- 1Department of Microbiology, University of Bergen, Bergen, Norway
- 2Department of Arctic and Marine Biology, UiT – The Arctic University of Tromsø, Tromsø, Norway
The warming of the Arctic causes increased riverine discharge, coastal erosion, and the thawing of permafrost. Together, this is leading to an increased wash out of terrestrial dissolved organic matter (tDOM) into the coastal Arctic ecosystems. This tDOM may be anticipated to affect both carbon and nutrient flow in the microbial food web and microbial community composition, but there are few studies detailing this in Arctic marine ecosystems. We tested the effects of tDOM on the bacterial community composition and net-growth by extracting DOM from the active layer of permafrost soil and adding the aged tDOM concentrate to a natural microbial fjord community (Kongsfjorden, NW Svalbard). This resulted in an increased carbon load of 128 μM DOC in the tDOM treatment relative to the control of 83 μM DOC. We observed changes in community composition and activity in incubations already within 12 h where tDOM was added. Flow cytometry revealed that predominantly large bacteria increased in the tDOM treated incubations. The increase of this group correlated with the increase in relative abundance of the genus Glaciecola (Gammaproteobacteria). Glaciecola were initially not abundant in the bacterial community (0.6%), but their subsequent increase up to 47% after 4 days upon tDOM addition compared to 8% in control incubations indicates that they are likely capable of degrading permafrost derived DOM. Further, according to our experimental results we hypothesize that the tDOM addition increased bacterivorous grazing by small protists and thus tDOM might indirectly also effect higher trophic levels of the microbial food web.
Introduction
The Arctic is experiencing a warming at nearly twice the global rate, with drastic changes for the ecosystem (Trenberth and Josey, 2007; Screen and Simmonds, 2010; Vincent, 2010). Higher sea surface temperatures, melting sea ice, and increased freshwater input from large Arctic rivers, transporting nutrients, and terrestrial organic matter into the ocean, have multiple implications for the marine carbon cycle (Li et al., 2009; Doney et al., 2012; Fichot et al., 2013; Holmes et al., 2013; El-Swais et al., 2015). Higher temperatures are on the one hand responsible for a decreasing sea ice cover, which in turn may enhance primary production and thus the biological carbon pump (CO2 burial), but on the other hand could also increase the rate of bacterial degradation of phytoplankton derived dissolved organic matter (DOM) (CO2 production) (Wohlers et al., 2009). This bacterial transformation of phytoplankton derived DOM might lead to the accumulation of more complex humic-like organic matter via the microbial carbon pump (Jiao et al., 2010).
In the Arctic, another source of DOM comes from permafrost soil organic matter and enters the Arctic Ocean via rivers (Feng et al., 2013; Holmes et al., 2013). Estimations show that mobilization of DOM has increased up to 6% from 1985 to 2004 (Feng et al., 2013) and will further increase under the current warming climate (Amon et al., 2012). Yearly, about 3,300 km3 of freshwater stream into the Arctic Ocean and influence stratification, light absorption, surface temperature, gas exchange, productivity, and carbon sequestration (Rachold et al., 2004). This input is often characterized by a high dissolved organic carbon (DOC) concentration, reaching more than 1,000 μmol kg−1, compared to open ocean concentrations of around 80 μmol kg−1 (Dittmar and Kattner, 2003; Hansell et al., 2009; Stedmon et al., 2011). The quality of the DOC has in some studies been described to be mainly refractory (Opsahl et al., 1999; Dittmar and Kattner, 2003; Xie et al., 2012), while other studies showed that up to 40% can be degraded within weeks up to months (Hansell, 2004; Holmes et al., 2008; Vonk et al., 2013; Sipler et al., 2017a). Thus, it is still disputed whether Arctic tDOM can represent an important carbon source for marine bacteria, leading to increased CO2 production and how this may affect the marine trophic network via the microbial loop.
Several studies have examined the ability of bacteria to degrade the seasonally available phytoplankton derived DOM and found that an increase of such carbon sources influences both the structure and the activity of the bacterial community (Pinhassi et al., 2004; Sapp et al., 2007; Teeling et al., 2012). Especially a versatile group of Gammaproteobacteria, belonging to the order Alteromonadales, responds immediately both in abundance and activity, when phytoplankton derived DOM becomes available (Eilers et al., 2000; McCarren et al., 2010; Pedler et al., 2014; Beier et al., 2015; von Scheibner et al., 2017). Only few studies have investigated the effects of terrestrial derived DOM on marine microbial community structure and activity (Herlemann et al., 2014, 2017; Blanchet et al., 2017; Traving et al., 2017), of which even less have been conducted in the coastal Arctic (Sipler et al., 2017a). Common for all studies is an observed shift in bacterial community structure due to the addition of tDOM. There is a need to understand how this community shift might affect higher trophic levels in order to better understand climate change impacts on the marine Arctic ecosystem. A higher bacterial activity due to the degradation of tDOM might cause a higher turnover within the microbial loop and therewith increased CO2 production, but ultimately depends on the bacterial growth efficiency. Increased carbon availability might also enhance the competition between bacteria and phytoplankton for inorganic nutrients and indirectly disadvantage larger phytoplankton (Thingstad et al., 2008; Sipler et al., 2017b). Thus, high tDOM input may decrease primary production in coastal Arctic areas.
We here studied the impact of permafrost-derived DOM on an Arctic fjord microbial community using 16S rRNA amplicon sequencing and followed the changes over the course of a nine-day incubation experiment. We hypothesized that the increased organic matter input, as a consequence of increased run-off from land, would provide a potential source of organic matter for fjord microbial communities. If bioavailable, this tDOM will stimulate the growth of some fast-responding bacterial groups that were initially underrepresented and increase in abundance over time. In particular, we were interested in answering two questions (1) how tDOM might alter the fjord bacterial community composition and (2) how tDOM might affect the growth and size of bacteria and subsequently protist grazers. This study thus aims to improve our understanding of the implications of a warmer Arctic, influenced by increased run-off from land, on coastal microbial communities.
Materials and Methods
Preparation of Aged Permafrost-Derived tDOM Stock Solution
Active layer permafrost soil from 50 cm depth, just above the frozen permafrost table, was sampled in Adventdalen, Svalbard (78.19 N, 15.89 E), and mixed with unfiltered water from the nearby Adventfjorden (1 m depth) in the ratio 600 g soil to 1 L water. The mix was stored in the dark for 30 days at 4°C to degrade the predominantly labile compounds, thus producing “aged tDOM” as has been done in similar studies (Eiler et al., 2003). The rationale behind using an aged tDOM stock was to increase resemblance to the organic matter that reaches the coastal systems, as the most labile compounds will be degraded during its transportation from soil to coastal waters (Lobbes et al., 2000). Before being added to tDOM incubations, the stock solution was filtered through 0.2 μm polycarbonate filters, ensuring that only dissolved organic matter was present in the tDOM stock solution. To test the character of DOM in the tDOM-solution relative to the control, the fluorescent properties were examined during an earlier analysis, performed in 2014. Here five fluorescence components (two humic-like and three amino-like) were described following the method explained in Stedmon and Markager (2005). The averaged intensity (given in Raman units) of these components are given in Table S1. The intensity of the humic-like substances was two-fold higher in the tDOM-stock relative to the control (0.2 μm filtered Atlantic water) and further one of the amino-like components was 100 times higher in the tDOM stock solution. Since we did not characterize the DOM composition at the end of the experiment, we cannot say what exact compounds were consumed or produced throughout our incubations. The rationale behind measuring the DOM components in the beginning was to ensure that more complex compounds were enriched in the tDOM-solution. The results strongly indicate that the character of the DOM was significantly different in the tDOM treatment compared to the control.
Study Site and Experimental Set-Up
Kongsfjorden is a 26 km long fjord, 6 to 14 km wide and includes two tidewater-glaciers, Kronebreen and Kongsvegen (Figure 1A). Water samples for incubations were collected on the 29th of June in 2015 from the center of the fjord near Kings Bay (78.95°N, 11.93°E) at 40 m depth (Figure 1A). The water was filtered through pre-combusted GFC filters (1.2 μm) to reduce the presence of protists. The tDOM-stock solution had a carbon concentration of 190 μM DOC and was mixed in the ratio of 1:2.5 with fjord water (83 μM DOC) and aliquoted into eight 1 L air-tight glass bottles (Figure 1B). Filtered (0.2 μm) fjord water was added in the same ratio to the eight control bottles. The final DOC concentration was 1.5 times higher in the tDOM treatment incubations (128 μM) than in the control incubations (83 μM). The elevated DOC concentration in the tDOM treatment reflects ranges of natural, elevated concentrations near the sample site in Kongsfjorden (Zhu et al., 2016). The 16 bottles, 8 treatment and 8 controls were incubated in the dark at 2°C. Samples for bacterial community composition were obtained after 0, 0.5, 1, 2, 3, 4, 5, and 9 days of incubation by harvesting one treatment and one control bottle, each representing an independent incubation, as described below. The rationale behind the experimental design and the statistical power of such an un-replicated time series sampling strategy are evaluated in the Supplementary Material (Figures S3, S4). Samples (6 mL) for measurements of bacterial abundance via flow-cytometry were collected as replicates according to the number of bottles remaining at each respective sampling point (e.g., 8 replicates at t0, 4 at d4 and 1 at d9). Both bacterial abundance and community composition were also analyzed for the untreated 40 m fjord sample and 0.2 μm filtered tDOM-stock solution.
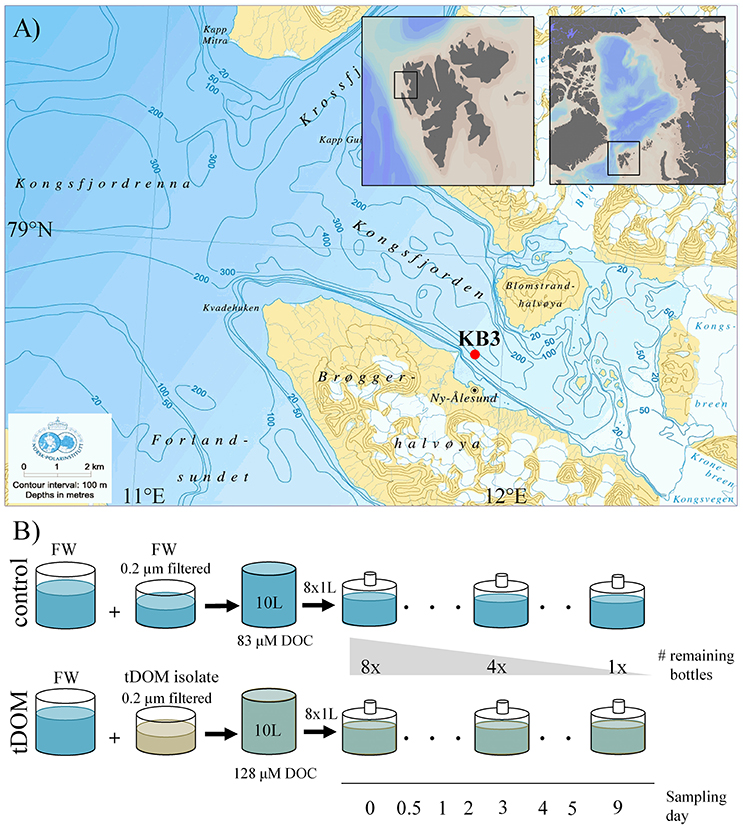
Figure 1. (A) Study area in northwest Svalbard (78.95°N, 11.93°E) showing the sampling location in Kongsfjorden (red circle). © Norwegian Polar Institute and small maps were created using Ocean Data View (Schlitzer, 2011). (B) Illustration of the experimental design showing that for both tDOM treatment and control eight bottles with the same starting condition were incubated for different periods (from 12 h up to 9 days). One bottle was harvested at each sampling point to analyse the bacterial community composition. Bacterial abundance was measured using flow cytometry for samples at the beginning of the experiment, including the fjord water (8.1 × 105 mL−1), the 0.2 μm filtered fjord water (2.8 × 104 mL−1), and the tDOM isolate (3.2 × 104 mL−1) and over the course of the incubation in both treatment and control bottles (number of replicates was dependent on the number of remaining bottles). FW, fjord water; tDOM, solution of terrigenous dissolved organic matter from permafrost; DOC, dissolved organic carbon.
Bacterial Enumeration Using Flow Cytometry
The abundance of bacteria, virus and heterotrophic nanoflagellates (HNF) were determined on an Attune® Acoustic Focusing Flow Cytometer (Applied Biosystems by Life technologies) with a syringe-based fluidic system and a 20 mW 488 nm (blue) laser. Triplicate samples of 2 mL were fixed with glutaraldehyde (0.5% final conc.) at 4°C for a minimum of 30 min, flash frozen in liquid nitrogen and stored at −80°C. Samples were first thawed and diluted x10 with 0.2 μm filtered TE buffer (Tris 10 mM, EDTA 1 mM, pH 8), stained with a green fluorescent nucleic acid dye (SYBR Green I; Molecular Probes, Eugene, Oregon, USA) and then incubated for 10 min at 80°C in a water bath (Marie et al., 1999). Samples were counted at a low flow rate of 25 μL min−1 and a minimum volume of 100 μL. Bacteria were discriminated on a biparametric plot of green florescence (BL1) vs. red florescence (BL3).
Additionally, these plots allowed to distinguish between low nuclear acid (LNA) and high nuclear acid (HNA) bacteria, virus, and a subgroup we here term “large bacteria.” Heterotrophic nanoflagellates (HNF) were measured at a high flow rate (500 μL min−1) according to Zubkov et al. (2007). Pico-and nano-sized phytoplankton were counted directly after thawing and the various groups discriminated based on their red fluorescence (BL3) vs. orange fluorescence (BL2) (Paulsen et al., 2016).
DNA Extraction, PCR Amplification, and Amplicon Sequencing
The bacterial biomass for molecular analysis was collected by filtering ca. 1 L onto 0.22 μm Millipore® Sterivex filters (Merck-Millipore), which were flash frozen in liquid nitrogen and stored at −80°C. DNA and RNA were simultaneously extracted from the Sterivex filters using the AllPrep DNA/RNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. In this study, only RNA was used in order to investigate changes in the active community. Before PCR amplification, RNA was treated with the DNA-free DNA Removal kit (Invitrogen, CA, USA). Subsequently, 10 ng of DNA-free RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen), according to the manufacturer's instructions. Amplification of cDNA (reverse transcribed RNA), targeting the bacterial/archaeal 16S rRNA gene V4 hypervariable region, was performed using a two-step nested PCR approach with primers 519F (CAGCMGCCGCGGTAA; Øvreås et al., 1997) and 806R (GGACTACHVGGGTWTCTAAT; Caporaso et al., 2011b). In brief, the first PCR step was performed in triplicates. Samples were amplified, comprising 10 ng cDNA, 10 μL HotStarTaq Master Mix (Qiagen), 0.5 μM of each primer and nuclease-free water. PCR reaction conditions were as follows: initial denaturation of 15 min at 95°C, followed by 25 cycles of 95°C for 20 s, 55°C for 30 s, and 72°C for 30 s and a final extension step of 72°C for 7 min. After triplicate PCR products were pooled, the DNA Clean & Concentrator-5 kit (Zymo Research Corporation, CA, USA) was used for purification. During the second PCR step, 10 ng of pooled PCR product, 25 μL HotStarTaq Master Mix, 0.5 μM of each nested primer (containing a unique eight-nucleotide barcode) were mixed with nuclease-free water to a reaction volume of 50 μL. PCR reaction conditions were as follows: initial denaturation of 15 min at 95°C, followed by 15 cycles of 95°C for 20 s, 62°C for 30 s, 72°C for 30 s, and a final extension step of 72°C for 7 min. Final PCR products were purified using Agencourt AMPure XP Beads (Beckman Coulter Inc., CA, USA) and pooled in equimolar amounts. Before sequencing, the quality, and concentration of the amplicon pool were assessed by agarose gel electrophoresis and a Qubit 3.0 Fluorometer, respectively. The final amplicon library was sequenced at the Norwegian Sequencing Centre (Oslo, Norway) using their MiSeq platform (MiSeq Reagent Kit v2, Illumina, CA, USA). All Illumina sequencing data is available at the European Nucleotide Archive (ENA) under study accession number PRJEB25031.
16S rRNA Gene Sequence Analysis
Illumina Paired-end sequence data was processed using different bioinformatic tools incorporated on a QIIME-processing platform (Caporaso et al., 2011a). In short, FASTQ files were quality end-trimmed at a phred quality score ≥ 24 using Trimmomatic (Bolger et al., 2014) and merged using PANDAseq (Masella et al., 2012), while all reads < 200 bp were removed. A total of 1,916,574 sequences were retrieved across 18 samples and two sequencing controls. Those sequences were used to select prokaryotic OTUs at a sequence similarity threshold of 97%, using a de novo uclust (Edgar, 2010) OTU clustering method and taxonomy assigned using the Silva 111 reference database (Quast et al., 2013). After removal of singletons and unassigned OTUs, sequences were rarefied to 10,000 reads per sample, with a total of 15,513 unique OTUs at 97% sequence identity. Rarefaction curves were calculated using QIIME's alpha rarefaction script and showed that sequencing coverage was sufficiently high, as samples approached an asymptote. The phylogenetic data was then used to calculate relative abundance at different taxonomical levels. When combined with absolute bacterial abundance data from flow cytometer measurements, the absolute abundance of taxa can be calculated. For this the bacterial abundance in cells per mL is multiplied with the relative abundance of the taxa of interest.
Indicator OTU Analysis
Calculations to identify indicator OTUs associated with the treatment of tDOM addition were performed using the “indicspecies” package (De Cáceres and Legendre, 2009) included in the statistical software R 3.2.3 (R Development Core Team, 2012) and the script “otu_category_significance.py” within the QIIME-processing platform (Caporaso et al., 2011a). Both tools can be used to assess statistically significant differences between OTU abundances and defined groups. We defined groups according to the experimental strategy in tDOM treatment and control. The analysis included only samples after 2 days of the experiment when abundances of Pseudoalteromonas sequences, an artifact of the experimental set-up, were greatly reduced.
Statistical Analysis
The statistical power of the experimental design is evaluated in detail in the Supplementary Material. The significance of changes in relative abundance of certain taxa can be calculated by comparing the slopes of regression analysis of treatment and control incubations. Correlations between bacterial abundance and community composition were calculated using the Pearson correlation coefficient (Pearson‘s r) and were carried out using GraphPad Prism v 6.01 for Windows (GraphPad Software, CA, USA).
Calculations to Estimate Bacterial and HNF Carbon Turnover
Calculations of carbon turnover are based on measurements of abundance and growth efficiency values for bacteria (B) and HNF (HNF) from literature. First carbon accumulation (CA) was calculated from the difference in cell abundance over time (ΔA) and values of fixed carbon content per cell for bacteria (0.02 pg C per cell; Lee and Fuhrman, 1987) and HNF (3.8 pg C per cell; Børsheim and Bratbak, 1987) from literature (1). The release of carbon as CO2 via respiration (R) is further calculated from the estimated CA values and expected growth efficiency of bacteria (10%; Kritzberg et al., 2010; Middelboe et al., 2012; Paulsen et al., 2017) and HNF (30%; Fenchel, 1982) (2).
Results
tDOM Effect on Bacterial Growth
During the 9-day incubation period the bacterial net-growth was documented (Figure 2). The initial fjord water contained 8.13 × 105 bacteria mL−1 and when mixed with either the aged tDOM-solution (tDOM treatment) or 0.22 μm filtered fjord water (control), this concentration was diluted to an average abundance of 4.32 × 105 or 4.19 × 105 mL−1, respectively (Figure 2A). After a lag phase during the first 24 h, we observed net-growth in both treatment and control. The bacterial abundance (BA) increased at twice the rate in the tDOM treatment between day 1 and 4 and the BA was on average 24% higher in the tDOM treatment than in the control during the first 4 days. After day 5, a different pattern emerged. While bacteria continued to grow in the control incubations reaching 4.32 × 106 mL−1 by day 9, we observed a significant decline of 63% in BA from 1.76 × 106 mL−1 (5 d) to 6.51 × 105 mL−1 (9 d) in the incubations with tDOM addition (Figure 2A).
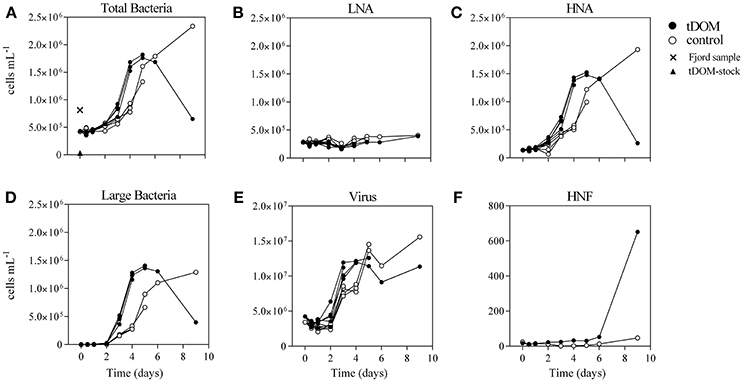
Figure 2. Flow cytometer counts over the course of the experiment in cells mL−1 of (A) Total bacteria; (B) low nucleic acid (LNA) containing bacteria; (C) high nucleic acid (HNA) containing bacteria; (D) a group of large bacteria; (E) Virus; (F) and heterotrophic nanoflagellates (HNF). Treatment incubations with added tDOM are illustrated as black circles and control incubations as open circles. The bacterial abundance of the untreated fjord sample and of the 0.2 μm filtered tDOM-stock solution is indicated as cross and triangle, respectively. The different lines represent the sample replicates, which declined over the course of the experiment depending on the number of remaining bottles (e.g., eight at t0, four at day 3 and one at day 9).
The group of total bacteria was divided into three subgroups, “LNA,” “HNA,” and “large bacteria” within the HNA group, to investigate whether a specific group is connected to the increase or decrease in BA (Figures 2B–D). The LNA group showed no differences in abundance between treatment and control over the 9 days and stayed overall stable, ranging between 1.78 × 105 and 4.04 × 105 mL−1 (Figure 2B). In contrary, the HNA group showed significant correlations (r = 0.99; p < 0.0001) with the increase in BA, including the same differences between treatment and control described earlier (Figure 2C). At day 3 we observed a new group on the flow cytometer plots within the HNA group, which we here term “large bacteria” (Figure 3). This group was well-defined in tDOM treatments where it started with low values of 1.5 × 104 mL−1 (day 2) and reached up to 1.36 × 106 mL−1 at day 5, thereby contributing to more than 77% of the BA (Figure 2D).
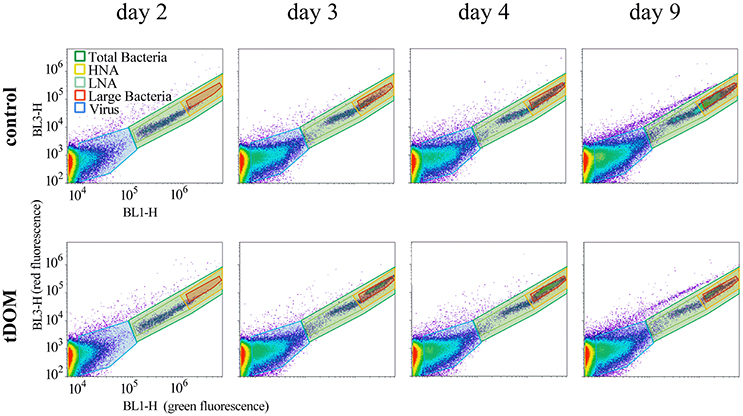
Figure 3. Flow cytometer plots of measurements from day 2 to 9 showing the changes in bacterial abundance and the subgroups within, illustrating the increase in large bacteria in the tDOM treatment. HNA, High nucleic acid containing bacteria; LNA, Low nucleic acid containing bacteria.
Virus abundance was on average one order of magnitude higher than bacteria ranging from 2.58 × 106 mL−1 to 1.56 × 107 mL−1 and followed the changes observed for BA in both treatment and control (Figure 2E). Due to the pre-filtration of the fjord water through 1.2 μm GFC filters, the abundance of small protists was substantially reduced from 600 mL−1 in the fjord water to 16 ± 2 mL−1 in both treatment and control until day 6 (Figure 2F). At day 9, the abundance of HNF reached 651 mL−1 in the incubation where tDOM was added, while it remained low in the control (47 mL−1). Picophytoplankton were additionally enumerated throughout the incubation period to confirm that autotrophic production did not contribute to the carbon pool. Abundances were reduced from 2,056 mL−1 in fjord water to < 100 mL−1 at the beginning of the experiment and remained low (< 150 mL−1).
tDOM Effect on Community Composition
The fjord water used to set up the incubations was taken from 40 m depth and was characterized as Atlantic water with a salinity of 34.6 and temperature of 4°C. The analysis of the untreated Atlantic water showed a high abundance of the phylum Proteobacteria (96.5%) (Figure 4A). Gammaproteopbacteria were dominating (±76.2%), followed by Alphaproteobacteria (±14.9%) and Betaproteobacteria (±3.0%). Due to the high diversity within the different Proteobacteria classes, 40% of all sequences at genus level were categorized as “Other” (Figure 4A).
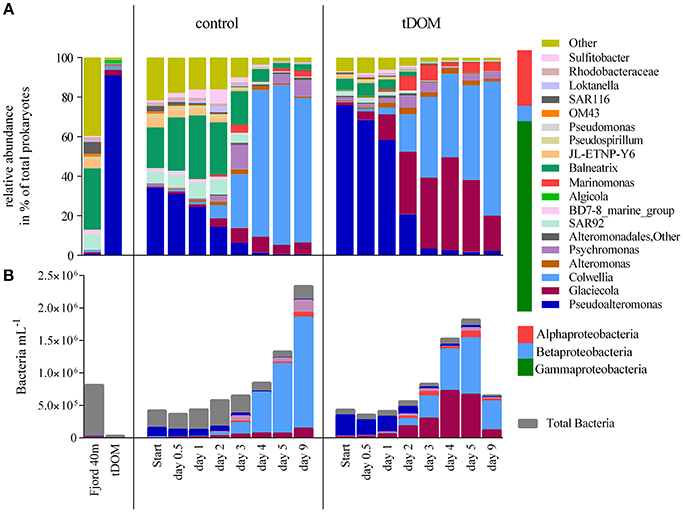
Figure 4. (A) Bacterial community composition derived from 16S rRNA sequencing data showing the relative abundance of the 20 most abundant taxa at genus level in the fjord sample from 40 m depth and during the 9 days of incubation in the control and treatment incubations where tDOM was added. Taxa comprising < 1% of the total number of sequences within a sample were summarized as “Other.” (B) Calculated absolute abundance of the most abundant genera in control and treatment incubations, based on absolute bacterial abundance measured on the flow cytometer and phylogenetic relative abundance.
Community composition at the beginning of the experiment was similar in the control and the tDOM treatment, with around 30–50% of the sequences resembling the in situ fjord community and all other sequences belonging to the genus Pseudoalteromonas. Sequences belonging to this genus, possibly introduced with the addition of 0.22 μm filtered tDOM solution and 0.22 μm filtered fjord water, decreased within 4 days from up to 70 to 4% in the treatment and from 30 to 3% in the control incubations. With increasing incubation time, changes could be attributed to the increase of certain genera. We observed a substantial increase in relative abundance of Glaciecola, Marinomonas, and Colwellia in the treatment experiments. In the control incubations, it was predominantly Colwellia that increased in relative abundance and to a lesser extend Glaciecola, Marinomonas, and Psychromonas. Glaciecola increased from 1.5 to 47.1% on day 5 in the incubations where tDOM was added, while the abundance in the controls increased only up to 7.9%. A Simper analysis showed in agreement, that predominantly the changes in Glaciecola relative abundance contributed, with up to 40% (at day 4), for the differences caused by the tDOM addition.
We analyzed the effect of tDOM addition on the community structure by combining the relative abundance of bacterial community composition and absolute abundance of bacterial counts obtained from sequencing data and flow cytometer counts, respectively (Figure 4B). Using this estimation of absolute species abundance, the Glaciecola abundance increased two-fold within the first 12 h and 138-fold after 4 days relative to the beginning (Figure 4B). At day 4, Glaciecola abundance was 90.6% higher in tDOM treatment incubations than in control incubations. The abundance of Marinomonas increased 92-fold after 5 days and was up to 93.8% higher in incubations with added tDOM than in controls (Figure 4B). Both genera, Glaciecola and Marinomonas, showed significant (p = 0.006 and p = 0.018) responses due to the addition of tDOM. Only in the tDOM treatment incubations Glaciecola abundance significantly correlated (r = 0.77; p = 0.03) with the abundance of large bacteria (Figure S1). In the control incubations it was only Colwellia abundance that correlated significantly (r = 0.83; p = 0.01) with the abundance of large bacteria. This genus however showed no significant difference in abundance between treatment and control. In the first days until day 3, the abundance was up to three times higher in the tDOM treatment than the control. This changed on day 4, when abundance in the control incubation was twice as high as in the treatment.
The observed changes for the different genera are based on cumulative abundances of several OTUs which were taxonomically assigned to these genera and grouped accordingly. In order to identify whether all or just some OTUs within each genus are causing the observed changes between treatment and controls, we performed an indicator OTU analysis (Figure 5). This analysis identified the OTUs that significantly contributed to the differences between tDOM treatment and control. Out of the 20 most significant OTUs, seven were significantly more abundant in incubations with tDOM addition and 13 OTUs had a significant higher abundance in control incubations. Taxonomically, the great majority of OTUs (19/20) belonged to the class of Gammaproteobacteria and within that class to genera including Glaciecola, Marinomonas, Colwellia, Balneatrix, SAR92, and Psychromonas. The overall most abundant OTU (33%) belonged to the genus Glaciecola and was at day 3 seven times more abundant in the tDOM treatment than the control incubations. Of all genera, Glaciecola was the genus with the highest number of OTUs (85%) that were positively associated with incubations where tDOM was added. Other genera, like Colwellia, with an overall high abundance had a more equal distribution of OTUs, which were higher in abundance in either treatment or control. 66% of Colwellia OTUs were significantly more abundant in control incubations and 33% more abundant in treatment incubations.
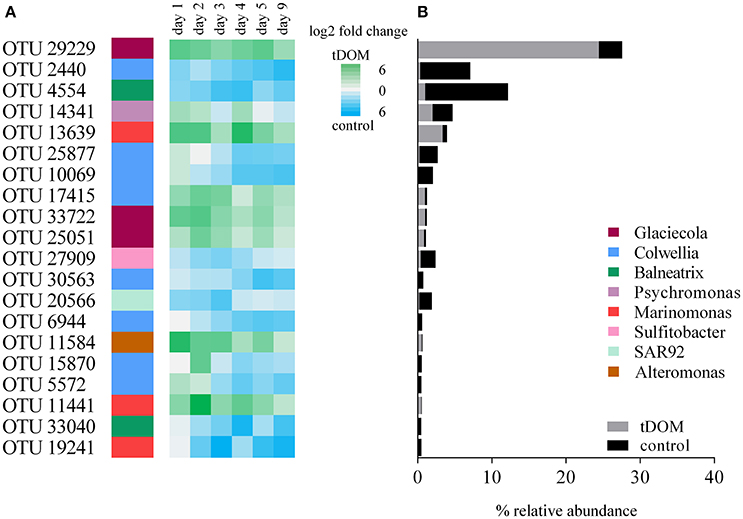
Figure 5. (A) The 20 most significant OTUs, identified by an indicator OTU analysis, contributing to the differences in community composition between control and treatment incubations. Relative abundance differences are visualized as 2-fold change from day1 until day 9. (B) Relative abundance (average values from d1 to d9) of the 20 most significant indicator OTUs visualized in the same order as in (A).
The results in this study are based on an experimental setup using an un-replicated sampling strategy. Using this strategy improves the probability of identifying trends in community composition change, and reduces the chance of making a type-2-error (finding a pattern that is not there) compared to a replicate sampling strategy. The statistical power of the results obtained from the incubation experiments is evaluated in the Supplementary Material (Figures S3, S4).
Discussion
Climate model predictions suggest a 30% increase of terrestrial run-off into the Arctic Ocean by the end of the century (Lehner et al., 2012). The tDOM in this run-off is originating from thawing Arctic soil and is modified during the transport into the Arctic Ocean (Lobbes et al., 2000; Serreze et al., 2000; Feng et al., 2013; Fichot et al., 2013; Holmes et al., 2013). It is uncertain how this will affect the marine microbial life and in particular the coastal communities. Our results indicated increased bacterial abundance (Figure 2), enlarged cell sizes (Figure 3) and changes in the community composition (Figure 4) as an immediate (within 3 days) response to tDOM addition. Together, this suggests that in the future the activity, the physiology and the structure of the fjord microbial community might be affected by increased tDOM rich run-off.
In situ Microbial Community Composition
Environmental conditions in the Arctic are highly affected by seasonality. In accordance, seasonal changes in microbial community composition have been reported for different parts of the Arctic Oceans, i.e., the increase of Gammaproteobacteria, in association to phytoplankton bloom dynamics and increased concentrations of dissolved organic matter in the summer months (Alonso-Sáez et al., 2008; Buchan et al., 2014; El-Swais et al., 2015; Wilson et al., 2017). The fjord water used for our incubation experiments, taken in June, was indicative for a post-bloom situation, with high relative abundance of the phylum Proteobacteria (96.5%) and in particular the class Gammaproteobacteria, with up to 76.1% of all proteobacterial reads (Figure 4), similar to reports from other studies (Piquet et al., 2010; Zeng et al., 2013). The largest contributor was the genus Balneatrix (30.9%), known to be associated with phytoplankton blooms and observed in other Arctic fjords (Nikrad et al., 2014; Paulsen et al., 2017). Other dominant taxa within the Gammaproteobacteria, such as SAR92 (10%) and OM182 (13%), are commonly associated with rather oligotrophic conditions (Cho and Giovannoni, 2004). Surprisingly, Bacteroidetes, commonly found in summer coastal Arctic communities, comprised only 0.3% in our samples (Nikrad et al., 2012; Sipler et al., 2017a). The low abundance of Betaproteobacteria, which comprised, with up to 3%, only a small proportion of all proteobacterial reads is characteristic for Atlantic water masses (Cottrell and Kirchman, 2003; Garneau et al., 2006).
tDOM Addition Induced Changes in Bacterial Community Composition
The large initial relative abundance of Pseudoalteromonas was likely an experimental artifact and rapidly decreased in abundance under both experimental conditions. Changes in community composition due to tDOM addition were already measurable after 12 h of incubation, for example the doubling of Glaciecola relative abundance (Figure 4). Glaciecola also increased in abundance in control incubations (4171% increase from t0 until d4), confirming both that this taxa is part of the in situ microbial community and able to grow using in situ carbon sources. The fact that Glaciecola grew faster and to a higher abundance in the treatment incubations (10781% increase from t0 until d4) indicates that this genus has the potential to degrade the introduced complex tDOM compounds. Other growth experiments with Glaciecola revealed both general phylotypes, capable of degrading a broad range of carbon compounds and specialized phylotypes, capable of degrading only specific carbon sources (Gómez-Consarnau et al., 2012).
Besides Glaciecola, Marinomonas, and Colwellia, two taxa known to degrade complex organic matter, also increased in abundance in the tDOM treatment incubations. Marinomonas and Colwellia had, similar to Glaciecola, low starting abundances and increased in both treatment and control incubations, with a stronger response under tDOM addition. It has been shown that a member of the genus Marinomonas is capable of catalyzing ring cleavage of aromatic compounds and correlates with lignocellulosic carbon uptake (Chandra and Chowdhary, 2015; Gontikaki et al., 2015). Also Colwellia has been considered to produce extracellular enzymes for the breakdown of high molecular-weight organic compounds (Huston et al., 2004; Methé et al., 2005). Glaciecola and Colwellia have also recently been shown to increase in abundance under the presence of tDOM derived from Arctic rivers (Sipler et al., 2017a). Interestingly, it was a different Glaciecola OTU that was dominating in their dataset. This OTU was also found in our data set, but is only one of the least abundant Glaciecola OTUs. It remains unclear whether this difference is caused by substrate specificity or simply which OTU is most abundant at in situ conditions. Sipler and colleagues used seven times higher DOC concentrations (400–500 μM) than in their control to stimulate a community response, while the DOC concentrations (128 μM) in our study were only 1.5 times higher than in the control incubations. Pulses of tDOM released via Arctic rivers can reach the DOC concentrations used by Sipler and colleagues (Benner et al., 2005), but at our sampling site in Kongsfjorden the DOC concentration is on average 109 μM (Zhu et al., 2016). Despite the relatively small increase in tDOM concentration in our study, we here stimulated faster growth of certain taxa than the change reported by Sipler and colleagues. This might be due to the fact that our incubations were conducted in the dark and therefore inhibited phototrophic processes.
OTU Specific Response to tDOM Addition and “the Bottle Effect”
We detected a significantly stronger increase of the genera Glaciecola and Marinomonas in incubations where we added tDOM compared to control incubations. The other genus found to increase, Colwellia, showed no significant difference between treatment and control. This is reflected in the differential response we observed at the taxonomic level of OTUs (Figure 5). Several OTUs were positively affected by tDOM addition and became more abundant during incubation, whereas other OTUs of the same genus decreased upon tDOM addition (Figure 5). This non-coherent tendency was found for all genera and indicates that strains within the same genus might have different functional roles. This was also observed by Sipler and colleagues, where they identified so called sentinels of increasing tDOM, OTUs that increased in abundance due to tDOM addition, while they did not identified genera where all OTUs were affected by the tDOM addition.
We compared changes in relative OTU abundance between treatment and control to differentiate between potential effects due to the tDOM input and effects caused by the experimental set-up, the so called “bottle effects.” The increase of a number of Colwellia OTUs was similar in both control and treatment incubations and is therefore likely to be attributed to the bottle effect, which is a well-known inherent concern in incubations studies (Lee and Fuhrman, 1991; Massana et al., 2001; Stewart et al., 2012). Several studies have suggested a combination of factors, including biofilm formation and the binding of nutrients, cells or carbon to the surface of the incubation container, as potential cause of bottle effects (Fogg and Calvario-Martinez, 1989; Fletcher, 1996; Eilers et al., 2000). It appears that the bottle effect predominantly leads to an increase in Gammaproteobacteria taxa, as documented in our and other studies (Eilers et al., 2000; Stewart et al., 2012; Dinasquet et al., 2013; Herlemann et al., 2014). While the bottle effect in the study from Stewart and colleagues and in our study can be attributed to an increase in Colwelliaceae, different families, such as Moraxellaceae (Herlemann et al., 2014), Pseudoalteromonadaceae (Dinasquet et al., 2013), or Oceanospirillaceae (Sipler et al., 2017a) were affected in other studies. This suggests that several different types of Gammaproteobacteria can benefit from a bottle effect and that the starting community composition might be the determining factor.
tDOM Effects on the Coastal Microbial Food Web
The increase of Gammaproteobacteria, in particular of taxa belonging to the order Alteromonadales including Glaciecola and Colwellia, has also been observed during marine phytoplankton spring blooms in lower latitudes (Tada et al., 2011; Teeling et al., 2012) and in the Arctic Ocean (Bano and Hollibaugh, 2002; Wilson et al., 2017). This suggests that they can rapidly proliferate in response to new carbon sources, including phytoplankton-derived organic carbon or tDOM as indicated in our study. A strong grazing pressure by bacterivorous protists has been shown to particularly affect Gammaproteobacteria of the order Alteromonadales (Beardsley et al., 2003; Allers et al., 2007). These studies demonstrated selective grazing on large metabolically active bacteria by heterotrophic flagellate grazers. Size-selective predator-prey interactions have also been shown for Glaciecola, that first became abundant upon rapid utilization of phytoplankton derived DOM and subsequently declined in their abundance due to grazing (von Scheibner et al., 2017). It was suggested that once abundant, Glaciecola became a target for size selective predation by protists, including heterotrophic nanoflagellates (HNF) due to their above-average cell size.
We also observed an increase in Glaciecola abundance upon tDOM addition, which correlated with the appearance of above-average large bacteria measured via flow cytometry (Figure 3 and Figure S1). Toward the end of the experiment, Glaciecola abundance declined by 84%, while at the same time the abundance of HNF increased substantially (from 32 to 651 cells mL−1). Interestingly, in the control incubations where Glaciecola abundance stayed low, HNF abundance remained unchanged at a low level and did not increase toward the end. This suggests that after Glaciecola, fueled by the tDOM addition, increased in abundance, size-selective HNF caused the decline in Glaciecola abundance. The specific predator-prey relation between Glaciecola and HNF might be an important link in the microbial food web of Arctic fjord systems, with cascading effects on higher trophic levels, including ciliates, copepods and up to the top level predators.
This link has also consequences for the coastal carbon budget. We calculated carbon turnover assuming a bacterial biomass of 0.02 pg per cell (Lee and Fuhrman, 1987) and a 10% growth efficiency for bacteria (Kritzberg et al., 2010; Middelboe et al., 2012; Paulsen et al., 2017). From day 2 to 4, bacterial growth resulted in the release of 188 μg C-CO2 L−1 in the tDOM treatment compared to 41 μg C-CO2 L−1 in the control. Based on these calculations, 30% of the added tDOM was already processed by the bacteria within 4 days. Since the increase in bacterial abundance after day 4 in the control incubations most likely was caused by the bottle effect, carbon turnover for the later period was not considered as representative for an in situ fjord community carbon turnover. The grazing and subsequent growth of HNF in the tDOM treatment caused a further transition of the bacterial biomass. The carbon turnover by HNF was calculated assuming a biomass of 3.8 pg (Børsheim and Bratbak, 1987) and 30% growth efficiency for HNF (Fenchel, 1982). The increase of HNF from day 5 to day 9 resulted in the incorporation of 2 μg C L−1 as biomass and an additional release of 6 μg C-CO2 L−1. Both the initial growth of Glaciecola and the subsequent grazing by HNF will thus affect the carbon turnover in Arctic coastal ecosystems with increased tDOM inputs. Based on our study design, we cannot fully predict such effects, but we can document that the addition of tDOM affected not only bacteria, but indirectly also the organisms grazing on bacteria. To our knowledge we here provide the first results on the effects of permafrost-derived tDOM input on fjord microbial communities and to understand the interactions at higher trophic levels, it is necessary to conduct further experiments with tDOM additions at larger scales, including more members of the marine food web.
Author Contributions
MP and OM led the planning of the study. LS collected and OM processed samples. MP did flow cytometric analysis. OM, GB, LS, and MP analyzed data, OM prepared figures and tables and led the writing of the paper. All authors contributed to discussing and interpreting data and writing the paper.
Funding
This study is part of the project MicroPolar (RCN 225956) funded by the Norwegian Research Council. LS participated as member of the Carbon Bridge project (RCN 226415). Parts of the study was funded by the project Microorganisms in the Arctic: Major drivers of biogeochemical cycles and climate change (RCN 227062).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank all colleagues in the UiB Marine Microbiology group and collaborators abroad who contributed to the research effort. We would like to thank Lise Øvreås for her assistance throughout the sampling. Also thank you Aud Larsen for facilitating the transport of samples and Colin Stedmon for performing the Parafac analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00263/full#supplementary-material
References
Allers, E., Gómez-Consarnau, L., Pinhassi, J., Gasol, J. M., Simek, K., and Pernthaler, J. (2007). Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ. Microbiol. 9, 2417–2429. doi: 10.1111/j.1462-2920.2007.01360.x
Alonso-Sáez, L., Sánchez, O., Gasol, J. M., Balagué, V., and Pedrós-Alio, C. (2008). Winter-to-summer changes in the composition and single-cell activity of near-surface Arctic prokaryotes. Environ. Microbiol. 10, 2444–2454. doi: 10.1111/j.1462-2920.2008.01674.x
Amon, R. M. W., Rinehart, A. J., Duan, S., Louchouarn, P., Prokushkin, A., Guggenberger, G., et al. (2012). Dissolved organic matter sources in large Arctic rivers. Geochim. Cosmochim. Acta 94, 217–237. doi: 10.1016/j.gca.2012.07.015
Bano, N., and Hollibaugh, J. T. (2002). Phylogenetic composition of bacterioplankton assemblages from the Arctic Ocean. Appl. Environ. Microbiol. 68, 505–518. doi: 10.1128/AEM.68.2.505-518.2002
Beardsley, C., Pernthaler, J., Wosniok, W., and Amann, R. (2003). Are readily culturable bacteria in coastal North Sea waters suppressed by selective grazing mortality? Appl. Environ. Microbiol. 69, 2624–2630. doi: 10.1128/AEM.69.5.2624-2630.2003
Beier, S., Rivers, A. R., Moran, M. A., and Obernosterer, I. (2015). The transcriptional response of prokaryotes to phytoplankton-derived dissolved organic matter in seawater. Environ. Microbiol. 17, 3466–3480. doi: 10.1111/1462-2920.12434
Benner, R., Louchouarn, P., and Amon, R. M. W. (2005). Terrigenous dissolved organic matter in the Arctic Ocean and its transport to surface and deep waters of the North Atlantic. Glob. Biogeochem. Cycles 19:GB2025. doi: 10.1029/2004GB002398
Blanchet, M., Pringault, O., Panagiotopoulos, C., Lefèvre, D., Charrière, B., Ghiglione, J. F., et al. (2017). When riverine dissolved organic matter (DOM) meets labile DOM in coastal waters: changes in bacterial community activity and composition. Aquat. Sci. 79, 27–43. doi: 10.1007/s00027-016-0477-0
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Børsheim, K., and Bratbak, G. (1987). Cell volume to cell carbon conversion factors for a bacterivorous Monas sp. enriched from seawater. Mar. Ecol. Prog. Ser. 36, 171–175. doi: 10.3354/meps036171
Buchan, A., LeCleir, G. R., Gulvik, C. A., and González, J. M. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Microbiol. 12, 686–698. doi: 10.1038/nrmicro3326
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2011a). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011b). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chandra, R., and Chowdhary, P. (2015). Properties of bacterial laccases and their application in bioremediation of industrial wastes. Environ. Sci. Process. Impacts 17, 326–342. doi: 10.1039/C4EM00627E
Cho, J. C., and Giovannoni, S. J. (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 70, 432–440. doi: 10.1128/AEM.70.1.432-440.2004
Cottrell, M. T., and Kirchman, D. L. (2003). Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware estuary. Limnol. Oceanogr. 48, 168–178. doi: 10.4319/lo.2003.48.1.0168
De Cáceres, M., and Legendre, P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90, 3566–3574. doi: 10.1890/08-1823.1
Dinasquet, J., Kragh, T., Schrøter, M. L., Søndergaard, M., and Riemann, L. (2013). Functional and compositional succession of bacterioplankton in response to a gradient in bioavailable dissolved organic carbon. Environ. Microbiol. 15, 2616–2628. doi: 10.1111/1462-2920.12178
Dittmar, T., and Kattner, G. (2003). The biogeochemistry of the river and shelf ecosystem of the Arctic Ocean: a review. Mar. Chem. 83, 103–120. doi: 10.1016/S0304-4203(03)00105-1
Doney, S. C., Ruckelshaus, M., Emmett Duffy, J., Barry, J. P., Chan, F., English, C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Eiler, A., Langenheder, S., Bertilsson, S., and Tranvik, L. J. (2003). Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 69, 3701–3709. doi: 10.1128/AEM.69.7.3701-3709.2003
Eilers, H., Pernthaler, J., and Amann, R. (2000). Succession of pelagic marine bacteria during enrichment: a close look at cultivation-induced shifts. Appl. Environ. Microbiol. 66, 4634–4640. doi: 10.1128/AEM.66.11.4634-4640.2000
El-Swais, H., Dunn, K. A., Bielawski, J. P., Li, W. K., and Walsh, D. A. (2015). Seasonal assemblages and short-lived blooms in coastal north-west Atlantic Ocean bacterioplankton. Environ. Microbiol. 17, 3642–3661. doi: 10.1111/1462-2920.12629
Fenchel, T. (1982). Ecology of heterotrophic microflagellates. IV quantitative occurrence and importance as bacterial consumers. Mar. Ecol. Prog. Ser. 9, 35–42. doi: 10.3354/meps009035
Feng, X., Vonk, J. E., van Dongen, B. E., Gustafsson, Ö., Semiletov, I. P., Dudarev, O. V., et al. (2013). Differential mobilization of terrestrial carbon pools in Eurasian Arctic river basins. Proc. Natl. Acad. Sci. U.S.A. 110, 14168–14173. doi: 10.1073/pnas.1307031110
Fichot, C. G., Kaiser, K., Hooker, S. B., Amon, R. M., Babin, M., Bélanger, S., et al. (2013). Pan-Arctic distributions of continental runoff in the Arctic Ocean. Sci. Rep. 3:1053. doi: 10.1038/srep01053
Fletcher, M. (1996). Bacterial Adhesion: Molecular and Ecological Diversity. Wiley. Available online at: https://books.google.com/books?hl=en&lr=&id=bYmsjh4ppZYC&pgis=1 (Accessed December 6, 2017).
Fogg, G. E., and Calvario-Martinez, O. (1989). Effects of bottle size in determinations of primary productivity by phytoplankton. Hydrobiologia 173, 89–94. doi: 10.1007/BF00015518
Garneau, M., Vincent, W., Alonso-Sáez, L., Gratton, Y., and Lovejoy, C. (2006). Prokaryotic community structure and heterotrophic production in a river-influenced coastal arctic ecosystem. Aquat. Microb. Ecol. 42, 27–40. doi: 10.3354/ame042027
Gómez-Consarnau, L., Lindh, M. V., Gasol, J. M., and Pinhassi, J. (2012). Structuring of bacterioplankton communities by specific dissolved organic carbon compounds. Environ. Microbiol. 14, 2361–2378. doi: 10.1111/j.1462-2920.2012.02804.x
Gontikaki, E., Thornton, B., Cornulier, T., and Witte, U. (2015). Occurrence of priming in the degradation of lignocellulose in marine sediments. PLoS ONE 10:e0143917. doi: 10.1371/journal.pone.0143917
Hansell, D. A. (2004). Degradation of terrigenous dissolved organic carbon in the Western Arctic Ocean. Science 304, 858–861. doi: 10.1126/science.1096175
Hansell, D. A., Carlson, C. A., Repeta, D. J., and Schlitzer, R. (2009). Dissolved organic matter in the ocean - a controversy stimulates new insights. Oceanography 22, 202–211. doi: 10.5670/oceanog.2009.109
Herlemann, D. P. R., Manecki, M., Dittmar, T., and Jürgens, K. (2017). Differential responses of marine, mesohaline and oligohaline bacterial communities to the addition of terrigenous carbon. Environ. Microbiol. 19, 3098–3117. doi: 10.1111/1462-2920.13784
Herlemann, D. P., Manecki, M., Meeske, C., Pollehne, F., Labrenz, M., and Schulz-Bull, D. (2014). Uncoupling of bacterial and terrigenous dissolved organic matter dynamics in decomposition experiments. PLoS ONE 9:e93945. doi: 10.1371/journal.pone.0093945
Holmes, R. M., Coe, M. T., Fiske, G. J., Gurtovaya, T., McClelland, J. W., Shiklomanov, A. I., et al. (2013). “Climate change impacts on the hydrology and biogeochemistry of Arctic rivers,” in Climatic Change and Global Warming of Inland Waters: Impacts and Mitigation for Ecosystems and Societies, eds C. R. Goldman, M. Kumagai, and R. D. Robarts (Oxford UK: John Wiley), 1–26.
Holmes, R. M., McClelland, J. W., Raymond, P. A., Frazer, B. B., Peterson, B. J., and Stieglitz, M. (2008). Lability of DOC transported by Alaskan rivers to the Arctic Ocean. Geophys. Res. Lett. 35:L03402. doi: 10.1029/2007GL032837
Huston, A. L., Methe, B., and Deming, J. W. (2004). Purification, characterization, and sequencing of an extracellular cold-active aminopeptidase produced by marine psychrophile Colwellia psychrerythraea strain 34H. Appl. Environ. Microbiol. 70, 3321–3328. doi: 10.1128/AEM.70.6.3321-3328.2004
Jiao, N., Herndl, G. J., Hansell, D. A., Benner, R., Kattner, G., Wilhelm, S. W., et al. (2010). Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8, 593–599. doi: 10.1038/nrmicro2386
Kritzberg, E. S., Duarte, C. M., and Wassmann, P. (2010). Changes in Arctic marine bacterial carbon metabolism in response to increasing temperature. Polar Biol. 33, 1673–1682. doi: 10.1007/s00300-010-0799-7
Lee, S., and Fuhrman, J. A. (1987). Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl. Environ. Microbiol. 53, 1298–1303.
Lee, S., and Fuhrman, J. A. (1991). Species composition shift of confined bacterioplankton studies at the level of community DNA. Mar. Ecol. Prog. Ser. 79, 195–201. doi: 10.3354/meps079195
Lehner, F., Raible, C. C., Hofer, D., and Stocker, T. F. (2012). The freshwater balance of polar regions in transient simulations from 1500 to 2100 AD using a comprehensive coupled climate model. Clim. Dyn. 39, 347–363. doi: 10.1007/s00382-011-1199-6
Li, W. K. W., McLaughlin, F. A., Lovejoy, C., and Carmack, E. C. (2009). Smallest algae thrive as the arctic ocean freshens. Science 326:539. doi: 10.1126/science.1179798
Lobbes, J. M., Fitznar, H. P., and Kattner, G. (2000). Biogeochemical characteristics of dissolved and particulate organic matter in Russian rivers entering the Arctic Ocean. Geochim. Cosmochim. Acta 64, 2973–2983. doi: 10.1016/S0016-7037(00)00409-9
Marie, D., Brussaard, C. P. D., Thyrhaug, R., Bratbak, G., and Vaulot, D. (1999). Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 65, 45–52.
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq : PAired-eND Assembler for Illumina sequences. BMC Bioinformatics 13:31. doi: 10.1186/1471-2105-13-31
Massana, R., Pedros-Alio, C., Casamayor, E. O., and Gasol, J. M. (2001). Changes in marine bacterioplankton phylogenetic composition during incubations designed to measure biogeochemically significant parameters. Limnol. Oceanogr. 46, 1181–1188. doi: 10.4319/lo.2001.46.5.1181
McCarren, J., Becker, J. W., Repeta, D. J., Shi, Y., Young, C. R., Malmstrom, R. R., et al. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc. Natl. Acad. Sci. U.S.A. 107, 16420–16427. doi: 10.1073/pnas.1010732107
Methé, B. A., Nelson, K. E., Deming, J. W., Momen, B., Melamud, E., Zhang, X., et al. (2005). The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc. Natl. Acad. Sci. U.S.A. 102, 10913–10918. doi: 10.1073/pnas.0504766102
Middelboe, M., Glud, R. N., and Sejr, M. K. (2012). Bacterial carbon cycling in a subarctic fjord: a seasonal study on microbial activity, growth efficiency, and virus-induced mortality in Kobbefjord, Greenland. Limnol. Oceanogr. 57, 1732–1742. doi: 10.4319/lo.2012.57.6.1732
Nikrad, M. P., Cottrell, M. T., and Kirchman, D. L. (2012). Abundance and single-cell activity of heterotrophic bacterial groups in the western Arctic Ocean in summer and winter. Appl. Environ. Microbiol. 78, 2402–2409. doi: 10.1128/AEM.07130-11
Nikrad, M. P., Cottrell, M. T., and Kirchman, D. L. (2014). Growth activity of gammaproteobacterial subgroups in waters off the west Antarctic Peninsula in summer and fall. Environ. Microbiol. 16, 1513–1523. doi: 10.1111/1462-2920.12258
Opsahl, S., Benner, R., and Amon, R. M. W. (1999). Major flux of terrigenous dissolved organic matter through the Arctic Ocean. Limnol. Oceanogr. 44, 2017–2023. doi: 10.4319/lo.1999.44.8.2017
Ovreås, L., Forney, L., and Daae, F. L. Torsvik, V. (1997). Distribution of bacterioplankton in meromictic lake sælenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63, 3367–3373.
Paulsen, M. L., Doré, H., Garczarek, L., Seuthe, L., Müller, O., Sandaa, R.-A., et al. (2016). Synechococcus in the Atlantic gateway to the Arctic Ocean. Front. Mar. Sci. 3:191. doi: 10.3389/fmars.2016.00191
Paulsen, M. L., Nielsen, S. E. B., Müller, O., Møller, E. F., Stedmon, C. A., Juul-Pedersen, T., et al. (2017). Carbon bioavailability in a high arctic fjord influenced by glacial meltwater, NE Greenland. Front. Mar. Sci. 4:176. doi: 10.3389/fmars.2017.00176
Pedler, B. E., Aluwihare, L. I., and Azam, F. (2014). Single bacterial strain capable of significant contribution to carbon cycling in the surface ocean. Proc. Natl. Acad. Sci. U.S.A. 111, 7202–7207. doi: 10.1073/pnas.1401887111
Pinhassi, J., Sala, M. M., Havskum, H., Peters, F., Guadayol, O., Malits, A., et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70, 6753–6766. doi: 10.1128/AEM.70.11.6753-6766.2004
Piquet, A. M.-T., Scheepens, J. F., Bolhuis, H., Wiencke, C., and Buma, A. G. J. (2010). Variability of protistan and bacterial communities in two Arctic fjords (Spitsbergen). Polar Biol. 33, 1521–1536. doi: 10.1007/s00300-010-0841-9
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project : improved data processing and web-based tools. Nucleic Acid Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rachold, V., Eicken, H., Gordeev, V. V., Grigoriev, M. N., Hubberten, H.-W., Lisitzin, A. P., et al. (2004). The Organic Carbon Cycle in the Arctic Ocean. Heidelberg: Springer, 33–55.
Sapp, M., Wichels, A., Wiltshire, K. H., and Gerdts, G. (2007). Bacterial community dynamics during the winter-spring transition in the North Sea. FEMS Microbiol. Ecol. 59, 622–637. doi: 10.1111/j.1574-6941.2006.00238.x
Schlitzer, R. (2011). Ocean Data View User's Guide, 1–156. Available online at: http://odv.awi.de
Screen, J. A., and Simmonds, I. (2010). The central role of diminishing sea ice in recent Arctic temperature amplification. Nature 464, 1334–1337. doi: 10.1038/nature09051
Serreze, M., Walsh, J., and Iii, F. C. (2000). Observational evidence of recent change in the northern high-latitude environment. Clim. Change 46, 159–207. doi: 10.1023/A:1005504031923
Sipler, R. E., Kellogg, C. T. E., Connelly, T. L., Roberts, Q. N., Yager, P. L., and Bronk, D. A. (2017a). Microbial community response to terrestrially derived dissolved organic matter in the coastal Arctic. Front. Microbiol. 8:1018. doi: 10.3389/fmicb.2017.01018
Sipler, R. E., Baer, S. E., Connelly, T. L., Frischer, M. E., Roberts, Q. N., Yager, P. L., et al. (2017b). Chemical and photophysiological impact of terrestrially-derived dissolved organic matter on nitrate uptake in the coastal western Arctic. Limnol. Oceanogr. 62, 1881–1894. doi: 10.1002/lno.10541
Stedmon, C. A., and Markager, S. (2005). Resolving the variability in dissolved organic matter fluorescence in a temperate estuary and its catchment using PARAFAC analysis. Limnol. Oceanogr. 50, 686–697. doi: 10.4319/lo.2005.50.2.0686
Stedmon, C. A., Amon, R. M. W., Rinehart, A. J., and Walker, S. A. (2011). The supply and characteristics of colored dissolved organic matter (CDOM) in the Arctic ocean: pan Arctic trends and differences. Mar. Chem. 124, 108–118. doi: 10.1016/j.marchem.2010.12.007
Stewart, F. J., Dalsgaard, T., Young, C. R., Thamdrup, B., Revsbech, N. P., Ulloa, O., et al. (2012). Experimental incubations elicit profound changes in community transcription in OMZ bacterioplankton. PLoS ONE 7:e37118. doi: 10.1371/journal.pone.0037118
Tada, Y., Taniguchi, A., Nagao, I., Miki, T., Uematsu, M., Tsuda, A., et al. (2011). Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl. Environ. Microbiol. 77, 4055–4065. doi: 10.1128/AEM.02952-10
Teeling, H., Fuchs, B. M., Becher, D., Klockow, C., Gardebrecht, A., Bennke, C. M., et al. (2012). Induced by a phytoplankton bloom. Science 336, 608–611. doi: 10.1126/science.1218344
Thingstad, T. F., Bellerby, R. G., Bratbak, G., Børsheim, K. Y., Egge, J. K., Heldal, M., et al. (2008). Counterintuitive carbon-to-nutrient coupling in an Arctic pelagic ecosystem. Nature 455, 387–390. doi: 10.1038/nature07235
Traving, S. J., Rowe, O., Jakobsen, N. M., Sørensen, H., Dinasquet, J., Stedmon, C. A., et al. (2017). The effect of increased loads of dissolved organic matter on estuarine microbial community composition and function. Front. Microbiol. 8:351. doi: 10.3389/fmicb.2017.00351
Trenberth, K. E., and Josey, S. A. (2007). Observations: surface and atmospheric climate change. Changes 164, 235–336. doi: 10.5194/cp-6-379-2010
Vincent, W. F. (2010). Microbial ecosystem responses to rapid climate change in the Arctic. ISME J. 4, 1087–1090. doi: 10.1038/ismej.2010.108
Vonk, J. E., Mann, P. J., Davydov, S., Davydova, A., Spencer, R. G. M., Schade, J., et al. (2013). High biolability of ancient permafrost carbon upon thaw. Geophys. Res. Lett. 40, 2689–2693. doi: 10.1002/grl.50348
von Scheibner, M., Sommer, U., and Jürgens, K. (2017). Tight coupling of Glaciecola spp. and diatoms during cold-water phytoplankton spring blooms. Front. Microbiol. 8:27. doi: 10.3389/fmicb.2017.00027
Wilson, B., Müller, O., Nordmann, E.-L., Seuthe, L., Bratbak, G., and Øvreås, L. (2017). Changes in marine prokaryote composition with season and depth over an arctic polar year. Front. Mar. Sci. 4:95. doi: 10.3389/fmars.2017.00095
Wohlers, J., Engel, A., Zöllner, E., Breithaupt, P., Jürgens, K., Hoppe, H. G., et al. (2009). Changes in biogenic carbon flow in response to sea surface warming. Proc. Natl. Acad. Sci. U.S.A. 106, 7067–7072. doi: 10.1073/pnas.0812743106
Xie, H., Bélanger, S., Song, G., Benner, R., Taalba, A., Blais, M., et al. (2012). Photoproduction of ammonium in the southeastern Beaufort Sea and its biogeochemical implications. Biogeosciences 9, 3047–3061. doi: 10.5194/bg-9-3047-2012
Zeng, Y. X., Zhang, F., He, J. F., Lee, S. H., Qiao, Z. Y., Yu, Y., et al. (2013). Bacterioplankton community structure in the Arctic waters as revealed by pyrosequencing of 16S rRNA genes. Antonie Van Leeuwenhoek 103, 1309–1319. doi: 10.1007/s10482-013-9912-6
Zhu, Z. Y., Wu, Y., Liu, S. M., Wenger, F., Hu, J., Zhang, J., et al. (2016). Organic carbon flux and particulate organic matter composition in Arctic valley glaciers: examples from the Bayelva River and adjacent Kongsfjorden. Biogeosciences 13, 975–987. doi: 10.5194/bg-13-975-2016
Keywords: dissolved organic matter, Arctic, terrestrial run-off, permafrost, tDOM, Kongsfjorden, microbial community composition, Glaciecola
Citation: Müller O, Seuthe L, Bratbak G and Paulsen ML (2018) Bacterial Response to Permafrost Derived Organic Matter Input in an Arctic Fjord. Front. Mar. Sci. 5:263. doi: 10.3389/fmars.2018.00263
Received: 28 February 2018; Accepted: 16 July 2018;
Published: 06 August 2018.
Edited by:
Veronica Molina, Universidad de Playa Ancha, ChileReviewed by:
Eva Ortega-Retuerta, UMR7621 Laboratoire d'Océanographie Microbienne (LOMIC), FranceClaudia Piccini, Instituto de Investigaciones Biológicas Clemente Estable (IIBCE), Uruguay
Copyright © 2018 Müller, Seuthe, Bratbak and Paulsen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oliver Müller, b2xpdmVyLm11bGxlckB1aWIubm8=
 Oliver Müller
Oliver Müller Lena Seuthe
Lena Seuthe Gunnar Bratbak
Gunnar Bratbak Maria L. Paulsen
Maria L. Paulsen