- 1Laboratory of Aquatic Animal Nutrition and Feed, Fisheries College, Guangdong Ocean University, Zhanjiang, China
- 2Aquatic Animals Precision Nutrition and High-Efficiency Feed Engineering Research Center of Guangdong Province, Zhanjiang, China
- 3Key Laboratory of Aquatic, Livestock and Poultry Feed Science and Technology in South China, Ministry of Agriculture, Zhanjiang, China
- 4Southern Marine Science and Engineering Guangdong Laboratory, Guangdong Ocean University, Zhanjiang, China
A 10-week feeding experiment was conducted to reveal the immune mechanism for soybean meal-induced enteritis (SBMIE) in hybrid grouper, Epinephelus fuscoguttatus ♀ × Epinephelus lanceolatus ♂. Four isonitrogenous and isolipidic diets were formulated by replacing 0, 10, 30, and 50% fish meal protein with soybean meal (namely FM, SBM10, SBM30, and SBM50, respectively). The weight gain rate of the SBM50 group was significantly lower than those of the other groups. Plica height, muscular layer thickness, and goblet cells of the distal intestine in the SBM50 group were much lower than those in the FM group. The intestinal transcriptomic data, including the transcriptome and miRNAome, showed that a total of 6,390 differentially expressed genes (DEGs) and 92 DEmiRNAs were identified in the SBM50 and FM groups. DEmiRNAs (10 known and 1 novel miRNAs) and their DE target genes were involved in immune-related phagosome, natural killer cell-mediated cytotoxicity, Fc gamma R-mediated phagocytosis, and the intestinal immune network for IgA production pathways. Our study is the first to offer transcriptomic and small RNA profiling for SBMIE in hybrid grouper. Our findings offer important insights for the understanding of the RNA profile and further elucidation of the underlying molecular immune mechanism for SBMIE in carnivorous fish.
Introduction
In recent years, the global contribution of fish meal to aquafeeds has sharply reduced (1). As a result, soybean meal, which is considered one of the most hopeful candidates for fish meal replacement, can partially or fully replace the fish meal but introduces many anti-nutritional factors. Consequently, fish enteritis induced by plant proteins has become one of the main challenges for sustainable aquaculture (2), and this occurs in a dose-dependent manner (1, 3).
Soybean meal-induced enteritis (SBMIE) has been found in many commercial fish species such as Atlantic salmon (Salmo salar) (4), grass carp (Ctenopharyngodon idella) (5), and turbot (Scophthalmus maximus L.) (6, 7), and mainly occurs in carnivorous fish (4, 6, 7). The symptoms of SBMIE are most apparent in the posterior/distal intestine of turbot (6, 7) and grass carp (5), the most important mucosal immune organ (8). Fish SBMIE has been found to be accompanied by a decrease in the height of villi and microvilli, downregulation of tight junction protein claudin-4, occluding, and ZO-1 mRNA levels, and upregulation of pro-inflammatory cytokine genes, including TNF-α, IL-1β, IL-8, and IL-16 (9–12), influencing the innate immune response. Zebrafish SBMIE is T cell-dependent and has a T helper (Th) 17 cytokine profile (13). What are the underlying immune mechanisms in fish SBMIE? The new omics technologies, including genomics, proteomics, and transcriptomics, have great potential for investigating and explaining the complex relationship between fish nutrition and immunity, both in intestine health and disease (14). At the genomic level, most components associated with T lymphocyte function have been identified in fish, suggesting that gut-associated lymphoid tissue has similar functionalities between fish and mammalian T lymphocytes (15). At the transcriptomic level, immune-related pathways of fish SBMIE have been gradually reported, showing that cytokine–cytokine receptor interaction, NOD-like receptor interaction, the intestinal immune network for IgA production, and the NF-kB, Jak-STAT, T-cell receptor, and TNF signaling pathways played key roles in response to SBM stress (3, 16). Fish meal replacement in fish diet by alternative protein sources could change the fish intestine proteome, including innate immune proteins (17).
The especially interesting fact is that miRNAs are involved in regulating intestine function, including epithelial cell growth (18), mucosal barrier function (19), and the development of gastroenteric diseases (20–22). An important aspect is that miRNA can also regulate mRNA expression in fish at a transcriptional level. Recently, studies of miRNAome in turbot intestinal function have reported that miRNAs contributed to the intestinal immune responses, preventing host infection, which the potential target genes of differentially expressed miRNAs were involved in multiple functional categories, including the RIG-I signaling pathway, immune defense/evasion, the toll-like receptor signaling pathway, and inflammatory responses (23). Also, it was found via small RNA sequencing that fish diet could affect the expression of intestinal miRNAs and target genes and immune-related pathways, including cell adhesion molecules, ECM–receptor interaction, the apoptosis signaling pathway, cytokine-cytokine receptor, and the VEGF signaling pathway (3, 24). However, there has been a lack of investigation of the underlying immune response by combining both transcriptomes and small RNA sequencing, and this requires further elucidation, especially in carnivorous fish.
Hybrid grouper (Epinephelus fuscoguttatus♀ × Epinephelus lanceolatus♂) is a carnivorous fish species that is the main farmed species in China due to its outstanding delicious taste and better growth rate and survival compared with the broodfish. Information on how nutrition influences the intestine health of hybrid grouper has been gradually accumulated in recent years (25–30). Our previous research used metabolomics technology to identify 17 potential markers of SBMIE in hybrid grouper (31). However, this study aims to reveal the immune-related miRNA-mRNA regulation network for SBMIE in hybrid grouper by integrative transcriptome and small RNA profiling from the perspective of molecular immunology and to provide another important insight to further solve the problem of fish intestinal health.
Materials and Methods
Experimental Diets
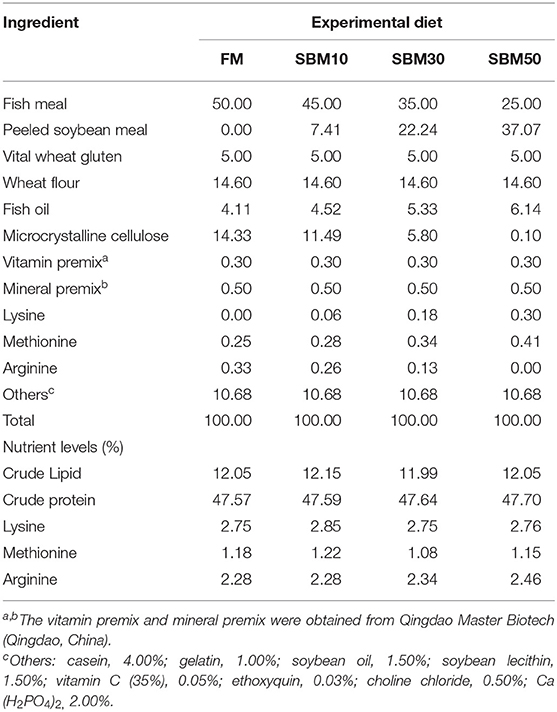
The use of hybrid grouper juveniles was approved by the Animal Research and Ethics Committees of Guangdong Ocean University, China. Four isonitrogenous and isolipidic diets were formulated to contain 0, 7.41, 22.24, and 37.07% of soybean meal (SBM) by replacing 0% (FM, control), 10% (SBM10), 30% (SBM30), and 50% (SBM50) of fish meal (FM) protein, respectively. The formulation of the basic experimental feeds is presented in Table 1. All ingredients were systematically mixed with lipid sources such as fish oil, soybean oil, and soybean lecithin and then purified water was added to produce a homogenous mixture. The dough was pelleted through a double helix extrusion mechanism (F-75, South China University of Technology, China). Feeds (2.5 mm diameter) were air-dried and then stored at −20°C until feeding.
Feeding Trial
Hybrid grouper Juveniles were obtained from a native species farm (Zhanjiang, China). All fish were adapted under the feeding system for 2 weeks by feeding with a commercial diet. Uniformly sized fish (mean initial weight ± SE = 17.01 ± 0.04 g) were randomly divided into four groups in triplicate, with 30 individuals in each fiberglass tank (300 L). The fish were slowly fed twice a day at 08:00 and 17:00 for 10 weeks. During the experiment, the water temperature fluctuated from 28 to 30°C, the dissolved oxygen concentration was kept at >7 mg/L, and ammonia and nitrate were kept at <0.03 mg/L.
Sample Collection
Before the termination of the 10-week feeding experiment, the fish per tank were fasted for 24 h before collecting samples and were then counted and weighed to determine growth indexes, including weight gain rate, feed conversion ratio, and survival rate. After weighing, distal intestines of two fish per tank were collected and instantly transferred to 4% paraformaldehyde solution for histological examination. At the same time, distal intestines of another three fish per tank were collected as a single sample and instantly frozen in liquid nitrogen, then stored at −80°C for RNA extraction. Based on growth performance (see section Growth Performance) and histological examination, small RNA and transcriptome analyses were performed on distal intestine samples of the FM and SBM50 groups to ensure the maximum difference between samples. Thereby, the probability of detecting differential expression was increased between samples.
Intestinal Morphology
The fixed distal intestine samples from FM and SBM50 groups were dehydrated in a series of graded ethanol and embedded in paraffin. Distal intestine sections (7 μm thick) from each sample were cut and then stained with hematoxylin/eosin. The sections were observed under an inverted microscope (Nikon, Japan), and 10 plicas and muscle layer thicknesses (MLT) were randomly selected per slice. Plica height (PH), plica width (PW), MLT, and the number of goblet cells (GC) per slice were measured using the image acquisition software (NIS Elements, version 4.60, Nikon, Japan).
Transcriptome Sequencing and de novo Assembly
One microgram total RNA from the FM and SBM50 treatment groups was used for transcriptome library preparation. Total RNA was purified by beads containing oligo (dT). First-strand cDNA was then generated in a First-Strand Reaction System by PCR, and the second-strand cDNA was generated as well. The cDNA fragments with adapters were amplified by PCR, and the products were purified using AMPure XP Beads. The library was validated on the Agilent Technologies 2100 bioanalyzer for quality control. Transcriptome sequencing was carried out on a BGISEQ-500 platform (BGI-Shenzhen, China). Trinity (32) was used to achieve de novo assembly with clean reads, and Tgicl (33) was then used to cluster transcripts to Unigenes. The expected number of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) was used to calculate the mRNA gene expression. Differentially expressed genes (DEGs) of two groups (SBM50 and FM) were quantified using the cutoff |log2FC| >1, P < 0.001 by DESeq R package (34, 35). After assembly, All-Unigenes were searched and annotated against the publicly available protein databases, including Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), KOG (EuKaryotic Orthologous Groups), Swiss-Prot, and GO (Gene Ontology). The pathway assignments were performed by sequence searches against the KEGG (Kyoto Encyclopedia of Genes and Genomes) database. KEGG terms with corrected P-values (Q-values) ≤ 0.05 were considered significant. Transcriptome (de novo assembly) sequencing data were deposited into the NCBI SRA database with the accession number SUB7020170.
Construction and Sequencing of Small RNA Libraries
Six small RNA libraries were constructed from the FM and SBM50 treatment groups. Total RNA extraction was performed from the distal intestine using Trizol Reagent (Invitrogen, USA). Subsequently, 1 μg of total RNA per sample was used for small RNA sequencing. The quality of RNA samples was evaluated using the Agilent 2100 Bioanalyzer. Small RNA fractions were ligated to 3′ and 5′ adapter. Quantitative reverse transcription PCR (RT-PCR) was carried out on the adaptor-ligated small RNAs. PCR products were purified by QIAquick Gel Extraction Kit (Qiagen, Germany) and used for sequencing on the BGISEQ-500 platform (BGI-Shenzhen, China). Clean reads were obtained by cleaning low-quality tags, removing adapter sequences, and filtering adaptor-ligated contaminants and sequences fewer than 18 nucleotides (nt). The final reads were mapped to the Hypoplectrus puella (GCA_900610375.1) reference genome by Bowtie2 (36). Clean reads were compared against small RNAs (rRNA, scRNA, snoRNA, snRNA, tRNA, and mRNA) using the Rfam database to annotate small RNA sequences. Finally, the miRBase20.0 was used to look for know miRNA. The hairpin structures were used to predict novel miRNAs using miRDeep2 software (37).
miRNA expression levels were compared between the SBM50 and FM groups to identify differentially expressed miRNAs (DEmiRNAs). Firstly, data were normalized to obtain transcripts per million (TPM) values using the following formula: normalized expression = actual miRNA count/(total reads) ×1,000,000 (38). Fold-change values were then calculated based on log2 (SBM50/FM) expression. The corrected P-value corresponds to the differential gene expression test using the Bonferroni method (39). Differential miRNA expression between two groups was analyzed with DESeq software based on the following thresholds: P-value ≤ 0.01 and |log2 ratio| ≥ 1. RNAhybrid (40) and miRanda (41) software predicted the potential target genes of miRNA candidates, as described previously elsewhere (42, 43). The DAVID gene annotation tool was used for the KEGG pathway annotation of predicted miRNA targets. Small RNA sequencing data were deposited into the NCBI SRA database with the accession number SUB7175134.
Network Analysis of DEmiRNA and DEG Interaction
Pearson's correlation coefficients between DEmiRNAs and their target genes were calculated using the correlation function in RStudio. To obtain the positive and negative correlations between two groups, the potential target genes of miRNAs were overlapped with the identified upregulated or downregulated DEGs, respectively. All of the relationship pairs between DEmiRNAs and their DE target genes were used to construct the interaction network using Cytoscape v.3.7.2 software.
Validation by Real-Time Quantitative PCR (RT-qPCR)
RT-qPCR validation was carried out on the same samples used for transcriptome sequencing (n = 3). Primers were designed from the candidate gene sequences by premier 5.0 software and the online Primer-BLAST program. Primers used in this study are provided in Table S1. One microgram total RNA for RNA sequencing was reverse transcribed into cDNA. Real-time PCR assays were conducted on a CFX96 real-time PCR Detection System (Bio-Rad, Hercules, CA) with 5 μL SYBR Green Master Mix (Takara, China). β-actin was selected as the reference gene according to a previous study (26). The small RNA of the same samples used for sequencing was extracted using an RNAiso for Small RNA Kit (Takara, China) according to the manufacturer's protocol. Subsequently, the first-strand cDNA was synthesized for mature miRNA expression analysis by a Mir-X™ miRNA First-Strand Synthesis Kit (Code No. 638315, Takara, China). The qPCR was carried out using a miRNA SYBR Green RT-qPCR Kit (Takara, China) with the provided miRNA reference gene (U6). Relative quantitative levels were calculated based on the 2−ΔΔCT method (44).
Statistical Analysis
The normal distribution and the homogeneity of the variance of growth indexes were tested, followed by one-way analysis of variance and Tukey s test. Morphological analysis between two groups was assessed by two-tailed unpaired Student's t-test (GraphPad). For statistically significant differences, P < 0.05 was required. All statistical analyses were carried out using SPSS 24.0 software. The barplot was generated by Graphpad Prism 8.0.1 software.
Results
Growth Performance
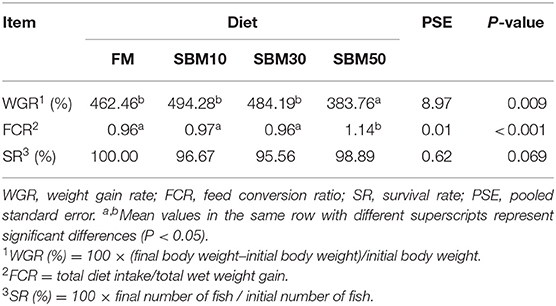
The survival rate (SR) was not affected by dietary treatment levels (P > 0.05, Table 2). The weight gain rate (WGR) of the SBM50 group was significantly lower than that of other groups (P < 0.05). The feed conversion ratio (FCR) of the SBM50 group was significantly higher than that of the other groups (P < 0.05).
Histological Examinations and Intestinal Morphometry
More swelling of the lamina propria (LP) of the SBM50 group was observed compared with the FM group, and the intestinal villi of the SBM50 group showed signs of shedding (Figures 1A,B). The plica height (PH), muscular layer thickness (MLT), and number of goblet cells (GC) of the SBM50 group were much lower than those of the FM group (Figure 1C, P < 0.01). There was no significant difference in plica width (PW) between the FM and SBM50 groups (P > 0.05).
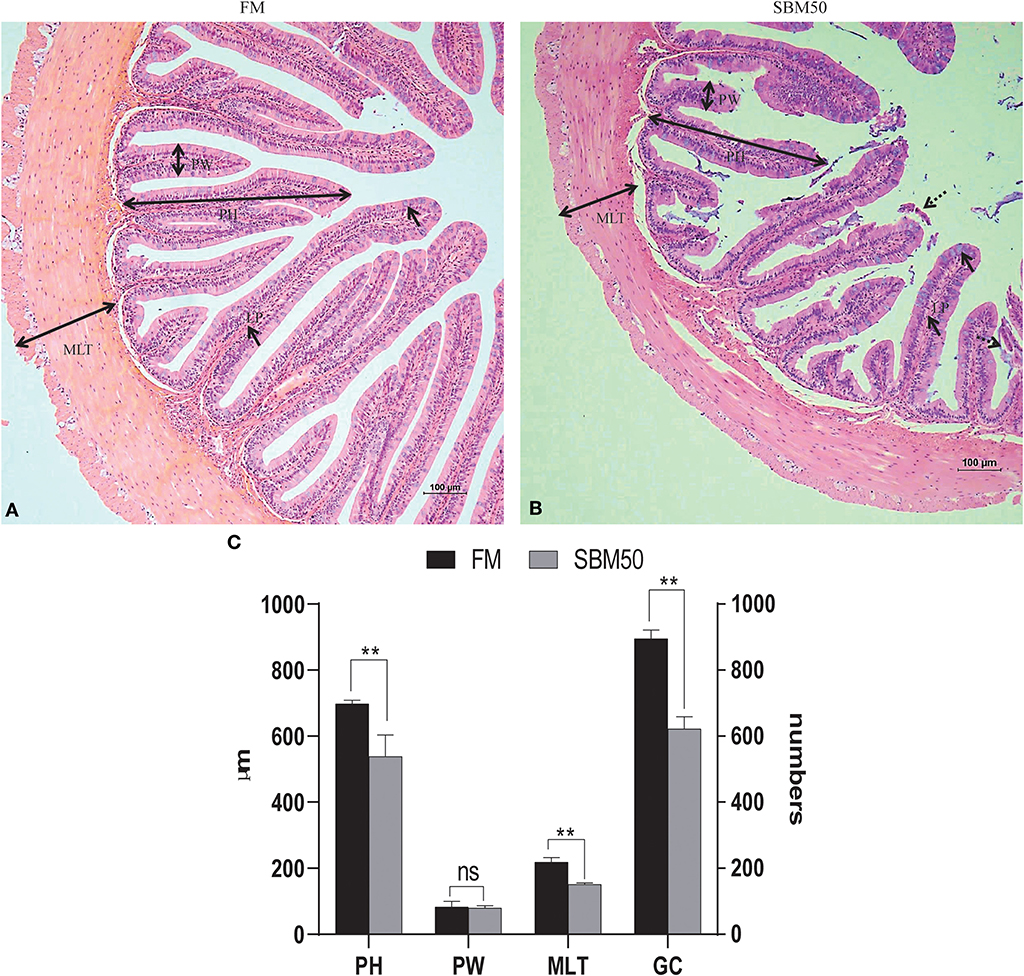
Figure 1. Histological analysis of the distal intestine in hybrid grouper. (A) HE staining of the FM group (100×). (B) HE staining of the SBM50 group (100×). (C) Intestinal morphometric measurements between FM and SBM50 groups. Mean ± S.E.M (n =10). LP, lamina propria; PH, Plica height; PW, plica width; MLT, muscular layer thickness; GC, goblet cells. **0.001<P<0.01 between two groups, NS, no significance.
Analysis of mRNA Sequencing
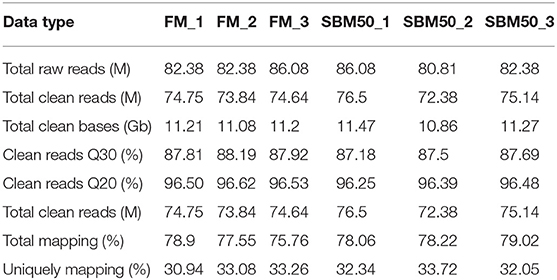
A total of six qualified libraries from the FM and SBM50 groups, with three biological replicates per treatment, were sequenced. An overview of the sequencing and assembly data is presented in Table 3. Approximately 33.49 and 33.6 Gb of clean reads were obtained in the FM and SBM50 groups. More than 87.1% of the reads had Q-scores at the Q30 level, and more than 75.8% of the clean reads were aligned. The length distribution of the Unigene in all six libraries is shown in Figure S1.
Differentially Expressed Genes and KEGG Pathway Analysis
In brief, 6,390 DEGs were identified in the FM and SBM50 groups, with 3,330 upregulated and 3,060 downregulated genes (Figure 2A). The results of clustered DEGs are shown in Figure 2B. Also, 184 and 146 genes were specifically expressed in the FM and SBM50 groups, respectively (Table S2). These total DEGs were enriched in 336 pathways, and each enriched pathway contained numbers of DEGs ranging from 1 to 150 (Figure 3A). From these, the top 20 KEGG pathways were mainly involved in disease process-related pathways, such as staphylococcus aureus infection (ko05150), systemic lupus erythematosus (ko05322), allograft rejection (ko05330), autoimmune thyroid disease (ko05320), leishmaniasis (ko05140), graft-vs.-host disease (ko05332), type I diabetes mellitus (ko04940), tuberculosis (ko05152), viral myocarditis (ko05416), and asthma (ko05310), and immune system-related pathways, such as natural killer cell-mediated cytotoxicity (ko04650), Fc gamma R-mediated phagocytosis (ko04666), and the intestinal immune network for IgA production (ko04672), were significantly enriched (Q < 0.0001). Also, upregulated genes were significantly enriched in immune and inflammatory-related pathways such as phagosome (ko04145), natural killer cell-mediated cytotoxicity, Fc gamma R-mediated phagocytosis, the intestinal immune network for IgA production, and NF-kappa B signaling pathways (ko04064) (Q < 0.001, Figure 3B). The downregulated genes were mainly involved in lipid metabolisms such as fat digestion and absorption (ko04975), biosynthesis of unsaturated fatty acids (ko01040), cholesterol metabolism (ko04979), fatty acid elongation (ko00062), and linoleic acid metabolism (ko00591) (Q < 0.01, Figure 3C).
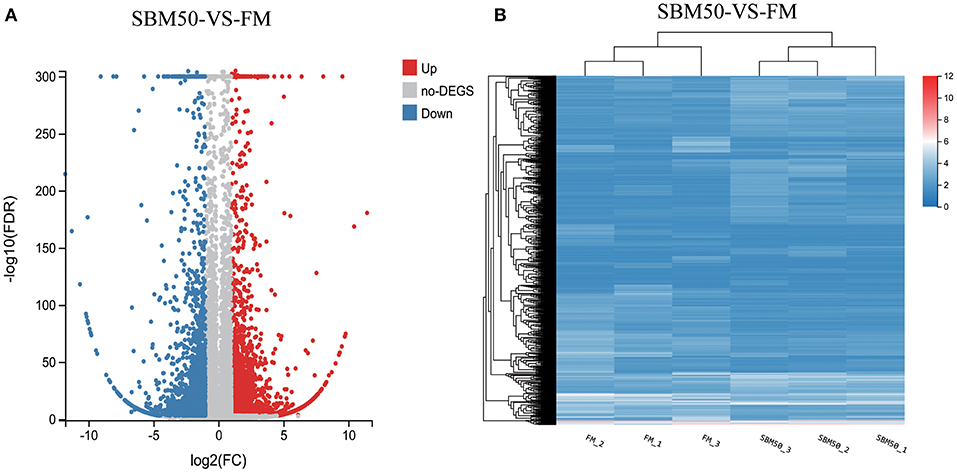
Figure 2. Volcano plot (A) and hierarchical cluster (B) analysis of differentially expressed genes (DEGs) in the FM and SBM50 groups. (A) Each point in the figure indicates a gene, the horizontal axis indicates the numerical value of DEGs, and the vertical axis indicates the negative logarithm of P-value-FDR. Red and blue dots represent the upregulation and downregulation of DEGs, respectively. Gray dots represent no differential expression of genes. (B) The color indicates the level of gene expression. Red and blue colors represent the high and low expression of genes, respectively.
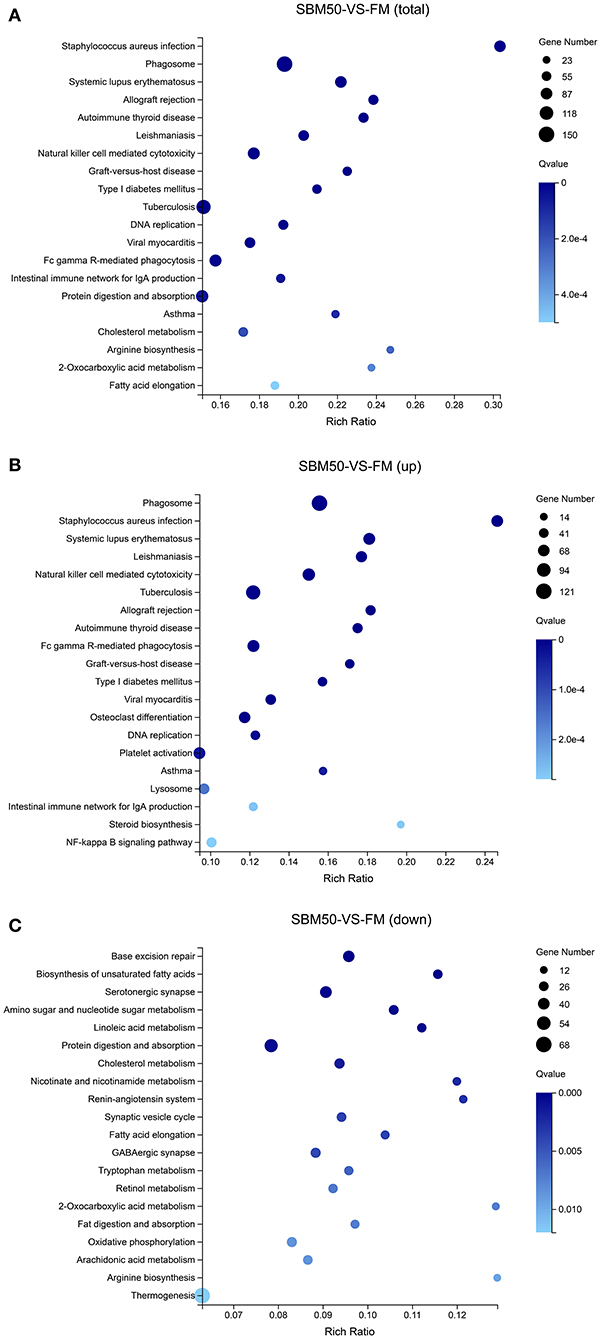
Figure 3. Enrichment analysis of the top 20 KEGG pathways for differentially expressed genes (DEGs). The horizontal axis represents the rich ratio, and the vertical axis represents the name of the KEGG pathway. The size of the bubble represents the number of genes annotated to the pathway. The color expresses the enriched Q-value. (A) KEGG pathways enriched by the total DEGs. (B) KEGG pathways enriched by the upregulated DEGs. (C) KEGG pathways enriched by the downregulated DEGs.
Analysis of Small RNA Sequencing
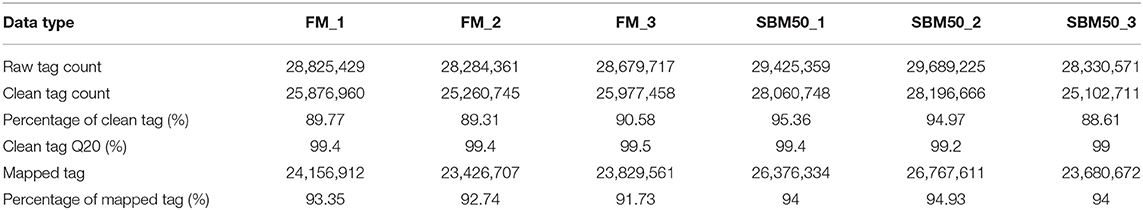
Six small RNA libraries were constructed, with three biological replicates per treatment. A total of 85,789,507 and 87,445,155 raw reads were found in the FM and SBM50 groups, respectively (Table 4). Also, 77,115,163 and 81,360,125 clean reads were found in the FM and SBM50 groups, respectively. The total mapped tags to the reference genome in the FM and SBM50 groups were 71,413,180 (92.60%) and 76,824,617 (94.43%), respectively. Most small RNAs were 21–23 nt in length in all six libraries, with 22 nt being the most frequent length (Figure S2), and more than 61.7% were miRNA in the catalog of small RNA in all six libraries (Figure S3). A total of 682 mature miRNAs (Table S3) and 29 novel miRNAs (Table S4) were identified in these six small RNA libraries.
Differential Expression of miRNAs and Potential Target Gene Enrichment
Concerning the miRNA sequencing data analysis, a total of 92 DEmiRNAs were identified in the FM and SBM50 groups, with 88 known and 4 novel miRNAs (Figure 4A, Table S5). Also, 55 and 37 miRNAs were upregulated and downregulated, respectively. Cluster analysis of the DEmiRNAs was shown in Figure 4B. It was further observed that 2 miRNAs (miR-143_1, miR-21-5p) and 4 miRNAs (miR-21-5p, miR-143_1, miR-143_2, and miR-194-5p) exhibited higher expression (TPM >10,000) in the FM and SBM50 groups, respectively. Furthermore, 5 miRNAs (miR-9-3p, miR-23a-3-5p, miR-9-4-5p, novel_mir29, miR-219-5p_1, and novel_mir21) were uniquely expressed in the SBM50 group (Table S6).
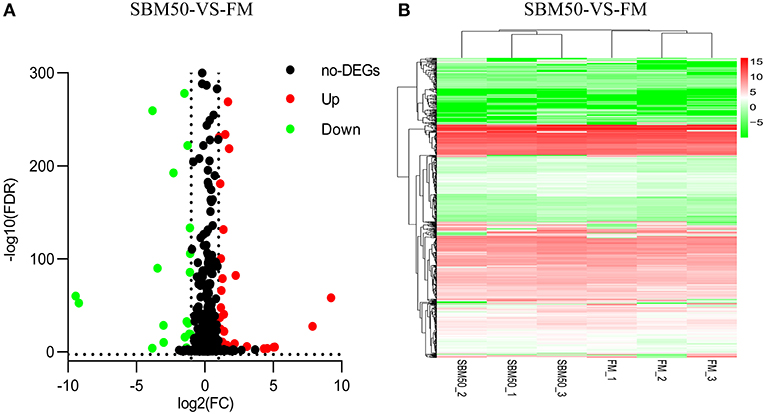
Figure 4. Volcano plot (A) and hierarchical cluster (B) analysis of differentially expressed miRNAs (DEmiRNAs) in the FM and SBM50 groups. (A) Each point in the figure indicates a gene, the horizontal axis represents the numerical value of gene expression, and the vertical axis represents the negative logarithm of P-value-FDR. Red and green dots indicate the upregulation and downregulation of DEmiRNAs, respectively. Black dots indicate no differential expression of genes. (B) The color indicates the level of gene expression. Red and green colors represent the high and low expression of genes, respectively.
In total, 15,317 target genes were predicted using the miRanda and RNAhybrid tools. The potential target genes were annotated to 340 pathways (Figure 5). The top 20 enriched pathways were most enriched in gastric cancer (ko05226), amoebiasis (ko05146), osteoclast differentiation (ko04380), ECM–receptor interaction (ko04512), and carbon metabolism (ko01200).
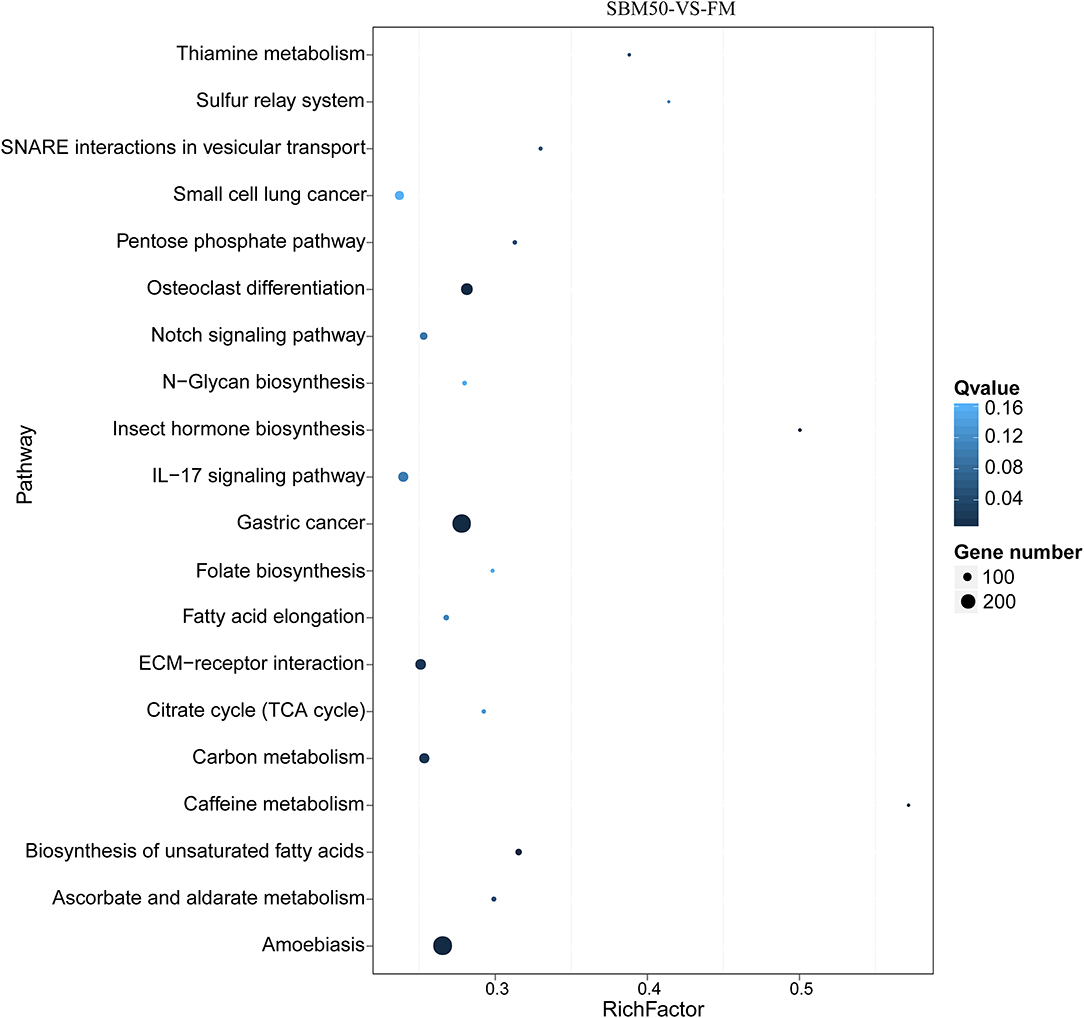
Figure 5. The top 20 KEGG pathways of predicted target genes in FM and SBM50 groups. The horizontal axis represents the rich ratio, and the vertical axis represents the name of the KEGG pathway. The size of the bubble represents the number of genes annotated to the pathway. The color represents the enriched Q-value.
Integration Analysis of the DEmiRNAs and DEGs
A total of 244 miRNA–mRNA interactions were identified in the FM and SBM50 groups, with the involvement of 92 DEmiRNAs and 211 DEGs (Figure 6, Table S7). A positively correlated expression pattern was seen for 180 mRNA–miRNA pairs. Most miRNAs had multiple possible target genes, while different miRNAs could regulate the same target. For instance, miR-196 was the regulator of CL4654.Contig2_All, CL7786.Contig3_All, and Unigene16944_All, whereas miR-20a-5p and miR-459-5p_1 could regulate the expression of CL8081.Contig7_All.
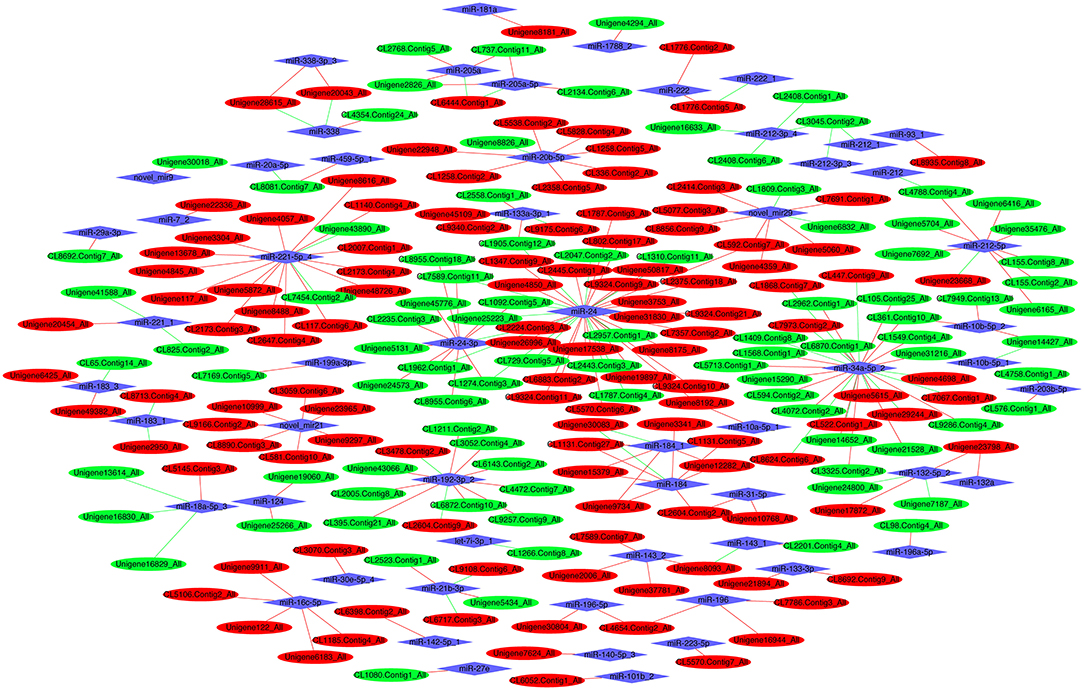
Figure 6. The interaction network of differentially expressed miRNAs (DEmiRNAs) combined with differentially expressed mRNAs (DEGs). Red and green ellipses represent upregulated and downregulated mRNA, respectively. Blue diamonds represent DEmiRNAs. Red and green lines represent a positive and negative correlation, respectively, between DEmiRNAs and DEGs.
KEGG Enrichment Analysis of Differential Target Genes
Differential target genes were predicted and annotated to 241 pathways. The top 5 of the top 20 KEGG pathways were phagosome, tuberculosis, osteoclast differentiation, natural killer cell-mediated cytotoxicity, and mineral absorption (ko04978), respectively (Figure 7, Table S8).
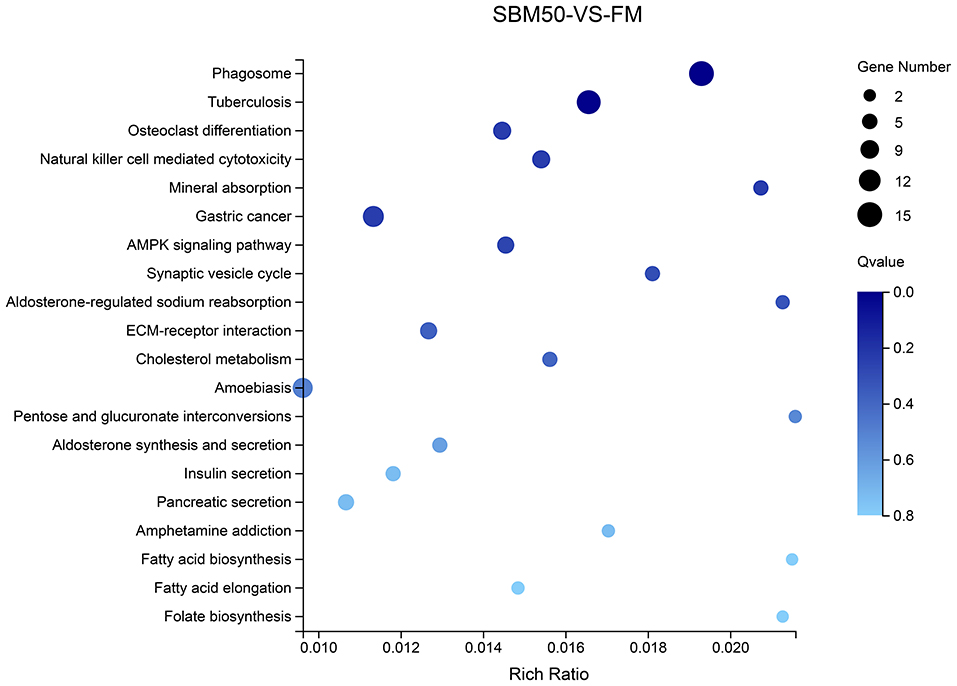
Figure 7. The top 20 KEGG pathways of differential target genes of differentially expressed miRNAs (DEmiRNAs) in the FM and SBM50 groups. The horizontal axis represents the rich ratio, and the vertical axis represents the name of the KEGG pathway. The size of the bubble represents the number of genes annotated to the pathway. The color indicates the enriched Q-value.
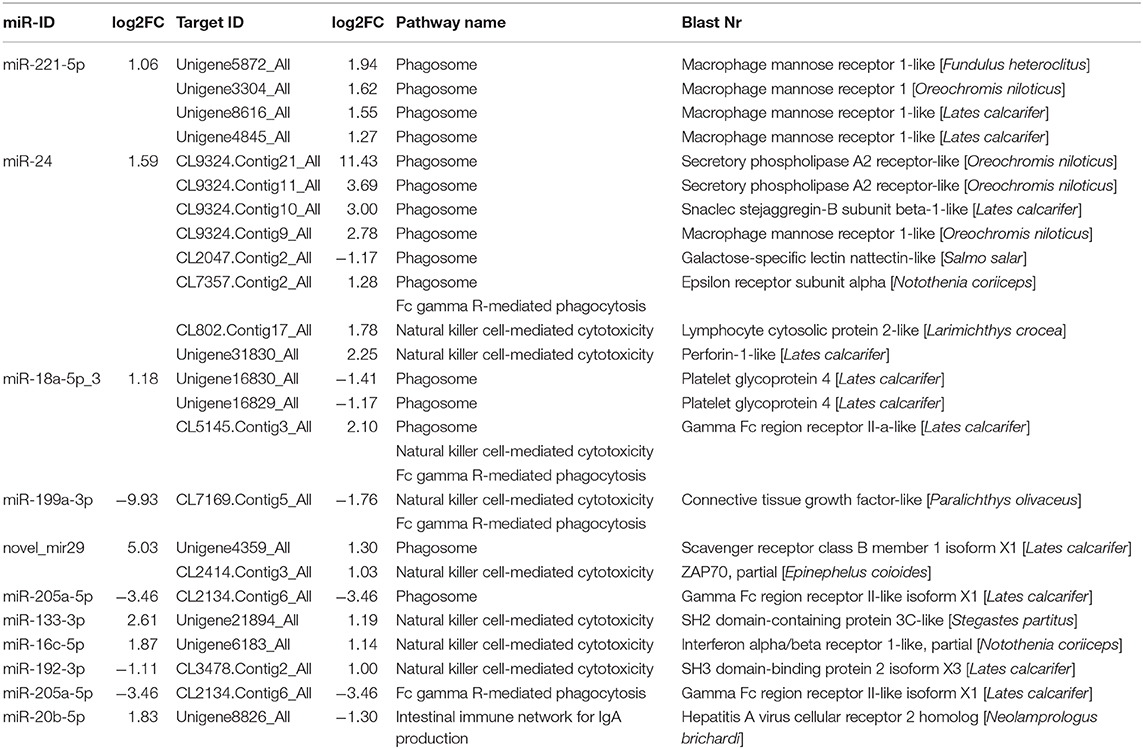
To further explain the possible immune response for SBMIE, combined with the KEGG enrichment pathway of DEGs found that 15 miRNA–mRNA pairs involved in the phagosome, 8 miRNA–mRNA pairs involved in natural killer cell-mediated cytotoxicity, 4 miRNA–mRNA pairs involved in Fc gamma R-mediated phagocytosis, and 1 miRNA–mRNA pair involved in the intestinal immune network for IgA. Details concerning the miRNA–mRNA pairs in immune-related pathways are provided in Table 5.
Real-Time Quantitative PCR Validation
In total, 10 miRNAs and 10 mRNAs were selected to test their expression and the results suggested that DEmiRNAs except for miR-194-5p and DEGs showed a similar expression pattern to the high-throughput sequencing data (Figure 8). There were no significant differences in expression levels of mRNA genes (Unigene1343_All and CL7662.Contig2_All) and miRNA (miR-27e, −23a-3-5p, and miR-194-5p) between the two groups (P > 0.05). Also, fold-changes between miRNA or mRNA and their respective qPCR results correlated well (Table S9).
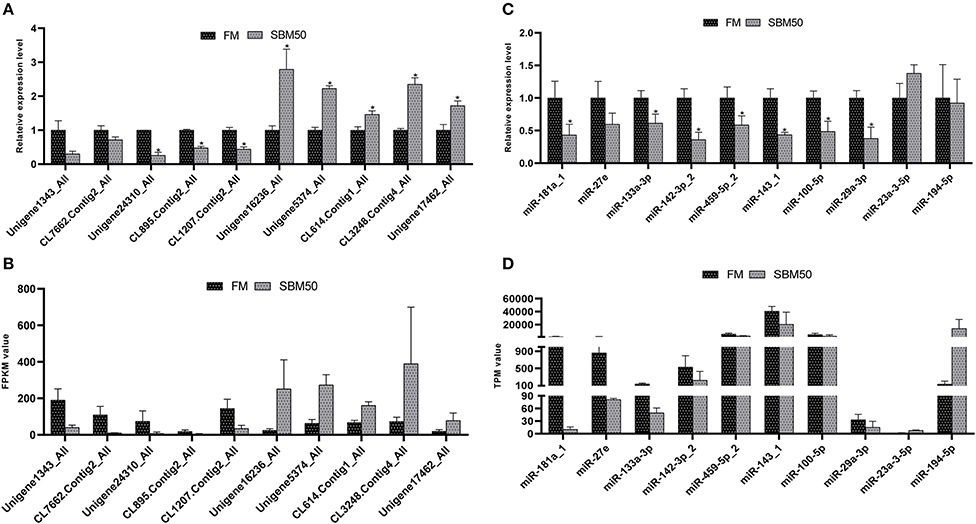
Figure 8. RT-qPCR validation of differentially expressed miRNAs (DEmiRNAs), and genes (DEGs) in the FM and SBM50 groups. (A) The relative expression levels of mRNA between the two groups. (B) The value of fragments per kilobase of transcript sequence per million base pairs sequenced (FPKM) of the DEGs between the two groups. (C) The relative expression levels of the DEmiRNAs between the two groups. (D) The transcripts per million (TPM) values of miRNA between the two groups. The results of (A,C) are presented as the mean ± S.E.M (n =3). The sequencing results of (B,D) are presented as the mean. *P < 0.05 between two groups.
Discussion
The nutritional trial of this study revealed that a 22.4% SBM, i.e., 30% substitution of FM protein, did not significantly influence the growth of hybrid grouper compared to the FM diet. However, the effect of SBM on growth was substitution-related, and the growth performance became progressively compromised with increasing SBM substitution level, which is consistent with the previous studies (45, 46). In this study, growth performance was significantly reduced when dietary SBM content reached 370 g/kg, i.e., 50% substitution of the FM protein. Hybrid grouper fed with SBM replacing 50% of the FM protein diet showed swelling of the lamina propria and reduction of plica height, muscular layer thickness, and number of goblet cells in the distal intestine of hybrid grouper, indicating that enteritis and intestinal injury had appeared. The severity of histopathological changes observed under SBM application relies on the level of soybean inclusion. SBMIE is now commonly used as a model for the study of intestinal inflammation in fish (6, 7, 47). The histopathological changes of SBMIE have been widely researched and are characterized by a swelling of the subepithelial mucosa and lamina propria, a reduced mucosal fold height, a profound infiltration of various inflammatory cells, and loss of normal enterocyte supranuclear absorptive vacuolization (13, 31, 48). Liu et al. (10) found that turbot fed with SBM replacing 40% of the fish meal protein diet showed obvious enteropathy, including a reduction of absorptive surface and obvious infiltration of mixed leukocytes in the lamina propria.
To reveal the underlying immune response of fish SBMIE, 6,390 DEGs in the distal intestine of hybrid grouper were identified in this study. Also, the enhanced gene expressions were found to involved in immune- and inflammatory-related pathways, including phagosome, natural killer cell-mediated cytotoxicity, the intestinal immune network for IgA production, Fc gamma R-mediated phagocytosis, and the NF-kappa B signaling pathway. The above results suggested that these pathways played a vital role in fish SBMIE. Similar immune-related pathways have been reported in SBMIE in other carnivorous fish, such as salmon (16, 49) and turbot (6, 7). It is worth noting that the downregulated gene expression was in KEGG pathways mainly involved in lipid metabolisms such as biosynthesis of unsaturated fatty acids, linoleic acid metabolism, cholesterol metabolism, fat digestion, absorption, and arachidonic acid metabolism. This may indicate that impaired lipid metabolism could be a consequence of “tissue malfunction” (47, 49). The results enable a better understanding of why LC-PUFA biosynthesis, cholesterol biosynthesis, lipid digestion, and the PPAR signaling pathway of distal intestine were influenced when Atlantic salmon ingested feed in which the fish meal was partially replaced by soybean meal (1, 16).
Despite the major contribution of mRNAs, miRNAs also play a key role during immune processes in fish SBMIE. In this study, predicted target genes for DEmiRNAs are annotated to 340 signaling pathways. Immune-related pathways, including ECM–receptor interaction, the NF-kappa B signaling pathway, and the IL-17 signaling pathway, were enriched. The downregulation of miR-192-3p and miR-212-5p expression involved in the regulation of the ECM–receptor interaction pathway. The down-regulation of genes involved in the ECM–receptor interaction pathway in response to SBM stress was also reported in Grass carp (5). To further introduce miRNAs in detail, enteritis-related miRNAs have been found both in human and mammals. The present miRNA results include inflammatory bowel diseases (IBD) related-miRNAs, such as miR-124, miR-24, miR-221, and miR-132. The results of this study showed that upregulation of miR-124 expression and downregulation of miR-24, miR-221, and miR-132 expression were observed in the SBM50 group (Table S5). Similar expression patterns were found in colon tissues of children with active ulcerative colitis (UC), where decreased levels of miR-124 appeared to enhance expression and activity of STAT3, which could induce inflammation and pathogenesis (50). There were elevated levels of miR-24, miR-221, and miR-132 in colonic biopsies from UC, which suggested that they are an important regulator of the intestine barrier that may be essential in the pathogenesis of IBD (51). The above results indicated that miR-124, miR-24, miR-221, and miR-132 may play important roles in SBMIE of hybrid grouper juveniles, which may suggest their therapeutic potential.
To further explain the possible immune response for SBMIE through integrative transcriptomic and small RNA sequencing, the KEGG enrichment pathways of differential target genes were analyzed. Combined with the KEGG enrichment pathways of DEGs, it was concluded that immune-related signaling pathways such as phagosome, natural killer cell-mediated cytotoxicity, Fc gamma R-mediated phagocytosis, and the intestinal immune network for IgA production were enriched. The most enhanced gene expression being in the phagosome pathway suggested the involvement of a macrophage as the main intestine phagocyte during enteritis (52). A total of 14 miRNA–mRNA pairs also suggested that the phagosome pathway could play a key role in intestinal inflammation. In addition, this study identified 92 DEGs related to the natural killer cell-cytotoxicity pathway. The role of this pathway in the immune response to pathogens has also been reported in different fish species such as large yellow croaker (Larimichthys crocea) (53) and half-smooth tongue sole (Cynoglossus semilaevis) (54). A previous study reported that mammalian natural killer (NK) cells mediated cytotoxic activity via two distinct pathways (55). NK cells can release cytotoxic granules, including perforin and granzymes, on the surface of diseased cells. Granzymes can then stimulate caspase activation, mitochondrial dysfunction, or apoptosis (55). In this study, the upregulation of granzyme (gene id: Unigene24888_All) and perforin (gene id: Unigene31830_All, Blast Nr annotated in Table 5) gene expressions indicated that granule-mediated cytotoxicity may be triggered by the targeted release of lytic granules toward a locally attached target cell. Wu et al. (5) reported that the intestinal immune network for the IgA production pathway was upregulated in the early stages of Grass carp in response to high-SBM-content stress. Similar results were also reported in the present study: most genes in pathways of Fc gamma receptor-mediated phagocytosis and the intestinal immune network for IgA production were significantly upregulated. The role of the two pathways in immune response to pathogens has also been reported in fish such as half-smooth tongue sole (53), large yellow croaker (54), and darkbarbel catfish (Pelteobagrus vachellii) (56). Previous studies reported that the target genes of the DEmiRNAs were involved in pathways of Fcγ R-mediated phagocytosis and the intestinal immune network for IgA production (57, 58). In this study, related miRNA–mRNA pairs were also enriched in the two pathways mentioned above (Table 5). Thus, our results suggested that phagosome, natural killer cell-mediated cytotoxicity, Fc gamma R-mediated phagocytosis, and the intestinal immune network for IgA production pathways may play a vital role in SBMIE of carnivorous fish.
In conclusion, our study is the first to offer the transcriptomic and small RNA profiles for SBMIE in hybrid grouper. Overall, 6,390 mRNAs and 92 miRNAs were differentially expressed under dietary SBM stress. Our findings support the notion that DEmiRNAs and their target mRNAs play an important role in immune regulation. Also, investigation of KEGG enrichment pathways by integrative transcriptomic and small RNA profiling revealed that the immune mechanism for SBMIE in hybrid grouper may be associated with the phagosome, natural killer cell-mediated cytotoxicity, Fc gamma R-mediated phagocytosis, and the intestinal immune network for IgA production pathways. Our findings offer important insights for the understanding of the RNA profiles and further elucidation of the underlying molecular immune mechanism for SBMIE in carnivorous fish.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SUB7020170; https://www.ncbi.nlm.nih.gov/, SUB7175134.
Ethics Statement
The animal study was reviewed and approved by The Animal Research and Ethics Committees of the Guangdong Ocean University, China.
Author Contributions
BT and SC conceived the project. GY performed animal experiments. XD bought feed ingredients. SZ bought sample analysis regent kits. HL and QY managed and repaired experimental instruments. YH did data analysis and wrote the manuscript. SC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was financially supported by the National Key R&D Program of China (2019YFD0900200), the China Agriculture Research System (CARS-47), the National Natural Science Foundation of China (NSFC 31772864), and the Fund of Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (ZJW-2019-06).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01502/full#supplementary-material
References
1. Krol E, Douglas A, Tocher DR, Crampton VO, Speakman JR, Secombes CJ, et al. Differential responses of the gut transcriptome to plant protein diets in farmed Atlantic salmon. BMC Genomics. (2016) 17:156. doi: 10.1186/s12864-016-2473-0
2. Hu H, Kortner TM, Karina G, Elvis C, John T, Ashild K. Intestinal fluid permeability in Atlantic Salmon (Salmo salar L.) is affected by dietary protein source. PLoS ONE. (2016) 11:e0167515. doi: 10.1371/journal.pone.0167515
3. Wu N, Wang B, Cui ZW, Zhang XY, Cheng YY, Xu X, et al. Integrative transcriptomic and microRNAomic profiling reveals immune mechanism for the resilience to soybean meal stress in fish gut and liver. Front Physiol. (2018) 9:1154. doi: 10.3389/fphys.2018.01154
4. Refstie S, Korsøen ØJ, Storebakken T, Baeverfjord G, Lein I, Roem AJ. Differing nutritional responses to dietary soybean meal in rainbow trout (Oncorhynchus mykiss) and Atlantic salmon (Salmo salar). Aquaculture. (2000) 190:49–63. doi: 10.1016/S0044-8486(00)00382-3
5. Wu N, Zhang XY, Huang B, Zhang N, Zhang XJ, Guo X, et al. Investigating the potential immune role of fish NCAMs: molecular cloning and expression analysis in mandarin fish. Fish Shellfish Immunol. (2015) 46:765–77. doi: 10.1016/j.fsi.2015.08.006
6. Liu Y, Chen Z, Dai J, Yang P, Xu W, Ai Q, et al. Sodium butyrate supplementation in high-soybean meal diets for turbot (Scophthalmus maximus L.): effects on inflammatory status, mucosal barriers and microbiota in the intestine. Fish Shellfish Immunol. (2019) 88:65–75. doi: 10.1016/j.fsi.2019.02.064
7. Gu M, Bai N, Xu B, Xu X, Jia Q, Zhang Z. Protective effect of glutamine and arginine against soybean meal-induced enteritis in the juvenile turbot (Scophthalmus maximus). Fish Shellfish Immunol. (2017) 70:95–105. doi: 10.1016/j.fsi.2017.08.048
8. Nowarski R, Jackson R, Flavell RA. The stromal intervention: regulation of immunity and inflammation at the epithelial-mesenchymal barrier. Cell. (2017) 168:362–75. doi: 10.1016/j.cell.2016.11.040
9. Wang YR, Wang L, Zhang CX, Song K. Effects of substituting fish meal with soybean meal on growth performance and intestinal morphology in orange-spotted grouper (Epinephelus coioides). Aquacult Rep. (2017) 5:52–7. doi: 10.1016/j.aqrep.2016.12.005
10. Liu Y, Chen Z, Dai J, Yang P, Hu H, Ai Q, et al. The protective role of glutamine on enteropathy induced by high dose of soybean meal in turbot, Scophthalmus maximus L. Aquaculture. (2018) 497:510–9. doi: 10.1016/j.aquaculture.2018.08.021
11. Tan C, Zhou H, Wang X, Mai K, He G. Resveratrol attenuates oxidative stress and inflammatory response in turbot fed with soybean meal based diet. Fish Shellfish Immunol. (2019) 91:130–5. doi: 10.1016/j.fsi.2019.05.030
12. Fehrmann-Cartes K, Coronado M, Hernandez AJ, Allende ML, Feijoo CG. Anti-inflammatory effects of aloe vera on soy meal-induced intestinal inflammation in zebrafish. Fish ShellFish Immunol. (2019) 95:564–73. doi: 10.1016/j.fsi.2019.10.075
13. Feijoo CG, Coronado M. Soybean meal-induced intestinal inflammation in Zebrafish is T cell-dependent and has a Th17 cytokine profile. Front Immunol. (2019) 10:610. doi: 10.3389/fimmu.2019.00610
14. Martin SA, Król E. Nutrigenomics and immune function in fish: new insights from omics technologies. Dev Comp Immunol. (2017) 75:86–98. doi: 10.1016/j.dci.2017.02.024
15. Tafalla C, Leal E, Yamaguchi T, Fischer U. T cell immunity in the teleost digestive tract. Dev Comp Immunol. (2016) 64:167–77. doi: 10.1016/j.dci.2016.02.019
16. De SC, Bartie KL, Olsen RE, Taggart JB, Tocher DR. Nutrigenomic profiling of transcriptional processes affected in liver and distal intestine in response to a soybean meal-induced nutritional stress in Atlantic salmon (Salmo salar). Comp Biochem Phys D. (2015) 15:1–11. doi: 10.1016/j.cbd.2015.04.001
17. Wulff T, Petersen J, Nørrelykke MR, Jessen F, Nielsen HH. Proteome analysis of pyloric ceca: a methodology for fish feed development? J Agric Food Chem. (2012) 60:8457–64. doi: 10.1021/jf3016943
18. Nguyen HTT, Dalmasso G, Yan Y, Laroui H, Dahan S, Mayer L, et al. MicroRNA-7 modulates CD98 expression during intestinal epithelial cell differentiation. J Biol Chem. (2010) 285:1479–89. doi: 10.1074/jbc.M109.057141
19. McKenna LB, Schug J, Vourekas A, McKenna JB, Bramswig NC, Friedman JR, et al. MicroRNAs control intestinal epithelial differentiation, architecture, and barrier function. Gastroenterology. (2010) 139:1654–64, 1664 e1651. doi: 10.1053/j.gastro.2010.07.040
20. Biton M, Levin A, Slyper M, Alkalay I, Horwitz E, Mor H, et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium–T cell crosstalk. Nat Immunol. (2011) 12:239. doi: 10.1038/ni.1994
21. Goto Y, Kiyono H. Epithelial cell microRNAs in gut immunity. Nat Immunol. (2011) 12:195. doi: 10.1038/ni0311-195
22. Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, et al. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell Res. (2008) 18:823. doi: 10.1038/cr.2008.81
23. Gao C, Cai X, Fu Q, Yang N, Song L, Su B, et al. Dynamics of miRNA transcriptome in turbot (Scophthalmus maximus L.) intestine following Vibrio anguillarum infection. Mar Biotechnol (NY). (2019) 21:550–64. doi: 10.1007/s10126-019-09903-z
24. Miao LH, Lin Y, Pan WJ, Huang X, Ge XP, Ren MC, et al. Identification of Differentially expressed micrornas associate with glucose metabolism in different organs of blunt snout bream (Megalobrama amblycephala). Int J Mol Sci. (2017) 18:E1161. doi: 10.3390/ijms18061161
25. Wang J, Liang D, Yang Q, Tan B, Dong X, Chi S, et al. The effect of partial replacement of fish meal by soy protein concentrate on growth performance, immune responses, gut morphology and intestinal inflammation for juvenile hybrid grouper (Epinephelus fuscoguttatus female symbol x Epinephelus lanceolatus male symbol). Fish ShellFish Immunol. (2019) 98:619–31. doi: 10.1016/j.fsi.2019.10.025
26. An W, Dong X, Tan B, Yang Q, Chi S, Zhang S, et al. Effects of dietary n-3 highly unsaturated fatty acids on growth, non-specific immunity, expression of some immune-related genes and resistance to Vibrio harveyi in hybrid grouper (female symbol Epinephelus fuscoguttatus x male symbol Epinephelus lanceolatu). Fish ShellFish Immunol. (2019) 96:86–96. doi: 10.1016/j.fsi.2019.11.072
27. Wu M, Wu X, Lu S, Gao Y, Yao W, Li X, et al. Dietary arginine affects growth, gut morphology, oxidation resistance and immunity of hybrid grouper (Epinephelus fuscoguttatus female symbolxEpinephelus lanceolatus male symbol) juveniles. Br J Nutr. (2018) 120:269–82. doi: 10.1017/S0007114518001022
28. Sun Z, Chen L, Liu Q, Mai K, Xu M, Zhou Y, et al. Effects of dietary Senecio scandens buch-ham extracts on growth performance, plasma biochemical, histology and the expression of immune-related genes in hybrid grouper (Epinephelus lanceolatusmale symbol x Epinephelus fuscoguttatus female symbol). Fish ShellFish Immunol. (2019) 98:681–90. doi: 10.1016/j.fsi.2019.11.002
29. Tan X, Sun Z, Ye C. Dietary Ginkgo biloba leaf extracts supplementation improved immunity and intestinal morphology, antioxidant ability and tight junction proteins mRNA expression of hybrid groupers (Epinephelus lanceolatus male symbol x Epinephelus fuscoguttatus female symbol) fed high lipid diets. Fish ShellFish Immunol. (2019) 98:611–8. doi: 10.1016/j.fsi.2019.09.034
30. Yin B, Liu H, Tan B, Dong X, Chi S, Yang Q, et al. Cottonseed protein concentrate (CPC) suppresses immune function in different intestinal segments of hybrid grouper female symbol Epinephelus fuscoguttatus x male symbol Epinephelus lanceolatu via TLR-2/MyD88 signaling pathways. Fish ShellFish Immunol. (2018) 81:318–28. doi: 10.1016/j.fsi.2018.07.038
31. Zhang W, Tan B, Ye G, Wang J, Dong X, Yang Q, et al. Identification of potential biomarkers for soybean meal-induced enteritis in juvenile pearl gentian grouper, Epinephelus lanceolatus♂ × Epinephelus fuscoguttatus♀. Aquaculture. (2019) 512:734337. doi: 10.1016/j.aquaculture.2019.734337
32. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. (2011) 29:644. doi: 10.1038/nbt.1883
33. Pertea G, Huang X, Liang F, Antonescu V, Sultana R, Karamycheva S, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. (2003) 19:651–2. doi: 10.1093/bioinformatics/btg034
34. Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. (2010) 26:136–8. doi: 10.1093/bioinformatics/btp612
35. Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. (2011) 11:R106. doi: 10.1186/gb-2010-11-10-r106
36. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. (2009) 10:R25. doi: 10.1186/gb-2009-10-3-r25
37. Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. (2012) 40:37–52. doi: 10.1093/nar/gkr688
38. Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE. (2010) 5:e15224. doi: 10.1371/journal.pone.0015224
39. Rasmussen K. Encyclopedia of Measurement and Statistics. Thousand Oaks, CA: Sage Publications. (2007).
40. Jan K, Marc R. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. (2006) 34:W451–4. doi: 10.1093/nar/gkl243
41. John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Correction: human microRNA targets. PLoS Biol. (2004) 3:e264. doi: 10.1371/journal.pbio.0030264
42. Li Y, Li RH, Ran MX, Zhang Y, Liang K, Ren YN, et al. High throughput small RNA and transcriptome sequencing reveal capacitation-related microRNAs and mRNA in boar sperm. BMC Genomics. (2018) 19:1–12. doi: 10.1186/s12864-018-5132-9
43. Zhao L, Lu H, Meng Q, Wang J, Wang W, Yang L, et al. Profilings of microRNAs in the liver of common carp (Cyprinus carpio) infected with Flavobacterium columnare. Int J Mol Sci. (2016) 17:566. doi: 10.3390/ijms17040566
44. Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression's CT difference” formula. J Mol Med. (2006) 84:901–10. doi: 10.1007/s00109-006-0097-6
45. Miao S, Zhao C, Zhu J, Hu J, Dong X, Sun L. Dietary soybean meal affects intestinal homoeostasis by altering the microbiota, morphology and inflammatory cytokine gene expression in northern snakehead. Sci Rep. (2018) 8:113. doi: 10.1038/s41598-017-18430-7
46. Liu H, Jin J, Zhu X, Han D, Yang Y, Xie S. Effect of substitution of dietary fish meal by soybean meal on different sizes of gibel carp (Carassius auratus gibelio): digestive enzyme gene expressions and activities, and intestinal and hepatic histology. Aquacult Nutr. (2017) 23:129–47. doi: 10.1111/anu.12375
47. Sahlmann C, Sutherland BJG, Kortner TM, Koop BF, Krogdahl s, Bakke AM. Early response of gene expression in the distal intestine of Atlantic salmon (Salmo salar L.) during the development of soybean meal induced enteritis. Fish Shellfish Immunol. (2013) 34:599–609. doi: 10.1016/j.fsi.2012.11.031
48. Gu M, Jia Q, Zhang Z, Bai N, Xu X, Xu B. Soya-saponins induce intestinal inflammation and barrier dysfunction in juvenile turbot (Scophthalmus maximus). Fish Shellfish Immunol. (2018) 77:264–72. doi: 10.1016/j.fsi.2018.04.004
49. Kortner TM, Skugor S, Penn MH, Mydland LT, Djordjevic B, Hillestad M, et al. Dietary soyasaponin supplementation to pea protein concentrate reveals nutrigenomic interactions underlying enteropathy in Atlantic salmon (Salmo salar). BMC Vet Res. (2012) 8:101. doi: 10.1186/1746-6148-8-101
50. Koukos G, Polytarchou C, Kaplan JL, Morley–Fletcher A, Gras–Miralles B, Kokkotou E, et al. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. (2013) 145:842–52. doi: 10.1053/j.gastro.2013.07.001
51. Soroosh A, Rankin CR, Polytarchou C, Lokhandwala ZA, Patel A, Chang L, et al. miR-24 is elevated in ulcerative colitis patients and regulates intestinal epithelial barrier function. Am J Pathol. (2019) 189:1763–74. doi: 10.1016/j.ajpath.2019.05.018
52. Grainger JR, Konkel JE, Zangerle-Murray T, Shaw TN. Macrophages in gastrointestinal homeostasis and inflammation. Pflug Arch Eur J Phy. (2017) 469:527–39. doi: 10.1007/s00424-017-1958-2
53. Wang P, Wang J, Su YQ, Mao Y, Zhang JS, Wu CW, et al. Transcriptome analysis of the Larimichthys crocea liver in response to Cryptocaryon irritans. Fish Shellfish Immunol. (2016) 48:1–11. doi: 10.1016/j.fsi.2015.11.011
54. Zhang X, Wang S, Chen S, Chen Y, Liu Y, Shao C, et al. Transcriptome analysis revealed changes of multiple genes involved in immunity in Cynoglossus semilaevis during Vibrio anguillarum infection. Fish Shellfish Immunol. (2015) 43:209–18. doi: 10.1016/j.fsi.2014.11.018
55. Prager I, Watzl C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J Leukocyte Biol. (2019) 105:1319–29. doi: 10.1002/JLB.MR0718-269R
56. Qin C, Gong Q, Wen Z, Yuan D, Shao T, Wang J, et al. Transcriptome analysis of the spleen of the darkbarbel catfish Pelteobagrus vachellii in response to Aeromonas hydrophila infection. Fish Shellfish Immunol. (2017) 70:498–506. doi: 10.1016/j.fsi.2017.09.042
57. Parashar D, Cherian S. RNAi agents as chikungunya virus therapeutics. Future Virol. (2016) 11:321–9. doi: 10.2217/fvl-2016-0010
Keywords: hybrid grouper, inflammation, RNA-seq, intestinal health, immune response
Citation: He Y, Ye G, Chi S, Tan B, Dong X, Yang Q, Liu H and Zhang S (2020) Integrative Transcriptomic and Small RNA Sequencing Reveals Immune-Related miRNA–mRNA Regulation Network for Soybean Meal-Induced Enteritis in Hybrid Grouper, Epinephelus fuscoguttatus♀ × Epinephelus lanceolatus♂. Front. Immunol. 11:1502. doi: 10.3389/fimmu.2020.01502
Received: 30 April 2020; Accepted: 09 June 2020;
Published: 06 August 2020.
Edited by:
Reinaldo B. Oria, Federal University of Ceara, BrazilReviewed by:
Byron Morales-Lange, Norwegian University of Life Sciences, NorwayYanjiao Zhang, Ocean University of China, China
Copyright © 2020 He, Ye, Chi, Tan, Dong, Yang, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuyan Chi, Y2hpc2h1eWFuNzdAMTYzLmNvbQ==; Beiping Tan, YnB0YW5AMTI2LmNvbQ==
 Yuanfa He
Yuanfa He Guanlin Ye
Guanlin Ye Shuyan Chi
Shuyan Chi Beiping Tan
Beiping Tan Xiaohui Dong
Xiaohui Dong Qihui Yang
Qihui Yang Hongyu Liu1,2,3
Hongyu Liu1,2,3