- 1Department of Biological Sciences, Virginia Institute of Marine Science, College of William & Mary, Gloucester Point, VA, United States
- 2Department of Ecology, Evolution and Marine Biology, University of California, Santa Barbara, Santa Barbara, CA, United States
- 3Department of Biological Sciences, Louisiana State University, Baton Rouge, LA, United States
- 4Whitney Laboratory for Marine Bioscience, University of Florida, St. Augustine, FL, United States
As global ocean change progresses, reef-building corals and their early life history stages will rely on physiological plasticity to tolerate new environmental conditions. Larvae from brooding coral species contain algal symbionts upon release, which assist with the energy requirements of dispersal and metamorphosis. Global ocean change threatens the success of larval dispersal and settlement by challenging the performance of the larvae and of the symbiosis. In this study, larvae of the reef-building coral Pocillopora damicornis were exposed to elevated pCO2 and temperature to examine the performance of the coral and its symbionts in situ and better understand the mechanisms of physiological plasticity and stress tolerance in response to multiple stressors. We generated a de novo holobiont transcriptome containing coral host and algal symbiont transcripts and bioinformatically filtered the assembly into host and symbiont components for downstream analyses. Seventeen coral genes were differentially expressed in response to the combined effects of pCO2 and temperature. In the symbiont, 89 genes were differentially expressed in response to pCO2. Our results indicate that many of the whole-organism (holobiont) responses previously observed for P. damicornis larvae in scenarios of ocean acidification and warming may reflect the physiological capacity of larvae to cope with the environmental changes without expressing additional protective mechanisms. At the holobiont level, the results suggest that the responses of symbionts to future ocean conditions could play a large role in shaping success of coral larval stages.
Introduction
Understanding the sensitivities of reef-building corals to the ongoing simultaneous shifts in temperature and pCO2 is imperative, as corals are the engineers of the coral reef ecosystem that provides habitat for an incredible diversity of species, as well as food, income, and coastline security to millions of humans. Significant changes in temperature and pCO2 have already been detected in the surface ocean (Feely, 2008) as anthropogenic fossil fuel use continues to cause ocean acidification (OA) and ocean warming. The increase in frequency and duration of periods of thermal stress over the past three decades has led to three pan-tropical mass coral bleaching events (Heron et al., 2016; Hughes et al., 2017), and sea surface temperature is projected to further increase by ~2°C by 2100 (IPCC, 2013). Concurrently, the surface ocean is absorbing about one-third of anthropogenic CO2 (Sabine et al., 2004), causing OA. Although OA and warming cause stress through different mechanisms, OA and warming may have synergistic effects for reef-building corals: increased physiological sensitivity to increased temperature and/or broader fitness-related consequences (Metzger et al., 2007; Walther et al., 2009; Pörtner et al., 2017; Prada et al., 2017).
The persistence of coral reefs is partially dependent on the successful dispersal and recruitment of early life history stages of coral, which may be particularly affected by OA and warming. While OA and warming are known to decrease calcification and cause bleaching in adult corals (Chan and Connolly, 2013; Hughes et al., 2017), their interactive effects on coral larvae are less understood. Dispersing early life stages have high energy requirements and sometimes limited physiological capacity. For example, coral larvae have high metabolic demands while they are actively swimming and locating optimal settlement sites (Okubo et al., 2008). High-pCO2 can cause a reduction in oxygen consumption rates in larvae (Pocillopora damicornis, Rivest and Hofmann, 2014), while rates in adults are not significantly affected (P. damicornis, Putnam and Gates, 2015). Consequently, larvae are especially vulnerable to changing environmental conditions (Pechenik, 1999; Kurihara, 2008; Byrne, 2011).
An understanding of the mechanistic underpinnings of the biological responses of larvae to OA and temperature will enable better predictions of success for dispersal and settlement of these life stages. Changes in environmental conditions can cause stress through molecular damage that requires energy to repair, as in the damage causing proteotoxic effects (e.g., Kültz, 2005). Environmental changes can also increase the maintenance requirements for homeostatic processes by disrupting or shifting acid-base balances and ionic gradients (Pörtner, 2008; Calosi et al., 2013; Tresguerres and Hamilton, 2017). Increased maintenance of homeostatic processes and up-regulation of stress response pathways could prevent mortality of coral larvae under stressful environmental conditions, though the energetic costs of stress response may slow developmental timing, reduce growth, and impair competency to settle (Albright et al., 2010; Albright and Langdon, 2011; Timmins-Schiffman et al., 2013; Ko et al., 2014). Physiological plasticity serves to minimize and repair damage in response to changing environmental conditions. A growing body of work describes biological responses of coral larvae to conditions of OA and warming (Putnam et al., 2010, 2013; Albright and Langdon, 2011; Cumbo et al., 2013a,b; Rivest and Hofmann, 2014, 2015; Putnam and Gates, 2015; Graham et al., 2017; Rivest et al., 2017). However, it is still unknown how coral larvae are physiologically plastic–what changes are made at the molecular level to preserve function and survival under environmental stress? Transcriptome sequencing can provide insight for the physiological mechanisms employed to maintain homeostasis in a new environment, but to date, changes in gene expression of larvae, particularly larvae that host algal symbionts, in response to OA and warming have not been characterized.
We present the first study to examine transcriptome-wide changes in gene expression in symbiotic coral larvae in response to projected ocean conditions, testing the response of the cauliflower coral, Pocillopora damicornis, and its dinoflagellate symbionts (genus Symbiodinium) to OA and temperature stress. Pocillopora damicornis broods lecithotrophic, free-swimming planula larvae, which are released monthly with the lunar cycle (e.g., Harriott, 1983; Richmond and Jokiel, 1984; Fan et al., 2002). The larvae receive endosymbiotic Symbiodinium from the parent through vertical transmission prior to release (Harrison and Wallace, 1990). Pocillopora damicornis larvae fuel their dispersal, settlement, and metamorphosis largely from stored lipid and translocated metabolites from Symbiodinium (Harii et al., 2010).
We assessed broad patterns in the larval transcriptome in response to laboratory exposures of pCO2 and temperature. Using next-generation high-throughput RNA sequencing (RNA-seq), a de novo transcriptome was generated, and differential gene expression between pCO2 and temperature levels were quantified in both the coral and Symbiodinium. Measurements of larval physiological condition, in this case, total protein, Symbiodinium density, and size, complement these transcriptomic data. Our exploration of responses to a changing environment at the level of the transcriptome yields valuable information about current physiological thresholds, mechanisms driving changes in biological processes, and the potential for physiological plasticity and tolerance (e.g., Hofmann et al., 2008; Todgham and Hofmann, 2009; Whitehead et al., 2010).
Materials and Methods
Collection of Coral Larvae
On January 22, 2012 (the new moon), eight colonies of adult P. damicornis were collected from a depth of ~1–3 m on a fringing reef (17.4803 S, 149.7989 W) on Moorea, French Polynesia. Average seawater temperature on the fringing reef was 28.19 ± 0.50°C during the week of coral collection (Rivest, 2014). Colonies were individually held in aquaria that received indirect natural sunlight and coarsely filtered seawater. Daytime temperatures in the aquaria varied between 26.0 and 29.5°C. Larvae were collected from adult colonies following Rivest and Hofmann (2014). The “peak” larvae, determined by the predicted relationship of larval release according to the lunar cycle (January 30, 2012; e.g., Fan et al., 2006; Rivest and Hofmann, 2014, 2015), were used in the experiment. Due to the smaller larval production by some colonies, all larvae released were pooled. A portion of larvae from this pool were immediately photographed and preserved to quantify larval quality metrics and gene expression (see below) that represented the condition of freshly-released larvae. The remaining larvae in the pool were randomly assigned to experimental treatments immediately after collection.
Experimental Incubations
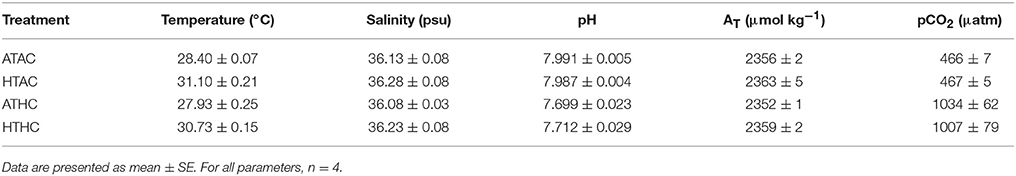
Two pCO2 levels were prescribed: ~465 μatm pCO2 and ~1,030 μatm pCO2. The control (ambient) treatment approximated the present-day environmental conditions for the adult coral (verified by environmental data; Rivest and Hofmann, 2015), while the high treatment represents a level of pCO2 expected in the surface ocean by year 2100 (RCP8.5 scenario; IPCC, 2013). Two experimental temperatures (T) were crossed with the pCO2 levels: 28 and 31°C. The control (ambient) temperature approximated the multi-year average temperature (20-min sampling frequency) for the fringing reefs close to the collection sites for adult P. damicornis (Leichter, 2015). The elevated temperature represents an average surface ocean temperature projected by year 2100 (RCP8.5 scenario, IPCC, 2013). The four treatments created by this experimental set-up are defined as ambient temperature–ambient pCO2 (ATAC), ambient temperature–high pCO2 (ATHC), high temperature–ambient pCO2 (HTAC), and high temperature–high pCO2 (HTHC).
Treatment conditions were generated in a Moorea Coral Reef Long-Term Ecological Research program (MCR LTER) facility as described previously (Rivest and Hofmann, 2015), with two aquaria for each combination of pCO2 and temperature treatment levels (8 aquaria total). Photosynthetically-active radiation (PAR) was provided at 1,800 ± 140 μmol m−2 s−1 on a 13h:11h: light:dark cycle (Sol LED Module, Aquaillumination, 75 W, Ames, IA, USA), an ecologically relevant level for shallow reef environments studied by MCR LTER (Carpenter, 2016). To verify and monitor the physical parameters of the pCO2 × temperature treatments, the carbonate chemistry (pH and total alkalinity), salinity, and temperature of the seawater in the aquaria was measured during the incubations. For a description of the methods, see Rivest and Hofmann (2015). pH and pCO2 of the treatments were calculated from pH at 25°C, total alkalinity (AT), temperature, and salinity using CO2calc (Robbins et al., 2010). CO2 constants K1 and K2 from Lueker et al. (2000) were used, and pH was expressed on the total scale (mol kg SW−1).
Larvae were incubated under experimental conditions for 24 h in two 400 mL containers per aquarium at 0.15 larva mL−1. Larval containers had 100 μm mesh sides and a PAR-transparent lid. Incubations were staggered by 1 h per aquarium, with the order randomized, due to the time needed to process the larvae at the end of the incubation. At 24 h, larvae within tanks were pooled, and 10 larvae were randomly selected for size measurements (n = 20). The remaining larvae were aliquoted and preserved at −80°C for downstream analyses of gene expression (25 larvae per aliquot), total protein content (5 larvae per aliquot), and symbiont density (5 larvae per aliquot). Therefore, for comparisons between treatments, n = 4 for gene expression, total protein, and symbiont density.
Larval Quality Metrics
Larval size, protein content, and Symbiodinium density were measured as described in Rivest and Hofmann (2015). Briefly, larval size was quantified through image analysis using ImageJ (Rasband, 1997). Protein content was measured using the Bradford assay (Bradford, 1976; Rivest and Hofmann, 2014). Symbiont density was quantified from larval homogenates using a haemocytometer.
Statistical Analyses for Physical Conditions and Larval Quality
All data were analyzed using R v3.0.1 (R Core Team, 2013). Effects of pCO2 and temperature on tank conditions and larval quality response variables were estimated using linear mixed-effect models (nlme package in R; Pinheiro and Bates, 2000) with pCO2 and temperature treatments as fixed factors and “tank” as a random factor (following Rivest and Hofmann, 2015). Statistical assumptions of normality and homogeneity of variance were met, sometimes following a power-based transformation of the response variable.
Transcriptome Sequencing
RNA was extracted from aliquots of 25 P. damicornis larvae homogenized in 0.5 mL TRIzol® using glass beads and a bead-beater. Homogenates were brought to a total volume of 2 mL TRIzol® and split into two microcentrifuge tubes. Proceeding with one half of the homogenate, RNA was isolated using the guanidinium thiocyanate method (Chomczynski and Sacchi, 1987). To improve the quality of the RNA extracts, a Qiagen RNeasy® MinElute® Cleanup Kit was used, following the manufacturer's instructions. After the clean-up, RNA quality was assessed using an Agilent RNA 6000 Pico Kit and a Bioanalyzer Agilent 2100 instrument, following the manufacturer's instructions. The electropherograms showed clean, resolved peaks with no obvious signs of RNA degradation and consistent quality across all extracts.
cDNA libraries were prepared from at least 1 μg total RNA from each extract using a TruSeq RNA Sample Prep Kit v2 (Illumina, Inc., San Diego, CA USA), following the manufacturer's instructions. From the two tubes of larvae preserved from each aquarium, one was used to prepare a library for paired-end RNA sequencing and the other was used to prepare a library for single-read RNA sequencing. A unique adapter index was ligated to the cDNA fragments in each library. The libraries contained cDNA sequences of insert size ~260 bp.
Pooled, barcoded libraries were sequenced at the UC Davis Genome Center DNA Technologies Core (http://dnatech.genomecenter.ucdavis.edu/) on the HiSeq 2500 Illumina sequencing platform. Nine libraries were pooled in each of 2 lanes. One lane was used to generate 100 bp paired end reads, and the second lane was used to generate 50 bp single reads, resulting in 187,925,419 paired-end and 157,325,043 single-end reads, respectively. Adapter contamination and bases with Phred quality scores < 20 were removed with Trim Galore v3.23 (Trim Galore!, Bioinformatics Group, Babraham Institute) with Cutadapt v1.2.1 (Martin, 2011) and FastQC v0.10.1 (Andrews, 2014). The reads generated were uploaded to the European Nucleotide Archive under the project accession PRJEB26650. The individual RNA-seq libraries are accessioned with numbers ERS2472791-ERS2472808.
Transcriptome Assembly
A de novo transcriptome was assembled using Trinity v2013-8-25 (Haas et al., 2013) implemented on the Knot computing cluster at the UCSB Center for Scientific Computing (http://csc.cnsi.ucsb.edu/clusters/knot). Subsequent bioinformatics analyses were performed using high performance computing resources provided by Louisiana State University (http://www.hpc.lsu.edu). Because P. damicornis harbors Symbiodinium zooxanthellae, host and symbiont mRNA was present in our samples. A holobiont transcriptome was assembled and then filtered to separate the host coral and symbiont zooxanthellae contigs. Contigs in the holobiont transcriptomes were collapsed using CD-HIT v4.6 (Fu et al., 2012) with a 0.98 cutoff, and contigs < 500 bp long were removed. The host and symbiont contigs were filtered from the holobiont transcriptome following the procedure outlined in Davies et al. (2016). Briefly, a series of blast searches were performed against four databases created from cnidarian and Symbiodinium genome and transcriptome sequences publicly available as of July 2016. The databases contained i) sequences from cnidarians (n = 28) that may contain Symbiodinium contamination, ii) sequences from cnidarians (n = 10) known to be free of Symbiodinium, iii) sequences from Symbiodinium that may contain cnidarian contamination (n = 19), and iv) sequences from Symbiodinium known to be free of cnidarian sequences (n = 13). The symbiont-free cnidarian database contained sequences obtained from aposymbiotic larvae and adult individuals (for example, deep sea corals). The cnidarian-free Symbiodinium databases contained sequences from free-living zooxanthellae or from lines cultured in the laboratory. A list of all genomes and transcriptomes used to construct the blast databases and their citation information is available in Supplementary Table 1. The databases were indexed, and translated blast searches (tblastx v2.4.0, Camacho et al., 2009) were performed on the coral holobiont transcriptome against all four databases. If a contig from the holobiont transcriptome had an amino acid overlap >100 bp with 80% identity cutoff to any cnidarian sequence, it was considered a host contig. However, if the same contig had a 100 bp amino acid overlap with 80% identity to a clean Symbiodinium sequence, it was removed from the coral host transcriptome. Symbiodinium contigs were identified in the same manner as described above, removing any contigs that were assigned to aposymbiotic cnidarian sequences with 100 bp amino acid overlap and 80% identity from the Symbiodinium transcriptome. After filtering, 1,200 contigs (1.5 and 2.6% of the coral and symbiont transcriptomes, respectively) that had equally good matches to both contaminant-free cnidarian and contaminant-free Symbiodinium blast databases were removed because it was not possible to determine whether they were of coral or symbiont origin. This conservative approach increases confidence that the findings for P. damicornis are based unambiguously on the response of the coral host and those for Symbiodinium are based unambiguously on the response of the symbiont.
Differential Gene Expression Analysis
To measure changes in gene expression associated with pCO2, temperature, and the interaction of these environmental stressors, reads were mapped against the coral and symbiont transcriptomes and counted using RSEM v1.2.31 (Li and Dewey, 2011). Read counts for each technical replicate (one set of 50 and 100 bp reads each per tank) were added together before testing differential gene expression among treatments. Effects of temperature, pCO2, and their interaction were tested via a likelihood ratio test in DESeq2 v1.12.4 (Love et al., 2014) in R v3.3.1 (R Core Team, 2013), and genes differentially expressed among treatments were identified with a p-value cutoff corresponding to an FDR of 0.05 (Benjamini and Hochberg, 1995). Overall patterns of gene expression were assessed by plotting a two-dimensional principal components analysis (PCA) of log-transformed gene counts. Two PCAs for both the coral and symbiont transcriptome data were performed: one comparing larvae among the experimental treatments and one comparing larvae from the experimental treatments to the initial sample of larvae collected immediately after release from the parents. Including the freshly-released larvae in our analysis allowed us to test the hypothesis that High-T and High-pCO2 treatments resulted in developmental delay, causing larvae from these treatments to exhibit a gene expression pattern more similar to freshly-released larvae.
Functional Enrichment Analysis
A rank-based gene ontology (GO) analysis was performed to identify GO terms that were significantly enriched by up or down-regulated genes following the method of Wright et al. (2015). First, coral and symbiont transcriptomes were translated into protein sequences in TransDecoder v3.0.1 (http://transdecoder.github.io/) using default parameters, and GO terms for the protein sequences were retrieved using InterProScan v5.22-61.0 (Jones et al., 2014). Next, the uncorrected p-values for differentially expressed genes in the DESeq2 analysis were converted by taking the log of the p-value and multiplying by −1 if the gene was down-regulated. After this conversion, highly down-regulated genes had highly negative values, and highly up-regulated genes had highly positive values. Using the GO terms list from InterProScan and the positive and negative log p-values for genes expressed under each condition, the Mann-Whitney U (MWU) test in R was used to determine functional enrichment of up and down-regulated genes. Scripts for this procedure and example files are available at https://github.com/z0on/GO_MWU.
Identification of Symbiodinium Clade
Because P. damicornis can host multiple clades of symbionts (Putnam et al., 2012), and the Symbiodinium community could shuffle in response to changes in seawater conditions (Jones et al., 2008), the clade(s) of Symbiodinium present were identified for freshly-released larvae and larvae exposed to experimental treatments. Symbiodinium form eight distinct clades, which can be definitively characterized based on chloroplast 23S-rDNA and nuclear 28S-rDNA sequences (Pochon et al., 2006). To identify which clade(s) of Symbiodinium were present in the coral larvae sampled in this experiment, a local blast search (Zhang et al., 2004) of the symbiont transcriptome was performed against a database created from 23S and 28S genes from each of the eight Symbiodinium clades (see Table 1 in Pochon et al., 2006).
Results
To assess how changes in the transcriptome might shape physiological plasticity in Pocillopora damicornis planulae, larvae were exposed to levels of pCO2 and temperature in laboratory microcosm experiments (Table 1). In general, pCO2 and T differed significantly between treatments, and temperatures in Ambient-pCO2 tanks were on average 0.4–0.5°C higher than in the High-pCO2 tanks (Supplementary Table 2). Salinity and total alkalinity did not differ significantly among treatments (Supplementary Table 2).
Changes in larval quality in response to pCO2 and temperature were measured in the form of total protein per larva, Symbiodinium density, larval length, and larval size. Larval total protein ranged from 9.07 μg larva−1 at ATAC to 29.00 μg larva−1 at HTAC (Figure 1A). At the end of the 24-h exposures, total protein did not differ significantly by pCO2, temperature (T) or their interaction (Supplementary Table 3). Varying between 6,188 and 14,357 cells larva−1, Symbiodinium density was lowest at ATAC and greatest at HTAC (Figure 1B). Symbiodinium density was significantly affected by T and pCO2xT. However, post-hoc analyses did not confirm significant differences (Supplementary Table 3). Larval area did not differ significantly by pCO2, T or their interaction. Larval length varied significantly by pCO2, though post-hoc analyses did not confirm the significant difference (Supplementary Table 3). Larvae had the smallest area and longest length at HTAC while having the biggest area at ATAC (Figures 1C,D).
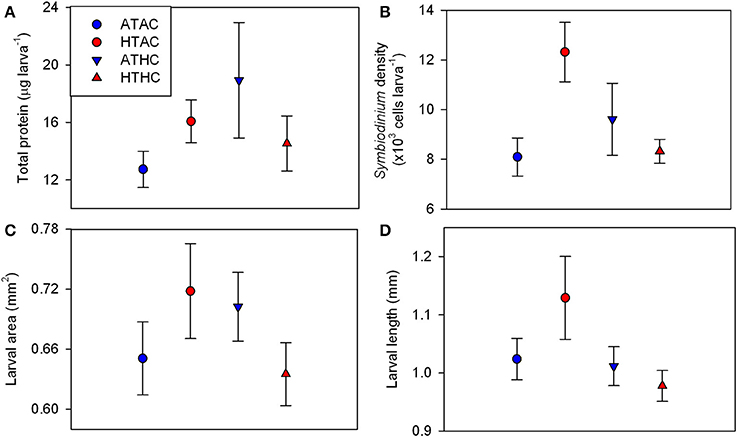
Figure 1. Larval quality traits for Pocillopora damicornis larvae following 24-h exposures to pCO2 and temperature. Mean ± SE changes in (A) total protein content (n = 4), (B) Symbiodinium density (n = 4), (C) larval area (n = 20), and (D) larval length (n = 20). Experimental treatments: Ambient-T, Ambient-pCO2 (ATAC), High-T, Ambient-pCO2 (HTAC), Ambient-T, High-pCO2 (ATHC), High-T, High-pCO2 (HTHC). Symbiont density was significantly affected by the interaction of pCO2 and temperature (X2 = 5.081, df = 1, p = 0.034) as well as temperature alone (X2 = 5.248, df = 1, p = 0.022). Larval length was significantly affected by pCO2 (X2 = 4.026, df = 1, p = 0.045). Refer to Table S3 for full statistical details.
Assembly of the Reference Transcriptome
To quantify changes in gene expression between high-temperature, high-pCO2 conditions and ambient conditions, a de novo reference transcriptome for P. damicornis larvae was assembled. The holobiont assembly yielded a transcriptome of 422.3 Mb with 169,472 contigs. The mean contig length was 1,279 bases, the N50 (the shortest sequence length such that 50% of the total sequence output is contained in sequences that are shorter) was 2,317 bases, and the GC content was 43.39%. The filtered reference transcriptomes contained 49,941 and 23,479 contigs for the coral and symbiont, respectively. The coral transcriptome had a N50 of 3,531 and a GC content of 40.65%, while the symbiont transcriptome had a N50 of 1,931 and a GC content of 55.41%.
Differential Gene Expression
After filtering, the total number of reads mapped per biological replicate ranged from 9.4 to 22.7 million for the coral transcriptome and from 1.7 to 5.4 million for the symbiont transcriptome. For the coral transcripts, only a small number of genes were differentially expressed (17 genes, FDR = 0.05, Supplementary Table 4), and a LRT indicated that the best-fit model for coral gene expression was one that included the interactive effects of temperature and pCO2, but no main effects of either treatment (0 differentially expressed genes for either temperature or pCO2 alone, FDR = 0.05). All 17 of these genes showed elevated expression in the ACAT and HCHT treatments, relative to the ACHT and HCAT treatments. Gene ontology categories for these genes (as determined by an InterProScan search) included transcription factor activity, ion and protein binding, nuclease and receptor activity, and regulation of apoptotic process (Supplementary Table 4).
Despite having fewer mapped reads (and thus lower power of detection), many more differentially expressed genes were identified in the symbiont transcriptome (89 genes, FDR = 0.05, Supplementary Table 5), consistent with a greater separation among pCO2 treatments in the principal components analysis (Figure 2). A LRT indicated that the best-fit model was for symbiont gene expression was one that included only the effects of CO2 (89 genes, FDR = 0.05), with no T or interaction effects (0 differentially expressed genes for either T or TxpCO2, FDR = 0.05).
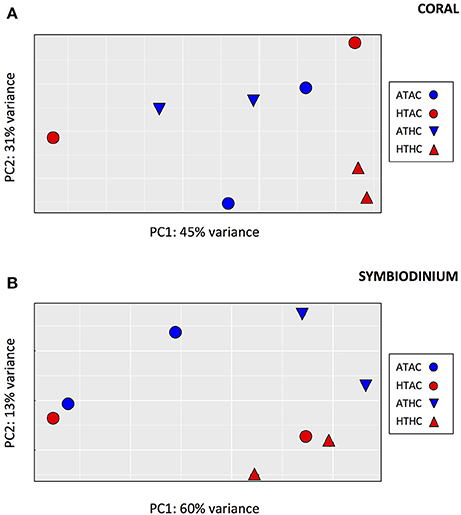
Figure 2. Principal components analysis depicting global pattern of gene expression for transcripts from (A) coral host tissue and (B) Symbiodinium for each of four experimental treatments: Ambient-T, Ambient-pCO2 (ATAC), High-T, Ambient-pCO2 (HTAC), Ambient-T, High-pCO2 (ATHC), High-T, High-pCO2 (HTHC).
When the freshly-released larvae were included in the PCA (Supplementary Figure 1), the first principal component separated the freshly-released larvae from the eight experimental samples. This axis accounted for 64 and 52% of the variance among samples for the coral and symbiont, respectively, with 5,772 transcripts being differentially expressed between time points in the coral and 796 transcripts being differentially expressed between time points in the symbionts. However, there was no evidence that any of the experimental treatments resulted in a gene expression pattern that was more similar to the freshly-released larvae than to any other treatment.
Functional Enrichment
For the coral transcriptome, only the Molecular Function category included enriched gene ontologies (Figure 3, Supplementary Table 6). Here, gene expression affected by the interaction of pCO2 and temperature was enriched for enzyme and binding activities, which were upregulated in the ACAT and HCHT treatments, relative to the others, while structural molecule and oxoreductase activities were downregulated in the ACAT and HCHT treatments.
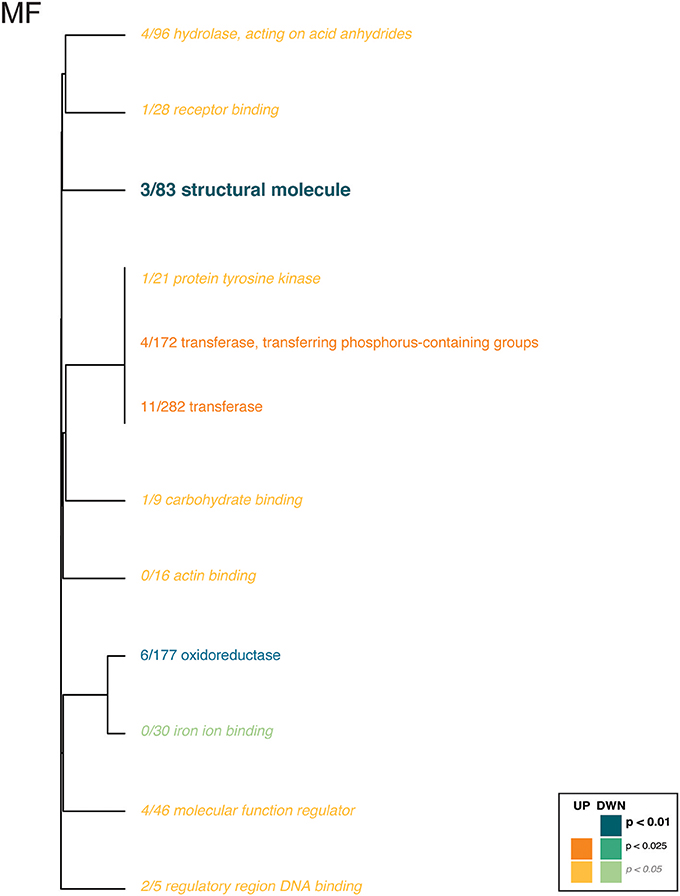
Figure 3. Gene ontology categories enriched by genes of Pocillopora damicornis larvae that are up-regulated (shades of orange) or down-regulated (shades of turquoise) in response to conditions of warming and ocean acidification. For coral host tissue, only the molecular function category (MF) contained any ontologies with significant enrichment. The size of the text and darkness of the color indicate the significance of the term as indicated by the inset key. The fraction preceding each GO term indicates the proportion of genes annotated with the term that pass an unadjusted p-value threshold of 0.05. The trees indicate sharing of genes among GO categories (the categories with no branch length between them are subsets of each other).
Several GO categories were functionally enriched for symbiont genes that were up- and down-regulated in response to High-pCO2 (Figure 4, Supplementary Table 7). Under the Biological Function class, the up-regulated genes were in metabolic process and ion transport categories, and the down-regulated genes had biosynthetic, metabolic process, and RNA modification categories. Under the Molecular Function class, up-regulated genes were largely in transporter and binding categories. Down-regulated genes were enriched for several enzyme-related categories (i.e. isomerases, peptidases, hydrolases). Under the Cellular Component class, up-regulated genes included cellular complexes, and down-regulated genes included ribosomal subunit complex and photosystem.
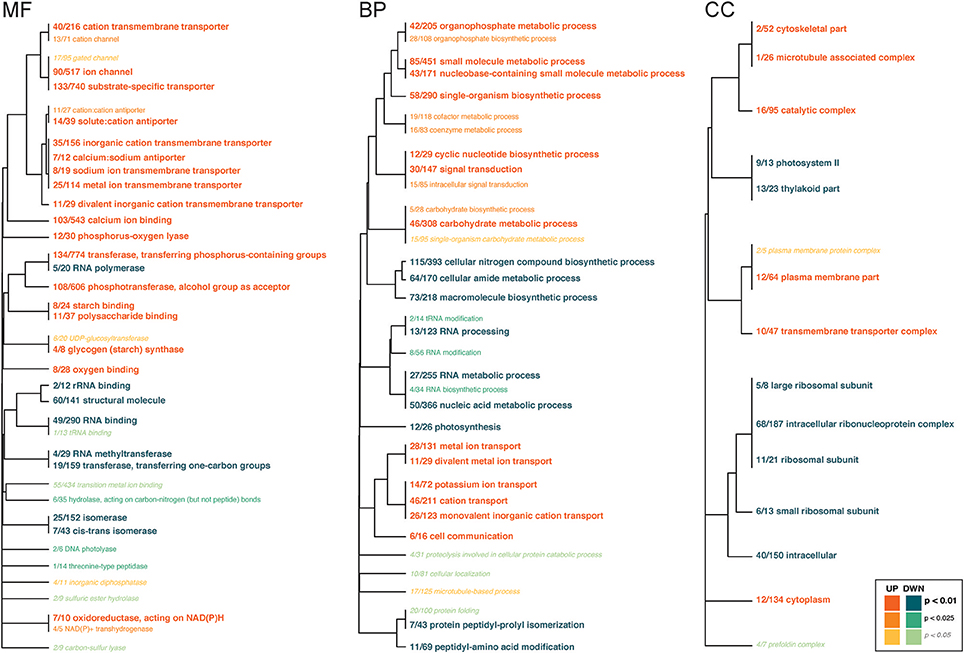
Figure 4. Gene ontology categories enriched by Symbiodinium genes up-regulated (shades of orange) or down-regulated (shades of turquoise) in response to OA, summarized by molecular function (MF), biological process (BP), and cellular component (CC). The size of the text and darkness of the color indicate the significance of the term as indicated by the inset key. The fraction preceding each GO term indicates the proportion of genes annotated with the term that pass an unadjusted p-value threshold of 0.05. The trees indicate sharing of genes among GO categories (the categories with no branch length between them are subsets of each other).
Identification of Symbiodinium Clade
Blast searches of the symbiont transcriptome against databases of Symbiodinium 28S nuclear and 23S chloroplast rDNA genes identified the symbiont clade present in coral larvae from this study as D1 (Pochon et al., 2006). Symbiont genes comp97910 and comp84390 had 99% and 96% sequence identity to clade D1 28S Genbank sequence AJ311948.1 and clade D1 23s Genbank sequence JN558001.1, respectively.
Discussion
Previous work examining effects of climate change stressors on gene expression in corals has tended to focus on the coral host (e.g., Voolstra et al., 2009; Kaniewska et al., 2012; Kenkel et al., 2013; Moya et al., 2015; Davies et al., 2016; Mass et al., 2016; González-Pech et al., in review). By examining coral and symbiont gene expression individually, differential effects of elevated pCO2 and temperature on the components of the holobiont were observed in this study. There was a greater effect of experimental conditions on the symbiont than on the coral, with 89 vs. 17 differentially expressed genes, respectively. The larval coral and the symbiont in the present study responded to different variables, with the symbiont responding only to elevated pCO2, and the coral responding only to the interaction between pCO2 and temperature. These patterns of gene expression are likely to be direct responses to temperature and pCO2 levels and not due to a development delay under particular treatment conditions. We observed a large number of transcripts that were differentially expressed between freshly released larvae and larvae from the experimental treatments, and larval development is commonly associated with large changes in gene expression across time points (e.g., Helm et al., 2013). Without field-caught 24-h-old larvae for comparison, the possibility that the differential gene expression observed is the result of acclimation to laboratory conditions cannot be excluded. However, all four experimental treatments were equally dissimilar to the gene expression profile of the freshly released larvae, even though the ATAC lab treatment was similar to pCO2 and temperature conditions in the field. Therefore, experimental treatment, not development, was the major driver of gene expression changes between treatment groups.
The gene expression response observed here is much smaller than in previous studies of the effects of climate change stressors on P. damicornis. For example, Vidal-Dupiol et al. (2013) observed that 16% of the holobiont transcriptome was affected by exposure to high pCO2 in adult P. damicornis. However, different outcomes in the present study are perhaps not surprising given the more extreme pH values (pH 7.4), longer duration of exposure (3 weeks), and different life history stage used by Vidal-Dupiol et al. (2013). Greater effects of high pCO2 on gene expression in adults may be more likely, given that Vidal-Dupiol et al. (2013) observed that many of the differentially expressed genes were involved in calcification. Altered expression of these genes would be less likely for P. damicornis larvae, given that they do not begin to actively calcify until settlement. Indeed, another recent study of gene expression in P. damicornis observed that expression of genes involved in the ‘biomineralization toolkit' is elevated upon settlement in P. damicornis (Mass et al., 2016).
Gene Expression in the Coral Animal
When examining compartments of the holobiont separately, conditions of OA and temperature had little effect on the gene expression profile of the coral animal, given the small number of differentially expressed genes and the greater power to detect differential expression given the higher number of mapped reads. These results provide insight for previous work showing that larvae released on the peak day of the spawning period, as were used in this study, are often larger in size with higher protein content and have different response phenotypes to high-pCO2, high-temperature conditions (i.e., changes in oxygen consumption rates, changes in lipid utilization) than larvae released on other days (Cumbo et al., 2012, 2013a; Rivest and Hofmann, 2014, 2015). The ability of these larvae to tolerate high temperature, high pCO2 conditions may be rooted at different levels of control other than gene expression: using existing enzymatic machinery or post-translational modification. For example, different oxygen consumption rates of P. damicornis larvae have been observed under different pCO2 levels despite the same maximum activity of aerobic metabolism enzymes (Rivest and Hofmann, 2014). Indeed, in Acropora millepora embryos, key components of DNA/RNA metabolism are down-regulated while heat shock proteins are upregulated in response to elevated temperatures, but responses to long-term exposures (>24 h) differ (Rodriguez-Lanetty et al., 2009; Meyer et al., 2011). Larvae released close to the peak day, which are generally larger and contain more symbionts and protein that were vertically transmitted from the parent colony (Cumbo et al., 2012), may also receive more mRNA, proteins, and hormones that enhance physiological plasticity immediately after release (McCormick, 1999; Place and Smith, 2012). Future studies should pair measurements of larval gene expression with enzyme and whole-organism measurements of phenotype to provide a comparative investigation of mechanisms of plasticity.
The subdued response of the coral animal to multistressor conditions is unique and may inform its relationship with the symbiont. In contrast to previous studies on the animal compartment of other species (Voolstra et al., 2009; Moya et al., 2015), here there was no evidence that exposure to either increased temperature or increased pCO2 led to upregulation of a generalized cellular stress response in P. damicornis larvae. We also observed no transcriptomic evidence for either depressed metabolism (as in Voolstra et al., 2009; Moya et al., 2012) or elevated metabolism (as in Davies et al., 2016) in response to high pCO2. These results support the above hypothesis that the coral larvae can tolerate high temperature and high pCO2 using mechanisms other than changes in gene expression. Additionally, the coral's subdued response in gene expression may indicate a lack of stress response on the part of the symbionts. Many previous studies have shown that photosynthetic dysfunction of symbionts during thermal stress produces reactive oxygen species (ROS), which elicit a stress response in the coral animal in the form of upregulation of antioxidant genes (Griffin et al., 2006; Merle et al., 2007; DeSalvo et al., 2008; Richier et al., 2008; Sunagawa et al., 2008). Our observed lack of change in expression in antioxidant genes may indicate that the symbionts in the coral larvae are not stressed by the elevated temperatures imposed during the experiment and are not producing a greater number of ROS. This hypothesis is supported by Rodriguez-Lanetty et al. (2009), which suggested that ROS from symbionts are required to stimulate upregulation of host oxidative stress genes.
Ours is the first study to compare transcriptome-wide expression patterns between host and symbiont compartments in coral larvae in response to temperature and pCO2 conditions. Although other studies have examined the expression patterns of candidate genes in larvae (e.g., Putnam et al., 2013), the full transcriptome provides a global view of all genes involved in a particular stress response. Previous studies examining gene expression in animal and symbiont compartments of adult coral provide an interesting comparison to our results with larvae. For adult Acropora millepora near CO2 seeps in Papua New Guinea, expression changes in Symbiodinium were also greater in response to pCO2 than in the coral host (Kenkel et al., 2017). In adult A. hyacinthus, heat stress did not change symbiont transcriptomes (Barshis et al., 2014) but did affect expression of hundreds of host coral genes (Barshis et al., 2013). Fold change differences in Symbiodinium gene expression were smaller than for coral host genes in A. aspera following heat exposure (Leggat et al., 2011). The relative role of symbiont physiology and performance, as influenced by gene expression, may differ among life stages and species of coral. While host maternal effects and host plasticity at levels other than transcription are likely important, our results indicate that the symbiont may be playing a relatively large role in shaping the overall performance of symbiotic larvae post-release.
Gene Expression in the Symbiont
The gene expression profile in the symbionts of P. damicornis larvae changed dramatically in response to conditions of OA and warming. Parsing out the symbiont-specific gene expression response is important in the effort to accurately predict effects of global ocean change on coral-dinoflagellate symbioses, as Symbiodinium and other dinoflagellates can display muted or dramatic responses in gene expression to elevated temperature and other stressors (Rosic et al., 2010, 2011; Guo and Ki, 2012). Our results—greater expression changes in Symbiodinium vs. host—agree with a recent study in Papua New Guinea using adult A. millepora (Kenkel et al., 2017). As ocean conditions continue to change, coral animal populations will need to acclimatize or locally adapt to changes in environmental conditions (direct effects on animal host) as well as changes in symbiont performance (Kenkel et al., 2017) and in composition of hosted Symbiodinium genotypes/clades/types (DeSalvo et al., 2010).
Although symbiont density was significantly affected by the interaction of pCO2 and temperature, symbiont gene expression was only affected by pCO2. One of the most prominent effects of high pCO2 observed in this study was the upregulation of ion transport genes in symbionts. Other studies of gene expression responses of coral holobionts to elevated pCO2 have observed increased expression of ion transporters in high pCO2 treatments (Kaniewska et al., 2012; Vidal-Dupiol et al., 2013; Davies et al., 2016). However, most of these previous studies have observed changes in ion transporter expression on the part of the coral host and have hypothesized that these changes in gene regulation allowed the host to maintain conditions necessary for calcification. Our results represent the first observation (of which we are aware) of altered ion transporter expression on the part of the symbiont. Observations of isolated Symbiodinium have shown that compatible intracellular ion composition with the host improves carbon assimilation by zooxanthellae (Seibt and Schlichter, 2001). Our observations suggest that symbiont gene regulation may be playing an important role in the maintenance of holobiont homeostasis. However, it is also important to note that ion regulation is often energetically costly (Seibt and Schlichter, 2001), so if symbionts are required to increase active ion regulation under high pCO2 as part of physiological maintenance, it may come at the cost of long-term carbon assimilation. Alternatively, increased ion regulation under high pCO2, may reflect the higher capacity of symbionts to enhance carbon metabolism under increased carbon availability.
The larvae in this experiment contained Clade D1 Symbiodinium, known for the high thermotolerance of its symbiosis with corals (Stat and Gates, 2011). Although symbiont density varied by temperature as a function of pCO2 level, the lack of significant differential expression of symbiont genes among temperature levels supports the idea that the symbionts can tolerate the range of temperatures imposed with minimal adjustment from the level of gene expression. In Moorea, Pocilloporid corals are symbiotically flexible, known to harbor 5 symbiont types from clades A, C, and D (Putnam et al., 2012). While Clade D1 has a global distribution in tropical corals, it is relatively rare and is found in higher abundance in reefs exposed to high sea surface temperatures (Stat and Gates, 2011). The exclusive presence of Clade D1 in the larvae from 8 P. damicornis colonies suggests that these colonies may have a history of exposure to elevated temperatures in their shallow fringing reef habitat. Acclimatization to an environmental history of elevated temperature exposure may also explain the muted gene expression response in the coral animal. Additionally, the clade or genotype of symbiont harbored by the larvae in this study may have influenced the gene expression response of the coral, as previous work with adult Montipora faveolata showed that host gene expression profiles were significantly distinct based on the clade of symbiont hosted, above and beyond differential effects of thermal stress on gene expression (DeSalvo et al., 2010). Consequently, P. damicornis larvae harboring other symbiont clades may have produced different responses to conditions of high temperature and pCO2. Indeed, the magnitude of host gene expression response to thermal stress differs between corals hosting different symbiont clades (DeSalvo et al., 2010).
Our observation of the plethora of differentially expressed genes in Symbiodinium as compared to the coral host offers important insight for future efforts to examine the response of the coral holobiont to future ocean conditions. Measurements of symbiont density alone are not enough to adequately describe or predict the functional change of symbiosis, as symbiont density and gene expression were affected by different environmental factors in our study. Furthermore, while a change in symbiont density would affect the coral host through changes in photosynthate delivery, ROS production, etc. (Cunning and Baker, 2014), clearly, the physiological function of symbionts, as controlled by their gene expression, is responsive to external seawater conditions. Corals whose symbiont densities do not change in response to environmental stress may still experience a fundamental functional shift in their symbiotic relationships.
The Holobiont Perspective
Our study on genome-wide changes in expression of P. damicornis larvae in response to multiple stressors provides new insight for functioning of the coral-algal symbiosis under high-pCO2, high-temperature conditions as well as the role of the gene expression patterns of each partner that correlate with responses observed at the level of the whole organism. Firstly, our results indicate that many of the whole-organism responses previously observed for P. damicornis larvae in scenarios of OA and warming may reflect physiological plasticity at levels of control other than gene expression. For example, multiple studies have observed significant changes in oxygen consumption rates in P. damicornis larvae in response to elevated temperature and pCO2 (Cumbo et al., 2013a,b; Putnam et al., 2013; Rivest and Hofmann, 2014). However, in this study, coral genes involved in metabolic processes were not among the few differentially expressed genes. Additionally, Rivest and Hofmann (2014) found that citrate synthase activity did not differ among larvae cultured under combinations of temperature and OA conditions. Taken together, these results suggest that plasticity in metabolic rate may be accomplished through changes in fluxes of pathways using existing biochemical machinery, rather than the synthesis of new enzymes, at least for the population and seawater conditions evaluated.
Much of the work to date on responses of coral to environmental conditions has measured biological and physiological traits at the level of the whole-organism holobiont. For example, rates of growth/calcification and respiration are measured for a nubbin, colony, or intact larva. These measurements are made on the performance of the holobiont, and it is assumed that both coral and Symbiodinium contributed in some way to the response measured. The results of this study reveal strong potential merit in examining the gene expression of both partners, rather than just focusing on the gene expression of the coral animal host as a way to inform control of broader biological performance. While symbiont density and clade/genotype are known to cause differences in whole-organism performance like calcification and oxygen fluxes (e.g., Jokiel and Coles, 1977; Goreau and Macfarlane, 1990; Goulet et al., 2005), our results indicate that the performance of an existing symbiont population can change significantly with potential ramifications for host physiology and holobiont performance. Future work is needed to link differential expression in host and symbiont compartments with changes in processes of larval or colony biology.
Finally, it is clear that the responses of symbionts to future ocean conditions could play a large role in shaping success of coral larval stages. The relative role of symbiont function could depend on the clade/genotype hosted. Robust differential gene expression was observed in Symbiodinium clade D1 in P. damicornis larvae in response to experimental treatments. Pocilliopora damicornis is able to host other clades of Symbiodinium (Putnam et al., 2012), so corals may respond differently to environmental conditions based on unique transcriptomic responses of their symbiont clades. If the clade or genotype of symbiont is an important factor in holobiont function (through transcriptomic responses), then the brooding reproductive mode (vs. broadcast spawning) may be advantageous, as it allows parents to curate the symbionts the offspring receive. Symbiont clades that performed well and proliferated under the environmental conditions experienced by the parent can be vertically transmitted to brooded larvae and may allow them to perform better under those same environmental conditions immediately upon release.
Author Contributions
ER and GH contributed substantially to the conception and design of the work. ER, MK, and MD contributed substantially to the acquisition, analysis, and interpretation of data for the work. ER, MK, MD, and GH drafted and critically revised the work for important intellectual content. ER, MK, MD, and GH provided final approval of the version to be published. ER, MK, MD, and GH agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
This work was supported by the U.S. National Science Foundation under Grant Nos. OCE 10-26851 and OCE12-26905 as well as a generous gift from the Gordon and Betty Moore Foundation. Research was completed under permits issued by the French Polynesian Government (Délégation à la Recherche) and the Haut-commissariat de la République en Polynésie Francaise (DTRT) (Protocole d'Accueil 2005–2018). This work represents a contribution of the Moorea Coral Reef (MCR) LTER Site. During the course of this research, ER was supported by a National Science Foundation Graduate Research Fellowship as well as a graduate fellowship from the Department of Ecology, Evolution and Marine Biology at UC Santa Barbara.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SF and handling Editor declared their shared affiliation.
Acknowledgments
The culturing experiments for this research were conducted at the Richard B. Gump South Pacific Research Station (UC Berkeley) in collaboration with the Moorea Coral Reef Long-Term Ecological Research (MCR LTER) program. We thank Dr. Peter Edmunds and members of his research group for his support of this work. We thank Dr. Brian Rivest for assistance with experiments and fieldwork in Moorea, as well as Keith Seydel, Vince Moriarty, Dr. Steeve Comeau, Nate Spindel, Chelsea Behymer, and Gump Station staff. Many thanks to undergraduates Joshua Hancock, Katrina Shao, Silke Bachhuber, Farallon Broughton, Fiona Luong, and Yana Nebuchina for assistance with sample analysis. Portions of this research were conducted with high performance computing resources provided by Louisiana State University (http://www.hpc.lsu.edu). This paper is Contribution No. 3748 of the Virginia Institute of Marine Science, College of William & Mary.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00186/full#supplementary-material
References
Albright, R., and Langdon, C. (2011). Ocean acidification impacts multiple early life history processes of the Caribbean coral Porites astreoides. Glob. Chang. Biol. 17, 2478–2487. doi: 10.1111/j.1365-2486.2011.02404.x
Albright, R., Mason, B., Miller, M., and Langdon, C. (2010). Ocean acidification compromises recruitment success of the threatened Caribbean coral Acropora palmata. Proc. Natl. Acad. Sci. U.S.A. 107, 20400–20404. doi: 10.1073/pnas.1007273107
Andrews, S. (2014). FastQC a quality-control tool for high-throughput sequence data. Babraham Bioinformat. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Barshis, D. J., Ladner, J. T., Oliver, T. A., and Palumbi, S. R. (2014). Lineage-specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol. Biol. Evol. 31, 1343–1352. doi: 10.1093/molbev/msu107
Barshis, D. J., Ladner, J. T., Oliver, T. A., Seneca, F. O., Traylor-Knowles, N., and Palumbi, S. R. (2013). Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 1387–1392. doi: 10.1073/pnas.1210224110
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300.
Bradford, M. M. (1976). Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Byrne, M. (2011). “Impact of ocean warming and ocean acidification on marine invertebrate life history stages: vulnerabilities and potential for persistence in a changing ocean,” in Oceanography and Marine Biology: An Annual Review, eds R. N. Gibson, R. J. A. Atkinson, J. D. M. Gordon, I. P. Smith, and D. J. Hughes (Boca Raton, FL: CRC Press), 1–42.
Calosi, P., Rastrick, S. P. S., Graziano, M., Thomas, S. C., Baggini, C., Carter, H. A., et al. (2013). Distribution of sea urchins living near shallow water CO2 vents is dependent upon species acid–base and ion-regulatory abilities. Mar. Pollut. Bull. 73, 470–484. doi: 10.1016/j.marpolbul.2012.11.040
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carpenter, R. and of Moorea Coral Reef, LTER. (2016). MCR LTER: Coral Reef: Benthic Photosynthetically Active Radiation (PAR), ongoing since 2009. knb-lter-mcr.4005.8
Chan, N. C., and Connolly, S. R. (2013). Sensitivity of coral calcification to ocean acidification: a meta-analysis. Glob. Chang. Biol. 19, 282–290. doi: 10.1111/gcb.12011
Chomczynski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. doi: 10.1016/0003-2697(87)90021-2
Cumbo, V. R., Edmunds, P. J., and Wall, C. B. (2013a). Brooded coral larvae differ in their response to high temperature and elevated pCO2 depending on the day of release. Mar. Biol. 160, 2903–2917. doi: 10.1007/s00227-013-2280-y
Cumbo, V. R., Fan, T.-Y., and Edmunds, P. J. (2012). Physiological development of brooded larvae from two pocilloporid corals in Taiwan. Mar. Biol. 159, 2853–2866. doi: 10.1007/s00227-012-2046-y
Cumbo, V. R., Fan, T.-Y., and Edmunds, P. J. (2013b). Effects of exposure duration on the response of Pocillopora damicornis larvae to elevated temperature and high pCO2. J. Exp. Mar. Biol. Ecol. 439, 100–107. doi: 10.1016/j.jembe.2012.10.019
Cunning, R., and Baker, A. C. (2014). Not just who, but how many: the importance of partner abundance in reef coral symbioses. Front. Microbiol. 5:400. doi: 10.3389/fmicb.2014.00400
Davies, S. W., Marchetti, A., Ries, J. B., and Castillo, K. D. (2016). Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 3:112. doi: 10.3389/fmars.2016.00112
DeSalvo, M. K., Sunagawa, S., Fisher, P. L., Voolstra, C. R., Iglesias-Prieto, R., and Medina, M. (2010). Coral host transcriptomic states are correlated with Symbiodinium genotypes. Mol. Ecol. 19, 1174–1186. doi: 10.1111/j.1365-294X.2010.04534.x
DeSalvo, M. K., Voolstra, C. R., Sunagawa, S., Schwarz, J. A., Stillman, J. H., Coffroth, M. A., et al. (2008). Differential gene expression during thermal stress and bleaching in the Caribbean coral Montastraea faveolata. Mol. Ecol. 17, 3952–3971. doi: 10.1111/j.1365-294X.2008.03879.x
Fan, T.-Y., Li, J.-J., Ie, S.-X., and Fang, L.-S. (2002). Lunar periodicity of larval release by Pocilloporid corals in southern Taiwan. Zool. Stud. 41, 288–294. Available online at: http://zoolstud.sinica.edu.tw
Fan, T.-Y., Lin, K.-H., Kuo, F.-W., Soong, K., Liu, L.-L., and Fang, L.-S. (2006). Diel patterns of larval release by five brooding scleractinian corals. Mar. Ecol. Prog. Ser. 321, 133–142. doi: 10.3354/meps321133
Feely, R. A. (2008).“Ocean acidification,” in State of the Climate in 2007, eds D. H. Levinson and J. H. Lawrimore (Boston, MA: Bulletin of the American Meteorological Society), S1–S179.
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Goreau, T. J., and Macfarlane, A. H. (1990). Reduced growth rate of Montastrea annularis following the 1987?1988 coral-bleaching event. Coral Reefs 8, 211–215. doi: 10.1007/BF00265013
Goulet, T. L., Cook, C. B., and Goulet, D. (2005). Effect of short-term exposure to elevated temperatures and light levels on photosynthesis of different host-symbiont combinations in the Aiptasia pallida/Symbiodinium symbiosis. Limnol. Oceanogr. 50, 1490–1498. doi: 10.4319/lo.2005.50.5.1490
Graham, E. M., Baird, A. H., Connolly, S. R., Sewell, M. A., and Willis, B. L. (2017). Uncoupling temperature-dependent mortality from lipid depletion for scleractinian coral larvae. Coral Reefs 36, 97–104. doi: 10.1007/s00338-016-1501-5
Griffin, S. P., Bhagooli, R., and Weil, E. (2006). Evaluation of thermal acclimation capacity in corals with different thermal histories based on catalase concentrations and antioxidant potentials. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 144, 155–162. doi: 10.1016/j.cbpa.2006.02.017
Guo, R., and Ki, J.-S. (2012). Differential transcription of heat shock protein 90 (HSP90) in the dinoflagellate Prorocentrum minimum by copper and endocrine-disrupting chemicals. Ecotoxicology 21, 1448–1457. doi: 10.1007/s10646-012-0898-z
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-Seq: reference generation and analysis with Trinity. Nat. Protoc. 8, 1494–1512. doi: 10.1038/nprot.2013.084
Harii, S., Yamamoto, M., and Hoegh-Guldberg, O. (2010). The relative contribution of dinoflagellate photosynthesis and stored lipids to the survivorship of symbiotic larvae of the reef-building corals. Mar. Biol. 157, 1215–1224. doi: 10.1007/s00227-010-1401-0
Harriott, V. J. (1983). Reproductive seasonality, settlement, and post-settlement mortality of Pocillopora damicornis (Linnaeus), at Lizard Island, Great Barrier Reef. Coral Reefs 2, 151–157. doi: 10.1007/BF00336721
Harrison, P. L., and Wallace, C. C. (1990). “Reproduction, dispersal and recruitment of scleractinian corals,” in Ecosystems of the World, Vol. 25, Coral Reefs, ed Z. Dubinsky (New York, NY: Elsevier), 133–207.
Helm, R. R., Siebert, S., Tulin, S., Smith, J., and Dunn, C. (2013). Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genomics 14:266. doi: 10.1186/1471-2164-14-266
Heron, S. F., Maynard, J. A., van Hooidonk, R., and Eakin, C. M. (2016). Warming trends and bleaching stress of the world's coral reefs 1985–2012. Sci. Rep. 6:38402. doi: 10.1038/srep38402
Hofmann, G. E., O'Donnell, M. J., and Todgham, A. E. (2008). Using functional genomics to explore the effects of ocean acidification on calcifying organisms. Mar. Ecol. Prog. Ser. 373, 219–225. doi: 10.3354/meps07775
Hughes, T. P., Kerry, J. T., Álvarez-Noriega, M., Álvarez-Romero, J. G., Anderson, K. D., Baird, A. H., et al. (2017). Global warming and recurrent mass bleaching of corals. Nature 543, 373–377. doi: 10.1038/nature21707
IPCC (2013). “Climate change 2013: the physical science basis, ” in Contribution of the Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds T. F. Stocker, D. Qin, G.-K. Plattner, M. M. B. Tignor, S. K. Allen, J. Boschung, A. Nauels, Y. Xia, V. Bex, and P. M. Midgley (New York, NY: Cambridge University Press).
Jokiel, P. L., and Coles, S. L. (1977). Effects of temperature on the mortality and growth of Hawaiian reef corals. Mar. Biol. 43, 201–208. doi: 10.1007/BF00402312
Jones, A. M., Berkelmans, R., Oppen, M. J., Mieog, J. C., and Sinclair, W. (2008). A community change in the algal endosymbionts of a scleractinian coral following a natural bleaching event: field evidence of acclimatization. Proc. Biol. Sci. 275, 1359–1365. doi: 10.1098/rspb.2008.0069
Jones, P., Binns, D., Chang, H.-Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kaniewska, P., Campbell, P. R., Kline, D. I., Rodriguez-Lanetty, M., Miller, D. J., Dove, S., et al. (2012). Major cellular and physiological impacts of ocean acidification on a reef building coral. PLoS ONE 7:e34659. doi: 10.1371/journal.pone.0034659
Kenkel, C. D., Meyer, E., and Matz, M. (2013). Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 22, 4322–34. doi: 10.1111/mec.12390
Kenkel, C. D., Moya, A., Strahl, J., Humphrey, C., and Bay, L. K. (2017). Functional genomic analysis of corals from natural CO2-seeps reveals core molecular responses involved in acclimatization to ocean acidification. Glob. Chang. Biol. 24, 158–171. doi: 10.1111/gcb.13833
Ko, G. W., Dineshram, R., Campanati, C., Chan, V. B., Havenhand, J., and Thiyagarajan, V. (2014). Interactive effects of ocean acidification, elevated temperature, and reduced salinity on early-life stages of the Pacific oyster. Environ. Sci. Technol. 48, 10079–10088. doi: 10.1021/es501611u
Kültz, D. (2005). Molecular and evolutionary basis of the cellular stress response. Annu. Rev. Physiol. 67, 225–257. doi: 10.1146/annurev.physiol.67.040403.103635
Kurihara, H. (2008). Effects of CO2-driven ocean acidification on the early developmental stages of invertebrates. Mar. Ecol. Prog. Ser. 373, 275–284. doi: 10.3354/meps07802
Leggat, W., Seneca, F., Wasmund, K., Ukani, L., Yellowlees, D., and Ainsworth, T. D. (2011). Differential responses of the coral host and their algal symbiont to thermal stress. PLoS ONE 6:e26687. doi: 10.1371/journal.pone.0026687
Leichter, J. of Moorea Coral Reef, LTER. (2015). MCR LTER: Coral Reef: Benthic Water Temperature, ongoing since 2005. knb-lter-mcr.1035.10
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 23:323. doi: 10.1186/1471-2105-12-323
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lueker, T. J., Dickson, A. G., and Keeling, C. D. (2000). Ocean pCO2 calculated from dissolved inorganic carbon, alkalinity, and equations for K1 and K2: validation based on laboratory measurements of CO2 in gas and seawater at equilibrium. Mar. Chem. 70, 105–119. doi: 10.1016/S0304-4203(00)00022-0
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 17:10. doi: 10.14806/ej.17.1.200
Mass, T., Putnam, H. M., Drake, J. L., Zelzion, E., Gates, R. D., Bhattacharya, D., et al. (2016). Temporal and spatial expression patterns of biomineralization proteins during early development in the stony coral Pocillopora damicornis. Proc. R. Soc. Lond. B Biol. Sci. 283:20160322. doi: 10.1098/rspb.2016.0322
McCormick, M. I. (1999). Experimental test of the effect of maternal hormones on larval quality of a coral reef fish. Oecologia 118, 412–422. doi: 10.1007/s004420050743
Merle, P.-L., Sabourault, C., Richier, S., Allemand, D., and Furla, P. (2007). Catalase characterization and implication in bleaching of a symbiotic sea anemone. Free Radic. Biol. Med. 42, 236–246. doi: 10.1016/j.freeradbiomed.2006.10.038
Metzger, R., Sartoris, F. J., Langenbuch, M., and Pörtner, H. O. (2007). Influence of elevated CO2 concentrations on thermal tolerance of the edible crab Cancer pagurus. J. Therm. Biol. 32, 144–151. doi: 10.1016/j.jtherbio.2007.01.010
Meyer, E., Aglyamova, G. V., and Matz, M. (2011). Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol. Ecol. 20, 3599–3616. doi: 10.1111/j.1365-294X.2011.05205.x
Moya, A., Ganot, P., Furla, P., and Sabourault, C. (2012). The transcriptomic response to thermal stress is immediate, transient and potentiated by ultraviolet radiation in the sea anemone Anemonia viridis. Mol. Ecol. 21, 1158–1174. doi: 10.1111/j.1365-294X.2012.05458.x
Moya, A., Huisman, L., Forêt, S., Gattuso, J.-P., Hayward, D. C., Ball, E. E., et al. (2015). Rapid acclimation of juvenile corals to CO2-mediated acidification by upregulation of heat shock protein and Bcl-2 genes. Mol. Ecol. 24, 438–452. doi: 10.1111/mec.13021
Okubo, N., Yamamoto, H. H., Nakaya, F., and Okaji, K. (2008). Oxygen consumption of a single embryo/planula in the reef-building coral Acropora intermedia. Mar. Ecol. Prog. Ser. 366, 305–309. doi: 10.3354/meps07562
Pechenik, J. A. (1999). On the advantages and disadvantages of larval stages in benthic marine invertebrate life cycles. Mar. Ecol. Prog. Ser. 177, 269–297. doi: 10.3354/meps177269
Pinheiro, J. C., and Bates, D. M. (2000). Mixed-effects Models in S and S-Plus. New York, NY: Springer.
Place, S. P., and Smith, B. W. (2012). Effects of seawater acidification on cell cycle control mechanisms in Strongylocentrotus purpuratus embryos. PLoS ONE 7:e34068. doi: 10.1371/journal.pone.0034068
Pochon, X., Montoya-Burgos, J. I., Stadelmann, B., and Pawlowski, J. (2006). Molecular phylogeny, evolutionary rates, and divergence timing of the symbiotic dinoflagellate genus Symbiodinium. Mol. Phylogenet. Evol. 38, 20–30. doi: 10.1016/j.ympev.2005.04.028
Pörtner, H.-O. (2008). Ecosystem effects of ocean acidification in times of ocean warming: a physiologist's view. Mar. Ecol. Prog. Ser. 373, 203–217. doi: 10.3354/meps07768
Pörtner, H.-O., Bock, C., and Mark, F. C. (2017). Oxygen- and capacity-limited thermal tolerance: bridging ecology and physiology. J. Exp. Biol. 220, 2685–2696. doi: 10.1242/jeb.134585
Prada, F., Caroselli, E., Mengoli, S., Brizi, L., Fantazzini, P., Capaccioni, B., et al. (2017). Ocean warming and acidification synergistically increase coral mortality. Sci. Rep. 7:40842. doi: 10.1038/srep40842
Putnam, H. M., Edmunds, P. J., and Fan, T.-Y. (2010). Effect of a fluctuating thermal regime on adult and larval reef corals. Invertebr. Biol. 129, 199–209. doi: 10.1111/j.1744-7410.2010.00199.x
Putnam, H. M., and Gates, R. D. (2015). Preconditioning in the reef-building coral Pocillopora damicornis and the potential for trans-generational acclimatization in coral larvae under future climate change conditions. J. Exp. Biol. 218, 2365–2372. doi: 10.1242/jeb.123018
Putnam, H. M., Mayfield, A. B., Fan, T.-Y., Chen, C.-S., and Gates, R. D. (2013). The physiological and molecular responses of larvae from the reef-building coral Pocillopora damicornis exposed to near-future increases in temperature and pCO2. Mar. Biol. 160, 2157–2173. doi: 10.1007/s00227-012-2129-9
Putnam, H. M., Stat, M., Pochon, X., and Gates, R. D. (2012). Endosymbiotic flexibility associates with environmental sensitivity in scleractinian corals. Proc. R. Soc. B Biol. Sci. 279, 4352–4361. doi: 10.1098/rspb.2012.1454
Rasband, W. S. (1997). ImageJ. Available online at: http://imagej.nih.gov/ij/
R Core Team (2013). R: A language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Richier, S., Cottalorda, J.-M., Guillaume, M. M. M., Fernandez, C., Allemand, D., and Furla, P. (2008). Depth-dependant response to light of the reef building coral, Pocillopora verrucosa: implication of oxidative stress. J. Exp. Mar. Biol. Ecol. 357, 48–56. doi: 10.1016/j.jembe.2007.12.026
Richmond, R. H., and Jokiel, P. L. (1984). Lunar periodicity in larva release in the reef coral Pocillopora damicornis at Enewetak and Hawaii. Bull. Mar. Sci. 34, 280–287.
Rivest, E. B., Chen, C.-S., Fan, T.-Y., Li, H.-H., and Hofmann, G. E. (2017). Lipid consumption in coral larvae differs among sites: a consideration of environmental history in a global ocean change scenario. Proc. Biol. Sci. 284:20162825. doi: 10.1098/rspb.2016.2825
Rivest, E. B., and Hofmann, G. E. (2014). Responses of the metabolism of the larvae of Pocillopora damicornis to ocean acidification and warming. PLoS ONE 9:e96172. doi: 10.1371/journal.pone.0096172
Rivest, E. B., and Hofmann, G. E. (2015). Effects of temperature and pCO2 on lipid use and biological parameters of planulae of Pocillopora damicornis. J. Exp. Mar. Biol. Ecol. 473, 43–52. doi: 10.1016/j.jembe.2015.07.015
Rivest, E. of Moorea Coral Reef, LTER. (2014). MCR LTER: Coral Reef: Seawater pH, Temperature and Depth Time Series from Bottom-mounted Sensors on the Fringing Reef, January-March 2012. knb-lter-mcr.5014.1.
Robbins, L. L., Hansen, M. E., Kleypas, J. A., and Meylan, S. C. (2010). CO2calc - a user-friendly seawater carbon calculator for Windows, Mac OS X and iOS (iPhone). US Geological Survey Open-File Report 2010–1280.
Rodriguez-Lanetty, M., Harii, S., and Hoegh-Guldberg, O. (2009). Early molecular responses of coral larvae to hyperthermal stress. Mol. Ecol. 18, 5101–5114. doi: 10.1111/j.1365-294X.2009.04419.x
Rosic, N. N., Pernice, M., Dove, S., Dunn, S., and Hoegh-Guldberg, O. (2011). Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: possible implications for coral bleaching. Cell Stress Chaperones 16, 69–80. doi: 10.1007/s12192-010-0222-x
Rosic, N. N., Pernice, M., Dunn, S., Dove, S., and Hoegh-Guldberg, O. (2010). Differential regulation by heat stress of novel cytochrome P450 genes from the dinoflagellate symbionts of reef-building corals. Appl. Environ. Microbiol. 76, 2823–2829. doi: 10.1128/AEM.02984-09
Sabine, C. L., Feely, R. A., Gruber, N., Key, R. M., Lee, K., Bullister, J. L., et al. (2004). The oceanic sink for anthropogenic CO2. Science 305, 367–371. doi: 10.1126/science.1097403
Seibt, C., and Schlichter, D. (2001). Compatible intracellular ion composition of the host improves carbon assimilation by zooxanthellae in mutualistic symbioses. Naturwissenschaften 88, 382–386. doi: 10.1007/s001140100240
Stat, M., and Gates, R. D. (2011). Clade D Symbiodinium in scleractinian corals: a “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J. Mar. Biol. 2011:730715. doi: 10.1155/2011/730715
Sunagawa, S., Choi, J., Forman, H. J., and Medina, M. (2008). Hyperthermic stress-induced increase in the expression of glutamate-cysteine ligase and glutathione levels in the symbiotic sea anemone Aiptasia pallida. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 151, 133–138. doi: 10.1016/j.cbpb.2008.06.007
Timmins-Schiffman, E., O'Donnell, M. J., Friedman, C. S., and Roberts, S. B. (2013). Elevated pCO2 causes developmental delay in early larval Pacific oysters, Crassostrea gigas. Mar. Biol. 160, 1973–1982. doi: 10.6084/M9.FIGSHARE.95828
Todgham, A. E., and Hofmann, G. E. (2009). Transcriptomic response of sea urchin larvae Strongylocentrotus purpuratus to CO2-driven seawater acidification. J. Exp. Biol. 212, 2579–2594. doi: 10.1242/jeb.032540
Tresguerres, M., and Hamilton, T. J. (2017). Acid-base physiology, neurobiology and behaviour in relation to CO2-induced ocean acidification. J. Exp. Biol. 220, 2136–2148. doi: 10.1242/jeb.144113
Vidal-Dupiol, J., Zoccola, D., Tambutté, E., Grunau, C., Cosseau, C., Smith, K. M., et al. (2013). Genes related to ion-transport and energy production are upregulated in response to CO2-driven pH decrease in corals: new insights from transcriptome analysis. PLoS ONE 8:e58652. doi: 10.1371/journal.pone.0058652
Voolstra, C. R., Schnetzer, J., Peshkin, L., Randall, C. J., Szmant, A. M., and Medina, M. (2009). Effects of temperature on gene expression in embryos of the coral Montastraea faveolata. BMC Genomics 10:627. doi: 10.1186/1471-2164-10-627
Walther, K., Sartoris, F. J., Bock, C., and Pörtner, H. O. (2009). Impact of anthropogenic ocean acidification on thermal tolerance of the spider crab Hyas araneus. Biogeosciences 6, 2207–2215. doi: 10.5194/bg-6-2207-2009
Whitehead, A., Galvez, F., Zhang, S., Williams, L. M., and Oleksiak, M. F. (2010). Functional genomics of physiological plasticity and local adaptation in killifish. J. Hered. 102, 499–511. doi: 10.1093/jhered/esq077
Wright, R. M., Aglyamova, G. V., Meyer, E., and Matz, M. V. (2015). Gene expression associated with white syndromes in a reef building coral, Acropora hyacinthus. BMC Genomics 16:371. doi: 10.1186/s12864-015-1540-2
Keywords: coral larvae, ocean acidification, ocean warming, holobiont, transcriptomics, Symbiodinium, moorea, multiple stressors
Citation: Rivest EB, Kelly MW, DeBiasse MB and Hofmann GE (2018) Host and Symbionts in Pocillopora damicornis Larvae Display Different Transcriptomic Responses to Ocean Acidification and Warming. Front. Mar. Sci. 5:186. doi: 10.3389/fmars.2018.00186
Received: 19 October 2017; Accepted: 09 May 2018;
Published: 30 May 2018.
Edited by:
Stefano Goffredo, Università degli Studi di Bologna, ItalyReviewed by:
Susana Enríquez, Universidad Nacional Autónoma de México, MexicoSilvia Franzellitti, Università degli Studi di Bologna, Italy
Copyright © 2018 Rivest, Kelly, DeBiasse and Hofmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emily B. Rivest, ZWJyaXZlc3RAdmltcy5lZHU=
 Emily B. Rivest
Emily B. Rivest Morgan W. Kelly
Morgan W. Kelly Melissa B. DeBiasse
Melissa B. DeBiasse Gretchen E. Hofmann
Gretchen E. Hofmann