Corrigendum: Vaccines Against Antimicrobial Resistance
- 1GSK, Siena, Italy
- 2Department of Pharmacy and Biotechnology (FaBiT), University of Bologna, Bologna, Italy
- 3vAMRes Lab, Toscana Life Sciences, Siena, Italy
- 4Faculty of Medicine, Imperial College London, London, United Kingdom
In the last century, life expectancy has increased considerably, thanks to the introduction of antibiotics, hygiene and vaccines that have contributed to the cure and prevention of many infectious diseases. The era of antimicrobial therapy started in the nineteenth century with the identification of chemical compounds with antimicrobial properties. However, immediately after the introduction of these novel drugs, microorganisms started to become resistant through different strategies. Although resistance mechanisms were already present before antibiotic introduction, their large-scale use and mis-use have increased the number of resistant microorganisms. Rapid spreading of mobile elements by horizontal gene transfer such as plasmids and integrative conjugative elements (ICE) carrying multiple resistance genes has dramatically increased the worldwide prevalence of relevant multi drug-resistant human pathogens such as Staphylococcus aureus, Neisseria gonorrhoeae, and Enterobacteriaceae. Today, antimicrobial resistance (AMR) remains one of the major global concerns to be addressed and only global efforts could help in finding a solution. In terms of magnitude the economic impact of AMR is estimated to be comparable to that of climate global change in 2030. Although antibiotics continue to be essential to treat such infections, non-antibiotic therapies will play an important role in limiting the increase of antibiotic resistant microorganisms. Among non-antibiotic strategies, vaccines and therapeutic monoclonal antibodies (mAbs) play a strategic role. In this review, we will summarize the evolution and the mechanisms of antibiotic resistance, and the impact of AMR on life expectancy and economics.
Introduction
Throughout the evolution of our species, life expectancy has generally been between 25 and 35 years with infection diseases being the balance needle. The living conditions of human beings progressively improved to the point that nowadays, a person born in a high-income country can expect to live more than 85 years (1). This result has been achieved primarily by improved hygiene, antibiotics and vaccination introduction. These three pillars have represented the most effective medical interventions to reduce mortality and morbidity caused by infectious diseases. In fact, in the poor areas of our planet where hygiene and the use of antibiotics and vaccination are not well-established, life expectancy is still lower than 50 years (2). Vaccination has protected humans by training the immune system to recognize and establish a rapid and effective response against a pathogen. In the 20th century, vaccines were introduced to large populations with the immunization practice developed by Edward Jenner. Since then, vaccination has become a widespread medical treatment determining the control of 16 major diseases, namely, smallpox, diphtheria, tetanus, yellow fever, pertussis, Haemophilus influenzae type b disease, poliomyelitis, measles, mumps, rubella, typhoid, rabies, rotavirus, hepatitis B, pneumococcal, and meningococcal diseases. Noteworthy is the complete or partial eradication achieved for smallpox and poliomyelitis, respectively (3).
To counteract infectious diseases, the discovery of antimicrobial treatment was another significant milestone that has dramatically reduced mortality. The modern era of antimicrobial therapy initiated in the 19th century with the identification of anti-syphilitic and anti-trypanosomal molecules derived from organic compounds chemically synthetized (4). In 1928, the discovery by Alexander Fleming of a new class of non-toxic antimicrobial agents derived from environmental fungi gave rise to the “golden era” of antibiotic discovery (1945–1960) (5). Conversely to active vaccination, drugs are therapeutics with different modes of action targeting the bacterial functions such as cell wall integrity, nucleic acid synthesis and repair, or protein biosynthesis. Moreover, drugs can be naturally produced by microorganisms (including environmental fungi and saprophytic bacteria), generated by chemical modifications of the natural antimicrobial agents or fully synthetized (6).
In combination with the vaccination practice, the discovery of antibiotics and their successful use in medicine is considered one the most relevant findings from a global health perspective (Figure 1). Nevertheless, the effectiveness of antibiotics has weakened to the point that our lives can be severely threatened. In fact, the antimicrobial resistance (AMR) is one of the most daunting problems that is causing the spread of infectious diseases and the increase in the number of deaths caused by infections that were previously considered uncomplicated (7). For example, the bloodstream infections caused by bacteria resistant to one or several drugs (multidrug-resistant; MDR) such as Pseudomonas aeruginosa are characterized by a 50% of mortality compared with the 24% of the non-multidrug-resistant infections (8). In addition, medical procedures such as surgeries, immunosuppressive chemotherapy and organ transplantation are becoming more critical and, in some cases, even prohibitive considering the need of effective antibiotics against multidrug-resistant pathogens. Therefore, the consequences of such microbial evolution can be dramatic with infectious diseases that could severely reduce our lifespan to an extent similar to the pre-antibiotic era. Globally, AMR pathogens are causing 700,000 deaths/year, and 10 million deaths/year are expected by 2050, a number even, higher than the 8.2 million caused by cancer today (9) (Figure 2).
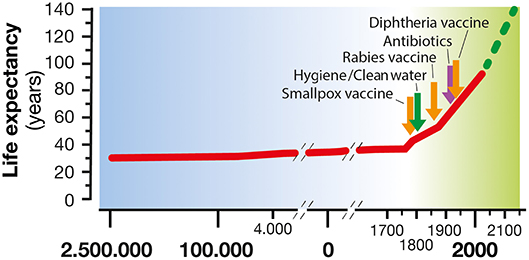
Figure 1. Life expectancy increase along human civilization. In the last century, life expectancy has increased considerably, thanks to the introduction of hygiene, clean water, antibiotics, and vaccines as a means of treatment and prevention of many infectious diseases.
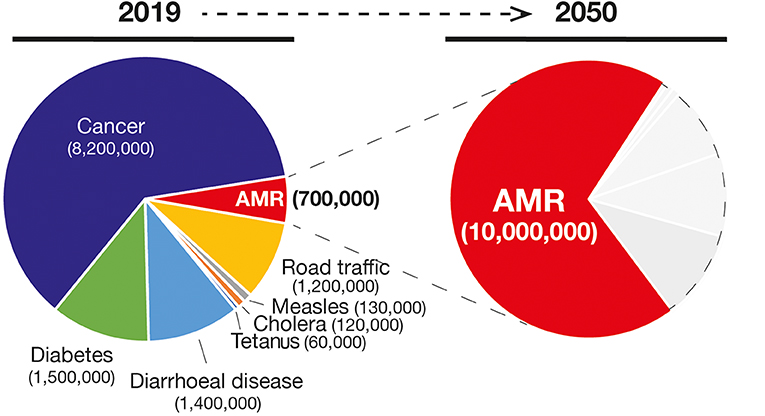
Figure 2. Number of deaths and the main causes (Left) in 2019 and the projection of number of deaths due to AMR infections in 2050 (in red in the Right). Gray areas represent other causes of deaths.
Antibiotic Resistance Mechanisms and Prioritization of Antibiotic Resistant Microorganisms
Antibiotic resistance is considered nowadays as one of the greatest threats to human health (10). Cases of antibiotic resistance are constantly reported, and the time needed for bacteria to become resistant to newly introduced antibiotics, is getting shorter. In fact, antimicrobial use exerts evolutionary pressure for the creation and transmission of resistant pathogens, thus reducing antimicrobial effectiveness and raising the incidence of severe disease (11). However, this is not a new phenomenon and is commonly observed as soon as the introduction of new classes of antibiotics occurs (12). In 1946, Alexander Fleming anticipated this global burden with the renowned sentence “There is probably no chemotherapeutic drug to which in suitable circumstances the bacteria cannot react by in some way acquiring “fastness” [resistance]” (13). In fact, penicillin became commercially available in 1943 and resistance was observed for Staphylococcus aureus by 1948. In this context, the discovery by Barbara McClintok that transposons play a major role in the genomic diversity and evolution paved the way for a deeper understanding of the genetic basis underlying the antimicrobial resistance dissemination. Transduction, conjugation, transformation and other mobile genetic materials (transposons and integrons) are all possible mechanisms for the transmission of genetic determinants involved in AMR (14, 15) and in the generation of “superbugs” (16). Worryingly, the transmission of multiple resistance genes is not only relatively common among organisms within the same genus but also among evolutionarily distant organisms.
A plethora of mechanisms has contributed to the emergence of antimicrobial resistant microorganisms and to their spread. The first classification is between natural and acquired resistance mechanisms. The natural resistance relies on the bacterial ability to block the antibiotic function as a result of inherent structural or functional characteristics. For example, vancomycin (glycopeptide antibiotic) which inhibits peptidoglycan crosslinking, is generally effective only against Gram-positive but not Gram-negative bacteria, due to its inability to cross the outer membrane to get access to the d-Ala-d-Ala peptides. By contrast, the acquired resistance is based on the gaining of new functions; bacteria that would be originally susceptible, became resistant to one or more antibiotics (17). The main mechanisms exploited by bacteria to tackle the action of antibiotics are based on (i) the inactivation of the drug by its hydrolysis or structural modification, (ii) the prevention of the access to the target by reducing membrane permeability or overexpression of efflux pumps, (iii) changes in the antibiotic targets by mutation or post-translational modifications (Figures 3A–C) (18). The mechanisms of antibiotics inactivation comprise both the production of enzymes that destroys the antibiotic itself (for example, β-lactamases) and the production of enzymes that modify its structure (for example, aminoglycoside-modifying enzymes). β-lactam antibiotics, such as penicillin and cephalosporins, are hydrolyzed by a wide variety of β-lactamases. Since the early discovery of these enzymes, bacteria resistant to all β-lactam antibiotics have emerged (19, 20). In this regard, the genes encoding for extended-spectrum β-lactamases (ESBLs with activity against oxyimino-cephalosporins) and carbapenemases (21), including the IMP (imipenemase), VIM (Verona integron encoded metallo β-lactamase), K. pneumoniae carbapenemase (KPC) and OXA (oxacillinase) have been identified not only on the chromosomes, but also on plasmids as in the case of Enterobacteriaceae (22, 23) and of P. aeruginosa (8). Among them, there is the New Delhi metallo-β- lactamase 1 (NDM-1), an enzyme that confers high level of resistance to multiple antibiotics, including penicillin, cephalosporins and carbapenems and that is capable of rapid worldwide dissemination (14). This is of high concern since there are few available treatment options beyond the carbapenems. The main producers of the NDM-1 enzyme are Enterobacteriaceae such as Klebsiella pneumoniae, Escherichia coli and Proteus mirabilis, but also Acinetobacter spp. and Pseudomonas aeruginosa (Figure 3A) (24). Lastly, aminoglycoside-modifying enzymes are able to modify the structure of the antibiotic molecule by the transfer of different chemical groups such as acyl, phosphate, nucleotidyl and ribitoyl groups. This modification prevents the antibiotic from binding to its target protein mainly due to the steric hindrance (Figure 3A). For example, in 2012 a novel genomic island encoding six aminoglycoside-modifying enzymes, including gentamycin, was identified in Campylobacter coli isolated from broiler chickens in China (25) and more recently in Group B streptococcus (26). Yet, the prevention of antibiotic access to a specific target can occur by reducing the permeability of the membrane and/or increasing efflux in order to reduce the intracellular concentration of a drug (27, 28). Differently from Gram-positive bacteria, Gram-negative bacteria are intrinsically less permeable to many antibiotics as their outer membrane forms a sort of permeability barrier. Generally, the permeability is reduced by the downregulation of the major porins, which results in a decrease in antibiotics entry into the bacterial cell. Porins are beta barrel proteins that cross the cellular membranes acting as channels for passive diffusion of certain molecules (29). The expression or the structure of a specific porin can be influenced by mutations that decrease membrane permeability, reducing the entry of antibiotics inside the cell. For instance, P. aeruginosa OprD-deficient strains are characterized by high level of resistance to carbapenems (30). Bacterial efflux pumps belonging to the resistance-nodulation-division (RND) family are not selective for a specific class of antibiotic and confer intrinsic resistance to multiple and structurally distinct classes of drugs, including the β-lactams, quinolones, and aminoglycosides (31). The high-level expression of efflux genes is often due to mutations in their regulatory network that generally occur within a local repressor, a global transcription factor, promoter regions or intergenic sites. For example, by mutations and gene duplication mechanisms the overexpression of MexAB-OprM multidrug efflux systems confers to some P. aeruginosa isolates the ability to became resistant to carbenicillin antibiotics. Moreover, Nicoloff et al. identified a large tandem duplication of the E. coli acrAB locus in a mutant isolated in the presence of tetracycline (32). Such a mutation was found to overexpress the AcrAB drug efflux pump, producing an MDR phenotype (33). In N. gonorrhoeae, a mutation in the consensus −10 sequence upstream of mtrC strengthens its promoter activity determining constitutive overexpression of the efflux pump (34), (Figure 3B).
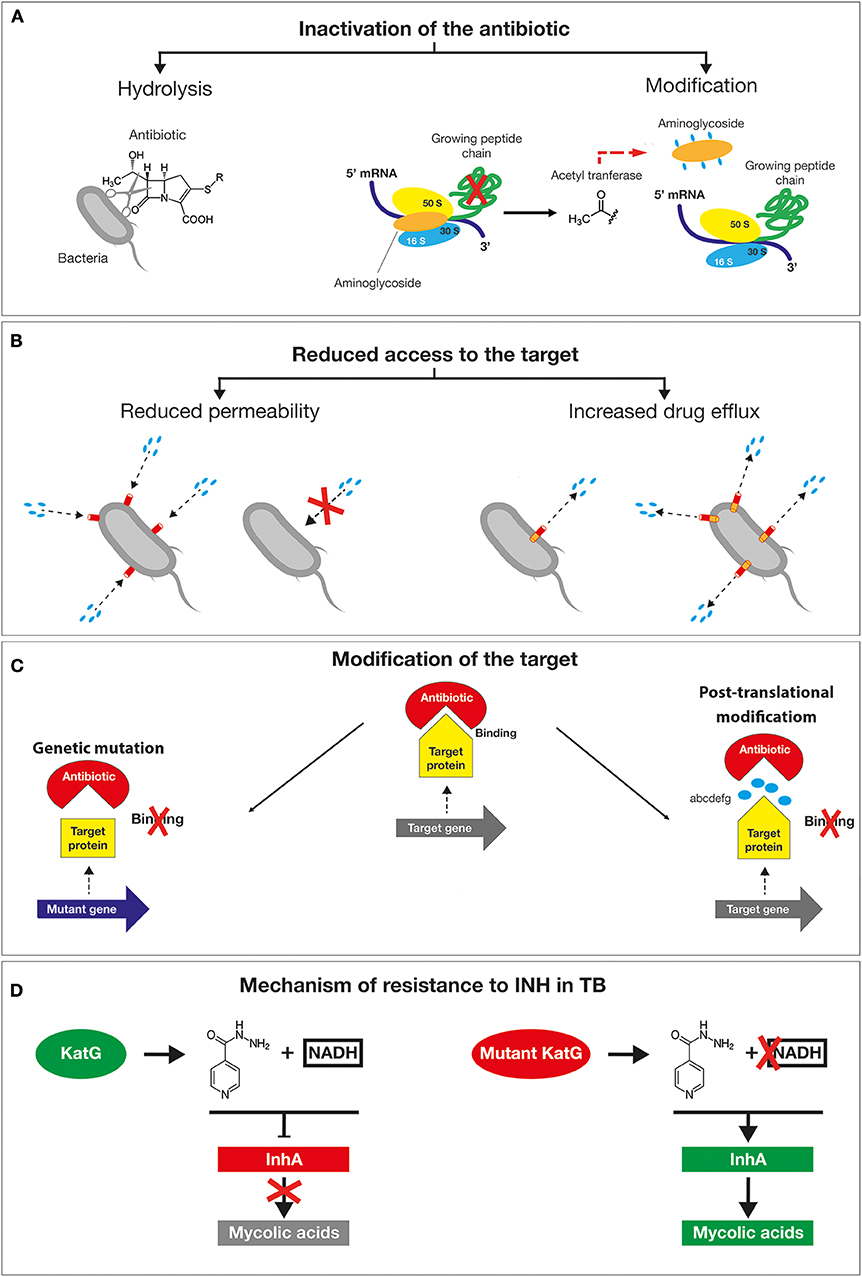
Figure 3. Schematic representation of the main mechanisms exploited by bacteria to tackle the action of antibiotics. (A) The inactivation of the drug by its hydrolysis or structural modification. (B) The prevention of the access to the target by reducing membrane permeability or overexpression of efflux pumps. (C) Changes in the antibiotic targets by mutation or post-translational modifications. (D) Resistance mechanism of Mycobacterium tuberculosis to isoniazid drug mutations in the katG gene causes the accumulation of the inactive form of the antibiotic inside the bacilli without the formation of active adduct.
The other mechanism responsible for drug resistance is represented by the modification of the target by genetic mutations or post-translational modifications. Mutations in target genes can enable bacteria to overcome the toxic effects of antibiotics. Examples are provided by mutations in bacterial DNA gyrases (e.g. gyrA) and topoisomerase (e.g. parC) for fluoroquinolones, the β subunit of RNA polymerase (rpoB) for rifampin, 16S rRNA (rrs) for tetracyclines and aminoglycosides and 23S rRNA (rrl) for linezolid (13). The clinical use of linezolid has selected S. pneumoniae (35) and S. aureus (36) resistant lineages by a mutation in one of the rrl copies. The high recombination frequency has subsequently enabled the allele enrichment in the bacterial population. Another example of a target change is the acquisition of specific genes by horizontal gene transfer (HGT), such as in methicillin-resistant S. aureus (MRSA). In this case, the methicillin resistance is obtained by the acquisition of the staphylococcal cassette chromosome mec (SCCmec) element. This cassette encodes for a mutated form of PBP2 that is not recognized as a target by the β-lactamase enzyme but is still able to mediate a regular cell wall biosynthesis. Additional antibiotic resistance mechanisms consist in the protection by modification of the target molecule. This is exemplified by the polymyxin resistance in Acinetobacter baumanni (polymyxin B and polymyxin E, also known as colistin). The polymyxin antibiotics, are cyclic antimicrobial peptides with long, hydrophobic tails that bind to the lipopolysaccharide (LPS) of Gram-negative bacteria disrupting cell membranes. This resistance is often associated with changes in the expression of regulators affecting LPS production. For example, the addition of phosphoethanolamine (PE) to lipid A due to overexpression of pmrC results in a substantial reduction of colistin binding as the negative charge of the LPS is hugely lowered (37).
Given the alarming increase in antibiotic resistance cases, several human pathogens have been classified as a priority for the design of appropriate preventive and therapeutic strategies. In 2017, WHO published a prioritization list of pathogens (classified as critical, high and medium priority) to guide discovery, research and development of new antibiotic (38). In 2019 an estimation performed by the Centers for Disease Control and Prevention (CDC) in the United States reported that more than 2.8 million antibiotic-resistant infections occur in the U.S. each year, and more than 35,000 people die as a result of these infections. The antibiotic-resistant strains were classified into four categories according to the urgency of intervention (urgent, serious, concerning and watch list) (39). These bacteria can cause severe and often deadly infections and have become resistant to a large number of antibiotics, including carbapenems and third generation cephalosporins (the best available antibiotics for treating multidrug resistant bacteria). A relevant example is provided by A. baumannii multi-drug resistant isolates, that frequently cause outbreaks in hospitals. These strains are characterized by an extraordinary ability to rapidly alter their genome surviving on inanimate surfaces and medical equipments even in the presence of disinfectants (40). In A. baumannii, “resistance islands” located on the bacterial chromosomes and/or on mobile genetic elements induce a drug-resistant phenotype. For example, AbaR1 is an 86kb resistance- island generated by recurrent insertions of different mobile genetic elements, which can spread by horizontal gene transfer (HGT) within the genera Pseudomonas, Salmonella, and Escherichia. Genomic islands like AbaR1 confer resistance to various antibiotics by spreading the cluster of genes encoding for OXA-type beta-lactamases, aminoglycoside-modifying enzymes, proteins responsible for the modification of the lipid A and overexpression of the AdeABC efflux system (41). Tuberculosis (TB), even if not initially included in the WHO list, was already considered one of the most urgent priority because of its growing resistance to traditional treatment. TB now kills more people than any other pathogen (1.8 million in 2015) (42). Unfortunately, the resistance to isoniazid (INH), one of the four antibiotics used for the treatment of TB, is constantly increasing. INH leads to the inhibition of mycolic acids (MA) synthesis, the major lipid component of the mycobacterial cell envelope (43). The mechanisms of action of isoniazid are diverse and not fully known due to the involvement of a number of different genes. However, several studies suggest that the catalase KatG activates INH enabling the binding to NAD and the inhibition of enoyl acyl carrier protein (ACP) reductase (InhA). As a result, MA synthesis is blocked leading to mycobacteria death. When mutations in the katG gene occur, INH remains in an inactive form, unable to bind NAD and to inhibit InhA activity (Figure 3D). Consequently, InhA accomplishes its function in cell wall synthesis determining the survival of TB. This type of resistance is of concern as it negatively affects the outcome of TB-treatment facilitating the dissemination of MDR-TB and reducing the efficacy of INH (44). The second and third tiers in the list—the high and medium priority categories—include other increasingly drug resistant bacteria that cause more common diseases, such as Neisseria gonorrhoeae (the causative agent of gonorrhea) (45, 46). Since the treatment of N. gonorrhoeae with sulfonamides, this bacterium has acquired genetic resistance determinants capable to inhibit the killing by all major classes of antibiotics that are used as first line methods of treatment against gonorrhea infection (47). The main lethal target of cephalosporins is the transpeptidase penicillin binding protein 2 (PBP2), encoded by penA. Most of the resistant isolates contain mosaic mutations in penA that confer resistance to cephalosporins. In addition, the overexpression of efflux pump, MtrCDE, contributes to resistance through an increase of the drug efflux. The simultaneous mutations in the major porins PorB and mtrR, the repressors of MtrCDE efflux pump, represent an important resistance determinant of gonococcus. Hence, the outstanding capacity of N. gonorrhoeae to rapidly acquire resistance has led the World Health Organization and Center for Disease Control (US) to classify this bacterium as “superbug.” As a result, if new prophylactic and/or therapeutic strategies will not be discovered soon, we may enter an era of incurable gonorrhea infections (48). Therefore, understanding the mechanisms underlying the AMR will help to rationally design new preventive strategies to effectively tackle these dangerous threats for the human health.
Evolution of Drug Resistance: What Did We Do Wrong?
Antibiotic resistance occurs when bacteria are not controlled nor killed by previously effective drugs through several mechanisms described above. Resistance is largely driven by the abuse of antibiotics in humans and animals, often with little or no therapeutic benefit. The use of antibiotics in viral respiratory infections or abuse in animal care and in agriculture to promote livestock growth, are common examples of the misuse (49). Additionally, the inability to rapidly identify the specific infecting pathogen at the point of care make the empiric treatment the only possible choice. In fact, the empiric treatment requires the use of broader spectrum antibiotics to cover all the key pathogens responsible for a particular infection (50). Moreover, the relationships between the human microbiome and the host immune system is hugely threatened. Antimicrobials alter the structure of the microbiota, expand the pool of antimicrobial-resistance genes, degrade the protective effects of the microbiota against invasion by pathogens, and may impair the efficacy of alternative medical treatments (51). Hence, the abuse of antibiotics has negatively affected the immune response making individuals more susceptible to infections. In fact, the pressure imposed by the antibiotic treatments on microbes other than the targeted pathogen (“bystander selection”) is hypothesized to be a major factor in the propagation of antibiotic resistance (11). International traveling is also a key factor in the dissemination of highly resistant strains. New Delhi metallo-β- lactamase 1 (NDM-1) and mcr-1 are the mobile resistance genes increasingly spreading worldwide and generating resistance to a variety of antibiotics including penicillins, cephalosporins and carbapenems. This is particularly relevant for infections caused by Gram-negative bacteria sensitive only to carbapenems, and for which only very few alternative possibilities are available. Furthermore, the NDM-1 resistance genes are transmitted very easily, as shown by the two Dutch travelers which were infected by a NDM-1-producing bacterial strain during their trip to India, even if not visiting any health centers (52). Enterobacteriaceae (Klebsiella pneumoniae, E. coli and Proteus mirabilis), Acinetobacter spp. and Pseudomonas aeruginosa are the most common NDM-1-producing bacteria for which colistin is considered the only possible therapeutic option. Nevertheless, with the spread of the mcr resistance gene (mcr-1) a generation of pan-resistant strains is arising. This has been probably due to the large use of colistin to increase pig growth in farms in China. In addition, to the raw meat, mcr-1 has been isolated also in samples from infected patients (53). The mcr-1 resistance determinant is able to easily spread in different bacterial species and also worldwide due to business globalization (24). Hence, the spread of resistant organisms, highly infectious and inducing high rate of morbidity and mortality has a strong economic impact, in terms of costs of treatment and long-term hospital stay, with a great burden on health systems.
Vaccine Against Antibiotic Resistance: A Valuable Weapon To Fight AMR
The rising of AMR is a threat to modern medicine and new measures to control antibiotic-resistant bacteria are desperately needed including the development of new antibiotics. Nevertheless, the discovery of new effective chemical compounds with an appropriate balance of antibacterial activity, drug metabolism, pharmacokinetics properties and safety it is a daunting task. In fact, the pipeline run dry ~40 years ago (54). In addition, challenging logistics and high costs of large clinical trials make them nearly impossible to bring to market. Even if successful, the clinical utility of antibiotics will decline as resistance to them inevitably rises (55). Improving hygiene and the correct use of antibiotics while expanding their access in low- and middle-income countries are important tools to limit the burden of AMR, despite the fact these measures alone are not enough (50). Hence, vaccines may become a valuable and effective weapon to fight AMR. An important aspect is that resistance mechanisms are of less concern in vaccination. As reported above the mechanisms of resistance to antibiotics are mediated by the generation of spontaneous mutations or the acquisition of mobile genetic element by HGT. These mechanisms confer to bacteria the ability to tackle the killing effect of a drug and survive. Antibiotics are therapeutic measures since generally prescribed after the settling down of an infection when hundreds of millions of bacteria are infecting the body. On the contrary, vaccines are designed to prevent diseases. Their prophylactic use allows the host to build an immune response before encountering the pathogen or even at the beginning of an infection when only a few hundred or thousands bacteria are present. As the occurrence of resistance mechanisms can stochastically arise among billions of bacteria, it is evident that this is less likely to occur following vaccination. Furthermore, most antibiotics have a single target while vaccines have multiple targets inducing host-specific antibody and/or T cell responses. Even in this case, more mutations are likely needed to confer resistance to vaccines making the development of microbial resistance even harder (56, 57). Therefore, vaccines can be effective against antimicrobial resistance in different ways:
(I) By lowering the inappropriate use of antimicrobial compounds. Perhaps counterintuitive is the evidence that viral vaccines are also very effective in reducing AMR. For istance, vaccines against influenza virus reduce the incidence of fever and sickness which affect a significant proportion of community-dwelling elderly population each year in the US (58). By preventing a proportion of these cases, vaccines can reduce both, the inappropriate use of antibiotics prescribed in case of viral infections and the need of antibiotic treatment to cure secondary bacterial infections (59).
(II) By reducing the insurgence of resistant serotypes. For example, the pneumococcal polysaccharide conjugate vaccines had an effect of direct protection of infants and of herd immunity in adults initially not targeted by routine immunization (60). Remarkably, also the antibiotic prescriptions and the prevalence of antibiotic resistant strains decreased. In the 1990s, before the introduction of a 7-valent pneumococcal conjugate vaccine (PCV7), more than 63,000 cases of invasive pneumococcal disease occurred each year in the US. Between 2000 and 2004, a 57% reduction in the incidence of penicillin-non-susceptible invasive pneumococcal disease (IPD) and 84% reduction in the rate of multidrug-resistant strains were achieved. These data indicate that vaccination is effective, regardless of the bacterial resistance phenotype. However, the universal use of PCV-7 led to increased prevalence of serotype 19A, a non-vaccine serotype with high rate of penicillin resistance (61). Introduction of 13-valent PCV in 2010, which contains 6 additional serotypes, including 19A, further reduced the incidence of IPD and of antibiotic-resistant pneumococci. Nevertheless, the risk of the evolution of AMR in pneumococcal serotypes not contained in the vaccine is still high. In this context, the design of a vaccine aimed to specifically target resistance determinants or resistant strains is highly valuable (62).
(III) By reducing infection rate of resistant strains in closely related species. Between 2004 and 2008, in New Zealand, 1 million people were vaccinated with the Outer Membrane Vesicles (OMV) based vaccine (MeNZB) to fight a meningococcus B outbreak. A retrospective case-control study has shown that the immunization with MeNZB resulted in 31% reduction of Neisseria gonorrhoeae infection (63). N. gonorrhoeae is one of the most common bacterial sexually transmitted diseases (STDs), with ~100 million cases worldwide. One of the main concerns of gonococcal infection is the emergence of strains resistant to nearly all classes of antibiotics including the expanded-spectrum cephalosporins and the lack of an effective vaccine. Hence, N. gonorrhoeae infections are becoming as the most prevalent and difficult to treat. Despite causing very different diseases, N. meningitidis and N. gonorrhoeae share 80–90% nucleotide identity at the genome level. Therefore, it is reasonable to assume that antibodies against common antigens could induce a cross-protective effect (64). Bioinformatic analyses has revealed that 57 OMPs are of N. meningitidis are conserved also in 970 N. gonorrhoeae strains isolated in US (65). Among them, PilQ, Omp85 (BamA), NspA, MtrE, MetQ, and LbpA show 93% amino acid sequence similarity to N. gonorrhoeae, suggesting their potential contribution to cross-protection (64). Further investigations are needed to evaluate the effectiveness of MenB-4C vaccine in preventing gonococcal infections (66).
(IV) By directly targeting antibiotic resistant microorganisms. Although many different vaccine formulations have been proposed to prevent infection by antimicrobial resistant pathogens, (such as S. aureus, E. coli, Clostridium difficile etc.) no successful phase III clinical trial data have been published yet (67, 68). A possible reason for the failure in developing vaccines against these pathogens are the multiple virulence mechanisms a vaccine should target, as well as the absence of animal models that are representative of human diseases (69, 70). A better understanding of host-pathogen interactions such as immune evasion, and increased knowledge in the epidemiology and variability of the main antigens, could help in the development of novel effective vaccine. However, new incentives may be necessary for the development of novel vaccines leading to the refinement of health care system against AMR infections and eventually saving millions of lives.
Modern Technologies and The Future of Vaccines Against AMR
Live-Attenuated or Killed Microorganisms and Subunit Vaccines
Attempts to “vaccinate” begin even before the Edward Jenner's smallpox vaccination was introduced. In the 7th century, some Indian Buddhists drank snake venom in an attempt to become immune to its effect. The process of variolation, consisting in the introduction of dried pus from smallpox pustules into the skin of a patient, was practiced at regular intervals by the Brahmin caste of Hindus in India in the 16th century (71). Nevertheless, Edward Jenner is considered the real father of vaccinology; in 1796 he tested the protective effect of a prior cowpox infection against smallpox by actively immunizing an eight-year-old-boy with cowpox and then carrying out smallpox challenge (variolation) (72). In the 19th century, Louis Pasteur gave a fundamental contribution in the field with the creation in laboratory of the first vaccine. The main principle of Pasteur was to “isolate, inactivate and inject” the causative agent of a disease. His rationale was to attenuate or inactivate the virulence of the disease-causing organisms while preserving their immunogenicity (5). Live attenuated vaccines (LAV) have been available since the last century and derive from viruses or bacteria that have been weakened under laboratory conditions, so that they induce immunity by simulating natural infections causing no or very mild disease (73). Moreover, they provide a recurrent antigenic stimulation that ensures memory cells production. An example is provided by the TB treatment with the Bacillus Calmette-Guérin (BCG) developed in 1922. Killed microorganisms are safer, do not induce disease and have an excellent stability profile, even if the immune response that they induce is reduced in terms of magnitude compared to LAV and several doses of vaccine may be required. For many viral diseases, the risk of reinfection is null due to the induction of antibodies directed against invariable antigens (e.g., smallpox, yellow fever, polio, rabies, mumps, measles, rubella, varicella, herpes zoster, hepatitis A, and Japanese encephalitis). Therefore, vaccines that were developed following the principles described in this paragraph have proven to be successful (74). A further development of Pasteur's strategy was the so-called “subunit” vaccine that contains only the antigenic determinants that are able to induce an effective immune response. This approach has been successfully applied to diphtheria and tetanus, two diseases caused by bacterial toxins (75).
Recombinant DNA
The development of safer live-attenuated vaccines and subunit vaccines through rational design has been possible when in the 1980s recombinant DNA techniques were introduced and revolutionized the field of vaccinology. This new approach enabled the generation of the genetically detoxified Bordetella pertussis toxin (76) and the recombinant hepatitis B surface antigen (HBsAg) (77). Genetic engineering has also been applied to generate attenuated viral and bacterial strains as antigen and delivery system for heterologous antigens, or to develop genetic “reassortants,” as in the case of the influenza attenuated vaccine or the rotavirus vaccine, which is based on a bovine rotavirus “reassorted” with 5 human rotavirus strains (78).
Glycoconjugate and Bioconjugate
In the previous decades, innovative polysaccharides-based vaccines have been developed to protect against infections caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae type b (Hib). However, these vaccines were not immunogenic in children below 2 years of age and they induced a short memory response. To enhance immunogenicity, an effective strategy was to covalently link the polysaccharide antigen to a carrier protein, thereby providing helper T-cell activation in addition to the B-cell dependent immune response. Examples of protein carriers that have been efficiently used for the chemical conjugation are tetanus toxoid (TT), diphtheria toxoid (DT), and a non-toxic cross-reacting mutant of DT (CRM197). The resulting glycoconjugate vaccines elicited a more potent functional immune response in all age groups. Nowadays, this type of vaccines are available for the meningococcus serogroups A, C, W, Y, and H. influenzae type b (Hib) and Pneumococcal vaccines. The glycoconjugate vaccines have been able to successfully prevent the deaths and hospitalization of millions of people during the past three decades (79, 80). However, the requirement of complex manufacturing makes the process of glycoconjugation expensive and unsuitable for very complex vaccines, or for their use in low-income countries. These limitations can be overcome by a process called “bioconjugation,” a new platform technology to generate glycoconjugate vaccines in vivo in a single step, drastically simplifying glycoconjugates development and manufacture. Bioconjugation is based on recombinant non-pathogenic Escherichia coli strain that co-express (i) a heterologous, antigenic polysaccharide, (ii) a carrier protein encoding the glycosylation sites, and (iii) an enzyme covalently coupling both substrates during bacterial growth. Clinical trials with either mono- and multi-valent vaccine candidates produced using the PglB conjugating enzyme showed safety, immunogenicity and functionality of the immune response (81, 82). Bioconjugates may play a fundamental role in the future to prevent infection caused by dangerous AMR pathogens expressing polysaccharide antigens, such as those reported in the WHO and CDC priority lists (38, 39).
Adjuvants
A limitation of many vaccines is the poorly immunogenic response they evoke. Thanks to the advances in the knowledge of mechanisms underlying innate immunity, effective vaccine adjuvants have been developed to enhance the immune systems' response. (83). These compounds are able to improve the speed, potency, and persistency of the immune response to vaccination. While aluminum was the first and only adjuvant routinely used in human vaccines for more than 50 years, novel adjuvanted system (AS) composed of the combination of two or more different immunostimulants were developed to achieve the desired response. For example, one of them, namely AS04, was used in 2005 for the hepatitis B (HBV) vaccine and in 2007 for the human papillomavirus HPV-16/18 vaccine (84). More recently, AS01, an adjuvant composed by two immunostimulants, (saponin QS21 and monophosphoryl lipid A, targeting TLR4), has been licensed for a vaccine against malaria and herpes zoster vaccine. Also, it is currently used in clinical trials for vaccines designed to prevent reactivation of tuberculosis, and for a vaccine against non-typeable H. influenzae and Moraxella catarrhalis, aimed to prevent exacerbation in chronic obstructive pulmonary disease. The TB vaccine phase 2b trial conducted in Kenya, South Africa, and Zambia provided 54.0% protection in M. tuberculosis infected adults against active pulmonary tuberculosis disease. This vaccine is composed of the M72 recombinant fusion protein including two immunogenic M. tuberculosis antigens (Mtb32A and Mtb39A), combined with the AS01E adjuvant system (85). Additional adjuvants targeting the innate immune receptors TLR9 (DNA oligonucleotides), TLR5 (Flagellin), TLR7, surface-exposed (TLR1/2), endosomal (TLR3), or cytoplasmic (RIG I, DNA sensors) innate immune receptors have been successfully tested (83).
Reverse Vaccinology, Generalized Modules for Membrane Antigens (GMMAs) and Nanoparticles
A new era in vaccine design is started through the use of the bacterial genomes to discover new vaccine candidates. “in silico” prediction tools are applied to identify genes encoding for surface-exposed or secreted antigens. Newly discovered antigens are expressed in E. coli as recombinant proteins, and their immunogenicity evaluated in preclinical models (86). This approach, named “reverse vaccinology” has allowed the development of the first multivalent protein-based vaccine against meningococcus B, 4CMenB, that is now approved in many countries worldwide. In addition to the recombinant proteins identified by the reverse vaccinology approach, the licensed 4CMenB vaccine contains the highly immunogenic outer membrane vesicles (OMV) component produced through detergent extraction (87). The same OMV component has been used in New Zealand to fight a MenB outbreak. An immunization campaign was implemented in all population starting from 5 weeks to 20 years of age in the period 2004–2006 (64, 88, 89). Interestingly, by using genetic approaches, it is possible to induce an over-blebbing phenotype without using detergents leading to a new generation of improved bacterial OMVs called generalized modules for membrane antigens (GMMAs). The vesicles are obtained by mutation in genes responsible for stabilizing the link between the bacterial outer membrane and peptidoglycan (e.g., tolR, tolB, nlpl). GMMAs can also be further engineered by targeting genes responsible for LPS acetylation, such as lpxlM and lpxL1 therefore reducing LPS reactogenicity (90). Yet, another class of vaccines proposed to overcome the limitations of conventional and subunit vaccines is based on nanoparticles. In this regard, thanks to advancement in structural biology the rational design of molecule structures with desired size, shape, stability, improved immunogenicity and functionality became feasible (91). Nanoparticles were initially discovered in virus such as those from Hepatitis B. Once produced recombinantly these particles were found to be composed either of the surface antigen (HBsAg) or the core antigen (HBcAg). The HBcAg was shown to be assembled into particles of 24–31 nm diameter resembling the highly immunogenic viral cores obtained from HBV-infected human liver. Other class of self-assembling protein nanoparticles has been identified from a wide variety of source. For example, one of these is represented by a 24 subunits molecule, each composed of a four-alpha-helix bundle, that self-assembles in a quaternary structure with octahedral symmetry named Ferritin. DNA technology can be also used to construct genes that encode for self-assembling polypeptides fused with the desired immunogenic epitope for expression of the chimeric molecule in a selected cell expression system. The chimeric polypeptide then self-assembles within the cell, with an ordered pattern of surface exposed epitopes (92). An important advantage of this approach is that proteins of interest are exposed on the surface of the nanoparticle in the correct conformation thus eliciting a high specific immune response.
Reverse Vaccinology 2.0 and Structural Vaccinology
Reverse Vaccinology 2.0 is aimed at the identification of antigens inducing high functional antibodies (93). By single B cells sorting and culturing, antibodies (Ab) with the desired functionality are selected and the corresponding Ig gene sequenced, and Abs produced as recombinant proteins. Three-dimensional structure resolution of the Ag–Ab (Fab) complex leads to a detailed definition of the protective epitope. Structural information and identification of the protective epitopes can drive the design of a novel optimized immunogens (“structure-based Ag design”). The new Ag can then be included in the best formulation or delivery system and tested in humans.
The Reverse Vaccinology 2.0 approach has allowed the identification of the cytomegalovirus CMV pentameric complex (94) and of the pre-fusion of F protein of the respiratory syncytial virus (RSV) (95). Although this approach has been exploited for viral pathogens, it is expected that the same technologies may also be applied to bacterial pathogens. Growing knowledge in this field could lead to rational design of new antigens more stable and able to elicit high level of functional antibodies. An example of structure-based design of bacterial antigens is the factor H binding protein (fHbp) of Neisseria meningitidis. This is a key meningococcal vaccine which exists as more than 300 peptides that can be grouped into three variants that do not elicit cross-protective immunity. Definition of the 3D structure and the mapping of all protective epitopes has guided the design of a new chimeric molecule containing combination of the main epitopes of the three variants but maintaining the overall conformation and the same structural features. The new chimeric fHbp was able to induce, in mice, cross-reactive immunity against the three variants (96).
Monoclonal Antibodies
An alternative strategy proposed to combat AMR infection is based on the use of monoclonal antibodies. The main modes of action of anti-bacterial antibodies are based on the neutralization of virulence factor activities and inhibition of complement-mediated bacterial lysis. An important aspect is that the efficacy of monoclonal antibodies is dependent upon the expression level and the degree of sequence conservation of the corresponding target epitope. Hence, the ideal epitope should be highly conserved in structure and to be easily accessible for antibody binding. This is the case of monoclonal antibodies against bacterial toxins approved for clinical use which include two antibodies against anthrax (97, 98) and one targeting the toxin B of Clostridium difficile, approved for the prevention of recurrent infections in high-risk patients (99). As described above, the new technologies could allow the analysis of immune response and the identification of human monoclonal antibodies naturally produced by infected or vaccinated human donors. This will guide the design of novel and highly immunogenic antigens and antigen derived peptides. Analyzing the Ab-Ag complex, it would be possible to design powerful antigen molecules with high neutralization activity as in the case of the RSV preF antigen (95). The outstanding power of these new technologies is boosting our ability to identify, design and stabilize antigens in a very short time frame and consequently starts clinical trials immediately after (100). Effective medical treatments are urgently needed to address major global threats, and in this context, monoclonal antibodies represent a unique valuable weapon. In developing human monoclonal antibodies careful attention should be paid to evaluate possible reactions with human tissue at early preclinical stages. Reactions can be limited by using fully human monoclonal antibodies and by an accurate selection of the targeted epitopes.
RNA Vaccines
RNA-based vaccines can be divided into non-replicating and self-amplifying mRNA vaccines (SAM) both of which make use of the host cell translational machinery to produce the selected antigens. More in detail, SAM-based vaccines consist of an engineered RNA viral genome encoding nonstructural proteins (nsPs) in which the genes encoding for the structural protein are replaced with the antigen of choice. RNA vaccines have shown to be effective in case of viral pathogens; however, this technology could be successfully applied also to bacterial AMR targets. Interestingly, a SAM vaccine encoding for a double-mutant of Streptolysin-O (SLOdm) and the backbone protein of pilus island 2a (BP-2a) from Group A (GAS, Streptococcus pyogenes) and Group B (GBS, Streptococcus agalactiae) was shown to be immunogenic in mice inducing both humoral and cellular immunity (101). Differently from viral vectors and DNA -based vaccine, RNA vaccines do not interact with the host-cell DNA, avoiding the potential risk of genomic integration and should not induce anti-vector immunity. Therefore, based on (i) their fully synthetic nature, (ii) relatively low production cost, and (iii) the possibility of expressing complex antigens in large amounts and in a short time, mRNA technologies could represent a rapid and valuable platform to be further exploited for vaccines against AMR infections.
Altogether, these new technologies may greatly advance the field for the control of AMR infections (Figure 4).
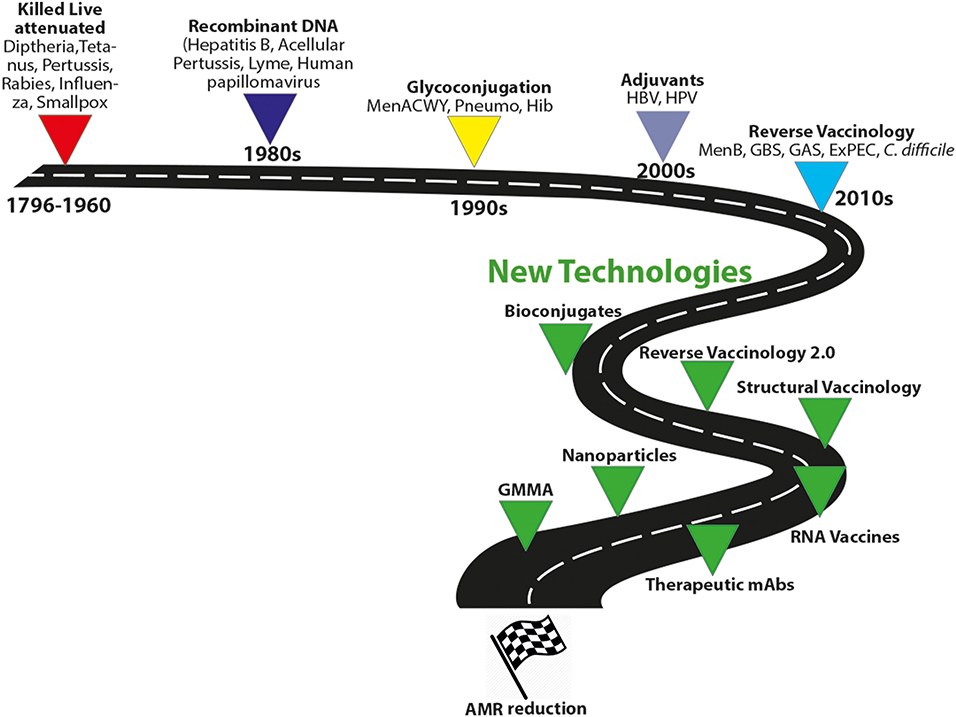
Figure 4. Evolution of vaccine technologies and platforms. MenACWY, meningococcus ACWY; Pneumo, pneumococcus; HBV, hepatitis B virus; Hib, type B Haemophilus influenzae; GBS, group B Streptococcus, GAS HPV, human papillomavirus; MenB, meningococcus B; GAS, group A streptococcus; ExPEC, extraintestinal pathogenic E. coli; Generalized Modules for Membrane Antigens, GMMA.
Conclusions and Future Perspectives
Rising antimicrobial resistance (AMR) is a threat to modern medicine and it also represents a major economic burden on healthcare systems. In the European Union, antibiotic resistant infections determine an economic burden of ~€1.5 billion per year. The US health system spends 20 billion USD in excess each year to treat resistant infections (102). By 2050, it is foreseen that USD100 trillion will be the economic damage due to AMR. According to the World Bank simulation, the global economy could lose as much as 3.8% of its annual gross domestic product by 2050 in a worst-case scenario. However, estimating the incidence and attributable mortality is challenging. This cost depends on a wide variety of factors: which drug and pathogen are involved, the mechanism of antibiotic resistance, the prevalence of that pathogen, the types of infections, the level of transmissibility, the health burden of those infections and whether alternative treatments are available. AMR can result in treatment failures and previously uncomplicated infections become more complex and severe (103, 104). It has recently been estimated that a 30% reduction in the efficacy of antibiotic prophylaxis for surgical procedures and chemotherapy would result in 120,000 additional infections and 6,300 infection-related deaths per year only in the United States. There is a risk of the cost being far higher than current best estimates. As already mentioned, vaccines represent a valuable and extraordinary tool to tackle the AMR problem. Besides being able to protect against serious diseases, vaccines may also reduce avoid mild disease episodes that may not receive medical attention, but which have important societal consequences. Vaccination decreases the disease incidence, the number of deaths, the longer hospital stays, and the need for more antibiotics if others fail owing to resistance. Unvaccinated individuals also benefit from wider societal benefits such as the lower transmission rates. This can be translated into a reduced risk of epidemics. In health economics, Incremental Cost–Utility Analysis (ICUA) is a method of financial analysis that can drive procurement decisions. The purpose of ICUA is to perform a cost-effectiveness analysis based on a careful evaluation of the ratio between the cost of an intervention and the benefit it generates in terms of increase in life expectancy. Value of vaccines and drugs is calculated based on the ICUA financial method resulting in an underestimation of the former. The added value of vaccines such as the induction of herd protection, reduction in time of hospitalization and reduction in anti-microbial resistance are difficult to measure in the short term. Therefore, since in the ICUA analysis, the benefits of medical interventions are evaluated during the period of medical observation, important benefits of vaccines which are only measurable in the long term are not considered. Although the ICUA analysis has generated more comprehensive models for financial evaluation, all the benefits of vaccination still need to be included in the analysis for the measurement of a full vaccine value (105). In 2015, the burden of resistant infections in countries of the European Economic Area (EEA) was measured in terms of number of cases, attributable deaths, and disability-adjusted life-years (DALYs) providing useful information for public health decision-makers prioritizing interventions for infectious diseases (106). The contribution of various antibiotic-resistant bacteria to the overall burden varies greatly between countries, thus highlighting the need for prevention and control strategies that are tailored to the needs of each country. Evaluation of the economic cost of AMR is important for decision-making and should be estimated accurately. Only combined efforts in the fields of antibiotics, new technologies for vaccine development and monoclonal antibodies will provide affordable solutions to tackle antimicrobial resistance.
Author Contributions
SN, RRo, MP, and RRa wrote the manuscript and contributed to the ideas and concepts it contains. All authors reviewed and approved the manuscript.
Funding
This work has received funding under the European Research Council (ERC) advanced grant agreement number 787552 (vAMRes).
Conflict of Interest
RRo, MP, and RRa are permanent employees of the GSK group of companies. RRo, MP, and RRa are listed as inventors on patents owned by the GSK group of companies. SN is a PhD Research Fellow of University of Bologna (IT). This work was sponsored by GlaxoSmithKline Biologicals SA.
Acknowledgments
The authors wish to thank Giorgio Corsi for artwork and Catherine Mallia for the editorial assistance.
References
1. Christensen K, Doblhammer G, Rau R, Vaupel JW. Ageing populations: the challenges ahead. Lancet. (2009) 374:1196–208. doi: 10.1016/S0140-6736(09)61460-4
2. Rappuoli R. Twenty-first century vaccines. Philos Trans R Soc Lond B Biol Sci. (2011) 366:2756–8. doi: 10.1098/rstb.2011.0075
3. Rappuoli R, Pizza M, Del Giudice G, De Gregorio E. Vaccines, new opportunities for a new society. Proc Natl Acad Sci USA. (2014) 111:12288–93. doi: 10.1073/pnas.1402981111
4. Rappuoli R, Bloom DE, Black S. Deploy vaccines to fight superbugs. Nature. (2017) 552:165–7. doi: 10.1038/d41586-017-08323-0
5. Plotkin SL, Plotkin SA. (2013). A short history of vaccination. Vaccines. 6 edn. 1–13. doi: 10.1016/B978-1-4557-0090-5.00017-3
6. Wohlleben W, Mast Y, Stegmann E, Ziemert N. Antibiotic drug discovery. Microb Biotechnol. (2016) 9:541–8. doi: 10.1111/1751-7915.12388
7. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. (2010) 74:417–33. doi: 10.1128/MMBR.00016-10
8. Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. Antibiotic resistance in Pseudomonas Aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv. (2019) 37:177–92. doi: 10.1016/j.biotechadv.2018.11.013
10. Delany I, Rappuoli R, Seib KL. Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harb Perspect Med. (2013) 3:a012476. doi: 10.1101/cshperspect.a012476
11. Tedijanto C, Olesen SW, Grad YH, Lipsitch M. Estimating the proportion of bystander selection for antibiotic resistance among potentially pathogenic bacterial flora. Proc Natl Acad Sci USA. (2018) 115:E11988–95. doi: 10.1073/pnas.1810840115
12. Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. (2015) 13:42–51. doi: 10.1038/nrmicro3380
13. Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. (2007) 128:1037–50. doi: 10.1016/j.cell.2007.03.004
14. Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. (2016) 4:1–37. doi: 10.1128/microbiolspec.VMBF-0016-2015
15. Baker KS, Dallman TJ, Field N, Childs T, Mitchell H, Day M, et al. Horizontal antimicrobial resistance transfer drives epidemics of multiple Shigella species. Nat Commun. (2018) 9:1462. doi: 10.1038/s41467-018-03949-8
16. Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist. (2018) 11:1645–58. doi: 10.2147/IDR.S173867
17. Blake KL, O'Neill AJ. Transposon library screening for identification of genetic loci participating in intrinsic susceptibility and acquired resistance to antistaphylococcal agents. J Antimicrob Chemother. (2013) 68:12–6. doi: 10.1093/jac/dks373
18. Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. (2007) 5:175–86. doi: 10.1038/nrmicro1614
19. Sampaio JL, Gales AC. Antimicrobial resistance in Enterobacteriaceae in Brazil: focus on beta-lactams and polymyxins. Braz J Microbiol. (2016) 47 (Suppl. 1):31–7. doi: 10.1016/j.bjm.2016.10.002
20. Pana ZD, Zaoutis T. Treatment of extended-spectrumβ-lactamase-producing Enterobacteriaceae (ESBLs) infections: what have we learned until now? F1000Res. (2018) 7:F1000. doi: 10.12688/f1000research.14822.1
21. Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. (2007) 20:440–58. doi: 10.1128/CMR.00001-07
22. Trecarichi EM, Tumbarello M. Therapeutic options for carbapenem-resistant Enterobacteriaceae infections. Virulence. (2017) 8:470–84. doi: 10.1080/21505594.2017.1292196
23. Lepuschitz S, Schill S, Stoeger A, Pekard-Amenitsch S, Huhulescu S, Inreiter N, et al. Whole genome sequencing reveals resemblance between ESBL-producing and carbapenem resistant Klebsiella pneumoniae isolates from Austrian rivers and clinical isolates from hospitals. Sci Total Environ. (2019) 662:227–35. doi: 10.1016/j.scitotenv.2019.01.179
24. Frost I, Van Boeckel TP, Pires J, Craig J, Laxminarayan R. Global geographic trends in antimicrobial resistance: the role of international travel. J. Travel Med. (2019) 26:taz036. doi: 10.1093/jtm/taz036
25. Qin S, Wang Y, Zhang Q, Chen X, Shen Z, Deng F, et al. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter Coli. Antimicrob Agents Chemother. (2012) 56:5332–9. doi: 10.1128/AAC.00809-12
26. Campisi E, Rosini R, Ji W, Guidotti S, Rojas-Lopez M, Geng G, et al. Genomic analysis reveals multi-drug resistance clusters in group B Streptococcus CC17 hypervirulent isolates causing neonatal invasive disease in southern mainland China. Front Microbiol. (2016) 7:1265. doi: 10.3389/fmicb.2016.01265
27. Nikaido H. Multidrug resistance in bacteria. Annu Rev Biochem. (2009) 78:119–46. doi: 10.1146/annurev.biochem.78.082907.145923
28. Lavigne JP, Sotto A, Nicolas-Chanoine MH, Bouziges N, Pages JM, Davin-Regli A. An adaptive response of Enterobacter Aerogenes to imipenem: regulation of porin balance in clinical isolates. Int J Antimicrob Agents. (2013) 41:130–6. doi: 10.1016/j.ijantimicag.2012.10.010
29. Welte W, Nestel U, Wacker T, Diederichs K. Structure and function of the porin channel. Kidney Int. (1995) 48:930–40. doi: 10.1038/ki.1995.374
30. Li H, Luo YF, Williams BJ, Blackwell TS, Xie CM. Structure and function of OprD protein in Pseudomonas Aeruginosa: from antibiotic resistance to novel therapies. Int J Med Microbiol. (2012) 302:63–8. doi: 10.1016/j.ijmm.2011.10.001
31. Blair JM, Richmond GE, Piddock LJ. Multidrug efflux pumps in gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. (2014) 9:1165–77. doi: 10.2217/fmb.14.66
32. Nicoloff H, Perreten V, McMurry LM, Levy SB. Role for tandem duplication and lon protease in AcrAB-TolC- dependent multiple antibiotic resistance (Mar) in an Escherichia Coli mutant without mutations in marRAB or acrRAB. J Bacteriol. (2006) 188:4413–23. doi: 10.1128/JB.01502-05
33. Ricci V, Attah V, Overton T, Grainger DC, Piddock LJV. CsrA maximizes expression of the AcrAB multidrug resistance transporter. Nucleic Acids Res. (2017) 45:12798–807. doi: 10.1093/nar/gkx929
34. Ohneck EA, Zalucki YM, Johnson PJ, Dhulipala V, Golparian D, Unemo M, et al. A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria Gonorrhoeae. MBio. (2011) 2:e00187–11. doi: 10.1128/mBio.00187-11
35. Cattoir V, Merabet L, Legrand P, Soussy CJ, Leclercq R. Emergence of a Streptococcus Pneumoniae isolate resistant to streptogramins by mutation in ribosomal protein L22 during pristinamycin therapy of Pneumococcal Pneumonia. J Antimicrob Chemother. (2007) 59:1010–2. doi: 10.1093/jac/dkm041
36. Ikeda-Dantsuji Y, Hanaki H, Nakae T, Takesue Y, Tomono K, Honda J, et al. Emergence of linezolid-resistant mutants in a susceptible-cell population of methicillin-resistant Staphylococcus Aureus. Antimicrob Agents Chemother. (2011) 55:2466–8. doi: 10.1128/AAC.01548-10
37. Gerson S, Betts JW, Lucassen K, Nodari CS, Wille J, Josten M, et al. Investigation of novel pmrB and eptA mutations in isogenic acinetobacter baumannii isolates associated with colistin resistance and increased virulence in vivo. Antimicrob Agents Chemother. (2019) 63:e01586–18. doi: 10.1128/AAC.01586-18
38. World Health Organization. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis. Geneva:WHO (2017).
39. CDC. Antibiotic Resistance Threats in the United States. Atlanta, GA: Department of Health and Human Services, CDC. (2019).
40. Michalopoulos A, Falagas ME. Treatment of acinetobacter infections. Expert Opin Pharmacother. (2010) 11:779–88. doi: 10.1517/14656561003596350
41. Nasr P. Genetics, epidemiology, and clinical manifestations of multidrug-resistant Acinetobacter Baumannii. J Hosp Infect. (2019) 104:4–11. doi: 10.1016/j.jhin.2019.09.021
42. Gygli SM, Borrell S, Trauner A, Gagneux S. Antimicrobial resistance in Mycobacterium Tuberculosis: mechanistic and evolutionary perspectives. FEMS Microbiol Rev. (2017) 41:354–73. doi: 10.1093/femsre/fux011
43. Marrakchi H, Laneelle MA, Daffe M. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol. (2014) 21:67–85. doi: 10.1016/j.chembiol.2013.11.011
44. Stagg HR, Lipman MC, McHugh TD, Jenkins HE. Isoniazid-resistant tuberculosis: a cause for concern? Int J Tuberc Lung Dis. (2017) 21:129–39. doi: 10.5588/ijtld.16.0716
45. Rice PA, Shafer WM, Ram S, Jerse AE. Neisseria Gonorrhoeae: drug resistance, mouse models, and vaccine development. Annu Rev Microbiol. (2017) 71:665–86. doi: 10.1146/annurev-micro-090816-093530
46. Sanchez-Buso L, Harris SR. Using genomics to understand antimicrobial resistance and transmission in Neisseria Gonorrhoeae. Microb Genom. (2019) 5:e000239. doi: 10.1099/mgen.0.000239
47. Quillin SJ, Seifert HS. Neisseria Gonorrhoeae host adaptation and pathogenesis. Nat Rev Microbiol. (2018) 16:226–40. doi: 10.1038/nrmicro.2017.169
48. Unemo M, Del Rio C, Shafer WM. Antimicrobial resistance expressed by neisseria gonorrhoeae: a major global public health problem in the 21st century. Microbiol Spectr. (2016) 4:1–18. doi: 10.1128/microbiolspec.EI10-0009-2015
49. McEwen SA, Collignon PJ. Antimicrobial resistance: a one health perspective. Microbiol Spectr. (2018) 6:iv36-iv41. doi: 10.1128/microbiolspec.ARBA-0009-2017
50. Bloom DE, Black S, Salisbury D, Rappuoli R. Antimicrobial resistance and the role of vaccines. Proc Natl Acad Sci USA. (2018) 115:12868–71. doi: 10.1073/pnas.1717157115
51. Relman DA, Lipsitch M. Microbiome as a tool and a target in the effort to address antimicrobial resistance. Proc Natl Acad Sci USA. (2018) 115:12902–10. doi: 10.1073/pnas.1717163115
52. Leverstein-Van Hall MA, Stuart JC, Voets GM, Versteeg D, Tersmette T, Fluit AC. Global spread of New Delhi metallo-β-lactamase 1. Lancet Infect Dis. (2010) 10:830–1. doi: 10.1016/S1473-3099(10)70277-2
53. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
54. Gupta SK, Nayak RP. Dry antibiotic pipeline: regulatory bottlenecks and regulatory reforms. J Pharmacol Pharmacother. (2014) 5:4–7. doi: 10.4103/0976-500X.124405
55. Richter MF, Drown BS, Riley AP, Garcia A, Shirai T, Svec RL, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. (2017) 545:299–304. doi: 10.1038/nature22308
56. Kennedy DA, Read AF. Why does drug resistance readily evolve but vaccine resistance does not? Proc Biol Sci. (2017) 284:20162562. doi: 10.1098/rspb.2016.2562
57. Tagliabue A, Rappuoli R. Changing priorities in vaccinology: antibiotic resistance moving to the top. Front Immunol. (2018) 9:1068. doi: 10.3389/fimmu.2018.01068
58. Nichol KL, Nordin JD, Nelson DB, Mullooly JP, Hak E. Effectiveness of influenza vaccine in the community-dwelling elderly. N Engl J Med. (2007) 357:1373–81. doi: 10.1056/NEJMoa070844
59. Kwong JC, Maaten S, Upshur RE, Patrick DM, Marra F. The effect of universal influenza immunization on antibiotic prescriptions: an ecological study. Clin Infect Dis. (2009) 49:750–6. doi: 10.1086/605087
60. Moore MR, Link-Gelles R, Schaffner W, Lynfield R, Lexau C, Bennett NM, et al. Effect of use of 13-valent pneumococcal conjugate vaccine in children on invasive pneumococcal disease in children and adults in the USA: analysis of multisite, population-based surveillance. Lancet Infect Dis. (2015) 15:301–9. doi: 10.1016/S1473-3099(14)71081-3
61. Cassiolato AP, Almeida SCG, Andrade AL, Minamisava R, Brandileone MCC. Expansion of the multidrug-resistant clonal complex 320 among invasive Streptococcus Pneumoniae serotype 19A after the introduction of a ten-valent pneumococcal conjugate vaccine in Brazil. PLoS ONE. (2018) 13:e0208211. doi: 10.1371/journal.pone.0208211
62. Gaviria-Agudelo CL, Jordan-Villegas A, Garcia C, McCracken GHJr. The effect of 13-valent pneumococcal conjugate vaccine on the serotype distribution and antibiotic resistance profiles in children with invasive pneumococcal disease. J Pediatric Infect Dis Soc. (2017) 6:253–9. doi: 10.1093/jpids/piw005
63. Goldstein E, Finelli L, O'Halloran A, Liu P, Karaca Z, Steiner CA, et al. Hospitalizations associated with respiratory syncytial virus and influenza in children, including children diagnosed with asthma. Epidemiology. (2019) 30:918–26. doi: 10.1097/EDE.0000000000001092
64. Semchenko EA, Tan A, Borrow R, Seib KL. The serogroup B meningococcal vaccine bexsero elicits antibodies to Neisseria Gonorrhoeae. Clin Infect Dis. (2019) 69:1101–11. doi: 10.1093/cid/ciy1061
65. Marjuki H, Topaz N, Joseph SJ, Gernert KM, Kersh EN Antimicrobial-Resistant Neisseria gonorrhoeae Working G. Genetic similarity of gonococcal homologs to meningococcal outer membrane proteins of serogroup B vaccine. MBio. (2019) 10:e01668–19. doi: 10.1128/mBio.01668-19
66. Rivero-Calle I, Raguindin PF, Gomez-Rial J, Rodriguez-Tenreiro C, Martinon-Torres F. Meningococcal group B vaccine for the prevention of invasive meningococcal disease caused by Neisseria Meningitidis serogroup B. Infect Drug Resist. (2019) 12:3169–88. doi: 10.2147/IDR.S159952
67. Leuzzi R, Adamo R, Scarselli M. Vaccines against Clostridium difficile. Hum Vaccin Immunother. (2014) 10:1466–77. doi: 10.4161/hv.28428
68. Poolman JT, Anderson AS. Escherichia Coli and Staphylococcus Aureus: leading bacterial pathogens of healthcare associated infections and bacteremia in older-age populations. Expert Rev Vaccines. (2018) 17:607–18. doi: 10.1080/14760584.2018.1488590
69. Lacey KA, Geoghegan JA, McLoughlin RM. The role of Staphylococcus aureus virulence factors in skin infection and their potential as vaccine antigens. Pathogens. (2016) 5. doi: 10.3390/pathogens5010022
70. Dayan GH, Mohamed N, Scully IL, Cooper D, Begier E, Eiden J, et al. Staphylococcus aureus: the current state of disease, pathophysiology and strategies for prevention. Expert Rev Vaccines. (2016) 15:1373–92. doi: 10.1080/14760584.2016.1179583
71. Plotkin SA, Plotkin SL. The development of vaccines: how the past led to the future. Nat Rev Microbiol. (2011) 9:889–93. doi: 10.1038/nrmicro2668
72. Riedel S. Edward Jenner and the history of smallpox and vaccination. Proc (Bayl Univ Med Cent). (2005) 18:21–5. doi: 10.1080/08998280.2005.11928028
73. Minor P. D. (2015). Live attenuated vaccines: historical successes and current challenges. Virology. 479–480, 379–392. doi: 10.1016/j.virol.2015.03.032
74. Finco O, Rappuoli R. Designing vaccines for the twenty-first century society. Front Immunol. (2014) 5:12. doi: 10.3389/fimmu.2014.00012
75. Glenny AT, Hopkins BE. Duration of passive immunity. J Hyg. (1923) 22:208–21. doi: 10.1017/S0022172400008172
76. Pizza M, Covacci A, Bartoloni A, Perugini M, Nencioni L, De Magistris MT, et al. Mutants of pertussis toxin suitable for vaccine development. Science. (1989) 246:497–500. doi: 10.1126/science.2683073
77. van den Ende C, Marano C, van Ahee A, Bunge EM, De Moerlooze L. The immunogenicity of GSK's recombinant hepatitis B vaccine in children: a systematic review of 30 years of experience. Expert Rev. Vaccines. (2017) 16:789–809. doi: 10.1080/14760584.2017.1338569
78. Nogales A, Martinez-Sobrido L. Reverse genetics approaches for the development of influenza vaccines. Int J Mol Sci. (2016) 18:20. doi: 10.3390/ijms18010020
79. Jefferies JM, Macdonald E, Faust SN, Clarke SC. 13-valent pneumococcal conjugate vaccine (PCV13). Hum Vaccin. (2011) 7:1012–8. doi: 10.4161/hv.7.10.16794
80. Klugman KP. Contribution of vaccines to our understanding of pneumococcal disease. Philos Trans R Soc Lond B Biol Sci. (2011) 366:2790–8. doi: 10.1098/rstb.2011.0032
81. Huttner A, Hatz C, van den Dobbelsteen G, Abbanat D, Hornacek A, Frolich R, et al. Safety, immunogenicity, and preliminary clinical efficacy of a vaccine against extraintestinal pathogenic Escherichia Coli. in women with a history of recurrent urinary tract infection: a randomised, single-blind, placebo-controlled phase 1b trial. Lancet Infect. Dis. 17:528–37. doi: 10.1016/S1473-3099(17)30108-1
82. Ravenscroft N, Braun M, Schneider J, Dreyer AM, Wetter M, Haeuptle MA, et al. Characterization and immunogenicity of a shigella flexneri 2a O-antigen bioconjugate vaccine candidate. Glycobiology. (2019) 29:669–80. doi: 10.1093/glycob/cwz044
83. Bastola R, Noh G, Keum T, Bashyal S, Seo JE, Choi J, et al. Vaccine adjuvants: smart components to boost the immune system. Arch Pharm Res. (2017) 40:1238–48. doi: 10.1007/s12272-017-0969-z
84. Laupeze B, Herve C, Di Pasquale A, Tavares Da Silva F. Adjuvant Systems for vaccines: 13years of post-licensure experience in diverse populations have progressed the way adjuvanted vaccine safety is investigated and understood. Vaccine. (2019) 37:5670–80. doi: 10.1016/j.vaccine.2019.07.098
85. Van Der Meeren O, Hatherill M, Nduba V, Wilkinson RJ, Muyoyeta M, Van Brakel E, et al. Phase 2b controlled trial of M72/AS01E vaccine to prevent tuberculosis. N Engl J Med. (2018) 379:1621–34. doi: 10.1056/NEJMoa1803484
86. Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, et al. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. (2000) 287:1816–20. doi: 10.1126/science.287.5459.1816
87. Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine. (2012) 30 (Suppl. 2):B87–97. doi: 10.1016/j.vaccine.2012.01.033
88. Unemo M, Shafer WM. Antimicrobial resistance in Neisseria Gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev. (2014) 27:587–613. doi: 10.1128/CMR.00010-14
89. Petousis-Harris H, Paynter J, Morgan J, Saxton P, McArdle B, Goodyear-Smith F, et al. Effectiveness of a group B outer membrane vesicle meningococcal vaccine against gonorrhoea in New Zealand: a retrospective case-control study. Lancet. (2017) 390:1603–10. doi: 10.1016/S0140-6736(17)31449-6
90. Baker SJ, Payne DJ, Rappuoli R, De Gregorio E. Technologies to address antimicrobial resistance. Proc Natl Acad Sci USA. (2018) 115:12887–95. doi: 10.1073/pnas.1717160115
91. Al-Halifa S, Gauthier L, Arpin D, Bourgault S, Archambault D. Nanoparticle-based vaccines against respiratory viruses. Front Immunol. (2019) 10:22. doi: 10.3389/fimmu.2019.00022
92. Lopez-Sagaseta J, Malito E, Rappuoli R, Bottomley MJ. Self-assembling protein nanoparticles in the design of vaccines. Comput Struct Biotechnol J. (2016) 14:58–68. doi: 10.1016/j.csbj.2015.11.001
93. Rappuoli R, Bottomley MJ, D'Oro U, Finco O, De Gregorio E. Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med. (2016) 213:469–81. doi: 10.1084/jem.20151960
94. Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. (2010) 84:1005–13. doi: 10.1128/JVI.01809-09
95. McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. (2013) 340:1113–7. doi: 10.1126/science.1234914
96. Scarselli M, Arico B, Brunelli B, Savino S, Di Marcello F, Palumbo E, et al. Rational design of a meningococcal antigen inducing broad protective immunity. Sci Transl Med. (2011) 3:91ra62. doi: 10.1126/scitranslmed.3002234
97. Tsai CW, Morris S. Approval of raxibacumab for the treatment of inhalation anthrax under the US food and drug administration “animal rule”. Front Microbiol. (2015) 6:1320. doi: 10.3389/fmicb.2015.01320
98. Hou AW, Morrill AM. Obiltoxaximab: adding to the treatment arsenal for Bacillus Anthracis infection. Ann Pharmacother. (2017) 51:908–13. doi: 10.1177/1060028017713029
99. Kufel WD, Devanathan AS, Marx AH, Weber DJ, Daniels LM. Bezlotoxumab: a novel agent for the prevention of recurrent Clostridium difficile infection. Pharmacotherapy. (2017) 37:1298–308. doi: 10.1002/phar.1990
100. Andreano E, D'Oro U, Rappuoli R, Finco O. Vaccine evolution and its application to fight modern threats. Front Immunol. (2019) 10:1722. doi: 10.3389/fimmu.2019.01722
101. Maruggi G, Chiarot E, Giovani C, Buccato S, Bonacci S, Frigimelica E, et al. Immunogenicity and protective efficacy induced by self-amplifying mRNA vaccines encoding bacterial antigens. Vaccine. (2017) 35:361–8. doi: 10.1016/j.vaccine.2016.11.040
102. Wozniak TM, Barnsbee L, Lee XJ, Pacella RE. Using the best available data to estimate the cost of antimicrobial resistance: a systematic review. Antimicrob Resist Infect Control. (2019) 8:26. doi: 10.1186/s13756-019-0472-z
103. Sevilla JP, Bloom DE, Cadarette D, Jit M, Lipsitch M. Toward economic evaluation of the value of vaccines and other health technologies in addressing AMR. Proc Natl Acad Sci USA. (2018) 115:12911–9. doi: 10.1073/pnas.1717161115
104. Roope LSJ, Smith RD, Pouwels KB, Buchanan J, Abel L, Eibich P, et al. The challenge of antimicrobial resistance: what economics can contribute. Science. (2019) 364:eaau4679. doi: 10.1126/science.aau4679
105. Standaert B, Rappuoli R. 2. How is the economic assessment of vaccines performed today? J Mark Access Health Policy. (2017) 5:1335163. doi: 10.1080/20016689.2017.1335163
106. Cassini A, Högberg LD, Plachouras D, Quattrocchi A, Hoxha A, Simonsen GS, et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis. (2019) 19:56–66. doi: 10.1016/S1473-3099(18)30605-4
Keywords: vaccines, infectious diseases, antimicrobial resistance (AMR), vaccine development, vaccinology, mechanisms of antibiotic resistance
Citation: Rosini R, Nicchi S, Pizza M and Rappuoli R (2020) Vaccines Against Antimicrobial Resistance. Front. Immunol. 11:1048. doi: 10.3389/fimmu.2020.01048
Received: 17 December 2019; Accepted: 30 April 2020;
Published: 03 June 2020.
Edited by:
Rita Carsetti, Bambino Gesù Children Hospital (IRCCS), ItalyReviewed by:
Shigeto Yoshida, Kanazawa University, JapanAnnie Searls De Groot, EpiVax, United States
Copyright © 2020 Rosini, Nicchi, Pizza and Rappuoli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rino Rappuoli, cmluby5yLnJhcHB1b2xpQGdzay5jb20=
 Roberto Rosini
Roberto Rosini Sonia Nicchi
Sonia Nicchi Mariagrazia Pizza
Mariagrazia Pizza Rino Rappuoli
Rino Rappuoli