- 1College of Forestry and Landscape Architecture, Anhui Agricultural University, Hefei, China
- 2Key Laboratory of Eutrophication and Red Tide Prevention of Guangdong Higher Education Institutes, College of Life Science, Jinan University, Guangzhou, China
- 3College of Horticulture, Anhui Agricultural University, Hefei, China
- 4Key Laboratory of High Magnetic Field and Ion Beam Physical Biology, Hefei Institutes of Physical Science, Chinese Academy of Sciences, Hefei, China
Burgeoning demand for long-chain polyunsaturated fatty acids (LC-PUFA) due to their established pharmacological and economic significances along with the declining sources of fish urge to explore the sustainable sources of LC-PUFA. Being the predominant LC-PUFA source for marine fishes at the base of the aquatic food web, microalgae has been hailed as a promising natural source for LC-PUFA. However, the potential of algal systems to overproduce LC-PUFA via metabolic engineering is warranted to meet the ever-increasing demand. In this study, we identified and overexpressed Δ6-desaturase, the key enzyme involved in fatty acid desaturation and exemplified its potential on elevating PUFA and lipid content in Nannochloropsis oceanica.Δ6-desaturase overexpression enhanced growth and photosynthetic efficiency. Transgenic cells exhibited a remarkable increase in EPA content and reached up to 62.35 mg/g DW, the highest EPA production in transgenic N. oceanica by expressing a single key enzyme without impeding growth. Total lipid content was significantly increased by 1.7-fold in the transgenic cell than WT. Together, these findings exemplify a potential candidate for LC-PUFA overproduction and also open a new avenue for sustainable production of microalgal PUFAs.
Introduction
Long-chain polyunsaturated fatty acids (LC-PUFA; ≥C20) are the predominant structural molecules of biological membranes and pivotal dietary compounds that play a potential role in human mental and physical developmental processes thereby garnered significant research attention (Kabeya et al., 2018). These PUFAs are primarily synthesized by marine photosynthetic microalgae and consequently accumulated into fish lipids by the aquatic food chain. Presently, marine fish are considered the primary sources for LC-PUFA, however, declining fisheries and contamination of toxic pollutants have provided the impetus to explore a sustainable alternative to depleting conventional sources (Wang et al., 2017). Earlier studies have shown that various photosynthetic microalgae naturally biosynthesize high amount of LC-PUFA (Khozin-Goldberg et al., 2011). In microalgae, LC-PUFA biosynthesis is initiated by Δ-9 desaturase to introduce first unsaturation in saturated stearic acid (C18:0) to yield oleic acid (18:1, n-9), which is subsequently unsaturated into LA by Δ-12 desaturase. Thereafter, the unsaturation of LA to produce γ-linolenic acid (GLA) is catalyzed by Δ6-desaturase, which could be further elongated to produce LC-PUFA. Despite the involvement of various key proteins, Δ6-desaturase of the ω-3 pathway has been considered as a unique rate-limiting enzyme in EPA biosynthesis that present in algae but not in plants (Zhu et al., 2017).
Among various algae, Nannochloropsis sps. are known to produce a remarkable amount of LC-PUFA and considered to be the predominant natural source of valuable PUFAs such as arachidonic acid (ARA; C20:4 Δ5,8,11,14), eicosapentaenoic acid (EPA, C20:5 Δ5,8,11,14,17) and docosahexaenoic acid (DHA; C22:6 Δ4,7,10,13,16,19) (Chen et al., 2013) and they thus garnered huge research attention for producing LC-PUFA. However, the LC-PUFA accumulation in native strains is incapable of meeting the burgeoning global demand for LC-PUFA which generate an impetus to develop a potential strategy to obviate this critical bottleneck to overproduce LC-PUFA in order to meet the contemporary LC-PUFA demand. Consequently, previous studies have employed nutrient limitation and sequential treatment to enhance the content of polyunsaturated fatty acids in N. oceanica (Wang et al., 2019; Yuan et al., 2019). Nevertheless, reduced overall yield and productivity under nutrient deprivation conditions and the cost account for establishing the combinatorial treatment strategy have provoked the necessities to genetically improve the algal strain to enhance lipid content without compromising growth and overall productivity. Genetic engineering has been hailed as the potential strategy for enhancing the titer of desired products by precisely perturb the target metabolic node(s), thereby rewire the metabolic circuit and thus increase the titer of the desired product without constraining the cellular physiological properties. Previous studies have attempted to increase LC-PUFA content (Chen et al., 2013; Kaye et al., 2015; Poliner et al., 2018). Nevertheless, promising strategies to overproduce LC-PUFA without impeding cellular biomass are yet to be explored and specifically, characterization of Δ6-desaturase is yet to be elucidated in Nannochloropsis, despite its functional significance. In this study, we identified and overexpressed the Δ6-desaturase and elucidated its crucial role in enhancing LC-PUFA and cellular physiological properties in N. oceanica.
Materials and Methods
Strain and Culture Conditions
Nannochloropsis oceanica CCAP 849/10 was purchased from NCMA (National Center for Marine Algae and Microbiota) and cultivated in the filter-sterilized f/2 medium at 22 ± 1°C with an irradiance of 200 μmol photons m–2s–1 under 12/12 h light/dark photoperiod (Li et al., 2016).
NoD6 Cloning and Construction of Recombinant Expression Vector
Total RNA was prepared from N. oceanica using RNA/DNA/Protein isolation kit (Omega, United States) and subsequently, retro-transcribed into cDNA using the Prime ScriptTM RT reagent kit (Takara, China). The full-length coding region of Δ6-desaturase (referred to as NoD6) was amplified using the primers NOD6-F and NOD6-R (Table 1) and the amplicon was purified using EZNA gel extraction kit (Omega, United States). The resultant NoD6 amplicon was cloned into pNa03 under the control of fcpC promoter and fcpA terminator using ClonExpress II One Step kit (Vazyme, China) according to the manufacturer’s specification (Figure 1A). Besides, an omega motif leader sequence was cloned between promoter and the transgene to facilitate its translation. The recombinant expression vector was linearized and electroporated into N. oceanica by using a Bio-Rad Gene Pulser Xcell electroporator following the protocol described by Li et al. (2016). Thereafter, the electroporated cells were harvested and preliminarily screened on the solid medium containing zeocin (5 μg/ml, Invitrogen) and the screened cells were cultivated in the selection liquid f/2 medium as described elsewhere (Li et al., 2016).
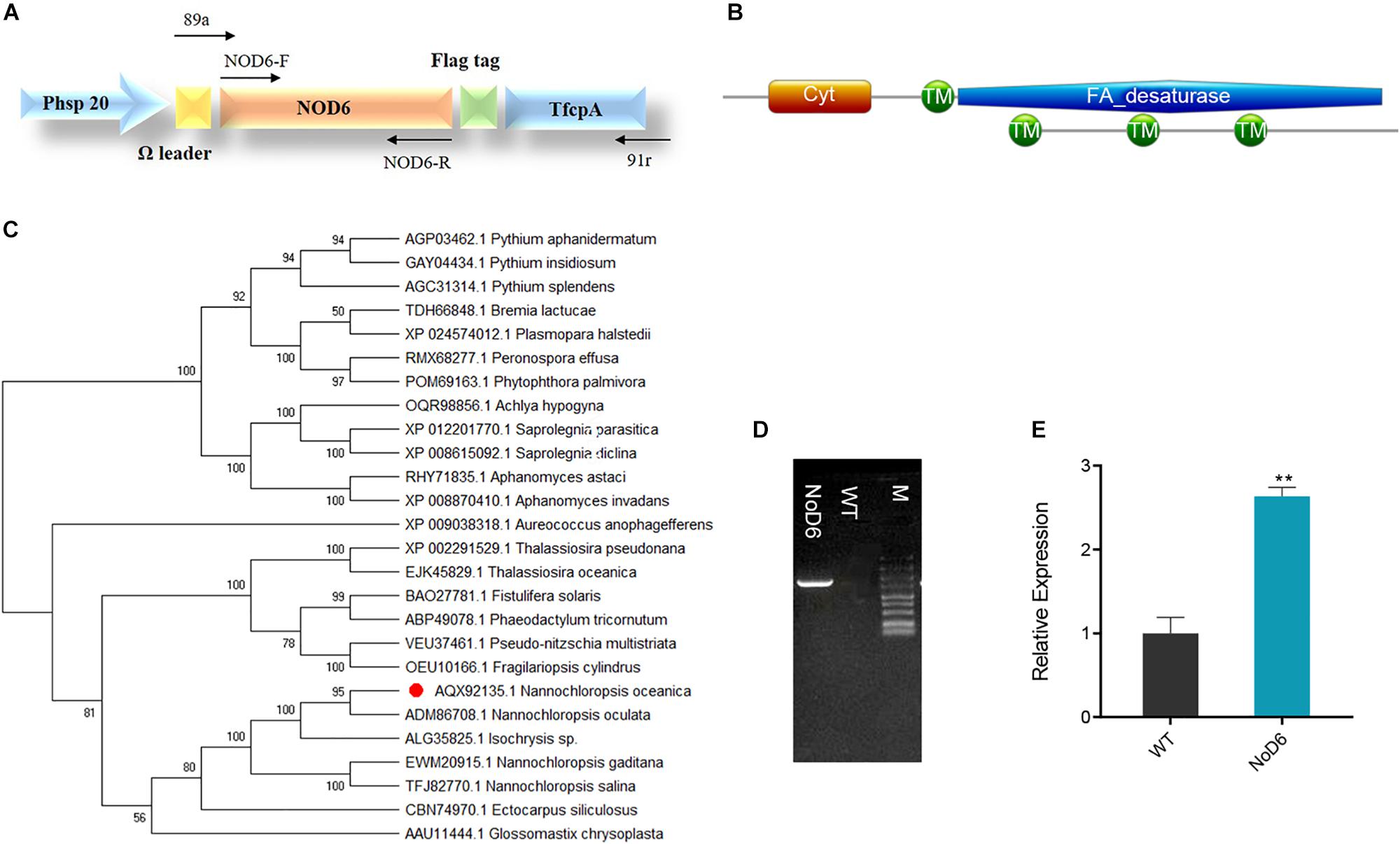
Figure 1. Sequence analyses of NoD6 and recombinant vector construction. (A) Schematic representation of recombinant expression. NoD6 was cloned into the pNa03 under the control of promoter fcpC and an omega leader motif was inserted in between the promoter and the transgene to enhance translation. (B) Conserved domain analysis of NoD6 as predicted by Pfam and SMART. (C) Phylogenetic tree was constructed from the deduced amino acid sequences of Δ6-desaturase from various organisms by MEGA using neighbor-joining method. The red dot denotes the D6 of Nannochloropsis oceanica. (D) Colony genomic PCR analysis. Lane NoD6, transgenic cell; Lane WT, wild type; M, marker. (E) Relative mRNA level of NoD6 as determined by qPCR. Data were normalized against tublin gene. Significant difference is indicated at ∗∗p < 0.01 level. Each value represents means ± SD (n = 3).
Evaluation of Transgenic Strain by Molecular Approaches
The putative transgenic strain was evaluated by single colony genomic PCR using the primers 89A and 91R that amplify the flanking region of the introduced NoD6 and the vector backbone as described previously (Li et al., 2016). Relative transcript level of NoD6 was determined by qPCR using a SYBR Green SuperMix for qPCR (Invitrogen, United States) on ABI Prism 7500 Sequence Detection System (Applied Biosystems, United States) as reported previously (Li et al., 2016). Briefly, total RNA was prepared from transgenics and WT using RNAiso Plus (Takara, Japan) and used as the template for the synthesis of first strand cDNA using the PrimeScript RT Reagent Kit (Takara, Japan) as per the provider’s instruction. The qPCR was performed in 96-well plates (20 μl reaction volume) as the manufacturer’s specifications (Applied Biosystems, United States). The threshold cycle value (Ct) for each reaction was determined and the relative mRNA abundance was determined after normalization to tublin gene.
Measurement of General Physiological Properties
The growth rate of transgenic and WT cells was determined by direct cell count method (Balamurugan et al., 2017) and estimating the specific growth rate (Sandnes et al., 2005). Photosynthetic efficiency of the cells was determined by measuring the maximum quantum yield of photosystem II (Fv/Fm) (Li et al., 2016).
Fluorometric, Gravimetric and Confocal Lipid Analyses
Relative neutral lipid content of transgenic cell was measured by qualitative Nile-red (Sigma, United States) fluorometric analysis as described previously (Li et al., 2016). Lipid content of transgenic and WT cells was further quantitatively determined by gravimetric method. Total lipids were extracted from the freeze-dried microalgal cells as described by Bligh and Dyer, 1959 and the extracted lipid content was determined gravimetrically (Li et al., 2016). Nile-red stained cells were observed under confocal microscope LSM 510 Meta (Zeiss, Germany) as described previously (Li et al., 2016).
Solid-Phase Extraction Mediated Lipid Fractionation and Fatty Acid Analysis
Total lipids were fractionated into phospholipid by solid-phase extraction (SPE) through gravity on a disposable pre-packed silica cartridges (500 mg, 6 cc Sep-Pak, Waters) conditioned with chloroform (5 ml). Thereafter, the extracted lipid sample (1 ml) was loaded on the chloroform-conditioned column and the PL was eluted into the pre-weighed glass vials with 10 ml of methanol. The fractionated lipids were transmethylated and the fatty acid composition of the extracted lipids was determined by using gas chromatography-mass spectrometry (GC-MS) as described elsewhere (Li et al., 2016).
Statistical Analysis
All experiments were carried out in biological triplicate. The data were represented as mean ± SD (n = 3). The data obtained were subjected to one−way analysis of variance (ANOVA) followed by Tukey’s test using GraphPad Prism 7.0 (GraphPad, San Diego, CA, United States).
Results and Discussion
Sequence and Phylogenetic Analyses of NoD6
BLAST analysis using P. tricornutum Δ6-desaturase against N. oceanica genome, identified Δ6-desaturase coding region (Accession number KY214451.1) with high sequence similarity (E-value 4e-175). Pfam and SMART tools (Letunic and Bork, 2018; El-gebali et al., 2019) predicted the presence of four transmembrane domains and cytochrome b5 domain (31–101 aa), which could facilitate the acquisition of electrons from NADH cytochrome b5 reductase (Meesapyodsuk and Qiu, 2012; Figure 1B). Phylogenetic analysis by MEGA showed that NoD6 exhibited high homology with Δ6-desaturase of N. oculata and both were clustered into the same clade, while distinctly separated from others (Figure 1C). To elucidate its role in LC-PUFA biosynthesis, we cloned NoD6 in pNa03 expression vector and overexpressed in N. oceanica.
Molecular Evaluation of Transgenic Cells
The putative transgenic algal cells were screened for zeocin resistance and cultivated on the selection medium supplemented with zeocin for at least four successive subcultures and thereafter, evaluated by molecular approaches. Genomic PCR was performed to identify the integration of transgene in the host, which resulted in the presence of 1.5-kb amplicon in transgenic cells, which is in accordance with the expected amplicon range, but no such band was detected in WT (Figure 1D). qPCR analysis showed that the relative transcript level of NoD6 was significantly increased by 2.6-fold in transgenic cells than WT (Figure 1E), however, enzymatic studies are needed to confirm the protein expression in transgenic cells. These results showed that NoD6 was successfully introduced and transcribed in the transgenic cells.
NoD6 Overexpression Enhanced Cellular Growth and Photosynthetic Efficiency
Concurrent enhancement of biomass and desired products is considered a critical parameter for commercial applications (Li et al., 2019). Hence, we examined the impact of NoD6 overexpression on cellular physiological parameters. Interestingly, NoD6 overexpression increased growth rate in transgenic cells compared to WT and it is worth mentioning that growth was significantly higher during stationary phase (Figure 2A), which is consistent with previous reports. Previous study has shown that expression of Δ12- and Δ5-desaturase enhanced growth in N. oceanica (Poliner et al., 2018). Similarly, expression of Δ5-elongase from Ostreococcus tauri enhanced growth in P. tricornutum during stationary phase. Thereafter, we examined the photosynthetic efficiency by determining Fv/Fm, which implied that NoD6 overexpression increased Fv/Fm during exponential phase, meanwhile Fv/Fm was reduced in transgenic cells during the stationary phase (Figure 2B). Besides, the specific growth rate was also found to be increased in transgenic cells (Figure 2C). Being the major membrane lipids of chloroplasts, PUFAs play a crucial role in maintaining plastidial membrane integrity and photosynthesis (Valentine and Valentine, 2004). Particularly, the potential of Nannochloropsis sp. to hyperaccumulate EPA in cellular membrane and thylakoids has been reported (Kaye et al., 2015). The abundance of PUFAs in plastids could enhance the rate of electron transport, thereby enhance photosynthetic efficiency (Chen et al., 2013). Consistently, our results showed that NoD6 overexpression enhanced photosynthetic efficiency, which enables the transgenic cells to harness the photosynthetic carbon sink toward enhanced growth and metabolite production during stationary phase which is in good agreement with previous studies (Thawechai et al., 2016).
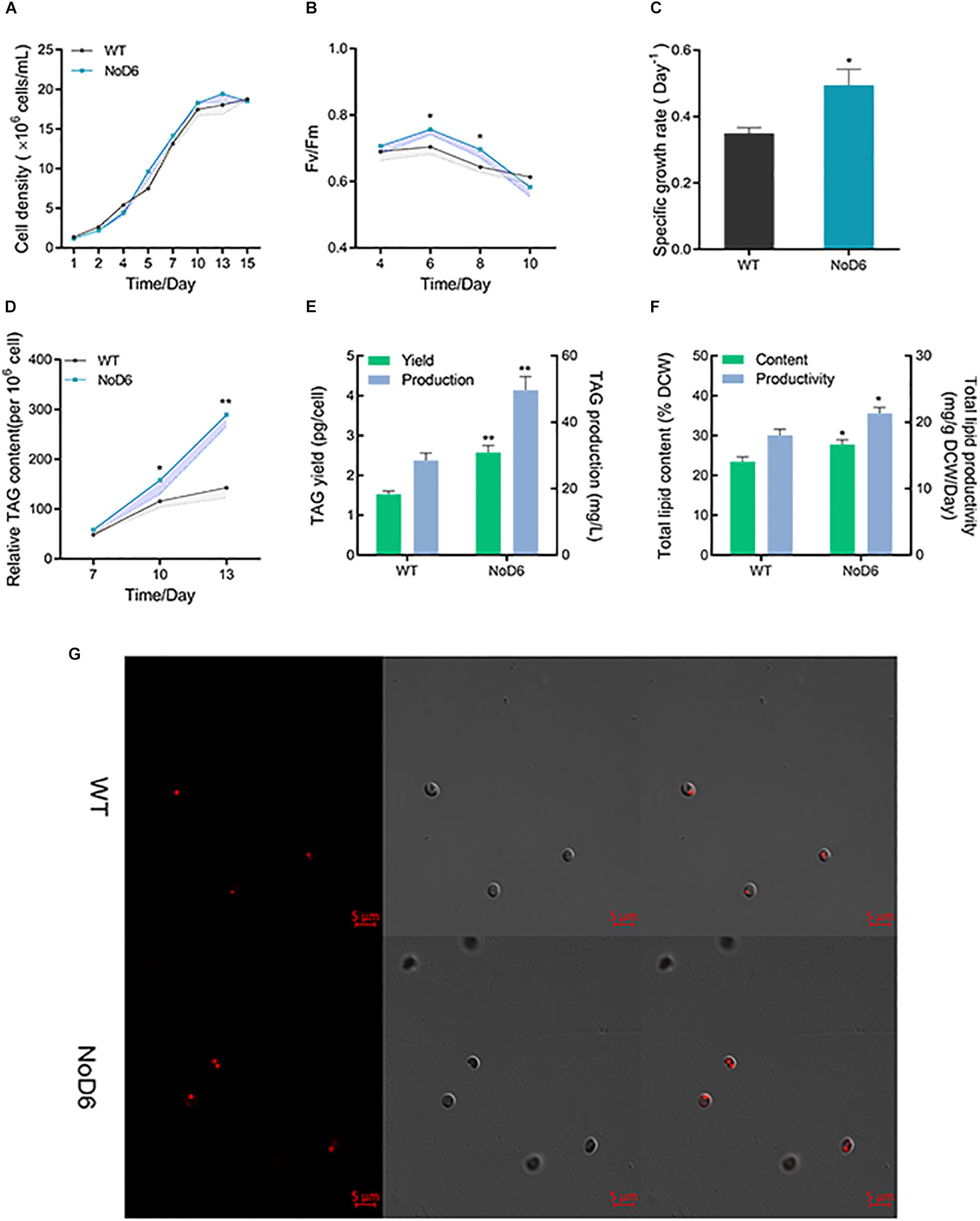
Figure 2. Physiological and lipidomic analyses of transgenic cell overexpressing NoD6. (A) Growth curves of transgenic and WT as determined by direct cell count methods. (B) Photosynthetic efficiency as reflected by Fv/Fm. (C) Specific growth rate. (D) Nile-red based fluorometric lipid analysis. (E) TAG yield (pg/cell). (F) Total lipid content (% DCW). (G) Laser scanning confocal microscopic observation of Nile-red stained cells. Bars = 5 μm. Left panel, lipid fluorescence; Middle panel, differential interference contrast (DIC); Right panel, overlay image. Significant difference is indicated at ∗p < 0.05 or ∗∗p < 0.01 level. Each value represents means ± SD (n = 3).
Lipidomic Studies Demonstrated That NoD6 Elevated Total Lipid Content
As fatty acids are the key precursors for lipogenesis, fatty acid hyperaccumulation could enhance lipid production (Peng et al., 2014). To assess the effect of NoD6 overexpression on lipid accumulation and to elucidate the lipogenic potential of NoD6, we determined lipid content by fluorometric and gravimetric analyses. Nile-red analysis showed that lipid fluorescence was gradually increased, particularly, lipid fluorescence was significantly higher in transgenic cells than WT at 13th day (Figure 2D). Congruently, the gravimetric analysis showed that TAG was significantly increased and reached up to 50 mg/L in transgenic cells (Figure 2E). Laser scanning confocal microscopic analysis of the Nile-red stained cells corroborated the fluorometric and gravimetric lipid analysis, which showed that volume of lipid droplets was increased in transgenic cells than WT (Figure 2G). Interestingly, total lipid productivity was remarkably increased in transgenic cells and reached up to 21 mg/gDCW/day (Figure 2F). Delta-6 fatty acid desaturase overexpression significantly elevated total lipid in P. tricornutum (Zhu et al., 2017). Elongase overexpression increased TAG in Thalassiosira pseudonana (Cook and Hildebrand, 2015). Simultaneous expression of MCAT and Δ5-desaturase increased total lipid content by 2.61-fold in P. tricornutum (Wang et al., 2017). Congruently, individual overexpression of Δ5-desaturase elevated neutral lipid up to 65% in transgenic P. tricornutum (Peng et al., 2014). Effective consumption of fatty acid moieties for the generation of PUFA by NoD6 could result in the activation of upstream fatty acid biogenesis and provide adequate fatty acid precursors for lipogenesis, thereby overproducing lipids in the transgenic cell which is consistent with the previous reports (Chen et al., 2017).
NoD6 Overexpression Significantly Elevated LC-PUFA in N. oceanica
Given the established health benefits of LC-PUFAs and their burgeoning demand, there exists a pressing need to develop a potential engineering strategy to overproduce algal LC-PUFAs (Martins et al., 2013). Consequently, tremendous efforts have been devoted to enhance algal PUFA content through various strategies (Yang et al., 2013; Hamilton et al., 2016). Stress treatments enhanced LC-PUFA content but impeded biomass and overall productivity, thereby obstructing the algal commercial potential (Mühlroth et al., 2013). On the other hand, microalgal genetic engineering yields mixed success owing to intricacies in LC-PUFA metabolism despite its potential (Peng et al., 2014). Hence, it is crucial to identify the key node to exploit its full potential (Li et al., 2019). To investigate the impact of NoD6 on LC-PUFA biosynthesis, we determined the fatty acid composition of total lipids and phospholipids during 10th day of cultivation, which showed that NoD6 overexpression significantly altered fatty acid composition (Figures 3D,E). Interestingly, fatty acids such as palmitic acid (PA), ARA and DHA were significantly increased. Total saturated and monounsaturated fatty acids were significantly reduced in transgenic cells, particularly, C16:1 was found to be significantly reduced. The reduced C16:1 content was is in accordance with the enhanced desaturation of oleic acid and subsequent desaturation of LA to GLA (Kaye et al., 2015). It is worth noting that LC-PUFAs such as ARA, EPA and DHA were increased by 2.04-, 1.53- and 2.32-fold, respectively in NoD6 overexpressing cells (Figures 3A–C), which accounts to 11.58, 62.35, and 5.80 μg/mg, respectively. Among the three hyperaccumulated LC-PUFAs, EPA was found to be predominant which constitutes up to 62.35 mg/g DCW at the 10th day of cultivation, however, EPA content was found to be decreased thereafter (Figure 3B). Meanwhile, DHA content was significantly higher in transgenic cells during 10th and 13th day, however, DHA content during the 13th day was found to be remarkably higher than that of the 10th day (Figure 3C).
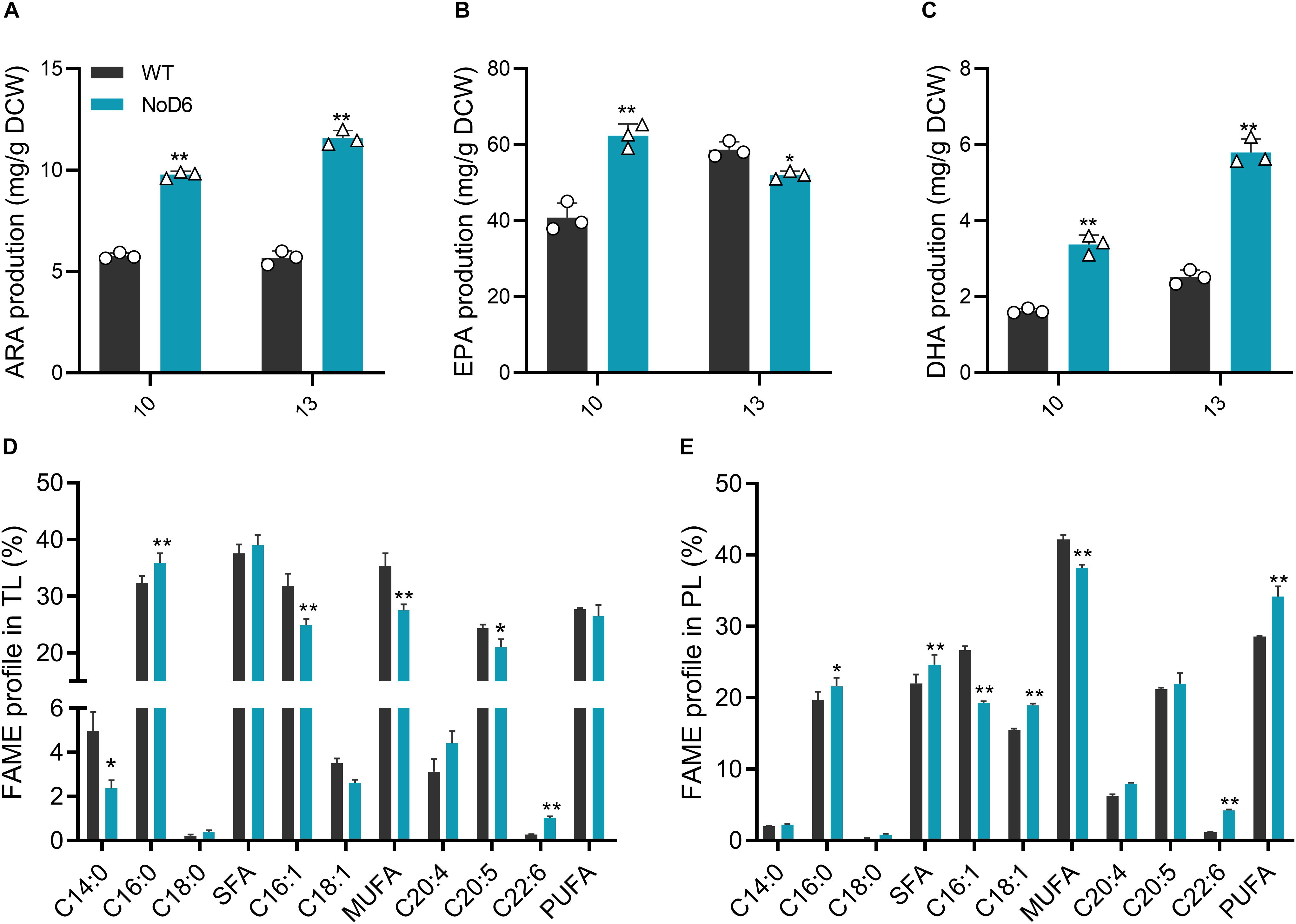
Figure 3. Fatty acid analysis. (A) Arachidonic acid content (mg/g DCW). (B) Eicosapentaenoic content (mg/g DCW). (C) Docosahexaenoic acid content (mg/g DCW). (D) Fatty acid composition in total lipids (%). (E) Fatty acid composition in total phospholipid (%). Significant difference is indicated at ∗p < 0.05 or ∗∗p < 0.01 level. Each value represents means ± SD (n = 3).
Given the crucial role of Δ6-desaturase in catalyzing the first committed step of generating EPA from ALA, it emerged as the potential metabolic target for LC-PUFA overproduction (Zhu et al., 2017). Various reports demonstrated the exploitation of elongases, desaturases, fatty acid synthase or combinatorial multiple gene expression to hyperaccumulate algal LC-PUFAs (Martins et al., 2013; Gong et al., 2014), however, promising strategies and molecular candidates are yet to be characterized (Domergue et al., 2002). Zhu et al. (2017) reported the overexpression of Δ6-desaturase enhanced EPA up to 38.101 mg/g DW in P. tricornutum (Zhu et al., 2017). Expression of multiple desaturase genes increased EPA by 25% in Nannochloropsis oceanica (Poliner et al., 2018). Similarly, overexpression of endogenous elongases overproduced EPA and DHA, by 1.4- and 4.5-fold, respectively in transgenic T. pseudonana (Cook and Hildebrand, 2015). In contrast, our study showed the potential of NoD6 gene on remarkably increasing EPA and DHA which reached the highest record up to 62.35 and 5.80 mg/g of DCW, respectively in transgenic N. oceanica. The increased EPA content enhanced physiological properties of the transgenic cells which is reflected in the enhanced photosynthetic efficiency and subsequent primary metabolite content (Thawechai et al., 2016). Taken together, these findings demonstrated the unprecedented regulatory role of NoD6 on enhancing LC-PUFAs, providing the balanced carbon flux in the form of acyl-CoA moieties. Previous studies have reported the abundance of PUFAs in the acyl-CoA pool could govern the adequate provision of carbon precursors for acylation into neutral lipids (Hamilton et al., 2014). Congruently, increased PUFA content has been reported to elicit the upstream metabolic pathways of fatty acid biosynthesis and enhance the availability of carbon metabolic precursors (Peng et al., 2014), which in turn adequately provide the carbon precursors for growth and lipogenesis.
Conclusion
Overexpression of Δ6-desaturase characterized its mechanistic role in LC-PUFA biogenesis and lipogenesis in oleaginous heterokont N. oceanica. NoD6 overexpression enhanced cellular growth and photosynthetic efficiency. Fluorometric and gravimetric lipidomic analyses showed that NoD6 significantly enhanced lipid content possibly by activating the upstream de novo fatty acid biosynthetic pathway, thereby facilitating the adequate provision of fatty acid precursors for lipogenesis. NoD6 overexpression significantly elevated the EPA and DHA content to 62.35 and 5.80 mg/g DCW, respectively, which has been recorded as the highest EPA production by engineering a key enzyme rather than multiple overexpression and stress treatments. Collectively, this report epitomizes a potential candidate for overproducing LC-PUFAs and lipids without hindering cellular physiological properties and provides a promising candidate for economically feasible production of microalgal LC-PUFAs.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
SF, H-YL, and LW conceived of and designed the study. FY, WY, YM, and BS carried out the experiments and analyzed the data. H-YL, SF, and LW prepared the manuscript and supervised the project.
Funding
This work was funded by the National Key R&D Program of China (2018YFD0502001 and 2018YFD0300901), the Science and Technology Service Program of Chinese Academy of Sciences (KFJ-STS-ZDTP-054), the Key Program of 13th Five-Year Plan, CASHIPS (No. KP-2019-21), Natural Science Foundation of Anhui Province (1808085QC73), and the supplementary program of Science and Technology Service Program of Chinese Academy of Sciences of Fujian Province (2019T3031).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Balamurugan, S., Wang, X., Wang, H. L., An, C. J., Li, H., Li, D. W., et al. (2017). Occurrence of plastidial triacylglycerol synthesis and the potential regulatory role of AGPAT in the model diatom Phaeodactylum tricornutum. Biotechnol. Biofuels 10, 1–14. doi: 10.1186/s13068-017-0786-0
Bligh, E. G., and Dyer, W. J. (1959). A rapid method of total lipid extraction, and purification. Can. J. Biochem. Physiol. 37, 911–917.
Chen, C., Chen, Y., Huang, H., Huang, C., and Lee, W. (2013). Bioresource Technology Engineering strategies for enhancing the production of eicosapentaenoic acid (EPA) from an isolated microalga Nannochloropsis oceanica CY2. Bioresour. Technol. 147, 160–167. doi: 10.1016/j.biortech.2013.08.051
Chen, J. W., Liu, W. J., Hu, D. X., Wang, X., Balamurugan, S., Alimujiang, A., et al. (2017). Identification of a malonyl CoA-acyl carrier protein transacylase and its regulatory role in fatty acid biosynthesis in oleaginous microalga Nannochloropsis oceanica. Biotechnol. Appl. Biochem. 64, 620–626. doi: 10.1002/bab.1531
Cook, O., and Hildebrand, M. (2015). Enhancing LC-PUFA production in Thalassiosira pseudonana by overexpressing the endogenous fatty acid elongase genes. J. Appl. Phycol. 28, 897–905. doi: 10.1007/s10811-015-0617-2
Domergue, F., Lerchl, J., Zähringer, U., and Heinz, E. (2002). Cloning and functional characterization of Phaeodactylum tricornutum front-end desaturases involved in eicosapentaenoic acid biosynthesis. Eur. J. Biochem. 269, 4105–4113.
El-gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Potter, S. C., Qureshi, M., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, 427–432. doi: 10.1093/nar/gky995
Gong, Y., Wan, X., Jiang, M., Hu, C., Hu, H., and Huang, F. (2014). Metabolic engineering of microorganisms to produce omega-3 very long-chain polyunsaturated fatty acids. Prog. Lipid Res. 56, 19–35. doi: 10.1016/j.plipres.2014.07.001
Hamilton, M. L., Haslam, R. P., Napier, J. A., and Sayanova, O. (2014). Metabolic engineering of Phaeodactylum tricornutum for the enhanced accumulation of omega-3 long chain polyunsaturated fatty acids. Metab. Eng. 22, 3–9. doi: 10.1016/j.ymben.2013.12.003
Hamilton, M. L., Powers, S., Napier, J. A., and Sayanova, O. (2016). Heterotrophic production of omega-3 long-chain polyunsaturated fatty acids by trophically converted marine diatom Phaeodactylum tricornutum. Mar. Drugs 14:E53. doi: 10.3390/md14030053
Kabeya, N., Fonseca, M. M., Ferrier, D. E. K., Navarro, J. C., Bay, L. K., Francis, D. S., et al. (2018). Genes for de novo biosynthesis of omega-3 polyunsaturated fatty acids are widespread in animals. Sci. Adv. 4:eaar6489. doi: 10.1126/sciadv.aar6849
Kaye, Y., Grundman, O., Leu, S., Zarka, A., Zorin, B., Didi-Cohen, S., et al. (2015). Metabolic engineering toward enhanced LC-PUFA biosynthesis in Nannochloropsis oceanica: overexpression of endogenous delta-12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG. Algal Res. 11, 387–398.
Khozin-Goldberg, I., Iskandarov, U., and Cohen, Z. (2011). LC-PUFA from photosynthetic microalgae: occurrence, biosynthesis, and prospects in biotechnology. Appl. Microbiol. Biotechnol. 91, 905–915. doi: 10.1007/s00253-011-3441-x
Letunic, I., and Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Li, D., Balamurugan, S., Yang, Y., Zheng, J., Huang, D., Zou, L., et al. (2019). Transcriptional regulation of microalgae for concurrent lipid overproduction and secretion. Sci. Adv. 5:eaau3795. doi: 10.1126/sciadv.aau3795
Li, D. W., Cen, S. Y., Liu, Y. H., Balamurugan, S., Zheng, X. Y., Alimujiang, A., et al. (2016). A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J. Biotechnol. 229, 65–71. doi: 10.1016/j.jbiotec.2016.05.005
Martins, D. A., Pereira, H., Ben-hamadou, R., Abu-salah, K. M., and Arabia, S. (2013). Alternative sources of n-3 long-chain polyunsaturated fatty acids in marine microalgae. Mar. Drugs 11, 2259–2281. doi: 10.3390/md11072259
Meesapyodsuk, D., and Qiu, X. (2012). The front-end desaturase: structure, function, evolution and biotechnological use. Lipids 47, 227–237. doi: 10.1007/s11745-011-3617-2
Mühlroth, A., Li, K., Røkke, G., Winge, P., Olsen, Y., Hohmann-Marriott, M. F., et al. (2013). Pathways of lipid metabolism in marine algae, co-expression network, bottlenecks and candidate genes for enhanced production of EPA and DHA in species of chromista. Mar. Drugs 11, 4662–4697. doi: 10.3390/md11114662
Peng, K. T., Zheng, C. N., Xue, J., Chen, X. Y., Yang, W. D., Liu, J. S., et al. (2014). Delta 5 fatty acid desaturase upregulates the synthesis of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum. J. Agric. Food Chem. 62, 8773–8776. doi: 10.1021/jf5031086
Poliner, E., Pulman, J. A., Zienkiewicz, K., Childs, K., Benning, C., and Farré, E. M. (2018). A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production. Plant Biotechnol. J. 16, 298–309. doi: 10.1111/pbi.12772
Sandnes, J. M., Källqvist, T., Wenner, D., and Gislerød, H. R. (2005). Combined influence of light and temperature on growth rates of Nannochloropsis oceanica: linking cellular responses to large-scale biomass production. J. Appl. Phycol. 17, 515–525.
Thawechai, T., Cheirsilp, B., Louhasakul, Y., Boonsawang, P., and Prasertsan, P. (2016). Mitigation of carbon dioxide by oleaginous microalgae for lipids and pigments production: effect of light illumination and carbon dioxide feeding strategies. Bioresour. Technol. 219, 139–149. doi: 10.1016/j.biortech.2016.07.109
Valentine, R. C., and Valentine, D. L. (2004). Omega -3 fatty acids in cellular membranes: a unified concept. Prog. Lipid. Res. 43, 383–402.
Wang, X., Fosse, H. K., Li, K., Chauton, M. S., Vadstein, O., and Inge, K. (2019). Influence of nitrogen limitation on lipid accumulation and EPA and DHA content in four marine microalgae for possible use in aquafeed. Front. Mar. Sci. 6:95. doi: 10.3389/fmars.2019.00095
Wang, X., Liu, Y. H., Wei, W., Zhou, X., Yuan, W., Balamurugan, S., et al. (2017). Enrichment of long-chain polyunsaturated fatty acids by coordinated expression of multiple metabolic nodes in the oleaginous microalga Phaeodactylum tricornutum. J. Agric. Food Chem. 65, 7713–7720. doi: 10.1021/acs.jafc.7b02397
Yang, Z. K., Niu, Y. F., Ma, Y. H., Xue, J., Zhang, M. H., Yang, W. D., et al. (2013). Molecular and cellular mechanisms of neutral lipid accumulation in diatom following nitrogen deprivation. Biotechnol. Biofuels 6, 1–14.
Yuan, W., Ma, Y., Wei, W., Liu, W., Ding, Y., and Balamurugan, S. (2019). Sequential treatment with bicarbonate and low-temperature to potentiate both biomass and lipid productivity in Nannochloropsis oceanica. 2019. J. Chem. Technol. Biotechnol. 94, 3413–3419. doi: 10.1002/jctb.6155
Keywords: Δ6-desaturase, docosahexaenoic acid, eicosapentaenoic acid, long-chain polyunsaturated fatty acids, Nannochloropsis oceanica
Citation: Yang F, Yuan W, Ma Y, Balamurugan S, Li H-Y, Fu S and Wu L (2019) Harnessing the Lipogenic Potential of Δ6-Desaturase for Simultaneous Hyperaccumulation of Lipids and Polyunsaturated Fatty Acids in Nannochloropsis oceanica. Front. Mar. Sci. 6:682. doi: 10.3389/fmars.2019.00682
Received: 23 August 2019; Accepted: 21 October 2019;
Published: 01 November 2019.
Edited by:
Periasamy Anbu, Inha University, South KoreaReviewed by:
Grigorios Krey, Hellenic Agricultural Organization – ELGO, GreeceChanghong Yao, Sichuan University, China
Copyright © 2019 Yang, Yuan, Ma, Balamurugan, Li, Fu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songling Fu, ZnVzb25nbGluZ0BhaGF1LmVkdS5jbg==; Lifang Wu, bGZ3dUBpcHAuYWMuY24=
†These authors have contributed equally to this work
 Fan Yang1†
Fan Yang1† Wasiqi Yuan
Wasiqi Yuan Srinivasan Balamurugan
Srinivasan Balamurugan Hong-Ye Li
Hong-Ye Li Lifang Wu
Lifang Wu