- 1Africa Health Research Institute, University of KwaZulu-Natal, Durban, South Africa
- 2Department of Infectious Diseases, University of KwaZulu-Natal, Durban, South Africa
- 3Vanderbilt Tuberculosis Center, Vanderbilt University School of Medicine, Nashville, TN, United States
- 4Global Division, The Aurum Institute, Johannesburg, South Africa
- 5Department of Infection and Immunity, University College London, London, United Kingdom
Tuberculosis remains a leading cause of death globally despite curative treatment, partly due to the difficulty of identifying patients who will not respond to therapy. Simple host biomarkers that correlate with response to drug treatment would facilitate improvement in outcomes and the evaluation of novel therapies. In a prospective longitudinal cohort study, we evaluated neutrophil count and phenotype at baseline, as well as during TB treatment in 79 patients [50 (63%) HIV-positive] with microbiologically confirmed drug susceptible TB undergoing standard treatment. At time of diagnosis, blood neutrophils were highly expanded and surface expression of the neutrophil marker CD15 greatly reduced compared to controls. Both measures changed rapidly with the commencement of drug treatment and returned to levels seen in healthy control by treatment completion. Additionally, at the time of diagnosis, high neutrophil count, and low CD15 expression was associated with higher sputum bacterial load and more severe lung damage on chest x-ray, two clinically relevant markers of disease severity. Furthermore, CD15 expression level at diagnosis was associated with TB culture conversion after 2 months of therapy (OR: 0.14, 95% CI: 0.02, 0.89), a standard measure of early TB treatment success. Importantly, our data was not significantly impacted by HIV co-infection. These data suggest that blood neutrophil metrics could potentially be exploited to develop a simple and rapid test to help determine TB disease severity, monitor drug treatment response, and identify subjects at diagnosis who may respond poorly to treatment.
Introduction
Though global tuberculosis (TB) disease incidence is falling at a rate of 1.5–2% per year, it remains one of the leading causes of death from a single infectious agent, and much greater declines in incidence are required to meet global TB targets (1). TB is a curable disease, but better tools to evaluate early TB treatment response would facilitate optimization of TB treatment, and cure rates (2, 3). Shorter TB treatment regimens are also needed as poor adherence to the current standard 6-months regimen can lead to treatment failure, relapse, or the development of drug resistant TB (4). Likewise, tools for identifying patients at the point of diagnosis who are at risk of responding poorly to treatment could help with clinical management. In addition, novel methods of evaluating response to treatment could expedite testing of new TB treatment regimens in clinical trials. Host immune responses are potential biomarkers for predicting response to anti-TB treatment (5, 6) as the current approach of evaluating culture conversion at 2 months has proved poorly predictive of cure (7–9). Typically, these biomarkers are measured in blood or urine, which also makes them an attractive option for groups that are inherently difficult to diagnose and monitor with conventional sputum samples, such as children, pregnant women, and people living with HIV (10–12).
As proof of this concept, Adekambi et al. showed that the expression of cellular activation markers on Mycobacterium tuberculosis (Mtb)-specific CD4+ T-cells was highly effective at distinguishing individuals with asymptomatic latent TB infection (LTBI) from those with untreated active TB (13). Furthermore, decreasing expression of these markers correlated with decreasing mycobacterial load during treatment. These observations were subsequently confirmed in HIV/TB co-infected individuals (14). The advantage of this biomarker is that it is highly TB disease specific. However, the assay requires an extended period of cell stimulation and also, by definition, is only useful in individuals with detectable Mtb responses. This last point is important, as up to 30% of people with active TB infection do not have a detectable Mtb-specific CD4 response at the time of diagnosis (15), and this proportion can be higher in individuals co-infected with HIV (16, 17).
Neutrophils are the most abundant immune cells in the blood, accounting for ~60% of circulating leukocytes, and are rapidly activated and recruited to sites of infection or tissue damage (18). Moreover, neutrophils have a lifespan in circulation of ~7 h, increasing on activation to 2–4 days. Consequently, the large pool of neutrophils in blood and tissue is continuously being replenished from precursors in the bone marrow (19–21). Therefore, neutrophil based biosignatures may be expected to change rapidly. The role neutrophils play during active TB is complex and depends on timing. Initially, following arrival at the site of infection, neutrophils can recognize Mtb through both opsonin-dependent and -independent pathways, resulting in phagocytosis (22, 23). This probably contributes to early immune control, as experimental depletion of neutrophils during initial infection in mice has a negative outcome for the host (24). In addition, low blood neutrophil count in household contacts of TB patients is an independent predictor of TB infection (25). However, if infection fails to resolve, neutrophils appear to be one of the main drivers of lung destruction (6, 26, 27). A detailed investigation of the neutrophil response to TB infection in humans may also, therefore, improve our understanding of the immunopathology of this disease.
Transcriptomic analysis of whole-blood from TB patients has revealed a 393-transcript signature for active TB disease that is dominated by a neutrophil-driven IFN-inducible gene profile, consisting of both IFN-γ and type I IFN-αβ signaling (28). This signature was subsequently confirmed in sorted neutrophils and found to correlate with radiographic extent of disease and could distinguish active TB disease from other respiratory diseases. In addition, this signature significantly diminished following 2 months of TB treatment (29). We, therefore, hypothesized that (1) active TB is associated with both increased blood neutrophil count and phenotypic changes that could be easily measured by flow cytometry, (2) neutrophil count and phenotype is correlated with disease severity and, (3) due to the lifespan of neutrophils, these changes rapidly respond to changing disease status.
Materials and Methods
Study Population
TB Cases
Study participants were recruited from the Regional Prospective Observational Research for Tuberculosis (RePORT)-South Africa, Durban study between December 2016 and December 2019. All participants were TB symptomatic and referred for TB testing by attending clinician. Enrolled subjects were GeneXpert and culture positive (solid or liquid media), had evidence of pulmonary involvement by chest X-ray (CXR), were 18 years or older and provided written informed consent. All RePORT study participants received standard 6-months treatment for TB, consisting of isoniazid, rifampicin, pyrazinamide, and ethambutol. Study participants were followed for the duration of TB treatment and up to 24 months post-enrolment. Enrolees found to have baseline drug resistance to any standard anti-TB drug (isoniazid, rifampicin, ethambutol, or pyrazinamide), who developed resistance during treatment, or who had concomitant extrapulmonary TB were excluded from the study.
Controls
TB symptomatic participants initially recruited for RePORT as GeneXpert positive, but subsequently found to be culture negative at baseline were included as Xpert-positive/culture-negative controls (n = 14) and were followed for 2 months from enrolment. An additional control group consisting of healthy asymptomatic individuals (n = 23) were recruited from the Collection of Urine, Blood, and Sputum (CUBS) cohort in Durban between January and December 2018. This group is referred to as healthy controls.
Descriptive Statistics
For TB cases and Xpert-positive/culture-negative controls age, sex, race, body mass index (BMI), self-reported alcohol use, tobacco use, and employment status were recorded. For persons living with HIV (PLWH), CD4 cell counts, HIV-1 RNA (viral load), and timing of antiretroviral therapy were also collected. Extent of lung disease at enrolment was determined from CXR, evaluated by a radiologist blinded to all study participant information, using a CXR score [% area of lung infected + 40 for the presence of cavities; giving a scale of 0–140 (30)] see Table S1. Bacterial burden was estimated from liquid culture using the BD BACTEC MGIT 960 mycobacterial detection system (Becton Dickinson Microbiology Systems, Sparks, Md.) and full blood counts were measured independently by a commercial laboratory. For the healthy controls age, sex, and race were recorded.
Sample Collection, Processing, and Analysis
Samples
Blood samples were collected from healthy controls at one time-point only, and blood and sputum samples from TB cases were collected at baseline (pre-treatment) and at post-treatment initiation study visits: week 1, week 3, month 2, and month 6. For all participants, fresh blood samples were collected in EDTA vacutainers (BD Biosciences) for cell phenotyping.
Neutrophil Isolation and Staining
A 100 μl of whole blood was stained with 13 surface markers and incubated for 30 min at 4°C in the dark. The phenotyping panel comprised of standard lineage markers V500 conjugated mAb anti-CD45 (clone HI30), BUV496 conjugated mAb anti-CD16 (clone 3G8), BUV395 conjugated mAb anti-CD11b (clone ICRF44), V450 conjugated mAb anti-CD66b (clone G10F5) all from BD Bioscience; BUV785 conjugated mAb anti-CD3 (clone OKT3) and BV711 conjugated mAb anti-CD14 (clone M5E2) from Biolegend. The following phenotype markers were used, PE conjugated mAb anti-CD177 (clone MEM-166) and BV605 conjugated mAb anti-HLA-DR (clone G46-6) from BD Bioscience; FITC conjugated mAb anti-CD15 (clone HI98) and PE-Cy7 conjugated mAb anti-CXCR4 (clone 12G5) from Biolegend; APC conjugated mAb anti-CD32 (clone FLI8.28) and PE-Cy5 conjugated mAb anti-CD10 (clone HI10a) from BD Pharmigen and Live dead stain (ThermoFischer Scientific). Phenotype markers were chosen based on preliminary data from our lab indicating differential expression between TB cases and HIV-TB- controls. Red blood cells were removed, and leukocytes fixed by adding 2 mls of FacsLyse solution (BD Bioscience) and incubating for 10 min in the dark at room temperature. Samples were then centrifuged at 500 g for 5 min and the supernatant decanted. Cells were washed with 2 mls of 1X PBS (Sigma Aldrich) for 5 min at 500 g and the cell pellet resuspended in 2% PFA (Sigma Aldrich). Cells were acquired on a 17 color FACSAria Fusion (BD Bioscience), with calibrating beads run before each acquisition to ensure consistent performance. Due to the longitudinal nature of the study we normalized the data based on a published method by Upreti et al. (31). Data normalization was performed by transforming the Mean Fluorescence Intensity (MFI) values of the test samples (TB cases, Xpert-positive/culture-negative, and healthy controls) to a common scale using the following equation: Final relative fluorescence intensity (RFI) = MFI of the test sample/MFI of the internal control. Gating strategy shown in Figure S1.
TB Treatment Outcomes
We examined two TB treatment outcomes; (1) culture status on solid media 2 months after initiation of anti-TB therapy; and (2) end of TB treatment outcome, as defined by World Health Organization (WHO) into two mutually-exclusive categories—treatment success (cure or treatment completion) vs. unsuccessful outcome (death, failure, loss to follow-up, or not evaluated) (32). Cure was defined as completing treatment with a negative culture result at the end of treatment and on at least one previous occasion. Treatment completion was based on completion of standard anti-TB therapy without evidence of failure, and without documentation of a negative culture in the last month of TB treatment and/or on at least one previous occasion (either because it was not done, or results are unavailable). Death included mortality from any cause during TB treatment. Treatment failure was defined as a positive culture at month 5 of treatment or later. Loss to follow-up was any participant who interrupted treatment for 2 consecutive months or more. Not evaluated included any participant for whom treatment outcome was unassigned, including cases who transferred out of care or whom treatment outcome was unknown.
In addition, for exploratory analyses, we categorized TB cases into four groups based on time to culture negativity. Individuals who were culture negative within 1 month of treatment initiation were defined as early responders, those who converted to culture-negative by 2 months as medium responders, those who converted to culture-negative at 6 months were slow responders, and treatment failures were those who remained culture positive during the full 6 months of treatment.
Statistical Analysis
Baseline characteristics were compared between TB cases, Xpert-positive/culture-negative, and healthy controls using Chi-square or Fisher's exact test for categorical variables and Kruskal-Wallis test for continuous variable. Correlations between CXR score, bacterial burden, blood counts, neutrophil phenotype, and demographic data were determined using the Mann-Whitney test and Spearman's correlation analysis. Differences in distributions and median values of neutrophil blood counts and CD15 expression were compared between cases and controls and also examined for changes from baseline during the course of TB treatment using the Wilcoxon rank sum test and Kruskal-Wallis test followed by Dunn's multiple comparison test. We graphically evaluated the impact of pre-treatment immune measures on patient response to treatment, defined by early, medium, slow and treatment failure responses. Additionally, we examined the association between baseline immune markers and culture status at month 2 (positive vs. negative) and end of treatment outcome (successful vs. unsuccessful treatment) using univariable and HIV-adjusted logistic regression. Confounders, including age, sex, baseline BMI, and HIV were considered a priori based on clinical expertise. Due to a limited number of outcome events, adjusted models controlled only for HIV co-infection, as it was believed to be most clinically relevant confounder. We also examined the association between the aforementioned exposures with treatment failure/death and with loss to follow-up/not evaluated. The referent group for these analyses was successful outcome (i.e., in analyses with the outcome of failure/death), persons who were lost to follow-up/not evaluated were excluded, and in analysis with the outcome of lost to follow-up/not evaluated, persons with failure/death were excluded. Statistical analysis was performed using StataIC, version 16 (StataCorp, College Station, TX, USA) and GraphPad PRISM, version 8.3.1 (GraphPad Software, San Diego, CA, USA).
Results
The characteristics of the study population are listed in Table 1. Of note, HIV co-infection was 63%, the majority of whom had not started antiretroviral therapy (ART) at baseline (67%). As expected, smoking was significantly higher in the TB subjects (22%) compared to healthy controls (4%). HIV co-infected individuals had significantly lower CXR scores, primarily due to fewer cavities (P = 0.0028); they also had lower bacterial burden than HIV uninfected individuals (median HIV- = 34.54 and HIV+ = 31.88; P = 0.0083) (Figure S2A). Consistent with these results, we observed a weak correlation between CD4 count and CXR score (r = 0.2370; P = 0.0268) and bacterial load (r = 0.2478; P = 0.0143, Figures S2D,E).
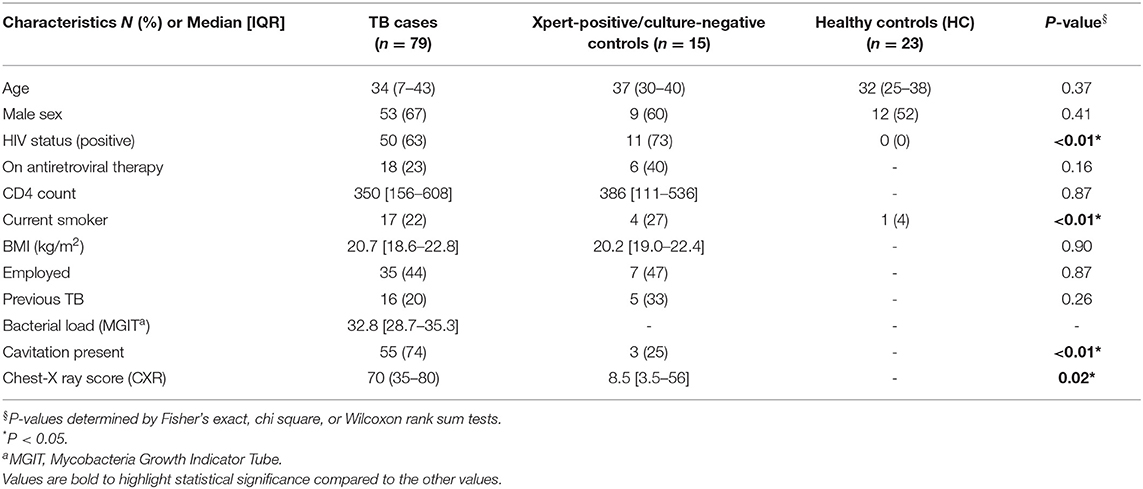
Table 1. Baseline clinical and demographic characteristics of culture positive, drug susceptible TB cases; Xpert-positive/culture-negative controls and healthy controls.
Neutrophil Blood Count and CD15 Expression Level Are Altered at Baseline and Associated With Disease Severity
Full blood counts were measured at baseline and during the course of treatment. Neutrophil count and neutrophil/lymphocyte ratio (NLR) were significantly higher in TB cases at diagnosis compared to healthy controls (HC) (Figures 1A,B). Interestingly, symptomatic Xpert-positive/culture-negative controls also had slightly higher neutrophil counts and NLR compared to healthy controls, but this did not reach statistical significance. Of the surface markers measured in this study, CD15 expression level showed the most consistent difference between TB cases and controls at baseline. As shown in Figure 1C, surface expression of CD15, a canonical neutrophil marker, was found to be highly down regulated at baseline in TB patients compared to healthy controls (P < 0.0001). In addition, TB symptomatic Xpert-positive/culture-negative controls displayed significantly higher CD15 expression levels than active TB cases (P = 0.01).
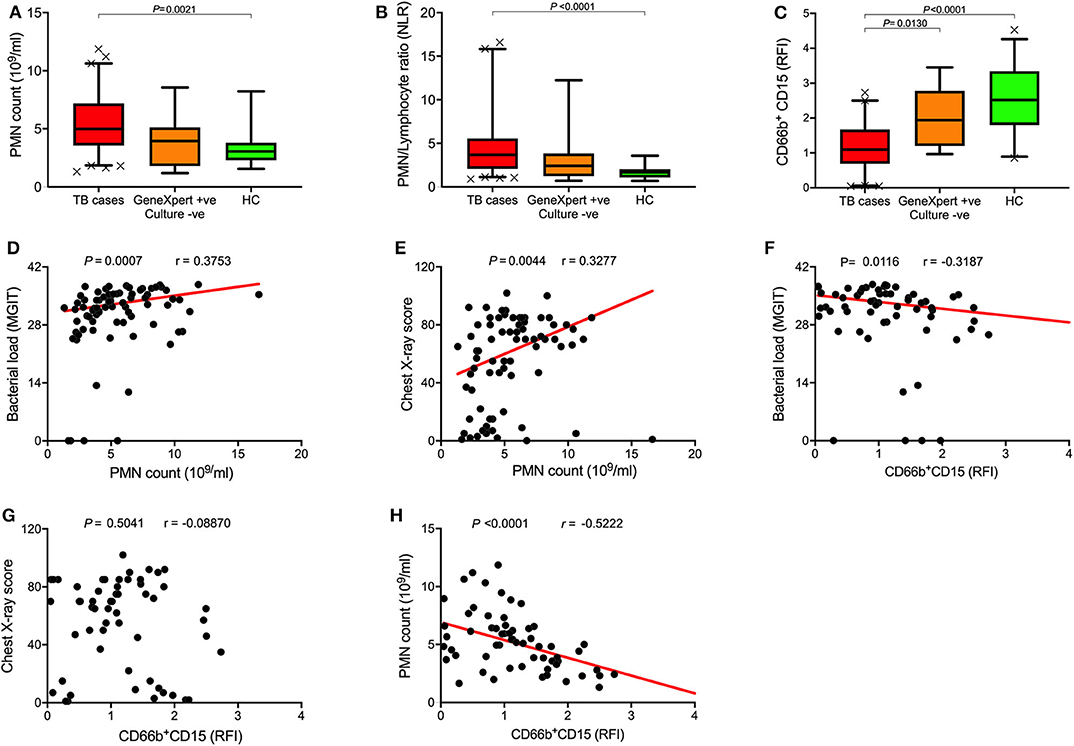
Figure 1. Baseline neutrophil blood counts and phenotypic changes in active TB disease. (A–C) Box plots of (A) baseline neutrophil blood count (PMN count), (B) PMN/Lymphocyte ratios, and (C) Surface CD15 expression levels of TB cases (n = 79), Xpert-positive/culture-negative controls (n = 15), and healthy controls (HC; n = 23), Data represents median values and was analyzed using Kruskal-Wallis test for Dunn's multiple comparisons. (D–H) Spearman's rank correlation analysis of neutrophil blood counts and CD15 expression with bacterial burden and chest X-ray score.
To explore the relationship between baseline neutrophils and disease severity, we performed univariate correlations with sputum bacterial load and CXR score, clinically important measures of TB disease severity. Despite the relatively small sample size, we observed significant correlations between neutrophil count and both bacterial load (r = 0.25, P = 0.034; Figure 1D) and CXR score (r = 0.41, P = 0.0004; Figure 1E), and CD15 expression and bacterial load (r = −0.3187; P = 0.01; Figure 1F). Together these data support the existence of a relationship between the blood neutrophil response to TB and disease pathogenesis in the lung, as suggested by several recent publications (6, 33, 34). Finally, we found CD15 expression level was inversely correlated with neutrophil count (r = −0.5222, P < 0.0001; Figure 1H) but had no correlation with CXR score (Figure 1G). Importantly these measurements were taken independently by our in-house flow cytometry assay and by an accredited commercial laboratory, thereby adding support to the observed phenotypic changes.
Neutrophil Count and CD15 Expression Resolved With TB Treatment
Neutrophil characteristics were examined longitudinally to determine how they respond to TB drug treatment. Baseline neutrophil count and NLR both significantly improved within the 1st month of TB treatment, and reached the level seen in healthy controls by month 6 (Figures 2A,B), irrespective of HIV status (Figures 3A,B). Additionally, surface expression of CD15 increased during TB treatment and expression level was no longer significantly different from healthy controls by 2 months of treatment (Figure 2C and Figure S1). CD15 expression was already elevated by week 1 (for which complete blood counts were not available). This supports our hypothesis that rapid turnover of blood neutrophils makes them extremely sensitive to changes in the lung environment. This is also supported by the fact that, following 3 weeks of drug treatment, neutrophil count and CD15 expression remain correlated with falling sputum bacterial load (r = 0.3929, P = 0.0006 and r = −0.3498, P = 0.0046, respectively; median bacterial load baseline vs. month 1 = 33 vs. 21 P < 0.0001), and each other (r = −0.5751, P < 0.0001) (Figures S4A–C). As with neutrophil count, changes in CD15 expression were not affected by HIV co-infection (Figure S3C). Interestingly, no changes in neutrophil count, NLR and CD15 expression were detected among the Xpert positive/culture negative controls following 2 months of treatment with anti-TB drugs (Figures 2D–F).
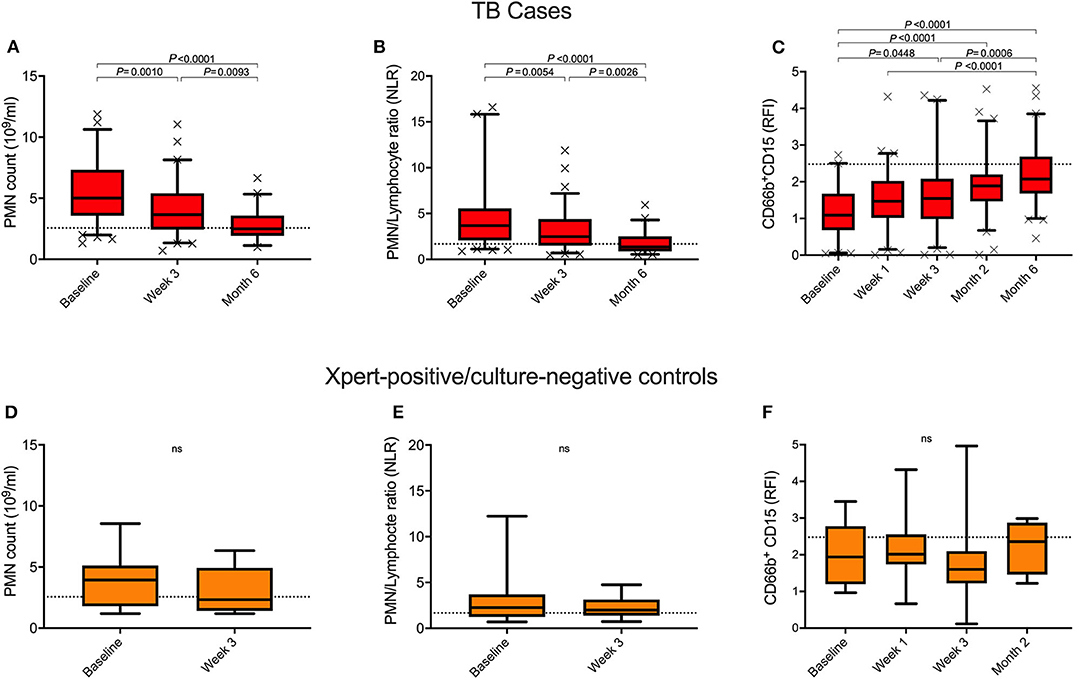
Figure 2. Longitudinal measurement of blood neutrophil count and CD15 expression during anti-TB treatment. Changes in neutrophil blood count (PMN), PMN/Lymphocyte ratio and surface CD15 expression levels over the course of treatment in TB cases (A–C) and Xpert-positive/culture-negative controls (D–F). P-values were obtained using the Wilcoxon rank sum test and Kruskal-Wallis tests for Dunn's multiple comparisons. Dotted line represents the median healthy control levels.
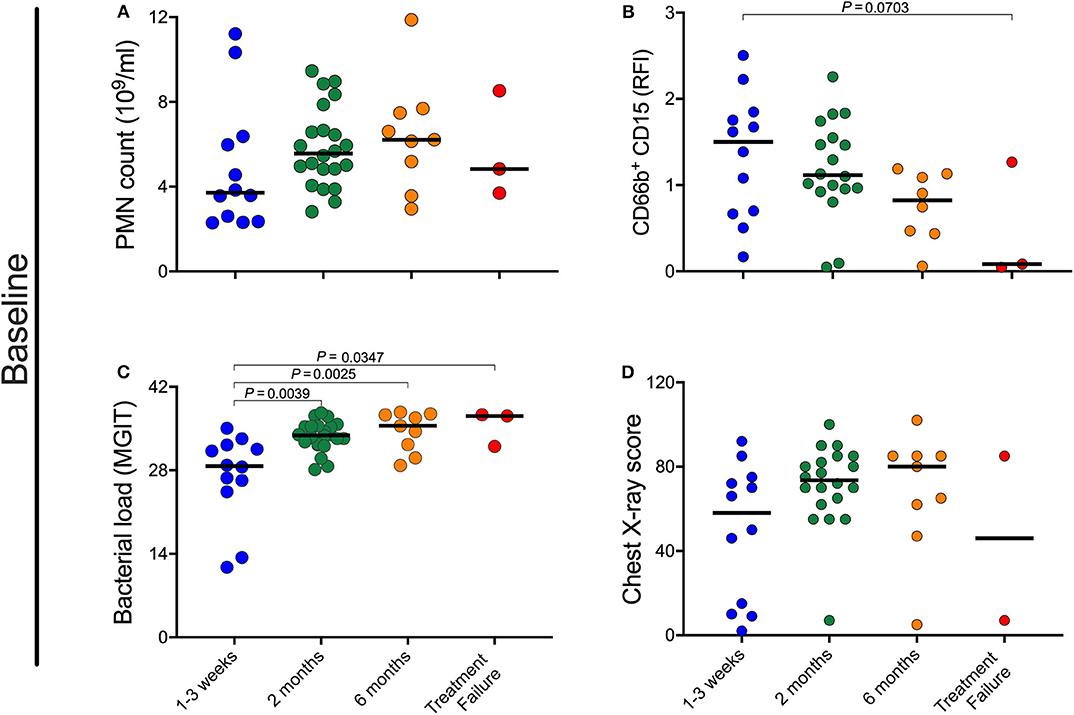
Figure 3. Impact of pre-treatment characteristics on rate of culture conversion. (A–D) Baseline neutrophil blood (PMN) count, CD15 expression, bacterial load, and chest X-ray scores were retrospectively separated into four groups based on time to MGIT culture negativity. Early responders became and remained culture negative within the 1st month, medium responders by 2 months, and slow responders by month 6 post-treatment initiation. In this analysis treatment failure are individuals who were culture positive at all time points. P-values were obtained using the Mann-Whitney test.
Pre-treatment Characteristics Are Associated With Rate of Culture Conversion
Given the correlation between neutrophil characteristics and clinical measures of disease severity at baseline, we sought to determine whether the baseline neutrophil characteristics, bacterial load, or chest X-ray changes correlated with treatment response kinetics. Initially, participants were grouped as being early, medium or slow responders as well as treatment failure, based on achieving and maintaining TB culture negative status at month 1, month 2, month 6, or never, respectively. As shown in Figure 3, there is a trend for individuals with a lower blood neutrophil counts and higher CD15 surface expression at baseline to become culture negative early compared to those who culture converted later or not at all (Figures 3A,B). These differences did not reach statistical significance by univariate analysis. Bacterial load at baseline, however, was significantly associated with treatment response, with early responders having significantly lower bacterial burden than all others (Figure 3C). CXR score, on the other hand, was not predictive of treatment response (Figure 3D).
Next, we examined the association between baseline neutrophil characteristics, CXR score, and bacterial load with sputum culture conversion at 2 months as an endpoint, a clinically relevant measure of TB treatment response (35). For this analysis, we excluded four participants whose baseline culture was positive on liquid media, but not solid media, and 8 participants who were missing culture results at month 2. Among 67 remaining participants with a solid culture at baseline, 7 (10%) remained culture positive after 2 months of anti-TB treatment. Results of this analysis are shown in Table 2. Interestingly, in this reduced dataset, the only baseline characteristic associated with remaining culture positive at month 2 was CD15 expression level (Odds Ratio (OR): 0.17, 95% Confidence Interval (CI): 0.03, 0.97). In other words, for every unit increase in baseline CD15, the odds of remaining culture positive after 2 months of TB treatment was reduced by 83%. After adjusting for HIV-status, the relationship appears slightly stronger (adjusted OR: 0.14, 95% CI: 0.02, 0.89).
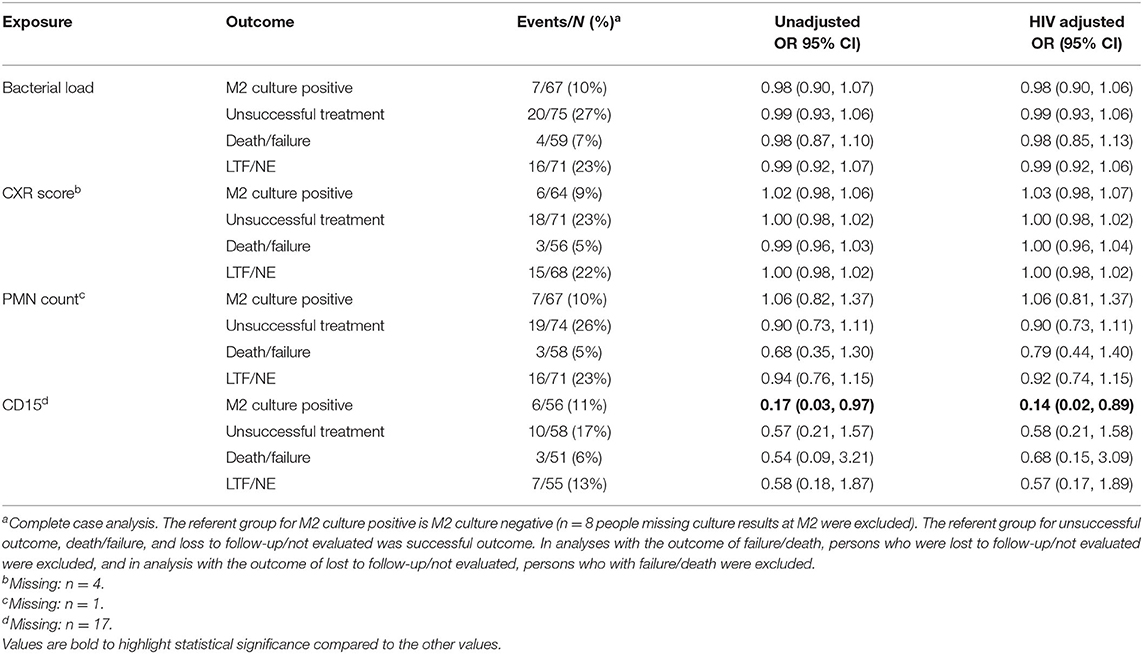
Table 2. Crude and HIV-adjusted effect measures for association between baseline bacterial load, chest x-ray score, neutrophil (PMN) count, and CD15 with TB treatment outcomes (M2 culture conversion, unsuccessful outcome, death/failure, and losses to follow-up/not evaluated), using solid culture.
TB Treatment Outcome
We also examined the association between bacterial load, CXR score, neutrophil count, and CD15 with TB treatment outcome, defined according to the WHO definitions. Among 75 TB cases who were positive for Mtb on solid culture, 53 (71%) were cured, 2 (3%) completed treatment, 2 (3%) failed treatment), 2 died (3%), 8 (11%) were lost to follow-up (LTF), and 8 (11%) were not evaluated (NE). Combining death, failure, loss to follow-up, and not evaluated, the unsuccessful outcome rate was 27%. However, none of baseline bacterial load, CXR score, neutrophil count, or CD15 were associated with TB treatment outcomes (Table 2).
Discussion
In this study we investigated changes in the frequency and phenotype of blood neutrophils associated with active TB at diagnosis and patient response to standard TB drug therapy. Overall, we find that active TB is associated with both an increase in neutrophil count and down-regulation of the canonical neutrophil marker CD15, irrespective of HIV co-infection. In addition, changes in neutrophil count and CD15 expression correlate with clinical markers of disease severity—sputum bacterial load, as measured by time to positivity in liquid culture, and extent of lung involvement, as determined by an independent radiology examination of the CXR. This suggests a potential direct relationship between the blood neutrophil response and the severity of lung disease. As neutrophils are short-lived and rapidly recruited to the site of infection, we hypothesized that any blood neutrophil signature would respond rapidly to clearance of the bacteria. Our data support that hypothesis, as both neutrophil count and CD15 expression level significantly increased by month 2. Moreover, CD15 expression, which was measured more frequently, was higher after only 1 week of drug treatment. The fact that the correlation between neutrophils and bacterial load is maintained after 1 month of treatment, when bacterial load has greatly reduced, also highlights the direct relationship between the blood neutrophil response and the kinetics of the response to drug therapy. Finally, in a multivariate analysis, baseline CD15 expression level was the only variable that predicted sputum culture conversion at 2 months.
The potential role of neutrophils in the immune response to TB is somewhat conflicting and has been well-reviewed elsewhere (36, 37). However, targeting these cells as potential biomarkers for diagnosing active TB disease and monitoring treatment outcomes has been less well-studied (38, 39). Abakay et al. found that an elevated neutrophil count and NLR were both associated with advanced TB disease; with increased NLR, in particular, associated with the most severe lung involvement (40). Disease severity was estimated from CXR and thus our findings are comparable, although in our study, NLR was less informative than neutrophil count alone or CD15 expression level. Furthermore, our data are consistent with clinical studies and experimental infections showing that acute pulmonary tuberculosis is accompanied by an influx of neutrophils (26, 41). In mice, for example, neutrophil influx is associated with mycobacterial burden and lung pathology, and their depletion in chronic infection leads to decreased CFU and improved survival (26, 42–44). It is important to note, however, that the influx of neutrophils is not unique to TB disease and has been well-described in other inflammatory conditions such as asthma, influenza and bacterial or viral pneumonia (45–47). Therefore, neutrophil characteristics alone would not be appropriate as a diagnostic test. None the less, studies have shown the usefulness of neutrophil phenotyping in distinguishing bacterial infection from other inflammatory conditions (48, 49). Therefore, together with other diagnostic tests, neutrophil monitoring could be valuable as a rule our test and for identifying patients at base line with the most severe disease. Such metrics could prove useful for testing treatment shortening interventions, for example.
Unlike other immune subsets, it is not possible to freeze neutrophils for future analysis. Consequently, few studies have examined neutrophil phenotypic changes in human TB cohorts. The phenotyping of blood neutrophils by flow cytometry in this study was undertaken with the aim of potentially identifying sensitive markers of the neutrophil response. Initially, this included eight neutrophil surface markers based on preliminary data from our lab showing differential expression between healthy individuals and TB cases. However, in this longitudinal study, only CD15 expression showed a consistent response during the course of TB treatment. CD15 is a carbohydrate antigen associated with cell surface of glycoproteins or glycolipids, including CD11b/CD18 (Mac-1) and CR3 β-chain (50, 51). It is highly expressed by mature neutrophils (52) and has been shown to affect a number of neutrophil functions including adhesion to endothelium, phagocytosis, degranulation, and the respiratory burst (50, 53–55). However, there are very few studies that have examined CD15 expression level ex vivo. CD15 expression is elevated on low density neutrophils (56), which can arise from activation both in vitro and in vivo (57). Conversely, expression of CD15 was found to dramatically decreases in asthmatic patients following acute allergen challenge (58), suggesting that activation per se does not elevate CD15 in general. Interestingly, treatment of healthy donors with Granulocyte-Colony Stimulating Factor (G-CSF) to induce neutrophil release from the bone marrow (59, 60), also leads to decreased CD15 expression on blood neutrophils (61). Therefore, both the elevated blood neutrophil numbers and low CD15 expression observed in active TB may relate to sustained recruitment of neutrophils from the bone marrow in response to cytokine release from Mtb infection. As neutrophils mature in the bone marrow they upregulate CD15 (62, 63), implying that the CD15 low neutrophils we observed in blood are not fully mature. In addition, when G-CSF treatment was ceased in the above studies, CD15 expression level in returned to normal, consistent with the rapid response kinetics observed in our study. Whether the influx of CD15 low neutrophils observed in TB directly relates to G-CSF, alone or in combination with other neutrophil related cytokines, is an interesting area for future study and might lead to alternative and potentially more sensitive measures of on-going lung involvement.
In the analysis we included individuals who were TB symptomatic and GeneXpert positive at diagnosis, but turned out to be culture negative. Discordant gene-expert and culture results have previously been linked, at least in part, to residual DNA in the lung from killed bacteria (64), poor sputum quality and low bacterial load as determined by CT cycle number (65). One third of these subjects reported previous TB episodes, slightly higher than the culture positive group (20%), but detail on sputum quality and GeneXpert quantitation data was not recorded. Nonetheless, the neutrophil data on this group were interesting and potentially informative. First, neutrophil counts and CD15 expression in this group were not significantly higher than healthy controls, and, second, neither neutrophil count nor CD15 expression level were changed by 2 months of TB treatment. This suggests that monitoring blood neutrophils during TB screening could help to inform classification and management of discordant subjects. In addition, to identify novel prognostic factors, we sought to determine whether neutrophil characteristics at TB diagnosis were predictive of response to treatment. We found that baseline CD15 expression level was predictive of culture positivity at month 2, suggesting potential value in helping triage subjects into those likely to respond well to treatment. In the univariate analysis, sputum bacterial load also appeared to distinguish subjects who became culture negative rapidly compared to those with a slower response to treatment, consistent with other studies (66, 67). However, neutrophil phenotype can be determined much faster than sputum bacterial load, and thus could be more relevant in the clinical setting.
There were some limitations of this study. First, sample size was relatively small. However, it was sufficiently large to detect significant correlations between neutrophil count, CD15 expression level, and TB treatment outcomes. Second, study participants were from one city in South Africa, and may not be generalizable to other settings. However, there was a high proportion of HIV-positive participants, and we were able to demonstrate that findings did not differ according to HIV status. Further work is planned to further explore the potential utility of neutrophil signatures in larger cohorts and with additional measures of neutrophil activity and neutrophil related cytokines.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Biomedical Research Ethics Committee of the University of Kwa-Zulu Natal. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
AL and LN conceived, designed the experiments, and wrote the paper. AP, TS, FK, and M-YM designed and managed the cohort. FK and MM enrolled patients and collected samples and data. KK and SM supervised laboratory collection of clinical samples and analysis. LN, SM, SN, and AN performed the experiments. TS, YH, FM, and LP made intellectual contributions. All authors contributed to the article and approved the submitted version.
Funding
This work was jointly sponsored by the South African National Science Foundation and the National Institute for Allergy and Infectious Diseases, National Institutes of Health, and administered by CRDF Global (OISE-16-62060-1).
Funding was also sourced from the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative [DEL-15-006]. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)'s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa's Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust [107752/Z/15/Z] and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust, or the UK government. This publication was also supported by CTSA Award No. TL1TR000447 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01872/full#supplementary-material
References
1. World Health Organisation. Tuberculosis. (2019). Available online at: https://www.who.int/news-room/fact-sheets/detail/tuberculosis (accessed March 23, 2020).
2. Centis R, D'Ambrosio L, Zumla A, Migliori GB. Shifting from tuberculosis control to elimination: where are we? What are the variables and limitations? Is it achievable? Int J Infect Dis. (2017) 56:30–3. doi: 10.1016/j.ijid.2016.11.416
3. Walzl G, Ronacher K, Hanekom W, Scriba TJ, Zumla A. Immunological biomarkers of tuberculosis. Nat Rev Immunol. (2011) 11:343–54. doi: 10.1038/nri2960
4. Naidoo P, Peltzer K, Louw J, Matseke G, McHunu G, Tutshana B. Predictors of tuberculosis (TB) and antiretroviral (ARV) medication non-adherence in public primary care patients in South Africa: a cross sectional study. BMC Public Health. (2013) 13:396. doi: 10.1186/1471-2458-13-396
5. Walzl G, McNerney R, du Plessis N, Bates M, McHugh TD, Chegou NN, et al. Tuberculosis: advances and challenges in development of new diagnostics and biomarkers. Lancet Infect Dis. (2018) 18:e199–210. doi: 10.1016/S1473-3099(18)30111-7
6. Friedland J, Ong C, Elkington P, Dos Santos Brilha S, Ugarte-Gill C, Sathyamoorthy T, et al. Neutrophil-derived MMP-8 drives AMPK-dependant matrix destruction in human pulmonary tuberculosis. PLoS Pathog. (2015) 11:e1004917. doi: 10.1371/journal.ppat.1004917
7. Johnson JL, Hadad DJ, Dietze R, Noia Maciel EL, Sewali B, Gitta P, et al. Shortening treatment in adults with noncavitary tuberculosis and 2-month culture conversion. Am J Respir Crit Care Med. (2009) 180:558–63. doi: 10.1164/rccm.200904-0536OC
8. Davies P, Pai M. The diagnosis and misdiagnosis of tuberculosis. Int J Tuberc Lung Dis. (2008) 12:1226–34.
9. Wallis RS, Perkins MD, Phillips M, Joloba M, Namale A, Johnson JL, et al. Predicting the outcome of therapy for pulmonary tuberculosis. Am J Respir Crit Care Med. (2000) 161:1076–80. doi: 10.1164/ajrccm.161.4.9903087
10. Elliott AM, Namaambo K, Allen BW, Luo N, Hayes RJ, Pobee JO, et al. Negative sputum smear results in HIV-positive patients with pulmonary tuberculosis in Lusaka, Zambia. Tuberc Lung Dis. (1993) 74:191–4. doi: 10.1016/0962-8479(93)90010-U
11. Zar HJ, Tannenbaum E, Apolles P, Roux P, Hanslo D, Hussey G. Sputum induction for the diagnosis of pulmonary tuberculosis in infants and young children in an urban setting in South Africa. Arch Dis Child. (2000) 82:305–8. doi: 10.1136/adc.82.4.305
12. Maddineni M, Panda M. Pulmonary tuberculosis in a young pregnant female: challenges in diagnosis and management. Infect Dis Obstet Gynecol. (2008) 2008:628985. doi: 10.1155/2008/628985
13. Adekambi T, Ibegbu CC, Cagle S, Kalokhe AS, Wang YF, Hu Y, et al. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J Clin Invest. (2015) 125:1827–38. doi: 10.1172/JCI77990
14. Wilkinson KA, Oni T, Gideon HP, Goliath R, Wilkinson RJ, Riou C. Activation profile of mycobacterium tuberculosis–specific CD4+ T cells reflects disease activity irrespective of HIV status. Am J Respir Crit Care Med. (2016) 193:1307–10. doi: 10.1164/rccm.201601-0116LE
15. Rozot V, Patrizia A, Vigano S, Mazza-Stalder J, Idrizi E, Day CL, et al. Combined use of mycobacterium tuberculosis–specific CD4 and CD8 T-cell responses is a powerful diagnostic tool of active tuberculosis. Clin Infect Dis. (2015) 60:432–7. doi: 10.1093/cid/ciu795
16. Dewan PK, Grinsdale J, Kawamura LM. Low sensitivity of a whole-blood interferon-γ release assay for detection of active tuberculosis. Clin Infect Dis. (2007) 44:69–73. doi: 10.1086/509928
17. Cattamanchi A, Smith R, Steingart KR, Metcalfe JZ, Date A, Coleman C, et al. Interferon-gamma release assays for the diagnosis of latent tuberculosis infection in HIV-infected individuals – a systematic review and meta-analysis. J Acquired Immune Defic Syndr. (2011) 56:230–8. doi: 10.1097/QAI.0b013e31820b07ab
18. Eruslanov EB, Lyadova IV, Kondratieva TK, Majorov KB, Scheglov IV, Orlova MO, et al. Neutrophil responses to Mycobacterium tuberculosis infection in genetically susceptible and resistant mice. Infect Immun. (2005) 73:1744–53. doi: 10.1128/IAI.73.3.1744-1753.2005
19. Laskay T, van Zandbergen G, Solbach W. Neutrophil granulocytes as host cells and transport vehicles for intracellular pathogens: apoptosis as infection-promoting factor. Immunobiology. (2008) 213:183–91. doi: 10.1016/j.imbio.2007.11.010
20. Korbel DS, Schneider BE, Schaible UE. Innate immunity in tuberculosis: myths and truth. Microb Infect. (2008) 10:995–1004. doi: 10.1016/j.micinf.2008.07.039
21. Summers C, Rankin SM, Condliffe AM, Singh N, Peters AM, Chilvers ER. Neutrophil kinetics in health and disease. Trends Immunol. 31:318–24. doi: 10.1016/j.it.2010.05.006
22. Seiler P, Aichele P, Raupach B, Odermatt B, Steinhoff U, Kaufmann SH. Rapid neutrophil response controls fast-replicating intracellular bacteria but not slow-replicating mycobacterium tuberculosis. J Infect Dis. (2000) 181:671–80. doi: 10.1086/315278
23. Alemán M, Schierloh P, de la Barrera SS, Musella RM, Saab MA, Baldini M, et al. Mycobacterium tuberculosis triggers apoptosis in peripheral neutrophils involving toll-like receptor 2 and p38 mitogen protein kinase in tuberculosis patients. Infect Immun. (2004) 72:5150–8. doi: 10.1128/IAI.72.9.5150-5158.2004
24. Pedrosa J, Saunders BM, Appelberg R, Orme IM, Silva MT, Cooper AM. Neutrophils play a protective nonphagocytic role in systemic mycobacterium tuberculosis infection of mice. Infect Immun. (2000) 68:577–83. doi: 10.1128/IAI.68.2.577-583.2000
25. Martineau AR, Newton SM, Wilkinson KA, Kampmann B, Hall BM, Nawroly N, et al. Neutrophil-mediated innate immune resistance to mycobacteria. J Clin Invest. (2007) 117:1988–94. doi: 10.1172/JCI31097
26. Yeremeev V, Linge I, Kondratieva T, Apt A. Neutrophils exacerbate tuberculosis infection in genetically susceptible mice. Tuberculosis. (2015) 95:447–51. doi: 10.1016/j.tube.2015.03.007
27. Nusbaum RJ, Calderon VE, Huante MB, Sutjita P, Vijayakumar S, Lancaster KL, et al. Pulmonary tuberculosis in humanized mice infected with HIV-1. Sci Rep. (2016) 6:21522. doi: 10.1038/srep21522
28. Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. (2010) 466:973–7. doi: 10.1038/nature09247
29. Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS One. (2013) 8:8. doi: 10.1371/annotation/7d9ec449-aee0-48fe-8111-0c110850c0c1
30. Ralph AP, Ardian M, Wiguna A, Maguire GP, Becker NG, Drogumuller G, et al. A simple, valid, numerical score for grading chest x-ray severity in adult smear-positive pulmonary tuberculosis. Thorax. (2010) 65:863–9. doi: 10.1136/thx.2010.136242
31. Upreti D, Pathak A, Kung SK. Development of a standardized flow cytometric method to conduct longitudinal analyses of intracellular CD3ζ expression in patients with head and neck cancer. Oncol Lett. (2016) 11:2199–206. doi: 10.3892/ol.2016.4209
32. World Health Organization. Definitions and Reporting Framework for Tuberculosis-2013 Revision. (2013). Available online at: https://www.who.int/tb/publications/definitions/en/ (accessed March 20, 2020).
33. Eum S-Y, Kong J-H, Hong M-S, Lee Y-J, Kim J-H, Barry CE, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. (2010) 137:122–8. doi: 10.1378/chest.09-0903
34. Panteleev AV, Nikitina IY, Burmistrova IA, Kosmiadi GA, Radaeva TV, Amansahedov RB, et al. Severe tuberculosis in humans correlates best with neutrophil abundance and lymphocyte deficiency and does not correlate with antigen-specific CD4 T-cell response. Front Immunol. (2017) 8:963. doi: 10.3389/fimmu.2017.00963
35. Rockwood N., du Bruyn E, Morris T, Wilkinson RJ. Assessment of treatment response in tuberculosis. Expert Rev Respir Med. (2016) 10: 643–54. doi: 10.1586/17476348.2016.1166960
36. Lowe DM, Redford PS, Wilkinson RJ, O'Garra A, Martineau AR. Neutrophils in tuberculosis: friend or foe? Trends Immunol. (2012) 33:14–25. doi: 10.1016/j.it.2011.10.003
37. Lyadova IV. Neutrophils in tuberculosis: heterogeneity shapes the way? Mediators Inflamm. (2017) 2017:1–11. doi: 10.1155/2017/8619307
38. Brahmbhatt S, Black GF, Carroll NM, Beyers N, Salker F, Kidd M, et al. Immune markers measured before treatment predict outcome of intensive phase tuberculosis therapy. Clin Exp Immunol. (2006) 146:243–52. doi: 10.1111/j.1365-2249.2006.03211.x
39. Yoon N-B, Son C, Um S-J. Role of the neutrophil-lymphocyte count ratio in the differential diagnosis between pulmonary tuberculosis and bacterial community-acquired pneumonia. Ann Lab Med. (2013) 33:105–10. doi: 10.3343/alm.2013.33.2.105
40. Abakay O., Abakay A, Sen HS, Tanrikulu AC. The relationship between inflammatory marker levels and pulmonary tuberculosis severity. Inflammation. (2015) 38: 691–6. doi: 10.1007/s10753-014-9978-y
41. Kerkhoff AD, Wood R, Lowe DM, Vogt M, Lawn SD. Blood neutrophil counts in HIV-infected patients with pulmonary tuberculosis: association with sputum mycobacterial load. PLoS ONE. (2013) 8:e67956. doi: 10.1371/journal.pone.0067956
42. Marzo E. Vilaplana C, Tapia G, Diaz J, Garcia V, Cardona P-J. Damaging role of neutrophilic infiltration in a mouse model of progressive tuberculosis. Tuberculosis. (2014) 94:55–64. doi: 10.1016/j.tube.2013.09.004
43. Gopal R, Monin L, Torres D, Slight S, Mehra S, McKenna KC, et al. S100A8/A9 proteins mediate neutrophilic inflammation and lung pathology during tuberculosis. Am J Respir Crit Care Med. (2013) 188:1137–46. doi: 10.1164/rccm.201304-0803OC
44. Almeida FM, Ventura TLB, Amaral EP, Ribeiro SCM, Calixto SD, Manhães MR, et al. Hypervirulent Mycobacterium tuberculosis strain triggers necrotic lung pathology associated with enhanced recruitment of neutrophils in resistant C57BL/6 mice. PLoS ONE. (2017) 12:e0173715. doi: 10.1371/journal.pone.0173715
45. Ciepiela O, Ostafin M, Demkow U. Neutrophils in asthma—a review. Respir Physiol Neurobiol. (2015) 209:13–6. doi: 10.1016/j.resp.2014.12.004
46. Pechous RD. With friends like these: the complex role of neutrophils in the progression of severe pneumonia. Front Cell Infect Microbiol. (2017) 7:160. doi: 10.3389/fcimb.2017.00160
47. Camp JV, Jonsson CB. A role for neutrophils in viral respiratory disease. Front Immunol. (2017) 8:550. doi: 10.3389/fimmu.2017.00550
48. Jalava-Karvinen P, Hohenthal U, Laitinen I, Kotilainen P, Rajamäki A, Nikoskelainen J, et al. Simultaneous quantitative analysis of FcγRI (CD64) and CR1 (CD35) on neutrophils in distinguishing between bacterial infections, viral infections, inflammatory diseases. Clin Immunol. (2009) 133:314–323. doi: 10.1016/j.clim.2009.08.003
49. Allen E, Bakke AC, Purtzer MZ, Deodhar A. Neutrophil CD64 expression: distinguishing acute inflammatory autoimmune disease from systemic infections. Ann Rheum Dis. (2002) 61:522–5. doi: 10.1136/ard.61.6.522
50. Stocks SC, Albrechtsen M, Kerr MA. Expression of the CD15 differentiation antigen (3-fucosyl-N-acetyl-lactosamine, LeX) on putative neutrophil adhesion molecules CR3 and NCA-160. Biochem J. (1990) 268:275–80. doi: 10.1042/bj2680275
51. van Gisbergen KP., Sanchez-Hernandez M, Geijtenbeek TB, van Kooyk Y. Neutrophils mediate immune modulation of dendritic cells through glycosylation-dependent interactions between Mac-1 and DC-SIGN. J Exp Med. (2005) 201:1281–92. doi: 10.1084/jem.20041276
52. Nakayama F, Nishihara S, Iwasaki H, Kudo T, Okubo R, Kaneko M, et al. CD15 expression in mature granulocytes is determined by α1, 3-fucosyltransferase IX, but in promyelocytes and monocytes by α1, 3-fucosyltransferase IV. J Biol Chem. (2001) 276:16100–6. doi: 10.1074/jbc.M007272200
53. Melnick DA, Nauseef WM, Markowitz SD, Gardner JP, Malech HL. Biochemical analysis and subcellular localization of a neutrophil-specific antigen, PMN-7, involved in the respiratory burst. J Immunol. (1985) 134:3346–55.
54. Melnick DA, Meshulam T, Manto A, Malech HL. Activation of human neutrophils by monoclonal antibody PMN7C3: cell movement and adhesion can be triggered independently from the respiratory burst. Blood. (1986) 67:1388–94. doi: 10.1182/blood.V67.5.1388.bloodjournal6751388
55. Skubitz KM, Snook RW. Monoclonal antibodies that recognize lacto-N-fucopentaose III (CD15) react with the adhesion-promoting glycoprotein family (LFA-1/HMac-1/gp 150, 95) and CR1 on human neutrophils. J Immunol. (1987) 139:1631–9.
56. Ssemaganda A, Kindinger L, Bergin P, Nielsen L, Mpendo J, Ssetaala A, et al. Characterization of neutrophil subsets in healthy human pregnancies. PLoS ONE. (2014) 9:e85696. doi: 10.1371/journal.pone.0085696
57. Hassani M, Hellebrekers P, Chen N, van Aalst C, Bongers S, Hietbrink F, et al. On the origin of low-density neutrophils. J Leukoc Biol. (2020) 107:809–18. doi: 10.1002/JLB.5HR0120-459R
58. Pelikan Z. Expression of surface markers on the blood cells during the delayed asthmatic response to allergen challenge. Allergy Rhinol. (2014) 5:96–109. doi: 10.2500/ar.2014.5.0087
59. Semerad CL, Liu F, Gregory AD, Stumpf K, Link DC. G-CSF is an essential regulator of neutrophil trafficking from the bone marrow to the blood. Immunity. (2002) 17:413–23. doi: 10.1016/S1074-7613(02)00424-7
60. Dale DC. Colony-stimulating factors for the management of neutropenia in cancer patients. Drugs. (2002) 62:1–15. doi: 10.2165/00003495-200262001-00001
61. Zarco MA, Ribera JM, Urbano-Ispizua A, Filella X, Arriols R, Martínez C, et al. Phenotypic changes in neutrophil granulocytes of healthy donors after G-CSF administration. Haematologica. (1999) 84:874–8.
62. Terstappen L, Safford M, Loken MR. Flow cytometric analysis of human bone marrow. III. Neutrophil maturation. Leukemia. (1990) 4:657–63.
63. Elghetany MT. Surface antigen changes during normal neutrophilic development: a critical review. Blood Cells Mol Dis. (2002) 28:260–74. doi: 10.1006/bcmd.2002.0513
64. Theron G, Venter R, Smith L, Esmail A, Randall P, Sood V, et al. False-positive Xpert MTB/RIF results in retested patients with previous tuberculosis: frequency, profile, and prospective clinical outcomes. J Clin Microbiol. (2018) 56:e01696–17. doi: 10.1128/JCM.01696-17
65. Shi J, Dong W, Ma Y, Liang Q, Shang Y, Wang F, et al. GeneXpert MTB/RIF outperforms Mycobacterial culture in detecting Mycobacterium tuberculosis. from salivary sputum. BioMed Res Int. (2018) 2018:1514381. doi: 10.1155/2018/1514381
66. Visser ME, Stead MC, Walzl G, Warren R, Schomaker M, Grewal HM, et al. Baseline predictors of sputum culture conversion in pulmonary tuberculosis: importance of cavities, smoking, time to detection and W-Beijing genotype. PLoS ONE. (2012) 7:e29588. doi: 10.1371/journal.pone.0029588
Keywords: tuberculosis biomarker, neutrophil phenotype, blood neutrophil count, tuberculosis treatment outcome, tuberculosis disease severity
Citation: Ndlovu LN, Peetluk L, Moodley S, Nhamoyebonde S, Ngoepe AT, Mazibuko M, Khan K, Karim F, Pym AS, Maruri F, Moosa M-YS, van der Heijden YF, Sterling TR and Leslie A (2020) Increased Neutrophil Count and Decreased Neutrophil CD15 Expression Correlate With TB Disease Severity and Treatment Response Irrespective of HIV Co-infection. Front. Immunol. 11:1872. doi: 10.3389/fimmu.2020.01872
Received: 10 May 2020; Accepted: 13 July 2020;
Published: 28 August 2020.
Edited by:
Abhay Satoskar, The Ohio State University, United StatesReviewed by:
Carmen Judith Serrano, Mexican Social Security Institute (IMSS), MexicoShashank Gupta, National Institutes of Health, United States
Copyright © 2020 Ndlovu, Peetluk, Moodley, Nhamoyebonde, Ngoepe, Mazibuko, Khan, Karim, Pym, Maruri, Moosa, van der Heijden, Sterling and Leslie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alasdair Leslie, YWwubGVzbGllQGFocmkub3Jn
 Lerato N. Ndlovu
Lerato N. Ndlovu Lauren Peetluk
Lauren Peetluk Sashen Moodley1
Sashen Moodley1 Abigail T. Ngoepe
Abigail T. Ngoepe Khadija Khan
Khadija Khan Fernanda Maruri
Fernanda Maruri Yuri F. van der Heijden
Yuri F. van der Heijden Alasdair Leslie
Alasdair Leslie