- 1Department of Biological Sciences, University of Bergen, Bergen, Norway
- 2Department of Earth Sciences, University of Bergen, Bergen, Norway
- 3K. G. Jebsen Centre for Deep-Sea Research, University of Bergen, Bergen, Norway
- 4NORCE Norwegian Research Centre, Bergen, Norway
- 5Department of Marine Sciences, University of Gothenburg, Gothenburg, Sweden
- 6Gothenburg Global Biodiversity Centre, Gothenburg, Sweden
Large parcels of organic matter in the deep sea, such as whale carcasses, harbor a very specialized fauna, most famously the bone-eating worms in the genus Osedax (Annelida, Siboglinidae). Although Osedax was first described only 15 years ago, there are already 26 described species from the Pacific, Atlantic, and Southern Oceans. The high discovery rate of new Osedax species indicates that there is still a lot of undescribed diversity. In this study we describe the most northerly species of Osedax to date, Osedax fenrisi sp. nov. from 73°N on the Arctic Mid-Ocean Ridge. We also present an updated molecular phylogeny of Osedax based on cytochrome oxidase subunit I and 18S rRNA, including all described species in the genus. The molecular results support that O. fenrisi sp. nov. is distinct from the previously known species of Osedax. Both morphological characters and the molecular phylogeny support the placement of O. fenrisi sp. nov. in clade V. The most striking morphological character shared with other described species in this clade (Osedax rubiplumus, Osedax roseus, and Osedax bryani) is the presence of long pinnules inserted on the outside of the palps. Nomenclatural act recorded in Zoobank. LSID: E55A5C87-0CB6-4146-B3D9-7E3B19B68628.
Introduction
Large parcels of organic matter in the deep sea such as whale carcasses or sunken wood logs provide nutrients both for opportunistic scavengers and a more specialized fauna (Smith and Baco, 2003; Bienhold et al., 2013). The most famous of the organic fall specialists is probably the bone-eating worms in the genus Osedax (Rouse et al., 2004). Osedax belongs to the annelid family Siboglinidae, which is characterized by adult worms lacking a functional digestive system, and instead relying on bacterial symbionts for nutrients (Schulze and Halanych, 2003). Siboglinids are found in reducing habitats such as organically enriched sediments, organic falls, cold seeps and hydrothermal vents, and most of the siboglinid taxa have chemoautotrophic symbionts that rely on sulfide or methane (Hilário et al., 2011). Within the siboglinids, Osedax is unique in having heterotrophic symbionts, which live in the posterior root-like extensions that protrude into the bone substratum and probably utilize collagen from the bones (Goffredi et al., 2005, 2007). Colonization experiments have demonstrated that Osedax are not limited to inhabiting the bones of marine mammals such as whales and seals, some species can also inhabit bones from turtles, birds, teleosts or from terrestrial mammals such as cows and pigs (Glover et al., 2008; Jones et al., 2008; Rouse et al., 2011, 2018).
Although Osedax was first described only 15 years ago (Rouse et al., 2004), there are already 26 described species and an additional 9 unnamed OTUs (Rouse et al., 2018; Fujiwara et al., 2019). The Monterey Bay in California, from where the first Osedax species was described, has the highest documented diversity with 18 named species (Rouse et al., 2018). Research on natural organic falls and colonization experiments in other oceans has, however, lead to the discovery of new Osedax species in most of the world oceans, and at depths from 10–4204 m (e.g., Glover et al., 2005, 2013; Fujikura et al., 2006). The continued high discovery rate of new Osedax species would indicate that more undescribed diversity remains, and that the high diversity observed along the California margin is a product of the intensity of deep-sea research in this region.
The co-occurrence of many species of Osedax in the Monterey Bay area, where the diversity of Osedax is best explored, could be partly explained by depth segregation. It appears that most species are restricted to a certain depth range, and many species are only known from a single depth (Rouse et al., 2018). It has been suggested that the Antarctic species of Osedax have a more eurybathic distribution due to the lack of strong temperature clines through the water column (Amon et al., 2014). However, depth ranges of over 1000 m are also known in three species from Monterey Bay (Lundsten et al., 2010), so the ability of some species to occupy wide depth ranges is not unique to the Antarctic. In addition to a species turnover with depth, some species are segregated by stage of decomposition of the bones, with early colonizers being replaced by other species after some time (Rouse et al., 2018).
Compared to the Pacific and the Antarctic, organic fall fauna from the Atlantic and Arctic are not very well known. The only described species of Osedax in the North Atlantic, Osedax mucofloris, was originally described from 125 m depth in a fjord on the west coast of Sweden (Glover et al., 2005), and has subsequently been reported from a similar depth on the west coast of Norway (Schander et al., 2010) and from 1000 m depth in the Setubal Canyon, off Portugal (Hilario et al., 2015). One new species of Osedax and four OTUs were recently recorded from the deep southwest Atlantic (4204 m depth off Brazil; Fujiwara et al., 2019; Shimabukuro and Sumida, 2019). In addition, there are two undescribed putative species: one from the Setubal Canyon off Portugal (1000 m depth; Hilario et al., 2015) and one from the Mediterranean (53 m depth; Taboada et al., 2015). The most northerly species of Osedax, O. mucofloris, has not been recorded further north than 60°N (Schander et al., 2010), and thus there are no species of Osedax previously known from within the Arctic circle.
Here we report on findings of a new species of Osedax from colonization experiments on the Arctic Mid-Ocean Ridge at 73° N and 2340 m depth, near the Loki’s Castle Vent Field (LCVF). To assess the phylogenetic position of the new species, we performed a phylogenetic analysis based on cytochrome oxidase subunit I (COI) and 18S rRNA (18S), including all described species of Osedax and known OTUs available in GenBank.
MATERIALS AND METHODS
Colonization Experiment
The colonization experiments were deployed and retrieved during the K. G. Jebsen Centre for Deep-Sea Research cruises to the Arctic mid-Ocean Ridge on the RV G. O. Sars in 2017 and 2018. Dried cow bones bought from a pet shop were used as substratum for the colonization experiment. The bones were enclosed in cages with holes of 4 mm in diameter to prevent them from being picked up and moved by larger animals, and to retain animals that had colonized the bones during retrieval of the experiments. Two bones, each in a separate cage, were deployed on (10.07.2017) on a sedimented plain at 2341 m depth near the LCVF using the ROV Ægir 6000. Although in close proximity to the active hydrothermal vent field, the sediments where the experiments were deployed were passive, without any signs of venting fluid or bacterial mats. The experiments were collected by ROV 1 year later (22.07.2018) at 73°34′01.6″N 8°09′31.7″E (experiment CB1) and at 73°34′01.2″N 8°09′29.9″E (experiment CB2). The bones were densely colonized by Osedax fenrisi sp. nov., and over 100 specimens were collected. The worms were carefully removed from the bones and fixed in 96% ethanol, RNA later or formalin, and some specimens were flash frozen in liquid nitrogen and stored at −80°C.
Morphological Analyses
In the lab, fixed specimens were examined using dissecting and compound microscopes. Scanning electron microscopy (SEM) using a Zeiss Supra 55VP was applied on two specimens that were critical point dried and mounted on stubs following a preparation of graded ethanol dehydration.
Molecular Analyses
For phylogenetic analysis the mitochondrial marker COI and the nuclear small ribosomal subunit 18S were sequenced. Tissue for DNA extraction was sampled from the palps of the worms. DNA was extracted using the Qiagen DNeasy Blood and Tissue kit, following the manufacturers protocol. PCR and sequencing primers can be found in Table 1. PCR reactions were run with TaKaRa taq and PCR protocols were as follows: COI – 4 min at 96°C, 45 cycles of 30 s at 95°C, 30 s at 48°C, and 1 min at 72°C, and finally 7 min at 72°C. 18S – 3 min at 94°C, 35 cycles with 1 min at 94°C, 1.5 min at 42°C, and 2 min at 72°C, and finally 7 min at 72°C. Quality and quantity of amplicons were assessed by gel electrophoresis imaging using a FastRuler DNA Ladder (Life Technologies) and GeneSnap and GeneTools (SynGene) for image capture and band quantification. Successful PCRs were purified using Exonuclease 1 (EXO, 10 U mL–1) and Shrimp 90 Alkaline Phosphatase (SAP, 10 U mL–1, USB Europe, Germany) in 10 μL reactions (0.1 mL EXO, 1 μL SAP, 0.9 μL ddH 2O, and 8 μL PCR product). Samples were incubated at 37°C for 15 min followed by an inactivation step at 80°C for 15 min. The purified PCR products were sequenced using BigDye v3.1 (Life Technologies) and run on an Automatic Sequencer 3730XL at the sequencing facility of the Institute of Molecular Biology, University of Bergen.
Forward and reverse sequences were assembled in Geneious (Biomatters Ltd.) and checked for contamination using BLAST (Altschul et al., 1990). Sequences of 35 known species of Osedax (26 described species and 9 undescribed putative species) and sequences of Sclerolinum brattstromi as an outgroup were downloaded from Genbank. The dataset was based on the phylogenetic dataset in Rouse et al. (2018), with some additional sequences of newly described species and OTUs added. For species with very wide distributions, one specimen from each region was included, and also two specimens of the new species. For Genbank accession numbers see Table 2. For the four OTUs and specimens of Osedax frankpressi recorded from the South-West Atlantic (Shimabukuro and Sumida, 2019), only sequences of COI were available. An initial attempt was made to include these in the combined analysis of COI + 18S with 18S as missing data, but this resulted in a poorly supported tree. Thus, these sequences were only included in the single-gene analysis of COI.
Sequences were aligned in Geneious using the MUSCLE algorithm for COI (Edgar, 2004), and MAFFT for 18S (Katoh and Standley, 2013). Alignments were trimmed and missing data at the ends were coded with question marks. Pairwise p-distances for COI were calculated in Geneious and can be found in Supplementary Table 1. Substitution saturation was tested for the first, second and third codon position of COI using the Xia method implemented in DAMBE6 (Xia and Lemey, 2009), which indicated high levels of saturation in the third codon position. This position was thus excluded from the alignment for the combined analysis with 18S. Single-gene analysis of COI was performed both with and without the third codon position.
The best partition scheme and substitution models were identified using Partition Finder v2.1.1 with the greedy algorithm and PhyML (Guindon et al., 2010; Lanfear et al., 2016). The best partition scheme was found to be one with the first and second codon position of COI as separate partitions, and 18S as one partition. For the first codon position of COI the SYM + G model was selected, GTR + I for the second codon position and GTR + I + G for 18S. The datasets were analyzed by Bayesian Inference in MrBayes v.3.2.7a (Ronquist and Huelsenbeck, 2003) and by Maximum Likelihood in RAxML v.8.2.10 (Stamatakis, 2014). The MrBayes analysis was run for 10 million generations for the analysis of the concatenated dataset, and 3 million generations for the single-gene analyses. Tracer v1.5 was used to check for convergence, and to determine the burnin, which was set to 10%. The RAxML analyses were run with the GTRGAMMA model and the program was allowed to halt bootstrapping automatically. All phylogenetic analyses were run on the CIPRES Science Gateway (Miller et al., 2010). The trees were converted to graphics using FigTree v1.4.0 (Rambaut, 2012) and final adjustments were made in Adobe Illustrator v24.1 (Adobe Systems, San Jose, CA, United States).
Results
Morphological Description
Annelida Lamarck, 1809, Siboglinidae Caullery, 1914.
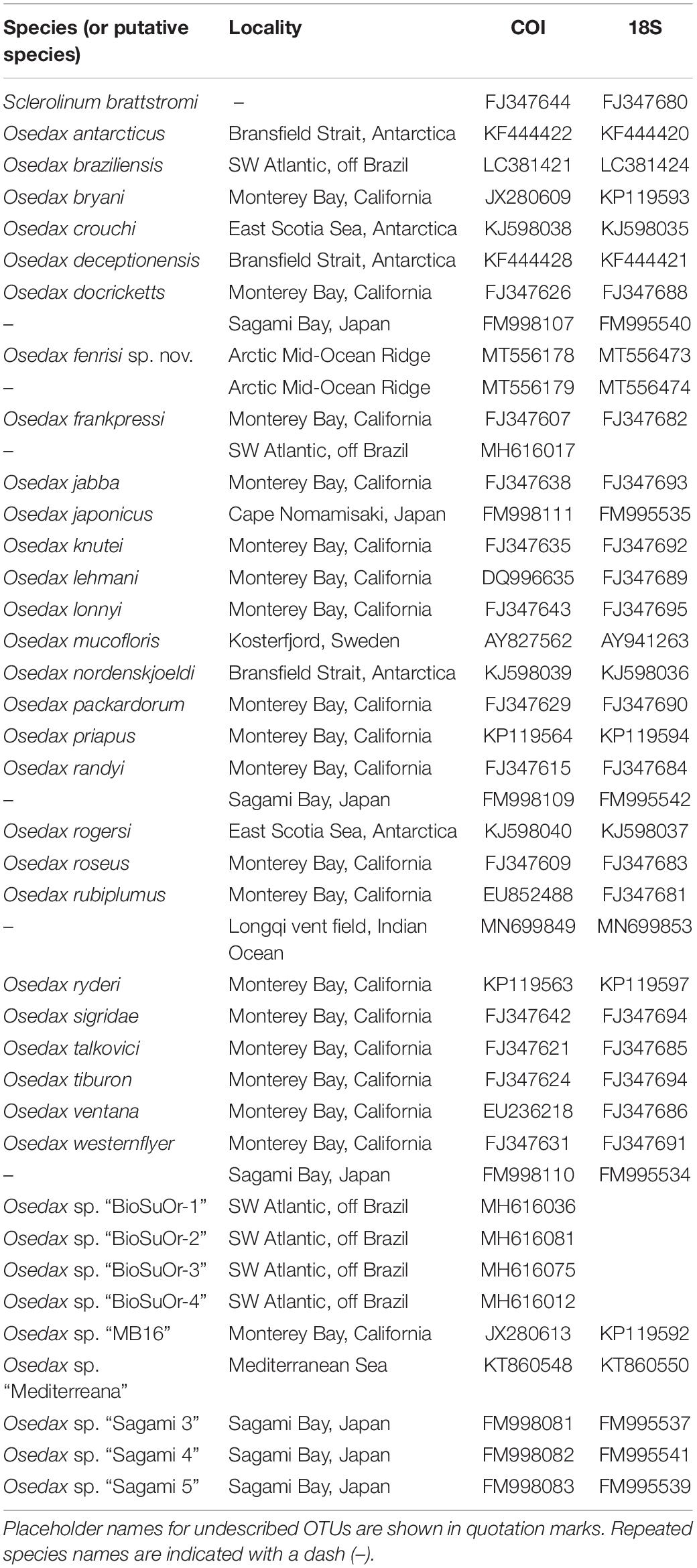
O. fenrisi sp. nov. Eilertsen, Dahlgren and Rapp, 2020 (Figures 1A–F).
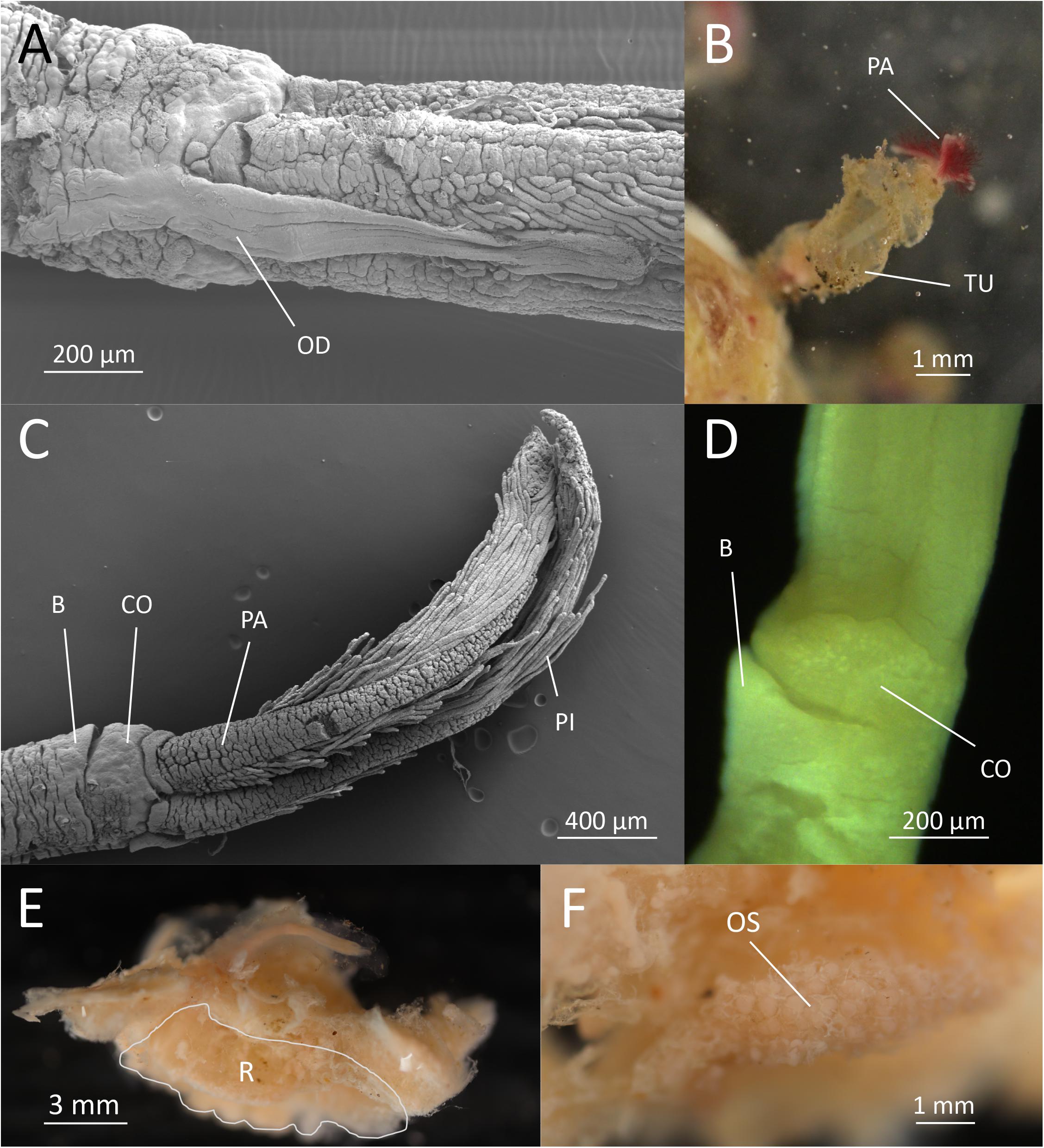
Figure 1. Osedax fenrisi sp. nov. (A) SEM. Dorsal view. Oviduct (OD) extending between the dorsal most palps. (B) Live image from tank. Red palps (PA) extending out from mucous tube (TU). (C) SEM. Lateral view. Ventral collar (CO) visible with a small bump (B). (D) Light microscopy. Ventral view. Bump (B) visible on the lower rim of collar (CO). (E) Formalin fixed female dissected out of bone with attached bone material. The root tissue is indicated (R). (F) Ovisac (OS) in formalin fixed root tissue.
Material Examined
Holotype: ZMBN 136747, Female, fixed in formalin preserved in ethanol, collected with ROV Ægir 6000 from cow bone at 2340 m depth at 73°34′01.6″N 8°09′31.7″E (experiment CB1) on 22.07.2018. Paratypes: ZMBN 136748, Females (2), fixed in formalin preserved in ethanol, collected with ROV Ægir 6000 from cow bone at 2340 m depth at 73°34′01.2″N 8°09′29.9″E (experiment CB2) on July 22, 2018.
Diagnosis and Description
Holotype female; preserved trunk 2.4 mm long, 0.3 mm wide; crown of palps 1.7 mm long. Pinnules long, some extending up to half the length of the palps, inserted dorsally, with a distal bulb or knob (Figure 1C). Oviduct short, emerging from the rim of the trunk, extending between the base of the dorsal palps a quarter of the length of palps (Figure 1A). Trunk with inconspicuous collar on anterior margin, thicker on the ventral side (opposite the oviduct) including a “bump” (Figures 1C,D). No demarcation of upper and lower trunk. Live specimen with red palps (Figure 1B). Color of trunk pale or white (Figure 1B). Color of the collar not observed. Preserved specimens without pigmentation. Tube cylindrical and not very gelatinous (Figure 1B). Root structure bulbous (Figure 1E). Ovisac an ellipsoidal mass (Figure 1F). Males not observed in 10 preserved specimens investigated with dissecting microscope.
Distribution
Known from LCVF at 2340 m depth in cow bone.
Etymology
This species is named after the Fenris wolf, son of the Norse god Loki, for whom the vent field where the species was discovered is named.
Remarks
Osedax fenrisi sp. nov. is part of Osedax clade V and is sister taxon to the O. rubiplumus, O. roseus, O. “sagami 3,” and O. “sagami 4” clade (Figure 2). O. fenrisi sp. nov. has palps with long pinnules inserted dorsally as in O. bryani. Like other Osedax in clade V, O. fenrisi sp. nov. has an oviduct that extend beyond the trunk into the crown. However, in O. fenrisi sp. nov. it is relatively short and like in Osedax randyi and Osedax braziliensis not extending beyond half the length of the palps. A weak collar present at crown base similar to O. roseus, including a bump that is less pronounced than in O. braziliensis. The root structure is bulbous but branched, while in O. rubiplumus and O. roseus the root is long. No comparable morphological data currently available for O. “sagami 3” or O. “sagami 4.”
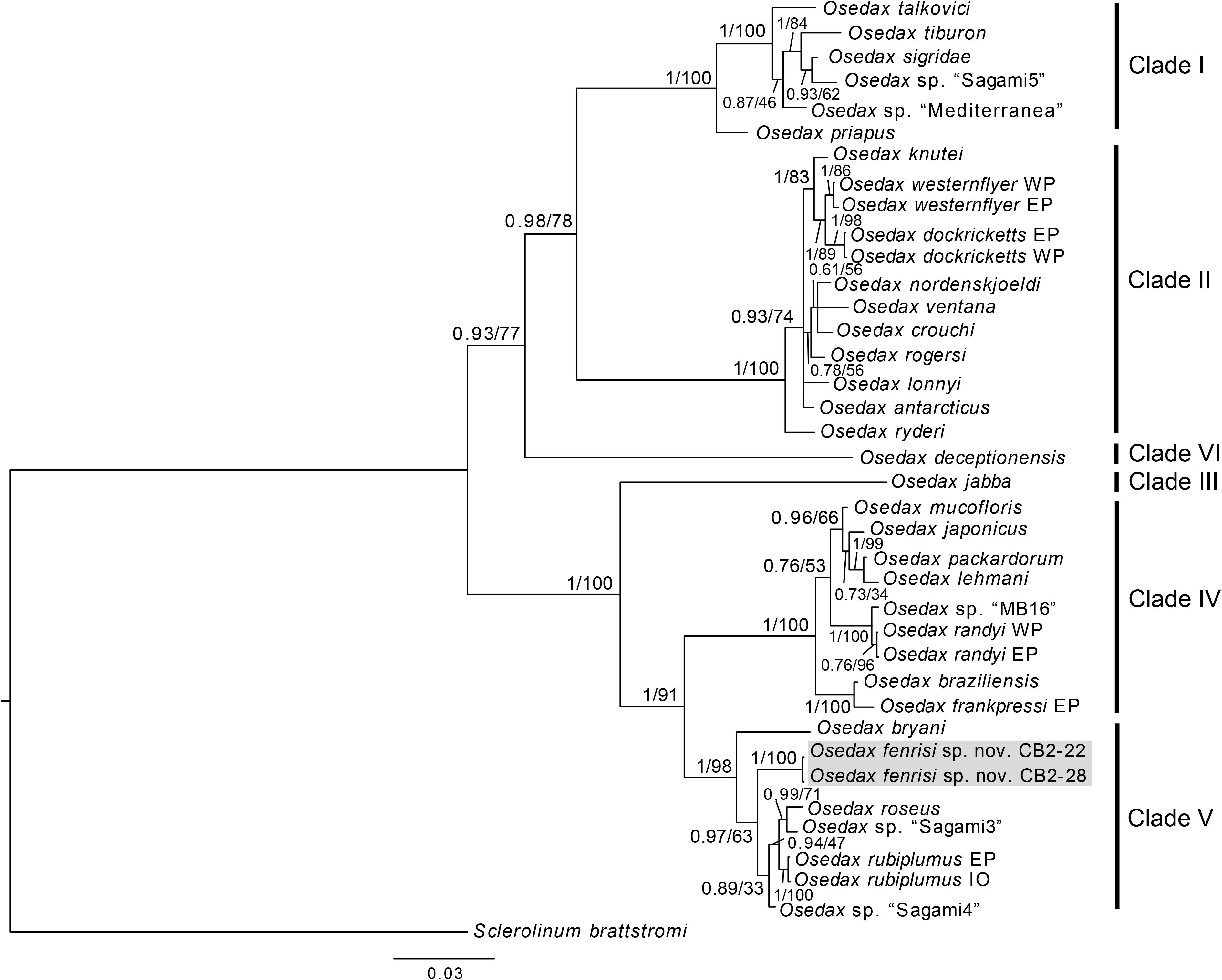
Figure 2. Phylogenetic reconstruction based on the concatenated dataset of COI and 18S. The tree is the summary tree from the MrBayes analysis, and node support values are given as PP/BS. For nodes that were not recovered in the tree from the maximum likelihood analysis, support is denoted as a hyphen (-). Placeholder names for undescribed OTUs are shown in quotation marks, while for species with more than one specimen is included, the region of the specimens is indicated following the species name (EP, East Pacific; WP, West Pacific; At, Atlantic; IO, Indian Ocean).
A peculiar observation and apparently unique (among known Osedax) is that the tubes are inhabited by a small nematode in very high abundancies. However, similar observations have been made in other polychaetes from the Loki’s Castle, where the nematode abundances were particularly high in Nicomache lokii (Kongsrud and Rapp, 2012).
Molecular Results
The COI sequences of O. fenrisi sp. nov. were 0.4% different from each other (2 bp difference), and 14.6–14.8% different from the most closely related species (Osedax sp. “Sagami 4”). A complete matrix of pairwise uncorrected p-distances can be found in Supplementary Table 1. The gene-trees for COI (including third codon position) and 18S can be found in Supplementary Figures 1, 2. The phylogenetic analyses of COI with and without the third codon position included yielded a similar topology, but with lower support when the third codon position was excluded. In general, the COI gene-tree is very poorly resolved (Supplementary Figure 1).
The phylogenetic analysis of the concatenated dataset of COI and 18S recovered the same well supported clades (clade I–V, see Figure 2) as described by Vrijenhoek et al. (2009), and with Osedax deceptionensis recovered as sister to clades I and II (clade VI; Amon et al., 2014; Rouse et al., 2018). O. fenrisi sp. nov. is recovered within clade V with high support (Figure 2). O. mucofloris, the only species previously known from the North Atlantic, is recovered within clade IV, which is the sister clade to clade V.
Discussion
In the present study, we described the most northerly species of Osedax to date, and confirmed the presence of Osedax in the Arctic. The molecular results supported that O. fenrisi sp. nov. was distinct from the previously known species of Osedax. Both morphological characters and the molecular phylogeny supported the placement of O. fenrisi sp. nov. in clade V. The most striking morphological character shared with other described species in this clade (O. rubiplumus, O. Roseus, and O. bryani) is the presence of long pinnules inserted dorsally on the palps. Males were not observed in the investigated material. This is most likely due to the small size of males, and that they are difficult to observe in preserved specimens. The OTU “BioSuOr 4” from the South-West Atlantic also most likely belongs to clade V, based on the presence of pinnules and a phylogenetic association with species in clade V (Shimabukuro and Sumida, 2019; see also Supplementary Figure 1). However, the lack of sequence data for other genes in addition to COI for this OTU, and other OTUs from the South-West Atlantic, prevents confident placement of these OTUs in the phylogeny of Osedax. The low resolution of the COI gene-tree (see Supplementary Figure 1) shows that although COI works well as a barcoding gene in Osedax, it has limited use as a phylogenetic marker. The phylogeny presented here based on COI and 18S mainly shows the same topology as previous phylogenies based on more markers (e.g., Rouse et al., 2018), but with some minor differences in the relationships between species within clades II and V. To produce a robust phylogeny of the genus including the new species would require a higher number of independent genetic markers, but that was outside the scope of this article.
The North-Atlantic species O. mucofloris was not found on the deployed bones. This could indicate that the experiments were outside of either the geographic range or the depth range of O. mucofloris. Several Osedax species are known to have wide geographic distributions, for example there are five species that have a trans-Pacific distribution (Rouse et al., 2018), a range of over 8000 km. Recently, two species have also been recorded from multiple oceans; O. rubiplumus from the Pacific, Antarctic, and Indian Oceans (Zhou et al., 2020), and O. frankpressi from the Pacific and the Atlantic Oceans (Shimabukuro and Sumida, 2019). O. mucofloris is known from the coast of Sweden, Norway and from the Setubal Canyon off Portugal (Glover et al., 2005; Dahlgren et al., 2006; Schander et al., 2010; Hilario et al., 2015), which is also a sizeable range. Therefore, it seems unlikely that distance alone could limit O. mucofloris from colonizing the deployed bones. The known depth range of O. mucofloris is 30–1000 m, which means that the Arctic bone deployments were over 1000 m outside the known depth range of O. mucofloris. However, it is more likely that the distribution is regulated by water mass and temperature. Shelf and slope occurrences of O. mucofloris are linked to NE Atlantic- and coastal waters with temperatures ranging from 7 to 10°C. Going deeper into the Nordic Seas and the Arctic the temperature decreases rapidly and is negative below 600–1000 m depth. The site of our deployment is characterized by Norwegian Sea deep water with a temperature of −0.5 to −0.9°C, which is likely to be too cold for O. mucofloris. It should also be noted that only cow bones were deployed in this experiment, and as Osedax species appear to have different preferences when it comes to the settlement substrate (Rouse et al., 2018), cow bones might not attract all Osedax species. Additional experiments with a wide range of bone substrates, both in terms of size and animal origin, would be desirable to capture the entire diversity of Osedax in the region.
The phylogeny shows that O. fenrisi sp. nov. belongs to a different clade than O. mucofloris, which refutes the hypothesis that these two species might be part of a regional North Atlantic/Arctic radiation. Most of the species in clade V are from the Pacific, which could indicate a Pacific origin of the clade. It has previously been suggested that some of the hydrothermal vent fauna of the LCVF may have originated from the Pacific (Pedersen et al., 2010). However, recent advances on the taxonomy and distributions of the annelids from LCVF has called this hypothesis into question (Kongsrud et al., 2017; Eilertsen et al., 2018). In other groups there are apparent links both to the Polar basin and the Pacific on one hand, and to the Mid-Atlantic Ridge on the other (Antje, 2015; Tandberg et al., 2017). It should be noted that there is an over-representation of Pacific Osedax species in the phylogeny compared to other oceans due to unbalanced sampling efforts, and the prevalence of Pacific species in clade V may thus be an artifact. A more complete sampling of the global diversity of Osedax is necessary to infer biogeographic patterns and evolutionary pathways.
Data Availability Statement
The datasets generated for this study can be found in GenBank and Figshare.
Author Contributions
ME participated in the deployment and retrieval of the colonization experiment, sampling of Osedax, performed the DNA analyses, and drafted the manuscript. TD did the morphological work and wrote the species descriptions. HR came up with the study design and coordinated the study. All authors contributed to the general text and read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the crew, scientists and students onboard the R/V G.O. Sars during the cruises in 2017 and 2018, and the operators of the ROV Ægir6000 for their assistance at sea. We are especially grateful to Tone Ulvatn and Anne Helene Tandberg for their help with deployment and retrieval of the colonization experiments. We thank David J. Rees for help with the molecular work, which was performed at the Biodiversity Laboratories, University of Bergen. We also thank Jon A. Kongsrud and Katrine Kongshavn, University Museum of Bergen for assisting with SEM images. Finally, we would like to thank the two reviewers for their feedback, which helped improve this manuscript. This work is a contribution from the KG Jebsen Centre for Deep Sea Research (funded through Stiftelsen Kristian Gerhard Jebsen).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00443/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1006/jmbi.1990.9999
Amon, D. J., Wiklund, H., Dahlgren, T. G., Copley, J. T., Smith, C. R., Jamieson, A. J., et al. (2014). Molecular taxonomy of Osedax (Annelida: Siboglinidae) in the Southern Ocean. Zool. Scr. 43, 405–417. doi: 10.1111/zsc.12057
Antje, B. (2015). The Expedition PS86 of the Research Vessel POLARSTERN to the Arctic Ocean in 2014. (Germany: Berichte zur Polar- und Meeresforschung = Reports on Polar and Marine Research), 685:133. doi: 10.2312/BzPM_0685_2015
Bienhold, C., Ristova, P. P., Wenzhofer, F., Dittmar, T., and Boetius, A. (2013). How deep-sea wood falls sustain chemosynthetic life. PLoS One 8:e53590. doi: 10.1371/journal.pone.0053590
Cohen, B. L., Gawthrop, A., and Cavalier-Smith, T. (1998). Molecular phylogeny of brachiopods and phoronids based on nuclear-encoded small subunit ribosomal RNA gene sequences. Philos. Trans. R. Soc. B 353, 2039–2061. doi: 10.1098/rstb.1998.0351
Dahlgren, T. G., Wiklund, H., Kallstrom, B., Lundalv, T., Smith, C. R., and Glover, A. G. (2006). A shallow-water whale-fall experiment in the north Atlantic. Cah. Biol. Mar. 47, 385–389.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Eilertsen, M. H., Georgieva, M. N., Kongsrud, J. A., Linse, K., Wiklund, H., Glover, A. G., et al. (2018). Genetic connectivity from the Arctic to the Antarctic: Sclerolinum contortum and Nicomache lokii (Annelida) are both widespread in reducing environments. Sci. Rep. 8:4810. doi: 10.1038/s41598-018-23076-0
Fujikura, K., Fujiwara, Y., and Kawato, M. (2006). A new species of Osedax (Annelida: Siboglinidae) associated with whale carcasses off Kyushu. Jpn. Zool. Sci. 23, 733–740. doi: 10.2108/zsj.23.733
Fujiwara, Y., Jimi, N., Sumida, P. Y. G., Kawato, M., and Kitazato, H. (2019). New species of bone-eating worm Osedax from the abyssal South Atlantic Ocean (Annelida: Siboglinidae). Zookeys 814, 53–69. doi: 10.3897/zookeys.814.28869
Glover, A. G., Källström, B., Smith, C. R., and Dahlgren, T. G. (2005). World-wide whale worms? A new species of Osedax from the shallow north Atlantic. Proc. Royal Soc. B 272, 2587–2592. doi: 10.1098/rspb.2005.3275
Glover, A. G., Kemp, K. M., Smith, C. R., and Dahlgren, T. G. (2008). On the role of bone-eating worms in the degradation of marine vertebrate remains. Proc. Royal Soc. B 275, 1959–1961. doi: 10.1098/rspb.2008.0177
Glover, A. G., Wiklund, H., Taboada, S., Avila, C., Cristobo, J., Smith, C. R., et al. (2013). Bone-eating worms from the Antarctic: the contrasting fate of whale and wood remains on the Southern Ocean seafloor. Proc. Royal Soc. B 280:1768. doi: 10.1098/rspb.2013.1390
Goffredi, S. K., Johnson, S. B., and Vrijenhoek, R. C. (2007). Genetic diversity and potential function of microbial symbionts associated with newly discovered species of Osedax polychaete worms. Appl. Environ. Microbiol. 73, 2314–2323. doi: 10.1128/aem.01986-06
Goffredi, S. K., Orphan, V. J., Rouse, G. W., Jahnke, L., Embaye, T., Turk, K., et al. (2005). Evolutionary innovation: a bone-eating marine symbiosis. Environ. Microbiol. 7, 1369–1378. doi: 10.1111/j.1462-2920.2005.00824.x
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Halanych, K. M., Bacheller, J. D., Aguinaldo, A. M., Liva, S. M., Hillis, D. M., and Lake, J. A. (1995). Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science 267, 1641–1643. doi: 10.1126/science.7886451
Hilário, A., Capa, M., Dahlgren, T. G., Halanych, K. M., Little, C. T. S., Thornhill, D. J., et al. (2011). New perspectives on the ecology and evolution of siboglinid tubeworms. PLoS One 6:e16309. doi: 10.1371/journal.pone.0016309
Hilario, A., Cunha, M. R., Génio, L., Marçal, A. R., Ravara, A., Rodrigues, C. F., et al. (2015). First clues on the ecology of whale falls in the deep Atlantic Ocean: results from an experiment using cow carcasses. Mar. Ecol. 36, 82–90. doi: 10.1111/maec.12246
Hillis, D. M., and Dixon, M. T. (1991). Ribosomal DNA: molecular evolution and phylogenetic inference Q. Rev. Biol. 66, 411–453. doi: 10.1086/417338
Jones, W. J., Johnson, S. B., Rouse, G. W., and Vrijenhoek, R. C. (2008). Marine worms (genus Osedax) colonize cow bones. Proc. Royal Soc. B 275, 387–391. doi: 10.1098/rspb.2007.1437
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kongsrud, J. A., Eilertsen, M. H., Alvestad, T., Kongshavn, K., and Rapp, H. T. (2017). New species of Ampharetidae (Annelida: Polychaeta) from the Arctic loki castle vent field. Deep Sea Res. Pt II 137, 232–245. doi: 10.1016/j.dsr2.2016.08.015
Kongsrud, J. A., and Rapp, H. T. (2012). Nicomache (Loxochona) lokii sp. nov. (Annelida, Polychaeta, Maldanidae) from the Loki’s Castle vent field – an important structure builder in an Arctic vent system. Pol. Biol. 35, 161–170. doi: 10.1007/s00300-011-1048-4
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Lundsten, L., Schlining, K. L., Frasier, K., Johnson, S. B., Kuhnz, L. A., Harvey, J. B. J., et al. (2010). Time-series analysis of six whale-fall communities in Monterey Canyon, California, USA. Deep Sea Res. Pt I 57, 1573–1584. doi: 10.1016/j.dsr.2010.09.003
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES science gateway for inference of large phylogenetic trees,” in Proceedings of the Gateway Computing Environments Workshop (GCE) (New Orleans, LA: IEEE), 1–8.
Passamaneck, Y. J., Schander, C., and Halanych, K. M. (2004). Investigation of molluscan phylogeny using large-subunit and small-subunit nuclear rRNA sequences. Mol. Phylogenet. Evol. 32, 25–38. doi: 10.1016/j.ympev.2003.12.016
Pedersen, R. B., Rapp, H. T., Thorseth, I. H., Lilley, M. D., Barriga, F., Baumberger, T., et al. (2010). Discovery of a black smoker vent field and vent fauna at the Arctic Mid-Ocean Ridge. Nat. Commun. 1:126. doi: 10.1038/ncomms1124
Rambaut, A. (2012). FigTree. Version 1.4.0. Available online at: http://tree.bio.ed.ac.uk/software/figtree (accessed October 1, 2019).
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Rouse, G. W., Goffredi, S. K., Johnson, S. B., and Vrijenhoek, R. C. (2011). Not whale-fall specialists, Osedax worms also consume fishbones. Biol. Lett. 7, 736–739. doi: 10.1098/rsbl.2011.0202
Rouse, G. W., Goffredi, S. K., Johnson, S. B., and Vrijenhoek, R. C. (2018). An inordinate fondness for Osedax (Siboglinidae: Annelida): fourteen new species of bone worms from California. Zootaxa 4377, 451–489. doi: 10.11646/zootaxa.4377.4.1
Rouse, G. W., Goffredi, S. K., and Vrijenhoek, R. C. (2004). Osedax: bone-eating marine worms with dwarf males. Science 305, 668–671. doi: 10.1126/science.1098650
Schander, C., Rapp, H. T., and Dahlgren, T. G. (2010). Osedax mucofloris (Polychaeta, Siboglinidae), a bone-eating marine worm new to Norway. Fauna Norv. 30, 5–8. doi: 10.5324/fn.v30i0.632
Schulze, A., and Halanych, K. M. (2003). Siboglinid evolution shaped by habitat preference and sulfide tolerance. Hydrobiologia 496, 199–205. doi: 10.1023/A:1026192715095
Shimabukuro, M., and Sumida, P. Y. G. (2019). Diversity of bone-eating Osedax worms on the deep Atlantic whale falls—bathymetric variation and inter-basin distributions. Mar. Biodivers. 49, 2587–2599. doi: 10.1007/s12526-019-00988-2
Smith, C. R., and Baco, A. R. (2003). Ecology of whale falls at the deep-sea floor. Oceanogr. Mar. Biol. 41, 311–354.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Struck, T., Hessling, R., and Purschke, G. (2002). The phylogenetic position of the Aeolosomatidae and Parergodrilidae, two enigmatic oligochaete-like taxa of the “Polychaeta”, based on molecular data from 18SrDNA sequences. J. Zool. Syst. Evolut. Res. 40, 155–163. doi: 10.1046/j.1439-0469.2002.00200.x
Struck, T. H., Purschke, G., and Halanych, K. M. (2005). A scaleless scale worm: molecular evidence for the phylogenetic placement of Pisione remota (Pisionidae: Annelida). Mar. Biol. Res. 1, 243–253. doi: 10.1080/17451000500261951
Taboada, S., Riesgo, A., Bas, M., Arnedo, M. A., Cristobo, J., Rouse, G. W., et al. (2015). Bone-eating worms spread: insights into shallow-water Osedax (Annelida, Siboglinidae) from Antarctic, Subantarctic, and Mediterranean waters. PLoS One 10:e0140341. doi: 10.1371/journal.pone.0140341
Tandberg, A. H. S., Olsen, B. R., and Rapp, H. T. (2017). Amphipods from the arctic hydrothermal vent field ≪Loki’s Castle≫, Norwegian Sea. Biodiver. J. 8, 553–554.
Vrijenhoek, R. C., Johnson, S. B., and Rouse, G. W. (2009). A remarkable diversity of bone-eating worms (Osedax; Siboglinidae; Annelida). BMC Biol. 7:74. doi: 10.1186/1741-7007-7-74
Xia, X., and Lemey, P. (2009). “Assessing substitution saturation with DAMBE,” in The Phylogenetic Handbook, eds P. Lemey, M. Salemi, and A. M. Vandamme (Cambridge: Cambridge University Press), 615–630. doi: 10.1017/cbo9780511819049.022
Keywords: deep sea, Osedax, organic falls, phylogeny, Siboglinidae
Citation: Eilertsen MH, Dahlgren TG and Rapp HT (2020) A New Species of Osedax (Siboglinidae: Annelida) From Colonization Experiments in the Arctic Deep Sea. Front. Mar. Sci. 7:443. doi: 10.3389/fmars.2020.00443
Received: 01 December 2019; Accepted: 20 May 2020;
Published: 16 June 2020.
Edited by:
Andrew Kvassnes Sweetman, Heriot-Watt University, United KingdomReviewed by:
Paulo Yukio Gomes Sumida, University of São Paulo, BrazilRobert C. Vrijenhoek, Monterey Bay Aquarium Research Institute (MBARI), United States
Copyright © 2020 Eilertsen, Dahlgren and Rapp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mari Heggernes Eilertsen, Mari.eilertsen@uib.no
†This article is dedicated to the memory of Professor Hans Tore Rapp who passed away on the 7th of March 2020
 Mari Heggernes Eilertsen
Mari Heggernes Eilertsen Thomas G. Dahlgren
Thomas G. Dahlgren Hans Tore Rapp
Hans Tore Rapp