- 1Climate Change Cluster (C3), University of Technology Sydney, Sydney, NSW, Australia
- 2School of Life and Environmental Sciences, The University of Sydney, Sydney, NSW, Australia
Biogenic volatile organic compounds (BVOCs) are a large group of molecules involved in trophic interactions, stress response and atmospheric chemistry. Although they have been extensively studied in terrestrial ecosystems, their identity and prevalence in the marine environment remains largely unexplored. Here we characterized the volatilome of two abundant marine bacteria that were previously identified as members of the core microbiome of Symbiodiniaceae (phylum: Dinoflagellata), the photosynthetic endosymbionts of reef building corals. To determine the influence of Symbiodiniaceae exudate on their associated bacteria, we incubated isolates of Marinobacter adhaerens HP15 and Labrenzia sp. 21p with Symbiodiniaceae culture filtrate or culture medium (control) and investigated their volatilomes using GC–MS. The volatilome of Labrenzia sp. incubated in Symbiodiniaceae filtrate was significantly different and more diverse relative to the control. In contrast, the overall composition of the M. adhaerens volatilomes were consistent between treatment and control. Among the 35 compounds detected in both bacterial species, the dominant chemical functional groups were halogenated hydrocarbons, aromatic hydrocarbons and organosulfurs, some of which are known to play roles in inter-organism signaling, to act as antioxidants and as antimicrobials. This study provides new insights into the potential sources and diversity of marine BVOCs, uncovering a wide range of molecules that may play important physiological and ecological roles for these organisms, while also revealing the role of Symbiodiniaceae-associated bacteria in the emission of important atmospheric gases.
Introduction
All organisms are capable of producing biogenic volatile organic compounds (BVOCs), a class of chemicals known for their ecological and physiological roles. BVOCs can function as infochemicals, allowing organisms to locate food sources (Runyon et al., 2006), warn conspecifics of danger (Frost et al., 2007) or provide defense from predators (De Moraes et al., 2001). These compounds can also act as antioxidants, preventing the build-up of damaging reactive oxygen species (Loreto and Velikova, 2001; Sunda et al., 2002), and upon their release, they can influence atmospheric chemistry by forming secondary organic aerosols (Hallquist et al., 2009; Fu et al., 2010), reacting with greenhouse gases (Atkinson and Arey, 2003) and thus impacting climate. Previous studies characterizing BVOC production and emission have largely focused on terrestrial organisms, and have established the physiological (e.g. stress response; Vickers et al., 2009), ecological (e.g. signaling; Runyon et al., 2006) and atmospheric (e.g. ozone depletion; Matsumoto, 2014) roles that BVOCs can play. However, the occurrence and potential roles of BVOCs in important marine ecosystems, such as coral reefs, has remained largely overlooked. Coral reefs are known to produce large concentrations of the sulfur BVOC, dimethyl sulfide (DMS), resulting in a large body of research on the potential antioxidant and atmospheric functions of this compound (Broadbent and Jones, 2004; Deschaseaux et al., 2014b; Hopkins et al., 2016). A recent study also identified the capacity of corals to produce other sulfur volatiles (methyl mercaptan, carbon disulfide, dimethyl disulfide, dimethyl trisulfide, thiirane, 2,4-dithiapentance, 1,2,4-trithiolane) as well as isoprene, a ubiquitous BVOC in terrestrial systems (Swan et al., 2016). However, the diversity of coral derived BVOCs remains to be fully explored and it is likely that the wide range of organisms living in symbioses with corals are partially responsible for the production of BVOCs across coral reef ecosystems.
Corals are now widely considered as holobionts – complex assemblages which include the cnidarian host, endosymbiotic algae (Symbiodiniaceae) and diverse microbial communities (Rohwer et al., 2002). Multiple mutualistic associations sustain the health of the coral holobiont (Muscatine and Porter, 1977; Rohwer et al., 2002; Little et al., 2004; Ziegler et al., 2019), with inter-organism chemical signaling likely to form an important foundation for these relationships (Koike et al., 2004; Tebben et al., 2011; Garren et al., 2014; Raina et al., 2016). Given their importance as signaling molecules in other environments (Ryu et al., 2003; Runyon et al., 2006; Sharifi and Ryu, 2018; Shan et al., 2019), BVOCs may play an important role in mediating interactions within the coral holobiont, by acting as infochemicals and influencing stress responses. We recently performed a characterization of the Symbiodiniaceae volatilome (total BVOCs), with a diverse suite of BVOCs identified, including compounds implicated in chemical signaling (e.g. benzaldehyde, methyl jasmonate) and stress tolerance (e.g. DMS) (Lawson et al., 2019). However, it is probable that these measurements have only scratched the surface of the full diversity of BVOCs produced by different members of the coral holobiont.
A central feature of the coral holobiont is the tripartite interaction between corals, their Symbiodiniaceae partners and associated bacterial assemblages. We recently identified putative ecological links between Symbiodiniaceae and bacterial partners by characterizing the core microbiome associated with 18 strains of Symbiodiniaceae (Lawson et al., 2018). We identified three bacterial operational taxonomic units (OTUs), belonging to the Labrenzia and Marinobacter genera and an unclassified Chromatiaceae, which occurred in all strains of Symbiodiniaceae (Lawson et al., 2018). Members of the Marinobacter and Labrenzia commonly develop associations with microalgae (Han et al., 2016; Sandhya et al., 2017), and Marinobacter species have previously been isolated from other Symbiodiniaceae cultures (Frommlet et al., 2015). Notably, Labrenzia species (which comprised up to 38.4% of the Symbiodiniaceae microbiome; Lawson et al., 2018) can synthesize the DMS precursor, dimethylsulfoniopropionate (DMSP) (Curson et al., 2017), a sulfur compound also abundantly produced by Symbiodiniaceae (Broadbent et al., 2002; Deschaseaux et al., 2014b).
Given the close associations between these bacteria and Symbiodiniaceae, we examined the impact of the microalgal exudates on the bacterial volatilomes. We specifically aimed to provide the first characterization of the Labrenzia sp. 21p and Marinobacter adhaerens HP15 (hereafter referred to as Labrenzia sp. and M. adhaerens) volatilomes and to investigate the presence of putative inter-organism signaling BVOCs. We hypothesized that when bacteria are incubated in Symbiodiniaceae exudate, they will produce a greater diversity of BVOCs compared to controls incubated in Symbiodiniaceae growth medium.
Materials and Methods
Bacterial Isolation
A non-axenic species of Symbiodiniaceae, Breviolum minutum (strain: UTSB; originally isolated from the coral Euphyllia glabrescens; Lawson et al., 2018), was grown in a sterile 1L Schott bottle in IMK medium (in artificial seawater; Berges et al., 2001) [ARALAB incubator; 26 ± 1.5°C; ca. 100 ± 10 μmol photons m–2 s–1 (LEDs) on a 12:12 light:dark cycle; as per Lawson et al., 2018]. Bacterial isolates from the B. minutum microbiome were subsequently obtained by plating 20 μL aliquots of B. minutum culture on Marine Agar (100%; Difco 2216). Plates were then incubated for 10 days at room temperature before visible individual colonies were picked, grown overnight in Marine Broth (Difco 2216) at 22°C under shaking (180 rpm; Ratek Orbital Mixer Incubator), and re-plated on Marine Agar until axenic. Subsequent individual colonies were picked and grown overnight in Marine Broth (22°C, 180 rpm). Following establishment of cultures, cells were preserved through the addition of glycerol (Sigma-Aldrich; 20% final concentration), snap frozen in liquid nitrogen and stored at −80°C. Small volumes of pure overnight cultures were centrifuged (1.5 mL at 7,000 g for 10 min), before the supernatant was discarded and the pellets frozen (−20°C) for subsequent DNA extraction.
DNA Extraction, 16S rRNA PCR, and Strain Selection
To identify the bacterial isolates (25 in total; Supplementary Table S1), their DNA was extracted and their 16S rRNA gene sequenced. Briefly, frozen bacterial pellets were resuspended in 200 μL of nuclease free water (Ambion, Austin, TX, United States), heated at 95°C for 10 min to break cell walls, and centrifuged to pellet debris (10,000 g for 5 min). The DNA in the resulting supernatant was amplified with PCR targeting the 16S rRNA gene using the primers: 27F-AGAGTTTGATCMTGGCTCAG and 907R-CCGTCAATTCMTTTRAGTTT. The PCR mixture comprised 1 μL DNA, 1 μL of each primer, 9.5 μL nuclease free water (Ambion, Austin, TX, United States) and 12.5 μL of GoTaq colorless master mix (Promega, Madison, WI, United States) for a total reaction volume of 25 μL. The PCR reaction conditions were as follows: 95°C for 3 min followed by 30 cycles of 95°C for 30 s, 50°C for 30 s, 72°C for 1.45 min, and a final extension of 72°C for 7 min. The PCR products were visualized with electrophoresis on a 1% agarose gel strained with Gel Red (Biotium, Fremont, CA, United States) and analyzed using Sanger sequencing at the Australian Genome Research Facility (AGRF). Sequences were identified using nucleotide BLAST1 (Supplementary Table S1). Isolate 21p was identified as Labrenzia sp. (GenBank Accession number: MN305698), and displayed 97% sequence identity (BLASTN 2.9.0) to the Labrenzia sp. OTU that was previously identified as a core member of the Symbiodiniaceae microbiome (Lawson et al., 2018). None of the other bacterial isolates matched members of the Symbiodiniaceae core microbiome as defined in Lawson et al. (2018); present in 100% of samples from 18 Symbiodiniaceae cultures (n = 3). In lieu of other core microbiome isolates, we used a laboratory culture of M. adhaerens, a model bacterium extensively studied in the context of its interactions with phytoplankton (Gärdes et al., 2010; Sonnenschein et al., 2012), and present in the Symbiodiniaceae core microbiome (Lawson et al., 2018). M. adhaerens displayed 97% sequence similarity to the Marinobacter sp. OTU previously identified as a core member of the Symbiodiniaceae microbiome (Lawson et al., 2018). To our knowledge, neither of these bacteria have previously had their volatilomes characterized and are currently not included in the microbial VOC database (mVOC; Lemfack et al., 2018).
Volatile Sampling of Labrenzia sp. and Marinobacter adhaerens
Glycerol stocks of Labrenzia sp. and M. adhaerens were replated onto Marine Agar (100%; Difco 2216) and all bacterial biomass was resuspended in 300 mL of Marine Broth (Difco 2216). Bacteria cultures were grown overnight in a shaking incubator (KuhnerShaker X; room temperature), before being centrifuged at 1,000 g for 10 min. The supernatant was discarded, and the pellet washed by resuspension in IMK medium (Diago, Japan) (in artificial seawater; Berges et al., 2001). The process was repeated, before the cells were resuspended in either IMK or non-axenic Symbiodiniaceae culture filtrate (B. minutum; grown in IMK, n = 3, as above) and incubated for 4.5 h prior to volatile sampling. This timing was selected based on a pilot study that identified additional compounds in bacterial cultures incubated for 4.5 h in IMK medium relative to axenic IMK medium. The IMK medium incubation was used as a reference to investigate the influence of Symbiodiniaceae culture filtrate on the bacterial volatilomes, hereafter the IMK incubation will be referred to as the “control.”
Symbiodiniaceae culture (B. minutum) was grown as described above, before triple filtration (5, 2, and 0.22 μm Isopore Membrane filters, Merck Millipore, Ltd., Tullagreen, Ireland) using a peristaltic pump (Watson Marlow Sci-Q 323) in order to remove all algal cells and bacteria. This filtration was carried out, on average, 3.5 h prior to its use in the experiment.
Sampling of volatiles was staggered so that all samples received a 4.5 h incubation period within either Symbiodiniaceae culture filtrate or the control. Following the incubation, technical duplicates were taken from each biological replicate for volatile sampling. Each technical duplicate (50 mL) was placed into a 100 mL gas tight vial and purged for 30 min (while under identical growth conditions) with instrument grade air (BOC Gases, Linde Group, Australia). Volatiles were captured on thermal desorption (TD) tubes (Markes Tenax TA) that were stored at 4°C until processing on GC–MS (<2 weeks later). For processing, TD tubes were desorbed (ULTRA 2 & UNITY 2; Markes International, Ltd., Llantrisant, United Kingdom) for 6 min at 300°C, then concentrated on a Tenax TA cold trap at −30°C. This concentrated sample was flash heated (300°C) and injected, at a flow rate of 2.3 mL/min, via a heated transfer line (150°C) onto a 7890A GC–MS (Agilent Technologies Pty, Ltd., Melbourne) fitted with a BP1 capillary column (60 m × 0.32 mm, 1 μm film thickness; SGE Analytical Science Pty, Ltd., Melbourne). Samples were run splitless to allow detection of trace compounds. The GC oven was heated at 35°C for 5 min, then ramped up 4°C min–1 to 160°C, then 20°C min–1 to 300°C for 10 min. The GC–MS was coupled to a mass-selective detector (Model 5975C; Agilent) that was set to a scanning range of 35–300 amu.
Flow Cytometry
Bacterial cell abundance was calculated in order to normalize BVOC concentrations. Aliquots (500 μL) of bacterial cultures were taken at the time of volatile sampling and fixed in glutaraldehyde (Sigma-Aldrich; 2% final concentration). Bacterial abundance was quantified by staining the cells with SYBR Green (1:10,000 final dilution; Applied Biosystems, Foster City, CA, United States) and analysis on a CytoFLEX S (Beckman Coulter, Brea, CA, United States) flow cytometer with filtered MilliQ water as the sheath fluid. For each sample, forward scatter (FSC), side scatter (SSC), and green fluorescence (SYBR Green) were recorded. The samples were analyzed at a flow rate of 25 μL min–1, with bacterial cells discriminated according to SSC and SYBR Green fluorescence.
Data Analysis
Chromatograms were stripped of common contaminating ions (73, 84, 147, 149, 207, 221, 262, and 281 m/z) using the Denoising function in OpenChrom (Wenig and Odermatt, 2010), before peak integration in ChemStation (Agilent Technologies Pty, Ltd., Melbourne) with an initial threshold of 15 and an initial peak width of 0.068. The chromatograms were then processed in the MSeasyTkGUI package (Nicolè et al., 2012) in R version 3.5.3 (R Core Team, 2017) to cluster all putative compounds. Peaks were identified by manually comparing mass spectra against a commercial library (NIST08 library in NIST MS Search v.2.2f; NIST, Gaithersburg, MD, United States). Blank media samples of both algal filtrate and IMK medium were run in conjunction with all analysis; the average values for contaminants present in the blanks were subtracted from all samples. Trimethyl silanol was removed from the analysis as it was likely contamination resulting from the hydrolysis of dimethylpolysiloxane (GC column material) (Cella and Carpenter, 1994). Any compound that did not occur in at least 4 of the 6 samples (2 technical replicates per biological replicate) was removed. All data were normalized to the bacterial cell concentration (cells mL–1).
PAST (PAleontological STatisitics; version 3.25) was used to generate principal component analysis (PCA) and non-metric multidimensional scaling (nMDS) plots on fourth root transformed data (Hammer et al., 2001). Primer v6.1.14 was used to perform a PERMANOVA+ (v1.0.4) analysis (Clarke and Gorley, 2006; Anderson et al., 2008) with p values based on the Monte Carlo statistic (Hope, 1968). All other statistical tests were run in IBM SPSS Statistic (version 25). UC denotes an unclassified compound (the number following UC indicates the retention time of the compound if the chemical functional group could not be determined). All mass spectra files are available (Accession number MSV000084431; MassIVE2), to help future studies as databases continue to improve.
Results
Across the two bacterial strains (Labrenzia sp. and M. adhaerens) tested here, a total of 35 volatile compounds were detected. Half of these compounds were successfully identified, and a further 11% were assigned to a chemical functional group. Only six of the compounds detected from the two Symbiodiniaceae-associated bacteria examined here have previously been reported from other bacteria. These included acetone, DMS, dimethyl trisulfide (DTS), toluene, camphor and 2-ethylhexanal (mVOC 2.0; Lemfack et al., 2018; as of 10/06/2019). The remaining 29 newly reported bacterial BVOCs included alcohols, aromatic hydrocarbons, esters, ethers, halogenated hydrocarbons, ketones, and organosulfurs. The volatilomes of the two bacterial isolates were not only distinguished from each other, but each bacterium generated a different volatilome when incubated in Symbiodiniaceae filtrate relative to the control (nMDS, Stress = 0.1259; Figure 1A).
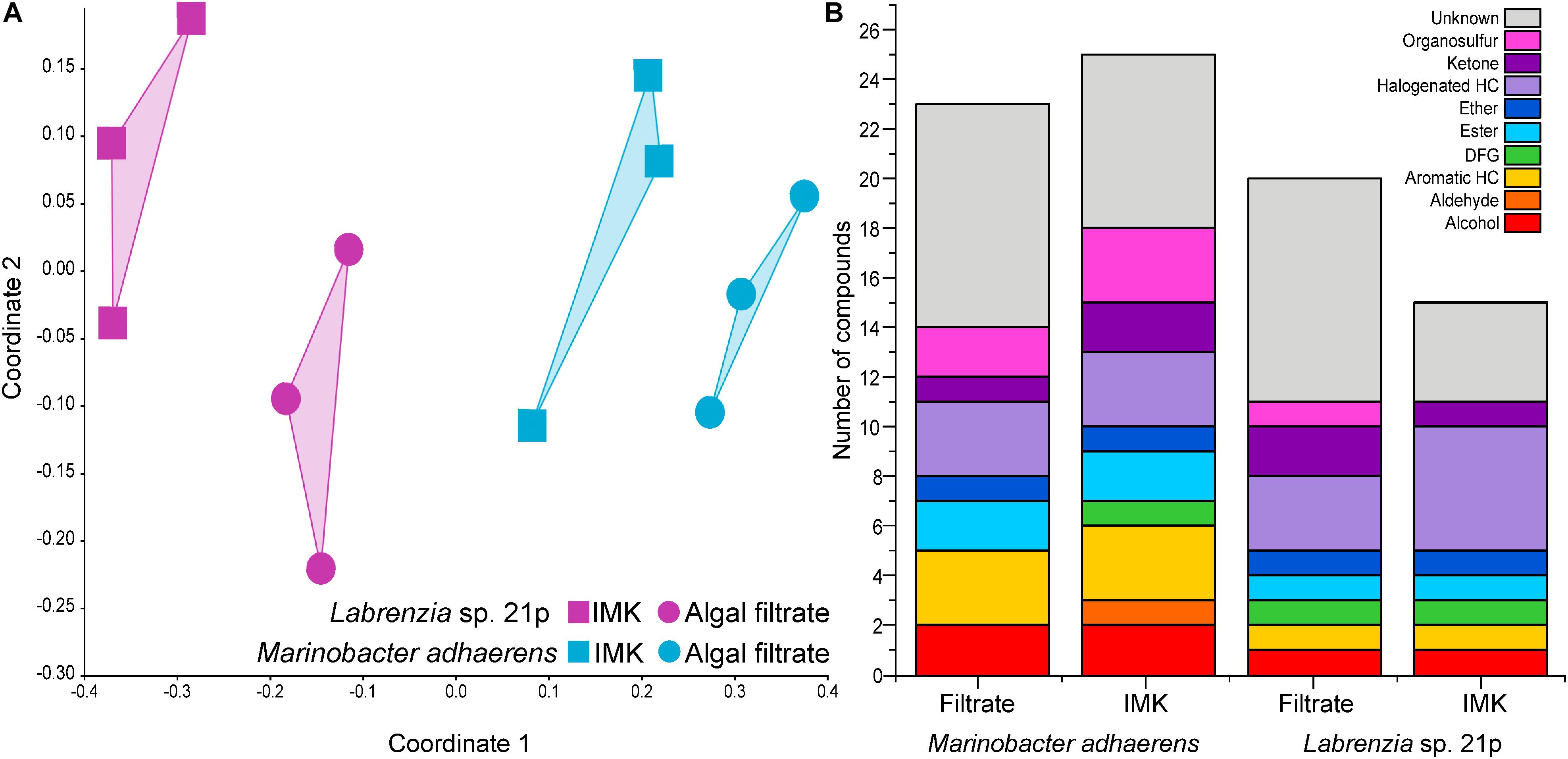
Figure 1. (A) Non-metric multidimensional scaling (nMDS; Bray–Curtis Similarity; Stress = 0.1055) analysis on the volatilomes of Labrenzia sp. 21p and Marinobacter adhaerens incubated either IMK medium or algal filtrate. (B) Chemical functional groups present in Labrenzia sp. 21p and Marinobacter adhaerens incubated in either IMK medium or algal filtrate (total number of volatile compounds present in at least two out of three replicates). HC, hydrocarbon; DFG, diverse functional groups.
The Labrenzia sp. volatilome was strongly influenced by the presence of Symbiodiniaceae filtrate. This induced significant differences in the overall structure of the volatilome (p = 0.014; PERMANOVA+; Supplementary Table S2) and more BVOCs present in the filtrate compared to the control incubations (20 vs. 15 compounds; Figure 1B). These differences were also confirmed using PCA, which identified the volatiles driving the strong separation between the two incubations. DMS was the leading cause of separation between the Labrenzia sp. control and filtrate volatilomes [Principal Component loading (PC)1 = 0.631], followed by acetone (PC1 = 0.405) and UC44.40 (PC2 = 0.409) (Figure 2A). Specifically, incubation of Labrenzia sp. in Symbiodiniaceae filtrate resulted in the production of one extra ketone and organosulfur, five additional unclassified compounds and two fewer halogenated hydrocarbons relative to the control (Figure 1B). Additionally, the relative quantities of nine compounds significantly differed among Labrenzia sp. cells incubated in the two media types, including DMS, acetone and 1,2-15,16-diepoxyhexadecane (Figure 3). Only one of these compounds, 2-ethyl-3-hydroxyhexyl-2-methylpropanoate, was detected in higher levels in the control incubation, while the other eight compounds were present in higher quantities in the Symbiodiniaceae culture filtrate (Figure 3).
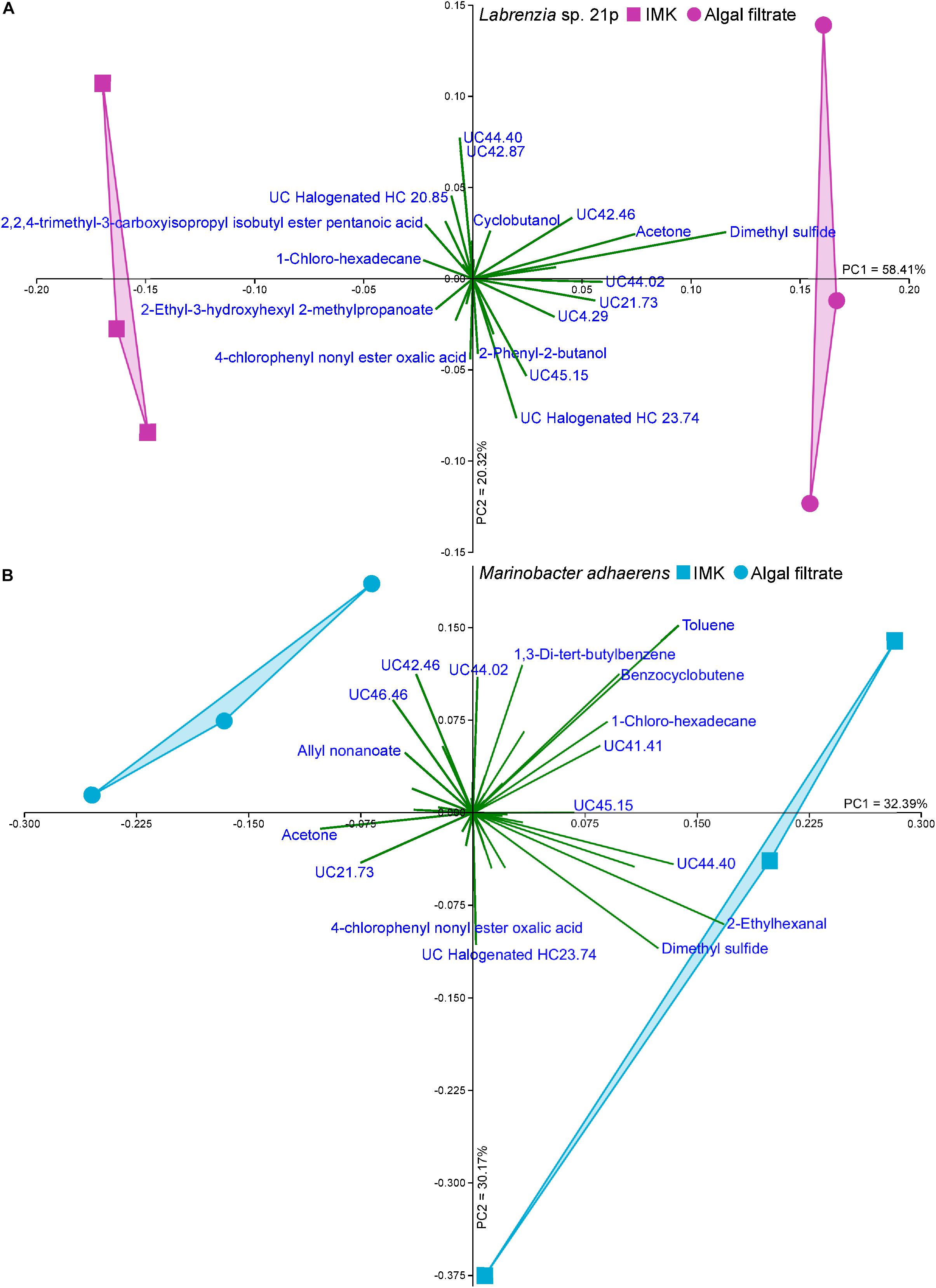
Figure 2. Principal component analysis (PCA; Bootstrap N = 1000) of (A) Labrenzia sp. 21p and (B) Marinobacter adhaerens incubated either IMK medium or Symbiodiniaceae filtrate.
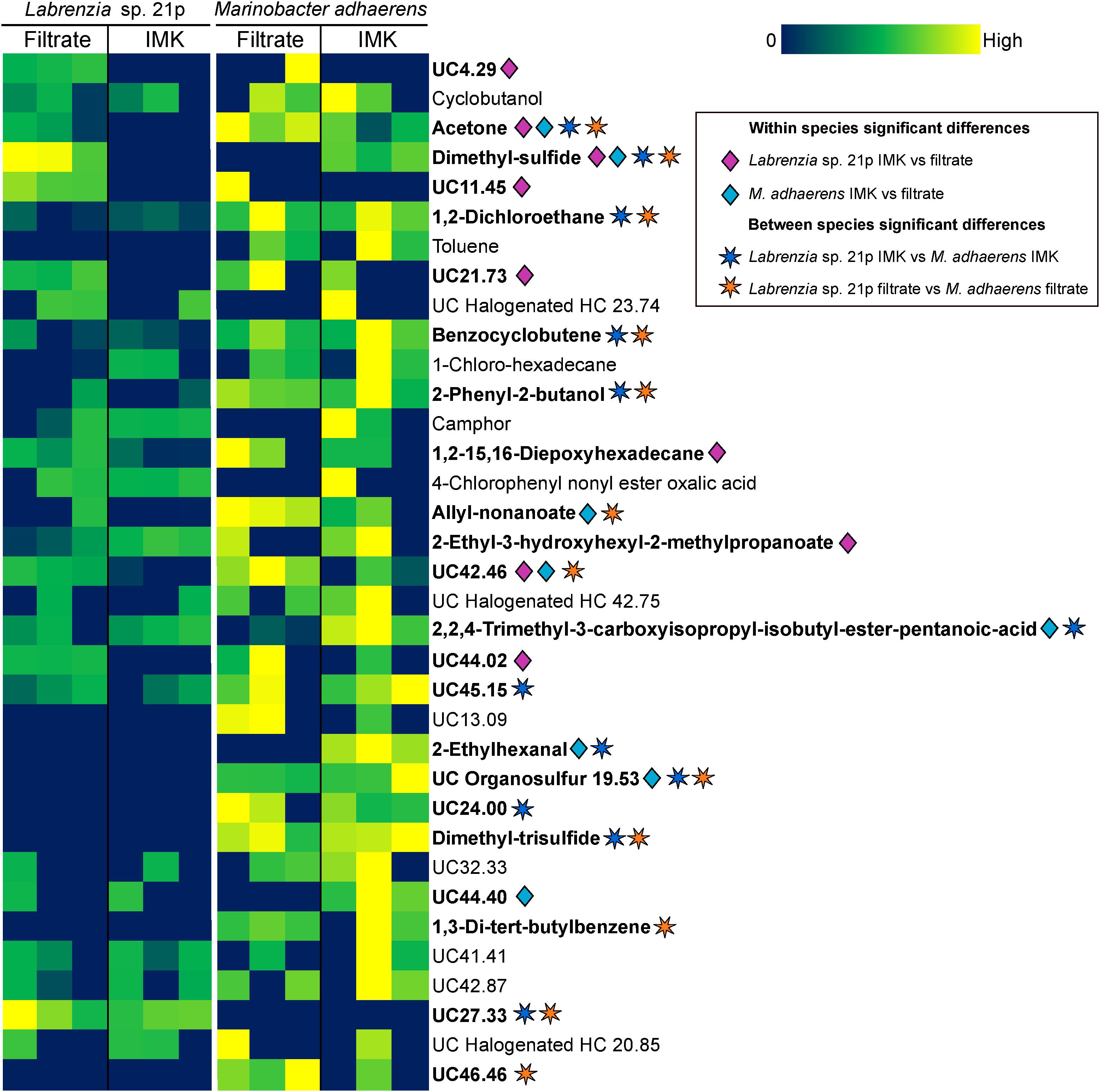
Figure 3. Heat map of all BVOCs present in at least two out of three replicates of Labrenzia sp. 21p and Marinobacter adhaerens HP15 incubated in either IMK medium or algal filtrate. Color scale is specific to each compound. Significant differences (p < 0.05; Kruskal–Wallis; IBM SPSS version 25) are denoted with diamonds for within species differences (pink for Labrenzia sp. 21p IMK vs. filtrate; blue for M. adhaerens IMK vs. filtrate) and stars for between species differences (dark blue for Labrenzia sp. 21p IMK vs. M. adhaerens IMK; orange for Labrenzia sp. 21p filtrate vs. M. adhaerens filtrate). HC, hydrocarbon; UC, unclassified (the number following UC indicates the retention time of the compound).
Incubation of M. adhaerens in Symbiodiniaceae filtrate did not result in significant differences in the overall structure of the M. adhaerens (p = 0.163; PERMANOVA+; Supplementary Table S2), nor a considerable change in the number of compounds detected relative to the control (23 BVOCs in filtrate, 25 in control) (Figure 1B). However, PCA did reveal some separation between the M. adhaerens volatilomes, with 2-ethylhexanal (PC1 = 0.445), UC44.40 (PC1 = 0.348), DMS (PC1 = 0.339, PC2 = −0.244) and toluene (PC1 = 0.307, PC2 = 0.421) driving this separation (Figure 2B). Moreover, there were specific differences in individual BVOCs, with the control incubation resulting in the production of the only aldehyde detected, an extra ketone and an extra organosulfur (Figure 1B). Additionally, the relative quantities of three BVOCs were significantly higher following incubation in the Symbiodiniaceae filtrate compared to the control, two of which were identified as acetone and allyl nonanoate (Figure 3). A further five compounds occurred in significantly lower quantities in the filtrate incubation, and included DMS, 2,2,4-trimethyl-3-carboxyisopropyl-isobutylester-pentanoic acid, 2-ethylhexanal, an UC organosulfur and UC44.40 (Figure 3). However, these differences in the quantities of individual BVOCs were not sufficient to significantly alter the overall structure of the M. adhaerens volatilome between culture filtrate and control.
The volatilomes of Labrenzia sp. and M. adhaerens cells also differed significantly to one another when incubated in Symbiodiniaceae culture filtrate (p = 0.007; PERMANOVA+; Supplementary Table S2). DMS and UC27.33 were significantly over-represented in Labrenzia sp. relative to M. adhaerens (Figure 3). However, 10 compounds were detected in significantly higher levels in M. adhaerens, including acetone, 1,2-dichloroethane, benzocyclobutene, 2-phenyl-2-butanol, allyl nonanoate, DTS, 1,3-di-tert-butylbenzene, UC organosulfur, UC42.46 and UC46.46 (Figure 3). Similarly, the control volatilomes of the Labrenzia sp. and M. adhaerens cells also differed significantly to one another (p = 0.014; PERMANOVA+; Supplementary Table S2). This difference was driven by significantly higher amounts of UC27.33 in the Labrenzia sp. relative to the control and a further 11 BVOCs over-represented in M. adhaerens, these encompassed acetone, DMS, 1,2-dichloroethane, benzocyclobutene, 2-phenyl-2-butanol, 2,2,4-trimethyl-3-carboxyisopropyl-isobutylester-pentanoic acid, 2-ethylhexanal, DTS, UC45.15, and UC24.00 (Figure 3). Neither species of bacteria showed any significant difference in cell abundance following incubation in either IMK medium or culture filtrate (Supplementary Figure S1).
Discussion
Despite the important roles that BVOCs can play in enhancing the growth and survival of microorganisms (Stotzky and Schenck, 1976; Ryu et al., 2003, 2004; Ramirez et al., 2010; Schmidt et al., 2015; Moore et al., 2019), bacterial BVOC production and emission remains relatively unexplored. Numerous BVOCs have been detected from a range of bacteria and while we are currently unable to identify or determine the ecological relevance of them all, they have the potential to strongly influence the growth and success of neighboring microorganisms. For example, the most abundant bacterium on Earth, Pelagibacter (SAR11), metabolizes diatom derived BVOCs, which constitute a significant portion of the carbon released by phytoplankton cells (Moore et al., 2019). Furthermore, bacteria often live within or associated with a host Raina et al. (2019) and as such, bacterial emissions can interact and react with host emissions (Schmidt et al., 2015; Kai et al., 2018). Importantly, such associations can potentially lead to BVOCs being falsely attributed to the host and can also lead to the formation of new compounds resulting from the interaction of different BVOCs (Kai et al., 2018). The multitude of bacterial BVOCs that have been studied in terrestrial ecosystems have also highlighted the signaling role of many of these BVOCs (Sharifi and Ryu, 2018). Following the characterization of the Symbiodiniaceae volatilome and microbiome (Lawson et al., 2018, 2019), here we investigated the potential influence of the exudates from non-axenic Symbiodiniaceae cultures on the volatilomes of two members of the Symbiodiniaceae core microbiome.
Between Labrenzia sp. and M. adhaerens, a total of 35 BVOCs were detected. This BVOC richness is comparable to observations in other marine bacterial species. For example, 69 different BVOCs were detected in a liquid culture of the Actinobacteria Streptomyces sp. (Dickschat et al., 2005a), while 38 BVOCs were detected from two Proteobacteria grown on agar plates, Loktanella BIO-204 and Dinoroseobacter shibae DFL-27 (Dickschat et al., 2005b).
The range of BVOCs detected in this study varied significantly between species and the substrates that the bacteria were incubated in. The Labrenzia sp. volatilome differed significantly between cells incubated in Symbiodiniaceae culture filtrate and the control, with more BVOCs detected when the bacteria were inoculated in the Symbiodiniaceae filtrate. The quantities of all but one of the BVOCs that differed significantly between the Labrenzia sp. volatilomes were higher within the Symbiodiniaceae filtrate incubation. Among these were DMS, acetone, 1,2-15,16-diepoxyhexadecane and five unclassified BVOCs. The extract of the herbaceous plant Artemisia annua has been shown to contain 1,2-15,16-diepoxyhexadecane, but this compound has no known functions in microorganisms (Hameed et al., 2016). Acetone has previously been reported in 13 bacterial genera (mVOC2.0; Lemfack et al., 2018) and is thought to inhibit fungal growth (Stotzky and Schenck, 1976; Amavizca et al., 2017). Notably, acetone was also present in a mixture of bacterial volatiles that was shown to promote the growth of the microalga Chlorella sorokiniana (Amavizca et al., 2017). The number of halogenated hydrocarbons decreased in the Labrenzia sp. filtrate relative to the control, with some of these compounds thought to function as signaling molecules (Ohsawa et al., 2001; Cabrita et al., 2010; Paul and Pohnert, 2011) and quorum sensing inhibitors (Hentzer et al., 2002). The higher total numbers of BVOCs detected in the Symbiodiniaceae filtrate incubation relative to the control could suggest that Labrenzia sp. is capable of utilizing Symbiodiniaceae-derived organic molecules to produce a wider diversity of volatile compounds. Given the dominance of this bacterial genus in the Symbiodiniaceae core microbiome (Lawson et al., 2018) and its culturability, future experiments should identify the nature of the Symbiodiniaceae-Labrenzia relationship and the role(s) that this bacterium could fulfill.
The number of M. adhaerens derived BVOCs detected were equivalent when incubated in either Symbiodiniaceae culture filtrate or control (23 and 25, respectively), and the structure of the M. adhaerens volatilome did not differ significantly between conditions. As such, the presence of the chemicals produced by the Symbiodiniaceae host appear to have had little to no effect on the M. adhaerens volatilome. Despite no apparent influence from the Symbiodiniaceae culture filtrate, the M. adhaerens volatilome was still diverse, comprising a range of alcohols (2), aromatic hydrocarbons (3), esters (2), ethers (1), halogenated hydrocarbons (3), ketones (1), and organosulfurs (2). The aromatic hydrocarbon toluene was detected in both M. adhaerens incubations. This compound has previously been detected in the volatilomes of 20 different bacterial genera (mVOC 2.0; Lemfack et al., 2018) and is also produced by both higher plants and microalgae (Misztal et al., 2015). The enzyme responsible for toluene biosynthesis (PhdB) was recently identified in bacteria (Beller et al., 2018), and while the biological function of this molecule remains uncertain, it has been proposed to serve as an infochemical and/or to play a role in stress response (Misztal et al., 2015). Dimethyl trisulfide (DTS) was also detected in all M. adhaerens samples under both incubation conditions. This compound has previously been detected in 34 bacterial genera (mVOC 2.0; Lemfack et al., 2018) and has been shown to have a strong antifungal activity (Fernando et al., 2005), while also playing a role in inter-organism signaling (Thibout et al., 1995). Marinobacter has previously been reported to consume volatile hydrocarbons (Ali et al., 2015), yet to the best of our knowledge no studies have reported BVOC production by this genus. Given the close association of this bacterial genus with phytoplankton, its production of the diverse range of BVOCs reported here may contribute to the complex inter-organism signaling underpinning these relationships.
Dimethyl sulfide is an important sulfur molecule known to act as antioxidant (Sunda et al., 2002) and as a signaling molecule (Seymour et al., 2010). DMS was a strong contributor to the differentiation between the volatilomes of Labrenzia sp. cells, as it was not detected in the control incubation. This is particularly interesting as Labrenzia aggregata is capable of synthesizing the DMS precursor, DMSP (Curson et al., 2017). Additionally, Symbiodiniaceae produce large quantities of both DMS and DMSP (Broadbent et al., 2002; Steinke et al., 2011; Deschaseaux et al., 2014a). Detection of DMS solely in the filtrate incubation may indicate that Labrenzia sp. was preferentially using Symbiodiniaceae-derived DMSP to form DMS. Surprisingly, DMS was detected in the M. adhaerens control volatilome. Given that IMK media only contains inorganic sulfur and no DMSP, the presence of DMS in this sample provides the first evidence that M. adhaerens may have the capacity to biosynthesize DMSP. Furthermore, the absence of the DMSP-producing gene dsyB in M. adhaerens genome (Supplementary Table S3) potentially points to the existence of alternative production pathways and a more widely distributed capacity to produce DMSP among marine bacteria.
Further comparing the BVOCs emissions of Labrenzia sp. and M. adhaerens with those previously identified from Symbiodiniaceae cultures (Lawson et al., 2019) demonstrated that only two compounds, DMS and toluene, were common to both bacteria and Symbiodiniaceae volatilomes (Figure 4). The ubiquitous nature of these BVOCs in the Labrenzia sp. and M. adhaerens volatilomes, and their similar putative functions, suggests that they might be critical for the physiology of both Symbiodiniaceae and its core bacteria. Excluding DMS and toluene, a further 33 BVOCs were only detected in the bacterial volatilomes, highlighting the capacity of these bacteria to produce a wide variety of BVOCs.
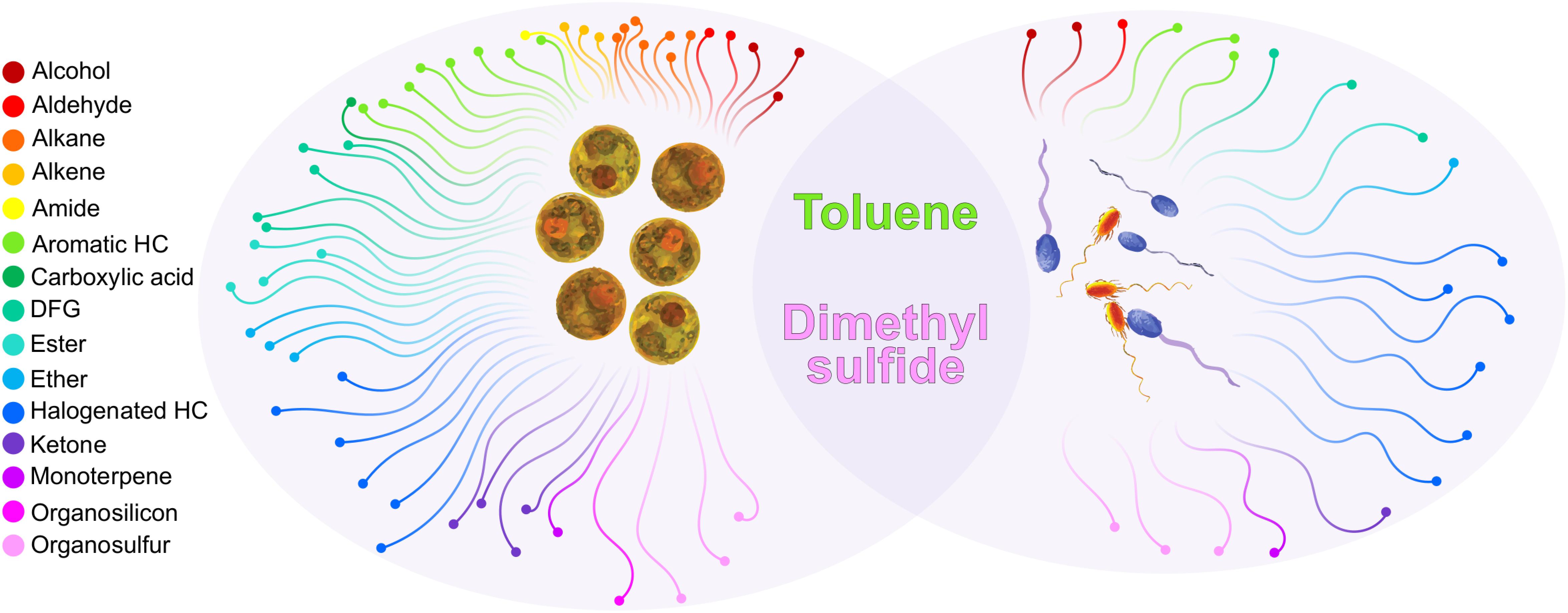
Figure 4. Functional chemical groups produced by Symbiodiniaceae (left oval) and Marinobacter adhaerens and Labrenzia sp. 21p (right oval) and the compounds that were present in both (overlap). Only compounds detected in Symbiodiniaceae during ambient conditions (26°C) were included. Compounds had to be present in at least four (out of six) technical replicates in at least one species. Unclassified BVOCs were excluded from this figure, see Supplementary Table S4 for a full list of BVOCs present in Symbiodiniaceae and bacteria.
In summary, we have demonstrated that two members of the Symbiodiniaceae core microbiome, M. adhaerens and Labrenzia sp. are capable of producing a diverse range of BVOCs, with the structure of the volatilome varying with the availability of Symbiodiniaceae culture-derived nutrients. Our experiments demonstrate the importance of marine bacteria as a source of BVOCs and provides evidence that their production may play a role in the complex chemical interplay between bacteria and their hosts – and thus between microbes that are critical to sustaining the health and productivity of coral reef ecosystems. Identification of the production of several key compounds such as DTS, toluene and a range of halogenated BVOCs, provides direction for future work specifically targeting and quantifying the occurrence and potential functions of these chemicals in marine ecosystems.
Data Availability Statement
The datasets generated for this study can be found in the GenBank MN305698, Mass Spectra data: https://massive. ucsd.edu.
Author Contributions
CL, J-BR, JS, and DS conceived and designed the project. CL performed the experiments, analyzed the data, produced the figures, and wrote the manuscript. MP provided technical and data analysis assistance. All authors edited the manuscript.
Funding
CL was supported by an Australian Government Research Training Program Scholarship. J-BR was supported by an Australian Research Council fellowship DE160100636. JS and J-BR were supported by an Australian Research Council grant DP180100838.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Axel Olander for his assistance transporting samples for analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00106/full#supplementary-material
Footnotes
References
Ali, N., Al-Awadhi, H., Dashti, N., Khanafer, M., El-Nemr, I., Sorkhoh, N., et al. (2015). Bioremediation of atmospheric hydrocarbons via bacteria naturally associated with leaves of higher plants. Int. J. Phytoremediation 17, 1160–1170. doi: 10.1080/15226514.2015.1045125
Amavizca, E., Bashan, Y., Ryu, C. M., Farag, M. A., Bebout, B. M., and De-Bashan, L. E. (2017). Enhanced performance of the microalga Chlorella sorokiniana remotely induced by the plant growth-promoting bacteria Azospirillum brasilense and Bacillus pumilus. Sci. Rep. 7, 1–11. doi: 10.1038/srep41310
Anderson, M., Gorley, R., and Clarke, K. (2008). PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods. Plymouth: PRIMER-E.
Atkinson, R., and Arey, J. (2003). Atmospheric degradation of volatile organic compounds. Chem. Rev. 103, 4605–4638. doi: 10.1021/cr0206420
Beller, H. R., Rodrigues, A. V., Zargar, K., Wu, Y. W., Saini, A. K., Saville, R. M., et al. (2018). Discovery of enzymes for toluene synthesis from anoxic microbial communities. Nat. Chem. Biol. 14, 451–457. doi: 10.1038/s41589-018-0017-4
Berges, J. A., Franklin, D. J., and Harrison, P. J. (2001). Evolution of an artificial seawater medium: improvements in enriched seawater, artificial water over the last two decades. J. Phycol. 37, 1138–1145. doi: 10.1046/j.1529-8817.2001.01052.x
Broadbent, A. D., and Jones, G. B. (2004). DMS and DMSP in mucus ropes, coral mucus, surface films and sediment pore waters from coral reefs in the great barrier reef. Mar. Freshw. Res. 55, 849–855. doi: 10.1071/MF04114
Broadbent, A. D., Jones, G. B., and Jones, R. J. (2002). DMSP in corals and benthic algae from the great barrier reef. Estuar. Coast. Shelf Sci. 55, 547–555. doi: 10.1006/ecss.2002.1021
Cabrita, M. T., Vale, C., and Rauter, A. P. (2010). Halogenated compounds from marine algae. Mar. Drugs 8, 2301–2317. doi: 10.3390/md8082301
Cella, J. A., and Carpenter, J. C. (1994). Procedures for the preparation of silanols. J. Organomet. Chem. 480, 23–26. doi: 10.1016/0022-328x(94)87098-5
Curson, A. R. J., Liu, J., Martínez, A. B., Green, R. T., Chan, Y., Carrión, O., et al. (2017). Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat. Microbiol. 2:17009. doi: 10.1038/nmicrobiol.2017.9
De Moraes, C. M., Mescher, M. C., and Tumlinson, J. H. (2001). Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 410, 577–580. doi: 10.1038/35069058
Deschaseaux, E. S. M., Beltran, V. H., Jones, G. B., Deseo, M. A., Swan, H. B., Harrison, P. L., et al. (2014a). Comparative response of DMS and DMSP concentrations in Symbiodinium clades C1 and D1 under thermal stress. J. Exp. Mar. Bio. Ecol. 459, 181–189. doi: 10.1016/j.jembe.2014.05.018
Deschaseaux, E. S. M., Jones, G. B., Deseo, M. A., Shepherd, K. M., Kiene, R. P., Swan, H. B., et al. (2014b). Effects of environmental factors on dimethylated sulfur compounds and their potential role in the antioxidant system of the coral holobiont. Limnol. Oceanogr. 59, 758–768. doi: 10.4319/lo.2014.59.3.0758
Dickschat, J. S., Martens, T., Brinkhoff, T., Simon, M., and Schulz, S. (2005a). Volatiles released by a Streptomyces species isolated from the North Sea. Chem. Biodivers. 2, 837–865. doi: 10.1002/cbdv.200590062
Dickschat, J. S., Wagner-Döbler, I., and Schulz, S. (2005b). The chafer pheromone buibuilactone and ant pyrazines are also produced by marine bacteria. J. Chem. Ecol. 31, 925–947. doi: 10.1007/s10886-005-3553-9
Fernando, W. G. D., Ramarathnam, R., Krishnamoorthy, A. S., and Savchuk, S. C. (2005). Identification and use of potential bacterial organic antifungal volatiles in biocontrol. Soil Biol. Biochem. 37, 955–964. doi: 10.1016/j.soilbio.2004.10.021
Frommlet, J. C., Sousa, M. L., Alves, A., Vieira, S. I., Suggett, D. J., and Serôdio, J. (2015). Coral symbiotic algae calcify ex hospite in partnership with bacteria. Proc. Natl. Acad. Sci. U.S.A. 112, 6158–6163. doi: 10.1073/pnas.1420991112
Frost, C. J., Appel, H. M., Carlson, J. E., De Moraes, C. M., Mescher, M. C., and Schultz, J. C. (2007). Within-plant signalling via volatiles overcomes vascular constraints on systemic signalling and primes responses against herbivores. Ecol. Lett. 10, 490–498. doi: 10.1111/j.1461-0248.2007.01043.x
Fu, P., Kawamura, K., Kanaya, Y., and Wang, Z. (2010). Contributions of biogenic volatile organic compounds to the formation of secondary organic aerosols over Mt. Tai, Central East China. Atmos. Environ. 44, 4817–4826. doi: 10.1016/j.atmosenv.2010.08.040
Gärdes, A., Kaeppel, E., Shehzad, A., Seebah, S., Teeling, H., Yarza, P., et al. (2010). Complete genome sequence of Marinobacter adhaerens type strain (HP15), a diatom-interacting marine microorganism. Stand. Genomic Sci. 3, 97–107. doi: 10.4056/sigs.922139
Garren, M., Son, K., Raina, J.-B., Rusconi, R., Menolascina, F., Shapiro, O. H., et al. (2014). A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J. 8, 999–1007. doi: 10.1038/ismej.2013.210
Hallquist, M., Wenger, J. C., Baltensperger, U., Rudich, Y., Simpson, D., Claeys, M., et al. (2009). The formation, properties and impact of secondary organic aerosol: current and emerging issues. Atmos. Chem. Phys. 9, 5155–5236.
Hameed, I. H., Altameme, H. J., and Idan, S. A. (2016). Artemisia annua: biochemical products analysis of methanolic aerial parts extract and anti-microbial capacity. Res. J. Pharm. Biol. Chem. Sci. 7, 1843–1868.
Hammer, Ø, Harper, D. A. T., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron 4:9.
Han, J., Zhang, L., Wang, S., Yang, G., Zhao, L., and Pan, K. (2016). Co-culturing bacteria and microalgae in organic carbon containing medium. J. Biol. Res. 23:8. doi: 10.1186/s40709-016-0047-6
Hentzer, M., Riedel, K., Rasmussen, T. B., Heydorn, A., Andersen, J. B., Parsek, M. R., et al. (2002). Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148, 87–102. doi: 10.1099/00221287-148-1-87
Hope, A. C. A. (1968). A simplified monte carlo significance test procedure. J. R. Stat. Soc. Ser. B 30, 582–598. doi: 10.1088/0031-9155/54/3/005
Hopkins, F. E., Bell, T. G., Yang, M., Suggett, D. J., and Steinke, M. (2016). Air exposure of coral is a significant source of dimethylsulfide (DMS) to the atmosphere. Sci. Rep. 6, 1–11. doi: 10.1038/srep36031
Kai, M., Effmert, U., Lemfack, M. C., and Piechulla, B. (2018). Interspecific formation of the antimicrobial volatile schleiferon. Sci. Rep. 8, 1–6. doi: 10.1038/s41598-018-35341-3
Koike, K., Jimbo, M., Sakai, R., Kaeriyama, M., Muramoto, K., Ogata, T., et al. (2004). Octocoral chemical signaling selects and controls dinoflagellate symbionts. Biol. Bull. 207, 80–86. doi: 10.2307/1543582
Lawson, C. A., Possell, M., Seymour, J. R., Raina, J.-B., and Suggett, D. J. (2019). Coral endosymbionts (Symbiodiniaceae) emit species-specific volatilomes that shift when exposed to thermal stress. Sci. Rep. 9:17395. doi: 10.1038/s41598-019-53552-0
Lawson, C. A., Raina, J. B., Kahlke, T., Seymour, J. R., and Suggett, D. J. (2018). Defining the core microbiome of the symbiotic dinoflagellate. Symbiodinium. Environ. Microbiol. Rep. 10, 7–11. doi: 10.1111/1758-2229.12599
Lemfack, M. C., Gohlke, B. O., Toguem, S. M. T., Preissner, S., Piechulla, B., and Preissner, R. (2018). MVOC 2.0: a database of microbial volatiles. Nucleic Acids Res. 46, D1261–D1265. doi: 10.1093/nar/gkx1016
Little, A. F., van Oppen, M. J. H., and Willis, B. L. (2004). Flexibility in algal endosymbioses shapes growth in reef corals. Science 304, 1492–1494. doi: 10.1126/science.1095733
Loreto, F., and Velikova, V. (2001). Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 127, 1781–1787. doi: 10.1104/pp.010497
Matsumoto, J. (2014). Measuring biogenic volatile organic compounds (BVOCs) from vegetation in terms of ozone reactivity. Aerosol Air Qual. Res. 14, 197–206. doi: 10.4209/aaqr.2012.10.0275
Misztal, P. K., Hewitt, C. N., Wildt, J., Blande, J. D., Eller, A. S. D., Fares, S., et al. (2015). Atmospheric benzenoid emissions from plants rival those from fossil fuels. Sci. Rep. 5, 1–10. doi: 10.1038/srep12064
Moore, E. R., Davie−Martin, C. L., Giovannoni, S. J., and Halsey, K. H. (2019). Pelagibacter metabolism of diatom−derived volatile organic compounds imposes an energetic tax on photosynthetic carbon fixation. Environ. Microbiol 78, 57–64. doi: 10.1111/1462-2920.14861
Muscatine, L., and Porter, J. W. (1977). Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27, 454–460. doi: 10.2307/1297526
Nicolè, F., Guitton, Y., Courtois, E. A., Moja, S., Legendre, L., and Hossaert-McKey, M. (2012). MSeasy: unsupervised and untargeted GC-MS data processing. Bioinformatics 28, 2278–2280. doi: 10.1093/bioinformatics/bts427
Ohsawa, N., Ogata, Y., Okada, N., and Itoh, N. (2001). Physiological function of bromoperoxidase in the red marine alga, Corallina pilulifera: production of bromoform as an allelochemical and the simultaneous elimination of hydrogen peroxide. Phytochemistry 58, 683–692. doi: 10.1016/S0031-9422(01)00259-X
Paul, C., and Pohnert, G. (2011). Production and role of volatile halogenated compounds from marine algae. Nat. Prod. Rep. 28, 186–195. doi: 10.1039/c0np00043d
Raina, J.-B., Fernandez, V. I., Lambert, B. S., Stocker, R., and Seymour, J. R. (2019). The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol. 17, 284–294. doi: 10.1038/s41579-019-0182-9
Raina, J.-B., Tapiolas, D., Motti, C. A., Foret, S., Seemann, T., Tebben, J., et al. (2016). Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ 4:e2275. doi: 10.7717/peerj.2275
R Core Team (2017). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: https://www.r-project.org/.
Ramirez, K. S., Lauber, C. L., and Fierer, N. (2010). Microbial consumption and production of volatile organic compounds at the soil-litter interface. Biogeochemistry 99, 97–107. doi: 10.1007/s10533-009-9393-x
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243, 1–10. doi: 10.3354/meps243001
Runyon, J. B., Mescher, M. C., and De Moraes, C. M. (2006). Chemical plants cues guide host location and host selection by parasitic plants. Science 313, 1964–1967. doi: 10.1126/science.1131371
Ryu, C.-M., Farag, M. A., Hu, C.-H., Reddy, M. S., Kloepper, J. W., and Pare, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026. doi: 10.1104/pp.103.026583
Ryu, C.-M., Farag, M. A., Hu, C.-H., Reddy, M. S., Wei, H.-X., Pare, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 100, 4927–4932. doi: 10.1073/pnas.0730845100
Sandhya, S., Preetha, K., Nair, A., and Antony, M. (2017). Isolation, characterisation and phylogenetic diversity of culturable bacteria associated with marine microalgae from saline habitats of south India. Aquat. Microb. 79, 21–30. doi: 10.3354/ame01815
Schmidt, R., Cordovez, V., De Boer, W., Raaijmakers, J., and Garbeva, P. (2015). Volatile affairs in microbial interactions. ISME J. 9, 2329–2335. doi: 10.1038/ismej.2015.42
Seymour, J. R., Simo, R., Ahmed, T., and Stocker, R. (2010). Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–346. doi: 10.1126/science.1188418
Shan, S., Wang, W., Song, C., Wang, M., Sun, B., Li, Y., et al. (2019). The symbiotic bacteria Alcaligenes faecalis of the entomopathogenic nematodes Oscheius spp. exhibit potential biocontrol of plant- and entomopathogenic fungi. Microb. Biotechnol. 12, 459–471. doi: 10.1111/1751-7915.13365
Sharifi, R., and Ryu, C. M. (2018). Sniffing bacterial volatile compounds for healthier plants. Curr. Opin. Plant Biol. 44, 88–97. doi: 10.1016/j.pbi.2018.03.004
Sonnenschein, E. C., Syit, D. A., Grossart, H.-P., and Ullrich, M. S. (2012). Chemotaxis of Marinobacter adhaerens and its impact on attachment to the diatom thalassiosira weissflogii. Appl. Environ. Microbiol. 78, 6900–6907. doi: 10.1128/aem.01790-12
Steinke, M., Brading, P., Kerrison, P., Warner, M. E., and Suggett, D. J. (2011). Concentrations of dimethylsulfoniopropionate and dimethyl sulfide are strain-specific in symbiotic dinoflagellates (symbiodinium sp., dinophyceae). J. Phycol. 47, 775–783. doi: 10.1111/j.1529-8817.2011.01011.x
Stotzky, G., and Schenck, S. (1976). Volatile organic compounds and microorganisms. Crit. Rev. Microbiol. 4, 333–382. doi: 10.3109/10408417609102303
Sunda, W., Kieber, D. J., Kiene, R. P., and Huntsman, S. (2002). An antioxidant function for DMSP and DMS in marine algae. Nature 418, 317–320. doi: 10.1038/nature00851
Swan, H. B., Crough, R. W., Vaattovaara, P., Jones, G. B., Deschaseaux, E. S. M., Eyre, B. D., et al. (2016). Dimethyl sulfide and other biogenic volatile organic compound emissions from branching coral and reef seawater: potential sources of secondary aerosol over the great barrier reef. J. Atmos. Chem. 73, 303–328. doi: 10.1007/s10874-016-9327-7
Tebben, J., Tapiolas, D. M., Motti, C. A., Abrego, D., Negri, A. P., Blackall, L. L., et al. (2011). Induction of larval metamorphosis of the coral Acropora millepora by tetrabromopyrrole isolated from a Pseudoalteromonas bacterium. PLoS One 6: e19082. doi: 10.1371/journal.pone.0019082
Thibout, E., Guillot, J. F., Ferary, S., Limouzin, P., and Auger, J. (1995). Origin and identification of bacteria which produce kairomones in the frass of Acrolepiopsis assectella (Lep. Hyponomeutoidea). Experientia 51, 1073–1075. doi: 10.1007/BF01946919
Vickers, C. E., Possell, M., Cojocariu, C. I., Velikova, V. B., Laothawornkitkul, J., Ryan, A., et al. (2009). Isoprene synthesis protects transgenic tobacco plants from oxidative stress. Plant, Cell Environ. 32, 520–531. doi: 10.1111/j.1365-3040.2009.01946.x
Wenig, P., and Odermatt, J. (2010). OpenChrom: a cross-platform open source software for the mass spectrometric analysis of chromatographic data. BMC Bioinformatics 11:405. doi: 10.1186/1471-2105-11-405
Keywords: volatilome, Labrenzia sp., Marinobacter adhaerens HP15, GC–MS, core microbiome, Symbiodiniaceae, algal–bacteria interactions
Citation: Lawson CA, Seymour JR, Possell M, Suggett DJ and Raina J-B (2020) The Volatilomes of Symbiodiniaceae-Associated Bacteria Are Influenced by Chemicals Derived From Their Algal Partner. Front. Mar. Sci. 7:106. doi: 10.3389/fmars.2020.00106
Received: 08 October 2019; Accepted: 10 February 2020;
Published: 25 February 2020.
Edited by:
Tony Gutierrez, Heriot-Watt University, United KingdomReviewed by:
Carl J. Carrano, San Diego State University, United StatesZongze Shao, Third Institute of Oceanography, China
Alex J. Poulton, The Lyell Centre, United Kingdom
Copyright © 2020 Lawson, Seymour, Possell, Suggett and Raina. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Caitlin A. Lawson, Y2FpdGxpbi5hbGlueWFAZ21haWwuY29t; Jean-Baptiste Raina, amVhbi1iYXB0aXN0ZS5yYWluYUB1dHMuZWR1LmF1
 Caitlin A. Lawson
Caitlin A. Lawson Justin R. Seymour
Justin R. Seymour Malcolm Possell
Malcolm Possell David J. Suggett
David J. Suggett Jean-Baptiste Raina
Jean-Baptiste Raina