- 1Kewalo Marine Laboratory, Pacific Biosciences Research Center, University of Hawai‘i at Mānoa, Honolulu, HI, United States
- 2Proteomics and Metabolomics Shared Resource, Duke University, Durham, NC, United States
- 3Duke University Marine Laboratory, Department of Marine Science and Conservation, Duke University, Beaufort, NC, United States
Growth, molting, and hardening cuticle are intertwined processes for arthropods and share common protein systems to execute these functions. For barnacles, these processes are also tied to adhesion, which is vital to their survival and under great selection pressure. Unlike other crustaceans, barnacle growth, deposition of adhesive material and the curing of glue and cuticle, though associated, are spatially distinct from molting and occur at the interface of the calcareous baseplate and substrata. Here, we use an ancient system, the innate immune response as context and use proteomics and enzyme activity assays to test the hypothesis that chemistries associated with barnacle fluids and glue curing have their origins in wound healing and the innate immune response. Glue precursor material was extracted from barnacles and manipulated to accelerate known trypsin activity, curing, and the effects of sequential sampling. Using non-trypsin digests and searching for tryptic peptides, new products of endogenous trypsin were detected. We found extensive evidence of a prophenoloxidase system, including activating factors, serine proteases, and serine protease inhibitors. Through enzyme-specific substrate assays, phenoloxidase-like activity was found in glue and at the periphery of the base. The enzymatic nature of the phenoloxidase-like activity was confirmed through chemical inhibition and through heating the glue material to denature the enzyme. Further evidence for antimicrobial activity mediated by reactive oxygen species previously described during larval settlement and metamorphosis was the presence of peroxidases in glue precursor material. Beyond clotting, numerous extracellular matrix proteins were identified, suggesting glue curing may lead to stabilization of the initial clot and wound repair. We propose a model of the emerging hypothesis and find exciting potential for further testing innate immune components in this evolutionary approach to barnacle glue curing.
Introduction
The evolutionarily ancient innate immune system is present in all metazoans and enables recognition of self versus non-self and defense responses, which is especially vital in occurrences of wounding (Janeway and Medzhitov, 2002; Medzhitov and Janeway, 2002; Kurtz and Franz, 2003; Litman et al., 2005). In all multicellular organisms, injury activates pathways that stabilize the site, limit infection and repair the injury (Nagai and Kawabata, 2000; Li et al., 2002; Theopold et al., 2002; Frantz et al., 2005; Jiravanichpaisal et al., 2006; Eming et al., 2007). Clotting limits fluid loss is driven by tightly controlled enzymatic cascades (Ravanti and Kähäri, 2000; Werner and Grose, 2003; Theopold et al., 2004; Braiman-Wiksman et al., 2007; Eming et al., 2007).
Convergent evolution has led to extensive overlap between enzyme cascades for blood and hemolymph coagulation and innate immune responses that target microorganisms (Krem and Di Cera, 2002). Three clotting pathways are the blood coagulation cascade (Durliat, 1985; Furie and Furie, 1988; Li et al., 2002), quinone tanning and melanization (Tomasetti et al., 1994; Waite, 1995; Sugumaran, 2002; Suderman et al., 2006), and the prophenoloxidase system (Aspan and Söderhäll, 1991; Sritunyalucksana and Söderhäll, 2000; Wang et al., 2001; Lai et al., 2002; Li et al., 2002; Cerenius et al., 2008; Amparyup et al., 2013). Central to proteolytic cascades is the use of serine proteases to cleave zymogens in activation and serine protease inhibitors to regulate activity, modulating responses with multiple feedback mechanisms (Davie and Ratnoff, 1964; Lee et al., 1998; Dahlbäck, 2000; Jiang and Kanost, 2000; Sritunyalucksana and Söderhäll, 2000; Wang et al., 2001; Yu et al., 2003; Kanost et al., 2004).
The open circulatory systems of invertebrates has extreme sensitivity to exogenous signals and increased clotting speed (Theopold et al., 2004). For arthropods to grow, they must undergo molting events, which can be challenging with an open circulatory system. Molting is complex and requires coordination for the living tissue to separate from the extracellular cuticle so that the exoskeleton can be shed and new material quickly hardened in replacement (Chang and Mykles, 2011). As the molting process unfolds, the internal circulating fluids in contact with tissue, or hemolymph, can be released as a byproduct. Hemolymph can be thought of as a dynamic tissue, the largest tissue of decapod crustaceans, and changes during the molt cycle (Chang, 1995; Chang and Mykles, 2011).
Crustaceans undergo molting in marine or semi-terrestrial environments replete with microbes, enhancing their need for quickly hardening new cuticle and limiting potential infection (Terwilliger, 1999; Taylor and Kier, 2003). One versatile proteolytic cascade that is highly conserved across arthropods, including crustaceans, is the prophenoloxidase system. Phenoloxidases often perform multiple functions, participating in wound healing and cuticle sclerotization via enzymatic oxidation of substrates (Sritunyalucksana and Söderhäll, 2000; Cerenius et al., 2008; Amparyup et al., 2013). Antimicrobial activity is inherent in the proPO cascade as cytotoxic by-products, including quinones and reactive oxygen species are generated in the phenoloxidase reactions. Components of the proPO cascade and quinone tanning mechanisms can be spatially separated with some in the cuticle and others in the hemolymph (Ashida and Brey, 1995; Suderman et al., 2006; Wu et al., 2013). Activation of these components during molting underscores the multifunctionality of phenoloxidases and the degree to which wound healing and cuticle hardening are intertwined, since simultaneous requirements are to harden new cuticle and prevent infection (Dillaman and Roer, 1980; Roer and Dillaman, 1984; Willis, 1999; Glazer et al., 2013).
Barnacles constitute the infraclass of Cirripedia and differ from other crustaceans in their sessile lifestyle and spatial separation of tissues that are shed during molting and the largely calcite shell that remains after molting (Crisp and Bourget, 1985). Planktonic larvae locate a suitable habitat, cued by environmental factors and adult pheromones, and then the cypris larva undergoes settlement and metamorphosis, where it transforms into the adult body plan and permanently adheres to substratum. From that point on, the soft tissue of the animal living within the shell is shed during molting, but the external calcareous shell remains. In acorn barnacles, like Amphibalanus (=Balanus) amphitrite, growth occurs through radial expansion of cuticle of the base and proteinaceous glue is deposited, both of which harden (or cure) and the tissue and glue material eventually becomes calcified into new shell (Bocquet-Vedrine, 1965; Walker, 1971; Bourget and Crisp, 1975; Fyhn and Costlow, 1976; Crisp and Bourget, 1985). The adhesive strength, difficulty to remove these calcareous platforms, and ability to recruit conspecifics and other biofouling organisms through pheromones and kairomones make barnacles particularly pernicious biofoulers on marine substrata. Biofouling comes at great economic cost to the United States Navy, through attempted prevention and removal as well as substantial increases of fuel by ships that become heavily fouled (Schultz et al., 2011). The use of non-toxic coatings on ships to reduce biofouling burden is of great interest to improve efficiency, lessen environmental impact of ships through decreasing fuel use and limit invasive species transport (Callow and Callow, 2002; Holm, 2012). Understanding their glues has gained attention over the last 50 years, both their role in adhesion and in mechanisms to disrupt the curing of them (Saroyan et al., 1968, 1970; Walker, 1971; Lindner and Dooley, 1974).
Barnacle secretions and hemolymph associated with cuticle expansion during molting must manage the same issues as in other arthropods, which is to harden new cuticle and kill microbes. As in all crustaceans, wounding is an inherent part of barnacle molting and growth and leaking of fluids between the base and the surface occurs (Bourget and Crisp, 1975; Crisp and Bourget, 1985; Burden et al., 2012, 2014; Essock-Burns et al., 2017). Barnacle fluid deposition is spatiotemporally dynamic and related to growth and molting. First a cyprid adhesive is secreted via antennules in a bi-phasic system of lipids and proteins, some of which are phosphorylated (Walker, 1971, 1973; Gohad et al., 2014), followed by complex biopolymeric materials associated with reactive oxygen species during the metamorphic molt (Essock-Burns et al., 2017). As juveniles and adults molt and grow, several distinct proteinaceous materials are released (Burden et al., 2012). Detailed work at the growth interface shows lipids are secreted beyond the base and act as a surface-cleaning fluid followed by the deposition of proteins some of which might be delivered through capillaries and ducts (Burden et al., 2012; Golden et al., 2016; Fears et al., 2018). Numerous cement proteins have been identified from solubilizing hardened glue, as well as two similar groups of cuticular proteins, settlement inducing protein complex (SIPC) and MULTIFUNCin (Kamino and Shizuri, 1998; Matsumura et al., 1998a; Kamino, 2001; Dreanno et al., 2006a; Ferrier et al., 2016).
The wound-healing hypothesis of barnacle glue curing proposed initial glue curing was related to blood clotting (Dickinson et al., 2009). Using precursor material collected from the periphery of barnacle bases, two enzymes, trypsin-like serine protease and transglutaminase, were isolated and characterized to participate in barnacle glue curing. Concurrent with a release of proteinaceous fluids under the cuticle during settlement and metamorphosis, reactive oxygen species are present, and contribute to observed antimicrobial activity (Essock-Burns et al., 2017; Fears et al., 2018). The ability to study glue curing as barnacle larvae undergo the metamorphic transition to the adult form and as they continue to grow and adhere, is a strength to this system of study.
In well-studied marine adhesives there are common themes. One theme is involvement of phosphoproteins and quinone tanning (Waite, 1990a, b, 1995). Another theme is in adhesives associated with calcareous structures, phosphorylated proteins within the extracellular matrix participate in the biomineralization (Flammang et al., 2009). Barnacle glue contains phosphoproteins, released during settlement and metamorphosis and during adult growth (Gohad et al., 2014; Dickinson et al., 2016; Essock-Burns et al., 2017). There is one report of evidence for quinone tanning in barnacle adhesive (Lindner and Dooley, 1974). Two known forms of enzymatic crosslinking in glue curing are oxidase activity (So et al., 2017) and transglutaminase (Dickinson et al., 2009).
Proteomics is a tool that has been used on barnacle secretions, cured glue, larvae, and adult tissues to probe mechanisms in this biological process (Thiyagarajan and Qian, 2008; Zhang et al., 2010; Han et al., 2013; Chen et al., 2014; Al-Aqeel et al., 2016). While many barnacle secretion proteins have been described, much of the focus has been on phylogenetic comparisons of those identified with great confidence and distinctions between barnacle groups (Kamino et al., 1996; Matsumura et al., 1998a; Kamino, 2001; He et al., 2013; Ferrier et al., 2016; So et al., 2016). Factors contributing to inter-study variability include working at the limits of the technology, the species and population of barnacle used, collection technique, sample preparation, mass spectrometry equipment sensitivity, databases and annotation used and scoring cutoffs accepted. Here, we report proteomic surveys on precursor material of glue, under different collection schemes and enzymatic treatment to target aspects of glue curing, enabling the discovery of proteins and enzymatic activity that may be relevant at the interface where adhesion occurs.
To assay enzyme activity suggested by proteomic evidence was present, we use enzyme-specific substrates to measure and test the ability to inhibit activity. The evidence suggests that a prophenoloxidase-like system is present in barnacle precursor material and this oxidase activity may contribute to barnacle glue curing. This enzyme system could function to aid in crosslinking of newly deposited glue and harden new cuticle while exhibiting antimicrobial activity to protect from microbial invasion as barnacles grow on biofilmed substrata. This work expands the hypothesis that barnacle glue has similarities to wound healing processes to include the prophenoloxidase system and additional innate immune system components that may also contribute to coagulation and extracellular matrix formation.
Materials and Methods
Barnacle Larval Culture, Settlement and Maintenance
Barnacles, Amphibalanus (=Balanus) amphitrite, were reared at the Duke University Marine Lab in Beaufort, NC from nauplii released from barnacles from the field to mature adults following established culture techniques (Rittschof et al., 1984, 1992, 2008; Holm et al., 2005). Barnacle larvae were settled on glass panels coated with T2® silicone (Dow Corning Silastic, Midland, MI, United States) and grown to a basal diameter of approximately 0.5 cm.
Barnacle Secretion Collection
Barnacle larvae were settled on T2® silicone panels, to allow for removal of the animal while maintaining the integrity of the baseplate (Rittschof et al., 2008). Prior to removal of barnacle from the surfaces were rinsed in deionized water and the surface around the barnacle and outer shell were swabbed with sterile Q-tips and ethanol to minimize biofilm. Using a sterile fine gauge needle (B-D, 22G1, Cat# 305155, Becton Dickinson Company, Franklin Lakes, NJ, United States), barnacles were gently pried from the surface. Immediately after removal, liquid glue was collected by pricking the outer edge of the base, the location of secretion release during cuticle expansion (Dickinson et al., 2009). One to four microliters of fluid were collected with either a glass capillary or a micropipette and sterile plastic tip.
Experimental Design
Precursor glue material was used rather than reworked insoluble cement material as has been done previously (Cheung et al., 1977; Dougherty, 1996; Dickinson et al., 2009). To accelerate the activity of known endogenous trypsin activity, we first treated glue precursor material with experimentally added trypsin (series 1). In a second approach, we allowed extracted precursor material to briefly cure in air, which would result in proteins that become a part of the coagulated material to be bound, and the ones identified to be those that escape the clot (series 2). The third collection scheme was three sequential extractions from the same set of barnacles, 24-h apart, to mimic periodic wounding associated with growth and evaluate the composition of the precursor material (series 3). The material in both series 2 and 3 was digested with a non-trypsin enzyme, a glutamyl endopeptidase, and when searching the database we looked for tryptic peptides assuming those would be from endogenous trypsin activity.
Series 1 Liquid Glue Without Curing
A glass capillary pipette was used to harvest 2 to 6 μl samples of fluids from each of 10 mature barnacles. Each sampled was pipetted immediately into a tube containing 60 μl of 50 mM ammonium bicarbonate buffer (pH 8) on ice and then frozen at −80°C. The total processing time was less than 15 s for each sample.
The Duke Proteomics and Metabolomics Shared Resource (DPMSR) received the ten barnacle samples and generated a master pool of 100 μL by combining 10 μL from each. From the master pool, 10 μL (30.5 μg) of the sample was removed and run on a 10-well, 1.5 mm 4–12% Bis-Tris gel (Life Sciences) with a 46-min run time. After following the vendor-recommended gel staining and destaining protocol, four gel bands were excised and subjected to in-gel trypsin digestion with reduction and alkylation using a standardized protocol found here: https://genome.duke.edu/sites/genome.duke.edu/files/In-gelDigestionProtocol_abbreviatedrevised_0.pdf. The bands were approximately 30 kDa, 90 kDa, 97 kDa, and the fourth band was protein >400 kDa, which remained at the top of gel (Supplementary Figure S1). The incubation fluid was transferred to new tubes and samples were spun to dryness in a vacuum centrifuge.
Series 2 Enrichment for Proteins That Escape Curing Glue
We collected 1 to 3 μl of curing glue from each of 30 adult barnacles using a plastic pipette tip and micropipettor. Each drop of glue extruded from the pipette and cured in air as a hanging drop at 25°C for 1 min and then deposited into one tube containing 200 μl of 30% acetic acid at room temperature. This pooled sample was incubated overnight at 4°C to allow diffusion out of the clots and then centrifuged at 10,000 rpm for 3 min. The supernate was transferred to a new tube. The pellet and supernate were lyophilized, then washed with deionized water, and lyophilized again before storing at −80°C. Samples were transported to the DPMSR facility in Durham, NC on dry ice and stored at −80°C until processing.
The freeze-dried glue sample was resuspended in 200 μL of 50 mM AmBic and subjected to bath sonication (4 rounds per sample at ∼30 s per round). A molecular weight cutoff was performed on the samples using a 0.5 mL 10 kDa Amicon filter (Millipore). The low molecular weight fraction of each sample was lyophilized and subjected to a C18 cleanup using a 1 cc C18 Sep-Pak column (Waters). After elution, the samples were spun to ∼50% dryness in the vacuum centrifuge and then lyophilized to full dryness. The high molecular weight fraction samples were supplemented with 1% ALS-I, reduced, alkylated and digested with GluC overnight at 37°C. The samples were then acidified and lyophilized to dryness.
Series 3 Three Sequential Sampling
To assay the effect of time on wounding and the healing response, in this series barnacles were removed from a surface, glue was collected, and wounds were allowed to heal. Thirty adult barnacles of comparable size were used. After collection they were placed upside down (baseplate facing up) on a bed of damp paper towels in a sealed plastic container, at 28°C to maintain the humidity. Then after 24 and 48 h we gently picked open the scab and collected fluid. At each time interval, 2 μl of fluid were collected with a micropipette and sterile plastic tip and immediately transferred into 200 μl of 30% acetic acid in a microcentrifuge tube. The collection procedure was done at time 0, 24, and 48 h. All thirty barnacles were alive at each time point. Barnacle fluid samples were lyophilized overnight and transported on dry ice to the Duke Proteomics Core Facility in Durham.
The samples were resuspended in 200 μL of 50 mM AmBic and subjected to bath sonication (4 rounds per sample at ∼30 s per round). A molecular weight cutoff was performed on the samples using a 0.5 mL 10 kDa Amicon filter (Millipore). The low molecular weight fraction of each sample was lyophilized and subjected to a C18 cleanup using a 1 cc C18 Sep-Pak column (Waters). After elution, the samples were spun to ∼50% dryness in the vacuum centrifuge and then lyophilized to full dryness. The high molecular weight fraction samples were supplemented with 1% ALS-I, reduced, alkylated and digested with the protease, GluC, overnight at 37°C. The samples were then acidified and lyophilized to dryness. GluC cleaves polypeptides at glutamates and asparagine amino acid residues and was used to avoid cross reactivity with endogenous trypsin in barnacle fluids (Dickinson et al., 2009).
Qualitative Analysis of Sample
Samples were resuspended in 12 μL of 0.2% formic acid/2% acetonitrile (series 1 and 2) or 20 μL of 1% trifluoroacetic acid/2% acetonitrile (series 3). Each sample was subjected to chromatographic separation on a Waters NanoAcquity UPLC equipped with a 1.7 μm BEH130 C18 75 μm I.D. X 250 mm reversed-phase column. The mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. Following a 5 μL injection, peptides were trapped for 5 min on a 5 μm Symmetry C18 180 μm I.D. X 20 mm column at 20 μl/min in 99.9% A. The analytical column was held at 5% B for 5 min then switched in-line and a linear elution gradient of 5% B to 40% B was performed over 60 min at 400 nL/min. The analytical column was connected to a fused silica PicoTip emitter (New Objective, Cambridge, MA, United States) with a 10 μm tip orifice and coupled to a Waters Synapt G2 QToF mass spectrometer through an electrospray interface operating in a data-dependent mode of acquisition. The instrument was set to acquire a precursor MS scan from m/z 50–2000 with MS/MS spectra acquired for the three most abundant precursor ions. For all experiments, charge dependent CID energy settings were employed and a 120 s dynamic exclusion was employed for previously fragmented precursor ions.
Data Analysis and Processing
LC-MS/MS
All MS/MS samples were analyzed using Mascot (Matrix Science, London, United Kingdom; version 2.4.1) and X! Tandem [The GPM, thegpm.org; version CYCLONE (2010.12.01.1)]. Mascot was set up to search the BALAM_adhesproteins_003 database (1492 entries) and a newly deposited transcriptome for A. amphitrite, deposited to NCBI, (Wang et al., 2015) assuming the digestion enzyme trypsin or GluC, depending on the experiment. X! Tandem was set up to search a subset of the BALAM database also assuming trypsin. The programs, Mascot and X! Tandem, were searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 0.80 Da. Deamidation of asparagine and glutamine, oxidation of methionine, acetylation of the n-terminus and carbamidomethyl cysteine were specified in Mascot as variable modifications. Glu->pyro-Glu of the n-terminus, ammonia-loss of the n-terminus, gln->pyro-Glu of the n-terminus, deamidated asparagine and glutamine, oxidation of methionine, acetylation of the n-terminus and carbamidomethyl of cysteine were specified in X! Tandem as variable modifications.
Scaffold (version Scaffold_4.2.1, Proteome Software Inc., Portland, OR, United States) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 99% probability by the Peptide Prophet algorithm (Keller et al., 2002) with Scaffold delta-mass correction. Protein identifications were accepted if they could be established at greater than 98% probability and contained at least 1 identified peptide. The peptide FDR and protein FDR for all experiments was 0.0%. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. Each cluster was counted as a discrete protein.
Proteome Database Treatment
The database used for matching mass spectrometry data to proteins was an amalgamation of previously published and accessible data. The database is largely comprised of sequences translated from the transcriptome of adult Amphibalanus (=Balanus) amphitrite, from the same population of barnacles used in this study, through different stages of molting (NCBI Bioproject PRJNA271096) (Wang et al., 2015). The database also includes two other proteomes from the same barnacle species, one of the barnacle shell (database available with Han et al., 2013) and one that includes tissues in different larval stages (NCBI Bioproject PRJNA79921) (Chen et al., 2014). Recent studies that characterized several novel proteins from barnacle secretions were included (So et al., 2016, 2017). Finally, the contaminant repository for affinity purification (the CRAPome) was added to ensure that background proteins were not incorrectly assigned to samples (Mellacheruvu et al., 2013).
The combined database from the published references was over 3 million sequences. To reduce the database size and only sequences with hits to the public databases were kept as reference. This was done using a BLASTp (Camacho et al., 2009) against NCBI non-redundant protein database was used to annotate the reference proteome. As much of the sequence data was from translated transcripts, asterisks within a sequence indicate a stop codon, sequences containing X’s, indicating unknown amino acids were not removed. A peptide containing a single X generated 20 peptides to be tested through the Mascot program. To avoid excessive search times, Mascot tests all possibilities up to a maximum of 3 B or Z residues and 1 X residue per peptide. Sequences were clustered to eliminate duplicate entries. Enzyme classifications were done using the KEGG pathway analysis (Kanehisa and Goto, 2000). For the functional annotation of the proteome, Gene Ontology (GO) mapping of the protein sequences was performed with Blast2go software (Götz et al., 2008). The final database (Supplementary Data Sheet S1) included 35,748 annotated sequences.
Enzyme Activity Assays
Substrates
Enzyme assays with colorimetric substrates assessed activity on monophenols and diphenols. All reagents were purchased from Sigma-Aldrich, Inc., (St. Louis, MO, United States). Substrates were either 1 or 10 mM in MES buffer (Wang and Stewart, 2013). To assay monophenolase activity, L-tyrosine was used. To assess diphenolase activity of PO L-dopa (3,4-dihydroxyphenylalanine), 4-methoxyphenol and DHPPA, an analog of dopamine, were used. ABTS was a third diphenolase substrate for laccase was also used. Also, 3-methyl-2-benzothiazolinone hydrazone (MBTH) was used in conjunction with DHPPA and L-dopa to trap the initial quinone product as an adduct, more stable than dopachrome (Morales et al., 1995; Espín et al., 1997). All substrates were freshly made for each assay and stored in the dark prior to use to limit the auto-oxidation of L-dopa.
Inhibitors
All inhibitors were made fresh to 10 mM in MES buffer for each assay (Wang and Stewart, 2013). Salicylhydroxamic acid (SHAM) is a copper chelator that inhibits catechol oxidase (diphenolase) activity but not laccase. Phenylthiourea (PTU) is a copper chelator that inhibits all phenoloxidase and laccase activity. Arbutin was used as an L-tyrosine analog that inhibits monophenolase activity. For glue assays, the inhibitor was incubated with the substrate and glue concurrently, for barnacle baseplate assays, the inhibitors were incubated overnight prior to exposure with substrates.
Assay and Analysis
Each assay was run with a standard curve, using 0, 1, 5, and 10 U of tyrosinase (from Mushroom, Cat No 9002-10-2). Each assay with glue used 10 μl of glue combined from 3 to 5 barnacles per well. Solutions were stored at 4°C until the start of the reaction. Total reaction volume was 250 μl (clear bottom 96-well plates), 100 μl of substrate with 50 μl of inhibitor, along with 1, 5, or 10 μl of tyrosinase (for standards) or 10 μl of glue and the rest of the volume with 0.22 μm filtered seawater. The change in absorbance over time was measured using a spectrophotometer Spectramax M2 Microplate Reader (Molecular Devices). Assays were run at 37°C, taking readings every minute and kept in the dark to prevent autooxidation. For L-dopa, the absorbance was read with 490 nm, for L-tyrosine, the wavelength was 405 nm and for ABTS, 280 nm was used. Co-incubation with chymotrypsin was done to test if enzymatic activation of an endogenous phenoloxidase was needed. For assays with heated glue, glue was boiled at 95°C for 10 min prior to incubation with substrates. Prism 7.0 for Mac, GraphPad Software, La Jolla, CA, United States, www.graphpad.com was used for analyses and figures. Statistical analyses conducted were a one-way ANOVA and Tukey’s multiple comparisons test to determine which treatments were significantly different.
Results
Characterization of the Barnacle Proteome Database
The filtering and refinement of the barnacle protein database resulted in 35,748 annotated sequences (Supplementary Data Sheet S1). The results from comparing our database of sequences to the NCBI non-redundant database using BLAST showed that the top hit matches 19,250 different taxa, including microbial and human proteins (Supplementary Data Sheet S2). The most represented invertebrate group was the Mediterranean mussel, Mytilus galloprovincialis (961 sequences, Supplementary Data Sheet S2). There were 19 taxa of barnacles representing 440 sequence entries, with Amphibalanus amphitrite making up 37% (164 sequences) of those. Enzyme classifications revealed 2,330 enzymes in the database, about 70% of which were hydrolases (1619 sequences) matching six EC subclasses (Supplementary Table S1). The most common hydrolase type was peptidases (1220 sequences). Another enzyme group of interest, oxidoreductases, had 283 sequences matching 15 EC subclasses.
Gene ontology classification enables the grouping of the barnacle protein database into three main categories: biological processes, cellular components, and molecular functions. Within the category of biological process, several salient subcategories were immune system process (GO:0002376), response to wounding (GO:0009611), regulation of bodily fluid levels (GO:0050878), and secretion (GO:0046903). Representative proteins from each subcategory include fibrinogen, prothrombin, serine peptidase inhibitor, and amyloid-beta A4 family. Cellular component subcategories like extracellular region (GO:0005576) and vesicle (GO:0031982) contained metalloproteases, serine protease inhibitors, coagulation factors, and plasminogen. Molecular function subcategories of interest were serine-type peptidase activity (GO:0008236), of which there were 353 entries, and endopeptidase activity (GO:0004175). Both subcategories included numerous forms of trypsin and other coagulation factors. Gene ontology terms and coagulation literature was used to group the proteins identified through peptide hits into the broad categories of clotting-related, extracellular matrix (ECM)-related, and other, which included vitellogenin, barnacle waterborne settlement pheromone and nucleosome assembly protein and other cell functions.
Clotting-Related Proteins Are Most Common in Barnacle Fluids
Across all experiments, 867 peptides were identified overall matching 73 proteins. Experimental series 1 (accelerated trypsin activity) yielded 544 peptides matching 38 proteins, while series 2 (proteins excluded from the clot) had 141 matching 16 proteins, and series 3 (three sequential wounds) had 182 peptides matching 19 proteins (Table 1). Certain proteins were only found when searching for trypsin specificity for samples digested with glutamyl endpeptidase (GluC), due to the activity of naturally occurring trypsin-like serine proteases in the glue (Table 1). The sampling strategy was the main defining factor between each experiment, but a secondary factor was the sample preparation strategy and two enzyme digests were used, trypsin or glutamyl endopeptidase. The searching strategy for peptides assuming both trypsin and endopeptidase digestion (those with GluC) revealed an additional 57 peptides in series 2 and 144 peptides in series 3 (Table 1).
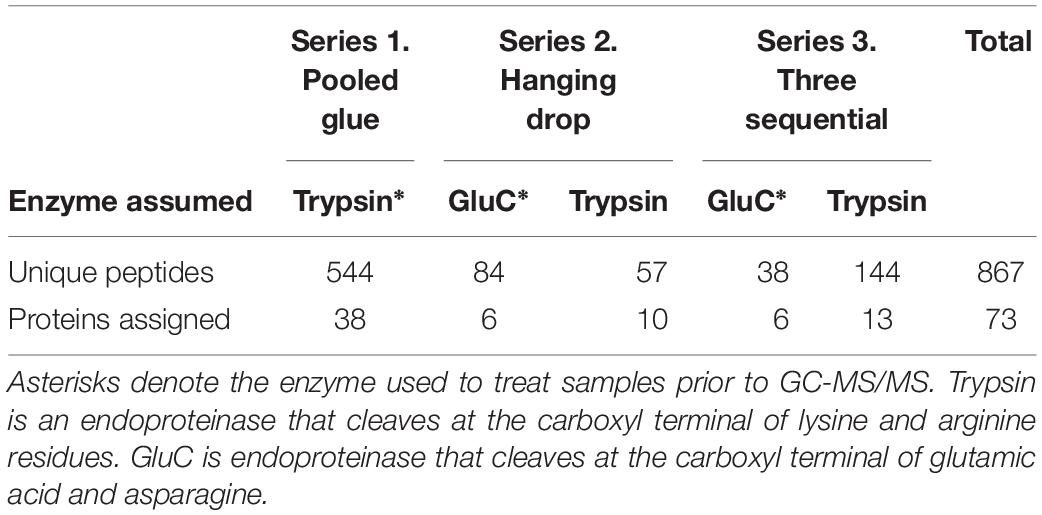
Table 1. Total unique peptides identified in barnacle secretions for each experimental series and enzyme-specificity search strategy.
The protein sequences that the peptide data recognized were grouped into three broad categories based on the putative function of the best-annotated hit (Tables with the transcript ID are in Supplementary Data Sheet S2). Hypothetical or uncharacterized proteins were classified based on the conserved domains reported or the second best hit if no domains were detected. The proteins identified were grouped into clotting-related, extracellular matrix-related, and other. Clotting included activators and regulators of clotting systems like serine proteases and inhibitors as well as proteins with domains like fibrinogen and C-type lectins. Clotting activators and regulators were in the gene ontology categories of response to wounding (GO:0009611), regulation of bodily fluid levels (GO:0050878) and secretion (GO:0046903). C-type lectins were included here because of their role as a pattern recognition protein in initiating innate immune responses (King et al., 2003; Robinson et al., 2006), and are one way to activate the prophenoloxidase system (Kanost et al., 2004; Wu et al., 2013; Wang and Wang, 2013).
The top seven identified clotting-related proteins found in two or more experiments accounted for 29% of all peptides found (254 unique peptides). These shared proteins included four clotting activators and one inhibitor (Table 2 and Figure 1). Clotting activators were prophenoloxidase activating factor, C-type lectin, heat shock protein 70, and serine proteases. The inhibitor represented was pacifastin-like (light chain), a serine protease inhibitor that regulates the prophenoloxidase cascade. The sampling strategy of pooled glue samples with in-gel trypsin digestion yielded the most clotting-related peptides, but the two others experimental series had representative peptides from all five of these proteins (Figure 1 and Table 2).
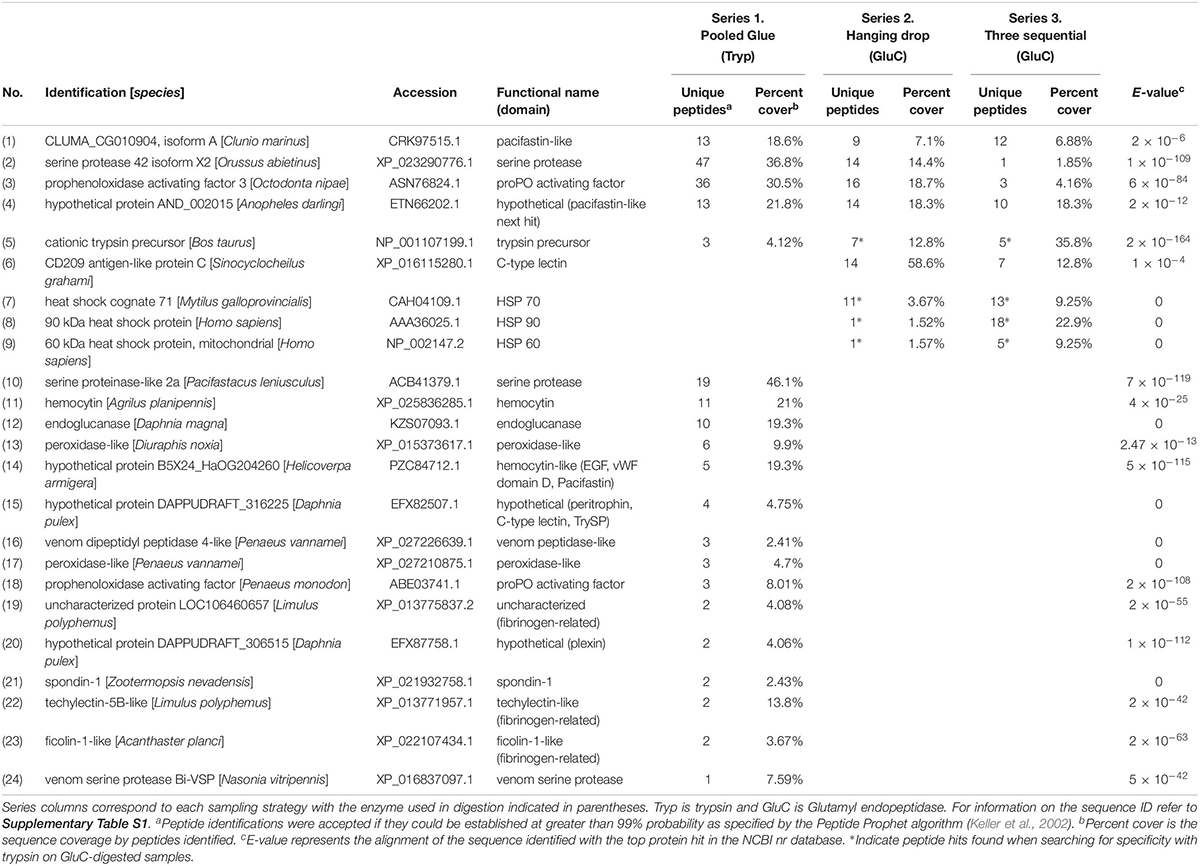
Table 2. Clotting-related proteins detected in barnacle secretions by LC-MS/MS based on peptides identified and sequence coverage. The protein identification corresponds to the annotation with the top hit in NCBI with the based on the E-value given for the match.
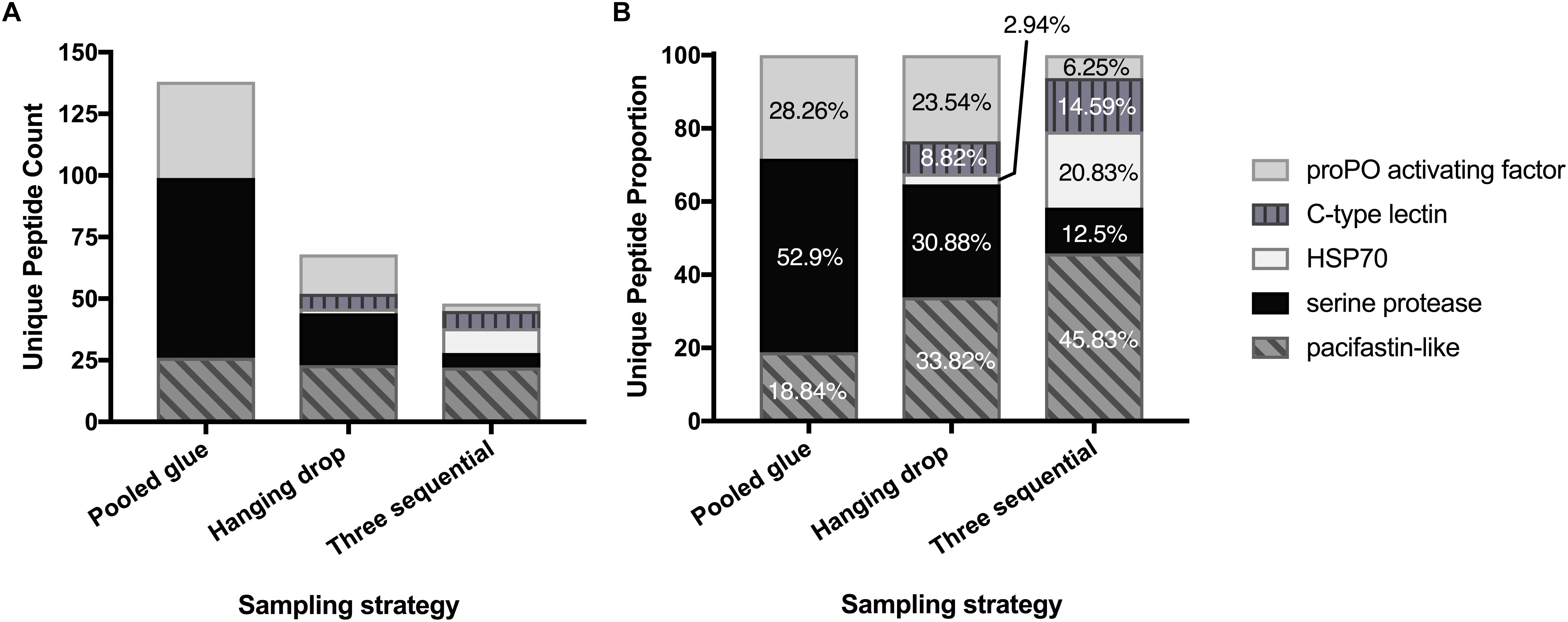
Figure 1. Enrichment of key clotting-related proteins across the three experimental series. Distribution of unique peptides counts (A) and proportion of each (B) for five prevalent clotting proteins shared across experimental series.
Extracellular matrix (ECM)-related proteins shared between experimental series were less prevalent than clotting proteins. The only shared extracellular protein shared in all three studies was a barnacle cuticle protein called settlement inducing protein complex (SIPC). SIPC was found in abundance (143 peptides and 55% sequence coverage) in the accelerated trypsin activity approach (series 1) and detected in lower abundance in series 2 and 3 (Table 3). The top shared ECM-related proteins between series 2 and 3 were actin, vimentin, and tubulin (131 peptides; Table 3). They were revealed when endogenous trypsin digestion was assumed during Mascot searching of the database.
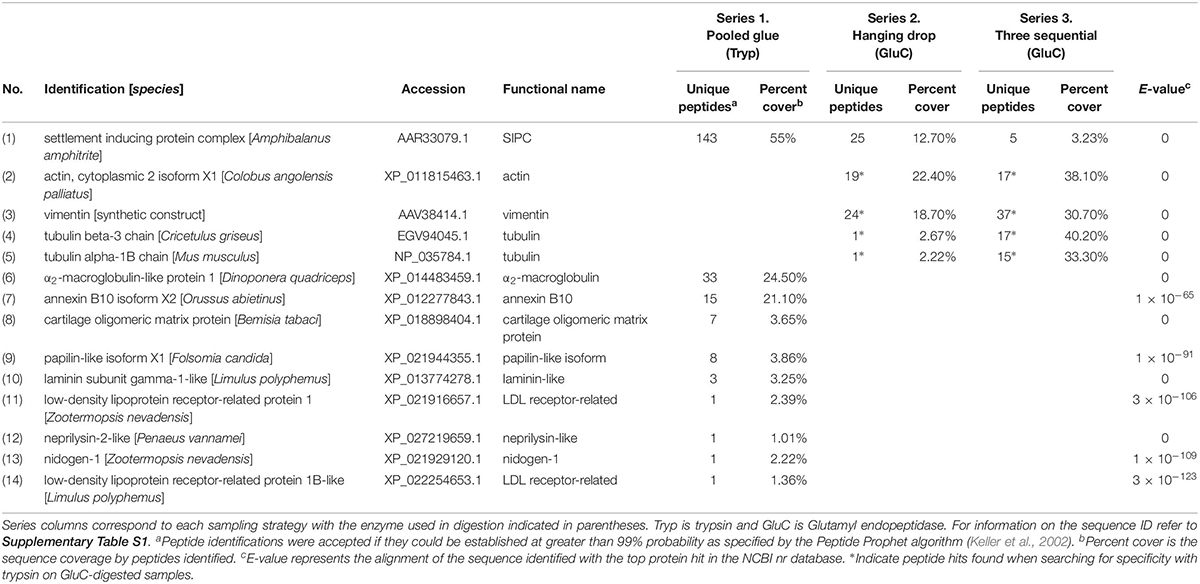
Table 3. Extracellular Matrix (ECM)-related proteins detected in barnacle secretions by LC-MS/MS based on peptides identified and sequence coverage. The protein identification corresponds to the annotation with the top hit in NCBI with the based on the E-value given for the match.
The trypsin-digest approach in experimental series 1 produced another 15 clotting proteins unique to it and 9 ECM-related proteins (Tables 2, 3). The most prevalent clotting-related proteins identified only in series 1 were hemocytin (16 peptides), endoglucanase (10 peptides) and peroxidase (9 peptides) (Table 2). The remaining clotting-related proteins had fewer peptide hits and were categorized based on conserved domains (Table 2). Common themes were hits to proteins that with domains like fibrinogen, von Willebrand factor domain D, trypsin inhibitor, and C-type lectin, totaling 23 unique peptides.
Experimental series 2 aimed to enrich for proteins that escape the clot during initial curing and series 3 for those that are upregulated after initial wounding by sampling. Unique to these experiments were several clotting-related proteins that initiate innate immune responses, CD209 antigen-like protein C, heat shock cognate 71, heat shock protein 90 and 60 (70 peptides; Table 2). The heat shock proteins were found when searching for tryptic specificity in GluC-digested samples. In series 3, the first collection had the most clotting-related proteins (20 peptides for 7 proteins) and the samples collected 24 h after the first wounding event had the most peptides identified (130 peptides), many of which were cytoskeletal and heat shock proteins. The third collection at 48 h, had the fewest peptides overall (18 peptides).
The proteins in the remaining group were comprised of additional previously described barnacle proteins and those with an array of cell functions (Table 4). The two most abundant in series 1 were vitellogenin (112 peptides) and waterborne settlement pheromone (14 peptides) (Table 4). The most abundant protein of this category in series 2 and 3 was a nucleosome assembly protein; histone H4-like, with 17 peptides and 52.4% sequence coverage.
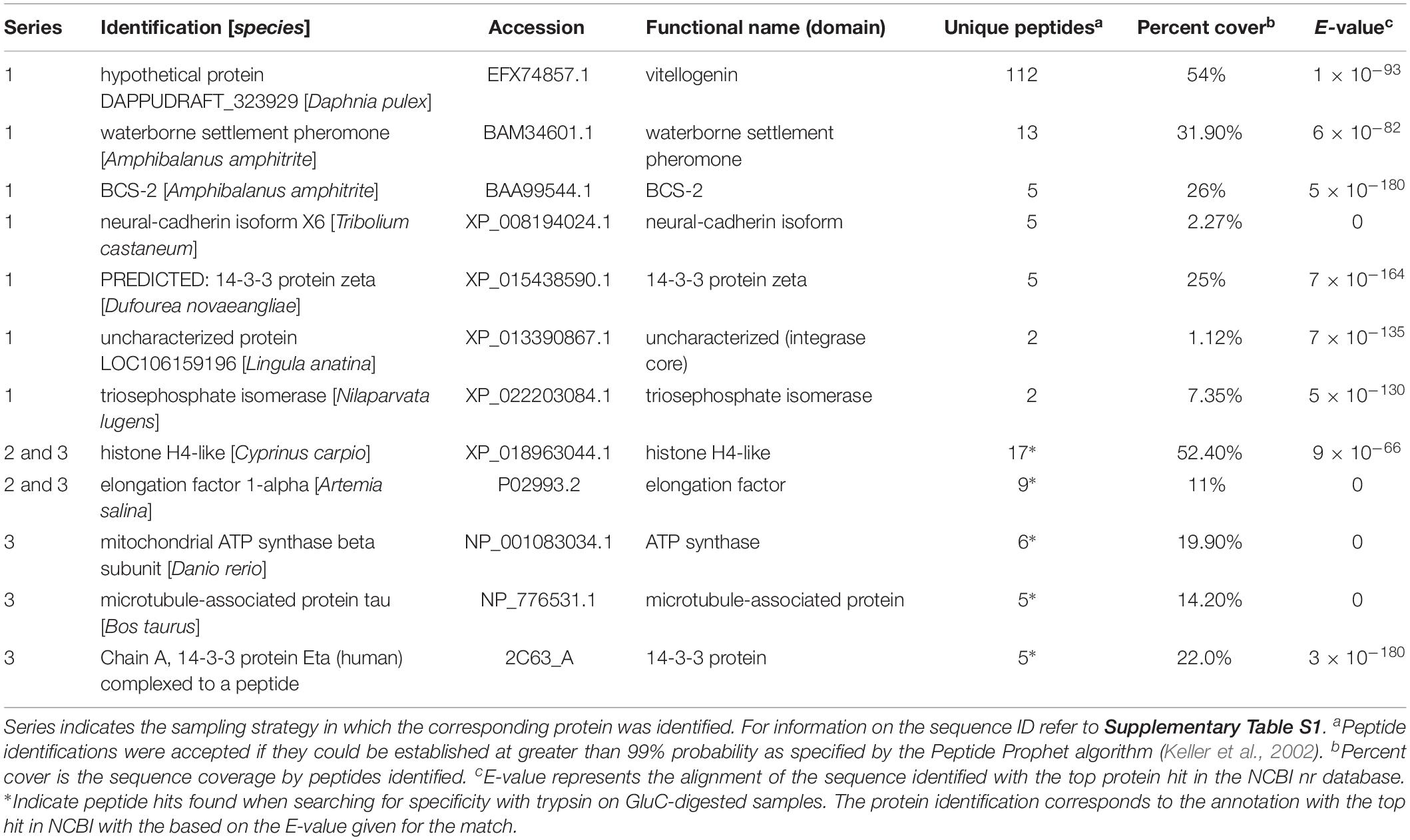
Table 4. Other proteins identified in barnacle secretions by LC-MS/MS based on peptides identified and sequence coverage.
Phenoloxidase Activity Is Detectable on Barnacle Bases and in Glue
To further explore the idea of clotting mediated through the prophenoloxidase (proPO) cascade, we probed barnacle fluid and bases with enzyme-specific substrates and inhibitors. We found that barnacle fluids contain enzyme activities resembling diphenolases in proPO as well as broader phenoloxidase activity in the barnacle basis as described below.
Barnacle baseplates showed colorimetric changes in response to L-dopa, L-tyrosine, and ABTS (Figure 2A). Exposure to inhibitors prior to the addition of substrate did not affect the reaction. A dose-response of color change was observed when comparing 1 to 10 mM substrates, especially L-dopa (Figure 2A). The regions of the observed color change were often at the edge of the base, where the parietal plates meet the baseplate, and found in concentric rings away from the edge. The phenoloxidase activity on the barnacle base resembles broader phenoloxidase activity as it reacts with monophenols (L-tyrosine), diphenols (L-dopa) and ABTS, a substrate for laccase, an enzyme with a similar role to diphenolases.
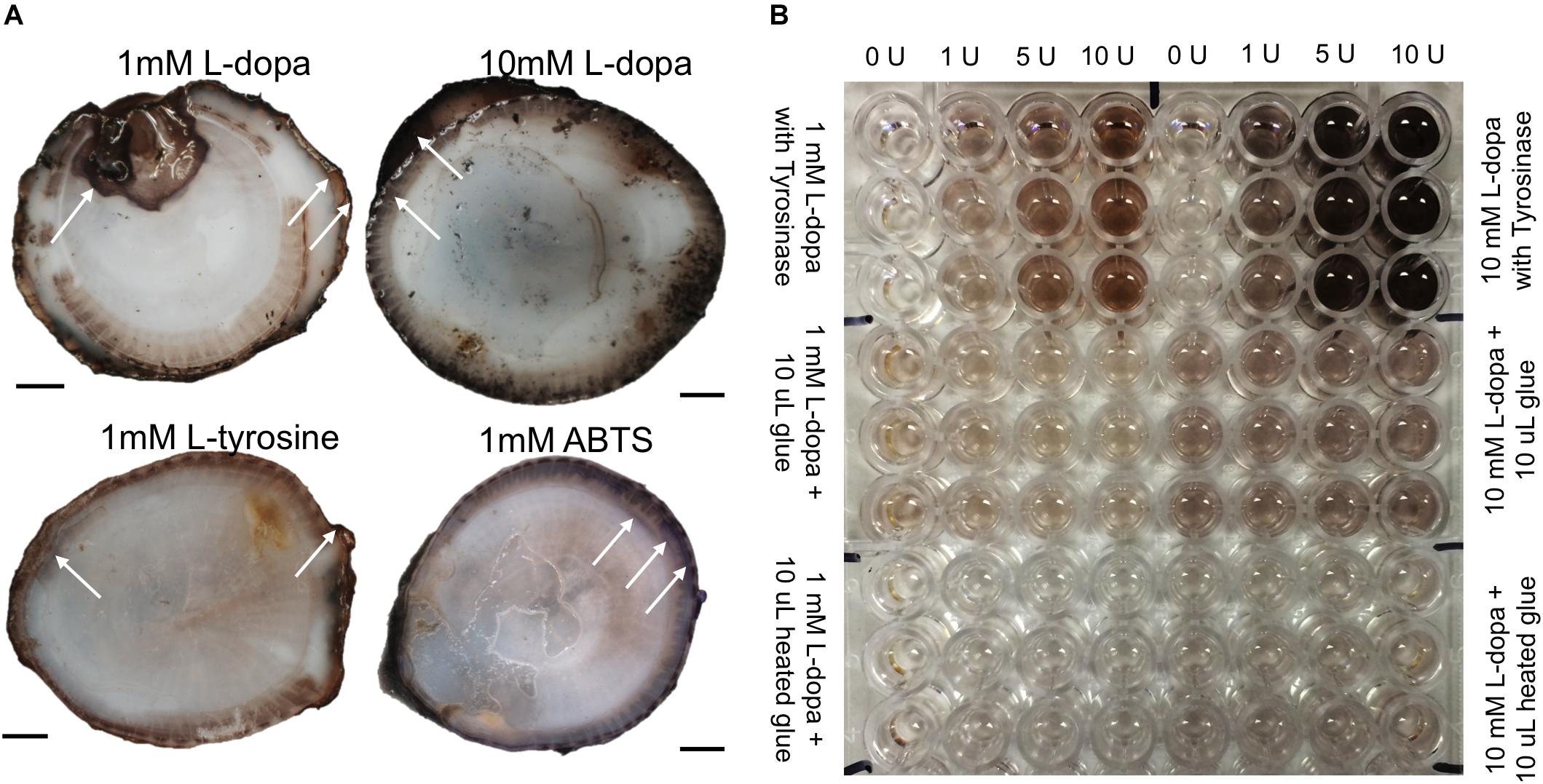
Figure 2. Colorimetric enzyme-specific substrate incubated on barnacle bases (A) and with liquid glue in spectrophotometric assays (B). (A) Arrows indicate example regions of observed color changes, L-dopa product was black (darker with higher concentration substrate), L-tyrosine product was lighter brown, and ABTS product was purple. (B) Top third of plate shows enzyme standards, run in triplicate with every assay and the corresponding units of tyrosinase added to generate the associated color change. Middle region shows liquid glue with L-dopa substrate (12 replicates for each concentration), and bottom third shows assays with heated glue. Scale bars are 1 cm.
The sole activity in glue assays was with L-dopa as a substrate (Figure 2B). There was no monophenolase-like activity with L-tyrosine and no diphenolase-like activity with the other substrates, DHPPA or ABTS. The addition of MBTH did not change activity with L-dopa, nor did co-incubation with chymotrypsin on the standards or the glue assays (data not shown). Glue showed diphenolase-like activity, acting with L-dopa significantly more than the L-dopa alone, for both 1 mM (p < 0.001) and 10 mM (p < 0.0001) concentrations (Figures 3, 4).
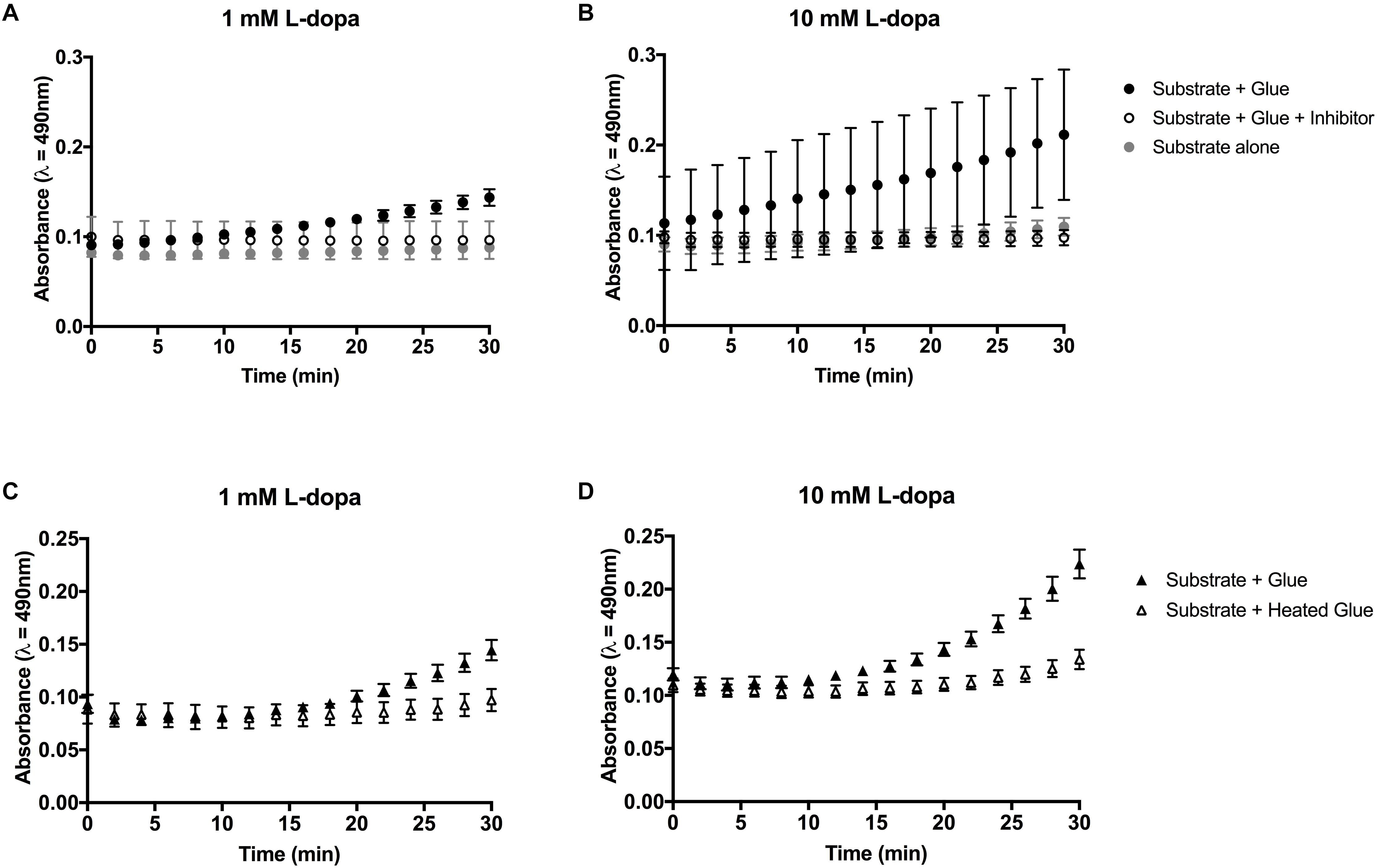
Figure 3. Colorimetric assays for diphenolase activity in barnacle glue with L-dopa as the substrate. (A,B) Diphenolase activity in glue is reduced in the presence of salicylhydroxamic acid (SHAM) inhibitor. (C,D) Heating the glue prior to the assay limits the diphenolase activity. Bars indicate the 95% confidence intervals of the mean at each time point. Non-overlapping data points are considered statistically different.
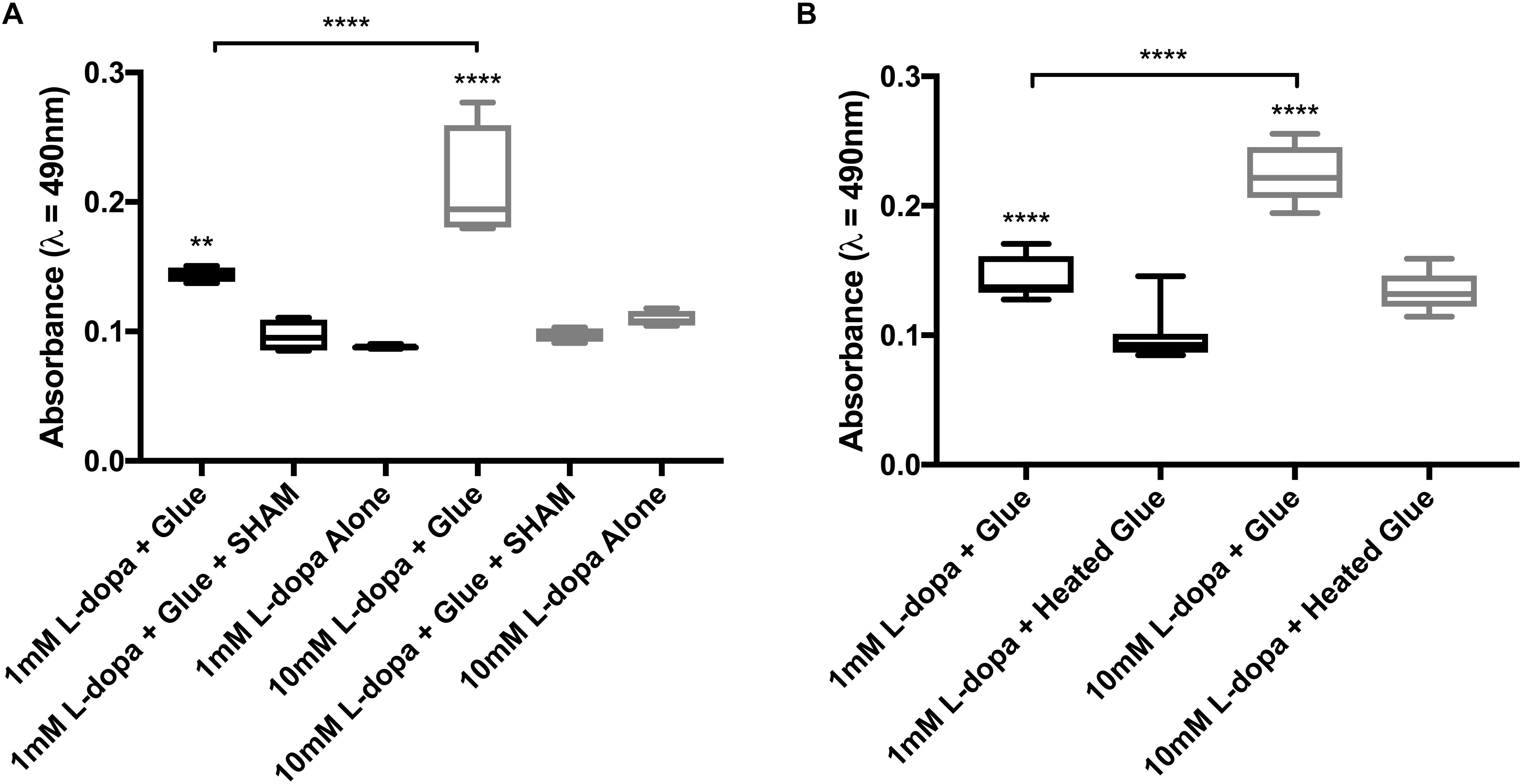
Figure 4. Endpoint data from colorimetric assays of diphenolase activity in barnacle glue, taken at 30 min. (A) Activity with 1 and 10 mM substrate (L-dopa), inhibitor (SHAM) and control with substrate alone. (B) Activity with both substrate concentrations and glue untreated as compared to heated prior to the assay. Significance is indicated by asterisks (∗∗p < 0.01 and ****p < 0.0001).
Activity in glue could be effectively inhibited by the addition of a copper chelating inhibitor SHAM (Figures 3A,B, 4A). At the endpoint of the L-dopa with glue enzyme assay, estimation of activity was significantly higher in treatments with substrate and glue over that with the SHAM inhibitor (p < 0.01) (Figure 4A). While diphenolase activity in the glue was greater in assays with a higher concentration of substrate (p < 0.0001), and SHAM was equally effective at inhibiting diphenolase activity at both concentrations (p < 0.0001) (Figures 3A,B, 4A). There was no significance difference between SHAM inhibitor and 1 or 10 mM L-dopa alone.
Further evidence of endogenous diphenolase activity was in the heated glue treatments. Heating the glue significantly eliminated diphenolase activity (p < 0.0001) (Figures 2B, 3C,D, 4B). This effect was equally pronounced at both concentrations of substrates (p < 0.0001) (Figure 4B). By inferring activity levels in glue from the standard curve of activity with tyrosinase, 10 μl barnacle glue has between 0.48 and 0.68 units of diphenolase-like activity (Supplementary Figure S2).
Discussion
Our goal was to probe aspects of barnacle glue curing and similarities to coagulation cascades using a broad proteomics approach with targeted enzymatic treatment of precursor material to selectively enrich rarer proteins swamped by the dominant ones. The three proteomic-series were designed to accelerate the effect of trypsin on precursor material to analyze the products of a known endogenous enzyme, which yielded more serine proteases than any other strategy. In addition to serine proteases detected in strategies that relied on endogenous trypsin, initiator proteins of the prophenoloxidase cascade were identified when searching for tryptic peptides, further supporting that trypsin-like serine proteases are key activators in precursor glue material (Dickinson et al., 2009). While we found previously described barnacle cement proteins, these were in the minority. This was expected as most of cement studies begin with reworked cured cement, and our strategies were designed to enrich for less abundant proteins and precursors in curing rather than the final product. As full curing and reworking may take many weeks especially for some oxidative chemistries to take effect, we expected the beginning material would be substantially richer in content than the final calcified cement products.
Clotting Systems Are Prevalent in Barnacle Secretions
Proteolytic clotting systems in arthropods contain diverse proteins to initiate cascades. Once the cascade is initiated, serine proteases sequentially activate other enzymes and serine protease inhibitors help to regulate the processes. Serine proteases, prophenoloxidase activating factor, and serine protease inhibitors were most common proteins across all experiments, which are major components of the clotting and prophenoloxidase cascades (Figure 5; Davie and Ratnoff, 1964; Jiang and Kanost, 2000; Wang et al., 2001; Kanost et al., 2004; Dickinson et al., 2009). Additional clotting activators identified include C-type lectins, heat shock protein 70, endoglucanase, and hemocytin, each of which are implicated in initiating clotting responses and defense systems in invertebrates (Ochiai and Ashida, 1988; Kotani et al., 1995; Cheng et al., 2003; Kikuchi et al., 2005; Cellura et al., 2006; Wu et al., 2013; Niksirat et al., 2014). Heat shock proteins have intracellular roles as chaperones, but have emerged as potent signals eliciting innate immune responses (Byrd et al., 1999; Milani et al., 2002; Srivastava, 2002a, b). In particular, heat shock protein 70 is an evolutionarily conserved molecule that can prime the prophenoloxidase system in crustacean (Artemia), inducing a protective effect to challenge with bacteria (Baruah et al., 2011). HSP90 was one of the most differentially expressed proteins identified in a proteomic study of shrimp hemocytes after infection (Somboonwiwat et al., 2010). While we also identified heat shock protein 60 and 90, only HSP70 was included in the clotting initiators because of its aforementioned connection with the proPO cascade, but it is possible that they all initiate immune responses.
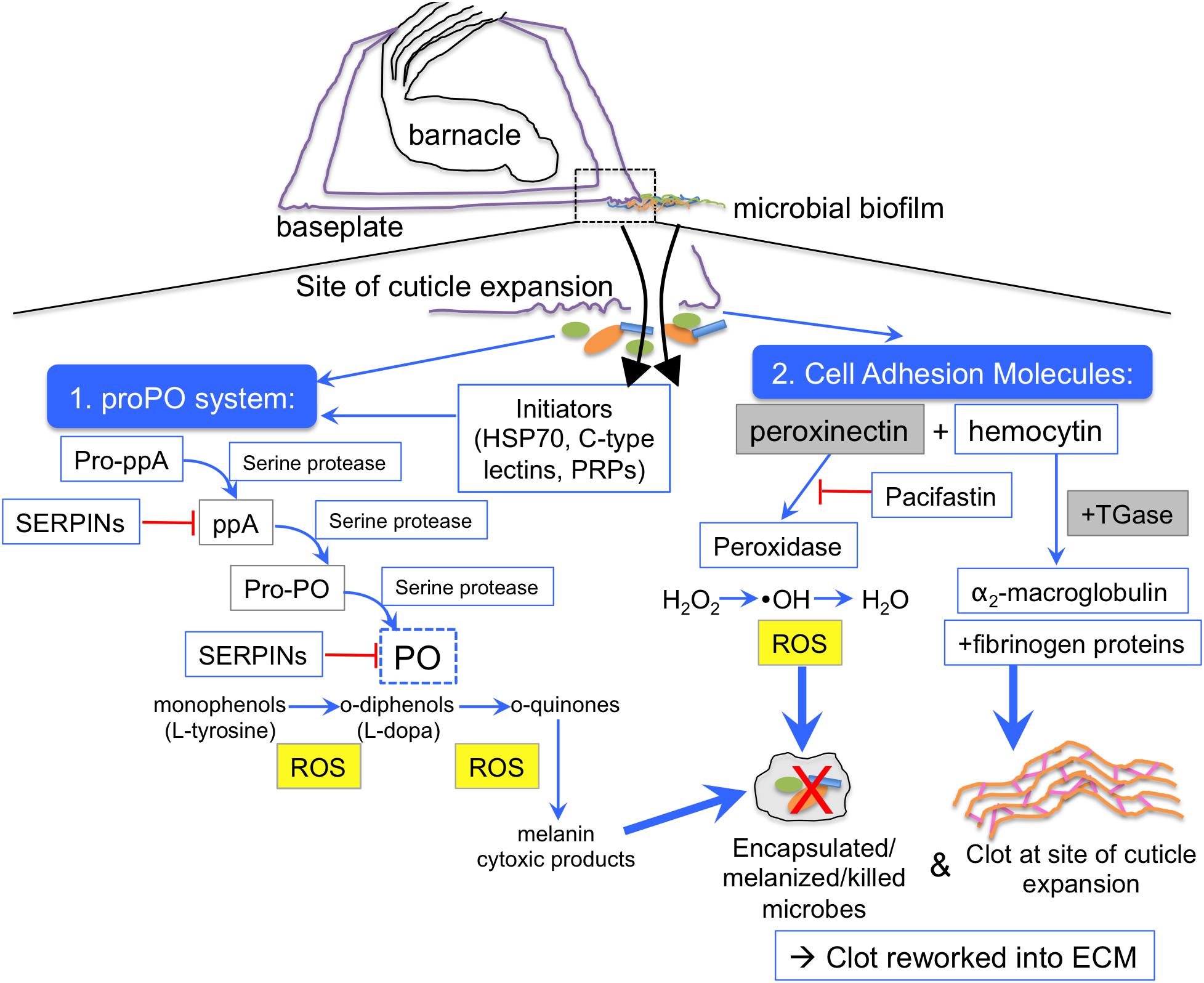
Figure 5. Emerging hypothesis of barnacle glue coagulation scheme mediated by the prophenoloxidase cascade. During barnacle growth cuticle is extended at the outer edge of the base over a biofilmed surface (dotted box). As the cuticle stretches (purple), there are tears, which release fluids (black arrows) that come into contact with microorganisms (orange/green/blue shapes), which present microbe-associated molecular patterns (MAMPs) to the host innate immune system. Two main clotting and defense responses are initiated, one mediated by the prophenoloxidase system (proPO) and one mediated by peroxinectin. ProPO can be activated by initiator proteins like C-type lectins and heat shock protein 70, and other pattern recognition proteins (PRPs) that recognize MAMPs, like endoglucanase. Activation of the phenoloxidase system is initiated via prophenoloxidase activating factor/enzyme (pro-ppA) and mediated by zymogen activation via serine proteases. After the activation of pro-ppA, intermediates like phenoloxidase activating enzyme (ppA), and prophenoloxidase (PPO) are cleaved until finally phenoloxidase (PO) is freed to act on monophenol and diphenol substrates. Serine protease inhibitors (SERPINs), including pacifastin, inhibit (red line) the action of serine proteases at any step of the cascade. Reactive oxygen species (ROS), quinones, and cytotoxic intermediates can all kill nearby microorganisms via encapsulation, melanization and direct oxidation. A concurrent pathway could occur through activation cell adhesion molecules like peroxinectin, a multifunctional protein with peroxidase activity, which converts hydrogen peroxide to a hydroxyl radical (a reactive oxygen species) that can kill microbes and also converts that to water, thus creating and neutralizing ROS in one enzyme. Peroxinectin and hemocytin can interact with clotting proteins (fibrinogen-like proteins) and the crosslinking enzyme transglutaminase (TGase), which crosslinks proteins. Transglutaminase also may act on α-2 macroglobulin, incorporating it into the clot to inhibit bacterial proteases. A crustacean serine protease inhibitor, Pacifastin, stops the phenoloxidase-like activity of peroxinectin (red line). Blue boxes indicate proteins identified through proteomics in this study, dotted blue box indicates confirmation through enzyme assays of this study, the yellow-filled gray box has been shown through microscopy of previous studies (Essock-Burns et al., 2017; So et al., 2017), and gray boxes indicate suspected proteins based on previous work, such as peroxinectin (So et al., 2016) and transglutaminase (Dickinson et al., 2009).
Previous work identified trypsin-like serine proteases and characterized their activity in barnacle glue (Dickinson et al., 2009). This allowed us to test the impact of endogenous trypsin activity on proteins in samples by searching for tryptic peptides in the experiments not digested with trypsin. Serine proteases were identified in all experiments, even those without trypsin digestion, demonstrating the prevalence of this endogenous proteolytic activity. Additional clotting enzymes demonstrated in glue include transglutaminase, a crosslinking enzyme conserved in fibrinogen-based blood clotting and peroxidases (Dickinson et al., 2009; So et al., 2017). Although we identified several peroxidase peptides, they did not match the recently described proteins from the same barnacle species and a similar database (So et al., 2017). Fibrinogen domains were prevalent in proteins identified like techylectin, ficolin, and uncharacterized proteins, which could be substrates for transglutaminase.
In addition to activators and inhibitors of prophenoloxidase, evidence of phenoloxidase-like activity was supported by the enzyme activity assays. Barnacle baseplates, particularly at the periphery where new cuticle growth occurs, converted many of the substrates, indicating substrate promiscuity of the active enzymes. In contrast, the extracted glue solely had diphenolase activity, which was effectively inhibited by a copper chelator and through heating the glue. The fact that the activity with L-dopa could be inhibited by heating the glue prior to the assay indicates that the activity is enzymatic and not due to auto-oxidation. We hypothesize that the cuticle and shells of barnacle bases provide a broader range of enzyme functions, including monophenolase and diphenolase activity as shown here, and peroxidase activity as previously described (Dickinson, 2008; So et al., 2017). This suggests that the components to initiate the prophenoloxidase cascade and the diphenolase activity of the phenoloxidase are mostly in the glue, but enzymes with monophenolase activity in the first part of the reaction are in the cuticle. Separation of quinone tanning components in hemolymph and cuticle is common in other arthropods (Ashida and Brey, 1995; Suderman et al., 2006; Gorman et al., 2007; Wu et al., 2013). As the growing cuticle at the periphery of the barnacle base is the site of expansion and release of glue fluids, all of the enzymatic components of this reaction occur in the same region. This interface between new barnacle growth, glue release, and phenoloxidase activity also include microbial biofilms on a natural surface in seawater. We hypothesize that the phenoloxidase activity in glue is involved with crosslinking, clotting and killing microorganisms and the phenoloxidase activity in base/cuticle assists in these aspects of cuticle sclerotization and biomineralization (Figure 5). The presence of phenoloxidase components and activity in barnacle glue has been debated for the past 50 years (Walker, 1971; Lindner and Dooley, 1974; Cheung et al., 1977; Lindner, 1984; Naldrett, 1993). We propose that the analysis of precursor material presented here presents a fuller picture of potential molecular players in the mechanisms contributing to glue curing.
Many of the oxidase enzymes can play interchangeable roles in coagulation and pathogen defense in arthropods. One such dual-functioning protein has been identified in barnacle, peroxinectin, has roles in defense via phenoloxidase activation leading to encapsulation and phagocytosis of invading microorganisms (Johansson, 1999; Lin et al., 2007) as well as peroxidase activity which generates reactive oxygen species, hydrogen peroxide (H2O2), at sites of injury (Johansson, 1999; Lin et al., 2007; Minibayeva et al., 2009). Phenoloxidases also generate ROS in the conversion of monophenols to quinones. ROS are present in barnacles at times of cuticle expansion and fluid release during barnacle metamorphosis (Golden et al., 2016; Essock-Burns et al., 2017; Fears et al., 2018). We suspect that the ROS found are involved in clotting and killing microorganisms on the surface, and then are neutralized by protective enzymes, like thioredoxin, previously found to be upregulated prior to molting (Wang et al., 2015). As defense and crosslinking mechanisms are intertwined, we expected that barnacle fluids would contain redundant mechanisms for antimicrobial defense. This would include antimicrobial peptides, which were not identified in the present study, but are common in crustaceans (Amparyup et al., 2008; Battison et al., 2008; Smith et al., 2008).
Regulation of proteolytic cascades by protease inhibitors is crucial to survival (Werner and Grose, 2003; Kanost et al., 2004). Our proteomic data suggest that the putative prophenoloxidase cascade in barnacle secretions is likely mediated by the serine protease inhibitor similar to pacifastin light chain, a well-described serine protease inhibitor first described in the plasma of freshwater crayfish Pacifastacus leniusculus, and involved in many arthropod prophenoloxidase cascade (Kopáček et al., 1993; Liang et al., 1997; Söderhäll and Cerenius, 1998; Simonet et al., 2002; Breugelmans et al., 2009). Another inhibitor of proteases that was identified is α2-macroglobulin, which can bind a broad range of proteases, physically trapping them, and marking them for removal through endocytotic pathways in innate immune systems, including arthropods (Orth et al., 1992; Armstrong and Qmoley, 1993; Armstrong et al., 1993; Armstrong and Quigley, 1999).
Extracellular Matrix Formation May Be a Precursor to Calcification
Due to the prevalence of structural, mediating, and reworking proteins common to extracellular matrices found in barnacle precursor material, this extended similarities of barnacle glue curing beyond initial clotting to incorporate aspects of wound repair. Similar to wound healing in barnacles, the initial clot formed through proteolytic cascades previously described becomes further crosslinked and reworked into an extracellular matrix (ECM). Wound repair involves the stabilization of tissue at the wound site and reworking the structural ECM (Gailit and Clark, 1994; Ravanti and Kähäri, 2000; Li et al., 2003), providing a structural scaffold with the flexibility to become degraded, reinforced, or calcified over time (Bosman and Stamenkovic, 2003; Badylak et al., 2009).
The cuticular protein complex with pheromone activity, named SIPC, was classified as ECM-related in this study because of its prevalence in glue and cuticle, and homology with α2-macroglobulin (Matsumura et al., 1998b; Dreanno et al., 2006b) and we also identified a distinct α2-macroglobulin annotation. Although the role of SIPC α2-macroglobulin has not been defined, it acts as a substrate for the crosslinking enzyme transglutaminase in crayfish and can mediate ECM development (Hall and Söderhäll, 1994; Narita et al., 1997). In addition to SIPC, a variety of potential ECM structural proteins are found in barnacle fluids, including vimentin and laminin, which participate in cardiac ECM formation (Speiser et al., 1992; Bosman and Stamenkovic, 2003; Li et al., 2003). Mediator proteins in barnacle secretions, like annexin B-10 and cartilage oligomeric matrix protein could interface with structural proteins like SIPC and cytoskeletal proteins (Adams, 2001; Werner and Grose, 2003), similar to mediator interaction with actin in crayfish (Niksirat et al., 2014).
Actin is another potential substrate for transglutaminase and was most abundant 24 h after the initial sampling in the sequential wounding in the present study (Nemes et al., 1997). Both actin and tubulin are upregulated after bacterial infection in shrimp Penaeus monodon hemocytes, and were most abundant 24 h after the first sampling in our sequential wounding experiment (Somboonwiwat et al., 2010). The release of cytoskeletal proteins in barnacle fluids may provide structural elements for the extracellular matrix that participate in binding of the calcium carbonate base plate to the substratum. We found very few constitutive proteins related to general cellular function and recovery, which reflects the relatively low density of cells, as compared to tissues, in the coagulating fluids.
Extracellular matrices (ECM) are central components in wound repair across all taxa, as they help form connective tissue (Montesano and Orci, 1988; Peterkofsky, 1991), calcify eggshells and bone (Arias et al., 1993; Arias and Fernández, 2003), and barnacle shell (Fernández et al., 2002; Li et al., 2003; Verderio et al., 2004; Saito and Marumo, 2010). Much of the proteomic evidence for the extracellular matrix was structural, rather than mediators and enzymes to coordinate and rework. Components to rework the clot into an ECM may be more prevalent later, and not detected in the time course of sequential wounding. Zinc metalloproteases participate in ECM remodeling by degrading fibrin, collagen and peptides and are a critical part of reworking extracellular matrices (Ravanti and Kähäri, 2000; Parks et al., 2004). Zinc metalloproteases were previously identified in liquid barnacle secretions and their gene expression is upregulated prior to molting (Dougherty, 1996; Wang et al., 2015).
A consequence of having growth and adhesion tied to the innate immune response is redundancy in the function of clotting components. Part of the reason why barnacle glue curing is a difficult process to study is that it is a dynamic process that occurs in the same spatial area, the periphery of the baseplate at the interface with the substratum. We propose that barnacle fluids released between the base and substrate provide material structural biopolymers precursors that become highly crosslinked as evidenced by the structure of cured glue (Kamino, 2006, 2013; Barlow et al., 2009, 2010; Sullan et al., 2009;So et al., 2016). Unlike mussel and tubeworm attachment, which employs specialized glands and applicators, juvenile and adult barnacle adhesion may be an extension of molting and related to arthropod clotting and cuticle hardening. We hypothesize that, as in other crustaceans, expansion associated with molting tears cuticle-releasing fluids and activates innate immune responses, including the prophenoloxidase system and cell adhesion molecules (Figure 5). Cell adhesion molecules, like peroxinectin, can have peroxidase activity as well as phenoloxidase activity in crustaceans (Johansson et al., 1999; Sritunyalucksana et al., 2001; Lin et al., 2007; Schmidt et al., 2010). Additional cell adhesion proteins found in precursor material, like hemocytin and others with fibrinogen domains can be further crosslinked by transglutaminase, and non-enzymatic means like reactive oxygen species and quinones. This initial clot can be eventually reworked through matrix metalloproteinases and other enzymes and the extracellular matrix becomes a stable structure that is calcified over time. The innate immune system hypothesis provides an experimental framework for probing barnacle glue curing, adhesion and calcification.
Conclusion
Ancient protein systems, like the innate immune response, that are common across taxa provide opportunities to understand their core components, functions, and evolution. Barnacle glue curing is external to the animal, but is driven by mechanisms related to molting, blood clotting and wound repair. Insight into potential coagulation and cross-linking mechanisms that may contribute to barnacle glue curing is interesting from a comparative biology perspective and may have roles in fouling management and medicine. Through an understanding of the biological processes that drive barnacle glue curing, we can better design modes of intervention with biological adhesion for economic applications such as foul-release coatings for ships and medical applications such as biocompatible surgical glues that cure in aqueous environments and potentially direct calcification.
Data Availability
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
TE-B and DR conceptualized and implemented the experimental design and writing. TE-B carried out all of the sample preparation for proteomics and enzyme assays and analyzed the enzyme assays. BO maintained barnacle stock and provided expertise with the glue extraction technique. ES and MM helped in design and carried out the proteomic sample preparation and analysis and provided conceptual feedback throughout the study.
Funding
This work was financially supported by the Office of Naval Research grant N00014-11-10180, the American Society for Engineering Education, and Naval Research Enterprise Internship Program. NIH grant 1S10RR026977-01, which enabled the purchase of the mass spectrometer by the DPMSR facility.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Silvia Moriano-Gutierrez for her assistance with the database used and the Naval Research Laboratory barnacle group for access to their version of the Wang et al. (2015) database.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00343/full#supplementary-material
FIGURE S1 | Pooled barnacle secretion sample and corresponding bands cut for the in-gel trypsin digestion in experimental series 1.
FIGURE S2 | Standard curves shown with 1 and 10 mM L-dopa and varying concentrations of tyrosinase for the two experiments, both with glue and inhibitors (A,B) and with heated glue (C,D). Error bars indicate the standard deviation of triplicate conditions.
TABLE S1 | Distribution of enzyme classes present in the proteome database, highlighting two subclasses of interest, oxidoreductases and hydrolases and number of sequences within each classification.
DATA SHEET S1 | Fasta file of the annotated, clustered, database used for proteomic searches.
DATA SHEET S2 | Frequency of database annotations by species.
References
Adams, J. C. (2001). Thrombospondins: multifunctional regulators of cell interactions. Annu. Rev. Cell Dev. Biol. 17, 25–51. doi: 10.1146/annurev.cellbio.17.1.25
Al-Aqeel, S., Ryu, T., Zhang, H., Chandramouli, K. H., and Ravasi, T. (2016). Transcriptome and proteome studies reveal candidate attachment genes during the development of the barnacle Amphibalanus amphitrite. Front. Mar. Sci. 3:171. doi: 10.3389/fmars.2016.00171
Amparyup, P., Charoensapsri, W., and Tassanakajon, A. (2013). Prophenoloxidase system and its role in shrimp immune responses against major pathogens. Fish Shellfish Immunol. 34, 990–1001. doi: 10.1016/j.fsi.2012.08.019
Amparyup, P., Donpudsa, S., and Tassanakajon, A. (2008). Shrimp single WAP domain (SWD)-containing protein exhibits proteinase inhibitory and antimicrobial activities. Dev. Comp. Immunol. 32, 1497–1509. doi: 10.1016/j.dci.2008.06.005
Arias, J. L., and Fernández, M. S. (2003). Biomimetic processes through the study of mineralized shells. Mater. Charact. 50, 189–195. doi: 10.1016/S1044-5803(03)00088-3
Arias, J. L., Fink, D. J., Xiao, S.-Q., Heuer, A. H., and Caplan, A. I. (1993). Biomineralization and eggshells: cell-mediated acellular compartments of mineralized extracellular matrix. Int. Rev. Cytol. 145, 217–250. doi: 10.1016/s0074-7696(08)60428-3
Armstrong, P. B., Armstrong, M. T., and Quigley, J. P. (1993). Involvement of a2-macroglobulin and C-reactive protein in a complement-like hemolytic system in the arthropod, Limulus polyphemus. Mol. Immunol. 30, 929–934. doi: 10.1016/0161-5890(93)90017-6
Armstrong, P. B., and Qmoley, J. P. (1993). Immune function of alpha 2-macroglobulin in invertebrates. Prog. Mol. Subcell. Biol. 15, 101–130. doi: 10.1007/978-3-642-79735-4_6
Armstrong, P. B., and Quigley, J. P. (1999). a2-macroglobulin: an evolutionarily conserved arm of the innate immune system. Dev. Comp. Immunol. 23, 375–390. doi: 10.1016/s0145-305x(99)00018-x
Ashida, M., and Brey, P. T. (1995). Role of the integument in insect defense: pro-phenol oxidase cascade in the cuticular matrix. Proc. Natl. Acad. Sci. U.S.A. 92, 10698–10702. doi: 10.1073/pnas.92.23.10698
Aspan, A., and Söderhäll, K. (1991). Purification of prophenoloxidase from crayfish blood cells, and its activation by an endogenous serine proteinase. Insect Biochem. 21, 363–373. doi: 10.1016/0020-1790(91)90002-v
Badylak, S. F., Freytes, D. O., and Gilbert, T. W. (2009). Extracellular matrix as a biological scaffold material: structure and function. Acta Biomater. 5, 1–13. doi: 10.1016/j.actbio.2008.09.013
Barlow, D. E., Dickinson, G. H., Orihuela, B., Kulp, J. L., Rittschof, D., and Wahl, K. J. (2010). Characterization of the adhesive plaque of the barnacle Balanus amphitrite: amyloid-like nanofibrils are a major component. J. Chem. Ecol. 26, 6549–6556. doi: 10.1021/la9041309
Barlow, D. E., Dickinson, G. H., Orihuela, B., Rittschof, D., and Wahl, K. J. (2009). In situ ATR-FTIR characterization of primary cement interfaces of the barnacle Balanus amphitrite. Biofouling 25, 359–366. doi: 10.1080/08927010902812009
Baruah, K., Ranjan, J., Sorgeloos, P., MacRae, T. H., and Bossier, P. (2011). Priming the prophenoloxidase system of Artemia franciscana by heat shock proteins protects against Vibrio campbellii challenge. Fish Shellfish Immunol. 31, 134–141. doi: 10.1016/j.fsi.2011.04.008
Battison, A. L., Summerfield, R., and Patrzykat, A. (2008). Isolation and characterisation of two antimicrobial peptides from haemocytes of the American lobster Homarus americanus. Fish Shellfish Immunol. 25, 181–187. doi: 10.1016/j.fsi.2008.04.005
Bocquet-Vedrine, J. (1965). Etude du tegument et de la mue chez le cirripede opercule Elminius modestus Darwin. Arch. Zool. Exp. Gen. 105, 30–76.
Bosman, F. T., and Stamenkovic, I. (2003). Functional structure and composition of the extracellular matrix. J. Pathol. 200, 423–428. doi: 10.1002/path.1437
Bourget, E., and Crisp, D. J. (1975). An analysis of the growth bands and ridges of barnacle shell plates. J. Mar. Biol. Assoc. U.K. 55, 439–461. doi: 10.1017/s0025315400016052
Braiman-Wiksman, L., Solomonik, I., Spira, R., and Tennenbaum, T. (2007). Novel insights into wound healing sequence of events. Toxicol. Pathol. 35, 767–779. doi: 10.1080/01926230701584189
Breugelmans, B., Simonet, G., van Hoef, V., Van Soest, S., and Vanden Broeck, J. (2009). Pacifastin-related peptides: structural and functional characteristics of a family of serine peptidase inhibitors. Peptides 30, 622–632. doi: 10.1016/j.peptides.2008.07.026
Burden, D. K., Barlow, D. E., Spillmann, C. M., Orihuela, B., Rittschof, D., Everett, R. K., et al. (2012). Barnacle Balanus amphitrite adheres by a stepwise cementing process. Langmuir 28, 13364–13372. doi: 10.1021/la301695m
Burden, D. K., Spillmann, C. M., Everett, R. K., Barlow, D. E., Orihuela, B., Deschamps, J. R., et al. (2014). Growth and development of the barnacle Balanus amphitrite: time- and spatially-resolved structure and chemistry of the base plate. Biofouling 30, 799–812. doi: 10.1080/08927014.2014.930736
Byrd, C. A., Bornmann, W., Erdjument-Bromage, H., Tempst, P., Pavletich, N., Rosen, N., et al. (1999). Heat shock protein 90 mediates macrophage activation by Taxol and bacterial lipopolysaccharide. Proc. Natl. Acad. Sci. U.S.A. 96, 5645–5650. doi: 10.1073/pnas.96.10.5645
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Cellura, C., Toubiana, M., Parrinello, N., and Roch, P. (2006). HSP70 gene expression in Mytilus galloprovincialis hemocytes is triggered by moderate heat shock and Vibrio anguillarum, but not by Vibrio splendidus or Micrococcus lysodeikticus. Dev. Comp. Immunol. 30, 984–997. doi: 10.1016/j.dci.2005.12.009
Cerenius, L., Lee, B. L., and Söderhäll, K. (2008). The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 29, 263–271. doi: 10.1016/j.it.2008.02.009
Chang, E. S., and Mykles, D. L. (2011). General and Comparative Endocrinology Regulation of crustacean molting: a review and our perspectives. Gen. Comp. Endocrinol. 172, 323–330. doi: 10.1016/j.ygcen.2011.04.003
Chang, S. (1995). Physiological and biochemical changes during the molt cycle in decapod crustaceans: an overview. J. Exp. Mar. Biol. Ecol. 193, 1–14. doi: 10.1016/0022-0981(95)00106-9
Chen, Z.-F., Zhang, H., Wang, H., Matsumura, K., Wong, Y. H., Ravasi, T., et al. (2014). Quantitative proteomics study of larval settlement in the Barnacle Balanus amphitrite. PLoS One 9:e88744. doi: 10.1371/journal.pone.0088744
Cheng, S. H., So, C. H., Chan, P. K., Cheng, C. W., and Wu, R. S. S. (2003). Cloning of the HSP70 gene in barnacle larvae and its expression under hypoxic conditions. Mar. Pollut. Bull. 46, 665–671. doi: 10.1016/S0025-326X(03)00059-6
Cheung, P. J., Ruggieri, G. D., and Nigrelli, R. F. (1977). A new method for obtaining barnacle cement in the liquid state for polymerization studies. Mar. Biol. 43, 157–163. doi: 10.1007/BF00391263
Crisp, D. J., and Bourget, E. (1985). “Growth in barnacles,” in Advances in Marine Biology, Vol. 22, eds J. H. S. Blaxter and F. S. Russel (London: Academic Press), 199–244. doi: 10.1016/s0065-2881(08)60052-8
Davie, E. W., and Ratnoff, O. D. (1964). Waterfall sequence for intrinsic blood clotting. Science 145, 1310–1312. doi: 10.1126/science.145.3638.1310
Dickinson, G. H. (2008). Barnacle Cement: A Polymerization Model Based on Evolutionary Concepts. Durham: Duke University.
Dickinson, G. H., Vega, I. E., Wahl, K. J., Orihuela, B., Beyley, V., Rodriguez, E. N., et al. (2009). Barnacle cement: a polymerization model based on evolutionary concepts. J. Exp. Biol. 212(Pt 21), 3499–3510. doi: 10.1242/jeb.029884
Dickinson, G. H., Yang, X., Wu, F., Orihuela, B., Rittschof, D., and Beniash, E. (2016). Localization of phosphoproteins within the barnacle adhesive interface. Biol. Bull. 230, 233–242. doi: 10.1086/BBLv230n3p233
Dillaman, R. M., and Roer, R. D. (1980). Carapace repair in the green crab, Carcinus maenas (L.). J. Morphol. 163, 135–155. doi: 10.1002/jmor.1051630203
Dougherty, W. J. (1996). Zinc metalloprotease activity in the cement precursor secretion of the barnacle, Chthamalus fragilis Darwin. Tissue Cell 28, 439–447. doi: 10.1016/s0040-8166(96)80029-2
Dreanno, C., Kirby, R. R., and Clare, A. S. (2006a). Locating the barnacle settlement pheromone: spatial and ontogenetic expression of the settlement-inducing protein complex of Balanus amphitrite. Proc. Biol. Sci. 273, 2721–2728. doi: 10.1098/rspb.2006.3649
Dreanno, C., Matsumura, K., Dohmae, N., Takio, K., Hirota, H., Kirby, R. R., et al. (2006b). An α2-macroglobulin-like protein is the cue to gregarious settlement of the barnacle Balanus amphitrite. Proc. Natl. Acad. Sci. U.S.A. 103, 14396–14401. doi: 10.1073/pnas.0602763103
Durliat, M. (1985). Clotting processes in crustacea decapoda. Biol. Rev. 60, 473–498. doi: 10.1111/j.1469-185x.1985.tb00620.x
Eming, S. A., Krieg, T., and Davidson, J. M. (2007). Inflammation in wound repair: molecular and cellular mechanisms. J. Investig. Dermatol. 127, 514–525. doi: 10.1038/sj.jid.5700701
Espín, J. C., Morales, M., García-Ruiz, P. A., Tudela, J., and García-Cánovas, F. (1997). Improvement of a continuous spectrophotometric method for determining the monophenolase and diphenolase activities of mushroom polyphenol oxidase. J. Agric. Food Chem. 45, 1084–1090. doi: 10.1021/jf960428a
Essock-Burns, T., Gohad, N. V., Orihuela, B., Mount, A. S., Spillmann, C. M., Wahl, K. J., et al. (2017). Barnacle biology before, during and after settlement and metamorphosis: a study of the interface. J. Exp. Biol. 220, 194–207. doi: 10.1242/jeb.145094
Fears, K. P., Orihuela, B., Rittschof, D., and Wahl, K. J. (2018). Acorn barnacles secrete phase-separating fluid to clear surfaces ahead of cement deposition. Adv. Sci. 5:1700762. doi: 10.1002/advs.201700762
Fernández, M. S., Vergara, I., Oyarzun, A., Arias, J. L., Rodriguez, R., Wiff, J. P., et al. (2002). Extracellular matrix molecules involved in barnacle shell mineralization. Mater. Res. Soc. Symp. Proc. 724, 3–9.
Ferrier, G. A., Kim, S. J., Kaddis, C. S., Loo, J. A., Ann Zimmer, C., and Zimmer, R. K. (2016). MULTIFUNCin: a multifunctional protein cue induces habitat selection by, and predation on, barnacles. Integr. Comp. Biol. 56, 901–913. doi: 10.1093/icb/icw076
Flammang, P., Lambert, A., Bailly, P., and Hennebert, E. (2009). Polyphosphoprotein-containing marine adhesives. J. Adh. 85, 447–464. doi: 10.1080/00218460902996358
Frantz, S., Vincent, K. A., Feron, O., and Kelly, R. A. (2005). Innate immunity and angiogenesis. Circ. Res. 96, 15–26. doi: 10.1161/01.RES.0000153188.68898.ac
Furie, B., and Furie, B. C. (1988). The molecular basis of blood coagulation. Cell 53, 505–518. doi: 10.1016/0092-8674(88)90567-3
Fyhn, U. E. H., and Costlow, J. D. (1976). A histochemical study of cement secretion during the intermolt cycle in barnacles. Biol. Bull. 150, 47–56. doi: 10.2307/1540588
Gailit, J., and Clark, R. A. (1994). Wound repair in the context of extracellular matrix. Curr. Opin. Cell Biol. 6, 717–725. doi: 10.1016/0955-0674(94)90099-X
Glazer, L., Tom, M., Weil, S., Roth, Z., Khalaila, I., Mittelman, B., et al. (2013). Hemocyanin with phenoloxidase activity in the chitin matrix of the crayfish gastrolith. J. Exp. Biol. 216(Pt 10), 1898–1904. doi: 10.1242/jeb.080945
Gohad, N. V., Aldred, N., Hartshorn, C. M., Jong Lee, Y., Cicerone, M. T., Orihuela, B., et al. (2014). Synergistic roles for lipids and proteins in the permanent adhesive of barnacle larvae. Nat. Commun. 5:4414. doi: 10.1038/ncomms5414
Golden, J. P., Burden, D. K., Fears, K. P., Barlow, D. E., So, C. R., Burns, J., et al. (2016). Imaging active surface processes in barnacle adhesive interfaces. Langmuir 32, 541–550. doi: 10.1021/acs.langmuir.5b03286
Gorman, M. J., An, C., and Kanost, M. R. (2007). Characterization of tyrosine hydroxylase from Manduca sexta. Insect Biochem. Mol. Biol. 37, 1327–1337. doi: 10.1016/j.ibmb.2007.08.006
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., et al. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Hall, M., and Söderhäll, K. (1994). Crayfish α-macroglobulin as a substrate for transglutaminases. Comp. Biochem. Physiol. 108, 65–72. doi: 10.1016/0305-0491(94)90166-x
Han, Z., Sun, J., Zhang, Y., He, F., Xu, Y., Matsumura, K., et al. (2013). iTRAQ-based proteomic profiling of the barnacle Balanus amphitrite in response to the antifouling compound meleagrin. J. Proteome Res. 12, 2090–2100. doi: 10.1021/pr301083e
He, L. S., Zhang, G., and Qian, P.-Y. (2013). Characterization of two 20kDa-cement protein (cp20k) homologues in Amphibalanus amphitrite. PLoS One 8:e64130. doi: 10.1371/journal.pone.0064130
Holm, E. R. (2012). Barnacles and biofouling. Integr. Comp. Biol. 52, 348–355. doi: 10.1093/icb/ics042
Holm, E. R., Orihuela, B., Kavanagh, C. J., and Rittschof, D. (2005). Variation among families for characteristics of the adhesive plaque in the barnacle Balanus amphitrite. Biofouling 21, 121–126. doi: 10.1080/08927010512331344188
Janeway, C. A., and Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. doi: 10.1146/annurev.immunol.20.083001.084359
Jiang, H., and Kanost, M. R. (2000). The clip-domain family of serine proteinases in arthropods. Insect Biochem. Mol. Biol. 30, 95–105. doi: 10.1016/s0965-1748(99)00113-7
Jiravanichpaisal, P., Lee, B. L., and Söderhäll, K. (2006). Cell-mediated immunity in arthropods: hematopoiesis, coagulation, melanization and opsonization. Immunobiology 211, 213–236. doi: 10.1016/j.imbio.2005.10.015
Johansson, M. W. (1999). Cell adhesion molecules in invertebrate immunity. Dev. Comp. Immunol. 23, 303–315. doi: 10.1016/s0145-305x(99)00013-0
Johansson, M. W., Holmblad, T., Thörnqvist, P. O., Cammarata, M., Parrinello, N., and Söderhäll, K. (1999). A cell-surface superoxide dismutase is a binding protein for peroxinectin, a cell-adhesive peroxidase in crayfish. J. Cell Sci. 112(Pt 6), 917–925.
Kamino, K. (2001). Novel barnacle underwater adhesive protein is a charged amino acid-rich protein constituted by a Cys-rich repetitive sequence. Biochem. J. 356(Pt 2), 503–507. doi: 10.1042/bj3560503
Kamino, K. (2006). “Barnacle underwater attachment,” in Biological Adhesives, eds A. M. Smith and J. A. Callow (Berlin: Springer-Verlag), 145–166. doi: 10.1007/978-3-540-31049-5_8
Kamino, K. (2013). Mini-review: barnacle adhesives and adhesion. Biofouling 29, 735–749. doi: 10.1080/08927014.2013.800863
Kamino, K., Odo, S., and Maruyama, T. (1996). Cement proteins of the acorn barnacle, Megabalanus rosa. Biol. Bull. 190, 403–409. doi: 10.2307/1543033
Kamino, K., and Shizuri, Y. (1998). “Structure and function of barnacle cement proteins,” in New Developments in Marine Biotechnology, eds Y. Le Gal and H. O. Halvorson (New York, NY: Plenum Press), 77–80. doi: 10.1007/978-1-4757-5983-9_16
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Kanost, M. R., Jiang, H., and Yu, X.-Q. (2004). Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 198, 97–105. doi: 10.1111/j.0105-2896.2004.0121.x
Keller, A., Nesvizhskii, A. I., Kolker, E., and Aebersold, R. (2002). Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392. doi: 10.1021/ac025747h
Kikuchi, T., Shibuya, H., and Jones, J. T. (2005). Molecular and biochemical characterization of an endo-β-1,3-glucanase from the pinewood nematode Bursaphelenchus xylophilus acquired by horizontal gene transfer from bacteria. Biochem. J. 389, 117–125. doi: 10.1042/bj20042042
King, N., Hittinger, C. T., and Carroll, S. B. (2003). Evolution of key cell signaling and adhesion protein families predates animal origins. Science 301, 361–363. doi: 10.1126/science.1083853
Kopáček, P., Hall, M., and Söderhäll, K. (1993). Characterization of a clotting protein, isolated from plasma of the freshwater crayfish Pacifastacus leniusculus. Eur. J. Biochem. 213, 591–597. doi: 10.1111/j.1432-1033.1993.tb17798.x
Kotani, E., Yamakawa, M., Iwamoto, S.-I., Tashiro, M., Mori, H., Sumida, M., et al. (1995). Cloning and expression of the gene of hemocytin, an insect humoral lectin which is homologous with the mammalian von Willebrand factor. Biochim. Biophys. Acta 1260, 245–258. doi: 10.1016/0167-4781(94)00202-e
Krem, M. M., and Di Cera, E. (2002). Evolution of enzyme cascades from embryonic development to blood coagulation. Trends Biochem. Sci. 27, 67–74. doi: 10.1016/s0968-0004(01)02007-2
Kurtz, J., and Franz, K. (2003). Innate defence: evidence for memory in invertebrate immunity. Nature 425, 37–38. doi: 10.1038/425037a
Lai, S.-C., Chen, C.-C., and Hou, R. F. (2002). Immunolocalization of prophenoloxidase in the process of wound healing in the mosquito Armigeres subalbatus (Diptera: Culicidae). J. Med. Entomol. 39, 266–274. doi: 10.1603/0022-2585-39.2.266
Lee, S. Y., Kwon, T. H., Hyun, J. H., Choi, J. S., Kawabata, S. I., Iwanaga, S., et al. (1998). In vitro activation of pro-phenol-oxidase by two kinds of pro-phenol-oxidase-activating factors isolated from hemolymph of coleopteran, Holotrichia diomphalia larvae. Eur. J. Biochem. 254, 50–57. doi: 10.1046/j.1432-1327.1998.2540050.x
Li, D., Scherfer, C., Korayem, A. M., Zhao, Z., Schmidt, O., and Theopold, U. (2002). Insect hemolymph clotting: evidence for interaction between the coagulation system and the prophenoloxidase activating cascade. Insect Biochem. Mol. Biol. 32, 919–928. doi: 10.1016/s0965-1748(02)00030-9
Li, J., Zhang, Y.-P., and Kirsner, R. S. (2003). Angiogenesis in wound repair: angiogenic growth factors and the extracellular matrix. Microsc. Res. Tech. 60, 107–114. doi: 10.1002/jemt.10249
Liang, Z., Sottrup-Jensen, L., Aspán, A., Hall, M., and Söderhäll, K. (1997). Pacifastin, a novel 155-kDa heterodimeric proteinase inhibitor containing a unique transferrin chain. Proc. Natl. Acad. Sci. U.S.A. 94, 6682–6687. doi: 10.1073/pnas.94.13.6682
Lin, X., Cerenius, L., Lee, B. L., and Söderhäll, K. (2007). Purification of properoxinectin, a myeloperoxidase homologue and its activation to a cell adhesion molecule. Biochim. Biophys. Acta 1770, 87–93. doi: 10.1016/j.bbagen.2006.06.018
Lindner, E. (1984). “The attachment of macrofouling invertebrates,” in Marine Biodeterioration: An Interdisciplinary Study, eds J. D. Costlow and R. C. Tipper (Annapolis, MD: Naval Institute Press), 183–201. doi: 10.1007/978-1-4615-9720-9_24
Lindner, E., and Dooley, C. A. (1974). “Chemical bonding in cirriped adhesive,” in Proceedings of the Third International Congress on Marine Corrosion and Fouling, eds R. F. Acker, B. F. Brown, J. R. de Palma, and W. R. Iverson (Evanston, IL: Northwestern University Press), 653–673.
Litman, G. W., Cannon, J. P., and Dishaw, L. J. (2005). Reconstructing immune phylogeny: new perspectives. Nat. Rev. Immunol. 5, 866–879. doi: 10.1038/nri1712
Matsumura, K., Nagano, M., and Fusetani, N. (1998a). Purification of a larval settlement-inducing protein complex (SIPC) of the barnacle, Balanus amphitrite. J. Exp. Zool. 281, 12–20. doi: 10.1080/08927010902875113
Matsumura, K., Nagano, M., Kato-Yoshinaga, Y., Yamazaki, M., Clare, A. S., and Fusetani, N. (1998b). Immunological studies on the settlement-inducing protein complex (SIPC) of the barnacle Balanus amphitrite and its possible involvement in larva-larva interactions. Proc. Biol. Sci. 265, 1825–1830. doi: 10.1098/rspb.1998.0508
Medzhitov, R., and Janeway, C. A. J. (2002). Decoding the patterns of self and nonself by the innate immune system. Science 296, 298–301.
Mellacheruvu, D., Wright, Z., Couzens, A. L., Lambert, J. P., St-Denis, N. A., Li, T., et al. (2013). The CRAPome: a contaminant repository for affinity purification – mass spectrometry data. Nat. Methods 10, 730–740. doi: 10.1038/nmeth.2557
Milani, V., Noessner, E., Ghose, S., Kuppner, M., Ahrens, B., Scharner, A., et al. (2002). Heat shock protein 70: role in antigen presentation and immune stimulation. Int. J. Hyperthermia 18, 563–575. doi: 10.1080/0265673021016614
Minibayeva, F., Kolesnikov, O., Chasov, A., Beckett, R. P., Lüthje, S., Vylegzhanina, N., et al. (2009). Wound-induced apoplastic peroxidase activities: their roles in the production and detoxification of reactive oxygen species. Plant Cell Environ. 32, 497–508. doi: 10.1111/j.1365-3040.2009.01944.x
Montesano, R., and Orci, L. (1988). Transforming growth factor beta stimulates collagen-matrix contraction by fibroblasts: implications for wound healing. Proc. Natl. Acad. Sci. U.S.A. 85, 4894–4897. doi: 10.1073/pnas.85.13.4894
Morales, M., Vardn, R., Tudela, J., and Bt, F. G. (1995). Monophenolase activity of polyphenol oxidase from verdedoncella apple. J. Agric. Food Chem. 43, 2807–2812. doi: 10.1021/jf00059a007
Nagai, T., and Kawabata, S. I. (2000). A link between blood coagulation and prophenol oxidase activation in arthropod host defense. J. Biol. Chem. 275, 29264–29267. doi: 10.1074/jbc.M002556200
Naldrett, M. J. (1993). The importance of sulphur cross-links and hydrophobic interactions in the polymerization of barnacle cement. J. Mar. Biol. Assoc. U.K. 73, 689–702. doi: 10.1017/S0025315400033221
Narita, M., Holtzman, D. M., Schwartz, A. L., and Bu, G. (1997). a2-macroglobulin complexes with and mediates the endocytosis of beta-amyloid peptide via cell surface low-density lipoprotein receptor-related protein. J. Neurochem. 69, 1904–1911. doi: 10.1046/j.1471-4159.1997.69051904.x
Nemes, Z. J., Adány, R., Balázs, M., Boross, P., and Fésüs, L. (1997). Identification of cytoplasmic actin as an abundant glutaminyl substrate for tissue transglutaminase in HL-60 and U937 cells undergoing apoptosis. J. Biol. Chem. 272, 20577–20583. doi: 10.1074/jbc.272.33.20577
Nesvizhskii, A. I., Keller, A., Kolker, E., and Aebersold, R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658. doi: 10.1021/ac0341261
Niksirat, H., Andersson, L., James, P., Kouba, A., and Kozák, P. (2014). Proteomic profiling of the signal crayfish Pacifastacus leniusculus egg and spermatophore. Anim. Reprod. Sci. 149, 335–344. doi: 10.1016/j.anireprosci.2014.07.024
Ochiai, M., and Ashida, M. (1988). Purification of a beta-1,3-glucan recognition protein in the prophenoloxidase activating system from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 263, 12056–12062.
Orth, K., Madison, E. L., Gething, M. J., Sambrook, J. F., and Herz, J. (1992). Complexes of tissue-type plasminogen activator and its serpin inhibitor plasminogen-activator inhibitor type 1 are internalized by means of the low density lipoprotein receptor-related protein/alpha 2-macroglobulin receptor. Proc. Natl. Acad. Sci. U.S.A. 89, 7422–7426. doi: 10.1073/pnas.89.16.7422
Parks, W. C., Wilson, C. L., and López-Boado, Y. S. (2004). Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat. Rev. Immunol. 4, 617–629. doi: 10.1038/nri1418
Peterkofsky, B. (1991). Ascorbate requirement for hydroxylation and secretion of procollagen: relationship to inhibition of collagen synthesis in scurvy. Am. J. Clin. Nutr. 54(6 Suppl.), 1135S–1140S. doi: 10.1093/ajcn/54.6.1135s
Ravanti, L., and Kähäri, V. M. (2000). Matrix metalloproteinases in wound repair (review). Int. J. Mol. Med. 6, 391–407.
Rittschof, D., Branscomb, E. S., and Costlow, J. D. (1984). Settlement and behavior in relation to flow and surface in larval barnacles, Balanus amphitrite Darwin. J. Exp. Biol. 82, 131–146. doi: 10.1016/0022-0981(84)90099-6
Rittschof, D., Clare, A. S., Gerhart, D. J., Mary, S. A., and Bonaventura, J. (1992). Barnacle in vitro assays for biologically active substances: toxicity and settlement inhibition assays using mass cultured Balanus amphitrite amphitrite Darwin. Biofouling 6, 115–122. doi: 10.1080/08927019209386217
Rittschof, D., Orihuela, B., Stafslien, S., Daniels, J., Christianson, D., Chisholm, B., et al. (2008). Barnacle reattachment: a tool for studying barnacle adhesion. Biofouling 24, 1–9. doi: 10.1080/08927010701784920
Robinson, M. J., Sancho, D., Slack, E. C., LeibundGut-Landmann, S., and Reis e Sousa, C. (2006). Myeloid C-type lectins in innate immunity. Nat. Immunol. 7, 1258–1265. doi: 10.1038/ni1417
Roer, R., and Dillaman, R. (1984). The structure and calcification of the crustacean cuticle. Am. Zool. 24, 893–909. doi: 10.1093/icb/24.4.893
Saito, M., and Marumo, K. (2010). Collagen cross-links as a determinant of bone quality: a possible explanation for bone fragility in aging, osteoporosis, and diabetes mellitus. Osteoporos. Int. 21, 195–214. doi: 10.1007/s00198-009-1066-z
Saroyan, J. R., Lindner, E., and Dooley, C. A. (1970). Repair and reattachment in the balanidae as related to their cementing mechanism. Biol. Bull. 139, 333–350. doi: 10.2307/1540088
Saroyan, J. R., Lindner, E., and Dooley, C. A. (1968). Attachment Mechanism of Barnacles: Fouling Prevention. The Study of Adhesion of Calcareous Types Attaching Marine Organisms. Report No. 68-1. First Progress Report. In-House Independent Research Program. NAVSHIPS Project No. SF020-99-02. Vallejo, CA.
Schmidt, O., Söderhäll, K., Theopold, U., and Faye, I. (2010). Role of adhesion in arthropod immune recognition. Annu. Rev. Entomol. 55, 485–504. doi: 10.1146/annurev.ento.54.110807.090618
Schultz, M. P., Bendick, J. A., Holm, E. R., and Hertel, W. M. (2011). Economic impact of biofouling on a naval surface ship. Biofouling 27, 87–98. doi: 10.1080/08927014.2010.542809
Simonet, G., Claeys, I., and Broeck, J. V. (2002). Structural and functional properties of a novel serine protease inhibiting peptide family in arthropods. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 132, 247–255. doi: 10.1016/S1096-4959(01)00530-9
Smith, V. J., Fernandes, J. M. O., Kemp, G. D., and Hauton, C. (2008). Crustins: enigmatic WAP domain-containing antibacterial proteins from crustaceans. Dev. Comp. Immunol. 32, 758–772. doi: 10.1016/j.dci.2007.12.002
So, C. R., Fears, K. P., Leary, D. H., Scancella, J. M., Wang, Z., Liu, J. L., et al. (2016). Sequence basis of barnacle cement nanostructure is defined by proteins with silk homology. Sci. Rep. 6:36219. doi: 10.1038/srep36219
So, C. R., Scancella, J. M., Fears, K. P., Essock-Burns, T., Haynes, S. E., Leary, D. H., et al. (2017). Oxidase activity of the barnacle adhesive interface involves peroxide-dependent catechol oxidase and Lysyl oxidase enzymes. ACS Appl. Mater. Interfaces 9, 11493–11505. doi: 10.1021/acsami.7b01185
Söderhäll, K., and Cerenius, L. (1998). Role of the prophenoloxidase-activating system in invertebrate immunity. Curr. Opin. Immunol. 10, 23–28. doi: 10.1016/s0952-7915(98)80026-5
Somboonwiwat, K., Chaikeeratisak, V., Wang, H.-C., Fang Lo, C., and Tassanakajon, A. (2010). Proteomic analysis of differentially expressed proteins in Penaeus monodon hemocytes after Vibrio harveyi infection. Proteome Sci. 8:39. doi: 10.1186/1477-5956-8-39
Speiser, B., Weihrauch, D., Riess, C. F., and Schaper, J. (1992). The Extracellular Matrix in human cardiac tissue. Part II: vimentin, laminin, and fibronectin. Cardioscience 3, 41–48.
Sritunyalucksana, K., and Söderhäll, K. (2000). The proPO and clotting system in crustaceans. Aquaculture 191, 53–69. doi: 10.1016/S0044-8486(00)00411-7
Sritunyalucksana, K., Wongsuebsantati, K., Johansson, M. W., and Söderhäll, K. (2001). Peroxinectin, a cell adhesive protein associated with the proPO system from the black tiger shrimp, Penaeus monodon. Dev. Comp. Immunol. 25, 353–363. doi: 10.1016/S0145-305X(01)00009-X
Srivastava, P. (2002a). Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu. Rev. Immunol. 20, 395–425. doi: 10.1146/annurev.immunol.20.100301.064801
Srivastava, P. (2002b). Roles of heat-shock proteins in innate and adaptive immunity. Nat. Rev. Immunol. 2, 185–194. doi: 10.1038/nri749
Suderman, R. J., Dittmer, N. T., Kanost, M. R., and Kramer, K. J. (2006). Model reactions for insect cuticle sclerotization: cross-linking of recombinant cuticular proteins upon their laccase-catalyzed oxidative conjugation with catechols. Insect Biochem. Mol. Biol. 36, 353–365. doi: 10.1016/j.ibmb.2006.01.012
Sugumaran, M. (2002). Comparative biochemistry of eumelanogenesis and the protective roles of phenoloxidase and melanin in insects. Pigment Cell Res. 15, 2–9. doi: 10.1034/j.1600-0749.2002.00056.x
Sullan, R. M., Gunari, N., Tanur, A. E., Chan, Y., Dickinson, G. H., Orihuela, B., et al. (2009). Nanoscale structures and mechanics of barnacle cement. Biofouling 25, 263–275. doi: 10.1080/08927010802688095
Taylor, J. R. A., and Kier, W. M. (2003). Switching skeletons: hydrostatic support in molting crabs. Science 301, 209–210. doi: 10.1126/science.1085987
Terwilliger, N. B. (1999). Hemolymph proteins and molting in crustaceans and insects. Integr. Comp. Biol. 39, 589–599. doi: 10.1093/icb/39.3.589
Theopold, U., Li, D., Fabbri, M., Scherfer, C., and Schmidt, O. (2002). The coagulation of insect hemolymph. Cell. Mol. Life Sci. 59, 363–372. doi: 10.1007/s00018-002-8428-4
Theopold, U., Schmidt, O., Söderhäll, K., and Dushay, M. S. (2004). Coagulation in arthropods: defence, wound closure and healing. Trends Immunol. 25, 289–294. doi: 10.1016/j.it.2004.03.004
Thiyagarajan, V., and Qian, P. Y. (2008). Proteomic analysis of larvae during development, attachment, and metamorphosis in the fouling barnacle, Balanus amphitrite. Proteomics 8, 3164–3172. doi: 10.1002/pmic.200700904
Tomasetti, M., Pinotti, M., Muzzarelli, R. A. A., Xia, W., and Ilari, P. (1994). Tyrosinase-mediated quinone tanning of chitinous materials. Carbohydr. Polym. 24, 295–300. doi: 10.1016/0144-8617(94)90074-4
Verderio, E. A., Johnson, T., and Griffin, M. (2004). Tissue transglutaminase in normal and abnormal wound healing: review article. Amino Acids 26, 387–404. doi: 10.1007/s00726-004-0094-4
Waite, J. H. (1990a). Marine adhesive proteins: natural composite thermosets. Int. J. Biol. Macromol. 12, 139–144. doi: 10.1016/0141-8130(90)90065-I
Waite, J. H. (1990b). The phylogeny and chemical diversity of quinone-tanned glues and varnishes. Comp. Biochem. Physiol. 97B, 19–29. doi: 10.1016/0305-0491(90)90172-p
Waite, J. H. (1995). Precursors of quinone tanning: dopa-containing proteins. Methods Enzymol. 258, 1–20. doi: 10.1016/0076-6879(95)58033-6
Walker, G. (1971). A study of the cement apparatus of the cypris larva of the barnacle Balanus balanoides. Mar. Biol. 212, 205–212. doi: 10.1007/bf00351380
Walker, G. (1973). The early development of the cement apparatus in the barnacle, Balanus Balanoides (L.) (Crustacea: Cirripedia). J. Exp. Mar. Biol. Ecol. 12, 305–314. doi: 10.1007/s00216-015-9253-6
Wang, C. S., and Stewart, R. J. (2013). Multipart copolyelectrolyte adhesive of the sandcastle worm, Phragmatopoma californica (Fewkes): catechol oxidase catalyzed curing through peptidyl-DOPA. Biomacromolecules 14, 1607–1617. doi: 10.1021/bm400251k
Wang, R., Lee, S. Y., Cerenius, L., and Söderhäll, K. (2001). Properties of the prophenoloxidase activating enzyme of the freshwater crayfish, Pacifastacus leniusculus. Eur. J. Biochem. 268, 895–902. doi: 10.1046/j.1432-1327.2001.01945.x
Wang, X.-W., and Wang, J.-X. (2013). Pattern recognition receptors acting in innate immune system of shrimp against pathogen infections. Fish Shellfish Immunol. 34, 981–989. doi: 10.1016/j.fsi.2012.08.008
Wang, Z., Leary, D., Liu, J., Mostaghim, A., Settlage, R., Fears, K. P., et al. (2015). Molt-dependent transcriptomic analysis of cement proteins in the barnacle Amphibalanus amphitrite. BMC Genomics 16:859. doi: 10.1186/s12864-015-2076-1
Werner, S., and Grose, R. (2003). Regulation of wound healing by growth factors and cytokines. Physiol. Rev. 83, 835–870. doi: 10.1152/physrev.2003.83.3.835
Willis, J. H. (1999). Cuticular proteins in insects and crustaceans. Integr. Comp. Biol. 39, 600–609. doi: 10.1093/icb/39.3.600
Wu, C., Charoensapsri, W., Nakamura, S., Tassanakajon, A., Söderhäll, I., and Söderhäll, K. (2013). An MBL-like protein may interfere with the activation of the proPO-system, an important innate immune reaction in invertebrates. Immunobiology 218, 159–168. doi: 10.1016/j.imbio.2012.02.011
Yu, X.-Q., Jiang, H., Wang, Y., and Kanost, M. R. (2003). Nonproteolytic serine proteinase homologs are involved in prophenoloxidase activation in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 33, 197–208. doi: 10.1016/S0965-1748(02)00191-1
Keywords: barnacle adhesion, innate immune, crustaceans, proteomics, prophenoloxidase, reactive oxygen species
Citation: Essock-Burns T, Soderblom EJ, Orihuela B, Moseley MA and Rittschof D (2019) Hypothesis Testing With Proteomics: A Case Study Using Wound Healing Mechanisms in Fluids Associated With Barnacle Glue. Front. Mar. Sci. 6:343. doi: 10.3389/fmars.2019.00343
Received: 04 April 2019; Accepted: 04 June 2019;
Published: 26 June 2019.
Edited by:
Andrew Stanley Mount, Clemson University, United StatesReviewed by:
Adam Michael Reitzel, The University of North Carolina at Charlotte, United StatesVittoria Roncalli, University of Barcelona, Spain
Copyright © 2019 Essock-Burns, Soderblom, Orihuela, Moseley and Rittschof. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tara Essock-Burns, RXNzb2NrQGhhd2FpaS5lZHU=; dGFyYS5lc3NvY2tidXJuc0BnbWFpbC5jb20=
 Tara Essock-Burns
Tara Essock-Burns Erik J. Soderblom
Erik J. Soderblom Beatriz Orihuela
Beatriz Orihuela M. Arthur Moseley
M. Arthur Moseley Daniel Rittschof
Daniel Rittschof