
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol. , 25 September 2020
Sec. Molecular Innate Immunity
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.569127
Toll-like receptors (TLRs) are at the forefront of pathogen recognition ensuring host fitness and eliciting protective cellular and humoral responses. Signaling pathways downstream of TLRs are tightly regulated for preventing collateral damage and loss of tolerance toward commensals. To trigger effective intracellular signaling, these receptors require the involvement of adaptor proteins. Among these, Toll/Interleukin-1 receptor domain containing adaptor protein (Tirap or MAL) plays an important role in establishing immune responses. Loss of function of MAL was associated with either disease susceptibility or resistance. These opposite effects reveal paradoxical functions of MAL and their importance in containing infectious or non-infectious diseases. In this review, we summarize the current knowledge on the signaling pathways involving MAL in different pathologies and their impact on inducing protective or non-protective responses.
Commensal and pathogenic microorganisms contain pathogen-associated molecular patterns (PAMPs) that are recognized by different families of pattern-recognition receptors (PRRs) (1). Mammals have distinct classes of PRRs, including Toll-like receptors (TLRs), that are patrolling both, extracellular and intracellular environments. They are expressed in innate immune cells, such as dendritic cells (DCs) and macrophages, but also in non-immune cells, such as fibroblasts and epithelial cells. TLRs are composed of the Toll/Interleukin-1 receptor homology (TIR) domain capable of binding to intracellular signaling adaptor proteins. For more details on downstream signaling cascades, we direct the reader to comprehensive reviews (2, 3).
Among a variety of TLRs adaptor proteins, TIR-containing adaptor protein (Tirap), also named MyD88 adaptor-like protein [MAL, also referred to as megakaryoblastic leukemia (translocation) 1], was mostly reported for its involvement in the regulation of signaling cascades downstream of TLR-2 and TLR-4 by bridging the adaptor protein myeloid differentiation primary response 88 (MyD88) (4–6). Human MAL consists of 221 amino acids (Figure 1A). At the N-terminus, there are a phosphatidylinositol 4,5-bisphosphate (PIP2) binding motif (PBM) and a putative proline, glutamic acid, serine and threonine (PEST) domain associated to short-lived proteins (7). At the C-terminus, a TIR domain extends from amino acid 84–221 and orchestrates the signal transduction pathways after TLR and Interleukin-1 (IL-1) receptor engagement. Because of its analogy with MyD88, MAL signaling was initially confused with that of MyD88. Actually, it impacts on inflammation and innate immune responses in a TLR- and MyD88-independent manner (8), pinpointing its role to other putative cellular mechanisms, such as vesicular trafficking. Heterozygosity and homozygosity for some inherited mutations in MAL are associated with different outcomes in patients, suggesting a paradoxical role in protection against diseases. This may reflect the influence of gene-gene and gene-environment interactions that vary across populations. In this review, we summarize the current knowledge on cellular mechanisms of MAL/Tirap and highlight its role in disease predisposition.
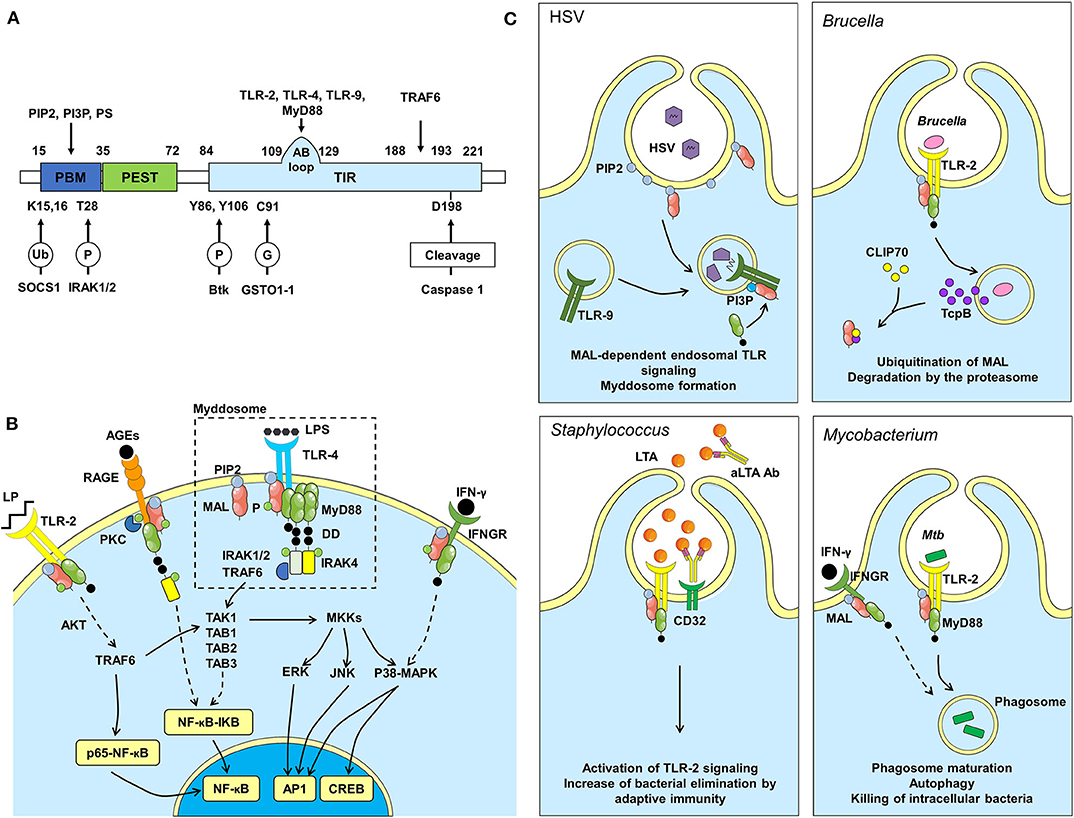
Figure 1. MAL structure, pathway and dependent immune responses during infections. (A) Protein structure of MAL, binding sites (above) and regulation sites (below). P, phosphorylation; Ub, ubiquitination; G, S-glutathionylation. (B) Among other receptors, MAL affects signals downstream of TLR-2, TLR-4, RAGE and IFNGR. When TLR-4 is activated by LPS, MAL is recruited by its PIP2 binding domain to TLR-4 rich regions of the plasma membrane. MAL then facilitates the recruitment of MyD88 and the formation of the myddosome, which is important for the activation of the NF-κB pathway, thus inducing inflammation. Signaling pathway activated downstream of RAGE can also induce NF-κB dependent inflammation. MAL induces the translocation of p65-NF-κB to the nucleus through AKT phosphorylation, independently from MyD88 signaling. Also, independently from MyD88, MAL can activate CREB via p38-MAPK and Mitogen-activated protein kinase kinase (MKK) signaling pathways. Another pathway involving MAL is downstream IFNGR, which leads to phosphorylation of p38-MAPK. AGEs, Advanced Glycation End products; DD, Death Domain; P, Phosphate group; LP, Lipopeptide. (C) The Herpes simplex virus (HSV) infection model was used to confirm MAL signaling from endosomes. TLRs found at the cell surface signal from a PIP2-rich subdomain, and MAL is recruited to that location via interactions with PIP2. TLRs found on endosomes (TLR-9) signal from a domain rich in PI3P. These lipids direct MAL to endosomes to promote TLR-9 signaling after viral DNA recognition. Brucella infection: Brucella interferes with immune responses by producing TcpB, which targets MAL to inhibit NF-κB activation. TcpB also targets CLIP70 inducing MAL ubiquitination and degradation by the proteasome. Staphylococcus infection: Lipoteichoic acid (LTA), a toxin produced by Staphylococcus bacteria, is recognized by TLR-2. TLR-2 activation induces MAL signaling to eliminate the toxin/bacteria. LTA is also recognized by antibodies (aLTA Ab), which are recognized by CD32 (Fcγ receptor II). In patients carrying the R121W mutation in MAL, adaptive immunity can compensate for defects in MAL function. Mycobacterium infection: Killing M. tuberculosis requires activation of TLR-2- and IFNGR-dependent signaling pathways within phagocytes to induce phagosome maturation and autophagy.
MAL recruitment at the plasma membrane occurs upon binding to PIP2 prior to its interaction with TLRs (Figure 1B) (9). MAL is then phosphorylated by Burton tyrosine kinase (Btk), which facilitates its interaction with the TIR domain of TLRs and consequently MyD88 to initiate the transduction signal (10). Recently, it has been shown that MAL is S-glutathionylated on Cys91 to facilitate the interaction with MyD88 (11). Notably, it is possible that the MAL-MyD88 tandem is prepositioned on the membrane awaiting ligand-induced TLR activation. Upon activation, MyD88 oligomerizes to form a large signaling platform called the “myddosome,” which also contains members of the Interleukin-1-Receptor-Associated Kinase (IRAK) family. The N-terminal Death Domain of MyD88 recruits firstly IRAK4 and then IRAK1 and IRAK2, which are phosphorylated to interact with the TNF receptor-associated factor 6 (TRAF6). TRAF6 is an E3 ubiquitin ligase able to generate K63-linked polyubiquitination chains. The linear ubiquitin assembly complex can bind these chains to recruit preassembled kinase complexes containing TAK1, TAB1, TAB2, and TAB3. This leads to nuclear factor kappa B (NF-κB) translocation to the nucleus after inhibitor of NF-κB (IKB) proteolytic degradation. Concomitantly, this complex controls the mitogen-activated protein kinase (MAPK) signaling that activates members of the activator protein-1 (AP1) transcription factor family, Jun and Fos, resulting in cytokine production, initiation of inflammation and metabolic cell polarization (6, 12).
Thus, MAL is an important actor in the establishment of inflammation. Over the last decade, a large number of clinical and experimental studies focused on the role of MAL in the control of infectious and non-infectious diseases (Table 1). Unexpectedly, as shown in Table 1, genetic variation in MAL was associated with either protection or susceptibility to diseases. Recent studies have shown that MAL is involved in other processes besides TLR-2 and TLR-4 making its biology more complex than expected. These new functions, discussed below, will make it possible to emit new hypotheses explaining the paradoxical behavior of MAL.
Several studies have investigated whether intracellular TLRs require MAL to signal. While PIP2 is required for MAL recruitment at TLR-2 and TLR-4 (10, 52), functional analysis by Kagan's group revealed that Phosphatidylinositol 3-phosphate (PI3P) and Phosphatidylserine (PS) are needed to recruit MAL to endosomal TLR-9 (21). The authors used MAL-deficient plasmacytoid DCs, known to respond exclusively via endosomal TLRs, to demonstrate that MAL is required for production of type I interferon (IFN) downstream TLR-9 stimulation with Herpes simplex Virus (HSV) (Figure 1C). Wild type and MAL-deficient primary bone marrow-derived macrophages were stimulated with substrains of HSV-1 showing that deficient cells presented a defective production of IL-1β and IL-6, specifically downstream of endocytic TLR-9 (21, 53). Since then, it became clear that multiple targets of the lipid-binding domain of MAL are functionally important and allow this adaptor to promote TLR signaling from both plasma membrane and endosomal compartments. Corroborating this, Shan et al., also demonstrated that MAL was recruited as an adaptor to endosomal compartments by TLR-8 (54). The complexity of the endosomal system fine-tunes the immune response by ensuring the proper compartmentalization of intracellular TLRs. The contribution of MAL downstream of intracellular TLRs increases the complexity of its biology and further investigations are needed to fully understand the regulation of endosomal signaling which could provide new hypotheses explaining the paradoxical role of MAL.
Besides the above-mentioned MAL-mediated pathways that are downstream of TLRs, Keane's group discovered in murine macrophages that MAL binds to IFN-γ receptor (IFNGR), which triggered its interaction with MyD88 (Figure 1B). In the context of Mycobacterium tuberculosis (Mtb) infection, authors reported that MAL leads to autophagy and vacuole acidification that kills the bacteria (Figure 1C). In addition, the S180L polymorphism (this single-nucleotide polymorphism (SNP) encodes a serine instead of a leucine), and its murine equivalent S200L, compromised IFNGR signaling impairing host responses to Mtb (8).
MAL was also described to interact with Receptor for Advanced Glycation End products (RAGE), a type I single-pass transmembrane protein belonging to the immunoglobulin superfamily (Figure 1B). Upon binding of the extracellular domain of RAGE to its ligands, the cytoplasmic domain of this receptor is phosphorylated at Ser391 by PKCζ. Consequently, this leads to the recruitment of MAL and MyD88, further inducing a recruitment of IRAK4, activation of the downstream effector kinases and finally production of inflammatory cytokines through activation of NF-κB. RAGE and TLR-2/4 partly share an intracellular signaling pathway. These receptors display a precise motif in the intracellular domain (Q residue followed by three successive negatively charged residues), which, upon phosphorylation, enhances its affinity to MAL (55, 56). During infection, such as tuberculosis (Tb), the RAGE pathway is modulated (57). Since MAL is implicated in this signaling pathway, mutations in the adaptor could certainly impact on disease severity. It was shown that PKCζ is upregulated during Tb progression, which increases effector killing functions (58). As PKCζ facilitates the recruitment of MAL to RAGE, it becomes an interesting way of investigation to decipher MAL's role during Mtb infection. Furthermore, co-morbidities such as diabetes are known to increase the amount of RAGE ligands, which could influence MAL functions during infection (59). Based on these observations, we can legitimately assume that MAL is implicated in other unknown pathways.
The PEST domain of MAL undergoes phosphorylation and polyubiquitination of lysine residues targeting degradation via the 26S proteasome. Mansell et al., demonstrated that stimulation of both TLR-2 and TLR-4 induced MAL degradation within 15–30 min after stimulation in order to avoid chronic inflammation. The degradation of MAL is a consequence of its polyubiquitination, which occurs via the SH2 domain of SOCS-1 and subsequent recruitment of the ubiquitin machinery (60). Tyrosine phosphorylation of MAL via Btk is necessary for the SOCS-1–mediated degradation. Moreover, MAL ubiquitination and degradation was also shown to be mediated by Cytoplasmic Linker Protein 170 (CLIP170) that is implicated in regulation of microtubule dynamics, cell migration and intracellular transport (7, 61). A study also demonstrated that MAL phosphorylation at Thr28 within its PBM reduces PI interactions and cell membrane targeting, leading to its ubiquitination and degradation (62).
Moreover, limiting the amount of PIP2 at the plasma membrane would prevent an exacerbated inflammation. Aksoy et al., showed in DCs that the interaction of MAL with phosphoinositol-3 kinases (PI3K) converts PIP2 to PIP3 and leads to the shedding of its membrane anchor sites. The change in the PIP2/PIP3 ratio favored the redistribution of MAL into the cytosol, where it was thus degraded. Moreover, TLR-4 is internalized, inactivating its downstream signaling pathway (1, 7).
In the context of whooping cough, a pulmonary infection caused by Bordetella pertussis, the absence of MAL induced susceptibility to the infection in mouse lungs with exacerbated dissemination leading to death (15). In this setting, MAL impacted on early local pro-inflammatory cytokine production by alveolar macrophages in lungs and prevented apoptosis-induced cell death and depletion of alveolar macrophages. As expected through this example, loss of function in MAL decreases the inflammatory response which leads to susceptibility to the infection. However, MAL biology is not as straightforward in other models for which the level of TLR/MAL-derived inflammation will strongly impact the severity of the disease. The impact of S180L polymorphism was deeply investigated. In a group of Pakistani population, it was observed that the 180L allele frequency is higher than that of the 180S allele in patients infected with Plasmodium, the causative agent of malaria, demonstrating that MAL deficiency enhances 3.000 times the chance of acquiring malaria caused by Plasmodium falciparum (35). In a cohort of adults from India, the heterozygous S180L mutation led to an optimal release of TNF-α that was shown to be protective against severe P. falciparum infection and mortality (36). Several studies also focused on the implication of MAL S180L polymorphism during Tb resulting in confusing conclusions. Indeed, S180L SNP has been often associated with protection. Capparelli et al., demonstrated that MAL S180L conferred resistance against Tb in heterozygous individuals, showing that those subjects displayed intermediate levels of IFN-γ, TNF-α or nitric oxide (NO), which helped to control the infection (29). Together, these results suggest that an intermediate level of inflammation decreases the severity of the pathology leading to a better protection to some pathogens that normally take advantage of an exacerbated inflammation.
Whereas MAL signaling was long considered important for antimicrobial immunity, recent studies demonstrated that genetic impairments in MAL are also associated with tumorigenesis. Interestingly, MAL overexpression was reported in ~20% of investigated lymphoma, and a whole exome sequencing in human recently revealed that MAL SNP R81C activated its downstream signaling to enhance NF-κB gene expression, whose constitutive activity is characteristic of B cell lymphoma (50, 63). It became clear that TLR/MAL-dependent inflammation strongly influences disease control with versatile consequences.
Following the hypothesis that negative feedback could avoid chronic inflammation and septic shock, a putative immunoregulatory role of MAL was investigated (20, 64). Mellett et al., demonstrated that MAL is the unique TIR adaptor protein capable of activating cyclic adenosine monophosphate (c-AMP) Response Element-binding proteins (CREB), a key transcription factor that mediates regulation of gene expression. They showed that MAL-induced phosphorylation of CREB was induced by LPS that stimulated TLR-4, suggesting a positive feedback system in dysregulated inflammatory responses, where MAL induces the production of IL-10 and cyclooxygenase 2 (COX-2). During Trypanosoma cruzi infection, the causative agent of Chagas disease, it was shown in a mouse model of infection that MAL deficiency is associated to exacerbated inflammation, similarly to TLR-2 deficient mice, leading to decreased parasitemia and delayed mortality (20). Moreover, authors distinguished between pro-inflammatory LyC6hiTLR2hi and anti-inflammatory Ly6CloTLR2hi splenic monocytes and demonstrated that MAL was associated with cytokine production by the immunosuppressive population after triggering TLR-2 or TLR-9.
The confusing role of MAL could be explained by the fact that MAL induces both pro-inflammatory and anti-inflammatory responses depending on the stimulated receptor and the targeted cell population. For example, Mtb is known to colonize different cell types and organs depending on the chronicity of the infection (65), possibly inducing different levels of inflammation. For instance, Russell's team demonstrated that Mtb grows differentially within interstitial macrophages compared to alveolar macrophages, which are more permissive to infection (66). The involvement of MAL in the immune response to Mtb infection in these two macrophage subtypes could provide new insights in the versatility of MAL functions.
During Staphylococcus aureus infection, the rare human SNP R121W was identified to impair the interaction of MAL with MyD88, TLR-2 and TLR-4 (43). The effect of this SNP, initially predicted deleterious, resulted in increased compensatory adaptive immune responses and decreased invasive hematogenous infections in children (Figure 1C). This demonstrated that MAL not only affects innate immune responses but also adaptive immunity through not yet understood mechanisms.
To counteract immune response activation triggered by PAMPs detection by PRRs, several pathogenic bacteria express virulence factors, such as TIR domain-containing proteins, to perturb TIR-dependent interactions, which are essential in the initiation of innate immune responses (67–69). Salcedo's team demonstrated that Pseudomonas aeruginosa PA7 has a TIR domain-containing protein called PumA (Pseudomonas UBAP1 modulator A) conferring the ability to downmodulate innate immune responses (70). Indeed, PumA was translocated into host cells during infection to directly interact with MAL at the plasma membrane controlling TLR signaling. Similarly, Brucella produces a TIR domain-containing protein (TcpB/Btp1) to selectively target MAL and inhibit NF-κB activation, which is essential for intracellular Brucella survival and replication (Figure 1C) (71). Moreover, TcpB/Btp1 was also described to target CLIP170 enhancing MAL proteasome-mediated degradation (61). Furthermore, a number of viral proteins were described to interfere with innate immune signaling, highlighting the implication of the TLR pathway in antiviral immunity (72). Among them, the poxvirus protein A46 was identified to inhibit TLR-4 signaling by interfering physically with MAL (73, 74). It is of interest to investigate if other pathogens are also able to physically modulate MAL signaling to determine whether its deficiency could interfere with the progression of infection. To answer this question, bioinformatics might be useful to identify potential effectors that may interact with MAL.
Epistasis is a gene-gene interaction that influences a phenotype. As MAL interacts with numerous proteins, it is possible that genetic variations in these partners could modulate its signaling. Fulgione et al., investigated an epistatic interaction between MyD88 and MAL during Helicobacter pylori infection. A cohort study revealed that heterozygosity for S180L confer increased resistance to infection, which was found associated with a low level of IL-6, COX-2, TNF-α, and IL-1β production. Regardless of MAL, polymorphism in MyD88 alone did not influence the infection. However, in some combinations with MAL, MyD88 has an effect on the risk of infection. Together with MAL S180L, certain polymorphisms in MyD88 confer higher protection providing evidence of an epistatic interaction occurring between the two genes (44). The same polymorphic sites have also been documented to act epistatically against Mtb infection as well (29). These results showed that epistasis could play a key role in MAL versatility.
Impairment of MAL expression in diseases result in some controversy. These discrepant findings about the differential effect of MAL during diseases could simply represent heterogeneity of association in different populations, which is well described for many immunogenetic polymorphisms (75). Frequencies of polymorphisms in MAL can vary among different races. This difference, along with gene-gene interactions, environmental and cultural factors, and variations in microbial strains make understanding the observed differences between ethnic groups even more complicated. Moreover, the size of the selected populations in different cohort studies can vary. This can impact the power of detection of small effects due to a rare mutation.
More investigations are still needed to characterize the contribution of MAL in each setting and to reach understanding of its impact on immune response to infection. In particular, the severity of the disease and its inflammatory status seem to have an essential impact. Deeper characterization of the local environments that are less favorable for the pathogen to survive may bring some cues.
In the emerging field of host-directed therapies to intracellular pathogens, TLRs and their adaptor proteins were proposed as putative targets for the treatment of inflammatory disorders and to overcome microbial resistance (76). For example, Gefitinib, Phycocianin and other peptides were recently studied for their inhibiting effect on MAL in the context of endotoxic injury, lung cancers and autoimmune diseases, respectively, showing promising results (77–80).
All authors participated in the concept, preparation, and writing of the manuscript.
Financial support for this work was provided by the European Community (CycloNHit no. 608407, ERC-STG INTRACELLTB no. 260901), the Agence Nationale de la Recherche (ANR-10-EQPX-04-01, ANR-14-CE08-0017, ANR-16-CE35-0009, ANR-16-IDEX-0004 ULNE), the Projet Transversal de l'Institut Pasteur (PTR441, PTR22-16), the EMBO Young Investigator Program, the Feder [12001407 (D-AL) Equipex Imaginex BioMed], the Région Hauts-de-France (convention no. 12000080), and the Fondation pour la Recherche Medicale (SPF20170938709).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Kieser KJ, Kagan JC. Multi-receptor detection of individual bacterial products by the innate immune system. Nat Rev Immunol. (2017) 17:376–90. doi: 10.1038/nri.2017.25
2. Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. (2010) 11:373–84. doi: 10.1038/ni.1863
3. Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. (2015) 33:257–90. doi: 10.1146/annurev-immunol-032414-112240
4. Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. (2002) 420:329–33. doi: 10.1038/nature01180
5. Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, Kaisho T, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. (2002) 420:324–9. doi: 10.1038/nature01182
6. Bernard NJ, O'neill LA. Mal, more than a bridge to MyD88. IUBMB Life. (2013) 65:777–86. doi: 10.1002/iub.1201
7. Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilagyi A, Banoczi Z, et al. On the sequential determinants of calpain cleavage. J Biol Chem. (2004) 279:20775–85. doi: 10.1074/jbc.M313873200
8. Ni Cheallaigh C, Sheedy FJ, Harris J, Munoz-Wolf N, Lee J, West K, et al. A common variant in the adaptor mal regulates interferon gamma signaling. Immunity. (2016) 44:368–79. doi: 10.1016/j.immuni.2016.01.019
9. Patra MC, Choi S. Insight into phosphatidylinositol-dependent membrane localization of the innate immune adaptor protein toll/interleukin 1 receptor domain-containing adaptor protein. Front Immunol. (2018) 9:75. doi: 10.3389/fimmu.2018.00075
10. Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. (2006) 125:943–55. doi: 10.1016/j.cell.2006.03.047
11. Hughes MM, Lavrencic P, Coll RC, Ve T, Ryan DG, Williams NC, et al. Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling. Proc Natl Acad Sci USA. (2017) 114:E6480–9. doi: 10.1073/pnas.1701868114
12. Gong L, Wang H, Sun X, Liu C, Duan C, Cai R, et al. Toll-Interleukin 1 Receptor domain-containing adaptor protein positively regulates BV2 cell M1 polarization. Eur J Neurosci. (2016) 43:1674–82. doi: 10.1111/ejn.13257
13. Jeyaseelan S, Manzer R, Young SK, Yamamoto M, Akira S, Mason RJ, et al. Toll-IL-1 receptor domain-containing adaptor protein is critical for early lung immune responses against Escherichia coli lipopolysaccharide and viable Escherichia coli. J Immunol. (2005) 175:7484–95. doi: 10.4049/jimmunol.175.11.7484
14. Jeyaseelan S, Young SK, Yamamoto M, Arndt PG, Akira S, Kolls JK, et al. Toll/IL-1R domain-containing adaptor protein (TIRAP) is a critical mediator of antibacterial defense in the lung against Klebsiella pneumoniae but not Pseudomonas aeruginosa. J Immunol. (2006) 177:538–47. doi: 10.4049/jimmunol.177.1.538
15. Bernard NJ, Finlay CM, Tannahill GM, Cassidy JP, O'neill LA, Mills KH. A critical role for the TLR signaling adapter Mal in alveolar macrophage-mediated protection against Bordetella pertussis. Mucosal Immunol. 8:982–92. doi: 10.1038/mi.2014.125
16. Jerke S, Srinivasan A, Mcsorley SJ. Expression of Toll/IL-1R domain-containing adaptor protein. (TIRAP) is detrimental to primary clearance of Salmonella and is not required for the generation of protective immunity. Immunol Lett. (2008) 116:64–71. doi: 10.1016/j.imlet.2007.11.007
17. Talbot S, Totemeyer S, Yamamoto M, Akira S, Hughes K, Gray D, et al. Toll-like receptor 4 signalling through MyD88 is essential to control Salmonella enterica serovar typhimurium infection, but not for the initiation of bacterial clearance. Immunology. (2009) 128:472–83. doi: 10.1111/j.1365-2567.2009.03146.x
18. Corr SC, Palsson-Mcdermott EM, Grishina I, Barry SP, Aviello G, Bernard NJ, et al. MyD88 adaptor-like. (Mal) functions in the epithelial barrier and contributes to intestinal integrity via protein kinase C. Mucosal Immunol. (2014) 7:57–67. doi: 10.1038/mi.2013.24
19. Fremond CM, Togbe D, Doz E, Rose S, Vasseur V, Maillet I, et al. IL-1 receptor-mediated signal is an essential component of MyD88-dependent innate response to Mycobacterium tuberculosis infection. J Immunol. (2007) 179:1178–89. doi: 10.4049/jimmunol.179.2.1178
20. Gravina HD, Goes AM, Murta SM, Ropert C. MyD88 adapter-like. (Mal)/TIRAP is required for cytokine production by splenic Ly6CloTLR2hi but not by Ly6ChiTLR2hi Monocytes during Trypanosoma cruzi infection. J Biol Chem. (2016) 291:23832–41. doi: 10.1074/jbc.M116.729509
21. Bonham KS, Orzalli MH, Hayashi K, Wolf AI, Glanemann C, Weninger W, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of toll-like receptor signal transduction. Cell. (2014) 156:705–16. doi: 10.1016/j.cell.2014.01.019
22. Aviello G, Corr SC, Johnston DG, O'neill LA, Fallon PG. MyD88 adaptor-like. (Mal) regulates intestinal homeostasis and colitis-associated colorectal cancer in mice. Am J Physiol Gastrointest Liver Physiol. (2014) 306:G769–78. doi: 10.1152/ajpgi.00399.2013
23. Hawn TR, Dunstan SJ, Thwaites GE, Simmons CP, Thuong NT, Lan NTN, et al. A polymorphism in Toll-interleukin 1 receptor domain containing adaptor protein is associated with susceptibility to meningeal tuberculosis. J Infect Dis. (2006) 194:1127–34. doi: 10.1086/507907
24. Khor CC, Chapman SJ, Vannberg FO, Dunne A, Murphy C, Ling EY, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. (2007) 39:523–8. doi: 10.1038/ng1976
25. Castiblanco J, Varela DC, Castano-Rodriguez N, Rojas-Villarraga A, Hincapie ME, Anaya JM. TIRAP (MAL) S180L polymorphism is a common protective factor against developing tuberculosis and systemic lupus erythematosus. Infect Genet Evol. (2008) 8:541–4. doi: 10.1016/j.meegid.2008.03.001
26. Zhang YX, Xue Y, Liu JY, Zhao MY, Li FJ, Zhou JM, et al. Association of TIRAP (MAL) gene polymorhisms with susceptibility to tuberculosis in a Chinese population. Genet Mol Res. (2011) 10:7–15. doi: 10.4238/vol10-1gmr980
27. Sanchez D, Lefebvre C, Rioux J, Garcia LF, Barrera LF. Evaluation of Toll-like receptor and adaptor molecule polymorphisms for susceptibility to tuberculosis in a Colombian population. Int J Immunogenet. (2012) 39:216–23. doi: 10.1111/j.1744-313X.2011.01077.x
28. Selvaraj P, Harishankar M, Singh B, Jawahar MS, Banurekha VV. Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis. (2010) 90:306–10. doi: 10.1016/j.tube.2010.08.001
29. Capparelli R, De Chiara F, Di Matteo A, Medaglia C, Iannelli D. The MyD88 rs6853 and TIRAP rs8177374 polymorphic sites are associated with resistance to human pulmonary tuberculosis. Genes Immun. (2013) 14:504–11. doi: 10.1038/gene.2013.48
30. Nejentsev S, Thye T, Szeszko JS, Stevens H, Balabanova Y, Chinbuah AM, et al. Analysis of association of the TIRAP (MAL) S180L variant and tuberculosis in three populations. Nat Genet. (2008) 40:261–2; author reply 262-263. doi: 10.1038/ng0308-261
31. Miao R, Li J, Sun Z, Xu F, Shen H. Meta-analysis on the association of TIRAP S180L variant and tuberculosis susceptibility. Tuberculosis. (2011) 91:268–72. doi: 10.1016/j.tube.2011.01.006
32. Liu Q, Li W, Li D, Feng Y, Tao C. TIRAP C539T polymorphism contributes to tuberculosis susceptibility: evidence from a meta-analysis. Infect Genet Evol. (2014) 27:32–9. doi: 10.1016/j.meegid.2014.06.025
33. Siebert JN, Hamann L, Verolet CM, Gameiro C, Grillet S, Siegrist CA, et al. Toll-interleukin 1 receptor domain-containing adaptor protein 180L single-nucleotide polymorphism is associated with susceptibility to recurrent pneumococcal lower respiratory tract infections in children. Front Immunol. (2018) 9:1780. doi: 10.3389/fimmu.2018.01780
34. Ramasawmy R, Cunha-Neto E, Fae KC, Borba SC, Teixeira PC, Ferreira SC, et al. Heterozygosity for the S180L variant of MAL/TIRAP, a gene expressing an adaptor protein in the Toll-like receptor pathway, is associated with lower risk of developing chronic Chagas cardiomyopathy. J Infect Dis. (2009) 199:1838–45. doi: 10.1086/599212
35. Nawaz SK, Ahmed B, Arshad N, Rani A, Rasool H, Arshad M. Role of S180L polymorphism in etiology of malaria caused by Plasmodium falciparum in a small group of Pakistani population. Bosn J Basic Med Sci. (2015) 15:20–3. doi: 10.17305/bjbms.2015.413
36. Panda AK, Das BK, Panda A, Tripathy R, Pattnaik SS, Mahto H, et al. Heterozygous mutants of TIRAP. (S180L) polymorphism protect adult patients with Plasmodium falciparum infection against severe disease and mortality. Infect Genet Evol. (2016) 43:146–50. doi: 10.1016/j.meegid.2016.04.035
37. Esposito S, Molteni CG, Zampiero A, Baggi E, Lavizzari A, Semino M, et al. Role of polymorphisms of toll-like receptor (TLR) 4, TLR9, toll-interleukin 1 receptor domain containing adaptor protein (TIRAP) and FCGR2A genes in malaria susceptibility and severity in Burundian children. Malar J. (2012) 11:196. doi: 10.1186/1475-2875-11-196
38. Zakeri S, Pirahmadi S, Mehrizi AA, Djadid ND. Genetic variation of TLR-4, TLR-9 and TIRAP genes in Iranian malaria patients. Malar J. (2011) 10:77. doi: 10.1186/1475-2875-10-77
39. Rani A, Nawaz SK, Irfan S, Arshad M, Bashir R, Shaheen N. Role of MyD88-adaptor-like gene polymorphism rs8177374 in modulation of malaria severity in the Pakistani population. Braz J Infect Dis. (2017) 21:418–23. doi: 10.1016/j.bjid.2017.04.002
40. Sheedy FJ, Marinou I, O'neill LA, Wilson AG. The Mal/TIRAP S180L and TLR4 G299D polymorphisms are not associated with susceptibility to, or severity of, rheumatoid arthritis. Ann Rheum Dis. (2008) 67:1328–31. doi: 10.1136/ard.2007.083337
41. Ladhani SN, Davila S, Hibberd ML, Heath PT, Ramsay ME, Slack MP, et al. Association between single-nucleotide polymorphisms in Mal/TIRAP and interleukin-10 genes and susceptibility to invasive haemophilus influenzae serotype b infection in immunized children. Clin Infect Dis. (2010) 51:761–7. doi: 10.1086/656236
42. Papadopoulos AI, Ferwerda B, Antoniadou A, Sakka V, Galani L, Kavatha D, et al. Association of Mal/TIRAP S180L variant polymorphism with decreased infection risk in patients with advanced HIV-1 infection. Cytokine. (2012) 60:104–7. doi: 10.1016/j.cyto.2012.05.008
43. Israel L, Wang Y, Bulek K, Della Mina E, Zhang Z, Pedergnana V, et al. Human adaptive immunity rescues an inborn error of innate immunity. Cell. (2017) 168:789–800 e710. doi: 10.1016/j.cell.2017.01.039
44. Fulgione A, Di Matteo A, Contaldi F, Manco R, Ianniello F, Incerti G, et al. Epistatic interaction between MyD88 and TIRAP against Helicobacter pylori. FEBS Lett. (2016) 590:2127–37. doi: 10.1002/1873-3468.12252
45. Rupasree Y, Naushad SM, Rajasekhar L, Uma A, Kutala VK. Association of TLR4 (D299G, T399I), TLR9−1486T>C, TIRAP S180L and TNF-alpha promoter (-1031,−863,−857) polymorphisms with risk for systemic lupus erythematosus among South Indians. Lupus. (2015) 24:50–7. doi: 10.1177/0961203314549792
46. Sacre SM, Andreakos E, Kiriakidis S, Amjadi P, Lundberg A, Giddins G, et al. The Toll-like receptor adaptor proteins MyD88 and Mal/TIRAP contribute to the inflammatory and destructive processes in a human model of rheumatoid arthritis. Am J Pathol. (2007) 170:518–25. doi: 10.2353/ajpath.2007.060657
47. Durrani O, Banahan K, Sheedy FJ, Mcbride L, Ben-Chetrit E, Greiner K, et al. TIRAP Ser180Leu polymorphism is associated with Behcet's disease. Rheumatology. (2011) 50:1760–5. doi: 10.1093/rheumatology/ker200
48. Turunc G, Coskun D, Alibaz-Oner F, Coit P, Duzgun N, Alpsoy E, et al. TIR-domain-containing adaptor protein gene TIRAP S180L polymorphism is not increased in Behcet's disease patients in two ethnic cohorts. Clin Exp Rheumatol. (2013) 31:54–6.
49. Castano-Rodriguez N, Kaakoush NO, Pardo AL, Goh KL, Fock KM, Mitchell HM. Genetic polymorphisms in the Toll-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer. Hum Immunol. (2014) 75:808–15. doi: 10.1016/j.humimm.2014.06.001
50. Burkhard R, Keller I, Arambasic M, Juskevicius D, Tzankov A, Lundberg P, et al. TIRAP p.R81C is a novel lymphoma risk variant which enhances cell proliferation via NF-kappaB mediated signaling in B-cells. Haematologica. (2019) 104:766–77. doi: 10.3324/haematol.2018.201590
51. An Y, Ohnishi H, Matsui E, Funato M, Kato Z, Teramoto T, et al. Genetic variations in MyD88 adaptor-like are associated with atopic dermatitis. Int J Mol Med. (2011) 27:795–801. doi: 10.3892/ijmm.2011.645
52. Aksoy E, Taboubi S, Torres D, Delbauve S, Hachani A, Whitehead MA, et al. The p110delta isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat Immunol. (2012) 13:1045–54. doi: 10.1038/ni.2426
53. Zyzak J, Mitkiewicz M, Leszczynska E, Reniewicz P, Moynagh PN, Siednienko J. HSV-1/TLR9-Mediated IFNbeta and TNFalpha induction is Mal-dependent in macrophages. J Innate Immunity. (2019) 12:387–98. doi: 10.1159/000504542
54. Shan S, Liu R, Jiang L, Zhu Y, Li H, Xing W, et al. Carp Toll-like receptor 8. (Tlr8): An intracellular Tlr that recruits TIRAP as adaptor and activates AP-1 pathway in immune response. Fish Shellfish Immunol. (2018) 82:41–9. doi: 10.1016/j.fsi.2018.08.001
55. Sakaguchi M, Murata H, Yamamoto K, Ono T, Sakaguchi Y, Motoyama A, et al. TIRAP, an adaptor protein for TLR2/4, transduces a signal from RAGE phosphorylated upon ligand binding. PLoS ONE. (2011) 6:e23132. doi: 10.1371/journal.pone.0023132
56. Teissier T, Boulanger E. The receptor for advanced glycation end-products. (RAGE) is an important pattern recognition receptor. (PRR) for inflammaging. Biogerontology. (2019) 20:279–301. doi: 10.1007/s10522-019-09808-3
57. Lui G, Wong CK, Ip M, Chu YJ, Yung IM, Cheung CS, et al. HMGB1/RAGE signaling and pro-inflammatory cytokine responses in non-HIV adults with active pulmonary tuberculosis. PLoS ONE. (2016) 11:e0159132. doi: 10.1371/journal.pone.0159132
58. Parihar SP, Ozturk M, Marakalala MJ, Loots DT, Hurdayal R, Maasdorp DB, et al. Protein kinase C-delta. (PKCδ), a marker of inflammation and tuberculosis disease progression in humans, is important for optimal macrophage killing effector functions and survival in mice. Mucosal Immunol. (2018) 11:496–511. doi: 10.1038/mi.2017.68
59. Kumar NP, Moideen K, Nancy A, Viswanathan V, Shruthi BS, Sivakumar S, et al. Systemic RAGE ligands are upregulated in tuberculosis individuals with diabetes co-morbidity and modulated by anti-tuberculosis treatment and metformin therapy. BMC Infect Dis. (2019) 19:1039. doi: 10.1186/s12879-019-4648-1
60. Mansell A, Smith R, Doyle SL, Gray P, Fenner JE, Crack PJ, et al. Suppressor of cytokine signaling 1 negatively regulates Toll-like receptor signaling by mediating Mal degradation. Nat Immunol. (2006) 7:148–55. doi: 10.1038/ni1299
61. Jakka P, Bhargavi B, Namani S, Murugan S, Splitter G, Radhakrishnan G. Cytoplasmic linker protein CLIP170 negatively regulates TLR4 signaling by targeting the TLR adaptor protein TIRAP. J Immunol. (2018) 200:704–14. doi: 10.4049/jimmunol.1601559
62. Zhao X, Xiong W, Xiao S, Tang TX, Ellena JF, Armstrong GS, et al. Membrane targeting of TIRAP is negatively regulated by phosphorylation in its phosphoinositide-binding motif. Sci Rep. (2017) 7:43043. doi: 10.1038/srep43043
63. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. (2018) 24:679–90. doi: 10.1038/s41591-018-0016-8
64. Mellett M, Atzei P, Jackson R, O'neill LA, Moynagh PN. Mal mediates TLR-induced activation of CREB and expression of IL-10. J Immunol. (2011) 186:4925–35. doi: 10.4049/jimmunol.1002739
65. Cohen SB, Gern BH, Delahaye JL, Adams KN, Plumlee CR, Winkler JK, et al. Alveolar macrophages provide an early Mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe. (2018) 24:439–46 e434. doi: 10.1016/j.chom.2018.08.001
66. Huang L, Nazarova EV, Tan S, Liu Y, Russell DG. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J Exp Med. (2018) 215:1135–52. doi: 10.1084/jem.20172020
67. Salcedo SP, Marchesini MI, Degos C, Terwagne M, Von Bargen K, Lepidi H, et al. BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front Cell Infect Microbiol. (2013) 3:28. doi: 10.3389/fcimb.2013.00028
68. Rosadini CV, Kagan JC. Microbial strategies for antagonizing Toll-like-receptor signal transduction. Curr Opin Immunol. (2015) 32:61–70. doi: 10.1016/j.coi.2014.12.011
69. Rosadini CV, Zanoni I, Odendall C, Green ER, Paczosa MK, Philip NH, et al. A single bacterial immune evasion strategy dismantles both MyD88 and TRIF signaling pathways downstream of TLR4. Cell Host Microbe. (2015) 18:682–93. doi: 10.1016/j.chom.2015.11.006
70. Imbert PR, Louche A, Luizet JB, Grandjean T, Bigot S, Wood TE, et al. A Pseudomonas aeruginosa TIR effector mediates immune evasion by targeting UBAP1 and TLR adaptors. EMBO J. (2017) 36:1869–87. doi: 10.15252/embj.201695343
71. Li W, Ke Y, Wang Y, Yang M, Gao J, Zhan S, et al. Brucella TIR-like protein TcpB/Btp1 specifically targets the host adaptor protein MAL/TIRAP to promote infection. Biochem Biophys Res Commun. (2016) 477:509–14. doi: 10.1016/j.bbrc.2016.06.064
72. Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. (2008) 8:911–22. doi: 10.1038/nri2436
73. Oda S, Franklin E, Khan AR. Poxvirus A46 protein binds to TIR domain-containing Mal/TIRAP via an alpha-helical sub-domain. Mol Immunol. (2011) 48:2144–50. doi: 10.1016/j.molimm.2011.07.014
74. Stack J, Bowie AG. Poxviral protein A46 antagonizes Toll-like receptor 4 signaling by targeting BB loop motifs in Toll-IL-1 receptor adaptor proteins to disrupt receptor:adaptor interactions. J Biol Chem. (2012) 287:22672–82. doi: 10.1074/jbc.M112.349225
75. Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. (2002) 186:1463–8. doi: 10.1086/344891
76. Kaufmann SHE, Dorhoi A, Hotchkiss RS, Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov. (2018) 17:35–56. doi: 10.1038/nrd.2017.162
77. Hao S, Li S, Wang J, Yan Y, Ai X, Zhang J, et al. Phycocyanin exerts anti-proliferative effects through down-regulating TIRAP/NF-kappaB activity in human non-small cell lung cancer cells. Cells. (2019) 8:588. doi: 10.3390/cells8060588
78. Srivastava M, Saqib U, Banerjee S, Wary K, Kizil B, Muthu K, et al. Inhibition of the TIRAP-c-Jun interaction as a therapeutic strategy for AP1-mediated inflammatory responses. Int Immunopharmacol. (2019) 71:188–97. doi: 10.1016/j.intimp.2019.03.031
79. Achek A, Kwon HK, Patra MC, Shah M, Hong R, Lee WH, et al. A peptide derived from the core beta-sheet region of TIRAP decoys TLR4 and reduces inflammatory and autoimmune symptoms in murine models. EBioMedicine. (2020) 52:102645. doi: 10.1016/j.ebiom.2020.102645
Keywords: TLRs, MAL/Tirap, chronic diseases, signaling pathways, inflammation
Citation: Belhaouane I, Hoffmann E, Chamaillard M, Brodin P and Machelart A (2020) Paradoxical Roles of the MAL/Tirap Adaptor in Pathologies. Front. Immunol. 11:569127. doi: 10.3389/fimmu.2020.569127
Received: 03 June 2020; Accepted: 24 August 2020;
Published: 25 September 2020.
Edited by:
Jagadeesh Bayry, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Catherine Ropert, Federal University of Minas Gerais, BrazilCopyright © 2020 Belhaouane, Hoffmann, Chamaillard, Brodin and Machelart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Priscille Brodin, cHJpc2NpbGxlLmJyb2RpbkBpbnNlcm0uZnI=; Arnaud Machelart, YXJuYXVkLm1hY2hlbGFydEBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.