- 1Microbial Systems Ecology, Department of Freshwater and Marine Ecology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, Amsterdam, Netherlands
- 2Marine Ecology and Evolution Research Group, CCMAR-CIMAR Centre for Marine Sciences, Universidade do Algarve, Faro, Portugal
Seagrasses play an important role as ecosystem engineers; they provide shelter to many animals and improve water quality by filtering out nutrients and by controlling pathogens. Moreover, their rhizosphere promotes a myriad of microbial interactions and processes, which are dominated by microorganisms involved in the sulfur cycle. This study provides a detailed insight into the metabolic sulfur pathways in the rhizobiome of the seagrass Zostera marina, a dominant seagrass species across the temperate northern hemisphere. Shotgun metagenomic sequencing revealed the relative dominance of Gamma- and Deltaproteobacteria, and comparative analysis of sulfur genes identified a higher abundance of genes related to sulfur oxidation than sulfate reduction. We retrieved four high-quality draft genomes that are closely related to the gill symbiont of the clam Solemya velum, which suggests the presence of putative free-living forms of symbiotic bacteria. These are potentially highly versatile chemolithoautotrophic bacteria, able to alternate their metabolism between parallel pathways of sulfide oxidation (via sqr and fcc), nitrate reduction (denitrification or DNRA) and carbon fixation (via CBB or TCA cycle), depending on the environmental availability of sulfide. Our results support the hypothesis that seagrass meadows might function as a source of symbionts for invertebrates that inhabit within or around seagrass meadows. While providing ideal conditions for the proliferation of these free-living forms of symbionts, seagrasses would benefit from their genetic versatility, which contributes to sulfide detoxification and ammonium production, the seagrasses' preferred nitrogen source.
Introduction
Global carbon and sulfur biogeochemical cycles are tightly coupled in marine sediments, mostly through sulfate reduction, which is responsible for approximately 50% of the organic carbon remineralization in anoxic coastal shelf sediments (Jørgensen, 1982). Although marine sediments in general harbor active biogeochemical cycles, these seem to be further boosted in seagrass-vegetated areas (Devereux, 2005). The rhizosphere of seagrasses is a thin layer of sediment surrounding the roots, which is highly enriched with dissolved organic matter (OM), mostly originated from photosynthetic products that are released through the roots (Holmer and Nielsen, 1997; Pérez et al., 2007). These root exudates create microniches that are distributed along the rhizosphere according to the availability of the most energetically favorable terminal electron acceptors for microbial respiration (Capone and Kiene, 1988; Devereux, 2005), creating hotspots of microbial activity (Holmer and Nielsen, 1997; Blaabjerg and Finster, 1998; Donnelly and Herbert, 1999). The high abundance of sulfate in seawater (Capone and Kiene, 1988) coupled with OM enrichment in the rhizosphere, enhance the activity of sulfate reducing bacteria (SRB, Holmer and Nielsen, 1997; Blaabjerg and Finster, 1998; Donnelly and Herbert, 1999). This results in the production of high levels of sulfide, which pose a strong threat to seagrass health and survival (Borum et al., 2005; Pérez et al., 2007). Detoxification of sulfide in the rhizosphere occurs by different chemical and biological processes. Chemical oxidation can occur by oxygen released from the seagrass roots during photosynthesis (Jørgensen and Nelson, 2004; Borum et al., 2005; Frederiksen and Glud, 2006; Holmer et al., 2006), or by binding to iron resulting in iron sulfide and pyrite (Jørgensen and Nelson, 2004). Although the majority of studies focus on the chemical oxidation of sulfide, its biological oxidation by sulfide-oxidizing bacteria (SOB) also plays an important role in coastal marine sediments (Jørgensen and Nelson, 2004; Frigaard and Dahl, 2008).
The microbial communities established in the root and rhizosphere of seagrasses are mainly dominated by members of the classes Alpha-, Delta-, Epsilon-, and Gammaproteobacteria, as well as Bacteroidia (Cifuentes et al., 2000; Jensen et al., 2007; Green-García and Engel, 2012; Cúcio et al., 2016; Mejia et al., 2016; Fahimipour et al., 2017). The most abundant bacteria identified on the root and rhizosphere have been related to sulfate reduction and sulfur oxidation (Cifuentes et al., 2000; Crump and Koch, 2008; Cúcio et al., 2016; Fahimipour et al., 2017), and their importance has been linked to nitrogen fixation and sulfide detoxification (Donnelly and Herbert, 1999; Cifuentes et al., 2000; Cúcio et al., 2016). Cifuentes et al. (2000) detected 16S rRNA genes of sulfur-oxidizing Gammaproteobacteria closely related to marine endosymbionts in sediments colonized by Zostera noltii, which highlights the potential for an intimate relationship between seagrasses and the bacteria in their rhizosphere. Several reports suggest that seagrass-inhabited sediments harbor hosts of sulfur-oxidizing and sulfate-reducing endosymbionts, such as bivalves (Gros et al., 2003; Van der Heide et al., 2012; Dmytrenko et al., 2014; König et al., 2016) and marine oligochaetes (Dubilier et al., 2001; Blazejak et al., 2005), as well as some free-living forms of these symbionts (e.g., Gros et al., 2003; König et al., 2016).
Zostera marina, also known as eelgrass, is the most widespread seagrass species in the world (Green and Short, 2003). It is found in subtidal areas up to 15 meters depth (Borum and Greve, 2004) along temperate coasts (Green and Short, 2003) throughout the northern hemisphere. Eelgrasses, as seagrasses in general, are ecosystem engineers. They provide valuable ecosystem services, such as a habitat, refuge and nursery ground for many animals, improve water quality through sediment and organic matter retention, and by filtering out nutrients and contaminants (Gacia et al., 1999; Short et al., 2000). Recently it was shown that seagrasses might even control the abundance of potential pathogens of marine animals and humans (Lamb et al., 2017).
In a previous study in which we used 16S rRNA gene sequencing, we found that bacteria involved in the sulfur cycle are abundant in the rhizosphere of different European seagrass species (Cúcio et al., 2016). Although the importance of biogeochemical sulfur cycling in coastal marine sediments has been widely studied, to our knowledge, no information is available about the molecular pathways involved in sulfate reduction and sulfide oxidation in the seagrass rhizosphere. In the present study, we investigated the diversity, structure and gene content of bacterial communities involved in dissimilatory sulfur cycling in the rhizosphere of Zostera marina, and hypothesized that the rhizosphere harbors a diverse community of bacteria involved in the sulfur cycle, which is enriched in sulfide oxidizers over sulfate reducers. Furthermore, we reconstructed and characterized four gammaproteobacterial draft genomes. Shotgun metagenomic sequencing allowed us to investigate for the first time in detail the molecular pathways involved in sulfate reduction and sulfide oxidation in the rhizosphere.
Materials and Methods
Sample Collection and Preparation
The rhizosphere of the seagrass Zostera marina was sampled at Culatra Island (ZmPt-Faro, Portugal, 36°59′56.0″ N 7°49′31.7″ W) and Pointe de Cléguer (ZmFr-Roscoff, France, 48°43′37.1″ N 3°58′35.9″ W), in 2013 as described and used in Cúcio et al. (2016). Briefly, sampling consisted of randomly collecting cores (15 cm diameter) of Z. marina (n = 5), from which 4–6 shoots were carefully separated and shaken to remove loose sediment. The rhizosphere was retrieved by washing the roots in 0.2 μm-filtered seawater, and was subsequently transported to the laboratory in cool boxes. Upon arrival in the laboratory, meiofauna and plant detritus were removed whenever present, and samples were treated in a homogenizer (Stomacher 80 Laboratory Blender, Seward Medical) to loosen the bacteria from the sediment particles. After 3 homogenizing cycles of 1 min at normal speed, the supernatant was centrifuged for 30 min at 10,000 g (Costa et al., 2006), and the resulting pellet was used for DNA extractions.
Metagenomic Sequencing
Extraction of genomic DNA from the rhizosphere bacteria was performed using the PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, CA, USA), according to manufacturer's instructions. Part of the DNA resulting from this extraction was used in our previous publication as 5 individual replicates per location (Cúcio et al., 2016), and due to the highly comparable community composition among the samples within location (Cúcio et al., 2016), all replicates of each location were pooled to create two rhizosphere samples for shotgun metagenomic analysis, one from Portugal (Pt) and another from France (Fr). The quality of the genomic DNA was determined by agarose electrophoresis, and the quantification was performed with a dsDNA HS Assay Kit on a Qubit 2.0 Fluorometer.
The genomic DNA of the two samples was sequenced by the Beijing Genomics Institute (BGI, Hong Kong, China) in one lane of paired-end sequences with an insert size of 170 bp, using an Illumina HiSeq 2000 PE 100 sequencer.
Metagenomic Analysis
The quality of the metagenomic reads was checked with FastQC (Babraham Bioinformatics, http://www.bioinformatics.babraham.ac.uk/projects/fastqc), and trimmed using CLC Genomics Workbench (v8.5.1, CLCbio) with a quality score limit of 0.05, and a maximum allowance of 2 ambiguous nucleotides.
Taxonomic assignment of metagenomic reads was carried out using Kaiju (http://kaiju.binf.ku.dk), using the non-redundant protein database NCBI BLAST nr as reference database (Menzel et al., 2016). “Greedy” run mode was applied with a minimum match score of 70, and an allowance of five mismatches (Menzel et al., 2016). High-quality metagenomic paired-end reads were assembled using IDBA-UD (v1.1.1, Peng et al., 2012), with pre-correction for improved efficiency of highly uneven sequencing depth, and k-mer sizes from 40 to 100. To obtain more insights into the eelgrass rhizobiome and to increase the amount of information for coverage and binning, both metagenomes were further co-assembled using MEGAHIT (v1.0.3-6, Li et al., 2015), using a minimum k-mer size of 25 incremented in steps of 10 on each iteration. Hereinafter, results obtained from the individually assembled metagenomes are referred to by the name of their sampling location (i.e., “Portugal” or “ZmPt”, and “France” or “ZmFr”), and results obtained from the co-assembled metagenomes are referred to as “co-assembly”.
Protein-coding genes were predicted with Prodigal (Hyatt et al., 2010), and tRNAs with tRNAscan-SE (Lowe and Eddy, 1997). Annotation of protein-coding genes was performed with BLAST against COGs (Tatusov et al., 2003), TIGRfams (Haft et al., 2001), and NCBI's non-redundant database. In parallel, annotation of functional genes was also performed using the KEGG (Kyoto Encyclopedia of Genes and Genomes) Orthologs (KO) database in the KEGG's online annotation tool, GhostKOALA (Kanehisa et al., 2016). For those genes that can participate in both oxidative and reductive pathways of the sulfur cycle (Table 1), the function was confirmed by phylogenetic placement in reference trees, as well as according to the percentage of similarity to the closest hit attributed by BLAST during annotation, using a minimum identity of 75%.
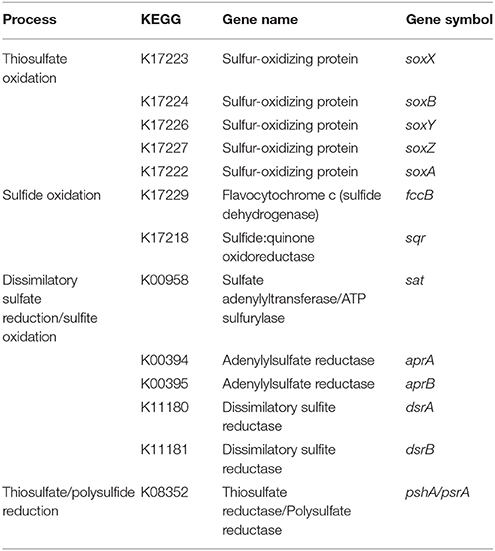
Table 1. Summary of marker genes for the (dissimilatory) sulfur metabolism present in the metagenomes of the rhizobiomes of Zostera marina.
The presence of specific eukaryotic sequences was investigated based on raw reads, using the MG-RAST server (v4, Meyer et al., 2008).
Phylogenetic Reconstruction of Sulfur Genes
Databases of the marker genes for dissimilatory sulfur processes (“sulfur” genes) adenylylsulfate reductase (aprA) and sulfate adenylyltransferase (sat) were built using reference alignments of the ortholog clusters ENOG4105CFK and ENOG4107RIQ, respectively, retrieved from the eggNOG phylogenomic database (v4.5, Huerta-Cepas et al., 2016). A database of sulfide:quinone oxidoreductase/flavocytochrome c (sqr/fcc) orthologs was generated with sequences used in previous work performed by Marcia et al. (2010) and Han and Perner (2015). To complement the databases with genetic information about symbiotic bacteria, amino acid sequences of the proteins aprA, sat, sqr and fccB from symbionts were obtained from the Nucleotide database of NCBI (https://www.ncbi.nlm.nih.gov) by searching the terms “<gene of interest> AND symbiont.” A reference alignment for these proteins was performed on the online multiple sequence alignment tool Clustal Omega (Sievers et al., 2011). To build the phylogenetic tree for the sulfur gene dissimilatory sulfite reductase dsrAB, we used the reference database published by Müller et al. (2015).
Contigs annotated as orthologs of each gene were manually aligned to the reference alignments of their respective database in the ARB alignment tool (Ludwig et al., 2004), and phylogenetic trees were calculated using an approximately maximum-likelihood method in FastTree v2.1.3 (Price et al., 2010). Contigs that did not align to the reference databases used were not included in this analysis.
Reconstruction and Analysis of Draft Genomes
Assembled contigs (>1 kb) from ZmPt, ZmFr and co-assembly, were binned according to their pentanucleotide frequency using VizBin (Laczny et al., 2015). Taxonomic affiliation, completeness, contamination, strain heterogeneity and coverage of draft genomes were determined with the software CheckM (v1.0.5, Parks et al., 2015). Furthermore, assembled genomes were manually inspected to verify that the operons/gene cassettes are consistent in their structure. We retrieved about 20 draft genomes, but only selected those that had a completeness of > 70% and a contamination of < 5%. To identify the potential for CO2 fixation, we searched for the carbon fixation marker genes cbbL, cbbS and cbbM from the Calvin-Benson-Bassham cycle (CBB), and korAB, frdA, porAB, and aclAB from the reverse tricarboxylic acid cycle (rTCA). The presence of nitrate-reducing and anammox bacteria was investigated through the presence/absence of genes involved in denitrification (narG, napAB, nirS, nirK, norC, and nosZ), dissimilatory nitrate reduction to ammonium (DNRA; nrfA), and anaerobic ammonium oxidation (Anammox; hzo, and hao). Nitrogenase-encoding genes nifH, nifD and nifK were also searched for, in order to identify nitrogen-fixing bacteria. Furthermore, sulfur metabolism was assessed by investigating the presence of the sulfur genes mentioned in the previous section, and summarized in Table 1. The search for these genes was performed based on their KO identifiers and gene/protein names.
Results
Metagenomic sequencing resulted in approximately 98 and 91 million high-quality paired-end reads from the rhizospheres sampled in Portugal (ZmPt) and France (ZmFr), respectively. The metagenome ZmPt yielded better quality results than the metagenome ZmFr, i.e., more than twice as many contigs larger than 1 kb and a N50 of 1,150 bp, compared to a N50 of 1,097 bp in ZmFr (Table 2). The co-assembly resulted in a total of 76,983 contigs larger than 1 kb and a N50 of 1,768 bp, higher values than those observed for ZmPt and ZmFr (Table 2). The average G+C content of the contigs from ZmPt, ZmFr and the co-assembly was 55.61, 50.79, and 54.20%, respectively (Table 2).
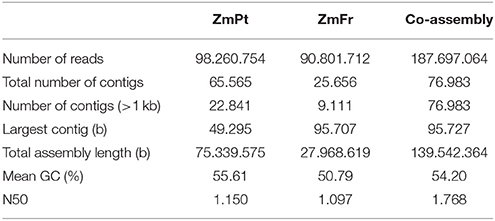
Table 2. Characteristics of the metagenomes from the rhizobiomes of Zostera marina sampled in Portugal (ZmPt), France (ZmFr), and the co-assembly of both.
Bacterial Community Structure
Based on the most abundant bacterial classes identified among classified reads, the community structure, as revealed by taxonomic assignment of raw sequence reads, was highly similar between Portugal and France. The percentage of unclassified reads was similar in both metagenomes (37.36% in ZmPt and 38.57% in ZmFr). Nearly 98% of the classified reads from both metagenomes could be assigned to Bacteria, whereas < 2% of the reads were assigned to Archaea, and 0.1–0.2% of the reads had a viral origin. Based on classified reads, the bacterial communities were dominated by Gammaproteobacteria (25.3% for ZmPt, and 21.6% for ZmFr), followed by Deltaproteobacteria (20.0% for ZmPt, and 16.3% for ZmFr), and Alphaproteobacteria (7.9% for ZmPt, and 12.2% for ZmFr) (Figure 1, Supplementary Table 1). The most abundant classes were comparable between sites. However, a larger difference was found in the number of reads from the less abundant classes, such as Flavobacteriia, Anaerolineae, Gemmatimonadetes, Epsilonproteobacteria, Caldinae, Acidimicrobiia, Dehalococcoidia, Ardenticatenia, Mollicutes, and Thermoflexia (Supplementary Table 1). The order Desulfobacterales equally dominated the Deltaproteobacteria in both sites, whereas Chromatiales, the most dominant order of the Gammaproteobacteria in the metagenomes, harbored nearly two-fold more reads in Portugal than in France (Figure 1, Supplementary Table 2).
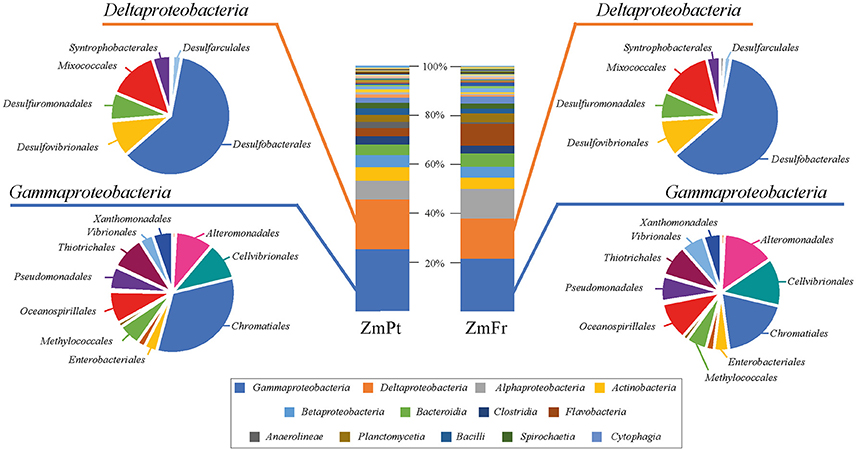
Figure 1. Community composition of the microbiomes from the rhizosphere of Zostera marina from Portugal (ZmPt) and France (ZmFr). Taxonomic classification of metagenomic reads of bacterial classes (bar charts), and orders of the two most abundant classes, the Gamma- and the Deltaproteobacteria (pie charts). All unassigned reads, as well as those unclassified at higher taxonomic ranks were excluded from the analysis. Taxonomy assignment was performed on raw reads in the software program Kaiju (Menzel et al., 2016).
To provide genome and ecosystem level insight in the general rhizosphere of Z. marina we co-assembled the metagenomes from Portugal and France. By mapping the reads from both metagenomes to the co-assembly, we identified a higher coverage in Portugal than in France (Figure 2). Furthermore, although most reads were not shared between sites, three clusters of the co-assembly formed a clear exception, as their coverage was high for both metagenomes (clusters 1, 2, and 3, highlighted in Figure 2). Assembly of the reads of each cluster followed by phylogenetic placement with CheckM allowed the identification of cluster 1 as a member of the family Desulfobacteraceae within the Deltaproteobacteria and cluster 2 and 3 as Gammaproteobacteria.
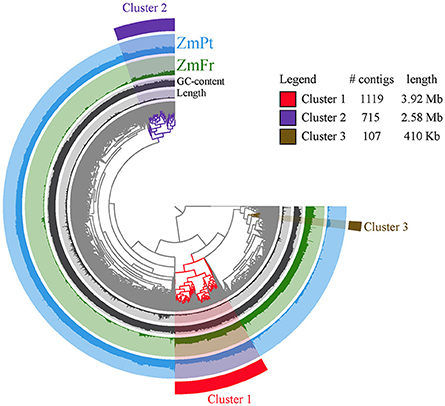
Figure 2. Mapping of reads to the co-assembled metagenomes. Visualization of the contigs from Portugal (blue, ZmPt) and France (green, ZmFr) mapped against the co-assembled metagenomes, represented in a log scale with a maximum of 2.2 for the former and 3.2 for the latter, respectively. The GC content is also represented, with a maximum of 73%. The dendogram was calculated based on sequence composition and differential coverage of raw reads.
Diversity of Sulfur Genes in the Rhizobiome of Z. marina
A total of 263 high-quality sulfur-related gene copies were retrieved from the rhizosphere of ZmPt, from which 90% were assigned to the oxidative sulfur cycle with an average total coverage of 125x, in contrast to reductive genes, which only had an average total coverage of 35x (Figure 3).
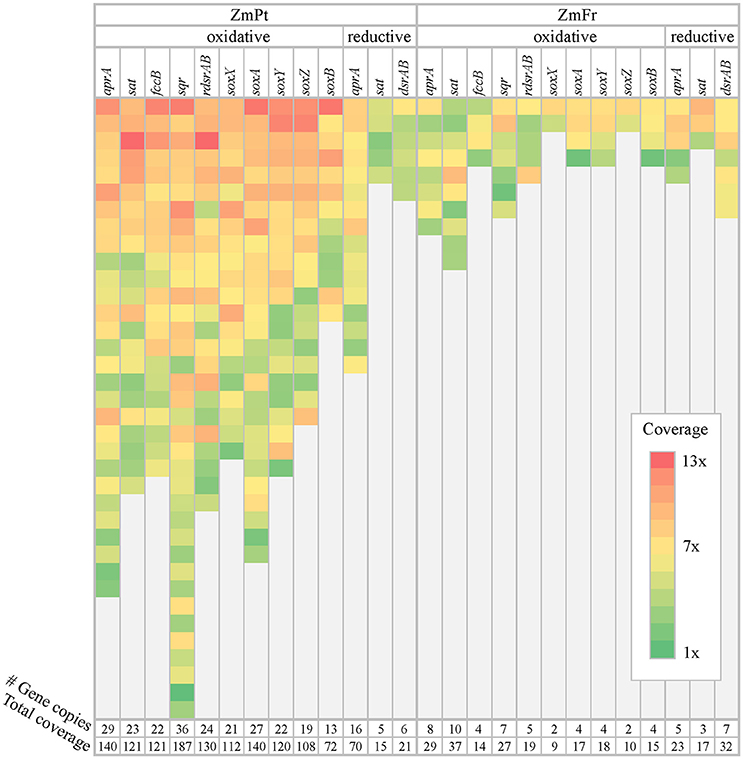
Figure 3. Diversity of sulfur genes in the rhizobiome of Zostera marina. Distribution of genes involved in the (dissimilatory) sulfur metabolism throughout the metagenomes of the rhizosphere of Z. marina sampled in Portugal (ZmPt) and France (ZmFr). The partial coverage of each gene copy of the marker genes is showed per cell according to the color label, and the total coverage of each marker gene in the metagenome is indicated at the bottom of the heat-map in absolute values.
The most abundant oxidation pathway in ZmPt was sulfide oxidation via sqr with a total coverage of 187x, followed by sulfite oxidation via aprAB, and thiosulfate oxidation via the sox complex (Figure 3). On the other hand, aprAB dominated the pool of reductive genes, with a total coverage of 70x (Figure 3). Only 65 sulfur-related gene copies were identified in the rhizosphere of ZmFr (Figure 3), of which 77% were assigned to genes involved in oxidative pathways. Nevertheless, reductive sulfur pathways dominated ZmFr in terms of coverage (average coverage of 20x vs. 24x for oxidative and reductive sulfur genes, respectively, Figure 3).
Phylogenetic analysis allowed the distinction of dsrAB (Figure 4), aprAB (Supplementary Figure 1) and sat (Supplementary Figure 2) genes from oxidative and reductive microorganisms. Contig sequences aligning to sequences in the reverse operating dsrAB database were mostly affiliated to the Gammaproteobacteria, whereas those aligning to the direct operating dsrAB were affiliated to the Deltaproteobacteria. Several contigs were grouping with symbiotic bacteria, such as a cluster of five contigs closely related to the endosymbiont of the polychaete Oligobrachia haakonmosbiensis (contigs FR-20896a, PT-3377ab, PT-61199b, PT-1051a, and PT-10561ab). Eight other contigs were directly related to endosymbionts of the oligochaete worms Olavius algarvensis and O. ilvae (contigs PT-42780ab, PT-47632ab, PT-7484ab, PT-55619ab as Gammaproteobacteria, and contigs FR-942b, FR-17686a, FR-8850b, and PT-27360ab as Deltaproteobacteria). Moreover, three other contigs were related to Candidatus Thiobios zoothamnicoli (contig PT-954ab, PT-562ab, and PT-16897a), an ectosymbiont of the ciliate Zoothamnium niveum (Figure 4).
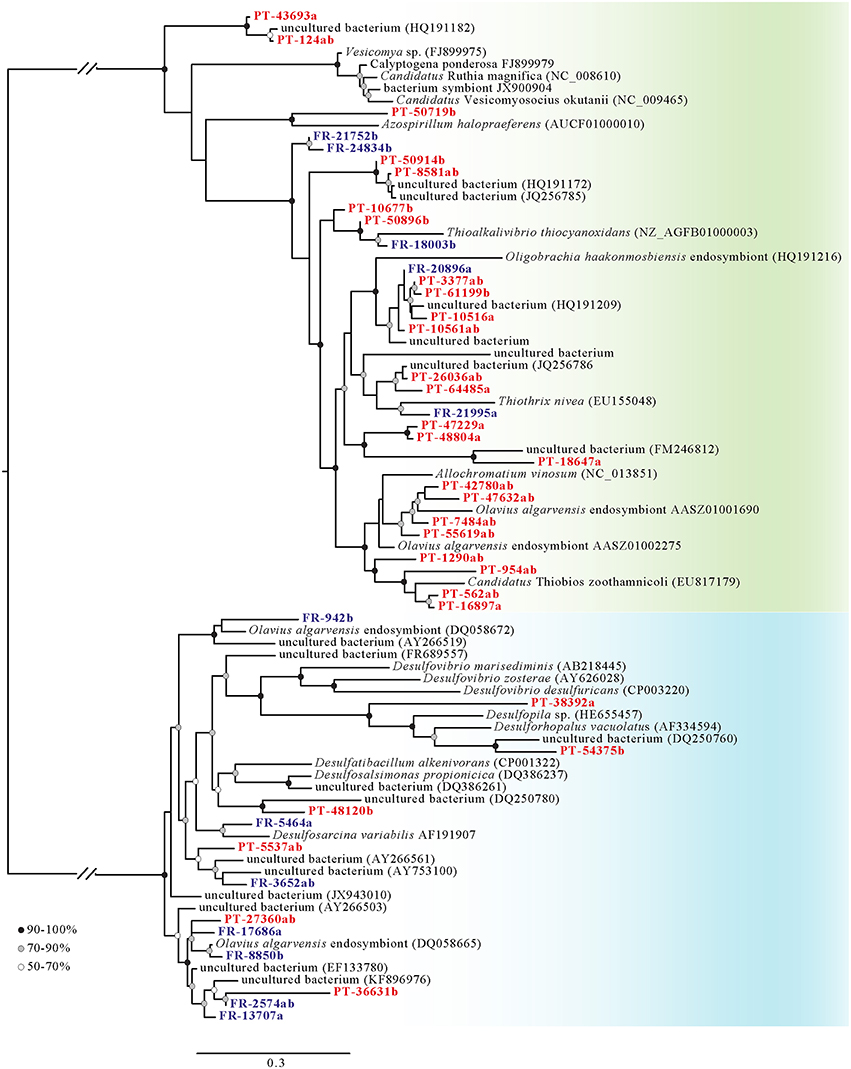
Figure 4. Diversity of dissimilatory sulfite reductase (dsrAB) genes. Phylogenetic tree of dsrAB marker genes present in the rhizobiomes of Zostera marina from Portugal (red, prefix PT) and France (blue, prefix FR). Contigs labeled with suffix “–a” correspond to sequences annotated to dsrA, “–b” to dsrB, and “–ab” to concatenated dsrAB. Sequences were concatenated whenever dsrA and dsrB were assembled in the same contig. Contigs were aligned at the protein level to the dsrAB reference alignment published by Müller et al. (2015) using the ARB alignment tool. The phylogenetic tree was inferred using the approximate maximum-likelihood method in FastTree2 (Price et al., 2010). The green background indicates sequences involved in oxidative processes, the blue background sequences involved in reductive processes. Bootstrap values are indicated. Scale bar indicates percentage sequence difference.
As the taxonomic resolution obtained from the dsrAB tree is not extremely high due to the large number of uncultured bacteria to which our sequences aligned, sister lineages containing species such as Thioalkalivibrio thiocyanoxidans, Thiothrix nivea, and Allochromatium vinosum, allowed the placement of our sequences in the orders Chromatiales and Thiotrichales (Figure 4). On the reductive side of the phylogeny, however, a more detailed identification was possible for members of the families Desulfovibrionaceae, Desulfobulbaceae and Desulfobacteraceae (Figure 4).
Reconstructed Draft Genomes
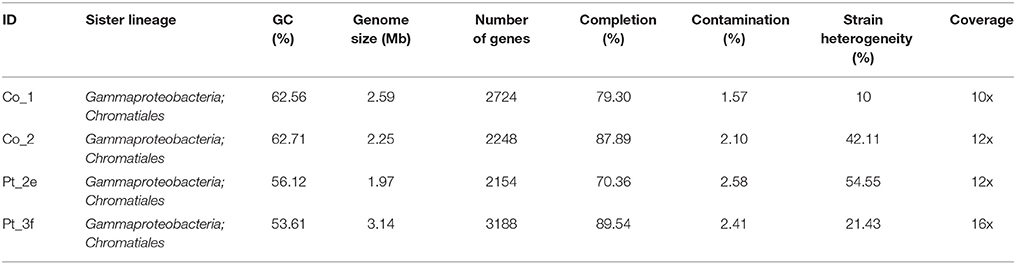
Metagenomic binning resulted in 4 draft genomes isolated from ZmPt, which ranged in genome size from ~1.9 to ~3.1 Mb, and GC content between 53.6 and 62.7% (Table 3). With a coverage of 16x, Pt_3f was the most abundant organism among the recovered draft genomes, whereas Co_1 was the least abundant with a 10x coverage (Table 3).
Genes for the oxidation of reduced sulfur compounds were identified in all draft genomes, and additionally Co_1 and Co_2 contained sgpA, one of the genes encoding proteins for sulfur globule envelopes. Co_1 and Pt_3f contained the genes necessary for the complete oxidation of sulfide to sulfate (Table 4). Furthermore, Co_1 was the draft genome with the highest number of genes involved in nitrogen cycling, including the following two pathways: i) denitrification to dinitrogen, and ii) dissimilatory nitrate reduction to ammonium (DNRA). Nitrogenase-encoding genes nifH, nifD and nifK were absent in all draft genomes. Genes for two carbon fixation pathways were identified in Co_1, Co_2, and Pt_2d (cbbL and cbbS from the CBB cycle, and korAB, frdA, sdhA, sdhC, and porAB from rTCA cycle), whereas in Pt_3f genes for the CBB cycle were absent (Table 4). Comparative sequence analysis attributed these draft genomes to the family Chromatiaceae, and placed them in close proximity to the Solemya velum gill symbiont (IMG_2502171186; Figure 5).
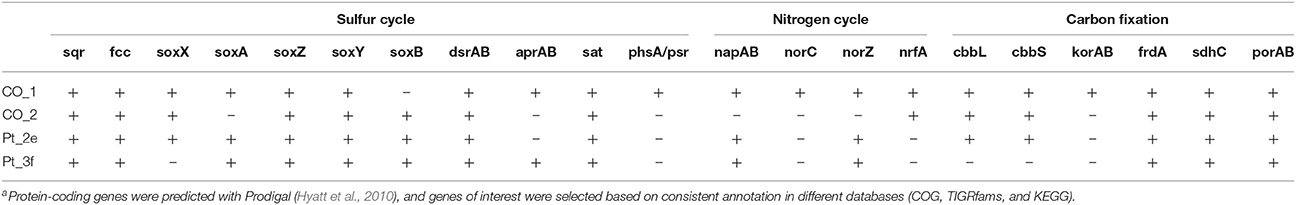
Table 4. Putative genes involved in sulfur, nitrogen and carbon cycling present in reconstructed draft genomesa.
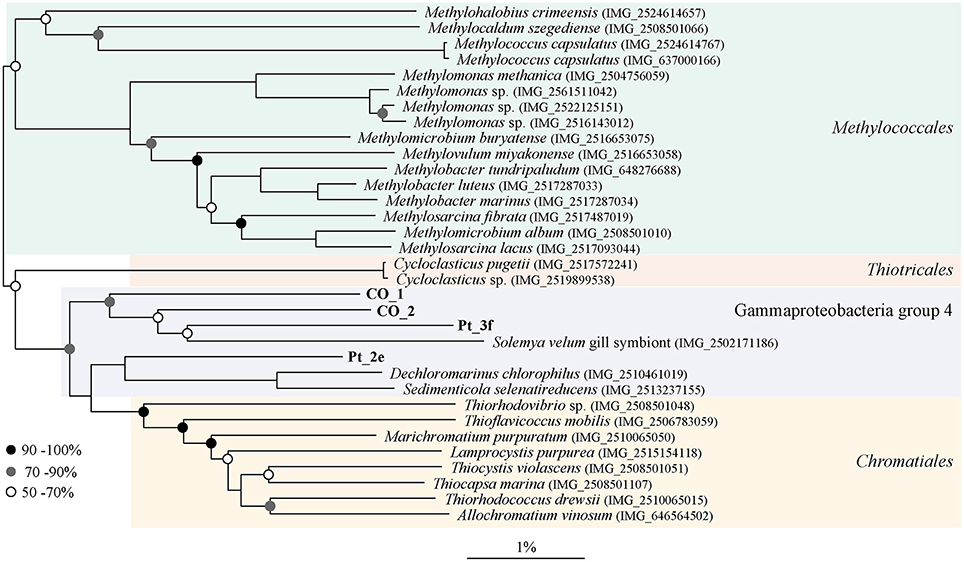
Figure 5. Phylogenetic affiliation of reconstructed draft genomes. Draft genomes were aligned to genomes in the CheckM reference tree (Parks et al., 2015). Subsequently, sequences were selected and the final phylogeny was calculated using the approximate maximum-likelihood method in FastTree2 (Price et al., 2010). Bootstrap values are indicated. Scale bar indicates percentage sequence difference.
Discussion
Bacterial Community Structure
Marine plants anchored in anoxic sediments harbor a steep redox gradient around their roots that creates microniches for different types of bacteria (Holmer and Nielsen, 1997; Blaabjerg and Finster, 1998; Devereux, 2005). By using 16S rRNA amplicon sequencing, we have found that the seagrass rhizosphere microbiome (“rhizobiome”) is enriched by Gamma- and Deltaproteobacteria (Cúcio et al., 2016). Although Gammaproteobacteria contains an extremely large and metabolically versatile group of bacteria, their importance in the sulfur cycle is indicated by the high abundance of members of the orders Chromatiales and Thiotrichales in the metagenomes. Members of the order Chromatiales are mainly phototrophic sulfur oxidizers (Imhoff, 2005). Even though most of these purple sulfur bacteria (PSB) require light as an energy source, some are able to grow chemotrophically under micro-oxic conditions in the absence of light (Imhoff, 2005). Chromatiales have been previously shown to be dominant in the rhizosphere of seagrasses (Cúcio et al., 2016) and salt marsh plants (Thomas et al., 2014). Some of the most abundant bacteria among the Chromatiales (Thiohalocapsa sp. and Lamprocystis purpurea) identified in the rhizosphere of Zostera marina from Portugal, are bacteria that can grow in the dark as chemolithoautotrophs or chemoorganotrophs using sulfide/thiosulfate and pyruvate, respectively, as electron donors (Eichler and Pfennig, 1988; Caumette et al., 1991). While profiting from the oxygen that is released from the roots during daytime (Borum et al., 2005; Frederiksen and Glud, 2006), these bacteria can actively consume the sulfide from the rhizosphere, and so contribute to a detoxification of the root area.
Nearly half of the class Deltaproteobacteria in our metagenomes was composed by members of the order Desulfobacterales. Ten percent of this order in ZmPt (corresponding to 5% of the Deltaproteobacteria and 2% of the Proteobacteria) were assigned to the species Desulfosarcina cetonica. The genus Desulfosarcina was among the most abundant taxa of the Deltaproteobacteria in the rhizosphere of the seagrasses Zostera marina, Z. noltii and Cymodocea nodosa (Cúcio et al., 2016). Desulfosarcina cetonica is a complete oxidizer that is able to utilize sulfate, thiosulfate and/or elemental sulfur as electron acceptors for the oxidation of a wide range of organic compounds, including acetone, benzoate, ethanol, lactate, and acetate (Galushko and Rozanova, 1991). As previously discussed by Cúcio et al. (2016), the presence of SRB capable of ethanol oxidation in the rhizosphere might represent a trade-off between plant and bacteria, in which the latter, although it produces sulfide, contributes to another mean of detoxification of the rhizosphere by consuming the ethanol released by the roots. Another deltaproteobacterium present in ZmPt was the (uncultured) PSCGC 5451, which was sequenced in a project targeting hydrocarbon degradation in marine sediments (GOLD–JGI). This strain, like several other SRB found (namely those belonging to the family Desulfobacteraceae), have been described to degrade hydrocarbons (Kleindienst et al., 2014). Desulfobacteraceae were also dominant among other Deltaproteobacteria on belowground compartments of the tropical seagrass Halophila stipulaceae (Mejia et al., 2016).
The overall community composition at the class level was similar in both locations, contradicting previous findings in salt marshes and seagrass meadows (Thomas et al., 2014; Cúcio et al., 2016). We consider that this discrepancy is likely to be associated with three reasons, i) different sequencing depth between the samples, as indicated by large variation in coverage (visible in Figure 2), (ii) biases inherent to PCR amplification in the former studies, and iii) to the high number (ca. 38%) of unclassified reads in the present study, which masks the presence, c.q. dominance of unknown bacteria. Furthermore, shotgun metagenomic sequencing does not feature the biases inherent to PCR amplification used in previous studies. Furthermore, the difference in the GC content found between both metagenomes (55.6 and 50.8% in ZmPt and ZmFr, respectively, Table 2), suggests that they harbor different microbial communities. Notwithstanding this fact, some sequences were abundant in both metagenomes (see clusters 1, 2, and 3, and encircled regions in Figure 2), highlighting an overrepresentation of some bacteria in these metagenomes, in particular in ZmFr. Cluster 1, in particular, was identified as a member of the Desulfobacteracae. Previous determination of the core rhizobiome of seagrasses (Cúcio et al., 2016) identified the Desulfobacteraceae as the most abundant family within the core rhizobiome; therefore it is not surprising that these bacteria are highly represented in both locations.
Diversity of Sulfur Genes in the Rhizobiome of Z. marina
Although the importance of biological oxidation of sulfide in the rhizosphere of seagrasses has not yet been fully understood (Sayama et al., 2005; Preisler et al., 2007; Lenk et al., 2011), Luther et al. (2011) have demonstrated that biological sulfide oxidation rates are three or more orders of magnitude higher than abiotic rates, indicating the importance of sulfide oxidizing bacteria in the rhizosphere of seagrasses.
Overall, we observed a higher diversity and abundance of sulfur genes involved in oxidation than in reduction, such as the gene sqr encoding the enzyme sulfide:quinone reductase (Figure 3, Supplementary Figure 3). The gene sqr and its homolog fccAB, are essential for sulfide oxidation. They can be present in one or multiple copies per genome (e.g., Reinartz et al., 1998; Chan et al., 2009), and are characterized and divided into six different types, which exhibit different affinities to sulfide (Marcia et al., 2010). According to phylogenetic placement of the contig sequences annotated as SQR, we identified a majority of sequences clustering with the SQR types VI, I, and III. (Supplementary Figure S3). SQR type VI, responsible for growth on sulfide as the only electron donor (Chan et al., 2009), was the most abundant sulfur gene in ZmPt. This type has been shown to be transcribed under sulfide concentrations up to 8 mM in Chlorobium tepidum (Chan et al., 2009), indicating that sulfide-oxidizing bacteria present in the rhizosphere of Z. marina might be adapted to perform sulfide oxidation under high sulfide concentrations (Han and Perner, 2015). Nevertheless, the presence of other types of SQR in the rhizosphere of Z. marina with affinity to sulfide at the micromolar level, as well as a high diversity of FCC (Brune, 1995; Marcia et al., 2010), suggest that sulfide oxidizing bacteria possess a diverse set of genes which allow them to thrive under a large range of sulfide concentrations. Such versatility enables these bacteria to successfully explore the complex redox gradient and settle in specific microniches of the rhizosphere.
The oxidation of sulfide via SQR and/or FCC, as well as the oxidation of thiosulfate via the Sox multi-enzyme complex, result in the formation of sulfur globules as an intermediate product (Dahl and Prange, 2006; Eichinger et al., 2014). Although sulfur globule formation is not yet fully understood, the pathway for thiosulfate oxidation via SoxB seems to result in the accumulation of sulfur intra- or extracellularly (Dahl and Prange, 2006; Hensen et al., 2006; Eichinger et al., 2014). A study on endosymbiotic bacteria suggested that the formation of sulfur globules actively contributes to the detoxification of sulfide from the cells of the host (Eichinger et al., 2014), and functions as a reservoir of energy for the bacterium itself. Furthermore, reduced sulfur compounds are preferentially utilized as electron donors rather than organic compounds (Grimm et al., 2011). Intra- and extracellular sulfur globules are oxidized via the reverse-operating dissimilatory sulfite reductase, encoded by the genes rdsrAB (Dahl et al., 2005; Dahl and Prange, 2006; Hensen et al., 2006; Müller et al., 2015), which were abundant in ZmPt (Figure 3). The well represented presence of all these genes allowed us to speculate that bacteria in the seagrass rhizosphere are well adapted to varying concentrations and different types of reduced sulfur compounds, and furthermore, the high abundance of rdsrAB genes might represent an advantageous side mechanism for SOB to obtain energy (Müller et al., 2015).
Another abundant gene involved in oxidation, as determined by phylogenetic analysis (Figure 3, Supplementary Figure S1), was the APS reductase-encoding gene aprA. This is the reverse-operating homologous gene of the aprA found in sulfate-reducing prokaryotes (Meyer and Kuever, 2007a,b), which is essential for the oxidation of sulfite to an APS intermediate and is further oxidized to sulfate by the ATP sulfurylase (Sat).
Symbiont-Related Bacteria in the Rhizosphere of Z. marina
The functional genes dsrAB, aprAB, and soxB are commonly used to infer the phylogenetic affiliation of SRB and SOB (Meyer and Kuever, 2007b; Meyer et al., 2007; Müller et al., 2015). Whereas aprAB and soxB genes have suffered several events of lateral gene transfer (LGT) among SRB and SOB (Meyer and Kuever, 2007b; Meyer et al., 2007), the dsrAB gene is highly conserved in both groups, and its phylogeny is congruent with that of the 16S rRNA gene (Müller et al., 2015). Despite that a majority of sequences of dsrAB clustered with those of uncultured bacteria (Figure 4), indicating the presence of a high diversity of unknown bacteria in the rhizosphere of Z. marina, another number of sequences analyzed in our study clustered in close proximity to those of well-known photo- and chemolithoautotrophic bacteria, such as Allochromatium vinosum (Kämpf and Pfennig, 1980), and (endo) symbionts of several marine invertebrates (Figure 4). Likewise, sequences of other target genes involved in oxidation processes, such as dsrAB, aprA, sat, sqr, and fcc also clustered closely with sequences from chemolithoautotrophic and endosymbiotic bacteria (Figure 4, Supplementary Figures S1–S3). In particular, several assembled sequences closely clustered with those of the gamma- and deltaproteobacterial endosymbionts of the gutless worm Olavius algarvensis (Figure 4). The gammaproteobacterial endosymbionts are SOB that have genes for autotrophic CO2 fixation and accumulation of sulfur globules, and provide their gutless host with their vital nutrition (Woyke et al., 2006).
Three out of the four draft genomes isolated from the rhizosphere of Z. marina clustered in proximity to a sulfur-oxidizing gill symbiont of the clam Solemya velum. Gros et al. (2003) also found multiple 16S rDNA sequences identical to Codakia orbicularis symbiont. Nevertheless, no eukaryotic genes from this clam or from O. algarvensis were detected in our metagenomes, suggesting that these bacteria might be free-living forms of these symbionts. Free-living forms of endosymbionts of the clam Codakia orbicularis (Lucinidae) were previously identified in seagrass meadows (Gros et al., 2003; König et al., 2016). Furthermore, Gros et al. (1996, 2012) demonstrated that this clam is able to acquire its chemoautotrophic gill endosymbionts from the environment, although the release of these bacteria from the host to the environment has, to our knowledge, never been observed for these clams (Brissac et al., 2009). Symbionts of members of the family Solemyidae were initially thought to be exclusively vertically transmitted (Krueger et al., 1996), nevertheless recent evidence suggests a very dynamic mode of transmission, which further includes horizontal/environmental acquisition (Dmytrenko et al., 2014). Though little information is available about the transmission of chemoautotrophic symbionts of gutless oligochaetes, such as O. algarvensis, some evidence supports both vertical (Giere and Langheld, 1987) and environmental transmission (Dubilier et al., 2006). Hence, there seem to be several indications suggesting that seagrass meadows could harbor a pool of fauna endosymbionts in a free-livingstate.
Some seagrasses can, at least partly, cope with high levels of sulfide through a three-stage symbiosis between a Lucinidae clam and their chemolithoautotrophic gill symbionts (Van der Heide et al., 2012). However, this does not include meadows devoid of clams. Due to the large proportion of reads annotated as symbiotic bacteria and the absence of invertebrate hosts in our metagenome, we support the hypothesis of Brissac et al. (2009), which states that seagrass meadows function as a source of endosymbionts. Horizontal (environmental) symbiont transmission requires an inoculum of free-living bacteria (reviewed in Bright and Bulgheresi, 2010), which might originate by detaching from their symbiotic association upon the host's death, as recently shown in Riftia pachyptila by Klose et al. (2015). We hypothesize that these bacteria could accumulate and proliferate in the rhizosphere of seagrasses, until they are brought to the water column by bioturbators, for instance, and inoculate putative hosts in the periphery of the seagrass meadow.
Reconstructed Symbiont-Related Draft Genomes
Like the gill symbiont of Solemya velum, all four draft genomes recovered in our study possessed genes necessary for carbon fixation, as well as a repertoire of genes for the complete oxidation of sulfide to its most oxidized form sulfate (Table 4; Stewart et al., 2011), thereby confirming that these organisms can function as chemolithoautotrophs. Furthermore, they are also genetically equipped to perform nitrate reduction (Table 4), indicating that they may be able to couple sulfide oxidation to nitrate reduction, as previously described for bacteria such as Thioploca sp. and Beggiatoa sp. (Fossing et al., 1995; Otte et al., 1999; Sayama, 2001; Sayama et al., 2005).
A vast range of genes involved in the sulfur and nitrogen cycles, as well as carbon fixation, were identified in Co_1 (Table 4). The presence of alternative pathways in sulfur reactions (sulfate, thiosulfate, sulfur oxidation), carbon fixation mechanisms (CO2 fixation via the CBB and carbon fixation via the TCA cycle) and nitrate reduction pathways (denitrification, DNRA) in this draft genome, are likely an advantageous trait in a dynamic environment as the seagrass rhizosphere. For this reason, we propose a conceptual model for the presence and function of Co_1-like free-living forms of symbiotic bacteria, in the rhizosphere of seagrasses (Figure 6). Having such a complete set of genes in its genome potentially gives this bacterium the ability to exploit a large variety of substrates, which is particularly important if it needs to thrive as free-living as well as in symbiotic associations. For instance, this bacterium is able to synthesize organic matter under oxic via the CBB cycle, and under micro-oxic/anoxic conditions via the TCA cycle (Figure 6). To our knowledge, the presence of these two pathways simultaneously has only been described in the endosymbionts of the hydrothermal vent worms Riftia pachyptila and Tevnia jerichonana, and represents an adaptation to transient conditions marked by the amount of energy available (Markert et al., 2007; Gardebrecht et al., 2012). Similar to the gill symbiont of Solemya velum (Stewart et al., 2011), this microorganism is capable of oxidizing sulfide at low and high environmental concentrations using FCC and SQR, respectively (Brune, 1995; Marcia et al., 2010). Moreoever, it might be able to store sulfur intracellularly as suggested by the presence of the gene sgpA. Sulfide concentrations in the rhizosphere of seagrasses can be very dynamic and particularly respond to light/dark regimes, mainly due to the lower oxygen release in the dark, compared to light conditions (e.g., Pagès et al., 2012). Given its ability to thrive in such conditions, this bacterium also has the genetic setup necessary to switch their usage between nitrogen compounds, according to the availability of sulfide. Brunet and Garcia-Gil (1996) found evidence that under low sulfide concentrations denitrification is favored, whereas under high levels of sulfide, DNRA is more likely to proceed (as sulfide inhibits the final two steps of denitrification, Figure 6). In this regard, high sulfide concentrations may benefit the seagrass (below the threshold of sulfide detrimental for the fitness of the plant), by inducing the production of ammonium, which is the seagrasses' preferred nitrogen source (Alexandre et al., 2015). Like sulfide, ammonium concentrations in the rhizosphere increase during dark periods (Pagès et al., 2012). Although these authors attributed higher concentrations to a decrease in seagrass uptake, according to our conceptual model, high sulfide concentrations sustain DNRA, which results in the release of ammonium during the night.
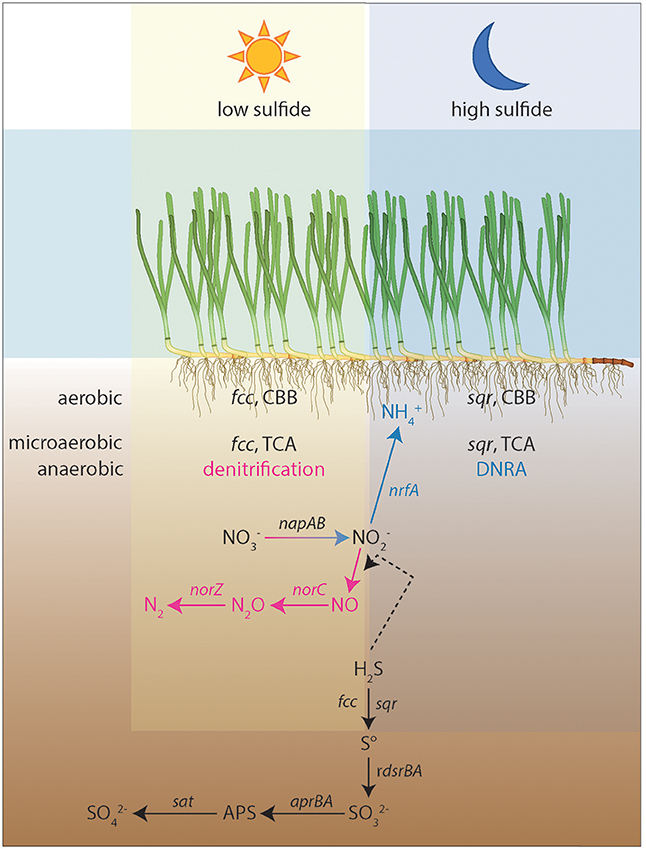
Figure 6. Conceptual model of sulfur, nitrogen and carbon fixation pathways in one of the reconstructed genomes, CO_1, obtained from the rhizosphere of Zostera marina. At aerobic conditions and low sulfide concentrations fcc is used to oxidize sulfide with oxygen, while at high sulfide concentrations sqr is used to oxidize sulfide. In both cases, the Calvin cycle (CBB) is used to fix CO2. However, at microaerobic and anaerobic conditions and low sulfide concentrations fcc is oxidizing sulfide via denitrification (purple), while at high sulfide concentrations sqr is oxidizing sulfide via DNRA (dissimilatory nitrate reduction to ammonium represented in blue). At these conditions carbon is fixed using the TCA-cycle. The ammonium produced in the DNRA is used as a nitrogen source by the seagrasses. The dashed arrow represents the proposed interactions between the sulfur and nitrogen cycle in which sulfide reduces nitrite to nitric acid.
Notwithstanding the fact that nitrate reduction via DNRA is favored by environmental conditions (Sayama, 2001), Co_1 possesses genes that encode nitric oxide and nitrous oxide reductases. The presence of these genes indicates that this organism could have the potential to completely denitrify nitrate to dinitrogen, however we did not find the genes that encode the nitrite reductase responsible for the reduction of nitrite to nitric oxide (nir, Figure 6). Either nir genes were not captured due to the limited sequencing depth (this genome is missing approximately 20% of its genes), or they are naturally not present in Co_1. While we cannot rule out the first possibility, complete denitrification with the concomitant absence of nir genes in Co_1 could be bridged by the reduction of nitrite to nitric oxide through interaction with sulfide (Figure 6, Grossi, 2009; Cortese-Krott et al., 2015). This abiotic reaction could be of major importance for the seagrasses, because it would allow the double detoxification of two toxic components, H2S and .
The present study unveiled the so-far unknown diversity of sulfur genes present in the rhizosphere of the seagrass Zostera marina, and supported the hypothesis that the rhizobiome is enriched with sulfur-oxidizing bacteria. We found indications supported by phylogenetic inference that the rhizosphere hosts highly versatile bacteria related to symbionts of marine invertebrates that are capable of exploiting a wide range of sulfur and nitrogen compounds, using alternate pathways that are favorable under different environmental conditions. We suggested a conceptual model for the sulfur, nitrogen and carbon fixation metabolism of these organisms in the rhizosphere of seagrasses. The draft genomes recovered further supported the presence of chemolithoautotrophic bacteria closely related to free-living forms of symbionts, that are able to couple the complete oxidation of sulfide to sulfate with nitrate reduction to ammonium. The dominance of these bacteria should be determined by other techniques, such as fluorescent in situ hybridisation (FISH) or qPCR, and their contribution to the well-being of seagrasses needs to be unraveled in future studies by characterization of their activity.
Data Availability
The raw sequence reads have been deposited as dataset SRP126211 in the NCBI Sequence Read Archive (SRA).
Author Contributions
CC, AE, and GM designed the study and collected the samples; CC performed all lab experiments, and data analysis together with LO; CC wrote the manuscript and all authors contributed to the discussion of the results and to the final version of the manuscript.
Funding
This work was supported by grants from the European Union (MaCuMBA and ASSEMBLE 8) to GM and CC, the ERC Advanced Grant PARASOL (No. 322551) to GM and LO, the European Science Foundation ConGenOmics program (No. 6349) to CC, and fellowships SFRH/BPD/63/03/2009 and SFRH/BPD/107878/2015 of Fundação para a Ciência e Tecnologia (FCT) to AE.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Francisco Rodríguez-Valera, Mário Lopez-Lopez, and Rohit Ghai for an introduction into metagenomic analysis, Emily D. Melton for helpful discussions about microbial sulfur metabolism, Tom Berben for very helpful contribution to scripting, and Nicole Dubilier for insightful comments on the final version of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00171/full#supplementary-material
References
Alexandre, A., Hill, P. W., Jones, D. L., and Santos, R. (2015). Dissolved organic nitrogen: a relevant complementary source of nitrogen for the seagrass Zostera marina. Limnol. Oceanogr. 60, 1477–1483. doi: 10.1002/lno.10084
Blaabjerg, V., and Finster, K. (1998). Sulphate reduction associated with roots and rhizomes of the marine macrophyte Zostera marina. Aquat. Microb. Ecol. 15, 311–314. doi: 10.3354/ame015311
Blazejak, A., Erséus, C., Amann, R., and Dubilier, N. (2005). Coexistence of bacterial sulfide oxidizers, sulfate reducer, and spirochetes in a gutless worm (Oligochaeta) from the Peru margin. Appl. Environ. Microbiol. 71, 1553–1561. doi: 10.1128/AEM.71.3.1553-1561.2005
Borum, J., and Greve, T. (2004). “The four European seagrass species”, in European Seagrasses: An Introduction to Monitoring and Management, eds J. Borum, C. M. Duarte, D. Krause-Jensen, and T. M. Greve (Copenhagen: The M&MS Project), 1–7.
Borum, J., Pedersen, O., Greve, T. M., Frankovich, T. A., Zieman, J. C., Fourqurean, F. W., et al. (2005). The potential role of plant oxygen and sulphide dynamics in die-off events in tropical seagrass, Thalassia testudinum. J. Ecol. 93, 148–158. doi: 10.1111/j.1365-2745.2004.00943.x
Bright, M., and Bulgheresi, S. (2010). A complex journey: transmission of microbial symbionts. Nat. Rev. Microbiol. 8, 218–230. doi: 10.1038/nrmicro2262
Brissac, T., Gros, O., and Merçot, H. (2009). Lack of endosymbiont release by two Lucinidae (Bivalvia) of the genus Codakia: consequences for symbiotic relationships. FEMS Microbiol. Ecol. 67, 261–267. doi: 10.1111/j.1574-6941.2008.00626.x
Brune, D. C. (1995). “Sulfur compounds as photosynthetic electron donors,” in Anoxygenic Photosynthetic Bacteria, eds R. E. Blankenship, M. T. Madigan, and C. E. Bauer (Dordrecht: Kluwer), 847–870.
Brunet, R. C., and Garcia-Gil, L. J. (1996). Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol. Ecol. 21, 131–138. doi: 10.1111/j.1574-6941.1996.tb00340.x
Capone, D. G., and Kiene, R. P. (1988). Comparison of microbial dynamics in marine and freshwater sediments: contrasts in anaerobic carbon metabolism. Limnol. Oceanogr. 33. 725–749. doi: 10.4319/lo.1988.33.4_part_2.0725
Caumette, O., Baulaigue, R., and Matheron, R. (1991). Thiocapsa halophile sp. nov., a new halophilic phototrophic purple sulfur bacterium. Arch. Microbiol. 155, 170–176. doi: 10.1007/BF00248613
Chan, L. K., Morgan-Kiss, R. M., and Hanson, T. E. (2009). Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum. J. Bacteriol. 191, 1026–1034. doi: 10.1128/JB.01154-08
Cifuentes, A., Antón, J., Benlloch, S., Donnelly, A., Herbert, R. A., and Rodríguez-Valera, F. (2000). Prokaryotic diversity in Zostera noltii-colonized marine sediments. Appl. Environ. Microbiol. 66, 1715–1719. doi: 10.1128/AEM.66.4.1715-1719.2000
Cortese-Krott, M. M., Fernandez, B. O., Kelm, M., Butler, A. R., and Feelisch, M. (2015). On the chemical biology of the nitrite/sulfide interaction. Nitric Oxide 46, 14–24. doi: 10.1016/j.niox.2014.12.009
Costa, R., Götz, M., Mrotzek, N., Lottmann, J., Berg, G., and Smalla, K. (2006). Effects of site and plant species on rhizosphere community structure as revealed by molecular analysis of microbial guilds. FEMS Microbiol. Ecol. 56, 236–249. doi: 10.1111/j.1574-6941.2005.00026.x
Crump, B. C., and Koch, E. W. (2008). Attached bacterial populations shared by four species of aquatic angiosperms. Appl. Environ. Microbiol. 74, 5948–5957. doi: 10.1128/AEM.00952-08
Cúcio, C., Engelen, A. E., Costa, R., and Muyzer, G. (2016). Rhizosphere microbiomes of European seagrasses are selected by the plant, but are not species specific. Front. Microbiol. 7:440. doi: 10.3389/fmicb.2016.00440
Dahl, C., Engels, S., Pott-Sperling, A. S., Schulte, A., Sander, J., Lübbe, Y., et al. (2005). Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187, 1392–1404. doi: 10.1128/JB.187.4.1392-1404.2005
Dahl, C., and Prange, A. (2006). “Bacterial sulfur globules: occurrence, structure and metabolism,” in Microbiology Monographs, Inclusions in Prokaryotes, Vol. 1, ed J. M. Shively (Berlin; Heidelberg: Springer), 21–51.
Devereux, R. (2005). “Seagrass rhizosphere microbial communities,” in Interactions Between Macro- and Microorganisms in Marine Sediments, eds E. Kristensen, R. R. Haese, and J. E. Kostka (Washington, DC: American Geophysical Union), 199–216.
Dmytrenko, O., Russel, S. L., Loo, W. T., Fontanez, K. M., Liao, L., Roeselers, G., et al. (2014). The genome of the intracellular bacterium of the coastal bivalve, Solemya velum: a blueprint for thriving in and out of symbiosis. BMC Genomics 15:924. doi: 10.1186/1471-2164-15-924
Donnelly, A. P., and Herbert, R. A. (1999). Bacterial interactions in the rhizosphere of seagrass communities in shallow coastal lagoons. J. Appl. Microbiol. 85, 151–160. doi: 10.1111/j.1365-2672.1998.tb05294.x
Dubilier, N., Blazejak, A., and Rühland, C. (2006). “Symbioses between bacteria and gutless marine oligochaetes,” in Molecular Basis of Symbiosis. Progress in Molecular and Subcellular Biology, Vol. 41, ed J. Overmann (Berlin; Heidelberg: Springer), 251–275.
Dubilier, N., Mülders, C., Ferdelman, T., de Beer, D., Pernthaler, A., Klein, M., et al. (2001). Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature 411, 298–302. doi: 10.1038/35077067
Eichinger, I., Schmitz-Esser, S., Schmid, M., Fisher, C. R., and Bright, M. (2014). Symbiont-driven sulfur crystal formation in a thiotrophic symbiosis from deep-sea hydrocarbon seeps. Env. Microbiol. Rep. 6, 364–372. doi: 10.1111/1758-2229.12149
Eichler, B., and Pfennig, N. (1988). A new purple sulfur bacterium from stratified freshwater lakes, Amoebobacter purpureus sp. nov. Arch. Microbiol. 149, 395–400. doi: 10.1007/BF00425577
Fahimipour, A. K., Kardish, M. R., Lang, J. M., Green, J. L., Eisen, J. A., and Stachowicz, J. J. (2017). Global-scale structure of the eelgrass microbiome. Appl. Environ. Microb. 83, e-03391–e-03316. doi: 10.1128/AEM.03391-16
Fossing, H., Gallardo, V. A., Jørgensen, B. B., Hüttel, M., Nielsen, L. P., Schulz, H., et al. (1995). Concentration and transport of nitrate by the mat-forming sulphur bacterium Thioploca. Nature 374, 713–715. doi: 10.1038/374713a0
Frederiksen, M. S., and Glud, R. N. (2006). Oxygen dynamics in the rhizosphere of Zostera marina: a two-dimensional planar optode study. Limnol. Oceanogr. 51, 1072–1083. doi: 10.4319/lo.2006.51.2.1072
Frigaard, N. U., and Dahl, C. (2008). Sulfur metabolism in phototrophic sulfur bacteria. Adv. Microb. Physiol. 54, 103–200. doi: 10.1016/S0065-2911(08)00002-7
Gacia, E., Granata, T. C., and Duarte, C. M. (1999). An approach to measurement of particle flux and sediment retention within seagrass (Posidonia oceanica) meadows. Aquat. Bot. 65, 255–268. doi: 10.1016/S0304-3770(99)00044-3
Galushko, A. S., and Rozanova, E. P. (1991). Desulfobacterium cetonicum sp. nov.: a sulfate-reducing bacterium which oxidizes fatty acids and ketones. Microbiology 60, 742–746.
Gardebrecht, A., Markert, S., Sievert, S. M., Felbeck, H., Thürmer, A., Albrecht, D., et al. (2012). Physiological homogeneity among the endosymbionts of Riftia pachyptila and Tevnia jerichonana revealed by proteogenomics. ISME J. 6, 766–776. doi: 10.1038/ismej.2011.137
Giere, O., and Langheld, C. (1987). Structural organisation, transfer and biological fate of endosymbiotic bacteria in gutless oligochaetes. Mar. Biol. 93, 641–650. doi: 10.1007/BF00392801
Green, E. P., and Short, F. T. (2003). World Atlas of Seagrasses. Berkeley, CA: University of California Press.
Green-García, A. M., and Engel, A. S. (2012). Bacterial diversity of siliciclastic sediments in a Thalassia testudinum meadow and the implications for Lucinisca nassula chemosymbiosis. Estuar. Coast. Shelf Sci. 112, 153–161. doi: 10.1016/j.ecss.2012.07.010
Grimm, F., Franz, B., and Dahl, C. (2011). Regulation of dissimilatory sulfur oxidation in the purple sulfur bacterium Allochromatium vinosum. Front. Microbiol. 2:51. doi: 10.3389/fmicb.2011.00051
Gros, O., Darrasse, A., Durand, P., Frenkiel, L., and Mouëza, M. (1996). Environmental transmission of a sulfur-oxidizing bacterial gill endosymbiont in the tropical lucinid bivalve Codakia orbicularis. Appl. Environ. Microb. 62, 2324–2330.
Gros, O., Elisabeth, N. H., Gustave, S. D., Caro, A., and Dubilier, N. (2012). Plasticity of symbiont acquisition throughout the life cycle of the shallow-water tropical lucinid Codakia orbiculata (Mollusca: Bivalvia). Environ. Microbiol. 14, 1584–1595. doi: 10.1111/j.1462-2920.2012.02748.x
Gros, O., Liberge, M., Heddi, A., Khatchadourian, C., and Felbeck, H. (2003). Detection of free-living forms of sulfide-oxidizing gill endosymbionts in the lucinid habitat (Thalassia testudinum environment). Appl. Environ. Microb. 69, 6264–6267. doi: 10.1128/AEM.69.10.6264-6267.2003
Grossi, L. (2009). Hydrogen sulfide induces nitric oxide release from nitrite. Bioorg. Med. Chem. Lett. 19, 6092–6094. doi: 10.1016/j.bmcl.2009.09.030
Haft, D. H., Loftus, B. J., Richardson, D. L., Yang, F., Eisen, J. A., Paulsen, I. T., et al. (2001). TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res. 29, 41–43. doi: 10.1093/nar/29.1.41
Han, T., and Perner, M. (2015). The globally widespread genus Sulfurimonas: versatile energy metabolisms and adaptations to redox clines. Front. Microbiol. 6:989. doi: 10.3389/fmicb.2015.00989
Hensen, D., Sperling, D., Trüper, H. G., Brune, D. C., and Dahl, C. (2006). Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 62, 794–810. doi: 10.1111/j.1365-2958.2006.05408.x
Holmer, M., and Nielsen, S. L. (1997). Sediment sulfur dynamics related to biomass-density patterns in Zostera marina (eelgrass) beds. Mar. Ecol. Prog. Ser. 146, 163–171. doi: 10.3354/meps146163
Holmer, M., Pedersen, O., and Ikejima, K. (2006). Sulfur cycling and sulfide intrusion in mixed Southeast Asian tropical seagrass meadows. Bot. Mar. 49, 91–102. doi: 10.1515/BOT.2006.013
Huerta-Cepas, J., Szklarczyk, D., Forslund, K., Cook, H., Heller, D., Walter, M. C., et al. (2016). eggNOG 4.5: a hierarchical orthology framework with improved functional annotation for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44, D286–D293. doi: 10.1093/nar/gkv1248
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Imhoff, J. F. (2005). “Chromatiales ord. nov.,” in Bergey's Manual of Systematic Bacteriology, The Proteobacteria Part B, The Gammaproteobacteria, eds D. J. Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (New York, NY: Springer), 1–59.
Jensen, S. I., Kühl, M., and Priem,é, A. (2007). Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina. FEMS Microbiol. Ecol. 62, 108–117. doi: 10.1111/j.1574-6941.2007.00373.x
Jørgensen, B. B. (1982). Mineralization of organic matter in the sea bed – the role of sulphate reduction. Nature 296, 643–645. doi: 10.1038/296643a0
Jørgensen, B. B., and Nelson, D. C. (2004). “Sulfide oxidation in marine sediments: Geochemistry meets microbiology,” in Sulfur Biogeochemistry-Past and Present, eds J. P. Amend, K. J. Edwards, and T. W. Lyons (Boulder, CO: Geological Society of America), 63–81.
Kämpf, C., and Pfennig, N. (1980). Capacity for Chromatiaceae for chemotrophic growth. Specific respiration rates of Thiocystis violacea and Chromatium vinosum. Arch. Microbiol. 127, 125–135. doi: 10.1007/BF00428016
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mar. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kleindienst, S., Herbst, F. A., Stagars, M., von Netzer, F., von Bergen, M., Seifert, J., et al. (2014). Diverse sulfate-reducing bacteria of the Desulfosarcine/Desulfococcus clade are the key alkane degraders at marine seeps. ISME J. 8, 2029–2044. doi: 10.1038/ismej.2014.51
Klose, J., Polz, M. F., Wagner, M., Schimak, M. P., Gollner, S., and Bright, M. (2015). Endosymbionts escape dead hydrothermal vent tubeworms to enrich the free-living population. Proc. Natl. Acad. Sci. U.S.A. 112, 11300–11305. doi: 10.1073/pnas.1501160112
König, S., Gros, O., Heiden, S. E., Hinzke, T., Thürmer, A., Poehlein, A., et al. (2016). Nitrogen fixation in a chemoautotrophic lucinid symbiosis. Nat. Microbiol. 2:16193. doi: 10.1038/nmicrobiol.2016.193
Krueger, D. M., Gustafson, R. G., and Cavanaugh, C. M. (1996). Vertical transmission of chemoautotrophic symbionts in the bivalve Solemya velum (Bivalvia: Protobranchia). Biol. Bull. 190, 195–202. doi: 10.2307/1542539
Laczny, C. C., Sternal, T., Plugaru, V., Gawron, P., Atashpendar, A., Matgossian, H. H., et al. (2015). VizBin – an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome 3:1. doi: 10.1186/s40168-014-0066-1
Lamb, J. B., van de Water, J. A., Bourne, D. G., Altier, C., Hein, M. Y., Fiorenza, E. A., et al. (2017). Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science 355, 731–733. doi: 10.1126/science.aal1956
Lenk, S., Arnds, J., Zerjatke, K., Musat, N., Amann, R., and Muβmann, M. (2011). Novel groups of Gammaproteobacteria catalyse sulfur oxidation and carbon fixation in a coastal, intertidal sediment. Environ. Microbiol. 13, 758–774. doi: 10.1111/j.1462-2920.2010.02380.x
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genome sequence. Nucl. Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.0955
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Luther, G. W. III., Findlay, A. J., Macdonald, D. J., Owings, S. M., Hanson, T. E., Beinart, R. A., et al. (2011). Thermodynamics and kinetics of sulfide oxidation by oxygen: a look at inorganically controlled reactions and biologically mediated processes in the environment. FEMS Microbiol. Ecol. 2:62. doi: 10.3389/fmicb.2011.00062
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2010). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78, 1073–1083. doi: 10.1002/prot.22665
Markert, S., Arndt, C., Felbeck, H., Becher, D., Sievert, S. M., Hügler, M., et al. (2007). Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science 315, 247–250. doi: 10.1126/science.1132913
Mejia, A. Y., Rotini, A., Lacasella, F., Bookman, R., Thaller, M., Shem-Tov, R., et al. (2016). Assessing the ecological status of seagrasses using morphology, biochemical descriptors and microbial community analysis. A study in Halophila stipulaceae (Forsk.) Aschers meadows in the northern Red Sea. Ecol. Indic. 60, 1150–1163. doi: 10.1016/j.ecolind.2015.09.014
Menzel, P., Ng, K. L., and Krogh, A. (2016). Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 7:11257. doi: 10.1038/ncomms11257
Meyer, B., Imhoff, J. F., and Kuever, J. (2007). Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria – evolution of the Sox sulfur oxidation enzyme system. Environ. Microbiol. 9, 2957–2977. doi: 10.1111/j.1462-2920.2007.01407.x
Meyer, B., and Kuever, J. (2007a). Phylogeny of the alpha and beta subunit of the dissimilatory adenosine-5'-phosphosulfate (APS) reductase from sulfate-reducing prokaryotes – origin and evolution of the dissimilatory sulfate-reduction pathway. Microbiology 153, 2026–2044. doi: 10.1099/mic.0.2006/003152-0
Meyer, B., and Kuever, J. (2007b). Molecular analysis of the distribution and phylogeny of dissimilatory adenosine-5'-phosphosulphate reductase-encoding genes (aprBA) among sulfur-oxidizing prokaryotes. Microbiology 153, 3478–3498. doi: 10.1099/mic.0.2007/008250-0
Meyer, F., Paarmann, D., D'Souza, M., Olson, R., Glass, E. M., Kubal, M., et al. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386
Müller, A. L., Kjeldsen, K. U., Rattei, T., Pester, M., and Loy, A. (2015). Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 9, 1152–1165. doi: 10.1038/ismej.2014.208
Otte, S., Kuenen, J. G., Nielsen, L. P., Paerl, H. W., Zopfi, J., Schulz, H. N., et al. (1999). Nitrogen, carbon, and sulfur metabolism in natural Thioploca samples. Appl. Environ. Microbiol. 65, 3148–3157.
Pagès, A., Welsh, D. T., Robertson, D., Panther, J. G., Schäfer, J., et al. (2012). Diurnal shifts in co-distributions of sulfide and iron(II) and profiles of phosphate and ammonium in the rhizosphere of Zostera capricorni. Estuar. Coast. Shelf Sci. 115, 282–290. doi: 10.1016/j.ecss.2012.09.011
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Peng, Y., Leung, H. C., Yiu, S. M., and Chin, F. Y. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. doi: 10.1093/bioinformatics/bts174
Pérez, M., Invers, O., Ruiz, J. M., Frederiksen, M. S., and Holmer, M. (2007). Physiological responses of the seagrass Posidonia oceanica to elevated organic matter content in sediments: an experimental assessment. J. Exp. Mar. Biol. Ecol. 344, 149–160. doi: 10.1016/j.jembe.2006.12.020
Preisler, A., de Beer, D., Lichtschlag, A., Lavik, G., Boetius, A., and Jørgensen, B. B. (2007). Biological and chemical sulfide oxidation in a Beggiatoa inhabited marine sediment. ISME J. 1, 341–353. doi: 10.1038/ismej.2007.50
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – Approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Reinartz, M., Tschäpe, J., Brüser, T., Trüper, H. G., and Dahl, C. (1998). Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch. Microbiol. 170, 59–68. doi: 10.1007/s002030050615
Sayama, M. (2001). Presence of nitrate-accumulating sulfur bacteria and their influence on nitrogen cycling in a shallow coastal marine sediment. Appl. Environ. Microbiol. 67, 3481–3487. doi: 10.1128/AEM.67.8.3481-3487.2001
Sayama, M., Risgaard-Petersen, N., Nielsen, L. P., Fossing, H., and Christensen, P. B. (2005). Impact of bacterial transport on sedimentary biogeochemistry. Appl. Environ. Microbiol. 71, 7575–7577. doi: 10.1128/AEM.71.11.7575-7577.2005
Short, F. T., Burdick, D. M., Short, C. A., Davis, R. C., and Morgan, P. A. (2000). Developing success criteria for restored eelgrass, saltmarsh and mud flat habitats. Ecol. Eng. 15, 239–252. doi: 10.1016/S0925-8574(00)00079-3
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Stewart, F. J., Dmytrenko, O., DeLong, E. F., and Cavanaugh, C. M. (2011). Metatranscriptomic analysis of sulfur oxidation genes in the endosymbiont of Solemya velum. Front. Microbiol. 2:134. doi: 10.3389/fmicb.2011.00134
Tatusov, R. L., Fedorova, N. D., Jackson, J. D., Jacobs, A. R., Kiryutin, B., Koonin, E. V., et al. (2003). The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. doi: 10.1186/1471-2105-4-41
Thomas, F., Giblin, A. E., Cardon, Z. G., and Sievert, S. M. (2014). Rhizosphere heterogeneity shapes abundance and activity of sulfur-oxidizing bacteria in vegetated salt marsh sediments. Front. Microbiol. 5:309. doi: 10.3389/fmicb.2014.00309
Van der Heide, T., Govers, L. L., de Fouw, J., Olff, H., van der Geest, M., van Katwijk, M. M., et al. (2012). A three-stage symbiosis forms the foundation of seagrass ecosystems. Science 336, 1432–1434. doi: 10.1126/science.1219973
Keywords: chemolithoautotrophs, metagenomics, plant-microbe interactions, rhizobiome, seagrass microbiome, sulfur bacteria, symbionts
Citation: Cúcio C, Overmars L, Engelen AH and Muyzer G (2018) Metagenomic Analysis Shows the Presence of Bacteria Related to Free-Living Forms of Sulfur-Oxidizing Chemolithoautotrophic Symbionts in the Rhizosphere of the Seagrass Zostera marina. Front. Mar. Sci. 5:171. doi: 10.3389/fmars.2018.00171
Received: 20 December 2017; Accepted: 30 April 2018;
Published: 28 May 2018.
Edited by:
Jillian Petersen, Universität Wien, AustriaReviewed by:
Stephanie Markert, Institut für Marine Biotechnologie, GermanyAnnette Summers Engel, University of Tennessee, Knoxville, United States
Copyright © 2018 Cúcio, Overmars, Engelen and Muyzer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerard Muyzer, Zy5tdWlqemVyQHV2YS5ubA==
 Catarina Cúcio
Catarina Cúcio Lex Overmars
Lex Overmars Aschwin H. Engelen
Aschwin H. Engelen Gerard Muyzer
Gerard Muyzer