- 1Department of Biological Sciences, The University of Illinois at Chicago, Chicago, IL, United States
- 2Environmental and Fisheries Sciences Division, Northwest Fisheries Science Center, National Marine Fisheries Service, NOAA, Port Orchard, WA, United States
Algae, or “greenwater,” is a traditional water additive used in finfish aquaculture but it is associated with high costs and potentially harmful bacterial growth. “Claywater,” a mix of clay and seawater, has been explored as a replacement for greenwater. In some fish species, however, claywater reduces survival rates, but the mechanisms are not understood. A link between water additive and microbial community composition may exist. In this study, the effects of different water additives on the microbial communities of larval sablefish were studied. Three treatments were evaluated: a traditional greenwater additive, a claywater additive, and a greenwater additive switched to claywater after 1 week. Microbial communities were characterized using 16S rRNA gene sequencing, and sablefish survival and growth were recorded. Tank seawater microbial communities were significantly influenced by water additive (treatment). Sablefish microbiomes were significantly but weakly influenced by treatment, and there were time-specific differences within the claywater treatment. Sablefish, from the treatment that was switched after 1 week, maintained microbiomes that were more similar to the initial greenwater treatment. In general, sablefish were dominated by Vibrionaceae operational taxonomic units (OTUs). Variability in the sablefish microbiomes between tanks from the same treatment was high, especially in the claywater treatment, which may have confounded treatment effects. Larvae in the claywater treatment had significantly lower survival rates compared to greenwater and greenwater-claywater treatments, but no treatment effect was observed on sablefish growth (length). Overall, results suggest that claywater does not negatively impact survival or the microbial community of sablefish when preceded by 1 week of greenwater.
Introduction
In aquaculture, most marine fish species require water additives, such as algae, to increase turbidity so larvae can feed successfully (Boehlert and Morgan, 1985; Utne-Palm, 2002; Carton, 2005). Water with this type of additive is known as “greenwater,” and while effective, it is also costly and contributes to excess organic material in tanks and subsequent bacterial proliferation, including pathogens (Attramadal et al., 2012). Alternatives such as clay (“claywater”) have been proposed as algae replacements and have been successful in rearing other fish species (Attramadal et al., 2012; Daugherty, 2013; Stuart et al., 2015). In sablefish, however, claywater results in lower survival rates compared to greenwater in the first week of rearing (Lee et al., 2017b), but the mechanisms responsible for differences in survival between claywater and traditional additives are not well understood.
One potential benefit of greenwater may be the chemically distinct environment provided by greenwater compared to clay additives related to the algal production of the organic sulfur compound dimethylsulfoniopropionate (DMSP). DMSP is released by many algal species, (Charlson et al., 1987; Vila-Costa et al., 2014; Zhou et al., 2017), during senescence, microbial and viral attack, and zooplankton grazing (Yoch, 2002; Reisch et al., 2011). DMSP can be an olfactory cue, indicating to fish that prey is nearby, and leading to increased fish aggregation in the wild (DeBose et al., 2008). Additionally, Lee et al. (2016) found that DMSP increased survival in larval sablefish, likely due to its ability to stimulate foraging. As DMSP degradation is microbially mediated, there may be a link between fish survival, water additives, and the microbial communities in larval sablefish and the surrounding seawater.
Furthermore, bacteria often have specific associations with algae, due in part to algal exudates such as DMSP (Nicholas et al., 2004; Grossart et al., 2005; Goecke et al., 2013, for reviews, see Hollants et al., 2013; Ramanan et al., 2016). Nannochloropsis sp., an algal species that is a common fish hatchery water additive, has been shown to be associated with Alpha- and Gammaproteobacteria and Bacteroidetes, especially those in the Cytophaga-Flavobacterium cluster (Nakase and Eguschi, 2007). Synergistic effects between bacteria and algae are possible, including the ability of Nannochloropsis sp. to enhance the probiotic inhibition of fish pathogens (Sharifah and Eguchi, 2011). Specifically, members of the Roseobacter clade found within the Alphaproteobacteria class are known probiotics (Porsby et al., 2008); these bacteria degrade DMSP and are commonly observed with algae (Buchan et al., 2005; Luo and Moran, 2014). The absence of these associations may be a potential factor in reduced fish survival when rearing with claywater instead of greenwater.
In this study, we investigated the effects of water additives on the microbial communities of sablefish larvae and seawater, as well as those of the tank and larval skin surfaces. The microbiome of sablefish as it related to rearing tank water additives, parental cross, and time (age) were all evaluated. By addressing differences between algae- and clay-based water additives on a microbial level, we hoped to better understand the underlying factors responsible for survival differences between additives. We hypothesized that greenwater would have a different microbial community structure compared to claywater, and that those differences would directly relate to sablefish growth and survival.
Materials and Methods
Experimental Design
Broodstock collection, spawning, and rearing are described in Cook et al. (2015). Tank setup, lighting, and feeding details are outlined in Lee et al. (2016). Briefly, fertilized eggs were initially kept in incubators, then transferred to silos. When yolk sacs became spent (approximately 0% yolk sac remaining), they were stocked into rearing tanks and fed live rotifers.
Claywater was made daily by mixing seawater with clay (12 mg per L seawater; Kentucky OM4 Ball Clay). Greenwater was made daily by hand-mixing algal paste (0.021 mL/L seawater, Nannochloropsis Instant Algae, 68 billion cells/mL, Reed Mariculture, Campbell, CA, United States) and green dye (0.005 mL per L seawater, “green shade color,” Esco Foods, San Francisco, CA, United States). Those concentrations of claywater and greenwater were achieved with peristaltic pumps that delivered more concentrated solutions of the additives to rearing tanks. Tanks were flow-through, with new seawater flowing into each tank at a rate of 270 mL per minute. Seawater was sourced from Puget Sound, UV sterilized, and filtered to 1 μm. Water temperature was 12°C at stocking on 7 March, gradually increased until reaching 15°C on 11 March and maintained at 15°C thereafter.
Eighteen 37-L tanks were divided among three treatments. Six tanks each were maintained on greenwater or claywater for the duration of the experiment. Another six tanks were started on greenwater and after 1 week were switched to claywater. Each tank contained larvae from only one parental cross. Three parental crosses were stocked at 300 larvae per tank into six tanks per cross on 8 March. The three treatments were divided equally among parental crosses, to control for possible parental cross effects.
Microbial Community Sampling
Samples were collected over the course of three weeks after initiation of feeding. Fish, seawater, and tank surfaces from the hatching silos were sampled 3 days prior to the start of the experiment. These pre-feeding larvae are henceforth referred to as “silo fish.” Rearing tanks were stocked out and sampled 3, 7, and 14 days post-stocking and post-first feed. The day-7 sampling was done before the greenwater-to-claywater switch was made. At each sampling time point, two 60-mL samples of seawater per tank were collected and filtered using Sterivex 0.22 μm filters (EMD Millipore Corp.) with syringes. The sides of the tanks were sampled with sterile cotton tip swabs and stored in transport tubes (Puritan Medical Products Co.). Eight to 12 larvae from each tank were captured and swabbed, with the exception of the final time point in claywater, when three tanks had only one survivor, one tank had two survivors, and two tanks had at least 10 survivors. After swabbing larval surfaces, whole fish surfaces were sterilized in a bath of 0.1% (v/v) benzalkonium chloride in sterile seawater. Fish were then rinsed twice with sterile seawater and twice in nuclease-free water before storing in cryotubes. Only rearing tank seawater and tank swabs were collected on stock out day (7 March). Tank seawater samples were also preserved for cell counts (1,125 μL seawater, 375 μL 4% paraformaldehyde). Additional samples including clay, algal paste, rotifer feed, and incoming seawater from Puget Sound were taken. All samples were frozen at -20°C until further processing.
Sablefish Survival and Growth
After microbial sampling on the final day of the experiment, surviving larvae were hand counted from each tank. The number of survivors was calculated as the number of larvae hand counted from each tank, plus the number of survivors removed from the tank for microbial sampling earlier that day. Prior to swabbing during microbial sampling on that final day, larvae were photographed with a ruler for scale. Total larval length was later measured from those images using image analysis software (Infinity Analyze, Lumenara Corporation, Ottawa, ON, Canada).
DNA Extractions and PCR
Bacterial DNA from water samples was extracted from the Sterivex filters using the PowerWater Sterivex DNA Isolation Kit (Mo Bio Laboratories, Inc.) while DNA from whole fish and swab samples was extracted using the PowerSoil and PowerSoil –htp 96-Well Soil DNA Isolation kits, respectively (Mo Bio Laboratories, Inc.). Whole fish (8–12 individuals) from each tank time point were extracted together. Swab extractions were modified to enhance yield by reducing the volume of reagents used for the IRT steps. Additional samples were also extracted using the PowerSoil kit. A targeted amplicon sequencing (TAS) protocol was employed as previously described by Green et al. (2015) with modifications to the 16S hypervariable region used and polymerase. We amplified the V3-V4 hyper-variable region of the 16S rRNA gene using previously designed 341F/806R primers with CS1/CS2 linkers, using GoTaq Hot Start Colorless Master Mix (Promega).
16S Amplicon Sequencing
Libraries were prepared and sequenced on the Illumina MiSeq platform at the DNA Services Facility at The University of Illinois at Chicago (V3 chemistry, 2 × 300 paired-end sequencing). Raw sequencing reads are available for download from the NCBI Sequence Read Archive, BioProject accession number PRJNA431797.
Read pairs were merged, trimmed, quality and length filtered, and chimera checked using PEAR (v. 0.9.10), CLC Workspace, and UCLUST, respectively. Sequences were then analyzed using the Quantitative Insights into Microbial Ecology (QIIME, v. 1.9.1) pipeline. Greengenes (13_8 release; DeSantis et al., 2006) was used as a reference database for the taxonomic assignment of operational taxonomic units (OTUs) based on 97% sequence similarity. Biom tables were filtered to remove singletons, doubletons, and OTUs accounting for less than 0.005% of the total abundance. Sequences identified as chloroplasts and mitochondria were removed prior to analysis. QIIME files were then analyzed using the phyloseq (v. 1.19.1) and DESeq2 (v. 1.16.1) packages in R (McMurdie and Holmes, 2013; Love et al., 2014). Samples were not rarefied (McMurdie and Holmes, 2014), reads per sample were between 10–30k, depending on sample type. Samples of the algal paste and powdered clay had fewer than 200 reads but were retained in order to use to compare to greenwater and claywater. OTUs of interest were checked with NCBI’s Nucleotide Basic Local Alignment Search Tool (BLAST) (Altschul et al., 1990; Zhang et al., 2000).
Cell Counts
Preserved samples were diluted 1:100 with 1X phosphate buffered saline (PBS). Dilutions were filtered onto black polycarbonate filters (0.2 μm, 25 mm; Millipore, Cork, Ireland) with 4′,6-diamidino-2-phenylindole (DAPI), to achieve a final concentration of 5 μg/mL DAPI. After staining, filters were rinsed with PBS and Milli-Q water and mounted on microscope slides. Counting was done according to the guidelines outlined in Muthukrishnan et al. (2017) with 30 random fields of view per sample. Slides were imaged using fluorescent microscopy (Leica DM5500 B, Leica Microsystems, Wetzlar, Germany; 100x/1.4 Oil Plan Apochromat objective lens; QImaging EXiAqua Bio-Imaging Microscopy camera) in ImagePro software (v. 7.0), and processed using ImageJ software (v. 1.51j8, Schneider et al., 2012). The effects of additive and date on cell counts were modeled in R statistical software (v. 3.4.4) using a generalized linear mixed effects model with a poisson family, log link function, and a random effect of tank.
Statistical Analyses
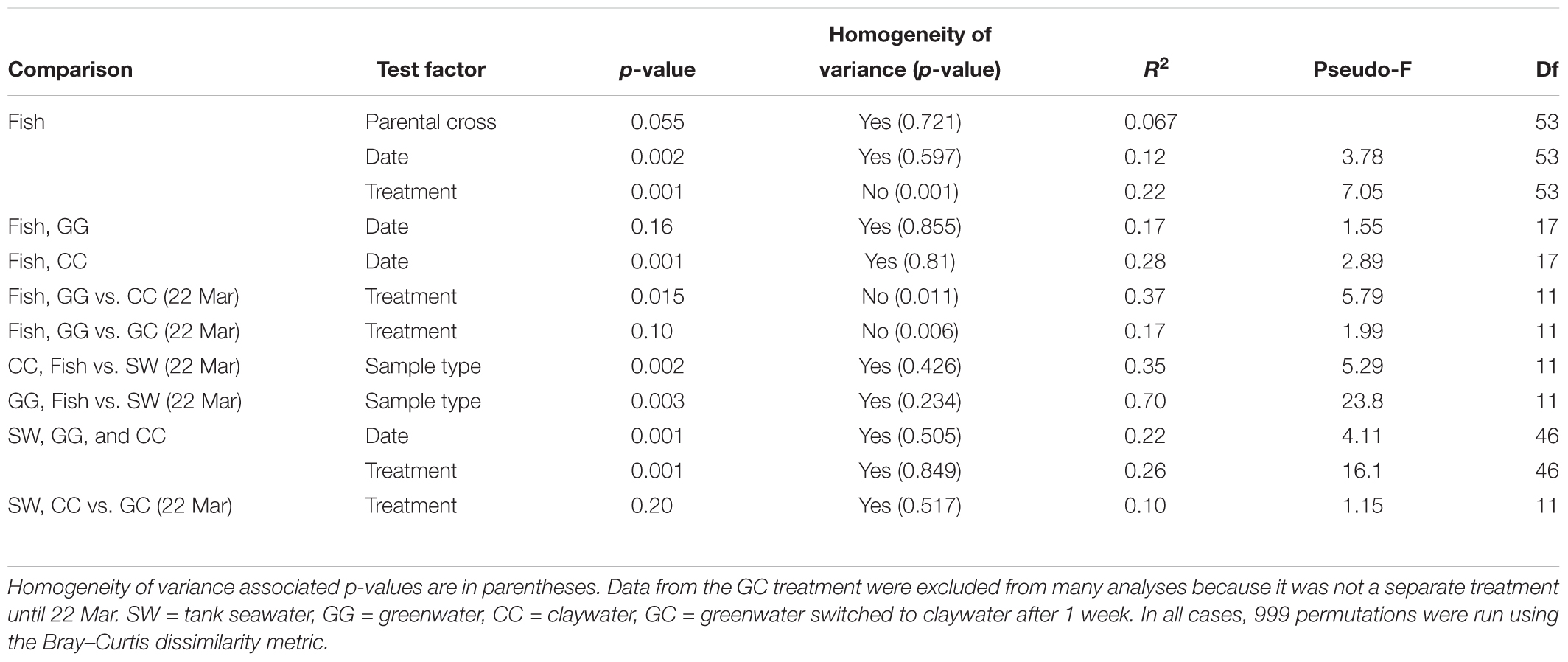
Permutational multivariate analysis of variance (PERMANOVA) were carried out in R (adonis function, vegan package v. 2.4-4) using Bray–Curtis OTU-based distance matrices to test the effect of the factors time (date), treatment, and where applicable, parental cross, on the microbial community structure of each sample type. Homogeneity of variance was evaluated with the betadisper function, which utilizes a multivariate analogue of Levene’s test. Results that did not violate the homogeneity assumption of the PERMANOVA test were interpreted as the most reliable. Those comparisons with a balanced design that were statistically heterogenous were also reported as they are still considered admissible as outlined in Anderson and Walsh (2013). DESeq2 analysis was carried out using code from the phyloseq tutorial “Using Negative Binomial in Microbiome Differential Abundance Testing,” including the calculation of geometric means prior to DESeq2 testing to account for zero values (McMurdie and Holmes, 2013). DESeq2, by default, calculates adjusted p-values using the Benjamini–Hochberg correction (Love et al., 2014). One-way analysis of variances (ANOVA) were run to test the effect of treatment on sablefish survival and length (GLM, Systat 13). All statistical analyses assessed significance using an alpha level of 0.05.
Results
Survival was low for all treatments, between 2.1–8.5%. There was a significant effect of treatment on survival, with both GG and GC treatments having significantly higher survival compared to the CC treatment (Figure 1, p < 0.05). No effect of treatment was seen on sablefish growth based on larval length (Figure 1, p > 0.05).
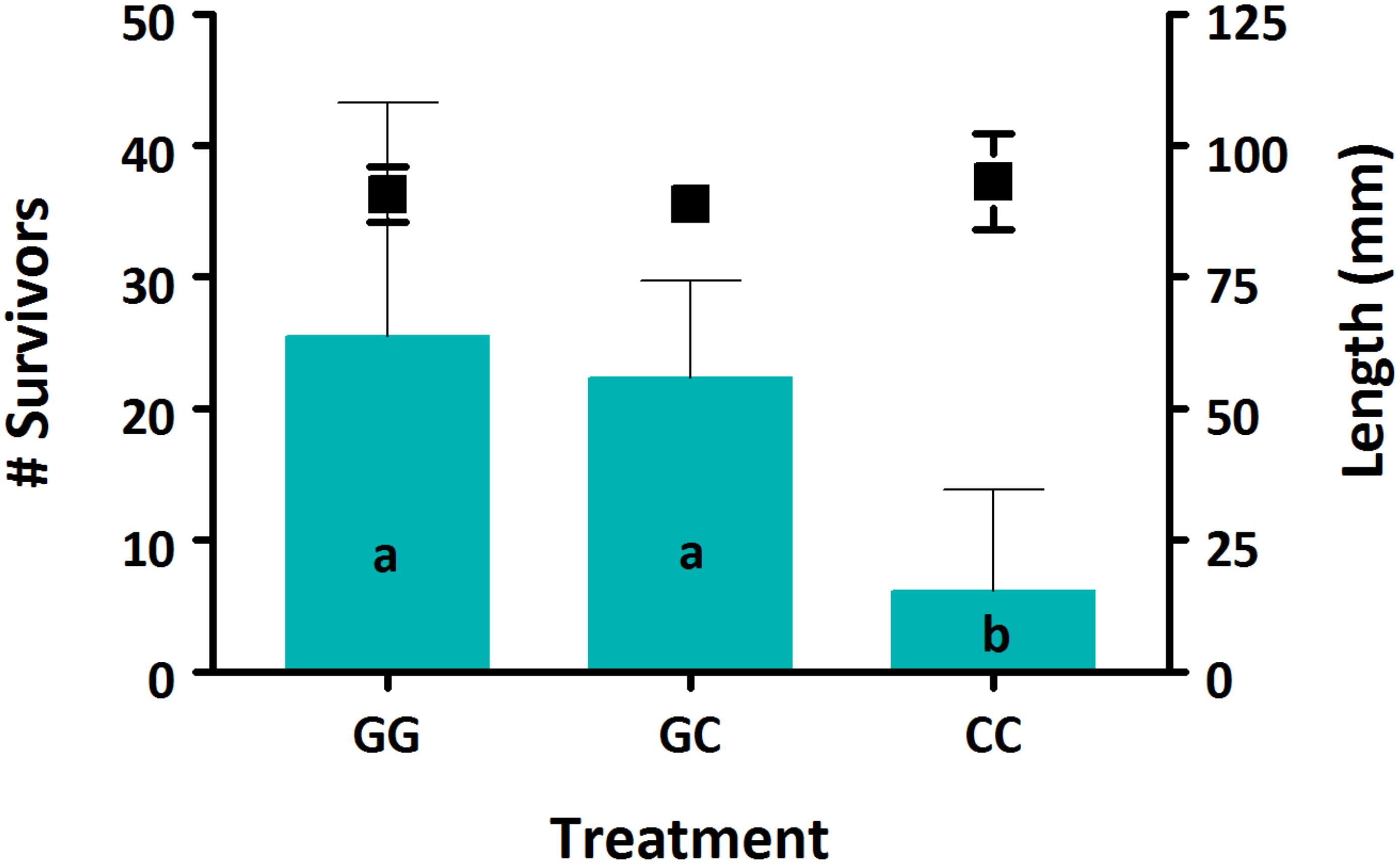
Figure 1. Survival (bars) and growth (markers) of sablefish larvae from each treatment. Data are presented as means (±SD). Different lowercase letters denote significant differences between treatments. Where there are no letters, no significance was found. A significant effect of treatment was seen on sablefish survival (p = 0.016) but not on length (p > 0.05).
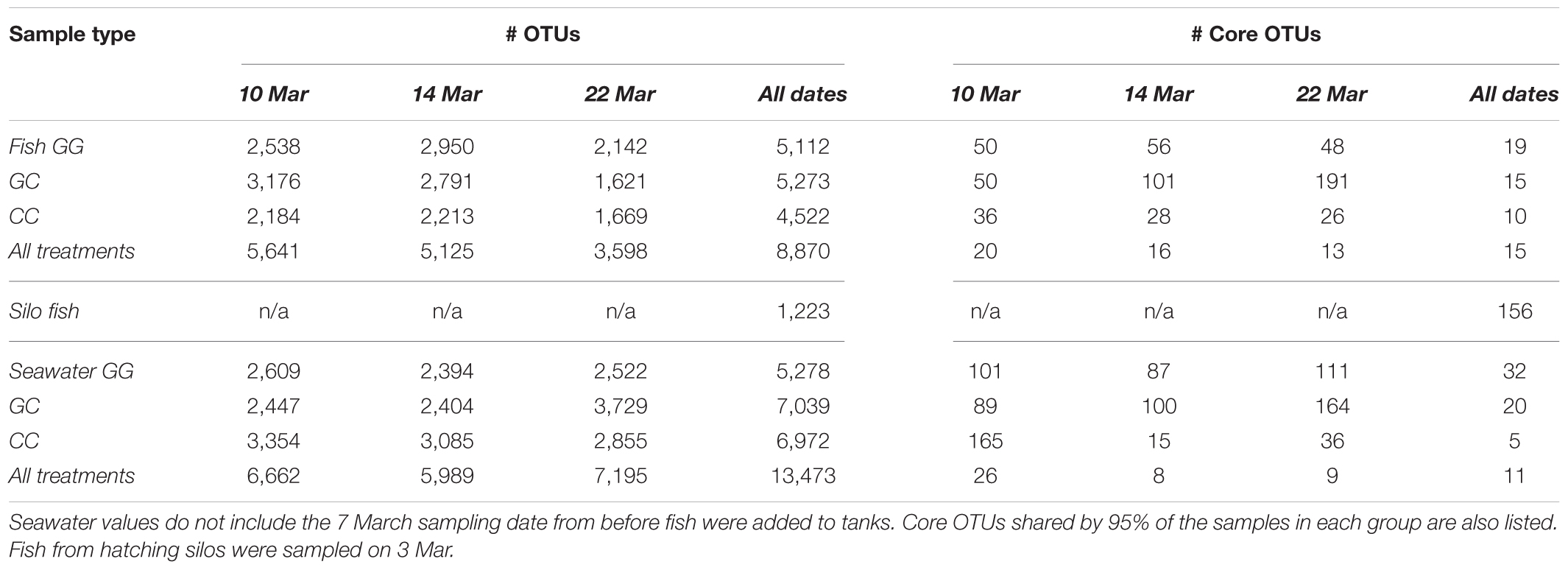
A total of 7.7 million sequences were analyzed from 371 samples, with a median of 19,120 reads per sample. After filtering, across all treatments and dates sampled, a total of 34,565 OTUs were observed. Seawater samples had 14,616 OTUs, whole fish had 8,870 OTUs, fish surfaces had 23,262 OTUs, and tank surfaces had 23,738 OTUs.
All sample types were dominated by Alpha- and Gamma proteobacteria, especially fish, which maintained OTUs belonging to the family Vibrionaceae at abundances of >70% in many cases (Figure 2). Relative abundances of Vibrionaceae were lower in fish and seawater samples from the claywater (CC) treatment, compared to those in the greenwater (GG) and greenwater-claywater (GC) treatments (Figure 2). The Vibrionaceae OTU that comprised the majority of fish communities was found in all treatments from all dates (core OTU, defined at 95% prevalence). BLAST results showed that this sequence had a 98% identity with Vibrio spp. isolated from other marine hosts. Other abundant families found to be associated with sablefish included Rhodobacteraceae, Colwelliaceae, and Oceanospirillaceae. Overall, fish samples were more similar to one another, based on composition (Bray–Curtis), than to any other sample type in the experiment, despite individual variation (Figure 3).
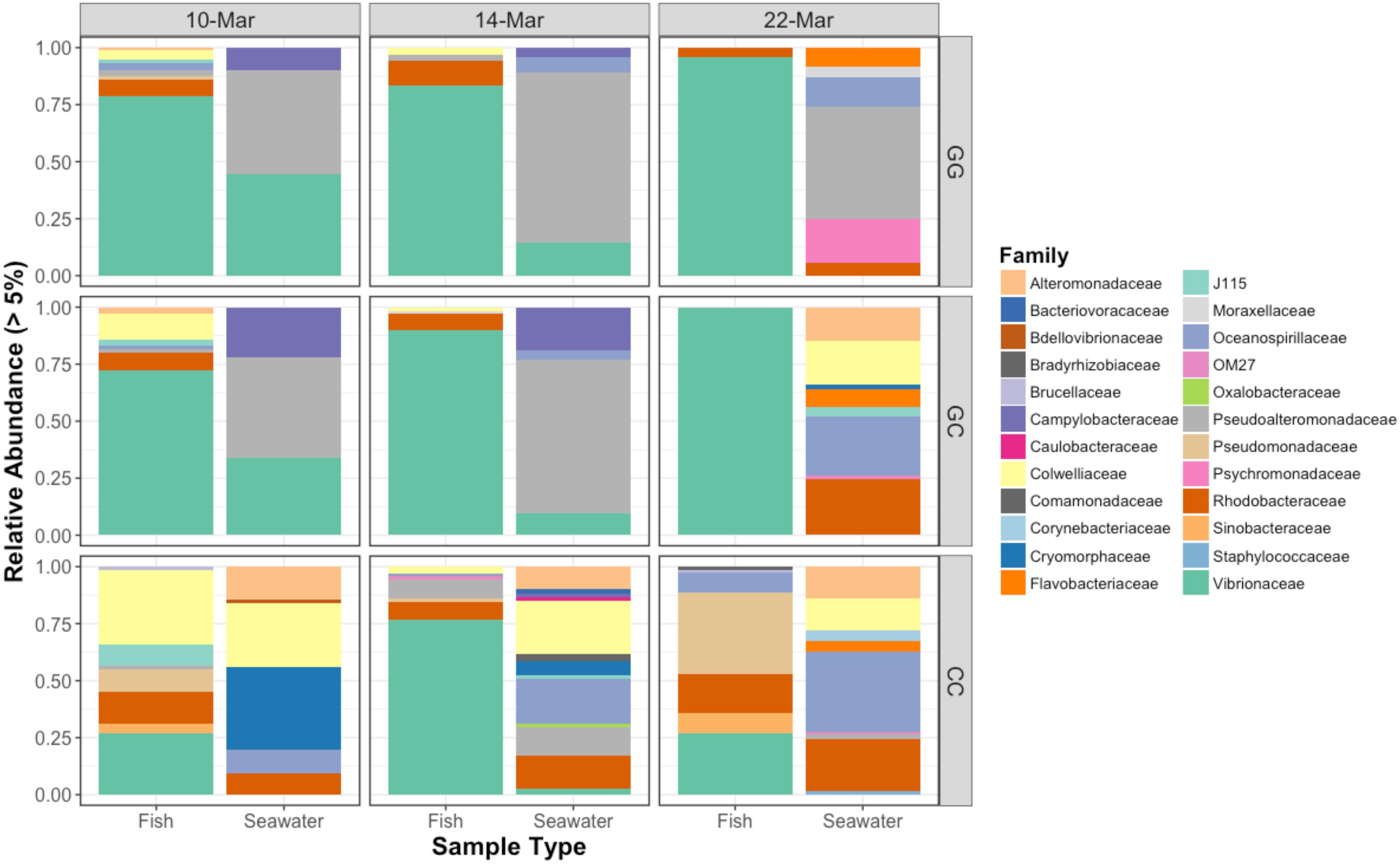
Figure 2. Relative abundances of bacterial families that represent greater than 5% of the total community for fish and seawater samples. GG = greenwater, CC = claywater, and GC = greenwater-claywater treatment. Fish microbial communities were dominated by Vibrionaceae OTUs throughout the experiment, especially in the GG and GC treatments.
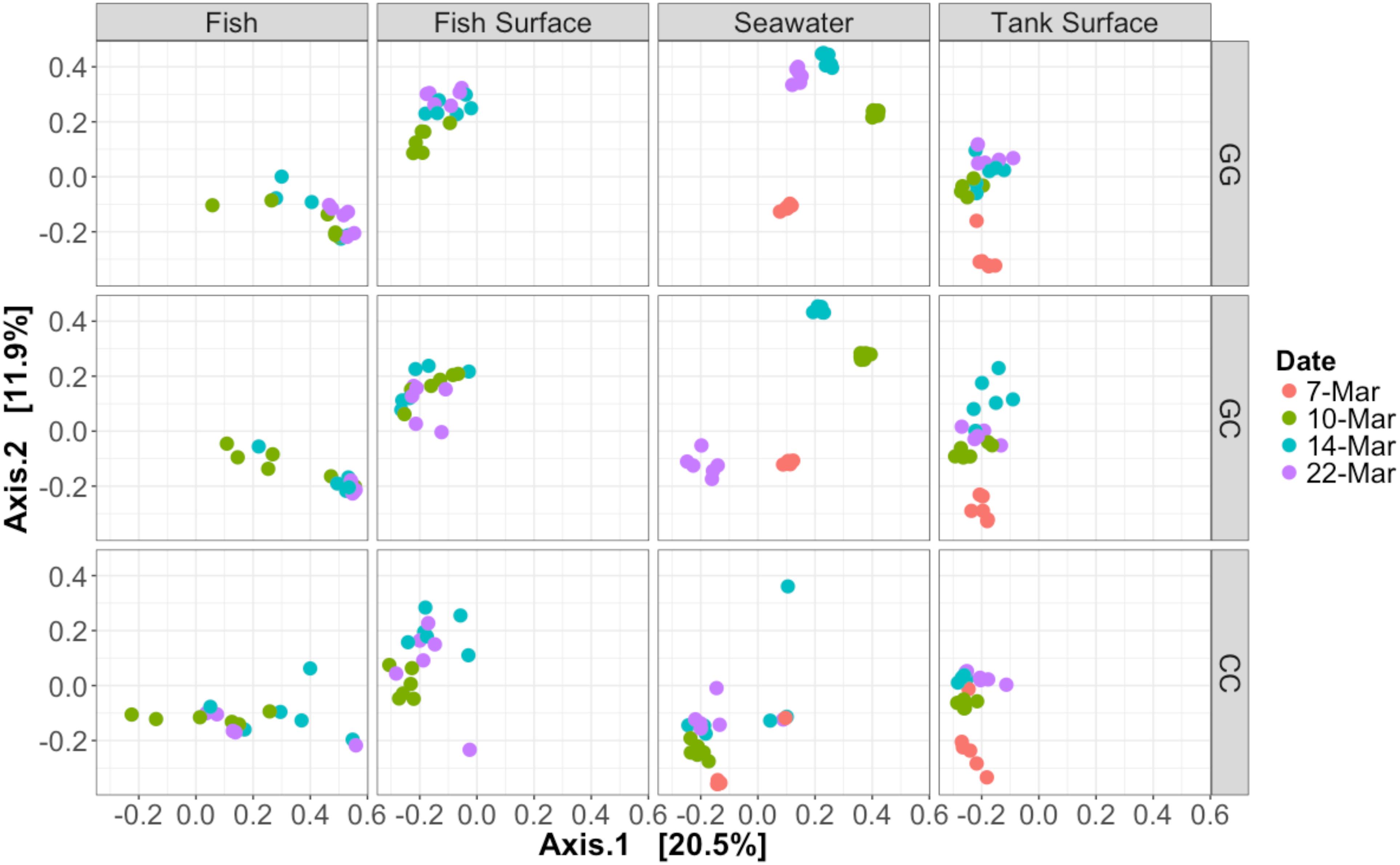
Figure 3. Bray–Curtis PCoA of all samples taken during the experiment. Samples are faceted according to type and treatment. Dates are represented by different colors. Percentage variation explained by each axis is given in brackets. Seawater microbial communities changed over time and were influenced by treatment. Fish samples did not cluster together closely, especially those in the claywater treatment.
The community composition of fish from the claywater treatment exhibited high variability and shifts in composition and abundance at each time point were observed during the experiment (Figures 2, 3 and Table 1). The microbial communities of whole fish from the greenwater treatment, however, exhibited lower variation between individuals and were not significantly influenced by time (Table 1, p = 0.16). Fish from the GC treatment maintained microbial communities that were most similar to those associated with fish in the GG treatment, even after being switched to the claywater additive (Figures 2, 3). On the final sampling date, when three true treatments were represented, fish from the GC treatment had microbiomes that were not significantly different from those in the GG treatment (Table 1, p > 0.05). At this time point, fish from the GG and GC treatment shared more OTUs than either shared with fish in the CC treatment (Figure 4A). Additionally, fish from the CC treatment maintained microbiomes which were significantly different from fish in the GG treatment (Table 1, p < 0.05). Although a significant effect of treatment was seen on the composition of the fish microbiome, the samples did not meet the homogeneity of variance assumption for the PERMANOVA.
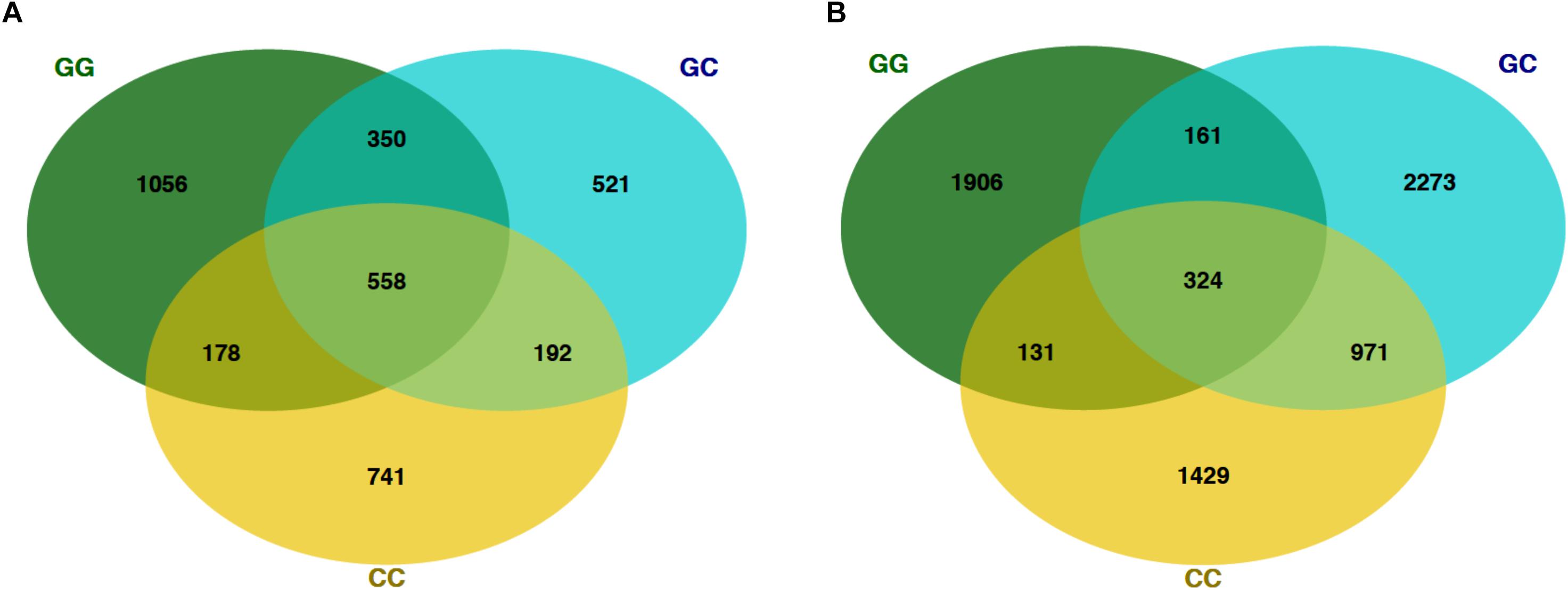
Figure 4. Venn diagrams showing the number of shared OTUs on the final sampling date (22 Mar) among fish and seawater samples. (A) OTUs from fish microbiomes of each treatment and (B) seawater microbiomes.
At the final time point, fish from GG and CC treatments shared 14 core OTUs, five of which were members of the top 10 most abundant OTUs in both treatments (Supplementary Figure S1). Fish from all treatments shared one-third of all the OTUs observed (Table 2 and Figure 4A, 558 shared/3,598 total OTUs). On this date, half the OTUs observed in fish from the GC treatment were also observed in fish from the GG treatment (Figure 4A, 908 shared/1,621 total OTUs in GC). DESeq2 analysis revealed that across all dates, 35 OTUs were significantly enriched in GG compared to CC fish microbiomes. Fish from the greenwater treatment had 1–3.0 log2 fold increases in OTUs from the Rhodobacteraceae, Campylobacteraceae, Vibrionaceae, and Pseudoalteromonadaceae families, among others (Figure 5B). Due to heterogeneity of variance of the samples, however, these results should be interpreted with caution as high within-group variability can artificially inflate the dispersion estimates for DESeq2.
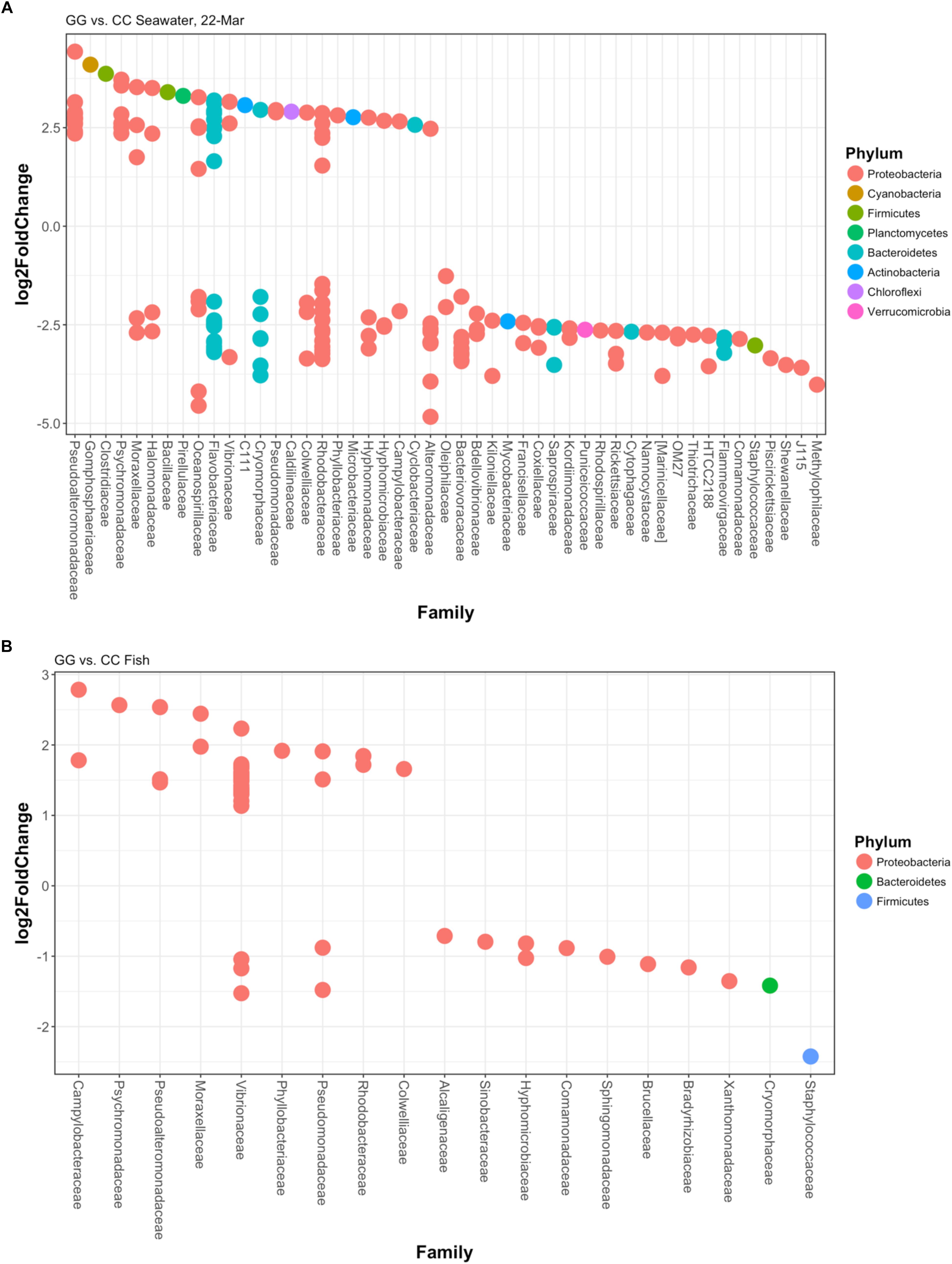
Figure 5. Log2fold changes in OTUs found in (A) greenwater compared to claywater on the final sampling date and (B) whole fish in the greenwater treatment compared to whole fish in the claywater treatment from all timepoints of the experiment. Each dot represents a different OTU. Colors designate the phylum each OTU belongs to, and their associated family is on the x-axis. All are significant (p < 0.05) using DESEq2’s built-in Benjamini–Hochberg correction for adjusted p-values.
There was no significant effect of parental cross on the OTU composition in fish (p > 0.05, Table 1). Date (time) from first feed did have a significant but weak influence on community structure (R2 = 0.12, Table 1). When treatments were analyzed individually, there was no significant effect of time on the GG treatment (p > 0.05), but there was a significant effect on the CC treatment (p < 0.05, R2 = 0.28). Thus, the microbial communities of fish changed over time in the CC but not the GG treatment.
Interestingly, fish in hatching silos were dominated (>90%) by the family Pseudomonadaceae (Figure 6), but by 3 days post-first feed, they were absent or reduced (Figure 2). Hatching silo fish had 156 core OTUs, 151 of which were Pseudomonadaceae (Table 2). There were no core Vibrionaceae OTUs in fish from silos. The diversity of these silo fish was low compared to the fish in rearing tanks (Shannon diversity indices, Figure 7).
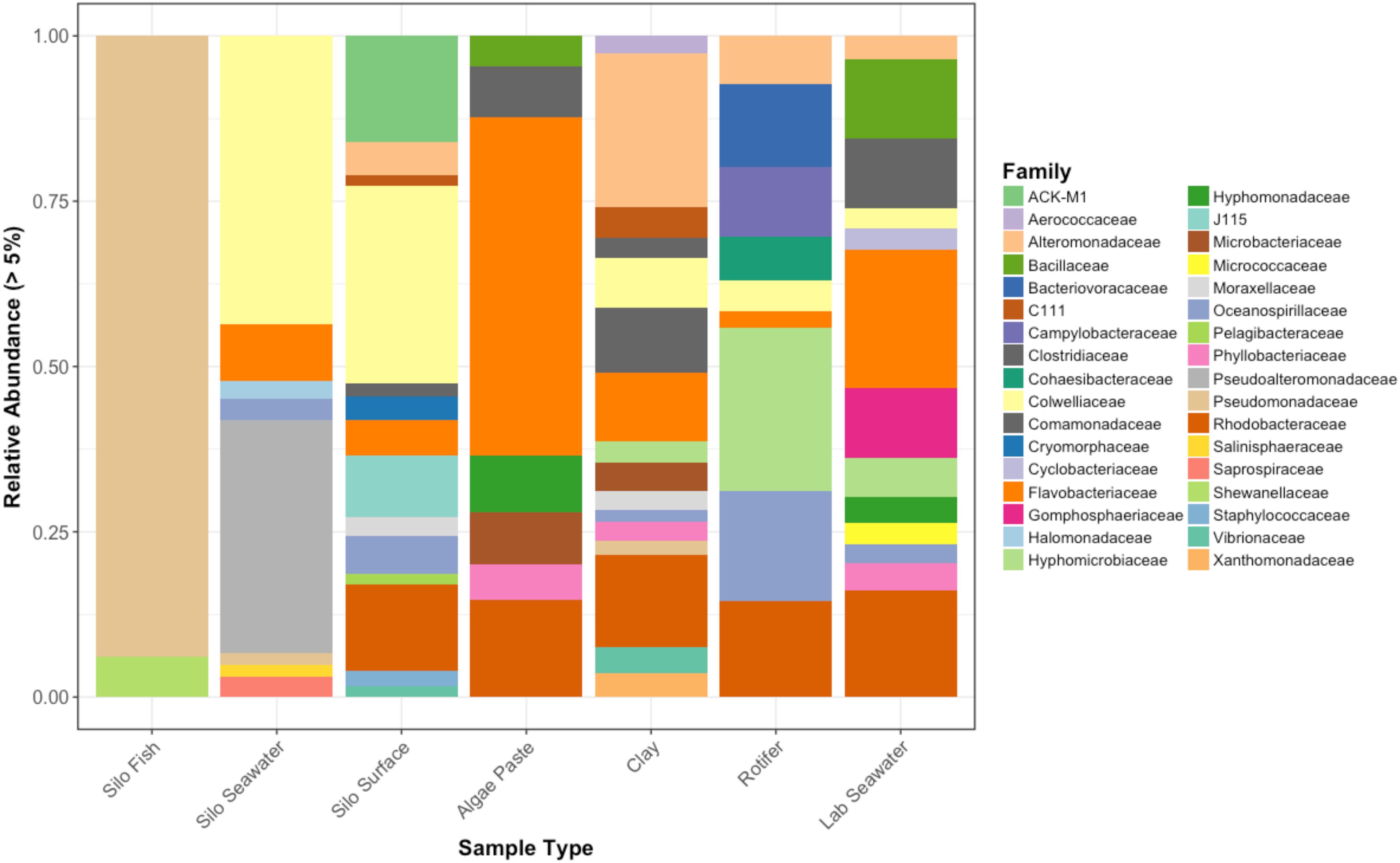
Figure 6. Relative abundances of bacterial families that represent greater than 5% of the total for each sample type. Pre-experimental samples taken from sablefish hatching silos, algal paste, clay, rotifer feed, and inflowing seawater from Puget Sound.
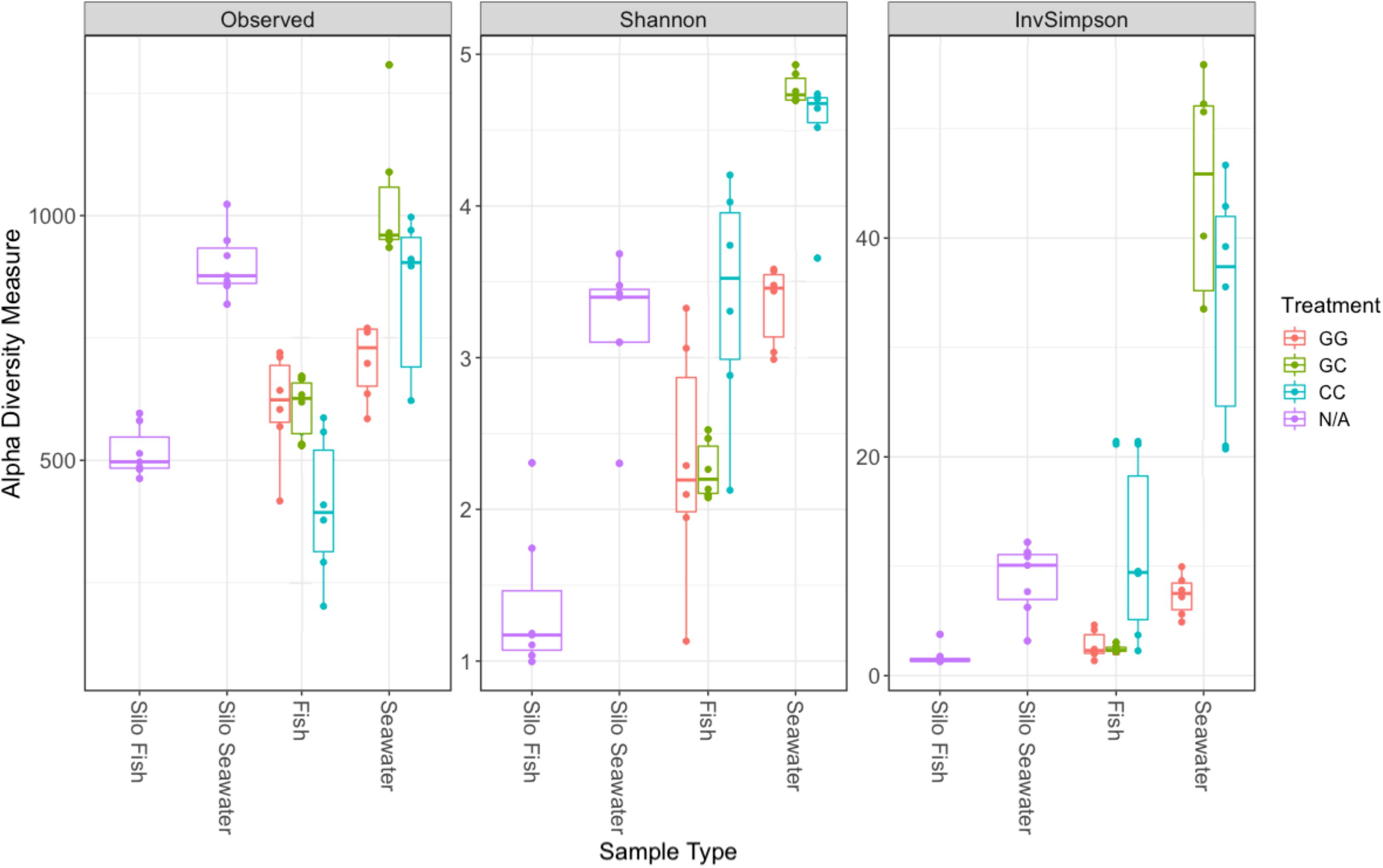
Figure 7. Alpha diversity metrics for samples collected on the final sampling date (fish, seawater) and samples collected prior to the start of the experiment (silo fish, silo seawater). Samples from hatching silos are designed “N/A” because they were not subjected to experimental treatments. Richness estimate means and theoretical standard error ranges are shown by the boxplot for each sample type and treatment.
Fish maintained microbial communities distinct from the surrounding seawater (Table 1). On the final sampling date, both GG and CC treatments showed significant differences between fish and their associated tank seawater (Table 1, p < 0.01). In the greenwater treatment especially, 70% of the variability in the microbial communities of fish and tank seawater was predicted by the sample type (R2 = 0.70, Table 1). Fewer than 400, or 11.5–19.7% of the total OTUs, were shared between fish and tank seawater for both greenwater and claywater treatments (data not shown). Fish microbial communities were also less diverse than seawater communities across all treatments (Figure 7).
The OTU composition of seawater was significantly affected by both treatment and time (Table 1, p < 0.001). When the GC treatment was switched from greenwater to claywater, the seawater microbiome shifted to resemble that of the CC treatment. One week post-water additive switch, the GC seawater microbiome was not significantly different from that of the CC seawater (Table 1, p > 0.05; Figure 2), but both were significantly different from the GG seawater microbiome. GG and GC shared a large portion of OTUs, nearly one-third of the total observed (971 shared OTUs/3,729 total GC), while all three treatments shared fewer than 400 OTUs (Figure 4B and Table 2). The CC and GC OTUs driving the differences between treatments belonged to the phyla Cyanobacteria, Proteobacteria, Chloroflexi, Actinobacteria, Firmicutes, Bacteroidetes, Verrucomicrobia, and Planctomycetes (Figure 5). Most of the OTUs that were significantly enriched in GG seawater compared to CC were from the families Pseudoalteromonadaceae, Rhodobacteraceae, Flavobacteriaceae, Moraxellaceae, and Psychromonadaceae (Figure 5, p < 0.05).
Vibrionaceae abundance decreased over time in GG and GC seawater (Figure 2). Pseudoalteromonadaceae and Camplyo bacteraceae were abundant in GG seawater at all sampling dates, while CC seawater microbial communities shifted for each date sampled (Figure 2). Colwelliaceae, Alteromonadaceae, Rhodobacteraceae, and Oceanospirillaceae, however, were regularly observed in the CC seawater. Samples from the rotifer feed, algae, and incoming seawater from Puget Sound were analyzed in order to pinpoint the contribution and potential source of Vibrio OTUs to the GG and GC seawater, but none of these contained Vibrionaceae OTUs in abundances higher than 5% of the total community (Figure 6). Microbes associated with the algal paste were mostly Flavobacteriaceae and Rhodobacteraceae, while clay-associated microbes were more diverse and included Colwelliaceae, and Alteromonadaceae, among others (Figure 6).
Cell counts showed significantly higher cell concentrations in CC seawater compared to GG seawater (Supplementary Figure S2). The GG treatment had significantly fewer bacterial cells (β = -2.190, SE = 0.204, z = -10.75, p < 0.001) than the CC treatment. After 1 week (14 March), cell counts from all treatments were significantly higher than the start date of the experiment (β = 0.474, SE = 0.017, z = 28.05, p < 0.001). This increase was especially evident in the GG seawater, where cell counts were 2.6 times higher on the final date (22 Mar) compared to the initial date (7 Mar) of the experiment (β = 0.582, SE = 0.208, z = 2.80, p < 0.01).
Discussion
The results of this experiment demonstrated that water additives result in different larval survival rates and microbial community compositions in sablefish rearing tanks. The microbial communities of fish and tank seawater were both affected by treatment and time. Treatment effects, however, were variable depending on the type of sample. Treatment had the greatest effect on seawater microbial communities, maintaining distinct compositions between the greenwater and claywater treatments.
Sablefish in the hatching silos harbored microbiomes, which were different from the silo seawater as well as the post-first feed larvae in rearing tanks. Silo fish may have derived their microbiome from the yolk sac, which could account for these differences. Because larvae were stocked out at approximately 0% yolk sac, it is likely that the shift in the fish microbiome happened rapidly and in response to first feeding. Other work has also shown a major shift in the salmon-associated microbiome during development from eggs to first-feeding (Romero and Navarrete, 2006). Our results indicate that intra-specific differences were not attributable to parental cross, further implicating the yolk sac and diet as factors in the microbial community structure of the larvae. Although treatment effects on the whole fish-associated microbial communities were seen, all larvae still maintained one Vibrionaceae OTU that dominated the community. Additionally, despite treatment differences, sablefish shared a large portion of OTUs, up to 33% of the total sequenced. Individual fish microbial communities were variable in composition and abundance, which obscured treatment-specific effects. Thus, even after accepting the statistical results based on the balanced design, R2 values associated with significant treatment effects were low in most cases. Less than 40% of the variability between fish microbiomes was explained by treatment. Further, fish from all treatments and dates shared many of the same core OTUs, which accounted for a large proportion of the microbial community in some cases.
The claywater fish microbiome was characterized by both homoscedasticity and shifts in composition and abundance over the course of the experiment. The use of the clay additive may have contributed to stress in the fish, resulting in dysbiosis and thus more within-group variability. Stress in these fish could further be evidenced by their significantly lower survival. Zaneveld et al. (2017) proposed applying the “Anna Karenina principle” to animal microbiomes; that is, perturbation induced changes in the microbiome are stochastic. The authors provide numerous examples supporting the idea that a healthy microbiome may be characterized by specific patterns in composition, but dysbiotic individuals do not display an “alternative” or unhealthy pattern of microbial community composition, they are simply random and variable (Zaneveld et al., 2017). If claywater additions are stressful to the fish, it may account for lower survival and the increased variability in the types and abundances of microbes between individuals. Interestingly, alpha diversity was highest in fish and seawater from the claywater treatment, which could reinforce the idea of an unstable, non-selective, stochastic colonization of these fish compared to those reared in greenwater. Further, other researchers have documented intraspecific variability in the fish microbiota, suggesting it may be a common feature (Givens et al., 2015; Bledsoe et al., 2016).
Overall, the effect of treatment on sablefish-associated microbial communities was difficult to interpret due to heterogeneity of variance, causing the data to fail the assumptions of the PERMANOVA. Through modeled datasets, Anderson and Walsh (2013) determined that with a balanced design, datasets failing to meet the homogeneity assumptions of the PERMANOVA were still statistically robust and acceptable. The concern with heterogeneity is that differences within a group influence the PERMANOVA’s interpretation of differences between groups. As such, the statistical results of sablefish microbial communities presented here should still be interpreted with some caution.
It is likely that internal fish microbiota are independent of the seawater microbial community, to some degree. A few results indicate that this may be the case. First, fish from different treatments had nearly double the OTUs in common with each other than with the seawater, even when comparing fish and seawater from the same water additive treatment. Second, fish in the claywater treatment maintained high relative abundances of Vibrionaceae, even when these bacteria were absent or in low abundances in the seawater. Finally, the sablefish-associated microbes in the greenwater-claywater treatment stayed similar to those in the initial greenwater treatment, even though the seawater microbial community shifted to be more similar to the claywater treatment. This further indicates that environment may not be the most important factor in determining the sablefish microbial community, at least not throughout the entire larval stage. Specifically, the environmental microbiome may impact the fish more or less at different stages in ontogeny. Additionally, survival in fish from the GC treatment was not significantly different from survival in the GG treatment. This result supports our research group’s previous findings that sablefish have poor survival if reared in claywater but can have better survival if reared in greenwater for the first week post-first feed, before switching to claywater (Lee et al., 2017b). It may be that after 1 week post-first feed, the window for microbial colonization has closed and water additive matters less. Alternatively, the fish may represent a more favorable environment for greenwater-associated microbes and thus, retain them. The interplay between water additive and ontogeny in microbiome succession is still unclear, and further research is needed.
Many studies on fish microbiota have shown that environmental recruitment was a major route of colonization of fish species (Nayak, 2010; Semova et al., 2012; Wu et al., 2013; Schmidt et al., 2015; Sullam et al., 2015). Although this is disputed by others (Roeselers et al., 2011; Li et al., 2012; Larsen et al., 2014; Ye et al., 2014; Li et al., 2015), who argue that host factors (i.e., species) are more influential in shaping fish microbial community structure. More likely, a combination of extrinsic and intrinsic factors together is responsible for shaping the fish microbiome. In a meta-analysis of 25 fish gut communities, Sullam et al. (2012) found that both environmental and ecological factors influenced microbial community composition and structure. Additionally, diet has been implicated as an important influence. In a comprehensive study on five different fish species, diet, but not host species, was the major determinant of fish microbiome composition (Dr. Konstantinos Kormas, personal communication). In first feeding rainbow trout, a marine-based fish meal and fish oil diet resulted in an abundance of Proteobacteria compared to a plant-based diet (Ingerslev et al., 2014), and shifts in their bacterial community were dependent on the ratio of dietary plant to animal protein source (Michl et al., 2017). In particular, animal protein-based diets promoted the growth of Vibrionales, among others, in the gut microbiome of fish (Michl et al., 2017). Similar work on another salmonid has also shown that animal vs. plant-based diet influences the microbial community structure of the intestines (Schmidt et al., 2016). The rotifer-based diet in the current study was not seen to contain vibrios in abundance on its own but may have promoted an environment which was favorable to their colonization and growth. Since all treatments received the same diet, rotifers may have contributed to the observed similarities among fish microbiomes. In addition to protein source, lipids, polysaccharides, iron, antibiotics, and nutraceuticals have all been demonstrated to impact the gut microbial communities of fish (for reviews, see Ringø et al., 2012; Romero et al., 2014). Taking these studies as a whole, many intrinsic and extrinsic factors, and not one, likely contribute to fish microbial community colonization, maintenance, and succession.
Our results showed low diversity at the phylum and even family level for fish microbial communities, which were dominated by the Gammaproteobacteria family Vibrionaceae. This is in line with what research on other fish species has shown. In a study of 12 species of bony fish and three species of sharks, Givens et al. (2015) observed that Proteobacteria accounted for 62.5% of all sequences acquired from the gut microbial communities of all species. Other fish species have also been characterized by low phylogenetic diversity at the phylum level during the larval stage (Bledsoe et al., 2016). It should be noted that most microbial community studies focus on adult fish, and due to shifts in the microbiome over ontogeny, comparisons are limited.
Greenwater-associated microbes were Pseudoalteromona-daceae, Flavobacteriaceae, Rhodobacteraceae, Moraxellaceae, Campylobacteraceae, and Oceanosprillaceae. These results are comparable to other research, which has shown Flavobacteriaceae and Rhodobacteraceae to be highly abundant algae-associated bacteria, especially with Nannochloropsis sp. (Goecke et al., 2013; Geng et al., 2016). Members of the Roseobacter clade, such as Rhodocateraceae, are not only regularly associated with microalgae, but are known DMSP metabolizers (Malmstrom et al., 2004; Luo and Moran, 2014). Their role in the conversion of DMSP, a foraging cue in fish, may contribute to the reason why greenwater is necessary during the first week of sablefish rearing. Porsby et al. (2008) found that two Roseobacter strains within the Rhodobacteraceae family conveyed a probiotic effect in turbot aquaculture system. Other researchers have shown higher survival rates with the addition of Nannochloropsis sp. to larval rearing tanks, correlating the results to the presence of the Cytophaga–Flavobacterium cluster, associated with algae and their subsequent suppression of pathogenic Gammaproteobacteria (Nakase et al., 2007). Additionally, Cytophaga-like bacteria, like the Roseobacters, have been implicated in DMSP metabolism (Malmstrom et al., 2004). Due to their functional metabolic roles and antagonism of pathogens, these algae-associated microbes represent potential probiotics that could supplement claywater. The claywater microbiome was characterized by high variability over time. Despite this, Colwelliaceae, Alteromonadaceae, Rhodobacteraceae, and Oceanospirillaceae, among others, were regular members of the claywater-associated microbial community. Overlapping OTUs between the two treatments was likely due to the incoming seawater from Puget Sound, which also harbored these groups in abundance, especially Flavobacteriaceae and Rhodobacteraceae. As was observed in Stuart et al. (2015), clay reduced vibrio relative abundances in seawater compared to greenwater. Unlike Stuart et al. (2015), however, in the current study of this reduction of vibrios did not equate to increased survival of sablefish. One reason for this may be the increased number of total bacterial cells in the claywater treatment, as indicated by the cell counts. Clay is sticky, negatively charged, and as such may be binding cells, leading to significantly higher counts. Attramadal et al. (2012) observed the opposite result with regards to cell counts in clay and greenwater, but the concentration of algae used was higher than in the current study.
Although overall survival was low, the trends with respect to water additive observed here are consistent with our research group’s previous work (Lee et al., 2017a,b). Further, tank size is known to influence survival (Cook et al., 2015), and the small tank size used in this study likely contributed to poor survival. Survival rates in these experiments were consistent with past survival rates in similar sized tanks. These results highlight the larval stage as a bottleneck for the aquaculture industry, and the importance of research aimed at improving methods during this critical phase.
The results presented here support the use of greenwater in the first week of sablefish rearing, then switching to claywater to reduce cost. Longer term experiments are needed to validate that microbial communities in those fish continue to remain stable post-switch. The connection between the larval host and microbial colonization is likely nuanced and affected by multiple factors such as fish ontogeny, health, species, environment, and diet. Further studies are needed to understand the mechanisms driving larval fish microbial colonization. This work, however, provides an important step in understanding how different water additives influence the survival and microbial communities of sablefish in a flow through aquaculture system.
Conclusion
Seawater microbial communities were most significantly influenced by water additive. Fish-associated microbial communities were influenced by treatment to some degree, but were characterized by high individual variation, especially with respect to beta diversity. Clay may be stressful, causing stochastic changes in the microbiome, and could explain lower survival rates in this treatment. Fish from all treatments shared one-third of their OTUs and were more similar to one another than to the seawater. Host parental cross did not appear to influence the microbial communities of sablefish. Date influenced sablefish microbial community structure, but its effects were different between treatments. A major shift in fish microbiomes was observed from the hatching silos to 3 days post-first feed. More research on the dynamics between fish ontogeny and diet are needed. Claywater replacement of greenwater after 1 week resulted in comparable survival rates and similar microbial communities as those sablefish reared solely on greenwater. The results presented here contribute to refining an alternative water additive to reduce costs and maintain or increase survival for the sablefish aquaculture community.
Ethics Statement
This study was carried out in accordance with the recommendations of the Institutional Animal Care and Use Committee (IACUC) animal use procedures. The protocol was approved by the University of Washington IACUC.
Author Contributions
JL and RP designed the study and collected samples. MP performed laboratory work and analysis of sequencing data as well as writing of the manuscript. ED performed cell count laboratory work and data analysis and aided in analysis of sequencing data. All authors contributed, read, edited, and approved the final manuscript.
Funding
Funding was provided by The Saltonstall-Kennedy Grant Program (National Oceanic and Atmospheric Administration), Award 15WCR040 to RP.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Stefan Green and the UIC DNAS facility for sequencing work. Rick Goetz and staff in the Marine Fish and Shellfish Biology Program at the Manchester Research Station (NOAA Fisheries) provided sablefish larvae and technical support. The views expressed herein are those of the authors and do not necessarily reflect the views of the National Oceanic and Atmospheric Administration or any of its sub-agencies.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2019.00203/full#supplementary-material
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Anderson, M. J., and Walsh, D. C. I. (2013). PERMANOVA, ANOSIM, and the mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol. Monogr. 83, 557–574. doi: 10.1890/12-2010.1
Attramadal, K. J. K., Tøndel, B., Salvesen, I., Øie, G., Vladstein, O., and Olsen, Y. (2012). Ceramic clay reduces the load of organic matter and bacteria in marine fish larval culture tanks. Aquacult. Eng. 49, 23–34. doi: 10.1016/j.aquaeng.2012.02.003
Bledsoe, J. W., Peterson, B. C., Swanson, K. S., and Small, B. C. (2016). Otogenetic characterization of the intestinal microbiota of channel catfish throuh 16S rRNA gene sequencing reveals insights on temporal shifts and the influence of environmental microbes. PLoS One 11:e0166379. doi: 10.1371/journal.pone.0166379
Boehlert, G. W., and Morgan, J. B. (1985). Turbidity enhances feeding abilities of larval pacific herring, clupea harengus pallasi. Hydrobiologia 123, 161–170. doi: 10.1007/BF00018978
Buchan, A., González, J. M., and Moran, M. A. (2005). Overview of the marine roseobacter lineage. Appl. Environ. Microbiol. 71, 5665–5677. doi: 10.1128/AEM.71.105665-5677.2005
Carton, A. G. (2005). The impact of light intensity and algal-induced turbidity on first-feeding Seriola lalandi larvae. Aquacult. Res. 36, 1588–1594. doi: 10.1111/j.1365-2109.2005.01383.x
Charlson, R. J., Lovelock, J. E., Andreae, M. O., and Warren, S. G. (1987). Oceanic phytoplankton, atmospheric Sulfur, cloud albedo and climate. Nature 326, 655–661. doi: 10.1038/326655a0
Cook, M. A., Massee, K. C., Wade, T. H., Oden, S. M., Jensen, C., Jasonowicz, A., et al. (2015). Culture of sablefish (Anoplopoma fimbria) larvae in four experimental tank designs. Aquacult. Eng. 69, 43–49. doi: 10.1007/s10886-016-0713-z
Daugherty, Z. N. (2013). Effects of Algae Paste Substitutes on the Larval Rearing Performance and Microbial Communities in the Culture of Cobia (Rachycentron canadum) and Yellowtail Kingfish (Seriola lalandi). Ph. D. thesis, University of Miami Miama, FL.
DeBose, J. L., Lema, S. C., and Nevitt, G. A. (2008). Dimethylsulfoniopropionate as a foraging cue for reef fishes. Science 319, 1356–1356. doi: 10.1126/science.1151109
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Geng, H., Tran-Gyamfi, M. B., Lane, T. W., Sale, K. L., and Yu, E. T. (2016). Changes in the structure of the microbial community associated with Nannochloropsis salina following treatments with antibiotics and bioactive compounds. Front. Microbiol. 7:1155. doi: 10.3389/fmicb.2016.01155
Givens, C. E., Ransom, B., Bano, N., and Hollibaugh, J. T. (2015). Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 518, 209–223. doi: 10.3354/meps11034
Goecke, F., Thiel, V., Wiese, J., Labes, A., and Imhoff, J. F. (2013). Algae as an important environment for bacteria – phylogenetic relationships among new bacterial species isolated from algae. Phycologia 52, 14–24. doi: 10.2216/12-24.1.s1
Green, S. J., Venkatramanan, R., and Naqib, A. (2015). Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS One 10:e0128122. doi: 10.1371/journal.pone.0128122
Grossart, H. P., Levold, F., Allgaier, M., Simon, M., and Brinkhoff, T. (2005). Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 2005, 860–873. doi: 10.1111/j.1462-2920.2005.00759.x
Hollants, J., Leliaert, F., De Clerck, O., and Willems, A. (2013). What we can learn from sushi: a review on seaweed-bacterial associations. FEMS Microbiol. Ecol. 83, 1–16. doi: 10.1111/j.1574-6941.2012.01446.x
Ingerslev, H. C., von Gersdorff Jørgensen, L., Lenz Strube, M., Larsen, N., Dalsgaard, I., Boye, M., et al. (2014). The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 424–425, 24–34. doi: 10.1016/j.aquaculture.2013.12.032
Larsen, A. M., Mohammed, H. H., and Arias, C. R. (2014). Characterization of the gut microbiota of three commercially valuable warmwater fish species. J. Appl. Microbiol. 116, 1396–1404. doi: 10.1111/jam.12475
Lee, J. S. F., Britt, L. L., Cook, M. A., Wade, T. H., Berejikian, B. A., and Goetz, F. W. (2017a). Effect of light intensity and feeding density on feeding behavior, growth, and survival of larval sablefish Anoplopoma fimbria. Aquacult. Res. 48, 4438–4448. doi: 10.1111/are.13269
Lee, J. S. F., Cook, M. A., Berejikian, B. A., and Goetz, F. W. (2017b). Temporal changes in the suitability of claywater as a greenwater substitute for rearing larval sablefish (Anoplopoma fimbria). Aquaculture 470, 11–16. doi: 10.1016/j.aquaculture.2016.12.011
Lee, J. S. F., Poretsky, R. S., Cook, M. A., Reyes-Tomassini, J. J., Berejikian, B. A., and Goetz, F. W. (2016). Dimethylsulfoniopropionate (DMSP) increases survival of larval sablefish, anoplopoma fimbria. J. Chem. Ecol. 42, 533–536. doi: 10.1007/s10886-016-0713-z
Li, T., Long, M., Gatesoupe, F. J., Zhang, Q., Li, A., and Gong, X. (2015). Comparative analysis of the intestinal bacterial communities in different species of carp by pyrosequencing. Microb. Ecol. 69, 25–36. doi: 10.1007/s00248-014-0480-8
Li, X., Yu, Y., Feng, W., Yan, Q., and Gong, Y. (2012). Host species as a strong determinant of the intestinal microbiota of fish larvae. J. Microbiol. 50, 29–37. doi: 10.1007/s12275-012-1340-1
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Luo, H., and Moran, M. A. (2014). Evolutionary ecology of the marine Roseobacter clade. Microbiol. Mol. Biol. Rev. 78, 573–587. doi: 10.1128/MMBR.00020-14
Malmstrom, R. R., Kiene, R. P., and Kirchman, D. L. (2004). Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 49, 597–606. doi: 10.4319/lo.2004.49.2.0597
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an r package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
McMurdie, P. J., and Holmes, S. (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS One 10:e1003531. doi: 10.1371/journal.pcbi.1003531
Michl, S. C., Ratten, J. M., Beyer, M., Hasler, M., LaRoch, J., and Schulz, C. (2017). The malleable gut microbiome of juvenile rainbow trout (Oncorhynchus mykiss): diet-dependent shifts of bacterial community structures. PLoS One 12:e0177735. doi: 10.1371/journal.pone.0177735
Muthukrishnan, T., Govender, A., Dobretsov, S., and Abed, R. M. M. (2017). Evaluating the reliability of counting bacteria using epifluorescence microscopy. J. Mar. Sci. Eng. 5:4. doi: 10.3390/jmse5010004
Nakase, G., and Eguschi, M. (2007). Analysis of bacterial communities in Nannochloropsis sp. Cultures used for larval fish production. Fish. Sci. 73, 543–549. doi: 10.1111/j.1444-2906.2007.01366.x
Nakase, G., Nakagawa, Y., Miyashita, S., Nasu, T., Senoo, S., Matsubara, H., et al. (2007). Association between bacterial community structures and mortality of fish larvae in intensive rearing systems. Fish. Sci. 73, 784–791. doi: 10.1111/j.1444-2906.2007.01397.x
Nayak, S. K. (2010). Role of gastrointestinal microbiota in fish. Aquacult. Res. 41, 1553–1573. doi: 10.1111/j.1365-2109.2010.02546.x
Nicholas, J. L., Corre, S., and Cochard, J. C. (2004). Bacterial population association with phytoplankton cultured in a bivalve hatchery. Microb. Ecol. 48, 400–413. doi: 10.1007/s00248-003-2031-6
Porsby, C. H., Nielsen, K. F., and Gram, L. (2008). Phaeobacter and ruegeria species of the roseobacter clade colonize separate niches in a danish turbot (Scophthalmus maximus)-rearing farm and antagonize Vibrio anguillarum under different growth conditions. Appl. Environ. Microbiol. 74, 7356–7364. doi: 10.1128/AEM.01738-08
Ramanan, R., Kim, B. H., Cho, D. H., Oh, H. M., and Kim, H. S. (2016). Algae-bacteria interactions: evolution, ecology and emerging applications. Biotechnol. Adv. 34, 14–29. doi: 10.1016/j.biotechadv.2015.12.003
Reisch, C. R., Moran, M. A., and Whitman, W. B. (2011). Bacterial catabolism of dimethylsulfoniopropionate (DMSP). Front. Microbiol. 2:172. doi: 10.3389/fmicb.2011.00172
Ringø, E., Zhou, Z., Olsen, R. E., and Song, S. K. (2012). Use of chitin and krill in aquaculture – effect on gut microbiota and the immune system: a review. Aquacult. Nutr. 18, 117–131. doi: 10.1111/j.1365-2095.2011.00919.x
Roeselers, G., Mittge, E. K., Stephens, W. Z., Parichy, D. M., Cavanaugh, C. M., Guillemin, K., et al. (2011). Evidence for a core gut microbiota in the zebrafish. ISME J. 5, 1595–1608. doi: 10.1038/ismej.2011.38
Romero, J., and Navarrete, P. (2006). 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch). Microb. Ecol. 51, 422–430. doi: 10.1007/s00248-006-9037-9
Romero, J., Ringø, E., and Merrifield, D. L. (2014). “The gut microbiota of fish,” in Aquaculture Nutrition: Gut Health, Probiotics and Prebiotics, ed. D. Merrifield (Oxford: Wiley-Blackwell Publishing), 75–100.
Schmidt, V. T., Amaral-Zettler, L. A., Davidson, J., Summerfelt, S., and Good, C. (2016). Influence of fishmeal-free diets on microbial communities in Atlantic salmon (Salmo salar) recirculation aquaculture systems. Appl. Environ. Microbiol. 82, 4470–4481. doi: 10.1128/AEM.00902-16
Schmidt, V. T., Smith, K. F., Melvin, D. W., and Amaral-Zettler, L. A. (2015). Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol. Ecol. 24, 2537–2550. doi: 10.1111/mec.13177
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Semova, I., Carten, J. D., Stombaugh, J., Mackey, L. C., Knight, R., Farber, S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Sharifah, E. N., and Eguchi, M. (2011). The phytoplankton nannochloropsis oculata enhances the ability of roseobacter clade bacteria to inhibit the growth of fish pathogen Vibrio anguillarum. PLoS One 6:e26756. doi: 10.1371/journal.pone.0026756
Stuart, K., Rotman, F., and Drawbridge, M. (2015). Methods of microbial control in marine fish larval rearing: clay-based turbidity and passive larval transfer. Aquac. Res. 47, 2470–2480. doi: 10.1111/are.12696
Sullam, K. E., Essinger, S. D., Lozupone, C. A., O’Connor, M. P., Rosen, G. L., Knight, R. O. B., et al. (2012). Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol. Ecol. 21, 3363–3378. doi: 10.1111/j.1365-294.X.2012.05552
Sullam, K. E., Rubin, B. E., Dalton, C. M., Kilham, S. S., Flecker, A. S., and Russell, J. A. (2015). Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. ISME J. 9, 1508–1522. doi: 10.1038/ismej.2014.231
Utne-Palm, A. C. (2002). Visual feeding of fish in a turbid environment: physical and behavioral aspects. Mar. Freshw. Behav. Physiol. 35, 111–128. doi: 10.1080/10236240290025644
Vila-Costa, M., Rinta-Kanto, J., Poretsky, R. S., Sun, S., Kiene, R. P., and Moran, M. A. (2014). Microbial controls on DMSP degradation and DMS formation in the Sargasso Sea. Biogeochemistry 120, 295–305. doi: 10.1007/s10533-014-9996-8
Wu, S. G., Tian, J. Y., Gatesoupe, F. J., Li, W. X., Zou, H., Yang, B. J., et al. (2013). Intestinal microbiota of gibel carp (Carassius auratus gibelio) and its origin as revealed by 454 pyrosequencing. World J. Microbiol. Biotechnol. 29, 1585–1595. doi: 10.1007/s11274-013-1322-4
Ye, L., Amberg, J., Chapman, D., Gaikowski, M., and Liu, W. T. (2014). Fish gut microbiota analysis differentiates physiology and behavior of invasive Asian carp and indigenous American fish. ISME J. 8, 541–551. doi: 10.1038/ismej.2013.181
Yoch, D. C. (2002). Dimethylsulfoniopropionate: it’s sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68, 5304–5315. doi: 10.1128/AEM.68.12.5804-5815.2002
Zaneveld, J. R., McMinds, R., and Thurber, R. V. (2017). Stress and stability: applying the anna karenina principle to animal microbiomes. Nat. Microbiol. 2:17121. doi: 10.1038/nmicrobiol.2017.121
Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. (2000). A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214. doi: 10.1089/10665270050081478
Keywords: aquaculture, microbial community, water additive, greenwater, fish, sablefish, 16S rDNA
Citation: Pierce ML, Lee JSF, Dodd E and Poretsky RS (2019) Algae and Clay Water Additives Differentially Impact Survival and Microbial Community Structure in Sablefish (Anoplopoma fimbria) Rearing Tanks. Front. Mar. Sci. 6:203. doi: 10.3389/fmars.2019.00203
Received: 29 October 2018; Accepted: 01 April 2019;
Published: 26 April 2019.
Edited by:
Suhelen Egan, University of New South Wales, AustraliaReviewed by:
Melissa Garren, California State University, Monterey Bay, United StatesAlexander Chong Shu-Chien, University of Science, Malaysia, Malaysia
Copyright © 2019 Pierce, Lee, Dodd and Poretsky. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melissa L. Pierce, bWxwMTZAdWljLmVkdQ==
 Melissa L. Pierce
Melissa L. Pierce Jonathan S. F. Lee2
Jonathan S. F. Lee2 Emily Dodd
Emily Dodd Rachel S. Poretsky
Rachel S. Poretsky