- 1Department of Neurosurgery, Xiangya Hospital, Central South University, Changsha, China
- 2National Clinical Research Center for Geriatric Disorders, Changsha, China
- 3Department of Neurosurgery, West China Hospital, Sichuan University, Chengdu, China
- 4Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha, China
The pentraxin is a superfamily of proteins with the same domain known as the pentraxin domain at C-terminal. This family has two subgroups, namely; short pentraxins (C-reactive protein and serum amyloid P component) and long pentraxins (neuronal pentraxin 1, neuronal pentraxin 2, neuronal pentraxin receptor, pentraxin 3 and pentraxin 4). Each group shares a similar structure with the pentameric complexes arranged in a discoid shape. Previous studies revealed the functions of different pentraxin family members. Most of them are associated with human innate immunity. Inflammation has commonly been associated with tumor progression, implying that the pentraxin family might also participate in tumor progression. Therefore, we reviewed the basic characteristics and functions of the pentraxin family and their role in tumor progression.
Introduction
The pentraxin family is a superfamily of protein that share the same domain and are made from monomers arranged in pentameric structures with a discoid shape (1). The members of the family are characterized by a 205 amino acids (AA) long conserved sequence located at C-terminal called the pentraxin domain. Members of the pentraxin family share a similar 8 AA (His-x-Cys-x-Ser/Thr-Trp-x-Ser, in which x represent any AA) long conserved sequence called the pentraxin signature within the pentraxin domain (2). Based on the length of the protein sequence, the pentraxin family can be classified into two subfamilies: the short and long pentraxins. The short pentraxins are comprised of C-reactive protein (CRP) and serum amyloid P component (SAP), whereas the long pentraxins are composed of neuronal pentraxin 1(NPTX1), neuronal pentraxin 2 (NPTX2), neuronal pentraxin receptor (NPTXR), pentraxin 3 (PTX3), and pentraxin 4 (PTX4) (3). The long pentraxins are approximately twice the size of the short pentraxins with an un-related long N-terminal sequence. This structure variability of family members could explain their function difference.
In the past decades, studies have revealed the functions of specific members of the pentraxin family. For example, the member of the short pentraxin family, CRP, and SAP were previously reported as mediators in human immune system regulation (4–6). Their functions in immune regulation include acting against pathogen invasion, removing mutant cells, and triggering inflammation. The neuronal pentraxins are involved in the development of the central nervous system and neurodegenerative diseases (7). PTX3 not only participates in immune system activation but also affects tumor progression (8, 9).
Chronic inflammations such as chronic atrophic gastritis and cervical intraepithelial neoplasia have been recognized as precancerous lesions (10). Therefore, inflammation is closely associated with tumor progression, including tumorigenesis, metastasis. Reactive oxygen/nitrogen species (ROS/RNS) produced by immune cells and epithelial cells fight against microbial invasion and eliminate the mutant cell. However, they can cause cell dysfunction and promote tumorigenesis (11, 12). Tumors induce inflammation by either producing antibodies or rejecting immunocytes infiltration to avoid immune system surveillance (13). Additionally, the tumor microenvironment facilitates the growth of cancer cells, metastasis, and enhances drug resistance (14). It is necessary to explore the mechanisms through which inflammation and tumor microenvironment enhances tumor progression.
Several reports have also been documented on the role of the pentraxin family in tumor progression. Likewise, several receptors and pathways have been proposed that could be associated with the mechanisms employed by the pentraxin family in mediating tumor progression. Most members of the pentraxin family can activate the PI3K/AKT/mTOR pathways, thereby interfering with the normal cell- cycle. The neuronal pentraxins and PTX3 possess specific unique receptors that mediates tumor progression (15). In this review, we have summarized the basic characteristics and functions of each pentraxin family member and highlighted the existing connection between their structures and specific roles in tumor progression.
C-Reactive Protein
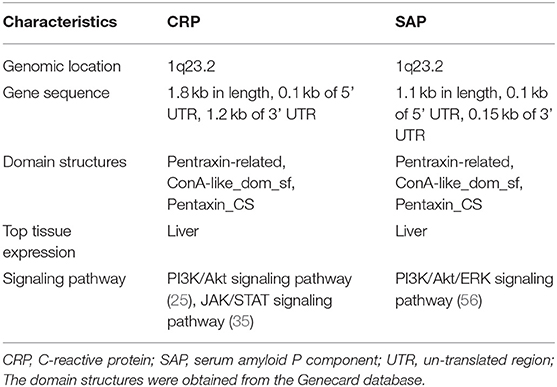
The human C-reactive protein (CRP) gene is located on the chromosome 1q23.2 (16), and has a length of 1.8-kb. It consists of 0.1 kb at untranslated region (UTR) in the 5′ terminal and a 1.2 kb pair UTR region in the 3′ terminal, with two exons separated by an intron. The first exon encodes 18 AA signal peptide and the first two amino acids, whereas, the second exon encodes the rest AA (16). The X-ray derived structures of CPR are pentameric with five subunits arranged in a discoid shape. Each unit contains 206 AA with two anti-parallel b-sheets appearing as a flattened b-barrel with a jellyroll topology. The two sides of its discoid have distinct functions. The Ca2+ binds to the “A” side and activates the classical complement pathway and phagocytosis by interacting with C1q and Fcγ receptor, respectively (17, 18). The “B” side, CRP recognizes phosphocholine (PCh), a bacterial cell wall component, and eliminates the pathogen (19). The CRP also binds to soluble control protein factor H regulating the alternative-pathway amplification and C3 convertase (18). The secretion of CRP by hepatocytes can be stimulated by the IL6 and IL1 (20), which enhances innate immunity by triggering inflammation and neutralizing pathogen (18, 21).
CRP and Tumor
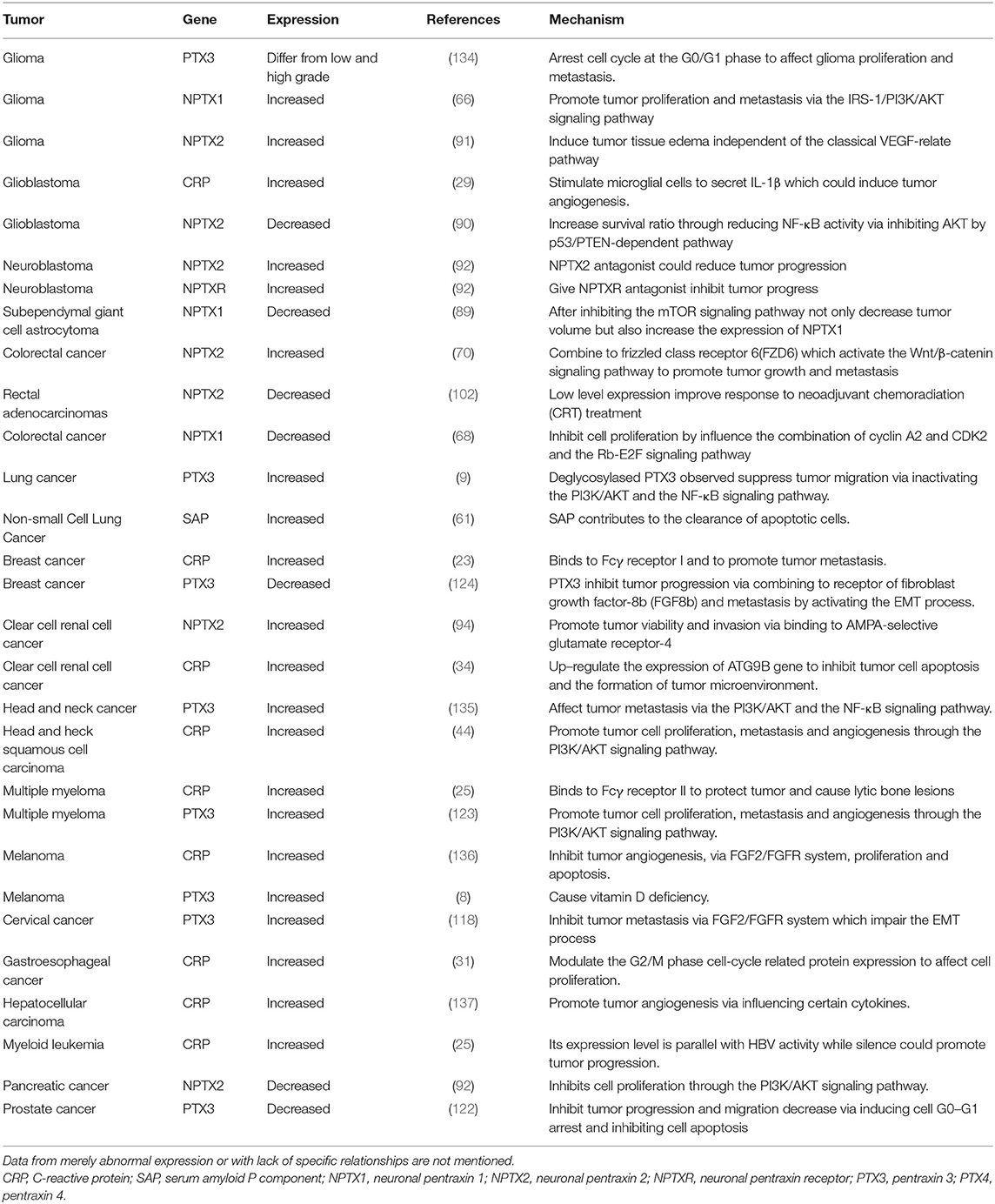
CRP activates various signaling pathways by binding to the Fcγ receptor (22, 23), which links it with inflammation (17). The PI3K/AKT/mTOR signaling pathway is associated with tumor cell proliferation, metabolic reprogramming, apoptosis, and metastasis (24). For instance, the CRP arrest cell-cycle at the sub G1 phase by negatively regulating the PI3K/AKT/mTOR signaling pathway in myeloid leukemia (25) and tongue squamous cell carcinoma (26) thus promoting tumor progression. It has been reported that interaction between CRP and Fcγ receptor I facilitate tumor cell metastasis in breast cancer (23). Furthermore, CRP targets to the p38/MAPK pathway causing lytic bone lesions (27) and activates the PI3K/AKT/mTOR and the ERK/NF-κB pathway thereby inhibiting tumor cell apoptosis via the Fcγ receptor II in multiple myeloma (22).
Previous reports showed that CRP is a clinical marker for infection and has a regulatory role in innate immunity (17, 28). The IL-6/JAK/STAT signaling pathway has been reported to enhance CRP expression in glioblastoma (29), clear cell renal cell carcinoma (30) and gastroesophageal cancers (31). Moreover, the pathway enhances the formation of CRP-mediated tumor microenvironment by activating tumor-associated macrophages (30) and tumor angiogenesis (32, 33). CRP regulates cell apoptosis and cell-cycle in clear cell renal cell cancer (34). Notably, inhibiting CRP expression by targeting IL-1 can prolong overall survival time for patients with multiple myeloma (35).
Multiple studies have, therefore, confirmed that CRP could be used as a prognosis factor (36–38). This has been reported in different tumors including breast cancer (39), prostate cancer (40), non-small cell lung cancer (41), hepatocellular carcinoma (42), cervical cancer (43), head and neck squamous cell carcinoma (44), diffuse large B-cell lymphoma (45) and osteosarcoma (46). The specific mechanisms involved are still lacking. Controversies have also emerged in determining the role of CRP in pancreatic cancer (47, 48) and colorectal cancer (49–51). Considering that CRP also affects tumor progression, more effort in explicating its role in the tumor might be beneficial in understanding the connection between tumor and human innate immunity (Figure 1).
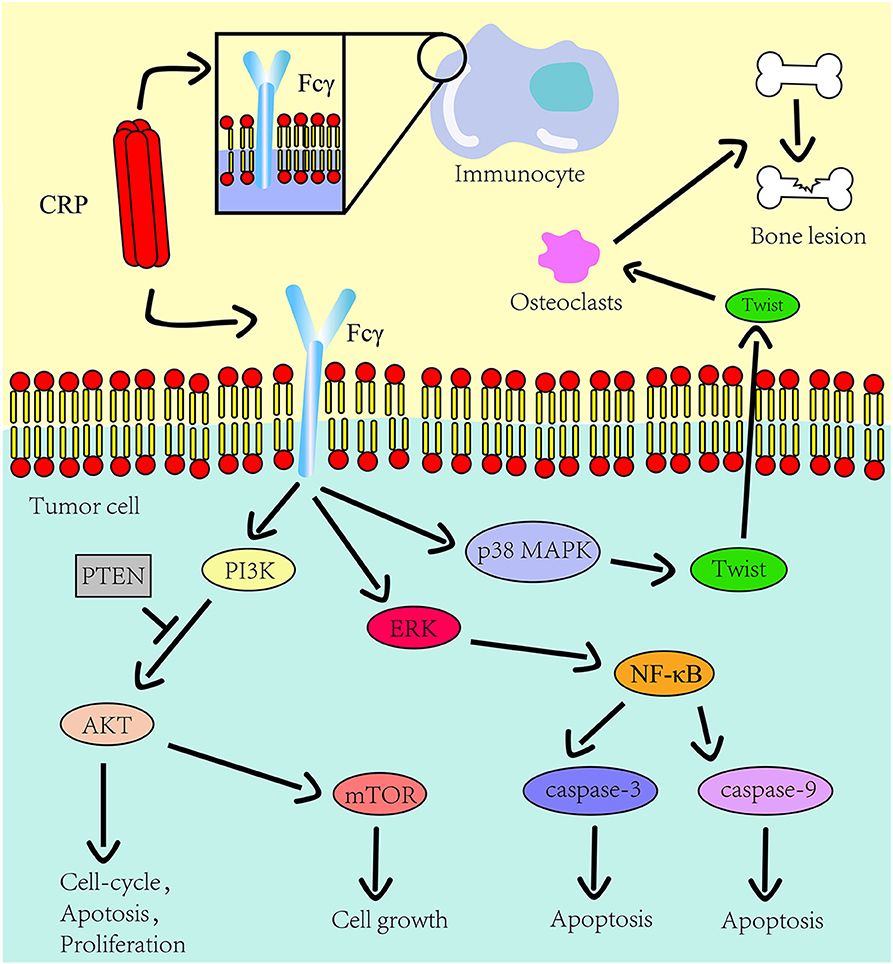
Figure 1. The role CRP on the innate immune system and tumor progression. The Fcγ receptor is expressed on the cell surface of immunocyte and multiple myeloma cells. CRP binds to Fcγ receptor to promote inflammation and tumor progression. The downstream pathways of this receptor in tumors include the PI3K/AKT/mTOR pathway, the ERK/NF-κB pathway and the p38/MAPK pathway. CRP regulates the expression of osteolytic cytokines in myeloma cells through p38 MAPK-Twist signaling.
Serum Amyloid P Component
The serum amyloid P component (SAP) gene, identified as a close CRP paralog, is also localized on the chromosome 1q23.2 and shares the same gene architecture (52). The gene is approximately 1.1 kb long with 0.1 kb 5′ UTR and 0.15 kb 3′ UTR. The SAP structure is similar to CRP except that its subunits consist of 204 AA and has a slight difference at the calcium-binding site (4, 53, 54). In the absence of calcium, SAP form decamers composed of two pentamers facing each other (4). The two ligands of SAP, deoxyadenosine 5′-monophosphate (dAMP), and the 4,6-pyruvate acetal of β-D-galactose (MoβDG), bind calcium and amyloid fibrils, respectively (55). To distinguish structure and gene sequence of short pentraxins, we archived data on the 3D structure for CRP and SAP from the PDB web portal (https://www.rcsb.org/, Figures 2A,B). The prediction of their domain was inferred from the Pfam database (http://pfam.xfam.org/, Figure 3) and the Genecard database (https://www.genecards.org/, Table 1).
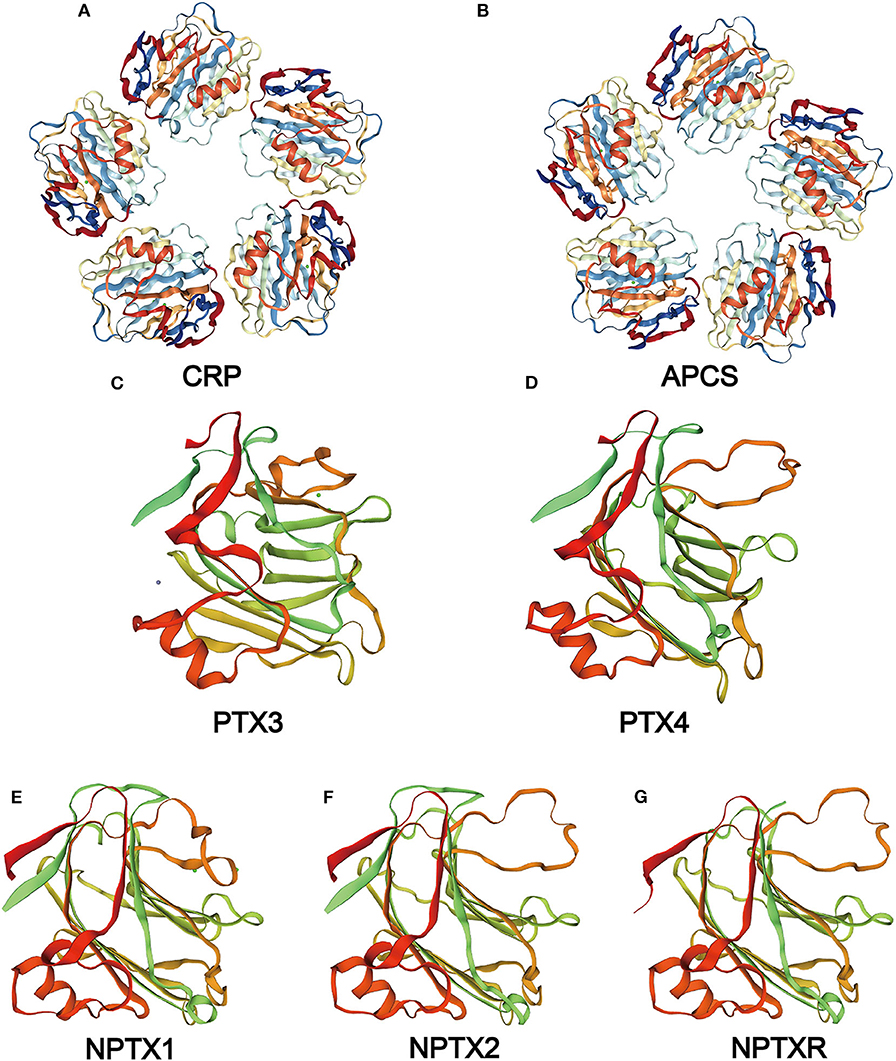
Figure 2. Structure of the pentraxin family members. Protein structure of CRP (A) and SAP (B) from the PDB website and the hypothetical structure of the full monomer (C-terminal domain and N-terminal domain) of PTX3 (C), PTX4 (D), NPTX1 (E), NPTX2 (F), NPTXR (G) from the Swiss database.
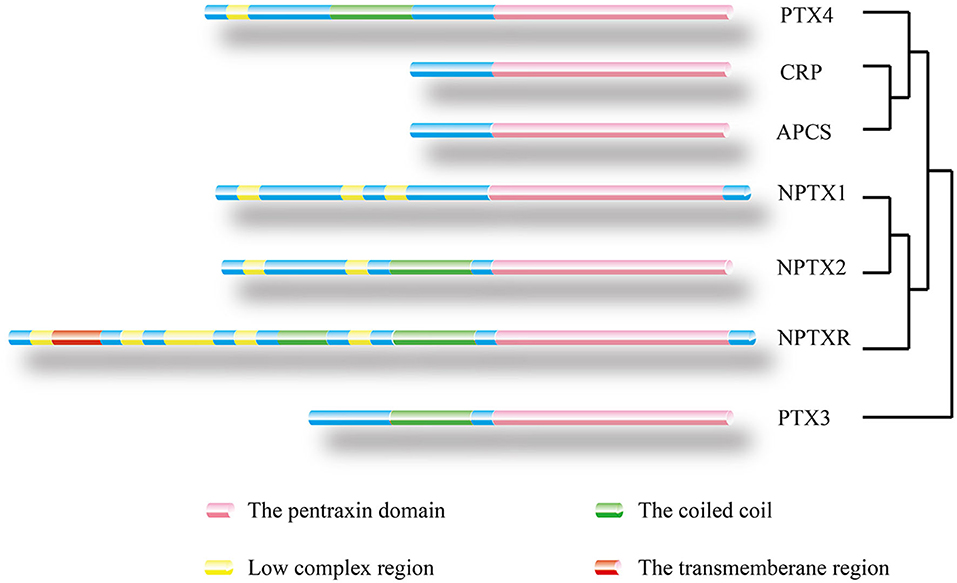
Figure 3. Location of protein domain for each family member and their sequence homology. The outcome was predicted by the Pfam database. Pink box: Pentraxin domain. Green box: Coiled coil. Yellow box: Low complex region. Red box: Transmembrane region. A homology tree based on the similarity of the protein sequence of members of the pentraxin family is generated.
Like CRP, SAP is also secreted by hepatocytes and mediates innate immunity by interacting with the complement system and the Fcγ receptor (17). This decreases neutrophil adhesion, inhibits neutrophil spreading, regulates macrophage activation (56), and inhibits fibrocyte differentiation (57). Besides, SAP is involved in immunological tolerance by binding to DNA or chromatin resulting from necrosis and apoptosis cell (58). Furthermore, SAP binds to amyloid fibrils through MoβDG, thereby causing amyloidosis disease (4–6). SAP is also associated with tuberculosis (59) and sickle cell disease (60), but the specific mechanisms are unknown. Despite limited reports on the role of SAP in tumors, SAP is considered a prognosis factor in non-small cell lung cancer (61). The highly structural homology between SAP and CRP suggests that SAP has the potential to mediate tumor progression through the Fcγ receptor.
The Neuronal Pentraxins
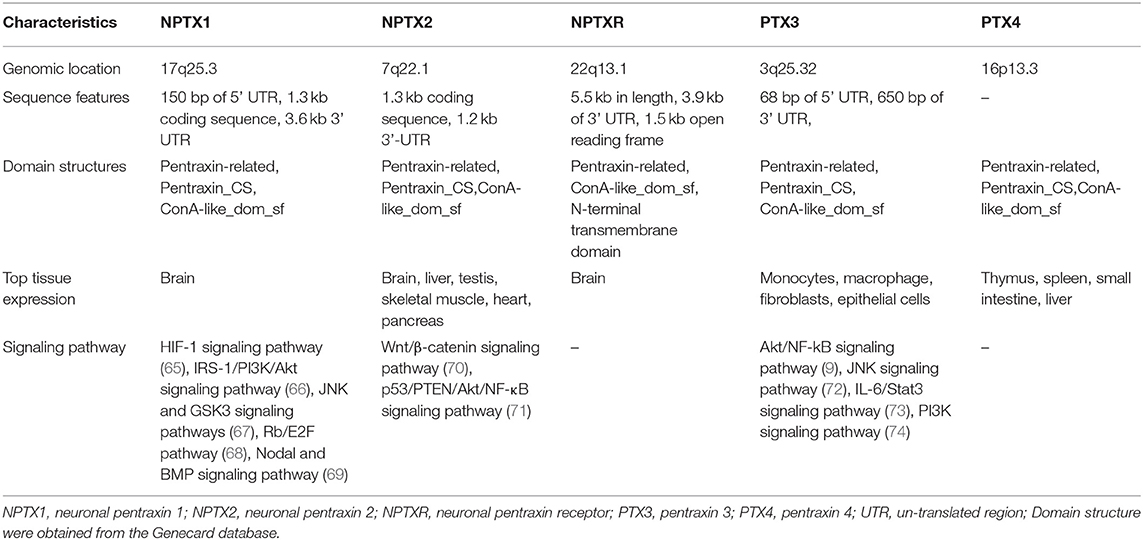
Neuronal pentraxin 1 (NPTX1 or NP1) is a 47–50 kDa secreted glycoprotein mainly expressed in neurons. The NPTX1 gene is located on chromosome 17q25.3. Its cDNA clones sequence is made up of a 150 bp 5′UTR, a 1.3 kb coding region, and a 3.6 kb 3′UTR with four introns (62). NPTX1 contains three main domains: a putative ligand- and calcium-binding site, the pentraxin domain, and an Asn-linked glycosylation site (62). Three NPTX1 domains, including the Pentraxin-related domain, the Pentraxin_CS (Pentraxin, conserved site), and glucanase domain superfamily (ConA-like_dom_sf) are speculated from the Genecard database.
Neuronal pentraxin 2 (NPTX2 or NP2, also known as apexin/p50 in guinea pig or narp in rat), is a ~47 kDa secretory glycoprotein with 431 AA. It is expressed in various tissues of the brain, testicle, pancreas, and skeletal muscle (1). The human NPTX2 gene is located on chromosome 7q22.1. Its cDNA sequence is made up of a 1.3 kb coding region and 1.2 kb 3′-UTR with four introns (1). Domains of NPTX2 are the same as those of NPTX1, according to the speculation from the Genecard website.
Neuronal pentraxin receptor (NPTXR or NPR) is an ~53 kDa type-II transmembrane protein with 500 AA and is mostly expressed in the brain. It is the only pentraxin family member anchored to the cell membrane by a putative N-terminal transmembrane domain. The receptor binds tightly to its ligands, such as taipoxin, TCBP49, NPTX1, and NPTX2, and activates different downstream signal transduction processes (63). In the human genome, the NPTXR gene is located on chromosome 22q13.1 and has the longest cDNA clones sequence containing a 3.9 kb 3′ UTR and a 1.5 kb open reading frame (63). From the Gencard database, this protein consists of two main domains, Pentraxin-related domain and glucanase domain superfamily (ConA-like_dom_sf). The N-terminal structure of the neuronal pentraxins is unrelated to other known human protein structures (64). Therefore, multiple online databases were used to generated detailed information about the structures of the neuronal pentraxins. The 3D structures of monomer were referenced from the Swiss database (https://www.swissmodel.expasy.org/, Figures 2E–G), and their domains were projected from the Pfam (Figure 3) and the Genecard databases (Table 2).
The Function of the Neuronal Pentraxins
The neuronal pentraxins have different functions in the development of the central nervous system (7) such as mediation of neural differentiation (69), synaptogenesis (75) and synapse plasticity (76, 77). Abnormal expression of the neuronal pentraxins has been reported in some mental diseases such as bipolar disorder (78), central precocious puberty (79), anxiety (80), depression (80), childhood-onset mood disorders (81) and schizophrenia (82). The neuronal pentraxins are associated with neurodegenerative diseases, including Alzheimer's disease (AD) (83, 84) and Parkinson's disease (PD) (85, 86). Researchers concluded that NPTXR protein in cerebrospinal fluid is a novel potential biomarker of AD progression and could have important utility in assessing treatment success in clinical trials (83), and NPTX1 could significantly contribute to the pathogenesis of PD (87). Moreover, only NPTX1 among all the neuronal pentraxins participates in inflammation by inducing mitochondria dysfunction (65, 67). Despite the neuronal pentraxins contains the pentraxin domain like the short pentraxins, few studies have classified their roles in human innate immunity which might result from the difference in their tertiary structure.
The Neuronal Pentraxins in Cancer
Several pathways were identified as potential mechanisms through which the neuronal pentraxins promote tumor progression. NPTX1 and NPTX2 were reported to cause dysfunction of the PI3K/AKT/mTOR pathway thereby affecting tumor progression in glioma (66), gastrointestinal stromal tumors (GIST) (88) and subependymal giant cell astrocytoma (89). Additionally, NPTX2 promotes tumor cell proliferation and metastasis by activating the NF-κB pathway (71, 90) and the Wnt/β-catenin pathway (70). It also induces tissue edema via an independent pathway from the classical VEGF-relate pathway (91). The dysfunction of the PI3K/AKT/mTOR pathway interferes with the normal cell-cycle and causes tumorigenesis. Similarly, NPTX1 and NPTX2 can inhibit cyclin A2 and CDK2 through the Rb/E2F signaling pathway (68), respectively, thus inducing G0/G1 arrest in pancreatic cancer (92, 93). Overexpression of NPTX2 has been identified as a prognosis factor in clear cell renal cell carcinoma, and its interaction with AMPA-selective glutamate receptor-4 affects tumor cell viability and metastasis (94, 95).
Abnormal expression of the neuronal pentraxins has been reported in different tumors such as cervical carcinoma (96), primary lung cancer (97), Ewing sarcoma (98), neuroblastoma (99), small cell lung cancer (100) and neuroblastoma (99). Another study has reported an increase in NPTX1 expression in pancreatic cancer after treatment with metformin and aspirin (101). On the contrary, a low level of NPTX2 showed better response to neoadjuvant chemoradiation (CRT) treatment in rectal adenocarcinomas (102).
The neuronal pentraxins are crucial in the central nervous system development when they interact with the AMPA receptor. The AMPA receptor has been proved to be associated with tumors (103, 104). Therefore, the AMPA receptor is a potential linkage between the neuronal pentraxins and tumor development as opposed to other members of the pentraxin family. Conclusively, the above-reviewed studies did not reveal the explicit mechanism through which the neuronal pentraxins affect tumor progression; however, they form the basis for in-depth studies on the existing association between the two.
Pentraxin 3 and Pentraxin 4
Pentraxin 3 (PTX3) and pentraxin 4 (PTX4) are characterized as long pentraxins. The PTX3 gene is located on chromosome 3q25 and has three exons and two introns (105). The three exons encode the leader peptide, an N-terminal domain, and the pentraxin domain, respectively (106). The N-terminal domain of PTX3 and PTX4 is not related to any known protein. However, based on previous research, PTX3 has a putative N-terminal domain that shows structural similarity to the mannose-binding protein and the surfactant proteins (107). PTX3 is secreted by different cell types that include dendritic cells, macrophages, and fibroblasts (108).
PTX4 gene is located on chromosome 16p13.3 and consists of three exons. However, the human PTX4 cDNA sequence and its first exon failed to amplify because it was different from the sequences in the various database (109). The sequence analysis of the protein showed a highly structural homology between PTX4 and the short pentraxins (Figure 2D). This indicates that PTX4 might be playing specific roles in innate immunity or tumor progression. To compare the PTX3 and PTX4 structures, their 3D structures were predicted from the Swiss database (https://www.swissmodel.expasy.org/, Figures 2C,D), and their domain predicted from the Pfam (Figure 3) and the Genecard databases (Table 2).
Functions of PTX3 and PTX4
From the previous study, an increase in blood PTX3 concentration serves as a monitor of inflammation initiation. The maximal PTX3 level increased slightly earlier than the CRP level (110), suggesting that PTX3 could be a highly sensitive inflammation-related factor. The interaction between PTX3 and the complement system has broad implications in host defense against microbial infections, regulation of the inflammatory reaction, and removal of dead cells. PTX3 is actively involved in the complement pathways activation (111). For example, the classical activation cascade may be initiated when PTX3 binds to C1q via the Fcγ receptor III once the latter is bound on a microbial surface (64, 112, 113). However, the process can be inhibited if the interaction occurs in the fluid-phase (114).
Furthermore, the N-terminal domain of PTX3 enhances tissue repair and remodeling functions (115). A study reported that PTX3 inhibited interstitial fibrosis in acute renal injury (73), indicating its role in an extracellular matrix formation. The N-terminal domain also combines with FGF2 to mediate angiogenic activity (116). For PTX4, there is currently no evidence supporting its functions in innate immunity or tumor progression.
PTX3 and Cancer
PTX3 interacts with the PI3K/AKT/mTOR signaling pathway to induce tumor cell proliferation, apoptosis and metastasis in lung cancer (9), head and neck squamous cell carcinoma (74) and breast cancer (117). PTX3 also inhibits cell proliferation and tumor metastasis by modulating the expression of protein related to the G2/M phase cell-cycle in cervical cancer (118). Furthermore, it arrests cell-cycle at the G0/G1 phase by stimulating the secretion of p21 protein in glioma (119).
Notably, we reported that PTX3 interacts with the fibroblast growth factor-2 (FGF2)/FGF receptor (FGFR) system that mediates the epithelial-mesenchymal transition (EMT) through its N-terminal domain (120). Through this system, PTX3 inhibits tumor metastasis, tumor growth and tumor angiogenesis in melanoma (8), breast cancer (121), prostate cancer (122) and multiple myeloma (123). Besides, PTX3 binds to fibroblast growth factor-8b receptor (FGF8b) and inhibits tumor cell proliferation in steroid hormone-regulated tumors (124). Compared to other pentraxin family members, the PTX3 is highly associated with the FGFR system.
Abnormal PTX3 expression was also observed in different tumors, including glioma (125), esophageal squamous cell carcinoma (126), pancreatic cancer (127), gastric cancer (128), colorectal cancer (129), leiomyosarcoma and desmoid tumors (130). Previous studies have shown that the activation of Fcγ receptor promotes tumor progression (131, 132). Of note, Fcγ receptor expression on NK cells modulates tumor response to immunotherapy (133), and PTX3 can exert its function in human immunity by interacting with this receptor. However, no current research has bridged PTX3 and tumor progression through the Fcγ receptor. Therefore, the multifaceted role of PTX3 in cancer requires further comprehensive study.
Overall Survival Analysis Prediction
We conducted the overall survival analysis of the pentraxin family to establish the relationship between the tumor and the pentraxin family members using the Gepia web portal (http://gepia.cancer-pku.cn/, Supplementary Table).
For short pentraxins, low expression of CRP showed better survival outcomes in kidney renal papillary cell carcinoma (KIRP) than high CRP expression (Figure 4A). There was no significant difference in survival outcome between the high and low expression of SAP. The expression levels of PTX4, with similar protein sequence as short pentraxins, showed a significant difference in survival outcome in adrenocortical carcinoma (ACC) and head and neck squamous cell carcinoma (HNSC) (Figures 4B,C).
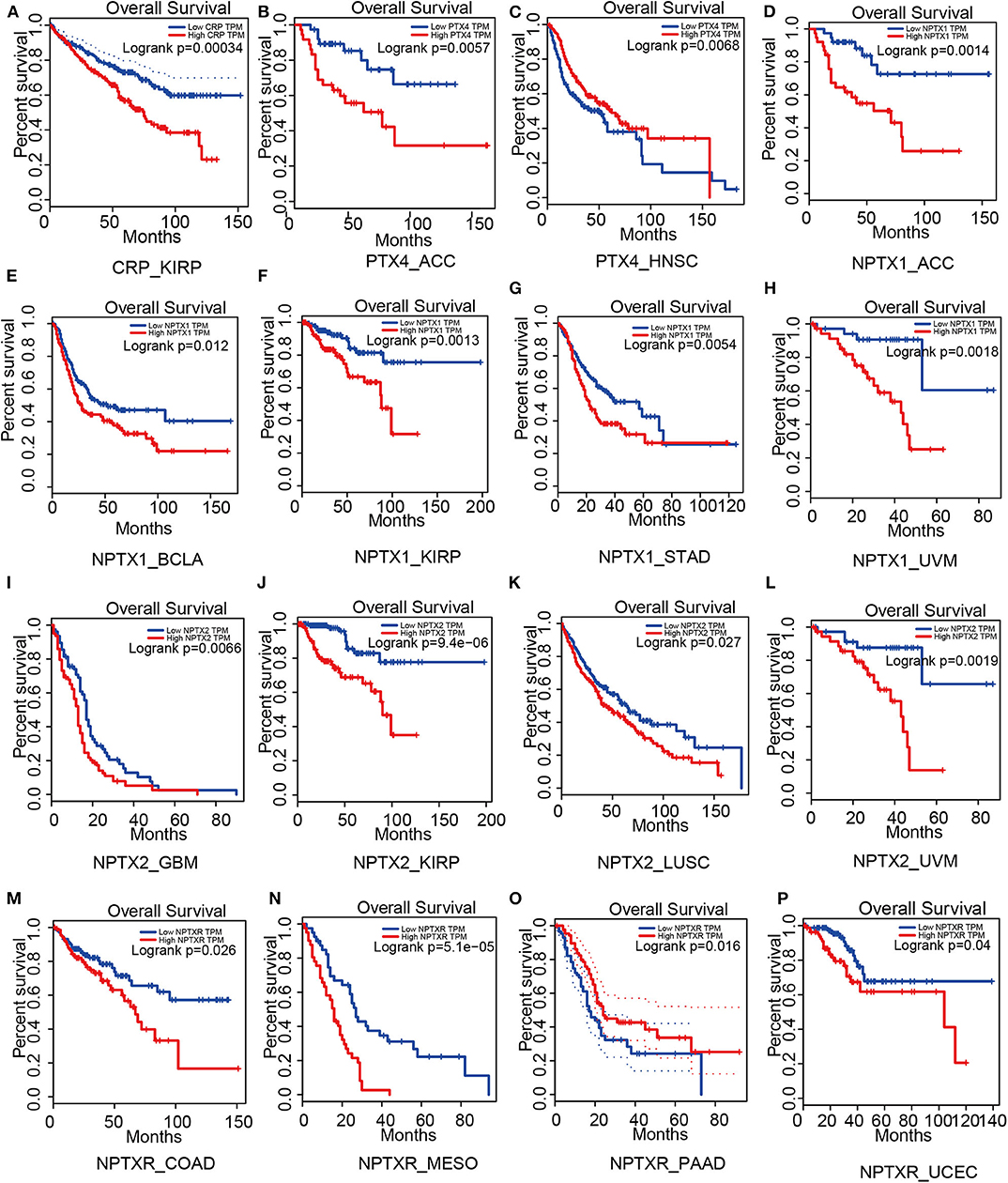
Figure 4. Survival analysis based on members of the pentraxin family from the Gepia website. Outcome of survival analysis predicted by the Gepia database including CRP (A), PTX4 (B,C) and the neuronal pentraxins (D–P).
We revealed that low NPTX1 expression improved the survival outcomes in patients with adrenocortical carcinoma (ACC), urothelial bladder carcinoma (BCLA), kidney renal papillary cell carcinoma (KIRP), stomach adenocarcinoma (STAD) and uveal melanoma (UVM), (Figures 4D–H). Patients that showed high expression of NPTX2 frequently exhibited worse survival outcomes for glioblastoma multiforme (GBM), kidney renal papillary cell carcinoma (KIRP), lung squamous cell carcinoma (LUSC) and uveal melanoma (UVM), (Figures 4I–L). On the other hand, overexpression of NPTXR predicted worse survival outcomes for colon adenocarcinoma (COAD), mesothelioma (MESO), and pancreatic adenocarcinoma (PAAD) and better survival outcome in uterine corpus endometrial carcinoma (UCEC) (Figures 4M–P).
Therefore, PTX3 is considered a promoter in tumor progression since its overexpression resulted in worse survival outcomes in invasive breast carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), brain lower-grade glioma (LGG), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA) and uterine corpus endometrial carcinoma (UCEC), (Figure 5).
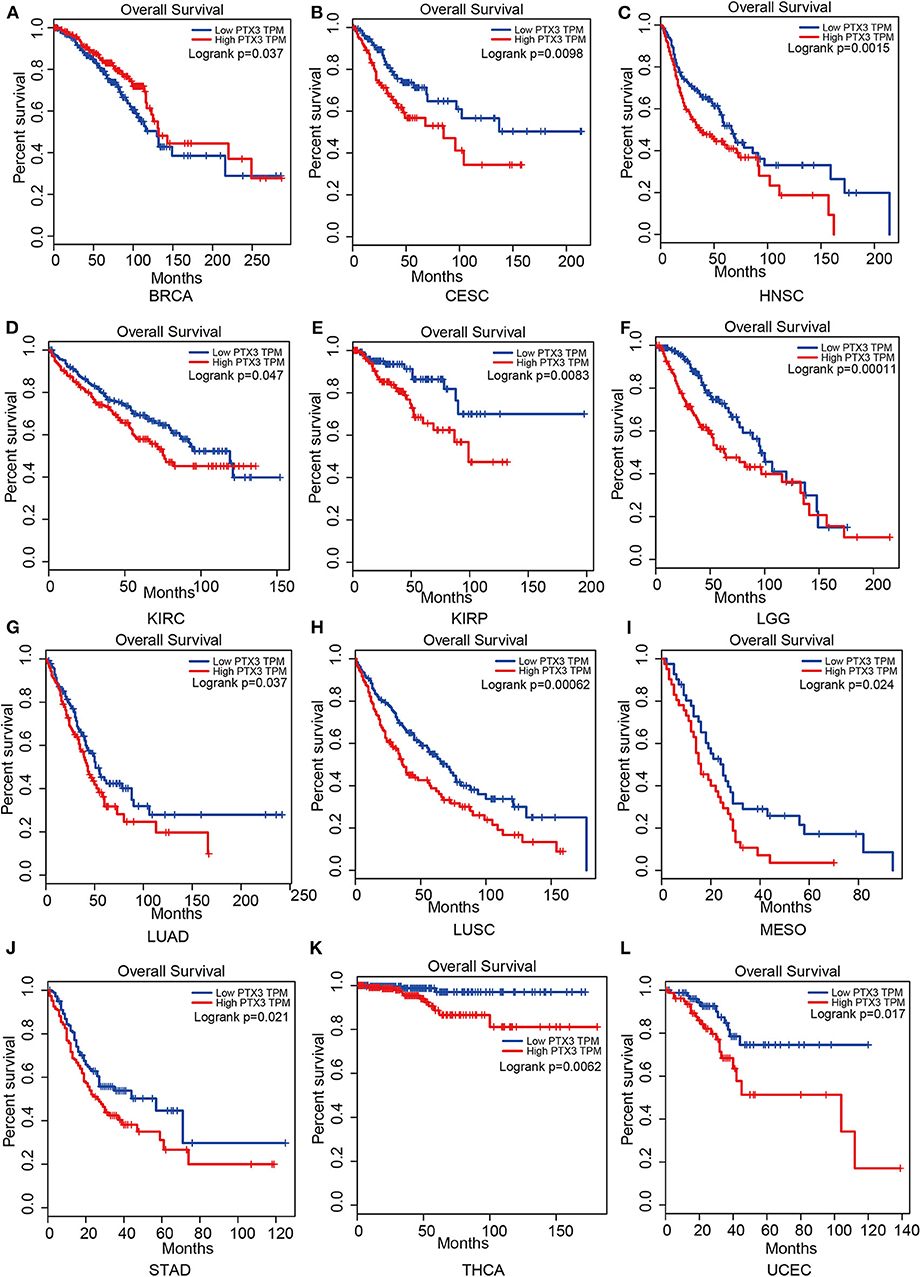
Figure 5. Impact of PTX3 on overall survival and tumor phenotypes based on the Gepia dataset. (A) Breast invasive carcinoma. (B) Cervical squamous cell carcinoma and endocervical adenocarcinoma. (C) Head and Neck squamous cell carcinoma. (D) Kidney renal clear cell carcinoma. (E) Kidney renal papillary cell carcinoma. (F) Brain Lower Grade Glioma. (G) Lung adenocarcinoma. (H) Lung squamous cell carcinoma. (I) Mesothelioma. (J) Stomach adenocarcinoma. (K) Thyroid carcinoma. (L) Uterine Corpus Endometrial Carcinoma. The overall survival of tumor that cannot be predicted by PTX3 expression is not listed.
The protein structure variability sheds light on the prognosis prediction ability of members of the pentraxin family. The expression of PTX3 affects multiple tumor types compared to other members of the pentraxin family, implying a potential association between its putative N-terminal domain and tumor progression. PTX4 shares a similar protein structure with the short pentraxins, and they are both poor predictors of survival outcomes of patients suggesting that they mainly concentrate on monitoring inflammation. However, this deduced relationship between protein structure and their prognosis capability requires further confirmation.
Conclusions
Emerging evidence confirms that the pentraxin family is associated with tumor progression by affecting tumor proliferation, mediating tumor cell apoptosis, inducing tumor metastasis, and promoting tumor tissue edema (Table 3, Figure 6). The tumor microenvironment is an extremely complex network consisting of cancer-associated fibroblasts, adipose cells, immunocytes, new-born vessels, and extracellular matrix. The pentraxin family promotes the formation of tumor microenvironment by facilitating macrophage infiltration and stimulating cytokines secretion. The pentraxin family, therefore, plays an essential role in tumor progression, although the exact mechanisms are still unknown and require further in-depth studies.
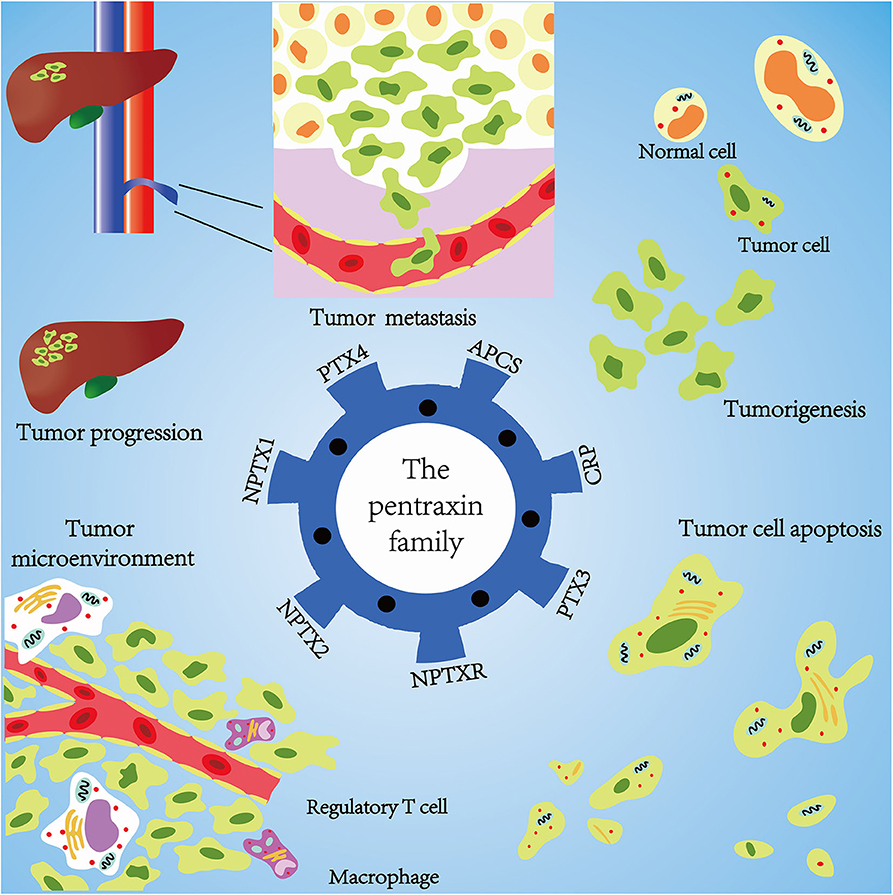
Figure 6. Mechanisms of pentraxin family in tumor progression. At the core is the pentraxin family and its seven members. The pentraxin family mainly participates in tumor metastasis, tumorigenesis, tumor cell apoptosis, tumor cell proliferation, and regulation of tumor microenvironment.
Protein structure of short pentraxins and their role in the immune system has highly been explored compared to the long pentraxins. It has been reported that PTX3 and NPTXR, for example, have an additional N-terminal domain other than the known pentraxin domain and exhibit different functions. Each pentraxin family member has their specific unique functions, for instance, SAP binds to amyloid fibrils and the neuronal pentraxins and extensively participates in the central nervous system development. Furthermore, the pentraxin family affects tumor progression through their unique receptors and pathways. Therefore, an in-depth analysis of the protein structure of the pentraxins family and their underlying mechanisms using advanced electron microscopy technologies, such as cryo-electron microscopy, is important for future research.
The PI3K/AKT/mTOR pathway is commonly associated with the pentraxin family in inducing tumor progression. Nevertheless, each pentraxin family member poses a specific receptor linked with tumor progression. Short pentraxins bind with the Fcγ receptor to activate different pathways. Studies have proposed that the AMPA receptor bridges the neuronal pentraxins and tumor by mediating intracellular free concentration of calcium known to be vital for various downstream pathways. PTX3 modulates tumor cell adhesion and metastasis by interacting with the FGFR system. SAP and PTX4 share highly structural homology with CRP, but limited research has focused on their association with the tumor, similarly with the neuronal pentraxins. Tumor microenvironment and inflammation are two crucial components that influence tumor progression through active communication with each other (138, 139). The pentraxin family can both initiate inflammation and promote tumor progression.
Author Contributions
QC, ZL, and GX: offered the idea of this review, and acted as the mentors and guarantors of the review. ZW and XW: wrote the review. ZW, XW, and ZD: modified the review. ZW: drew the figures. HZ, MZ, and SF: offered modification advice and checked grammar. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Nature Science Foundation of China (Nos. 81703622 and 81803582); the China Postdoctoral Science Foundation (No. 2018M63302); Hunan Provincial Natural Science Foundation of China (Nos. 2018JJ3838, 2019JJ50949, and 2020JJ8111); Grants from Xiangya Hospital Central South University postdoctoral foundation; Grants from Clinical medical technology innovation guidance program of Hunan Province (No. 2017SK50109); and grants from the research projects of Hunan Provincial Health Commission of China (Nos. C2019186 and B2019187).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01757/full#supplementary-material
References
1. Hsu YC, Perin MS. Human neuronal pentraxin II (NPTX2): conservation, genomic structure, and chromosomal localization. Genomics. (1995) 28:220–7. doi: 10.1006/geno.1995.1134
2. Liu Q, Tu T, Bai Z, Liu Z, Zhou S. Elevated plasma pentraxin 3: a potential cardiovascular risk factor? Med Hypotheses. (2011) 77:1068–70. doi: 10.1016/j.mehy.2011.09.004
3. Gonzalez OA, Kirakodu S, Novak MJ, Stromberg AJ, Orraca L, Gonzalez-Martinez J, et al. Comparative analysis of microbial sensing molecules in mucosal tissues with aging. Immunobiology. (2018) 223:279–87. doi: 10.1016/j.imbio.2017.10.034
4. Thompson D, Pepys MB, Tickle I, Wood S. The structures of crystalline complexes of human serum amyloid P component with its carbohydrate ligand, the cyclic pyruvate acetal of galactose. J Mol Biol. (2002) 320:1081–6. doi: 10.1016/S0022-2836(02)00514-4
5. Richards DB, Cookson LM, Berges AC, Barton SV, Lane T, Ritter JM, et al. Therapeutic clearance of amyloid by antibodies to serum amyloid P component. N Engl J Med. (2015) 373:1106–14. doi: 10.1056/NEJMoa1504942
6. Richards DB, Cookson LM, Barton SV, Liefaard L, Lane T, Hutt DF, et al. Repeat doses of antibody to serum amyloid P component clear amyloid deposits in patients with systemic amyloidosis. Sci Transl Med. (2018) 10:eaan3128. doi: 10.1126/scitranslmed.aan3128
7. Farhy-Tselnicker I, van Casteren ACM, Lee A, Chang VT, Aricescu AR, Allen NJ. Astrocyte-secreted glypican 4 regulates release of neuronal pentraxin 1 from axons to induce functional synapse formation. Neuron. (2017) 96:428–45.e13. doi: 10.1016/j.neuron.2017.09.053
8. Ronca R, Di Salle E, Giacomini A, Leali D, Alessi P, Coltrini D, et al. Long pentraxin-3 inhibits epithelial-mesenchymal transition in melanoma cells. Mol Cancer Ther. (2013) 12:2760–71. doi: 10.1158/1535-7163.MCT-13-0487
9. Ahmmed B, Khan MN, Nisar MA, Kampo S, Zheng Q, Li Y, et al. Tunicamycin enhances the suppressive effects of cisplatin on lung cancer growth through PTX3 glycosylation via AKT/NF-kappaB signaling pathway. Int J Oncol. (2019) 54:431–42. doi: 10.3892/ijo.2018.4650
10. Landskron G, De la Fuente M, Thuwajit P, Thuwajit C, Hermoso MA. Chronic inflammation and cytokines in the tumor microenvironment. J Immunol Res. (2014) 2014:149185. doi: 10.1155/2014/149185
11. Duan Y, Li Z, Cheng S, Chen Y, Zhang L, He J, et al. Nasopharyngeal carcinoma progression is mediated by EBER-triggered inflammation via the RIG-I pathway. Cancer Lett. (2015) 361:67–74. doi: 10.1016/j.canlet.2015.02.037
12. Murata M. Inflammation and cancer. Environ Health Prev Med. (2018) 23:50. doi: 10.1186/s12199-018-0740-1
13. Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. (2015) 348:74–80. doi: 10.1126/science.aaa6204
14. Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, et al. Role of tumor microenvironment in tumorigenesis. J Cancer. (2017) 8:761–73. doi: 10.7150/jca.17648
15. Ronca R, Tamma R, Coltrini D, Ruggieri S, Presta M, Ribatti D. Fibroblast growth factor modulates mast cell recruitment in a murine model of prostate cancer. Oncotarget. (2017) 8:82583–92. doi: 10.18632/oncotarget.19773
16. Woo P, Korenberg JR, Whitehead AS. Characterization of genomic and complementary DNA sequence of human C-reactive protein, and comparison with the complementary DNA sequence of serum amyloid P component. J Biol Chem. (1985) 260:13384–8.
17. Lu J, Marjon KD, Mold C, Du Clos TW, Sun PD. Pentraxins and Fc receptors. Immunol Rev. (2012) 250:230–8. doi: 10.1111/j.1600-065X.2012.01162.x
18. Vilahur G, Badimon L. Biological actions of pentraxins. Vascul Pharmacol. (2015) 73:38–44. doi: 10.1016/j.vph.2015.05.001
19. Clark SE, Weiser JN. Microbial modulation of host immunity with the small molecule phosphorylcholine. Infect Immun. (2013) 81:392–401. doi: 10.1128/IAI.01168-12
20. Ridker PM. From C-reactive protein to interleukin-6 to interleukin-1: moving upstream to identify novel targets for atheroprotection. Circ Res. (2016) 118:145–56. doi: 10.1161/CIRCRESAHA.115.306656
21. Zhang YA, Salinas I, Li J, Parra D, Bjork S, Xu Z, et al. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat Immunol. (2010) 11:827–35. doi: 10.1038/ni.1913
22. Yang J, Wezeman M, Zhang X, Lin P, Wang M, Qian J, et al. Human C-reactive protein binds activating Fcgamma receptors and protects myeloma tumor cells from apoptosis. Cancer Cell. (2007) 12:252–65. doi: 10.1016/j.ccr.2007.08.008
23. Kim ES, Kim SY, Koh M, Lee HM, Kim K, Jung J, et al. C-reactive protein binds to integrin alpha2 and Fcgamma receptor I, leading to breast cell adhesion and breast cancer progression. Oncogene. (2018) 37:28–38. doi: 10.1038/onc.2017.298
24. Aoki M, Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. (2017) 407:153–89. doi: 10.1007/82_2017_6
25. Chen W, Pilling D, Gomer RH. C-reactive protein (CRP) but not the related pentraxins serum amyloid P and PTX3 inhibits the proliferation and induces apoptosis of the leukemia cell line Mono Mac 6. BMC Immunol. (2017) 18:47. doi: 10.1186/s12865-017-0230-z
26. Du J, Hu W, Yang C, Wang Y, Wang X, Yang P. C-reactive protein is associated with the development of tongue squamous cell carcinoma. Acta Biochim Biophys Sin. (2018) 50:238–45. doi: 10.1093/abbs/gmy004
27. Yang J, Liu Z, Liu H, He J, Yang J, Lin P, et al. C-reactive protein promotes bone destruction in human myeloma through the CD32-p38 MAPK-Twist axis. Sci Signal. (2017) 10:eaan6282. doi: 10.1126/scisignal.aan6282
28. Morrison L, Laukkanen JA, Ronkainen K, Kurl S, Kauhanen J, Toriola AT. Inflammatory biomarker score and cancer: a population-based prospective cohort study. BMC Cancer. (2016) 16:80. doi: 10.1186/s12885-016-2115-6
29. Nijaguna MB, Schroder C, Patil V, Shwetha SD, Hegde AS, Chandramouli BA, et al. Definition of a serum marker panel for glioblastoma discrimination and identification of Interleukin 1beta in the microglial secretome as a novel mediator of endothelial cell survival induced by C-reactive protein. J Proteomics. (2015) 128:251–61. doi: 10.1016/j.jprot.2015.07.026
30. Nakayama T, Saito K, Kumagai J, Nakajima Y, Kijima T, Yoshida S, et al. Higher serum C-reactive protein level represents the immunosuppressive tumor microenvironment in patients with clear cell renal cell carcinoma. Clin Genitourin Cancer. (2018) 16:e1151–8. doi: 10.1016/j.clgc.2018.07.027
31. Xu BB, Lu J, Zheng ZF, Xie JW, Wang JB, Lin JX, et al. The predictive value of the preoperative C-reactive protein-albumin ratio for early recurrence and chemotherapy benefit in patients with gastric cancer after radical gastrectomy: using randomized phase III trial data. Gastric Cancer. (2019) 22:1016–28. doi: 10.1007/s10120-019-00936-w
32. Krzystek-Korpacka M, Matusiewicz M, Diakowska D, Grabowski K, Blachut K, Kustrzeba-Wojcicka I, et al. Acute-phase response proteins are related to cachexia and accelerated angiogenesis in gastroesophageal cancers. Clin Chem Lab Med. (2008) 46:359–64. doi: 10.1515/CCLM.2008.089
33. Kuribayashi K, Motoyama S, Sasaki T, Sato Y, Yoshino K, Wakita A, et al. C-reactive protein reduces the relative number of tumor-associated M2 macrophages and intratumoral angiogenesis in mice. Tohoku J Exp Med. (2014) 233:249–55. doi: 10.1620/tjem.233.249
34. Ma Z, Qi Z, Shan Z, Li J, Yang J, Xu Z. The role of CRP and ATG9B expression in clear cell renal cell carcinoma. Biosci Rep. (2017) 37:BSR20171082. doi: 10.1042/BSR20171082
35. Lust JA, Lacy MQ, Zeldenrust SR, Witzig TE, Moon-Tasson LL, Dinarello CA, et al. Reduction in C-reactive protein indicates successful targeting of the IL-1/IL-6 axis resulting in improved survival in early stage multiple myeloma. Am J Hematol. (2016) 91:571–4. doi: 10.1002/ajh.24352
36. Saito K, Tatokoro M, Fujii Y, Iimura Y, Koga F, Kawakami S, et al. Impact of C-reactive protein kinetics on survival of patients with metastatic renal cell carcinoma. Eur Urol. (2009) 55:1145–53. doi: 10.1016/j.eururo.2008.10.012
37. Xia WK, Wu X, Yu TH, Wu Y, Yao XJ, Hu H. Prognostic significance of lymphocyte-to-monocyte ratio and CRP in patients with nonmetastatic clear cell renal cell carcinoma: a retrospective multicenter analysis. Onco Targets Ther. (2016) 9:2759–67. doi: 10.2147/OTT.S101458
38. Dalpiaz O, Luef T, Seles M, Stotz M, Stojakovic T, Pummer K, et al. Critical evaluation of the potential prognostic value of the pretreatment-derived neutrophil-lymphocyte ratio under consideration of C-reactive protein levels in clear cell renal cell carcinoma. Br J Cancer. (2017) 116:85–90. doi: 10.1038/bjc.2016.393
39. Kaur RP, Rubal Banipal RPS, Vashistha R, Dhiman M, Munshi A. Association of elevated levels of C-reactive protein with breast cancer, breast cancer subtypes, and poor outcome. Curr Probl Cancer. (2019) 43:123–9. doi: 10.1016/j.currproblcancer.2018.05.003
40. Liu ZQ, Chu L, Fang JM, Zhang X, Zhao HX, Chen YJ, et al. Prognostic role of C-reactive protein in prostate cancer: a systematic review and meta-analysis. Asian J Androl. (2014) 16:467–71. doi: 10.4103/1008-682X.123686
41. Muller DC, Larose TL, Hodge A, Guida F, Langhammer A, Grankvist K, et al. Circulating high sensitivity C reactive protein concentrations and risk of lung cancer: nested case-control study within Lung Cancer Cohort Consortium. BMJ. (2019) 364:k4981. doi: 10.1136/bmj.k4981
42. Kinoshita A, Onoda H, Imai N, Iwaku A, Oishi M, Tanaka K, et al. The C-reactive protein/albumin ratio, a novel inflammation-based prognostic score, predicts outcomes in patients with hepatocellular carcinoma. Ann Surg Oncol. (2015) 22:803–10. doi: 10.1245/s10434-014-4048-0
43. He X, Li JP, Liu XH, Zhang JP, Zeng QY, Chen H, et al. Prognostic value of C-reactive protein/albumin ratio in predicting overall survival of Chinese cervical cancer patients overall survival: comparison among various inflammation based factors. J Cancer. (2018) 9:1877–84. doi: 10.7150/jca.23320
44. Park MJ, Roh JL, Kim SB, Choi SH, Nam SY, Kim SY. Prognostic value of circulating biomarker score in advanced-stage head and neck squamous cell carcinoma. Eur J Cancer. (2018) 92:69–76. doi: 10.1016/j.ejca.2018.01.069
45. Qin W, Yuan Q, Wu J, Yu H, Wang Y, Chen Q. Prognostic value of pre-therapy C-reactive protein level in diffuse large B-cell lymphoma: a meta-analysis. Leuk Lymphoma. (2019) 60:358–66. doi: 10.1080/10428194.2018.1482540
46. Li W, Luo X, Liu Z, Chen Y, Li Z. Prognostic value of C-reactive protein levels in patients with bone neoplasms: A meta-analysis. PLoS ONE. (2018) 13:e0195769. doi: 10.1371/journal.pone.0195769
47. Grote VA, Kaaks R, Nieters A, Tjonneland A, Halkjaer J, Overvad K, et al. Inflammation marker and risk of pancreatic cancer: a nested case-control study within the EPIC cohort. Br J Cancer. (2012) 106:1866–74. doi: 10.1038/bjc.2012.172
48. Szkandera J, Stotz M, Absenger G, Stojakovic T, Samonigg H, Kornprat P, et al. Validation of C-reactive protein levels as a prognostic indicator for survival in a large cohort of pancreatic cancer patients. Br J Cancer. (2014) 110:183–8. doi: 10.1038/bjc.2013.701
49. Chan AT, Ogino S, Giovannucci EL, Fuchs CS. Inflammatory markers are associated with risk of colorectal cancer and chemopreventive response to anti-inflammatory drugs. Gastroenterology. (2011) 140:799–808, quiz e711. doi: 10.1053/j.gastro.2010.11.041
50. Nimptsch K, Aleksandrova K, Boeing H, Janke J, Lee YA, Jenab M, et al. Association of CRP genetic variants with blood concentrations of C-reactive protein and colorectal cancer risk. Int J Cancer. (2015) 136:1181–92. doi: 10.1002/ijc.29086
51. Farsad-Naeimi A, Alizadeh M, Esfahani A, Darvish Aminabad E. Effect of fisetin supplementation on inflammatory factors and matrix metalloproteinase enzymes in colorectal cancer patients. Food Funct. (2018) 9:2025–31. doi: 10.1039/C7FO01898C
52. Xi D, Luo T, Xiong H, Liu J, Lu H, Li M, et al. SAP: structure, function, and its roles in immune-related diseases. Int J Cardiol. (2015) 187:20–6. doi: 10.1016/j.ijcard.2015.03.179
53. Emsley J, White HE, O'Hara BP, Oliva G, Srinivasan N, Tickle IJ, et al. Structure of pentameric human serum amyloid P component. Nature. (1994) 367:338–45. doi: 10.1038/367338a0
54. Mikolajek H, Kolstoe SE, Pye VE, Mangione P, Pepys MB, Wood SP. Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component. J Mol Recognit. (2011) 24:371–7. doi: 10.1002/jmr.1090
55. Hind CR, Collins PM, Renn D, Cook RB, Caspi D, Baltz ML, et al. Binding specificity of serum amyloid P component for the pyruvate acetal of galactose. J Exp Med. (1984) 159:1058–69. doi: 10.1084/jem.159.4.1058
56. Zhang W, Xu W, Xiong S. Macrophage differentiation and polarization via phosphatidylinositol 3-kinase/Akt-ERK signaling pathway conferred by serum amyloid P component. J Immunol. (2011) 187:1764–77. doi: 10.4049/jimmunol.1002315
57. Cox N, Pilling D, Gomer RH. Serum amyloid P: a systemic regulator of the innate immune response. J Leukoc Biol. (2014) 96:739–43. doi: 10.1189/jlb.1MR0114-068R
58. Bickerstaff MC, Botto M, Hutchinson WL, Herbert J, Tennent GA, Bybee A, et al. Serum amyloid P component controls chromatin degradation and prevents antinuclear autoimmunity. Nat Med. (1999) 5:694–7. doi: 10.1038/9544
59. Jacobs R, Tshehla E, Malherbe S, Kriel M, Loxton AG, Stanley K, et al. Host biomarkers detected in saliva show promise as markers for the diagnosis of pulmonary tuberculosis disease and monitoring of the response to tuberculosis treatment. Cytokine. (2016) 81:50–6. doi: 10.1016/j.cyto.2016.02.004
60. Chantrathammachart P, Mackman N, Sparkenbaugh E, Wang JG, Parise LV, Kirchhofer D, et al. Tissue factor promotes activation of coagulation and inflammation in a mouse model of sickle cell disease. Blood. (2012) 120:636–46. doi: 10.1182/blood-2012-04-424143
61. Zhao J, Chi A, Mao R, Hu G, Ji M. Serum amyloid P-component level may be a biomarker for lung toxicities and overall survival after thoracic radiotherapy for non-small cell lung cancer. Clin Lab. (2016) 62:2183–90. doi: 10.7754/Clin.Lab.2016.160411
62. Omeis IA, Hsu YC, Perin MS. Mouse and human neuronal pentraxin 1 (NPTX1): conservation, genomic structure, and chromosomal localization. Genomics. (1996) 36:543–5. doi: 10.1006/geno.1996.0503
63. Dodds DC, Omeis IA, Cushman SJ, Helms JA, Perin MS. Neuronal pentraxin receptor, a novel putative integral membrane pentraxin that interacts with neuronal pentraxin 1 and 2 and taipoxin-associated calcium-binding protein 49. J Biol Chem. (1997) 272:21488–94. doi: 10.1074/jbc.272.34.21488
64. Du Clos TW. Pentraxins: structure, function, and role in inflammation. ISRN Inflamm. (2013) 2013:379040. doi: 10.1155/2013/379040
65. Thatipamula S, Al Rahim M, Zhang J, Hossain MA. Genetic deletion of neuronal pentraxin 1 expression prevents brain injury in a neonatal mouse model of cerebral hypoxia-ischemia. Neurobiol Dis. (2015) 75:15–30. doi: 10.1016/j.nbd.2014.12.016
66. Huo L, Wang B, Zheng M, Zhang Y, Xu J, Yang G, et al. miR-128-3p inhibits glioma cell proliferation and differentiation by targeting NPTX1 through IRS-1/PI3K/AKT signaling pathway. Exp Ther Med. (2019) 17:2921–30. doi: 10.3892/etm.2019.7284
67. Guzeloglu-Kayisli O, Basar M, Shapiro JP, Semerci N, Huang JS, Schatz F, et al. Long-acting progestin-only contraceptives enhance human endometrial stromal cell expressed neuronal pentraxin-1 and reactive oxygen species to promote endothelial cell apoptosis. J Clin Endocrinol Metab. (2014) 99:E1957–66. doi: 10.1210/jc.2014-1770
68. Peng X, Pan K, Zhao W, Zhang J, Yuan S, Wen X, et al. NPTX1 inhibits colon cancer cell proliferation through down-regulating cyclin A2 and CDK2 expression. Cell Biol Int. (2018) 42:589–97. doi: 10.1002/cbin.10935
69. Boles NC, Hirsch SE, Le S, Corneo B, Najm F, Minotti AP, et al. NPTX1 regulates neural lineage specification from human pluripotent stem cells. Cell Rep. (2014) 6:724–36. doi: 10.1016/j.celrep.2014.01.026
70. Xu C, Tian G, Jiang C, Xue H, Kuerbanjiang M, Sun L, et al. NPTX2 promotes colorectal cancer growth and liver metastasis by the activation of the canonical Wnt/beta-catenin pathway via FZD6. Cell Death Dis. (2019) 10:217. doi: 10.1038/s41419-019-1467-7
71. Shukla S, Pia Patric IR, Thinagararjan S, Srinivasan S, Mondal B, Hegde AS, et al. A DNA methylation prognostic signature of glioblastoma: identification of NPTX2-PTEN-NF-kappaB nexus. Cancer Res. (2013) 73:6563–73. doi: 10.1158/0008-5472.CAN-13-0298
72. Liu H, Jiang Q, Ju Z, Guan S, He B. Pentraxin 3 promotes cardiac differentiation of mouse embryonic stem cells through JNK signaling pathway. Cell Biol Int. (2018) 42:1556–63. doi: 10.1002/cbin.11049
73. Xiao Y, Yang N, Zhang Q, Wang Y, Yang S, Liu Z. Pentraxin 3 inhibits acute renal injury-induced interstitial fibrosis through suppression of IL-6/Stat3 pathway. Inflammation. (2014) 37:1895–901. doi: 10.1007/s10753-014-9921-2
74. Chan SH, Tsai JP, Shen CJ, Liao YH, Chen BK. Oleate-induced PTX3 promotes head and neck squamous cell carcinoma metastasis through the up-regulation of vimentin. Oncotarget. (2017) 8:41364–78. doi: 10.18632/oncotarget.17326
75. Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. (2007) 55:87–102. doi: 10.1016/j.neuron.2007.06.020
76. Cummings DM, Benway TA, Ho H, Tedoldi A, Fernandes Freitas MM, Shahab L, et al. Neuronal and peripheral pentraxins modify glutamate release and may interact in blood-brain barrier failure. Cereb Cortex. (2017) 27:3437–48. doi: 10.1093/cercor/bhx046
77. Lee SJ, Wei M, Zhang C, Maxeiner S, Pak C, Calado Botelho S, et al. Presynaptic neuronal pentraxin receptor organizes excitatory and inhibitory synapses. J Neurosci. (2017) 37:1062–80. doi: 10.1523/JNEUROSCI.2768-16.2016
78. Rajkumar AP, Christensen JH, Mattheisen M, Jacobsen I, Bache I, Pallesen J, et al. Analysis of t(9;17)(q33.2;q25.3) chromosomal breakpoint regions and genetic association reveals novel candidate genes for bipolar disorder. Bipolar Disord. (2015) 17:205–11. doi: 10.1111/bdi.12239
79. Liu H, Kong X, Chen F. Mkrn3 functions as a novel ubiquitin E3 ligase to inhibit Nptx1 during puberty initiation. Oncotarget. (2017) 8:85102–9. doi: 10.18632/oncotarget.19347
80. Li T, Mamillapalli R, Ding S, Chang H, Liu ZW, Gao XB, et al. Endometriosis alters brain electrophysiology, gene expression and increases pain sensitization, anxiety, and depression in female mice. Biol Reprod. (2018) 99:349–59. doi: 10.1093/biolre/ioy035
81. Strauss J, McGregor S, Freeman N, Tiwari A, George CJ, Kovacs M, et al. Association study of early-immediate genes in childhood-onset mood disorders and suicide attempt. Psychiatry Res. (2012) 197:49–54. doi: 10.1016/j.psychres.2011.11.022
82. Manchia M, Piras IS, Huentelman MJ, Pinna F, Zai CC, Kennedy JL. Pattern of gene expression in different stages of schizophrenia: down-regulation of NPTX2 gene revealed by a meta-analysis of microarray datasets. Eur Neuropsychopharmacol. (2017) 27:1054–63. doi: 10.1016/j.euroneuro.2017.07.002
83. Begcevic I, Tsolaki M, Brinc D, Brown M, Martinez-Morillo E, Lazarou I, et al. Neuronal pentraxin receptor-1 is a new cerebrospinal fluid biomarker of Alzheimer's disease progression. F1000Res. (2018) 7:1012. doi: 10.12688/f1000research.15095.1
84. Goetzl EJ, Abner EL, Jicha GA, Kapogiannis D, Schwartz JB. Declining levels of functionally specialized synaptic proteins in plasma neuronal exosomes with progression of Alzheimer's disease. FASEB J. (2018) 32:888–93. doi: 10.1096/fj.201700731R
85. Charbonnier-Beaupel F, Malerbi M, Alcacer C, Tahiri K, Carpentier W, Wang C, et al. Gene expression analyses identify Narp contribution in the development of L-DOPA-induced dyskinesia. J Neurosci. (2015) 35:96–111. doi: 10.1523/JNEUROSCI.5231-13.2015
86. Li J, Sun Y, Chen J. Transcriptome sequencing in a 6-hydroxydopamine rat model of Parkinson's disease. Genes Genet Syst. (2019) 94:61–9. doi: 10.1266/ggs.18-00036
87. Qu Z, D'Mello SR. Proteomic analysis identifies NPTX1 and HIP1R as potential targets of histone deacetylase-3-mediated neurodegeneration. Exp Biol Med. (2018) 243:627–38. doi: 10.1177/1535370218761149
88. Zook P, Pathak HB, Belinsky MG, Gersz L, Devarajan K, Zhou Y, et al. Combination of imatinib mesylate and AKT inhibitor provides synergistic effects in preclinical study of gastrointestinal stromal tumor. Clin Cancer Res. (2017) 23:171–80. doi: 10.1158/1078-0432.CCR-16-0529
89. Tyburczy ME, Kotulska K, Pokarowski P, Mieczkowski J, Kucharska J, Grajkowska W, et al. Novel proteins regulated by mTOR in subependymal giant cell astrocytomas of patients with tuberous sclerosis complex and new therapeutic implications. Am J Pathol. (2010) 176:1878–90. doi: 10.2353/ajpath.2010.090950
90. Skiriute D, Vaitkiene P, Asmoniene V, Steponaitis G, Deltuva VP, Tamasauskas A. Promoter methylation of AREG, HOXA11, hMLH1, NDRG2, NPTX2 and Tes genes in glioblastoma. J Neurooncol. (2013) 113:441–9. doi: 10.1007/s11060-013-1133-3
91. Carlson MR, Pope WB, Horvath S, Braunstein JG, Nghiemphu P, Tso CL, et al. Relationship between survival and edema in malignant gliomas: role of vascular endothelial growth factor and neuronal pentraxin 2. Clin Cancer Res. (2007) 13:2592–8. doi: 10.1158/1078-0432.CCR-06-2772
92. Zhang L, Gao J, Li L, Li Z, Du Y, Gong Y. The neuronal pentraxin II gene (NPTX2) inhibit proliferation and invasion of pancreatic cancer cells in vitro. Mol Biol Rep. (2011) 38:4903–11. doi: 10.1007/s11033-010-0632-y
93. Zhang L, Gao J, Li Z, Gong Y. Neuronal pentraxin II (NPTX2) is frequently down-regulated by promoter hypermethylation in pancreatic cancers. Dig Dis Sci. (2012) 57:2608–14. doi: 10.1007/s10620-012-2202-8
94. von Roemeling CA, Radisky DC, Marlow LA, Cooper SJ, Grebe SK, Anastasiadis PZ, et al. Neuronal pentraxin 2 supports clear cell renal cell carcinoma by activating the AMPA-selective glutamate receptor-4. Cancer Res. (2014) 74:4796–810. doi: 10.1158/0008-5472.CAN-14-0210
95. Schrodter S, Braun M, Syring I, Klumper N, Deng M, Schmidt D, et al. Identification of the dopamine transporter SLC6A3 as a biomarker for patients with renal cell carcinoma. Mol Cancer. (2016) 15:10. doi: 10.1186/s12943-016-0495-5
96. Yang N, Eijsink JJ, Lendvai A, Volders HH, Klip H, Buikema HJ, et al. Methylation markers for CCNA1 and C13ORF18 are strongly associated with high-grade cervical intraepithelial neoplasia and cervical cancer in cervical scrapings. Cancer Epidemiol Biomarkers Prev. (2009) 18:3000–7. doi: 10.1158/1055-9965.EPI-09-0405
97. Zhou C, Qin Y, Xie Z, Zhang J, Yang M, Li S, et al. NPTX1 is a novel epigenetic regulation gene and associated with prognosis in lung cancer. Biochem Biophys Res Commun. (2015) 458:381–6. doi: 10.1016/j.bbrc.2015.01.124
98. Alholle A, Brini AT, Gharanei S, Vaiyapuri S, Arrigoni E, Dallol A, et al. Functional epigenetic approach identifies frequently methylated genes in Ewing sarcoma. Epigenetics. (2013) 8:1198–204. doi: 10.4161/epi.26266
99. Bartolini A, Di Paolo D, Noghero A, Murgia D, Sementa AR, Cilli M, et al. The neuronal pentraxin-2 pathway is an unrecognized target in human neuroblastoma, which also offers prognostic value in patients. Cancer Res. (2015) 75:4265–71. doi: 10.1158/0008-5472.CAN-15-0649
100. Poulsen TT, Pedersen N, Perin MS, Hansen CK, Poulsen HS. Specific sensitivity of small cell lung cancer cell lines to the snake venom toxin taipoxin. Lung Cancer. (2005) 50:329–37. doi: 10.1016/j.lungcan.2005.06.011
101. Yue W, Wang T, Zachariah E, Lin Y, Yang CS, Xu Q, et al. Transcriptomic analysis of pancreatic cancer cells in response to metformin and aspirin: an implication of synergy. Sci Rep. (2015) 5:13390. doi: 10.1038/srep13390
102. Karagkounis G, Thai L, DeVecchio J, Gantt GA, Duraes L, Pai RK, et al. NPTX2 is associated with neoadjuvant therapy response in rectal cancer. J Surg Res. (2016) 202:112–7. doi: 10.1016/j.jss.2015.12.042
103. de Groot J, Sontheimer H. Glutamate and the biology of gliomas. Glia. (2011) 59:1181–9. doi: 10.1002/glia.21113
104. Radin DP, Patel P. A current perspective on the oncopreventive and oncolytic properties of selective serotonin reuptake inhibitors. Biomed Pharmacother. (2017) 87:636–9. doi: 10.1016/j.biopha.2017.01.024
105. Breviario F, d'Aniello EM, Golay J, Peri G, Bottazzi B, Bairoch A, et al. Interleukin-1-inducible genes in endothelial cells. Cloning of a new gene related to C-reactive protein and serum amyloid P component. J Biol Chem. (1992) 267:22190–7.
106. Lee GW, Lee TH, Vilcek J. TSG-14, a tumor necrosis factor- and IL-1-inducible protein, is a novel member of the pentaxin family of acute phase proteins. J Immunol. (1993) 150:1804–12.
107. Presta M, Camozzi M, Salvatori G, Rusnati M. Role of the soluble pattern recognition receptor PTX3 in vascular biology. J Cell Mol Med. (2007) 11:723–38. doi: 10.1111/j.1582-4934.2007.00061.x
108. Moalli F, Jaillon S, Inforzato A, Sironi M, Bottazzi B, Mantovani A, et al. Pathogen recognition by the long pentraxin PTX3. J Biomed Biotechnol. (2011) 2011:830421. doi: 10.1155/2011/830421
109. Martinez de la Torre Y, Fabbri M, Jaillon S, Bastone A, Nebuloni M, Vecchi A, et al. Evolution of the pentraxin family: the new entry PTX4. J Immunol. (2010) 184:5055–64. doi: 10.4049/jimmunol.0901672
110. Mantovani A, Garlanda C, Doni A, Bottazzi B. Pentraxins in innate immunity: from C-reactive protein to the long pentraxin PTX3. J Clin Immunol. (2008) 28:1–13. doi: 10.1007/s10875-007-9126-7
111. Ma YJ, Doni A, Hummelshoj T, Honore C, Bastone A, Mantovani A, et al. Synergy between ficolin-2 and pentraxin 3 boosts innate immune recognition and complement deposition. J Biol Chem. (2009) 284:28263–75. doi: 10.1074/jbc.M109.009225
112. Bottazzi B, Vouret-Craviari V, Bastone A, De Gioia L, Matteucci C, Peri G, et al. Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component. J Biol Chem. (1997) 272:32817–23. doi: 10.1074/jbc.272.52.32817
113. Inforzato A, Peri G, Doni A, Garlanda C, Mantovani A, Bastone A, et al. Structure and function of the long pentraxin PTX3 glycosidic moiety: fine-tuning of the interaction with C1q and complement activation. Biochemistry. (2006) 45:11540–51. doi: 10.1021/bi0607453
114. Nauta AJ, Bottazzi B, Mantovani A, Salvatori G, Kishore U, Schwaeble WJ, et al. Biochemical and functional characterization of the interaction between pentraxin 3 and C1q. Eur J Immunol. (2003) 33:465–73. doi: 10.1002/immu.200310022
115. Bottazzi B, Inforzato A, Messa M, Barbagallo M, Magrini E, Garlanda C, et al. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J Hepatol. (2016) 64:1416–27. doi: 10.1016/j.jhep.2016.02.029
116. Presta M, Foglio E, Churruca Schuind A, Ronca R. Long pentraxin-3 modulates the angiogenic activity of fibroblast growth factor-2. Front Immunol. (2018) 9:2327. doi: 10.3389/fimmu.2018.02327
117. Thomas C, Henry W, Cuiffo BG, Collmann AY, Marangoni E, Benhamo V, et al. Pentraxin-3 is a PI3K signaling target that promotes stem cell-like traits in basal-like breast cancers. Sci Signal. (2017) 10:eaah4674. doi: 10.1126/scisignal.aah4674
118. Ying TH, Lee CH, Chiou HL, Yang SF, Lin CL, Hung CH, et al. Knockdown of Pentraxin 3 suppresses tumorigenicity and metastasis of human cervical cancer cells. Sci Rep. (2016) 6:29385. doi: 10.1038/srep29385
119. Pfeifer GP. Defining driver DNA methylation changes in human cancer. Int J Mol Sci. (2018) 19:1166. doi: 10.3390/ijms19041166
120. Giacomini A, Ghedini GC, Presta M, Ronca R. Long pentraxin 3: A novel multifaceted player in cancer. Biochim Biophys Acta Rev Cancer. (2018) 1869:53–63. doi: 10.1016/j.bbcan.2017.11.004
121. Kampo S, Ahmmed B, Zhou T, Owusu L, Anabah TW, Doudou NR, et al. Scorpion venom analgesic peptide, BmK AGAP inhibits stemness, and epithelial-mesenchymal transition by down-regulating PTX3 in breast cancer. Front Oncol. (2019) 9:21. doi: 10.3389/fonc.2019.00021
122. Ronca R, Alessi P, Coltrini D, Di Salle E, Giacomini A, Leali D, et al. Long pentraxin-3 as an epithelial-stromal fibroblast growth factor-targeting inhibitor in prostate cancer. J Pathol. (2013) 230:228–38. doi: 10.1002/path.4181
123. Basile A, Moschetta M, Ditonno P, Ria R, Marech I, De Luisi A, et al. Pentraxin 3. (PTX3) inhibits plasma cell/stromal cell cross-talk in the bone marrow of multiple myeloma patients. J Pathol. (2013) 229:87–98. doi: 10.1002/path.4081
124. Leali D, Alessi P, Coltrini D, Ronca R, Corsini M, Nardo G, et al. Long pentraxin-3 inhibits FGF8b-dependent angiogenesis and growth of steroid hormone-regulated tumors. Mol Cancer Ther. (2011) 10:1600–10. doi: 10.1158/1535-7163.MCT-11-0286
125. Locatelli M, Ferrero S, Martinelli Boneschi F, Boiocchi L, Zavanone M, Maria Gaini S, et al. The long pentraxin PTX3 as a correlate of cancer-related inflammation and prognosis of malignancy in gliomas. J Neuroimmunol. (2013) 260:99–106. doi: 10.1016/j.jneuroim.2013.04.009
126. Ma D, Zong Y, Zhu ST, Wang YJ, Li P, Zhang ST. Inhibitory role of pentraxin-3 in esophageal squamous cell carcinoma. Chin Med J. (2016) 129:2233–40. doi: 10.4103/0366-6999.189921
127. Zambirinis CP, Pushalkar S, Saxena D, Miller G. Pancreatic cancer, inflammation, and microbiome. Cancer J. (2014) 20:195–202. doi: 10.1097/PPO.0000000000000045
128. Choi B, Lee EJ, Park YS, Kim SM, Kim EY, Song Y, et al. Pentraxin-3 silencing suppresses gastric cancer-related inflammation by inhibiting chemotactic migration of macrophages. Anticancer Res. (2015) 35:2663–8. Available online at: http://ar.iiarjournals.org/content/35/5/2663.long
129. Tsuji S, Midorikawa Y, Seki M, Takayama T, Sugiyama Y, Aburatani H. Network-based analysis for identification of candidate genes for colorectal cancer progression. Biochem Biophys Res Commun. (2016) 476:534–40. doi: 10.1016/j.bbrc.2016.05.158
130. Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, et al. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. (2015) 160:700–14. doi: 10.1016/j.cell.2015.01.004
131. Dahan R, Sega E, Engelhardt J, Selby M, Korman Alan J, Ravetch Jeffrey V. FcγRs modulate the anti-tumor activity of antibodies targeting the PD-1/PD-L1 axis. Cancer Cell. (2015) 28:285–95. doi: 10.1016/j.ccell.2015.08.004
132. Brandsma AM, Bondza S, Evers M, Koutstaal R, Nederend M, Jansen JHM, et al. Potent Fc receptor signaling by IgA leads to superior killing of cancer cells by neutrophils compared to IgG. Front Immunol. (2019) 10:704. doi: 10.3389/fimmu.2019.00704
133. Sanseviero E. NK cell-Fc receptors advance tumor immunotherapy. J Clin Med. (2019) 8:1667. doi: 10.3390/jcm8101667
134. Liu Q, Wang XY, Qin YY, Yan XL, Chen HM, Huang QD, et al. SPOCD1 promotes the proliferation and metastasis of glioma cells by up-regulating PTX3. Am J Cancer Res. (2018) 8:624–35. Available online at: http://www.ajcr.us/files/ajcr0067117.pdf
135. Chang WC, Wu SL, Huang WC, Hsu JY, Chan SH, Wang JM, et al. PTX3 gene activation in EGF-induced head and neck cancer cell metastasis. Oncotarget. (2015) 6:7741–57. doi: 10.18632/oncotarget.3482
136. Fang S, Sui D, Wang Y, Liu H, Chiang YJ, Ross MI, et al. Association of vitamin D levels with outcome in patients with melanoma after adjustment for C-reactive protein. J Clin Oncol. (2016) 34:1741–7. doi: 10.1200/JCO.2015.64.1357
137. She S, Xiang Y, Yang M, Ding X, Liu X, Ma L, et al. C-reactive protein is a biomarker of AFP-negative HBV-related hepatocellular carcinoma. Int J Oncol. (2015) 47:543–54. doi: 10.3892/ijo.2015.3042
138. Huang A, Cao S, Tang L. The tumor microenvironment and inflammatory breast cancer. J Cancer. (2017) 8:1884–91. doi: 10.7150/jca.17595
Keywords: C-reactive protein, serum amyloid P component, the long pentraxins, pentraxin 3, tumor
Citation: Wang Z, Wang X, Zou H, Dai Z, Feng S, Zhang M, Xiao G, Liu Z and Cheng Q (2020) The Basic Characteristics of the Pentraxin Family and Their Functions in Tumor Progression. Front. Immunol. 11:1757. doi: 10.3389/fimmu.2020.01757
Received: 16 March 2020; Accepted: 30 June 2020;
Published: 18 August 2020.
Edited by:
Silvano Sozzani, Sapienza University of Rome, ItalyCopyright © 2020 Wang, Wang, Zou, Dai, Feng, Zhang, Xiao, Liu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quan Cheng, Y2hlbmdxdWFuQGNzdS5lZHUuY24=; Zhixiong Liu, emhpeGlvbmdsaXVAY3N1LmVkdS5jbg==; Gelei Xiao, eGlhb2dlbGVpQGNzdS5lZHUuY24=
‡ORCID: Quan Cheng orcid.org/0000-0003-2401-5349
†These authors have contributed equally to this work and share first authorship
 Zeyu Wang1,2†
Zeyu Wang1,2† Xing Wang
Xing Wang Ziyu Dai
Ziyu Dai Gelei Xiao
Gelei Xiao Zhixiong Liu
Zhixiong Liu Quan Cheng
Quan Cheng