- 1Department of Allergy and Rheumatology, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
- 2Department of Functional Genomics and Immunological Diseases, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
- 3Laboratory for Autoimmune Diseases, RIKEN Center for Integrative Medical Sciences, Yokohama, Japan
- 4Division of Molecular Cell Signaling, Cancer Research Institute, Kanazawa University, Kanazawa, Japan
Peptidylarginine deiminase 4 (PAD4), encoded by PADI4, plays critical roles in the immune system; however, its contribution to the pathogenesis of lupus nephritis remains controversial. The pathological roles of PAD4 were investigated in lupus model mice. An imiquimod (IMQ)-induced lupus model was analyzed in wild-type (WT) and Padi4-knockout (KO) mice. Proteinuria, serum anti-double stranded DNA (anti-dsDNA) antibody, and renal infiltrated cells were evaluated. Neutrophil migration and adhesion were assessed using adoptive transfer and adhesion assay. PAD4-regulated pathways were identified by RNA-sequencing of Padi4 KO neutrophils. Padi4 KO mice exhibited significant improvements in proteinuria progression compared with WT mice, whereas, serum anti-dsDNA antibody and immune complex deposition in the glomeruli showed no difference between both mice strains. Padi4 KO mice showed decreased neutrophil infiltration in the kidneys. Adoptively transferred Padi4 KO neutrophils showed decreased migration to the kidneys of IMQ-treated WT mice, and adhesion to ICAM-1 was impaired in Padi4 KO neutrophils. Padi4 KO neutrophils exhibited reduced upregulation of p38 mitogen-activated protein kinase (MAPK) pathways. Toll-like receptor 7 (TLR7)-primed Padi4 KO neutrophils demonstrated reduced phosphorylation of p38 MAPK and lower expression of JNK-associated leucine zipper protein (JLP), a p38 MAPK scaffold protein. Neutrophils from heterozygous Jlp KO mice showed impaired adhesion to ICAM-1 and decreased migration to the kidneys of IMQ-treated WT mice. These results indicated a pivotal role of PAD4-p38 MAPK pathway in renal neutrophil infiltration in TLR7 agonist-induced lupus nephritis, and the importance of neutrophil-mediated kidney inflammation. Inhibition of the PAD4-p38 MAPK pathway may help in formulating a novel therapeutic strategy against lupus nephritis.
Introduction
Systemic lupus erythematosus (SLE) is a systemic autoimmune disease characterized by multi-organ inflammation with immune complex deposition and leukocyte infiltration. The treatment of SLE remains a huge challenge for clinicians considering the unfavorable prognosis associated with the side effects of steroids and immunosuppressive drugs. Genetic studies revealed the susceptibility of genes associated with the Toll-like receptor (TLR) pathway, interferon pathway, immune complex processing, and immune signal transduction in SLE, suggesting that the pathogenesis of SLE is based on immunological abnormalities (1). Autoantibodies are typically present many years before the diagnosis of SLE, consistent with the observation by Gower et al. (2) that deposition of immune complexes and complement precedes the inflammatory infiltrate. Therefore, leukocyte infiltration after immune complex deposition is the key process of organ damage in SLE and is thought to be a promising therapeutic target.
Peptidylarginine deiminase 4 (PAD4) is an enzyme that converts arginine residues to citrulline through post-translational modifications (3). Peptidylarginine deiminase 4 (PADI4), which encodes PAD4, was initially identified as a non-HLA susceptibility gene in rheumatoid arthritis (4). PADI4 is primarily expressed in neutrophils (5). Subsequently, extensive research revealed the contribution of PADI4 to the pathogenesis of diverse diseases including inflammatory arthritis (3, 5), myocardial ischemia (6), and deep vein thrombosis (7). Several physiological roles associated with PADI4, such as pluripotency (8), p53 target gene repression (9), and regulation of the proliferation of multipotent hematopoietic cells (10) have been identified. The importance of PAD4 for innate immunity was underscored by the observation that the formation of neutrophil extracellular traps (NETs) requires PAD4 (11). In addition, citrullination regulates the functions of histones and gene transcripts. A previous study reported that PAD4 binds to transcription factors to regulate numerous gene transcriptions (12). Considering the multiple roles of PAD4 in the regulation of gene expression and immunological functions, PAD4 may be a potential target for the therapy of autoimmune diseases. However, the precise contribution of PAD4 to the pathogenesis of lupus nephritis remains unclear.
The aim of the present study was to elucidate the pathological roles of PAD4 in lupus nephritis. A TLR7 agonist, Imiquimod (IMQ)-induced SLE mouse model (13) was used to explore the pathogenesis of immune complex-mediated lupus nephritis in several previous reports (14, 15). In this model, topical treatment with IMQ cream on the ear skin induced lupus-like phenotypes, such as elevated levels of serum anti-dsDNA IgG and multi-organ injury, including glomerulonephritis with immune complex deposition (13). We studied IMQ-induced lupus model mice in Padi4 KO background to explore the pathological roles of PAD4 in lupus nephritis.
Materials and Methods
Mice
Padi4 KO mice were generated by deletion of Padi4 exon 1 in C57BL/6 (B6) background mice (3). Heterozygous Jlp KO mice (Jlp +/– mice) were described previously (16). All mice were bred in a specific pathogen-free facility.
IMQ-Treated Mice Experiments
IMQ cream (5%, Mochida Pharmaceutical) was administered on the skin of the left ear of age-matched 8–9-week-old female B6 WT and Padi4 KO mice every alternate day up to 8 weeks as reported previously (13). Degree of proteinuria was evaluated semi-quantitatively using Albustix (Siemens Healthineers) every week. Serum anti-dsDNA IgG titers were measured using an anti-dsDNA antibody mouse ELISA kit (Shibayagi). Serum anti-Sm (IgG, IgA, and IgM) antibodies were detected using mouse anti-Sm Total IgG ELISA kit (Alpha Diagnostic). Matrix metalloproteinase-9 (MMP-9) concentrations in kidney supernatants were measured using a Mouse Total MMP-9 Quantikine ELISA kit (R&D Systems). Serum BUN and Creatinine was measured using QuantiChrom Urea Assay Kit (BioAssay) and Serum Creatinine Detection Kit (Arbor Assays), respectively. Renal histopathology was analyzed by hematoxylin-eosin (HE) staining and immunohistochemistry. Myeloid lineage cells in the kidneys and spleen were isolated and analyzed by flow cytometry (MoFlo XDP, Beckman Coulter).
Histological Assessment of the Ear Skin and Kidneys
The ear skin and kidneys were excised from sacrificed IMQ-treated mice 8 weeks after the first IMQ treatment, fixed with 4% paraformaldehyde, followed by embedding in paraffin. Paraffin-embedded fragments were stained with H&E. For kidney immunohistochemistry, paraffin-embedded sections were immunostained for 1 h at 4°C with goat antiserum to mouse complement C3 primary antibody (ICN/CAPPEL), followed by staining for 1 h at room temperature with Alexa Fluor 594 Donkey anti-goat IgG secondary antibody (Invitrogen). The, slides were also immunostained with 1 h at 4°C with rabbit F (ab') 2 anti-mouse IgG primary antibody (BIO-RAD), followed by staining for 1 h at room temperature with Alexa Fluor 488 goat anti-rabbit IgG secondary antibody (Invitrogen). DAPI (Invitrogen) was used for nuclear staining.
Inflammatory cells in the ear skin were counted per high-power field. Glomerular score represents the sum of scores for glomerular inflammation, proliferation, crescent formation, and necrosis as described previously (17). Each score was graded from 0 to 4. For assessing immune complex deposition in the kidneys, fluorescence intensity was scored semiquantitatively (0: no staining, 1+: mild staining, 2+: moderate staining, 3+: high staining) and average scores were calculated as described previously (17). At least 60 glomeruli per animal were assessed by two independent investigators.
Bone Marrow Neutrophil Isolation
Bone marrow-derived neutrophils were isolated by density gradient centrifugation. For transcriptome analysis, bone marrow neutrophils from 8-week-old female B6 WT and Padi4 KO mice were isolated magnetically using a Neutrophil isolation kit (Miltenyi Biotec). CD11b+ Ly6G+ neutrophils were isolated at higher than 85% purity by density gradient isolation, and > 97% by magnetic isolation.
Neutrophil Adoptive Transfer
Bone marrow-derived neutrophils extracted from age-matched female B6 WT, Padi4 KO and Jlp +/– mice were stained with CellTracker Green CMFDA Dye (Thermo Fisher Scientific), and 2.5 × 106 neutrophils were adoptively transferred to B6 WT control mice and B6 WT mice after IMQ treatment for 4 weeks as described previously (18). The frequencies of CellTracker Green-labeled neutrophils in the kidneys and spleen of the recipients were analyzed by flow cytometry 4 h after adoptive transfer.
Neutrophil Adhesion Assay
Bone marrow-derived neutrophils from age-matched female B6 WT, Padi4 KO, and Jlp +/– mice were isolated, and 3 × 106/ml of neutrophils in Hank's balanced salt solution (HBSS; with Ca2+ and Mg2+) containing 20 mM HEPES and 0.1% bovine serum albumin (BSA) were incubated with or without 1 μg/ml of R848 for 1 h. In some wells, p38 MAPK inhibitor SB203580 (Sigma-Aldrich) was added 30 min prior to R848 stimulation. Furthermore, neutrophils were plated in 96-well plates coated with recombinant mouse ICAM-1/CD54 Fc chimeric protein (R&D systems) for 30 min. Supernatants was removed, and analyzed for the neutrophil adhesion to ICAM-1 using CytoTox 96 non-radioactive cytotoxicity assay (Promega) as described previously (19). Neutrophil adhesion to ICAM-1 was calculated using the ratio of neutrophils adhering to ICAM-1-coated plates to neutrophils adhering to 96-well poly-L-lysine plates (20).
Assessment of Neutrophil Extracellular Traps
Bone marrow-derived neutrophils from age-matched female B6 WT and Padi4 KO mice after IMQ treatment for 8 weeks were resuspended in NET medium containing RPMI 1,640, 1% BSA (Cayman Chemical), 1 mM CaCl2 (Cayman Chemical), 10 mM HEPES (Invitrogen), and 0.2 μM SytoxGreen (Thermo Fisher Scientific) and plated at a concentration of 3 × 105/ml. Neutrophils were incubated without any stimulants overnight 37°C in 5% CO2. Supernatants were removed and plates were washed with PBS twice. SytoxGreen positive cells, which included NETs in the extracellular parts, were counted using a fluorescence microscope in a blinded fashion. The percentage of SytoxGreen positive cells were calculated as the average of 5 fields (×100) per well.
Flow Cytometric Analysis
Cell staining was performed in the presence of purified rat anti-mouse CD16/32 antibody (BD). Antibodies used for staining were as follows; anti-Ly6G-PE (1A8, BD), anti-CD11b-APC (M1/70, eBioscience), anti-Ly6G-APC-Cy7 (1A8, BD), anti-Ly6C-APC-Cy7 (AL-21, BD), mouse biotin-conjugated anti-CD45 (30-F11, eBioscience), anti-streptavidin-V500 (BD), and anti-7-AAD (BioLegend).
For intracellular staining for phosphorylated p38 MAPK, 1 × 106/ml of bone marrow-derived neutrophils were stimulated with 1 μg/ml of R848 (Enzo Life Sciences) for 30 min in RPMI 1640 (Invitrogen) containing 10% fetal bovine serum (FBS), 100 μg/ml of L-glutamine (Sigma-Aldrich), 100 U/ml of penicillin (Sigma-Aldrich), 100 μg/ml of streptomycin (Sigma-Aldrich), and 50 μM 2-mercaptoethanol (Sigma-Aldrich). Stimulated neutrophils were stained with anti-CD11b-APC (M1/70, eBioscience) and anti-Ly6G-APC-Cy7 (1A8, BD) antibodies in the presence of purified rat anti-mouse CD16/32 antibody (BD), fixed with Lyse/Fix Buffer (BD), and permeabilized by BD Phosflow Perm Buffer II (BD). Then, neutrophils were intracellularly stained with Alexa Fluor 488 Mouse IgG1κ isotype control (BD) or Alexa Fluor 488 mouse anti-p38 MAPK (pT180/pY182) (BD).
Flow cytometric analysis was performed using MoFlo XDP and data were analyzed using FlowJo Software (Tree star).
Transwell Assays
Transwell assays were performed as described previously (21). Bone marrow-derived neutrophils from age-matched female B6 WT and Padi4 KO mice were resuspended in HBSS (with Ca2+ and Mg2+) (Wako) containing 0.25% fatty acid free, low endotoxin BSA (Sigma-Aldrich), and 14 mM HEPES at a concentration of 5 × 106/ml. Then, 300 μl of 1 nM recombinant mouse CXCL2 (carrier-free) (BioLegend) was loaded at the bottom of the 24-well plate (ultra-low attachment; Costar, Corning). Neutrophils in 200 μl of the above-mentioned buffer were put on the Transwell filter (polycarbonate, 3 μm pores; Millipore) and placed at the top of the plate. The plates were incubated in 37°C, in 5% CO2. After 1 h, the Transwell filter and medium in the lower chamber were collected. Lower chambers were filled with 300 μl of HBSS (without Ca2+ and Mg2+) (Wako) containing 5 mM EDTA and put on ice for 30 min. Additionally, 300 μl of HBSS (without Ca2+ and Mg2+) containing 5 mM EDTA was added again and the cells were collected. Cell numbers were counted and relative migration was calculated for each well.
RNA Sequencing
Neutrophils at a concentration of 1 × 106/ml were incubated in 24-well plates and left unstimulated or stimulated with 1 μg/ml of R848 for 6 h in RPMI 1,640 containing 10% FBS, 100 μg/ml of L-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 50 μM 2-mercaptoethanol. Each sample was analyzed in duplicate. After collecting the cells and removal of the supernatant, 250 μl of PBS was added, followed by pipetting with 750 μl of TRIzol LS reagent (Life Technologies) in accordance with the manufacturer's instructions. RNA extraction was performed using an RNeasy micro kit (Qiagen) in accordance with the manufacturer's instructions. The quality of extracted RNA was confirmed using a 2,100 Bioanalyzer (Agilent) and quantified with a Qubit RNA HS assay kit (Thermo Fisher Scientific). RNA sequencing libraries were prepared using a TrueSeq RNA sample preparation kit v2 (Illumina) in accordance with the manufacturer's instructions. Paired-end sequencing was performed using a MiSeq Reagent V2 kit (Illumina) on the MiSeq system.
Sequence Data Analysis
Raw read quality was evaluated using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) (v0.11.7). After trimming off the adaptors using cutadapt (22) (v1.16), reads containing many low quality bases (Phred quality score <20 in >20% of the bases) or very low quality bases (Phred quality score <10) were removed using the FASTX toolkit. Mean of the total read count after QC was 1.02 × 106. Filtered reads were mapped to the UCSC mm10 using STAR (v2.5.3a) (23), and the read count was calculated using HTSeq (v0.6.1p1) (24). Genes expressed in less than two samples were omitted from further analysis. We compared changes in mRNA expression before and after R848 stimulation in WT and Padi4 KO neutrophils. Differentially expressed genes before and after R848 stimulation were analyzed using the nbinomWaldTest function in the DESeq2 (25) package (v1.16.1) for R (v3.4.0) in WT and Padi4 KO neutrophils. Differentially expressed genes were filtered by 2-fold increase or decrease and false discovery rate <0.1, then analyzed using core analysis in Ingenuity® Pathway Analysis (IPA). We conducted comparison analysis in IPA between the results of the core analysis of WT and Padi4 KO neutrophils. Activation Z scores were calculated using IPA comparison analysis between WT and Padi4 KO neutrophils. Read counts were normalized as count per million and scaled by low Z-score and were plotted as a heatmap with hierarchical clustering based on Ward's Method.
PAD4-binding genes were adopted from a report that involved ChIP-chip analysis in MCF-7 (12). Gene names were converted to mouse homologs using biomaRt (26). Downregulated genes in TLR 7-primed Padi4 KO neutrophils were defined as differentially upregulated genes (more than 2-fold increase and false discovery rate <0.1) in TLR 7-primed WT neutrophils but not in Padi4 KO neutrophils. Overlapping genes were selected between PAD4-binding genes and downregulated genes of TLR7-primed Padi4 KO neutrophils.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
Total RNA from neutrophils was extracted using an RNeasy Micro kit as described in the manufacturer's instructions. RNA was reverse-transcribed to cDNA with random primers (Invitrogen) and SuperScript III (Invitrogen). To measure gene expression, quantitative real-time PCR was performed by using a CFX connect real-time PCR detection system (Bio-Rad) with a QuantiTect SYBR Green PCR kit (QIAGEN). Gene expression was normalized to β-actin expression levels as a control. Sequences of the PCR primers were as follows: Actb (5′- AGAGGGAAATCGTGCGTGAC-3′ and 5′- CAATAGTGATGACCTGGCCGT-3′), Padi4 (5′-CTCTCCAGGAGTCATCGTAG-3′ and 5′-CCAACACCAGCTGATACTTT-3′), Jlp (5′-GAGCATGTGTTTACAGATCCACTG-3′ and 5′-CATTTTCTGAGCTTCTTCTCTCGC-3′).
Western Blotting
Bone marrow-derived neutrophils (3 × 106/ml) from age-matched female B6 WT, Padi4 KO mice were incubated in 24-well plates and stimulated with 1 μg/ml of R848 in RPMI 1640 containing 10% FBS, 100 μg/ml of L-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 50 μM 2-mercaptoethanol. After collecting cells and removal of the supernatant, neutrophils were homogenized in RIPA buffer (Sigma-Aldrich) containing Halt phosphatase inhibitor cocktail (Thermo Fisher Scientific) and Halt protease inhibitor (Thermo Fisher Scientific). The protein samples were heated to 95°C for 5 min in Laemmli sample buffer (Bio-Rad) containing 2-mercaptoethanol and loaded onto Mini-PROTEAN TGX precast gels (Bio-Rad). Following electrophoretic transfer of proteins to an Immobilon transfer membrane (Millipore) and membrane was blocked with PVDF blocking reagent for Can Get Signal® (Toyobo). The membrane was incubated overnight with primary antibodies as follows; anti-JLP (D72F4, Cell Signaling) and anti-GAPDH (D16H11, Cell Signaling). The membrane was then incubated with a secondary goat anti-rabbit IgG-HRP (Invitrogen) antibody, and specific proteins were visualized using ImageQuant LAS4010 (GE Healthcare) with ECL Select (GE Healthcare).
Statistical Analysis
GraphPad Prism7 (GraphPad Software) was used for statistical analysis except in sequencing data. Two-tailed unpaired t-test was used for comparison between two groups. Kruskal–Wallis test with Dunn's multiple comparisons test for proteinuria, and one-way ANOVA with Tukey's multiple comparison test for others were used for comparison among three or more groups. P < 0.05 was considered statistically significant.
Results
IMQ-Induced Lupus Like Phenotypes Were Ameliorated in Padi4 KO Mice
First, we compared the lupus-like phenotypes induced by topical treatment of IMQ between WT and Padi4 KO mice. The spleen weight was almost equivalent between untreated WT and Padi4 KO mice (Figure S1). IMQ-treated Padi4 KO (Padi4 KO-IMQ) mice showed a significant reduction in splenomegaly compared with IMQ-treated WT (WT-IMQ) mice (Figure 1A). Epidermal hyperplasia and dermal immune cell infiltration in the ear skin of WT-IMQ mice were minimized, and cellular infiltrations were rarely observed in Padi4 KO-IMQ mice (Figure 1B). Moreover, proteinuria was significantly ameliorated in Padi4 KO-IMQ mice compared with WT-IMQ mice (Figure 1C). The serum BUN and creatinine showed no difference between IMQ-treated WT and Padi4 KO mice (Figure S2). The concentrations of MMP-9, a candidate biomarker of renal damage (27), in kidney extracts of Padi4 KO-IMQ mice were significantly decreased compared with WT-IMQ mice (Figure 1D). Spontaneous NETosis was observed in the bone marrow neutrophils of WT-IMQ mice, and was totally abrogated in Padi4 KO-IMQ mice (Figure S3). Taken together, in Padi4 KO-IMQ mice, lupus-like organ damage was ameliorated compared with WT-IMQ mice.
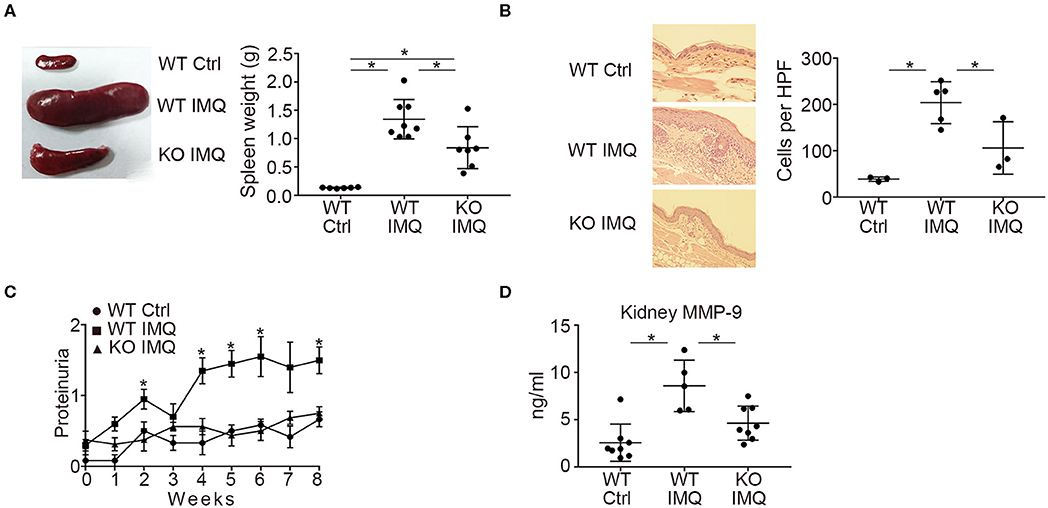
Figure 1. Organ involvement of an imiquimod-treated lupus model in Padi4 KO mice. Organ involvement was analyzed in IMQ-treated WT (WT IMQ), and IMQ-treated Padi4 KO (KO IMQ) mice 8 weeks after IMQ treatment. (A) Representative pictures of the spleen of WT Ctrl, WT IMQ, and Padi4 KO IMQ mice (Left), and the spleen weights of these mice (Right) (WT Ctrl, n = 6; WT IMQ, n = 8; KO IMQ, n = 7). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). (B) Representative pictures of the HE staining of the ear skin (Left), and the number of inflammatory cell infiltrates in the dermis counted per high-power field from these mice (Right) (WT Ctrl, n = 3; WT IMQ, n = 5; KO IMQ, n = 3). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). (C) Chronological progression of proteinuria (Left) (WT Ctrl, n = 6; WT IMQ, n = 10, KO IMQ, n = 8). *p < 0.05 (Kruskal–Wallis test with Dunn's multiple comparisons test, *indicates a significant difference in proteinuria between WT IMQ and KO IMQ mice). (D) MMP-9 concentrations in kidney supernatants (WT Ctrl, n = 8; WT IMQ, n = 5; KO IMQ, n = 8). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). Data are representative of at least three independent experiments. All error bars represent ± SD except for proteinuria progression (±SEM).
Neutrophil Infiltration in the Kidneys Was Decreased in Padi4 KO-IMQ Mice
Pertaining to the immunological characteristics, there was no difference in serum anti-dsDNA IgG and anti-Sm titers between WT-IMQ and Padi4 KO-IMQ mice (Figures 2A, S4). Immunohistochemical analysis of the kidneys revealed that the degree of immune complex depositions were identical in WT-IMQ and Padi4 KO-IMQ mice (Figures 2B,C). In contrast, Padi4 KO-IMQ mice exhibited reduced glomerular scores, especially in view of cellular infiltration, compared with WT-IMQ mice (Figures 2B,D). In both WT-IMQ and Padi4 KO-IMQ mice, cellular infiltration into the glomeruli was predominant, and infiltration into the tubulointerstitium was hardly observed (data not shown).
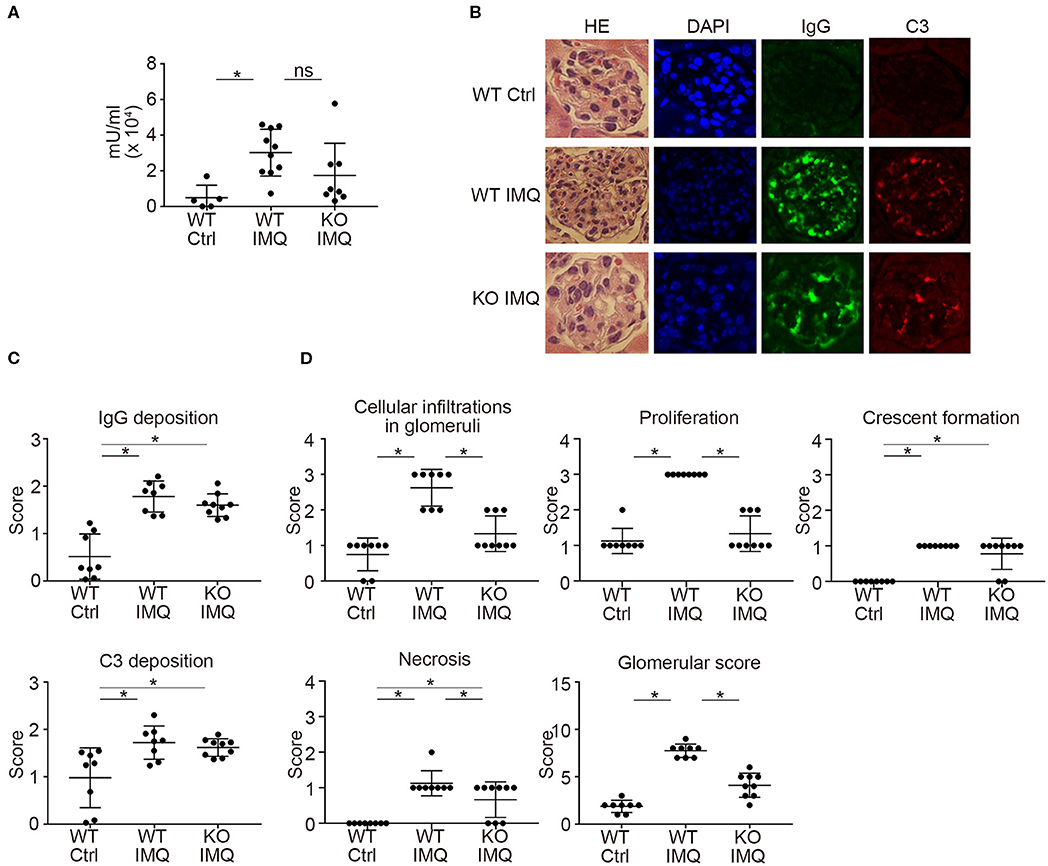
Figure 2. Histological analysis of the glomeruli in IMQ-treated Padi4 KO mice. (A) Serum titers of anti-dsDNA antibody in the serum of WT Ctrl, WT IMQ, and KO IMQ mice for 4 weeks are indicated (WT Ctrl, n = 5; WT IMQ, n = 10; KO IMQ, n = 8). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test); ns, non-significant. (B–D) The glomeruli of the kidneys of WT Ctrl, WT IMQ, and KO IMQ mice were analyzed histologically 8 weeks after IMQ treatment. (B) Representative pictures of HE, DAPI, IgG, and C3 staining. (C) Scores of IgG deposition and C3 deposition in the kidneys. Fluorescence intensity was scored semiquantitatively (0: no staining, 1+: mild staining, 2+: moderate staining, 3+: high staining) and average scores of at least 60 glomeruli per animal were calculated. *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). (D) Glomerular score consisting of glomerular inflammation (cellular infiltration in glomeruli), proliferation, crescent formation, and necrosis of the kidneys (WT Ctrl, n = 8; WT IMQ, n = 8; KO IMQ, n = 9). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). All error bars represent ± SD.
Flow cytometry analysis revealed a significant reduction in the numbers of myeloid cells, such as neutrophils (CD11b+Ly6G+) and CD11b+CD11c+ cells in the kidneys of Padi4 KO-IMQ mice (Figures 3A,B). Although the frequencies of these myeloid cells in the spleens were similar between WT-IMQ and Padi4 KO-IMQ mice, the numbers of CD11b+CD11c+ cells and monocytes (CD11b+Ly6Chigh) decreased in the spleen of Padi4 KO-IMQ mice (Figures 3A–C), which may partially explain the improvement of splenomegaly in Padi4 KO-IMQ mice shown in Figure 1A. The frequencies of CD11b+CD11c+ cells and monocyte were similar between untreated WT and Padi4 KO mice (Figure S5). Even though the frequency of neutrophils was increased in Padi4 KO mice (Figure S5), the range of the frequencies was much lower. The frequencies of neutrophils and monocytes infiltrating the kidney were higher in the group of the high levels of proteinuria (Figure 3D). Considering neutrophils were decreased in the kidney of the Padi4 KO-IMQ mice in Figure 3A, neutrophils were thought to play pivotal roles in the pathogenesis of lupus nephritis. Kidney neutrophils highly expressed Padi4 mRNA, whereas CD11b+CD11c+ cells showed limited Padi4 expression (Figure 3E). Interestingly, Padi4 mRNA expression was significantly decreased in the kidney neutrophils of WT-IMQ mice compared with those of WT-control mice (Figure 3E). Taken together, the significant decrease in renal neutrophil infiltration was closely associated with the amelioration of proteinuria in Padi4 KO-IMQ mice.
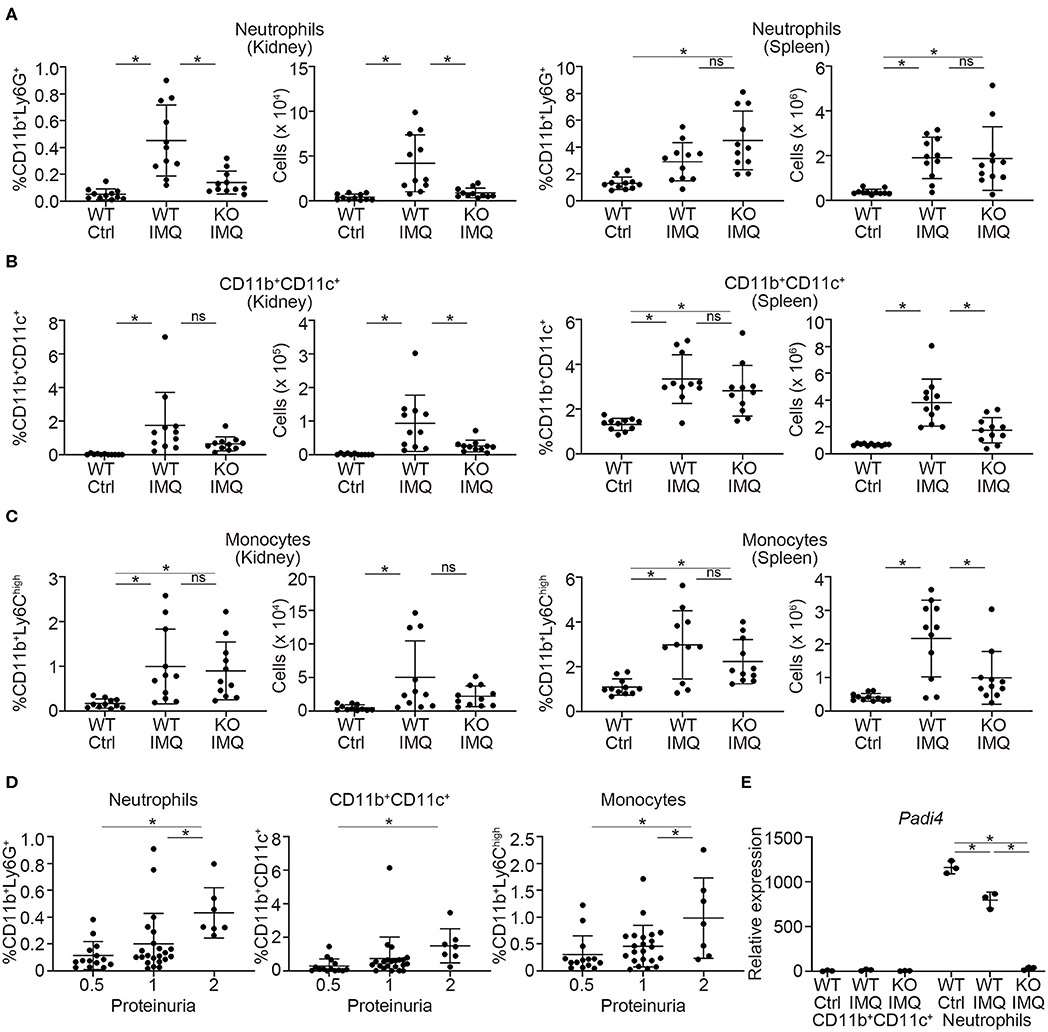
Figure 3. Myeloid cell infiltration in the kidneys of IMQ-treated Padi4 KO mice. (A–C) The frequencies (Left) and the numbers (Right) of CD11b+Ly6G+ cells (neutrophils) (A), CD11b+CD11c+ cells (B), and CD11b+Ly6high cells (monocytes). (C) in the kidneys and spleen 7AAD− cells 8 week after IMQ treatment (WT Ctrl, n = 11; WT IMQ, n = 11; KO IMQ, n = 11). Data were analyzed by flow cytometry. *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test); ns, non-significant. (D) The percentage of myeloid cells infiltrating the kidney in each semi-quantitatively measured proteinuria (0.5, 1, and 2) (WT Ctrl, n = 14; WT IMQ, n = 14; KO IMQ, n = 15). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). (E) Real-time PCR analysis of the expression of Padi4 mRNA in CD11b+CD11c+ cells (Left), and neutrophils (Right) in the kidneys (n = 3 per group). Results were normalized to Actb. *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). Data are representative of at least three independent experiments. All error bars represent ± SD.
Padi4 KO Neutrophils Reduced the Migration Capacity to the Kidneys
Next, we investigated the role of Padi4 in neutrophil migration to the kidneys using adoptive transfer experiments (Figure 4A). There was a significant decrease in Padi4 KO neutrophil migration to the kidneys compared with WT neutrophils, whereas there was no difference between WT and Padi4 KO neutrophil migration to the spleen (Figure 4B). ICAM-1 is a representative adhesive molecule in neutrophil recruitment to the kidneys and is associated with lupus nephritis (28, 29). Padi4 KO neutrophils showed decreased adhesion to ICAM-1 after TLR7 simulation (Figure 4C). CXCL2 is a representative renal neutrophil-attracting chemokine (30). Cxcl2 mRNA was expressed equivalently in the kidneys of WT-IMQ and Padi4 KO-IMQ mice (data not shown). In Transwell assays, there were no differences in chemotaxis between WT and Padi4 KO neutrophils toward CXCL2 in vitro (Figure 4D). Therefore, neutrophil adhesion and migration capacity to the kidneys were regulated by Padi4, consistent with the decreased renal neutrophil infiltration in Padi4 KO-IMQ mice.
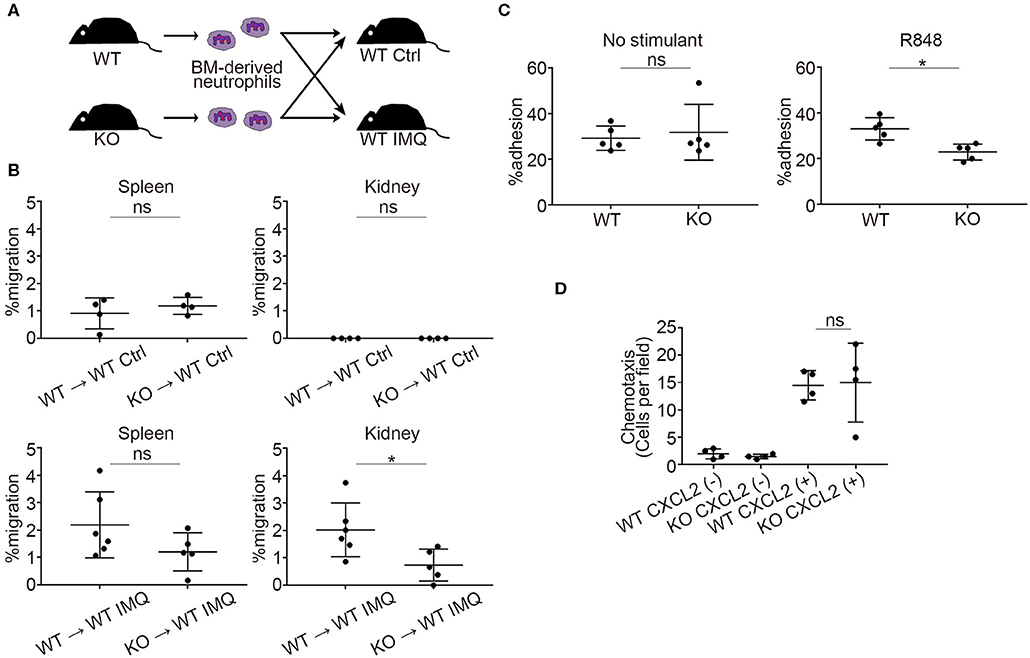
Figure 4. Padi4 KO neutrophil migratory capacity in vitro and in vivo. (A) A schematic explanation of the adoptive transfer experiments. Bone marrow-derived neutrophils (2.5 × 106) from WT and Padi4 KO mice stained with Cell Tracker Green were adoptively transferred to WT Ctrl and WT IMQ mice 4 weeks after IMQ treatment. (B) The frequencies of migrated Cell Tracker Green-positive neutrophils of 7AAD−CD45+CD11b+Ly6G+ in the spleen (Left) and the kidneys (Right) (WT Ctrl for WT neutrophils and KO neutrophils, n = 4 for each; WT IMQ, n = 6 for WT neutrophils and n = 5 for KO neutrophils). *p < 0.05 (two-tailed unpaired t-test); ns, non-significant. (C) Adhesion of bone marrow-derived WT and Padi4 KO neutrophils (3 × 106/ml) to plates coated in ICAM-1 molecules with 1 μg/ml of R848. Cell adhesion was evaluated using the ratio of cells adhered to 96-well poly-L-lysine plates (n = 5 for each group). *p < 0.05 (two-tailed unpaired t-test); ns, non-significant. (D) Chemotaxis of bone marrow-derived neutrophils (1 × 106) from WT and Padi4 KO mice was assessed to CXCL2 (1 nM) (n = 4 per group) (one-way ANOVA with Tukey's multiple comparisons test, only the result of the analysis between WT CXCL2(+) and KO CXCL2(+) is shown.); ns, non-significant. Data are representative of at least three independent experiments. All error bars represent ± SD.
Padi4 Promoted the Neutrophil p38 MAPK Pathway After TLR7 Stimulation
Next, we investigated the transcriptome of TLR7-stimulated neutrophils by RNA-sequencing. We focused on the changes in mRNA expression before and after R848 stimulation in WT and Padi4 KO neutrophils, because there was a decrease in the adhesion of Padi4 KO neutrophils to ICAM-1 after TLR7 stimulation. Padi4 KO neutrophils showed impaired upregulation of several signaling pathways after TLR7 stimulation (Figure 5A). In particular, p38 mitogen-activated protein kinase (MAPK) signaling pathway exhibited a clear reduction in Padi4 KO neutrophils (Figures 5A, S6). The p38 MAPK pathway regulates various neutrophil functions including adhesion (31). Indeed, the phosphorylation of p38 MAPK was significantly decreased in Padi4 KO neutrophils compared with WT neutrophils following TLR7 stimulation (Figure 5B). Consistent with this, the α and β isoforms of the p38 MAPK inhibitor SB203580 inhibited the adhesion of WT neutrophils to ICAM-1 following TLR7 stimulation (Figure 5C). To summarize, Padi4 promoted upregulation of the p38 MAPK pathway after TLR7 stimulation, and the p38 MAPK pathway contributed to neutrophil adhesion after TLR7 stimulation.
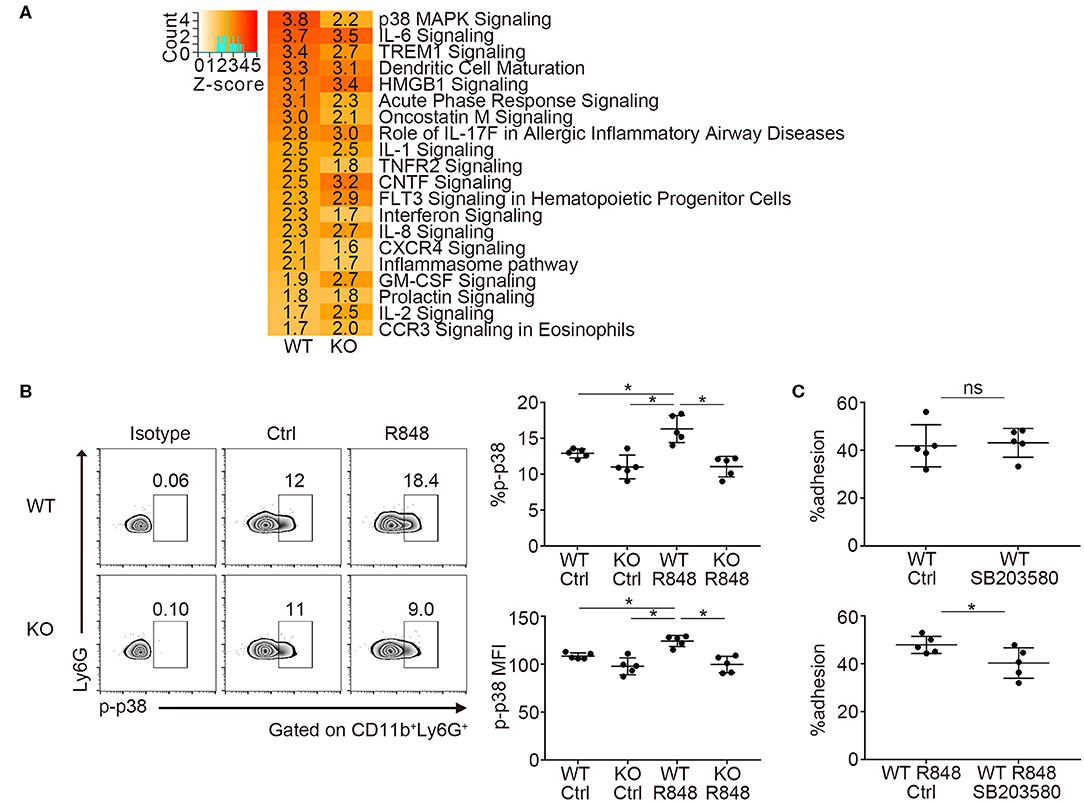
Figure 5. Transcriptome analysis of Padi4 KO neutrophils. (A) Bone marrow-derived neutrophils were incubated with or without 1 μg/ml of R848 for 6 h. We detected differentially expressed genes before and after R848 stimulation in WT and Padi4 KO neutrophils, respectively. The activation Z-scores in the canonical pathway analysis were calculated using IPA software. The top 20 pathways are listed. Activation Z-scores are displayed in each column. (B) Bone marrow-derived neutrophils (3 × 106/ml) were stimulated with 1 μg/ml of R848 for 30 min and were intracellularly stained with isotype control or anti-phosphorylated p38 MAPK (p-p38 MAPK) antibodies and analyzed using a flow cytometer (n = 5 for each group). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). (C) R848-stimulated bone marrow-derived WT neutrophils (3 × 106/ml) were incubated on ICAM-1-coated plates with or without SB203580 prior to R848 stimulation. Neutrophil adhesion to ICAM-1 was calculated using the ratio of cells adhered to 96-well poly-L-lysine plates (n = 5 for each group). *p < 0.05 (two-tailed unpaired t-test). Data are representative of at least three independent experiments. All error bars represent ± SD.
Padi4 Regulated the Expression of JLP, a p38 MAPK Scaffold Protein
Previous reports demonstrated that PAD4 binds to the histone and transcriptional factor complex at the promoter regions and works as a transcriptional coactivator (12). We examined the overlap between PAD4-binding genes detected by chromatin immunoprecipitation coupled with a promoter tiling array (ChIP-chip) analysis reported in MCF-7 cells (12) and downregulated genes of TLR7-primed Padi4 KO neutrophils. Nineteen overlapping genes were identified, and the Jlp gene, encoding JNK-associated leucine zipper protein (JLP), was included among these (Figure 6A). JLP is a key scaffold protein for the activation of the p38 MAPK pathway (32). Indeed, mRNA expression of Jlp and protein expression of JLP were reduced in Padi4 KO neutrophils after TLR7 stimulation (Figures 6B,C). mRNA expression of Jlp was also suppressed following LPS stimulation (Figure S7). In Jlp +/– neutrophils (Figure S8), phosphorylation of p38 MAPK (Figure 6D) and neutrophil adhesion to ICAM-1 in vitro (Figure 6E) were significantly decreased. To analyze the in vivo migratory capacity of the Jlp +/– neutrophils to the kidneys, WT and Jlp +/– neutrophils were adoptively transferred to WT-IMQ mice (Figure 6F). There was a significant decrease in Jlp +/– neutrophil migration to the kidneys compared with WT neutrophils, whereas there was no difference between WT and the Jlp +/– neutrophil migration to the spleen (Figure 6G). Taken together, Padi4 promoted JLP expression after TLR7 stimulation, which led to activation of the p38 MAPK pathway and neutrophil adhesion to ICAM-1.
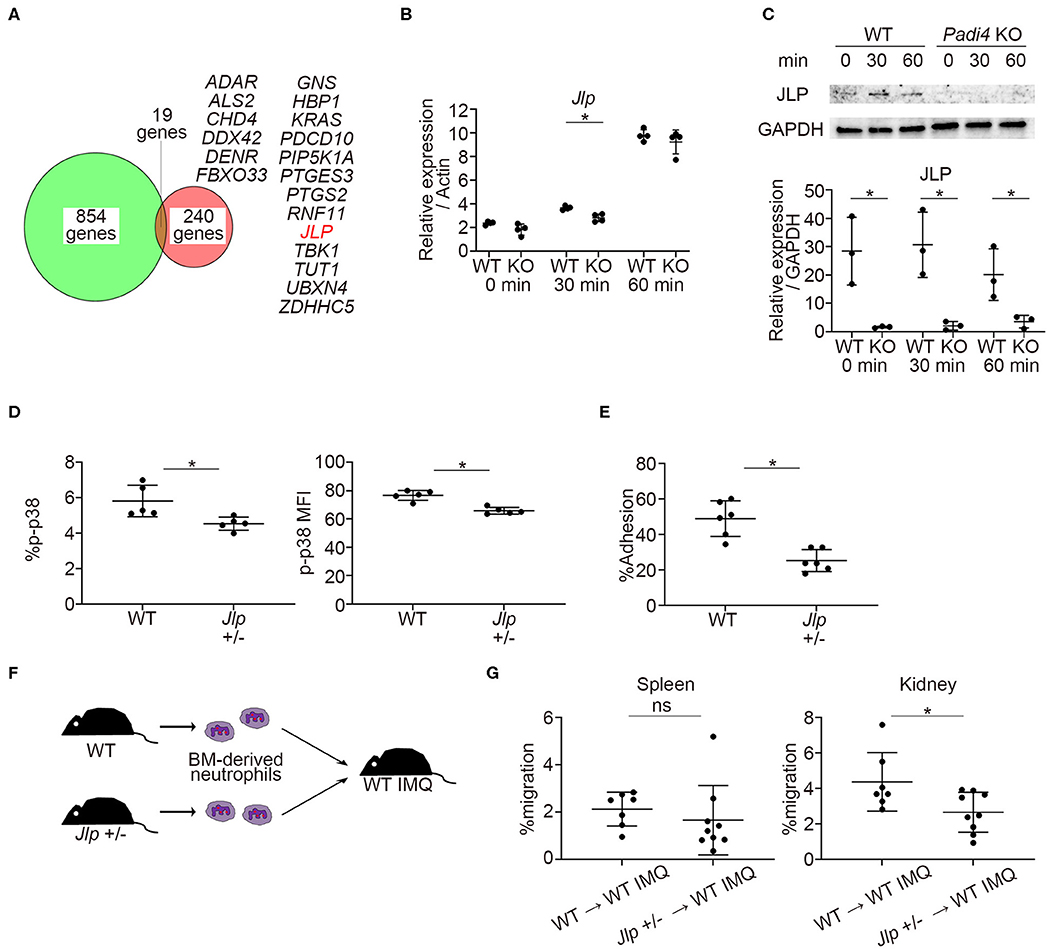
Figure 6. Identification of PAD4-dependent gene, Jlp, which encodes JLP, a scaffold protein of p38 MAPK. (A) Venn diagram of PAD4 Chip-chip binding genes and downregulated genes of TLR7-primed Padi4 KO neutrophils. Downregulated genes of TLR7-primed Padi4 KO neutrophils were defined as differentially upregulated genes (more than 2-fold increase and false discovery rate <0.1) in TLR7-primed WT neutrophils but not in Padi4 KO neutrophils. Nineteen overlapping genes are listed in alphabetical order. (B) Real-time PCR analysis of the expression of Jlp mRNA in WT and Padi4 KO neutrophils at 0, 30, and 60 min after R848 stimulation (n = 4 per group). Results were normalized to Actb. *p < 0.05 (two-tailed unpaired t-test) (C) Western blotting of JLP and GAPDH in WT and Padi4 KO neutrophils at 0, 30, and 60 min after R848 stimulation. Relative expression of JLP protein (normalized to GAPDH) was analyzed between WT and Padi4 KO neutrophils (n = 3 per group). *p < 0.05 (two-tailed unpaired t-test) (D) The bone marrow-derived neutrophils (3 × 106/ml) of WT and Jlp +/– mice were intracellularly stained with isotype control or anti-phosphorylated p38 MAPK (p-p38 MAPK) antibodies and analyzed using flow cytometry (n = 5 for each group). *p < 0.05 (one-way ANOVA with Tukey's multiple comparisons test). (E) The adhesion of bone marrow-derived WT and Jlp +/– neutrophils (3 × 106/ml) to plate-bound ICAM-1 molecules. Cell adhesion was evaluated using the ratio of cells adhered to 96-well poly-L-lysine plates (n = 5 for each group). *p < 0.05 (two-tailed unpaired t-test). (F) A schematic explanation of the adoptive transfer experiments. Bone marrow-derived neutrophils (2.5 × 106) from WT and Jlp +/– mice stained with Cell Tracker Green were adoptively transferred to WT IMQ mice 4 weeks after IMQ treatment. (G) The frequencies of migrated Cell Tracker Green-positive neutrophils of 7AAD−CD45+CD11b+Ly6G+ in the spleen (Left) and the kidneys (Right) (n = 7 for WT neutrophils and n = 9 for Jlp +/– neutrophils). *p < 0.05 (two-tailed unpaired t-test); ns, non-significant. Data are representative of at least three independent experiments. All error bars represent ±SD.
Discussion
Our study demonstrated the pivotal role of PAD4 in neutrophil infiltration to the kidneys with immune complex deposition. In lupus nephritis, immune complex deposition triggers several immunological reactions, such as cellular infiltration and the activation of immune cells, including neutrophils (33). Neutrophils stimulated with immune complexes injure renal tissue via the production of reactive oxygen species, cytokines, and chemokines along with degranulation and the release of MMPs and elastase (33). Our study demonstrated that neutrophil infiltration to the kidneys was significantly suppressed in Padi4 KO IMQ-mice, even though immune complexes were equivalently deposited. Consistent with our observations, lupus-prone MRL-Faslpr/lpr (MRL/lpr) mice lacking ICAM-1 or lymphocyte function-associated antigen-1 showed amelioration of glomerulonephritis, which was correlated with a decrease in neutrophil accumulation in the glomeruli (34, 35). In humans, neutrophils are one of the major infiltrated cells in the kidneys in lupus nephritis (36). In addition, the blood transcriptome profiles of adult lupus patients revealed an association between neutrophil signatures and lupus nephritis (37). Our observations provide an experimental evidence for the linkage between neutrophil and lupus nephritis. In our data, concentrations of MMP-9, a candidate biomarker of renal damage (27), in kidney extracts of Padi4 KO-IMQ mice were significantly decreased compared with WT-IMQ mice. In a rat model of membranous nephropathy, a correlation between increased expression of MMP-9 within the visceral epithelial cells and the proteinuria was reported (38). Also in NZBxNZW F1 lupus mice, MMP-9 contributed to increase glomerular proteolytic activity around the onset of proteinuria (39). Additionally, glomerular MMP-9 staining were increased in the mesangial region and its levels correlated the glomerular cell proliferation in human lupus nephritis (40). Thus, MMP-9 is associated with renal impairment, especially with glomerular inflammation and proliferation. As neutrophils are known as a source of MMP-9 (41), the result might be due to a decrease in infiltration of neutrophils into the kidney of Padi4 KO-IMQ mice. In fact, neutrophil extracellular trap-bound MMP-9 induces endothelial cells death and vascular dysfunction (42). Therefore, we selected MMP-9 as renal damage marker representing the degree of proteinuria and the renal infiltrations of myeloid cells that are characteristic of this mice model of lupus. Further investigation is needed to determine whether MMP-9 secreted from neutrophils directly contributes to renal damage in this mouse model.
The roles of PAD4 in neutrophils have been investigated. A previous study showed that there was no difference in neutrophil functions, such as phagocytosis capability, generation of reactive oxygen species, or in absolute neutrophil counts between WT and Padi4 KO mice (43). Some recent reports suggested an association between NETosis and lupus nephritis (44, 45). NETs in SLE patients contain autoantigens, such as self-DNA, and form immune complexes to activate plasmacytoid dendritic cells (46). Because PAD4 is required for NETosis (11), the amelioration of lupus-like phenotypes in Padi4 KO mice can be partially explained by the lack of NETosis. Nevertheless, the pathological roles of NETosis in this IMQ-induced model of lupus remains unclear. Apart from NETosis, our study demonstrated that adhesion was impaired in Padi4 KO neutrophils, especially after TLR7 stimulation. In addition, Padi4 KO neutrophils reduced the expression of JLP and impaired upregulation of the p38 MAPK pathway after TLR7 stimulation. Several reports suggested an alteration of integrin affinity via p38 MAPK signaling. Mice with aged neutrophils had altered integrin affinity in p38 MAPK-dependent pathways (47). In human neutrophils, p38α specifically contributes to LPS-induced adhesion (31). Moreover, p38 MAPK inhibitors decreased proteinuria in MRL/lpr mice (48). In this way, we supposed that the PAD4-p38 MAPK pathway and regulation of neutrophil adhesion may be promising therapeutic targets against lupus nephritis. Further study is needed to elucidate the connection between p38 MAPK pathway in neutrophils and the defect in adhesion to ICAM-1, including the possibility that expression of activated LFA-1 or Mac-1 is diminished on the surface of Padi4 KO neutrophils. Also, further in vivo study is needed using conditional knockout mice in which p38α expression is specifically ablated in neutrophils. Padi4 mRNA expression was significantly decreased in the kidney neutrophils of WT-IMQ mice compared with those of WT-control mice. Some negative feedback system may exist in the regulation of Padi4 expression, however, additional investigation is needed to unveil its detailed mechanism. Among the identified nineteen genes, the other genes could be related to the activation of MAPK pathway. According to the literature review, four candidate genes were listed; Kras, Rnf11, Hbp1, and Ptgs2. Kras encodes RAS, a guanosine triphosphate (GTP)-binding protein. When stimulated by binding GTP, various signaling pathways are activated (49). Of interest, KRAS mutation was reported in SLE patient (50). Rnf11 encodes RING finger protein 11 (RNF11), involved in various pathways, such as regulation of signal transduction, trafficking and modulation of the transcriptional activity (51). miR-200a-5p and its target gene Rnf11 is involved in selenium deficiency-induced cardiac necrosis via MAPK activation (52). Hbp1 encodes HMG-box transcription factor 1 (HBP1), which is involved in transcriptional repressor and cell cycle inhibitor (53). HBP1 is downstream target in p38 MAPK pathway (53). Ptgs2 encodes cyclooxygenase 2 (COX2), and COX2 is also downstream target in p38 MAPK pathway (54). We selected Jlp for the further study because Jlp extensively regulated p38 MAPK-related genes compared to other candidate genes. Additional investigation is required to determine whether the other candidate genes control MAPK pathway in the PAD4-dependent manner.
The effectiveness of PAD4 inhibition for lupus models remains controversial. These conflicting data were due to the usage of a pan-PAD inhibitor, which also inhibits all PADs. Another important reason was the difference in the models of lupus mice and mice strains. A previous study showed that Padi4-deficiency and the pan-PAD inhibitor Cl-amidine (55) did not ameliorate nephritis in MRL/lpr mice and another lupus model induced by the transfer of SLE sera to FcγRIIAγ−/−Mac1−/− mice, respectively (56). Additionally, Kienhöfer et al. (57) reported nephritis aggravated in a pristane model in the absence of PAD4. In contrast, Cl-amidine exhibits clear therapeutic efficacy in two lupus-prone strains, MRL/lpr mice (44) and New Zealand Mixed 2328 mice (45). Particularly, a recent study demonstrated that Padi4 deficiency ameliorated immunological and clinical phenotypes of IMQ-treated mice in the FVB background (58). They attributed less kidney injury to reduced NETosis and associated vascular damage. They also observed an abrogated T cell response and a decrease of autoantibodies in Padi4 KO mice, which was different form our observation. However, IMQ induces strain dependent responses, at least in dermatitis, and CD4+ T cell-associated gene expression was weaker in the B6 background (59). Therefore, we speculated that the controversial results depended on the different characteristics of the mice strains. We thought that each result reflected the various aspects of lupus nephritis. Of note, there was no difference of BUN and creatinine between IMQ-treated WT and Padi4 KO mice, even though there was a significant difference in the proteinuria. One possible explanation is that PAD4-dependent neutrophil infiltration could be principally associated with the pathogenesis of proteinuria, not the reduction of renal functions. In human lupus nephritis, the poor outcome of renal sufficiency is associated with tubular intestinal damage (60). Since neutrophil infiltration was mainly found in glomerular lesions in IMQ-induced mice, we supposed that PAD4 would principally contribute to the glomerular lesions and proteinuria in this model. Further studies are required to explore the function of PAD4 in human lupus nephritis. In addition, the role of PAD4 in neutrophils and renal proximal tubular cells has been reported in acute kidney injury (61). Therefore, future studies with conditional KO mice specific for either neutrophils or tubular cells are also needed to dissect the cell-specific effect of PAD4 in lupus nephritis.
The importance of PADI4 in the pathogenesis of SLE remains unclear. A genome wide association study (GWAS) revealed a number of critical genes and pathways for SLE (1). Although PADI4 has not been identified as a GWAS risk gene for SLE, PADI4 gene polymorphisms were reported to be associated with lupus nephritis (62). This may be due to the SLE case population with nephritis constitutes only a proportion of patients. With regard to immune-related nephritis, the fact that PADI4 has been identified as a GWAS risk gene for IgA nephropathy supports the possibility that PADI4 plays a critical role in immune complex-mediated nephritis (63). Recently, genetic variations in the TNFAIP3 de-ubiquitinase domain, which is known to be associated with an increased risk of SLE, upregulates PADI4 expression (64). These genetic studies supported the importance of PADI4 in the pathogenesis of human lupus nephritis, and further human studies are required for unveiling the precise roles of PADI4 in human diseases.
In summary, PAD4 promoted neutrophil infiltration into the kidneys and the development of nephritis in TLR7 agonist-induced lupus model mice. PAD4 was implicated in regulating the p38 MAPK pathway, which affects neutrophil adhesion to the kidneys. Our study demonstrated the importance of neutrophils in the pathogenesis of lupus nephritis, and the suppression of neutrophil adhesion and inhibition of the PAD4-p38 MAPK pathway may be a unique and promising therapeutic strategy against lupus nephritis.
Data Availability Statement
The datasets generated for this study can be found in the GEO, GSE145422, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE145422.
Ethics Statement
The animal study was reviewed and approved by the ethics committee of The University of Tokyo Institutional Animal Care and Use Committee (15-P-069) and (P18-098).
Author Contributions
HS, TK, TO, KYa, and KF designed experiments. NH conducted the experiments and acquired data. NH, HH, YN, and AS analyzed the data. AS, IG, and KYo provided animals. NH, HS, and KF wrote the manuscript. NH, HH, and YN prepared figures. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Ministry of Health, Labor and Welfare, Ministry of Education, Culture, Sports, Science and Technology KAKENHI Grant-in-Aid for Scientific Research (C) (17K09971) and the GSK Japan Research Grant 2019.
Conflict of Interest
NH has received grants from GlaxoSmithKline. HS has received speaking fees from Takeda, BMS, Asahi Kasei, Sanofi, Eli Lilly, Janssen, Astellas, Pfizer, Chugai, UCB, and Daiichi-Sankyo. YN has received financial support or fees from BMS, Kissei, Mitsubishi Tanabe, and Pfizer. TK has received grants from GlaxoSmithKline. TO has received grants from Chugai. KYa has received fees for lecture from AbbVie, Ayumi, BMS, Chugai, Eisai, Janssen, Mitsubishi Tanabe, Ono, and UCB. KF has received grants, consulting fees, speaking fees, and/or honoraria from Mitsubishi Tanabe, BMS, Eli Lilly, Chugai, Janssen, Pfizer, Ono, AbbVie, Ayumi, Astellas, Sanofi, Novartis, Daiichi-Sankyo, Eisai, Asahi Kasei, MSD, and Japan Blood Products Organization.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2020.01095/full#supplementary-material
References
1. Bentham J, Morris DL, Graham DSC, Pinder CL, Tombleson P, Behrens TW, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. (2015) 47:1457–64. doi: 10.1038/ng.3434
2. Gower RG, Sams WM, Thorne EG, Kohler PF, Claman HN. Leukocytoclastic vasculitis: sequential appearance of immunoreactants and cellular changes in serial biopsies. J Invest Dermatol. (1977) 69:477–84. doi: 10.1111/1523-1747.ep12511631
3. Seri Y, Shoda H, Suzuki A, Matsumoto I, Sumida T, Fujio K, et al. Peptidylarginine deiminase type 4 deficiency reduced arthritis severity in a glucose-6-phosphate isomerase-induced arthritis model. Sci Rep. (2015) 5:13041. doi: 10.1038/srep13041
4. Suzuki A, Yamada R, Chang X, Tokuhiro S, Sawada T, Suzuki M, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. (2003) 34:395–402. doi: 10.1038/ng1206
5. Suzuki A, Kochi Y, Shoda H, Seri Y, Fujio K, Sawada T, et al. Decreased severity of experimental autoimmune arthritis in peptidylarginine deiminase type 4 knockout mice. BMC Musculoskelet Disord. (2016) 17:205. doi: 10.1186/s12891-016-1055-2
6. Savchenko AS, Borissoff JI, Martinod K, De Meyer SF, Gallant M, Erpenbeck L, et al. VWF-mediated leukocyte recruitment with chromatin decondensation by PAD4 increases myocardial ischemia/reperfusion injury in mice. Blood. (2014) 123:141–8. doi: 10.1182/blood-2013-07-514992
7. Martinod K, Demers M, Fuchs TA, Wong SL, Brill A, Gallant M, et al. Neutrophil histone modification by peptidylarginine deiminase 4 is critical for deep vein thrombosis in mice. Proc Natl Acad Sci USA. (2013) 110:8674–9. doi: 10.1073/pnas.1301059110
8. Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, Loos R, Radzisheuskaya A, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. (2014) 507:104–8. doi: 10.1038/nature12942
9. Li P, Yao H, Zhang Z, Li M, Luo Y, Thompson PR, et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. (2008) 28:4745–58. doi: 10.1128/MCB.01747-07
10. Nakashima K, Arai S, Suzuki A, Nariai Y, Urano T, Nakayama M, et al. PAD4 regulates proliferation of multipotent haematopoietic cells by controlling c-myc expression. Nat Commun. (2013) 4:1836. doi: 10.1038/ncomms2862
11. Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. (2010) 207:1853–62. doi: 10.1084/jem.20100239
12. Zhang X, Gamble MJ, Stadler S, Cherrington BD, Causey CP, Thompson PR, et al. Genome-wide analysis reveals PADI4 cooperates with Elk-1 to activate c-Fos expression in breast cancer cells. PLoS Genet. (2011) 7:e1002112. doi: 10.1371/journal.pgen.1002112
13. Yokogawa M, Takaishi M, Nakajima K, Kamijima R, Fujimoto C, Kataoka S, et al. Epicutaneous application of toll-like receptor 7 agonists leads to systemic autoimmunity in wild-type mice: a new model of systemic Lupus erythematosus. Arthritis Rheumatol. (2014) 66:694–706. doi: 10.1002/art.38298
14. Greiling TM, Dehner C, Chen X, Hughes K, Iñiguez AJ, Boccitto M, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med. (2018) 10:eaan2306. doi: 10.1126/scitranslmed.aan2306
15. Brightbill HD, Suto E, Blaquiere N, Ramamoorthi N, Sujatha-Bhaskar S, Gogol EB, et al. NF-κB inducing kinase is a therapeutic target for systemic lupus erythematosus. Nat Commun. (2018) 9:179. doi: 10.1038/s41467-017-02672-0
16. Iwanaga A, Wang G, Gantulga D, Sato T, Baljinnyam T, Shimizu K, et al. Ablation of the scaffold protein JLP causes reduced fertility in male mice. Transgenic Res. (2008) 17:1045–58. doi: 10.1007/s11248-008-9191-6
17. Atkinson C, Qiao F, Song H, Gilkeson GS, Tomlinson S. Low-dose targeted complement inhibition protects against renal disease and other manifestations of autoimmune disease in MRL/lpr mice. J Immunol. (2008) 180:1231–8. doi: 10.4049/jimmunol.180.2.1231
18. Swamydas M, Lionakis MS. Isolation, purification and labeling of mouse bone marrow neutrophils for functional studies and adoptive transfer experiments. J Vis Exp. (2013) 77:e50586. doi: 10.3791/50586
19. Watanabe M, Terasawa M, Miyano K, Yanagihara T, Uruno T, Sanematsu F, et al. DOCK2 and DOCK5 act additively in neutrophils to regulate chemotaxis, superoxide production, and extracellular trap formation. J Immunol. (2014) 193:5660–7. doi: 10.4049/jimmunol.1400885
20. Weckbach LT, Gola A, Winkelmann M, Jakob SM, Groesser L, Borgolte J, et al. The cytokine midkine supports neutrophil trafficking during acute inflammation by promoting adhesion via β2 integrins (CD11/CD18). Blood. (2014) 123:1887–96. doi: 10.1182/blood-2013-06-510875
21. Suire S, Condliffe AM, Ferguson GJ, Ellson CD, Guillou H, Davidson K, et al. Gbetagammas and the Ras binding domain of p110gamma are both important regulators of PI(3)Kgamma signalling in neutrophils. Nat Cell Biol. (2006) 8:1303–9. doi: 10.1038/ncb1494
22. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. (2011)17:10–12. doi: 10.14806/ej.17.1.200
23. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
24. Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. (2015) 31:166–9. doi: 10.1093/bioinformatics/btu638
25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
26. Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. (2009) 4:1184–91. doi: 10.1038/nprot.2009.97
27. Faber-Elmann A, Sthoeger Z, Tcherniack A, Dayan M, Mozes E. Activity of matrix metalloproteinase-9 is elevated in sera of patients with systemic lupus erythematosus. Clin Exp Immunol. (2002) 127:393–8. doi: 10.1046/j.1365-2249.2002.01758.x
28. Rossaint J, Zarbock A. Tissue-specific neutrophil recruitment into the lung, liver, and kidney. J Innate Immun. (2013) 5:348–57. doi: 10.1159/000345943
29. Guan J, Wang G, Tam LS, Kwan BC, Li EK, Chow KM, et al. Urinary sediment ICAM-1 level in lupus nephritis. Lupus. (2012) 21:1190–5. doi: 10.1177/0961203312451334
30. Chung AC, Lan HY. Chemokines in renal injury. J Am Soc Nephrol. (2011) 22:802–9. doi: 10.1681/ASN.2010050510
31. Nick JA, Avdi NJ, Young SK, Lehman LA, McDonald PP, Frasch SC, et al. Selective activation and functional significance of p38alpha mitogen-activated protein kinase in lipopolysaccharide-stimulated neutrophils. J Clin Invest. (1999) 103:851–8. doi: 10.1172/JCI5257
32. Lee CM, Onésime D, Reddy CD, Dhanasekaran N, Reddy EP. JLP: a scaffolding protein that tethers JNK/p38MAPK signaling modules and transcription factors. Proc Natl Acad Sci USA. (2002) 99:14189–94. doi: 10.1073/pnas.232310199
33. Flores-Mendoza G, Sansón SP, Rodríguez-Castro S, Crispín JC, Rosetti F. Mechanisms of Tissue Injury in Lupus Nephritis. Trends Mol Med. (2018) 24:364–78. doi: 10.1016/j.molmed.2018.02.003
34. Bullard DC, King PD, Hicks MJ, Dupont B, Beaudet AL, Elkon KB. Intercellular adhesion molecule-1 deficiency protects MRL/MpJ-Fas(lpr) mice from early lethality. J Immunol. (1997) 159:2058–67.
35. Kevil CG, Hicks MJ, He X, Zhang J, Ballantyne CM, Raman C, et al. Loss of LFA-1, but not Mac-1, protects MRL/MpJ-Fas(lpr) mice from autoimmune disease. Am J Pathol. (2004) 165:609–16. doi: 10.1016/S0002-9440(10)63325-1
36. Villanueva E, Yalavarthi S, Berthier CC, Hodgin JB, Khandpur R, Lin AM, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol. (2011) 187:538–52. doi: 10.4049/jimmunol.1100450
37. Panousis NI, Bertsias GK, Ongen H, Gergianaki I, Tektonidou MG, Trachana M, et al. Combined genetic and transcriptome analysis of patients with SLE: distinct, targetable signatures for susceptibility and severity. Ann Rheum Dis. (2019) 78:1079–89. doi: 10.1136/annrheumdis-2018-214379
38. McMillan JI, Riordan JW, Couser WG, Pollock AS, Lovett DH. Characterization of a glomerular epithelial cell metalloproteinase as matrix metalloproteinase-9 with enhanced expression in a model of membranous nephropathy. J Clin Invest. (1996) 97:1094–101. doi: 10.1172/JCI118502
39. Tveita AA, Rekvig OP, Zykova SN. Increased glomerular matrix metalloproteinase activity in murine lupus nephritis. Kidney Int. (2008) 74:1150–8. doi: 10.1038/ki.2008.308
40. Urushihara M, Kagami S, Kuhara T, Tamaki T, Kuroda Y. Glomerular distribution and gelatinolytic activity of matrix metalloproteinases in human glomerulonephritis. Nephrol Dial Transplant. (2002) 17:1189–96. doi: 10.1093/ndt/17.7.1189
41. Chakrabarti S, Zee JM, Patel KD. Regulation of matrix metalloproteinase-9 (MMP-9) in TNF-stimulated neutrophils: novel pathways for tertiary granule release. J Leukoc Biol. (2006) 79:214–22. doi: 10.1189/jlb.0605353
42. Carmona-Rivera C, Zhao W, Yalavarthi S, Kaplan MJ. Neutrophil extracellular traps induce endothelial dysfunction in systemic lupus erythematosus through the activation of matrix metalloproteinase-2. Ann Rheum Dis. (2015) 74:1417–24. doi: 10.1136/annrheumdis-2013-204837
43. Biron BM, Chung CS, Chen Y, Wilson Z, Fallon EA, Reichner JS, et al. PAD4 deficiency leads to decreased organ dysfunction and improved survival in a dual insult model of hemorrhagic shock and sepsis. J Immunol. (2018) 200:1817–28. doi: 10.4049/jimmunol.1700639
44. Knight JS, Subramanian V, O'Dell AA, Yalavarthi S, Zhao W, Smith CK, et al. Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann Rheum Dis. (2015) 74:2199–206. doi: 10.1136/annrheumdis-2014-205365
45. Knight JS, Zhao W, Luo W, Subramanian V, O'Dell AA, Yalavarthi S, et al. Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest. (2013) 123:2981–93. doi: 10.1172/JCI67390
46. Lande R, Ganguly D, Facchinetti V, Frasca L, Conrad C, Gregorio J, et al. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci Transl Med. (2011) 3:73ra19. doi: 10.1126/scitranslmed.3001180
47. Uhl B, Vadlau Y, Zuchtriegel G, Nekolla K, Sharaf K, Gaertner F, et al. Aged neutrophils contribute to the first line of defense in the acute inflammatory response. Blood. (2016) 128:2327–37. doi: 10.1182/blood-2016-05-718999
48. Iwata Y, Wada T, Furuichi K, Sakai N, Matsushima K, Yokoyama H, et al. p38 Mitogen-activated protein kinase contributes to autoimmune renal injury in MRL-Fas lpr mice. J Am Soc Nephrol. (2003) 14:57–67. doi: 10.1097/01.ASN.0000037402.83851.5F
49. Thatcher JD. The Ras-MAPK signal transduction pathway. Sci Signal. (2010) 3:tr1. doi: 10.1126/scisignal.3119tr1
50. Ragotte RJ, Dhanrajani A, Pleydell-Pearce J, Del Bel KL, Tarailo-Graovac M, van Karnebeek C, et al. The importance of considering monogenic causes of autoimmunity: a somatic mutation in KRAS causing pediatric Rosai-Dorfman syndrome and systemic lupus erythematosus. Clin Immunol. (2017) 175:143–6. doi: 10.1016/j.clim.2016.12.006
51. Santonico E, Belleudi F, Panni S, Torrisi MR, Cesareni G, Castagnoli L. Multiple modification and protein interaction signals drive the Ring finger protein 11 (RNF11) E3 ligase to the endosomal compartment. Oncogene. (2010) 29:5604–18. doi: 10.1038/onc.2010.294
52. Yang T, Cao C, Yang J, Liu T, Lei XG, Zhang Z, et al. miR-200a-5p regulates myocardial necroptosis induced by Se deficiency via targeting RNF11. Redox Biol. (2018) 15:159–69. doi: 10.1016/j.redox.2017.11.025
53. Zhang X, Kim J, Ruthazer R, McDevitt MA, Wazer DE, Paulson KE, et al. The HBP1 transcriptional repressor participates in RAS-induced premature senescence. Mol Cell Biol. (2006) 26:8252–66. doi: 10.1128/MCB.00604-06
54. Suzuki R, Nemoto E, Shimauchi H. Cyclic tensile force up-regulates BMP-2 expression through MAP kinase and COX-2/PGE2 signaling pathways in human periodontal ligament cells. Exp Cell Res. (2014) 323:232–41. doi: 10.1016/j.yexcr.2014.02.013
55. Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, et al. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. (2010) 49:4852–63. doi: 10.1021/bi100363t
56. Gordon RA, Herter JM, Rosetti F, Campbell AM, Nishi H, Kashgarian M, et al. Lupus and proliferative nephritis are PAD4 independent in murine models. JCI Insight. (2017) 2:e92926. doi: 10.1172/jci.insight.92926
57. Kienhöfer D, Hahn J, Stoof J, Csepregi JZ, Reinwald C, Urbonaviciute V, et al. Experimental lupus is aggravated in mouse strains with impaired induction of neutrophil extracellular traps. JCI Insight. (2017) 2:e92920. doi: 10.1172/jci.insight.92920
58. Liu Y, Lightfoot YL, Seto N, Carmona-Rivera C, Moore E, Goel R, et al. Peptidylarginine deiminases 2 and 4 modulate innate and adaptive immune responses in TLR-7-dependent lupus. JCI Insight. (2018) 3:e124729. doi: 10.1172/jci.insight.124729
59. Swindell WR, Michaels KA, Sutter AJ, Diaconu D, Fritz Y, Xing X, et al. Imiquimod has strain-dependent effects in mice and does not uniquely model human psoriasis. Genome Med. (2017) 9:24. doi: 10.1186/s13073-017-0415-3
60. Yu F, Wu LH, Tan Y, Li LH, Wang CL, Wang WK, et al. Tubulointerstitial lesions of patients with lupus nephritis classified by the 2003 international society of nephrology and renal pathology society system. Kidney Int. (2010) 77:820–9. doi: 10.1038/ki.2010.13
61. Li H, Han SJ, Kim M, Cho A, Choi Y, D'Agati V, et al. Divergent roles for kidney proximal tubule and granulocyte PAD4 in ischemic AKI. Am J Physiol Renal Physiol. (2018) 314:F809–19. doi: 10.1152/ajprenal.00569.2017
62. Massarenti L, Enevold C, Damgaard D, Ødum N, Nielsen CH, Jacobsen S. Peptidylarginine deiminase-4 gene polymorphisms are associated with systemic lupus erythematosus and lupus nephritis. Scand J Rheumatol. (2019) 48:133–40. doi: 10.1080/03009742.2018.1488273
63. Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. (2014) 46:1187–96. doi: 10.1038/ng.3118
Keywords: peptidylarginine deiminase 4, lupus nephritis, neutrophil, p38 mitogen-activated protein kinase, JNK-associated leucine zipper protein
Citation: Hanata N, Shoda H, Hatano H, Nagafuchi Y, Komai T, Okamura T, Suzuki A, Gunarta IK, Yoshioka K, Yamamoto K and Fujio K (2020) Peptidylarginine Deiminase 4 Promotes the Renal Infiltration of Neutrophils and Exacerbates the TLR7 Agonist-Induced Lupus Mice. Front. Immunol. 11:1095. doi: 10.3389/fimmu.2020.01095
Received: 09 January 2020; Accepted: 06 May 2020;
Published: 23 June 2020.
Edited by:
Carlo Riccardi, University of Perugia, ItalyCopyright © 2020 Hanata, Shoda, Hatano, Nagafuchi, Komai, Okamura, Suzuki, Gunarta, Yoshioka, Yamamoto and Fujio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirofumi Shoda, c2hvZGEtdGt5JiN4MDAwNDA7dW1pbi5hYy5qcA==; Keishi Fujio, a2Z1amlvLXRreSYjeDAwMDQwO3VtaW4uYWMuanA=
 Norio Hanata
Norio Hanata Hirofumi Shoda
Hirofumi Shoda Hiroaki Hatano
Hiroaki Hatano Yasuo Nagafuchi
Yasuo Nagafuchi Toshihiko Komai
Toshihiko Komai Tomohisa Okamura
Tomohisa Okamura Akari Suzuki3
Akari Suzuki3 Keishi Fujio
Keishi Fujio