- 1Department of Marine Organism Taxonomy and Phylogeny, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Key Laboratory of Marine Ecosystem Dynamics, Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China
- 3Museums Victoria, Melbourne, VIC, Australia
- 4Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
- 5Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 6College of Marine Science, University of Chinese Academy of Sciences, Beijing, China
- 7School of Oceanography, Shanghai Jiao Tong University, Shanghai, China
Eiconaxius Bate, 1888 is a genus of axiid shrimps exclusively associated with deep-sea hexactinellid sponges. Due to its special morphology and habitat, Eiconaxius is taxonomically and ecologically controversial. Based on material recently collected from seamounts in the northwestern Pacific, a new species of Eiconaxius is described. Intraspecific morphological and genetic variation and host specificity were evaluated. The complete mitochondrial genome of the new species was sequenced to explore the systematic status of Eiconaxius and some other axiidean taxa. Our analyses showed that differentiation of the new species occurs both allopatrically and sympatrically, probably resulting from the interaction of geographical isolation and deep water current movement, rather than from adaptation to different hosts. In addition, species of Eiconaxius are suggested to have wider ranges of distribution and host than expected. The reconstructed mitogenomic phylogeny supported merging Eiconaxius into Axiidae, and recognized most axiidean families, except that Strahlaxiidae was suggested to be paraphyletic. However, more comprehensive taxon sampling is still needed to resolve the explicit internal relationships among Axiidea.
Introduction
Eiconaxius Bate, 1888 is an axiidean genus (Crustacea: Decapoda: Axiidae) comprising more than 30 species confined to deep waters in all except polar oceans (Sakai, 2011; WoRMS, 2020; Poore, in press). Like few other axiid genera, such as Montanaxius Dworschak, 2016 and Spongiaxius Sakai and de Saint Laurent, 1989, Eiconaxius is associated with deep-sea hexactinellid sponges and frequently found as monogamous pairs in the internal cavity of sponge hosts (Ortmann, 1891; Faxon, 1893, 1896; Bouvier, 1925; De Man, 1925; Kensley, 1996; Komai, 2011; Sakai, 2011, 2014; Komai and Tsuchida, 2012; Poore, 2017, in press; Poore and Dworschak, 2018). Many species descriptions are based on few specimens, but some samples contain hundreds of individuals such as those in the collections of the Muséum nationale d’Histoire naturelle, Paris (GCBP observations). Differences between species are slight (Kensley, 1996; Poore, in press). The known distributional range of most species is restricted, but others are apparently widely distributed (Sakai, 2011; Komai and Tsuchida, 2012; Poore, in press), which raises the question of whether restricted distributions are real or can be attributed to limited sampling. Some species of Eiconaxius have been reported to be sympatric (Sakai, 1992, 2011; Komai et al., 2010; Komai, 2011; Poore, 2018, in press). Increasing evidence has shown that host selection could drive speciation in diverse shallow water marine organisms (Munday et al., 2004; Morrison et al., 2004; Faucci et al., 2007; Tsang et al., 2009; Hurt et al., 2013; Kou et al., 2013, 2015; Horká et al., 2016). Perhaps host specificity also plays a key role in speciation of Eiconaxius and some other deep-sea crustaceans.
The classification and systematic status of Eiconaxius within Axiidae has been controversial. Molecular phylogenetic analyses using three ribosomal RNA genes (16, 18S, 28S rRNA; Tsang et al., 2008a) and two nuclear protein coding genes (NaK, PEPCK; Tsang et al., 2008b; Chu et al., 2009) suggested that a family erected for Eiconaxius, Eiconaxiidae Sakai and Ohta, 2005, was embedded within the clade of Axiidae, a conclusion supported by Robles et al. (2009), Poore and Collins (2009), Komai and Tsuchida (2012), and Poore (2017, in press) but not by Sakai (2011, 2014).
Mitochondrial genomes have been widely used to resolve deeper phylogenetic relationships and are promising to settle systematic disputes. The typical metazoan mitochondrial genome is a closed circular molecule of 15–20 kb, usually comprising 37 genes, including 13 protein coding genes (PCGs), two ribosomal RNA (rRNA) genes, and 22 transfer RNA (tRNA) genes (Boore, 1999). Mitochondrial genomes include more genetic information than incomplete mitochondrial (e.g., COI, 12S rRNA, 16S rRNA) and nuclear genes (e.g., 18S rRNA, 28S rRNA, H3, NaK, PEPCK) and have the advantages of rather conserved gene content, easily accessible nature, and diverse evolutionary rates among different segments (Shen et al., 2017; Sun et al., 2018b; Li et al., 2019). Besides, the gene order and the RNA secondary structure can provide additional useful evolutionary information (Macey et al., 1997; Roehrdanz et al., 2002). As the technology and affordability of next-generation sequencing (NGS) matures, phylogenetic studies of decapods based on whole mitochondrial genome sequences have recently become popular (Tan et al., 2015, 2017, 2018a,b, 2019; Basso et al., 2017; Cheng et al., 2018; Sun et al., 2018a, b, 2019a,b). Currently, mitochondrial genomes of 11 species of Axiidae from five families, Axiidae Huxley, 1879, Callianassidae Dana, 1852, Callichiridae Manning and Felder, 1991, Eucalliacidae Manning and Felder, 1991, and Strahlaxiidae Poore, 1994, are available. However, only a single representative of Axiidae Calocaris macandreae Bell, 1846 has been reported, and no mitochondrial genome of the genus Eiconaxius has been determined. This makes evaluation of the monophyly of Axiidae and the systematic status of Eiconaxiidae using mitochondrial genomes unachievable.
During recent cruises to seamounts in the northwestern Pacific, conducted by the Second Institute of Oceanography, Ministry of Natural Resources (SIOMNR) and Institute of Oceanology, Chinese Academy of Sciences (IOCAS), five shrimps of the genus Eiconaxius and their sponge hosts were collected from Caiwei and Weijia Guyots in the Magellan Seamount Chain and from one unnamed seamount on the Caroline Ridge. This new material provides an opportunity to present: (1) descriptions of a new species of Eiconaxius; (2) assessment of the intraspecific divergence of the new species between populations using two mitochondrial (16S rRNA and COI) genes; (3) discussion of host specificity and the role it plays in the diversification of deep-sea commensal axiid shrimps; (4) evaluation of the systematic status of Eiconaxius and other families of Axiidea based on mitogenomic analyses. Our study provides new insights into the distribution, speciation, and phylogeny of axiidean shrimps and other deep-sea crustaceans.
Materials and Methods
Specimen Collection
Three specimens of Eiconaxius were collected during survey cruises near Caiwei Guyot, Magellan Seamount Chain in 2014 (one ovigerous female), and near Weijia Guyot, Magellan Seamount Chain in 2016 (one ovigerous female, one mature male), using RV Xiangyanghong 9. These three axiid shrimps and their sponge hosts were captured by the Chinese manned submersible Jiaolong. Two more ovigerous females were collected during a survey cruise from two sites on an unnamed seamount on the Caroline Ridge in 2019 using the RV Kexue. These two and their sponge hosts were captured by the remotely operated vehicle (ROV) Faxian. All specimens were immediately fixed and preserved in 75% ethanol after being photographed on board. When the specimens were unloaded and carried to the laboratory, fresh 75% ethanol was replaced. The specimens are deposited in the Sample Repository of the Second Institute of Oceanography (SRSIO), Ministry of Natural Resources, Hangzhou, China, and the Marine Biological Museum, Chinese Academy of Sciences (MBMCAS), Qingdao, China.
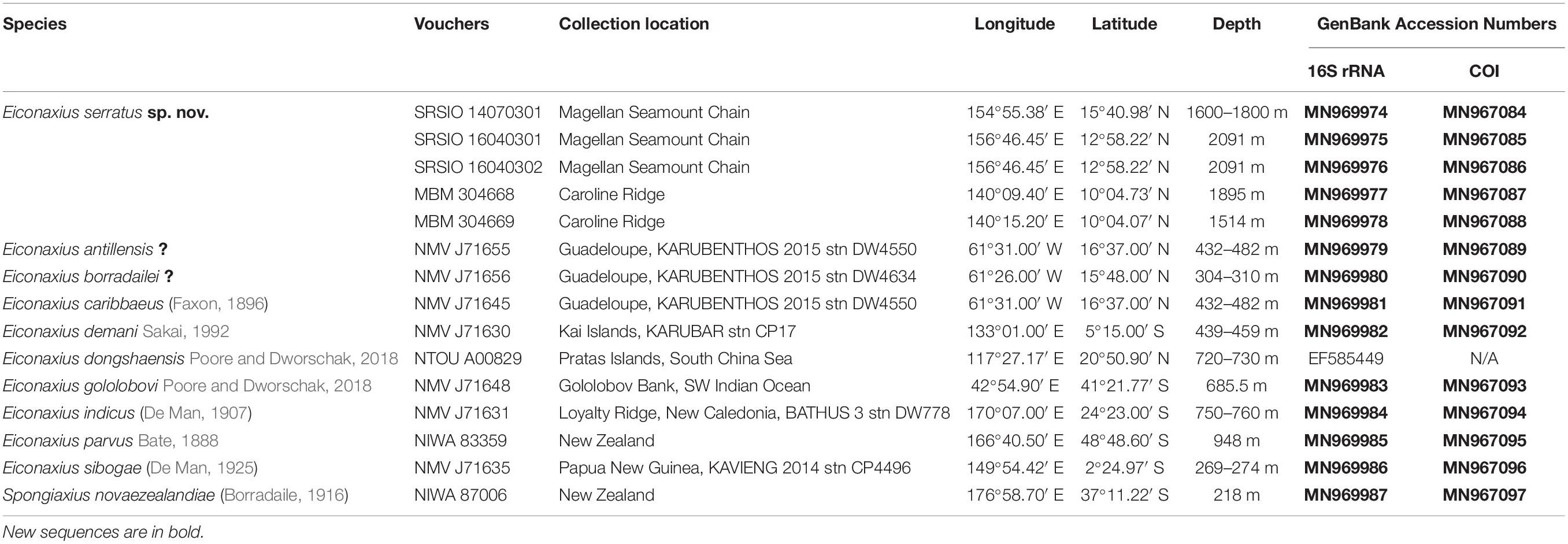
To resolve the taxonomic status of the undescribed Eiconaxius and to explore interspecific genetic divergence, tissue of nine other species of Eiconaxius was sampled from collections in Museums Victoria, Melbourne, Australia (NMV), and National Institute of Water and Atmospheric Research, Wellington, New Zealand (NIWA). One individual from another deep-sea sponge associated axiid species, Spongiaxius novaezealandiae (Borradaile, 1916), was included in the 16S rRNA dataset, with an aligned length of 559 bp, and in the COI dataset with an aligned length of 591 bp (Table 1). The collection locations of all the axiid specimens used in the present study are shown in Figure 11A.
Morphology Observation
On board, the shrimps and their sponge hosts were photographed using a Canon EOS-1D Digital Single Lens Reflex camera. In the laboratory, the specimens were measured and illustrated under a Zeiss SteREO Discovery V8 stereomicroscope. Carapace length (cl) was measured from the tip of the rostrum to the posterior end of the carapace. Total length (tl) was measured from the tip of the rostrum to the posterior end of the telson, with body stretched. The diagnosis was derived from the same DELTA database (Dallwitz, 2018) of species and characters of Eiconaxius as that used by Poore (in press).
DNA Extraction, PCR Amplification, and Sanger Sequencing
Total genomic DNA of the specimens was extracted from a small piece of muscle tissue (5∼10 mg). DNA was extracted using a QIAamp DNA Micro Kit (Qiagen, Hilden, Germany) and then eluted in 50 μL of sterile distilled H2O (RNase free), and stored at −20°C. Polymerase chain reaction (PCR) amplification was carried out in a reaction mix containing 5 μL of template DNA, 25 μL of Premix TaqTM (Takara, Otsu, Shiga, Japan), 1 μL of each primer (10 mM), and sterile distilled H2O to a total volume of 50 μL. Mitochondrial 16S rRNA and COI genes were amplified using the primers 16S-ar/br (Simon et al., 1994) and LCO1490/HCO2198 (Folmer et al., 1994), respectively, with the following thermal profile: initial denaturation for 3 min at 94°C, followed by 35–40 cycles of denaturation at 94°C for 30 s, annealing at 48°C for 40 s, extension at 72°C for 30 s, and a final extension at 72°C for 10 min. PCR products were purified using the WizardTM SV Gel and PCR Clean-UP System (Promega, Madison, WI, United States) before sequencing. The purified PCR products were bidirectionally sequenced using the same forward and reverse primers for PCR amplification with ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, United States).
Next-Generation Sequencing, Mitochondrial Genome Assembly, and Annotation
One μg of purified total genomic DNA of the holotype was fragmented and used to construct a paired-end library (insert size 300∼500 bp) using TruSeqTM Nano DNA Sample Prep Kit (Illumina, United States). The library was sequenced on the Illumina Hiseq 4000 platform (2 × 150 bp paired-end reads) by BIOZERON Co., Ltd (Shanghai, China).
Prior to assembly, raw reads were filtered to remove the reads with adaptors, the reads with a quality score below 20 (Q < 20), the reads containing a percentage of uncalled based (“N” characters) equal or greater than 10% and the duplicated sequences. Then the mitochondrial genome was reconstructed using a combination of de novo and reference-guided assemblies, and the following three steps were used to assemble mitochondria genomes. First, the filtered reads were assembled into contigs using SOAPdenovo 2.04 (Luo et al., 2012). Second, contigs were aligned to the reference mitochondrial genome of Calocaris macandreae (KC107812) using BLAST, and aligned contigs (≥80% similarity and query coverage) were ordered according to the reference genome. Third, clean reads were mapped to the assembled draft mitochondrial genome to correct the wrong bases, and the majority of gaps were filled through local assembly.
The mitochondrial genome was annotated using the MITOS2 webserver (Bernt et al., 2013). Locations and sizes of the protein-coding genes (PCGs) were identified by Open Reading Frame Finder (ORFfinder) available on NCBI with the invertebrate mitochondrial code. Transfer RNA (tRNA) genes were identified by tRNAscan-SE 2.0 webserver (Chan and Lowe, 2019), and their secondary structures were predicted and visualized using Forna (Kerpedjiev et al., 2015). Ribosome RNA (rRNA) genes were delineated by rRNAmmer 1.2 webserver (Lagesen et al., 2007). All the gene predictions were reconfirmed by comparing nucleotide or amino acid sequences with those of published mitochondrial genomes of Axiidae using the Basic Local Alignment Search Tool (BLAST). The frequencies of both amino acids and codons, and the relative synonymous codon usage (RSCU), were calculated using MEGA 6.06 (Tamura et al., 2013). The assembly of the mitochondrial genome was verified by comparison with the 16S rRNA and COI sequence obtained from the foregoing Sanger sequencing. The circular mitochondrial genome map of the new axiid species was generated by CGView Server (Grant and Stothard, 2008).
Phylogenetic Analysis
For the 16S rRNA and COI gene sequences obtained from Sanger sequencing, sequence chromatograms were checked using CHROMAS 2.23 (Technelysium Pty Ltd) by eye. The forward and reverse sequence fragments were assembled by CONTIG EXPRESS (a component of Vector NTI Suite 6.0, Life Technologies, Carlsbad, CA, United States). The homologous sequences were aligned by MAFFT version 7 webserver (Katoh et al., 2019) with default parameters, and manually trimmed to the same length for all the taxa. The Kimura’s 2-parameter genetic distances were calculated using MEGA 6.06 (Tamura et al., 2013).
Phylogeny was reconstructed based on the mitochondrial genomes of the new species and those of 59 decapod species belonging to two suborders, 10 infraorders, with three species of Stomatopoda and one species of Euphausiacea as outgroups (Supplementary Table S1). The nucleotide sequences of 13 PCGs were aligned by MAFFT version 7 webserver (Katoh et al., 2019) with default parameters. GBlocks v0.91b (Castresana, 2000) was used to eliminate the highly divergent and poorly aligned segments of each gene (GBlocks parameters: minimum length of a block = 5; allowed gap positions = with half). Then the trimmed alignments were concatenated into a single dataset consisting of 13 PCGs using Sequence Matrix 1.8 (Vaidya et al., 2011), and each gene was treated as separate data partition in the subsequent analyses. Phylogenetic relationships were inferred from the concatenated dataset using both maximum likelihood (ML) and Bayesian inference (BI) methods. For ML analyses, the best-fit substitution models (including FreeRate heterogeneity models) and partition schemes were inferred by ModelFinder (Kalyaanamoorthy et al., 2017) implemented in IQ-TREE 1.6.10 (Nguyen et al., 2015). The ML tree was reconstructed using IQ-TREE, and branch support was assessed by performing Bayesian-like transformation of aLRT (aBayes) test (Anisimova et al., 2011) as well as ultrafast bootstrap (BP) with 1,000 replicates (Hoang et al., 2018). For BI analyses, the best-fit substitution models and partition schemes were inferred by PartitionFinder 2.1.1 (Lanfear et al., 2017) with the ‘greedy’ algorithm according to the Bayesian information criterion. Bayesian analysis was conducted using MrBayes 3.2.7a (Ronquist et al., 2012). Two independent runs were carried out with four Markov Chains for 20,000,000 generations started from a random tree, with sampling every 1,000 generations. The average standard deviation of split frequencies and the likelihood values were monitored, to determine whether the two runs converged onto the stationary distribution. The first 25% (5,000) trees generated prior to the achievement of stationarity of the log-likelihood values were discarded as burn-in. The remaining trees were used to construct the 50% majority rule consensus tree and to estimate the posterior probabilities (PP). The effective sample size (ESS) values for all sampled parameters were diagnosed by Tracer 1.7.1 (Rambaut et al., 2018) to ensure convergence. The phylogenetic trees and node labels were visualized using FigTree 1.4.3 (Rambaut, 2016). Finally, all newly obtained 16 rRNA, COI, and mitochondrial genome sequences were submitted to the GenBank database.
Gene Order Analysis
We mapped all mitochondrial gene orders on to the phylogeny for comparison. Additionally, the putative ancestral state of the pancrustacean ground pattern and the mitochondrial genome order of the new species were pairwise compared to predict the mitochondrial genome rearrangement events (e.g., gene reversals, transpositions, reverse transpositions, tandem duplication random loss [TDRL]) using Common interval Rearrangement Explorer, heuristically exploring mitochondrial rearrangements based on common intervals (CREx; Bernt et al., 2007).
Results
Taxonomy
Axiidae Huxley, 1879.
Eiconaxius Bate, 1888.
Remarks
For the most recent diagnosis of Eiconaxius see Poore (2017).
Eiconaxius serratus sp. nov. (Figures 1–6).

Figure 1. Eiconaxius serratus sp. nov. Holotype, SRSIO 14070301 (ovigerous female, cl 9.0 mm). Scale: 5 mm.
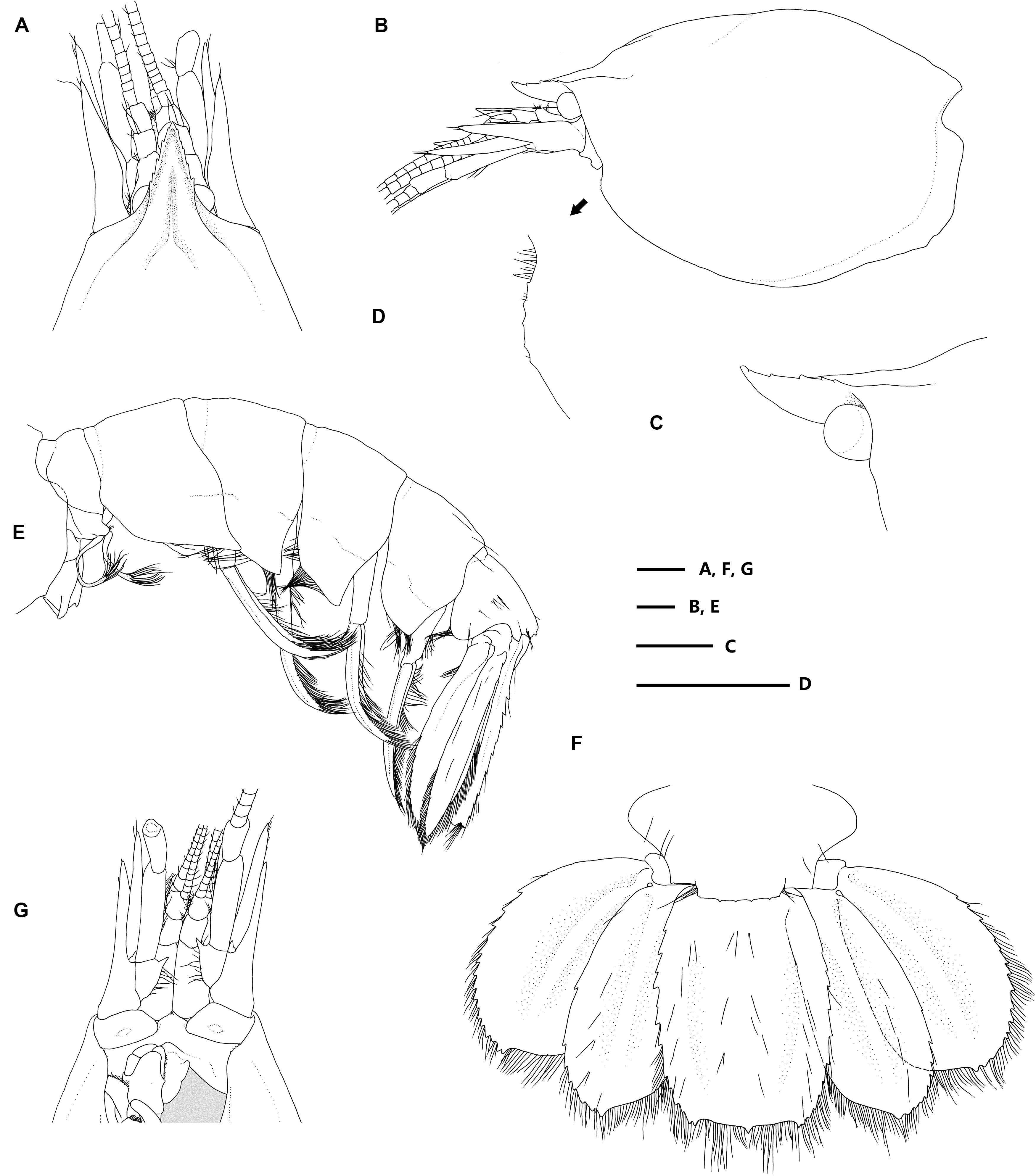
Figure 2. Eiconaxius serratus sp. nov. Holotype, SRSIO 14070301 (ovigerous female, cl 9.0 mm). (A) anterior carapace, antenna, antennule, dorsal view; (B) carapace, antenna, antennule, lateral view; (C) rostrum, anterior median carina, lateral view; (D) pterygostomial angle, lateral view; (E) pleon, pleopods, telson, uropods, lateral view; (F) pleomere 6, telson, uropods, dorsal view; (G) anterior carapace, antenna, antennule, ventral view. Scales: 1 mm.
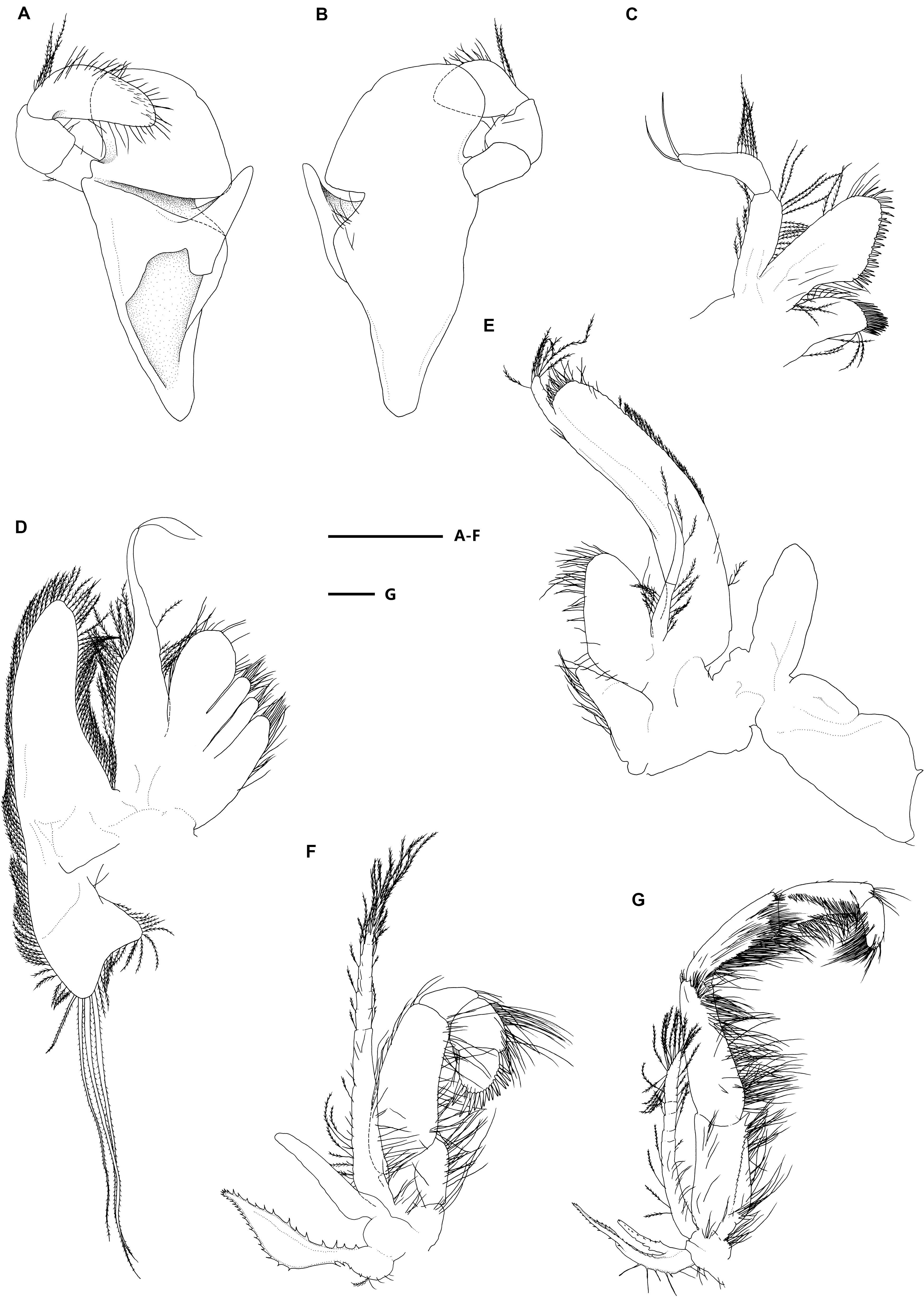
Figure 3. Eiconaxius serratus sp. nov. Holotype, SRSIO 14070301 (ovigerous female, cl 9.0 mm). (A) Mandible, left, inner view; (B) same, outer view; (C) maxillule, left, inner view; (D) maxilla, left, inner view; (E) maxilliped 1, right, inner view; (F) maxilliped 2, left, inner view; (G) maxilliped 3, left, inner view. Scales: 1 mm.
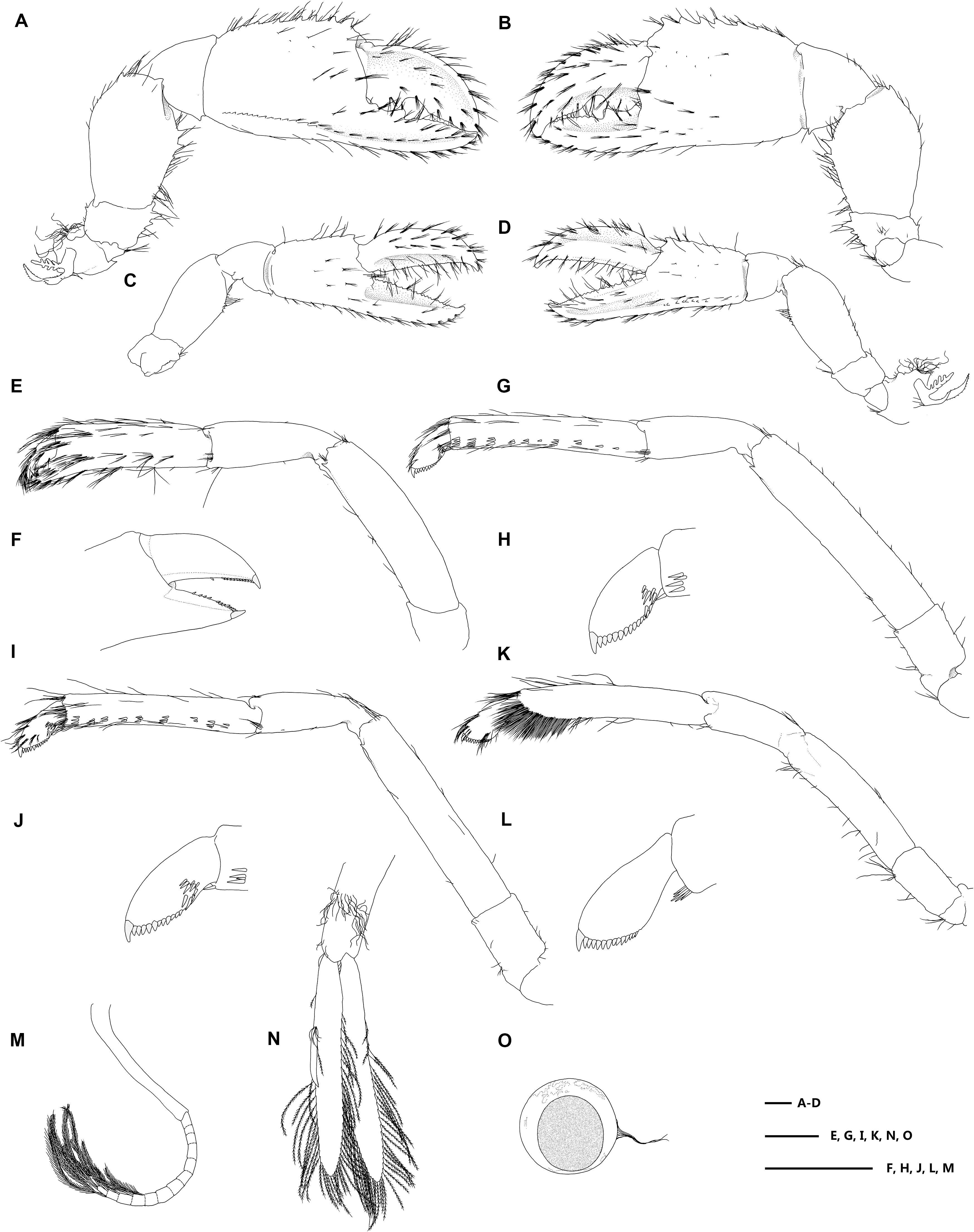
Figure 4. Eiconaxius serratus sp. nov. Holotype, SRSIO 14070301 (ovigerous female, cl 9.0 mm). (A) major cheliped, right, lateral view; (B) same, inner view; (C) minor cheliped, left, inner view; (D) same, lateral view; (E) pereopod 2, left, lateral view; (F) same, fingers, mesial view; (G) pereopod 3, left, lateral view; (H) same, dactylus, lateral view; (I) pereopod 4, left, lateral view; (J) same, dactylus, lateral view; (K) pereopod 5, left, lateral view; (L) same, dactylus, lateral view; (M) pleopod 1, right; (N) pleopod 2, right; (O) egg, with ovipositing setae. Scales: 1 mm.
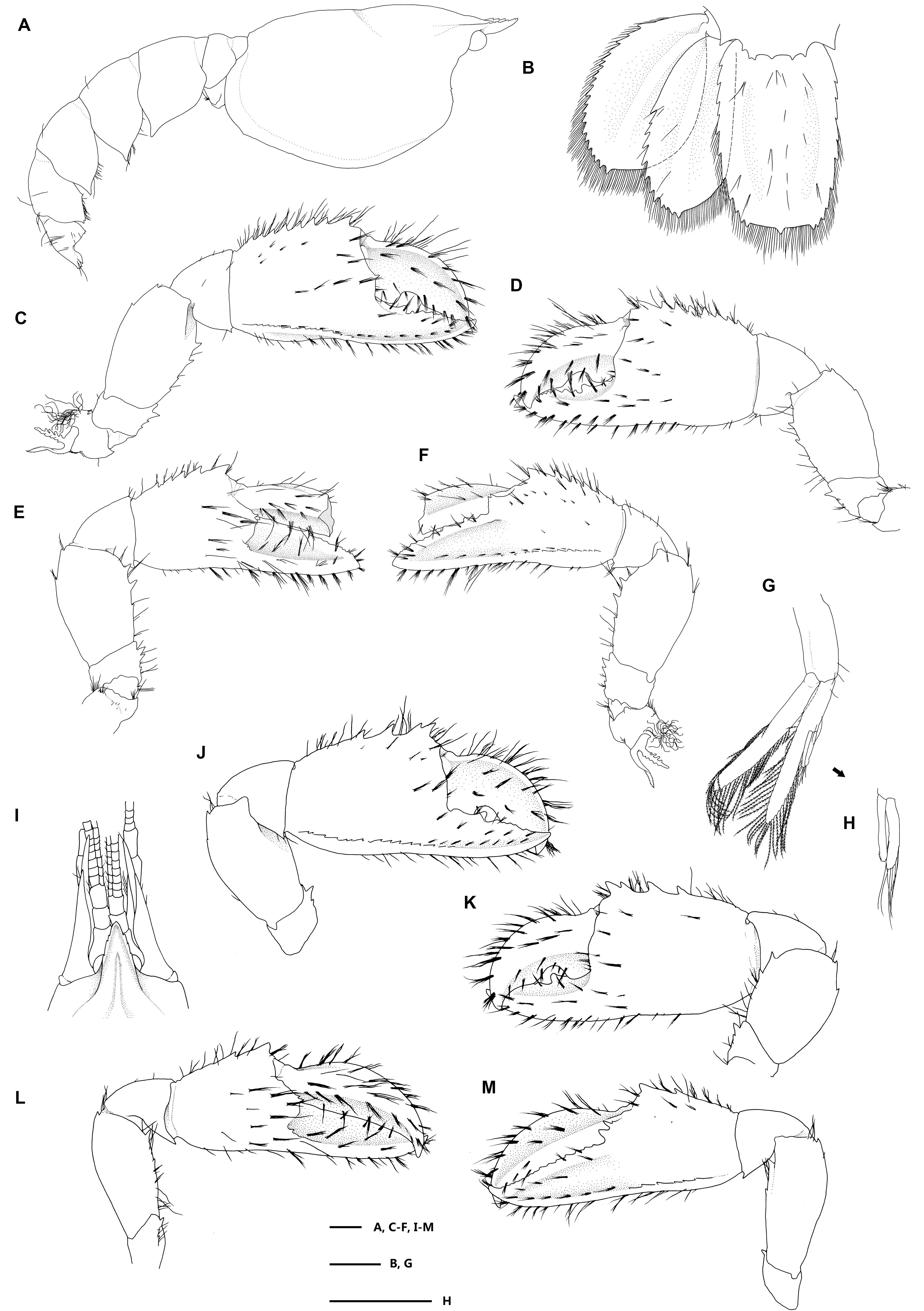
Figure 5. Eiconaxius serratus sp. nov. Paratype, SRSIO 16040302 (male, cl 7.3 mm). (A) Carapace, pleon, lateral view; (B) pleomere 6, telson, left uropod, dorsal view; (C) major cheliped, right, lateral view; (D) same, inner view; (E) minor cheliped, left, inner view; (F) same, lateral view; (G) pleopod 2, left; (H) appendices interna and masculine, right. Paratype, SRSIO 16040301 (ovigerous female, cl 7.1 mm); (I) anterior carapace, antenna, antennule, dorsal view; (J) major cheliped, right, lateral view; (K) same, inner view; (L) minor cheliped, left, inner view; (M) same, lateral view. Scales: 1 mm.
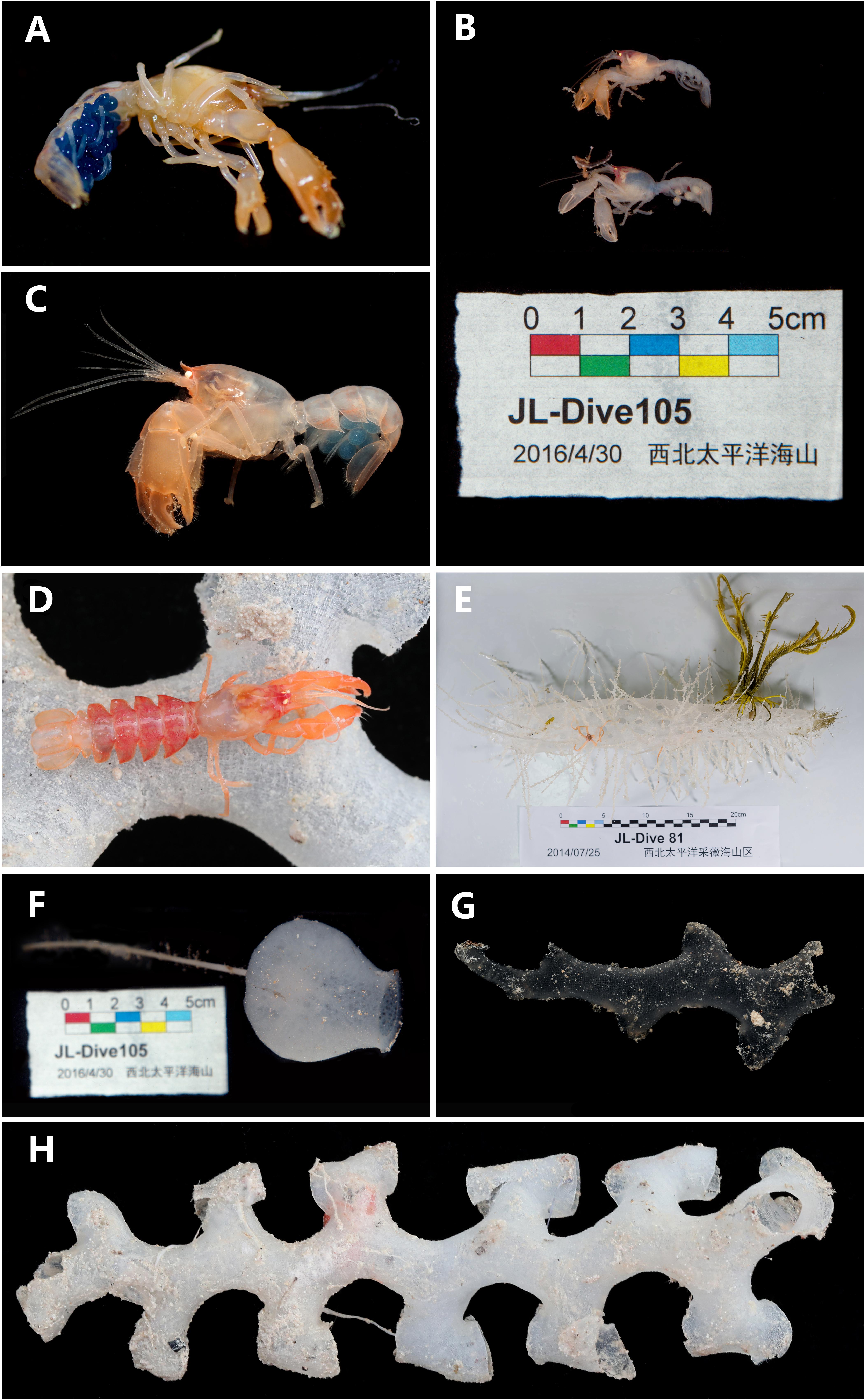
Figure 6. (A) Eiconaxius serratus sp. nov. Holotype, SRSIO 14070301 (ovigerous female, cl 9.0 mm), color in life; (B) Eiconaxius serratus sp. nov. Paratypes, SRSIO 16040301 (ovigerous female, cl 7.1 mm) and SRSIO 16040302 (male, cl 7.3 mm), color in life; (C) Eiconaxius serratus sp. nov. Non-type, MBM 304669 (ovigerous female, cl 7.8 mm), color in life; (D) Eiconaxius serratus sp. nov. Non-type, MBM 304668 (ovigerous female, cl 10.0 mm), color in life; (E) host sponge of holotype (SRSIO 14070301), Corbitellinae gen. sp. (Lyssacinosida, Euplectellidae); (F) host sponge of paratypes (SRSIO 16040301 and 16040301), Amphidiscella sp. (Lyssacinosida, Euplectellidae, Bolosominae); (G) host sponge of non-type (MBM 304668), Farrea sp. (Sceptrulophora, Farreidae); (H) host sponge of non-type (MBM 304669), Farrea sp. (Sceptrulophora, Farreidae).
ZooBank registration LSID
urn:lsid:zoobank.org:act:908113CF-494B-4B87-BABF-D37DD4055BEC.
Material Examined
Holotype
Caiwei Guyot, Magellan Seamount Chain, North West Pacific, 15°40.98′ N, 154°55.38′ E, 1600–1800 m, RV Xiangyanghong 9, collected by crew of manned submersible Jiaolong, stn DY35-I, JL-Dive81, 25 July 2014, SRSIO 14070301 (ovigerous female, cl 9.0 mm, tl 31.2 mm).
Paratypes
Weijia Guyot, Magellan Seamount Chain, North West Pacific, 12°58.22′ N, 156°46.45′ E, 2091 m, RV Xiangyanghong 9, collected by crew of manned submersible Jiaolong, stn DY37-I, JL-Dive105, 30 April 2016, SRSIO 16040301 (1 ovigerous female, cl 7.1 mm, tl 24.4 mm); SRSIO 16040302 (1 male, cl 7.3 mm, tl 25.0 mm).
Other material
Unnamed seamount on the Caroline Ridge, North West Pacific, 10°04.73′ N, 140°09.40′ E, 1895 m, RV Kexue, collected by crew of RV Kexue, stn FX-Dive 222, 10 June 2019, MBM 304668 (ovigerous female, cl 10.0 mm, tl 33.1 mm). Unnamed seamount on the Caroline Ridge, North West Pacific, 10°04.07′ N, 140°15.20′ E, 1514 m, RV Kexue, collected by crew of RV Kexue, stn FX-Dive 223, 11 June 2019, MBM 304669 (ovigerous female, cl 7.8 mm, tl 25.2 mm).
Diagnosis
Rostrum tapering evenly, 1.3 times as long as wide. Submedian gastric carinae U-shaped, diverging widely from base of median carina. Major cheliped, merus lower margin with 2 prominent teeth in distal half; palm wider distally than at midpoint; palm upper margin denticulate; fixed finger cutting edge with notch and blunt tooth in proximal half; dactylus cutting edge with basal molar, notch and straight beyond. Minor cheliped, palm upper margin denticulate, fingers almost as long to longer than upper margin of palm; fixed finger cutting edge denticulate.
Description of Female Holotype
Body robust, integument solid, surface generally glabrous. Rostrum triangular in dorsal view, 0.15 times as long as carapace, 1.25 times as long as wide at base, apex acute, slightly directed upwards, overreaching antennular peduncle article 2; lateral margins each with 5 small, shallow teeth, continuous with lateral carinae; ventral margin unarmed. Carapace glabrous, with gastric region inflated; cervical groove faint; median carina shallow, entire, unarmed, reaching middle of rostrum; submedian gastric carinae U-shaped, diverging widely from base of median carina, short; lateral carina unarmed, diverging posteriorly, extending over anterior 0.1 of carapace length; pterygostomian angle rounded, with five obsolete marginal denticles.
Pleomeres smooth, pleura without anteroventral tooth; pleomere 1 0.45 times as long as pleomere 2, pleuron posteroventrally subacute; pleomeres 2–4 subequal in length, pleura posteroventrally acute; pleomere 5 0.9 times as long as pleomere 4, pleuron posteroventrally subacute, dorsal surface with several long simple setae; pleomere 6 subequal in length to pleomere 5, pleuron extended laterally, lateral surface with scattered simple setae, posterodorsal margin with 8 widely spaced denticles.
Telson sub-oval, 0.75 times as long as maximum width at midlength, lateral margin arcuate, each side armed with 12 uneven teeth, posterolateral angle obtuse, defined by 2 small teeth, posterior margin truncate, with median tooth; dorsal surface concave, with scattered long simple setae, posterolateral and posterior margins setose.
Eyestalk well developed, not reaching middle of rostrum; cornea globular, without pigment. Antennular peduncle article 2 half-length of article 1, almost reaching end of rostrum; articles 2, 3 subequal in length, 1.2 times as long as wide; flagella 1.5 times as long as carapace. Antennal peduncle article 1 unarmed; article 2 with blunt distal tooth on ventral surface, triangular blade elongate, acute, reaching distal margin of article 4; article 3 with acute ventromesial tooth; article 4 4.0 times as long as wide; article 5 half-length of article 4; scaphocerite slender, acute, reaching middle of article 5; flagellum 0.7 times as long as carapace.
Mandible with triangular molar and incisor processes separated by deep groove; incisor margin entire; palp article 3 tapering, 1.5 times as long as article 2, lateral surface with long plumose setae proximally, dense simple setae distally. Maxillule proximal endite short, with numerous long distal complex setae; mesial margin of distal endite with numerous simple setae, 2 rows of about 30 stout setae; palp 2-articled, margins with plumose setae, with 2 long apical setae. Maxilla distal and proximal endites bilobed, mesial margins with long simple setae; endopod, tapering, with 2 long, apical setae; scaphognathite 5.0 times as long as wide, outer margin setose, proximal corner with 3 elongate setae. Maxilliped 1 proximal endite bilobed, mesial margin setose; distal endite sub-oval, mesial margin setose; endopod 2-articled, with few plumose setae; article 2 slender, tapering, 1.4 times as long as article 1; exopod broad, lateral margin with short plumose setae, tip with dense simple setae, with distal 4-segmented process tipped with several long plumose setae; epipod bilobed. Maxilliped 2 endopod ischium and basis not fused, both 0.4 times as long as merus, with long simple setae on mesial margin; merus straight, 2.8 times as long as carpus; carpus subequal in length to dactylus; propodus sub-quadrangular, 1.2 times as long as dactylus, with dense simple setae on lateral margin; dactylus sub-oval, distal margin armed with 15 stout setae; exopod distal half segmented, tip with dense long plumose setae; podobranch not branched; epipod leaf-like, outer margins serrated. Maxilliped 3 coxa with leaf-like epipod and podobranch, both with outer margins serrated; basis unarmed, 0.25 times length of ischium; ischium 0.85 times as long as merus, mesial surface with crista dentata of 17 small teeth; merus unarmed, 1.3 times as long as carpus; carpus unarmed, 1.2 times as long as propodus; propodus unarmed, 2.0 times as long as dactylus, with oblique row of long simple setae on dorsal surface; dactylus cylindrical, 2.2 times as long as broad; exopod distal half segmented, reaching middle of merus, tip with dense long plumose setae.
Major (right) cheliped 1.8 times as long as carapace; coxa with distal tooth on mesial margin; basis unarmed; ischium compressed laterally, 1.4 times as long as broad, with 4 teeth on lower margin, increasing in size, small tooth on upper margin; merus 3.2 times as long as ischium, compressed laterally, upper margin arcuate, with curved, distal tooth, lower margin with 2 prominent teeth in distal half, plus 2 more proximal denticles; carpus 0.5 times as long as merus, lower margin with subdistal tooth, upper margin unarmed; propodal palm inflated, slightly longer than merus, upper margin 1.1 times as long as wide, carinate, with 8 teeth, increasing in size distally, lateral and mesial surfaces with few scattered tubercles in distal half, mesial face with oblique row of 4 tufts of setae, lower-lateral carina sharp, extending to tip of fixed finger, with 15 teeth proximally, distolateral and distomesial margins oblique, unarmed; fingers forming deep sub-oval cavity defined by sharp longitudinal carina on mesial surface when closed; fixed finger 0.8 times as long as palm, nearly straight, distally slightly upturned, opposable margin with shallow concavity over proximal third, prominent triangular tooth, triangular notch followed by several weak teeth distally; dactylus as long as palm, distally curved, upper margin carinate, unarmed, lateral surface slightly concave, mesial surface flattened, opposable margin with large blunt proximal cusp and several weak teeth distally, separated by semicircular, prominent notch.
Minor (left) cheliped shorter, more slender than major, 1.4 times as long as carapace, palm 0.6 times width of major; coxa with distal tooth on mesial margin; basis unarmed; ischium compressed laterally, 1.2 times as long as broad, with prominent triangular tooth, 4 denticles on lower margin, upper margin unarmed; merus 2.4 times as long as ischium, compressed laterally, upper margin slightly arcuate, unarmed, lower margin with 3 small teeth at mid-length; carpus 0.6 times as long as merus, lower distal angle unarmed; palm slightly inflated, subequal in length to merus, upper margin 1.2 times as long as wide, sharply carinate, with 8 subequal obsolete teeth, lateral and mesial surfaces with few scattered tubercles in distal half, lower lateral carina sharp, extending to tip of fixed finger, with row of 8 teeth proximally, distolateral and distomesial margins oblique, unornamented; fingers forming deep sub-triangular cavity defined by sharp longitudinal carina on mesial surface when closed; fixed finger as long as palm, nearly straight, distally slightly upturned, opposable margin with row of small triangular teeth along whole length; dactylus 1.2 times as long as palm, distally curved, upper margin sharply carinate, unarmed, lateral surface with blunt longitudinal carina on midline, opposable margin with several tiny denticles in distal half, small proximal notch.
Pereopod 2 almost reaching distal margin of carpus of pereopod 1; coxa with distomesial tooth; basis unarmed; ischium 1.4 times as long as broad, unarmed; merus 4.8 times as long as ischium, upper margin slightly arcuate, unarmed, lower margin with acute distal tooth; carpus 0.6 times as long as merus, unarmed; chela setose; palm unarmed, slightly longer than carpus, 3.2 times as long as broad; fixed finger and dactylus terminating in corneous claw, cutting edge with row of tiny corneous spinules. Pereopod 3 coxa with distomesial tooth; basis unarmed; 1.3 times as long as broad, unarmed; merus 3.5 times as long as ischium, upper margin unarmed, lower margin with distal acute tooth; carpus 0.5 times as long as merus, unarmed; propodus 1.6 times as long as carpus, with 10 (left), 9 (right) sets of spinules on lower lateral surface (each set of 1–4 spinules), 1 spinule at lower distal margin; dactylus sub-oval, with unguis, flexor margin with 12 stout spinules, lateral face with 7 (left), 10 (right) submarginal proximal spinules. Pereopod 4 similar to pereopod 3, slightly thinner; propodus with 9 (left), 10 (right) sets of spinules (each set of 1–5 spinules); dactylus with unguis, flexor margin with 11 (left), 14 (right) stout spinules, lateral face with 10 (left), 9 (right) submarginal proximal spinules. Pereopod 5 coxa to carpus unarmed; propodus distal lower-lateral face densely setose, 5 slender spinules at lower distal angle; dactylus flexor margin with unguis, 13 (left), 14 (right) stout spinules.
Pleopod 1 slender, 2 articles of similar length; article 2 segmented. Pleopod 2 peduncle shorter than rami, bearing long, curly simple ovipositing setae on mesial margin and lower surface; endopod with rod-like appendix interna, tapering, tip slightly curved, bearing cluster of small hooks; exopod slightly longer than endopod, both with dense long plumose setae along margins. Pleopods 3–5 similar to pleopod 2, decreasing in length posteriorly.
Uropod as long as telson; peduncle stout, unarmed; endopod 2.3 times as long as broad, lateral margin armed with 8 (left), 10 (right) teeth over distal half, distal angle slightly produced, posterior margin straight, dorsal surface with faint longitudinal ridge and scattered long simple setae; exopod 1.7 times as long as broad, shorter than endopod, lateral margin armed with row of 18 (left), 16 (right) teeth over distal two-thirds, distal angle bifid or almost so, posterior margin almost straight, distally strongly convex, dorsal surface with faint longitudinal ridge; exopod and endopod both with long plumose setae along mesial and posterodistal margins.
About 50 eggs, average diameter 1.4 mm.
Description of Male Paratype
Generally similar to holotype. Rostrum lateral margins each with 4 small, shallow teeth. Carapace gastric region slightly inflated; cervical groove faint; pterygostomian angle with 9 (left), 7 (right) tiny marginal denticles. Pleura 2–5 more acute than in female; pleuron 5 with additional tooth and small anteroventral tooth; pleuron 6 lateral process with 2 ventral teeth, posterodorsal margin convex, with more prominent teeth than in female. Telson lateral margin armed with 11 (left), 10 (right) teeth subequal in size. Major (right) cheliped ischium lower margin with 3 teeth, increasing in size distally, upper margin unarmed; merus lower margin with 4 sharp teeth, upper margin with small distal tooth and midlength tooth; carpus lower margin with distal tooth, upper margin unarmed; palm upper margin 1.1 times greatest width, with 8 sharp subequal teeth, lower-lateral carina with row of 11 teeth proximally, distolateral margin with tooth at midlength, distomesial margin unarmed; fixed finger 0.8 times as long as palm upper margin, opposable margin with prominent rectangular tooth at proximal third, 8 triangular teeth and several tiny denticles distally. Minor (left) cheliped ischium lower margin with 3 teeth, increasing in size distally, upper margin unarmed; merus lower margin with 3 sharp teeth, upper margin with 2 small teeth distal to midlength; carpus lower margin with distal tooth, upper margin unarmed; palm width 0.8 that of major cheliped, upper margin as long as greatest width, with 5 sharp subequal teeth, lower-lateral carina with row of 12 teeth proximally, distolateral margin with prominent triangular tooth, bifid at tip, distomesial margin with 2 small teeth; fixed finger (tip broken), with 2 teeth at base of lower mesial ridge, opposable margin with 10 small irregular teeth along whole length; dactylus distal half broken off. Pereopod 3 propodus with 9 sets of spinules on lower-lateral surface (each set of 1–4 spinules), spinule at lower-distal margin; dactylus with 12 accessory teeth on flexor margin and 6 (left), 7 (right) submarginal spinules proximally. Pereopod 4 propodus with 9 (left), 8 (right) sets of spinules (each set of 1–5 spinules); dactylus with 13 (left), 12 (right) accessory teeth on flexor margin with 7 (left), 8 (right) submarginal spinules proximally. Pereopod 5 dactylus with 11 (left), 10 (right) accessory teeth on flexor margin. Pleopod 1 absent. Pleopod 2 appendix interna, appendix masculina subequal in length; appendix masculina quarter length of endopod.
Description of Other Specimens
Non-type ovigerous female (MBM 304668)
Generally similar to holotype. Rostrum slightly directed upwards, reaching to middle of antennular peduncle article 2, lateral margins each with 2 small, shallow teeth. Telson lateral margin armed with 9 (left), 12 (right) teeth subequal in size. Major (right) cheliped ischium lower margin with 2 teeth, upper margin unarmed; merus lower margin with 2 sharp teeth, upper margin with small distal tooth and midlength tooth; carpus lower margin with distal tooth, upper margin unarmed; palm upper margin with 7 sharp subequal teeth, lower-lateral carina with row of 6 teeth proximally, distolateral and distomesial margins unarmed; dactylus upper margin with tooth at proximal quarter. Minor (left) cheliped ischium lower margin with tooth, upper margin unarmed; merus lower margin with 2 sharp teeth, upper margin with 2 small teeth distal to midlength; carpus lower margin with distal tooth, upper margin unarmed; palm with 5 sharp subequal teeth, lower-lateral carina with row of 5 teeth proximally, distolateral margin with prominent bifid triangular tooth and small additional tooth, distomesial margin with 3 small teeth; fixed finger with lower mesial ridge unarmed; dactylus upper margin unarmed.
Non-type ovigerous female (MBM 304669)
Generally similar to holotype. Rostrum slightly directed upwards, reaching to middle of antennular peduncle article 2, lateral margins each with 2 small, shallow teeth. Telson lateral margin armed with 11 (left), 13 (right) teeth subequal in size. Major (left) cheliped ischium lower margin with 3 teeth, upper margin unarmed; merus lower margin with 3 sharp teeth, upper margin with small distal tooth and midlength tooth; carpus lower margin with distal tooth, upper margin unarmed; palm upper margin with 7 sharp subequal teeth, lower-lateral carina with row of 5 teeth proximally, distolateral and distomesial margins unarmed; dactylus upper margin with tooth at proximal third. Minor (right) cheliped ischium lower margin with 3 teeth, upper margin unarmed; merus lower margin with 3 sharp teeth, upper margin with 2 small teeth distal to midlength; carpus lower margin with distal tooth, upper margin unarmed; palm with 6 sharp subequal teeth, lower-lateral carina with row of 5 teeth proximally, distolateral margin with prominent bifid triangular tooth and small additional tooth, distomesial margin with 3 small teeth; fixed finger with lower mesial ridge unarmed; dactylus upper margin with tooth at half length.
Color in Life
Body and appendages orangish translucent, tips of rostrum and chelipeds darker; cornea of eye faint yellow, opaque; mature female with ovary light blue; embryos sapphire at early embryonic stage, whitish at late embryonic stage.
Host
The specimen from Caiwei Guyot was found in the cavity of a hexactinellid sponge belonging to the subfamily Corbitellinae (Lyssacinosida, Euplectellidae) (Figure 6E); the specimens from Weijia Guyot were found in the cavity of the hexactinellid sponge Amphidiscella sp. (Lyssacinosida, Euplectellidae, Bolosominae) (Figure 6F), and the specimens from an unnamed seamount on the Caroline Ridge were found in the cavity of the hexactinellid sponge Farrea sp. (Sceptrulophora, Farreidae) (Figures 6G,H).
Etymology
From Latin serratus, meaning serrated, referring to the strongly serrated upper margin of the cheliped palm.
Distribution
Northwestern Pacific Ocean: Caiwei Guyot, Magellan Seamount Chain, 1600–1800 m; Weijia Guyot, Magellan Seamount Chain, 2091 m; unnamed seamount on the Caroline Ridge, 1514–1895 m.
16S rRNA and COI Genetic Distances
The 16S rRNA sequences of the three individuals of E. serratus sp. nov. from Magellan Seamount Chain were identical. Similarly, the two individuals from the seamount on the Caroline Ridge had the same 16S rRNA sequences. The 16S genetic divergence between them was 0.5% (Table 2). Furthermore, the interspecific genetic divergence between E. serratus sp. nov. and other congeneric species ranged from 5.2% (E. caribbaeus) to 12.6% (E. sibogae). The averaged intrageneric genetic divergence was 9.5% (3.2–14.4%) and the averaged intergeneric genetic divergence (between Eiconaxius and Spongiaxius novaezealandiae) was 19.6% (15.0–22.5%) (Table 2).
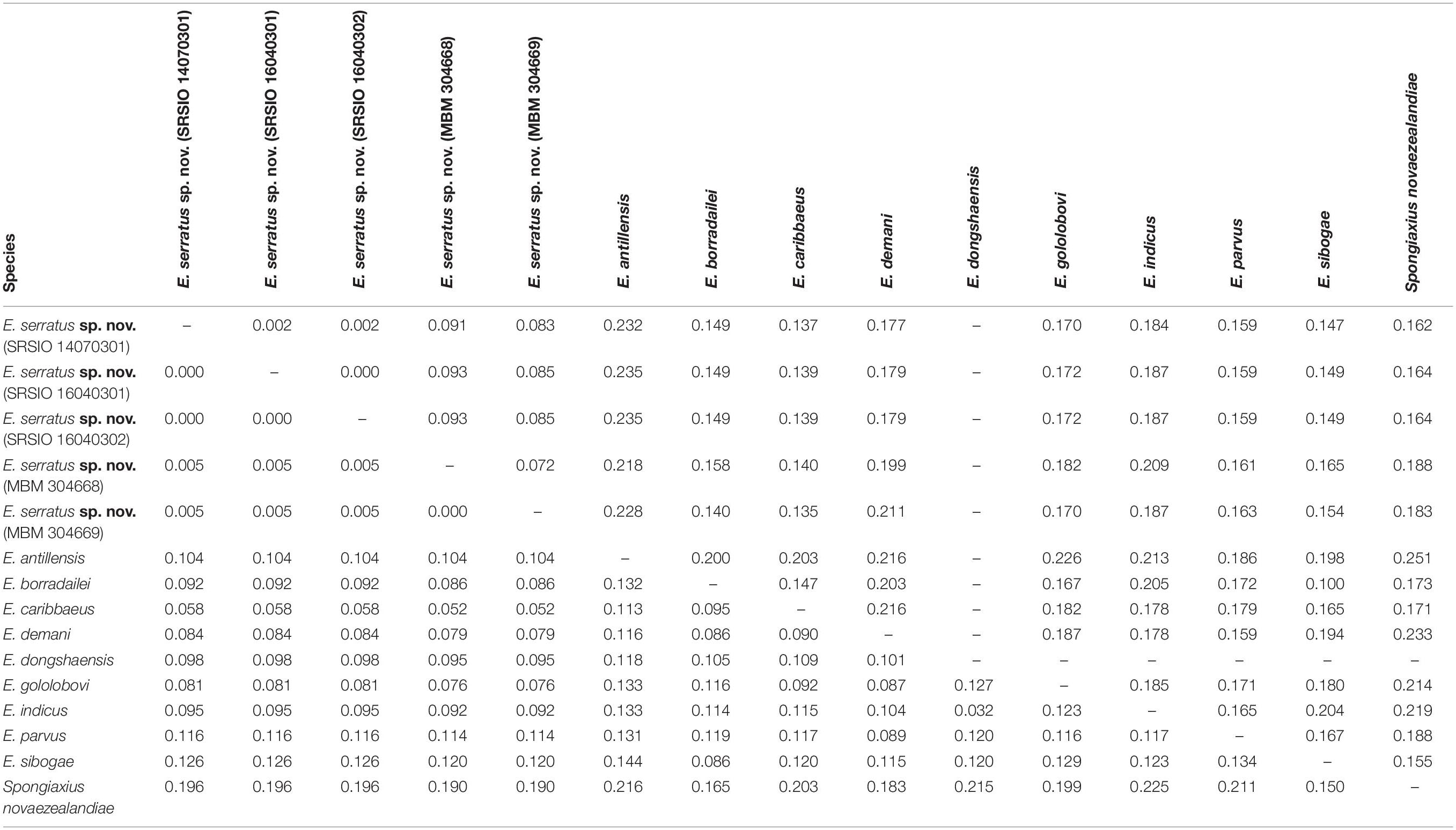
Table 2. Kimura’s 2-parameter pair-wise genetic distances of 16S rRNA (below diagonal) and COI (above diagonal) gene sequences among species studied.
COI divergence between the individuals of E. serratus sp. nov. from Caiwei Guyot and Weijia Guyot was close to zero (0.2%). However, the genetic divergence between the individuals from two sampled sites from the unnamed seamount on the Caroline Ridge reached to 7.2%. The COI averaged intraspecific divergence of E. serratus sp. nov. was 4.7% (0–9.3%) (Table 2). The interspecific genetic divergence between E. serratus sp. nov. and other congeneric species ranged from 13.5% (E. caribbaeus) to 23.5% (E. antillensis). The averaged intrageneric genetic divergence was 16.2% (10.0–23.5%) and the averaged intergeneric genetic divergence (between Eiconaxius and Spongiaxius) was 19.0% (15.5–25.1%) (Table 2).
Organization and Characterization of Mitochondrial Genome
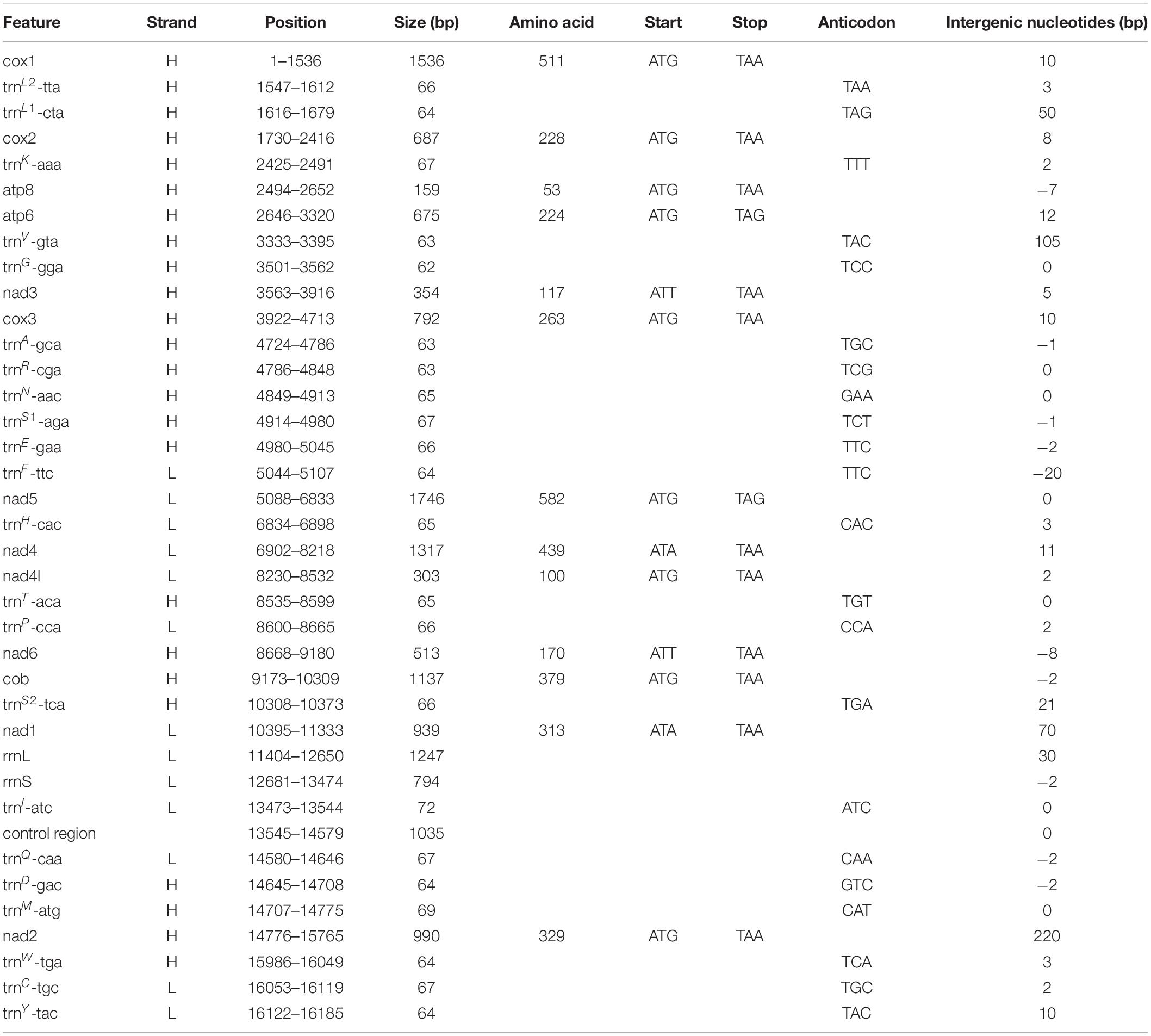
A total of 51,653,517 clean reads (7,578,946,749 bp) were generated by Illumina HiSeq sequencing with an insert size of approximate 450 bp. After assembling, a 16,195 bp circular molecule was obtained (Figure 7), which represented the complete mitochondrial genome of Eiconaxius serratus sp. nov. This length is comparable to that of the complete mitochondrial genomes of other axiideans, which range from 16,899 bp [Filhollianassa ceramica (Fulton and Grant, 1906)] to 14,909 bp [Neaxius glyptocercus (von Martens, 1868)] (Supplementary Table S1).
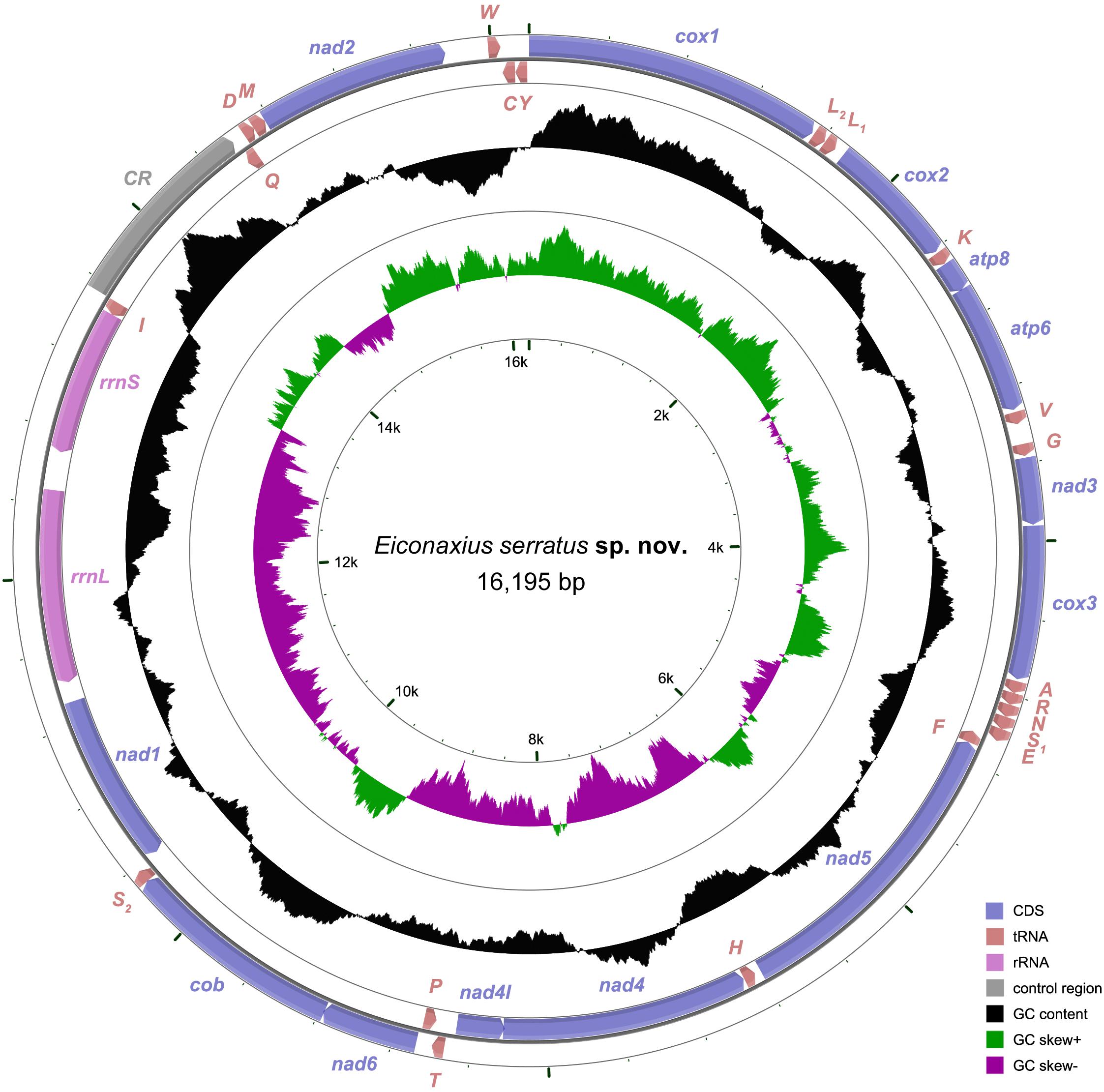
Figure 7. The organization of the mitochondrial genome of Eiconaxius serratus sp. nov. The full names of protein coding genes, rrnS and rrnL, are listed under abbreviations rrnS and rrnL, 12S and 16S ribosomal RNA genes, respectively; atp6 and atp8, ATPase subunit 6 and 8 genes, respectively; cox1–cox3, cytochrome c oxidase subunits I–III genes, respectively; cob, cytochrome b gene; nad1–6 and 4l, NADH dehydrogenase subunit 1–6 and 4l genes, respectively. One uppercase letter amino acid abbreviations are used to label the corresponding tRNA genes. The position of control region (CR) is indicated in the figure.
The complete mitochondrial genome encodes 37 genes, including 13 PCGs, 2 rRNA genes, and 22 tRNA genes (duplication of trnL and trnS), which exhibits the same components as most other decapod mitochondrial genomes. Twenty-four genes (9 PCGs and 15 tRNAs) are encoded on the heavy (H) strand, while other 13 genes (4 PCGs, 7 tRNAs, and 2 rRNAs) are encoded on the light (L) strand (Table 3). The base composition of the heavy strand is A = 34.69%, T = 38.01%, C = 14.26%, and G = 13.04%. The A + T content (72.70%) is distinctly higher than the G + C content (27.30%). A total of 1,614 bp of non-coding nucleotides are scattered among 19 intergenic regions varying from 2 to 1,035 bp (Table 3). The largest non-coding region (1,035 bp) located between trnI and trnQ is identified as the putative control region (CR) according to its location in the mitochondrial genome (Figure 7). Furthermore, there are 11 overlaps between adjacent genes in the new species with a size range of 2 to 20 bp (Table 3). The combined length of 13 PCGs was 11,148 bp, accounting for 68.83% of the entire mitochondrial genome. All PCGs started with ATD as initiation codons (9 with ATG, 2 with ATT, and 2 with ATA) and end with two conventional stop codons (TAA and TAG) (Table 3). Among 13 PCGs, Leu (15.31%) and Cys (1.11%) are the most and the least frequently used amino acids, respectively (Supplementary Figure S1A). RSCU analysis shows that UUA (Leu, 3.80%) is the most and CGC (Arg, 0.00%) the least frequently used codons. In addition, NNW codons have a higher abundance than NNS codons (Supplementary Figure S1B). The lengths of 22 tRNA genes range from 62 to 72 bp, and all tRNA genes can be folded into classic clover leaf structures (Supplementary Figure S2). Two ribosomal RNA genes (rrnS and rrnL) are located on the L strand between nad1 and trnI, with lengths of 794 bp and 1247 bp, respectively (Figure 7 and Table 3).
Phylogenetic Analysis
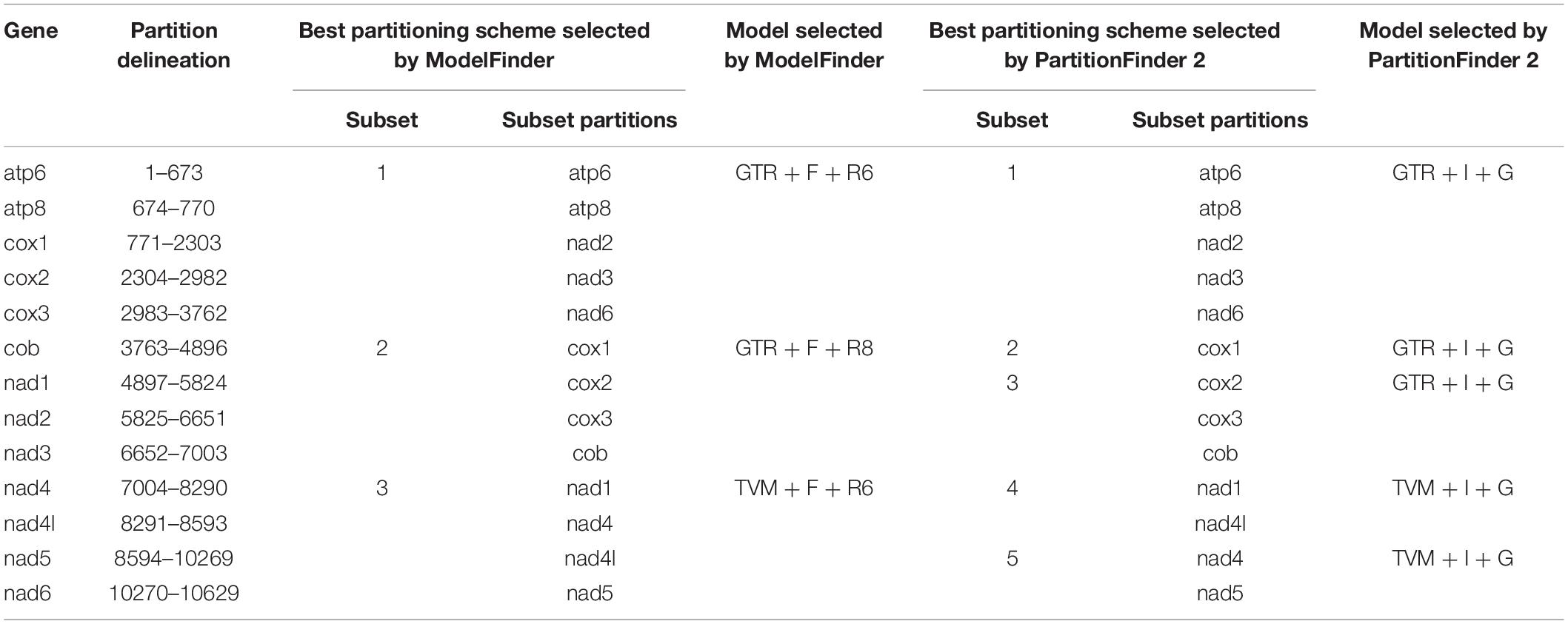
The final concatenated dataset consisted of 10,629 bp (∼95.34% of the original 11,148 bp alignment) after the poorly aligned positions and the hypervariable regions were removed with GBlocks. We found that the BIC scores of edge-linked partition scheme were always better than that of edge-unlinked partition scheme. Therefore, we chose the best-fit substitution models and partition schemes selected by edge-linked partition scheme findings for subsequent phylogenetic analyses (Table 4).
Tree topologies resulting from the BI and ML analyses were highly congruent and generally well supported, except for a few internal nodes in the clades of Caridea, Axiidae, and Brachyura (Figure 8 and Supplementary Figure S3). At the order level, both Stomatopoda and Decapoda were found to be monophyletic with high support values (BP = 100%, aBayes = 1.00, PP = 1.00). At the suborder level, both Dendrobranchiata and Pleocyemata were monophyletic (BP = 100%, aBayes = 1.00, PP = 1.00) and Dendrobranchiata located at the basal position of decapods with strong support (BP = 98%, aBayes = 1.00, PP = 1.00). At the infraorder level, the monophyly of each infraorder was consistently well supported in all analyses (BP ≥ 95%, aBayes ≥ 0.95, PP ≥ 0.95). Furthermore, apart from the grouping of Polychelida and Glypheidea (BP = 50%, aBayes = 0.63, PP = 0.63) and the position of Stenopodidea (BP = 83%, aBayes = 0.99, PP = 0.88), the relationships between infraorders were mostly well resolved. Although this study was not intended to investigate the familial relationships within decapod infraorders, these too were generally well supported in both analyses. Axiidae (represented by two genera) were monophyletic (BP = 89%, aBayes = 1.00, PP = 1.00) and basally positioned within Axiidea in all analyses with high support values (BP = 100%, aBayes = 1.00, PP = 1.00). Strahlaxiidae were paraphyletic, Neaxius and Strahlaxius basal to a clade consisting of Eucalliacidae, Callichiridae, and Callianassidae (BP = 100%, aBayes = 1.00, PP = 1.00). The relationships among the last three families were obscure, as the topologies derived from ML and BI analyses were different yet well supported. In the ML tree, Eucalliacidae and Callichiridae (represented by one species each) formed a sister group to Callianassidae with high support values (BP = 90%, aBayes = 1.00). While in the BI tree, Callichiridae and Callianassidae grouped together first (PP = 1.00), and then clustered with Eucalliacidae (PP = 1.00). Callianassidae (five species) were always recognized as a monophyletic group with high support values (BP = 100%, aBayes = 1.00, PP = 1.00) in our analyses.
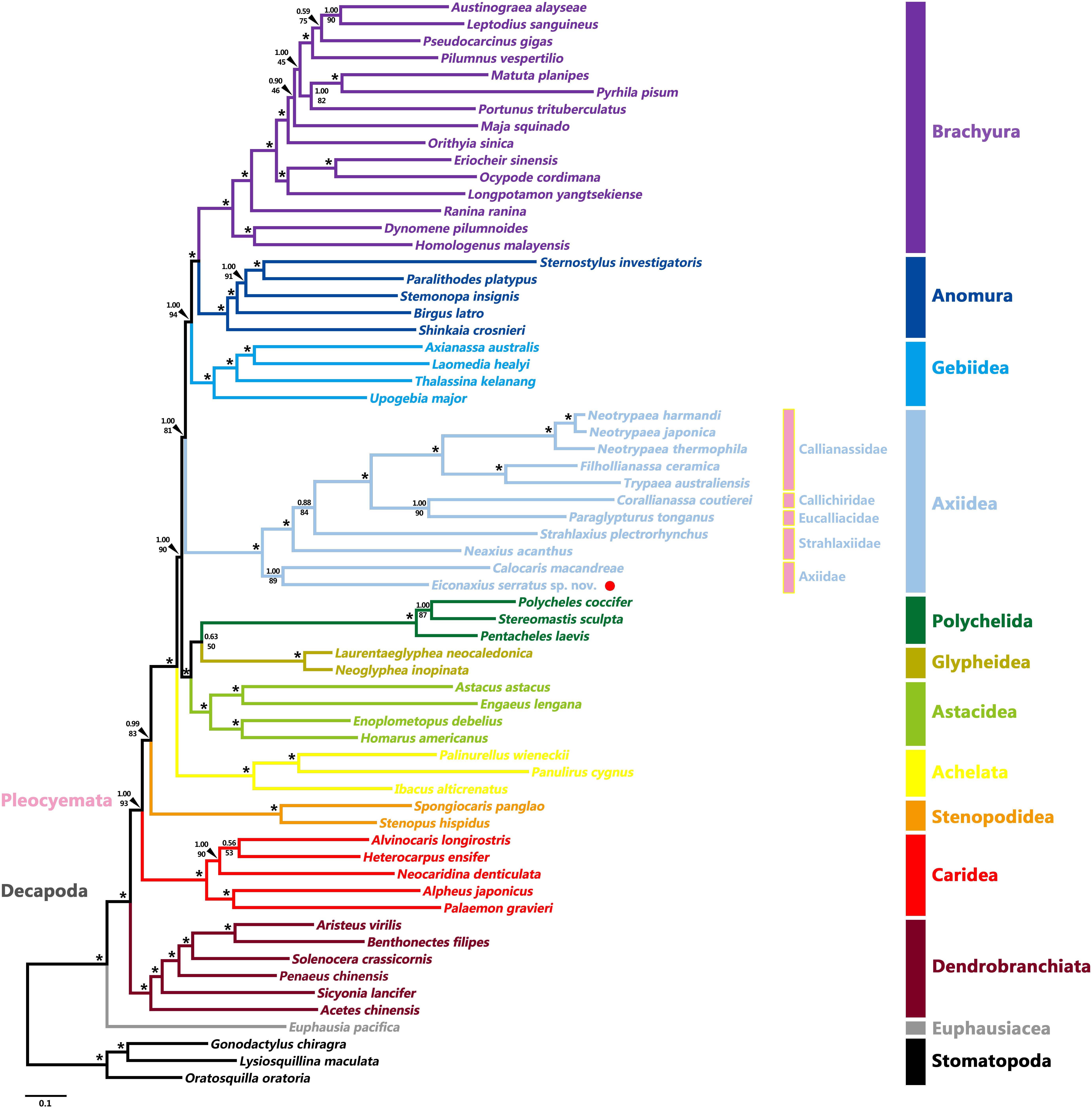
Figure 8. Maximum likelihood tree inferred from the concatenated 13 mitochondrial PCGs nucleotide sequences partitioned by gene. Nodal supports are denoted on the corresponding branches, and the black asterisk (*) indicates both BS and aBayes test value ≥ 0.95 for the node.
Mitochondrial Gene Order and Rearrangements
The gene orders of mitochondrial genomes were mapped onto the phylogeny based on analyses of 13 PCG nucleotide sequences (Figure 9). Within Axiidea, four unique gene arrangements were identified. Among them, the mitochondrial gene order of Eiconaxius serratus sp. nov. was identical to that of the other axiid species Calocaris macandreae.
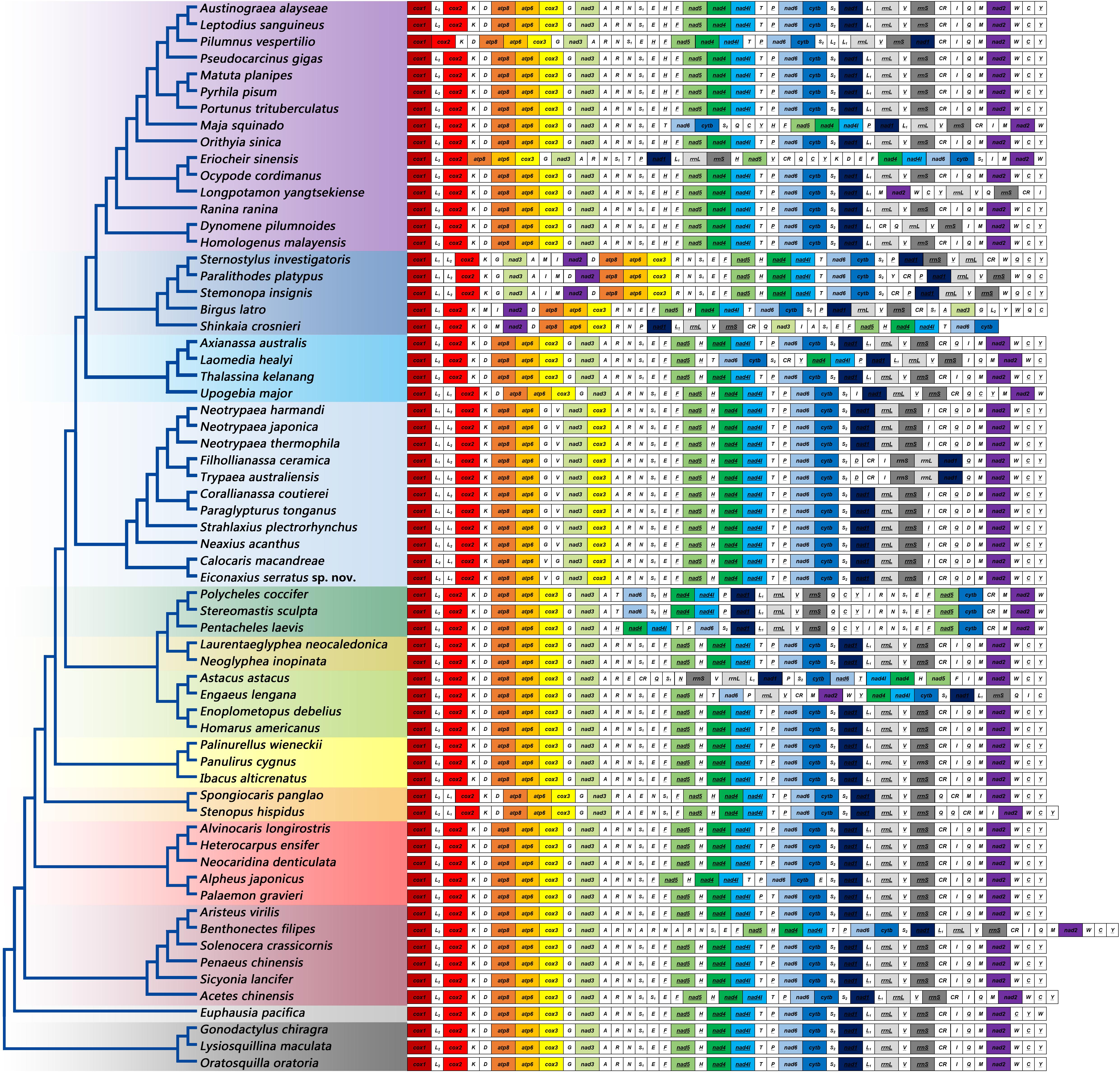
Figure 9. The gene orders of mitochondrial genomes of the studied species mapped onto the ML tree. Genes encoded by the light strand are underlined. Protein-encoding and rRNA genes are color coded to emphasize patterns and gene rearrangements.
Compared with the pancrustacean ground pattern, at least five genes were rearranged in the mitochondrial genome of E. serratus sp. nov. The trnL (CUN) (trnL1), which is located between nad1 and rrnL in the mitochondrial genomes of more primitive taxa, was found between trnL (UUR) (trnL2) and cox2 in E. serratus sp. nov. Besides, the rearrangement of trnL1 also involved shifting between two strands, which can find parallels in Stenopodidea and Anomura. Similarly, the trnI relocated between rrnS and CR, and the trnV moved to upstream of trnG, both with a reversal. In addition, the trnD moved to upstream of trnM, while the PCG cox3 relocated between nad3 and trnA.
Based on the analysis of CREx, two alternative rearrangement scenarios were inferred, from the putative ancestral state of the pancrustacean ground pattern to E. serratus sp. nov., as a result of successive events of transposition/reverse transposition, reversal, and TDRL (Figure 10).
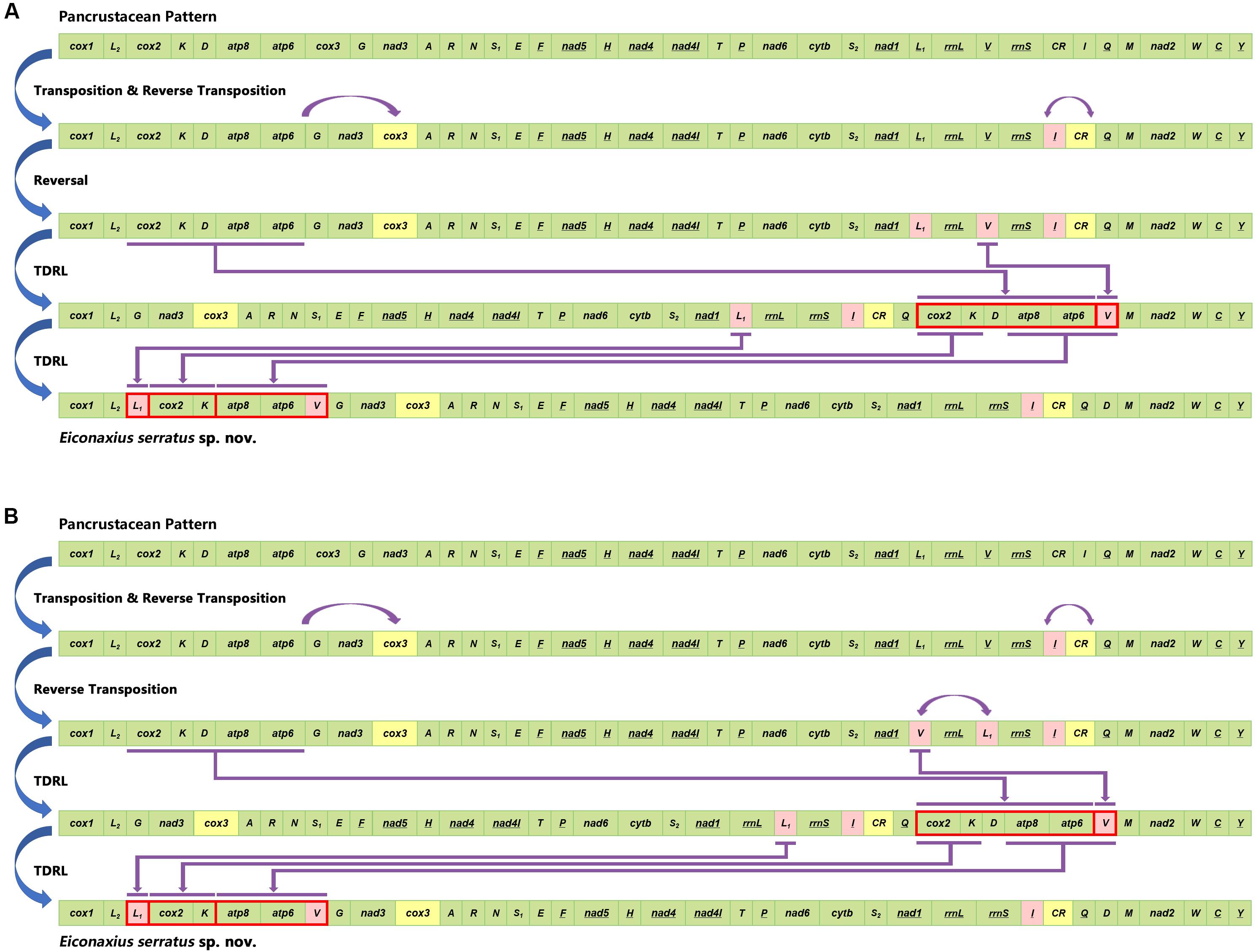
Figure 10. Two alternative rearrangement scenarios (A,B) inferred from the putative ancestral state of the pancrustacean ground pattern to Eiconaxius serratus sp. nov. by CREx analysis.
Discussion
Morphological Differences
Eiconaxius serratus sp. nov. is uniquely diagnosed by the combination of a relatively narrow acute rostrum and two prominent distal teeth on the lower margin of the merus of the major cheliped. Many species have a similar rostrum but no others have such prominent meral teeth. Most species have a serrated meral lower margin while a few have one or two teeth or tubercles just beyond the midpoint. E. spinigera (MacGilchrist, 1905), E. rubrirostris Komai et al., 2010, and E. albatrossae Kensley, 1996 are similar (differing in having much smaller meral teeth). While the upper margin of the chelipeds in several species are obscurely denticulate, none has the strong erect teeth possessed by E. serratus sp. nov.
Of the nine species of Eiconaxius for which 16S and COI genes are available, E. serratus sp. nov. has the least genetic divergence from E. caribbaeus (Faxon, 1896) (Table 2). E. caribbaeus differs in having a rounded rostrum, weakly serrated cheliped meral margin, and without strong tooth on the upper margin of the palm of the minor cheliped.
We noted several obsolete marginal denticles along the pterygostomian angle in E. serratus sp. nov., a feature easily missed by earlier authors.
Distribution and Differentiation
The 16S rRNA genetic divergence between specimens of Eiconaxius serratus sp. nov. from Magellan Seamount Chain and the seamount on the Caroline Ridge was 0.5%. Although 16S rRNA tends to have slower rates of substitution than other mitochondrial genes (Toon et al., 2009), normally a genetic divergence of less than 1% in the 16S rRNA gene indicates conspecificity for decapod crustaceans (Cabezas et al., 2009; Matzen da Silva et al., 2011; Lavery et al., 2014). Besides, this value was significantly lower than the averaged intrageneric genetic divergence (9.5%) of Eiconaxius. We recognize all individuals as belonging to the same species.
It is notable that high COI intraspecific divergence of E. serratus sp. nov. was observed. The divergence between the specimens from Magellan Seamount Chain and the unnamed seamount on the Caroline Ridge ranged from 8.3 to 9.3% (Table 2), indicating low connectivity between the two populations. Because adults of Eiconaxius are restricted to the cavity of deep-sea hexactinellid sponges, gene flow between populations must be mediated through larval dispersal rather than migration of adults. For one thing, the long distance (>1700 km) across the East Mariana Basin is a barrier leading to potential geographical isolation. For another, two deep-water currents in the Northwest Pacific could play an important role directing larval dispersal. The Northwest Pacific (5°–15° N) contains two main westward oxygen-rich water currents at 2000–3000 m depth (Kawabe et al., 2003; Kawabe and Fujio, 2010) (Figure 11B). The southern current which flows westwards south of the Carolina Seamounts toward the Yap Trench does not concern us. The northern one bifurcates at 150° E, just north of the Caroline Seamounts, its major branch flowing through the vicinity of the Challenger Deep and proceeding westward into the West Mariana Basin, while the minor branch swerves along west of the East Mariana Basin and flows eastward near the Magellan Seamounts. Consequently, the two current branches flowing in opposite directions could confine larval dispersal, promoting the divergence of E. serratus sp. nov. populations. For the same reason, the low divergence between the two populations from the Magellan Seamount Chain (0.2% for COI) might because the sampled regions are influenced by same branch. Interestingly, the specimens collected from two very close sites on the unnamed seamount on the Caroline Ridge showed significant COI genetic divergence (7.2%). Because these two specimens came from similar depths and hosts, neither bathymetric segregation nor host shift explains their divergence but could result from their separation at two sites on opposite flanks of the steep seamount (summit < 800 m). We speculate the seamount itself between them is a physical barrier preventing larval dispersal and connectivity.
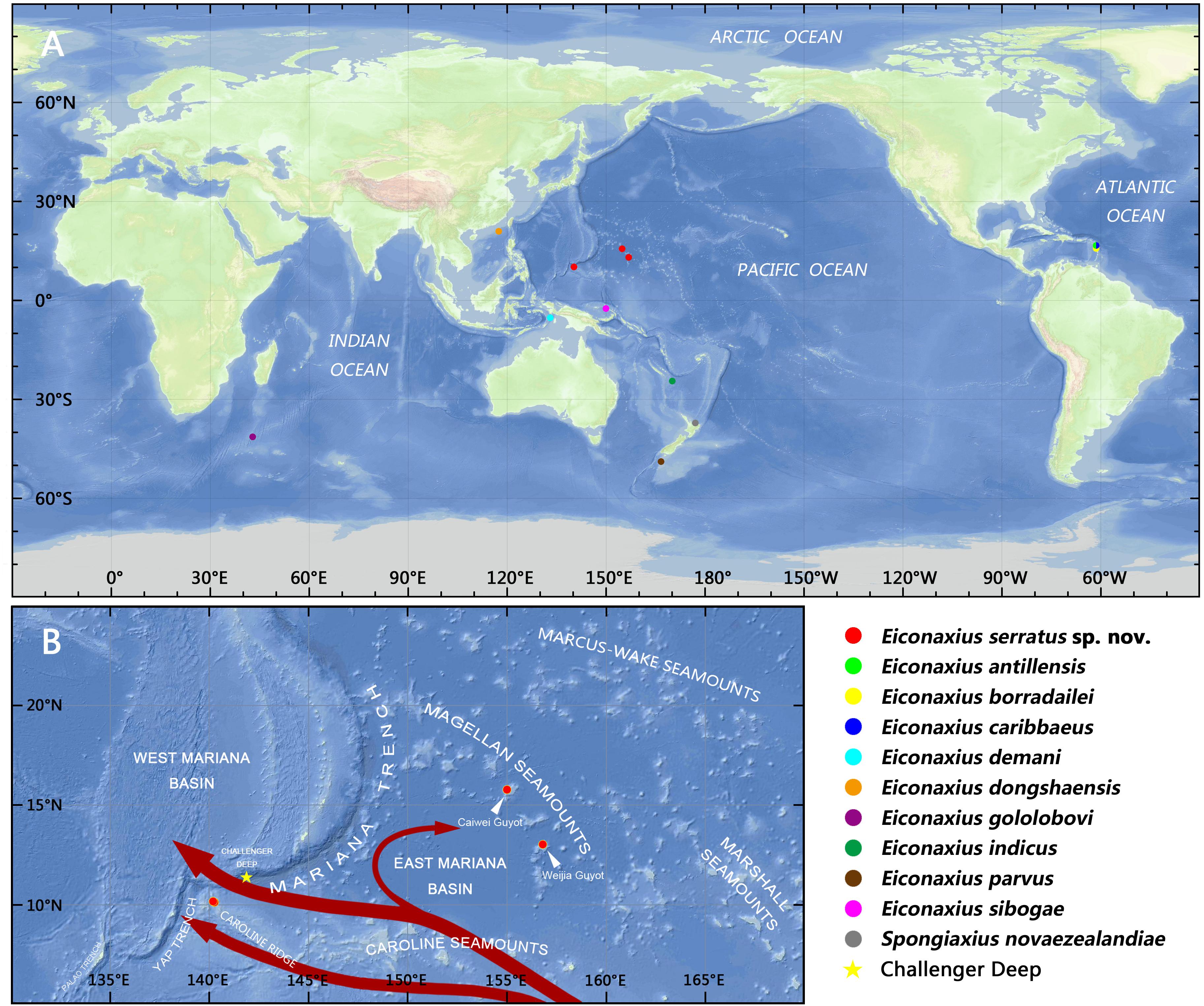
Figure 11. (A) Collection locations of the axiid specimens in the present study; (B) collection locations of Eiconaxius serratus sp. nov., with schematic diagram of the deep water currents at low latitudes in the West Pacific in the upper deep layer (marked in red arrows, from Kawabe et al., 2003; Kawabe and Fujio, 2010).
In summary, we can confirm that this species of Eiconaxius ranges over 1500 km despite genetic discontinuity. The interaction of geographical isolation and deep-water currents contribute to this intraspecific differentiation. The high COI intraspecific divergence between specimens of E. serratus sp. nov. from two very close sites is consistent with sympatric speciation in Eiconaxius. Poore (2018) recorded four species from the eastern Caribbean Sea, and Poore (in press) recorded several species from limited areas in the southwest Pacific, even two species from the same dredge sample. While most species have been described from few specimens from small areas, others such as E. demani Sakai, 1992, E. indicus (De Man, 1907), and E. sibogae (De Man, 1925) have been recorded over wide distances, up to 6000 km, in the southwest Pacific (Poore and Dworschak, 2018; Poore, in press).
Host Specificity and Speciation
Host-mediated speciation is common among shallow-water symbiotic marine invertebrates, having been documented in many taxonomic groups, such as coral-eating snails (Simmonds et al., 2018), nudibranchs (Faucci et al., 2007; Fritts-Penniman et al., 2020), barnacles (Tsang et al., 2009, 2014), snapping shrimps (Duffy, 1996; Morrison et al., 2004; Hurt et al., 2013), and palaemonid shrimps (Kou et al., 2013, 2015; Horká et al., 2016). However, this type of ecological speciation is poorly studied in deep water.
Hitherto, little has been known about host specificity of species of Eiconaxius (Ortmann, 1891; Faxon, 1893, 1896; Bouvier, 1925; De Man, 1925; Kensley, 1996; Komai, 2011; Komai and Tsuchida, 2012; Poore and Dworschak, 2018). The reasons for this are various. Specimens are often collected in large numbers without their hosts, presumably because their sponge homes have been destroyed by destructive dredge sampling. Shrimps found inside hexactinellids have been identified by decapod taxonomists who have not consulted sponge taxonomists for precise identification of the host.
Encouragingly, recent collections by deep-sea submersibles and ROVs have provided a better picture of the relationship between deep-sea decapods and their sponge hosts (Saito et al., 2006; Komai and Tsuchida, 2012; Komai, 2013; Jiang et al., 2015; Komai et al., 2016; Dworschak, 2016; Xu et al., 2016, 2017; Kou et al., 2018; Poore and Dworschak, 2018). Komai and Tsuchida (2012) found Eiconaxius acutifrons Bate, 1888 associated with sponges of the family Farreidae Gray, 1872 (Hexactinellida, Sceptrulophora). Interestingly, two males were found inside the internal cavity of the same sponge. In our study, submersible and ROV successfully collected intact sponge hosts of E. serratus sp. nov. belonging to two orders (Lyssacinosida, Sceptrulophora) and at least three genera, indicating E. serratus sp. nov. has low host specificity. The diversity and number of sponges decrease significantly with greater depth, narrowing the choice of hosts for Eiconaxius. The two populations from the Magellan Seamount Chain with low genetic divergence have distinct hosts while the two populations from the Caroline Ridge with high genetic divergence share the same host. We postulate that species of Eiconaxius could associate with a variety of sponges, or even other organisms. Accordingly, we consider host shift may not play a key role in speciation of Eiconaxius, as was found for some spongicolid shrimps (Saito and Komai, 2008; Goy, 2010; Kou et al., 2018).
Mitogenomic Phylogeny
On the whole, our phylogenetic reconstruction based on nucleotide sequences of 13 PCGs recovered topologies consistent with recent phylogenomic studies in supporting Axiidea as the sister group of the Gebiidea-Anomura-Brachyura clade (Shen et al., 2013; Tan et al., 2019; Wolfe et al., 2019). Within Axiidea, Axiidae were recovered as monophyletic (albeit with only two species) and at the most basal position, consistent with previous inferences based on morphology (Poore, 1994), Sanger sequencing (Tsang et al., 2008a; Felder and Robles, 2009; Robles et al., 2009), and genomic results (Shen et al., 2013; Tan et al., 2015; Wolfe et al., 2019). In addition, the two species of Axiidae shared the same mitochondrial gene order pattern which differs from that in other axiidean families, an observation strengthening the monophyly of Axiidae. However, the only two axiid genera included, Calocaris and Eiconaxius, have been placed in other families in the past, Calocarididae and Eiconaxiidae, respectively, both of which have been synonymized with Axiidae (Poore and Collins, 2009; Robles et al., 2009). Hence, more mitochondrial genomes of taxa from Axiidae sensu lato are needed to clearly demonstrate the monophyly of Axiidae.
Previous phylogenetic studies have been unable to clarify the validity of Strahlaxiidae due to the limited taxon representatives (Tsang et al., 2008a; Bracken et al., 2009; Robles et al., 2009; Lin et al., 2012; Shen et al., 2013; Tan et al., 2015, 2017; Sun et al., 2018b, 2019a). Tan et al. (2019) reconstructed the phylogeny of Decapoda based on mitochondrial genomes and found that two representative genera of Strahlaxiidae were paraphyletic, as did we. These questions about the monophyly of Strahlaxiidae are also reflected in different mitochondrial gene orders. Morphological differences between the three strahlaxiid genera need to be further investigated.
The phylogeny of higher axiidean genera, so-called “Callianassoidea” was reviewed by Robles et al. (2020) who offered a new phylogeny based on analysis of four genes, mitochondrial 16S and 12S rRNA along with nuclear histone 3 and 18S rRNA, and morphology of about half the known species. The family Eucalliacidae was found to be polyphyletic in the molecular analysis but the family and its genera had well defined morphological synapomorphies. Callichiridae has ambiguous molecular support but good morphological support. Callianassidae sensu stricto and four smaller families were well supported. Our study based on few representatives does not contradict these findings. We have provided new data and highlighted the potential of mitochondrial genomes to robustly resolve the deeper relationships among Axiidea, while undeniably the present taxon coverage of this mitogenomic tree is still limited. Therefore, more comprehensive taxon samplings in future will be necessary to lead us closer to the goal of reconstructing the natural evolutionary history of axiidean shrimps.
Data Availability Statement
The datasets generated for this study can be found in the GenBank (http://www.ncbi.nlm.nih.gov/genbank). ZooBank registration publication LSID: urn:lsid:zoobank.org:pub:B6A819C8-6B78-47B1-978F-54CA771B82FF.
Author Contributions
XL and CW designed the study and edited the several versions of the manuscript. PX and CW collected the samples and provided the color photos. QK and GP described and illustrated the species. QK conducted the molecular experiments and data analyses. QK, PX, and GP prepared the first draft of the manuscript. All the authors contributed to the final version of the manuscript.
Funding
This work was supported by the China Ocean Mineral Resources Research and Development Association Program (Nos. DY135-E2-1-02, DY135-E2-2-03, DY135-E2-2-06, and DY135-E2-3-04), Science & Technology Basic Resources Investigation Program of China (2017FY100804 and 2017FY201404), the National Natural Science Foundation of China (Nos. 41876178 and 41930533), Scientific Research Fund of the Second Institute of Oceanography, MNR (Nos. JG1418 and JG1613), and National Programme on Global Change and Air-Sea Interaction (No. 03-01-07-02).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Dr. Kuidong Xu (Institute of Oceanology, Chinese Academy of Sciences, Qingdao) for providing us with the deep-sea specimens collected during the M5 seamount cruise in 2019. Drs. Lin Gong (Institute of Oceanology, Chinese Academy of Sciences, Qingdao) and Chengcheng Shen (Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou) for identifying the sponge host of the new axiid, and Dr. Xuwen Wu (Institute of Oceanology, Chinese Academy of Sciences, Qingdao) for offering us color photos of the new species and its sponge hosts taken shortly after collection. We are grateful to the crews of the RV Xiangyanghong 9 and manned submersible Jiaolong for their help in sampling during the Magellan Seamount Chain cruises in 2014 and 2016, and the crews of the RV Kexue for their support in sampling with ROV Faxian during the M5 seamount cruise in 2019. We want to express our appreciation to Dr. Kareen Schnabel (National Institute of Water & Atmospheric Research, New Zealand) for sending us tissue samples. Comparative material was also obtained from specimens collected during the KAVIENG 2014, BATHUS 3 and KARUBAR expeditions to the deep waters in the southwest Pacific, and the KARUBENTHOS 2015 to Guadeloupe, organized by the Muséum national d’Histoire naturelle, Paris (MNHN) and associated organizations. We also thank all the organizers, cruise leaders, scientific and technical crews, and sorters involved to making these collections available. We especially thank Philippe Bouchet, Laure Corbari, Paula Lefèvre-Martin, and Anouchka Krygelmans for help in making the collections available at MNHN and sharing them with Museums Victoria.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00469/full#supplementary-material
FIGURE S1 | (A) Codon usage of the mitochondrial genome of Eiconaxius serratus sp. nov. Numbers to the left refer to the total number of codons; (B) the relative synonymous codon usage (RSCU) of the mitochondrial genome of Eiconaxius serratus sp. nov. Numbers to the left refer to the total number of the RSCU values. Codon families are provided on the X axis.
FIGURE S2 | Putative secondary structures for the 22 transfer RNAs of the of mitochondrial genome of Eiconaxius serratus sp. nov.
FIGURE S3 | Bayesian tree inferred from the concatenated 13 mitochondrial pro PCGs nucleotide sequences partitioned by gene. Nodal supports are denoted on the corresponding branches, and the black asterisk (∗) indicates PP ≥ 0.95 for the node.
TABLE S1 | The mitochondrial genomes sequences used in this study.
References
Anisimova, M., Gil, M., Dufayard, J. F., Dessimoz, C., and Gascuel, O. (2011). Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699. doi: 10.1093/sysbio/syr041
Basso, A., Babbucci, M., Pauletto, M., Riginella, E., Patarnello, T., and Negrisolo, E. (2017). The highly rearranged mitochondrial genomes of the crabs Maja crispata and Maja squinado (Majidae) and gene order evolution in Brachyura. Sci. Rep. 7, 4096. doi: 10.1038/s41598-017-04168-9
Bate, C. S. (1888). Report on the Crustacea macrura collected by the challenger during the years 1873–76. Rep. Sci. Challenger Zool. 24, 1873–1876.
Bell, T. (1846). “Part V,” in A History of the British Stalk-Eyed Crustacea (London: John van Voorst), 193–240.
Bernt, M., Donath, A., Jühling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313–319. doi: 10.1016/j.ympev.2012.08.023
Bernt, M., Merkle, D., Ramsch, K., Fritzsch, G., Perseke, M., Bernhard, D., et al. (2007). CREx: inferring genomic rearrangements based on common intervals. Bioinformatics 23, 2957–2958. doi: 10.1093/bioinformatics/btm468
Boore, J. L. (1999). Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767–1780. doi: 10.1093/nar/27.8.1767
Borradaile, L. A. (1916). Crustacea. Part I. Decapoda. Brit. Ant. Terra Nova Exp. Nat. Hist. Rep. Zool. 3, 75–110.
Bouvier, E. L. (1905). Sur les Thalassinidés recueillis par le Blake dans la mer des Antilles et le golfe du Mexique. Comp. Rend. Hebd. S. Acad. Sci. 141, 802–806.
Bouvier, E. L. (1925). Les Macroures marcheurs. Reports on the results of dredging under the supervision of Alexander Agassiz in the Gulf of Mexico (1877–78), in the Caribbean Sea (1878–79), and along the Atlantic coast of the United States (1880), by the U. S. Coast Survey steamer “Blake”. Mem. Mus. Comp. Zool. Harv. Univ. 47, 401–472.
Bracken, H. D., Toon, A., Felder, D. L., and Martin, J. W. (2009). The decapod tree of life: compiling the data and moving toward a consensus of decapod evolution. Arthropod Syst. Phylogeny 67, 99–116.
Cabezas, P., Macpherson, E., and Machordom, A. (2009). Morphological and molecular description of new species of squat lobster (Crustacea: Decapoda: Galatheidae) from the Solomon and Fiji Islands (South-West Pacific). Zool. J. Linn. Soc. 156, 465–493. doi: 10.1111/j.1096-3642.2008.00492.x
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Cheng, J., Chan, T. Y., Zhang, N., Sun, S., and Sha, Z. (2018). Mitochondrial phylogenomics reveals insights into taxonomy and evolution of Penaeoidea (Crustacea: Decapoda). Zool. Scr. 47, 582–594. doi: 10.1111/zsc.12298
Chu, K. H., Tsang, L. M., Ma, K. Y., Chan, T. Y., and Ng, P. K. L. (2009). “Decapod phylogeny: what can protein-coding genes tell us?,” in Crustacean Issues Vol. 18, Decapod Crustacean Phylogenetics, eds J. W. Martin, K. A. Crandall, and D. L. Felder (Bocan Raton, FL: CRC Press), 89–99. doi: 10.1201/9781420092592-c6
Dallwitz, M. J. (2018). Overview of the DELTA System. Available online at: http://delta-intkey.com/www/overview.htm (accessed June 22, 2020).
Dana, J. D. (1852). Conspectus crustaceorum, & c. Conspectus of the Crustacea of the Exploring Expedition under Capt. C. Wilkes, U. S. N. Proc. Acad. Nat. Sci. Phila. 6, 10–28.
De Man, G. J. (1907). Diagnoses of new species of macrurous decapod Crustacea from the “Siboga-Expedition”. Notes Leiden Mus. 29, 127–147.
De Man, G. J. (1925). The Decapoda of the Siboga Expedition. Part VI. The Axiidae collected by the Siboga-Expedition. Siboga Expéd. Monogr. 39a5, 1–127.
Duffy, J. E. (1996). Species boundaries, specialization, and the radiation of sponge-dwelling alpheid shrimp. Biol. J. Linnean Soc. 58, 307–324. doi: 10.1111/j.1095-8312.1996.tb01437.x
Dworschak, P. C. (2016). A new genus and species of axiid shrimp (Crustacea, Decapoda) from a southwestern Indian Ocean seamount. Eur. J. Taxon. 229, 1–11.
Faucci, A., Toonen, R. J., and Hadfield, M. G. (2007). Host shift and speciation in a coral-feeding nudibranch. Proc. R. Soc. B-Biol. Sci. 274, 111–119. doi: 10.1098/rspb.2006.3685
Faxon, W. (1893). Reports on the dredging operations off the west coast of Central America to the Galapagos, to the west coast of Mexico, and in the Gulf of California, in charge of Alexander Agassiz, carried on by the U. S. Fish Commission Steamer Albatross during 1891, lieut. Commander Z.L. Tanner, U.S.N., commanding, 6: preliminary description of new species of Crustacea. Bull. Mus. Comp. Zool. Harv. Coll. 24, 149–220.
Faxon, W. (1896). Reports on the results of dredging, under the supervision of Alexander Agassiz, in the Gulf of Mexico and the Caribbean Sea, and on the east coast of United States, 1877–1880, by the U. S. coast survey steamer Blake, Lieut-Commander C. D. Sigsbee, U. S. N., and Commander J.R. Bartlett, U. S. N., commanding, 37: supplementary notes on the Crustacea. Bull. Mus. Comp. Zool. Harv. Coll. 30, 153–168.
Felder, D. L., and Robles, R. (2009). “Molecular phylogeny of the family Callianassidae based on preliminary analysis of two mitochondrial genes,” in Crustacean Issues Vol. 18, Decapod Crustacean Phylogenetics, eds J. W. Martin, K. A. Crandall, and D. L. Felder (Bocan Raton, FL: CRC Press), 319–342.
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299.
Fritts-Penniman, A. L., Gosliner, T. M., Mahardika, G. N., and Barber, P. H. (2020). Cryptic ecological and geographic diversification in coral-associated nudibranchs. Mol. Phylogenet. Evol. 144:106698. doi: 10.1016/j.ympev.2019.106698
Fulton, S. W., and Grant, F. E. (1906). Some little known Victorian decapod Crustacea with descriptions of new species. Proc. R. Soc. Vic. 19, 5–15.
Goy, J. W. (2010). “Infraorder Stenopodidea Claus, 1872,” in Treatise on Zoology – Anatomy, Taxonomy, Biology – The Crustacea, Decapoda, Volume 9 Part A. Eucarida: Euphausiacea, Amphionidacea, and Decapoda (partim), eds F. R. Schram, J. C. von Vaupel Klein, M. Charmantier-Daures, and J. Forest (Leiden: Brill), 215–265. doi: 10.1163/9789004187801_009
Grant, J. R., and Stothard, P. (2008). The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36, 181–184.
Gray, J. E. (1872). Notes on the classification of the sponges. Ann. Mag. Nat. Hist. Ser. 4 9, 442–461. doi: 10.1080/00222937208696616
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Horká, I., De Grave, S., Fransen, C. H. J. M., Petrusek, A., and Duriš, Z. (2016). Multiple host switching events shape the evolution of symbiotic palaemonid shrimps (Crustacea: Decapoda). Sci. Rep. 6:26486.
Hurt, C., Silliman, K., Anker, A., and Knowlton, N. (2013). Ecological speciation in anemone-associated snapping shrimps (Alpheus armatus species complex). Mol. Ecol. 22, 4532–4548. doi: 10.1111/mec.12398
Huxley, T. H. (1879). On the classification and the distribution of the crayfishes. Proc. Zool. Soc. Lond. 1878, 752–788.
Jiang, Q., Kou, Q., and Li, X. (2015). New species of Globospongicola Komai and Saito, 2006 (Decapoda: Stenopodidea: Spongicolidae) from the South China Sea with discussion of its phylogenetic position. J. Crust. Biol. 35, 271–281. doi: 10.1163/1937240x-00002310
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kawabe, M., and Fujio, S. (2010). Pacific Ocean circulation based on observation. J. Oceanogr. 66, 389–403. doi: 10.1007/s10872-010-0034-8
Kawabe, M., Fujio, S., and Yanagimoto, D. (2003). Deep-water circulation at low latitudes in the western North Pacific. Deep Sea Res. Part I Oceanogr. Res. Pap. 50, 631–656. doi: 10.1016/s0967-0637(03)00040-2
Kensley, B. (1996). New thalassinidean shrimp from the Pacific Ocean (Crustacea: Decapoda: Axiidae and Calocarididae). Bull. Mar. Sci. 59, 469–489.
Kerpedjiev, P., Hammer, S., and Hofacker, I. L. (2015). Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics 31, 3377–3379. doi: 10.1093/bioinformatics/btv372
Komai, T. (2011). Deep-sea shrimps and lobsters (Crustacea: Decapoda: Dendrobranchiata and Pleocyemata) from the Sagami Sea and Izu Islands, central Japan. Mem. Natl. Mus. Nat. Sci. 47, 279–337.
Komai, T. (2013). A new species of the hippolytid genus Paralebbeus Bruce & Chace, 1986 (Crustacea: Decapoda: Caridea) from the Coral Seamount, southwestern Indian Ocean. Zootaxa 3646, 171–179.
Komai, T., De Grave, S., and Saito, T. (2016). Two new species of the stenopodidean shrimp genus Spongiocaris Bruce & Baba, 1973 (Crustacea: Decapoda: Spongicolidae) from the Indo-West Pacific. Zootaxa 4111, 421–447.
Komai, T., Lin, F. J., and Chan, T. Y. (2010). Five new species of Axiidae (Crustacea: Decapoda: Axiidea) from deep-water off Taiwan, with description of a new genus. Zootaxa 2352, 1–28.
Komai, T., and Tsuchida, S. (2012). Rediscovery and redescription of a sponge-associated axiid shrimp, Eiconaxius acutifrons Bate, 1888 (Crustacea: Decapoda: Axiidea). Zootaxa 3393, 27–40.
Kou, Q., Gong, L., and Li, X. (2018). A new species of the deep-sea spongicolid genus Spongicoloides (Crustacea, Decapoda, Stenopodidea) and a new species of the glass sponge genus Corbitella (Hexactinellida, Lyssacinosida, Euplectellidae) from a seamount near the Mariana Trench, with a novel commensal relationship between the two genera. Deep Sea Res. Part I Oceanogr. Res. Pap. 135, 88–107. doi: 10.1016/j.dsr.2018.03.006
Kou, Q., Li, X., Chan, T. Y., and Chu, K. H. (2015). Divergent evolutionary pathways and host shifts among the commensal pontoniine shrimps: a preliminary analysis based on selected Indo-Pacific species. Org. Divers. Evol. 15, 369–377. doi: 10.1007/s13127-014-0198-y
Kou, Q., Li, X., Chan, T. Y., Chu, K. H., Huang, H., and Gan, Z. (2013). Phylogenetic relationships among genera of the Periclimenes complex (Crustacea: Decapoda: Pontoniinae) based on mitochondrial and nuclear DNA. Mol. Phylogenet. Evol. 68, 14–22. doi: 10.1016/j.ympev.2013.03.010
Lagesen, K., Hallin, P., Rodland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2017). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773.
Lavery, S. D., Farhadi, A., Farahmand, H., Chan, T. Y., Azhdehakoshpour, A., Thakur, V., et al. (2014). Evolutionary divergence of geographic subspecies within the scalloped spiny lobster Panulirus homarus (Linnaeus 1758). PLoS One 9:e97247. doi: 10.1371/journal.pone.0097247
Li, J. Y., Zeng, C., Yan, G. Y., and He, L. S. (2019). Characterization of the mitochondrial genome of an ancient amphipod Halice sp. mt-2017 (Pardaliscidae) from 10,908 m in the Mariana Trench. Sci. Rep. 9:2610.
Lin, F. J., Liu, Y., Sha, Z. L., Tsang, L. M., Chu, K. H., Chan, T. Y., et al. (2012). Evolution and phylogeny of the mud shrimps (Crustacea: Decapoda) revealed from complete mitochondrial genomes. BMC Genomics 13:631. doi: 10.1186/1471-2164-13-631
Luo, R. B., Liu, B. H., Xie, Y. L., Li, Z. Y., Huang, W. H., Yuan, J. Y., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1, 18–23.
Macey, J. R., Larson, A., Ananjeva, N. B., Fang, Z., and Papenfuss, T. J. (1997). Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 14, 91–104. doi: 10.1093/oxfordjournals.molbev.a025706
MacGilchrist, A. C. (1905). Natural history notes from the R. I. M. S. “Investigator”, Capt. T. H. Heming, R. N. (retired), commanding, (3) 6. An account of the new and some of the rarer decapod Crustacea obtained during the surveying seasons 1901–1904. Ann. Mag. Nat. Hist. Ser. 7 15, 233–268. doi: 10.1080/03745480509443038
Manning, R. B., and Felder, D. L. (1991). Revision of the American Callianassidae (Crustacea: Decapoda: Thalassinidea). Proc. Biol. Soc. Wash. 104, 764–792.
Matzen da Silva, J., dos Santos, A., Cunha, M. R., Costa, F. O., Creer, S., and Carvalho, G. R. (2011). Multigene molecular systematics confirm species status of morphologically convergent Pagurus hermit crabs. PLoS One 6:e28233. doi: 10.1371/journal.pone.0028233
Morrison, C., Rios, R., and Duffy, E. (2004). Phylogenetic evidence for an ancient rapid radiation of Caribbean sponge-dwelling snapping shrimps (Synalpheus). Mol. Phylogenet. Evol. 30, 563–581. doi: 10.1016/s1055-7903(03)00252-5
Munday, P. L., Van Herwerden, L., and Dudgeon, C. L. (2004). Evidence for sympatric speciation by host shift in the sea. Curr. Biol. 14, 1498–1504. doi: 10.1016/j.cub.2004.08.029
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Ortmann, A. (1891). Die Decapoden-Krebse des Strassburger Museums, mit besonderer Berücksichtigung der von Herrn Dr. Döderlein bei Japan und bei den Liu-Kiu-Inseln gesammelten und zur Zeit im Strassburger Museum aufbewahrten Formen. III. Die Abtheilungen der Reptantia Baos: homaridea, Loricata und Thalassinidea. Zool. Jahrb. Abtheilung Syst. Geogr. Biol. Thiere 6, 1–58.
Poore, G. C. B. (1994). A phylogeny of the families of Thalassinidea (Crustacea: Decapoda) with keys to the families and genera. Mem. Mus. Vic. 54, 79–120. doi: 10.24199/j.mmv.1994.54.03
Poore, G. C. B. (2017). Synonymy and problematic species of Eiconaxius Spence Bate, 1888, with descriptions of new species (Crustacea: Decapoda: Axiidea: Axiidae). Zootaxa 4231, 364–376.
Poore, G. C. B. (2018). Caribbean species of Eiconaxius (Decapoda: Axiidea: Axiidae). Zootaxa 4524, 139–146.
Poore, G. C. B. (in press). “Axiid and micheleid lobsters from Indo-West Pacific deep-sea environments (Crustacea: Decapoda: Axiidea: Axiidae, Micheleidae),” in Tropical Deep-Sea Benthos, Vol. 31. Papua New Guinea, eds L. Corbari, T.-Y. Chan, and S. T. Ahyong (Paris: Mémoires du Muséum National d’Histoire Naturelle), 213.
Poore, G. C. B., and Collins, D. J. (2009). Australian Axiidae. Mem. Mus. Vic. 66, 221–287. doi: 10.24199/j.mmv.2009.66.20
Poore, G. C. B., and Dworschak, P. C. (2018). The Eiconaxius cristagalli species complex (Decapoda, Axiidea, Axiidae). Mem. Mus. Vic. 77, 105–120. doi: 10.24199/j.mmv.2018.77.06
Rambaut, A. (2016). FigTree 1.4.3. Available online at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed June 22, 2020).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., and Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Robles, R., Dworschak, P. C., Felder, D. L., Mantelatto, F. L., and Poore, G. C. B. (2020). A new molecular phylogeny of the Callianassoidea (Crustacea: Decapoda: Axiidea) with morphological support. Invertebr. Syst. 34, 113–132.
Robles, R., Tudge, C. C., Dworschak, P. C., Poore, G. C. B., and Felder, D. (2009). “Molecular phylogeny of the Thalassinidea based on nuclear and mitochondrial genes,” in Crustacean Issues Vol. 18, Decapod Crustacean Phylogenetics, eds J. W. Martin, K. A. Crandall, and D. L. Felder (Bocan Raton, FL: CRC Press), 309–326. doi: 10.1201/9781420092592-c15
Roehrdanz, R. L., Degrugillier, M. E., and Black, W. C. (2002). Novel rearrangements of arthropod mitochondrial DNA detected with Long-PCR: applications to arthropod phylogeny and evolution. Mol. Biol. Evol. 19, 841–849. doi: 10.1093/oxfordjournals.molbev.a004141
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Saito, T., and Komai, T. (2008). A review of species of the genera Spongicola de Haan, 1844 and Paraspongicola de Saint Laurent & Cleva, 1981 (Crustacea, Decapoda, Stenopodidea, Spongicolidae). Zoosystema 30, 87–147.
Saito, T., Tsuchida, S., and Yamamoto, T. (2006). Spongicoloides iheyaensis, a new species of deep-sea sponge-associated shrimp from the Iheya Ridge, Ryukyu Islands, southern Japan (Decapoda: Stenopodidea: Spongicolidae). J. Crust. Biol. 26, 224–233. doi: 10.1651/c-2650.1
Sakai, K. (1992). Axiid collections of the Zoological Museum, Copenhagen, with the description of one new genus and six new species (Axiidae, Thalassinidea, Crustacea). Zool. Scr. 21, 157–180. doi: 10.1111/j.1463-6409.1992.tb00318.x
Sakai, K. (2011). Axioidea of the world and a reconsideration of the Callianassoidea (Decapoda, Thalassinidea, Callianassida). Crust. Monogr. 13, 1–10.
Sakai, K. (2014). Axioidea material collected by the Japanese RV-s “Hakuhou-maru” and “Nagasaki-maru” in the East China Sea with a revised key to the families of the superfamily Axioidea Huxley, 1879 (Decapoda, Callianassidea). Crustaceana 87, 609–626. doi: 10.1163/15685403-00003302
Sakai, K., and de Saint Laurent, M. (1989). A check list of Axiidae (Decapoda, Crustacea, Thalassinidea, Anomura), with remarks and in addition descriptions of one new subfamily, eleven new genera and two new species. Naturalists Shikoku Univ. 3, 1–104.
Sakai, K., and Ohta, S. (2005). Some thalassinid collections by RV “Hakuhou-Maru” and RV “Tansei-Maru”, University of Tokyo, in the Sulu Sea, Philippines, and in Sagami Bay and Suruga Bay, Japan, including two new species, one new genus, and one new family (Decapoda, Thalassinidea). Crustaceana 78, 67–93. doi: 10.1163/1568540054024619
Shen, H., Braband, A., and Scholtz, G. (2013). Mitogenomic analysis of decapod crustacean phylogeny corroborates traditional views on their relationships. Mol. Phylogenet. Evol. 66, 776–789. doi: 10.1016/j.ympev.2012.11.002
Shen, Y., Kou, Q., Zhong, Z., Li, X., He, L., He, S., et al. (2017). The first complete mitogenome of the South China deep-sea giant isopod Bathynomus sp. (Crustacea: Isopoda: Cirolanidae) allows insights into the early mitogenomic evolution of isopods. Ecol. Evol. 7, 1869–1881. doi: 10.1002/ece3.2737
Simmonds, S. E., Chou, V., Cheng, S. H., Rachmawati, R., Calumpong, H. P., Mahardika, G. N., et al. (2018). Evidence of host-associated divergence from coral-eating snails (genus Coralliophila) in the Coral Triangle. Coral Reefs 37, 355–371. doi: 10.1007/s00338-018-1661-6
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., and Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 87, 651–701. doi: 10.1093/aesa/87.6.651
Sun, S., Hui, M., Wang, M., and Sha, Z. (2018a). The complete mitochondrial genome of the alvinocaridid shrimp Shinkaicaris leurokolos (Decapoda, Caridea): insight into the mitochondrial genetic basis of deep-sea hydrothermal vent adaptation in the shrimp. Comp. Biochem. Physiol. D Genomics Proteomics 25, 42–52. doi: 10.1016/j.cbd.2017.11.002
Sun, S., Sha, Z., and Wang, Y. (2018b). Complete mitochondrial genome of the first deep-sea spongicolid shrimp Spongiocaris panglao (Decapoda: Stenopodidea): novel gene arrangement and the phylogenetic position and origin of Stenopodidea. Gene 676, 123–138. doi: 10.1016/j.gene.2018.07.026
Sun, S., Sha, Z., and Wang, Y. (2019a). Divergence history and hydrothermal vent adaptation of decapod crustaceans: a mitogenomic perspective. PLoS One 14:e0224373. doi: 10.1371/journal.pone.0224373
Sun, S., Sha, Z., and Wang, Y. (2019b). The complete mitochondrial genomes of two vent squat lobsters, Munidopsis lauensis and M. verrilli : novel gene arrangements and phylogenetic implications. Ecol. Evol. 9, 12390–12407. doi: 10.1002/ece3.5542
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, M. H., Gan, H. M., Dally, G., Horner, S., Moreno, P. A. R., Rahman, S., et al. (2018a). More limbs on the tree: mitogenome characterisation and systematic position of ‘living fossil’ species Neoglyphea inopinata and Laurentaeglyphea neocaledonica (Decapoda: Glypheidea: Glypheidae). Invertebr. Syst. 32, 448–456.
Tan, M. H., Gan, H. M., Lee, Y. P., Bracken-Grissom, H., Chan, T. Y., Miller, A. D., et al. (2019). Comparative mitogenomics of the Decapoda reveals evolutionary heterogeneity in architecture and composition. Sci. Rep. 9:10756.
Tan, M. H., Gan, H. M., Lee, Y. P., Linton, S., and Austin, C. M. (2018b). Order within the chaos: insights into phylogenetic relationships within the Anomura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Mol. Phylogenet. Evol. 127, 320–331. doi: 10.1016/j.ympev.2018.05.015
Tan, M. H., Gan, H. M., Lee, Y. P., Poore, G. C. B., and Austin, C. M. (2017). Digging deeper: new gene order rearrangements and distinct patterns of codons usage in mitochondrial genomes among shrimps from the Axiidea, Gebiidea and Caridea (Crustacea: Decapoda). PeerJ 5:e2982. doi: 10.7717/peerj.2982
Tan, M. H., Gan, H. M., Schultz, M. B., and Austin, C. M. (2015). MitoPhAST, a new automated mitogenomic phylogeny tool in the postgenomic era with a case study of 89 decapod mitogenomes including eight new freshwater crayfish mitogenomes. Mol. Phylogenet. Evol. 85, 180–188. doi: 10.1016/j.ympev.2015.02.009
Toon, A., Finley, M., Staples, J., and Crandall, K. A. (2009). “Decapod phylogenetics and molecular evolution,” in Crustacean Issues Vol. 18, Decapod Crustacean Phylogenetics, eds J. W. Martin, K. A. Crandall, and D. L. Felder (Bocan Raton, FL: CRC Press), 9–24.
Tsang, L. M., Chan, B. K. K., Shih, F. L., Chu, K. H., and Chen, C. A. (2009). Host-associated speciation in the coral barnacle Wanella milleporae (Cirripedia: Pyrgomatidae) inhabiting the Millepora coral. Mol. Ecol. 18, 1463–1475. doi: 10.1111/j.1365-294x.2009.04090.x
Tsang, L. M., Chu, K. H., Nozawa, Y., and Chan, B. K. K. (2014). Morphological and host specificity evolution in coral symbiont barnacles (Balanomorpha: Pyrgomatidae) inferred from a multi-locus phylogeny. Mol. Phylogenet. Evol. 77, 11–22. doi: 10.1016/j.ympev.2014.03.002
Tsang, L. M., Lin, F. J., Chu, K. H., and Chan, T. Y. (2008a). Phylogeny of Thalassinidea (Crustacea, Decapoda) inferred from three rDNA sequences: implications for morphological evolution and superfamily classification. J. Zool. Syst. Evol. Res. 46, 216–223. doi: 10.1111/j.1439-0469.2008.00459.x
Tsang, L. M., Ma, K. Y., Ahyong, S. T., Chan, T. Y., and Chu, K. H. (2008b). Phylogeny of Decapoda using two nuclear protein-coding genes: origin and evolution of the Reptantia. Mol. Phylogenet. Evol. 48, 359–368. doi: 10.1016/j.ympev.2008.04.009
Vaidya, G., Lohman, D. J., and Meier, R. (2011). SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Wolfe, J. M., Breinholt, J. W., Crandall, K. A., Lemmon, A. R., Lemmon, E. M., Timm, L. E., et al. (2019). A phylogenomic framework, evolutionary timeline and genomic resources for comparative studies of decapod crustaceans. Proc. R. Soc. B Biol. Sci. 286:20190079. doi: 10.1098/rspb.2019.0079
WoRMS (2020). Eiconaxius Bate, 1888. Available online at: http://www.marinespecies.org/aphia.php?p=taxdetails&id=390862 (accessed June 22, 2020).
Xu, P., Liu, F., Ding, Z., and Wang, C. (2016). A new species of the thorid genus Paralebbeus Bruce & Chace, 1986 (Crustacea: Decapoda: Caridea) from the deep sea of the Northwestern Pacific Ocean. Zootaxa 4085, 119–126.
Keywords: biodiversity, taxonomy, host specificity, isolation, divergence, mitochondrial genome
Citation: Kou Q, Xu P, Poore GCB, Li X and Wang C (2020) A New Species of the Deep-Sea Sponge-Associated Genus Eiconaxius (Crustacea: Decapoda: Axiidae), With New Insights Into the Distribution, Speciation, and Mitogenomic Phylogeny of Axiidean Shrimps. Front. Mar. Sci. 7:469. doi: 10.3389/fmars.2020.00469
Received: 27 January 2020; Accepted: 26 May 2020;
Published: 07 July 2020.
Edited by:
Zhijun Dong, Yantai Institute of Coastal Zone Research (CAS), ChinaReviewed by:
Hongying Sun, Nanjing Normal University, ChinaXiaoshou Liu, Ocean University of China, China
Copyright © 2020 Kou, Xu, Poore, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinzheng Li, bGl4emhAcWRpby5hYy5jbg==; Chunsheng Wang, d2FuZ3Npb0BzaW8ub3JnLmNu
†These authors have contributed equally to this work
 Qi Kou
Qi Kou Peng Xu
Peng Xu Gary C. B. Poore
Gary C. B. Poore Xinzheng Li1,4,5,6*
Xinzheng Li1,4,5,6*