that the first linear discriminant function (LD1) accounted for 62.17% of the variability, while the 2nd linear discriminant function score (LD2) explained for 37.83% of the variation (Figure 3B). For the PCA and LDFA analyses of morphometric dataset of different salinity gradients habitats, the sea and estuarine populations were appeared to form overlapping clusters with the riverine populations, but both populations were sufficiently distinctive from their freshwater counterparts (Figures 3A,B). Interestingly, higher body depth related morphometric distances such as D2-9, D2-10, D3-9, D3-10, D3-8, D4-9, and D4-8 mostly discriminated the riverine population from the estuarine and marine population (Figure 3A), indicating the riverine Hilsa shad population were morphometrically wider than the marine and brackishwater population. Interestingly, the PCA and LDFA analyses using putative adaptive SNP loci of Hilsa shad also revealed consistent results with the above morphometric outcomes (Figures 3C,D). Moreover, the CVA analysis based on 22 truss morphometric distances and putatively adaptive panels of SNP loci, displayed the similar trend as those in PCA and LDFA analyses (Figure 4). The outcomes of various multivariate approaches were also in complete concurrence with the results attained using the clustering analysis by the NJ trees derived based on Nei’s genetic distances for putatively adaptive SNP loci dataset, and Euclidean distances between the clusters of centroids applying an UPGMA for morphometric dataset (Figure 5). Based on the results of different multivariate and cluster analyses, it was confirmed that the majority of the Hilsa shad population collected from the freshwater riverine habitats displayed pronounced morpho-genetic divergence than the marine and estuarine populations.
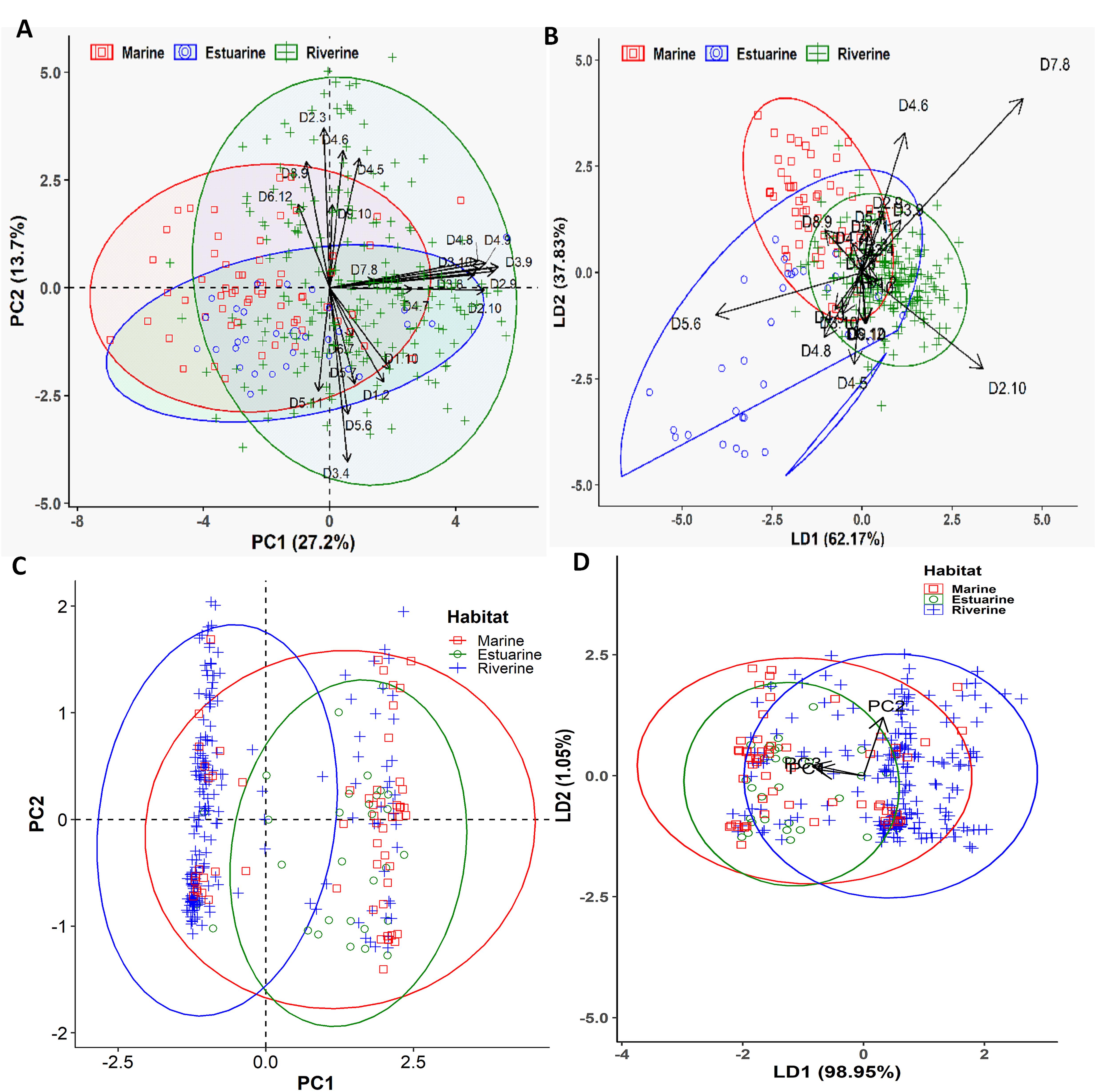
Figure 3. The Principal Component Analysis (PCA) and Linear Discriminant Function Analysis (LDFA) showing the morphogenetic variation of Hilsa shad Tenualosa ilisha collected from the marine (salinity 30.3–33.4 ppt), estuarine (salinity 2.5 to 20.6 ppt) and riverine (salinity < 1.0 ppt) habitats. (A)- biplots of the PCA using 22 morphometric distances of Hilsa shad; (B)- biplots of the LDFA using 22 truss morphometric distances of Hilsa shad; (C)- biplots of the PCA using 74 putatively adaptive panels of SNP loci identified by the FST OutFLANK approach; (D)- biplots of the LDFA using 74 putatively adaptive panels of SNP loci identified by the FST OutFLANK approach.
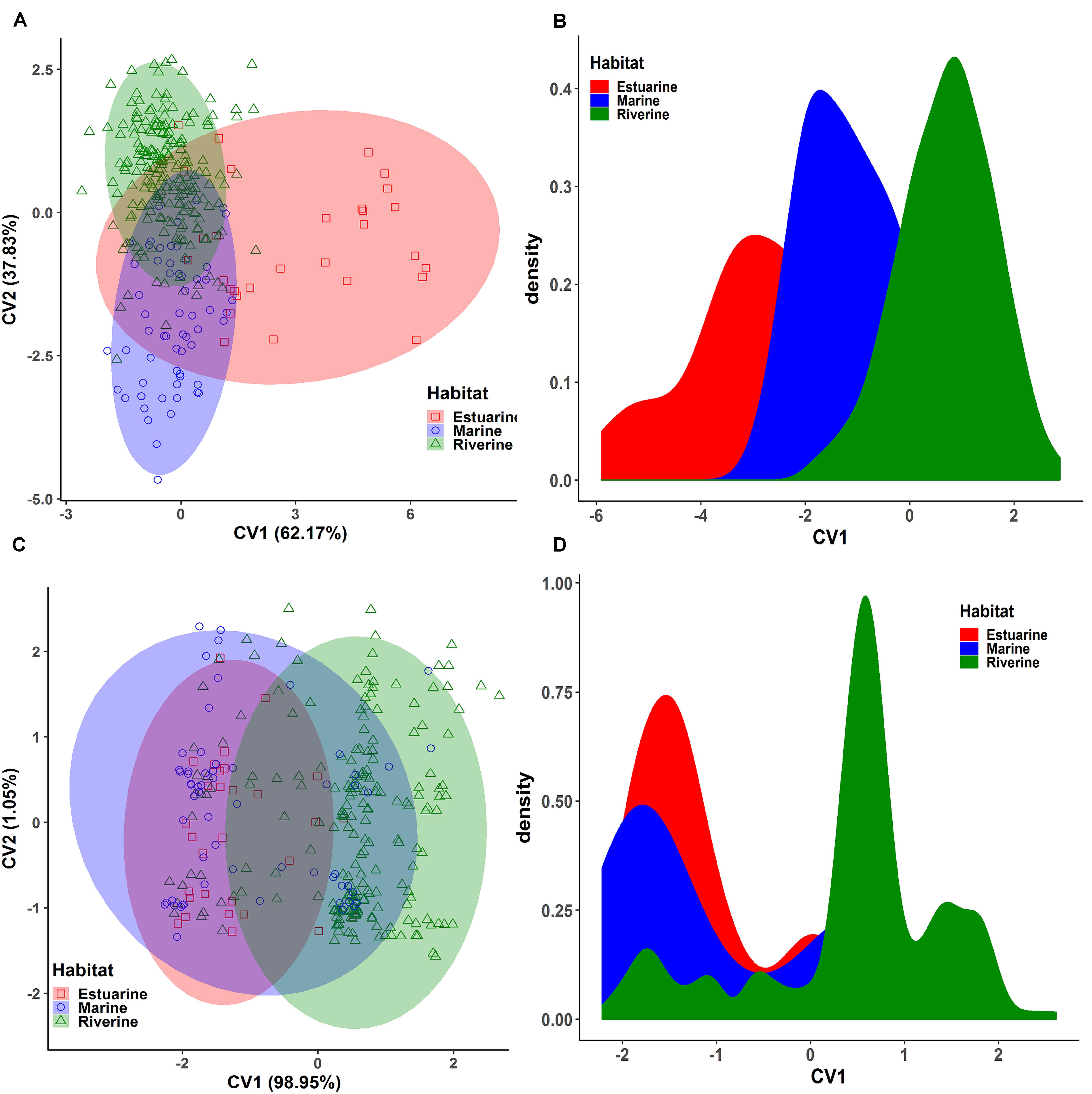
Figure 4. The Canonical Variate Analysis (CVA) showing the morpho-genetic variation of Hilsa shad Tenualosa ilisha collected from the marine (salinity 30.3–33.4 ppt), estuarine (salinity 2.5 to 20.6 ppt) and riverine (salinity < 1.0 ppt) habitats. Biplots of sample centroids of the canonical variates scores are shown based on the 22 truss morphometric length (A,B) and 74 putatively adaptive panels of SNP loci identified by the FST OutFLANK approach (C,D).
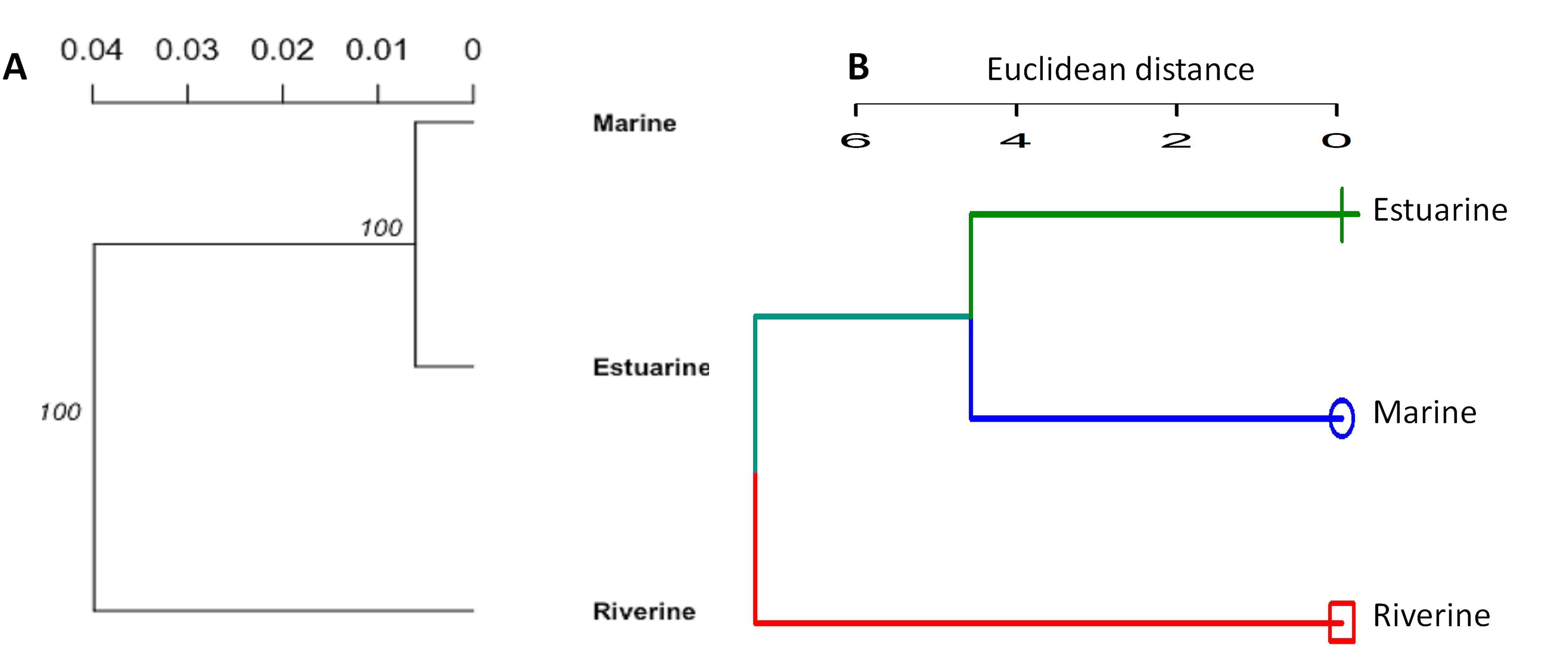
Figure 5. The cluster analysis showing the morpho-genetic variation of Hilsa shad Tenualosa ilisha collected from the marine (salinity 30.3–33.4 ppt), estuarine (salinity 2.5 to 20.6 ppt) and riverine (salinity < 1.0 ppt) habitats. (A)- neighbor-joining trees, based on the Nei’s genetic distances using the 74 putatively adaptive panel of SNP loci identified by the FST OutFLANK approach. (B)- Unweighted pair group method with arithmetic mean (UPGMA) dendrogram of hierarchical cluster analysis based on the Euclidean distance using the 22 truss morphometric distances of Hilsa shad population collected from their different habitats.
The correlation plot (Figure 6) revealed that geographical distances had only one significant negative correlation with the morphometric distance of D8-9 (r = -0.22; p < 0.001). Most body depth related morphometric distances provided significant and strong positive correlations with geographical distance such as D2-9 (r = 0.55; p < 0.001), D2-10 (r = 0.73; p < 0.05), D3-8 (r = 0.51; p < 0.001), D3-9 (r = 0.57; p < 0.001), and D4-9 (r = 0.55; p < 0.001). Other morphometric distances were observed positively correlated with the geographical distance, but with lower r-values such as D1-2 (r = 0.26; p < 0.001), D1-10 (r = 0.34; p < 0.001), D3-10 (r = 0.43; p < 0.001), D4-8 (r = 0.38; p < 0.01), and D7-8 (r = 0.37, p < 0.001). We also investigated the correlation of genetic discrimination between the pairwise FST value and geographical distance (Figure 7). The Isolation-by-Distance (IBD) analysis using the Mantel test revealed a significant association between increasing the distance of geographical separation and increasingly larger the FST value for the putatively adaptive SNP loci, in which the slope of IBD was significantly higher with R2 values equal to 0.4871 (p < 0.001) (Figure 7).
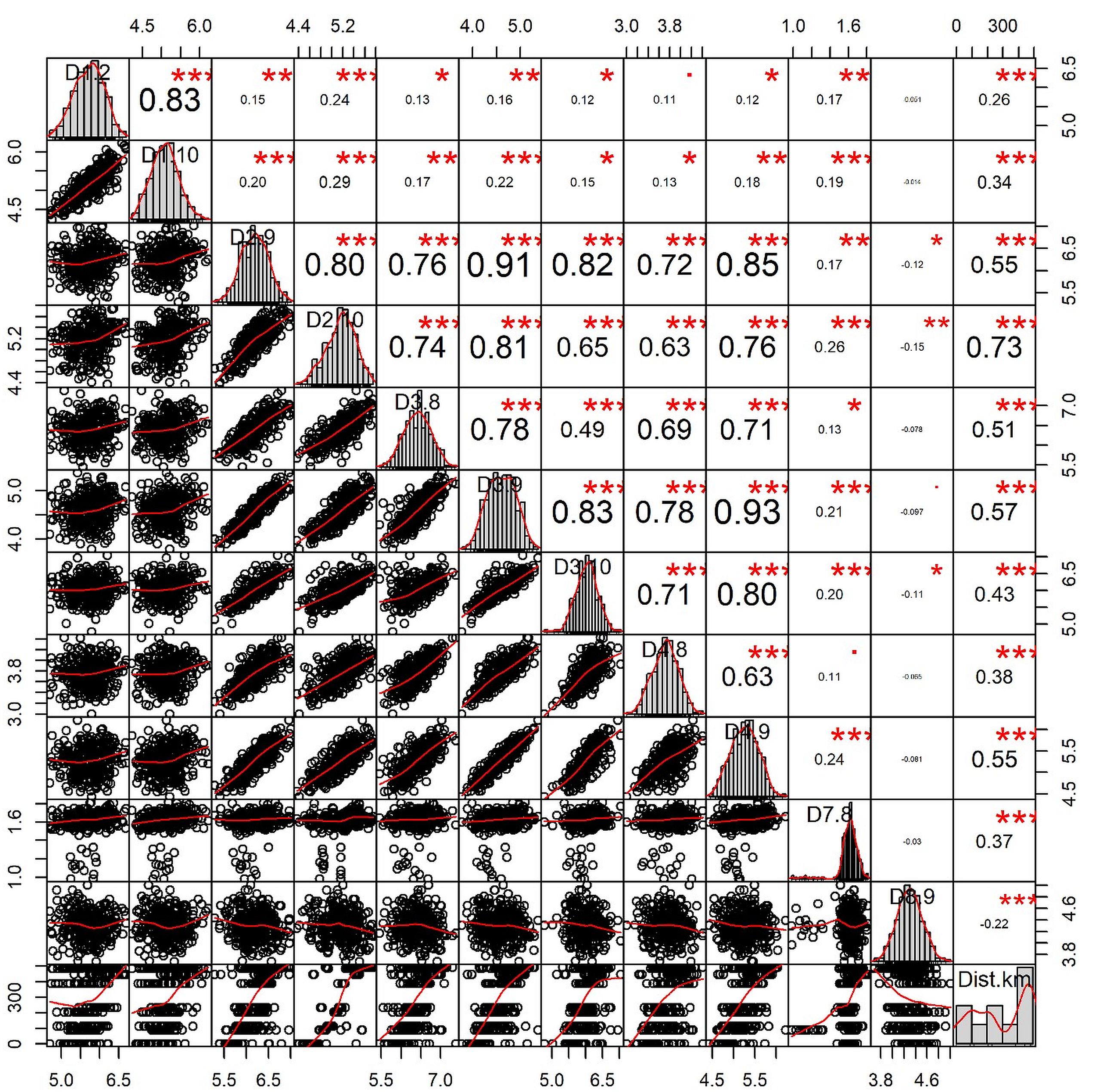
Figure 6. The correlation plot among the geographic distances of collection sites from the deep sea and morphometric distances of Hilsa shad Tenualosa ilisha collected from heterogeneous habitats including the sea, estuary and different rivers. The values given around all the axes are the range of each individual parameter’s measured unit values (cm). Correlation coefficients (r) are indicated with numeric values, while significance levels (p) are denoted by asterisks (*<0.05, **<0.01, ***<0.001).
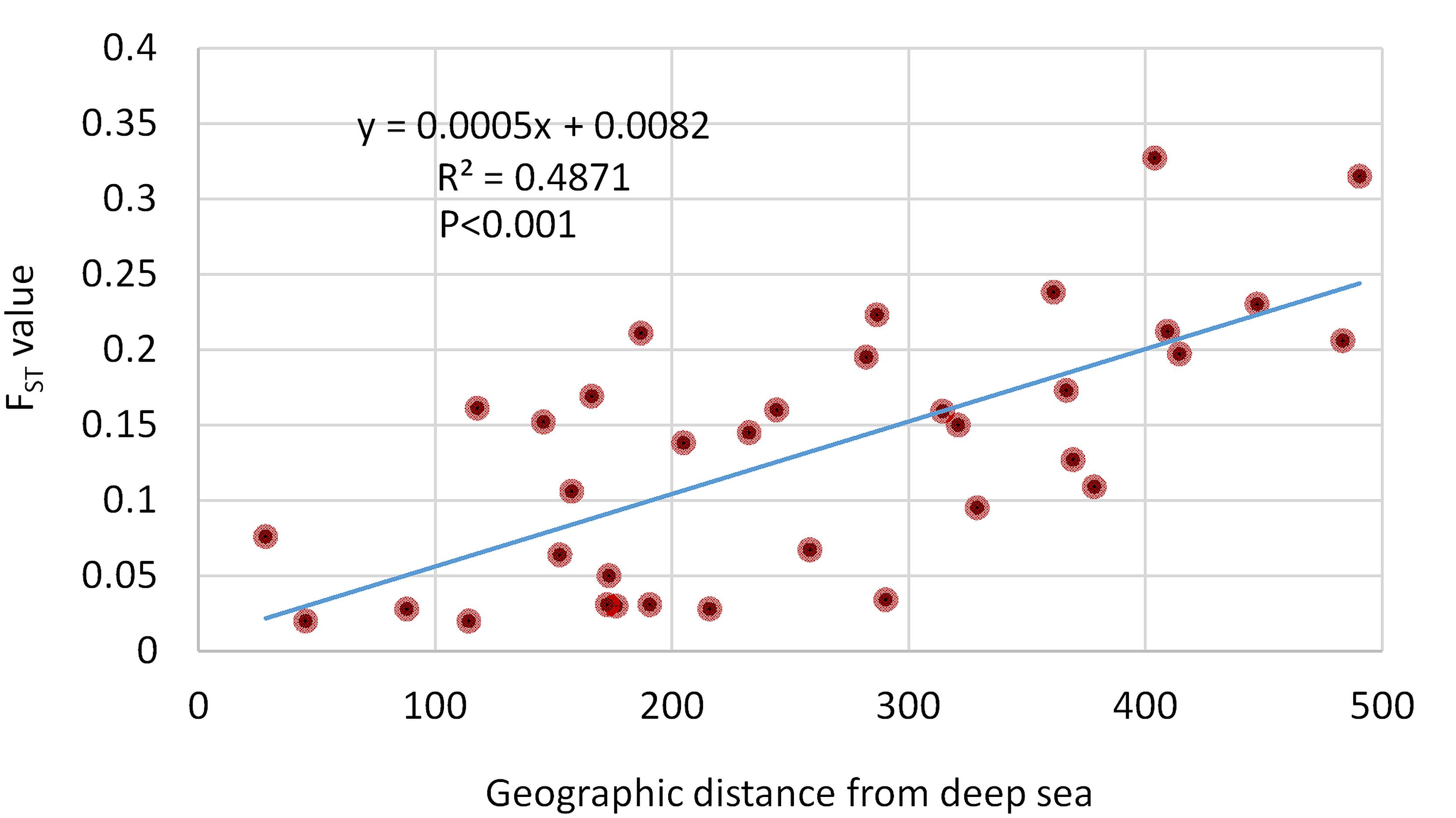
Figure 7. The regression plot between the pairwise FST values of 74 putatively adaptive SNP loci and the geographical distances of Hilsa shad Tenualosa ilisha collection sites associated with the spatial environmental heterogeneity. The Mantel test showed highly significant effects based on 9999 replicates.
Six collection sites of three rivers were further grouped into the western turbid rivers (Padma and Jamuna) and the eastern clear river (Meghna) based on the annual variation of turbidity levels (Supplementary Table S1). Similarly, the multivariate analyses were executed to condense the dimensionality of the morphogenetic data sets into few number of components that summarized the information of the overall data set. Applying PCA to the morphometric data set, it was possible to extract four significant and cross-validated PCs having eigenvalues > 1 (Supplementary Table S5). Together, they accounted for 64.67% of the variation in the original dataset. The PC1 accounted for 26.58% of the variability and mostly associated with the body depth related loading variables such as D3-9 (0.91), D2-9 (0.87), D2-10 (0.83), D4-9 (0.83), D3-10 (0.80), D4-8 (0.80), and D3-8 (0.75). The PC2 accounted for 15.63% of the variability and mostly dominated by the body length related loading variables such as D3-4 (-0.78), D2-3 (0.69), D5-6 (-0.59), and D8-9 (0.51). Interestingly, the PCA outcomes further demonstrated that the higher value of both the body depth (D2-9, D3-9, D4-9, D4-8, D2-10, and D3-10) and body length (D1-2, D1-10, D3-4, D5-6, D6-7, and D7-8) has triggered the morphometric discrimination (Figure 8A), indicating that the eastern turbid riverine population were morphometrically wider than the eastern clear river population. The biplots of PC1 and PC2 scores also showed a clear distinctive clustering pattern between the eastern clear water and the western turbid water populations (Figure 8A). Like PCA biplot, the density plot of CVA analysis also showed a clear morphometric divergence between the eastern clear water and the western turbid water populations of Hilsa shad. Interestingly, the outcomes of both PCA and CVA analyses of putatively adaptive panels of SNP loci were in complete agreement with the results obtained from the morphometric dataset (Figures 8C,D).
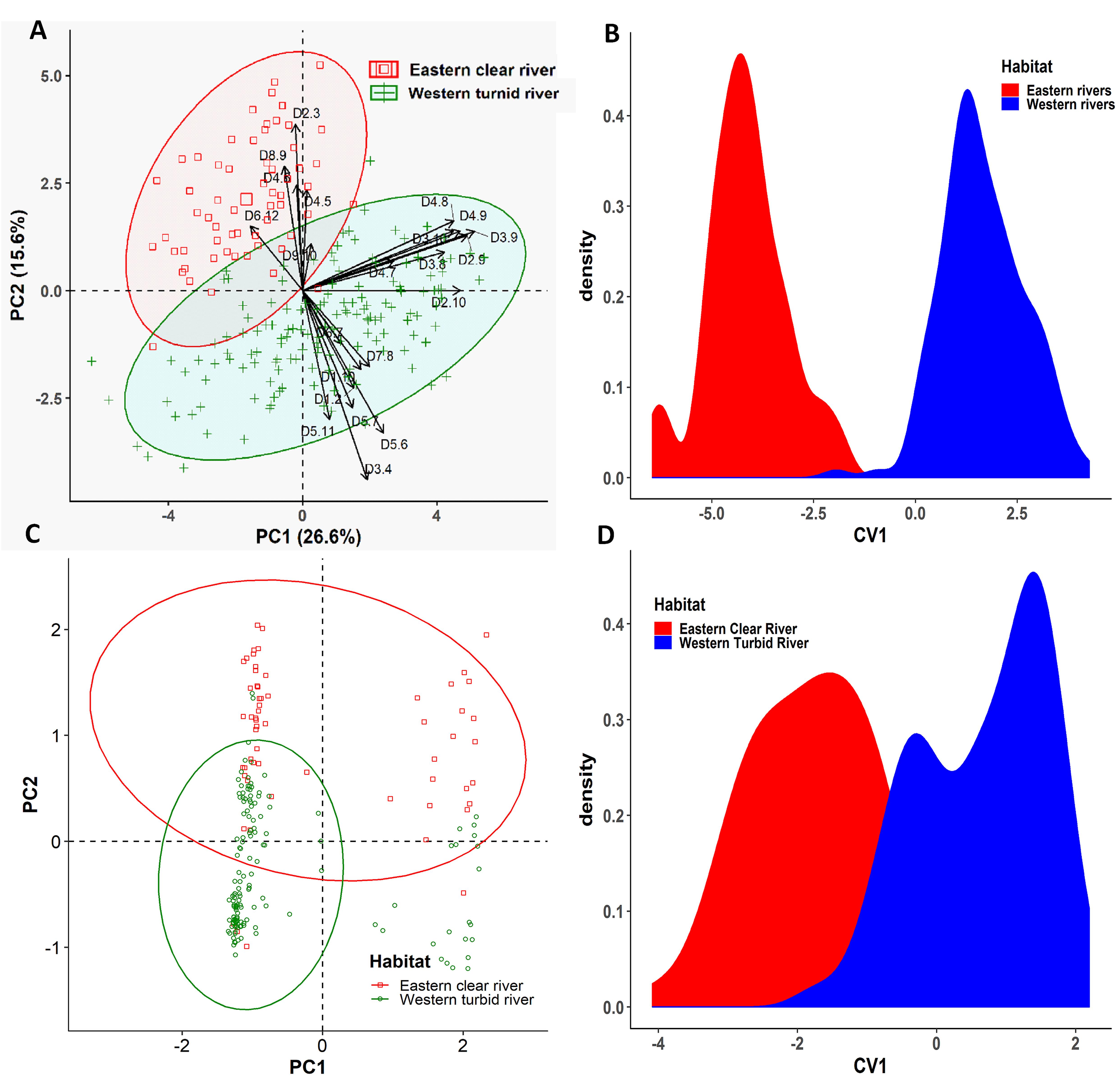
Figure 8. The Principal Component Analysis (PCA) and Canonical Variate Analysis (CVA) showing the morpho-genetic variation of Hilsa shad Tenualosa ilisha collected from the eastern clear water (total suspended sediment 20–750 mg/l) and the western turbid water (total suspended sediment 190–1600 mg/l). (A) Biplots of the PCA using 22 truss morphometric distances of Hilsa shad; (B) biplots of sample centroids of the canonical variate scores are shown based on the 22 truss morphometric distances; (C) biplots of the PCA using 74 putatively adaptive panels of SNP loci identified by the FST OutFLANK approach; (D) biplots of sample centroids of the canonical variate scores are shown based on 74 putatively adaptive panels of SNP loci identified by the FST OutFLANK approach.
The annotated 36 genes (Supplementary Table S6) were subjected to GO enrichment analysis and three functional groupings were considered such as biological process, cellular component and molecular function. For gene function prediction, we considered GO biological terms and observed that these genes perform diverse function in anadromous Hilsa shad including growth, metabolism, developmental process, body homeostasis, osmoregulation and so on. It was noticed that the majority of the genes were involved in growth and metabolism related physiological processes including MTHFD1, AADAT, PGM1, GMPS, PDHA1, BLMH, PLCB1, ITPKB, PPP1R9B, EXTL3, UBR3, GABBR1, ARAP1, FBXO11, TUBGCP6, ELFN2, CCNF, KDM6A, ACIN1, MYO6, EPR1, and SFI1 (Table 1). It was also discovered that many genes were actively involved in maintaining homeostasis and osmoregulation such as PrRPR, SLC7A10, SIDT2, TRIM67, nrxn3b, Grik2, MYO6, PLCB1, EXOC8, SRP68, GABBR1, MYRF, NFIB, PLCB1, PPP1R9B, TUBGCP6, HERC1, and ARAP1 (Table 2). Furthermore, the Enriched KEGG pathways additionally demonstrated that PLCB2, GRIK2, GABBR1, and SPR68 genes were found to be involved in various signal transduction pathways, which are important for maintaining the osmotic balance and homeostasis (Table 3). Besides, some genes participated in other functions like embryonic development (UBR3), immunity (MPEG1, PDHA1, ACIN1, and PLCB1) and epigenetic control (KDM6A).
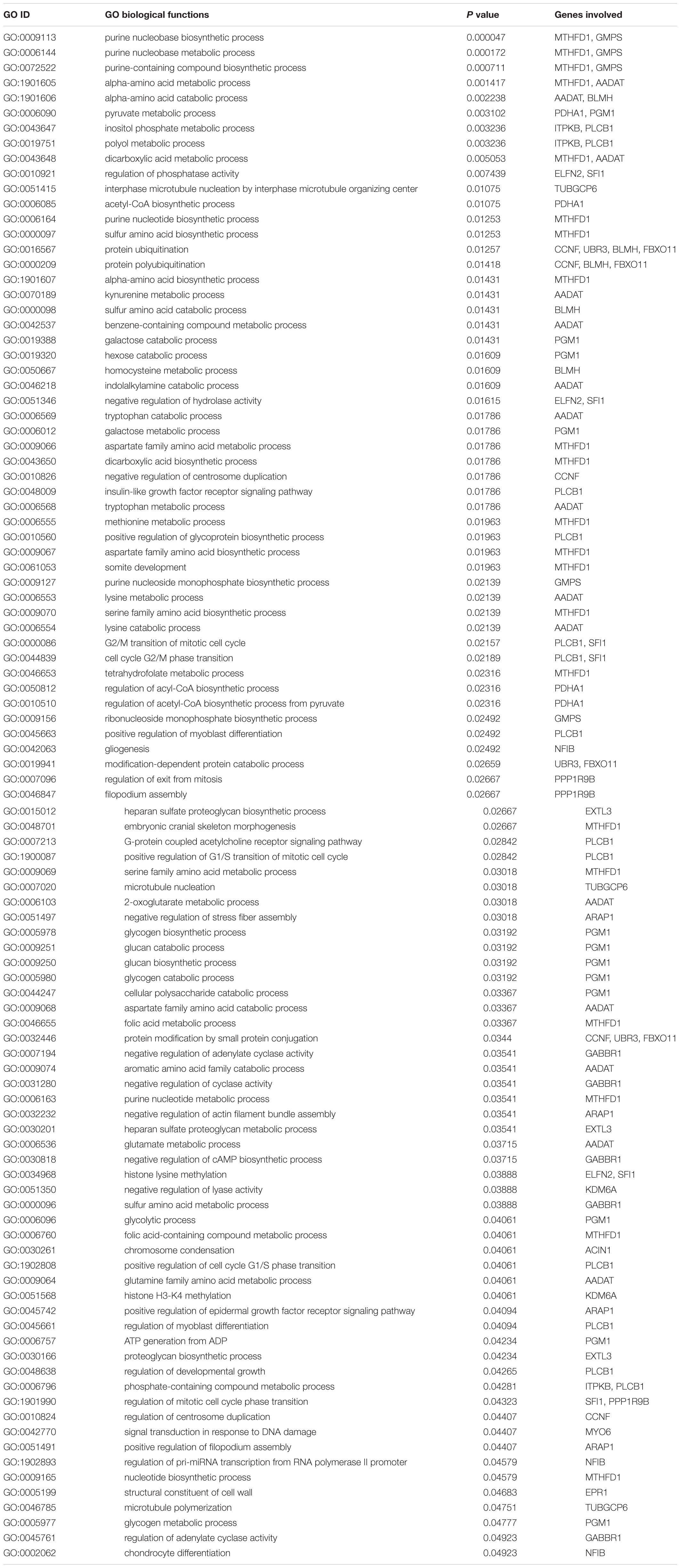
Table 1. Gene Ontology (GO) terms pertaining to growth and metabolism related functions of the genes encoded by the putatively adaptive panel of the SNP loci of T. ilisha populations.
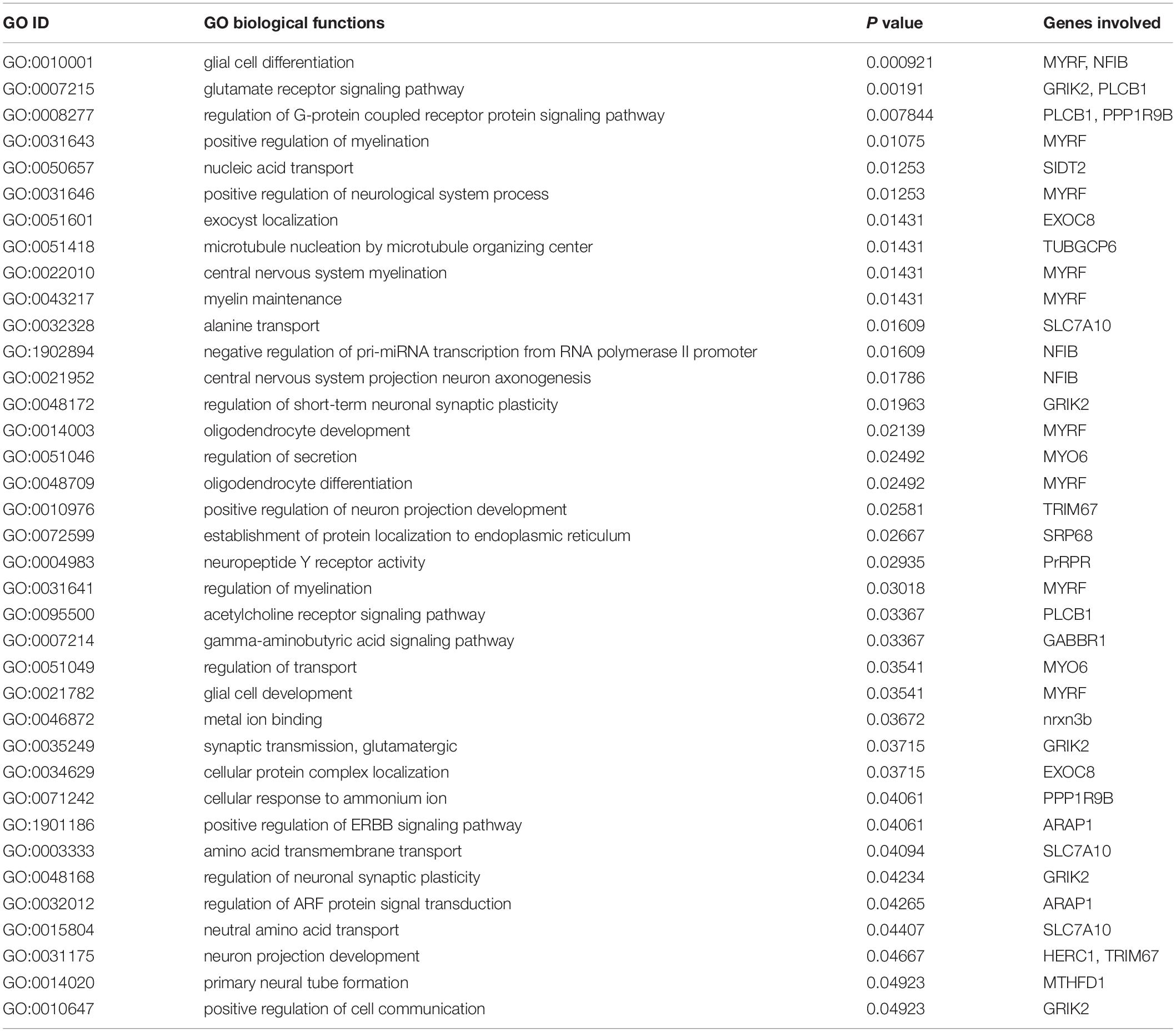
Table 2. Gene Ontology (GO) terms pertaining to the homeostasis and osmoregulation related functions of the genes encoded by the putatively adaptive panel of the SNP loci of T. ilisha populations.
This study so far represents the first ever efforts made to combine both nextRAD genotypes and morphometric analyses of anadromous Hilsa shad populations collected from a broader sampling locations covering all types of their natural habitats. The results generated from this study would help to determine among and within-site divergence and the extent of parallelism at both phenotypic plasticity and genomic levels between the Hilsa shad populations residing in diversified habitats. In general, phenotypic divergence and genetic differences were estimated by outlier loci, which were more pronounced among ecotypes than among sites, suggesting that ecotypic differentiation was more dominant than local adaptation among sites. Although it was not fully understood, whether the Hilsa shad morpho-genetically pre-adapted or modified (phenotypic plasticity) their shape according to the environmental heterogeneity, several non-mutually exclusive hypotheses may elucidate the observed continuum differentiation at both phenotypic and genotypic levels. In the present study, hence, we hypothesized that: (i) the morphometric discrimination are largely determined by the genetic variation, (ii) the interactive evolutionary processes and salinity gradients predominantly contribute to the morphogenetic divergence between the marine-estuarine and freshwater riverine populations, (iii) spatial salinity gradients influences the genotypes leading to phenotypic plasticity, and (iv) the local environmental discrimination may contribute to morphogenetic divergence among the riverine populations.
Environmental heterogeneity is one of the key factors shaping the genetic and phenotypic variation. As Hilsa shad is a widely distributed species with large effective population size, local population often experience heterogenic environmental conditions to evolve in such kind of diversified environments for thousands of years. Under these heterogeneous environmental conditions, they may have evolved a distinct genetic composition, and genetically based phenotypic traits through local adaptations. Therefore, we hypothesized that the observed morphometric body shape divergence of Hilsa shad are largely determined by the genetic adaptation to the heterogeneous environmental conditions. In support of our hypothesis, we observed that the outcomes of different spatial multivariate analyses of morphometric dataset in different heterogenic habitats were consistent with the results obtained using putatively adaptive panels of SNP loci dataset. Moreover, the linear model outcomes of morphometric dataset (Figure 6) corroborated with the Mantel test outcomes of putatively adaptive SNP loci (Figure 7), indicating the morphogenetic differentiation observed in this study may have evolved in parallel with the geographical distance. Furthermore, the GO enrichment analysis demonstrated that most of the genes encoded by the putative adaptive loci were involved in growth and metabolism related functions (Table 1). Since metabolism is very essential for cell growth and proliferation, the differences in body shape and size of Hilsa shad in various habitats may likely link to growth supported by different metabolic pathways. Therefore, the present findings demonstrated that the variations in morphometric shape, observed among Hilsa populations in different habitats, might possibly be regulated by the mutation of these genes. Involvement of these genes in different metabolic pathways like purine nucleobase metabolic process (MTHFD1, GMPS), folic acid metabolic process (MTHFD1), galactose and glycogen metabolic process (PGM1), dicarboxylic acid metabolic process (MTHFD1, AADAT), insulin-like growth factor receptor signaling pathway (PLCB1), regulation of acyl-CoA biosynthetic process (PDHA1) and alpha-amino acid biosynthetic process (MTHFD1) most probably indicate the crucial role of these genes in growth and development of Hilsa shad in respect to their habitat differences. The association of MTHFD1 in folate metabolic pathway was found to be essential for early development of zebrafish and mice, and the deficiency of this protein resulted in embryonic defects (Christensen et al., 2016; Lee et al., 2012). PGM1 plays a significant role in cell proliferation and cell survival by maintaining metabolic carbohydrate homeostasis through the inter-conversion of glucose-1-phosphate (G-1-P) and glucose-6-phosphate (G-6-P) (Bae et al., 2014; Nolting et al., 2017). Moreover, PGM1 may also increase the efficiency of dietary carbohydrate resulting in higher growth rate in rainbow trout (Allendorf et al., 1983). PDHA1 is also very essential for growth and proliferation of mammalian cells that regulates glucose homeostasis by converting pyruvate to acetyl-CoA (Rajagopalan et al., 2015). However, we suggested that the variation in body shape in migratory Hilsa shad i.e., the deeper body in the riverine populations than their counterparts in seawater is likely to be related to the polymorphism of these genes throughout the adaptive SNP loci.
Understanding the evolutionary processes that generate the morphometric divergence of anadromous fish is important as body shape can often significantly affects most of the physiological and fitness traits. The cluster and spatial analyses on morphometric dataset showed that the marine and estuarine populations adapted to maintain streamlined body (narrow and slender type) than their counterpart freshwater populations. Prior to migration, a streamlined body was maintained by the marine and estuarine populations to obtain a range of evolutionary benefits for fast swimming performance during their long distance migration against the water current (Chapman et al., 2015). In general, migration against water flow and gravitational force to upstream rivers from marine habitat is energetically costly (Quinn, 2005; Skov et al., 2008; Hulthen et al., 2014). In a range of fish species, many findings support the link between increased thin body shape with streamlining body and reduced energy expenditure to maintain stable and fast swimming performance for long distance migration (e.g., Vogel, 1994; Langerhans and Reznick, 2010). Moreover, a significant number of studies investigated the morphometric phenotypic discrimination of migrant and local residents of anadromous fish (Boily and Magnan, 2002; Morinville and Rasmussen, 2003, 2006, 2008). All of these studies reported that migratory anadromous fish adopting a specialized life history strategy (prior to migration) to have less robust, streamlined and narrower body shape than those adopting the residential life-history in rivers. Beside the anadromous nature of Hilsa shad, studies have also demonstrated -a fluvial potamodromous type, which inhabit the freshwater riverine systems throughout their entire life cycle (Blaber et al., 2003; Milton and Chenery, 2001; Hossain et al., 2014). Although the phenotypic discrimination is very challenging, further studies should need to consider the body shape variation associated with the migratory anadromous (migrants) and fluvial potamodromous type (local residents) of Hilsa shad in upstream riverine habitats.
We also observed subtle shifts on body shape between Hilsa shad populations across habitat of different salinities, but this phenotypic shift does not seem to be under selection. Natural selection may determine local adaptation, especially in fragmented and heterogeneous habitats (Kawecki and Ebert, 2004). However, the anadromous lifestyle of Hilsa shad requires movement between variable environments, does not conform to this evolutionary mechanism, which might result in speciation (Kirkpatrick and Barton, 2006). Although adaptive loci have been detected, the functions of these loci from gene annotation approach could not fully determine the connection of genotypes with the phenotypic effects for these loci. Moreover, phenotypes may be highly polygenic, and several phenotypes may contribute to a multifaceted adaptive process, which is especially common for adaptation to various environment (Rockman, 2012). This is particularly true for Hilsa shad migrating from the marine to freshwater gradients, with varying levels of salinity and multiple environmental parameters such as oxygen, pH, turbidity, food, predators, microbes, etc. (Austin et al., 2012; Ali et al., 2014; Osborne et al., 2015; Ou et al., 2015; Hossain et al., 2016). The presence of multiple genes in Hilsa shad suggested the expansion of these genes in developing physiological adaptations between hypo-osmotic freshwater and hyper-osmotic seawater conditions throughout the life cycle of this anadromous fish. This adaptive process to large salinity variation is energetically costly and is highly dependent on carbohydrate and lipid metabolic pathways involving homeostasis genes for sufficient energy supply. In fact, changes in body shape and size in Hilsa shad may be likely linked to growth supported by these metabolic pathways. Given that lower energy may be required by fish to acclimatize to the freshwater habitat rather than the sea and estuarine habitats (Sangiao-Alvarellos et al., 2005), fish from the freshwater rivers have deeper body than their counterparts in environment with higher salinity. Some other studies have reported the similar results of relationship between salinity and body shape. For example, sticklebacks reared in saline conditions tend to develop a more streamlined body shape than those of freshwater conditions (Mazzarella et al., 2015). Moreover, our GO and KEGG pathways results supported the presence of multiple genes involved in the homeostasis and osmoregulation to enable this euryhaline fish to adapt to varying environmental salinity (Tables 2, 3). The changing osmotic conditions become the stimulus that trigger the activation of several crucial gene families in Hilsa shad such as SLC7A10, SIDT2, GRIK2, SRP68, PPP1R9B, nrxn3b, and MYO6 which are actively involved in the ion secretion and transportation for the osmotic balance maintenance. PLCB1 and ARAP1 are highly involved in signal transduction pathway whereas MYRF, NFIB, GRIK2, PrRPR, nxrn3b, and HERC1 are involved in neuronal function regulation that might be important to maintain body homeostasis and facilitate the Hilsa shad to adapt in different environments throughout their life cycle. Our result also discovered another secondary transporter, SLC7A10 which control the trans-membrane movement of amino acid in cellular homeostasis maintenance of Hilsa shad. Other SLCs have also been found to be involved in regulating salinity stress and osmoregulation in fish species such as spotted seabass and freshwater eel (Nakada et al., 2005; Zhang et al., 2017). The significant regulation of molecular activities by these trans-membrane and signaling pathways prove their importance in up-keeping the intracellular homeostasis of Hilsa shad under various salinity and osmotic conditions. Very recently, the association of SLCs, GRIKs, and MYOs has also been observed in C. harengus in maintaining ionic homeostasis (Mohindra et al., 2019). PrRPR was found to be reported to involve in neuromodulation in the brain of the guppy as well as in tilapia (Amano et al., 2007; Watanabe and Kaneko, 2010). As an anadromous fish, the osmoregulation-related genes play a pivotal role in maintaining body homeostasis while Hilsa shad are migrating from sea to freshwater bodies or vice-versa. The KEGG pathways additionally demonstrated the significant involvement of PLCB1, Grik2, GABBR1 and SRP68 in different signaling pathways like phosphatidylinositol signaling system, glucagon signaling pathway, glutamatergic synapse, estrogen signaling pathway, protein export and calcium signaling pathway (Table 3) that might play significant role in energy homeostasis and maintaining the osmotic balance (Edwards and Michel, 2002; Yan et al., 2016). The functional analysis of these genes, therefore, will be helpful to understand migratory behavior of Hilsa shad and how they adapt between different environments during their life cycle.
The anadromous Hilsa shad, as like most teleosts, excrete water and ions passively using their skin, and thus osmoregulation efficiency may influence the body shape variation (phenotypic plasticity) during migration between the high and low salinity environments. Spatial homogeneity promotes local adaptations and divergent selection, while temporarily heterogeneous environments results in phenotypic plasticity (Endler, 1977; Hedrick, 1986; Meyers and Bull, 2002). Although local adaptation may play a role in population differentiation, we hypothesized that association of phenotype to habitat in Hilsa shad are result of plasticity of salinity tolerance, which are also found in other anadromous fish such as salmonids (Klemetsen, 2010; Valiente et al., 2010; Stelkens et al., 2012). Phenotypic plasticity is a phenomenon of a genotype exhibiting different phenotypes in response to heterogenic environmental conditions, whereby in this context, the salinity variation (West-Eberhard, 2003). The body shape and size of Hilsa shad appear to be differed by habitat types, some of which may have heritable components to be passed on to the next generations. Furthermore, habitat type discrimination of PC values was found to be significant, suggesting that habitat types of different salinities rather than specific habitat are determining these divergences. Therefore, it is not known whether the Hilsa shad at each stage and heterogeneous habitats are capable to modulate phenotypic traits or harbor enough standing genetic variation to cope with changes in salinity associated with their anadromous life cycle. Besides, our GO analysis has also identified two genes which are crucial for Hilsa shad developmental process, notably UBR3 and MTHFD1 gene. UBR3 is a novel regulator of Hh signaling, which regulates numerous developmental processes in vertebrates (Varjosalo and Taipale, 2008; Gulino et al., 2012). On the other hand, MTHFD1, which is involved in the folate metabolic pathway, was shown to be essential for early development of zebrafish, and the deficiency of this protein resulted in embryonic defects (Lee et al., 2012). Considering the annotated genes, we further assume that adaptive developmental plasticity may have allowed the genotype of Hilsa shad to have a broader salinity throughout different life stages as cued by signaling pathways since embryonic stage. As a strong determinant factor, salinity gradients have also been reported to predominantly influence the plankton diversity and species richness (Larson and Belovsky, 2013). Since body depth in fishes can be strongly influenced by foraging conditions in a water body, it is distinctly possible that differing planktonic food resources may also play a predominant role in the observed phenotypic plasticity of planktivorous Hilsa shad among different salinity gradient habitats, as reported in another clupeid species Gilchristella aestuaria (Blaber et al., 1981). However, reports on the feeding behavior of pre-spawning adults during migration are rather mixed. Pillay and Rosa (1963), Quereshi (1968) and Shafi et al. (1977) observed that the matured adult ceased eating during upstream migration, and they were depending on accumulated fat deposits for energy. In contrast, Pillay (1958) and Halder (1968) found no evidence of cessation or even any significant decrease in the feed uptake during upstream spawning migration. Moreover, the overall size of Hilsa shad depicted by body depth, head length and anal fin length, are highly dependent on the seasonal migratory behavior (Dwivedi, 2019b). During monsoon season (July to October), fish with deeper body profile are found commonly at mature adult stage, dominate the freshwater habitats, while individuals with slender body profile remain in estuarine and marine environments (Borah et al., 2019; Dwivedi, 2019b). These morphogenetic differences among heterogeneous habitats are confounded by variation of environmental parameters and seasonal life-stages, preventing us from concluding firmly as to whether these differences are of exclusively plastic. Thus, these findings provide us an avenue for follow-up studies, notably to further investigate the developmental time points capable of capturing the possible timing of development important for migratory behavior.
Our results identified different morphologies and genetic structure between the north-eastern clear riverine (the Meghna and its upper tributaries) and the north-western turbid riverine (the Padma and Jamuna) systems. Different multivariate approaches revealed that the north-western turbid riverine population were morphometrically broader than the north-eastern clear river population (Figure 8). Previous studies also reported that Hilsa shad inhabiting the north-eastern riverine (the Meghna) is of “slender” type, while those in the north-western riverine system (Padma-Jamuna) is a “broad” type (Ghosh et al., 1968; Shafi et al., 1977). This morphological variation observed between the riverine habitats, that varied by turbidity differences, could be attributed by developmental plasticity, specifically for individuals of the north-western riverine system which experience extremely higher turbidity of the Gangetic water flows throughout their development stages. Although turbidity deteriorates visuality which negatively impacts the aquatic organisms relying on sight (Gregory and Northcote, 1993), Hilsa shad in such turbid systems may compensate this limitation through the sensory system and maintain sufficient foraging rates. Given that Hilsa shad is a plankton feeder, the turbid waters offer refuge from the predation, higher primary productivity and planktonic food availability for these individuals (Gardner, 1981; Roy et al., 2004; Lehtiniemi et al., 2005; Sonin et al., 2007; Bhaumik, 2015). In another clupeid fish G. aestuaria, body shape differences (wider vs. slender) were found to be linked with the feeding strategies in which filter feeding was the norm in turbid system, whereas individual selection of scarce planktonic prey items appeared to be the norm in clear systems (Blaber et al., 1981). Therefore, similar differences in foraging behavior may also be applicable to Hilsa shad in the north-eastern clear riverine and the north-western turbid riverine systems which, requires further study. Under turbidity influence, the growth of Hilsa shad varies between these two riverine systems due to changes in environmental parameters such as sediment input, the upstream runoff, food availability, population size and density-dependent growth factors, which will affect diet and trophic-related traits. Consistently, plankton composition and diversity were also found to be distinctly different in the migratory rivers of Hilsa shad in which Bacillariophyceae (diatom) were mostly dominated in the turbid north-western rivers, whereas the north-eastern rivers were dominated by the Chlorophyceae (Ahmed et al., 2005; Ahsan et al., 2012). Total dissolved solid (TDS) in the north-eastern riverine system ranged from 72–130 mg/L, while the range of TDS in the north-western riverine system was 255–370 mg/L (Rikta et al., 2016; Bhuyan et al., 2017). Previous studies have also revealed plasticity in morphology in fish in response to food type (Lucek et al., 2014; Svanback and Schluter, 2012), environmental heterogeneity (Garduno-Paz et al., 2010), and a combination of these two cues (Wund et al., 2012). Thus, the north-western riverine Padma Hilsa shad of turbid waters are broader and thicker size than those from the clearer north-eastern riverine Hilsa shad of Meghna river. Adaptive plasticity indeed represents the initial step in adaptation to the new environment, decreasing the cost of selection force, and generating enough time for populations to be established in multiple environments (Snell-Rood, 2013). Importantly, body shape differentiation between heterogeneous environments most likely results from an interaction of genetic and environmental factors (Pfenning et al., 2010; Hendry et al., 2011).
The integration of genomic and morphometric approaches have been demonstrated to be a powerful tool to resolve population structure and short-term variation induced by environmental conditions, respectively. We observed a pronounced pattern of phenotypic parallelism with the nextRAD genotypes between nine sampling sites for Hilsa shad throughout the marine, coastal and freshwater river waters of Bangladesh. The results support the view that adaptive phenotypic plasticity to salinity variation has driven the divergence between populations residing in the sea and estuarine with the freshwater riverine systems (Meghna, Padma, and Jamuna). Focusing on multiple traits, particularly those involved in the metabolic and signaling pathways have further contributed to inheritable components of this fish in developing local adaptation without directional selection process at different life stages and habitats of varying salinity levels. Hilsa shad, inhabiting the coastal areas in the south, are commonly adult fish with slender body type that resist higher water current, while those in the freshwater habitat are deep bodied matured adults ready for spawning. Together, we have also found that adaptive phenotypic plasticity potentially promotes local adaptation for Hilsa shad to persist as “broad” type in northwestern riverine with turbid water and “slender” type in northeastern riverine with clear water, whereby these plastic traits become genetically assimilated. Finally, we concluded that genetic adaptation, phenotypic plasticity and evolutionary constraints jointly determine the morphogenetic divergence of Hilsa shad across their heterogenic migratory habitats. However, the interactions of these forces and relative strengths across heterogenic environments may be one reason that made it has been so difficult to determine the underlying selective pressures that have shaped the Hilsa shad morphogenetic divergence along their diversified migratory habitats. The influence of different landscape parameters including monsoon patterns on genetic connectivity should need to be investigated to attain a deeper knowledge on morphogenetic adaptation of Hilsa shad along their diverse migratory habitats. The pronounced genetic structures paralleled with the phenotypic differences indicate that the studied populations should be managed as independent units. While these findings could be further refined, it is imperative to investigate functionally informative population proxies to design conservation efforts based upon functional genetic diversity. This study also account for life stage-specific habitat suitability of Hilsa, provides insight for planning specific measures to augment natural recruitment for sustainable Hilsa shad fishery stocks in Bangladesh waters.
The datasets generated for this study can be found in the DDBJ (https://ddbj.nig.ac.jp), with the DRA accession number: DRA009185.
The animal study was reviewed and approved by Chittagong Veterinary and Animal Sciences University.
MA conceived and designed the experiments. MA and LW performed the molecular and nextRAD genotyping experiments. MA and BR performed the landmark based morphometric measurements. MN and MJR carried out the sampling work in the field. MA, M, and MMR performed the bioinformatics analysis, gene ontology study, multivariate analyses, and prepared the figures and tables. MA, M, and LW wrote the draft of the manuscript. MA, LW, MW, and MP managed funding for this study. MW, MMR, and MP reviewed and edited the manuscript. All authors have approved the final version.
This research was carried out under a sub-project of Enhanced Coastal Fisheries in Bangladesh (ECOFISH-BD) activity, which is funded by the United States Agency for International Development (USAID) and jointly implemented by the WorldFish, Bangladesh and South Asia Office, and the Department of Fisheries (DOF), Bangladesh. This sub-project was under the collaborative agreement among WorldFish, Chattogram Veterinary and Animal Sciences University and Universiti Malaysia Terengganu and a part of the ECOFISH-BD project activity.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work is a part of the CGIAR Research Program on Fish Agri-Food Systems (FISH). We would like to express our gratitude toward the administrative support of University Malaysia Terengganu (UMT) and Chittagong Veterinary and Animal Sciences University (CVASU) for supporting the administrative and financial management activities of this sub-project. We would also like to express our gratefulness to research assistants and research associates of the ECOFISH-BD Project for their hard works and efforts in hiring the Hilsa shad fishers’ facilities and assisting us during the samples collection. We would also like to acknowledge Dr. Yoji Igarashi, Laboratory of Aquatic Molecular Biology and Biotechnology, Department of Aquatic Bioscience, The University of Tokyo for his kind help during bioinformatics analysis of this works.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00554/full#supplementary-material
FIGURE S1 | The Principal Component Analysis (PCA) and Linear Discriminant Function Analysis (LDFA) showing the morpho-genetic variation of Hilsa shad Tenualosa ilisha among heterogeneous habitats including the sea, estuary and different rivers. (A) Biplots of the PCA using 22 truss morphometric distances of Hilsa shad; (B) biplots of the LDFA using 22 truss morphometric distances of Hilsa shad; (C) biplots of the PCA using 74 putatively adaptive panels of SNP loci identified by the FST Outflank approach; (D) biplots of the LDFA using 74 putatively adaptive panels of SNP loci identified by the FST OutFLANK approach.
FIGURE S2 | The cluster analysis showing morpho-genetic variation of Hilsa shad Tenualosa ilisha among heterogeneous habitats including the sea, estuary and different rivers. (A) Neighbor-joining trees, based on the Nei’s genetic distances using the 74 putatively adaptive panel of SNP loci identified by the FST OutFLANK approach. (B) Unweighted pair group method with arithmetic mean (UPGMA) dendrogram of hierarchical cluster analysis based on Euclidean distance using the 22 truss morphometric distances of Hilsa shad population collected from their different habitats.
Ahmed, A. S. I., Islam, M. S., Azam, M. S., Khan, M. M. R., and Alam, M. S. (2004). RFLP analysis of the mtDNA D-loop region in Hilsa shad (Tenualosa ilisha) population from Bangladesh. Indian J. Fish. 51, 25–31.
Ahmed, K. K. U., Ahmed, S. U., Haldar, G. C., Hossain, M. R. A., and Ahmed, T. (2005). Primary production and fish yield estimation in the Meghna river system, Bangladesh. Asian Fish. Sci. 18, 95–105.
Ahsan, D., Kabir, A., Rahman, M., Mahabub, S., Yesmin, R., Faruque, M., et al. (2012). Plankton composition, abundance and diversity in Hilsa (Tenualosa ilisha) migratory rivers of Bangladesh during spawning season. Dhaka Univ. J. Biol. Sci. 21, 177–189. doi: 10.3329/dujbs.v21i2.11516
Ali, A. D., Naser, M. N., Bhaumik, U., Hazra, S., and Bhattacharya, S. B. (2014). Migration, Spawning Patterns and Conservation of Hilsa Shad (Tenualosa ilisha) in Bangladesh and India. Gland: International Union for Conservation of Nature and Natural Resources (IUCN), 95.
Allendorf, F. W., Knudsen, K. L., and Leary, R. F. (1983). Adaptive significance of differences in the tissue-specific expression of a phosphoglucomutase gene in rainbow trout. Proc. Natt. Acad. Sci. U.S.A. 80, 1397–1400. doi: 10.1073/pnas.80.5.1397
Allison, M. A. (1998). Geologic framework and environmental status of the Ganges-Brahmaputra Delta. J. Coast. Res. 14, 826–836.
Amano, M., Ok, Y., Amiya, N., and Yamamori, K. (2007). Immunohistochemical localization and ontogenic development of prolactin-releasing peptide in the brain of the ovoviviparous fish species Poecilia reticulata (guppy). Neurosci. Lett. 413, 206–209. doi: 10.1016/j.neulet.2006.10.011
AnvariFar, H., Khyabani, A., Farahmand, H., Vatandoust, S., AnvariFar, H., and Jahageerdar, S. (2011). Detection of morphometric differentiation between isolated up- and downstream populations of siah mahi (Capoeta capoeta gracilis) (Pisces: Cyprinidae) in the Tajan River (Iran). Hydrobiologia 673, 41–52. doi: 10.1007/s10750-011-0748-7
Arai, T., and Amalina, R. (2014). New record of a tropical shad Tenualosa ilisha (Teleostei: Clupeidae) in Malaysian waters. Mar. Biodivers. Rec. 7, 1–4. doi: 10.1017/S1755267214000736
Asaduzzaman, M., Igarashi, Y., Wahab, M. A., Nahiduzzaman, M., Rahman, M. J., Phillips, M. J., et al. (2020). Population genomics of an anadromous Hilsa Shad Tenualosa ilisha species across its diverse migratory habitats: discrimination by fine-scale local adaptation. Genes 11:46. doi: 10.3390/genes11010046
Asaduzzaman, M., Wahab, M. A., Rahman, M. J., Nahiduzzaman, M., Dickson, M. W., Igarashi, Y., et al. (2019). Fine-scale population structure and ecotypes of anadromous Hilsa shad (Tenualosa ilisha) across complex aquatic ecosystems revealed by NextRAD genotyping. Sci. Rep. 9:16050. doi: 10.1038/s41598-019-52465-2
Austin, B., Austin, D. A., Austin, B., and Austin, D. A. (2012). Bacterial Fish Pathogens. Dordrecht: Springer Netherlands, doi: 10.1007/978-1-4020-6069-4
Bae, E., Kim, H. E., Koh, E., and Kim, K. S. (2014). Phosphoglucomutase1 is necessary for sustained cell growth under repetitive glucose depletion. FEBS Lett. 588, 3074–3080. doi: 10.1016/j.febslet.2014.06.034
Barua, D. K. (1990). Suspended sediment movement in the estuary of the Ganges-Brahmaputra-Meghna River system. Mar. Geol. 91, 243–253. doi: 10.1016/0025-3227(90)90039-m
Behera, B. K., Singh, N. S., Paria, P., Sahoo, A. K., Panda, D., Meena, D. K., et al. (2015). Population genetic structure of Indian shad, Tenualosa ilisha Inferred from variation in mitochondrial DNA sequences. J. Environ. Biol. 2015, 1193–1197.
Bernatchez, L. (2016). On the maintenance of genetic variation and adaptation to environmental change: considerations from population genomics in fishes. J. Fish Biol. 89, 2519–2556. doi: 10.1111/jfb.13145
Bhaumik, U. (2015). Migration of Hilsa shad in the Indo-Pacific Region – a review. Int. J. Curr. Res. Acad. Rev. 3, 139–155.
Bhuyan, M. S., Bakar, M. A., Akhtar, A., Hossain, M. B., and Islam, M. S. (2017). Analysis of water quality of the Meghna River using multivariate analyses and RPI. J. Asiat. Soc. Bangladesh Sci. 43, 23–35.
Blaber, S. J. M. (2000). Tropical Estuarine Fishes: Ecology, Exploitation and Conservation. Oxford: Blackwell Science.
Blaber, S. J. M., Cyrus, D. P., and Whitfield, A. K. (1981). The influence of zooplankton food resources on the morphology of the estuarine clupeid Gilchristella aestuarius (Gilchrist, 1914). Environ. Biol. Fishes 6, 351–355. doi: 10.1007/bf00005764
Blaber, S. J. M., Milton, D. A., Chenery, S. R., and Fry, G. C. (2003). New insights into the life history of Tenualosa ilisha and fishery implications. Am. Fish. Soc. Symp. 35, 223–240.
BOBLME (2012). Management Advisory for the Bay of Bengal Hilsa Fishery. Regional Fisheries Management Advisory Committee. Available online at: http://www.boblme.org/documentRepository/BOBLME-2012-Brochure-02.pdf (accessed December 12, 2015).
Boily, P., and Magnan, P. (2002). Relationship between individual variation in morphological characters and swimming costs in brook charr (Salvelinus fontinalis) and yellow perch (Perca flavescens). J. Exp. Biol. 205, 1031–1036.
Borah, S., Vaisakh, G., Jaiswar, A. K., Bhattacharjya, B. K., Sahoo, A. K., Deshmukhe, G., et al. (2019). Association pattern between dimensions of fish and otolith to expedite morphometric variations of three geographically isolated stocks of Tenualosa ilisha (Hamilton, 1822) from diverse ecosystems. Indian J. Fish. 66, 48–54. doi: 10.21077/ijf.2019.66.3.91245-06
Borges, R. M. (2008). Plasticity comparisons between plants and animals: concepts and mechanisms. Plant Signal Behav. 3, 367–375. doi: 10.4161/psb.3.6.5823
Bradshaw, W. E., and Holzapfel, C. M. (2006). Evolutionary response to rapid climate change. Science 312, 1477–1478. doi: 10.1126/science.1127000
Brahmane, M. P., Kundu, S. N., Das, M. K., and Sharma, A. P. (2013). Low genetic diversity and absence of population differentiation of Hilsa (Tenualosa ilisha) revealed by mitochondrial DNA cytochrome b region in Ganga and Hooghly rivers. Afr. J. Biotechnol. 12, 3383–3389. doi: 10.5897/AJB2013.12364
Cadrin, S. X. (2000). Advances in morphometric analysis of fish stock structure. Rev. Fish Biol. Fish. 10, 91–112. doi: 10.1023/A:1008939104413
Carreras, C., Ordóñez, V., Zane, L., Kruschel, C., Nasto, I., Macpherson, E., et al. (2016). Population genomics of an endemic Mediterranean fish: differentiation by fine scale dispersal and adaptation. Sci. Rep. 7:43417. doi: 10.1038/srep43417
Cavalcanti, M. J., Monteiro, L. R., and Lopez, P. R. D. (1999). Landmark based morphometric analysis in selected species of Serranid fishes (Perciformes: Teleostei). Zool. Stud. 38, 287–294.
Chapman, B. B., Hulthen, K., Bronmark, C., Nilsson, A., Skov, C., Hansson, L., et al. (2015). Shape up or ship out: migratory behaviour predicts morphology across spatial scale in a freshwater fish. J. Animal Ecol. 84, 1187–1193. doi: 10.1111/1365-2656.12374
Christensen, K. E., Hou, W., Bahous, R. H., Deng, L., Malysheva, O. V., Arning, E., et al. (2016). Moderate folic acid supplementation and MTHFD1-synthetase deficiency in mice, a model for the R653Q variant, result in embryonic defects and abnormal placental development. Am. J. Clin. Nutr. 104, 1459–1469. doi: 10.3945/ajcn.116.139519
Department of Fisheries [DOF] (2018). Yearbook of Fisheries Statistics of Bangladesh 2017–18. Dhaka: Ministry of Fisheries and Livestock and Department of Fisheries.
Des Roches, S., Post, D. M., Turley, N. E., Bailey, J. K., Hendry, A. P., Kinnison, M. T., et al. (2018). The ecological importance of intraspecific variation. Nat. Ecol. Evol. 2, 57–64. doi: 10.1038/s41559-017-0402-5
Drinan, D. P., Gruenthal, K. M., Canino, M. F., Lowry, D., Fisher, M. C., and Hauser, L. (2018). Population assignment and local adaptation along an isolation-by-distance gradient in Pacific cod (Gadus macrocephalus). Evol. Appl. 11, 1448–1464. doi: 10.1111/eva.12639
Dutton, I. M., Hossain, M. S., and Kabir, H. (2018). Enhanced Coastal Fisheries in Bangladesh Mid-Term Performance Evaluation Report: USAID/ACME Contract No. AID-388-C-14-00001. Vienna, VA: International Business & Technical Consultants, Inc., 99.
Dwivedi, A. K. (2019a). Differentiating three Indian shads by applying shape analysis from digital images. J. Fish Biol. 2019, 1–11. doi: 10.1111/jfb.14074
Dwivedi, A. K. (2019b). Morphometric variations between seasonal migrants of anadromous shad Tenualosa ilisha (Hamilton, 1822) from Hooghly Estuary. India Mar. Freshw. Res. 70, 1427–1435. doi: 10.1071/MF19004
Edwards, J. G., and Michel, W. C. (2002). Odor-stimulated glutamatergic neurotransmission in the zebrafish olfactory bulb. J. Comp. Neurol. 454, 294–309. doi: 10.1002/cne.10445
Elliot, N. G., Haskard, K., and Koslow, J. A. (1995). Morphometric analysis of orange roughy (Hoplostethus atlanticus) off the continental slope of Southern Australia. J. Fish Biol. 46, 202–220. doi: 10.1111/j.1095-8649.1995.tb05962.x
Endler, J. A. (1977). Geographic Variation, Speciation and Clines. Princeton, NJ: Princeton University Press, 262.
Food and Agricultural Organization [FAO] (2017). FishStat J Database. Rome: Food and Agriculture Organization.
Forsman, A., and Wennersten, L. (2016). Inter-individual variation promotes ecological success of populations and species: evidence from experimental and comparative studies. Ecography 39, 630–648. doi: 10.1111/ecog.01357
Fox, J., and Weisberg, S. (2011). An {R} Companion to Applied Regression, 2nd Edn. Thousand Oaks, CA: Sage.
Galili, T. (2015). dendextend: an R package for visualizing, adjusting, and comparing trees of hierarchical clustering. Bioinformatics 31, 3718–3720. doi: 10.1093/bioinformatics/btv428
Gardner, M. B. (1981). Effects of turbidity on feeding rates and selectivity of bluegills. Trans. Am. Fish. Soc. 110, 446–450. doi: 10.1577/1548-86591981110<446:EOTOFR<2.0.CO;2
Garduno-Paz, M. V., Couderic, S., and Adams, C. E. (2010). Habitat complexity modulates phenotype expression through developmental plasticity in the three spine stickleback. Biol. J. Linn. Soc. 100, 407–413. doi: 10.1111/j.1095-8312.2010.01423.x
Ghosh, A. N., Bhattacharya, R. K., and Rao, K. V. (1968). On the identification of the sub-populations of Hilsa ilisha (Ham.) in the Gangetic system with a note on their distribution. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 34, 44–57.
Gregory, R. S., and Northcote, T. G. (1993). Surface, planktonic, and benthic foraging by juvenile chinook salmon (Oncorhynchus tshawytscha) in turbid laboratory conditions. Can. J. Fish. Aquat. Sci. 50, 233–220. doi: 10.1139/f93-026
Gulino, A., Di Marcotullio, L., Canettieri, G., De Smaele, E., and Screpanti, I. (2012). Hedgehog/Gli control by ubiquitination/acetylation interplay. Vitam. Horm. 88, 211–227. doi: 10.1016/B978-0-12-394622-5.00009-2
Halder, D. D. (1968). Observations on the food of young Hilsa ilisha (Hamilton) around Nabadwip in the Hooghly estuary. J. Bombay Nat. Hist. Soc. 65, 796–798.
Hedrick, P. W. (1986). Genetic polymorphism in heterogeneous environments: a decade later. Ann. Rev. Ecol. Syst. 17, 535–566. doi: 10.1146/annurev.es.17.110186.002535
Hendry, A. P., Kinnison, M. T., Heino, M., Day, T., Smith, T. B., Fitt, G., et al. (2011). Evolutionary principles and their practical application. Evol. Appl. 4, 159–183. doi: 10.1111/j.1752-4571.2010.00165.x
Hess, J. E., Campbell, N. R., Close, D. A., Docker, M. F., and Narum, S. R. (2012). Population genomics of Pacific lamprey: adaptive variation in a highly dispersive species. Mol. Ecol. 22, 2898–2916. doi: 10.1111/mec.12150
Hoffmann, A., Griffin, P., Dillon, S., Catullo, R., Rane, R., Byrne, M., et al. (2015). A framework for incorporating evolutionary genomics into biodiversity conservation and management. Clim. Change Response 2, 1–23. doi: 10.1186/s40665-014-0009-x
Hossain, M. S., Sarker, S., Sharifuzzaman, S. M., and Chowdhury, S. R. (2014). Discovering spawning ground of Hilsa Shad (Tenualosa ilisha) in the coastal waters of Bangladesh. Ecol. Model. 282, 59–68. doi: 10.1016/j.ecolmodel.2014.03.001
Hossain, M. S., Sharifuzzaman, S. M., Chowdhury, S. R., and Sarker, S. (2016). Habitats across the lifecycle of hilsa shad (Tenualosa ilisha) in aquatic ecosystem of Bangladesh. Fish. Manag. Ecol. 23, 450–462. doi: 10.1111/fme.12185
Hossain, M. S., Sharifuzzaman, S. M., Rouf, M. A., Pomeroy, R. S., Hossain, M. D., Chowdhury, S. R., et al. (2019). Tropical hilsa shad (Tenualosa ilisha): biology, fishery and management. Fish Fish. 20, 44–65. doi: 10.1111/faf.12323
Hothorn, T., Bretz, F., and Westfall, P. (2008). Simultaneous inferene in general parametric models. Biom. J. 50, 346–363. doi: 10.1002/bimj.200810425
Hulthen, K., Chapman, B. B., Nilsson, P. A., Hansson, L. A., Skov, C., Baktoft, H., et al. (2014). Sex identification and PIT-tagging: tools and prospects for studying intersexual differences in freshwater fishes. J. Fish Biol. 84, 503–512. doi: 10.1111/jfb.12300
Islam, M. M., Essam, Y. M., and Ali, L. (2016). Economic incentives for sustainable Hilsa fishing in Bangladesh: an analysis of the legal and institutional framework. Mar. Policy 68, 8–22. doi: 10.1016/j.marpol.2016.02.005
Jombart, T. (2008). adegenet: an R package for the multivariate analysis of genetic markers. Bioinformatics 22, 1403–1405. doi: 10.1093/bioinformatics/btn129
Kawecki, T. J., and Ebert, D. (2004). Conceptual issues in local adaptation. Ecol. Lett. 7, 1225–1241. doi: 10.1111/j.1461-0248.2004.00684.x
Kirkpatrick, M., and Barton, N. (2006). Chromosome inversions, local adaptation and speciation. Genetics 173, 419–434. doi: 10.1534/genetics.105.047985
Klemetsen, A. (2010). The charr problem revisited: exceptional phenotypic plasticity promotes ecological speciation in postglacial lakes. Freshw. Rev. 3, 49–75. doi: 10.1608/FRJ-3.1.3
Kuparinen, A., and Merilä, J. (2007). Detecting and managing fisheries induced evolution. Trends Ecol. Evol. 22, 652–659. doi: 10.1016/j.tree.2007.08.011
Langerhans, R. B., and Reznick, D. N. (2010). Ecology and Evolution of Swimming Performance in Fishes: Predicting Evolution with Biomechanics. Fish Locomotion: An Eco-Ethological Perspective, eds P. Domenici and B. G. Kapoor (Enfield, NH: Science Publishers), 200–248.
Larson, C. A., and Belovsky, G. E. (2013). Salinity and nutrients influence species richness and evenness of phytoplankton communities in microcosm experiments from Great Salt Lake, Utah, USA. J. Plankton Res. 35, 1154–1166. doi: 10.1093/plankt/fbt053
Lee, C. E., and Gelembiuk, G. W. (2008). Evolutionary origins of invasive populations. Evol. App. 1, 427–448. doi: 10.1111/j.1752-4571.2008.00039.x
Lee, M. S., Bonner, J. R., Bernard, D. J., Sanchez, E. L., Sause, E. T., Prentice, R. R., et al. (2012). Disruption of the folate pathway in zebrafish causes developmental defects. BMC Dev. Biol. 12:12. doi: 10.1186/1471-213X-12-12
Lehtiniemi, M., Engström-Öst, J., and Viitasalo, M. (2005). Turbidity decreases anti-predator behavior in pike larvae. Esox lucius. Environ. Biol. Fish. 73, 1–8. doi: 10.1007/s10641-004-5568-4
Lowerre-Barbieri, S., DeCelles, G., Pepin, P., Catalán, I. A., Muhling, B., Erisman, B., et al. (2017). Reproductive resilience: a paradigm shift in understanding spawner-recruit systems in exploited marine fish. Fish Fish. 18, 285–312. doi: 10.1111/faf.12180
Lucek, K., Sivasundar, A., and Seehausen, O. (2014). Disentangling the role of phenotypic plasticity and genetic divergence in contemporary ecotype formation during a biological invasion. Evolution 68, 2619–2632. doi: 10.1111/evo.12443
Mazumder, S. K., and Alam, M. S. (2009). High levels of genetic variability and differentiation in Hilsa shad, Tenualosa ilisha (Clupeidae, Clupeiformes) populations revealed by PCR-RFLP analysis of the mitochondrial DNA D-loop region. Genet. Mol. Biol. 32, 190–196. doi: 10.1590/S1415-47572009005000023
Mazzarella, A. B., Voje, K. L., Hansson, T. H., Taugbøl, A., and Fischer, B. (2015). Strong and parallel salinity-induced phenotypic plasticity in one generation of threespine stickleback. J. Evol. Biol. 28, 667–677. doi: 10.1111/jeb.12597
Meyers, L. A., and Bull, J. J. (2002). Fighting change with change: adaptive variation in an uncertain world. Trends Ecol. Evol. 17, 551–557. doi: 10.1016/S0169-5347(02)02633-2
Miah, M. S. (2015). Climatic and anthropogenic factors changing spawning pattern and production zone of hilsa fishery in the Bay of Bengal. Weather Clim. Extrem. 7, 109–115. doi: 10.1016/j.wace.2015.01.001
Milton, D. A., and Chenery, S. R. (2001). Can otolith chemistry detect the population structure of the shad hilsa Tenualosa ilisha? Comparison with genetic and morphological studies. Mar. Ecol. Prog. Ser. 222, 239–251. doi: 10.3354/meps222239
Mohindra, V., Dangi, T., Tripathi, R. K., Kumar, R., Singh, R. K., Jena, J. K., et al. (2019). Draft genome assembly of Tenualosa ilisha, Hilsa shad, provides resource for osmoregulation studies. Sci. Rep. 9:16511. doi: 10.1038/s41598-019-52603-w
Morinville, G., and Rasmussen, J. (2008). Distinguishing between juvenile anadromous and resident brook trout (Salvelinus fontinalis) using morphology. Environ. Biol. Fish. 81, 171–184. doi: 10.1007/s10641-007-9186-9
Morinville, G. R., and Rasmussen, J. B. (2003). Early juvenile bioenergetic differences between anadromous and resident brook trout (Salvelinus fontinalis). Can. J. Fish. Aquat. Sci. 60, 401–410. doi: 10.1139/f03-036
Morinville, G. R., and Rasmussen, J. B. (2006). Does life-history variability in salmonids affect habitat use by juveniles? A comparison among streams open and closed to anadromy. J. Animal Ecol. 75, 693–704. doi: 10.1111/j.1365-2656.2006.01090.x
Nakada, T., Zandi-Nejad, K., Kurita, Y., Kudo, H., Broumand, V., Kwon, C. Y., et al. (2005). Roles of Slc13a1 and Slc26a1 sulfate transporters of eel kidney in sulfate homeostasis and osmoregulation in freshwater. Am. J. Physiol. Regul. Integr. Comp. Physiol. 289, R575–R585. doi: 10.1152/ajpregu.00725.2004
Nolting, K., Park, J. H., Tegtmeyer, L. C., Zühlsdorf, A., Grüneberg, M., Rust, S., et al. (2017). Limitations of galactose therapy in phosphoglucomutase 1 deficiency. Mol. Genet. Metab. Rep. 13, 33–40. doi: 10.1016/j.ymgmr.2017.07.010
Osborne, R. I., Bernot, M. J., and Findlay, S. E. G. (2015). Changes in nitrogen cycling processes along a salinity gradient in tidal wetlands of the Hudson River, New York, USA. Wetlands 35, 323–334. doi: 10.1007/s13157-014-0620-4
Ou, M., Hamilton, T. J., Eom, J., Lyall, E. M., Gallup, J., Jiang, A., et al. (2015). Responses of pink salmon to CO2-induced aquatic acidification. Nat. Clim. Change 5, 950–955. doi: 10.1038/nclimate2694
Paez, D. J., Morrissey, M., Bernatchez, L., and Dodson, J. J. (2010). The genetic basis of early-life morphological traits and their relation to alternative male reproductive tactics in Atlantic salmon. J. Evol. Biol. 23, 757–768. doi: 10.1111/j.1420-9101.2010.01941.x
Palstra, F. P., Connell, M. F. O., and Ruzzante, D. E. (2007). Population structure and gene flow reversals in Atlantic salmon (Salmo salar) over contemporary and long-term temporal scales: effects of population size and life history. Mol. Ecol. 16, 4504–4522. doi: 10.1111/j.1365-294X.2007.03541.x
Peterson, B. G., and Carl, P. (2019). Performance Analytics: Econometric Tools for Performance and Risk Analysis. R Package Version 1.5.3. 2019. Available online at: https://cran.r-project.org/web/packages/PerformanceAnalytics/PerformanceAnalytics.pdf (accessed September 04, 2019).
Pfenning, D. W., Wund, M. A., Snell-Rood, E. C., Cruickshank, T., Schlichting, C. D., and Moczek, A. P. (2010). Phenotypic plasticity’s impact on diversification and speciation. Trends Ecol. Evol. 25, 459–467. doi: 10.1016/j.tree.2010.05.006
Pillay, S. R., and Rosa, H. (1963). Synopsis of biological data on Hilsa ilisha (Hamilton). FAO Fish. Biol. Synop. 25, 1–6.
Pillay, T. V. R. (1958). Biology of the hilsa, Hilsa ilisha (Hamilton) of the river Hooghly. Indian J. Fish. 5, 201–257.
Quddus, M. A., Shimizu, M., and Nose, Y. (1984). Meristic and morphometric differences in two types of Hilsa Ilisha in Bangladesh waters. Bull. Jpn. Soc. Sci. Fish. 50, 43–49. doi: 10.2331/suisan.50.43
Quinn, T. P. (2005). The Behavior and Ecology of Pacific Salmon and Trout. Seattle, WA: University of British Columbia Press.
R Development Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R foundation for statistical computing.
Rahman, M., and Naevdal, G. (2000). Population genetics studies of Hilsa shad, Tenualosa ilisha (Hamilton), in Bangladesh waters: evidence for the existence of separate gene pools. Fish. Manag. Ecol. 7, 401–412. doi: 10.1046/j.1365-2400.2000.00211.x
Rahman, M. A., Miah, M. S., Rahman, M. J., Haldar, G. C., and Mazid, M. A. (1997). Application of biometric and electrophoretic methods for the stock discrimination of hilsa (Tenualosa ilisha) in Bangladesh water. Ind. J. An. Sci. 67, 1022–1027.
Rahman, M. J., Wahab, M. A., Amin, S. M. N., Nahiduzzaman, M., and Romano, N. (2018). Catch trend and stock assessment of Hilsa Tenualosa ilisha using digital image measured length-frequency data. Mar. Coast. Fish. 10, 386–401. doi: 10.1002/mcf2.10034
Rajagopalan, K. N., Egnatchik, R. A., Calvaruso, M. A., Wasti, A. T., Padanad, M. S., Boroughs, L. K., et al. (2015). Metabolic plasticity maintains proliferation in pyruvate dehydrogenase deficient cells. Cancer Metab. 3:7. doi: 10.1186/s40170-015-0134-4
Rikta, S. Y., Rahaman, M. S., Mehjabin, J. J., Uddin, M. K., Kabir, M. M., and Tareq, S. M. (2016). Evaluation of water quality parameters and Humic substance status of Bangshi, Dhaleshwari and Padma Rivers in Bangladesh. Int. J. Environ. Sci. 6, 1129–1139. doi: 10.6088/ijes.6018
Rockman, M. V. (2012). The QTN program and the alleles that matter for evolution: all that’s gold does not glitter. Evolution 66, 1–17. doi: 10.1111/j.1558-5646.2011.01486.x
Roy, D., Docker, M. F., Hehanussa, P., Heath, D. D., and Haffner, G. D. (2004). Genetic and morphological data supporting the hypothesis of adaptive radiation in the endemic fish of Lake Matano. J. Evol. Biol. 17, 1268–1276. doi: 10.1111/j.1420-9101.2004.00783.x
Salini, J. P., Milton, D. A., Rahman, M. J., and Hussain, M. G. (2004). Allozyme and morphological variation throughout the geographic range of the tropical shad, Hilsa Tenualosa ilisha. Fish. Res. 66, 53–69. doi: 10.1016/S0165-7836(03)00124-3
Sangiao-Alvarellos, S., Arjona, F. J., Martin del Rio, M. P., Miquez, J. M., Mancera, J. M., and Soengas, J. L. (2005). Time course of osmoregulatory and metabolic changes during osmotic acclimation in Sparus auratus. J. Exp. Bio. 208, 4291–4304. doi: 10.1242/jeb.01900
Sarker, M. H., Huque, I., Alam, M., and Koudstaal, R. (2003). Rivers, chars and char dwellers of Bangladesh. Int. J. River Basin Manag. 1, 61–80. doi: 10.1080/15715124.2003.9635193
Sebastien, L., Josse, J., and Husson, F. (2008). FactoMineR: an R package for multivariate analysis. J. Stat. Softw. 25, 1–18. doi: 10.18637/jss.v025.i01
Sen, S., Jahageerdara, S., Jaiswara, A. K., Chakrabortya, S. K., Sajinab, A. M., and Dash, G. R. (2011). Stock structure analysis of Decapterus russelli (Ruppell, 1830) from east and westcoast of India using truss network analysis. Fish. Res. 112, 38–43. doi: 10.1016/j.fishres.2011.08.008
Shafi, M., Quddus, M. M. A., and Hossain, H. A. (1977). Morphometric study of the population of Hilsa ilisha (Hamilton-Buchanan) from the river Meghna. Proc. Second Bangladesh Sci. Conf. 2, 22–22.
Skov, C., Brodersen, J., Nilsson, P. A., Hansson, L. A., and Bronmark, C. (2008). Inter- and size-specific patterns of fish seasonal migration between a shallow lake and its streams. Ecol. Freshw. Fish 17, 406–415. doi: 10.1111/j.1600-0633.2008.00291.x
Snell-Rood, E. C. (2013). An overview of the evolutionary causes and consequences of behavioral plasticity. Anim. Behav. 85, 1004–1011. doi: 10.1016/j.anbehav.2012.12.031
Sonin, O., Spanier, E., Levi, D., Patti, B., Rizzo, P., and Andreoli, M.-G. (2007). Nanism (dwarfism) in fish: a comparison between red mullet Mullus barbatus from the southeastern and the central Mediterranean. Mar. Ecol. Prog. Ser. 343, 221–228. doi: 10.3354/meps06917
Stelkens, R. B., Jaffuel, G., Escher, M., and Wedekind, C. (2012). Genetic and phenotypic population divergence on a microgeographic scale in brown trout. Mol. Ecol. 21, 2896–2915. doi: 10.1111/j.1365-294X.2012.05581.x
Svanback, R., and Schluter, D. (2012). Niche specialization influences adaptive phenotypic plasticity in the threespine stickleback. Am. Nat. 180, 50–59. doi: 10.1086/666000
Tamario, C., Sunde, J., Petersson, E., Tibblin, P., and Forsman, A. (2019). Ecological and evolutionary consequences of environmental change and management actions for migrating fish. Front. Ecol. Evol. 7:271. doi: 10.3389/fevo.2019.00271
Turan, C. (1999). A note on the examination of morphometric differentiation among fish populations: the truss system. Turk. J. Zool. 23, 259–263.
Tzeng, T. D. (2004a). Morphological variation between populations of spotted mackerel (Scomber australasicus) off Taiwan. Fish. Res. 68, 45–55. doi: 10.1016/j.fishres.2004.02.011
Tzeng, T. D. (2004b). Stock identification of sword prawn Parapenaeopsis hardwickii in the East China Sea and Taiwan Strait inferred by morphometric variation. Fish. Sci. 70, 758–764. doi: 10.1111/j.1444-2906.2004.00868.x
Valiente, A. G., Juanes, F., Nuñez, P., and Garcia-Vazquez, E. (2010). Brown trout (Salmo trutta) invasiveness: plasticity in life-history is more important than genetic variability. Biol. Invasions 12, 451–462. doi: 10.1007/s10530-009-9450-3
Varjosalo, M., and Taipale, J. (2008). Hedgehog: functions and mechanisms. Genes Dev. 22, 2454–2472. doi: 10.1101/gad.1693608
Veasey, E. A., Schammas, E. A., Vencovsky, R., Martins, P. S., and Bandel, G. (2001). Germplasm characterization of Sesbania accessions based on multivariate analyses. Genet. Resour. Crop Evol. 48, 79–90. doi: 10.1023/A:1011238320630
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S, 4th Edn. New York, NY: Springer.
Vera, M., Diez-del-Molino, D., and Garcia-Marin, J. L. (2016). Genomic survey provides insights into the evolutionary changes that occurred during European expansion of the invasive mosquitofish (Gambusia holbrooki). Mol. Ecol. 25, 1089–1105. doi: 10.1111/mec.13545
Watanabe, S., and Kaneko, T. (2010). Prolactin-releasing peptide receptor expressed in the pituitary in Mozambique tilapia Oreochromis mossambicus: an aspect of prolactin regulatory mechanisms. Gen. Comp. Endocrinol. 167, 27–34. doi: 10.1016/j.ygcen.2010.03.004
West-Eberhard, M. J. (2003). Developmental Plasticity and Evolution. New York, NY: Oxford University Press.
Wund, M. A., Valena, S., Wood, S., and Baker, J. A. (2012). Ancestral plasticity and allometry in three spine stickleback reveal phenotypes associated with derived, freshwater ecotypes. Biol. J. Linn. Soc. 105, 573–583. doi: 10.5061/dryad.hb824gd4
Yakubu, A., and Okunsebor, S. A. (2011). Morphometric differentiation of two Nigerian fish species (Oreochromis niloticus and Lates niloticus) using principal components and discriminant analysis. Int. J. Morphol. 29, 1429–1434. doi: 10.4067/s0717-95022011000400060
Yan, A., Chen, T., Chen, S., Tang, D., Liu, F., Jiang, X., et al. (2016). Signal transduction mechanism for glucagon-induced leptin gene expression in goldfish liver. Int. J. Biol. Sci. 12, 1544–1554. doi: 10.7150/ijbs.16612
Keywords: Hilsa shad (Tenualosa ilisha), anadromous fish, morphogenetic divergence, environmental heterogeneity, nextRAD genotyping, local adaptation
Citation: Asaduzzaman M, Wahab MA, Rahman MM, Mariom, Nahiduzzaman M, Rahman MJ, Roy BK, Phillips MJ and Wong LL (2020) Morpho-Genetic Divergence and Adaptation of Anadromous Hilsa shad (Tenualosa ilisha) Along Their Heterogenic Migratory Habitats. Front. Mar. Sci. 7:554. doi: 10.3389/fmars.2020.00554
Received: 28 February 2020; Accepted: 16 June 2020;
Published: 07 July 2020.
Edited by:
Ciro Rico, The University of the South Pacific, FijiReviewed by:
Alan Whitfield, South African Institute for Aquatic Biodiversity, South AfricaCopyright © 2020 Asaduzzaman, Wahab, Rahman, Mariom, Nahiduzzaman, Rahman, Roy, Phillips and Wong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Md Asaduzzaman, YXNhZHV6emFtYW5AY3Zhc3UuYWMuYmQ=; YV96YW1hbmJhdUB5YWhvby5jb20=; Li Lian Wong, bGlsaWFuQHVtdC5lZHUubXk=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.