- 1Department of Applied Biological Chemistry, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
- 2Mie Prefecture Fisheries Research Institute, Shima, Japan
- 3Marine Genomics Unit, Okinawa Institute of Science and Technology Graduate University, Onna, Japan
The mollusk class, Bivalvia, plays an important role in the formation of calcium carbonate in the ocean. The bivalve hinge ligament is a hard but pliant tissue that resists the stress placed on the hinge during opening and closing. The ligament comprises a fine microstructure of fibrous aragonite crystals surrounded by a dense organic matrix. The matrix consists of organic fibers that are aligned with the fibrous aragonite crystals. Previous studies identified a tissue inhibitor of metalloproteinase (Pf-TIMP: Pinctada fucata-Tissue Inhibitor of Metalloproteinase) in the organic fibers of P. fucata ligaments. This enzyme exhibited strong inhibition of matrix metalloproteinase (MMP) activity in vitro, suggesting that MMPs also play a role in formation of the organic fibers of the ligament. Using transcriptome data, we identified MMP genes from the mantle isthmus, which is a soft tissue attached to the ligament. To investigate the function of MMP genes in vivo, we performed RNA interference experiments. The expression of MMP14973 and MMP07860 genes was inhibited after injection of each dsRNA. Cumulative injection of MMP07860 dsRNA induced aggregated aragonite fiber orientation, whereas the injection of MMP14973 showed minor effects. When the decalcified ligament was incubated in a solution saturated with calcium carbonate, aragonite fibers aligned along the surface. When the decalcified ligament was treated with recombinant human (hr) MMP-13, the precipitation of calcium carbonate was inhibited. To investigate general MMP functions in calcium carbonate crystallization in detail, we precipitated aragonite crystals in collagen gels treated with or without recombinant human (hr) MMP-1. Treatment with hrMMP-1 increased the interaction between collagen gels and calcium carbonate. These results imply that Pinctada fucata (Pf)-MMPs degrade extracellular matrices in the ligament to produce the fine organic fibers that regulate the orientation of fibrous aragonite crystals.
Introduction
The precipitation of calcium carbonate in the ocean is a source of significant carbon translocation. Mollusk shells consist of calcium carbonate and small amounts of organic matrices. The crystallization of calcium carbonate is regulated by these organic matrices to produce the fine microstructures of the shell. Bivalve shells are made of two valves that protect the two sides of the animal. These two shells are connected by the hinge ligament, which functions as a joint allowing the shells to open and close. The hinge exists in the bottom of the bivalve shells and consists of a fibrous microstructure to resist the mechanical pressure. A previous study identified a positive correlation between nacre deposition capability and shell closing strength in the Japanese pearl oyster Pinctada fucata (Aoki et al., 2012). It has also been reported that the photonic structure of the ligament was determined by the humidity, because the water absorption changed the structural color of the ligament (Zhang and Zhang, 2015). This report suggested that the ligament is an interesting biomaterial for development of biomimetic material. Therefore, an investigation of the mechanism of ligament formation would be of interest for understanding bivalve biomineralization processes, and may provide useful insight into the efficient development of high-quality pearl production methods or functional materials.
The components of the ligament are secreted by the mantle isthmus, which is part of the soft mantle tissue attached to the shell hinge. Small, fibrous aragonite crystals, and organic frameworks are secreted from mantle isthmus cells and then align. The small crystals connect to form long aragonite crystals within the organic fibrous matrices (Bevelander and Nakahara, 1969). Short aragonite rods extend vertically toward the growing surface of the ligament. Transmission electron microscopy (TEM) observations revealed a pseudohexagonal aragonite crystal in the cross-section of a ligament (Kahler et al., 1976; Marsh and Sass, 1980) that reflects the euhedral shape of the aragonite crystal. The diameters of the aragonite crystals are ca. 50–100 nm, and the c axis is parallel to the morphological long axis. In contrast, the a and b axes are oriented randomly between crystals. The contrast due to the {110} twinning defects were observed in the center of the aragonite crystals (Kahler et al., 1976; Marsh and Sass, 1980).
The crystals and the fibers of the matrix are aligned in in the growth direction and the ligament in this direction is strengthened by the embedded crystals (Ono et al., 1990). Millon and xanthoprotein staining methods were used to demonstrate that the Anodonta ligament contains proteins tanned by quinone (Trueman, 1950). Amino acid composition analysis showed that the unmineralized outer layer of the ligament contains a high concentration of glycine, whereas the crystalized inner layer of the ligament is dominated by methionine and proline (Hare, 1963). A comparison of the amino acid compositions of the crystalized inner layer of the ligament and those of other calcified microstructures in bivalve shells showed that the ratio of acidic residues in the ligament was lower than those in other microstructures of the shell. One previous study suggested that the ligament of Pecten consists of a rubber-like protein, abductin (Kelly and Rice, 1967). Although the physical characteristics of abductin are similar to those of elastin and resilin, the protein sequence of abductin has not yet been determined. Kikuchi et al. (1987) discovered that the Pseudocardiurn sachalinensis ligament contains components that form a connection between the proteins desmosine and isodesmosine, which are related to elastin formation. Another study reported that aragonite crystals in the ligament of P. fucata contain a ligament intra-crystalline peptide (Suzuki et al., 2015), which inhibited the growth of the first deposited crystals along the c axis, keeping the aragonite crystals small. These small aragonite crystals were aligned in the same direction and stacked to form long aragonite fibers within the organic framework. However, the mechanism by which these small crystals are assembled in the organic matrix of the ligament is still unknown.
Recently, a tissue inhibitor of metalloproteinase (Pf-TIMP) was identified from organic fibers in the P. fucata ligament (Kubota et al., 2017). As the previous research revealed that TIMP inhibited the activity of matrix metalloproteinases (MMPs) in vitro, the authors investigated if the purified Pf-TIMP from the ligament has the true function of inhibitory activity using human MMPs. However, the functions of MMPs and TIMP in P. fucata are unknown, because no other publications about TIMP in the hinge ligament of bivalves have not been reported yet. The results of knockdown experiments using a double-stranded RNA (dsRNA) targeting MMP54089, which was identified from the P. fucata genome database, showed that the disordered aragonite fibers in the ligament were observed indicating that MMP54089 protein plays an important role in the formation of organic fibers between aragonite crystals.
The periodontal ligament in the tooth of mammals has similar structure of the hinge ligament of bivalves. Similar function of human MMPs has been reported in human bones and teeth (Sasano et al., 2012). MMPs degrade extracellular matrices which inhibit the growth of hydroxyapatite to make some space and hydroxyapatite crystals can then be formed in this space. Our first discovery of Pf-MMPs and Pf-TIMP functions in the hinge ligament means the convergent evolution of two similar structures in mammals and mollusks. Then, we assumed that the combination of human MMPs and collagen is a useful model system to study molluscan biomineralization in the hinge ligament. Because many other Pinctada fucata (Pf)-MMP genes are encoded by the P. fucata genome, the functions of Pf-MMPs and organic fibers in the ligament needs detailed clarification.
The objectives of this study are described below.
(1) Identification of MMP genes from transcriptome data.
(2) Functional analyses of MMP genes for the formation of the ligament using RNAi technique.
(3) Functional analyses of MMP proteins using in vitro calcium carbonate crystallization.
We analyzed transcriptome data from the P. fucata mantle isthmus and identified the expressed Pf-MMP genes. Knockdown and in vitro calcium carbonate crystallization experiments demonstrated the function of organic fibers in ligament formation. To gain further insight into possible ways that the matrix can influence aragonite formation, we also carried out experiments on Pf-MMP treated collagen matrices in which aragonite was induced to form. Knockdown and in vitro experiments revealed the formation process of organic fibers by Pf-MMP functions. Lastly, calcium carbonate crystallization experiment showed that human MMP treatments increased the interaction between aragonite crystals and organic fibers suggesting that Pf-MMP have some functions for calcification in the ligament.
Materials and Methods
Collection of Shells and Living Tissues
The Japanese pearl oyster P. fucata was cultured in Ago Bay (Mie Prefecture, Japan) by the Fisheries Research Division, Mie Prefectural Science and Technology Promotion Center, Japan (latitude, 34.3 degree and longitude, 136.8 degree), and sent to The University of Tokyo, where P. fucata was cultured for 1–2 weeks before collecting the shells by removing the soft bodies. Live P. fucata were cultured in 10 L aquarium filled with natural seawater (natural seawater from outside of Tokyo bay was kept in 10 ton tank) at 20°C. We changed the natural seawater every 2 days. Other shells were washed, dried, and separated by MIKIMOTO COSMETICS (Mie, Japan) and sent to the University of Tokyo. Living mantle isthmus tissue was collected and stored in a freezer at -80°C.
Transcriptome Analysis by Next-Generation Sequencing
Total RNA was extracted from the mantle isthmus of three individuals at the age of 2 years using Sepasol-RNA I Super G (Nacalai), following the manufacturer’s instructions. We chose the P. fucata which kept tightly closed of bivalve shells. Transcriptome libraries were prepared using the TruSeq RNA Library Preparation Kit v2 (Illumina). RNA was sequenced using an Illumina Miseq (Illumina), following the manufacturer’s instructions. Only high-quality sequences (quality value ≥ 20) were assembled using Trinity ver. 2.2.11 (Grabherr et al., 2011). Read preprocessing, assembly, and mapping were performed in the read annotation pipeline of the DNA Databank of Japan (DDBJ) using the default parameters2.
Identification of Pf-MMP Genes From Transcriptome Data
To identify the Pf-MMP genes, we performed a homology search in transcriptome data from the mantle isthmus using the basic local alignment search tool (BLAST). Identified Pf-MMP sequences were combined with those of Pf-MMP genes from the P. fucata genome database (Takeuchi et al., 2012) to determine the entire sequences. The sequences of Pf-MMP genes were registered to the DDBJ and assigned accession numbers (MMP21914: LC388937, MMP07860: LC388938, MMP60936: LC388939, MMP32404: LC388940, MMP23659: LC388941, MMP54089: LC388942, MMP14973: LC388943).
RNA Interference (RNAi)
To reveal the function of other Pf-MMPs other than MMP54089 (Kubota et al., 2017), we chose MMP07860, which showed a similar expression pattern to MMP54089, and MMP14973, which showed lower expression level than MMP54089. A dsRNA targeting MMP07860 or MMP14973 was generated to observe the growing end of the ligament in the absence of MMP07860 or MMP14973 using a scanning electron microscope (SEM). To prepare the template to construct dsRNAs targeting MMP07860 and MMP149739, we conducted polymerase chain reaction (PCR) using cDNA prepared from oyster mantle isthmus RNA. PCR was performed using the following cycling conditions: 30 cycles of 30 s at 94°C (3 min 30 s for the first cycle), 30 s at 55°C, and 30 s at 72°C. The PCR product was diluted with distilled water to 100× the volume and used as a template for the following nested PCR. The primers used in the nested PCR contained a T7 polymerase recognition sequence, enabling T7 polymerase to recognize the sequence and start transcription. For MMP14973, the 5′ primer sequence was GGGTAATACGACTCACTATAGGGTCAAAGAATATCAAAAGAGG and that of the 3′ primer GGGTAATACGACTCACTATAGGGTCACCATTTCCAGATGTCAT (Under bars mean the sequence of T7 promoter). For MMP07860, the 5′ primer sequence was GGGTAATACGACTCACTATAGGGAGTACTGGTCTGACGTCACT and that of the 3′ primer GGGTAATACGACTCACTATAGGGAGTTGGTGACACTGGGGTTA. The nested PCR was performed under the same conditions, and the resulting product was used as a template to produce dsRNA. The in vitro Transcription T7 Kit for siRNA synthesis (TaKaRa) was used to produce dsRNA according to the manufacturer’s instructions. A dsRNA targeting enhanced green fluorescence protein (EGFP) was also prepared as a control. We dissolved 30 μg each of dsRNA in 50 μL phosphate-buffered saline and injected the dsRNA into the adductor muscle of P. fucata. We used more than three individuals of young oysters (shell size: 7 cm) in this experiment, because it was necessary to observe the growth process of the ligament, which can be difficult to observe in mature oysters. We extracted total RNA from the mantle isthmus of each sample 4 days post-injection using the method described above; reverse transcription was performed using the PrimeScript Perfect Real Time Reagent Kit (TaKaRa) according to the manufacturer’s instructions.
SEM Observations of the Ligament
Fresh ligaments were collected from living pearl oysters following dsRNA injection. The ligament was washed with distilled water and dried on an aluminum stage. The growing end of the ligament in each sample (more than three individuals in each condition) was observed using an SEM (Hitachi S-4800, Tokyo, Japan). The surface of the ligament was observed via secondary electron images at an acceleration voltage of 2 kV. The samples were coated with Pt–Pd for high-resolution secondary electron image recording.
Real-Time PCR
Real-time quantitative PCR (qPCR) was performed to quantity gene expression levels of MMP14973 and MMP07860. qPCR primers were designed using the Primer3Plus software. For MMP14973, the 5′ primer sequence was AGGTCGGTGCAATGATAGCA and that of the 3′ primer CGTTTTCGTCTTTCCTCTTCTGT. For MMP07860, the 5′ primer sequence was CGACACACATTTCGATGACG and that of the 3′ primer TTTGGGCTCACTTGAGTGTC. Actin was used as an internal control gene for qPCR (5′ primer: CAGAAGGAAATCACCGCACT; 3′ primer: AGAGGGAAGCAAGGATGGAG). We performed qPCR using the Applied Biosystems 7300 Fast Real-Time PCR System with SYBR Premix Ex Taq II (Takara). To compare gene expression levels, we used the ΔΔCT method. The qPCR reaction mixture (10 μl) was adjusted according to the manufacturer’s instructions. The cycling parameters were: 1 cycle of 0.5 min at 95°C, 40 cycles of 5 s at 95°C, and 34 s at 60°C. Dissociation curves were analyzed at the end of each run to determine the purity of the product and the amplification specificity.
Calcium Carbonate Crystallization in the Decalcified Ligament
The ligaments were decalcified individually in 15 ml tubes with 5 ml of 1 M acetic acid solution for 24 h. We collected the decalcified ligament after centrifugation to remove the acetic acid. The decalcified ligament was washed with distilled water using centrifugation. The ligaments were collected from more than three individuals for each condition. The experimental treatments were described below.
(1) 20 μl of human recombinant MMP-13 (hrMMP-13) (340 mU; MMP inhibitor profiling kit, fluorometric red; ENZO) was mixed with the ligament from one individual in the 1.5 ml tube.
(2) The solution of (1) was mixed with 20 μl of assay buffer from MMP inhibitor profiling kit and 60 μl of 10 mM Tris-HCl (pH 8.0) for 3 h at 37°C.
(3) The ligament treated with or without human MMP13 in 20 mM CaCl2/5 mM MgCl2/25 mM Tris-HCl (pH 7.4) solution was then placed in a desiccator with 5 g ammonium carbonate powder in 50 ml beaker.
(4) After 24 h, the recalcified ligament was placed on the sample stage for SEM observation (Hitachi, S-4800) and coated with Pt–Pd at 20 mA for 60 s (Hitachi, E-1030).
In vitro Calcium Carbonate Crystallization in a Collagen Gel
To synthesize collagen gels, we followed the method of Nudelman et al. (2010) with modifications. Collagen type I from bovine skin (Nippi, Inc., Tokyo, Japan) was dissolved in 1 M acetic acid at 4°C. The collagen solution was then dialyzed in 67 mM phosphate buffer (pH 6.0) using a dialysis membrane for 3 h at 4°C; the buffer was replaced three times (every 3 h) at 4°C. The solution was then placed in a dish to produce gels. These collagen gels were treated with 734.4 U of the hrMMP-1 described above for 14 h at 37°C. The collagen gels treated with or without human MMP13 in the 20 mM CaCl2/5 mM MgCl2/25 mM Tris-HCl (pH 7.4) solution were then placed in the desiccator. The calcification experiment was performed as described above. After calcification, the collagen gel was degraded with 18% sodium hypochlorite solution to purify the calcium carbonate precipitates, which were then observed by SEM. The Hitachi FB-2100 focused ion beam (FIB) was used to cut electron-transparent cross-sections from synthetic precipitate aggregates. The cross-sections were transferred to a JEOL JEM-2010 TEM operating at 200 kV for imaging and electron diffraction.
The collagen gels and calcium carbonate crystals were mounted on an aluminum platelet with a cavity of 200 μm depth and filled with 1-hexadecene (Sigma -Aldrich, St. Louis, MO, United States) for cryo-SEM. The samples were then cryo-immobilized in the HPM10 high-pressure freezing device (Bal-Tec AG, Balzers, Liechtenstein). The frozen samples were mounted on a holder and transferred to the BAF 60 FF device (Bal-Tec) using the VCT 100 vacuum cryo-transfer device (Bal-Tec). Frozen samples were fractured at 120°C and etched for 5 min at 105°C. Samples were then transferred to the Ultra 55 SEM using the VCT 100 cryo-transfer device and observed using secondary electron and backscattered electron in-lens detectors at 120°C. The backscattered electron in-lens detector was operated at 2 kV using an energy selective backscattered detector (ESB) grid voltage of 300 V. Stronger signals in the backscattered images were obtained from elements with higher atomic masses, i.e., calcium atoms within the mineral phase, which provided information on the distribution of the mineral relative to the organic material.
Results and Discussion
Identification of Pf-MMPs in P. fucata
In a previous study (Kubota et al., 2017), we identified seven Pf-MMP genes in the P. fucata genome database (Takeuchi et al., 2012). However, the assembled genome sequence in the genome database was insufficient to reveal the entire sequence of each Pf-MMP gene. To determine the whole sequence of Pf-MMP genes expressed in the mantle isthmus, we performed a homology search of the amino acid sequence of the MMP activity domain using transcriptome data from mantle isthmus tissue. We identified seven Pf-MMP genes (MMP21914, 07860, 60936, 32404, 23659, 54089, and 14973) from the transcriptome data (Figure 1 and Supplementary Figure S1). All Pf-MMPs in the transcriptome database except for MMP32404 contained a signal peptide, an MMP superfamily domain, a putative peptidoglycan-binding domain, and hemopexin-like repeats. As signal peptide sequences in Pf-MMP genes were not identified in our previous study, we extended the sequences used for analysis using the transcriptome data. Signal peptides and hemopexin-like repeats were not found in MMP32404, suggesting that MMP32404 expression was too low to obtain many fragments from the mantle isthmus. We also checked that the whole sequence of Pf-TIMP was included in the transcriptome data.
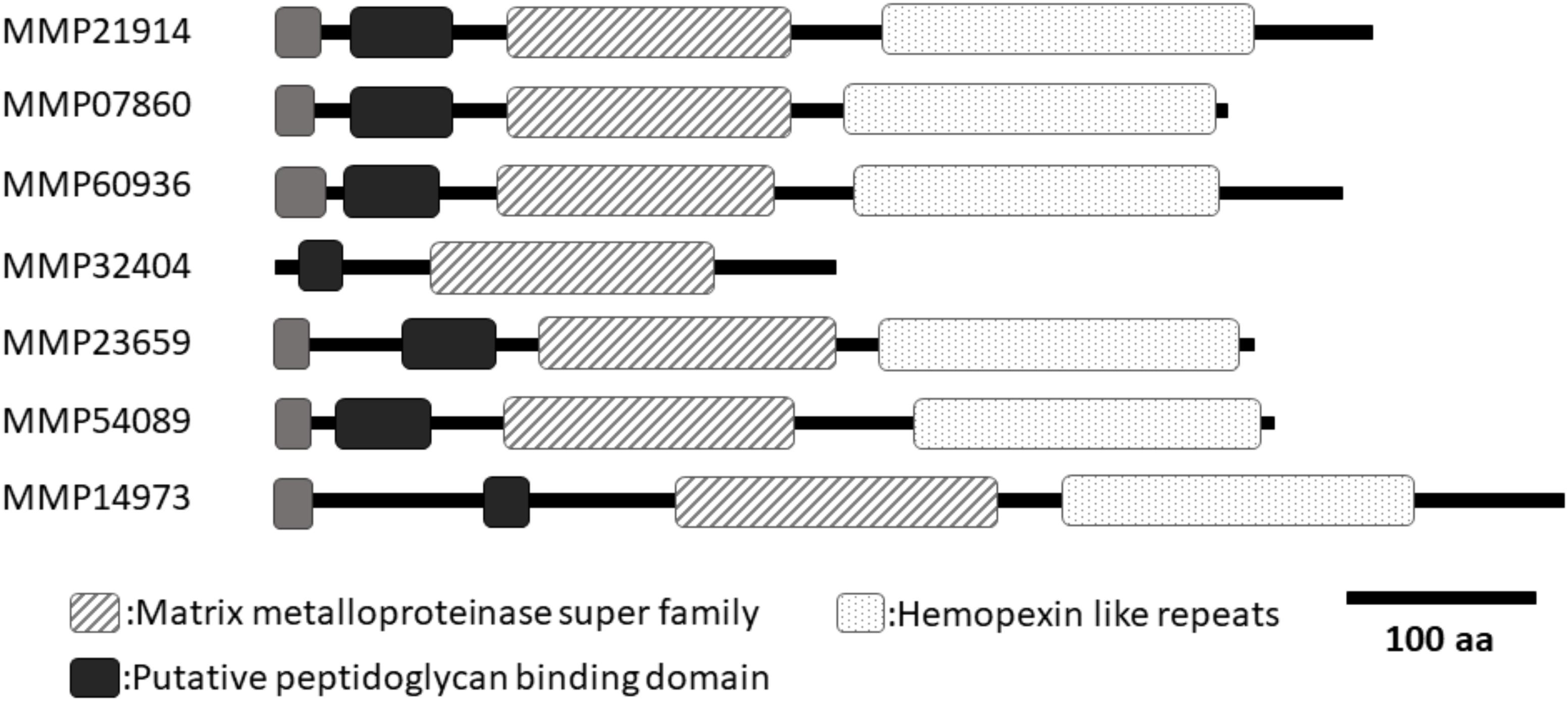
FIGURE 1. A schematic of the domain structures of seven MMPs from Pinctada fucata. All MMPs, except for MMP32404, had similar lengths and domain structures. Only MMP32404 did not contain hemopexin like repeats in the C-terminal region. Black boxes indicate the putative peptidoglycan-binding domain. Dotted boxes indicate hemopexin-like repeats. Hashed boxes indicate the MMP superfamily domain. Gray boxes indicate the signal peptide. DDBJ accession numbers (MMP21914: LC388937, MMP07860: LC388938, MMP60936: LC388939, MMP32404: LC388940, MMP23659: LC388941, MMP54089: LC388942, MMP14973: LC388943).
Peptidoglycan-binding domain and hemopexin-like repeats have also been identified in human MMPs (Sekhon, 2010). Previous studies have shown that these regions are related to the interaction between human MMPs and their substrates (Gillian and Vera, 1997). A cysteine switch (PRCGVP) occurs after the peptidoglycan-binding domain binds to zinc in the human MMP activity domain, thereby inhibiting the activity (Springman et al., 1990; Van Wart and Hansen, 1990). Processing of the propeptide in human MMPs is necessary to activate human MMPs. All pf-MMPs in P. fucata exhibit conservation of this cysteine switch, suggesting that Pf-MMP activity in P. fucata may require this processing step.
RNAi of Pf-MMPs
In the previous study, a knockdown experiment using a dsRNA targeting MMP54089 affected the growth direction of aragonite fibers in the ligament. To investigate the functions of MMP07860 and MMP14973 in the ligament, we performed RNAi experiments in the current study. The growing edge of the ligament after RNAi experiments was observed by SEM 4 days after injection (Figure 2). In the control treatment, the tips of aragonite crystals were observed as white dots along the surface of the growing edge. Similar microstructures were observed after injection of EGFP-specific dsRNA that is a negative control of RNAi experiment. Some aggregation of the aragonite fibers was observed after MMP07860-specific dsRNA injection. The knockdown of MMP14973 increases the diameter of each aragonite tips in the ligament. However, the effect of MMP14973 is small, because the expression level of MMP14973 is too low in the ligament to cause a marked impact by knockdown. MMP07860 expression after injection was measured using qPCR (Figure 3). Expression was lower 12 h after injection, suggesting that MMP07860 production was inhibited by MMP07860-specific dsRNA injection. However, it was difficult to detect MMP14973 PCR products using qPCR (data not shown), preventing us from assessing the efficacy of the RNAi procedure.
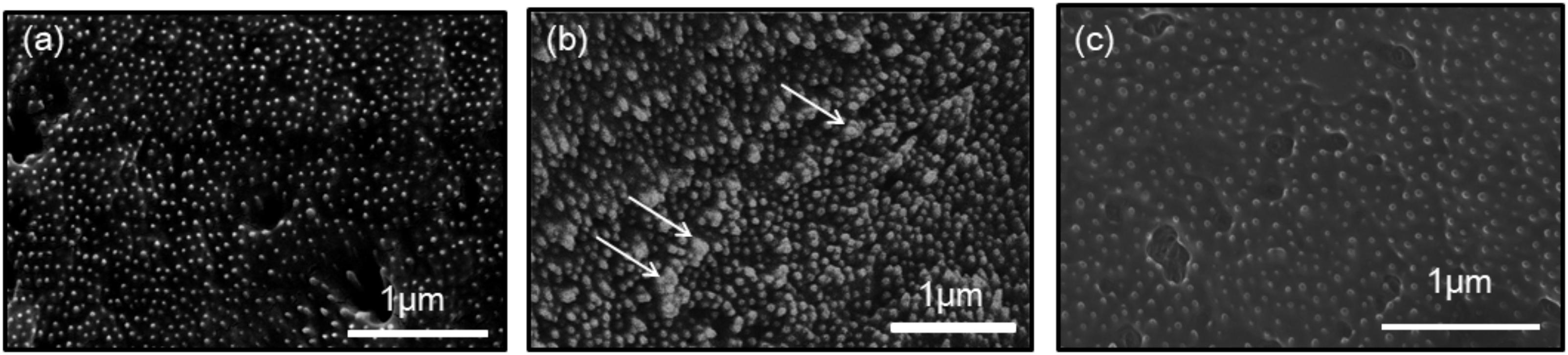
FIGURE 2. Scanning electron microscopy (SEM) observations of the surface of the growing edge of the ligament in the samples injected with dsRNA targeting (a) enhanced green fluorescence protein (EGFP), (b) MMP07860, and (c) MMP14973. The injection of EGFP and MMP14073 showed normal surface, while the injection of MMP07860 showed small aggregation of aragonite crystals. White arrows indicate the aggregation of aragonite crystals.
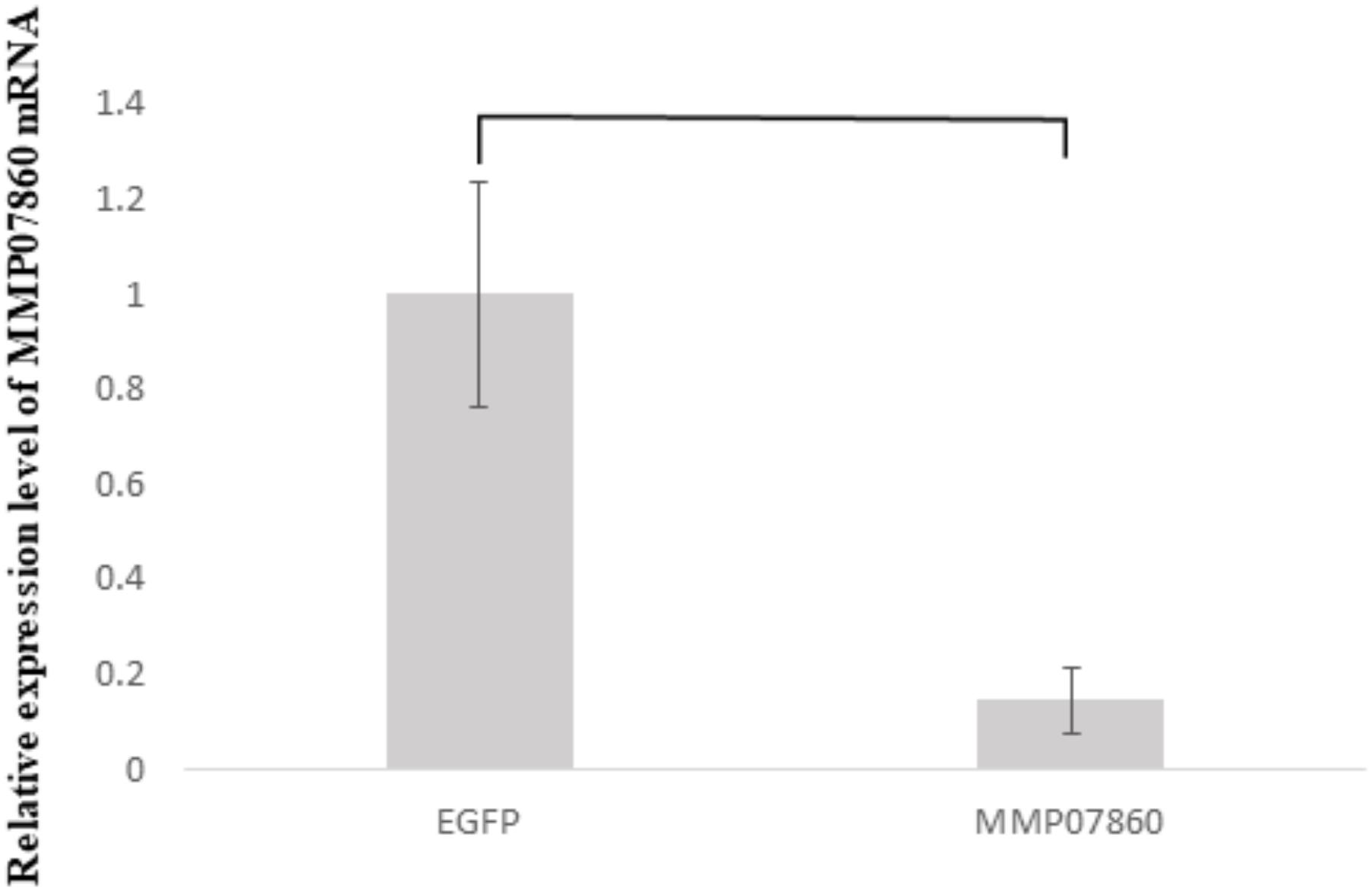
FIGURE 3. Expression levels of MMP07860 mRNA in the mantle isthmus, determined using real-time quantitative polymerase chain reaction (qPCR) 12 h after injection. The injection of dsRNA (MMP07860) depressed the expression level of MMP07860. The MMP07860 mRNA expression in the EGFP group (EGFP–dsRNA injected) was given a relative value of 1.0. Five oysters (n = 5) were used. Statistically significant differences were determined by t-test (p < 0.05).
The RNAi knockdown experiment conducted in our previous study showed that MMP54089 regulates the organic framework to align aragonite fiber orientation in the ligament (Kubota et al., 2017). MMP07860 knockdown led to minor aggregation of aragonite fibers, whereas knockdown of MMP14973 had small effect, because the expression level of MMP14973 was very low. Although the expression level and pattern of MMP07860 were similar to those of MMP54089, the results of RNAi experiments were different. The knockdown of MMP54089 disordered the orientation of aragonite fibers and disrupted the organic matrices (Kubota et al., 2017), while the knockdown of MMP07860 resulted in small aggregations of aragonite fibers. These results suggest that MMP07860 has few effects for the ligament formation. The knockdown effects of these Pf-MMP genes were dramatically different, suggesting that each Pf-MMP has a specific function in the formation of the ligament.
Previous work has reported on the functions of human MMPs related to biomineralization. In the process of tooth formation, expression of human MMP 20 was detected. Human MMP20 degrades amelogenin to induce calcification of hydroxyapatite in tooth enamel (Uskokovic et al., 2011). Pf-MMP proteins in P. fucata probably have similar functions during ligament formation.
Organic Fibers in the Decalcified Ligament Treated With Recombinant MMP-13
To investigate Pf-MMP function in the ligament, we investigated if organic fibers in the ligament were degraded by human MMP activity, or not. The decalcified ligament was washed with distilled water and observed by SEM (Figure 4). The dissolution of aragonite fibers created holes in its surface (Figure 4a). Some aragonite fibers remained after decalcification. The decalcified ligament was treated with hrMMP-13, because hrMMP-13 activity was most inhibited by Pf-TIMP. Treatment with hrMMP-13 resulted in alteration of the surface structure; the holes disappeared, and the hexagonal shape of the organic framework appeared (Figure 4b).
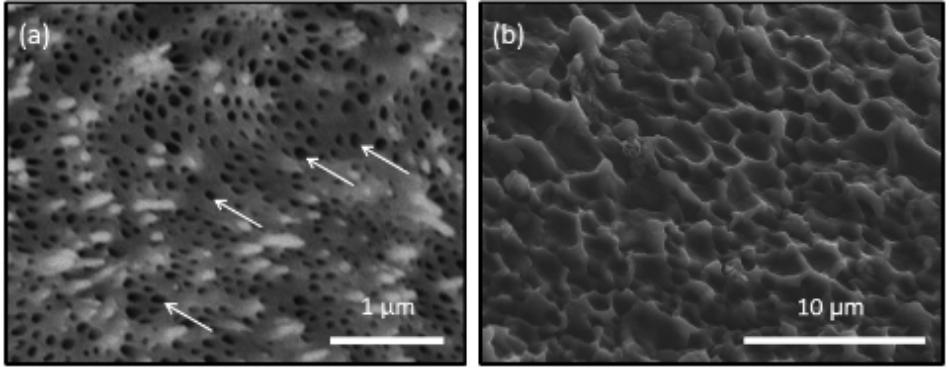
FIGURE 4. Scanning electron microscopy observations of the surface of the growing edge of the ligament (a) after decalcification using 1 M CH3COOH and (b) after treatment with hrMMP-13. The decalcified treatment showed many holes on the surface of the ligament. Treatment of hrMMP-13 degraded the microstructure of the ligament. White arrows indicate the holes.
Many small holes (diameter ca. 50–100 nm) were observed following decalcification treatment, which were the sites of the aragonite fibers. These holes disappeared in the decalcified ligament after hrMMP-13 treatment, and a large, hexagonal organic framework (diameter ca. 1–3 μm) appeared, suggesting that large (micrometer order) organic frameworks formed following the formation of a small (nanometer order) space for aragonite fibers. Calcium carbonate crystallization on the decalcified ligament following hrMMP-13 treatment indicated that hrMMP-13 indirectly prevented calcium carbonate formation by destroying the surface which promoted mineral formation; thus, excessive hrMMP-13 degradation depressed calcification in the framework. During the biomineralization process, fibrous organic matrices such as chitin play an important role as a framework for crystals (Kintsu et al., 2017).). As human MMPs are essential for the ordered assembly of collagen, it is conceivable that the hinge ligament protein contains a collagen-like protein. Collagen in various mineralized tissues forms a fibrous framework.
Calcium Carbonate Crystallization on the Decalcified Ligament
The existence of TIMP in the ligament (Kubota et al., 2017) suggested that excess MMP activity may inhibit the formation of the ligament. The effect for calcification of the ligament after hrMMP-13 treatment was investigated. The decalcified ligament was incubated in a calcium carbonate-supersaturated solution containing 50 mM MgCl2 to induce the formation of aragonite crystals on the surface. Many fibrous aragonite crystals were observed on the surface (Figures 5a,b). Each fiber was aligned, and large grains were formed. In contrast, no crystals formed on the surface treated with hrMMP-13, indicating that degraded organic frameworks cannot induce the formation of aragonite crystals (Figures 5c,d). These results suggested that the excess treatment of hrMMP-13 changed a good balance of MMP and TIMP functions in the ligament to inhibit the calcification.

FIGURE 5. Scanning electron microscopy observations of the surface of the growing edge of the ligament after crystallization. (a,c) hrMMP-13 treatment and (b,d) no MMP treatment (decalcification only). (a,b) Low magnification; (c,d) high magnification. The treatment of hrMMP-13 inhibited the calcification of the decalcified ligament. White arrows indicate the aragonite crystals.
Calcium Carbonate Crystallization in the Collagen Gels Treated With hrMMP-1
To observe the organic fibers after MMP treatment, we used collagen fiber and hrMMP-1 as in vitro model system. The components of the organic framework in the ligament are similar to those of extracellular matrices such as collagen proteins. Aragonite crystals were precipitated in the collagen fibers to mimic ligament formation. We dissolved type 1 bovine collagen in acid and transferred to phosphate buffer at a neutral pH to induce collagen gel formation. Then, the collagen gel was treated with hrMMP-1. TEM observations showed that the untreated collagen gels retained the same thickness and elongated separately (Figure 6a). Collagen gels treated with hrMMP-1 became short, thin fibers (Figure 6b); the tips of some of these fibers aggregated to form a spherical shape.
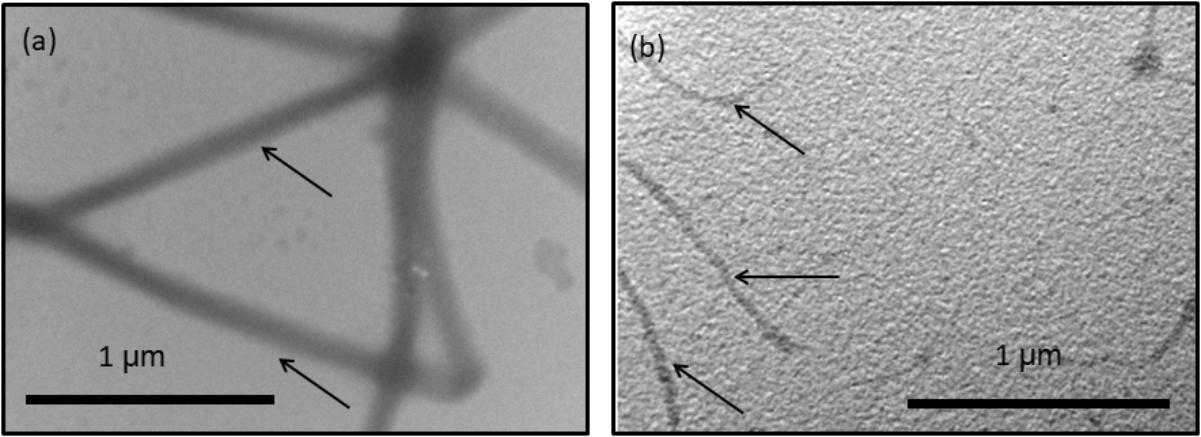
FIGURE 6. Transmission electron microscopy (TEM) observation of collagen fibers treated with (a) no hrMMP-1 and (b) hrMMP-1. The treatment of hrMMP-1 degraded the collagen fibers and made the fibers thin. Black arrows indicate the collagen fibers.
The collagen gels with or without treatment of hrMMP-1 with aragonite crystals were observed by SEM. A ball shape covered by needles was observed under both conditions. The needle shape is typical of aragonite crystals. Each needle was clearly observed in the collagen gels that were not treated with hrMMP-1 (Figures 7a–d), whereas the surface of the ball in the collagen gels treated with hrMMP-1 was covered with organic substances (Figures 7e–h), which were attached to the needles to form a complex shape, suggesting that degraded collagen fibers enhanced the interaction with the calcium carbonate surface.
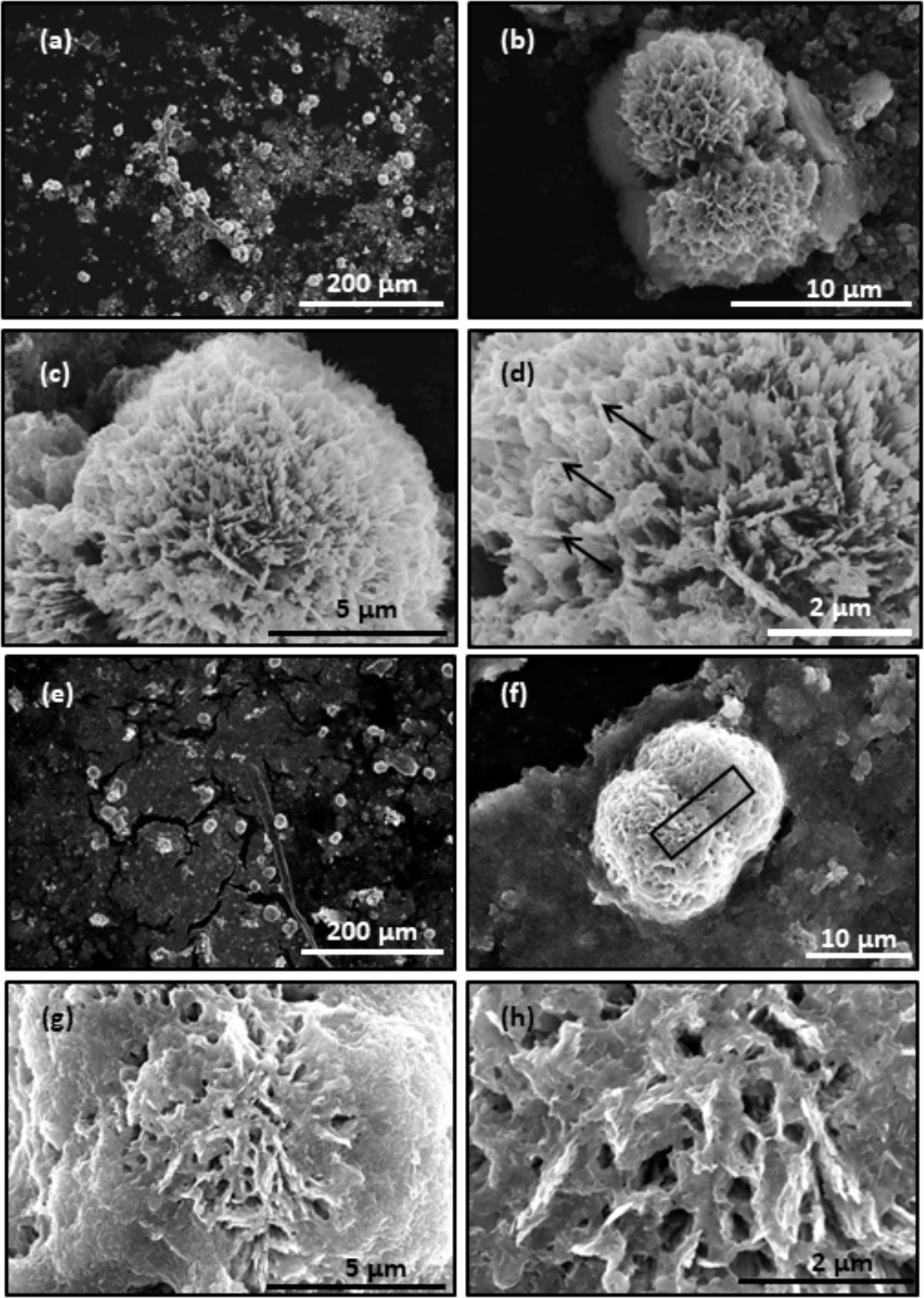
FIGURE 7. Scanning electron microscopy observation of CaCO3 crystals in the collagen gel. (a–d) No hrMMP-1 treatment; same region, different magnification. (e–h) hrMMP-1 treatment; same region, different magnification. The treatment of hrMMP-1 increased the interaction between collagen fibers and aragonite crystals. Black arrows show the needles covering the ball crystal. Black rectangle in (f) indicates the region where the cross-section was made for FIB-TEM analysis.
To observe the interface between crystal and collagen fibers treated with hrMMP-1, a crystal cross-section was prepared by FIB and freeze fracturing. TEM observations showed that the organic layer covered the spiky aragonite surface (Figure 8). Cryo-SEM observations also showed that the collagen fibers interacted with the aragonite surface (Figure 9). Back-scattered images clearly revealed the boundary between calcium carbonate and the collagen fibers (Figures 9b,d).
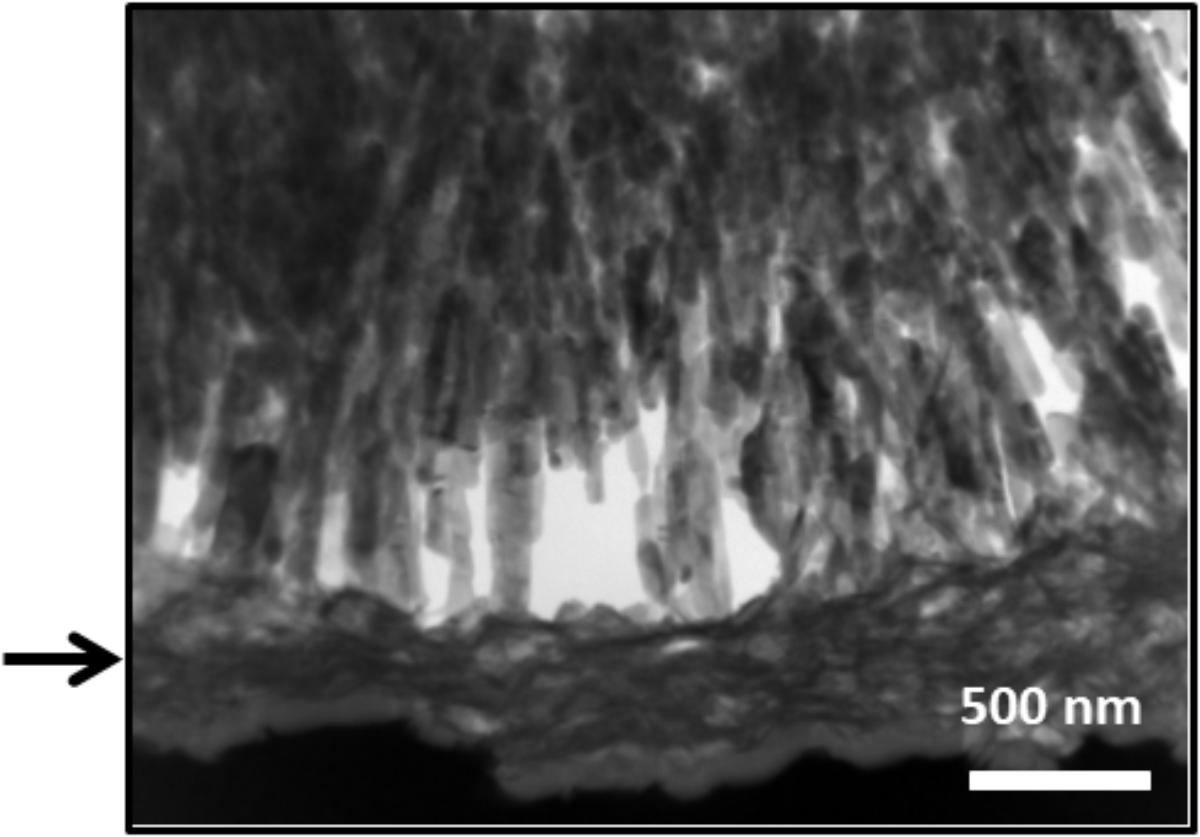
FIGURE 8. Transmission electron microscopy images of the FIB cross-section. The collagen fibers covered the aragonite needles. Crystal surface with organic fibers covering the calcium carbonate crystal. Arrow indicated the localization of collagen fibers.
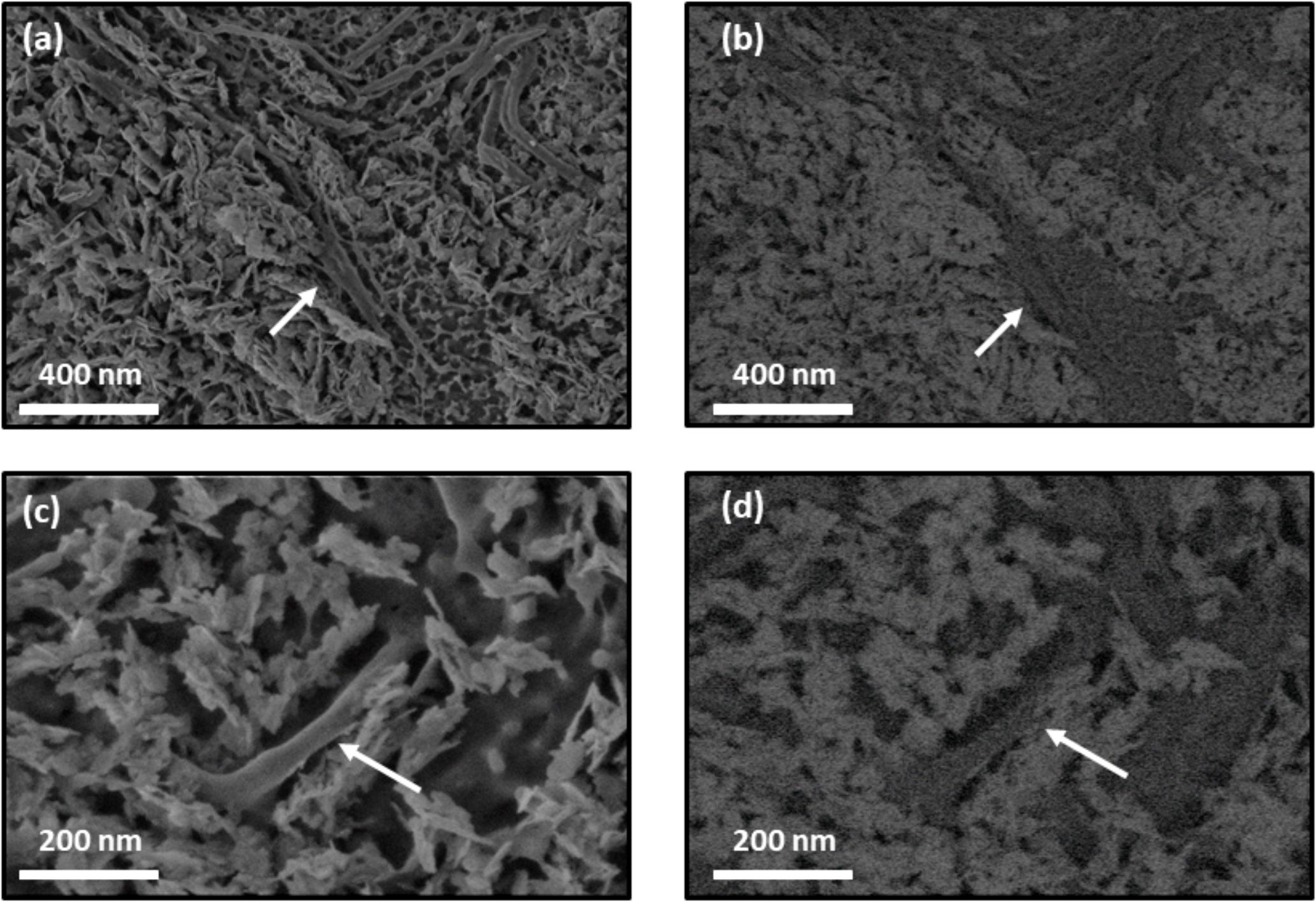
FIGURE 9. Cryo-SEM images of collagen gel with calcium carbonate crystals. Collagen fibers treated with hrMMP-1 tightly interacted with aragonite crystals. (a,c) Secondary electron images of calcium carbonate with collagen fibers. (b,d) Back-scattered images of (a,c), respectively. White arrows indicate collagen fibers.
Our results showed that the degradation process of organic fibers increased the interaction of calcium carbonate and collagen fibers. The contents of organic compounds will be regulated by this technique. The organic content affects the toughness and stiffness of the organic-inorganic hybrid materials.
Conclusion
We identified seven Pf-MMP genes from the P. fucata genome database in a previous study (Kubota et al., 2017). Because MMP is a proteinase for extracellular matrix proteins, MMP is secreted from the cell and must contain an N-terminal signal peptide to do so. However, the length of the nucleotide sequence was insufficient to reveal the entire gene sequence of each Pf-MMP, and the N-terminal signal peptide was not detected in almost all Pf-MMP genes. In the P. fucata genome database, the intron–exon prediction may have misidentified the Pf-MMPs open reading frame region, or whole Pf-MMP gene sequences may have been separated into different scaffolds. The mantle isthmus transcriptome data in this study was used to extend the nucleotide sequences of the Pf-MMP genes successfully. All Pf-MMP proteins, except for MMP32404, had similar lengths and domain structures. Only MMP32404 did not contain hemopexin like repeats in the C-terminal region.
Pf-TIMP plays an important role in regulating human MMP activity (Kubota et al., 2017). Calcium carbonate crystallization on the decalcified ligament that was not treated with hrMMP-13 induced the orientation of bundled aragonite crystals. Normal aragonite crystals in the calcium carbonate-supersaturated solution containing magnesium ions were covered by radiating needles. The components of the ligament may have a role in regulating the growth direction of aragonite crystals.
To investigate general MMP functions during calcification in detail, we performed in vitro calcium carbonate crystallization experiments with collagen gels. TEM observations showed that fine collagen fibers covered the surface of the aragonite needles. Cryo-SEM showed the boundary between collagen fibers and aragonite needles. These results suggest that hrMMP-1 degraded the collagen fibers and increased the interaction between aragonite surface and collagen fibers. The previous work also showed the collagen fibers regulated the orientation of aragonite crystals (Falini et al., 1997). In this study, we revealed that hrMMP-13 functions in the matrix calcification of the P. fucata ligament. This information provides insight into the biomineralization mechanism and will contribute to the development of novel applications in material science.
Author Contributions
KK did the experiments and described the images of figures. HK supported the experiments and paper preparation. AM did the experiments of next generation sequencer. YT cultured and sent the living P. fucata. TT and NS supported to do the next generation sequencer. MS organized this project and wrote the manuscript.
Funding
This work was supported by a Grant-in-Aid for Young Scientists B (JP25850078 and JP16K20995) (to MS) from Japan Society for the Promotion of Science (JSPS). This study was also supported by the JSPS-ISF (Israel Science Foundation) Joint Academic Research Program.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Toshihiro Kogure in The University of Tokyo, Prof. Stephen Weiner and Prof. Lia Addadi in Weizmann Institute of Science for their invaluable helps.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00373/full#supplementary-material
Footnotes
References
Aoki, H., Tanaka, S., Atsumi, T., Abe, H., Fujiwara, T., Kamiya, N., et al. (2012). Correlation between Nacre-deposition ability and shell-closing strength in Japanise Pearl Oyster Pinctada fucata. Aquac. Sci. 60, 451–457.
Bevelander, G., and Nakahara, H. (1969). An electron microscope study of the formation of the ligament of Mytilus edulis and Pinctada radiata. Calcif. Tissue Res. 4, 101–112. doi: 10.1007/BF02279112
Falini, G., Fermani, S., Gazzano, M., and Ripamonti, A. (1997). Biomimetic crystallization of calcium carbonate polymorphs by means of collagenous matrices. Chem. Eur. J. 3, 1807–1814. doi: 10.1002/chem.19970031113
Gillian, M., and Vera, K. (1997). Relating matrix metalloproteinase structure to function: why the ‘hemopexin’ domain? Matrix Boil. 15, 511–518.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hare, P. (1963). Amino acids in the proteins from aragonite and calcite in the shells of Mytilus californianus. Science 139, 216–217. doi: 10.1126/science.139.3551.216
Kahler, G., Sass, R., and Fisher, F. (1976). The fine structure and crystallography of the hinge ligament of Spisula solidissima (Mollusca: Bivalvia: Mactridae). J. Comp. Physiol. 109, 209–220. doi: 10.1007/BF00689419
Kelly, R., and Rice, R. (1967). Abductin: a rubber-like protein from the internal triangular hinge ligament of Pecten. Science 155, 208–210. doi: 10.1126/science.155.3759.208
Kikuchi, Y., Tsuchiura, O., Hirama, M., and Tamiya, N. (1987). Desmosine and isodesmosine as cross-links in the hinge-ligament protein of bivalves. 3,3’-Methylenebistyrosine as an artifact. Eur. J. Biochem. 164, 397–402. doi: 10.1111/j.1432-1033.1987.tb11071.x
Kintsu, H., Okumura, T., Negishi, L., Ifuku, S., Kogure, T., Sakuda, S., et al. (2017). Crystal defects induced by chitin and chitinolytic enzymes in the prismatic layer of Pinctada fucata. Biochem. Biophys. Res. Commun. 489, 89–95. doi: 10.1016/j.bbrc.2017.05.088
Kubota, K., Tsuchihashi, Y., Kogure, T., Maeyama, K., Hattori, F., Kinoshit, S., et al. (2017). Structural and functional analyses of a TIMP and MMP in the ligament of Pinctada fucata. J. Struct. Biol. 199, 216–224. doi: 10.1016/j.jsb.2017.07.010
Marsh, M., and Sass, R. (1980). Aragonite twinning in the molluscan bivalve hinge ligament. Science 208, 1262–1263. doi: 10.1126/science.208.4449.1262
Nudelman, F., Pieterse, K., George, A., Bomans, P. H., Friedrich, H., and Brylka, L. J. (2010). The role of collagen in bone apatite formation in the presence of hydroxyapatite nucleation inhibitors. Nat. Mater. 9, 1004–1009. doi: 10.1038/nmat2875
Ono, K., Kikuchi, Y., Higashi, K., and Tamiya, N. (1990). Elastic anisotropy of bivalve hinge-ligament. J. Biomech. 23, 307–312. doi: 10.1016/0021-9290(90)90058-B
Sasano, Y., Nakamura, M., Okata, H., Henmi, A., and Mikami, Y. (2012). Remodeling of extracellular matrices initiates and advances calcification during development and healing of bones and teeth. J. Oral Biosci. 54, 25–29. doi: 10.1016/j.job.2011.06.001
Springman, E. B., Angleton, E. L., Birkedal-Hansen, H., and Van Wart, H. E. (1990). Multiple modes of activation of latent human fibroblast collagenase: evidence for the role of a Cys73 active-site zinc complex in latency and a “cysteine switch” mechanism for activation. Proc. Natl. Acad. Sci. U.S.A. 87, 364–368. doi: 10.1073/pnas.87.1.364
Suzuki, M., Kogure, T., Sakuda, S., and Nagasawa, H. (2015). Identification of Ligament Intra-Crystalline Peptide (LICP) from the Hinge Ligament of the Bivalve, Pinctada fucata. Mar. Biotechnol. 17, 153–161. doi: 10.1007/s10126-014-9603-y
Takeuchi, T., Kawashima, T., Koyanagi, R., Gyoja, F., Tanaka, M., Ikuta, T., et al. (2012). Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res. 19, 117–130. doi: 10.1093/dnares/dss005
Uskokovic, V., Khan, F., Liu, H. C., Witkowska, H. E., Zhu, L., Li, W., et al. (2011). Hydrolysis of amelogenin by matrix metalloprotease-20 accelerates mineralization in vitro. Arch. Oral Biol. 56, 1548–1559. doi: 10.1016/j.archoralbio.2011.06.016
Van Wart, H. E., and Hansen, B. H. (1990). The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix matelloproteinase gene family. Proc. Natl. Acad. Sci. U.S.A. 87, 5578–5582. doi: 10.1073/pnas.87.14.5578
Keywords: aragonite, biomineralization, ligament, matrix metalloproteinase (MMP), Pinctada fucata
Citation: Kubota K, Kintsu H, Matsuura A, Tsuchihashi Y, Takeuchi T, Satoh N and Suzuki M (2018) Functional Analyses of MMPs for Aragonite Crystal Formation in the Ligament of Pinctada fucata. Front. Mar. Sci. 5:373. doi: 10.3389/fmars.2018.00373
Received: 16 April 2018; Accepted: 26 September 2018;
Published: 16 October 2018.
Edited by:
Vera Bin San Chan, Clemson University, United StatesReviewed by:
Takashi Toyofuku, Japan Agency for Marine-Earth Science and Technology, JapanVengatesen Thiyagarajan, University of Hong Kong, Hong Kong
Gary H. Dickinson, The College of New Jersey, United States
Copyright © 2018 Kubota, Kintsu, Matsuura, Tsuchihashi, Takeuchi, Satoh and Suzuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michio Suzuki, YW1pY2hpd29AbWFpbC5lY2MudS10b2t5by5hYy5qcA==
 Kazuki Kubota1
Kazuki Kubota1 Hiroyuki Kintsu
Hiroyuki Kintsu Noriyuki Satoh
Noriyuki Satoh Michio Suzuki
Michio Suzuki