- 1Department of Biology, University of Louisiana at Lafayette, Lafayette, LA, United States
- 2Smithsonian Marine Station, Fort Pierce, FL, United States
Endolithic, red unicells residing in the interior of Lithothamnion rhodoliths, collected offshore the NW Gulf of Mexico in mesophotic rhodolith beds at ∼54–55 m depth and maintained in closed microcosms, were used to establish cultures following their isolation. These endolithic unicells subsequently developed into amorphous blobs of palmelloid cell colonies. Each cell contains unstacked, 2–5 lobed parietal chloroplasts, one prominent central pyrenoid, and have a thin or thick cell wall. Single cells, or cell clusters (in pairs, tetrads, or up to 12) are embedded inside an extracellular matrix whose boundaries remain closely appressed to neighboring cell clusters. Cell division by concavo-convex division resulted in hemispherical cells subsequently expanding in size. Plastid tufA, psbA and 16S rDNA sequence analyses confirmed that the colonies are Rhodosorus marinus Geitler. This is the first report of a unicellular red alga spending part of its life history endolithically inside biogenic rhodoliths.
Introduction
The unicellular marine coccoid red algal genus Rhodosorus was described by Geitler (1930, p. 633, Figure 15) from seawater cultures originating from Las Palmas, Canary Islands. Rhodosorus marinus Geitler was the only species in the genus until Fresnel and Billard (1995) described a second species, Rhodosorus magnei, isolated from the French West Indies (Isle de St Barthélemy). The current distribution of Rhodosorus (Guiry and Guiry, 2019) indicates that these unicellular red algae predominantly inhabit warm and coastal waters worldwide (West and Calumpong, 1990; Fresnel and Billard, 1995; Zuccarello et al., 2008). The two species are distinguished by their color, cell size and number of chloroplast lobes (Fresnel and Billard, 1995). Pickett-Heaps et al. (2001) reported that the four strains of Rhodosorus investigated displayed continuous cytoplasmic rotation within the wall, and Wilson et al. (2002) documented chloroplast rotation and morphological plasticity in R. marinus. Previously placed in the Porphyridiales (e.g., West and Calumpong, 1990), the genus currently belongs in the family Stylonemataceae, order Stylonematales, in the class Stylonematophyceae (Yoon et al., 2006, 2010; Yang et al., 2010, 2016).
Whereas R. marinus and R. magnei typically grow epiphytically on the surface of seaweeds (Fresnel and Billard, 1995), we recently found a member of Rhodosorus growing inside (endolithically) calcified Lithothamnion (Hapalidiaceae, Hapalidiales) rhodoliths collected in mesophotic rhodolith beds in the northwestern Gulf of Mexico. Rhodoliths are free-living marine benthic spheroidal nodules predominantly accreted by crustose coralline red algae (CCA) precipitating CaCO3 within their organic cell walls (Foster, 2001; Krayesky-Self et al., 2016, 2017; Fredericq et al., 2019; Spalding et al., 2019). Two major rhodolith categories can be found in the northwestern Gulf of Mexico (NWGMx), i.e., biogenic and autogenic rhodoliths. Biogenic rhodoliths (Figure 1a) are formed by the non-geniculate CCA themselves, e.g., Lithothamnion sp. In contrast, autogenic rhodoliths are derived from already existing calcium carbonate rubble established by differential erosion processes of the caprock (Gore, 1992), with the rubble becoming secondarily covered by various encrusting and fleshy algae (Felder et al., 2014; Fredericq et al., 2014; Richards et al., 2016; Krayesky-Self et al., 2017; Schmidt et al., 2017). Autogenic rhodoliths are viewed as a specific type of nucleated rhodoliths (sensu Freiwald and Henrich, 1994) in which the core derives from calcium carbonate rubble as opposed to other materials. These two categories of rhodoliths co-inhabit the same rhodolith beds but the internal (endolithic) microbiome of each category may differ with regard to the diversity of taxa (biogenic: Krayesky-Self et al., 2017, and autogenic: Sauvage et al., 2016a; Fredericq et al., 2019).
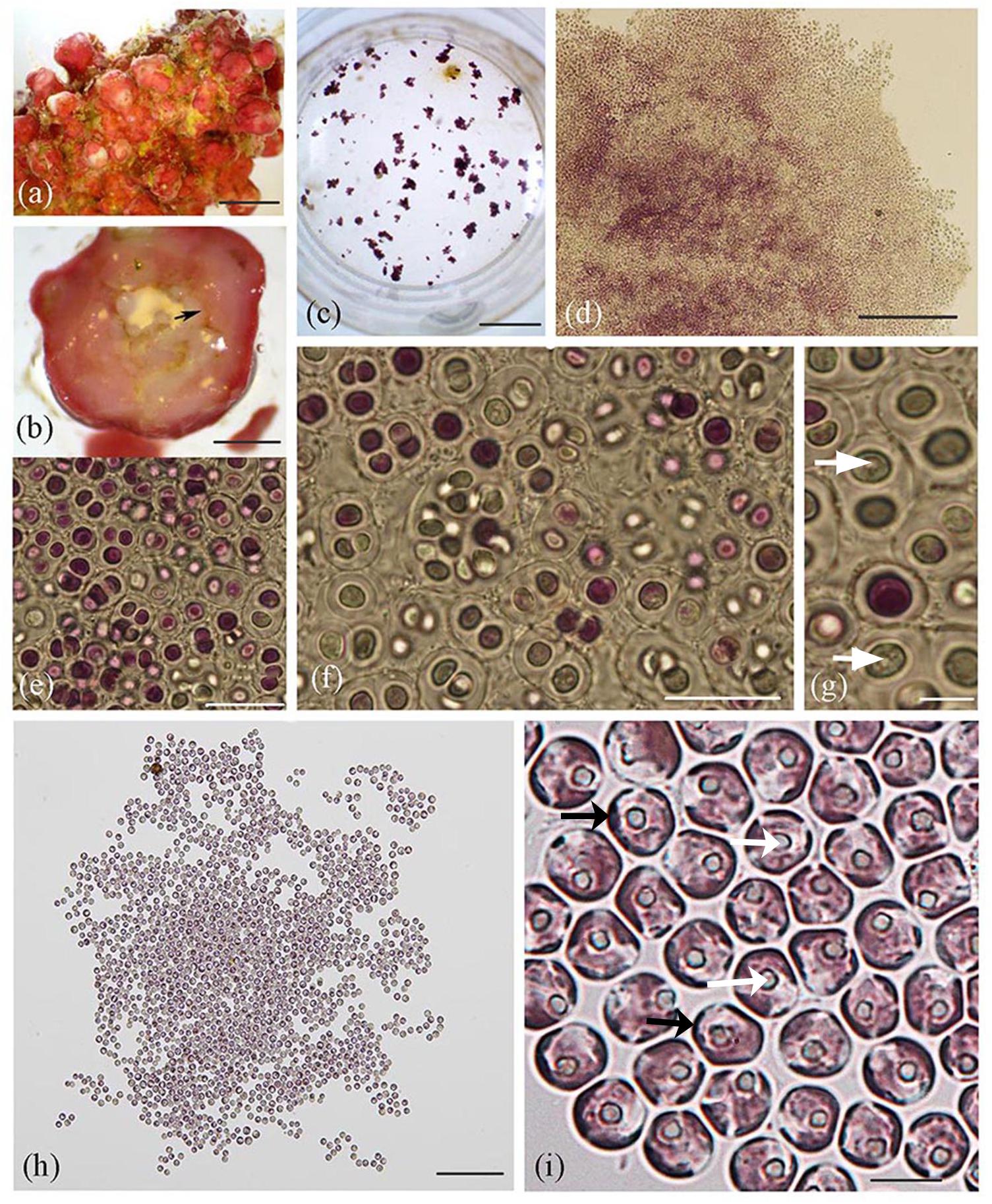
Figure 1. Lithothamnion sp. rhodolith collected from Ewing Bank, northwestern Gulf of Mexico, and light microscopy images of Rhodosorus marinus cells. (a) Surface view of habit of Lithothamnion (LAF6573b); (b) cross section through rhodolith protuberance displaying internal area where endolithic Rhodosorus cells were found (arrow); (c) culture well plate viewed under dissecting scope, with cultured Rhodosorus cells and Ochrosphaera haptophyte cells (brown clusters); (d) magnified view of cell clusters seen in Figure 3c); (e–g) light micrographs of single cells or cell clusters (in pairs, tetrads, or up to 12 cells). Cell division by concavo-convex division resulted in hemispherical cells subsequently expanding in size, each with a prominent central pyrenoid (white arrows); (h) Rhodosorus subculture; (i) close-up of cells shown in (3 h) showing parietal chloroplast lobes (black arrows) and pyrenoid (white arrows). Scale bars: (a) = 0.5 mm, (b) = 2 mm, (c) = 15.5 mm, (d) = 200 μm, (e) = 20 μm, (f) = 20 μm, (g) = 5 μm, (h) = 150 μm, (i) = 6 μm.
This endolithic taxon retrieved from a biogenic rhodolith is herein characterized on the basis of anatomical evidence via light, phase contrast, fluorescent and TEM microscopy, and also on the basis of DNA sequence analyses of plastid tufA (plastid-encoded protein chain elongation factor EF-Tu), rbcL (encodes the large subunit of the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase), psbA (photosystem II reaction center protein D1 gene), and 16S rDNA.
Materials and Methods
Study Area and Sample Collection
Mesophotic rhodolith collections representing Lithothamnion sp., a currently undescribed species of CCA (Figure 1a) (Hapalidiaceae, Hapalidiales, Rhodophyta) (J. Richards, unpublished data), were collected at Ewing Bank (27°57.08N, 92°01.03.′W, coll. S. Fredericq, depth 54–55 m, 26.viii.2012, LAF6573b) offshore Louisiana in the northwestern Gulf of Mexico aboard the R/V Pelican, a UNOLS research vessel operated out of LUMCON, Cocodrie, LA. Rhodoliths were retrieved using a Hourglass-design box dredge (Joyce and Williams, 1969) with minimum tows (usually 8 min or less) (Felder et al., 2014; Fredericq et al., 2014, 2019). Rhodoliths were initially stored on-site by location in containers filled with seawater collected in situ from the same depth and site of the sampled rhodoliths using the onboard CTD water-sampling rozette. Samples were kept aerated on board ship for the duration of the trip (4 days) and immediately transferred into microcosms, filled with in situ collected seawater, located in our laboratory at UL Lafayette 2–5 h upon return to the laboratory. The Lithothamnion rhodolith investigated in the present study is part of the same voucher that included endolithic Ochrosphaera (haptophyte) cellular inclusions housed within the rhodolith’s interior (Krayesky-Self et al., 2017).
Establishment of Microcosm
A 75-L closed microcosm tank, established from a subset of samples from Ewing Bank, was equipped with a SeaClone 100 protein skimmer (Instant Ocean, Blacksburg, VA, United States), water jet (MJ2000) and 600 lumen light (FugeRay Unibody). The protein skimmer provided filtration and a flow of 1,200 L/h (Krayesky-Self et al., 2017; Fredericq et al., 2019). The LED photosynthetically available radiation (PAR) in the microcosm was about 30 μmol photons m–2 s–1, a measurement approximating in situ light PAR or irradiance levels measured with a LI-COR Biosciences (Lincoln, NE, United States) biospherical PAR sensor incorporated in a CTD (for conductivity, temperature, and depth) rosette and water sampler. The closed microcosm was filled with in situ collected water with CTD rosette Niskin bottles and the systems was maintained at approximately 10 h light/14 h dark cycle at 24°C, the same temperature measured in the field at 55 m depth in late summer. Deionized water was used to counteract evaporation within the microcosm. Rhodolith vouchers are deposited in the Algal Herbarium of the University of Louisiana at Lafayette (LAF).
Light Microscopy and Phase Contrast Microscopy
Rhodoliths were cross-sectioned with straight-edged razor blades, and cultured cells were viewed under a Zeiss Stemi 2000-C (Oberkochen, Germany), Olympus SZ61 stereomicroscope (Waltham, MA, United States), and with phase contrast using a Motic BA300 microscope (Carlsbad, CA, United States).
Fluorescence Microscopy
Specimens from the culture wells were pipetted onto microscope slides following the protocol of Krayesky-Self et al. (2017). A Nikon E600FN epifluorescence microscope (Melville, NY, United States) was used to visualize autofluorescing cells with blue light. Photographs were taken with an Olympus digital camera.
Transmission Electron Microscopy
A subsample of the Rhodosorus cells was fixed in Trumps fixative (Electron Microscopy Science) for 1 h at ∼22°C, then fixed in 2% OsO4 for 15 min, dehydrated, embedded in Spurr’s resin, cut into 1 nm sections, stained using uranyl acetate and lead citrate and viewed on a Hitachi 7600 TEM microscope (Dallas, TX, United States) following the procedure in Krayesky-Self et al. (2017).
Establishment of Cell Cultures
With a sterile razor blade, a rhodolith nodule was sectioned into thin pieces and examined under an Olympus BX60 compound microscope with a SLMPlan 50X/0.45 M-plain objective attached to a Canon PowerShot A330 camera (Melville, NY, United States). A Sutter Ultra-micropipette puller produced fine ultra-micropipette tips that were used with a mechanical manipulator to remove single cells from within the coralline cells. The pipettes were controlled using a micro-manipulator and suction was controlled using a one-way valve and a transfer pipette. Following the procedure of Krayesky-Self et al. (2017), cells retrieved from the inside of the rhodoliths were cultured, and subcultured into 24-well culture plates. The isolated cells grew within the well-plates which contained 50% filtered seawater with 50% K-Media nutrients at room temperature, following the procedure of Krayesky-Self et al. (2017). Cells were placed in filtered microwaved-sterilized natural seawater for 5–7 days and this seawater was refreshed with modified K-media every 2 weeks after 7 days. The cultures have been maintained since 2012. Larger cultures were then established in Corning 25 cm2 tissue culture flasks at 19°C and periodically checked for growth.
Single Cell DNA Amplification
Single cells were isolated from the inside of Lithothamnion sp. (LAF6573b) and transferred into culture plates. The DNA of the isolated cells was then amplified by whole genome amplification using a Phi29 REPLI-g single cell kit (Qiagen, Valencia, CA, United States) to produce sufficient DNA for subsequent polymerase chain reaction (PCR) with primers for genes of interest. The manufacturer’s REPLI-g protocol was modified by adding a 5-min incubation period at 95°C to lyse the cells before the denaturing buffer incubation step (65°C for 10 min). All other steps occurred following the manufacturer’s protocol.
DNA Amplification and Sequencing
The genomic DNA from the individual cells was PCR-amplified and sequenced using four different molecular markers. Plastid tufA was amplified and sequenced using the methods and primer combination designed by Sauvage et al. (2016a), plastid rbcL using primers listed in Schmidt et al. (2016), plastid psbA using primers designed by Yoon et al. (2002), and 16S rDNA using primers listed in Olsen et al. (2004). The resulting PCR products of the four genes were gel-purified and cycle sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies, Grand Island, NY, United States). The cycle sequencing reactions were then purified using Ethanol/EDTA precipitation. The resulting dried precipitated DNA was then resuspended in HiDiTM formamide (Life Technologies, Grand Island, NY, United States), heat-denatured and sequenced on the ABI 3130xl Genetic Analyzer at UL Lafayette. Chromatograms were assembled in Sequencher v5.1 (GeneCodes®, Ann Arbor, MI, United States).
Metabarcode Database
A tufA Illumina-metabarcoding (amplicon-based environmental sequencing) framework on environmental samples of various limestone fragments established by Sauvage et al. (2016a, b), provided us with reference tufA metabarcodes of cryptic (hidden) phototrophs retrieved from four small CaCO3 substrata collected from the Ryukyu archipelago, Japan; the NW Gulf of Mexico; and the Florida Keys. The Sanger tufA sequence of the endolithic tufA sequence from Ewing Bank, NW Gulf of Mexico, was approximately 900 bp long and was BLASTed against Sauvage et al.’s (2016b) tufA barcode reference dataset (with each barcode approximately 375 bp long) and against available tufA GenBank sequences. This was done in order to possibly link the taxonomic identity of any of the tufA metabarcodes with that of the tufA Sanger sequence.
Tree Building
The assembled sequences were run through BLASTn on GenBank1 and the nearest hits were downloaded from the public NCBI database and used to establish the data sets. The available resulting tufA dataset of Rhodosorus sequences consisted of a GenBank-downloaded R. marinus sequence from Venice, Italy (AF545599), Core OTU 374 retrieved within endolithic samples (reef rubble) originating from Japan (Ryukyus archipelago)2 (Sauvage et al., 2016a, b) and a newly generated R. marinus (LAF7199 S13) sequence from offshore LA, NW Gulf of Mexico (MN808825), and of a R. magnei sequence from Guadeloupe, F.W. I. (EF660206). The psbA sequence of the endolithic taxon (MN808824) was compared to that of R. marinus from Venice, Italy (AY119744), the Florida Keys (EF660237) and the Maldives (EF660236), and to R. magnei from Guadeloupe F.W.I. (EF660268). The 16S (MN808823) and rbcL sequence of the endolithic taxon (MN808826) was compared to R. marinus from Italy (AF170719, AY119778, respectively).
The sequences of each gene dataset were then aligned manually in Mega v5.2.2 (Tamura et al., 2011). The resulting alignments were analyzed using PartitionFinder2 (Lanfear et al., 2016) to determine the best fitting model of evolution and the optimum data partition. The analyses resulted in the selection of the General Time Reversible model with a proportion of invariable sites and a gamma distribution applied separately to each codon position of the three protein-encoding genes, and as a single partition for the non-protein-coding gene (16S) on the basis of the three information criteria, i.e., Akaike information criterion corrected (AICc), Akaike information criterion (AIC), and Bayesian information criterion (BIC). The alignments were analyzed by Maximum likelihood (ML) as implemented by RAXML (Stamatakis, 2014) with the above model and partition scheme with 1000 restarts to find the tree with the lowest likelihood score and 1000 Bootstrap (BS) replications.
Results
Non-flagellated unicells were captured from the rhodolith’s (Figure 1a) interior (Figure 1b) using a micromanipulator and microscope and used to establish cell cultures (Figure 1c) following their isolation. These cells subsequently developed into free-living amorphous blobs of palmelloid cell colonies (Figures 1c–i) that consistently grew along with brown cell clusters (Figure 1c). Unicells were either purple, pink or greenish and appeared to divide by vegetative cell division to produce two-to-four cells in compact (Figures 1e–g) or loose (Figures 1h,i, 2a) cell clusters (in pairs, groups of three, tetrads, or up to 12 cells) that were embedded inside a cell wall whose boundaries remain closely appressed to neighboring clusters (Figures 1e–g, 2b) or more spatially isolated from one another (Figure 1h). Cell walls were either thick (Figures 1e,g, 2a) or thin (Figure 1i). Vegetative concavo-convex cell division resulted in hemispherical cells (Figure 1f) subsequently expanding in size. Each cell contained one prominent central or off-centric pyrenoid (Figures 1g,i), parietal chloroplasts with 2–5 peripheral plastid lobes barely extending from the cell surface and visible using different focus levels (Figure 1i). Other cellular details were not perceptible with conventional brightfield microscopy.
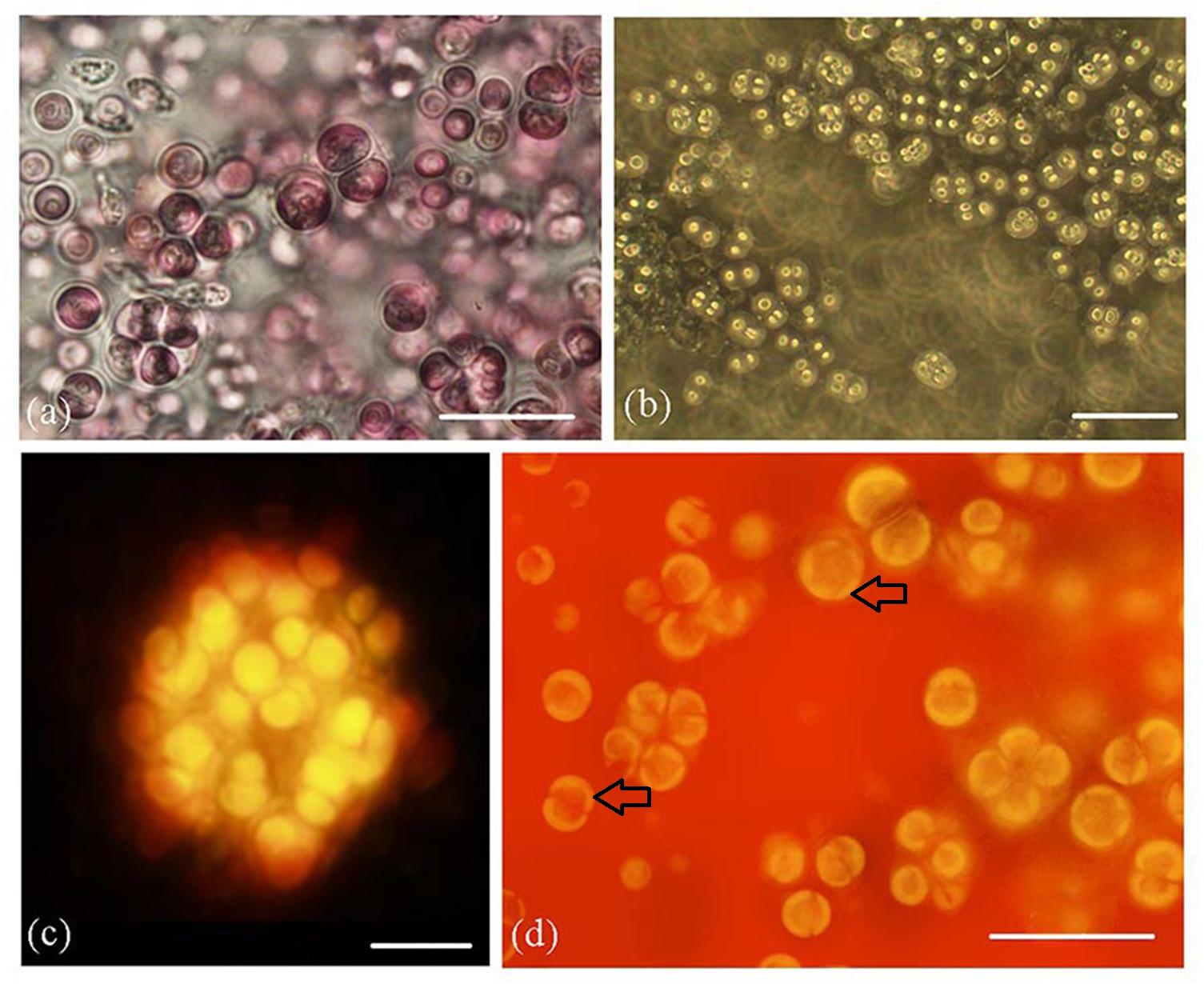
Figure 2. Rhodosorus marinus cultures. (a) light photomicrograph; (b) phase-contrast photomicrograph; (c) auto-fluorescence of cluster of cells shown from above in (2a); (d) autofluorescence of cells, showing 2–5 chloroplast lobes (black arrows). Scale bars: (a) = 20 μm, (b) = 35 μm, (c) = 10 μm, (d) = 20 μm.
Phase contrast (Figure 2a) and autofluorescence (Figure 2b) also showed the palmelloid organization of cultured cells. Autofluorescence micrographs (Figure 2c) clearly documented 2–5 plastid lobes per cell (Figure 2d).
Transmission electron microscopy (TEM) observations confirmed that the Rhodosorus cells contained crescent-shaped to oval plastids and one prominent pyrenoid (Figures 3a–c). The pyrenoid extends toward the interior of the cell and depending on the plane of the section are shown to be connected to a plastid lobe appearing stalked (Figure 3a). One-to-five plastid lobes per cell were seen in various planes of sectioning (Figures 3a–c) and follow the contour of the cells. Each plastid lobe contains parallel, evenly spaced, unstacked thylakoids (Figures 3a–c) and does not abut the cell wall but remains separated from it by an evenly distributed intracellular space immediately below the cell wall. Phycobilisomes were not seen on the thylakoids. Floridean starch was not abundant in young, actively growing cultured cells (Figure 3a) but formed extensive floridean starch sheaths (Figures 3b,c) surrounding the pyrenoid in cells from mature cultures, i.e., in non-actively growing cells that were maintained in culture for a period of 2 months (Figure 3b) and 2 years (Figure 3c), respectively.
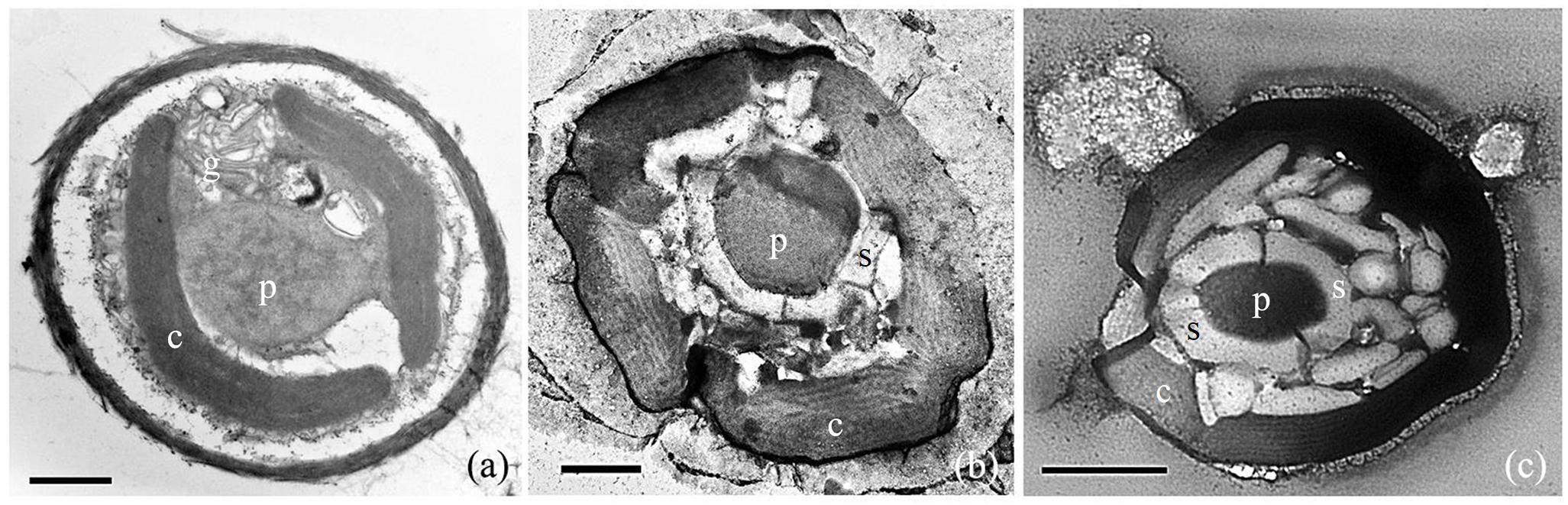
Figure 3. TEM photomicrographs of Rhodosorus marinus cells taken from cultures established from individual cells removed from Lithothamnion sp. rhodolith. (a) Two crescent-shaped chloroplast lobes (c), a stalked pyrenoid (p), and golgi apparatus (g); (b) five chloroplast lobes (c) and a central pyrenoid (p) surrounded by floridean starch (s); (c) copious amounts of floridean starch granules (s) surrounding the pyrenoid (p), and chloroplast lobes (c). Scale bars: (a) = 1 μm, (b) = 0.5 μm, (c) = 1 μm.
The small branch length between the psbA (Figure 4a), tufA (Figure 4b), 16S (Figure 4c) and rbcL (Figure 4d) sequences of the endolithic Rhodosorus taxon from the NW Gulf of Mexico and downloaded GenBank sequences of R. marinus worldwide indicate that they do not represent separate species. tufA metabarcoding enabled us to link the taxonomic identity of a hidden (cryptic) ∼375 tufA metabarcode from reef rubble from Okinawa, Ryukyu archipelago, Japan with the ∼900 bp tufA sequence of the endolithic Louisiana taxon (Figure 4b).
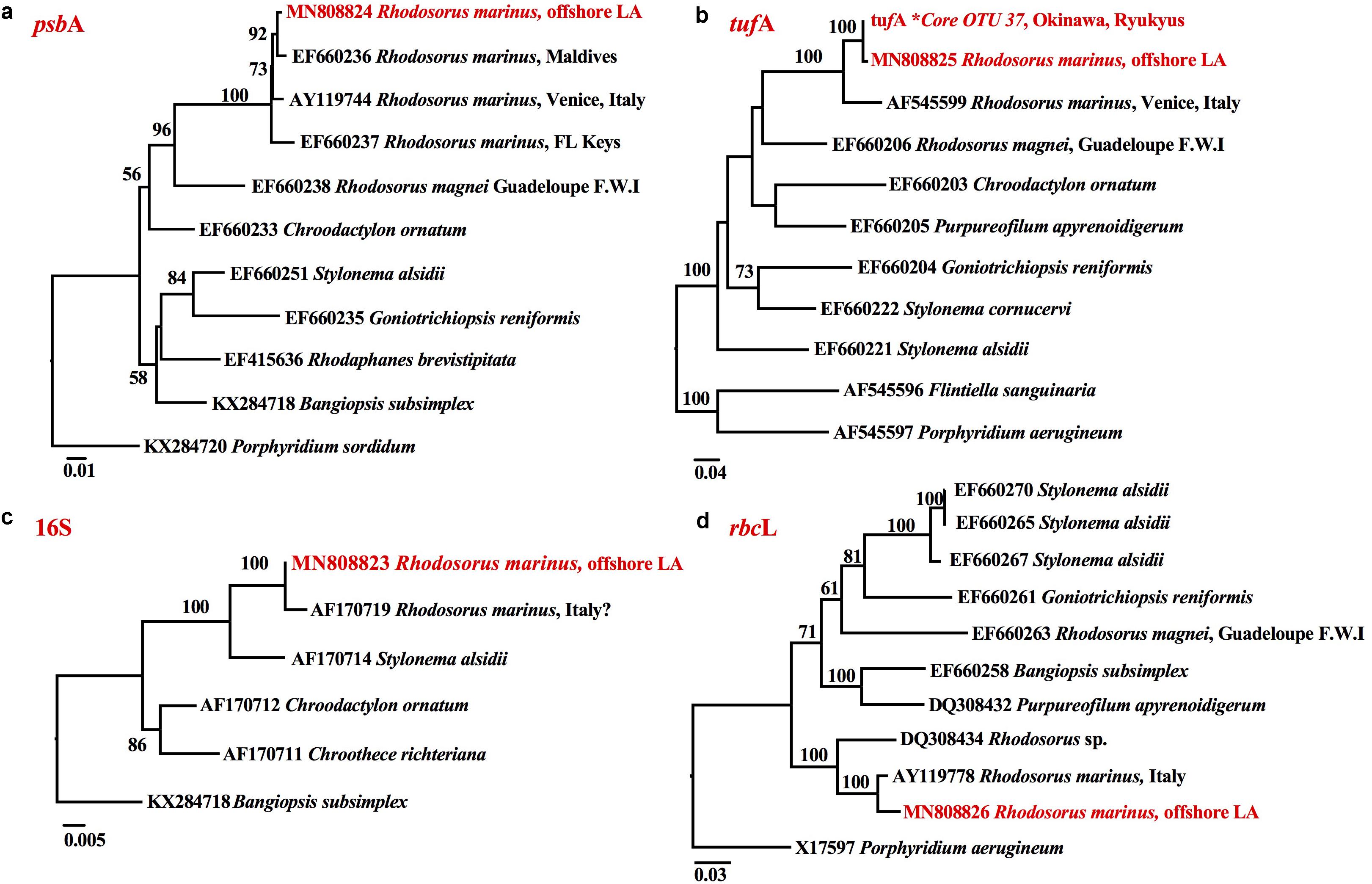
Figure 4. RaxML phylograms of (a) psbA, (b) tufA, (c) 16S, and (d) rbcL sequences showing that the endolithic Rhodosorus species from offshore Louisiana (highlighted in red) corresponds to the R. marinus from (a) the Maldives, Venice, Italy, and the Florida Keys, (b) Venice, Italy, and a Core OTU∗ from Okinawa, Ryukyus, Japan, (c) Italy? (SAG.116.79), and (d) Italy.
Discussion
Morphologically, each of the cultured cells originating from endolithic cells inhabiting NW Gulf of Mexico Lithothamnion rhodoliths conforms to the concept of R. marinus Geitler, a species that has fewer chloroplast lobes (∼2–5) than R. magnei (8–11) (Fresnel and Billard, 1995); however, it should be noted that these characteristics were found to be quite variable and dependent on culture conditions and that they cannot be used to differentiate these two species (Wilson et al., 2002). Our observations coincide with those by Giraud (1976) who noted that the width of the cell wall in R. marinus varies greatly depending on culture conditions. Our observations of the NW Gulf of Mexico cultured cells also resemble aspects of cultures of R. marinus from Hawaii in that they may form loose cell packets enclosed in a poorly defined mucilage (West, 1969). Pyrenoids encased by prominent cytoplasmic starch as shown in R. marinus by Giraud (1976), Lee (1974), and Fresnel and Billard (1995) were not observed in the actively dividing Louisiana vouchers but were prominent in cultures that were maintained for up to 2 years. Perhaps the fact that such prominent starch cells were not found in young cultures may indicate that cells were still actively growing and not storing energy in the form of floridean starch. Molecularly, comparative plastid tufA, psbA and 16S sequence analysis also confirmed that the endolithic Rhodosorus taxon from the NW Gulf of Mexico is R. marinus.
Rhodosorus marinus may be more ubiquitous than the present records indicate. It may be overlooked in collections because of its very small size and because the distribution record of the genus is patchy (West and Calumpong, 1990). Rhodosorus was originally described from the Canary Islands (Geitler, 1930) and subsequently recorded in France (Giraud, 1958), Italy, and the Florida Keys (Ott, 1967), and Hawaii (West, 1969). More recent reports, listed in Guiry and Guiry (2019) include a European distribution for France (Billard and Gayral, 1972; Anon, 2017), Spain (incl. Canary Is., Gallardo et al., 2016), the Canary Islands (John et al., 1979; Gil-Rodríguez and Afonso-Carrillo, 1980; Haroun et al., 2002; Gil-Rodríguez et al., 2003; John et al., 2004; Afonso-Carrillo, 2014), in addition to being distributed in British Columbia (Scagel et al., 1989), the tropical and subtropical western Atlantic (Wynne, 2017), Japan (Yoshida et al., 1990, 2015; Yoshida, 1998), and the Philippines (Ang et al., 2014).
Rhodosorus marinus typically grows epiphytically on macroalgae, mostly siphonous green algae, and co-occurs with chrysophytes and haptophytes (e.g., Ochrosphaera) (West and Calumpong, 1990; Fresnel and Billard, 1995). West (1969) isolated R. marinus and Ochrosphaera verrucosa from Porites coral fragments which he maintained for 7 years in aerated seawater cultures from Coconut Island, Kaneohe Bay, Oahu, Hawaii. Just as O. verrucosa endolithic life history stages were detected by tufA metabarcoding in calcium carbonate substrata from geographically isolated reef habitats in southern Japan, the Florida Keys, and the NWGMx (Sauvage et al., 2016a, b), so too is the O. verrucosa metabarcode a perfect match to the tufA sequence retrieved from cells in cultures co-occurring with Rhodosorus and isolated from Lithothamnion rhodoliths. Besides O. verrucosa, we have previously documented the presence of other microalgal cells growing endolithically within biogenic Lithothamnion rhodoliths from the NW Gulf of Mexico, i.e., the dinoflagellate Prorocentrum lima (Krayesky-Self et al., 2017).
This is the first report of a unicellular species of red algae, i.e., R. marinus, that grows inside biogenic rhodoliths indicating that this species spends part of its life history endolithically inside mesophotic rhodoliths, co-habiting with the haptophyte Ochrosphaera (Coccolithales, Prymnesiophycidae Coccolithophyceae) a taxon occasionally referred to as Hymenomonas globosa (Magne) Gayral and Fresnel, a common coastal haptophyte (Fresnel and Probert, 2005). Both taxa thus appear to have a wide distribution in the CaCO3 endolithic niche worldwide. R. marinus was not observed growing as free-living cells in our laboratory microcosms.
This study adds to our previous discovery that the interior of rhodoliths are marine biodiversity hotspots (Sauvage et al., 2016a; Krayesky-Self et al., 2017; Fredericq et al., 2019) for previously unknown endolithic stages in the life history of ecologically important and diverse microalgae encompassing phyla as diverse as dinoflagellates (i.e., Prorocentrum), haptophytes (i.e., Ochrosphaera), and, as noted herein, red algae (i.e., Rhodosorus) as well. The metabarcoding approach may reveal additional unknown biodiversity that can form the basis for species description through careful culturing efforts and anatomical observations, as was performed here.
Data Availability Statement
This manuscript contains previously unpublished data. The repository of the tufA metabarcode (part of the Core OTU 374) is DRYAD, 10.5061/dryad.6cj8h (Sauvage et al., 2016b). All DNA sequences are available from GenBank, and newly generated DNA sequences represent GenBank numbers MN808824 (psbA), MN808825 (tufA), MN808823 (16S), and MN808826 (rbcL).
Author Contributions
SF, SK-S, TS, and WS conceived the study and collected the samples. SK-S, DP, TS, LB, and WS conducted the laboratory work. WS, TS, and SK-S conducted the data analyses. SF, SK-S, and WS wrote the manuscript with contributions from TS. All authors edited the manuscript before submission.
Funding
This work was funded by NSF DEB-1754504.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We greatly acknowledge the support from NSF research grants DEB1754504 and DEB-0315995 and a Coastal Water Consortium grant of the BP/The Gulf of Mexico Research Initiative (GoMRI-I) following the 2010 Macondo oil spill to SF. We also thank the Louisiana Board of Regents Supervised Undergraduate Research Experience (SURE) program [LEQSF-EPS(2015)-SURE-168] funded by NSF (EPS-1003897), the LAMP and McNair Scholars Programs and a UL Lafayette Undergraduate Research Grant to DP, and Faculty Improvement Grant to SK-S. We thank Tom Pesacreta and Mike Purpura at the UL Lafayette Microscopy Center for advice with the TEM and Fluorescence Microscopy protocols.
Footnotes
References
Afonso-Carrillo, J. (2014). Lista actualizada de las algas marinas de las islas Canarias. Las Palmas: Elaborada para la Sociedad Española de Ficología (SEF) 2014, 1–64.
Ang, P. O. Jr., Leung, S. M., and Choi, M. M. (2014). A verification of reports of marine algal species from the Philippines. Philippine J. Sci. 142, 5–49.
Anon (2017). Inventaire National du Patrimoine Naturel. Website. Paris: Muséum National d’Histoire Naturelle.
Billard, C., and Gayral, P. (1972). Two new species of Isochrysis with remarks on the genus Ruttnera. Br. Phycol. J. 7, 289–297.
Felder, D. L., Thoma, B. P., Schmidt, W. E., Sauvage, T., Self-Krayesky, S., Chistoserdov, A., et al. (2014). Seaweeds and decapod crustaceans on Gulf deep banks after the Macondo Oil Spill. Bioscience 64, 808–819. doi: 10.1093/biosci/biu119
Foster, M. S. (2001). Rhodoliths: between rocks and soft places – Minireview. J. Phycol. 37, 659–667. doi: 10.1046/j.1529-8817.2001.00195.x
Fredericq, S., Arakaki, B., Camacho, O., Gabriel, D., Krayesky, D., Self-Krayesky, S., et al. (2014). A dynamic approach to the study of rhodoliths: a case study for the Northwestern Gulf of Mexico. Cryptog. Algol. 35, 77–98. doi: 10.7872/crya.v35.iss1.2014.77
Fredericq, S., Krayesky-Self, S., Sauvage, S., Richards, J., Kittle, R., Arakaki, N., et al. (2019). The critical importance of rhodoliths in the life cycle completion of both macro- and microalgae, and as holobionts for the establishment and maintenance of marine biodiversity. Front. Mar. Sci. 5:502. doi: 10.3389/fmars.2018.00502
Freiwald, A., and Henrich, R. (1994). Reefal coralline algal build-ups within the arctic circle: morphology and sedimentary dynamics under extreme environmental seasonality. Sediment 41, 963–984. doi: 10.1111/j.1365-3091.1994.tb01435.x
Fresnel, J., and Billard, C. (1995). A new marine unicellular red alga Rhodosorus magnei sp. nov. (Rhodophyceae. Porphyridiales). Cryptog. Algol. 16, 63–75.
Fresnel, J., and Probert, I. (2005). The ultrastructure and life cycle of the coastal coccolithophorid Ochrosphaera neapolitana (Prymnesiophyceae). Eur. J. Phycol. 40, 105–122. doi: 10.1080/09670260400024659
Gallardo, T., Bárbara, I., Afonso-Carrillo, J., Bermejo, R., Altamirano, M., Gómez Garreta, A., et al. (2016). Nueva lista crítica de las algas bentónicas marinas de España. a new checklist of benthic marine algae of Spain. Algas. Boletín Informativo de la Sociedad Española de Ficología 51, 7–52.
Geitler, L. (1930). Ein grünes filarplasmodium und andere neue Protisten. Archiv für Protistenkunde 69, 614–636.
Gil-Rodríguez, M. C., Haroun, R., Ojeda Rodríguez, A., Berecibar Zugasti, E., Domínguez Santana, P., and Herrera Morán, B. (2003). “Proctoctista,” in Lista de Especies Marinas de Canarias (algas, hongos, plantas y animales), eds L. Moro, J. L. Martín, M. J. Garrido, and I. Izquierdo, (Las Palmas: Consejería de Política Territorial y Medio Ambiente del Gobierno de Canarias), 5–30.
Gil-Rodríguez, M. C., and Afonso-Carrillo, J. (1980). Adiciones al la flora y catálogo ficológico para la isla de Lanzarote. Vieraea 10, 59–70.
Giraud, G. (1958). Sur la vitesse de croissance d’une Rhodophycée monocellulaire marine, le Rhodosorus marinus cultivé en milieu synthétique. Compt. Rend. Hebd. Séances Acad. Sci. T. 247, 3501–3504.
Giraud, G. (1976). Some aspects of cell-wall secretion in red algae. Bull. Soc. Bot. France 123, 73–80.
Guiry, M. D., and Guiry, G. M. (2019). AlgaeBase. World-wide electronic publication, National University of Ireland, Galway 2019. Available from: http://www.algaebase.org. (accessed January 19, 2019).
Haroun, R. J., Gil-Rodríguez, M. C., Díaz de Castro, J., and Prud’homme van Reine, W. F. (2002). A checklist of the marine plants from the Canary Islands (central eastern Atlantic Ocean). Bot. Mar. 45, 139–169.
John, D. M., Price, J. H., Maggs, C. A., and Lawson, G. W. (1979). Seaweeds of the western coast of tropical Africa and adjacent islands: a critical assessment. III. Rhodophyta (Bangiophyceae). Bull. British Mus. (Nat. Hist.) Bot. 7, 69–82.
John, D. M., Prud’homme van Reine, W. F., Lawson, G. W., Kostermans, T. B., and Price, J. H. (2004). A taxonomic and geographical catalogue of the seaweeds of the western coast of Africa and adjacent islands. Beih. Nova Hedwigia 127:139.
Joyce, A. E., and Williams, J. (1969). Rationale and pertinent data. Mem. Hourglass Cruises 1, 11–50.
Krayesky-Self, S., Richards, J. L., Rahmatian, M., and Fredericq, S. (2016). Aragonite infill in overgrown conceptacles of coralline Lithothamnion spp. (Hapalidiaceae, Hapalidiales, Rhodophyta): new insights in biomineralization and phylomineralogy. J. Phycol. 52, 161–173. doi: 10.1111/jpy.12392
Krayesky-Self, S., Schmidt, W. E., Phung, D., Henry, C., Sauvage, T., Camacho, O., et al. (2017). Eukaryotic life inhabits rhodolith-forming coralline algae (Hapalidiales, Rhodophyta), remarkable marine benthic microhabitats. Sci. Rep. 7:45850. doi: 10.1038/srep45850
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., and Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Lee, R. E. (1974). Chloroplast structure and starch grain production as phylogenetic indicators in the lower Rhodophyceae. Br. Phycol. J. 9, 291–295. doi: 10.1080/00071617400650351
Olsen, K. N., Melton, R. S., Yaudes, K. M., Norwood, K. G., and Freshwater, D. W. (2004). Characteristics and utility of plastid-encoded 16S rRNA gene sequence data in phylogenetic studies of red algae. J. N. C. Acad. Sci. 120, 143–151.
Ott, F. D. (1967). Rhodosorus marinus geitler: a new addition to the marine algal flora of the western hemisphere. J. Phycol. 3, 158–159. doi: 10.1111/j.1529-8817.1967.tb04651.x
Pickett-Heaps, J. D., West, J. A., Wilson, S. M., and McBride, D. L. (2001). Time-lapse videomicroscopy of cell (spore) movement in red algae. Eur. J. Phycol. 36, 9–22. doi: 10.1080/09670260110001735148
Richards, J. L., Vieira-Pinto, T., Schmidt, W. E., Sauvage, T., Gabrielson, P. W., Oliveira, M. C., et al. (2016). Molecular and morphological diversity of Lithothamnion spp. rhodoliths (Hapalidiaceae, Hapalidiales) from deepwater rhodolith beds in the northwestern Gulf of Mexico. Phytotaxa 278, 81–114.
Sauvage, T., Schmidt, W. E., Suda, S., and Fredericq, S. (2016a). A metabarcoding framework for facilitated survey of coral reef and rhodolith endolithic communities with tufA. BMC Ecol. 16:8. doi: 10.1186/s12898-016-0068-x
Sauvage, T., Schmidt, W. E., Suda, S., and Fredericq, S. (2016b). Data from: a metabarcoding framework for facilitated survey of endolithic phototrophs with tufA. Dryad Digit. Repos. 16:8. doi: 10.5061/dryad.6cj8h
Scagel, R. F., Gabrielson, P. W., Garbary, D. J., Golden, L., Hawkes, M. W., Lindstrom, S. C., et al. (1989). A synposis of the benthic marine algae of British Columbia, southeast Alaska, Washington and Oregon. Phycol. Contr., Univ. British Columbia 3:532.
Schmidt, W. E., Gurgel, C. F. D., and Fredericq, S. (2016). Taxonomic transfer of the red algal genus Gloiosaccion to Chrysymenia (Rhodymeniaceae, Rhodymeniales), including the description of a new species, Chrysymenia pseudoventricosa, for the Gulf of Mexico. Phytotaxa 243, 54–70.
Schmidt, W. E., Lozada-Troche, C., Ballantine, D. L., Arakaki, N., Norris, J. N., Gabriel, D., et al. (2017). Taxonomic transfer of Chrysymenia enteromorpha and C. wrightii to Botryocladia (Rhodymeniaceae. Rhodymeniales, Rhodophyta). Phytotaxa 324, 122–138.
Spalding, H. L., Amado-Filho, G. M., Bahia, R. G., Ballantine, D. L., Fredericq, S., Leichter, J. J., et al. (2019). “Macroalgae,” in Mesophotic Coral Ecosystems, Coral Reefs of the World 12, eds Y. Loya, A. K. Puglise, and T. C. L. Bridge, (Switzerland: Springer), 507–536.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
West, J. A. (1969). Observations on four rare marine microalgae from Hawaii. Phycologia 8, 187–192. doi: 10.2216/i0031-8884-8-3-187.1
West, J. A., and Calumpong, H. P. (1990). New records of marine algae from the Philippines. Micronesica 23, 181–190.
Wilson, S., West, J., Pickett-Heaps, J., Yokoyama, A., and Yoshisaki, H. (2002). Chloroplast rotation and morphological plasticity of the unicellular alga Rhodosorus (Rhodophyta. Stylonematales). Phycol. Res. 50, 183–191. doi: 10.1111/j.1440-1835.2002.tb00150.x
Wynne, M. J. (2017). A checklist of benthic marine algae of the tropical and subtropical Western Atlantic: fourth revision. Nova Hedwigia, Beih. 145:202.
Yang, C. Y., Boo, S. M., Bhattacharya, D., Saunders, G. W., Knoll, A. H., Fredericq, S., et al. (2016). Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci. Rep. 6:21361. doi: 10.1038/srep21361
Yang, E. C., Scott, J., West, J. A., Yoon, H. S., Yokoyama, A., Karsten, A., et al. (2010). New taxa of the Porphyridiophyceae (Rhodophyta): Timspurckia oligopyrenoides gen. et sp. nov. and Erythrolobus madagascariensis sp. nov. Phycologia 49, 604–616. doi: 10.2216/09-105.1
Yoon, H. S., Hackett, J. D., Pinto, G., and Bhattacharya, D. (2002). The single, ancient origin of chromist plastids. PNAS 99, 15507–15512. doi: 10.1073/pnas.242379899
Yoon, H. S., Müller, K. M., Sheath, R. G., Ott, F. D., and Bhattacharya, D. (2006). Defining the major lineages of red algae (Rhodophyta). J. Phycol. 42, 482–492. doi: 10.1111/j.1529-8817.2006.00210.x
Yoon, H. S., Zuccarello, G. C., and Bhattacharya, D. (2010). “Evolutionary history and taxonomy of red algae,” in Red Algae in the Genomic Age, eds J. Seckbach and D. J. Chapman, (Berlin: Springer), 27–42.
Yoshida, T., Nakajima, Y., and Nakata, Y. (1990). Check-list of marine algae of Japan (revised in 1990). Jap. J. Phycol. 38, 269–320.54.
Yoshida, T., Suzuki, M., and Yoshinaga, K. (2015). Checklist of marine algae of Japan (Revised in 2015). Jap. J. Phycol. 63, 129–189.
Keywords: CCA, coralline algae, Gulf of Mexico, mesophotic, metabarcoding, rhodoliths, Rhodophyta, Rhodosorus
Citation: Krayesky-Self S, Phung D, Schmidt W, Sauvage T, Butler L and Fredericq S (2020) First Report of Endolithic Members of Rhodosorus marinus (Stylonematales, Rhodophyta) Growing Inside Rhodoliths Offshore Louisiana, Northwestern Gulf of Mexico. Front. Mar. Sci. 7:7. doi: 10.3389/fmars.2020.00007
Received: 11 July 2019; Accepted: 08 January 2020;
Published: 23 January 2020.
Edited by:
Joshua D. Voss, Florida Atlantic University, United StatesReviewed by:
Conxi Rodríguez-Prieto, University of Girona, SpainDennis Hanisak, Florida Atlantic University, United States
Copyright © 2020 Krayesky-Self, Phung, Schmidt, Sauvage, Butler and Fredericq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzanne Fredericq, c2xmOTIwOUBsb3Vpc2lhbmEuZWR1
 Sherry Krayesky-Self
Sherry Krayesky-Self Delena Phung1
Delena Phung1 William Schmidt
William Schmidt Thomas Sauvage
Thomas Sauvage Luke Butler
Luke Butler Suzanne Fredericq
Suzanne Fredericq