- 1Institute of Marine Biology, Biotechnology & Aquaculture, Hellenic Centre for Marine Research, Heraklion, Greece
- 2Life Sciences Department, Natural History Museum, London, United Kingdom
- 3Institut National de la Recherche Agronomique (INRA), Génétique et Amélioration des Fruits et Légumes (GALF), Montfavet Cedex, France
- 4Norwegian Sequencing Centre, Centre for Ecological and Evolutionary Synthesis, University of Oslo, Oslo, Norway
The emblematic sponge Spongia officinalis is currently threatened by recurrent mortality incidents in its native habitats. Elevated temperature has been indicated as a major triggering factor, but the molecular mechanisms recruited for the organism’s response to thermal shifts are yet unknown. Here, we experimentally tested the effect of exposure to temperatures of varying intensity and span on its gene expression profile, replicating gradients encountered in the species’ native habitat. Analysis revealed major shifts in the organism’s transcriptomic profile induced by temperatures corresponding to the standard seasonal maximum, triggering processes related to signal transduction, inflammation, and apoptotic pathway. Further elevation of temperature corresponding to local extremes activated further the immune response of the sponge along with protein ubiquitination. Following prolonged exposure, activation of endoplasmic reticulum stress related to accumulation of misfolded proteins and signs of resilience were observed. In the latter condition, categories such as cellular response to stress, wound repair, and diminution of pathological inflammation as also genes related to cell regeneration and cell growth were upregulated. Our results highlight the acknowledged sensitivity of S. officinalis to environmental shifts, providing an insight into the molecular mechanisms involved in the process. Furthermore, they suggest innate capacity for resilience at the current thermal extremes, implying a combination of factors and not temperature per se as the lethal agent. This sheds light on the mechanisms of pressure induced by the ongoing ocean warming trend to coastal sessile invertebrates.
Introduction
Global warming, one of the most challenging drivers of change with massive impacts on biodiversity and ecosystem functioning, is expected to influence dramatically coastal sedentary organisms. Temperatures in the oceans are predicted to rise by up to 4°C by 2100 (IPCC, 2014), causing disease outbreaks and mass mortality events on benthic communities (Harvell et al., 2002; Sutherland and Porter, 2004; Webster, 2007). The Mediterranean Sea, in particular, is warming up much faster than the oceans and is hence acknowledged as a climate change hotspot (Giorgi, 2006). Much effort has been spent to understand the effect of increasing seawater temperatures on the biological processes of marine invertebrates and how organisms react to this stress (Dove et al., 2005; Duckworth and Peterson, 2013; Runzel, 2016; Lathlean et al., 2017). To shed light on the latter, the focus has been put on the molecular aspects of such a response, and more particularly on the various molecular mechanisms triggered in animal tissues during stressful conditions. To date, various sessile marine invertebrates in different developmental stages have been experimentally exposed to a diverse range of temperature regimes and exposure times: molluscs (Place et al., 2012; Li et al., 2017), cnidarians (Richier et al., 2006; Desalvo et al., 2008; Kenkel et al., 2013; Mayfield et al., 2014; Seneca and Palumbi, 2015; Ryu et al., 2016), and sponges (Muller et al., 1995; Bachinski et al., 1997; Krasko et al., 1997; López-Legentil et al., 2008; Pantile and Webster, 2011; Fan et al., 2013; Webster et al., 2013; Guzman and Conaco, 2016), manifesting the sensitivity of some species to thermal stress, as well as the molecular resilience and potential adaptation of others.
Sponges are key species in their ecosystems, performing essential functions in terms of microhabitat formation –e.g., via provision of shelter to other animals–, nutrient recycling, enhancement of primary production, and clearing of the water column, among others (Bell, 2008). Like other sessile marine invertebrates, they are highly dependent on the environmental traits of their habitat, lacking the ability to escape abiotic stressors during their adult life stages. Changes in seawater temperature can affect vital biological processes, including reproduction (Asa et al., 2000; Ettinger-Epstein et al., 2007), filtration rate (Massaro et al., 2012), and dispersal capacity. Thus, those changes can directly impact their physiological fitness and survival. Prolonged exposure to increased temperature can provoke alterations to the relationship between a sponge and its symbionts, thus inducing expansion of opportunistic bacteria and virulence of pathogens (Webster et al., 2008; Cebrian et al., 2011). Several studies have correlated increased pathogenesis and mortality events in sponges with environmental temperature anomalies; for instance, mortalities of species of the genera Spongia and Hippospongia in the Caribbean (Vicente, 1989), and mass mortality events in Spongia, Hippospongia, and Cacospongia spp. in the Mediterranean (Gaino et al., 1992; Cerrano et al., 2000). Despite a strong correlation between ocean warming and sponge mortality events manifested in the studies above, there are also contrasting experimental studies showing evidence that thermal fluctuations do not affect fitness. For instance, Kelmo et al. (2013) have found that sponges, compared to other benthic communities, showed resilience to large-scale thermal events, maintaining their population density and biodiversity stable in the tested habitats. The deep-sea sponge Geodia barretti has shown thermal resistance, maintaining stable balance with its symbionts even after a 5°C rise in the surrounding medium (Strand et al., 2017). Similarly, a transcriptomic study of the sponge Haliclona tubifera (Guzman and Conaco, 2016) showed thermal tolerance and adaptation potential when subjected to sublethal thermal increases.
The sponge Spongia officinalis, the common bath sponge and archetype of the poriferan phylum, has attracted the interest of Mediterranean civilizations since ancient times due to its remarkable properties as a cleansing tool, but also due to its therapeutic value (Voultsiadou, 2007). It is native in the temperate Mediterranean Sea (Carballo et al., 1994), with denser populations in its eastern basin where they form sponge grounds (Voultsiadou, 2005, 2009). This species occupies the shallow sublittoral shelf and therefore is subjected to broad annual seawater temperature variations. In the last few decades, apart from the intensive and unregulated harvesting, especially in Greece (Voultsiadou et al., 2013), outbreaks of sponge disease have also affected its abundance locally (Gaino et al., 1992; Webster, 2007; Pronzato and Manconi, 2008) and consequently it has recently been proposed as an endangered species (Gerovasileiou et al., 2018). Importantly, some of the above mass mortality events have been positively linked to elevated temperatures (Vicente, 1989; Cerrano et al., 2000; Perez et al., 2000). Despite the severity of these incidences, no study has yet investigated the molecular response of S. officinalis to adverse environmental conditions.
Given the imperiled state of S. officinalis, as well as the important ecological and socioeconomic value of this species, the goal of this study was to provide the first evidence on the molecular mechanisms activated in response to thermal stress. A transcriptomic analysis was subsequently performed, to identify the effects of the induced thermal stress to the functional molecular traits of the sponge. This study provides a detailed snapshot of the molecular toolkit deployed by a sessile marine invertebrate in response to a prominent environmental stressor that already actively affects its ecology and is anticipated to gain additional importance in the face of an escalating ocean warming.
Materials and Methods
Experimental Design
Individuals of S. officinalis originating from a single wild population (35.57 N, 23.77 E) previously shown to exhibit high genetic diversity (Dailianis et al., 2011) were used in the experiments. They were translocated and reared at the Underwater Biotechnological Park of Crete (UBPC) (35.35 N, 25.28 E), a monitored open-sea underwater facility operated by the Hellenic Centre for Marine Research. Four individuals (biological replicates s1, s2, s3, s4) were cut into five clonal fragments each (genotypically identical clones A, B, C, D, E) and attached to cultivation structures at 20 m depth, where they stayed for regeneration and acclimation for 4 months prior to transportation to indoor, condition-controlled tanks (Figures 1A–C). The tanks were filled with natural seawater directly transported from the site of the open-sea cultivation facility, with no adjacent contaminant or pollution sources.
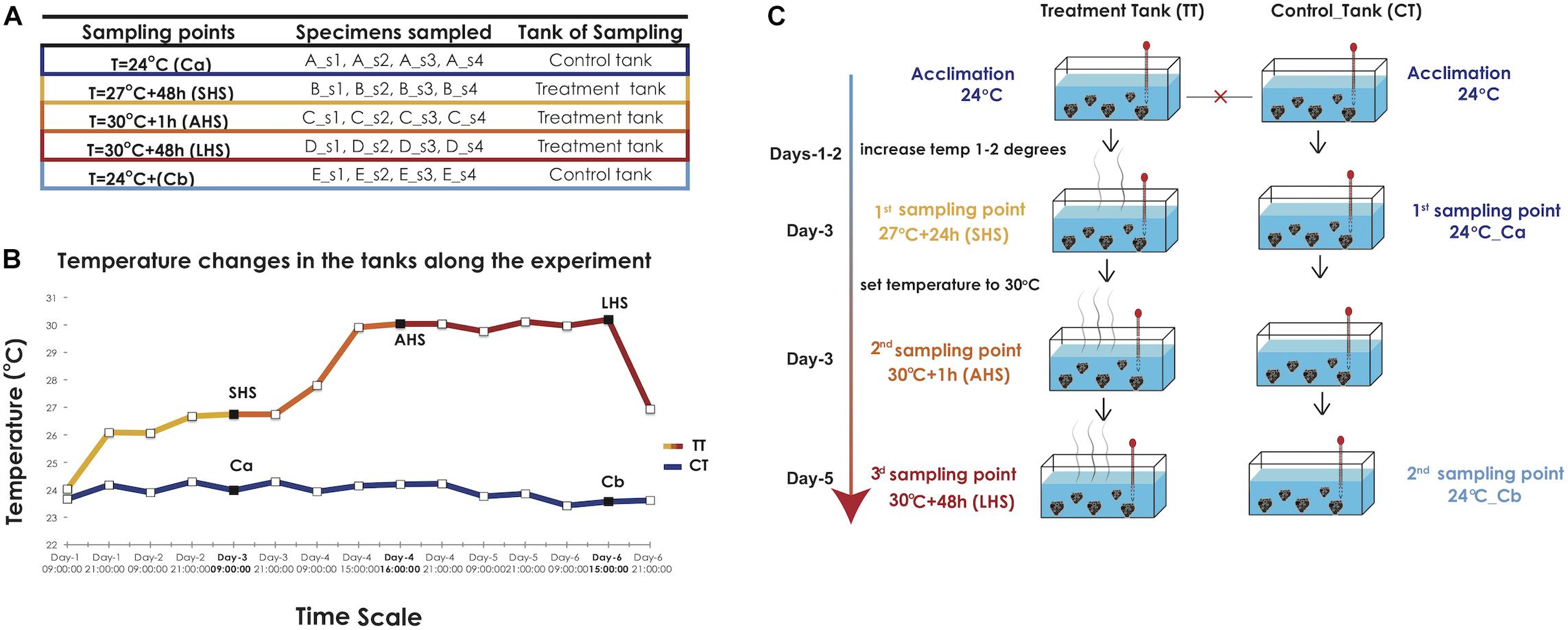
Figure 1. Experimental design. (A) The number and labeling of biological replicates and clones collected in each sampling point from the treatment and the control tank. (B) Graph indicating the temperature conditions in the TT and CT along the time of the experiment, as recorded by HOBO loggers. The three sampling points in the TT curve and the two sampling points in the CT, respectively, are indicated with black squares. SHS, Short-term Heat Shock; AHS, Acute Heat Shock; LHS, Long-term Heat Stress, CT, Control Tank; TT, Treatment tank. (C) The diagram depicts the different stages of temperature increases at the treatment tank (TT) and the sampling points at the control and treatment tank (CT, TT) along the duration of the experiment.
In our experimental approach, we attempted to replicate conditions prevailing in the species’ natural habitat. In the wild, the sponge is subjected to broad annual seawater temperature variations commonly ranging from 15 to 27°C with some sporadic heatwave events (29–30°C) that are encountered in Mediterranean coastal areas during periods of wind calmness which can last from several hours to a few days. Our experimental setting is summarized in Figures 1A–C: two 50 m3 experimental tanks, a treatment tank (TT) and a control tank (CT) were filled with natural seawater and were interconnected via a central open recirculation system. The temperature in the tanks was adjusted with thermostats (Eheim) and recorded at 30-min intervals with HOBO U23 Pro v2 data loggers (Onset) (Figure 1B). Salinity during the experiment was kept at the local value of 39 ppt, while day and night cycles were adjusted to reflect the local photoperiod at the time of the experiment (i.e., 12-h daytime).
The starting temperature in both tanks was 24°C, identical to the Sea Surface Temperature (SST) in situ at the time of sampling (September) and corresponding to actual local average summer SST. After a 5-day acclimation period, tanks were isolated from each other and therefore considered as separate systems from now on (Figure 1C). The temperature in the CT was maintained at 24°C throughout the experiment. In the TT, we increased the water temperature to 27°C over the course of 3 days, provoking an initial Short-term Heat Shock (SHS). The first tissue sampling was conducted after 48 h in the TT (27°C + 48 h) and in CT (Ca) After the first sampling point, the water temperature in the TT was set to increase rapidly (8 h) to 30°C in order to induce an Acute Heat Shock (AHS). The second sampling was done only in the TT 1 h after achieving 30°C (30°C + 1 h). This temperature was maintained constant for the rest of the experiment in the TT. The third and final sampling took place 48 h later in both TT (30°C + 48 h; Long-term Heat Stress, LHS) and CT (Cb), which was still at 24°C (Figures 1B,C). For each experimental condition (5 in total), we sampled tissue from 4 distinct clones corresponding to the original individuals (four biological replicates) (Figure 1A). The used clones were consequently removed from the TT and CT tanks. Tissue samples were fragments of ca. 2 cm3, flash frozen in liquid nitrogen prior to RNA extraction and consequent transcriptomic analyses. The live sponge explants used in the experimental setup were kept for a further 7 days in the closed cultivation system at 24°C, prior to being transferred to the open-sea cultivation, where they were monitored for survival and growth.
RNA Extraction and Sequencing
Total RNA was extracted from tissue samples with TRIZOL (TRIzolTM Reagent, Ambion) according to the protocol provided by the manufacturer. RNA quantity and quality were assessed using NanoDropTM (Thermo Scientific NanoDrop 2000) and electrophoresis through a 2% agarose gel, respectively. Quality of the extracted RNA (RIN number) was further assessed using a HS DNA assay in a Bioanalyzer 2100 (Agilent 2100 Bioanalyzer) at the Norwegian Sequencing Centre (NSC) where the library preparation and sequencing took place. Twenty cDNA libraries were prepared using the TruSeq Stranded Total RNA Library Prep Kit with Ribo-ZeroTM (Illumina), which is based on a polyA selection method (keeping the eukaryotic mRNA and excluding the majority of prokaryotic mRNA) and further sequenced with an Illumina HiSeq4000 using a paired-end (150 bp length) sequencing strategy.
Bioinformatic Analysis
Read Pre-processing
We assessed the quality of the sequencing reads with FastQC1. The fastq raw data were subjected to a thorough filtering with a pipeline including an array of software packages (Ilias et al., 2015). Briefly, we used Scythe2 for adapter trimming, Sickle3 for low quality read trimming, Trimmomatic (Bolger et al., 2014) to further remove 5′ and 3′ adaptor sequences and apply extra filtering steps and finally Prinseq4 lite to filter out low complexity sequences (threshold entropy value of 30) and perform poly A/T 5′ tail trimming (minimum of five A/T).
Reads of bacterial origin were removed from our data using two different approaches. First, we downloaded all bacterial REFSEQ sequences from NCBI and mapped the filtered reads against them using riboPicker standalone-0.4.3 version (Schmieder et al., 2012). The unmapped reads were kept and used for the transcriptome assembly. Secondly, following transcriptome assembly, all transcripts were used in a similarity search through blastx using NOBLAST (Lagnel et al., 2009) against the Swiss-Prot database (e-value: 1.0E-5). The transcripts that had as best hit sequences of prokaryotic origin were eliminated. Details of the pipeline are given in Manousaki et al. (in press).
Assembly
The filtered paired pre-processed reads (excluding singletons) were assembled de novo using the Trinity v2.1.1 package (Haas, 2013) with default parameters (kmer = 25, and minimum contig length to 200 bp). To assess the quality of the transcriptome we performed several steps. First, we checked the percentage of paired-end reads that were properly aligned to the assembly with Bowtie2 (Langmead and Salzberg, 2012) Finally, we assessed the transcriptome completeness with Benchmarking Universal Single-Copy Orthologs (Busco v2) (Simão et al., 2015) using the program gVolante5 selecting as reference gene set the Metazoa.
Annotation
The assembled transcripts were annotated through a blastx search using BLAST + 2.2.23 + (Camacho et al., 2009) against the Swiss-Prot database (e-value: 1.0E-5; Blast hits:20). The taxonomic distribution of blast hits and bacterial/viral contamination were assessed qualitatively and quantitatively using an in-house Perl script. Contaminant bacterial and viral contigs and their mapped reads were removed from the assembly using another in-house Perl script. The Swiss-Prot hits were further annotated by Blast2GO (Conesa et al., 2005) to retrieve the gene ontology (GO) terms and the functional information of those transcripts.
Differential Gene Expression
The transcript quantification was conducted using the alignment-based quantification method RSEM (Li and Dewey, 2011), supported by the Trinity package. The paired reads of each sample were aligned to the reference transcriptome with Bowtie2 (Langmead and Salzberg, 2012) and the read abundance was estimated with RSEM. The estimated expected counts for each sample at the gene level were extracted and used for the analysis of differential expression.
The differential gene expression was conducted with DESeq2 (Love et al., 2014) with parameters FDR 0.01 and log2-fold change differences >2 to determine and discuss the number of genes that show a greater magnitude of change among the conditions. Pairwise comparisons were applied between all condition pairs. For our analysis, we focused on pairwise comparisons between the samples collected at the control condition during the first sampling point (24°C, Ca) and samples of the treatment conditions SHS and AHS (27°C + 48 h and 30°C + 1 h). The first control condition (Ca) was collected simultaneously with SHS and was also closer to the collection time point of the AHS. The last thermal condition (LHS, 30°C + 48 h) was compared with the second control condition Cb as they were collected at the same time point. Furthermore, we made comparisons between the pairs SHS-AHS and AHS-LHS to monitor step-by-step the changes in the molecular responses of the sponges during the progress of thermal shock. Finally, we compared gene expression between the two control conditions (Ca and Cb) to assess potential “tank effect,” i.e., potential molecular response induced by other abiotic stressors not accounted for (e.g., lack of nutrients, bacterial, or viral contamination) over the course of the experiment.
Functional Enrichment Analysis
The upregulated genes in each comparison were subjected to GO descriptive analysis and illustration using the REVIGO (Supek et al., 2011). We restricted the GO terms to the biological processes to mitigate the amount of information. A further GO enrichment analysis was conducted on the upregulated genes with the Fisher’s Exact Test (FDR < 0.1) in the Blast2GO platform (Conesa et al., 2005) in order to observe the most enriched GO categories.
Results
Sequencing and Processing
The sequencing of 20 cDNA libraries yielded a total of 258,664,633 reads, an average of 12,933,232 raw paired reads per library (stdev 1.5 million). After the filtering process, 11,241,585 paired reads per library remained on average. The number of reads was uniformly distributed among the libraries (Supplementary Figure S1).
The final assembly of the de novo reference transcriptome included 581,983 putative transcripts (N50: 605 bp, mean contig length: 308 bp and total assembled bases: 176,505,435). The majority (94.8%) of the reads were represented in the de novo reference transcriptome. The analysis of BUSCO suggested 93.6% transcriptome completeness. Finally, about 1% of all genes (56,877 genes) were annotated (with a blastx hit) and had associated GO terms (Supplementary Figure S2).
Differential Gene Expression Analysis
All comparisons between the three thermal conditions and the controls (Ca and Cb) clearly differed in expression patterns, as observed in the general heatmap (Figure 2A) and the sample correlation matrix (Supplementary Figure S3A). All four biological replicates of each condition clustered together, forming independent groups (Figure 2A). The SHS treatment (27°C + 48 h) appeared as outgroup in the heatmap dendrogram and had the most distinctive expression profile when compared to the control conditions (Figure 2A and Supplementary Figures S3A,B).
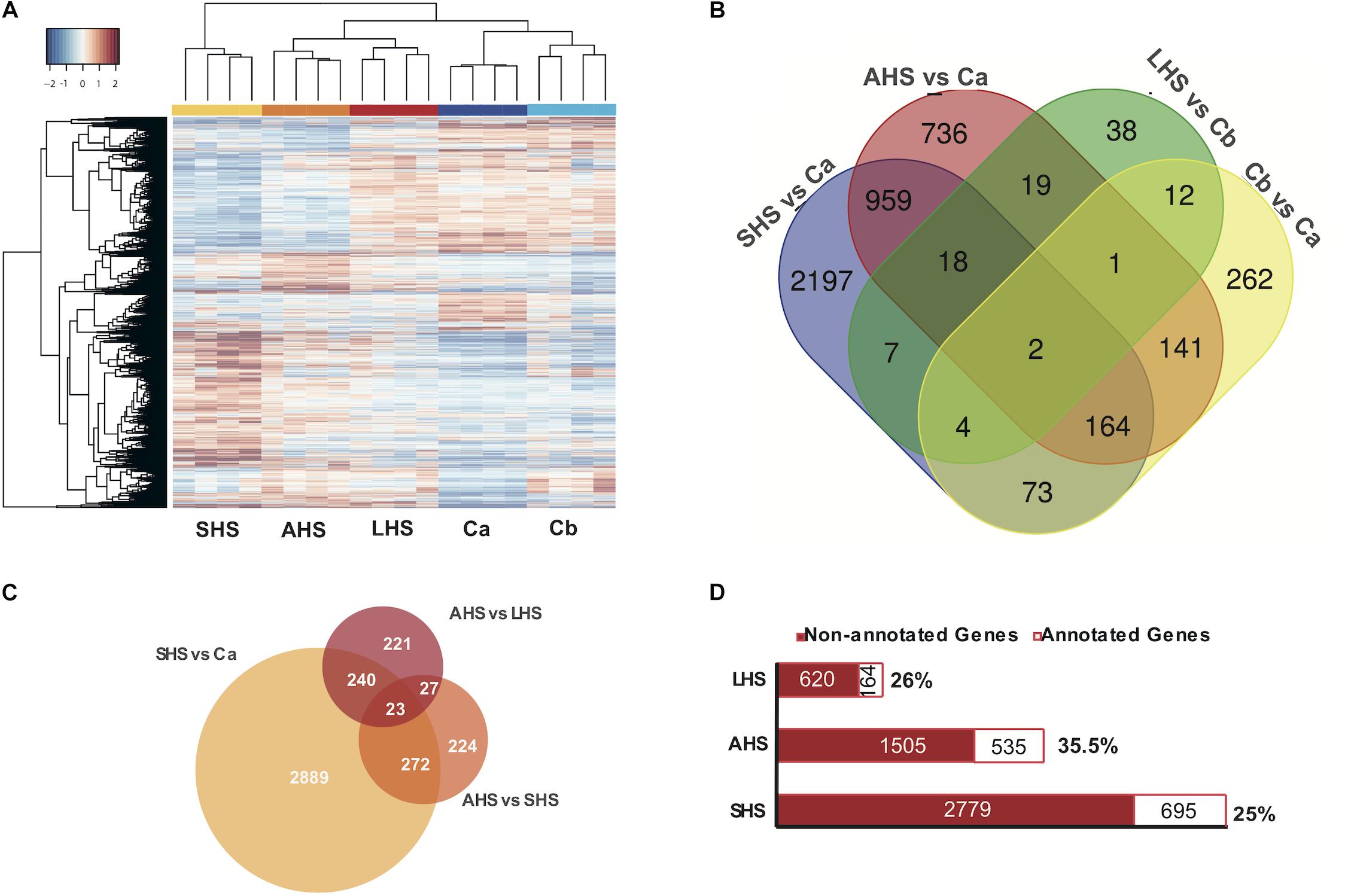
Figure 2. Differential Expression Analysis. (A) Heatmap depicts the clustering of all the differentially expressed genes, based on FDR < 0.01 along the different conditions. Colors from blue to red indicate the increasing expression. (B) Venn diagram presenting the amount of differentially expressed genes in each treatment condition (SHS, AHS, LHS) compared to the corresponding control condition Ca, Cb as also in the comparison between the two control conditions. (C) Venn diagram representing the differentially expressed genes in each treatment condition when compared among themselves in a gradual increase from Ca to SHS, from SHS to AHS, and from AHS to LHS. (D) Bar graph depicting the amount of differentially expressed genes in each thermal condition that are annotated and have an associated GO term. SHS, Short heat Shock; AHS, Acute Heat Shock; LHS, Long Heat Shock; CT, Control tank; TT, treatment tank.
During the exposure to the initial SHS treatment compared to the control Ca, we found 3424 differentially expressed (DE) genes (1973 upregulated, 1451 downregulated) (Figure 2B and Supplementary Table S4A). In the AHS treatment vs. Ca, 2040 DE genes were registered (1105 upregulated, 935 downregulated), i.e., 60% of the number of DE genes found in the comparison between SHS and Ca (Figure 2B and Supplementary Table S4B). However, after 48 h at 30°C (LHS), the number of DE genes compared to the control Cb dropped notably compared to the previous conditions. In the LHS treatment, only 101 genes were DE (40 upregulated and 61 downregulated) compared to the second control condition, Cb (Figure 2B and Supplementary Table S4C). In the comparison of gene expression between the two control conditions (Ca and Cb) to account for the “tank effect,” we found 659 DE genes (456 upregulated and 203 downregulated in Cb vs. Ca) (Figure 2B and Supplementary Table S4D). When we compared the differential gene expression patterns during the transition from SHS to AHS, we observed 546 DE genes (260 upregulated and 286 downregulated) (Figure 2C and Supplementary Table S4E), and from AHS to LHS we found 511 DE genes (350 upregulated and 161 downregulated in the latest condition) (Figure 2C and Supplementary Table S4F). Out of the total DE genes, 28% had an associated GO term (Figure 2D and Supplementary Table S4G). Non-annotated genes included both non-coding genes and genes with currently unknown function.
Upregulated Genes Under Exposure to the Elevated Temperatures
Studying the DE genes several groups were chosen for individual study, based on their high expression level in the thermal treatments. Environmental sensors such as G-protein coupled (GPCRs) and Scavenger receptors were upregulated mainly in SHS and AHS treatments compared to control conditions (Figure 3A and Supplementary Tables S4A,B). Moreover, upregulation of genes coding for glutamate receptors and secretin relating pathways was identified in our thermal treatments. For instance, the gene coding for Calcium-binding mitochondrial carrier protein Aralar1 (slc25a12) with glutamate transmembrane transporter activity, was upregulated during AHS (Figure 3A and Supplementary Table S4B). In SHS, we found genes coding for cAMP kinases (e.g., cAMP-dependent protein kinase type i-alpha regulatory subunit, camp-specific 3 -cyclic phosphodiesterase 4b) and the gene coding for cAMP-responsive element modulator (crem) (Supplementary Table S4A). After 48 h at the highest treatment temperature (LHS), we observed the circularly permutated RAS protein 1 (cpras1) related with signal transduction when compared with Cb (Supplementary Table S4C). We also observed overexpressed genes involved in calcium regulation during thermal treatments (Figure 3A), including the Two Pore Calcium Channel Protein 1 gene (tpcn1) and several copine genes (CPN); specifically, tpcn1 was upregulated in SHS and cpne9 in AHS (Figure 3A and Supplementary Table S4A). In addition, we confirmed upregulation of genes with antioxidant activity while exposed to thermal treatment in S. officinalis (Figure 3B), including the gene cytochrome b5 reductase 4 (CYB5R4), overexpressed at SHS and AHS (Figure 3B and Supplementary Tables S4A,B).
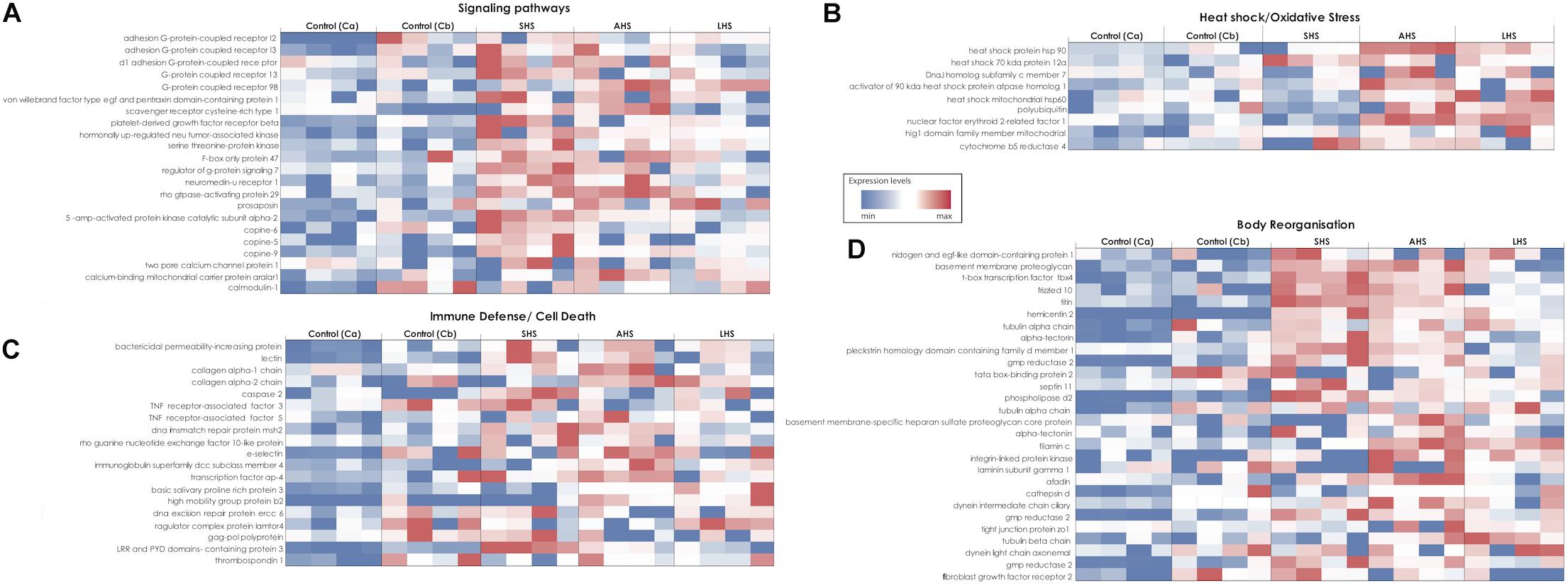
Figure 3. Heatmaps showing the relative expression level of target genes in all the different conditions of the experiment. Expression levels increase from blue to red. The different heatmaps (A–D) represent different groups of genes according to their function. (A) Signaling pathways. (B) Heat shock and oxidative stress. (C) Immune response and Cell death. (D) Skeleton formation and body organization. SHS, Short heat Shock; AHS, Acute Heat Shock; LHS, Long Heat Shock; Ca, control treatment at the first sampling point; Cb, control treatment at the last sampling point.
Heat shock protein genes (Hsp) generally showed higher relative expression levels in all three experimental conditions compared to both control conditions (Figure 3B and Supplementary Tables S4A–C). Specifically, the Hsp70 kda protein 12a (HSPA12A) was upregulated in SHS (Supplementary Table S4A) and Hsp90-alpha (HSP90AA1), Hsp70 kda protein 12a (HSPA12A), the activator of 90 kda Hsp, atpase homolog 1 (AHSA1) and the DnaJ homolog subfamily C member 7 (DNAJC7) in AHS (Figure 3B and Supplementary Table S4B), when compared to control Ca. During LHS, the 60 kda heat shock mitochondrial (HSP60) was upregulated when compared with control Cb (Figure 3B and Supplementary Table S4C).
Genes involved in defense against pathogenic bacteria, innate immune response and apoptosis, such as lectins and Bactericidal Permeability-Increasing protein (BPI), were consistently found upregulated in the SHS and AHS conditions relative to control Ca (Figure 3C and Supplementary Tables S4A,B). In the same comparisons, genes related to programed cell death were overexpressed, such as caspase-2 (CASP2) and Tumor Necrosis Factor Receptor-Associated Factor-3 (TRAF-3) (Figure 3C and Supplementary Tables S4A,B). At the transition from AHS to LHS, genes with negative regulation of cell death, including the thioredoxin domain-containing protein 5 (txndc5) and the gene Ragulator complex protein (lamtor4) were upregulated (Figure 3C and Supplementary Table S4F).
Finally, we detected more than 15 upregulated genes related to “body reorganization” processes during exposure to increased thermal conditions (Figure 3D). For instance, the frizzled-10 factor, which is a receptor in the Wnt signaling pathway, was upregulated in sponges during SHS treatment (Figure 3D and Supplementary Table S4A). In addition, genes that have a role in cell-to-cell adhesion and in the extracellular matrix such as the filamin-c, adhesion g protein-coupled receptor, basement membrane proteoglycan, and laminin were over-expressed in SHS and AHS (Figure 3D and Supplementary Tables S4A,B). Also, several genes involved in the production of lipids responsible for stabilization of the membrane, such as phosphatidylserine decarboxylase, 1-phosphatidylinositol-bisphosphate phosphodiesterase epsilon-1, and n-acetylglucosaminyl-phosphatidylinositol de-n-acetylase were upregulated in sponges during SHS and LHS treatment, and membrane-associated phosphatidylinositol transfer protein during AHS (Supplementary Table S4B). In LHS, we observed overexpression of the gene DNAJ homolog subfamily b member 6 (DNAJB6), which is active during extracellular matrix reorganization and actin cytoskeleton organization (Supplementary Table S4C).
Gene Ontology Analysis
A GO term descriptive analysis and a further GO enrichment analysis of the upregulated genes was assessed in order to identify the most important biological processes induced in each comparison (Supplementary Tables S5,S6). At the initial SHS treatment compared to control Ca, sponges had upregulated many stress-related mechanisms including signaling pathways with cell surface and transmembrane receptors (transmembrane receptor protein tyrosine kinase signaling pathway, G-protein coupled receptor signaling pathway), production of molecular mediator of immune response, polyubiquitinated misfolded protein transport, microtubule-based process, cortical actin cytoskeleton organization, membrane lipid metabolic process, phagocytosis, stress-activated protein kinase (MAPK) cascade, cellular response to DNA damage stimulus, fibroblast growth factor receptor apoptotic signaling pathway and positive regulation of cytokine secretion, among others (Figure 4 and Supplementary Tables S5A, S6A). Sponges in AHS had still activated signaling pathways, cytoskeleton reorganization and membrane lipid processes as also response to misfolded protein- autoubiquitination and phagocytosis when compared to Ca. In this condition, sponges also expressed cellular response to heat and further immune activation with the overexpression of Toll-like receptor 9 signaling pathway and cellular response to interleukin-4 (Figure 4 and Supplementary Tables S5B, S6B).
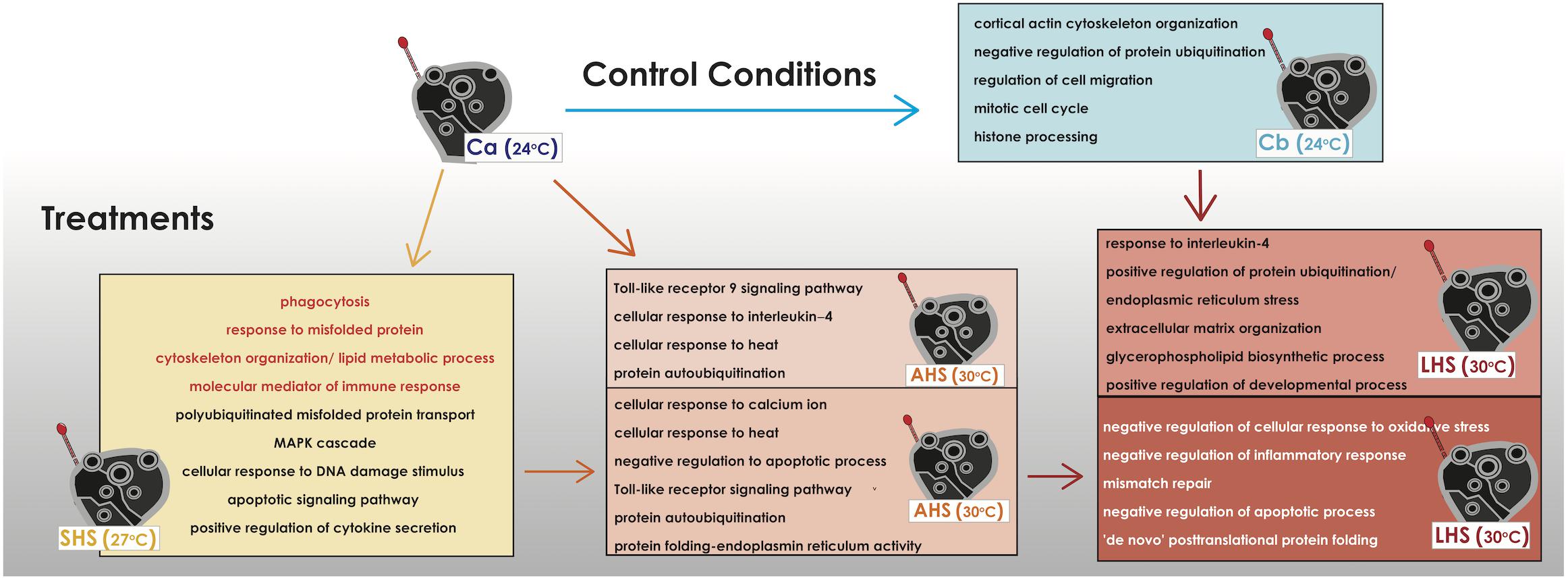
Figure 4. Depiction of the overrepresented functional categories in each thermal condition when compared to the control condition or with other thermal conditions. The present functional categories of the last control condition (Cb) when compared to the former control condition (Ca) are also depicted. The functional categories come from the GO terms extracted from the upregulated genes in each comparison and/or the further enrichment of GO categories (data available in Supplementary Tables S5, S6). In orange color are categories that are find common in the comparisons SHS vs. Ca and AHS vs. Ca.
In the pairwise comparisons between the treatments themselves (AHS vs. SHS), within big functional groups, GO categories falling into “cellular response to calcium ion,” “regulation of cellular response to heat,” “negative regulation of apoptotic process,” “leukocyte mediated immunity” including the toll-like receptor signaling pathway, and protein auto-ubiquitation were retrieved from the annotation of the DE genes when comparing SHS to AHS (Figure 4 and Supplementary Tables S5C, S6C). Comparing the LHS treatment to control Cb, main GO categories of the upregulated genes implicated in biological processes were positive regulation of protein ubiquitination, regulation of cellular response to stress and cellular response to interleukin-4, positive regulation of the anti-inflammatory interleukin-6 production and biological processes related to body reorganization/extracellular matrix and glycerophospholipid biosynthetic process (Figure 4 and Supplementary Table S5D). Positive regulation of developmental process was also present the most dominant biological process happening during this comparison was the endoplasmic reticulum stress related to regulation of protein misfolding (Supplementary Table S6D). When comparing LHS to the previous AHS condition, GO categories such as G-protein coupled receptor signaling pathway, negative regulation of inflammatory response, and negative regulation of cellular response to oxidative stress and mismatch repair were overexpressed (Figure 4 and Supplementary Table S5E). Another important, among others, functional-GO group found in the same comparison was the positive regulation of DNA binding which included the negative regulation of apoptotic process and (Figure 4 and Supplementary Table S5E). Finally, we observed categories which included mitotic cell cycle and metabolic processes, such as organic hydroxy compound biosynthesis (transcription and translation, cellular biosynthetic process, lipid and steroid metabolic process) and “de novo” post-translational protein folding (Figure 4 and Supplementary Tables S5E, S6E).
Tank Effect
When observing the overexpressed genes in the control treatment at the end of the experiment (Cb) compared to the starting point (Ca), upregulation was recorded for genes related to extracellular matrix reorganization [e.g., collagen alpha-2 chain (let-2), thrombospondin-1 (thbs1), tubulin alpha chain, tata box-binding protein 2 (tbpl2)], progress of cell cycle [e.g., g2 mitotic-specific cyclin-b2 (ccnb2), cyclin-dependent-like kinase 5 (cdk5) and histone deacetylase 4 (hdac4)], as well as for genes involved in inflammation DNA damage-regulated autophagy modulator (dram1), DNA excision repair protein (ercc6), e-selectin (sele), arachidonate (alox5) (Supplementary Table S4D). According to the GO terms, the overexpressed biological processes belonged to few broader categories, such as cortical actin cytoskeleton organization and negative regulation of protein ubiquitination, as well as some smaller such as cell proliferation and cell-to-matrix adhesion (Figure 4 and Supplementary Tables S5F, S6F).
Discussion
In our experimental setup, we attempted to assess the response of S. officinalis to conditions approximating those encountered in the species’ natural habitat during the peak of the warm season (summer), including the sporadic anomalous temperature incidences that are occasionally manifested in Mediterranean coastal areas during periods of wind calmness (Baldi et al., 2006). The gene expression of S. officinalis, as expected, varied considerably in response to our experimental thermal treatment. A pronounced response was evidenced during the initial “short-term heat shock”; this is notable, since the temperature selected for this treatment (27°C) can be often encountered as a seasonal maximum in the species’ native habitats during the peak of summer. A further steep rise of the temperature to 30°C in the “acute heat shock” treatment –corresponding to annual extremes occurring during the summer season and lasting from several hours to days– induced a further important molecular response of the sponge. However, this response was moderated following prolonged exposure at the same temperature.
The studied organism clearly demonstrated responsiveness to temperature shifts in its environment, activating relevant molecular mechanisms. In other studies on cnidarians and sponges, a temperature increase provoked several downstream negative effects such as oxidative stress (Richier et al., 2006; Desalvo et al., 2008; Voolstra et al., 2009), malfunction and misfolding of proteins, and increase of pathogens (Webster et al., 2008; Cebrian et al., 2011). This caused the activation of several defense molecular mechanisms, including the innate immune system, apoptotic processes, and protein ubiquitination. Genes and GO categories responding to the above stimuli have been previously mentioned to be activated in sponges during thermal stress (Wagner et al., 1998; Pålsson-McDermott and O’Neill, 2004; Wiens et al., 2005, 2007; Takeda, 2005; Wiens and Müller, 2006; Morizot and Saleh, 2012; Guzman and Conaco, 2016) and were also upregulated during SHS and AHS, suggesting that the studied individuals experienced an environmental stress during our experiment. Combining the known molecular pathways activated during thermal or other stressogenic factors in other metazoans with genes -as well as functional categories- DE in our study, we gained insight into the functional framework in response to elevated environmental temperatures in S. officinalis (Figure 5). According to this hypothetical scenario, the first response of the sponge included sensing the temperature increase via thermosensors, which activated downstream pathways of inflammation. The calcium channel detector is clearly indicated as a candidate thermosensor in the molecular toolkit of S. officinalis. Structural genes for Ca2+ channels were found mainly upregulated in our SHS treatment, indicating potential thermosensibility of calcium mobilizers in S. officinalis. In sponges, calcium mobilizers are ion channels with environmental detecting properties and act as a signaling molecules for diverse functions (Leys and Meech, 2006). In a similar manner, increase in intracellular Ca2+ was apparent only 10 min after a thermal shock of 6°C in the sponge Axinella polypoides (Zocchi et al., 2001). Moreover, GPCRs and Scavenger receptors are known as significant groups of environmental sensors in sponges (Srivastava et al., 2010; Conaco et al., 2012). Genes from both groups were upregulated in the two first thermal treatments, rendering them potential candidates as thermosensors in our species as well. Similarly, Guzman and Conaco (2016) found upregulation of rhodopsin, glutamate and secretin GPCRs in H. tubifera during exposure to thermal stress, explaining cellular stimulation by external stimuli.
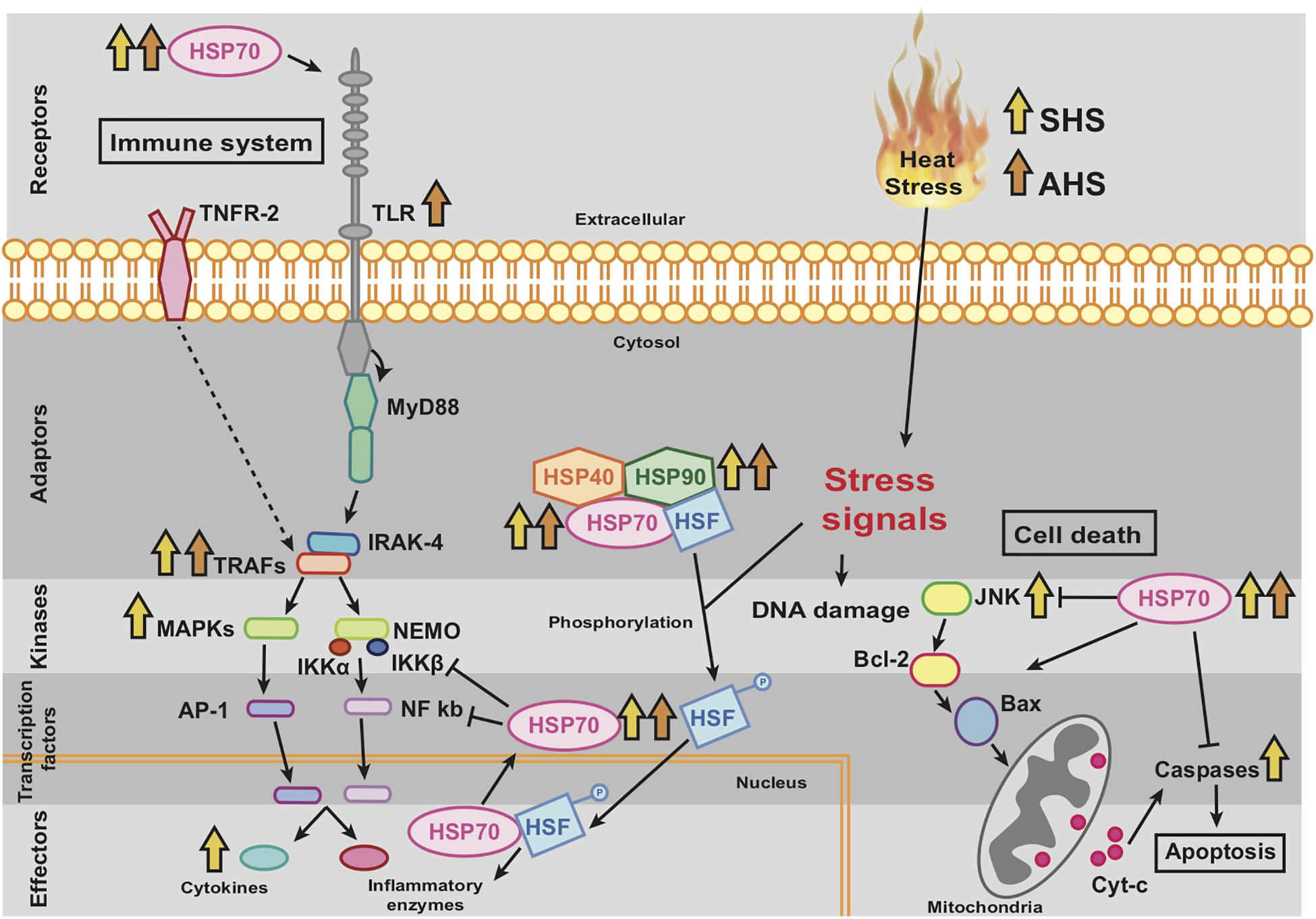
Figure 5. Theoretical representation and interconnection of molecular pathways related to thermal stress which have been appeared partially in our experiment. The arrows indicate which of the genes, included in those pathways, were upregulated. The orange arrows refer to genes upregulated in SHS (Short-term Heat Stress) and red arrows to genes upregulated in AHS (Acute Heat Stress).
The activation of thermosensors consequently induced the transcription of an array of hsp genes or other signal genes, which may have triggered the activation of the apoptotic pathway via caspases and the immune system via TRAFs, MAP kinases and inflammatory cytokines during SHS. Extracellular release of HSPs further induced the activation of the immune system via the TLR receptor at AHS, as it has also been shown to happen in other stress scenarios known from more developmentally complex animals (Kim and Yenari, 2013). However, the overexpression of HSPs may additionally be involved in the suppression of cell death and inflammation, sustaining the cell survival after damaging stimuli (Beere, 2004). The present upregulation of HSPs mainly in AHS and partially in LHS with parallel absence of any further upregulation of apoptotic pathway (no more upregulation of pro-apoptotic elements such as JNK or caspases), indicates that at this stage (AHS), high intracellular amounts of HSPs are functionally supressing the apoptotic pathway protecting the sponge from cell death. In LHS, the survival of the sponge has already been achieved as no upregulation of genes related to either the immune system or the cell death pathway was detected. Indeed, LHS was found to promote overexpression of genes related to regulation of cellular response to stress, wound repair, and diminution of pathological inflammation. Accordingly, the gene hsp70 has been isolated from the deep-water sponge Geodia cydonium under thermal stress (Koziol et al., 1996, 1997) and was also found upregulated during thermal stress in other marine and freshwater sponges (e.g., Muller et al., 1995; Guzman and Conaco, 2016).
Among the inflammation pathways, genes involved in the TLR pathway –TLR receptor, TRAFs and MAPKs– were DE, eventually promoting the overexpression of cytokines and triggering the phagocytic activity of the sponge (Pålsson-McDermott and O’Neill, 2004; Takeda, 2005). In sponges, the role of TLRs is to detect foreign antigens and trigger signaling pathways, activating the innate immune system (Wiens et al., 2005, 2007). Phagocytic activity is usually upregulated during sponge host defense in the presence of foreign cells or self-damaged tissue (Bayne, 1990), and has also been shown to be activated during heat shock in vertebrates (Vega and De Maio, 2005). Several kinases and transcription factors of the MAPK family are known to be induced by cellular stresses such as UV radiation, osmotic and heat shock (Kyriakis and Avruch, 1996). Furthermore, the cell death pathway was activated mainly at SHS, as indicated by upregulation of several caspases and TRAF proteins, responsible for regulation of cell death (Bradley and Pober, 2001; Wiens et al., 2003; Wiens and Müller, 2006; McIlwain et al., 2013). These genes were also found upregulated in the sponge H. tubifera (Guzman and Conaco, 2016) and the sea anemone Anemonia viridis (Richier et al., 2006) during exposure to thermal treatment. Induction of programed cell death as a defense mechanism against heat shock and pathogen infections has been described previously in sponges (Wagner et al., 1998; Wiens and Müller, 2006; Morizot and Saleh, 2012).
Notably, the prolonged exposure to extreme temperature (LHS) was characterized by a distinctly different transcriptomic profile. The main activated category during this treatment was the endoplasmic reticulum (ER) stress (Figure 6), which occurs when a high number of unfolded proteins is accumulated in the respective organelle (Kadowaki and Nishitoh, 2013). The initial exposure to elevated temperatures (SHS and AHS) apparently had already caused misfolding of proteins as manifested through overrepresentation of protein folding and transport of polyubiquitinated proteins. The sponge shows the capacity to compensate with this only during the LHS via the upregulation of ER stress. An array of genes related to unfolded protein response (UPR) in ER were found upregulated in this treatment, in particular, genes for the PERK, IRE, ATF6 receptors of the ER signaling pathway. These receptors detect misfolded proteins and activate chaperones and transcription factors in the nucleus to act in the ER for quality control to ensure survival of the cell. The misfolded proteins which cannot be corrected, pass through the endoplasmic-reticulum-associated protein degradation (ERAD) pathway to the proteasome for degradation (Kadowaki and Nishitoh, 2013). This pathway, in combination with death factors such as TRAFs, may lead to cell apoptosis in case of irreversible damage. However, no genes related to immune response and cell death were present in this condition (LHS). On the contrary, only during the transition from AHS to LHS, S. officinalis upregulated thioredoxin; the latter plays an essential role in eliminating dead cells after induced cell apoptosis and promotes programmed cell regeneration in sponges (Wiens et al., 1999). Furthermore, genes that make part of a cell growth pathway (lamtor4; Bar-Peled et al., 2012) were upregulated during LHS. Genes with similar function were found upregulated during exposure to sublethal temperatures (32°C + 12 h) in H. tubifera, suggesting that acclimatization to thermal stress enables sponges to mitigate further environmental stress and improve their chances of surviving (Guzman and Conaco, 2016). In an attempt to understand the transcriptomic response of the sponges to the maintenance in experimental tanks, we compared the two control conditions at the very beginning and at the end of the experiment to ultimately find no indication of stress. Interestingly, a high number of genes from those upregulated in Cb was related to body organization, as also observed in all other treatments when compared to their relevant control conditions. Potentially, the sponge employs this structural response to avoid any possible abiotic stress, presumably by removing some damaged tissue. Reorganization of the body plan and epithelial morphogenesis is a common strategy during reparative regeneration in sponges to isolate damaged areas or recover from tissue wounds (Ereskovsky et al., 2015; Runzel, 2016). This strategy has been previously reported for individuals of S. officinalis when suffering disease outbreaks (Gaino and Pronzato, 1989). However, no visible signs of tissue damage were manifested over the course of our experiment. On the other hand, in all comparisons between conditions spanning over extended periods (Cb vs. Ca, LHS vs. Ca, and LHS vs. AHS), we observed an overexpression of genes related to cell growth and positive regulation of the cell cycle, which does not indicate any negative impact of the cultivation regime over the studied period.
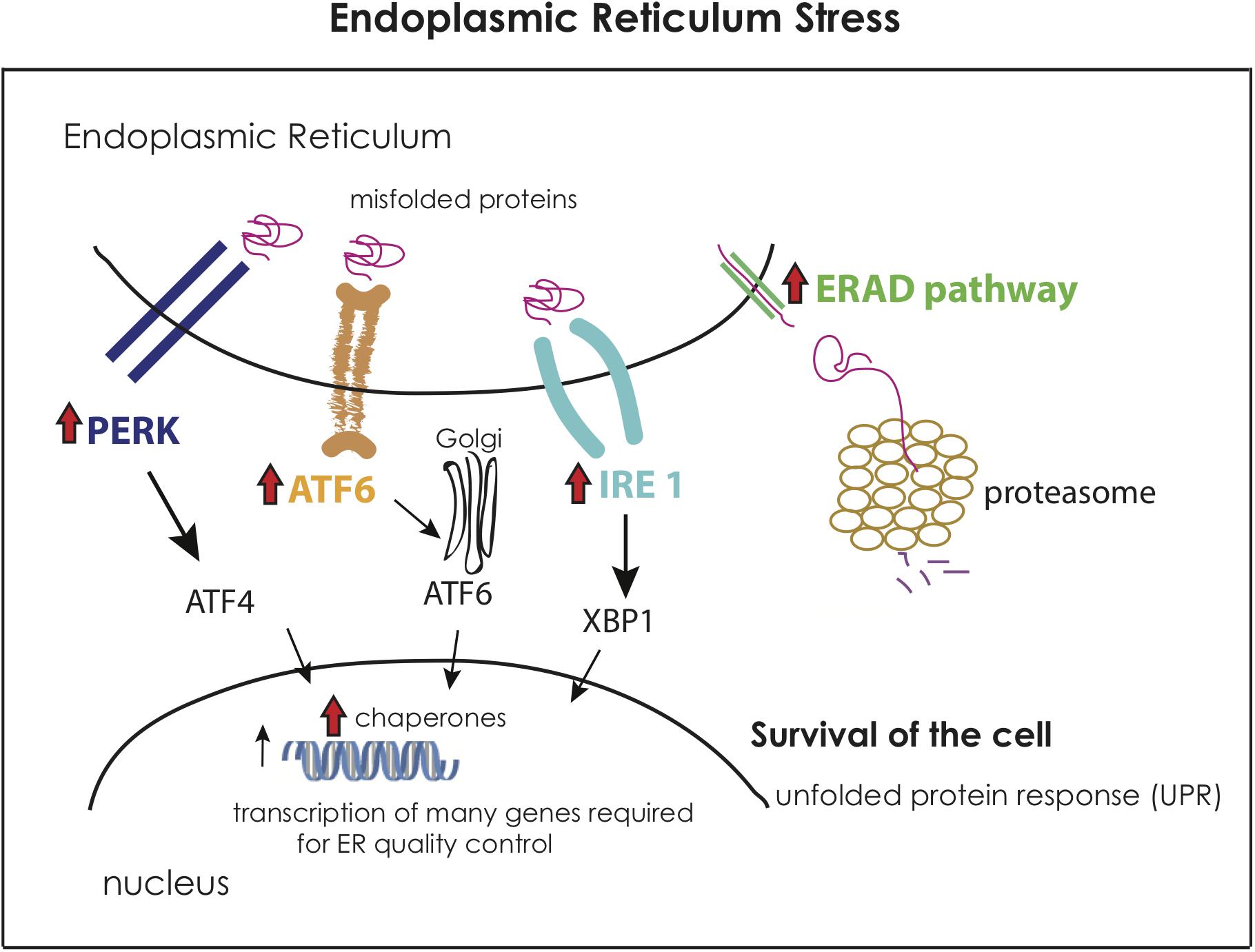
Figure 6. Representation of the molecular activation of the Endoplasmic Reticulum Stress (ER stress) based on theoretical data and the activated genes in the Long-term Heat Shock (LHS) condition compared to the control Cb. The red arrow indicates the overexpression of this gene or functional category in LHS when compared to Cb. The figure is adapted from Kadowaki and Nishitoh (2013).
From the above we can extract the conclusions that this sponge species has a very well-developed mechanism for reacting even to mild heat shocks, since its main transcriptional response occurred at the first thermal shock (AHS). When exposed to the following thermal condition (SHS), given the sponge already deployed a molecular response to face the adverse conditions, it expressed less numbers of genes. After few days enduring high thermal stress (LHS), it already started to acclimatize to the new abiotic conditions of its surroundings. We observed that during prolonged exposure to extreme temperatures, S. officinalis demonstrated recovering from a stress-inducing condition and initiated upregulation of common physiological functions that were paused during the initial heat shock.
In conclusion, this study paves the way to understanding the molecular response of this emblematic species to thermal shift. The results of the present study are based solely on transcriptomic data, which do not always coincide with the translated proteins (e.g., Peng et al., 2015; Mayfield et al., 2018). Nevertheless, the fact that many genes involved in known pathways responding to thermal stress were found upregulated in our study provides some evidence toward the conservation of the molecular machinery of thermal shock response that appeared early in evolution. The activation of the described pathways and the actual physiological status of the sponge during exposure to different thermal treatments along time remains to be tested using alternative techniques. Moreover, additional experiments are required to further address the proteomic response of this species to thermal shift, as well as to explore the effect of extended temperature increases and/or additional stressors; those should ideally include physiological measurements and experiments in natural settings. This is especially important under the light of the anticipated climatic changes in the Mediterranean environment in the following years (Darmaraki et al., 2019). Our study provides a sound baseline on the gene expression response of S. officinalis to elevated temperatures, indicating which genes are overexpressed under the current thermal thresholds and predicting the molecular pathways that will likely be targeted by selective forces in near future climate change scenarios. This is essential information to better tune the management and conservation of this emblematic and economically important organism, whose populations have faced a dramatic decline in the past years and is also relevant to other sublittoral sessile species that have recently undergone mass mortality incidents.
Data Availability Statement
The datasets generated for this study can be found in the Sequence Read Archive (SRA) with accession number SRP150632.
Author Contributions
TD conceived and managed the study and provided the live samples. TD, VK, and TM designed the experiments. VK and TD performed the experimental treatments. VK carried out laboratory analyses and wrote the first draft of the manuscript. SK performed library preparation and sequencing. VK, JL, TM, and AR analyzed the data. CD, CT, CA, and AM provided access to research infrastructures. AR, TM, and TD made major contributions to the writing. All authors reviewed and contributed to the final version of the manuscript.
Funding
This research was co-financed by Greece and the European Union (European Social Fund-ESF) through the Operational Programme “Human Resources Development, Education and Lifelong Learning” in the context of the project “Reinforcement of Postdoctoral Researchers” (MIS-5001552), implemented by the State Scholarships Foundation (IKY).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Julius Glampedakis for support in the collection and rearing of S. officinalis in the open sea, and Grigorios Skouradakis for setup of the experimental aquaria and continuous technical support during the experiments. The authors also thank the technicians and other researchers of the molecular laboratory of the institute Vasso Terzoglou, Katerina Oikonomaki, and Eliza Kaitetzidou for important advice during RNA extraction and Alexandros Tsakogiannis for useful discussions on transcriptome analysis. Finally, thanks are due to Sergio Taboada, Nathan Kenny, Bruna Plese, Cristina Diez, and Giorgos Kotoulas for important comments and advice during the data analysis and writing of the manuscript. The sequencing service was provided by the Norwegian Sequencing Centre (www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00786/full#supplementary-material
Footnotes
- ^ http://www.bioinformatics.babraham.ac.uk/services.html
- ^ https://github.com/vsbuffalo/scythe
- ^ https://github.com/najoshi/sickle
- ^ http://prinseq.sourceforge.net/
- ^ gvolante.riken.jp/analysis.html
References
Asa, S., Yeemin, T., Chaitanawisuti, N., and Kritsanapuntu, S. (2000). “Sexual reproduction of a marine sponge, petrosia sp. from coral communities in the Gulf of Thailand,” in Proceedings of the 9th International Coral Reef Symposium, Bali.
Bachinski, N., Koziol, C., Batel, R., Labura, Z., Schröder, H. C., and Müller, W. E. G. (1997). Immediate early response of the marine sponge Suberites domuncula to heat stress: reduction of trehalose and glutathione concentrations and glutathione S-transferase activity. J. Exp. Mar. Bio. Ecol. 210, 129–141. doi: 10.1016/S0022-0981(96)02705-2700
Baldi, M., Dalu, G., Maracchi, G., Pasqui, M., and Cesarone, F. (2006). Heat waves in the Mediterranean: a local feature or a larger-scale effect? Int. J. Climatol. 26, 1477–1487. doi: 10.1002/joc.1389
Bar-Peled, L., Schweitzer, L. D., Zoncu, R., and Sabatini, D. M. (2012). An expanded ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208. doi: 10.1016/j.cell.2012.07.032
Bayne, C. J. (1990). Phagocytosis and non-self recognition in invertebrates. Bioscience 40, 723–731. doi: 10.2307/1311504
Beere, H. M. (2004). ‘The stress of dying’: the role of heat shock proteins in the regulation of apoptosis. J. Cell Sci. 117, 2641–2651. doi: 10.1242/jcs.01284
Bell, J. J. (2008). The functional roles of marine sponges. Estuar. Coast. Shelf Sci. 79, 341–353. doi: 10.1016/j.ecss.2008.05.002
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bradley, J. R., and Pober, J. S. (2001). Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20, 6482–6491. doi: 10.1038/sj.onc.1204788
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Carballo, J. L., Sánchez-Moyano, J. E., and Carballo, J. C. (1994). Esponjas del estrecho de gibraltar. I. Esponjas corneas. Graellsia 50, 25–56.
Cebrian, E., Uriz, M. J., Garrabou, J., and Ballesteros, E. (2011). Sponge mass mortalities in a warming mediterranean sea: are cyanobacteria-harboring species worse off? PLoS One 6:e20211. doi: 10.1371/journal.pone.0020211
Cerrano, C., Bavestrello, G., Bianchi, C. N., Cattaneo-Vietti, R., Bava, S., Morganti, C., et al. (2000). A catastrophic mass-mortality episode of gorgonians and other organisms in the Ligurian Sea (North-western Mediterranean), summer 1999. Ecol. Lett. 3, 284–293. doi: 10.1046/j.1461-0248.2000.00152.x
Conaco, C., Neveu, P., Zhou, H., Arcila, M. L., Degnan, S. M., Degnan, B. M., et al. (2012). Transcriptome profiling of the demosponge Amphimedon queenslandica reveals genome-wide events that accompany major life cycle transitions. BMC Genom. 13:209. doi: 10.1186/1471-2164-13-209
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Dailianis, T., Tsigenopoulos, C. S., Dounas, C., and Voultsiadou, E. (2011). Genetic diversity of the imperilled bath sponge Spongia officinalis linnaeus, 1759 across the mediterranean Sea: patterns of population differentiation and implications for taxonomy and conservation. Mol. Ecol. 20, 3757–3772. doi: 10.1111/j.1365-294X.2011.05222.x
Darmaraki, S., Somot, S., Sevault, F., Nabat, P., Cabos Narvaez, W. D., Cavivvhia, L., et al. (2019). Future evolution of marine heatwaves in the mediterranean sea. Clim. Dyn. 53:1371. doi: 10.1007/s00382-019-04661-z
Desalvo, M. K., Voolstra, C. R., Sunagawa, S., Schwarz, J. A., Stillman, J. H., Coffroth, M. A., et al. (2008). Differential gene expression during thermal stress and bleaching in the caribbean coral Montastraea faveolata. Mol. Ecol. 17, 3952–3971. doi: 10.1111/j.1365-294X.2008.03879.x
Dove, A. D. M., All Am, B., Powers, J. J., and Sokolowski, M. S. (2005). A prolonged thermal stress experiment on the american lobster, Homarus americanus. J. Shellfish Res. 24, 761–765. doi: 10.2983/0730-8000(2005)24%5B761:aptseo%5D2.0.co;2
Duckworth, A. R., and Peterson, B. J. (2013). Effects of seawater temperature and pH on the boring rates of the sponge Cliona celata in scallop shells. Mar. Biol. 160, 27–35. doi: 10.1007/s00227-012-2053-z
Ereskovsky, A. V., Borisenko, I. E., Lapébie, P., Gazave, E., Tokina, D. B., and Borchiellini, C. (2015). Oscarella lobularis (Homoscleromorpha, Porifera) regeneration: epithelial morphogenesis and metaplasia. PLoS One 10:e0134566. doi: 10.1371/journal.pone.0134566
Ettinger-Epstein, P., Whalan, S. W., Battershill, C. N., and De Nys, R. (2007). Temperature cues gametogenesis and larval release in a tropical sponge. Mar. Biol. 153, 171–178. doi: 10.1007/s00227-007-0793-y
Fan, S., Ren, H., Wang, R., Pan, C., Huang, G., Chen, Y., et al. (2013). Vegetation restoration and conservation plans for integrated coastal management in Hengqin China. J. Coast. Res. 29, 1297–1308. doi: 10.2112/JCOASTRES-D-12-00243.1
Gaino, E., and Pronzato, R. (1989). Ultrastructural evidence of bacterial damage to Spongia officinalis fibres (Porifera, Demospongiae). Dis. Aquat. Org. 6, 67–74. doi: 10.3354/dao006067
Gaino, E., Pronzato, R., Corriero, G., and Buffa, P. (1992). Mortality of commercial sponges: Incidence in two mediterranean areas. Boll. di Zool. 59, 79–85. doi: 10.1080/11250009209386652
Gerovasileiou, V., Dailianis, T., Sini, M., Otero, M., del, M., Numa, C., et al. (2018). Assessing the regional conservation status of sponges (Porifera): the case of the Aegean ecoregion. Mediterr. Mar. Sci. 19, 526–537.
Giorgi, F. (2006). Climate change hot-spots. Geophys. Res. Lett. 33:L08707. doi: 10.1029/2006GL025734
Guzman, C., and Conaco, C. (2016). Gene expression dynamics accompanying the sponge thermal stress response. PLoS One 11:e0165368. doi: 10.1371/journal.pone.0165368
Haas, B. J. (2013). De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Harvell, C. D., Mitchell, C. E., Ward, J. R., Altizer, S., Dobson, A. P., Ostfeld, R. S., et al. (2002). Climate warming and disease risks for terrestrial and marine biota. Science 296, 2158–2162. doi: 10.1126/science.1063699
Ilias, A., Lagnel, J., Kapantaidaki, D. E., Roditakis, E., Tsigenopoulos, C. S., Vontas, J., et al. (2015). Transcription analysis of neonicotinoid resistance in Mediterranean (MED) populations of B. tabaci reveal novel cytochrome P450s, but no nAChR mutations associated with the phenotype. BMC Genom. 16:939. doi: 10.1186/s12864-015-2161-2165
IPCC (2014). “Climate change 2014: synthesis report,” in Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds R. Core Writing Team, and L. Meyer, (Geneva: IPCC), 151.
Kadowaki, H., and Nishitoh, H. (2013). Signaling pathways from the endoplasmic reticulum and their roles in disease. Genes 4, 306–333. doi: 10.3390/genes4030306
Kelmo, F., Bell, J. J., and Attrill, M. J. (2013). Tolerance of sponge assemblages to temperature anomalies: resilience and proliferation of sponges following the 1997-8 el-niño southern oscillation. PLoS One 8:e76441. doi: 10.1371/journal.pone.0076441
Kenkel, C. D., Meyer, E., and Matz, M. V. (2013). Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 22, 4322–4334. doi: 10.1111/mec.12390
Kim, J. Y., and Yenari, M. A. (2013). The immune modulating properties of the heat shock proteins after brain injury. Anat. Cell Biol. 46, 1–7. doi: 10.5115/acb.2013.46.1.1
Koziol, C., Batel, R., Arinc, E., Schröder, H. C., and Müller, W. E. G. (1997). Expression of the potential biomarker heat shock protein 70 and its regulator, the metazoan DnaJ homolog, by temperature stress in the sponge Geodia cydonium. Mar. Ecol. Prog. Ser. 154, 261–268. doi: 10.3354/meps154261
Koziol, C., Wagner-ulsmann, C., Koc, A. I., Gamulin, V., Krusel, M., Pancerl, Z., et al. (1996). Cloning of a heat-inducible biomarker, the cDNA encoding the 70 kDa heat shock protein, from the marine sponge Geodia cydonium: response to natural stressors. Mar. Ecol. Prog. Ser. 136, 153–161. doi: 10.3354/meps136153
Krasko, A., Scheffer, U., Koziol, C., Pancer, Z., Batel, R., Badria, F., et al. (1997). Diagnosis of sublethal stress in the marine sponge Geodia cydonium: application of the 70 kDa heat-shock protein and a novel biomarker, the Rab GDP dissociation inhibitor, as probes. Aquat. Toxicol. 37, 157–168. doi: 10.1016/S0166-445X(96)00822-3
Kyriakis, J. M., and Avruch, J. (1996). Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays 18, 567–577. doi: 10.1002/bies.950180708
Lagnel, J., Tsigenopoulos, C. S., and Iliopoulos, I. (2009). NOBLAST and JAMBLAST: new options for BLAST and a Java application manager for BLAST results. Bioinformatics 25, 824–826. doi: 10.1093/bioinformatics/btp067
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lathlean, J. A., Seuront, L., and Ng, T. P. T. (2017). On the edge: the use of infrared thermography in monitoring responses of intertidal organisms to heat stress. Ecol. Indic. 81, 567–577. doi: 10.1016/j.ecolind.2017.04.057
Leys, S. P., and Meech, R. W. (2006). Physiology of coordination in sponges. Can. J. Zool. 84, 288–306. doi: 10.1139/z05-171
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12:323. doi: 10.1186/1471-2105-12-323
Li, J., Zhang, Y., Mao, F., Tong, Y., Liu, Y., Zhang, Y., et al. (2017). Characterization and identification of differentially expressed genes involved in thermal adaptation of the Hong Kong Oyster Crassostrea hongkongensis by digital gene expression profiling. Front. Mar. Sci. 4:112. doi: 10.3389/fmars.2017.00112
López-Legentil, S., Song, B., Mcmurray, S. E., and Pawlik, J. R. (2008). Bleaching and stress in coral reef ecosystems: hsp70 expression by the giant barrel sponge Xestospongia muta. Mol. Ecol. 17, 1840–1849. doi: 10.1111/j.1365-294X.2008.03667.x
Love, M. I., Huber, W., Anders, S., Lönnstedt, I., Speed, T., Robinson, M., et al. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom. Biol. 15:550. doi: 10.1186/s13059-014-0550-558
Manousaki, T., Koutsouveli, V., Lagnel, J., Kollias, S., Tsigenopoulos, C. S., Arvanitidis, C., et al. (in press). A de novo transcriptome assembly for the bath sponge Spongia officinalis, adjusting for microsymbionts. BMC Res. Notes. doi: 10.1186/s13104-019-4843-6
Massaro, A. J., Weisz, J. B., Hill, M. S., and Webster, N. S. (2012). Behavioral and morphological changes caused by thermal stress in the great barrier reef sponge Rhopaloeides odorabile. J. Exp. Mar. Bio. Ecol. 41, 55–60. doi: 10.1016/j.jembe.2012.02.008
Mayfield, A. B., Chen, Y. J., Lu, C. Y., and Chen, C. S. (2018). The proteomic response of the reef coral Pocillopora acuta to experimentally elevated temperatures. PLoS One 13:e0192001. doi: 10.1371/journal.pone.0192001
Mayfield, A. B., Wang, Y. B., Chen, C. S., Lin, C. Y., and Chen, S. H. (2014). Compartment-specific transcriptomics in a reef-building coral exposed to elevated temperatures. Mol. Ecol. 23, 5816–5830. doi: 10.1111/mec.12982
McIlwain, D. R., Berger, T., and Mak, T. W. (2013). Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 5:a008656. doi: 10.1101/cshperspect.a008656
Morizot, A., and Saleh, M. (2012). Non-apoptotic functions of cell death effectors in inflammation and innate immunity. Microb. Infect. 14, 1241–1253. doi: 10.1016/j.micinf.2012.06.005
Muller, W. E. G., Koziol, C., Kurelec, B., Dapper, J., Batel, R., and Rinkevich, B. (1995). Combinatory effects of temperature stress and nonionic organic pollutants on stress protein (Hsp70) gene-expression in the fresh-water sponge Ephydatia-Fluviatilis. Environ. Toxicol. Chem. 14, 1203–1208. doi: 10.1002/etc.5620140712
Pålsson-McDermott, E. M., and O’Neill, L. A. J. (2004). Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology 113, 153–162. doi: 10.1111/j.1365-2567.2004.01976.x
Pantile, R., and Webster, N. (2011). Strict thermal threshold identified by quantitative PCR in the sponge Rhopaloeides odorabile. Mar. Ecol. Prog. Ser. 431, 97–105. doi: 10.3354/meps09128
Peng, X., Qin, Z., Zhang, G., Guo, Y., and Huang, J. (2015). Integration of the proteome and transcriptome reveals multiple levels of gene regulation in the rice dl2 mutant. Front. Plant Sci. 6:351. doi: 10.3389/fpls.2015.00351
Perez, T., Garrabou, J., Sartoretto, S., Harmelin, J.-G., Francour, P., and Vacelet, J. (2000). Mortalité massive d’invertébrés marins: un événement sans précédent en Méditerranée nord-occidentale. C. R. Acad. Sci. III 323, 853–865. doi: 10.1016/s0764-4469(00)01237-3
Place, S. P., Menge, B. A., and Hofmann, G. E. (2012). Transcriptome profiles link environmental variation and physiological response of Mytilus californianus between Pacific tides. Funct. Ecol. 26, 144–155. doi: 10.1111/j.1365-2435.2011.01924.x
Pronzato, R., and Manconi, R. (2008). Mediterranean commercial sponges: over 5000 years of natural history and cultural heritage. Mar. Ecol. 29, 146–166. doi: 10.1111/j.1439-0485.2008.00235.x
Richier, S., Sabourault, C., Courtiade, J., Zucchini, N., Allemand, D., and Furla, P. (2006). Oxidative stress and apoptotic events during thermal stress in the symbiotic sea anemone Anemonia viridis. FEBS J. 273, 4186–4198. doi: 10.1111/j.1742-4658.2006.05414.x
Runzel, C. C. (2016). Sponge physiology: the effects of temperature on the regeneration and reaggregation of sponges (Haliclona reniera). PeerJ Prepr. 4:e2654v1. doi: 10.7287/PEERJ.PREPRINTS.2654V1
Ryu, T., Seridi, L., Moitinho-Silva, L., Oates, M., Liew, Y. J., Mavromatis, C., et al. (2016). Hologenome analysis of two marine sponges with different microbiomes. BMC Genom. 17:158. doi: 10.1186/s12864-016-2501-2500
Schmieder, R., Lim, Y. W., and Edwards, R. (2012). Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics 28, 433–435. doi: 10.1093/bioinformatics/btr669
Seneca, F. O., and Palumbi, S. R. (2015). The role of transcriptome resilience in resistance of corals to bleaching. Mol. Ecol. 24, 1467–1484. doi: 10.1111/mec.13125
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Srivastava, M., Simakov, O., Chapman, J., Fahey, B., Gauthier, M. E. A., Mitros, T., et al. (2010). The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466, 720–726. doi: 10.1038/nature09201
Strand, R., Whalan, S., Webster, N. S., Kutti, T., Fang, J. K. H., Luter, H. M., et al. (2017). The response of a boreal deep-sea sponge holobiont to acute thermal stress. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-01091-x
Supek, F., Bošnjak, M., Škunca, N., and Šmuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800. doi: 10.1371/journal.pone.0021800
Sutherland, K. P., and Porter, J. W. (2004). Disease and immunity in caribbean and indo-pacific zooxanthellate corals. Mar. Ecol. Prog. Ser. 266, 273–302. doi: 10.3354/meps266273
Takeda, K. (2005). Evolution and integration of innate immune recognition systems: the Toll-like receptors. J. Endotoxin Res. 11, 51–55. doi: 10.1177/09680519050110011101
Vega, V. L., and De Maio, A. (2005). Increase in phagocytosis after geldanamycin treatment or heat shock: role of heat shock proteins. J. Immunol. 175, 5280–5287. doi: 10.4049/JIMMUNOL.175.8.5280
Vicente, V. P. (1989). Regional commercial sponge extinctions in the west indies: are recent climatic changes responsible? Mar. Ecol. 10, 179–191. doi: 10.1111/j.1439-0485.1989.tb00073.x
Voolstra, C. R., Schnetzer, J., Peshkin, L., Randall, C. J., Szmant, A. M., and Medina, M. (2009). Effects of temperature on gene expression in embryos of the coral Montastraea faveolata. BMC Genom. 10:627. doi: 10.1186/1471-2164-10-627
Voultsiadou, E. (2005). Demosponge distribution in the eastern Mediterranean: a NW-SE gradient. Helgol. Mar. Res. 59, 237–251. doi: 10.1007/s10152-005-0224-228
Voultsiadou, E. (2007). Sponges: an historical survey of their knowledge in Greek antiquity. J. Mar. Biol. Assoc. U. K. 87, 1757–1763. doi: 10.1017/S0025315407057773
Voultsiadou, E. (2009). Reevaluating sponge diversity and distribution in the Mediterranean Sea. Hydrobiologia 628, 1–12. doi: 10.1007/s10750-009-9725-97290
Voultsiadou, E., Gerovasileiou, V., and Dailianis, T. (2013). Extinction trends of marine species and populations in the Aegean Sea and adjacent ecoregions. CIESM Work Monogr. 45, 58–73.
Wagner, C., Steffen, R., Koziol, C., Batel, R., Lacorn, M., Steinhart, H., et al. (1998). Apoptosis in marine sponges: a biomarker for environmental stress (cadmium and bacteria). Mar. Biol. 131, 411–421. doi: 10.1007/s002270050334
Webster, N., Pantile, R., Botté, E., Abdo, D., Andreakis, N., and Whalan, S. (2013). A complex life cycle in a warming planet: gene expression in thermally stressed sponges. Mol. Ecol. 22, 1854–1868. doi: 10.1111/mec.12213
Webster, N. S. (2007). Sponge disease: a global threat? Environ. Microbiol. 9, 1363–1375. doi: 10.1111/j.1462-2920.2007.01303.x
Webster, N. S., Cobb, R. E., and Negri, A. P. (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2, 830–842. doi: 10.1038/ismej.2008.42
Wiens, M., Korzhev, M., Krasko, A., Thakur, N. L., Perović-Ottstadt, S., Breter, H. J., et al. (2005). Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway: induction of a perforin-like molecule. J. Biol. Chem. 280, 27949–27959. doi: 10.1074/jbc.M504049200
Wiens, M., Korzhev, M., Perović-Ottstadt, S., Luthringer, B., Brandt, D., Klein, S., et al. (2007). Toll-like receptors are part of the innate immune defense system of sponges (Demospongiae: Porifera). Mol. Biol. Evol. 24, 792–804. doi: 10.1093/molbev/msl208
Wiens, M., Krasko, A., Perovic, S., and Müller, W. E. G. (2003). Caspase-mediated apoptosis in sponges: cloning and function of the phylogenetic oldest apoptotic proteases from Metazoa. Biochim. Biophys. Acta Mol. Cell Res. 1593, 179–189. doi: 10.1016/S0167-4889(02)00388-389
Wiens, M., and Müller, W. E. G. (2006). Cell death in porifera: molecular players in the game of apoptotic cell death in living fossils. Can. J. Zool. 84, 307–321. doi: 10.1139/z05-165
Wiens, M., Seack, J., Koziol, C., Hassanein, H. M. A., Steffen, R., Korzhev, M., et al. (1999). 17β-estradiol-dependent regulation of chaperone expression and telomerase activity in the marine sponge Geodia cydonium. Mar. Biol. 133, 1–10. doi: 10.1007/s002270050436
Keywords: heat stress, gene expression, RNAseq, sponges, porifera, climate change
Citation: Koutsouveli V, Manousaki T, Riesgo A, Lagnel J, Kollias S, Tsigenopoulos CS, Arvanitidis C, Dounas C, Magoulas A and Dailianis T (2020) Gearing Up for Warmer Times: Transcriptomic Response of Spongia officinalis to Elevated Temperatures Reveals Recruited Mechanisms and Potential for Resilience. Front. Mar. Sci. 6:786. doi: 10.3389/fmars.2019.00786
Received: 20 August 2019; Accepted: 06 December 2019;
Published: 10 January 2020.
Edited by:
Jose M. Eirin-Lopez, Florida International University, United StatesReviewed by:
Maha Joana Cziesielski, King Abdullah University of Science and Technology, Saudi ArabiaHeather Diana Veilleux, University of Alberta, Canada
Copyright © 2020 Koutsouveli, Manousaki, Riesgo, Lagnel, Kollias, Tsigenopoulos, Arvanitidis, Dounas, Magoulas and Dailianis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thanos Dailianis, dGhhbm9zZEBoY21yLmdy
 Vasiliki Koutsouveli
Vasiliki Koutsouveli Tereza Manousaki
Tereza Manousaki Ana Riesgo2
Ana Riesgo2 Costas S. Tsigenopoulos
Costas S. Tsigenopoulos Christos Arvanitidis
Christos Arvanitidis Costas Dounas
Costas Dounas Thanos Dailianis
Thanos Dailianis