- Key Laboratory of Eutrophication and Red Tide Prevention of Guangdong Higher Education Institute, College of Life Science and Technology, Jinan University, Guangzhou, China
Though microalgae have been considered the potential resource for lipid production, native strains are unable to meet the industrial demand. Here, we aim to uncover the complex molecular relationship between algal growth and lipid accumulation. Transcriptomic analysis revealed the crucial role of plastidial fatty acid and triacylglycerol (TAG) biosynthetic machinery in lipid overproduction. The expression of key fatty acid biosynthetic genes such as acetyl-CoA carboxylase (ACCase), malonyl CoA−acyl carrier protein transacylase (MCAT), 3-ketoacyl synthase (KAS), 3-ketoacyl-ACP reductase (KAR) increased during day 10–13 of cultivation, particularly plastidial TAG biosynthetic genes substantially increased. However, expression of genes involved in ER TAG biosynthesis increased only in the stationary phase, which implied the potential of plastidial TAG biosynthesis. This report provides a novel insight into the growth-phase dependent lipogenic orchestration, and also uncovers the signature genes and plastidial TAG biosynthesis that might be extrapolated for improving lipogenic traits of algae.
Introduction
Microalgae have emerged as the potential biological feedstocks for the sustainable production of a wide range of biocomponents including lipids, polyunsaturated fatty acids, vitamins, carotenoids, and recombinant proteins (Mata et al., 2010). Considering the negative environmental and economic impact of fossil fuels combustion and rapidly depleting reserves have further attracted the research attention toward commercial exploitation of microalgal biofuel owing to their inherited beneficial characteristics (Chisti, 2007). Particularly, oleaginous microalgal species are gaining huge research interests due to their promising oleaginicity and possibilities of wide commercial applications (Han et al., 2017). However, native strains do not hyperaccumulate biocomponents under optimal conditions, which extremely incumbers their commercial applications (Chisti, 2013). For instance, algal cells have been shown to overproduce triacylglycerol (TAG), the critical precursor for biofuels under stress conditions which constrain the biomass and growth, thereby imped the overall productivity and commercial viability of the whole production process (Yang et al., 2013). Thus, it is of paramount significance to improve the genetic traits to produce algal biocomponents in a commercially feasible manner.
Genetic engineering provides a promising strategy to precisely perturb the crucial metabolic node to enhance the titer of desired products, possibly without impeding the cellular physiological characteristics. However, previous algal genetic engineering studies have yielded inconsistent success (Li et al., 2010). Among the factors governing algal genetic engineering, judicious selection of potential metabolic targets has been considered crucial (Li et al., 2019). With the advancement of genomics, development of genetic tool kits and characterization of potential genetic elements have paved the way for designing proficient genetic transformation (Zou et al., 2018). Various studies have interrogated the differential expression pattern of key lipogenic genes under nutrient stress that could elicit algal lipid accumulation potential (Yang et al., 2013; Li et al., 2014). These studies have examined the mechanistic role of nutrient limitation in orchestrating the key metabolic circuits in various microalgae at the cost of cellular growth and altered metabolic activities.
Meanwhile, the growth phase of the algal life cycle possesses crucial points that govern the growth during log phase and accumulation of energy reserves for the cellular requirements during late-growth phase (Yang et al., 2013; Yuan et al., 2019). However, data availability on the expression pattern of key genes during algal growth is limited, which has been the critical point in governing the biomass and lipid accumulation. There are few reports that report the complex and contradictory relationships between growth and lipid accumulation during the growth phase. To this end, we sought to investigate the transcriptome to uncover the expression pattern of key genes involved in lipid metabolic pathway in the oleaginous heterokont Nannochloropsis oceanica, which might provide some valuable molecular cues that underlie the concurrent biomass and lipid overproduction.
Materials and Methods
Algal Strain and Cultivation
Nannochloropsis oceanica CCAP 849/10 (formerly CCMP1779) was procured from NCMA (National Center for Marine Algae and Microbiota, United States). The algal cells were cultivated in f/2 liquid medium at 25°C as reported previously (Li et al., 2016).
Process Monitoring of N. oceanica Cells
Algal cultivation was monitored by evaluating certain crucial parameters. Cell growth was determined by direct cell count method under a light microscope (Li et al., 2016). Photosynthetic efficiency was monitored by measuring the chlorophyll fluorescence parameter Fv/Fm, the maximum photochemical quantum yield of photosystem II as described previously (Li et al., 2016). Total phosphorus concentration in the culture medium was determined by using inductively coupled plasma analyzer iCAP 7400 (Thermo Scientific, United States) and the total nitrogen concertation was determined using a Dionex ICS-5000 ion chromatography system with suppressed conductivity detector (Thermo Scientific).
Analyses of Lipids and Fatty Acid Composition
Relative neutral lipid content was fluorometrically measured by Nile red staining as described (Li et al., 2016). The total fatty acid composition was determined as fatty acid methyl esters by gas chromatography-mass spectrometry (GC-MS) as described (Li et al., 2016). Lipid saturation index was calculated as described previously by using the formulae below (Chandra et al., 2019).
Transcriptome Sequencing and Analyses
Total RNA was extracted from the cells at Day 7, 10, and 13 of cultivation using RNA Iso plus kit (Takara, Japan), with three replicates each treatment. Illumina sequencing was conducted at the Gene de novo company (Guangzhou, China). The prepared mRNA samples were enriched with oligo (dT) beads and added with fragmentation buffer and the mRNA was fragmented. Thereafter, the mRNA was retroscribed using a random hexamer primer and subsequently, the second strands were synthesized. The double-stranded cDNA was purified using the QIAGEN quick PCR extraction kit and the purified cDNAs were subjected to end repair and single adenine (A) addition. Finally, sequencing adaptors were ligated to the cDNA fragments, and the resultant samples were resolved in agarose gel electrophoresis for gel-purification and then enriched by PCR amplification. The library products were sequenced using the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, United States). The default parameters were used to pass reads using the Illumina quality-control tools (Li et al., 2014). The relative transcript abundance of key fatty acid and lipid biosynthetic genes was quantified by real-time quantitative PCR (qPCR) using a SYBR green qPCR SuperMix (Invitrogen, United States) on ABI PRISM 7500 Sequence Detection System (Applied Biosystems, United States) as per the protocol described elsewhere (Li et al., 2016).
RNA-seq Data Processing and Pathway Analysis
Nannochloropsis oceanica reference genome was retrieved (Vieler et al., 2017) and used for annotation. We aligned our RNA short reads under optimized conditions to the reference genome using SOAP 2.21 (BGI, Shenzhen, China). The identified gene with a minimum of one read of lower than this was considered as non-confident and thus discarded. The RPKM value was estimated and log2-transformed for each gene defined in the Nannochloropsis database, to estimate the differential expression pattern between three different cultivation phases (Li et al., 2014). The KEGG pathway database was downloaded, and all annotated genes of the N. oceanica genome were obtained from the KEGG database, with an e-value cut-off of 1e-5. BLAST (Basic Local Alignment Search Tool1) search was carried out for the Nannochloropsis genes as the query against all available annotated KEGG genes from other species.
Subcellular Localization Prediction of Key Proteins Involved in Lipogenesis
The subcellular localization of key proteins involved in TAG biosynthesis with significant differential expression was predicted by using online tools including SignalP (Petersen et al., 2011), ChloroP (Emanuelsson et al., 1999), Mitoprot (Claros, 1995), and HECTAR (Artzi et al., 2008).
Ultrastructural Analysis by TEM
Ultrastructural analyses of the N. oceanica cells at 7th, 10th, and 13th day of cultivation were analyzed by transmission electron microscopy as described previously (Li et al., 2019) and ultrathin sections were cut made on an ultramicrotome LKB 8800 (LKB Instruments, United States). Thereafter, sections were stained and observed under a JEM-1200EX transmission electron microscope (JEOL, Japan), and images were recorded on EM film 4489 (Eastman-Kodak, NY, United States) (Li et al., 2019).
Results and Discussion
Bioprocess Monitoring and Lipidomic Analyses Revealed the Potential Growth Phase for Lipid Accumulation
Growth rate and algal biomass are regarded as the crucial parameters that determine the economic feasibility of the algal industrial applications (Li et al., 2018). Growth rate of the photosynthetic microalgae is regulated by various factors such as nutrient, light and temperature and thus, monitoring the microalgal cultivations has been considered crucial for assessing algal commercial potential (Havlik et al., 2013a, b). Thus, we attempted to monitor the process of microalgal cultivation during the study by assessing growth rate, biomass content, maximum quantum yield of photosystem II (Fv/Fm), determination of nitrogen and phosphorus content and cellular morphology. It is well known that algal cells involve in growth during log phase, whereas cells involve in energy metabolism in the stationary phase to meet the energy demand and for the cell survival under nutrient-deprived conditions (Yang et al., 2013). Consequently, various strategies such as nutrient deprivation, stress treatments, etc. have been used to increase algal lipid accumulation, however, these sub-optimal treatment methods resulted in reduced growth and impaired biomass accumulation. Thus, concurrent biomass and lipid accumulation during the microalgal cultivation phase has been considered the most contradictory parameters to be achieved (Wang et al., 2019). However, large-scale production of algal biocomponents in a commercially viable manner warrants simultaneous overproduction of biomass and lipids. We thus evaluated the growth and lipid accumulation properties of N. oceanica cells during the whole 14-day cultivation phase.
As shown in Figures 1A,B, the growth rate was gradually increased and reached the stationary phase on day 10 of cultivation and similar trend was observed in terms of Fv/Fm. We then determined total nitrogen and phosphorus content in the cultivation medium, which showed that both nitrogen and phosphorus content was found to be reduced during cultivation (Figure 1C). It is well known that nitrogen and phosphorus are the indispensable nutrient elements required for the biosynthesis of biological macromolecules, photosynthesis, and energy transfer, thereby govern the crucial role in cellular metabolism and growth (Singh et al., 2015; Kokabi et al., 2019). Consistently, adequate availability of the nitrogen and phosphorus content during log phase of the microalgal cultivation facilitated the growth rate of the cells. Meanwhile, effective consumption of these nutrient molecules by the algal cells during log phase resulted in reduced availability of N and P content during the stationary phase, thereby resulted in induced lipid accumulation (de Alva et al., 2018). This observation is in accordance with the previous report which implied that reduced nutrient concentration during the stationary phase induced lipid accumulation (Ren et al., 2019). However, the phenomenon of lipid increment during the stationary phase in the optimized cultivation medium is different from that cultivated in nutrient deprived conditions (Yang et al., 2013). Under nutrient deprivation the routine cellular metabolic activities were altered and physiological characteristics were impaired, which in turn led to accumulation of energy rich molecules at the cost of algal growth, whereas under optimized cultivation conditions the cellular physiological parameters and metabolic activities would not be impaired, thereby cells would enter the lipid accumulation phase without hindered growth activities during the stationary phase (Adams et al., 2013). Similarly, the lipidomic analysis revealed that lipid content was lower during the log phase and gradually increased from day 10, and reached the peak level on day 13 (Figures 1D,E), and lipid productivity was 5.93 mg/L/Day (Figure 1E). Besides, lipid saturation index was calculated and lipid unsaturation was found to be increased on day 10, whereas lipid saturation was found to be increased on day 7 and 13 (Figure 1F). During day 10 and 13, both growth and lipid content were significantly altered, which revealed that the lipid accumulation is highly associated with the growth phase (Chiu et al., 2009). In this regard, it is of paramount importance to scrutiny the potential phase that elicited both biomass and lipid accumulation in microalgae (Klok et al., 2013). Zheng et al. (2016) identified that cells hyperaccumulated biomass and lipids after day 6 of cultivation under high-temperature treatment in Scenedesmus quadricauda. Congruently, Chlorella ellipsoidea accumulated both biomass and lipid on day 9 of cultivation which was between the log and stationary phase (Yang et al., 2011). Besides, the scrutinization of the cultivation phase for simultaneous accumulation of lipids and biomass is considered crucial and this time point can exemplify the critical time point for analyzing the expression pattern of key genes that underpin the concurrent enhancement of lipids and biomass (Msanne et al., 2012). To further gain insights into the fatty acid composition of the lipids under this cultivation phase, we determined fatty acid composition by GC-MS on day 7, 10, and 13 of cultivation. Interestingly, fatty acid content was comparable on day 13 than other days (Supplementary Table S1). Besides, Our previous study has reported the significance of chloroplast localized TAG biosynthetic pathway on governing the fatty acid composition in TAG pool, specifically C16:0 at the sn-2 position was found to be remarkably increased over C18:0 fatty acids (Balamurugan et al., 2017). Fan et al. (2011) demonstrated that nitrogen starved Chlamydomonas cells accumulated significantly higher amount of C16 fatty acid moieties, which is accounted for up to 90% of the total TAG implied the substrate specificity of plastidial LPAAT toward C16:0 fatty acid moieties rather than ER-localized LPAAT (Fan et al., 2011). These studies have provided the crucial role of subcellular organelles in governing the TAG biogenesis and our results which depicted that C16:0-fatty acids were the predominant fatty acid composition compared to other fatty acids, thereby provoking the necessity to interrogate the role of subcellular compartments in governing lipogenesis in a growth-phase dependent manner (Kim et al., 2018).
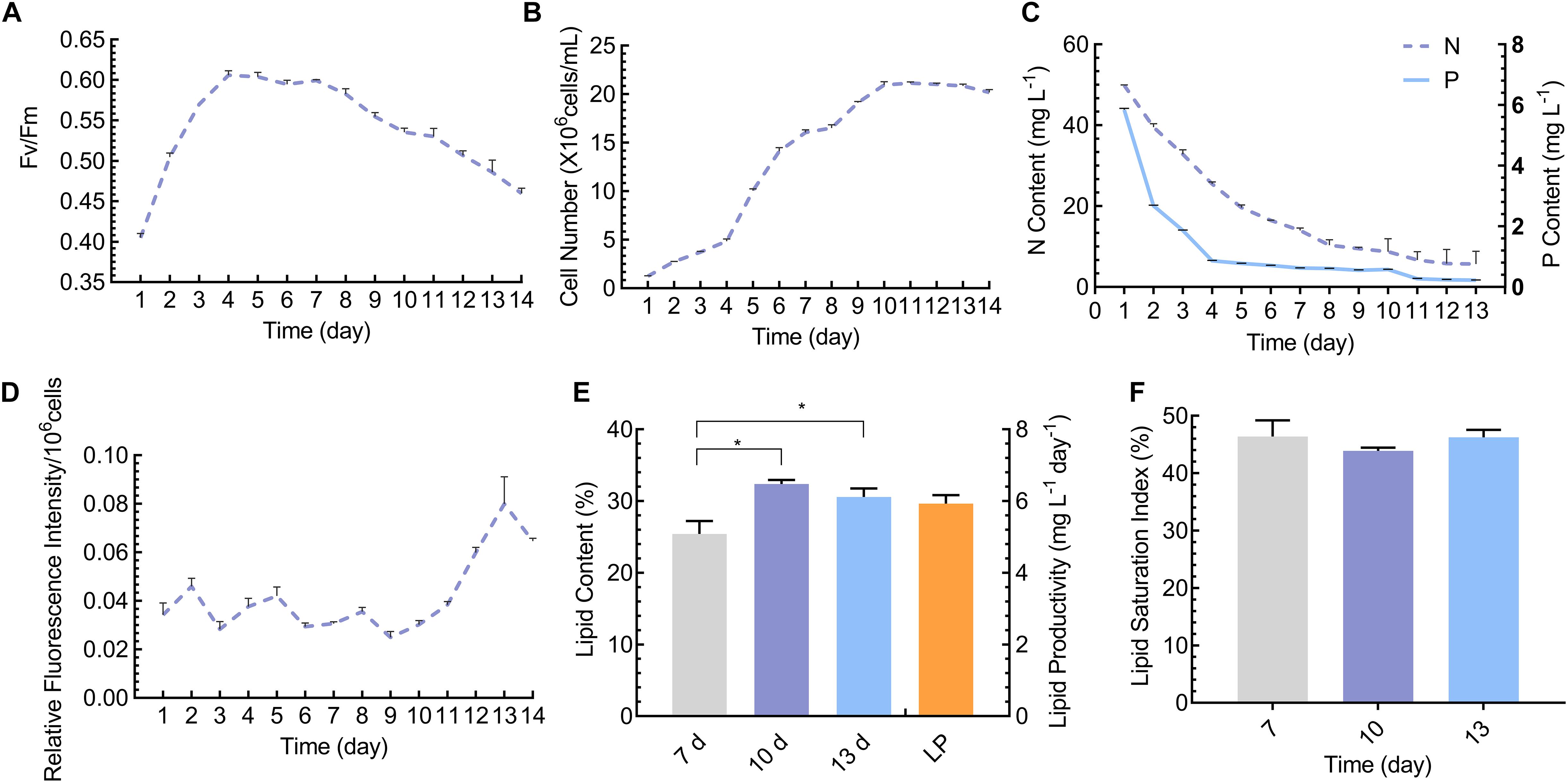
Figure 1. Bioprocess monitoring and lipidomic analyses throughout the cultivation phase. (A) Photosynthetic performance as determined by Fv/Fm; (B) Cell growth curve as measured by direct count method; (C) Determination of nitrogen and phosphorus concentration in the growth medium; (D) Determination of relative neutral lipid content by Nile-red fluorometric method per ml of culture; (E) Total lipid content (DCW) and total lipid productivity, LP- lipid productivity (mg/L/Day); (F) Lipid saturation index (%). Significant difference is indicated at P < 0.05 (*) level. Each value represents mean ± SD (n = 3).
RNA-seq Analyses of Transcriptional Modifications and Their Functional Annotation
Thereafter, transcriptomic analysis was carried out at different cultivation phases. Totally, 10939 of genes were expressed in all experimental sets, i.e. day 7, 10, and 13 of the cultivation. Among them, 340 were differentially expressed at a significant level (p > 0.05) between day 7 and 10. Meanwhile, 1283 genes were differentially expressed in the set compared between day 13 and 7 of cultivation. To investigate the characteristics of the transcriptome pattern during various phases of the growth curve, we carried out the gene ontology analysis of the genes expressed on day 7, 10, and 13 and the expression pattern was compared among these cultivation phases and defined as three categories such as day 7 versus day 10, day 7 versus day 13 and day 10 versus day 13 (Supplementary Figure S1). In the first category, i.e. day 7 versus day 10, most of the GO terms under all the “Biological Process,” “Molecular Function” and “Cellular Component” were down-regulated. In the second category, most of the GO terms under all three categories were found to be differentiated. Particularly, “death” GO term under “biological process,” “virion” GO term under the cellular component and “transporter” and “translation regulator activity” under “molecular function” category were downregulated. In the third category, except “localization,” “biological regulation” GO terms under “biological process” category and “catalytic activity” GO term under “molecular function” all the GO terms were up- and down-regulated. The differentially expressed genes (10393) were clustered into the 8 major clusters according to their differential expression patterns (Supplementary Figure S2). The transcriptional patterns of key lipogenic genes including GPAT, LPAAT, PAP, and DGAT were measured quantitatively (Supplementary Dataset S1), which corroborated the above-mentioned growth phase-dependent lipogenic phenomenon. In addition, we have determined the relative transcript abundance of the genes LPAAT, PAP, and ACBP by qPCR, which showed that plastidial localized enzymes were highly dynamic during the growth-phase dependent TAG biosynthesis and ACBP did not exhibit any difference during growth phases, which corroborate our transcriptome data (Supplementary Figure S3). These results implied the importance of the cultivation phase on governing the biochemical potential including lipogenesis and provided the necessity to uncover the expression pattern of the key metabolic pathways that underlie the lipogenic property.
Expression Pattern of Key Lipogenic Genes Was Orchestrated in a Growth Phase-Dependent Manner
Given the interesting lipidomic and growth data during different growth phases, we further sought to investigate the expression pattern of key lipogenic genes under these growth phases. Generally, ER-localized TAG biosynthesis has been considered as the traditional pathway for TAG biogenesis, however, previous studies reported the occurrence of plastidial localized TAG biosynthesis (Frentzen, 1998; Fan et al., 2011). However, such data remain largely obscured in N. oceanica. Thus, we examined the crucial role of growth phase-dependent lipid accumulation and the underlying TAG biogenesis. Intriguingly, we found that genes involved in plastidial TAG pathway such as GPAT, LPAAT, and DGAT were remarkably higher on both day 10 and 13 (Figure 2A). On the other hand, the expression pattern of key genes involved in ER-localized TAG pathway did not display significant increment on day 10, however, expression of these genes was drastically increased on day 13. These data uncovered the unique expression pattern of these key lipogenic genes in a growth-phase dependent manner.
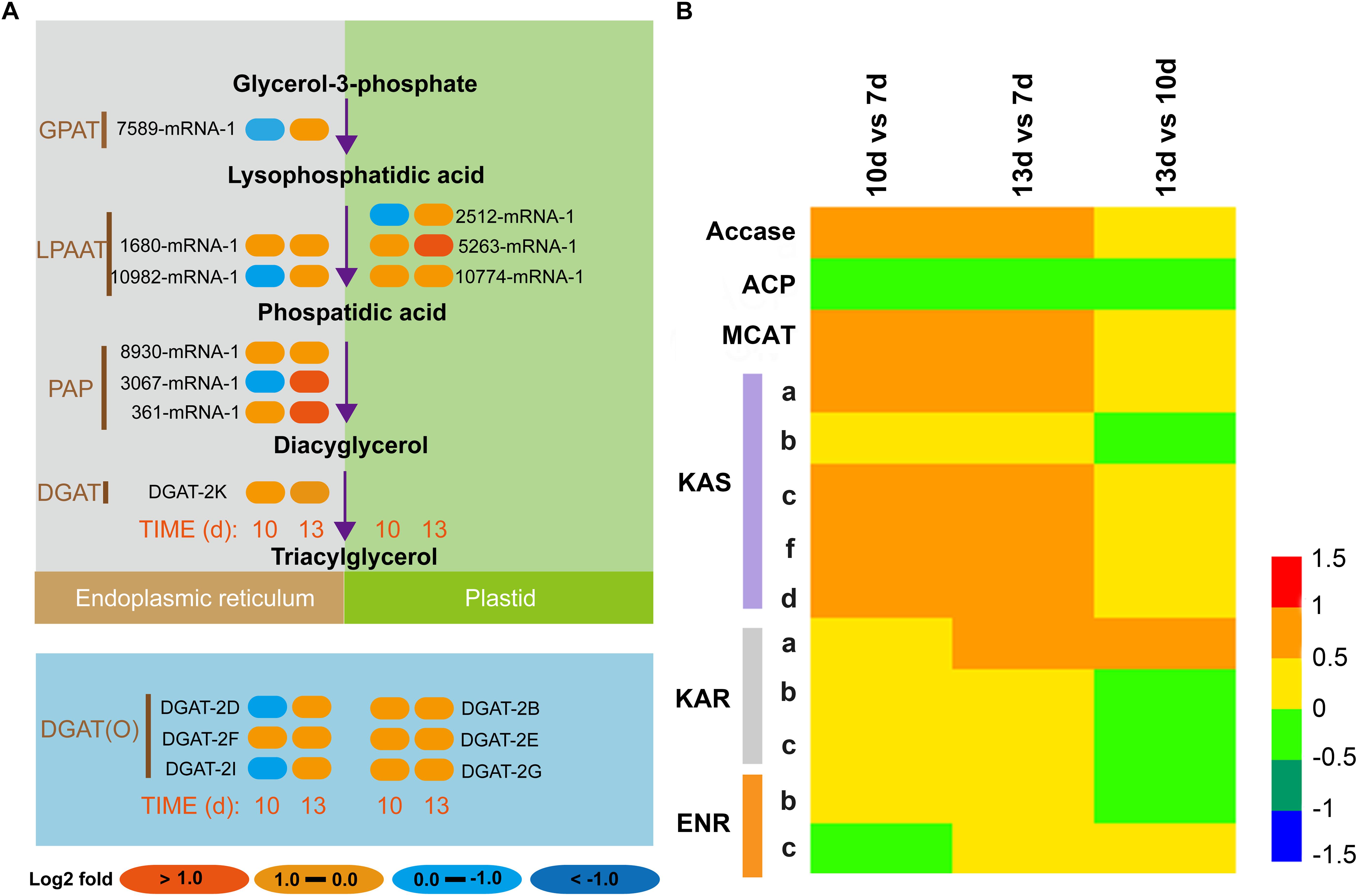
Figure 2. Regulation of the expression of key genes involved in ER and plastidial TAG pathways and fatty acid metabolism. (A) Heat map indicates the fold changes of relative transcripts of key lipogenic genes during various growth phases in the chloroplast and ER; (B) Heat map depicts the transcriptional pattern of key genes involved in fatty acid metabolism. Values of the differential gene expression were calculated as the log2-fold changes.
Prokaryotic TAG Biosynthetic Pathway Could Be the Master Regulator in Growth-Phase Dependent Lipid Accumulation
It is worth mentioning that plastidial TAG biosynthetic machinery has been found to be highly active during the late-log and early stationary phase compared to the ER-localized enzymes, which were increased during the stationary phase. To reinforce this notion, we predicted the subcellular localization of the enzymes using various bioinformatic tools such as SignalP, ChloroP, Mitoprot and HECTAR which corroborated the possible plastidial localization of these enzymes such as LPAAT (2512-mRNA, 5263-mRNA and 10774-mRNA) and DGAT (DGAT-2K) was predicted to be localized in the ER (Supplementary Dataset S2). Microalgal TAG has been considered to be synthesized via acetyl-CoA dependent and/or acetyl-CoA independent phospholipid:diacylglycerol acyltransferase (PDAT) TAG pathway (Yang et al., 2013). TAG is synthesized by sequential acylation of glycerol backbone at sn-1, sn-2, and sn-3 positions by glycerol-3-phosphate acyltransferase (GPAT), lysophosphatidate acyltransferase (LPAAT), and diacylglycerol acyltransferase (DGAT), respectively, to form TAG (Li et al., 2016). Among these key enzymes, LPAAT plays a crucial role in determining the fatty acid composition of TAGs via their stringent substrate specificity toward certain acetyl-CoAs (Balamurugan et al., 2017). Substrate specificity of these LPAATs has been determined by its subcellular localization (Allen et al., 2015), plastidial LPAAT has been demonstrated to possess prokaryotic enzymatic activity which prefers C16 acyl-CoA instead of ER-localized LPAAT that prefers C18 acyl-CoA moieties, referred to as the eukaryotic pathway (Wang et al., 2017). Besides, the potential role of LPAAT in governing the fatty acid profile has received research interests owing to the importance of fatty acid composition that elevate the functional and economic features of biofuels. The previous report has shown that overexpression of plastidial LPAAT elevated saturated and polyunsaturated fatty acids, and decreased monounsaturated fatty acids with particular increment and decrement of C16:0 and C16:1 fatty acids, respectively (Balamurugan et al., 2017). Similarly, dual overexpression of plastidial GPAT and LPAAT resulted in remarkable lipid enhancement with an altered fatty acid profile in Phaeodactylum tricornutum (Wang et al., 2018). In addition, DGAT, which catalyzes the final committed step of TAG biosynthesis, was remarkably increased during the late growth phase. Various previous studies have demonstrated the pivotal role of DGAT in elevating the TAG content of microalga, which is in accordance with our lipidomic data (Li et al., 2016; Klaitong et al., 2017; Wei et al., 2017).
Given these observations which highlighted the differential expression of key lipogenic genes, we further investigated the ultrastructural changes in the cells by transmission electron microscopy. There were no physical aberrations observed in terms of cellular structural, while at the same time lipid droplets accumulated more in the cytosol compared to the chloroplast (Supplementary Figure S4). It has been well documented that TAG biosynthetic mechanisms warrant intricate trafficking of fatty acid precursors between ER and chloroplasts (Andersson et al., 2007; Sparkes et al., 2009). Being the preliminary site for fatty acid biosynthesis, chloroplasts play a crucial role in providing fatty acid precursors, which could be exported to ER for TAG biosynthesis (Ohlrogge and Browse, 1995). Previous reports have demonstrated the transport of various lipid molecules between chloroplasts and ER by the involvement of various membrane transporters (Mehrshahi et al., 2014). Besides, the structural modification of organelle membranes in response to the interaction of lipid molecules with the membrane proteins could facilitate lipid transportation across membranes (Goren et al., 2014). Phosphatidic acid has also been reported to play a pivotal role in plastid-ER lipid transportation by modulating membrane curvature and enzymatic activity (Block and Jouhet, 2015). Collectively, our observations implied that the overproduced fatty acids and lipids could be exported to the cytosol for lipid droplet formation, which is in accordance with the reports mentioned above.
Our data showed that a series of transporter genes was upregulated during the stationary phase, particularly, the expression pattern of the identified transporters was significantly increased on day 13 (Figure 3). Various studies have demonstrated the crucial role of these transporters in inter-organelle lipid trafficking (Mehrshahi et al., 2013), however, there are few reports available on their molecular characterization, particularly such data are yet to be explored in Nannochloropsis. Interestingly, the expression of phosphate translocators was significantly increased along with the transporters such as CCMP1779| 10107-mRNA-1, CCMP1779| 11843-mRNA-1. Sharkey et al. (2019) reported that transcriptional activation of phosphate translocator 2 resulted in the transportation of reduced carbon molecules across plastidial membranes, which enhanced photosynthesis and carbon metabolism. Similarly, loss of phosphate transporters resulted in serious growth and photosynthesis impairment in Arabidopsis (Hilgers et al., 2018). These data are consistent with our findings, which imply that expressed transporters might play a crucial role in lipid trafficking, thereby plastidial fatty acids could be transported to the cytosol to form cytosolic lipid droplets (Block et al., 2007). Together, these data provide a novel insight into the mechanistic role of plastidial TAG biosynthesis in Nannochloropsis for the first time, which warrants further in-depth molecular characterization.
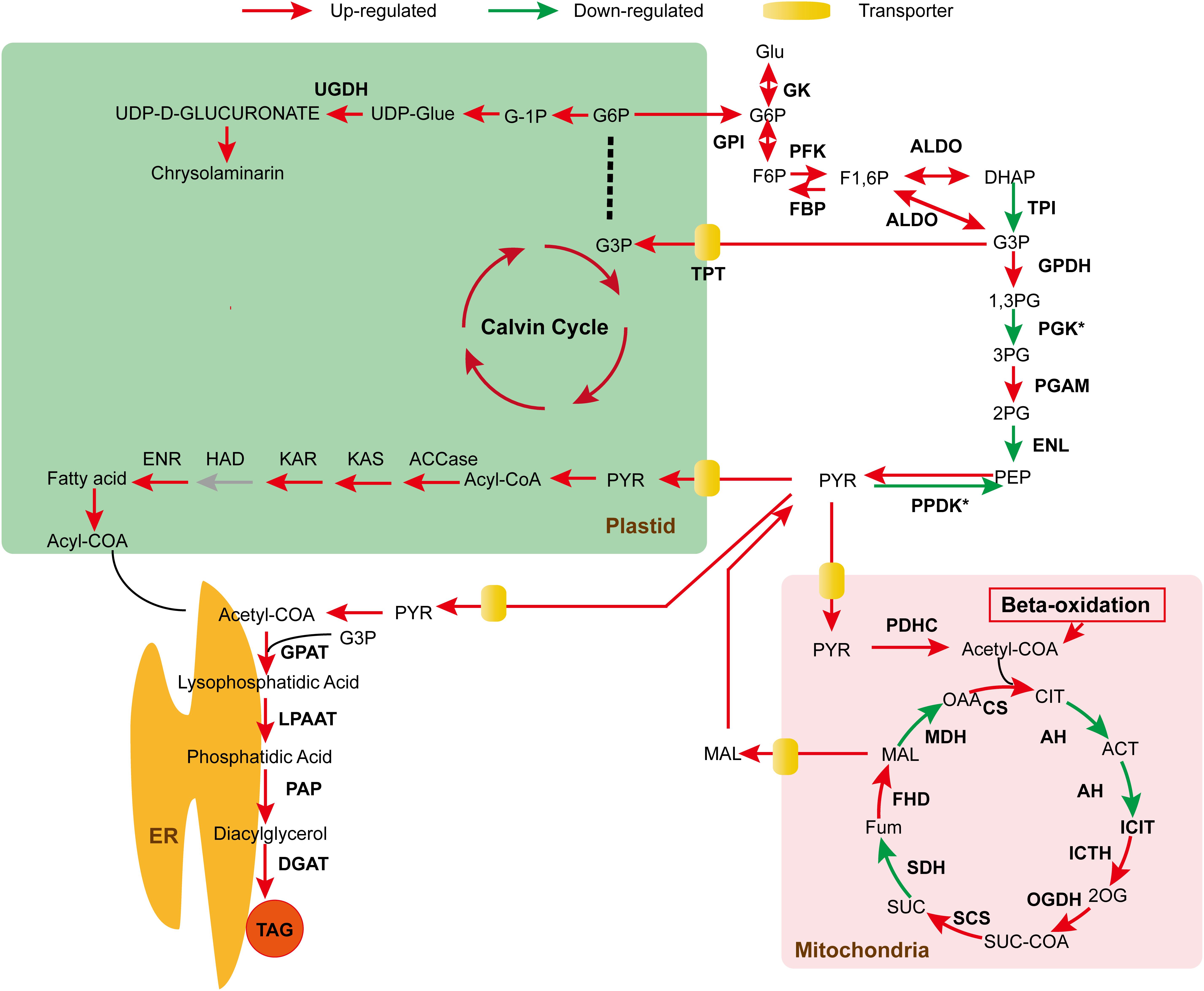
Figure 3. Schematic representation of the expression pattern of key genes on day 13 of cultivation. Genes that are up- and down- regulated are marked by red and green arrows, respectively. Yellow boxes denote the transporter genes.
Analyses of Fatty Acid Biosynthetic Genes Under Different Cultivation Phases
Reasoning the remarkable alterations in the lipid content and the expression pattern of key lipogenic genes, particularly the genes involved in plastidial TAG biogenesis, we sought to explore the expression mechanism of key genes involved in fatty acid biosynthesis. As shown in Figure 2B, key genes such as acetyl-CoA carboxylase (ACCase), MCAT, KAS, KAR, and ENR involved in fatty acid biosynthesis were significantly increased on day 10 and 13. Particularly, expression of these genes was remarkably higher on day 10, which implied the crucial role of these genes in providing fatty acid moieties for lipogenesis.
Lipid biosynthesis is critically regulated by various regulatory factors such as the provision of metabolic carbon precursors, reducing equivalents and reducing the competing metabolic pathways. Amongst, provision of fatty acids as the key precursors has been considered crucial for fatty acid biosynthesis. Overexpression of plastidial ACCase resulted in significant elevation of fatty acid and lipid content in P. tricornutum (Li et al., 2018). Similarly, overexpression of MCAT resulted in the remarkable elevation of fatty acids and lipids by providing adequate fatty acid precursors for lipogenesis in Nannochloropsis (Chen et al., 2017). KAS, KAR, and ENR have also been shown to play a role in lipid metabolism in various organisms (Ajjawi et al., 2017). Altogether, our data demonstrated the potential of plastidial lipogenic machinery on substantially improving algal lipid content.
In summary, we have elucidated the mechanisms of lipid biosynthesis and uncovered the signature genes that underpin the intricate lipogenesis without impairing cellular physiological properties. Our data demonstrated that plastidial fatty acid biosynthetic machinery played a promising role not only in providing adequate fatty acid precursors but also in TAG biosynthesis in the heterokont Nannochloropsis (Figure 3). These results are of paramount importance which exemplify the orchestration of key metabolic circuits under optimized conditions instead of previous stress treatment. Moreover, our data provide a novel insight into the crucial lipid trafficking, particularly from plastids to ER, which raises an interesting topic for further study.
Data Availability Statement
The datasets generated for this study can be found in the https://dataview.ncbi.nlm.nih.gov/object/PRJNA558252?reviewer=vbp72kf5odbpjlgqgfi6lvq2g1.
Author Contributions
DH and H-YL conceived and designed the experiments. DH, D-WL, SB, W-JL, and L-GZ performed the experiments. DH and J-WZ contributed to the bioinformatics analysis. DH and W-DY analyzed the data. D-WL, J-SL, and H-YL wrote the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31870027 and 41576132) and the China Postdoctoral Science Foundation (2017M612838).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2020.00104/full#supplementary-material
FIGURE S1 | Gene ontology annotation. Distribution of the relative transcripts with GO involved in Biological Process, Molecular Function, and Cellular Component categories.
FIGURE S2 | Transcript abundance of various genes under seven clusters differentially expressed during various growth phases.
FIGURE S3 | Relative transcript abundance of key fatty acid and lipid biosynthetic genes. Significant difference is indicated at P < 0.05 (∗) level. Each value represents mean ± SD (n = 3).
FIGURE S4 | Ultrastructural analyses of the cells by TEM on (A) day 7; (B) 10, and (C) 13 of cultivation. Cp, Chloroplast; OB, Oil Bodies. Bars = 0.5 μm.
TABLE S1 | Fatty acid composition of N. oceanica cell during cultivation.
DATASET S1 | Transcriptional dynamics of key genes involved in central carbon metabolism.
DATASET S2 | Bioinformatic prediction of the subcellular localization of the key proteins.
Footnotes
References
Adams, C., Godfrey, V., Wahlen, B., Seefeldt, L., and Bugbee, B. (2013). Understanding precision nitrogen stress to optimize the growth and lipid content tradeoff in oleaginous green microalgae. Bioresour. Technol. 131, 188–194. doi: 10.1016/j.biortech.2012.12.143
Ajjawi, I., Verruto, J., Aqui, M., Soriaga, L. B., Coppersmith, J., Kwok, K., et al. (2017). Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol. 35:647. doi: 10.1038/nbt.3865
Allen, J. W., DiRusso, C. C., and Black, P. N. (2015). Triacylglycerol synthesis during nitrogen stress involves the prokaryotic lipid synthesis pathway and acyl chain remodeling in the microalgae Coccomyxa subellipsoidea. Algal Res. 10, 110–120. doi: 10.1016/j.algal.2015.04.019
Andersson, M. X., Goksör, M., and Sandelius, A. S. (2007). Membrane contact sites: physical attachment between chloroplasts and endoplasmic reticulum revealed by optical manipulation. Plant Signal. Behav. 2, 185–187. doi: 10.4161/psb.2.3.3973
Artzi, S., Kiezun, A., and Shomron, N. (2008). miRNAminer: a tool for homologous microRNA gene search. BMC Bioinformatics 9:39. doi: 10.1186/1471-2105-9-39
Balamurugan, S., Wang, X., Wang, H. L., An, C. J., Li, H., Li, D. W., et al. (2017). Occurrence of plastidial triacylglycerol synthesis and the potential regulatory role of AGPAT in the model diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 10:97. doi: 10.1186/s13068-017-0786-0
Block, M. A., Douce, R., Joyard, J., and Rolland, N. (2007). Chloroplast envelope membranes: a dynamic interface between plastids and the cytosol. Photosynth. Res. 92, 225–244. doi: 10.1007/s11120-007-9195-8
Block, M. A., and Jouhet, J. (2015). Lipid trafficking at endoplasmic reticulum–chloroplast membrane contact sites. Curr. Opin. Cell Biol. 35, 21–29. doi: 10.1016/j.ceb.2015.03.004
Chandra, R., Das, P., Vishal, G., and Nagra, S. (2019). Factors affecting the induction of UV protectant and lipid productivity in Lyngbya for sequential biorefinery product recovery. Bioresour. Technol. 278, 303–310. doi: 10.1016/j.biortech.2019.01.084
Chen, J. W., Liu, W. J., Hu, D. X., Wang, X., Balamurugan, S., Alimujiang, A., et al. (2017). Identification of a malonyl CoA−acyl carrier protein transacylase and its regulatory role in fatty acid biosynthesis in oleaginous microalga Nannochloropsis oceanica. Biotechnol. Appl. Biochem. 64, 620–626. doi: 10.1002/bab.1531
Chisti, Y. (2013). Constraints to commercialization of algal fuels. J. Biotechnol. 167, 201–214. doi: 10.1016/j.jbiotec.2013.07.020
Chiu, S.-Y., Kao, C.-Y., Tsai, M.-T., Ong, S.-C., Chen, C.-H., and Lin, C.-S. (2009). Lipid accumulation and CO2 utilization of Nannochloropsis oculata in response to CO2 aeration. Bioresour. Technol. 100, 833–838. doi: 10.1016/j.biortech.2008.06.061
Claros, M. G. (1995). MitoProt, a macintosh application for studying mitochondrial proteins. Bioinformatics. 11, 441–447. doi: 10.1093/bioinformatics/11.4.441
de Alva, M. S., Pabello, V. M. L., Ledesma, M. T. O., and Gómez, M. J. C. (2018). Carbon, nitrogen, and phosphorus removal, and lipid production by three saline microalgae grown in synthetic wastewater irradiated with different photon fluxes. Algal Res. 34, 97–103. doi: 10.1016/j.algal.2018.07.006
Emanuelsson, O., Nielsen, H., and Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Fan, J., Andre, C., and Xu, C. (2011). A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett. 585, 1985–1991. doi: 10.1016/j.febslet.2011.05.018
Frentzen, M. (1998). Acyltransferases from basic science to modified seed oils. Lipid/Fett 100, 161–166. doi: 10.1002/(sici)1521-4133(19985)100:4/5<161::aid-lipi161>3.0.co;2-p
Goren, M. A., Morizumi, T., Menon, I., Joseph, J. S., Dittman, J. S., Cherezov, V., et al. (2014). Constitutive phospholipid scramblase activity of a G protein-coupled receptor. Nat. Commun. 5:5115. doi: 10.1038/ncomms6115
Han, D., Jia, J., Li, J., Sommerfeld, M., Xu, J., and Hu, Q. (2017). Metabolic remodeling of membrane glycerolipids in the microalga Nannochloropsis oceanica under nitrogen deprivation. Front. Mar. Sci. 4:242.
Havlik, I., Lindner, P., Scheper, T., and Reardon, K. F. (2013a). On-line. (monitoring)of large cultivations of microalgae and cyanobacteria. Trends Biotechnol. 31, 406–414. doi: 10.1016/j.tibtech.2013.04.005
Havlik, I., Reardon, K. F., Ünal, M., Lindner, P., Prediger, A., Babitzky, A., et al. (2013b). Monitoring of microalgal cultivations with on-line, flow-through microscopy. Algal Res. 2, 253–257. doi: 10.1016/j.algal.2013.04.001
Hilgers, E. J., Schöttler, M. A., Mettler-Altmann, T., Krueger, S., Doermann, P., Eicks, M., et al. (2018). The combined loss of triose phosphate and xylulose 5-phosphate/phosphate translocators leads to severe growth retardation and impaired photosynthesis in Arabidopsis thaliana tpt/xpt double mutants. Front. Plant Sci. 9:1331. doi: 10.3389/fpls.2018.01331
Kim, Y., Terng, E. L., Riekhof, W. R., Cahoon, E. B., and Cerutti, H. (2018). Endoplasmic reticulum acyltransferase with prokaryotic substrate preference contributes to triacylglycerol assembly in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 115, 1652–1657. doi: 10.1073/pnas.1715922115
Klaitong, P., Fa-Aroonsawat, S., and Chungjatupornchai, W. (2017). Accelerated triacylglycerol production and altered fatty acid composition in oleaginous microalga Neochloris oleoabundans by overexpression of diacylglycerol acyltransferase 2. Microb. Cell Fact. 16:61. doi: 10.1186/s12934-017-0677-x
Klok, A. J., Martens, D. E., Wijffels, R. H., and Lamers, P. P. (2013). Simultaneous growth and neutral lipid accumulation in microalgae. Bioresour. Technol. 134, 233–243. doi: 10.1016/j.biortech.2013.02.006
Kokabi, K., Gorelova, O., Ismagulova, T., Itkin, M., Malitsky, S., Boussiba, S., et al. (2019). Metabolomic foundation for differential responses of lipid metabolism to nitrogen and phosphorus deprivation in an arachidonic acid-producing green microalga. Plant Sci. 283, 95–115. doi: 10.1016/j.plantsci.2019.02.008
Li, D. W., Balamurugan, S., Yang, Y. F., Zheng, J. W., Huang, D., Zou, L. G., et al. (2019). Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci. Adv. 5:eaau3795. doi: 10.1126/sciadv.aau3795
Li, D. W., Cen, S. Y., Liu, Y. H., Balamurugan, S., Zheng, X. Y., Alimujiang, A., et al. (2016). A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol. 229, 65–71. doi: 10.1016/j.jbiotec.2016.05.005
Li, D. W., Xie, W. H., Hao, T. B., Cai, J. X., Zhou, T. B., Balamurugan, S., et al. (2018). Constitutive and chloroplast targeted expression of Acetyl-CoA carboxylase in oleaginous microalgae elevates fatty acid biosynthesis. Mar. Biotechnol. 20, 566–572. doi: 10.1007/s10126-018-9841-5
Li, J., Han, D., Wang, D., Ning, K., Jia, J., Wei, L., et al. (2014). Choreography of transcriptomes and lipidomes of Nannochloropsis reveals the mechanisms of oil synthesis in microalgae. Plant Cell 26, 1645–1665. doi: 10.1105/tpc.113.121418
Li, Y., Han, D., Hu, G., Sommerfeld, M., and Hu, Q. (2010). Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol. Bioeng. 107, 258–268. doi: 10.1002/bit.22807
Mata, T. M., Martins, A. A., and Caetano, N. S. (2010). Microalgae for biodiesel production and other applications: a review. Renew. Sust. Energ. Rev. 14, 217–232. doi: 10.1016/j.rser.2009.07.020
Mehrshahi, P., Johnny, C., and DellaPenna, D. (2014). Redefining the metabolic continuity of chloroplasts and ER. Trends Plant Sci. 19, 501–507. doi: 10.1016/j.tplants.2014.02.013
Mehrshahi, P., Stefano, G., Andaloro, J. M., Brandizzi, F., Froehlich, J. E., and DellaPenna, D. (2013). Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 110, 12126–12131. doi: 10.1073/pnas.1306331110
Msanne, J., Xu, D., Konda, A. R., Casas-Mollano, J. A., Awada, T., Cahoon, E. B., et al. (2012). Metabolic and gene expression changes triggered by nitrogen deprivation in the photoautotrophically grown microalgae Chlamydomonas reinhardtii and Coccomyxa sp. C-169. Phytochemistry 75, 50–59. doi: 10.1016/j.phytochem.2011.12.007
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8:785. doi: 10.1038/nmeth.1701
Ren, H.-Y., Xiao, R.-N., Kong, F., Zhao, L., Xing, D., Ma, J., et al. (2019). Enhanced biomass and lipid accumulation of mixotrophic microalgae by using low-strength ultrasonic stimulation. Bioresour. Technol. 272, 606–610. doi: 10.1016/j.biortech.2018.10.058
Sharkey, T. D., Weise, S. E., Childs, K., Preiser, A. L., Katulski, H. M., Perrin-Porzondek, C., et al. (2019). Transcriptional regulation of the glucose-6-phosphate/Phosphate translocator 2 is related to carbon exchange across the chloroplast envelope. Front. Plant Sci. 10:827. doi: 10.3389/fpls.2019.00827
Singh, P., Guldhe, A., Kumari, S., Rawat, I., and Bux, F. (2015). Investigation of combined effect of nitrogen, phosphorus and iron on lipid productivity of microalgae Ankistrodesmus falcatus KJ671624 using response surface methodology. Biochem. Eng. J. 94, 22–29. doi: 10.1016/j.bej.2014.10.019
Sparkes, I. A., Frigerio, L., Tolley, N., and Hawes, C. (2009). The plant endoplasmic reticulum: a cell-wide web. Biochem. J. 423, 145–155. doi: 10.1042/BJ20091113
Vieler, A., Wu, G., Tsai, C. H., Bullard, B., Cornish, A. J., Harvey, C., et al. (2017). Correction: genome, functional gene annotation, and nuclear transformation of the heterokont oleaginous alga Nannochloropsis oceanica CCMP1779. PLoS Genet. 13:e1006802. doi: 10.1371/journal.pgen.1006802
Wang, N., Ma, J., Pei, W., Wu, M., Li, H., Li, X., et al. (2017). A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: organization, expression, sequence variation, and association with seed oil content and fiber quality. BMC Genomics 18:218. doi: 10.1186/s12864-017-3594-9
Wang, X., Dong, H.-P., Wei, W., Balamurugan, S., Yang, W.-D., Liu, J.-S., et al. (2018). Dual expression of plastidial GPAT1 and LPAT1 regulates triacylglycerol production and the fatty acid profile in Phaeodactylum tricornutum. Biotechnol. Biofuels 11:318. doi: 10.1186/s13068-018-1317-3
Wang, X., Luo, S. W., Luo, W., Yang, W. D., Liu, J. S., and Li, H. Y. (2019). Adaptive evolution of microalgal strains empowered by fulvic acid for enhanced polyunsaturated fatty acid production. Bioresour. Technol. 277, 204–210. doi: 10.1016/j.biortech.2018.12.116
Wei, H., Shi, Y., Ma, X., Pan, Y., Hu, H., Li, Y., et al. (2017). A type-I diacylglycerol acyltransferase modulates triacylglycerol biosynthesis and fatty acid composition in the oleaginous microalga, Nannochloropsis oceanica. Biotechnol. Biofuels. 10:174. doi: 10.1186/s13068-017-0858-1
Yang, J., Li, X., Hu, H., Zhang, X., Yu, Y., and Chen, Y. (2011). Growth and lipid accumulation properties of a freshwater microalga, Chlorella ellipsoidea YJ1, in domestic secondary effluents. Appl. Energ. 88, 3295–3299. doi: 10.1016/j.apenergy.2010.11.029
Yang, Z., Niu, Y. F., Ma, Y. H., Xue, J., Zhang, M. H., Yang, W. D., et al. (2013). Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 6:67. doi: 10.1186/1754-6834-6-67
Yuan, W., Ma, Y., Wei, W., Liu, W., Ding, Y., and Balamurugan, S. (2019). Sequential treatment with bicarbonate and low-temperature to potentiate both biomass and lipid productivity in Nannochloropsis oceanica. J. Chem. Technol. Biotechnol. 94, 3413–3419. doi: 10.1002/jctb.6155
Zheng, J. W., Li, D. W., Lu, Y., Chen, J., Liang, J. J., Zhang, L., et al. (2016). Molecular exploration of algal interaction between the diatom Phaeodactylum tricornutum and the dinoflagellate Alexandrium tamarense. Algal Res. 17, 132–141. doi: 10.1016/j.algal.2016.04.019
Zou, L. G., Chen, J. W., Zheng, D. L., Balamurugan, S., Li, D. W., Yang, W. D., et al. (2018). High-efficiency promoter-driven coordinated regulation of multiple metabolic nodes elevates lipid accumulation in the model microalga Phaeodactylum tricornutum. Microb. Cell Fact. 17:54. doi: 10.1186/s12934-018-0906-y
Keywords: growth phase, lipogenesis, triacylglycerol assembly, fatty acid biosynthesis, Nannochloropsis oceanica
Citation: Huang D, Li D-W, Balamurugan S, Zheng J-W, Liu W-J, Zou L-G, Yang W-D, Liu J-S and Li H-Y (2020) Plastidial and ER Triacylglycerol Biosynthesis in a Growth Phase-Dependent Manner in the Heterokont Nannochloropsis oceanica. Front. Mar. Sci. 7:104. doi: 10.3389/fmars.2020.00104
Received: 03 August 2019; Accepted: 10 February 2020;
Published: 21 February 2020.
Edited by:
Sachin Kumar, Sardar Swaran Singh National Institute of Renewable Energy, IndiaReviewed by:
Rashmi Chandra, University of Waterloo, CanadaLi Wei, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2020 Huang, Li, Balamurugan, Zheng, Liu, Zou, Yang, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Ye Li, dGh5bGlAam51LmVkdS5jbg==
†These authors have contributed equally to this work
 Dan Huang
Dan Huang Da-Wei Li
Da-Wei Li Srinivasan Balamurugan
Srinivasan Balamurugan Jian-Wei Zheng
Jian-Wei Zheng Hong-Ye Li
Hong-Ye Li