- 1Tohoku National Fisheries Research Institute, Japan Fisheries Research and Education Agency, Shiogama, Japan
- 2Faculty of Environmental & Symbiotic Sciences, Prefectural University of Kumamoto, Kumamoto, Japan
- 3Faculty of Marine Bioscience, Fukui Prefectural University, Obama, Japan
- 4UMR7144, Station Biologique de Roscoff, CNRS – Sorbonne Université, Roscoff, France
- 5GEMA Center for Genomics, Ecology and Environment, Universidad Mayor, Huechuraba, Chile
- 6Asian School of the Environment, Nanyang Technological University, Singapore, Singapore
Pico- and nano-phytoplankton (respectively, 0.2–2 and 2–20 μm in cell size) play a key role in many marine ecosystems. In this size range, Bolidophyceae is a group of eukaryotes that contains species with cells surrounded by 5 or 8 silica plates (Parmales) as well as naked flagellated species (formerly Bolidomonadales). Bolidophyceae share a common ancestor with diatoms, one of the most successful groups of phytoplankton. This review summarizes the current information on taxonomy, phylogeny, ecology, and physiology obtained by recent studies using a range of approaches including metabarcoding. Despite their rather small contribution to the phytoplankton communities (on average less than 0.1%), Bolidophyceae are very widespread throughout marine systems from the tropics to the pole. This review concludes by discussing similarities and differences between Bolidophyceae and diatoms.
Introduction
Following the appearance of oxygenic photosynthesis in the ancestors of cyanobacteria, this complex process was distributed across all eukaryotic lineages via permanent primary, secondary, and tertiary endosymbioses (Not et al., 2012). Ocean photosynthesis is dominated by phytoplankton, a functional group of single cell organisms including prokaryotes and eukaryotes. In the late 70’s, early 80’s the work of Waterbury et al. (1979) and Johnson and Sieburth (1982) revealed the importance of very small cells, some below one micron in size, for primary productivity, which importance was formalized with the concept of the microbial loop by Azam et al. (1983). However, it was only in the mid 90′s, when researchers began to investigate the eukaryotic compartment of picophytoplankton, and realized that while cyanobacteria are very little diversified, at the least at the genus level with a couple of taxa (Prochlorococcus, Synechococcus) dominating, eukaryotes turned out to be very diverse with picoplankton taxa distributed widely across several branches of eukaryotic tree of life (Vaulot et al., 2008).
Microphytoplankton such as diatoms, dinoflagellates or coccolithophorids that produce short lived blooms such as Emiliania huxleyi, have been extensively investigated, in contrast to other members of the picoplanktonic community. A large number of photosynthetic picoeukaryotes species (and clades) described to date belong to Stramenopiles (also called Heterokonts), which are characterized by flagellated cells, produced at least at some point of their life cycles, with two unequal flagella (heterokont), one being ornamented with hair-like structures called mastigonemes. Their plastids are thought to have been acquired through secondary endosymbiosis and typically contain chlorophylls a and c. Stramenopiles besides include diatoms, a very successful lineage which cells are encased in ornamented silica structures, Dictyochophyceae, also called silicoflagellates, and Pelagophyceae, well known because of the toxic algal blooms of the brown tide species Aureococcus anophagefferens and Aureoumbra lagunensis (Gobler and Sunda, 2012) or Pelagomonas calceolata, frequently isolated from sea water. Although often seen as less diverse, some of these groups play important ecological roles in marine ecosystems are fundamental for our understanding of the evolution of algae.
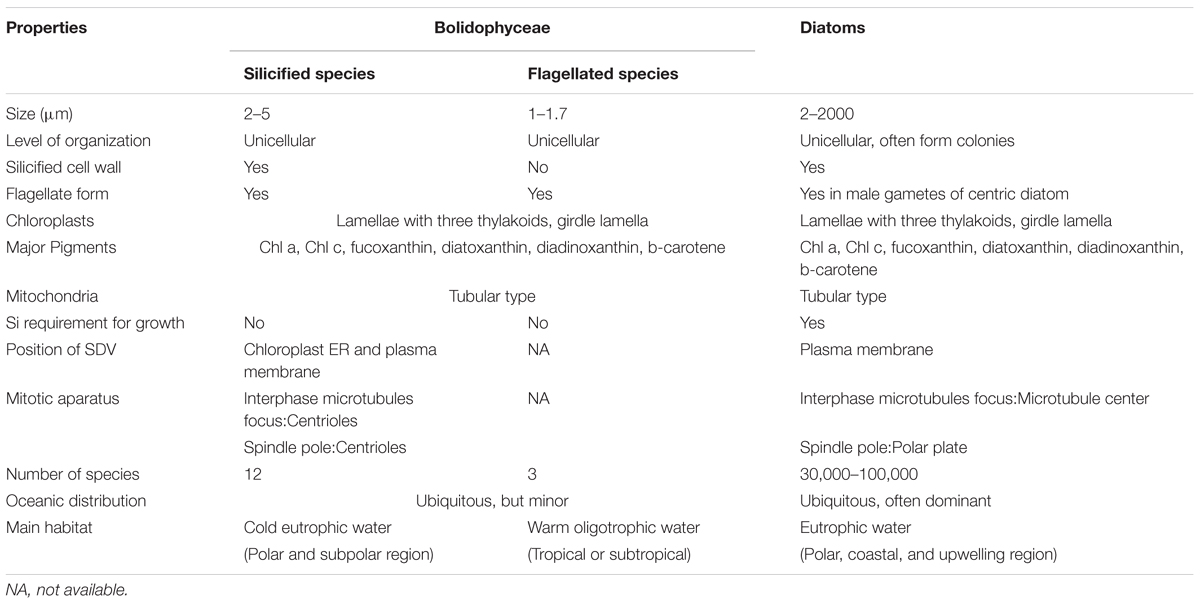
Bolidophyceae, a class created by Guillou et al. (1999a) is the Stramenopiles group phylogenetically nearest to the diatoms. They are often detected in molecular surveys, although in low abundance. We now know that they can occur as two distinct forms, either silicified pico-sized (2–5 μm) or non-silicified flagellated (1–1.7 μm) cells (Guillou et al., 1999a; Ichinomiya et al., 2011, 2016) and cultures from both forms have been isolated from the marine environment. This review summarizes the discovery of Bolidophyceae and current information of phylogeny, ecology, and physiology obtained by recent studies using a range of approaches. We discuss similarities and differences between Bolidophyceae and diatoms to explore the evolutionary link between these silicified algal groups.
Discovery and Taxonomy
Well before the creation of the class Bolidophyceae, their silicified forms (Figure 1) were first reported from scanning electron microscopy (SEM) images in oceanic samples from the North Pacific (Iwai and Nishida, 1976). Initially, they were thought to be resting cysts of silicified loricate choanoflagellates (Silver et al., 1980). However, the observation of red auto fluorescence indicating the presence of chlorophyll and the existence of a chloroplast in sectioned cells observed by transmission electron microscopy revealed that they were active phytoplankton cells (Marchant and McEldowney, 1986). Marchant and McEldowney (1986) could not establish their taxonomic position, although they suggested some morphological similarities with other algae groups such as Bacillariophyceae and Chrysophyceae.
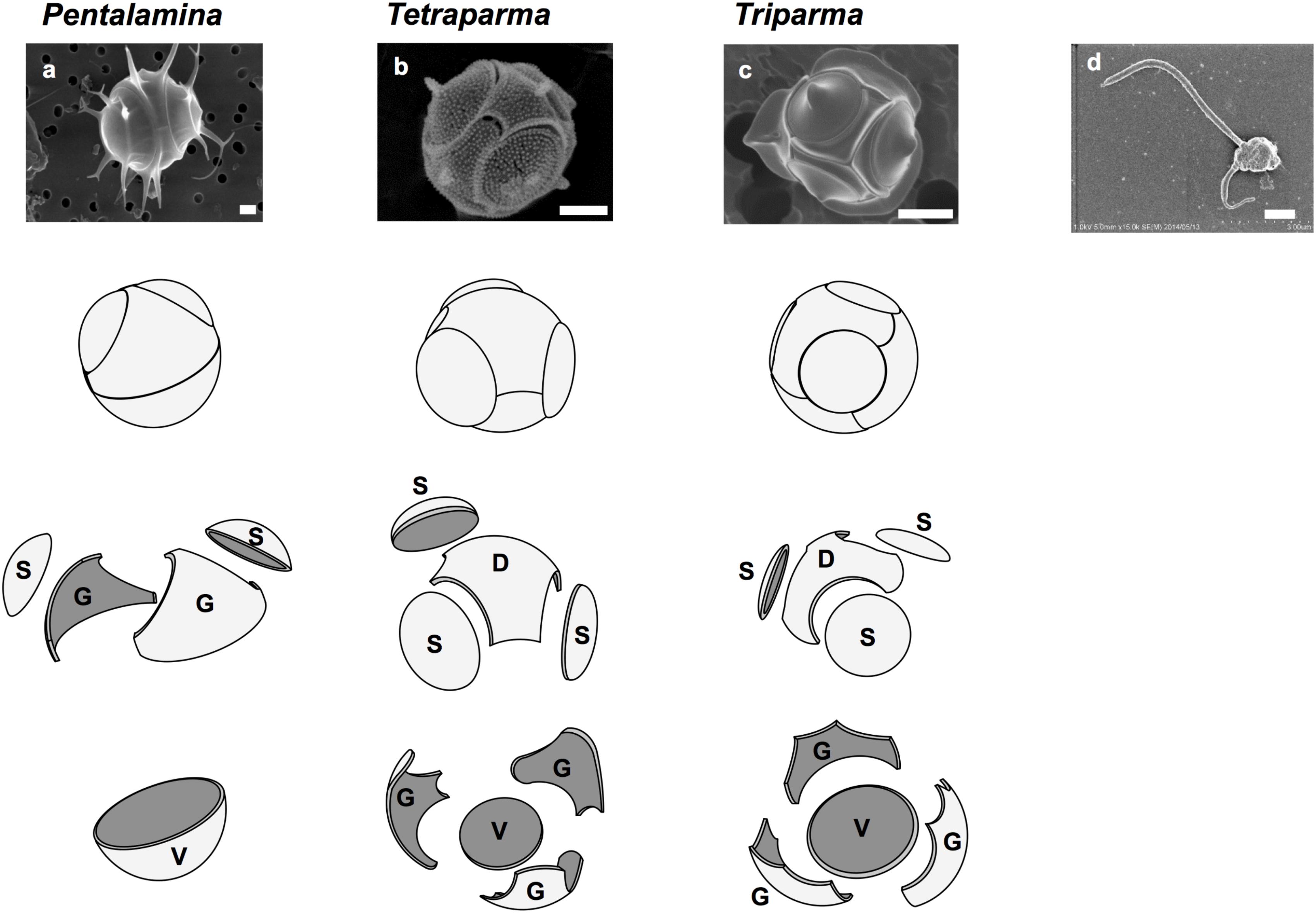
FIGURE 1. Silicified and flagellated species of Bolidophyceae. (a) Pentalamina corona, (b) Tetraparma pelagica, (c) Triparma laevis f. inornata, and (d) Triparma eleuthera. Scale bars = 1 μm. Diagrams of silica plates in the three genera of silicified species (Parmales); D, dorsal plate; G, girdle plate; S, shield plate; and V, ventral plate. Redrawn from Booth and Marchant (1987) with permission.
Booth and Marchant (1987) tentatively established Parmales (Latin: small round shields) as a new order within Chrysophyceae. The taxonomy of Parmales was based on the morphological features of silica plates that can only be observed with SEM. Two families and three genera were established: Pentalaminaceae with one genus Pentalamina (Latin: five plates) and Triparmaceae with two genera, Tetraparma (Latin: four small round shields) and Triparma (Latin: three small round shields) (Figures 1a–c). Pentalamina has 2 circular shield plates of equal size, a larger ventral plate and 2 triradiate girdle plates. Tetraparma has 3 shield plates of equal size, a smaller ventral plate, a triradiate dorsal plate and 3 girdle plates. Triparma has 3 shield plates of equal size, a larger ventral plate, a triradiate dorsal plate and 3 girdle plates (Booth and Marchant, 1987, 1988; Kosman et al., 1993; Bravo-Sierra and Hernández-Becerril, 2003; Konno and Jordan, 2007; Konno et al., 2007).
The flagellated forms of Bolidophyceae (Figure 1d) were originally isolated from the Pacific Ocean and Mediterranean Sea, and described as two flagellated species, Bolidomonas pacifica and B. mediterranea, differing in the angle of the insertion of the two flagellum, swimming patterns as well as in 18S rRNA gene signatures (Guillou et al., 1999a). The name “Bolidomonas” refers to the rapid swimming behavior of the cells remembering a racing car. A variety, B. pacifica var. eleuthera, was later proposed based on both cultures and environmental sequences (Guillou et al., 1999b). Analyses of photosynthetic pigments as well as nuclear 18S rRNA and plastid RubisCO large subunit (rbcL) sequences (Guillou et al., 1999a; Daugbjerg and Guillou, 2001) demonstrated the sister relationship between Bolidomonas and diatoms, although Bolidomonas are flagellates and lack the siliceous frustule characteristic of diatoms. Bolidophyceae were thus proposed to be an intermediate group between diatoms and all other Stramenopiles (Guillou et al., 1999a).
For more than 24 years, Parmales escaped isolation. These silicified cells are small and difficult to distinguish them from other small phytoplankton in field samples under the light microscope. To overcome this problem, Ichinomiya et al. (2011) used the fluorescence dye PDMPO (2-(4-pyridyl)-5-((4-(2-dimethylaminoethylaminocarbamoyl) methoxy)phenyl) oxazole) (Shimizu et al., 2001), which is co-deposited with silicon into the solid silica matrix of the newly produced cell walls and fluoresces under UV excitation whenever silicic acid is polymerized forming biogenic silica. Using PDMPO staining and a serial dilution technique, the first Parmales strain was established from Oyashio region of the western North Pacific (Ichinomiya et al., 2011).
Scanning electron microscopy established that this strain belonged to the species Triparma laevis and transmission electron microscope observations showed the typical ultrastructure of photosynthetic Stramenopiles, with two endoplasmic reticulate membranes surrounding the chloroplast, a girdle and two to three thylakoid lamellae as well as a mitochondrion with tubular cristae. Phylogenetic analyses based on 18S ribosomal rRNA sequences from the new strain demonstrated that T. laevis was closely related to Bolidophyceae (Ichinomiya et al., 2011), rather than part of Chrysophyceae as hypothesized initially (Booth and Marchant, 1987). Phylogenetic analyses using plastidial and mitochondrial encoded genes from T. laevis also confirmed its sistership with Bolidophyceae and diatoms (Tajima et al., 2016).
Recent phylogenetic analyses using nuclear, plastidial, and mitochondrial genes from several novel strains, including a flagellate form very closely related to the silicified strains, led to a taxonomic revision (Table 1) in which the order Parmales was included within the class Bolidophyceae and Bolidomonas species were transferred to the genus Triparma (Ichinomiya et al., 2016).
Genetic Diversity
Clade Diversity
The analysis of full-length nuclear 18S rRNA gene sequences from public databases revealed the existence of two environmental clades (Env. clade I and II) in addition to the group corresponding to the genus Triparma (Ichinomiya et al., 2016). These clades are only formed by environmental sequences and no sequences from cultures or isolates are available. Within the Triparma group, sub-clades formed by sequences from strains and the environment corresponded to the species Triparma eleuthera, Triparma pacifica, and Triparma mediterranea. The “T. laevis” sub-clade, including the species T. laevis f. inornata, T. laevis f. longispina, Triparma strigata, Triparma aff. verrucosa and the flagellated strain Triparma sp. RCC1657. Other molecular markers (plastid 16S rRNA and rbcL, nuclear ITS rRNA and mitochondrial nad1) revealed the presence of two distinct sub-clades within the “T. laevis” sub-clade, hereafter called for convenience Triparma clade I with the two forms of T. laevis (f. inornata, and longispina) and Triparma clade II with T. strigata, T. aff. verrucosa and the flagellated strain RCC1657.
In order to review the current state of the diversity of Bolidophyceae, we analyzed existing GenBank sequences as well as metabarcodes obtained from a range of recent studies (Table 2) focusing on the V4 region of the 18S rRNA gene (see Supplementary Material for Methodology). The phylogenetic analysis of the newly obtained V4 sequences (Figure 2) recovered the two major environmental clades previously described (Env. clade I and II, Ichinomiya et al., 2016), but also revealed the existence of a third environmental clade (called Env. clade III) within which two sub-clades IIIA and IIIB can be clearly separated. Each environmental clade contained sequences from clone libraries (GenBank) as well as identical or nearly identical metabarcode sequences from different surveys suggesting that these environmental clades are not artefactual. These environmental clades may, for some of them, correspond to species of Parmales (e.g., from genera Tetraparma or Pentalamina) that have not yet been isolated in cultures.
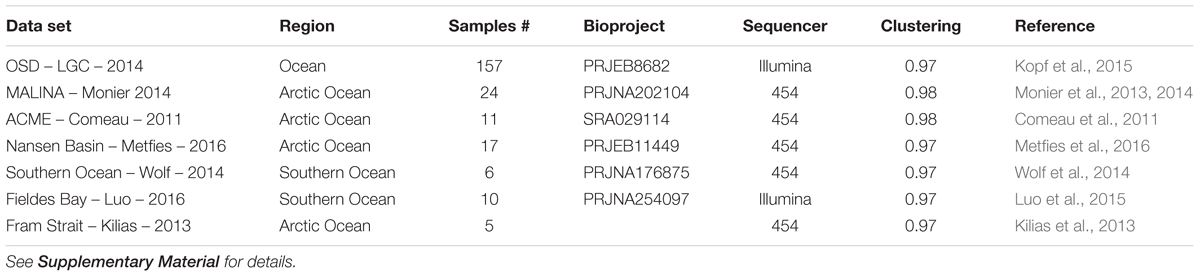
TABLE 2. List of metabarcoding studies using the V4 region of the 18S rRNA genes that have been used for the phylogenetic (Figure 2) and the biogeography analyses (Figures 6, 7).
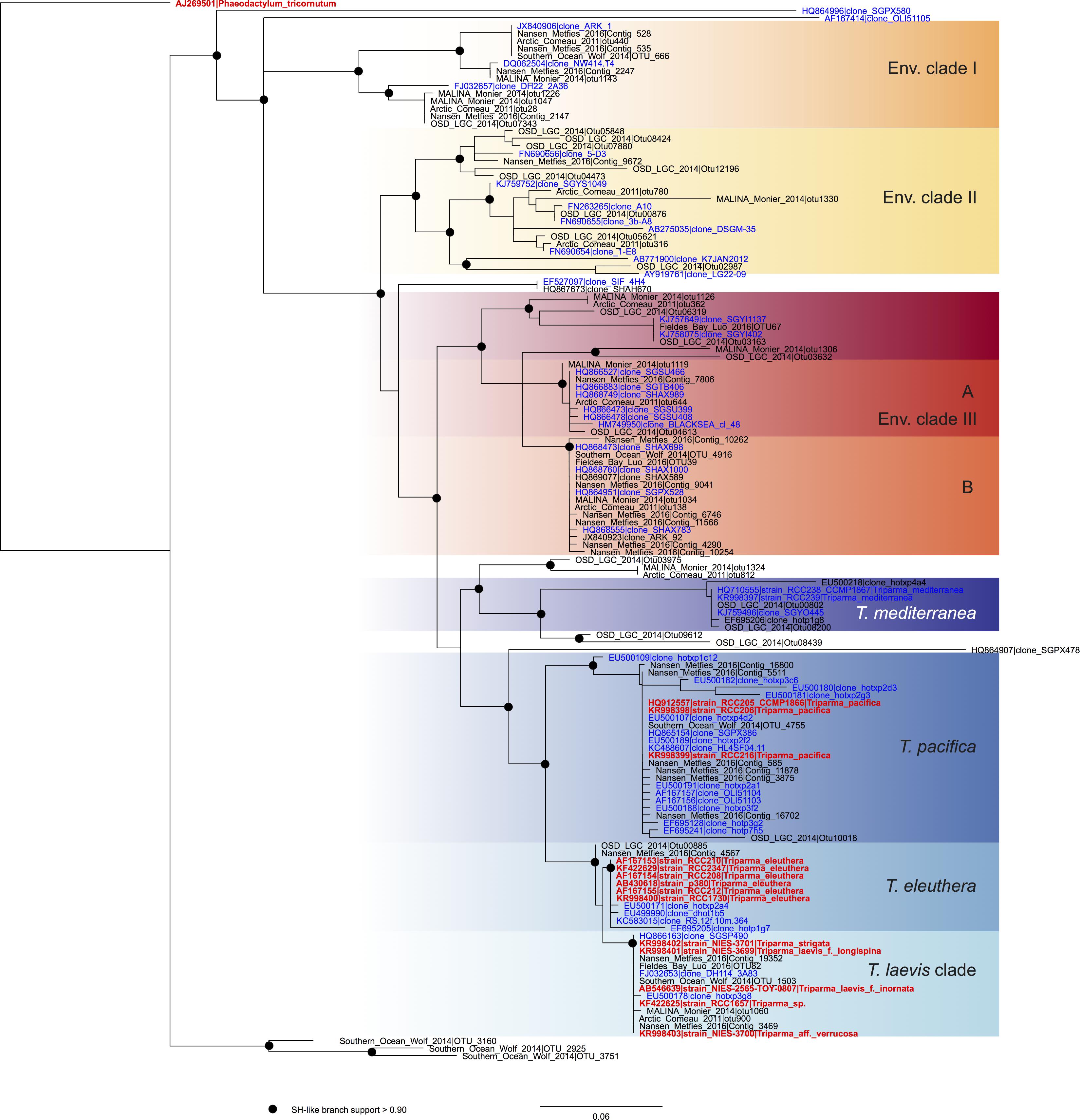
FIGURE 2. ML phylogenetic tree based on the V4 region of the 18S rRNA gene based both on GenBank sequences available from the PR2 database (Guillou et al., 2013) and on metabarcodes OTUs obtained from the studies listed in Table 2 (see Methodology in Supplementary Material). The tree was constructed with phyML and dots correspond to nodes with SH-like branch support >0.90. GenBank sequences from environmental samples are colored in blue and those from cultures, in red.
Diversity Within the Genus Triparma
We explored the level of inter- and intra-clade diversity within the genus Triparma by analyzing the folding pattern of the ITS2 (see Supplementary Material for Methodology) from 14 strains previously described in Ichinomiya et al. (2016). The general ITS2 secondary structure of Bolidophyceae proposed contains the four-helices domains known in many eukaryotic taxa in addition to helix B9 (Figure 3). We located in Helices II and III the universal hallmarks proposed by Mai and Coleman (1997) and Müller et al. (2007): the pyrimidine–pyrimidine (Y–Y) mismatch in helix II and YRRY (pyrimidine – purine – pyrimidine) motif on the 5′ side of Helix III, respectively, at alignment positions 95 and 148 and between nucleotides 205 and 208 (Figures 3, 4). In all strains analyzed, the Y–Y mismatch was represented by the pair U x C, with the exception of T. pacifica strains (U x U), and the YRRY motif of helix III by the sequence UGGU (Figure 3).
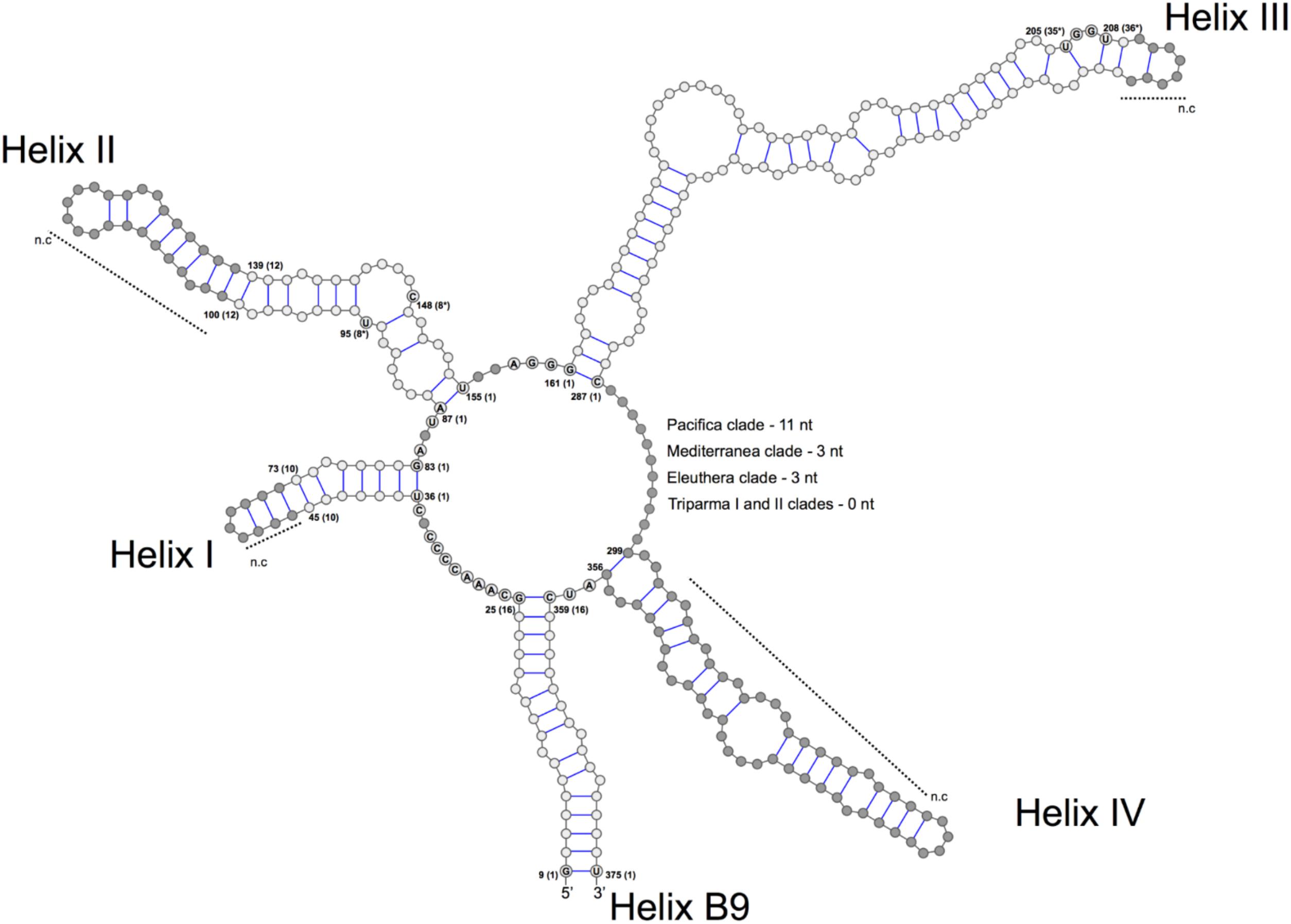
FIGURE 3. General structure model of the ITS2 molecule of the Triparma clades. The four major helices are labeled as Helix I – Helix IV and the interaction region of 5.8S and 28S rRNA as B9. Numbers refer to the alignment positions and those between brackets to the compared positions in each helix. The nucleotides that are 100% conserved in the helices spacers, hallmarks positions in helices II and III and first two base pairs of the helices are shown. The gray dots represent segments that display length and sequence variation (see Methodology in Supplementary Material).
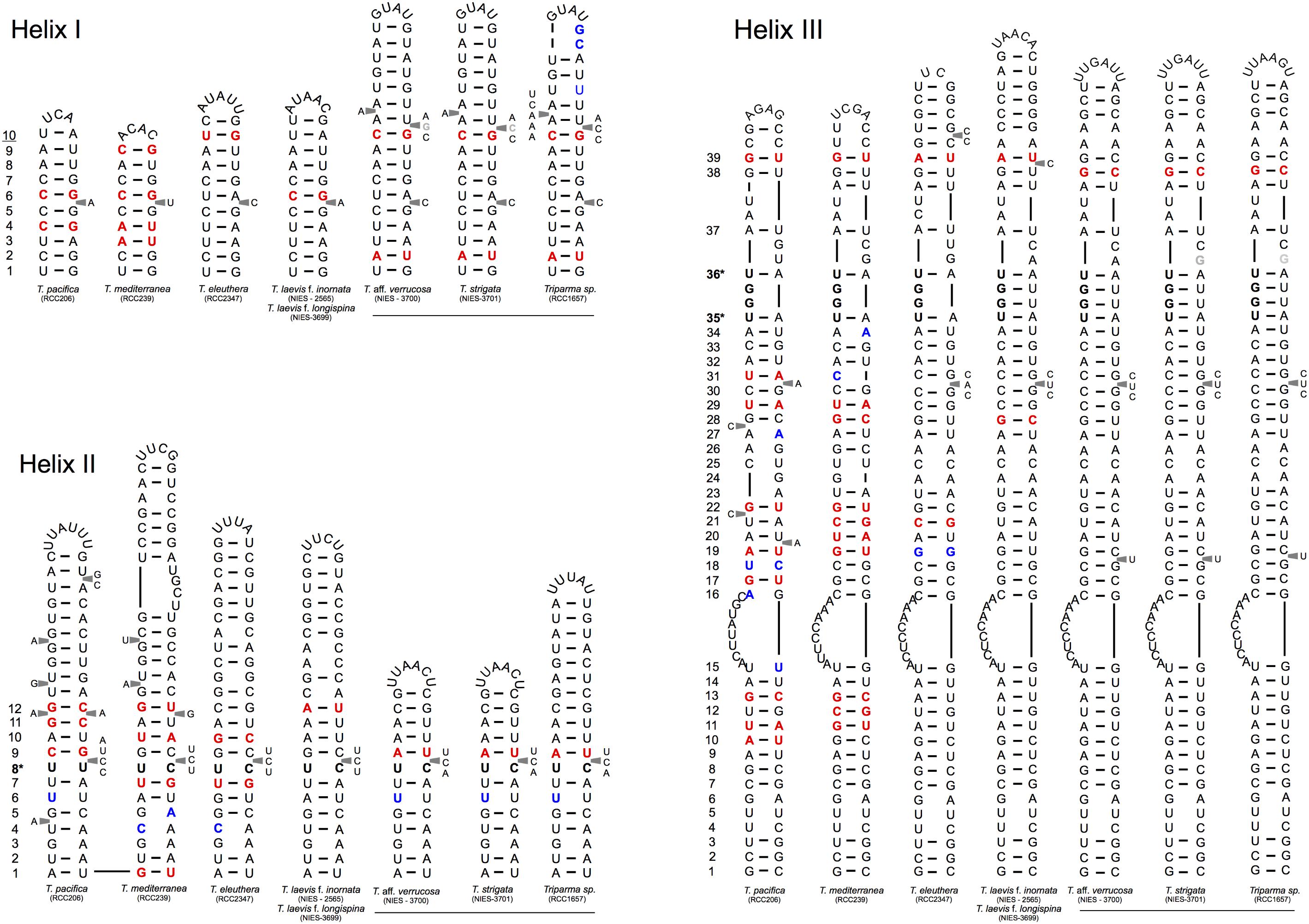
FIGURE 4. Simplified secondary structural diagram of helices I, II, and III among the different Triparma clades. The conserved base pair among the different clades are numbered. Double-sided CBCs (compensatory base changes) and hemi-CBCs are highlighted by bold red nucleotides. Non-CBCs (N – N ↔ N × N) are represented by bold blue nucleotides. The pyrimidine-pyrimidine (Y–Y) mismatch in helix II and the YRRY (pyrimidine – purine – pyrimidine) motif on the 5′ side arm of the helix III are showed by bold nucleotides and by ∗ positions. Single nucleotide substitution between T. aff. verrucosa and T. strigata is showed by gray nucleotides (see Methodology in Supplementary Material).
The spacers between helices B9 and I, I and II, II and III, and IV and B9 were conserved in length and sequence among the clades (Figure 3), as well as the first two base pairs of helices I, II, and III. In contrast, the spacer between helices III and IV showed greater variation between Bolidophyceae clades but it was conserved at the intra-clade level (Figure 3). Helices B9 (a region of the 5.8S and 28S rRNA interaction) and III showed good intra and inter-clade conservation (Figure 3 and Supplementary Figure 1). The ITS2 sequence from T. aff. verrucosa is incomplete and the 3′side arm of helix B9 could not be determined (Supplementary Figure 1).
The identification of CBCs in Bolidophyceae ITS2 secondary structure was based on the phenetic approach which relies on a base pair sequence comparison of all CBCs between two sequences without direct reference to their evolutionary origin (Müller et al., 2007; Coleman, 2009). The phylogenetic approach method which considers the status of a given base pair in the ancestor of two sister taxon could not be applied for Bolidophyceae given the conflicting branching pattern among phylogenies (for more details see Ichinomiya et al., 2016).
Putative CBCs, hCBCs, and non-CBCs type changes were identified in the conserved regions of the helices B9, I, II, and III within each clade and between clades (Figure 4 and Supplementary Figure 1). Helix IV (Figure 4) was not included in the inter-clade analysis given its known variable nature (Coleman, 2007). Several CBCs and hCBCs were identified at inter-clade level suggesting that each clade within Triparma genus (T. pacifica, T. mediterranea, T. eleuthera, Triparma I and II, sensu Ichinomiya et al., 2016) is composed by at least one species (Figure 4).
At the intra -clade level, no CBC, nor hCBCs were identified between the two forms of T. laevis, f. inornata and f. longispina (Triparma clade I), that differ by the plate morphology, suggesting that these two forms may belong to the same species, although the absence of CBCs is not an absolute indicator that two organisms belong to the same species (Müller et al., 2007; Caisová et al., 2011, 2013). However, at least one CBC is a good indicator (93.1% of confidence for plants and fungi) that in most of the cases, two organisms represent distinct species (Müller et al., 2007). For Triparma clade II (T. aff. verrucosa, T. strigata, and Triparma sp. RCC1657), no CBC or hCBCs were identified between the morpho species T. aff. verrucosa and T. strigata. The ITS operon sequences from these two strains, including the two internal transcribed spacers and 5.8S rRNA, are nearly identical (99.1%) differing only by six nucleotides. Although three of these substitutions are within the ITS2, none correspond to a nucleotide pair in the ITS2 secondary structure (Figure 4). However, between Triparma sp. RCC1657 on the one side and T. aff. verrucosa plus T. strigata on the other side, 1 hCBC (helix B9, position 6, Supplementary Figure 1) and 1 CBC (helix IV, box, Supplementary Figure 1) were identified, suggesting that Triparma clade II is composed by at least two species, one corresponding to Triparma sp. RCC1657 and the other by T. aff. verrucosa and T. strigata.
Ecology
Oceanic Distribution
We explored the distribution of Bolidophyceae in the ocean using data obtained by SEM observation, environmental sequencing and metabarcoding. By compiling available records of observed silicified morphological species of Bolidophyceae in natural waters (Supplementary Table 1), we mapped the geographical and specific distribution pattern of each morphological species (Figure 5). Tetraparma pelagica, and the Triparma species, T. laevis, T. columnacea, T. retinervis, and T. strigata are widely distributed from polar to subtropical regions. In contrast, Pentalamina corona, Tetraparma gracilis, Tetraparma catinifera, and Triparma verrucosa are restricted to polar or subpolar regions. T. gracilis was observed in both, Arctic and Antarctic regions while P. corona seems endemic to the Antarctic and, T. verrucosa and T. catinifera to the subarctic region. Tetraparma insecta and the recently described species Tetraparma silverae and Tetraparma trullifera seem to be restricted so-far to subtropical regions.
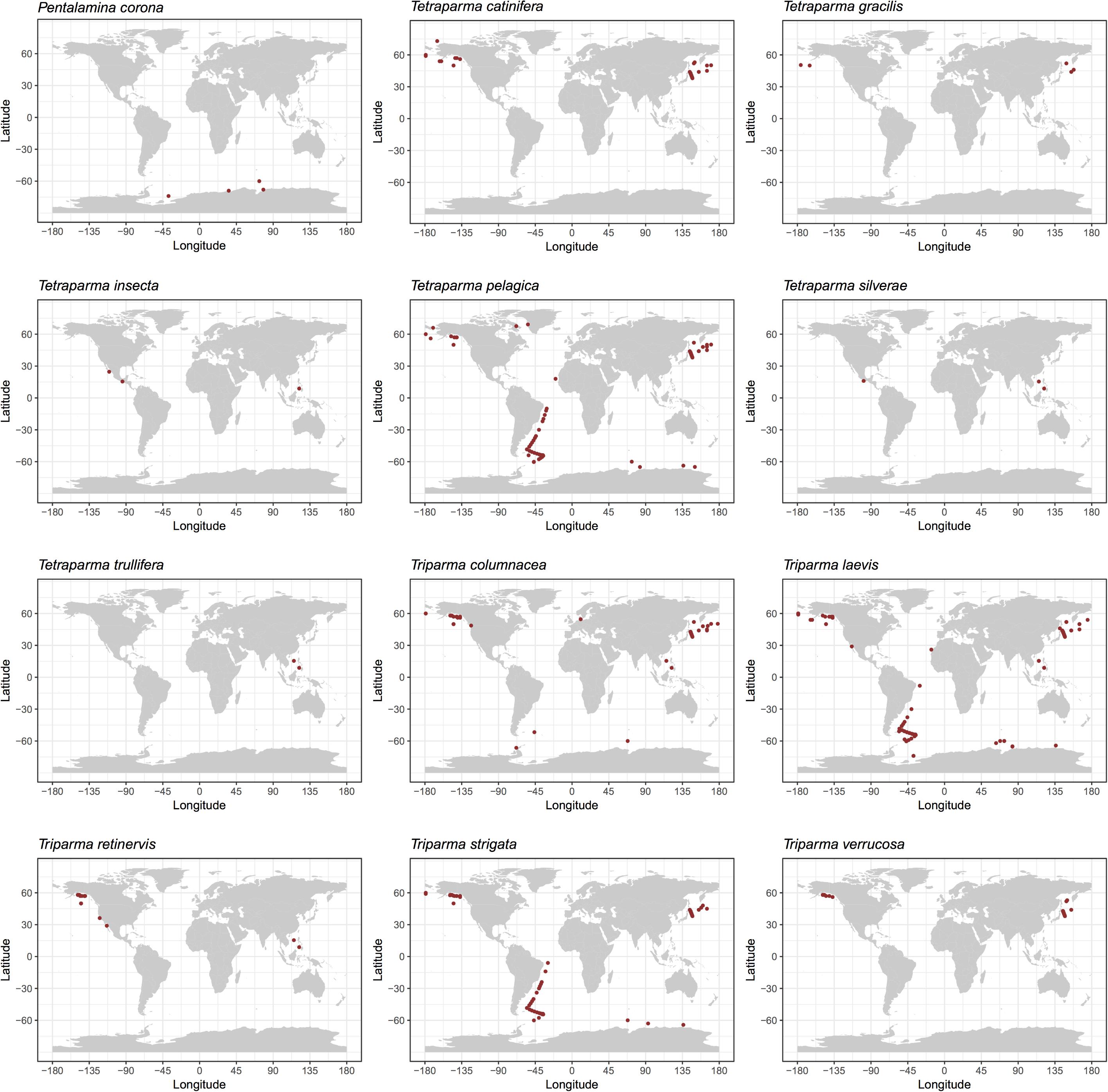
FIGURE 5. Distribution of silicified Bolidophyceae species based on literature records of SEM observations (see Supplementary Table 1).
Using both available environmental GenBank sequences and 18S rRNA V9 metabarcodes acquired during the Tara Oceans expedition, Ichinomiya et al. (2016) established the oceanic distribution of the major Triparma species and of environmental clades. T. mediterranea metabarcodes dominated in the Mediterranean Sea while T. pacifica and T. eleuthera were co-dominant in the tropical and sub-tropical oceans. The T. laevis clade was clearly associated with cold Antarctic waters but was also found near the Costa Rica dome. Bolidophyceae sequences were most abundant in the picoplanktonic fraction (0.8–5 μm) of the Tara Oceans samples and represented at most 4% of the photosynthetic reads and less than 1% on average (Ichinomiya et al., 2016).
In order to obtain a more complete image of the Bolidophyceae distribution, we used the large data set of 18S rRNA V4 metabarcodes described above. This data set includes a range of studies (Table 2) including OSD (Ocean Sampling Day) that sampled an extensive set of coastal stations (Kopf et al., 2015) and several from Arctic and Antarctic waters that were not covered by the Tara expedition.
Among these metabarcodes, the Triparma clade was slightly dominating in terms of total reads followed by the three environmental clades III, I, and II, respectively, in this order (Figure 6A). Within Triparma, T. pacifica was most abundant followed by T. mediterranea. One environmental subclade (IIIA) was also particularly abundant. The relative contribution of Bolidophyceae to total metabarcodes at each station varied widely with an average of 0.23% (Figure 6B). It was highest, up to 12%, in both Arctic and Antarctic regions as well as around the European coast. In contrast Bolidophyceae were absent at several stations along the East coast of North America and in the Eastern Mediterranean Sea (Figure 6C).
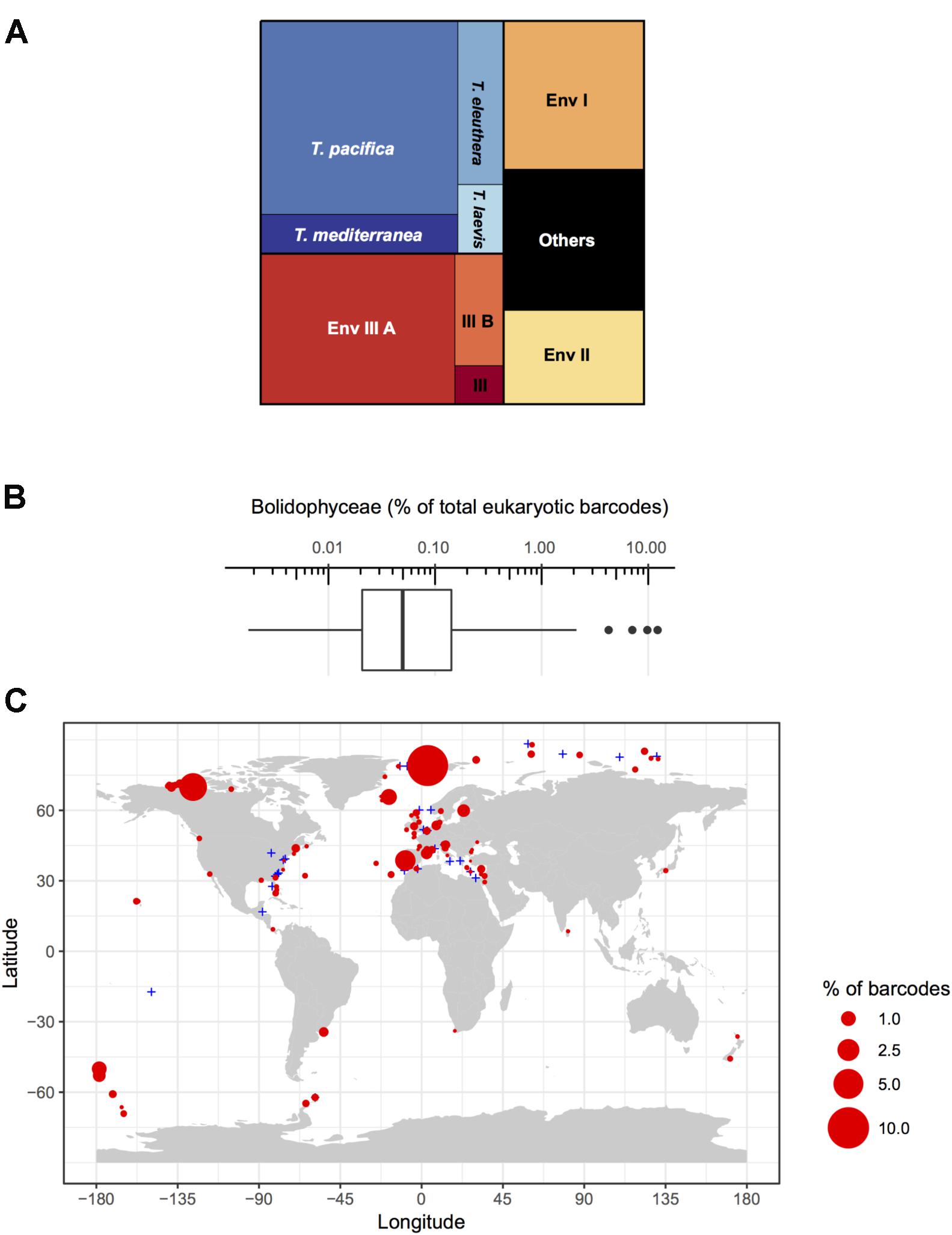
FIGURE 6. (A) Distribution of read abundance among the different environmental Bolidophyceae clades and Triparma species in the metabarcoding studies based on the 18S rRNA V4 region listed in Table 2. (B) Overall statistics of the proportion of Bolidophyceae reads related to the total number of reads from studies listed in Table 2. Only surface samples were considered. (C) Oceanic distribution of Bolidophyceae. Circle size is proportional to the number of Bolidophyceae reads related to the total number of reads at a given station.
The distribution of individual Triparma species and environmental clades confirmed some of the trends observed in the Tara Oceans data (Ichinomiya et al., 2016) but also revealed new features (Figure 7). Among the Triparma species, T. eleuthera and T. pacifica were the most ubiquitous and often co-occurred at the same stations, suggesting that their ecological niches are very close. They did not seem to be present though in really polar waters such as in the Beaufort Sea. In contrast, it was confirmed that T. mediterranea was indeed mostly restricted to the Mediterranean Sea while the T. laevis clade was only found at high latitudes both in the Arctic and Antarctic. Some environmental clades had clear biogeographic distributions such as clade IIIA found mostly in temperate latitudes and IIIB only in the Arctic and Antarctic regions. The latter clade seemed particularly prevalent in the high Arctic Ocean. Clade I was also mostly observed at high latitudes, although not restricted to polar waters, in contrast to clade II which was more widespread. These distribution patterns may reflect the genetic diversity within these clades. Clades IIIA and B have very low genetic diversity (Figure 2) in contrast clades I or II. The former may therefore correspond to a single species with a narrow niche and the latter to several species or even genera, explaining their wide distribution.
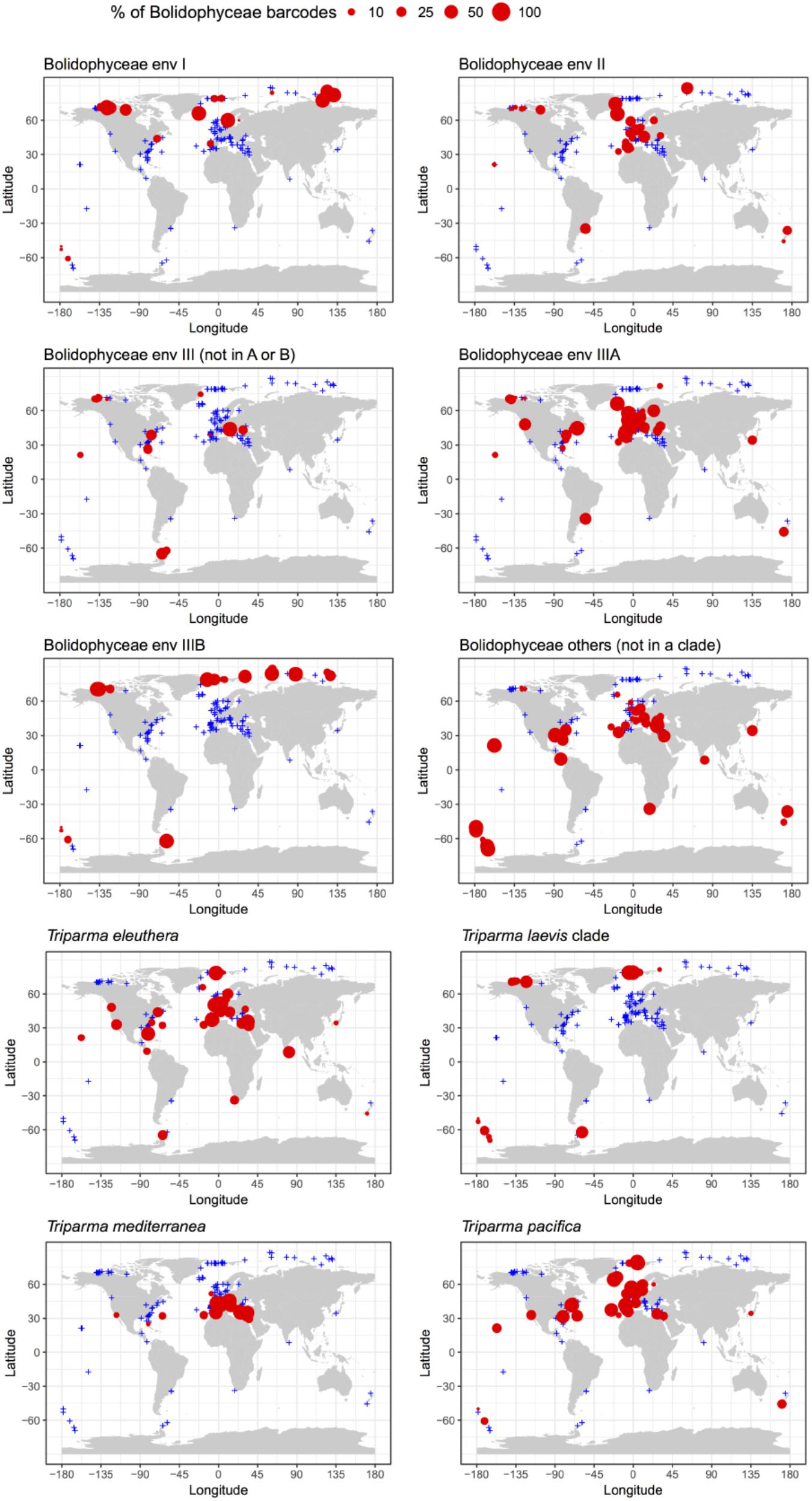
FIGURE 7. Percentage relative to the total number of Bolidophyceae reads of the different environmental Bolidophyceae clades and Triparma species in the metabarcoding studies based on the 18S rRNA V4 region listed in Table 2. Only surface samples were considered.
Seasonal Cycle
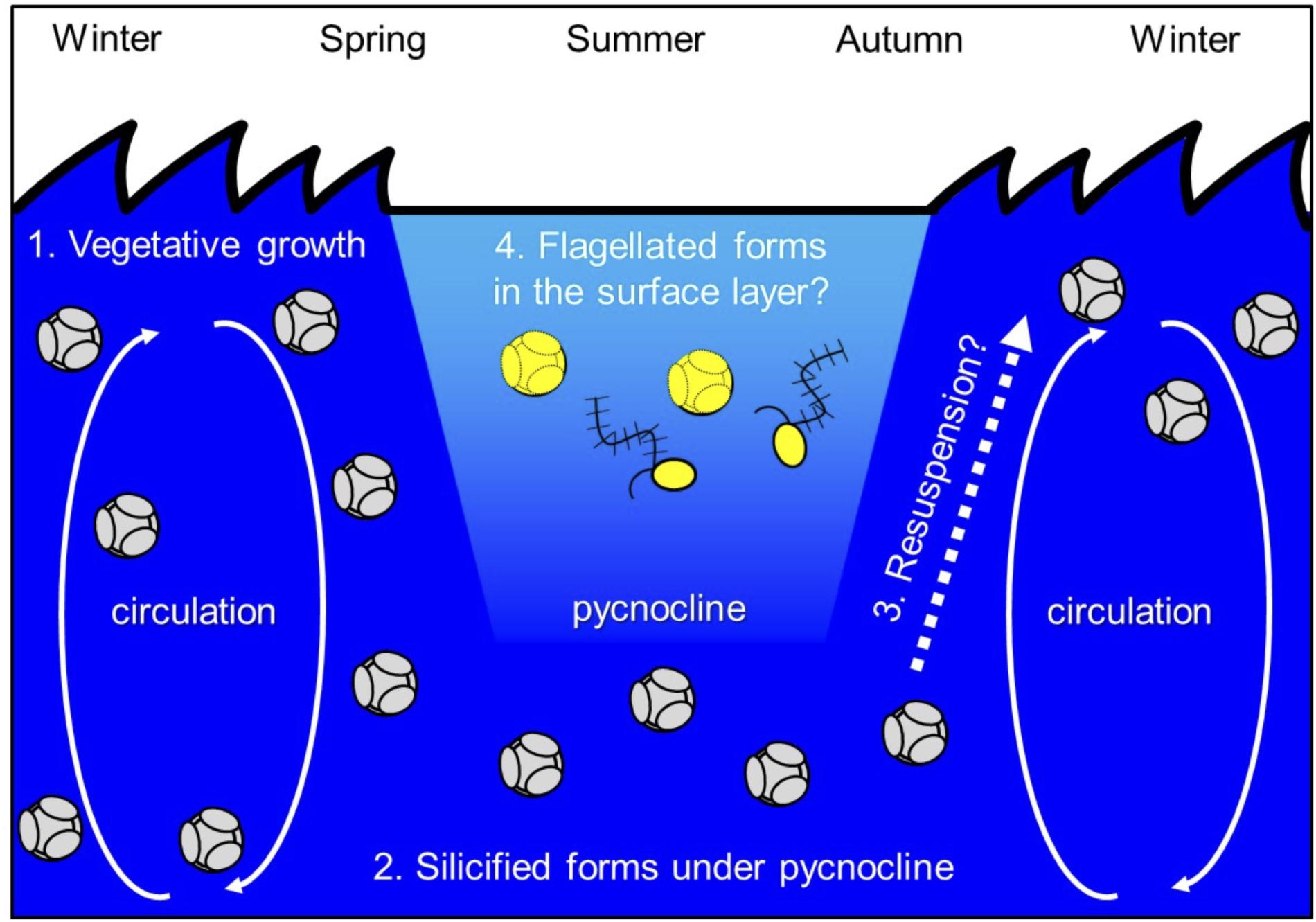
Ichinomiya and Kuwata (2015) investigated the seasonal influence in the abundance and vertical distribution of the silicified forms of Bolidophyceae in the western North Pacific using SEM. The area investigated is surrounded by the cold Oyashio current with water temperature below 5–8°C at 100 m (Shimizu et al., 2009). The Bolidophyceae community was mainly composed of T. laevis (64 ± 22%) with only small regional and seasonal differences in contrast to diatoms that display clear seasonality patterns (Takahashi et al., 2008; Suzuki et al., 2011). The vertical distribution of the silicified Bolidophyceae community changed seasonally according to the hydrographic condition. Silicified Bolidophyceae had a wide vertical distribution between 0 and 100 m with high abundance of 10–102 cells mL-1 in March and May at stations where the water column was well mixed or weakly stratified (Figure 8). In contrast, from May to October at stations where the water was stratified Bolidophyceae were absent from the surface layer, but mainly distributed under the pycnocline from 20 to 50 m with lower abundance of <0.1–10 cells mL-1. Komuro et al. (2005) also reported similar seasonal variations in depth distributions of the silicified form at Station KNOT (44°N, 155°E) in the western North Pacific. They extended from 0 to 100 m in January and May, but were restricted to the subsurface layer from 30 to 100 m in August. Silicified Bolidophyceae have optimal growth temperatures below 10°C, but do not grow above 15°C (see section “Cell Physiology”). These data suggest that silicified Bolidophyceae actively grow during the cold mixing season and maintain their population under the pycnocline during the warm stratified season (Figure 9). Flagellated forms of Bolidophyceae may also be present in the surface layer during stratification since they have been reported in the surface layer during the summer season in the English Channel (Not et al., 2002) and northern South China Sea (Wu et al., 2017) using 18S rRNA-targeted oligonucleotide probes specific of Bolidophyceae detected by in situ hybridization and tyramide signal amplification (FISH-TSA). 18S rDNA sequences of Bolidophyceae have also been detected using high-throughput sequencing (Kataoka et al., 2017) at 10 m in summer and autumn in the Oyashio region when silicified forms were absent.
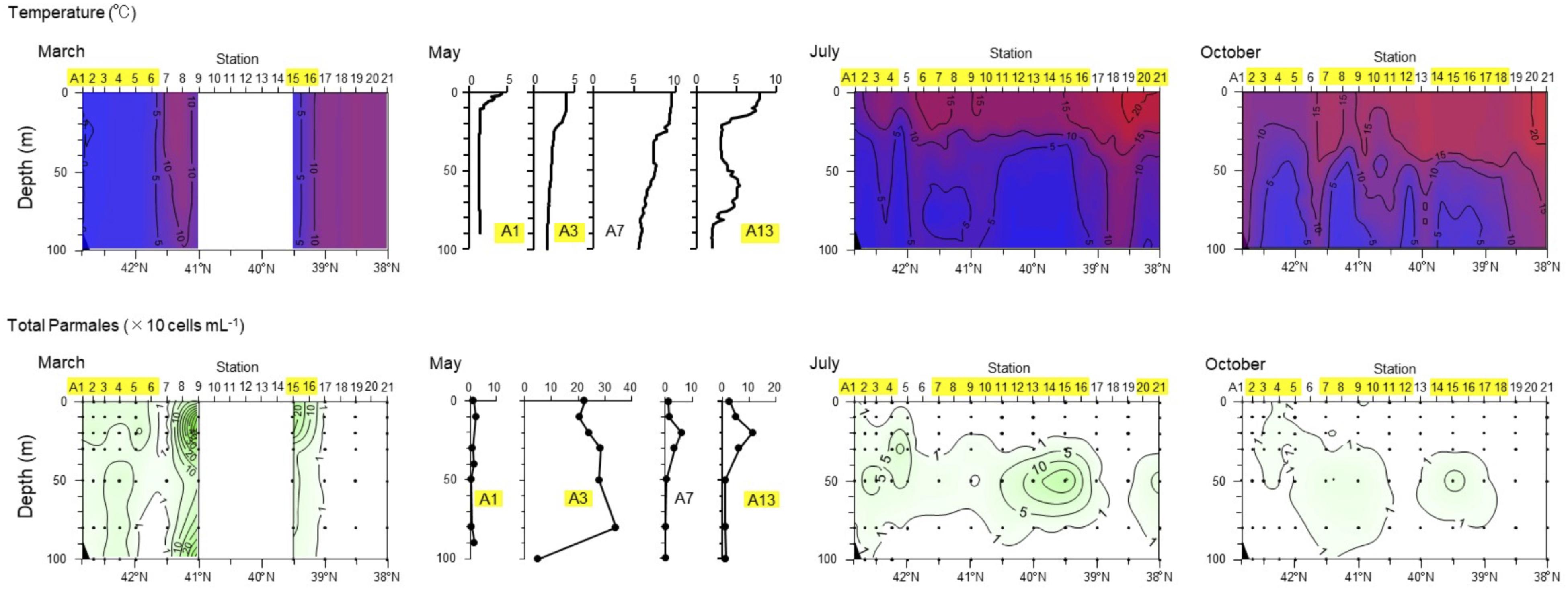
FIGURE 8. Seasonal variation in vertical distributions of temperature and abundance of the Parmales community along the A-line in March, May, July, and October 2009. Station numbers shaded in yellow indicate the Oyashio region. Redrawn from Ichinomiya and Kuwata (2015) with permission.
Role in Food Webs
It is not clear how Bolidophyceae contribute to the microbial food web. Materials resembling silicified Bolidophyceae have been reported in fecal pellets of copepods (Booth et al., 1980; Urban et al., 1993) and Antarctic krill (Marchant and Nash, 1986), indicating that they can be grazed by larger predators (Kosman et al., 1993). Bolidophyceae main grazers are expected to be small protozoans, such as choanoflagellates (Taniguchi et al., 1995) although there is no evidence of their direct ingestion by protists.
Cell Physiology
Temperature
Ichinomiya et al. (2013) and Ichinomiya and Kuwata (2015) conducted growth experiments at various temperature ranging from 0 to 15°C, using three silicified strains: T. laevis f. inornata, T. laevis f. longispina, and T. strigata. These silicified Bolidophyceae species were able to grow at 0 to 10°C (T. laevis f. inornata and T. strigata) and 5 to 10°C (T. laevis f. longispina) but not over 15°C (Figure 10). The optimal growth temperatures were 5°C for T. laevis f. inornata and 10°C for T. laevis f. longispina and T. strigata, with growth rates of 0.35 d-1, 0.50 d-1, and 0.69 d-1, respectively. In contrast, strains of naked flagellated forms have higher growth rates and grow at higher temperatures (Figure 10). T. eleuthera showed positive growth at 16–24°C with the maximum growth rate of 1.7 d-1 at 22°C (Stawiarski et al., 2016) while T. pacifica and Triparma sp. RCC201 had growth rates of 0.91 d-1 and 0.51 d-1 at 20°C, respectively (Jacquet et al., 2001; Thomas and Campbell, 2013).
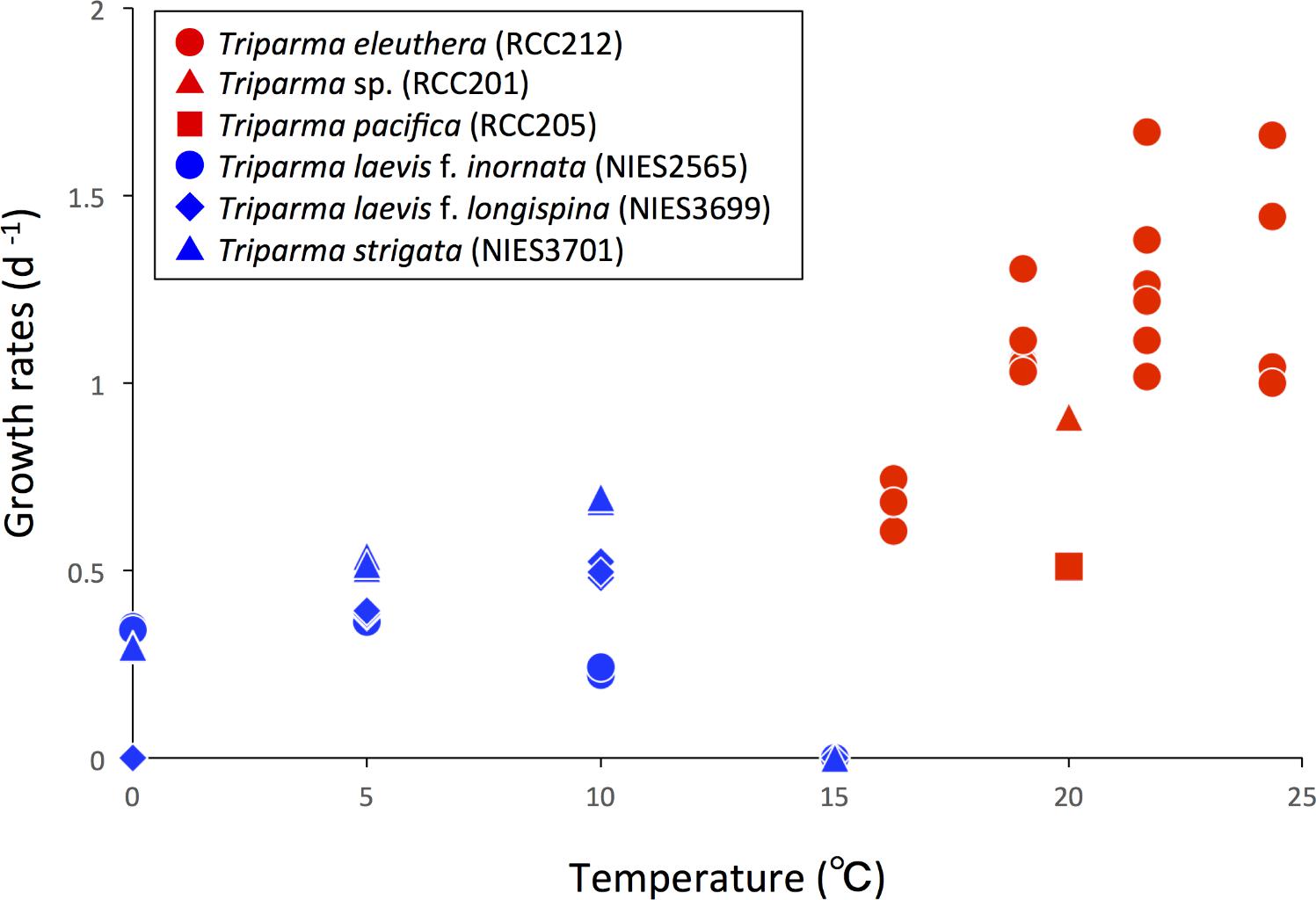
FIGURE 10. Growth rates of the silicified (blue symbols) and naked flagellated (red symbols) Triparma strains. Data redrawn from Stawiarski et al. (2016) for T. eleuthera (RCC212), Jacquet et al. (2001) for Triparma sp. (RCC201), Thomas and Campbell (2013) for T. pacifica (RCC205) and Ichinomiya and Kuwata (2015) for T. laevis f. inornata (NIES-2565), T. laevis f. longispina (NIES-3699), and T. strigata (NIES-3701).
Silica
The growth of diatoms is limited by dissolved silicate (Martin-Jézéquel et al., 2000; Sarthou et al., 2005). Diatoms cell cycle is controlled by silica and silica limitation arrests cells at the G1–S boundary (Darley and Volcani, 1969; Okita and Volcani, 1978; Vaulot et al., 1987) and during the G2–M transition due to silica requirement for DNA replication and cell wall formation, respectively (Vaulot et al., 1987; Brzezinski et al., 1990).
In contrast, Bolidophyceae despite possessing silica plates can grow in the absence of silica (Yamada et al., 2014). T. laevis f. inornata cells growing under sufficient silicate (100 μM) are surrounded by eight plates, rounded shield and ventral plates, as well as non-rounded dorsal and girdle plates. However, plate formation becomes incomplete and the fraction of cells lacking dorsal and girdle plates increases at low silicate concentration (10 μM). Cells finally loose almost all plates at silicate concentrations lower than 1 μM (Yamada et al., 2014). Other silicified Bolidophyceae strains, T. laevis f. longispina and T. strigata, can also grow under silicate depletion without formation of a silica cell wall (unpublished data). Cell wall is restored within a day in about 40% of the naked cells after replenishment of silicate (Yamada et al., 2014). Direct observation of regeneration of the silica cell wall in naked cells after re-supply of silicate using transmission and SEM revealed that shield plates appear first, followed by ventral, dorsal, and girdle plates, in this order. The dorsal and girdle plates are inserted into the space between the previously secreted shield and ventral plates to complete cell wall (Yamada et al., 2016). Similar uncoupling between the formation of silica structures and cell growth has also been observed in other silicified Stramenopiles such as Dictyochales (Henriksen et al., 1993) and Synurales (Leadbeater and Barker, 1995; Sandgren et al., 1996).
Rounded plates of silicified Bolidophyceae have a structure similar to the valves and scales of auxospores from centric diatoms (Mann and Marchant, 1989; Yamada et al., 2014). In other groups that display silica structures such as diatoms, chrysophytes, synurophytes, and dictyochophytes (Simpson and Volcani, 1981; Knoll and Kotrc, 2015; Finkel, 2016; Marron et al., 2016), silica formation takes place within a specialized membrane-bound compartment termed the Silica Deposition Vesicle (SDV) (Simpson and Volcani, 1981; Preisig, 1994). The origin and location of SDV differ among taxa. Diatoms SDVs for the development of valve, girdle bands of vegetative cells and auxospores scales are formed adjacent to the plasma membrane (Stoermer et al., 1965; Edgar and Pickett-Heaps, 1984; Lee and Li, 1992; Idei et al., 2012), possibly generated from the Golgi body (Lee and Li, 1992). Synurophytes and chrysophytes SDVs are located in the cytoplasmic or chloroplast endoplasmic reticulum. Bolidophyceae SDVs forming the shield and ventral plates are initially produced around the chloroplast and moving toward the plasma membrane like synurophytes and chrysophytes (Yamada et al., 2016). In contrast, SDVs for dorsal and girdle plates are formed adjacent to the plasma membrane like in diatoms. Such differentiation in the development site of SDVs depending on the type of silica plates within a single species has not been previously reported in other organisms (Yamada et al., 2016).
Mitotic Nuclear Division
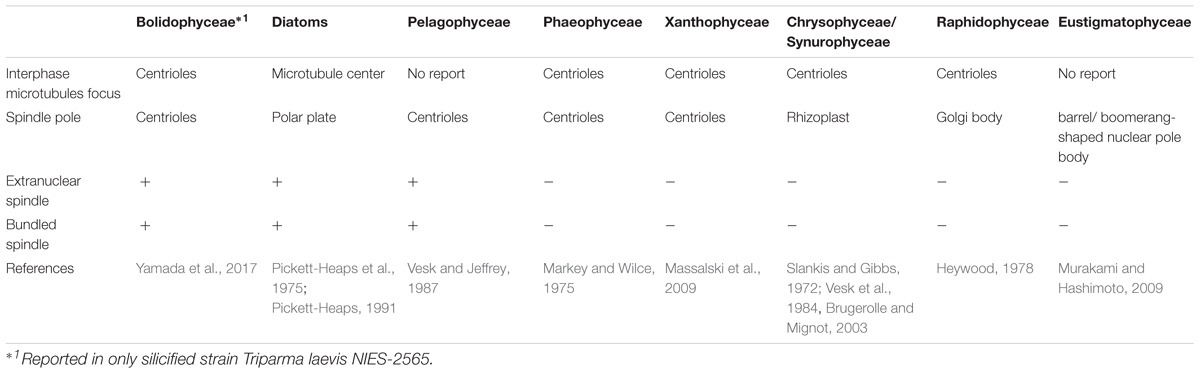
In eukaryotes, cell division, mitotic process, and related apparatus are often well conserved within high phylogenetic levels (e.g., at the class or phylum levels, Heath, 1980; Schmit and Nick, 2008; De Martino et al., 2009). Among Stramenopiles, the organelles related to the focus of interphase microtubules and spindle poles are the centrioles, like recently observed in Bolidophyceae (Yamada et al., 2017, Table 3). However, some classes have unique organelles (Table 3), such as the Microtubule Center (MC) and Polar Plate (PP) in diatoms (Pickett-Heaps et al., 1975; Tippit and Pickett-Heaps, 1977; Edgar and Pickett-Heaps, 1984; Pickett-Heaps, 1991; De Martino et al., 2009), the rhizoplast in chrysophytes and synurophytes (Slankis and Gibbs, 1972; Vesk et al., 1984; Brugerolle and Mignot, 2003) or a barrel/boomerang-shaped nuclear pole body in eustigmatophytes (Murakami and Hashimoto, 2009).
During the mitosis of T. laevis f. inornata, the interphase cell has more than four very short centrioles (ca. 80 nm in contrast to 150–500 nm of typical mature centrioles in other Stramenopiles, (Figure 11A). In prophase, the spindle bundle forms at in the extranuclear region (Figure 11B), the centrioles move to the spindle poles (Figure 11C) and then it moves to the cytoplasmic tunnel of the nucleus (Figure 11D). All along metaphase, the kinetochore microtubules elongate from the spindle poles to the condensed chromatin through the region of partially disintegrated nuclear envelope (Figure 11D). Finally, the chromatin is separated to both sides of the cell.
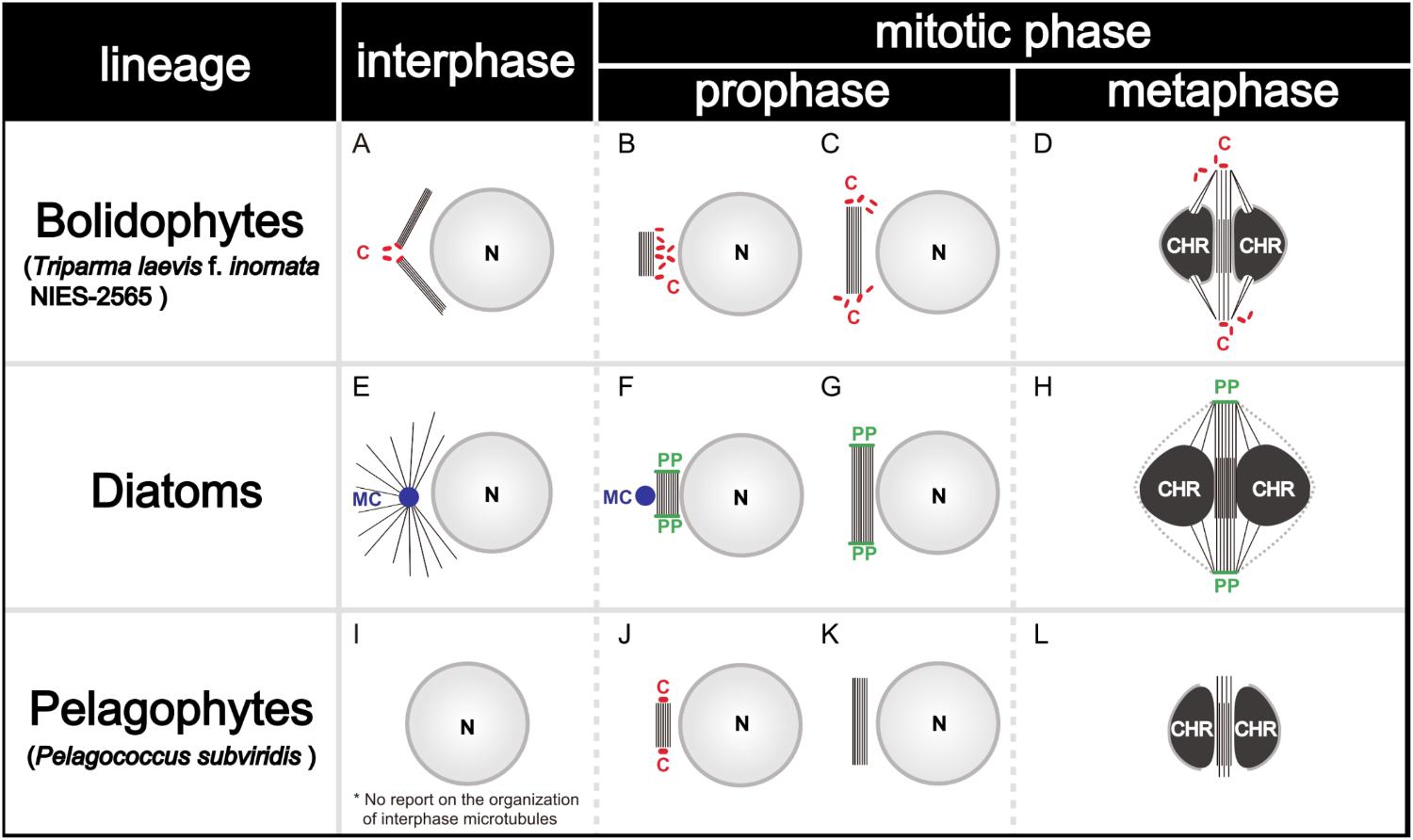
FIGURE 11. Graphical scheme of the interphase microtubule nucleation and the spindle formation in bolidophytes (A–D), diatoms (E–H), and pelagophytes (I–L). See text for details. Figures (A–D) and (E–H) are adapted from Yamada et al. (2017) and De Martino et al. (2009) with permission.
Spindle configuration and formation of T. laevis f. inornata are very similar to the process found among diatoms and pelagophytes (Table 3). They share two conspicuous characters: extranuclear spindle formation (Figures 11B,C,F,G) and the bundling of the interpolar microtubules (Figures 11D,H). However, the organelle serving as a Microtubule Organizing Center (MTOC) and its behavior differ. T. laevis f. inornata and pelagophytes have centrioles while diatoms have the specialized MC and PP (Figures 11E–H). The centrioles of pelagophytes (reported only in one species, Pelagococcus subviridis) appear only during the spindle initiation phase (Vesk and Jeffrey, 1987) while those of T. laevis f. inornata occur after central spindle formation in the extranuclear region. Since centriole is the most common organelle serving as MTOC in Stramenopiles (Table 3), the mitotic apparatus of T. laevis f. inornata shows more ancestral features than the diatoms.
Conclusion – the Evolutionary Relationships Between Diatoms and Bolidophyceae
Diatoms are highly diverse with 30,000 to 100,000 species (Mann and Vanormelingen, 2013) and constitute one of the top group of primary producers, contributing to about 20% of the photosynthesis on Earth, the equivalent of terrestrial rainforests (Nelson et al., 1995; Falkowski et al., 1998; Mann, 1999). They cover a wide size range from 2 μm to 2 mm and form large blooms in high-nutrient coastal and upwelling systems (Margalef, 1978; Hasle and Syvertsen, 1997). They are the main prey for zooplankton and the carbon that they fix through photosynthesis is efficiently transferred to higher trophic levels highlighted by their role in fish production (Ryther, 1969; Cushing, 1989). From an evolutionary point of view, the appearance of this highly productive group and the resulting increase in oceanic primary production may have driven the evolution of crustaceans, pelagic fish, and whales and shaped modern marine pelagic ecosystems (Parsons, 1979). Although, the origin and early evolution of diatoms remain controversial, the first well–preserved diatom fossils have been dated from ∼110 Myr ago, in the early Cretaceous (Gersonde and Harwood, 1990; Harwood and Gersonde, 1990), while molecular-clock-based estimations suggest that the origin of diatoms may have occurred 180 – 250 Myr ago (Medlin, 2011).
Recent multigene phylogenetic analyses suggest that bolidophytes, diatoms, pelagophytes, and dictyochophytes form a monophyletic lineage (Riisberg et al., 2009; Yang et al., 2012; Ševèíková et al., 2015; Derelle et al., 2016). This lineage, called Diatomiista (Derelle et al., 2016) or Khakista (Riisberg et al., 2009), originally only included diatoms and bolidophytes (Cavalier-Smith and Chao, 2006).
Recent success in the isolation of strains of both silicified and naked flagellated Bolidophyceae species allow detailed phylogenetic studies, clarifying the taxonomic position of this group as a sister group of diatoms and revealing the close relationship between silicified and naked strains. Cell wall formation and mitotic division in the silicified species T. laevis f. inornata have intermediate features between diatom and more ancient stramenopiles (Figure 11 and Tables 3, 4). Analysis of organellar genomes of this species also suggested that it displays more ancestral characteristics than diatoms (Tajima et al., 2016). Gene contents of the plastid and mitochondrial genomes are similar between T. laevis f. inornata and diatoms whereas the gene order of the mitochondrial genome is different. The structure of the mitochondrial genome is also more compact in T. laevis f. inornata than in diatoms since the latter species has no introns or repeat regions which are often observed in some diatoms species (Oudot-Le Secq and Green, 2011).
The phylogenetically close relationship between silicified and naked Bolidophyceae strains and recent occasional observation of flagellated cells in cultures of T. laevis f. inornata (Ichinomiya et al., 2016) suggest that bolidophytes may have a life cycle that switches between silicified non-flagellated and naked flagellate stages. This hypothetical life cycle has similarities to centric diatoms for which the diploid vegetative stage produces haploid naked flagellated cells (male gametes or spermatozoa) during sexual reproduction (Drebes, 1977; Vaulot and Chisholm, 1987). The origin, early evolution and key processes for the acquisition of the silica cell wall are not yet been fully understood and Bolidophyceae could play a key role in answering these questions. More comprehensive analyses, including whole genome sequences, observations of the life-cycle, and fossil records, would lead a deeper understanding of the evolutionary relationships between Bolidophyceae and diatoms.
Author Contributions
DV analyzed GenBank and metabarcode sequences and draw distribution maps. MT analyzed the OSD metabarcoding data. ALdS performed ITS secondary structure. MI and AK analyzed the distribution of silicified species. AK, KY, MI, SY, DV, and ALdS wrote the manuscript and contributed to the final editing.
Funding
Financial support for this work was provided by the following projects: Grants-in-Aid for Scientific Research 22657027, 23370046, 26291085, 15K14784, and 17H03724 from the Japan Society for the Promotion of Science (JSPS), the Canon Foundation, ANR PhytoPol (ANR-15-CE02-0007), ECCOS MicrAntar (C16B02), TaxMArc (Research Council of Norway, 268286/E40), and CONICYT/FONDECYT regular PiSCOSouth (1171802). MT was supported by a Ph.D. fellowship from the Université Pierre et Marie Curie and the Région Bretagne.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to Adam Monier, Katja Metfies, Estelle Kilias, and Wei Luo for communicating to us their raw metabarcoding data, and Mary-Hélène Noël for providing us with a SEM image of T. eleuthera. We would also like to thank the Ocean Sampling Day consortium for providing sequence data and the ABIMS platform in Roscoff for access to bioinformatics resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00370/full#supplementary-material and https://doi.org/10.6084/m9.figshare.5549638.
References
Azam, F., Fenchel, T., Field, J., Gray, J., Meyer-Reil, L., and Thingstad, F. (1983). The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10, 257–263. doi: 10.3354/meps010257
Booth, B. C., Lewin, J., and Norris, R. E. (1980). Siliceous nanoplankton I. Newly discovered cysts from the Gulf of Alaska. Mar. Biol. 58, 205–209. doi: 10.1007/BF00391877
Booth, B. C., and Marchant, H. J. (1987). Parmales, a new order of marine chrysophytes, with descriptions of three new genera and seven new species. J. Phycol. 23, 245–260. doi: 10.1111/j.1529-8817.1987.tb04132.x
Booth, B. C., and Marchant, H. J. (1988). Triparmaceae, a substitute name for a family in the order Parmales (Chrysophyceae). J. Phycol. 24:124. doi: 10.1111/j.1529-8817.1988.tb04467.x
Bravo-Sierra, E., and Hernández-Becerril, D. U. (2003). Parmales (Chrysophyceae) from the Gulf of Tehuantepec, Mexico, including the description of a new species, Tetraparma insecta sp Nov., and a proposal to the taxonomy of the group. J. Phycol. 39, 577–583. doi: 10.1046/j.1529-8817.2003.02181.x
Brugerolle, G., and Mignot, J. P. (2003). The rhizoplast of chrysomonads, a basal body-nucleus connector that polarises the dividing spindle. Protoplasma 222, 13–21. doi: 10.1007/s00709-003-0016-4
Brzezinski, M., Olson, R., and Chisholm, S. (1990). Silicon availability and cell-cycle progression in marine diatoms. Mar. Ecol. Prog. Ser. 67, 83–96. doi: 10.3354/meps067083
Caisová, L., Marin, B., and Melkonian, M. (2011). A close-up view on ITS2 evolution and speciation - a case study in the Ulvophyceae (Chlorophyta, Viridiplantae). BMC Evol. Biol. 11:262. doi: 10.1186/1471-2148-11-262
Caisová, L., Marin, B., and Melkonian, M. (2013). A consensus secondary structure of ITS2 in the Chlorophyta identified by phylogenetic reconstruction. Protist 164, 482–496. doi: 10.1016/j.protis.2013.04.005
Cavalier-Smith, T., and Chao, E. E. Y. (2006). Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J. Mol. Evol. 62, 388–420. doi: 10.1007/s00239-004-0353-8
Coleman, A. W. (2007). Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 35, 3322–3329. doi: 10.1093/nar/gkm233
Coleman, A. W. (2009). Is there a molecular key to the level of “biological species” in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 50, 197–203. doi: 10.1016/j.ympev.2008.10.008
Comeau, A. M., Li, W. K. W., Tremblay, J. -É., Carmack, E. C., and Lovejoy, C. (2011). Arctic Ocean microbial community structure before and after the 2007 record sea ice minimum. PLoS One 6:e27492. doi: 10.1371/journal.pone.0027492
Cushing, D. H. (1989). A difference in structure between in strongly stratified waters and in those that are only weakly stratified. J. Plankton. Res. 11, 1–13. doi: 10.1093/plankt/11.1.1
Darley, W. M., and Volcani, B. E. (1969). Role of silicon in diatom metabolism: a silicon requirement for deoxyribonucleic acid synthesis in the diatom Cylindrotheca fusiformis Reimann and Lewin. Exp. Cell Res. 58, 334–342. doi: 10.1016/0014-4827(69)90514-x
Daugbjerg, N., and Guillou, L. (2001). Phylogenetic analyses of Bolidophyceae (Heterokontophyta) using rbcL gene sequences support their sister group relationship to diatoms. Phycologia 40, 153–161. doi: 10.2216/i0031-8884-40-2-153.1
De Martino, A., Amato, A., and Bowler, C. (2009). Mitosis in diatoms: rediscovering an old model for cell division. Bioessays 31, 874–884. doi: 10.1002/bies.200900007
Derelle, R., López-García, P., Timpano, H., and Moreira, D. (2016). A phylogenomic framework to study the diversity and evolution of stramenopiles (=Heterokonts). Mol. Biol. Evol. 33, 2890–2898. doi: 10.1093/molbev/msw168
Drebes, G. (1977). “Sexuality,” in The Biology of Diatoms, ed. D. Werner (Oxford: Blackwell Scientific Publications), 250–283.
Edgar, L. A., and Pickett-Heaps, J. D. (1984). Valve morphogenesis in the pennate diatom Navicula cuspidata. J. Phycol. 20, 47–61. doi: 10.1111/j.0022-3646.1984.00047.x
Falkowski, P. G., Barber, R. T., and Smetacek, V. (1998). Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206. doi: 10.1126/science.281.5374.200
Finkel, Z. V. (2016). “Silicification in the microalgae,” in The Physiology of Microalgae, eds M. A. Borowitzka, J. Beardall, and J. A. Raven (Basel: Springer International Publishing), 289–300. doi: 10.1007/978-3-319-24945-2_13
Fujita, R., and Jordan, R. W. (2017). Tropical Parmales (Bolidophyceae) assemblages from the Sulu Sea and South China Sea, including the description of five new taxa. Phycologia 56, 499–509. doi: 10.2216/16-128.1
Gersonde, R., and Harwood, D. M. (1990). “Lower Cretaceous diatoms from ODP Leg 113 Site 693 (Weddell Sea). Part 1: vegetative cells,” in Proceedings of the Ocean Drilling Program, Scientific Results, College Station, TX, 365–402.
Gobler, C. J., and Sunda, W. G. (2012). Ecosystem disruptive algal blooms of the brown tide species, Aureococcus anophagefferens and Aureoumbra lagunensis. Harmful Algae 14, 36–45. doi: 10.1016/j.hal.2011.10.013
Guillou, L., Bachar, D., Audic, S., Bass, D., Berney, C., Bittner, L., et al. (2013). The protist ribosomal reference database (PR2): a catalog of unicellular eukaryote Small SubUnit rRNA sequences with curated taxonomy. Nucleic Acids Res. 41, D597–D604. doi: 10.1093/nar/gks1160
Guillou, L., Chrétiennot-Dinet, M.-J., Medlin, L. K., Claustre, H., Loiseaux-de Goër, S., and Vaulot, D. (1999a). Bolidomonas: a new genus with two species belonging to a new algal class, the Bolidophyceae (Heterokonta). J. Phycol. 35, 368–381. doi: 10.1046/j.1529-8817.1999.3520368.x
Guillou, L., Moon-van der Staay, S. Y., Claustre, H., Partensky, F., and Vaulot, D. (1999b). Diversity and abundance of Bolidophyceae (Heterokonta) in two oceanic regions. Appl. Environ. Microbiol. 65, 4528–4536.
Harwood, D. M., and Gersonde, R. (1990). “Lower cretaceous diatoms from ODP Leg 113 site 693 (Weddell Sea). Part 2?: resting spores, chrysophycean cysts, an endoskeletal dinoflagellate, and notes on the origin of diatoms,” in Proceedings of the Ocean Drilling Program, Scientific Results, Vol. 113, College Station, TX, 403–425. doi: 10.2973/odp.proc.sr.113.201.1990
Hasle, G. R., and Syvertsen, E. E. (1997). “Marine diatoms,” in Identifying Marine Phytoplankton, ed. C. R. Tomas (San Diego, CA: Academic Press), 5–385.
Heath, I. B. (1980). Variant mitoses in lower eukaryotes: indicators of the evolution of mitosis? Int. Rev. Cytol. 64, 1–80. doi: 10.1016/S0074-7696(08)60235-1
Henriksen, P., Knipschildt, F., Moestrup,Ø., and Thomsen, H. A. (1993). Autecology, life history and toxicology of the silicoflagellate Dictyocha speculum (Silicoflagellata, Dictyochophyceae). Phycologia 32, 29–39. doi: 10.2216/i0031-8884-32-1-29.1
Heywood, P. (1978). Ultrastructure of mitosis in the chloromonadophycean alga Vacuolaria virescens. J. Cell Sci. 31, 37–51.
Ichinomiya, M., dos Santos, A. L., Gourvil, P., Yoshikawa, S., Kamiya, M., Ohki, K., et al. (2016). Diversity and oceanic distribution of the Parmales (Bolidophyceae), a picoplanktonic group closely related to diatoms. ISME J. 10, 2419–2434. doi: 10.1038/ismej.2016.38
Ichinomiya, M., and Kuwata, A. (2015). Seasonal variation in abundance and species composition of the Parmales community in the Oyashio region, western North Pacific. Aquat. Microb. Ecol. 75, 207–223. doi: 10.3354/ame01756
Ichinomiya, M., and Kuwata, A. (2017). Establishment of first ever culture revealed an unidentified algal taxa: the Parmales (in Japanese). Jpn. J. Phycol. 65, 153–158.
Ichinomiya, M., Nakamachi, M., Shimizu, Y., and Kuwata, A. (2013). Growth characteristics and vertical distribution of Triparma laevis (Parmales) during summer in the Oyashio region, western North Pacific. Aquat. Microb. Ecol. 68, 107–116. doi: 10.3354/ame01606
Ichinomiya, M., Yoshikawa, S., Kamiya, M., Ohki, K., Takaichi, S., and Kuwata, A. (2011). Isolation and characterization of Parmales (Heterokonta/Heterokontophyta/Stramenopiles) from the Oyashio region, Western North Pacific. J. Phycol. 47, 144–151. doi: 10.1111/j.1529-8817.2010.00926.x
Idei, M., Osada, K., Sato, S., Toyoda, K., Nagumo, T., and Mann, D. G. (2012). Gametogenesis and auxospore development in Actinocyclus (Bacillariophyta). PLoS One 7:e41890. doi: 10.1371/journal.pone.0041890
Iwai, T., and Nishida, S. (1976). The distribution of modern coccolithophores in the North Pacific. News Osaka Micropaleontol. 5, 1–11.
Jacquet, S., Partensky, F., Lennon, J. F., and Vaulot, D. (2001). Diel patterns of growth and division in marine picoplankton in culture. J. Phycol. 37, 357–369. doi: 10.1046/j.1529-8817.2001.037003357.x
Johnson, P. W., and Sieburth, J. M. N. (1982). In-situ morphology and occurrence of eucaryotic phototrophs of bacterial size in the picoplankton of estuarine and oceanic waters. J. Phycol. 18, 318–327. doi: 10.1111/j.1529-8817.1982.tb03190.x
Kataoka, T., Yamaguchi, H., Sato, M., Watanabe, T., Taniuchi, Y., Kuwata, A., et al. (2017). Seasonal and geographical distribution of near-surface small photosynthetic eukaryotes in the western North Pacific determined by pyrosequencing of 18S rDNA. FEMS Microbiol. Ecol. 93:fiw229. doi: 10.1093/femsec/fiw229
Kilias, E., Wolf, C., Nöthig, E.-M., Peeken, I., and Metfies, K. (2013). Protist distribution in the Western Fram Strait in summer 2010 based on 454-pyrosequencing of 18S rDNA. J. Phycol. 49, 996–1010. doi: 10.1111/jpy.12109
Knoll, A. H., and Kotrc, B. (2015). “Protistan skeletons: a geologic history of evolution and constraints,” in Evolution of Lightweight Structures, ed. C. Hamm (Dordrecht: Springer), 1–16. doi: 10.1007/978-94-017-9398-8
Komuro, C., Narita, H., Imai, K., Nojiri, Y., and Jordan, R. W. (2005). Microplankton assemblages at Station KNOT in the subarctic western Pacific, 1999-2000. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 52, 2206–2217. doi: 10.1016/j.dsr2.2005.08.006
Konno, S., and Jordan, R. W. (2007). An amended terminology for the Parmales (Chrysophyceae). Phycologia 46, 612–616. doi: 10.2216/07-29.1
Konno, S., Ohira, R., Harada, N., and Jordan, R. W. (2007). Six new taxa of subarctic Parmales (Chrysophyceae ). J. Nannoplankt. Res. 29, 108–128.
Kopf, A., Bicak, M., Kottmann, R., Schnetzer, J., Kostadinov, I., Lehmann, K., et al. (2015). The ocean sampling day consortium. Gigascience 4:27. doi: 10.1186/s13742-015-0066-5
Kosman, C. A., Thomsen, H. A., and Østergaard, J. B. (1993). Parmales (Chrysophyceae) from Mexican, Californian, Baltic, Arctic and Antarctic waters with the description of a new subspecies and several new forms. Phycologia 32, 116–128. doi: 10.2216/i0031-8884-32-2-116.1
Leadbeater, B. S. C., and Barker, D. A. N. (1995). “Biomineralization and scale production in the Chrysophyta,” in Chrysophyte Algae: Ecology, Phylogeny and Development, eds C. D. Sandgren, J. P. Smol, and J. Kristiansen (Cambridge: Cambridge University press), 141–164. doi: 10.1017/CBO9780511752292.008
Lee, M., and Li, C. W. (1992). The origin of the silica deposition vesicle of diatoms. Bot. Bull. Acad. Sin. 33, 317–325.
Luo, W., Li, H., Gao, S., Yu, Y., Lin, L., and Zeng, Y. (2015). Molecular diversity of microbial eukaryotes in sea water from Fildes Peninsula, King George Island, Antarctica. Polar Biol. 39, 605–616. doi: 10.1007/s00300-015-1815-8
Mai, J. C., and Coleman, A. W. (1997). The internal transcribed spacer 2 exhibits a common secondary structure in green algae and flowering plants. J. Mol. Evol. 44, 258–271. doi: 10.1007/PL00006143
Mann, D. G. (1999). The species concept in diatoms. Phycologia 38, 437–495. doi: 10.2216/i0031-8884-38-6-437.1
Mann, D. G., and Marchant, H. J. (1989). “The origins of the diatom and its life cycle,” in The Chromophyte Algae: Problems and Perspectives, eds J. C. Green, B. S. C. Leadbeater, and W. L. Diver (Oxford: Clarendon Press), 307–323.
Mann, D. G., and Vanormelingen, P. (2013). An inordinate fondness? The number, distributions, and origins of diatom species. J. Eukaryot. Microbiol. 60, 414–420. doi: 10.1111/jeu.12047
Marchant, H., and Nash, G. (1986). Electron microscopy of gut contents and faeces of Euphausia superba Dana. Mem. Natl. Inst. Polar Res. Spec. Issue 40, 167–177.
Marchant, H. J., and McEldowney, A. (1986). Nanoplanktonic siliceous cysts from Antarctica are algae. Mar. Biol. 92, 53–57. doi: 10.1007/BF00392745
Margalef, R. (1978). Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanol. Acta 1, 493–509. doi: 10.1007/BF00202661
Markey, D. R., and Wilce, R. T. (1975). The ultrastructure of reproduction in the brown alga Pylaiella littoralis. Protoplasma 85, 219–241. doi: 10.1007/BF01567948
Marron, A. O., Ratcliffe, S., Wheeler, G. L., Goldstein, R. E., King, N., Not, F., et al. (2016). The evolution of silicon transport in eukaryotes. Mol. Biol. Evol. 33, 3226–3248. doi: 10.1093/molbev/msw209
Martin-Jézéquel, V., Hildebrand, M., and Brzezinski, M. A. (2000). Silicon metabolism in diatoms: implications for growth. J. Phycol. 36, 821–840. doi: 10.1046/j.1529-8817.2000.00019.x
Massalski, A., Kostikov, I., Olech, M., and Hoffmann, L. (2009). Mitosis, cytokinesis and multinuclearity in a Xanthonema (Xanthophyta) isolated from Antarctica. Eur. J. Phycol. 44, 263–275. doi: 10.1080/09670260802636274
Medlin, L. K. (2011). “A review of the evolution of the diatoms from the origin of the lineage to their populations,” in The Diatom World, eds J. Seckbach and P. Kociolek (Dordrecht: Springer), 94–118. doi: 10.1007/978-94-007-1327-7_4
Metfies, K., von Appen, W.-J., Kilias, E., Nicolaus, A., and Nöthig, E.-M. (2016). Biogeography and photosynthetic biomass of arctic marine pico-eukaroytes during summer of the record sea ice minimum 2012. PLoS One 11: e0148512. doi: 10.1371/journal.pone.0148512
Monier, A., Comte, J., Babin, M., Forest, A., Matsuoka, A., and Lovejoy, C. (2014). Oceanographic structure drives the assembly processes of microbial eukaryotic communities. ISME J. 9, 990–1002. doi: 10.1038/ismej.2014.197
Monier, A., Terrado, R., Thaler, M., Comeau, A., Medrinal, E., and Lovejoy, C. (2013). Upper Arctic Ocean water masses harbor distinct communities of heterotrophic flagellates. Biogeosciences 10, 4273–4286. doi: 10.5194/bg-10-4273-2013
Müller, T., Philippi, N., Dandekar, T., Schultz, J., and Wolf, M. (2007). Distinguishing species. RNA 13, 1469–1472. doi: 10.1261/rna.617107
Murakami, R., and Hashimoto, H. (2009). Unusual nuclear division in Nannochloropsis oculata (Eustigmatophyceae, Heterokonta) which may ensure faithful transmission of secondary plastids. Protist 160, 41–49. doi: 10.1016/j.protis.2008.09.002
Nelson, D. M., Tréguer, P., Brzezinski, M. A., Leynaert, A., and Quéguiner, B. (1995). Production and dissolution of biogenic silica in the ocean: revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Glob. Biogeochem. Cycles 9, 359–372. doi: 10.1029/95GB01070
Not, F., Siano, R., Kooistra, W. H. C. F., Simon, N., Vaulot, D., and Probert, I. (2012). Diversity and ecology of eukaryotic marine phytoplankton. Adv. Bot. Res. 64, 1–53. doi: 10.1016/B978-0-12-391499-6.00001-3
Not, F., Simon, N., Biegala, I., and Vaulot, D. (2002). Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat. Microb. Ecol. 28, 157–166. doi: 10.3354/ame028157
Okita, T. W., and Volcani, B. E. (1978). Role of silicon in diatom metabolism IX. Differential synthesis of DNA polymerases and DNA-binding proteins during silicate starvation and recovery in Cylindrotheca fusiformis. Biochem. Biophys. Acta 519, 76–86.
Oudot-Le Secq, M. P., and Green, B. R. (2011). Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 476, 20–26. doi: 10.1016/j.gene.2011.02.001
Parsons, T. R. (1979). Some ecological, experimental and evolutionary aspets of the upwelling ecosystem. S. Afr. J. Sci. 75, 536–540.
Pickett-Heaps, J. D. (1991). Cell division in diatoms. Int. Rev. Cytol. 128, 63–108. doi: 10.1016/S0074-7696(08)60497-0
Pickett-Heaps, J. D., McDonald, K. L., and Tippit, D. H. (1975). Cell division in the pennate diatom Diatoma vulgare. Protoplasma 86, 205–242. doi: 10.1007/BF01275633
Preisig, H. R. (1994). Siliceous structures and silicification in flagellated protists. Protoplasma 181, 29–42. doi: 10.1007/BF01666387
Riisberg, I., Orr, R. J. S., Kluge, R., Shalchian-Tabrizi, K., Bowers, H. A., Patil, V., et al. (2009). Seven gene phylogeny of Heterokonts. Protist 160, 191–204. doi: 10.1016/j.protis.2008.11.004
Ryther, J. H. (1969). Photosynthesis and fish production in the sea. Science 166, 72–76. doi: 10.1126/science.166.3901.72
Sandgren, C. D., Hall, S. A., and Barlow, S. B. (1996). Siliceous scale production in chrysophyte and synurophyte algae. I. Effects of silica-limited growth on cell silica content, scale morphology, and the construction of the scale layer of Synura petersenii. J. Phycol. 32, 675–692. doi: 10.1111/j.0022-3646.1996.00675.x
Sarthou, G., Timmermans, K. R., Blain, S., and Tréguer, P. (2005). Growth physiology and fate of diatoms in the ocean: a review. J. Sea Res. 53, 25–42. doi: 10.1016/j.seares.2004.01.007
Schmit, A. C., and Nick, P. (2008). Microtubules and the evolution of mitosis. Plant Cell Monogr. 11, 233–266. doi: 10.1007/7089_2007_161
Ševèíková, T., Horák, A., Klimeš, V., Zbránková, V., Demir-Hilton, E., Sudek, S., et al. (2015). Updating algal evolutionary relationships through plastid genome sequencing: did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 5:10134. doi: 10.1038/srep10134
Shimizu, K., Del Amo, Y., Brzezinski, M. A., Stucky, G. D., and Morse, D. E. (2001). A novel fluorescent silica tracer for biological silicification studies. Chem. Biol. 8, 1051–1060. doi: 10.1016/S1074-5521(01)00072-2
Shimizu, Y., Takahashi, K., Ito, S. I., Kakehi, S., Tatebe, H., Yasuda, I., et al. (2009). Transport of subarctic large copepods from the oyashio area to the mixed water region by the coastal oyashio intrusion. Fish. Oceanogr. 18, 312–327. doi: 10.1111/j.1365-2419.2009.00513.x
Silver, M. W., Mitchell, J. G., and Ringo, D. L. (1980). Siliceous nanoplankton. II. Newly discovered cysts and abundant choanoflagellates from the Weddell Sea, Antarctica. Mar. Biol. 58, 211–217. doi: 10.1007/BF00391878
Simpson, T. L., and Volcani, B. E. (1981). Silicon and Siliceous Structures in Biological Systems. Hamburg: Springer Verlag Gmbh, doi: 10.1017/CBO9781107415324.004
Slankis, T., and Gibbs, S. P. (1972). The fine structure of mitosis and cell division in the chrysophycean alga Ochromonas danica. J. Phycol. 8, 243–256. doi: 10.1111/j.1529-8817.1972.tb04035.x
Stawiarski, B., Buitenhuis, E. T., and Le Quéré, C. (2016). The physiological response of picophytoplankton to temperature and its model representation. Front. Mar. Sci. 3:164. doi: 10.3389/fmars.2016.00164
Stoermer, E. F., Pankratz, H. S., and Bowen, C. C. (1965). Fine structure of the diatom Amphipleura pellucida. II. Cytoplasmic fine structure and frustule formation. Am. J. Bot. 52, 1067–1078. doi: 10.1002/j.1537-2197.1965.tb07286.x
Suzuki, K., Kuwata, A., Yoshie, N., Shibata, A., Kawanobe, K., and Saito, H. (2011). Population dynamics of phytoplankton, heterotrophic bacteria, and viruses during the spring bloom in the western subarctic Pacific. Deep Sea Res. Part 1 Oceanogr. Res. Pap. 58, 575–589. doi: 10.1016/j.dsr.2011.03.003
Tajima, N., Saitoh, K., Sato, S., Maruyama, F., Ichinomiya, M., Yoshikawa, S., et al. (2016). Sequencing and analysis of the complete organellar genomes of Parmales, a closely related group to Bacillariophyta (diatoms). Curr. Genet. 62, 887–896. doi: 10.1007/s00294-016-0598-y
Takahashi, K., Kuwata, A., Saito, H., and Ide, K. (2008). Grazing impact of the copepod community in the Oyashio region of the western subarctic Pacific Ocean. Prog. Oceanogr. 78, 222–240. doi: 10.1016/j.pocean.2008.06.002
Taniguchi, A., Suzuki, T., and Shimada, S. (1995). Growth characteristics of Parmales (Chrysophyceae) observed in bag cultures. Mar. Biol. 123, 631–638. doi: 10.1007/BF00349241
Thomas, S. L., and Campbell, D. A. (2013). Photophysiology of Bolidomonas pacifica. J. Plankton Res. 35, 260–269. doi: 10.1093/plankt/fbs105
Tippit, D. H., and Pickett-Heaps, J. D. (1977). Mitosis in the pennate diatom Surirella ovalis. J. Cell Biol. 73, 705–727. doi: 10.1083/jcb.73.3.705
Urban, J. L., McKenzie, C. H., and Deibel, D. (1993). Nanoplankton found in fecal pellets of macrozooplankton in coastal Newfoundland waters. Bot. Mar. 36, 267–281. doi: 10.1515/botm.1993.36.4.267
Vaulot, D., and Chisholm, S. W. (1987). Flow cytometric analysis of spermatogenesis in the diatom Thalassiosira weissflogii (Bacillariophyceae). J. Phycol. 23, 132–137. doi: 10.1111/j.1529-8817.1987.tb04435.x
Vaulot, D., Eikrem, W., Viprey, M., and Moreau, H. (2008). The diversity of small eukaryotic phytoplankton ( ≤ 3 μm) in marine ecosystems. FEMS Microbiol. Rev. 32, 795–820. doi: 10.1111/j.1574-6976.2008.00121.x
Vaulot, D., Olson, R. J., Merkel, S., and Chisholm, S. W. (1987). Cell-cycle response to nutrient starvation in two phytoplankton species, Thalassiosira weissflogii and Hymenomonas carterae. Mar. Biol. 95, 625–630. doi: 10.1007/BF00393106
Vesk, M., Hoffman, L. R., and Pickett-Heaps, J. D. (1984). Mitosis and cell division in Hydrurus foetidus (Chrysophyceae). J. Phycol. 20, 461–470. doi: 10.1111/j.0022-3646.1984.00461.x
Vesk, M., and Jeffrey, S. W. (1987). Ultrastructure and pigments of two strains of the picoplanktonic alga Pelagococcus subviridis (Chrysophyceae). J. Phycol. 23, 322–336. doi: 10.1111/j.1529-8817.1987.tb04141.x
Waterbury, J. B., Watson, S. W., Guillard, R. R. L., and Brand, L. E. (1979). Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature 277, 293–294. doi: 10.1038/277293a0
Wolf, C., Frickenhaus, S., Kilias, E. S., Peeken, I., and Metfies, K. (2014). Protist community composition in the Pacific sector of the Southern Ocean during austral summer 2010. Polar Biol. 37, 375–389. doi: 10.1007/s00300-013-1438-x
Wu, W., Wang, L., Liao, Y., Xu, S., and Huang, B. (2017). Spatial and seasonal distributions of photosynthetic picoeukaryotes along an estuary to basin transect in the northern South China Sea. J. Plankton Res. 39, 423–435. doi: 10.1093/plankt/fbx017
Yamada, K., Nagasato, C., Motomura, T., Ichinomiya, M., Kuwata, A., Kamiya, M., et al. (2017). Mitotic spindle formation in Triparma laevis NIES-2565(Parmales, Heterokontophyta). Protoplasma 254, 461–471. doi: 10.1007/s00709-016-0967-x
Yamada, K., Yoshikawa, S., Ichinomiya, M., Kuwata, A., Kamiya, M., and Ohki, K. (2014). Effects of silicon-limitation on growth and morphology of Triparma laevis NIES-2565 (Parmales, Heterokontophyta). PLoS One 9:e103289. doi: 10.1371/journal.pone.0103289
Yamada, K., Yoshikawa, S., Ohki, K., Ichinomiya, M., Kuwata, A., Motomura, T., et al. (2016). Ultrastructural analysis of siliceous cell wall regeneration in the stramenopile Triparma laevis (Parmales, Bolidophyceae). Phycologia 55, 602–609. doi: 10.2216/16-32.1
Keywords: bolidophyceae, parmales, diatoms, genetic diversity, mitosis, geographical distribution, seasonal dynamics and silicification
Citation: Kuwata A, Yamada K, Ichinomiya M, Yoshikawa S, Tragin M, Vaulot D and Lopes dos Santos A (2018) Bolidophyceae, a Sister Picoplanktonic Group of Diatoms – A Review. Front. Mar. Sci. 5:370. doi: 10.3389/fmars.2018.00370
Received: 05 December 2017; Accepted: 25 September 2018;
Published: 29 October 2018.
Edited by:
Brivaela Moriceau, UMR6539 Laboratoire des Sciences de L’environnement Marin (LEMAR), FranceReviewed by:
Jun Sun, Tianjin University of Science and Technology, ChinaYantao Liang, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), China
Copyright © 2018 Kuwata, Yamada, Ichinomiya, Yoshikawa, Tragin, Vaulot and Lopes dos Santos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akira Kuwata, YWt1d2F0YUBhZmZyYy5nby5qcA==
 Akira Kuwata
Akira Kuwata Kazumasa Yamada
Kazumasa Yamada Mutsuo Ichinomiya2
Mutsuo Ichinomiya2 Shinya Yoshikawa
Shinya Yoshikawa Margot Tragin
Margot Tragin Daniel Vaulot
Daniel Vaulot Adriana Lopes dos Santos
Adriana Lopes dos Santos