- 1Institute for Marine and Antarctic Studies, University of Tasmania, Hobart, TAS, Australia
- 2Antarctic Climate and Ecosystems Cooperative Research Centre, University of Tasmania, Hobart, TAS, Australia
Marine host-associated microbiomes can strongly influence their host’s function and are shaped by selection, dispersal, diversification, and drift. These processes can lead to spatially structured microbiomes, with potential implications for host fitness in different locations. We review the literature on marine host-associated microbiomes to identify if spatially structured microbiomes are more prevalent in certain taxonomic groups, are linked to species traits, or sampling design and methodology. The 28 papers analyzed represented 38 host species, with spatial structure detected in 75% of species, increasing to 83% when restricted to studies using high-throughput DNA sequencing. Spatial structure was detected in all coral and marine mammal microbiomes, but was less common in fish (69%) and sponges (46%). Mobile species and external tissues were more likely to show spatially structured microbiomes than sessile species and internal tissues. We found no relationship between spatial structuring and maximum distance between sampling sites, with studies on large (>1000 km) and small spatial scales (<100 km) almost as likely to show spatial structure (87% vs. 79%). Our results support using high-throughput sequencing for studying marine host-associated microbiomes due to better taxonomic resolution compared to other methods. Given the observed generality of spatially structured microbiomes, future studies should test whether microbiome variation between locations affects host fitness. Researchers should include sufficient environmental microbiome sampling and host data to distinguish host and environmental effects. This will help resolve the relative importance of selection, dispersal, diversification and drift in shaping marine host-associated microbiomes.
Introduction
Host-associated microbiomes (including bacteria, unicellular eukaryotes and fungi) can strongly influence their host’s function (O’Brien et al., 2019). For example, corals depend on symbiotic relationships with photosynthetic Symbiodiniaceae dinoflagellates, and heterotrophic foraminifera and radiolarians also house endosymbiotic microalgae. Microbial influence on host function can include semi-permanent microbially mediated adaptation (Correa and Baker, 2011; Sison-Mangus et al., 2014) or acclimation (Dittami et al., 2016; Röthig et al., 2016) of the host to their environment (e.g., to temperature and salinity). Additionally, host-associated microbiomes can also influence and reflect the host’s health (e.g., skin microbiome of marine mammals, Bierlich et al., 2018). The green alga and major marine primary producer Ostreococcus exchanges B vitamins with bacterial partners, highlighting the importance of mutualistic interactions between the microbiome and the host (Cooper et al., 2019). Sponge and coral microbiomes also play important roles in cycling key nutrients including phosphorus, carbon, nitrogen, and sulfur (Raina et al., 2009; Pita et al., 2018).
Marine host-associated microbiomes vary between genera (Pita et al., 2013a), species (Reveillaud et al., 2014), and between individuals (Datta et al., 2018). The spatial structure of host-associated microbial communities is crucial for understanding potential effects on the host’s ecology and physiology (Mark Welch et al., 2016). For the purposes of this review, we define spatially structured microbiomes in terms of biogeography, i.e., if individuals from a given geographic location share more similar microbiomes than with individuals from other locations, the species shows a spatially structured microbiome, as opposed to micro-scale or tissue/niche-based spatial structuring. There is still a knowledge gap regarding large-scale patterns of microbial distribution among ecosystems (Nemergut et al., 2013), including host-associated microbiomes. Host-associated microbes from different geographic areas might have a different function despite similar environmental conditions (Martiny et al., 2006). Therefore, microbial biogeography studies of host-associated communities are key for predicting effects on both the host organism and ecosystem (Martiny et al., 2006).
Host-associated microbial communities are shaped by four ecological processes, consisting of selection, dispersal, diversification, and ecological drift (Hanson et al., 2012; Zhou and Ning, 2017). Selection shapes communities due to fitness differences, including survival, growth and reproduction, between community members in a given environment (Vellend, 2010; Stegen et al., 2015; Zhou and Ning, 2017). Selection can be influenced by abiotic and biotic factors on local and regional scales (Zhou and Ning, 2017). Additionally, the host organism itself can exert selection pressure on the microbiome, for example through the immune system (Müller and Müller, 2003; Sipkema et al., 2015). Dispersal describes movement and successful colonization across space, which can be passive or active (Vellend, 2010; Hanson et al., 2012; Zhou and Ning, 2017). Marine microbes are considered to disperse passively (e.g., by ocean currents, Troussellier et al., 2017) due to their restricted ability to move large distances. Marine host-associated microbes can be dispersed either with a mobile host or separate from the host in the water column. High dispersal rates can decrease the difference between microbiomes in different locations, reducing spatial structure through homogenizing dispersal, while low dispersal rates, interacting with other processes such as drift and selection, can increase differentiation between locations, known as dispersal limitation (Stegen et al., 2013, 2015). Oceanographic barriers (e.g., fronts) influence dispersal ability of marine microbes (Martiny et al., 2006). Diversification involves new genetic variation arising from mutations, which for bacteria includes horizontal gene transfer and recombination in general. Diversification typically affects the species pool over large spatial and temporal scales, although evolution through mutation can be much faster, and even actively promoted, within microbial communities (Rensing et al., 2002; Vellend, 2010; Nemergut et al., 2013; Zhou and Ning, 2017). The fourth process, drift, is due to random fluctuations in abundance, which is more important when the community is small and other processes (e.g., selection) are weak (Chase and Myers, 2011; Zhou and Ning, 2017). Ecological drift, however, requires individuals of different species to be demographically identical, which is extremely unlikely (Vellend, 2010). The interaction of these four processes determines whether host-associated microbiomes are spatially structured.
Many studies have examined the spatial structure of marine host-associated microbiomes, mostly within a single or a few related species. This approach has left a knowledge gap as to whether host-associated microbiomes are influenced by host taxonomy, species traits or study design (e.g., tissue sampled, spatial separation of samples, and sequencing method). First, we describe the ecological processes structuring marine host-associated microbiomes. We then compile the results from the studies to date to identify if spatially structured microbiomes are more prevalent in (i) certain taxonomic groups (e.g., fish, corals, sponges, and marine mammals), (ii) species with certain traits (mobile or sessile species), (iii) certain host tissues, (iv) studies with a broader spatial scale of sampling, or (v) studies using high-throughput DNA sequencing compared to non-sequencing approaches (e.g., DGGE/TRFLP). Factors known to influence host-associated microbiomes of fish, coral, sponge, and marine mammals are described. Based on these findings, we provide recommendations for future studies of marine host-associated microbiomes.
Materials and Methods
We searched the scientific literature for studies that examined marine host-associated microbiomes for one or more species in multiple locations, and tested for significant differences between locations for individual species. Initially papers were chosen based on our knowledge of the literature, as well as papers cited in or citing the most relevant papers. We also used Google Scholar and Scopus to find papers with the keywords “microbiome,” “biogeography,” and/or “spatial structure” as well as each of the key taxonomic groups (“coral,” “fish,” “sponge,” “whale,” and “dolphin” etc.). Studies with only one sampling site, or with only two samples per site (hence low statistical power), or no species-specific data were excluded from our analysis. For each host species, we scored the presence or absence (1 or 0) of statistically significant microbiome spatial structure, with a P-value threshold of 0.05. One species where the skin microbiome showed spatial structure but the gut microbiome did not (the fish Elacatinus prochilos) was scored as 0.5 for presence and 0.5 for absence. Additionally, we noted the taxonomy and mobility of the host species, the tissue type examined, the number of locations and individuals sampled, the maximum geographic distance between sites and whether high-throughput sequencing was used to characterize the microbiome.
Results
Taxonomy and Species Traits
We found 28 papers (representing 38 species) that examined marine host-associated microbiomes for one or more species in multiple locations, and tested for significant differences between locations for individual species. Spatially structured microbiomes were detected in the majority of species studied (28.5 species, 75%). Within taxonomic groups, spatial structure was detected in all coral and marine mammal microbiomes, but was less common in fish (69%) and sponges (46%, Figure 1A). Different species within a genus did not necessarily show the same pattern. For example, the sponges Ircinia strobilina (Pita et al., 2013a) and I. campana (Griffiths et al., 2019) both showed spatially structured microbiomes, but I. felix (Pita et al., 2013a), I. fasciculata, I. variabilis and I. oros did not (Pita et al., 2013b; Table 1). Studies on sessile species showed 70% spatially structured microbiomes, while mobile species showed 84% spatially structured microbiomes (Figure 1B).
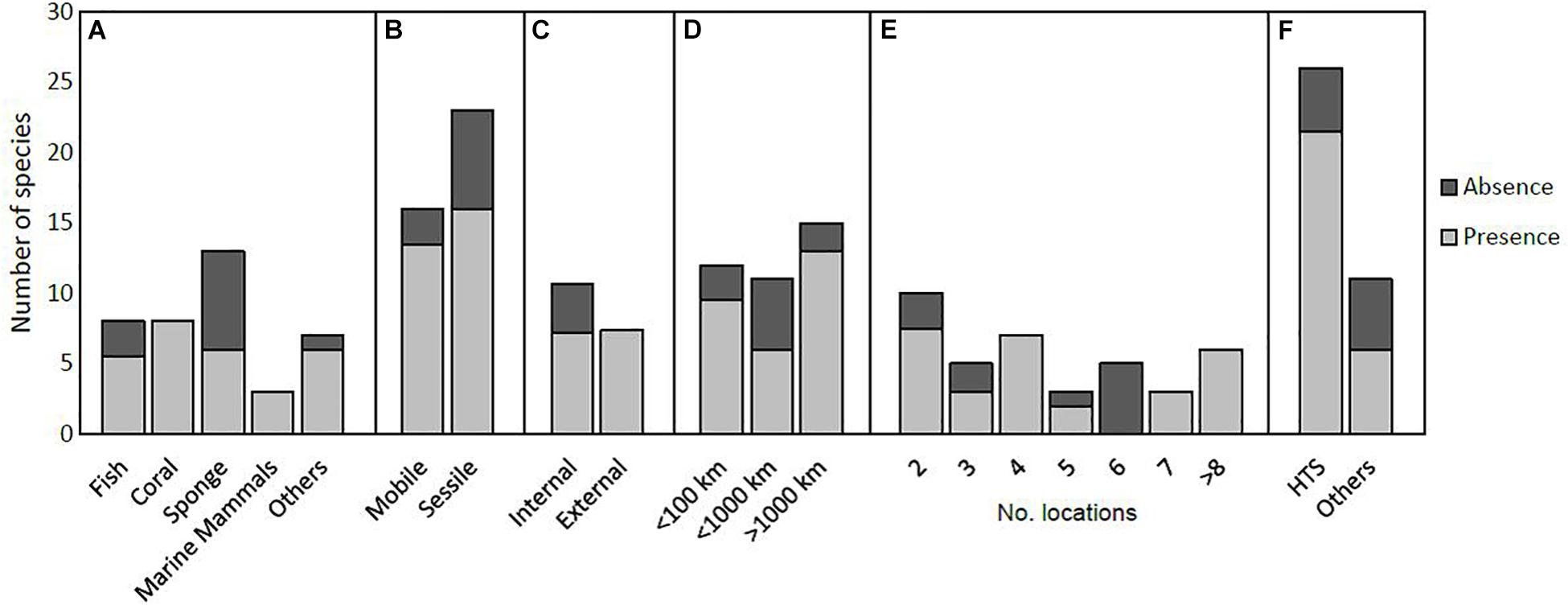
Figure 1. Relationship between detecting presence or absence of species’ microbiome spatial structure and (A) taxonomic group, (B) species traits (mobile vs. sessile species), (C) host tissue sampled (internal vs. external), (D) maximum distance between sampled individuals, (E) number of locations sampled, and (F) analysis method (high-throughput sequencing (HTS) and others). Three species’ microbiomes were tested for more than one tissue type; for example one species where the skin microbiome showed spatial structure but the gut microbiome did not (the fish Elacatinus prochilos) 0.5 was scored for presence and 0.5 for absence.
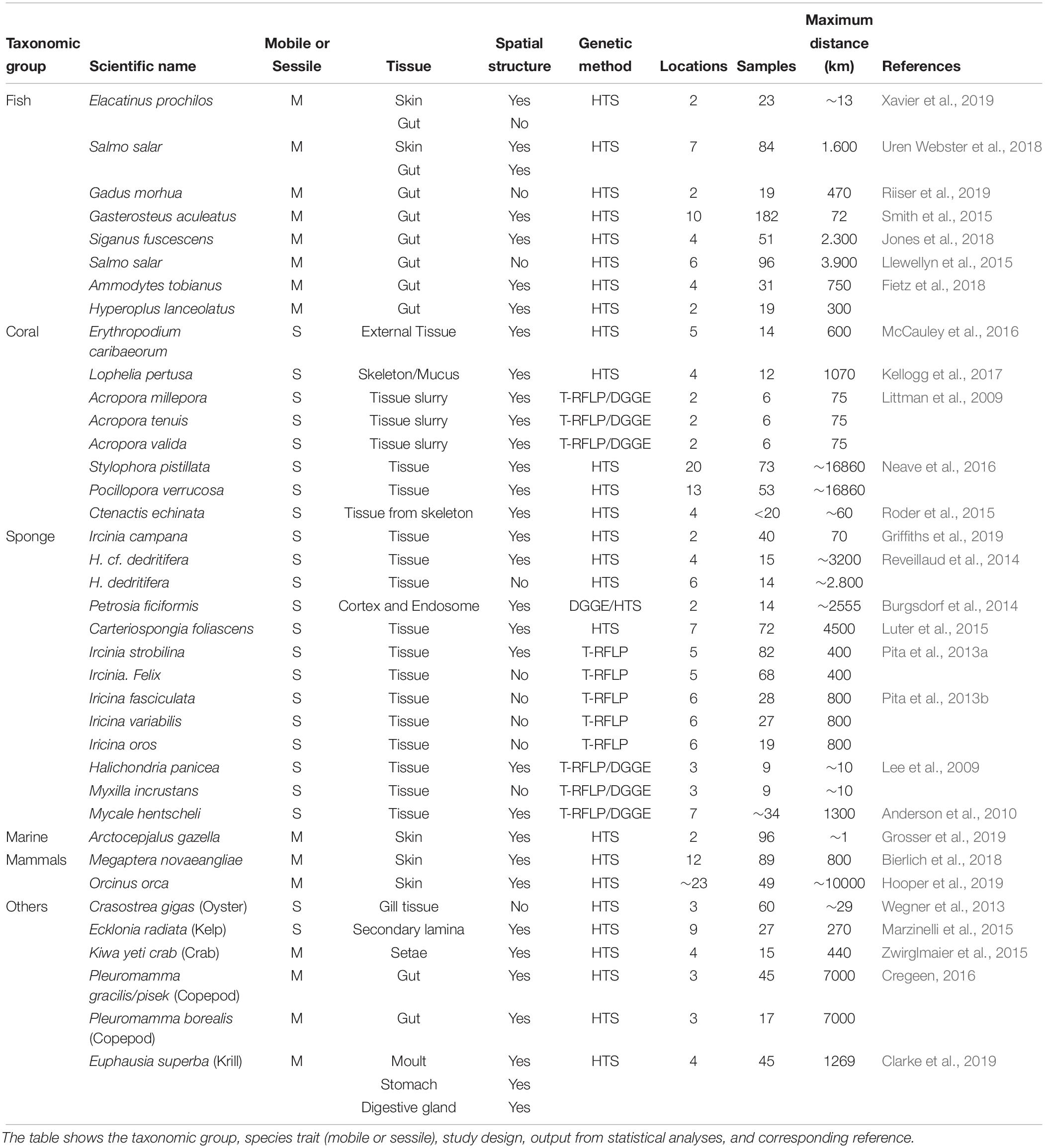
Table 1. Data on presence or absence of spatial structure in marine host-associated microbiomes for 38 species extracted from 28 research papers.
Study Design
We examined whether the microbiome of internal (e.g., gut and coral skeleton) or external (e.g., skin and coral mucus) host tissues were more likely to show spatial structure. Spatial structure was detected in all external microbiomes, in comparison to 67% of internal microbiomes (Figure 1C). Sponges were excluded from this analysis due to issues differentiating between external and internal tissue, as well as coral species where tissue sample type was not specified.
We examined whether the maximum distance between sampled individuals influenced the chance of detecting a spatially structured microbiome. Spatially structured microbiomes were most likely to be detected in studies over the largest spatial scales (>1000 km, 87%, Figure 1D). Interestingly, the next highest proportion was for studies covering <100 km (79% presence). These results suggest detection of spatially structured microbiomes was not dependent on the spatial scale of the study. However, spatially structured microbiomes were detected in all nine species where seven or more locations were sampled, suggesting the number of sites sampled influences the chance of detecting spatial structure (Figure 1E).
Studies using high-throughput DNA sequencing were more likely to detect spatial structure than non-sequencing methods such as terminal restriction fragment length polymorphism (TRFLP) or denaturing gradient gel electrophoresis (DGGE) (81% vs. 55%, Figure 1F). This difference in detection may reflect lack of resolution for TRFLP and DGGE, as these methods are not able to detect species below 1% of the community composition (Muyzer et al., 1993; Taylor et al., 2007).
Fish
Fish microbiome research has primarily focused on commercial and farmed fish species (reviewed by Legrand et al., 2019), with most studying the bacterial microbiome of the skin and the gut (Table 1). The fish gut microbiome is related to their food sources and trophic level (Egerton et al., 2018). The absence of spatial structure in the Atlantic cod gut microbiome suggests colonization by a limited number of bacterial species (Riiser et al., 2019), and may reflect similar food sources across their distribution. The skin, on the other hand, is strongly influenced by both host and environmental factors (Larsen et al., 2013; Xavier et al., 2019). Both location and seasonal environmental changes significantly influence fish skin microbiomes (Larsen et al., 2013). The strong interaction between these two factors makes it difficult to distinguish the importance of each parameter separately. Interestingly, a reciprocal transplant experiment showed Atlantic salmon skin and gut microbiomes are strongly influenced by environmental conditions, but that developmental history also influences microbiome structure (Uren Webster et al., 2019). Similarly, returning adult Atlantic salmon in Canadian and Irish sites shared similar gut microbiomes to oceanic adult salmon from Greenland, but adult microbiomes were distinct from those of juvenile freshwater life stages (Llewellyn et al., 2015). The anadromous life history of salmon highlights the impact of environment, developmental stage, and diet on fish gut microbiomes. Lab experiments showed that interhost dispersal can overwhelm host genotype in (freshwater) zebrafish gut microbiomes, demonstrating the importance of metacommunity dynamics (Burns et al., 2017). Although the experiment was performed in much smaller volumes than experienced by marine fish, interhost dispersal may be important for schooling fish species.
Corals
Coral microbiomes gained a lot of interest over the last few years in regard to their influence on host fitness and survival in the face of ongoing environmental changes [e.g., ocean warming, reviewed by van Oppen and Blackall (2019)]. Coral microbiomes consist of three essential elements (1) a conserved core microbiome, (2) regional bacteria specific to the geographic area, and (3) a set of environmentally variable bacteria (Hernandez-Agreda et al., 2016; Kellogg et al., 2017; van de Water et al., 2017).
The coral microbial community changes across anatomy and therefore the coral tissue sampled affects the likelihood of detecting a spatially structured microbiome (Pollock et al., 2018). Pollock et al. (2018) investigated the microbial community differences between skeleton, mucus, and tissue in 36 coral species over 21 sites. The mucus was strongly influenced by environmental factors, while the skeleton microbiome was the most diverse and most likely to show phylogenetic structure, reflecting the influence of host traits. The influence of phylosymbiosis, defined as “microbial community relationships that recapitulate the phylogeny of their host” (Brucker and Bordenstein, 2013), was higher than regional dispersal or environmental heterogeneity and differed across anatomy, being stronger in the skeleton than in tissue or mucus (Pollock et al., 2018; Dunphy et al., 2019). Regarding the prevalence of spatial structure, Pollock et al. (2018) showed that the external mucus microbiome is 1.15-fold more influenced by collection site than coral tissue and 1.28-fold more than skeleton communities (Pollock et al., 2018). This differences in spatial structuring across anatomy is also observed in the krill Euphausia superba, where the exoskeleton (moult) microbiome showed stronger spatial structuring than the gut microbiome (Clarke et al., 2019).
The reproductive mode of corals also appears to influence spatial structuring of their microbiomes. Neave et al. (2016) showed the microbiome of the brooding species Stylophora pistillata is strongly spatially structured, whereas the microbiome of the broadcast spawning Pocillopora verrucosa has a much weaker spatial structure. As a brooder, S. pistillata uses vertical transmission to control the larval microbiome, resulting in a high structuring due to the location (Hall and Hughes, 1996; Shlesinger et al., 1998; Sharp et al., 2011; Neave et al., 2016). In contrast, the sterile larvae of P. verrucosa gain microbes from seawater resulting in a weak spatial structuring (Sharp et al., 2010; Ceh et al., 2013; Pinzón et al., 2013; Neave et al., 2016).
The spatial structure of coral microbiomes could be influenced by regional processes including dispersal limitation and spatiotemporal environmental heterogeneity even at small spatial distances (Dunphy et al., 2019). Nevertheless, these spatial differences are limited in comparison to differences between coral genera or species (Dunphy et al., 2019).
Sponges
The microbiome of sponges can contribute up to 35% of their entire mass, with the diversity and core functions of the sponge microbiome reviewed by Pita et al. (2018). Due to its filter-feeding activity, sponges have a diverse and abundant microbial community, approximately three to four times greater than surrounding seawater (Taylor et al., 2007; Hentschel et al., 2012), with most microbial organisms inhabiting the sponge mesophyll tissue (Hentschel et al., 2012). The bacterial community among sponges is widely thought to be a result of both vertical and horizontal transmission (Taylor et al., 2007; Sipkema et al., 2015).
Approximately half the sponge species studied to date show spatially structured microbiomes. Most sponge microbiome studies that did not detect spatial structure (5/7) used non-sequencing based methods (Table 1). Other studies found differences in occurrence of spatial structure not only within but also between different genera (Lee et al., 2009; Pita et al., 2013a; Reveillaud et al., 2014). These studies were carried out over spatial scales from 10 km (Lee et al., 2009) to over 4000 km (Luter et al., 2015). Nevertheless, there was no clear influential pattern of this factor. For example, Griffiths et al. (2019) reported spatial structure within 70 kilometers (Ircinia campana), while Pita et al. (2013b) found no spatial structure at 80 up to 800 km in congeneric species (I. fasciculata, I. variabilis, and I. oros).
The sponge microbiome is also influenced by the host’s innate immune system and metabolism strongly due to the production of both antimicrobial compounds and nutrients (Müller and Müller, 2003; Wiens et al., 2006; Gauthier et al., 2010; Srivastava et al., 2010; Blunt et al., 2011; Hentschel et al., 2012). The sponge host can thus exert selection pressure on its microbiome at an individual level (Sipkema et al., 2015).
Marine Mammals
The three studies of spatial structuring of marine mammal microbiomes (all examining the skin microbiome), all showed significant spatial structure (Bierlich et al., 2018; Grosser et al., 2019; Hooper et al., 2019).
Several factors influence marine mammal microbial diversity, including horizontal transmission of bacteria due to social interactions (Hooper et al., 2019), or vertical transmission between mother and offspring resulting in similar microbial patterns between them (Grosser et al., 2019). Additionally, there are complex interactions between environmental and host genetic effects (Grosser et al., 2019). Grosser et al. (2019) found no significant influence on Antarctic fur seal microbial community structure of age, gender, or the proximity of mother to their offspring, as well as no relationship between microbial similarity and host genetic traits. However, humpback microbial diversity is affected by seasonal change and foraging even at the core microbiome level (Bierlich et al., 2018).
Conclusion
We have shown that 75% of marine species studied to date show spatially structured microbiomes, with the proportion increasing to more than 80% when restricted to studies using modern high-throughput sequencing technology. These results suggest spatially structured microbiomes are common in marine taxa, with absence of spatial structure the exception. Although less than 50% of sponge microbiomes were spatially structured, most sponge microbiome studies that did not detect spatial structure (5/7) used non-sequencing based methods. Given the connection between the microbiome and host health, future studies should investigate whether microbiome variations between locations affect host fitness. We are unaware of similar quantitative reviews for freshwater or terrestrial host-associated microbiomes. Future studies along these lines would demonstrate whether spatially structured host-associated microbiomes are common in all environments, and whether the increased prevalence of spatial structure in external microbiomes is a general rule. Future studies should aim for a high taxonomic resolution of the microbial β-diversity and distinguish between effects of environment and the respective host. High-throughput sequencing is recommended for studying marine host-associated microbiomes due to better taxonomic resolution compared to other methods (e.g., DGGE/TRFLP). We also recommend researchers test the statistical power of their study design (number of sites, samples and sequencing depth) using tools especially developed for microbiome studies (e.g., Kelly et al., 2015).
Geographic distance alone is likely not generating differences in marine host-associated microbiomes, but acting in concert with environmental or host factors. Future biogeographical studies of host-associated microbiomes should therefore aim to distinguish between effects of environment and the host. The influence of host effects require data about the host itself, such as size, age or developmental stage, and genotype (e.g., Pollock et al., 2018). The influence of environment on host-associated microbiomes can be examined by measuring environmental variables (e.g., temperature, salinity, pH, depth, chlorophyll a), and helps to unravel the extent to which environmental and spatial variability are confounded. We recommend researchers collect extensive metadata to facilitate these analyses [e.g., the MIMARKS (Minimum Information about a Marker gene Sequence) checklist, Yilmaz et al., 2011]. However, a more direct test is to sample the relevant environmental microbiome (e.g., seawater, sediment, or biofilm bacterial communities) and compare variation in the environmental microbiome to that observed in the host (e.g., Fietz et al., 2018). We did not explicitly examine the effect of environmental variability in this review given the difficulty of generalizing across studies, and the inclusion of mobile species like fish and marine mammals that can traverse large environmental gradients. Future review studies could explore relationships between environmental and microbiome variability, particularly for sessile species, taking into account temporal variability in both the environment and microbiome.
The relative importance of selection, dispersal, diversification and drift on structuring host-associated microbiomes needs further investigation. For example, the selective influence of various environmental and host parameters can be tested as in Pollock et al. (2018) by analyzing factors affecting the microbial composition (e.g., host species, typical growth form, geographic region, and light availability). Additionally, aquarium experiments allow environmental conditions to be tightly controlled, and can also be used to control interhost dispersal (Burns et al., 2017). Furthermore, microbial phylogenetic approaches can give further information about the host-associated microbiome pattern and its processes (Pollock et al., 2018). Null modeling approaches developed by Stegen et al. (2013, 2015) allow the relative contributions of variable selection, homogenizing selection, dispersal limitation, and homogenizing dispersal in shaping host-associated microbiomes to be estimated. Yan et al. (2016) used this approach to show shifts in the processes shaping fish gut microbiomes during development. Combining estimates of the importance of selection with environmental and host data could be used to test whether host or environmental factors drive selection in marine host-associated microbiomes.
Author Contributions
LS and LC conceived the idea for this review and wrote the manuscript. LS compiled the data.
Funding
This work was supported by the Australian Government’s Business Cooperative Research Centres Programme through the Antarctic Climate and Ecosystems Cooperative Research Centre (ACE CRC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Anderson, S. A., Northcote, P. T., and Page, M. J. (2010). Spatial and temporal variability of the bacterial community in different chemotypes of the New Zealand marine sponge Mycale hentscheli. FEMS Microbiol. Ecol. 72, 328–342. doi: 10.1111/j.1574-6941.2010.00869.x
Bierlich, K. C., Miller, C., DeForce, E., Friedlaender, A. S., Johnston, D. W., and Apprill, A. (2018). Temporal and regional variability in the skin microbiome of humpback whales along the Western Antarctic Peninsula. Appl. Environ. Microbiol. 84:e02574-17. doi: 10.1128/aem.02574-17
Blunt, J. W., Copp, B. R., Munro, M. H. G., Northcote, P. T., and Prinsep, M. R. (2011). Marine natural products. Nat. Prod. Rep. 28, 196–268. doi: 10.1039/C005001F
Brucker, R. M., and Bordenstein, S. R. (2013). The hologenomic basis of speciation: gut bacteria cause hybrid lethality in the genus Nasonia. Science 341, 667–669. doi: 10.1126/science.1240659
Burgsdorf, I., Erwin, P. M., López-Legentil, S., Cerrano, C., Haber, M., Frenk, S., et al. (2014). Biogeography rather than association with cyanobacteria structures symbiotic microbial communities in the marine sponge Petrosia ficiformis. Front. Microbiol. 5:529. doi: 10.3389/fmicb.2014.00529
Burns, A. R., Miller, E., Agarwal, M., Rolig, A. S., Milligan-Myhre, K., Seredick, S., et al. (2017). Interhost dispersal alters microbiome assembly and can overwhelm host innate immunity in an experimental zebrafish model. Proc. Natl. Acad. Sci. U.S.A. 114, 11181–11186. doi: 10.1073/pnas.1702511114
Ceh, J., van Keulen, M., and Bourne, D. G. (2013). Intergenerational transfer of specific bacteria in corals and possible implications for offspring fitness. Microb. Ecol. 65, 227–231. doi: 10.1007/s00248-012-0105-z
Chase, J. M., and Myers, J. A. (2011). Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B Biol. Sci. 366, 2351–2363. doi: 10.1098/rstb.2011.0063
Clarke, L. J., Suter, L., King, R., Bissett, A., and Deagle, B. E. (2019). Antarctic krill are reservoirs for distinct southern ocean microbial communities. Front. Microbiol. 9:3226. doi: 10.3389/fmicb.2018.03226
Cooper, M. B., Kazamia, E., Helliwell, K. E., Kudahl, U. J., Sayer, A., Wheeler, G. L., et al. (2019). Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J. 13, 334–345. doi: 10.1038/s41396-018-0274-y
Correa, A. M. S., and Baker, A. C. (2011). Disaster taxa in microbially mediated metazoans: how endosymbionts and environmental catastrophes influence the adaptive capacity of reef corals. Glob. Chang. Biol. 17, 68–75. doi: 10.1111/j.1365-2486.2010.02242.x
Cregeen, S. J. J. (2016). Microbiota of Dominant Atlantic Copepods: Pleuromamma sp. As a Host to a Betaproteobacterial Symbiont. Doctoral thesis, University of Southampton, Southampton.
Datta, M. S., Almada, A. A., Baumgartner, M. F., Mincer, T. J., Tarrant, A. M., and Polz, M. F. (2018). Inter-individual variability in copepod microbiomes reveals bacterial networks linked to host physiology. ISME J. 12, 2103–2113. doi: 10.1038/s41396-018-0182-1
Dittami, S. M., Duboscq-Bidot, L., Perennou, M., Gobet, A., Corre, E., Boyen, C., et al. (2016). Host–microbe interactions as a driver of acclimation to salinity gradients in brown algal cultures. ISME J. 10, 51–63. doi: 10.1038/ismej.2015.104
Dunphy, C. M., Gouhier, T. C., Chu, N. D., and Vollmer, S. V. (2019). Structure and stability of the coral microbiome in space and time. Sci. Rep. 9:6785. doi: 10.1038/s41598-019-43268-6
Egerton, S., Culloty, S., Whooley, J., Stanton, C., and Ross, R. P. (2018). The gut microbiota of marine fish. Front. Microbiol. 9:873. doi: 10.3389/fmicb.2018.00873
Fietz, K., Rye Hintze, C. O., Skovrind, M., Kjaergaard Nielsen, T., Limborg, M. T., Krag, M. A., et al. (2018). Mind the gut: genomic insights to population divergence and gut microbial composition of two marine keystone species. Microbiome 6:82. doi: 10.1186/s40168-018-0467-7
Gauthier, M. E. A., Du Pasquier, L., and Degnan, B. M. (2010). The genome of the sponge Amphimedon queenslandica provides new perspectives into the origin of Toll-like and interleukin 1 receptor pathways. Evol. Dev. 12, 519–533. doi: 10.1111/j.1525-142X.2010.00436.x
Griffiths, S. M., Antwis, R. E., Lenzi, L., Lucaci, A., Behringer, D. C., Butler, M. J., et al. (2019). Host genetics and geography influence microbiome composition in the sponge Ircinia campana. J. Anim. Ecol. 88, 1684–1695. doi: 10.1111/1365-2656.13065
Grosser, S., Sauer, J., Paijmans, A. J., Caspers, B. A., Forcada, J., Wolf, J. B. W., et al. (2019). Fur seal microbiota are shaped by the social and physical environment, show mother–offspring similarities and are associated with host genetic quality. Mol. Ecol. 28, 2406–2422. doi: 10.1111/mec.15070
Hall, V. R., and Hughes, T. P. (1996). Reproductive strategies of modular organisms: comparative studies of reef- building corals. Ecology 77, 950–963. doi: 10.2307/2265514
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C., and Martiny, J. B. H. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10:497. doi: 10.1038/nrmicro2795
Hentschel, U., Piel, J., Degnan, S. M., and Taylor, M. W. (2012). Genomic insights into the marine sponge microbiome. Nat. Rev. Microbiol. 10:641. doi: 10.1038/nrmicro2839
Hernandez-Agreda, A., Leggat, W., Bongaerts, P., and Ainsworth, T. D. (2016). The microbial signature provides insight into the mechanistic basis of coral success across reef habitats. mBio 7:e00560-16. doi: 10.1128/mBio.00560-16
Hooper, R., Brealey, J. C., van der Valk, T., Alberdi, A., Durban, J. W., Fearnbach, H., et al. (2019). Host-derived population genomics data provides insights into bacterial and diatom composition of the killer whale skin. Mol. Ecol. 28, 484–502. doi: 10.1111/mec.14860
Jones, J., DiBattista, J. D., Stat, M., Bunce, M., Boyce, M. C., Fairclough, D. V., et al. (2018). The microbiome of the gastrointestinal tract of a range-shifting marine herbivorous fish. Front. Microbiol. 9:2000. doi: 10.3389/fmicb.2018.02000
Kellogg, C. A., Goldsmith, D. B., and Gray, M. A. (2017). Biogeographic comparison of lophelia-associated bacterial communities in the Western Atlantic reveals conserved core microbiome. Front. Microbiol. 8:796. doi: 10.3389/fmicb.2017.00796
Kelly, B. J., Gross, R., Bittinger, K., Sherrill-Mix, S., Lewis, J. D., Collman, R. G., et al. (2015). Power and sample-size estimation for microbiome studies using pairwise distances and PERMANOVA. Bioinformatics 31, 2461–2468. doi: 10.1093/bioinformatics/btv183
Larsen, A., Tao, Z., Bullard, S. A., and Arias, C. R. (2013). Diversity of the skin microbiota of fishes: evidence for host species specificity. FEMS Microbiol. Ecol. 85, 483–494. doi: 10.1111/1574-6941.12136
Lee, O. O., Wong, Y. H., and Qian, P.-Y. (2009). Inter- and intraspecific variations of bacterial communities associated with marine sponges from San Juan Island, Washington. Appl. Environ. Microbiol. 75, 3513–3521. doi: 10.1128/aem.00002-09
Legrand, T. P. R. A., Wynne, J. W., Weyrich, L. S., and Oxley, A. P. A. (2019). A microbial sea of possibilities: current knowledge and prospects for an improved understanding of the fish microbiome. Rev. Aquacul. 1–34. doi: 10.1111/raq.12375
Littman, R. A., Willis, B. L., Pfeffer, C., and Bourne, D. G. (2009). Diversities of coral-associated bacteria differ with location, but not species, for three Acroporid corals on the Great Barrier Reef. FEMS Microbiol. Ecol. 68, 152–163. doi: 10.1111/j.1574-6941.2009.00666.x
Llewellyn, M. S., McGinnity, P., Dionne, M., Letourneau, J., Thonier, F., Carvalho, G. R., et al. (2015). The biogeography of the atlantic salmon (Salmo salar) gut microbiome. ISME J. 10:1280. doi: 10.1038/ismej.2015.189
Luter, H. M., Widder, S., Botté, E. S., Abdul Wahab, M., Whalan, S., Moitinho-Silva, L., et al. (2015). Biogeographic variation in the microbiome of the ecologically important sponge, Carteriospongia foliascens. PeerJ 3:e01435. doi: 10.7717/peerj.1435
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E., and Borisy, G. G. (2016). Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800. doi: 10.1073/pnas.1522149113
Martiny, J. B. H., Bohannan, B. J. M., Brown, J. H., Colwell, R. K., Fuhrman, J. A., Green, J. L., et al. (2006). Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112. doi: 10.1038/nrmicro1341
Marzinelli, E. M., Campbell, A. H., Zozaya Valdes, E., Vergés, A., Nielsen, S., Wernberg, T., et al. (2015). Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 17, 4078–4088. doi: 10.1111/1462-2920.12972
McCauley, E. P., Haltli, B., Correa, H., and Kerr, R. G. (2016). Spatial and temporal investigation of the microbiome of the caribbean octocoral Erythropodium caribaeorum. FEMS Microbiol. Ecol. 92:fiw147. doi: 10.1093/femsec/fiw147
Müller, W. E. G., and Müller, I. M. (2003). Origin of the metazoan immune system: identification of the molecules and their functions in sponges. Integ. Comp. Biol. 43, 281–292. doi: 10.1093/icb/43.2.281
Muyzer, G., de Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/aem.59.3.695-700.1993
Neave, M. J., Rachmawati, R., Xun, L., Michell, C. T., Bourne, D. G., Apprill, A., et al. (2016). Differential specificity between closely related corals and abundant Endozoicomonas endosymbionts across global scales. ISME J. 11:186. doi: 10.1038/ismej.2016.95
Nemergut, D. R., Schmidt, S. K., Fukami, T., O’Neill, S. P., Bilinski, T. M., Stanish, L. F., et al. (2013). Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 77, 342–356. doi: 10.1128/mmbr.00051-12
O’Brien, P. A., Webster, N. S., Miller, D. J., and Bourne, D. G. (2019). Host-microbe coevolution: applying evidence from model systems to complex marine invertebrate holobionts. mBio 10, 1–14. doi: 10.1128/mBio.02241-18
Pinzón, J. H., Sampayo, E., Cox, E., Chauka, L. J., Chen, C. A., Voolstra, C. R., et al. (2013). Blind to morphology: genetics identifies several widespread ecologically common species and few endemics among Indo-Pacific cauliflower corals (Pocillopora, Scleractinia). J. Biogeogr. 40, 1595–1608. doi: 10.1111/jbi.12110
Pita, L., López-Legentil, S., and Erwin, P. M. (2013a). Biogeography and host fidelity of bacterial communities in Ircinia spp. from the Bahamas. Microb. Ecol. 66, 437–447. doi: 10.1007/s00248-013-0215-2
Pita, L., Turon, X., López-Legentil, S., and Erwin, P. M. (2013b). Host rules: spatial stability of bacterial communities associated with marine sponges (Ircinia spp.) in the Western Mediterranean Sea. FEMS Microbiol. Ecol. 86, 268–276. doi: 10.1111/1574-6941.12159
Pita, L., Rix, L., Slaby, B. M., Franke, A., and Hentschel, U. (2018). The sponge holobiont in a changing ocean: from microbes to ecosystems. Microbiome 6:46. doi: 10.1186/s40168-018-0428-1
Pollock, F. J., McMinds, R., Smith, S., Bourne, D. G., Willis, B. L., Medina, M., et al. (2018). Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 9:4921. doi: 10.1038/s41467-018-07275-x
Raina, J.-B., Tapiolas, D., Willis, B. L., and Bourne, D. G. (2009). Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl. Environ. Microbiol. 75, 3492–3501. doi: 10.1128/aem.02567-08
Rensing, C., Newby, D. T., and Pepper, I. L. (2002). The role of selective pressure and selfish DNA in horizontal gene transfer and soil microbial community adaptation. Soil Biol. Biochem. 34, 285–296. doi: 10.1016/S0038-0717(01)00183-3
Reveillaud, J., Maignien, L., Eren, A. M., Huber, J. A., Apprill, A., Sogin, M. L., et al. (2014). Host-specificity among abundant and rare taxa in the sponge microbiome. ISME J. 8:1198. doi: 10.1038/ismej.2013.227
Riiser, E. S., Haverkamp, T. H. A., Varadharajan, S., Borgan, Ø, Jakobsen, K. S., Jentoft, S., et al. (2019). Switching on the light: using metagenomic shotgun sequencing to characterize the intestinal microbiome of Atlantic cod. Environ. Microbiol. 21, 2576–2594. doi: 10.1111/1462-2920.14652
Roder, C., Bayer, T., Aranda, M., Kruse, M., and Voolstra, C. R. (2015). Microbiome structure of the fungid coral Ctenactis echinata aligns with environmental differences. Mol. Ecol. 24, 3501–3511. doi: 10.1111/mec.13251
Röthig, T., Ochsenkühn, M. A., Roik, A., van der Merwe, R., and Voolstra, C. R. (2016). Long-term salinity tolerance is accompanied by major restructuring of the coral bacterial microbiome. Mol. Ecol. 25, 1308–1323. doi: 10.1111/mec.13567
Sharp, K. H., Distel, D., and Paul, V. J. (2011). Diversity and dynamics of bacterial communities in early life stages of the Caribbean coral Porites astreoides. ISME J. 6:790. doi: 10.1038/ismej.2011.144
Sharp, K. H., Ritchie, K. B., Schupp, P. J., Ritson-Williams, R., and Paul, V. J. (2010). Bacterial acquisition in juveniles of several broadcast spawning coral species. PLoS One 5:10898. doi: 10.1371/journal.pone.0010898
Shlesinger, Y., Goulet, T. L., and Loya, Y. (1998). Reproductive patterns of scleractinian corals in the northern Red Sea. Mar. Biol. 132, 691–701. doi: 10.1007/s002270050433
Sipkema, D., de Caralt, S., Morillo, J. A., Al-Soud, W. A., Sørensen, S. J., Smidt, H., et al. (2015). Similar sponge-associated bacteria can be acquired via both vertical and horizontal transmission. Environ. Microbiol. 17, 3807–3821. doi: 10.1111/1462-2920.12827
Sison-Mangus, M. P., Jiang, S., Tran, K. N., and Kudela, R. M. (2014). Host-specific adaptation governs the interaction of the marine diatom, Pseudo-nitzschia and their microbiota. ISME J. 8, 63–76. doi: 10.1038/ismej.2013.138
Smith, C. C. R., Snowberg, L. K., Gregory Caporaso, J., Knight, R., and Bolnick, D. I. (2015). Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J. 9:2515. doi: 10.1038/ismej.2015.64
Srivastava, M., Simakov, O., Chapman, J., Fahey, B., Gauthier, M. E. A., Mitros, T., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720. doi: 10.1038/nature09201
Stegen, J. C., Lin, X., Fredrickson, J. K., Chen, X., Kennedy, D. W., Murray, C. J., et al. (2013). Quantifying community assembly processes and identifying features that impose them. ISME J. 7:2069. doi: 10.1038/ismej.2013.93
Stegen, J. C., Lin, X., Fredrickson, J. K., and Konopka, A. E. (2015). Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 6:370. doi: 10.3389/fmicb.2015.00370
Taylor, M. W., Radax, R., Steger, D., and Wagner, M. (2007). Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol. Mol. Biol. Rev. 71, 295–347. doi: 10.1128/mmbr.00040-06
Troussellier, M., Escalas, A., Bouvier, T., and Mouillot, D. (2017). Sustaining rare marine microorganisms: macroorganisms as repositories and dispersal agents of microbial diversity. Front. Microbiol. 8:947. doi: 10.3389/fmicb.2017.00947
Uren Webster, T., Rodriguez Barreto, D., Castaldo, G., Taylor, J., Gough, P., Consuegra, S., et al. (2019). Environmental plasticity and colonisation history in the Atlantic salmon microbiome: a translocation experiment. bioRxiv [Preprint], doi: 10.1101/564104
Uren Webster, T. M., Consuegra, S., Hitchings, M., and Garcia de Leaniz, C. (2018). Interpopulation variation in the atlantic salmon microbiome reflects environmental and genetic diversity. Appl. Environ. Microbiol. 84:e00691-18. doi: 10.1128/aem.00691-18
van de Water, J. A. J. M., Melkonian, R., Voolstra, C. R., Junca, H., Beraud, E., Allemand, D., et al. (2017). Comparative assessment of mediterranean gorgonian-associated microbial communities reveals conserved core and locally variant bacteria. Microb. Ecol. 73, 466–478. doi: 10.1007/s00248-016-0858-x
van Oppen, M. J. H., and Blackall, L. L. (2019). Coral microbiome dynamics, functions and design in a changing world. Nat. Rev. Microbiol. 17, 557–567. doi: 10.1038/s41579-019-0223-4
Vellend, M. (2010). Conceptual synthesis in community ecology. Q. Rev. Biol. 85, 183–206. doi: 10.1086/652373
Wegner, K. M., Volkenborn, N., Peter, H., and Eiler, A. (2013). Disturbance induced decoupling between host genetics and composition of the associated microbiome. BMC Microbiol. 13:252. doi: 10.1186/1471-2180-13-252
Wiens, M., Korzhev, M., Peroviæ-Ottstadt, S., Luthringer, B., Brandt, D., Klein, S., et al. (2006). Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol. 24, 792–804. doi: 10.1093/molbev/msl208
Xavier, R., Mazzei, R., Pérez-Losada, M., Rosado, D., Santos, J. L., Veríssimo, A., et al. (2019). A risky business? Habitat and social behavior impact skin and gut microbiomes in caribbean cleaning gobies. Front. Microbiol. 10:716. doi: 10.3389/fmicb.2019.00716
Yan, Q., Li, J., Yu, Y., Wang, J., He, Z., Van Nostrand, J. D., et al. (2016). Environmental filtering decreases with fish development for the assembly of gut microbiota. Environ. Microbiol. 18, 4739–4754. doi: 10.1111/1462-2920.13365
Yilmaz, P., Kottmann, R., Field, D., Knight, R., Cole, J. R., Amaral-Zettler, L., et al. (2011). Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420. doi: 10.1038/nbt.1823
Zhou, J., and Ning, D. (2017). Stochastic community assembly: does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 81:e00002-17. doi: 10.1128/mmbr.00002-17
Zwirglmaier, K., Reid, W. D. K., Heywood, J., Sweeting, C. J., Wigham, B. D., Polunin, N. V. C., et al. (2015). Linking regional variation of epibiotic bacterial diversity and trophic ecology in a new species of kiwaidae (Decapoda, Anomura) from East Scotia Ridge (Antarctica) hydrothermal vents. Microbiol. Open 4, 136–150. doi: 10.1002/mbo3.227
Keywords: host-associated microbiome, biogeography, high-throughput sequencing, fish, coral, sponge, selection, dispersal
Citation: Schellenberg L and Clarke LJ (2020) Spatial Structure of Marine Host-Associated Microbiomes: Effect of Taxonomy, Species Traits, and Study Design. Front. Mar. Sci. 7:146. doi: 10.3389/fmars.2020.00146
Received: 15 November 2019; Accepted: 25 February 2020;
Published: 11 March 2020.
Edited by:
Simon Bahrndorff, Aalborg University, DenmarkReviewed by:
Lucia Pita, GEOMAR Helmholtz Centre for Ocean Research Kiel, GermanyRyan Christopher McMinds, Oregon State University, United States
Robert W. Thacker, Stony Brook University, United States
Mengfei Ho, University of Illinois at Urbana-Champaign, United States
Copyright © 2020 Schellenberg and Clarke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lisa Schellenberg, lisa.schellenberg@utas.edu.au
 Lisa Schellenberg
Lisa Schellenberg Laurence J. Clarke
Laurence J. Clarke