
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 22 July 2020
Sec. Cancer Immunity and Immunotherapy
Volume 11 - 2020 | https://doi.org/10.3389/fimmu.2020.01214
This article is part of the Research TopicRoles of Tumor-Recruited Myeloid Cells in Immune Evasion in CancerView all 13 articles
 Elena Daveri†
Elena Daveri† Elisabetta Vergani†
Elisabetta Vergani† Eriomina Shahaj*
Eriomina Shahaj* Laura Bergamaschi
Laura Bergamaschi Stefano La Magra
Stefano La Magra Michela Dosi
Michela Dosi Chiara Castelli
Chiara Castelli Monica Rodolfo
Monica Rodolfo Licia Rivoltini
Licia Rivoltini Viviana Vallacchi‡
Viviana Vallacchi‡ Veronica Huber‡
Veronica Huber‡Immunotherapy with immune checkpoint inhibitors can achieve long-term tumor control in subsets of patients. However, its effect can be blunted by myeloid-induced resistance mechanisms. Myeloid cells are highly plastic and physiologically devoted to wound healing and to immune homeostasis maintenance. In cancer, their physiological activities can be modulated, leading to an expansion of pro-inflammatory and immunosuppressive cells, the myeloid-derived suppressor cells (MDSCs), with detrimental consequences. The involvement of MDSCs in tumor development and progression has been widely investigated and MDSC-induced immunosuppression is acknowledged as a mechanism hindering effective immune checkpoint blockade. Small non-coding RNA molecules, the microRNAs (miRs), contribute to myeloid cell regulation at different levels, comprising metabolism and function, as well as their skewing to a MDSC phenotype. miR expression can be indirectly induced by cancer-derived factors or through direct miR import via extracellular vesicles. Due to their structural stability and their presence in body fluids miRs represent promising predictive biomarkers of resistance, as we recently found by investigating plasma samples of melanoma patients undergoing immune checkpoint blockade. Dissection of the miR-driven involved mechanisms would pave the way for the identification of new druggable targets. Here, we discuss the role of these miRs in shaping myeloid resistance to immunotherapy with a special focus on immunosuppression and immune escape.
Myeloid cells are involved in inflammatory processes, including cancer, and their accrual to the tumor microenvironment (TME) leads to immunosuppression and angiogenesis, thereby promoting tumor growth. Thanks to their plasticity, they are acknowledged cancer allies, negative prognostic factors, and pharmacological targets. Low/negative HLA-DR expression (1) defines monocytic myeloid-derived suppressor cells (CD14+HLA-DRlow/neg; M-MDSCs), which influence cancer aggressiveness and resistance to immune checkpoint inhibitors (ICIs) (2). We focused on myeloid cells for more than a decade and first defined M-MDSCs in melanoma patients (3). We dissected underlying mechanisms via an in vitro tumor extracellular vesicle (EV)-healthy donor monocyte-MDSC model and identified a set of causally involved microRNAs (miRs), the “MDSC-miRs.” miRs are small non-coding RNAs of ~22 nucleotides, which modulate biological processes by mostly interacting with the 3′-untranslated region (UTR) of the target messenger RNA (mRNA). An imperfect base-pair interaction induces translational repression, while a perfectly base-paired miR directly cleaves the mRNA (4, 5). However, some miRs can also bind the 5′-UTR of mRNA, upregulating its translation (6). We measured increased MDSC-miR levels in circulating CD14+ cells and lesions of melanoma patients in association with myeloid infiltrates and peripheral blood MDSC accrual (7, 8). Matching of MDSC-miR predicted target genes with EV-MDSC transcriptional profile revealed miR involvement in chemotaxis, adhesion, and differentiation of myeloid cells. The upregulation of MDSC-miRs, including miR-146a, miR-146b, miR-155, miR-125b, miR-100, let-7e, miR-125a, and miR-99b, in baseline plasma predicted resistance to ICIs (8). In vitro, MDSC-miR antagonists relieved the suppressive potential of patients' monocytes leading to autologous T cell reactivation. Thus, MDSC-miRs could account for myeloid deregulation, implying an involvement of blood factors in the epigenetic control of MDSC functions. A higher MDSC frequency is associated with poor prognosis, even upon immunotherapy, anticipating a reduced treatment efficacy. Pharmacological MDSC reduction, inhibition of their suppressive activities or promotion of their differentiation are under testing at preclinical and clinical levels (9, 10). The functional roles of miR expression by immune cells remain controversial. In case of MDSC-miRs, their overexpression impacts myeloid cell differentiation and polarization by participating in immunosuppressive pathways. Like other miRs, also MDSC-miRs are detectable in EVs, whose size correlates with M-MDSC frequency (8). Tumor and immune cell EVs attracted interest as reservoirs of functional messages exchanged between adjacent cells in the TME and at a distance. EV membrane guarantees content integrity, enabling safe traveling of proteins, lipids, and genetic material to interaction-prone cells. Major efforts are dedicated to investigate EVs as biomarkers of response or drivers of resistance mechanisms to ICIs.
This review discusses the role of MDSC-miRs in shaping myeloid resistance to immunotherapy with a focus on immunosuppression and escape.
As oncomiRs or tumor-suppressors, miRs can promote or inhibit cancer development. They directly target cell proliferation and apoptosis genes, thus being involved in chemotherapy resistance, drug target deregulation, and drug metabolism mechanisms (11). In immunotherapy, miRs can control the success of ICIs by targeting PD-1 and PD-L1, MHC-antigen presentation machinery, and TLR signaling (12). Among MDSC-miRs, miR-155 suppresses PD-L1 through directly binding the 3′-UTR of PD-L1 in human lymphatic endothelial cells (13). The reduction of PD-L1 expression and the consequent disruption of the PD-L1/PD-1 axis may contribute to sustaining T cell antitumor responses, thereby synergizing with ICIs to improve cancer immunotherapy outcome. This miR contributes essentially to mounting of CD8+ T cell responses by restraining T cell senescence and exhaustion through epigenetic silencing of transcription factors determining their terminal differentiation (14). Moreover, miR-155 expression correlates with TCR stimulation of tumor-infiltrating T cells in melanoma patients (15). The MDSC-miR-146a, 146b, 155, and let-7, bind to the 3′-UTR of TLRs or TLR-associated genes resulting in post-translational TLR signaling repression and inflammatory response modulation (16). Similarly, the MDSC-miR-125a and let-7e regulate the inflammatory response and the IL-10-mediated tolerance to LPS, by targeting the TLR4 pathway in monocytes (17). TLR4 can promote expansion of PD-L1+ MDSCs, an effect mediated via HSP86-TLR4 signaling pathway activation (18). Since specific miRs can directly activate TLRs expressed at endosomal level (19, 20), MDSC-miRs might target these proteins and contribute to expanding PD-L1 expressing MDSCs. Thus, targeting MDSC-miRs might potentiate ICI-based immunotherapy.
The development of therapeutic antagomiRs and miR mimics have entered phase I and II clinical studies (21). The DNA-single strand antagomiRs are usually designed on first-generation antisense oligonucleotides or modified with locked nucleic acids to reduce the oncomiR activity by competition with the native cancer-suppressing target transcripts (22). MiR mimics are double strand oligonucleotides that enter the native cellular process mimicking pre-miR duplex (23). Cobomarsen (MRG-106), a miR-155 inhibitor, has entered phase I trials to study safety and potential efficacy following local or intravenous administration in lymphoma and leukemia patients (24).
Despite the therapeutic potential of miRs, their delivery remains challenging, due to undesired off-target effects, hindered cell uptake, and short circulation half-life (25). Synthetic nanoparticles (NPs) mediate specific cell uptake and prevent miR clearance (26). In preclinical models, effective miR supply was obtained via neutral lipid emulsion-based approach for miRs of the let-7 family, as well as neutral liposomes and synthetic polyethylenimine-based nanocarriers for miR-145. Lastly, pH low insertion peptide-modified antagomiRs were able to inhibit the oncomiR miR-155 (27). Otherwise, miR-155-loaded NPs can repolarize tumor-associated macrophages (TAMs) from pro-tumorigenic M2 to anti-inflammatory M1-like phenotype, reversing the immunosuppressive TME (28). In clinical setting, NP-based miR manipulation comprises liposomal (DOPC)-encapsulated siRNAs targeting EphA2 in solid tumors (29), bacterial derived nanocells EDVs (EnGeneIC Delivery Vehicle), or TargomiRs, for miR-16 mimic delivery (30). EVs may be also suitable for miR delivery (31). Healthy donor plasma miR-loaded EVs promoted apoptosis in HCC cells (31), while miR-sponge engineered EVs reduced glioblastoma volume in rats (32). Finally, the natural exchange of endogenous miRs between immune cells, such as miR-155 and miR-146a carried by dendritic cell EVs, controls inflammatory gene expression or promotes apoptotic cell clearance, as in case of endothelial cell EVs transferring miR-125a to macrophages (33).
Specific miR inhibition is accomplished by Small Molecule Inhibitors of miRs (SMIRs), which target synergistically tumor cells and oncomiRs, such as linifanib. This VEGF- and PDGF-receptor tyrosine kinase-inhibitor effectively inhibits the oncogenic function of miR-10b in preclinical cancer models (34). Finally, several miRs are related to tumor radioresistance management, where, thanks to the inhibition of ATM protein, they can modulate DNA damage response sensitizing tumor cells to radiotherapy (35).
The upregulation of miR-146a, miR-146b, miR-155, miR-125b, miR-100, let-7e, miR-125a, and miR-99b can skew immune cells into inhibitors of response to immune and other cancer therapies (Table 1). Of note, five miRs out of eight show a coordinated expression pattern due to their transcription as clusters. The miR-125a~99b~let-7e cluster is hosted in the first intron of the long non-coding RNA NCRNA00085, whereas miR-125b, miR-100, and let-7a are hosted in MIR100HG (17, 58). A clear association of miR-125a~99b~let-7e cluster and acquisition of a myeloid immunosuppressive phenotype has been demonstrated (17, 36, 59, 60). In particular, stimulation of monocytes with GM-CSF, IL-4, and R848 TLR7/8 agonist upregulates the miR-125a~99b~let-7e cluster, activates STAT3, and induces the acquisition of an immunosuppressive phenotype. Conversely, the depletion of the cluster reverses immunosuppressive functions and MDSC phenotype hallmarks, by downregulating PD-L1 and IDO, while increasing HLA-DR expression. This contributes to STAT3 stabilization through downregulation of TRIB2, a suppressor of MAPK signaling, and SOCS1, a key regulator of cytokine signaling and STAT3 inhibitor. The miR-125a~99b~let-7e cluster is negatively regulated by IFNγ, while it is induced by STAT3 and SMAD3, in turn activated by IL-10 and TGFβ immunoregulatory cytokines. miR-125a and let-7e also exert their anti-inflammatory activity by targeting the TLR signaling pathway molecules TLR4, CD14, and IRAK1, leading to decreased pro-inflammatory cytokine release by myeloid cells (17, 59).
MIR100HG and its encoded miR-125b and miR-100 are induced by TGFβ, the main cytokine released by M2 macrophages (61). TGFβ promotes cancer epithelial-to-mesenchymal transition (EMT) through MIR100HG induction and SMAD2/3 transcription factor activation. The dysregulation of this cluster is also causally linked with drug resistance in several tumor types (58, 62). In immune cells, miR-125b expression is usually linked to antitumor M1-like macrophages, whereas in T cells it inhibits CD4 T cell differentiation and γδ T cell activation (37). In contrast, little is known about miR-100 expression and function in immune cells. In regulatory T cells (Tregs) increased levels of the edited variant of miR-100 changes its target gene from MTOR to SMAD2, resulting in limited differentiation and increase of Treg plasticity (38).
Under physiological conditions the miR-146 family (miR-146a and miR-146b) and miR-155 actively control innate immunity, whereas in cancer these miRs have gained attention for their deregulation and acquisition of oncogenic roles. Both are transcriptionally regulated by NFkB, but with opposite functions: miR-146 represents the anti-inflammatory and miR-155 the pro-inflammatory counterpart. miR-146a/b act as negative feedback regulators of TLR signaling through inhibition of the NFkB pathway by downregulation of TRAF6 and IRAK1 (63), thereby dampening the production of pro-inflammatory mediators (64). On the other hand, miR-146b is also induced by TLR4 signaling via an IL-10-mediated STAT3-dependent loop (65), and it inhibits macrophage activation by targeting IRF5 (39). miR-146a is an essential regulator of immune cell activation and malignant transformation (64), and knockout mice are affected by chronic NFkB dysregulation and myeloid malignancies (40, 41). Several studies proposed miR-146a as an immunotherapeutic target: its overexpression reduces the metastatic potential of breast cancer (BC) cell lines through NFkB inhibition (42), whereas it supports the M2-like phenotype of TAMs in endometrial cancer (43). In a preclinical model of HCC, miR-146a inhibition alters the STAT3 activation-associated cytokine profile improving the anti-tumor effect of lymphocytes (44). Mastroianni et al. identified miR-146a as a central negative regulator of the STAT1/IFNγ axis, affecting migration, proliferation, and inducing PD-L1 expression. Combined PD-1 blockade and miR-146a antagomiR improve survival of melanoma-bearing mice (45). We found that high miR-146a levels, concomitantly with the other MDSC-miRs, are associated with MDSC induction and ICI resistance (8). In myeloid leukemia, miR-146a mimics can inhibit tumorigenic NFkB activity (46). Finally, miR-146a is also involved in ICI-mediated immune-related adverse events (irAEs), as shown by knockout mice exhibiting increased T cell activity and inflammation during ICI intake. These effects could be restrained by miR-146a mimics (47).
The pro-inflammatory miR-155 is induced upon TLR/IFNγ stimulation in monocyte/macrophages and drives their response by regulating mRNA targets with inhibitory effects on innate immune cell activation (66). In tumor cells, intrinsic miR-155 mediates pro- or anti-tumor effects (67). Similarly to miR-146a, miR-155 upregulation promotes cell proliferation, colony formation, and xenograft tumor growth in BC models by negative regulation of SOCS1 and SHIP1, leading to constitutive STAT3 activation and pro-tumor inflammation (48). Deficiency of miR-155 can also foster tumor growth through MDSC recruitment and potentiation of their tumor promoting functions, as demonstrated in BC. Here, miR-155 loss in myeloid cells impairs TAM activation, while in tumor cells it stimulates C/EBP-β-mediated cytokine production in turn stimulating tumor-infiltrating MDSCs (49, 50). Similar results were obtained in mouse models of melanoma and lung cancer (51). As for other miRs also miR-155 appears to cover apparently contradictory roles depending on the expressing cell or the setting. Li et al. showed that upregulated miR-155 together with miR-21 led to MDSC expansion, whereas their loss reversed this effect. In particular, by targeting SHIP1 and PTEN these miRs synergistically increase STAT3 activity, promoting MDSCs (52). In this line, loss of miR-155 can enhance antitumor T cell activity by reducing MDSC immunosuppression and tumor infiltration (53). In contrast, miR-155 expression by T cells promotes antitumor immunity and ICIs hinder miR-155-deficiency-induced immune escape (54). We found that miR-146a and miR-155 along with the other MDSC-miRs contribute to MDSC induction (8), suggesting that the expression levels of different miRs can influence the fine-tuning of pro- or anti-inflammatory pathways depending on the cell type. Interestingly, in tumors with high mutational burden, such as melanoma and lung cancer, miR-155 was associated with a strong immune signature and improved clinical outcomes (55). Likewise, miR-155 potentiates immunotherapy through epigenetic regulation of CD8+ T cell differentiation via PRC2/Phf19 signaling (14). Yan and coworkers demonstrated that miR-155-induced downregulation of TIM3, a negative immune checkpoint, enhanced the cytolytic activity of anti-HCC CD8+ T cells (56). Finally, miR-155 overexpression can optimize CD8+ T cell antitumor activity and improve adoptive-transfer in low-affinity antigen tumors (57).
All cell types release EVs, membrane-surrounded structures devoted to intercellular communication. EVs are present in body fluids including plasma, serum, lymph, urine, saliva, tears, and milk (68). Their content of proteins, nucleic acids, lipids, and their stability, make EVs potential biomarkers and therapeutic targets of disease (69, 70). Recent evidence shows the ability of tumor-derived EVs to blunt anti-tumor immunity at multiple levels (71). They can operate within the TME or at a distance by boosting angiogenesis, triggering tumor cell EMT, activating cancer-associated fibroblasts, and shaping the immune environment toward a condition of immune escape. Tumor EVs can induce myeloid cell dysfunctions and increase MDSC expansion (7, 8, 18, 72, 73). Indeed, EVs derived from melanoma cell cultures as well as those from plasma of melanoma patients contain the MDSC-miRs and promote the acquisition of MDSC characteristics by healthy donors' monocytes (8). The potential of these miRs to induce such dramatic changes might depend on their integrity, protected by the EV membrane, as well as on their way of transfer. In fact, the internalization of whole EVs carrying different miRs or EV receptor-ligand interaction might account for diverse effects (74). In support of our findings, Gerloff et al. (75) found that miR-125b encapsulated in melanoma EVs promotes a TAM phenotype in macrophages through targeting of lysosomal acid lipase (LIPA). In fact, LIPA deficiency stimulates MDSC expansion in mice, and their tumor promoting functions are driven by mTOR pathway overactivation (76).
As a major obstacle to immunotherapy, it is crucial to study MDSC effects in the TME (77). Myeloid EVs may support immune activation or tolerance (78). Like other cells, also MDSCs release EVs, taking part in intercellular communication. Proteomics of MDSC EVs of BALB/c mice bearing 4T1 or 4T1-IL-1β+ mammary carcinoma showed a higher expression of 63 pro-inflammatory proteins in 4T1-IL-1β+ mice, due to a more inflammatory environment. The MDSC chemotactics S100A8 and S100A9 are abundant in MDSC EVs and polarize macrophages into M2 phenotype (79). These pro-inflammatory proteins are characterized by multiple ubiquitination sites, and MDSC EVs were identified as carriers of enzymes catalyzing ubiquitination (80). Interestingly, EVs from TME-resident MDSC display a stronger immunosuppressive potential than those deriving from spleen or bone marrow MDSCs, suggesting the existence of different phenotypes and functions (81). In contrast, the miR content of MDSC EVs is still elusive, but the dissection could contribute to targeting MDSC EVs release and spreading (82). EVs modulate innate and adaptive immune responses via ligand-receptor interaction or via miRs (83, 84). Indeed, a set of miRs including miR-155, regulate PD-L1 protein expression (85) and induce MDSCs if released via EVs by CLL cells (86). EVs also express immune checkpoints. Actually, PD-L1 carried by EVs was investigated for its role as biomarker and functional inducer of PD-1-mediated immunosuppression (87–90). TIM3 and GAL9 bound to EVs were proposed as prognostic biomarkers in NSCLC patients (91).
Large scale profiling studies demonstrated the association of specific circulating miRs with certain types of human cancer, proposing miRs as biomarkers (92). Their detection could contribute to early cancer diagnosis, patient stratification, and evaluation of therapy outcomes (93). miRs can be found free or EV-bound in peripheral blood or other body fluids (94). Among MDSC-miRs, the miR-125a~99b~let-7e cluster was identified as a potential diagnostic biomarker in many tumor types. Colorectal and ovarian cancer patients display lower levels of EV-bound miR-99b compared to healthy controls (95, 96). Dysregulated levels of free circulating let-7e were observed in retinoblastoma, papillary thyroid carcinoma, lung, and prostate cancer (97–100), whereas altered levels of EV-bound let-7 characterize esophageal adenocarcinoma and lung cancer patients (101, 102). miR-125a also represents a potential biomarker of treatment outcome for HCC patients (103) and altered levels of this miR were detected in certain blood malignancies (104, 105), where they predicted response to chemotherapy, as demonstrated in patients with myelodysplastic syndromes (106). In serum, increased miR-146b levels correlate with papillary thyroid carcinoma recurrence (92), while elevated miR-146a is associated with higher overall response rate and survival in NSCLC (107). Furthermore, lower EV-bound miR-146a levels correlate with cisplatin resistance and shorter progression-free survival in NSCLC patients (108). BC patients display high plasma miR-155 levels and in the absence of disease, its increase is associated with treatment failure (109). Interestingly, also urinary miR-155 can be correlated with BC development (93, 110). In NSCLC, an increase of miR-155 in plasma identifies stage I-II patients, implying this miR as diagnostic tool (93, 111), although it was not suitable as a prognostic biomarker (112). miR-155 expression is also related to risk of relapse in colorectal cancer patients and chemoresistance in pancreatic ductal adenocarcinoma, where anti-apoptotic mechanisms are driven by tumor cell exchange of miR-155 containing EVs (93, 113, 114). Concerning MIR100HG, reduced miR-100 expression coincides with diagnosis and prognosis of bladder cancer (115). In contrast, higher circulating miR-100 levels were found in HCC and esophageal squamous cell carcinoma patients and predicted poor survival (116, 117). Lastly, circulating miR-125b was identified as a biomarker of diagnosis and poor prognosis in NSCLC, BC, colorectal, and epithelial ovarian cancer patients, also during chemotherapy or after surgery (118–123).
Despite major advances, the role of miRs, including MDSC-miRs, expressed by immune cells remains controversial. For instance, both pro and antitumoral potentials are ascribed to miR-155, depending on its expression levels (124). Of interest is also their interplay: miR-146a−/− mice succumb to chronic inflammation and miR-155 expressed by T cells contributes to shortening lifespan by activating autoimmunity (125). The continuous technical improvement will facilitate in-depth investigations of the finely-tuned mechanisms governing the miR balance, expression levels, and consequent repression/overexpression of target genes to clarify the mechanisms governing myeloid cell dysfunctions and MDSC activity. This will be of major relevance also for cancer therapies. In fact, similarly to SMIRs, also ICIs may induce changes in myeloid MDSC-miR expression potentially related to clinical responses. The complex tumor-immune relationship regulated by miRs and the miR-based therapeutic approaches are summarized in Figure 1. Thus, the dissection of therapy-induced miR modulation in immune cells may contribute to decipher and antagonize resistance mechanisms.
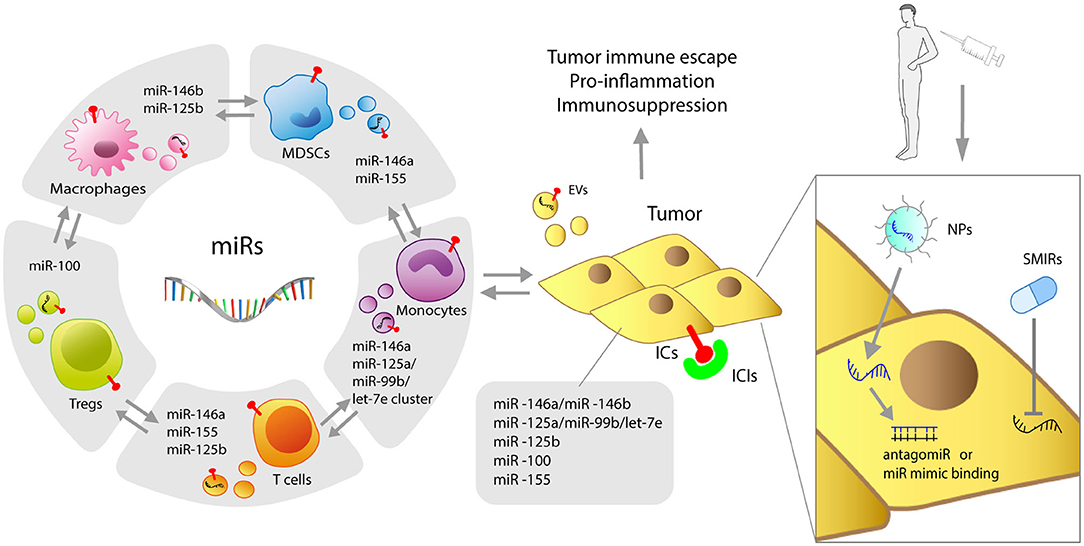
Figure 1. Graphic representation of miRs shaping myeloid resistance to ICIs. Immune and tumor cells, both expressing miRs, mutually interact through miR-loaded EVs. MDSC-derived miRs, including miR-146a, miR-146b, miR-155, miR-125b, miR-100, let-7e, miR-125a, and miR-99b can intervene in cancer progression and interfere with the success of cancer immunotherapy by regulating immune checkpoints (ICs) and different molecular immune targets. Delivery of antagomiRs or miR mimics with NPs, as well as SMIR drugs, represents the current therapeutic strategies to overcome resistance to ICIs induced by miRs. NPs, nanoparticles; SMIR, small molecule inhibitor of miR; ICIs, immune checkpoint inhibitors; EVs, extracellular vesicles.
All authors wrote and revised the manuscript. LB made the figure. All authors have read and agreed to the submitted version of the manuscript.
This work was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) Special Program Innovative Tools for Cancer Risk Assessment and early Diagnosis 5X1000 (no. 12162 to LR), by the Cariplo Foundation (2015-0911 to VV), by Fondi 5x1000 Ministero della Salute 2015 (D/17/1VH to VH), and by PRECIOUS Project-Horizon 2020, Grant Agreement no. 686089.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res. (2017) 5:3–8. doi: 10.1158/2326-6066.CIR-16-0297
2. Tesi RJ. MDSC; the most important cell you have never heard of. Trends Pharmacol Sci. (2019) 40:4–7. doi: 10.1016/j.tips.2018.10.008
3. Filipazzi P, Valenti R, Huber V, Pilla L, Canese P, Iero M, et al. Identification of a new subset of myeloid suppressor cells in peripheral blood of melanoma patients with modulation by a granulocyte-macrophage colony-stimulation factor-based antitumor vaccine. J Clin Oncol. (2007) 25:2546–53. doi: 10.1200/JCO.2006.08.5829
5. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. (2005) 433:769–73. doi: 10.1038/nature03315
6. Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. (2007) 318:1931–4. doi: 10.1126/science.1149460
7. Valenti R, Huber V, Filipazzi P, Pilla L, Sovena G, Villa A, et al. Human tumor-released microvesicles promote the differentiation of myeloid cells with transforming growth factor-beta-mediated suppressive activity on T lymphocytes. Cancer Res. (2006) 66:9290–8. doi: 10.1158/0008-5472.CAN-06-1819
8. Huber V, Vallacchi V, Fleming V, Hu X, Cova A, Dugo M, et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J Clin Invest. (2018) 128:5505–16. doi: 10.1172/JCI98060
9. Anani W, Shurin MR. Targeting myeloid-derived suppressor cells in cancer. Adv Exp Med Biol. (2017) 1036:105–28. doi: 10.1007/978-3-319-67577-0_8
10. Fleming V, Hu X, Weber R, Nagibin V, Groth C, Altevogt P, et al. Targeting myeloid-derived suppressor cells to bypass tumor-induced immunosuppression. Front Immunol. (2018) 9:398. doi: 10.3389/fimmu.2018.00398
11. Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. (2019) 11:25. doi: 10.1186/s13148-018-0587-8
12. Omar HA, El-Serafi AT, Hersi F, Arafa E-SA, Zaher DM, Madkour M, et al. Immunomodulatory MicroRNAs in cancer: targeting immune checkpoints and the tumor microenvironment. FEBS J. (2019) 286:3540–557. doi: 10.1111/febs.15000
13. Yee D, Shah KM, Coles MC, Sharp TV, Lagos D. MicroRNA-155 induction via TNF-α and IFN-γ suppresses expression of programmed death ligand-1 (PD-L1) in human primary cells. J Biol Chem. (2017) 292:20683–93. doi: 10.1074/jbc.M117.809053
14. Ji Y, Fioravanti J, Zhu W, Wang H, Wu T, Hu J, et al. miR-155 harnesses Phf19 to potentiate cancer immunotherapy through epigenetic reprogramming of CD8+ T cell fate. Nat Commun. (2019) 10:1–2. doi: 10.1038/s41467-019-09882-8
15. Martinez-Usatorre A, Sempere LF, Carmona SJ, Carretero-Iglesia L, Monnot G, Speiser DE, et al. MicroRNA-155 expression is enhanced by t-cell receptor stimulation strength and correlates with improved tumor control in melanoma. Cancer Immunol Res. (2019) 7:1013–24. doi: 10.1158/2326-6066.CIR-18-0504
16. Bayraktar R, Bertilaccio MTS, Calin GA. The interaction between two worlds: microRNAs and toll-like receptors. Front Immunol. (2019) 10:1053. doi: 10.3389/fimmu.2019.01053
17. Curtale G, Renzi TA, Mirolo M, Drufuca L, Albanese M, De Luca M, et al. Multi-step regulation of the TLR4 pathway by the miR-125a~99b~let-7e cluster. Front Immunol. (2018) 9:2037. doi: 10.3389/fimmu.2018.02037
18. Fleming V, Hu X, Weller C, Weber R, Groth C, Riester Z, et al. Melanoma extracellular vesicles generate immunosuppressive myeloid cells by upregulating PD-L1 via TLR4 signaling. Cancer Res. (2019) 79:4715–28. doi: 10.1158/0008-5472.CAN-19-0053
19. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. (2012) 109:E2110–6. doi: 10.1073/pnas.1209414109
20. Abdi J, Rashedi I, Keating A. Concise Review: TLR pathway-miRNA interplay in mesenchymal stromal cells: regulatory roles and therapeutic directions. Stem Cells. (2018) 36:1655–62. doi: 10.1002/stem.2902
21. Hanna J, Hossain GS, Kocerha J. The potential for microRNA therapeutics and clinical research. Front Genet. (2019) 10:478. doi: 10.3389/fgene.2019.00478
22. Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. (2014) 13:622–638. doi: 10.1038/nrd4359
23. Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. (2013) 31:577. doi: 10.1038/nbt0713-577
24. Safety Tolerability Pharmacokinetics of MRG-106 in Patients with Mycosis Fungoides (MF) CLL DLBCL or ATLL. Available at: https://clinicaltrials.gov/ct2/show/NCT02580552 (Accessed April 6, 2020).
25. Lee SWL, Paoletti C, Campisi M, Osaki T, Adriani G, Kamm RD, et al. MicroRNA delivery through nanoparticles. J Control Release. (2019) 313:80–95. doi: 10.1016/j.jconrel.2019.10.007
26. Chaudhary V, Jangra S, Yadav NR. Nanotechnology based approaches for detection and delivery of microRNA in healthcare and crop protection. J Nanobiotechnology. (2018) 16:40. doi: 10.1186/s12951-018-0368-8
27. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. (2017) 16:203–222. doi: 10.1038/nrd.2016.246
28. Zang X, Zhang X, Zhao X, Hu H, Qiao M, Deng Y, et al. Targeted delivery of miRNA 155 to tumor associated macrophages for tumor immunotherapy. Mol Pharm. (2019) 16:1714–22. doi: 10.1021/acs.molpharmaceut.9b00065
29. EphA2 siRNA in Treating Patients With Advanced or Recurrent Solid Tumors. Available online at: https://clinicaltrials.gov/ct2/show/NCT01591356 (Accessed April 6, 2020).
30. MesomiR 1: A Phase I Study of TargomiRs as 2nd or 3rd Line Treatment for Patients With Recurrent MPM and NSCLC. Available online at: https://clinicaltrials.gov/ct2/show/NCT02369198 (Accessed April 6, 2020).
31. Pomatto MAC, Bussolati B, D'Antico S, Ghiotto S, Tetta C, Brizzi MF, et al. Improved loading of plasma-derived extracellular vesicles to encapsulate antitumor miRNAs. Mol Ther Methods Clin Dev. (2019) 13:133–44. doi: 10.1016/j.omtm.2019.01.001
32. Monfared H, Jahangard Y, Nikkhah M, Mirnajafi-Zadeh J, Mowla SJ. Potential therapeutic effects of exosomes packed with a miR-21-sponge construct in a rat model of glioblastoma. Front Oncol. (2019) 9:782. doi: 10.3389/fonc.2019.00782
33. Chen J, Hu C, Pan P. Extracellular vesicle microRNA transfer in lung diseases. Front Physiol. (2017) 8:1028. doi: 10.3389/fphys.2017.01028
34. Monroig-Bosque PDC, Shah MY, Fu X, Fuentes-Mattei E, Ling H, Ivan C, et al. OncomiR-10b hijacks the small molecule inhibitor linifanib in human cancers. Sci Rep. (2018) 8:13106. doi: 10.1038/s41598-018-30989-3
35. Rezaeian A-H, Khanbabaei H, Calin GA. Therapeutic Potential of the miRNA-ATM axis in the management of tumor radioresistance. Cancer Res. (2020) 80:139–50. doi: 10.1158/0008-5472.CAN-19-1807
36. Hildebrand D, Eberle M-E, Wölfle SM, Egler F, Sahin D, Sähr A, et al. Hsa-miR-99b/let-7e/miR-125a cluster regulates pathogen recognition receptor-stimulated suppressive antigen-presenting cells. Front Immunol. (2018) 9:1224. doi: 10.3389/fimmu.2018.01224
37. Wang JK, Wang Z, Li G. MicroRNA-125 in immunity and cancer. Cancer Lett. (2019) 454:134–45. doi: 10.1016/j.canlet.2019.04.015
38. Negi V, Paul D, Das S, Bajpai P, Singh S, Mukhopadhyay A, et al. Altered expression and editing of miRNA-100 regulates iTreg differentiation. Nucleic Acids Res. (2015) 43:8057–65. doi: 10.1093/nar/gkv752
39. Peng L, Zhang H, Hao Y, Xu F, Yang J, Zhang R, et al. Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine. (2016) 14:83–96. doi: 10.1016/j.ebiom.2016.10.041
40. Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, et al. MiR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. (2011) 208:1189–201. doi: 10.1084/jem.20101823
41. Zhao JL, Rao DS, Boldin MP, Taganov KD, O'Connell RM, Baltimore D. NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc Natl Acad Sci U S A. (2011) 108:9184–9. doi: 10.1073/pnas.1105398108
42. Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-κB activity with reduction of metastatic potential in breast cancer cells. Oncogene. (2008) 27:5643–7. doi: 10.1038/onc.2008.171
43. Zhou Y-X, Zhao W, Mao L-W, Wang Y-L, Xia L-Q, Cao M, et al. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int J Biochem Cell Biol. (2018) 104:25–33. doi: 10.1016/j.biocel.2018.08.017
44. Sun X, Zhang J, Hou Z, Han Q, Zhang C, Tian Z. MiR-146a is directly regulated by STAT3 in human hepatocellular carcinoma cells and involved in anti-tumor immune suppression. Cell Cycle. (2015) 14:243–52. doi: 10.4161/15384101.2014.977112
45. Mastroianni J, Stickel N, Andrlova H, Hanke K, Melchinger W, Duquesne S, et al. MiR-146a controls immune response in the melanoma microenvironment. Cancer Res. (2019) 79:183–95. doi: 10.1158/0008-5472.CAN-18-1397
46. Su Y-L, Wang X, Mann M, Adamus TP, Wang D, Moreira DF, et al. Myeloid cell-targeted miR-146a mimic inhibits NF-κB-driven inflammation and leukemia progression in vivo. Blood. (2020) 135:167–80. doi: 10.1182/blood.2019002045
47. Marschner D, Falk M, Javorniczky NR, Hanke-Müller K, Rawluk J, Schmitt-Graeff A, et al. MicroRNA-146a regulates immune-related adverse events caused by immune checkpoint inhibitors. JCI Insight. (2020) 5:e132334. doi: 10.1172/jci.insight.132334
48. Jiang S, Zhang H-W, Lu M-H, He X-H, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. (2010) 70:3119–27. doi: 10.1158/0008-5472.CAN-09-4250
49. Zonari E, Pucci F, Saini M, Mazzieri R, Politi LS, Gentner B, et al. A role for miR-155 in enabling tumor-infiltrating innate immune cells to mount effective antitumor responses in mice. Blood. (2013) 122:243–52. doi: 10.1182/blood-2012-08-449306
50. Kim S, Song JH, Kim S, Qu P, Martin BK, Sehareen WS, et al. Loss of oncogenic miR-155 in tumor cells promotes tumor growth by enhancing C/EBP-β-mediated MDSC infiltration. Oncotarget. (2016) 7:11094–112. doi: 10.18632/oncotarget.7150
51. Wang J, Yu F, Jia X, Iwanowycz S, Wang Y, Huang S, et al. MicroRNA-155 deficiency enhances the recruitment and functions of myeloid-derived suppressor cells in tumor microenvironment and promotes solid tumor growth. Int J Cancer. (2015) 136:E602–E13. doi: 10.1002/ijc.29151
52. Li L, Zhang J, Diao W, Wang D, Wei Y, Zhang C-Y, et al. MicroRNA-155 and microRNA-21 promote the expansion of functional myeloid-derived suppressor cells. J Immunol. (2014) 192:1034–43. doi: 10.4049/jimmunol.1301309
53. Chen S, Wang L, Fan J, Ye C, Dominguez D, Zhang Y, et al. Host miR155 promotes tumor growth through a myeloid-derived suppressor cell-dependent mechanism. Cancer Res. (2015) 75:519–31. doi: 10.1158/0008-5472.CAN-14-2331
54. Huffaker TB, Lee S-H, Tang WW, Wallace JA, Alexander M, Runtsch MC, et al. Antitumor immunity is defective in T cell-specific microRNA-155-deficient mice and is rescued by immune checkpoint blockade. J Biol Chem. (2017) 292:18530–41. doi: 10.1074/jbc.M117.808121
55. Ekiz HA, Huffaker TB, Grossmann AH, Stephens WZ, Williams MA, Round JL, et al. MicroRNA-155 coordinates the immunological landscape within murine melanoma and correlates with immunity in human cancers. JCI Insight. (2019) 4:e126543. doi: 10.1172/jci.insight.126543
56. Yan K, Fu Y, Zhu N, Wang Z, Hong J-L, Li Y, et al. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8+T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Int J Biochem Cell Biol. (2019) 110:1–8. doi: 10.1016/j.biocel.2019.01.019
57. Monnot GC, Martinez-Usatorre A, Lanitis E, Lopes SF, Cheng W-C, Ho P-C, et al. miR-155 overexpression in OT-1 CD8+ T cells improves anti-tumor activity against low-affinity tumor antigen. Mol Ther Oncolytics. (2020) 16:111–23. doi: 10.1016/j.omto.2019.12.008
58. Vallacchi V, Rodolfo M. MiR-100 and miR-125b regulate epithelial-mesenchymal transition and drug resistance in tumors. Noncoding RNA investig. (2018) 2:57. doi: 10.21037/ncri.2018.09.05
59. Curtale G, Rubino M, Locati M. MicroRNAs as molecular switches in macrophage activation. Front Immunol. (2019) 10:799. doi: 10.3389/fimmu.2019.00799
60. Curtale G. MiRNAs at the crossroads between innate immunity and cancer: focus on macrophages. Cells. (2018) 7:12. doi: 10.3390/cells7020012
61. Batlle E, Massagué J. Transforming growth factor-β signaling in immunity and cancer. Immunity. (2019) 50:924–940. doi: 10.1016/j.immuni.2019.03.024
62. Vergani E, Di Guardo L, Dugo M, Rigoletto S, Tragni G, Ruggeri R, et al. Overcoming melanoma resistance to vemurafenib by targeting CCL2-induced miR-34a, miR-100 and miR-125b. Oncotarget. (2016) 7:4428–41. doi: 10.18632/oncotarget.6599
63. Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. (2006) 103:12481–6. doi: 10.1073/pnas.0605298103
64. Iacona JR, Lutz CS. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip Rev RNA. (2019) 10:e1533. doi: 10.1002/wrna.1533
65. Lee H-M, Kim TS, Jo E-K. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. (2016) 49:311–8. doi: 10.5483/BMBRep.2016.49.6.056
66. Zhang L, Xu X, Su X. Noncoding RNAs in cancer immunity: functions, regulatory mechanisms, and clinical application. Mol Cancer. (2020) 19:48. doi: 10.1186/s12943-020-01154-0
67. Mahesh G, Biswas R. MicroRNA-155: A master regulator of inflammation. J Interferon Cytokine Res. (2019) 39:321–30. doi: 10.1089/jir.2018.0155
68. Maas SLN, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol. (2017) 27:172–88. doi: 10.1016/j.tcb.2016.11.003
69. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. (2008) 10:1470–6. doi: 10.1038/ncb1800
70. Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. (2012) 18:883–91. doi: 10.1038/nm.2753
71. Li X, Wang Y, Wang Q, Liu Y, Bao W, Wu S. Exosomes in cancer: small transporters with big functions. Cancer Lett. (2018) 435:55–65. doi: 10.1016/j.canlet.2018.07.037
72. Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. (2014) 26:707–21. doi: 10.1016/j.ccell.2014.09.005
73. Tuccitto A, Shahaj E, Vergani E, Ferro S, Huber V, Rodolfo M, et al. Immunosuppressive circuits in tumor microenvironment and their influence on cancer treatment efficacy. Virchows Arch. (2019) 474:407–20. doi: 10.1007/s00428-018-2477-z
74. French KC, Antonyak MA, Cerione RA. Extracellular vesicle docking at the cellular port: extracellular vesicle binding and uptake. Semin Cell Dev Biol. (2017) 67:48–55. doi: 10.1016/j.semcdb.2017.01.002
75. Gerloff D, Lützkendorf J, Moritz RKC, Wersig T, Mäder K, Müller LP, et al. Melanoma-derived exosomal miR-125b-5p educates tumor associated macrophages (TAMs) by targeting lysosomal acid lipase A (LIPA). Cancers (Basel). (2020) 12:464. doi: 10.3390/cancers12020464
76. Zhao T, Du H, Ding X, Walls K, Yan C. Activation of mTOR pathway in myeloid-derived suppressor cells stimulates cancer cell proliferation and metastasis in lal(-/-) mice. Oncogene. (2015) 34:1938–48. doi: 10.1038/onc.2014.143
77. Groth C, Hu X, Weber R, Fleming V, Altevogt P, Utikal J, et al. Immunosuppression mediated by myeloid-derived suppressor cells (MDSCs) during tumour progression. Br J Cancer. (2019) 120:16–25. doi: 10.1038/s41416-018-0333-1
78. Colombo F, Bastoni M, Nigro A, Podini P, Finardi A, Casella G, et al. Cytokines stimulate the release of microvesicles from myeloid cells independently from the P2X7 receptor/acid sphingomyelinase pathway. Front Immunol. (2018) 9:204. doi: 10.3389/fimmu.2018.00204
79. Burke M, Choksawangkarn W, Edwards N, Ostrand-Rosenberg S, Fenselau C. Exosomes from myeloid-derived suppressor cells carry biologically active proteins. J Proteome Res. (2014) 13:836–43. doi: 10.1021/pr400879c
80. Adams KR, Chauhan S, Patel DB, Clements VK, Wang Y, Jay SM, et al. Ubiquitin conjugation probed by inflammation in myeloid-derived suppressor cell extracellular vesicles. J Proteome Res. (2018) 17:315–24. doi: 10.1021/acs.jproteome.7b00585
81. Rashid MH. The critical immunosuppressive effect of MDSC-derived exosomes in the tumor microenvironment. bioRxiv [Preprint]. (2020). doi: 10.1101/2020.03.05.979195
82. Zöller M. Janus-faced myeloid-derived suppressor cell exosomes for the good and the bad in cancer and autoimmune disease. Front Immunol. (2018) 9:137. doi: 10.3389/fimmu.2018.00137
83. Whiteside TL. Exosomes carrying immunoinhibitory proteins and their role in cancer. Clin Exp Immunol. (2017) 189:259–67. doi: 10.1111/cei.12974
84. Eichmüller SB, Osen W, Mandelboim O, Seliger B. Immune modulatory microRNAs involved in tumor attack and tumor immune escape. J Natl Cancer Inst. (2017) 109:djx034. doi: 10.1093/jnci/djx034
85. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. (2018) 48:434–52. doi: 10.1016/j.immuni.2018.03.014
86. Bruns H, Böttcher M, Qorraj M, Fabri M, Jitschin S, Dindorf J, et al. CLL-cell-mediated MDSC induction by exosomal miR-155 transfer is disrupted by vitamin D. Leukemia. (2017) 31:985–8. doi: 10.1038/leu.2016.378
87. Theodoraki M-N, Yerneni SS, Hoffmann TK, Gooding WE, Whiteside TL. Clinical significance of PD-L1+ exosomes in plasma of head and neck cancer patients. Clin Cancer Res. (2018) 24:896–905. doi: 10.1158/1078-0432.CCR-17-2664
88. Del Re M, Marconcini R, Pasquini G, Rofi E, Vivaldi C, Bloise F, et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br J Cancer. (2018) 118:820–4. doi: 10.1038/bjc.2018.9
89. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. (2018) 560:382–6. doi: 10.1038/s41586-018-0392-8
90. Daassi D, Mahoney KM, Freeman GJ. The importance of exosomal PDL1 in tumour immune evasion. Nat Rev Immunol. (2020) 20:209–15. doi: 10.1038/s41577-019-0264-y
91. Gao J, Qiu X, Li X, Fan H, Zhang F, Lv T, et al. Expression profiles and clinical value of plasma exosomal Tim-3 and Galectin-9 in non-small cell lung cancer. Biochem Biophys Res Commun. (2018) 498:409–15. doi: 10.1016/j.bbrc.2018.02.114
92. Filipów S, Łaczmanski Ł. Blood circulating miRNAs as cancer biomarkers for diagnosis and surgical treatment response. Front Genet. (2019) 10:169. doi: 10.3389/fgene.2019.00169
93. Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin Epigenetics. (2018) 10:59. doi: 10.1186/s13148-018-0492-1
94. Wu Q, Yu L, Lin X, Zheng Q, Zhang S, Chen D, et al. Combination of serum miRNAs with serum exosomal miRNAs in early diagnosis for non-small-cell lung cancer. Cancer Manag Res. (2020) 12:485–95. doi: 10.2147/CMAR.S232383
95. Zhao YJ, Song X, Niu L, Tang Y, Song X, Xie L. Circulating exosomal miR-150-5p and miR-99b-5p as diagnostic biomarkers for colorectal cancer. Front Oncol. (2019) 9:1129. doi: 10.3389/fonc.2019.01129
96. Zhang H, Xu S, Liu X. MicroRNA profiling of plasma exosomes from patients with ovarian cancer using high-throughput sequencing. Oncol Lett. (2019) 17:5601–7. doi: 10.3892/ol.2019.10220
97. Liu S-S, Wang Y-S, Sun Y-F, Miao L-X, Wang J, Li Y-S, et al. Plasma microRNA-320, microRNA-let-7e and microRNA-21 as novel potential biomarkers for the detection of retinoblastoma. Biomed Rep. (2014) 2:424–8. doi: 10.3892/br.2014.246
98. Yu S, Liu Y, Wang J, Guo Z, Zhang Q, Yu F, et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab. (2012) 97:2084–92. doi: 10.1210/jc.2011-3059
99. Kumar S, Sharawat SK, Ali A, Gaur V, Malik PS, Kumar S, et al. Identification of differentially expressed circulating serum microRNA for the diagnosis and prognosis of Indian non-small cell lung cancer patients. Curr Probl Cancer. (2020). doi: 10.1016/j.currproblcancer.2020.100540. [Epub ahead of print].
100. Cochetti G, Poli G, Guelfi G, Boni A, Egidi MG, Mearini E. Different levels of serum microRNAs in prostate cancer and benign prostatic hyperplasia: evaluation of potential diagnostic and prognostic role. Onco Targets Ther. (2016) 9:7545–53. doi: 10.2147/OTT.S119027
101. Chiam K, Wang T, Watson DI, Mayne GC, Irvine TS, Bright T, et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J Gastrointest Surg. (2015) 19:1208–15. doi: 10.1007/s11605-015-2829-9
102. Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. (2017) 23:5311–9. doi: 10.1158/1078-0432.CCR-17-0577
103. Oura K, Fujita K, Morishita A, Iwama H, Nakahara M, Tadokoro T, et al. Serum microRNA-125a-5p as a potential biomarker of HCV-associated hepatocellular carcinoma. Oncol Lett. (2019) 18:882–90. doi: 10.3892/ol.2019.10385
104. Ahmadvand M, Eskandari M, Khakpour G, Pashaiefar H, Manoochehrabadi S, Yaghmaie M, et al. Identification of MiR-125a as a novel plasma diagnostic biomarker for chronic lymphoblastic leukemia. Clin Lab. (2019) 65. doi: 10.7754/Clin.Lab.2018.180815
105. Khare D, Goldschmidt N, Bardugo A, Gur-Wahnon D, Ben-Dov IZ, Avni B. Plasma microRNA profiling: exploring better biomarkers for lymphoma surveillance. PLoS One. (2017) 12:e0187722. doi: 10.1371/journal.pone.0187722
106. Hrustincova A, Krejcik Z, Kundrat D, Szikszai K, Belickova M, Pecherkova P, et al. Circulating small noncoding RNAs have specific expression patterns in plasma and extracellular vesicles in myelodysplastic syndromes and are predictive of patient outcome. Cells. (2020) 9:794. doi: 10.3390/cells9040794
107. Wu C, Cao Y, He Z, He J, Hu C, Duan H, et al. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J Exp Med. (2014) 232:85–95. doi: 10.1620/tjem.232.85
108. Yuwen D-L, Sheng B-B, Liu J, Wenyu W, Shu Y-Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci. (2017) 21:2650–58.
109. Khalighfard S, Alizadeh AM, Irani S, Omranipour R. Plasma miR-21, miR-155, miR-10b, and Let-7a as the potential biomarkers for the monitoring of breast cancer patients. Sci Rep. (2018) 8:17981. doi: 10.1038/s41598-018-36321-3
110. Erbes T, Hirschfeld M, Rücker G, Jaeger M, Boas J, Iborra S, et al. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer. (2015) 15:193. doi: 10.1186/s12885-015-1190-4
111. Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. (2014) 15:149. doi: 10.1186/s12931-014-0149-3
112. Shao C, Yang F, Qin Z, Jing X, Shu Y, Shen H. The value of miR-155 as a biomarker for the diagnosis and prognosis of lung cancer: a systematic review with meta-analysis. BMC Cancer. (2019) 19:1103. doi: 10.1186/s12885-019-6297-6
113. Monzo M, Santasusagna S, Moreno I, Martinez F, Hernáez R, Muñoz C, et al. Exosomal microRNAs isolated from plasma of mesenteric veins linked to liver metastases in resected patients with colon cancer. Oncotarget. (2017) 8:30859–69. doi: 10.18632/oncotarget.16103
114. Mikamori M, Yamada D, Eguchi H, Hasegawa S, Kishimoto T, Tomimaru Y, et al. MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci Rep. (2017) 7:42339. doi: 10.1038/srep42339
115. Motawi TK, Rizk SM, Ibrahim TM, Ibrahim IA-R. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem Funct. (2016) 34:142–8. doi: 10.1002/cbf.3171
116. Jin Y, Wong YS, Goh BKP, Chan CY, Cheow PC, Chow PKH, et al. Circulating microRNAs as potential diagnostic and prognostic biomarkers in hepatocellular carcinoma. Sci Rep. (2019) 9:1–2. doi: 10.1038/s41598-019-46872-8
117. Wu C, Wang C, Guan X, Liu Y, Li D, Zhou X, et al. Diagnostic and prognostic implications of a serum miRNA panel in oesophageal squamous cell carcinoma. PLoS One. (2014) 9:e92292. doi: 10.1371/journal.pone.0092292
118. Yue H, He J, Zhang H, Wang C, Hu W, Gu J, et al. Contribution of Myostatin gene polymorphisms to normal variation in lean mass, fat mass and peak BMD in Chinese male offspring. Acta Pharmacol Sin. (2012) 33:660–7. doi: 10.1038/aps.2012.12
119. Ponomaryova AA, Morozkin ES, Rykova EY, Zaporozhchenko IA, Skvortsova TE, Dobrodeev AY, et al. Dynamic changes in circulating miRNA levels in response to antitumor therapy of lung cancer. Exp Lung Res. (2016) 42:95–102. doi: 10.3109/01902148.2016.1155245
120. Liu B, Su F, Chen M, Li Y, Qi X, Xiao J, et al. Serum miR-21 and miR-125b as markers predicting neoadjuvant chemotherapy response and prognosis in stage II/III breast cancer. Hum Pathol. (2017) 64:44–52. doi: 10.1016/j.humpath.2017.03.016
121. Kassem NM, Makar WS, Kassem HA, Talima S, Tarek M, Hesham H, et al. Circulating miR-34a and miR-125b as promising non invasive biomarkers in Egyptian locally advanced breast cancer patients. Asian Pac J Cancer Prev. (2019) 20:2749–55. doi: 10.31557/APJCP.2019.20.9.2749
122. Zhu T, Gao W, Chen X, Zhang Y, Wu M, Zhang P, et al. A pilot study of circulating microRNA-125b as a diagnostic and prognostic biomarker for epithelial ovarian cancer. Int J Gynecol Cancer. (2017) 27:3–10. doi: 10.1097/IGC.0000000000000846
123. Yamada A, Horimatsu T, Okugawa Y, Nishida N, Honjo H, Ida H, et al. Serum miR-21, miR-29a, and miR-125b are promising biomarkers for the early detection of colorectal neoplasia. Clin Cancer Res. (2015) 21:4234–42. doi: 10.1158/1078-0432.CCR-14-2793
124. Michaille J-J, Awad H, Fortman EC, Efanov AA, Tili E. miR-155 expression in antitumor immunity: the higher the better? Genes Chromosomes Cancer. (2019) 58:208–218. doi: 10.1002/gcc.22698
Keywords: microRNAs, myeloid-derived suppressor cells, immunotherapy, immune checkpoints, therapy resistance, extracellular vesicles
Citation: Daveri E, Vergani E, Shahaj E, Bergamaschi L, La Magra S, Dosi M, Castelli C, Rodolfo M, Rivoltini L, Vallacchi V and Huber V (2020) microRNAs Shape Myeloid Cell-Mediated Resistance to Cancer Immunotherapy. Front. Immunol. 11:1214. doi: 10.3389/fimmu.2020.01214
Received: 10 April 2020; Accepted: 15 May 2020;
Published: 22 July 2020.
Edited by:
Sergei Kusmartsev, University of Florida, United StatesReviewed by:
Viktor Umansky, German Cancer Research Center, GermanyCopyright © 2020 Daveri, Vergani, Shahaj, Bergamaschi, La Magra, Dosi, Castelli, Rodolfo, Rivoltini, Vallacchi and Huber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eriomina Shahaj, ZXJpb21pbmEuc2hhaGFqQGlzdGl0dXRvdHVtb3JpLm1pLml0
†These authors share first authorship
‡These authors share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.