- 1Department of Earth and Planetary Sciences, University of California, Davis, Davis, CA, United States
- 2Genome Center, University of California, Davis, Davis, CA, United States
Some oyster species precipitate a soft, friable form of calcite that is occluded within their shells, often referred to as chalk or chalky deposits. Because of the unusual microstructure of this shell feature, it has been proposed that chalk is the result of microbial involvement in the calcification process. Specifically, chalk has been hypothesized to be induced or influenced by microbial sulfate-reduction, and therefore chalk formation may not be under direct control of the oysters themselves. Specimens of the Pacific oyster, Magallana gigas, well-known for chalk deposition within shells, were grown in Bodega Harbor, Bodega Bay, California, and exposed monthly to treatments that altered the abundance of sulfate-reducing bacteria within oysters. The four treatment conditions evaluated included a control group (not exposed to any reagents), as well as oysters exposed to ciprofloxacin (a broad spectrum antibiotic), sodium molybdate (a compound known to inhibit bacterial sulfate reduction), and an inoculum of a sulfate-reducing bacterium isolated for this study. At the end of a 7 month growing period, specimens were culled and shells from treatment groups were assessed for chalk content by measuring bulk shell density and percentage of chalk in a cross sectional area. While analyses show that treatment conditions were successful with respect to altering abundances of sulfate-reducing bacteria in oysters, increasing SRB populations did not correlate to enhanced chalk expression in oyster shells. Interestingly, control oysters produced more chalk than the other treatment groups, according to both bulk shell density and percent chalk measured in cross section. Given that control oysters represent the wild type for chalk expression in shells, it is inferred that the decreased formation of chalk in the other groups was due to a perturbation of the microbiome in the oyster calcifying fluid. However, the methods used here only quantify the presence of sulfate-reducing bacteria in oysters, and therefore, additional work is necessary to evaluate the role of the microbiome in oyster calcification.
1. Introduction
The process of biomineralization is defined as the selective uptake of elements that are incorporated into a defined mineral structure under strict biological control (Dupraz et al., 2009). However, mineralization does not always occur under strict genetic (i.e., metazoan) control. Microbially-induced mineralization occurs when microbial metabolism creates a chemical environment that favors mineral precipitation (Dupraz et al., 2009). This type of mineralization is exemplified by the formation of stromatolites (e.g., Baumgartner et al., 2006) and marine peloids (Chafetz, 1986). Microbially-induced calcification may also take place when carbonate-precipitating microbes colonize a shell-secreting organism and enhance the ability of the host to build shell material. These microbes alter the internal chemical environment resulting in biomineralization that is not directly controlled by the metazoan host. Cases of microbially-induced mineralization in a metazoan host include the development of polychaete tubes (Guido et al., 2014), formation of rhodolith carbonate (Cavalcanti et al., 2014), precipitation of calcium carbonate in modern stromatoporid sponges (Jackson et al., 2010), growth of metal rich (pyrite or greigite) scales on a hydrothermal vent gastropod (Goffredi et al., 2004; Nakagawa et al., 2014), and specific structures found in bivalve mollusc shell carbonate (e.g., Braithwaite et al., 2000; Checa, 2000; Glover and Taylor, 2010).
Another potential case of microbially-induced mineralization in bivalve skeletons is chalky calcite found in the shells of certain oyster species (Chinzei and Seilacher, 1993; Chinzei, 1995; Vermeij, 2014). Chalky deposits, or chalk, is a form of porous, chalky calcite that may be irregularly interspersed throughout the shells of species (both fossil and extant) that display this feature. A number of hypotheses have been put forward for both the ecological function and mechanism of formation of this trait (e.g., Chinzei, 1995; Higuerea-Ruiz and Elorza, 2009; Vermeij, 2014; Checa et al., 2018). The oyster shell, including chalky deposits, have also been well-characterized in terms of structural and chemical properties (e.g., MacDonald et al., 2009; Lee et al., 2011b; Meng et al., 2018a,b), and recent efforts have also sought to evaluate the organic components of this biomineral structure (Dauphin et al., 2013; Mouchi et al., 2016).
The goal of the present study was to manipulate the populations of sulfate-reducing bacteria (SRB) in oysters to determine whether there was an associated effect on the expression of chalk in shells. For example, if SRB populations in oysters are supplemented, it is hypothesized that a higher amount of chalk will be expressed in the shell. SRB are of particular interest because they play a central role in carbonate precipitation in microbial mats via their ability to manipulate carbonate chemistry (Visscher et al., 2000; Baumgartner et al., 2006). It has also been hypothesized that chalk formation in oysters is the result of bacterial sulfate-reduction that occurs in the calcifying fluid, found in the extrapallial space between the mantle and the shell (Chinzei and Seilacher, 1993; Vermeij, 2014; Banker and Vermeij, 2018). Individuals of Magallana gigas (also known as Crassostrea gigas) were grown in Bodega Harbor, Bodega Bay, California for approximately 7 months, and were exposed periodically to treatments designed to alter SRB populations and activity within oysters. At the end of the experimental period, oysters were sacrificed and the amount of SRB in oysters was quantified, as was the amount of chalk in shells. Comparing SRB abundance to chalk expression in oysters across treatment, and assessing whether or not these variables correlate, will provide insight into whether or not SRB are involved in oyster chalk formation.
2. Methods
2.1. Study Organism and Environmental Setting
For this study, chalky deposit formation was investigated using M. gigas as a representative for organisms that display this shell feature. Oysters were grown out for a period of approximately 7 months in Bodega Harbor (38°19'20.6544" N, 123°2'48.6708"W) and exposed to experimental treatments in order to characterize the relationship between chalky deposit formation and SRB abundance in oysters in M. gigas. M. gigas is a fast growing species that displays ample chalky deposit growth after the juvenile stage. Shell structure for this oyster has also been relatively well-characterized (e.g., Higuerea-Ruiz and Elorza, 2009; MacDonald et al., 2009; Lee et al., 2011a,b; Dauphin et al., 2013; Mouchi et al., 2016; Checa et al., 2018), making it ideal for this work. Approximately 200 specimens of M. gigas of the same cohort (i.e., same age) were obtained for this experiment from Starbird Mariculture, Inc. facility in Bodega Harbor, Bodega Bay, California (38°19'41.56"N, 123°03'22.61"W). Oysters were grown in the Floating Commercial Upweller System (FLUPSY) at Starbird Mariculture Inc., which supplies the juvenile oysters at the facility with ample nutrients, until they reached a size of 4–5 mm (approximately 6 months old). At this time, oysters used for this experiment were separated and moved to mesh cages that were secured to a dock immediately adjacent to the FLUPSY, and thus remained in Bodega Harbor seawater for the duration of the experiment, except when oysters were brought to BML for treatments.
Bodega Harbor, within Bodega Bay, is a shallow portion of the bay that is largely protected from wave action by two parallel jetties located at its entrance. The harbor is almost completely flushed during each tidal cycle, and receives little freshwater input from April to November. However, the harbor may receive nutrient rich water during strong upwelling events (Olyarnik and Stachowicz, 2012). Bodega Bay seawater temperature during the course of the experiment (June 20, 2018 to January 24, 2019) was 13.3 ± 1.4°C as measured by Station BDXC1, a monitoring buoy owned and maintained by the UC Davis Bodega Marine Laboratory. Buoy data from Station BDXC1 was downloaded from the National Ocean Atmospheric Administration (https://www.ndbc.noaa.gov/), data from the experimental period was spliced together, and the average and standard deviation of temperature were calculated. Data for December 8, 2018 to December 11, 2018 were removed because temperature measurements for those days were erroneously recorded as 99°C. Because the Harbor receives little freshwater, temperature is generally within 5°C of the ocean temperature. Therefore, the temperature range experienced by the oysters during the experimental interval was approximately 8 to 18°C.
2.2. Oyster Cultivation and Experimental Treatments
After obtaining specimens from Starbird Mariculture, Inc. on June 20, 2018, oysters were split into 4 equal groups and placed in separate cages that were deployed in Bodega Harbor. Therefore, for each of the four treatments there was one cage that contained 50 individuals. On treatment dates, beginning July 6, 2018, all four cages were removed from the harbor and moved to the UC Davis Bodega Marine Laboratory (BML). Treatments occurred approximately 3–4 weeks apart, except for the final two dates which took place 5–6 weeks apart (Table 1). This was due to inclement weather, which prevented safe access to the outplant location, and resulted in delays in exposure to treatments. Oysters were then rinsed with seawater to remove sediment and pseudofeces, and a brush was used to remove epibionts from all individuals. Each treatment group was then placed in a 10-gallon bucket with filtered seawater (FSW). During the treatment period, oysters supplied a diet of 20,000 algal cells mL−1 (calculated using a hemocytometer), composed of Isochrysis galbana in each of the four separate buckets.

Table 1. Table summarizing dates on which oysters were removed from Bodega Harbor and transported to BML for treatment.
Experimental conditions were intended to either supplement SRB populations in oysters, or reduce the SRB microbial load, by adding specific reagents (or an inoculum) to the buckets of each of the four experimental groups. The first group, or the control, had no additional reagents added to the bath of FSW and algae. The second group was exposed to ciprofloxacin, a broad spectrum antibiotic that was selected for this study to reduce the microbial load of SRB within oysters. Although the hypothesis was that an antibiotic would decrease the prevalence of SRB in oysters, other research suggests that it may have the opposite effect by removing competitors of SRB, thus promoting SRB growth (Córdova-Kreylos and Scow, 2007). In this study, ciprofloxacin was added to FSW for a final concentration of 200 mg L−1, based on the protocol described in Córdova-Kreylos and Scow (2007). Because it was unclear how ciprofloxacin would affect the oyster microbiome, particularly SRB within the oysters, the third treatment group was exposed to sodium molybdate. This compound was selected because sodium molybdate, when applied at a concentration of 0.08 mM (12.8 mg L−1), can inhibit the activity of SRB (de Jesus et al., 2015). Therefore, sodium molybdate was added to the treatment bucket for the third group at a concentration of 12.8 mg L−1. The fourth and final treatment group was exposed to an inoculum of SRB (Desulfovibrio sp.), that was intended to supplement existing populations of SRB in oysters of this treatment group. A liquid culture of the inoculum was added to buckets at a concentration of approximately 20,000 cells mL−1. Details on SRB isolation, cultivation, and genome analysis are described below. The permits necessary for outplanting oysters and using the following protocols were obtained from the California Department of Fish and Wildlife.
2.3. Sulfate-Reducing Bacteria Isolation and Cultivation
The permit issued for this work specified that any bacterial cultures used here needed to be acquired from local sources. Therefore, sulfate-reducing bacteria were isolated and cultured in the lab specifically for this study. An intact seagrass plant (Zostera marina) was collected from near the BML within the harbor. Small pieces of root were vortexed in 1X Phosphate Buffered Saline (PBS), which was then plated onto Marine Broth 2216 (Difco) agar. Plates were incubated anaerobically in a GasPak™EZ Container System (Becton, Dickinson, and Company) at room temperature. Subsequent dilution streaking took place in a pure N2 atmosphere in a glove box. Liquid cultures were maintained in stoppered vials sparged with pure N2. Genomic DNA extraction was performed with a Wizard Genomic DNA Purification Kit (Promega) following the manufacturer's instructions. The isolate was identified by Sanger sequencing of the extracted DNA using the 27F (Lane, 1991) and 1391R (Turner et al., 1999) primers to isolate the entire 16S rRNA gene. When amplifying the bacterial 16S rRNA gene, PCR was performed with the following protocol: 95°C for 3 min, 40 cycles at 95°C for 15 s, 54°C for 30 s, 72°C for 1 min and 30 s, and a final extension at 72°C for 5 min (modified from Dunitz et al., 2015). The resulting 16S rRNA consensus sequence was queried using BLAST (Altschul et al., 1990) against the nr database at NCBI. The isolate, Desulfovibrio sp. strain UCD-KL4C, was >97% identical (95–97% query coverage, E = 0) to D. ferrireducens (NCBI Reference: 043581.1), D. lacusfryxellense (NCBI Reference: 115861.1), and D. frigidus (NCBI Reference: 043580.1).
2.4. Desulfovibrio sp. Strain UCD-KL4C Genome Sequencing
Full genome sequencing was performed by SNPsaurus (www.snpsaurus.com/). Ten nanograms of genomic DNA was used in a 1:10 reaction of the Nextera DNA Flex Library Prep protocol. Tagmented DNA was amplified with Phusion DNA polymerase and 12 PCR cycles with 1 min extension time. Samples were sequenced on an Illumina HiSeq4000 (University of Oregon GC3F) with paired-end 150 bp reads. Quality filtering, adaptor and PhiX removal were performed with BBMap (Bushnell, 2014), and A5-miseq was used to assemble contaminated reads with the metagenomic flag enabled (Coil et al., 2014). The assembly was then fed to metagenomic binners, MaxBin (Wu et al., 2014) and Metabat (Kang et al., 2015) targeting contigs 300 bp or longer. Bins were reconciled using DASTool (Sieber et al., 2018), which provided 2 clean bins. Completeness and contamination were evaluated using CheckM (Parks et al., 2015).
A phylogenetic tree of 16S rRNA sequences was produced following the protocol in Dunitz et al. (2015). First, the Ribosomal Database Project (RDP) was used to produce an alignment of the isolate used here, closely related sequences (i.e., members of Desulfovibrio), and an archean outgroup (Maidak et al., 1994). Tree-building was accomplished with FastTree using default parameters (Price et al., 2009), and was then visualized using Dendroscope (Huson and Scornavacca, 2012). FastANI was used to compute pairwise Average Nucleotide Identity (ANI) values, a whole genome similarity metric, between Desulfovibrio sp. strain UCD-KL4C and other closely related isolates (Jain et al., 2018). A 95% similarity cutoff was used to determine species as this has been proposed to be a valid classifier for delineating currently named prokaryotic species (Jain et al., 2018). Rapid Annotation using Subsystem Technology (RAST) was used to obtain functional annotations for Desulfovibrio sp. strain UCD-KL4C (Aziz et al., 2008; Overbeek et al., 2013; Brettin et al., 2015).
2.5. Sample Collection and Quantification of Sulfate-Reducing Bacteria
After 8 treatment cycles, or approximately 7 months, (section 3.2), 20 oysters from each of the four treatment groups were sacrificed on January 24, 2019. Oysters were grown out for 7 months to allow oysters to reach a size that provided ample shell material for subsequent analyses. This grow out period also allowed time for potential differences in chalk expression to develop between groups. Because chalk expression is associated with rapid growth and large size (Kirby, 2001), the 20 largest specimens, ranging from 7 to 10 cm (umbo to commissure), were selected from each treatment group to control for size as a factor affecting chalk expression. At the time cages were collected from Bodega Harbor, four seawater samples were collected in 20-mL scintillation vials immediately adjacent to the oyster cages to enable comparison of SRB populations between treatment groups and seawater.
Sani-Check SRB Kits (Biosan Laboratory, Inc.) were used to quantify the amount of SRB contained in oysters from each treatment group in order to determine whether or not experimental treatments (i.e., control, ciprofloxacin, sodium-molybdate, and the Desulfovibrio inoculum) had an affect on SRB populations within oysters. These kits were also applied to seawater samples taken from the oyster grow-out locality. The SRB Kits include tubes of culture media designed to promote the growth of sulfate-reducing bacteria, as well as applicator swabs that resemble small pipe cleaners. To use the kit, the applicator can be applied to any liquid or surface of interest, and is then inserted into the growth medium in sample tubes. Drops of mineral solution are added before the vial is closed and then incubated according to manufacturer instructions. When SRB present on the applicator release sulfide, it reacts with iron in the culture medium and forms iron sulfide, a distinct black precipitate. The amount of black sulfide produced, and the length of time it takes to form, is proportional to the amount of SRB in the sample, and can thus be used to approximate SRB counts from the original sample (Table 2). It is important to note that the Sani-Check SRB Kits used here take advantage of SRB activity in culture (i.e., test kits), that has been calibrated as a proxy for SRB abundance in the original sample. Given that the goal of this experiment was to assess whether or not SRB activity within oysters affects chalk formation, this is an appropriate method for quantifying SRB abundance in oysters.
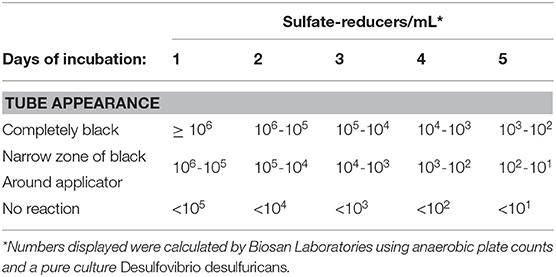
Table 2. Table reproduced from manufacturer guidelines on quantification of SRB in samples based on amount of black sulfide present after incubation periods ranging from 1 to 5 days.
Prior to use of the SRB Kits, oysters were rinsed and scrubbed with filtered seawater to remove sediment and epibionts, and were then opened dorsally at the umbo. The right valve was removed, leaving the soft tissue of the animal in the left, cupped valve. The test kit applicator was inserted between the mantle and the left valve, thus immersing it in the oyster calcifying fluid left in the extrapallial space. Test kits were applied to 10 of the 20 oysters collected from each treatment group, as well as to the 4 seawater samples collected in vials (Table 3).
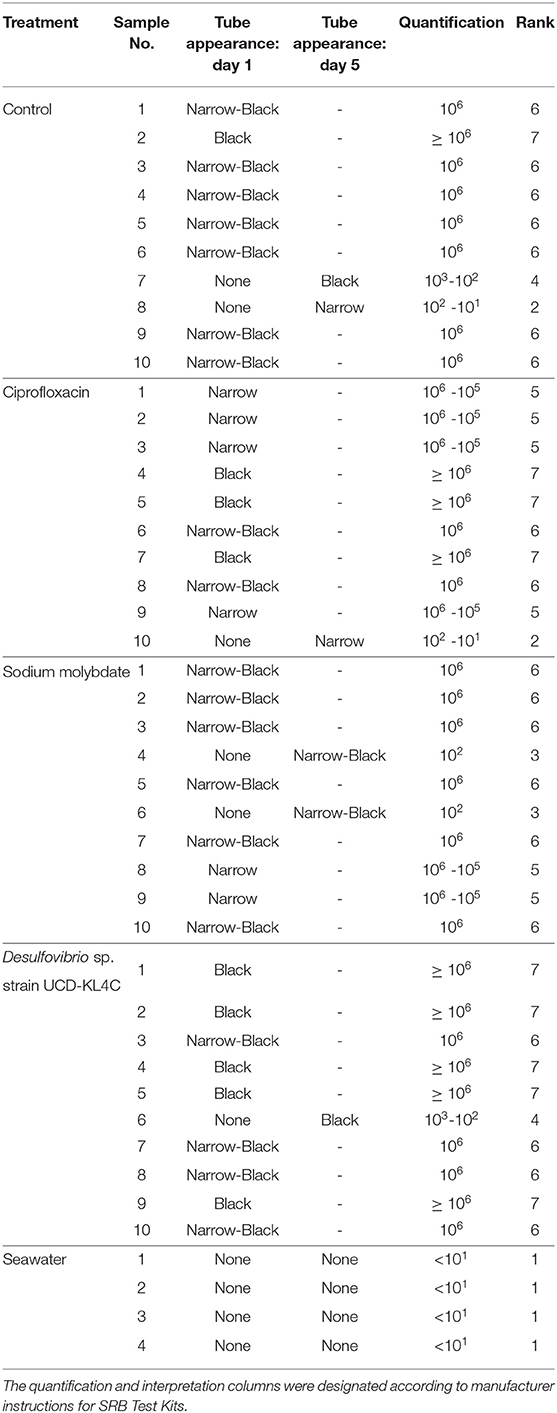
Table 3. Table detailing the results of SanBio SRB Test Kits applied to oyster and seawater samples.
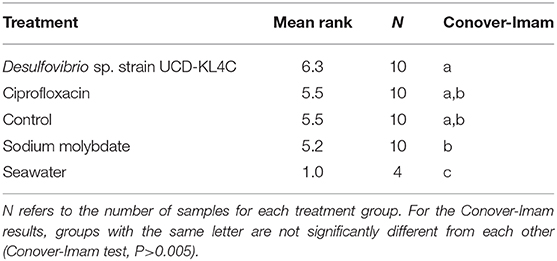
Because the SanBio test kits used to assess levels of SRB did not yield exact cell counts, treatment groups were compared statistically by assigning ranks to each sample based on the kit results (Table 3). The Kruskal–Wallis test was applied to rank data in R (Version 3.5.3) using the kruskal.test function, and significance was determined using a confidence level of 0.05. This test is typical for datasets that have one nominal and one ranked variable, as is the case with the SRB quantification here. The Conover-Imam test was used as a post-hoc analysis to perform pairwise comparisons of all treatment groups, and was implemented using conover.test in the conover.test package (version 1.1.5). A Bonferroni correction was used to reduce the rate of Type I error (i.e., the rate of incorrectly rejecting a true null hypothesis). The Bonferroni correction indicates significance for a post hoc test when the P value is less than 0.05/k, where k is the number of comparisons being made. For this study, the five treatment groups were compared, including the control, resulting in ten pairwise comparisons (k = 10). Thus a second confidence level, when P was less than 0.05/10(= 0.005), was used.
2.6. Quantification of Chalk in Oyster Shells
Chalk expression in oysters was assessed in two ways: First, the average density of shells (both valves) was measured for each of the 20 specimens taken from the four treatment groups (i.e., control, ciprofloxacin, sodium-molybdate, and the Desulfovibrio inocolum). This was done by taking the dry weight of both valves, then measuring water displacement when both shells were placed in a beaker.
The second method to quantify chalk expression in oysters was to measure the percent of cross-sectional area occupied by chalk. After mass and volume measurements were taken for each specimen to calculate shell density, the left valves from these same oysters (20 specimens in each of the four treatment groups) were cut along the axis of maximum growth using a BuehlerTM IsoMet® low-speed saw (Figure S1). The cut surface was ground using a 600 grit diamond wheel, and the shell-half was mounted onto a large slide (51 × 75 mm) using Hillquist AB thin section epoxy. Shells were then re-sectioned using BuehlerTM PetroThin thin section saw, resulting in mounted shell sections that were approximately 500–700 μm thick. Sections were digitized using a scanner and the proportion of chalk in each specimen was calculated as a percentage of cross-sectional area using Image J open-source software (https://imagej.nih.gov/ij/).
A one-way analysis of variance (ANOVA), was employed to compare the means of each treatment group for both shell density and percent chalk of cross-sectional area. This test is typical for datasets that have one nominal and one measurement variable, and evaluates whether means of the measurement variable are different between groups as determined by the nominal variable. The Tukey–Kramer test was used as a post-hoc test to make pairwise comparisons between each treatment group for both parameters. This test includes an inherent P-value adjustment to account for multiple comparisons. Mean shell volume was also compared between treatment groups, as a proxy for overall shell growth, using a one-way ANOVA and the Tukey–Kramer test for post-hoc analysis.
3. Results
3.1. Desulfovibrio sp. Strain UCD-KL4C Genome Analysis
A preliminary assembly determined that the genome of Desulfovibrio sp. strain UCD-KL4C was contaminated with reads from Bacillus. 4,853,674 raw pairs of reads were received. After quality filtering, adaptor and PhiX removal, 4,854,199 pairs of reads (bbduk) remained (Bushnell, 2014). Of these, 92% of reads belonged to Desulfovibrio sp. strain UCD-KL4C. A5-miseq (Coil et al., 2014) assembled the contaminated reads with the metagenomic flag enabled (this prevents overcorrecting the reads to a single isolate) into 288 contigs. Only 3.8% of contigs (11 out of the 288) were contamination attributed to Bacillus. MaxBin produced 2 bins with near perfect completeness and minimal contamination. Metabat produced 4 bins, with only 1 with 100% completeness. Two 16S rRNA sequences (of length > 1kb) were identified using ssu-align (Nawrocki, 2009) of which only one was assigned to the bins (the second one belonged to a contig that was not binned). Eleven contigs were binned into the bin of interest and included all the single copy marker genes used by CheckM and the 16S rRNA sequence, which blasted to Desulfovibrio lacusfryxellense at 97.85% identity over 99% of 1402 nucleotides.
A phylogenetic tree, using 16S rRNA sequences, was used to confirm the taxonomic identity of the isolate Desulfovibrio sp. strain UCD-KL4C. Desulfovibrio sp. strain UCD-KL4C fell within a well-supported (96% bootstrap support) clade that contained only Desulfovibrio isolates (Figure S2). However, the only members of this clade with designated taxonomy were Desulfovibrio ferrireducens and Desulfovibrio frigidus (Figure S2). Pairwise ANI values were computed to compare our isolate to these organisms (Table 4), though values fell below 95% similarity, indicating that Desulfovibrio sp. strain UCD-KL4C is potentially a separate species.
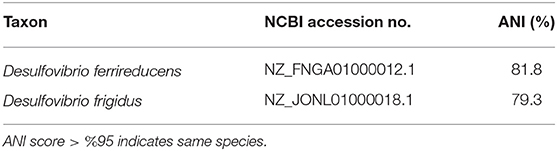
Table 4. Table summarizing Average Nucleotide Identity percent similarities between Desulfovibrio sp. strain UCD-KL4C.
Results from the RAST annotation of the Desulfovibrio sp. strain UCD-KL4C genome indicated that this isolate contains 3,627 protein coding sequences and 63 non-coding RNAs. This includes genes for sulfur metabolism, such as the sulfite reduction-associated complex DsrMKJOP, and several other dissimilatory sulfite reductase (Dsr) subunits.
3.2. Abundance of Sulfate-Reducing Bacteria in Oysters
Samples were visually assessed for black residue after 1 day of incubation. At this point, most test kits showed significant sulfide production, and these results were recorded (Table 3). Samples that did not produce black residue over the first 24-h were monitored for an additional 4 days, as per manufacturer instructions. At this point, most samples that did not have black residue after 1 day of incubation had developed some black sulfides. However, the seawater samples never developed any sulfide deposits. As per the manufacturer guidlines (Table 2), no sulfide production (no reaction) indicates that a given sample very few sulfate-reducers per milliliter, but does not mean that there were no sulfate-reducers present in the original sample. The designation “Narrow-Black” was applied to samples that showed more sulfide production than a narrow band of black around the applicator, but less black residue than could be described as completely black (Table 3). Frequency distributions of the number of samples from each treatment group based on the rank, or amount of SRB present, is shown in Figure 1.
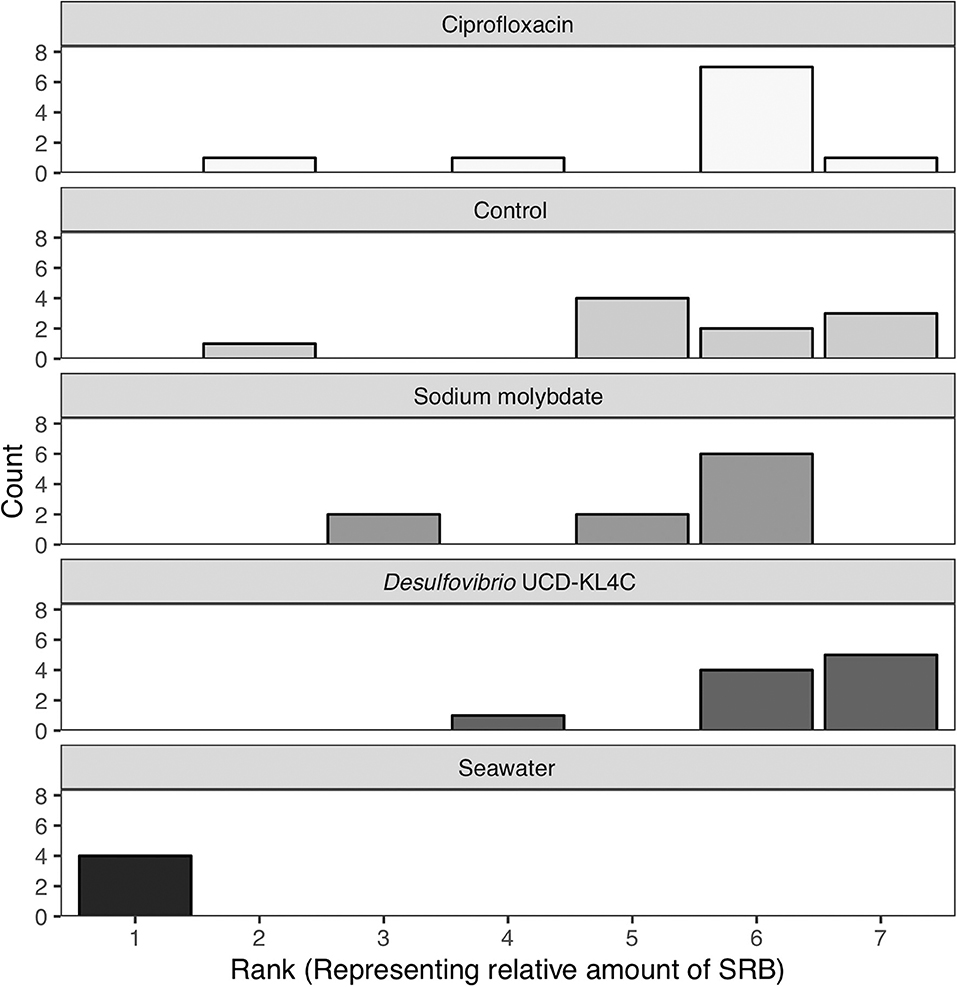
Figure 1. Counts of the number of samples from each treatment group with a certain rank representing the amount of sulfate-reducing bacteria (SRB) based on SanBio Test Kit results (Table 3). For example, all four seawater samples displayed sulfide formation after 5 days of incubation. According to the manufacturer guidelines (Table 3), no reaction indicates <101 sulfate-reducers per milliliter in the original seawater sample. Because this was the lowest concentration of sulfate-reducing bacteria for any sample, these four seawater samples were assigned a rank of 1 (Table 3). Therefore, the seawater sample panel (N = 4) shows a count of 4 at a rank of 1, indicating that all four samples from this group had a rank of 1 (Table 3). N = 10 for each oyster treatment group; N = 4 for seawater.
The Kruskal–Wallis test yielded a significant result, which indicates that there is a significant different in values among groups (H = 16.521, df = 4, P = 0.002394). Because the null hypothesis of the Kruskal–Wallis test was rejected, it was appropriate to use the Conover-Imam method as a post-hoc test to evaluate pairwise differences between groups (Conover and Imam, 1979; Conover, 1999). Results from the Conover-Imam test showed that there was a significant difference between each oyster treatment group when compared to seawater samples. Furthermore, the only two groups that were distinct from one another were those exposed to sodium molybdate and the inoculum of Desulfovibrio UCD-KL4C, otherwise all samples can be sorted into homogenous subsets (P > 0.005) (Table 5).
3.3. Shell Chalk Content
Tests performed on shell density measurements indicated that there was a significant difference between treatment groups for this parameter [one-way ANOVA, F(3, 76) = 4.636 P = 0.00496]. The Tukey–Kramer test, applied to discern pairwise differences in shell density, indicated that groups exposed to ciprofloxacin, sodium molybdate, and Desulfovibrio sp. strain UCD-KL4C, were not statistically different from one another, but that these three groups were all distinct from the control group (Figure 2).
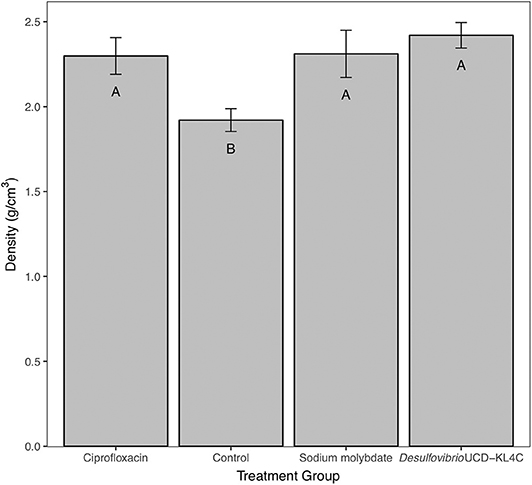
Figure 2. Bulk density of shells from M. gigas. Means ± one standard error are shown for the four treatment groups. Ciprofloxacin: = 2.29, SE = 0.11, N = 20; Control = 1.92, SE = 0.07, N = 20; Sodium molybdate: = 2.31, SE = 0.14, N = 20; Desulfovibrio sp. strain UCD-KL4C: = 2.42, SE = 0.08, N = 20. Means with the same letter are not significantly different from each other (Tukey–Kramer test, P > 0.05).
For percent of chalk in cross-section, treatment groups were not statistically different from one another [one-way ANOVA, F(3, 76) = 1.883, P = 0.14]. However, even though differences across groups is not statistically significant, means for percent chalk content in shells follow the same pattern for shell density. Results show that the control group had a higher average of chalk as a percentage of the cross sectional area as compared to all other treatment groups (Figure 3). Comparison of mean shell volume amongst groups also yielded a significant result [one-way ANOVA, F(3, 76) = 3.037, P = 0.0327], though post-hoc analysis indicated that only the control and Desulfovibrio sp. strain UCD-KL4C groups were distinct from one another (Figure 4). Overall, although results indicate treatment conditions successfully altered SRB counts in oysters, the abundance of SRB did not correlate to the amount of chalk in shells.
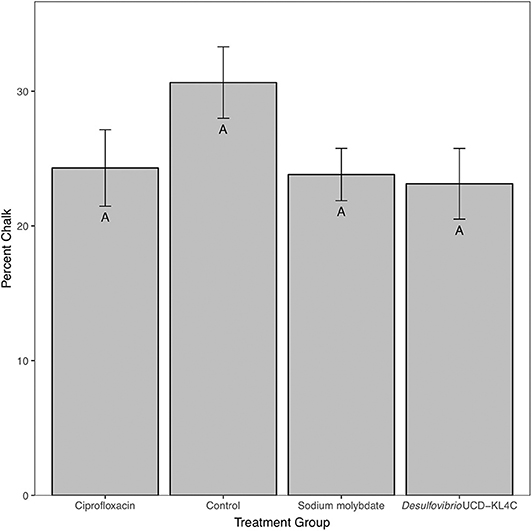
Figure 3. Percent chalk (as a percentage of the cross sectional area) of shells from M. gigas. Means ± one standard error are shown for the four treatment groups. Ciprofloxacin: = 24.30, SE = 2.84, N = 20; Control = 30.65, SE = 2.65, N = 20; Sodium molybdate: = 23.82, SE = 1.94, N = 20; Desulfovibrio sp. strain UCD-KL4C: = 23.12, SE = 2.62, N = 20. Means with the same letter are not significantly different from each other (Tukey–Kramer test, P > 0.05).
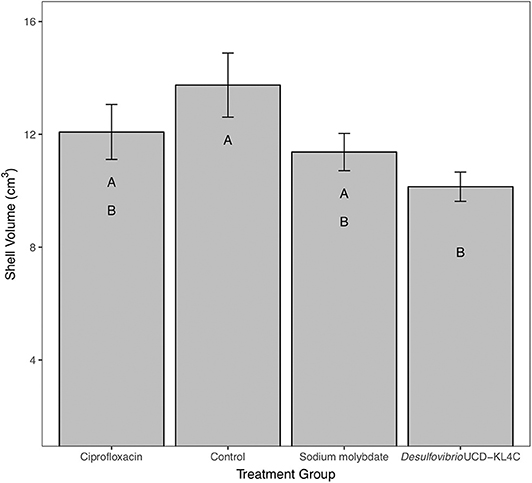
Figure 4. Shell volume of shells from M. gigas. Means ± one standard error are shown for the four treatment groups. Ciprofloxacin: = 12.08, SE = 0.97, N = 20; Control = 13.75, SE = 1.14, N = 20; Sodium molybdate: = 11.37, SE = 0.66, N = 20; Desulfovibrio sp. strain UCD-KL4C: = 10.14, SE = 0.52, N = 20. Means with the same letter are not significantly different from each other (Tukey–Kramer test, P > 0.05).
4. Discussion
4.1. Potential for Desulfovibrio sp. Strain UCD-KL4C to Contribute to Calcification
The primary goal of this study was to test the hypothesis that microbial activity, particularly bacterial sulfate-reduction, induces or influences the formation of chalky deposits found in oyster shells. Sulfate-reducing bacteria are of particular interest because they are known to contribute to calcium carbonate precipitation in microbial mats due to their ability to manipulate carbonate chemistry and promote mineral nucleation (Visscher et al., 2000; Baumgartner et al., 2006). Depending on the organic compound consumed for sulfate reduction, SRB are able to raise the saturation index of calcium carbonate (Dupraz and Visscher, 2005), increase local alkalinity (Gallagher et al., 2012), and create active sites that promote crystal growth by altering organic substrates (Dupraz et al., 2009; Bontognali et al., 2014). The proposed relationship between oysters and SRB is similar: SRB activity in the calcifying fluid will affect oyster shell formation by influencing carbonate chemistry, thus promoting rapid shell growth in the form of chalky deposits.
Although further analyses to precisely resolve the taxonomy of the Desulfovibrio sp. strain UCD-KL4C are beyond the scope of this study, phylogenetic analysis reveals that this isolate is definitively a member of the genus Desulfovibrio (Figure S2). Furthermore, genome annotation indicates that the isolate has genes that are integral to bacterial sulfate-reduction, including several Dsr subunits and the DsrMKJOP complex, which catalyze the reduction of sulfite to sulfide (Pires et al., 2006; Grein et al., 2013; Fike et al., 2016). In fact, the presence of Dsr genes are used to assess microspatial distributions of SRB in microbial mats because they are diagnostic of microbes that can perform sulfate-reduction (Minz et al., 1999; Dar et al., 2007; Petrisor et al., 2014).
4.2. Effect of Treatments on Abundance of Sulfate-Reducing Bacteria
Overall, the treatments selected for this study to manipulate internal oyster populations of SRB were successful. Although not all sample pairs revealed significant statistical differences, mean ranks (representing relative amounts of SRB across samples) showed trends that align with expectations of how treatment conditions would affect the populations of sulfate-reducing bacteria. The group of oysters inoculated with the Desulfovibrio sp. strain UCD-KL4C had the highest mean rank. This was expected because test kits assayed for sulfate-reduction, which increase when SRB populations are supplemented. In contrast, amounts of sulfate-reducing bacteria in the group exposed to sodium molybdate, an agent that inhibits sulfate-reduction (de Jesus et al., 2015), had the lowest mean rank value (Table 5). Given that oysters in the control were not exposed to any reagents intended to alter SRB populations, one would expect that the control would have intermediate population sizes of SRB, which is what was observed here. Interestingly, oysters from the control and the group exposed to ciprofloxacin had identical mean ranks for the number of SRB, indicating that the antibiotic had little or no effect on SRB. It is possible that ciprofloxacin only affected other bacterial taxa, as was found in Córdova-Kreylos and Scow (2007) when this antibiotic applied to salt marsh sediments, though characterization of the whole oyster microbiome would be required to confirm whether this was the case in the present study.
It is important to note that not all trends are statistically supported. However, the lack of statistical significance may be a function of the time elapsed between the final treatment that occurred on December 11, 2018, and the date when samples for SRB test kits were taken on January 24, 2019. In another study that investigated bacterial uptake dynamics in bivalves, results showed that retention of Escherichia coli in the oyster Ostrea edulis was primarily driven by the concentration of E. coli in seawater, as well as by temperature and salinity (Jozić et al., 2012). Furthermore, research on the uptake of Vibrio vulnificus in oysters has indicated that although oysters may be quick to accumulate this pathogen, it is also rapidly depurated when oysters are replaced in pathogen free water (Froelich and Noble, 2014). This effect is attributed to the endogenous oyster microbiome inhibiting the establishment of external bacteria (Froelich and Noble, 2014). All oysters used in this study were brought to the Bodega Marine Laboratory each month and exposed to reagents over a discrete period of time (3 h) before being returned to Bodega Harbor to grow in natural conditions. Prolonged exposure to the ambient environment in Bodega Harbor likely provided time for SRB populations in oysters to return to pre-treatment levels, resulting in a lack of significance for statistical tests. Last, even though there were very few SRB in seawater according to test kits (Table 3), the ambient environment is still the source of oyster microbial communities (Murchelano and Bishop, 1969; Kueh and Chan, 1985; Beleneva et al., 2003; Lokmer et al., 2016; Pierce et al., 2016; Banker and Vermeij, 2018; Pierce and Ward, 2018).
4.3. Sulfate-Reducing Bacteria Influence on Chalk Expression
If sulfate-reducing bacteria are able to induce or enhance the precipitation of chalky material in shells (Vermeij, 2014), then oysters with more SRB should exhibit increased chalk expression. However, analyses comparing shell parameters (i.e., density and percent chalk) amongst treatment groups revealed that oysters from the control group precipitated more chalk than all other groups. Moreover, comparisons of overall shell volume between treatment group indicates that control oysters experienced overall more shell growth over the course of the experiment. It makes sense that the group with the most chalk, the control group, also displays the most overall shell growth, as enhanced chalk expression has been linked to fast growth rates for oysters in previous studies (Kirby, 2000, 2001).
Although the pattern of enhanced chalk formation and overall growth in the control group is only statistically significant for shell density, and not for percent chalk or volume measurements, the fact that all analyses yielded the same pattern indicates that this is a true signal. The lack of the significance between treatment groups for percent chalk is likely due to the fact that chalk is not perfectly evenly distributed throughout the shell, which may have skewed the results of this measurement slightly. Overall, the fact that there is no correlation between the abundance of SRB in oysters and the amount of chalk in shells illustrates that SRB activity in oysters is not responsible for inducing the formation of chalky deposits in the Pacific oyster.
4.4. Microbiome and Shell Formation
Although results here show that sulfate-reducing bacteria do not directly cause chalk precipitation, the pattern of chalk expression across oyster groups suggests that treatments did affect shell formation. For example, even though there were differences in SRB populations between the oysters exposed to sodium molybdate vs. those exposed to Desulfovibrio sp. strain UCD-KL4C, these two groups expressed similar amounts of chalk in their shells. Furthermore, even though the control and the group exposed to ciprofloxacin had very similar populations of SRB, they exhibited varying amounts of chalk in their shells. Overall, the fact that all treatment groups produced less chalk compared to control-group oysters, and that the control group showed overall more growth according to volume measurements, indicates that treatment conditions did have an effect on shell formation.
We hypothesize that changes to the microbiome, that could not be detected with the SRB test kit, induced changes to the microbiome within oysters that ultimately affected chalk formation and overall shell growth. Mounting evidence indicates that the relationship between metazoa and their resident microbial communities are central to host health and functioning (e.g., Bourne et al., 2016; Gould et al., 2018; Pierce and Ward, 2018; Vezzulli et al., 2018; van Oppen and Blackall, 2019). This is also true for marine taxa, and recent research shows that the microbiome can affect disease resistance of seaweeds (Longford et al., 2019), sponges (Slaby et al., 2019), as well as resistance to symbiont loss and bleaching in corals (Bourne et al., 2013; Rosado et al., 2019). Additional work shows that the coral skeletal microbiome is highly structured (Marcelino et al., 2018) and that disruption to the microbiome may also result in reduced calcification rates in corals (Grottoli et al., 2018); moreover, microbial communities have also been shown to be important for calcification of rhodoliths (Cavalcanti et al., 2018). Taken together, this evidence supports the idea that the microbiome is critical for skeletal formation in marine taxa. Therefore, it is proposed that oysters in this study were similarly affected by a dysbiosis caused by treatment conditions in all but the control group, which resulted in reduced shell growth for non-control oysters. However, evaluation of the whole microbiome is required to substantiate this hypothesis. Because the data collected here only address SRB abundance, further work utilizing next generation sequencing to evaluate microbial community composition overall will be central to resolving questions regarding the relationship between marine taxa, their microbiota, and skeletal formation. This is particularly true for non-coral groups that have received relatively less attention in regards to how the microbiome affects calcification.
5. Conclusion
Different types of treatments (an antimicrobial reagent, sodium molybdate, and a biological agent, the Desulfovibrio culture) affected abundances of sulfate-reducing bacteria within oysters examined here. This indicates that internal microbial flora are affected not only by the ambient external environment, but may be modified by targeted mechanisms as well. This in turn may have important implications for aquaculture, particularly for engineering the oyster microbiome to promote resilience to disease and other stressors that are expected to increase under future climate scenarios. Even though treatments were successful, abundance of SRB did not correlate with chalk expression. These results suggest that SRB alone are not responsible for chalk formation. However, all treatment groups displayed less chalk formation than the control oysters. It is hypothesize that this is caused by other changes in the oyster microbiome, induced by treatment conditions, that could not be detected with the methods used here. Mounting evidence indicates that the microbiome is important for healthy organism functioning, including calcification, in various marine taxa. Thus, treatments used here disrupted the wild-type microbiome, which ultimately affected organism health and shell formation in the form of chalk expression. Future work should utilize next generation sequencing to assess the whole microbiome and community composition to more precisely characterize how the microbiome affects organismal processes, such as shell formation.
Data Availability Statement
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession VCNC00000000. The version described in this paper is version VCNC01000000.
Author Contributions
RB conceived, designed, and conducted the experiment and collection of material, performed analysis and interpretation of data, and drafted the manuscript. DC consulted on experimental design, oversaw culturing of the bacterial strain used in this work, and reviewed the article for intellectual content.
Funding
Funding for this work came from the Melbourne R. Carriker Student Research Award from the American Malacological Society, the Ernest E. Hill Fellowship for Environmental Sciences, and the Durrell Award for Graduate Research via the Earth and Planetary Sciences Department at UC Davis.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
First, we would like to thank Dr. Jonathan Eisen for providing laboratory space and materials for this work. Thanks also to Guillaume Jospin for his expertise and assistance performing bioinformatic analyses. We would also like to thank Dr. Geerat Vermeij for providing feedback on earlier iterations of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00407/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The rast server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Banker, R. M., and Vermeij, G. J. (2018). Oyster microbial communities and implications for chalky deposit formation. Hydrobiologia 816, 121–135. doi: 10.1007/s10750-018-3569-0
Baumgartner, L., Reid, R., Dupraz, C., Decho, A., Buckley, D., Spear, J., et al. (2006). Sulfate reducing bacteria in microbial mats: Changing paradigms, new discoveries. Sed. Geol. 185, 131–145. doi: 10.1016/j.sedgeo.2005.12.008
Beleneva, I. A., Zhukova, N. V., and Maslennikova, E. F. (2003). Comparative study of microbial communities from cultured and natural populations of the mussel Mytilus trossulus in Peter the Great Bay. Microbiology 72, 528–535. doi: 10.1023/A:1025005025620
Bontognali, T. R. R., McKenzie, J. A., Warthamm, R. J., and Vasconcelos, C. (2014). Microbiall influenced formation of Mg-calcite and ca-dolomite in the presence of exopolymeric substances produced by sulphate-reducing bacteria. Terra Nova 26, 72–77. doi: 10.1111/ter.12072
Bourne, D. G., Dennis, P. G., Uthicke, S., Soo, R. M., Tyson, G. W., and Webster, N. (2013). Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J. 7, 1751–7362. doi: 10.1038/ismej.2013.71
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Ann. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Braithwaite, C. J. R., Taylor, J. D., and Glover, E. A. (2000). Marine carbonate cements, biofilms, biomineralization, and skeletogenesis: some Bivalves do it all. J. Sed. Res. 70, 1129–1138. doi: 10.1306/091699701129
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). Rasttk: a modular and extensible implementation of the rast algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Cavalcanti, G. S., Gregoracci, G. B., dos Santos, E. O., Silveira, C. B., Meirelles, P. M., Longo, L., et al. (2014). Physiologic and metagenomic attributes of the rhodoliths forming the largest CaCO3 bed in the South Atlantic Ocean. ISME J. 8, 52–62. doi: 10.1038/ismej.2013.133
Cavalcanti, G. S., Shukla, P., Morris, M., Ribeiro, B., Foley, M., Doane, M. P., et al. (2018). Rhodoliths holobionts in a changing ocean: host-microbes interactions mediate coralline algae resilience under ocean acidification. BMC Genomics 19:701. doi: 10.1186/s12864-018-5064-4
Chafetz, H. (1986). Marine peloids: a product of bacterially induced precipitation of calcite. SEPM J. Sed. Res. 56, 812–817. doi: 10.1306/212F8A58-2B24-11D7-8648000102C1865D
Checa, A. G. (2000). Remote biomineralization in divaricate ribs of Strigilla and Solecurtus (Tellinoidea: Bivalvia). J. Mollus Stud. 66, 457–466. doi: 10.1093/mollus/66.4.457
Checa, A. G., Harper, E. M., and González-Segura, A. (2018). Structure and crystallography of foliated and chalk shell microstructures of the oyster Magallana: the same materials grown under different conditions. Sci. Rep. 8, 1–12. doi: 10.1038/s41598-018-25923-6
Chinzei, K. (1995). Adaptive significant of lightweight shell structure in soft bottom oysters. N. J. Geol. Palaont. Abh. 195, 217–227. doi: 10.1127/njgpa/195/1995/217
Chinzei, K., and Seilacher, A. (1993). Remote Biomineralization I: fill skeletons in vesicular oyster shells. N. J. Geol. Palaont. Abh. 190, 349–361.
Coil, D., Jospin, G., and Darling, A. E. (2014). A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31, 587–589. doi: 10.1093/bioinformatics/btu661
Conover, W. J., and Imam, R. L. (1979). On Multiple-Comparisons Procedures. Technical Report LA-7677-MS, Los Alamos Scientific Laboratory.
Córdova-Kreylos, A. L., and Scow, K. M. (2007). Effects of ciprofloxacin on salt marsh sediment microbial communities. ISME J. 1, 585–595. doi: 10.1038/ismej.2007.71
Dar, S. A., Yao, L., van Dongen, U., Kuenen, J. G., and Muyzer, G. (2007). Analysis of diversity and activity of sulfate-reducing bacterial communities in sulfidogenic bioreactors using 16s rRNA and DSRB genes as molecular markers. Appl. Environ. Microbiol. 73, 594–604. doi: 10.1128/AEM.01875-06
Dauphin, Y., Ball, A. D., Castillo-Michel, H., Chevallard, C., Cuif, J.-P., Farre, B., et al. (2013). In situ distribution and characterization of the organic content of the oyster shell Crassostrea gigas (Mollusca, Bivalvia). Micron 44, 373–83. doi: 10.1016/j.micron.2012.09.002
de Jesus, E., R P Lima, L., Bernardez, L., and Almeida, P. (2015). Inhibition of microbial sulfate reduction by molybdate. Braz. J. Petrol. Gas 9:95. doi: 10.5419/bjpg2015-0010
Dunitz, M. I., Lang, J. M., Jospin, G., Darling, A. E., Eisen, J. A., and Coil, D. A. (2015). Swabs to genomes: a comprehensive workflow. PeerJ 3:e960. doi: 10.7717/peerj.960
Dupraz, C., Reid, R. P., Braissant, O., Decho, A. W., Norman, R. S., and Visscher, P. T. (2009). Processes of carbonate precipitation in modern microbial mats. Earth Sci. Rev. 96, 141–162. doi: 10.1016/j.earscirev.2008.10.005
Dupraz, C., and Visscher, P. T. (2005). Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 13, 429–438. doi: 10.1016/j.tim.2005.07.008
Fike, D. A., Bradley, A. S., and Leavitt, W. D. (2016). “Geomicrobiology of sulfur,” in Ehrlich's Geomicrobiology, eds H. L. Ehrlich, D. K. Newman, A. Kappler (Boca Raton, FL Taylor Francis Group), 479–515. doi: 10.1201/b19121-21
Froelich, B. A., and Noble, R. T. (2014). Factors affecting the uptake and retention of Vibrio vulnificus in oysters. Appl. Environ. Microbiol. 80, 7454–7459. doi: 10.1128/AEM.02042-14
Gallagher, K. L., Kading, T. J., Braissant, O., Dupraz, C., and Visscher, P. T. (2012). Inside the alkalinity engine: the role of electron donors in the organomineralization potential of sulfate-reducing bacteria. Geobiology 10, 518–30. doi: 10.1111/j.1472-4669.2012.00342.x
Glover, E. A., and Taylor, J. D. (2010). Needles and pins: acicular crystalline periostracal calcification in venerid bivalves (Bivalvia: Veneridae). J. Mollus Stud. 76, 157–179. doi: 10.1093/mollus/eyp054
Goffredi, S. K., Warén, A., Orphan, V. J., Dover, C. V., and Vrijenhoek, R. C. (2004). Novel forms of structural integration between microbes and a hydrothermal vent gastropod from the Indian Ocean. Appl. Environ. Microbiol. 70, 3082–3090. doi: 10.1128/AEM.70.5.3082-3090.2004
Gould, A. L., Zhang, V., Lamberti, L., Jones, E. W., Obadia, B., Korasidis, N., et al. (2018). Microbiome interactions shape host fitness. Proc. Natl. Acad. Sci. U.S.A. 115, E11951–E11960. doi: 10.1073/pnas.1809349115
Grein, F., Ramos, A. R., Venceslau, S. S., and Pereira, I. A. (2013). Unifying concepts in anaerobic respiration: insights from dissimilatory sulfur metabolism. Biochim. Biophys. Acta Bioenerg. 1827, 145–160. doi: 10.1016/j.bbabio.2012.09.001
Grottoli, A. G., Dalcin Martins, P., Wilkins, M. J., Johnston, M. D., Warner, M. E., Cai, W.-J., et al. (2018). Coral physiology and microbiome dynamics under combined warming and ocean acidification. PLoS ONE 13:e0191156. doi: 10.1371/journal.pone.0191156
Guido, A., Mastandrea, A., Rosso, A., Sanfilippo, R., Tosti, F., Riding, R., et al. (2014). Commensal symbiosis between agglutinated polychaetes and sulfate-reducing bacteria. Geobiology 12, 265–275. doi: 10.1111/gbi.12084
Higuerea-Ruiz, R., and Elorza, J. (2009). Estuarine, Coastal and Shelf Science Biometric, microstructural, and high-resolution trace element studies in Crassostrea gigas of Cantabria (Bay of Biscay, Spain): anthropogenic and seasonal influences. Estuar. Coast. Shelf 82, 201–213. doi: 10.1016/j.ecss.2009.01.001
Huson, D. H., and Scornavacca, C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61, 1061–1067. doi: 10.1093/sysbio/sys062
Jackson, D. J., Thiel, V., and Wörheide, G. (2010). An evolutionary fast-track to biocalcification. Geobiology 8, 191–196. doi: 10.1111/j.1472-4669.2010.00236.x
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90k prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jozić, S., Šolić, M., and Krstulović, N. (2012). The accumulation of the indicator bacteria Escherichia coli in mussels (Mytilus galloprovincialis) and oysters (Ostrea edulis) under experimental conditions. Acta Adriatica 53, 353–362.
Kang, D. D., Froula, J., Egan, R., and Wang, Z. (2015). Metabat, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165. doi: 10.7717/peerj.1165
Kirby, M. X. (2000). Paleoecological differences between tertiary and quaternary crassostrea oysters, as revealed by stable isotope sclerochronology. Palaios 15, 132–141. doi: 10.1669/0883-1351(2000)015<0132:PDBTAQ>2.0.CO;2
Kirby, M. X. (2001). Differences in growth rate and environment between tertiary and quaternary crassostrea oysters. Paleobiology 27, 84–103. doi: 10.1666/0094-8373(2001)027<0084:DIGRAE>2.0.CO;2
Kueh, C. S., and Chan, K. Y. (1985). Bacteria in bivalve shellfish with special reference to the oyster. J. Appl. Bacteriol. 59, 41–47. doi: 10.1111/j.1365-2672.1985.tb01773.x
Lane, D. (1991). “Nucleic acid techniques in bacterial systematics,” in 16S/23S rRNA Sequencing, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley and Sons), 115–175.
Lee, S.-W., Jang, Y.-N., and Kim, J.-C. (2011a). “Characteristics of the aragonitic layer in adult oyster shells, Crassostrea gigas: structural study of Myostracum including the adductor muscle scar,” in Evidence-Based Complementary and Alternative Medicine (eCAM). doi: 10.1155/2011/742963
Lee, S.-W., Jang, Y.-N., Ryu, K.-W., Chae, S.-C., Lee, Y.-H., and Jeon, C.-W. (2011b). Mechanical characteristics and morphological effect of complex crossed structure in biomaterials: fracture mechanics and microstructure of chalky layer in oyster shell. Micron 42, 60–70. doi: 10.1016/j.micron.2010.08.001
Lokmer, A., Kuenzel, S., Baines, J. F., and Wegner, K. M. (2016). The role of tissue-specific microbiota in initial establishment success of Pacific oysters. Environ. Microbiol. 18, 970–987. doi: 10.1111/1462-2920.13163
Longford, S. R., Campbell, A. H., Nielsen, S., Case, R. J., Kjelleberg, S., and Steinberg, P. D. (2019). Interactions within the microbiome alter microbial interactions with host chemical defences and affect disease in a marine holobiont. Sci. Rep. 9:1363. doi: 10.1038/s41598-018-37062-z
MacDonald, J., Freer, A., and Cusack, M. (2009). Alignment of crystallographic c -axis throughout the four distinct microstructural layers of the oyster Crassostrea gigas. Cryst. Growth Des. 10, 1243–1246. doi: 10.1021/cg901263p
Maidak, B. L., Larsen, N., McCaughey, M. J., Overbeek, R., Olsen, G. J., Fogel, K., et al. (1994). The ribosomal database project. Nucleic Acids Res. 22, 3485–3487. doi: 10.1093/nar/22.17.3485
Marcelino, V. R., van Oppen, M. J., and Verbruggen, H. (2018). Highly structured prokaryote communities exist within the skeleton of coral colonies. ISME J. 12, 300–303. doi: 10.1038/ismej.2017.164
Meng, Y., Fitzer, S., Chung, P., Li, C., Thiyagarajan, V., and Cusack, M. (2018a). Crystallographic interdigitation in oyster shell folia enhances material strength. Cryst. Growth Des. 18, 3753–3761. doi: 10.1021/acs.cgd.7b01481
Meng, Y., Guo, Z., Fitzer, S. C., Upadhyay, A., Chan, V., Li, C., et al. (2018b). Ocean acidification reduces hardness and stiffness of the Portuguese oyster shell with impaired microstructure: a hierarchical analysis. Biogeosciences 15, 6833–6846. doi: 10.5194/bg-15-6833-2018
Minz, D., Flax, J. L., Green, S. J., Muyzer, G., Cohen, Y., Wagner, M., et al. (1999). Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65, 4666–4671. doi: 10.1128/AEM.65.10.4666-4671.1999
Mouchi, V., Lartaud, F., Guichard, N., Immel, F., de Rafélis, M., Broussard, C., et al. (2016). Chalky versus foliated: a discriminant immunogold labelling of shell microstructures in the edible oyster Crassostrea gigas. Mar. Biol. 163, 1–15. doi: 10.1007/s00227-016-3040-6
Murchelano, R., and Bishop, J. (1969). Bacteriological Study of Laboratory-Reared Juvenile American Oysters (Crassostrea virginica). J. Invertebr. Pathol. 14, 321–327. doi: 10.1016/0022-2011(69)90158-X
Nakagawa, S., Shimamura, S., Takaki, Y., Suzuki, Y., Murakami, S.-I., Watanabe, T., et al. (2014). Allying with armored snails: the complete genome of gammaproteobacterial endosymbiont. ISME J. 8, 40–51. doi: 10.1038/ismej.2013.131
Nawrocki, E. P. (2009). Structural RNA homology search and alignment using covariance models (Ph.D. Thesis). Washington University in Saint Louis School of Medicine, St. Louis, MO.
Olyarnik, S. V., and Stachowicz, J. J. (2012). Multi-year study of the effects of Ulva sp. blooms on eelgrass Zostera marina. Mar. Ecol. Prog. Ser. 468, 107–117. doi: 10.3354/meps09973
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2013). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). Checkm: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Petrisor, A. I., Szyjka, S., Kawaguchi, T., Visscher, P. T., Norman, R. S., and Decho, A. W. (2014). Changing microspatial patterns of sulfate-reducing microorganisms (SRM) during cycling of marine stromatolite mats. Int. J. Mol. Sci. 15, 850–877. doi: 10.3390/ijms15010850
Pierce, M. L., and Ward, E. J. (2018). Microbial ecology of the bivalvia, with an emphasis on the family ostreidae. J. Shellfish Res. 37, 793–806. doi: 10.2983/035.037.0410
Pierce, M. L., Ward, J. E., Holohan, B. A., Zhao, X., and Hicks, R. E. (2016). The influence of site and season on the gut and pallial fluid microbial communities of the eastern oyster, Crassostrea virginica (Bivalvia, Ostreidae): community-level physiological profiling and genetic structure. Hydrobiologia 765, 97–113. doi: 10.1007/s10750-015-2405-z
Pires, R. H., Venceslau, S. S., Morais, F., Teixeira, M., Xavier, A. V., and Pereira, I. A. C. (2006). Characterization of the Desulfovibrio desulfuricans ATCC 27774 DsrMKJOP complexa membrane-bound redox complex involved in the sulfate respiratory pathway. Biochemistry 45, 249–262. doi: 10.1021/bi0515265
Price, M. N., Dehal, P. S., and Arkin, A. P. (2009). FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 26, 1641–1650. doi: 10.1093/molbev/msp077
Rosado, P. M., Leite, D. C. A., Duarte, G. A. S., Chaloub, R. M., Jospin, G., Nunes da Rocha, U., et al. (2019). Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J. 13, 921–936. doi: 10.1038/s41396-018-0323-6
Sieber, C. M. K., Probst, A. J., Sharrar, A., Thomas, B. C., Hess, M., Tringe, S. G., et al. (2018). Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 3, 836–843. doi: 10.1038/s41564-018-0171-1
Slaby, B. M., Franke, A., Rix, L., Pita, L., Bayer, K., Jahn, M. T., et al. (2019). “Symbiotic microbiomes of coral reefs sponges and corals,” in Marine Sponge Holobionts in Health and Disease, ed S. Li (Zhiyong: Springer), 29–41. doi: 10.1007/978-94-024-1612-1_7
Turner, S., Pryer, K., Miao, V., and Palmer, J. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46, 327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x
van Oppen, M. J. H., and Blackall, L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Vermeij, G. J. (2014). The oyster enigma variations: a hypothesis of microbial calcification. Paleobiology 40, 1–13. doi: 10.1666/13002
Vezzulli, L., Stagnaro, L., Grande, C., Tassistro, G., Canesi, L., and Pruzzo, C. (2018). Comparative 16srdna gene-based microbiota profiles of the pacific oyster (Crassostrea gigas) and the Mediterranean mussel (Mytilus galloprovincialis) from a shellfish farm (Ligurian sea, Italy). Microb. Ecol. 75, 495–504. doi: 10.1007/s00248-017-1051-6
Visscher, P. T., Reid, R. P., and Bebout, B. M. (2000). Microscale observations of sulfate reduction: correlation of microbial activity with lithified micritic laminae in modern marine stromatolites. Geology 28, 919–923. doi: 10.1130/0091-7613(2000)28<919:MOOSRC>2.0.CO;2
Keywords: pacific oyster, chalk, sulfate-reducing bacteria, calcification, shell formation
Citation: Banker RMW and Coil D (2020) Inoculation With Desulfovibrio sp. Does Not Enhance Chalk Formation in the Pacific Oyster. Front. Mar. Sci. 7:407. doi: 10.3389/fmars.2020.00407
Received: 05 December 2020; Accepted: 11 May 2020;
Published: 10 June 2020.
Edited by:
Punyasloke Bhadury, Indian Institute of Science Education and Research Kolkata, IndiaReviewed by:
Oumar Sadio, Institut de Recherche pour le Développement, SenegalVengatesen Thiyagarajan Rajan, The University of Hong Kong, Hong Kong
Copyright © 2020 Banker and Coil. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Roxanne M. W. Banker, cmJhbmtlciYjeDAwMDQwO3VjYXZpcy5lZHU=
 Roxanne M. W. Banker
Roxanne M. W. Banker David Coil2
David Coil2