Corrigendum: In the Wake of the Ongoing Mass Mortality Events: Co-occurrence of Mycobacterium, Haplosporidium and Other Pathogens in Pinna nobilis Collected in Italy and Spain (Mediterranean Sea)
- 1Department of Biology, University of Naples Federico II, Naples, Italy
- 2Department of Veterinary Medicine, University of Sassari, Sassari, Italy
- 3Mediterranean Center for Disease Control (MCDC), Sassari, Italy
- 4IMC–International Marine Centre, Oristano, Italy
- 5Fish Disease and Aquaculture Center, IZS of Sardinia, Oristano, Italy
- 6IRTA-Sant Carles de la Ràpita, Tarragona, Spain
- 7Dipartimento di Medicina Veterinaria, Università degli Studi di Bari, Valenzano, Italy
- 8Department of Chemical, Biological, Pharmaceutical and Environmental Sciences, University of Messina, Messina, Italy
- 9IZS delle Venezie, Viale dell’Università, Legnaro, Italy
Following the Mass Mortality Events (MMEs) of the pen shell P. nobilis in Campania region and Sicily, a survey of moribund P. nobilis specimens was also conducted in other Italian regions (Campania, Tuscany, Sardinia, and Apulia) and Spain (Catalunya). Histopathological and molecular examination of 27 specimens of P. nobils revealed different types of pathogens associated with tissue lesions, morbidity and mortality. Presence of Mycobacterium, Vibrio species, Haplosporidium pinnae and Perkinsus sp. were detected, differently distributed into the areas. The Mycobacterium sp., previously reported in Campania and Sicily samples, was observed in all the analyzed areas and individuals, associated to systemic inflammatory lesions. In Spain, H. pinnae was observed in 36% of cases, always associated to the Mycobacterium sp. Molecular study using hsp65 genes and Internal Transcriber Spacer ITS support that a new species of Mycobacteria is infecting P. nobilis, close to M. triplex and belonging to the group of M. simiae complex with M. sherrisi. Presence of Perkinsus spp. resembling P. mediterraneus was observed in 2 out of 13 Italian individuals whose presence should be addressed as potential risk for shellfish aquaculture of the area. Vibrio spp. were also detected in some case. The preliminary results of this study suggest that Mycobacterium sp., Vibrio spp., H. pinnae and Perkinsus sp. cooperate to disease pathogenesis, being Mycobacterium and Haplosporidium most of the time involved. Vigilant inspection of those areas where MME is now starting, along with continuous systematic surveys, are crucial to define the spatiotemporal progress of mortality and the role of every single pathogen in the disease outcome.
Introduction
The co-occurrence of different pathogens in the context of aquatic animals disease has received little attention so far, even considering that this condition is common in human and fish pathology and critical to understand disease dynamic (Bakaletz, 2004; Griffiths et al., 2011; Johnson and Hoverman, 2012). The interactions in the same host of different protozoan species, along with bacteria and viruses, are reported in literature with a dramatic effect in disease susceptibility (Cox, 2001; Lassalle et al., 2007; Arzul et al., 2012). Co-infection can in fact alter the progression and the severity of disease pathogenesis and clinical outcomes, also influence the spread of infections at population level and bring to animal outbreaks, as described in coral, fish and bivalves (Lassalle et al., 2007; Arzul et al., 2012; Kotob et al., 2016; Tracy et al., 2018). The little consideration given to this aspect is due by the fact that the interaction between pathogens is complex and pathogenesis hard to understand. Moreover, even if MMEs in aquatic animals are increasing, like in the case of sea star wasting disease impacted marine communities recently, studies on marine disease still results as a neglected and emerging field (Hewson et al., 2014; Carella et al., 2015; Fey et al., 2015). During episodes of co-infection, interactions between the infectious agents can bring to varied outcomes: the weight of one or both the pathogens may be amplified, one or both may be suppressed or one may be amplified and the other suppressed (Johnson and Hoverman, 2012).
Since 2016, numerous mortality events of P. nobilis have been reported in the Mediterranean Sea reaching up to 100% of the population in many areas like Italy, Greece and Spain (Catanese et al., 2018; Carella et al., 2019; Katsanevakis et al., 2019). At first, an Haplosporidian parasite, named Haplosporidium pinnae, was designated as the solely responsible for the mortality (Catanese et al., 2018). After that, a mycobacterial infection has been also associated to the disease outcome (Carella et al., 2019), while Vibrio mediterranei appears to cause major mortality among stabled individuals in Spain, possibly associated to animal stress in captivity (Prado et al., 2019). Mycobacteriosis is a chronic, systemic, progressive and frequently lethal disease affects a wide range of terrestrial and aquatic teleost fishes (Ross, 1970; Gauthier and Rhodes, 2009). The mycobacteriaceae family includes severalgroups of species: (1) the Mycobacterium tuberculosis complex (MTBC) including species that are pathogenic to humans, wild and domesticated mammals (M. tuberculosis, M. bovis, M. africanum) which cause Tuberculosis (TB); and (2) Non-Tuberculous Mycobacteria (NTM), consisting on free-living organisms, ubiquitous in the environment and saprophytic habit and that can cause a wide range of infections both in humans and animals from different environments (Porvaznik et al., 2017; Honda et al., 2018). NTM infections are among the most common chronic disease of aquatic animals (Gcebe et al., 2018). In the aquatic environment, at present, there are over 167 species of freshwater and marine fishes described as susceptible to Mycobacteria (Bruno et al., 1998; Jacobs et al., 2009). Haplosporidian parasites (phylum Haplosporidia) are a small group of four genera of pathogenic protozoan parasites usually with uninucleated spores. They infect molluscs including commercially important bivalves and other molluscs, annelids, crustaceans, ascidians, trematodes, turbellarians (Arzul and Carnegie, 2015). This phylum contains 52 described species and several unnamed species reported in the four genera (Burreson and Ford, 2004).
After a recent study reporting mortality episodes of P. nobilis in the Campania region associated to a mycobacterial disease (Carella et al., 2019), the aim of this work is to evaluate the geographic spreading of the mycobacteriosis and its co-occurrence with H. pinnae and other pathogens in other parts of Mediterranean Sea, in particular in different regions of Italy (Campania, Tuscany, Apulia, and Sardinia), and one region in Spain (Ebro Delta, Catalunya).
Moreover, in order to characterize the mycobacteria species, we used the conserved genes hsp65 and the 16S–23S internal transcribed spacer region (ITS) (Kim et al., 2005a; Mohamed et al., 2005). Primary surveillance systems based on a rapid identification of pathogens are critical for effective disease control and for improving epidemiological assessments (De Vico and Carella, 2019). The conventional detection of mycobacteria can be done through microscopic examination for acid-fast bacilli by Ziehl–Neelsen (ZN) stain in smear samples and histopathology (Koch and Cote, 1965). In this study, we also investigated the application of an in situ hybridization (ISH) as diagnostic assay and discriminating it from other acid-fast mycobacteria in aquatic environment.
Materials and Methods
Animal Collection and Histopathology
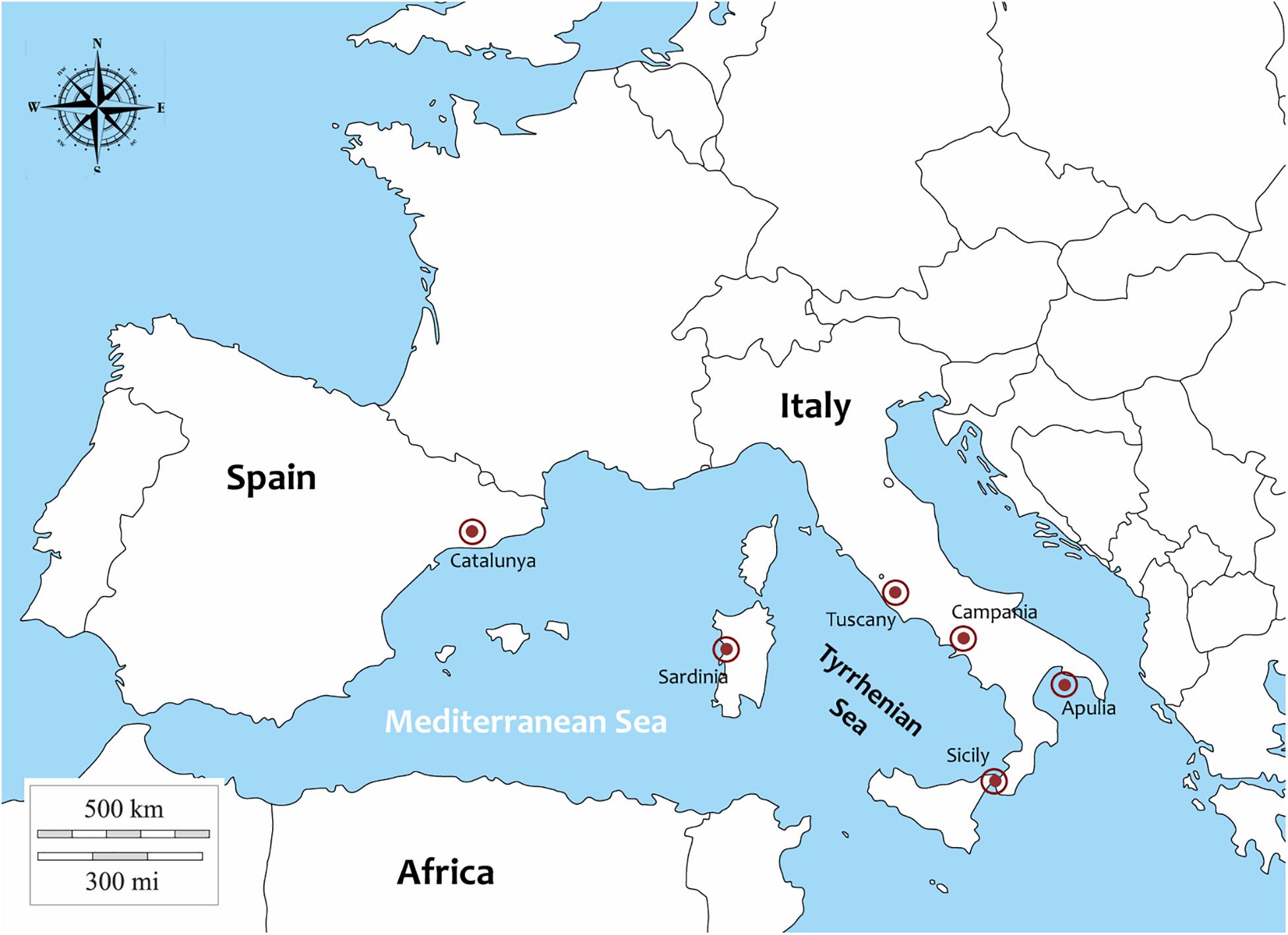
Samples of P. nobilis (27) were collected from different Italian regions and in one Spanish region (Catalunya) between 2018 and 2019 (Figure 1). In particular: 1 animal from Campania (Cilento area), 1 from Sicily (Lago Faro), 3 from Sardinia (Oristano Gulf), 1 from Tuscany (Isola di Gorgona), 3 from Apulia from Mare Piccolo di Taranto, published by Panarese et al. (2019) and 4 form Porto Cesareo. Moreover 14 animals were collected in Catalunya (Ebro Delta) in 2018.
In Campania Region, in the context of the Marine Strategy of Marine Protected Areas (D.M. 22 del 11/02/2015) (Italy) the percentage of death, moribund and live animals in the field was also evaluated and recorded along the coast form scuba divers. A permit for collection of P. nobilis individuals in Campania was issued by the ISPRA Istituto Superiore per la Protezione e la Ricerca Ambientale (Ref.: Prot. 25888 5 April 2018). In Spain the animals were collected following a permission of the Catalonian government in the context of a rescue project of Pinna nobilis individuals (Generalitat de Catalunya, permission of the 17th November 2017). In the other areas moribund individuals were collected in order to define the animal’s health status. In every region, animal valve length was measured and opened with a blade to cut the adductor muscles. In each area, from each animals portions of tissues were collected (digestive glands, mantle, labial palps, gills, gonads, adductor and retractor muscles) and were fixed in Davidson’s solution and stored for 48 h at room temperature. Complementary tissue used for histopathology were also fixed in absolute ethanol for DNA isolation. For histopathology, tissue samples were embedded in paraffin blocks and sectioned at 5 μm with a rotary microtome (Bioptica, Naples, Italy). Tissue sections were deparaffinised, stained with Carazzi haematoxylin and eosin and special stain like V.O.F. (Verde Luz-orange G-acid fuchsin) stain (Gutierrez, 1977) and examined by light microscopy (Zeiss Axioscope A1). In order to define the Mycobacteria presence and distribution, in every tissue per samples Ziehl-Neelsen (Mazzi, 1977) stain was also performed.
DNA Isolation and PCR Diagnosis of Common P. nobilis Pathogens
DNA was isolated from tissues using the Qiagen Blood and Tissue Kit (Qiagen). DNA quality and quantity were checked with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Inc.). In order to define the presence of Mycobacterium sp. and the possible co-existence of Haplosporidium pinnae in the same individual, in all the animals belonging to the different locations DNA was amplified by PCR using the 16S rRNA for Mycobacterium (Böddinghaus et al., 1990), and for and the small subunit ribosomal DNA (SSU rDNA) for H. pinnae (Catanese et al., 2018; Table 1). Moreover, in Catalunya samples, specific primers for V. mediterranei were also used (Andree et al., under review). The PCR was performed in 25 μl of reaction volume containing 1 μl of genomic DNA (100 ng/μl), 12.5 μl of GoTaq MasterMix (Promega) at 1x concentration, 6.5 μl of water and 2.5 μl of each primer (10 μM). PCR products were electrophoresed on 2% agarose gels in 1x TAE buffer. Negative controls were also included in all the PCR reactions.
Amplification of Mycobacterial hsp65, ITS and Phylogenetic Analysis
The 65 kDa heat-shock protein family hsp65 has been considered as useful phylogenetic markers for Mycobacteria along with the internal transcriber spacer ITS. Both hsp65 and ITS were used to characterize the Mycobacterium species in all the Italian regions and in Spain. The sequence of the primers and the reference are reported in the Table 1. The amplified fragments were gel eluted and directly sequenced. The chromatograms were analyzed using the program ChromasPro version 2.6.4 (Technelysium). Regions of similarity between edited and published biological sequences were compared using the BLAST-Basic Local Alignment Search Tool1. The pairwise p-distances were calculated among the hsp65 and ITS nucleotide sequences of Mycobacterium sp. obtained in the present study and those of different Mycobacterium species present in GenBank, selected on the basis of the highest BLASTN score. The nucleotide alignment was constructed using ClustalW and the neighbor-joining tree was obtained using the Maximum Composite Likelihood model implemented in MEGA X with 1000 bootstrap replicates (Kumar et al., 2018).
In situ Hybridizationfor the Detection of P. nobilis Mycobacterium
In order to confirm that the amplified region corresponded to the Mycobacteria detected by histology, a specific probe (326bp) was generated by PCR with primers MycPn7F (5′-GTGGAAAGCTTTTGCGGTGT-3′) and MycPn7R (5′-CGCTCGCACCCTACGTATTA-3′) constructed on 16S sequences (Table 1) using digoxigenin dUTP 25 mM in the reaction mix following a modification of the method described by Lipart and Renault (2002). Briefly, sections of different tissues of P. nobilis (7 μm thick) from different Italian regions and from Spain, were placed on salinized slides (Sigma), dewaxed with xylene and treated with Proteinase K (100 μg ml–1). After dehydration with 95% ethanol and absolute ethanol, tissue sections were air dried and a prehybridization step was carried out with prehybridization buffer (50% formamide, 10% dextran sulphate, 4X SSC, 250 μg ml–1 yeast tRNA) for 30 min at 42°C in a humid chamber. Slides were incubated overnight at 42°C with 100 μl of the prehybridization buffer containing the specific probe (20 ng μl–1). After hybridization, sections were washed with 2X SSC and 0.4X SSC and the detection of the digoxigenin-labeled probe was conducted. Samples were incubated with anti-DIG-AP antibody solution DIG-1 (1:500 in Tris–HCl buffer, pH 7.5) for 1 h at RT. After that, sections are incubated in nitroblue tetrazolium/bromochloroindolyl phosphate (NBT/BCIP, in Tris–HCl based buffer, pH 9.5) for 1 h in the dark to visualize the hybridized products. The slides were finally counter-stained with Bismarck yellow, dehydrated and mounted using Eukitt (Bioptica, Italy). In order to define the specificity of the probe, in situ hybridization was conducted in tissue sections from aquatic animals infected by other mycobacteria: a zebrafish infected by M. tuberculosis and the common flathead mullet Mugil cephalus infected by M. marinum. Negative controls comprehend samples incubated without the probe were also included.
Results
Mortality Data, Light Microscopy and in situ Hybridization
From north to the south of the Campania Region, including the islands of Ischia and Procida, the monitoring activity of the scuba divers of AMPs revealed a high rate of mortality of P. nobilis ranging from 65 to 100% by the end of December 2017 to May 2018 (Figure 2). The other Italian regions reported a similar situation (personal communication). In Italian samples, both histopathological and molecular analysis revealed the presence of Mycobacterium sp., Vibrio species, H. pinnae and other parasites like Perkinsus sp. (Table 2). In particular, in Italian samples from Tuscany, Sardinia, Sicily, Apulia and Campania the co-occurrence of H. pinnae and Mycobacterium was observed. The Haplosporidium was always detected at the level of digestive tubules in advanced phases of development and scarce plasmodium phases, with instead well visible sporocysts accompanied by light interstitial inflammation (Figure 3A). In Sardinia, all the three specimens showed both the presence of Mycobacterium sp. and H. pinnae with a co-occurrence of a Perkinsus species in two out of three samples analyzed (Table 2, Figure 3B). The trophozoites of Perkinsus sp. were detected in the interstitial of connective tissue of inter tubular spaces of digestive gland, constituted by densely packed spherical trophozoites ranged from 10–25 μm in size. They showed a large vacuole, sometimes enclosing a vacuoplast and peripheral nucleus and a clear nucleolus. In the specimen from December 18, a massive digestive gland necrosis was also observed.
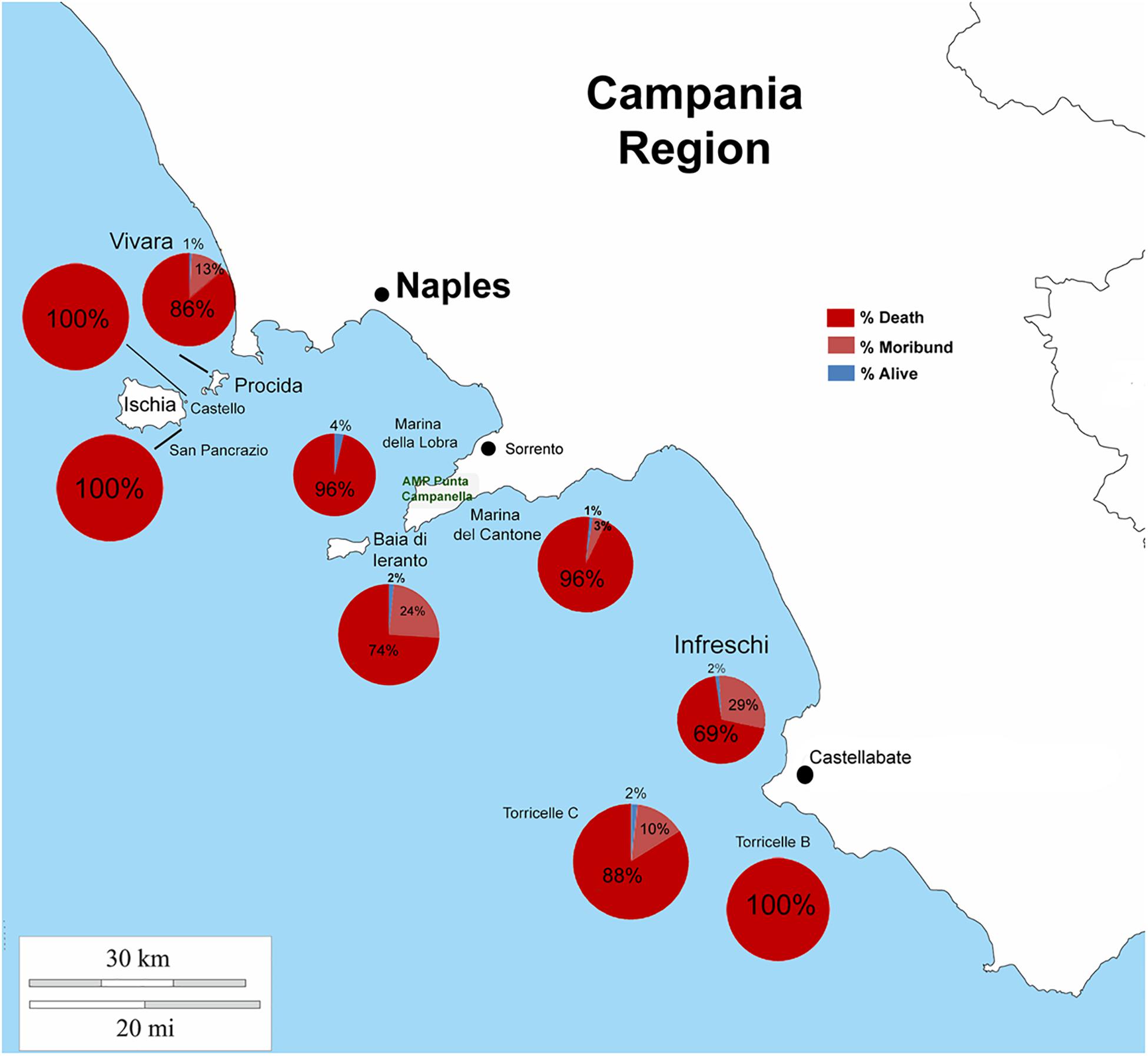
Figure 2. Percentage of death, moribund and survived individuals of P. nobilis in Campania Region in period 2017–2018.
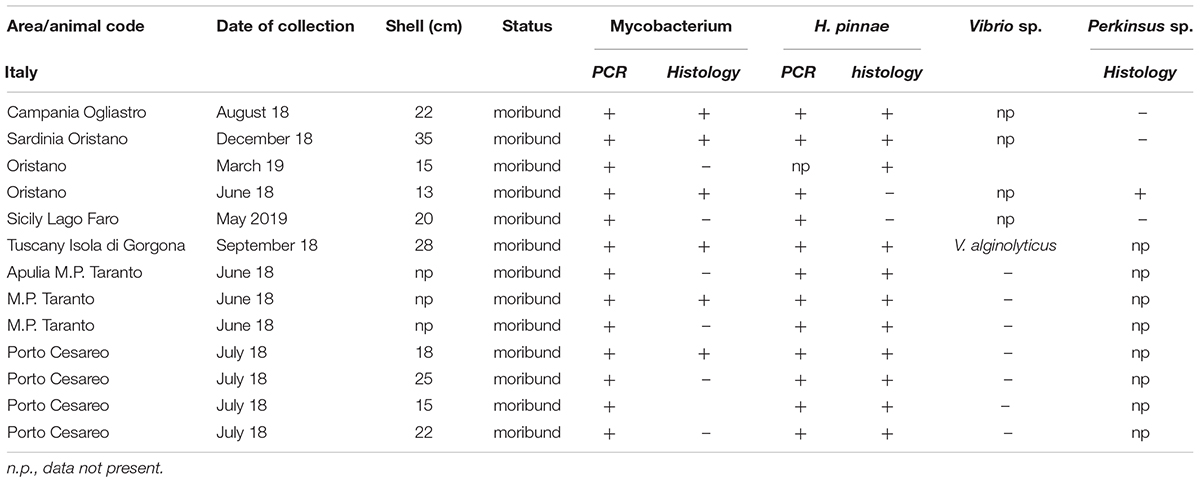
Table 2. Sampling data of the 13 Pinna nobilis specimens from Italy analyzed in the study and results of the detection of Mycobacterium, H. pinnae and other pathogens by histology and molecular diagnostic.
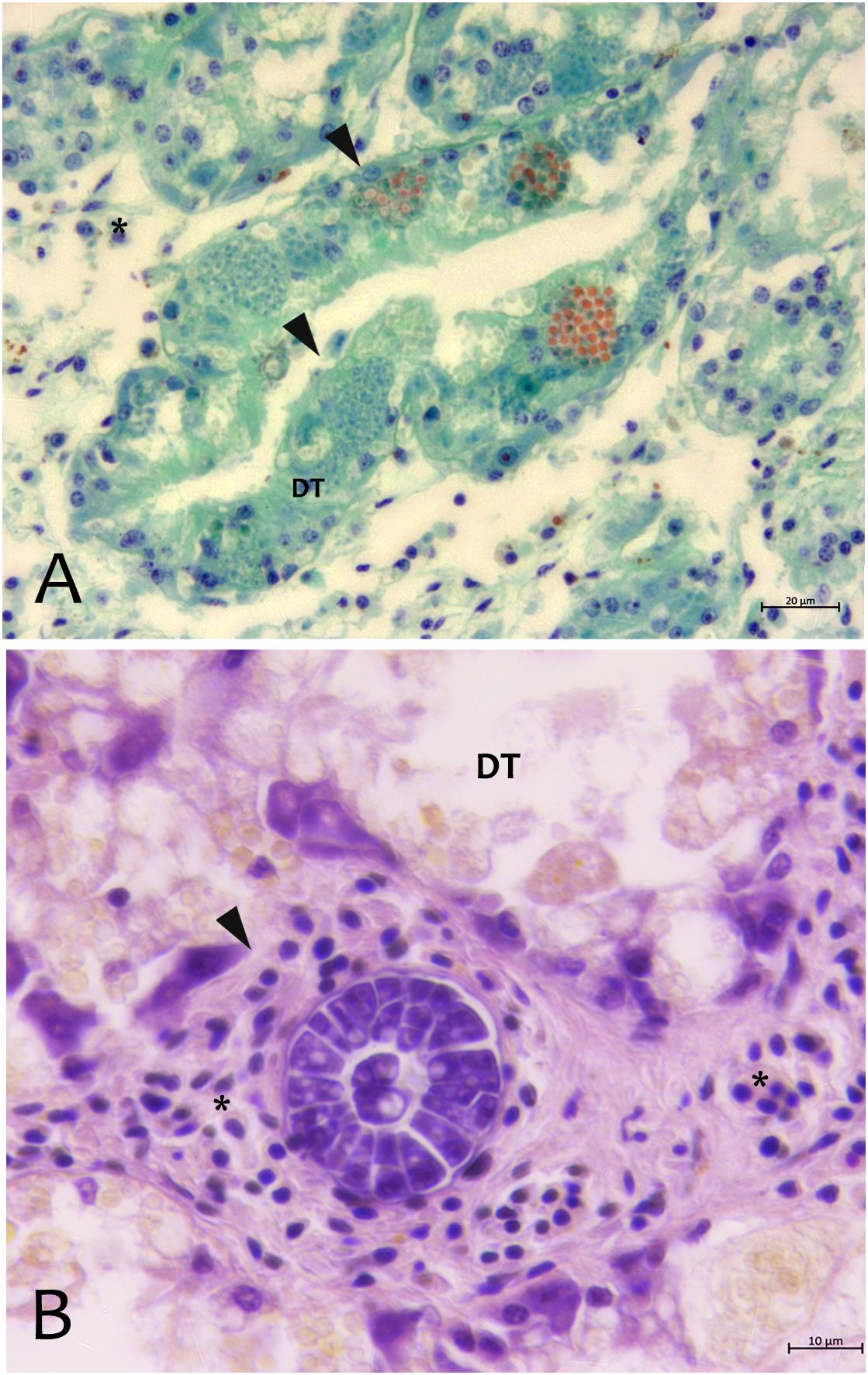
Figure 3. Parasites observed in Campania (A) and Sardinia samples (B) of P. nobilis. (A) Digestive gland of Pinna nobilis infected with by H. pinnae in the digestive tubules (DT) showing sporocysts with deeply stained plasmodia in green, and mature spore in bright orange stained with V.O.F. stain (arrows). (B) Perkinsus sp. with encapsulated multicellular trophozoites (rosettes) in the interstitial space of digestive gland. ∗haemocytes; DT, digestive tubules.
In Spain, at the molecular diagnostic, three moribund specimens, three recently death animals and 1 in decomposition resulted positive at the detection of Mycobacterium (7/14–50%) (Table 3). In three sample, the presence of V. mediterranei was also observed. Haplosporidium was also present in 1 individual in decomposition, in two moribund individuals and two recently death animals (5/14–36%). The samples positive to both H. pinnae and Mycobaterium sp., histologically showed few sparse plasmodium phases of the H. pinnae distributed in the interstitium of the digestive tissue or of the gonad, with Mycobacterium present within scattered immune cells or forming inflammatory nodules (Figure 4).
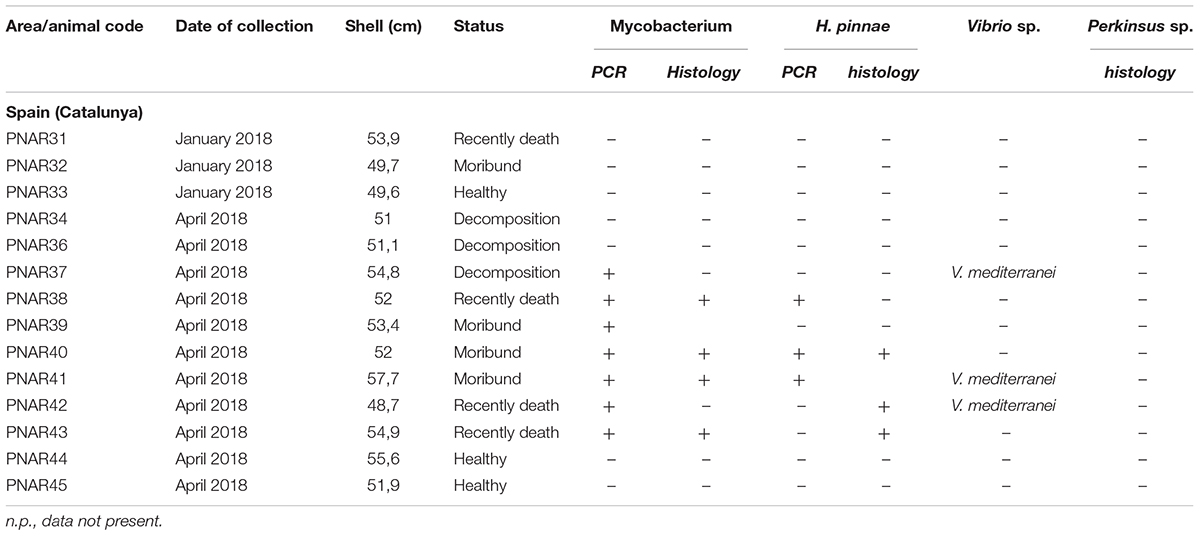
Table 3. Sampling data of the 14 Pinna nobilis specimens from Catalunya analyzed in the study and results of the detection of Mycobacterium, H. pinnae and V. mediterranei by histology and/or molecular diagnostic.
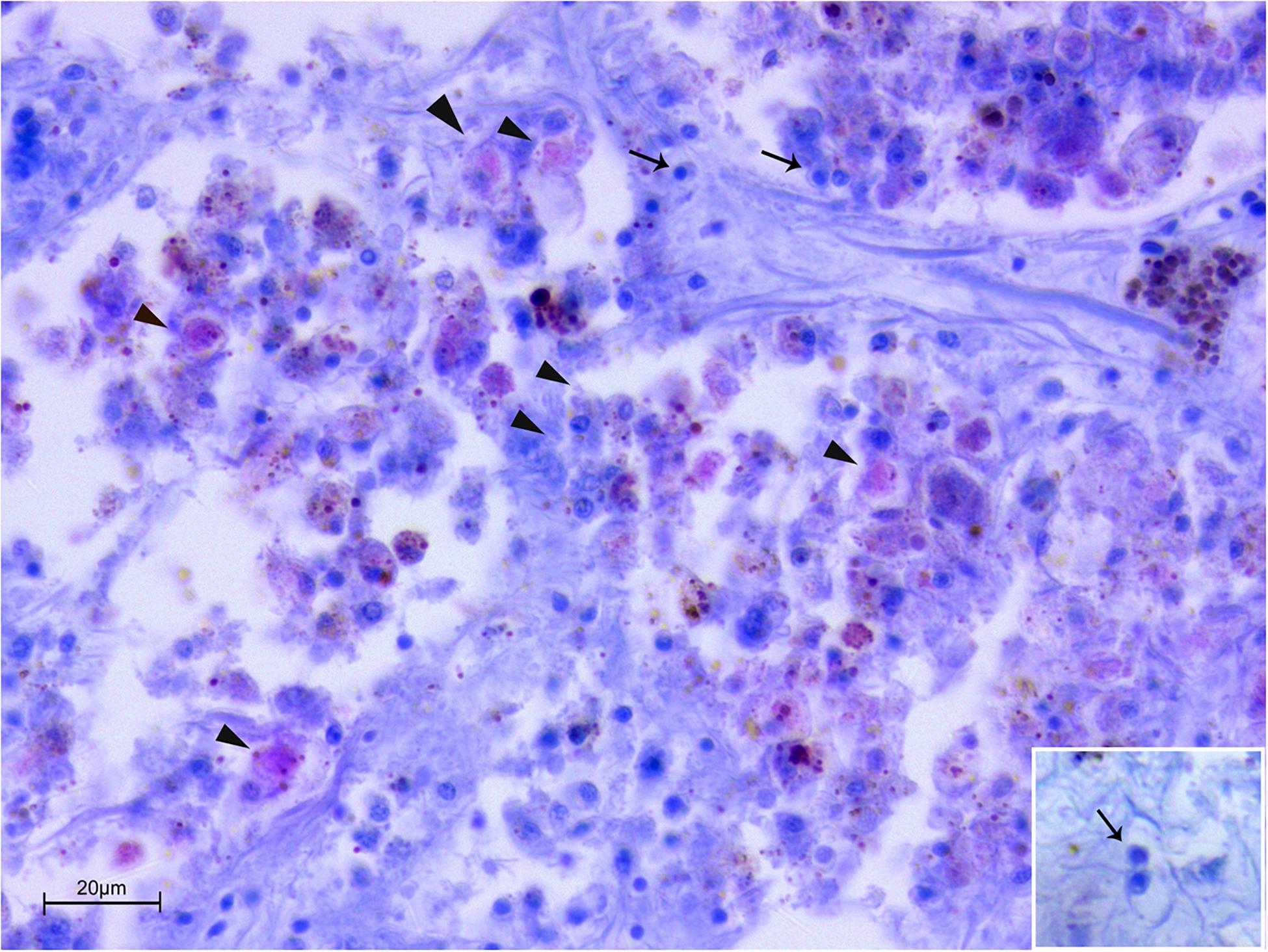
Figure 4. Co-infection by Mycobacterium sp. and H. pinnae in samples of P. nobilis from Catalunya (ZN stain). Presence of Mycobacterium filling immune cells (arrowheads) forming nodules accompanied by the presence of plasmodium phases of H. pinnae (arrows) and insert.
About the Mycobacterium tropism, it was mainly observed at level of the connective tissue of mantle as well as in the fibrous capsule surrounding gonad and digestive gland (Figure 5). Mycobacteria were present filling one or few aggregates of immune cell expanding within the areas around the gonadal follicles or infiltrating digestive tubules (Figures 5C,D), and as single or multiple immune cells aggregate rich regions coupled with Brown Cells (Figures 5E,F).
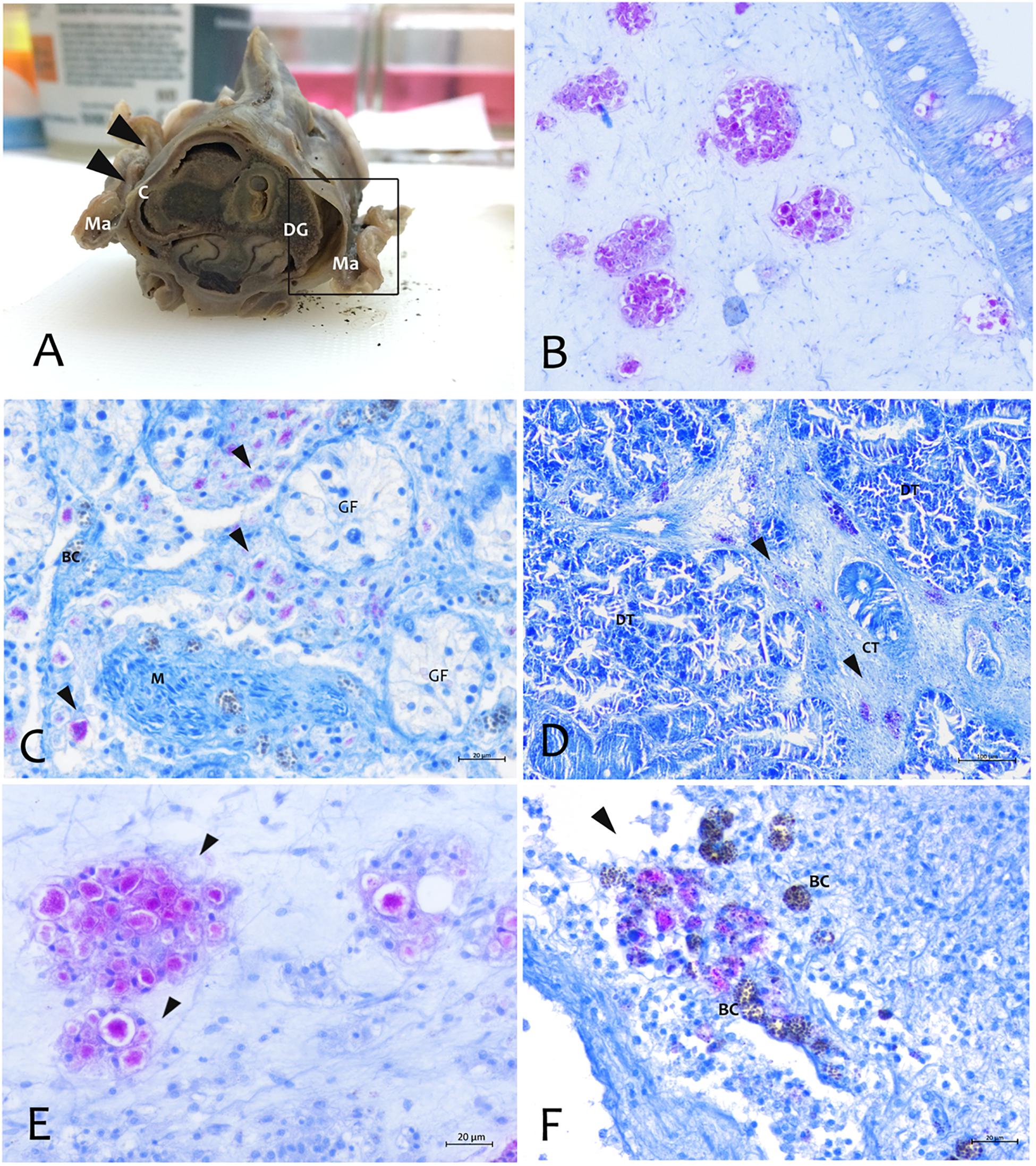
Figure 5. Distribution of the Mycobacterium at tissue and organs level by Zhiel-Neelsen stain in P. nobilis specimens. (A) Gross observation of digestive system and mantle in transverse section showing tropism of M. at fibrous capsule circumscribing digestive gland (DG) and image at microscopic level (B). (C) Tropism in the interstitial space of gonadal follicles (GF) and in the connective tissue of digestive gland (black arrows) (D). (E) Mycobacteria present as aggregates filling immune cells (arrowheads) or present in few cells, and recalling a strong inflammatory response of haemocyte in the site of infection associated with Brown Cells (BC) (F) at mantle level; Ma, Mantle; Ct, Connective tissue; M, muscle; DT, Digestive tubules; C, connective capsule.
By using the in situ hybridization technique, the amplified sequence, used as DNA probe, showed to be specific to this Mycobacterium. The probe showed strong to mild label within immune cells in different tissues like mantle epithelium and the underline connective tissue (Figure 6). This signal was not detected in negative controls or uninfected tissues. Moreover, there was no cross-reactivity with other mycobacteria since the mycobacterial probe did not hybridize with M. tuberculosis in zebrafish and M. marinum in common flathead gray mullet.
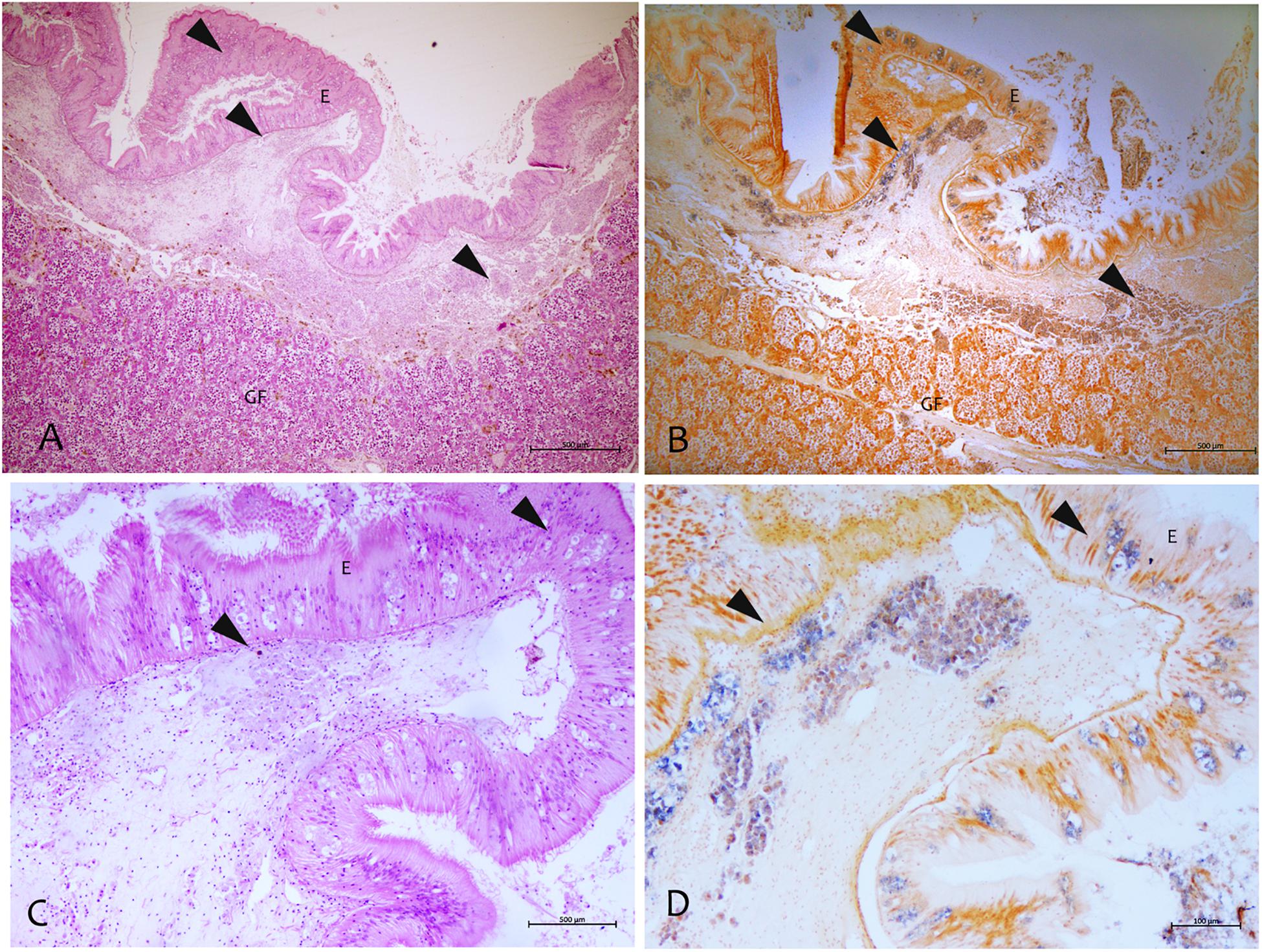
Figure 6. Mycobacterium identified by the in situ hybridization technique in samples of P. nobilis in H&E (A–C) compared with the in situ (B–D). (A,B) General view of the gonad with signal in the connective tissue and epithelia (arrowheads) (B) and detail showing the labeled mycobacterium cells (arrowheads). (C,D) Detection of mycobacteria in the immune cells aggregates of mantle underline connective and epithelia using the digoxigenin-labeled probe. GF, Gonadal Follicle; E, epithelium.
PCR Amplification of hsp65 and ITS Sequences of Mycobacterium
Sequences of 441 bp and 289 bp were obtained for the hsp65 and ITS loci, respectively. For the hsp65, BLAST analysis revealed that the nucleotide sequences of these fragments were homologs of the hsp65 region presenting the 97–98% of identity with Mycobacterium sp. isolated in clinical non-tuberculous isolates (Accession number: AY379077). The hsp65 Mycobacterium sequences isolated from all the areas were very similar, showing a p-distance ranging from 0 to 0.0053. The neighbor-joining tree constructed from the nucleotide alignment of the sequences obtained in the present study and those of different hsp65 Mycobacterium species downloaded from Gene Bank are shown in Figure 7. The six sequences of the different areas were deposited. Accession number: (MN854405-MN854410).

Figure 7. Neighbor-joining tree of the hsp65 sequences. The analysis involved 22 nucleotide sequences and 882 positions in the final dataset. The percentages of bootstrap replicates are shown next to the branches.
On the other side, the ITS sequences of Mycobacterium in all the areas showed a p-distance ranging from 0 to 0.0044 showing and identity of 95–96% with a M. triplex isolated in human pulmonary infection (Figure 8). The six sequences of the different areas were deposited. Accession number: (MN637877-MN637882).
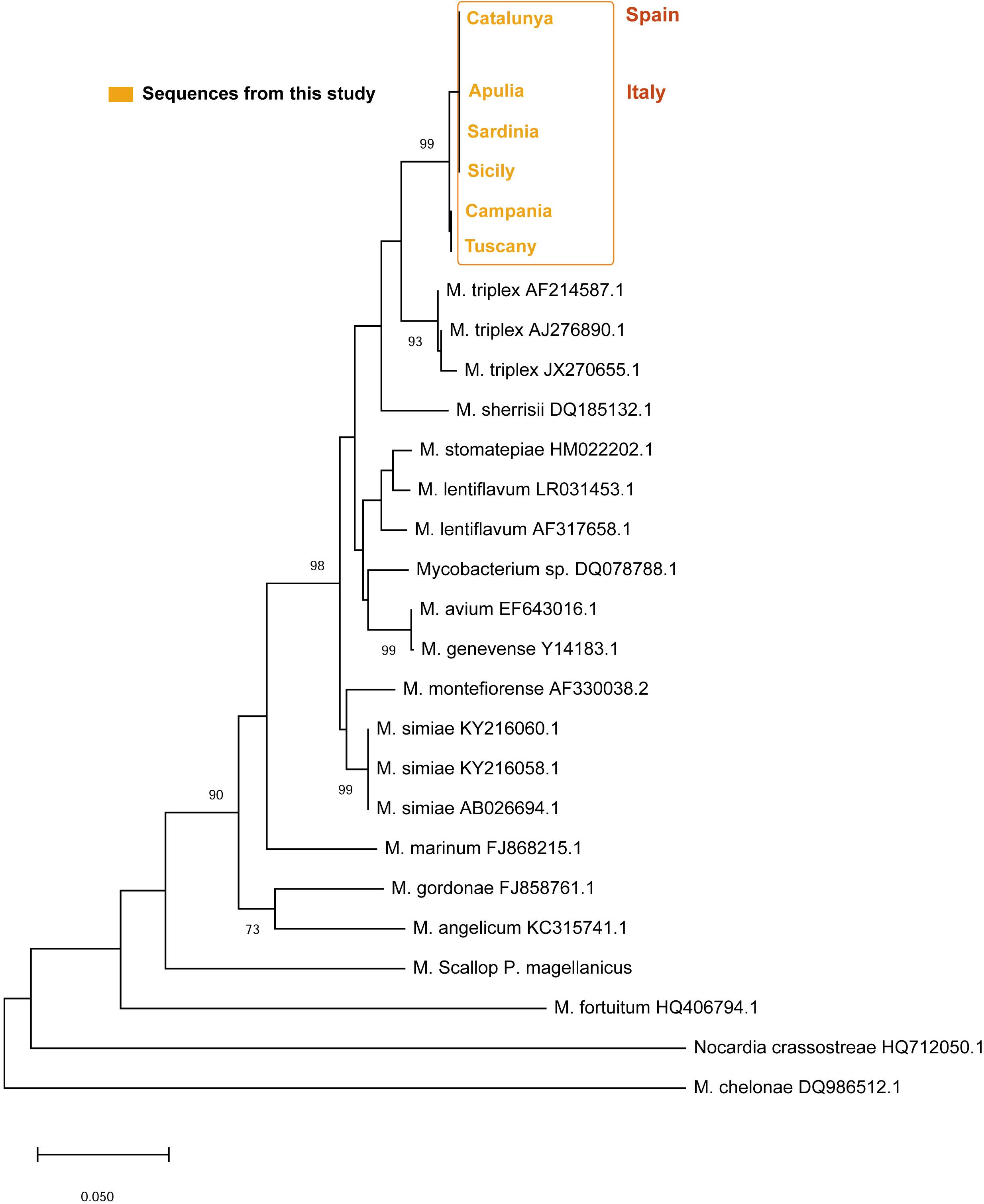
Figure 8. Neighbor-joining tree of the ITS rDNA sequences. The analysis involved 21 nucleotide sequences and a total of 303 positions in the final dataset. The percentages of bootstrap replicates are shown next to the branches.
Discussion
The present study describe for the first time the presence of different pathogens, comprehending bacteria and parasites of different taxonomy, co-occurring over P. nobilis MMEs in two different areas of Mediterranean Sea, Italy and Spain, collected in 2018/2019. Concurrent infections by multiple pathogens species are common in human and animals, but how this component affects host morbidity remains mainly theoretical (Griffiths et al., 2011; Johnson and Hoverman, 2012; Groner et al., 2016). This condition may be more likely happen in immune depressed individuals, more disposed to multiple opportunistic infections, or co-infecting pathogens can interact synergistically with host’s immune system, and consequently the presence of one enhances the abundance and/or virulence of the other, with many examples in human disease. Co-infection of mycobacteriosis and parasitic diseases in humans is an important problem in developing countries, where parasite species are simultaneous with Mycobacterium sp. at multiple organ level, also related to the age of the host (Petney and Andrews, 1998; Fenton et al., 2014).
Mycobacterium species have been documented as an important source of morbidity and mortality in fish aquaculture, as well as in wild animals (Gauthier and Rhodes, 2009), but few descriptions exist in bivalves (Grimm et al., 2016; Carella et al., 2019). Showing to be present at the level of phagocytes as intracellular pathogens (Carella et al., 2019). The behavior of mycobacteria within the immune cells of molluscs has not been characterized yet, but previous study in P. nobilis showed granulocyte tropism and no evidence of phago-lysosomal membranes to enclose it (Carella et al., 2019). Mycobacteria are generally visualized in tissue sections using the Ziehl–Neelsen stain, which is based on acid-fastness of the mycobacterial cell wall to acid–alcohol decolorization after staining with carbolfuchsin (Mazzi, 1977). In this study, apart from Ziehl–Neelsen stain, we also tested a specific probe based on sequences of P. nobilis mycobacterium. In addition to their known infectivity to fishes, aquatic mycobacteria pose significant zoonotic concerns (De Vico and Carella, 2019). Molecular markers have been used to elucidate the taxonomy of the Mycobacterium of P. nobilis. Although the hsp65 gene has been reported to be the most suitable target for the differentiation of mycobacteria species (Kim et al., 2005b; Mun et al., 2007), it has also been reported that some discrepancies can exist between hsp65, internal transcribed spacer 1 (ITS1) and 16S rDNA results for species differentiation. When comparing the hsp65 and ITS, we noticed that the 16S–23S internal transcribed spacer region (ITS) had better identification ability. First of all, nor hsp65 or ITS showed 100% of similarity of any type of mycobacteria in Blast associated to the obtained sequences. The hsp65 sequence analysis associates the Mycobacterium to an unknown Mycobacterium species located in the group of M. lentiflavum. On the other side, ITS sequences locate the bacterium close to another human mycobacterium species M. triplex responsible of disease in both immunocompromised and immunocompetent individuals (Piersimoni et al., 2004; Caruso et al., 2013) and reported as causing pulmonary infections in human from different countries (Dias-Campos et al., 2016). Molecular results support likely a new species of Mycobacteria infecting P. nobilis, close to M. triplex and belonging to the group of M. simiae complex with M. sherrisi.
Two different Vibrio species have been isolated in two geographic areas. Vibrio alginolyticus in Tuscany and Vibrio mediterranei (V. shiloi) in Catalunya. Vibrio mediterranei has been found associated to some disease condition of aquatic animals such as corals and scallops (Rubio-Portillo et al., 2014; Serrano et al., 2018) and associated to P. nobilis mortality in stabled individuals (Prado et al., 2019). No Vibrio mediterranei have been observed in Apulia (Panarese et al., 2019) and still lack data from Campania Sardinia and Sicily. V. alginolyticus is associated disease of marine animals including fish, crustaceans and molluscs (Balebona et al., 1998; Gómez-León et al., 2005; Ben kahla-Nakbi et al., 2006).
haplosporidian parasites have been major pathogens of concern for aquatic animal health managers and shellfish industries around the world (Arzul and Carnegie, 2015). The phylum Haplosporidium contains spore-forming protozoan parasites responsible of mortality episodes of both freshwater and marine invertebrates including bivalves, crustaceans, and polychaetes (Arzul and Carnegie, 2015). In brackish environments the causative agent of MSX disease Haplosporidium nelsoni has contributed to major mortalities in the eastern oyster populations (Crassostrea virginica) along the coast of the United States (Burreson and Ford, 2004). The new haplosporidian species, H. pinnae is reported associated to mass mortality events of P. nobilis in Balearic Islands (Catanese et al., 2018), Greece (Lesvos Island) (Katsanevakis et al., 2019) also reported in Italy (Panarese et al., 2019).
In our study, all the Italian regions resulted positive to both Mycobacterium and Haplosporidium. Few months before, in Campania and Sicily, Carella et al. (2019) reported the main representation of Mycobacterium associated to a strong and systemic inflammatory response. H. pinnae was instead detected in only one sample. Few months later, however, we identified the two pathogens in both the areas. It is interesting to note that, similarly to early report from Campania region in Italy (Carella et al., 2019), in Catalunya, where the mortality was at the beginning, we found samples affected by the mycobacteriosis and H. pinnae was less represented. Interestingly, parasites belonging to the Perkinsus genus has been observed in two over three analyzed samples of Sardinia. The genus Perkinsus has been associated with mortalities of molluscs around the world, including oysters, clams, abalones and scallops (Soudant et al., 2013). The pathogen observed resembled P. mediterraneus, a parasite firstly detected in oysters like Ostrea edulis from Mahon (Minorca, Balearic Islands, Spain) in 2004 and in other mollusc species. Several years later it was also found in Andratx Harbor (Majorca, Balearic Islands) (Casas et al., 2004) in oysters from the Gulf of Manfredonia (SE Italy), in V. verrucosa and Arca noae from Balearic Islands and Chlamys varia from Balearic Islands, Alicant and Delta de l’Ebre (Ramilo et al., 2015). In some case, simultaneous presence of P. mediterraneus and other important parasites like Marteilia in the natural bed of C. gallina have also been described (Valencia et al., 2014).
Conclusion
Our data underline that many pathogens are associated to moribund animals, among which Mycobacterium sp. and H. pinnae seems to be the most relevant. About possible disease pathogenesis, it is important to emphasize that the data collected in some areas of Catalonia, in accordance with the data of Carella et al. (2019), show also morbidity/mortality in the presence of Mycobacterium and absence of H. pinnae, suggesting that the mycobacteriosis is most often associated to the condition. Moreover, the samples of Sardinia showed the co-occurrence of other opportunistic pathogens like Perkinsus sp., that can also contribute to animal morbidity. Given the great number of pathogens detected, this study also underline that in future research, the study of a single agent possibly associate to the disease, should be replaced by a diagnostic panel aimed at verifying the simultaneous presence and quantity of multiple pathogens potentially involved in disease pathogenesis. Furthermore, continuous sampling on time scale, rather than punctual sampling, is necessary, to define the role of every single pathogen. Unfortunately, continuous data for the other Italian regions are lacking, and no solid conclusions can be based on single reports. For that reason, the areas where the clinical signs of the disease and the first cases of mortality are displaying, are central to understand disease aetio-pathogenesis. Finally, the multitude of pathogens associated to the disease, as observed in this study, also suggests that the immune status of the animals could be involved in P. nobilis mortality. The inter relationships between multiple pathogens within a host are very complex. The result of our study supports the view recently exposed by Carella et al. (2019), that there may be a common primary cause, not yet identified, which favors the above condition.
Data Availability Statement
The datasets generated for this study can be found in the accession numbers from: MN637877–MN637882 and MN854405–MN854410.
Author Contributions
FC and GD conceived the study, formally analyzed the data, and wrote the original draft. FC investigated the results and developed the methodology. EA, SF, FS, DM, FM, PP, and RP helped in the analysis of the samples. TP and EF provided part of the biomolecular diagnostic.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer PM declared a shared affiliation, with no collaboration, with several of the authors, FC and GD, to the handling Editor at the time of review.
Acknowledgments
The authors are very grateful to Dr. Domenico Sgambati and the group of SCUBA divers from Area Marina Protetta (AMP) Punta Campanella and Dr. Francesco Pollaro form CEESM. The authors also thank Dr. Chiara Miele and Dr. Grazia Villari for the technical support in histopathology.
Footnotes
References
Arzul, I., and Carnegie, R. B. (2015). New perspective on the haplosporidian parasites of molluscs. J. Invertebr. Pathol. 131, 32–42. doi: 10.1016/j.jip.2015.07.014
Arzul, I., Chollet, B., Michel, J., Robert, M., Garcia, C., Joly, J. P., et al. (2012). One Perkinsus species may hide another: characterization of Perkinsus species present in clam production areas of France. Parasitology 139, 1757–1771. doi: 10.1017/S0031182012001047
Bakaletz, L. O. (2004). Developing animal models for polymicrobial diseases. Nat. Rev. Microbiol. 2, 552–568. doi: 10.1038/nrmicro928
Balebona, M. C., Andreu, M. J., Bordas, M. A., Zorrilla, I., Morinigo, M. A., and Borrego, J. J. (1998). Pathogenicity of Vibrio alginolyticus from cultured gilt-head sea bream (Sparus aurata, L.). Appl. Environ. Microbiol. 64, 4269–4275. doi: 10.1128/aem.64.11.4269-4275.1998
Ben kahla-Nakbi, A., Chaieb, K., Besbes, A., Zmantar, T., and Bakhrouf, A. (2006). Virulence and enterobacterial repetitive intergenic consensus PCR of Vibrio alginolyticus strains isolated from Tunisian cultured gilthead sea bream and sea bass outbreaks. Vet. Microbiol. 117, 321–327. doi: 10.1016/j.vetmic.2006.06.012
Böddinghaus, B., Rogall, T., Flohr, T., Blöcker, H., and Böttger, E. C. (1990). Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28, 1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990
Bruno, D. W., Griffiths, J., Mitchell, C. G., Wood, B. P., Fletcher, Z. J., Drobniewski, F. A., et al. (1998). Pathology attributed to Mycobacteria chelonae infection among farmed and laboratory-infected Atlantic salmon Salmo salar. Dis. Aquat. Org. 33, 101–109. doi: 10.3354/dao033101
Burreson, E. M., and Ford, S. E. (2004). A review of recent information on the Haplosporidia, with special reference to Haplosporidium nelsoni (MSX disease). Aquat. Living Resour. 17, 499–517. doi: 10.1051/alr:2004056
Carella, F., Aceto, S., Pollaro, F., Miccio, A., Iaria, C., Carrasco, N., et al. (2019). A mycobacterial disease is associated with the silent mass mortality of the pen shell Pinna nobilis along the tyrrhenian coastline of Italy. SREP 9:2725. doi: 10.1038/s41598-018-37217-y
Carella, F., Feist, S. W., Bignell, J. P., and De Vico, G. (2015). Comparative pathology in bivalves: aetiological agents and disease processes. J. Invertebr. Pathol. 131, 107–120. doi: 10.1016/j.jip.2015.07.012
Caruso, G., Angotti, R., Molinaro, F., Benicchi, E., Cerchia, E., and Messina, M. (2013). Cervical lymphadenitis by Mycobacterium triplex in an immunocompetent child: case report and review Indian. J. Microbiol. 53, 241–244. doi: 10.1007/s12088-013-0367-2
Casas, S. M., Grau, A., Reece, K. S., Apakupakul, K., Azevedo, C., and Villalba, A. (2004). Perkinsus mediterraneus n. sp., a protistan parasite of the European flat oyster Ostrea edulis from the Balearic Islands, Mediterranean Sea. Dis. Aquat. Org. 58, 231–244. doi: 10.3354/dao058231
Catanese, G., Grau, A., Valencia, J. M., Garcia-March, J. R., Vázquez-Luis, M., Alvarez, E., et al. (2018). Haplosporidium pinnae sp. nov., a haplosporidan parasite associated with mass mortalities of the fan mussel, Pinna nobilis, in the Western Mediterranean Sea. J. Invert. Pathol. 157, 9–24. doi: 10.1016/j.jip.2018.07.006
Cox, F. E. G. (2001). Concomitant infections, parasites and immune responses. Parasitology 122, S23–S38.
De Vico, G., and Carella, F. (2019). Nocardiosis and mycobacteriosis of bivalves: “Yet-to emerge” zoonoses of public concern? Zoonoses Public Health 66, 559–561. doi: 10.1111/zph.12614
Dias-Campos, C. E., Fontoura Dias, C., Unis, G., de Souza Caldas, P. C., Redner, P., and Distásio, et al. (2016). First detection of mycobacterium triplex in Latin America. Int. J. Mycobact. 5, 89–91. doi: 10.1016/j.ijmyco.2015.09.004
Fenton, A., Knowles, S. C., Petchey, O. L., and Pedersen, A. B. (2014). The reliability of observational approaches for detecting interspecific parasite interactions: comparison with experimental results Int. J. Parasitol. 44, 437–445. doi: 10.1016/j.ijpara.2014.03.001
Fey, S. B., Siepielski, A. M., Nusslé, S., Cervantes-Yoshida, K., Hwan, J. L., Huber, E. R., et al. (2015). Recent shifts in the occurrence, cause, and magnitude of animal mass mortality events. PNAS 112, 1083–1088. doi: 10.1073/pnas.1414894112
Gauthier, D. T., and Rhodes, M. W. (2009). Mycobacteriosis in fishes: a review. Vet. J. 180, 33–47. doi: 10.1016/j.tvjl.2008.05.012
Gcebe, N., Michel, A. L., and Hlokwe, T. M. (2018). Non-tuberculous Mycobacterium species causing mycobacteriosis in farmed aquatic animals of South Africa. BMC Microbiol. 18:32. doi: 10.1186/s12866-018-1177-9
Gómez-León, J., Villamil, L., Lemos, M. L., Novoa, B., and Figueras, A. (2005). Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. App. Environ. Microbiol. 71, 98–104. doi: 10.1128/aem.71.1.98-104.2005
Griffiths, E. C., Pedersen, A. B., Fenton, A., and Petchey, O. (2011). The nature and consequences of coinfection in humans. J. Infect. 63, 200–206. doi: 10.1016/j.jinf.2011.06.005
Grimm, C., Huntsberger, C., Markey, K., Inglis, S., and Smolowitz, R. (2016). Identification of a Mycobacterium sp. as the causative agent of orange nodular lesions in the Atlantic sea scallop Placopecten magellanicus. Dis. Aquat. Org. 118, 247–258. doi: 10.3354/dao02961
Groner, M. L., Maynard, J., Breyta, R., Carnegie, R. B., Dobson, A., Friedman, C. S., et al. (2016). Managing marine disease emergencies in an era of rapid change. Philos. Trans. R. Soc. Lond. B Biol. Sci. 5:20150364. doi: 10.1098/rstb.2015.0364
Gutierrez, M. (1977). Técnicas de coloración del agente de la enfermedad de la glándula digestiva de la ostra, Ostrea edulis L. Invest. Pesqueras 41, 643–645.
Hewson, I., Button, J. B., Gudenkauf, B. M., Miner, B., Newton, A. L., Gaydos, J. K., et al. (2014). Densovirus associated with sea-star wasting disease and mass mortality. Proc. Natl. Acad. Sci. U.S.A. 111, 17278–17283. doi: 10.1073/pnas.1416625111
Honda, J. R., Virdi, R., and Chan, E. D. (2018). Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front. Microbiol. 9:2029. doi: 10.3389/fmicb.2018.02029
Jacobs, J. M., Stine, C. B., Baya, A. M., and Kent, M. L. (2009). A review of Mycobacteriosis in marine fish. J. Fish Dis. 32, 119–130. doi: 10.1111/j.1365-2761.2008.01016.x
Johnson, P. T., and Hoverman, J. T. (2012). Parasite diversity and coinfection determine pathogen infection success and host fitness. Proc. Natl. Acad. Sci. U.S.A. 109, 9006–9011. doi: 10.1073/pnas.1201790109
Katsanevakis, S., Tsirintanis, K., Tsaparis, D., Doukas, D., Sini, M., Athanassopoulou, F., et al. (2019). The cryptogenic parasite Haplosporidium pinnae invades the Aegean Sea and causes the collapse of Pinna nobilis populations. Aquat. Invas. 14, 150–164. doi: 10.3391/ai.2019.14.2.01
Kim, H., Kim, S. H., Shim, T. S., Kim, M. N., Bai, G. H., Park, Y. G., et al. (2005a). Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Syst. Evol. Microbiol. 55, 1649–1656. doi: 10.1099/ijs.0.63553-0
Kim, H., Kim, S. H., Shim, T. S., Kim, M. N., Bai, G. H., Park, Y. G., et al. (2005b). PCR restriction fragment length polymorphism analysis (PRA)-algorithm targeting 644 bp heat shock protein 65 (hsp65) gene for differentiation of Mycobacterium spp. J. Microbiol. Methods 62, 199–209. doi: 10.1016/j.mimet.2005.02.010
Koch, M. L., and Cote, R. A. (1965). Comparison of fluorescence microscopy with Ziehl-Neelsen stain for demonstration of acid-fast bacilli in smear preparations and tissue sections. Am. Rev. Respir. Dis. 91, 283–284. doi: 10.1164/arrd.1965.91.2.283
Kotob, M. H., Menanteau-Ledouble, S., Kumar, G., Abdelzaher, M., and El-Matbouli, M. (2016). The impact of co-infections on fish: a review. Vet. Res. 47:98. doi: 10.1186/s13567-016-0383-4
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lassalle, G., de Montaudouin, X., Soudant, P., and Paillard, C. (2007). Parasite co-infection of two sympatric bivalves, the Manila clam (Ruditapes philippinarum) and the cockle (Cerastoderma edule) along a latitudinal gradient. Aquat. Liv. Res. 20, 33–42. doi: 10.1051/alr:2007013
Lipart, C., and Renault, T. (2002). Herpes-like virus detection in infected Crassostrea gigas spat using DIG-labelled probes. J. Virol. Methods 101, 1–10. doi: 10.1016/s0166-0934(01)00413-x
Mohamed, A. M., Kuyper, D. J., Iwen, P. C., Ali, H. H., Bastola, D. R., and Hinrichs, S. H. (2005). Computational approach involving use of the internal transcribed spacer 1 region for identification of Mycobacterium species. J. Clin. Microbiol. 43, 3811–3817. doi: 10.1128/jcm.43.8.3811-3817.2005
Mun, H. S., Kim, H. J., Oh, E. J., Kim, H., Park, Y. G., Bai, G. H., et al. (2007). Direct application of AvaII PCR restriction fragment length polymorphism analysis (AvaII PRA) targeting 644 bp heat shock protein 65 (hsp65) gene to sputum samples. Microbiol. Immunol. 51, 105–110. doi: 10.1111/j.1348-0421.2007.tb03880.x
Panarese, R., Tedesco, P., Chimienti, G., Latrofa, M. S., Quaglio, F., Passantino, G., et al. (2019). Haplosporidium pinnae associated with mass mortality in endangered Pinna nobilis (Linnaeus 1758) fan mussels. J. Invertebr. Pathol. 164, 32–37. doi: 10.1016/j.jip.2019.04.005
Petney, T. N., and Andrews, R. H. (1998). Multiparasite communities in animals and humans: frequency, structure and pathogenic significance. Int. J. Parasitol. 28:377e.93.
Piersimoni, C., Zitti, P., Mazzarelli, G., Mariottini, A., Nista, D., and Zallocco, D. (2004). Mycobacterium triplex pulmonary disease in immunocompetent host. Em. Inf. Dis. 10, 1859–1862.
Porvaznik, I., Solovic, I., and Mokry, J. (2017). Non-tuberculous mycobacteria: classification, diagnostics, and therapy. Adv. Exp. Med. Biol. 944, 19–25. doi: 10.1007/5584_2016_45
Prado, P., Carrasco, N., Catanese, G., Grau, A., Cabanes, P., Carella, F., et al., (2019). Presence of Vibrio mediterranei associated to major mortality in stabled individuals of Pinna nobilis L. Aquaculture 519:734899. doi: 10.1016/j.aquaculture.2019.734899
Ramilo, A., Carrasco, N., Reece, K., Valencia, J. M., Grau, A., Aceituno, P., et al. (2015). Update of information on perkinsosis in NW Mediterranean coast: identification of Perkinsus spp. (Protista) in new locations and hosts. J. Invertebr. Pathol. 125, 37–41. doi: 10.1016/j.jip.2014.12.008
Ross, A. J. (1970). “Mycobacteriosis among Pacific salmonid fishes,” in A Symposium on Diseases of Fishes and Shellfishes, Special Publication nr. 5. ed. S. F. Snieszko (Washington, DC: American Fisheries Society).
Rubio-Portillo, E., Yarza, P., Peñalver, C., Ramos-Esplá, A. A., and Antón, J. (2014). New insights into Oculina patagonica coral diseases and their associated Vibrio spp. communities. ISME J. 8, 1794–1807. doi: 10.1038/ismej.2014.33
Serrano, W., Tarazona, U. I., Olaechea, R. M., and Friedrich, M. W. (2018). Draft genome sequence of a new Vibrio strain with the potential to produce bacteriocin-like inhibitory substances, isolated from the gut microflora of scallop (Argopecten purpuratus). Genome Announc. 6:e00419-18. doi: 10.1128/genomeA.00419-18
Soudant, P., Chu, F.-L. E., and Volety, A. (2013). Host–parasite interactions: marine bivalve molluscs and protozoan parasites, Perkinsus species. J. Invertebr. Pathol. 114, 196–216. doi: 10.1016/j.jip.2013.06.001
Steingrube, V. A., Gibson, J. L., Brown, B. A., Zhang, Y., Wilson, R. W., Rajagopalan, M., et al. (1995). PCR amplification and restriction endonuclease analysis of a 65-kilodalton heat shock protein gene sequence for taxonomic separation of rapidly growing mycobacteria. J. Clin. Microbiol. 33, 149–153. doi: 10.1128/jcm.33.1.149-153.1995
Tracy, A. M., Weil, E., and Harvell, C. D. (2018). Octocoral co-infection as a balance between host immunity and host environment. Oecologia 186, 743–753. doi: 10.1007/s00442-017-4051-9
Keywords: Pinna nobilis, Mycobacterium sp., mass mortality, co-infection, invertebrate pathology
Citation: Carella F, Antuofermo E, Farina S, Salati F, Mandas D, Prado P, Panarese R, Marino F, Fiocchi E, Pretto T and De Vico G (2020) In the Wake of the Ongoing Mass Mortality Events: Co-occurrence of Mycobacterium, Haplosporidium and Other Pathogens in Pinna nobilis Collected in Italy and Spain (Mediterranean Sea). Front. Mar. Sci. 7:48. doi: 10.3389/fmars.2020.00048
Received: 06 November 2019; Accepted: 27 January 2020;
Published: 13 February 2020.
Edited by:
Rubina Sirri, University of Bologna, ItalyReviewed by:
Youji Wang, Shanghai Ocean University, ChinaPaola Maiolino, University of Naples Federico II, Italy
Alfina Grasso, University of Catania, Italy
Copyright © 2020 Carella, Antuofermo, Farina, Salati, Mandas, Prado, Panarese, Marino, Fiocchi, Pretto and De Vico. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Carella, ZnJhbmNlc2NhLmNhcmVsbGFAdW5pbmEuaXQ=
 Francesca Carella
Francesca Carella Elisabetta Antuofermo2,3
Elisabetta Antuofermo2,3 Patricia Prado
Patricia Prado Fabio Marino
Fabio Marino Eleonora Fiocchi
Eleonora Fiocchi Gionata De Vico
Gionata De Vico