
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 22 April 2020
Sec. Marine Megafauna
Volume 7 - 2020 | https://doi.org/10.3389/fmars.2020.00253
This article is part of the Research Topic Advances in Understanding Sea Turtle Use of the Gulf of Mexico View all 17 articles
Reptile growth is influenced by many ecological processes that can cumulatively give rise to divergent somatic growth rates within spatially structured populations. As somatic growth variation can strongly influence a species’ population dynamics, identifying proximate drivers can be critical to the conservation and management of protected species. Kemp’s ridley sea turtles (Lepidochelys kempii) exhibit spatial variation in both diet composition and growth, but whether components of this variation are linked has not been evaluated. Through an integration of skeletochronological and stable isotope analyses of stranded turtle humerus bones we characterized regional variation in Kemp’s ridley diet composition and potential relationships with somatic growth rates. Turtles were divided among five regions within the United States Gulf of Mexico (GoM) and Atlantic Coast based on location of stranding, and humerus bones were sampled for stable carbon (δ13C) and nitrogen (δ15N) isotope ratios. These data were combined with region-specific prey stable isotope data sourced from the primary literature into Bayesian stable isotope mixing models (MixSIAR) to estimate the proportional contribution of five prey groups (crustaceans, bivalves, gastropods, fish, and macroalgae/seagrass) to Kemp’s ridley diets. Our analysis revealed strong regional differences in mixing model-derived diet composition estimates that closely tracked published records of Kemp’s ridley diet. Invertebrates generally comprised the largest proportion (43.5–97.7%) of turtle diets. However, we also observed high proportional contributions of fish (42.6–43.1%) to western GoM turtle diets and macroalgae/seagrass (42.4–47.8%), or isotopically similar prey resources (e.g., tunicates), to eastern GoM turtle diets. Growth rates were poorly correlated with δ15N values—a proxy for trophic level—and diet composition estimates, suggesting that diet composition alone may not explain the regional differences in somatic growth observed in this species. This study highlights the value of complementary skeletal and isotopic analyses to understanding regional diet variation in sea turtles as well as the importance of continued collection of isotopic data for both sea turtles and their prey. These results also help fill critical knowledge gaps pertaining to the relationship between sea turtle foraging ecology and somatic growth dynamics, a topic of high importance to sea turtle conservation and management.
Somatic growth variation manifests from the cumulative effect of multiple biological, ecological, and environmental processes (Congdon, 1989; Stearns, 1992). Environmental effects on growth rates are particularly strong in ectothermic reptiles, such as sea turtles, where resource use, quality, and availability interact with temperature to determine how much of an individuals’ total energy budget is devoted to growth, maintenance, storage, and reproduction (Gibbons, 1967; Dunham et al., 1989). Spatiotemporal variation in energy allocation to growth within and among individuals can have profound effects on individual fitness and species population dynamics through influences on key life history features such as time to maturity (Frazer et al., 1993; Bjorndal et al., 2013), size-dependent mortality (Werner and Gilliam, 1984; O’Brien et al., 2005), and fecundity (Berry and Shine, 1980; Frazer and Richardson, 1986). Sea turtle somatic growth rates are highly variable within and among species and life stages, and a suite of environmental factors are thought to contribute to this variability (e.g., temperature, density-dependence, prey dynamics, diet quality, and individual behavior; Balazs, 1982; Bjorndal et al., 2003; Balazs and Chaloupka, 2004; Hatase et al., 2010; Peckham et al., 2011). Disentangling the myriad potential drivers of sea turtle somatic growth variation is challenging given the logistical limitations associated with studying highly migratory species (Omeyer et al., 2017). Nevertheless, identifying factors influential to sea turtle growth rates is of high importance to their conservation and management given that their population dynamics are sensitive to changes in demographic rates (Crouse et al., 1987; Gerber and Heppell, 2004).
Variation in resource use and availability is a primary driver of somatic growth variation within animal populations. For example, it is well-established that fish growth and population dynamics are strongly influenced by zooplankton composition, abundance, and distribution (Cushing, 1990; Brodeur and Ware, 1992; Durant et al., 2007). Similarly, variation in multiple seabird demographic rates, including growth, have been linked to differences in prey availability, composition, and energy density (Cairns, 1988; Abraham and Sydeman, 2004; Hennicke and Culik, 2005; Piatt et al., 2007). For loggerhead sea turtles (Caretta caretta), geographic variation in resource availability is thought to contribute to differences in somatic growth rates among life stages and breeding populations (Bjorndal et al., 2003; Piovano et al., 2011). These differences may relate to divergent prey energy densities or geographic differences in primary productivity (Bosc et al., 2004; Peckham et al., 2011). Observations of compensatory and density-dependent growth in loggerhead and green sea turtles (Chelonia mydas) provide further support for the importance of resource use in shaping sea turtle growth rates (Bjorndal et al., 2000, 2003). Within the Gulf of Mexico, factors that affect foraging resources for sea turtles include fisheries (Robinson et al., 2015), seasonal hypoxic zones (Craig et al., 2001), oil spills (Wallace et al., 2017), red tides (Dupont et al., 2010), hurricane activity (Engle et al., 2009), and climate change (Sanchez-Rubio et al., 2011), among others.
Kemp’s ridley sea turtles (Lepidochelys kempii) display distinct regional differences in somatic growth rates that may be linked to differences in diet composition. During neritic life stages, this species occupies nearshore marine habitats throughout the Gulf of Mexico (GoM) and United States Atlantic (NMFS and USFWS, 2015). Comparative studies prior to the year 2000 suggest juvenile Atlantic Kemp’s ridley sea turtles exhibit slower grow rates than conspecifics in the GoM (Caillouet et al., 1995; Zug et al., 1997; NMFS and USFWS, 2015; Avens et al., 2017), though causal mechanisms remain unknown. In contrast, juvenile Kemp’s ridley growth rates do not appear to vary substantially within the United States GoM and Atlantic (Ramirez, 2019). Although crabs are generally thought to constitute the bulk of their diet across their range, regional differences in Kemp’s ridley foraging patterns have been observed that may influence their somatic growth rates (Shaver, 1991; Burke et al., 1993, 1994; Seney and Musick, 2005; Schmid and Tucker, 2018). Diets are particularly variable among Kemp’s ridleys that inhabit the GoM. For example, tunicates are a common prey item for turtles in southwest Florida (Witzell and Schmid, 2005), whereas fish—likely sourced as discards from shrimp fisheries—are often consumed by turtles in the northern and western GoM (Werner, 1994; Cannon, 1998; Stacy, 2015). Shrimp fisheries are the overwhelmingly dominant source of fish discards throughout the Kemp’s ridleys’ range but the availability of this potential resource is an order of magnitude higher in the western and northern GoM than in the eastern GoM and United States Atlantic (Diamond, 2004; Harrington et al., 2005; Scott-Denton et al., 2012). In contrast to their GoM counterparts, Atlantic Kemp’s ridleys appear less likely to deviate from the traditional diet of crabs and molluscs (Burke et al., 1993, 1994; Frick and Mason, 1998; Seney and Musick, 2005). Ultimately, whether this spatial variability in diet composition correlates with regional differences in growth rates has yet to be evaluated.
As the isotopic composition of consumer tissues closely tracks that of their assimilated diet, stable isotope analyses provide a means of characterizing intra-population variation in diet composition over space and time (Newsome et al., 2007; Katzenberg, 2008). Importantly, the proportional contribution of different resources to a consumer’s diet can be quantified using mass-balance stable isotope mixing models when isotopic data are available for both consumers and potential prey (Phillips, 2001). While many environmental and physiological processes can influence stable isotope deposition rates into consumer tissues, the latest generation of mixing models allows for incorporation of various sources of uncertainty through Bayesian inference to improve estimations of diet composition (Phillips and Koch, 2002; Semmens et al., 2009; Parnell et al., 2010; Stock and Semmens, 2016). This approach in turn yields source contribution estimates that are accompanied by probability distributions that more accurately reflect model uncertainties. Kemp’s ridley humerus bones contain annual records of somatic growth that can be revealed through histological processing and analysis (Snover and Hohn, 2004; Avens et al., 2017). Combining skeletochronological and stable isotope analyses within a mixing model framework may thus provide a means of investigating the influence of diet composition on sea turtle growth rates across multiple spatiotemporal scales. The integration of these tools has already shed valuable insight into sea turtle ontogenetic growth dynamics and resource shifts (Snover et al., 2010; Avens et al., 2013; Ramirez et al., 2017, 2019; Turner Tomaszewicz et al., 2017a).
In this study we integrated skeletochronological and stable isotope analyses of Kemp’s ridley humerus bones to (1) characterize regional variation in diet composition and (2) quantify the relationship between diet composition and somatic growth rates. To reduce biases associated with translating isotopic data to diet composition estimates for a highly mobile species, our analysis assesses diet composition at a broad taxonomic level (e.g., % fish, % invertebrate, and % macroalgae/seagrass). We specifically investigated if turtles inhabiting areas where fish discards are prevalent (western and northern GoM) showed evidence of consuming greater proportions of fish relative to turtles from other regions (eastern GoM and United States Atlantic). Because the energy density of fish is generally higher than that of crustaceans (Doyle et al., 2007; Peckham et al., 2011; Schaafsma et al., 2018), we also investigated whether fish subsidies to turtle diets enhance somatic growth rates, thereby contributing to regional differences in somatic growth. This investigation presents one of the first studies explicitly linking sea turtle foraging ecology to their somatic growth dynamics.
Variation in Kemp’s ridley diet composition and growth was evaluated by dividing turtle and prey data among five geographic regions within the species’ range (Figure 1): (1) western Gulf of Mexico (wGoM, n = 44 turtles; Texas/Mexico border to Vermilion Bay, LA), (2) northern Gulf of Mexico (nGoM, n = 28 turtles; Vermilion Bay, LA, to Mobile Bay, AL), (3) eastern Gulf of Mexico (eGoM, n = 24 turtles; Apalachicola Bay to Florida Bay, FL), (4) North Carolina (NC, n = 32 turtles; Long Bay to Albemarle Sound, NC), and (5) Virginia (VA, n = 25 turtles; North Carolina/Virginia border to lower Chesapeake Bay). These breakpoints were primarily determined based on known or presumed spatial variation in ocean chemistry. We explored using smaller geographic areas to more closely link turtle and prey stable isotope data in space. However, there was generally insufficient prey data for one or more prey groups to use smaller regional units for this analysis (see below).
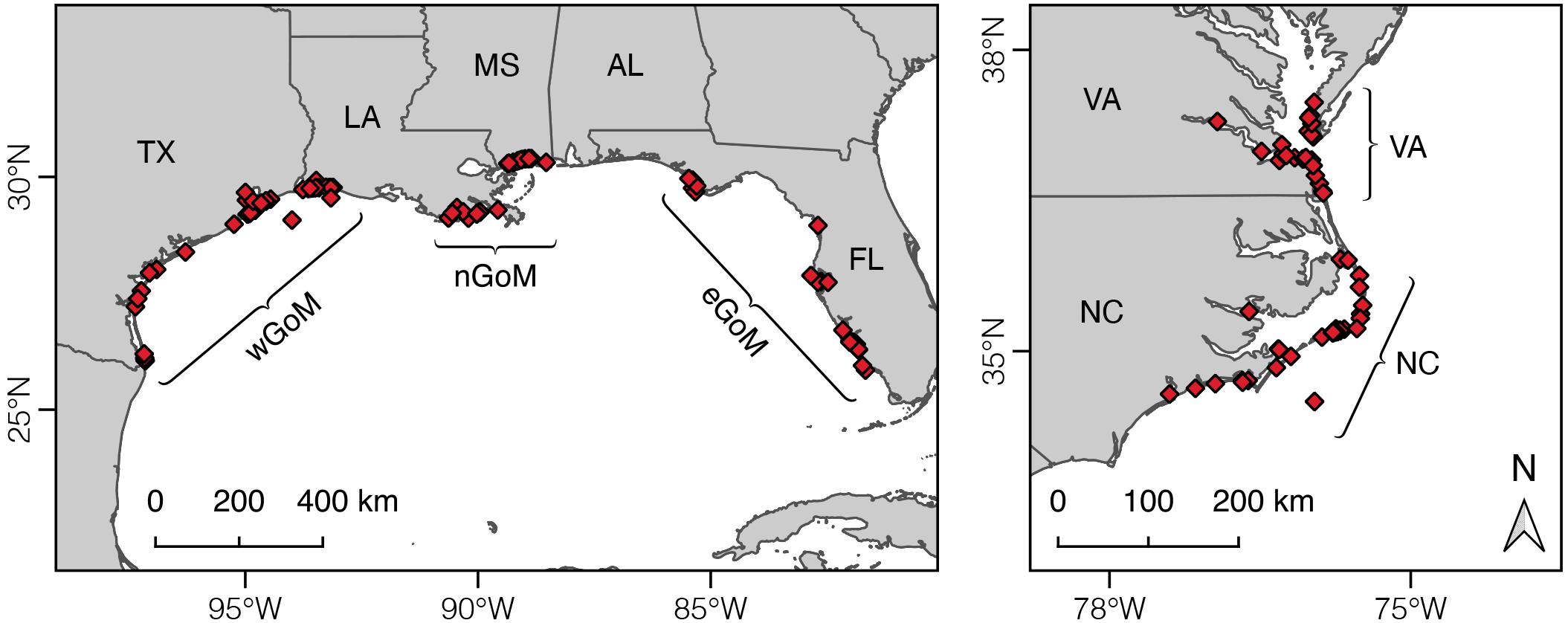
Figure 1. Map of Kemp’s ridley sea turtle stranding locations for the humerus bones used in this study and geographic breakpoints used to cluster turtles and prey groups. wGoM, western Gulf of Mexico; nGoM, northern Gulf of Mexico; eGoM, eastern Gulf of Mexico; NC, North Carolina; VA, Virginia.
Within the GoM, the West Florida Shelf is characterized by relatively low stable nitrogen isotope ratios (δ15N) due to the presence of Trichodesmium (Lenes et al., 2001; Mulholland et al., 2006; Vander Zanden et al., 2015), a N2-fixing cyanobacteria; N2-fixation reduces δ15N values (Montoya et al., 2002). Marine organisms occupying the West Florida Shelf thereby may have lower δ15N values than conspecifics elsewhere due to chemical differences at the base of the food web. In contrast, the nGoM and Virginia regions may have relatively high δ15N values and low stable carbon isotope (δ13C) values than adjacent regions due to high nitrogen loading from agricultural runoff (i.e., high nitrogen content; Black et al., 2017; Fritts et al., 2017) and freshwater influences, respectively—freshwater systems have distinctly lower δ13C values than marine systems (Fry and Sherr, 1989).
Kemp’s ridley sea turtles are generalist carnivores, consuming primarily invertebrates (crustaceans, gastropods, bivalves, and tunicates) but also variable amounts of fish, macroalgae, and seagrasses (Shaver, 1991; Seney and Musick, 2005; Witzell and Schmid, 2005). Although regional differences in foraging patterns have been observed for this species, such as increased consumption of fish in turtles from Louisiana and Texas (Werner, 1994; Cannon, 1998; Stacy, 2015) and tunicates in turtles from Southwest Florida (Witzell and Schmid, 2005), crabs have generally constituted > 75% of their observed diet by weight (Shaver, 1991; Burke et al., 1993, 1994; Seney and Musick, 2005; Schmid and Tucker, 2018). Given the spatiotemporal extent and retrospective nature of this study, we relied on the primary literature to source stable isotope data of representative prey species for our mixing model.
We first performed a structured literature search in Web of Science and Google Scholar using the following Boolean search terms: stable isotope, crustacean, crab, shrimp, mollusc, arthropod, gastropod, sea snail, bivalve, clam, oyster, mussel, fish, tunicate, seagrass, and macroalgae. We then performed an unstructured literature search using the reference lists of relevant publications found in the structured search. Following exclusion of studies performed outside the focal geographic areas, the literature search yielded 86 studies from which stable carbon and nitrogen isotope ratios were collated. If studies reported multiple stable isotope values for a single prey species, a weighted mean and pooled standard deviation (SD) were calculated to collapse the reported data into one estimate per species per study. Tunicates, though potentially an important Kemp’s ridley prey group, were excluded from our analysis given their poor representation in the literature (n = 2 studies) and overlap in isotopic values with macroalgae and seagrass. The final prey stable isotope dataset comprised 552 isotopic records (see Table 1 for summary and Supplementary Table S1 for full dataset). Original collection dates spanned 1975 to 2016, but primarily encompassed the years 1990 to 2016—pre-1990 data were included in some instances to fill important data gaps for poorly represented taxa within each region, namely bivalves and gastropods.

Table 1. Taxonomic and geographic summaries of Kemp’s ridley sea turtle prey stable isotope data collated from the published literature.
Prey stable isotope data were grouped into five primary prey groups (crustaceans, bivalves, gastropods, fish, and macroalgae/seagrass) within each of the five geographic regions (Supplementary Figures S1, S2). For all animal prey groups, a simple mean and pooled SD were calculated for each region using the 552 isotopic values from the published literature. Although isotopically distinct, macroalgae and seagrass were grouped to reduce the number of sources in the mixing model. As with the other prey groups, we first calculated a simple mean and pooled SD for macroalgae and seagrass separately and then calculated a simple mean of these estimates to yield final estimates for the macroalgae/seagrass prey group, thereby weighting each prey type equally in the models. Final means and SDs for all prey groups used in the mixing model are presented in Table 2. We assume that isotopic data collated from the published literature accurately reflect the means and variances of these prey groups.
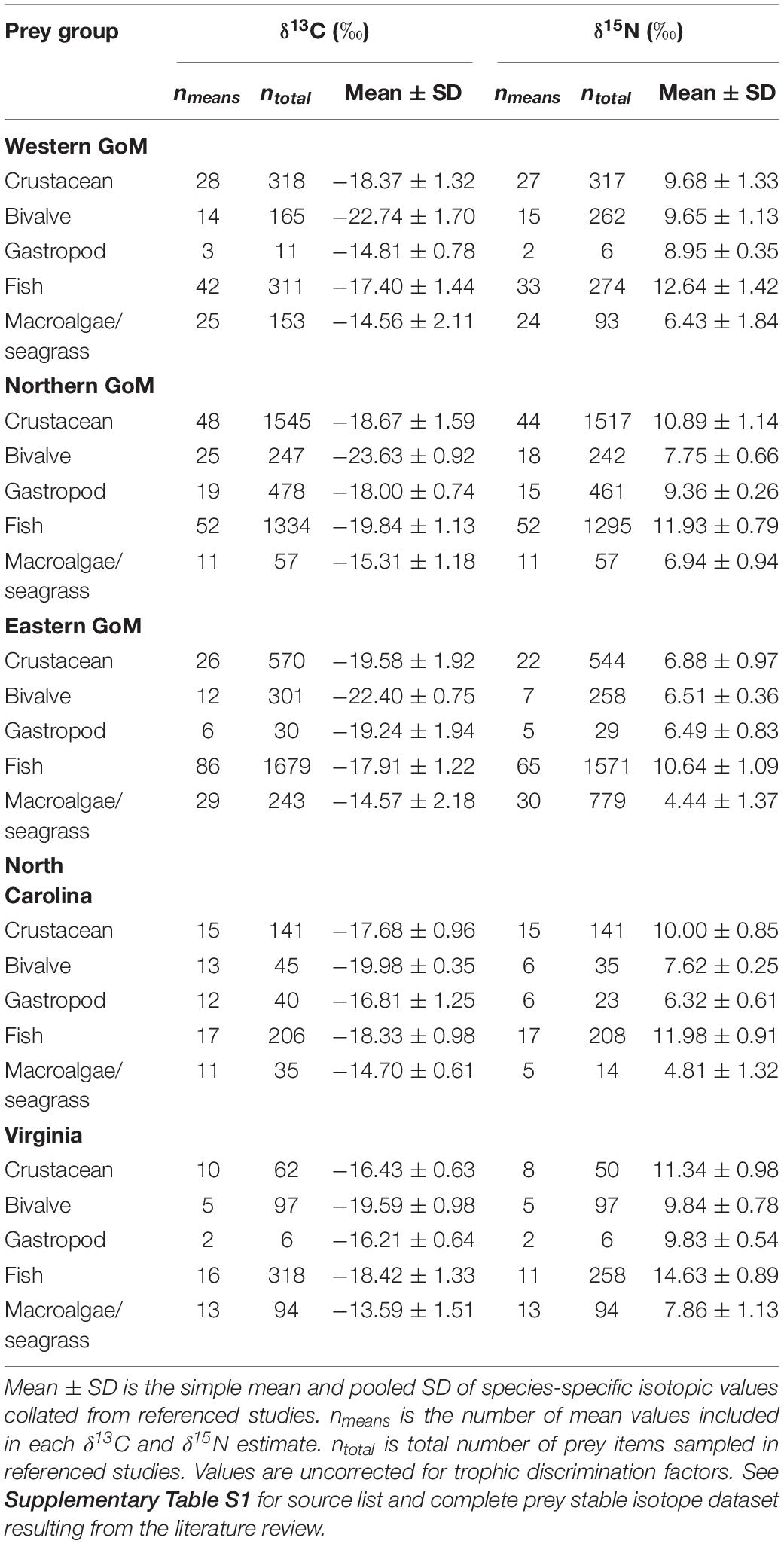
Table 2. Mean ± SD stable carbon (δ13C) and nitrogen (δ15N) isotope ratios collated from the primary literature for Kemp’s ridley sea turtle prey groups by geographic region.
Given uncertainties in the types of fish consumed by sea turtles, potential fish prey for our analysis included species previously observed in Kemp’s ridley gut and fecal contents (e.g., mullet, croaker, weakfish, menhaden, sea catfish, flatfish, and lizardfish; see Shaver, 1991; Werner, 1994; Cannon, 1998; Witzell and Schmid, 2005; Stacy, 2015; Seney, 2016) as well as ecologically similar species abundant in shrimp fishery discards (e.g., porgy, pinfish, herring, and searobin; Harrington et al., 2005; Benaka et al., 2019). When possible, fish isotopic data were restricted to specimens <30 cm in length to align with those likely to be consumed by Kemp’s ridleys. However, only 45% of studies reported fish lengths, so this was not always possible. Fish stable isotope data were initially grouped based on feeding mode (e.g., piscivorous, benthophagous, and planktivorous) to evaluate trophic differences. However, isotopic data for these three fish groups tended to overlap extensively in isospace within each region and were thus collapsed to reduce the number of sources in the mixing model.
Kemp’s ridley humerus bones utilized in this study were originally collected as whole front flippers from 153 turtles stranded dead along the United States Gulf and Atlantic Coasts between 1993 and 2015 by participants of the Sea Turtle Stranding and Salvage Network. At time of stranding, carapace length (notch to tip), calendar date, and stranding location (state, latitude, and longitude) were recorded for each turtle. Body size was typically measured as straightline carapace length (SCL), but in cases where only curved carapace length was recorded measurements were converted to SCL following Avens et al. (2017). Sea turtle diets and growth rates vary throughout their lifetime (Bjorndal, 1997; Chaloupka and Musick, 1997). To reduce the potential for ontogenetic effects to bias our results we only sampled bone growth layers from juvenile Kemp’s ridleys corresponding to their benthic life stage (i.e., age ≥ 0.75 and δ15N values ≥ 10.7‰; Ramirez, 2019; Ramirez et al., 2019).
Prior to sampling, each humerus bone was cleaned of soft tissue using a knife and then boiled. To perform complementary growth and stable isotope analyses, two sequential 2–3 mm thick cross-sections were cut from each humerus bone distal to the site of the deltopectoral muscle insertion scar using a low-speed isomet saw (Buehler). One section was histologically processed using standard methods to reveal the annual growth layers contained within each bone and estimate sea turtle growth rates (see below), whereas the second was reserved for complementary stable isotope analysis. Methods for histologically processing sea turtle bones are detailed in Avens and Snover (2013) but are briefly outlined here. First, humerus bone sections were decalcified over multiple days using a fixative/decalcifier (Cal-Ex II or RDO). Then, bone sections were thin sectioned using a freezing-stage microtome or cryostat, stained using Ehrlich’s hematoxylin, and finally mounted onto microscope slides and digitally imaged. Two or three independent readers (among L. Avens, L. Goshe, M. Ramirez, and M. Snover) then analyzed the bone images to determine the number and placement of lines of arrested growth (LAGs), which delimit the outer edges of each skeletal growth mark.
To characterize resource use, ∼1.5 mg of bone dust was milled from the most recently deposited growth layer of each sea turtle bone cross-section reserved for stable isotope analysis (ESI New Wave Research MicroMill). This time period represents the geochemical history within 1 year of death, dependent on individual stranding date. A 0.3 mm diameter carbide drill bit (Brasseler) was used in conjunction with transparencies of the digital skeletochronology images to guide precision drilling to a depth of ≤1.0 mm for each sample. Bulk bone dust samples were analyzed for δ13C and δ15N values via continuous-flow isotope-ratio mass spectrometry at the Oregon State University Stable Isotope Lab (Corvallis, OR, United States). The system consists of a Carlo Erba NA1500 elemental analyzer interfaced with a DeltaPlusXL isotope-ratio mass spectrometer (Finnigan MAT, Bremen, Germany). The standards used for δ13C and δ15N were Vienna Pee Dee Belemnite (VPDB) and atmospheric N2, respectively. The internal standard IAEA-600 (Caffeine; isotopic composition of δ15N = 1.00‰) was calibrated at regular intervals and used to correct for instrument drift and linearity. Analytical precision was 0.08‰ for δ13C values and 0.05‰ for δ15N values. In addition to stable isotope ratios, %N and %C were calculated using mass 28 and mass 44 peak areas, respectively, with a precision of 0.55% for %N and 0.28% for %C. C:N ratios (%C divided by %N) for all samples were below 3.5, characteristic of unaltered protein with low lipid content (Post et al., 2007). Following stable isotope analysis, bulk bone δ13C values were mathematically corrected to account for carbonate-derived carbon as recommended by Turner Tomaszewicz et al. (2015). Using their approach, we developed a δ13C conversion equation (δ13Ccollagen = 0.975 ∗ δ13Cbulk − 1.126, F1,42 = 550.1, P < 0.001, adjusted R2 = 0.93) that was used to mathematically correct bulk bone δ13C values (see Supplementary Mateiral for details).
We assumed that stranding location was reflective of recent foraging location based on two lines of evidence. First, while we did not know precise locations of death in the ocean for turtles herein, it is likely that most turtles included in this study died relatively close to their stranding locations as ocean conditions were likely favorable for short carcass drift distances. The majority of turtles included in our study stranded in the spring, summer, and fall when sea surface temperatures, and thereby decomposition rates, would have been relatively high (Higgins et al., 2007). Therefore, in order for stranding to occur before carcasses dissociated due to decomposition, drift times and distances would have needed to be low (∼2–5 days, 15–30 km; Nero et al., 2013; Santos et al., 2018). Second, Kemp’s ridleys display relatively high intra- and inter-annual site fidelity to nearshore, shallow (<50 m depth) foraging areas (generally <1,000 km2) that are well constrained spatially within our defined geographic regions (Renaud and Williams, 2005; Schmid and Witzell, 2006; Shaver and Rubio, 2008; Seney and Landry, 2011; Coleman et al., 2017). Therefore, turtles that stranded within each geographic area are likely to have been foraging within their stranding location-assigned geographic area prior to death. As Kemp’s ridleys have been occasionally documented migrating >1,000 km in a single year (Renaud and Williams, 2005), we acknowledge that some of our turtles may be misclassified geographically, particularly those that stranded near the edges of our pre-defined geographic areas.
We implemented multiple Bayesian hierarchical mixing models using the MixSIAR package (v 3.1.10, Stock et al., 2018) in R (v 3.5.3, R Core Team, 2019) to estimate the proportional contribution of five prey groups (crustaceans, bivalves, gastropods, fish, and macroalgae/seagrass) to Kemp’s ridley diets. MixSIAR uses Markov chain Monte Carlo (MCMC) procedures to estimate posterior probability distributions of plausible proportional contributions of prey groups to consumer diets (Moore and Semmens, 2008), while accounting for uncertainty associated with trophic discrimination factors (TDFs; Parnell et al., 2010), concentration dependence (Phillips and Koch, 2002), fixed and random effects (Semmens et al., 2009), and variability in the predation process (i.e., error structure) (Parnell et al., 2010; Stock and Semmens, 2016). Initial investigations using a hierarchical structure that nested individuals within regions in a single modeling framework failed to converge after running for multiple days due to model complexity and size. Therefore, we implemented separate mixing models for each region.
To characterize inter- and intra-regional differences in diet composition, we implemented four mixing models for each region in a 2 × 2 factorial design that included one of two prior distributions (uninformative vs. informative prior; Supplementary Figure S3) for each prey group and one of two model configurations (null model vs. individual random effect model). We first ran the models using uninformative priors that assumed a generalist diet and weighted prey groups equally (α = 1, 1, 1, 1, 1). We then ran the model using an informative/specialist prior that weighted the prey group prior distributions using published diet composition data. Taking a weighted average of taxon-specific diet composition estimates from six Kemp’s ridley diet studies (Supplementary Table S2), we constructed the informative priors assuming diet compositions (by dry mass) of 76.74% for crustaceans, 2.12% for bivalves, 2.12% for gastropods, 5.97% for fish, and 2.13% for macroalgae/seagrass—10.92% of diet contents were categorized as Other/Unidentified, which was excluded from this analysis. As recommended by Stock et al. (2018), the hyperparameters (α) for the informative priors were scaled to have a total weight equal to the number of sources (α = 4.31, 0.12, 0.12, 0.34, 0.12). Between region diet variation was assessed using null models, whereas within region diet variation was assessed using models that included individual as a random effect. In all models, the invertebrate prey groups were aggregated a posteriori (Phillips et al., 2005). All models included multiplicative error (process × residual error) and were run using the “extreme” MCMC settings (chain length = 3,000,000 iterations; burn-in = 1,500,000; posterior thinning = 500; 3 chains). Convergence was assessed using Gelman–Rubin (Rc < 1.01) and Geweke diagnostics (Gelman and Rubin, 1992; Geweke, 1992). Most models that included an informative prior and individual random effects failed to converge with these settings. Convergence was achieved after re-running them using a chain length of 6,000,000 and burn-in of 3,000,000.
Prior to model implementation all source and consumer δ13C values were corrected for the Suess Effect, the global decrease in atmospheric δ13C values driven by the combustion of fossil fuels over the past 150 years (Keeling et al., 1979; Francey et al., 1999). We followed Chamberlain et al. (2005) and Fox-Dobbs et al. (2007) in applying a linear correction to standardize our data. To develop a δ13C correction factor we analyzed the atmospheric δ13C data for Maua Loa and La Jolla available on the Scripps CO2 Program website1 (Keeling et al., 2001), which indicated that atmospheric δ13C values declined by ∼0.025 ‰ per year since 1978. We used this rate of δ13C change to correct turtle and prey δ13C values to modern values (modern = 2016; i.e., δ13C values were reduced by 0.025‰ in 2015, 0.050‰ in 2014, and so on). Concentrations of carbon and nitrogen for each prey group, derived from the literature (Supplementary Table S3), were also included in the models to account for taxon-specific differences in digestibility (Phillips and Koch, 2002).
Stable isotope mixing models require estimates of diet-tissue trophic discrimination factors (TDFs; Δ)—the difference in isotopic ratios between consumers and their diet—to estimate the proportional contribution of different prey groups to consumer diets. As diet-bone TDFs have not been quantified for Kemp’s ridleys or other primarily carnivorous sea turtles, we used diet-bone TDFs estimated from dead, captive, juvenile green sea turtles (Chelonia mydas) (Δ13C = 2.1 ± 0.6, Δ15N = 5.1 ± 1.1) (Turner Tomaszewicz et al., 2017b). Although these turtles were maintained on omnivorous diets composed of ∼56% animal matter (squid, shrimp, and fish) and ∼43% plant matter (lettuce) by weight, percent digestible N and C from animal protein was estimated to be 96.8 and 81.9%, respectively. Even though Bayesian stable isotope mixing models account for uncertainty in TDFs, their outputs are still highly sensitive to variation in TDFs (Bond and Diamond, 2011). Given uncertainty in the diet-bone TDFs for sea turtles, we used a sensitivity analysis to characterize the influence of varying TDFs on diet composition estimates that encompass the range of diet-bone TDFs reported for sea turtles and other animal species maintained on carnivorous diets (∼2–6‰; e.g., Ambrose and DeNiro, 1986; Hobson and Clark, 1992; Fox-Dobbs et al., 2007; Borrell et al., 2012; Kim et al., 2012; Cloyed et al., 2015; Webb et al., 2016; Matsubayashi et al., 2017).
To examine the influence of sea turtle trophic ecology on somatic growth rates, we compared complementary diet composition data generated from the stable isotope mixed models with annual somatic growth rate data generated through skeletochronology for each stranded turtle. The somatic growth rate data presented herein are a combination of newly collected (n = 58 turtles stranded 2010–2015) and previously collected data (n = 95 turtles stranded 1993–2009) originally presented in Snover et al. (2007) and Avens et al. (2017). We followed Avens et al. (2017) to calculate growth rates for the newly processed turtles.
First, LAG diameter and humerus section diameter (HSD) were measured using image analysis software (Olympus Microsuite and cellSens) for each histologically prepared bone cross-section. The body proportional hypothesis back-calculation technique (BPH; Francis, 1990) was then used to estimate SCL for every measurable LAG, adjusted for turtle–specific SCL and HSD at death (Snover and Hohn, 2004; Avens et al., 2017). Annual somatic growth rates were calculated by taking the difference between SCL estimates of successive LAGs. However, given that LAGs are deposited in the spring and we sampled turtles that died throughout the year, only 73/153 turtles had true annual growth rate estimates.
To extend the growth dataset we calculated marginal growth rates for the 36 turtles that stranded between November and March by taking the difference between SCL at stranding and the SCL estimate of the most external LAG. While these marginal growth rates are considered minimum estimates of annual somatic growth, Kemp’s ridleys likely grow little during the boreal winter when temperatures are cooler and sea turtle metabolic rates and activity patterns are reduced (Balazs and Chaloupka, 2004; Hochscheid et al., 2007; McMichael et al., 2008). Indeed, skeletal growth appears to asymptote in November (Snover and Hohn, 2004). The 44 turtles that stranded between June and October were excluded from the growth analysis, highlighting a potential disconnect in data availability for linking sea turtle growth and diet that could be overcome in future analyses through targeted sampling of only turtles that stranded in the spring.
To examine the influence of sea turtle trophic ecology on somatic growth rates, we implemented a series of Generalized Linear Models (GLMs) that included somatic growth as the response variable, age as a fixed effect, and either δ15N value or estimated diet composition as a fixed effect. Separate GLMs that included δ15N values as a fixed effect were implemented for each region, whereas GLMs that included estimated diet composition as a fixed effect were only implemented for regions with considerable intra-population variation in diet composition. As sea turtle growth rates change throughout their ontogeny, age was included in the model to account for ontogenetic effects on growth and diet. Age was chosen over body size to account for ontogenetic effects as models that included age had consistently lower AIC values than models that included body size. All GLMs included a Gamma distribution and were implemented in R (version 3.5.3) using the mgcv package (Wood, 2006; R Core Team, 2019).
Prey δ13C and δ15N values were significantly different both within and among regions (Kruskal–Wallis rank sum tests, P < 0.05; see Supplementary Table S4). Gastropod was the only prey group that did not exhibit significant regional differences for both stable isotopes examined, although differences in δ13C were sometimes evident. Despite this regional variation in isotopic composition within prey groups, the relative positioning of prey groups in bivariate isospace was similar across regions (Figure 2). As expected, fish δ15N values were greater than the other prey groups in all cases, with mean values ranging between 10.64 and 14.63‰ (Table 2). Similarly, the macroalgae/seagrass group exhibited the lowest δ15N values (mean range 4.44–7.86 ‰) and highest δ13C values of all prey groups (mean range −15.31 to −13.59‰), reflective of their position at the base of coastal benthic food webs. Bivalves, which tended to be sampled in closest proximity to coastlines and freshwater inputs, had the lowest δ13C values (mean range −23.63 to −19.59‰). Crabs and gastropods displayed the greatest variability in isospace positioning of the five prey groups but generally fell within the polygon formed by macroalgae/seagrass, bivalves, and fish (Figure 2). Within regions, fish, crustaceans, bivalves, and macroalgae/seagrass differed statistically for at least one stable isotope (Wilcoxon rank sum tests, P < 0.05; see Supplementary Table S5). However, gastropods tended to share isospace with at least one other prey group in each region, likely due in part to small sample sizes—gastropod stable isotope values are poorly represented in the primary literature (see Supplementary Figure S1).
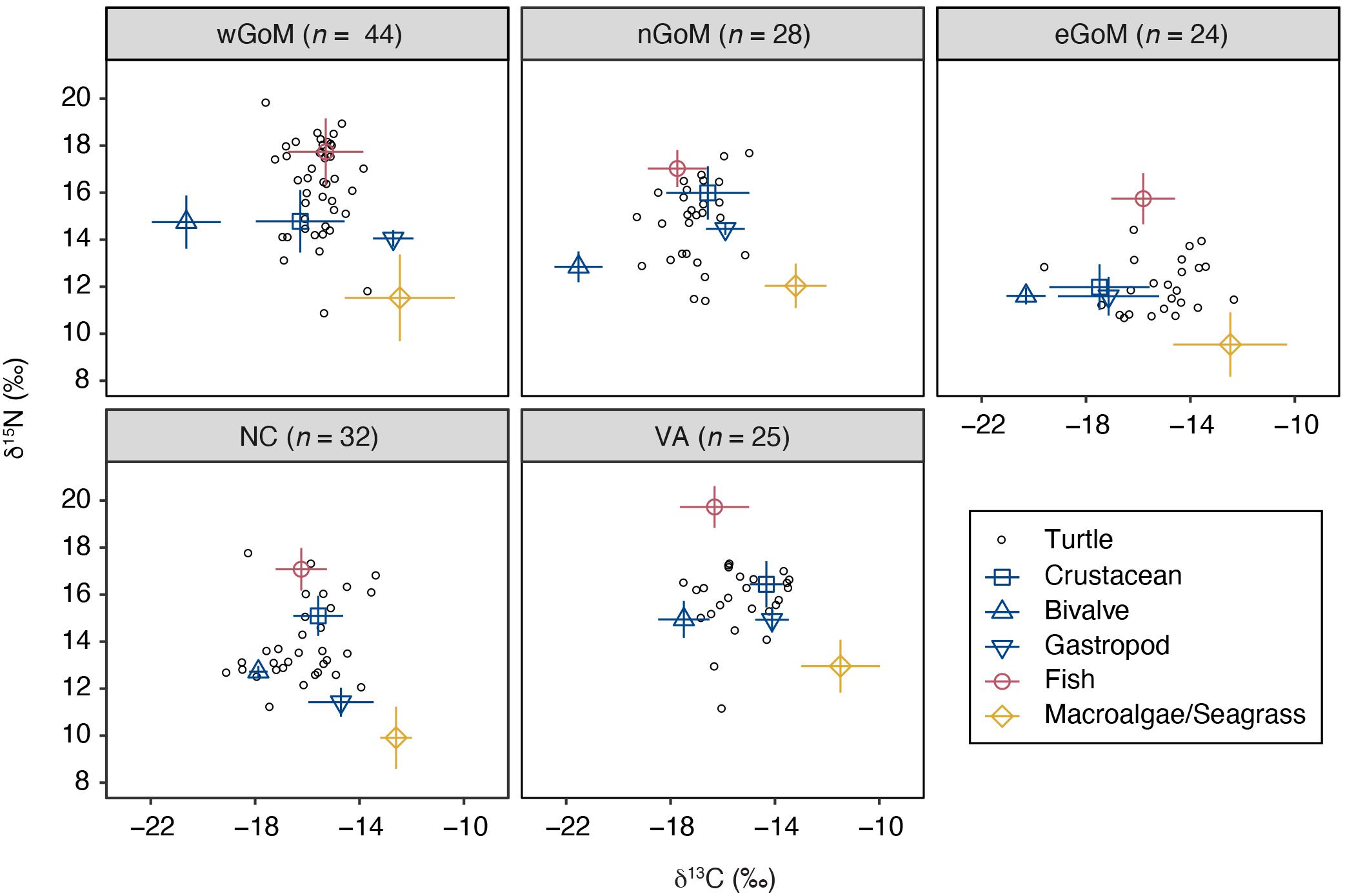
Figure 2. Biplots of δ13C and δ15N values for Kemp’s ridley sea turtles (open circles) and their potential prey groups (mean ± SD) by geographic region. Turtle sample sizes are presented at the top of each plot. Data are for the most recently deposited growth layer prior to death only. Prey values are corrected for trophic discrimination factors (Δ13C = 2.1 ‰, Δ15N = 5.1 ‰; see Table 2 for uncorrected values). δ13C values are corrected for carbonate carbon and the Suess effect.
Kemp’s ridley bone stable isotope values were generally constrained by the prey stable isotope data (Figure 2). Summary characteristics of bone growth layers sampled for stable isotope ratios are presented in Table 3. An analysis of variance on these data showed there was significant variation among regions for both δ13C (F4,148 = 11.68, P < 0.001) and δ15N (F4,148 = 129.19, P < 0.001) values. A post hoc Tukey test determined that turtle bone δ13C values were significantly lower in turtles stranded in the nGoM relative to all other regions (P < 0.05; Supplementary Table S6), possibly a result of influences of the Mississippi River, as freshwater systems generally have distinctly lower δ13C values than marine systems (Fry and Sherr, 1989). In addition, δ15N values were significantly higher in turtles from the wGoM and lower in turtles from the eGoM relative to all other regions (P < 0.05). Differences in δ15N values between turtles in the eGoM and other regions may be driven by regional differences in nitrogen cycling or trophic ecology. The West Florida Shelf is an area of high N2-fixation due to the presence of the cyanobacteria Trichodesmium (Lenes et al., 2001; Mulholland et al., 2006; Vander Zanden et al., 2015), which reduces δ15N values (Montoya et al., 2002). Similarly, Kemp’s ridleys in southwest Florida are known to eat tunicates, a low trophic level marine species with characteristically low δ15N values (Williams et al., 2014). Along the United States Atlantic Coast, δ15N values were significantly higher and less variable in turtles from Virginia relative to turtles in North Carolina, tracking differences in prey isotopic composition, which is possibly due to nutrient loading by anthropogenic activities in the Chesapeake Bay.
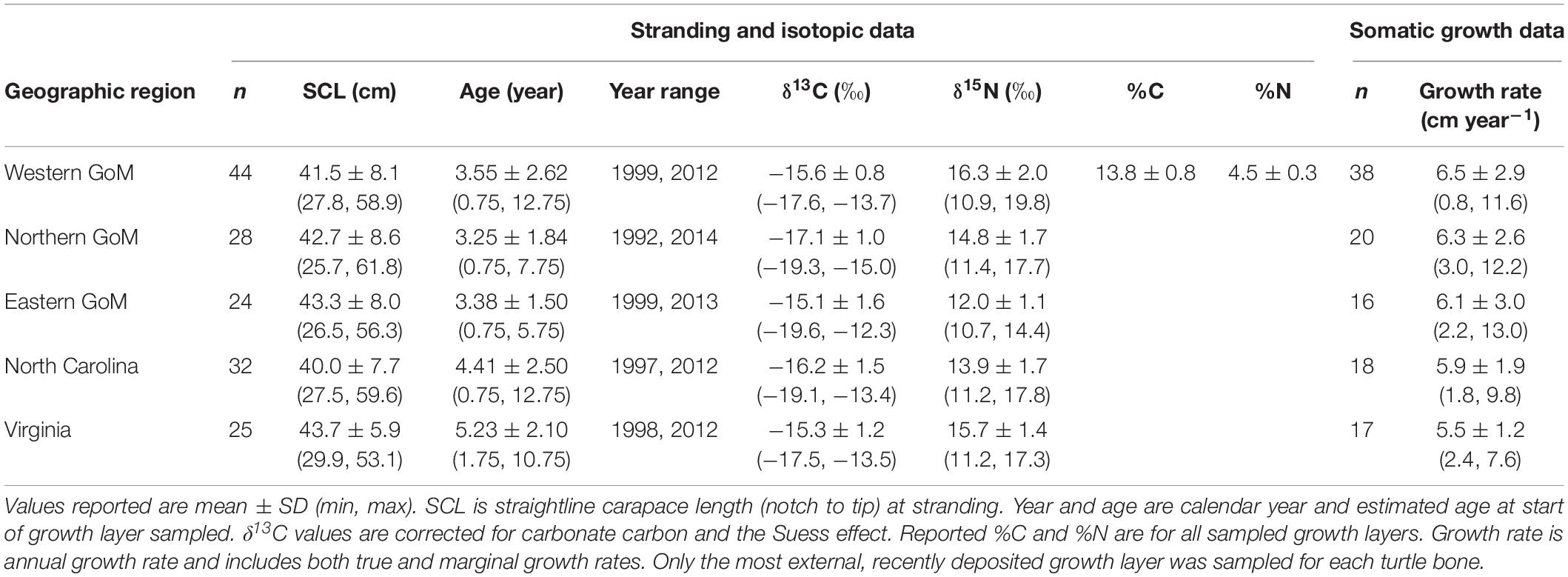
Table 3. Summary characteristics for Kemp’s ridley sea turtle bone growth layers sampled for stable isotope values.
We observed distinct regional differences in diet composition (% fish vs. % invertebrate vs. % macroalgae/seagrass) for Kemp’s ridleys (Figure 3 and Table 4). Diet proportion estimates derived from mixing models that included both uninformative and informative priors indicated that Kemp’s ridley diets were dominated by invertebrates in the nGoM, North Carolina, and Virginia (65.6–97.7%). In contrast, diets in the wGoM and eGoM were more evenly divided between invertebrates (43.6–54.5%) and fish (42.6–43.1%) or invertebrates (43.5–53.6%) and macroalgae/seagrass (42.4–47.8%), respectively. As it is unlikely that Kemp’s ridleys would consume such high proportions of macroalgae/seagrass, the eGoM results likely reflect consumption of an isotopically similar benthic resource, such as tunicates (∼5.5‰; Williams et al., 2014), or incorrect parameterization of the model. Within the wGoM and eGoM regions, individual variation in turtle diets was high for wGoM turtles but low for eGoM turtles. The proportional contribution of fish and invertebrates to individual wGoM turtle diets ranged between 12 and 60% and 36 and 85%, respectively, whereas the proportional contribution of macroalgae/seagrass and invertebrates to individual eGoM turtle diets ranged between 32 and 48% and 49 and 63%.
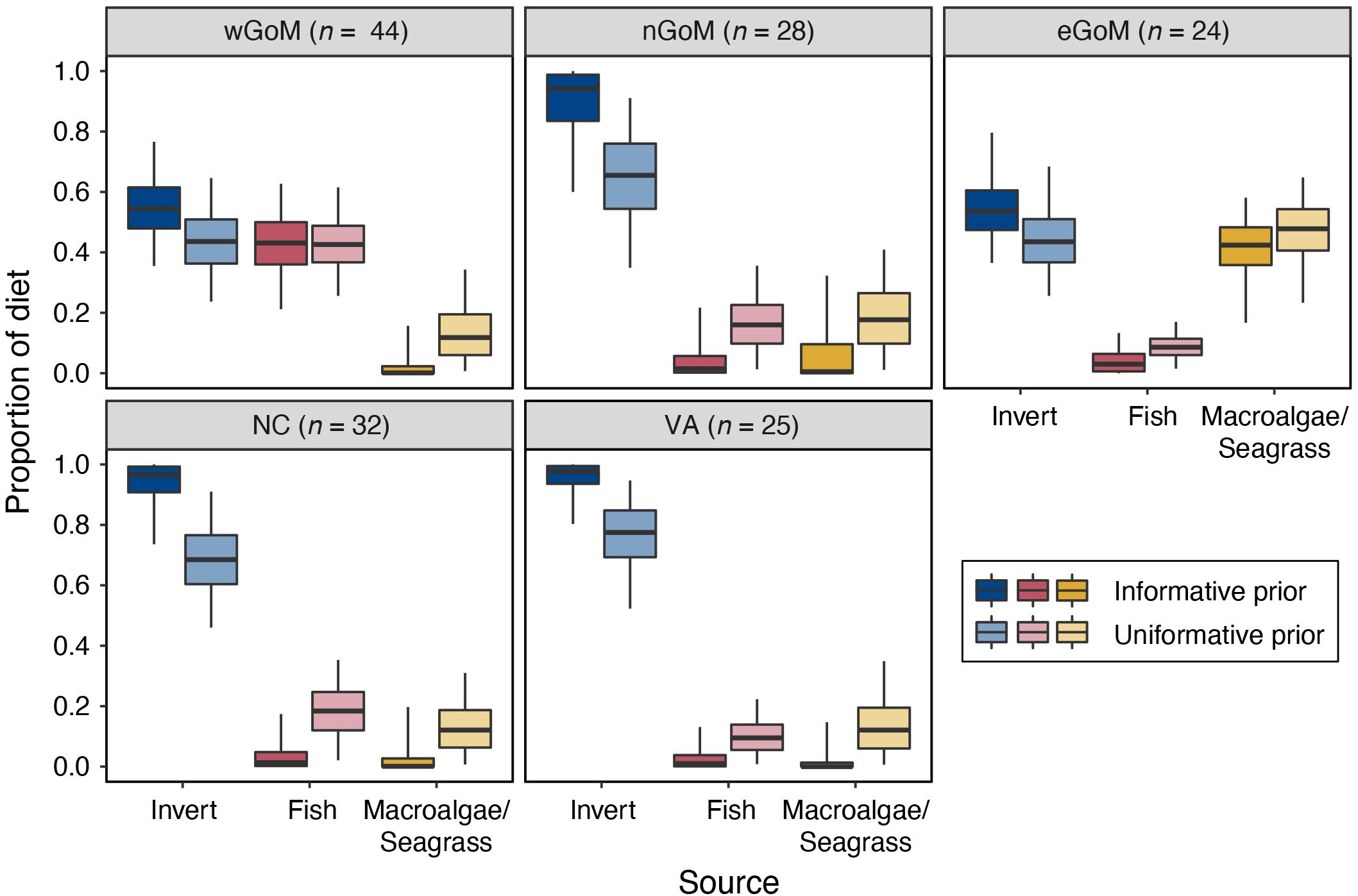
Figure 3. Proportional contribution of each prey group to Kemp’s ridley sea turtle diets by geographic region based on MixSIAR models that included an informative prior constructed from published diet proportion data and an uninformative prior that assigned equal probability to all prey groups. Turtle sample sizes are presented at the top of each plot. Data for invertebrate prey groups (crustacean, bivalve, and gastropod) were aggregated a posteriori. Lines in boxes are medians, boxes are 50% credible intervals, error bars are 95% credible intervals. See Table 4 for samples sizes, medians, and credible interval values.
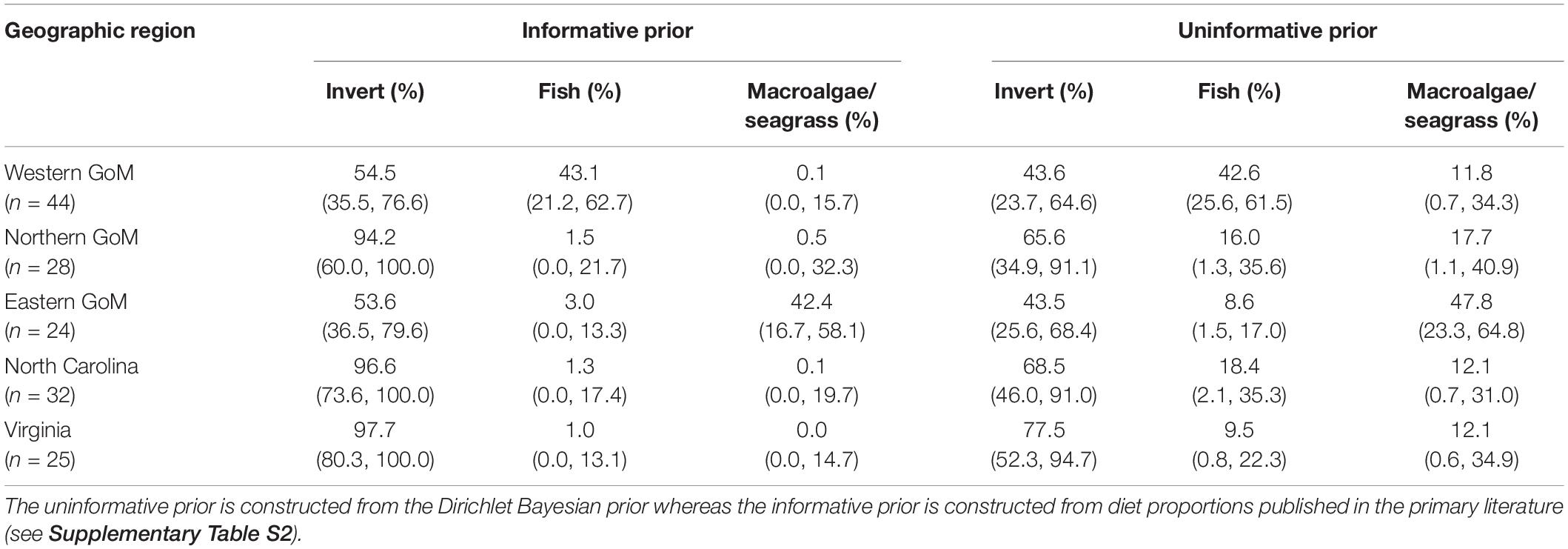
Table 4. Median (95% CI) posterior Bayesian mixing model estimates of diet proportion by geographic region for Kemp’s ridley sea turtles (n = 153).
In most cases, models that included uninformative priors estimated slightly greater contribution of fish and macroalgae/seagrass prey groups to Kemp’s ridley diets relative to models with informative priors. However, posterior distributions and 95% credible intervals overlapped extensively between each set of models (Figure 3 and Supplementary Figure S5). Larger differences between these model sets were evident in the pre-aggregated invertebrate data, where mixing models with uninformative priors estimated more even contribution of crustaceans, bivalves, and gastropods to Kemp’s ridley diets relative to models with the informative priors (Supplementary Figure S6).
As expected for Bayesian stable isotope mixing models (Bond and Diamond, 2011), sensitivity analyses performed on the null mixing model with informative priors for wGoM turtles showed that changes in diet-bone TDFs affected estimated proportional contribution of prey groups to Kemp’s ridley diets (Supplementary Figure S6). Specifically, the median estimated proportional contribution of fish and invertebrate prey to wGoM turtle diets was highly sensitive to changes in Δ15N but less sensitive to changes in Δ13C, unsurprising given that these prey groups primarily differ in δ15N values (Figure 2). Diet estimates within one standard deviation of the Δ15N mean ranged between 7.9 and 66.9% for fish and 30.7 and 79.2% for invertebrates, whereas estimates within one standard deviation of the Δ13C mean ranged between 35.7.and 45.4% for fish and 41.7 and 62.6% for invertebrates. Mixing model estimates for proportional contribution of individual invertebrate groups to turtle diets displayed greater sensitivity to changes in Δ13C values. Bivalve and gastropod diet composition estimates were more sensitive to changes in Δ13C than Δ15N, although their relative contribution to turtle diets remained low within one standard deviation of the mean Δ13C value (0–7.4% for bivalve, 0–11.9% for gastropod). Crustacean estimates were equally sensitive to both changes in Δ13C and Δ15N values, with bivariate changes in both TDFs resulting in estimates ranging from 19.5 to 90.7%.
After controlling for the influence of age on somatic growth rates, our GLMs revealed no significant relationships between δ15N values and somatic growth rates across most regions (Table 5 and Figure 4). The only exception was for nGoM turtles, where there was a weakly negative relationship between δ15N values and somatic growth rates (P = 0.07). This negative trend was still evident when marginal growth rates were excluded from the analysis, but the relationship became non-significant (P = 0.11). When marginal growth rates were excluded, trends across the other regions remained the same, exhibiting a shallow, non-significant decline in somatic growth rates with increasing δ15N values. These patterns ran counter to our expectation of higher growth rates with increasing δ15N values (i.e., foraging higher in food web), and could indicate that turtles consuming proportionally higher amounts of fish bycatch might be growing slower than conspecifics feeding primarily on invertebrates, or that physiological processes related to changes in size/age are influencing δ15N values. In nGoM and VA turtles, δ15N values and age exhibited a weakly positive relationship (Figure 4). However, across all regions, turtles with the highest δ15N values tended to span a wide range of ages, suggesting that larger/older turtles are generally not any more likely than smaller/younger turtles to feed higher in the food web.
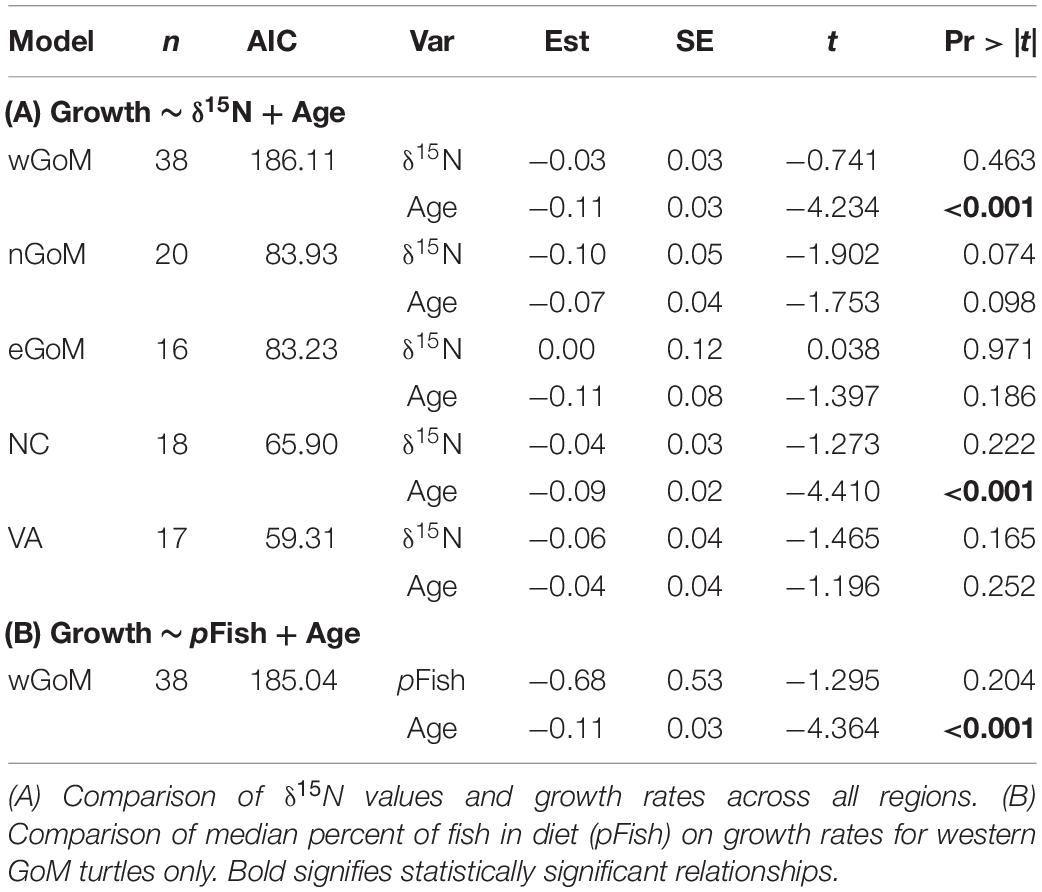
Table 5. Summary of statistical output for Generalized Linear Models used to evaluate the influence of diet (composition and δ15N values) on Kemp’s ridley sea turtle annual growth rates.
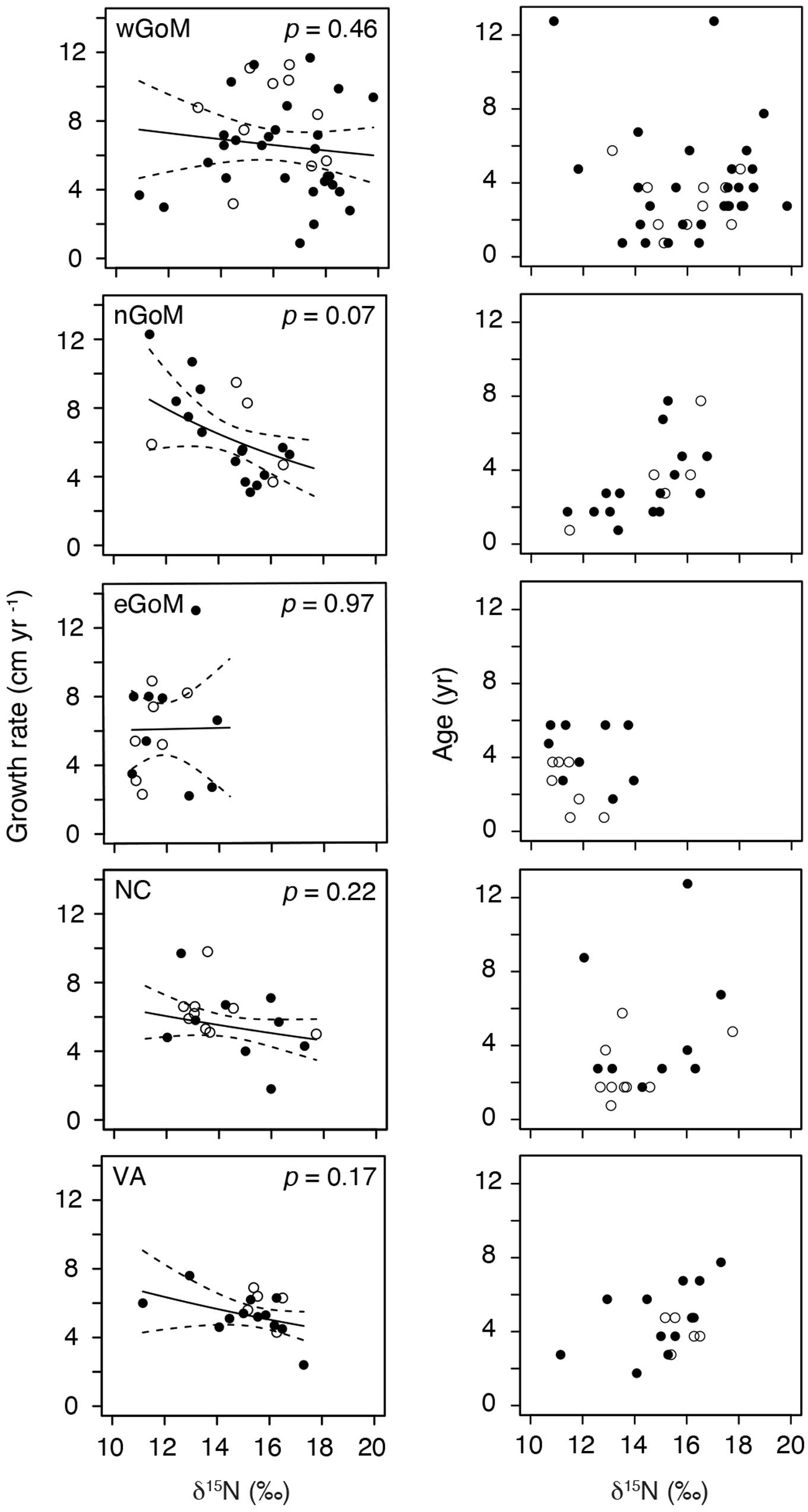
Figure 4. Generalized Linear Model results examining the relationships between annual Kemp’s ridley sea turtle growth rates and δ15N values, and age and δ15N values, for individual turtles by geographic region. Data are for the most recently deposited growth layer prior to death only. Closed circles are true annual growth rates (i.e., turtle stranded in spring, yielding a complete growth interval). Open circles are estimated annual growth rates for turtles that stranded during the winter (November–March); we assumed that annual skeletal growth asymptotes in November (see section “Somatic Growth Rates in Materials and Methods”).
Given the low intra-regional variation in diet composition for most regions, we only examined relationships between estimated diet composition and growth rates for turtles from the wGoM (Figure 5). For these turtles, growth rates were not strongly related to the proportion of fish in turtle diets (P = 0.20). Again, a shallow, non-significant, negative trend was evident in this relationship that did not change following exclusion of marginal growth rates from the analysis. Similar to covariate relationships with δ15N values, the proportional contribution of fish was not strongly related to age (Figure 5).
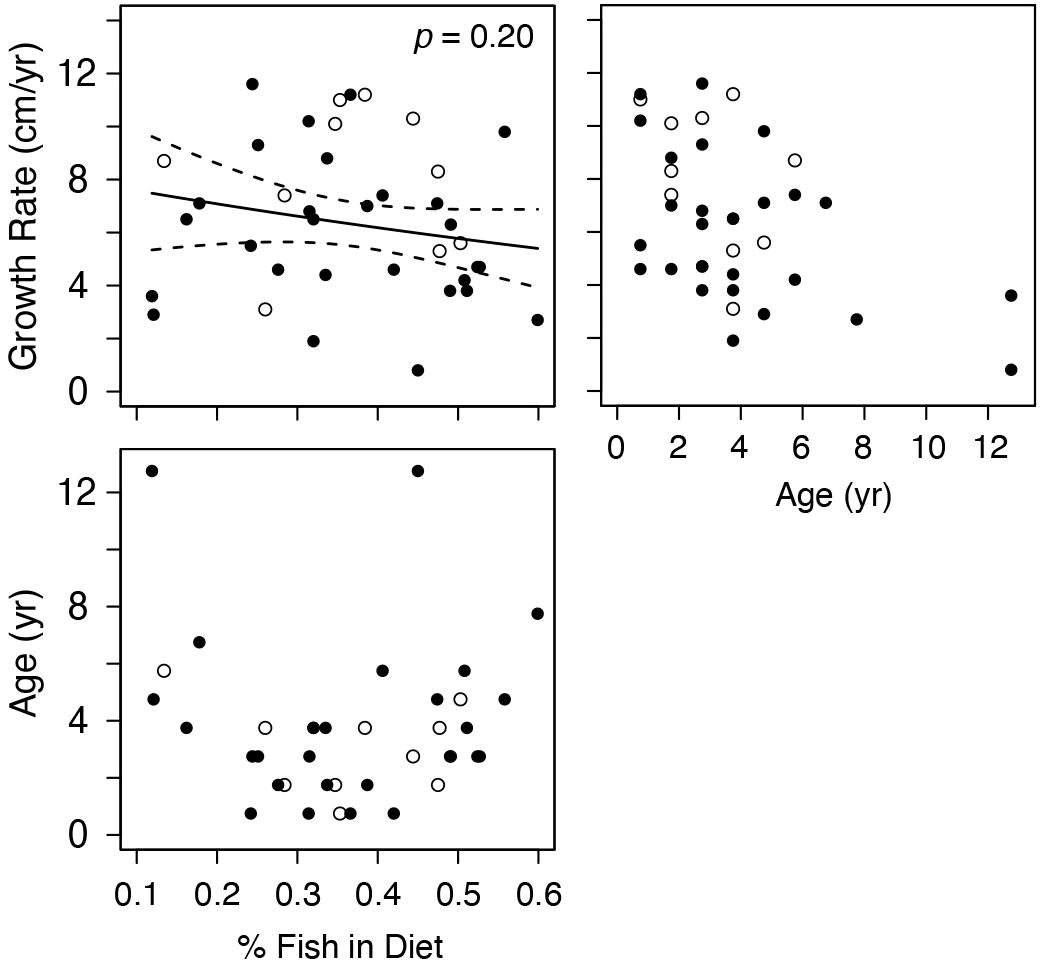
Figure 5. Generalized Linear Model results examining the relationship between annual Kemp’s ridley sea turtle growth rates and proportional contribution of fish to western Gulf of Mexico turtle diets. Diet estimates are derived from a stable isotope mixing model that included informative priors. Data are for the most recently deposited growth layer prior to death only. Closed circles are true annual growth rates (i.e., turtle stranded in spring, yielding a complete growth interval). Open circles are estimated annual growth rates for turtles that stranded during the winter (November–March); we assumed that annual skeletal growth asymptotes in November (see section “Somatic Growth Rates in Materials and Methods”).
Through an integration of multiple skeletal analyses, we provide the first population-level evaluation of Kemp’s ridley diet composition and investigation into the relationship between individual foraging ecology and somatic growth. Our stable isotope mixing models revealed strong regional differences in the proportional contribution of different prey groups to turtle diets that generally followed findings of published gut and fecal content studies. We specifically observed greater contribution of fish to turtle diets in the western GoM and greater contribution of macroalgae/seagrass—or other isotopically similar benthic resources—to turtle diets in the eastern GoM, whereas invertebrates dominated turtle diets in other regions. Through comparative analyses of somatic growth rates, stable isotope values, and mixing model-derived diet composition estimates, we found that individual Kemp’s ridley somatic growth rates were generally poorly correlated with stable isotope-based evidence of turtle trophic ecology within regions. Turtles that foraged higher in the food web (i.e., more fish in diet, higher δ15N values) grew at the same rate as or slower than conspecifics foraging lower in the food web, even after accounting for ontogenetic effects on growth rates. Our results suggest that diet composition alone is not a primary determinant of Kemp’s ridley growth rates, which may be more strongly influenced by other factors such as prey availability, foraging rate and efficiency, and nutritional condition.
Kemp’s ridleys are opportunistic foragers, naturally feeding on a wide range of invertebrate species (Shaver, 1991). Various crab species generally constitute > 75% of total dietary dry mass, whereas molluscs and vegetation generally make up <5–10% (Shaver, 1991; Burke et al., 1993, 1994; Seney and Musick, 2005; Servis et al., 2015; Schmid and Tucker, 2018). In the western and northern GoM, Kemp’s ridleys also consume a significant amount of fish and shrimp. Fish can comprise up to 13.7% of total dietary dry mass and have been reported in 40.1–76.1% of stranded turtle gastrointestinal tracts in these regions (Werner, 1994; Cannon, 1998; Stacy, 2015). Fish prey are most likely obtained as discarded bycatch or bait from fisheries given that Kemp’s ridleys are thought to lack the speed necessary to catch them live (Shoop and Ruckdeschel, 1982; National Research Council, 1990). This conclusion has been supported by the co-occurrence of Nassarius species—molluscs that scavenge dead animal tissues—in turtle stomachs that also contain fish (Shaver, 1991; Bjorndal, 1997). In contrast, fish are an uncommon prey item for Kemp’s ridleys along the United States Atlantic Coast, occurring in a maximum of 16.7% of sampled turtles (Burke et al., 1993, 1994; Seney and Musick, 2005).
Results of our Bayesian isotope mixing models largely follow these patterns, with invertebrates comprising 68.5–97.7% of turtle diets along the United States Atlantic Coast but smaller and more variable proportions within the GoM. In the western GoM, where shrimp fishing effort is relatively high (Scott-Denton et al., 2012), we estimated the region-level contribution of fish to turtle diets was 42.6–43.1%. The similarity in posterior distribution estimates for models with informative and uninformative priors suggests our stable isotope data were highly informative and that these estimates are relatively robust (Moore and Semmens, 2008). Kemp’s ridleys display remarkable plasticity in diet that appears largely driven by local availability rather than preferences for specific prey species (Bjorndal, 1997). Importantly, even with the implementation of bycatch reduction devices, shrimp fishery discard rates are high in the GoM, accounting for ∼50% of total United States fishery discards (Diamond, 2004; Harrington et al., 2005; Scott et al., 2012). It is thus probable that consumption of fish bycatch discarded by shrimp trawlers is a facultative response to local availability in addition to ease of acquisition.
Diet composition estimates for turtles in the northern GoM were similar to those for turtles along the United States Atlantic Coast, with estimated contributions of invertebrates to diets ranging between 65.6 and 94.2%. These results were unexpected given our hypothesis regarding the spatial relationship between shrimp trawl activity and fish consumption, and contrast with recent necropsy results for the region which suggest higher contributions of fish to turtle diets (Stacy, 2015). Even though fishery discard rates are high in the northern GoM, natural prey availability is also high in this region and may be sufficient to support the Kemp’s ridley population. Indeed, blue crab landings in Louisiana represent > 75% of all landings in the Gulf of Mexico, whereas those in Texas comprise only 7% (GSMFC, 2015). The negligible estimated contribution of fish to northern GoM turtle diets may also be due to the close proximity of fish and crustaceans in isospace (Figure 2). Mixing models require sources to be sufficiently separated in order for the model to be able to differentiate them (Parnell et al., 2013). It is thus possible that fish contribute more to Kemp’s ridley diets in this region than our mixing models indicate. Further refinement of the prey stable isotope data to more accurately reflect fish (species and size) and invertebrate species consumed by Kemp’s ridleys may improve mixing model-derived diet estimations for this and other regions.
Within the eastern GoM, we estimated Kemp’s ridley diets primarily comprise invertebrates (43.5–53.6%) and macroalgae/seagrass (42.4–47.8%). These results do not align with the current understanding of Kemp’s ridley diet composition and are likely due to two factors. First, the invertebrate prey groups in the eastern GoM are the most clustered in isospace relative to other regions, with δ13C values for crustaceans and gastropods being particularly low (Figure 2). Such a δ13C mismatch could arise if the eastern GoM crustaceans and gastropods included in our study derived a greater proportion of their carbon from terrestrial vs. marine sources relative to the other regions (Michener and Schell, 1994). This, combined with slightly higher turtle δ13C values in this region, resulted in the largest isotopic mismatch between invertebrates and turtles of all regions after accounting for trophic enrichment. It is thus possible that the prey data included in our mixing model did not accurately reflect those prey groups or turtle diets in this region. Second, it is also possible that our mixing model is missing a key prey source. Notably, tunicates are thought to be an important prey source for Kemp’s ridleys in southwest Florida, occurring in 83.3% of fecal samples and constituting 38.6% of fecal dry mass (n = 64 turtles; Witzell and Schmid, 2005). A dearth of tunicate stable isotope data prevented their inclusion in our mixing models. However, two tunicates sampled in Saint Joseph’s Bay, Florida, had δ15N values of 5.51 and 5.56‰ and δ13C values of −12.72 and −12.78‰ (Williams et al., 2014), which fall within the range of seagrass and macroalgae stable isotope values included in our study. Therefore, our results may in fact reflect consumption of this or another similar benthic resource rather than macroalgae/seagrass.
While isotopic mixing models have greatly advanced our ability to discern diets from isotopic data, their utility and accuracy still rely on substantial ecological knowledge for proper parameterization—these models will always attempt to fit the data, even if the consumers fall outside the mixing space (Phillips and Koch, 2002; Parnell et al., 2010). Given the spatiotemporal scale of this study it was necessary to rely on prey isotopic data from the primary literature, which may have inserted certain biases into the analysis. We ameliorated temporal effects to the best of our abilities by using time-corrected δ13C values. However, it was not possible to overcome spatial biases in sample collection and as a result this may represent the greatest source of bias in our analysis. Kemp’s ridley sea turtles forage in a wide range of shallow, benthic marine habitats, including a substantial part of the continental shelf (Shaver et al., 2013; Hart et al., 2018). Unfortunately, few studies have characterized invertebrate stable isotope values for continental shelf habitats resulting in greater prevalence of estuarine and coastal organisms in our prey isotopic dataset. Given the growing application of stable isotopes to the study of sea turtle foraging and spatial ecology (Pearson et al., 2017; Figgener et al., 2019), quantifying means and variances in known prey stable isotope values across sea turtle ranges should be a high-priority research area. Future analyses using compound-specific isotope analysis of amino acids, which can more accurately estimate consumer trophic position, may also greatly aid in understanding diet variation in sea turtles (Evershed et al., 2007; McMahon and Newsome, 2018).
The lack of strong relationships among bone δ15N values, mixing model-derived diet composition estimates, and somatic growth rates suggests that within-population variation in diet composition may not be a primary determinant of Kemp’s ridley somatic growth variation, and that diet composition may not be a strong driver of the regional (Atlantic vs. GoM) somatic growth differences observed in this species. However, we measured only one component of a sea turtles’ diet—composition—and foraging rate, nutrient assimilation rate, and nutritional status can also strongly influence animal growth rates. Unfortunately, these factors are difficult to study in sea turtles due to their high mobility and conservation status, which has thus far limited investigations into relationships between sea turtle trophic ecology and growth. Wallace et al. (2009) provides the only other comparison of sea turtle trophic ecology and somatic growth where they compared blood plasma δ15N and δ13C values with growth rates of recaptured loggerhead turtles from North Carolina, United States. They found no strong relationships between these covariates and hypothesized that intra-population growth variation may instead be driven by alternative habitat use (coastal vs. oceanic habitat; McClellan and Read, 2007). However, recent research suggests that loggerhead growth dynamics are similar between coastal and oceanic life stages and foragers (Ramirez et al., 2017), indicating that perhaps other factors underlie the observed variability in growth.
Surprisingly, our results suggest that turtles foraging at higher trophic levels may in fact exhibit lower growth rates than conspecifics foraging at lower trophic levels. Our study does not shed light on underlying mechanisms for this pattern, but these findings suggest that foraging strategies that rely on higher trophic level prey may not be energetically optimal for sea turtles. For example, that this energy rich (Williams et al., 2014; Schaafsma et al., 2018), yet presumably similarly digestible (Tibbetts et al., 2006; Peckham et al., 2011), prey does not infer a growth advantage may indicate that the energetic costs associated with searching for and consuming fish (discards) outweigh energetic gains. Similarly, Kemp’s ridleys may not be well adapted to consume fish given that fish are considered an unnatural prey item. Our understanding of sea turtle nutritional ecology is poor for omnivorous species (Bjorndal, 1997), but it is plausible that sea turtles may less efficiently assimilate nutrients from fish relative to invertebrates due to evolutionary constraints.
However, it is also possible that the conditions that lead Kemp’s ridleys to consume fish also contribute to reduced growth rates. If Kemp’s ridleys consume fish due to low natural prey availability or poor condition, turtles may consume fewer resources overall or be nutritionally stressed which would lead to reduced growth rates. Additionally, the tissues of nutritionally stressed animals tend to have higher δ15N values because they catabolize their own tissues for energy (Hobson et al., 1993; Fuller et al., 2005). Given the retrospective nature of our study, we were not able to evaluate the nutritional condition at stranding for sampled turtles. However, necropsies of Kemp’s ridleys stranded in the northern GoM (Louisiana, Mississippi, and Alabama) between 2010 and 2014 suggest there was a decline in stranded turtle nutritional condition during this period (Stacy, 2015). As all but one of the northern GoM humerus bones we sampled were from turtles stranded between 2010 and 2014, the apparent decline in growth rates with increasing δ15N values for this region may be attributed in part to this general decline in turtle nutritional condition in the region. Future studies combining stranded turtle nutritional assays, skeletochronology, and stable isotope analyses would greatly aid in identifying factors underpinning the observed growth patterns.
An important source of uncertainty in our growth analysis is the potential influence of growth rates on isotopic signatures and trophic discrimination factors (TDFs). For neonate loggerhead sea turtles (Caretta caretta), somatic growth can explain up to half of the total rate of isotopic incorporation into blood, skin, and scute tissues, and likely explains age-related differences in nitrogen TDFs (Reich et al., 2008). Indeed, multiple studies have demonstrated that faster growth can reduce Δ15N values because nitrogen input greatly exceeds nitrogen loss—more 14N is retained in the body which lowers δ15N values and reduces isotopic differences between consumers and their prey (Fuller et al., 2004; Martinez del Rio and Wolf, 2005; Reich et al., 2008; Kurle et al., 2014). Such physiological effects, if not accounted for in stable isotope-based studies, can lead to spurious conclusions, particularly in species with distinct ontogenetic changes in size and growth (Villamarín et al., 2018). For our study, a growth-induced decline in Δ15N values may have caused us to underestimate the proportional contribution of fish to turtle diets for faster growing individuals. In contrast, animals that consume large amounts of animal-derived proteins typically have higher Δ15N values (Vander Zanden et al., 2012; Kurle et al., 2014; Turner Tomaszewicz et al., 2017b). A diet-induced increase in Δ15N would therefore potentially have the opposite effect as growth on TDFs, causing an overestimation of the proportional contribution of fish to turtle diets for individuals that forage higher in the food web. Given the sensitivity of our results to changes in Δ15N values, more studies are needed that characterize isotopic routing within sea turtle tissues and effects of diet type and physiology on TDFs, particularly for bone tissue (e.g., Turner Tomaszewicz et al., 2017b).
The integration of skeletal growth and stable isotope analysis provides a powerful tool to reconstruct sea turtle trophic ecology while simultaneously investigating relationships between diet composition and somatic growth rates. Using this approach, we elucidated substantial regional variation in Kemp’s ridley diet composition that aligns with results of site-specific studies of their foraging ecology. This study also provides one of the few quantitative assessments of the relationship between sea turtle trophic ecology and somatic growth. While we present a promising new approach for studying drivers of somatic growth variation in sea turtles, our analysis was limited due to critical data and knowledge gaps. Greater characterization of sea turtle prey stable isotope values throughout the western North Atlantic Ocean, and diet-tissue isotopic discrimination factors, would substantially improve the stable isotope mixing models herein and allow for more robust isotope-based investigations into sea turtle foraging ecology (Pearson et al., 2017; Figgener et al., 2019). Additionally, applications of stable isotope mixing models to Kemp’s ridleys at narrower spatiotemporal scales (e.g., specific foraging grounds, ages, and years) and using greater taxonomic specificity for prey groupings may help reduce sources of uncertainty, improve model estimates, and clarify relationships between diet composition and growth rates (e.g., Wallace et al., 2009; Lemons et al., 2011; Goodman Hall et al., 2015). Integrating additional data gleaned from dead stranded turtles (e.g., gut contents, nutritional condition, and parasite load) into these analyses may also be informative. Ultimately, our analysis further highlights the unique importance of stranded and salvaged turtles to investigating otherwise intractable questions in sea turtle ecology.
All prey isotope datasets, and turtle isotope and growth metadata, generated for this study are included in the article/Supplementary Material. Full turtle isotope and growth datasets are available upon request to the corresponding author.
Ethical review and approval was not required for the animal study because this study only utilized samples opportunistically collected from animals that died naturally and washed up on beaches.
MR, LA, and SH contributed to the conception and design of the study. LA and MC provided samples for analysis. MR, LA, LG, and MS performed skeletochrological analyses. MR performed stable isotope and statistical analyses and wrote the first draft of the manuscript. All authors contributed to manuscript revision and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
MR was funded by the NSF Graduate Research Fellowship Program, National Oceanic and Atmospheric Administration National Marine Fisheries Service (NOAA-NMFS), and NOAA Office of Education Educational Partnership Program (award # NA16SEC4810007). This publications content is solely the responsibility of the award recipient and does not necessarily represent the official views of the United States Department of Commerce, National Oceanic and Atmospheric Administration. Research support was also provided by the NOAA-NMFS and PADI Foundation (award # 28838).
We thank all past and present participants of the Sea Turtle Stranding and Salvage Network whose dedicated work made this study possible. We also thank K. Magnusson for providing lab space and equipment for skeletochronological analyses and J. McKay for assistance with stable isotope analyses. Thank you to J. Cordeiro, M. Davis, H. Hagler, K. McNeely, N. Owen, and M. VanBemmel for assistance with laboratory analyses. Thank you to B. Stacy and J. Keene for collection of humeri associated with the DWH Natural Resource Damage Assessment, and to J. Miller, A. Shiel, J. McKay, H. Haas, C. Allen, and the two reviewers for comments on this manuscript. Research was conducted under USFWS permit number TE-676379-5 issued to the NMFS Southeast Fisheries Science Center.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00253/full#supplementary-material
Abraham, C. L., and Sydeman, W. J. (2004). Ocean climate, euphausiids and auklet nesting: inter-annual trends and variation in phenology, diet and growth of a planktivorous seabird, Ptychoramphus aleuticus. Mar. Ecol. Prog. Ser. 274, 235–250. doi: 10.3354/meps274235
Ambrose, S. H., and DeNiro, M. J. (1986). The isotopic ecology of East African mammals. Oecologia 69, 395–406. doi: 10.1007/BF00377062
Avens, L., Goshe, L. R., Coggins, L., Shaver, D. J., Higgins, B., Landry, A. M., et al. (2017). Variability in age and size at maturation, reproductive longevity, and long-term growth dynamics for Kemp’s ridley sea turtles in the Gulf of Mexico. PLoS One 12:e0173999. doi: 10.1371/journal.pone.0173999
Avens, L., Goshe, L. R., Pajuelo, M., Bjorndal, K. A., MacDonald, B. D., Lemons, G. E., et al. (2013). Complementary skeletochronology and stable isotope analyses offer new insight into juvenile loggerhead sea turtle oceanic stage duration and growth dynamics. Mar. Ecol. Prog. Ser. 491, 235–251. doi: 10.3354/meps10454
Avens, L., and Snover, M. L. (2013). “Age and age estimation in sea turtles,” in The Biology of Sea Turtles, eds J. Wyneken, K. J. Lohmann, and J. A. Musick (Boca Raton, FL: CRC Press), 97–134. doi: 10.1201/b13895-6
Balazs, G. H. (1982). “Growth rates of immature green turtles in the Hawaiian Archipelago,” in Biology and Conservation of Sea Turtles, ed. K. A. Bjorndal (Washington, D.C: Smithsonian Institution Press), 117–125.
Balazs, G. H., and Chaloupka, M. (2004). Spatial and temporal variability in somatic growth of green sea turtles (Chelonia mydas) resident in the Hawaiian Archipelago. Mar. Biol. 145, 1043–1059. doi: 10.1007/s00227-004-1387-6
Benaka, L. R., Bullock, D., Hoover, A. L., and Olsen, N. A. (2019). U.S. National Bycatch Report First Edition Update 3. Silver Spring, MD: U.S. Department of Commerce.
Berry, J. F., and Shine, R. (1980). Sexual size dimorphism and sexual selection in turtles (order testudines). Oecologia 44, 185–191. doi: 10.1007/BF00572678
Bjorndal, K. A. (1997). “Foraging ecology and nutrition of sea turtles,” in The Biology of Sea Turtles, eds P. L. Lutz and J. A. Musick (Boca Raton, FL: CRC Press), 199–231.
Bjorndal, K. A., Bolten, A. B., and Chaloupka, M. Y. (2000). Green turtle somatic growth model: evidence for density dependence. Ecol. Appl. 10, 269–282. doi: 10.2307/2641001
Bjorndal, K. A., Bolten, A. B., Dellinger, T., Delgado, C., and Martins, H. R. (2003). Compensatory growth in oceanic loggerhead sea turtles: response to a stochastic environment. Ecology 84, 1237–1249. doi: 10.1890/0012-9658(2003)084
Bjorndal, K. A., Parsons, J., Mustin, W., and Bolten, A. B. (2013). Threshold to maturity in a long-lived reptile: interactions of age, size, and growth. Mar. Biol. 160, 607–616. doi: 10.1007/s00227-012-2116-1
Black, H. D., Andrus, C. F. T., Lambert, W. J., Rick, T. C., and Gillikin, D. P. (2017). δ15N values in Crassostrea virginica shells provides early direct evidence for nitrogen loading to Chesapeake Bay. Sci. Rep. 7:44241. doi: 10.1038/srep44241
Bond, A. L., and Diamond, A. W. (2011). Recent Bayesian stable-isotope mixing models are highly sensitive to variation in discrimination factors. Ecol. Appl. 21, 1017–1023. doi: 10.1890/09-2409.1
Borrell, A., Abad−Oliva, N., Gómez−Campos, E., Giménez, J., and Aguilar, A. (2012). Discrimination of stable isotopes in fin whale tissues and application to diet assessment in cetaceans. Rapid Commun. Mass Spectrom. 26, 1596–1602. doi: 10.1002/rcm.6267
Bosc, E., Bricaud, A., and Antoine, D. (2004). Seasonal and interannual variability in algal biomass and primary production in the Mediterranean Sea, as derived from 4 years of SeaWiFS observations. Glob. Biogeochem. Cycles 18:GB1005. doi: 10.1029/2003GB002034
Brodeur, R. D., and Ware, D. M. (1992). Long-term variability in zooplankton biomass in the subarctic Pacific Ocean. Fish. Oceanogr. 1, 32–38. doi: 10.1111/j.1365-2419.1992.tb00023.x
Burke, V. J., Morreale, S. J., and Standora, E. A. (1994). Diet of the Kemp’s ridley sea turtle, Lepidochelys kempii, in New York waters. Fish. Bull. 92, 26–32.
Burke, V. J., Standora, E. A., and Morreale, S. J. (1993). Diet of juvenile Kemp’s ridley and loggerhead sea turtles from Long island, New York. Copeia 1993, 1176–1180. doi: 10.2307/1447107
Caillouet, C. W., Fontaine, C. T., Manzella-Tirpak, S. A., and Williams, T. D. (1995). Growth of head-started Kemp’s ridley sea turtles (Lepidochelys kempii) following release. Chelonian Conserv. Biol. 1, 231–234.
Cairns, D. K. (1988). Seabirds as indicators of marine food supplies. Biol. Oceanogr. 5, 261–271. doi: 10.1080/01965581.1987.10749517
Cannon, A. C. (1998). Gross Necropsy Results Of Sea Turtles Stranded On The Upper Texas And Western Louisiana Coasts, 1 January-31 December 1994. Silver Spring, MD: U.S. Department of Commerce.
Chaloupka, M., and Musick, J. A. (1997). “Age, growth, and population dynamics,” in The Biology of Sea Turtles, eds P. L. Lutz and J. A. Musick (Boca Raton, FL: CRC Press), 233–376.
Chamberlain, C. P., Waldbauer, J. R., Fox-Dobbs, K., Newsome, S. D., Koch, P. L., Smith, D. R., et al. (2005). Pleistocene to recent dietary shifts in California condors. PNAS 102, 16707–16711. doi: 10.1073/pnas.0508529102
Cloyed, C. S., Newsome, S. D., and Eason, P. K. (2015). Trophic discrimination factors and incorporation rates of carbon- and nitrogen-stable isotopes in adult green frogs, Lithobates clamitans. Physiol. Biochem. Zool. 88, 576–585. doi: 10.1086/682576
Coleman, A. T., Pitchford, J. L., Bailey, H., and Solangi, M. (2017). Seasonal movements of immature Kemp’s ridley sea turtles (Lepidochelys kempii) in the northern Gulf of Mexico. Aquat. Conserv. Mar. Freshwater Ecosyst. 27, 253–267. doi: 10.1002/aqc.2656
Congdon, J. D. (1989). Proximate and evolutionary constraints on energy relations of reptiles. Physiol. Zool. 62, 356–373. doi: 10.1086/physzool.62.2.30156175
Craig, J. K., Crowder, L. B., Gray, C. D., McDaniel, C. J., Kenwood, T. A., and Hanifen, J. G. (2001). “Ecological effects of hypoxia on fish, sea turtles, and marine mammals in the northwestern Gulf of Mexico,” in Coastal and Estuarine Studies, eds N. N. Rabalais and R. E. Turner (Washington, DC: American Geophysical Union), 269–291. doi: 10.1029/CE058p0269
Crouse, D. T., Crowder, L. B., and Caswell, H. (1987). A stage-based population model for loggerhead sea turtles and implications for conservation. Ecology 68, 1412–1423. doi: 10.2307/1939225
Cushing, D. H. (1990). “Plankton production and year-class strength in fish populations: an update of the match/mismatch hypothesis,” in Advances in Marine Biology, eds J. H. S. Blaxter and A. J. Southward (Cambridge, MA: Academic Press), 249–293. doi: 10.1016/S0065-2881(08)60202-3
Diamond, S. L. (2004). Bycatch quotas in the Gulf of Mexico shrimp trawl fishery: can they work? Rev. Fish Biol. Fish. 14, 207–237. doi: 10.1007/s11160-004-7121-0
Doyle, T. K., Houghton, J. D. R., McDevitt, R., Davenport, J., and Hays, G. C. (2007). The energy density of jellyfish: estimates from bomb-calorimetry and proximate-composition. J. Exp. Mar. Biol. Ecol. 343, 239–252. doi: 10.1016/j.jembe.2006.12.010
Dunham, A. E., Grant, B. W., and Overall, K. L. (1989). Interfaces between biophysical and physiological ecology and the population ecology of terrestrial vertebrate ectotherms. Physiol. Zool. 62, 335–355. doi: 10.1086/physzool.62.2.30156174
Dupont, J., Hallock, P., and Jaap, W. (2010). Ecological impacts of the 2005 red tide on artificial reef epibenthic macroinvertebrate and fish communities in the eastern Gulf of Mexico. Mar. Ecol. Prog. Ser. 415, 189–200. doi: 10.3354/meps08739
Durant, J., Hjermann, D., Ottersen, G., and Stenseth, N. (2007). Climate and the match or mismatch between predator requirements and resource availability. Clim. Res. 33, 271–283. doi: 10.3354/cr033271
Engle, V. D., Hyland, J. L., and Cooksey, C. (2009). Effects of Hurricane Katrina on benthic macroinvertebrate communities along the northern Gulf of Mexico coast. Environ. Monit. Assess. 150, 193–209. doi: 10.1007/s10661-008-0677-8
Evershed, R. P., Bull, I. D., Corr, L. T., Crossman, Z. M., van Dongen, B. E., Evans, C. J., et al. (2007). “Compound-specific stable isotope analysis in ecology and paleoecology,” in Stable Isotopes in Ecology and Environmental Science, eds R. H. Michener and K. Lajtha (Malden, MA: Blackwell Publishing), 480–540. doi: 10.1002/9780470691854.ch14
Figgener, C., Bernardo, J., and Plotkin, P. T. (2019). Beyond trophic morphology: stable isotopes reveal ubiquitous versatility in marine turtle trophic ecology. Biol. Rev. 94, 1947–1973. doi: 10.1111/brv.12543
Fox-Dobbs, K., Bump, J. K., Peterson, R. O., Fox, D. L., and Koch, P. L. (2007). Carnivore-specific stable isotope variables and variation in the foraging ecology of modern and ancient wolf populations: case studies from Isle Royale, Minnesota, and La Brea. Can. J. Zool. 85, 458–471. doi: 10.1139/Z07-018
Francey, R. J., Allison, C. E., Etheridge, D. M., Trudinger, C. M., Enting, I. G., Leuenberger, M., et al. (1999). A 1000-year high precision record of δ13C in atmospheric CO2. Tellus Bull. 51, 170–193. doi: 10.3402/tellusb.v51i2.16269
Francis, R. (1990). Back-calculation of fish length: a critical review. J. Fish Biol. 36, 883–902. doi: 10.1111/j.1095-8649.1990.tb05636.x
Frazer, N. B., Greene, J. L., and Gibbons, J. W. (1993). Temporal variation in growth rate and age at maturity of male painted turtles, Chrysemys picta. Am. Midl. Nat. 130, 314–324. doi: 10.2307/2426130
Frazer, N. B., and Richardson, J. I. (1986). The relationship of clutch size and frequency to body size in loggerhead turtles, Caretta caretta. J. Herpetol. 20, 81–84. doi: 10.2307/1564129
Frick, M. G., and Mason, P. A. (1998). Lepidochelys kempi (Kemp’s ridley sea turtle) diet. Herpetol. Rev. 29, 166–168.
Fritts, A. K., Fritts, M. W., Haag, W. R., DeBoer, J. A., and Casper, A. F. (2017). Freshwater mussel shells (Unionidae) chronicle changes in a North American river over the past 1000 years. Sci. Total Environ. 575, 199–206. doi: 10.1016/j.scitotenv.2016.09.225
Fry, B., and Sherr, E. B. (1989). “13C measurements as indicators of carbon flow in marine and frehswater ecosystems,” in Stable Isotopes in Ecological Research, eds P. W. Rundel, J. R. Ehleringer, and K. A. Nagy (New York, NY: Springer), 196–229. doi: 10.1007/978-1-4612-3498-2_12
Fuller, B. T., Fuller, J. L., Sage, N. E., Harris, D. A., O’Connell, T. C., and Hedges, R. E. M. (2004). Nitrogen balance and δ15N: why you’re not what you eat during pregnancy. Rapid Commun. Mass Spectrom. 18, 2889–2896. doi: 10.1002/rcm.1708
Fuller, B. T., Fuller, J. L., Sage, N. E., Harris, D. A., O’Connell, T. C., and Hedges, R. E. M. (2005). Nitrogen balance and δ15N: why you’re not what you eat during nutritional stress. Rapid Commun. Mass Spectrom. 19, 2497–2506. doi: 10.1002/rcm.2090
Gelman, A., and Rubin, D. B. (1992). Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 457–472. doi: 10.1214/ss/1177011136
Gerber, L. R., and Heppell, S. S. (2004). The use of demographic sensitivity analysis in marine species conservation planning. Biol. Conserv. 120, 121–128. doi: 10.1016/j.biocon.2004.01.029
Geweke, J. (1992). “Evaluating the accuracy of sampling-based approaches to the calculation of posterior moments,” in Bayesian Statistics 4, eds J. M. Bernardo, J. O. Berger, A. P. Dawid, and A. F. M. Smith (Oxford: Claredon Press).
Gibbons, J. W. (1967). Variation in growth rates in three populations of the painted turtle, Chrysemys picta. Herpetologica 23, 296–303.
Goodman Hall, A. G., Avens, L., McNeill, J. B., Wallace, B., and Goshe, L. R. (2015). Inferring long-term foraging trends of individual juvenile loggerhead sea turtles using stable isotopes. Mar. Ecol. Prog. Ser. 537, 265–276. doi: 10.3354/meps11452
GSMFC (2015). The Blue Crab Fishery of the Gulf of Mexico: A Regional Management Plan. Ocean Springs, MS: Gulf State Marine Fisheries Commission.
Harrington, J. M., Myers, R. A., and Rosenberg, A. A. (2005). Wasted fishery resources: discarded by-catch in the USA. Fish Fish. 6, 350–361. doi: 10.1111/j.1467-2979.2005.00201.x
Hart, K. M., Iverson, A. R., Fujisaki, I., Lamont, M. M., Bucklin, D., and Shaver, D. J. (2018). Marine threats overlap key foraging habitat for two imperiled sea turtle species in the Gulf of Mexico. Front. Mar. Sci. 5:336. doi: 10.3389/fmars.2018.00336
Hatase, H., Omuta, K., and Tsukamoto, K. (2010). Oceanic residents, neritic migrants: a possible mechanism underlying foraging dichotomy in adult female loggerhead turtles (Caretta caretta). Mar. Biol. 157, 1337–1342. doi: 10.1007/s00227-010-1413-9
Hennicke, J. C., and Culik, B. M. (2005). Foraging performance and reproductive success of Humboldt penguins in relation to prey availability. Mar. Ecol. Prog. Ser. 296, 173–181. doi: 10.3354/meps296173
Higgins, B., Cannon, A., and Gitschlag, G. (2007). Sea Turtle Decomposition Study. Unpublished Report. Galveston, TX: National Marine Fisheries Service.
Hobson, K. A., Alisauskas, R. T., and Clark, R. G. (1993). Stable-nitrogen isotope enrichment in avian tissues due to fasting and nutritional stress: implications for isotopic analyses of diet. Condor 95, 388–394. doi: 10.2307/1369361
Hobson, K. A., and Clark, R. G. (1992). Assessing avian diets using stable isotopes II: factors influencing diet-tissue fractionation. Condor 94, 189–197. doi: 10.2307/1368808
Hochscheid, S., Bentivegna, F., Bradai, M., and Hays, G. (2007). Overwintering behaviour in sea turtles: dormancy is optional. Mar. Ecol. Prog. Ser. 340, 287–298. doi: 10.3354/meps340287
Katzenberg, M. A. (2008). “Stable isotope analysis: a tool for studying past diet, demography, and life history,” in Biological Anthropology of the Human Skeleton, eds M. A. Katzenberg and S. R. Saunders (Hoboken, NJ: John Wiley & Sons, Inc), 413–441.
Keeling, C. D., Mook, W. G., and Tans, P. P. (1979). Recent trends in the 13C/12C ratio of atmospheric carbon dioxide. Nature 277, 121–122. doi: 10.1038/277121a0
Keeling, C. D., Piper, S. C., Bacastow, R. B., Wahlen, M., Whorf, T. P., Heimann, M., et al. (2001). Exchanges of Atmospheric CO2 and 13CO2 with the Terrestrial Biosphere And Oceans from 1978 to 2000. I. Global Aspects. SIO Reference Series, No. 01-06. San Diego: Scripps Institution of Oceanography.
Kim, S. L., Tinker, M. T., Estes, J. A., and Koch, P. L. (2012). Ontogenetic and among-individual variation in foraging strategies of Northeast Pacific white sharks based on stable isotope analysis. PLoS One 7:e45068. doi: 10.1371/journal.pone.0045068
Kurle, C. M., Koch, P. L., Tershy, B. R., and Croll, D. A. (2014). The effects of sex, tissue type, and dietary components on stable isotope discrimination factors (Δ13C and Δ15N) in mammalian omnivores. Isotopes Environ. Health Stud. 50, 307–321. doi: 10.1080/10256016.2014.908872
Lemons, G., Lewison, R., Komoroske, L., Gaos, A., Lai, C.-T., Dutton, P., et al. (2011). Trophic ecology of green sea turtles in a highly urbanized bay: insights from stable isotopes and mixing models. J. Exp. Mar. Biol. Ecol. 405, 25–32. doi: 10.1016/j.jembe.2011.05.012
Lenes, J. M., Darrow, B. P., Cattrall, C., Heil, C. A., Callahan, M., Vargo, G. A., et al. (2001). Iron fertilization and the Trichodesmium response on the West Florida shelf. Limnol. Oceanogr. 46, 1261–1277. doi: 10.4319/lo.2001.46.6.1261
Martinez del Rio, C., and Wolf, B. O. (2005). “Mass-balance models for animal isotopic ecology,” in Physiological and Ecological Adaptations to Feeding in Vertebrates, eds J. M. Starck and T. Wang (Enfield: Science Publishers), 141–174.
Matsubayashi, J., Saitoh, Y., Osada, Y., Uehara, Y., Habu, J., Sasaki, T., et al. (2017). Incremental analysis of vertebral centra can reconstruct the stable isotope chronology of teleost fishes. Methods Ecol. Evol. 8, 1755–1763. doi: 10.1111/2041-210X.12834
McClellan, C. M., and Read, A. J. (2007). Complexity and variation in loggerhead sea turtle life history. Biol. Lett. 3, 592–594. doi: 10.1098/rsbl.2007.0355
McMahon, K. W., and Newsome, S. D. (2018). “Amino acid isotope analysis: a new frontier in studies of animal migration and foraging ecology,” in Tracking Animal Migration with Stable Isotopes, eds K. A. Hobson and L. I. Wassenaar (London, UK: Academic Press), 173–190. doi: 10.1016/b978-0-12-814723-8.00007-6
McMichael, E., Seminoff, J., and Carthy, R. (2008). Growth rates of wild green turtles, Chelonia mydas, at a temperate foraging habitat in the northern Gulf of Mexico: assessing short−term effects of cold−stunning on growth. J. Nat. Hist. 42, 2793–2807. doi: 10.1080/00222930802357335
Michener, R. H., and Schell, D. M. (1994). “Stable isotope ratios as tracers in marine aquatic food webs,” in Stable Isotopes in Ecology and Environmental Science, eds K. Lajtha and R. H. Michener (Oxford: Blackwell), 138–157.
Montoya, J. P., Carpenter, E. J., and Capone, D. G. (2002). Nitrogen fixation and nitrogen isotope abundances in zooplankton of the oligotrophic North Atlantic. Limnol. Oceanogr. 47, 1617–1628. doi: 10.4319/lo.2002.47.6.1617
Moore, J. W., and Semmens, B. X. (2008). Incorporating uncertainty and prior information into stable isotope mixing models. Ecol. Lett. 11, 470–480. doi: 10.1111/j.1461-0248.2008.01163.x
Mulholland, M. R., Bernhardt, P. W., Heil, C. A., Bronk, D. A., and O’Neil, J. M. (2006). Nitrogen fixation and release of fixed nitrogen by Trichodesmium spp. in the Gulf of Mexico. Limnol. Oceanogr. 51, 1762–1776. doi: 10.4319/lo.2006.51.4.1762
National Research Council (1990). Decline of the Sea Turtles: Causes and Prevention. Washington, DC: National Academy Press.
Nero, R., Cook, M., Coleman, A., Solangi, M., and Hardy, R. (2013). Using an ocean model to predict likely drift tracks of sea turtle carcasses in the north central Gulf of Mexico. Endanger. Species Res. 21, 191–203. doi: 10.3354/esr00516
Newsome, S. D., Martinez del Rio, C., Bearhop, S., and Phillips, D. L. (2007). A niche for isotopic ecology. Front. Ecol. Environ. 5:429–436. doi: 10.1890/060150.1
NMFS, and USFWS (2015). Kemp’s Ridley Sea Turtle (Lepidochelys kempii) 5-Year Review: Summary and Evaluation. Silver Spring, MD: National Marine Fisheries Service.
O’Brien, S., Robert, B., and Tiandray, H. (2005). Hatch size, somatic growth rate and size-dependent survival in the endangered ploughshare tortoise. Biol. Conserv. 126, 141–145. doi: 10.1016/j.biocon.2005.03.022
Omeyer, L., Godley, B., and Broderick, A. (2017). Growth rates of adult sea turtles. Endanger. Species Res. 34, 357–371. doi: 10.3354/esr00862
Parnell, A. C., Inger, R., Bearhop, S., and Jackson, A. L. (2010). Source partitioning using stable isotopes: coping with too much variation. PLoS One 5:e9672. doi: 10.1371/journal.pone.0009672
Parnell, A. C., Phillips, D. L., Bearhop, S., Semmens, B. X., Ward, E. J., Moore, J. W., et al. (2013). Bayesian stable isotope mixing models. Environmetrics 24, 387–399. doi: 10.1002/env.2221
Pearson, R., van de Merwe, J., Limpus, C., and Connolly, R. (2017). Realignment of sea turtle isotope studies needed to match conservation priorities. Mar. Ecol. Prog. Ser. 583, 259–271. doi: 10.3354/meps12353
Peckham, S. H., Maldonado Diaz, D., Tremblay, Y., Ochoa, R., Polovina, J., Balazs, G., et al. (2011). Demographic implications of alternative foraging strategies in juvenile loggerhead turtles Caretta caretta of the North Pacific Ocean. Mar. Ecol. Prog. Ser. 425, 269–280. doi: 10.3354/meps08995
Phillips, D. L. (2001). Mixing models in analyses of diet using multiple stable isotopes: a critique. Oecologia 127, 166–170. doi: 10.1007/s004420000571
Phillips, D. L., and Koch, P. L. (2002). Incorporating concentration dependence in stable isotope mixing models. Oecologia 130, 114–125. doi: 10.1007/s004420100786
Phillips, D. L., Newsome, S. D., and Gregg, J. W. (2005). Combining sources in stable isotope mixing models: alternative methods. Oecologia 144, 520–527. doi: 10.1007/s00442-004-1816-8
Piatt, J. F., Harding, A. M. A., Shultz, M., Speckman, S. G., van Pelt, T. I., Drew, G. S., et al. (2007). Seabirds as indicators of marine food supplies: cairns revisited. Mar. Ecol. Prog. Ser. 352, 221–234. doi: 10.3354/meps07078
Piovano, S., Clusa, M., Carreras, C., Giacoma, C., Pascual, M., and Cardona, L. (2011). Different growth rates between loggerhead sea turtles (Caretta caretta) of Mediterranean and Atlantic origin in the Mediterranean Sea. Mar. Biol. 158, 2577–2587. doi: 10.1007/s00227-011-1759-7
Post, D. M., Layman, C. A., Arrington, D. A., Takimoto, G., Quattrochi, J., and Montaña, C. G. (2007). Getting to the fat of the matter: models, methods and assumptions for dealing with lipids in stable isotope analyses. Oecologia 152, 179–189. doi: 10.1007/s00442-006-0630-x
R Core Team (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Ramirez, M., Miller, J., Parks, E., Avens, L., Goshe, L., Seminoff, J., et al. (2019). Reconstructing sea turtle ontogenetic habitat shifts through trace element analysis of bone tissue. Mar. Ecol. Prog. Ser. 608, 247–262. doi: 10.3354/meps12796
Ramirez, M. D. (2019). It’s in Their Bones: Ecological drivers of Kemp’s Ridley Sea Turtle (Lepidochelys kempii) Somatic Growth and Population Dynamics, Ph.D. Dissertation, Oregon State University, Corvallis, OR.
Ramirez, M. D., Avens, L., Seminoff, J. A., Goshe, L. R., and Heppell, S. S. (2017). Growth dynamics of juvenile loggerhead sea turtles undergoing an ontogenetic habitat shift. Oecologia 183, 1087–1099. doi: 10.1007/s00442-017-3832-5
Reich, K. J., Bjorndal, K. A., and Martínez del Rio, C. (2008). Effects of growth and tissue type on the kinetics of 13C and 15N incorporation in a rapidly growing ectotherm. Oecologia 155, 651–663. doi: 10.1007/s00442-007-0949-y
Renaud, M. L., and Williams, J. A. (2005). Kemp’s ridley sea turtle movements and migrations. Chelonian Conserv. Biol. 4, 808–816.
Robinson, K. L., Ruzicka, J. J., Hernandez, F. J., Graham, W. M., Decker, M. B., Brodeur, R. D., et al. (2015). Evaluating energy flows through jellyfish and gulf menhaden (Brevoortia patronus) and the effects of fishing on the northern Gulf of Mexico ecosystem. ICES J. Mar. Sci. 72, 2301–2312. doi: 10.1093/icesjms/fsv088
Sanchez-Rubio, G., Perry, H. M., Biesiot, P. M., Johnson, D. R., and Lipcius, R. N. (2011). Climate-related hydrological regimes and their effects on abundance of juvenile blue crabs (Callinectes sapidus) in the northcentral Gulf of Mexico. Fish. Bull. 109, 139–146.
Santos, B. S., Friedrichs, M. A. M., Rose, S. A., Barco, S. G., and Kaplan, D. M. (2018). Likely locations of sea turtle stranding mortality using experimentally-calibrated, time and space-specific drift models. Biol. Conserv. 226, 127–143. doi: 10.1016/j.biocon.2018.06.029
Schaafsma, F. L., Cherel, Y., Flores, H., van Franeker, J. A., Lea, M.-A., Raymond, B., et al. (2018). Review: the energetic value of zooplankton and nekton species of the Southern Ocean. Mar. Biol. 165, 129. doi: 10.1007/s00227-018-3386-z
Schmid, J. R., and Tucker, A. D. (2018). Comparing diets of Kemp’s ridley sea turtles (Lepidochelys kempii) in mangrove estuaries of Southwest Florida. J. Herpetol. 52, 252–258. doi: 10.1670/16-164
Schmid, J. R., and Witzell, W. N. (2006). Seasonal migrations of immature Kemp’s ridley turtles (Lepidochelys kempii) along the West coast of Florida. Gulf Mex. Sci. 24, 28–40.
Scott, R., Marsh, R., and Hays, G. C. (2012). Life in the really slow lane: loggerhead sea turtles mature late relative to other reptiles. Funct. Ecol. 26, 227–235. doi: 10.1111/j.1365-2435.2011.01915.x
Scott-Denton, E., Cryer, P. F., Duffy, M. R., Gocke, J. P., Harrelson, M. R., Kinsella, D. L., et al. (2012). Characterization of the U.S. Gulf of Mexico and South Atlantic Penaeid and rock shrimp fisheries based on observer data. Mar. Fish. Rev. 74, 1–27.
Semmens, B. X., Ward, E. J., Moore, J. W., and Darimont, C. T. (2009). Quantifying inter- and intra-population niche variability using hierarchical Bayesian stable isotope mixing models. PLoS One 4:e6187. doi: 10.1371/journal.pone.0006187
Seney, E., and Landry, A. M. Jr. (2011). Movement patterns of immature and adult female Kemp’s ridley sea turtles in the northwestern Gulf of Mexico. Mar. Ecol. Prog. Ser. 440, 241–254. doi: 10.3354/meps09380
Seney, E. E. (2016). Diet of Kemp’s ridley sea turtles incidentally caught on recreational fishing gear in the Northwestern Gulf of Mexico. Chelonian Conserv. Biol. 15, 132–137. doi: 10.2744/CCB-1191.1
Seney, E. E., and Musick, J. A. (2005). Diet analysis of Kemp’s ridley sea turtles (Lepidochelys kempii) in Virginia. Chelonian Conserv. Biol. 4, 864–871.
Servis, J. A., Lovewell, G., and Tucker, A. D. (2015). Diet analysis of subadult Kemp’s ridley (Lepidochelys kempii) turtles from West-central Florida. Chelonian Conserv. Biol. 14, 173–181. doi: 10.2744/ccb-1177.1
Shaver, D. J. (1991). Feeding ecology of wild and head-started Kemp’s ridley sea turtles in South Texas waters. J. Herpetol. 25, 327–334. doi: 10.2307/1564592
Shaver, D. J., Hart, K. M., Fujisaki, I., Rubio, C., Sartain, A. R., Peña, J., et al. (2013). Foraging area fidelity for Kemp’s ridleys in the Gulf of Mexico. Ecol. Evol. 3, 2002–2012. doi: 10.1002/ece3.594
Shaver, D. J., and Rubio, C. (2008). Post-nesting movement of wild and head-started Kemp’s ridley sea turtles Lepidochelys kempii in the Gulf of Mexico. Endanger. Species Res 4, 43–55. doi: 10.3354/esr00061
Shoop, C. R., and Ruckdeschel, C. (1982). Increasing turtle strandings in the southeast United States: a complicating factor. Biol. Conserv. 23, 213–215. doi: 10.1016/0006-3207(82)90076-3
Snover, M. L., Avens, L., and Hohn, A. A. (2007). Back-calculating length from skeletal growth marks in loggerhead sea turtles Caretta caretta. Endanger. Species Res. 3, 95–104. doi: 10.3354/esr003095
Snover, M. L., and Hohn, A. A. (2004). Validation and interpretation of annual skeletal marks in loggerhead (Caretta caretta) and Kemp’s ridley (Lepidochelys kempii) sea turtles. Fish. Bull. 102, 682–692.
Snover, M. L., Hohn, A. A., Crowder, L. B., and Macko, S. A. (2010). Combining stable isotopes and skeletal growth marks to detect habitat shifts in juvenile loggerhead sea turtles Caretta caretta. Endanger. Species Res. 13, 25–31. doi: 10.3354/esr00311
Stacy, B. A. (2015). Summary of Necropsy Findings for Non-Visibly Oiled Sea Turtles Documented by Stranding Response in Alabama, Louisiana, and Mississippi 2010 through 2014. DWH Sea Turtles NRDA Technical Working Group Report. Silver Spring, MD: U.S. Department of Commerce.
Stock, B. C., Jackson, A. L., Ward, E. J., Parnell, A. C., Phillips, D. L., and Semmens, B. X. (2018). Analyzing mixing systems using a new generation of Bayesian tracer mixing models. PeerJ 6:e5096. doi: 10.7717/peerj.5096
Stock, B. C., and Semmens, B. X. (2016). Unifying error structures in commonly used biotracer mixing models. Ecology 97, 2562–2569. doi: 10.1002/ecy.1517
Tibbetts, S. M., Milley, J. E., and Lall, S. P. (2006). Apparent protein and energy digestibility of common and alternative feed ingredients by Atlantic cod, Gadus morhua (Linnaeus, 1758). Aquaculture 261, 1314–1327. doi: 10.1016/j.aquaculture.2006.08.052
Turner Tomaszewicz, C. N., Seminoff, J. A., Peckham, S. H., Avens, L., and Kurle, C. M. (2017a). Intrapopulation variability in the timing of ontogenetic habitat shifts in sea turtles revealed using δ15N values from bone growth rings. J. Anim. Ecol. 86, 694–704. doi: 10.1111/1365-2656.12618
Turner Tomaszewicz, C. N., Seminoff, J. A., Price, M., and Kurle, C. M. (2017b). Stable isotope discrimination factors and between-tissue isotope comparisons for bone and skin from captive and wild green sea turtles (Chelonia mydas). Rapid Commun. Mass Spectrom. 31, 1903–1914. doi: 10.1002/rcm.7974
Turner Tomaszewicz, C. N., Seminoff, J. A., Ramirez, M. D., and Kurle, C. M. (2015). Effects of demineralization on the stable isotope analysis of bone samples. Rapid Commun. Mass Spectrom. 29, 1879–1888. doi: 10.1002/rcm.7295
Vander Zanden, H. B., Bjorndal, K. A., Mustin, W., Ponciano, J. M., and Bolten, A. B. (2012). Inherent variation in stable isotope values and discrimination factors in two life stages of green turtles. Physiol. Biochem. Zool. 85, 431–441. doi: 10.1086/666902
Vander Zanden, H. B., Tucker, A. D., Hart, K. M., Lamont, M. M., Fujisaki, I., Addison, D. S., et al. (2015). Determining origin in a migratory marine vertebrate: a novel method to integrate stable isotopes and satellite tracking. Ecol. Appl. 25, 320–335. doi: 10.1890/14-0581.1
Villamarín, F., Jardine, T. D., Bunn, S. E., Marioni, B., and Magnusson, W. E. (2018). Body size is more important than diet in determining stable-isotope estimates of trophic position in crocodilians. Sci. Rep. 8:2020. doi: 10.1038/s41598-018-19918-6
Wallace, B. P., Avens, L., Braun-McNeill, J., and McClellan, C. M. (2009). The diet composition of immature loggerheads: insights on trophic niche, growth rates, and fisheries interactions. J. Exp. Mar. Biol. Ecol. 373, 50–57. doi: 10.1016/j.jembe.2009.03.006
Wallace, B. P., Brosnan, T., McLamb, D., Rowles, T., Ruder, E., Schroeder, B., et al. (2017). Effects of the Deepwater Horizon oil spill on protected marine species. Endanger. Species Res. 33, 1–7. doi: 10.3354/esr00789
Webb, E. C., Stewart, A., Miller, B., Tarlton, J., and Evershed, R. P. (2016). Age effects and the influence of varying proportions of terrestrial and marine dietary protein on the stable nitrogen-isotope compositions of pig bone collagen and soft tissues from a controlled feeding experiment. Sci. Technol. Archaeol. Res. 2, 54–66. doi: 10.1080/20548923.2015.1133121
Werner, E. E., and Gilliam, J. F. (1984). The ontogenetic niche and species interactions in size-structured populations. Annu. Rev. Ecol. Syst. 15, 393–425. doi: 10.1146/annurev.es.15.110184.002141
Werner, S. A. (1994). Feeding Ecology of Wild and Head Stared Kemp’s Ridley Sea Turtles. Ph.D. Thesis Texas A&M University, College Station, TX.
Williams, N. C., Bjorndal, K. A., Lamont, M. M., and Carthy, R. R. (2014). Winter diets of immature green turtles (Chelonia mydas) on a northern feeding ground: integrating stomach contents and stable isotope analyses. Estuar. Coast. 37, 986–994. doi: 10.1007/s12237-013-9741-x
Witzell, W. N., and Schmid, J. R. (2005). Diet of immature Kemp’s ridley turtles (Lepidochelys kempii) from Gullivan Bay, ten thousand islands, southwest Florida. Bull. Mar. Sci. 77, 191–200.
Wood, S. N. (2006). Generalized Additive Models: An Introduction with R. Boca Raton, FL: Chapman and Hall.
Keywords: stable isotope analysis, skeletochronology, mixing model, growth rates, diet analysis, foraging ecology
Citation: Ramirez MD, Avens L, Goshe LR, Snover ML, Cook M and Heppell SS (2020) Regional Variation in Kemp’s Ridley Sea Turtle Diet Composition and Its Potential Relationship With Somatic Growth. Front. Mar. Sci. 7:253. doi: 10.3389/fmars.2020.00253
Received: 10 January 2020; Accepted: 30 March 2020;
Published: 22 April 2020.
Edited by:
Donna Jill Shaver, National Park Service, United StatesReviewed by:
Federico Alberto Abreu-Grobois, National Autonomous University of Mexico, MexicoCopyright © 2020 Ramirez, Avens, Goshe, Snover, Cook and Heppell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Matthew D. Ramirez, bWRyYW1pcmV6QHVyaS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.