- 1Key Laboratory of Marine Genetics and Breeding (OUC), Ministry of Education, Qingdao, China
- 2College of Marine Life Sciences, Ocean University of China, Qingdao, China
- 3Laboratory for Marine Biology and Biotechnology, Pilot Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4Key Laboratory of Utilization and Conservation for Tropical Marine Bioresources (Hainan Tropical Ocean University), Ministry of Education, Sanya, China
Pyropia haitanensis, one of the most economically important marine crops worldwide, can recover from up to 80% water loss during emersion in the intertidal zone. However, little is currently known regarding the molecular mechanism involved in the response to osmotic stress at the post-transcriptional level. In this study, we profiled the transcriptional patterns of microRNAs (miRNAs) in P. haitanensis under the conditions of dehydration and succeeding rehydration. Forty-seven miRNAs were sequenced and characterized, which included two evolutionarily conserved and 45 novel miRNAs. Among these, 12 miRNAs were differentially expressed under osmotic stress conditions. The target genes were predicted to be related to catalytic activity, binding, transporter activity, transcription factor activity, signal transducer activity, and antioxidant activity. The miRNAPC-3p-99769_194 exhibited constant upregulation during dehydrated stress, whereas its target gene, a carbonic anhydrase, was downregulated as detected via qPCR. Furthermore, we identified genes that encode Dicer and Argonaute (AGO) proteins. The Dicer protein is encoded by two genes, each partially harboring several conserved domains. In this study, we discovered that an inverse expression profiles of miRNAPC-3p-99769_194 and its target CA under osmotic stress, which revealed the essential role of miRNAs in the suppression of carbon fixation to maintain basic living conditions. Additionally, the Pyropia-specific organization of Dicer genes may be indicative of the distinctive structural features of miRNAs. This study provided novel insights into the miRNA-mediated regulatory mechanisms that are essential for desiccation-tolerance in P. haitanensis.
Introduction
Pyropia, a genus of red algae, is one of the most economically important marine crops worldwide, especially in East Asian countries such as China, South Korea, and Japan (Sahoo et al., 2002). Its annual fresh weight can reach 1,806,000 tons according to statistics provided by the Food and Agriculture Organization in 20141. P. haitanensis is a native species that is distributed and intensively farmed in southern China. The production of P. haitanensis represents 75–80% of the total production of cultivated Pyropia in China (Zhang et al., 2005).
Most Pyropia species are naturally distributed in the intertidal zone, with a drastically changing environment (Nakamura et al., 2013). Due to their living environment, they are periodically exposed to an environment in which they experience a variety of potentially stressful environmental conditions, which include nutrient limitation, high and low temperature, intense light, and osmotic stress caused by tidal changes (Davison and Pearson, 1996). The stress caused by environmental factors and long-term evolution make Pyropia highly environmentally tolerant, especially toward osmotic stress (Hwang et al., 1997). For example, many species of Pyropia (or species related to Porphyra) can survive after they lose 85–95% of their cellular water content during low tide in the daytime (Blouin et al., 2011). Therefore, Pyropia is an ideal model organism for the study of the molecular mechanism involved in the tolerance of seaweed to osmotic stress in the intertidal zone (Cock and Coelho, 2011).
Studies show that Pyropia adapt and tolerate osmotic stress via many physiological and molecular mechanisms, including signal transduction systems, antioxidant systems, molecular chaperones, protective proteins, soluble permeants, and changes to cell wall components (Davison and Pearson, 1996). For example, “floridoside” protects the cell when the gametophytes of Porphyra purpurea are exposed to the stress of salinity (Reed et al., 1980). Additionally, putative fucosyltransferase is located in the nucleus and confers tolerance to desiccation and osmotic stress in P. tenera (Wi et al., 2018). In addition, the genes that change under natural hydration and desiccation conditions are involved in protein synthesis, processing, and degradation, photosynthesis, and respiration in P. columbina (Contreras-Porcia et al., 2013). Using dynamic stochastic general equilibrium modeling (DGE) to characterize the tolerance mechanism of P. haitanensis under dehydration and rehydration conditions, it is clear that these differential genes are related to the synthesis of unsaturated fatty acids, transcription factor families, and molecular chaperones (Wang et al., 2015). Unigenes related to glutathione transferase, superoxide dismutase, heat shock proteins, serine/threonine kinases, and phospholipases under osmotic stress are upregulated significantly, which implies that these genes play important roles in the prevention of damage from osmotic stress (Sun et al., 2019). However, little is currently known regarding the molecular mechanisms involved in the response to osmotic stress at the post-transcriptional level.
MicroRNAs (miRNAs) are endogenous small RNAs that are 21–25 nucleotides (nt) long. They play an important role in the regulation of animal and plant growth and development, hormone signaling, programmed cell death, and disease infection (Bartel, 2004; Lin and Gregory, 2015; Thomson and Dinger, 2016; Gao et al., 2019). Recent studies show that miRNAs play important roles in a variety of stress conditions. For example, the expression of miR169, miR394, miR159, miR395, and miR160 is induced when plants are subjected to drought stress, some of which are involved in plant root development and transcriptional regulation; thus, allow the plant to tolerate water stress (Ni et al., 2013). Additionally, the expression of several miRNAs, such as miR396, miR168, miR167, miR165, miR319, miR159, miR394, miR156, miR393, miR171, and miR158 is altered in response to salinity and consequently affects the expression of their targets in Arabidopsis (Liu et al., 2008) and rice (Zhao et al., 2009). Drought stress in wheat enhances protein storage via the regulation of the expression of miRNAs and their target genes (Chen et al., 2017). In Yu et al. (2019), overexpressed miRNA169c in Arabidopsis and discovered that the transgenic strain is more sensitive to drought stress. However, little is known about the osmotic stress response and regulatory molecular mechanisms in intertidal seaweeds compared with that in higher plants.
To understand the molecular mechanisms underlying osmotic acclimation and adaptation to intertidal environments at the post-transcriptional level, a comprehensive analysis of miRNA expression in response to dehydration and rehydration in P. haitanensis based on the genome was performed. This study was undertaken to identify conserved and novel miRNAs expressed in P. haitanensis and characterize osmotic-regulated miRNAs. The results of this study will lay the foundation for the elucidation of the molecular mechanisms involved in osmotic acclimation.
Materials and Methods
Seaweed Culture and Osmotic Treatments
A lab-cultured pure line PH-40 of P. haitanensis was used in the experiments to eliminate the interference caused by genotypic differences. The sporophytes (conchocelis) of this pure line were directly developed from a single somatic cell that was isolated using 2% snail enzyme from a farmed gametophyte collected from Putian, Fujian Province, China. The gametophytes of P. haitanensis were cultured in a light incubator under the following conditions: 20 ± 1°C with 50–60 μmol photons⋅m–2⋅s–1 of illumination during a 12: 12 h light: dark cycle. Provasoli-Enriched Seawater (PES) medium was changed every 3 days. Material processing was performed when the blade was 10–15 cm long. For miRNA identification, gametophytes were exposed to dehydration and rehydration. The gametophytes were harvested in the following conditions: normal conditions as control (CON), 30 ± 5% water loss rate (moderate water loss, MWL), 50% ± 5% (high water loss, HML), and 80% ± 5% (severe water loss, SWL). The gametophytes from SWL were then put back into seawater to rehydrate for 30 min (REH) according to a previous study (Wang et al., 2015). The different water loss levels were determined according to a previous study (Kim et al., 2009). Three biological replicates were performed for each treatment. After treatment, all samples were stored in liquid nitrogen for subsequent RNA extraction.
RNA Extraction and miRNA Library Construction
Total RNA was separately isolated from gametophytes (control and osmotic-treated samples) using Trizol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s protocol. Concentrations and qualities of the isolated RNAs were measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Approximately 2.5 μg of total RNA was used to prepare a small RNA library, according to the protocol of TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, CA, United States). Then, the libraries were sequenced by Illumina Hiseq2500 50SE (single-end) at LC-Bio (Hangzhou, China).
Data Processing
The raw reads were processed using Illumina’s Genome Analyzer Pipeline software (Solexa 0.3) and the ACGT V3.1 program developed by LC Sciences (LC Sciences, Houston, TX, United States). After removing adapter dimers, junk, and low complexity sequences from the small RNA reads, total read counts for the small RNAs were established. Then, small RNAs that mapped to common RNA families [ribosomal RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), and small nucleolar RNA (snoRNA)] were discarded. Subsequently, the remaining sequences were mapped to the assembled genome of P. haitanensis using the Bowtie software (Langmead et al., 2009; Cao et al., 2019). For alignment, one mismatch in the first 16 nt was allowed. Mapped reads were used for further analysis. The mapped sequences that were 18–25 nt long were mapped to specific species precursors in miRBase 21.0 by Bowtie search to identify known miRNAs and novel 3p- and 5p- derived miRNAs. Length variation at 3′ and 5′ ends and one mismatch inside of the sequence were allowed in the alignment. Meanwhile, the remaining sequences that did not match known miRNAs were used to identify novel miRNAs in P. haitanensis.
Differential miRNA Expression
miRNA differential expression based on normalized deep sequencing counts was analyzed by selectively using DeSeq. The miRNA expression level fold-change was calculated using the formula mentioned in a previous report (Marsit et al., 2006): Fold-change = log2 of RPKM value. The significance threshold (p-value) was set at 0.01 and 0.05 for each test. The fold-change (>1) and p-values (p < 0.05) were combined to identify differentially expressed genes, which were defined as osmotic stress-responsive miRNAs.
Target Prediction and Annotation of Differentially Expressed miRNAs
To further understand the function of the differentially expressed miRNAs, computational target prediction algorithms (TargetScan5.0 and miRanda 3.3a) were used to predict the miRNA binding sites (potential target genes) (Yang et al., 2012). The function of the target genes of these differentially expressed miRNAs were annotated using the Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways2 databases.
Verification of miRNA PC-3p-99769 and Its Target CA Using qPCR
To validate the osmotic stress-responsive miRNAs identified by high-throughput sequencing and expression trends, the differentially expressed miRNA and its target gene was confirmed by qPCR (PC-3p-99769_194-ph08980.t1). The gametophytes were collected as described for miRNA library preparation and sequencing. Then, small RNAs (<200 nt) were extracted from the samples following the manufacturer’s instructions for the RNA isolation for Small RNA Kit (TaKaRa). Equal amounts of high-quality small RNAs were reverse transcribed to complementary DNA (cDNA). Reverse transcription (RT) was performed using the PrimeScript RT reagent Kit (TaKaRa, China) according to the manufacturer’s instructions but with specific stem-loop RT primers for miRNAs (Supplementary Table S1). The 5.8S and 18S subunits were used as internal controls for miRNA and mRNAs, respectively. The expression levels of these genes were analyzed by qRT-PCR with a Light-Cycle® 480 Real-Time PCR System. The reactions were performed using 20 μL volumes containing 10 μL of SYBR® Premix Ex Taq (TaKaRa Biotech Co., Dalian, China), 0.6 μL of each primer (forward and reverse primer), 2 μL of diluted cDNA, and 6.8 μL of RNA-free water using the following protocol: 95°C for 30 s, 40 cycles at 95°C for 5 s, 57°C for 30 s, and 72°C for 30 s. The relative expression levels were calculated using the comparative Ct (2–ΔΔCt) method.
Results
Small RNA Sequencing
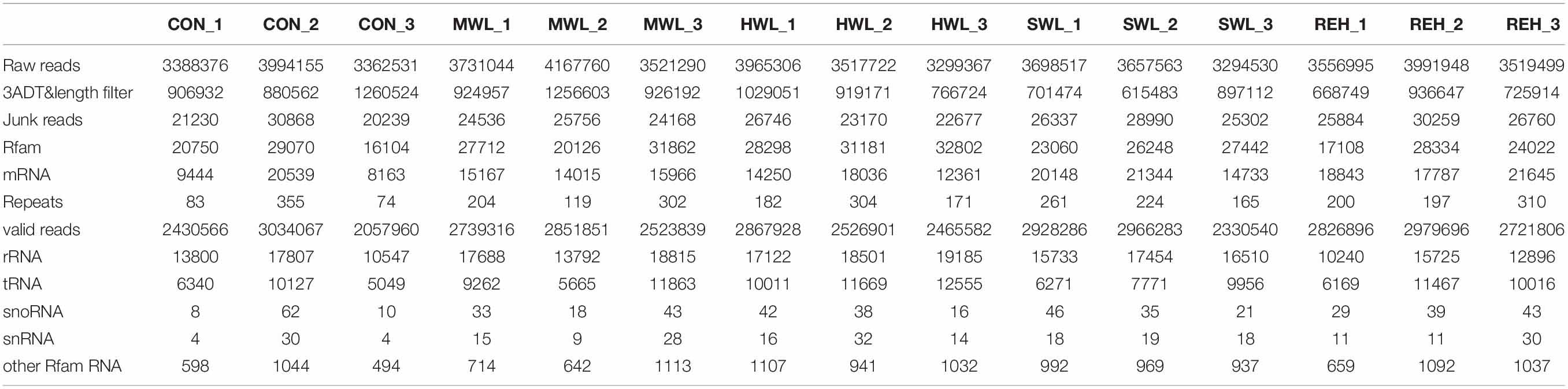
To profile small RNAs and identify the osmotic stress-responsive miRNAs in P. haitanensis, small RNA sequencing for Pyropia gametophytes subjected to varying degrees of dehydration and rehydration (CON, MWL, HWL, SWL, and REH) were performed. After filtering low-quality raw reads and mRNA, Rfam, and Repbase mappable reads, the average reads of these samples were 2,507,531, 2,705,002, 2,620,137, 2,741,703, and 2,842,799 bp, respectively (Table 1). These reads were used for further miRNA identification and prediction.
Identification and Analysis of miRNAs in P. haitanensis
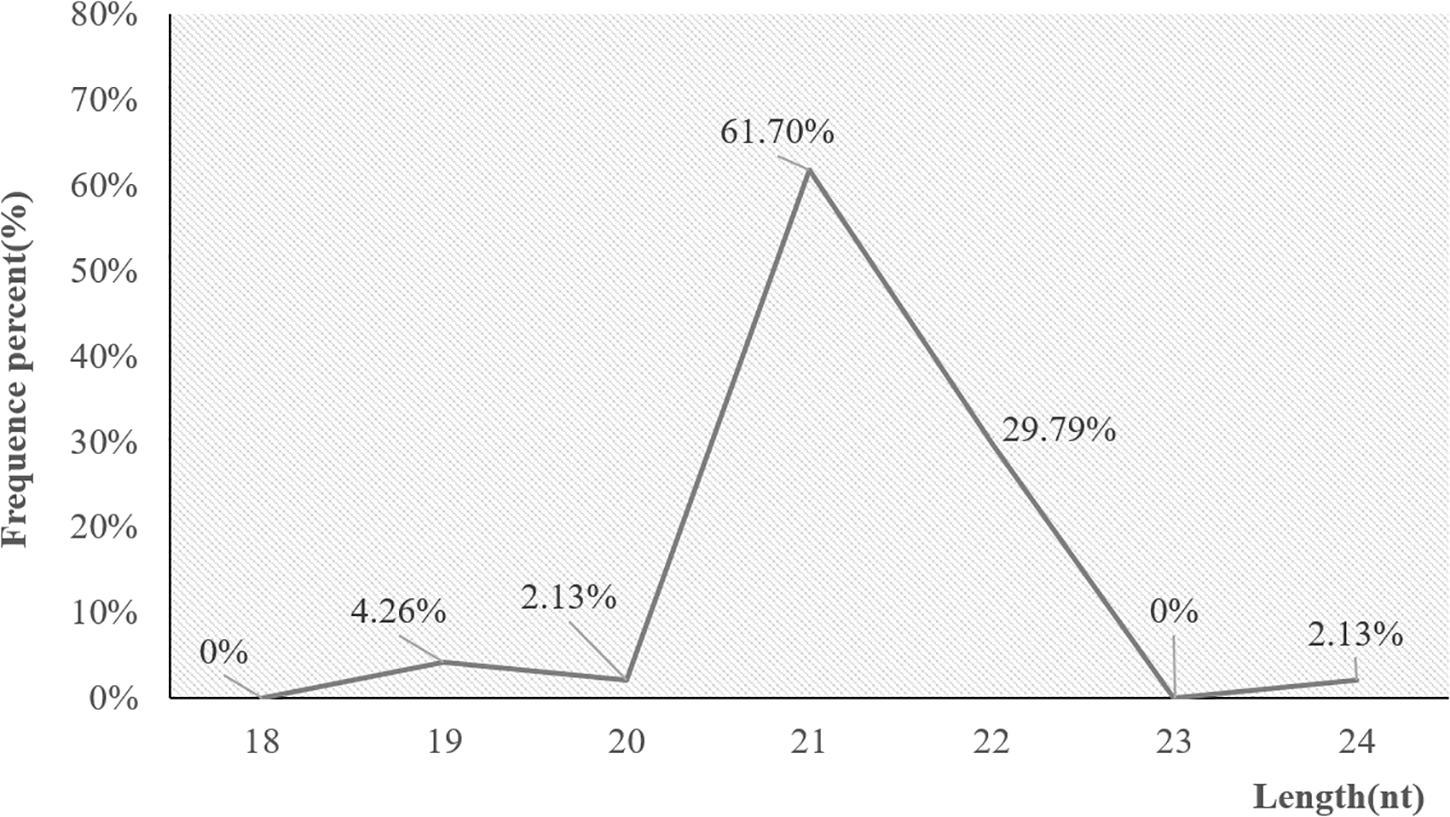
Small RNA sequences were mapped to miRBase and the P. haitanensis genome to identify conserved and novel miRNAs. Forty-seven miRNAs were predicted and 95.74% of these were 20–24 nt long, with the 21 nt as the dominant length (Figure 1). In the first nucleotide bias analysis of miRNAs in P. haitanensis, the first residue of the 19 nt miRNAs was either predominantly uridine (U) or cytosine (C). The first residue of the 20, 21, and 22 nt miRNAs was predominantly U, whereas the 24 nt miRNAs predominantly started with guanine (G).
Among the 47 miRNAs, 2 could be mapped to specific miRNA sequences in miRBase and the extended Pyropia genome sequences from their genomic loci could form hairpins. Thus, they were defined as conserved miRNAs for this study. The other 45 are novel miRNAs (Table 2).
To trace the evolutionary scenario of miRNAs in the red algae lineage, we downloaded the published miRNA data for the Pyropia species P. yezoensis and the sister red algal clade Floriceadens Chondrus crispus for phylogenetic comparative analysis. We discovered that the pre-miRNA sequences of 15 miRNAs could be aligned to both P. yezoensis and Porphyra umbilicalis, which indicated that these sequences were well conserved in Pyropia species. Only two were also observed in C. crispus (Supplementary Table S2). The target genes of these conserved miRNAs are mainly involved in DNA synthesis (DNA repair proteins and DNA mismatch repair proteins), signal pathways (mitogen-activated protein kinase and serine/threonine protein kinase), transporters (ABC transport system and zinc transporter proteins), and some transcriptional regulators.
Differential Expression Analysis of miRNAs
Many genes undergo significant expressional variation in response to osmotic stress (Wang et al., 2015). Our identification of miRNAs in the P. haitanensis genome caused us to investigate whether miRNAs contribute to the post-transcriptional regulation of key genes that lead to physiological modulation to adapt to stressful conditions. Therefore, we further investigated the expression levels of miRNAs under different degrees of dehydration and subsequent rehydration. Twelve miRNAs exhibited significant variation in at least one dehydrated or rehydrated condition (Figure 2). According to their transcriptional patterns along the time course of osmotic stresses, they can be classified into three clusters (Figure 2). In cluster I, three miRNAs exhibited slight variations in response to most of the stress conditions, while the levels of PC-3P-45934_389 and PC-3P-75147_255 were transiently down-regulated in HWL and SWL, respectively. There were four miRNAs in cluster II. Although there was no significant change in their expression under dehydrated conditions, they were induced when Pyropia thalli were rehydrated after severe water loss. The final five miRNAs were grouped in cluster III. All of which showed a sharp increase in expression when Pyropia underwent water loss, except for PC-5P-180717_97, which was upregulated only until HWL. When rehydrated, the levels of most of the miRNAs in cluster III barely changed, and only PC-5p-210644_78 showed marked downregulation.
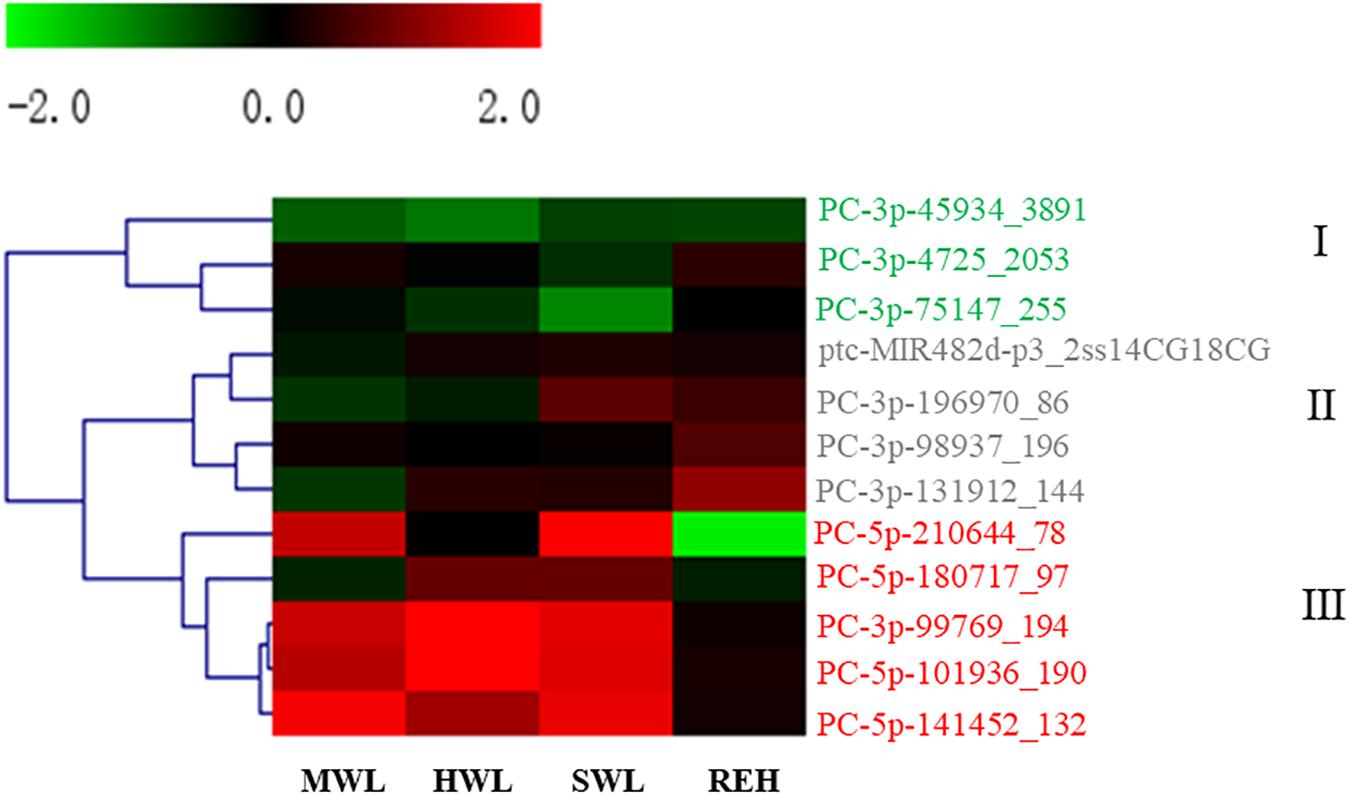
Figure 2. Expressional patterns of miRNAs during desiccated stresses. CON, Normal conditions (control); MWL, 30 ± 5% water loss rate (moderate water loss); HML, 50% ± 5% (severe water loss); SWL, 80% ± 5% (Severe water loss); REH, the gametophytes from SWL were put back into seawater to rehydrate for 30 min. The expressional variation was represented by fold-change = log2(FPKM under stress/FPKM under control). The miRNAs in cluster I, II, and III have been depicted in green, gray and red, respectivley.
PC-3p-99769_194 Regulated Photosynthetic Carbon Fixation Under Dehydration via the Repression of the Expression of Carbonic Anhydrase (CA)
To understand the specific functions of these differentially expressed miRNAs under osmotic stress in P. haitanensis, we predicted the target genes of differentially expressed miRNAs and analyzed the enriched functional categories that they encoded in terms of GO and KEGG pathways. Overall, 60 target genes were predicted for 12 miRNAs. According to the GO classification system, these genes were classified into molecular function categories related to catalytic activity, binding, transporter activity, transcription factor activity, signal transducer activity, and antioxidant activity (Supplementary Table S3). The results of KEGG metabolic pathway annotation showed that cell cycle, meiosis, nucleotide excision repair, DNA replication, glycerolipid metabolism, and base excision repair pathways were significantly enriched (P < 0.01) (Supplementary Table S3). Nucleotide excision repair, base excision repair, cell cycle, and DNA replication were also significantly enriched in differentially transcribed genes under osmotic stress in P. haitanensis as previously described (Wang et al., 2015). The consistency between the two datasets supports the regulatory role of miRNAs in response to osmotic stress.
miRNA PC-3p-99769_194 from cluster III was predicted to bind to the 523–542 bp region of the CA gene and may regulate its expression by degrading mRNA. The miRNA PC-3p-99769_194 was upregulated during the whole dehydrated process. Accordingly, the target CA gene had declined in transcriptional activity as observed in the published transcriptomic data under dehydration in P. haitanensis. The CA gene exhibited 0.77- and 0.44-fold downregulation under 30 and 80% dehydration and 1.3-fold upregulation under rehydration conditions.
PC-5p-335121_37 and PC-5p-350045_35 were highly expressed in samples under dehydration stress. The target genes of PC-5p-335121_37 are ph07921.t1 and ph07931.t1 which encode the U5 snRNP complex subunit and superoxide dismutase, respectively. PC-5p-350045_35 targets cid-thiol ligase and glycosyltransferase (ph09862.t1 and ph00598.t1), respectively. PTC-MIR482D-P3_2SS14CG18CG exhibited upregulated expression in rehydration samples. The target gene of ptc-MIR482D-P3_2SS14CG18CG is ph04985.t1, which encodes epoxyqueuosine reductase (Supplementary Table S3).
Validation of the Expression of PC-3p-99769_194 and Its Target CA Using qPCR
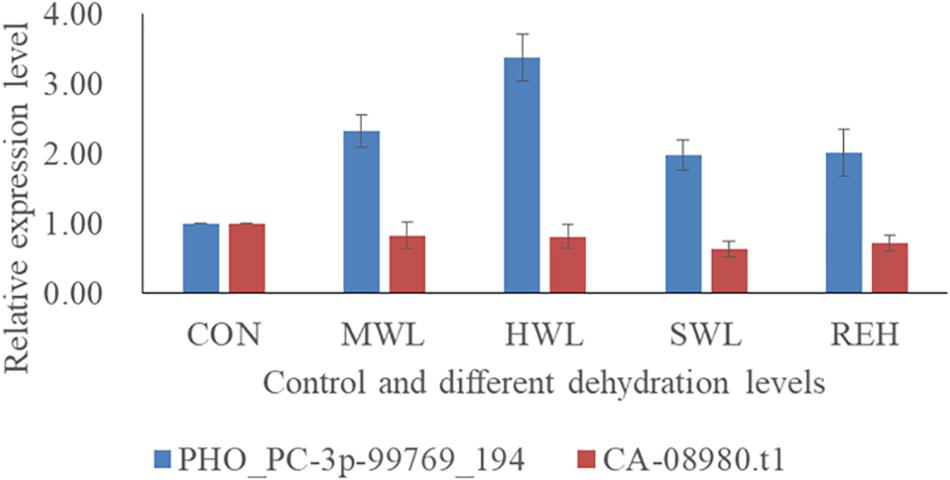
To validate the expression of miRNAs obtained from the high throughput sequencing approach, qPCR was performed on three miRNAs among samples of CON, MWL, HWL, SWL, and REH. PC-3p-99769_194 exhibited 2. 21-, 3. 46-, 1. 71-, and 1.83-fold upregulation in P. haitanensis after MWL, HWL, SWL, and REH, respectively. The expression of PC-5p-350045_35 in dehydration and rehydration samples were higher than those in the control (Figure 3). Its target gene CA (ph0980.t1) was downregulated 0. 87-, 0. 84-, and 0. 51-fold during MWL, HWL, SWL, respectively. And underwent a slight 1.39-fold increase during REH. These inverse transcriptional profiles suggest that PC-3p-99769_194A might function as a negative regulator of CA expression.
Small RNA Biogenesis and Function Apparatus in P. haitanensis
To unravel the RNA silencing machinery for miRNA generation and function in Pyropia, we identified genes encoding the two proteins that mediate the central activities required for miRNA and small RNA biogenesis and functioning, Dicer and Argonaute (AGO), respectively. Dicer proteins are characterized in many organisms, have multiple functional domains from the N- to C-terminus, and are approximately 2000 amino acids (aa) long. In P. haitanensis, we performed a Basic Local Alignment Search Tool (BLAST) search against the genome using Dicer proteins in model organisms as the query and discovered two candidate genes that exhibited high sequence similarity (ph07342.t1 and ph05047.t1 at 562 aa and 1163 aa long, respectively). Ph07342.t1 contains a DEAD-like helicase domain (DEXDc) and a helicase C (HELIc) domain and was aligned to the N-terminal part of reference Dicer. Ph05047.t1 has two parallel RIBO (ribonuclease III) domains. The two genes are located in two separate loci and are not adjacent to each other, which is different from the gene arrangement in other species (Figure 4). This phenomenon was verified in Po. umbilicalis. However, red macroalgae C. crispus and Gracilariopsis chorda, as well as the unicellular red algae Porphyridium purpureum, have one full-length canonical Dicer gene with multiple functional domains. To trace the evolutionary history of the two segmented Dicer genes, we identified Dicer homologs in available species and performed phylogenetic analyses. In the evolutionary trees constructed based on model Dicer sequences for ph07342.t1 and ph05047.t1, both closely clustered with the two Po. umbilicalis proteins and formed a unique branch, whereas the Dicer proteins from other species united as a single group (Supplementary Figure S1). For AGO proteins, there were 1, 2, 1, 3, and 3 candidate genes found in P. haitanensis, Po. umbilicalis, G. chorda, C. crispus, and P. purpureum genomes, respectively. All had canonical structures with an AGO-specific domain, a PAZ domain, and a PIWI domain at the C-terminal. P. haitanensis AGO exhibited close phylogeny with homologs in other red algae (Supplementary Figure S2). Conserved amino acids in the PIWI domain that facilitate siRNA binding were observed in P. haitanensis AGO (Figure 5). Moreover, it harbored a classical glutamate as a key residue of the catalytic DDE motif in RNase H. The DDH motif is required for metal ion coordination in the catalytic center, albeit “H” residue was not found in the corresponding locus (Figure 5). The identification of the key components of miRNA biogenesis machinery in P. haitanensis, as well as their expressional variation under desiccation, indicated the potential involvement of an miRNA-based regulatory mechanism in response to osmotic stress and gene engineering via artificial miRNA and siRNA. Although AGO that exhibited contractive gene copies, albeit well-conserved sequences, the distinctive structure of Dicer proteins lets us envisage some unique features possibly existing in Pyropia and Porphyra. In addition, we searched the transcriptional levels of two Dicer genes and the AGO gene in the published transcriptomic data under dehydration in P. haitanensis and discovered that the two Dicer genes were increased under dehydration and rehydration conditions. The AGO gene was 1.14-fold upregulated under medium dehydration, whereas AGO was 0.69- and 0.52-fold downregulated under severe dehydration and rehydration conditions.
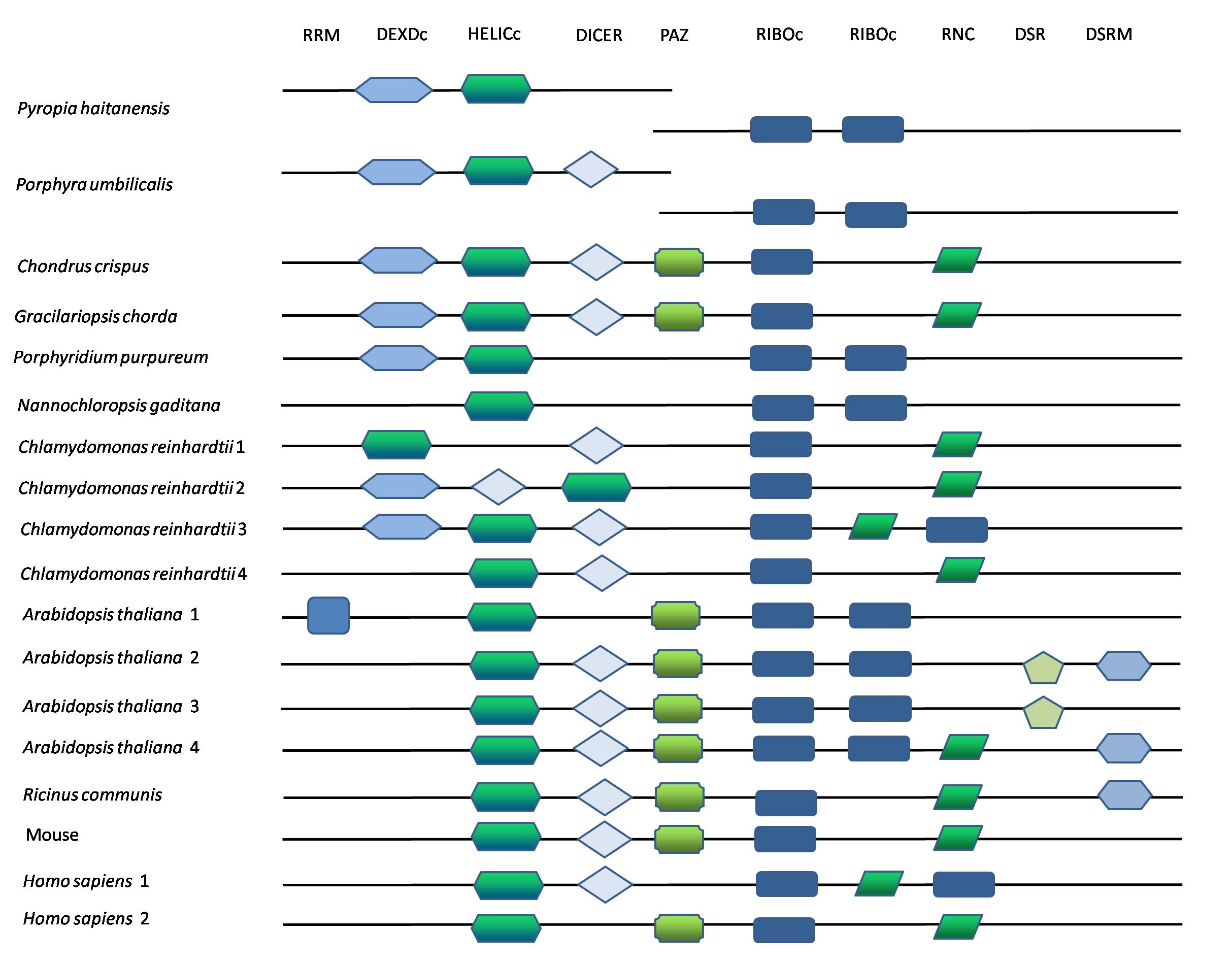
Figure 4. Structural features of Dicers in P. haitanensis and other species. Each conserved domains were represented as different shapes. RRM, RNA recognition motif; DEXDc, DEAD-like helicase domain; HELICc, helicase C; RIBOc, Ribonuclease III C terminal domain; RNC, dsRNA-specific ribonuclease.

Figure 5. Sequence alignment of Argonaute PIWI domains. Accession numbers of AGOs in P. haitanensis and other species were listed in Supplementary Table S4. Residues highlighted in dark gray are shared ≥75% similarities, media gray are those shared ≥50% similarities, light gray are those shared ≥30% similarities in all sequences. Conserved amino acids were marked as N/Q, D/E, R/K, S/T, F/Y, A/G, and V/I/L/M in this figure. D, D and H amino acid residues in bold letters represent the DDH motif. Italicized residules are class-switch residues that attribute a given protein either to the (p)iwi or to the (a)rgonaute subfamily.
Discussion
miRNAs Regulate Carbon Fixation by Downregulating CA Under Osmotic Stress
The expression of miRNAs, an important class of gene regulators, is altered by abiotic stress treatment such as light, temperature, moisture, salt, and heavy metal ions (Jia et al., 2009; Lv et al., 2010). However, as most of these studies were performed using model organisms, little is known about seaweeds. CA catalyzes the interconversion between carbon dioxide and dissolved bicarbonate. It is the key component of the carbon concentration mechanism in aquatic photosynthetic organisms by facilitating the accumulation of sufficient carbon dioxide in the vicinity of rubisco (Badger and Price, 1994). However, the exact molecular mechanism involved in the control of the expression of CA is unclear. In this study, we demonstrated a post-transcriptional regulatory mechanism of CA and identified an miRNA as the key element involved in the regulation of the expression of the CA gene. The raised level of expression of PC-3p-99769_194 under dehydration stress suggested its increased negative regulation of the target CA gene. This is further supported by the decreased transcriptional level of CA gene in our previous study (Wang et al., 2015). Three enzymes involved in photosynthesis (rubisco, fumarate and nitrate reductase, and CA are downregulated in desiccated individuals compared to naturally hydrated controls by 2DE and LC-MS/MS analyses (López-Cristoffanini et al., 2015). Under high salinity treatment, the algae Nitzschia closterium f. minutissima adjusts its photosynthetic pigment content to decrease photosynthesis and inhibits the activity of CA (Yu et al., 2011). Combined with our study, we speculate that Pyropia reduce their photosynthetic activities in response to osmotic stress via miRNA-based regulatory mechanisms.
Rehydration can induce various metabolic processes that responding to osmotic stress in seaweed. The target gene of upregulated ptc-MIR482D-P3_2SS14CG18CG is related to epoxyqueuosine reductase, which catalyzes the last step of the two-electron reduction of epoxyqueuosine to queuosine during the synthesis of tRNA (Nishimura, 1983) and plays an important role in translational efficiency, accuracy, and structural stabilization of tRNA (El Yacoubi et al., 2012). In our study, genes encoding epoxyqueuosine reductase showed high expression when P. haitanensis was subjected to osmotic stress and its expression was suppressed during rehydration. Salt treatment in Anabaena fertilissima leads to the hyperaccumulation of epoxyqueuosine reductase (Rai and Swapnil, 2019). Therefore, we speculate that suppressed miRNA leads to the increased expression of epoxyqueuosine reductase in response to osmotic stress in Pyropia.
The miRNA Biogenesis in P. haitanensis
Dicer and AGO are the key components in miRNA biogenesis and are characterized in almost all organisms that have miRNA regulatory mechanisms (Bernstein et al., 2003; Yang et al., 2013). Both have well-conserved homologs with similarly organized functional domains and motifs. However, in this study, we identified that in Pyropia, Dicer was divided into two genes, ph07342.t1 that harbors the N-terminal functional domains and an open reading frame (ORF) that contains the other domains. The segmentation of Dicer was also observed in other Bangiales including P. yezoensis and Po. umbilicalis, whereas in the Florideophyceae genomes, such as C. crispus, G. chorda, and P. purpureum, full-length canonical Dicer was present. This particular distribution of segmented Dicer protein suggested that it might be generated after the evolutionary diversification of Bangiales. The length of a small RNA is mainly related to the type of enzymes involved in its processing. For example, a 21 nt segment is generally generated by Dicer-like 1 (DCL1), a 22 nt segment by RNase endonuclease, and a 24 nt segment by Dicer-like 3 (DCL3) (Xie et al., 2004; Vazquez, 2006; Ou et al., 2012). The characteristic length distribution of small RNAs, with 21 nt as the dominant length in Pyropia, suggests that Pyropia Dicer protein might function in a DCL1-like way in miRNA biogenesis.
Partial and truncated Dicer with a C-terminal fragment was observed in Caenorhabditis elegans before and characterized to be a negative regulator of miRNA biogenesis via its competitive interaction with AGO proteins (Sawh and Duchaine, 2013). However, the truncated small Dicer was generated by in vivo proteolytic cleavage, rather than by alternative splicing or individual “truncated” genes as in Pyropia. Thus, we envisage that they might function in a different way. The action mode is “two-peptides,” which means that the two peptides encoded by the two ORFs serve as the “subunits” and form a heterodimer. In Pyropia AGO, although a close phylogenetic relationship with its red algal counterparts was observed, it was encoded by only one gene, whereas three were observed in other red algae. Therefore, the segmented structure of Dicer and the contracted copy numbers of AGO represented the distinctive feature of miRNA biogenesis in Pyropia and imply a diverse miRNA regulatory mechanism. The latter was further reinforced by the dominant length of miRNA and a relative lack of universally conserved miRNAs.
Conclusion
To investigate the miRNA-based regulatory mechanisms in response to desiccation stress in Pyropia, we profiled the miRNAs from a series of dehydrated and rehydrated Pyropia thalli by high-throughput sequencing. Analysis of their transcriptional variations, as well as target genes, revealed that miRNAs play essential roles in suppressing carbon fixation by downregulating the CA gene to survive in the intertidal zone. The genetic structural and phylogenetic analysis of Dicer and AGO proteins disclosed the distinctive features of miRNA-mediated mechanism in Pyropia. Our discoveries provide valuable information for further functional characterization of miRNAs in response to abiotic stress in Pyropia and shed light on the evolution of miRNA-mediated mechanism in algae.
Data Availability Statement
The sequencing data have been deposited into the NCBI Sequence Read Archive under the BioProject: PRJNA573823.
Author Contributions
YM conceived the study. MC performed the experiments. KX, FK, and JW analyzed the results. MC and DW wrote the manuscript.
Funding
This work was financially supported by the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2018SDKJ0302-4), the National Key R&D Program of China (No. 2018YFD0900106), and the Shandong Natural Science Foundation (No. ZR2019MC012).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00766/full#supplementary-material
FIGURE S1 | Phylogenetic relationships of Dicer genes.
FIGURE S2 | Phylogenetic relationships of AGO genes.
TABLE S1 | Specific primers for miRNAs and mRNAs.
TABLE S2 | Conserved miRNAs in Pyropia and C. crispus.
TABLE S3 | Significant expressed miRNAs and the function of their target genes.
TABLE S4 | The AGO and Dicer sequences used in this study.
Footnotes
References
Badger, M. R., and Price, G. D. (1994). The role of carbonic anhydrase in photosynthesis. Annu Rev. Plant Biol. 45, 369–392. doi: 10.1146/annurev.arplant.45.1.369
Bernstein, E., Kim, S. Y., Carmell, M. A., Murchison, E. P., Alcorn, H., Li, M. Z., et al. (2003). Dicer is essential for mouse development. Nat. Genet. 35, 215–217. doi: 10.1038/ng1253
Blouin, N. A., Brodie, J. A., Grossman, A. C., Xu, P., and Brawley, S. H. (2011). Porphyra: a marine crop shaped by stress. Trends Plant Sci. 16, 29–37. doi: 10.1016/j.tplants.2010.10.004
Cao, M., Xu, K., Yu, X., Bi, G., Liu, Y., Kong, F., et al. (2019). A chromosome-level genome assembly of Pyropia haitanensis (Bangiales, Rhodophyta). Mol. Ecol. Resour. doi: 10.1111/1755-0998.13102 [Epub ahead of print].
Chen, X. Y., Yang, Y., Ran, L. P., Dong, Z. D., Zhang, E. J., Yu, X. R., et al. (2017). Novel insights into miRNA regulation of storage protein biosynthesis during wheat caryopsis development under drought stress. Front. Plant Sci. 8:1707. doi: 10.3389/fpls.2017.01707
Cock, J. M., and Coelho, S. M. (2011). Algal models in plant biology. J. Exp. Bot. 62, 2425–2430. doi: 10.1093/jxb/err117
Contreras-Porcia, L., López-Cristoffanini, C., Lovazzano, C., Flores-Molina, M. R., Thomas, D., Núñez, A., et al. (2013). Differential gene expression in Pyropia columbina (Bangiales, Rhodophyta) under natural hydration and desiccation conditions. Lat. Am. J. Aquat. Res. 41, 933–958. doi: 10.3856/vol41-issue5-fulltext-13
Davison, I. R., and Pearson, G. A. (1996). Stress tolerance in intertidal seaweeds. J. Phycol. 32, 197–211. doi: 10.1111/j.0022-3646.1996.00197.x
El Yacoubi, B., Bailly, M., and de Crecy-Lagard, V. (2012). Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Ann. Rev. Genet. 46, 69–95. doi: 10.1146/annurev-genet-110711-155641
Gao, C., Cai, X., Fu, Q., Yang, N., Song, L., Su, B., et al. (2019). Dynamics of MiRNA Transcriptome in Turbot (Scophthalmus maximus L.) Intestine Following Vibrio anguillarum Infection. Mar. Biotechnol. 21, 550–564. doi: 10.1007/s10126-019-09903-z
Hwang, M. S., Chung, I. K., and Oh, Y. S. (1997). Temperature responses of Porphyra tenera Kjellman and P. yezoensis Ueda (Bangiales. Rhodophyta) from Korea. Algae 12, 207–207.
Jia, X., Wang, W. X., Ren, L., Chen, Q. J., Mendu, V., Willcut, B., et al. (2009). Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol. Biol. 71, 51–59. doi: 10.1007/s11103-009-9508-8
Kim, J. K., Kraemer, G. P., and Yarish, C. (2009). Comparison of growth and nitrate uptake by New England Porphyra species from different tidal elevations in relation to desiccation. Phycological Res. 57, 152–157. doi: 10.1111/j.1440-1835.2009.00533.x
Langmead, B., Trapnell, C., Pop, M., and Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. doi: 10.1186/gb-2009-10-3-r25
Lin, S., and Gregory, R. I. (2015). MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 15, 321–333. doi: 10.1038/nrc3932
Liu, H. H., Tian, X., Li, Y. J., Wu, C. A., and Zheng, C. C. (2008). Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843. doi: 10.1261/rna.895308
López-Cristoffanini, C., Zapata, J., Gaillard, F., Potin, P., Correa, J. A., and Contreras-Porcia, L. (2015). Identification of proteins involved in desiccation tolerance in the red seaweed Pyropia orbicularis (Rhodophyta, Bangiales). Proteomics 15, 3954–3968. doi: 10.1002/pmic.201400625
Lv, D. K., Bai, X., Li, Y., Ding, X. D., Ge, Y., Cai, H., et al. (2010). Profiling of cold-stress-responsive miRNAs in rice by microarrays. Gene 459, 39–47. doi: 10.1016/j.gene.2010.03.011
Marsit, C. J., Eddy, K., and Kelsey, K. T. (2006). MicroRNA responses to cellular stress. Cancer Res. 66, 10843–10848. doi: 10.1158/0008-5472.can-06-1894
Nakamura, Y., Sasaki, N., Kobayashi, M., Ojima, N., Yasuike, M., Shigenobu, Y., et al. (2013). The first symbiont-free genome sequence of marine red alga. Susabi-nori (Pyropia yezoensis). PloS One 8:e57122. doi: 10.1371/journal.pone.0057122
Ni, Z., Hu, Z., Jiang, Q., and Zhang, H. (2013). GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. doi: 10.1007/s11103-013-0040-5
Nishimura, S. (1983). “Structure, biosynthesis, and function of queuosine in transfer RNA,” in Progress in Nucleic Acid Research and Molecular Biology, Vol. 28, ed. W. E. Cohn, (New York, NY: Academic Press), 49–73. doi: 10.1016/s0079-6603(08)60082-3
Ou, J., Meng, Q., Li, Y., Xiu, Y., Du, J., Gu, W., et al. (2012). Identification and comparative analysis of the Eriocheir sinensis microRNA transcriptome response to Spiroplasma eriocheiris infection using a deep sequencing approach. Fish Shellfish Immunol. 32, 345–352. doi: 10.1016/j.fsi.2011.11.027
Rai, A. K., and Swapnil, P. (2019). Proteomic analysis of the salt-adapted and directly salt-(NaCl and NaCl+Na2SO4 mixture) stressed cyanobacterium Anabaena fertilissima. J. Appl. Phycol. 31, 1185–1196. doi: 10.1007/s10811-018-1607-y
Reed, R. H., Collins, J. C., and Russell, G. (1980). The effects of salinity upon galactosyl-glycerol content and concentration of the marine red alga Porphyra purpurea (Roth) C. Ag. J. Exp. Bot. 31, 1539–1554. doi: 10.1093/jxb/31.6.1539
Sahoo, D., Tang, X., and Yarish, C. (2002). Porphyra–the economic seaweed as a new experimental system. Curr. Sci. 83, 1313–1316.
Sawh, A. N., and Duchaine, T. F. (2013). A truncated form of dicer tilts the balance of RNA interference pathways. Cell Rep. 4, 454–463. doi: 10.1016/j.celrep.2013.07.013
Sun, P., Tang, X., Bi, G., Xu, K., Kong, F., and Mao, Y. (2019). Gene expression profiles of Pyropia yezoensis in response to dehydration and rehydration stresses. Mar. Genom. 43, 43–49. doi: 10.1016/j.margen.2018.09.005
Thomson, D. W., and Dinger, M. E. (2016). Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17, 272–283. doi: 10.1038/nrg.2016.20
Vazquez, F. (2006). Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 11, 460–468. doi: 10.1016/j.tplants.2006.07.006
Wang, L., Mao, Y., Kong, F., Cao, M., and Sun, P. (2015). Genome-wide expression profiles of Pyropia haitanensis in response to osmotic stress by using deep sequencing technology. BMC Genom. 16:1012. doi: 10.1186/s12864-015-2226-5
Wi, J., Jung, H. S., Im, S., Yang, S., Park, E. J., Hwang, M. S., et al. (2018). A nuclear fucosyltransferase-like protein, PtFUT, from marine red alga Pyropia tenera (Rhodophyta) confers osmotic stress tolerance. J. Appl. Phycol. 30, 717–727. doi: 10.1007/s10811-017-1241-0
Xie, Z., Johansen, L. K., Gustafson, A. M., Kasschau, K. D., Lellis, A. D., Zilberman, D., et al. (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2:E104. doi: 10.1371/journal.pbio.0020104
Yang, G., Yang, L., Zhao, Z., Wang, J., and Zhang, X. (2012). Signature miRNAs involved in the innate immunity of invertebrates. PloS One 7:e39015. doi: 10.1371/journal.pone.0039015
Yang, Y., Zhong, J., Ouyang, Y. D., and Yao, J. (2013). The integrative expression and co-expression analysis of the AGO gene family in rice. Gene 528, 221–235. doi: 10.1016/j.gene.2013.07.002
Yu, J. L., Xia, J. R., and Zou, Y. D. (2011). Response of carbonic anhydrase activity and photosynthesis to high salinity stress in Nitzschia closterium f. minutissima. J. Fisheries China 35, 515–523.
Yu, Y., Ni, Z., Wang, Y., Wan, H., Hu, Z., Jiang, Q., et al. (2019). Overexpression of soybean miR169c confers increased drought stress sensitivity in transgenic Arabidopsis thaliana. Plant Sci. 285, 68–78. doi: 10.1016/j.plantsci.2019.05.003
Zhang, X. C., Qin, S., Ma, J. H., and Xu, P. (2005). “Genetics of marine red algae — Gracilaria,” In The Genetics of Marine Algae[J]. ed. Z Y Lin, (Beijing: China Agriculture Press), 459–464.
Keywords: Pyropia haitanensis, genomics, miRNA, osmotic stress, Dicer, AGO
Citation: Cao M, Wang D, Kong F, Wang J, Xu K and Mao Y (2019) A Genome-Wide Identification of Osmotic Stress-Responsive MicroRNAs in Pyropia haitanensis (Bangiales, Rhodophyta). Front. Mar. Sci. 6:766. doi: 10.3389/fmars.2019.00766
Received: 24 September 2019; Accepted: 27 November 2019;
Published: 20 December 2019.
Edited by:
Jianhua Fan, East China University of Science and Technology, ChinaReviewed by:
Jin Sun, Hong Kong University of Science and Technology, Hong KongXiaodong Wang, Jinan University, China
Copyright © 2019 Cao, Wang, Kong, Wang, Xu and Mao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongmei Wang, d2FuZ2RtQG91Yy5lZHUuY24=; Yunxiang Mao, eXhtYW9AaG50LmVkdS5jbg==
†These authors have contributed equally to this work
 Min Cao
Min Cao Dongmei Wang
Dongmei Wang Fanna Kong1,2
Fanna Kong1,2 Junhao Wang
Junhao Wang Kuipeng Xu
Kuipeng Xu