- 1Medical Research Center, Taizhou Hospital of Zhejiang Province, Wenzhou Medical University, Linhai, China
- 2Biological Resource Center, Taizhou Hospital of Zhejiang Province, Wenzhou Medical University, Linhai, China
Human leukocyte antigen (HLA)-G, a non-classical HLA-class I molecule, has a low polymorphism frequency, restricted tissue distribution and immunoinhibitory property. HLA-G expression in tumor cells and cells chronically infected with virus may enable them to escape from host immune surveillance. It is well-known that the HLA-G molecule is a novel biomarker and potential therapeutic target that is relevant in various types of cancers, but its role in cervical cancer has not been fully explored. In this review, we aim to summarize and discuss the immunologic role of the HLA-G molecule in the context of HPV infections and the process of cervical cancer carcinogenesis. A better understanding of the potential impact of HLA-G on the clinical course of persistent HPV infections, cervical epithelial cell transformation, tumor growth, recurrence and metastasis is needed to identify a novel diagnostic/prognostic biomarker for cervical cancer, which is critical for cervical cancer risk screening. In addition, it is also necessary to identify HLA-G-driven immune mechanisms involved in the interactions between host and virus to explore novel immunotherapy strategies that target HLA-G/immunoglobulin-like transcript (ILT) immune checkpoints.
Introduction
Cervical cancer ranks as the fourth most common female cancer worldwide, with an estimated 569,847 new cases and 311,365 deaths in 2018 (1). Persistent infection with high-risk human papillomavirus (hrHPV) is necessary but not sufficient to induce cervical cancer (2). Most HPV infections are transient and are cleared within months by host innate and adaptive immune responses (3). Failure to clear the virus leads to infection persistence, and only a minority of HPV-infected and transformed cells eventually avoid host immune surveillance, which leads to tumor growth and lymph node metastasis (4, 5). This host-dependent immunological status and HPV-induced immune escape are reflected in persistent infection and the subsequent progression of precancerous lesions to invasive cervical cancer, which indicates the complexity of host-virus interactions. Therefore, the roles of the immune system, not only in viral elimination but also in tumor antigen recognition, are extremely relevant in the process of cervical cancer carcinogenesis.
Accumulating evidence has supported the idea of a critical role for immunosuppressive mechanisms in promoting HPV-induced carcinogenesis, either by suppressing the capacity of the host to overcome HPV infection or by preventing the elimination of HPV-transformed epithelial cells (3–7). Human leucocyte antigen (HLA) complex is located on chromosome 6p21.3. Several HLA molecules with different functions can be broadly divided into classical HLA-class I (HLA-A, -B, -C), non-classical HLA-class I (HLA-E, -F, -G), classical HLA-class II (HLA-DR, -DQ, -DP), and classical HLA-class III (8). The HLA system influences the host immune response by mediating antigen presentation (9). HLA-G has been termed “non-classical” due to its low frequency of polymorphisms, restricted tissue distribution and immunoinhibitory properties, which are different from the properties of classical HLA-class I molecules (10, 11). It has become increasingly evident that the HLA-G molecule is involved in modulating both innate and adaptive immune responses and in promoting immune escape in various types of cancers (10–13) and infectious diseases (14–16). However, to date, the possibility that HLA-G gene polymorphisms and/or protein expression affecting HPV infection persistence and cervical cancer risk remains to be explored.
Molecular Structure of Human Leukocyte Antigen-G
The HLA-G gene consists of eight exons, seven introns, a 5′upstream regulatory region (URR) that extends at least 1,400 bp from the initial ATG start codon, and a 3′untranslated region (UTR), with a total length of 6,000 bp (12, 17). It is widely accepted that the HLA-G primary transcript is alternatively spliced into seven mRNAs, which encode four membrane-bound (HLA-G1, -G2, -G3, -G4) and three soluble (HLA-G5, -G6, -G7) protein isoforms (18, 19). Each unique HLA-G isoform contains one to three extracellular globular domains (α1, α2, α3) encoded by exon 2, exon 3, and exon 4, whereas the presence of intronic sequences are variable (IMGT/HLA Database).
The overall structure of HLA-G1 and that of its soluble counterpart HLA-G5 is similar to the structure of the classical HLA-class I antigens, which contain a heavy chain non-covalently bound to β2-microglobulin (β2m) (18). Peptide is bound in the antigen-binding cleft formed by the α1 and α2 domains (11, 20), whereas the α3 domain can bind co-receptors such as CD8 (21). Both HLA-G1 and HLA-G5 isoforms can also exist as β2m-free antigens (22). Other HLA-G isoforms lacking one or two extracellular globular domains (α2, or α3, or both) are smaller than HLA-G1/-G5 isoforms and are not associated with β2m (23). HLA-G1 to HLA-G4 are membrane-bound isoforms due to the presence of the transmembrane region encoded by exon 5 and a short cytoplasmic tail encoded by exon 6, which contains a stop codon. HLA-G5 and HLA-G6 are soluble isoforms due to the presence of intron 4, which contains a premature stop codon to prevent the translation of the transmembrane and cytoplasmic tail. HLA-G7 is a soluble isoform due to the presence of intron 2, which contains a premature stop codon and results in the expression of a soluble protein (18–20). All seven reported HLA-G isoforms contain the extracellular α1 domain.
In addition to the seven HLA-G monomers reported, the molecular structure of HLA-G is even more complex. A study on its crystal structure demonstrated that HLA-G can exist as a dimer with the intermolecular Cys42-Cys42 disulphide bond (24). In vitro and in vivo studies have shown that HLA-G dimers are observed for all isoforms except HLA-G3 (25). Moreover, β2m-associated and β2m-free dimers of HLA-G1 or HLA-G5 also exist (26–28). Dimer formation affects the specificity of receptor-HLA-G binding, as dimers exhibit a higher overall affinity to immunoglobulin-like transcript (ILT)2/4 receptors than monomers due to significant avidity effects (24, 28, 29).
Notably, unidentified HLA-G isoforms without an α1 domain were predicted based on RNA sequencing (RNA-seq), and several previously undescribed HLA-G isoforms have been identified in renal cancer samples (30). According to the nucleotide sequence of the HLA-G gene listed in the Ensembl database (ENST00000376828), this gene may possess a supplementary exon at the 5′-end, but this is absent from the sequence in the IMGT/HLA database. A novel HLA-G isoform named HLA-G1L was predicted by Tronik-Le Roux et al. (30); this isoform has five additional amino acids (MKTPR) located at the N-terminal end. Analysis of RNA-seq data indicates that some sequence reads may be initiated at exon 4, and thus could predict the existence of novel α1-deleted HLA-G isoforms that contain α2 and α3 domains or only the α3 domain. Other novel soluble HLA-G isoforms can be generated by the skipping of exon 6 coding for the transmembrane domain (30, 31). Lin et al. (32) indicated the existence of novel α1-deleted HLA-G isoforms containing intron 4 in 11.6% (44/379) of colorectal cancer lesions that exhibited negative staining with mAb 4H84 but that exhibited positive staining with mAb 5A6G7 (4H84neg5A6G7pos). Moreover, patients with 4H84neg5A6G7pos HLA-G isoforms had a better survival than patients with 4H84pos5A6G7neg, and thus suggests a functional role for the novel α1-deleted HLA-G isoforms (31). However, the specific function of these novel HLA-G isoforms remains to be determined. The development of specific antibodies for these novel HLA-G isoforms is urgently needed and even inevitable (33).
HLA-G-Mediated Immune Suppression
HLA-G expression was initially observed on cytotrophoblasts at the maternal-fetal interface (34), where HLA-G modulates the response of maternal immune cells that contribute to maintenance of tolerance to the fetus (35–37). HLA-G has a physiological tissue-restricted distribution property, as it is expressed by cytotrophoblasts (34), cornea (38), thymus (39), nail matrix (40), pancreatic islets (41), and erythroblasts (42). However, aberrant upregulated expression of HLA-G molecules has been detected in pathological conditions such as malignancies (43–45), infections and inflammatory diseases (14, 46–49), transplant grafts (50, 51), and autoimmune disorders (16, 52–54). In malignancies, aberrant HLA-G expression was preferentially detected in tumor tissues but was rarely detected in normal or adjacent non-tumorous tissues, which indicates that HLA-G might play a key role in tumor development (44).
Functionally, HLA-G has comprehensive immunosuppressive properties exerted in multiple steps to weaken anti-tumor immune responses by acting on immune cells through its inhibitory receptors: ILT2(CD85j/LILRB1), ILT4(CD85d/LILRB2), and KIR2DL4(CD158d) (11, 12, 55–59) (Figure 1). HLA-G inhibits the cytolytic function of natural killer (NK) cells (60, 61), cytotoxic T lymphocyte (CTL)-mediated cytolysis (62), macrophage-mediated cytotoxicity (63), allo-proliferative response of CD4+ T cells (64, 65), maturation and function of dendritic cells (DCs) or B lymphocytes (66–69), stimulation of antigen-presenting cells (APCs) to secrete functional cytokines TGF-β and IL-10, and induction of apoptosis of CD8+ T cells and CD8+ NK cells (70, 71). In addition, HLA-G-receptor interactions could also exert long-term immunomodulatory effects by inducing immune suppressor/regulatory cells, such as regulatory T cells (Tregs) (72, 73), tolerogenic DCs (tDCs) (74, 75), mesenchymal stem cells (MSCs) (76), and myeloid-derived suppressor cells (MDSCs) (77, 78), among others. In addition to the interactions between HLA-G and its receptors, HLA-G-mediated immunosuppression by intercellular transfer mechanisms such as trogocytosis, exosomes, or tunneling nanotubes (TnTs) also represents another important complementary mechanism through which cancer cells escape destruction by the host immune system (11, 12, 79–81).
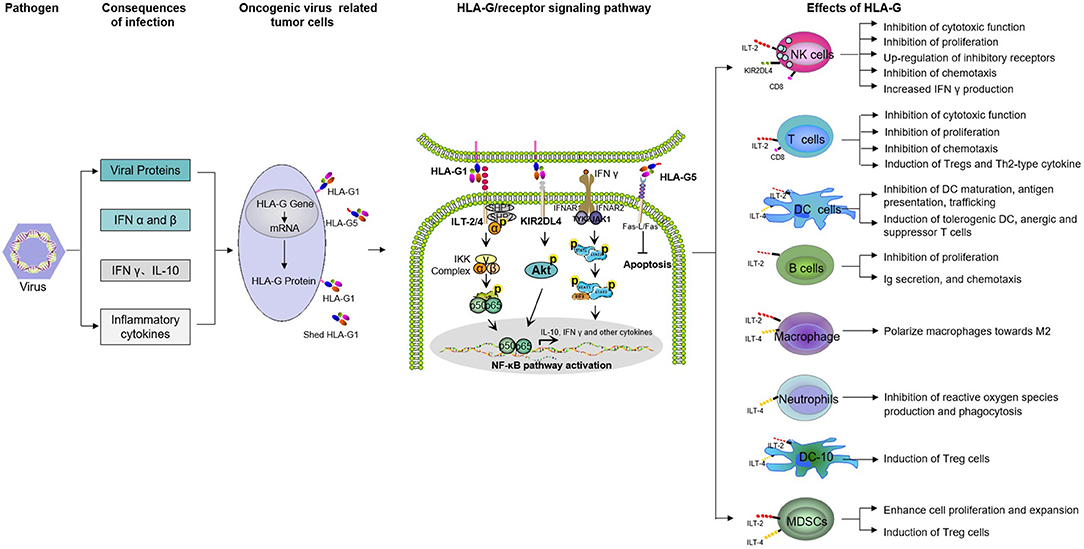
Figure 1. Mechanisms of both membrane-bound and soluble HLA-G-mediated immune suppression in tumor immune evasion. NK, natural killer cells; DC, dendritic cells; MDSC, myeloid-derived suppressor cells; Tregs, regulatory T cells; DC-10, IL-10-differentiated dendritic cells.
HLA-G Polymorphisms in the Context of HPV Infections
To date, 69 alleles that encode 19 proteins have been discovered (IMGT/HLA Database, February 2020). Polymorphic sites along the HLA-G gene may change the affinity of gene-targeted sequences for transcriptional or post-transcriptional factors (82, 83). In particular, the 14bp Insertion/Deletion (Ins/Del) (rs66554220) in the 3′UTR is associated with HLA-G alternative splicing and mRNA stability (84, 85). The +3142C/G (rs1063320) located 167 bp downstream from the 14bp Ins/Del polymorphic site may be a target for HLA-G-specific miRNAs (86), which could directly downregulate HLA-G expression through post-transcriptional regulation (87, 88).
Accumulating evidence has supported the concept that HLA-G polymorphisms are genetic susceptibility and/or protection-relevant factors for cervical HPV infections and viral persistence (89–101). Many studies have primarily focused on polymorphisms in the 3′UTR of the HLA-G gene (89–95), while few have assessed its promoter region (96). Studies by Xu et al. (89, 90) showed that HLA-G 14bp Ins or +3142G alleles are risk factors for HPV infections, especially hrHPV infections, compared with the alleles found in healthy women and that these alleles affect the progression of HPV18-associated cervical lesions in Chinese women. A similar finding was reported in a study performed in Brazilian women from São Paulo, Brazil; this study showed that the HLA-G 14bp Ins/+3142G haplotype was related to increased risk of high-grade cervical lesions, especially in smokers (91). Inconsistent results were obtained in Italy (92) and Taiwan (93), where increased risk for squamous cell carcinoma (SCC) was found to be associated with the 14bp del or +3142C alleles, especially in SCC patients infected with the HPV16 genotype (93). Moreover, some have focused on the association between HLA-G 3′UTR polymorphisms and HPV infection among HIV-positive women who have a higher risk of developing HPV co-infection. The combination of the +3142CX (CC or CG) and +3187AA genotypes conferred the highest risk of HPV-induced aneuploidy in cervical cells among Brazilian women with HIV/HPV co-infections (94).A SNP (rs1633038) in the 3′UTR of the HLA-G gene was significantly related to higher HPV clearance rates among African-Americans with HIV/HPV co-infection, but this association was not observed in Hispanics or European-Americans (95).
Further evidence for the role of genetic factors in HPV infections and the carcinogenic process was provided by studies that showed an association with specific HLA-G coding region polymorphisms (96–104). Among the Canadian population, the HLA-G*01:01:02 and HLA-G*01:03 alleles were found to be related to an increased risk of HPV16 infection and persistent infections (96), while the HLA-G*01:01:03 and HLA-G*01:01:05 alleles were identified as significant predictors of cumulative coinfections over the follow-up period (97). In the same cohort, the HLA-G*01:01:02, HLA-G*01:04:01 and HLA-G*01:06 alleles were related to high-grade cervical intraepithelial neoplasia (HG-CIN) (98). The HLA-G*01:01:02, HLA-G*01:06 and 3′UTR 14bp Ins alleles were associated with disease progression from preinvasive to invasive cervical cancer among HPV-positive Canadian women (99). The homozygous HLA-G*01:04:01 genotype was related to a significantly decreased risk of HPV infection (98), and the heterozygotic form of the HLA-G*01:01:01 allele conferred significant protection against cancer (99). Among Brazilian women, the HLA-G*01:04/14bp Ins haplotype as well as HPV16 and HPV18 co-infection were preferentially related to HG-CIN, while the HLA-G*01:03 allele was related to protection against HPV-related cervical lesions (100). Among HPV-positive pregnant women in Brazil, a protective effect of the HLA-G*01:01:02 allele against the occurrence of CIN was observed in a cohort of HPV/HIV co-infected pregnant women (101). In a study that focused on the role of host factors in the vertical transmission of HPV infection from mother to offspring, the results showed that the HLA-G*01:01:01/01:04:01 genotype increased the risk of hrHPV infection in both cord blood and the infant's oral mucosa; moreover, the mother-child concordance of HLA-G*01:01:02/01:01:02 increased the risk of oral hrHPV infection both in the mother and offspring (102). In addition, a pilot analysis of HLA-G promoter methylation and HPV infection status showed no association between HLA-G methylation and HPV infections in healthy women (103, 104).
Overall, the discrepancy among these studies could be explained by differences in the study designs, ethnicity, sample sizes, and cancer types. Current data suggested that HLA-G gene polymorphisms (mainly located in the coding region or 3′UTR region) appear to be independent risk factors for HPV infection and cervical carcinogenesis, which supports the biological role of HLA-G molecules in shaping the tumor microenvironment (105).
HLA-G Expression in Cervical Carcinogenesis
HLA-G expression may be induced after HPV infection, which leads to escape from host immunosurveillance. This evidence is derived from the results of a study that showed that HLA-G expression was significantly higher in CIN and cancer patients with HPV16/18 infections than in CIN patients without HPV infection (106). Several studies have investigated the relationship between HLA-G isoform expression and clinicopathologic features in patients with precancerous lesions and invasive cervical cancer (45, 90, 106–117).
Another study focused on HLA-G mRNA expression in cervical cancer in Korean patients using RT-PCR (15 normal tissues and 40 cervical cancer tissues) and found that high HLA-G mRNA expression was related to the early stages of cervical cancer (108). These results are consistent with the report by Rodriguez et al. (109), which showed upregulation of HLA-G protein expression in the early stages of cervical cancer in Colombian patients using immunohistochemistry (IHC) with mAb 4H84 (9 CIN III and 54 cervical cancer cases). Both studies supported a possible role for the HLA-G molecule in early cervical carcinogenesis (108, 109). The results of both studies further showed that Interleukin-10 (IL-10) expression was also significantly increased in cervical cancer tissues (108, 109), which supports a shift toward a Th2 cytokine microenvironment; this in turn may promote local immunosuppression by upregulating HLA-G expression (111, 118). Consistent with this, the results also revealed the inverse relationship among HLA-G expression levels and estimated numbers of tumor infiltrating lymphocytes (TILs) and CD57+ NK cells, which favors an escape from host anti-tumor activity (115). Moreover, three independent studies have reported evidence of a positive correlation between HLA-G expression and cervical carcinogenesis in a Chinese population (45, 106, 107). The results of all three studies indicated that HLA-G expression was negative in normal or adjacent non-tumorous tissues but was significantly increased along with CIN grade and cervical cancer metastasis. HLA-G expression may play an important role in determining the risk for cervical carcinogenesis. These clinical studies also analyzed clinicopathological parameters and demonstrated significant correlations between HLA-G expression and unfavorable prognosis, poor overall survival, and lymph node metastasis. However, inconsistent results were obtained in only two studies that showed that HLA-G expression was not related to cervical carcinogenesis (112, 113). Zhou et al. (112) described that in all normal epithelium, HLA-G expression was strong and uniform but was statistically down-regulated in CIN and SCC. Gonçalves et al. (113) reported that HLA-G was not expressed in any CIN or SCC. Futhermore, in experimental model of cervical cancer research, Real et al. (119) reported that low expression of HLA-G in Hela cell line (HPV18 infection). Thus, the role of HLA-G in malignancies has gained considerable clinical interest due to the possibility of exploiting it as a novel diagnostic/prognostic biomarker to identify cervical cancer and to monitor disease stage.
Additionally, three studies focused on soluble HLA-G (sHLA-G) isoform expression using different detection technologies (107, 110, 114). Guimarães et al. (110) analyzed sHLA-G expression in cervical cancer tissues from Brazilian patients using IHC with the specific mAb 5A6G7 (27 with metastasis and 52 without metastasis). Low expression of sHLA-G isoforms was detected in all HPV-positive tissues, and the sHLA-G expression level was similar in both groups (110). Zheng et al. (107) investigated the sHLA-G expression level in the plasma of patients with cervical lesions using ELISA kit (sHLA-G, Exbio) with mAb MEM-G/9 (20 normal cervical tissues, 15 CIN I, 22 CIN II, 35 CIN III, and 80 cervical cancer tissues). sHLA-G expression levels in the plasma were significantly increased in CIN II-III and SCC patients, and their expression levels were also associated with differentiation and metastasis. Therefore, sHLA-G molecules may have significance in early cervical cancer screening (107). However, inconsistent results obtained in the Netherlands (366 cervical cancer) using ELISA kit (sHLA-G, Exbio) reported that sHLA-G levels were not associated with clinicopathological parameters or survival (114).
Overall, the discrepancies in these studies that examined HLA-G expression in cervical cancer patients are partly due to tumor heterogeneity (31). In the future, there will be a need for additional studies to obtain deeper insight into the association between HLA-G expression levels and advanced cervical cancer.
HLA-G as a Novel Target for Immunotherapies
Cervical cancer accounts for 6.6% of all female cancers and is thus a major global health challenge, as ~90% of cervical cancer deaths occur in less developed countries (1). High-risk HPV causes almost all invasive cervical cancers, and therefore, HPV screening and vaccination are needed to improve cervical cancer control (2). Despite significant advances in effective screening and preventive vaccination during the past decade, substantial regional and global disparities in the prognosis of cervical cancer patients still exist (120). Unfortunately, ~30% of patients experience recurrence and metastasis after primary treatment, with an expected 5-year survival of <10%. Few effective therapeutic strategies have been developed that specifically target recurrent or metastatic cervical cancer, particularly advanced-stage disease. Thus, novel therapeutic strategies, such as immunotherapy, are urgently needed in clinical settings (121, 122).
In recent years, an improved understanding of the molecular mechanisms of the interactions between HPV-associated cervical cancer and the host immune responses has driven the exploration of immunotherapy as one of the new therapeutics targeting immune checkpoints (123). HLA-G has comprehensive immunosuppressive properties that are exerted in multiple steps to weaken the anti-tumor immune responses by acting on immune cells through its inhibitory receptors. Fortunately, HLA-G expression can be downregulated through RNA interference or antibody blockade, which can allow recovery of the functions of immune effectors and prevent tumor reoccurrence. Thus, HLA-G could serve as a novel immune checkpoint molecule and play a key role in novel immunotherapy approaches that offer a promising perspective for tumor progression and advanced- stage cervical cancer.
It has been confirmed that miR-148a negatively regulates HLA-G expression by binding to the 3′UTR of the HLA-G gene (88). The long non-coding RNA HOX transcript antisense RNA (HOTAIR) may also serve as a competing endogenous RNAs (ceRNAs) to regulate HLA-G expression by sponging miR-148a in cervical cancer cells (116). Targeting the HOTAIR-miR-148a-HLA-G axis or HLA-G-specific miRs could represent a novel therapeutic strategy in cervical cancer. Intra-tumor heterogeneity of checkpoint molecule expression in cervical cancer is related to a poor chemo/radio-therapy response, lymph node metastasis and tumor recurrence. HLA-G has been identified as a cervical cancer stem cell (CCSC)-specific marker, and targeting HLA-G and its related signaling pathways may offer a novel strategy for CCSC-targeted therapy (124). Moreover, the HLA-G/ILTs axis has been recently recognized as a new immune checkpoint in addition to other immune checkpoints such as programmed cell death 1 (PD-1)/PD-L1 and cytotoxic T lymphocyte-associated protein 4 (CTLA-4)/B7 (125). Different responses to checkpoint inhibitor therapy could be a consequence of heterogeneous intra- and inter-tumor expression of different types of checkpoint molecules, although data on the expression status of HLA-G, CTLA-4 and PD-L1 in cancers are rather limited (13, 125, 126). PD-1 is a major immunotherapeutic checkpoint target in various cancer types, but until now, few data have been available on the clinical efficacy of blocking this checkpoint protein in cervical cancer (2, 126). The expression of PD-1 was found to be heterogeneous in tumors and could be co-expressed with the immune checkpoint protein HLA-G (127). A recent study focused on tumor-infiltrating CD8+ T cells that express the HLA-G receptor ILT2 in renal-cell carcinoma (RCC), and the results emphasize the potential of therapeutically targeting the HLA-G/ILT2 checkpoint in HLA-G+ tumors (127). Overexpression of the immune checkpoint HLA-G molecule by tumor cells profoundly affects tumor-specific T cell immunity in the cancer microenvironment. In this regard, targeting multiple checkpoints, especially potential antagonists of the HLA-G/ILT-2/4 pathway, is urgently needed to target the entire tumor.
Conclusions
Considering the above studies that were reviewed, we propose that HLA-G gene polymorphisms have an impact on the immune response and likely determine those in specific populations who are at higher risk for cervical HPV infections and viral persistence. Aberrant HLA-G expression in cervical lesions could generate inhibitory signals in the cancer microenvironment, which would ultimately help tumor cells escape from immunosurveillance and reshape tumor progression and metastasis. The checkpoint molecule HLA-G with immune tolerance contribute to cervical carcinogenesis, but HLA-G could also represent a good immunotherapeutic target for cervical cancer treatment.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by grants from National Natural Science Foundation of China (81901625) and by Science and Technology Bureau of Taizhou (1901ky05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
2. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. (2019) 393:169–82. doi: 10.1016/S0140-6736(18)32470-X
3. Sasagawa T, Takagi H, Makinoda S. Immune responses against human papillomavirus (HPV) infection and evasion of host defense in cervical cancer. J Infect Chemother. (2012) 18:807–15. doi: 10.1007/s10156-012-0485-5
4. Steinbach A, Riemer AB. Immune evasion mechanisms of human papillomavirus: an update. Int J Cancer. (2018) 142:224–9. doi: 10.1002/ijc.31027
5. Graham SV. The human papillomavirus replication cycle, and its links to cancer progression: a comprehensive review. Clin Sci. (2017) 131:2201–21. doi: 10.1042/CS20160786
6. Kuss-Duerkop SK, Westrich JA, Pyeon D. DNA Tumor virus regulation of host DNA methylation and its implications for immune evasion and oncogenesis. Viruses. (2018) 10: E82. doi: 10.3390/v10020082
7. Litwin TR, Clarke MA, Dean M, Wentzensen N. Somatic host cell alterations in HPV carcinogenesis. Viruses. (2017) 9: E206. doi: 10.3390/v9080206
8. Paaso A, Jaakola A, Syrjänen S, Louvanto K. From HPV infection to lesion progression: the role of HLA alleles and host immunity. Acta Cytol. (2019) 63:148–58. doi: 10.1159/000494985
9. Garrido F, Aptsiauri N. Cancer immune escape: MHC expression in primary tumours versus metastases. Immunology. (2019) 158:255–66. doi: 10.1111/imm.13114
10. Lin A, Yan WH. HLA-G as an inhibitor of immune responses. Methods Mol Biol. (2016) 1371:3–9. doi: 10.1007/978-1-4939-3139-2_1
11. Carosella ED, Rouas-Freiss N, Tronik-Le Roux D, Moreau P, LeMaoult J. HLA-G: an immune checkpoint molecule. Adv Immunol. (2015) 127:33–144. doi: 10.1016/bs.ai.2015.04.001
12. Lin A, Yan WH. Intercellular transfer of HLA-G: its potential in cancer immunology. Clin Transl Immunology. (2019) 8:e1077. doi: 10.1002/cti2.1077
13. Własiuk P, Putowski M, Giannopoulos K. PD1/PD1L pathway, HLA-G and T regulatory cells as new markers of immunosuppression in cancers. Postepy Hig Med Dosw. (2016) 70:1044–58. doi: 10.5604/17322693.1220994
14. Amiot L, Vu N, Samson M. Immunomodulatory properties of HLA-G in infectious diseases. J Immunol Res. (2014) 2014:298569. doi: 10.1155/2014/298569
15. Catamo E, Zupin L, Crovella S, Celsi F, Segat L. Non-classical MHC-I human leukocyte antigen (HLA-G) in hepatotropic viral infections and in hepatocellular carcinoma. Hum Immunol. (2014) 75:1225–31. doi: 10.1016/j.humimm.2014.09.019
16. Rizzo R, Bortolotti D, Bolzani S, Fainardi E. HLA-G molecules in autoimmune diseases and infections. Front Immunol. (2014) 5:592. doi: 10.3389/fimmu.2014.00592
17. Geraghty DE, Koller BH, Orr HT. A human major histocompatibility complex class I gene that encodes a protein with a shortened cytoplasmic segment. Proc Natl Acad Sci USA. (1987) 84:9145–9. doi: 10.1073/pnas.84.24.9145
18. Ishitani A, Geraghty DE. Alternative splicing of HLA-G transcripts yields proteins with primary structures resembling both class I and class II antigens. Proc Natl Acad Sci USA. (1992) 89:3947–51. doi: 10.1073/pnas.89.9.3947
19. Paul P, Cabestre FA, Ibrahim EC, Lefebvre S, Khalil-Daher I, Vazeux G, et al. Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum Immunol. (2000) 61:1138–49. doi: 10.1016/S0198-8859(00)00197-X
20. Clements CS, Kjer-Nielsen L, McCluskey J, Rossjohn J. Structural studies on HLA-G: implications for ligand and receptor binding. Hum Immunol. (2007) 68:220–6. doi: 10.1016/j.humimm.2006.09.003
21. Gao GF, Willcox BE, Wyer JR, Boulter JM, O'Callaghan CA, Maenaka K, et al. Classical and nonclassical class I major histocompatibility complex molecules exhibit subtle conformational differences that affect binding to CD8alphaalpha. J Biol Chem. (2000) 275:15232–8. doi: 10.1074/jbc.275.20.15232
22. Poláková K, Kuba D, Russ G. The 4H84 monoclonal antibody detecting beta2m free nonclassical HLA-G molecules also binds to free heavy chains of classical HLA class I antigens present on activated lymphocytes. Hum Immunol. (2004) 65:157–62. doi: 10.1016/j.humimm.2003.10.005
23. Alegre E, Rizzo R, Bortolotti D, Fernandez-Landázuri S, Fainardi E, González A. Some basic aspects of HLA-G biology. J Immunol Res. (2014) 2014:657625. doi: 10.1155/2014/657625
24. Shiroishi M, Kuroki K, Ose T, Rasubala L, Shiratori I, Arase H, et al. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J Biol Chem. (2006) 281:10439–47. doi: 10.1074/jbc.M512305200
25. HoWangYin KY, Loustau M, Wu J, Alegre E, Daouya M, Caumartin J, et al. Multimeric structures of HLA-G isoforms function through differential binding to LILRB receptors. Cell Mol Life Sci. (2012) 69:4041–9. doi: 10.1007/s00018-012-1069-3
26. Morales PJ, Pace JL, Platt JS, Langat DK, Hunt JS. Synthesis of beta(2)-microglobulin-free, disulphide-linked HLA-G5 homodimers in human placental villous cytotrophoblast cells. Immunology. (2007) 122:179–88. doi: 10.1111/j.1365-2567.2007.02623.x
27. Boyson JE, Erskine R, Whitman MC, Chiu M, Lau JM, Koopman LA, et al. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proc Natl Acad Sci USA. (2002) 99:16180–5. doi: 10.1073/pnas.212643199
28. Kuroki K, Matsubara H, Kanda R, Miyashita N, Shiroishi M, Fukunaga Y, et al. Structural and functional basis for LILRB immune checkpoint receptor recognition of HLA-G isoforms. J Immunol. (2019) 203:3386–94. doi: 10.4049/jimmunol.1900562
29. Nardi Fda S, Konig L, Wagner B, Giebel B, Santos Manvailer LF, Rebmann V. Soluble monomers, dimers and HLA-G-expressing extracellular vesicles: the three dimensions of structural complexity to use HLA-G as a clinical biomarker. HLA. (2016) 88:77–86. doi: 10.1111/tan.12844
30. Tronik-Le Roux D, Renard J, Verine J, Renault V, Tubacher E, LeMaoult J, et al. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol Oncol. (2017) 11:1561–78. doi: 10.1002/1878-0261.12119
31. Lin A, Yan WH. Heterogeneity of HLA-G expression in cancers: facing the challenges. Front Immunol. (2018) 9:2164. doi: 10.3389/fimmu.2018.02164
32. Lin A, Zhang X, Zhang RL, Zhang JG, Zhou WJ, Yan WH. Clinical significance of potential unidentified HLA-G isoforms without a1 domain but containing intron 4 in colorectal cancer patients. Front Oncol. (2018) 8:361. doi: 10.3389/fonc.2018.00361
33. Swets M, Wouters A, Krijgsman D, van Vlierberghe RLP, Boot A, van Eendenburg JD, et al. HLA-G protein expression in colorectal cancer evaluated by immunohistochemistry and western blot analysis: its expression characteristics remain enigmatic. Clin Immunol. (2018) 194:80–6. doi: 10.1016/j.clim.2018.07.005
34. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. (1990) 248:220–3. doi: 10.1126/science.2326636
35. Carosella ED, Moreau P, Le Maoult J, Le Discorde M, Dausset J, Rouas-Freiss N. HLA-G molecules: from maternal-fetal tolerance to tissue acceptance. Adv Immunol. (2003) 81:199–252. doi: 10.1016/S0065-2776(03)81006-4
36. Rebmann V, da Silva Nardi F, Wagner B, Horn PA. HLA-G as a tolerogenic molecule in transplantation and pregnancy. J Immunol Res. (2014) 2014:297073. doi: 10.1155/2014/297073
37. Ferreira LMR, Meissner TB, Tilburgs T, Strominger JL. HLA-G: at the interface of maternal-fetal tolerance. Trends Immunol. (2017) 38:272–86. doi: 10.1016/j.it.2017.01.009
38. Le Discorde M, Moreau P, Sabatier P, Legeais JM, Carosella ED. Expression of HLA-G in human cornea, an immune-privileged tissue. Hum Immunol. (2003) 64:1039–44. doi: 10.1016/j.humimm.2003.08.346
39. Mallet V, Blaschitz A, Crisa L, Schmitt C, Fournel S, King A, et al. HLA-G in the human thymus: a subpopulation of medullary epithelial but not CD83(+) dendritic cells expresses HLA-G as a membrane-bound and soluble protein. Int Immunol. (1999) 11:889–98. doi: 10.1093/intimm/11.6.889
40. Ito T, Ito N, Saathoff M, Stampachiacchiere B, Bettermann A, Bulfone-Paus S, et al. Immunology of the human nail apparatus: the nail matrix is a site of relative immune privilege. J Invest Dermatol. (2005) 125:1139–48. doi: 10.1111/j.0022-202X.2005.23927.x
41. Cirulli V, Zalatan J, McMaster M, Prinsen R, Salomon DR, Ricordi C, et al. The class I HLA repertoire of pancreatic islets comprises the nonclassical class Ib antigen HLA-G. Diabetes. (2006) 55:1214–22. doi: 10.2337/db05-0731
42. Menier C, Rabreau M, Challier JC, Le Discorde M, Carosella ED, Rouas-Freiss N. Erythroblasts secrete the nonclassical HLA-G molecule from primitive to definitive hematopoiesis. Blood. (2004) 104:3153–60. doi: 10.1182/blood-2004-03-0809
43. Paul P, Rouas-Freiss N, Khalil-Daher I, Moreau P, Riteau B, Le Gal FA, et al. HLA-G expression in melanoma: a way for tumor cells to escape from immunosurveillance. Proc Natl Acad Sci USA. (1998) 95:4510–5. doi: 10.1073/pnas.95.8.4510
44. Lin A, Yan WH. HLA-G expression in cancers: roles in immune evasion, metastasis and target for therapy. Mol Med. (2015) 21:782–91. doi: 10.2119/molmed.2015.00083
45. Li XJ, Zhang X, Lin A, Ruan YY, Yan WH. Human leukocyte antigen-G (HLA-G) expression in cervical cancer lesions is associated with disease progression. Hum Immunol. (2012) 73:946–9. doi: 10.1016/j.humimm.2012.07.041
46. Souto FJ, Crispim JC, Ferreira SC, da Silva AS, Bassi CL, Soares CP, et al. Liver HLA-G expression is associated with multiple clinical and histopathological forms of chronic hepatitis B virus infection. J Viral Hepat. (2011) 18:102–5. doi: 10.1111/j.1365-2893.2010.01286.x
47. Amiot L, Vu N, Rauch M, L'Helgoualc'h A, Chalmel F, Gascan H, et al. Expression of HLA-G by mast cells is associated with hepatitis C virus-induced liver fibrosis. J Hepatol. (2014) 60:245–52. doi: 10.1016/j.jhep.2013.09.006
48. Rizzo R, Bortolotti D, Baricordi OR, Fainardi E. New insights into HLA-G and inflammatory diseases. Inflamm Allergy Drug Targets. (2012) 11:448–63. doi: 10.2174/187152812803590037
49. Carosella ED, Moreau P, Aractingi S, Rouas-Freiss N. HLA-G: a shield against inflammatory aggression. Trends Immunol. (2001) 22:553–5. doi: 10.1016/S1471-4906(01)02007-5
50. Lazarte J, Adamson MB, Tumiati LC, Delgado DH. 10-year experience with HLA-G in heart transplantation. Hum Immunol. (2018) 79:587–93. doi: 10.1016/j.humimm.2018.05.003
51. Deschaseaux F, Delgado D, Pistoia V, Giuliani M, Morandi F, Durrbach A. HLA-G in organ transplantation: towards clinical applications. Cell Mol Life Sci. (2011) 68:397–404. doi: 10.1007/s00018-010-0581-6
52. Rizzo R, Hviid TV, Govoni M, Padovan M, Rubini M, Melchiorri L, et al. HLA-G genotype and HLA-G expression in systemic lupus erythematosus: HLA-G as a putative susceptibility gene in systemic lupus erythematosus. Tissue Antigens. (2008) 71:520–9. doi: 10.1111/j.1399-0039.2008.01037.x
53. Beneventi F, Badulli C, Locatelli E, Caporali R, Ramoni V, Cavagnoli C, et al. Soluble HLA-G in pregnancies complicated by autoimmune rheumatic diseases. J Reprod Immunol. (2015) 110:67–73. doi: 10.1016/j.jri.2015.04.005
54. Brenol CV, Veit TD, Chies JA, Xavier RM. The role of the HLA-G gene and molecule on the clinical expression of rheumatologic diseases. Rev Bras Reumatol. (2012) 52:82–91. doi: 10.1590/S0482-50042012000100009
55. Zhang F, Zheng J, Kang X, Deng M, Lu Z, Kim J, et al. Inhibitory leukocyte immunoglobulin-like receptors in cancer development. Sci China Life Sci. (2015) 58:1216–25. doi: 10.1007/s11427-015-4925-1
56. Rouas-Freiss N, LeMaoult J, Verine J, Tronik-Le Roux D, Culine S, Hennequin C, et al. Intratumor heterogeneity of immune checkpoints in primary renal cell cancer: focus on HLA-G/ILT2/ILT4. Oncoimmunology. (2017) 6:e1342023. doi: 10.1080/2162402X.2017.1342023
57. Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G alpha1 domain involve in KIR2DL4 recognition. Cell Res. (2005) 15:176–82. doi: 10.1038/sj.cr.7290283
58. Fons P, Chabot S, Cartwright JE, Lenfant F, L'Faqihi F, Giustiniani J, et al. Soluble HLA-G1 inhibits angiogenesis through an apoptotic pathway and by direct binding to CD160 receptor expressed by endothelial cells. Blood. (2006) 108:2608–15. doi: 10.1182/blood-2005-12-019919
59. Hofmeister V, Weiss EH. HLA-G modulates immune responses by diverse receptor interactions. Semin Cancer Biol. (2003) 13:317–23. doi: 10.1016/S1044-579X(03)00022-1
60. Rouas-Freiss N, Goncalves RM, Menier C, Dausset J, Carosella ED. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc Natl Acad Sci USA. (1997) 94:11520–5. doi: 10.1073/pnas.94.21.11520
61. Lin A, Xu HH, Xu DP, Zhang X, Wang Q, Yan WH. Multiple steps of HLA-G in ovarian carcinoma metastasis: alter NK cytotoxicity and induce matrix metalloproteinase-15 (MMP-15) expression. Hum Immunol. (2013) 74:439–46. doi: 10.1016/j.humimm.2012.11.021
62. Le Gal FA, Riteau B, Sedlik C, Khalil-Daher I, Menier C, Dausset J, et al. HLA-G-mediated inhibition of antigen-specific cytotoxic T lymphocytes. Int Immunol. (1999) 11:1351–6. doi: 10.1093/intimm/11.8.1351
63. Esquivel EL, Maeda A, Eguchi H, Asada M, Sugiyama M, Manabe C, et al. Suppression of human macrophage-mediated cytotoxicity by transgenic swine endothelial cell expression of HLA-G. Transpl Immunol. (2015) 32:109–15. doi: 10.1016/j.trim.2014.12.004
64. Naji A, Durrbach A, Carosella ED, Rouas-Freiss N. Soluble HLA-G and HLA-G1 expressing antigen-presenting cells inhibit T-cell alloproliferation through ILT-2/ILT-4/FasL-mediated pathways. Hum Immunol. (2007) 68:233–9. doi: 10.1016/j.humimm.2006.10.017
65. Lila N, Rouas-Freiss N, Dausset J, Carpentier A, Carosella ED. Soluble HLA-G protein secreted by allo-specific CD4+ T cells suppresses the allo-proliferative response, a CD4+ T cell regulatory mechanism. Proc Natl Acad Sci USA. (2001) 98:12150–5. doi: 10.1073/pnas.201407398
66. Ristich V, Liang S, Zhang W, Wu J, Horuzsko A. Tolerization of dendritic cells by HLA-G. Eur J Immunol. (2005) 35:1133–42. doi: 10.1002/eji.200425741
67. Gros F, Cabillic F, Toutirais O, Maux AL, Sebti Y, Amiot L. Soluble HLA-G molecules impair natural killer/dendritic cell crosstalk via inhibition of dendritic cells. Eur J Immunol. (2008) 38:742–9. doi: 10.1002/eji.200736918
68. Naji A, Menier C, Morandi F, Agaugué S, Maki G, Ferretti E, et al. Binding of HLA-G to ITIM-bearing Ig-like transcript 2 receptor suppresses B cell responses. J Immunol. (2014) 192:1536–46. doi: 10.4049/jimmunol.1300438
69. Naji A, Menier C, Maki G, Carosella ED, Rouas-Freiss N. Neoplastic B-cell growth is impaired by HLA-G/ILT2 interaction. Leukemia. (2012) 26:1889–92. doi: 10.1038/leu.2012.62
70. Contini P, Ghio M, Poggi A, Filaci G, Indiveri F, Ferrone S, et al. Soluble HLA-A,-B,-C and -G molecules induce apoptosis in T and NK CD8+ cells and inhibit cytotoxic T cell activity through CD8 ligation. Eur J Immunol. (2003) 33:125–34. doi: 10.1002/immu.200390015
71. Fournel S, Aguerre-Girr M, Huc X, Lenfant F, Alam A, Toubert A, et al. Cutting edge, soluble HLA-G1 triggers CD95/CD95 ligand-mediated apoptosis in activated CD8+ cells by interacting with CD8. J Immunol. (2000) 164:6100–4. doi: 10.4049/jimmunol.164.12.6100
72. Selmani Z, Naji A, Zidi I, Favier B, Gaiffe E, Obert L, et al. Human leukocyte antigen-G5 secretion by human mesenchymal stem cells is required to suppress T lymphocyte and natural killer function and to induce CD4+CD25highFOXP3+ regulatory T cells. Stem Cells. (2008) 26:212–22. doi: 10.1634/stemcells.2007-0554
73. Naji A, Le Rond S, Durrbach A, Krawice-Radanne I, Creput C, Daouya M, et al. CD3+CD4low and CD3+CD8low are induced by HLA-G: novel human peripheral blood suppressor T-cell subsets involved in transplant acceptance. Blood. (2007) 110:3936–48. doi: 10.1182/blood-2007-04-083139
74. Liang S, Horuzsko A. Mobilizing dendritic cells for tolerance by engagement of immune inhibitory receptors for HLA-G. Hum Immunol. (2003) 64:1025–32. doi: 10.1016/j.humimm.2003.08.348
75. Liang S, Ristich V, Arase H, Dausset J, Carosella ED, Horuzsko A. Modulation of dendritic cell differentiation by HLA-G and ILT4 requires the IL-6–STAT3 signaling pathway. Proc Natl Acad Sci USA. (2008) 105:8357–62. doi: 10.1073/pnas.0803341105
76. Nasef A, Mathieu N, Chapel A, Frick J, François S, Mazurier C, et al. Immunosuppressive effects of mesenchymal stem cells: involvement of HLA-G. Transplantation. (2007) 84:231–7. doi: 10.1097/01.tp.0000267918.07906.08
77. Agaugue S, Carosella ED, Rouas-Freiss N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood. (2011) 117:7021–31. doi: 10.1182/blood-2010-07-294389
78. Carosella ED, HoWangYin KY, Favier B, LeMaoult J. HLA-G-dependent suppressor cells, diverse by nature, function, and significance. Hum Immunol. (2008) 69:700–7. doi: 10.1016/j.humimm.2008.08.280
79. Brown R, Kabani K, Favaloro J, Yang S, Ho PJ, Gibson J, et al. CD86+ or HLA-G+ can be transferred via trogocytosis from myeloma cells to T cells and are associated with poor prognosis. Blood. (2012) 120:2055–63. doi: 10.1182/blood-2012-03-416792
80. Alegre E, Rebmann V, Lemaoult J, Rodriguez C, Horn PA, Diaz-Lagares A, et al. In vivo identification of an HLA-G complex as ubiquitinated protein circulating in exosomes. Eur J Immunol. (2013) 43:1933–9. doi: 10.1002/eji.201343318
81. Rebmann V, Konig L, Nardi Fda S, Wagner B, Manvailer LF, Horn PA. The potential of HLA-G-bearing extracellular vesicles as a future element in HLA-G immune biology. Front Immunol. (2016) 7:173. doi: 10.3389/fimmu.2016.00173
82. Dias FC, Castelli EC, Collares CV, Moreau P, Donadi EA. The role of HLA-G molecule and HLA-G gene polymorphisms in tumors, viral hepatitis, and parasitic diseases. Front Immunol. (2015) 6:9. doi: 10.3389/fimmu.2015.00009
83. Donadi EA, Castelli EC, Arnaiz-Villena A, Roger M, Rey D, Moreau P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell Mol Life Sci. (2011) 68:369–95. doi: 10.1007/s00018-010-0580-7
84. Rousseau P, Le Discorde M, Mouillot G, Marcou C, Carosella ED, Moreau P. The 14 bp deletion-insertion polymorphism in the 3' UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol. (2003) 64:1005–10. doi: 10.1016/j.humimm.2003.08.347
85. Hviid TV, Hylenius S, Rorbye C, Nielsen LG. HLA-G allelic variants are associated with differences in the HLA-G mRNA isoform profile and HLA-G mRNA levels. Immunogenetics. (2003) 55:63–79. doi: 10.1007/s00251-003-0547-z
86. Porto IO, Mendes-Junior CT, Felício LP, Georg RC, Moreau P, Donadi EA, et al. MicroRNAs targeting the immunomodulatory HLA-G gene: a new survey searching for microRNAs with potential to regulate HLA-G. Mol Immunol. (2015) 65:230–41. doi: 10.1016/j.molimm.2015.01.030
87. Veit TD, Chies JA. Tolerance versus immune response–microRNAs as important elements in the regulation of the HLA-G gene expression. Transpl Immunol. (2009) 20:229–31. doi: 10.1016/j.trim.2008.11.001
88. Seliger B. Role of microRNAs on HLA-G expression in human tumors. Hum Immunol. (2016) 77:760–3. doi: 10.1016/j.humimm.2016.04.006
89. Xu HH, Shi WW, Lin A, Yan WH. HLA-G 3' untranslated region polymorphisms influence the susceptibility for human papillomavirus infection. Tissue Antigens. (2014) 84:216–22. doi: 10.1111/tan.12359
90. Xu HH, Zhang X, Zheng HH, Han QY, Lin AF, Yan WH. Association of HLA-G 3' UTR polymorphism and expression with the progression of cervical lesions in human papillomavirus 18 infections. Infect Agent Cancer. (2018) 13:42. doi: 10.1186/s13027-018-0217-2
91. Silva ID, Muniz YC, Sousa MC, Silva KR, Castelli EC, Filho JC, et al. HLA-G 3′UTR polymorphisms in high grade and invasive cervico-vaginal cancer. Hum Immunol. (2013) 74:452–8. doi: 10.1016/j.humimm.2012.11.025
92. Bortolotti D, Gentili V, Rotola A, Di Luca D, Rizzo R. Implication of HLA-G 3′ untranslated region polymorphisms in human papillomavirus infection. Tissue Antigens. (2014) 83:113–8. doi: 10.1111/tan.12281
93. Yang YC, Chang TY, Chen TC, Lin WS, Chang SC, Lee YJ. Human leucocyte antigen-G polymorphisms are associated with cervical squamous cell carcinoma risk in Taiwanese women. Eur J Cancer. (2014) 50:469–74. doi: 10.1016/j.ejca.2013.10.018
94. Medeiros FS, Martins AES, Gomes RG, de Oliveira SAV, Welkovic S, Maruza M, et al. Variation sites at the HLA-G 3' untranslated region confer differential susceptibility to HIV/HPV co-infection and aneuploidy in cervical cell. PLoS ONE. (2018) 13:e0204679. doi: 10.1371/journal.pone.0204679
95. Sudenga SL, Wiener HW, King CC, Rompalo AM, Cu-Uvin S, Klein RS, et al. Dense genotyping of immune related loci identifies variants associated with clearance of HPV among HIV positive women in the HIV epidemiology research study (HERS). PLoS ONE. (2014) 9:e99109. doi: 10.1371/journal.pone.0099109
96. Ferguson R, Ramanakumar AV, Richardson H, Tellier PP, Coutlée F, Franco EL, et al. Human leukocyte antigen (HLA)-E and HLA-G polymorphisms in human papillomavirus infection susceptibility and persistence. Hum Immunol. (2011) 72:337–41. doi: 10.1016/j.humimm.2011.01.010
97. Smith MA, Tellier PP, Roger M, Coutlée F, Franco EL, Richardson H. Determinants of human papillomavirus coinfections among Montreal university students: the influence of behavioral and biologic factors. Cancer Epidemiol Biomarkers Prev. (2014) 23:812–22. doi: 10.1158/1055-9965.EPI-13-1255
98. Metcalfe S, Roger M, Faucher MC, Coutlée F, Franco EL, Brassard P. The association between human leukocyte antigen (HLA)-G polymorphisms and human papillomavirus (HPV) infection in Inuit women of northern Quebec. Hum Immunol. (2013) 74:1610–5. doi: 10.1016/j.humimm.2013.08.279
99. Ferguson R, Ramanakumar AV, Koushik A, Coutlée F, Franco E, Roger M, et al. Human leukocyte antigen G polymorphism is associated with an increased risk of invasive cancer of the uterine cervix. Int J Cancer. (2012) 131:E312–9. doi: 10.1002/ijc.27356
100. Simões RT, Gonçalves MA, Castelli EC, Júnior CM, Bettini JS, Discorde ML, et al. HLA-G polymorphisms in women with squamous intraepithelial lesions harboring human papillomavirus. Mod Pathol. (2009) 22:1075–82. doi: 10.1038/modpathol.2009.67
101. Alves BM, Prellwitz IM, Siqueira JD, Meyrelles ÂR, Bergmann A, Seuánez HN, et al. The effect of human leukocyte antigen G alleles on human papillomavirus infection and persistence in a cohort of HIV-positive pregnant women from Brazil. Infect Genet Evol. (2015) 34:339–43. doi: 10.1016/j.meegid.2015.06.027
102. Louvanto K, Roger M, Faucher MC, Syrjänen K, Grenman S, Syrjänen S. HLA-G and vertical mother-to-child transmission of human papillomavirus infection. Hum Immunol. (2018) 79:471–6. doi: 10.1016/j.humimm.2018.03.002
103. Gillio-Tos A, Bicalho Mda G, Fiano V, Grasso C, Tarallo V, De Marco L, et al. Case-control study of HLA-G promoter methylation status, HPV infection and cervical neoplasia in Curitiba, Brazil: a pilot analysis. BMC Cancer. (2012) 12:618. doi: 10.1186/1471-2407-12-618
104. Cordeiro JC, da Silva JS, Roxo VS, da Graça Bicalho M. A pilot study on Hla-G locus control region haplotypes and cervical intraepithelial neoplasias. Hum Immunol. (2017) 78:281–6. doi: 10.1016/j.humimm.2016.12.004
105. Gimenes F, Teixeira JJ, de Abreu AL, Souza RP, Pereira MW, da Silva VR, et al. Human leukocyte antigen (HLA)-G and cervical cancer immunoediting: a candidate molecule for therapeutic intervention and prognostic biomarker? Biochim Biophys Acta. (2014) 1846:576–89. doi: 10.1016/j.bbcan.2014.10.004
106. Dong DD, Yang H, Li K, Xu G, Song LH, Fan XL, et al. Human leukocyte antigen-G (HLA-G) expression in cervical lesions: association with cancer progression, HPV 16/18 infection, and host immune response. Reprod Sci. (2010) 17:718–23. doi: 10.1177/1933719110369183
107. Zheng N, Wang CX, Zhang X, Du LT, Zhang J, Kan SF, et al. Up-regulation of HLA-G expression in cervical premalignant and malignant lesions. Tissue Antigens. (2011) 77:218–24. doi: 10.1111/j.1399-0039.2010.01607.x
108. Yoon BS, Kim YT, Kim JW, Kim SH, Kim JH, Kim SW. Expression of human leukocyte antigen-G and its correlation with interleukin-10 expression in cervical carcinoma. Int J Gynaecol Obstet. (2007) 98:48–53. doi: 10.1016/j.ijgo.2007.03.041
109. Rodriguez JA, Galeano L, Palacios DM, Gómez C, Serrano ML, Bravo MM, et al. Altered HLA class I and HLA-G expression is associated with IL-10 expression in patients with cervical cancer. Pathobiology. (2012) 79:72–83. doi: 10.1159/000334089
110. Guimarães MC, Soares CP, Donadi EA, Derchain SF, Andrade LA, Silva TG, et al. Low expression of human histocompatibility soluble leukocyte antigen-G (HLA-G5) in invasive cervical cancer with and without metastasis, associated with papilloma virus (HPV). J Histochem Cytochem. (2010) 58:405–11. doi: 10.1369/jhc.2009.954131
111. Ferns DM, Heeren AM, Samuels S, Bleeker MCG, de Gruijl TD, Kenter GG, et al. Classical and non-classical HLA class I aberrations in primary cervical squamous and adenocarcinomas and paired lymph node metastases. J Immunother Cancer. (2016) 4:78. doi: 10.1186/s40425-016-0184-3
112. Zhou JH, Ye F, Chen HZ, Zhou CY, Lu WG, Xie X. Altered expression of cellular membrane molecules of HLA-DR, HLA-G and CD99 in cervical intraepithelial neoplasias and invasive squamous cell carcinoma. Life Sci. (2006) 78:2643–9. doi: 10.1016/j.lfs.2005.10.039
113. Gonçalves MA, Le Discorde M, Simões RT, Rabreau M, Soares EG, Donadi EA, et al. Classical and non-classical HLA molecules and p16(INK4a) expression in precursors lesions and invasive cervical cancer. Eur J Obstet Gynecol Reprod Biol. (2008) 141:70–4. doi: 10.1016/j.ejogrb.2008.06.010
114. Samuels S, Ferns DM, Meijer D, van Straalen JP, Buist MR, Zijlmans HJ, et al. High levels of soluble MICA are significantly related to increased disease-free and disease-specific survival in patients with cervical adenocarcinoma. Tissue Antigens. (2015) 85:476–83. doi: 10.1111/tan.12562
115. Fahim NM, Shehata IH, Taha SE, Fahmy RA, Elsayed MS. Human Leukocyte Antigen-G (HLA-G) expression in precancerous and cancerous cervical lesions: association with human papilloma virus infection and host immune response. Egypt J Immunol. (2018) 25:125–34.
116. Sun J, Chu H, Ji J, Huo G, Song Q, Zhang X. Long non-coding RNA HOTAIR modulates HLA-G expression by absorbing miR-148a in human cervical cancer. Int J Oncol. (2016) 49:943–52. doi: 10.3892/ijo.2016.3589
117. Miranda LN, Reginaldo FP, Souza DM, Soares CP, Silva TG, Rocha KB, et al. Greater expression of the human leukocyte antigen-G (HLA-G) and interleukin-17 (IL-17) in cervical intraepithelial neoplasia: analytical cross-sectional study. São Paulo Med J. (2015) 133:336–42. doi: 10.1590/1516-3180.2013.7170009
118. Wang Y, Liu XH, Li YH, Li O. The paradox of IL-10-mediated modulation in cervical cancer. Biomed Rep. (2013) 1:347–51. doi: 10.3892/br.2013.69
119. Real LM, Cabrera T, Collado A, Jimenez P, Garcia A, Ruiz-Cabello F, et al. Expression of HLA G in human tumors is not a frequent event. Int J Cancer. (1999) 81:512–8. doi: 10.1002/(SICI)1097-0215(19990517)81:4<512::AID-IJC2>3.0.CO;2-O
120. Cibula D, Pötter R, Planchamp F, Avall-Lundqvist E, Fischerova D, Haie-Meder C, et al. The European Society of Gynaecological Oncology/European Society for Radiotherapy and Oncology/European Society of Pathology Guidelines for the management of patients with cervical Cancer. Virchows Arch. (2018) 472:919–36. doi: 10.1007/s00428-018-2362-9
121. Zagouri F, Sergentanis TN, Chrysikos D, Filipits M, Bartsch R. Molecularly targeted therapies in cervical cancer. A systematic review. Gynecol Oncol. (2012) 126:291–303. doi: 10.1016/j.ygyno.2012.04.007
122. Menderes G, Black J, Schwab CL, Santin AD. Immunotherapy and targeted therapy for cervical cancer: an update. Expert Rev Anticancer Ther. (2016) 16:83–98. doi: 10.1586/14737140.2016.1121108
123. Eskander RN, Tewari KS. Beyond angiogenesis blockade: targeted therapy for advanced cervical cancer. J Gynecol Oncol. (2014) 25:249–59. doi: 10.3802/jgo.2014.25.3.249
124. Sato M, Kawana K, Adachi K, Fujimoto A, Yoshida M, Nakamura H, et al. Regeneration of cervical reserve cell-like cells from human induced pluripotent stem cells (iPSCs): a new approach to finding targets for cervical cancer stem cell treatment. Oncotarget. (2017) 8:40935–45. doi: 10.18632/oncotarget.16783
125. Carosella ED, Ploussard G, LeMaoult J, Desgrandchamps F. A systematic review of immunotherapy in urologic cancer: evolving roles for targeting of CTLA-4, PD-1/PD-L1, and HLA-G. Eur Urol. (2015) 68:267–79. doi: 10.1016/j.eururo.2015.02.032
126. Saglam O, Conejo-Garcia J. PD-1/PD-L1 immune checkpoint inhibitors in advanced cervical cancer. Integr Cancer Sci Ther. (2018) 5. doi: 10.15761/ICST.1000272
Keywords: human leukocyte antigen G, human papillomavirus, viral infection, carcinogenesis, cervical cancer, immunotherapy
Citation: Xu H-H, Yan W-H and Lin A (2020) The Role of HLA-G in Human Papillomavirus Infections and Cervical Carcinogenesis. Front. Immunol. 11:1349. doi: 10.3389/fimmu.2020.01349
Received: 24 March 2020; Accepted: 27 May 2020;
Published: 25 June 2020.
Edited by:
Joao P. B. Viola, National Cancer Institute (INCA), BrazilReviewed by:
Ana Paula Lepique, University of São Paulo, BrazilMarcelo A. Soares, National Cancer Institute (INCA), Brazil
Copyright © 2020 Xu, Yan and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aifen Lin, YWlmZW5saW5AeWFob28uY29t
 Hui-Hui Xu
Hui-Hui Xu Wei-Hua Yan
Wei-Hua Yan Aifen Lin
Aifen Lin