- 1Department of Marine Science, Bharathidasan University, Tiruchirappalli, India
- 2Department of Marine Systems, Tallinn University of Technology, Tallinn, Estonia
- 3Biological Oceanography Division, CSIR-National Institute of Oceanography, Dona Paula, India
Skeletal growth anomalies (SGA) are easily recognizable disease in reef-forming corals which drain energy from the host and end up with mortality. We collected mucus samples of healthy and SGA affected colonies of Acropora cytherea from Palk Bay, the southeast coast of India and investigated the taxonomic composition of mucus-associated bacterial communities using full-length 16S rRNA gene nanopore sequencing. Metagenomic analysis revealed the dominance of Proteobacteria, Firmicutes, and Actinobacteria in both healthy and diseased samples. The proportion of Proteobacteria in diseased samples was almost doubled compared to the healthy one, whereas Acidobacteria (fourth dominant phyla in healthy sample) and Actinobacteria were significantly low in diseased samples. Planctomycetes, Cyanobacteria, and Chloroflexi were some of the other major phyla found in healthy samples which were low in diseased corals. Kruskal-Wallis test indicated high number of putative pathogenic bacteria group viz. Staphylococcus, Salmonella, Klebsiella, and Escherichia in diseased samples compared to healthy coral. The corals perhaps have acquired potential pathogens belong to the phylum Proteobacteria during SGA formation.
Introduction
Coral diseases become a rising issue over several decades, yet fundamental knowledge of their causes is not known in many cases. Skeletal growth anomalies (SGAs) are a group of coral diseases characterized by reduced colony growth, absence of polyp formation, decreased skeleton density and reduced numbers of zooxanthellae (Cheney, 1975; Bak, 1983) and sources confirm that SGAs are not transmissible (Peters et al., 1986). SGA on coral reefs was first witnessed in Madrepora kauaiensis from Hawaii (Squires, 1965) since then it has been reported in several species of reef-building corals including Acropora, Montipora, Platgyra, Turbinaria, and Porites (Burns and Takabayashi, 2011; Irikawa et al., 2011; Williams, 2013; Ng et al., 2015; Tavakoli-Kolour et al., 2015; Hussain et al., 2016). Though the etiology of many coral diseases remains unknown, majority of the diseases reported are recognized or suspected to be of microbial origin (Richardson and Aronson, 2002). However, previous studies on reef coral Acropora cytherea revealed that the formation of SGA is potentially intensified by environmental stresses such as increased sea temperature (Irikawa et al., 2011). While hitherto no report of microbial community variations from healthy and SGA affected A. cytherea are available.
Microorganisms are important diverse component of the coral tissues as well as surface mucus layer and plays a major role in nutrient cycling (Shashar et al., 1994; Ainsworth et al., 2010; Thompson et al., 2014), response to environmental stress (Bourne et al., 2008), coral health (Bourne et al., 2016; Peixoto et al., 2017) and defense against diseases (Reshef et al., 2006; Ritchie, 2006; Rosenberg et al., 2007; Kvennefors et al., 2012). Coral-associated microbial communities are host-specific (Rohwer et al., 2002; Medina, 2011) even if the same coral species from geographically different locations (Rohwer et al., 2001). Yet, many reports revealed that healthy and diseased corals have different microbial associations (Reis et al., 2009; de Castro et al., 2010; Sere et al., 2013). A substantial number of coral diseases i.e., coral bleaching, white-band syndrome, white-plague, white-pox, were demonstrated to have microbial origins (Harvell et al., 2007). In recent years, culture-independent and metagenomics techniques paved way to identify a wide array of microorganisms that are associated with coral surface mucus and tissues of healthy and diseased corals. To date, only few studies are available on microbial community composition from SGA affected reef corals Platgyra carnosus (Chiu et al., 2012; Ng et al., 2015). For better understanding the future health of coral reefs, it is important to be aware of the microbial communities that are associated with corals and how they shift with response to a disease or environmental stress.
Palk Bay is a fringing reef, 25–30 km long and 300 m wide, located on the southeast coast of India. The fringing reef runs parallel to land in east to west direction with an average water depth of 9 meters (Ravindran et al., 2012). In Palk Bay region, scleractinian diversity studies showed a reduction in species richness from >60 species (Pillai, 1971) to 34 species in 40 years of time (Venkataraman and Rajan, 2013). The dominant genera recorded along the Palk Bay regions were Porites sp., Favia sp., Acropora sp., Platygyra sp., Goniastrea sp., Favites sp. and Siderastrea sp. (Marimuthu et al., 2016). Earlier studies documented various diseases including bleaching, black band, white band, white spot, pink spot, white plaque and yellow band diseases (Thinesh et al., 2011). During a coral survey in the Palk Bay region in 2018, we observed SGA affected colonies of A. cytherea for the first time. With the aim of characterizing bacterial community composition, we collected healthy and the SGA affected A. cytherea colonies from Palk Bay region. This investigation presents the first comprehensive metagenomic analysis (using nanopore sequencing) on the comparison of bacterial communities between healthy and SGA affected A. cytherea colonies from Palk Bay, southeast coast of India.
Materials and Methods
Study Area and Sample Details
Samples were collected from healthy and SGA reef coral A. cytherea from Palk Bay region, Tamil Nadu during June 2018 (Figure 1A). The SGA affected colonies are characterized by poorly developed branches with a small portion of the colony showing abnormal skeletal mass (≤5 cm diameter) (Figure 1B). Mucus samples were collected from the abnormal portion of the colony using sterile syringes. Approximately 10 cm2 area of the tissue was sampled for mucus in both healthy and SGA affected corals. Collected samples were transported to the laboratory in ice and stored at −20°C until DNA extraction.
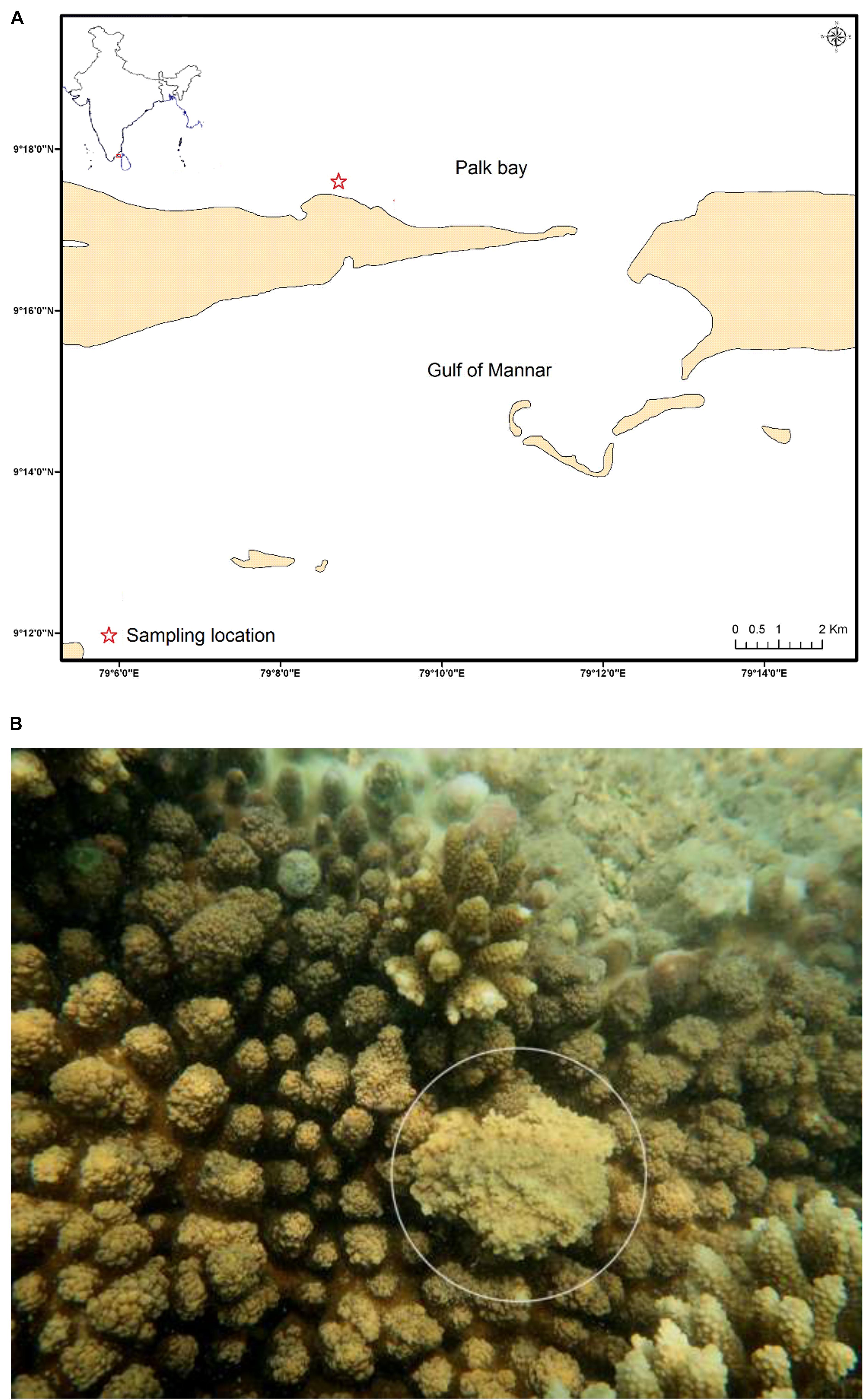
Figure 1. (A) Sampling location of the Palk Bay coral reef ecosystem, Southeast coast of India (Asterisk indicates the sampling site); (B) Reef coral Acropora cytherea (white circle indicates the SGA).
DNA Extraction and PCR Amplification
Total DNA was extracted from both the mucus samples using a DNA isolation kit (Bogar Biobee) following the manufacturer’s protocol. PCR amplification of 16S rRNA gene was performed using consensus primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) (Lane, 1991). PCR products were purified by using Montage PCR Clean-up kit according to manufacturer’s instructions. The quality, quantity (approx. 100 ng/μl) and formulation of the PCR product were checked using Qubit Fluorometer 3.0.
Sequencing and Bioinformatics Analysis
Library was prepared using 1 μg of the purified DNA template. The library preparation was carried out using NEB End repair/dA-tailing (EXP-PBC001) and ligation sequencing kit (SQK-LSK109) as per the manufacturer’s protocol. Libraries were purified using AMPure XP beads and loaded on to the Nanopore MinION SpotON flow cell (R9.4.1). Reads containing raw reads were base called using MinKNOW (Oxford Nanopore Technologies). The raw reads were further quality checked using FastQC Version 0.11.8 (Andrews, 2010). Further, poor quality reads were trimmed using Trimmomatic with default parameters (Bolger et al., 2014). The taxonomic abundance profile of healthy and diseased coral was classified using Kraken classification tool (Wood and Salzberg, 2014) in OmicsBox server1. The raw sequence data obtained from this study are available from the GenBank BioProject under the accession number PRJNA576005.
Statistical Analysis
Alpha diversity measures (Chao-1, Shannon and Simpson indices) were calculated using EstimateS v9.1.0 (Colwell, 2013) followed by Principal coordinate analysis (PCoA) and Kruskal-Wallis test based on the classified species in healthy and diseased samples (PAST software 3.26). Rarefaction analysis was performed by plotting the number of species against the total number of reads using EcoSim700 (Gotelli and Entsminger, 2004). The community composition between healthy and diseased was also tested using non-parametric Wilcoxon rank-sum test, p-value was calculated using GraphPad 8.2.1 Software (GraphPad Int, United States). The significant level was determined as p < 0.05.
Results and Discussion
Skeletal growth anomalies (SGA) is an easily identifiable disease on the basis of irregular or spherical protuberant masses, reduced growth rates and decreased skeletal density (Burns and Takabayashi, 2011) and has been reported in several scleractinian corals, including 11 species of Acropora (Irikawa et al., 2011). However, no reports have been recorded in the aspect of microbial contribution or microbial role in the SGA affected Acropora reef corals. This study provides detailed amplicon metagenomic analysis into the coral mucus associated bacterial communities based on 16S rRNA gene of the healthy and diseased coral A. cytherea.
A total of 104652 sequences of bacteria were qualified from the OmicsBox tool in which 34998 sequences for the SGA samples and 69654 sequences for healthy samples of the reef coral A. cytherea. Sequences were classified into 38 phyla, 71 classes, and 926 genera for the healthy samples, and 34 phyla, 61 classes, and 697 genera for the diseased samples. Based on our study, the bacterial community associated with the healthy coral was dominated by Proteobacteria (31.6%), followed by Firmicutes (23.6%), Actinobacteria (18.0%), Acidobacteria (6.6%), Chloroflexi (5.6%), Bacteroidetes (5.1%), and Planctomycetes (2.1%). SGA affected corals exhibited high diversity of Proteobacteria (57.3%), followed by Firmicutes (26.9%), Actinobacteria (6.5%), Bacteroidetes (4.8%) and Planctomycetes (2.7%) (p-value <0.0001). Acidobacteria and Chloroflexi were at lower abundance (0.1 and 0.2% respectively) of total sequences (Figure 2A). Among the Proteobacteria, the Gammaproteobacteria sub-group was predominant in both healthy and SGA corals respectively.
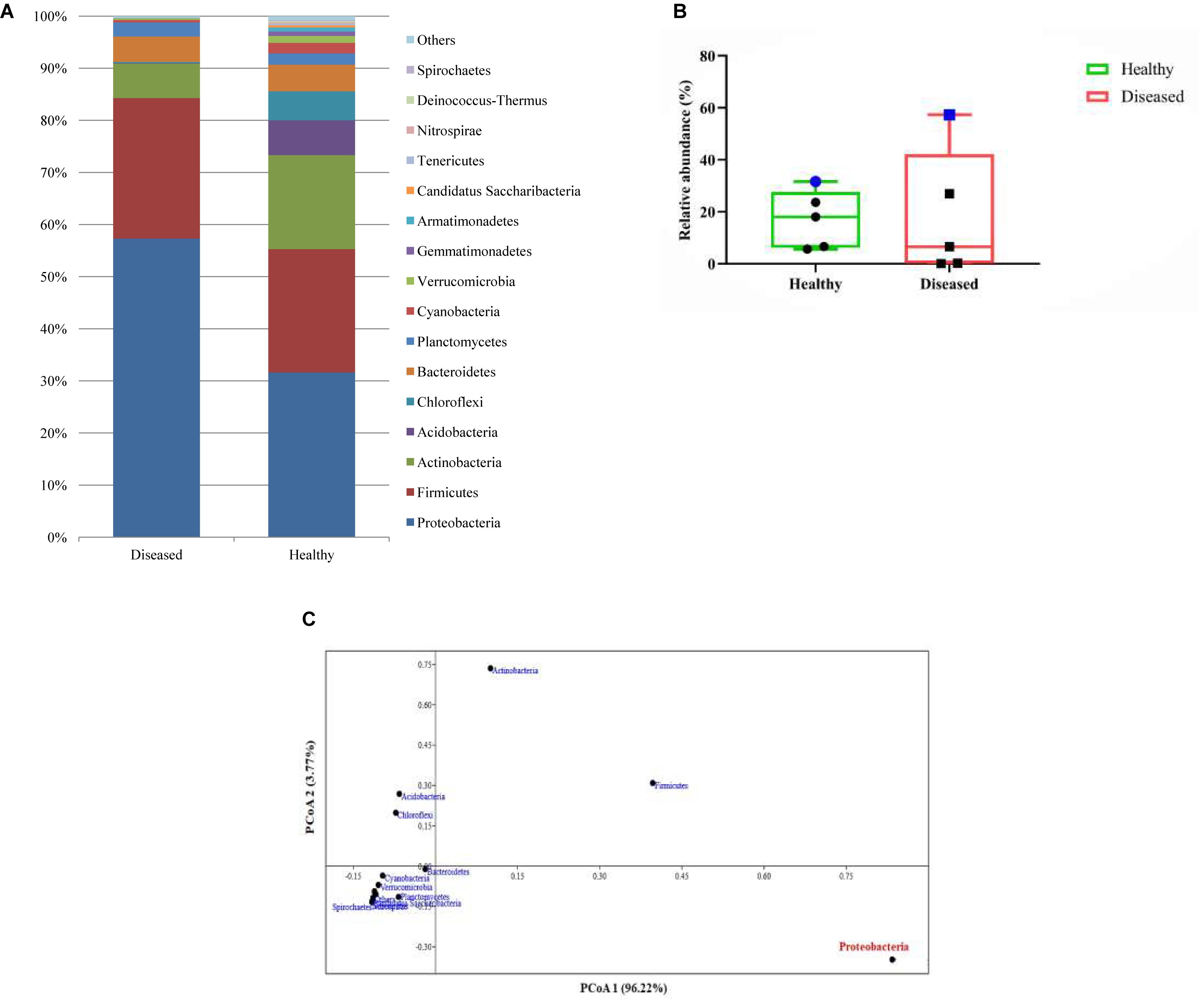
Figure 2. (A) Relative abundance (%) identified based on the metagenomic reads of major bacterial communities at the phylum level; (B) Box and whisker plots of relative abundance of the top 5 abundant phyla. Blue symbol represents the variation of proteobacteria abundance in healthy and diseased coral; (C) Principal coordinate analysis (PCoA) based on the abundance of bacterial phyla.
Several studies have reported the variations of microbial communities associated between the healthy and reef corals with variety of diseases such as white patch syndrome (Godwin et al., 2012; Wilson et al., 2012; Sere et al., 2013), paling and necrosis (de Castro et al., 2010), dark spot-affected (Kellogg et al., 2014), black band disease (Sato et al., 2010; Miller and Richardson, 2011), and white plague disease (Sunagawa et al., 2009; Cárdenas et al., 2011; Roder et al., 2014). There are also fewer reports microbial communities of SGA affected reef coral Platgyra carnosus (Chiu et al., 2012; Ng et al., 2015) and Porites compressa (Breitbarta et al., 2005). Overall, all the studies reported the dominance of Proteobacteria which is coinciding with the present report.
Based on the Box-Plot (Figure 2B) and PCoA (Figure 2C) at the phyla level, we found that the Proteobacteria groups were the major contributor to the variations of microbial abundance between the healthy and the diseased coral. This result is in accordance with the previous reports on bacterial communities associated with various diseases (Sunagawa et al., 2009; Sato et al., 2010; Cárdenas et al., 2011; Miller and Richardson, 2011; Sere et al., 2013; Ng et al., 2015). Whereas, members of the Acidobacteria were scarce in the diseased samples which is contradictory to previous reports (Roder et al., 2014; Ng et al., 2015). Within the Proteobacteria, Gammaproteobacteria was identified as the predominant class, which is almost equal to the percentage of Bacilli. These findings contradict previous findings that have revealed the predominance of Alphaproteobacteria in SGA affected corals and other diseases as well (Pantos et al., 2003; Godwin et al., 2012; Ng et al., 2015).
The association of Vibrio spp. have been well documented with high abundance in various species of diseased reef corals (Cervino et al., 2004; Sussman et al., 2008; Luna et al., 2010; Ushijima et al., 2016; Hadaidi et al., 2018). Whereas, in our study Vibrio spp. represented only 0.14 and 0.18% of the analyzed sequences in healthy and diseased corals respectively. This clearly pointed out that Vibrio spp. was not associated with the progress of SGA in A. cytherea (present study) and Platgyra carnosus as well (Ng et al., 2015).
The association of cyanobacteria (Miller and Richardson, 2011; Hadaidi et al., 2018), Thalassomonas (Thompson et al., 2006), Arcobacter, Desulfovibrio (Hadaidi et al., 2018), Roseovarius, Hoeflea, and Cytophaga (Ng et al., 2015), as main contributors of diseases have been recorded in various coral species. Even though none of the above-mentioned groups of bacteria observed in this study, we identified high abundance of Staphylococcus (4.79%), Salmonella (5.58%), Klebsiella (4.21%), and Escherichia (7.98%) in SGA affected corals (Figure 3). These groups were closely related to pathogens of other terrestrial and as well as marine organisms (Ayulo et al., 1994; Wang et al., 2009; Boss et al., 2016), which indicates the role of opportunistic exogenous pathogens. Besides, an increase of Caulobacter abundance (8.09%) has also been associated with SGA coral which is comparable to the study of Pootakham et al. (2018) where they reported the predominance of Caulobacterales during bleaching of Porites lutea. In addition, A high abundance of Candidatus Solibacter and Candidatus Koribacter were observed in healthy samples (4.2 and 2.2% respectively) when compared to diseased ones (<0.01%).
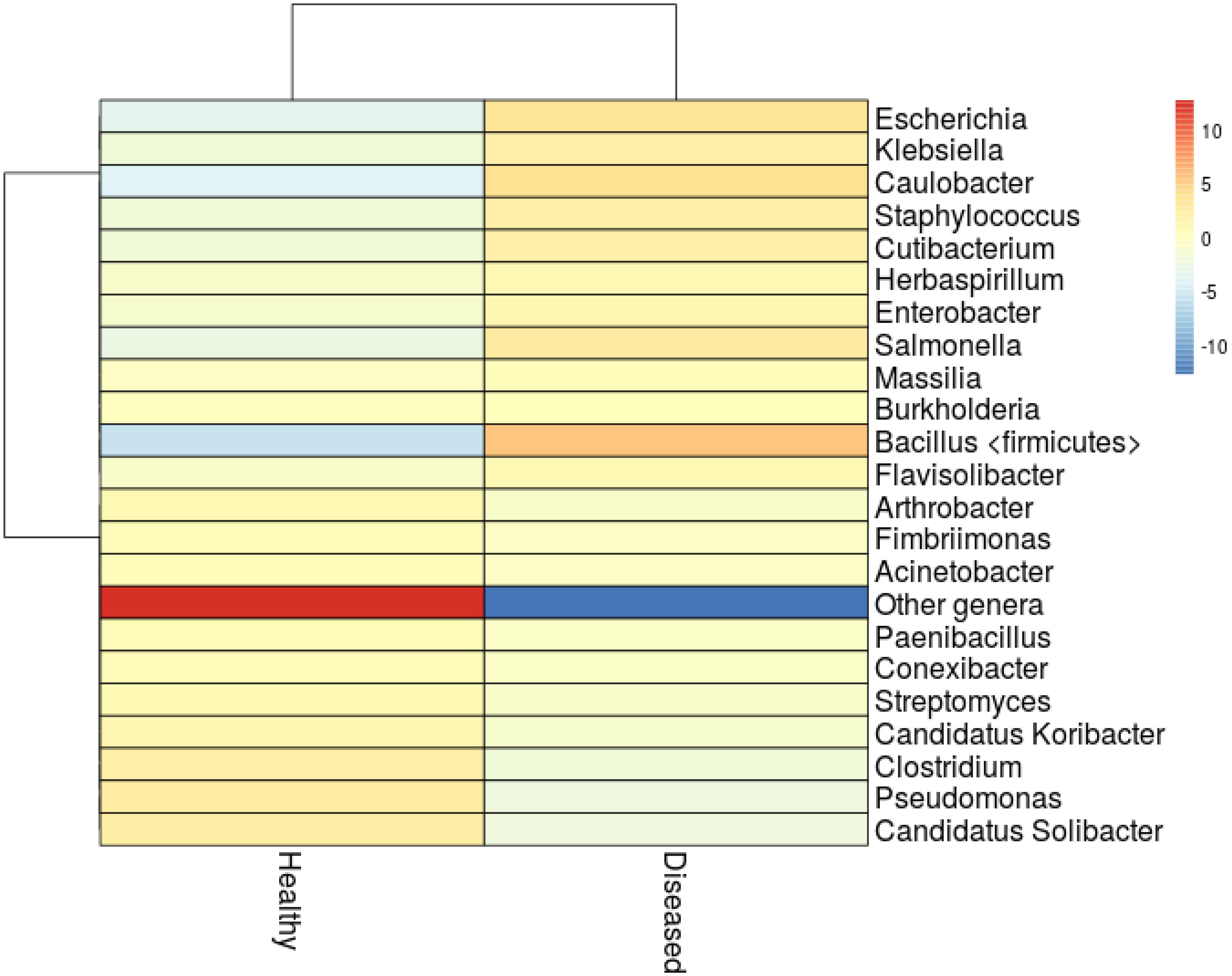
Figure 3. Heatmap analysis showing clustering of the major genera from healthy and SGA samples. Colors represent correlation between samples.
Marine bacteria belonging to Actinobacteria are well documented in the aspect of antibiotics or secondary metabolite producers (e.g., Blunt et al., 2013; Abdelmohsen et al., 2014; Kamjam et al., 2017). In this study, Actinobacteria was found to be the predominant phyla with 18.0% in healthy corals whereas its abundance severely affected in the SGA corals with just 6.4%. Reis et al. (2009) in his study revealed that loss of beneficial microbes (e.g., Actinobacteria and Pseudoalteromonas), in diseased coral might have resulted in increased susceptibility to diseases by opportunistic bacteria. We also found that the abundance of Pseudomonas spp. declined greatly in the diseased sample (Figure 3) which might affect the metabolism of dimethylsulfoniopropionate (DMSP) in A. cytherea (Peixoto et al., 2017). This clearly indicated that the natural bacterial communities are gradually replaced by a succession of opportunistic microorganisms including potential pathogens.
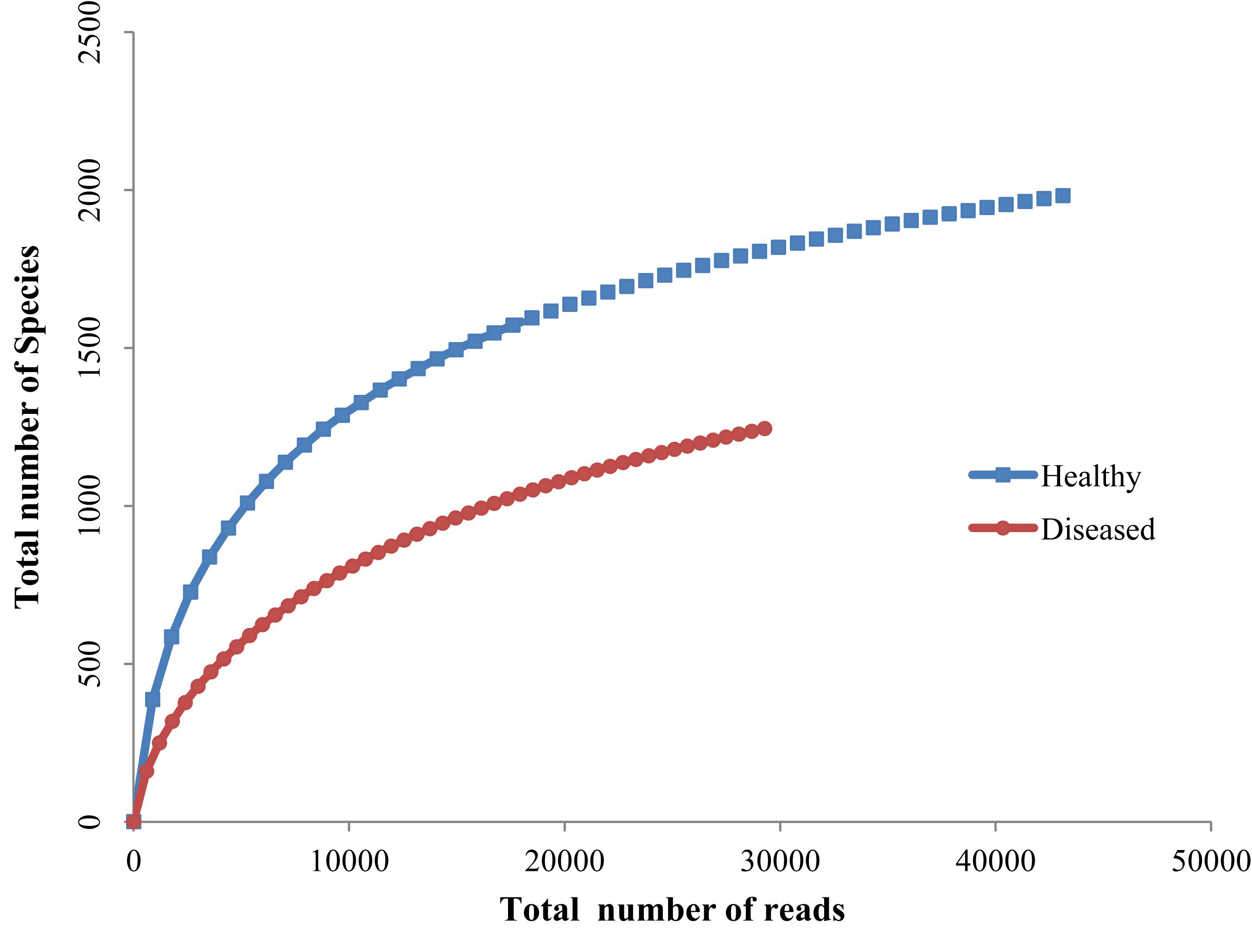
Rarefaction analyses of healthy and SGA affected corals indicated that the healthy coral was hosted with more species and also the rarefaction curve reached a clear saturation (Figure 4). A high alpha-diversity (Shannon Index) and richness estimate (Chao1) were observed in both the healthy mucus (5.75 and 2542 respectively) and SGA affected coral (4.55 and 1668 respectively).
Conclusion
In conclusion, we established that the bacterial community structure is greatly varied between healthy and SGA affected corals and found Proteobacteria groups were the major contributor to these variations. Several bacterial genera affiliated to putative pathogens were highly recovered and at the same time, beneficial groups such as Actinobacteria were drastically reduced in SGA affected coral. We believe that SGA formation perhaps caused by environmental stress but the coral health potentially aggravated by the shift in the bacterial community. This is the first study to provide detailed metagenomic analysis of bacterial communities associated with SGA affected A. cytherea based on 16S rRNA gene amplicon sequencing.
Data Availability Statement
The datasets generated for this study can be found in the GenBank BioProject under the accession number PRJNA576005.
Author Contributions
RR and BM designed the research work and carried out the sampling. RR and KR performed all the experiments, analyzed all the data, and wrote the entire manuscript. RA and CM contributed in designing the research work and edited the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
RR acknowledges University Grants Commission, India for granting Dr. D. S. Kothari Postdoctoral Fellowship (BL/15-16/0370). BM acknowledges Science and Engineering Research Board, Department of Science and Technology, Government of India for sanction of funds under Early Career Research Grant (ECR/2017/002533).
Footnotes
References
Abdelmohsen, U. R., Yang, C., Horn, H., Hajjar, D., Ravasi, T., and Hentschel, U. (2014). Actinomycetes from red sea sponges: sources for chemical and phylogenetic diversity. Mar. Drugs 12, 2771–2789. doi: 10.3390/md12052771
Ainsworth, T. D., Thurber, R. V., and Gates, R. D. (2010). The future of coral reefs: a microbial perspective. Trends Ecol. Evol. 25, 233–240. doi: 10.1016/j.tree.2009.11.001
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed November 2, 2019).
Ayulo, A. M., Machado, R. A., and Scussel, V. M. (1994). Enterotoxigenic Escherichia coli and Staphylococcus aureus in fish and seafood from the southern region of Brazil. Int. J. Food. Microbiol. 24, 171–178. doi: 10.1016/0168-1605(94)90116-3
Bak, R. P. M. (1983). Neoplasia, regeneration and growth in the reef-building coral Acropora palmata. Mar. Biol. 77, 221–227. doi: 10.1007/bf00395810
Blunt, J. W., Copp, B. R., Keyzers, R. A., Munro, M. H., and Prinsep, M. R. (2013). Marine natural products. Nat. Prod. Rep. 30, 237–323. doi: 10.1039/c2np20112g
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boss, R., Overesch, G., and Baumgartner, A. (2016). Antimicrobial resistance of Escherichia coli, Enterococci, Pseudomonas aeruginosa, and Staphylococcus aureus from Raw fish and seafood imported into Switzerland. J. Food. Prot. 79, 1240–1246. doi: 10.4315/0362-028X.JFP-15-463
Bourne, D. G., Iida, Y., Uthicke, S., and Smith-Keune, C. (2008). Changes in coral-associated microbial communities during a bleaching event. ISME J. 2, 350–363. doi: 10.1038/ismej.2007.112
Bourne, D. G., Morrow, K. M., and Webster, N. S. (2016). Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Annu. Rev. Microbiol. 70, 317–340. doi: 10.1146/annurev-micro-102215-095440
Breitbarta, M., Bhagoolib, R., Griffin, S., Johnstond, I., and Rohwera, F. (2005). Microbial communities associated with skeletal tumors on Porites compressa. FEMS Microbiol. Lett. 243, 431–436. doi: 10.1016/j.femsle.2005.01.004
Burns, J. H. R., and Takabayashi, M. (2011). Histopathology of growth anomaly affecting the coral, Montipora capitata: implications on biological functions and population viability. PLoS One 6:e28854. doi: 10.1371/journal.pone.0028854
Cárdenas, A., Rodriguez, R. L. M., Pizarro, V., Cadavid, L. F., and Arévalo-Ferro, C. (2011). Shifts in bacterial communities of two caribbean reef-building coral species affected by white plague disease. ISME J. 6, 502–512. doi: 10.1038/ismej.2011.123
Cervino, J. M., Hayes, R. I, Polson, S. W., Polson, S. C., Goreau, T. J., Martinez, R. J., et al. (2004). Relationship of Vibrio species infection and elevated temperature to yellow blotch/band disease in Caribbean corals. Appl. Environ. Microbiol. 70, 6855–6864. doi: 10.1128/AEM.70.11.6855-6864.2004
Cheney, D. P. (1975). Hard tissue tumors of scleractinian corals. Adv. Exp. Med. Biol. 64, 77–87. doi: 10.1007/978-1-4684-3261-9_9
Chiu, J. M. Y., Li, S., Li, A., Po, B., Zhang, R., Shin, P. K., et al. (2012). Bacteria associated with skeletal tissue growth anomalies in the coral Platygyra carnosus. FEMS Microbiol. Ecol. 79, 380–391. doi: 10.1111/j.1574-6941.2011.01225.x
Colwell, R. K. (2013). EstimateS: Statistical Estimation of Species Richness and Shared Species from Samples. Version 9. User’s Guide and Application. Available at: http://purl.oclc.org/estimates (accessed October 4, 2019).
de Castro, A. P., Araújo, S. D. Jr., Reis, A. M. M., Moura, R. L., Francini-Filho, R. B., and Pappas, G. Jr., et al. (2010). Bacterial community associated with healthy and diseased reef coral Mussismilia hispida from Eastern Brazil. Microb. Ecol. 59, 658–667. doi: 10.1007/s00248-010-9646-1
Godwin, S., Bent, E., Borneman, J., and Pereg, L. (2012). The role of coral-associated bacterial communities in australian subtropical white syndrome of Turbinaria mesenterina. PLoS One 7:e44243. doi: 10.1371/journal.pone.0044243
Gotelli, N. J., and Entsminger, G. L. (2004). Ecosim: Null Models Software for Ecology, Version 7. Burlington, VT: Acquired Intelligence, Inc.
Hadaidi, G., Ziegler, M., Shore-Maggio, A., Jensen, T., Aeby, G., and Voolstra, C. R. (2018). Ecological and molecular characterization of a coral black band disease outbreak in the Red Sea during a bleaching event. PeerJ 6:e5169. doi: 10.7717/peerj.5169
Harvell, D., Jordán-Dahlgren, E., Merkel, S., Rosenberg, E., Raymundo, L., Smith, G., et al. (2007). Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography 20, 58–81. doi: 10.5670/oceanog.2007.91
Hussain, A., De, K., Thomas, L., Nagesh, R., Mote, S., and Ingole, B. (2016). Prevalence of skeletal tissue growth anomalies in a scleractinian coral: Turbinaria mesenterina of malvan marine sanctuary, eastern Arabian Sea. Dis. Aquat. Org. 121, 79–83. doi: 10.3354/dao03038
Irikawa, A., Casareto, B. E., Suzuki, Y., Agostini, S., Hidaka, M., and van Woesik, R. (2011). Growth anomalies on Acropora cytherea corals. Mar. Poll. Bull. 62, 1702–1707. doi: 10.1016/j.marpolbul.2011.05.033
Kamjam, M., Sivalingam, P., Deng, Z., and Hong, K. (2017). Deep sea actinomycetes and their secondary metabolites. Front. Microbiol. 8:760. doi: 10.3389/fmicb.2017.00760
Kellogg, C. A., Piceno, Y. M., Tom, L. M., DeSantis, T. Z., Gray, M. A., and Anderson, G. L. (2014). Comparing bacterial community composition of healthy and dark spot-affected Siderastrea siderea in Florida and the Caribbean. PLoS One 9:e108767. doi: 10.1371/journal.pone.0108767
Kvennefors, E. C. E., Sampayo, E., Kerr, C., Vieira, G., Roff, G., and Barnes, A. C. (2012). Regulation of bacterial communities through antimicrobial activity by the coral holobiont. Microb. Ecol. 63, 605–618. doi: 10.1007/s00248-011-9946-0
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, N.Y: John Wiley & Sons, Inc), 115–176. doi: 10.12691/jaem-2-4-11
Luna, G. M., Bongiorni, L., Gili, C., Biavasco, F., and Danovaro, R. (2010). Vibrio harveyi as a causative agent of the white syndrome in tropical stony corals. Environ. Microbiol. Rep. 2, 120–127. doi: 10.1111/j.1758-2229.2009.00114.x
Marimuthu, N., Ramachandran, P., Robin, R. S., Tudu, D., Hariharan, G., and Ramesh, R. (2016). Spatial variation in the health of coral reef communities of Palk Bay, southeast coast of India. Aquat. Ecosyst. Health Manag. 19, 360–367. doi: 10.1080/14634988.2016.1258925
Medina, M. (2011). Analyzing coral reefs and their microbial assemblages. Microbe Mag. 6, 226–232. doi: 10.1128/microbe.6.226.1
Miller, A. W., and Richardson, L. L. (2011). A meta-analysis of 16S rRNA gene clone libraries from the polymicrobial black band disease of corals. FEMS Microbiol. Ecol. 75, 231–241. doi: 10.1111/j.1574-6941.2010.00991.x
Ng, J. C. Y., Chan, Y., Tun, H. M., Leung, F. C. C., Shin, P. K. S., and Chiu, J. M. (2015). Pyrosequencing of the bacteria associated with Platygyra carnosus corals with skeletal growth anomalies reveals differences in bacterial community composition in apparently healthy and diseased tissues. Front. Microbiol. 6:1142. doi: 10.3389/fmicb.2015.01142
Pantos, O., Cooney, R. P., Le Tissier, M. D. A., Barer, M. R., O’Donnell, A. G., and Bythell, J. C. (2003). The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environ. Microbiol. 5, 370–382. doi: 10.1046/j.1462-2920.2003.00427.x
Peixoto, R. S., Rosado, P. M., Leite, D. C. A., Rosado, A. S., and Bourne, D. G. (2017). Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience. Front. Microbiol. 8:341. doi: 10.3389/fmicb.2017.00341
Peters, E. C., Halas, J. C., and McCarty, H. B. (1986). Calicoblastic neoplasms in Acropora palmata with a review of reports on anomalies of growth and form in corals. J. Natl. Cancer Inst. 76, 895–912.
Pillai, C. S. G. (1971). “Composition of the coral fauna of the southeastern coast of India,” in Regional Variation in Indian Ocean Coral Reefs, Vol. 28, eds D. R. Stoddart and C. M. Young (London: Symposia of the Zoological society of London), 301–325.
Pootakham, W., Mhuantong, W., Putchim, L., Yoocha, T., Sonthirod, C., Kongkachana, W., et al. (2018). Dynamics of coral- associated microbiomes during a thermal bleaching event. Microbiol. Open 7:e604. doi: 10.1002/mbo3.604
Ravindran, J., Kannapiran, E., Manikandan, B., Mani Murali, R., and Joseph, A. (2012). Bleaching and secondary threats on the corals of Palk Bay: a survey and proactive conservation needs. Ind. J. Geo Mar. Sci. 41, 19–26.
Reis, A. M. M., Araújo, S. D., Moura, R. L., Francini-Filho, R. B., Pappas, G., Coelho, A. M. A., et al. (2009). Bacterial diversity associated with the Brazilian endemic reef coral Mussismilia braziliensis. J. Appl. Microbiol. 106, 1378–1387. doi: 10.1111/j.1365-2672.2008.04106.x
Reshef, L., Koren, O., Loya, Y., Zilber-Rosenberg, I., and Rosenberg, E. (2006). The coral probiotic hypothesis. Environ. Microbiol. 8, 2068–2073. doi: 10.1111/j.1462-2920.2006.01148.x
Richardson, L. L., and Aronson, R. B. (2002). “Infectious diseases of reef corals,” in Proceedings 9th International Coral Reef Symposium, Bali.
Ritchie, K. B. (2006). Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar. Ecol. Progr. Ser. 322, 1–14. doi: 10.3354/meps322001
Roder, C., Arif, C., Daniels, C., Wil, E., and Voolstra, C. R. (2014). Bacterial profiling of white plague disease across corals and oceans indicates a conserved and distinct disease microbiome. Mol. Ecol. 23, 965–974. doi: 10.1111/mec.12638
Rohwer, F., Breitbart, M., Jara, J., Azam, F., and Knowlton, N. (2001). Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20, 85–91. doi: 10.1007/s003380100138
Rohwer, F., Seguritan, V., Azam, F., and Knowlton, N. (2002). Diversity and distribution of coral-associated bacteria. Mar. Ecol. Progr. Ser. 243, 1–10. doi: 10.3354/meps243001
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease, and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
Sato, Y., Willis, B. L., and Bourne, D. G. (2010). Successional changes in bacterial communities during the development of black band disease on the reef coral, Montipora hispida. ISME J. 4, 203–214. doi: 10.1038/ismej.2009.103
Sere, G. M., Tortosa, P., Chabanet, P., Turquet, J., Quod, J.-P., and Schleyer, M. H. (2013). Bacterial communities associated with Porites white patch syndrome (PWPS) on three Western Indian Ocean (WIO) coral reefs. PLoS One 8:e83746. doi: 10.1371/journal.pone.0083746
Shashar, N., Cohen, Y., Loya, Y., and Sar, N. (1994). Nitrogen-fixation (acetylene reduction) in stony corals: evidence for coral-bacteria interactions. Mar. Ecol. Progr. Ser. 111, 259–264. doi: 10.3354/meps111259
Squires, E. J. (1965). Neoplasia in a coral? Science 148, 503–505. doi: 10.1126/science.148.3669.503
Sunagawa, S., DeSantis, T. Z., Piceno, Y. M., Brodie, E. L., DeSalvo, M. K., Voolstra, C. R., et al. (2009). Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3, 512–521. doi: 10.1038/ismej.2008.131
Sussman, M., Willis, B. L., Victor, S., and Bourne, D. G. (2008). Coral pathogens identified for white syndrome (WS) epizootics in the Indo-Pacific. PLoS One 3:e2393. doi: 10.1371/journal.pone.0002393
Tavakoli-Kolour, P., Kavousi, J., and Rezai, H. (2015). Outbreak of growth anomalies in coral communities of Qeshm Island, Persian Gulf. Int. Aquat. Res. 7, 151–156. doi: 10.1007/s40071-015-0100-3
Thinesh, T., Mathews, G., and Edward, J. K. P. (2011). Coral disease prevalence in the Palk Bay, Southeastern India - with special emphasis to black band. Ind. J. Geo Mar. Sci. 40, 813–820. doi: 10.3354/dao03155
Thompson, F. L., Barash, Y., Sawabe, T., Sharon, G., Swings, J., and Rosenberg, E. (2006). Thalassomonas loyana sp. nov., a causative agent of the white plaguelike disease of corals on the Eilat coral reef. Int. J. Syst. Evol. Microbiol. 56, 365–368. doi: 10.1099/ijs.0.63800-0
Thompson, J. R., Rivera, H. E., Closek, C. J., and Medina, M. (2014). Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front. Cell Infect. Microbiol. 4:176. doi: 10.3389/fcimb.2014.00176
Ushijima, B., Videau, P., Poscablo, D., Stengel, J. W., Beurmann, S., Burger, A. H., et al. (2016). Mutation of the toxR or mshA genes from Vibrio coralliilyticus strain OCN014 reduces infection of the coral Acropora cytherea. Environ. Microbiol. 18, 4055–4067. doi: 10.1111/1462-2920.13428
Venkataraman, K., and Rajan, R. (2013). Status of coral reefs in Palk Bay. Rec. Zool. Surv. India 113, 01–11. doi: 10.1007/s10661-014-3835-1
Wang, G., Zhang, S., and Wang, Z. (2009). Responses of alternative complement expression to challenge with different combinations of Vibrio anguillarum, Escherichia coli and Staphylococcus aureus: evidence for specific immune priming in amphioxus Branchiostoma belcheri. Fish Shellfish Immunol. 26, 33–39. doi: 10.1016/j.fsi.2008.09.018
Williams, G. J. (2013). Contrasting recovery following removal of growth anomalies in the corals Acropora and Montipora. Dis. Aquat. Org. 106, 181–185. doi: 10.3354/dao02652
Wilson, B., Aeby, G. S., Work, T. M., and Bourne, D. G. (2012). Bacterial communities associated with healthy and Acropora white syndrome-affected corals from American Samoa. FEMS Microbiol. Ecol. 80, 509–520. doi: 10.1111/j.1574-6941.2012.01319.x
Keywords: coral reef, Acropora cytherea, growth anomalies, bacterial community, nanopore sequencing
Citation: Rajasabapathy R, Ramasamy KP, Manikandan B, Mohandass C and Arthur James R (2020) Bacterial Communities Associated With Healthy and Diseased (Skeletal Growth Anomaly) Reef Coral Acropora cytherea From Palk Bay, India. Front. Mar. Sci. 7:92. doi: 10.3389/fmars.2020.00092
Received: 12 November 2019; Accepted: 04 February 2020;
Published: 21 February 2020.
Edited by:
Sami Souissi, Lille University of Science and Technology, FranceReviewed by:
Vaikundamoorthy Ramalingam, Chonbuk National University, South KoreaRonaldo Bastos Francini-Filho, Federal University of Paraíba, Brazil
Copyright © 2020 Rajasabapathy, Ramasamy, Manikandan, Mohandass and Arthur James. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raju Rajasabapathy, c2FiYWp1bHkwNkBnbWFpbC5jb20=; Rathinam Arthur James, amFtZXMubXNiZHVAZ21haWwuY29t
 Raju Rajasabapathy
Raju Rajasabapathy Kesava Priyan Ramasamy
Kesava Priyan Ramasamy Balakrishnan Manikandan
Balakrishnan Manikandan Chellandi Mohandass
Chellandi Mohandass Rathinam Arthur James
Rathinam Arthur James