- 1Environment, Ecology and Energy Program, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 2Department of Biology, The University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Seafood mislabeling is a widely documented problem that has significant implications for human and environmental health. Defined as when seafood is sold under something other than its true species name, seafood fraud allows less-desired or illegally caught species to be marketed as one recognizable to consumers. Red snapper is one of the most frequently mislabeled species, with previous studies showing mislabeling rates as high as 77%. We assessed whether red snapper mislabeling rates varied among states or vendor type. We also determined the IUCN Red List designation of substituted species to assess whether frequently substituted stocks were more or less at-risk than red snapper stocks. We used standard DNA barcoding protocols to determine the identity of products labeled as “red snapper” from sushi restaurants, seafood markets, and grocery stores in the Southeastern United States. Overall, 72.6% of samples (out of 62) were mislabeled, with sushi restaurants mislabeling samples 100% of the time. Out of 13 substituted species (including samples that were indistinguishable between two species), seven (53.8%) were not native to the United States of the 12 substituted species assessed by the IUCN Red List, 11 (91.6%) were listed as less threatened than red snapper. These results contribute to a growing body of mislabeling research that can be used by government agencies trying to develop effective policies to combat seafood fraud and consumers hoping to avoid mislabeled products.
Introduction
Seafood mislabeling in the United States has been recognized for decades: a 1997 press release by the United States National Seafood Inspection Laboratory stated 37% of fish tested between 1988 and 1997 were mislabeled (Ropicki et al., 2010). Over a decade and a half later, the mislabeling rate remained at over 30%, with one-third of over 1,200 samples nationwide mislabeled according to Food and Drug Administration (FDA) guidelines (Warner et al., 2013). Recent assessment of sushi restaurants in Los Angeles found mislabeling rates as high as 47%, with some species mislabeled up to 77% of the time (Willette et al., 2017). Despite growing public awareness about the practice of seafood fraud, rates of mislabeling remain high, indicating that there is still economic incentive to mislabel along the supply chain, while lack of awareness and/or enforcement allows the practice to continue.
Labeling of seafood is dependent on species’ identity, country of origin, production method, and potential eco-labels (Buck, 2010). Each of these factors presents an opportunity for mislabeling as consumers, especially in the United States, are generally unfamiliar with the seafood production process (Jacquet and Pauly, 2008). Restaurants and businesses can exclude information about the origin of the product, which can lead to consumers receiving a product that is of lesser value than the desired species (Stiles et al., 2013; Khaksar et al., 2015). Seafood filets can be extremely similar in taste, texture, and appearance, allowing fraud to pass undetected by the consumer (Ropicki et al., 2010).
Unintentional mislabeling occurs when species are misidentified or when information is lost along the supply chain. One example is accidental assignment to a species with a common vernacular name, such as labeling a red-colored vermillion snapper (Rhomboplites aurorubens), as “red snapper,” which is a different species (Lutjanus campechanus) according to FDA guidelines (Willette et al., 2017). Intentional mislabeling allows retailers to label less-desirable species as more profitable ones, or to mask the sale of illegally captured species (Jacquet and Pauly, 2008). In 2009, Florida restaurants sold imported catfish as grouper, one of the most popular finfishes in the state. The restaurants paid only $2.50 per pound for the catfish, whereas domestic grouper cost $11 to $12 per pound (Vasquez, 2009).
Seafood fraud, whether intentional or unintentional, weakens public trust, compromises consumers’ ability to adhere to dietary restrictions, and poses public health concerns (Ling et al., 2008; Miller and Mariani, 2014). Mislabeling makes it impossible for consumers, especially children and pregnant women, to monitor their intake of high-trophic level species that could contain elevated levels of mercury (Marko et al., 2014). A previous study found tilefish, a species that the FDA warns consumers against eating due to its high mercury content, substituted for red snapper (Warner et al., 2012). Additionally, a fish that seems to be readily available but is actually mislabeled leads the public to believe the fish stock is plentiful, regardless of the true state of the stock (Marko et al., 2004). This is particularly critical for popular seafood like red snapper, where the South Atlantic stock is considered overfished and is undergoing overfishing (SEDAR, 2016). If mislabeling occurs before landing data is collected, commercial landing data could be artificially inflated for in-demand species, and artificially low for substituted species (Di Pinto et al., 2015a). This could affect management efforts by potentially allowing unregulated overharvesting of substitute species (Carvalho et al., 2011; Cox et al., 2012; Cawthorn et al., 2018).
Lastly, mislabeling undermines efforts to promote consumption of sustainable seafood. Increasing education and awareness about the decline of wild-caught fisheries has led to a rise in consumers wanting to make environmentally sustainable choices when buying seafood (Marko et al., 2011). A number of seafood certification and education programs have arisen worldwide, including the Global Sustainable Seafood Initiative, Seafood Watch, Seafood Choice Alliance, and the Marine Stewardship Council. Seafood certification programs are a way for people to engage in marine conservation initiatives, and 72% of respondents in a United States survey said they would be more likely to purchase seafood labeled as “environmentally responsible” (Logan et al., 2008). However, the success of certification programs depends on the integrity of labeling: seafood substitution can undermine initiatives intended to provide sustainable seafood options to consumers (Gulbrandsen, 2009; Stawitz et al., 2017).
Although seafood fraud is widely documented in the literature, many studies are limited by small sample sizes or restricted to small geographic regions, such as a single city. Additionally, many studies analyze a few samples from many different species, making it difficult to draw conclusions about mislabeling rates of a single species.
We measured the frequency and distribution of red snapper mislabeling and assessed how mislabeling rates vary between vendor type and state in the Southeastern United States. Red snapper is one of the most widely mislabeled species in the United States and one of the most popular and controversial fisheries in the South Atlantic and Gulf of Mexico (Cowan et al., 2011). Despite being declared overfished in the late 1980s, red snapper remains among the most valued fisheries in the South Atlantic and Gulf of Mexico, and the stock is currently managed by a rebuilding plan to restore stocks to sustainable levels (Goodyear, 1988; SEDAR, 2016). In the study region in 2016 alone, commercial red snapper landings were valued at $2,565,290 dockside (NOAA, 2017). But somehow, the red snapper on the dock is not ending up on plates at the same rate: red snapper is mislabeled up to 77% of the time (Marko et al., 2004; Warner et al., 2012). According to the U.S. Food and Drug Administration, only L. campechanus can legally be marketed as “red snapper,” but previous studies have found the name colloquially used for a wide range of other fish, including species outside the snapper family (Marko et al., 2004).
We measured red snapper mislabeling throughout the Southeastern coast of the United States to test the hypotheses that there are differences in mislabeling rates among states and vendor types, and that substituted species typically have healthier stocks than red snapper.
Materials and Methods
Collection
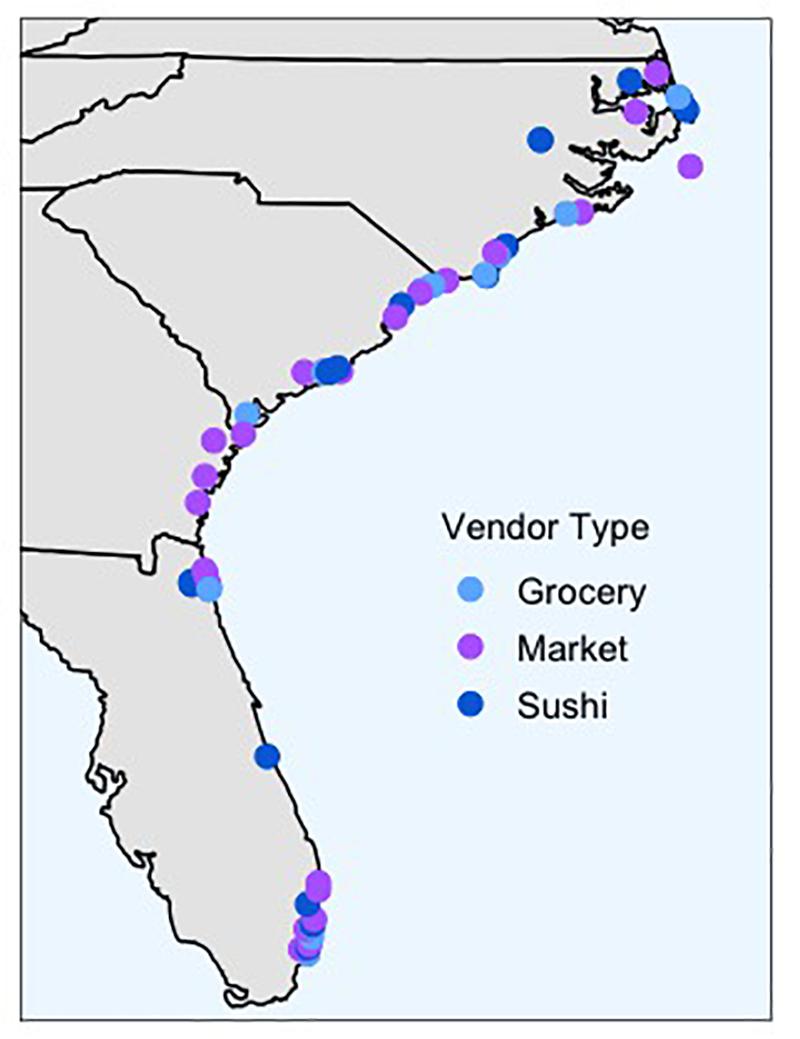
Seafood labeled as red snapper was collected from March–May 2018 from sushi restaurants, fish markets, and grocery stores along the coastline of North Carolina, South Carolina, Georgia, and Florida (Figure 1). For a holistic view of regional mislabeling, the number of samples collected from each state was loosely proportional to the length of the coastline. We collected 66 samples, with 20 samples from North Carolina, 15 from South Carolina, 4 from Georgia, and 27 from Florida. 22 samples were from grocery stores, 25 from fish markets, and 19 from sushi restaurants. Sites were sampled only once, with the exception of two vendors who sold both fileted and whole “red snapper,” in which case both products were collected and tested. Our study defined mislabeling in terms of incorrect identification of the species, but the scope of mislabeling can extend to other information like country of origin, farmed or wild caught, and more (Di Pinto et al., 2015b). Samples either needed to be physically labeled “red snapper,” or verbally confirmed as “red snapper” by a vendor employee. Vendors were not aware that samples were being collected for this study. To simulate the experience of a consumer, if we asked an employee for red snapper and the employee indicated a specific product, it was included as a sample regardless of whether it was physically labeled “red snapper.” For example, when asked for red snapper, one grocery store employee indicated a filet was red snapper, so that sample was collected despite it being physically labeled as mutton snapper. In sushi restaurants, only sashimi or rolls specifically marketed as “red snapper” were included. No samples only labeled as “snapper” were included unless an employee confirmed it was red snapper. A small piece of each sample was preserved in 95% ethanol and stored for processing in the lab. The specific location of vendors sampled varied within each state due to availability of “red snapper” products for sale.
DNA Extraction and Polymerase Chain Reaction (PCR)
For each sample, we extracted genomic DNA from thawed fish tissue using the Qiagen DNeasy Blood and Tissue Kit Protocol (Qiagen, Inc.). Each DNeasy kit included proteinase K, spin columns, and buffers used in the protocol. First, 20 mg of fish tissue was placed in a 1.5 mL microcentrifuge tube with 180 μl ATL Buffer and 20 μl proteinase K and incubated at 65°C for 1 h. Samples were vortexed approximately every 10 min during incubation. We added 200 μl AL Buffer, vortexed, incubated at 55°C for 10 min, then added 200 μl ethanol. After transferring the resulting liquid to a DNAeasy Mini spin column, samples were placed in a centrifuge at 8,000 rpm for 1 min. Samples were run in the centrifuge twice more: first after adding 500 μl Buffer AW1 at 8,000 rpm for 1 min, then after adding 500 μl of Buffer AW2 at 14,000 rpm for 3 min. Flow through was discarded from spin columns after each centrifuge run. Spin columns were then transferred to a new microcentrifuge tube, eluted with 20 μl of diH20, incubated at room temperature for 5 min, then centrifuged at 8,000 rpm for 1 min.
Polymerase Chain Reaction was used to amplify a fragment of the cytochrome c oxidase 1 (CO1) gene, which has been shown to be a strong diagnostic marker of fish identification to the species level (Wong and Hanner, 2008; Willette et al., 2017). We used a primer cocktail designed in Ivanova et al. (2007) (C_FishF1t1 and C_FishR1t1). We added 1 μl of each sample’s DNA to separate 0.2 ml illustra puReTaq Ready-To-Go PCR bead tubes, along with the primer cocktail consisting of 1.3 μl each of CO1_F1, CO1_F2, CO1_R1, and CO1_R2 PCR primers. A control PCR bead tube was used to ensure primers were not contaminated with DNA. To bring the overall volume to 25 μl, 19 μl of distilled water was added to the PCR bead tubes (20 μl was added for the control). After ensuring the PCR beads in the tubes were dissolved, tubes were placed in a Bio-Rad T100 Thermal Cycler using the following protocol adapted from Willette et al. (2017).
(1) Initial denaturing: 95°C for 5 min
(2) Denature: 94°C for 30 s
(3) Annealing: 50°C for 45 s
(4) Extension: 72°C for 60 s
(5) Final extension: 72°C for 10 min
(where steps 2–4 were repeated for 35 cycles)
Analysis of PCR Amplified Products and Sequence Analysis
We used gel electrophoresis to assess the results of PCR processing. We mixed and heated 50 mL of 1X TAE Buffer and 0.5 g of agarose powder until the agarose was fully dissolved. We added 3 μl of ethidium bromide before pouring the mixture into the gel tray. We mixed 1 μl of 6X loading dye with 5 μl of each sample, added each sample to the gel, and ran the chamber for 30 min at 100 V. If the PCR reaction was determined successful, PCR products were shipped to Eton Bioscience in Durham, North Carolina, for purification and sequencing using the M13 forward primer from Ivanova et al. (2007). Using 4Peaks software (version 1.8, developed by Nucleobytes) we selected at least 300 base pairs and identified each sample to the species level with the Basic Local Alignment Search Tool (BLAST) on the National Center for Biotechnology Information (NCBI) website1. Every identity with 98% confidence or above was considered a positive identification. Of 66 total samples, four were contaminated with bacteria and were unable to be identified. Contaminated samples were collected from different vendor types on different days and we were unable to determine the source of contamination. We determined positive identifications for the remaining 62 samples. Sometimes the CO1 gene is not enough to differentiate two species, specifically between rose and lane snapper (Lutjanus guttatus/Lutjanus synagris) and Malabar blood and crimson snapper (Lutjanus malabaricus/Lutjanus erythropterus). In these cases, samples were noted as being either species. Chi square tests and two-proportions z tests in R Studio were used to see if the proportion of mislabeled samples was significantly different between vendor and state.
Results
Identity of Substituted Species
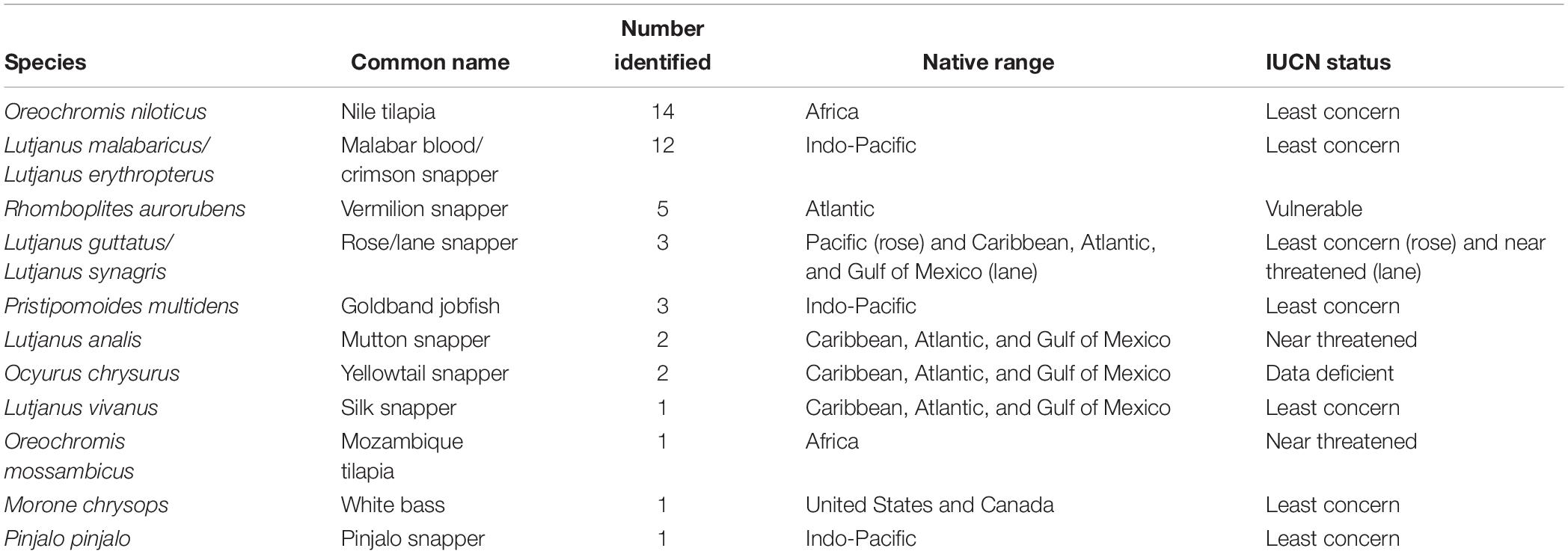
Of 62 samples, 45 (72.6%) were mislabeled. Vendors substituted 11 different species for red snapper (Table 1) and 29 of the mislabeled samples (64.4%) were another species of the family Lutjanidae. Of mislabeled samples, one third were identified as Oreochromis sp. (tilapia). For a full list of sample identities, please see Supplementary Table 1.
Mislabeling by Vendor Type and State
Mislabeling rates were 55.0%, 66.7%, and 100% for grocery stores, seafood markets, and sushi restaurants, respectively, and varied significantly among vendor types (Chi square test, p = 0.006, α = 0.05). A total of six species, including red snapper, were represented in grocery store samples. Seafood markets sold nine unique species (including red snapper) as red snapper, the most common of which was vermilion snapper (31.2% of mislabeled market samples). Of samples collected in markets and grocery stores, filets had a marginally higher mislabeling rate than whole fish (two proportions z-test, p = 0.046, α = 0.05). Of 12 whole fish collected from grocery stores and super markets, eight were correctly labeled (66.7%), compared to only nine of 32 filets (28.1%).
Every sample (n = 18) from sushi restaurants was mislabeled, with five different species being sold as red snapper. Sushi restaurants were the only vendors to substitute red snapper with tilapia, and 83.3% of sushi samples were tilapia.
Of the states, North Carolina had a mislabeling rate of 90.0%, Georgia and South Carolina both had a rate of 75.0%, and Florida had a rate of 57.7%. Although Florida had the lowest rate of mislabeling, there was not a statistically significant difference in mislabeling rates among states (Chi square test, p = 0.112, α = 0.05).
Discussion
Our findings were consistent with studies that assessed red snapper mislabeling rates in other parts of the United States. Marko et al.’s (2004) study across eight states found 17 of 22 samples mislabeled (77%), commonly replaced by lane or vermilion snapper. Despite extensive media coverage of the topic and presumably increased public awareness, 14 years later the rate of red snapper mislabeling is still over 70%. Like Marko et al. (2004), we found that about half of all samples were species not native to North America. Of 45 mislabeled samples, 68.9% were species native to other parts of the world. The mislabeling rate of sushi restaurants in our study (100%) was concordant with results from Willette et al. (2017), which reported all red snapper sushi samples mislabeled.
Florida had the lowest rate of mislabeling, and if Florida samples were removed from analysis, the overall mislabeling rate would jump from 72.6% to 83.3%. This regional trend is similar to Warner et al. (2013), who found that Miami, Florida had lower rates of red snapper mislabeling (38%) than the United States West Coast (100%), which is geographically further from a commercial red snapper fishery. While the South Atlantic commercial red snapper fishery was closed during the sampling period, the primary commercial red snapper fishery in the Gulf of Mexico was open at the time of collection. Ease of accessibility to fresh fish from the Gulf of Mexico could account for lower rates of mislabeling in Florida.
Out of 13 substituted species (if we consider those that are genetically indistinguishable using CO1 – L. guttatus/L. synagris and L. malabaricus/L. erythropterus – as separate species), six were not native to the continental United States (46.2%) (Table 1). Of the 12 substituted species assessed by the IUCN Red List, 11 were listed as less threated than red snapper. Vermilion snapper is the only species that is considered as at risk (“Vulnerable”) as red snapper.
When mislabeling occurred, grocery stores were most likely to sell species closely related to red snapper. Of mislabeled grocery samples, 81.8% came from the same genus as red snapper (Lutjanus), compared to 43.8% of market samples. Only 11.1% of sushi samples were species of genus Lutjanus.
Filets were more likely to be mislabeled than whole fish, likely because it is easier to pass off a variety of species as plain white filets rather than whole fish with distinguishing morphological features. Of the four whole fish that were mislabeled, one was a rose/lane snapper, one was a silk snapper, and two were vermilion snapper. All three species have roughly similar coloring and body shape to red snapper which could decrease the likelihood that consumers would detect fraud (Figure 2).

Figure 2. Many snapper species are difficult to tell apart, even as whole fish. For example, lane snapper (top) resembles red snapper (middle). There is also variation in coloration of red snapper, as seen in the two red snapper samples (middle and bottom), which makes positive identification even more challenging.
All substituted species, with the exception of vermilion snapper, were considered less threatened than red snapper by the International Union for Conservation of Nature (IUCN) (IUCN, 2019). Both red and vermilion snapper are considered “Vulnerable,” which means the IUCN considers the species threatened with extinction. Despite a similar IUCN listing, there are differences in the stock status of red and vermilion snapper. According to stock assessments in 2015 and 2016, red snapper is overfished in the South Atlantic and Gulf of Mexico, and are undergoing overfishing in the South Atlantic (SEDAR, 2016). In contrast, a 2012 stock assessment found that vermilion snapper is not overfished and are not undergoing overfishing in the Gulf of Mexico or the South Atlantic (SEDAR, 2012). Compared to red snapper, which is significantly below its target population, vermilion snapper is close to its target levels, suggesting that they are currently a more sustainable seafood option than red snapper.
However, if mislabeling of vermilion snapper occurs before the fish reaches the dock, it could lead to artificially low estimates of vermilion snapper catch, and therefore the population status designation may not be accurate. Artificially high population estimates for vermilion snapper could lead to catch limits that are too high for the population to sustain, therefore putting the stock at risk for overexploitation. A 2009 study assessed this problem in two North American species of hake: they found that offshore hake (Merluccius albidus) was being sold as the morphologically similar silver hake (M. bilinearis), and unreported offshore hake could make up as much as 12% of exported hake to Spain, one of the largest markets for hake (Garcia-Vazquez et al., 2009). At that rate, over 11,000 metric tons of offshore hake could have been caught and labeled as silver hake over the last decade (Garcia-Vazquez et al., 2009).
Substituting tilapia for red snapper, as seen in one third of the mislabeled samples in this study, has health implications for consumers. Compared to snapper species, tilapia species are lower in nutrient content in a number of categories, including protein, calcium, magnesium, phosphorous, potassium, and vitamin B-12 (USDA, 2019). Tilapia is also lower in omega-3 polyunsaturated fatty acids, which is associated with positive health effects such as reduced risk of stroke, cardiovascular disease, and diabetes (Smith and Guentzel, 2010). The nutritional value of a fish is cited as a reason why some people choose to consume one type of fish over another, and substitution undermines the consumer’s ability to purchase fish based on its nutritional benefit (Oken et al., 2012).
Our results suggest that purchasing whole fish from grocery stores is the best way to avoid red snapper mislabeling. There was some redundancy, however, in the grocery store chains that were sampled. Although samples came from different geographic locations, some grocery store chains were sampled repeatedly. For example, of the 22 grocery store samples, seven were from a regional grocery chain (five were correctly labeled), and four were from a nation-wide grocery chain (all correctly labeled) – both of which are chains that emphasize seafood sustainability in their marketing materials. Disproportionate sampling of grocery stores that have better seafood traceability could result in artificially lower mislabeling rates.
We were also limited by the availability of samples – sometimes it was difficult to find vendors who sold red snapper. Some markets, especially in North Carolina, who advertised selling local seafood stressed that they did not carry red snapper because it was not in season in the South Atlantic. Multiple employees in fish markets in the study region explained why it was important to eat local fish that were in season, and suggested we try a different species of fish that was similar in texture and taste to red snapper. Therefore, our sampling was limited to vendors who carry red snapper in the off season, which is a subsection that might not be reflective of the average red snapper mislabeling rate across all vendors when red snapper is in season. Also, we were not able to sample all vendors types from all states – in Georgia, we could only find markets who sold red snapper, so there are no samples from sushi restaurants or grocery stores in Georgia.
Although isolated, there were examples of either misidentification or overt deception when purchasing samples for this study. A North Carolina seafood market employee said they caught the whole “red snapper” off the dock that morning, even though commercial red snapper season was closed in North Carolina at the time of collection. An employee at another market assured us that a fish labeled as vermilion snapper was red snapper, then pulled a different fish from under the table to wrap up for purchase that was later identified as silk snapper.
Further research into mislabeling rates at each stage of the supply chain (fisher, distributor, or vendor) would help determine at which stage the mislabeling occurs. Although our study assesses mislabeling rates by vendor, we were unable to account for retailers that had the same distributor. It is possible, for example, that two sushi restaurants could unknowingly receive mislabeled “red snapper” from the same food provider, or that a seafood market could receive mislabeled products from a fisher.
Increased transparency and traceability throughout the seafood supply chain and across vendors is imperative to not only preventing mislabeling, but also ensuring seafood is sustainable and safe for consumers. Traceability, which is defined as the ability to follow a food item through all stages of production, processing, and distribution, allows consumers and vendors to confirm a seafood product was harvested legally, from a sustainable source, and without the use of forced or illegal labor (Leal et al., 2015; Marschke and Vandergeest, 2016). Traceability requires accurate recordkeeping from players at all levels of the supply chain, as well as accountability tools, such as DNA testing, that authorities can use to test for compliance (Leal et al., 2015).
Understanding the scope, scale, and trends of seafood mislabeling is important for consumers, fisheries managers, and participants in the seafood supply chain. Testing large sample sizes of commercially popular seafood species could indicate whether the economic value of those fisheries is inflated by the inclusion of artificially inflated seafood sales. Regular, strategic testing (and retesting) could also point to seasonal trends, such as whether a species is more likely to be mislabeled when commercial seasons are closed. Disseminating mislabeling data could also encourage vendors and consumers to more closely assess where their fish is coming from and could motivate vendors to test their own products to check that they are not receiving mislabeled products from their suppliers. Additionally, encouraging consumers to learn what to look and ask for in their seafood incentivizes vendors to ensure they are not selling mislabeled products.
Data Availability
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
Both authors conceived the project based on previous mislabeling work done by JB and the Seafood Forensics class at The University of North Carolina at Chapel Hill. ES collected and processed the samples, analyzed the data with statistical help from JB, and wrote the manuscript with editing assistance from JB.
Funding
This work was supported by funds from National Geographic Early Career Grant (#EC-428R-18); the Department of Biology, The University of North Carolina at Chapel Hill; the QEP program within College of Arts and Sciences, The University of North Carolina at Chapel Hill; and by the National Science Foundation (OCE #1737071 to JB).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank C. Moscarito for assisting in processing the samples, and the Bruno Lab at The University of North Carolina at Chapel Hill for their helpful feedback on manuscript drafts.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00513/full#supplementary-material
Footnotes
References
Buck, E. H. (2010). Seafood Marketing: Combating Fraud and Deception. Report RL34124. Washington, DC: Congressional Research Service.
Carvalho, D. C., Neto, D. A. P., Brasil, B. S. A. F., and Oliveira, D. A. A. (2011). DNA barcoding unveils a high rate of mislabeling in a commercial freshwater catfish from Brazil. Mitochondrial DNA 22, 97–105. doi: 10.3109/19401736.2011.588219
Cawthorn, D.-M., Baillie, C., and Mariani, S. (2018). Generic names and mislabeling conceal high species diversity in global fisheries markets. Conserv. Lett. 2018:e12573. doi: 10.1111/conl.12573
Cowan, J. H., Grimes, C. B., Patterson, W. F., Walters, C. J., Jones, A. C., Lindberg, W. J., et al. (2011). Red snapper management in the Gulf of Mexico: science- or faith-based? Rev. Fish Biol. and Fish. 21, 187–204. doi: 10.1007/s11160-010-9165-7
Cox, C. E., Jones, C. D., Wares, J. P., Castillo, K. D., Mcfield, M. D., and Bruno, J. F. (2012). Genetic testing reveals some mislabeling but general compliance with a ban on herbivorous fish harvesting in Belize. Conserv. Lett. 6, 132–140. doi: 10.1111/j.1755-263X.2012.00286.x
Di Pinto, A., Marchetti, P., Mottola, A., Bozzo, G., Bonerba, E., Ceci, E., et al. (2015a). Species identification in fish fillet products using DNA barcoding. Fish. Res. 170, 9–13. doi: 10.1016/j.fishres.2015.05.006
Di Pinto, A., Mottola, A., Marchetti, P., Bottaro, M., Terio, V., Bozzo, G., et al. (2015b). Packaged frozen fishery products: species identification, mislabeling occurrence and legislative implications. Food Chem. 194, 279–283. doi: 10.1016/j.foodchem.2015.07.135
Garcia-Vazquez, E., Horreo, J. L., Campo, D., Machado-Schiaffino, G., Bista, I., Triantafyllidis, A., et al. (2009). Mislabeling of two commercial North American hake species suggests underreported exploitation of offshore hake. Trans. Am. Fish. Soc. 138, 790–796. doi: 10.1577/T08-169.1
Goodyear, C. P. (1988). Recent Trends in the Red Snapper Fishery of the Gulf of Mexico. Miami: National Marine Fisheries Service. Technical Report CRD 87/88-16.
Gulbrandsen, L. H. (2009). The emergence and effectiveness of the marine stewardship council. Mar. Policy 33, 654–660. doi: 10.1016/j.marpol.2009.01.002
IUCN (2019). The IUCN Red List of Threatened Species. Version 2019-2. Available at: http://www.iucnredlist.org (accessed March 26, 2019).
Ivanova, N. V., Zemlak, T. S., Hanner, R. H., and Hebert, P. D. N. (2007). Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 7, 544–548. doi: 10.1111/j.1471-8286.2007.01748.x
Jacquet, J. L., and Pauly, D. (2008). Trade secrets: renaming and mislabeling of seafood. Mar. Policy 32, 309–318. doi: 10.1016/j.marpol.2007.06.007
Khaksar, R., Carlson, T., Schaffner, D. W., Ghorashi, M., Best, D., Jandhyala, S., et al. (2015). Unmasking seafood mislabeling in U.S. markets: DNA barcoding as a unique technology for food authentication and quality control. Food Control 56, 71–76. doi: 10.1016/j.foodcont.2015.03.007
Leal, M. C., Pimentel, T., Ricardo, F., Rosa, R., and Calado, R. (2015). Seafood traceability: current needs, available tools, and biotechnological challenges for origin certification. Trends Biotechnol. 33, 331–336. doi: 10.1016/j.tibtech.2015.03.003
Ling, K. H., Cheung, C. W., Cheng, S. W., Cheng, L., Li, S. L., Nichols, P. D., et al. (2008). Rapid detection of oilfish and escolar in fish steaks: a tool to prevent keriorrhea episodes. Food Chem. 110, 538–546. doi: 10.1016/j.foodchem.2008.02.066
Logan, C. S., Alter, E., Haupt, A. J., Tomalty, K., and Palumbi, S. R. (2008). An impediment to consumer choice: overfished species are sold as Pacific red snapper. Biol. Conserv. 141, 1591–1599. doi: 10.1016/j.biocon.2008.04.007
Marko, P. B., Lee, S., Rice, A. M., Gramling, J. M., Fitzhenry, T. M., McAlister, J. S., et al. (2004). Mislabeling of a depleted reef fish. Nature 430, 309–310. doi: 10.1038/nature02689
Marko, P. B., Nance, H. A., and Guynn, K. D. (2011). Genetic detection of mislabeled fish from a certified sustainable fishery. Curr. Biol. 21, 621–622. doi: 10.1016/j.cub.2011.07.006
Marko, P. B., Nance, H. A., and Van Den Hurk, P. (2014). Seafood substitutions obscure patterns of mercury contamination in patagonian toothfish (Dissostichus eleginoides) or “Chilean sea bass”. PLoS One 9:e104140. doi: 10.1371/journal.pone.0104140
Marschke, M., and Vandergeest, P. (2016). Slavery scandals: unpacking labour challenges and policy responses within the off-shore fisheries sector. Mar. Policy 68, 39–46. doi: 10.1016/j.marpol.2016.02.009
Miller, D. D., and Mariani, S. (2014). Smoke, mirrors, and mislabeled cod: poor transparency in the European cod seafood industry. Ecol. Environ. 8, 517–521. doi: 10.1890/090212
NOAA (2017). Specification of Annual Catch Limits for Red Snapper (Lutjanus campechanus) in the South Atlantic Region. Silver Spring, MD: National Oceanic And Atmospheric Administration.
Oken, E., Choi, A. L., Karagas, M. R., Mariën, K., Rheinberger, C. M., Schoeny, R., et al. (2012). Which fish should I eat? Perspectives influencing fish consumption choices. Environ. Health Perspect. 120, 790–798. doi: 10.1289/ehp.1104500
Ropicki, A. J., Larkin, S. L., and Adams, C. M. (2010). Seafood substitution and mislabeling: WTP for a locally caught grouper labeling program in Florida. Mar. Resour. Econ. 25, 77–92. doi: 10.5950/0738-1360-25.1.77
SEDAR (2012). Stock Assessment of Vermilion Snapper off the Southeastern United States. North Charleston, SC: Southeast Data, Assessment, and Review.
SEDAR (2016). SEDAR 41 – South Atlantic Red Snapper Assessment Report. North Charleston, SC: Southeast Data, Assessment, and Review.
Smith, K. L., and Guentzel, J. L. (2010). Mercury concentrations and omega-3 fatty acids in fish and shrimp: preferential consumption for maximum health benefits. Mar. Pollut. Bull. 60, 1615–1618. doi: 10.1016/j.marpolbul.2010.06.045
Stawitz, C. C., Siple, M. C., Munsch, S. H., Lee, Q., and Safs Research Derby. (2017). Financial and ecological implications of global seafood mislabeling. Conserv. Lett. 10, 681–689. doi: 10.1111/conl.12328
Stiles, M. L., Kagan, A., Lahr, H. J., Pullekines, E., and Walsh, A. (2013). Seafood Sticker Shock Why You May Be Paying Too Much for Your Fish Americans Are Eating More Seafood. New York, NY: Oceana.
USDA (2019). Nutrient Report on 15101, Fish, Snapper, Mixed Species, Raw” and “Nutrient Report on 15261, Fish, Tilapia, Raw. Washington, DC: United States Department of Agriculture.
Warner, K., Timme, W., and Lowell, B. (2012). Widespread Seafood Fraud Found in New York City. New York, NY: Oceana.
Warner, K., Timme, W., Lowell, B., and Hirshfield, M. (2013). Oceana Study Reveals Seafood Fraud Nationwide. New York, NY: Oceana.
Willette, D. A., Simmonds, S. E., Cheng, S. H., Esteves, S., Kane, T. L., Nuetzel, H., et al. (2017). Using DNA barcoding to track seafood mislabeling in Los Angeles restaurants. Conserv. Biol. 31, 1076–1085. doi: 10.1111/cobi.12888
Keywords: red snapper, seafood mislabeling, DNA barcoding, Southeastern United States, marine fisheries, seafood
Citation: Spencer ET and Bruno JF (2019) Fishy Business: Red Snapper Mislabeling Along the Coastline of the Southeastern United States. Front. Mar. Sci. 6:513. doi: 10.3389/fmars.2019.00513
Received: 12 February 2019; Accepted: 05 August 2019;
Published: 27 August 2019.
Edited by:
Marty Riche, Florida Atlantic University, United StatesReviewed by:
Ricardo Calado, University of Aveiro, PortugalPaul S. Wills, Florida Atlantic University, United States
Copyright © 2019 Spencer and Bruno. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin T. Spencer, ZXRzcGVuY2VyMTRAZ21haWwuY29t
 Erin T. Spencer
Erin T. Spencer John F. Bruno
John F. Bruno